Abstract
Formation of neutrophil extracellular traps was first described in 2004, showing that NETs are composed of decondensed chromatin fibers and nuclear and granule components. Free DNA is often used to quantify NETs, but to differentiate NETosis from necrotic DNA-release, immunofluorescence microscopy with NET-specific markers is required. Although evaluation by hand is time-consuming and difficult to standardize, it is still widespread. Unfortunately, no standardized method and only limited software tools are available for NET evaluation. This study provides an overview of recent techniques in use and aims to compare two published computer-based methods with hand counting. We found that the selected semi-automated quantification method and fully automated quantification via NETQUANT differed significantly from results obtained by hand and exhibited problems in detection of complex NET structures with partially illogical results. In contrast to that, trained persons were able to adapt to varying settings. Future approaches aimed at developing deep-learning algorithms for fast and reproducible quantification of NETs are needed.
Keywords: NETs, ImageJ, Quantification, Cell counting, NETQUANT, In vitro
1. Introduction
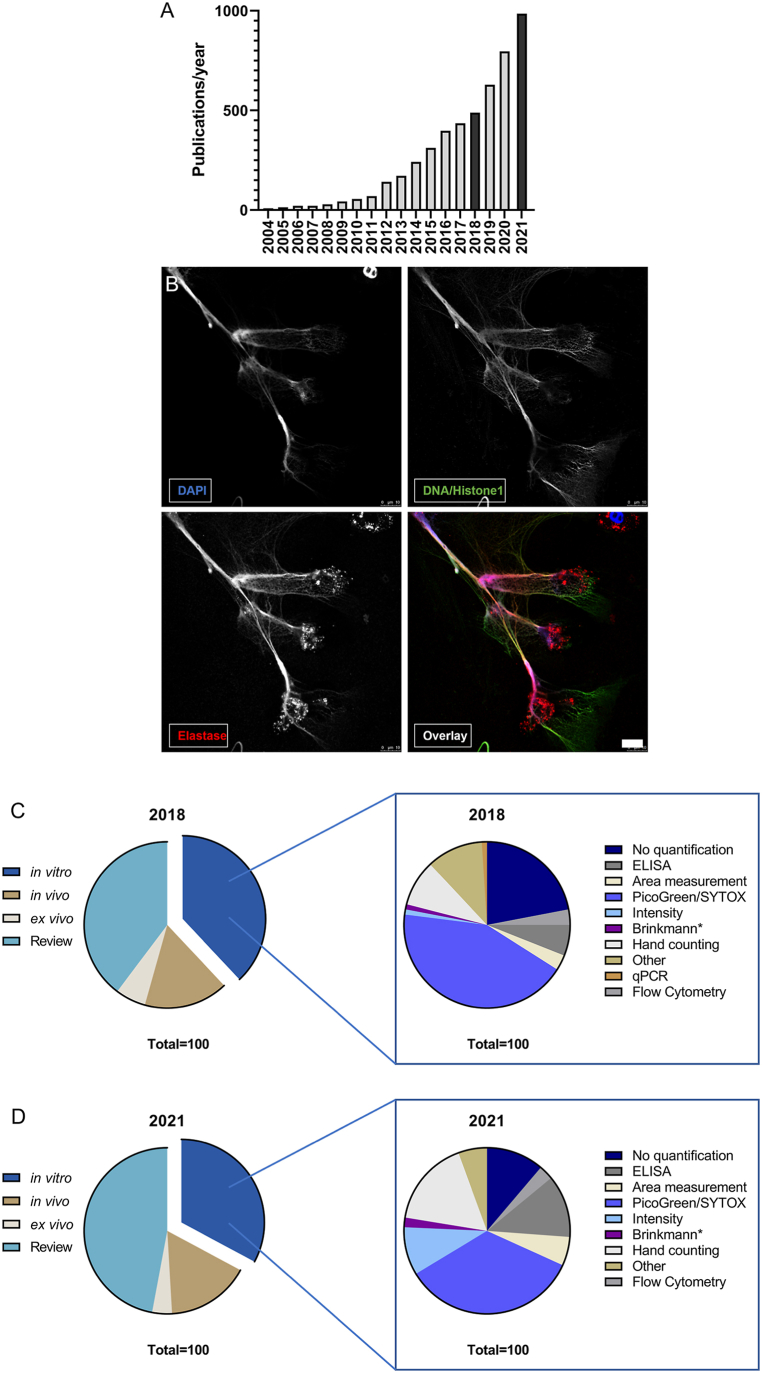
Neutrophils are the most abundant cells of the innate immune system which are known for their diverse repertoire of mechanisms like phagocytosis, degranulation, or reactive oxygen species production [1]. Additionally, the mechanism of neutrophil extracellular trap (NET) formation was described in the early 2000s [2]. NET formation is a response to invading pathogens described as a cell death mechanism, NETosis, which is distinct from necrosis and apoptosis [3]. Neutrophils release their decondensed DNA into the extracellular space to entrap pathogens, and these NETs are decorated with histones and host defense peptides (HDPs) like neutrophil elastase (NE), myeloperoxidase (MPO), and LL-37 [4,5]. NET formation can be triggered by different kinds of proinflammatory stimuli or pathogens [[2], [3], [4],6]. Since its discovery in 2004, NET formation has been intensively studied and was shown to be involved in defense against various pathogens such as viruses [7,8], fungi [9,10], and bacteria [2,6]. However, also detrimental effects against the host were observed and accounted for exceeding NET formation or malfunctioning clearance of NET structures like in thrombosis, cystic fibrosis, autoimmune or chronic diseases [[11], [12], [13], [14], [15]]. Recently, the role of NETs during SARS-CoV-2 infection has become of huge interest. Early in the pandemic, increased NET marker levels in patient serum was found as a marker for a poor disease outcome. Studies revealed that increased NET formation and elevated presence of NET markers contributed to thrombosis, vascular leakage, worsening oxygenation, acute respiratory distress syndrome, and generally poor prognosis [[16], [17], [18], [19], [20]]. Hence, the phenomenon of NET formation is still a growing research field with an increasing number of publications each year (Fig. 1A). However, to study both the beneficial and detrimental role of NETs, the in vitro visualization of NETs is a common and important tool to investigate their properties and mechanisms in detail (reviewed in: [21,22]. One possible and widely used approach to visualize NETs is the use of DNA intercalating dyes which bind to the anionic DNA fibers due to their cationic properties, such as PicoGreen® and SYTOX® staining. Nonetheless, this method cannot discriminate between DNA derived from NET formation or DNA release by necrosis [23]. The usage of immunofluorescence (IF) staining overcomes these difficulties by the specificity of the primary antibodies, which are directed against NET components and subsequent fluorophore-conjugated secondary antibodies and image acquisition by immunofluorescence microscopy. Such an example is shown in Fig. 1B. Commonly used antibodies to specifically stain NETs are antibodies against DNA-histone 1-complexes, which stain delobulated and decondensed nuclei after activation for NET formation. This antibody is often used in combination with neutrophil-specific granule components, e.g., elastase or myeloperoxidase that translocates to the nuclei during NET formation and that are abundantly found associated with extracellular NET fibers [21,22,24].
Fig. 1.
Increasing interest in NET research and the associated techniques used for NET quantification.
A shows the number of publications listed by Pubmed under the term “neutrophil extracellular trap”. Since the first description in 2004, NETs are a research topic with increasing representation in literature. B Exemplary immunofluorescence micrograph of NET formation by human neutrophils stimulated with Simvastatin (10 μM, 3 h) and stained with DAPI and antibodies against DNA/Histone1 complex (green) and neutrophil elastase (red). The image is presented with the individual channels and an overlay. Individual channels highlight the advantage of antibody staining over pure DNA intercalating dyes by exposing more details of NET structures. The image was acquired as stated in the methods section with the following modifications: additional staining against neutrophil elastase (primary antibody: anti-neutrophil elastase, rabbit, polyclonal, Millipore, Cat# 3864, secondary antibody: Alexa Fluor 633, goat anti-rabbit, Thermo Scientific Cat#: A21070), and image acquisition with HCX PL APO lambda blue 63.0×1.40 OIL UV objective. Scale bar 10 μm. C provides an overview of NET research and the proportion of research categories in 2018. Most publications were reviews (39.72%), followed by in vitro research (37.98%). In vivo and ex vivo studies were described to a lesser extent. Within the group of in vitro studies, the NET quantification methods were counted. A total of 22% of the studies did not show quantification data. Most publications used tools for quantification with DNA intercalating dyes, followed by hand counting of NETs on micrographs by immunofluorescence microscopy (9%) and ELISA (6%). D shows the same analysis for the year 2021 where reviews made up 47.01% of the published articles, while in vitro research still accounted for 32.83% of the screened articles. In the group of in vitro studies, the number of studies which did not quantify NET formation decreased to 11.21%. Analysis of NET formation via DNA intercalating dyes was still the most favored method (34.58%), but other techniques were used more frequently. Hand counting increased to 16.82% of the studies and ELISA techniques were used in 12.15% of these. Also, area and intensity measurements were used more frequently, while flow cytometry and program-based analysis of micrographs only played a minor role in both years.
However, there is still a need for standardized quantification methods of NETs derived from images by immunofluorescence microscopy. Counting NETs by hand is still performed in many groups. Nevertheless, this requires training and is highly time consuming and prone to personal bias. To date, only a limited amount of publications on such automated programs are available that, for example, quantify the area covered with NETs [25,26] or the amount of NET-releasing cells [[25], [26], [27], [28]], some of these based on freely available software and others relying on fee-based platforms like MATLAB. For in vitro studies that aim for a better understanding of processes associated with NET formation, a valid quantification tool for the amount of NET releasing cells is needed. Therefore, in the present study, we aimed to compare the evaluation of two sets of immunofluorescence images using three different approaches: (1) hand counting, (2) semi-automated quantification [27], and (3) the MATLAB-based automated NETQUANT application [28]. This comparison enables advantages and problems to be identified which can occur using those techniques and to highlight the need for more research not only on NET formation processes and the development thereof, but also research on quantification of the same.
2. Results
2.1. Actual situation of NET quantification in the literature
To first gain an overview of recently used techniques for NET quantification, we screened all hits occurring under the term “neutrophil extracellular trap” from 2004 until 2021 in PubMed (https://pubmed.ncbi.nlm.nih.gov/), showing an increased publication rate over the years (Fig. 1 A). As a following step, the search terms “neutrophil extracellular trap” was expanded to determine the number of publications in the selected respective categories: in vitro, in vivo, ex vivo, or review. We investigated the years 2018 as the last complete year before the COVID-19 pandemic and 2021 as the most recent complete year to determine changes in application of NET quantification methods. The analysis revealed that in 2018 as well as 2021, most publications were reviews (39.72% in 2018 and 47.07% in 2021), followed by in vitro research (37.98% in 2018 and 32.83% in 2021). In vivo and ex vivo studies were described to a lesser extent (Fig. 1C and D). All in vitro studies were scrutinized for the applied NET-quantification methods. Data are shown in Fig. 1C for 2018 and 1D for 2021. In most cases, simple extracellular DNA quantification with PicoGreen or SYTOX staining was used for NET quantification (43% in 2018 and 34.58% in 2021), while hand counting of immunofluorescence micrographs was used as a second priority (9% in 2018 and 16.82% in 2021). Notably, 22% of in vitro studies conducted on the topic of NETs in 2018 did not quantify NET formation at all, a number which decreased in the year 2021 to 11.21%. Overall, in 2021, the number of quantification methods like hand counting of immunofluorescence images or ELISA-based techniques targeting NET complexes like MPO-DNA increased. Also, area and intensity measurements were used more frequently, while flow cytometry and software-based analysis of micrographs only played a minor role in both years.
Remarkably, hand counting of NET-releasing cells is still one of the most frequently used techniques in laboratories conducting NET research (Fig. 1C + D), although this is quite a time-consuming technique which is difficult to be standardized. This finding highlights the need for a reliable quantification method for immunofluorescence micrographs.
2.2. Quantification of image set 1 by different methods
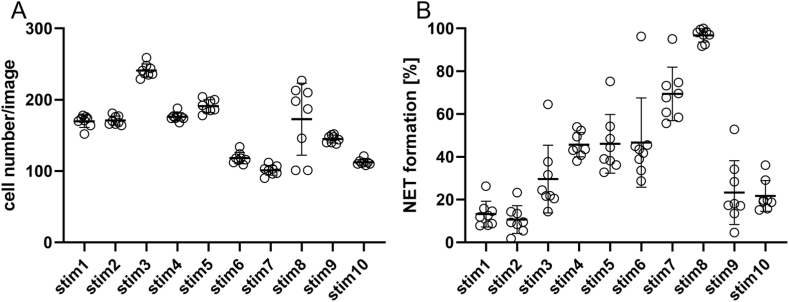
For a better understanding of the advantages and challenges in the usage of NET quantification methods, in this study, three different approaches for evaluating NET formation on immunofluorescence microscopy images were applied and compared: on the one hand, commonly used hand counting of NET-active cells, on the other hand, a semi-automated and a fully automated software. Image sets of different origin and preparation techniques were used: “Image set 1” consisted of images with either control or stimulated neutrophils acquired by the authors. Images were analyzed for total cell number per image (independent of their activation status) as well as determining the number of cells that release NETs (NET formation). First, quantification of NET-releasing cells was scrutinized by eight of the authors of this manuscript who counted 10 images each with different rates of NET formation (named “stim 1–10”). All individuals were introduced to defined NET-criteria: the occurrence of a distinct extracellular off-shoot was defined as a NET-positive criterion as was any cell touching an off-shoot (Supplementary Fig. 1). Moreover, cells where the nucleus lost its shape and density as well as increased in size were defined as NET-positive (Supplementary Fig. 1). Each individual examiner determined the total cell number and NET formation rate for each image (Fig. 3A+B; raw data of each single image evaluation is shown in Supplementary Fig. 2). It was observed that the values for NET formation were not completely consistent between individuals. However, with one exception (Fig. 3A stim8), all other images showed a standard deviation of a maximum of 10 cells between individual examiners when counting the total cell number per image. Regarding the number of NET-positive cells per image, the results showed a standard deviation of 6.4% between the individual examiners (Fig. 3B). Furthermore, reliable quantification depended on experience and training of the respective individuals. In our case, one person had received no training and had less than 3 months’ experience in working with NET visualization using immunofluorescence microscopy, which resulted in higher deviations compared to the rest of the group. This highlights the need for thorough training of staff but also points out the weakness of this method.
Fig. 3.
Manual quantification of cell number and NET formation derived from image set 1.
The graph depicts values for each image derived from the individual examiners (circles) as well as mean and standard deviation for each image (bars). A: Cell counts determined by the individual examiners. B: Determination of NET formation by the individual examiners.
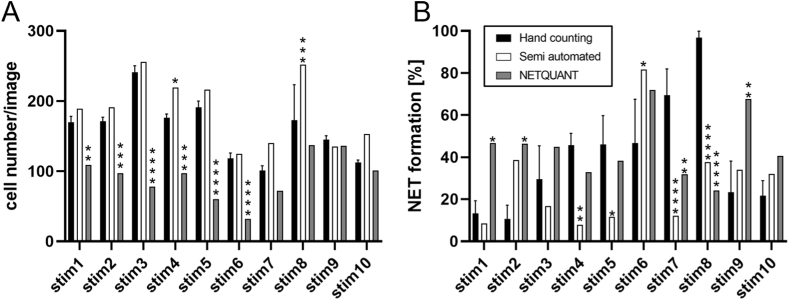
On comparing the average result from hand counting with those of the software-based evaluations, a discrepancy was observed between NETQUANT and both the other techniques (Fig. 4). NETQUANT determined significantly lower numbers of cells compared to hand counting in six images, while the semi-automated methods detected significantly more cells in two images (Fig. 4A). In the case of NET formation, further differences in the results of each technique became evident (Fig. 4B). NETQUANT determined a NET formation rate significantly different from hand counting in five images, and the semi-automated method differed significantly in five images as well. For the other remaining images, the differences were not statistically significant. However, the partly different outcome was visible to the naked eye. Since NETQUANT required a set of control images for calibration, image set 1 was supplemented with six images with control conditions with unstimulated neutrophils (Supplementary Fig. 3).
Fig. 4.
Comparison of NET quantification by hand counting semi-automated quantification and NETQUANT.
A: Cell numbers for Image set 1 determined by all three methods, showing significantly fewer cell counts by NETQUANT in the majority of the images. B: NET formation determined by all methods revealed a significant difference between hand counting and NETQUANT as well as the semi-automated method. Statistical analysis was performed by Two-Way ANOVA with Dunnett correction for multiple comparison. Semi-automated and NETQUANT values were compared with hand counting. Data are given as mean ± SD. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
2.3. Quantification of image set 2 using different methods
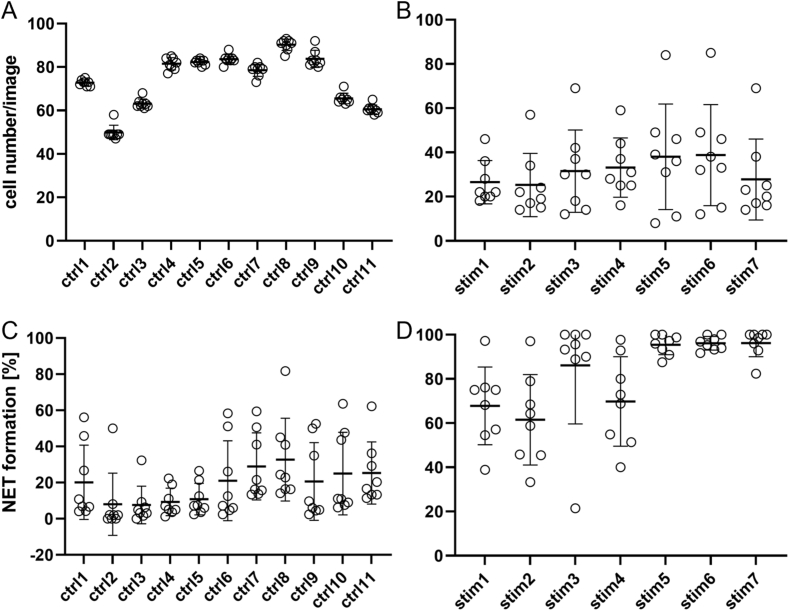
Similar to image set 1, the same analysis pattern was applied to image set 2. The set consisted of the available test images provided by the programmers of the NETQUANT software [28]. Eight of the authors evaluated the images of the control group and stimulated cells (Fig. 5). Subsequently, the average outcome of hand counting was compared to the semi-automated method and NETQUANT results (Fig. 6). For the control group, the number of cells were determined at a similar value, resulting in a small standard deviation (Fig. 5A), while the stimulated group showed higher differences in the cell count with consequently higher standard deviations (Fig. 5B). In terms of NET formation, the control and stimulated groups showed a heterogeneous outcome (Fig. 5C + D).
Fig. 5.
Cell number and NET formation of control and stimulated groups determined by hand counting from image set 2.
Eight individual examiners (circles) determined the cell number and NET formation. Averages of all values (vertical bars) are given with respective standard deviation. A shows the cell number in the control group and B the cell number of the stimulated group. C shows the NET formation determined in the control group and D the stimulated group; both groups show differences in the individual values for NET formation.
Fig. 6.
Comparison of hand counting, NETQUANT, and semi-automated quantification for image set 2.
A shows the cell count for the control group with the semi-automated methods giving unrealistically high values for some images. Significant differences by both semi-automated and NETQUANT compared to hand counting were apparent. In B, the pattern remained similar to A, where the semi-automated method showed remarkably higher values of cell counts in several cases. C depicts NET formation values by all three methods, with the semi-automated method showing significantly higher results than both other methods in the control group, even exceeding 100% NET formation. D shows NET formation of the stimulated group where NETQUANT exhibited values within the range of hand counting, while the semi -automated method revealed significant differences. Statistical analysis was performed by Two-Way ANOVA with Dunnett correction for multiple comparison. Semi -automated and NETQUANT values were compared to hand counting. Data are given as mean ± SD. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
When comparing the results acquired by hand counting with the software-based results, differences in the outcome became clear between the techniques (Fig. 6). The semi-automated evaluation was significantly higher in cell count determination in nine images, and NETQUANT significantly different in eight images. Remarkably, the semi-automated method showed illogical values of 1000 to almost 3000 cells for some images (Fig. 6A). In the case of the stimulated group, values of NETQUANT and hand counting were found to occur within a similar range without significant differences (Fig. 6B), whereas the semi -automated method showed results significantly above what was determined by hand counting in four images (Fig. 6B). For NET formation, the semi-automated method detected significantly higher values close to 100% in seven images, while NETQUANT and hand counting were shown to be at low levels. Noteworthy, despite lacking significance, hand counting and NETQUANT values differed significantly as seen for the images ctrl6-11 (Fig. 6C). In the stimulated group, the semi-automated method again differed significantly from hand counting, while NETQUANT was at a similar level in five images. Interestingly, for the control and stimulated images, the semi-automated method calculated illogical values of more than 100% NET formation in some cases (Fig. 6D). This issue will be addressed in the discussion section below.
In summary, the data showed that semi-automated quantification and fully automated quantification via NETQUANT produced mostly different results from hand counting with an overall heterogenous pattern, ranging from highly significant to similar levels.
3. Discussion
Missing a standard quantification technique makes comparisons of results between different research groups, which are working on the topic of NET formation, a challenging task. As the interest in this subject has been growing over the last years (Fig. 1A), it remains an urgent future goal to develop and establish standardized methods. With an increasing number of publications on the role of NETs in COVID-19, more quantification techniques based on ELISA and also IF have been performed. Nevertheless, hand counting of NET formation on immunofluorescence images is still highly relevant. However, multiple tools to ease the workload and reduce hand counting bias were already developed and published, as reviewed in van Breda et al., 2019., although none of them seemed to prevail and gain general acceptance among groups working on NET formation. Notably, the NETQUANT method described by Ref. [28] has been recently recommended by van Breda et al. [29], but so far, this method has only been rarely applied [30]. In comparison, the semi-automated quantification described by Brinkmann [28] has been used more frequently (Fig. 1C and D).
Overall, as seen in the literature screening (Fig. 1), automated or semi-automated techniques were not considered as valid options by the majority of research groups and related publications in international peer-reviewed journals in 2018 and 2021 (Fig. 1C and D). Nonetheless, hand counting as a technique was seen to increase, despite being a time-consuming and to a certain extent biased technique, resulting in discrepancies between different individuals evaluating the same images, as seen for both image sets here (Fig. 3, Fig. 5). Clearly defined criteria for NET formation and training can result in lower deviations between persons and allow partial standardization. As seen in the control group (Fig. 5D), five individuals determined NET formation rates at similar levels of under 20%, whereas the other three individuals detected high rates of NET formation in most of the control images. However, discrepancies were not great enough to define a significant outlier. This highlights the need for extensive training, as the staining technique applied in image set 2 was not frequently used within the group at that time point.
Nevertheless, a standardized quantification via automated or semi-automated programs could help to enhance the quality of studies on NET formation and thus help to improve comparability of results.
The advantage of hand counting is that well-trained persons can more easily adapt to variations in staining and NET phenotypes. The diversity in nuclear morphology could turn out to be challenging for programs that rely on the definition of, for example, size and morphology of inactive cells. It is important to realize that control samples of healthy patients may provide a different composition of neutrophils than those of the diseased group [31]. This is also apparent in the images used in the present study. Based on morphological alteration, challenges for software tools occurred in the case of false positive or negative results and fusion of cells within a NET fiber. Examples are given in Supplementary Fig. 4. Finally, also the manual settings of thresholds and cut-offs used in the automated software provide the possibility of bias or even manipulation, similar to the initial criticism about hand counting.
Furthermore, we believe that the respective staining techniques are one of the major problems for the software-based evaluation, as the semi-automated quantification struggled with image set 2 as did the NETQUANT software with image set 1. In our opinion, using NE as classical granule protein as the only NET marker bears the risk of false statements about colocalization, even if signals may be lying in different focal planes of the sample, e.g., in simple non-activated granule components (Supplementary Fig. 5). Moreover, not all NETs always show NE staining in all structures, possibly leading to underestimated NETs when applying this method. The criteria of DNA/NET area used by the program may cause problems, as the area covered by NE staining would vary during the NET formation process. Non-active cells would give a strong signal due to NE localized in neutrophil granules that are present in the cytosol. Subsequent NET formation would be initialized by granular content transmigrating to the nucleus [4,5], resulting in a smaller area covered by the signal.
In contrast to that, using DNA/Histone1 complex as a NET marker appears more specific, as only NETs should be stained by this method. The DNA/Histone1 complexes only become accessible for the antibody during the NET formation process, as the histone packing, and intact nuclear membrane hinder a binding in NET negative cells. However, especially NETQUANT results differed distinctly from the two other quantification techniques for this staining (image set 1; Fig. 4). In the same line, semi-automated quantification was unable to deliver logical results for images stained with NE as NET marker, as NET formation of more than 100% was reported for several images (Fig. 6). This was, on the one hand, due to NE as NET marker being present in NET negative and positive cells, and, on the other hand, due to the threshold setting during image processing. As described in the original publication, the threshold was set on one image with a low number of NETs and applied to all other images of the set. This led to some images showing events on the DNA channel, also not excluded by size, leading to increased counts in the numerator without corresponding counts in the denominator, yielding NET formation rates of over 100% (Supplementary Fig. 5).
A more basic problem for the semi-automated method was identified in the cell fusion, which occurred when several cells were in contact with a larger NET structure. Each cell would have to be counted as a single event, but the program created an outline, combining multiple cells to a single event, thus leading to wrong cell counts and a subsequently altered calculation of NET formation (Supplementary Fig. 4C+D). Both programs had in common that during processing, cells were lost, and hence, were not included in the cell count. This consequently led to a wrong calculation of NET formation rates (Supplementary Fig. 4).
In our opinion, a fundamental problem of fully or semi-automated quantification of NET occurs within the heterogeneous character of NETs themselves. This factor makes it very difficult to mathematically define criteria, which, on the one hand, are strict enough to give clear cut-offs and, on the other hand, leave enough room for the diverse morphology of NETs. It even increases this difficulty when NETs of different species arise, as nucleus morphology can differ [32].
In general, the tested programs did not achieve similar values as the results derived from hand counting. The need for an unbiased, standardized and ideally faster evaluation of immunofluorescence microscopy images of NET formation has yet to be fulfilled. A promising approach could be programs based on machine learning with a specific deep-learning algorithm as previously shown for quantification of NET-degradation by quantifying the area of NET surfaces [26]. Such programs are trained on the basis of multiple events with various phenotypes and learn to differentiate NET-forming cells from inactive ones. The more intense the training of the program, the better and more specific is the outcome of the data. However, many groups do not yet consider this approach, as those programs are complicated to operate, especially for non-IT-experienced users, and often expensive. Nevertheless, for now, until those deep-learning algorithm tools are used more frequently, the evaluation of visualized NETs by hand counting seems to be the most reliable technique, even despite the required time and possible personal bias. To overcome this, combining this with other methods, e.g., ELISA-based techniques, can strengthen the validity of results found by hand counting and vice versa, as is often seen in the literature and reflected by increasing application of both methods (Fig. 1C + D). Additionally, facilitation of highly NET-specific markers will be helpful, as the recently described specific antibody against the cleavage site of histone 3 [33]. These will enable a more secure identification of NETs and, thus, in combination with a subsequent standardized quantification of NETs, this would allow for robust and more importantly, reproducible results.
In summary, this work points out a need for future approaches which aim for the development of deep learning algorithms that enable fast and reproducible quantification of NETs.
4. Limitations of the study
In future experiments, the obtained data need to be extended to in vivo images. However, since in vivo images of NET formation are even more complex and difficult for separation of single cells and NET structures from simple DNA release during necrosis, this study focused on in vitro NET assays only. Evaluation of NET structures in in vivo lung samples derived from equine asthma and human COVID-19 patients were shown to be possible via NET volume determination. The volume of NET positive structures was evaluated relative to the sample volume [34,35]. A comparison of the above methods in a similar manner to those performed in this manuscript might help to explain differences between different projects or laboratories with NET formation as a crucial readout.
Funding
This work was supported by R2N Project under Grant [74ZN1574], which is funded by the Federal State of Lower Saxony. This Open Access publication was funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) within the program LE 824/10-1 “Open Access Publication Costs” and the University of Veterinary Medicine Hannover, Foundation.
Author contribution statement
Timo Henneck: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Christina Krüger: Performed the experiments; Analyzed and interpreted the data.
Andreas Nerlich and Sönke von den Berg: Contributed reagents, materials, analysis tools or data.
Melissa Langer, Leonie Fingerhut, Marta C. Bonilla, Marita Meurer, Nicole de Buhr and Katja Branitzki-Heinemann: Performed the experiments.
Maren von Köckritz-Blickwede: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Data availability statement
Data included in article/supp. material/referenced in article.
Experimental model and subject details (STAR methods)
Literature research on NET quantification techniques
First, the number of publications given by Pubmed under the search term “neutrophil extracellular trap” was determined from the year 2004, the date of discovery of NETs, until 2021. Highlighted years 2018 and 2021 were scrutinized. The search terms “neutrophil extracellular trap, x” were used to determine the number of publications in the respective categories (for x either of the following terms was inserted: in vitro, in vivo, ex vivo, or review). The resulting number of publications for each search was added up and the percentage of the respective categories was calculated.
To determine the NET quantification methods applied for in vitro research, the papers found under “neutrophil extracellular trap, in vitro” for the years 2018 were manually screened in the method sections for their NET quantification technique. For 2018, a total of 109 papers were displayed in the search. After excluding those papers which were solely in vivo topics, reviews, or did not focus on NETs, 91 paper were left for analysis. The same approach was performed for 2021, where 196 papers showed up in the search. Of those, the first 109 were manually screened for the respective methods. After excluding papers according to the same criteria as for 2018, a total of 96 papers were used in the analysis.
Generation of image sets for quantification
Isolation of human neutrophils
Primary neutrophils were collected from fresh blood of voluntary, healthy donors after informed consent in accordance with the Declaration of Helsinki and in agreement with the local ethical board. The study was approved by the Ethical Committee of Hannover Medical School, Hannover, Germany No: 3295-2016. Neutrophils were isolated from voluntary healthy donors as previously described [36]. Briefly, heparin-blood was carefully layered onto the same amount of separating solution Polymorphprep (Progen), and after centrifugation, a layer of neutrophils was visible. Neutrophils were transferred to a fresh 50 mL falcon tube and topped with LPS-free 1× PBS (Sigma). After centrifugation, supernatant was discarded and remaining erythrocytes were eradicated by water lysis with LPS free water (Roth) and immediately filled up with LPS-free PBS again. Centrifugation sedimented neutrophils, the supernatant was discarded and remaining neutrophils were resuspended in RPMI (RPMI Medium 1640 without phenol red, Gibco) at room temperature.
NET induction
Neutrophils were seeded at 5 × 105 cells/mL in 24-well suspension plates (Greiner bio-one), on 12 mm glass slides which were coated with poly-L-lysin one day prior to the experiment. Cells in RPMI medium served as negative control (Supplementary Fig. 3), whereas PMA (25 nM, phorbol 12-myristate 13-acetate), methyl-β-cyclodextrin (10 mM Sigma) or LL-37 (5 μm, Annaspec) were added as NET inducer (Fig. 3, Fig. 4). Cells were incubated at 37 °C and 5% CO2 for 2–4 h and afterwards fixed in PFA 4% final concentration.
Immunofluorescence staining and confocal microscopy
Cells were washed, permeabilized, and incubated in blocking buffer (2% BSA, 0.2% TritonX100 in PBS) for 20 min at room temperature. Supernatant was removed and cells were incubated with primary antibody (1:1000, mouse monoclonal anti-DNA/Histone1, Millipore, MAB 3864, Stock 2.2 mg/mL) diluted in blocking buffer for 1 h. After washing, secondary antibody (1:1000, Dylight 488, goat anti-mouse IgG, Thermo Fisher, 35502) was added and incubated for 45 min at room temperature in the dark. Subsequently, cells were washed and mounted on ProlongGold antifade + DAPI (Thermo Fisher). Slides were dried over night at 4 °C and were sealed the following morning. Images were acquired using a confocal laser scanning microscope (Leica TCS SP5 AOBS) with an HCX PL APO 40 × 0.75–1.25 oil immersion objective and Leica Application Suite (LAS) software. Each coverslip was screened without selection of certain areas to acquire six images per stimulus for later evaluation. Of the different experiments performed, a total of 10 images (Supplementary Fig. 2) were selected for testing the deviation between those results obtained by 1) hand counting by the individual examiners and also by the aforementioned software-based quantification methods.
In addition to the above mentioned images, a set of representative images provided by the Nordenfelt laboratory, Lund University, Lund, Sweden (https://nordlab.med.lu.se/?page_id=34) to test their NETQUANT software was used for all three quantification methods. Isolation, stimulation, and imaging were described previously in Ref. [28].
Quantification of NET formation by hand counting
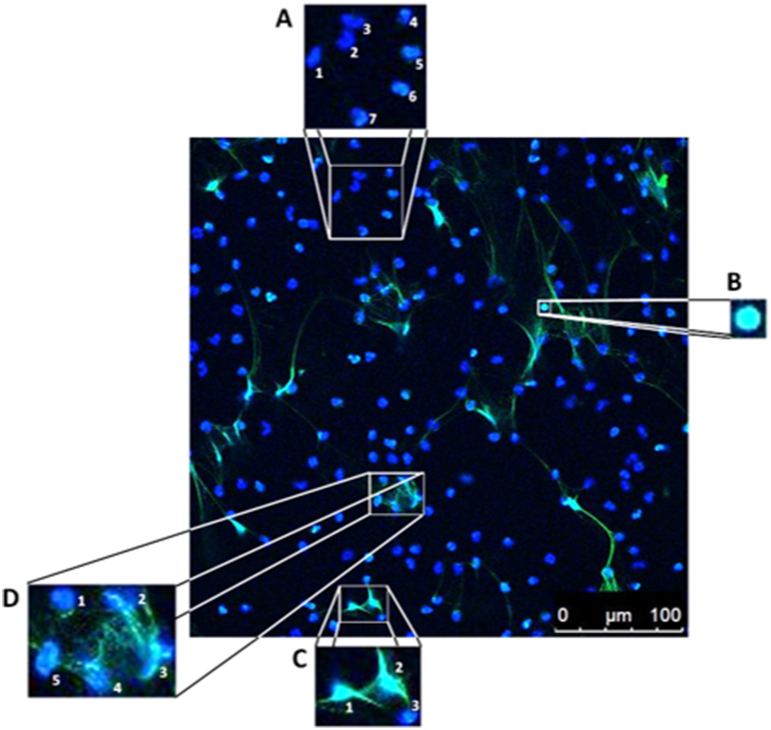
Criteria for evaluating the NET formation were defined beforehand and later applied for generating the results via hand counting. Microscopic images had more than 50, but fewer than 350 cells per image. NET-negative cells were defined as showing small, round, or lobulated nuclei (stained blue with DAPI) with a clear sharp rim. The occurrence of a distinct extracellular off-shoot was defined as a NET-positive criterion as was any cell touching an off-shoot. Moreover, cells where the nucleus had lost its shape and density as well as increased in size were defined as NET-positive (Supplementary Fig. 1).
Hand counting
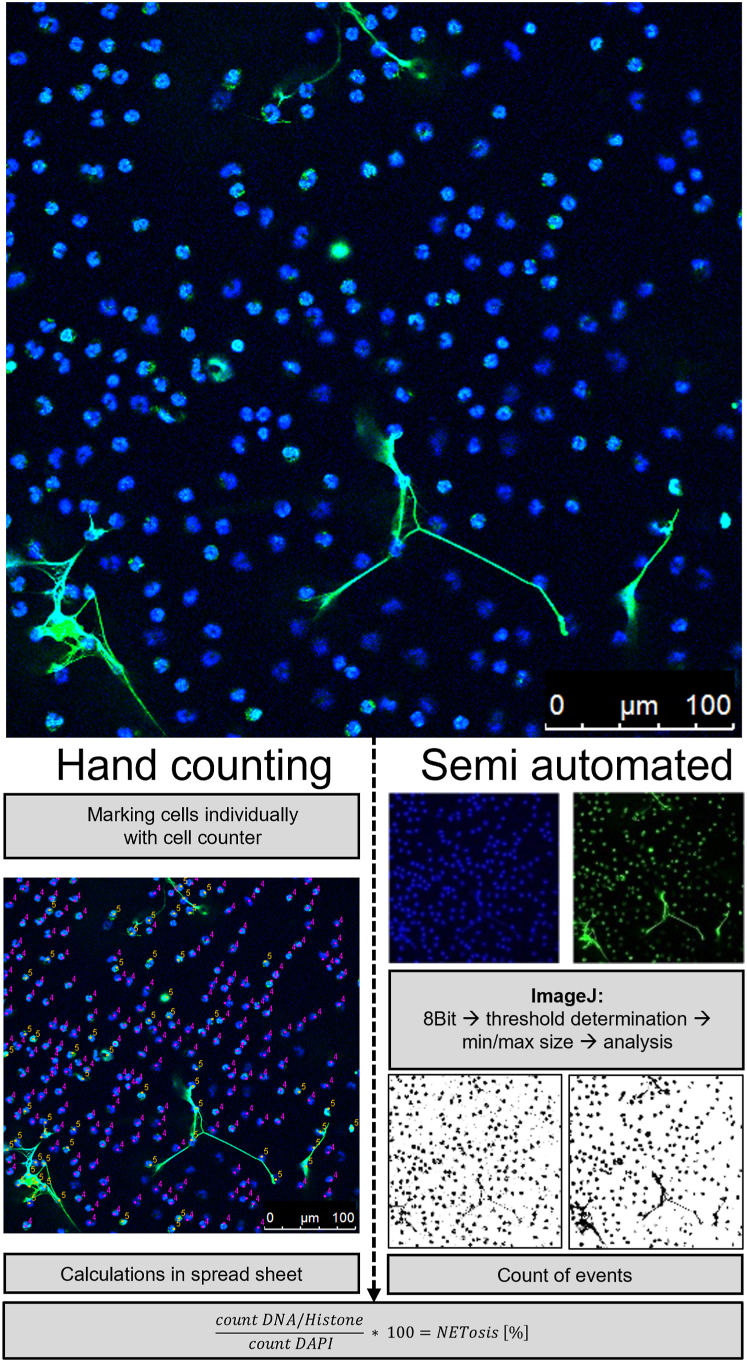
Manual quantification of NET formation also requires image processing programs with the ability of individually marking single cells. Here, we used ImageJ (National Institutes of Health) with additionally installed Cell Counter plugin (De Vos, University of Sheffield, England, https://imagej.nih.gov/ij/plugins/cell-counter.html). Images were loaded into the Cell counter plugin and initialized. If desired, options like keeping the original image without markers are available. Afterwards, each single nucleus was determined as either NET-negative or NET-positive and marked accordingly by applying a marker of choice for that nucleus (Fig. 2B). After counting, the respective values for NET-positive and NET-negative markers were transferred to a spreadsheet and the respective NET formation rate was calculated for each image as follows: , and finally averaged for each stimulus.
Fig. 2.
Overview of fluorescence images of NETs with subsequent evaluation by hand counting or semi-automated quantification.
NET-negative cells are morphologically seen as small, round, or lobulated nuclei that are stained with DAPI (a blue-fluorescent DNA stain). NET-positive cells show green extracellular off-shoots including any cells that attach to the NET or are distinguished by their blurry rim, enlarged and decondensed (puffy) green nucleus. Green indicates positive staining for DNA-histone-1-complexes as marker for NETs. For hand counting, all cells were counted with the Cell Counter plugin for ImageJ for individually marking each cell. Here, NET-negative cells were labeled in magenta with the number 4, and NET-positive cells were marked in yellow with the number 5. The workflow of NET quantification with the semi-automated approach is shown in ImageJ. Images were separated into respective channels for DAPI (blue) and DNA/Histone complex (green). After converting to the 8-Bit format, values for the background threshold and minimal size were determined. The program determined the events for each channel and respective values were used for manual calculation of NET formation rates.
Semi-automated quantification with Brinkmann method using ImageJ
Semi-automated quantification of NET formation by ImageJ was described [27] as s standardized and unbiased approach for better comparison of results coming from different laboratories, based on freely available ImageJ software. Images were imported separately as a series of each respective channel (here blue for DAPI staining, green for DNA/Histone complex, Fig. 2C). For each channel, the images were converted to 8-bit format for further processing. Afterwards, a threshold for the background was set by hi-pass filtering to separate the stained structures from the background. Therefore, the picture with the least amount of NET-positive cells was chosen as reference and settings were fixed at that point, where all impurities were removed but all cells were still visible. For subsequent analysis of particles, first, a value for minimal particle size for both channels was defined. The following particle analysis calculated the number of events for each channel, which then had to be manually set in relation with the aforementioned formula to determine the amount of NET formation for the respective image (Fig. 2C).
NETQUANT (Loading, analysis, output)
The quantification of NET formation by NETQUANT was automatically performed by the program. For this study, the NETQUANT software user manual version 1.21 was used for application (available in the supplementary material of the original manuscript [28]). NETQUANT separates the images, defines background levels, and compares the channels on its own. Nevertheless, previous arrangements were necessary. As described in Ref. [28], the program is based on fee-based MATLAB and requires a subset of toolboxes. Different kinds of bioformats were selectable to be run through the program. Here, .tiff and.nd2 format were used. The.nd2 images offered by the publishers were obtained from their website (https://nordlab.med.lu.se/?page_id=34) and loaded to the program as described by the respective authors. Here, it was important to have the images precisely saved as described in the manual, saving the images directly in a folder on the drive (e.g., C:\samples) and not in personal subfolders, which may result in error messages. The image set was processed with default settings and outcomes were similar to those described in the original publication. It was noticed that.tif files ideally had to be presented in a way where each channel of the same image was separately saved directly during image acquisition. Later generation of stacks or separate channel images from overlays in ImageJ led to errors or extremely prolonged and error prone processing. Settings were adjustable in terms of sensitivity for the respective channels as well as segmentation methods and minimal areas, whereas changing of segmentation method was not recommended by the authors and not used in our case. Since NETQUANT requires a set of unstimulated control images for each adjustment (as described in Chapter 2.7 of the NETQUANT user manual, which is published in supplementary material of the original publication [28], additional control images of unstimulated control neutrophils treated with RPMI medium alone were used for this purpose in the case of image set 1 (Supplementary Fig. 3).
Statistical analysis
Analysis was performed in Excel (Microsoft) and GraphPad Prism 9.1.0 (GraphPad Software, San Diego, CA, USA). Data are presented as mean ± SD, statistical analysis is indicated in the figure legends.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
Acknowledgments
The authors wish to thank Silke Akhdar for her excellent technical assistance.
TH is a PhD student at the University of Veterinary Medicine Hannover, Foundation, Hannover, Germany. This work is submitted in partial fulfillment of the requirement for the PhD.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e16982.
Contributor Information
Timo Henneck, Email: timo.henneck@tiho-hannover.de.
Maren von Köckritz-Blickwede, Email: Maren.von.Koeckritz-Blickwede@tiho-hannover.de.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
figs1.
figs2.
figs3.
figs4.
figs5.
References
- 1.Mayadas T.N., Cullere X., Lowell C.A. The multifaceted functions of neutrophils. Annu. Rev. Pathol. 2014;9:181–218. doi: 10.1146/annurev-pathol-020712-164023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D.S., Weinrauch Y., Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science (1979) 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 3.Fuchs T.A., Abed U., Goosmann C., Hurwitz R., Schulze I., Wahn V., Weinrauch Y., Brinkmann V., Zychlinsky A. Novel cell death program leads to neutrophil extracellular traps. JCB (J. Cell Biol.) 2007;176:231–241. doi: 10.1083/jcb.200606027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Papayannopoulos V., Metzler K.D., Hakkim A., Zychlinsky A. Neutrophil elastase and myeloperoxidase regulate the formation of neutrophil extracellular traps. JCB (J. Cell Biol.) 2010;191:677–691. doi: 10.1083/jcb.201006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neumann A., Berends E.T.M., Nerlich A., Molhoek E.M., Gallo R.L., Meerloo T., Nizet V., Naim H.Y., von Köckritz-Blickwede M. The antimicrobial peptide LL-37 facilitates the formation of neutrophil extracellular traps. Biochem. J. 2014;464:3–11. doi: 10.1042/BJ20140778. [DOI] [PubMed] [Google Scholar]
- 6.Kenny E.F., Herzig A., Krüger R., Muth A., Mondal S., Thompson P.R., Brinkmann V., von Bernuth H., Zychlinsky A. Diverse stimuli engage different neutrophil extracellular trap pathways. Elife. 2017;6 doi: 10.7554/eLife.24437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wardini A.B., Guimarães-Costa A.B., Nascimento M.T.C., Nadaes N.R., Danelli M.G.M., Mazur C., Benjamin C.F., Saraiva E.M., Pinto-da-Silva L.H. Characterization of neutrophil extracellular traps in cats naturally infected with feline leukemia virus. J. Gen. Virol. 2010;91:259–264. doi: 10.1099/vir.0.014613-0. [DOI] [PubMed] [Google Scholar]
- 8.Saitoh T., Komano J., Saitoh Y., Misawa T., Takahama M., Kozaki T., Uehata T., Iwasaki H., Omori H., Yamaoka S., Yamamoto N., Akira S. Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe. 2012;12:109–116. doi: 10.1016/j.chom.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 9.Urban C.F., Reichard U., Brinkmann V., Zychlinsky A. Neutrophil extracellular traps capture and kill Candida albicans and hyphal forms. Cell Microbiol. 2006;8:668–676. doi: 10.1111/j.1462-5822.2005.00659.x. [DOI] [PubMed] [Google Scholar]
- 10.McCormick A., Heesemann L., Wagener J., Marcos V., Hartl D., Loeffler J., Heesemann J., Ebel F. NETs formed by human neutrophils inhibit growth of the pathogenic mold Aspergillus fumigatus. Microb. Infect. 2010;12:928–936. doi: 10.1016/j.micinf.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 11.Fuchs T.A., Brill A., Duerschmied D., Schatzberg D., Monestier M., Myers D.D., Wrobleski S.K., Wakefield T.W., Hartwig J.H., Wagner D.D. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. USA. 2010;107:15880–15885. doi: 10.1073/pnas.1005743107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knight J.S., Kaplan M.J. Lupus neutrophils: “NET” gain in understanding lupus pathogenesis. Curr. Opin. Rheumatol. 2012;24:441–450. doi: 10.1097/BOR.0b013e3283546703. [DOI] [PubMed] [Google Scholar]
- 13.Hakkim A., Furnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V., Herrmann M., Voll R.E., Zychlinsky A. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. USA. 2010;107:9813–9818. doi: 10.1073/pnas.0909927107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Manzenreiter R., Kienberger F., Marcos V., Schilcher K., Krautgartner W.D., Obermayer A., Huml M., Stoiber W., Hector A., Griese M., Hannig M., Studnicka M., Vitkov L., Hartl D. Ultrastructural characterization of cystic fibrosis sputum using atomic force and scanning electron microscopy. J. Cyst. Fibros. 2012;11:84–92. doi: 10.1016/j.jcf.2011.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Papayannopoulos V., Staab D., Zychlinsky A. Neutrophil elastase enhances sputum solubilization in cystic fibrosis patients receiving dnase therapy. PLoS One. 2011;6:1–7. doi: 10.1371/journal.pone.0028526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ackermann M., Anders H.-J., Bilyy R., Bowlin G.L., Daniel C., de Lorenzo R., Egeblad M., Henneck T., Hidalgo A., Hoffmann M., Hohberger B., Kanthi Y., Kaplan M.J., Knight J.S., Knopf J., Kolaczkowska E., Kubes P., Leppkes M., Mahajan A., Manfredi A.A., Maueröder C., Maugeri N., Mitroulis I., Muñoz L.E., Narasaraju T., Naschberger E., Neeli I., Ng L.G., Radic M.Z., Ritis K., Rovere-Querini P., Schapher M., Schauer C., Simon H.-U., Singh J., Skendros P., Stark K., Stürzl M., van der Vlag J., Vandenabeele P., Vitkov L., von Köckritz-Blickwede M., Yanginlar C., Yousefi S., Zarbock A., Schett G., Herrmann M. Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ. 2021;28:3125–3139. doi: 10.1038/s41418-021-00805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Englert H., Rangaswamy C., Deppermann C., Sperhake J.-P., Krisp C., Schreier D., Gordon E., Konrath S., Haddad M., Pula G., Mailer R.K., Schlüter H., Kluge S., Langer F., Püschel K., Panousis K., Stavrou E.X., Maas C., Renné T., Frye M. Defective NET clearance contributes to sustained FXII activation in COVID-19-associated pulmonary thrombo-inflammation. EBioMedicine. 2021;67 doi: 10.1016/j.ebiom.2021.103382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hidalgo A. A NET-thrombosis axis in COVID-19. Blood. 2020;136:1118–1119. doi: 10.1182/blood.2020007951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppkes M., Knopf J., Naschberger E., Lindemann A., Singh J., Herrmann I., Stürzl M., Staats L., Mahajan A., Schauer C., Kremer A.N., Völkl S., Amann K., Evert K., Falkeis C., Wehrfritz A., Rieker R.J., Hartmann A., Kremer A.E., Neurath M.F., Muñoz L.E., Schett G., Herrmann M. Vascular occlusion by neutrophil extracellular traps in COVID-19. EBioMedicine. 2020;58 doi: 10.1016/j.ebiom.2020.102925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Middleton E.A., He X.-Y., Denorme F., Campbell R.A., Ng D., Salvatore S.P., Mostyka M., Baxter-Stoltzfus A., Borczuk A.C., Loda M., Cody M.J., Manne B.K., Portier I., Harris E.S., Petrey A.C., Beswick E.J., Caulin A.F., Iovino A., Abegglen L.M., Weyrich A.S., Rondina M.T., Egeblad M., Schiffman J.D., Yost C.C. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Buhr N., von Köckritz-Blickwede M. How neutrophil extracellular traps become visible. J Immunol Res. 2016;2016 doi: 10.1155/2016/4604713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Buhr N., von Köckritz-Blickwede M. Methods in Molecular Biology. Humana Press Inc.; 2020. Detection, visualization, and quantification of neutrophil extracellular traps (NETs) and NET markers; pp. 425–442. [DOI] [PubMed] [Google Scholar]
- 23.Choi J.J., Reich C.F., Pisetsky D.S. Release of DNA from dead and dying lymphocyte and monocyte cell lines in vitro. Scand. J. Immunol. 2004;60:159–166. doi: 10.1111/j.0300-9475.2004.01470.x. [DOI] [PubMed] [Google Scholar]
- 24.Metzler K.D., Goosmann C., Lubojemska A., Zychlinsky A., Papayannopoulos V. Myeloperoxidase-containing complex regulates neutrophil elastase release and actin dynamics during NETosis. Cell Rep. 2014;8:883–896. doi: 10.1016/j.celrep.2014.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rebernick R., Fahmy L., Glover C., Bawadekar M., Shim D., Holmes C.L., Rademacher N., Potluri H., Bartels C.M., Shelef M.A. DNA area and NETosis analysis (DANA): a high-throughput method to quantify neutrophil extracellular traps in fluorescent microscope images. Biol. Proced. Online. 2018;20:1–9. doi: 10.1186/s12575-018-0072-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coelho L.P., Pato C., Friães A., Neumann A., von Köckritz-Blickwede M., Ramirez M., Carriço J.A. Automatic determination of NET (neutrophil extracellular traps) coverage in fluorescent microscopy images. Bioinformatics. 2015;31:2364–2370. doi: 10.1093/bioinformatics/btv156. [DOI] [PubMed] [Google Scholar]
- 27.Brinkmann V., Goosmann C., Kühn L.I., Zychlinsky A. Automatic quantification of in vitro NET formation. Front. Immunol. 2012;3:1–8. doi: 10.3389/fimmu.2012.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohanty T., Sørensen O.E., Nordenfelt P. NETQUANT: automated quantification of neutrophil extracellular traps. Front. Immunol. 2018;8:1–10. doi: 10.3389/fimmu.2017.01999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Breda S.v., Vokalova L., Neugebauer C., Rossi S.W., Hahn S., Hasler P. Computational methodologies for the in vitro and in situ quantification of neutrophil extracellular traps. Front. Immunol. 2019;10:1–7. doi: 10.3389/fimmu.2019.01562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher J., Mohanty T., Karlsson C.A.Q., Khademi S.M.H., Malmström E., Frigyesi A., Nordenfelt P., Malmstrom J., Linder A. Proteome profiling of recombinant DNase therapy in reducing NETs and aiding recovery in COVID-19 patients. Mol. Cell. Proteomics. 2021;20 doi: 10.1016/J.MCPRO.2021.100113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silvin A., Chapuis N., Dunsmore G., Goubet A.G., Dubuisson A., Derosa L., Almire C., Hénon C., Kosmider O., Droin N., Rameau P., Catelain C., Alfaro A., Dussiau C., Friedrich C., Sourdeau E., Marin N., Szwebel T.A., Cantin D., Mouthon L., Borderie D., Deloger M., Bredel D., Mouraud S., Drubay D., Andrieu M., Lhonneur A.S., Saada V., Stoclin A., Willekens C., Pommeret F., Griscelli F., Ng L.G., Zhang Z., Bost P., Amit I., Barlesi F., Marabelle A., Pène F., Gachot B., André F., Zitvogel L., Ginhoux F., Fontenay M., Solary E. Elevated calprotectin and abnormal myeloid cell subsets discriminate severe from mild COVID-19. Cell. 2020;182:1401–1418.e18. doi: 10.1016/j.cell.2020.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fingerhut L., Dolz G., de Buhr N. What is the evolutionary fingerprint in neutrophil granulocytes? Int. J. Mol. Sci. 2020;21:1–37. doi: 10.3390/ijms21124523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tilley D.O., Abuabed U., Arndt U.Z., Schmid M., Florian S., Jungblut P.R., Brinkmann V., Herzig A., Zychlinsky A. Histone H3 clipping is a novel signature of human neutrophil extracellular traps. Elife. 2022;11 doi: 10.7554/ELIFE.68283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Janssen P., Tosi I., Hego A., Maréchal P., Marichal T., Radermecker C. Neutrophil extracellular traps are found in bronchoalveolar lavage fluids of horses with severe asthma and correlate with asthma severity. Front. Immunol. 2022;13 doi: 10.3389/FIMMU.2022.921077/FULL. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Radermecker C., Detrembleur N., Guiot J., Cavalier E., Henket M., d'Emal C., Vanwinge C., Cataldo D., Oury C., Delvenne P., Marichal T. Neutrophil extracellular traps infiltrate the lung airway, interstitial, and vascular compartments in severe COVID-19. J. Exp. Med. 2020;217 doi: 10.1084/JEM.20201012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Köckritz-Blickwede M., Chow O., Ghochani M., Nizet V. Visualization and functional evaluation of phagocyte extracellular traps. Methods Microbiol. 2010;37:139–160. doi: 10.1016/S0580-9517(10)37007-3. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.