Abstract
The Chiehyuan herbal oral protection solution (GB-2) is a herbal mixture commonly utilized in Taiwan for combating severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) as per traditional Chinese medicine practices. This study assessed the clinical impact of GB-2 through prospective clinical trials. With twice-daily use for a week, GB-2 was shown to diminish the expression of angiotensin-converting enzyme 2 (ACE2) in oral mucosal cells. Moreover, after two weeks of use, it could reduce transmembrane protease, serine 2 (TMRPSS2) expression in these cells. Additionally, in vitro experiments demonstrated that GB-2 lessened the entry efficiency of the Omicron, L452R–D614G, T478K–D614G, and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus. It also impeded the interaction between ACE2 and the receptor-binding domain (RBD) presenting N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R and L452R–T478K mutations. Glycyrrhizic acid, a major compound in GB-2, also hindered the entry of the Omicron variant (BA.1) of the SARS-CoV-2 pseudotyped lentivirus by obstructing the binding between ACE2 and the RBD presenting the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation. To sum up, these findings suggest that GB-2 can decrease ACE2 and TMPRSS2 expression in oral mucosal cells. Both glycyrrhizic acid and GB-2 were found to reduce the entry efficiency of the Omicron variant (BA.1) of the SARS-CoV-2 pseudotyped lentivirus and block the binding between ACE2 and the RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation. This evidence implies that GB-2 might be a potential candidate for further study as a preventative measure against SARS-CoV-2 infection.
Keywords: SARS-CoV-2, Spike protein, ACE2, Omicron variant, Mouth washing, TMPRSS2
Abbreviations
- ACE2
angiotensin converting enzyme 2
- TMPRSS2
Transmembrane protease, serine 2
- SARS-CoV-2
The severe acute respiratory syndrome coronavirus 2
- COVID-19
coronavirus disease 2019
- RBD
receptor binding domain
- T3G
Theaflavin 3-gallate
- GB-2
Chiehyuan herbal oral protection solution
- SEM
standard error of mean
- HPLC
High-performance Liquid Chromatography
- 293T-ACE2 cells
293T cells with over-expression of ACE2
- IHC assay
immunohistochemistry assay
- PBS
phosphate buffered saline
- BSA
bovine serum albumin
1. Introduction
COVID-19 has globally affected more than 576 million individuals, leading to 6.4 million fatalities. The responsible pathogen, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is characterized by a spike protein on its envelope that facilitates host cell infection [1]. The receptor-binding domain (RBD) within this spike protein directly engages with the peptidase domain of the angiotensin-converting enzyme 2 (ACE2) on host cells [2,3]. Once bound to ACE2, transmembrane proteases, serine 2 (TMPRSS2), and furin within host cells cleave and activate the spike protein, allowing SARS-CoV-2 to invade the host cells [[1], [2], [3], [4]].
SARS-CoV-2, an RNA virus, holds a higher mutation rate leading to a steadily increasing number of variants [[5], [6], [7]]. The World Health Organization has identified variants such as Alpha (B.1.1.7), Beta (B.1.351), Gamma (P.1), Delta (B.1.617.2), and Omicron (B.1.1.529) as variants of concern due to their increased transmissibility and resistance to neutralizing antibodies [8]. The Omicron and Delta variants have gained global dominance owing to their enhanced transmissibility [[9], [10], [11], [12], [13]]. In the Omicron variant, 30 mutations in the spike protein, predominantly located in the RBD, have been discovered [11,14]. These mutations in the Omicron variant relate to increased infectivity and a certain level of evasion from therapeutic antibodies [[15], [16], [17]]. In contrast, the Delta variant carries spike protein mutations T19R, L452R, T478K, D614G, P681R, D960N, and deletions at positions 157 and 158 [18]. The L452R mutation increases transmissibility and infectivity and reduces susceptibility to antibody neutralization [[19], [20], [21], [22], [23], [24], [25]]. These findings suggest that these variants threaten the protective effects of existing SARS-CoV-2 vaccines and treatments [26].
Various traditional medicinal herbs have been employed to mitigate COVID-19 via their immunomodulatory properties [[27], [28], [29]]. Chiehyuan herbal oral protection solution (GB-2), sourced from Tian Shang Sheng Mu of Chiayi Puzi Peitian Temple in Taiwan (嘉義朴子配天宮-天上聖母), is a prevalent herbal formulation in Taiwan and comprises Glycyrrhiza uralensis Fisch. and Camellia sinensis var. assamica (J.W.Mast.) Kitam. The therapeutic benefits of these herbal formulations have been highlighted in past traditional Chinese medicine studies, including the “Supplement to Compendium of Materia Medica” [30] in the context of infectious diseases. As stated in the Brush Records of Chouchi, oral rinsing with C. sinensis was employed to manage oral diseases [31]. Past research showed that GB-2 decreased both ACE2 and TMPRSS2 protein expression in cellular and animal models [32]. Moreover, a report detailed the inhibitory effect of GB-2 on the ACE2 and RBD binding with K417N–E484K–N501Y, K417N, N501Y, and L452R mutations [33]. However, the therapeutic impact of GB-2 remains undetermined. There is also ambiguity regarding how GB-2 interacts with various variants, including the Omicron and Delta variants. This research scrutinized the clinical implications of GB-2 on the expression of ACE2 and TMPRSS2 in oral mucosal cells via a clinical trial. Additionally, this investigation analyzed the suppressive effect of GB-2 and its principal compound on the entry competence of the Omicron variant (BA.1) and the L452R–D614G, T478K–D614G, and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus, alongside the interaction of ACE2 and RBD with N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R and L452R–T478K mutations.
2. Methods and materials
2.1. Preparation of Chiehyuan herbal oral protection solution (GB-2)
The Chiehyuan herbal oral protection solution, sourced from the Tian Shang Sheng Mu of Chiayi Puzi Peitian Temple in Taiwan (嘉義朴子配天宮-天上聖母), was procured from the Chinese pharmacy department of Chiayi Chang Gung Memorial Hospital. To briefly describe the process, 10 g of Glycyrrhiza uralensis Fisch., originating from Hangjinqi, Inner Mongolia (voucher reference: No.7H-E014, obtained from Chang Gung Memorial Hospital), and 25 g of Camellia sinensis var. assamica (J.W. Mast.) Kitam., originating from Chiayi city, Taiwan (voucher reference: No.7H-Y059, acquired from Chang Gung Memorial Hospital and Qingheyu Biological Technology Co., Ltd), were measured. Dr. Yu-Shih Lin, a pharmacist at the Chinese pharmacy department of Chiayi Chang Gung Memorial Hospital, authenticated all herbs. These raw herbs were then immersed in 2000 mL of water and boiled for 25 min in thermal flasks. The resulting water extracts (1500 mL) were then filtered using filter paper to eliminate any particulate matter, before being packaged for the clinical trial. For the in vitro experiments, the water extracts (1500 mL) were subjected to pressure reduction to create a viscous mass (6 g), which was then stored at −80 °C. For all experiments, the final concentrations of the tested compound were obtained by diluting the stock with water.
2.2. Quality control of GB-2
The Agilent 1100 High-performance Liquid Chromatography (HPLC) system was utilized to conduct the quality control of GB-2. The fingerprint chromatography for the approved formulation was performed using an HPLC method with a C18 column (4.6 mm × 150 mm, 5 μM, Discovery®). Mobile phase A consisted of 0.3% phosphoric acid, while mobile phase B was 100% acetonitrile. The mobile phase was programmed as follows: 90:10 at 0 min, 80:20 at 16.2 min, 40:60 at 23.4 min, and returning to 90:10 at 23.76 min. The flow rate was set at 0.7 mL/min with a detection wavelength of 278 nm and a column temperature of 30 °C. Theaflavin 3-gallate (acquired from ChromaDex, Irvine, CA, USA, Lot Number: 000258-841) and (+)-catechin (obtained from Sigma-Aldrich, Batch Number: BCCC3128) were employed as index compounds. For glycyrrhizic acid (sourced from ChromaDex, Irvine, CA, USA), a C18 column (4.6 mm × 150 mm, 5 μM, Discovery®) was used. The mobile phase for detection constituted 100% acetonitrile and 2% acetic acid in a ratio of 36% and 64% respectively, with a flow rate of 0.6 mL/min, detection wavelength of 278 nm, and column temperature of 25 °C.
2.3. Ethical approval and consent to participate
This forward-looking clinical study adhered to both ethical principles and the standards established for carrying out medical research in Taiwan. Approval for the study protocol was granted by the Chang Gung Memorial Hospital’s research ethics review committee, located in the Chia-Yi Branch in Taiwan, with Ethics Code: 202001919A3. The study has been recorded on the ClinicalTrials.gov site under trial number NCT05010928. All participants voluntarily joined the study and provided written informed consent prior to any data collection. Compliance with the Declaration of Helsinki and Tokyo guidelines for human research was ensured in all conducted experiments.
2.4. Subject enrollment and protocol of clinical trial
A total of 35 participants were engaged in the clinical trial from September 1, 2021, to October 31, 2021. Healthy adults, aged 20 years and above, were consecutively enrolled from the outpatient clinic of Otolaryngology-head and neck surgery at Chiayi Chang Gung Memorial Hospital. To minimize confounding factors, individuals with unhealed oral trauma, laceration, or wounds from recent oral surgery, as well as those undergoing radiotherapy or chemotherapy for oral tumors, were excluded from the study. Participants voluntarily chose to join either the experimental group, who used GB-2, or the control group, who used water as mouthwash. All participants underwent oral mucosal cell collection at the time of enrollment and again at one week, two weeks, and four weeks, to analyze the expression levels of ACE2 and TMPRSS2.
2.5. The protocol for using Chiehyuan herbal oral protection solution
The clinical impact of the Chiehyuan herbal oral protection solution was evaluated by implementing a modified version of the previously described clinical trial protocols [34,35]. Each time, one week of Chiehyuan herbal oral protection solution (14 packs in total, two packs per day, 100 mL per pack) was prepared to be sent to the patients, and the subjects were asked to store them in the refrigerator at 4 °C, and took them out of the refrigerator at 4 °C every day. Used mouthwash twice a day, one pack each morning and evening. The subjects followed the following steps: Pour the mouthwash into the mouth, fully gargle up and down, left, right, and left for 30–60 s, then spit out the mouthwash.
2.6. Oral mucosal cell collection and immunohistochemistry (IHC) assessment of ACE2 and TMPRSS2
To examine the effect of GB-2 on the expression of ACE2 and TMPRSS2 in human oral mucosal cells, these cells were collected and tested via IHC assessment. The process for mucosal cell collection was conducted as outlined in earlier works [36,37], while the IHC assessment followed procedures from a previous study [32,38]. Dr. Ming-Shao Tsai performed all mucosal collection and followed the steps below: a. Subject did not eat within 30 min before sampling. b. Rinsed mouth with boiling water for 10 s before sampling and vomited dry as much as possible. c. Used the sterile cotton swab in the package, lightly touched the inner wall of the cheek. d. Scraped the oral mucosa cells in a rotating manner and wiped them repeatedly for more than 20 times. e. Put the oral mucosal cell sample on the cotton stick on the glass slide f. Waited for the oral mucosal cells to dry naturally, soaked the slide in 95% alcohol, and let it rest for 5 min to fix. g. Washed the slide with 1xPBS for three times. h. Took 200 μl blocking buffer and gently dropped it on the slide, put it in a dark box and let it stand for 1 h in the dark. i. Took out the slide and wash three times with 1xPBS. j. Stained with ACE2 primary antibody (1:100 dilution; Bioss Antibodies, bs-1004R) or TMPRSS2 primary antibody (1:500 dilution; Abcam, ab92323). k. Peroxidase-linked goat anti-rabbit secondary antibodies were used. Photomicrographs were observed using Nikon TE3000 microscope. ImageJ (1.50d, USA) system was used to quantify integrated optical density per stained area (IOD/area) for ACE2 and TMPRSS2 staining.
2.7. Statistical analyses for clinical trial
All values were the means ± standard error of mean (SEM) (experimental group (n = 20) and control group (n = 13)). Differences between experimental group and control group were assessed using the unpaired two-tailed Student’s t-test. For testing the significance of pairwise group comparisons, the Tukey test was used. For all comparisons, P values of <0.05 were considered statistically significant. SPSS version 13.0 (SPSS Inc., Chicago, IL, USA) was used for all calculations.
2.8. Cell culture and treatment
293T cells (human embryonic kidney cell line) were obtained from the Bioresource Collection and Research Center, Taiwan. 293T cells were cultured in Dulbecco's Modified Eagle’s medium (DMEM, Invitrogen Corp., Carlsbad, Cat. Number: 11965-048), supplemented with 10% FBS at 37 °C and 5% CO2. pCEP4-myc-ACE2 plasmid was a gift from Erik Procko (Addgene plasmid # 141185; http://n2t.net/addgene:141185;RRID:Addgene_141185). pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmid was a gift from Erik Procko (Addgene plasmid # 141184; http://n2t.net/addgene:141184;RRID:Addgene_141184) [39]. pLENTI_hACE2_PURO was a gift from Raffaele De Francesco (Addgene plasmid # 155295; http://n2t.net/addgene:155295;RRID:Addgene_155295). psPAX2 was a gift from Didier Trono (Addgene plasmid # 12260; http://n2t.net/addgene:12260; RRID:Addgene_12260). pLV-eGFP was a gift from Pantelis Tsoulfas (Addgene plasmid # 36083; http://n2t.net/addgene:36083;RRID:Addgene_36083). HDM_SARS2_Spike_del21_D614G was a gift from Jesse Bloom (Addgene plasmid # 158762; http://n2t.net/addgene:158762; RRID:Addgene_158762) [40]. pTwist-SARS-CoV-2 Δ18 B.1.1.529 was a gift from Alejandro Balazs (Addgene plasmid # 179907; http://n2t.net/addgene:179907; RRID:Addgene_179907) [41]. Before treatment, 293T cells were cultured to 60–70% confluence. Then, cultured medium was replaced with fresh medium containing indicated compounds in water at the indicated concentrations. 293T cells treated with water were used as controls. 293T cells without treatment were used as blank control.
2.9. XTT assay (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide assay)
293T cells or 293T-ACE2 cells were propagated in DMEM supplemented with 10% FBS at a concentration of 1 × 103 cells per well in a 96-well plate. Post attachment, the culture medium was refreshed with new DMEM medium containing 10% FBS. Subsequently, the cells underwent treatment with specified drugs for a period of 24 h. The resultant absorbance was measured utilizing an XTT assay kit (Roche, catalog number: 11465015001) as directed by the manufacturer. The XTT formazan complex was quantified at 492 nm using an ELISA reader from Bio-Rad Laboratories, Inc.
2.10. Site-directed mutagenesis
Amino acid substitutions (G339D, K417N, L452R, T478K, E484K, Q493R, G496S, Q498R or N501Y) were induced in the pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmid or the HDM_SARS2_Spike_del21_D614G plasmid using the QuikChange Lightning Site-Directed Mutagenesis kit (Agilent Technologies Inc., Santa Clara, CA, Cat. Number: 210519). This was accomplished following the instructions provided by the manufacturer.
2.11. Flow cytometry analysis of ACE2-spike protein binding
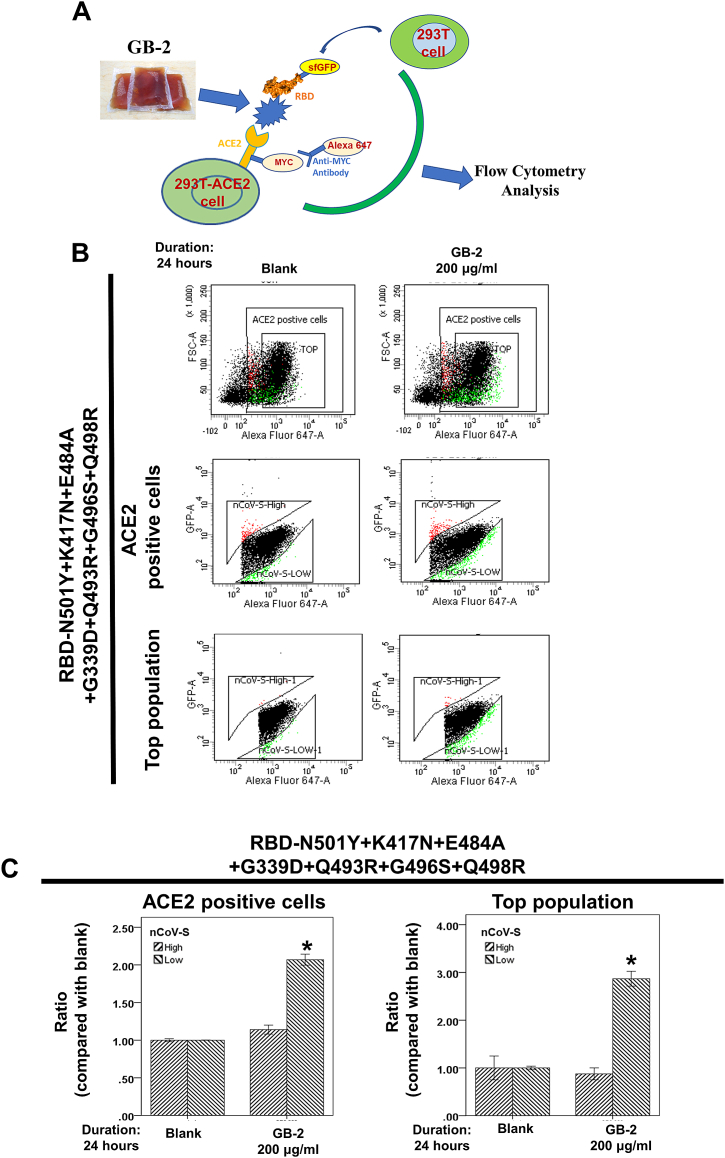
ACE2-Spike protein binding analysis via flow cytometry was conducted as outlined in prior studies [33,39,42]. A schematic diagram of this dual-color flow cytometry analysis is presented inFig. 6A. Initially, 293T cells were transfected with pCEP4-MYC-ACE2 or pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmids with specific mutations (500 ng DNA per mL of culture at 2 × 106/mL) using lipofectamine 2000 (ThermoFisher, Cat. Number: 11668-019). The medium containing RBD-sfGFP with specific mutations was gathered from 293T cells transfected with the pcDNA3-SARS-CoV-2-S-RBD-sfGFP plasmid with respective mutations after a 48-hour period. Subsequent to a 24-hour pretreatment with certain drugs, 293T cells transfected with the pCEP4-MYC-ACE2 plasmid were rinsed with ice-cold PBS-BSA and incubated for 30 min on ice with a medium composed of RBD-sfGFP and anti-MYC Alexa 647 (clone 9B11, Cell Signaling Technology, Cat. Number: 2233S). Lastly, the cells were washed twice with PBS-BSA and analyzed by a flow cytometer BD FACSCanto (Becton Dickinson) as described in prior research [39] (Fig. 6A).
Fig. 6.
Effect of GB-2 on interaction between ACE2 and SARS-CoV-2 spike with N501Y-K417N-E484A-G339D-Q493R-G496S-Q498R mutation. (A) The schematic diagram of this dual-color flow cytometry analysis. (B) Flow cytometry analysis of ACE2-Spike protein binding. 293T cells with pCEP4-MYC-ACE2 plasmid were incubated with RBD (N501Y-K417N-E484A-G339D-Q493R-G496S-Q498R)-sfGFP-containing medium and co-stained with anti-MYC Alexa 647 to detect surface ACE2 by flow cytometry. During analysis, the top population were chosen from the ACE2-positive population. Then, two subsets of the ACE2-positive population were collected: the top population (nCoV-S-High sort, red gate) and the bottom population (nCoV-S-Low sort, green gate) based on the fluorescence of bound RBD-sfGFP relative to ACE2 surface expression. (C) Quantitative results of nCoV-S-High sort and nCoV-S-low sort, which were presented as ratio compared with blank, in the top population or ACE2-positive population. All the results are representative of at least three independent experiments. (Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01, n = 3).
2.12. Creation of 293T cells with over-expression of ACE2 (293T-ACE2 cells) and Western’s blotting assay
Lentivirus carrying the human ACE2 was synthesized by co-transfecting 293T cells with the pLENTI_hACE2_PURO plasmid and lentiviral assistant plasmids (psPAX2). Produced lentivirus was utilized to infect 293T cells in the presence of 5 μg/mL polybrene. The transduced cells underwent selection via 3 μg/mL puromycin for 7 days. The clone demonstrating high ACE2 expression was further developed and named as 293T-ACE2 cells. Thereafter, the expression of ACE2 in 293T-ACE2 cells was confirmed using Western blotting, following the procedures described previously [32,38]. Protein extracts from 293T-ACE2 or 293T cells were prepared as indicated in past studies [43,44]. Proteins of equal amounts were separated on a 10% SDS-PAGE gel and transported to polyvinylidene difluoride membranes. These membranes were then blocked with 5% nonfat dried milk followed by a 12-hour incubation with primary antibodies at room temperature. The enlisted primary antibodies included: anti-ACE2 antibody (1:1000, Cell Signaling, #4355) and anti-GAPDH antibody (1:10000, GeneTex, GTX100118). Both primary and secondary antibodies were diluted in 1% nonfat dried milk in Tris-buffered saline with 0.1% Tween 20 detergent. Subsequently, the membranes were rinsed with 0.1% Tris-Buffered Saline Tween-20 and incubated with horseradish peroxidase-conjugated secondary anti-mouse or anti-rabbit antibodies (Santa Cruz, ratio: 1:5000) for 1 h at room temperature. Following this, the membranes were washed and protein signal was detected through chemiluminescence using the SuperSignal West Pico PLUS chemiluminescent substrate (Pierce, catalog number: 34087).
2.13. Generation of SARS-CoV-2 pseudotyped lentivirus
The production of SARS-CoV-2 pseudotyped lentivirus was conducted following established methods [40]. Twenty-four hours post-seeding, 293T cells were transfected with lentiviral backbone–pLV-eGFP plasmids (9 μg DNA per ml of culture at 2 × 106/ml), viral entry protein–either pTwist-SARS-CoV-2 Δ18 B.1.1.529 (Omicron variant) or HDM_SARS2_Spike_del21_D614G (3 μg DNA per ml of culture at 2 × 106/ml) with L452R–D614G, T478K–D614G or L452R–T478K–D614G mutations, pMD.G plasmid (VSV-G, positive control, supplied by Core Facility for Manipulation of Gene Function by RNAi, miRNA, miRNA sponges, and CRISPR, Academia Sinica, GB-2) and helper plasmids–psPAX2 (6 μg DNA per ml of culture at 2 × 106/ml) utilizing lipofectamine 2000 (ThermoFisher). After 24 h of transfection, the culture medium was refreshed. The virus was gathered 48 h post-transfection by procuring the supernatant and filtering it through a 0.45 μm low protein-binding filter. Subsequently, the virus could be stored at 4 °C for immediate application.
2.14. SARS-CoV-2 pseudotyped lentivirus infectivity assay
The infectivity assay of pseudotyped SARS-CoV-2 lentivirus was carried out following previously established methods [40]. 293T-ACE2 cells were exposed to the specified compounds for a duration of 24 h before the infection process. The supernatants that contained SARS-CoV-2 pseudotyped lentivirus were combined with these specified compounds for 30 min. Subsequently, an equivalent volume of supernatants encompassing SARS-CoV-2 pseudotyped lentivirus was introduced to the 293T-ACE2 cells, allowing for a 3-hour incubation period. The culture medium was then substituted with a fresh one, inclusive of the indicated drugs. Following a 48-hour incubation at 37 °C, the treated 293T-ACE2 cells underwent a double rinse process with PBS (phosphate buffered saline) - BSA (bovine serum albumin). Centrifugation of the treated 293T-ACE2 cells took place at 300 × g for 4 min, followed by a double rinse with 3% BSA in PBS. Post final rinse, these cells were resuspended in 1% BSA in PBS. Quantification of 293T-ACE2 cells infected by the GFP backbone SARS-CoV-2 pseudotyped lentivirus was achieved through enumeration of green cells, made possible by the flow cytometer BD FACSCanto (Becton Dickinson).
2.15. Statistical analyses
All reported values represent the means ± standard error of mean (SEM) from replicate samples (n = 3, based on the experiment). These experiments were conducted a minimum of three times. The unpaired two-tailed Student’s t-test was employed to examine differences between two groups. The Tukey test was utilized to assess the significance of pairwise group comparisons. A P value of <0.01 was deemed statistically significant in all comparisons. All computations were carried out using SPSS version 13.0 (SPSS Inc., Chicago, IL, USA).
3. Results
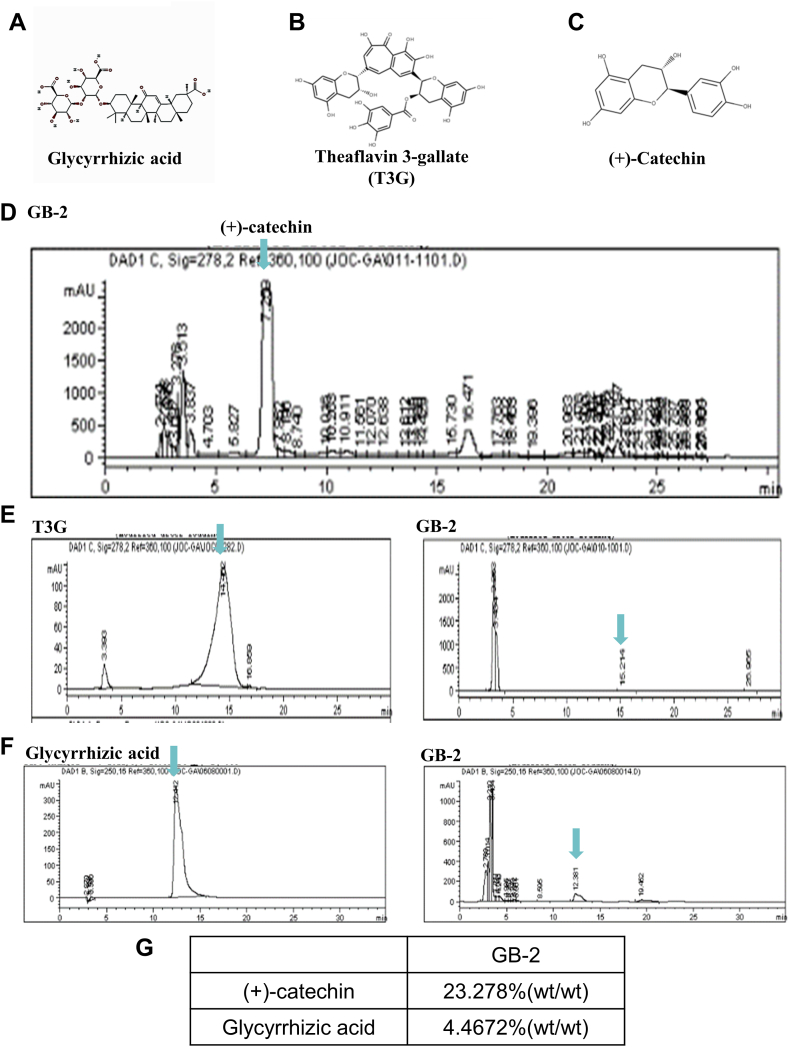
3.1. Identification of reference compounds in GB-2 by HPLC analysis
Glycyrrhizic acid (Fig. 1A), theaflavin 3-gallate (Fig. 1B), and (+)-Catechin (Fig. 1C) were utilized as reference standards to confirm the primary components within GB-2. The approved GB-2 fingerprint chromatography identified (+)-catechin as a reference compound in GB-2 using high-performance liquid chromatographic analysis (Fig. 1D). The concentration of (+)-catechin in GB-2 was recorded as 23.278 % (wt/wt) (Fig. 1G). Theaflavin 3-gallate was identified within Camellia sinensis var. assamica (J.W.Mast.) (Fig. 1E). Additionally, glycyrrhizic acid, present in Glycyrrhiza uralensis Fisch., was validated as a reference compound in GB-2 through high-performance liquid chromatographic analysis (Fig. 1F), and its concentration in GB-2 was determined to be 4.4672 % (wt/wt) (Fig. 1G).
Fig. 1.
The fingerprint chromatography of GB-2 by HPLC analysis. (A) The structure of glycyrrhizic acid. (B) The structure of theaflavin 3-gallate (T3G). (C) The structure of (+)-catechin. (D) The fingerprint chromatography of GB-2 and (+)-catechin were used as reference compounds. (E) HPLC chromatograms of theaflavin 3-gallate (T3G). (F) HPLC chromatograms of glycyrrhizic acid. (G) The concentrations of (+)-catechin and glycyrrhizic acid in GB-2.
3.2. The expression ratio of ACE2 and TMPRSS2 in oral mucosal cells after treated with GB-2
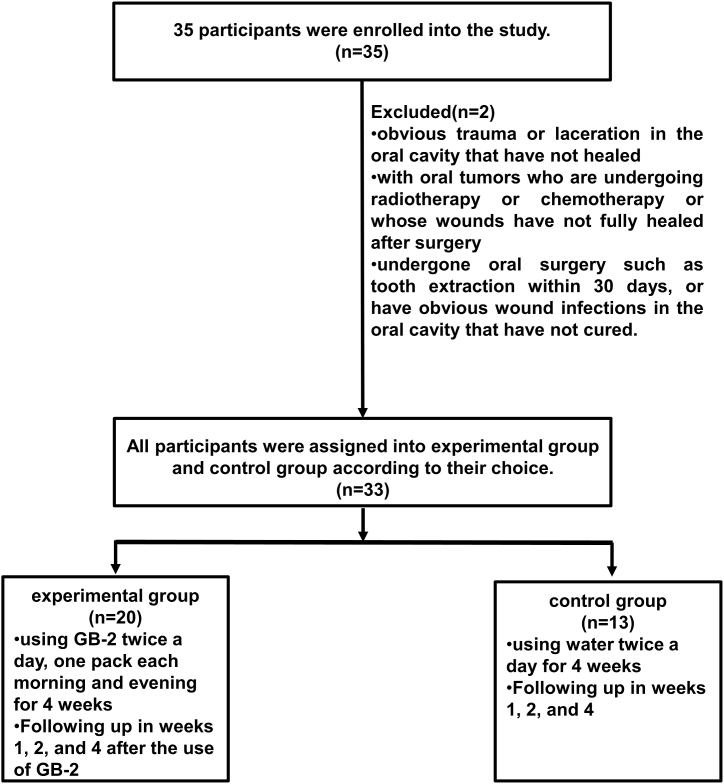
The prior research has indicated that GB-2 can suppress ACE2 mRNA expression along with ACE2 and TMPRSS2 protein expression in cellular and animal models [32]. To explore the clinical effects of GB-2, a prospective clinical trial was carried out. This trial involved 35 consecutive participants. However, due to the development of oral ulcers and bacterial infections, two participants were excluded. Hence, 33 participants, 20 in the experimental group using GB-2 and 13 in the control group not using any mouthwash, were included in the study (Fig. 2). The experimental group had 9 males and 11 females, aged between 24 and 56 years (average: 39.8, standard error [SEM]: 3.4). The control group consisted of 7 males and 6 females, aged between 20 and 50 years (average: 31.5, SEM: 2.2). No significant difference in age and gender distribution was found between the two groups.
Fig. 2.
Disposition of the subjects in this study.
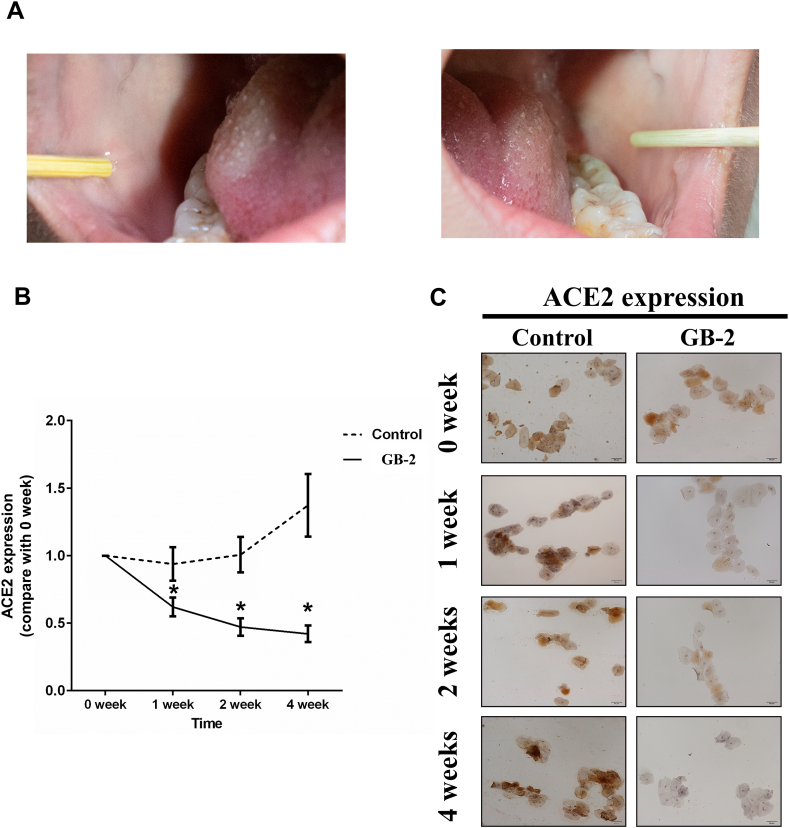
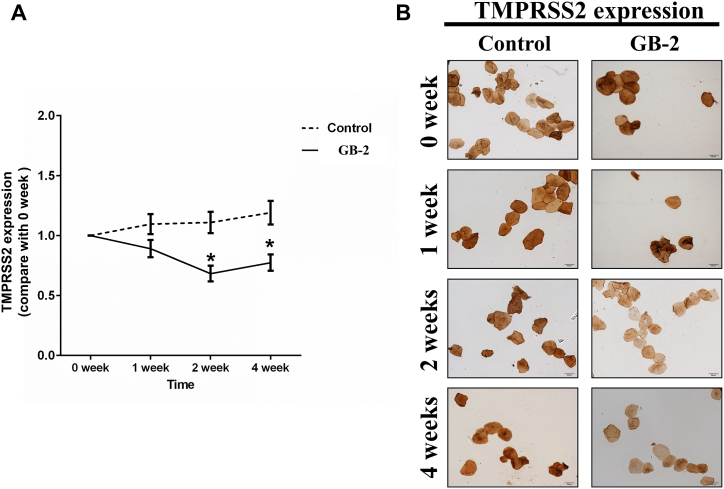
The outcome of the study showed a significant decrease in ACE2 expression in oral mucosal cells in the experimental group in weeks 1, 2, and 4 following the use of GB-2, as compared to the baseline (week 0, without GB-2). Conversely, ACE2 expression in oral mucosal cells of the control group remained virtually unchanged from baseline to weeks 1, 2, or 4 (Fig. 3B, C). After using the mouthwash, the TMPRSS2 ratio in the experimental group significantly reduced in weeks 2 and 4 compared to the baseline (week 0, without GB-2). The TMPRSS2 ratio in the control group showed no notable change from baseline at weeks 1, 2, or 4 (Fig. 4A, B). These findings indicate that gargling with GB-2 might inhibit the protein expression of ACE2 and TMPRSS2.
Fig. 3.
Effect of GB-2 on ACE2 expression of oral mucosal cells on clinical trial. (A) the oral mucosa cells on both sides of the buccal areas were collected, fixed and stained, and the results were analyzed. (B) Quantitative results of IHC staining, which were presented as IOD/area and were proportional to the level of ACE2 in oral mucosal cells. (C) Representative IHC staining photomicrographs of oral mucosal cells. (n = 20 in the experimental group and n = 13 in the control group, Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01).
Fig. 4.
Effect of GB-2 on TMPRSS2 expression of oral mucosal cells on clinical trial. (A) Quantitative results of IHC staining, which were presented as IOD/area and were proportional to the level of TMPRSS2 in oral mucosal cells. (B) Representative IHC staining photomicrographs of oral mucosal cells. (n = 20 in the experimental group and n = 13 in the control group, Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01).
3.3. Effect of GB-2 on entry efficiency of Omicron variant (BA.1) of SARS-CoV-2 pseudotyped lentivirus
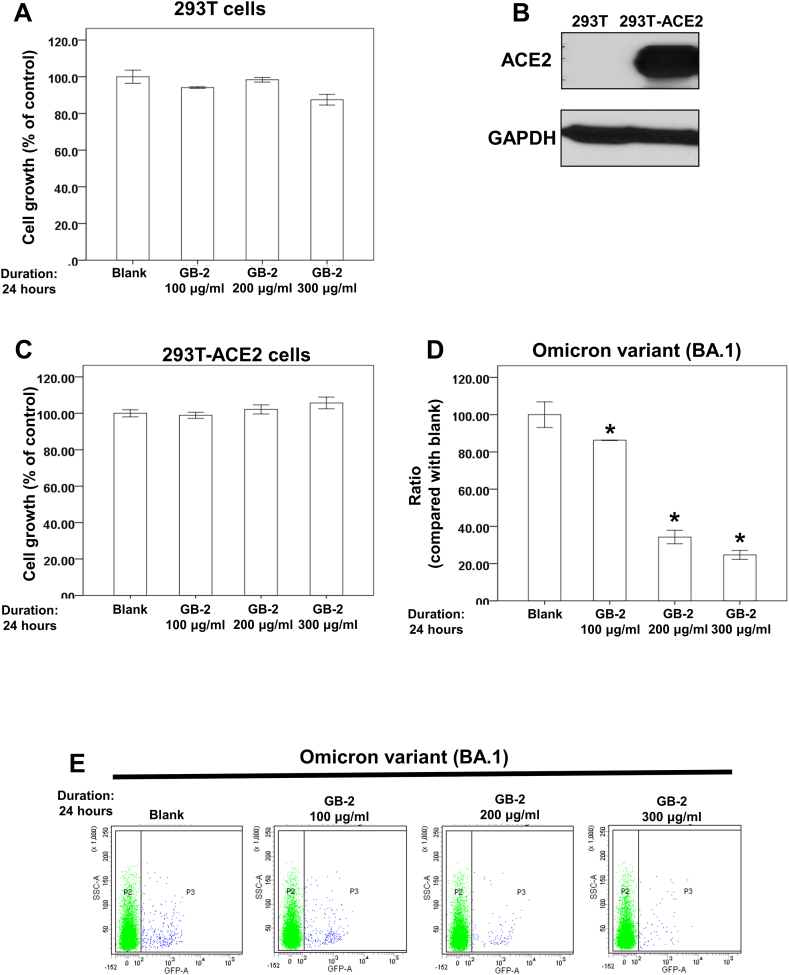
The impact of GB-2 on the entry efficiency of the Omicron variant (BA.1) was studied using 293T cells as the experimental model. An initial XTT assay was performed to determine the cytotoxicity of GB-2 on these cells. The findings indicated that within a concentration range of 100–300 μg/mL, GB-2 didn’t significantly hinder the proliferation of 293T cells after 24 h (Fig. 5A). This suggested that these concentrations of GB-2 do not induce notable cytotoxicity on 293T cells. Subsequently, the anti-infectivity of GB-2 on the Omicron variant of SARS-CoV-2 was examined using a SARS-CoV-2 pseudotyped lentivirus [40] in the model. 293T cells overexpressing ACE2 (293T-ACE2 cells) were included in the SARS-CoV-2 pseudotyped lentivirus infectivity test (Fig. 5B). It was observed that GB-2, in the range of 100–300 μg/mL, didn’t restrict the growth of the 293T-ACE2 cells after a 24-hour period (Fig. 5C). Finally, the influence of GB-2 on the entry efficiency of the Omicron variant (BA.1) of SARS-CoV-2 pseudotyped lentivirus was assessed using the infectivity assay. Following the outlined treatment, the inhibitory effect of GB-2 at concentrations between 100 and 300 μg/mL on the quantity of 293T-ACE2 cells infected with the Omicron variant (BA.1) was observed (Fig. 5C, D D and E). These observations imply that GB-2 has the ability to decrease the entry efficiency of the Omicron variant (BA.1) of SARS-CoV-2 pseudotyped lentivirus without causing substantial cytotoxicity.
Fig. 5.
Effect of GB-2 on entry efficiency of Omicron variant (BA.1) of SARS-CoV-2 pseudotyped lentivirus. (A) 293T cells were measured by XTT assay after 24 h of culturing in the presence of GB-2. (B) Total cell extracts of 293T-ACE2 or 293T cells were harvested. The protein was immunoblotted with polyclonal antibodies specific for ACE2. GAPDH was used as an internal loading control. (C) 293T-ACE2 cells were measured by XTT assay after 24 h of culturing in the presence of GB-2. (D) The entry efficiency of the GFP-backbone Omicron variant of SARS-CoV-2 pseudotyped lentivirus were quantified by counting 293T-ACE2 cells with GFP expression (blue spot) via flow cytometry analysis. (E) Quantitative results of 293T-ACE2 cells infected by Omicron variant of SARS-CoV-2 pseudotyped lentivirus. (Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01, n = 3).
3.4. Effect of GB-2 on the interaction between ACE2 and RBD with the N501Y-K417N-E484A-G339D-Q493R-G496S-Q498R mutation
Previous research has identified significant mutations (N501Y, K417N, and E484A) in the Omicron variant [11]. A docking analysis has also indicated that mutations Q493R, N501Y, and Q498R exhibit a heightened binding affinity to human ACE2, and mutations K417N, Q493R, and Q498R can enhance disease susceptibility in the Omicron variant [45]. An earlier study showed that GB-2 can impede the interaction between ACE2 and RBD with mutations K417N–E484K–N501Y, K417N, N501Y, and L452R in a dose-responsive manner [33]. The study subsequently examined the influence of GB-2 on the ACE2–mutated spike protein interaction in 293T cells, utilizing dual-color flow cytometry [39]. The schematic diagram of this dual-color flow cytometry analysis can be seen in Fig. 6A. 293T cells, expressing MYC-tagged ACE2, were incubated in a medium that contained the RBD fused with the superfolder green fluorescent protein and bearing the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation. Following treatment with specified concentrations of GB-2, the investigation examined cell populations expressing MYC-tagged ACE2 on the cell surface, which exhibited high or low binding to the RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation, using fluorescence-activated cell sorting. The analysis revealed a significant rise in the number of 293T cells with low binding to the RBD in both ACE2-positive and top cell populations post treatment with 200 μg/mL of GB-2 (Fig. 6B, C). These findings suggest that GB-2 inhibits the interaction between ACE2 and the RBD harboring the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation.
3.5. Effect of GB-2 on the entry efficiency of the L452R–D614G, T478K–D614G, and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus
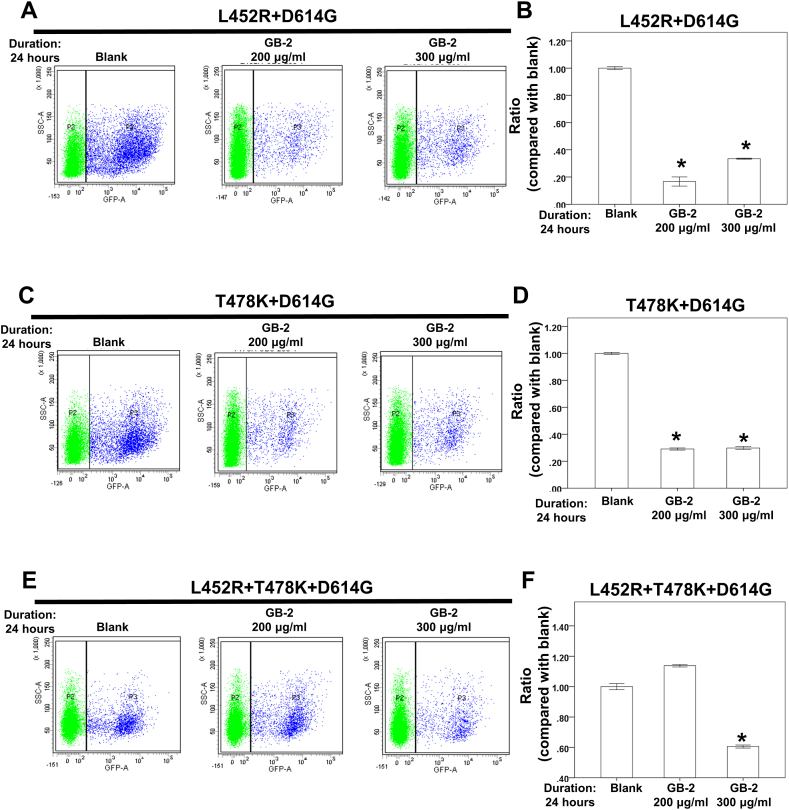
The Delta variant has been reported to exhibit greater infectivity and immune evasion capabilities [8], with L452R and T478K identified as the two critical mutations in the variant’s RBD [8]. Subsequently, the effect of GB-2 on the entry efficiency of L452R–D614G, T478K–D614G, and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus was examined by utilizing the SARS-CoV-2 pseudotyped lentivirus infectivity assay. It was observed that 200–300 μg/mL of GB-2 lessened the number of cells infected with the L452R–D614G (F. 7A, B) or T478K–D614G variants (Fig. 7C, D). Additionally, 300 μg/mL of GB-2 diminished the number of cells infected with the L452R–T478K–D614G variant (Fig. 7E, F). These observations suggest that GB-2 decreased the entry efficiency of the L452R–D614G, T478K–D614G, and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus.
Fig. 7.
Effect of GB-2 on entry efficiency of L452R–D614G, T478K–D614G and L452R–T478K–D614G variants of SARS-CoV-2 pseudotyped lentivirus. (A, C, E) The entry efficiency of the GFP-backbone L452R–D614G (A), T478K–D614G (C) and L452R–T478K–D614G (E) variants of SARS-CoV-2 pseudotyped lentivirus were quantified by counting 293T-ACE2 cells with GFP expression (blue spot) via flow cytometry analysis. (B, D, F) Quantitative results of 293T-ACE2 cells infected by L452R–D614G (B), T478K–D614G (D) and L452R–T478K–D614G (F) variants of SARS-CoV-2 pseudotyped lentivirus. (Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01, n = 3).
3.6. Effect of GB-2 on the interaction between ACE2 and RBD with the L452R–T478K mutation
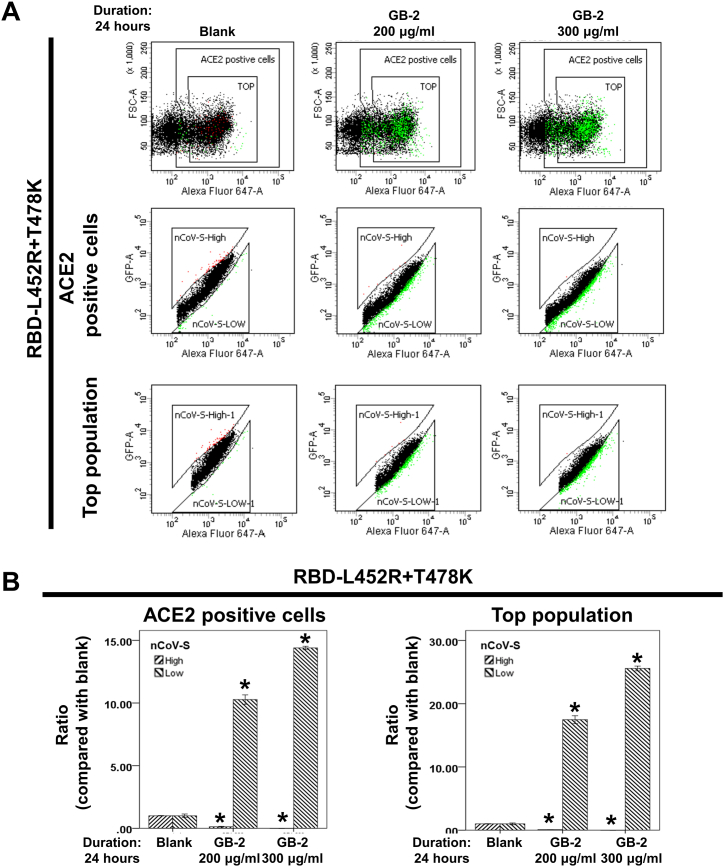
Observing that GB-2 reduces the entry efficiency of the L452R–T478K–D614G variant of the SARS-CoV-2 pseudotyped lentivirus prompted an investigation into GB-2's ability to inhibit the interaction between ACE2 and the RBD with the L452R–T478K mutation. This investigation was conducted using dual-color flow cytometry. The findings showed a substantial increase in the number of 293T-ACE2 cells displaying low binding to the RBD, seen in both the ACE2-positive and top cell populations, with this effect increasing in a dose-dependent manner (Fig. 8A, B). This provides evidence that GB-2 inhibits the binding between ACE2 and RBD with the L452R–T478K mutation.
Fig. 8.
Effect of GB-2 on interaction between ACE2 and SARS-CoV-2 spike with L452R–T478K mutation. (A) Flow cytometry analysis of ACE2-Spike protein binding. 293T cells with pCEP4-MYC-ACE2 plasmid were incubated with RBD (L452R–T478K)-sfGFP-containing medium and co-stained with anti-MYC Alexa 647 to detect surface ACE2 by flow cytometry. During analysis, the top population were chosen from the ACE2-positive population. Then, two subsets of the ACE2-positive population were collected: the top population (nCoV-S-High sort, red gate) and the bottom population (nCoV-S-Low sort, green gate) based on the fluorescence of bound RBD-sfGFP relative to ACE2 surface expression. (B) Quantitative results of nCoV-S-High sort and nCoV-S-low sort, which were presented as ratio compared with blank, in the top population or ACE2-positive population. All the results are representative of at least three independent experiments. (Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01, n = 3).
3.7. Effect of glycyrrhizic acid on the entry efficiency of the Omicron (BA.1) and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus
Glycyrrhizic acid (Fig. 1A), a principal compound in the G. uralensis Fisch. extract of GB-2, has been demonstrated to inhibit the interaction between ACE2 and the RBD with Wuhan strain of SARS-CoV-2, as the earlier findings [42]. To investigate further, the influence of glycyrrhizic acid on the entry efficiency of the Omicron (BA.1) and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus was assessed by performing the SARS-CoV-2 pseudotyped lentivirus infectivity assay. The assay revealed that 25–50 μg/mL of glycyrrhizic acid decreased the quantity of cells infected with the Omicron variant (Fig. 9A, B). In addition, it was observed that a concentration of 50 μg/mL of glycyrrhizic acid minimally reduced the number of cells infected with the L452R–T478K–D614G variant (Fig. 9C, D).
Fig. 9.
Effect of glycyrrhizic acid on entry efficiency of Omicron (BA.1) or L452R–T478K–D614G variants of SARS-CoV-2 pseudotyped lentivirus. (A, C) The effect of glycyrrhizic acid on the entry efficiency of the GFP-backbone Omicron (A) or L452R–T478K–D614G (B) variants of SARS-CoV-2 pseudotyped lentivirus were quantified by count-ing 293T-ACE2 cells with GFP expression (blue spot) via flow cytometry analysis. (B, D) Quantitative results of glycyrrhizic acid on 293T-ACE2 cells infected by Omicron (B) or L452R–T478K–D614G (D) variants of SARS-CoV-2 pseudotyped lentivirus. (Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01, n = 3).
3.8. Effect of glycyrrhizic acid on interaction between ACE2 and the RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation
The prior research highlighted the ability of glycyrrhizic acid to hinder the interaction between ACE2 and the RBD of the SARS-CoV-2 Wuhan strain, as evidenced by flow cytometry results [42]. The outcomes of the research showed that glycyrrhizic acid had a substantial inhibitory impact on the entry efficiency of the Omicron variant (BA.1). Further investigation was carried out to determine the effect of glycyrrhizic acid on the interaction between ACE2 and the RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation using dual-color flow cytometry [33,42]. The data indicated an increase in the number of 293T-ACE2 cells displaying low binding to the RBD in both the ACE2-positive and top cell populations following treatment with 25 μg/mL of glycyrrhizic acid (Fig. 10A, B). This infers that glycyrrhizic acid plays a role in the inhibition of the entry efficiency of the Omicron variant of the SARS-CoV-2 pseudotyped lentivirus. Furthermore, the interaction between ACE2 and RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation was found to be blocked by glycyrrhizic acid.
Fig. 10.
Effect of glycyrrhizic acid on interaction between ACE2 and SARS-CoV-2 spike with N501Y-K417N-E484A-G339D-Q493R-G496S-Q498R mutation. (A) Flow cytometry analysis of ACE2-Spike protein binding. 293T cells with pCEP4-MYC-ACE2 plasmid were incubated with RBD (N501Y-K417N-E484A-G339D-Q493R-G496S-Q498R)-sfGFP-containing medium and co-stained with anti-MYC Alexa 647 to detect surface ACE2 by flow cytometry. During analysis, the top population were chosen from the ACE2-positive population. Then, two subsets of the ACE2-positive population were collected: the top population (nCoV-S-High sort, red gate) and the bottom population (nCoV-S-Low sort, green gate) based on the fluorescence of bound RBD-sfGFP relative to ACE2 surface expression. (B) Quantitative results of nCoV-S-High sort and nCoV-S-low sort, which were presented as ratio compared with blank, in the top population or ACE2-positive population. All the results are representative of at least three independent experiments. (Error bars = mean ± S.E.M. Asterisks (*) mark samples significantly different from control group with p < 0.01, n = 3).
4. Discussion
Numerous researches have established that airborne transmission via aerosols is a critical pathway for SARS-CoV-2 infection [[46], [47], [48]]. One proposed method of safeguarding healthcare providers who carry out aerosol-generating procedures is to have patients perform pre-procedural rinses or mouthwashes to diminish the viral load in their saliva [49,50]. Compounds like povidone-iodine, chlorhexidine, and cetylpyridinium chloride have been demonstrated in several studies to have significant virucidal capabilities against SARS-CoV-2 in the saliva of COVID-19 patients [48,[51], [52], [53], [54], [55], [56]]. Prophylactic rinses or mouthwashes for healthcare providers, however, are still lacking. Previous studies have shown that GB-2 can inhibit ACE2 and TMPRSS2 protein expression in human cellular models and decrease ACE2 expression levels in lung and kidney tissues in animal models [32]. This study found that GB-2, when used twice daily for 1 week, can reduce ACE2 expression in oral mucosal cells. After 2 weeks of use, it can also reduce TMPRSS2 expression in oral mucosal cells. This suggests that GB-2 might be beneficial in reducing the risk of SARS-CoV-2 infection in healthcare providers over the long term.
The Omicron variant (B.1.1.529 variant) was designated as a variant of concern by the World Health Organization on November 26, 2021. This variant, with its increased transmissibility and potential for reinfection, has spread to over 80 countries worldwide. Some clinical antibodies and vaccines may be less effective against this variant due to its mutations [41,57]. A docking study indicated that Q493R, Q498R, T478K, and N501Y mutations play a crucial role in the variant’s high binding affinity to human ACE2 [45]. This study shows that both GB-2 and glycyrrhizic acid can inhibit the binding between ACE2 and the RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation. This may explain the capacity of GB-2 and glycyrrhizic acid to reduce the entry efficiency of the Omicron variant (BA.1) of the SARS-CoV-2 pseudotyped lentivirus. The impact of GB-2 and glycyrrhizic acid on the other mutations of the Omicron variant is still unclear, warranting further investigation.
In 2021, the Delta variant spread to over 100 countries. The key mutations in the RBD of this variant are L452R and T478K [8]. The L452R mutation has shown increased resistance to various neutralizing monoclonal antibodies (mAbs) and convalescent and vaccine plasma samples [[22], [23], [24], [25]]. Nonetheless, most mAbs and convalescent and vaccine plasma samples still show susceptibility to the T478K mutation [[58], [59], [60]]. SARS-CoV-2 pseudoviruses carrying the L452R mutation showed an increased entry efficiency into lung organoids compared to other SARS-CoV-2 pseudoviruses carrying the D614G mutation [19]. The D614G mutation started spreading in 2020, achieving global prevalence within several months [61]. In animal and cellular models, the virus with the D614G mutation enhanced SARS-CoV-2 infectivity and transmission [62]. Given that T478K–L452R–D614G mutations are the key mutations in the Delta variant, this study investigated the effect of GB-2 on these mutations. Results showed that 200–300 μg/mL of GB-2 reduced the entry efficiency of the L452R–D614G and T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus. Nevertheless, only a concentration of 300 μg/mL of GB-2 had a significant inhibitory effect on the entry efficiency of the L452R–D614G and T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus. The influence of GB-2 on viral entry efficiency might be affected by conformational alterations in the L452R–T478K-D614G variant.
The G. uralensis Fisch. extract is renowned for its medicinal uses across several nations. Different viruses, including the vesicular stomatitis virus from the Rhabdoviridae family, Epstein–Barr virus, Newcastle disease virus from the Paramyxoviridae family, human respiratory syncytial virus, herpes simplex virus 1, pseudorabies, and varicella zoster virus, have been shown to be susceptible to glycyrrhizin and licorice extracts [[63], [64], [65], [66]]. According to a docking study, glycyrrhizic acid could interact with the binding pocket of the SARS-CoV-2 spike glycoprotein (Protein Data Bank identifier 6VSB), suggesting its potential in preventing SARS-CoV-2 entry [67]. Additionally, glycyrrhizic acid was found to obstruct SARS-CoV-2 lentivirus infection by hindering the binding of the spike protein to host cells [68]. Previous findings also revealed that glycyrrhizic acid could disrupt the interaction between ACE2 and the RBD of the SARS-CoV-2 Wuhan strain as determined by flow cytometry [42]. It was also found that glycyrrhizic acid could be compatible with the RBD of both the Wuhan strain and Omicron variant [69]. The evidence in this study suggests that glycyrrhizic acid could limit the entry of the Omicron variant of the SARS-CoV-2 pseudotyped lentivirus and inhibit the binding of ACE2 with the RBD featuring the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation. These findings contribute to understanding the mechanism behind the anti-SARS-CoV-2 effect of glycyrrhizic acid. However, it was noted that only a partial inhibition of the entry of the L452R–T478K–D614G variant of the SARS-CoV-2 pseudotyped lentivirus was achieved with 50 μg/mL of glycyrrhizic acid. Conformational changes in these variants could potentially affect the impact of glycyrrhizic acid on viral entry efficiency.
The extract of C. sinensis var. assamica (J.W. Mast.) Kitam is a major component of GB-2. It was previously found that theaflavin-3-gallate, a compound of C. sinensis var. assamica (J.W. Mast.) Kitam, could interrupt the interaction between ACE2 and the RBD of the Wuhan-type SRAS-CoV-2 [33]. Another docking study indicated that theaflavin, another compound in C. sinensis var. assamica (J.W. Mast.) Kitam, could attach to SARS-CoV-2 RNA-dependent RNA polymerase with a low binding energy [70]. Catechin has been demonstrated to inactivate SARS-CoV-2 in vitro [71,72] and in silico studies [73,74]. These results suggest that the extract of C. sinensis var. assamica (J.W. Mast.) Kitam or its reference compounds might have a virucidal effect. Prior research has indicated that numerous traditional medicinal herbs containing potent bioactive compounds could prevent COVID-19 disease progression [27]. Future investigations should focus on the anti-infective capability of the extract of C. sinensis var. assamica (J.W. Mast.) Kitam and its reference compounds against different SARS-CoV-2 variants.
5. Conclusions
To sum up, using GB-2 as an oral rinse markedly diminished the expression of both ACE2 and TMPRSS2 in human oral mucosa cells without any adverse effects. GB-2 has demonstrated the ability to disrupt the binding between ACE2 and the RBD with N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R and L452R–T478K mutations. It has also shown potential in decreasing the entry efficiency of the Omicron (BA.1) and L452R–D614G, T478K–D614G, and L452R–T478K–D614G variants of the SARS-CoV-2 pseudotyped lentivirus. Furthermore, glycyrrhizic acid has been found to inhibit the entry of the Omicron variant (BA.1) of the SARS-CoV-2 pseudotyped lentivirus and to block the interaction between ACE2 and the RBD with the N501Y–K417N–E484A–G339D–Q493R–G496S–Q498R mutation.
Funding
Financial support was obtained from grant MOST 109-2320-B-182-021-MY3 from Ministry of Science and Technology (TW) and CORPG6K0211, CORPG6K0212 and CMRPG6L0241 from Chiayi Chang Gung Memorial Hospital.
Availability of data and materials
All data generated or analyzed during this study are indicated in this article (with no patient data). The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
Not applicable (not contain any individual person’s data).
Author contribution statement
Ching-Yuan Wu: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper; Contributed reagents, materials, analysis tools or data.
Yu-Shih Lin, Li-Hsin Shu, Yu-Ching Cheng, Pei-Rung Yang, Ying-Ying Tsai, Rou-Chen Shen and Hung-Te Liu: Performed the experiments; Ming-Shao Tsai: Analyzed and interpreted the data; Wrote the paper; Performed the experiments.
Yin-Yin Lin, I-Yun Lee, Wei-Tai Shih, Cheng-Ming Hsu, Reming-Albert Yeh, Yu-Huei Wu and Yu-Heng Wu: Analyzed and interpreted the data.
Yao-Hsu Yang and Geng-He Chang: Conceived and designed the experiments.
Pei-Rung Yang, Ying-Ying Tsai, Yu-Huei Wu and Yu-Heng Wu: Wrote the paper.
Institutional review board statement
This prospective clinical trial study was conducted in accordance with the ethical principles and the national norms and standards for conducting medical research in Taiwan. The study protocol was approved by the research ethics review committee of Chang Gung Memorial Hospital, Chia-Yi Branch, Taiwan (Ethics Code: 202001919A3). This study has been registered on the Clinical-Trials.gov website, and the trial number is NCT05010928. Prior to any data collection, all the subjects who voluntarily participated in this study and signed their written in-formed consent for inclusion. All experiments were performed in compliance with the research followed guidelines of the Declaration of Helsinki and Tokyo for humans.
Informed consent statement
Informed consent was obtained from all subjects involved in the study. Written informed consent has been obtained from the patient(s) to publish this paper.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgements
The authors acknowledge the Health Information and Epidemiology Labor-atory at the Chiayi Chang Gung Memorial Hospital for the comments and assistance in data analysis.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17701.
Contributor Information
Ching-Yuan Wu, Email: smbepigwu77@gmail.com.
Ming-Shao Tsai, Email: smbepig77work@gmail.com.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292 e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280 e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann M., Kleine-Weber H., Pohlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78(4):779–784 e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Duffy S. Why are RNA virus mutation rates so damn high? PLoS Biol. 2018;16(8) doi: 10.1371/journal.pbio.3000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L., Qin H., Wang M., Lu Q., Li X., Sun Q., Liu J., Zhang L., Li X., Huang W., Wang Y. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294 e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavda V.P., Patel A.B., Vaghasiya D.D. SARS-CoV-2 variants and vulnerability at the global level. J. Med. Virol. 2022;94(7):2986–3005. doi: 10.1002/jmv.27717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao K., Tzou P.L., Nouhin J., Gupta R.K., de Oliveira T., Kosakovsky Pond S.L., Fera D., Shafer R.W. The biological and clinical significance of emerging SARS-CoV-2 variants. Nat. Rev. Genet. 2021;22(12):757–773. doi: 10.1038/s41576-021-00408-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J., Xiao T., Cai Y., Lavine C.L., Peng H., Zhu H., Anand K., Tong P., Gautam A., Mayer M.L., Walsh R.M., Jr., Rits-Volloch S., Wesemann D.R., Yang W., Seaman M.S., Lu J., Chen B. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science. 2021 doi: 10.1126/science.abl9463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes Q., Inchakalody V.P., Merhi M., Mestiri S., Taib N., Moustafa Abo El-Ella D., Bedhiafi T., Raza A., Al-Zaidan L., Mohsen M.O., Yousuf Al-Nesf M.A., Hssain A.A., Yassine H.M., Bachmann M.F., Uddin S., Dermime S. Emerging COVID-19 variants and their impact on SARS-CoV-2 diagnosis, therapeutics and vaccines. Ann. Med. 2022;54(1):524–540. doi: 10.1080/07853890.2022.2031274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Araf Y., Akter F., Tang Y.D., Fatemi R., Parvez M.S.A., Zheng C., Hossain M.G. Omicron variant of SARS-CoV-2: genomics, transmissibility, and responses to current COVID-19 vaccines. J. Med. Virol. 2022;94(5):1825–1832. doi: 10.1002/jmv.27588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiecco G., Storti S., Degli Antoni M., Foca E., Castelli F., Quiros-Roldan E. Omicron genetic and clinical peculiarities that may overturn SARS-CoV-2 pandemic: a literature review. Int. J. Mol. Sci. 2022;23(4) doi: 10.3390/ijms23041987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Team C.-F. Past SARS-CoV-2 infection protection against re-infection: a systematic review and meta-analysis. Lancet. 2023;401 doi: 10.1016/S0140-6736(22)02465-5. 833-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daria S., Bhuiyan M.A., Islam M.R. Detection of highly muted coronavirus variant Omicron (B.1.1.529) is triggering the alarm for South Asian countries: associated risk factors and preventive actions. J. Med. Virol. 2022;94(4):1267–1268. doi: 10.1002/jmv.27503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shah M., Woo H.G. Omicron: a heavily mutated SARS-CoV-2 variant exhibits stronger binding to ACE2 and potently escapes approved COVID-19 therapeutic antibodies. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.830527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adhikari P., Jawad B., Podgornik R., Ching W.Y. Mutations of Omicron variant at the interface of the receptor domain motif and human angiotensin-converting enzyme-2. Int. J. Mol. Sci. 2022;23(5) doi: 10.3390/ijms23052870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar R., Murugan N.A., Srivastava V. Improved binding affinity of omicron’s spike protein for the human angiotensin-converting enzyme 2 receptor is the key behind its increased virulence. Int. J. Mol. Sci. 2022;23(6) doi: 10.3390/ijms23063409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shiehzadegan S., Alaghemand N., Fox M., Venketaraman V. Analysis of the Delta variant B.1.617.2 COVID-19. Clin. Pract. 2021;11(4):778–784. doi: 10.3390/clinpract11040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng X., Garcia-Knight M.A., Khalid M.M., Servellita V., Wang C., Morris M.K., Sotomayor-Gonzalez A., Glasner D.R., Reyes K.R., Gliwa A.S., Reddy N.P., Sanchez San Martin C., Federman S., Cheng J., Balcerek J., Taylor J., Streithorst J.A., Miller S., Sreekumar B., Chen P.Y., Schulze-Gahmen U., Taha T.Y., Hayashi J.M., Simoneau C.R., Kumar G.R., McMahon S., Lidsky P.V., Xiao Y., Hemarajata P., Green N.M., Espinosa A., Kath C., Haw M., Bell J., Hacker J.K., Hanson C., Wadford D.A., Anaya C., Ferguson D., Frankino P.A., Shivram H., Lareau L.F., Wyman S.K., Ott M., Andino R., Chiu C.Y. Transmission, infectivity, and neutralization of a spike L452R SARS-CoV-2 variant. Cell. 2021;184(13):3426–3437 e8. doi: 10.1016/j.cell.2021.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCallum M., Bassi J., De Marco A., Chen A., Walls A.C., Di Iulio J., Tortorici M.A., Navarro M.J., Silacci-Fregni C., Saliba C., Sprouse K.R., Agostini M., Pinto D., Culap K., Bianchi S., Jaconi S., Cameroni E., Bowen J.E., Tilles S.W., Pizzuto M.S., Guastalla S.B., Bona G., Pellanda A.F., Garzoni C., Van Voorhis W.C., Rosen L.E., Snell G., Telenti A., Virgin H.W., Piccoli L., Corti D., Veesler D. SARS-CoV-2 immune evasion by the B.1.427/B.1.429 variant of concern. Science. 2021;373(6555):648–654. doi: 10.1126/science.abi7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreira I., Kemp S.A., Datir R., Saito A., Meng B., Rakshit P., Takaori-Kondo A., Kosugi Y., Uriu K., Kimura I., Shirakawa K., Abdullahi A., Agarwal A., Ozono S., Tokunaga K., Sato K., Gupta R.K., I.S.-C.-G.C. Citiid-Nihr BioResource Covid-19 Collaboration. C. Genotype to Phenotype Japan SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J. Infect. Dis. 2021;224(6):989–994. doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Copin R., Baum A., Wloga E., Pascal K.E., Giordano S., Fulton B.O., Zhou A., Negron N., Lanza K., Chan N., Coppola A., Chiu J., Ni M., Wei Y., Atwal G.S., Hernandez A.R., Saotome K., Zhou Y., Franklin M.C., Hooper A.T., McCarthy S., Hamon S., Hamilton J.D., Staples H.M., Alfson K., Carrion R., Jr., Ali S., Norton T., Somersan-Karakaya S., Sivapalasingam S., Herman G.A., Weinreich D.M., Lipsich L., Stahl N., Murphy A.J., Yancopoulos G.D., Kyratsous C.A. The monoclonal antibody combination REGEN-COV protects against SARS-CoV-2 mutational escape in preclinical and human studies. Cell. 2021;184(15):3949–3961 e11. doi: 10.1016/j.cell.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., Alford B., Buchser W.J., Ellebedy A.H., Fremont D.H., Diamond M.S., Whelan S.P.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29(3):477–488 e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greaney A.J., Loes A.N., Gentles L.E., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci. Transl. Med. 2021;13(600) doi: 10.1126/scitranslmed.abi9915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferreira I., Kemp S.A., Datir R., Saito A., Meng B., Rakshit P., Takaori-Kondo A., Kosugi Y., Uriu K., Kimura I., Shirakawa K., Abdullahi A., Agarwal A., Ozono S., Tokunaga K., Sato K., Gupta R.K., I.S.-C.-G.C. Citiid-Nihr BioResource Covid-19 Collaboration, C Genotype to phenotype Japan, SARS-CoV-2 B.1.617 mutations L452R and E484Q are not synergistic for antibody evasion. J. Infect. Dis. 2021;224(6):989–994. doi: 10.1093/infdis/jiab368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chavda V.P., Kapadia C., Soni S., Prajapati R., Chauhan S.C., Yallapu M.M., Apostolopoulos V. A global picture: therapeutic perspectives for COVID-19. Immunotherapy. 2022;14(5):351–371. doi: 10.2217/imt-2021-0168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Al-Kuraishy H.M., Al-Fakhrany O.M., Elekhnawy E., Al-Gareeb A.I., Alorabi M., De Waard M., Albogami S.M., Batiha G.E. Traditional herbs against COVID-19: back to old weapons to combat the new pandemic. Eur. J. Med. Res. 2022;27(1):186. doi: 10.1186/s40001-022-00818-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chavda V.P., Patel A.B., Vihol D., Vaghasiya D.D., Ahmed K., Trivedi K.U., Dave D.J. Herbal remedies, nutraceuticals, and dietary supplements for COVID-19 management: an update. Clin Complement Med Pharmacol. 2022;2(1) doi: 10.1016/j.ccmp.2022.100021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malapela R.G., Thupayagale-Tshweneagae G., Baratedi W.M. Use of home remedies for the treatment and prevention of coronavirus disease: an integrative review. Health Sci Rep. 2023;6(1) doi: 10.1002/hsr2.900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xuemin Z. Supplement to Compendium of Materia Medica. 1765. [Google Scholar]
- 31.S. Shi, Brush Records of Chouchi, The Complete Library in Four Sections (960-1126).
- 32.Wu C.Y., Lin Y.S., Yang Y.H., Shu L.H., Cheng Y.C., Liu H.T. GB-2 inhibits ACE2 and TMPRSS2 expression: in vivo and in vitro studies. Biomed. Pharmacother. 2020;132 doi: 10.1016/j.biopha.2020.110816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsai M.S., Yang Y.H., Lin Y.S., Chang G.H., Hsu C.M., Yeh R.A., Shu L.H., Cheng Y.C., Liu H.T., Wu Y.H., Wu Y.H., Shen R.C., Wu C.Y. GB-2 blocking the interaction between ACE2 and wild type and mutation of spike protein of SARS-CoV-2. Biomed. Pharmacother. 2021;142 doi: 10.1016/j.biopha.2021.112011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Khan F.R., Kazmi S.M.R., Iqbal N.T., Iqbal J., Ali S.T., Abbas S.A. A quadruple blind, randomised controlled trial of gargling agents in reducing intraoral viral load among hospitalised COVID-19 patients: a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):785. doi: 10.1186/s13063-020-04634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrouel F., Viennot S., Valette M., Cohen J.M., Dussart C., Bourgeois D. Salivary and Nasal Detection of the SARS-CoV-2 Virus after Antiviral Mouthrinses (BBCovid): a structured summary of a study protocol for a randomised controlled trial. Trials. 2020;21(1):906. doi: 10.1186/s13063-020-04846-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Theda C., Hwang S.H., Czajko A., Loke Y.J., Leong P., Craig J.M. Quantitation of the cellular content of saliva and buccal swab samples. Sci. Rep. 2018;8(1):6944. doi: 10.1038/s41598-018-25311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang N., Perez P., Kato T., Mikami Y., Okuda K., Gilmore R.C., Conde C.D., Gasmi B., Stein S., Beach M., Pelayo E., Maldonado J.O., Lafont B.A., Jang S.I., Nasir N., Padilla R.J., Murrah V.A., Maile R., Lovell W., Wallet S.M., Bowman N.M., Meinig S.L., Wolfgang M.C., Choudhury S.N., Novotny M., Aevermann B.D., Scheuermann R.H., Cannon G., Anderson C.W., Lee R.E., Marchesan J.T., Bush M., Freire M., Kimple A.J., Herr D.L., Rabin J., Grazioli A., Das S., French B.N., Pranzatelli T., Chiorini J.A., Kleiner D.E., Pittaluga S., Hewitt S.M., Burbelo P.D., Chertow D., Consortium N.C.-A., Oral H.C.A., Craniofacial Biological N., Frank K., Lee J., Boucher R.C., Teichmann S.A., Warner B.M., Byrd K.M. SARS-CoV-2 infection of the oral cavity and saliva. Nat. Med. 2021;27(5):892–903. doi: 10.1038/s41591-021-01296-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu C.Y., Lin Y.S., Yang Y.H., Shu L.H., Cheng Y.C., Liu H.T. Potential simultaneous inhibitors of angiotensin-converting enzyme 2 and transmembrane protease, serine 2. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.584158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chan K.K., Dorosky D., Sharma P., Abbasi S.A., Dye J.M., Kranz D.M., Herbert A.S., Procko E. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science. 2020;369(6508):1261–1265. doi: 10.1126/science.abc0870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford K.H.D., Eguia R., Dingens A.S., Loes A.N., Malone K.D., Wolf C.R., Chu H.Y., Tortorici M.A., Veesler D., Murphy M., Pettie D., King N.P., Balazs A.B., Bloom J.D. Protocol and reagents for pseudotyping lentiviral particles with SARS-CoV-2 spike protein for neutralization assays. Viruses. 2020;12(5) doi: 10.3390/v12050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia-Beltran W.F., St Denis K.J., Hoelzemer A., Lam E.C., Nitido A.D., Sheehan M.L., Berrios C., Ofoman O., Chang C.C., Hauser B.M., Feldman J., Roederer A.L., Gregory D.J., Poznansky M.C., Schmidt A.G., Iafrate A.J., Naranbhai V., Balazs A.B. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell. 2022;185(3):457–466 e4. doi: 10.1016/j.cell.2021.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai M.S., Shih W.T., Yang Y.H., Lin Y.S., Chang G.H., Hsu C.M., Yeh R.A., Shu L.H., Cheng Y.C., Liu H.T., Wu Y.H., Wu Y.H., Shen R.C., Wu C.Y. Potential inhibitor for blocking binding between ACE2 and SARS-CoV-2 spike protein with mutations. Biomed. Pharmacother. 2022;149 doi: 10.1016/j.biopha.2022.112802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lin Y.Y., Lee I.Y., Huang W.S., Lin Y.S., Kuan F.C., Shu L.H., Cheng Y.C., Yang Y.H., Wu C.Y. Danshen improves survival of patients with colon cancer and dihydroisotanshinone I inhibit the proliferation of colon cancer cells via apoptosis and skp2 signaling pathway. J. Ethnopharmacol. 2017;209:305–316. doi: 10.1016/j.jep.2017.08.011. [DOI] [PubMed] [Google Scholar]
- 44.Lee I.Y., Lin Y.Y., Yang Y.H., Lin Y.S., Lin C.L., Lin W.Y., Cheng Y.C., Shu L.H., Wu C.Y. Dihydroisotanshinone I combined with radiation inhibits the migration ability of prostate cancer cells through DNA damage and CCL2 pathway. BMC Pharmacol Toxicol. 2018;19(1):5. doi: 10.1186/s40360-018-0195-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumar S., Thambiraja T.S., Karuppanan K., Subramaniam G. Omicron and Delta variant of SARS-CoV-2: a comparative computational study of spike protein. J. Med. Virol. 2022;94(4):1641–1649. doi: 10.1002/jmv.27526. [DOI] [PubMed] [Google Scholar]
- 46.Morawska L., Milton D.K. It is time to address airborne transmission of coronavirus disease 2019 (COVID-19) Clin. Infect. Dis. 2020;71(9):2311–2313. doi: 10.1093/cid/ciaa939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng X., Xu X., Li Y., Cheng L., Zhou X., Ren B. Transmission routes of 2019-nCoV and controls in dental practice. Int. J. Oral Sci. 2020;12(1):9. doi: 10.1038/s41368-020-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Garcia-Sanchez A., Pena-Cardelles J.F., Salgado-Peralvo A.O., Robles F., Ordonez-Fernandez E., Ruiz S., Vegh D. Virucidal activity of different mouthwashes against the salivary load of SARS-CoV-2: a narrative review. Healthcare (Basel) 2022;10(3) doi: 10.3390/healthcare10030469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marui V.C., Souto M.L.S., Rovai E.S., Romito G.A., Chambrone L., Pannuti C.M. Efficacy of preprocedural mouthrinses in the reduction of microorganisms in aerosol: a systematic review. J. Am. Dent. Assoc. 2019;150(12):1015–1026 e1. doi: 10.1016/j.adaj.2019.06.024. [DOI] [PubMed] [Google Scholar]
- 50.Carrouel F., Conte M.P., Fisher J., Goncalves L.S., Dussart C., Llodra J.C., Bourgeois D. COVID-19: a recommendation to examine the effect of mouthrinses with beta-cyclodextrin combined with citrox in preventing infection and progression. J. Clin. Med. 2020;9(4) doi: 10.3390/jcm9041126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Sanchez A., Pena-Cardelles J.F., Ruiz S., Robles F., Ordonez-Fernandez E., Salgado-Peralvo A.O., Balloch J., Simon J.C. Efficacy of pre-procedural mouthwashes against SARS-CoV-2: a systematic review of randomized controlled trials. J. Clin. Med. 2022;11(6) doi: 10.3390/jcm11061692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jain A., Grover V., Singh C., Sharma A., Das D.K., Singh P., Thakur K.G., Ringe R.P. Chlorhexidine: an effective anticovid mouth rinse. J. Indian Soc. Periodontol. 2021;25(1):86–88. doi: 10.4103/jisp.jisp_824_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu C., Wang A., Hoskin E.R., Cugini C., Markowitz K., Chang T.L., Fine D.H. Differential effects of antiseptic mouth rinses on SARS-CoV-2 infectivity in vitro. Pathogens. 2021;10(3) doi: 10.3390/pathogens10030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Seneviratne C.J., Balan P., Ko K.K.K., Udawatte N.S., Lai D., Ng D.H.L., Venkatachalam I., Lim K.S., Ling M.L., Oon L., Goh B.T., Sim X.Y.J. Efficacy of commercial mouth-rinses on SARS-CoV-2 viral load in saliva: randomized control trial in Singapore. Infection. 2021;49(2):305–311. doi: 10.1007/s15010-020-01563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaudhary P., Melkonyan A., Meethil A., Saraswat S., Hall D.L., Cottle J., Wenzel M., Ayouty N., Bense S., Casanova F., Chaney M., Chase H., Hermel R., McClement M., Sesson C., Woolsey B., Kumar P. Estimating salivary carriage of severe acute respiratory syndrome coronavirus 2 in nonsymptomatic people and efficacy of mouthrinse in reducing viral load: a randomized controlled trial. J. Am. Dent. Assoc. 2021;152(11):903–908. doi: 10.1016/j.adaj.2021.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Eduardo F.P., Correa L., Heller D., Daep C.A., Benitez C., Malheiros Z., Stewart B., Ryan M., Machado C.M., Hamerschlak N., Rebello Pinho J.R., Bezinelli L.M. Salivary SARS-CoV-2 load reduction with mouthwash use: a randomized pilot clinical trial. Heliyon. 2021;7(6) doi: 10.1016/j.heliyon.2021.e07346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.VanBlargan L.A., Errico J.M., Halfmann P.J., Zost S.J., Crowe J.E., Jr., Purcell L.A., Kawaoka Y., Corti D., Fremont D.H., Diamond M.S. An infectious SARS-CoV-2 B.1.1.529 Omicron virus escapes neutralization by therapeutic monoclonal antibodies. Nat. Med. 2022;28(3):490–495. doi: 10.1038/s41591-021-01678-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Greaney A.J., Loes A.N., Crawford K.H.D., Starr T.N., Malone K.D., Chu H.Y., Bloom J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe. 2021;29(3):463–476 e6. doi: 10.1016/j.chom.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371(6531):850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D., Dingens A.S., Nargi R.S., Sutton R.E., Suryadevara N., Rothlauf P.W., Liu Z., Whelan S.P.J., Carnahan R.H., Crowe J.E., Jr., Bloom J.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29(1):44–57 e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield C.-G.G., McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827 e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hou Y.J., Chiba S., Halfmann P., Ehre C., Kuroda M., Dinnon K.H., 3rd, Leist S.R., Schafer A., Nakajima N., Takahashi K., Lee R.E., Mascenik T.M., Graham R., Edwards C.E., Tse L.V., Okuda K., Markmann A.J., Bartelt L., de Silva A., Margolis D.M., Boucher R.C., Randell S.H., Suzuki T., Gralinski L.E., Kawaoka Y., Baric R.S. SARS-CoV-2 D614G variant exhibits efficient replication ex vivo and transmission in vivo. Science. 2020;370(6523):1464–1468. doi: 10.1126/science.abe8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Diomede L., Beeg M., Gamba A., Fumagalli O., Gobbi M., Salmona M. Can antiviral activity of licorice help fight COVID-19 infection? Biomolecules. 2021;11(6) doi: 10.3390/biom11060855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sui X., Yin J., Ren X. Antiviral effect of diammonium glycyrrhizinate and lithium chloride on cell infection by pseudorabies herpesvirus. Antivir. Res. 2010;85(2):346–353. doi: 10.1016/j.antiviral.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Feng Yeh C., Wang K.C., Chiang L.C., Shieh D.E., Yen M.H., San Chang J. Water extract of licorice had anti-viral activity against human respiratory syncytial virus in human respiratory tract cell lines. J. Ethnopharmacol. 2013;148(2):466–473. doi: 10.1016/j.jep.2013.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pompei R., Flore O., Marccialis M.A., Pani A., Loddo B. Glycyrrhizic acid inhibits virus growth and inactivates virus particles. Nature. 1979;281(5733):689–690. doi: 10.1038/281689a0. [DOI] [PubMed] [Google Scholar]
- 67.Sinha S.K., Prasad S.K., Islam M.A., Gurav S.S., Patil R.B., AlFaris N.A., Aldayel T.S., AlKehayez N.M., Wabaidur S.M., Shakya A. Identification of bioactive compounds from Glycyrrhiza glabra as possible inhibitor of SARS-CoV-2 spike glycoprotein and non-structural protein-15: a pharmacoinformatics study. J. Biomol. Struct. Dyn. 2020:1–15. doi: 10.1080/07391102.2020.1779132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li J., Xu D., Wang L., Zhang M., Zhang G., Li E., He S. Glycyrrhizic acid inhibits SARS-CoV-2 infection by blocking spike protein-mediated cell attachment. Molecules. 2021;26(20) doi: 10.3390/molecules26206090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vardhan S., Sahoo S.K. Computational studies on the interaction of SARS-CoV-2 Omicron SGp RBD with human receptor ACE2, limonin and glycyrrhizic acid. Comput. Biol. Med. 2022;144 doi: 10.1016/j.compbiomed.2022.105367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lung J., Lin Y.S., Yang Y.H., Chou Y.L., Shu L.H., Cheng Y.C., Liu H.T., Wu C.Y. The potential chemical structure of anti-SARS-CoV-2 RNA-dependent RNA polymerase. J. Med. Virol. 2020;92(6):693–697. doi: 10.1002/jmv.25761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nishimura H., Okamoto M., Dapat I., Katsumi M., Oshitani H. Inactivation of SARS-CoV-2 by catechins from green tea. Jpn. J. Infect. Dis. 2021;74(5):421–423. doi: 10.7883/yoken.JJID.2020.902. [DOI] [PubMed] [Google Scholar]
- 72.Ohgitani E., Shin-Ya M., Ichitani M., Kobayashi M., Takihara T., Kawamoto M., Kinugasa H., Mazda O. Significant inactivation of SARS-CoV-2 in vitro by a green tea catechin, a catechin-derivative, and black tea galloylated theaflavins. Molecules. 2021;26(12) doi: 10.3390/molecules26123572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mishra C.B., Pandey P., Sharma R.D., Malik M.Z., Mongre R.K., Lynn A.M., Prasad R., Jeon R., Prakash A. Identifying the natural polyphenol catechin as a multi-targeted agent against SARS-CoV-2 for the plausible therapy of COVID-19: an integrated computational approach. Briefings Bioinf. 2021;22(2):1346–1360. doi: 10.1093/bib/bbaa378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jha R.K., Khan R.J., Parthiban A., Singh E., Jain M., Amera G.M., Singh R.P., Ramachandran P., Ramachandran R., Sachithanandam V., Muthukumaran J., Singh A.K. Identifying the natural compound Catechin from tropical mangrove plants as a potential lead candidate against 3CL(pro) from SARS-CoV-2: an integrated in silico approach. J. Biomol. Struct. Dyn. 2021:1–20. doi: 10.1080/07391102.2021.1988710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are indicated in this article (with no patient data). The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.