Abstract
Background
Cervical cancer (CC) is the second most common type of female malignancy in Bangladesh. Polymorphisms in the CYP1A1 gene have been reported to be associated with CC in different populations. This case-control study with meta-analysis was undertaken to assess the relation of CYP1A1 rs4646903 and rs1048943 polymorphisms with the susceptibility of CC.
Methods
A total of 185 CC patients and 220 controls were recruited, and the PCR-RFLP (Polymerase chain reaction-restriction fragment length polymorphism) technique was applied for genotyping. Again, 42 eligible studies (24 with rs4646903 and 18 with rs1048943) were included for meta-analysis, and RevMan 5.3 and the MetaGenyo web-based tool were used.
Results
The rs4646903 polymorphism was significantly linked with CC in all association models, namely, additive 1, additive 2, dominant, recessive, overdominant, and allele models (OR = 2.41, 4.75, 2.67, 3.61, 2.13, and 2.44 with corresponding 95% CI = 1.55–3.76, 1.81–12.45, 1.75–4.07, 1.39–9.35, 1.38–3.30, and 1.71–3.48, respectively). On the contrary, rs1048943 showed no association (p > 0.05) with CC. Haplotype analysis revealed AT and AC haplotypes significantly decreased (OR = 0.45) and increased (OR = 4.86) CC risk, respectively, and SNPs are in strong linkage disequilibrium (D’ = 0.912, r2 = 0.448). Again, rs4646903 carriers with a contraception history and >5 years of taking contraceptives showed an enhanced risk of CC (OR = 2.39, OR = 3.05). Besides, rs1048943 carriers aged >40 years (OR = 0.44), conceived first child aged ≤18 years (OR = 3.45), and history of contraceptives (OR = 2.18) were significantly linked with CC. Our meta-analysis found that for CYP1A1 rs4646903 codominant 1 (COD 1), codominant 2 (COD 2), codominant 3 (COD 3), dominant model (DM), recessive model (RM), and allele model (AM) in Caucasians and overdominant model (OD) in the overall population are associated with an elevated risk of CC, whereas rs1048943 is also associated with CC in overall, Caucasians and Asians in some genetic models.
Conclusion
Our case-control study and meta-analysis summarize that CYP1A1 rs4646903 and rs1048943 polymorphisms may be correlated with cervical cancer.
Keywords: CYP1A1, rs4646903, rs1048943, PCR-RFLP, Contraceptives, Haplotype
1. Introduction
Cancer is a pathological condition or disease in which an abnormal and uncontrolled multiplication and spread of cells occur within the body. According to 2020 GLOBOCAN statistics, about 19 million new cases were identified, and more than 10 million patients died worldwide due to cancer, making it the second major cause of mortality [1]. On the basis of mortality and incidence, breast cancer is the most frequent [2] and cervical cancer (CC) is the second most common female (aged 15–49 years) malignancy in the world [1]. Notably, the rate of mortality due to CC in low- and middle-income countries is comparably higher (almost 18 times) than in high-income countries. Again, a maximum number of women suffered from squamous cell carcinoma (70%) rather than from adenocarcinoma (25%) [3]. At present, CC holds second place among Southeast Asian females [4]. In South Asia, the age-standardized incidence rate of CC is 22, 19.2, 13, and 2.8 in India, Bangladesh, Sri Lanka, and Iran, respectively, per 100,000 women [5]. In Bangladesh, CC is the second (25–30 cases/100,000) most important cause in terms of incidence, morbidity, and mortality [6].
Human papillomavirus (HPV) infection is mainly responsible for CC, along with many other etiological factors, such as environmental and socioeconomic factors [7]. Different types of HPV are found in the world, among which type 16 and type 18 are the most dangerous types for developing CC (about 75%) [8]. Genetic polymorphisms are inherent changes in the sequence of DNA that are present in at least 1% of specific populations. Almost 90% of genetic variations are single nucleotide polymorphisms (SNPs), and insertions, deletions, sequence repeats, and recombination are responsible for these genetic variations. Several host genetic factors dominate to regulate gene expression as well as disease pathogenesis and progression [7]. Numerous epidemiological studies revealed the relationship between host genetic makeup and cervical cancer. Cervical tumors have been determined to be around 27% in females by birth [9], and various types of strategies are used for cancer treatment, among which nanotechnology is extensively researched for selective cancer treatment [10,11].
The variation in responses to different xenobiotic-metabolizing enzymes occurs due to an individual's genetic variation. Cytochrome P450 1A1 (CYP1A1) gene located on the 15q22-24 chromosomal region consists of 5987 bp and codes for a protein (512 amino acids) [12]. A lot of endogenous agents, toxins, hormones and environmental pro-carcinogens (polyaromatic hydrocarbons, polyaromatic amines, dibenzofurans and biphenyls) are metabolized in the body by this enzyme, but cells are damaged if CYP1A1 enzyme activities are changed. Inducing agents commonly bind with specific receptors (such as the aryl hydrocarbon receptor), which are then translocated into the nucleus by AHR nuclear translocator (ARNT), and this heterodimer (AHR/ARNT) is then conjugated with a response element called xenobiotic response element (XRE) of CYP1A1 gene leading to transcription. Besides activating xenobiotics, CYP1A1 also has an important role in estrogen metabolism. It catalyzes the hydroxylation of 17-β estradiol [13].
So far, a total of 11 variant alleles have been recognized for CYP1A1, named from CYP1A1*1 to CYP1A1*11. Among these variants, four alleles, CYP1A1*2A (3698 T > C) or m1, CYP1A1*2C (2454 A > G) or m2, CYP1A1*3 (3204 T > C) or m3 and CYP1A1*4 (2452C > A) or m4 have been studied mostly for the association with cancer [14]. Moreover, CYP1A1*2A (rs4646903) and CYP1A1*2C (rs1048943) are the most common SNPs, and their genetic expressions are being frequently investigated. CYP1A1 rs1048943, situated near a heme-binding region of the protein, changes isoleucine to valines and modulates the effect of tobacco carcinogens metabolizing enzyme [15]. There is a strong relationship between rs4646903 and rs1048943 SNPs and the risk of CC in Indians [16,17] and Chinese women [18]. In contrast, some studies have reported that rs4646903 and rs1048943 polymorphisms have no relationship with the susceptibility of CC in Chhattisgarh women [19] and the Jewish population [20]. Scientists have found that many inherited genetic materials are responsible for cancer development, transmitted from generation to generation, and about 4% of all cancer in the world are heterogeneous [21]. It has been reported that 40% of human CYP450 isoforms that are involved in xenobiotic metabolism are highly polymorphic [22].
Different types of cancer, such as breast cancer [23,24], cervical cancer [25], prostate cancer [7,26], colorectal cancer [7,27] and lung cancer [27], are vulnerable to abnormalities in the actions of different genes. To date, genetic polymorphisms in human CYP1A1 have been widely investigated for the susceptibility of different cancers (e.g., cancer of the lung, oral, larynx, breast, thyroid, prostate, renal, cervix, gastric, and colon) in different ethnicity [28,29]. Unfortunately, the rate of CC incidence and death are increasing rapidly in Bangladesh day by day. Although previous studies investigated the role of multiple genetic polymorphisms of ECCR1, ECCR4, GCNT1P5, HLA-DRB1, interleukins (IL1β, IL4R, IL6 and IL10), INSIG2, and XPC with the risk of CC in the Bangladeshi patients, no previous study was conducted to evaluate the association of CYP1A1 polymorphisms [30]. Considering the above-mentioned facts, the current case-control study with meta-analysis was aimed to assess the potential relationship of CC susceptibility with two common CYP1A1 polymorphisms (rs4646903 and rs1048943).
2. Methods and materials
2.1. Recruitment of study population
Our study recruited patients suffering from CC, confirmed by the National Institute of Cancer Research and Hospital (NICRH), Dhaka-1212, Bangladesh. The controls were also recruited from the same institute at the same time as the patients. The study includes 185 CC respondents and 220 normal controls. All the relevant information of cases and controls, including demographic features, clinicopathological features, list of medicines taken, diagnostic, treatment and perceived features, were collected through a detailed preformed questionnaire by authors in the presence of the physicians. Each and every volunteer were specifically informed of the study intent before blood collection, and each of the participants/or legal guardians signed a written consent regarding the study (available upon request from the corresponding author). NICRH ethical committee approved ethical clearance (NICRH/Ethics/2019/447) for this study to ensure ethical issues. Helsinki Declaration and its further correction were followed during the study. We confirmed that no patients had severe diseases such as pulmonary disease, kidney disorder, and liver disease.
2.2. Sample preparation and primer design
About 3 ml of blood was collected from every participant by a 3 ml disposable syringe and quickly transferred into an EDTA (Ethylene Diamine Tetra Acetic acid) containing tube, and −80 °C freezer was used for storage of the collected blood till DNA extraction. DNA extraction was done by a chemical method described previously [31]. Micro-volume Spectrophotometer (Genova Nano, Jenway) was used to measure the concentration and purity of DNA, setting the absorbance ratio at 260/280 nm. Primer sequences were designed from the published paper by Islam et al., 2013 [31], shown in Table S1.
2.3. Method validation and SNP genotyping
To complete the genotyping of rs4646903 and rs1048943 SNPs, the PCR-based restriction fragment length polymorphism (RFLP) technique was used. Briefly, a working mix for PCR was prepared by adding EmeraldAmp GT PCR Master Mix (2x) (Takara Bio USA, Inc.) and two designed complementary primers (forward and reverse) at a suitable concentration. Then, 20 μl of the working mix was taken in a PCR tube, and 1 μl DNA sample was mixed in it, making a final volume of 21 μl. PCR conditions to amplify the rs1048943 and rs4646903 of CYP1A1 gene and the respective length of PCR products are given in Table S1. The gel electrophoresis (1% agarose) method was used to analyze the presence of PCR products at desired fragments. After the confirmation of desired PCR fragments, the respective PCR products were digested with specific restriction endonucleases for RFLP. To detect the CYP1A1 rs4646903 (T > C) and rs1048943 (A > G) alleles in the CC patients and controls, the digestion enzyme, condition of digestion, and the expected fragment lengths of both SNPs are listed in Table S1. After digestion, electrophoresis was done for the digested products using 1.3% agarose gel to get the RFLP products of rs1048943 and rs1048943. A 100 bp DNA ladder was used for size estimation of all RE digestion fragments, allowing for accurate and reliable genotyping of samples.
2.4. Meta-analysis
Two independent authors (MAB and MAA) systematically searched online platforms, including PubMed, BMC, Wiley, Springer, Web of Science, Cochrane Library, and ScienceDirect, to retrieve the eligible studies. The search keywords included: CYP1A1 and cervical cancer, rs4646903 and cervical cancer, rs108943 and cervical cancer, CYP1A1 polymorphisms and cervical cancer. The articles were evaluated based on the following inclusion criteria - i) Articles reporting the association of CYP1A1 polymorphisms (rs4646903 and rs1048943) with CC; ii) Design was case-control analysis; iii) Complete dataset to calculate OR and 95% CI value; iv) Human-sample-based studies; v) No language and geographical restriction. Besides, the exclusion criteria followed are - i) Overlapping or duplicate publications; ii) Studies with incomplete data; iii) Literature reviews, expert reviews, letters to the editor, and commentary articles; iv) Animal model studies, and v) Studies except for CYP1A1 polymorphisms (rs4646903 and rs1048943). To conduct the meta-analysis, the following information was gathered and recorded: first author's name, year of publication, study country, ethnic group, genotyping method, case/control quantity, and genotyping distribution. Another investigator (MSI) resolved any anomalies or conflicting results.
2.5. Statistical analysis
MedCalc (v19.0.7) was used for the estimation of ORs and associated 95% CIs to assess genotype and allele frequency variability with the Chi-square goodness of fit test. Moreover, the SHEsis online application was applied to determine the link between linkage disequilibrium (LD) and haplotypes with rs4646903 and rs1048943 for CC risk. All p < 0.05 was set as statistically significant. Using statistical software SPSS, version 25.0 for Windows (IBM), all statistical calculation was completed. For correcting the p-values, Bonferroni correction was performed, setting the significance level p < 0.017 [32]. The statistical power of the sample size in this case-control correlation investigation was determined using Online Sample Size Estimator (OSSE). Calculations of SNP-SNP interactions were also performed. To do the meta-analysis and assess heterogeneity between the included trials, we used Review Manager 5.3 (RevMan 5.3, The Cochrane Collaboration, Oxford, UK) and the MetaGenyo web tool. If there was significant heterogeneity (PH-value less than 0.1 or I2 > 50%), a random-effect model was used; otherwise, a fixed-effect model was utilized. The association of CYP1A1 polymorphisms (rs4646903 and rs1048943) has been examined in seven genetic models. In addition, Egger's regression test and Begg-Mazumdar's test were used to investigate publication biases [33]. The level of significance was fixed at p < 0.05 for publication bias, and values higher than this were projected to have no publishing bias. Finally, we conducted a sensitivity analysis by excluding each of the studies one by one to test the stability of the overall analysis.
2.6. False-positive report probability (FPRP) analysis
Analysis of the false-positive report probability (FPRP) was applied to analyze the notable linkages of the significant results and used a threshold of 0.2 and a prior probability of 0.25 for a correlation with the genotypes under study. In FPRP calculations, we recommended utilizing statistical power to determine an odds ratio of 2.
3. Results
3.1. Demographic and clinicopathological features
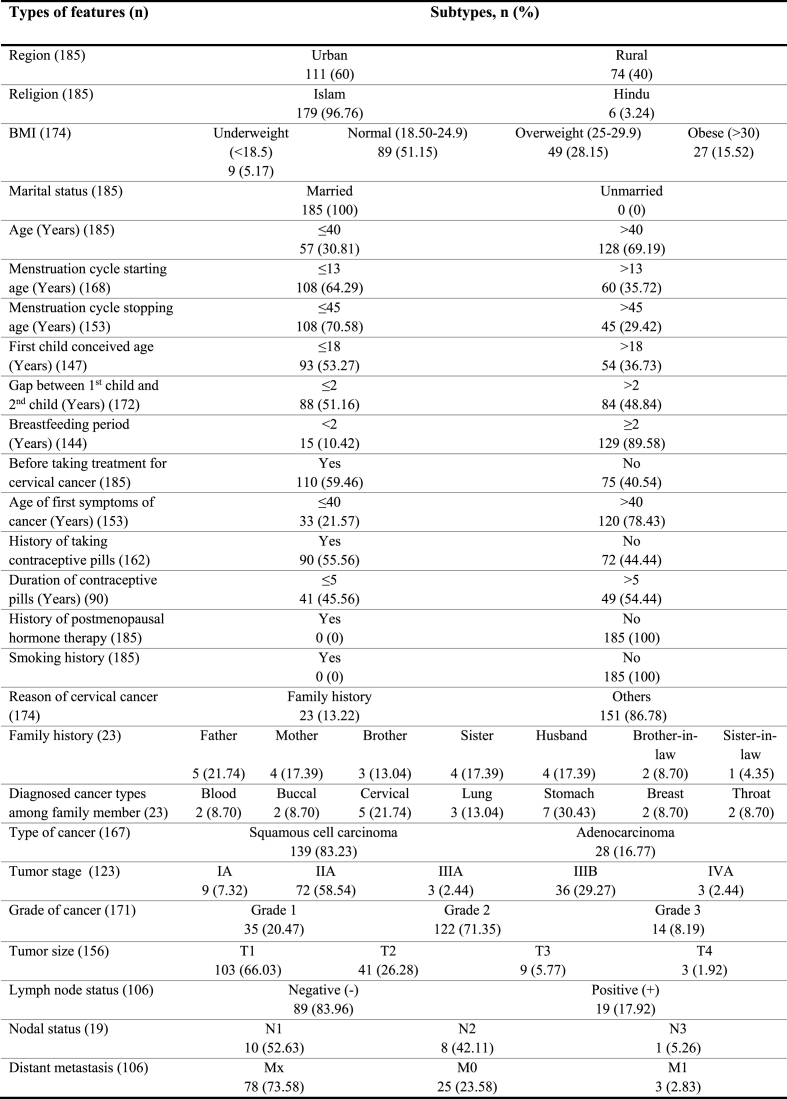
Among 185 CC patients, 60% were urban, and 40% were rural, whereas 96.76% of patients were Muslims and others were Hindus. Again, 51.15% of patients had normal BMI, while 28.15% and 15.52% were overweight and obese, respectively. All the patients were married, and 69.19% of them were aged more than 40 years. In 64.29% of cases, the menstrual cycle started at the age of 13 years or below 13 years, and in 70.58% of cases, the menstrual cycle stopped before 45 years or 45 years. Maximum women conceived their first child at ≤18 years (53.27%), and 51.16% of cases conceived their second child within 2 years. 89.58% of patients breastfed for at least or equal to 2 years. 59.46% of patients had experience taking treatment for CC during blood sample collection, and 78.43% faced the first symptoms of cancer after 40 years. 55.56% of patients took contraceptive pills, among which 54.44% took pills for more than 5 years. Only 13.22% of patients had a family history of CC, and 83.23% suffered from squamous cell carcinoma. Moreover, 58.54% were with IIA tumor stage, 71.35% were with grade 2, 66.03% were at T1 stage, and 83.96% had negative lymph node status. The details of demographic and clinicopathological features of CC cases are described in Table 1.
Table 1.
Demographic and clinicopathological features of cervical cancer patients.
3.2. List of different medicines taken by CC patients
From the available prescriptions of 157 patients, it was reported that 36.94% omeprazole (stomach acid-reducing agents), 14.65% levocetirizine (antihistamines), 34.39% ciprofloxacin (antibiotics), 17.83% multivitamin (vitamins), 31.85% metronidazole (antidiarrheals), 11.46% paracetamol or paracetamol + tramadol (analgesics), 20.38% ondansetron (anti-vomiting), 24.20% tranexamic acid (antifibrinolytics), 3.82% dexamethasone (corticosteroids), 6.37% lactulose (laxatives), 1.91% fluconazole (antifungals), and 3.82% amitriptyline (anxiolytics) were mostly prescribed medicines. The full list of different prescribed medicines taken by CC patients is shown in Table S2.
3.3. Diagnostic, treatment and perceived features of CC patients
The diagnostic, treatment and perceived features of CC patients are described in Table S3. Multiple diagnostic and treatment techniques were used and applied to CC patients. Most of the patients were diagnosed using pelvic examination (91.62%), pap test (98.80%), biopsy (97%), X-ray (86.235), and CT scan (73.65%). Our study found that 78.50%, 56.07%, and 29.91% of patients received chemotherapy, radiotherapy and surgery, respectively. Again, 79.76% of patients took ≤3 chemotherapy cycles and 5-FU (46.84%), cisplatin (53.16%), carboplatin (36.71%) and paclitaxel (44.30%) injections were mainly used during chemotherapy.
Patients with CC faced different adverse reactions during chemotherapy cycles, among which pain (16.67%), diarrhea (40.47%), constipation (28.57%), fever (42.86%), headache (30.95%), leg pain (35.71%), weakness (47.62%), irritation (21.43%), sleep disturbance (23.81%), and abdominal pain (19.05%) were common. Maximum patients experienced nausea & vomiting (73.27%), anorexia (69.31%), and alopecia (67.33%) with other side effects. 63.10% of patients showed no improvements after chemotherapy, and 36.90% showed improvements like decreased pain (67.74%), improved eating (29.03%), reduced irritation (19.35%), and stopped bleeding (77.42%).
3.4. Genotypic and haplotypic linkage of CYP1A1 polymorphisms with CC
The distribution of genotypes in both studied CYP1A1 SNPs (rs4646903 and rs1048943) was found to be consistent with HWE in terms of CC cases and controls (p > 0.05) (Supplementary Table S4). The association of different genotypes and their combinations with CC risk are described in Table 2. In terms of rs4646903, both TC (OR = 2.41, p = 0.0001, 95% CI = 1.55–3.76) and CC (OR = 4.75, p = 0.002, 95% CI = 1.81–12.45) carriers showed significantly increased association with CC risk. An increased risk of CC also found with the dominant (TC + CC vs. TT: OR = 2.67, p < 0.0001, 95% CI = 1.75–4.07), recessive (CC vs. TT + TC: OR = 3.61, p = 0.008, 95% CI = 1.39–9.35), over-dominant (TC vs. TT + CC: OR = 2.13, p = 0.0007, 95% CI = 1.38–3.30) and allele (C vs. T: OR = 2.44, p < 0.0001, 95% CI = 1.71–3.48) models that were statistically significant. The significance level of all associations remained stable after conducting the Bonferroni correction (p < 0.017). In the case of rs1048943, no genetic models were found to be significantly linked with CC susceptibility (p > 0.05) in the studied population. The statistical power for rs4646903 and rs1048943 SNPs were 93.4 and 22% obtained from the OSSE web tool (Table 2).
Table 2.
Genotypic and haplotypic linkage of CYP1A1 rs4646903 and rs1048943 polymorphisms with cervical cancer.
| SNPs | Model | Genotype/Allele | Case (%) | Control (%) | Crude analysis |
Statistical Power (%) | |
|---|---|---|---|---|---|---|---|
| OR (95% Cl) | p-value | ||||||
| rs4646903 | TT | 99 (53.51%) | 166 (75.45%) | 1 | 93.4 | ||
| Additive model 1 (TC vs. TT) | TC | 69 (37.30%) | 48 (21.82%) | 2.41 (1.55–3.76) | 0.0001 | ||
| Additive model 2 (CC vs. TT) | CC | 17 (9.19%) | 6 (2.73%) | 4.75 (1.81–12.45) | 0.002 | ||
| Dominant model (TC + CC vs. TT) | TT | 99 (53.51%) | 166 (75.45%) | 1 | |||
| TC + CC | 86 (46.49%) | 54 (24.55%) | 2.67 (1.75–4.07) | <0.0001 | |||
| Recessive model (CC vs. TT + TC) | TT + TC | 168 (90.81%) | 214 (97.27%) | 1 | |||
| CC | 17 (9.19%) | 6 (2.73%) | 3.61 (1.39–9.35) | 0.008 | |||
| Over-dominant model (TC vs TT + CC) | TT + CC | 116 (62.70%) | 172 (78.18) | 1 | |||
| TC | 69 (37.30%) | 48 (21.82%) | 2.13 (1.38–3.30) | 0.0007 | |||
| Allele | T | 267 (72.16%) | 380 (86.36%) | 1 | |||
| C | 103 (27.84%) | 60 (13.64%) | 2.44 (1.71–3.48) | <0.0001 | |||
| rs1048943 | AA | 138 (74.59%) | 174 (79.09%) | 1 | |||
| Additive model 1 (AG vs. AA) | AG | 40 (21.62%) | 40 (18.18%) | 1.26 (0.77–2.06) | 0.356 | ||
| Additive model 2 (GG vs. AA) | GG | 7 (3.78%) | 6 (2.73%) | 1.47 (0.48–4.48) | 0.497 | ||
| Dominant model (AG + GG vs. AA) | AA | 138 (74.59%) | 174 (79.09%) | 1 | |||
| AG + GG | 47 (25.41%) | 46 (20.91%) | 1.29 (0.81–2.05) | 0.285 | 22 | ||
| Recessive model (GG vs. AA + AG) | AA + AG | 178 (96.22%) | 214 (97.27%) | 1 | |||
| GG | 7 (3.78%) | 6 (2.73%) | 1.40 (0.46–4.25) | 0.550 | |||
| Over-dominant model (AG vs AA + GG) | AA + GG | 145 (78.38%) | 120 (54.55%) | 1 | |||
| AG | 40 (21.62%) | 40 (18.18%) | 0.83 (0.50–1.37) | 0.459 | |||
| Allele | A | 316 (85.41%) | 388 (88.18%) | 1 | |||
| G | 54 (14.59%) | 52 (11.82%) | 1.28 (0.85–1.92) | 0.244 | |||
| rs4646903 and rs1048943 |
Haplotypes | χ2 | Cases | Controls | OR (95% CI) | p-value | |
| AC | 30.61 | 0.137 | 0.031 | 4.86 (2.65–8.94) | 9.47x10−8 | ||
| AT | 21.60 | 0.716 | 0.85 | 0.45 (0.32–0.63) | 6.72x10−6 | ||
| GC | 2.45 | 0.14 | 0.104 | 1.4 (0.92–2.14) | 0.117 | ||
*Bold values indicate statistically significant (p < 0.05).
Linkage analysis using different haplotypes of rs4646903 and rs1048943 SNPs with the risk of CC showed that AC haplotype was significantly linked with increased risk (OR = 4.86, p = 9.47 × 10−8). In contrast, AT haplotype revealed a significantly reduced chance of CC (OR = 0.45, p = 6.72 × 10−6) (Table 2). Besides, the constructed LD block for rs4646903 and rs1048943 demonstrated that both SNPs are in strong linkage disequilibrium (D’ = 0.912 and r2 = 0.448) in the CC cases and controls (Fig. 1).
Fig. 1.
LD block between CYP1A1 rs4646903 and rs1048943 in the cases-controls (D’ = 0.912 and r2 = 0.448).
3.5. Risk of CYP1A1 SNPs carriers for CC respective to demographic and clinicopathological features
The risk of CYP1A1 SNPs (rs4646903 and rs1048943) carriers respective to demographic and clinicopathological features is shown in Table 3. As the table shows, rs4646903 carrier patients were found to be linked with an increased risk of CC in the dominant model when they took contraceptive pills (OR = 2.39, p = 0.008, 95% CI = 1.26–4.54) and the duration of taking contraceptive pills was more than 5 years (OR = 3.05, p = 0.012, 95% CI = 1.28–7.23). Again, the dominant model in rs1048943 polymorphism carrier CC patients was significantly correlated with age of >40 years (OR = 0.44, p = 0.019, 95% CI = 0.22–0.87), the first child conceived before 18 years (OR = 0.29, p = 0.003, 95% CI = 0.13–0.66), history of taking contraceptive pills (OR = 2.18, p = 0.036, 95% CI = 1.05–4.51), and the duration of taking contraceptive pills for more than 5 years (OR = 3.80, p = 0.004, 95% CI = 1.53–9.44).
Table 3.
Genotype distribution of CYP1A1 rs4646903 and rs1048943 polymorphisms with respect to demographic and clinicopathological features of cervical cancer patients.
| Variables (n) | rs4646903 |
Total | OR (95% CI) | p-value | rs1048943 |
Total | OR (95% CI) | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| TT | TC + CC | AA | AG + GG | |||||||
| Region (185) | ||||||||||
| Rural | 43 | 31 | 74 | 1 | 54 | 20 | 74 | 1 | ||
| Urban | 56 | 55 | 111 | 1.36 (0.75–2.47) | 0.307 | 84 | 27 | 111 | 0.87 (0.44–1.70) | 0.679 |
| Age (Years) (185) | ||||||||||
| ≤40 | 34 | 23 | 57 | 1 | 36 | 21 | 57 | 1 | ||
| >40 | 65 | 63 | 128 | 1.43 (0.76–2.70) | 0.265 | 102 | 26 | 128 | 0.44 (0.22–0.87) | 0.019 |
| BMI (174) | ||||||||||
| Underweight (<18.5) | 5 | 4 | 9 | 0.98 (0.25–3.89) | 0.977 | 7 | 2 | 9 | 0.69 (0.13–3.56) | 0.660 |
| Normal (18.50–24.9) | 49 | 40 | 89 | 1 | 63 | 26 | 89 | 1 | ||
| Overweight (25.00–29.9) | 27 | 22 | 49 | 1.00 (0.50–2.01) | 1.00 | 40 | 9 | 49 | 0.55 (0.23–1.28) | 0.165 |
| Obese (>30) | 14 | 13 | 27 | 1.14 (0.48–2.70) | 0.770 | 21 | 6 | 27 | 0.69 (0.25–1.91) | 0.478 |
| First child conceived age (147) | ||||||||||
| >18 | 24 | 30 | 54 | 1 | 45 | 9 | 54 | 1 | ||
| ≤18 | 41 | 52 | 93 | 1.01 (0.52–1.99) | 0.966 | 55 | 38 | 93 | 3.45 (1.51–7.89) | 0.003 |
| History of taking contraceptive pills (162) | ||||||||||
| No | 48 | 24 | 72 | 1 | 58 | 14 | 72 | 1 | ||
| Yes | 41 | 49 | 90 | 2.39 (1.26–4.54) | 0.008 | 59 | 31 | 90 | 2.18 (1.05–4.51) | 0.036 |
| Duration of taking contraceptive pills (90) | ||||||||||
| ≤5 | 27 | 14 | 41 | 1 | 31 | 10 | 41 | 1 | ||
| >5 | 19 | 30 | 49 | 3.05 (1.28–7.23) | 0.012 | 22 | 27 | 49 | 3.80 (1.53–9.44) | 0.004 |
| Type of cancer (167) | ||||||||||
| Adenocarcinoma | 18 | 10 | 28 | 1 | 21 | 7 | 28 | 1 | ||
| Squamous cell carcinoma | 67 | 72 | 139 | 1.93 (0.83–4.49) | 0.124 | 102 | 37 | 139 | 1.09 (0.43–2.77) | 0.859 |
| Tumor stage (123) | ||||||||||
| I | 6 | 3 | 9 | 1 | 4 | 5 | 9 | 1 | ||
| IIB | 39 | 33 | 72 | 1.69 (0.39–7.30) | 0.480 | 46 | 26 | 72 | 0.45 (0.11–1.83) | 0.267 |
| IIIA | 2 | 1 | 3 | 1.00 (0.06–15.99) | 1.00 | 2 | 1 | 3 | 0.40 (0.03–6.18) | 0.512 |
| IIIB | 16 | 20 | 36 | 2.50 (0.54–11.59) | 0.14 | 27 | 9 | 36 | 0.27 (0.06–1.21) | 0.087 |
| IVA | 3 | 0 | 3 | 0.27 (0.01–6.74) | 0.421 | 2 | 1 | 3 | 0.40 (0.03–6.18) | 0.512 |
| Histologic grade of cancer (171) | ||||||||||
| Grade 1 | 20 | 15 | 35 | 1 | 25 | 10 | 35 | 1 | ||
| Grade 2 | 62 | 60 | 122 | 1.29 (0.60–2.75) | 0.510 | 90 | 32 | 122 | 0.89 (0.38–2.05) | 0.783 |
| Grade 3 | 10 | 4 | 14 | 0.53 (0.14–2.03) | 0.358 | 11 | 3 | 14 | 0.68 (0.16–2.97) | 0.610 |
| Tumor size (156) | ||||||||||
| T1 | 48 | 55 | 103 | 1 | 76 | 27 | 103 | 1 | ||
| T2 | 17 | 24 | 41 | 1.23 (0.59–2.56) | 0.576 | 28 | 13 | 41 | 1.31 (0.59–2.88) | 0.507 |
| T3 | 6 | 3 | 9 | 0.44 (0.10–1.84) | 0.259 | 7 | 2 | 9 | 0.80 (0.16–4.11) | 0.794 |
| T4 | 2 | 1 | 3 | 0.44 (0.04–4.96) | 0.504 | 2 | 1 | 3 | 1.41 (0.12–16.15) | 0.784 |
| Lymph node status (106) | ||||||||||
| Negative (−) | 53 | 36 | 89 | 1 | 62 | 27 | 89 | 1 | ||
| Positive (+) | 10 | 9 | 19 | 1.33 (0.49–3.58) | 0.579 | 12 | 7 | 19 | 1.34 (0.48–3.77) | 0.580 |
| Nodal status (106) | ||||||||||
| N0 | 53 | 36 | 89 | 1 | 62 | 27 | 89 | 1 | ||
| N1 | 6 | 4 | 10 | 0.98 (0.26–3.73) | 0.978 | 6 | 3 | 9 | 1.15 (0.27–4.93) | 0.853 |
| N2 | 3 | 5 | 8 | 0.88 (0.19–3.93) | 0.871 | 4 | 3 | 7 | 1.72 (0.36–8.23) | 0.496 |
| N3 | 1 | 0 | 1 | 0.49 (0.02–12.33) | 0.66 | 2 | 1 | 3 | 1.15 (0.10–13.21) | 0.912 |
| Distant metastasis (106) | ||||||||||
| Mx | 49 | 29 | 78 | 1 | 48 | 30 | 78 | 1 | ||
| M0 | 16 | 9 | 25 | 0.95 (0.37–2.43) | 0.915 | 19 | 6 | 25 | 0.51 (0.18–1.41) | 0.192 |
| M1 | 2 | 1 | 3 | 0.84 (0.07–9.73) | 0.892 | 2 | 1 | 3 | 0.80 (0.07–9.21) | 0.858 |
*Bold values indicate statistically significant (p < 0.05).
3.6. FPRP analysis and SNP-SNP of interaction CYP1A1 rs4646903 and rs1048943 polymorphisms
False-positive report probability (FPRP) values for the association between CYP1A1 rs4646903 and rs1048943 with cervical cancer risk are described in Table 4. Additive model 1, Additive model 2, DM, RM, OD, and AM showed substantial correlation for rs4646903 (FPRP = 0.001, 0.105, 0.000, 0.180, 0.005, 0.000, respectively), but no genetic model demonstrated the relationship for rs1048943 at the prior probability threshold of 0.25. Table 5 shows the SNP-SNP interaction of CYP1A1 rs4646903 and rs1048943 polymorphisms with cervical cancer. The interaction of rs4646903 CT with rs1048943 AA potentially increased the risk of CC (OR = 4.69, 95% CI: 2.38–9.27, p < 0.0001).
Table 4.
False-positive report probability values for the association between CYP1A1 rs4646903 and rs1048943 with cervical cancer risk.
| Model | Crude analysis |
Statistical Power | Prior probability |
|||||
|---|---|---|---|---|---|---|---|---|
| OR (95% Cl) | p-value | 0.25 | 0.1 | 0.01 | 0.001 | 0.0001 | ||
| rs4646903 | ||||||||
| TC vs. TT | 2.41 (1.55–3.76) | 0.0001 | 0.206 | 0.002 | 0.005 | 0.049 | 0.340 | 0.838 |
| CC vs. TT | 4.75 (1.81–12.45) | 0.002 | 0.039 | 0.105 | 0.259 | 0.794 | 0.975 | 0.997 |
| TC + CC vs. TT | 2.67 (1.75–4.07) | <0.0001 | 0.090 | 0.000 | 0.000 | 0.005 | 0.053 | 0.357 |
| CC vs. TT + TC | 3.61 (1.39–9.35) | 0.008 | 0.112 | 0.180 | 0.397 | 0.879 | 0.987 | 0.999 |
| TC vs. TT + CC | 2.13 (1.38–3.30) | 0.0007 | 0.389 | 0.005 | 0.016 | 0.153 | 0.646 | 0.948 |
| Allele | 2.44 (1.71–3.48) | <0.0001 | 0.136 | 0.000 | 0.000 | 0.001 | 0.006 | 0.059 |
| rs1048943 | ||||||||
| AG vs. AA | 1.26 (0.77–2.06) | 0.356 | 0.967 | 0.525 | 0.769 | 0.973 | 0.997 | 1.000 |
| GG vs. AA | 1.47 (0.48–4.48) | 0.497 | 0.706 | 0.679 | 0.864 | 0.986 | 0.999 | 1.000 |
| AG + GG vs. AA | 1.29 (0.81–2.05) | 0.285 | 0.968 | 0.466 | 0.723 | 0.966 | 0.997 | 1.000 |
| (GG vs. AA + AG) | 1.40 (0.46–4.25) | 0.550 | 0.736 | 0.693 | 0.871 | 0.987 | 0.999 | 1.000 |
| (AG vs AA + GG) | 0.83 (0.50–1.37) | 0.459 | 0.976 | 0.589 | 0.811 | 0.979 | 0.998 | 1.000 |
| Allele | 1.28 (0.85–1.92) | 0.244 | 0.985 | 0.415 | 0.680 | 0.959 | 0.996 | 1.000 |
| Haplotypes | ||||||||
| AC | 4.86 (2.65–8.94) | 9.47 × 10−8 | 0.002 | 0.000 | 0.001 | 0.012 | 0.109 | 0.550 |
| AT | 0.45 (0.32–0.63) | 6.72 × 10−6 | 0.270 | 0.000 | 0.000 | 0.001 | 0.012 | 0.109 |
| GC | 1.4 (0.92–2.14) | 0.117 | 0.950 | 0.275 | 0.532 | 0.926 | 0.992 | 0.999 |
Table 5.
SNP-SNP interaction of CYP1A1 rs4646903 and rs1048943 polymorphisms with cervical cancer.
| SNP-SNP Interaction | SNP Genotype | Cases | Controls | OR | 95% CI | p-value | |
|---|---|---|---|---|---|---|---|
| rs4646903 & rs1048943 |
m1 | m2 | |||||
| TT | AA | 97 | 160 | 1 | – | – | |
| CT | AA | 37 | 13 | 4.69 | 2.38–9.27 | <0.0001 | |
| CT | AG | 32 | 35 | 1.50 | 0.88–2.59 | 0.137 | |
| CC | GG | 7 | 6 | 1.92 | 0.63–5.89 | 0.252 | |
| TT | AG | 2 | 6 | 0.55 | 0.11–2.78 | 0.469 | |
| CC | AG | 6 | 0 | – | – | – | |
| CC | AA | 4 | 0 | – | – | – | |
3.7. Meta-analysis results
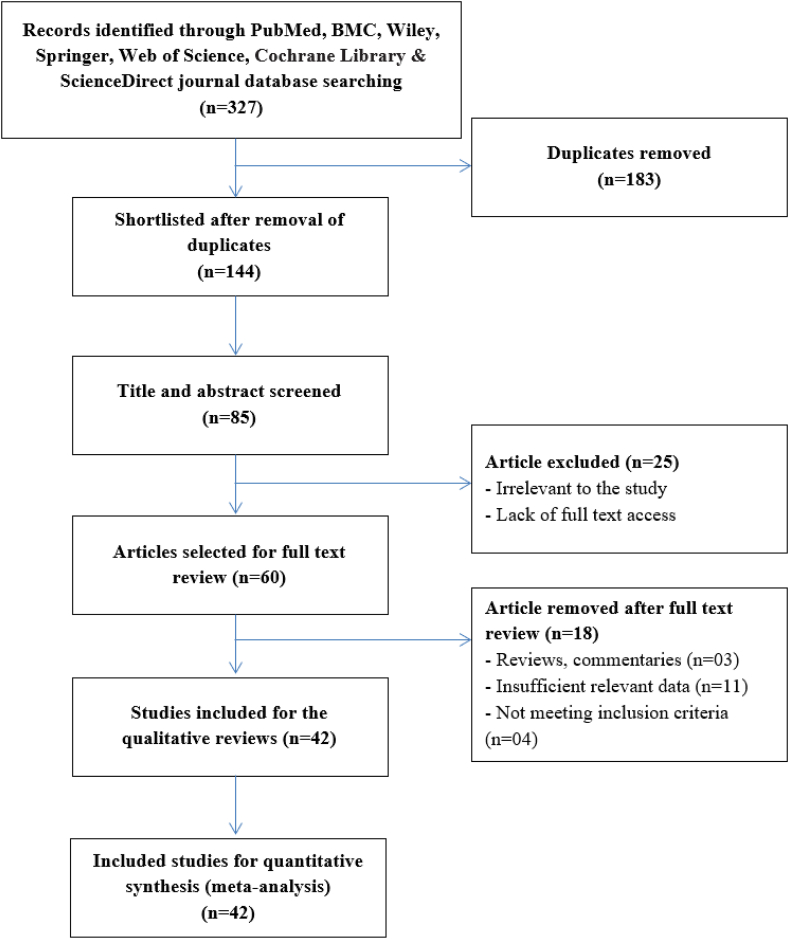
At first, 327 articles were identified from seven databases (PubMed, CNKI, Wiley, Springer, Web of Science, and ScienceDirect) during the preliminary retrieval. Due to duplication, a total of 183 records have been excluded. After that, 59 research articles were eliminated based on their titles and abstracts, while another 25 papers were removed due to inadequate data. Finally, 24 studies were accepted for rs4646903, with 3385 patients and 3425 controls, and 18 studies for rs1048943, with 2648 patients and 2914 controls. For the meta-analysis of these two SNPs, the procedure of searching, filtering, and selecting relevant publications is shown in Fig. 2, and the outcomes are described in Table S5.
Fig. 2.
PRISMA flow chart for the assessment of selected studies for the systemic review.
Table 6 and Fig. 3 demonstrate the relationship of CC with rs4646903 and rs1048943. Only the OD genetic model of rs4646903 was observed to be substantially linked with an elevated risk of CC in the overall population (TC vs. CC + TT: OR = 1.21, 95% CI = 1.04–1.41, Pz (p-value in meta-analysis) = 0.015). Other six genetic models showed a strong association with CC in Caucasian populations (COD1 (TC vs. TT): OR = 1.75, 95% CI = 1.03–2.97, Pz = 0.038; COD2 (CC vs. TT): OR = 8.36, 95% CI = 4.60–15.19, Pz < 0.0001; COD3 (CC vs. TC): OR = 3.39, 95% CI = 1.86–6.17, Pz = 6.76 × 10−5; DM (CC + TC vs. TT): OR = 2.02, 95% CI = 1.13–3.61, Pz = 0.018; RM (CC vs. TC + TT): OR = 5.54, 95% CI = 3.11–9.85, Pz = 5.8 × 10−9; and AM (C vs. T): OR = 1.96, 95% CI = 1.24–3.10, Pz = 0.004)).
Table 6.
Meta-analysis of the link between CYP1A1 rs4646903 and rs1048943 polymorphisms with cervical cancer.
| Comparison | Subgroup | N | OR | 95% Cl | PZ | Model | I2 | PH | Publication bias |
|
|---|---|---|---|---|---|---|---|---|---|---|
| Egger's test | Begg-Mazumdar's test | |||||||||
| CYP1A1 rs4646903 | ||||||||||
| COD1 (TC vs. TT) | Overall | 24 | 1.15 | 0.85–1.55 | 0.361 | Random | 83.58 | <0.0001 | 0.677 | 0.655 |
| Asian | 19 | 1.02 | 0.72–1.45 | 0.897 | Random | 83.7 | <0.0001 | 0.708 | 0.421 | |
| Caucasian | 5 | 1.75 | 1.03–2.97 | 0.038 | Random | 77.9 | 0.001 | 0.816 | 1.00 | |
| COD2 (CC vs. TT) | Overall | 24 | 1.43 | 0.74–2.80 | 0.290 | Random | 91.43 | <0.0001 | 0.394 | 0.941 |
| Asian | 19 | 1.03 | 0.51–2.10 | 0.931 | Random | 91.7 | <0.0001 | 0.691 | 0.529 | |
| Caucasian | 5 | 8.36 | 4.60–15.19 | <0.0001 | Fixed | 0 | 0.481 | 0.174 | 0.142 | |
| COD3 (CC vs. TC) | Overall | 24 | 1.19 | 0.79–1.80 | 0.412 | Random | 81.45 | <0.0001 | 0.030 | 0.298 |
| Asian | 19 | 0.98 | 0.64–1.50 | 0.907 | Random | 81.65 | <0.0001 | 0.104 | 0.506 | |
| Caucasian | 5 | 3.39 | 1.86–6.17 | 6.76x10−5 | Fixed | 2.26 | 0.394 | 0.917 | 0.624 | |
| DM (CC + TC vs. TT) | Overall | 24 | 1.15 | 0.81–1.6 | 0.432 | Random | 89.79 | <0.0001 | 0.572 | 0.254 |
| Asian | 19 | 0.99 | 0.66–1.51 | 0.974 | Random | 89.92 | <0.0001 | 0.615 | 0.278 | |
| Caucasian | 5 | 2.02 | 1.13–3.61 | 0.018 | Random | 83.19 | 0.0001 | 0.897 | 0.624 | |
| RM (CC vs. TC + TT) | Overall | 24 | 1.36 | 0.81–2.30 | 0.247 | Random | 90.1 | <0.0001 | 0.049 | 0.487 |
| Asian | 19 | 1.04 | 0.61–1.80 | 0.876 | Random | 90.35 | <0.0001 | 0.148 | 0.868 | |
| Caucasian | 5 | 5.54 | 3.11–9.85 | 5.8x10−9 | Fixed | 0 | 0.520 | 0.543 | 0.327 | |
| OD (TC vs. CC + TT) | Overall | 24 | 1.21 | 1.04–1.41 | 0.015 | Random | 50.42 | 0.003 | 0.796 | 0.710 |
| Asian | 19 | 1.15 | 0.98–1.36 | 0.088 | Random | 46.38 | 0.014 | 0.718 | 0.834 | |
| Caucasian | 5 | 1.44 | 0.98–2.12 | 0.066 | Random | 61.45 | 0.035 | 0.987 | 1.00 | |
| AM (C vs. T) | Overall | 24 | 1.15 | 0.80–1.66 | 0.445 | Random | 95.23 | <0.0001 | 0.127 | 0.823 |
| Asian | 19 | 1.01 | 0.68–1.51 | 0.956 | Random | 95.37 | <0.0001 | 0.150 | 1.00 | |
| Caucasian | 5 | 1.96 | 1.24–3.10 | 0.004 | Random | 81.35 | 0.0003 | 0.623 | 0.624 | |
| CYP1A1 rs1048943 | ||||||||||
| COD1 (AG vs. AA) | Overall | 18 | 1.48 | 1.16–1.90 | 0.002 | Random | 69.46 | <0.0001 | 0.766 | 0.520 |
| Asian | 15 | 1.37 | 1.06–1.77 | 0.015 | Random | 64.4 | 0.0003 | 0.762 | 0.805 | |
| Caucasian | 3 | 2.10 | 1.04–4.27 | 0.039 | Random | 77.38 | 0.012 | 0.770 | 0.602 | |
| COD2 (GG vs. AA) | Overall | 16 | 1.99 | 1.22–3.24 | 0.006 | Random | 66.09 | 0.0001 | 0.230 | 0.910 |
| Asian | 14 | 2.04 | 1.21–3.44 | 0.007 | Random | 68.83 | 0.0001 | 0.133 | 0.458 | |
| Caucasian | 2 | 2.09 | 0.72–6.01 | 0.173 | Fixed | 54.77 | 0.137 | 0.405 | 0.602 | |
| COD3 (GG vs. AG) | Overall | 16 | 1.22 | 0.78–1.93 | 0.386 | Random | 63.12 | 0.0004 | 0.160 | 0.970 |
| Asian | 14 | 1.34 | 0.82–2.19 | 0.243 | Random | 66.85 | 0.0002 | 0.066 | 0.656 | |
| Caucasian | 2 | 0.59 | 0.20–1.70 | 0.328 | Fixed | 0 | 0.439 | 0.603 | 0.602 | |
| DM (GG + AG vs. AA) | Overall | 18 | 1.51 | 1.16–1.97 | 0.002 | Random | 75.57 | <0.0001 | 0.882 | 0.970 |
| Asian | 15 | 1.43 | 1.08–1.90 | 0.014 | Random | 74.22 | <0.0001 | 0.866 | 0.961 | |
| Caucasian | 3 | 1.98 | 0.96–4.11 | 0.065 | Random | 79.26 | 0.008 | 0.655 | 0.602 | |
| RM (GG vs. AG + AA) | Overall | 16 | 1.59 | 0.99–2.55 | 0.057 | Random | 70.08 | <0.0001 | 0.088 | 0.910 |
| Asian | 14 | 1.67 | 1.00–2.80 | 0.049 | Random | 73.31 | <0.0001 | 0.047 | 0.520 | |
| Caucasian | 2 | 1.25 | 0.45–3.51 | 0.670 | Fixed | 27.73 | 0.240 | 0.493 | 0.602 | |
| OD (AG vs. GG + AA) | Overall | 18 | 1.38 | 1.09–1.75 | 0.007 | Random | 69.83 | <0.0001 | 0.739 | 0.850 |
| Asian | 15 | 1.27 | 1.00–1.61 | 0.050 | Random | 64.41 | 0.0003 | 0.807 | 0.520 | |
| Caucasian | 3 | 2.09 | 1.11–3.94 | 0.023 | Random | 72.58 | 0.026 | 0.740 | 0.602 | |
| AM (G vs. A) | Overall | 18 | 1.38 | 1.10–1.74 | 0.005 | Random | 81.66 | <0.0001 | 0.414 | 0.677 |
| Asian | 15 | 1.34 | 1.04–1.71 | 0.021 | Random | 82.24 | <0.0001 | 0.402 | 0.586 | |
| Caucasian | 3 | 1.65 | 0.95–2.88 | 0.076 | Random | 72.55 | 0.026 | 0.141 | 0.117 | |
COD1-Codominant 1; COD2-Codominant 2; COD3-Codominant 3; DM-Dominant model; RM-Recessive model; OD-Overdominant model; AM-Allele model; N-Number of studies.
Fig. 3.
(A) Forest plot (B) Funnel plot and (C) Sensitivity analysis plot for the meta-analysis of the link between CYP1A1 rs4646903 and rs1048943 polymorphisms and cervical cancer in codominant 1 model.
Again, rs1048943 revealed a significant correlation with CC in five association models for overall population, namely COD1 (AG vs. AA): OR = 1.48, 95% CI = 1.16–1.90, Pz = 0.002; COD2 (GG vs. AA): OR = 1.99, 95% CI = 1.22–3.24, Pz = 0.006; DM (GG + AG vs. AA): OR = 1.51, 95% CI = 1.16–1.97, Pz = 0.002; OD (AG vs. GG + AA): OR = 1.38, 95% CI = 1.09–1.75, Pz = 0.007; and AM (G vs. A): OR = 1.38, 95% CI = 1.10–1.74, Pz = 0.005) and for Asians, namely COD1 (AG vs. AA): OR = 1.37, 95% CI = 1.06–1.77, Pz = 0.015; COD2 (GG vs. AA): OR = 2.04, 95% CI = 1.21–3.44, Pz = 0.007; DM (GG + AG vs. AA): OR = 1.43, 95% CI = 1.08–1.90, Pz = 0.014; RM (GG vs. AG + AA): OR = 1.67, 95% CI = 1.00–2.80, Pz = 0.049; and AM (G vs. A): OR = 1.34, 95% CI = 1.04–1.71, Pz = 0.021). Besides, only COD1 (AG vs. AA: OR = 2.10, 95% CI = 1.04–4.27, Pz = 0.039) and OD (AG vs. GG + AA: OR = 2.09, 95% CI = 1.11–3.94, Pz = 0.023) of rs1048943 depicted substantial association with CC in the Caucasian population.
For the heterogeneity test, we applied the random-effect model for both SNPs except for COD2, COD3 and RM in Caucasians (Table 6). We used Begg's funnel plot and Egger's test to assess the publication bias as a part of our systemic evaluation, and both tests confirmed that there was no publication bias except Egger's test in COD3 and RM (Egger's value = 0.030 and 0.049, respectively) (Table 6 and Fig. 3). Each article in the meta-analysis was excluded one by one in sensitivity analysis to evaluate if individual data altered the pooled ORs. Using the step-by-step exclusion method, we were able to show that the findings of this meta-analysis were consistent among genetic models (Fig. 3).
4. Discussion
CC is a complex heterogeneous disease and the second most prevalent gynecological cancer behind breast carcinoma [34]. However, HPV infection acts as the primary etiological factor for the progression of CC; other environmental and socioeconomic factors like tobacco smoke, age of marriage and first pregnancy, use of contraceptives, and having more sexual partners also modify the cancer risk [35]. The immune system is well equipped to counter the viral attack, and in most infected individuals, the viral load is cleared out or becomes dormant by the immune responses. However, persistent HPV infection and other risk modifiers synergistically cause cellular transformations that lead to CC. Studies reported that inherent host genetic features have a relationship with the pathogenesis of CC and its progressions [36]. Our case-control study provides clear evidence of the link between CYP1A1 gene polymorphisms (rs4646903 and rs1048943) with CC in Bangladeshi females.
A lot of carcinogens (such as biphenyls, polyaromatic hydrocarbons and amines, and dibenzofurans) are metabolized by CYP1A1 (Phase I metabolizing enzyme) and subsequently metabolized by GSTM1 and GSTT1 (phase II detoxifying enzymes) [37]. But, various metabolites are not detoxified by enzymes, resulting in DNA damaging agents, which may cause cell damage and lead to mutations causing cancer [38]. Recent candidate gene-based investigations show how genetic polymorphisms alter enzymatic activities and predispose the risk of CC. The relationship between different SNPs in the CYP1A1 gene with different types of malignancies, such as pulmonary, bladder, oral, breast, and gynecological cancer in several ethnicities, has been evaluated rigorously [29].
In the present study, we found that women from urban areas are more susceptible to CC (60%) than rural women, and the majority of patients are Muslims (96.76%). Besides, patients over 40 years are more susceptible to CC, and a maximum number of women conceived their first child at 18 or below (63.27%). A case-control study in South India found that women married below 18 years old were more likely to be susceptible to CC [39]. Again, 78.43% of patients experienced the first symptom after 40 years of age. 55.56% of patients took contraceptive pills, among which 45.56% took them for at least 5 years. More than 210 million females currently take birth control pills or injections to control their pregnancy [40]. Another World Health Organization (WHO) report showed a relationship between CC risk and long-time intake of birth-control pills [41]. According to TNM, the maximum number of patients were with T1 tumor size (66.03%), negative lymph nodal status (83.96%), N1 nodal status (52.63%), and Mx distant metastasis (73.58%). We found that 13.22% of patients carried the familial risk of CC, and a maximum number of patients were suffering from squamous cell carcinoma (83.23%), stage IIB tumors (58.54%), and grade 2 (71.35%). A PCR-based line blot study in India found that the maximum number of cervical cancer patients (87.8%) carried squamous cell carcinomas [42]. Another study in the Indian population also showed similar findings [43].
Moreover, we traced the maximum prescribed medicines for CC patients in Bangladesh, which will be helpful in finding the best-acting medicines in the studied population. The diagnostic and treatment features of the study population were also recorded. Common drugs used during chemotherapy were 5-FU (46.84%), cisplatin (53.16%), carboplatin (36.71%), and paclitaxel injection (44.30%). Experts also recommend some common anti-cancer drugs, such as paclitaxel, cisplatin, carboplatin, and 5-FU to treat CC [44]. We have reported the common side effects and adverse reactions due to chemotherapy, including alopecia, anorexia, diarrhea, nausea, vomiting, fever, fatigue, irritations, pain etc., which were also reported by previous studies [45].
Genetic association analysis of CYP1A1 polymorphisms revealed that rs4646903 is significantly correlated with the risk of CC in all evaluated genetic association models, including TC (OR = 2.41) and CC (OR = 4.75) genotypes and dominant (OR = 2.67), over-dominant (OR = 2.13), recessive (OR = 3.61), and allele (OR = 2.44) models. After performing the Bonferroni correction (p < 0.017), the significance level of all associations was found to be stable. Again, the meta-analysis revealed that the OD genetic model of rs4646903 was substantially linked with an elevated risk of CC in the overall population (OR = 1.21). Besides, other six genetic models showed a significant link with CC in Caucasian populations (COD1: OR = 1.75, COD2: OR = 8.36, COD3: OR = 3.39, DM: OR = 2.02, RM: OR = 5.54, and AM: OR = 1.96). Several studies also found that rs4646903 SNP of CYP1A1 gene has an inter-relationship with CC in different ethnicities [17,18]. However, some studies failed to report any significant association in Chhattisgarh women [19], Jewish women [20], and Northeast Thai women [46] with CC recommending careful evaluation in the future.
Again, in the case of rs1048943, our study failed to demonstrate any notable link with CC susceptibility (p > 0.05) in the Bangladeshi population. However, meta-analysis revealed that COD1: OR = 1.48, COD2: OR = 1.99, DM: OR = 1.51, OD: OR = 1.38, and AM: OR = 1.38) are correlated with CC for the overall population and for Asians, COD1: OR = 1.37, COD2: OR = 2.04, DM: OR = 1.43, RM: OR = 1.67, and AM: OR = 1.34). Again, COD1 (OR = 2.10) and OD (OR = 2.09) of rs1048943 depicted a significant association with CC in Caucasians. Previous studies evaluated the relationship between CYP1A1 rs1048943 SNP and the development of different cancers like cervical, pharyngeal, prostate, pulmonary, oral, ovary, bladder, and colorectal carcinoma [47]. Only one study on Chinese ethnicity has found a relationship between rs1048943 and the risk of CC progression [48]. In line with our present outcomes, previous studies conducted in Polish [49], Jewish women [20], and the Japanese population [50] also did not find an association of rs1048943 with CC risk.
Importantly, we have also evaluated the risk association of CYP1A1 polymorphisms carriers respective to their demographic and clinicopathological features, which showed that rs4646903 carriers taking contraceptive pills (OR = 2.39) and for more than 5 years of pills intake (OR = 3.05) were linked with an increased risk of CC in dominant model. Again, rs1048943 carriers were significantly correlated with CC when they were >40 years (OR = 0.44), conceived their first child before 18 years (OR = 0.29), took contraceptive pills (OR = 2.18), or the duration of taking pills were more than five years (OR = 3.80). Evidence shows that women who had taken birth control pills for 5–9 years and who had taken these for more than 10 years were at three- and four-times higher risk of CC progression, respectively [41]. Studies on Australian [51], Thai [52], Jamaican [53], Iranian [54], and American women [55] proved that taking contraceptive pills for a prolonged time is a threat to developing CC. Again, first sexual intercourse at a premature age, more sexual partners, early pregnancy, family history, and multiparity are major threats to CC in developing countries [56]. Active or passive smoking is also a risk factor for CC, and studies found that married smoker couples are more prone to develop CC [57]. Though no history was found of smoking in our study population, we have found that older age, premature marriage and pregnancy, family history, and long-term contraceptive pill use are risk factors for CC.
Haplotype analysis of rs4646903 and rs1048943 with the risk of CC also demonstrated that AC haplotype significantly increased the risk of CC (OR = 4.86) but AT haplotype significantly reduced the risk (OR = 0.45). Moreover, both SNPs are found to be in strong linkage disequilibrium (D’ = 0.912 and r2 = 0.448). Besides, The SNP-SNP interaction of CYP1A1 rs4646903 CT with CYP1A1 rs1048943 AA significantly increased 4.69 times risk of CC (p < 0.0001). In addition to these, there was no substantial heterogeneity in our analysis, and sensitivity analysis confirmed the stability of the findings of the meta-analysis. The inclusion of meta-analysis with the case-control study has increased the acceptability as well as established the theme that the outcomes of genetic association studies may vary due to differences in ethnicity or geographical location.
Our study has some shortcomings that must be acknowledged to overcome in future research. Firstly, we only selected publicly available SNPs of the CYP1A1 gene other than novel SNPs. Secondly, the total number of participants is relatively small with low statistical power, especially for rs1048943 SNP, and the samples might not reveal the original scheme of the studied population. Thirdly, there was a lack of complete demographic and clinicopathological data, HPV information and survival data. This current study has provided a clear indication between CYP1A1 gene polymorphisms and the risk of CC development in Bangladeshi females without these limitations. The rapidly increasing prevalence and sufferings of women in our country are due to a lack of precaution and health care measures. Bangladesh lacks cutting-edge research and sufficient information for the proper treatment and management of cervical cancer. Our research aims to develop effective and affordable approaches to assess women's genetic variation-based risk of cervical cancer development, and we have already published six articles [30,[58], [59], [60], [61], [62]] to find the connection between different genetic variants with CC in our population. Our previous studies found significant associations between different genetic variants and CC. We hope our outcome will help clinicians manage early detection, ensuring better treatment options for CC patients. Furthermore, as this is the first study with these SNPs in Bangladeshi patients, it will also be helpful to make a cervical cancer biobank of this population.
5. Conclusion
Our case-control study and meta-analysis summarize that CYP1A1 rs4646903 and rs1048943 polymorphisms are correlated with CC risk. Moreover, carriers of these polymorphisms have a higher susceptibility to developing CC due to increased age, early pregnancy, or a history of contraceptives for more than five years. Haplotyping analysis revealed that rs4646903 and rs1048943 are in strong linkage disequilibrium, and AT and AC haplotypes are associated with cervical cancer in our study population. Future replication studies on these SNPs are recommended to get a more comprehensive association result on a large scale.
Declarations
Author contribution statement
Md Abdul Barek: Mohammad Anwarul Basher: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper
Md. Abdul Aziz: Md. Shafiul Hossen: Nusrat Jahan: Nahida Afroz: Mobashera Begum: Performed the experiments; Wrote the paper.
Sarah Jafrin: Mohammad Sarowar Uddin: Md. Shalahuddin Millat: Md. Mahmudul Hoque: Analyzed and interpreted the data; Wrote the paper.
Mohammad Safiqul Islam: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data will be made available on request.
Author contribution statement
Md Abdul Barek: Mohammad Anwarul Basher: Performed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Md. Abdul Aziz: Md. Shafiul Hossen: Nusrat Jahan: Nahida Afroz: Mobashera Begum: Performed the experiments; Wrote the paper.
Sarah Jafrin: Mohammad Sarowar Uddin: Md. Shalahuddin Millat: Md. Mahmudul Hoque: Analyzed and interpreted the data; Wrote the paper.
Mohammad Safiqul Islam: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data will be made available on request.
Research funding
Research Cell, Noakhali Science and Technology University and National Science and Technology Fellowship 2019, Ministry of Science and Technology, Bangladesh, have partially funded for conducting of this research work.
Ethics approval
The ethical committee of the National Institute of Cancer Research and Hospital (NICRH) reviewed the research protocol and consent form and approved the ethical clearance of the clinical part of this study. The ethical approval ID was NICRH/Ethics/2019/447.
Informed consent
Each and every volunteer were specifically informed of the study intent before blood collection, and each of the participants/or legal guardians signed a written consent regarding the study.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the authority of the National Institute of Cancer Research and Hospital, Dhaka, Bangladesh, for their cooperation and help during the collection of blood samples. We are also thankful to the Research Cell of Noakhali Science and Technology University and the Ministry of Science and Technology, Bangladesh, for funding partially.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17712.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Sargazi S., Kooshkaki O., Reza J.Z., et al. Mild antagonistic effect of Valproic acid in combination with AZD2461 in MCF-7 breast cancer cells. Med. J. Islam. Repub. Iran. 2019;33:29. doi: 10.34171/mjiri.33.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Small W., Bacon M.A., Bajaj A., et al. Cervical cancer: a global health crisis. Cancer. 2017;123:2404–2412. doi: 10.1002/cncr.30667. [DOI] [PubMed] [Google Scholar]
- 4.Shrestha A.D., Neupane D., Vedsted P., et al. Cervical cancer prevalence, incidence and mortality in low and middle income countries: a systematic review. Asian Pac. J. Cancer Prev. APJCP. 2018;19:319–324. doi: 10.22034/APJCP.2018.19.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sreedevi A., Javed R., Dinesh A. Epidemiology of cervical cancer with special focus on India. Int. J. Wom. Health. 2015;7:405–414. doi: 10.2147/IJWH.S50001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haque N., Uddin A., Dey B.R., et al. Challenges to cervical cancer treatment in Bangladesh: the development of a women's cancer ward at Dhaka Medical College Hospital. Gynecol. Oncol. Rep. 2017;21:67–72. doi: 10.1016/j.gore.2017.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harati-Sadegh M., Sargazi S., Saravani M., et al. Relationship between miR-143/145 cluster variations and cancer risk: proof from a meta-analysis. Nuc. Nucl. Nucl. 2021;40:578–591. doi: 10.1080/15257770.2021.1916030. [DOI] [PubMed] [Google Scholar]
- 8.Arbyn M., Weiderpass E., Bruni L., et al. Estimates of incidence and mortality of cervical cancer in 2018: a worldwide analysis. Lancet Global Health. 2020;8:e191–e203. doi: 10.1016/S2214-109X(19)30482-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Magnusson P.K., Lichtenstein P., Gyllensten U.B. Heritability of cervical tumours. Int. J. Cancer Res. 2000;88:698–701. doi: 10.1002/1097-0215(20001201)88:5<698::aid-ijc3>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.Sargazi S., Hajinezhad M.R., Rahdar A., et al. CoNi alloy nanoparticles for cancer theranostics: synthesis, physical characterization, in vitro and in vivo studies. Appl. Phys. A. 2021;127:1–12. [Google Scholar]
- 11.Mohammadzadeh V., Rahiman N., Hosseinikhah S.M., et al. Novel EPR-enhanced strategies for targeted drug delivery in pancreatic cancer: an update. J. Drug Deliv. Sci. Technol. 2022;73 [Google Scholar]
- 12.Vadlamuri S.V., Glover D.D., Turner T., et al. Regiospecific expression of cytochrome P4501A1 and 1B1 in human uterine tissue. Cancer Lett. 1998;122:143–150. doi: 10.1016/s0304-3835(97)00382-0. [DOI] [PubMed] [Google Scholar]
- 13.Lu J., Shang X., Zhong W., et al. New insights of CYP1A in endogenous metabolism: a focus on single nucleotide polymorphisms and diseases. Acta Pharm. Sin. B. 2020;10:91–104. doi: 10.1016/j.apsb.2019.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bag A., Jyala N.S., Bag N. Cytochrome P450 1A1 genetic polymorphisms as cancer biomarkers. Indian J. Cancer. 2015;52:479–489. doi: 10.4103/0019-509X.178380. [DOI] [PubMed] [Google Scholar]
- 15.Autrup H. Genetic polymorphisms in human xenobiotica metabolizing enzymes as susceptibility factors in toxic response. Mutat. Res. 2000;464:65–76. doi: 10.1016/s1383-5718(99)00167-9. [DOI] [PubMed] [Google Scholar]
- 16.Joseph T., Chacko P., Wesley R., et al. Germline genetic polymorphisms of CYP1A1, GSTM1 and GSTT1 genes in Indian cervical cancer: associations with tumor progression, age and human papillomavirus infection. Gynecol. Oncol. 2006;101:411–417. doi: 10.1016/j.ygyno.2005.10.033. [DOI] [PubMed] [Google Scholar]
- 17.Abbas M., Srivastava K., Imran M., et al. Association of CYP1A1 gene variants rs4646903 (T>C) and rs1048943 (A>G) with cervical cancer in a North Indian population. Eur. J. Obstet. Gynecol. Reprod. Biol. 2014;176:68–74. doi: 10.1016/j.ejogrb.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 18.Li S., Li G., Kong F., et al. The association of cyp1a1 gene with cervical cancer and additional snp-snp interaction in Chinese women. J. Clin. Lab. Anal. 2016;30:1220–1225. doi: 10.1002/jcla.22006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain V., Ratre Y.K., Amle D., et al. Polymorphism of CYP1A1 gene variants rs4646903 and rs1048943 relation to the incidence of cervical cancer in Chhattisgarh. Environ. Toxicol. Pharmacol. 2017;52:188–192. doi: 10.1016/j.etap.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 20.Gutman G., Morad T., Peleg B., et al. CYP1A1 and CYP2D6 gene polymorphisms in Israeli Jewish women with cervical cancer. Int. J. Gynecol. Cancer. 2009;19:1300–1302. doi: 10.1111/IGC.0b013e3181b9fa5d. [DOI] [PubMed] [Google Scholar]
- 21.Sloan F.A., Gelband H. National Academies Press; Washington (DC): 2007. Cancer Control Opportunities in Low- and Middle-Income Countries. [DOI] [PubMed] [Google Scholar]
- 22.Ingelman-Sundberg M. Human drug metabolising cytochrome P450 enzymes: properties and polymorphisms. Naunyn-Schmiedeb Arch Pharmacol. 2004;369:89–104. doi: 10.1007/s00210-003-0819-z. [DOI] [PubMed] [Google Scholar]
- 23.Harati-Sadegh M., Mohammadoo-Khorasani M., Sargazi S., et al. Quantitative assessment of the effects of IL-1ß-511 C> T variant on breast cancer risk: an updated meta-analysis of 3331 cases and 3609 controls. Lab. Med. 2021;52:36–46. doi: 10.1093/labmed/lmaa055. [DOI] [PubMed] [Google Scholar]
- 24.Sargazi S., Heidari Nia M., Mirinejad S., et al. Association of a novel KIF26B gene polymorphism with susceptibility to schizophrenia and breast cancer: a case-control study. Iran. J. Public Health. 2021;50:397–406. doi: 10.18502/ijph.v50i2.5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rezai M., Saravani R., Sargazi S., et al. Achillea Wilhelmsii C. Koch hydroalcoholic extract induces apoptosis and alters Lin28b and p53 gene expression in hela cervical cancer cells. Rep. Biochem. Mol. Biol. 2019;8:318–325. [PMC free article] [PubMed] [Google Scholar]
- 26.Sargazi S., Saravani R., Reza J.Z., et al. Novel Poly (Adenosine Diphosphate-Ribose) Polymerase (PARP) inhibitor, AZD2461, down-regulates VEGF and induces apoptosis in prostate cancer cells. Iran. Biomed. J. 2019;23:312–323. doi: 10.29252/.23.5.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sargazi S., Abghari A.Z., Sarani H., et al. Relationship between CASP9 and CASP10 gene polymorphisms and cancer susceptibility: evidence from an updated meta-analysis. Appl. Biochem. Biotechnol. 2021;193:4172–4196. doi: 10.1007/s12010-021-03613-w. [DOI] [PubMed] [Google Scholar]
- 28.Cordero K., Espinoza I., Caceres D., et al. Oral cancer susceptibility associated with the CYP1A1 and GSTM1 genotypes in Chilean individuals. Oncol. Lett. 2010;1:549–553. doi: 10.3892/ol_00000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H., Xiao D., Hu L., He T. Association of CYP1A1 polymorphisms with prostate cancer risk: an updated meta-analysis. Mol. Biol. Rep. 2012;39:10273–10284. doi: 10.1007/s11033-012-1904-5. [DOI] [PubMed] [Google Scholar]
- 30.Muhammad S.B., Hassan F., Bhowmik K.K., et al. Detection of association of IL1beta, IL4R, and IL6 gene polymorphisms with cervical cancer in the Bangladeshi women by tetra-primer ARMS-PCR method. Int. Immunopharm. 2021;90 doi: 10.1016/j.intimp.2020.107131. [DOI] [PubMed] [Google Scholar]
- 31.Islam M.S., Ahmed M.U., Sayeed M.S., et al. Lung cancer risk in relation to nicotinic acetylcholine receptor, CYP2A6 and CYP1A1 genotypes in the Bangladeshi population. Clin. Chim. Acta. 2013;416:11–19. doi: 10.1016/j.cca.2012.11.011. [DOI] [PubMed] [Google Scholar]
- 32.Morgan J.F. p Value fetishism and use of the Bonferroni adjustment. Evid. Base Ment. Health. 2007;10(2):34–35. doi: 10.1136/ebmh.10.2.34. [DOI] [PubMed] [Google Scholar]
- 33.Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 34.Wang L., Zhao W., Hong J., et al. Association between IL1B gene and cervical cancer susceptibility in Chinese Uygur Population: a case–control study. Mol. Genet. Gen. Med. 2019;7:e779. doi: 10.1002/mgg3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.National Center for Chronic Disease Prevention and Health Promotion (US) Office on Smoking and Health. Centers for Disease Control and Prevention (US); Atlanta (GA): 2014. http://purl.fdlp.gov/GPO/gpo45352 [Google Scholar]
- 36.Chen X., Jiang J., Shen H., et al. Genetic susceptibility of cervical cancer. J. Biomed. Res. 2011;25:155–164. doi: 10.1016/S1674-8301(11)60020-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pliarchopoulou K., Voutsinas G., Papaxoinis G., et al. Correlation of CYP1A1, GSTP1 and GSTM1 gene polymorphisms and lung cancer risk among smokers. Oncol. Lett. 2012;3:1301–1306. doi: 10.3892/ol.2012.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Crofts F., Taioll E., Trachman J., et al. Functional significance of different human CYPlAl genotypes. Carcinogenesis. 1994;15:2961–2963. doi: 10.1093/carcin/15.12.2961. [DOI] [PubMed] [Google Scholar]
- 39.Reichheld A., Mukherjee P.K., Rahman S.M., et al. Prevalence of cervical cancer screening and awareness among women in an urban community in South India-a cross sectional study. Ann. Glob. Heal. 2020;86 doi: 10.5334/aogh.2735. 30-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Urban M., Banks E., Egger S., et al. Injectable and oral contraceptive use and cancers of the breast, cervix, ovary, and endometrium in black South African women: case-control study. PLoS Med. 2012;9 doi: 10.1371/journal.pmed.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dyer O. WHO links long term pill use to cervical cancer. BMJ. 2002;324 doi: 10.1136/bmj.324.7341.808/a. 808-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sowjanya A.P., Jain M., Poli U.R., et al. Prevalence and distribution of high-risk human papilloma virus (HPV) types in invasive squamous cell carcinoma of the cervix and in normal women in Andhra Pradesh, India. BMC Infect. Dis. 2005;5 doi: 10.1186/1471-2334-5-116. 116-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sengupta D., Guha U., Mitra S., et al. Meta-analysis of polymorphic variants conferring genetic risk to cervical cancer in Indian women SupportsnCYP1A1 as an important associated locus. Asian Pac. J. Cancer Prev. APJCP. 2018;19(8):2071–2081. doi: 10.22034/APJCP.2018.19.8.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsuda N., Watari H., Ushijima K. Chemotherapy and molecular targeting therapy for recurrent cervical cancer. Chin. J. Cancer Res. 2016;28:241–253. doi: 10.21147/j.issn.1000-9604.2016.02.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saraswat N., Chopra A., Sood A., et al. A Descriptive study to analyze chemotherapy-induced hair loss and its psychosocial impact in adults: our experience from a tertiary care hospital. Ind. Dermat. Online J. 2019;10:426–430. doi: 10.4103/idoj.IDOJ_471_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wongpratate M., Ishida W., Phuthong S., et al. Genetic polymorphisms of the human cytochrome p450 1a1 (cyp1a1) and cervical cancer susceptibility among northeast Thai women. Asian Pac. J. Cancer Prev. APJCP. 2020;21:243–248. doi: 10.31557/APJCP.2020.21.1.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jin J.Q., Hu Y.Y., Niu Y.M., et al. CYP1A1 Ile462Val polymorphism contributes to colorectal cancer risk: a meta-analysis. World J. Gastroenterol. 2011;17:260–266. doi: 10.3748/wjg.v17.i2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geng J., Shi Y., Wang H., et al. Research of cytochrome p450 i a1 lle/val polymorphism and genetic susceptibility in cervical cancer. J. Bengbu Med. Coll. 2010;35:762. [Google Scholar]
- 49.Roszak A., Lianeri M., Sowinska A., et al. CYP1A1 Ile462Val polymorphism as a risk factor in cervical cancer development in the Polish population. Mol. Diagn. Ther. 2014;18:445–450. doi: 10.1007/s40291-014-0095-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugawara T., Nomura E., Sagawa T., et al. CYP1A1 polymorphism and risk of gynecological malignancy in Japan. Int. J. Gynecol. Cancer. 2003;13:785–790. doi: 10.1111/j.1525-1438.2003.13607.x. [DOI] [PubMed] [Google Scholar]
- 51.Chih H.J., Lee A.H., Colville L., et al. Condom and oral contraceptive use and risk of cervical intraepithelial neoplasia in Australian women. J. Gynecol. Oncol. 2014;25:183–187. doi: 10.3802/jgo.2014.25.3.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marks M., Gravitt P.E., Gupta S.B., et al. Combined oral contraceptive use increases HPV persistence but not new HPV detection in a cohort of women from Thailand. J. Infect. Dis. 2011;204:1505–1513. doi: 10.1093/infdis/jir560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McFarlane-Anderson N., Bazuaye P.E., Jackson M.D., et al. Cervical dysplasia and cancer and the use of hormonal contraceptives in Jamaican women. BMC Wom. Health. 2008;8:9. doi: 10.1186/1472-6874-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vaisy A., Lotfinejad S., Zhian F. Risk of cancer with combined oral contraceptive use among Iranian women. Asian Pac. J. Cancer Prev. APJCP. 2014;15:5517–5522. doi: 10.7314/apjcp.2014.15.14.5517. [DOI] [PubMed] [Google Scholar]
- 55.Ursin G., Peters R.K., Henderson B.E., et al. Oral contraceptive use and adenocarcinoma of cervix. Lancet. 1994;344:1390–1394. doi: 10.1016/s0140-6736(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 56.World Health Organization . Comprehensive Cervical Cancer Control. A Guide to Essential Practice; Geneva: 2014. https://www.ncbi.nlm.nih.gov/books/NBK269619/ [PubMed] [Google Scholar]
- 57.Trimble C.L., Genkinger J.M., Burke A.E., et al. Active and passive cigarette smoking and the risk of cervical neoplasia. Obstet. Gynecol. 2005;105:174–181. doi: 10.1097/01.AOG.0000148268.43584.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ivy S.C., Shabnaz S., Shahriar, et al. Association of RAD51 and XRCC2 gene polymorphisms with cervical cancer risk in the Bangladeshi women. Asian Pac. J. Cancer Prev. APJCP. 2021;22(7):2099–2107. doi: 10.31557/APJCP.2021.22.7.2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hasan M.E., Matin M., Haque M.E., et al. Polymorphic variants INSIG2 rs6726538, HLA-DRB1 rs9272143, and GCNT1P5 rs7780883 contribute to the susceptibility of cervical cancer in the Bangladeshi women. Cancer Med. 2021;10(5):1829–1838. doi: 10.1002/cam4.3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das S., Naher L., Aka T.D., et al. The ECCR1 rs11615, ERCC4 rs2276466, XPC rs2228000 and XPC rs2228001 polymorphisms increase the cervical cancer risk and aggressiveness in the Bangladeshi population. Heliyon. 2021;7(1) doi: 10.1016/j.heliyon.2021.e05919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Datta A., Zahora F.T., Aziz M.A., et al. Association study of IL10 gene polymorphisms (rs1800872 and rs1800896) with cervical cancer in the Bangladeshi women. Int. Immunopharm. 2020;89(Pt B) doi: 10.1016/j.intimp.2020.107091. [DOI] [PubMed] [Google Scholar]
- 62.Nazneen F., Millat M.S., Barek M.A., et al. Genetic polymorphism of miR-218-2 (rs11134527) in cervical cancer: a case-control study on the Bangladeshi women. MicroRNA. 2021;10(3):219–224. doi: 10.2174/2211536610666210715102554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.
Data will be made available on request.