Key Points
Question
What are the diagnostic yield and clinical utility of genetic sequencing for patients with unexplained pediatric epilepsy?
Findings
This cohort study of 522 children with previously unexplained epilepsy used exome sequencing to identify and clinically confirm diagnostic results for 100 children, including 89 with single nucleotide variants and 11 with copy number variants. Individuals with earlier seizure onset, intellectual disability, and motor impairment were more likely to have diagnostic results, and at least 29 patients had changes in treatment, surveillance, or prognosis based on their genetic diagnoses.
Meaning
These findings suggest that for children with unexplained epilepsy, genetic evaluation yielded precise diagnoses with direct clinical implications.
This cohort study examines the genetic landscape of pediatric epilepsy and clinical utility of genetic diagnoses for patients with epilepsy.
Abstract
Importance
Genomic advances inform our understanding of epilepsy and can be translated to patients as precision diagnoses that influence clinical treatment, prognosis, and counseling.
Objective
To delineate the genetic landscape of pediatric epilepsy and clinical utility of genetic diagnoses for patients with epilepsy.
Design, Setting, and Participants
This cohort study used phenotypic data from medical records and treating clinicians at a pediatric hospital to identify patients with unexplained pediatric-onset epilepsy. Exome sequencing was performed for 522 patients and available biological parents, and sequencing data were analyzed for single nucleotide variants (SNVs) and copy number variants (CNVs). Variant pathogenicity was assessed, patients were provided with their diagnostic results, and clinical utility was evaluated. Patients were enrolled from August 2018 to October 2021, and data were analyzed through December 2022.
Exposures
Phenotypic features associated with diagnostic genetic results.
Main Outcomes and Measures
Main outcomes included diagnostic yield and clinical utility. Diagnostic findings included variants curated as pathogenic, likely pathogenic (PLP), or diagnostic variants of uncertain significance (VUS) with clinical features consistent with the involved gene’s associated phenotype. The proportion of the cohort with diagnostic findings, the genes involved, and their clinical utility, defined as impact on clinical treatment, prognosis, or surveillance, are reported.
Results
A total of 522 children (269 [51.5%] male; mean [SD] age at seizure onset, 1.2 [1.4] years) were enrolled, including 142 children (27%) with developmental epileptic encephalopathy and 263 children (50.4%) with intellectual disability. Of these, 100 participants (19.2%) had identifiable genetic explanations for their seizures: 89 participants had SNVs (87 germline, 2 somatic mosaic) involving 69 genes, and 11 participants had CNVs. The likelihood of identifying a genetic diagnosis was highest in patients with intellectual disability (adjusted odds ratio [aOR], 2.44; 95% CI, 1.40-4.26), early onset seizures (aOR, 0.93; 95% CI, 0.88-0.98), and motor impairment (aOR, 2.19; 95% CI 1.34-3.58). Among 43 patients with apparently de novo variants, 2 were subsequently determined to have asymptomatic parents harboring mosaic variants. Of 71 patients who received diagnostic results and were followed clinically, 29 (41%) had documented clinical utility resulting from their genetic diagnoses.
Conclusions and Relevance
These findings suggest that pediatric-onset epilepsy is genetically heterogeneous and that some patients with previously unexplained pediatric-onset epilepsy had genetic diagnoses with direct clinical implications.
Introduction
Epilepsy, defined by recurrent unprovoked seizures or a single seizure with risk factors for developing others,1 is a common disorder often presenting in infancy or childhood2,3 and associated with comorbid conditions, including intellectual disability and autism spectrum disorder (ASD). Approximately 1 in 3 individuals with epilepsy have medically refractory seizures.4 Accordingly, patients, families, and clinicians seek underlying explanations and potentially etiologically specific treatments. Recent studies have demonstrated that a substantial proportion of nonacquired epilepsy is caused by inherited and de novo variants in several brain-expressed genes,5,6 providing insight into developmental and epileptic encephalopathies (DEE), genetic generalized epilepsy (GGE), and nonacquired focal epilepsy (NAFE), sometimes involving the same genes.5,7,8,9,10
Even in the research setting, only 30% to 50% of individuals with presumed genetic epilepsy have known genetic explanations.11 The discrepancy between presumed vs identified molecular diagnoses highlights a gap in understanding of the genetic causes of epilepsies. Furthermore, millions of individuals with presumed genetic epilepsy do not have identified genetic conditions, in part due to limited access to sequencing and challenges in interpretation of findings in many settings.12
Increasing potential for precision diagnosis has fueled a growing focus on precision medicine for the epilepsies.13,14,15 A genetic diagnosis provides an end to the diagnostic odyssey for patients and families and may inform prognosis, recurrence risk, and screening for additional clinical features.12 These latter aspects, and the knowledge that a search for a cause of the epilepsy has been attempted, reflect the potential for clinical and personal utility, which has not been systematically studied, to our knowledge.16,17
Leveraging a prospectively ascertained, single-institution cohort of 522 individuals with a range of pediatric-onset epilepsy phenotypes and performing exome sequencing (ES), we report diagnostic results and their clinical utility.
Methods
Study Cohort
This cohort study was approved by the Boston Children’s Hospital (BCH) institutional review board. All participants provided consent and assent when able. We enrolled biological parents and affected siblings whenever possible. Data were analyzed using descriptive statistics and reported according to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) reporting guideline.
Between August 2018 and October 2021, we recruited individuals from the BCH Department of Neurology and Division of Epilepsy and Clinical Neurophysiology inpatient and outpatient units. Patients with nonacquired epilepsy with unknown genetic etiology were eligible. We did not exclude patients with nonspecific brain magnetic resonance imaging abnormalities, focal cortical dysplasia, or nodular heterotopia. We included patients with abnormal electroencephalogram findings without clinical seizures (eg, DEE with spike-wave activation in sleep), as their genetic causes are expected to overlap with clinical epilepsy. We excluded patients with events suspicious for seizure without definitive epilepsy. DNA was collected as previously described,18,19 with details provided in the eMethods in Supplement 1).
Phenotypic Assessment
We reviewed clinical data from the BCH electronic medical record (EMR) and referring clinicians. We categorized age of seizure onset as neonatal (<1 month), infantile (1 month to <12 months), early childhood (1 year to <6 years), school-aged (6 years to <14 years), or adolescent (≥14 years). Seizure and epilepsy types were classified according to the International League Against Epilepsy classification by treating physicians and confirmed or reclassified by study epileptologists (K.N.W., N.C., C.M.E.A., H.E.O., and A.H.P.).20,21,22,23,24 Each patient was categorized as DEE vs non-DEE, and the non-DEE group divided into GGE (with specific idiopathic generalized epilepsy [IGE] syndromes noted), NAFE, or combined generalized and focal epilepsy. We assessed for the presence of intellectual disability, classified intellectual disability as borderline, mild, moderate, severe, or profound, based on reported IQ in neuropsychological evaluations (when available) or documentation by neurologists of developmental skills and supports needed, classified using standardized published criteria.25 We reviewed the description of the motor portion of the neurological examination and descriptions of motor function (eg, motor milestones, activities of daily living). We assessed for evidence of abnormalities in tone (hypotonia, hypertonia), movement disorder, cerebral palsy, and other diagnoses. We noted relevant neurological family history (including febrile seizures). As variants were identified, we reassessed clinical data relevant to the specific gene.
Variant Identification and Classification
We identified rare, predicted damaging, and clinically relevant variants using standard variant calling and analyses (eMethods in Supplement 1).19 Variants were reviewed by a multidisciplinary team of pediatric neurologists, epileptologists (C.M.E.A., C.J.Y., H.E.O., and A.H.P.), genetic counselors (L.S., S.M. and B.R.S.), and additional researchers (H.Y.K. and A.M.D.) with expertise in epilepsy genetics. We classified variants as pathogenic (P), likely pathogenic (LP), or variants of uncertain significance (VUS) according to the American College of Medical Genetics and Genomics/Association for Molecular Pathology guidelines.26 Variants were deemed diagnostic if they were P or LP in a gene associated with the patient’s phenotype or VUS in a gene associated with the phenotype but with unavailable parental segregation data.
Return of Results
All families opted to receive results. Clinical confirmation was conducted using original samples maintained at GeneDx’s Clinical Laboratory Improvement Amendments–certified laboratory. Clinical reports were issued and families notified of results by treating neurologists and/or qualified clinicians from the study team through the BCH Epilepsy Genetics Clinic.
Assessment of Clinical Utility
For participants receiving diagnostic results at BCH, we evaluated notes for data regarding clinical utility16: impact on treatment or clinical management and/or change in prognosis. We noted mention of personal utility (eg, relief, referral to gene-specific advocacy organizations). For participants with results communicated outside BCH, we assessed whether management recommendations would have been warranted based on the genes involved.
Statistical Analysis
To identify phenotypic factors associated with diagnostic findings, we performed a bivariate analysis for each variable, including sex, age at seizure onset, DEE or intellectual disability, ASD, attention deficit hyperactivity disorder (ADHD), motor impairment (eg, cerebral palsy, hypertonia, hypotonia), and history of afebrile seizure in a parent. We included these variables in a multivariable logistic regression model using R statistical software version 3.2.3 (R Project for Statistical Computing) and SPSS statistical software version 27.0 (IBM) with 2-sided P < .05 as the statistical significance threshold. Data were analyzed on a rolling basis through December 2022.
Results
Cohort Characterization
We enrolled 522 individuals, including 269 (51.5%) male patients, with a mean (SD) age at epilepsy onset of 1.2 (1.4) years and a mean (SD) age at assessment of 9.6 (6.7) years (Table 1). We classified 142 individuals (27.2%) as DEE. Individuals without DEE included 127 individuals (24.3%) with GGE, 53 individuals (10.2%) with specific IGE syndromes, 152 individuals (29.1%) with NAFE, and 48 individuals (9.2%) with combined generalized and focal epilepsy. The most frequently observed syndrome was infantile epileptic spasms syndrome (IESS), reported in 46 individuals (8.8%). Other diagnoses included childhood absence epilepsy (34 individuals [6.5%]), Lennox-Gastaut syndrome (24 individuals [4.6%]), self-limited epilepsy with centrotemporal spike (16 individuals [3.1%]), juvenile myoclonic epilepsy (14 individuals [2.7%]), and epilepsy with myoclonic–atonic seizures (11 individuals [2.1%]). Individuals with seizure onset in early childhood represented the largest subset (229 individuals [43.9%]). Seizures were reported to be refractory to antiseizure medications at last follow-up in 281 participants (53.5%). Comorbidities were common, including intellectual disability in 263 individuals (50.4%). In addition, 75 individuals (14.4%) had ASD and 71 individuals (13.6%) had ADHD. A total of 99 individuals (18.9%) had had previous nondiagnostic clinical genetic testing (ie, panel or chromosomal microarray analysis).
Table 1. Demographic Characteristics and Epilepsy Phenotypes.
| Characteristic | No. (%) (N = 522) |
|---|---|
| Sex | |
| Male | 269 (51.5) |
| Female | 253 (48.5) |
| Age at seizure onset | |
| Mean (SD), y | 1.2 (1.4) |
| Neonatal (<1 mo) | 20 (3.8) |
| Infantile (1 to <12 mo) | 105 (20.1) |
| Early childhood (1 to <6 y) | 229 (43.9) |
| School-aged (6 to <14 y) | 135 (25.9) |
| Adolescent (≥14 y) | 33 (6.3) |
| Epilepsy type | |
| DEE | 142 (27.2) |
| Non-DEE | |
| GGE | 127 (24.3) |
| IGE | 53 (10.2) |
| NAFE | 152 (29.1) |
| Combined generalized and focal | 48 (9.2) |
| Epilepsy syndrome diagnoses | |
| Syndromes associated with refractory seizures or developmental comorbidities | |
| Any | 118 (22.8) |
| Infantile epileptic spasms syndrome | 46 (8.8) |
| Lennox-Gastaut syndrome | 24 (4.6) |
| Epilepsy with myoclonic-atonic seizures | 11 (2.1) |
| Epilepsy with eyelid myoclonia | 9 (1.7) |
| Spike-and-wave activation in sleep | 7 (1.3) |
| Landau-Kleffner syndrome | 5 (1) |
| Sleep-related hypermotor epilepsy | 4 (0.8) |
| Myoclonic epilepsy in infancy | 4 (0.8) |
| Dravet syndrome | 3 (0.6) |
| Epilepsy of infancy with migrating focal seizures | 2 (0.4) |
| Febrile infection-related epilepsy syndrome | 2 (0.4) |
| Hemiconvulsion-hemiplegia epilepsy syndrome | 1 (0.2) |
| Syndromes associated with milder prognosis | |
| Any | 75 (14.4) |
| Childhood absence epilepsy | 34 (6.5) |
| Self-limited epilepsy with centrotemporal spikes | 16 (3.1) |
| Juvenile myoclonic epilepsy | 14 (2.7) |
| Juvenile absence epilepsy | 3 (0.6) |
| Self-limited infantile epilepsy | 3 (0.6) |
| Self-limited epilepsy with autonomic seizures | 2 (0.4) |
| Epilepsy with generalized tonic-clonic seizures alone | 1 (0.2) |
| Self-limited focal epilepsy | 1 (0.2) |
| Photosensitive occipital lobe epilepsy | 1 (0.2) |
| Responsive to ASMs (seizure-free) | 222 (46.4) |
| Intellectual disability | |
| None | 259 (49.6) |
| Borderline | 74 (14.2) |
| Mild | 79 (15.1) |
| Moderate | 59 (11.3) |
| Severe | 38 (7.3) |
| Profound | 13 (2.5) |
| Other neurodevelopmental diagnoses | |
| Presence of ASD | 75 (14.4) |
| Presence of ADHD | 71 (13.6) |
Abbreviations: ADHD, attention deficit hyperactivity disorder; ASD, autism spectrum disorder; ASM, antiseizure medication; DEE, developmental and epileptic encephalopathy; GGE, genetic generalized epilepsy; IGE, idiopathic generalized epilepsy; NAFE, nonacquired focal epilepsy.
Summary of Genetic Diagnoses
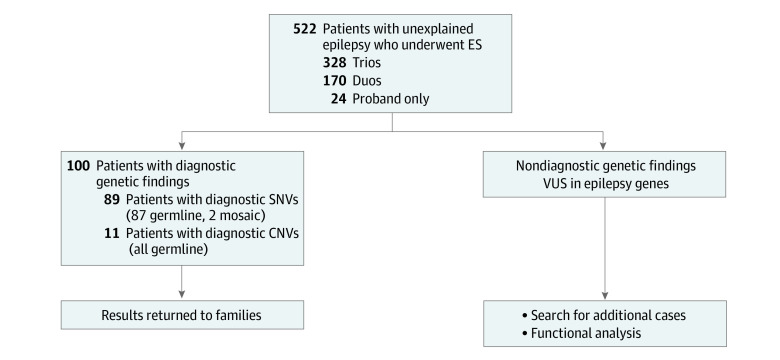
Sequencing was conducted on 328 trios (17 individuals with siblings), 170 duos with only 1 biological parent available (8 individuals with siblings), and 24 singletons (4 individuals with siblings) (Figure 1). We identified diagnostic genetic etiologies in 100 of 522 individuals (19.2%): 89 single nucleotide variants (SNVs) (17.0%) and 11 CNVs (2.1%). Among 142 individuals with DEE, we identified genetic etiologies in 45 individuals (31.7%). Of 180 individuals with GGE including IGE, we identified genetic etiologies in 26 individuals (14.4%); genetic etiologies were also identified in 22 of 152 individuals (14.5%) with NAFE and 7 of 48 individuals (14.6%) with combined focal and generalized epilepsy (eFigure 1 in Supplement 1). There was genetic heterogeneity in all groups, and some genes were identified in multiple groups (eFigure 1 in Supplement 1). Diagnostic yield among individuals with prior clinic testing was 18 of 99 individuals (18.2%), similar to those who had no prior testing (82 of 423 individuals [19.4%]; P = .79).
Figure 1. Summary of Diagnostic Yield From Exome Sequencing (ES) Patients With Unexplained Epilepsy.
A total of 522 patients with previously unexplained epilepsy were enrolled and underwent ES, with 1 or both parents as available. We identified diagnostic single nucleotide variants (SNVs) in 89 individuals. These pathogenic or likely pathogenic variants and diagnostic variants of uncertain significance (VUS) were clinically confirmed and returned to patients and families. Dedicated copy number variant (CNV) analysis of the ES data identified an additional 11 diagnostic CNVs, which were also returned to patients and families. Candidate gene findings and VUS in epilepsy-associated genes that were not determined to be diagnostic were not returned to families but will be reevaluated as additional data emerges or in the eventual emergence of functional data supporting pathogenesis.
Diagnostic SNVs
We initially identified 317 individuals with rare, potentially damaging SNVs. Manual filtering resulted in 89 individuals (17.0%) with a total of 96 variants (including compound heterozygous or homozygous variants) that we classified as diagnostic and returned to families (81 P or LP variants, 15 VUS) (Figure 2 and Table 2). These 89 participants harbored variants (including 43 previously reported) in 69 genes established as associated with epilepsy and neurodevelopmental disorders (eFigure 2 and eTable 1 in Supplement 1). Nine patients had diagnostic variants in SCN1A (OMIM: 182389), 3 patients each in DEPDC5 (OMIM: 614191) and PRRT2 (OMIM: 614386), 2 patients each in ANKRD11 (OMIM: 611192), CHD2 (OMIM: 602119), GABRG2 (OMIM: 137164), KCNMA1 (OMIM: 600150), PCDH19 (OMIM: 300460), SCN1B (OMIM: 600235), STXBP1 (OMIM: 602926), and SYNGAP1 (OMIM: 603384), and 1 patient each in 58 other genes. Variant types included missense (54 variants [50.5%]), nonsense (16 variants [15.0%]), frameshift (13 variants [12.2%]), and splice site affecting (9 variants [8.4%]) (eFigure 3 in Supplement 1). Two variants were present in mosaic form, 1 in SCN1A and 1 in NEXMIF. Heterozygous apparently de novo variants in genes associated with autosomal dominant conditions and mechanisms comprised 39 variants (40.2%) of our diagnostic variants (eFigure 3 in Supplement 1); 2 of these variants were subsequently identified to be mosaic in a parent. We identified inherited variants in 24 individuals, including 14 autosomal dominant conditions and 15 autosomal recessive conditions.
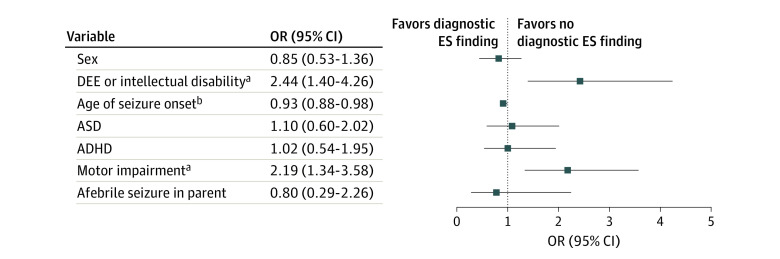
Figure 2. Clinical Features in Patients With Diagnostic Variants.
Multiple logistic regression analysis of 7 phenotypic variables found that developmental epileptic encephalopathy (DEE) or diagnosis of intellectual disability and history of motor impairment were the strongest factors associated with identifying a diagnostic Exome Sequencing (ES) finding. ADHD indicates attention deficit hyperactivity disorder; ASD, autism spectrum disorder; and OR, odds ratio.
aP < .001.
bP < .05.
Table 2. Diagnostic Genetic SNVs and CNVs and Their Associated Phenotypes, From a Cohort of 522 Patients With Unexplained Pediatric Epilepsya.
| Participant ID | Gene or coordinate | Variant or size/type of CNV | Inheritance | ACMG/AMP for SNVs or syndrome/involved genes for CNVs | Age at seizure onset | Sex | Epilepsy type | Syndrome |
|---|---|---|---|---|---|---|---|---|
| SNVs | ||||||||
| 1 | ANKRD11 | c.3039_3045del, p.Asp1013GlufsTer303 | De novo | P | 14 y | Female | Combined | NR |
| 2 | c.7535G>A, p.Arg2512Gln | Unknown | P | 1 y | Female | DEE | LGS | |
| 3 | ARHGEF9 | c.1285del, p.Glu429LysfsTer19 | Unknown | LP | 4 mo | Female | DEE | NR |
| 4 | ARID1B | c.3586C>T, p.Gln1196Ter | De novo | P | 8 y | Female | NAFE | NR |
| 5 | ATN1 | c.3193C>T, p.Gln1065Ter | De novo | VUS | 10 y | Male | GGE | NR |
| 6 | BCL11A | c.198C>G, p.His66Gln | Unknown | LP | 2 y | Female | GGE | EEM |
| 7 | BRAF | c.770A>G, p.Gln257Arg | De novo | P | 1 y 6 mo | Male | DEE | NR |
| 8 | BRAT1 | c.1925C>A, p.Ala642Glu | Paternal | LP | 6 wk | Female | DEE | NR |
| c.294dupA, p.Leu99ThrfsTer92 | Maternal | P | ||||||
| 9 | BRWD3 | c.4080+1G>A | Maternal | LP | 2 y | Male | NAFE | NR |
| 10 | CACNA1A | c.601C>T, p.Arg201Trp | Unknown | LP | 4 y | Female | IGE | CAE |
| 11 | CACNA1G | c.3568C>T, p.Arg1190Ter | De novo | VUS | 5 y | Female | IGE | CAE |
| 12 | CHD2 | c.2876+3_2876+6delAAGT | De novo | LP | 1 y | Female | DEE | NR |
| 13 | c.3895_3896insC, p.Val1299AlafsTer5 | Unknown | P | 1 y 10 mo | Male | DEE | NR | |
| 14 | CLN8 | c.784G>A, p.Asp262Asn | Unknown | LP | 5 y 6 mo | Female | DEE | NR |
| c.610C>T, p.Arg204Cys | Maternal | P | ||||||
| 15 | CREBBP | c.5315T>A, p.Ile1772Asn | Unknown | LP | Infantile | Male | DEE | NR |
| 16 | CSNK2A1 | c.921T>G, p.Tyr307Ter | De novo | LP | 9 m | Female | DEE | NR |
| 17 | CSNK2B | c.557+1G>A | Unknown | LP | 2 m | Male | DEE | NR |
| 18 | CYFIP2 | c.2542A>G, p.Met848Val | De novo | LP | 4 m | Male | DEE | IESS |
| 19 | DEPDC5 | c.363+1G>A | Paternal | LP | 3 y | Male | NAFE | NR |
| 20 | c.667A>G, p.Arg223Gly | Maternal | VUS | 8 y | Male | NAFE | NR | |
| 21 | c.1459C>T, p.Arg487Ter | Unknown | P | 10 y | Female | GGE | NR | |
| 22 | DYNC1H1 | c.5864G>T, p.Gly1955Val | De novo | LP | 2 mo | Male | DEE | IESS |
| 23 | EEF1A2 | c.364G>A, p.Glu122Lys | De novo | P | Infantile | Male | DEE | NR |
| 24 | FRRS1L | c.737_739del, p.Gly246del | Unknown | P | 11 mo | Female | DEE | NR |
| 25 | FOXP1 | c.-448G>C | Unknown | VUS | 4 y | Male | DEE | LGS |
| 26 | GABRA5 | c.902C>T, p.Thr301Met | De novo | LP | 11 y | Female | Combined | NR |
| 27 | GABRG2 | c.1087C>T, p.Arg363Trp | Unknown | LP | 1 y | Male | GGE | NR |
| 28 | c.542C>A, p.Thr181Asn | Maternal | LP | 7 y | Female | NAFE | NR | |
| 29 | GFAP | c.882C>A, p.Cys294Ter | Unknown | VUS | 2 y | Male | GGE | NR |
| 30 | GPHN | c.1471A>T, p.Arg491Ter | Unknown | P | 1 y | Male | GGE | NR |
| 31 | GRIA3 | c.1580C>A, p.Ser527Arg | Maternal | VUS | 1 y 6 mo | Male | GGE | NR |
| 32 | GRIN2A | c.1122+1G>C | Unknown | LP | 3 y | Male | NAFE | NR |
| 33 | GRIN2B | c.1843A>T, p.Asn615Tyr | De novo | P | 2 mo | Female | DEE | IESS |
| 34 | KCNA2 | c.217C>T, p.Arg73Ter | Unknown | LP | 9 y | Male | Combined | NR |
| 35 | KCNMA1 | c.1918C>T, p.Arg640Ter | Paternal | LP | 8 y | Female | Combined | NR |
| 36 | c.3199A>G, p.Lys1067Glu | De novo | LP | 14 y | Female | IGE | JME | |
| 37 | KCNQ2 | c.365C>T, p.Ser122Leu | De novo | P | 3 d | Female | DEE | NR |
| 38 | KCNQ3 | c.688C>T, p.Arg230Cys | De novo | P | 5 y | Female | DEE | NR |
| 39 | KDM4B | c.719G>A, p.Arg240Gln | De novo | LP | 1 y 6 mo | Male | DEE | NR |
| 40 | KDM6B | c.40C>G, p.Arg14Gly | De novo | VUS | 4 mo | Female | GGE | NR |
| 41 | KMT2E | c.1097_1116del20, p.Glu366ValfsTer4 | De novo | P | 4 y | Female | IGE | CAE |
| 42 | LGI1 | c.757G>A, p.Ala253Thr | Maternal | VUS | 4 y 6 mo | Male | NAFE | SELECTS |
| 43 | MECP2 | c.1200_1243del, p.Pro401Ter | De novo | P | 3 y | Female | NAFE | SHE |
| 44 | MTR | c.2411T>C, p.Ile804Thr | Paternal | VUS | 5 mo | Female | DEE | IESS |
| c.2472A>T, p.Ala824= | Maternal | VUS | ||||||
| 45 | NBEA | c.4702dup, p.Val1568GlyfsTer14 | De novo | P | 1 y 6 mo | Male | DEE | EMATS |
| 46 | NEXMIF | c.846_849delTGTC, p.V283tfsX20 | De novo mosaic | P | 5 y | Female | GGE | NR |
| 47 | NPRL2 | c.323_339+19del | Paternal | P | 3 mo | Male | NAFE | IESS |
| 48 | OTUD6B | c.433C>T, p.Arg145Ter | Both | P | 9 mo | Male | DEE | IESS |
| 49 | PCDH19 | c.811_825del, p.Gly271_Tyr275del | De novo | LP | 9 mo | Female | DEE | NR |
| 50 | c.1335C>A, p.Asp445Glu | Maternal | LP | 6 mo | Female | DEE | NR | |
| 51 | POLR2A | c.3281C>T, p.Ser1094Phe | De novo | LP | 2 y | Male | DEE | NR |
| 52 | PGAP2 | c.823A>G, p.Met275Val | Unknown | LP | 5 mo | Male | DEE | IESS |
| c.1040C>T, p.Ala347Val | Maternal | LP | ||||||
| 53 | POLG | c.1760C>T/c.752C>T, p.Pro587Leu/p.Thr251Ile | Maternal | P | 5 y | Male | GGE | NR |
| c.1703G>C, p.Gly568Ala | Paternal | VUS | ||||||
| 54 | PPP2R5D | c.592G>A, p.Glu198Lys | Unknown | P | 2 y | Male | DEE | NR |
| 55 | PRRT2 | c.870delT, p.Tyr290Ter | De novo | P | 5 mo | Female | NAFE | SELIE |
| 56 | c.649dup, p.Arg217ProfsTer8 | Paternal | P | 3 mo | Female | NAFE | NR | |
| 57 | c.649dup, p.Arg217ProfsTer8 | Maternal | P | 5 mo | Male | GGE | SELIE | |
| 58 | RORA | c.680del, p.Thr227ArgfsTer80 | De novo | P | 6 y | Male | Combined | NR |
| 59 | SCN1A | c.5066T>C, p.Met1689Thr | De novo | LP | 3 y | Male | Combined | NR |
| 60 | c.5495C>A, p.Ala1832Glu | Maternal mosaic | P | 6 mo | Male | DEE | DS | |
| 61 | c.3429G>C, p.Glu1143Asp | De novo | LP | 6 mo | Male | DEE | DS | |
| 62 | c.664C>T, p.Arg222Ter | Maternal mosaic | P | 9 mo | Male | DEE | DS | |
| 63 | c.4634T>G, p.Ile1545Arg | De novo | P | 3 mo | Male | GGE | NR | |
| 64 | c.2955T>G, p.Asn985Lys | Unknown | LP | 6 mo | Female | NAFE | NR | |
| 65 | c.332T>A, p.Leu111Ter | De novo mosaic | P | 11 mo | Male | NAFE | GEFS+ | |
| 66 | c.4057G>A, p.Val1353Ile | Unknown | P | 6 mo | Male | DEE | NR | |
| 67 | c.5606T>C, p.Phe1869Ser | Maternal | VUS | 2 y | Male | GGE | NR | |
| 68 | SCN1B | c.363C>G, p.Cys121Trp | Maternal | LP | 1 y 6 mo | Female | NAFE | NR |
| 69 | c.1A>C, p.Met1? | Paternal | LP | 2 y | Female | GGE | NR | |
| 70 | SCN8A | c.3955G>T, p.Ala1319Ser | De novo | P | 3 wk | Male | DEE | NR |
| 71 | SETD1A | c.4268A>G, p.Gln1423Arg | De novo | LP | 11 mo | Female | DEE | HHE |
| 72 | SETD1B | c.5726T>C, p.Ile1909Thr | De novo | LP | 5 y | Female | IGE | CAE |
| 73 | SHANK3 | c.3949dupG, p.Val1317GlyfsX28 | De novo | P | 8 y | Female | DEE | NR |
| 74 | SLC12A5 | c.1052A>G, p.Asn351Ser | Both | LP | 3 m | Female | DEE | EIMFS |
| 75 | SON | c.6888T>G, p.Asp2296Glu | De novo | LP | 8 y | Male | DEE | NR |
| 76 | SPATA5 | c.2045C>T, p.Ala682Val | Maternal | LP | 3.5 y | Male | DEE | NR |
| c.1883A>G, p.Asp628Gly | Paternal | LP | ||||||
| 77 | SPTAN1 | c.6589_6594dupGAGCT, p.Glu2197_Leu2198dup | De novo | P | 2 y | Male | DEE | NR |
| 78 | SRCAP | c.8919del, p.Leu2975Ter | Unknown | P | 5 y | Female | GGE | NR |
| 79 | STAG1 | c.1145C>T, p.Thr382Ile | De novo | VUS | 1 y 3 mo | Female | NAFE | NR |
| 80 | STXBP1 | c.1652G>A, p.Arg551His | Unknown | P | 1 m | Female | DEE | IESS |
| 81 | c.847G>A, p.Glu283Lys | Unknown | P | 1 y 6 mo | Male | NAFE | NR | |
| 82 | SYNGAP1 | c.403C>T, p.Arg135Ter | De novo | P | 3 y | Female | DEE | NR |
| 83 | c.1630C>T, p.Arg544Ter | Unknown | P | 2 y | Male | DEE | NR | |
| 84 | TANC2 | c.2326G>T, p.Glu776Ter | De novo | P | 8 mo | Female | DEE | IESS, LGS |
| 85 | TCF4 | c.1486+5delG | De novo | LP | 11 y | Female | DEE | NR |
| 86 | TRIT1 | c.967C>T, p.Arg323Trp | Both | LP | 1 y 6 mo | Female | DEE | NR |
| 87 | UBA5 | c.829G>A, p.Gly277Ser | Paternal | LP | 1 y 11 mo | Male | NAFE | NR |
| c.1111G>A, p.Ala371Thr | Maternal | P | ||||||
| 88 | WDR26 | c.706C>G, p.Leu236Val | Unknown | VUS | 3 y | Male | DEE | NR |
| 89 | ZEB2 | c.3135C>G, p.His1045Gln | Unknown | VUS | 2 y 6 mo | Female | GGE | NR |
| CNVs | ||||||||
| 90 | chr2:166847505-167334456 | 487 kb deletion | De novo | SCN1A | 1 y | Male | Combined | NR |
| 91 | chr3:11058648-11060634 | 2 kb deletion | Unknown27 | SLC6A1 | 1 y 10 mo | Female | GGE | NR |
| 92 | chr16:138446-140150 | 1.7 kb deletion | Unknown | NPRL3 | 4 mo | Male | NAFE | IESS |
| 93 | chr22:32121274-32302733 | 181 kb deletion | Maternal | DEPDC5 | 6 y | Male | NAFE | NR |
| 94 | chr22:32193336-32194893 | 1.5 kb deletion | Maternal | DEPDC5 | 3 y | Male | NAFE | NR |
| 95 | chr1:146630894-147415874 | 785 kb deletion | Paternal | 1q21.1 recurrent microdeletion | 5 y | Female | IGE | CAE |
| 96 | chr15:30896079-32404350 | 1.5 Mb deletion | De novo | 15q13.3 recurrent microdeletion | 6 y | Female | GGE | EMA |
| 97 | chr16:29674800-30199626 | 525 kb duplication | De novo | 16p11.2 recurrent microduplication | 8 mo | Male | NAFE | NR |
| 98 | chr16:21964495-22385880 | 421 kb deletion | Maternal | 16p12.1 recurrent microdeletion | 4 y | Male | GGE | NR |
| 99 | chr16:14960162-16297720 | 1.3 Mb deletion | Maternal | 16p13.11 recurrent microdeletion | 14 y | Male | NAFE | NR |
| 100 | chr22:18893638-21386351 | 2.5 Mb deletion | De novo | 22q11 deletion syndrome | 6 y | Female | GGE | NR |
Abbreviations: ACMG/AMP, American College of Medical Genetics and Genomics/Association for Molecular Pathology; ASM, antiseizure medication; CAE, childhood absence epilepsy; CNV, copy number variant; DEE, developmental and epileptic encephalopathy; DS, Dravet syndrome; EEM, epilepsy with eyelid myoclonia; EIMFS, epilepsy of infancy with migrating focal seizures; EMA, epilepsy with myoclonic absences; EMATS, epilepsy with myoclonic-atonic seizures; GEFS+, generalized epilepsy with febrile seizure plus; GGE, genetic generalized epilepsy; HHE, hemiconvulsion-hemiplegia epilepsy syndrome; ID, identification number; IESS, infantile epileptic spasm syndrome; IGE, idiopathic generalized epilepsy; JME, juvenile myoclonic epilepsy; LGS, Lennox-Gastaut syndrome; LP, likely pathogenic; NAFE, nonacquired focal epilepsy; NR, not reported; P, pathogenic; SELECTS, self-limited epilepsy with centrotemporal spikes; SELIE, self-limited infantile epilepsy; SHE, sleep-related hypermotor epilepsy; SNV, single nucleotide variant; VUS, variants of uncertain significance.
Gene, variant, and ACMG/AMP classification are given for SNVs. Coordinate, size/type of CNV, and syndrome or genes involved are given for CNVs.
Nondiagnostic SNVs
An additional 161 individuals (30.8%) had VUS in known epilepsy genes (eTable 2 in Supplement 1) not considered diagnostic due to phenotypic or disease mechanism inconsistency (eTable 3 in Supplement 1). A total of 93 individuals had variants (72 de novo variants) in 101 candidate genes (eFigure 2 and eTable 4 in Supplement 1) not yet implicated in epilepsy but with experimental evidence suggesting a role in brain development (eg, neuronal migration, signaling, or hyperexcitability).28
Genetics Related to Syndromes
We identified genetic diagnoses in 9 of 46 individuals (20%) with IESS. Of 9 individuals with SCN1A P, LP, or VUS variants, 3 had clinical Dravet syndrome; notably, 2 parents harbored these variants in mosaic form. The others had DEE (1 individual), GGE (2 individuals), NAFE (1 individual with a germline variant and 1 individual with a mosaic variant in the proband), and combined epilepsy (1 individuals), all with seizures in the setting of fever or illness. Notably, we observed a range of epilepsy phenotypes for those genes responsible for more than 1 condition, with SCN1A associated with all 4 of the aforementioned categories, DEPDC5 with GGE and NAFE and PRRT2 with GGE and NAFE.
We iteratively interrogated phenotypic data in our interpretation of VUS, accounting for clinical features relevant to the implicated genes. For example, following detection of compound heterozygous VUS in PGAP2, we confirmed hyperphosphatasia through clinical biochemical testing. A homozygous VUS in SLC12A5 was identified in a patient with epilepsy of infancy with migrating focal seizures.29,30 Finally, a VUS in CLN8 provided an early diagnosis of neuronal ceroid lipofuscinosis, allowing for anticipatory guidance. We designated these VUS as likely diagnostic, given phenotypic features closely associated with the relevant genes.
Diagnostic CNVs
We identified diagnostic CNVs in 11 individuals (2.1%) (Table 2; eTable 5 in Supplement 1), none of whom had a diagnostic SNV. Four variants were de novo, 5 variants were inherited, and 2 variants were unknown. In a child with refractory epilepsy and intellectual disability, we identified an 181 kb deletion in DEPDC5 (chromosome 22) inherited from a parent with well-controlled epilepsy without intellectual disability. For the 4 other inherited CNVs, the parents bearing the CNVs were unaffected, consistent with variable penetrance associated with many CNVs.27,31,32
Phenotypes Associated With Genetic Diagnoses
Multivariable analysis demonstrated higher diagnostic yield among individuals with DEE or intellectual disability (adjusted odds ratio [aOR], 2.44 [95% CI, 1.40-4.26]) and motor impairment (aOR, 2.19 [95% CI, 1.34-3.58]) (Figure 2; eTable 6 in Supplement 1). Patients with younger age at onset had more genetic diagnoses: each year of increasing age conferred a 7% reduction in the likelihood of identifying a genetic cause (aOR per 1-year, 0.93 [95% CI, 0.88-0.98]).
Clinical Utility of Genetic Diagnoses
Data were available to assess clinical utility for 71 of 100 participants with genetic diagnoses (Table 3). For 29 of these patients (40.8%), we observed evidence of impact in a change in treatment or management or a change in prognosis. Genetic diagnosis led to either a discussion of or actual change in treatment, including change in antiseizure medication or implementation of the ketogenic diet, in 27 patients (38.0%). Other management changes included referrals to other specialties due to risk of nonneurological manifestations and weaning of an antiseizure medication after genetic diagnosis suggested a self-resolving epilepsy (eg, PRRT2). Five patients (7%) had a striking change in prognosis, including an early and unsuspected diagnosis of neuronal ceroid lipofuscinosis (CLN8).
Table 3. Clinical Utility of Genetic Diagnoses for Individuals With Pediatric-Onset Epilepsya.
| Gene | Actions | Epilepsy type |
|---|---|---|
| Change in treatment and management | ||
| ANKRD11 | Referred to cardiovascular genetics, who recommended ECHO with follow-up in 1 y | DEE |
| Referred to audiology for hearing evaluation | ||
| Referred to endocrinology for short stature and bone mineralization seen in KBG syndrome | ||
| Referred to orthopedic surgery for evaluation for potential vertebral abnormalities and scoliosis | ||
| ARID1B | Referred for renal ultrasound and hypothyroidism screening | NAFE |
| Referred for evaluation for ASD | ||
| BCL11A | Referred to hematology for blood smear to assess for possible BCL11A-associated bone marrow abnormalities | GGE |
| BRAF | Referred to cardiology for evaluation due to high rate of cardiac abnormalities associated with BRAF | DEE |
| CREBBP | Recommended screening for cataracts, kidney, and thyroid abnormalities | DEE |
| CSNK2A1, CSNK2B | Referred to cardiology for baseline evaluation | DEE |
| DEPDC5 | Referred for epilepsy surgical evaluation due to high success rate with focal epilepsy in the setting of mTORopathies | NAFE |
| Counseled regarding increased risk for SUDEP and discussed monitoring devices | ||
| KCNMA1 | Monitoring for symptoms of movement disorders (paroxysmal dyskinesia and ataxia) | Combined |
| MECP2 | Annual EKG to evaluate for long QT | NAFE |
| Regular spine examinations for scoliosis | ||
| Monitoring for GERD, constipation, signs and symptoms of gallstones | ||
| Regular cholesterol screening | ||
| NPRL2 | Referred for epilepsy surgical evaluation due to high success rate with focal epilepsy in the setting of mTORopathies | NAFE |
| PGAP2 | Discussed reports of patients who benefit with vitamin B6 and recommended to monitoring for worsening symptoms if vitamin B6 were discontinued; referred to cardiology for EKG | DEE |
| Referred to endocrinology | ||
| SCN1A | Started cannabidiol and fenfluramine | DEE |
| Recommended temperature management (eg, cooling vest when playing outside) | ||
| Counseled regarding increased risk for SUDEP; family obtained a monitor for sleeping | ||
| SCN1B | Referred to cardiology for evaluation for arrhythmias (due to prior association with Brugada syndrome) | NAFE |
| SHANK3 | Referred to cardiology for baseline evaluation | DEE |
| Referred to nephrology for baseline evaluation | ||
| Recommended routine ophthalmology evaluations | ||
| SLC6A1 | Discussion of medications reported effective in this condition for seizures (ie, valproic acid) | GGE |
| TCF4 | Discussion of medications reported effective in this condition for seizures (ie, lamotrigine). | DEE |
| TRIT1 | Discussion of treatments reported effective in this condition for seizures (ie, ketogenic diet) | DEE |
| Referred to cardiology for evaluation. | ||
| Change in prognosis | ||
| CLN8 | Provided diagnosis and counseling regarding the presence of a neurodegenerative disorder. | DEE |
| PPP2R5D | Patient is substantially delayed and not yet walking; counseled that individuals with this diagnosis can develop skills much later than typical | NAFE |
| PRRT2 | Change in prognosis: confidence regarding weaning seizure medication and anticipatory guidance regarding possible movement disorder | NAFE |
| Explains family history of paroxysmal kinesigenic dyskinesia that was previously undiagnosed/unexplained | ||
Abbreviations: ASD, autism spectrum disorder; DEE, developmental and epileptic encephalopathy; ECHO, echocardiography; EKG, electrocardiography; GGE, genetic generalized epilepsy; GERD, gastroesophageal reflux disease; NAFE, nonacquired focal epilepsy; SUDEP, sudden unexpected death in epilepsy.
Includes only patients for whom such discussion is explicitly documented.
Genetic counseling, including reproductive risk counseling, was offered to all families seen at BCH with diagnostic results. This was particularly relevant for families with inherited variants and for the 2 unaffected parents with low-level mosaic variants.
For the 29 participants for whom follow-up data after genetic diagnosis were not available, we noted that 8 (27.6%) had diagnoses across 5 genes associated with clinical management recommendations: SCN1A (3 patients), DEPDC5 (2 patients), POLG, STXBP1, and MTR.
Discussion
In this cohort study, we report the genetic results for a large, clinically ascertained cohort with previously unexplained pediatric-onset epilepsy. Our research-based ES data analysis included evaluation for both SNVs and CNVs, the latter from ES data, and evaluation for somatic mosaic variants. The overall yield of 19% (17% SNVs, 2% CNVs) encompasses patients with diverse epilepsy syndromes and varying severity. Equally diverse are the 69 genes identified and the pathways implicated for both early and later onset epilepsies, demonstrating the strength of genomewide approaches.33,34 SCN1A represented the most commonly identified gene, accounting for 9% of our diagnostic findings and associated with a range of phenotypes.
Consistent with prior reports,16,35 our diagnostic yield was highest in patients with higher severity, with earlier age at onset, DEE or intellectual disability, and motor impairment. Specifically, diagnostic yield for DEE (32%) was more than twice that for the other groups (14%-15%). IESS was the most common DEE syndrome in our cohort, which may reflect hospitalization rates16,36,37 or referral bias. We also demonstrate genetic diagnoses in patients with non-DEE epilepsies, for whom there may be lower suspicion of genetic epilepsy and less clinical urgency. While non-DEE epilepsies (eg, GGE, NAGE) are considered influenced by polygenic factors,10,38 we found single-gene explanations for some patients and overlap of genes implicated in the DEE and non-DEE groups.
The ability to return clinically significant results allowed direct translation of our research into the clinical realm and allowed clinicians to conduct follow-up biochemical and imaging studies, as needed, to provide evidence supporting variant pathogenicity. This is imperative, particularly in the interpretation of VUS, which can benefit from additional phenotypic information. We highlight the importance of including all clinically relevant variants, including VUS that may warrant formal reclassification, and the important dynamic aspect of variant classification that incorporates emerging phenotypic, segregation, and functional data.39,40
Our continued access to enrolled patients and longitudinal EMR data enabled us to evaluate clinical utility. Delineation of clinical utility of genetic diagnoses in epilepsy is projected to increase testing by clinicians and support reimbursement from payers, thus increasing access to testing for patients. In contrast, we included a patient population with broad epilepsy phenotypes, some of whose insurance had denied coverage for clinical testing and some with low suspicion of genetic etiology. Our evidence of clinical utility in treatment, clinical management, and prognosis support clinical testing for a broad range of epilepsies.16 Beyond clinical utility, we noted anecdotally that several families expressed reduced guilt or shame after genetic diagnosis, relief at the end of a diagnostic odyssey that in some cases had lasted several years, and hope for still undiagnosed families that answers were still being explored through research. We advocate for prospective studies of the clinical and personal utility of genetic diagnoses among cohorts with epilepsy to more comprehensively demonstrate their impact.
We enrolled individuals from an academic hospital where patients seek care for new-onset epilepsy as well as long-term care for refractory epilepsy. While there were no overtly unusual features for patients with IGE or self-limited focal epilepsy, it is possible that patients with refractory seizures, who are seen more frequently, had more opportunities to enroll or more questions raised regarding etiology. This may bias our sample toward individuals with identifiable genetic diagnoses, although conversely, our overall yield may have been diminished by inclusion of patients with milder epilepsy phenotypes, such as GGE and NAFE.7,10 During the course of this study, we observed variability in genetic testing approval by insurance payers. Increased access to clinical ES, especially for DEE, may have reduced the seizure severity for patients referred for research ES, possibly accounting for our diagnostic yield of 19% being lower than some previous reports.41
We recognize the importance of continued evaluation of patients for whom genetic causes were not found. Exome reanalysis should include evaluation for novel genes and mosaic variants. For some patients, particularly those with syndromes suggesting 1 or more specific genes, genetic diagnoses may be identifiable through targeted deep sequencing of specific genes to assess for mosaic variants, or trio genome sequencing to evaluate for intronic, structural, or other types of variants undetectable with standard ES.42 Identification of additional patients and ongoing functional studies may ultimately lead to increased certainty regarding candidate genes. While the time required to scrutinize each variant of potential interest may be prohibitive in some settings, we demonstrate the merits of this approach, with referrals to neurogenetics specialists or subspecialty clinics as needed for variant assessment and explanation of results and their implications to patients and families. As technologies continue to evolve, we advocate for continued harmonization between the research and clinical realms for variant interpretation and translation of research findings to achieve diagnostic precision and clinical utility for all patients with unexplained epilepsy. Finally, future research concerning the psychological effects of these sometimes early genetic diagnoses on families will be important to inform future neurogenetics practice.
Limitations
This study has some limitations. We used exome capture and undertook CNV analysis of the resulting data to identify deletions or duplications. We acknowledge that exome capture may not detect all CNVs and that genome sequencing might be needed to detect variants beyond SNVs. We were also limited in our interpretation by lack of parental data for some patients, but we chose to include all individuals regardless of parental availability. Furthermore, as with a recent study reporting clinical utility of epilepsy panels,43 our EMR-based assessment could not accurately determine impact on hospitalization rates, morbidity or mortality, clinical trials eligibility, and avoidance of testing procedures (such as lumbar puncture, magnetic resonance imaging, or electroencephalography).
Conclusions
In this cohort study, we illustrated the diverse genetic landscape of pediatric-onset epilepsy in a hospital-based cohort, leveraging research-clinical partnerships to incorporate evolving clinical data in phenotyping, implement the most current guidelines with expertise in genomic analysis and variant interpretation, and increase diagnostic yield and clinical utility.
eMethods.
eTable 1. Clinical and Diagnostic Genetic Findings in 89 Patients With SNVs
eTable 2. A Phenotype-Driven Epilepsy Gene List
eTable 3. Nondiagnostic VUS in Epilepsy-Related Genes
eTable 4. Novel Candidate Epilepsy Genes and Their Associated Phenotypes
eTable 5. Clinical and Diagnostic Genetic Findings in 11 Cases With CNVs
eTable 6. Bivariate and Multivariate Regression Model of Phenotypic Variables to Diagnostic Yield by ES
eFigure 1. Diagnostic Yield and Age of Seizure Onset According to Epilepsy Type
eFigure 2. Established and Candidate Epilepsy Genes in the Study Cohort
eFigure 2. Variant Types Among the Diagnostic SNVs and CNVs
eFigure 3. Clinical Features in Patients With Epilepsy Associated With Diagnostic Variants
Members of the BCH Neurology Referral and Phenotyping Group
Data Sharing Statement
References
- 1.Fisher RS, Acevedo C, Arzimanoglou A, et al. ILAE official report: a practical clinical definition of epilepsy. Epilepsia. 2014;55(4):475-482. doi: 10.1111/epi.12550 [DOI] [PubMed] [Google Scholar]
- 2.Jacobs M, Jensen FE. Introduction to institute of medicine report: epilepsy across the spectrum: promoting health and understanding. Epilepsy Curr. 2012;12(6):243-244. doi: 10.5698/1535-7511-12.6.243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fiest KM, Sauro KM, Wiebe S, et al. Prevalence and incidence of epilepsy: a systematic review and meta-analysis of international studies. Neurology. 2017;88(3):296-303. doi: 10.1212/WNL.0000000000003509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Löscher W. Fit for purpose application of currently existing animal models in the discovery of novel epilepsy therapies. Epilepsy Res. 2016;126:157-184. doi: 10.1016/j.eplepsyres.2016.05.016 [DOI] [PubMed] [Google Scholar]
- 5.Helbig I, Lowenstein DH. Genetics of the epilepsies: where are we and where are we going? Curr Opin Neurol. 2013;26(2):179-185. doi: 10.1097/WCO.0b013e32835ee6ff [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas RH, Berkovic SF. The hidden genetics of epilepsy—a clinically important new paradigm. Nat Rev Neurol. 2014;10(5):283-292. doi: 10.1038/nrneurol.2014.62 [DOI] [PubMed] [Google Scholar]
- 7.Epi4K consortium; Epilepsy Phenome/Genome Project . Ultra-rare genetic variation in common epilepsies: a case-control sequencing study. Lancet Neurol. 2017;16(2):135-143. doi: 10.1016/S1474-4422(16)30359-3 [DOI] [PubMed] [Google Scholar]
- 8.Epi4K Consortium; EuroEPINOMICS-RES Consortium; Epilepsy Phenome Genome Project . Application of rare variant transmission disequilibrium tests to epileptic encephalopathy trio sequence data. Eur J Hum Genet. 2017;25(7):894-899. doi: 10.1038/ejhg.2017.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.EuroEPINOMICS-RES Consortium; Epilepsy Phenome/Genome Project; Epi4K Consortium . De novo mutations in synaptic transmission genes including DNM1 cause epileptic encephalopathies. Am J Hum Genet. 2014;95(4):360-370. doi: 10.1016/j.ajhg.2014.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koko M, Motelow JE, Stanley KE, Bobbili DR, Dhindsa RS, May P; Canadian Epilepsy Network; Epi4K Consortium; Epilepsy Phenome/Genome Project; EpiPGX Consortium; EuroEPINOMICS-CoGIE Consortium . Association of ultra-rare coding variants with genetic generalized epilepsy: a case-control whole exome sequencing study. Epilepsia. 2022;63(3):723-735. doi: 10.1111/epi.17166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McTague A, Howell KB, Cross JH, Kurian MA, Scheffer IE. The genetic landscape of the epileptic encephalopathies of infancy and childhood. Lancet Neurol. 2016;15(3):304-316. doi: 10.1016/S1474-4422(15)00250-1 [DOI] [PubMed] [Google Scholar]
- 12.Poduri A, Sheidley BR, Shostak S, Ottman R. Genetic testing in the epilepsies—developments and dilemmas. Nat Rev Neurol. 2014;10(5):293-299. doi: 10.1038/nrneurol.2014.60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Consortium E; EpiPM Consortium . A roadmap for precision medicine in the epilepsies. Lancet Neurol. 2015;14(12):1219-1228. doi: 10.1016/S1474-4422(15)00199-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demarest ST, Brooks-Kayal A. From molecules to medicines: the dawn of targeted therapies for genetic epilepsies. Nat Rev Neurol. 2018;14(12):735-745. doi: 10.1038/s41582-018-0099-3 [DOI] [PubMed] [Google Scholar]
- 15.Krueger DA, Sadhwani A, Byars AW, et al. Everolimus for treatment of tuberous sclerosis complex–associated neuropsychiatric disorders. Ann Clin Transl Neurol. 2017;4(12):877-887. doi: 10.1002/acn3.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sheidley BR, Malinowski J, Bergner AL, et al. Genetic testing for the epilepsies: a systematic review. Epilepsia. 2022;63(2):375-387. doi: 10.1111/epi.17141 [DOI] [PubMed] [Google Scholar]
- 17.Hayeems RZ, Luca S, Assamad D, Bhatt A, Ungar WJ. Utility of genetic testing from the perspective of parents/caregivers: a scoping review. Children (Basel). 2021;8(4):259. doi: 10.3390/children8040259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith LA, Ullmann JF, Olson HE, et al. A model program for translational medicine in epilepsy genetics. J Child Neurol. 2017;32(4):429-436. doi: 10.1177/0883073816685654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rockowitz S, LeCompte N, Carmack M, et al. Children’s rare disease cohorts: an integrative research and clinical genomics initiative. NPJ Genom Med. 2020;5:29. doi: 10.1038/s41525-020-0137-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zuberi SM, Wirrell E, Yozawitz E, et al. ILAE classification and definition of epilepsy syndromes with onset in neonates and infants: position statement by the ILAE Task Force on Nosology and Definitions. Epilepsia. 2022;63(6):1349-1397. doi: 10.1111/epi.17239 [DOI] [PubMed] [Google Scholar]
- 21.Specchio N, Wirrell EC, Scheffer IE, et al. International League Against Epilepsy classification and definition of epilepsy syndromes with onset in childhood: position paper by the ILAE Task Force on Nosology and Definitions. Epilepsia. 2022;63(6):1398-1442. doi: 10.1111/epi.17241 [DOI] [PubMed] [Google Scholar]
- 22.Riney K, Bogacz A, Somerville E, et al. International League Against Epilepsy classification and definition of epilepsy syndromes with onset at a variable age: position statement by the ILAE Task Force on Nosology and Definitions. Epilepsia. 2022;63(6):1443-1474. doi: 10.1111/epi.17240 [DOI] [PubMed] [Google Scholar]
- 23.Hirsch E, French J, Scheffer IE, et al. ILAE definition of the idiopathic generalized epilepsy syndromes: position statement by the ILAE Task Force on Nosology and Definitions. Epilepsia. 2022;63(6):1475-1499. doi: 10.1111/epi.17236 [DOI] [PubMed] [Google Scholar]
- 24.Scheffer IE, Berkovic S, Capovilla G, et al. ILAE classification of the epilepsies: position paper of the ILAE Commission for Classification and Terminology. Epilepsia. 2017;58(4):512-521. doi: 10.1111/epi.13709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boat TF, Wu JT; Committee to Evaluate the Supplemental Security Income Disability Program for Children with Mental Disorders; Board on the Health of Select Populations; Board on Children, Youth, and Families . Mental Disorders and Disabilities Among Low-Income Children. The National Academies Press; 2015. doi: 10.17226/21780 [DOI] [PubMed] [Google Scholar]
- 26.Richards S, Aziz N, Bale S, et al. ; ACMG Laboratory Quality Assurance Committee . Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405-424. doi: 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X, Huang Z, Liu J, et al. Phenotypic and genotypic characterization of DEPDC5-related familial focal epilepsy: case series and literature review. Front Neurol. 2021;12:641019. doi: 10.3389/fneur.2021.641019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Consortium GT; GTEx Consortium . The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580-585. doi: 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burgess R, Wang S, McTague A, et al. ; EIMFS Consortium . The genetic landscape of epilepsy of infancy with migrating focal seizures. Ann Neurol. 2019;86(6):821-831. doi: 10.1002/ana.25619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stödberg T, McTague A, Ruiz AJ, et al. Mutations in SLC12A5 in epilepsy of infancy with migrating focal seizures. Nat Commun. 2015;6:8038. doi: 10.1038/ncomms9038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mefford HC, Sharp AJ, Baker C, et al. Recurrent rearrangements of chromosome 1q21.1 and variable pediatric phenotypes. N Engl J Med. 2008;359(16):1685-1699. doi: 10.1056/NEJMoa0805384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Girirajan S, Rosenfeld JA, Cooper GM, et al. A recurrent 16p12.1 microdeletion supports a two-hit model for severe developmental delay. Nat Genet. 2010;42(3):203-209. doi: 10.1038/ng.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sánchez Fernández I, Loddenkemper T, Gaínza-Lein M, Sheidley BR, Poduri A. Diagnostic yield of genetic tests in epilepsy: a meta-analysis and cost-effectiveness study. Neurology. 2019;92(5):e418-e428. doi: 10.1212/WNL.0000000000006850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howell KB, Eggers S, Dalziel K, et al. ; Victorian Severe Epilepsy of Infancy Study Group . A population-based cost-effectiveness study of early genetic testing in severe epilepsies of infancy. Epilepsia. 2018;59(6):1177-1187. doi: 10.1111/epi.14087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Boonsimma P, Ittiwut C, Kamolvisit W, et al. Exome sequencing as first-tier genetic testing in infantile-onset pharmacoresistant epilepsy: diagnostic yield and treatment impact. Eur J Hum Genet. 2023;31(2):179-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chourasia N, Yuskaitis C, Zhang B, Poduri A, Harini C. Etiology of Infantile spasms and yield of genetic testing: a tertiary center study (2825). Neurology. 2021;96(15 suppl):2825. [Google Scholar]
- 37.Allen AS, Berkovic SF, Cossette P, et al. ; Epi4K Consortium; Epilepsy Phenome/Genome Project . De novo mutations in epileptic encephalopathies. Nature. 2013;501(7466):217-221. doi: 10.1038/nature12439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Epi25 Collaborative . Ultra-rare genetic variation in the epilepsies: a whole-exome sequencing study of 17,606 individuals. Am J Hum Genet. 2019;105(2):267-282. doi: 10.1016/j.ajhg.2019.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Preston CG, Wright MW, Madhavrao R, et al. ; Clinical Genome Resource (ClinGen) . ClinGen Variant Curation Interface: a variant classification platform for the application of evidence criteria from ACMG/AMP guidelines. Genome Med. 2022;14(1):6. doi: 10.1186/s13073-021-01004-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehm HL, Berg JS, Brooks LD, et al. ; ClinGen . ClinGen—the Clinical Genome Resource. N Engl J Med. 2015;372(23):2235-2242. doi: 10.1056/NEJMsr1406261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Olson H, Shen Y, Avallone J, et al. Copy number variation plays an important role in clinical epilepsy. Ann Neurol. 2014;75(6):943-958. doi: 10.1002/ana.24178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Palmer EE, Sachdev R, Macintosh R, et al. Diagnostic yield of whole genome sequencing after nondiagnostic exome sequencing or gene panel in developmental and epileptic encephalopathies. Neurology. 2021;96(13):e1770-e1782. doi: 10.1212/WNL.0000000000011655 [DOI] [PubMed] [Google Scholar]
- 43.McKnight D, Morales A, Hatchell KE, et al. ; ELEVIATE Consortium . Genetic testing to inform epilepsy treatment management from an International Study of Clinical Practice. JAMA Neurol. 2022;79(12):1267-1276. doi: 10.1001/jamaneurol.2022.3651 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eMethods.
eTable 1. Clinical and Diagnostic Genetic Findings in 89 Patients With SNVs
eTable 2. A Phenotype-Driven Epilepsy Gene List
eTable 3. Nondiagnostic VUS in Epilepsy-Related Genes
eTable 4. Novel Candidate Epilepsy Genes and Their Associated Phenotypes
eTable 5. Clinical and Diagnostic Genetic Findings in 11 Cases With CNVs
eTable 6. Bivariate and Multivariate Regression Model of Phenotypic Variables to Diagnostic Yield by ES
eFigure 1. Diagnostic Yield and Age of Seizure Onset According to Epilepsy Type
eFigure 2. Established and Candidate Epilepsy Genes in the Study Cohort
eFigure 2. Variant Types Among the Diagnostic SNVs and CNVs
eFigure 3. Clinical Features in Patients With Epilepsy Associated With Diagnostic Variants
Members of the BCH Neurology Referral and Phenotyping Group
Data Sharing Statement