Abstract
Food borne pathogens are one of the most common yet concerning cause of illnesses around the globe. These microbes invade the body via food items, through numerous mediums of contamination and it is impossible to completely eradicate these organisms from food. Extensive research has been made regarding their treatment. Unfortunately, the only available treatment currently is by antibiotics. Recent exponential increase in antibiotic resistance and the side effect of synthetic compounds have established a need for alternate therapies that could be utilized either on their own or along with antibiotics to provide protection against food-borne diseases. The aim of this review is to provide information regarding some common food borne diseases, their current and possible natural treatment. It will include details regarding some common foodborne pathogens, the disease they cause, prevalence, manifestations and treatment of the respective disease. Some natural modes of potential treatment will be summarized, which including phytochemicals, derived from plants either as crude extracts or as purified form and Bacteriocins as microbial based treatment, obtained from various types of bacteria. The paper will describe their mechanism of action, classification, susceptible organisms, some antimicrobial compounds and producing organisms, application in food systems and as potential treatment. Along with that, synthetic treatment i.e., antibiotics will be discussed including the first-line treatment of some common food borne infections, prevalence and mechanism of resistance against antibiotics in the pathogens.
Keywords: Food borne pathogens, Food pathogens, Natural antimicrobials, Food borne disease, Bacteriocin, Phytochemical
Graphical abstract
List of Abbreviations
- LAB
Lactic Acid Bacteria
- FDA
Food and Drug Administration
- MDR
Multi Drug Resistance
- GRAS
Generally Regarded As Safe
- STEC
Shiga Toxin Producing Escherichia coli
- LPS
Lipopolysaccharides
1. Introduction
There are several microorganisms which possess pathogenic traits. When these pathogenic microorganisms travel from their source of origin to humans via different food products and by travelling through different routes, they cause several diseases in humans known as foodborne illness and these particular pathogenic microorganisms are termed as foodborne pathogens [1]. Among various foodborne pathogens bacterial pathogens are the most common cause of foodborne illness [2].
Food borne pathogens come from different sources, most commonly found in plant, soil, animal and water. Zoonotic transmission occurs mostly due their residence in animal products such as eggs, dairy products, meat, poultry, etc. They can also contaminate the food products while processing due to unhygienic handling by the workers and malpractice can result in cross contamination [2]. Many pathogenic bacteria have unique and different properties and they survive on different conditions. Some of the most common food borne pathogens are Listeria monocytogens, Escherichia coli, Campyloacter jejuni, Clostridium botulinum, Bacillus subtilis, Salmonella typhi, and Staphylococcus aureus, resulting in foodborne illnesses like campylobacteriosis, listeriosis, salmonellosis and E. coli infections.
Once these pathogens are consumed with any of the food products, the onset of action occurs after the incubation period. It results in many infections, sometimes life threatening, immunocompromised people are majorly affected. The symptoms depends on the consumption amount of the microorganism nevertheless there are some symptoms similar to one and other which occurs in the illness caused by all of these organisms however they mostly tend to alter the gut microflora causing nausea, diarrhea, abdominal pain etc. [3].
Due to the contamination of these pathogenic microorganisms, food safety is becoming a major concern on a global level. 33 million people are affected by foodborne illness annually across the globe [4]. Many of the pathogenic microbes are heat resistant, spore forming, mesophilic and psychotropic, due to these exceptional surviving conditions they have a potential risk of surviving even after cleaning and sterilization process and becomes a great a risk of illness [2,5]. Although there is a continuous development in food technology but the morbidity caused by food borne illness is still prevailing specially in developing countries [5]. Both health and economic sector of the countries worldwide suffers due to this threat.
For the safety of food products from pathogenic microbes, there are several preservation techniques that has been developed however some negative effects of synthetic preservatives including formaldehyde, sorbates, sulfites, nitrates have been observed which causes allergic reactions and leads to a number of health problems [6,7]. Therefore there is an increasing demand of natural compounds and due to their effectiveness against various pathogens they are being considered as a safer option over synthetic ones.
Bio-preservation is a technique in which food products are preserved by using natural compounds. Several natural compounds have antibacterial properties which help to sustain and ensure the food safety [7]. Many natural preservatives and antimicrobial compounds are being used to counter numerous pathogenic microorganisms. These compounds are obtained from different sources including bacteria, plants, animals, fungi, mushrooms, virus, etc.
Among various natural preservatives there are some novel compounds which have been produced by particular species of Lactic Acid Bacteria (LAB) producers, these novel compounds are proteinaceous in nature and are known as bacteriocins [8]. They have an effective ability of inhibiting bacteria by several mechanisms and are regarded as safe to use in food products. A commercially used bacteriocin, used a preservative and approved by Food and Drug Administration (FDA) is Nisin [9]. Bacteriocins have inhibited the activity of many foodborne pathogens through different mechanism of actions and by different way of usage in food products [9,10].
Another antibacterial alternative which is of a great focus of attention and is covered in this article in detail are the antimicrobial compounds obtained from plants. Different compounds having antimicrobial properties such as phenols, terpenoids and alkaloids are achieved from different parts of the plant extracts as well as the secondary metabolites. All of them perform their action by certain mechanisms and are founds to be helpful in the treatment of food borne illness [11,12].
Multiple antibiotics are being used against several foodborne illnesses such as Fluroquinolones against salmonellosis [13]. Macrolides against campylobacteriois [14], β-lactam and aminoglycosides against listeriosis [15], Ciprofloxacin against E. coli infections [16]. However there is an emerging resistance of these pathogens against the conventional antibiotics. Through different mechanisms, microbes gets resistant to the antibiotics [17]. The insufficient use of antibiotics is leading towards multi drug resistance. Therefore, to combat this threat, the use of natural compounds is a focus of attention by many researchers and continuous studies are being done to discover novel compounds, which would help decreasing the risk of food related diseases [7].
This review article provided an insight of the food borne diseases, their causative agents, prevalence, and the different treatments to combat these pathogens. It also involves information about synergistic antimicrobial activity of synthetic and natural antimicrobials along with details about antibiotic resistance and their mode of actions. Finally, the study also entails a comparative analysis between the synthetic and natural antibiotics to overcome the knowledge gap of comparative analysis of efficiency of antibiotics i.e. synthetic antimicrobials, and that of bacteriocins and phytochemicals, which are considered as natural antimicrobials. The review article covers data from articles, which are focused on the food pathogens and the diseases caused by them, the treatments used for the diseases, both natural and synthetic. Articles, which focused on the sampling, isolation, and molecular identification of the food borne pathogens were excluded as this review article focuses on the comparative analysis of the treatments used against them. To best of our knowledge, this review article is a comparative analysis of the synthetic and natural treatment methods against food borne pathogens till date.
2. Food borne diseases
2.1. Listeriosis
The genus of Listeria is comprised of facultative anaerobic gram positive bacteria. The pathogenic species in this genus are L. ivanovii and L. monocytogenes. These two species have been responsible for the severe diseases caused in animals as well as humans, however only L. monocytogenes is known to infect humans, causing listeriosis, a disease with mortality rate of around 30% [18].
Listeriosis can be either invasive or noninvasive gastroenteritis [19]. The more commonly people affected by listeriosis are immunocompromised, pregnant women and older people. Once the listeriosis gets invasive it leads to further complications such as meningitis, sepsis and neonatal infections along with miscarriage [20]. The classification of invasive listeriosis is done in three categories including maternal neonatal infection, bacteraemia and neurolisteriosis [21].
Listeria can survive freezing temperatures and grow at temperatures as low as 0 °C. The primary habitat of Listeria monocytogenes is soil, from where it can spread to various food items grown, cattle or farmers and other workers, and enters the food chain. There are certain properties of Listeria monocytogenes which make it highly resilient. The properties includes: i) ability to survive freezing environment, ii) tolerating high salt concentration (up to 20% w/v) iii) biofilm production on surfaces of metals and plastics providing protection against environmental stress [15,22]. Due to the presence of these properties L. monocytogens is not prevented properly while food processing, and contaminate different types of food products such as meat, sausages, fish, dairy products, fruits and vegetables [23,24].
It gets difficult to control listeriosis outbreak because of the longer incubation period and widespread food trade connections [25]. Presence of this pathogen in food items have been reported multiple times suggesting the importance of contamination control [26,27].
2.2. Escherichia coli infections
Escherichia coli belongs to the family Enterobacteriaceae. The bacterium is gram negative, facultative anaerobe. Escherichia coli is a versatile microorganism which has both pathogenic and non-pathogenic variants. The non-pathogenic variants are found in the normal human gut flora. However, a number of pathogenic variants have been responsible to cause different types of intestinal or extra-intestinal infections in animals and humans [28]. Moreover, some of the strains of E. coli are commensal as they do not possess any virulence factors and do not harm the host [29].
Intestinal pathogenic E. coli (IPEC) are identified as obligate pathogens. Diseases like colitis or gastroenteritis is caused by IPEC. Six categories of diarrheagenic E. coli pathotypes which belongs to the intestinal pathogenic E. coli have been identified including, enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enterotoxigenic E. coli (ETEC), enteroaggregative E. coli (EAEC), enteroinvasive E. coli (EIEC) and diffusely adherent E. coli (DAEC) [30]. Enteropathogenic E. coli strain is a producer of shiga toxin (STEC) which is responsible for various diseases such as hemorrhagic colitis, bloody diarrhea and severe hemolytic uremic syndrome which can be life threatening [31].
Certain isolates such as uropathogenic E. coli (UPEC), neonatal meningitis associated E. coli (NMEC) and sepsis causing E. coli (SEPEC) have been isolated from outside of the intestinal tract and they are classified as Extra-intestinal Pathogenic E. coli (ExPEC). They are facultative pathogens [32] and can cause various infections at non-intestinal site such as prostate infections, bloodstream infections and most commonly urinary tract infection [33].
Number of different food items such as vegetables, meat, and raw milk is contaminated by E. coli and it is one of the major microorganism, which causes food borne diseases [34]. Pathogenic E. coli reside in the animal feces and are mostly found in soil and water due to animal fecal contamination [35]. The diseases can also reach the host through different routes which gets contaminated by feces [36].
2.3. Campylobacteriosis
Campylobacteriosis is any disease caused by the bacteria of genus Campylobacter. Campylobacter spp. Are Gram negative, spiral or rod shaped, motile, non-endospore forming bacteria. Motility contributes to virulence of pathogens as it helps in adherence to the intestinal walls. Campylobacter species are micro-aerophiles i.e., they require 5% Oxygen, 10% CO2 and 85% Nitrogen to survive. They are thermophiles i.e. can grow between 30°C and 46 °C (optimally at 40–42 °C) [37]. The size of Campylobacter ranges between 0.2 and 0.8 μm in width and 0.5–5 μm length. There is a total of 32 species and 12 sub species of Campylobacter but the infections in animals and humans are mainly caused by C. jejuni (95%) and C. coli (5%) [37,38]. Campylobacteriosis in humans is one of the most prevalent cause of Gastroenteritis. Depending on the severity of infection, the symptoms can range from mild diarrhea and vomiting to bloody diarrhea, abdominal cramps, fever and in very severe cases, it can lead to Guillain-Barre syndrome, an autoimmune condition that can ultimately lead to paralysis [38,39].
Zoonotic transmission by farm animals is one of the major sources of Campylobacter spread [40]. Natural habitat of campylobacter is in the GI tract of animals and birds and therefore, the infection can spread via feces contaminating food and water or poor food handling during slaughtering or food processing. Farm animals are one of the major sources of Campylobacter spread. The foods associated with Campylobacteriosis are mainly poultry and meat products such as eggs, chicken, turkey, beef and many other [41,42]. A study has been reported related to Campylobacter contamination in various food sources such as chicken, beef, meat, raw milk, and cheese, ready to eat foods and vegetables over a period of three years in Pakistan. It showed that raw chicken was the most frequently contaminated meat product (48%) and vegetables and salads (40.9%) were the most contaminated among other foods [43]. According to WHO, 550 million people are sick from diarrhea diseases with 33 million deaths due to food-borne illness, campylobacter being 1 of the 4 key global cause of Gastroenteritis worldwide signifies the importance of intervention against Campylobacter related food safety [44].
2.4. Salmonellosis
Salmonellosis is any disease caused by species belonging to Salmonella genus. The bacteria belonging to this group are Gram negative, facultative anaerobe, non-spore forming, motile (peritrichous flagella), straight bacilli and facultative pathogens. The bacteria size ranges between 2 and 3 μm, belonging to the Enterobacteriaceae family and are divided into two species, S. bongori and S. enterica, the latter specie is further divided into seven sub species and over 2600 serotypes. They are ubiquitous, and have the ability to grow over a wide range of conditions. Temperature usually range between 8 and 45 °C with 37 °C as optimum. However, as there are numerous bacteria in this genus, temperature ranges between 2 and 54 °C, pH of 4–9.5 with 6.5–7.5 being optimum, and can also grow in low moisture environments where water activity is 0.94 or above [37,45].
There are two types of salmonellosis: typhoidal and non-typhoidal salmonellosis. Non-typhoidal salmonellosis is further divided into invasive and non-invasive salmonellosis. Non-invasive Non-typhoidal salmonellosis causes gastroenteritis, usually the diarrhea is self-limiting and patient does not require any antibiotic treatment. Invasive non-typhoidal salmonellosis is characterized by invading sites that are otherwise free of microbes leading to systemic infections followed by localized infections such as meningitis [46,47]. Fatality rate of invasive salmonellosis is greater than non-invasive salmonellosis [47]. Serovar typhimurium and entriditis mainly cause invasive salmonellosis [48,49].
Typhoidal Salmonella is caused by Salmonella enterica serovars typhi and paratyphi A, B and C. Rest of the S. enterica species are responsible for non-typhoidal salmonellosis. Typhoidal strains are responsible for causing typhoid and paratyphoid fever collectively called as enteric fever [48,50]. The enteric fever may be presented with GIT disturbances such as diarrhea, abdominal pain, nausea, vomiting and cold-like symptoms such as sore throat, dry cough, headache and weakness [50].
Salmonellosis is one of the major food-borne disease, spreads via contaminated food such as beef, chicken/broilers, eggs pork [49,51,52]. Fecal contamination by animals or humans can spread it to vegetables/fruits, milk and water [52]. GBD (2017) described the prevalence of non-invasive and non-typhoidal salmonellosis globally from the year 1990–2017. The estimated cases ranged between 409.1000 and 622, 000 cases each year (2010–2017) with an average mortality rate of 14.5%. Majority of the infected population being children under the age of five [47]. The globally estimated cases of typhoid (76.3%) and paratyphoid fever are 14·3 million with a fatality rate of 0·95% [53]. These massive incidence rates indicates that variable intervention and control strategies must be adapted in order to reduce food-borne diseases’ incidence and mortality rate.
3. Natural modes of treatment
3.1. Bacteriocins
Contamination by pathogens is an ongoing and a very major concern in food industry which causes many food borne diseases. Regardless of the use of recent food preservation techniques, there is an increase in food related illness especially in those countries which has a poor monitoring system of food safety. A population of one third across the globe is suffering because of consuming intoxicated or contaminated food products such as meat, poultry, and dairy products [54].
The demand for safe food with minimum chemical additives is increasing. Biopreservation involves improvement of microbiological safety of foods and extend their shelf life through the use of non-pathogenic microorganisms [55]. Due to the safe history of use in fermented food products, lactic acid bacteria (LAB) has been of great importance and are attractive to be utilized as biopreservative [56]. Many new novel peptides having antimicrobial properties have been characterized by the isolates of LAB producers [57].
Bacteriocins are antimicrobial peptides which are synthesized ribosomally. Bacteriocins can be produced by both gram negative and gram positive bacteria as it has been suggested that this proteinaeous substance can be produced by all bacterial species [58,59]. However they are mostly produced by a wide range of LAB [60]. A large number of bacteriocins have been isolated from different strains of LAB such as L. lactis, L. lactis, L. plantarus, L. bulgaricus and L. acidophilus [61].
As bacteriocins have been isolated from different strains of LAB and Lactobacillus species, their isolation is based on the inhibitory activity against pathogenic bacteria, which is used as an indicator strain. Strains such as L. bavaricus, L. brevis, L. plantarum, and L. lactis have been used to check the inhibitory activity against L. monocytogenes, S. aureus, E. coli. [62].
Bacteriocins have the ability to kill closely related species (narrow spectrum), and in some cases they are able to kill the species of broad spectrum as well [63]. The growth of closely related species is inhibited by several mechanisms [64]. Microorganisms of different species become sensitive because of the interaction of cell membrane or cell surface with bacteriocin and the major mechanisms by which the targeted bacterial cell is inhibited are cell permeabilization, and pore formation [65].
Due to the toxic property of bacteriocins, they also become lethal to the bacteria through which they are produced. However, they get protected by a set of immunity proteins [66]. An operon cluster of genes that encode the bacteriocin, group of immunity proteins along with some additional proteins is present either in plasmid or genome [67].
Some of the infectious diseases have been prevented by bacterioicins, which were caused by gram-positive bacterial pathogenic strains such as Streptococci, Staphylococci and Micrococci. Along with these, bacterial strains by Gram-negative bacteria such as Salmonella, Shigella, Vibrio and Listeria have also been tested against the antibacterial activity of bacteriocin protein [68].
Some of the very useful and important characteristics found in bacteriocins produced by LAB are: a) being able to be active over a wide range of pH b) able to tolerate high thermal stress c) easily degradable by proteolytic enzymes d) highly specific in terms of employing receptors. Due to these characteristics, bacteriocins have been considered ideal as a useful component in food preservation [69,70].
Over the past years, a wide range of pathogenic bacteria is getting resistant to the conventional antibiotics and this emergence is leading towards the research and screening of natural compounds having effective mechanism of inhibition against the resistant bacteria. There are several bacterial species, which are getting resistant to the conventional antibiotics. Bacterial strains that show resistance to one or more antimicrobial agents are known as Multi Drug Resistant (MDR) bacteria. The use of such novel compounds would be helpful to replace antibiotics, which already exist, or contributes to make antibiotics efficacious [69,71]. A bacteriocin named as Nisin is produced by the specie Lactococcus lactis. It has been approved and has been granted Generally Regarded as Safe (GRAS) status by the FDA and is commercially used as a food preservative in more than 60 countries [72]. It belongs to Class I of bacteriocins which is known as lantibiotics, and is active against many gram positive bacteria which leads to their extensive use in food industry [60]. Thus, due to the stable property of bacteriocins, they are considered as a favorable alternatives in food preservations. Other than nisin, several bacteriocins have been isolated from probiotic organisms and have been found to be very effective against the multi drug resistant pathogens. One of them is reported as KAE01, isolated from Enterococcus faecium, has been found to have antimicrobial activity against Pseudomonas aeruginosa [55]. KAE01 was found to be stable at different physical parameters of temperature, pH and in presence of enzymes. These findings lead to the fact that bacteriocins can be used as food preservatives.
Due to the fast acting mechanism of bacteriocins, they are able to make pores in the target membrane even at very low concentrations. This reason is of great importance as it leads to lower chances of resistant development in bacteria. Whereas conventional antibiotics gets easily resistant in comparison of a specific and potent bacteriocin. Another advantage of bacteriocin being replaced by the antibiotics is the nature of these peptides, which is a primary metabolite and antibiotics are the secondary metabolites. The biosynthetic mechanism of a primary metabolite is comparatively simpler than the secondary metabolite and they can easily go through the bioengineering process to become more specific or active against a target microorganism [65]. Summarized data of the bacteriocins, their producers and the organisms against which they have antimicrobial activity have been summarized in Table 1.
Table 1.
Some of the bacteriocins, their producer species and the pathogenic species against which they are active.
| Bacteriocin | Produced by | Active against | Reference |
|---|---|---|---|
| Enterocin KAE01 | Enterococcus faecium | Pseudomonas aeruginosa | [55] |
| Lacticin 3147 | Lactococcus lactis | Staphylococcus aureus | [73] |
| Mutacin B-Ny266 | Streptococcus mutans | Staphylococcus aureus | [74] |
| Mutacin B-Ny266 | Streptococcus mutans | Escherichia coli | [75] |
| Pediocin PA-1 | Listeria monocytogenes | [76] | |
| ST151BR | Lactobacillus pentosus | Escherichia coli | [77] |
| Thermophylin | Streptococcus thermophiles | Listeria monocytogenes Salmonella typhimurium | [78] |
| B602 | Paenibacillus polymyxa | Campylobacter jejuni | [79] |
| OR-7 | Lactococcus salivarius | Campylobacter jejuni | [80] |
| Enterocin E−760 | Enterococcus sp. | Campylobacter jejuni | [81] |
| Albusin B | Ruminococcus albus 7 | Salmonella | [82] |
In some approaches, two different genes of bacteriocins are integrated together through recombinant PCR technique such as enterocin CL35 and microcin V genes which results in a combined bacteriocin named as Ent35-MccV. This combined bacteriocin is active against Listeria and Enterohemmorhagic E. coli [61].
Initially, these antimicrobial peptides (bacteriocins) produced by Gram positive bacteria were classified broadly into two categories: 1) Peptides which are ribosomally synthesized and post translationally modified, and 2) peptides which are unmodified [65]. As bacteriocins are produced by both Gram positive and Gram negative bacteria, therefore they are classified into different categories. Bacteriocins produced by Gram negative bacteria predominantly arise from Enterobacteriaceae and are divided into two classes: 1) colicins which are larger peptides and have high molecular mass; and 2) microcins which are smaller peptides and have relatively low molecular mass [83]. Due to the production of more prominent and potent bacteriocins by different species of LAB, they are considered as true bacteriocins and based on their genetic characteristics, molecular weight and presence of amino acid sequence, they were initially divided into four classes [84] along with some sub divisions depending on the taxonomy of the microorganism [85]. However, due to the composition of bacterioicins with large complexes of lipids and carbohydrates in class IV, it has been terminated and has been named as bacteriolysins [86]. Therefore, bacteriocins are majorly divided into three classes [87].
Bacteriocins in class I are broadly post translationally modified peptides and are further divided into two classes. Class Ia containing dehydrated amino acids and β-methyl lanthionine. They are known as lantibiotics. The peptides are linear, positively charged typically <5 kDa in size. Class Ib peptides are negatively charged and globular in shape [88].
Class II bacteriocins are relatively smaller in size with unmodified peptides. It is further divided into four sub classes:
IIa) This class include pediocin like petides.
IIb) This class have unmodified bacteriocins formed by two peptides.
IIc) This class has circular peptides.
IId) This class has linear peptides which are unmodified and are non pediocin like bacteriocins.
Among all of the four classes, pediocin like bacteriocins are the most dominant ones because of their distinctive features [66,89,90]. Bacteriocins of Class III are heat labile are larger peptides typically >30 kDa in size. An example of this class is colicin [91]. Recently, a novel bacteriocin helviticin M produced by Lactobacillus helveticus has been characterized. It is known to be effective against both Gram negative and Gram-positive bacteria due to the disruption of their cell wall [92].
3.2. Plant-based antimicrobials
Phytochemicals have gotten a lot of attention by researchers due to their diversity and numerous beneficial properties. Many researches have been done on the antimicrobial effects of various plant products. There are numerous plant species/varieties and even more products that could be used to treat food-borne diseases. Plant extracts of different parts such as leaves, stems, flowers, seeds, roots etc. Are used to obtain purified secondary metabolites, such as essential oils which are being tested for their antimicrobial potential. For the sake of simplicity, plant antimicrobials could be divided into three categories: 1st generation: Crude antimicrobials; 2nd generation: purified active compounds; and 3rd generation: Purified compounds with detailed pharmacological and biochemical assessment [93].
3.2.1. Mechanism of action
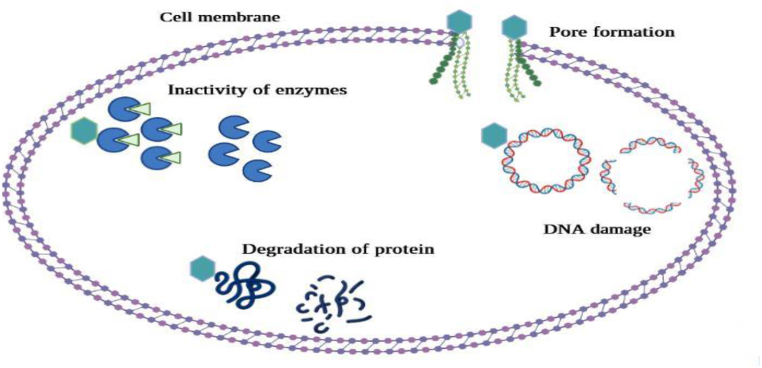
Mechanism of action of various phytochemicals is dependent upon their chemical nature. These compounds have the ability to either be bacteriostatic or bactericidal. Generally, there are many mechanisms by which plants exhibit the antimicrobial activity such as destabilizing plasma membrane, inhibition of biofilm synthesis, inhibition of intracellular enzymes, neutralization of bacterial toxins such as enterotoxins released by food borne pathogens responsible for causing diarrhea [94]. The phytochemicals inhibit cell to cell signaling in the neighboring bacteria. The signaling has the significance of stimulating various functions such as toxin production and biofilm production, which helps the bacteria in the invasion of the host and resist the environmental factors. Other mechanisms involve inhibition of efflux pumps, which are responsible for multidrug resistance in bacteria, inhibition of DNA synthesis, inhibition of cell wall synthesis or impairment of energy metabolism [12,93]. Different mechanisms of actions have been illustrated in Fig. 1.
Fig. 1.
Mechanism of action of natural antimicrobials.
3.2.2. Classification of phytochemicals and their antimicrobial properties
There are hundreds of plant-based compounds, each differing in its properties. Primarily, plant secondary metabolites are divided into three classes: 1) Alkaloids 2) Phenols 3) Terpenoids. Alkaloids are diverse group of compounds characterized by being basic, nitrogen containing compounds. The compounds belonging to this class are variable in their structures and possess a broad range of antimicrobial activity. They are further divided into two classes. Typical alkaloids which contain heterocyclic ring with nitrogen attached and atypical alkaloids which do not possess the heterocyclic ring. Typical alkaloids are much more commonly found in nature such as morphine, codeine. Typical alkaloids are further divided into many categories [12,95]: Erythromycin, a common macrolide antibiotic belongs to atypical class of alkaloids. Some of the typical alkaloids having antimicrobial activity against food borne pathogens are Piperine, a piperidine type alkaloid, isolated from black pepper. Berberine, is an isoquinoline alkaloid isolated from Berberis species, reserpine isolated from tropical plant Rauwolfia, tomatidine isolated from the leaves of tomatoes and many more, summarized by Khameneh et al.
The other class of secondary metabolites is Phenols (phenyl ring attached to a hydroxyl group), are compounds usually released as antimicrobials for defense against microbes, pests or animals. Phenols having strong reducing power are potent antioxidants, scavenges free radicals and can chelate metal ions therefore, prevent lipid or protein oxidation. Phenols are divided into many sub divisions based on their chemical nature such as simple phenols, phenolic acids, flavonoids, tannins, coumarins, quinines, lignans, stilbenes and xanthones [96]. Some of the phenolic compounds having antimicrobial activity are Resveratrol, effective against Campylobacter jejuni possessing cmeABC efflux pump [97]. Coumarin is effective against various food borne pathogens as shown in Table 2A, Table 2Ba and 2b along with Quercetin and Luteolin, plant flavonoids found in many fruits and vegetables [95].
Table 2A.
Some phytochemicals with antimicrobial properties.
| Compound | Susceptible pathogen | Zone of inhibition | MIC (mg/ml) | MBC (mg/ml) | Reference |
|---|---|---|---|---|---|
| Coumarin Extraction in DMSO (100 mg/ml) 10 μl solution per disc | E. coli | 25.5 ± 2.12 | 1.25 | >5 | [100] |
| S. typhimurium | 14 ± 1.41 | 2.5 | >5 | ||
| S. infantis | 10.5 ± 2.12 | 5 | >5 | ||
| Enterobacter aerogenes | 19.5 ± 0.7 | 0.625 | 5 | ||
| Blueberry (Vaccinium corymbosum L.) extract 75% ethanol (20 g/10 ml final concentration) | L. monocytogenes | NA | 300–750 | 450–900 | [14] |
| S.Enteritidis | 450–1200 | 600–1800 | |||
| Flowers of Punica granatum L. var. Pleniflora (Ethanol extract) | Staphylococcus aureus | 32 -21 ± 0.6 (500–1.95 mg/ml) | 3.12–0.19 | 0.78–12.50 | [101] |
| Bacillus cereus | 28 ± 0.6–11 ± 0 | 3.12–0.19 | 1.56–12.50 | ||
| Listeria monocytogenes | 32 ± 1–11 ± 0.6 | 6.25–1.56 | 6.25–25.00 | ||
| Escherichia coli | 22 ± 1–10 ± 0 (500–62.5 mg/ml) | 12.5–3.12 | 12.50–50.00 | ||
| Shigella dysantriae | 30 ± 1–18 ± 0 | 6.25–0.39 | 1.56–25.00 | ||
| Salmonella typhi | 27 ± 0.6–11 ± 0.6 (500–7.8 mg/ml) | 1.56–6.25 | 6.25–25.00 | ||
| Clove (Syzygium aromaticum) (Aqueous extract, ethanol extract, Essential oil) | L. monocytogenes | 11.0–26.6 | 1.0–2.5 | 1.5–5.0 | [102] |
| S. aureus | 14.0–25.5 | 1.5–2.5 | 2.0–5.0 | ||
| V. parahaemolyticus | 11.3–21.5 | 1–5.5 | 1.5–10.0 | ||
| E.coli | 13.0–18.0 | 2.5 | 5.0 | ||
| S. Enteritidis | 14.2–17.0 | 2.5 | 5.0 | ||
| Bacillus cereus | 12.5–14.2 | 2.5 | 5.0 | ||
| P. aeruginosa | 11.5–15.4 | 5.0–5.5 | 6.0–10.0 | ||
| P. putida | 13.4–22.0 | 5.0–5.5 | 6.0–10.0 | ||
| A. faecalis | 19.8–22.5 | 0.5–5 | 0.6–10.0 | ||
| A. hydrophila | 9.7–32.0 | 0.8–5.0 | 0.6–5.5 |
Table 2B.
Some phytochemicals with antimicrobial properties.
| Compound | Susceptible pathogen | Zone of inhibition | MIC (mg/ml) | MBC (mg/ml) | Reference |
|---|---|---|---|---|---|
| Cinnamon (Cinnamomum cassia) (Aqueous extract, ethanol extract, Essential oil) | L. monocytogenes | 27.3–38.4 | 1.25–2.5 | 2.5 | [102] |
| S. aureus | 24.0–44.0 | 2.0–2.5 | 2.0–5.0 | ||
| V. parahaemolyticus | 11.4–20.5 | 1.0–5.0 | 1.5–5.0 | ||
| E.coli | 20.0–21.5 | 2.5 | 5.0 | ||
| S. Enteritidis | 19.8–23.1 | 2.5 | 5.0 | ||
| Bacillus cereus | 46.5 | 1.25 | 2.5 | ||
| P. aeruginosa | 12.0 | 5.0 | 10.0 | ||
| P. putida | 11.0 | 5.0 | 10.0 | ||
| A. faecalis | 22.2 | 1.25 | 2.5 | ||
| A. hydrophila | 29.5–31.9 | 1.25 | 2.5 | ||
| Epigallocatechin gallate Carnosic acid (Pure phenols/plant phenolic extracts) | C.coli, C | – | 78 μg/ml | – | [97] |
| jejuni | 19.5 μg/ml | ||||
| S. officinalis | [103] | ||||
| T. capitatus | E. coli, S. | 7.0–17.3 | 22.78 | 22.78–45.55 | |
| R. officinalis | enterica, P. | 15.0–30.0 | 0.73–2.94 | 0.73–2.94 | |
| O. majorana | aeruginosa, B. | 8.3–25.3 | 11.38–91.00 | 22.75–182.0 | |
| (Essential oil) | subtilis, S. aureus | 6.0–14.0 | 22.50–45.0 | 22.50–45.0 | |
| Cardamom (Amomum subulatum) (Essential oil) | S. typhi | – | 3.7–6.6 | – | [104] |
| S. paratyphi | 4.1 | ||||
| E. coli | 2.83 | ||||
| S. aureus | 9.4 | ||||
| B. licheniformis | 4.7 | ||||
| P. fluorescens | 7.5 | ||||
| Cumin (Cuminum cyminum) (Essential oil) | S. typhi | – | 3.4–6.1 | – | [104] |
| S. paratyphi | 14 | ||||
| E. coli | 3.4 | ||||
| S. aureus | 29.7 | ||||
| B. licheniformis | 4.3 | ||||
| P. fluorescens | 12.2 | ||||
| Salvia officinalis | L. monocytogenes | – | – | [105] | |
| Thymus vulgaris | 2–16 μl/ml | ||||
| Thymus vulgaris | 0.5–4 μl/ml | ||||
| Salvia officinalis (Essential oil) | 0.312–0.5 μl/ml |
Terpenes are a diverse group of compounds present in various parts of plants such as flowers, fruits, vegetables etc. Characterized by the presence of isoprene units (C5H8). C5HB is a major component of resins and essentials oils, responsible for the unique fragrance of the plant. They are also responsible for providing defense against biotic stress to the plant [95]. Essential oils contain significant quantity of terpenes but also contain other compounds that may have to do with their antimicrobial properties. Along with the isoprene units, terpenes have other chemical moieties attached such as alcohols, aldehydes, phenols, ketones, ether etc. Based on the chemical composition the properties of the molecule and essential oil will vary. In a study, antimicrobial activity of thirty-three free terpenes, commonly present in essential oils, was identified against common food pathogens E. coli, S. Typhimurium, S. aureus and B. cereus. Sixteen terpenes had possessed antimicrobial activity against the pathogens, six being bactericidal [98].
3.2.3. Some antimicrobial compounds
Chakraborty et al., reported the antimicrobial activity of Guava leaves against many diarrhea causing food pathogens including E. coli, Salmonella, Shigella and more. 7–9% concentration of the ethanol and aqueous extracts were found to be either significantly or moderately effective against majority of the pathogens [99]. Another study carried out by Simpore and Dianou utilized isolated phenolic compounds, Coumarin and quercetin against enteropathogens. Coumarins are composed of benzene rings fused with á-pyrone rings, and quercetin is made up of aromatic ring with multiple hydroxyl groups attached [100]. Quercetin did not possess any antimicrobial properties but coumarin was effective in controlling the bacterial growth. Klančnik et al., identified two phenolic compounds to be active against both antibiotic susceptible and resistant strains of Campylobacter indicating that the mechanism of action of the tested compounds is different, and it possess potential in treating drug resistant pathogens [97].
Effects of some of the phytochemicals on food-borne pathogens is summarized in Table 2A, Table 2Ba and 2b These phytochemicals can be used solely or in combination with the available antibiotics against the food pathogens.
It is important to know that the studies summarized above were conducted with different methods of extraction such as aqueous, ethanol, methanol, chloroform etc. With variable composition and concentration of the active compound, all of which may affect the end results. Along with that, the pH of the tested compound may also play a significant role in the antimicrobial activity as identified by Hoque et al. (2008), that the optimum activity of clove and cinnamon essential oil was noted at the pH of 7 [102].
3.2.4. Synergism between plant-based microbials and natural/synthetic antimicrobials
Along with the use of plant extracts and its isolated compounds, these antimicrobials can also be used as supportive therapy to provide a synergistic action when used in combination with other natural or synthetic antimicrobials such as bacteriocins or antibiotics. Iseppi et al., carried out a study to check the antimicrobial activity of sage (phenolic acids, flavonoids and terpenes), thyme (carvacrol and thymol as active components) essential oils and bacteriocin Lp17 extracted from E. mundtii against Listeria monocytogenes [105]. All three were effective in controlling L. monocytogenes and biofilm production. However, synergism was observed in both combination of the essential oils, and the combination of each essential oil with bacteriocin. Lowest MIC was for the combination of T. vulgaris EO/bacLP17 and antibiofilm activity of both T. vulgaris EO/bacLP17 and bacLP17/S. officinalis was significantly greater than individual activity of each [105]. Another study performed by Abdollahzadeh et al., also demonstrated the synergistic action of bacteriocin Nisin with essential oils of thyme against L. monocytogenes in minced fish meat [106]. A number of essential oils were tested along with nisin and pediocin against common food pathogens by Turgis et al., [107]. Nisin plus oregano EO showed synergism against L. monocytogenes, Thyme EO plus nisin against S. typhimurium, and pediocin plus Satureja montana against E. coli. [107]. Another study reports the activity of bacteriocin and plant extract as food additive [108]. Lemongrass and hot pepper extract along with bacteriocin isolated from Bacillus velezensis showed bactericidal activity similar to commercially available Nisin against spoilage organisms in dried squid. In a nut shell, nisin can be used in solo as well as in combination with other phytochemicals as antimicrobial agent against the food pathogens. Using in combination will have an increased antimicrobial efficacy of phytochemicals along with nisin.
Rakholiya & Chanda identified that methanolic extracts of Carica papaya leaves were inactive on its own against multiple Gram positive and negative bacteria but also showed that synergism with different type of antibiotics, enhanced their activity against these pathogens [109]. Garvey et al. identified the ability of Levisticum officinale and other plant extracts to inhibit the activity of efflux pumps in gram-negative bacteria such as Enterobacteriaceae and S. typhimurium, thus providing a promising solution towards the control of resistant pathogens [110]. Sanhueza et al. investigated the antimicrobial activity of grape pomace extract, which is rich in phenolic compounds against S. aureus, and E. coli, and the extract reduced the MIC levels of all different class of antibiotics [111]. The extract was also non-toxic to HeLa cell lines, thus, it can further be tested on animal models as it has the potential of becoming cheap and harmless candidate as supportive antimicrobial against pathogenic bacteria [[112], [113], [114]].
3.2.5. Phytochemicals as food preservatives
Along-with the potential of phytochemicals to be used as treatment against food borne pathogens, these can also be utilized to prevent the contamination of pathogens in food, in order to enhance food safety and reduce the prevalence of food borne diseases. Secondary metabolites are also potent antioxidants, preventing oxidative deterioration of food [115]. Hoque et al. identified clove essential oil (10%) to be effective in controlling Listeria monocytogenes population within a single day of inoculation in ground chicken meat and then throughout 15 days of incubation [102]. Cinnamon oil was also tested, which decreased the cell count but was not successful in completely eliminating the pathogen. Pomegranate peel extract has shown anti-bacterial properties, and active coatings of it have increased the shelf-life of pork by 3 days [116]. Kanatt et al. carried out another study on pomegranate peel extract; the extract showed antimicrobial and antioxidant properties, and increased the shelf life of chicken products by 2–3 weeks when stored under refrigeration [117]. Combination of carvacrol and 1, 8-cineole, which are the major constituents of Rosemary, reduced the total viable count of Listeria monocytogenes, in fresh cut vegetables and vegetable broth. However, it was ineffective against Aeromonas hydrophila and Pseudomonas fluorescens [118]. On the other hand, rosemary extract was useful in reducing C. jejuni contamination in chicken meat juice, but only if the sample was frozen before inoculation to reduce the viable count of bacteria [119].
By the above-mentioned data, we can conclude that the plant-based preservatives could possibly be used in food products, however there are certain drawbacks to it as well. Essential oils have strong aroma and may disrupt the flavor of the food [120]. For that problem, edible coatings could be used so that the compound is not incorporated into the food. These phytochemicals could also be used along with other commercially available preservatives or bacteriocins to provide a synergistic effect, thus will reduce its concentration. Other factors such as storage time and conditions of food such as pH, temperature, chemical nature, processing can also affect their mainstream use as a food preservative [120].
4. Antibiotic resistance
Antibiotic resistance is a term used when microorganisms such as bacteria grow resistant to antimicrobial substances that they were once susceptible to. It is one of the most pressing health problems due to rapid increase in antibiotic resistance in the recent years. Excessive use of antibiotics to treat human and animal infections is the major reason behind this emerging resistance [121]. There are numerous classes of antibiotics which act on different part of the microbial cells such as cell membrane, cell wall, protein or nucleic acid synthesis. Fluroquinolones and macrolides are used as first line treatment for Campylobacter species, gentamycin and erythromycin can be used to treat systemic infections [122]. For Salmonella species, enteric fever could be treated by fluroquinolones or azithromycin [13]. For Listeria infections, the drug of choice usually is β-lactam and aminoglycosides. Other antibiotics such as erythromycin, chloramphenicol or sulfonamides may also be used based on the severity of infection and other factors [15]. For diarrhea caused by Shiga toxin producing Escherichia. Coli (STEC), antibiotic therapy is usually not recommended, however for severe systemic infections, bactericidal antibiotics are preferred over bacteriostatic antibiotics [123].
4.1. Mechanism of resistance
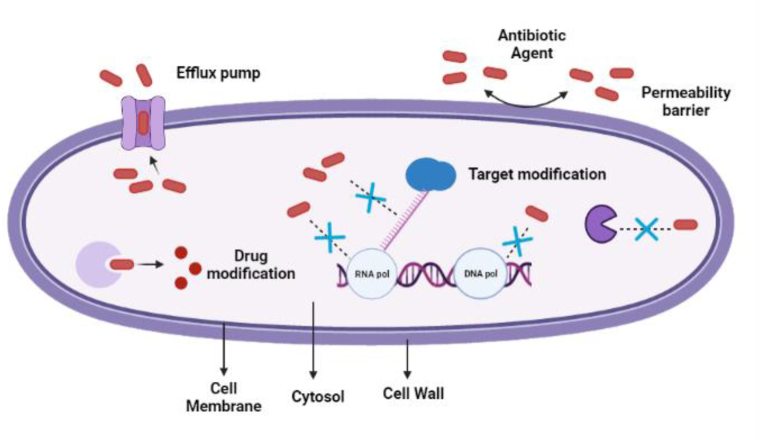
Microorganisms confers resistance to a single or multiple class of antibiotics usually by three basic ways: 1) Modification of the target site; 2) Modification of the antibiotic; 3) Reduced internalization of antibiotics either by decreased membrane permeability or expression of efflux pumps [17]. These mechanisms have been comprehensively demonstrated in Fig. 2.
Fig. 2.
Mechanism of antibiotic resistance in bacteria.
Throughout the years all the food pathogens have various mechanisms of antimicrobial resistance. For Campylobacter infections, as mentioned above, macrolide and fluroquinolones are used. Resistance against macrolides is due to rRNA methylase Erm (B) gene or point mutations in the 23sRNA, both of which modifies the ribosomal target site. Synergism in resistance is identified in efflux pump cmeABC which is responsible for multi drug resistancent. Mutations in the gene encoding the target site of fluroquinolones (gyrA) along with the expression of an efflux pump (cmeABC) confers resistance to quinolones. Other mechanisms of resistance include Tetracycline resistance by cmeABC and cmeG efflux pumps or tet(O) protein which provides ribosomal protection [122]. In Listeria monocytogenes, resistance to β-lactams can be achieved either by reduced membrane permeability or by PASTA domains (PBPs associated with serine threonine kinases) (PrkA) which binds to the beta lactam ring of the β-lactams inactivating the antibiotic [124]. Multi drug resistance is caused by the presence of efflux pump coded by MdrL and Lde genes. Fluroquinolone's resistance can be due to mutations in gyrA and parC genes coding for topoisomerase ii and iv subunits. Macrolide resistance such as clindamycin is due to the presence of Isa(A) and lnu(A) genes which inactivates the drug [125], and erythromycin resistance is by ermC genes. Whereas tetracycline resistance is exhibited by tet(A), tet(L) genes which code for efflux proteins, and tet(M) and tet(S) are responsible for providing ribosomal protection [126]. Escherichia. Coli strains can poses genes coding for β-lactamases such as blaTEM-1/ESBLs or cephalosporinases (ampCs) and carbapenemases. Other gene include 16 S rRNA methylases (against aminoglycosides), mcr genes (polymyxins) and PMQR genes (plasmid-mediated quinolone resistance) (fluoroquinolones) [127]. In general, resistance in Salmonella can be due to genes such as floR, cat 1 and cat 2 (chloramphenicol), tetC (tetracyclines), BlaTEM (ampicillin) strA, strB, aaddA1 and aaddA2 (streptomycin), and sul 1, sul 2 and sul 3 (sulphamethoxazole) [128]. Table 3 entails about the various sources of food borne pathogens which are antibiotic resistant and the frequency of their occurrence.
Table 3.
Frequency and sources of antibiotic resistant food-borne pathogens.
| Antibiotics | Organism | Percentage of resistant strains | Source of Sample | Reference |
|---|---|---|---|---|
| Ciprofloxacin | C. jejuni | 1111/1997 (55.63%) | Feces/blood (human) | [16] |
| 168/200 (84.0%) | Broiler meat | [129] | ||
| C. Coli | 270/419 (64.44%) | Feces/blood (human) | [16] | |
| 12/27 (44.4%) | Broiler meat | [129] | ||
| S. enteritidis/S. typhimurium | 3/11 (27.27%) | Raw chicken meat | [130] | |
| Erythromycin | C. jejuni | 9/1197 (0.45%) | Feces/blood (human) | [16] |
| 76/200 (38.0%) | Broiler meat | [129] | ||
| C. Coli | 49/419 (9.3%) | Feces/blood (human) | [16] | |
| 9/27 (33.0%) | Broiler meat | [129] | ||
| C. fetus | 1/100 (1%) | Feces/blood (human) | [16] | |
| Tetracycline | C. jejuni | 944/1197 (47.29%) | Feces/blood (human) | [16] |
| 181/200 (90.5%) | Broiler meat | [129] | ||
| C. Coli | 294/419 (70.17%) | Feces/blood (human) | [16] | |
| 17/27 (63.0%) | Broiler meat | [129] | ||
| C. fetus | 17/100 (17%) | Feces/blood (human) | [16] | |
| L. monocytogenes | 9/24 (34.7%) | RTE animal-based foods | [131] | |
| Salmonella spp. | 17/53 (32.1%) | Meat/butcher shops | [131] | |
| S. typhimurium | 9/11 (81.8%) | Meat products | [132] | |
| --- | 22/49 (44.9%) | Danish pigs | [133] | |
| E. coli | 15/48 (31.3%) | – | – | |
| Ampicillin | C. jejuni | 695/1197 (34.80%) | Feces/blood (human) | [16] |
| 13/200 (6.5%) | Broiler meat | [129] | ||
| C. Coli | 124/419 (9.59%) | Feces/blood (human) | [16] | |
| 3/27 (11.1%) | Broiler meat | [129] | ||
| Salmonella spp. | 47/53 (88.7) | Meat/butcher shops | [131] | |
| S.typhimurium | 58/58 (100%) | Chicken farms | [128] | |
| S. enteritidis/S. typhimurium | 8/11 (72.73%) | Raw chicken meat | [130] | |
| E. coli | 13/48 (27.1) | Danish pigs | [133] | |
| Amoxicillin | Salmonella spp. | 35/53 (62.3) | Meat/butcher shops | [131] |
| S. enteritidis/S. typhimurium | 3/8 (3 (27.27) | Raw chicken meat | [130] | |
| Nalidixic acid | L. monocytogenes | 12/24 (50%) | RTE animal-based foods | [131] |
| S. enteritidis | 20/37 (54%) | Meat products | [132] | |
| S. infantis | 11/15 (73.3%) | – | – | |
| S. hadar | 5/7 (71.4%) | – | – | |
| Gentamycin | S. typhimurium | 2/11 (18.2%) | Meat products | [132] |
| Clindamycin | L. monocytogenes | 72/206 (35%) | RTE meat products/surfaces with food contact | [134] |
| Penicillin | L. monocytogenes | 16/24 (66.7%) | RTE animal based foods | [131] |
| S. enteritidis/S. typhimurium | 11/11 (100%) | Raw chicken meat | [130] | |
| Sulfamethoxazole-Trimethoprim | Salmonella spp. | 16/53 (30.2) | Meat/butcher shops | [131] |
| S.typhimurium | 3/58 (5.2%) | Chicken farms | [128] | |
| 26/49 (53.1%) | Danish pigs | [133] | ||
| E. coli | 13/48 (27.1%) | – | ||
| Chloramphenicol | S.typhimurium | 58/58 (100%) | Chicken farms | [128] |
| 2/49 (4.3) | Danish pigs | [133] | ||
| E. coli | 5/48 (10.4) | – | – | |
| Streptomycin | E. coli | 25/48 (52.1%) | Danish pigs | [133] |
| S.typhimurium | 25/49 (51%) | – | – |
5. Membrane proteins in food pathogens
Membranous proteins of pathogens play essential roles such as adhesion, invasion, toxin release and other interactions of pathogen to the host in order to cause infections [[135], [136], [137]]. A pathogen such as Cronobacter sakazakii utilizes transmembrane regulatory proteins present in gram negative bacteria for its virulence which helps in production of biofilm and also evading the immune system [138]. Along with transmembrane proteins, outer membrane proteins including OmpA and OmpX are involved in the process of adhesion and invasion of the human cells [[139], [140], [141]]. It was reported by Kothary et al. that the protein responsible for the spread of C. sakazakii in the bloodstream might be due to the cell bound zinc containing protein metalloprotease, encoded by zpx gene [142]. However concrete evidence is yet to be established regarding the membrane proteins responsible for virulence in various C. sakazakii strains. Proteomics has emerged as one of the vital fields of science, can be used to identify proteomic virulence factors, by comparing pathogenic and non-pathogenic forms of these bacteria [[143], [144], [145]].
5.1. Outer membrane
All Gram-negative bacteria have an additional outer membrane surrounding the peptidoglycan cell wall, and is absent in Gram positive bacteria. The outer membrane is asymmetric and is composed of phospholipids on the inside, and the outer portion mainly constitutes of lipopolysaccharides. The lipopolysaccharide layer of Gram-negative bacteria consists of 3 parts: I) A conserved hydrophobic membrane made up of Lipid A, ii) A phosphorylated, anionic, core oligosaccharide, and iii) O-antigen, a polysaccharide made up of different type of sugar monomers providing a hydrophilic surface and is highly variable among different bacteria [[146], [147]].
In E. coli, the core-OS is divided into two regions. Outer region being made of various sugar residues and the inner region is composed of 2-keto-3- deoxy-d-manno-octulosonic acid (Kdo) and l-glycero-d-manno-heptose. The inner region is highly conserved among the family of Enterobacteriaceae providing stability to its outer membrane [148]. In E. coli, further modifications within the inner region of core-OS, depending upon its types can be achieved by variable non-stoichiometric substitutions [149]. Unlike the conserved inner region, the outer region of the core-OS shows variability. There are five types of cores in E. coli based on this variability including R1, R2, R3, R4 and K12. All these are composed of a (hexose)3 Carbohydrate backbone with two side chains. These are categorized based upon their difference in the order and position of the carbohydrate backbone along with the chemical nature and type of linkage of the side chain with the backbone. Smooth lipopolysaccharide (LPS) which contains three components is more prevalent in as clinical isolates, than rough LPS. E. Coli lacks the O-antigen and may have a shortened core-OS.
Outer membrane is crucial for pathogenic gram-negative bacteria to maintain cell viability, and protects it by acting as a permeability barrier [147,150,151]. The outer membrane being crucial, is highly conserved among various species of gram-negative bacteria with very little variation (Vaara, 1992). LPS plays a significant role in maintaining its integrity and selective permeability. This selective diffusion allows limited entry of hydrophobic molecules and inability of charged molecules to enter the prokaryotic cells [151]. The core-OS of LPS is mainly responsible for the semi permeability against non-polar molecules due to the presence of anionic phosphate groups. These phosphate residues bind to other core-OS molecules via divalent cations usually Magnesium or Calcium and form intermolecular electrostatic bonds between the neighboring LPS [152]. This interlinking provides a barrier against harmful hydrophobic molecules. Negatively charged lipid A and the inner core-OS play vital roles to maintain the integrity of the outer membrane [153].
5.2. LPS and other PAMPs
Based on the experimental results, it is indicated that the innate immune response against the outer membrane vesicles is due to the recognition of the combination of LPS and vesicle pathogen-associated molecular patterns (PAMPs). Proteins and lipoproteins of the outer membrane are active biological molecules having the tendency to activate immune system and initiate leukocyte migration [154]. Investigation of mutants lacking particular proteins or other components in the outer membrane can be beneficial for the identification immune stimulators. LPS is the most potent and abundant immune cell activator of the outer membrane. LPS is found in high amount in the membrane vesicles in comparison to other proteins even when combined. Toll-like receptor 4 (TLR4) complex senses LPS and induces a pro-inflammatory response in the body. High levels of LPS in the form of endotoxins and excess inflammation due to the TLR4 can lead to LPS toxicity and in severe cases, septic shock [155]. Due to abundancy of LPS in the outer membrane vesicles, all studies focused on host immune system activation must focus on its contribution. These vesicles deliver LPS and can increase the bacterial clearance but may also cause tissue damage in the host due to excessive inflammation.
For studying these immune regulations in-vivo, it is important to know that LPS that is purified and LPS that is bound to outer membrane vesicles are not the same in terms of chemical nature and immune system activation. These differ in terms of distribution and clearance in the host tissues. Vesicles provides amphipathic and heterogenous environment rich in proteins which enhances LPS diffusion across tissues. Pure LPS is much more hydrophobic and easily enters lipid bilayers of host cells. The vesicle bound LPS can penetrate deeper into the tissues where phagocytes are present. Therefore, the vesicles may also cause increased recognition and clearance by these phagocytes. Size of the LPS complexes is directly proportional to the immune response in host cells. A study found direct correlation between the size of LPS aggregated and LPS internalization by CD14 system [156].
5.3. Effector proteins in enterohemorrhagic Escherichia coli (EHEC)
There are a number of diseases which are caused by a commonly known pathogen called Enterohemorrhagic Escherichia coli (EHEC), diseases include colitis, diarrhea along with some severe infections which leads to acute kidney failure, hemolytic uremic syndrome and acute encephalopathy [157]. Shiga toxins produced by E. coli and effector proteins are major proteins which are held accountable for the pathogenicity of EHEC. Cell death of eukaryotic host cells occurs due to the impairment of ribosomal activity which leads to inhibition of protein synthesis caused by shiga toxin [158]. In epithelial cells A/E lesions are induced to attach due to the binding of some host proteins to effector proteins [159,160]. The characterization of A/E lesions are done by certain factors and under certain conditions: i) when gut epithelial microvilli gets disrupted, ii) when bacterial attaches to the plasma membrane of host cell, and iii) when actin gets accumulated in the host cells [161]. A structure which is needle like and is termed as type III secretion system (T3SS) secretes effector proteins through a transport complex. Host cells are targeted via EspA, EspB and EspD which are the translocator proteins [160]. A polymeric filamentous structure is formed by EspA proteins which facilitate effector proteins to translocate into the host cells [162,163]. EspA also delivers EspB and EspD and a pore structure complex is formed by them on the cell membrane of host [164]. First effector protein known as Intimin receptor (Tir) is transported into the host cell and once it reaches to the host plasma membrane, it gets re-translocated [161]. The family of outer membrane proteins is comprised of β-barrel which is eight stranded along with membrane spanning regions, main members of the family are OmpA, OmpW and OmpX [165]. High conservation of these proteins have been found in several Gram-negative bacteria, and they play a major role in bacterial pathogenesis. Their characterization have been observed in various pathogens such as Vibrio cholera, Yersinia pestis, E. coli, Salmonella enteria, Klebsiella pnueumoniae and Pseudomonas aeruginosa [166,167]. Pathogenesis of various strains of E. coli is associated with the interference of OmpA and OmpX. By deleting the OmpA gene of meningitic strains, bacterial invasion is decreased in microvascular cells of brain while surviving within the macrophages [166,168]. Defect in adhesion to the epithelial cells of colon and mouse bladder colonization is exhibited by certain mutants such as ompA in uropathogenic E. coli (UPEC) and EHEC [169,170]. Meanwhile, in comparison with the ompX mutants in UPEC and pig lung disease related strain, it has been observed that there is lower mortality rate of mouse and reduced flagellar production and colonization in the mouse kidney is exhibited by these wild type mutants [171,172].
5.4. Spore forming food pathogens
There are many causative agents which causes food borne illness, Bacillus cereus is one of them. It is a spore forming organism and a human pathogen. Once it is consumed, emetic and diarrheal syndromes occur due to certain toxins which are synthesized by this organism [173]. In the manufacture of safe and stable food, B. cereus, which are spore forming bacteria are a major challenge to be controlled. During the production of food, inactivation of these spore forming bacteria is hard as the spores which are formed by B. cereus are highly resistant to radiation, heat, UV, desiccation and chemicals. Dipicolinic acid is present in the core therefore there is thermal resistance for a part and the dipocolinic acid is replaced into water to a major extent [174]. A structure in which a spore forms multi layers also results due to the high resistance, each layer helps to protect the core and localizes the macromolecules which are essential to live, inner membrane surrounds the macromolecules. Layers starts from the spore core, and then comes towards the germ cell wall, after that the outer membrane and the coat is assembled. The outermost layer is the exosporium.
Spores return to the vegetative stage once they sense favorable environmental condition though germination and their high stress resisting properties gets released. Germinant receptors which are present on the inner membrane of spore give out the signals of germination for transduction. The inner membrane of the spore acts as a strong permeability barrier to small molecules which are hydrophobic and hydrophilic in nature [175]. In order to deal with the spore stress resistance, it is required that the inner membrane should have strong integrity. There are chances of inner membrane to expand approximately up to two folds in the initial stage of spore germination process. This happens without the production of ATP and without the formation of new membrane [176,177]. Several types of membrane proteins are assembled and supported by the inner membrane of spore. Not only germinant receptor but there are other localized proteins as well which are involved in the process of germination. These localized proteins play a crucial role in certain processes such as environmental cues transduction, information flow is an example. They are also crucial for metabolite transportation and for the catalysis of reactions [178]. There are some inner membrane proteins which are critical to the spore's life cycle, specifically for germination. Furthermore, up till now 60% of the drug targets are made up by membrane proteins [178].
Development of inner membrane from the spore to vegetative cell membrane takes places after the process of germination. By attaining a deeper understanding about formation of spore inner membrane proteins and how the comparison can be done with protein composition of the vegetative cell membrane, new insights can be gained into how survival of B. cereus spore in adverse conditions is done and how the germination is regulated.
5.5. Bacterial small proteins
Small open reading frames (sORFs) encodes bacterial small proteins, and it is a class of functional molecules which is recognized as an emerging class. These molecules were largely unnoticed in the past. Some of them were uncovered accidently. Recently, in order to detect these small opening reading frames approaches have been made across the globe. Most part of the small proteins seems to be hydrophobic in nature, and are placed in the bacterial membrane [179]. In this review, it is described that in pathogenic bacteria, some functional small hydrophobic proteins have been discovered, and there are also some new advances which tends to discover the additional ones. The ability of bacteria to adapt to changing environmental conditions is contributed by small membrane proteins. Either the stability or the function of larger membrane proteins gets modulated, and these small membrane proteins tend to implicate in negative feedback regulation loops. Toxin-antitoxin modules comprise a subset of these proteins. Small proteins that are characterized as hydrophobic and functional small proteins between novel sORFs are discovered. New and unique therapeutic interventions can be obtained through the identification of naturally occurring small hydrophobic proteins of pathogenic bacteria as recently displayed with synthetic peptides and their development which exhibit antibacterial properties [180].
6. Conclusion
In conclusion, the use of synthetic and natural antimicrobials as a control against foodborne pathogens is a promising approach in the food industry. This review has shown that both synthetic and natural antimicrobials have their own advantages and limitations. Synthetic antimicrobials i.e. antibiotics are effective, and have a longer shelf life but are becoming narrow spectrum as well due to the problem of multi drug resistance. It has been found that Campylobacter sp. Have become resistant to several antibiotics such as ciprofloxacin, erythromycin, tetracycline and ampicillin. Other antibiotics such as gentamicin, clindamycin and penicillin are effective against certain microbes such as L. monocytogenes and S. typhimurium. On the other hand, natural antimicrobials which include plant extracts and bacteriocins are generally safer for consumption, and have a better potential as a food preservative or as a substitute for antibiotics. As per the findings, cardamom, cumin and cinnamon have been found to have broad spectrum antimicrobial activity as compared to that of other phytochemicals. Bacteriocins extracted from probiotics are gaining importance as they have been found to be effective against many Gram positive and Gram negative organisms. Some of them are Enterocin KAE01, Pediocin PA, Lp17, Thermophylin and Mutacin B-Ny266. Natural antimicrobials despite having substantial research, rarely get attention for commercial application as drugs or food preservatives. This is due to several facts such as: limited antimicrobial activity against specific microbes only, limited data on the in-vivo effects of natural compounds against food borne pathogens, and impact of variable environmental and experimental factors affecting the activity and stability of the compound. Along with that, further research is needed to determine the optimal conditions and concentrations for their use, as well as to evaluate their safety and potential side effects. Additionally, the combination of different types of antimicrobials may be a promising approach to enhance the efficacy of both the synthetic and natural antimicrobials. This is evident from a study which reports the increased antimicrobial activity of sage, thyme essential oils and bacteriocin Lp17 extracted from E. mundtii against Listeria monocytogenes. Nisin has also been reported to show increased efficacy when used with different antimicrobials. Along with this, methods to ensure safe delivery of these compounds could be explored, such as edible coatings and nanoparticles/nanovesicles which are made safe to consume. Thus, the use of synthetic and natural antimicrobials as a control against foodborne pathogens has the potential to improve food safety and reduce the risk of foodborne illness. Careful consideration should be given to the selection and application of antimicrobials, based on their effectiveness, safety, and potential impact on product quality and consumer acceptance. Overall, this review highlights the importance of ongoing research into the development and application of safe and effective antimicrobial strategies to ensure the safety and quality of our food supply.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Data availability statement
Data will be made available on request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper
References
- 1.Bhunia A.K. 2018. Introduction to Foodborne Pathogens; pp. 1–23. [DOI] [Google Scholar]
- 2.Bintsis T. Foodborne pathogens. AIMS Microbiol. 2017;3:529–563. doi: 10.3934/microbiol.2017.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gursoy D. Foodborne illnesses: an overview of hospitality operations liability. J. Hospit. 2019;1(1):41–49. [Google Scholar]
- 4.Sharif M.K., Javed K., Nasir A. Foodborne Diseases. Elsevier; 2018. Foodborne illness: threats and control; pp. 501–523. [DOI] [Google Scholar]
- 5.Akhtar S., Sarker M.R., Hossain A. Microbiological food safety: a dilemma of developing societies. Crit. Rev. Microbiol. 2014;40:348–359. doi: 10.3109/1040841X.2012.742036. [DOI] [PubMed] [Google Scholar]
- 6.Pisoschi A.M., Pop A., Georgescu C., Turcuş V., Olah N.K., Mathe E. An overview of natural antimicrobials role in food. Eur. J. Med. Chem. 2018;143:922–935. doi: 10.1016/j.ejmech.2017.11.095. [DOI] [PubMed] [Google Scholar]
- 7.Gyawali R., Ibrahim S.A. Natural products as antimicrobial agents. Food Control. 2014;46:412–429. doi: 10.1016/j.foodcont.2014.05.047. [DOI] [Google Scholar]
- 8.Alvarez-Sieiro P., Montalbán-López M., Mu D., Kuipers O.P. Bacteriocins of lactic acid bacteria: extending the family. Appl. Microbiol. Biotechnol. 2016;100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu M., Ma Y., Dou X., Aslam M.Z., Liu Y., Xia X., Yang S., Wang X., Qin X., Hirata T., Dong eQ. A review of potential antibacterial activities of nisin against Listeria monocytogenes: the combined use of nisin shows more advantages than single use. Food Res. Int. 2022 doi: 10.1016/j.foodres.2022.112363. [DOI] [PubMed] [Google Scholar]
- 10.Kuniyoshi T.M., O'Connor P.M., Lawton E., Thapa D., Mesa-Pereira B., Abulu S., Hill C., Ross R.P., Oliveira R.P., Cotter P.D. An oxidation resistant pediocin PA-1 derivative and penocin A display effective anti-Listeria activity in a model human gut environment. Gut Microb. 2022;14(1) doi: 10.1080/19490976.2021.2004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazir F., Rehana Salim I., Nargis Yousf I., Bashir M., Naik I.H., Syed Zameer Hussain I., Correspondence Fiza Nazir I., Salim R., Yousf N., Naik H., Zameer Hussain S. Natural antimicrobials for food preservation. J. Pharmacogn. Phytochem. 2017;6:2078–2082. [Google Scholar]
- 12.McClements D.J., Das A.K., Dhar P., Nanda P.K., Chatterjee N. Nanoemulsion-based technologies for delivering natural plant-based antimicrobials in foods. Front. Sustain. Food Syst. 2021;5 doi: 10.3389/fsufs.2021.643208. [DOI] [Google Scholar]
- 13.Jin C., Gibani M.M., Pennington S.H., Liu X., Ardrey A., Aljayyoussi G., Moore M., Angus B., Parry C.M., Biagini G.A., Feasey N.A., Pollard A.J. Treatment responses to azithromycin and ciprofloxacin in uncomplicated Salmonella typhi infection: a comparison of clinical and microbiological data from a controlled human infection model. PLoS Neglected Trop. Dis. 2019;13 doi: 10.1371/journal.pntd.0007955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen X., Sun X., Xie Q., Liu H., Zhao Y., Pan Y., Hwang C.-A., Wu V.C.H. Antimicrobial effect of blueberry (Vaccinium corymbosum L.) extracts against the growth of Listeria monocytogenes and Salmonella enteritidis. Food Control. 2014;35:159–165. doi: 10.1016/j.foodcont.2013.06.040. [DOI] [Google Scholar]
- 15.Olaimat A.N., Al-Holy M.A., Shahbaz H.M., Al-Nabulsi A.A., Abu Ghoush M.H., Osaili T.M., Ayyash M.M., Holley R.A. Emergence of antibiotic resistance in Listeria monocytogenes isolated from food products: a comprehensive review. Compr. Rev. Food Sci. Food Saf. 2018;17:1277–1292. doi: 10.1111/1541-4337.12387. [DOI] [PubMed] [Google Scholar]
- 16.Sifré E., Salha B.A., Ducournau A., Floch P., Chardon H., Mégraud F., Lehours P. EUCAST recommendations for antimicrobial susceptibility testing applied to the three main Campylobacter species isolated in humans. J. Microbiol. Methods. 2015;119:206–213. doi: 10.1016/j.mimet.2015.10.018. [DOI] [PubMed] [Google Scholar]
- 17.Tang Y., Fang L., Xu C., Zhang Q. Antibiotic resistance trends and mechanisms in the foodborne pathogen, Campylobacter. Anim. Health Res. Rev. 2017;18:87–98. doi: 10.1017/S1466252317000135. [DOI] [PubMed] [Google Scholar]
- 18.Cocolin L., Stella S., Nappi R., Bozzetta E., Cantoni C., Comi G. Analysis of PCR-based methods for characterization of Listeria monocytogenes strains isolated from different sources. Int. J. Food Microbiol. 2005;103:167–178. doi: 10.1016/j.ijfoodmicro.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 19.Halbedel S., Prager R., Banerji S., Kleta S., Trost E., Nishanth G., Alles G., Hölzel C., Schlesiger F., Pietzka A., Schlüter D., Flieger A. A Listeria monocytogenes ST2 clone lacking chitinase ChiB from an outbreak of non-invasive gastroenteritis. Emerg. Microb. Infect. 2019;8:17–28. doi: 10.1080/22221751.2018.1558960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allerberger F., Wagner M. Listeriosis: a resurgent foodborne infection. Clin. Microbiol. Infection. 2010;16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 21.Charlier C., Perrodeau É., Leclercq A., Cazenave B., Pilmis B., Henry B., Lopes A., Maury M.M., Moura A., Goffinet F., Dieye H.B., Thouvenot P., Ungeheuer M.-N., Tourdjman M., Goulet V., de Valk H., Lortholary O., Ravaud P., Lecuit M. MONALISA study group, Clinical features and prognostic factors of listeriosis: the MONALISA national prospective cohort study. Lancet Infect. Dis. 2017;17:510–519. doi: 10.1016/S1473-3099(16)30521-7. [DOI] [PubMed] [Google Scholar]
- 22.Hamidiyan N., Salehi-Abargouei A., Rezaei Z., Dehghani-Tafti R., Akrami-Mohajeri F. The prevalence of Listeria spp. food contamination in Iran: a systematic review and meta-analysis. Food Res. Int. 2018;107:437–450. doi: 10.1016/j.foodres.2018.02.038. [DOI] [PubMed] [Google Scholar]
- 23.Stephan R., Althaus D., Kiefer S., Lehner A., Hatz C., Schmutz C., Jost M., Gerber N., Baumgartner A., Hächler H., Mäusezahl-Feuz M. Foodborne transmission of Listeria monocytogenes via ready-to-eat salad: a nationwide outbreak in Switzerland. Food Control. 2015;57:14–17. doi: 10.1016/j.foodcont.2015.03.034. 2013–2014. [DOI] [Google Scholar]
- 24.Smith A.M., Tau N.P., Smouse S.L., Allam M., Ismail A., Ramalwa N.R., Disenyeng B., Ngomane M., Thomas J. Outbreak of Listeria monocytogenes in South Africa, 2017–2018: laboratory activities and experiences associated with whole-genome sequencing analysis of isolates. Foodb. Pathog. Dis. 2019;16:524–530. doi: 10.1089/fpd.2018.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heiman K.E., Garalde V.B., Gronostaj M., Jackson K.A., Beam S., Joseph L., Saupe A., Ricotta E., Waechter H., Wellman A., Adams-Cameron M., Ray G., Fields A., Chen Y., Datta A., Burall L., Sabol A., Kucerova Z., Trees E.…Silk B.J. Multistate outbreak of listeriosis caused by imported cheese and evidence of cross-contamination of other cheeses, USA, 2012. Epidemiol. Infect. 2016;144(13):2698–2708. doi: 10.1017/S095026881500117X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gebretsadik S., Kassa T., Alemayehu H., Huruy K., Kebede N. Isolation and characterization of Listeria monocytogenes and other Listeria species in foods of animal origin in Addis Ababa, Ethiopia. J. Infect. Public Health. 2011;4:22–29. doi: 10.1016/j.jiph.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Faeji C.O., Fasoro A.A., O Oni I., Akingbade A.M. Assessment of Listeria monocytogenes in unpasteurized milk obtained from cattle in northern Nigeria. J. Microbiol. Res. 2016;6:23–27. [Google Scholar]
- 28.Tenaillon O., Skurnik D., Picard B., Denamur E. The population genetics of commensal Escherichia coli. Nat. Rev. Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 29.Escobar-Páramo P., Grenet K., Le Menac’h A., Rode L., Salgado E., Amorin C., Gouriou S., Picard B., Rahimy M.C., Andremont A., Denamur E., Ruimy R. Large-scale population structure of human commensal Escherichia coli isolates. Appl. Environ. Microbiol. 2004;70:5698–5700. doi: 10.1128/AEM.70.9.5698-5700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaper J.B., Nataro J.P., Mobley H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 31.Smith J.L., Fratamico P.M., Gunther N.W. 2014. Shiga Toxin-Producing Escherichia coli; pp. 145–197. [DOI] [PubMed] [Google Scholar]
- 32.Russo T.A., Johnson J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 33.Nielubowicz G.R., Mobley H.L.T. Host–pathogen interactions in urinary tract infection. Nat. Rev. Urol. 2010;7:430–441. doi: 10.1038/nrurol.2010.101. [DOI] [PubMed] [Google Scholar]
- 34.Havelaar A.H., Kirk M.D., Torgerson P.R., Gibb H.J., Hald T., Lake R.J., Praet N., Bellinger D.C., de Silva N.R., Gargouri N., Speybroeck N., Cawthorne A., Mathers C., Stein C., Angulo F.J., Devleesschauwer B. World health organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 2015;12 doi: 10.1371/journal.pmed.1001923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAuley C.M., McMillan K., Moore S.C., Fegan N., Fox E.M. Prevalence and characterization of foodborne pathogens from Australian dairy farm environments. J. Dairy Sci. 2014;97:7402–7412. doi: 10.3168/jds.2014-8735. [DOI] [PubMed] [Google Scholar]
- 36.Julian T.R. Environmental transmission of diarrheal pathogens in low and middle income countries. Environ. Sci. Process Impacts. 2016;18:944–955. doi: 10.1039/C6EM00222F. [DOI] [PubMed] [Google Scholar]
- 37.Chlebicz A., Śliżewska K. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: a review. Int. J. Environ. Res. Publ. Health. 2018;15:863. doi: 10.3390/ijerph15050863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Modi S., Brahmbhatt M.N., Chatur Y.A., Nayak J.B. Prevalence of Campylobacter species in milk and milk products, their virulence gene profile and antibiogram. Vet. World. 2015;8:1–8. doi: 10.14202/vetworld.2015.1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorthi S.P., Behari M., Perez-Perez G.I. 1997. Guillain-Barré Syndrome: Association with Campylobacter Jejuni and Mycoplasma Pneumoniae Infections in India. [PubMed] [Google Scholar]
- 40.de Vries S.P.W., Vurayai M., Holmes M., Gupta S., Bateman M., Goldfarb D., Maskell D.J., Matsheka M.I., Grant A.J. Phylogenetic analyses and antimicrobial resistance profiles of Campylobacter spp. from diarrhoeal patients and chickens in Botswana. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alaboudi A.R., Malkawi I.M., Osaili T.M., Abu-Basha E.A., Guitian J. Prevalence, antibiotic resistance and genotypes of Campylobacter jejuni and Campylobacter coli isolated from chickens in Irbid governorate, Jordan. Int. J. Food Microbiol. 2020;327 doi: 10.1016/j.ijfoodmicro.2020.108656. [DOI] [PubMed] [Google Scholar]
- 42.Lone J.A., Kotwal S.K., Rehman M.U., Wani N., Malik M.A., Singh M. Prevalence and antimicrobial resistance pattern of Campylobacter species among poultry and poultry handlers of Jammu. J. Anim. Res. 2016;6:351. doi: 10.5958/2277-940X.2015.00189.8. [DOI] [Google Scholar]
- 43.Hussain I., Shahid Mahmood M., Akhtar M., Khan A. Prevalence of Campylobacter species in meat, milk and other food commodities in Pakistan. Food Microbiol. 2007;24:219–222. doi: 10.1016/j.fm.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 44.WHO Campylobacter. 2020. https://www.who.int/news-room/fact-sheets/detail/campylobacter Retrieved from World Health Organisation.
- 45.Ryan M.P., O'Dwyer J., Adley C.C. Evaluation of the complex nomenclature of the clinically and veterinary significant pathogen Salmonella. BioMed Res. Int. 2017;2017:1–6. doi: 10.1155/2017/3782182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balasubramanian R., Im J., Lee J.-S., Jeon H.J., Mogeni O.D., Kim J.H., Rakotozandrindrainy R., Baker S., Marks F. The global burden and epidemiology of invasive non-typhoidal Salmonella infections. Hum. Vaccines Immunother. 2019;15:1421–1426. doi: 10.1080/21645515.2018.1504717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stanaway J.D., Parisi A., Sarkar K., Blacker B.F., Reiner R.C., Hay S.I., Crump J.A. The global burden of non-typhoidal Salmonella invasive disease: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019;19:1312–1324. doi: 10.1016/S1473-3099(19)30418-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feasey N.A., Dougan G., Kingsley R.A., Heyderman R.S., Gordon M.A. Invasive non-typhoidal Salmonella disease: an emerging and neglected tropical disease in Africa. Lancet. 2012;379:2489–2499. doi: 10.1016/S0140-6736(11)61752-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Friesema I.H.M., Schimmer B., Ros J.A., Ober H.J., Heck M.E.O.C., Swaan C.M., de Jager C.M., Peran i Sala R.M., van Pelt W. A regional Salmonella enterica Serovar typhimurium outbreak associated with raw beef products. Foodb. Pathog. Dis. 2012;9:102–107. doi: 10.1089/fpd.2011.0978. The Netherlands, 2010. [DOI] [PubMed] [Google Scholar]
- 50.Crump J.A., Sjölund-Karlsson M., Gordon M.A., Parry C.M. Epidemiology, clinical presentation, laboratory diagnosis, antimicrobial resistance, and antimicrobial management of invasive Salmonella infections. Clin. Microbiol. Rev. 2015;28:901–937. doi: 10.1128/CMR.00002-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mughini-Gras L., Enserink R., Friesema I., Heck M., van Duynhoven Y., van Pelt W. Risk factors for human salmonellosis originating from pigs, cattle, broiler chickens and egg laying hens: a combined case-control and source attribution analysis. PLoS One. 2014;9 doi: 10.1371/journal.pone.0087933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou M., Li X., Hou W., Wang H., Paoli G.C., Shi X. Incidence and characterization of Salmonella isolates from raw meat products sold at small markets in hubei province, China. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.02265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanaway J.D., Reiner R.C., Blacker B.F., Goldberg E.M., Khalil I.A., Troeger C.E., Hay S.I. The global burden of typhoid and paratyphoid fevers: a systematic analysis for the Global Burden of Disease Study 2017. Lancet Infect. Dis. 2019;19:369–381. doi: 10.1016/S1473-3099(18)30685-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akhtar S., Sarker M.R., Hossain A. Microbiological food safety: a dilemma of developing societies. Crit. Rev. Microbiol. 2014;40:348–359. doi: 10.3109/1040841X.2012.742036. [DOI] [PubMed] [Google Scholar]
- 55.Bashir A., Ali K., Bux K., Farid N., Khaireabadi M., Hassan K.A., Hussain A., Fatima K., Mehmood S., Haider S.A., Herwig R. Molecular characterization, purification, and mode of action of enterocin KAE01 from lactic acid bacteria and its in silico analysis against MDR/ESBL Pseudomonas aeruginosa. Genes. 2022;13(12):2333. doi: 10.3390/genes13122333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gholami-Shabani M., Shams-Ghahfarokhi M., Razzaghi-Abyaneh M. 2023. Food Microbiology: Application of Microorganisms in Food Industry, Intechopen.https://www.intechopen.com/online-first/86078 [Google Scholar]
- 57.Moslem M.N., Ghasemi M.F., Amirmozafari N., Ranji N. Antimicrobial activity of Pediococcus acidilactici PTCC 1954 and Leuconostoc mesenteroides PTCC 1953 strains isolated from organic meat sausages. Biol. J. Microorg. 2023;11(44) [Google Scholar]
- 58.Majeed H., Lampert A., Ghazaryan L., Gillor O. The weak shall inherit: bacteriocin-mediated interactions in bacterial populations. PLoS One. 2013;8 doi: 10.1371/journal.pone.0063837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O'Connor P.M., O'Shea E.F., Guinane C.M., O'Sullivan O., Cotter P.D., Ross R.P., Hill C. Nisin H is a new nisin variant produced by the gut-derived strain Streptococcus hyointestinalis DPC6484. Appl. Environ. Microbiol. 2015;81:3953–3960. doi: 10.1128/AEM.00212-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chikindas M.L., Weeks R., Drider D., Chistyakov V.A., Dicks L.M. Functions and emerging applications of bacteriocins. Curr. Opin. Biotechnol. 2018;49:23–28. doi: 10.1016/j.copbio.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang S.-C., Lin C.-H., Sung C.T., Fang J.-Y. Antibacterial activities of bacteriocins: application in foods and pharmaceuticals. Front. Microbiol. 2014;5 doi: 10.3389/fmicb.2014.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.A I S., A A.O. Characterization of bacteriocin produced by Lactobacillus plantarum F1 and Lactobacillus brevis OG1. Afr. J. Biotechnol. 2003;2:219–227. doi: 10.5897/AJB2003.000-1045. [DOI] [Google Scholar]
- 63.Gálvez A., López R.L., Pulido R.P., Burgos M.J.G. 2014. Natural Antimicrobials for Food Biopreservation; pp. 3–14. [DOI] [Google Scholar]
- 64.Inglis R.F., Bayramoglu B., Gillor O., Ackermann M. The role of bacteriocins as selfish genetic elements. Biol. Lett. 2013;9 doi: 10.1098/rsbl.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cotter P.D., Ross R.P., Hill C. Bacteriocins — a viable alternative to antibiotics? Nat. Rev. Microbiol. 2013;11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 66.Oppegård C., Rogne P., Emanuelsen L., Kristiansen P.E., Fimland G., Nissen-Meyer J. The two-peptide class II bacteriocins: structure, production, and mode of action. Microb. Physiol. 2007;13:210–219. doi: 10.1159/000104750. [DOI] [PubMed] [Google Scholar]
- 67.Noda M., Miyauchi R., Danshiitsoodol N., Matoba Y., Kumagai T., Sugiyama M. Expression of genes involved in bacteriocin production and self-resistance in Lactobacillus brevis 174A is mediated by two regulatory proteins. Appl. Environ. Microbiol. 2018;84 doi: 10.1128/AEM.02707-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nigam A., Gupta D., Sharma A. Treatment of infectious disease: beyond antibiotics. Microbiol. Res. 2014;169:643–651. doi: 10.1016/j.micres.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 69.Daba G.M., Ishibashi N., Zendo T., Sonomoto K. Functional analysis of the biosynthetic gene cluster required for immunity and secretion of a novel Lactococcus -specific bacteriocin, lactococcin Z. J. Appl. Microbiol. 2017;123:1124–1132. doi: 10.1111/jam.13564. [DOI] [PubMed] [Google Scholar]
- 70.Daba G.M., Ishibashi N., Gong X., Taki H., Yamashiro K., Lim Y.Y., Zendo T., Sonomoto K. Characterisation of the action mechanism of a Lactococcus-specific bacteriocin. Lactococcin Z, J Biosci. Bioeng. 2018;126:603–610. doi: 10.1016/j.jbiosc.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 71.Zendo T. Screening and characterization of novel bacteriocins from lactic acid bacteria. Biosci. Biotechnol. Biochem. 2013;77:893–899. doi: 10.1271/bbb.130014. [DOI] [PubMed] [Google Scholar]
- 72.de Arauz L.J., Jozala A.F., Mazzola P.G., Vessoni Penna T.C. Nisin biotechnological production and application: a review. Trends Food Sci. Technol. 2009;20:146–154. doi: 10.1016/j.tifs.2009.01.056. [DOI] [Google Scholar]
- 73.Piper C., Casey P.G., Hill C., Cotter P.D., Ross R.P. The lantibiotic lacticin 3147 prevents systemic spread of Staphylococcus aureus in a murine infection model. Internet J. Microbiol. 2012;2012:1–6. doi: 10.1155/2012/806230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mota-Meira M., Morency H., Lavoie M.C. In vivo activity of mutacin B-Ny266. J. Antimicrob. Chemother. 2005;56:869–871. doi: 10.1093/jac/dki295. [DOI] [PubMed] [Google Scholar]
- 75.Ghodhbane H., Elaidi S., Sabatier J.-M., Achour S., Benhmida J., Regaya I. Bacteriocins active against multi-resistant gram negative bacteria implicated in nosocomial infections. Infect. Disord.: Drug Targets. 2015;15:2–12. doi: 10.2174/1871526514666140522113337. [DOI] [PubMed] [Google Scholar]
- 76.Settanni L., Corsetti A. Application of bacteriocins in vegetable food biopreservation. Int. J. Food Microbiol. 2008;121:123–138. doi: 10.1016/j.ijfoodmicro.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 77.Caridi A. Selection of Escherichia coli -inhibiting strains of Lactobacillus paracasei subsp. paracasei. J. Ind. Microbiol. Biotechnol. 2002;29:303–308. doi: 10.1038/sj.jim.7000300. [DOI] [PubMed] [Google Scholar]
- 78.Ivanova I., Miteva V., Stefanova T., Pantev A., Budakov I., Danova S., Moncheva P., Nikolova I., Dousset X., Boyaval P. Characterization of a bacteriocin produced by Streptococcus thermophilus 81. Int. J. Food Microbiol. 1998;42:147–158. doi: 10.1016/S0168-1605(98)00067-1. [DOI] [PubMed] [Google Scholar]
- 79.Cole K., Farnell M.B., Donoghue A.M., Stern N.J., Svetoch E.A., Eruslanov B.N., Volodina L.I., Kovalev Y.N., Perelygin V.V., Mitsevich E.V., Mitsevich I.P., Levchuk V.P., Pokhilenko V.D., Borzenkov V.N., Svetoch O.E., Kudryavtseva T.Y., Reyes-Herrera I., Blore P.J., de los Santos F.S., Donoghue D.J. Bacteriocins reduce Campylobacter colonization and alter gut morphology in Turkey poults. Poultry Sci. 2006;85:1570–1575. doi: 10.1093/ps/85.9.1570. [DOI] [PubMed] [Google Scholar]
- 80.Stern N.J., Svetoch E.A., Eruslanov B.V., Perelygin V.V., Mitsevich E.V., Mitsevich I.P., Pokhilenko V.D., Levchuk V.P., Svetoch O.E., Seal B.S. Isolation of a Lactobacillus salivarius strain and purification of its bacteriocin, which is inhibitory to Campylobacter jejuni in the chicken gastrointestinal system. Antimicrob. Agents Chemother. 2006;50:3111–3116. doi: 10.1128/AAC.00259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Line J.E., Svetoch E.A., Eruslanov B.V., Perelygin V.V., Mitsevich E.V., Mitsevich I.P., Levchuk V.P., Svetoch O.E., Seal B.S., Siragusa G.R., Stern N.J. Isolation and purification of enterocin E-760 with broad antimicrobial activity against gram-positive and gram-negative bacteria. Antimicrob. Agents Chemother. 2008;52:1094–1100. doi: 10.1128/AAC.01569-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang H.-T., Yu C., Hsieh Y.-H., Chen S.-W., Chen B.-J., Chen C.-Y. Effects of albusin B (a bacteriocin) of Ruminococcus albus 7 expressed by yeast on growth performance and intestinal absorption of broiler chickens-its potential role as an alternative to feed antibiotics. J. Sci. Food Agric. 2011;91:2338–2343. doi: 10.1002/jsfa.4463. [DOI] [PubMed] [Google Scholar]
- 83.Rebuffat S. Prokaryotic Antimicrobial Peptides. Springer; New York, NY: 2011. Bacteriocins from gram-negative bacteria: a classification? pp. 55–72. New York. [DOI] [Google Scholar]
- 84.Rea M.C., Ross R.P., Cotter P.D., Hill C. Prokaryotic Antimicrobial Peptides. Springer; New York, NY: 2011. Classification of bacteriocins from gram-positive bacteria; pp. 29–53. New York. [DOI] [Google Scholar]
- 85.Cui Y., Zhang C., Wang Y., Shi J., Zhang L., Ding Z., Qu X., Cui H. Class IIa bacteriocins: diversity and new developments. Int. J. Mol. Sci. 2012;13:16668–16707. doi: 10.3390/ijms131216668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mokoena M.P. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules. 2017;22:1255. doi: 10.3390/molecules22081255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu W., Pang H., Zhang H., Cai Y. Lactic Acid Bacteria. Springer Netherlands; Dordrecht: 2014. Biodiversity of lactic acid bacteria; pp. 103–203. [DOI] [Google Scholar]
- 88.Ekblad B., Kyriakou P.K., Oppegård C., Nissen-Meyer J., Kaznessis Y.N., Kristiansen P.E. Structure–function analysis of the two-peptide bacteriocin plantaricin EF. Biochemistry. 2016;55:5106–5116. doi: 10.1021/acs.biochem.6b00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drider D., Fimland G., Héchard Y., McMullen L.M., Prévost H. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 2006;70:564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Belguesmia Y., Naghmouchi K., Chihib N.-E., Drider D. Prokaryotic Antimicrobial Peptides. Springer; New York, NY: 2011. Class IIa bacteriocins: current knowledge and perspectives; pp. 171–195. New York. [DOI] [Google Scholar]
- 91.Nilsen T., Nes I.F., Holo H. Enterolysin A, a cell wall-degrading bacteriocin from Enterococcus faecalis LMG 2333. Appl. Environ. Microbiol. 2003;69:2975–2984. doi: 10.1128/AEM.69.5.2975-2984.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sun Z., Wang X., Zhang X., Wu H., Zou Y., Li P., Sun C., Xu W., Liu F., Wang D. Class III bacteriocin Helveticin-M causes sublethal damage on target cells through impairment of cell wall and membrane. J. Ind. Microbiol. Biotechnol. 2018;45:213–227. doi: 10.1007/s10295-018-2008-6. [DOI] [PubMed] [Google Scholar]
- 93.Savoia D. Plant-derived antimicrobial compounds: alternatives to antibiotics. Future Microbiol. 2012;7:979–990. doi: 10.2217/fmb.12.68. [DOI] [PubMed] [Google Scholar]
- 94.Takó M., Kerekes E.B., Zambrano C., Kotogán A., Papp T., Krisch J., Vágvölgyi C. Plant phenolics and phenolic-enriched extracts as antimicrobial agents against food-contaminating microorganisms. Antioxidants. 2020;9:165. doi: 10.3390/antiox9020165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Khameneh B., Iranshahy M., Soheili V., Fazly Bazzaz B.S. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob. Resist. Infect. Control. 2019;8:118. doi: 10.1186/s13756-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vuolo M.M., Lima V.S., Maróstica Junior M.R. Bioactive Compounds. Elsevier; 2019. Phenolic compounds; pp. 33–50. [DOI] [Google Scholar]
- 97.Klančnik A., Šikić Pogačar M., Trošt K., Tušek Žnidarič M., Mozetič Vodopivec B., Smole Možina S. Anti- Campylobacter activity of resveratrol and an extract from waste Pinot noir grape skins and seeds, and resistance of Camp. jejuni planktonic and biofilm cells, mediated via the CmeABC efflux pump. J. Appl. Microbiol. 2017;122:65–77. doi: 10.1111/jam.13315. [DOI] [PubMed] [Google Scholar]
- 98.Guimarães A.C., Meireles L.M., Lemos M.F., Guimarães M.C.C., Endringer D.C., Fronza M., Scherer R. Antibacterial activity of terpenes and terpenoids present in essential oils. Molecules. 2019;24:2471. doi: 10.3390/molecules24132471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chakraborty R., Pusapati B.R. 2011. Screening for Antidiarrheal Activity of Psidium Guajava: A Possible Alternative in the Treatment against Diarrhea Causing Enteric Pathogens.www.jocpr.com [Google Scholar]
- 100.Nitiema L.W., Savadogo A., Simpore J., Dianou D., Traore A.S. In vitro antimicrobial activity of some phenolic compounds (coumarin and quercetin) against gastroenteritis bacterial strains. Int. J. Microbiol. Res. 2012;3(3):183–187. doi: 10.5829/idosi.ijmr.2012.3.3.6414. [DOI] [Google Scholar]
- 101.Mahboubi A., Asgarpanah J., Sadaghiyani P.N., Faizi M. Total phenolic and flavonoid content and antibacterial activity of Punica granatum L. var. pleniflora flowers (Golnar) against bacterial strains causing foodborne diseases. BMC Compl. Alternative Med. 2015;15:366. doi: 10.1186/s12906-015-0887-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hoque M., Inatsu M., Juneja V., Kawamoto S. Antimicrobial activity of cloves and cinnamon extracts against food borne pathogens and spoilage bacteria and inactivation of Listeria monocytogenes in ground chicken meat with their essential oils. Rep. Natl. Food Res. Inst. 2008;72 [Google Scholar]
- 103.Moumni S., Elaissi A., Trabelsi A., Merghni A., Chraief I., Jelassi B., Chemli R., Ferchichi S. Correlation between chemical composition and antibacterial activity of some Lamiaceae species essential oils from Tunisia. BMC Compl. Med. Ther. 2020;20 doi: 10.1186/s12906-020-02888-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Naveed R., Hussain I., Tawab A., Tariq M., Rahman M., Hameed S., Mahmood M.S., Siddique A.B., Iqbal M. Antimicrobial activity of the bioactive components of essential oils from Pakistani spices against Salmonella and other multi-drug resistant bacteria. BMC Compl. Alternative Med. 2013;13 doi: 10.1186/1472-6882-13-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Iseppi R., Camellini S., Sabia C., Messi P. Combined antimicrobial use of essential oils and bacteriocin bacLP17 as seafood biopreservative to control Listeria monocytogenes both in planktonic and in sessile forms. Res. Microbiol. 2020;171:351–356. doi: 10.1016/j.resmic.2020.07.002. [DOI] [PubMed] [Google Scholar]
- 106.Abdollahzadeh E., Rezaei M., Hosseini H. Antibacterial activity of plant essential oils and extracts: the role of thyme essential oil, nisin, and their combination to control Listeria monocytogenes inoculated in minced fish meat. Food Control. 2014;35:177–183. doi: 10.1016/j.foodcont.2013.07.004. [DOI] [Google Scholar]
- 107.Turgis M., Vu K.D., Dupont C., Lacroix M. Combined antimicrobial effect of essential oils and bacteriocins against foodborne pathogens and food spoilage bacteria. Food Res. Int. 2012;48:696–702. doi: 10.1016/j.foodres.2012.06.016. [DOI] [Google Scholar]
- 108.Soodsawaeng P., Butkhot N., Boonthai T., Vuthiphandchai V., Nimrat S. Antibacterial activity of bacteriocin produced by Bacillus velezensis BUU004, herb extracts and their combination for controlling spoilage and pathogenic bacteria in dried, seasoned and crushed squid. Srinakharinwirot Sci. J. 2021;37(1):1–20. [Google Scholar]
- 109.Rakholiya K., Chanda S. In vitro interaction of certain antimicrobial agents in combination with plant extracts against some pathogenic bacterial strains. Asian Pac. J. Trop. Biomed. 2012;2:S1466–S1470. [Google Scholar]
- 110.Garvey M.I., Rahman M.M., Gibbons S., Piddock L.J.V. Medicinal plant extracts with efflux inhibitory activity against Gram-negative bacteria. Int. J. Antimicrob. Agents. 2011;37:145–151. doi: 10.1016/j.ijantimicag.2010.10.027. [DOI] [PubMed] [Google Scholar]
- 111.Sanhueza L., Melo R., Montero R., Maisey K., Mendoza L., Wilkens M. Synergistic interactions between phenolic compounds identified in grape pomace extract with antibiotics of different classes against Staphylococcus aureus and Escherichia coli. PLoS One. 2017;12 doi: 10.1371/journal.pone.0172273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bag A., Chattopadhyay R.R. Synergistic antibacterial and antibiofilm efficacy of nisin in combination with p -coumaric acid against food-borne bacteria Bacillus cereus and Salmonella typhimurium. Lett. Appl. Microbiol. 2017;65:366–372. doi: 10.1111/lam.12793. [DOI] [PubMed] [Google Scholar]
- 113.Chandar B., Poovitha S., Ilango K., MohanKumar R., Parani M. Inhibition of New Delhi metallo-β-lactamase 1 (NDM-1) producing Escherichia coli IR-6 by selected plant extracts and their synergistic actions with antibiotics. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lacmata S.T., Kuete V., Dzoyem J.P., Tankeo S.B., Teke G.N., Kuiate J.R., Pages J.-M. Antibacterial activities of selected Cameroonian plants and their synergistic effects with antibiotics against bacteria expressing MDR phenotypes. Evid. base Compl. Alternative Med. 2012;2012:1–11. doi: 10.1155/2012/623723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Pisoschi A.M., Pop A., Georgescu C., Turcuş V., Olah N.K., Mathe E. An overview of natural antimicrobials role in food. Eur. J. Med. Chem. 2018;143:922–935. doi: 10.1016/j.ejmech.2017.11.095. [DOI] [PubMed] [Google Scholar]
- 116.Hu S., Wang H., Han W., Ma Y., Shao Z., Li L. Development of double-layer active films containing pomegranate peel extract for the application of pork packaging. J. Food Process. Eng. 2017;40 doi: 10.1111/jfpe.12388. [DOI] [Google Scholar]
- 117.Kanatt S.R., Chander R., Sharma A. Antioxidant and antimicrobial activity of pomegranate peel extract improves the shelf life of chicken products. Int. J. Food Sci. Technol. 2010;45:216–222. doi: 10.1111/j.1365-2621.2009.02124.x. [DOI] [Google Scholar]
- 118.de Oliveira K.Á.R., de Sousa J.P., da Costa Medeiros J.A., de Figueiredo R.C.B.Q., Magnani M., de Siqueira J.P., de Souza E.L. Synergistic inhibition of bacteria associated with minimally processed vegetables in mixed culture by carvacrol and 1,8-cineole. Food Control. 2015;47:334–339. doi: 10.1016/j.foodcont.2014.07.014. [DOI] [Google Scholar]
- 119.Piskernik S., Klančnik A., Riedel C.T., Brøndsted L., Možina S.S. Reduction of Campylobacter jejuni by natural antimicrobials in chicken meat-related conditions. Food Control. 2011;22:718–724. doi: 10.1016/j.foodcont.2010.11.002. [DOI] [Google Scholar]
- 120.Martínez-Graciá C., González-Bermúdez C.A., Cabellero-Valcárcel A.M., Santaella-Pascual M., Frontela-Saseta C. Use of herbs and spices for food preservation: advantages and limitations. Curr. Opin. Food Sci. 2015;6:38–43. doi: 10.1016/j.cofs.2015.11.011. [DOI] [Google Scholar]
- 121.Bilal H., Khan M.N., Rehman T., Hameed M.F., Yang X. Antibiotic resistance in Pakistan: a systematic review of past decade. BMC Infect. Dis. 2021;21:244. doi: 10.1186/s12879-021-05906-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Shen Z., Wang Y., Zhang Q., Shen J. vol. 6. 2018. (Antimicrobial Resistance in Campylobacter Spp, Microbiol Spectr). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.McGannon C.M., Fuller C.A., Weiss A.A. Different classes of antibiotics differentially influence shiga toxin production. Antimicrob. Agents Chemother. 2010;54:3790–3798. doi: 10.1128/AAC.01783-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pensinger D.A., Aliota M.T., Schaenzer A.J., Boldon K.M., Ansari I.H., Vincent W.J.B., Knight B., Reniere M.L., Striker R., Sauer J.-D. Selective pharmacologic inhibition of a PASTA kinase increases Listeria monocytogenes susceptibility to β-lactam antibiotics. Antimicrob. Agents Chemother. 2014;58:4486–4494. doi: 10.1128/AAC.02396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Moreno L.Z., Paixão R., Gobbi D.D.S., Raimundo D.C., Ferreira T.P., Moreno A.M., Hofer E., Reis C.M.F., Matté G.R., Matté M.H. Characterization of antibiotic resistance in Listeria spp. isolated from slaughterhouse environments, pork and human infections. J. Infect. Dev. Ctries. 2014;8:416–423. doi: 10.3855/jidc.4188. [DOI] [PubMed] [Google Scholar]
- 126.Lungu B., O'Bryan C.A., Muthaiyan A., Milillo S.R., Johnson M.G., Crandall P.G., Ricke S.C. Listeria monocytogenes : antibiotic resistance in food production. Foodb. Pathog. Dis. 2011;8:569–578. doi: 10.1089/fpd.2010.0718. [DOI] [PubMed] [Google Scholar]
- 127.Poirel L., Madec J.-Y., Lupo A., Schink A.-K., Kieffer N., Nordmann P., Schwarz S. Antimicrobial resistance in Escherichia coli. Microbiol. Spectr. 2018;6 doi: 10.1128/microbiolspec.ARBA-0026-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.El-Sharkawy H., Tahoun A., El-Gohary A.E.-G.A., El-Abasy M., El-Khayat F., Gillespie T., Kitade Y., Hafez H.M., Neubauer H., El-Adawy H. Epidemiological, molecular characterization and antibiotic resistance of Salmonella enterica serovars isolated from chicken farms in Egypt. Gut Pathog. 2017;9:8. doi: 10.1186/s13099-017-0157-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zendehbad B., Khayatzadeh J., Alipour A. Prevalence, seasonality and antibiotic susceptibility of Campylobacter spp. isolates of retail broiler meat in Iran. Food Control. 2015;53:41–45. doi: 10.1016/j.foodcont.2015.01.008. [DOI] [Google Scholar]
- 130.Thung T.Y., Mahyudin N.A., Basri D.F., Wan Mohamed Radzi C.W.J., Nakaguchi Y., Nishibuchi M., Radu S. Prevalence and antibiotic resistance of Salmonella enteritidis and Salmonella typhimurium in raw chicken meat at retail markets in Malaysia. Poultry Sci. 2016;95:1888–1893. doi: 10.3382/ps/pew144. [DOI] [PubMed] [Google Scholar]
- 131.Garedew L., Taddese A., Biru T., Nigatu S., Kebede E., Ejo M., Fikru A., Birhanu T. Prevalence and antimicrobial susceptibility profile of Listeria species from ready-to-eat foods of animal origin in Gondar Town, Ethiopia. BMC Microbiol. 2015;15:100. doi: 10.1186/s12866-015-0434-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Mąka Ł., Maćkiw E., Ścieżyńska H., Pawłowska K., Popowska M. Antimicrobial susceptibility of Salmonella strains isolated from retail meat products in Poland between 2008 and 2012. Food Control. 2014;36:199–204. doi: 10.1016/j.foodcont.2013.08.025. [DOI] [Google Scholar]
- 133.Zankari E., Hasman H., Kaas R.S., Seyfarth A.M., Agerso Y., Lund O., Larsen M.V., Aarestrup F.M. Genotyping using whole-genome sequencing is a realistic alternative to surveillance based on phenotypic antimicrobial susceptibility testing. J. Antimicrob. Chemother. 2013;68:771–777. doi: 10.1093/jac/dks496. [DOI] [PubMed] [Google Scholar]
- 134.Gómez D., Azón E., Marco N., Carramiñana J.J., Rota C., Ariño A., Yangüela J. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014;42:61–65. doi: 10.1016/j.fm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 135.Allaoui A., Sansonetti P.J., Parsot C. MxiD, an outer membrane protein necessary for the secretion of the Shigella flexneri Ipa invasins. Mol. Microbiol. 1993;7:59–68. doi: 10.1111/j.1365-2958.1993.tb01097.x. [DOI] [PubMed] [Google Scholar]
- 136.Monteville M.R. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology (N. Y.) 2003;149:153–165. doi: 10.1099/mic.0.25820-0. [DOI] [PubMed] [Google Scholar]
- 137.Ellis T.N., Kuehn M.J. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Krachler A.M., Ham H., Orth K. Outer membrane adhesion factor multivalent adhesion molecule 7 initiates host cell binding during infection by Gram-negative pathogens. Proc. Natl. Acad. Sci. USA. 2011;108:11614–11619. doi: 10.1073/pnas.1102360108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kumar M., Nair M., Venkitanarayanan K. 2007. Role of Bacterial OmpA and Host Cytoskeleton in the Invasion of Human Intestinal Epithelial Cells by Enterobacter Sakazakii. [DOI] [PubMed] [Google Scholar]
- 140.Mohan Nair M.K., Venkitanarayanan K., Silbart L.K., Kim K.S. Outer membrane protein A (OmpA) of cronobacter sakazakii binds fibronectin and contributes to invasion of human brain microvascular endothelial cells. Foodb. Pathog. Dis. 2009;6:495–501. doi: 10.1089/fpd.2008.0228. [DOI] [PubMed] [Google Scholar]
- 141.Kim K., Kim K.-P., Choi J., Lim J.-A., Lee J., Hwang S., Ryu S. Outer membrane proteins A (OmpA) and X (OmpX) are essential for basolateral invasion of Cronobacter sakazakii. Appl. Environ. Microbiol. 2010;76:5188–5198. doi: 10.1128/AEM.02498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kothary M.H., McCardell B.A., Frazar C.D., Deer D., Tall B.D. Characterization of the zinc-containing metalloprotease encoded by zpx and development of a species-specific detection method for Enterobacter sakazakii. Appl. Environ. Microbiol. 2007;73:4142–4151. doi: 10.1128/AEM.02729-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Srinivasa Rao P.S., Yamada Y., Tan Y.P., Leung K.Y. Use of proteomics to identify novel virulence determinants that are required for Edwardsiella tarda pathogenesis. Mol. Microbiol. 2004;53:573–586. doi: 10.1111/j.1365-2958.2004.04123.x. [DOI] [PubMed] [Google Scholar]
- 144.Hecker M., Becher D., Fuchs S., Engelmann S. A proteomic view of cell physiology and virulence of Staphylococcus aureus. Int. J. Med. Microbiol. 2010;300:76–87. doi: 10.1016/j.ijmm.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 145.Niemann G.S., Brown R.N., Gustin J.K., Stufkens A., Shaikh-Kidwai A.S., Li J., McDermott J.E., Brewer H.M., Schepmoes A., Smith R.D., Adkins J.N., Heffron F. Discovery of novel secreted virulence factors from Salmonella enterica Serovar typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 2011;79:33–43. doi: 10.1128/IAI.00771-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Erridge C., Bennett-Guerrero E., Poxton I.R. Structure and function of lipopolysaccharides. Microb. Infect. 2002;4:837–851. doi: 10.1016/s1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- 147.Raetz C.R.H., Whitfield C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Caroff M., Karibian D. Structure of bacterial lipopolysaccharides. Carbohydr. Res. 2003;338:2431–2447. doi: 10.1016/j.carres.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 149.Heinrichs D.E., Yethon J.A., Whitfield C. Molecular basis for structural diversity in the core regions of the lipopolysaccharides of Escherichia coli and Salmonella enterica. Mol. Microbiol. 1998;30:221–232. doi: 10.1046/j.1365-2958.1998.01063.x. [DOI] [PubMed] [Google Scholar]
- 150.Savage P.B. Multidrug-resistant bacteria: overcoming antibiotic permeability barriers of Gram-negative bacteria. Ann. Med. 2001;33:167–171. doi: 10.3109/07853890109002073. [DOI] [PubMed] [Google Scholar]
- 151.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol. Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Schneck E., Schubert T., Konovalov O.V., Quinn B.E., Gutsmann T., Brandenburg K., Oliveira R.G., Pink D.A., Tanaka M. Quantitative determination of ion distributions in bacterial lipopolysaccharide membranes by grazing-incidence X-ray fluorescence. Proc. Natl. Acad. Sci. USA. 2010;107:9147–9151. doi: 10.1073/pnas.0913737107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Whitmire W.M., Garon C.F. Specific and nonspecific responses of murine B cells to membrane blebs of Borrelia burgdorferi. Infect. Immun. 1993;61:1460–1467. doi: 10.1128/iai.61.4.1460-1467.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Opal S. The host response to endotoxin, antilipopolysaccharide strategies, and the management of severe sepsis. Int. J. Med. Microbiol. 2007;297:365–377. doi: 10.1016/j.ijmm.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 156.Kitchens R.L., Munford R.S. CD14-Dependent internalization of bacterial lipopolysaccharide (LPS) is strongly influenced by LPS aggregation but not by cellular responses to LPS. J. Immunol. 1998;160:1920–1928. doi: 10.4049/jimmunol.160.4.1920. [DOI] [PubMed] [Google Scholar]
- 157.Tarr P.I., Gordon C.A., Chandler W.L. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. 2005;365:1073–1086. doi: 10.1016/S0140-6736(05)71144-2. [DOI] [PubMed] [Google Scholar]
- 158.Nataro J.P., Kaper J.B. Diarrheagenic Escherichia coli. Clin. Microbiol. Rev. 1998;11:142–201. doi: 10.1128/CMR.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Hueck C.J. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 1998;62:379–433. doi: 10.1128/MMBR.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Galán J.E., Wolf-Watz H. Protein delivery into eukaryotic cells by type III secretion machines. Nature. 2006;444:567–573. doi: 10.1038/nature05272. [DOI] [PubMed] [Google Scholar]
- 161.Kenny B., DeVinney R., Stein M., Reinscheid D.J., Frey E.A., Finlay B.B., coli (Epec Enteropathogenic E. Transfers its receptor for intimate adherence into mammalian cells. Cell. 1997;91:511–520. doi: 10.1016/S0092-8674(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 162.Daniell S.J., Kocsis E., Morris E., Knutton S., Booy F.P., Frankel G. 3D structure of EspA filaments from enteropathogenic Escherichia coli. Mol. Microbiol. 2003;49:301–308. doi: 10.1046/j.1365-2958.2003.03555.x. [DOI] [PubMed] [Google Scholar]
- 163.Knutton S. A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J. 1998;17:2166–2176. doi: 10.1093/emboj/17.8.2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ide T., Laarmann S., Greune L., Schillers H., Oberleithner H., Schmidt M.A. Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol. 2001;3:669–679. doi: 10.1046/j.1462-5822.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- 165.Horne J.E., Brockwell D.J., Radford S.E. Role of the lipid bilayer in outer membrane protein folding in Gram-negative bacteria. J. Biol. Chem. 2020;295:10340–10367. doi: 10.1074/jbc.REV120.011473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.V Prasadarao N., Wass C.A., Weiser J.N., Stins M.F., Huang S.H., Kim K.S. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 1996;64:146–153. doi: 10.1128/iai.64.1.146-153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Kolodziejek A.M., Schnider D.R., Rohde H.N., Wojtowicz A.J., Bohach G.A., Minnich S.A., Hovde C.J. Outer membrane protein X (ail) contributes to Yersinia pestis virulence in pneumonic plague and its activity is dependent on the lipopolysaccharide core length. Infect. Immun. 2010;78:5233–5243. doi: 10.1128/IAI.00783-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Sukumaran S.K., Shimada H., Prasadarao N.V. Entry and intracellular replication of Escherichia coli K1 in macrophages require expression of outer membrane protein A. Infect. Immun. 2003;71:5951–5961. doi: 10.1128/IAI.71.10.5951-5961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Torres A.G., Kaper J.B. Multiple elements controlling adherence of enterohemorrhagic Escherichia coli O157:H7 to HeLa cells. Infect. Immun. 2003;71:4985–4995. doi: 10.1128/IAI.71.9.4985-4995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Nicholson T.F., Watts K.M., Hunstad D.A. OmpA of uropathogenic Escherichia coli promotes postinvasion pathogenesis of cystitis. Infect. Immun. 2009;77:5245–5251. doi: 10.1128/IAI.00670-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meng X., Liu X., Zhang L., Hou B., Li B., Tan C., Li Z., Zhou R., Li S. Virulence characteristics of extraintestinal pathogenic Escherichia coli; deletion of gene encoding the outer membrane protein X. J. Vet. Med. Sci. 2016;78:1261–1267. doi: 10.1292/jvms.16-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hirakawa H., Suzue K., Takita A., Kamitani W., Tomita H. Roles of OmpX, an outer membrane protein, on virulence and flagellar expression in uropathogenic Escherichia coli. Infect. Immun. 2021;89 doi: 10.1128/IAI.00721-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Abee T., Groot M.N., Tempelaars M., Zwietering M., Moezelaar R., van der Voort M. Germination and outgrowth of spores of Bacillus cereus group members: diversity and role of germinant receptors. Food Microbiol. 2011;28:199–208. doi: 10.1016/j.fm.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 174.Setlow P. Germination of spores of Bacillus species: what we know and do not know. J. Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 176.Cowan A.E., Olivastro E.M., Koppel D.E., Loshon C.A., Setlow B., Setlow P. Lipids in the inner membrane of dormant spores of Bacillus species are largely immobile. Proc. Natl. Acad. Sci. USA. 2004;101:7733–7738. doi: 10.1073/pnas.0306859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Laue M., Han H.M., Dittmann C., Setlow P. Intracellular membranes of bacterial endospores are reservoirs for spore core membrane expansion during spore germination. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-29879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kang M.K., Tullman-Ercek D. Engineering expression and function of membrane proteins. Methods. 2018;147:66–72. doi: 10.1016/j.ymeth.2018.04.014. [DOI] [PubMed] [Google Scholar]
- 179.Cassidy L., Kaulich P.T., Tholey A. Proteoforms expand the world of microproteins and short open reading frame-encoded peptides. iScience. 2023;26(2) doi: 10.1016/j.isci.2023.106069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Garai P., Blanc‐Potard A. Uncovering small membrane proteins in pathogenic bacteria: regulatory functions and therapeutic potential. Mol. Microbiol. 2020;114(5):710–720. doi: 10.1111/mmi.14564. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.