Abstract
Because nonopioid analgesics are much sought after, we computationally docked more than 301 million virtual molecules against a validated pain target, the α2A-adrenergic receptor (α2AAR), seeking new α2AAR agonists chemotypes that lack the sedation conferred by known α2AAR drugs, such as dexmedetomidine. We identified 17 ligands with potencies as low as 12 nanomolar, many with partial agonism and preferential Gi and Go signaling. Experimental structures of α2AAR complexed with two of these agonists confirmed the docking predictions and templated further optimization. Several compounds, including the initial docking hit ‘9087 [mean effective concentration () of 52 nanomolar] and two analogs, ‘7075 and PS75 ( 4.1 and 4.8 nanomolar), exerted on-target analgesic activity in multiple in vivo pain models without sedation. These newly discovered agonists are interesting as therapeutic leads that lack the liabilities of opioids and the sedation of dexmedetomidine.
Epidemics in pain (1) and in opioid-use disorder (2, 3) have inspired a search for nonopioid analgesics (1, 4). The α2A-adrenergic receptor (α2AAR) is a nonopioid receptor targeted by dexmedetomidine, a sedative that also has strong analgesic activity (5). Although dexmedetomidine has many advantages in emergency room and intensive care settings, its strong sedative effects (6, 7) and its lack of an oral formulation (8) have limited its broad use as an analgesic. These properties are barriers for future therapeutics targeting this receptor.
Most α2AAR analgesics are chemically related, and the relationship of their sedative to their analgesic properties is unclear. To find therapeutics with new pharmacology, we sought new α2AAR chemotypes, topologically unrelated to known α2AAR agonists. The α2B-adrenergic receptor (α2BAR) active-state structure (9) became available, and its binding site is highly conserved compared with that of α2AAR (fig. S1); therefore, it should be possible to identify new α2AAR agonists by structure-based docking. Meanwhile, the advent of readily accessible make-on-demand (“tangible”) molecules (10-12) ranging from hundreds of millions (10, 13, 14) to more than a billion molecules (15, 16) has vastly increased the chemotypes available for ligand discovery. Docking these libraries has revealed new chemotypes with 20 to 60% hit rates (13, 14, 17-20) and sometimes nanomolar potencies for a growing range of targets (10, 13, 14, 18, 21-24), often with new pharmacology (10, 13, 17, 25). Therefore, we targeted the α2BAR with an ultralarge library docking screen.
Docking 301 million molecules versus the α2BAR
The ZINC15 and ZINC20 virtual libraries are composed of millions to billions of tangible molecules, depending on the molecular property range targeted, and are accessed by combining hundreds of thousands of diverse building blocks through hundreds of well-characterized reactions (10-12). Most of the molecules have not previously been synthesized and range in mass, calculated LogP (cLogP) values (a measure of hydrophobicity), and formal charge. Given the small size of the α2BAR orthosteric site, we docked both the 20 million fragment-like [compounds with smaller masses of <250 atomic mass units (amu)] and 281 million lead-like (compounds with larger masses of 250 to 350 amu) molecules from the ZINC15 library (both sets having cLogP ≤ 3.5) (11) (Fig. 1A). More than 233 trillion complexes, an average of 452,000 per molecule, were sampled by DOCK3.7 and scored with its physics-based energy function (26) across three separate screens (two fragment screens with different variables and one lead-like screen; see Materials and methods). For each screen, the top 300,000 docking-ranked compounds were clustered for topological similarity and then filtered to identify scaffolds dissimilar to known agonists using an extended connectivity fingerprint (ECFP4). These known agonists were drawn from the International Union of Basic and Clinical Pharmacology (IUPHAR)–British Pharmacological Society (BPS) database (27) and from the literature (28-31). Ligands with internal torsional strain were deprioritized (32). An additional novelty filter removed molecules similar to annotated α2AAR compounds in CHEMBL29 (28). Of the remaining top-ranked cluster representatives, 5000 from each fragment screen and 20,000 for the lead-like screen were manually evaluated in UCSF Chimera (https://rbvi.ucsf.edu/chimera) for key polar and nonpolar interactions with α2BAR (9), including with D923.32, F4127.39, F3876.51, Y3916.55, and F3886.52 [residues conserved in α2AAR: D1283.32, F4277.39, F4056.51, Y4096.55, and F4066.52; superscripts use Ballesteros-Weinstein and the G protein–coupled receptor database (GPCRdb) nomenclature (33)]. Most α2AAR agonists, and certainly the clinically used dexmedetomidine and clonidine, are fragments (27), and the docking results reflected this. The docked fragment molecules fit in the orthosteric site, making key contacts with the receptor, whereas molecules in the lead-like screen generally did not fit in the small cavity (Fig. 1A). Accordingly, most selected ligands came from the fragment docking screens.
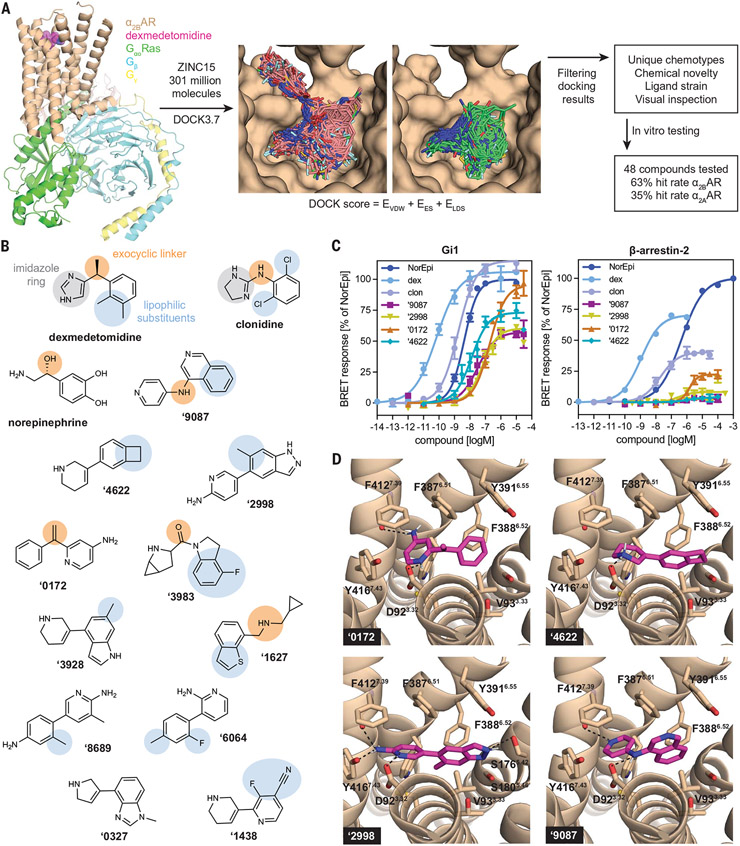
Fig. 1. Newly discovered a2AAR agonists from ultralarge library docking.
(A) 301 million molecules were docked against the active state of a2BAR (PDB 6K41). Lead-like molecules (pink carbons) often spilled out of the orthosteric site, whereas fragment molecules (green carbons) are well complemented by that site. Hit rates were determined with a Ki cutoff of 10 μM. EVDW, van der Waals; EES, electrostatic; ELDS, ligand desolvation. (B) The αAR pharmacophore model (9) overlaid on known α2AAR agonists dexmedetomidine, clonidine, and norepinephrine and new agonists from docking (colors represent the different moieties fulfilling the same role). (C) Gi activation and β-arrestin-2 recruitment for norepinephrine (NorEpi), dexmedetomidine (dex), clonidine (clon), and several of the newly discovered docking agonists. (D) Docked poses of these new agonists with hydrogen bonds to key recognition residues of α2BAR shown as black dashed lines. For (C), data are means ± SEMs of normalized results (n = 4 to 17 measurements for Gi and n = 3 to 8 measurements for β-arrestin-2). Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
From the 64 high-ranking docked compounds prioritized by visual inspection and purchased for in vitro testing, 48 were successfully synthesized—44 fragments and 4 lead-like molecules. Compounds were first tested for binding to the human α2BAR receptor, the structure used in docking screens. Thirty molecules of the 48 tested had binding constant (Ki) values less than 10 μM (table S1). This 63% hit rate is among the highest for a docking campaign to date (10, 14, 21, 23, 34). In radioligand competition assays, compound ZINC1173879087 (from here on referred to as ‘9087) had a Ki of 1.7 nM; the remaining 29 had Ki values ranging from 60 nM to 9.4 μM, which is relatively potent for initial docking hits. Ten compounds (21%) had Ki values below 1 μM (table S1). The compounds were then tested for binding to the murine α2AAR, again by radioligand competition. Of these, 17 (35%) had a Ki better than 10 μM, with affinities ranging from 72 nM to 9.4 μM; five compounds (10%) had Ki values below 1 μM (table S1). Against human α2AAR, the highest affinity was 12 nM (table S2).
Discovery of new α2AAR, partial Gi and Go (Gi/o) agonists
In functional assays, most of the potent binders were partial or full agonists for α2AAR and α2BAR (Fig. 1, B to D; tables S1 and S2; and figs. S2 to S4); few antagonists were found among the more potent docking hits. This reflects the targeting of the activated state of the receptor (35, 36) and was a goal of the screen. The best four agonists from the docking screen include ‘9087 as well as ZINC1240664622, ZINC1242282998, and ZINC001242890172 (from here on referred to as ‘4622, ‘2998, and ‘0172, respectively), with the α2AAR-mediated Gi activation maximum effect () ranging from 60 to 95% of norepinephrine response and mean effective concentration () values of 9.7 to 210 nM in Gαi bioluminescence resonance energy transfer (BRET) assays (Fig. 1C and table S2). We tested the effect of receptor expression in cells on the functional properties of the partial agonist, ‘9087, and ultimately of two optimized analogs, ‘7075 and PS75; all three remained potent Gi partial agonists, with decreasing with receptor expression (fig. S5). In an orthogonal cyclic adenosine monophosphate (cAMP) assay, ‘9087 was a partial agonist with an of 87 nM and of 42%, which is broadly consistent with the BRET assay (from here on, Gi activities are the Gαi BRET assay values, unless otherwise noted) (Fig. 1C, fig. S4, and table S2).
The docking agonists had strong differential activity for Gi activation compared with recruitment of β-arrestin-2. Although this was also true of the known agonists, dexmedetomidine and clonidine, for the new agonists the difference was accentuated so that arrestin recruitment was almost completely eliminated at relevant concentrations. Of the four best docking agonists, only ‘0172 had a measurable efficacy for β-arrestin-2 recruitment, but only with 22% of the of norepinephrine and with weak potency ( 1.7 μM); for the other three, β-arrestin-2 recruitment was negligible (Fig. 1C and table S2). We note that this lack of arrestin recruitment could reflect the partial agonism of the new agonists combined with the weaker coupling of the arrestin pathway versus the well-coupled Gi pathway, as indicated by the differences in potency and efficacy of the reference agonists, dexmedetomidine and clonidine.
Agonists of α2AAR, including its endogenous ligand norepinephrine, also activate other G protein pathways (37). Accordingly, we used the enhanced bystander BRET (ebBRET)–based effector membrane translocation assay (EMTA) (38) to test ‘9087 and its analogs, ‘7075 and PS75, against a more expansive panel of G protein and β-arrestin subtypes. The docking compounds preferentially activated Gi, Go, and Gz (Gi/o/z) signaling, whereas known agonists norepinephrine, dexmedetomidine, and brimonidine strongly activated multiple additional G proteins and β-arrestins (figs. S6 and S7 and tables S3 and S4). Receptor internalization after treatment with compound was also investigated by monitoring disappearance of α2AARs from the plasma membrane (α2AAR-RlucII/rGFP-CAAX biosensor) and relocalization of the receptors in endosomes (α2AAR-RlucII/rGFP-FYVE biosensor) (39). Known agonists brimonidine and norepinephrine show comparable responses for both biosensors, whereas dexmedetomidine has about half of this response. Consistent with their absence of β-arrestin recruitment, we found no effect of ‘9087, ‘7075, and PS75 on disappearance from the plasma membrane and marginal effect at the highest concentrations on endosomal relocalization (fig. S8). Although such functional selectivity was not explicitly modeled in the docking, it likely results from the new chemistry, which was explicitly required (13, 14, 17).
Comparing the new agonists with dexmedetomidine, clonidine, norepinephrine, and a previously described pharmacophore model for αAR selective agonists (9), both similar and distinct features emerge (Fig. 1B). The pharmacophore model for known agonists and the new docking compounds both have basic, nitrogen-containing rings. However, known agonists are dominated by imidazoles (unsaturated or partially saturated), whereas the docking compounds have diverse nonimidazole rings. Both sets of compounds contain additional moieties off of a second aryl ring, typically two substituents for the known agonists; however, for the docking-derived compounds, these vary from bulky hydrophobic rings, to hydrophilic rings, to single substituents, to having no substituents off of the aryl ring at all. Not all of the docking compounds have an exocyclic linker as described in the pharmacophore model. The protonated imidazole of known agonists ion pairs with D923.32 and hydrogen bonds to the backbone of F4127.39 of α2BAR (9, 40) (fig. S1). Although several of the docking-derived compounds also interacted with both D923.32 and F4127.39, they did so with different heterocyclic rings (Fig. 1D).
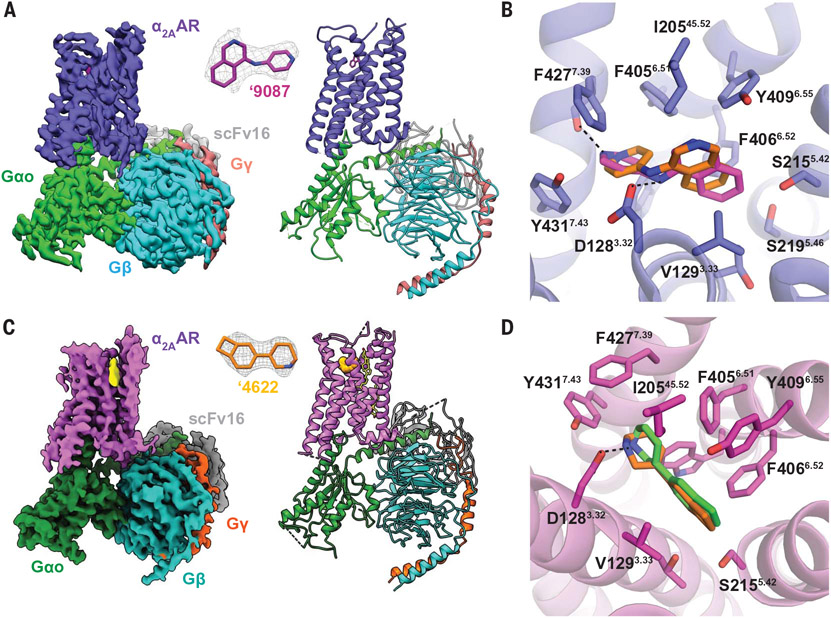
To test the docking model and to template structure-based optimization, we determined the structure of the ‘9087-α2AAR-GoA and ‘4622-α2AAR-GoA complexes at a nominal resolution of 3.47 and 3.38 Å, respectively, using single-particle cryo–electron microscopy (cryo-EM) (Fig. 2, A to D, and figs. S9 and S10). The predicted docked pose superimposes on the cryo-EM result of ‘9087 with a 1.14-Å all-atom root mean square deviation (RMSD) of the agonist; the docking-predicted interactions are recapitulated in the experimental structure (Fig. 2B). The interactions between ‘9087 and α2AAR differ from that of norepinephrine but resemble those of imidazoline-containing agonists (9, 40). ‘9087 interacts with α2AAR mainly through van der Waals and aromatic interactions to transmembrane helices (TM) 3, 5, 6, and 7 and I20545.52 of extracellular loop 2 (ECL2). It also forms an ionic interaction with the conserved D1283.32, and although this interaction is relatively distant at 3.6 Å, it is similar to those of norepinephrine (40) and dexmedetomidine (9) that are 3.0 and 3.7 Å from D1283.32, respectively (Fig. 2B). As in the docking prediction, the basic, formally cationic nitrogen of ‘9087 is not oriented toward D1283.32 to form a salt bridge (Fig. 2B and fig. S11), as seen in norepinephrine, but instead hydrogen bonds with the backbone carbonyl of F4277.39, as it does in dexmedetomidine (9). The bridging exocyclic and formally neutral amine of the ‘9087 ion pairs with D1283.32. Typically for aminergic G protein–coupled receptors (GPCRs), the conserved hydrogen bond with D1283.32 would be made by the stronger base (9, 17, 20, 40). In fact, the formal charge of ‘9087 after protonation of the pyridine moiety is almost equally shared between the two nitrogens, as calculated by semiempirical quantum mechanics and as reflected in the docking model (fig. S11). For ‘4622, the docked pose is also in good agreement with the cryo-EM result with an all-atom RMSD of 1.14 Å; ‘4622 forms a 3.4-Å hydrogen bond to D1283.32 and makes several hydrophobic interactions (Fig. 2D). Both ‘4622- and ‘9087-bound structures have similar receptor-GoA interfaces to other ligand-bound α2AR–G protein complexes (figs. S9 and S10).
Fig. 2. Docking-predicted poses of ‘9087 and ‘4622 superpose well on the cryo-EM structures of the ‘9087-α2AAR-GoA and ‘4622-α2AAR-GoA complexes.
(A and C) Cryo-EM structure of the ‘9087-α2AAR-GoA (A) and ‘4622-α2AAR-GoA (C) complexes. (B) Experimental ‘9087 structure (pink carbons) superposed on the docked pose (orange carbons) (PDB 7W6P; RMSD 1.14 Å). Hydrogen bonds and ion pairs are shown with dashed black lines to F4277.39 and D1283.32, respectively. (D) Experimental ’4622 structure (green carbons) superposed on the docked pose (orange carbons) (PDB 7W7E; RMSD 1.14 Å). Hydrogen bond shown with dashed black lines to D1283.32. For (B) and (D), side chains of α2AAR residues within 4 Å of ligands are shown as sticks.
The interactions observed in the ‘9087 and ‘4622 receptor complexes, and in the modeled pose of analog ‘7075, were tested by residue substitution for effects on Gi activation and β-arrestin recruitment (figs. S12 and S13 and tables S6 and S7). Consistent with the observed ion pair with D1283.32, norepinephrine and dexmedetomidine are highly sensitive to substitutions to D1283.32, with an almost complete loss of Gi activation and β-arrestin recruitment. For dexmedetomidine, the is 170,000-fold higher for activation of D1283.32A. Although the Gi activity of ‘9087 and ‘7075 is also diminished in the D1283.32 mutant receptors, potency only falls by ~200- to 1600-fold. By contrast, the Gi activity of ‘9087, ‘7075, and ‘4622 is eliminated in the F4277.39 A mutant. The backbone carbonyl of F4277.39 hydrogen bonds with ‘9087, whereas its aromatic side chain stacks with the agonist in the cryo-EM structure, perhaps indicating formation of a cation-pi interaction between the pyridine of ‘9087 and F4277.39, as previously suggested for agonist-induced α2AR activation (41). Meanwhile, dexmedetomidine and especially norepinephrine, which lack these interactions, are less affected by this mutant (Fig. 2B). Mutations of Y4096.55 greatly affect norepinephrine, increasing (weakening) values 500- to 10,000-fold, likely disrupting a key hydrogen bond (40, 41); the importance of position 6.55 was previously observed in agonist-induced β2AR activation (42-44). The potencies of ‘9087 and dexmedetomidine are only modestly worse in Y4096.55 mutants, and for ‘9087, the is even slightly increased. For ‘4622 and ‘7075, most substitutions diminished activity, with the exception of S2155.42 A, which slightly increased the agonist activity of ‘7075 and ‘4622 in the Gi activation and β-arrestin recruitment assays and hardly influenced ‘9087 and dexmedetomidine. By contrast, S2155.42A negatively affected norepinephrine-induced receptor activation, consistent with previous studies on direct interactions of full agonists and S2155.42 (40, 41). The Y4317.43A and F mutations overall influenced β-arrestin recruitment of norepinephrine more than Gi signaling. This has been previously observed, leading to the proposal that direct hydrogen bonding between the agonist and the residue at position 7.43 could more tightly couple TM7 and thereby play a role in β-arrestin signaling (40). Taken together, the differential responses to the residue substitutions supports suggestions from the structures that the new agonists, although binding in the same overall site as the canonical agonists, interact in meaningfully different ways, with potential implications for differential receptor signaling.
Optimization of docking hit ‘9087
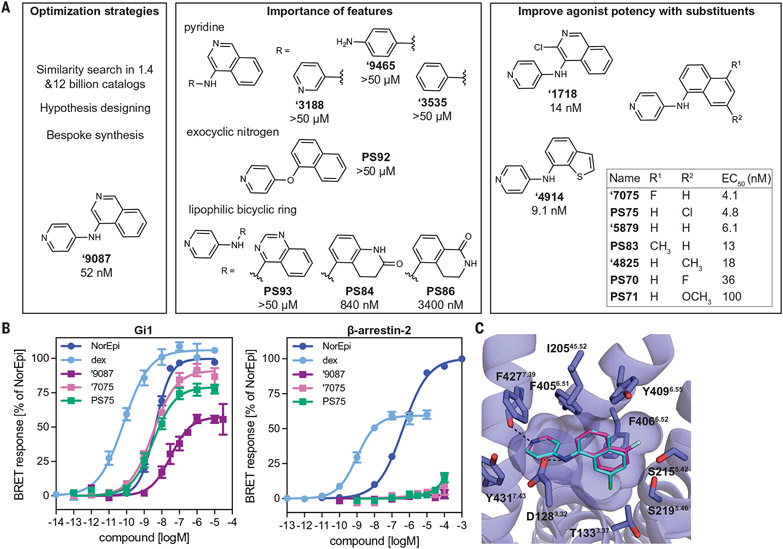
To optimize ‘9087, we adopted two strategies. First, we used classic medicinal chemistry hypothesis-testing and generation of analogs to investigate and improve key recognition features. We looked for possible analogs by similarity-searching among 1.4 billion and 12 billion tangible molecules using the Arthor and SmallWorld programs (12) (NextMove Software, UK). From these searches, we docked prioritized subsets, leading to 13 of the 19 ‘9087 analogs that we ultimately selected (Fig. 3A and table S5). Another six analogs were designed to probe particular protein-ligand interactions. Analogs were also investigated around compounds ‘2998, ‘4622, and ‘0172 (table S5). In the ‘9087 series, perturbing the pyridine eliminated activity and confirmed the importance of the cationic character and the importance of the hydrogen bond with F4277.39 (Fig. 3A, figs. S14 and S15, and table S8).
Fig. 3. Structure-based optimization of ‘9087.
(A) Strategies for analoging ‘9087 (left). Analogs of the pyridine, exocyclic nitrogen, and lipophilic nature of the bicyclic ring revealed their importance for ‘9087 activity (middle). Sampling alternate lipophilic bicyclic rings and modifying their substituents identified eight more potent agonists (right). values are shown for Gi activation. (B) Gi and β-arrestin-2 recruitment for ‘9087 and its two most potent analogs, ‘7075 and PS75. (C) Modeled poses of ‘7075 (pink carbons) and PS75 (blue carbons) based on the ‘9087-α2AAR cryo-EM structure with substituents oriented toward open space in the orthosteric site. Hydrogen bonds and ionic interactions are shown with dashed black lines to F4277.39 and D1283.32, respectively. For (A), Gi and β-arrestin-2 recruitment data for analogs are shown in figs. S14 and S15 and table S8. For (B), data are means ± SEMs of normalized results (n = 7 to 17 measurements for Gi and n = 4 to 8 measurements for β-arrestin-2).
The most potent analogs emerged from variations of the isoquinoline ring in ‘9087. Proximal hydrophobic residues F4056.51, F4066.52, Y4096.55, and I20545.52 do not make obvious polar interactions with the isoquinoline nitrogen of ‘9087 (Fig. 2B). Accordingly, we added small nonpolar groups, like the chlorine in ‘1718, or changed the isoquinoline to a different bicyclic system, like the benzothiophene in ‘4914 or naphthalene in ‘5879. A set of analogs also had the isoquinoline-to-naphthalene change but with a single substituent added at two different positions of the naphthalene, as in ‘4825 and PS83. Overall, this set of analogs resulted in five potent agonists ( 4.1 nM to 18 nM) (data supporting the ‘9087 optimization are summarized in Fig. 3, A and B; figs. S14 and S15; and tables S2, S5, and S8). ‘7075 was the most potent full agonist in the ‘9087 series with 13-fold increased potency in the BRET Gi activation ( 4.1 nM, 93%) and cAMP assays ( 18 nM, 96%) (Fig. 3B, fig. S4, and table S2). It preferentially activated Gi/o signaling over other G protein subtypes and β-arrestins and caused no receptor internalization (figs. S6 to S8 and tables S3 and S4).
Our second strategy for ligand optimization was purely structure based, using the newly determined ‘9087-α2AAR complex. Molecules were prioritized for their favorable docked pose in a ligand-free version of the α2AAR-’9087 structure or were designed to improve protein-ligand interactions based on the ‘9087-alpha2a cryo-EM structure, leading us to synthesize eight further analogs. Two derivatives of ‘9087 (PS84 and PS86) confirm the importance of the lipophilic and aromatic properties of the bicyclic moiety for α2AAR binding and activation, facilitating favorable interactions with the aromatic residues F4056.51, F4066.52, and Y4096.55 (Fig. 2B). Other substitutions confirmed the importance of the ion pair to D1283.32 and of the interaction between the protonated pyridine of ‘9087 and the backbone carbonyl atom of F4277.39 (PS92, PS93), as also shown by the mutational analysis (figs. S12 and S13 and tables S6 and S7).
Seeking more potent analogs, we focused on derivatizing the lipophilic substituents of ‘9087, building off analogs ‘5879 and ‘7075. Assuming the same binding mode for the naphthalene derivative ‘5879 as for ‘9087 in the cryo-EM structure, unexploited space between the ligand and the receptor in the orthosteric site was revealed in positions 5 and 7 of the bicyclic moiety of ‘5879 (R1 and R2 in Fig. 3A). We first tried to fill available space with substituents of different sizes at the R1 position of ‘5879. From largest to smallest, substituents were methoxy (PS71), chlorine (PS75), and fluorine (PS70). The potency of PS75 for Gi activation was similar to that of ‘5879 ( values of 4.8 nM and 6.1 nM, respectively), PS70 ( 36 nM) did not improve activity, and PS71 had 16-fold worse activity compared with ‘5879; for PS71, this may reflect entropic and desolvation penalties and a repulsive interaction for the bulky methoxy substituent that the receptor was unable to accommodate. Similar to ‘7075 with a substituent at the R2 position of the bicyclic moiety, addition of a methyl (PS83) had a similar of 13 nM to that of ‘7075 and ‘5879 ( values of 4.1 nM and 6.1 nM, respectively).
PS75 was the most potent analog to arise from the second round of optimization. The molecule was a full agonist with 11-fold improved potency ( 4.8 nM, 82%) for Gi activation compared with ‘9087, and it more potently activated Gi/o/z subtypes than ‘9087. Meanwhile, PS75 still retained the preferential signaling through the Gi/o versus other G protein families and β-arrestins and again led to negligible receptor internalization (figs. S5 to S8 and tables S3 and S4). In the modeled pose of PS75, the chlorine substituent is oriented toward the open space below its naphthalene ring toward T1333.37 (Fig. 3C). Its potency and efficacy make PS75 a lead molecule for treatment in pain.
Newly discovered α2AAR agonists are analgesic with reduced side effects
In preparation for in vivo studies, we investigated the selectivity and pharmacokinetic properties of our most potent agonists. ‘9087 activated only a few of the 320 GPCRs screened (45) (fig. S16A). Only the dopamine D2 receptor (D2R) had weak activity in secondary assays, with values of 4.5 μM and 16 μM in G protein signaling and β-arrestin recruitment, respectively (fig. S16B). ‘9087 did not measurably activate the μ-opioid receptor (μOR) nor did it inhibit the human ether-à-go-go-related gene (hERG) at concentrations below 10 μM (fig. S16, C and D). In binding experiments to other adrenergic receptors, ‘9087 bound to the α2C-subtype at mid-nanomolar concentration and to other α1-subtypes in the 1-to-10-μM range (table S9). The molecule had no measurable binding for β-adrenergic receptors up to 10 μM. Against the imidazoline-2 receptor (I2R), a common off-target of α2AAR agonists, ‘9087 bound with a Ki of 300 nM, showing a modest sixfold selectivity for the α2AAR, whereas a few docking-derived compounds actually had higher affinities for I2R than for the α2AAR (fig. S16E).
Computational models suggested that ‘9087, ‘4622, ‘7075, and ‘2998 would all have good physiologic permeability, consistent with their small size, low topological polar surface area, and weakly basic character (table S10). Consistent with this prediction, on 10-mg/kg intra-peritoneal (i.p.) injection in mice, the first three compounds, especially, had high brain and cerebrospinal fluid (CSF) exposure, indicating that the compounds are likely to reach centrally acting α2AARs (table S10). ‘9087 reached a similar maximum concentration () in the CSF as did ‘7075, both of which were fourfold greater than the of PS75, and ‘9087 had a 12- to 20-fold higher area under the concentration-time curve (AUC) in the CSF compared with either ‘7075 or PS75; CSF concentrations are often used as a proxy for fraction unbound in the brain (46). Encouragingly, ‘9087 reached high brain exposure after both intravenous (i.v.) and oral (p.o.) administration, with AUC values of 420,000 ng min mL−1 and 2,540,000 ng min mL−1 (table S10). The oral bioavailability was higher than 100%, which may reflect metabolic saturation at nonequal i.v. and p.o. doses, or enterohepatic recirculation (table S10); this merits further investigation. PS75 was fully bio-available, though, as an analog of ‘9087, the same caveats apply. We also investigated the metabolic stability of ‘9087, ‘7075, and PS75 in male rat liver microsomes. All three compounds remain largely unmodified alter 1 hour, with ‘9087 having lower clearance and a higher half-life than its two analogs (fig. S17).
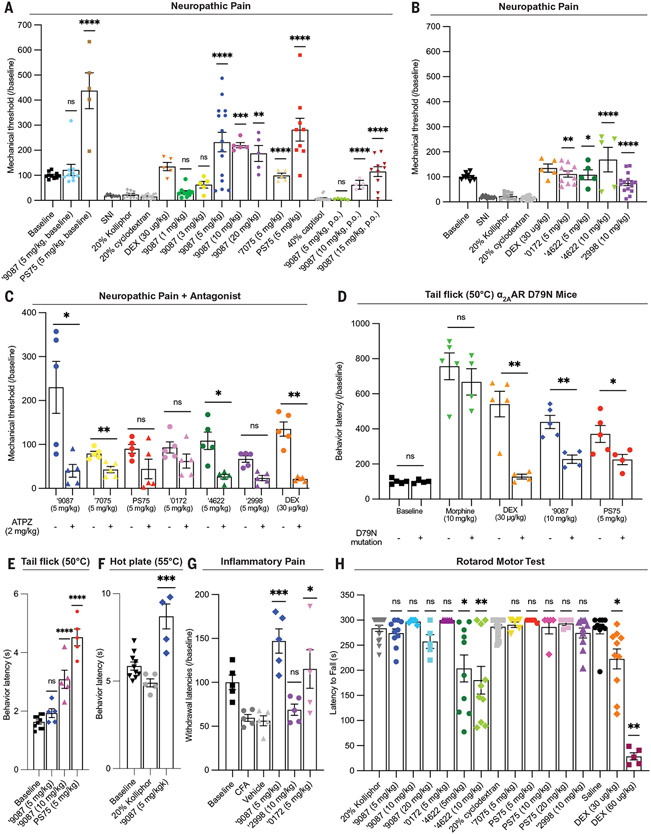
Given their selectivity and high brain exposures, we tested the more potent agonists for pain-relief after systemic dosing (Fig. 4, A to G). Initial doses were chosen to be less than the 10 mg/kg dose used in pharmacokinetics studies owing to favorable CSF and brain properties. We started with ‘9087, the initial docking hit, and evaluated analogs as they emerged from compound optimization. With naïve (uninjured) mice, ‘9087 did not increase baseline mechanical withdrawal thresholds, something observed with many antipain medications, which often only have an antinociceptive effect in the presence of pain. We then investigated the activity of ‘9087 in a mouse model of neuropathic pain, in which partial peripheral nerve injury invokes profound mechanical hypersensitivity (47). Systemic subcutaneous (s.c.) injections of ‘9087 dose-dependently increased the mechanical thresholds of spared nerve injury (SNI) mice, with a sharp increase in activity from 3 mg/kg to 5 mg/kg, at which point the effects plateaued (Fig. 4A). Lower doses were antiallodynic, returning mechanical thresholds to preinjury levels, whereas the higher doses were genuinely analgesic, generating mechanical thresholds substantially higher than baseline, preinjury levels. ‘9087 also increased thermal latencies in the complete Freund’s adjuvant (CFA)–mediated inflammatory pain model, which indicates that the molecule is effective in both tissue and nerve injury–induced pain models (Fig. 4G). ‘9087 also increased withdrawal latencies in the hot plate (55°C) and tail flick (50°C) assays of acute thermal (heat) pain (Fig. 4, E and F). Consistent with its relatively high exposure on oral dosing, this molecule also conferred a dose-dependent antiallodynic effect when delivered orally in the SNI neuropathic pain model (Fig. 4A). Doses up to 20 mg/kg of ‘9087 did not reduce the ability of the mice to perform in the rotarod test, which contrasts with the complete sedation of a dexmedetomidine dose of 60 μg/kg (Fig. 4H). This finding is an important differentiator for the new series and indicates that the analgesic effects of ‘9087 are not the result of motor impairment.
Fig. 4. The docking-derived agonists are antinociceptive in neuropathic, inflammatory, and acute thermal pain through the α2AAR but are not sedating.
(A to C) Effect of new α2AAR agonists in neuropathic pain model in mice after SNI with mechanical allodynia. (A) The new agonists ‘9087 and PS75 administered in naïve mice (baseline versus ‘9087, 5 mg/kg; baseline versus PS75, 5 mg/kg; one-way ANOVA; ns, not significant; ****P < 0.0001), dose response of ‘9087 in SNI mice and analogs ‘7075 and PS75 compared with their vehicles (20% kolliphor versus all ‘9087 doses; 20% cyclodextran versus ‘7075 and PS75; one-way ANOVA; **P < 0.01; ***P < 0.001; ****P < 0.0001) with positive control dexmedetomidine (DEX), and ‘9087 administered orally (p.o.) compared with its vehicle (40% captisol versus ‘9087 doses; one-way ANOVA; ****P < 0.0001). (B) Effect of additional agonists ‘4622, ‘0172, and ‘2998 compared with their vehicles (20% kolliphor versus ‘4622, 5 mg/kg; ‘4622, 10 mg/kg; and ‘0172, 5 mg/kg; one-way ANOVA; 20% cyclodextran versus ‘2998; two-tailed t test; ns = *P < 0.05; **P < 0.01; ****P < 0.0001) and positive control DEX. (C) Administration of α2AR antagonist atipamezole (ATPZ, 2 mg/kg i.p.) to block agonist efficacy in neuropathic pain model (‘9087 versus ‘9087 with ATPZ; ‘7075 versus ‘7075 with ATPZ; PS75 versus PS75 with ATPZ; ‘0172 versus ‘0172 with ATPZ; ‘4622 versus ‘4622 with ATPZ; ‘2998 versus ‘2998 with ATPZ; DEX versus DEX with ATPZ; two-tailed t test; *P < 0.05; **P < 0.01). (D) Diminished analgesia in α2AAR D79N mice in the 50°C tail flick assay for acute thermal (heat) pain. The mutation does not affect morphine analgesia but substantially decreases the analgesia by DEX, ‘9087, and PS75 (baseline WT versus D79N; morphine WT versus D79N; DEX WT versus D79N; ‘9087 WT versus D79N; PS75 WT versus D79N; two-tailed t test; *P < 0.05; **P < 0.01). (E) Analgesia of ‘9087 and PS75 in 50°C tail flick assay for acute thermal (heat) pain compared with its vehicle (20% Kolliphor versus ‘9087 and PS75; one-way ANOVA; ****P < 0.0001). (F) Analgesia of ‘9087 in 55°C hot plate assay for acute thermal (heat) pain compared with its vehicle (20% Kolliphor versus ‘9087; two-tailed t test; ***P < 0.001). (G) Efficacy of newly characterized agonists in CFA-induced hyperalgesia compared with the vehicle (vehicle versus ‘9087, ‘2998, and ‘0172; one-way ANOVA; *P < 0.05; ***P < 0.001). (H) Evaluating motor impairment and sedation of newly characterized agonists in the rotarod motor test. Only ‘4622 causes slight motor impairment, whereas other agonists do not. DEX causes significant impairment and complete sedation at higher doses. All compounds compared with their vehicles (20% Kolliphor versus ‘9087, ‘0172, and ‘4622; 20% cyclodextran versus ‘2298, ‘7075, and PS75; saline versus DEX; one-way ANOVA; *P < 0.05; **P < 0.01; ****P < 0.0001). For (A) to (G), all compounds were administered s.c., unless otherwise indicated. Data are shown as individual data points and means ± SEMs (n = 4to 25).
We also investigated the mechanistic bases for the analgesia of the new α2AAR agonists, both pharmacologically and genetically. Pharmacologically, the analgesic effect of ‘9087 was reversed by a systemic injection of the well-known α2AR antagonist, atipamezole (2 mg/kg; administered 15 min before ‘9087) (Fig. 4C). Because atipamezole has broad activity against the α2AR receptor subtypes and imidazoline receptors (48), we also tested ‘9087 in mice that express an inactive form of the α2AAR (point mutation D79N) (5, 49-51). D79N mutant mice were tested in the tail flick (50°C) assays. As previously reported, dexmedetomidine no longer induced analgesia in the mutant mice (52), and, as a control, the analgesia conferred by a 10-mg/kg dose of morphine was not significantly altered by the mutation (Fig. 4D). Consistent with activity through α2AAR, the analgesia conferred by ‘9087 was reduced by >50% back to baseline in the D79N mutant mice (Fig. 4D). Although most of the antinociceptive activity appears to derive from activity at the α2AAR, we cannot discount contributions from other receptors.
Five other of the new, docking-derived α2AAR agonists (‘2998, ‘4622, ‘0172, ‘7075, and PS75) also exhibited antiallodynic effects in the SNI mice (Fig. 4, A and B). The ‘9087 analogs, ‘7075 and PS75, completely reversed the mechanical hypersensitivity in the neuropathic pain model, with PS75 being more effective than ‘9087 (Fig. 4A). In contrast to ‘9087, PS75 did increase the mechanical thresholds of naïve (uninjured) mice (Fig. 4A). The antiallodynic effects of ‘4622 and ‘7075 were reversed by atipamezole (48); although this antagonist also partially reversed the antiallodynia of PS75, ‘2998, and ‘0172, these effects did not reach statistical significance at the small numbers of mice tested (Fig. 4C). PS75 also increased withdrawal latencies in the tail flick (50°C) acute thermal pain assay, and when tested in the D79N mutant mice, its analgesic effect was reduced by >50% (Fig. 4, D and E). Compounds ‘0172 and ‘4622 also exhibited antihyperalgesic effects in the CFA inflammatory pain model (Fig. 4G); ‘2998 did not, which may reflect the reduced brain penetration of this molecule (table S10). Only ‘4622 caused slight motor impairment at its equianalgesic dose in the rotarod test; however, the effect did not reach the full sedation observed with dexmedetomidine (Fig. 4H). As with ‘9087, increased dosing up to 20 mg/kg PS75 did not have an effect on the rotarod test (Fig. 4H). Taken together, these pharmacological and chemical-genetic epistasis experiments support a mechanism of action primarily through the α2AAR receptor, though a lesser contribution of other α2AR subtypes cannot be ruled out.
Some α2AAR agonists can produce changes in feeding, weight gain, and hyperglycemia (53, 54) as side effects. Accordingly, we evaluated the effect of compound treatment on body weight >48 hours after injection while allowing the mice to freely feed. We found no effect on body weight for ‘9087 dosed at 5 mg/kg, 10 mg/kg, or 20 mg/kg, nor for dexmedetomidine dosed at 30 μg/kg (fig. S18). We also investigated whether ‘9087 induced constipation, a side effect well-known for opioids and other classes of analgesics, comparing it with dexmedetomidine and with morphine tested at analgesic doses (30 μg/kg and 10 mg/kg, respectively). The number of accumulated pellets over 6 hours alter vehicle or compound injection was measured. As expected, morphine induced constipation when compared with vehicle at the 1-, 2-, and 3-hour marks. By contrast, although pellet number did decrease modestly with ‘9087 and dexmedetomidine, neither effect differed significantly from vehicle (fig. S18). We recognize that other possible side effects remain untested in this study; we return to these in the Discussion section.
Discussion
Three key observations emerge from this study. First, multiple chemotypes, unrelated to known agonists, discovered directly from large-library docking are efficacious in neuropathic, inflammatory, and acute pain models through α2AAR agonism (Fig. 4, A to F). In docking, as in other target-based screens, the initial goal is to identify molecules with in vitro activity; these are then optimized for in vivo activity through extensive structure-activity optimization (13, 14, 17, 20). Although it may be rare that direct hits from a docking screen are themselves in vivo active, such activity of the direct docking hits in this study does speak to the strengths of interrogating vast virtual libraries (11, 12). Second, functional assays reveal preferential Gi/o/z activation versus other G protein subtypes, no β-arrestin activity, and no receptor internalization for ‘9087 and its analogs, ‘7075 and PS75, compared with the established therapeutic α2AAR agonists, like dexmedetomidine and brimonidine, that also activate multiple other G protein and β-arrestin signaling pathways (Fig. 1C, Fig. 3B, table S2, and figs. S6 to S8). Although the high rate of agonist discovery was an intended outcome of docking against the activated state of the α2BAR, the functional selectivity was not designed and can be attributed to the chemotypes they explore and, by extension, their use of both canonical and noncanonical receptor interactions (Fig. 2 and figs. S12 and S13). There is no logical requirement that new chemotypes lead to new signaling pharmacology, though this has often been observed for other receptors (10, 13, 17, 25). Third, unlike dexmedetomidine and clonidine (6, 7, 55), ‘9087 and its analogs, PS75 and ‘7075, do not cause sedation or motor impairment at analgesic doses, potentially enabling broader applications to pain treatment and attesting to the ability to differentiate these two effects with α2AAR agonists (Fig. 4H).
The new α2AAR chemotypes explored (Fig. 1B) reflect the size and diversity of the docked libraries. Most of the actives emerged from the fragment-like library in ZINC, which covers a much greater portion of the chemical universe in its size range than do the lead-like or druglike libraries. This is akin to physical fragment libraries, which typically might include ~1500 molecules (56) but are thought to cover more chemotypes than high-throughput screening libraries that are 1000-fold larger. Meanwhile, the virtual fragment library in ZINC enumerates more than 20 million molecules (11, 12), ~10,000-fold more than in most physical fragment libraries. With more than 800,000 Bemis-Murcko scaffolds (18), there are 500-fold more fragment scaffolds in the docked library than there are molecules in most physical fragment libraries. From this great chemotype diversity springs opportunities for ligands with distinct pharmacology.
How the new chemotypes confer new pharmacology is uncertain. From the cryo-EM cocomplex with ‘9087 and substitution of binding site residues, ‘9087 and ‘7075 appear to make weaker interactions with D1283.32 and an apparently stronger interaction with F4277.39 compared with the canonical agonists (Fig. 2B and figs. S12 and S13). These structural differences may contribute to the unusual functional Gi/o/z selectivity over other G protein subtypes and β-arrestins as compared with known agonists, and to the lack of receptor internalization (Fig. 3B and figs. S6 to S8). In turn, this unusual signaling may play a role in conferring analgesia without sedation (Fig. 4). Admittedly, as the engagement of transducing G proteins and β-arrestins occurs 35 Å away from the orthosteric site, other mechanisms may be involved (57). Moreover, the physiological impact of the selective signaling will be entangled with the pharmacokinetics of the molecules. Regardless, what should be clear is that the analgesic potential of α2AAR agonists may be disentangled from their sedative effect, which is important for future drug development.
Several cautions merit re-emphasis. ‘9087 and its analogs are not as potent as dexmedetomidine. The action of ‘9087 in vivo is blocked by the α2AR antagonist atipamezole (48) and much reduced in D79N α2AAR mice, which indicates that the α2AAR is the primary receptor mediating activity in vivo (Fig. 4, C and D). However, especially for the mutant mice, we note that although most efficacy above baseline was reduced, it was not fully reversed (Fig. 4, C and D), and other targets may also play a role, including the I2R and other α2AR subtypes. We also have not extensively evaluated side effects common for α2AAR agonists, especially cardiovascular effects mediated by α2AAR and α2BAR activation (6). Finally, we do not anticipate that the ability to translate directly from docking hits to in vivo activity, as we saw in this study, will be common. In this case, it was helped by the small size of the α2BAR and α2AAR orthosteric sites and correspondingly the high potencies and good physical properties of the docking hits (fig. S1 and tables S2 and S10).
These caveats should not obscure the key observations of this study. From an ultralarge library docking screen emerged low-nanomolar α2AAR partial agonists, topologically unrelated to previously known ligands, making previously unobserved interactions with the receptor that appear to confer new pharmacology (Fig. 1; Fig. 2B; table S2; and figs. S6 to S8, S12, and S13). Several of the new α2AAR agonists were antiallodynic and analgesic in neuropathic and inflammatory pain models and against acute nociception in naïve animals (Fig. 4, A to G). Among the most promising are ’9087 and PS75, both of which are strongly analgesic without the sedative effects of dexmedetomidine (Fig. 4) and are orally bio-available (table S10). These properties make the compounds plausible therapeutic leads for new nonopioid pain therapeutics without the sedation of classic α2AAR drugs.
Materials and methods
Molecular docking
The α2BAR receptor with dexmedetomidine and GoA [Protein Data Bank (PDB) ID: 6K41] (9) was used for docking calculations before the determination of the α2BAR dexmedeto-midine-bound structure (PDB 7EJA; comparison shown in fig. S1). Three screens of the ZINC15 database were run, two for fragment molecules (<250 amu, cLogP ≤ 3.5) and one for lead-like (250 to 350 amu, cLogP ≤ 3.5). Docking was performed with DOCK3.7 (26). For the first screen, 45 matching spheres(26) were used, 15 from the docked pose of dexmedetomidine and 30 from SPHGEN-generated spheres (58). The receptor structure was protonated using REDUCE (59), and AMBER united atom charges were assigned (60). In control calculations (61) with 15 known agonists from the IUPHAR-BPS database(27) and from the literature (29-31), balanced against 1800 property matched decoys (62), docking parameters were optimized based on adjusted logAUC (61) and based on recapitulation of ligand interactions with residues α2BAR D923.32, F4127.39, F3876.51, Y3916.55, and F3886.52 (residues conserved in α2AAR: D1283.32, F4277.39, F4056.51, Y4096.55, and F4066.52). An “extrema” set was used to evaluate cationic charge preference, as described (18, 62). The protein low dielectric and desolvation regions, defined by pseudoatoms calculated with SPHGEN (58), were extended as previously described (63), based on the control calculations, by a radius of 1 and 0.3 Å, respectively (10, 64). Energy potential grids were calculated using CHEMGRID for AMBER-based van der Waals potential, QNIFFT (65) for Poisson-Boltzmann–based electrostatic potentials, and SOLVMAP (66) for context-dependent ligand desolvation. In the second and third docking screens, differences included modified matching spheres (added rigid fragments of xylazine docked-pose, only used 40 matching spheres) and extension of the desolvation pseudoatoms by a radius of 0.2 A.
For the first screen, 20 million molecules from the ZINC15 (https://zinc15.docking.org/) fragments subset were docked in 3008 core hours or ~6 wall-clock hours on a 500-core cluster. Almost 5 trillion complexes were sampled, on average each molecule sampled 2405 orientations and 202 conformations. Only ~8 of 20 million could be sterically accommodated in the orthosteric site, reflecting its small size. For the second screen, the same 20 million fragments were docked in 3830 core hours or 7.7 hours on 500-core cluster, sampling more than 6 trillion complexes; on average, each molecule sampled 3122 orientations and 203 conformations. About 9 million molecules were accommodated in the site. For the third screen, 281 million molecules from ZINC15 lead-like subset were screened in 71,625 core hours or ~1 week on 500 cores. More than 222 trillion complexes were sampled with an average of 4553 orientations and 469 conformations per molecule, though ultimately only 13.5 million could sterically fit in the site.
For the first and second screens, the top 161,055 scored compounds were clustered by ECFP4-based Tanimoto coefficient (Tc) of 0.5 to identify distinct chemotypes, resulting in 37,150 and 33,378 clusters. For the third screen, the top 300,000 scored compounds were clustered in the same manner, resulting in 57,168 clusters. Molecules were filtered for novelty, removing those with Tc > 0.35 to 15 α2AAR agonists used in control calculations. The top 5000 ranked molecules remaining were visually filtered for interactions at α2BAR residues D923.32, F4127.39, F3876.51, Y3916.55, and F3886.52 for the first and second screens; for the third screen, the top 20,000 molecules were examined by the same criteria. Lastly, prioritized molecules were also filtered for internal torsional strain; this was done visually for the first screen, whereas the second and third screens used a method drawing on CSD torsion populations cutting off at a total energy of 2 torsion energy units (32). An additional novelty filter was performed removing molecules with TC > 0.35 to CHEMBL29(28) α2AAR compounds. Sixty-four molecules were selected for purchasing: 33, 26, and 5 from the first, second, and third screens, respectively. Ten were sourced from WuXi and another 54 from Enamine, of which 8 and 40 were successfully synthesized, respectively. Most of these compounds have not previously been synthesized before, to the best of our knowledge, except for some of the smaller fragments, which have been previously used as building blocks (table S5).
Synthesis of tangible molecules
Forty-eight molecules prioritized for purchasing were synthesized by Enamine and Wuxi for a total fulfilment rate of 75%. Compounds were sourced from the Enamine REAL database (https://enamine.net/compound-collections/real-compounds) or the WuXi GalaXi virtual library. The purities of active molecules synthesized by Enamine and WuXi were at least 90% and typically above 95%. For bespoke compound synthesized in house purities were at least 96% and typically above 99%. The purity of compounds tested in vivo were >95% and typically above 98% (table S5 and figs. S19 and S20).
Ligand optimization
Analogs for four docking hits (‘9087, ‘2998, ‘0172, and ‘4622) were queried in Arthor and SmallWorld 1.4 and 12 Billion tangible libraries (https://sw.docking.org/, https://arthor.docking.org), the latter primarily containing Enamine REAL Space compounds (https://enamine.net/compound-collections/real-compounds/real-space-navigator). Results from SmallWorld, Bemis-Murcko framework, and substructure queries were pooled, docked into the α2BAR site before ‘9087-α2AAR structure being determined. Compounds with favorable interactions in the orthosteric site were prioritized, leading to 13 analogs for ‘9087 (table S5). Also, for the four docking hits, analogs were designed by modifying the two-dimensional (2D) chemical structure to test specific hypotheses, adding another six analogs for ‘9087 (table S5). The second round of analogs for ‘9087 were designed and prioritized for bespoke synthesis. Some were docked to a preliminary cryo-EM model of the ‘9087-α2AAR structure, whereas several were designed and synthesized regardless of docked pose to test specific hypotheses; in total, eight of these were synthesized and tested (table S5). Calculation of the contact areas was performed by means of UCSF Chimera (https://rbvi.ucsf.edu/chimera).
Bespoke synthesis
‘7075 was designed for hypothesis testing and was bespoke synthesized by Enamine (fig. S19).
Molecular modeling of ‘7075 and PS75
Maestro (version 2019-4, Schrödinger, Inc.) was used to manually change the chemical structure of ‘9087 to ‘7075 or PS75 in a preliminary model of the ‘9087-α2AAR complex cryo-EM structure. The isoquinoline nitrogen was changed to a carbon and the fluorine or chlorine substituent was added to the naphthalene ring for ‘7075 and PS75, respectively. The resulting complex of ‘7075 or PS75 and α2AAR coupled to the G protein but without scFv16 was energy minimized following the Protein Preparation Wizard protocol using the OPLS3e force field. The maximum heavy atom deviation from the initial model was 0.3 Å.
Passive membrane permeability prediction
Ligand structures were converted from SMILES strings to 3D structures using LigPrep (version 53013, Schrödinger, Inc.). For the passive membrane permeability prediction (67, 68), we retained only neutral form for each ligand. Passive membrane permeability of a ligand is predicted from the free-energy of insertion (), i.e., from the energy difference between a conformer in low and high dielectric media. Therefore, we generated conformations of each ligand using ConfGen software (version 5.1, Schrödinger, Inc.). We minimized each conformer in a low dielectric medium (chloroform) to mimic the membrane dielectric using Protein Local Optimization Program (PLOP) (69). After finding the lowest energy conformer in the low dielectric medium, we calculated the energy of that energy-minimized conformer in water. We subtracted the energy of the ligand in the high-dielectric water from the low-dielectric medium. We further added a deionization penalty term to account for transforming ionized form of the ligand in water to its neutral form in membrane. We computed the deionization penalty energy using the empirical pKa (where Ka is the acid dissociation constant) prediction software Epik (version 5.1013, Schrödinger, Inc.). We rank-ordered the ligands based on their free-energy of insertion.
Radioligand binding experiments
Receptor binding affinities for the α2AAR receptor and to α2BAR as well as the related adrenergic subtypes α1A, α1B, α2C, β1, and β2 were determined as described previously (44, 70). In brief, membranes were prepared from HEK293T cells transiently transfected with the cDNA for human α2AAR, murine α2AAR (provided by D. Calebiro, Birmingham, UK), human α2BAR (obtained from the cDNA resource center, www.cdna.org), or with the cDNAs for the human α1A, α1B, α2C, β2 (all from cDNA resource center), and β1 (provided by R. Sunahara, UCSD). Receptor densities ( value) and specific binding affinities (Kd value) for the radioligand [3H]RX82,1002 (specific activity 52 Ci/mmol, Novandi, Södertälje, Sweden) were determined as 1400 ± 210 fmol/mg protein and 0.54 ± 0.024 nM for human α2AAR, 4000 ± 720 fmol/mg protein and 1.8 ± 0.61 nM for murine α2AAR, and 3400 ± 580 fmol/mg protein and 2.3 ± 0.52 nM for α2BAR, respectively. Further values are 3200 ± 1900 fmol/mg protein and 0.58 ± 0.11 nM for α2C, 2000 ± 950 fmol/mg protein and 0.70 ± 0.13 nM for α1A, and 4000 fmol/mg protein and 0.11 nM for α1B, both determined with [3H]prazosin (51 Ci/mmol, PerkinElmer, Rodgau, Germany), respectively, and 1400 ± 360 fmol/mg protein and 0.070 ± 0.006 nM for β1 and 1300 ± 230 fmol/mg protein and 0.074 ± 0.012 nM for β2, both determined with [3H]CGP12,188 (52 Ci/mmol, PerkinElmer).
Competition binding with α2AR subtypes were performed by incubating membranes in buffer A (50 mM TRIS at pH 7.4) at final protein concentrations of 3 to 10 μg per well with the radioligand (final concentration 0.5 to 2.0 nM according to the appropriate Kd and ) and varying concentrations of the competing ligands for 60 min at 37°C. Binding to α1A and α1B was measured with buffer B (50 mM TRIS, 5 mM MgCl2, 1 mM EDTA, 100 μg/mL bacitracin and 5 μg/mL soybean trypsin inhibitor at pH 7.4) at 2 to 6 μg per well (radioligand at 0.2 to 0.3 nM) and binding to β1 and β2 was measured with buffer C (25 mM HEPES, 5 mM MgCl2, 1 mM EDTA, and 0.006% bovine serum albumin at pH 7.4) at 4 to 8 μg per well (radioligand 0.2 nM). Nonspecific binding was determined in the presence of unlabeled ligand at 10 μM. Protein concentration was measured using the method of Lowry (71).
The resulting competition curves were analyzed by nonlinear regression using the algorithms implemented in Prism 8.0 (GraphPad Software, San Diego, CA) to provide mean inhibitory concentration (IC50) values, which were subsequently transformed into a Ki values applying the equation of Cheng and Prusoff (72). Mean Ki values (±SEM for n ≥ 3 or ±SD for n = 2) were derived from two to seven experiments each performed in triplicates.
Functional assays
Plasmids
The human wild-type (WT) α2AAR, its respective receptor mutants (73) and the murine α2AAR, all carrying an N-terminal HA-signal sequence and a FLAG-tag, as well as the human adrenergic receptor subtypes α1A, α1B, α2C, β1, and β2 and the dopamine receptor D2long were cloned to pCDNA3.1 for G protein activation assays [BRET, inositol phosphate (IP) accumulation]. Human α2AAR and α2BAR were fused to the ARMS2-PK2 sequence and cloned to pCMV (DiscoverX, Eurofins) for β-arrestin-2 recruitment assays, respectively, using polymerase chain reaction (PCR) and Gibson Assembly (New England Biolabs) (70). Sequence integrity was verified by DNA sequencing (Eurofins Genomics).
Bioluminescence resonance energy transfer
G protein activation by human α2AAR and D2long was monitored with Gαi1-RLucII (74, 75) together with Gβ1 and Gγ2-GFP10. Assessment of arrestin recruitment was performed by enhanced bystander BRET using CAAX-rGFP and β-arrestin-2–RLucII as biosensors (39, 74) in the presence of GRK2, as described (44, 76). In brief, HEK293T cells (gift from the Chair of Physiology, FAU Erlangen-Nürnberg) were transfected with 200 ng receptor plasmid for G protein activation (receptor:Gα:Gβ:Gγ ratio 2:0.5:1:4) or 100 ng receptor plasmid for β-arrestin recruitment (receptor: β-arrestin: GRK2:CAAX ratio 1:0.2:1:3) using linear poly-ethyleneimine (PEI, Polysciences, 3:1 PEI:DNA ratio). The DNA was complemented to a total amount of 1 μg DNA per 3 × 105 cells with single-stranded DNA (ssDNA) (Sigma Aldrich) and 10,000 cells per well were transferred into 96-well half-area plates (Greiner, Frickenhausen, Germany). Additional experiments were performed using the same amount of G protein plasmids as described above but 50 ng or 10 ng α2AAR plasmid instead. 48 hours after transfection, the cell medium was exchanged with PBS (phosphate buffered saline), and cells were stimulated with ligands at 37°C for 10 min. Coelenterazine 400a (abcr GmbH, Karlsruhe, Germany) at a final concentration of 2.5 μM was added 5 min before measurement. BRET was monitored on a Clariostar plate reader (BMG, Ortenberg, Germany) with the appropriate filter sets (donor 410/80 nm, acceptor 515/30 nm) and was calculated as the ratio of acceptor emission to donor emission. BRET ratio was normalized to the effect of buffer (0%) and the maximum effect of norepinephrine (100%) for adrenergic receptors and quinpirole (100%) for D2long. For each compound, 3 to 17 individual experiments were performed, each done in duplicates.
Surface expression of the α2AAR in the G protein activation assays was monitored applying an enzyme-linked immunosorbent assay (ELISA) directed against the N-terminal FLAG tag. HEK293T cells were transfected with the cDNAs encoding α2AAR, Gαi1-RLucII, Gβ1, Gγ2-GFP10, and ssDNA, as described above. As a control, cells transfected with only α2AAR or mock pcDNA3.1 plasmid and ssDNA were used. Immediately after transfection, 50,000 cells per well were transferred to a 48-well plate (Greiner) pretreated with poly-D-lysine (Sigma Aldrich) and incubated at 37°C and 5% CO2 for 48h. The medium was removed, cells were treated with 4% paraformaldehyde for 10 min, washed once (wash buffer, 150 mM NaCl, 25 mM Tris, pH 7.5), and blocked for 60 min (30 g/liter skim milk powder in wash buffer, all steps carried out at room temperature). After incubation with anti-FLAG M2 mouse IgG (F3165, Sigma Aldrich, 1:4000 in blocking solution) for 60 min, cells were washed twice, blocked again for 60 min and incubated with antimouse rabbit IgG-HPR (A9044, Sigma Aldrich, 1:20,000 in blocking solution) for 60 min. Cells were washed thrice, before 200 μL substrate buffer was added (2.8 mM o-phenylenediamine, 35 mM citric acid, 66 mM Na2HPO4, pH 5.0). Reactions were kept in the dark for 5 to 15 min and stopped by addition of 1 M H2SO4 (200 μL). Resulting mixtures were transferred to a 96-well plate and absorption was determined with the Clariostar microplate reader at 492 nm. Data were normalized using cells transfected with only α2AAR (100%) and mock pcDNA3.1 (0%), respectively. n = 4 independent experiments were performed with each condition in triplicate.
The sensitivity of selected ligands to the receptor mutants α2AAR-D1283.32A, α2AAR-D1283.32T, α2AAR-D1283.32L, α2AAR-S2155.42A, α2AAR-Y4096.55A, α2AAR-Y4096.55T, α2AAR- Y4096.55F, α2AAR-F4277.39F, α2AAR-Y4317.43A, and α2AAR-Y4317.43F was monitored by G protein activation as described above, transfecting the appropriate receptor together with Gαi1-RLucII and Gβ1/Gγ2-GFP10. Data were analyzed as ligand-induced changes in BRET compared with vehicle (deltaBRET), and normalization was done according to the effect of buffer (0%) and norepinephrine (100%) with the exception of α2AAR-D1283.32A (dexmedetomidine = 100%), α2AAR-D1283.32T, and α2AAR-D1283.32L (‘7075 = 100%). Similarly, the effect of the α2AAR-D1283.32A, α2AAR-S2155.42A, α2AAR-Y4096.55A, α2AAR-Y4096.55F, α2AAR-F4277.39F, α2AAR-Y4317.43A, and α2AAR-Y4317.43F mutations on arrestin recruitment was evaluated as described above, transfecting the appropriate receptor together with CAAX-rGFP, GRK2, and β-arrestin-2–RLucII. Data were analyzed as ligand-induced changes in BRET compared with vehicle (deltaBRET). Three to seven experiments were done in duplicate.
IP accumulation assay
Determination of G protein–mediated signaling by human α2AAR, murine α2AAR, and human α2BAR was performed applying an IP accumulation assay (IP-One HTRF, Cisbio, Codolet, France) according to the manufacturer’s protocol and in analogy to previously described protocols (77, 78). In brief, HEK 293T cells were cotransfected with the cDNA for a receptor and the hybrid G protein Gαqi (Gαq protein with the last five amino acids at the C terminus replaced by the corresponding sequence of Gαi) (gift from The J. David Gladstone Institutes, San Francisco, CA), respectively, in a ratio of 1:2. After 1 day, cells were transferred into 384-well microplates (Greiner) and incubated for further 24 hours. On the day of the experiment, cells were incubated with test compounds for 90 min (α2AAR) or 120 min (α2BAR), and accumulation of second messenger was stopped by adding detection reagents (IP1-d2 conjugate and Anti-IP1cryptate TB conjugate). After 60 min, TR-FRET was monitored with a Clariostar plate reader. FRET-signals were normalized to buffer (0%) and the maximum effect of norepinephrine (100%). Three to nine (murine α2AAR, α2BAR) or 4 to 11 repeats (human α2AAR), respectively, were performed for each test compound all done in duplicate.
PathHunter arrestin recruitment assay
Investigation of α2AAR and α2BAR stimulated β-arrestin-2 recruitment was performed applying an assay which is based on fragment complementation of β-galactosidase (PathHunter assay, DiscoverX, Birmingham, UK) as described (79). In detail, HEK293T cells stably expressing the enzyme acceptor (EA) tagged β-arrestin-2 were cotransfected with human α2AAR or α2BAR each fused to the ProLink-ARMS2-PKS2 fragment for enzyme complementation and GRK2 (cDNA Resource Center) at equal amounts and subsequently transferred into 384 well microplates (Greiner) after 1 day. After incubation for further 24 hours cells were incubated with test compounds for 60 min (α2AAR) or 90 min (α2BAR), arrestin recruitment was stopped by adding detection regent and the resulting chemoluminescence was monitored with a Clariostar plate microreader. Data were normalized relative to buffer (0%) and the maximum effect of norepinephrine (100%). Three to nine repeats for α2AAR (three to six for α2BAR) in duplicate were measured.
DiscoverX HitHunter cAMP G protein activation assay
Dexmedetomidine, brimonidine, ‘9087, and ‘7075 were tested by DiscoverX (catalog item 86-0007P-2270AG; Eurofins; CA, USA) in their HitHunter XS+ assay. Freezer stock cAMP Hunter cell lines were expanded, then seeded in a total volume of 20 μL into white walled, 384-well microplates and incubated at 37°C before testing. For agonist determination, cells were incubated with compound samples in the presence of EC80 forskolin to induce response. Media was aspirated from cells and replaced with 15 μL 2:1 HBSS/10 mM Hepes: cAMP XS+ Ab reagent. Intermediate dilution of sample stocks was performed to generate 4X sample in assay buffer containing 4X EC80 forskolin. 5 μL of 4X sample was added to cells and incubated at 37°C or room temperature for 30 to 60 min. Finally assay vehicle concentration was 1%. After sample incubation assay signal was generated through incubation with 20 μL cAMP XS+ ED/CL lysis cocktail for one hour followed by incubation with 20 μL cAMP XS+ EA reagent for three hours at room temperature. Microplates were read after signal generation with a PerkinElmer Envision Instrument for chemiluminescent signal detection. Compound activity was analyzed using CBIS data analysis suite. For Gi agonist mode, percentage activity is calculated using the following formula: percentage activity = 100% × [1 − (mean RLU of test sample − mean RLU of MAX control) / (mean RLU of vehicle control − mean RLU of MAX control)]. Brimonidine was used as the control agonist. Each measurement was done in duplicate.
EMTA coupling panel for α2AAR
The ebBRET-based effector membrane translocation assay (EMTA) allows detection of each Ga protein subunit activation. Upon receptor activation, G protein–effector proteins fused at their C terminus to Renilla luciferase (RlucII) translocate from cytoplasm to the plasma membrane to selectively bind activated Gα proteins (p63-RhoGEF-RlucII with Gq/11 family, Rap1-GAP-RlucII with Gi/o family and PDZ-RhoGEF-RlucII with G12/13 family), thus leading to an increase in ebBRET by becoming in close proximity to the plasma membrane targeted energy acceptor, Renilla green fluorescent protein (rGFP-CAAX). The heterologous coexpression of each Gα subunits allow to identify which specific members of each G protein families (i.e.: Gi1, Gi2, Gi3, GoA, GoB, Gz, Gq, G11, G14, G15, G12, and G13) is activated by a receptor. The assay is also sensitive enough to detect responses elicited by endogenous Gi/o protein families in the absence of heterologously expressed G protein. The same plasma membrane translocation principle is used to measure β-arrestin-1 or -2 recruitment (39) using β-arrestin–RlucII/rGFP-CAAX biosensors.
Cell culture
HEK293 clonal cell line (HEK293SL cells), hereafter referred to as HEK293 cells, were a gift from S. Laporte (McGill University, Montreal, Quebec, Canada) and previously described (39). Cells were cultured in DMEM medium (Wisent; St-Jean-Baptiste, QC, Canada) supplemented with 10% newborn calf serum iron fortified (NCS) (Wisent). Cells were passaged weekly and incubated at 37°C in a humidified atmosphere with 5% CO2 and checked for mycoplasma contamination.
Transfection
HEK293 cells (1.2 mL at 3.5 × 105 cells per mL) were transfected with a fixed final amount of premixed biosensor-encoding DNA (0.57 μg, adjusted with salmon sperm DNA; Invitrogen) and human α2AAR DNA for Gs, Gi/o, Gq/11 and β-arrestins experiments. For G12/13 experiments, cells were transfected with 1 μg of total DNA (adjusted with salmon sperm DNA; Invitrogen), including empty pCDNA3.1 vector or human α2AAR DNA. Transfections were performed using linear polyethylenimine (PEI, 1 mg/mL; Polysciences) diluted in NaCl (150 mM, pH 7.0) as a transfecting agent (3:1 PEI/DNA ratio). Cells were immediately seeded (3.5 × 104 cells per well) into 96-well white microplates (PerkinElmer), maintained in culture for the next 48 hours and BRET experiments carried out. ebBRET (38) was used to monitor the activation of each Gα protein, as well as β-arrestin-1 and -2 recruitment to the plasma membrane. Gαs protein engagement was measured between the plasma membrane marker rGFP-CAAX and human Gαs-RlucII in presence of human Gβ1, Gγ9, and α2AAR. Gα12 or Gα13 protein family activation was assessed using the selective-G12/13 effector PDZ-RhoGEF-RlucII and rGFP-CAAX coexpressed with Gβ1, Gγ1, and either Gα12 or Gα13, in presence of α2AAR. Gαi/o protein family activation was followed using the selective-Gi/o effector Rap1GAP-RlucII and rGFP-CAAX along with the human Gαi1, Gαi2, GαoA, GαoB, or Gαz subunits and α2AAR. Gαq/11 protein family activation was determined using the selective-Gq/11 effector p63-RhoGEF-RlucII and rGFP-CAAX along with the human Gαq, Gα11, Gα14, or Gα15 subunits and α2AAR. β-arrestin recruitment to the plasma membrane was determined using DNA mix containing rGFP-CAAX and β-arrestin-1–RlucII or β-arrestin-2–RlucII in presence of α2AAR.
Bioluminescence resonance energy transfer measurement
The day of the experiment, cells were washed with phosphate-buffered saline (PBS) and incubated in Tyrode Hepes buffer (137 mM NaCl, 0.9 mM KCl, 1 mM MgCl2, 11.9 mM NaHCO3, 3.6 mM NaH2PO4, 25 mM HEPES, 5.5 mM D-Glucose and 1 mM CaCl2, pH 7.4) for 1 hour at 37°C. Cells were then treated with increasing concentrations of compounds for 10 min at 37°C. The luciferase substrate Prolume purple (1 μM, NanoLight Technologies) was added during the last 6 min before the reading. Plates were read on the TriStar2 LB 942 Multimode Microplate Reader (Berthold Technologies) with the energy donor filter (410 ± 80 nm; RlucII) and energy acceptor filter (515 ± 40 nm; GFP10 and rGFP CAAX). BRET signal (BRET2) was determined by calculating the ratio of the light intensity emitted by the acceptor (515 nm) over the light intensity emitted by the donor (410 nm) and data were normalized in percentage of the maximal response elicited by the reference compound norepinephrine. The data were analyzed in GraphPad Prism 9.1 using “dose–response-stimulation log(agonist) versus response (four parameters)” and data were presented as means ± SEMs of at least three different experiments each done in simplicate. and values were determined from dose-response curves to calculate the value for each pathway and each compound. To determine the relative efficacy of the compounds to activate the different signaling pathways, the difference between the values was calculated using the following equation
| (1) |
The compounds’ efficacy toward each pathway, relative to norepinephrine, were calculated as the inverse logarithm of the using the following equation
| (2) |
The SEM was calculated for the ratios using the following equation
| (3) |
where is the standard deviation, and is the number of experiments.
The SEM was calculated for the ratios using the following equation
| (4) |
Statistical analysis was performed using a two-tailed unpaired t test on the ratios to make pairwise comparisons between tested compounds and norepinephrine for a given pathway, where P < 0.05 was considered statistically significant.
Internalization assay with rGFP-CAAX and rGFP-FYVE biosensors
Plasmids
Human α2AAR sequence was fused to RlucII by cloning between the NheI and BamHI sites of pCDNA3.1/Zeo(+)-RlucII vector, using PCR (Q5 Hot Start High-Fidelity DNA Polymerase from NEB), enzymatic digestion (NEB) and ligation (Anza T4 DNA Ligase Master Mix; Invitrogen).
Transfection
The protocol used for transfection is the same as for G12/13 EMTA experiments [i.e., cells were transfected with 1 μg of total DNA (adjusted with salmon sperm DNA; Invitrogen)]. Transfections were performed using linear polyethylenimine (PEI, 1 mg/mL; Polysciences) diluted in NaCl (150 mM, pH 7.0) as a transfecting agent (3:1 PEI/DNA ratio). Cells were immediately seeded (3.5 × 104 cells per well) into 96-well white microplates (PerkinElmer), maintained in culture for the next 48 hours and BRET experiments carried out. Human α2AAR internalization was evaluated by measuring the disappearance of hα2AAR-RlucII from the plasma membrane labeled with rGFP-CAAX and its relocalization in endosome labeled with rGFP-FYVE (39).
Bioluminescence resonance energy transfer measurement
The day of the experiment, cells were washed with phosphate-buffered saline (PBS) and incubated in Tyrode Hepes buffer for 1 hour at 37°C. Cells were incubated during 6 min with the luciferase substrate Prolume purple (1 μM, NanoLight Technologies) before addition of the indicated compound (0 or 100 μM) and kinetics were recorded during 30 min. For concentration-response curves, BRET signal was measured after 30 min incubation. Plates were read on a Spark microplate reader (Tecan; Mannedorf, Switzerland) using the BRET2 manufacturer settings. BRET signal (BRET2) was determined by calculating the ratio of the light intensity emitted by the acceptor (515 nm) over the light intensity emitted by the donor (410 nm) and for concentration-response curves, data were normalized in percentage of the maximal response elicited by the reference compound norepinephrine. The data were analyzed in GraphPad Prism 9.1 using “log(agonist) versus response–Variable slope (four parameters)” and data were presented as means ± SEMs of three experiments performed in triplicate for kinetics or in simplicate for concentration-response curves.
Cryo-EM sample preparation and structure determination
Preparation of the ‘9087-α2AAR-GoA-scFv16 and ‘4622-α2AAR-GoA-scFv16 complexes
The human WT α2AAR was cloned to pFastBac vector with a N-terminal FLAG tag and a C-terminal histidine Tag. This construct was expressed in Sf9 insect cells using the pfastBac baculovirus system (Expression Systems). Cells were infected at a density of 4 × 106 cells per ml and then incubated for 48 hours at 27°C. Receptor was extracted and purified following the protocol described previously for α2BAR (9). Briefly, receptor was purified by Ni-NTA chromatography, Flag affinity chromatography and size exclusion chromatography in the presence of 100 μM ‘9087 or ‘4622. The monomeric peak fractions of receptor were collected and concentrated to ~20 mg/mL. The freshly purified ‘9087-bound or ‘4622-bound α2AAR was used for complex formation with the G protein. GoA heterotrimers were expressed and purified as previously described with minor modifications (78). Briefly, Hi5 cells were grown to a density of 3 million per mL and then infected with Gαo and Gβ1γ2 baculovirus at a ratio of 10 to 20 mL/L and 1 to 2 mL/L, respectively, and then incubated for 48 hours at 27°C. Cells were solubilized with 1% (w/v) sodium cholate and 0.05% (w/v) DDM. After centrifugation, the supernatant was loaded onto Ni-NTA column and then exchanged to 0.05% DDM. The eluted GoA heterotrimer was dephosphorylated by lambda phosphatase (homemade) and further purified through ion exchange using a Mono Q 10/100 GL column (GE Healthcare) and the peak fractions were collected and flash frozen in liquid nitrogen until use. The scFv16 (80) protein was expressed in insect Sf9 cells and purified with Ni-NTA column followed by the Superdex 200 Increase 10/300 GL column (GE Healthcare) with a buffer composed of 20 mM HEPEs, pH 7.5 and 100 mM NaCl. The monomeric peak fractions of receptor were collected and concentrated and stored at −80°C until use. The complex formation process is same as described. Briefly, the complex of α2AAR with heterotrimeric GoA was formed in a buffer containing 20 mM HEPEs pH 7.5, 100 mM NaCl, 0.1% DDM, 1 mM MgCl2, 10 mM GDP, and 100 mM ‘9087 or ‘4622. The α2AAR-GoA complex was then treated with 50 units of apyrase (NEB) on ice overnight, and exchanged on an anti-Flag M1 column into a buffer containing 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.0075% lauryl maltose neopentyl glycol (MNG, NG310 Anatrace), 0.0025% GDN (GDN101, Anatrace), and 0.001% CHS, 100 μM ‘9087 or ‘4622, and 2 mM CaCl2 in a stepwise manner. After elution by adding 5 mM EDTA and 0.2 mg/mL Flag peptide, the complex was concentrated and incubated with 1.5x molar excess scFv16 for 1 hour on ice, then further purified using Superdex 200 Increase 10/300 GL column (GE Healthcare) with a running buffer of 20 mM HEPES, pH 7.5, 100 mM NaCl, 0.00075% MNG, 0.00025% GDN and 0.0001% CHS, 100 μM ‘9087 or ‘4622. The monomeric peak fraction of α2AAR-GoA complex was collected and concentrated to ~5 mg/mL for cryo-EM.
Cryo-EM data collection, processing, and model building
3 μL of purified complex sample was applied onto the grid (CryoMatrix nickel titanium alloy film, R1.2/1.3, Zhenjiang Lehua Electronic Technology Co., Ltd.) (81) glow discharged at Tergeo-EM plasma cleaner and then blotted for 3 s with blotting force of 0 and quickly plunged into liquid ethane cooled by liquid nitrogen using Vitrobot Mark IV (Thermo Fisher Scientific, USA) at 10°C and with 95% humidity. Cryo-EM data were collected on a 300 kV Titan Krios Gi3 microscope. The raw movies were recorded by Gatan K3 BioQuantum Camera at the magnification of 105,000 and the corresponding pixel size is 0.85 Å. Inelastically scattered electrons were excluded by a GIF Quantum energy filter (Gatan, USA) using a slit width of 20 eV. The movie stacks were acquired with the defocus range of −1.0 to −1.6 μm with total exposure time 2.5 s fragmented into 50 frames (0.05 s per frame) with the dose rate of 22.0 e per pixel per second. The imaging mode is super resolution with 2-time hardware binning. The semiautomatic data acquisition was performed using SerialEM (82).
For the ‘9087-α2AAR-GoA-scFv16 complex, raw movie frames were aligned with MotionCor2 (83) using a 9 × 7 patch and the contrast transfer function (CTF) parameters were estimated using Gctf and ctf in JSPR (84). Micrographs with consistent CTF values including defocus and astigmatism parameter were kept for the following image processing, which kept 3768 micrographs from 4217 raw movies. Templates for particle auto-picking were generated by projecting the 3D volume of norepinephrine-bound α2AAR-GoA complex (40). The 2,137,146 particles picked from template picking was subjected 2D classification in cryoSPARC (85) and 3D classication in Relion3.1 (86). The sorted 321,762 particles were then subjected to homogeneous reconstruction in cryoSPARC, yielding a 3.57-Å map. Further 3D ab initio reconstruction reduced the particles number to 287,431, which was subjected to CTF refinement and nonuniform refinement after extracting with larger particle box size, and finally yield the 3.47-Å map.
For the ‘4622-α2AAR-GoA-scFv16 complex, 6983 raw movies were collected and subjected for motion correction using MotionCor2 (83). Contrast transfer function parameters were estimated by CTFFIND4, implemented in Relion3.1 (86). 2,593,747 particles were auto-picked using the templates in RELION3.1 and then subjected to 2D classification using cryoSPARC. Selected particles with appropriate 2D average from 2D classification were further subjected to ab initio reconstruction. Particles with appropriate initial model were selected from ab initio followed by heterogeneous refinement in cryoSPARC. The particles kept to 563,506 particles were subjected to nonuniform refinement and local refinement and yield a 3.38-Å reconstruction determined by gold standard Fourier shell correlation using the 0.143 criterion.
The norepinephrine-α2AAR-GoA complex structure (PDB 7EJ0) (40) was used as the initial template for model building. The model was docked into the cryo-EM density map using UCSF Chimera (https://rbvi.ucsf.edu/chimera), followed by iterative manual building in Coot (87) and real space refinement in Phenix. The statistics of the final models were validated by Molprobity. Cryo-EM data collection, refinement, and validation statistics are summarized in table S11. The ligand symmetry accounted RMSDs between the docked pose and cryo-EM pose of ‘9087 and ‘4622 were calculated by the Hungarian algorithm in DOCK6 (88).
pKa determination for ‘9087
The pKa of ‘9087 (2.90 mg, 0.013 mmol) was determined by potentiometric titration using a Metrohm pH Meter 632 equipped with a glass electrode (Metrohm 6.0259.100). The compound was dissolved in 15 mL of 10% methanol aqueous solution, at an ionic strength of I = 0.15 M using KCl. The resulting solution was stirred throughout the experiment using a magnetic stir bar and a magnetic agitator. The compound was titrated with 0.01 M HCl (Titrisol) using an automatic burette (Metrohm Dosimat Plus 876). The titrant was added to the analyte stepwise (0.024 to 2.87 mL). The resulting graph for pKa determination is presented in dependence of and . The pKa value was then determined using a simplified Henderson-Hasselbalch equation. The data from the titration experiment were evaluated with Origin 9.60.
Off-target activity
GPCRome
10 μM ‘9087 was tested for off-target activity at a panel of 320 nonolfactory GPCRs using PRESTO-Tango GPCRome arrestin-recruitment assay as described (45). Receptors with at least threefold increased relative luminescence over corresponding basal activity are potential positive hits. Screening was performed by the National Institutes of Mental Health Psychoactive Drug Screen Program (PDSP). Detailed experimental protocols are available on the NIMH PDSP website at https://pdsp.unc.edu/pdspweb/content/PDSP%20Protocols%20II%202013-03-28.pdf.
D2R activation
D2R was selected after the GPCRome panel, and ‘9087 was retested for full dose-response to determine G protein and arrestin recruitment (see above).
I2R binding
Top docking compounds (‘9087, ‘2998, ‘4622, and ‘0172) were tested for I2R binding, performed by Eurofins Cerep (France; catalog no. 81) as described (78). For compound ‘2998, no binding was seen in a single point radioligand competition experiment tested at 500 nM and the compound is not shown.
μOR competition binding
Equilibrium [3H] Diprenorphine competition and saturation binding were carried out in membranes prepared from Chinese Hamster Ovary (CHO-K1) cells stably expressing human μ-opioid receptor, as previously described (89-91). Briefly, binding was performed at 25°C for 90 min in the dark. Binding in μOR/CHO-K1 cells was carried out in a buffer consisting of 50 mM HEPES-base pH 7.4 (pH adjusted with KOH), 10 mM MgCl2, 0.1 mM EDTA, and 0.1% (w/v) bovine serum albumin with membranes containing ~40 μg/mL protein. After incubation with radioligand (1 pM to 10 nM for saturation, 500 pM for competition), drugs (33 μM to 3.3 pM) and/or 20 μM cold competitor naloxone, the reaction was rapidly filtered onto GF/B (PerkinElmer no. 1450-521) glass fiber filtermats which were equilibrated for 1 hour in binding buffer supplemented with 0.3% (v/v) polyethyleneimine. The filtermats were washed five times in ice-cold 50 mM HEPES-base pH 7.4 using a PerkinElmer semiautomated cell harvester (PerkinElmer FilterMate Harvester). The filtermats were dried and Meltilex solid scintillant (PerkinElmer no. 1450-442) was melted onto the mats for 10 min at 60°C. The scintillant was allowed to resolidify before disintegrations were quantified with a Wallac MicroBeta Scintillation counter using an integration time of 1 min. Nonspecific binding, total binding, the number of receptor binding sites, and the Kd of the radiotracer were determined from saturation binding experiments. Protein concentrations were determined using the microBCA method with BSA as the standard. Ki values were calculated by nonlinear regression analysis and application of the Cheng-Prusoff correction in GraphPad Prism 9.0.
hERG inhibition assays
‘9087 was tested for hERG inhibition in the FluxOR assay as described (92). hERG experiments used the National Institutes of Mental Health (NIMH) Psychoactive Drug Screening Program (PDSP). Experimental protocols are available on the NIMH PDSP website at https://pdsp.unc.edu/pdspweb/content/PDSP%20Protocols%20II%202013-03-28.pdf.
Metabolic stability studies
Metabolic stability of the test compounds was assessed using male pooled rat liver microsomes (Sprague Dawley, Sigma Aldrich) as previously described (93, 94). The reactions were carried out in 2 mL polyethylene tubes on a rotator carousel (Stuart SB3) in an incubator at 37°C. The incubation mixture contained ‘9087, ‘7075, PS75, or the positive controls rotigotine or imipramine (final concentration 20 μM), and pooled rat liver microsomes (0.25 mg protein per tube) in Tris-MgCl2 buffer (50 mM Tris, 5 mM MgCl2, pH 7.4, final volume 500 μL). Transformation reactions were initiated by the addition of 50 μL of cofactor solution (NADPH, Carl Roth, final concentration 1 mM). After time intervals of 0, 30, and 60 min for ‘9087, ‘7075, and PS75 or 0, 15, 30, and 60 min for rotigotine and imipramine, respectively, 100 μL aliquots of the reaction mixtures were added to 100 μL ice-cold acetonitrile (containing 10 μM chlorpromazine as internal standard) to terminate enzymatic reactions. Precipitated protein was removed by centrifugation (1 min, 16,000 relative centrifugal force) and the supernatants were analyzed by high-performance liquid chromatography–mass spectrometry (HPLC/MS) on a Thermo Scientific Dionex Ultimate 3000 HPLC system equipped with a Zorbax Eclipse XDB-C8 column (4.6 × 150 mm, 5 βm), a DAD detector (210 nm, 230 nm, 254 nm, 310 nm), and a BRUKER amaZon SL mass spectrometer with ESI source. The following binary eluent system (methanol in water + 0.1% (v/v) formic acid) was used: 10% for 1 min, 10 to 100% in 20 min, 100% for 5 min, 100 to 10% in 2 min, 10% for 2 min, flow 0.4 mL/min. Per compound, four (rotigotine, imipramine) or five (‘9087, ‘7075, PS75) independent experiments were performed. Control experiments were conducted in the absence of cofactor solution to determine non-specific binding to matrix. The integral (AUC) of the extracted ion chromatograms (EIC) was used to analyze the concentration of the remaining substrates. Concentrations were plotted in their logarithmic form as a function of the incubation time (in minutes) to calculate the elimination rate constant () and to determine the half-life () and intrinsic hepatic clearance () for each compound with the following equations (95)
| (5) |
| (6) |
In vivo methods
Animals and ethical compliance
Animal experiments were approved by the UCSF Institutional Animal Care and Use Committee and were conducted in accordance with the NIH Guide for the Care and Use of Laboratory animals (protocol no. AN181214). Adult (8 to 10 weeks old) male C56BL/6 mice (strain no. 664) were purchased from the Jackson Laboratory. Mice were housed in cages on a standard 12:12 hour light-dark cycle with food and water ad libitum. The α2AAR D79N mutant mice were purchased from Jackson (stock no. 2777), and 7- to 8-week-old females were used. Sample sizes were modeled on our previous studies and on studies using a similar approach, which were able to detect significant changes (96, 97). The animals were randomly assigned to treatment and control groups. Animals were initially placed into one cage and allowed to freely run for a few minutes. Then each animal was randomly picked up, injected with compound treatment or vehicle, and placed into a separate cylinder before the behavioral test.
In vivo compound preparation
All ligands were synthesized by Enamine (‘2998 and ‘7075) or WuXi (‘9087, ‘4622, and ‘0172) or in-house (PS75) and dissolved 30 min before testing. Available salt forms were used to aid solubility: HCl for ‘9087 and ‘7075, TFA for ‘4622, ‘0172, and PS75. ‘9087, ‘4622, and ‘0172 were resuspended in 20% Kolliphor (Sigma-Aldrich; cat. no. C5135) for s.c. and i.p. injections. ‘2998, ‘7075, and PS75 were resuspended in 20% cyclodextran (Sigma-Aldrich; cat. no. H107) for s.c. and i.p. injections. Atipamezole (Cayman Chemical Company; cat. no. 9001181) and Dexmedetomidine (Cayman Chemical Company; cat. no. 15581) were resuspended with NaCl 0.9% (Teknova; cat. no. S5819) for s.c. and i.p. injections. ‘9087 was formulated with 40% Captisol (Carbosynth; cat. no. OC15979) for p.o. dosing.
Behavioral analyses
For all behavioral tests, the experimenter was always blind to treatment. Animals were first habituated for 1 hour in Plexiglas cylinders and then tested 30 min after s.c. injection of the α2AAR compounds. The α2AAR antagonist atipamezole (2 mg/kg, i.p.) was injected 15 min before s.c. injection of the α2AAR agonists. The mechanical (von Frey), thermal (Hargreaves, hot plate, and tail flick) and ambulatory (rotarod) tests were conducted as described previously (98). Hindpaw mechanical thresholds were determined with von Frey filaments using the up-down method(99). Hindpaw thermal sensitivity was measured with a radiant heat source (Hargreaves) or a 55°C hot plate. For the tail flick assay, sensitivity was measured by immersing the tail into a 50°C water bath for both WT and D79N mutant mice. For the ambulatory (rotarod) test, mice were first trained on an accelerating rotating rod, three times for 5 min, before testing with any compound.
SNI model of neuropathic pain
Under isoflurane anesthesia, two of the three branches of the sciatic nerve were ligated and transected distally, leaving the sural nerve intact. Behavior was tested 7 to 14 days after injury.
CFA
The CFA model of chronic inflammation was induced as described previously (100). Briefly, CFA (Sigma) was diluted 1:1 with saline and vortexed for 30 min. When fully suspended, we injected 20 μL of CFA into one hindpaw. Heat thresholds were measured before the injection (baseline) and 3 days after using the Hargreaves test.
Constipation assay
Mice had access to food and water ad libitum before the test. On the test day, mice received an i.p. injection of a solution (100 μL) containing saline, 10 mg/kg morphine, 30 μg/kg dexmedetomidine, or 5 mg/kg ‘9087 and then individually placed in a clean cage, with no access to food or water. Fecal pellets were collected and counted every hour, up to 6 hours.
Body weight measurement
The body weights were measured before, 24 hours after, and 48 hours after mice received an i.p. injection of a solution (100 μL) containing dexmedetomidine (30 μg/μg) or ‘9087 (5, 10, or 20 mg/kg).
Pharmacokinetics
Pharmacokinetic experiments were performed by Bienta (Enamine Biology Services) in accordance with Enamine pharmacokinetic study protocols and Institutional Animal Care and Use Guidelines (protocol number 1-2/2020). Plasma pharmacokinetics and brain distribution for ‘9087, ‘2998, ‘4622, ‘7075, PS75, and CSF distribution for ‘7075, PS75, ‘9087, and ‘4622, were measured after a 10-mg/kg (i.p.) dose. Plasma and brain samples were also collected for ‘9087 after a 10-mg/kg i.v. and 30-mg/kg p.o. (oral) dose to determine oral bioavailability. In each compound study, nine time points (5, 15, 30, 60, 120, 240, 360, 480, and 1440 min) were collected, each of the time point treatment group included three animals. There was also a control group of one animal. In the ‘9087, ‘7075, and ‘4622 studies, male C57BL/6N mice were used, for PS75 CD-1 mice, and for ‘2998 male Balb/cAnN mice. For all compound studies the animals were randomly assigned to the treatment groups before the pharmacokinetic study; all animals were fasted for 4 hours before dosing. For injections, ‘9087 was dissolved in Captisol – water (40%:60%, w/v), ‘4622 was dissolved in a 20% Kolliphor HS – physiological saline solution, and ‘7075, PS75, and ‘2998 were dissolved in a 20% 2-HPBCD–aqueous solution. The batches of working formulations were prepared 10 min before the in vivo study.
Mice were injected i.p. with 2,2,2- tribromoethanol at 150 mg/kg before drawing CSF and blood. CSF was collected under a stereomicroscope from cisterna magna using 1-mL syringes. Blood collection was performed from the orbital sinus in microtainers containing K3EDTA Animals were euthanized by cervical dislocation after the blood samples collection. Blood samples were centrifuged 10 min at 3000 rpm. Brain samples (right lobe) were weighed and transferred into 1.5 mL tubes. All samples were immediately processed, flash-frozen and stored at −70°C until subsequent analysis.
Plasma samples (40 μL) were mixed with 200 μL of internal standard (IS) solution. After mixing by pipetting and centrifuging for 4 min at 6000 rpm, 4 μL of each supernatant was injected into the liquid chromatography–tandem mass spectrometry (LC-MS/MS) system. Solutions of internal standards were used to quantify compounds in the plasma samples. Brain samples (weight 200 ± 1 mg) were homogenized with 800 μL of an internal stock solution using zirconium oxide beads (115 ± 5 mg) in a Bullet Blender homogenizer for 30 s at speed 8. After this, the samples were centrifuged for 4 min at 14,000 rpm, and supernatant was injected into LC-MS/MS system. CSF samples (2 μL) were mixed with 40 μL of an internal stock solution. After mixing by pipetting and centrifuging for 4 min at 6000 rpm, 5 μL of each supernatant was injected into LC-MS/MS system.
Analyses of plasma, brain, and CSF samples were conducted at Enamine/Bienta. The concentrations of compounds in plasma and brain samples were determined using HPLC-MS/MS. Data acquisition and system control were performed using Analyst 1.5.2 software (AB Sciex, Canada). The concentrations of the test compound below the lower limit of quantitation (LLOQ = 10 ng/mL for plasma, 20 ng/g for brain, and 5 ng/mL for CSF samples) were designated as zero. Pharmacokinetic data analysis was performed using noncompartmental, bolus injection or extravascular input analysis models in WinNonlin 5.2 (PharSight). Data below LLOQ were presented as missing to improve validity of T1/2 calculations.
Additional pharmacokinetic experiments were performed by Sai Life Sciences (Hyderabad, India) in accordance with the Sai Study Protocol SAIDMPK/PK-22-04-0340. Brain distribution of dexmedetomidine was measured after a 30-μg/kg i.p. dose, using normal saline 0.9% as its vehicle. Plasma distributions were also collected for PS75 after 10-mg/kg i.v. and 30-mg/kg p.o. (oral) dosing to determine oral bioavailability; both doses were formulated in 20% v/v HβCD in saline. Testing was done in C57BL/6 mice. For PS75, 24 mice were divided into four groups: nine mice for i.v. dosing of the compound, three mice for i.v. dosing of the vehicle, nine mice for p.o. dosing with the compound, and three mice for p.o. vehicle dosing; sparse sampling of three mice per time point for compound-treated groups and one mouse per time point for vehicle groups was performed. For dexmedetomidine, 36 mice were included and split into two groups: three mice per time point for compound dosing, and one mouse per time point for vehicle-only dosing. For PS75, blood samples (60 μL) were collected under light isoflurane anesthesia (Surgivet) from retro orbital plexus from a set of three mice at 0.083, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 hours. Immediately after blood collection, plasma was harvested by centrifugation at 4000 rpm, 10 min at 4°C. For dexmedetomidine, brain samples were collected at the same time points indicated above. Animals were euthanized at respective time points and brain samples were isolated and homogenized in ice-cold phosphate buffer saline (pH 7.4). Total homogenate volume was three times the tissue weight. Samples were stored at −70°C until bioanalysis. All samples were processed for analysis by protein precipitation method and analyzed with fit-for-purpose LC-MS/MS method (LLOQ = 3.61 ng/mL for plasma for PS75, LLOQ =0.86 ng/mL for brain for dexmedetomidine). The pharmacokinetic parameters were estimated using the noncompartmental analysis tool of Phoenix WinNonlin software (version 8.0).
Statistical analyses
Data from functional experiments of adrenergic and D2long receptors were analyzed applying the algorithms for four-parameter nonlinear regression implemented in Prism 8.0 (GraphPad, San Diego, CA) to get dose-response curves representing and values. Mean values were derived by summarizing the results from each individual experiment to provide ± SEM and ± SEM (or SD where indicated). Additional statistical analyses for Fig. 4; fig. S4; fig. S16, A and C to E; and fig. S18 were performed with GraphPad Prism 9.0 (GraphPad Software Inc., San Diego). Data reported are means ± SEMs or, in Fig. 4 and fig. S18, single data points with means ± SEMs. Experiments of the compounds in the in vivo neuropathic, inflammatory, hot plate, tail flick, and rotarod models were evaluated using unpaired two-tailed Student’s t test or one-way analysis of variance (ANOVA) with Dunnett’s multiple comparison post hoc test to determine differences between groups. Experiments for body weight and constipation were analyzed with a two-way ANOVA. Details of the analyses, including groups compared in statistical sets, number of animals per group, and P values, can be found in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. Kobilka for sharing the structure of PDB 6K41 before publication. We thank T. Tummino and J. Lyu for reading this manuscript. We also thank WuXi AppTec for providing information on compounds sourced from the WuXi GalaXi virtual library. We gratefully acknowledge OpenEye Software for Omega and related tools and Schrödinger, Inc. for the Maestro package.
Funding:
This work is supported by DARPA grant HR0011-19-2-0020 (B.K.S., A.I.B., J.J.I., and M.P.J.), DFG grant GRK 1910 (P.G.), US NIH grant R35GM122481 (B.K.S.), US NIH grant R01GM133836 (J.J.I.), US R35 NS097306 (A.I.B.), Open Philanthropy (A.I.B.), the Facial Pain Research Foundation (A.I.B.), and CIHR Foundation grant FN-148431 (M.B.). C.M.W. is supported by an NSF Graduate Research Fellowship. Y.D. is supported by a grant from the Science, Technology and Innovation Commission of Shenzhen Municipality (project JCYJ20200109150019113) and in part by the Kobilka Institute of Innovative Drug Discovery and Shenzhen Key Lab (ZDSYS20190902093417963). X.-P.H. is supported by NIMH PDSP (HHSN-271-2018-00023-C), directed by B. Roth. M.B. holds the Canada Research Chair in Signal Transduction and Molecular Pharmacology.
Footnotes
Competing interests: B.K.S. and P.G. are founders of Epiodyne. B.K.S. is a founder of BlueDolphin and of Deep Apple Therapeutics and consults in docking and in the GPCR space.J.J.I. is a cofounder of BlueDolphin and Deep Apple Therapeutics. M.P.J. is a consultant to Deep Apple Therapeutics and to Schrödinger, Inc. M.B. is the chair of the scientific advisory board of Domain Therapeutics, to which some of the BRET-based biosensors used are licensed for commercial use. Y.S.M. is a CEO of Chemspace LLC and a scientific advisor at Enamine, Ltd. B.K.S., E.A.F., P.G., H.H., P.S., A.I.B., J.M.B., Y.D., and J.X. are authors of patents on the discovery of new pain modulators acting through the α2AAR. The authors declare no other competing interests.
Data and materials availability: All data are available in the main text, the supplementary materials, the listed Protein Data Bank (PDB) files, the Electron Microscopy Data Bank (EMDB) files, or at https://github.com/efink14/ADRA2AR_docking_results. The 3D cryo-EM density maps of 9087-α2AAR-GoA and ‘4622-α2AAR-GoA generated in this study have been deposited with accession codes EMD-32331 and EMD-32342, respectively. The coordinates of ‘9087-α2AAR-GoA and ‘4622-α2AAR-GoA have been deposited with PDB accession codes 7W6P and 7W7E, respectively. The identities of compounds docked in this study are freely available from the ZINC15 and ZINC20 databases https://zinc15.docking.org/ and https://zinc20.docking.org/), and active compounds may be purchased from Enamine and WuXi AppTec or are available from the authors. The docking results, including ZINC number, SMILES, and docking score, are located at https://github.com/efink14/ADRA2AR_docking_results. DOCK3.7 is freely available for noncommercial research (https://dock.compbio.ucsf.edu/DOCK3.7/). A web-based version is freely available to all (https://blaster.docking.org/). The biosensors used for generating the data in tables S3 and S4 and figures S6 to S8 are protected by a patent but are available from M.B. for noncommercial research without restrictions under a regular academic Material Transfer Agreement with the Université de Montréal.
SUPPLEMENTARY MATERIALS
REFERENCES AND NOTES
- 1.Yekkirala AS, Roberson DP, Bean BP, Woolf CJ, Breaking barriers to novel analgesic drug development. Nat. Rev. Drug Discov 16, 545–564 (2017). doi: 10.1038/nrd.2017.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manchikanti L et al. , Opioid epidemic in the United States. Pain Physician 15, ES9–ES38 (2012). doi: 10.36076/ppj.2012/15/ES9 [DOI] [PubMed] [Google Scholar]
- 3.Strang J et al. , Opioid use disorder. Nat. Rev. Dis. Primers 6, 3 (2020). doi: 10.1038/s41572-019-0137-5 [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen K, White DA, Acri JB, NIDA’s medication development priorities in response to the Opioid Crisis: Ten most wanted. Neuropsychopharmacology 44, 657–659 (2019). doi: 10.1038/s41386-018-0292-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malmberg AB, Hedley LR, Jasper JR, Hunter JC, Basbaum AI, Contribution of α2 receptor subtypes to nerve injury-induced pain and its regulation by dexmedetomidine. Br. J. Pharmacol 132, 1827–1836 (2001). doi: 10.1038/sj.bjp.0704032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lakhlani PP et al. , Substitution of a mutant α2a-adrenergic receptor via “hit and run” gene targeting reveals the role of this subtype in sedative, analgesic, and anesthetic-sparing responses in vivo. Proc. Natl. Acad. Sci. U.S.A 94, 9950–9955 (1997). doi: 10.1073/pnas.94.18.9950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buerkle H, Yaksh TL, Pharmacological evidence for different alpha2-adrenergic receptor sites mediating analgesia and sedation in the rat. Br J. Anaesth 81, 208–215 (1998). doi: 10.1093/bja/81.2.208 [DOI] [PubMed] [Google Scholar]
- 8.Anttila M, Penttilä J, Helminen A, Vuorilehto L, Scheinin H, Bioavailability of dexmedetomidine after extravascular doses in healthy subjects. Br J. Clin. Pharmacol 56, 691–693 (2003). doi: 10.1046/j.1365-2125.2003.01944.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan D et al. , Activation of the α2B adrenoceptor by the sedative sympatholytic dexmedetomidine. Nat. Chem. Biol 16, 507–512 (2020). doi: 10.1038/s41589-020-0492-2 [DOI] [PubMed] [Google Scholar]
- 10.Lyu J et al. , Ultra-large library docking for discovering new chemotypes. Nature 566, 224–229 (2019). doi: 10.1038/s41586-019-0917-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sterling T, Irwin JJ, ZINC 15 – Ligand discovery for everyone. J. Chem. Inf. Model 55, 2324–2337 (2015). doi: 10.1021/acs.jcim.5b00559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Irwin JJ et al. , ZINC20—A free ultralarge-scale chemical database for ligand discovery. J. Chem. Inf. Model 60, 6065–6073 (2020). doi: 10.1021/acs.jcim.0c00675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein RM et al. , Virtual discovery of melatonin receptor ligands to modulate circadian rhythms. Nature 579, 609–614 (2020). doi: 10.1038/s41586-020-2027-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alon A et al. , Structures of the receptor enable docking for bioactive ligand discovery. Nature 600, 759–764 (2021). doi: 10.1038/s41586-021-04175-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gorgulla C et al. , An open-source drug discovery platform enables ultra-large virtual screens. Nature 580, 663–668 (2020). doi: 10.1038/s41586-020-2117-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadybekov AA et al. , Synthon-based ligand discovery in virtual libraries of over 11 billion compounds. Nature 601, 452–459 (2022). doi: 10.1038/s41586-021-04220-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manglik A et al. , Structure-based discovery of opioid analgesics with reduced side effects. Nature 537, 185–190 (2016). doi: 10.1038/nature19112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuller M et al. , Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking. Sci. Adv 7, eabf8711 (2021). doi: 10.1126/sciadv.abf8711; [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rudling A et al. , Fragment-based discovery and optimization of enzyme inhibitors by docking of commercial chemical space. J. Med. Chem 60, 8160–8169 (2017). doi: 10.1021/acs.jmedchem.7b01006 [DOI] [PubMed] [Google Scholar]
- 20.Levit Kaplan A et al. , Structure-based design of a chemical probe set for the 5-HT5A Serotonin Receptor. J. Med. Chem 65, 4201–4217 (2022). doi: 10.1021/acs.jmedchem.1c02031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Graaf C et al. , Crystal structure-based virtual screening for fragment-like ligands of the human histamine H1 receptor. J. Med. Chem 54, 8195–8206 (2011). doi: 10.1021/jm2011589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss R et al. , Discovery of novel human histamine H4 receptor ligands by large-scale structure-based virtual screening. J. Med. Chem 51, 3145–3153 (2008). doi: 10.1021/jm7014777 [DOI] [PubMed] [Google Scholar]
- 23.Patel N et al. , Structure-based discovery of potent and selective melatonin receptor agonists. eLife 9, e53779 (2020). doi: 10.7554/eLife.53779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead CJ et al. , Identification of novel adenosine A2A receptor antagonists by virtual screening. J. Med. Chem 55, 1904–1909 (2012). doi: 10.1021/jm201455y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S et al. , D4 dopamine receptor high-resolution structures enable the discovery of selective agonists. Science 358, 381–386 (2017). doi: 10.1126/science.aan5468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coleman RG, Carchia M, Sterling T, Irwin JJ, Shoichet BK, Ligand pose and orientational sampling in molecular docking. PLOS ONE 8, e75992 (2013). doi: 10.1371/journal.pone.0075992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altosaar K et al. , Adrenoceptors (version 2019.4) in the IUPHAR/BPS Guide to Pharmacology Database (IUPHAR/BPS Guide to Pharmacology, 2019); doi: 10.2218/gtopdb/F4/2019.4 [DOI] [Google Scholar]
- 28.Gaulton A et al. , ChEMBL: A large-scale bioactivity database for drug discovery. Nucleic Acids Res. 40, D1100–D1107 (2012). doi: 10.1093/nar/gkr777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blaxall HS, Murphy TJ, Baker JC, Ray C, Bylund DB, Characterization of the Alpha-2C adrenergic receptor subtype in the opossum kidney and in the OK cell line. J. Pharmacol. Exp. Ther 259, 323–329 (1991). [PubMed] [Google Scholar]
- 30.Timmermans PBMWM, van Zwieten PA, α2 adrenoceptors: Classification, localization, mechanisms, and targets for drugs. J. Med. Chem 25, 1389–1401 (1982). doi: 10.1021/jm00354a001 [DOI] [PubMed] [Google Scholar]
- 31.Clineschmidt BV, Flataker LM, Faison EP, Haubrich DR, alpha-methyldopa reduces locomotor activity in rats via its metabolite, alpha-methylnorepinephrine, acting on alpha 2-adrenoceptors. Arch. Int. Pharmacodyn. Ther 244, 231–243 (1980). [PubMed] [Google Scholar]
- 32.Gu S, Smith MS, Yang Y, Irwin JJ, Shoichet BK, Ligand strain energy in large library docking. J. Chem. Inf. Model 61, 4331–4341 (2021). doi: 10.1021/acs.jcim.1c00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Isberg V et al. , Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol. Sci 36, 22–31 (2015). doi: 10.1016/j.tips.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Congreve M et al. , Discovery of 1,2,4-triazine derivatives as adenosine A2A antagonists using structure based drug design. J. Med. Chem 55, 1898–1903 (2012). doi: 10.1021/jm201376w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bhattacharya S, Hall SE, Li H, Vaidehi N, Ligand-stabilized conformational states of human β2 adrenergic receptor: Insight into G-protein-coupled receptor activation. Biophys. J 94, 2027–2042 (2008). doi: 10.1529/biophysj.107.117648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weiss DR et al. , Conformation guides molecular efficacy in docking screens of activated β-2 adrenergic G protein coupled receptor. ACS Chem. Biol 8, 1018–1026 (2013). doi: 10.1021/cb400103f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eason MG, Kurose H, Holt BD, Raymond JR, Liggett SB, Simultaneous coupling of α2-adrenergic receptors to two G-proteins with opposing effects: Subtype-selective coupling of α2C10, α2C4, and α2C2 adrenergic receptors to Gi and Gs. J. Biol. Chem 267, 15795–15801 (1992). doi: 10.1016/S0021-9258(19)49605-1 [DOI] [PubMed] [Google Scholar]
- 38.Avet C et al. , Effector membrane translocation biosensors reveal G protein and βarrestin coupling profiles of 100 therapeutically relevant GPCRs. eLife 11, e74101 (2022). doi: 10.7554/eLife.74101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Namkung Y et al. , Monitoring G protein-coupled receptor and β-arrestin trafficking in live cells using enhanced bystander BRET. Nat. Commun 7, 12178 (2016). doi: 10.1038/ncomms12178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xu J et al. , Structural insights into ligand recognition, activation, and signaling of the α2A adrenergic receptor. Sci. Adv 8, eabj5347 (2022). doi: 10.1126/sciadv.abj5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qu L et al. , Structural basis of the diversity of adrenergic receptors. Cell Rep. 29, 2929–2935.e4 (2019). doi: 10.1016/j.celrep.2019.10.088 [DOI] [PubMed] [Google Scholar]
- 42.Heydenreich FM et al. , Dissecting the allosteric networks governing agonist efficacy and potency in G protein-coupled receptors. bioRxiv2021.09.14.460253 [Preprint] (2021). doi: 10.1101/2021.09.14.460253 [DOI] [Google Scholar]
- 43.Masureel M et al. , Structural insights into binding specificity, efficacy and bias of a β2AR partial agonist. Nat. Chem. Biol 14, 1059–1066 (2018). doi: 10.1038/s41589-018-0145-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stanek M et al. , Hybridization of β-Adrenergic agonists and antagonists confers G protein bias. J. Med. Chem 62, 5111–5131 (2019). doi: 10.1021/acs.jmedchem.9b00349 [DOI] [PubMed] [Google Scholar]
- 45.Kroeze WK et al. , PRESTO-Tango as an open-source resource for interrogation of the druggable human GPCRome. Nat. Struct. Mol. Biol 22, 362–369 (2015). doi: 10.1038/nsmb.3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kodaira H et al. , Quantitative evaluation of the impact of active efflux by p-glycoprotein and breast cancer resistance protein at the blood-brain barrier on the predictability of the unbound concentrations of drugs in the brain using cerebrospinal fluid concentration as a surrogate. J. Pharmacol. Exp. Ther 339, 935–944 (2011). doi: 10.1124/jpet.111.180398 [DOI] [PubMed] [Google Scholar]
- 47.Shields SD, Eckert WA 3rd, Basbaum AI, Spared nerve injury model of neuropathic pain in the mouse: A behavioral and anatomic analysis. J. Pain 4, 465–470 (2003). doi: 10.1067/S1526-5900(03)00781-8 [DOI] [PubMed] [Google Scholar]
- 48.Virtanen R, Savola JM, Saano V, Highly selective and specific antagonism of central and peripheral alpha 2-adrenoceptors by atipamezole. Arch. Int. Pharmacodyn. Ther 297, 190–204 (1989). [PubMed] [Google Scholar]
- 49.MacMillan LB, Hein L, Smith MS, Piascik MT, Limbird LE, Central hypotensive effects of the α2a-adrenergic receptor subtype. Science 273, 801–803 (1996). doi: 10.1126/science.273.5276.801 [DOI] [PubMed] [Google Scholar]
- 50.Lakhlani PP, Lovinger DM, Limbird LE, Genetic evidence for involvement of multiple effector systems in alpha 2A-adrenergic receptor inhibition of stimulus-secretion coupling. Mol. Pharmacol 50, 96–103 (1996). [PubMed] [Google Scholar]
- 51.Ceresa BP, Limbird LE, Mutation of an aspartate residue highly conserved among G-protein-coupled receptors results in nonreciprocal disruption of α2-adrenergic receptor-G-protein interactions: A negative charge at amino acid residue 79 forecasts α2A-adrenergic receptor sensitivity to allosteric modulation by monovalent cations and fully effective receptor/G-protein coupling. J. Biol. Chem 269, 29557–29564 (1994). doi: 10.1016/S0021-9258(18)43916-6 [DOI] [PubMed] [Google Scholar]
- 52.Hunter JC et al. , Assessment of the role of α2-adrenoceptor subtypes in the antinociceptive, sedative and hypothermic action of dexmedetomidine in transgenic mice. Br. J. Pharmacol 122, 1339–1344 (1997). doi: 10.1038/sj.bjp.0701520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Metz SA, Halter JB, Robertson RP, Induction of defective insulin secretion and impaired glucose tolerance by clonidine. Selective stimulation of metabolic alpha-adrenergic pathways. Diabetes 27, 554–562 (1978). doi: 10.2337/diab.27.5.554 [DOI] [PubMed] [Google Scholar]
- 54.Lichtenstein SS, Marinescu C, Leibowitz SF, Chronic infusion of norepinephrine and clonidine into the hypothalamic paraventricular nucleus. Brain Res. Bull 13, 591–595 (1984). doi: 10.1016/0361-9230(84)90042-X [DOI] [PubMed] [Google Scholar]
- 55.Stone LS, German JP, Kitto KF, Fairbanks CA, Wilcox GL, Morphine and clonidine combination therapy improves therapeutic window in mice: Synergy in antinociceptive but not in sedative or cardiovascular effects. PLOS ONE 9, e109903 (2014). doi: 10.1371/journal.pone.0109903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Erlanson DA, Fesik SW, Hubbard RE, Jahnke W, Jhoti H, Twenty years on: The impact of fragments on drug discovery. Nat. Rev. Drug Discov 15, 605–619 (2016). doi: 10.1038/nrd.2016.109 [DOI] [PubMed] [Google Scholar]
- 57.Von Moo E et al. , Ligand-directed bias of G protein signaling at the dopamine D2 receptor. Cell Chem. Biol 29, 226–238.e4 (2022). doi: 10.1016/j.chembiol.2021.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE, A geometric approach to macromolecule-ligand interactions. J. Mol. Biol 161, 269–288 (1982). doi: 10.1016/0022-2836(82)90153-X [DOI] [PubMed] [Google Scholar]
- 59.Word JM, Lovell SC, Richardson JS, Richardson DC, Asparagine and glutamine: Using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol 285, 1735–1747 (1999). doi: 10.1006/jmbi.1998.2401 [DOI] [PubMed] [Google Scholar]
- 60.Weiner SJ et al. , A new force field for molecular mechanical simulation of nucleic acids and proteins. J. Am. Chem. Soc 106, 765–784 (1984). doi: 10.1021/ja00315a051 [DOI] [Google Scholar]
- 61.Bender BJ et al. , A practical guide to large-scale docking. Nat Protoc. 16, 4799–4832 (2021). doi: 10.1038/s41596-021-00597-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein RM et al. , Property-unmatched decoys in docking benchmarks. J. Chem. Inf. Model 61, 699–714 (2021). doi: 10.1021/acs.jcim.0c00598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mysinger MM et al. , Structure-based ligand discovery for the protein-protein interface of chemokine receptor CXCR4. Proc. Natl. Acad. Sci. U.S.A 109, 5517–5522 (2012). doi: 10.1073/pnas.1120431109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shoichet BK, Leach AR, Kuntz ID, Ligand solvation in molecular docking. Proteins 34, 4–16 (1999). doi: [DOI] [PubMed] [Google Scholar]
- 65.Gallagher K, Sharp K, Electrostatic contributions to heat capacity changes of DNA-ligand binding. Biophys. J 75, 769–776 (1998). doi: 10.1016/S0006-3495(98)77566-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mysinger MM, Shoichet BK, Rapid context-dependent ligand desolvation in molecular docking. J. Chem. Inf. Model 50, 1561–1573 (2010). doi: 10.1021/ci100214a [DOI] [PubMed] [Google Scholar]
- 67.Rezai T et al. , Conformational flexibility, internal hydrogen bonding, and passive membrane permeability: Successful in silico prediction of the relative permeabilities of cyclic peptides. J. Am. Chem. Soc 128, 14073–14080 (2006). doi: 10.1021/ja063076p [DOI] [PubMed] [Google Scholar]
- 68.Leung SSF, Mijalkovic J, Borrelli K, Jacobson MP, Testing physical models of passive membrane permeation. J. Chem. Inf. Model 52, 1621–1636 (2012). doi: 10.1021/ci200583t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jacobson MP, Kaminski GA, Friesner RA, Rapp CS, Force field validation using protein side chain prediction. J. Phys. Chem. B 106, 11673–11680 (2002). doi: 10.1021/jp021564n [DOI] [Google Scholar]
- 70.Gibson DG et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345 (2009). doi: 10.1038/nmeth.1318 [DOI] [PubMed] [Google Scholar]
- 71.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ, Protein measurement with the Folin phenol reagent. J. Biol. Chem 193, 265–275 (1951). doi: 10.1016/S0021-9258(19)52451-6 [DOI] [PubMed] [Google Scholar]
- 72.Cheng Y-C, Prusoff WH, Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem. Pharmacol 22, 3099–3108 (1973). doi: 10.1016/0006-2952(73)90196-2 [DOI] [PubMed] [Google Scholar]
- 73.Guan XM, Kobilka TS, Kobilka BK, Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J. Biol. Chem 267, 21995–21998 (1992). doi: 10.1016/S0021-9258(18)41623-7 [DOI] [PubMed] [Google Scholar]
- 74.Quoyer J et al. , Pepducin targeting the C-X-C chemokine receptor type 4 acts as a biased agonist favoring activation of the inhibitory G protein. Proc. Natl. Acad. Sci. U.S.A 110, E5088–E5097 (2013). doi: 10.1073/pnas.1312515110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galés C et al. , Probing the activation-promoted structural rearrangements in preassembled receptor-G protein complexes. Nat. Struct. Mol. Biol 13, 778–786 (2006). doi: 10.1038/nsmb1134 [DOI] [PubMed] [Google Scholar]
- 76.Möller D et al. , Discovery of G protein-biased dopaminergics with a pyrazolo[1,5-a]pyridine substructure. J. Med. Chem 60, 2908–2929 (2017). doi: 10.1021/acs.jmedchem.6b01857 [DOI] [PubMed] [Google Scholar]
- 77.Liu H et al. , Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists. Proc. Natl. Acad. Sci. U.S.A 115, 12046–12050 (2018). doi: 10.1073/pnas.1813988115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu J et al. , Conformational complexity and dynamics in a muscarinic receptor revealed by NMR spectroscopy. Mol. Cell 75, 53–65.e7 (2019). doi: 10.1016/j.molcel.2019.04.028 [DOI] [PubMed] [Google Scholar]
- 79.Liu X et al. , An allosteric modulator binds to a conformational hub in the β2 adrenergic receptor. Nat. Chem. Biol 16, 749–755 (2020). doi: 10.1038/s41589-020-0549-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maeda S et al. , Development of an antibody fragment that stabilizes GPCR/G-protein complexes. Nat. Commun 9, 3712 (2018). doi: 10.1038/s41467-018-06002-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang X et al. , Amorphous nickel titanium alloy film: A new choice for cryo electron microscopy sample preparation. Prog. Biophys. Mol. Biol 156, 3–13 (2020). doi: 10.1016/j.pbiomolbio.2020.07.009 [DOI] [PubMed] [Google Scholar]
- 82.Mastronarde DN, Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol 152, 36–51 (2005). doi: 10.1016/j.jsb.2005.07.007 [DOI] [PubMed] [Google Scholar]
- 83.Zheng SQ et al. , MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017). doi: 10.1038/nmeth.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jiang W, Guo F, Liu Z, A graph theory method for determination of cryo-EM image focuses. J. Struct. Biol 180, 343–351 (2012). doi: 10.1016/j.jsb.2012.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA, cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017). doi: 10.1038/nmeth.4169 [DOI] [PubMed] [Google Scholar]
- 86.Scheres SHW, RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol 180, 519–530 (2012). doi: 10.1016/j.jsb.2012.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Emsley P, Lohkamp B, Scott WG, Cowtan K, Features and development of Coot. Acta Cryst D66, 486–501 (2010). doi: 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Allen WJ, Rizzo RC, Implementation of the Hungarian algorithm to account for ligand symmetry and similarity in structure-based design. J. Chem. Inf. Model 54, 518–529 (2014). doi: 10.1021/ci400534h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Flanagan CA, in Methods in Cell Biology, Shukla AK, Ed. (Academic Press, 2016), pp. 191–215. [DOI] [PubMed] [Google Scholar]
- 90.DeHaven RN, DeHaven-Hudkins DL, Characterization of opioid receptors. Curr. Protocols Pharmacol 8, 1.4.1–1.4.12 (2000). doi: 10.1002/0471141755.ph0104s08 [DOI] [Google Scholar]
- 91.Hulme EC, Trevethick MA, Ligand binding assays at equilibrium: Validation and interpretation. Br. J. Pharmacol 161, 1219–1237 (2010). doi: 10.1111/j.1476-5381.2009.00604.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang X-P, Mangano T, Hufeisen S, Setola V, Roth BL, Identification of human Ether-à-go-go related gene modulators by three screening platforms in an academic drug-discovery setting. Assay Drug Dev Technol. 8, 727–742 (2010). doi: 10.1089/adt.2010.0331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hiller C et al. , Functionally selective dopamine D2/D3 receptor agonists comprising an enyne moiety. J. Med. Chem 56, 5130–5141 (2013). doi: 10.1021/jm400520c [DOI] [PubMed] [Google Scholar]
- 94.Hellmann J et al. , Structure-based development of a subtype-selective orexin 1 receptor antagonist. Proc. Natl. Acad. Sci U.S.A 117, 18059–18067 (2020). doi: 10.1073/pnas.2002704117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Di L et al. , Mechanistic insights from comparing intrinsic clearance values between human liver microsomes and hepatocytes to guide drug design. Eur. J. Med. Chem 57, 441–448 (2012). doi: 10.1016/j.ejmech.2012.06.043 [DOI] [PubMed] [Google Scholar]
- 96.Scherrer G et al. , Dissociation of the opioid receptor mechanisms that control mechanical and heat pain. Cell 137, 1148–1159 (2009). doi: 10.1016/j.cell.2009.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muralidharan A et al. , Identification and characterization of novel candidate compounds targeting 6- and 7-transmembrane μ-opioid receptor isoforms. Br. J. Pharmacol 178, 2709–2726 (2021). doi: 10.1111/bph.15463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Alvaro CG et al. , Hippocalcin-like 4, a neural calcium sensor, has a limited contribution to pain and itch processing. PLOS ONE 15, e0226289 (2020). doi: 10.1371/journal.pone.0226289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chaplan SR, Bach FW, Pogrel JW, Chung JM, Yaksh TL, Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994). doi: 10.1016/0165-0270(94)90144-9 [DOI] [PubMed] [Google Scholar]
- 100.Cao YQ et al. , Primary afferent tachykinins are required to experience moderate to intense pain. Nature 392, 390–394 (1998). doi: 10.1038/32897 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.