Abstract
Candida auris is a serious health concern of the current world that possesses a serious global health threat and is emerging at a high rate. Available antifungal drugs are failing to combat this pathogen as they are growing resistant to those drugs and some strains have already shown resistance to all three available antifungal drugs in the market. Hence, finding alternative therapies is essential for saving lives from this enemy. To make the development of new treatments easier, we conducted some in silico study of this pathogen to discover possible targets for drug design and also recommended some possible metabolites to test in vivo circumstances. The complete proteome of the representative strain was retrieved, and the duplicate, non-essential, human homologous, non-metabolic, and druggable proteins were then eliminated. As a result, out of a total of 5441 C. auris proteins, we were able to isolate three proteins (XP 028890156.1, XP 028891672.1, and XP 028891858.1) that are crucial for the pathogen's survival as well as host-non-homolog, metabolic, and unrelated proteins to the human microbiome. Their subcellular locations and interactions with a large number of proteins (10 proteins) further point to them being good candidates for therapeutic targets. Following in silico docking of 29 putative antifungals of plant origin against the three proteins we chose, Caledonixanthone E, Viniferin, Glaucine, and Jatrorrhizine were discovered to be the most effective means of inhibiting those proteins since they displayed higher binding affinities (ranging from −28.97 kcal/mol to −51.99 kcal/mol) than the control fluconazole (which ranged between −28.84 kcal/mol and −41.15 kcal/mol). According to the results of MD simulations and MM-PBSA calculations, Viniferin and Caledonixanthone E are the most effective ligands for the proteins XP 028890156.1, XP 028891672.1, and XP 028891858.1. Furthermore, they were predicted to be safe and also showed proper ADME properties.
Keywords: Candida auris, In silico approach, Drug targets, Potential drugs, Molecular docking, MD simulation
1. Introduction
Since 2009, one of the rarest fungal pathogens, C. auris, has piqued the interest of the global health community due to an increase in mortality due to severe invasive candidiasis and recurrent outbreaks [1,2]. It was first detected from the ear canal discharge of a patient in Japan and hence introduced “auris” in its nomenclature. However, it is believed that it was present long before it was identified as a misdiagnosed human pathogen. The dermal sites, respiratory, urogenital tract and the blood circulatory system are the potential sites of fatal invasive C. auris infection in the human body [3,4]. It is a budding yeast (ovoid to elongate structure), which may form rudimentary pseudohyphae. However, these phenotypes are not distinct enough to assist in the identification of C. auris. The whole genome sequencing, a VITEK 2 version 8.01 and mass spectrometry have shown promising results in identifying and differentiating C. auris from other closely related Candida species and also in characterizing the dynamics of its transmission [5]. The unique traits of C. auris like; nosocomial transmission in clinical settings, misidentification, colonizing ability, multi-drug resistance (MDR), high virulence and mortality rates, long-term persistence on healthcare environmental surfaces (both on the human host and inanimate surfaces) separate it from other Candida species and justifies its potential to generate pandemic situation [1,[5], [6], [7]]. Besides, the fungus has a high spreadability and can infect all types of patients, regardless of age or immune-suppression disease [8]. Along with the development of colonies at multiple sites of the human body, this fungus is found to persist and be isolated even after 3 months of primary detection and treatment (using Echinocandin) (Vallabhaneni et al., 2016; Schelenz et al., 2016). Moreover, recent investigations on COVID-19 conditions indicate a higher probability of C. auris outbreaks in the ICUs of overburdened hospitals. Hence extra caution is demanded regarding the identification and treatment of C. auris infections among COVID-19 patients [2,9].
The genome (haploid) of C. auris comprises 7 chromosomes, having a genomic size ranging from 12.1 Mb to 12.7 Mb (with approximately 45% GC content). The analysis of the genome estimates about 6500–8500 protein-coding sequences and around 5500 genes; that code different virulent proteins in Candida species which develops virulent characters of the fungus like biofilm formation. Besides, different kinase proteins and transporter genes have been sorted out suspecting that they might assist the drug resistance property of the fungus C. auris. The mortality rates at varied geographical locations portrayed significant differences. In Asia, the Far East, and the United States the mortality rate exceeds 50% whereas, in Venezuela and Colombia, the rate is 72% and 35.2% respectively [[10], [11], [12], [13]].
Till now three major classes of antifungal drugs i.e. azoles, polyenes and echinocandins are largely in practice regarding the treatment of invasive fungal infections or candidiasis. However, the multidrug resistance property of C. auris limits the options for the treatment of invasive infections caused by the pathogen. Different research groups and CDC (The Centers for Disease Control and Prevention) analyzed the resistance percentage of C. auris against different antifungals. For example; in a study, 93% of the fungus among 54 internationally collected isolates showed resistance against fluconazole, voriconazole (>50%), amphotericin B (35%), echinocandins (7%) and two antifungal classes i.e. azoles and polyenes (41%). Besides 4% isolates demonstrated raised MICs (minimum inhibitory concentration) towards the 3 major antifungal classes and thus interpreting dangerous situations regarding the control and treatment of multidrug resistant fungal pathogen, C. auris [[14], [15], [16]].
Different side-effects and multi-drug resistance generated due to different antifungal treatments, point out the significance of novel drug discovery, evaluating the antifungal properties of diverse natural and synthetic/semi-synthetic chemical compounds [17]. These compounds target host proteins and genes, disease-associated biomarkers, microRNAs and phenotypes, various biological pathways, and the connections between different biological networks and molecular functions in order to generate treatment measures. Multiple computational and in silico methods are being used nowadays in assisting the drug discovery process and at the same time reducing the risk factors, time consumed, cost and raw materials, unlike the traditional drug discovery approaches [18,19]. Besides this, the computer aided drug design (CADD) methodologies limit the pharmacological practices on animal models, designs safe and novel drug candidates, and thus broaden the field for potential antifungal treatments [20].
The multi-drug resistant condition and development of toxicity and side-effects in the host, developed as a result of the current antifungal therapeutics application, stimulates the research for novel drug development. Therefore, in this study, the subtractive proteomics approach was utilized to identify suitable drug targets of C. auris and reverse or inverse virtual screening or inverse docking technique was used that helped identify the probable relationships among targeted proteins via chemical probing. The reverse docking approach screens different protein targets (ligands/compounds) in order to find the potent binding partners via statistical analysis, unlike the conventional forward docking where a variety of ligands docks to a particular target only [21]. The plant secondary metabolites having antimicrobial and antifungal activities are being tested for this purpose. More than 50% of the Food and Drug Administration (FDA) approved drugs are derived from plant metabolites [22]. This validates the wide range of therapeutic potential of the natural products to be used as the basis of the innovation of new drugs. The recent advancement and interference of bioinformatics and in silico screening have largely enhanced the proportion of detecting potent plant metabolites with therapeutic properties. The researchers have developed multiple computerized databases containing detailed information about various plant metabolites for this purpose. This information helps virtual screening and selection of the potential natural products as novel drugs against the pathogen C. auris. Thus, this study aims at screening the best drug target candidates and bioactive compounds from plant origin and utilizing them as the building blocks for the development of novel drugs against the deadly fungal pathogen C. auris.
2. Material and methods
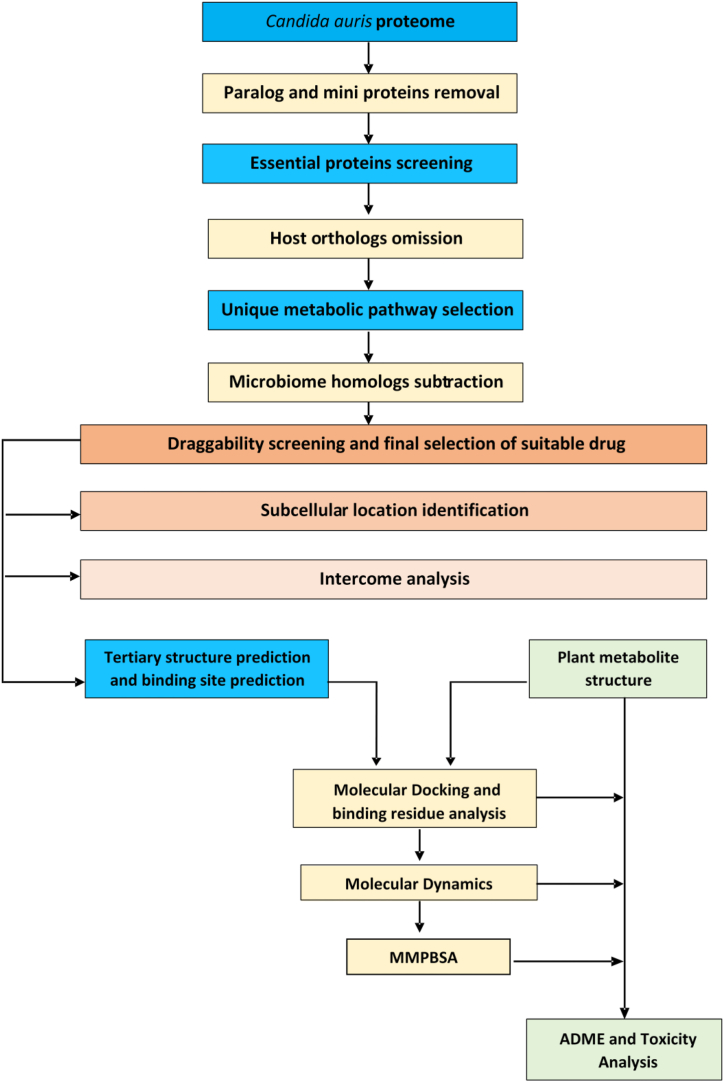
Several online servers and bioinformatics tools were utilized to scrutinize the unique drug targets of Candida auris and potential drugs to block those unique targets shown in Fig. 1.
Fig. 1.
Schematic pipeline of the process.
2.1. Retrieval of Candida auris proteome
Representative Candida auris (assembly Cand_auris_B11221_V1) proteome was retrieved from the National Center for Biotechnology Information Genome database (https://www.ncbi.nlm.nih.gov/genome).
2.2. Omission of paralog and mini proteins
In order to eliminate duplicate protein sequences, the proteome file was submitted to the CD-HIT server (http://cd-hit.org/) with the “sequence identity cut-off” set to 0.6 (60% identity) [23,24]. Mini proteins sequences of less than 100 amino acids play a significant role in a variety of regulatory activities and biological processes. Moreover, these proteins are less likely to be a suitable candidate for a therapeutic intervention. Therefore, the “length of the sequence to skip” was set at 100 to omit these sequences [25]. Amino acids having longer sequences, on the other hand, are frequently discovered to be engaged in important metabolic processes. Again, Haag et al., 2012, revealed metabolic pathway proteins as more appropriate for drug design as attacking bacterial energy-production machinery avoids the usual drug resistance sites, paving the way for multifaceted antibiotic resistance approaches [26]. Since our goal was to identify metabolic proteins and reduce the likelihood of medication resistance, we removed mini proteins from the investigation.
2.3. Finding essential proteins and screening orthologs in host
The Non-paralog protein sequences were subjected to BLAST against the DEG database (www.essentialgene.org/) to identify essential proteins [27] considering a cut-off e-value of 1e-100 and per identity of 25%. BLASTp was then run against the human genome in the NCBI database to separate non-paralog essential proteins from their ortholog counterparts in the host. Everything was left the same, with the exception of Homo sapiens (taxid: 9606) in the “organisms” box, and the threshold e-value was set to 0.0001. Only queries that resulted in no significant hits were retained for further investigation with the objective of developing a pathogen-specific remedy [28].
2.4. Selection of unique metabolic pathway proteins
By entering the respective three-letter KEGG organism codes in the organism box -"caur” for C. auris and “hsa” for H. sapiens-of the KEGG PATHWAY database (http://www.genome.jp/kegg/pathway.html) metabolic pathways for both species were obtained [29]. The fungus's unique pathways were then screened by manual comparison. The KO numbers of host nonhomologous proteins were obtained from the KEEG database's KAAS server (https://www.genome.jp/kaas-bin/kaas_main) using the BLASTp program. The KO number was then used to search the KO server (KEGG ORTHOLOGY) (https://www.genome.jp/kegg/ko.html) for protein pathways, and it was determined which proteins are solely engaged in the pathogen's unique metabolic pathways [30]. Proteins found to be involved in common pathways were excluded from the study.
2.5. Human microbiome non-homology analysis
An overwhelming number of beneficial microorganisms in the human body protect the body from diseases and potentially harmful external objects. The objective of the non-homology investigation on the human microbiome was to determine whether the distinct metabolic pathway sequences of C. auris correspond to the protein sequences of those helpful bacteria found in the human body. Therefore, the NCBI BLAST software was used to run unique metabolic pathway protein sequences against Bioproject-43021, with 0.005 as the cut-off score [31]. Bioproject-43021 is the Human Microbiome Project that explores the possible relationships between particular human diseases and the microbiome and it has collected and sequenced microbiome samples from volunteers [32]. Hence, using BLAST against this bioproject allowed for the identification of C. auris proteins that are identical to proteins found in the human microbiome and their subsequent deletion. Sequences that share fewer than 45% of their similarities are retained for the further analysis.
2.6. Finding novel drug targets using Drugbank Database
To determine the uniqueness of the chosen proteins as therapeutic targets, selected proteins from the preceding steps were examined in the DrugBank database (https://www.drugbank.ca/structures/search/bonds/sequence) [33]. The presence of targets demonstrates their draggability, but the lack of targets demonstrates the originality of the proteins categorized as “new targets” [34]. During the action, all of the parameters were left at their default values.
2.7. Analysis of cellular localization
Proteins located in membranes can be used as both drug targets and vaccination, where cytoplasmic proteins can be used as drug targets [35]. CELLO v.2.5 (http://cello.life.nctu.edu.tw/) server was utilized to forecast the subcellular localization of the unique drug targets [36].
2.8. Interactome analysis
The significance of the targeted proteins in the bacterial metabolic system can be figured out based on the number of interacting proteins (nodes) and interactions (edges) disrupted by its removal [37]. The number of interacting proteins (nodes) and interactions (edges) will indicate when the query protein will be blocked what effects it will put on the fungal metabolic system. Thus to determine the significance of the target protein in the metabolic system, the protein-protein interaction network of the unique drug targets was analyzed using the STRING 11.5 (https://string-db.org/) server [38]. To eliminate misleading negative and positive results, the Protein network includes high confidence interactors with scores ≥0.700.
2.9. Prediction of 3D structures and binding sites of the proteins
The Protein Data Bank (RCSB PDB) did not contain structures for the selected proteins. Therefore, molecular models of distinct proteins were predicted using the I-TASSER server (https://zhanggroup.org/I-TASSER/), an online resource for automated protein structure prediction and structure-based function annotation [39]. These molecular models were then refined using the GalaxyWEB server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) [40]. To evaluate the quality of the models, the Ramachandan plot and Errat quality score were studied using Saves server (https://saves.mbi.ucla.edu/) and RaptorX Binding site (http://raptorx.uchicago.edu/BindingSite/) prediction server was used to estimate ligand binding sites of the proteins [41,42].
2.10. Collection of metabolites structure
Following a review of the available research, a list of 29 plant metabolites from various species that have been shown to have antifungal properties was generated. The PubChem website (https://pubchem.ncbi.nlm.nih.gov/) provided the 3D structures of those metabolites in SDF (3D) format [43], which were then converted to PDB format using the Open Babel v2.3 program [44].
2.11. Docking analysis
The interaction between tiny ligands and macromolecules may be predicted using molecular docking in drug development [45]. PatchDock Server (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) was employed for docking purposes [46] since docking offers interactions between drug targets and prospective therapies [47]. Proteins were used as the receptors, while the metabolites were used as the ligands. Fluconazole, a widely prescribed antibiotic to treat C. auris, was chosen as the reference metabolite for molecular docking in order to conduct a more comprehensive investigation of the drug's effectiveness. The docked complexes were then refined using the FireDock refinement program [48]. The PyMOL v2.0 program was then used to visualize and analyze the binding sites of the metabolites [49].
2.12. Molecular dynamics and MM-PBSA calculations of the docked complex
The stability of three best docked complexes was evaluated through molecular dynamic simulations (MDS). The drug target proteins XP_028890156.1, XP_028891672.1 and XP_028891858.1 and three best ligand bounded target protein complexes for each protein docked complexes were subjected to MD simulations. The molecular dynamics was performed with Gromacs 2020.4 [50] MD simulation package on HPC cluster at Bioinformatics Resources and Applications Facility (BRAF), C-DAC, Pune. The protein topology was obtained using CHARMM-36 force field [51,52]; while the ligand topologies were generated using the CHARMM General Force Field [52,53] on CGenFF server. The systems were then solvated with TIP3P water models [54] in a dodecahedron unit cell. Subsequently the solvated systems were neutralized with addition of appropriate counter-ions where the bare protein and protein complexes of XP_028890156.1 required 20 chloride ions, XP_028891672.1 required 4 chloride ions and XP_028891858.1 required 10 sodium ions respectively. These solvated and neutralized systems were energy minimized with steepest descent criteria until the threshold (Fmax< 10 kJ mol−1 or emtol = 10) reaches and later equilibrated at constant volume and temperature conditions of 300 K using a modified Berendsen thermostat [55] and then at constant volume and pressure Berendsen barostat [56] for 1 ns each. The production phase unrestrained 100 ns MDS was performed at constant temperature and constant pressure conditions using the modified Berendsen thermostat and Parrinello-Rahman barostat [56]. During production phase MDS the covalent bonds were restrained with the LINCS algorithm [57]. The long-range electrostatic energies were measured with Particle Mesh Ewald (PME) method [58] with a cut-off of 1.2 nm. The stability and binding affinities of each ligand in respective protein complexes was analyzed in terms of root mean square deviations (RMSD) in backbone atoms, ligand atoms, root mean square fluctuations (RMSF) in the side-chain atoms of residues, hydrogen bonds analysis between respective ligand and proteins. The most dominant conformations which represent the stable conformations of proteins and protein-ligand complexes were evaluated with principal component analysis (PCA) and Gibb's free energy landscape analysis [59]. The MD trajectories of ligand bound systems isolated at each 1000 ps from the simulation period 75 ns till 100 ns were subjected to Poisson Boltzmann surface area continuum solvation (MM-PBSA) calculations [60,61] to derive the binding free energy estimates. Heavy atoms and the trajectories sampled from slightly unstable or fluctuating time steps could result in spurious and uncertain MM-PBSA energy calculations. Thus, we chose the reasonably stable simulation period for each system, which is 75–100 ns, for sampling the trajectories.
2.13. Pharmacoinformatics studies and toxicity analysis
The features of adsorption, distribution, metabolism, and excretion (ADME) are primarily linked to the kinetics of drug exposure to tissue. Analyzing ADME during the discovery phase will lower the chance of pharmacokinetics-related clinical failure [62]. The ADME characteristics of the top four metabolites were evaluated using the SwissADME server (http://www.swissadme.ch/) [63]. The medications were uploaded in SDF format to the server, transformed to SMILES, and then run to obtain predictions. Blood-brain barrier (BBB) of the top metabolites were then evaluated using the BOILED-Egg model [64]. To forecast the comparative harmful effects of leading medications, researchers used pkCSM (http://biosig.unimelb.edu.au/pkcsm/), an online program [65] that requires SMILES for the top four metabolites, which were obtained from PubChem database.
3. Results
3.1. Removal of duplicate proteins
The Candida auris representative proteome (assembly Cand_auris_B11221_V1), which has a total of 5441 proteins, was obtained from the 112 Candida auris genomes available at NCBI. After eliminating the paralogous sequences, 5217 non-duplicate large proteins were left since the CD-HIT service found 5217 clusters at 60% identity (Supplementary file 1).
3.2. Screening essential proteins and removal of orthologs in host
Proteins that had a significant hit in the DEG server under the defined parameters were considered to be essential for Candida auris. It was revealed that 795 out of 5217 non-paralog proteins were vital for the organisms and these were chosen for further analysis (Supplementary file 2). Drugs and therapeutic compounds should be designed in such a way that they do not unintentionally block host proteins. Therefore, to decrease unfavorable binding of drug to the active sites of the host homologous proteins, the human non-homology step is carried out [66] and orthologs were removed from the list as a result. In this stage, 66 non-paralog essential proteins showed no similarity with human proteins which were regarded as non-orthologous to the host (Supplementary file 3).
3.3. Finding unique metabolic pathway proteins
The KEGG server had 122 C. auris metabolic pathways and 345 human metabolic pathways, with 18 metabolic pathways (Supplementary file 4) being specific only for C. auris. Proteins involved in these unique pathways can be used as therapeutic targets. Following a BLAST at the KAAS server, 52 out of 66 non-homolog essential proteins were discovered to have both KO orthology and metabolic pathway involvement. These 52 proteins are essential for the proper functioning of metabolism, and 19 of them were found to be solely associated with C. auris unique pathways (Table 1).
Table 1.
Unique metabolic pathway proteins.
| SL No | Accession number | KO number | Protein (gene name) | Pathways |
|---|---|---|---|---|
| 1 | XP_028888316.1 | K01624 | fructose-bisphosphate aldolase, class II [EC:4.1.2.13] (FBA, fbaA) | -Methane metabolism |
| - Biosynthesis of secondary metabolites | ||||
| 2 | XP_028889164.1 | K09827 | 3-keto steroid reductase [EC:1.1.1.270] (ERG27) | - Biosynthesis of secondary metabolites |
| 3 | XP_028889502.1 | K13830 | pentafunctional AROM polypeptide [EC:4.2.3.4 4.2.1.10 1.1.1.25 2.7.1.71 2.5.1.19] (ARO1) | -Biosynthesis of various plant secondary metabolites |
| -Biosynthesis of secondary metabolites | ||||
| 4 | XP_028889622.1 | K00697 | trehalose 6-phosphate synthase [EC:2.4.1.15 2.4.1.347] (otsA) | - Biosynthesis of secondary metabolites |
| 5 | XP_028889979.1 | K01610 | phosphoenolpyruvate carboxykinase (ATP) [EC:4.1.1.49] (E4.1.1.49, pckA) | - Biosynthesis of secondary metabolites |
| 6 | XP_028890017.1 | K00641 | homoserine O-acetyltransferase/O-succinyltransferase [EC:2.3.1.31 2.3.1.46] (metX) | - Biosynthesis of secondary metabolites |
| 7 | XP_028890156.1 | K13507 | glycerol-3-phosphate O-acyltransferase/dihydroxyacetone phosphate acyltransferase [EC:2.3.1.15 2.3.1.42] (GAT) | -Biosynthesis of secondary metabolites |
| 8 | XP_028892424.1 | K01687 | dihydroxy-acid dehydratase [EC:4.2.1.9] (ilvD) | -Biosynthesis of secondary metabolites |
| 9 | XP_028890656.1 | K01694 | tryptophan synthase [EC:4.2.1.20] (TRP) | - Biosynthesis of secondary metabolites |
| 10 | XP_028890742.1 | K00053 | ketol-acid reductoisomerase [EC:1.1.1.86] (ilvC) | - Biosynthesis of secondary metabolites |
| 11 | XP_028891161.1 | K01663 | imidazole glycerol-phosphate synthase [EC:4.3.2.10] (HIS7) | -Biosynthesis of secondary metabolites |
| 12 | XP_028891194.1 | K01656 | anthranilate synthase/indole-3-glycerol phosphate synthase [EC:4.1.3.27 4.1.1.48] (TRP3) | -Biosynthesis of secondary metabolites |
| 13 | XP_028893153.1 | K16055 | trehalose 6-phosphate synthase/phosphatase [EC:2.4.1.15 3.1.3.12] (TPS) | -Biosynthesis of secondary metabolites |
| 14 | XP_028893355.1 | K00706 | 1,3-beta-glucan synthase [EC:2.4.1.34] (E2.4.1.34) | -MAPK signaling pathway - yeast |
| 15 | XP_028893411.1 | K00706 | 1,3-beta-glucan synthase [EC:2.4.1.34] (E2.4.1.34) | -MAPK signaling pathway - yeast |
| 16 | XP_028891587.1 | K01497 | GTP cyclohydrolase II [EC:3.5.4.25] (ribA, RIB1) | -Biosynthesis of secondary metabolites |
| 17 | XP_028891672.1 | K13507 | glycerol-3-phosphate O-acyltransferase/dihydroxyacetone phosphate acyltransferase [EC:2.3.1.15 2.3.1.42] (GAT) | -Biosynthesis of secondary metabolites |
| 18 | XP_028891801.1 | K00872 | homoserine kinase [EC:2.7.1.39] (thrB) | -Biosynthesis of secondary metabolites |
| 19 | XP_028891858.1 | K00938 | phosphomevalonate kinase [EC:2.7.4.2] (E2.7.4.2, mvaK2) | -Biosynthesis of secondary metabolites |
3.4. Human microbiome non-homology analysis and drug bank result
Microbiome analysis revealed that 6 pathogen proteins (Supplementary file 5) shared less than 45% similarity with human microflora, three of which shared similarity with approved, investigational, and experimental drug targets of Drugbank Database (Supplementary file 6). We ended up taking these three proteins off the list because we aimed to find novel targets. Three proteins did not show any resemblance to being novel targets. As a result, they were nominated (Table 2) as the unique drug targets of the pathogen.
Table 2.
Unique drug targets for C. auris.
| Sl No | Accession number | Subcellular Location |
|---|---|---|
| 01 | XP_028890156.1 | PlasmaMembrane |
| 02 | XP_028891672.1 | PlasmaMembrane |
| 03 | XP_028891858.1 | Cytoplasmic |
3.5. Identification of subcellular location
The selected proteins were found to be located either in the cytoplasm or the inner membrane. XP 028890156.1 and XP 028891672.1 are plasma membrane proteins, whereas XP 028891858.1 is located in the cytoplasm (Table 2). Both cytoplasmic and membrane proteins are potential therapeutic targets, therefore we retained all of them for additional research.
3.6. Protein-protein interaction network analysis
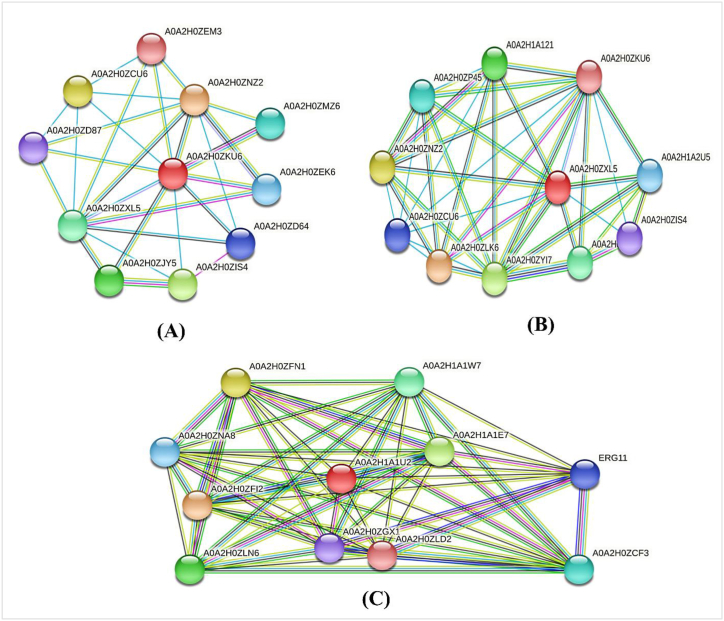
Proteins that interact with multiple proteins are thought to be metabolically active and they are suitable as a drug target [37,67]. STRING revealed that XP_028890156.1, XP_028891672.1 and XP_028891858.1 each exhibit interactions with 10 proteins as shown in Fig. 2(A–C).
Fig. 2.
Interaction of (A) XP_028890156.1, (B) XP_028891672.1 and (C) XP_028891858.1 with other proteins where red color balls represent the target proteins. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. Molecular modelling, quality assessment and binding site prediction
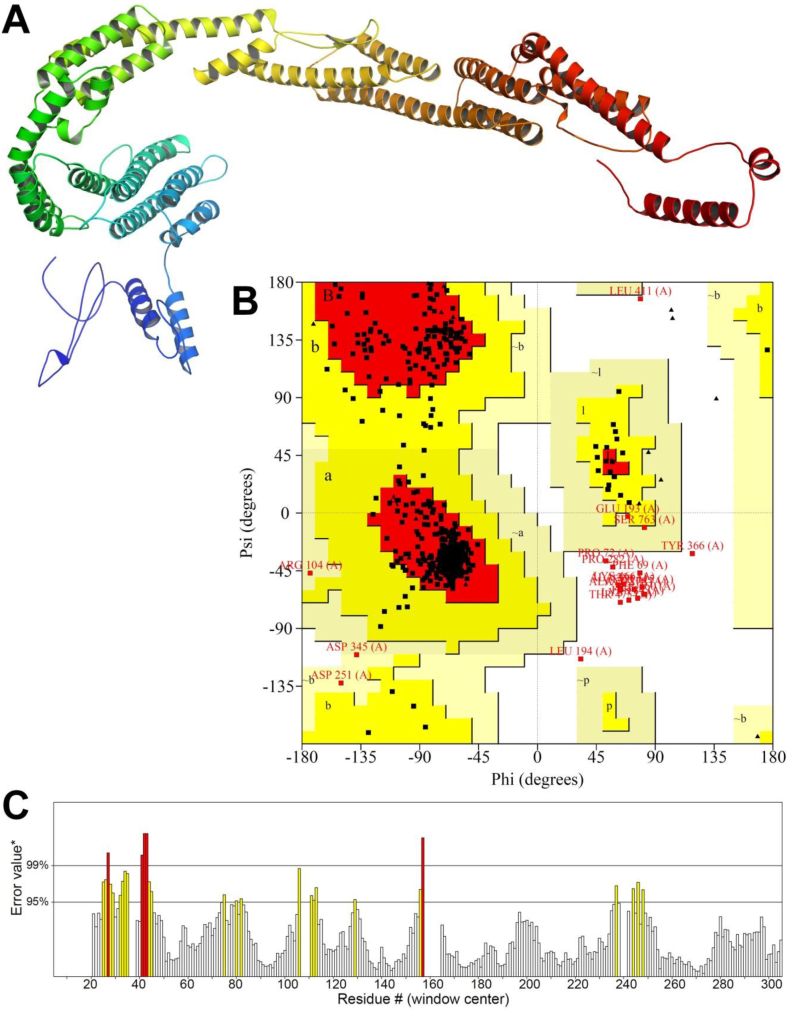
I-TASSER server uses multiple threading approaches to recognize the structural templates from the PDB. Then the full-length structure models are developed by iterative fragment assembly simulations. The functional insights are finally derived by matching the predicted structure models with known proteins in the function databases [68]. For each protein, there were 5 models predicted. Following the assessment of the ERRAT value and Ramachandan plot, the top candidate was then refined using the GalaxyWEB server. For each protein, this server also generated 5 improved models. By examining their ERRAT quality score and Ramachandan plot analysis, we ultimately chose the best 3D model. Table 3, Fig. 3, Fig. 4, Fig. 5 demonstrate the ERRAT value, the Ramachandan plot result, and the binding sites residues predicted by RaptorX Binding site server.
Table 3.
Refined protein model with their ERRAT value, Ramachandan plot result and binding sites residues.
| Protein | Errat value | Ramachandan plot result |
Ligand | Predicted ligand binding sites residues | |
|---|---|---|---|---|---|
| Favored region | Disallowed region | ||||
| >XP_028890156.1 | 92.6273 | 90.4% | 1.8% | PLM | L71 V80 A82 H84 D90 P91 L94 M95 I106 F108 I110 S124 G242 F244 C283 I299 |
| GOL | F88 K113 S114 G144 K145 G248 S249 D251 | ||||
| >XP_028891672.1 | 95.2035 | 91.4% | 1.4% | GOL | F80 G104 K105 S106 L168 G251 S252 D254 |
| PLM | S57 V72 A74 H76 D82 P83 V86 M87 I98 L100 T102 A116 G245 F247 G250 V304 | ||||
| >XP_028891858.1 | 78.2297 | 79.7% core | 2.4% | MG | K10 M35 S53 Q55 S80 F83 L107 S109 K143 G145 M146 G147 S148 S149 L152 Q188 T265 |
| GLA | K10 L12 G16 Y17 V19 I20 H184 G192 S193 F195 D196 T265 V269 G373 A374 | ||||
Fig. 3.
(A) 3D model (B) Ramachandan plot and (C) Errat quality value of XP_028890156.1.
Fig. 4.
(A) 3D model (B) Ramachandan plot and (C) Errat quality value of XP_028891672.1.
Fig. 5.
(A) 3D model (B) Ramachandan plot and (C) Errat quality value of XP_028891858.1.
3.8. Molecular docking and binding site analysis
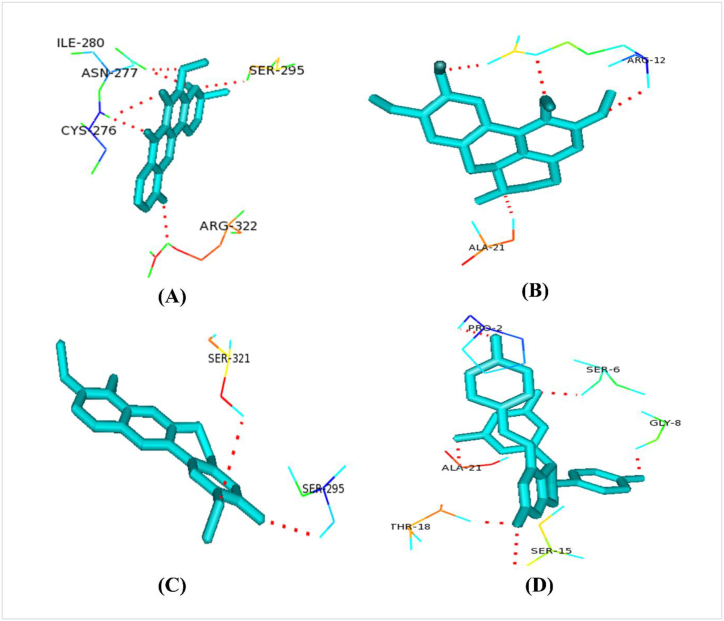
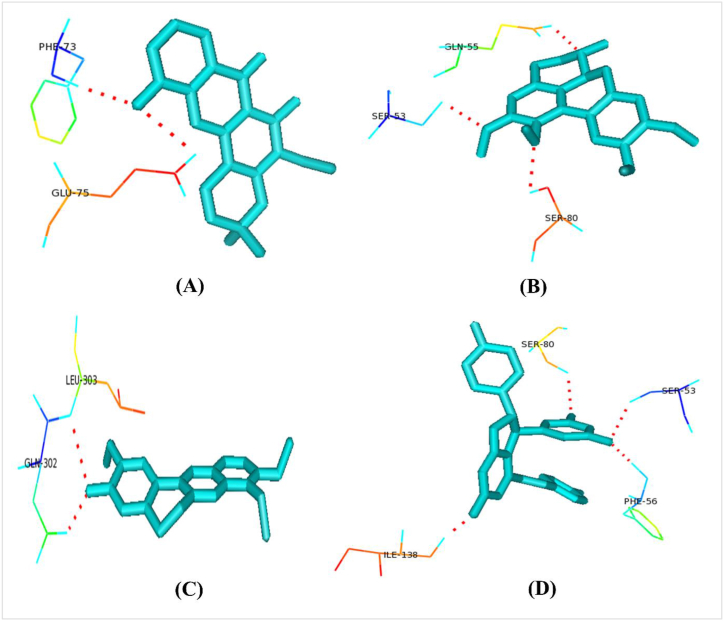
A list of collected plant metabolites is given in supplementary file 7. All of our selected unique drug targets (macromolecules) docked against all plant metabolites (ligands) (Supplementary file 8). Caledonixanthone E, Viniferin, Glaucine and Jatrorrhizine docked superiorly with the selected drug targets than that of Fluconazole. Against all the 3 proteins, all these four metabolites docked with lower binding energy than Fluconazole (Table 4). Therefore, we further analyzed the binding residues of those metabolites using the Pymol tool and enlisted in Table 4, Fig. 6, Fig. 7, Fig. 8.
Table 4.
Binding sites of Caledonixanthone E, Viniferin, Glaucine, Jatrorrhizine.
| Protein ID | Metabolites Name | Global Binding Energy (kcal/mol) | Polar Binding Residues |
|---|---|---|---|
| >XP_028890156.1 | Caledonixanthone E | −44.89 | CYS276,ASN277,ILE280,SER295,ARG322 |
| Viniferin | −51.99 | PRO2,SER6,GLY8,SER15,THR18,ALA21 | |
| Glaucine | −41.94 | ARG12,ALA21 | |
| Jatrorrhizine | −44.49 | SER295,SER321 | |
| Fluconazole | −41.15 | ||
| >XP_028891672.1 | Caledonixanthone E | −39.57 | LEU9,ASP12 |
| Viniferin | −48.28 | GLN11,ASP12,GLU125,ARG126 | |
| Glaucine | −43.88 | SER16,TYR18 | |
| Jatrorrhizine | −39.89 | GLU26 | |
| Fluconazole | −34.73 | ||
| >XP_028891858.1 | Caledonixanthone E | −33.76 | PHE73,GLU75 |
| Viniferin | −35.36 | SER53,PHE56,SER80,ILE138 | |
| Glaucine | −28.97 | SER53,GLN55, SER80 | |
| Jatrorrhizine | −31.94 | GLN302,LEU303 | |
| Fluconazole | −28.84 |
Fig. 6.
Polar binding residues of >XP_028890156.1with (A) Caledonixanthone E, (B) Glaucine, (C) Jatrorrhizine and (D) Viniferin.
Fig. 7.
Polar binding residues of >XP_028891858.1 with (A) Caledonixanthone E, (B) Glaucine, (C) Jatrorrhizine and (D) Viniferin.
Fig. 8.
Polar binding residues of >XP_028890156.1 with (A) Caledonixanthone E, (B) Glaucine, (C) Jatrorrhizine and (D) Viniferin.
3.9. Molecular dynamics simulation studies and MM-PBSA calculations
MD simulations of target proteins and ligand bound protein complexes for the top three ligands were undertaken. Specifically, in the case of XP_028890156.1 protein a bare protein and the complexes with the ligands Caledonixanthone E, Jatrorrhizine and Viniferin were taken for MD simulations. Similarly, in the case of XP_028891858.1 same ligands bound to it was taken up for MD simulations. While in the case of XP_028891672 the complex of Glaucine along with complexes of Jatrorrhizine and Viniferin were taken..
The results of RMSD in backbone atoms of XP_028890156.1 bare protein and ligand bound XP_028890156.1 complexes showed that the Viniferin bound XP_028890156.1 having the larger magnitude of RMSD with an average around 2.5 nm (Fig. 9(A–C). The Jatrorrhizine bound XP_028890156.1 has the lowest RMSD which is stabilized after around 20 ns with an average of around 1 nm. The Caledonixanthone E bound XP_028890156.1 is also having a lower RMSD compared to the RMSD of bare protein with an average of around 1.2 nm. The results of RMSD XP_028890156.1 backbone atoms corroborate with the lower RMSD in atoms of Jatrorrhizine and Caledonixanthone E and slightly higher RMSD in Viniferin. On the other hand, the RMSD in backbone atoms of XP_028891672.1 showed that Viniferin bound XP_028891672.1 has the lower RMSD which is stabilized after around 30 ns to an average of 1.5 nm. The Jatrorrhizine bound XP_028891672.1 has almost similar RMSD as that of bare protein with an average around 1.75 nm, while Glaucine bound XP_028891672.1 has a slightly larger magnitude of RMSD which apparently deviated till the end of MD simulation. However, the RMSD in atoms of Viniferin was seen slightly deviated during 30–40 ns simulation period. The RMSD in atoms of Jatrorrhizine is the lowest with an average of around 0.75 nm. The Jatrorrhizine bound XP_028891858.1 has the lowest RMSD with an average of around 0.41 nm which is stabilized after around 10 ns. The XP_028891858.1 bare protein, Viniferin and Caledonixanthone bound XP_028891858.1 stabilizes after around 25 ns with slightly higher RMSD. Similarly, the RMSD in atoms of Viniferin showed slightly higher RMSD compared to RMSD in atoms of Jatrorrhizine and Caledonixanthone E.
Fig. 9.
RMSD in protein backbone atoms and ligand atoms (A) XP_028890156.1 system, (B) XP_028891672.1, and (C) XP_028891858.1 system.
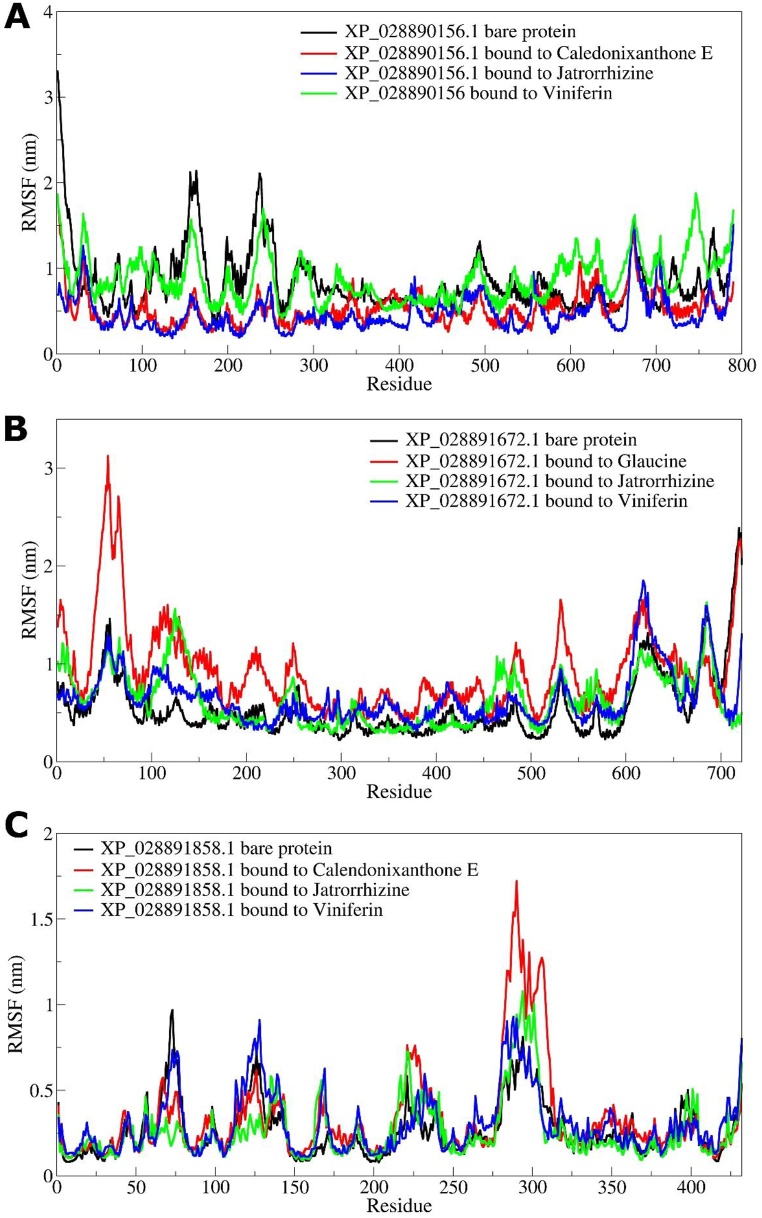
The results of RMSF analysis showed that XP_028890156.1 the bare protein having larger magnitude of fluctuations in the side chain atoms of residues in the range 150–250 (Fig. 10(A–C)). The Viniferin bound XP_028890156.1 also showed larger fluctuations however in a slightly extended region of residues in the range 100–250. The Jatrorrhizine and Caledonixanthone E bound XP_028890156.1 showed lower RMSF compared to the former two systems. In the case of XP_028891672.1 systems, the RMSF in side chain atoms of bare protein and Viniferin bound XP_028891672.1 has the lower RMSF with slight fluctuations in terminal residues in the range 0–100 and 600–720. The Glaucine bound XP_028891672.1 showed the largest deviations in the residues in the range 25–100, while Viniferin bound XP_028891672.1 showed larger deviations in the residues 50–150. The RMSF in XP_028891858.1 is almost similar in all the systems. However, the major fluctuations were seen in the residues 275–325 having larger magnitude with Caledonixanthone E bound XP_028891858.1 residues. The XP_028891858.1 bare protein residues showed the lowest RMSF.
Fig. 10.
RMSF analysis in side chain atoms of residues of (A) XP_028890156.1, (B) XP_028891672.1, and (C) XP_028891858.1 proteins in bare and bound form.
The hydrogen bonds formed over the simulation period between the target protein and respective ligands were analyzed. The results of hydrogen bonds formed between XP_028890156.1 and Caledonixanthone E showed a consistent 1 hydrogen bond being formed with residue Asp273 (Fig. 11(A–C)). Maximum 3 hydrogen bonds were seen during the simulation period 65–70 ns with residues Arg292 and Phe288. Jatrorrhizine scarcely formed one and maximum 4 hydrogen bonds with XP_028890156.1 where no hydrogen bonds were seen formed during the simulation period 20 ns–60 ns? Viniferin, on the other hand, formed more number of hydrogen bonds where two hydrogen bonds with residues Arg12 and Ile5 were seen consistently formed reaching maximum 3 in few trajectories. In the case of XP_028891672.1 complexes, Viniferin formed maximum 4 consistent hydrogen bonds with residues Ala30, Leu10, Ala15 and Met34. Jatrorrhizine occasionally formed one hydrogen bond, while Glaucine scarcely formed hydrogen bonds with XP_028891672.1. In the case of XP_028891858.1 complexes Caledonixanthone E formed two consistent hydrogen bonds throughout the simulation with Lys189 and Glu186 residues. Similarly, Viniferin formed around 3 hydrogen bonds consistently with Ser-53, Phe-56 and Asn-81 residues and on many occasions 4 hydrogen bonds with Ser-53, Phe-56, Asn-81 and Gln135 residues. Jatrorrhizine forms occasional one hydrogen bond with Glu61 residue.
Fig. 11.
Hydrogen bond analysis (A) XP_028890156.1 system, (B) XP_028891672.1 system, and (C) XP_028891858.1 system with Caledonixanthone E (red color), Glaucine (orange color) Jatrorrhizine (blue color) and viniferin (green color). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Further, in order to identify the stable lowest energy states of complexes and to have insights into the path of motion the principal component analysis and Gibb's free energy analysis were performed as given in Fig. 12(A–L). Here, the eigenvectors corresponding to each principal component give insights into the path of the motion along with the energy states of that system. Gibb's free energy analysis was performed with the first two principal components (PC1 and PC2) where the lower the value of Gibb's free energy better is the stability of the corresponding system. In the case of XP_028890156.1 systems, the bare protein showed a large number of conformations in the energy basin 20 to −60 kJ mol−1 on PC1 and 20 to −30 kJ.mol−1on PC2 (Fig. 12(A–L)). The Caledonixanthone bound system showed the lowest energy state conformations in the energy basin 0–10 kJ mol−1 on PC1 and 0 to −5 kJ mol−1 on PC2. A very small number of low energy conformations were observed for Jatrorrhizine bound system with the energy around 20 kJ mol−1 on PC1 and 0 to −20 kJ mol−1 on PC2. The largest low energy stable conformations were observed for Viniferin bound to XP_028890156.1 complex in the large energy basin 0 to −25 kJ mol−1 on PC1 and 10 to −20 kJ mol−1 on PC2. In the case of XP_028891672.1 systems, the bare protein showed low energy conformations in an energy basin 15 to −10 kJ mol−1 on PC1 and 15 to −10 kJ mol−1 on PC2. The Glaucine complex showed many low energy conformations with apparently overall low system energy. Jatrorrhizine complex showed a large energy basin with low energy conformation in the range 0 to −15 kJ mol−1 on PC1 and 15 to −10 kJ mol−1 on PC2. The Viniferin complex also showed a unique energy basin with low energy conformations in the range 10 to −10 kJ mol−1 on PC1 and PC2. In the case of XP_028891858.1 systems the bare protein showed a small low energy basin with few conformations in the range 4–5 kJ mol−1 on PC1 and 0 to −5 kJ mol−1 on PC2. The Caledonixanthone E complex showed the lowest energy conformations in the energy basin −2.5 to −7.5 kJ mol−1 on PC1 and 0 to −2 kJ mol−1 on PC2. A very small number of conformations of Jatrorrhizine complex was seen occupying low energy basin around −10 kJ mol−1 on PC1 and around 0 to −10 kJ mol−1 on PC2. Two distinct energy basins were observed for Viniferin complex amongst which the energy basin with energy −5 to −6 kJ mol−1 on PC1 and 2 to −2 kJ mol−1 being the largest and with the lowest energy.
Fig. 12.
Gibb's free energy landscape (A) XP_028890156.1bare protein, (B) XP_028890156.1-Caledonixanthone E complex, (C) XP_028890156.1-Jatrorrhizine complex, (D) XP_028890156.1-Viniferin complex, (E) XP_028891672.1 bare protein, (F) XP_028891672.1-Glaucine complex, (G) XP_028891672.1-Jatrorrhizine complex, (H) XP_028891672.1-Viniferin complex, (I) XP_028891858.1 bare protein, (J) XP_028891858.1-Caledonixanthone E complex, (K) XP_028891858.1-Jatrorrhizine complex, and (L) XP_028891858.1-Viniferin complex.
Further, the binding free energies of ligands bound to XP_028890156.1, XP_028891672.1 and XP_028891858.1 were obtained through MM-PBSA calculations. Particularly, various interaction energies such as van der Waal energy, electrostatic energy, polar solvation energy, SASA energy were calculated to obtain final binding free energy (ΔGbinding) between the protein and ligand. During MM-PBSA calculations the trajectories isolated at each 1 ns simulation step from the reasonably stable simulation period (75 ns–100 ns) were employed. The detail results of MM-PBSA calculations are given in Table 5. The results show that the XP_028890156.1 bound to Caledonixanthone E having the most favorable binding free energy (−103.015 ± 8.312 kJ mol−1), compared to Jatrorrhizine and Viniferin complexes. In the case of XP_028891672.1 complexes the Glaucine has lowest binding free energy of −78.537 ± 3.618 kJ mol−1. Other two ligands, Jatrorrhizine and Viniferin have comparatively higher binding free energies of −25.206 ± 8.493 and −59.571 ± 3.927 kJ mol−1, respectively. In the case of XP_028891858.1 complex with Viniferin, Viniferin has the lowest binding free energy of −128.250 ± 8.558 kJ mol−1, compared to Caledonixanthone E and Jatrorrhizine.
Table 5.
Results of MM-PBSA calculations.
| van der Waal energy (kJ mol−1) | Electrostatic energy (kJ mol−1) | Polar solvation energy (kJ mol−1) | Solvent accessible surface area energy (kJ mol−1) | Binding energy (ΔGbinding) (kJ mol−1) | |
|---|---|---|---|---|---|
| XP_028890156.1 systems | |||||
| Caledonixanthone E | −166.739 (8.663) | −4.074 (1.128) | 85.197 (3.114) | −18.418 (1.003) | −103.015 (8.312) |
| Jatrorrhizine | −107.026 (3.375) | −4.129 (1.208) | 91.060 (7.524) | −16.758 (7.161) | −36.098 (3.428) |
| Viniferin | −108.862 (2.851) | −4.142 (1.165) | 102.055 (3.542) | 0.213 (0.223) | −13.561 (2.541) |
| XP_028891672.1 systems | |||||
| Glaucine | −103.096 (5.158) | −10.747 (4.783) | 48.966 (5.910) | −13.808 (0.482) | −78.537 (3.618) |
| Jatrorrhizine | −99.955 (5.798) | −7.097 (8.199) | 94.626 (12.960) | −12.312 (0.701) | −25.206 (8.493) |
| Viniferin | −186.191 (12.860) | −28.795 (3.377) | 174.697 (0.651) | −21.154 (1.222) | −59.571 (3.927) |
| XP_028891858.1 systems | |||||
| Caledonixanthone E | −184.030 (2.563) | −23.914 (0.834) | 123.448 (3.282) | −17.604 (0.336) | −101.658 (0.864) |
| Jatrorrhizine | −73.723 (6.001) | −57.541 (2.756) | 54.951 (5.226) | −8.367 (0.295) | −84.886 (3.808) |
| Viniferin | −186.175 (7.123) | −8.871 (3.867) | 88.356 (5.032) | −21.403 (0.588) | −128.250 (8.558) |
Standard deviations are given in parentheses.
3.10. Pharmacoinformatics studies
ADME properties of top medications were assessed in order to define the pharmacologic profiles of the selected top antifungal drugs (Table 6). All of the metabolites had a high rate of absorption in the gastrointestinal tract and no BBB was found in cases of Caledonixanthone E and Viniferin. Apart from Viniferin, all of the medications had a modest amount of water solubility, and none of the metabolites suggested that they would cause pain (Table 6).
Table 6.
SwissADME properties of top metabolites.
| Parameter | Metabolites |
||||
|---|---|---|---|---|---|
| Caledonixanthone E | Viniferin | Glaucine | Jatrorrhizine | ||
| Physicochemical parameters | Formula | C19H16O6 | C28H22O6 | C21H25NO4 | C20H20NO4 |
| Molecular weight gm/mole | 340.33 | 454.47 | 355.43 | 338.38 | |
| Num. Heavy atoms | 25 | 34 | 26 | 25 | |
| Num. H-bond acceptors | 6 | 6 | 5 | 4 | |
| Num. H-bond donors | 2 | 5 | 0 | 1 | |
| Molar Refractivity | 94.65 | 130.24 | 104.94 | 97.33 | |
| Lipophilicity | LogPo/w(iLOGP) | 2.96 | 2.41 | 3.72 | −0.15 |
| LogPo/w(XLOGP3) | 3.7 | 5.4 | 3.37 | 3.42 | |
| LogPo/w(WLOGP) | 3.44 | 5.11 | 2.77 | 3.08 | |
| Consensus LogPo/w | 2.93 | 4.05 | 3.22 | 2.31 | |
| Pharmacokinetics | GI absorption | High | High | High | High |
| BBB permeant | No | No | Yes | Yes | |
| CYP1A2 inhibitor | Yes | No | No | Yes | |
| CYP2C19 inhibitor | No | No | No | No | |
| CYP2C9 inhibitor | Yes | Yes | No | No | |
| CYP2D6 inhibitor | Yes | No | Yes | Yes | |
| CYP3A4 inhibitor | Yes | No | Yes | Yes | |
| LogKp(skin permeation) (cm/s) | −5.75 | −5.24 | −6.08 | −5.94 | |
| Water solubility | LogS(ESOL) | −4.63 | −6.32 | −4.24 | −4.37 |
| Solubility (mg/ml) | 7.99E-03 | 2.18E-04 | 2.03E-02 | 1.45E-02 | |
| Solubility (mol/l) | 2.35E-05 | 4.81E-07 | 5.70E-05 | 4.28E-05 | |
| Class | Moderately soluble | Poorly soluble | Moderately soluble | Moderately soluble | |
| Medicinal chemistry | PAINS | 0 alert | 0 alert | 0 alert | 0 alert |
| Brenk | 1 alert: polycyclic_aromatic_hydrocarbon_2 | 1 alert: stilbene | 0 alert | 1 alert: quaternary_nitrogen_1 | |
| Leadlikeness | No; 1 violation: XLOGP3>3.5 | No; 2 violations: MW.350, XLOGP3.3.5 | No; 1 violation: MW>350 | yes | |
| Synthetic accessibility | 3.85 | 4.45 | 3.8 | 3.06 | |
3.11. Toxicity analysis of the selected metabolites
The toxicity profiles of top metabolites are enlisted in Table 7. The LD50 values for the top drugs ranged from 2.131 to 3.219 mol/kg and they displayed negative skin sensitization results and oral rat acute toxicity. Minnow Toxicity values of all drugs were more than −0.3 log mM, proving them non-toxic. Again, negative hepatotoxicity results, except for Jatrorrhizine, of all drugs indicate that the normal function of the liver will not be disrupted through these top drugs.
Table 7.
Toxicity parameter of selected metabolites.
| Toxicity parameter | Metabolites name |
|||
|---|---|---|---|---|
| Caledonixanthone E | Viniferin | Glaucine | Jatrorrhizine | |
| AMES Toxicity | Yes | Yes | No | No |
| Max. Tolerated Dose (log mg/kg/day) | 0.339 | 0.304 | 0.167 | −0.468 |
| hERG I inhibitor | No | No | No | No |
| hERG II inhibitors | Yes | Yes | Yes | Yes |
| Oral Rat Acute Toxicity, LD50 (mol/kg) | 2.131 | 2.441 | 3.219 | 3.089 |
| Oral Rat Chronic Toxicity, LOAEL (log mg/kg_bw/day) | 1.589 | 1.579 | 1.527 | 1.146 |
| Hepatotoxicity | No | No | No | Yes |
| Skin Sensitization | No | No | No | No |
| Minnow Toxicity (log mM) | 0.158 | 0.861 | 0.038 | 0.453 |
4. Discussion
Candida auris is a pathogenic yeast that has developed as a multidrug-resistant organism. In many countries, community health experts have identified that C. auris has affected hospitalized patients with serious illness. Diseases commenced by C. auris frequently do not react to regularly consuming antifungal medications, rendering them tough to cure effectively. Hence, we investigated the proteome of this yeast to anticipate the prospective drug targets and drugs to deal with this pathogenic yeast.
The paralog proteins (60% matching) were also excluded from the total representative proteome of C. auris since their motifs, domains, active sites, and other characteristics might be similar to those of paralogs. Since most therapeutics tends to bind with essential gene products, essential proteins are considered to be among the most effective drug targets. A prospective drug target has to be an essential protein as it has a vital attribute for the existence of the pathogen [66] and our target pathogen yeast contains 795 such essential proteins specifying that drugs will act out in opposition to those proteins will lead the pathogen to death. Subtraction of host-homolog proteins is regarded as the notable step of in-silico drug target determination [66,69]. Following the investigation, 66 non-orthologs were discovered, which can be targeted to develop new medications that might not cause a cross reactivity [66]. Fifty-two (52) proteins among those non-orthologs were predicted to be involved in yeast metabolism, however only 19 of them were shown to be exclusively involved in the distinctive metabolic pathways of C. auris (Table 1). Our digestive system is home to a large and diverse population of microbes, and if that “gut microbiome” is well populated with “good guy” strains, we enjoy several health benefits, including improved digestion, enhanced brain function, and increased immunological response. The protection of those beneficial microorganisms from unwanted blockage is confirmed by the assessment of host microbiome non-similarity. As a result, 13 unique metabolic pathway proteins that exhibited more alignment with the helpful bacteria were eliminated. The Drug-Bank database was inspected for evading mutational alterations as well as to prevent the progress of study bacteria by the broad-spectrum therapeutic drugs. Three of the six non-similar microbiome proteins were druggable and may be turned inactive by commercially available and approved medications. Thus, the remaining three proteins were kept as the novel therapeutic targets (Table 2). Although proteins are found to be located in five diverse sites of the cells, membrane and cytoplasmic one is regarded as the elite group for drug targets [70]. Each of the novel proteins was found to be located either in the cytoplasm or in the membrane showing their capability to act as a drug target (Table 2). Protein-protein interactions (PPIs) for all the distinctive proteins were discovered (Fig. 2(A–C)) as it allocates us to comprehend the function of a protein in a precise pathway and changes in the protein-protein network interrupt the regular flow of activities of the cell [71]. The discovery of interactions between all selected three proteins with ten other proteins demonstrates the unintended function interruption of those ten proteins caused by the blocking of the drug targets.
By serving as a key molecule in the discovery of suitable therapeutic candidates, plant metabolites play a vital role [72]. Thus, based on their potential to bind to specific therapeutic targets, a few inhibitory metabolites from plant sources were taken into consideration for C. auris. The results of the docking have demonstrated that four therapeutic molecules Caledonixanthone E, Viniferin, Glaucine, and Jatrorrhizine had strong affinity for all three proteins with the lowest global binding energy. Among them, Viniferin exhibited maximum binding affinity with each of the three drug targets (Table 4). The structural configurations of the docked protein complex were scrutinized to interpret the exterior drug hotspot of the targeted distinctive proteins. The binding arrangement of ligands and interrelating residues with their corresponding locations were inspected (Table 4) (Fig. 6, Fig. 7, Fig. 8). This inspection leads to the finding of the amino acids from 02 to 21 and 276–322 positions that were fundamental to the binding interactions of >XP_028890156.1. However, residues 9–26 were discovered as essential for the binding of >XP_028891672.1, the binding hotspot of ligands for protein > XP_028891858.1 that lies between 53 and 80 positions.
The RMSD analysis of protein backbone atoms and ligand atoms of XP_028890156.1 systems suggest that the Caledonixanthone E and Jatrorrhizine in bound form considerably stabilized the conformation of XP_028890156.1 (Fig. 9(A)). However, the XP_028890156.1 complex with Viniferin deviates considerably from the equilibrated state signifying unfavorable affinity of Viniferin towards XP_028890156.1. The RMSD in backbone atoms of XP_028891672.1 which is reasonably stabilized after around 25 ns in all the systems suggested that Jatrorrhizine and Viniferin stabilize to a stable conformation of XP_028891672.1 compared to Glaucine. In the case of XP_028891858.1 systems, the RMSD analysis suggests that Jatrorrhizine is the best ligand compared to Caledonixanthone E and Viniferin.
The RMSF analysis of XP_028890156.1 protein with 790 residues revealed that the binding site residues in the range 200–300 undergo major fluctuations (Fig. 10(A)). Viniferin causing the major fluctuations in the binding site residues may be slightly more unfavorable than Caledonixanthone E and Jatrorrhizine. Similarly, the complexes of XP_028891672.1 showed that the binding site residues in the range 25–125 undergo major fluctuations in Glaucine bound state signifying its unfavorable interactions. However, Jatrorrhizine and Viniferin stabilize the XP_028891672.1 as evidenced by the lower fluctuations in the corresponding binding site residues. The XP_028891858.1 complexes with Jatrorrhizine and Viniferin are comparably seen as more favorable and stable as there are lower fluctuations in the binding site residues in the range 275–325. Notably, Caledonixanthone E caused major fluctuation in the binding site residues when bound to XP_028891858.1 suggesting its unfavorable interactions at the binding site.
More the number of non-bonded interactions such as hydrogen bonds between protein and ligand, better is the binding affinity conferred to the ligand with consequent stability of protein-ligand complex. Caledonixanthone E and Viniferin formed consistent hydrogen bonds with XP_028890156.1 protein, where Viniferin formed slightly more number of hydrogen bonds compared to Caledonixanthone E (Fig. 11(A)). Since hydrogen bonds are continuously formed throughout the MD simulations, it is evident that these ligands remained in the binding pocket throughout the simulation. However, Jatrorrhizine showing no hydrogen bonds during the 20–60 ns simulation period showed it moved out of the binding pocket signifying its less favorable binding affinity. The corresponding RMSD evaluation for Jatrorrhizine bound XP_028890156.1 protein suggested lower RMSD which suggests XP_028890156.1 protein stabilizes without Jatrorrhizine in the binding pocket. However, the results of RMSD and RMSF corroborate well for Caledonixanthone E which augments that Caledonixanthone E could be the best ligand for XP_028890156.1 protein. In the case of XP_02889672.1 protein only Viniferin showed consistent hydrogen bonds, and this result corroborates well with the results of RMSD and RMSF evaluation. While Glaucine and Jatrorrhizine showed occasional hydrogen bonds and Glaucine is mostly seen coming out of binding pocket in visual inspection of its trajectories. In the case of XP_028891858.1 protein the ligands Caledonixanthone E and Viniferin have more favorable binding affinity as they show consistent and more number of hydrogen bonds. Further, the RMSD and RMSF analysis also supports the favorable binding affinity of these ligands. However, Jatrorrhizine which showed occasional hydrogen bonds may have a slightly unfavorable binding affinity.
The results of PC analysis and Gibb's free energy analysis suggest complexes of XP_028890156.1 protein have more number of low energy stable conformations for Caledonixanthone E and Viniferin than Jatrorrhizine (Fig. 12(A–L)). Similarly, Viniferin and Jatrorrhizine complexes with XP_028890156.1 protein having more number of low energy stable conformations may have more favorable binding affinity compared to Glaucine. The XP_028891858.1 complexes with Caledonixanthone E and Viniferin with more number of low energy stable conformations may have better binding affinity compared to XP_028891858.1 Jatrorrhizine complex.
The MM-PBSA calculations suggest that the Caledonixanthone E bound to XP_028890156.1 has most favorable binding free energy (−103.015 ± 8.312 kJ mol−1). The lower binding free energy may be due to lowest van der Waal energy and SASA energy of −166.739 ± 8.663 and −18.418 ± 1.003 kJ mol−1. The corresponding stabilization aspects such as RMSD, RMSF, hydrogen bond formation and Rg for Caledonixanthone E bound to XP_028890156.1 is also reasonably good. Glaucine bound to XP_028891672.1 was found having lower polar solvation energy of 48.966 ± 5.910 kJ mol−1, which resulted in lowest binding free energy of −78.537 ± 3.618 kJ mol−1. Although, Viniferin bound to XP_028891672.1 has favorable van der Waal energy, electrostatic energy and SASA energy, the slightly higher polar solvation energy resulted in slightly higher binding free energy (−59.571 ± 3.927 kJ mol-1) than Glaucine. In the case of XP_028891858.1 complexes, Viniferin showed least van der Waal energy, electrostatic energy, polar solvation energy and SASA energy, which resulted in the lowest binding free energy of −128.250 ± 8.558 kJ mol−1. The MM-PBSA studies points out favorable binding energies of Caledonixanthone E, Glaucine, and Viniferin against the XP_028890156.1, XP_028891672.1 and XP_028891858.1, respectively.
Inadequate ADME data is frequently linked with the malfunction of clinical trials in the course of several drug development projects [73]. Thus, ADME data analysis is crucial for those types of projects which can be accomplished by in-vitro or in-vivo or in-silico approaches. The main four drug contenders demonstrated no adverse effects in in-silico ADME study which may perhaps lessen the drug related properties (Table 5). Each of the potential drugs displayed water solubility and absorption in GI. The prediction of toxicity revealed that all of the four drug contenders are non-carcinogenic, non-mutagenic and impervious to skin and non-hepatotoxic. On the whole, the toxicity analysis of the drugs made known that the predicted novel drugs are harmless to conduct operations and can be used as therapeutic medications to treat C. auris.
5. Conclusion
This study of finding suitable drug targets and screening potential drugs will speed up the process to get remedies with fewer trials and error repeats of assays against C. auris. Pathogen protein XP_028890156.1, XP_028891672.1, and XP_028891858.1 were found as suitable drug targets and the metabolites Caledonixanthone E, Viniferin, Glaucine and Jatrorrhizine can be grater alternatives to get rid of this deadly pathogen. Moreover, the results from MD simulations and MM-PBSA calculations suggest that Caledonixanthone E and Viniferin are the best ligands for XP_028890156.1, XP_028891672.1, and XP_028891858.1 proteins. Finally selected proteins and metabolites to block those proteins may be greater foresteps of drug discovery against the emerging pathogen, C. auris. However, in vivo and clinical trials are highly recommended to validate the outcome of the study.
Author contribution statement
Md. Nazmul Islam Bappy, Tanjin Barketullah Robin: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed materials, analysis tools or data; Wrote the paper. Anindita Ash Prome, Rajesh B. Patil, Abu Tayab Moin, Rupali Akter, Fayeza Sadia Laskar, Anindita Roy, Hafsa Akter: Performed the experiments; Analyzed and interpreted the data; Wrote the paper. Kazi Md. Ali Zinnah: Conceived and designed the experiments; Analyzed and interpreted the data; Wrote the paper.
Data availability statement
Data will be made available on request.
Resources that were used in the study
-
1.
National Center for Biotechnology Information Genome database (https://www.ncbi.nlm.nih.gov/genome).
-
2.
CD-HIT server (http://weizhong-lab.ucsd.edu/cdhit_suite/cgi-bin/index. cgi?cmd = cd-hit) (Huang, Niu, Gao, Fu, & Li, 2010)
-
3.
DEG database (www.essentialgene.org/) (Luo, Lin, Gao, Zhang, & Zhang, 2014).
-
4.
KEGG PATHWAY database (https://www.genome.jp/kegg/pathway.html) (Ogata et al., 1999).
-
5.
KO server (KEGG ORTHOLOGY) (https://www.genome.jp/kegg/ko.html) (Damte et al., 2013).
-
6.
DrugBank database (https://www.drugbank.ca/structures/search/bonds/sequence) (Wishart et al., 2018)
-
7.
CELLO v.2.5 server (http://cello.life.nctu.edu.tw/) (Yu, Chen, Lu, & Hwang, 2006).
-
8.
STRING 11.5 server (https://string-db.org/) (Szklarczyk et al., 2019).
-
9.
I-TASSER server (https://zhanggroup.org/I-TASSER/) (Roy, Kucukural, & Zhang, 2010)
-
10.
GalaxyWEB server (http://galaxy.seoklab.org/cgi-bin/submit.cgi?type=REFINE) (Ko, Park, Heo, & Seok, 2012)
-
11.
RaptorX Binding site prediction server (http://raptorx.uchicago.edu/BindingSite/) (Colovos & Yeates, 1993)
-
12.
Open Babel v2.3 software (O'Boyle et al., 2011)
13. PatchDock Server (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) (Schneidman-Duhovny, Inbar, Nussinov, & Wolfson, 2005).
-
14.
PyMOL v2.0 (Wang et al., 2015).
-
15.
SwissParam server (https://www.swissparam.ch/) (Zoete et al., 2011).
-
16.
SwissADME server (http://www.swissadme.ch/) (Daina, Michielin, & Zoete, 2017)
-
17.
pkCSM (http://biosig.unimelb.edu.au/pkcsm/) (Pires, Blundell, & Ascher, 2015)
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17026.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Du H., et al. Candida auris: epidemiology, biology, antifungal resistance, and virulence. PLoS Pathog. 2020;16(10) doi: 10.1371/journal.ppat.1008921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson B.R., et al. On the origins of a species: what might explain the rise of Candida auris? J. Fungi. 2019;5(3) doi: 10.3390/jof5030058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arensman K., et al. Clinical outcomes of patients treated for Candida auris infections in a multisite health system, Illinois, USA. Emerg. Infect. Dis. 2020;26(5):876–880. doi: 10.3201/eid2605.191588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hata D.J., et al. Candida auris: an emerging yeast pathogen posing distinct challenges for laboratory diagnostics, treatment, and infection prevention. Arch. Pathol. Lab Med. 2020;144(1):107–114. doi: 10.5858/arpa.2018-0508-RA. [DOI] [PubMed] [Google Scholar]
- 5.Chow N.A., et al. Tracing the evolutionary history and global expansion of Candida auris using population genomic analyses. mBio. 2020;11(2) doi: 10.1128/mBio.03364-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsberg K., et al. Candida auris: the recent emergence of a multidrug-resistant fungal pathogen. Med. Mycol. 2018;57(1):1–12. doi: 10.1093/mmy/myy054. [DOI] [PubMed] [Google Scholar]
- 7.Welsh R.M., et al. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J. Clin. Microbiol. 2017;55(10):2996–3005. doi: 10.1128/JCM.00921-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saris K., et al. Candida auris. Curr. Opin. Infect. Dis. 2018;31(4) doi: 10.1097/QCO.0000000000000469. [DOI] [PubMed] [Google Scholar]
- 9.Chowdhary, A. and A. Sharma, The lurking scourge of multidrug resistant Candida auris in times of COVID-19 pandemic. J. Global Antimicrob. Resist.. 222020, p. 175-176. [DOI] [PMC free article] [PubMed]
- 10.Lee W.G., et al. First three reported cases of nosocomial fungemia caused by Candida auris. J. Clin. Microbiol. 2011;49(9):3139–3142. doi: 10.1128/JCM.00319-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morales-López S.E., et al. Invasive infections with multidrug-resistant yeast Candida auris, Colombia. Emerg. Infect. Dis. 2017;23(1):162–164. doi: 10.3201/eid2301.161497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarma S., et al. Candidemia caused by amphotericin B and fluconazole resistant Candida auris. Indian J. Med. Microbiol. 2013;31(1):90–91. doi: 10.4103/0255-0857.108746. [DOI] [PubMed] [Google Scholar]
- 13.Vallabhaneni S., et al. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States. Am. J. Transplant. 2017;17(1):296–299. doi: 10.1111/ajt.14121. May 2013-August 2016. [DOI] [PubMed] [Google Scholar]
- 14.Chowdhary A., et al. New clonal strain of Candida auris, Delhi, India. Emerg. Infect. Dis. 2013;19(10):1670–1673. doi: 10.3201/eid1910.130393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clancy C.J., Nguyen M.H. Emergence of Candida auris: an international call to arms. Clin. Infect. Dis. 2017;64(2):141–143. doi: 10.1093/cid/ciw696. [DOI] [PubMed] [Google Scholar]
- 16.Lockhart S.R., et al. Simultaneous emergence of multidrug-resistant Candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin. Infect. Dis. 2016;64(2):134–140. doi: 10.1093/cid/ciw691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fuentefria A.M., et al. Antifungals discovery: an insight into new strategies to combat antifungal resistance. Lett. Appl. Microbiol. 2018;66(1):2–13. doi: 10.1111/lam.12820. [DOI] [PubMed] [Google Scholar]
- 18.Agamah F.E., et al. Computational/in silico methods in drug target and lead prediction. Briefings Bioinf. 2020;21(5):1663–1675. doi: 10.1093/bib/bbz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhong F., et al. Artificial intelligence in drug design. Sci. China Life Sci. 2018;61(10):1191–1204. doi: 10.1007/s11427-018-9342-2. [DOI] [PubMed] [Google Scholar]
- 20.Brogi, S., et al., Editorial: in silico methods for drug design and discovery. Front. Chem.. 82020. [DOI] [PMC free article] [PubMed]
- 21.Lee M., Kim D. Large-scale reverse docking profiles and their applications. BMC Bioinf. 2012;13(17):S6. doi: 10.1186/1471-2105-13-S17-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shital S Chavan M.G.D., Devanand B Shinde, Sangshetti Jaiprakash N. vol. 1. Natural Products in Clinical Trials Bentham Science Publisher; 2018. Antibacterial and Antifungal Drugs from Natural Source: A Review of Clinical Development. [Google Scholar]
- 23.Huang Y., et al. CD-HIT Suite: a web server for clustering and comparing biological sequences. Bioinformatics. 2010;26(5):680–682. doi: 10.1093/bioinformatics/btq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutta A., et al. In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. Silico Biol. 2006;6(1–2):43–47. [PubMed] [Google Scholar]
- 25.Gupta, S., et al., Definition of potential targets in mycoplasma pneumoniae through subtractive genome analysis. J. Antivir. Antiretrovir.. 22010.
- 26.Haag N.L., et al. Potential antibacterial targets in bacterial central metabolism. Int. J. Adv. Life Sci. 2012;4(1–2):21–32. [PMC free article] [PubMed] [Google Scholar]
- 27.Luo H., et al. DEG 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42(Database issue):D574–D580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pourhajibagher M., Bahador A. Designing and in silico analysis of PorB protein from Chlamydia trachomatis for developing a vaccine candidate. Drug Res. 2016;66(9):479–483. doi: 10.1055/s-0042-110319. [DOI] [PubMed] [Google Scholar]
- 29.Ogata H., et al. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27(1):29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Damte D., et al. Putative drug and vaccine target protein identification using comparative genomic analysis of KEGG annotated metabolic pathways of Mycoplasma hyopneumoniae. Genomics. 2013;102(1):47–56. doi: 10.1016/j.ygeno.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 31.Turnbaugh P.J., et al. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Robin, T.B., et al., Identification of novel drug targets and screening potential drugs against Cryptococcus gattii: an in silico approach. Inform. Med. Unlocked. 382023, p. 101222.
- 33.Wishart D.S., et al. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46(D1):D1074–d1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knox C., et al. DrugBank 3.0: a comprehensive resource for 'omics' research on drugs. Nucleic Acids Res. 2011;39(Database issue):D1035–D1041. doi: 10.1093/nar/gkq1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahmud, A., et al., Identification of novel drug targets for humans and potential vaccine targets for cattle by subtractive genomic analysis of Brucella abortus strain 2308. Microb. Pathog.. 1372019, p. 103731. [DOI] [PubMed]
- 36.Yu C.S., et al. Prediction of protein subcellular localization. Proteins. 2006;64(3):643–651. doi: 10.1002/prot.21018. [DOI] [PubMed] [Google Scholar]
- 37.Kushwaha S.K., Shakya M. Protein interaction network analysis--approach for potential drug target identification in Mycobacterium tuberculosis. J. Theor. Biol. 2010;262(2):284–294. doi: 10.1016/j.jtbi.2009.09.029. [DOI] [PubMed] [Google Scholar]
- 38.Szklarczyk D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roy A., et al. I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko J., et al. GalaxyWEB server for protein structure prediction and refinement. Nucleic Acids Res. 2012;40:W294–W297. doi: 10.1093/nar/gks493. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colovos C., Yeates T.O. Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci. 1993;2(9):1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laskowski R.A., et al. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR. 1996;8(4):477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 43.Kim S., et al. PubChem substance and compound databases. Nucleic Acids Res. 2016;44(D1):D1202–D1213. doi: 10.1093/nar/gkv951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O'Boyle, N.M., et al., Open Babel: an open chemical toolbox. J. Cheminf.. 32011, p. 33. [DOI] [PMC free article] [PubMed]
- 45.Kitchen D.B., et al. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat. Rev. Drug Discov. 2004;3(11):935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 46.Schneidman-Duhovny D., et al. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W367. doi: 10.1093/nar/gki481. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meng X.Y., et al. Molecular docking: a powerful approach for structure-based drug discovery. Curr. Comput. Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mashiach E., et al. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:W229–W232. doi: 10.1093/nar/gkn186. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Q., et al., Interaction of α-cyperone with human serum albumin: determination of the binding site by using Discovery Studio and via spectroscopic methods. J. Lumin.. 1642015.
- 50.Berendsen H., van der Spoel D., van Drunen R. Gromacs: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91(1–3):43–56. [Google Scholar]
- 51.Best R.B., et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone φ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theor. Comput. 2012;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vanommeslaeghe K., et al. CHARMM general force field: a force field for drug‐like molecules compatible with the CHARMM all‐atom additive biological force fields. J. Comput. Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu W., et al. Extension of the CHARMM general force field to sulfonyl‐containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012;33(31):2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jorgensen W.L. Optimized intermolecular potential functions for liquid alcohols. J. Phys. Chem. 1986;90(7):1276–1284. [Google Scholar]
- 55.Brand W., et al. Interaction of hesperetin glucuronide conjugates with human BCRP, MRP2 and MRP3 as detected in membrane vesicles of overexpressing baculovirus‐infected Sf9 cells. Biopharm Drug Dispos. 2011;32(9):530–535. doi: 10.1002/bdd.780. [DOI] [PubMed] [Google Scholar]
- 56.Parrinello M., Rahman A. Strain fluctuations and elastic constants. J. Chem. Phys. 1982;76(5):2662–2666. [Google Scholar]
- 57.Hess B., et al. LINCS: a linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18(12):1463–1472. doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 58.Petersen H.G. Accuracy and efficiency of the particle mesh Ewald method. J. Chem. Phys. 1995;103(9):3668–3679. [Google Scholar]
- 59.Maisuradze G.G., et al. Relation between free energy landscapes of proteins and dynamics. J. Chem. Theor. Comput. 2010;6(2):583–595. doi: 10.1021/ct9005745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kumari R., et al. g_mmpbsa A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- 61.Baker N.A., et al. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U. S. A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hay M., et al. Clinical development success rates for investigational drugs. Nat. Biotechnol. 2014;32(1):40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 63.Daina, A., et al., SwissADME: a free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep.. 72017, p. 42717. [DOI] [PMC free article] [PubMed]
- 64.Daina A., Zoete V. A BOILED-egg to predict gastrointestinal absorption and brain penetration of small molecules. ChemMedChem. 2016;11(11):1117–1121. doi: 10.1002/cmdc.201600182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pires D.E., et al. pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J. Med. Chem. 2015;58(9):4066–4072. doi: 10.1021/acs.jmedchem.5b00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sarkar M., et al. In silico quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J. Mol. Model. 2012;18(5):1855–1866. doi: 10.1007/s00894-011-1204-3. [DOI] [PubMed] [Google Scholar]
- 67.Cui, T., et al., Uncovering new signaling proteins and potential drug targets through the interactome analysis of Mycobacterium tuberculosis. BMC Genomics. 102009, p. 118. [DOI] [PMC free article] [PubMed]
- 68.Yang J., Zhang Y. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res. 2015;43(W1):W174–W181. doi: 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Anishetty S., et al. Potential drug targets in Mycobacterium tuberculosis through metabolic pathway analysis. Comput. Biol. Chem. 2005;29(5):368–378. doi: 10.1016/j.compbiolchem.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 70.Michael, C.A., et al., The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health. 22014, p. 145. [DOI] [PMC free article] [PubMed]
- 71.Rabbani G., et al. Protein-protein interactions and their role in various diseases and their prediction techniques. Curr. Protein Pept. Sci. 2018;19(10):948–957. doi: 10.2174/1389203718666170828122927. [DOI] [PubMed] [Google Scholar]
- 72.Joseph J., et al. Molecular Docking of Phytoligands to the viral protein receptor P. monodon Rab7. Bioinformation. 2017;13(4):116–121. doi: 10.6026/97320630013116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shin H.K., et al. In: Handbook of Computational Chemistry. Leszczynski J., editor. Springer Netherlands; Dordrecht: 2016. Predicting ADME properties of chemicals; pp. 1–37. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.