Abstract
A database search revealed extensive sequence similarity between Streptomyces lividans plasmid pIJ101 and Streptomyces plasmid pSB24.2, which is a deletion derivative of Streptomyces cyanogenus plasmid pSB24.1. The high degree of relatedness between the two plasmids allowed the construction of a genetic map of pSB24.2, consisting of putative transfer and replication loci. Two pSB24.2 loci, namely, the cis-acting locus for transfer (clt) and the transfer-associated korB gene, were shown to be capable of complementing the pIJ101 clt and korB functions, respectively, a result that is consistent with the notion that pIJ101 and the parental plasmid pSB24.1 encode highly similar, if not identical, conjugation systems.
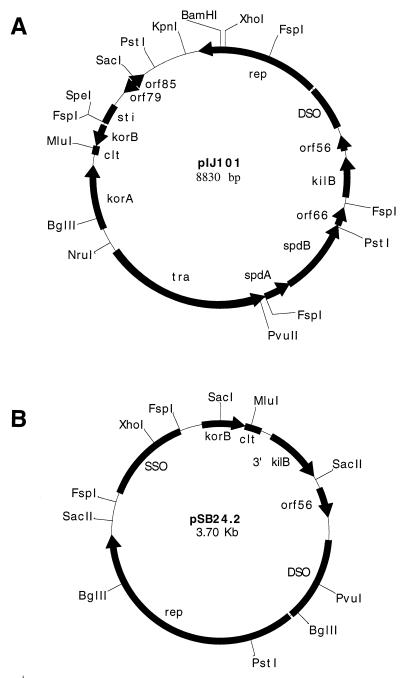
The conjugative plasmid pIJ101 is one of the most extensively studied of the Streptomyces extrachromosomal elements. Originally purified from Streptomyces lividans ISP5434 (14), pIJ101 (Fig. 1A) is an 8,830-bp (11), circular, high-copy-number plasmid which possesses a broad host range among actinomycete bacteria (14). To produce copies of itself, pIJ101 uses rolling-circle replication (RCR) (6), a process that involves the 456-amino-acid Rep protein encoded by the rep gene of pIJ101 (11), a double-strand origin (DSO) of replication where Rep-mediated single-stranded nicking and initiation of first-strand synthesis occurs, and a likely single-strand origin (SSO) termed sti, where second-strand synthesis is normally initiated (4). Certain motifs present within its Rep protein, along with conserved sequences present at the site of Rep nicking, have allowed for the classification of pIJ101 as a member of the pIJ101-pJV1 family of RCR plasmids (20), which is comprised mostly of Streptomyces replicons (13). The DSO of pIJ101, which has been mapped (3) to a 517-bp region that includes the likely nicking site (20) as well as other sequences required for plasmid replication and copy number control (3), is located between the rep gene and orf56, a small open reading frame (ORF) of undetermined function (11). The sti locus of pIJ101 (4) includes sequences that may form an extensive secondary structure and may contain consensus SSO hexanucleotide base positions (28).
FIG. 1.
Physical and genetic maps of pIJ101 (A) and pSB24.2 (B). Filled arrows represent ORFs which were determined from the published pIJ101 (11) or pSB24.2 (2) sequence or from the pSB24.2 sequence reported here. Genetic functions were determined for either pIJ101 genes (12, 14) or loci (solid bars with no arrows) (4, 17) as described previously. Small pIJ101 ORFs of undetermined genetic function (i.e., orf56, orf66, orf79, and orf85) are indicated. Certain pSB24.2 genetic functions (i.e., korB and clt) were determined here; other genetic functions (i.e., DSO and SSO) or small ORFs of unknown function (i.e., orf56) were assigned based on their similarity to corresponding pIJ101 sequences or in the case of the rep gene on the similarity of the deduced Rep protein to Rep proteins of other Streptomyces plasmids (9, 13).
The efficient transfer of pIJ101 is dependent on both the tra gene (12, 14), which encodes a trans-acting membrane protein with an as yet undetermined function (16), and a cis-acting transfer locus (clt), which appears to represent a site or sequence that participates in some unknown manner in the transfer event (17). The clt locus was originally subcloned on a 142-bp fragment of pIJ101 that spans the 3′ end of the korB gene, although sequences essential for clt function may be present only downstream of the korB ORF (17). Although its exact sequence determinants remain to be elucidated, clt does not appear to resemble cis-acting transfer sequences found on other bacterial plasmids (27), including the only other Streptomyces plasmid clt function so far identified (19).
The korB gene encodes a repressor protein that binds within its own promoter region as well as that of kilB (21, 22), a gene that is involved in, although not absolutely required for, pIJ101 transfer (12). The ability of the KorB protein to repress or override kilB is essential, since the unregulated expression of the kilB gene on pIJ101 is lethal in Streptomyces (12). Like kilB, the genes spdA and spdB contribute to plasmid transfer in an as yet undetermined ancillary manner (11, 12), while similar to korB control, the pIJ101 korA gene codes for a repressor (21) that appears to negatively regulate the expression of both korA and a transcript that includes tra, spdA, and spdB (8).
pSB24.1 is a conjugative plasmid isolated from Streptomyces cyanogenus (2), a species notable for its production of the antitumor agent landomycin A (26). Upon introduction of pSB24.1 into S. lividans, a stable, transfer-defective, deletion derivative designated pSB24.2 forms (Fig. 1B), which has a reported size of 3,706 bp (2). The deduced Rep protein of pSB24.2 (9) and potential site of Rep-mediated nicking (5) show homology to those of the other members of the pIJ101-pJV1 family of RCR plasmids (13). Here we show that a database search for sequences showing homology to the pIJ101 clt region (and thus plasmids that may share the same transfer mechanism as pIJ101) subsequently revealed extensive similarity between both transfer and replication loci of pIJ101 and corresponding previously unidentified loci on pSB24.2. We further show that the clt and korB functions encoded by pSB24.2 can complement their pIJ101 counterparts. These results support the hypothesis that pIJ101 and pSB24.1 share the same conjugation system and that pIJ101-like conjugative plasmids may be dispersed throughout at least a portion of the Streptomyces genus.
pIJ101 and pSB24.2 show extensive sequence similarity.
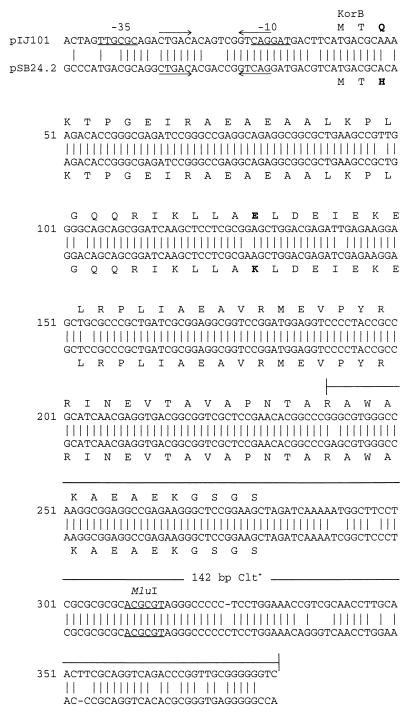
By using the BLASTN algorithm (1) and the 142-bp clt+ region of plasmid pIJ101 (Fig. 2) as a query sequence, a database search revealed only one other highly similar sequence, which belonged to Streptomyces plasmid pSB24.2. Subsequent BLASTN alignment of the entire pIJ101 and pSB24.2 sequences revealed extensive similarity between them (i.e., approximately 77% of the pSB24.2 sequence aligned to pIJ101 with 89% identity and 1% gaps by this method) and allowed the identification of putative transfer and replication loci on pSB24.2 (Fig. 1B) as detailed below. To resolve discrepancies arising through probable errors, both strands of the korB-clt region of pSB24.2 were sequenced by using relevant primers and an ABI Prism dye terminator cycle sequencing ready reaction kit (PE Applied Biosystems, Inc., Foster City, Calif.), followed by analysis of reactions on a model 310 genetic analyzer (PE Applied Biosystems, Inc.).
FIG. 2.
Alignment of the korB-clt regions of plasmids pIJ101 and pSB24.2. The revised korB-clt sequence of pSB24.2 is shown, aligned with the previously described korB-clt region of pIJ101 (11). One-letter amino acid designations are shown above (for pIJ101) or below (for pSB24.2) the corresponding korB ORF. Amino acid differences between the pIJ101 KorB protein and the deduced KorB protein of pSB24.2 are in bold. Putative −35 and −10 promoter determinants for the pIJ101 korB gene, which are based on previous transcriptional mapping (22), are indicated. Inverted repeats on both sequences are designated by arrows. The 142-bp region of pIJ101 that is known to contain clt activity (17) and that was used as a query sequence here for database searches is indicated.
The alignment of the revised korB-clt sequence of pSB24.2 with the corresponding region of pIJ101 revealed several features (Fig. 2). The 240-bp korB ORFs of the two plasmids are 97.5% identical, and the putative 80-amino-acid pSB24.2 KorB protein is identical to the pIJ101 KorB protein (11), with the exception of a histidine residue in place of a glutamine at amino acid position 3 and a lysine residue instead of a glutamic acid at position 30. The latter amino acid position is within the possible DNA binding domain of the mature pIJ101 KorB repressor protein (11, 24), although not at one of the most conserved positions of this motif (15). A comparison of the promoter region of the pIJ101 korB gene (which was identified following the demonstration that transcription initiates at the A residue of the start codon) (22) with the region immediately upstream of the pSB24.2 korB gene revealed that pSB24.2 contains an identical potential −10 but not a completely conserved possible −35 promoter determinant. Also within this region of pSB24.2 are inverted repeat sequences that are identical to those found in the pIJ101 korB promoter which were shown previously to be important for the autoregulation of korB gene expression (25). Downstream of the korB ORFs on both plasmids, there is complete conservation of sequences in the immediate vicinity of the MluI restriction site, a location at which the insertion of DNA into pIJ101 was previously shown to eliminate clt function (17).
The alignment of the two plasmids’ sequences also revealed that pSB24.2 contains an additional transfer-related sequence: in this case, a partial copy of a putative kilB ORF (Fig. 1B). The partial kilB ORF of pSB24.2 aligns with the 147-codon kilB ORF of pIJ101 (11) beginning one base prior to codon 42, extends to the end of the ORF, and is 95 and 75% identical at the respective nucleotide and deduced amino acid levels to the corresponding portion of the pIJ101 kilB gene (data not shown).
While sequences related to pIJ101 and other Streptomyces plasmids were previously identified for both the deduced Rep protein of pSB24.2 (9, 13), as well as for its putative site of single-stranded nicking (5), our analysis here identifies additional potential replication determinants (i.e., DSO and SSO sequences) present within the published pSB24.2 nucleotide sequence (2). The 480-bp region immediately 5′ to the pSB24.2 rep gene (designated DSO in Fig. 1B) aligned with 79% identity and 7% gaps (data not shown) to the corresponding 517-bp region of pIJ101 shown previously to contain all essential cis-acting sequences necessary for plasmid replication and copy number control (3). Included within this region of pSB24.2 is a stretch of 224 bp that is 100% identical to pIJ101 and includes the putative nicking site at the end proximal to the rep gene. The extended similarity between the pIJ101 DSO and the corresponding region of pSB24.2 may indicate that essential replication sequences, besides the site of Rep-mediated nicking, have remained conserved between the two plasmids.
Between its putative DSO region and partial kilB gene, pSB24.2 also contains a copy of orf56 (Fig. 1B) that is 99% identical at the nucleotide level and 96% identical at the deduced amino acid level (data not shown) to orf56 of pIJ101 (11). The significance, however, of this sequence conservation for orf56, an ORF that has been shown to be dispensable for pIJ101 replication (29) and copy number regulation (3), remains unknown.
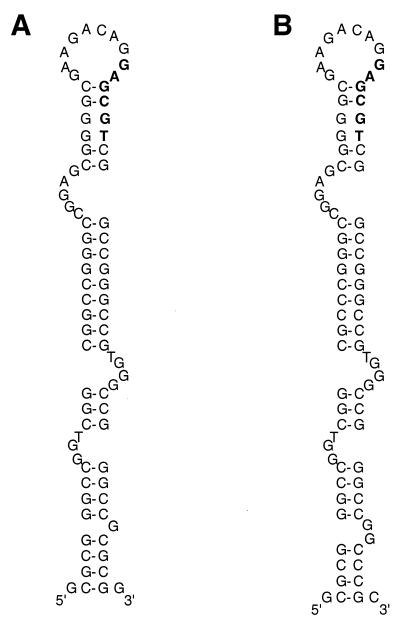
While DSO regions within families of RCR plasmids display some sequence similarity, SSO sequences are typically not well conserved, although they generally show the potential for an extensive secondary structure (13). In pIJ101, the SSO locus has been termed sti and appears to confer a strong incompatibility phenotype on pIJ101, such that Sti− plasmids cannot coexist with Sti+ plasmids (4). The sti locus (Fig. 1A) has been mapped to a point within roughly 200 bp (4) upstream of the korB promoter (22). A 485-bp region of pSB24.2 located upstream of its korB gene was found to align with 90% identity and 1% gaps (data not shown) to a 491-bp region of pIJ101 that contains the pIJ101 sti locus and extends from the korB promoter to a point within the small overlapping ORFs of unknown function (11), orf79 and orf85 (Fig. 1A). Within this region of pIJ101 exists a previously identified (28) potential stem-and-loop motif that contains five of six bases of the consensus hexanucleotide sequence 5′ TAGCGT 3′ (Fig. 3A), which is present in many SSOs (13). A nearly identical motif including the same hexanucleotide sequence was also found within the corresponding region of pSB24.2 (Fig. 3B). Because of the close similarity between the sequences located upstream of korB on the two plasmids, we have tentatively identified this region of pSB24.2 as the SSO (Fig. 1B); however, the exact determinants required for the efficient conversion of single-strand plasmid intermediates into double-stranded molecules as well as their potential to elicit strong incompatibility effects similar to the pIJ101 sti locus remain to be determined.
FIG. 3.
Potential stem-and-loop motifs within the SSO (i.e., sti) region of plasmid pIJ101 (A) and the SSO region of pSB24.2 (B). The pIJ101 structure was previously identified (28), while the pSB24.2 structure corresponds to base positions 3330 to 3405 of the published sequence (2). Hexanucleotide sequences that show similarity to the consensus sequence 5′ TAGCGT 3′ found in many SSO determinants (13) are indicated in bold.
The 1,311-bp rep gene (2) of pSB24.2 (Fig. 1B) and the 1,356-bp rep gene (11) of pIJ101 (Fig. 1A) were also very similar (43% identity at the nucleotide level and 68% identity at the amino acid level) (data not shown); however, as noted previously (10), the pIJ101 rep gene possesses striking homology (i.e., 92 and 95% identity at the nucleotide and deduced amino acid levels, respectively) to the rep gene of another well-studied Streptomyces conjugative plasmid, namely, pSN22 from Streptomyces nigrifaciens. This high degree of relatedness between the rep genes of pIJ101 and pSN22 is interesting, given that in all other respects pSN22 appears to be less related to pIJ101 than does the pSB24.2 plasmid; pSN22, for example, shows overall less similarity to the DSO region of pIJ101 than does pSB24.2 (data not shown), may have SSO determinants very different from those of pIJ101 (23), and also is unrelated to pIJ101 in its transfer region (10).
Possible physical derivation of pSB24.2 from the parental plasmid pSB24.1.
The genetic organization and nucleotide sequence of plasmid pSB24.2 thus strongly resemble the corresponding region of pIJ101 that extends from the clt locus clockwise to 3′ sequences within the kilB gene (Fig. 1A). It therefore seems reasonable to speculate that the parental plasmid pSB24.1 may be organized similarly to pIJ101 throughout and that upon passage through S. lividans, pSB24.1, for reasons unknown, undergoes a spontaneous deletion of sequences (including its transfer-essential tra gene) that correspond to the portion of pIJ101 extending from within kilB clockwise to just beyond the korA gene (Fig. 1A); such a deletion would thereby result in the formation of the transfer-defective pSB24.2 derivative (Fig. 1B).
pSB24.2 encodes a functional clt locus that complements the pIJ101 clt locus.
Our interest in conjugation in Streptomyces prompted us to examine whether pSB24.2 encodes a functional clt locus, and if so, whether this pSB24.2 locus can complement the clt function of the closely related pIJ101 plasmid. Such complementation could potentially allow for the subsequent identification of conserved functional sequences that are important for clt activity. To test these possibilities, we used a previously described mating system (16, 17) involving an S. lividans donor strain (TK23.42), which contains chromosomally integrated copies of the pIJ101 tra and korA genes, and an S. lividans recipient strain (TK23) containing pHYG1 (12), a transfer-defective, hygromycin-resistant pIJ101 derivative. TK23.42 was transformed with pSB24.202 (Table 1), a thiostrepton-resistant derivative of pSB24.2, and plasmid-containing spores of this strain were mixed with TK23(pHYG1) recipient spores, plated on nonselective MY (malt extract-yeast extract) agar (17), and then quantified following mating for donor, recipient, and transconjugant cell types as described previously (17).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| S. lividans | ||
| TK23 | spc-1 | 7 |
| TK23.42 | TK23 tra and korA genes of pIJ101 chromosomally integrated | 17 |
| Escherichia coli DH10B | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara leu)7697 galU galK λ− rpsL nupG | Gibco BRL |
| Plasmids | ||
| pGSP149 | pIJ350 derivative with pSP72 at the PstI site | 17 |
| pGSP263 | pIJ101 Clt+ derivative of pGSP149 | 17 |
| pGSP290 | KilB+ derivative of pSP72 containing the 1.0-kb PstI-BalI region of pIJ101 cloned as a blunt-end fragment in the EcoRV site (kilB is transcribed towards the T7 promoter) | This study |
| pGSP312 | pSP72 derivative containing the 142-bp Clt+ region of pSB24.2 cloned as a BamHI fragment in the BamHI site | This study |
| pGSP313 | pIJ350 derivative containing pGSP312 at the PstI site | This study |
| pGSP317 | Derivative of pHYG1 with pGSP290 at the PstI site (pIJ101 rep gene is transcribed towards the SP6 promoter) | This study |
| pGSP321 | KorB+ derivative of pGSP317 containing the 0.3-kb SpeI-MluI region of pIJ101 cloned as a blunt-end fragment in the PvuII site (korB is transcribed towards the SP6 promoter) | This study |
| pGSP322 | KorB+ derivative of pGSP317 containing the 0.4-kb FspI-MluI region of pSB24.2 cloned as a blunt-end fragment in the PvuII site (korB is transcribed towards the SP6 promoter) | This study |
| pHYG1 | Conjugally deficient Hygr derivative of pIJ101 lacking all transfer functions | 12 |
| pIJ101 | Conjugative S. lividans plasmid | 14 |
| pIJ350 | Conjugally deficient Tsrr derivative of pIJ101 lacking all transfer functions | 14 |
| pSB24.2 | Conjugally deficient Clt+ KorB+ deletion derivative of S. cyanogenus plasmid pSB24.1 | 2; obtained from the Russian National Collection of Industrial Microorganisms |
| pSB24.202 | Tsrr derivative of pSB24.2 with the 1.0-kb tsr gene (7) cloned as an XhoI fragment at the XhoI site (tsr gene transcribed away from korB) | This study |
| pSP72 | AprE. coli cloning vector | Promega Biotech |
Hygr, Tsrr, and Apr indicate hygromycin, thiostrepton, and ampicillin resistance, respectively.
As shown in Table 2, pSB24.202 transferred at a frequency (28%) that was nearly identical to that seen for a Clt+ pIJ101 derivative (i.e., pGSP263 at 29%) and that was 35-fold higher than the transfer frequency (0.80%) observed for pGSP149, a pIJ101-derived plasmid that lacks clt. This result strongly suggested that a clt function present on pSB24.2 can function in conjunction with the pIJ101 tra gene to promote the efficient transfer of pSB24.2. To show directly that the suspected pSB24.2 clt region can complement the pIJ101 clt function, the 142-bp Clt+ region of pIJ101 present on pGSP263 was replaced with the equivalent 142-bp sequence of pSB24.2 (Fig. 2), which had been PCR amplified with Pfu DNA polymerase (Stratagene, La Jolla, Calif.) and relevant primers prior to cloning, and the transfer frequency of the resulting plasmid (pGSP313) was then tested as before. As with pSB24.202, the transfer of pGSP313 from strain TK23.42 also occurred at a frequency (24%) comparable to that of pGSP263.
TABLE 2.
Transfer frequencies of pSB24.2 Clt+ derivatives from S. lividans TK23.42a
| Plasmid
|
Avg no. of:
|
Transfer frequencyb (%) | |||
|---|---|---|---|---|---|
| Donor (relevant genotype) | Recipient | Donors | Recipients | Transconjugants | |
| pGSP263 (clt+ [pIJ101]) | pHYG1 | 3.8 × 105 | 2.1 × 107 | 1.1 × 105 | 29 |
| pGSP149 (clt) | pHYG1 | 9.0 × 104 | 1.1 × 107 | 7.2 × 102 | 0.80 |
| pSB24.202 (clt+ [pSB24.2]) | pHYG1 | 9.7 × 105 | 3.9 × 107 | 2.7 × 105 | 28 |
| pGSP313 (clt+ [pSB24.2]) | pHYG1 | 2.6 × 105 | 2.6 × 107 | 6.3 × 104 | 24 |
Matings were performed and quantified as described previously (17). Numbers represent the averages obtained from four independent mating assays.
Transfer frequency is calculated as the ratio of transconjugants to donors expressed as a percentage.
The pSB24.2 korB gene can complement the pIJ101 korB function.
We also examined whether the pSB24.2 korB gene could functionally substitute for its pIJ101 counterpart to override the lethal effects of unregulated kilB gene expression in Streptomyces. When 100 ng of plasmid pGSP317, a pHYG1 derivative containing the unregulated pIJ101 kilB gene, was used to transform S. lividans TK23 according to the method of Hopwood et al. (7), a relatively low number of slow-growing transformants were observed after 4 days (i.e., an average of 2.8 × 102 transformants with a diameter of 1 mm or less was obtained in three independent assays). These transformants eventually grew to normal size and were therefore likely to be equivalent to similar transformants obtained previously (12) in which incoming plasmids carrying the unregulated kilB gene had subsequently deleted their kilB sequences. When TK23 was transformed with an equimolar amount of plasmid pGSP322, which is a derivative of pGSP317 containing the pSB24.2 korB gene, an average of 1.0 × 103 faster-growing transformants (i.e., with a diameter of greater than or equal to 2 mm after 4 days) was obtained in three independent assays; such numbers and growth phenotypes were similar to the results seen (i.e., an average of 1.8 × 103 faster-growing transformants) when TK23 was transformed with an equimolar amount of plasmid pGSP321, a pGSP317 derivative containing the pIJ101 korB gene.
Results here demonstrating that two important conjugation functions (i.e., clt and korB) have been conserved between plasmids pIJ101 and pSB24.2 are consistent with the hypothesis that pIJ101 and the parental plasmid pSB24.1 most likely encode the same conjugation system. Given the high degree of overall sequence relatedness between pIJ101 and pSB24.2, plasmids pIJ101 and pSB24.1 (along with its deletion derivative pSB24.2) may also encode highly similar, if not identical, replication systems. The possibility, however, that pIJ101 and pSB24.1 instead represent separate replicons possessing the same conjugation system is intriguing, as we are unaware of any example from other bacterial conjugation systems in which specific cis-acting transfer sequences (e.g., oriT sequences) (27) encoded by distinct replicons are functionally interchangeable.
The hosts of pIJ101 and pSB24.1 (i.e., S. lividans and S. cyanogenus, respectively) have 99.8% similarity in their 16S rRNA gene sequences, indicating that they are closely related species (18). Our results are therefore consistent with the notion that conjugative pIJ101-like plasmids may be dispersed throughout at least a portion of the Streptomyces genus. Such dispersal has the potential to be even more widespread, however, given the extremely high, albeit limited, degree of relatedness between pIJ101 and pSN22, two plasmids that are derived from species (i.e., S. lividans and S. nigrifaciens, respectively) that have only 95.7% homology in their 16S rRNA genes and thus fall into different phylogenetic clusters (18).
Nucleotide sequence accession number.
The revised pSB24.2 korB-clt sequence determined in this study has been deposited in the GenBank database under accession no. AF133709.
Acknowledgments
We are very grateful to Alexei Sorokin and Sergey Sineoky, for their assistance in obtaining plasmid pSB24.2, and to Fred A. Rainey for assistance with nucleotide sequencing. We thank Naomi Ward-Rainey, John Battista, and Matthew Ducote for critical reading of the manuscript.
This work was supported by grant MCB-9604879 from the National Science Foundation (to G.S.P.).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bolotin A P, Sorokin A V, Aleksandrov N N, Danilenko V N, Kozlov Y I. Nucleotide sequence of DNA of the actinomycete plasmid pSB24.2. Dokl Biochem. 1986;283:260–263. [PubMed] [Google Scholar]
- 3.Brasch M A, Cohen S N. Sequences essential for replication of plasmid pIJ101 in Streptomyces lividans. Plasmid. 1995;33:191–197. doi: 10.1006/plas.1995.1020. [DOI] [PubMed] [Google Scholar]
- 4.Deng Z, Kieser T, Hopwood D A. “Strong incompatibility” between derivatives of the Streptomyces multi-copy plasmid pIJ101. Mol Gen Genet. 1988;214:286–294. doi: 10.1007/BF00337723. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Gonzalez C, Cadenas R F, Noirot-Gros M F, Martin J F, Gil J A. Characterization of a region of plasmid pBL1 of Brevibacterium lactofermentum involved in replication via the rolling circle model. J Bacteriol. 1994;176:3154–3161. doi: 10.1128/jb.176.11.3154-3161.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gruss A, Ehrlich S D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989;53:231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 8.Hopwood D A, Kieser T. Conjugative plasmids of Streptomyces. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 293–311. [Google Scholar]
- 9.Ilyina T V, Koonin E V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kataoka M, Kiyose Y M, Michisuji Y, Horiguchi T, Seiki T, Yoshida T. Complete nucleotide sequence of the Streptomyces nigrifaciens plasmid, pSN22: genetic organization and correlation with genetic properties. Plasmid. 1994;32:55–69. doi: 10.1006/plas.1994.1044. [DOI] [PubMed] [Google Scholar]
- 11.Kendall K J, Cohen S N. Complete nucleotide sequence of the Streptomyces lividans plasmid pIJ101 and correlation of the sequence with genetic properties. J Bacteriol. 1988;170:4634–4651. doi: 10.1128/jb.170.10.4634-4651.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kendall K J, Cohen S N. Plasmid transfer in Streptomyces lividans: identification of a kil-kor system associated with the transfer region of pIJ101. J Bacteriol. 1987;169:4177–4183. doi: 10.1128/jb.169.9.4177-4183.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan S A. Rolling-circle replication of bacterial plasmids. Microbiol Mol Biol Rev. 1997;61:442–455. doi: 10.1128/mmbr.61.4.442-455.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieser T, Hopwood D A, Wright H M, Thompson C J. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185:223–238. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- 15.Pabo C O, Sauer R T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- 16.Pettis G S, Cohen S N. Plasmid transfer and expression of the transfer (tra) gene product of plasmid pIJ101 are temporally-regulated during the Streptomyces lividans life cycle. Mol Microbiol. 1996;19:1127–1135. doi: 10.1046/j.1365-2958.1996.493986.x. [DOI] [PubMed] [Google Scholar]
- 17.Pettis G S, Cohen S N. Transfer of the pIJ101 plasmid in Streptomyces lividans requires a cis-acting function dispensable for chromosomal gene transfer. Mol Microbiol. 1994;13:955–964. doi: 10.1111/j.1365-2958.1994.tb00487.x. [DOI] [PubMed] [Google Scholar]
- 18.Rainey, F. A. Personal communication.
- 19.Servín-González L. Identification and properties of a novel clt locus in the Streptomyces phaeochromogenes plasmid pJV1. J Bacteriol. 1996;178:4323–4326. doi: 10.1128/jb.178.14.4323-4326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Servín-González L. Relationship between the replication functions of Streptomyces plasmids pJV1 and pIJ101. Plasmid. 1993;30:131–140. doi: 10.1006/plas.1993.1040. [DOI] [PubMed] [Google Scholar]
- 21.Stein D S, Cohen S N. Mutational and functional analysis of the korA and korB gene products of Streptomyces plasmid pIJ101. Mol Gen Genet. 1990;222:337–344. doi: 10.1007/BF00633838. [DOI] [PubMed] [Google Scholar]
- 22.Stein D S, Kendall K J, Cohen S N. Identification and analysis of transcriptional regulatory signals for the kil and kor loci of Streptomyces plasmid pIJ101. J Bacteriol. 1989;171:5768–5775. doi: 10.1128/jb.171.11.5768-5775.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suzuki I, Kataoka M, Seki T, Yoshida T. Three single-strand origins located on both strands of the Streptomyces rolling circle plasmid pSN22. Plasmid. 1997;37:51–64. doi: 10.1006/plas.1996.1269. [DOI] [PubMed] [Google Scholar]
- 24.Tai J T-N, Cohen S N. The active form of the KorB protein encoded by the Streptomyces plasmid pIJ101 is a processed product that binds differentially to the two promoters it regulates. J Bacteriol. 1993;175:6996–7005. doi: 10.1128/jb.175.21.6996-7005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tai J T-N, Cohen S N. Mutations that affect regulation of the korB gene of Streptomyces lividans plasmid pIJ101 alter plasmid transmission. Mol Microbiol. 1994;12:31–39. doi: 10.1111/j.1365-2958.1994.tb00992.x. [DOI] [PubMed] [Google Scholar]
- 26.Westrich L, Domann S, Faust B, Bedford D, Hopwood D A, Bechthold A. Cloning and characterization of a gene cluster from Streptomyces cyanogenus S136 probably involved in landomycin biosynthesis. FEMS Microbiol Lett. 1999;170:381–387. doi: 10.1111/j.1574-6968.1999.tb13398.x. [DOI] [PubMed] [Google Scholar]
- 27.Wilkins B, Lanka E. DNA processing and replication during plasmid transfer between Gram-negative bacteria. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 105–136. [Google Scholar]
- 28.Zaman S, Radnedge L, Richards H, Ward J M. Analysis of the site for second-strand initiation during replication of the Streptomyces plasmid pIJ101. J Gen Microbiol. 1993;139:669–676. doi: 10.1099/00221287-139-4-669. [DOI] [PubMed] [Google Scholar]
- 29.Zaman S, Richards H, Ward J. Identification of the minimal replicon of the streptomycete plasmid pIJ101. Plasmid. 1993;29:57–62. doi: 10.1006/plas.1993.1007. [DOI] [PubMed] [Google Scholar]