Abstract
ςB, the general stress response ς factor of Bacillus subtilis, is activated when intracellular ATP levels fall or the bacterium experiences environmental stress. Stress activates ςB by means of a collection of regulatory kinases and phosphatases (the Rsb proteins), which catalyze the release of ςB from an anti-ς factor inhibitor. By using the yeast dihybrid selection system to identify B. subtilis proteins that could interact with Rsb proteins and act as mediators of stress signaling, we isolated the GTP binding protein, Obg, as an interactor with several of these regulators (RsbT, RsbW, and RsbX). B. subtilis depleted of Obg no longer activated ςB in response to environmental stress, but it retained the ability to activate ςB by the ATP responsive pathway. Stress pathway components activated ςB in the absence of Obg if the pathway’s most upstream effector (RsbT) was synthesized in excess to the inhibitor (RsbS) from which it is normally released after stress. Thus, the Rsb proteins can function in the absence of Obg but fail to be triggered by stress. The data demonstrate that Obg, or a process under its control, is necessary to induce the stress-dependent activation of ςB and suggest that Obg may directly communicate with one or more ςB regulators.
ςB is a transcription factor that directs RNA polymerase (RNAP) to promoters of the Bacillus subtilis general stress regulon (14). Induction of the regulon occurs when ςB is activated by either a decline in ATP levels or the onset of any of a number of environmental stresses (e.g., heat shock, acid salt, and ethanol) (5, 7, 14, 18, 34, 38). ςB is present in the prestressed cell, but it is held inactive in a complex with an anti-ςB protein, RsbW (6, 9). ςB is released from RsbW when a second protein (RsbV) binds to RsbW (9). The genes for RsbV, RsbW, ςB, and five additional ςB regulatory proteins (RsbR, -S, -T, -U, and -X) are cotranscribed as an eight-gene operon from a promoter (PA) that is recognized by the B. subtilis housekeeping ς factor, ςA (40). An internal ςB-dependent promoter (PB) enhances expression of the downstream four genes during periods of ςB activity (i.e., PA rsbR rsbS rsbT rsbU PB rsbV rsbW sigB rsbX) (4, 5, 7). A likely mechanism for ςB control by the Rsb proteins is illustrated in Fig. 1. In unstressed cells, RsbV, the effector of ςB release, is inactive due to RsbW-catalyzed phosphorylation (9, 37). Under conditions of low ATP (e.g., entry into the stationary phase of growth) this phosphorylation reaction is thought to be inefficient, and ςB remains active (2, 37, 38). In addition to this ATP-responsive activation, diverse environmental stresses initiate a sequence of Rsb-dependent processes which reactivate RsbV (18, 37, 38, 40, 41). In stressed B. subtilis, RsbT, normally inactive and complexed to RsbS, phosphorylates RsbS and becomes free to activate the RsbV-P phosphatase, RsbU (12, 41). RsbU then dephosphorylates RsbV-P, allowing RsbV to displace ςB from RsbW. Negative regulation is reestablished when RsbX, a RsbS-P phosphatase, dephosphorylates RsbS-P (41). This enables RsbS to again bind and inactivate RsbT. Although much has been learned about how the Rsb proteins function to alternatively silence or activate ςB, the mechanism by which stress communicates with the Rsb proteins is unknown. Neither the signal that is generated by environmental stress nor the regulator that is its target has been identified. In E. coli, protein denaturation and chaperone activation play key roles in communicating environmental stress to stress response transcription factors (8, 13, 43). Although similar processes appear to control chaperone expression in B. subtilis (23, 24, 27), we and others have not found an obvious correlation between chaperone activity and ςB activity in B. subtilis (23, 29). In addition, the known Rsb proteins appear to be inadequate to respond to environmental stress and activate ςB when they are expressed in Escherichia coli (29). Taken together, these results argue that the signal communicating environmental stress to the Rsb proteins is likely to be novel and bacillus specific. Assuming that an unidentified B. subtilis protein communicates stress signals to the Rsb phosphatase/kinase cascade, we attempted to identify candidate proteins, based on their ability to physically interact with key Rsb proteins in the yeast Gal4 dihybrid system. By this approach, several B. subtilis genes were identified as Rsb interactors. Although many of the interactions appear to be fortuitous, a number of biologically relevant associations were found. These included several known interacting Rsb proteins, as well as Obg, an essential GTP-binding protein (21, 31, 32) which proved to be needed for stress activation of ςB. The pattern of ςB induction in various mutant B. subtilis strains indicated that Obg, or a process under its control, plays a role in activating RsbT, the most upstream effector of ςB’s stress-induced pathway.
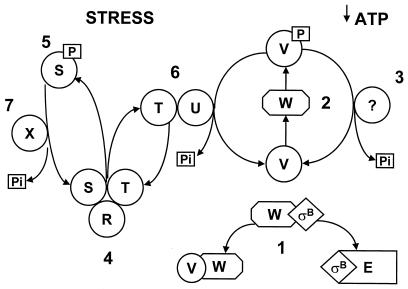
FIG. 1.
Model of ςB regulation. (Step 1) The anti-ςB protein RsbW (W) can form a mutually exclusive complex with either ςB or RsbV (V) (6, 9). When bound to RsbW, ςB is unavailable to RNA polymerase (E) (6). RsbV binding to RsbW allows the release of ςB and the formation of a ςB RNAP polymerase holoenzyme (E-ςB) (6). (Step 2) With ATP as a phosphate donor, RsbW can phosphorylate RsbV (2, 9). RsbV-P (V-P) is inactive as a ςB release factor (2, 9). During growth, relatively high ATP levels favor the phosphorylation and inactivation of RsbV, leaving ςB bound to RsbW (2, 38). If ATP levels fall, as when B. subtilis enters stationary phase, the phosphorylation of RsbV may be inefficient, leading to the persistence of active RsbV, the formation of RsbV-RsbW complexes, and the release of ςB (2, 38). (Step 3) The magnitude of the low-ATP activation of ςB is enhanced by dephosphorylation of a portion of the RsbV-P by an unknown mechanism (37). (Step 6) Environmental stress (e.g., heat shock, osmotic shock, and ethanol treatment) activates an RsbV-P phosphatase, RsbU (U), which creates active RsbV, regardless of ATP levels (37). (Step 4) RsbT (T), the activator of RsbU, is normally inactive due to an association with a negative regulator RsbS (S) (41). RsbR (R), an additional regulatory protein, binds to RsbS and RsbT (31) and is believed to play a structural role and to facilitate RsbT-RsbS interactions (1, 12). (Step 5) Upon exposure to stress, RsbT phosphorylates and inactivates RsbS and activates the RsbU phosphatase (12, 31). (Step 7) RsbS-P is dephosphorylated and reactivated by a phosphatase, RsbX (X) (31), which is encoded by one of the genes downstream of the sigB operon’s ςB-dependent promoter (17). RsbX levels increase with increasing ςB activity (10). This may serve to limit the activation process and return RsbT to an inactive complex with RsbS.
MATERIALS AND METHODS
Bacterial strains, plasmids, and cultivation.
All of the strains and plasmids used here are listed in Table 1. All BSJ and BSA strains are derivatives of PY22. Bacteria were grown in Luria-Bertani medium (LB [22]) at 37°C. The cells were exposed to ethanol or sodium azide during exponential growth at final concentrations of 4% or 2 mM, respectively. Pxyl and Pspac were induced by xylose (0.5%) or IPTG (isopropyl-β-d-thiogalactopyranoside; 1 or 0.1 mM), respectively. E. coli TG-2 was used as the host for cloning (26).
TABLE 1.
B. subtilis strains and plasmids used
| Strain or plasmid | Relevant genotype | Reference, source, or construction |
|---|---|---|
| Bascillus strains | ||
| SJV6 | Pspac::obg | A. Grossman |
| PY22 | trpC2 | P. Youngman |
| AG514 | spo0A::cat | A. Grossman |
| BSA46 | trpC2 Spβ ctc::lacZ | 3 |
| XS15 | trpC2 rsbX::spc rsbU194VA Spβ ctc::lacZ | 30 |
| BSA400 | trpC2 rsbT::pHV501T (Pspac::rsbT) | pHV501T→PY22 |
| BSA419 | trpC2 Pspac::rsbT Spβ ctc::lacZ | BSA46 →BSA400 |
| BSJ-5 | trpC2 Pspac::obg | SJV6→PY22 |
| BSJ-6 | trpC2 Pspac::obg Spβ ctc::lacZ | BSA46→BSJ-5 |
| BSJ-9 | trpC2 Pspac::obg (pJM2) Spβ ctc::lacZ | pJM2→BSA46 |
| BSJ-10 | trpC2 Pspac::obg rsbX::spc rsbU194VA Spβ ctc::lacZ | XS15→BSJ-6 |
| BSJ-11 | trpC2 Pxyl::obg | pJM4→PY22 |
| BSJ-12 | trpC2 Pxyl::obg Spβ ctc::lacZ | BSA46→BSJ-11 |
| BSJ-13 | trpC2 Pxyl::obg Pspac::rsbT Spβ ctc::lacZ | BSJ-11→BSA419 |
| BSJ-16 | trpC2 spo0A::cat | AG514→PY22 |
| BSJ-17 | trpC2 spo0A::cat Spβ ctc::lacZ | BSA46→BSJ-16 |
| Plasmids | ||
| pUS19 | Apr Specr | 4 |
| pUSX19 | Apr SpecrPxyl xylR | This study |
| pACT-2 | Apr vector with AD of GAL4 | Clontech |
| pGAD-10 | Apr vector with AD of GAL4 | Clontech |
| pGAD-GL | Apr vector with AD of GAL4 | Clontech |
| pX-2 | AprPxyl xylR | 20 |
| pUV31 | Apr protein fusion of RsbX with BD of GAL4 | 36 |
| pUV76 | Apr protein fusion of RsbT with BD of GAL4 | 36 |
| pUV145 | Apr protein fusion of RsbU with BD of GAL4 | 36 |
| pJM2 | Apr KanrPspac::obg | This study |
| pJM4 | Apr SpecrPxyl::obg150 xylR | This study |
| pDG148 | Apr KanrPspac lacI | P. Stragier |
| pHV501 | Apr ErmrPspac lacI lacZ | S. D. Ehrlich |
| pHV501T | Apr ErmrPspac::rsbT lacI | This study |
| pDT11 | Apr KanrPspac::rsbT lacI | 30 |
| pBluescript | Apr | 26 |
Library construction and yeast transformation.
Libraries of B. subtilis DNA::GAL4 activator domain (AD) fusions were constructed from PY22 chromosomal DNA that had been partially digested with either SauIIIA or Tsp509I and ligated into either the BamHI sites in pACT-2, pGAD-GL, and pGAD-10 (Clontech Laboratories, Inc., Palo Alto, Calif.) or the EcoRI sites of pACT-2 and pGAD-GL, respectively. These clonings created potential translational fusions between the SauIIIA fragments and the Gal4 AD in all three reading frames and between the Tsp509I fragments and the Gal4-AD in two reading frames. The libraries were transformed, according to established protocols (Clontech Laboratories) into yeast strain Y190 containing resident plasmids that encoded binding domain (BD) fusions to RsbU, RsbT, and RsbX (pUV31, pUV76, or pUV145) (36). Plasmids that encoded interacting proteins were selected by GAL4-dependent histidine prototrophy and screened for GAL4-dependent β-galactosidase activity (36). The B. subtilis DNA fragments of interest were amplified by PCR from positive colonies by using primers specific for the sequences immediately outside of the multiple cloning sites of the vectors. These DNAs were verified as the coding elements for proteins that interact with the rsb fusions by recloning into the AD vector and transformation into yeast cells carrying the appropriate rsb fusion. The DNA sequences that encoded interacting peptides were determined and their predicted protein sequences compared to the B. subtilis genome database (30a) to identify the genes from which they were derived.
Construction of plasmids for integration into target genes.
pUSX19 was created by cloning a 1.7-kb BamHI-SphI DNA fragment containing the xylose promoter and repressor from pX-2 (20) into similar sites of pUS19 (4). The 5′ ends (500 to 700 bp) of the genes that encoded protein fragments, which interacted with the Rsb proteins in the yeast two-hybrid system, were PCR amplified and cloned into the BamHI and SacI sites of pUSX19 by using sites that had been introduced during the amplification. This cloning placed the 5′ ends of the genes downstream of Pxyl in pUSX19. When these plasmids were transformed into PY22, a Campbell-like integration of the plasmid into the B. subtilis chromosome disrupted the resident copy of the gene and positioned the sole intact copy of the target gene downstream of Pxyl. These strains were then grown with or without xylose to determine whether the isolated genes were essential for growth. A ctc::lacZ reporter system was introduced into these strains by transformation with BSA46 (3) chromosomal DNA and selection for a ctc::lacZ-linked antibiotic resistance (erm and cat). To assess the possible need for each Rsb interacting protein in ςB stress activation, strains containing the Pxyl fusions were grown in LB with or without xylose and exposed to stress (4% ethanol) during the exponential phase of growth (optical density at 540 nm of 0.2). The ςB activity was monitored by reporter gene (ctc::lacZ) expression. To determine the effects of enhanced levels of the interacting proteins on stress activation of ςB, the cultures were grown in the presence of 2% xylose, which induces Pxyl several 100-fold (20), and stressed as described above.
Construction of plasmids and strains for Obg and RsbT manipulations.
pJM2, a Pspac::obg fusion plasmid, was constructed by cloning the obg gene, including its ribosomal binding site, which had been amplified by PCR into pBluescript (26), by using BamHI sites that had been placed at each end during the amplification. The orientation of the fragment in the plasmid was determined by restriction endonuclease digestion analysis. The obg segment was excised with HindIII and XbaI and cloned into the multiple cloning site of pDG148. pJM4 was constructed by PCR amplification of a 250-bp DNA fragment containing 150 bp of the 5′ end of obg and 100 bp of upstream DNA. BamHI and BglII sites, introduced into the ends of the fragment during amplification, were used to clone the fragment into the BamHI site of pUSX19, creating a Pxyl::obg150 fusion. pHV501T contains the HindIII/BamHI piece of rsbT from pDT11 (30). The fragment was cloned into pHV501 downstream of Pspac. The resulting plasmid was cut with ClaI and BamHI to remove lacZ, made blunt with the Klenow fragment of DNA polymerase, and religated. Transformation of PY22 with pHV501T places Pspac between rsbS and rsbT within the sigB operon (BSA400), due to a Campbell-like integration of the plasmid into rsbT.
General methods.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis, Western blotting, and β-galactosidase activity assays were performed as previously described (10, 19). DNA manipulations were performed according to standard protocols (26). Transformation of naturally competent B. subtilis cells was carried out as described by Yasbin et al. (42). Yeast β-galactosidase assays were performed as done previously (36). Predicted protein translations of the DNA fragments were determined by using DNAMAN software (Lynnon Biosoft Co.). Sequencing of DNA fragments identified in the yeast two-hybrid system screen was performed by the University of Texas Health Science Center at San Antonio Center for Advanced DNA Technologies.
RESULTS
Identification of Rsb interactors.
We considered RsbT, RsbU, and RsbX as the most likely of the Rsb proteins to be targeted by stress-generated signals. To identify possible protein mediators of such signals, we screened B. subtilis chromosomal DNA for genes whose products could be shown to physically interact with RsbT, -U, or -X in the yeast dihybrid system. As described in Materials and Methods, yeast strains carrying Gal4-activatable his and lacZ genes, as well as plasmids encoding rsbT, rsbU, or rsbX fused to the Gal4 DNA BD, were transformed with plasmid libraries of B. subtilis chromosomal DNA. The plasmid libraries contain potential translational fusions between various B. subtilis genes and the Gal4 AD. Transformants in which the cloned B. subtilis DNA encoded a Gal4-AD protein which could associate with an Rsb/Gal4-BD and create a functional gene activator protein were selected as histidine prototrophs and screened for a Lac+ phenotype. After we verified that the His+ Lac+ activities of the positive clones were due to an interaction between the AD fusion proteins and the Rsb-BD proteins (Materials and Methods), the AD fusion plasmids were recovered from the yeast cells, transformed into E. coli, and screened by restriction endonuclease-Southern blot analyses to eliminate duplicate clones. A total of 31 unique B. subtilis DNA inserts were isolated from approximately 400 His+ Lac+ transformants. The insert DNA was sequenced by using DNA primers complementary to the vectors’ translational fusion junctions. The identity of the B. subtilis element that had become fused to the Gal4-AD was determined by searching the B. subtilis genome sequence database (30a) for the sequences contained at the Gal4-AD fusion junctions. The results of this exercise are listed in Table 2. Eleven predicted intergenic regions were among the B. subtilis chromosome segments that displayed Gal4 activity when cloned into the AD vector. Presumably, these represent fortuitously generated AD fusion proteins that could pair with particular Rsb::Gal4-BD chimeras. In previous studies, where interactions between the various rsb genes were examined in the yeast dihybrid system, no RsbX interactions were detected with any of the other Rsb genes, but interactions were found between RsbT and RsbS/-R/-U and between RsbU and RsbV/-T (36). Our current screening of the B. subtilis genome detected two of these four associations: the interactions of RsbT with both RsbR and RsbS (Table 2).
TABLE 2.
Interacting proteins and intergenic regions identified in the yeast two-hybrid systema
| Bait protein | Interacting protein | Predicted function | Protein size (no. of amino acids) | Amino acid segment in BD fusion |
|---|---|---|---|---|
| RsbT | ProB | γ-Glutamyl kinase | 354 | 321–354 |
| RsbR | Regulator of ςB | 274 | 116–256 | |
| RsbS | Regulator of ςB | 121 | 8–117 | |
| YfmR | Similar to ABC transporter | 629 | 572–629 | |
| YhcH | Similar to ABC transporter | 305 | 113–203 | |
| YorL | Similar to DNA polymerase III, α subunit | 1,305 | 1,211–1,240 | |
| RsbU | PpsD | Peptide synthetase | 3,603 | 1,261–1,351 |
| RpoC | RNA polymerase, B subunit | 1,200 | 671–811 | |
| SpsB | Spore coat polysaccharide biosynthesis | 472 | 53–100 | |
| SpsJ | Spore coat polysaccharide biosynthesis | 315 | 7–47 | |
| YjlD | Similar to NADH dehydrogenase | 392 | 1–23, 1–46 | |
| YlaG | Similar to GTP binding elongation factor | 612 | 267–372 | |
| YoaR | Unknown, no similarity to known proteins | 303 | 222–303 | |
| YorL | Similar to DNA polymerase III, α subunit | 1,305 | 1,211–1,240 | |
| RsbX | Obg | GTP binding protein | 428 | 285–328 |
| YrkN | Unknown, no similarity to known proteins | 185 | 32–185 | |
| YtgA | Similar to ABC transporter | 307 | 140–296 | |
| YvgO | Unknown, no similarity to known proteins | 161 | 29–100, 55–100 |
The table lists each of the Rsb proteins fused to the Gal4::BD (bait protein), the identity of the B. subtilis genes that were isolated against them as interacting Gal4::AD fusions (interacting protein), and the predicted function of the genes. The predicted size of the gene products (protein size) and the particular amino acid segment of the proteins that were present as part of the fusion proteins are also listed. The nucleotide designations of the B. subtilis intergenic regions which yield “fortuitous” fusion proteins are noted as follows: for RsbT, 1886017–1885806; for RsbU, 3334901–3335145, 4094152–4094628, 521710–521628, 1298509–1298682, 3990625–3990466, 135661–135552, 3648551–3648811, 262440–262510, 2564095–2564127, and 2359371–2359314.
The remaining 18 cloned segments defined portions of 16 different predicted or known genes (Table 2). Two of these genes (yvgO and yjlD) were detected twice in overlapping fragments. Four of the genes were RsbT interactors, eight were RsbU interactors, and four were found in pairings with RsbX. As described in Materials and Methods, we attempted to distinguish biologically relevant associations from fortuitous biochemical interactions by reducing or elevating the expression of the identified genes in B. subtilis and then asking whether either of these manipulations would affect ςB activity during growth or stress (data not shown). Only obg, a gene isolated in the RsbX pairing, was found to have a significant effect on ςB’s activity in this analysis. It remains possible that one or more of the other B. subtilis genes could have a less obvious role in Rsb function; however, our inability to readily demonstrate this prompted us to focus on obg.
Obg is necessary for stress activation of ςB.
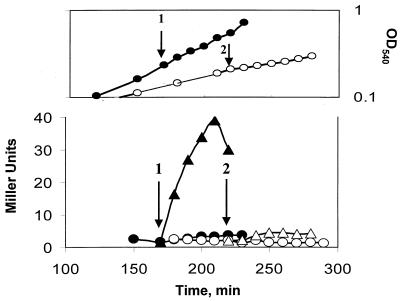
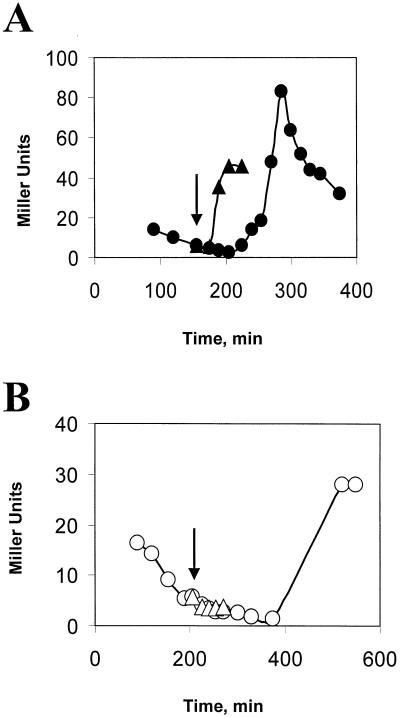
Obg is an essential B. subtilis protein (21, 31, 32). In order to study its possible role in ςB activation, we used a B. subtilis strain (BSJ-6) in which obg’s expression was under the control of an IPTG-inducible promoter (32). Withholding IPTG from this strain leads to a depletion of Obg and a concomitant cessation of growth (32). Cultures of BSJ-6 were resuspended in LB with or without IPTG and monitored until growth had slowed in the culture lacking IPTG. Portions of the cultures were then stressed by treatment with ethanol. The ςB activity in both cultures was estimated by using a reporter gene fused to a ςB-dependent promoter (i.e., ctc::lacZ). B. subtilis with Obg (i.e., the IPTG-containing culture) immediately activated ςB after ethanol treatment; however, the strain in which the Obg levels had been depleted failed to activate ςB in response to ethanol stress (Fig. 2). Similar results were obtained with heat shock as the stress and Western blot analyses to judge the inducibility of the Rsb genes that are downstream of the sigB operon’s ςB-dependent promoter (i.e., rsbV and -W, sigB, and rsbX) (data not shown).
FIG. 2.
Effect of Obg depletion on ethanol induction of ςB. (Top panel) Growth of BSJ-6 (Pspac::obg) in LB with (●) or without (○) IPTG (100 μM). The arrows represent the points during growth when ethanol (4% final concentration) was added to the IPTG-induced (1) and the IPTG− (2) cultures. The curves display the untreated cultures; the ethanol-treated cultures had their growth reduced by approximately 50% (not shown). (Bottom panel) ctc::lac expression in BSJ-6. Samples of the cultures represented in the top panel were analyzed for ςB-dependent β-galactosidase activity. The arrows depict the times of ethanol addition in the IPTG+ (1) and IPTG− (2) cultures. Closed symbols represent IPTG+ cultures with (▴) or without (●) ethanol; open symbols depict the IPTG− culture with (▵) or without (○) ethanol.
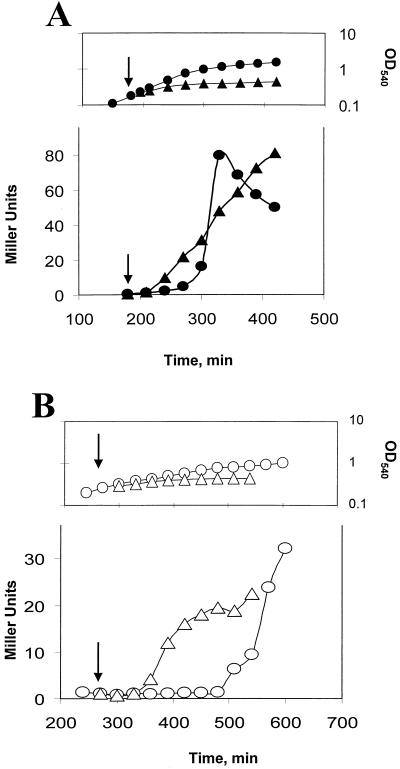
We had previously characterized two mechanisms by which ςB is activated (38). One relies on the above-described stress induction pathway. A second type of ςB activation occurs under culture conditions where ATP levels are low (e.g., glucose limitation, Mn2+ treatment, and entry into stationary phase) (38). Presumably, under low ATP conditions RsbV remains active due to ineffective phosphorylation by RsbW (Fig. 1) (2, 38). To determine whether Obg is needed by the ATP responsive pathway, we treated BSJ-6 (Pspac::obg), which was either growing in LB with IPTG or had ceased growth due to Obg depletion in LB lacking IPTG, with sodium azide, a compound which blocks oxidative phosphorylation and lowers ATP levels in B. subtilis (16). Azide addition resulted in enhanced ςB activity regardless of whether or not the culture was depleted of Obg (Fig. 3). Although ςB activity increased in both cultures, the level of reporter gene activity in the Obg-depleted culture (Fig. 3B) was approximately one-half of that seen in the culture in which Obg was not limiting (Fig. 3A).
FIG. 3.
ATP-responsive induction of ςB in Obg-depleted cells. BSJ-6 (Pspac::obg) was grown in the presence (A) or absence (B) of IPTG. The top portions of panels A and B illustrate the growth of the cultures without (○ and ●) or in the presence of 2 mM sodium azide (▵) and ▴), which was added at the times indicated by the arrows. The lower portions of panels A and B display ςB-dependent β-galactosidase activity of the cultures represented in the panels above them, with the arrows depicting the time of sodium azide addition. The triangles represent azide-treated cultures; the circles represent untreated cultures.
Continued incubation of Obg-induced and -depleted cultures led to the expected activation of ςB as the Obg+ culture entered stationary phase (Fig. 3A). ςB was also activated in the Obg− culture; however, a longer incubation period was required for its onset (Fig. 3B). Presumably, the delayed ςB activation in the Obg-depleted culture is due to the growth-inhibited culture requiring more time to reach the point where ATP levels fall sufficiently to activate ςB. As was the case with azide treatment, the stationary-phase induction of ςB in the Obg-depleted cells was approximately one-half of that seen in the control culture (Fig. 3). We have found that Obg-depleted cells, pulse labeled with 35S-labeled methionine-cysteine under conditions similar to those used in our experiments, incorporate only 60% of the label which is incorporated by an Obg+ culture (data not shown). Thus, although we cannot exclude the possibility that the reduced level of ςB activation represents a residue role for Obg in the ATP-responsive pathway, it is more likely to be the result of a general effect of Obg depletion on the biosynthetic capacity of B. subtilis. The activation of ςB in an Obg-depleted culture by conditions that trigger the ATP-responsive pathway, but not by those that induce the stress-dependent pathway, indicates that Obg’s role in ςB activation is largely limited to the stress activation process.
Obg effects on ςB are independent of Spo0A.
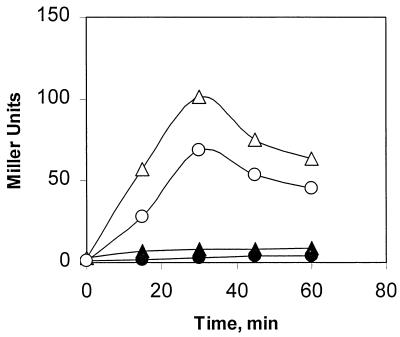
Obg is essential for both growth and sporulation (21, 31, 32). During studies of the sporulation requirement for Obg, it was discovered that Obg is needed, either directly or indirectly, for the phosphorylation reaction that activates the transition-phase regulatory protein Spo0A (32). Given that both the activation of Spo0A and the stress induction of ςB require Obg, we asked whether the stress activation of ςB could be a Spo0A-mediated process. Wild-type and spo0A::cat strains of B. subtilis were exposed to ethanol stress and examined for ςB activation. ςB induction occurred regardless of the presence or absence of Spo0A (Fig. 4). Thus, the need for Obg in ςB activation is not likely to be due to an Obg effect on Spo0A.
FIG. 4.
Activation of ςB in Spo0A− B. subtilis. B. subtilis BSA46 (wild type, circles) and BSJ-17 (spo0A::cat, triangles) were grown without stress in LB or exposed to ethanol (4%) during an early stage of exponential growth. Samples were taken from control (solid symbols) and ethanol-treated (open symbols) cultures at the times indicated and analyzed for ςB-dependent (ctc::lacZ) β-galactosidase activity.
Obg interacts with RsbT and RsbW in the yeast dihybrid system.
Knowing that high-molecular-weight complexes containing multiple Rsb proteins and unknown additional components are readily detectable in B. subtilis extracts (10) and that our initial screening of the B. subtilis library had not been exhaustive, we revisited the yeast dihybrid system to test whether Obg could interact with additional Rsb components. Specifically, we paired both the entire obg gene and the obg fragment which had interacted with RsbX with other rsb genes. rsbS was not included in the analyses due to its ability to activate Gal4-dependent promoters independently when fused to Gal4-BD.
No interactions were detected between either the Obg fusion and RsbR, -U, or -V (i.e., yeast strains expressing the fusion pairs failed to activate the lacZ reporter gene and turn blue in X-Gal [5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid] filter assays [36]); however, we observed reporter gene activity when the carboxy-terminal fragment of Obg was paired with RsbW or when the entire obg::Gal4-AD fusion was paired with either RsbW or RsbT. The RsbX-Obg interaction, although obvious when RsbX was paired with the Obg fragment, was barely detectable when RsbX was paired with the full-length Obg fusion protein. The relative reporter gene activity for each of the reactive pairings is given in Table 3.
TABLE 3.
Interactions of Obg and Rsb proteins by yeast two-hybrid systema
| BD fusion | AD fusion (ratio)
|
|
|---|---|---|
| Obg′ | Obg | |
| Vector, pAS2-1 | 1 | 1 |
| X | 1.88 | 1.15 |
| T | 0.525 | 4.41 |
| W | 5.52 | 33.86 |
The horizontal rows represent the Gal4 DNA BD vector (pAS2-1) either without insert DNA (row 1) or with the sigB gene depicted by a single letter (e.g., X = rsbX). The vertical rows depict the AD vector (pACT-2) encoding either the 285- to 328-amino-acid fragment of Obg, originally identified in the library screen (Obg′) or else intact Obg. Intersecting rows represent reporter gene activity (i.e., β-galactosidase expression) in a yeast strain cotransformed with both plasmids indicated by row. β-Galactosidase assays were performed on two clones from each pairing in duplicate. The results are presented as a ratio of the Rsb protein-AD fusion value from the indicated pairing divided by the value obtained from the AD fusion pairing with the pAS2-1 vector.
The level of reporter gene activity in the yeast dihybrid system can be a reflection of the affinity of the interacting proteins for each other (11). Based on reporter gene activity, relative to the vector pairings, the interaction between RsbT and the full-length Obg protein is approximately 2.5-fold that observed in a similar assay where RsbT was paired with RsbU, the phosphatase that it activates (36). The interaction between RsbW and the full-length Obg is more than three times the reporter gene response seen with RsbW paired with its antagonist RsbV (36). The degree of reporter gene activation that occurs in yeast cells when Obg is paired with RsbW or RsbT indicates that significant interactions are possible between these proteins. Although the relevance of these interactions in yeast cells to ςB control in B. subtilis is unclear, the fact that they occur suggest that RsbT and RsbW, as well as RsbX, are possible targets for Obg-essential steps in the stress activation of ςB.
Obg’s essential role in ςB activation is independent of RsbX.
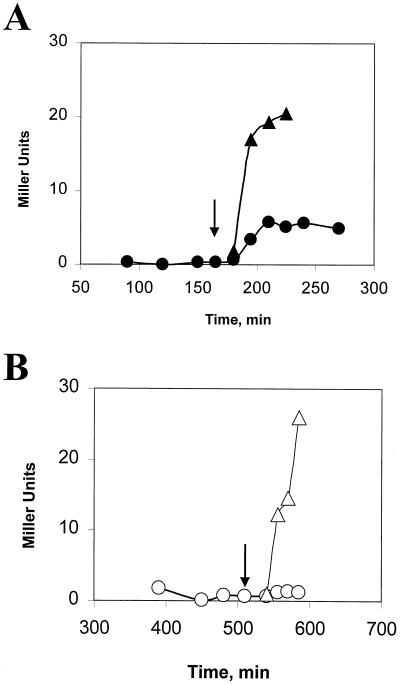
Obg was initially identified as a potential ςB regulator on the basis of an interaction between its carboxy terminus and RsbX in the yeast dihybrid system. RsbX is a negative regulator of the stress-inducible pathway for ςB activation (3, 15, 17, 30, 33, 35). Thus, if Obg’s essential role in ςB activation entails an effect on RsbX, we would expect this to involve a lifting of RsbX’s negative control. If this is so, Obg should not be essential for the stress activation of ςB in the absence of RsbX.
The loss of RsbX normally causes a toxic activation of ςB (3, 15, 17, 33); however, there are several B. subtilis strains in which the loss of RsbX is tolerated due to suppressor mutations in either RsbT, -U, or -V, that reduce their activities as positive regulators (30). We placed an inducible obg operon into one of these strains, XS15 (rsbU194VA rsbX::spec), and examined the effects of Obg depletion on ethanol-induced ςB activity in this RsbX− background. As was the case in the RsbX+ strains, ςB activity was induced after both ethanol treatment, and entry into stationary phase when Obg was present (Fig. 5A), but increased only upon entry into stationary phase in the culture depleted of Obg (Fig. 5B). Thus, Obg is needed for the stress activation of ςB in the absence of RsbX. This result does not exclude the possibility that Obg can modulate RsbX activity, but it does demonstrate that Obg provides an essential function for ςB activation aside from any putative RsbX interaction.
FIG. 5.
Effects of Obg on activation of ςB in RsbX− B. subtilis. BSJ-10 (Pspac::obg rsbX::spec rsbU194VA) was grown in LB with (A) or without (B) IPTG (0.1 mM). As in Fig. 2, cells were treated with ethanol (4%) at the times indicated by the arrows either during growth (A) or after the culture slowed due to Obg depletion (B). Symbols: triangles, ethanol-treated cultures; circles, untreated cultures.
RsbT can activate the ςB stress pathway in the absence of Obg.
Stress is believed to set in motion a process (Fig. 1) whereby RsbT frees itself from its inhibitor and then binds and activates RsbU, the RsbV-P phosphatase (41). RsbV then interacts with RsbW to free ςB (2, 6, 9). There are several points at which an Obg-dependent function might be needed in this process. To determine whether Obg is required for the activation of RsbT or a downstream event, we took advantage of the observation that the induced expression of RsbT, in the absence of a corresponding synthesis of its inhibitor (RsbS), can drive activation of ςB in unstressed bacteria (18, 28). If the essential Obg-dependent function is limited to the activation of RsbT, then the induced synthesis of RsbT should activate ςB in Obg-depleted cells. Conversely, if Obg has a critical role in RsbT’s ability to activate RsbU, RsbU’s capacity to dephosphorylate of RsbV-P, or RsbV’s ability to displace ςB from RsbW, then providing additional RsbT should not activate ςB in Obg-depleted cells.
To independently control both the levels of Obg and the induction of rsbT, we constructed a B. subtilis strain in which obg expression was driven from a xylose-inducible promoter (Pxyl::obg), and an IPTG-inducible promoter was placed immediately upstream of rsbT in the sigB operon. Growth and stress induction of this strain is xylose dependent due to Obg depletion in the absence of the Pxyl inductant. When IPTG was added to cultures of this strain, ςB was activated regardless of whether or not Obg had been depleted (Fig. 6). We conclude that Obg’s essential role in activation of ςB is upstream of RsbT’s activation of RsbU in the stress pathway and is likely in the activation of RsbT itself.
FIG. 6.
Activation of ςB by RsbT overexpression in Obg-depleted B. subtilis. BSJ-13 (Pxyl::obg Pspac::rsbT) was grown in LB with or without xylose (0.5%) to maintain Obg levels (A) or to deplete the cell of Obg (B), respectively. At the times indicated by the arrows, IPTG (1 mM) was added to the exponentially growing cells (A), and the culture that had slowed due to Obg depletion (B) to induce Pspac upstream of rsbT. Samples were taken from IPTG-induced (▴) and ▵) and control (● and ○) cultures and were analyzed for ςB-dependent β-galactosidase activity as in Fig. 2.
DISCUSSION
The general stress regulon of B. subtilis is induced when environmental stress communicates with a kinase/phosphatase pathway to trigger the activation of the ςB transcription factor (41). Neither the stress-generated signal that activates ςB nor the component within the pathway that initially responds to the signal are known. Previous studies indicated that the known ςB regulators are insufficient to detect and respond to stress and that the inducing signal is likely to be novel and Bacillus specific (29). This suggested that additional B. subtilis gene products are needed to communicate the presence of stress to the ςB activation cascade. In the current study, we attempted to identify such gene products by using the yeast dihybrid system to isolate B. subtilis proteins that could interact with important members of the ςB activation cascade and, as such, might be involved in this signaling. As is evident from our results, the dihybrid selection system readily detects peptides that can interact with Rsb fusion proteins to activate the yeast selection system. Although a few of these associations are clearly biologically relevant, i.e., known Rsb-Rsb interactions were detected, most of the isolates likely represent fortuitous interactions between domains on the fusion proteins. Fortunately, access to the B. subtilis genome database and our ability to readily alter the expression of the genes for these putative interactors allowed us to test whether the abundance or absence of their products could affect ςB stress activation and also allowed us to put aside unpromising candidates. By this approach, obg was detected in our current screening as a gene whose product influences ςB activity. If B. subtilis is depleted of Obg, stress activation of ςB, but not its ATP-responsive activation, is blocked. Overexpression of Obg from a high-copy-number plasmid had no effect on ςB activity (data not shown).
Obg is the second gene in the operon that encodes Spo0B, a critical protein in the phosphorelay which modulates the activity of the sporulation-transition state regulatory protein Spo0A (31). Obg is essential for both B. subtilis growth and sporulation (31, 32). It is a GTP binding protein (39) whose explicit function is unknown; however, there is evidence that Obg activity influences the initiation of chromosome replication (21) and the phosphorylation state of the postexponential-phase gene regulator, Spo0A (32). It is unclear whether Obg’s activities in either of these processes is related to its effects on ςB activity. We note however, that the Obg-dependent activation of Spo0A is not needed for the ςB activation process, since stress activation of ςB occurs in the absence of Spo0A function.
Obg is a member of a unique family of small GTP binding proteins that have been identified in diverse organisms from bacteria and mammals (reviewed in reference 25). In bacteria, the Obg subfamily is speculated to monitor the state of intracellular GTP levels and to serve as a switch to promote growth when bound to GTP, but not when associated with GDP (25). The actual targets for this switch protein are unknown. Our yeast dihybrid data suggest that Obg may directly interact with a number of the ςB Rsb regulators; however, even if these interactions ultimately prove to be biologically relevant, the Rsb proteins cannot be Obg’s only targets. Obg is essential for growth and sporulation, while neither the Rsb proteins nor ςB are essential for either process. Perhaps Obg, as a general sensor of intracellular GTP levels and a regulatory switch, has several targets within B. subtilis, allowing it to coordinate diverse functions in response to nucleotide changes.
Although it is clear that Obg is essential for stress activation of ςB, it remains possible that the apparent interaction of Obg with particular Rsb proteins is accidental and that the requirement for Obg in ςB stress activation is indirect. The putative RsbT-Obg interaction is, however, intriguing given that one function of the Obg class of proteins is intracellular signaling and that stress activation of RsbT fails to occur in its absence. Determining whether Obg directly communicates the existence of environmental stress to RsbT or is only indirectly involved in this process will likely require in vitro biochemical analyses.
ACKNOWLEDGMENTS
We thank Alan Grossman and Wolfgang Schulmann for strains and plasmids and Jim Hoch for advice and unpublished data.
This work was supported by NIH grant GM48220.
REFERENCES
- 1.Akbar S, Kang C M, Gaidenko T A, Price C W. Modulator protein RsbR regulates environmental signaling in the general stress pathway of Bacillus subtilis. Mol Microbiol. 1997;24:567–578. doi: 10.1046/j.1365-2958.1997.3631732.x. [DOI] [PubMed] [Google Scholar]
- 2.Alper S, Dufour A, Garsin D A, Duncan L, Losick R. Role of adenosine nucleotides in the regulation of a stress-response transcription factor in Bacillus subtilis. J Mol Biol. 1996;260:165–177. doi: 10.1006/jmbi.1996.0390. [DOI] [PubMed] [Google Scholar]
- 3.Benson A K, Haldenwang W G. Characterization of a regulatory network that controls ςB expression in Bacillus subtilis. J Bacteriol. 1992;174:749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson A K, Haldenwang W G. Regulation of ςB levels and activity in Bacillus subtilis. J Bacteriol. 1993;175:2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benson A K, Haldenwang W G. The ςB-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993;175:1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benson A K, Haldenwang W G. Bacillus subtilis ςB is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci USA. 1993;90:2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boylan S A, Redfield A R, Brody M S, Price C W. Stress-induced activation of the ςB transcription factor of Bacillus subtilis. J Bacteriol. 1993;175:7931–7937. doi: 10.1128/jb.175.24.7931-7937.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bukau B, Walker G C. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–4036. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dufour A, Haldenwang W G. Interactions between a Bacillus subtilis anti-ς factor (RsbW) and its antagonist (RsbV) J Bacteriol. 1994;176:1813–1820. doi: 10.1128/jb.176.7.1813-1820.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dufour A, Voelker U, Voelker A, Haldenwang W G. Relative levels and fractionation properties of Bacillus subtilis ςB and its regulators during balanced growth and stress. J Bacteriol. 1996;178:3701–3709. doi: 10.1128/jb.178.13.3701-9sigma.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Estojak J, Brent R, Golemis E A. Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol. 1995;15:5820–5829. doi: 10.1128/mcb.15.10.5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaidenko, T. A., X. Yang, Y. M. Lee, and C. W. Price. Threonine phosphorylation of modulator protein RsbR governs its ability to regulate a serine kinase in the stress signaling pathway of Bacillus subtilis. J. Mol. Biol., in press. [DOI] [PubMed]
- 13.Gross C A. Function and regulation of the heat shock proteins. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B Jr, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology, 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1382–1399. [Google Scholar]
- 14.Hecker M, Schumann W, Voelker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 15.Igo M, Lampe M, Ray C, Schaefer W, Moran C P, Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolliffe L K, Doyle R J, Streips U N. The energized membrane and cellular autolysis in Bacillus subtilis. Cell. 1981;25:753–763. doi: 10.1016/0092-8674(81)90183-5. [DOI] [PubMed] [Google Scholar]
- 17.Kalman S, Duncan M L, Thomas S M, Price C W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990;172:5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang C M, Brody M S, Akbar S, Yang X, Price C W. Homologous pairs of regulatory proteins control activity of Bacillus subtilis transcription factor ςB in response to environmental stress. J Bacteriol. 1996;178:3846–3853. doi: 10.1128/jb.178.13.3846-3853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kenney T J, Moran C P., Jr Organization and regulation of an operon that encodes a sporulation-essential sigma factor in Bacillus subtilis. J Bacteriol. 1987;169:3329–3339. doi: 10.1128/jb.169.7.3329-3339.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim L, Mogk A, Schumann W. A xylose-inducible Bacillus subtilis integration vector and its application. Gene. 1996;181:71–76. doi: 10.1016/s0378-1119(96)00466-0. [DOI] [PubMed] [Google Scholar]
- 21.Kok J, Trach K A, Hoch J A. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. J Bacteriol. 1994;176:7155–7160. doi: 10.1128/jb.176.23.7155-7160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 23.Mogk A, Voelker A, Engelmann S, Hecker M, Schumann W, Voelker U. Nonnative proteins induce expression of the Bacillus subtilis CIRCE regulon. J Bacteriol. 1998;180:2895–2900. doi: 10.1128/jb.180.11.2895-2900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mogk A, Homuth G, Scholz C, Kim L, Schmid F X, Schumann W. The GroE chaperonin machine is a major modulator of the CIRCE heat shock regulon of Bacillus subtilis. EMBO J. 1997;16:4579–4590. doi: 10.1093/emboj/16.15.4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okamoto S, Ochi K. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Mol Microbiol. 1998;30:107–119. doi: 10.1046/j.1365-2958.1998.01042.x. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Schulz A, Tzschaschel B, Schumann W. Isolation and analysis of mutants of the dnaK operon of Bacillus subtilis. Mol Microbiol. 1995;15:421–429. doi: 10.1111/j.1365-2958.1995.tb02256.x. [DOI] [PubMed] [Google Scholar]
- 28.Scott, J. M., and W. G. Haldenwang. Unpublished results.
- 29.Scott J M, Smirnova N, Haldenwang W G. A Bacillus specific factor is needed to trigger the stress-activated phosphatase/kinase cascade of ςB induction. Biochem Biophys Res Commun. 1999;257:106–110. doi: 10.1006/bbrc.1999.0418. [DOI] [PubMed] [Google Scholar]
- 30.Smirnova N, Scott J, Voelker U, Haldenwang W G. Isolation and characterization of Bacillus subtilis sigB operon mutations that suppress the loss of the negative regulators RsbX. J Bacteriol. 1998;180:3671–3680. doi: 10.1128/jb.180.14.3671-3680.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30a.The Bacillus subtilis Genome Database. 1999, copyright date. [Online.] Institut Pasteur. http://www.pasteur.fr/Bio/SubtiList. [18 June 1999, last date accessed.]
- 31.Trach K, Hoch J A. The Bacillus subtilis spo0B stage 0 sporulation operon encodes an essential GTP-binding protein. J Bacteriol. 1989;171:1362–1371. doi: 10.1128/jb.171.3.1362-1371.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vidwans, S. J., K. Ireton, and A. D. Grossman. 1995. Possible role for the essential GTP-binding protein Obg in regulating the initiation of sporulation in Bacillus subtilis. 177:3308–3311. [DOI] [PMC free article] [PubMed]
- 33.Voelker U, Dufour A, Haldenwang W G. The Bacillus subtilis rsbU gene product is necessary for RsbX-dependent regulation of ςB. J Bacteriol. 1995;177:114–122. doi: 10.1128/jb.177.1.114-122.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Voelker U, Engelmann S, Maul B, Riethdorf S, Voelker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 35.Voelker U, Luo T, Smirnova N, Haldenwang W G. Stress activation of Bacillus subtilis ςB can occur in the absence of the ςB negative regulator RsbX. J Bacteriol. 1997;179:1980–1984. doi: 10.1128/jb.179.6.1980-1984.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Voelker U, Voelker A, Haldenwang W G. The yeast two-hybrid system detects interactions between Bacillus subtilis ςB regulators. J Bacteriol. 1996;178:7020–7023. doi: 10.1128/jb.178.23.7020-7023.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Voelker U, Voelker A, Haldenwang W G. Reactivation of the Bacillus subtilis anti-ςB antagonist, RsbV, by stress or starvation-induced phosphatase activities. J Bacteriol. 1996;178:5456–5463. doi: 10.1128/jb.178.18.5456-5463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Voelker U, Voelker A, Maul B, Hecker M, Dufour A, Haldenwang W G. Separate mechanisms activate ςB of Bacillus subtilis in response to environmental and metabolic stresses. J Bacteriol. 1995;177:3771–3780. doi: 10.1128/jb.177.13.3771-3780.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Welsh K, Trach K A, Folger C, Hoch J A. Biochemical characterization of the essential GTP-binding protein Obg of Bacillus subtilis. J Bacteriol. 1994;176:7161–7168. doi: 10.1128/jb.176.23.7161-7168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wise A A, Price C W. Four additional genes in the sigB operon of Bacillus subtilis that control activity of the general stress factor ςB in response to environmental signals. J Bacteriol. 1995;177:123–133. doi: 10.1128/jb.177.1.123-133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang X, Kang C M, Brody M S, Price C W. Opposing pairs of serine protein kinases and phosphatases transmit signals of environmental stress to activate a bacterial transcription factor. Genes Dev. 1996;10:2265–2275. doi: 10.1101/gad.10.18.2265. [DOI] [PubMed] [Google Scholar]
- 42.Yasbin R E, Wilson G A, Young F E. Transformation and transfection of lysogenic strains of Bacillus subtilis 168. J Bacteriol. 1973;113:540–548. doi: 10.1128/jb.113.2.540-548.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]