Abstract
Millions of people are suffering from Alzheimer’s disease globally, but there is still no effective treatment for this neurodegenerative disease. Thus, novel therapeutic approaches for Alzheimer’s disease are needed, which requires further evaluation of the regulatory mechanisms of protein aggregate degradation. Lysosomes are crucial degradative organelles that maintain cellular homeostasis. Transcription factor EB-mediated lysosome biogenesis enhances autolysosome-dependent degradation, which subsequently alleviates neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and Huntington’s disease. In this review, we start by describing the key features of lysosomes, including their roles in nutrient sensing and degradation, and their functional impairments in different neurodegenerative diseases. We also explain the mechanisms — especially the post-translational modifications — which impact transcription factor EB and regulate lysosome biogenesis. Next, we discuss strategies for promoting the degradation of toxic protein aggregates. We describe Proteolysis-Targeting Chimera and related technologies for the targeted degradation of specific proteins. We also introduce a group of LYsosome-Enhancing Compounds, which promote transcription factor EB-mediated lysosome biogenesis and improve learning, memory, and cognitive function in APP-PSEN1 mice. In summary, this review highlights the key aspects of lysosome biology, the mechanisms of transcription factor EB activation and lysosome biogenesis, and the promising strategies which are emerging to alleviate the pathogenesis of neurodegenerative diseases.
Key Words: Alzheimer’s disease, degradation, lysosome biogenesis, LYsosome-Enhancing Compounds, neurodegenerative diseases, post-translational modifications, protein aggregates, transcription factor EB
Introduction
The pathogenesis of Alzheimer’s disease (AD) is characterized by the accumulation of protein aggregates, including amyloid-β (Aβ) and MAPT/Tau, which subsequently results in the loss of neuronal cells and dysfunction of the human brain. According to a recent study, dysfunction of the autophagy-lysosome system aggravates the pathogenesis of AD. In Tg2576/TRGL mice, an animal model of AD, the impairment of autolysosomal acidification occurs before Aβ accumulation in the brain (Lee et al., 2022). These results suggest that autolysosomal dysfunction further aggravates toxic protein accumulation. In this review, we summary the mechanisms of transcription factor EB activation and lysosome biogenesis, and the promising strategies which are emerging to alleviate the pathogenesis of neurodegenerative diseases.
Search Strategy and Selection Criteria
PubMed and Web of Science were searched to retrieve papers. During searching, all publications reviewed were in the English language, and no filters were applied such as restricting the date, publication type, or subjects. Literature retrieval was performed using the following keywords: lysosome biogenesis, small molecule compounds, TFEB, neurodegenerative disease, Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis, protein aggregates, degradation, phosphorylation, acetylation, ubiquitination, SUMOylation, oxidation, glycosylation, and PARsylation. The last search was conducted on December 29, 2022.
Structure and Degradative Function of Lysosomes
Lysosomes are major catabolic organelles, which were discovered by Christian de Duve and his colleagues in the 1950s (De Duve et al., 1955). They are usually found in all mammalian cells, except mature red blood cells, and they are located in the cytoplasm with the highest concentration in the perinuclear region. The number of lysosomes in each mammalian cell is between 50 and 1000 (Ballabio and Bonifacino, 2020), and the diameter of lysosomes typically ranges from 0.2 to 0.8 μm, with a minimum of 0.05 μm and a maximum of several microns. Lysosomes contain more than 70 acid hydrolases for the degradation of proteins, nucleic acids, lipids, and other biological macromolecules (Maxfield et al., 2016). The degradative capacity of lysosomes is critical for removing damaged organelles or recycling metabolites within cells via the autophagy pathway (Levine and Kroemer, 2019; Mizushima and Levine, 2020).
Notably, lysosomes are acidic organelles. The maintenance of an acidic lumenal microenvironment, with the pH between 4.5 and 5.0, is essential for lysosomal functions (Ballabio and Bonifacino, 2020). However, this does not mean that a lower pH is better. Lysosomal hyperacidification impairs lysosomal proteolytic activity and further leads to lysosome dysfunction (Hu et al., 2022). The acidic pH of the lysosome is maintained mainly by the vacuolar ATPase in the lysosomal membrane. However, the electrochemical gradient generated by the vacuolar ATPase must be dissipated to enable sustained proton import into the lysosomal lumen. ClC-7, a Cl–/H+ antiporter located in the lysosomal membrane, is responsible for lysosomal Cl– permeability (Leray et al., 2022). It imports 2 Cl– into the lysosomal lumen while ejecting 1 H+ into the cytoplasm to dissipate the electrochemical gradient across the lysosome membrane (Jentsch and Pusch, 2018). Previous studies have found that TMEM175, a genetic risk factor for PD, encodes a proton-activated/proton-selective channel in the lysosomal membrane which regulates the lysosomal H+ leak (Cang et al., 2015; Hu et al., 2022). Based on the highly acidic microenvironment of lysosomes, many lysosomal probes have been developed to monitor the acidity and function of lysosomes (Lukinavicius et al., 2016; Farfel-Becker et al., 2019; Lee et al., 2022). Besides proton transporters, there are other ion channels in the lysosomal membrane. For example, the transient receptor potential mucolipin 1 (TRPML1, also named as MCOLN1) and the two-pore channel proteins serve as lysosomal Ca2+ channels which release Ca2+ to the cytoplasm (Dong et al., 2008; Schieder et al., 2010). The voltage-activated BK channel, a potassium channel, delivers K+ into the lysosomal lumen from the cytosol. Dysregulation of the flux of these ions impairs lysosome function, which further leads to lysosome-related diseases.
Furthermore, there are many other types of transporters in the lysosomal membranes (Maxfield et al., 2016). After the damaged organelles and macromolecules are delivered to lysosomes and degraded in lysosomes, their products, including amino acids, sugars, and nucleosides, are selectively transferred from lysosomes to the cytoplasm via amino acid transporters, sugar transporters, and nucleoside transporters, respectively. These catabolites then become the building blocks for further cellular biosynthesis and metabolic processes.
Lysosomes Function as a Signaling Hub
Lysosomes also serve as nutrient sensors of amino acids, glucose, and cholesterol. Lysosomal nutrient sensing is related to the control of metabolic signal transduction via the key regulators mTORC1 and AMP-activated protein kinase (AMPK) (Zoncu et al., 2011; Wolfson et al., 2017; Ma et al., 2022; Shin et al., 2022; Zhang et al., 2022a). Lysosomes can sense amino acid concentrations and regulate mTORC1 activity. Under the condition of amino acid sufficiency, Rag guanosine triphosphatases (GTPases) recruit mTORC1 to the lysosomal surface from the cytoplasm and activate mTORC1 (Liu and Sabatini, 2020). Arginine binds to SLC38A9 and promotes the interaction of SLC38A9 with the Rag-Ragulator complex on the lysosome surface, then recruits mTORC1 anchored on Rag GTPases (Wyant et al., 2017). When leucine is sufficient, SAR1B, a small GTPase, directly binds to leucine and dissociates from GATOR2, resulting in mTORC1 activation. However, when leucine is deficient, SAR1B binds to GATOR2 and inhibits the activity of mTORC1 (Chen et al., 2021). Lysosomes can also sense the glucose concentration and regulate AMPK activity. Notably, lysosomal vacuolar ATPase functions as a crucial switch to regulate AMPK and mTORC1 activation and signal transduction via protein-protein interactions (Zhang et al., 2014; Zhang et al., 2017; Li et al., 2019). Furthermore, cholesterol transporters are also localized in the lysosomal membrane (Trinh et al., 2018). Cholesterol can be transported from the lysosomal lumen to the cytoplasm via its transporters, such as NPC1 (Niemann Pick type C1), LIMP-2 (lysosomal integral membrane protein 2), and PTCH1 (protein patched homolog 1) (Chu et al., 2015; Trinh et al., 2017; Lim et al., 2019; Meng et al., 2020; Qian et al., 2020). This subsequently drives mTORC1 recruitment and activation at the lysosomal surface via the Rag GTPases (Castellano et al., 2017; Shin et al., 2022).
Although lysosomes are located in the cytoplasm, especially in the perinuclear region, they also have crosstalk and/or contacts with other organelles, including perinuclear endoplasmic reticulum (Boutry and Kim, 2021; Tan and Finkel, 2022), peroxisomes (Chu et al., 2015), and mitochondria (Boutry and Kim, 2021). Lim et al. (2019) found that oxysterol binding protein is a key regulator that mediates cholesterol transfer from the endoplasmic reticulum to the lysosomal cholesterol pool via the endoplasmic reticulum anchor proteins VAPA and VAPB. This process subsequently activates mTORC1 on lysosome membranes (Lim et al., 2019). Lysosomes also transfer free cholesterol to peroxisomes through dynamic membrane contacts, which are regulated by the binding between lysosomal synaptotagmin VII and peroxisomal PI(4,5)P2 (Chu et al., 2015). Lysosome-mitochondrion contracts have also been found in mammalian cells and play important roles in maintaining metabolite homeostasis. For instance, the lysosomal cation channel TRPML1 mediates the efflux of intracellular calcium to mitochondria through voltage-dependent anion channel 1 (VDAC1) and mitochondrial calcium uniporter (MCU) (Peng et al., 2020). In neurons, lysosome-mitochondrion contacts exist in soma, axons, and dendrites. Dysregulation of lysosome-mitochondrion contacts may be related to multiple neurodegenerative diseases (Cioni et al., 2019; Kim et al., 2021a).
Lysosomes and Neurodegenerative Diseases
Lysosomal dysfunction aggravates many human diseases (Maxfield et al., 2016). One group of lysosome-related diseases is known as lysosomal storage disorders (LSDs) (Parenti et al., 2015; Platt et al., 2018). Lysosomal storage disorders are usually caused by mutations in the genes encoding lysosomal hydrolases or lysosomal membrane proteins. Since lysosomes are major catabolic organelles in almost all mammalian cells, they play a critical role in regulating cell homeostasis. In the nervous system, neuronal cells also contain a lot of lysosomes in both soma and neurites, and lysosomes play important roles in neural metabolic homeostasis and cargo delivery under physiological conditions (Iacoangeli et al., 2019; Roney et al., 2021). Therefore, the other group is age-related neurodegenerative diseases (Saftig and Klumperman, 2009), which also showed lysosomal function impaired. Under pathological conditions of neurodegenerative diseases lysosomal functions are impaired by many factors, including abnormality of lysosomal acidification, mutations of lysosomal genes, and calcium (Ca2+) dysregulations.
Alzheimer’s disease
AD is the most common neurodegenerative disease in China and is characterized by the abnormal accumulation of protein aggregates (Aβ and/or Tau). Presenilin-1 (PSEN1) and presenilin 2 (PSEN2) are catalytic subunits of the gamma (γ)-secretase enzyme complex and are responsible for the hydrolysis of Aβ. Lee et al. (2010) found that autophagy requires PSEN1, and the depletion or mutation of PSEN1 results in impaired lysosomal acidification, protease inactivation, and lysosomal proteolytic dysfunction. PSEN2 plays a critical role in modulating intracellular Ca2+ homeostasis independently from γ-secretase activity. In AD, mutant PSEN2 causes impaired lysosomal Ca2+ signaling and abnormal autolysosome degradation (Fedeli et al., 2019). Mutation of TMEM106B is also identified in AD patients, which results in hyperfunction of the encoded protein. TMEM106B, a single-pass transmembrane protein, is located on endosomal and lysosomal membranes. Its elevated expression causes lysosomal hyperacidification and trafficking disorders, which lead to the impairment of lysosome-dependent degradation (Root et al., 2021).
Parkinson’s disease
Parkinson’s disease (PD) is characterized by neurological and motor dysfunction. Several genes related to PD progression have been identified, for instance, TMEM175, GBA1, and VPS13C (Alcalay et al., 2014; Darvish et al., 2018; Wie et al., 2021). TMEM175, regulating potassium (K+) leak in the lysosomal membrane is also identified as mediating lysosomal H+ leak, as mentioned above. TMEM175 deficiency results in lysosomal hyperacidification and diminished catalytic activity, which leads to α-synuclein aggregation (Hu et al., 2022). Interestingly, Lee et al. (2022) discovered that reduced acidification of autolysosomes occurs before Aβ accumulation. This indicates that the functional impairment of autolysosomes is not completely caused by deposits of toxic proteins, and the defective autolysosomes will further aggravate the accumulation of protein aggregates. Therefore, enhancement of lysosomal function may be a better therapeutic strategy to improve PD.
Huntington’s disease
Huntington’s disease (HD) is a neurodegenerative disease characterized by striatal neurodegeneration resulting from mutation of the huntingtin (HTT) gene. The mutant HTT (mHTT) knock-in HD-like mouse model shows abnormal cargo loading into autophagosomes and abnormal fusion between autophagosomes and lysosomes (Heng et al., 2010), which lead to reduced autophagic clearance of mHTT aggregates by lysosomes. The enzymatic activity of Glutamine Synthetase 1 is reduced in HD patients. Glutamine synthetase 1 deficiency causes autophagy inactivation, subsequent accumulation of toxic Htt-Q93 aggregates, and neuron loss in a Drosophila model of HD (Vernizzi et al., 2020). In addition, the level of the protease Cathepsin D is reduced in lysosomes in HD because mHTT disrupts the interaction between optineurin and Rab8 within the Golgi, which subsequently prevents efficient trafficking of Cathepsin D to the lysosomes (Liang et al., 2011).
Amyotrophic lateral sclerosis
In recent studies, more and more mutations of lysosomal genes, such as C9orf72 (chromosome 9 open reading frame 72), have been identified to be associated with the pathogenesis of ALS. Iacoangeli et al. (2019) discovered that the C9orf72 gene contains a hexanucleotide repeat expansion in amyotrophic lateral sclerosis (ALS) patients. The hexanucleotide motif GGGGCC (G4C2) is repeated about 24 to 30 times (Iacoangeli et al., 2019). C9orf72 deficiency in mouse or human cells leads to lysosomal swelling, impairment of mTORC1 activation and reduced lysosomal degradation (Amick et al., 2016; Shao et al., 2020).
Lysosome biogenesis and function are deficient in neurodegenerative diseases (Bajaj et al., 2019). The impaired lysosome biogenesis and function will accelerate the accumulation of protein aggregates, which will further aggravate neurodegeneration. Enhancement of lysosome biogenesis will increase cargo degradation via lysosomes, reduce neuronal impairment, and ameliorate neurodegenerative diseases. Therefore, it is critical to uncover methods for promoting lysosome biogenesis, especially in diseases.
Transcription Factor EB and Lysosome Biogenesis
To ameliorate the pathogenesis of neurodegenerative diseases, we need to promote lysosome biogenesis and enhance the degradative functions of lysosomes. Transcription factor EB (TFEB) belongs to the microphthalmia-associated transcription factor (MiTF) family, which contains TFEB, TFE3, TFEC and MiTF. These proteins form homo/heterodimers and function as transcription factors to promote the expression of their target genes. In fed, unstressed cells, TFEB is inactive and sequestered by 14-3-3 proteins in the cytosol. During starvation or treatment with certain small-molecule compounds, TFEB becomes active and translocates into the nucleus to promote the expression of autophagic/lysosomal genes. During the activation and translocation of TFEB, many changes in post-translational modifications occur, including phosphorylation (Li et al., 2016; Puertollano et al., 2018; Yin et al., 2022), acetylation (Zhang et al., 2018; Wang et al., 2020b; Li et al., 2022), ubiquitination (Sha et al., 2017; Li et al., 2022), oxidation (Martina et al., 2021) and alkylation (Zhang et al., 2022b).
Phosphorylation
The phosphorylated sites on TFEB have been identified by recent studies (Sardiello et al., 2009; Settembre et al., 2011; Ferron et al., 2013; Li et al., 2016; Palmieri et al., 2017; Vega-Rubin-de-Celis et al., 2017; Hsu et al., 2018; Martina and Puertollano, 2018; Iacoangeli et al., 2019; Contreras et al., 2020; Yin et al., 2020, 2022; Paquette et al., 2021; Figure 1). In normal conditions, Rag GTPase recruits mTORC1 and TFEB onto lysosome membranes, where active mTORC1 phosphorylates TFEB at S122, S142, and S211 (Sardiello et al., 2009; Settembre et al., 2011; Vega-Rubin-de-Celis et al., 2017; Iacoangeli et al., 2019). Then, phosphorylated TFEB binds with 14-3-3 proteins and is sequestered in the cytosol. When amino acids are deficient, mTORC1 is inactivated. Thus, TFEB is dephosphorylated and activated, and it translocates into the nucleus to promote lysosome biogenesis. Based on a screen of small-molecule compounds, we found that HEP14 (a naturally occurring plant compound; see below) activated the PKCα/δ-GSK3β-TFEB axis to promote lysosome biogenesis independently of mTORC1 signaling (Li et al., 2016). In untreated cells, GSK3β phosphorylates TFEB at S134 and S138 on the lysosome membranes. After HEP14 treatment, activated PKCα/δ translocates onto lysosome membranes, where PKCα/δ inhibits GSK3β activity. Thus, inactivated GSK3β and dephosphorylated TFEB are released from lysosome membranes. Subsequently, the dephosphorylated TFEB translocates into the nucleus and promotes lysosome biogenesis. Recently, we also identified that inhibition of the dopamine transporter (DAT) promotes lysosome biogenesis (Yin et al., 2022). After treatment of cells with the small-molecule compound LH2-051, DAT translocates from the plasma membrane onto lysosome membranes, where DAT regulates the DAT-CDK9-TFEB axis and induces dephosphorylation of TFEB at S97, T99, S109, S144, S132, and T331. Then, dephosphorylated TFEB translocates into the nucleus and promotes lysosome biogenesis. Interestingly, CDK4/6 also phosphorylate TFEB at S142, which occurs in the nucleus and regulates TFEB export from the nucleus into the cytosol (Yin et al., 2020). Recent studies also demonstrate that TFEB phosphorylation is regulated by several kinases and phosphatases, including MAP4K3, PKCβ, Akt, c-Abl, ERK2, AMPK, PP2A and calcineurin (Settembre et al., 2011; Ferron et al., 2013; Palmieri et al., 2017; Hsu et al., 2018; Martina and Puertollano, 2018; Contreras et al., 2020; Paquette et al., 2021; Figure 1). Notably, phosphorylation of TFEB can increase or decrease its activity, especially phosphorylation at S467 in the C-terminus of TFEB.
Figure 1.

Post-translational modifications of TFEB.
The key sites of TFEB modifications are shown in the left panels, including phosphorylation, acetylation, ubiquitination, oxidation, and other modifications (SUMOylation, PARsylation, and glycosylation). The related effectors of TFEB modifications are shown in the right panels, including kinases, phosphatases, HDACs, KATs, and E3 ligases. Ac: Acetylation; AD: N-terminal transcriptional activation domain; bHLH: basic helix-loop-helix region; C: cysteine; E: glutamic acid; G: glycine; Gl: glycosylation; Gln rich: glutamine-rich domain; K: lysine; KATs: lysine acetyltransferases; NLS: nuclear localization signal; Ox: oxidation; P: phosphorylation; Pa: PARsylation; Pro rich: proline-rich domain; S: serine; Su: SUMOylation; T: threonine; TFEB: transcription factor EB; Ub: ubiquitination; V: valine; Zip: leucine zipper. Created with Adobe Illustrator CC 2018.
Acetylation
The mechanisms of TFEB acetylation have already been reported by several groups. Zhang et al. (2018) reported that treatment with SAHA, an inhibitor of histone deacetylases (HDACs), induces lysosome biogenesis via promoting TFEB acetylation at four sites, including K91, K103, K116, and K430. By screening small-molecule compounds, we found that trichostatin A (TSA), the pan-inhibitor of HDACs, also promotes TFEB activation and lysosome biogenesis (Li et al., 2022). Four acetylated lysine residues in TFEB, including K116, K236, K237, and K431, are important for TFEB nuclear translocation and lysosome biogenesis after TSA treatment. Notably, K236 and K237 localize in the NLS region of TFEB (Figure 1), suggesting that the acetylation at K236/K237 is crucial for unmasking the TFEB NLS region and interrupting the binding between TFEB and 14-3-3 proteins. We also found that TFEB acetylation is regulated by HDACs (HDAC5, HDAC6, and HDAC9) and lysine acetyltransferases, including ELP3, CREBBP, and HAT1 (Figure 1). Surprisingly, during TSA-induced TFEB activation, acetylation of TFEB is independent of TFEB dephosphorylation. On the other hand, acetyltransferase GCN5 promotes the acetylation of nuclear TFEB at K274 and K279, which subsequently disrupts nuclear TFEB dimerization and its transcriptional activity (Wang et al., 2020b).
Ubiquitination
A novel ubiquitination site on TFEB at K347 has been identified (Figure 1), which is evolutionarily conserved. After TSA treatment, a turnover between ubiquitination and acetylation occurs on K347: the ubiquitination on K347 is decreased, while the acetylation is increased. The reduced level of K347 ubiquitination allows TFEB to avoid proteasome-mediated degradation (Li et al., 2022). However, the E3 ligases which regulate K347 ubiquitination are still unknown. STUB1, an E3 ligase, has been reported to regulate TFEB protein levels and lysosome biogenesis. STUB1 can interact with phosphorylated TFEB (p-S142/p-S211), and dephosphorylation of TFEB (S142A and/or S211A) rescues the ubiquitination of TFEB and restores lysosome biogenesis (Sha et al., 2017). However, the STUB1-regulated ubiquitination sites of TFEB are still unknown. Further assays using mass spectrometry are needed to dissect the ubiquitination sites of phosphorylated TFEB (p-S142 or p-S211). STUB1 activates TFEB and its mutation is an effective method to explore the TFEB ubiquitination regulated by STUB1. However, one possibility needs to concerned: mutant STUB1 results in the accumulation of misfolded proteins, which also in turn promotes TFEB dephosphorylation and lysosome biogenesis for the autophagic clearance of these misfolded proteins. To exclude the above possibility, it will be important to mutate the STUB1-related ubiquitinated sites on TFEB after they have been found by mass spectrometry as mentioned above instead of STUB1 mutation. Functional analysis of the mutated TFEB will reveal the mechanisms of STUB1-related TFEB ubiquitination and lysosome biogenesis.
Oxidation and Other Modifications
TFEB contains a cysteine residue at site 212 (C212), which can be oxidized by reactive oxygen species. Cysteine oxidation promotes the formation of disulfide bonds and TFEB oligomers, which abolish the interaction between TFEB and Rag GTPases, thus inhibiting the phosphorylation of TFEB mediated by mTORC1 on lysosomal membranes (Wang et al., 2020a; Martina et al., 2021). Furthermore, TFEB can be alkylated at C212. Under normal conditions, mTORC1 phosphorylates TFEB at S211 and subsequently inactivates TFEB. After itaconate (an anti-inflammatory metabolite) treatment, TFEB is alkylated at C212, which attenuates the phosphorylation of TFEB at S211 by mTORC1. This in turn blocks the binding between 14-3-3 proteins and TFEB, which results in TFEB activation and enhanced lysosome biogenesis (Zhang et al., 2022b).
SUMOylation is a post-translational modification that adds a small ubiquitin-like-modifier (SUMO) to amino acid residues of target proteins. TFEB is SUMOylated at K316 by SUMO1 (Miller et al., 2005). There is no direct evidence showing whether SUMOylation affects the activity of TFEB. SUMOylation of MiTF does not affect MiTF localization, but it reduces the capacity of MiTF to bind to the promoter region of its target genes (Miller et al., 2005). Thus, an important future goal is to evaluate whether TFEB SUMOylation affects TFEB activity.
TFEB glycosylation occurs when the host is infected with Legionella pneumophila. SetA, a glucosyltransferase from Legionella pneumophila, glycosylates TFEB at S195 or S196, T201 or S203, and T208, which disrupts the binding between TFEB and 14-3-3 proteins. This molecular process promotes the nuclear translocation of TFEB (Beck et al., 2020). Meanwhile, SetA also directly glycosylates TFEB at S138 and blocks GSK3β-mediated phosphorylation of S138, which impairs TFEB nuclear export (Beck et al., 2020).
TFEB can also be modified by the addition of ADP-ribose polymers, which is called PARsylation. TNKS1 (Tankyrase 1) PARsylates TFEB at V65, G67 and E68 when the WNT/β-catenin signaling pathway is activated. The PARsylated TFEB translocates into the nucleus. However, the Wnt signaling-dependent PARsylation of TFEB does not induce the expression of lysosomal genes (Kim et al., 2021b).
LYsosome-Enhancing Compounds and Targeted Protein Degradation Technology
Toxic protein aggregates are the pathological characteristics of neurodegenerative diseases, and enhanced clearance of aggregated proteins is a direct therapeutic approach to increase the survival of neurons, restore the function of the central nervous system, and ameliorate neurodegenerative diseases. Protein quality control is dependent on two systems: the ubiquitin-proteasome system (UPS) and the autophagy-lysosome system. In the UPS system, misfolded proteins are targeted by ubiquitinating enzymes, then recognized and degraded by proteasomes. The UPS machinery is present in most mammalian cells, and the impairment of UPS has been reported during the pathogenesis of neurodegeneration. This suggests the possibility of treating neurodegenerative diseases through targeting UPS. A new technology named Proteolysis-Targeting Chimera (PROTAC) has been developed to enhance the specificity of the proteasomal pathway (Alabi and Crews, 2021). The PROTAC system consists of three parts: a ligand that binds the protein of interest (POI), a linker, and an E3 ligase-targeting ligand. Thus, the PROTAC molecule acts as a bridge between the POI and the E3 ligase, and the E3-PROTAC-POI complex increases the degradation of the POI by UPS.
Dysregulation of the autophagy-lysosome system has also been identified as a defining feature of neurodegenerative diseases, whereas enhancement of the autolysosome pathway is able to improve AD, PD, and HD. Based on knowledge of autophagosomes and lysosomes, new targeted protein degradation strategies have been developed to hijack the lysosomal degradation pathway. These include Autophagy-TArgeting Chimera (AUTAC), AuTophagosome-TEthering Compound (ATTEC), AUTOphagy-TArgeting Chimera (AUTOTAC), LYsosome-TArgeting Chimera (LYTAC), GlueTAC, and AbTAC (Ding et al., 2022) technologies. AUTAC incorporates the FBnG tag which mimics S-guanylation and binds to the POI; thus, the POI can be poly-ubiquitinated on K63. The POI is then recognized by SQSTM1/p62 and degraded by lysosomes (Takahashi et al., 2019). ATTEC directly combines POI and LC3; thus, the POI can be tethered by autophagosomes and subsequently degraded via the autophagy pathway. AUTOTAC is different from ATTEC: it combines SQSTM1/p62 and the POI, then the AUTOTAC system is recognized by LC3 rather than binding directly to LC3. Subsequently, the POI is tethered to the phagophores for degradation (Ji et al., 2022). The LYTAC system can degrade extracellular and membrane proteins via lysosomes. It is composed of an antibody or small molecule, which binds to receptors on the surface of the lysosome (e.g., cation-independent mannose-6-phosphate receptor, CI-M6PR) via a ligand (Banik et al., 2020). Notably, using the specific receptor on the surface of the lysosome can achieve tissue-specific targeting in human or mouse tissues. The POI will be degraded by lysosomes, and the receptor on the surface of the lysosome will be recycled to the plasma membrane for reuse. GlueTAC consists of three parts: a covalently-modified nanobody, a cell-penetrating peptide, and a lysosome-sorting sequence. The nanobody is responsible for recognizing and binding to the POI, and the cell-penetrating peptide promotes endocytosis of the POI-GlueTAC complex. The lysosome-sorting sequence then enhances lysosomal degradation (Zhang et al., 2021). AbTAC consists of a recombinant bispecific antibody. The recombinant bispecific antibody has two functions: one is targeting the POI and the other is targeting RNF43 (an E3 ligase). Subsequently, the POI is degraded in a lysosome-dependent manner (Cotton et al., 2021). The advantage of AbTAC is that it can target membrane proteins.
The above strategies are designed to enhance the recognition of cargos. During the pathogenesis of neurodegenerative diseases and aging, the functions of lysosomes are impaired, and the lysosomal degradative capacity is consequently reduced (Lee et al., 2022). Therefore, the enhancement of cargo degradation by lysosome biogenesis is a powerful strategy. As mentioned above, TFEB is one of the key regulators that promote lysosome biogenesis. Hence, small-molecule compounds that enhance TFEB activity and lysosome biogenesis are potential therapeutic agents for ameliorating AD (Table 1). In recent years, based on screening of small-molecule compounds, we identified a series of LYsosome-Enhancing Compounds (LYECs) that promote TFEB activation and lysosome biogenesis in cells stably expressing TFEB-EGFP. The previously published LYECs include HEP14, LH2-051, and TSA (Li et al., 2016, 2022; Yin et al., 2022). HEP14 (5β-O-angelate-20-deoxyingenol) is a natural compound isolated from Euphorbia peplus Linn. It promotes lysosome biogenesis via the PKC-GSK3β-TFEB pathway and the PKC-JNK/p38-ZKSCAN3 pathway (Li et al., 2016). Recently, we identified the LYEC LH2-051, which directly binds with and inhibits DAT. LH2-051 promotes TFEB activation and lysosome biogenesis via the DAT-CDK9-TFEB axis (Yin et al., 2022). Using APP-PSEN1 mice (an AD-like rodent model), we evaluated the effects of LYECs on cognitive and learning functions. Treatment with LYECs significantly decreased the accumulation of Aβ in APP-PSEN1 mice in a manner dependent on TFEB-mediated lysosome biogenesis. The performance of the AD mice was improved in the Morris water maze and Y maze tests (Li et al., 2016, 2022; Yin et al., 2022).
Table 1.
Small-molecule compounds enhance TFEB activity and lysosome biogenesis
| Compounds | Structure | Mechanisms | Animal models of neurodegenerative diseases | Effects on neurodegenerative diseases | References |
|---|---|---|---|---|---|
| Catalpol |

|
Activates AMPK and inhibits mTOR | APP/PS1-AD mice | Reduces Aβ load; alleviates cognitive impairments | Ren et al., 2019; Du et al., 2022 |
| Curcumin |

|
Inhibits mTORC1 | 5xFAD-AD mice; 3xTg-AD mice | Attenuates both APP and Tau pathology; improves cognitive deficits | Song et al., 2020 |
| Compound E4 |

|
Inhibits the Akt-mTORC1 pathway | N.A. | N.A. | Wang et al., 2020c |
| HEP14 |
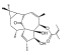
|
Directly binds to PKCα and PKCδ, thus activating PKCα and PKCδ | APP/PS1-AD mice | Reduces Aβ plaques; decreases Aβ40 and Aβ42 | Li et al., 2016 |
| Itaconate |

|
Enhances the alkylation of TFEB at site C212 | N.A. | N.A. | Zhang et al., 2022b |
| LH2-051 |
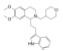
|
Directly binds to dopamine transporter (DAT) and inhibits DAT function | APP/PS1-AD mice | Reduces Aβ plaques; decreases Aβ40 and Aβ42; alleviates cognitive impairments | Yin et al., 2022 |
| MSL |
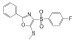
|
Directly binds to and stabilizes calcineurin A | N.A. | N.A. | Lim et al., 2018 |
| Oblongifolin C |

|
Inhibits mTORC1 | N.A. | N.A. | Wu et al., 2019 |
| Procyanidin B2 |

|
Potentially binds to TFEB | N.A. | N.A. | Su et al., 2018 |
| Quercetin |

|
Directly inhibits mTORC1 | 3xTg-AD mice; PD rats (rats were injected with 6-OHDA in striatum); HD rats (rats were injected with 3-NP) | Decreases Aβ and protects cognitive function; increases BDNF level and enhances cognitive effects; corrects movement disturbances and anxiety | Chakraborty et al., 2014; Huang et al., 2018; Paula et al., 2019; Naghizadeh et al., 2021 |
| Rapamycin |

|
Directly binds to mTOR and inhibits mTORC1 | 3xTg-AD mice; PD mice (C57BL/6 mice were intraperitoneally injected with MPTP) | Reduces Aβ plaques and Tau pathology; protects substantia nigral neurons | Malagelada et al., 2010; Majumder et al., 2011; Battaglioni et al., 2022 |
| SAHA |

|
Enhances the acetylation of TFEB at sites K91, K103, K116, and K430 | Mice with insulin resistance-induced cognitive deficit; R6/2-HD mice | Improves cognitive functions; reduces SDS-insoluble aggregate load | Mielcarek et al., 2011; Sharma and Taliyan, 2016; Zhang et al., 2018 |
| Siramesine |

|
Inhibits mTORC1 | N.A. | N.A. | Zhitomirsky et al., 2018 |
| Sulforaphane |
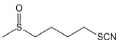
|
Increases intracellular ROS, then stimulates Ca2+ release | PS1V97L-AD mice | Inhibits the generation of Aβ; alleviates the cognitive deficits | Hou et al., 2018; Li et al., 2021 |
| Sunitinib |

|
Inhibits mTORC1 | AD2576APPSwe-AD mice; 3xTg-AD mice | Reduces Aβ; improves cognitive function | Grammas et al., 2014; Zhitomirsky et al., 2018 |
| Torin-1 |

|
Effectively blocks phosphorylation of mTOR (both mTORC1 and mTORC2) | PD mice (C57BL/6J female mice were injected with 6-OHDA in striatum) | Alleviates loss of dopaminergic neurons | Thoreen et al., 2009; Zhuang et al., 2020 |
| Trehalose |

|
Increases transient lysosomal membrane permeability, and releases Ca2+ from the lysosomes to activate the phosphatase PPP3/calcineurin | APP23-AD mice; R6/2-HD mice | Reduces Aβ generation and plaque formation; decreases polyglutamine aggregates improves motor function | Tanaka et al., 2004; Rusmini et al., 2019; Liu et al., 2020 |
| Trichostatin A (TSA) |
|
Enhances the acetylation of TFEB at sites K116, K236, K237, and K431, and decreases TFEB ubiquitination at site K347 | APP/PS1-AD mice | Reduces Aβ plaques; decreases Aβ40 and Aβ42; improves learning, memory, and cognitive functions | Li et al., 2022 |
| Tripterine |
|
Inhibits mTORC1, thus inducing TFEB dephosphorylation at sites S142 and S211 | P301S Tau-AD mice; 3xTg-AD mice | Attenuates Tau pathology and cognitive deficits | Yang et al., 2022 |
6-OHDA: 6-Hydroxydopamine; AD: Alzheimer’s disease; AMPK: AMP-activated protein kinase; APP: amyloid precursor protein; Aβ: amyloid beta; BDNF: brain-derived neurotrophic factor; HD: Huntington’s disease; MPTP: 1-Methyl-4-Phenyl-1,2,3,6-Tetrahydropyridine; mTOR: mammalian target of rapamycin; N.A.: not available; PD: Parkinson’s disease; PKC: protein kinase C; ROS: reactive oxygen species; SDS: sodium dodecyl sulfate; TFEB: transcription factor EB.
LYECs mainly enhance lysosome-dependent cargo degradation, while ATTEC, AUTAC, LYTAC, AUTOTAC, GlueTAC, and AbTAC technologies are used to enhance cargo recognition. In the future, if LYECs are combined with ATTEC, AUTAC, LYTAC, AUTOTAC, GlueTAC, or AbTAC strategies, they could have synergetic effects for the enhancement of both cargo recognition and cargo degradation by autolysosome-dependent systems. Drug cocktail therapy targeting protein aggregates will be a potential approach to enhance the degradation efficiency of autolysosomes and ameliorate neurodegenerative diseases (Figure 2).
Figure 2.

A schematic illustration showing how a combination of LYECs with ATTEC, AUTAC, LYTAC, AUTOTAC, GlueTAC, or AbTAC can be used as a potential therapeutic approach to ameliorate neurodegenerative diseases.
AbTAC: Antibody-based PROTAC; ATTEC: AuTophagosome-TEthering Compound; AUTAC: Autophagy-TArgeting Chimera; AUTOTAC: AUTOphagy-TArgeting Chimera; CPP: cell-penetrating peptide; GlueTAC: nanobody-based PROTAC; LSS: lysosome-sorting sequence; LYECs: LYsosome-Enhancing Compounds; LYTAC: LYsosome-TArgeting Chimera; M6P: mannose 6-phosphate; POI: protein of interest. Created with Adobe Illustrator CC 2018.
Additional file: Open peer review report 1 (73.6KB, pdf) .
Acknowledgments:
We thank Dr. Isabel Hanson for proofreading and editing.
Footnotes
Funding: This work was supported by the STI2030-Major Projects, No. 2022ZD0213000; the National Natural Science Foundation of China, Nos. 92057103 and 31872820; Shanghai Basic Research Program, No. 18ZR1404000; State Key Laboratory of Drug Research, No. SIMM2004KF-09 (all to YL).
Author contributions: Review design, figure preparation: WX and YL; data collection: WX and JZ; manuscript draft, manuscript revision: WX, JZ, and YL. All authors approved the final version of the manuscript.
Conflicts of interest: The authors declare no conflicts of interest.
Data availability statement: The data are available from the corresponding author on reasonable request.
Open peer reviewers: Jian Luo, Palo Alto Veterans Institute for Research, VA Palo Alto Health Care System, USA; Jonathan Martinez-Fabregas, Universidad de Sevilla, Spain.
P-Reviewers: Martinez-Fabregas J, Luo J; C-Editors: Zhao M, Liu WJ, Qiu Y; T-Editor: Jia Y
References
- 1.Alabi SB, Crews CM. Major advances in targeted protein degradation:PROTACs, LYTACs, and MADTACs. J Biol Chem. 2021;296:100647. doi: 10.1016/j.jbc.2021.100647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcalay RN, Dinur T, Quinn T, Sakanaka K, Levy O, Waters C, Fahn S, Dorovski T, Chung WK, Pauciulo M, Nichols W, Rana HQ, Balwani M, Bier L, Elstein D, Zimran A. Comparison of Parkinson risk in Ashkenazi Jewish patients with Gaucher disease and GBA heterozygotes. JAMA Neurol. 2014;71:752–757. doi: 10.1001/jamaneurol.2014.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amick J, Roczniak-Ferguson A, Ferguson SM. C9orf72 binds SMCR8, localizes to lysosomes, and regulates mTORC1 signaling. Mol Biol Cell. 2016;27:3040–3051. doi: 10.1091/mbc.E16-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bajaj L, Lotfi P, Pal R, Ronza AD, Sharma J, Sardiello M. Lysosome biogenesis in health and disease. J Neurochem. 2019;148:573–589. doi: 10.1111/jnc.14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ballabio A, Bonifacino JS. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21:101–118. doi: 10.1038/s41580-019-0185-4. [DOI] [PubMed] [Google Scholar]
- 6.Banik SM, Pedram K, Wisnovsky S, Ahn G, Riley NM, Bertozzi CR. Lysosome-targeting chimaeras for degradation of extracellular proteins. Nature. 2020;584:291–297. doi: 10.1038/s41586-020-2545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Battaglioni S, Benjamin D, Walchli M, Maier T, Hall MN. mTOR substrate phosphorylation in growth control. Cell. 2022;185:1814–1836. doi: 10.1016/j.cell.2022.04.013. [DOI] [PubMed] [Google Scholar]
- 8.Beck WHJ, Kim D, Das J, Yu H, Smolka MB, Mao Y. Glucosylation by the Legionella effector SetA promotes the nuclear localization of the transcription factor TFEB. iScience. 2020;23:101300. doi: 10.1016/j.isci.2020.101300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boutry M, Kim PK. ORP1L mediated PI(4)P signaling at ER-lysosome-mitochondrion three-way contact contributes to mitochondrial division. Nat Commun. 2021;12:5354. doi: 10.1038/s41467-021-25621-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cang C, Aranda K, Seo YJ, Gasnier B, Ren D. TMEM175 is an organelle K(+) channel regulating lysosomal function. Cell. 2015;162:1101–1112. doi: 10.1016/j.cell.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 11.Castellano BM, Thelen AM, Moldavski O, Feltes M, van der Welle RE, Mydock-McGrane L, Jiang X, van Eijkeren RJ, Davis OB, Louie SM, Perera RM, Covey DF, Nomura DK, Ory DS, Zoncu R. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science. 2017;355:1306–1311. doi: 10.1126/science.aag1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chakraborty J, Singh R, Dutta D, Naskar A, Rajamma U, Mohanakumar KP. Quercetin improves behavioral deficiencies, restores astrocytes and microglia, and reduces serotonin metabolism in 3-nitropropionic acid-induced rat model of Huntington's disease. CNS Neurosci Ther. 2014;20:10–19. doi: 10.1111/cns.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Ou Y, Luo R, Wang J, Wang D, Guan J, Li Y, Xia P, Chen PR, Liu Y. SAR1B senses leucine levels to regulate mTORC1 signalling. Nature. 2021;596:281–284. doi: 10.1038/s41586-021-03768-w. [DOI] [PubMed] [Google Scholar]
- 14.Chu BB, Liao YC, Qi W, Xie C, Du X, Wang J, Yang H, Miao HH, Li BL, Song BL. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell. 2015;161:291–306. doi: 10.1016/j.cell.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Cioni JM, Lin JQ, Holtermann AV, Koppers M, Jakobs MAH, Azizi A, Turner-Bridger B, Shigeoka T, Franze K, Harris WA, Holt CE. Late endosomes act as mRNA translation platforms and sustain mitochondria in axons. Cell. 2019;176:56–72. doi: 10.1016/j.cell.2018.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Contreras PS, Tapia PJ, Gonzalez-Hodar L, Peluso I, Soldati C, Napolitano G, Matarese M, Heras ML, Valls C, Martinez A, Balboa E, Castro J, Leal N, Platt FM, Sobota A, Winter D, Klein AD, Medina DL, Ballabio A, Alvarez AR, et al. c-Abl inhibition activates TFEB and promotes cellular clearance in a lysosomal disorder. iScience. 2020;23:101691. doi: 10.1016/j.isci.2020.101691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cotton AD, Nguyen DP, Gramespacher JA, Seiple IB, Wells JA. Development of antibody-based PROTACs for the degradation of the cell-surface immune checkpoint protein PD-L1. J Am Chem Soc. 2021;143:593–598. doi: 10.1021/jacs.0c10008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Darvish H, Bravo P, Tafakhori A, Azcona LJ, Ranji-Burachaloo S, Johari AH, Paisan-Ruiz C. Identification of a large homozygous VPS13C deletion in a patient with early-onset Parkinsonism. Mov Disord. 2018;33:1968–1970. doi: 10.1002/mds.27516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.De Duve C, Pressman BC, Gianetto R, Wattiaux R, Appelmans F. Tissue fractionation studies. 6. Intracellular distribution patterns of enzymes in rat-liver tissue. Biochem J. 1955;60:604–617. doi: 10.1042/bj0600604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding Y, Xing D, Fei Y, Lu B. Emerging degrader technologies engaging lysosomal pathways. Chem Soc Rev. 2022;51:8832–8876. doi: 10.1039/d2cs00624c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong XP, Cheng X, Mills E, Delling M, Wang F, Kurz T, Xu H. The type IV mucolipidosis-associated protein TRPML1 is an endolysosomal iron release channel. Nature. 2008;455:992–996. doi: 10.1038/nature07311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du J, Liu J, Huang X, Li Y, Song D, Li Q, Lin J, Li B, Li L. Catalpol ameliorates neurotoxicity in N2a/APP695swe cells and APP/PS1 transgenic mice. Neurotox Res. 2022;40:961–972. doi: 10.1007/s12640-022-00524-4. [DOI] [PubMed] [Google Scholar]
- 23.Farfel-Becker T, Roney JC, Cheng XT, Li S, Cuddy SR, Sheng ZH. Neuronal soma-derived degradative lysosomes are continuously delivered to distal axons to maintain local degradation capacity. Cell Rep. 2019;28:51–64. doi: 10.1016/j.celrep.2019.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedeli C, Filadi R, Rossi A, Mammucari C, Pizzo P. PSEN2 (presenilin 2) mutants linked to familial Alzheimer disease impair autophagy by altering Ca(2+) homeostasis. Autophagy. 2019;15:2044–2062. doi: 10.1080/15548627.2019.1596489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferron M, Settembre C, Shimazu J, Lacombe J, Kato S, Rawlings DJ, Ballabio A, Karsenty G. A RANKL-PKCbeta-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 2013;27:955–969. doi: 10.1101/gad.213827.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grammas P, Martinez J, Sanchez A, Yin X, Riley J, Gay D, Desobry K, Tripathy D, Luo J, Evola M, Young A. A new paradigm for the treatment of Alzheimer's disease:targeting vascular activation. J Alzheimers Dis. 2014;40:619–630. doi: 10.3233/JAD-2014-132057. [DOI] [PubMed] [Google Scholar]
- 27.Heng MY, Detloff PJ, Paulson HL, Albin RL. Early alterations of autophagy in Huntington disease-like mice. Autophagy. 2010;6:1206–1208. doi: 10.4161/auto.6.8.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hou TT, Yang HY, Wang W, Wu QQ, Tian YR, Jia JP. Sulforaphane inhibits the generation of amyloid-beta oligomer and promotes spatial learning and memory in Alzheimer's disease (PS1V97L) transgenic mice. J Alzheimers Dis. 2018;62:1803–1813. doi: 10.3233/JAD-171110. [DOI] [PubMed] [Google Scholar]
- 29.Hsu CL, Lee EX, Gordon KL, Paz EA, Shen WC, Ohnishi K, Meisenhelder J, Hunter T, La Spada AR. MAP4K3 mediates amino acid-dependent regulation of autophagy via phosphorylation of TFEB. Nat Commun. 2018;9:942. doi: 10.1038/s41467-018-03340-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu M, Li P, Wang C, Feng X, Geng Q, Chen W, Marthi M, Zhang W, Gao C, Reid W, Swanson J, Du W, Hume RI, Xu H. Parkinson's disease-risk protein TMEM175 is a proton-activated proton channel in lysosomes. Cell. 2022;185:2292–2308. doi: 10.1016/j.cell.2022.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang Y, Chen Y, Shaw AM, Goldfine H, Tian J, Cai J. Enhancing TFEB-mediated cellular degradation pathways by the mTORC1 inhibitor quercetin. Oxid Med Cell Longev. 2018;2018:5073420. doi: 10.1155/2018/5073420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iacoangeli A, Al Khleifat A, Jones AR, Sproviero W, Shatunov A, Opie-Martin S, Alzheimer's Disease Neuroimaging I. Morrison KE, Shaw PJ, Shaw CE, Fogh I, Dobson RJ, Newhouse SJ, Al-Chalabi A. C9orf72 intermediate expansions of 24-30 repeats are associated with ALS. Acta Neuropathol Commun. 2019;7:115. doi: 10.1186/s40478-019-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jentsch TJ, Pusch M. CLC chloride channels and transporters:structure, function, physiology, and disease. Physiol Rev. 2018;98:1493–1590. doi: 10.1152/physrev.00047.2017. [DOI] [PubMed] [Google Scholar]
- 34.Ji CH, Kim HY, Lee MJ, Heo AJ, Park DY, Lim S, Shin S, Ganipisetti S, Yang WS, Jung CA, Kim KY, Jeong EH, Park SH, Bin Kim S, Lee SJ, Na JE, Kang JI, Chi HM, Kim HT, Kim YK, et al. The AUTOTAC chemical biology platform for targeted protein degradation via the autophagy-lysosome system. Nat Commun. 2022;13:904. doi: 10.1038/s41467-022-28520-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim S, Wong YC, Gao F, Krainc D. Dysregulation of mitochondria-lysosome contacts by GBA1 dysfunction in dopaminergic neuronal models of Parkinson's disease. Nat Commun. 2021a;12:1807. doi: 10.1038/s41467-021-22113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S, Song G, Lee T, Kim M, Kim J, Kwon H, Kim J, Jeong W, Lee U, Na C, Kang S, Kim W, Seong JK, Jho EH. PARsylated transcription factor EB (TFEB) regulates the expression of a subset of Wnt target genes by forming a complex with beta-catenin-TCF/LEF1. Cell Death Differ. 2021b;28:2555–2570. doi: 10.1038/s41418-021-00770-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, Wolfe DM, Martinez-Vicente M, Massey AC, Sovak G, Uchiyama Y, Westaway D, Cuervo AM, Nixon RA. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010;141:1146–1158. doi: 10.1016/j.cell.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JH, Yang DS, Goulbourne CN, Im E, Stavrides P, Pensalfini A, Chan H, Bouchet-Marquis C, Bleiwas C, Berg MJ, Huo C, Peddy J, Pawlik M, Levy E, Rao M, Staufenbiel M, Nixon RA. Faulty autolysosome acidification in Alzheimer's disease mouse models induces autophagic build-up of Abeta in neurons, yielding senile plaques. Nat Neurosci. 2022;25:688–701. doi: 10.1038/s41593-022-01084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leray X, Hilton JK, Nwangwu K, Becerril A, Mikusevic V, Fitzgerald G, Amin A, Weston MR, Mindell JA. Tonic inhibition of the chloride/proton antiporter ClC-7 by PI(3,5)P2 is crucial for lysosomal pH maintenance. Elife. 2022;11 doi: 10.7554/eLife.74136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Levine B, Kroemer G. Biological functions of autophagy genes:a disease perspective. Cell. 2019;176:11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, Shao R, Wang N, Zhou N, Du K, Shi J, Wang Y, Zhao Z, Ye X, Zhang X, Xu H. Sulforaphane activates a lysosome-dependent transcriptional program to mitigate oxidative stress. Autophagy. 2021;17:872–887. doi: 10.1080/15548627.2020.1739442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li M, Zhang CS, Zong Y, Feng JW, Ma T, Hu M, Lin Z, Li X, Xie C, Wu Y, Jiang D, Li Y, Zhang C, Tian X, Wang W, Yang Y, Chen J, Cui J, Wu YQ, Chen X, et al. Transient receptor potential V channels are essential for glucose sensing by aldolase and AMPK. Cell Metab. 2019;30:508–524. doi: 10.1016/j.cmet.2019.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li T, Yin L, Kang X, Xue W, Wang N, Zhang J, Yuan P, Lin L, Li Y. TFEB acetylation promotes lysosome biogenesis and ameliorates Alzheimer's disease–relevant phenotypes in mice. J Biol Chem. 2022;298:102649. doi: 10.1016/j.jbc.2022.102649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Y, Xu M, Ding X, Yan C, Song Z, Chen L, Huang X, Wang X, Jian Y, Tang G, Tang C, Di Y, Mu S, Liu X, Liu K, Li T, Wang Y, Miao L, Guo W, Hao X, et al. Protein kinase C controls lysosome biogenesis independently of mTORC1. Nat Cell Biol. 2016;18:1065–1077. doi: 10.1038/ncb3407. [DOI] [PubMed] [Google Scholar]
- 45.Liang Q, Ouyang X, Schneider L, Zhang J. Reduction of mutant huntingtin accumulation and toxicity by lysosomal cathepsins D and B in neurons. Mol Neurodegener. 2011;6:37. doi: 10.1186/1750-1326-6-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lim CY, Davis OB, Shin HR, Zhang J, Berdan CA, Jiang X, Counihan JL, Ory DS, Nomura DK, Zoncu R. ER-lysosome contacts enable cholesterol sensing by mTORC1 and drive aberrant growth signalling in Niemann-Pick type C. Nat Cell Biol. 2019;21:1206–1218. doi: 10.1038/s41556-019-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim H, Lim YM, Kim KH, Jeon YE, Park K, Kim J, Hwang HY, Lee DJ, Pagire H, Kwon HJ, Ahn JH, Lee MS. A novel autophagy enhancer as a therapeutic agent against metabolic syndrome and diabetes. Nat Commun. 2018;9:1438. doi: 10.1038/s41467-018-03939-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y, Wang J, Hsiung GR, Song W. Trehalose inhibits abeta generation and plaque formation in Alzheimer's disease. Mol Neurobiol. 2020;57:3150–3157. doi: 10.1007/s12035-020-01942-1. [DOI] [PubMed] [Google Scholar]
- 50.Lukinavicius G, Reymond L, Umezawa K, Sallin O, D'Este E, Gottfert F, Ta H, Hell SW, Urano Y, Johnsson K. Fluorogenic probes for multicolor imaging in living cells. J Am Chem Soc. 2016;138:9365–9368. doi: 10.1021/jacs.6b04782. [DOI] [PubMed] [Google Scholar]
- 51.Ma T, Tian X, Zhang B, Li M, Wang Y, Yang C, Wu J, Wei X, Qu Q, Yu Y, Long S, Feng JW, Li C, Zhang C, Xie C, Wu Y, Xu Z, Chen J, Yu Y, Huang X, et al. Low-dose metformin targets the lysosomal AMPK pathway through PEN2. Nature. 2022;603:159–165. doi: 10.1038/s41586-022-04431-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Majumder S, Richardson A, Strong R, Oddo S. Inducing autophagy by rapamycin before, but not after, the formation of plaques and tangles ameliorates cognitive deficits. PLoS One. 2011;6:e25416. doi: 10.1371/journal.pone.0025416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Malagelada C, Jin ZH, Jackson-Lewis V, Przedborski S, Greene LA. Rapamycin protects against neuron death in in vitro and in vivo models of Parkinson's disease. J Neurosci. 2010;30:1166–1175. doi: 10.1523/JNEUROSCI.3944-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Martina JA, Puertollano R. Protein phosphatase 2A stimulates activation of TFEB and TFE3 transcription factors in response to oxidative stress. J Biol Chem. 2018;293:12525–12534. doi: 10.1074/jbc.RA118.003471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martina JA, Guerrero-Gomez D, Gomez-Orte E, Antonio Barcena J, Cabello J, Miranda-Vizuete A, Puertollano R. A conserved cysteine-based redox mechanism sustains TFEB/HLH-30 activity under persistent stress. EMBO J. 2021;40:e105793. doi: 10.15252/embj.2020105793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maxfield FR, Willard JM, Lu S. Lysosomes: biology, diseases, and therapeutics. Hoboken, New Jersey: Wiley; 2016. [Google Scholar]
- 57.Meng Y, Heybrock S, Neculai D, Saftig P. Cholesterol handling in lysosomes and beyond. Trends Cell Biol. 2020;30:452–466. doi: 10.1016/j.tcb.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 58.Mielcarek M, Benn CL, Franklin SA, Smith DL, Woodman B, Marks PA, Bates GP. SAHA decreases HDAC 2 and 4 levels in vivo and improves molecular phenotypes in the R6/2 mouse model of Huntington's disease. PLoS One. 2011;6:e27746. doi: 10.1371/journal.pone.0027746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller AJ, Levy C, Davis IJ, Razin E, Fisher DE. Sumoylation of MITF and its related family members TFE3 and TFEB. J Biol Chem. 2005;280:146–155. doi: 10.1074/jbc.M411757200. [DOI] [PubMed] [Google Scholar]
- 60.Mizushima N, Levine B. Autophagy in human diseases. N Engl J Med. 2020;383:1564–1576. doi: 10.1056/NEJMra2022774. [DOI] [PubMed] [Google Scholar]
- 61.Naghizadeh M, Mirshekar MA, Montazerifar F, Saadat S, Shamsi Koushki A, Jafari Maskouni S, Afsharfar M, Arabmoazzen S. Effects of quercetin on spatial memory, hippocampal antioxidant defense and BDNF concentration in a rat model of Parkinson's disease:an electrophysiological study. Avicenna J Phytomed. 2021;11:599–609. doi: 10.22038/AJP.2021.18526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Palmieri M, Pal R, Nelvagal HR, Lotfi P, Stinnett GR, Seymour ML, Chaudhury A, Bajaj L, Bondar VV, Bremner L, Saleem U, Tse DY, Sanagasetti D, Wu SM, Neilson JR, Pereira FA, Pautler RG, Rodney GG, Cooper JD, Sardiello M. mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat Commun. 2017;8:14338. doi: 10.1038/ncomms14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Paquette M, El-Houjeiri L, C Zirden L, Puustinen P, Blanchette P, Jeong H, Dejgaard K, Siegel PM, Pause A. AMPK-dependent phosphorylation is required for transcriptional activation of TFEB and TFE3. Autophagy. 2021;17:3957–3975. doi: 10.1080/15548627.2021.1898748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Parenti G, Andria G, Ballabio A. Lysosomal storage diseases:from pathophysiology to therapy. Annu Rev Med. 2015;66:471–486. doi: 10.1146/annurev-med-122313-085916. [DOI] [PubMed] [Google Scholar]
- 65.Paula PC, Angelica Maria SG, Luis CH, Gloria Patricia CG. Preventive effect of quercetin in a triple transgenic Alzheimer's disease mice model. Molecules. 2019;24:2287. doi: 10.3390/molecules24122287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Peng W, Wong YC, Krainc D. Mitochondria-lysosome contacts regulate mitochondrial Ca(2+) dynamics via lysosomal TRPML1. Proc Natl Acad Sci U S A. 2020;117:19266–19275. doi: 10.1073/pnas.2003236117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Platt FM, d'Azzo A, Davidson BL, Neufeld EF, Tifft CJ. Lysosomal storage diseases. Nat Rev Dis Primers. 2018;4:27. doi: 10.1038/s41572-018-0025-4. [DOI] [PubMed] [Google Scholar]
- 68.Puertollano R, Ferguson SM, Brugarolas J, Ballabio A. The complex relationship between TFEB transcription factor phosphorylation and subcellular localization. EMBO J. 2018;37:e98804. doi: 10.15252/embj.201798804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian H, Wu X, Du X, Yao X, Zhao X, Lee J, Yang H, Yan N. Structural basis of low-pH-dependent lysosomal cholesterol egress by NPC1 and NPC2. Cell. 2020;182:98–111. doi: 10.1016/j.cell.2020.05.020. [DOI] [PubMed] [Google Scholar]
- 70.Ren H, Wang D, Zhang L, Kang X, Li Y, Zhou X, Yuan G. Catalpol induces autophagy and attenuates liver steatosis in ob/ob and high-fat diet-induced obese mice. Aging (Albany NY) 2019;11:9461–9477. doi: 10.18632/aging.102396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roney JC, Li S, Farfel-Becker T, Huang N, Sun T, Xie Y, Cheng XT, Lin MY, Platt FM, Sheng ZH. Lipid-mediated motor-adaptor sequestration impairs axonal lysosome delivery leading to autophagic stress and dystrophy in Niemann-Pick type C. Dev Cell. 2021;56:1452–1468. doi: 10.1016/j.devcel.2021.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Root J, Merino P, Nuckols A, Johnson M, Kukar T. Lysosome dysfunction as a cause of neurodegenerative diseases:lessons from frontotemporal dementia and amyotrophic lateral sclerosis. Neurobiol Dis. 2021;154:105360. doi: 10.1016/j.nbd.2021.105360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rusmini P, Cortese K, Crippa V, Cristofani R, Cicardi ME, Ferrari V, Vezzoli G, Tedesco B, Meroni M, Messi E, Piccolella M, Galbiati M, Garre M, Morelli E, Vaccari T, Poletti A. Trehalose induces autophagy via lysosomal-mediated TFEB activation in models of motoneuron degeneration. Autophagy. 2019;15:631–651. doi: 10.1080/15548627.2018.1535292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saftig P, Klumperman J. Lysosome biogenesis and lysosomal membrane proteins:trafficking meets function. Nat Rev Mol Cell Biol. 2009;10:623–635. doi: 10.1038/nrm2745. [DOI] [PubMed] [Google Scholar]
- 75.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, Banfi S, Parenti G, Cattaneo E, Ballabio A. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 76.Schieder M, Rotzer K, Bruggemann A, Biel M, Wahl-Schott CA. Characterization of two-pore channel 2 (TPCN2)-mediated Ca2+currents in isolated lysosomes. J Biol Chem. 2010;285:21219–21222. doi: 10.1074/jbc.C110.143123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Settembre C, Di Malta C, Polito VA, Garcia Arencibia M, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, Sardiello M, Rubinsztein DC, Ballabio A. TFEB links autophagy to lysosomal biogenesis. Science. 2011;332:1429–1433. doi: 10.1126/science.1204592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sha Y, Rao L, Settembre C, Ballabio A, Eissa NT. STUB1 regulates TFEB-induced autophagy-lysosome pathway. EMBO J. 2017;36:2544–2552. doi: 10.15252/embj.201796699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shao Q, Yang M, Liang C, Ma L, Zhang W, Jiang Z, Luo J, Lee JK, Liang C, Chen JF. C9orf72 and smcr8 mutant mice reveal MTORC1 activation due to impaired lysosomal degradation and exocytosis. Autophagy. 2020;16:1635–1650. doi: 10.1080/15548627.2019.1703353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma S, Taliyan R. Epigenetic modifications by inhibiting histone deacetylases reverse memory impairment in insulin resistance induced cognitive deficit in mice. Neuropharmacology. 2016;105:285–297. doi: 10.1016/j.neuropharm.2016.01.025. [DOI] [PubMed] [Google Scholar]
- 81.Shin HR, Citron YR, Wang L, Tribouillard L, Goul CS, Stipp R, Sugasawa Y, Jain A, Samson N, Lim CY, Davis OB, Castaneda-Carpio D, Qian M, Nomura DK, Perera RM, Park E, Covey DF, Laplante M, Evers AS, Zoncu R. Lysosomal GPCR-like protein LYCHOS signals cholesterol sufficiency to mTORC1. Science. 2022;377:1290–1298. doi: 10.1126/science.abg6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Song JX, Malampati S, Zeng Y, Durairajan SSK, Yang CB, Tong BC, Iyaswamy A, Shang WB, Sreenivasmurthy SG, Zhu Z, Cheung KH, Lu JH, Tang C, Xu N, Li M. A small molecule transcription factor EB activator ameliorates beta-amyloid precursor protein and Tau pathology in Alzheimer's disease models. Aging Cell. 2020;19:e13069. doi: 10.1111/acel.13069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Su H, Li Y, Hu D, Xie L, Ke H, Zheng X, Chen W. Procyanidin B2 ameliorates free fatty acids-induced hepatic steatosis through regulating TFEB-mediated lysosomal pathway and redox state. Free Radic Biol Med. 2018;126:269–286. doi: 10.1016/j.freeradbiomed.2018.08.024. [DOI] [PubMed] [Google Scholar]
- 84.Takahashi D, Moriyama J, Nakamura T, Miki E, Takahashi E, Sato A, Akaike T, Itto-Nakama K, Arimoto H. AUTACs:cargo-specific degraders using selective autophagy. Mol Cell. 2019;76:797–810. doi: 10.1016/j.molcel.2019.09.009. [DOI] [PubMed] [Google Scholar]
- 85.Tan JX, Finkel T. A phosphoinositide signalling pathway mediates rapid lysosomal repair. Nature. 2022;609:815–821. doi: 10.1038/s41586-022-05164-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tanaka M, Machida Y, Niu S, Ikeda T, Jana NR, Doi H, Kurosawa M, Nekooki M, Nukina N. Trehalose alleviates polyglutamine-mediated pathology in a mouse model of Huntington disease. Nat Med. 2004;10:148–154. doi: 10.1038/nm985. [DOI] [PubMed] [Google Scholar]
- 87.Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Trinh MN, Brown MS, Seemann J, Goldstein JL, Lu F. Lysosomal cholesterol export reconstituted from fragments of Niemann-Pick C1. Elife. 2018;7:e38564. doi: 10.7554/eLife.38564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trinh MN, Lu F, Li X, Das A, Liang Q, De Brabander JK, Brown MS, Goldstein JL. Triazoles inhibit cholesterol export from lysosomes by binding to NPC1. Proc Natl Acad Sci U S A. 2017;114:89–94. doi: 10.1073/pnas.1619571114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vega-Rubin-de-Celis S, Pena-Llopis S, Konda M, Brugarolas J. Multistep regulation of TFEB by MTORC1. Autophagy. 2017;13:464–472. doi: 10.1080/15548627.2016.1271514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Vernizzi L, Paiardi C, Licata G, Vitali T, Santarelli S, Raneli M, Manelli V, Rizzetto M, Gioria M, Pasini ME, Grifoni D, Vanoni MA, Gellera C, Taroni F, Bellosta P. Glutamine synthetase 1 increases autophagy lysosomal degradation of mutant huntingtin aggregates in neurons, ameliorating motility in a drosophila model for Huntington's disease. Cells. 2020;9:196. doi: 10.3390/cells9010196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang H, Wang N, Xu D, Ma Q, Chen Y, Xu S, Xia Q, Zhang Y, Prehn JHM, Wang G, Ying Z. Oxidation of multiple MiT/TFE transcription factors links oxidative stress to transcriptional control of autophagy and lysosome biogenesis. Autophagy. 2020a;16:1683–1696. doi: 10.1080/15548627.2019.1704104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Huang Y, Liu J, Zhang J, Xu M, You Z, Peng C, Gong Z, Liu W. Acetyltransferase GCN5 regulates autophagy and lysosome biogenesis by targeting TFEB. EMBO Rep. 2020b;21:e48335. doi: 10.15252/embr.201948335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang Z, Yang C, Liu J, Chun-Kit Tong B, Zhu Z, Malampati S, Gopalkrishnashetty Sreenivasmurthy S, Cheung KH, Iyaswamy A, Su C, Lu J, Song J, Li M. A curcumin derivative activates TFEB and protects against parkinsonian neurotoxicity in vitro. Int J Mol Sci. 2020c;21:1515. doi: 10.3390/ijms21041515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wie J, Liu Z, Song H, Tropea TF, Yang L, Wang H, Liang Y, Cang C, Aranda K, Lohmann J, Yang J, Lu B, Chen-Plotkin AS, Luk KC, Ren D. A growth-factor-activated lysosomal K(+) channel regulates Parkinson's pathology. Nature. 2021;591:431–437. doi: 10.1038/s41586-021-03185-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wolfson RL, Chantranupong L, Wyant GA, Gu X, Orozco JM, Shen K, Condon KJ, Petri S, Kedir J, Scaria SM, Abu-Remaileh M, Frankel WN, Sabatini DM. KICSTOR recruits GATOR1 to the lysosome and is necessary for nutrients to regulate mTORC1. Nature. 2017;543:438–442. doi: 10.1038/nature21423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wu M, Lao YZ, Tan HS, Lu G, Ren Y, Zheng ZQ, Yi J, Fu WW, Shen HM, Xu HX. Oblongifolin C suppresses lysosomal function independently of TFEB nuclear translocation. Acta Pharmacol Sin. 2019;40:929–937. doi: 10.1038/s41401-018-0167-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wyant GA, Abu-Remaileh M, Wolfson RL, Chen WW, Freinkman E, Danai LV, Vander Heiden MG, Sabatini DM. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell. 2017;171:642–654. doi: 10.1016/j.cell.2017.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Yang C, Su C, Iyaswamy A, Krishnamoorthi SK, Zhu Z, Yang S, Tong BC, Liu J, Sreenivasmurthy SG, Guan X, Kan Y, Wu AJ, Huang AS, Tan J, Cheung K, Song J, Li M. Celastrol enhances transcription factor EB (TFEB)-mediated autophagy and mitigates Tau pathology:Implications for Alzheimer's disease therapy. Acta Pharm Sin B. 2022;12:1707–1722. doi: 10.1016/j.apsb.2022.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yin L, Zhou J, Li T, Wang X, Xue W, Zhang J, Lin L, Wang N, Kang X, Zhou Y, Liu H, Li Y. Inhibition of the dopamine transporter promotes lysosome biogenesis and ameliorates Alzheimer's disease-like symptoms in mice. Alzheimers Dement. 2022 doi: 10.1002/alz.12776. doi:10.1002/alz.12776. [DOI] [PubMed] [Google Scholar]
- 101.Yin Q, Jian Y, Xu M, Huang X, Wang N, Liu Z, Li Q, Li J, Zhou H, Xu L, Wang Y, Yang C. CDK4/6 regulate lysosome biogenesis through TFEB/TFE3. J Cell Biol. 2020;219:e201911036. doi: 10.1083/jcb.201911036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zhang CS, Hawley SA, Zong Y, Li M, Wang Z, Gray A, Ma T, Cui J, Feng JW, Zhu M, Wu YQ, Li TY, Ye Z, Lin SY, Yin H, Piao HL, Hardie DG, Lin SC. Fructose-1,6-bisphosphate and aldolase mediate glucose sensing by AMPK. Nature. 2017;548:112–116. doi: 10.1038/nature23275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang CS, Jiang B, Li M, Zhu M, Peng Y, Zhang YL, Wu YQ, Li TY, Liang Y, Lu Z, Lian G, Liu Q, Guo H, Yin Z, Ye Z, Han J, Wu JW, Yin H, Lin SY, Lin SC. The lysosomal v-ATPase-Ragulator complex is a common activator for AMPK and mTORC1, acting as a switch between catabolism and anabolism. Cell Metab. 2014;20:526–540. doi: 10.1016/j.cmet.2014.06.014. [DOI] [PubMed] [Google Scholar]
- 104.Zhang CS, Li M, Wang Y, Li X, Zong Y, Long S, Zhang M, Feng JW, Wei X, Liu YH, Zhang B, Wu J, Zhang C, Lian W, Ma T, Tian X, Qu Q, Yu Y, Xiong J, Liu DT, et al. The aldolase inhibitor aldometanib mimics glucose starvation to activate lysosomal AMPK. Nat Metab. 2022a;4:1369–1401. doi: 10.1038/s42255-022-00640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhang H, Han Y, Yang Y, Lin F, Li K, Kong L, Liu H, Dang Y, Lin J, Chen PR. Covalently engineered nanobody chimeras for targeted membrane protein degradation. J Am Chem Soc. 2021;143:16377–16382. doi: 10.1021/jacs.1c08521. [DOI] [PubMed] [Google Scholar]
- 106.Zhang J, Wang J, Zhou Z, Park JE, Wang L, Wu S, Sun X, Lu L, Wang T, Lin Q, Sze SK, Huang D, Shen HM. Importance of TFEB acetylation in control of its transcriptional activity and lysosomal function in response to histone deacetylase inhibitors. Autophagy. 2018;14:1043–1059. doi: 10.1080/15548627.2018.1447290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang Z, Chen C, Yang F, Zeng YX, Sun P, Liu P, Li X. Itaconate is a lysosomal inducer that promotes antibacterial innate immunity. Mol Cell. 2022b;82:2844–2857. doi: 10.1016/j.molcel.2022.05.009. [DOI] [PubMed] [Google Scholar]
- 108.Zhitomirsky B, Yunaev A, Kreiserman R, Kaplan A, Stark M, Assaraf YG. Lysosomotropic drugs activate TFEB via lysosomal membrane fluidization and consequent inhibition of mTORC1 activity. Cell Death Dis. 2018;9:1191. doi: 10.1038/s41419-018-1227-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhuang XX, Wang SF, Tan Y, Song JX, Zhu Z, Wang ZY, Wu MY, Cai CZ, Huang ZJ, Tan JQ, Su HX, Li M, Lu JH. Pharmacological enhancement of TFEB-mediated autophagy alleviated neuronal death in oxidative stress-induced Parkinson's disease models. Cell Death Dis. 2020;11:128. doi: 10.1038/s41419-020-2322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zoncu R, Bar-Peled L, Efeyan A, Wang S, Sancak Y, Sabatini DM. mTORC1 senses lysosomal amino acids through an inside-out mechanism that requires the vacuolar H(+)-ATPase. Science. 2011;334:678–683. doi: 10.1126/science.1207056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


