Abstract
Circular RNAs (circRNAs) are endogenous, covalently circularised, non-protein-coding RNAs generated from back-splicing. Most circRNAs are very stable, highly conserved, and expressed in a tissue-, cell- and developmental stage-specific manner. circRNAs play a significant role in various biological processes, such as regulation of gene expression and protein translation via sponging of microRNAs and binding with RNA-binding proteins. circRNAs have become a topic of great interest in research due to their close link with the development of various diseases. Their high stability, conservation and abundance in body fluids make them promising biomarkers for many diseases. A growing body of evidence suggests that aberrant expression of circRNAs and their targets plays a crucial role in pulmonary vascular remodelling and pulmonary arterial hypertension (group 1) as well as other forms (groups 3 and 4) of pulmonary hypertension (PH). Here we discuss the roles and molecular mechanisms of circRNAs in the pathogenesis of pulmonary vascular remodelling and PH. We also highlight the therapeutic and biomarker potential of circRNAs in PH.
Shareable abstract (@ERSpublications)
This review evaluates the role and molecular mechanisms of circular RNAs in pulmonary hypertension with a view to research opportunities, clinical study and therapeutic potential https://bit.ly/3MTHrdd
Introduction
Pulmonary hypertension (PH) is a complex disorder with abnormally elevated mean pulmonary arterial pressure (mPAP). Pulmonary arterial hypertension (PAH), group 1 PH as classified by the World Health Organization, is a rare but fatal progressive cardiopulmonary condition defined by abnormally enhanced mPAP and elevated pulmonary vascular resistance (PVR), which ultimately causes right ventricular failure (RVF) and death. PAH can be heritable (family history of the disease), idiopathic (IPAH; without a family history, unknown cause), associated (linked to congenital heart disease, interstitial lung disease, autoimmune disease, etc.) or drug- and toxin-induced. The key histopathological manifestations of the disease include dysfunction of pulmonary arterial endothelial cells (PAECs), smooth muscle cells (SMCs), fibroblasts, immune cells and pericytes, which all collaboratively induce pulmonary vascular remodelling (e.g. endothelial dysfunction, medial hypertrophy, neointima formation, plexiform lesion formation, perivascular inflammation and fibrosis). These manifestations narrow the pulmonary artery lumen and increase PVR and mPAP that define the disease [1]. While the exact cause of the disease is unknown, several genetic/epigenetic factors (e.g. loss-of-function mutations in the bone morphogenic protein receptor 2 gene (BMPR2), dysregulation of microRNAs (miRNAs) and long non-coding RNAs (lncRNAs)), immune and inflammatory triggers, and altered metabolic and environmental factors (e.g. hypoxia, infections and toxins) contribute to the occurrence and progression of the disease [2, 3]. Despite existing medical treatment options that improve survival and quality of life, the disease remains incurable. Thus, there is an urgent need to develop novel, effective therapies to treat this disease.
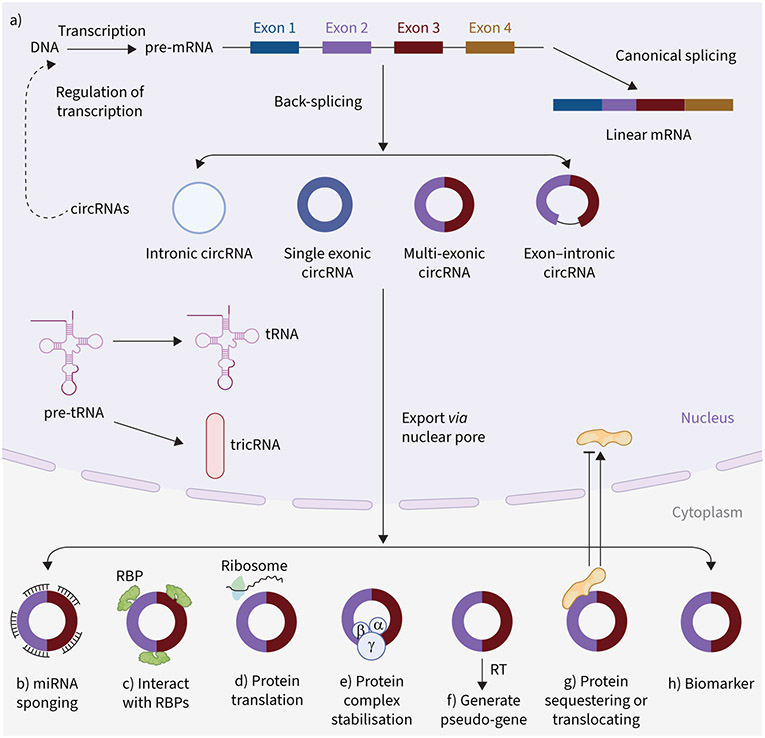
Circular RNAs (circRNAs) are single-stranded, covalently circularised endogenous non-coding RNAs lacking the 5′ cap or 3′ poly(A) tail. A non-canonical splicing process called back-splicing produces circRNAs from a single pre-mRNA of protein-coding genes, antisense transcripts, the intergenic spacer in tRNA or lncRNAs (figure 1) [4-6]. circRNAs were long thought of as transcriptional errors, but recent advances in RNA sequencing (RNAseq) technology and bioinformatics suggest that they play a significant role in health and disease. They are stable, highly conserved, and abundant in blood and tissues; their expression is greatly cell/tissue- and developmental stage-specific. circRNAs can be exonic (ecircRNA), exon–intronic (EIcircRNA) or intronic (ciRNA) (figure 1). Most circRNAs are ecircRNAs and predominantly exist in the cytoplasm. In contrast, ciRNAs and EIcircRNAs are generally located in the nucleus. Moreover, another type of circRNA, tricRNA, arises from cytoplasmic tRNA precursors [7]. Several factors regulate the biogenesis of circRNA, such as the canonical spliceosomal machinery, trans-factors (RNA-binding proteins (RBPs)) as well as cis-elements (intronic complementary sequences) [8, 9].
FIGURE 1.
Biogenesis and molecular mechanisms of circular RNAs (circRNAs) in health and disease. a) Generally, circRNAs (intronic, single exonic, multiple exonic and exon–intronic circRNAs) are produced by back-splicing from a single pre-mRNA of protein-coding genes, antisense transcripts or long non-coding RNAs. Another type of circRNA, tricRNA, is generated via a 3′-5′ phosphodiester bond between termini of introns that are removed from pre-tRNA by tRNA splicing enzymes. b–h) circRNAs can regulate transcription and translation, and also play important roles in different biological functions in health and disease. circRNAs can b) function as microRNA sponges, c) interact with RNA-binding proteins (RBPs) to regulate transcription of target mRNAs, d) be translated into peptides/proteins, e) stabilise protein complexes, f) produce pseudo-genes by reverse transcription (RT), g) translocate proteins to the nucleus or sequester them in the cytoplasm and h) serve as a molecular biomarker. Through these molecular mechanisms, circRNAs can influence different cellular physiology, such as proliferation and apoptosis.
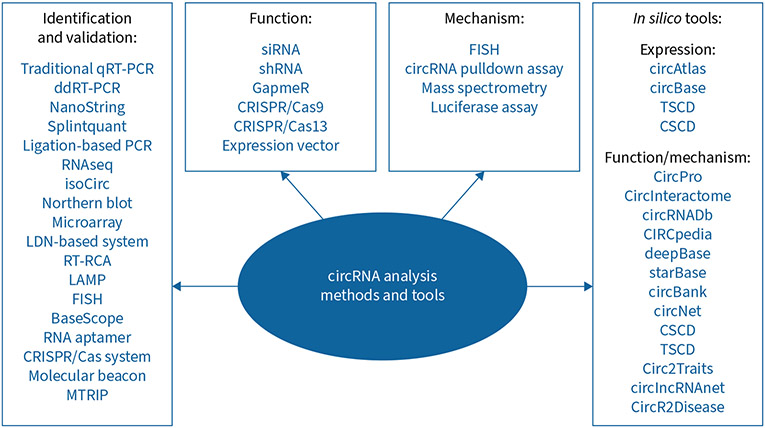
circRNAs regulate different physiological and biological processes, including proliferation, apoptosis, migration and angiogenesis, through various molecular mechanisms. miRNA sponging is a mechanism by which circRNAs competitively interact with miRNAs and affect their mRNA target expression. Further mechanisms include binding to RBPs and other proteins to regulate target gene expression, acting as a translational regulator and protein scaffold, and being associated with N6-methyladenosine (m6A) modification of RNAs (figure 1) [4, 10]. In the past few years, advances in RNAseq and bioinformatic approaches have allowed us to identify and characterise a long list of circRNAs that play a significant role in health and disease. For a detailed and precise understanding of circRNA, it is crucial to use tools and methods that accurately pinpoint circRNA sequence, length, abundance, subcellular localisation, function, mechanism, disease implications, biochemical interactions, biomarkers and therapeutic potential. A list of tools and methods for studying circRNAs is summarised in figure 2. Many circRNA annotation tools exist to identify circRNA from RNAseq data, including CIRCexplorer, CIRI2, DCC, Find_circ, CircSeq and UROBORUS [11]. These algorithm-based annotation tools identify circRNA mostly based on identifying back-splicing junction sites from RNAseq data. Additionally, circRNAs have also been identified using microarray techniques, but unlike RNAseq, this method uses probes to target the back-splice junction of known circRNAs. To validate circRNAs, in addition to traditional techniques, several state-of-the-art techniques are available. These include circRNA rolling circle amplification, which uses primers on the junction sequence of circRNAs to amplify their full-length sequence; isoCirc, a nanopore-based circRNAseq technique that can also capture full-length circRNA; and RNA imaging techniques, such as BaseScope and single-molecule RNA fluorescence in situ hybridisation (FISH) for fixed cells and fluorescent RNA aptamers for live cells [12, 13]. Recently, researchers have also developed a new imaging technique called circFISH that can simultaneously detect both linear RNAs and circRNAs [14]. Moreover, circRNAs may be visualised using molecular beacons, Cas-derived systems or multiply labelled tetravalent RNA imaging probes [12]. CRISPR/Cas9-, CRISPR/Cas13- and small interfering RNA (siRNA)/short hairpin RNA (shRNA)-mediated circRNA depletion approaches, the Cre-dependent conditional circRNA depletion approach, and circRNA overexpression plasmids can be used to investigate the function of circRNAs [15]. For mechanistic studies, bioinformatic prediction, luciferase reporter assays, RNA immunoprecipitation and RNA pulldown assays followed by mass spectrometry are conducted to explore circRNA–miRNA and circRNA–protein interactions. Finally, several online circRNA databases have been developed, such as circBase, CSCD (Cancer-Specific CircRNA Database), TSCD (Tissue-Specific CircRNA Database), circAtlas, CIRCpedia, CircInteractome, circBank, circRNADb and circNet, which are valuable in circRNA prediction, identification, evolutionary conservation, expression, subcellular localisation, function, mechanisms, protein-coding potential, design of silencing molecules and investigation of their associations with other molecules, such as miRNA response elements and RBPs (figure 2) [15]. Even though all of these databases assume circRNAs are functional in cells due to their interaction with cellular factors, they still need to be experimentally validated.
FIGURE 2.
Methods and tools for studying circular RNAs (circRNAs). qRT-PCR: quantitative reverse transcriptase-PCR; ddRT-PCR: droplet digital reverse transcriptase-PCR; RNAseq: RNA sequencing; RT-RCA: reverse transcriptase-rolling circle amplification; LAMP: loop-mediated isothermal amplification assay; FISH: fluorescence in situ hybridisation; MTRIP: multiply labelled tetravalent RNA imaging probes; siRNA: small interfering RNA; shRNA: short hairpin RNA.
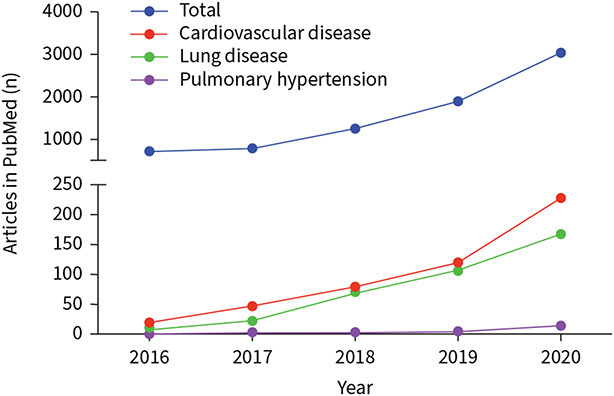
Dysregulation of circRNA expression occurs in many human diseases, including cancer, diabetes, and neurodegenerative and cardiovascular disorders [16, 17]. Recently, an expanded body of evidence suggests that circRNAs also play critical roles in pulmonary vascular remodelling and PAH [18-24]. circRNA research in PAH is an emergent field (figure 3). This review evaluates circRNAs in PH with a view to research opportunities, clinical study and therapeutic potential.
FIGURE 3.
Circular RNA (circRNA)-related publications in different disease conditions in PubMed: 2016–2020. Numbers of articles were retrieved from PubMed (September 2021) using key words searching “circRNA”, “circRNA AND cardiovascular disease”, “circRNA AND lung disease”, “circRNA AND pulmonary hypertension”, “circRNA AND pulmonary arterial hypertension”.
Overview of the role of circRNAs in PH
Several circRNA profiling studies of blood and lungs of PH patients, as well as rodent experimental PH models, have identified differentially expressed circRNAs associated with PH [21, 25-27]. For example, a prior study found 122 upregulated and 229 downregulated circRNAs in the blood of patients with chronic thromboembolic PH (CTEPH) (n=5) compared with healthy controls (n=5) using an Agilent circRNA chip [25]. By performing more advanced circRNA/miRNA/mRNA cross-talk analyses, the authors identified the hsa_circ_0002062/hsa-miR-942-5P/CDK6 and hsa_circ_0022342/hsa-miR-940/CRKL–ERBB signalling pathways that play a vital role in the development of CTEPH.
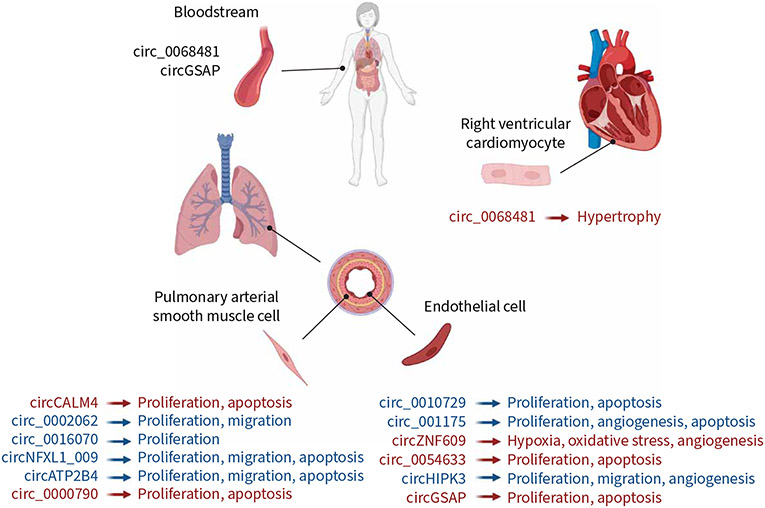
Zhou et al. [24] performed a circRNA microarray analysis in COPD patients with and without PH, and found that hsa_circ_0016070 expression was significantly increased in the COPD-PH condition. This study also showed that hsa_circ_0016070 promotes pulmonary arterial SMC (PASMC) proliferation by regulating miR-942/CCND1. Microarray analysis showed 23 upregulated and 41 downregulated circRNAs in the lung of hypoxia-induced PH mouse models [27]. Further quantitative PCR (qPCR) validation, miRNA prediction, and Gene Ontology and KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis identified the two most promising circRNAs, i.e. mmu_circRNA_018351 and mmu_circRNA_004592, to play essential roles in the pathogenesis of PH [27]. RNAseq analysis of the lungs of hypoxic PH mice by a different group, Zhang et al. [23], revealed 33 upregulated and 34 downregulated circRNAs, and among these 67 differentially expressed circRNAs, through qPCR validation and bioinformatic screening, and confirmed a novel circRNA, i.e. circCALM4, that regulated PASMC proliferation through the miR-337-3p/MYO10 signal transduction axis. Moreover, by carrying out a circRNA/miRNA/mRNA competitive endogenous RNA (ceRNA) regulatory network analysis in the lungs of hypoxic PH rat models, Xu et al. [21] identified that circ_002723, circ_016925, circ_020581 and circ_008021 regulated their downstream target genes by binding with miR-21 or miR-23a, potentially acting as ceRNA-mediated regulatory mechanisms and thereby contributing to the hypoxic PH. In addition, several candidate-based approaches have also revealed many circRNAs that play critical roles in PH [19, 28]. circRNAs regulate the function of the critical pulmonary vascular cells, such as in PASMCs and PAECs [18-20, 22-24, 28], and contribute to cardiac remodelling [29-34]. Therefore, it is likely that circRNAs play crucial roles in the pathogenesis of PH. In the following sections, we discuss the role and molecular mechanisms of circRNAs involved in the onset and progression of PH (figure 4).
FIGURE 4.
Circular RNAs (circRNAs) involved in biological processes in pulmonary hypertension (PH). Blue: in vitro findings; red: in vivo findings. circATP2B4, circ_0068481 and circGSAP findings are from studies related to pulmonary arterial hypertension; other circRNA findings represent other types of PH.
PASMC dysfunction
PASMCs are the predominant cell type involved in pulmonary vascular remodelling. PASMC dysfunction, including excessive proliferation and migration, and hypertrophy are considered the causes of pulmonary vascular remodelling and are instrumental for developing PH [2]. Therefore, addressing these dysfunctional PASMCs could be beneficial for treating PAH. Mounting evidence suggests that circRNAs are critical for regulating proliferation, apoptosis and migration of PASMCs in PH initiation [35].
circCALM4, generated from alternative splicing of the calmodulin 4 (CALM4) gene, is upregulated in the pulmonary vessels and lung of a murine model of hypoxic PH and hypoxia-exposed PASMCs [23]. Adenovirus-mediated knockdown of circCALM4 in the mouse improved hypoxia-induced PH features, as measured by right ventricular systolic pressure, Fulton Index (weight of the right ventricle/(left ventricle+ septum)), pulmonary artery acceleration time, pulmonary artery velocity–time integral and pulmonary vascular remodelling (SMC layer thickening) [23]. In addition, circCALM4 knockdown inhibited hypoxia-induced increases in PASMC proliferation through modulating cell cycle progression. Mechanistically, the circCALM4/miR-337-3p/MYO10 signalling axis regulates PASMC proliferation via regulating cell cycle progression. As the pathogenesis of PH has previously been linked to pyroptosis, another type of programmed cell death, which is different from apoptosis and necrosis [36], Jiang et al. [18] demonstrated that circCALM4 regulates hypoxia-induced pyroptosis of PASMCs potentially via the circCALM4/miR-124-3p/PDCD6 signalling axis. Together, these findings suggest that circCALM4 plays a significant role in the pathogenesis of hypoxic PH characterised by an increase in medial hypertrophy, and targeting circCALM4 could be an effective strategy to target PASMC proliferation and reduce medial hypertrophy in human PH.
The expression level of hsa_circ_0002062 was higher in the blood of patients with CTEPH and hypoxic PASMCs [20, 25]. In vitro studies revealed that hypoxia increased PASMC proliferation and migration, and was associated with induction of hsa_circ_0002062, while knockdown of hsa_circ_0002062 with shRNA reversed hypoxia-induced increases in proliferation and migration of PASMCs. Notably, the hsa_circ_0002062/hsa-miR-942-5P/CDK6 signalling pathway axis modulated PASMC proliferation [20]. Overall, these findings suggested that targeting hsa_circ_0002062 could effectively inhibit hypoxia-induced pulmonary vascular remodelling in PH. Considering the importance of hsa_circ_0002062 in hypoxic PH regulation, it would be critical to examine its downstream targets in detail to gain further insights into the mechanisms of hsa_circ_0002062 in the PH-regulating effect. A third circRNA, hsa_circ_0016070, has been reported to be significantly upregulated in COPD-PH patients compared with patients with COPD without PH [24], indicating that hsa_circ_0016070 may be associated with PH risk among COPD patients.
Additionally, hsa_circ_0016070 stimulated pulmonary vascular remodelling by increasing the proliferation of PASMCs through the miR-942/CCND1 pathway [24]. Analysing RNAseq data from blood collected from five patients with CTEPH and five healthy controls, Miao et al. [37] identified a different key circRNA/miRNA/mRNA regulatory axis: hsa_circ_0046159/miR-1226-3p/ATP2A2. Further qPCR validation also confirmed the regulatory axis in PH samples. One circRNA profiling study of an Arraystar circRNA microarray of COPD-PH patients identified a key circRNA, hsa_circNFXL1_009, that sponged with miR-29b-2-5p to potentially regulate proliferation, migration, apoptosis and potassium channel activation in PASMCs [26]. Another study suggested that increased levels of circATP2B4 in the serum of PAH patients were associated with SMC proliferation and migration. The subsequent in vitro studies demonstrated that circATP2B4 could sponge miR-223, increase the ATR serine/threonine kinase (ATR) protein level, promote PASMC proliferation/migration and decrease apoptosis [28]. In addition, a recent study by Sun et al. [38] showed that circGRM1 regulated PASMC functions through the GRM1/RAP1/ERK signalling axis. Recent studies by Steffes et al. [39] showed that neointima cells have a SMC origin. Therefore, identifying a novel way to control proliferation and induce apoptosis in smooth muscle-like cells might be beneficial to reduce medial hypertrophy but also vascular occlusions caused by neointima formation. Studies suggest that circRNAs can be a diagnostic biomarker and a therapeutic target for pulmonary vascular remodelling by regulating PASMC function in PH.
Endothelial cell dysfunction
The dysfunction of endothelial cells, such as abnormal proliferation and apoptosis, impaired and or inappropriate angiogenesis, and endothelial-to-mesenchymal transition, is believed to be one of the initial triggers for aberrant pulmonary vascular remodelling in PH [40]. Thus, treatments and studies targeting vascular endothelial dysfunction are of great therapeutic interest in treating and preventing aberrant vascular remodelling and PH. Increasing evidence suggests that circRNAs play a critical regulatory role in endothelial dysfunction in addition to their role in SMC biology.
circ_0010729 regulated proliferation and apoptosis of human umbilical vein endothelial cells (HUVECs) through targeting the miR-186/HIF1A axis [41]. circ_001175 stimulated cell proliferation and angiogenesis and attenuated apoptosis of HUVECs treated with high glucose, a risk factor for cardiovascular diseases and a positive regulator of endothelial dysfunction [42]. circZNF609, produced from zinc finger protein ZNF609, was upregulated in the fibrovascular membranes of diabetic patients and downregulated in the plasma of patients with coronary artery disease and hypertension [43]. Further cell culture studies suggested that circZNF609 protects against hypoxia and oxidative stress in HUVECs [43]. Moreover, knockdown of circZNF609 diminished retinal vessel loss and attenuated angiogenesis in pathological murine models. Mechanistically, circZNF609 regulated the vascular dysfunction via the miR-615-5p/MEF2A signalling axis [43].
Yang et al. [44] found that lipopolysaccharide (LPS)-treated rats and mouse pulmonary microvascular endothelial cells (PMVECs) showed higher expression levels of circ_0054633, interleukin-17A and tumour necrosis factor-α, and LPS treatment resulted in increased mouse PMVEC apoptosis and proliferation. Remarkably, circ_0054633 knockdown reversed these effects [44]. Another important circRNA that modulates endothelial dysfunction is circHIPK3, mostly generated from exon 2 of the homeodomain interacting protein kinase 3 gene (HIPK3), and abundant in the lung, liver and brain. circHIPK3 regulates cardiac vascular angiogenesis via miR-29a [33, 45]. circHIPK3 modulated retinal vascular function in diabetes mellitus through miR-30a function [46]. Hong et al. [47] have shown that circHIPK3 expression is higher in platelet-derived growth factor (PDGF)-stimulated human PAECs, and modulated PAEC proliferation, migration and angiogenesis potentially via the miR-328-3p/STAT3 signalling axis. However, further studies are needed to confirm these in vitro findings in PH pre-clinical animal models and clinical samples. Additional studies on mechanisms are also needed to clarify how other miRNAs and gene targets contribute to the action of this circRNA in PH.
Recently, Yuan et al. [22] performed whole-transcriptome sequencing of peripheral blood mononuclear cells (PBMCs) collected from IPAH patients and healthy controls, and identified a novel circRNA, circGSAP, that was lowly expressed in PBMCs of both discovery and validation cohorts of IPAH patients compared with controls. More importantly, circGSAP was also downregulated in lungs of patients with IPAH, SU5416/hypoxia- and monocrotaline-induced PH rats models, as well as hypoxia-treated PMVECs [22]. circGSAP overexpression, on the other hand, inhibited proliferation and promoted apoptosis of PMVECs under hypoxia [22]. However, this study did not investigate the mechanism how circGSAP regulates PAEC functions and therefore future studies are needed to better understand the role of circGSAP in PAEC biology. Together, circRNAs play a regulatory role in endothelial dysfunction and targeting the circRNAs could effectively modulate endothelial cell dysfunction in PH.
circRNAs in immune regulation in PH
circRNAs may play a key role in the immune dysregulation observed in PH [48-51]. Miao et al. [52] analysed a publicly available microarray dataset comprised of four IPAH, 14 CTEPH and four healthy controls to assess the molecular mechanisms of PH. Using protein–protein interaction analysis, they identified five immune-related genes that could be critical for developing PH: SLC2A3, C5AR1, CD83, PFN2 and ACKR1. Of these genes, MAN2A1 and SLC2A3 were associated with T-regulatory cells and interact with eosinophils. In the subsequent analysis, they identified several circRNA/miRNA/mRNA candidates, including circ_0022342/miR-503-5p/SLC2A3 and circ_0002062/miR-92b-3p/miR-92a-3p/MAN2A1 [52]. A previous study by the same research team also found that circ_0022342 and circ_002062 are downregulated in the peripheral blood of patients with CTEPH compared with healthy controls [25]. In summary, circ_0022342 and circ_0002062 may be relevant for the development and progression of CTEPH, possibly through immune regulation. However, although the authors correlate these findings with their previous study, verifying the accuracy of these results to a certain degree, further studies are needed to confirm these solely in silico predicted signalling pathways in animal models and human clinical samples.
circRNAs in right ventricular remodelling in PH
As a response to increased PVR and right ventricular pressure overload, several complex changes occur in the right ventricle, such as increased cardiomyocyte size, enhanced capillary angiogenesis, increased contractility and changes in the extracellular matrix composition [1, 53-56]. These changes ultimately lead to right ventricular dysfunction and failure, and eventually death. While different mechanisms and stimuli, including neurohormonal, metabolic and angiogenic factors, are involved in right ventricular remodelling in PH, no therapy directly targets the right ventricle and RVF remains the predominant cause of death in PH [1]. In general, currently available treatments aim to reduce PVR, thereby relieving symptoms and slowing clinical disease progression. However, as current medical therapies to lower the PVR long term are unsatisfactory, there is an unmet medical need to identify new targets that can help improve right ventricular remodelling and help the right ventricle adapt to a higher afterload. Emerging evidence suggests that circRNAs are associated with right ventricular remodelling [30]. For example, in a recent interesting study by Guo and Liu [30], expression of circ_0068481 and miR-646/miR-570/miR-885 was measured by qPCR in controls with neither PAH nor right ventricular hypertrophy (RVH) (n=52), patients with PAH and RVH (n=38), and patients with PAH yet without RVH (n=42). The study’s authors did not mention how they divided patients into the RVH(+) and RVH(−) groups, either by echocardiography or magnetic resonance imaging. Significantly higher expression levels of circ_0068481 and lower levels of miR-646 and miR-570, but no changes in miR-885 expression, were found in the patients with PAH with RVH compared with the patients with PAH without RVH and control patients. Remarkably, using a receiver operating characteristic curve analysis, the authors demonstrated that expression of circ_0068481, miR-646 and miR-570 predicted RVH in patients with PAH, suggesting that these non-coding RNAs could serve as a potential diagnostic biomarker for RVH in PAH. Subsequently, in vitro luciferase assay and alterations of circ_0068481 studies identified a regulatory network of circ_0068481/miR646/miR-570/miR-885 in the AC16 human cardiomyocyte cell line [30]. Another recent study identified dysregulation of several circRNAs (TTN, AKAP9, CACNA1C, CDC14B, SPECC1L, DENNDRA, DENNDRC, NAP1L4 and ZHX1) in right ventricular tissues of human RVH due to high-pressure load (compensated RVH, no RVF, non-failing heart (tetralogy of Fallot/pulmonary stenosis) subjects versus controls with ventricular septal defect (VSD) without pulmonary stenosis) [57]. using in silico analysis, the authors also predicted miRNA/mRNA targets of these dysregulated circRNAs. However, the study had several limitations. First, the authors utilised VSD tissues as control, which is not ideal. Second, the authors did not validate the identified circRNA/miRNA/mRNA signalling pathways in their samples. Thus, further studies are needed to confirm these findings in appropriate samples. It would also be interesting to investigate whether dysregulation of these circRNAs and the predicted signalling pathway axis in fact occurs also in the right ventricle of PAH patients. Several other circRNAs have been identified to play a role in cardiac remodelling, but none of the studies investigated the role of the circRNAs in the context of PAH [29, 31, 32, 34, 58, 59]. circ_000203 promoted cardiac hypertrophy through miR-140-3p/miR-26b-5p/GATA4 [31]. circYAP inhibited cardiac fibrosis by regulating actin polymerisation [34]. Heart-related circRNA (HRCR) inhibited cardiac hypertrophy and heart failure by the HRCR/miR-223/ARC axis [32]. Lim et al. [58] demonstrated a highly abundant circRNA, circSLC8A1, in cardiomyocytes attenuates pressure overload-induced hypertrophy in mice potentially through regulating miR-133/CTGF/SRF/ADCY6/ADRB1. The circ_0068655/miR-498/PAWR axis promoted apoptosis in cardiomyocytes [29]. A recent study by Du et al. [60] showed that circNLGN plays a significant role in cardiac fibrosis and heart failure in cardiac overload-induced remodelling. Together, these findings indicate that circRNAs influence cardiac remodelling. Therefore, circRNAs might be a promising tool to identify pathways and candidates that could improve adaptive cardiac remodelling, such as RVH in the context of PH.
Therapeutic potential of circRNAs in PH
In recent years, researchers have made significant efforts to develop RNA-based therapeutics for clinical application, using siRNAs and antisense oligonucleotides, several of which have been approved by the US Food and Drug Administration [61]. As therapeutic approaches, siRNAs, miRNA mimics or inhibitors (antagomiRs), CRISPR-mediated RNA/gene manipulation techniques and the delivery of RNA/genes to the body by adenovirus-mediated plasmids are becoming increasingly popular. The therapeutic approaches targeting non-coding RNAs, especially lncRNAs and miRNAs, have provided promising results for cancers, as well as many other diseases [61]. circRNAs are an emerging yet under-studied therapeutic target in treating various diseases, including cardiovascular disease [17]. circRNAs have attractive characteristics, such as expression stability and stage- and cell-type specificity. There is evidence that some circRNAs can be directly translated into proteins [62, 63]. Furthermore, the molecular mechanisms and biological functions of circRNAs and lncRNAs usually function as upstream regulators of miRNAs [64, 65]. All these characteristics make circRNAs an exciting research subject.
circRNAs play a significant role in PH (table 1). The majority of current circRNA PH studies have been conducted in vitro and only a few studies have translated their findings into animal models or validated them in PH clinical samples. Furthermore, to the best of our knowledge, no clinical trials have investigated the role of circRNAs in PH. A previous study showed increased expression of circCALM4 in the lung of hypoxia-induced PH in mice and PASMCs exposed to hypoxia [18]. Notably, on a pre-clinical level, targeting circCALM4 with intranasal administration of adenoviral particles carrying circCALM4 shRNA ameliorates hypoxia-induced PH features in mice potentially through inhibiting the pyroptosis mechanism [18].
TABLE 1.
List of circular RNAs (circRNAs) involved in pulmonary hypertension (PH)
| circRNA | Nature of study |
Expression in PH | Regulation | Function/mechanism | Reference |
|---|---|---|---|---|---|
| circCALM4 | In vitro, in vivo | Lung of HPH mice | ↑ | Regulated PASMC proliferation through miR-337-3p/MYO10 signalling axis; regulated hypoxia-induced PASMC pyroptosis by circCALM4/miR-124-3p/PDCD6 signalling | [18, 23] |
| hsa_circ_0016070 | In vitro, clinical | Lung of COPD±PAH patients | ↑ | Promoted PASMC proliferation through miR-942/CCND1 | [24] |
| circRNA CDR1 | In vitro | Hypoxic PASMCs | ↑ | Hypoxia-induced human PASMC osteoblastic differentiation and calcification were regulated via CDR1as/miR-7-5p/CNN3 and CAMK2D regulatory axis | [19] |
| circATP2B4 | In vitro, clinical | Serum of PAH patients and hypoxia-exposed PASMCs | ↑ | Stimulated hypoxia-induced proliferation and migration of PASMC by miR-223/ATR axis | [28] |
| mmu_circ_0000790 | In vitro, in vivo | Lung of HPH mouse models and hypoxic PASMCs | ↑ | Promoted pulmonary vascular remodelling in HPH mice via miR-374c-mediated FOXC1 | [66] |
| hsa_circ_0046159 | In vitro, clinical | Blood of CTEPH patients | ↑ | hsa_circ_0046159/miR-1226-3p/ATP2A2 axis; the mechanism was predicted in silico but not validated experimentally | [37] |
| hsa_circNFXL1_009 | In vitro, clinical | Blood of COPD-PAH patients and hypoxic PASMCs | ↓ | Regulated proliferation, migration, apoptosis and potassium channel activation of PASMCs | [26] |
| circ_0068481 | In vitro, clinical | Serum of IPAH patients | ↑ | Had higher sensitivity and specificity for predicting IPAH, correlated with haemodynamic parameters, poor clinical outcomes, discriminated survivors from non-survivors | [71] |
| circHIPK3 | In vitro | PDGF-activated PAECs | ↑ | Regulated PDGF-induced PAEC proliferation, migration and angiogenesis via circHIPK3/miR-328-3p/STAT3 axis | [47] |
| circGSAP | In vitro, in vivo, clinical | PBMCs and lung of IPAH patients, lung tissues of monocrotaline- and SU5416/hypoxia-induced PAH rats, and hypoxic PMECs | ↓ | Correlated with occurrence and poor outcomes of IPAH; overexpression stimulated hypoxia-induced apoptosis and decreased proliferation in PMVECs | [22] |
| hsa_circ_0002062 | In vitro, clinical | Blood of CTEPH patients and hypoxic PASMCs | ↑ | Promoted hypoxia-induced PASMC proliferation by hsa-miR-942-5p/CDK6 signalling axis | [20] |
| hsa_circ_0022342 | Clinical | Blood of CTEPH patients | ↑ | hsa_circ_0022342/hsa-miR-940/CRKL–ERBB signalling pathways; the mechanism was predicted in silico but not validated experimentally | [25] |
| circ_002723, circ_008021 | In vivo | Lung of HPH rats | ↑ | Potential mechanisms regulated by sponging miR-23a to control downstream target | [21] |
| circ_020581 | In vivo | Lung of HPH rats | ↓ | Potential mechanisms regulated by sponging miR-21 to control downstream target | [21] |
| m6A circXpo6 | In vitro, in vivo | Lung of HPH rats, hypoxic PASMCs and PAECs | ↓ | [77] | |
| m6A circTmtc3 | In vitro, in vivo | Lung of HPH rats, hypoxic PASMCs and PAECs | ↓ | [77] |
PAH: pulmonary arterial hypertension; HPH: hypoxic pulmonary hypertension; PASMC: pulmonary arterial smooth muscle cell; hsa_: Homo sapiens; mmu_: Mus musculus; CTEPH: chronic thromboembolic pulmonary hypertension; IPAH: idiopathic pulmonary arterial hypertension; PDGF: platelet-derived growth factor; PAEC: pulmonary arterial endothelial cell; PBMC: peripheral blood mononuclear cell; PMVEC: pulmonary microvascular endothelial cell; m6A; N6-methyladenosine.
Similarly, another pre-clinical study revealed a significant upregulation of mmu_circ_0000790 in pulmonary arteries of hypoxia-induced PH in mice and hypoxic mouse PASMCs, and targeting the circRNA with siRNA disrupted the hypoxia-induced PH manifestations such as haemodynamic parameters and pulmonary vascular remodelling in mice [66]. Although targeting circRNAs with siRNA is a convenient, time-saving and effective strategy to improve disease outcomes, several challenges limit their clinical translation, such as poor absorption into cells and off-target effects as administration of these exogenous RNA molecules can trigger an innate immune response. Chemical modification of siRNAs could overcome these barriers, making them potentially suitable for clinical translation [67]. In contrast to targeting upregulated circRNAs, a recent study demonstrated lower levels of circGSAP in IPAH patients and animal and cell culture PAH models, and overexpression via plasmid delivery of the circRNA resulted in inhibition of cell proliferation and promotion of apoptosis in PMVECs cultured under hypoxic conditions [22]. However, these in vitro findings need to be shown in vivo as well as in patient samples to confirm whether targeting the circRNA improves pulmonary vascular remodelling and PAH or not. In addition, targeting circRNAs with expression vehicle or viral vectors could produce unknown side-effects. There are thus still many challenges to overcome in this field. A deeper understanding of circRNA functional mechanisms and how to target circRNAs in vivo will play a crucial role in advancing the clinical potential of circRNA-based therapeutics in the coming years.
Biomarker potential of circRNAs in PAH
To date, a wealth of potential PH biomarkers have been described, such as N-terminal pro-brain natriuretic peptide (NT-proBNP), endothelin peptides, brain natriuretic peptide (BNP), galectin 3, ST-2, osteopontin and troponins [68]. NT-proBNP and BNP are two biomarkers routinely used in clinical trials associated with PH prognosis and mortality. Most biomarkers have not been rigorously tested as disease-specific surrogate markers for assessing the severity, prognosis or clinical response to therapy in PH [68]. Thus, identifying and applying novel biomarkers to diagnose and monitor PAH patients, provide prognostic information, guide therapy, and serve as surrogate end-points is a desirable goal. circRNAs exhibit several key characteristics that make them potentially ideal biomarkers for various diseases, including cancers, diabetes, and neurodegenerative and cardiovascular disorders [69, 70]. The features of circRNAs include that they are stable due to their closed-loop structures, highly conserved, with tissue- and developmental stage-specific expression patterns, and abundantly found in the extracellular spaces and body fluids such as blood, saliva, exosomes and urine, and are therefore easily detectable with high sensitivity and specificity. circRNAs have thus great potential as biomarkers in PH. For example, in the serum of 82 IPAH patients, increased levels of circ_0068481 compared with 82 normal controls were specific and sensitive to predicting PAH [71]. In addition, serum levels of circ_0068481 significantly correlated with clinicopathological manifestations of IPAH, such as 6-min walk distance (6MWD), heart function, RVF and death.
Furthermore, compared with IPAH patients without RVF (n=53), circ_0068481 levels were significantly higher in patients with RVF (n=29) and were able to predict RVF [71]. Serum circ_0068481 levels were also higher in patients who died with IPAH and could predict poorer clinical outcomes. While these findings suggest that circ_0068481 has potential as a non-invasive diagnostic and prognostic biomarker for PAH, the results are subject to several limitations. In this study, it is unclear whether circ_0068481 drives IPAH prognosis or is the consequence of the disease, or whether treatments had any effect on the expression of circ_0068481 that interfered with disease prognosis. Also, there was a relatively shorter follow-up period and a small sample size to assess the circRNAs and IPAH prognosis. The prognostic capability of circ_0068481 in IPAH will need to be confirmed by large-scale prospective studies. A different study recently showed that expression of circ_0068481, miR-570 and miR-646 could be a potential diagnostic biomarker to predict RVH in patients with PAH [30].
Another potential circRNA biomarker in PH, circGSAP, was lowly expressed in the PBMCs of IPAH patients; circGSAP expression was positively associated with cardiac output, cardiac index and 6MWD, while negatively associated with NT-proBNP, and discriminated survivor from non-survivor IPAH patients using Cox regression and receiver operating characteristic curve analysis [22]. Together, these findings suggest that the circGSAP expression levels could predict the occurrence of PAH and the survival of patients with IPAH. There are, however, several limitations to consider when interpreting these results. The discovery cohorts were small and recruited from one centre. However, the authors reduced the bias by validating their findings on a larger cohort. Collectively, these findings suggest that circRNAs could serve as biomarkers for PAH diagnosis and prognosis (table 2).
TABLE 2.
Circular RNAs (circRNAs) that have biomarker potential in pulmonary arterial hypertension (PAH)
| circRNA | Expression level | Study size | Biomarker role | Biomarker type | Reference |
|---|---|---|---|---|---|
| circ_0068481 | ↑ in serum of IPAH patients | Healthy controls (n=82); IPAH patients (n=82) | Had higher specificity and sensitivity for predicting IPAH; associated with poor clinical outcomes | Diagnostic and prognostic | [71] |
| circ_0068481 | ↑ in serum of PAH patients | PAH(−) RVH(−) (n=52); PAH(+) RVH(−) (n=42); PAH(+) RVH(+) (n=38) | Correlated with prediction of RVH diagnosis in PAH patients | Diagnostic | [30] |
| circGSAP | ↓ in PBMCs of IPAH patients | Healthy controls (n=30); IPAH patients (n=60) | Correlated with occurrence and poor outcomes of IPAH | Diagnostic and prognostic | [22] |
IPAH: idiopathic pulmonary arterial hypertension; RVH: right ventricular hypertrophy; PBMC: peripheral blood mononuclear cell.
Concluding remarks and future roadmap
circRNAs play essential roles in regulating different signalling pathways that modulate key pulmonary vascular cell functions (mostly tested in PAECs and PASMCs), mediating pulmonary vascular remodelling and PH. To the best of our knowledge, there are no studies investigating circRNAs in fibroblasts, pericytes and immune cells in PH. Future studies are needed to explore the role of circRNAs in these cell types in PH. Although several circRNAs play an important role in cardiomyocyte hypertrophy, fibrosis and right ventricular remodelling, none of the studies were conducted in the right ventricle under pressure, as seen in PH. As these features are common in PH patients, it is crucial to assess the role of circRNAs in cardiac remodelling in PH models and clinical samples. Emerging evidence also advocates that targeting circRNAs could be a promising approach to improve disease outcomes.
circRNAs play a crucial role in metabolic regulation in several diseases, including cancers [72, 73]. Moreover, as PH is associated with metabolic dysfunction [2], it is critical to investigate the role of circRNAs in metabolic regulation in PH. Future studies will need to address other research questions as well. These include, but are not limited to: Are there yet unknown factors influencing the production of circRNAs in PH? What are other functions and mechanisms of circRNAs that remain unknown in PH? What role do circRNAs play in regulating well-known molecular pathways involved in PAH, such as BMP/TGF-β2, WNT, mTOR, NOTCH, P53, HIF1, HIPPO, ERK, PDGF, EGFR and Rho-kinase signalling pathways [2, 74-76]?
As mentioned earlier, a clear limitation of the currently available knowledge of the role of circRNA in PH is that most data are derived from in vitro studies of pulmonary vascular cells. While the role of circRNAs in PASMCs has been relatively well documented, there are significantly less data on PAEC biology. Thus, we summarise some of that is known of circRNAs and endothelial cells from other vascular beds (most frequently HUVECS), that potentially some findings could be extrapolated from other vascular beds.
Aberrant expression of circRNAs has been implicated in a few in vivo pre-clinical models of PH, the majority of which are chronically hypoxic mice. Very little data to demonstrate the involvement of circRNAs in the development and progression of PH are derived from more robust experimental PH models, such as monocrotaline- and SU5416/hypoxia-induced PH, that more closely recapitulate the pulmonary vasculopathy characteristic of human PAH. Furthermore, the only attempts to therapeutically target circRNAs to reverse PH have been undertaken in a mouse model of chronically hypoxic PH by Zhang et al. [23]. No other studies have addressed the utility of targeting circRNAs to reverse PAH in the more severe models of pre-clinical PAH.
Moreover, current clinical observations have attempted to characterise circRNA expression in human PH, most notably CTEPH (group 4), COPD-PH (group 3) and IPAH (group 1). Yet, importantly, while the studies reviewed here report altered expression relative to typically healthy controls and, in some cases, associations with mortality, almost all the studies are limited by small numbers, cross-sectional design, single institution experiences, no external validation and insufficient numbers to appropriately address recognised covariates when building prognostic models. Future studies should focus on reproducible validation of biomarker datasets, larger studies to address confounding variables as they relate to the prognostic utility of these markers, human data addressing temporality (i.e. most studies compare disease with healthy controls, which has little clinical utility, and do not compare at-risk patients at different stages of the disease, longitudinal changes in biomarkers or response to therapy) and description of the tissue expression of circRNAs in diseased human tissues. Thus, the current understanding of any potential role for circRNAs in the pathogenesis of PH or PAH, their utility in diagnosis and prognosis, and finally, the ability to target them therapeutically are clearly at their infancy.
In summary, circRNAs have emerged as a promising yet under-studied, additional level of gene and pathway regulation in many diseases, and hold promise in becoming valuable tools in the diagnosis, prognosis and potential treatment of PH.
Support statement:
This work was supported by grants from the National Institutes of Health (R01 HL128734), Dept of Defense (PR161256) and Vera Moulton Wall Center for Pulmonary Vascular Diseases. Funding information for this article has been deposited with the Crossref Funder Registry.
Footnotes
Conflict of interest: The authors have no conflicts of interest to declare.
References
- 1.Ali MK, Ichimura K, Spiekerkoetter E. Promising therapeutic approaches in pulmonary arterial hypertension. Curr Opin Pharmacol 2021; 59: 127–139. [DOI] [PubMed] [Google Scholar]
- 2.Humbert M, Guignabert C, Bonnet S, et al. Pathology and pathobiology of pulmonary hypertension: state of the art and research perspectives. Eur Respir J 2019; 53: 1801887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dannewitz Prosseda S, Ali MK, Spiekerkoetter E. Novel advances in modifying BMPR2 signaling in PAH. Genes 2020; 12; 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci 2019; 26: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quan G, Li J. Circular RNAs: biogenesis, expression and their potential roles in reproduction. J Ovarian Res 2018; 11: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu KS, Pan F, Mao XD, et al. Biological functions of circular RNAs and their roles in occurrence of reproduction and gynecological diseases. Am J Transl Res 2019; 11: 1–15. [PMC free article] [PubMed] [Google Scholar]
- 7.Li J, Xu Q, Huang ZJ, et al. CircRNAs: a new target for the diagnosis and treatment of digestive system neoplasms. Cell Death Dis 2021; 12: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X, Yang L, Chen LL. The biogenesis, functions, and challenges of circular RNAs. Mol Cell 2018; 71: 428–442. [DOI] [PubMed] [Google Scholar]
- 9.Starke S, Jost I, Rossbach O, et al. Exon circularization requires canonical splice signals. Cell Rep 2015; 10: 103–111. [DOI] [PubMed] [Google Scholar]
- 10.Liu C, Li N, Dai G, et al. A narrative review of circular RNAs as potential biomarkers and therapeutic targets for cardiovascular diseases. Ann Transl Med 2021; 9: 578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L, Wang C, Sun H, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform 2021; 22: 1706–1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bejugam PR, Das A, Panda AC. Seeing is believing: visualizing circular RNAs. Noncoding RNA 2020; 6: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xin R, Gao Y, Gao Y, et al. isoCirc catalogs full-length circular RNA isoforms in human transcriptomes. Nat Commun 2021; 12: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koppula A, Abdelgawad A, Guarnerio J, et al. CircFISH: a novel method for the simultaneous imaging of linear and circular RNAs. Cancers 2022; 14: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang X, Ren H, Guo M, et al. Review on circular RNAs and new insights into their roles in cancer. Comput Struct Biotechnol J 2021; 19: 910–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee ECS, Elhassan SAM, Lim GPL, et al. The roles of circular RNAs in human development and diseases. Biomed Pharmacother 2019; 111: 198–208. [DOI] [PubMed] [Google Scholar]
- 17.Altesha MA, Ni T, Khan A, et al. Circular RNA in cardiovascular disease. J Cell Physiol 2019; 234: 5588–5600. [DOI] [PubMed] [Google Scholar]
- 18.Jiang Y, Liu H, Yu H, et al. Circular RNA Calm4 regulates hypoxia-induced pulmonary arterial smooth muscle cells pyroptosis via the Circ-Calm4/miR-124-3p/PDCD6 axis. Arterioscler Thromb Vasc Biol 2021; 41: 1675–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma C, Gu R, Wang X, et al. circRNA CDR1as promotes pulmonary artery smooth muscle cell calcification by upregulating CAMK2D and CNN3 via sponging miR-7-5p. Mol Ther Nucleic Acids 2020; 22: 530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Y, Tan X, Wu Y, et al. hsa_circ_0002062 promotes the proliferation of pulmonary artery smooth muscle cells by regulating the hsa-miR-942-5p/CDK6 signaling pathway. Front Genet 2021; 12: 673229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu SL, Deng YS, Liu J, et al. Regulation of circular RNAs act as ceRNA in a hypoxic pulmonary hypertension rat model. Genomics 2021; 113: 11–19. [DOI] [PubMed] [Google Scholar]
- 22.Yuan P, Wu WH, Gong SG, et al. Impact of circGSAP in peripheral blood mononuclear cells on idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med 2021; 203: 1579–1583. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Li Y, Qi J, et al. Circ-calm4 serves as an miR-337-3p sponge to regulate Myo10 (myosin 10) and promote pulmonary artery smooth muscle proliferation. Hypertension 2020; 75: 668–679. [DOI] [PubMed] [Google Scholar]
- 24.Zhou S, Jiang H, Li M, et al. Circular RNA hsa_circ_0016070 is associated with pulmonary arterial hypertension by promoting PASMC proliferation. Mol Ther Nucleic Acids 2019; 18: 275–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miao R, Wang Y, Wan J, et al. Microarray expression profile of circular RNAs in chronic thromboembolic pulmonary hypertension. Medicine 2017; 96: e7354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin X, Xu Y, Guo M, et al. hsa_circNFXL1_009 modulates apoptosis, proliferation, migration, and potassium channel activation in pulmonary hypertension. Mol Ther Nucleic Acids 2021; 23: 1007–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang J, Zhu MC, Kalionis B, et al. Characteristics of circular RNA expression in lung tissues from mice with hypoxia-induced pulmonary hypertension. Int J Mol Med 2018; 42: 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo J, Zhang L, Lian L, et al. CircATP2B4 promotes hypoxia-induced proliferation and migration of pulmonary arterial smooth muscle cells via the miR-223/ATR axis. Life Sci 2020; 262: 118420. [DOI] [PubMed] [Google Scholar]
- 29.Chai Q, Zheng M, Wang L, et al. Circ_0068655 promotes cardiomyocyte apoptosis via miR-498/PAWR axis. Tissue Eng Regen Med 2020; 17: 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo HM, Liu ZP. Up-regulation of circRNA_0068481 promotes right ventricular hypertrophy in PAH patients via regulating miR-646/miR-570/miR-885. J Cell Mol Med 2021; 25: 3735–3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Xu JD, Fang XH, et al. Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR-26b-5p and miR-140-3p binding to Gata4. Cardiovasc Res 2020; 116: 1323–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang K, Long B, Liu F, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J 2016; 37: 2602–2611. [DOI] [PubMed] [Google Scholar]
- 33.Wang Y, Zhao R, Shen C, et al. Exosomal circHIPK3 released from hypoxia-induced cardiomyocytes regulates cardiac angiogenesis after myocardial infarction. Oxid Med Cell Longev 2020; 2020: 8418407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu N, Xu J, Du WW, et al. YAP circular RNA, circYap, attenuates cardiac fibrosis via binding with tropomyosin-4 and gamma-actin decreasing actin polymerization. Mol Ther 2021; 29: 1138–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang J, Zhu M, Pan J, et al. Circular RNAs: a rising star in respiratory diseases. Respir Res 2019; 20: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M, Xin W, Yu Y, et al. Programmed death-ligand 1 triggers PASMCs pyroptosis and pulmonary vascular fibrosis in pulmonary hypertension. J Mol Cell Cardiol 2020; 138: 23–33. [DOI] [PubMed] [Google Scholar]
- 37.Miao R, Gong J, Zhang C, et al. hsa_circ_0046159 is involved in the development of chronic thromboembolic pulmonary hypertension. J Thromb Thrombolysis 2020; 49: 386–394. [DOI] [PubMed] [Google Scholar]
- 38.Sun S, Kong Q, Cai Z, et al. circGrm1 promotes pulmonary artery smooth muscle cell proliferation and migration via suppression of GRM1 expression by FUS. Int J Mol Med 2021; 48: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Steffes LC, Froistad AA, Andruska A, et al. A Notch3-marked subpopulation of vascular smooth muscle cells is the cell of origin for occlusive pulmonary vascular lesions. Circulation 2020; 142: 1545–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kurakula K, Smolders VFED, Tura-Ceide O, et al. Endothelial dysfunction in pulmonary hypertension: cause or consequence? Biomedicines 2021; 9: j5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dang RY, Liu FL, Li Y. Circular RNA hsa_circ_0010729 regulates vascular endothelial cell proliferation and apoptosis by targeting the miR-186/HIF-1alpha axis. Biochem Biophys Res Commun 2017; 490: 104–110. [DOI] [PubMed] [Google Scholar]
- 42.Pei X, Ye S, Jin G, et al. Overexpression of circRNA-001175 promotes proliferation and angiogenesis and inhibits apoptosis of the human umbilical vein endothelial cells (HUVECs) induced by high glucose. Int J Clin Exp Pathol 2018; 11: 359–366. [PMC free article] [PubMed] [Google Scholar]
- 43.Liu C, Yao MD, Li CP, et al. Silencing of circular RNA-ZNF609 ameliorates vascular endothelial dysfunction. Theranostics 2017; 7: 2863–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang CL, Yang WK, He ZH, et al. Quietness of circular RNA circ_0054633 alleviates the inflammation and proliferation in lipopolysaccharides-induced acute lung injury model through NF-kappaB signaling pathway. Gene 2021; 766: 145153. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y, Zhao R, Liu W, et al. Exosomal circHIPK3 released from hypoxia-pretreated cardiomyocytes regulates oxidative damage in cardiac microvascular endothelial cells via the miR-29a/IGF-1 pathway. Oxid Med Cell Longev 2019; 2019: 7954657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shan K, Liu C, Liu BH, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 2017; 136: 1629–1642. [DOI] [PubMed] [Google Scholar]
- 47.Hong L, Ma X, Liu J, et al. Circular RNA-HIPK3 regulates human pulmonary artery endothelial cells function and vessel growth by regulating microRNA-328-3p/STAT3 axis. Pulm Circ 2021; 11: 20458940211000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jafri S, Ormiston ML. Immune regulation of systemic hypertension, pulmonary arterial hypertension, and preeclampsia: shared disease mechanisms and translational opportunities. Am J Physiol Regul Integr Comp Physiol 2017; 313: R693–R705. [DOI] [PubMed] [Google Scholar]
- 49.Rabinovitch M, Guignabert C, Humbert M, et al. Inflammation and immunity in the pathogenesis of pulmonary arterial hypertension. Circ Res 2014; 115: 165–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L, Fu J, Zhou Y. Circular RNAs and their emerging roles in immune regulation. Front Immunol 2018; 9: 2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li I, Chen YG. Emerging roles of circular RNAs in innate immunity. Curr Opin Immunol 2021; 68: 107–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Miao R, Dong X, Gong J, et al. Possible immune regulation mechanisms for the progression of chronic thromboembolic pulmonary hypertension. Thromb Res 2021; 198: 122–131. [DOI] [PubMed] [Google Scholar]
- 53.Boehm M, Tian X, Ali MK, et al. Improving right ventricular function by increasing BMP signaling with FK506. Am J Respir Cell Mol Biol 2021; 65: 272–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernardo RJ, Haddad F, Couture EJ, et al. Mechanics of right ventricular dysfunction in pulmonary arterial hypertension and heart failure with preserved ejection fraction. Cardiovasc Diagn Ther 2020; 10: 1580–1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kocken JMM, da Costa Martins PA. Epigenetic regulation of pulmonary arterial hypertension-induced vascular and right ventricular remodeling: new opportunities? Int J Mol Sci 2020; 21: 8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Amsallem M, Sweatt AJ, Arthur Ataam J, et al. Targeted proteomics of right heart adaptation to pulmonary arterial hypertension. Eur Respir J 2021; 57: 2002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chouvarine P, Photiadis J, Cesnjevar R, et al. RNA expression profiles and regulatory networks in human right ventricular hypertrophy due to high pressure load. iScience 2021; 24: 102232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lim TB, Aliwarga E, Luu TDA, et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc Res 2019; 115: 1998–2007. [DOI] [PubMed] [Google Scholar]
- 59.Abbas N, Perbellini F, Thum T. Non-coding RNAs: emerging players in cardiomyocyte proliferation and cardiac regeneration. Basic Res Cardiol 2020; 115: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Du WW, Xu J, Yang W, et al. A neuroligin isoform translated by circNlgn contributes to cardiac remodeling. Circ Res 2021; 129: 568–582. [DOI] [PubMed] [Google Scholar]
- 61.Winkle M, El-Daly SM, Fabbri M, et al. Noncoding RNA therapeutics – challenges and potential solutions. Nat Rev Drug Discov 2021; 20: 629–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Legnini I, Di Timoteo G, Rossi F, et al. circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol Cell 2017; 66: 22–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pamudurti NR, Bartok O, Jens M, et al. Translation of circRNAs. Mol Cell 2017; 66: 9–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol 2016; 238: 42–51. [DOI] [PubMed] [Google Scholar]
- 65.Song X, Cao G, Jing L, et al. Analysing the relationship between lncRNA and protein-coding gene and the role of lncRNA as ceRNA in pulmonary fibrosis. J Cell Mol Med 2014; 18: 991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang L, Liang H, Meng X, et al. mmu_circ_0000790 is involved in pulmonary vascular remodeling in mice with HPH via microRNA-374c-mediated FOXC1. Mol Ther Nucleic Acids 2020; 20: 292–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sajid MI, Moazzam M, Kato S, et al. Overcoming barriers for siRNA therapeutics: from bench to bedside. Pharmaceuticals 2020; 13: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kanwar MK. Biomarkers in pulmonary arterial hypertension: moving closer toward precision medicine? J Heart Lung Transplant 2020; 39: 287–288. [DOI] [PubMed] [Google Scholar]
- 69.Verduci L, Tarcitano E, Strano S, et al. CircRNAs: role in human diseases and potential use as biomarkers. Cell Death Dis 2021; 12: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ren S, Lin P, Wang J, et al. Circular RNAs: promising molecular biomarkers of human aging-related diseases via functioning as an miRNA sponge. Mol Ther Methods Clin Dev 2020; 18: 215–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhang Y, Chen Y, Yao H, et al. Elevated serum circ_0068481 levels as a potential diagnostic and prognostic indicator in idiopathic pulmonary arterial hypertension. Pulm Circ 2019; 9: 2045894019888416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang L, Zhang W, Li H, et al. Five circular RNAs in metabolism pathways related to prostate cancer. Front Genet 2021; 12: 636419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeng Y, Zheng Z, Liu F, et al. Circular RNAs in metabolism and metabolic disorders. Obes Rev 2021; 22: e13220. [DOI] [PubMed] [Google Scholar]
- 74.Southgate L, Machado RD, Gräf S, et al. Molecular genetic framework underlying pulmonary arterial hypertension. Nat Rev Cardiol 2020; 17: 85–95. [DOI] [PubMed] [Google Scholar]
- 75.Atkinson C, Stewart S, Upton PD, et al. Primary pulmonary hypertension is associated with reduced pulmonary vascular expression of type II bone morphogenetic protein receptor. Circulation 2002; 105: 1672–1678. [DOI] [PubMed] [Google Scholar]
- 76.Chen NY, Collum SD, Luo F, et al. Macrophage bone morphogenic protein receptor 2 depletion in idiopathic pulmonary fibrosis and Group III pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol 2016; 311: L238–L254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Su H, Wang G, Wu L, et al. Transcriptome-wide map of m6A circRNAs identified in a rat model of hypoxia mediated pulmonary hypertension. BMC Genomics 2020; 21: 39. [DOI] [PMC free article] [PubMed] [Google Scholar]