Abstract
Ubiquitous antibiotic resistance genes (ARGs) is a significant global human health concern. Surfactants have been extensively used worldwide, and the consumption of surfactants containing hygiene, cleaning agents and disinfectants was multiplied during COVID-19 pandemic, which have caused significantly increased pollution of surfactants in aquatic environment. Whether such ever-increasing surfactant concentration boost dissemination risk of ARGs still remains unknown. Here the effects of three typical surfactants such as sodium dodecyl sulfate, cetyltrimethylammonium bromide and benzalkonium chloride on the transformation of pUC19 plasmid (2686 bp)-borne ARGs to recipient bacteria E. coli DH5ɑ were investigated. It was found that these surfactants at environmental concentrations facilitated horizonal gene transfer (HGT) via transformation. The transformation triggering concentrations for the three surfactants were 0.25–0.34 mg/L with a maximum increased transformation frequency of 13.51–22.93-fold. The mechanisms involved in activated HGT of ARGs via transformation triggered by surfactants could be mainly attributed to the increased production of reactive oxygen species, which further enhanced cell membrane permeability. These findings provide new sights for understanding of ARG propagation and also imply that the drastic rise of surfactant concentration in aquatic environment may significantly increase the dissemination risk of antibiotic resistance.
Keywords: Antibiotic resistance genes, Surfactants, Environmental concentration, Transformation, Dissemination risk
1. Introduction
Antibiotic resistance genes (ARGs) and the induced transmission of antibiotic resistant bacteria (ARB) was considered as emerging global contaminant, which even listed as one of the most worldwide ecological and human health concern in the coming decades [[1], [2], [3]]. Apart from spontaneous mutations, horizontal gene transfer (HGT) including conjugation, transformation and transduction prominently contributes to the spread of antibiotic resistance [4,5]. Transformation, exogenous DNA uptaken by competent bacteria from their surroundings, is a significant pathway of HGT, which can facilitate the dissemination of ARGs and also a significant evolution pattern of sensitive bacteria to ARB [6,7].
Antibiotics and non-antibiotics have been demonstrated to drive the spread of existing and newly emerging ARGs [8,9]. However, the role of surfactants at environmental concentrations in the dissemination of ARGs through transformation in aquatic environment has not been well studied. Surfactants, composed of both hydrophobic and hydrophilic groups, have been widely used as wetting agents, foaming agents, emulsifiers, dispersants, detergents and co-formulant in products used in agriculture, because of their enrichment at interfaces, lowering of interfacial tension and micelle formation trait [[10], [11], [12], [13]]. The global market size for surfactants was $41.3 billion in 2019, and is predicted to reach $58.5 billion by 2027 on basis of Allied Market Research. Moreover, the ongoing global public health emergency, COVID-19 pandemic, has conspicuously boosted the usage of surfactants containing products, such as hygiene, cleaning agents and disinfectants [[14], [15], [16], [17], [18]]. For instance, the sales of hand soaps, surfactants served as major active constituent, have been doubled since the declaration of COVID-19 pandemic in Britain, and the global soap market is expected to grow from $180.99 billion in 2020 to $302.25 billion by the end of 2030 [19]. A total of 430 commercial disinfectants were recommended by U.S Environmental Protection Agency to combat SARS-CoV-2 virus currently, and the dominate active ingredients for 52.1% of them are associated with surfactants like sodium dodecyl sulfate (SDS) and quaternary ammonium compounds (QACs) such as benzalkonium chloride (BAC) and cetyltrimethylammonium bromide (CTAB) (Fig. 1a).
Fig. 1.
(a) The classification of commercial disinfectants on the U.S EPA list to combat SARS-CoV-2 virus currently based on dominate active ingredients, which was redrawn from a previous study [14]. The type of various disinfectant and their proportion were displayed. The concentrations of surfactants in surface water (b) and wastewater treatment plants sewage (c) were re-displayed based on data from 55 published literature in the range of 1992 to 2020, and the detailed information are shown in supporting information (Table S1).
The broad application coupled with the ever-increasing surfactant consumption amid COVID-19 pandemic inevitably aggravate surfactant pollution. According to our summary, surfactants were common in surface water all over the world, and its concentration for 26.4% of the observations is above 250 μg/L (Fig. 1b), and the surfactants in municipal or industrial wastewater effluents can reach levels up to mg/L (Fig. 1c). For example, the average concentrations of QACs in surface water and wastewater in Turkey have been revealed to be around 40 μg/L and 500 μg/L, respectively [20]. Comparatively, COVID-19 pandemic has resulted in much more discharge of surfactants into aquatic environment and significant increase of surfactant concentration in surface water can be surely expected [19]. According to a survey conducted in Slovakia, the surfactant contaminated surface water monitoring sites, where the surfactant content higher than 1 mg/L, had an increase of 24.5–36.0% in 2020 in contrast to 2019 [21]. Furthermore, the concentrations of surfactants in wastewater are estimated to increase by several times, since about 60% of the used surfactants were discharged into aquatic environment [[22], [23], [24]]. It has been demonstrated that the concentration of QACs in wastewater treatment plant of Athens had a 331% increase in the lockdown conditions in 2020 compared with the data acquired under non-COVID-19 conditions [25].
A few recent studies showed that surfactants, similar to antibiotics, also have antimicrobial effects because of their solubilization of lipid membranes through electrostatic interaction, hydrophobic interaction as well as production of ROS [[26], [27], [28]]. In addition, surfactants can affect the mobility, stability, aggregation and vitality of bacteria as well as bacterial community structure [11,29]. The interactions between surfactants and bacteria make it a potential influence factor for gene transfer. We hypothesize that the prevalent and drastic rise of surfactants concentration during COVID-19 pandemic may exert significant effect on dissemination risk of ARGs in aquatic environment. A few relevant scientific questions need to be answered. (1) Whether the surfactants at environmental concentrations favor the HGT of ARGs via transformation or not? (2) If yes, what is the starting concentrations of surfactants that triggers the transformation? (3) Will the chargeability of surfactants impact the transformation of ARGs, and what is the dominant mechanism involved in effects of surfactants on ARG transformation?
In order to clarify the above-mentioned questions, herein, the transformation experiments in the presence of typical surfactants (BAC, CTAB and SDS) were carried out using E. coli DH5ɑ and free plasmid pUC19 containing ampicillin resistance gene ampR as the competent recipients and the extracellular ARG donor, respectively. The bacterial transformation frequency and efficiency were calculated to test if surfactants at environmental concentrations could significantly promote the transformation of ARGs. The underlying mechanisms were also explored through analysis of reactive oxygen species (ROS), cell membrane permeability and confocal laser scanning microscope (CLSM). The present work may advance our understanding of the influence of universal surfactants on the spread of ARGs through transformation in the aquatic environment.
2. Materials and methods
2.1. Materials
The competent E. coli DH5ɑ without resistance to ampicillin purchased from BeNa Culture Collection (BNCC353719) was used in this study, and pUC19 plasmid (2686 bp) having ampicillin resistance gene ampR, extracted from E. coli DH5ɑ (BNCC353848) performed by Genewiz Inc, was selected as extracellular ARG (eARG) donor. Three typical surfactants namely benzalkonium chloride (BAC), sodium dodecyl sulfate (SDS) and cetyltrimethylammonium bromide (CTAB) were chosen because they were widely used in commercial disinfectants recommended by U.S EPA to fight against COVID-19, and the specific information of these surfactants are listed in supporting information (Table S2). DCF-DA (2′7′-dichlorofluorescein diacetate) ROS detection assay kit was purchased from Nanjing Jiancheng Bioengineering Institute. Propidium iodide (PI) and SOC medium were obtained from Sangon Biotech (Shanghai) Co., Ltd. pBBR1MCS2-Tac-EGFP plasmid was kindly provided by Dr Bingshen Liu. The transformation test was conducted in the filter (0.22 μm pore size membrane) sterilized surface water sampled from Shangtang River (120.1711°E, 30.2986°N), Hangzhou, China, with the chemical properties showed in Table S3.
2.2. Effects of surfactants on bacterial growth
E. coli DH5ɑ (BNCC353719), was inoculated into lysogeny broth (LB), incubated at 30 °C with shaking at 200 rpm, and the pre-cultured bacteria (OD600 = 0.6) was inoculated (3%, v/v) into fresh LB medium in order to record the growth curve of bacteria to illustrate the effects of surfactants on bacteria as described previously [30]. Briefly, 50 μL of bacterial cells at stationary phase was added to 5 ml of LB medium with various concentration of surfactants and the absorbance at 600 nm of the culture (30 °C, 200 rpm) under 24 h exposure were measured.
2.3. Effects of surfactants on PCR amplification of ampR
Amplicons of the ampR gene in pUC19 plasmid with the final DNA concentration of 12 ng/μL, in the presence of surfactants (0, 0.05 μg/L, 1 mg/L, 25 mg/L), were quantified by means of QuantStudio 5 real-time PCR platform (Applied Biosystems, Massachusetts, USA), and the primers used for qPCR analysis of target ampR are given in Table S4. The threshold cycle (CT) of ampR gene under different concentration of surfactants were obtained to reveal their effects on amplification efficiencies.
2.4. Zeta potential
The zeta potential of pUC19 plasmid and bacterial cells in the absence/presence of surfactants were investigated by Zetasizer Nano ZS 90 (Malvern, Worcestershire, UK).
2.5. Transformation assays
Transformation assays were performed according to published protocols [31]. In general, 0.5 ml of E. coli DH5ɑ (BNCC353719) which has been cultured to stationary phase (∼10 h) was taken and went through 10 fold dilution, after another cultural period of 3 h, 2 ml liquid was sampled and chill-held for 10 min on ice, followed by 5 min centrifugation (4000 rpm, 4 °C). The obtained pellets were re-suspend in CaCl2 (0.1 M, ice-cold) and chilled for 20–25 min. Then the process of centrifugation and suspension were repeated. After that, 460 μL of the competent bacteria was mixed with 20 μL of pUC19 plasmid (10 ng/μL), 20 μL of surfactants, and the final concentration of plasmid was set as 0.4 ng/μL. The final concentration of surfactant was 0, 0.01, 0.05, 0.1, 0.25, 0.5, 0.75, 1, 2, 10 and 25 mg/L, which cover the environmentally relevant concentrations in surface water and comparative higher concentrations occur in wastewater [32,33]. Thereafter, the reaction tubes were exposed to a 42 °C heat-shock for 90 s and a chilling-shock for 5 min. Then, the tubes were incubated at room temperature for 6 h. Following that, the mixtures were diluted using sterile PBS buffer solution and spread onto LB agar plates containing 10 mg/L ampicillin, and then incubated for 48 h to record the number of transformants. The number of recipient bacteria was obtained through spreading diluted mating mixtures onto non-ampicillin containing LB agar plates likewise. In addition, a negative control without introduction of pUC19 plasmid was also carried out following same protocols. The transformation frequency was calculated as follows [34]:
2.6. ROS assessment
The generated intracellular ROS such as hydrogen peroxide, hydroxyl radicals, peroxynitrite after exposure to surfactants was detected using a DCF-DA ROS detection assay kit through BD Accuri C6 Plus flow cytometer (BD Biosciences, San Jose, USA) with excitation at 488 nm and emission at 525 nm [35,36]. Boiled competent bacterial cells were used as negative control in the ROS measurement.
2.7. Cell permeability assessment
The cell membrane permeability were detected by staining with 30 μM PI and the fluorescence was quantified using BD Accuri C6 Plus flow cytometer (BD Biosciences, San Jose, USA) with excitation at 488 nm and emission at 630 nm [37]. Boiled and untreated competent bacterial cells were used as controls for damaged and intact cell, respectively.
2.8. CLSM observation
A wide-host vector pBBR1MCS2-Tac-EGFP plasmid (5915 bp) with kanamycin resistance, was applied as cell free plasmid DNA to test the impacts of surfactants on plasmid transfer to E. coli DH5ɑ via transformation [38]. Because the green fluorescent protein (GFP) expression in pBBR1MCS2-Tac-EGFP contained E. coli DH5ɑ could emit GFP fluorescence under 440 nm excitation light. The obtained pBBR1MCS2-Tac-EGFP plasmid (3.55 μL, 36.47 ng/L) was mixed with 100 μL competent E. coli DH5ɑ induced by CaCl2 treatment to reach a final plasmid concentration of 1.25 ng/L. About 20 μL of surfactants was then added into the mixture to obtain solution with surfactants at concentrations of 0, 0.1 and 1 mg/L. The mixture was subsequently subjected to a 42 °C heat-shock (90 s) and a chilling-shock (5 min), and 200 μL of SOC sterile liquid medium was then added. After that, the mixture was cultured at 30 °C, 120 rpm for 6 h. At the endpoint of transformation assay, the mating mixtures at random locations was scanned using a CLSM (Zeiss LSM780, Jena, German) to reveal the occurrence of plasmid transfer. The acquired images were processed using Image-J (v1.52, NIH Image, US).
2.9. Statistical analysis
The similarity analysis (ANOSIM) with 999 permutations between groups was carried out using R 4.1.1 [39]. Principal coordinate analysis (PCoA) was performed using the phyloseq package in R 4.1.1 based on Bray-Curtis distance [40]. Bubble chart was obtained using origin 2021 (OriginLab Corporation, Massachusetts, USA). One-way analysis of variance (ANOVA) was conducted using IBM SPSS Version 26 to test the differences of transformation frequency, ROS generation and bacterial membrane permeability for surfactants treated samples under various concentration, as well as the calculation of pearson correlation coefficient. Concentration response relationships were modeled with Weibull-2 functions using origin 2021.
3. Results
3.1. Concentration-dependent transformation induced by surfactants
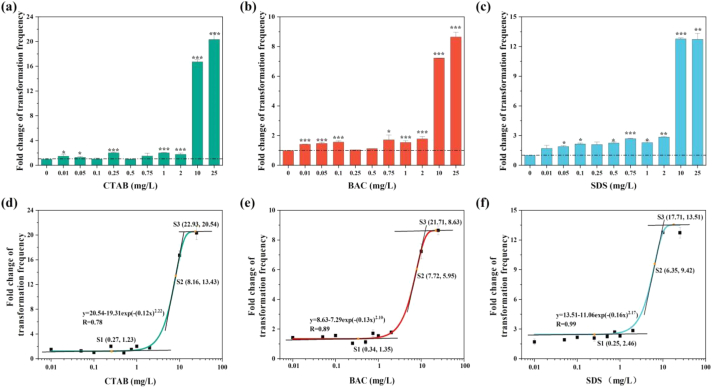
The transformation frequency of pUC19 plasmid as eDNA under surfactants (CTAB, BAC and SDS) exposure in the range of 0–25 mg/L was displayed in Fig. S1a, and the fold change of transformation frequency was then calculated to compare the effects of surfactants on ARG transformation. The transformation frequency was significantly increased (p < 0.05) in the presence of 1–25 mg/L surfactants (Fig. 2a, b, c). Moreover, the increased transformation frequency displayed a concentration-dependent pattern, which is in accordance with Weibull-2 model (R = 0.78–0.99) (Fig. 2d, e, f). In this study, the turning points labelled as S1 and S3 refer to the object with slope of 0.05, and S2 is the point with maximum slope. The increase in transformation frequency started from 0.27 mg/L for CTAB, 0.34 mg/L for BAC and 0.25 mg/L for SDS, respectively. The maximum increase of transformation frequency reached 20.54-fold for CTAB, 8.63-fold for BAC and 13.51-fold for SDS, respectively in comparison with the spontaneous transformation frequency of the control. However, the transformation was not promoted with increasing surfactants after the concentration arrived at 17.71–22.93 mg/L. The effects of surfactants on transformation are type-dependent (ANOSIM, R = 0.925, p = 0.001). PCoA based on the Bray-Curtis distances also support this idea, since the distribution of transformants under different type of surfactants were distinctly separated at the first axis (PC1), which explained 75.0% of the total variation (Fig. S1b).
Fig. 2.
Fold changes of transformation frequency under CTAB (a), BAC (b) and SDS (c), respectively. The simulated changes in the fold change of transformation frequency with the increase of surfactant concentration are showed in (d), (e) and (f) respectively.
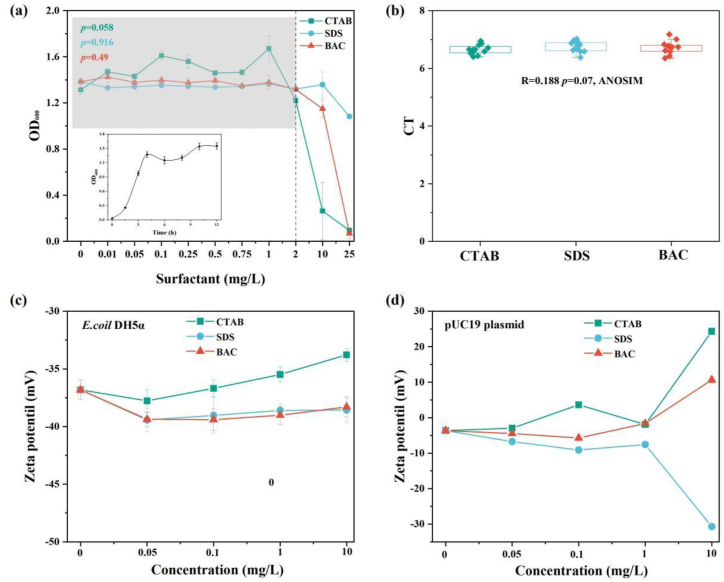
In addition, the effects of surfactants on bacteria and plasmid were also considered. The growth of E. coli DH5ɑ was not significantly affected for surfactants ranging from 0 to 2 mg/L. For surfactants at concentration of 2–25 mg/L, the transformation frequency showed a significantly increase trend in spite of the inhibited growth of E. coli DH5ɑ, which imply the horizontal transfer rather than vertical transfer contributed to the accumulation of transformants (Fig. 3a). The PCR amplification of ampR was not significantly influenced by various concentration of surfactants (p > 0.05) as well as surfactant type (ANOSIM, R = 0.188, p = 0.07), which exclude the damage of surfactants on plasmid to a certain extent (Fig. 3b). The stability of E. coli DH5ɑ and pUC19 plasmid were estimated based on zeta potential. Although, the negative surface charge of bacteria was slightly neutralized by cationic surfactants, the zeta potential of E. coli DH5ɑ under these three surfactants were in the range from −30 to −40 mV, which revealed the relatively homogeneous state of bacteria instead of aggregation (Fig. 3c). For pUC19 plasmid, the zeta potential of less negative charged plasmid was not significantly affected in the range of 0–1 mg/L (Fig. 3d). In case of 10 mg/L SDS, the negative charged plasmid comparable to bacteria could co-exist with bacteria stably through electrostatic repulsion, which also demonstrate the increased transformation was not driven by aggregation-mediated enhanced contact frequency between bacteria and plasmid.
Fig. 3.
The effects of surfactants on the growth of bacteria (a) and amplification of ampR gene embed in pUC19 plasmid (b). (c) and (d) are zeta potential of E. coli DH5ɑ and pUC19 plasmid in the presence of different concentration of surfactants, respectively.
The successful uptake of plasmid by bacteria was confirmed through CLSM using plasmid pBBR1MCS2-Tac-EGFP and E. coli DH5ɑ, since the expression of acquired GFP in E. coli DH5ɑ could present bacterial GFP fluorescence in green. Compared with control (Fig. 4a), the increased bacterial GFP fluorescence indicated abundant transformants in the presence of surfactants, and the increase of fluorescent intensity of transformants reached 1.29–1.45-fold and 1.69–3.32-fold under 0.1 and 1 mg/L surfactants exposure, respectively (Fig. 4a–d).
Fig. 4.
Typical CLSM images of E. coli DH5ɑ in the absence (a) or presence (b–d) of 1 mg/L surfactants, (b) CTAB, (c) BAC (d) SDS. E. coli DH5ɑ is fluorescence free under 440 nm excitation light, however, the GFP expression in pBBR1MCS2-Tac-EGFP contained transformants could emit GFP fluorescence. The normalized relative fluorescent intensity of transformants after addition of 0.1 and 1 mg/L surfactants are also presented (a).
3.2. Surfactants mediate the transformation via ROS and cell membrane permeability
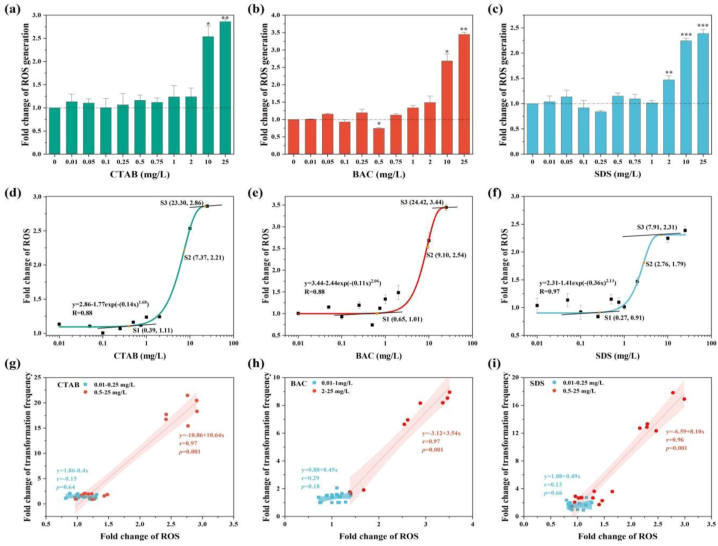
To explore the underlying mechanisms behind the surfactants mediated transformation, the ROS response and cell membrane permeability were determined. Generous generated ROS was detected after addition of 10–25 mg/L CTAB, BAC and SDS, respectively (Fig. 5a, b, c). According to the results of Weibull-2 fitting, the over-production of ROS happened from 0.39 mg/L for CTAB, 0.65 mg/L for BAC and 0.27 mg/L for SDS, respectively (Fig. 5d, e, f). At the concentration of 23.30 and 24.42 mg/L surfactatns, the maximum increase of ROS generation reached 2.86-fold and 3.44-fold for CTAB and BAC, respectively. In contrast, the maximum increase of ROS production (2.31-fold) arrived at 7.91 mg/L for SDS. The surfactants mediated increases in transformation frequency were significantly related to the surfactant mediated increases of ROS generation (Fig. 5g, h, i). For CTAB treatment, the turning point for ROS over-production is around 0.39 mg/L, and ROS had little correlation (r = 0.15, p = 0.64) with transformation frequency in the presence of 0.01–0.25 mg/L CTAB. Nevertheless, the induced ROS exposed to 0.5–25 mg/L of CTAB significantly contributed to the enhanced transformation (r = 0.97, p = 0.001).
Fig. 5.
Fold change of ROS content under a variety of concentrations of CTAB (a), BAC (b) and SDS (c), d-f are the fitting results of concentration-ROS generation effects, g-f displays the relationship between ROS and transformation frequency.
The bacteria at competence state or with higher cell membrane permeability, which in favor of the uptake of plasmid is another widely reported mechanism for enhanced transformation. Here, the bacterial membrane permeability was not significantly affected for surfactants below 10 mg/L (Fig. 6a, b, c). This is similar with the results of Weibull-2 fitting. The increase of cell membrane permeability induced by surfactants displayed a hysteresis effect compared to ROS, until the dose increased to 1.36 mg/L for CTAB, 4.19 mg/L for BAC and 0.83 mg/L for SDS, respectively, the bacterial membrane permeability was significantly influenced (Fig. 6d, e, f). A maximum of 35.23-fold under CTAB, 10.64-fold under BAC and 2.79-fold under SDS of cell membrane permeability was recorded, respectively. Similar to ROS over-production, the pearson correlation coefficient demonstrate that surfactants mediated increases in bacterial membrane permeability also significantly contributed to the increases in transformation (Fig. 6g, h, i).
Fig. 6.
Fold change of cell membrane permeability under different concentrations of surfactants is present in a-c, d-f are the relationship between the fold change of cell membrane permeability and concentrations and their fitting, g-f shows the correlation between cell membrane permeability and transformation frequency.
In addition, the relationship for ROS generation and cell membrane permeability was also explored in the present work (Fig. 7a–c). ROS response was illustrated to have a quick feedback for surfactants than cell membrane permeability, and the enriched ROS could result in the change of permeability of bacterial membrane. For instance, when the concentration of SDS increased to 0.27 mg/L, richer ROS was formed, but the bacterial membrane permeability was not significantly impacted. After the concentration of SDS increased to 0.83 mg/L, the membrane permeability of bacteria started to raise. Moreover, the increased bacterial membrane permeability was significantly related to the over-production of ROS (p = 0.001) (Fig. 7c).
Fig. 7.
The relationship between ROS generation and bacterial membrane permeability change in the presence of various concentration of CTAB (a), BAC (b) and SDS (c), respectively.
4. Discussion
Transformation has been clarified to be an important pathway of HGT [41]. As we know, the ubiquitous cell free DNA released from cell lysis is prone to bound with natural organic matter or minerals to stably exist for up to several months [42,43], and more than 80 competent bacterial species, even between taxnomically different bacteria, are capable of in-taking extracellular DNA as nutrition or for DNA repair [31,44,45]. To the best of our knowledge, naturally transformable bacteria are widespread in the environment, wastewater, surface water and soil are all included [36,43]. In addition, some pathogens have been revealed with natural competence [46]. Unfortunately, the role of transformation in ARG dissemination is underappreciated, and this process is still not fully understood [47]. In terms of the widely used surfactants during COVID-19 pandemic, little is known whether the prevalent presented surfactant in the aquatic environment could accelerate natural transformation.
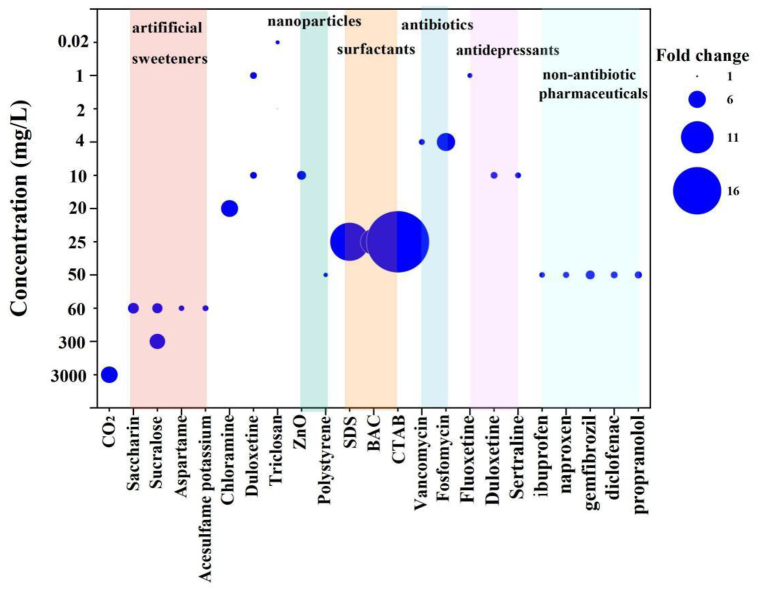
In the present work, three surfactants (CTAB, BAC and SDS) ranging from 0.01 to 25 mg/L, were illustrated to significantly facilitate the transformation frequency of plasmid-borne ARGs into E. coli DH5ɑ. This is in contradiction with a previous report, where the ability of Acinetobacter baylyi BD413 to acquire extracellular DNA did not result prominently different in the absence and presence of heptamethyltrisiloxane on leaf disc [13] Furthermore, the fold change of transformation frequency induced by surfactants is up to 20.54-fold increase for CTAB, 8.63-fold increase for BAC, and 13.51-fold increase for SDS at 25 mg/L, respectively (Fig. 8). Surfactants evoked transformation is comparable to that of antibiotics such as vancomycin and fosfomycin (2.0–6.0-fold increase) at 4 mg/L, and relatively higher than that caused by 60 mg/L of artificial sweeteners, 20 mg/L of chloramine, 10 mg/L of duloxetine, 0.02 mg/L of triclosan, 10 mg/L of ZnO nanoparticles, 50 mg/L of polystyrene nanoparticles, 50 mg/L of non-antibiotic pharmaceuticals and 1–10 mg/L of antidepressants [35,36,[48], [49], [50], [51], [52], [53]]. Even contrast to considerable high concentration of reagents, like 300 mg/L of sucralose (5.1-fold increase) and 3000 mg/L of CO2 (5.5-fold increase), surfactants could trigger greater transformation [54,55]. The concentration-transformation effects were further investigated with the help of Weibull-2 model. At concentration of 0.25–0.34 mg/L, the transformation frequency was significantly enhanced by surfactants. It should be noted that, surfactants at sub-microgram per liter concentration are common in wastewater. According to our summary based on the data collected from published papers as shown in Fig. 1c, the concentrations of surfactans for 65.4% of the observations are higher than 0.25 mg/L. For instance, the average concentration of QACs in wastewater from Turkey sewage treatment plant is about 0.5 mg/L, which is adequate to initiate promoted transformation of ARGs [20]. It is also applicable for the strong contaminated surface water. It was found that, 26.9% of the surveyed wastewater had surfactant concentration of >0.25 mg/L.
Fig. 8.
Comparison of the fold change of transformation frequency induced by studied surfactants (CTAB, BAC and SDS) with those of reported chemicals from previous research.
The underlying mechanisms for the promotion of transformation were explored in this work. The results of flow cytometer emphasize that, the surfactants mediated transformation is significantly associated with the surfactant mediated increases of ROS generation and cell membrane permeability (Fig. 5, Fig. 6). ROS response is relative rapid process (triggered by surfactants in range of 0.27–0.65 mg/L) and the accumulated ROS would eventually leading to the change of cell membrane permeability (triggered by surfactants in range of 0.83–4.19 mg/L). This is in line with previous research, the induced ROS overproduction (oxidative stress) is a major reason for the increased membrane permeability and bacterial competent state, which together significantly contribute to increased transformation [56,57].
Surfactants (CTAB, SDS and BAC) have been demonstrated to induce oxidative stress like ROS production in treated bacteria, and the caused production of ROS plays vital role in ARG transformation, the possible mechanisms were summarized as follows. First, the competence state of bacteria involved in ROS production, which favor the uptake of eDNA by crossing the cell outer membrane barrier. Bacterial DNA is susceptible to damage under extensive ROS exposure, competence-proficient bacteria can uptake DNA to repair the ROS-induced DNA damages in order to adapt better for survival [52,58]. The development of genetic competence is therefore proposed as a significant bacterial surviving strategy in response to stress [59]. Second, the increased uptake of plasmid and eDNA for E. coli are generally attributed to the appearance of pores on cell outer membrane and enhanced membrane permeability, which also aid in promoted ARG transformation [31,60]. The surfactants-increased cell membrane permeability and porosity could be associated with the oxidation of membrane lipids due to over-generation of ROS [61].
The chargeability of surfactants were assumed to linked with the transformation of ARGs. Whereas, the ARG transformation was both significantly enhanced in the presence of cationic surfactants such as BAC and CTAB and anionic surfactant SDS, and the facilitated transformation was not driven by aggregation-mediated enhanced contact frequency between bacteria and plasmid as demonstrated by zeta potential analysis. In summary, The amphotericit of surfactants seems to drive the transformation of ARGs, and further attention should be paid on the interfacial interaction between different functional groups of surfactants and bacteria, in order to decipher the influence mechanism of surfactants function groups on transformation of ARGs.
It was worth noting that sub-microgram per liter concentration of surfactants are particularly common in developing countries. For instance, the Ganga river in India is extremely contaminated owing to laundry, bathing, sewage disposal, industrial waste discharge, with the concentration of surfactants ranging from 0.31 to 3.20 mg/L [62,63]. According to previous report, the concentration of surfactant in surface water in Bangladesh can reach 0.4–0.8 mg/L [64]. It should be noticed that, the surveyed surfactants in previous research are usually chosen from limited candidates. The number of surfactant type that exists in surface water in prevalence and their abundance is still not perfectly clear, and the content of surfactants are suggested to be seriously underestimated due to lacking of thoroughgoing analysis focusing on complete type of surfactants. Several reason are associated with the ubiquitous surfactants pollution for low-income countries. First, The discharge of laundry detergents into water is a considerable reason that increases the content of surfactant in aquatic environment, because laundry washing is usually conducted on river banks for rural areas in developing countries such as Africa [65]. Second, large amount of wastewater are discharged into surface water without treatment coupled with deficient wastewater treatment plants [[66], [67], [68]]. In fact, the removal of surfactants from wastewater is still a growing concern when take the prevalence and high toxicity of surfactants into consideration [69]. Third, prevailing wastewater irrigation in developing countries facilitate the severe water pollution, surfactants are also included [70].
Furthermore, the surfactants contamination seems to become aggravate under covid-19 pandemic in low-income countries, because of the increasingly consumed surfactants contained detergents and disinfectants [71]. In this instance, the surfactants induced HGT of ARGs via transformation could significantly speed up the dissemination of ARGs, which may not only result in the formation of new ARB but also ARG harbouring pathogens or superbug. Bacterial antimicrobial resistance (AMR) is considered as a leading global health issue, which is responsible for the less effective performance of drugs for infections treatment [72]. About 4.95 million deaths were linked with bacterial AMR, and 1.27 million deaths were directly attributable to resistance globally in 2019 [73]. More strikingly, AMR is a particularly serious problem for some of the developing countries in the world given their poor medical conditions. For example, the highest rates of AMR burden was found in sub-Saharan Africa in 2019. If effective efforts can not be taken immediately, the growing AMR would aggravate the burden for fragile medical conditions. As a consequence, surfactants contamination and the effects of environmental surfactants on ARG propagation should be paid adequate attention, particularly in low-income countries.
Here, the influence of surfactants on HGT via transformation was investigated based on pure culture experiment in lab based on the calculation of fold change of transformation frequency, and heat-shock induced competent bacteria was used as recipient. The specific physiological state of prokaryotes capable of taking up genetic material from their surroundings was defined as natural competence for transformation [74]. To the best of our knowledge, eARGs can be taken up by both competent and non-competent bacteria, and the higher cell membrane permeability and porosity for competent bacteria favors the uptake of eARGs [75]. It should be recognized that, the natural transformation occurred in the environment is distinguished from the present artificial interfered laboratory settings. Firstly, the effects of surfactants on non-competent recipient were not concerned in this study, however, non-competent and competent bacteria are both associated with transformation. Secondly, the present transformation assays only focused on one competent species, however, the aim of competence likely differs from one organism to another, and the influence of the ambient bacterial was also neglected. Thirdly, CaCl2 treatment and heat-shock was selected to produce competent bacterial as receptor, while this condition for this artificial mediated competence of bacteria is not prevalent in the environment. Therefore, further research are required to test whether surfactants can result in promoted transformation in naturally existing non-competent and competent bacteria community under environmental conditions.
5. Conclusions
The present study firstly investigated the effects of surfactatns (CTAB, BAC and SDS) at environmental concentration on transformation frequency of plasmid-borne ARGs to E. coli DH5ɑ, and Weibull-2 model can provide further insights into the concentration-effect relationship for surfactants and transformation frequency, surfactants and ROS generation, surfactants and cell membrane permeability change. It was found that, surfactants at the commonly detected sub-microgram per liter concentration (0.25–0.34 mg/L) triggered HGT of ARGs via transformation, with a maximum increased transformation frequency of 13.51–22.93-fold. The underlying mechanism of enhanced transformation frequency is associated with surfactants induced ROS production as well as ROS-mediated promoted cell membrane permeability. This study highlight the critical role of widespread and increasingly consumed surfactants played in spread of ARGs through transformation. Our study implies that the widespread of surfactants in surface water may significantly speed up the propagation of antibiotic resistance, and these findings also extend our knowledge on the potential dissemination risk of ARGs.
Author contribution statement
Xiaonan Wang: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Gaoquan Du: Performed the experiments; Analyzed and interpreted the data.
Zhuang Qiao: Performed the experiments.
Yixuan Yang: Performed the experiments.
Huimin Shi: Performed the experiments.
Daoyong Zhang: Contributed reagents, materials, analysis tools or data.
Xiangliang Pan: Conceived and designed the experiments.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Xiaonan Wang reports financial support was provided by National Natural Science Foundation of China. Xiaonan Wang reports financial support was provided by Zhejiang Provincial Natural Science Foundation of China. Xiaonan Wang reports financial support was provided by Key Laboratory of Pollution Exposure and Health Intervention of Zhejiang Province.
Acknowledgements
This work was financially supported by the National Natural Science Foundation of China (42207441) and the Zhejiang Provincial Natural Science Foundation of China (LQ22D010008). Xiaonan Wang also gratefully acknowledges financial support from the Key Laboratory of Pollution Exposure and Health Intervention of Zhejiang Province (Grant No. 20220102).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17034.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Qiao M., Ying G., Singer A.C., Zhu Y. Review of antibiotic resistance in China and its environment. Environ. Int. 2018;110:160–172. doi: 10.1016/j.envint.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Amarasiri M., Sano D., Suzuki S. Understanding human health risks caused by antibiotic resistant bacteria (ARB) and antibiotic resistance genes (ARG) in water environments: current knowledge and questions to be answered. Crit. Rev. Environ. Sci. Technol. 2020;50(19):2016–2059. [Google Scholar]
- 3.Li S., Zhang C., Li F., Hua T., Zhou Q., Ho S.H. Technologies towards antibiotic resistance genes (ARGs) removal from aquatic environment: a critical review. J. Hazard Mater. 2021;411 doi: 10.1016/j.jhazmat.2021.125148. [DOI] [PubMed] [Google Scholar]
- 4.Hernando-Amado S., Coque T.M., Baquero F., Martínez J.L. Defining and combating antibiotic resistance from one health and global health perspectives. Nat. Microb. 2019;4(9):1432–1442. doi: 10.1038/s41564-019-0503-9. [DOI] [PubMed] [Google Scholar]
- 5.Feng G., Huang H., Chen Y. Effects of emerging pollutants on the occurrence and transfer of antibiotic resistance genes: a review. J. Hazard Mater. 2021;420 doi: 10.1016/j.jhazmat.2021.126602. [DOI] [PubMed] [Google Scholar]
- 6.Zainab S.M., Junaid M., Xu N., Malik R.N. Antibiotics and antibiotic resistant genes (ARGs) in groundwater: a global review on dissemination, sources, interactions, environmental and human health risks. Water Res. 2020;187 doi: 10.1016/j.watres.2020.116455. [DOI] [PubMed] [Google Scholar]
- 7.Larsson D.G., Flach C.F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 2022;20(5):257–269. doi: 10.1038/s41579-021-00649-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang Q., Feng M., Ye C., Yu X. Effects and relevant mechanisms of non-antibiotic factors on the horizontal transfer of antibiotic resistance genes in water environments: a review. Sci. Total Environ. 2022;806 doi: 10.1016/j.scitotenv.2021.150568. [DOI] [PubMed] [Google Scholar]
- 9.Shi X., Xia Y., Wei W., Ni B. Accelerated spread of antibiotic resistance genes (ARGs) induced by non-antibiotic conditions: roles and mechanisms. Water Res. 2022;224 doi: 10.1016/j.watres.2022.119060. [DOI] [PubMed] [Google Scholar]
- 10.Gu G., Hu J., Cevallos-Cevallos J.M., Richardson S.M., Bartz J.A., van Bruggen A.H. Internal colonization of Salmonella enterica serovar Typhimurium in tomato plants. PLoS One. 2011;6(11) doi: 10.1371/journal.pone.0027340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y., Li H., Gu J., Qian X., Yin Y., Li Y., Zhang R., Wang X. Effects of adding different surfactants on antibiotic resistance genes and intI1 during chicken manure composting. Bioresour. Technol. 2016;219:545–551. doi: 10.1016/j.biortech.2016.06.117. [DOI] [PubMed] [Google Scholar]
- 12.Myers D. John Wiley & Sons; 2020. Surfactant Science and Technology. [Google Scholar]
- 13.Riva V., Patania G., Riva F., Vergani L., Crotti E., Mapelli F. Acinetobacter baylyi strain BD413 can acquire an antibiotic resistance gene by natural transformation on lettuce phylloplane and enter the endosphere. Antibiotics. 2022;11(9):1231. doi: 10.3390/antibiotics11091231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hora P.I., Pati S.G., McNamara P.J., Arnold W.A. Increased use of quaternary ammonium compounds during the SARS-CoV-2 pandemic and beyond: consideration of environmental implications. Environ. Sci. Technol. Lett. 2020;7(9):622–631. doi: 10.1021/acs.estlett.0c00437. [DOI] [PubMed] [Google Scholar]
- 15.Israel S., Harpaz K., Radvogin E., Schwartz C., Gross I., Mazeh H., Cohen M.J., Benenson S. Dramatically improved hand hygiene performance rates at time of coronavirus pandemic. Clin. Microbiol. Infect. 2020;26(11):1566–1568. doi: 10.1016/j.cmi.2020.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pradhan D., Biswasroy P., Naik P.K., Ghosh G., Rath G. A review of current interventions for COVID-19 prevention. Arch. Med. Res. 2020;51(5):363–374. doi: 10.1016/j.arcmed.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brandtner P., Darbanian F., Falatouri T., Udokwu C. Impact of COVID-19 on the customer end of retail supply chains: a big data analysis of consumer satisfaction. Sustainability. 2021;13(3):1464. [Google Scholar]
- 18.Donde O.O., Atoni E., Muia A.W., Yillia P.T. COVID-19 pandemic: water, sanitation and hygiene (WASH) as a critical control measure remains a major challenge in low-income countries. Water Res. 2021;191 doi: 10.1016/j.watres.2020.116793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chirani M.R., Kowsari E., Teymourian T., Ramakrishna S. Environmental impact of increased soap consumption during COVID-19 pandemic: biodegradable soap production and sustainable packaging. Sci. Total Environ. 2021;796 doi: 10.1016/j.scitotenv.2021.149013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ertekin E., Hatt J.K., Konstantinidis K.T., Tezel U. Similar microbial consortia and genes are involved in the biodegradation of benzalkonium chlorides in different environments. Environ. Sci. Technol. 2016;50(8):4304–4313. doi: 10.1021/acs.est.5b05959. [DOI] [PubMed] [Google Scholar]
- 21.Hybská H., Lobotková M., Turčániová E., Salva J., Hýrošová T. 2022. Monitoring of the Surfactants in Surface Waters in Slovakia and the Impact COVID-19 Pandemic for Their Presence. Research Square. [Google Scholar]
- 22.Pradhan A., Bhattacharyya A. Quest for an eco-friendly alternative surfactant: surface and foam characteristics of natural surfactants. J. Clean. Prod. 2017;150:127–134. [Google Scholar]
- 23.Daverey A., Dutta K. COVID-19: eco-friendly hand hygiene for human and environmental safety. J. Environ. Chem. Eng. 2021;9(2) doi: 10.1016/j.jece.2020.104754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teymoorian T., Teymourian T., Kowsari E., Ramakrishna S. Direct and indirect effects of SARS-CoV-2 on wastewater treatment. J. Water Process Eng. 2021;42 doi: 10.1016/j.jwpe.2021.102193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alygizakis N., Galani A., Rousis N.I., Aalizadeh R., Dimopoulos M.A., Thomaidis N.S. Change in the chemical content of untreated wastewater of Athens, Greece under COVID-19 pandemic. Sci. Total Environ. 2021;799 doi: 10.1016/j.scitotenv.2021.149230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falk N.A. Surfactants as antimicrobials: a brief overview of microbial interfacial chemistry and surfactant antimicrobial activity. J. Surfactants Deterg. 2019;22(5):1119–1127. doi: 10.1002/jsde.12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L., Sha Y., Wu D., Wei Q., Chen D., Yang S., Jia F., Yuan Q., Han X., Wang J. Surfactant induces ROS-mediated cell membrane permeabilization for the enhancement of mannatide production. Process Biochem. 2020;91:172–180. [Google Scholar]
- 28.Sharma P., Vaiwala R., Parthasarathi S., Patil N., Verma A., Waskar M., Raut J.S., Basu J.K., Ayappa K.G. Interactions of surfactants with the bacterial cell wall and inner membrane: revealing the link between aggregation and antimicrobial activity. Langmuir. 2021;38(50):15714–15728. doi: 10.1021/acs.langmuir.2c02520. [DOI] [PubMed] [Google Scholar]
- 29.Zhong H., Liu G., Jiang Y., Yang J., Liu Y., Yang X., Liu Z., Zeng G. Transport of bacteria in porous media and its enhancement by surfactants for bioaugmentation: a review. Biotechnol. Adv. 2017;35(4):490–504. doi: 10.1016/j.biotechadv.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 30.Wang X., He Z., Luo H., Zhang M., Zhang D., Pan X., Gadd G.M. Multiple-pathway remediation of mercury contamination by a versatile selenite-reducing bacterium. Sci. Total Environ. 2018;615:615–623. doi: 10.1016/j.scitotenv.2017.09.336. [DOI] [PubMed] [Google Scholar]
- 31.Lu J., Wang Y., Zhang S., Bond P., Yuan Z., Guo J. Triclosan at environmental concentrations can enhance the spread of extracellular antibiotic resistance genes through transformation. Sci. Total Environ. 2020;713 doi: 10.1016/j.scitotenv.2020.136621. [DOI] [PubMed] [Google Scholar]
- 32.Siyal A.A., Shamsuddin M.R., Low A., Rabat N.E. A review on recent developments in the adsorption of surfactants from wastewater. J. Environ. Manag. 2020;254 doi: 10.1016/j.jenvman.2019.109797. [DOI] [PubMed] [Google Scholar]
- 33.Zhu X., Wang Z., Sun Y., Gu L., Zhang L., Wang J., Huang Y., Yang Z. Surfactants at environmentally relevant concentrations interfere the inducible defense of Scenedesmus obliquus and the implications for ecological risk assessment. Environ. Pollut. 2020;261 doi: 10.1016/j.envpol.2020.114131. [DOI] [PubMed] [Google Scholar]
- 34.Hu X., Sheng X., Zhang W., Lin Z., Gao Y. Nonmonotonic effect of montmorillonites on the horizontal transfer of antibiotic resistance genes to bacteria. Environ. Sci. Technol. Lett. 2020;7(6):421–427. [Google Scholar]
- 35.Jin M., Liu L., Wang D., Yang D., Liu W.L., Yin J., Yang Z., Wang H., Qiu Z., Shen Z., Shi D., Li H., Guo J., Li J. Chlorine disinfection promotes the exchange of antibiotic resistance genes across bacterial genera by natural transformation. ISME J. 2020;14(7):1847–1856. doi: 10.1038/s41396-020-0656-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Lu J., Engelstädter J., Zhang S., Ding P., Mao L., Yuan Z., Bond P.L., Guo J. Non-antibiotic pharmaceuticals enhance the transmission of exogenous antibiotic resistance genes through bacterial transformation. ISME J. 2020;14(8):2179–2196. doi: 10.1038/s41396-020-0679-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y., Gu A.Z., He M., Li D., Chen J. Subinhibitory concentrations of disinfectants promote the horizontal transfer of multidrug resistance genes within and across genera. Environ. Sci. Technol. 2017;51(1):570–580. doi: 10.1021/acs.est.6b03132. [DOI] [PubMed] [Google Scholar]
- 38.Xiao X., Lin W., Chen Z., Zou C., Jin H. Wide-host vector pBBR1MCS2-tac-EGFP suitable for the labeling of Ralstonia solanacearum. Chin. J. Top. Crops. 2020;42(6):1700. [Google Scholar]
- 39.Dixon P. VEGAN, a package of R functions for community ecology. J. Veg. Sci. 2003;14(6):927–930. [Google Scholar]
- 40.McMurdie P.J., Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4) doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas C.M., Nielsen K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005;3:711. doi: 10.1038/nrmicro1234. [DOI] [PubMed] [Google Scholar]
- 42.Nielsen K.M., Johnsen P.J., Bensasson D., Daffonchio D. Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 2007;6(1–2):37–53. doi: 10.1051/ebr:2007031. [DOI] [PubMed] [Google Scholar]
- 43.Mao D., Luo Y., Mathieu J., Wang Q., Feng L., Mu Q., Feng C., Alvarez P.J.J. Persistence of extracellular DNA in river sediment facilitates antibiotic resistance gene propagation. Environ. Sci. Technol. 2014;48(1):71–78. doi: 10.1021/es404280v. [DOI] [PubMed] [Google Scholar]
- 44.Dubnau D. DNA uptake in bacteria. Annu. Rev. Microbiol. 1999;53(1):217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- 45.Chang P.H., Juhrend B., Olson T.M., Marrs C.F., Wigginton K.R. Degradation of extracellular antibiotic resistance genes with UV254 treatment. Environ. Sci. Technol. 2017;51(11):6185–6192. doi: 10.1021/acs.est.7b01120. [DOI] [PubMed] [Google Scholar]
- 46.Dai K., He L., Chang Y.F., Cao S., Zhao Q., Huang X., Wu R., Huang Y., Yan Q., Han X., Ma X., Wen X., Wen Y. Basic characterization of natural transformation in a highly transformable Haemophilus parasuis strain SC1401. Front. Cell. Infect. Microbiol. 2018;8:32. doi: 10.3389/fcimb.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Winter M., Buckling A., Harms K., Johnsen P.J., Vos M. Antimicrobial resistance acquisition via natural transformation: context is everything. Curr. Opin. Microbiol. 2021;64:133–138. doi: 10.1016/j.mib.2021.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Wang X., Yang F., Zhao J., Xu Y., Mao D., Zhu X., Luo Y., Alvarez P.J.J. Bacterial exposure to ZnO nanoparticles facilitates horizontal transfer of antibiotic resistance genes. NanoImpact. 2018;10:61–67. [Google Scholar]
- 49.Wang X., Li H., Chen Y., Meng X., Dieketseng M.Y., Wang X., Yan S., Wang B., Zhou L., Zheng G. A neglected risk of nanoplastics as revealed by the promoted transformation of plasmid-borne ampicillin resistance gene by Escherichia coli. Environ. Microbiol. 2022;24(10):4946–4959. doi: 10.1111/1462-2920.16178. [DOI] [PubMed] [Google Scholar]
- 50.Zhang S., Wang Y., Lu J., Yu Z., Song H., Bond P.L., Guo J. Chlorine disinfection facilitates natural transformation through ROS-mediated oxidative stress. ISME J. 2021;15(10):2969–2985. doi: 10.1038/s41396-021-00980-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hu X., Waigi M.G., Yang B., Gao Y. Impact of plastic particles on the horizontal transfer of antibiotic resistance genes to bacterium: dependent on particle sizes and antibiotic resistance gene vector replication capacities. Environ. Sci. Technol. 2022;56(21):14948–14959. doi: 10.1021/acs.est.2c00745. [DOI] [PubMed] [Google Scholar]
- 52.Lu J., Ding P., Wang Y., Guo J. Antidepressants promote the spread of extracellular antibiotic resistance genes via transformation. ISME Commun. 2022;2:63. doi: 10.1038/s43705-022-00147-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Z., Wang Y., Henderson I.R., Guo J. Artificial sweeteners stimulate horizontal transfer of extracellular antibiotic resistance genes through natural transformation. ISME J. 2022;16(2):543–554. doi: 10.1038/s41396-021-01095-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liao J., Chen Y., Huang H. Effects of CO2 on the transformation of antibiotic resistance genes via increasing cell membrane channels. Environ. Pollut. 2019;254 doi: 10.1016/j.envpol.2019.113045. [DOI] [PubMed] [Google Scholar]
- 55.Yu Z., Wang Y., Lu J., Bond P.L., Guo J. Nonnutritive sweeteners can promote the dissemination of antibiotic resistance through conjugative gene transfer. ISME J. 2021;15(7):2117–2130. doi: 10.1038/s41396-021-00909-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen I., Dubnau D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004;2:241–249. doi: 10.1038/nrmicro844. [DOI] [PubMed] [Google Scholar]
- 57.Lim Y., Su C.H., Liao Y.C., Lee S.Y. Impedimetric analysis on the mass transfer properties of intact and competent E. coli cells. Biochim. Biophys. Acta Biomembr. 2019;1861(1):9–16. doi: 10.1016/j.bbamem.2018.10.010. [DOI] [PubMed] [Google Scholar]
- 58.Haijema B.J., Hahn J., Haynes J., Dubnau D. A ComGA-dependent checkpoint limits growth during the escape from competence. Mol. Microbiol. 2001;40(1):52–64. doi: 10.1046/j.1365-2958.2001.02363.x. [DOI] [PubMed] [Google Scholar]
- 59.De Furio M., Ahn S.J., Burne R.A., Hagen S.J. Oxidative stressors modify the response of Streptococcus mutans to its competence signal peptides. Appl. Environ. Microbiol. 2017;83(22) doi: 10.1128/AEM.01345-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Panja S., Aich P., Jana B., Basu T. How does plasmid DNA penetrate cell membranes in artificial transformation process of Escherichia coli? Mol. Membr. Biol. 2008;25(5):411–422. doi: 10.1080/09687680802187765. [DOI] [PubMed] [Google Scholar]
- 61.Stark G.J. Functional consequences of oxidative membrane damage. J. Membr. Biol. 2005;205(1):1–16. doi: 10.1007/s00232-005-0753-8. [DOI] [PubMed] [Google Scholar]
- 62.Naresh C.G., Dipankar S., Anjali G. Synthetic detergents (surfactants) and organochlorine pesticide signatures in surface water and groundwater of greater Kolkata, India. J. Water Resour. Protect. 2009;1:9. [Google Scholar]
- 63.Seth R., Singh P., Mohan M., Singh R., Aswal R.S. Monitoring of phenolic compounds and surfactants in water of Ganga Canal, Haridwar (India) Appl. Water Sci. 2013;3(4):717–720. [Google Scholar]
- 64.Islam M.M., Islam M.N., Rima F.R., Islam M.N. Quality analysis, miceller behavior, and environmental impact of some laundry detergents available in Bangladesh. Environ. Sci. Pollut. Res. 2016;23(6):5468–5476. doi: 10.1007/s11356-015-5724-8. [DOI] [PubMed] [Google Scholar]
- 65.Gordon A.K., Blatch G.L., Daniel S., Muller W.J. Stress protein responses in South African freshwater invertebrates exposed to detergent surfactant linear alkylbenzene sulfonate (LAS) Water Air Soil Pollut. 2008;193(1):123–130. [Google Scholar]
- 66.Mara D. Routledge; 2013. Domestic Wastewater Treatment in Developing Countries. [Google Scholar]
- 67.Afzal M., Arslan M., Müller J.A., Shabir G., Islam E., Tahseen R., Anwar-ul-Haq A., Hashmat A.J., Iqbal S., Khan Q.M. Floating treatment wetlands as a suitable option for large-scale wastewater treatment. Nat. Sustain. 2019;2(9):863–871. [Google Scholar]
- 68.Xu Z., Xu J., Yin H., Jin W., Li H., He Z. Urban river pollution control in developing countries. Nat. Sustain. 2019;2(3):158–160. [Google Scholar]
- 69.Palmer M., Hatley H. The role of surfactants in wastewater treatment: impact, removal and future techniques: a critical review. Water Res. 2018;147:60–72. doi: 10.1016/j.watres.2018.09.039. [DOI] [PubMed] [Google Scholar]
- 70.Zhang Y., Shen Y. Wastewater irrigation: past, present, and future. WIREs Water. 2019;6(3):e1234. [Google Scholar]
- 71.Bandala E.R., Kruger B.R., Cesarino I., Leao A.L., Wijesiri B., Goonetilleke A. Impacts of COVID-19 pandemic on the wastewater pathway into surface water: a review. Sci. Total Environ. 2021;774 doi: 10.1016/j.scitotenv.2021.145586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Temkin E., Fallach N., Almagor J., Gladstone B.P., Tacconelli E., Carmeli Y. Estimating the number of infections caused by antibiotic-resistant Escherichia coli and Klebsiella pneumoniae in 2014: a modelling study. Lancet Global Health. 2018;6(9):e969–e979. doi: 10.1016/S2214-109X(18)30278-X. [DOI] [PubMed] [Google Scholar]
- 73.Murray C.J., Ikuta K.S., Sharara F., Swetschinski L., Aguilar G.R., Gray A., Naghavi M. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Blokesch M. Natural competence for transformation. Curr. Biol. 2016;26(21):1126–1130. doi: 10.1016/j.cub.2016.08.058. [DOI] [PubMed] [Google Scholar]
- 75.Liu S., Qu H., Yang D., Hu H., Liu W., Qiu Z., Hou A., Guo J., Li J., Shen Z., Jin M. Chlorine disinfection increases intracellular and extracellular antibiotic resistance genes in a full-scale wastewater treatment plant. Water Res. 2018;136:131–136. doi: 10.1016/j.watres.2018.02.036. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.