Abstract
In the modern developing society, application of radiation has increased extensively. With significant improvement in the radiation protection practices, exposure to human could be minimized substantially, but cannot be avoided completely. Assessment of exposure is essential for regulatory decision and medical management as applicable. Until now, cytogenetic changes have served as surrogate marker of radiation exposure and have been extensively employed for biological dose estimation of various planned and unplanned exposures. Dicentric Chromosomal Aberration (DCA) is radiation specific and is considered as gold standard, micronucleus is not very specific to radiation and is considered as an alternative method for biodosimetry. In this study dose response curves were generated for X-ray induced “dicentric + ring” and micronuclei, in lymphocytes of three healthy volunteers [2 females (age 22, 23 years) and 1 male (24 year)]. The blood samples were irradiated with X-ray using LINAC (energy 6 MV, dose rate 6 Gy/min), in the dose range of 0–5Gy. Irradiated blood samples were cultured and processed to harvest metaphases, as per standard procedures recommended by International Atomic Energy Agency. Pooled data obtained from all the three volunteers, were in agreement with Poisson distribution for “dicentric + ring”, however over dispersion was observed for micronuclei. Data (“dicentric + ring” and micronuclei) were fitted by linear quadratic model of the expression Y C + αD + βD2 using Dose Estimate software, version 5.2. The data fit has resulted in linear coefficient α = 0.0006 (±0.0068) “dicentric + ring” cell−1 Gy−1 and quadratic coefficient β = 0.0619 (±0.0043) “dicentric + ring” cell−1 Gy−2 for “dicentric + ring” and linear coefficient α = 0.0459 ± (0.0038) micronuclei cell−1 Gy−1 and quadratic coefficient β = 0.0185 ± (0.0010) micronuclei cell−1 Gy−2 for micronuclei, respectively. Background frequencies for “dicentric + ring” and micronuclei were 0.0006 ± 0.0004 and 0.0077 ± 0.0012 cell−1, respectively. Established curves were validated, by reconstructing the doses of 8 dose blinded samples (4 by DCA and 4 by CBMN) using coefficients generated here. Estimated doses were within the variation of 0.9–16% for “dicentric + ring” and 21.7–31.2% for micronuclei respectively. These established curves have potential to be employed for biodosimetry of occupational, clinical and accidental exposures, for initial triage and medical management.
Keywords: Biodosimetry, Dicentrics chromosomal aberrations, Micronuclei, Dose-response curve, Dose blinded samples
1. Introduction
Biological dosimetry refers to estimation of absorbed dose in individuals with unintentional exposure to ionizing radiation when the physical dosimeter is unavailable or in dispute [1,2]. Biodosimetry classically relies on cytogenetic techniques where chromosomes are involved such as, Dicentric Chromosome Assay (DCA), Cytokinesis Blocked Micronuclei (CBMN) assays and Chromosomal Translocations using Fluorescence In Situ Hybridization (FISH) [[3], [4], [5], [6]]. The DCA was the only available cytogenetic technique intended for biodosimetry for many years, either after accidental or occupational radiation exposures. It has been used in various radiation accidents such as the Chernobyl disaster in 1986, Tokaimura nuclear accident in 1999 and recently in Fukushima accident in 2011 [[7], [8], [9]]. Based on ISO standards and IAEA recommendations, DCA has become a routine component of many radiological protection programs and its methodological standard has been accepted internationally [4,10]. DCA has also been applied in triage situation with specially designed scoring criteria wherein about 20–50 cells are analyzed instead of 500–1000 as in standard method. This has helped to achieve needed throughput and repurpose the assay for management of radiological emergencies involving large number of individuals with suspected exposures.
Fenech and Morley in 1985, developed CBMN assay, to detect micronuclei [[11], [12], [13]]. Micronuclei, are small extra-nuclear bodies produced from chromosome breaks or whole chromosomes lagging behind during anaphase of the cell cycle [[14], [15], [16]]. It is not very specific to radiation exposure as dicentrics are since it may be induced/influenced by some life style factors too [17,18]. Micronuclei are scored in peripheral blood lymphocytes with cytokinesis blocking (binucleated cell) in first division of cell cycle. Binucleated (BN) cells can be obtained by addition of cytokinesis inhibitor, Cytochalasin B during cell culturing. Micronuclei assay has been employed for biological dose estimation in various past radiological incidences [[19], [20], [21]]. The assay is easy to perform and scoring of aberrations (micronucleus), needs less effort and set of skills in comparison to DCA. Scoring of events is also amenable to automation. At moderate and high doses, the CBMN assay gives fairly accurate results. This assay is usually performed as a supplementary assay to support the finding of DCA, during triage management.
Dicentrics and micronuclei, are categorized as unstable chromosomal aberrations predominantly induced by ionizing radiation. They are considered as post repair events, since they are produced as a result of DSB mis-repair [22,23]. The number of cells possessing unstable chromosomal aberrations, declines with time, due to 1) Death of the cells after completion of their life span and 2) Selective apoptosis of aberration bearing cells during cell proliferation/division [24,25]. Therefore, analysis of the dicentric and micronuclei for biological dose estimation should be performed with in few months (as early as possible) after radiation exposure, for precised dose estimation [26,27].
In addition, some DSB (DNA double strand break) repair proteins like γH2AX and 53BP1 are presenting their candidature, as rapid biodosimetry tools, which can facilitate dosimetry in few hours [28,29]. Though, signal of these DSB-repair proteins decays with time, as DBS get repaired [30,31].
Application of radiation in the diagnostics and treatment of different diseases like malignancies are increasing exceptionally [32,33]. Radiotherapy (in combination with chemotherapy) contribute extensively in cancer treatment, approximately 50% of cancer patients under go radiotherapy during the period of their ailment [34,35]. Aforesaid, cytogenetic and protein markers are not only employed in dosimetry of occupational/accidental exposures but in clinical (diagnostics and therapeutics) exposures too [36].
As per standard recommendations, every biodosimetry lab need to establish their own dose-response curve to demonstrate the quality of preparations and validate the techniques and protocols deployed. Besides, dose estimation by employing coefficients generated from different laboratory may introduce extra uncertainties, which is undesirable [37,38]. Even with the advancements in the laboratory techniques and the practices, the differences in coefficients generated varies from lab to lab. This can be attributed to various factors, like varied culture conditions, micro-environment, instrumental sensitivity, imaging and personal scoring experiences etc [39,40]. Laboratories that perform biological dose assessment using cytogenetic markers must establish their own dose-response curve for different types and energies of radiation, as per their requirements [37,38]. Earlier reports have indicated that the yield of low LET-radiation induced dicentric and micronuclei follow linear-quadratic pattern. However, high-LET radiations exhibit linear pattern, with no quadratic component [41,42]. Lower limit of dose detection by dicentric and micronuclei for low LET radiation is ∼0.1 and ∼0.25 Gy respectively. Background frequency for dicentric and micronuclei varies from 0.5 to 1.0/1000 cells and 7.73 ± 3.91/1000 cells, respectively [43,44].
There is a dire need for every country to establish a biodosimetry network with sufficient nodal centres, ready with their own established dose-response curve for most of the recommended markers with demonstrated capability of handling multiple sample as per the need of time [[45], [46], [47]]. In the present study, we have established dose-response curve for X-ray induced dicentric and micronuclei in the blood sample of three volunteers in the dose range of 0–5 Gy. Values of the coefficients generated were in agreement with the values published in the scientific literatures. Coefficients generated were further validated by estimating the doses of 8 dose blinded samples, 4 (DCA-1 to 4) by dicentric chromosomal aberration assay and remaining 4 (MN-5 to 8) by cytokinesis blocked micronuclei assay. Estimated doses were well within the acceptable limits.
Biodosimetry laboratory at Bhabha Atomic Research centre (BARC), is the central facility in India and serving as reference biodosimetry laboratory for the nation [48,49]. Current work was carried out in Department of Human Genetics, Sri Ramachandra Institute of Higher Education and Research (Deemed to be University) [SRIHER (DU)], Chennai, India. SRIHER (DU) is the part of national biodosimetry network in India. This study was conducted in association with biodosimetry laboratory of BARC. The coefficients generated for dicentrics and micronuclei, would be employed for dose estimation of planned and unplanned radiation exposures in the country.
2. Materials and methods
2.1. Study design
The study was designed to establish a calibration curve for X-ray induced, “dicentric + ring” and micronuclei, in the blood sample of three volunteers (dose range, 0–5Gy), with manual scoring of the aberrations (“dicentric + ring” and micronuclei). Coefficients generated, was planned to validate with 8 dose blinded samples, 4 with “dicentric + ring” and remaining 4 with micronuclei.
This study can be considered as an inter-laboratory exercise with Radiological Physics and Advisory Division of BARC and extension of the part of biodosimetry network in India. The project was funded by Board of Research in Nuclear Sciences (BRNS), with sanction no. 54/14/03/2021-BRNS/10239.
2.2. Chemicals
l-glutamine and Giemsa were procured from Sigma-Aldrich, USA. Roswell Park Memorial Institute Medium 1640 (RPMI 1640), Phytohemagglutinin (PHA), Cyto-B Foetal Calf Serum (FCS), Streptomycin, Penicillin and Colcemid were procured from Gibco Life Technologies, USA. DPX (Distyrene, Plasticizer and Xylene) was purchased from Merck, USA.
2.3. Ethical approval donor selection and blood sampling
The study was approved by the Institutional Ethics committee (Ref: IEC-NI/21/OCT/80/128) and all experiments were conducted as per ethical guidelines provided. Three healthy volunteers [2 females (age 22, 23 years) and 1 male (24 year)], were recruited in this study, to construct the dose response curve for X-ray induced dicentric and micronuclei. Selected volunteers were with no recent history of, alcohol consumption, smoking, medication and radiation exposure (medical or occupational). 10 ml blood samples were collected in the lithium heparinized vacutainer by venepuncture, from each volunteer by expert phlebotomist.
2.4. Irradiation
The blood samples were aliquoted in 10 parts and irradiated with 0, 0.1, 0.25, 0.5, 0.75, 1, 2, 3, 4 and 5 Gy, using VERSA HD digital LINAC from Elekta [50], UK in the energy of 6 MeV at the dose rate of 6 Gy per minute with a SAD of 100 cm at the field size of 10 × 10 cm. Irradiation was carried out at the Department of Radiation Oncology, Sri Ramachandra Medical Centre, Chennai, India.
2.5. Dicentric chromosomal aberration assay
The irradiated blood samples were incubated in optimum conditions (37 °C temperature, 5% CO2 and 95% relative humidity) for 2 h to provide a time interval to DNA repair [51,52]. The methodology was framed according to International Atomic Energy Agency (IAEA, 2011); IAEA Technical Report Series, No.405 and ISO standard [4,48,49,53]. The cultures were set up with 8 ml RPMI-1640, 2 ml FBS, 0.2 ml Phytohemagglutinin (PHA) and 1 ml heparinized peripheral blood. The flasks were incubated in optimum conditions (37 °C temperature, 5% CO2 and 95% relative humidity) for 48 h. Long term (added at 24 h of cell culturing) colcemid (200 μl of 1 mg/ml) treatment was given to arrest the cell in metaphase of first division cycle [4,53]. The cells were treated with 8 ml of freshly prepared pre-warmed hypotonic solution (0.075 M potassium chloride) and incubated for 20 min in optimum conditions (37 °C temperature, 5% CO2 and 95% relative humidity). Cells were fixed with Carnoy's fixative (Methanol: Glacial acetic acid, 3:1), 3 times with repeated centrifugation and finally cell pellet was suspended in 0.2 ml of Carnoy's fixative. Cell suspension was dropped on top of a clean pre-chilled microscopic glass slide and placed on slide warmer to get it dried. The casted slides were stained with 10% Giemsa for 15 min and mounted with DPX [54]. About 800 to ∼12000 metaphases were scored to estimate yield of dicentrics and rings for each dose (0–5Gy) point.
2.6. Cytokinesis blocked micronuclei assay
Following irradiation, blood samples were incubated in optimum conditions and cultures were setup as described in section 2.5 (for Dicentric Chromosomal Aberration Assay) [4,51,55]. To arrest the cells in cytokinesis stage of the cell cycle, 200 μl (of 250 μg/ml) cytochalasin B was added to each culture flask at 44thh of cell culturing. To harvest binucleated cells, whole culture was transferred to 15 ml centrifuge tube, and pellet was obtained by centrifugation (at 1000 rpm, for 10 min). To the cell pellet, 8 ml of pre-chilled hypotonic solution (0.075 M potassium chloride) was added and incubated for 20 min at room temperature. Again, cells were pellet down by centrifugation (at 1000 rpm, for 10 min) and fixed with freshly prepared, chilled Carnoy's fixative (5:1:1, methanol: acetic acid: formaldehyde, 8 ml per tube). Fixation step was repeated three times, finally cell pellet was suspended in 0.3 ml of fixative solution. Slides were casted by dropping the cell suspension on top of the glass slide from the height of 10–15 cm and allowed it to dry on a hot plate (temperature, 40 °C). Slides bearing binucleated cells, were stained with 10% Giemsa (prepared in distilled water) for 20 min, followed by a brief rinse with DW, air-dried and mounted with DPX. For each sample, 5000 binucleated cells were scored manually, using an optical microscope, in bright field mode, at 40× magnification. The nuclear division index was calculated employing following formula [4].
where M1, M2, M3 and M4 are the number of lymphocytes with 1, 2, 3 and 4 nuclei, respectively and N is the total number of lymphocytes scored.
2.7. Dose blinded samples
To validate the established dose response curve, 8 ml peripheral blood sample was obtained from an informed male volunteer (age 32 years) without any history of occupational or medical radiation exposure. Blood sample was aliquoted in 8 different tubes (1ml/tube) and irradiated with different dose-blinded doses. As described in section 2.6 and 2.7, cultures were setup for DCA (4 cultures) and CBMN (4 cultures), metaphases and binucleated cells were harvested at 48 and 72 h of incubations, respectively. Slides were prepared, aberrations were scored and doses were reconstructed by employing coefficients generated for calibration curves (for “dicentric + ring” and micronuclei) established.
2.8. Statistical analysis
Dose-response curves were constructed using Dose Estimate software, version 5.2 [ 56]. The corresponding 95% confidence intervals were calculated assuming a Poisson distribution, as per IAEA recommendations [4,57]. To determine whether dicentrics with centric rings and micronuclei followed, a Poisson distribution, dispersion index (u) was used [4]. The goodness of fit for homogeneity were performed with the Dose Estimate and CABAS software [ 56]. All curves were fitted following least square method.
3. Results
3.1. Establishment of dose response curve for X-ray induced “dicentric + ring”
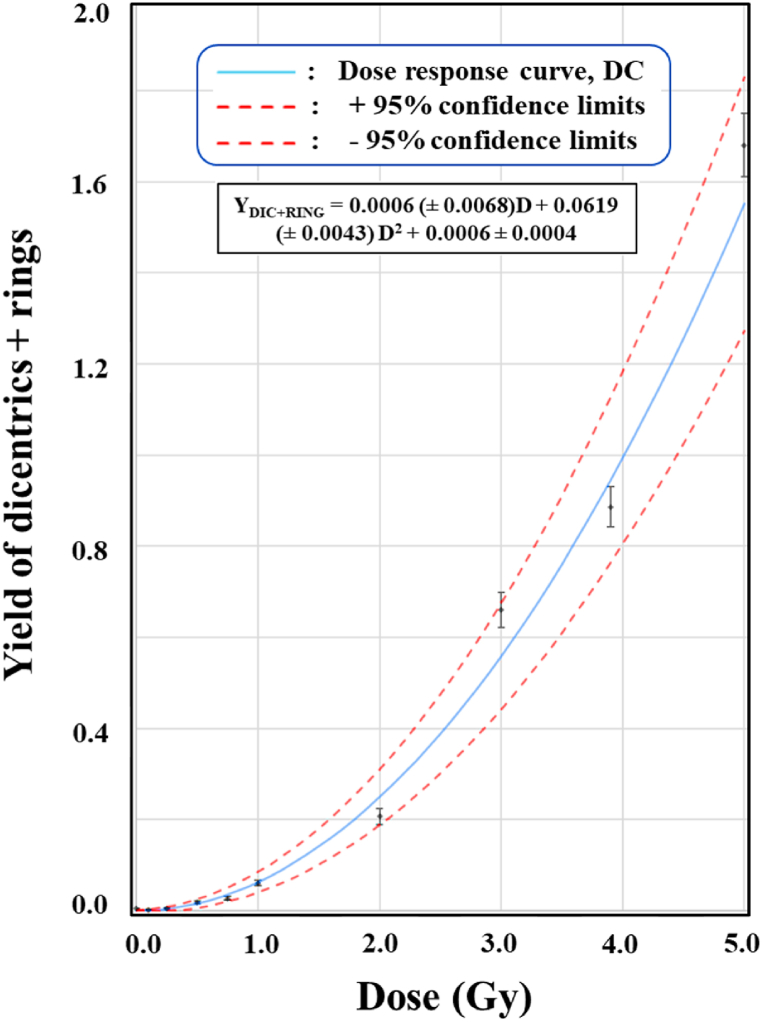
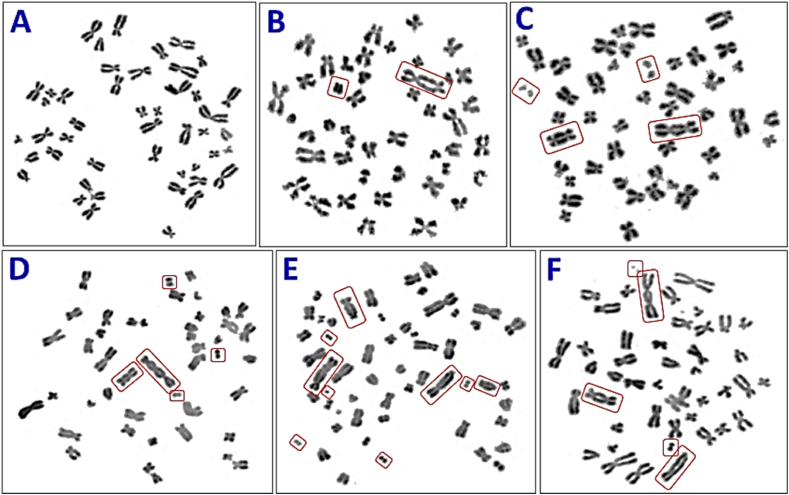
Dose-response curve was established for X-ray induced cytogenetic markers, “dicentric + ring”, in the peripheral blood samples of these volunteers, 2 females, ages 22 and 23 years and one 24 years old male (Fig. 1). A total of 25,104 metaphase spreads were analyzed and 1666 “dicentric + ring” were detected. Data were pooled from all the three volunteers for each dose points. Inclusion and exclusion criteria for metaphase-selections, were followed as per IAEA and ISO recommendations [4,53]. Fig. 2 (A-F), illustrates the representative metaphases that are included and excluded from the scoring. Metaphases bearing dicentric and fragment or ring and fragment were included in scoring however metaphases with fragment alone or with translocation were excluded from the scoring. It is evident from Table 1, that the number of cells with more than one “dicentric + ring”, increases with increase in dose. Up to 0.5 Gy, none of the cell with two or more “dicentric + ring” was observed. The first cell with two “dicentric + ring” was observed at 0.75 Gy. Statistical analysis revealed that distribution of “dicentric + ring” followed Poisson distribution for all thedoses, (Table 1). Table 1 also shows the number of metaphases analyzed, the number of “dicentric + ring” detected and their distribution for all dose points. Additionally, the dispersion index (u) and relative variance (σ2/Y) (values calculated as per IAEA recommendations) for all dose points [4] are also depicted in Table 1.
Fig. 1.
Establishment of dose response curve for 6 MeV, X-ray induced “dicentric + ring” in the dose range of 0–5 Gy. Curve followed linear-quadratic fit with the coefficients α = 0.0006 (±0.0068) “dicentric + ring” cell−1 Gy−2 and β = 0.0619 (±0.0043) “dicentric + ring” cell−1 Gy−2. C represents, background frequency (0.0006 ± 0.0004 “dicentric + ring” chromosomes cell−1) of “dicentric + ring” chromosomes, observed in the pooled data of all volunteers.
Fig. 2.
Representative metaphases with (A) No dicentric-control metaphase, (B) One dicentric accompanied with one fragment, (C) Two dicentrics accompanied with two fragments, (D) One dicentrics and one tricentric accompanied with three fragments, (E) Three dicentrics and one tricentric accompanied with five fragments and (F) Two dicentrics accompanied with two fragments and one translocation at the bottom - such metaphases were excluded from the scoring.
Table 1.
Yield and distribution of “dicentric + ring”, following ex vivo irradiation of 0–5 Gy (6 MeV, X-ray). All sets of data were in agreement with Poisson distribution (u values were within the range of ±1.96) and were fitted by a linear quadratic model (Y C + αD + βD2) using Dose Estimate software, version 5.2. D0, D1, D2, D3, D4, D5, D6 and D7, represents metaphases with 0, 1, 2, 3, 4, 5, 6 and 7 “dicentric + ring”, respectively.
| Dose (Gy) | Cells Scored |
D + R (No) | Distribution of “Dicentric + ring” |
Yield (Y) | Relative Variance σ2/Y |
Dispersion Index (u) | Poisson Distribution | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D1 | D2 | D3 | D4 | D5 | D6 | D7 | |||||||
| 0 | 12654 | 6 | 12648 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0005 | 1.000 | −0.034 | P |
| 0.1 | 2500 | 6 | 2494 | 6 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0024 | 0.998 | −0.077 | P |
| 0.25 | 2000 | 11 | 1989 | 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0055 | 0.995 | −0.166 | P |
| 0.5 | 2000 | 37 | 1963 | 37 | 0 | 0 | 0 | 0 | 0 | 0 | 0.0185 | 0.982 | −0.577 | P |
| 0.75 | 2000 | 55 | 1946 | 53 | 1 | 0 | 0 | 0 | 0 | 0 | 0.0275 | 1.009 | 0.299 | P |
| 1 | 2000 | 122 | 1884 | 111 | 4 | 1 | 0 | 0 | 0 | 0 | 0.0610 | 1.054 | 1.723 | P |
| 2 | 700 | 145 | 569 | 118 | 12 | 1 | 0 | 0 | 0 | 0 | 0.2071 | 1.001 | 0.022 | P |
| 3 | 450 | 297 | 236 | 155 | 41 | 12 | 6 | 0 | 0 | 0 | 0.6600 | 1.103 | 1.552 | P |
| 4 | 450 | 399 | 185 | 175 | 61 | 18 | 8 | 2 | 1 | 0 | 0.8867 | 1.108 | 1.624 | P |
| 5 | 350 | 588 | 28 | 189 | 56 | 46 | 15 | 9 | 5 | 2 | 1.6800 | 0.993 | −0.094 | P |
The data for “dicentric + ring” chromosomes, were fitted by linear quadratic model of the expression Y C + αD + βD2. The curve fitting yields, linear coefficient α = 0.0006 (±0.0068) “dicentric + ring” cell−1 Gy−1 and quadratic coefficient β = 0.0619 (±0.0043) “dicentric + ring” cell−1 Gy−2. ‘C’ represents, background frequency (0.0006 ± 0.0004) “dicentric + ring” chromosomes cell−1), in the pooled data of three volunteers. As shown in Table 3, pooled data followed a good statistical fit with the correlation coefficient (r) 0.9939, degrees of freedom7 and p value for goodness of fit0.0002.
Table 3.
Degrees of freedom (df), p values for goodness of fit and Correlation coefficient (r), for “dicentric + ring” and micronuclei, after fitting the pooled data obtained from all three volunteers.
| Radiation marker studied | Degrees of freedom | p value for goodness of fit | Correlation coefficient (r) |
|---|---|---|---|
| “Dicentric + Ring” | 7 | 0.0002 | 0.9939 |
| Micronuclei | 7 | 0.1955 | 0.9996 |
3.2. Establishment of dose response curve for X-ray induced micronuclei
As illustrated in Fig. 3, dose response curve was established for X-ray induced micronuclei in the blood sample of three volunteers (as detailed in section 3.1) using cytokinesis blocked micronuclei (CBMN) assay. To generate the curve, a total of 50,000 binucleated cells were analyzed and 9356 micronuclei were detected. Manual scoring was performed following the recommendations of IAEA and ISO [4,53,58]. Inclusion and exclusion criteria for scoring of micronuclei are shown in Fig. 4 (A-H). Only, binucleated cells (with or without micronuclei) were included in the scoring, however mono, tri and tetra nucleated-cells were excluded from the scoring. Pooled data of these volunteers, with statistical parameters like; distribution and yield of MN, dispersion index (u) and relative variance (σ2/Y), are shown in Table 4.
Fig. 3.
Establishment of calibration curve for X-ray induced micronuclei in the dose range of 0–5 Gy. Curve followed linear-quadratic fit with the coefficients α = 0.0459 ± 0.0038 micronuclei cell−1 Gy−1 and β = 0.0185 ± 0.0010 micronuclei cell−1 Gy−2. C represents, background frequency (0.0077 ± 0.0012 MN cell−1) of micronuclei, in the pooled data of all volunteers.
Fig. 4.
Representative (A) Mononucleated cell without MN, (B) Mononucleated cell with one MN, (C) Mononucleated cell with two MN, (D) Binucleated cell without MN, (E) Binucleated cell with one MN, and (F) Binucleated cell with one MN and a nucleoplasmic-bridge, (G and H) Tri and tetra-nucleated cells without MN. Only binucleated cells (without nucleoplasmic-bridge) were taken into account for construction of the dose response curve. Mono, tri and tetra nucleated-cells were not accounted in dose-response curve generation.
Table 4.
Yield and distribution of micronuclei, following ex vivo irradiation of 0–5 Gy (6 MeV, X-ray). All sets of data were over dispersed and were fitted by a linear quadratic model (Y C + αD + βD2) using Dose Estimate software, version 5.2. D0, D1, D2, D3, D4, D5 and D6, represents binucleated cells with 0, 1, 2, 3, 4, 5 and 6 micronuclei, respectively.
| Dose (Gy) | Cells scored | MN (No) | Distribution of Micronuclei (MN) |
Yield (Y) | Relative Variance σ2/Y |
Dispersion Index (u) | Poisson Distribution | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D0 | D1 | D2 | D3 | D4 | D5 | D6 | |||||||
| 0 | 5000 | 36 | 4966 | 32 | 2 | 0 | 0 | 0 | 0 | 0.007 | 0.02 | 5.28 | OD |
| 0.1 | 5000 | 72 | 4934 | 60 | 6 | 0 | 0 | 0 | 0 | 0.014 | 0.02 | 7.68 | OD |
| 0.25 | 5000 | 92 | 4918 | 72 | 10 | 0 | 0 | 0 | 0 | 0.018 | 0.02 | 10 | OD |
| 0.5 | 5000 | 174 | 4844 | 139 | 16 | 1 | 0 | 0 | 0 | 0.035 | 0.02 | 9.22 | OD |
| 0.75 | 5000 | 248 | 4777 | 201 | 19 | 3 | 0 | 0 | 0 | 0.05 | 0.02 | 8.84 | OD |
| 1 | 5000 | 400 | 4650 | 307 | 38 | 3 | 2 | 0 | 0 | 0.08 | 0.02 | 10.8 | OD |
| 2 | 5000 | 855 | 4275 | 622 | 79 | 21 | 3 | 0 | 0 | 0.171 | 0.02 | 10.2 | OD |
| 3 | 5000 | 1502 | 3809 | 933 | 213 | 38 | 6 | 1 | 0 | 0.3 | 0.02 | 9.83 | OD |
| 4 | 5000 | 2500 | 3146 | 1378 | 352 | 88 | 28 | 6 | 2 | 0.5 | 0.02 | 9.97 | OD |
| 5 | 5000 | 3477 | 2470 | 1881 | 424 | 168 | 44 | 10 | 3 | 0.695 | 0.02 | 3.69 | OD |
Distribution of micronuclei was tested for compliance with Poisson distribution for each dose point, since curve fitting was based on Poisson statistics [56]. As shown in Table 4, all sets of data were showing over-dispersion (u values were exceeding ±1.96 for all dose points). To generate the dose-response curve, data were fitted with linear quadratic fit. Data fitting resulted in, linear coefficient α = 0.0459 ± 0.0038 micronuclei cell−1 Gy−2 and quadratic coefficient β = 0.0185 ± 0.0010 micronuclei cell−1 Gy−2 (Table 2). C represents, background frequency of micronuclei, which was found to be, 0.0077 ± 0.0012 MN cell−1, in the pooled data of all the volunteers. Statistical parameters of data fitting are shown in Table 3, data showed a good statistical fit with the correlation coefficient (r) 0.9996, χ29.88, degrees of freedom 7 and p value for goodness of fit 0.1955.
Table 2.
Values of the coefficients (alpha and beta) generated for “dicentric + ring” and micronuclei, after fitting the pooled data obtained from all three volunteers.
| Radiation marker studied | Linear Coefficient (α) (Unit: DC or MN cell−1 Gy−2) |
Quadratic Coefficient (β) (Unit: DC or MN cell−1 Gy−2) |
Background Frequency (C) (Unit: DC or MN cell−1) |
|---|---|---|---|
| “Dicentric + Ring” | 0.0006 ± 0.0068 | 0.0619 ± 0.0043 | 0.0006 ± 0.0004 |
| Micronuclei | 0.0459 ± 0.0038 | 0.0185 ± 0.0010 | 0.0077 ± 0.0012 |
The nuclear division index measures the cell proliferation rate in CBMN assay [ 4,10]. As shown in Table 5 and Fig. 5, nuclear division index was found to be decreasing (from 2.06 to 1.09) with the increase in dose. These findings, clearly indicating the cyto-inhibitory effects of the increasing dose of radiation [ 11].
Table 5.
Nuclear division index (NDI) for all dose points (0–5Gy) taken into account (pooled data from 3 volunteers).
| S. No. | Dose | NDI (Pooled data of 3 volunteers) |
|---|---|---|
| 1 | 0 | 2.06 |
| 2 | 0.1 | 1.94 |
| 3 | 0.25 | 1.81 |
| 4 | 0.5 | 1.76 |
| 5 | 0.75 | 1.68 |
| 6 | 1 | 1.57 |
| 7 | 2 | 1.49 |
| 8 | 3 | 1.46 |
| 9 | 4 | 1.34 |
| 10 | 5 | 1.09 |
Fig. 5.
Nuclear division index (NDI±SD) for 10 dose points (0–5Gy), obtained from pooled data of 3 volunteers.
3.3. Validation of established dose response curves by estimating the doses of dose blinded samples
Generated coefficients for the established dose-response curves were validated by estimating doses of 8 dose blinded samples, using Dose Estimate and CABAS software [56]. Sample DCA-1 to 4 were analyzed by DCA and sample MN-5 to 8, were analyzed by CBMN-assays (Table 6). All the doses estimated by DCA, were within the limits of 95% lower and upper confidence intervals, except for 5Gy (Sample DCA-1). However, for the delivered dose of 5 Gy there was an overestimation of 16%. Two (Sample MN-2 and MN-4) out of 4 doses, estimated by CBMN, were within the limits of 95% lower and upper confidence intervals. Sample MN-1 (corresponding to the delivered dose of 1Gy) overestimatedthe dose by 31.2%. Similarly, sample MN-3 (corresponding to the delivered dose of 3Gy) underestimated the dose by21.7%. All estimated and delivered doses are shown in Table 6 and Fig. 6. It can be seen that the variation between estimated and delivered doses were within −21.7% to +31.2%. These estimates, validates the suitability of the coefficients generated (for DCA and CBMN) for biodosimetry.
Table 6.
Estimation of biological dose of the eight dose blinded samples (1–8) based on the yields of “dicentric + ring” and micronuclei. Four dose blinded samples were analyzed for scoring of “dicentric + ring” and remaining four were analyzed for micronuclei. 120 to 750 metaphases were analyzed for dicentrics (as per the yields obtained during the scoring) and more than 1000 binucleated cells were analyzed for micronuclei.
| S. No. | End Point | Blinded Sample | Dose Delivered (Gy) | Aberrations/Cells Scored | Aberration per Cell (Yield) | Estimated doses (Gy) | % variation in estimated doses |
|---|---|---|---|---|---|---|---|
| 1. | DCA | DCA-1 | 5.0 | 169/120 | 1.4083 | 5.79 {95% CI: 5.29–6.31} |
+16 |
| 2. | DCA | DCA-2 | 1.0 | 46/500 | 0.0920 | 1.01 {95% CI: 0.79–1.25} |
+0.9 |
| 3. | DCA | DCA-3 | 2.0 | 71/310 | 0.2290 | 1.91 {95% CI: 1.62–2.23} |
- 4.4 |
| 4. | DCA | DCA-4 | 0.5 | 4/750 | 0.0333 | 0.45 {95% CI: 0.31–0.62} |
- 9.94 |
| 5. | MN | MN-1 | 1.0 | 106/1071 | 0.0990 | 1.31 {95% CI: 1.16–1.47} |
+31.2 |
| 6. | MN | MN-2 | 0.25 | 17/1068 | 0.0153 | 0.31 {95% CI: 0.11–0.50} |
+22.8 |
| 7. | MN | MN-3 | 3.0 | 279/1037 | 0.2726 | 2.35 {95% CI: 2.20–2.51} |
- 21.7 |
| 8. | MN | MN-4 | 0.0 | 2/1120 | 0.0018 | 0 {95% CI: 0.0–0.0} |
0 |
Fig. 6.
Graphical illustration of the estimated doses of eight dose blinded samples. Four dose blinded samples (DCA-1 to 4) were analyzed for “dicentric + ring” and remaining 4 dose blinded samples (MN-5 to MN-8) were analyzed for micronuclei.
4. Discussion
Several approaches for biological dosimetry after exposure to low and high doses of radiation have been documented [27,[59], [60], [61], [62]]. DCA is most reliable and standard biological assay that is globally accepted [4,[63], [64], [65], [66], [67], [68]]. Recent studies suggest that chromosomal aberrations such as dicentrics can be employed for dose estimation following chronic or low-dose radiation exposures, too [69,70]. In this study, X-ray induced calibration curves for the “dicentric + ring” and micronuclei were generated as per the methods documented in IAEA manual, 2011 [4]. The coefficients were generated using the Dose Estimate statistical analysis software, which uses weighted fitting in the maximum-likelihood framework [56].
Before plotting the data, distribution patterns of “dicentric + ring” were subjected to dispersion analysis to ensure the Poisson distribution. As, shown in Table 1, dispersion index (u) for all dose points (0–5 Gy), were in the range of −1.96 to + 1.96, demonstrating a Poisson distribution. Distribution pattern was found to be in accordance with the previously published data [40,44,71,72]. Similarly, distribution pattern for micronuclei was also examined, as shown in Table 4, dispersion index (u) for all dose points (0–5 Gy), were exceeding to ±1.96, demonstrating an over-dispersion of all the data points. As per the literature, distribution of micronuclei should not necessarily follow Poisson distribution for all the dose points [4,73,74]. Hence over dispersion on MN is justified.
Coefficients generated for the dose response curves for “dicentric + ring” and micronuclei, are shown in Table 2. Taking into account pooled data from all three volunteers, the resultant fitted curve for “dicentric + ring” was; YDIC + RING = 0.0006 (±0.0068) D + 0.0619 (±0.0043) D2 + 0.0006 ± 0.0004 and for micronuclei; YMN = 0.0459 ± (0.0038)D + 0.0185 ± (0.0010)D2 + 0.0077 (±0.0012). The goodness of fit test for the “dicentric + ring” calibration curve indicated that the data were well adjusted by the linear quadratic model (degrees of freedom df = 7, p value for goodness of fit = 0.0002 and Correlation coefficient (r) = 0.9939). The same conclusion can be derived for the micronuclei calibration curve, which was also well fitted with a linear quadratic function (degrees of freedom (df) = 7, p value for goodness of fit = 0.1955 and Correlation coefficient (r) = 0.9996). Considering the points where over dispersion occurred, confidence intervals were corrected according to the IAEA recommendations [4].
It was interesting to note that higher alpha coefficient for MN but lower beta coefficient compared to DCA was found while both the assays followed linear quadratic responses. Breaks are associated with every dicentrics during their formation, which eventually lead to formation of micronuclei. Besides the associated breaks, interstitial breaks, acentric rings and telomere stabilized breaks also lead to formation of micronuclei. Sometimes, even whole chromosomes can lead to micronuclei albeit they are rare events. Such events are more prominent at low and moderate doses, which yields higher alpha value of response curve. Also, the fit coefficient of alpha is likely to weigh down beta at higher doses leading to reduced quadratic component compared to DCA.
The generated α and β coefficients for the “dicentric + ring” and micronuclei, dose response curve in the present study is comparable with previously reported values. Hence, it can be inferred that frequency of “dicentric + ring” and micronuclei, for all the donors and dose points, were consistent and were in accordance with the previously published data [39,40,72,[75], [76], [77], [78]].
Though micronuclei are not very specific to radiation exposure, it is imperative to established dose response curve, because it may be quickly examined in samples and scoring doesn't require much training. Micronuclei has potential application in biodosimetry, when large scale screening is required during radiological emergencies [[79], [80], [81]]. During dose-response curve establishment it was observed that, micronuclei exhibited zero-inflation at many dose-points accounted. Since some micronuclei are created when acentric fragments are not getting integrated into the daughter cell nuclei during cell division, the overdispersion and zero-inflation seen with micronuclei looked to be connected to acentric fragments (IAEA 2011) [4]. In general, a little higher variation in the yield of micronuclei, among the individuals is reported in the literature, this might be due to contribution of the factors like age, gender, diet and lifestyle [11,[82], [83], [84]]. Though in this study variation in the yield of micronuclei among the individuals was significantly low, this may be due to volunteers narrow age range and their healthy life style factor (no one was on any xenobiotic drug, alcohol consumption and smoking). A large-scale biomonitoring studies have shown that the spontaneous yield of micronuclei increases systematically with age. The micronuclei values between 0.24 and 0.44 micronuclei/1000 binucleated cells per year were observed in healthy male populations in the frame of studies performed by the Ghent group on nuclear power plant and hospital workers [39,85]. It is always recommended to establish the calibration curve, recruiting higher number of volunteers of different age groups and the gender [4,37]. In this study, dose-response curve is generated with the blood samples of three volunteers in the age range of 20–30 years. To strengthen the established calibration curve, volunteers of different age groups and the gender would be included in future. It will help to gain better confidence limits in dose estimations, when the test samples obtained will belong to different age groups and the gender.
Dicentrics are radiation specific, and their presence alone designates ionizing radiation exposure, whereas the presence of micronuclei is indicative and not very specific to the radiation exposure [4,86]. Though, in the mass exposure scenario, CBMN assay still has a time advantage for the analysis/scoring, over the DC-assay. To increase reliability, it is necessary to use biodosimetry together with the different assays, in addition to all the clinical and physical parameters. When all the approaches are used together, we might overcome the gaps inherent in each technique [26,87].
Established dose-response curves were validated by estimating the dose of 8 dose blinded samples (4 by DCA and rest 4 by CBMN), shown in Table 6. Out of 4 blind doses, DCA estimated 3 doses within the ±95% confidence limits, however CBMN estimated 1 (within the ±95% confidence limits) out of 3 and remaining estimate varied up to 31.2%. Such large errors were not surprising in case of CBMN for low doses owing to large uncertainties and inter-individual variation in responses (4th dose blinded sample was 0 Gy). Estimated dose for 5 Gy (sample DCA-1) by DCA was, 6% over-estimated from the ±95% confidence limits. Estimated doses for samples MN-1 (corresponds to 1Gy) and MN-3 (corresponds to 3 Gy) were found to be over and under-estimated by 16% and 16.3% from the ±95% confidence limits, respectively. Over all, DCA gave comparatively more precised dose estimation over CBMN, here in this study conducted with 8 dose blinded samples. “Constant upsurge in the application of radiation, poses the chances of exposures and growing threat to public health. The crucial medical support demands reconstruction of the dose received by the victims, concerning medical triage and proper treatment decision [88]. This study has helped to construct dose response curves (for dicentric and micronucleus), which can be directly employed in dose estimations and facilitating in treatment decisions. Established dose-response curve is the prerequisite for any biodosimetry laboratory, which is dedicated to biological dose estimations of exposed victims. The established dose-response curve for 6 MV (LINAC) X-ray induced cytogenetic markers is a value addition to the existing pool of other response curves such as low energy X-ray, gamma, Tritium beta and criticality neutrons by the Indian network of Biodosimetry labs. This study has given an effort to fulfil the gap and provided a tool to reconstruct the doses of victims exposed with 6 MV, X-ray. Majorly, 6 MV, X-ray is used in radiotherapy of cancer [89,90]. Cancer is a leading cause of death globally, according to estimates from the International Agency for Research on Cancer (IARC) approximately 10 million people died in 2020, (one in six death) [91]. In next 15–20 years, global cancer incidences (new) are anticipated to increase up to ∼27 million with ∼16 million deaths owing to growth and aging of the population [91,92]. These anticipations clearly indicate the increased application of radiotherapy units, hence increased chances of over exposures too. Dose-response curves established in this study, may have greater utility in the coming years”. Besides its role in radiation protection, these response curves can also be utilized as research tools to explore the feasibility of patient specific dosimetry and estimation of scattered dose or blood dose.
In spite of direct application, certain limitations of the study and the cytogenetic assays cannot be ignored. All the dose-response curves were generated through ex vivo experimentations and hence, in vivo validation of the results is warranted. Nonetheless, the response curves generated in vitro in other studies have shown clear matching with in vivo irradiation when clear physical dosimetry information were available. Generated dose response curves can be inter-compared and validated with calibration curves established in other cytogenetic biodosimetry laboratories (inter-laboratory comparison exercises) through dose blinded samples.
5. Conclusion
Dose-response curves were established for X-ray induced “dicentric + ring” and micronuclei, in the blood sample of three volunteers. Coefficients generated were in agreement with the data published in the literature. Established curves were validated by estimating the doses of 8 dose blinded samples. Estimated doses were within the variation of −21.7% to +31.2%. Closest estimate was made for sample DCA-2 (variation was just 0.9%, from the delivered dose of 1Gy) by dicentric chromosome assay. Farthest estimate was made for sample MN-1 (variation was +31.2%, from the delivered dose of 1Gy) by micronuclei assay. These outcomes validate the suitability of the coefficients generated (for DCA and CBMN) for biodosimetry.
The biodosimetry lab at SRIHER (DU), is a part of biodosimetry network in India, with the nodal centre at BARC, Mumbai. As per IAEA recommendations, these two assays and their in-house established calibration curves, can serve as tool for bi-parametric dose estimation, to gain better statistical confidence.
Author contribution statement
J. Vijayalakshmi, Rajesh Kumar Chaurasia, Solomon F.D. Paul, N.N. Bhat, B.K. Sapra: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper. ⟨/p⟩
K. Satish Srinivas, K. Vijayalakshmi: Contributed reagents, materials, analysis tools or data. ⟨/p⟩
Funding information
J. Vijayalakshmi was supported by Board of Research in Nuclear Sciences {54/14/03/2021-BRNS/10239}.
Data availability statement
Data included in article/supp. material/referenced in article.
Declaration of interest's statement
Authors declare no known conflict of interest.
Additional information
None.
Declaration of competing interest
Authors declare that, they do not have any conflict of interests.
Acknowledgement
The authors of the article would like to thank technical supports and encouragement received from Mr. Kapil B. Shirsath, Mr. Shrikant Jagtap and all other lab-mates of biodosimetry lab of SRIHER and BARC.
Contributor Information
J. Vijayalakshmi, Email: vijayalakshmi.j@sriramachandra.edu.in.
Rajesh Kumar Chaurasia, Email: rajeshc@barc.gov.in.
K. Satish Srinivas, Email: dr_satishsrinivas@srramachandra.edu.in.
K. Vijayalakshmi, Email: vijayalakshmi.k@sriramachandra.edu.in.
Solomon F.D. Paul, Email: paul@sriramachandra.edu.in.
N.N. Bhat, Email: nageshnb@barc.gov.in.
B.K. Sapra, Email: bsapra@barc.gov.in.
References
- 1.Martins V., Antunes A.C., Gil O.M. Implementation of a dose–response curve for γ-radiation in the Portuguese population by use of the chromosomal aberration assay. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2013;750(1–2):50–54. doi: 10.1016/j.mrgentox.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 2.Rojas-Palma C., Woda C., Discher M., Steinhäusler F. On the use of retrospective dosimetry to assist in the radiological triage of mass casualties exposed to ionising radiation. J. Radiol. Prot. 2020;40(4):1286. doi: 10.1088/1361-6498/abc181. [DOI] [PubMed] [Google Scholar]
- 3.Ainsbury E., Barquinero J.F., Beinke C., Blakely W.F., Braselmann H., Carr Z., Di Giorgio M., Fenech M., Garcia L.O., Kodama Y., Lindholm C. 2011. Cytogenetic Dosimetry: Applications in Preparedness for and Response to Radiation Emergencies. [Google Scholar]
- 4.Cytogenetic Dosimetry I.A.E.A. EPR‐Biodose; Vienna: 2011. Applications in Preparedness for and Response to Radiation Emergencies. [Google Scholar]
- 5.Tucker, J D. Fish cytogenetics and the future of radiation biodosimetry. Radiat. Protect. Dosim. 2001;97(1):55–60. doi: 10.1093/oxfordjournals.rpd.a006638. [DOI] [PubMed] [Google Scholar]
- 6.Grégoire E., Roy L., Buard V., Delbos M., Durand V., Martin-Bodiot C., Voisin P., Sorokine-Durm I., Vaurijoux A., Voisin P., Baldeyron C. Twenty years of FISH-based translocation analysis for retrospective ionizing radiation biodosimetry. Int. J. Radiat. Biol. 2018;94(3):248–258. doi: 10.1080/09553002.2018.1427903. [DOI] [PubMed] [Google Scholar]
- 7.Salassidis K., Schmid E., Peter R.U., Braselmann H., Bauchinger M. Dicentric and translocation analysis for retrospective dose estimation in humans exposed to ionising radiation during the Chernobyl nuclear power plant accident. Mutat. Res. Fund Mol. Mech. Mutagen. 1994;311(1):39–48. doi: 10.1016/0027-5107(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 8.Wojcik A., Gregoire E., Hayata I., Roy L., Sommer S., Stephan G., Voisin P. Cytogenetic damage in lymphocytes for the purpose of dose reconstruction: a review of three recent radiation accidents. Cytogenet. Genome Res. 2004;104(1–4):200–205. doi: 10.1159/000077489. [DOI] [PubMed] [Google Scholar]
- 9.Suto Y., Hirai M., Akiyama M., Kobashi G., Itokawa M., Akashi M., Sugiura N. Biodosimetry of restoration workers for the Tokyo electric power company (TEPCO) Fukushima Daiichi nuclear power station accident. Health Phys. 2013;105(4):366–373. doi: 10.1097/HP.0b013e3182995e42. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida K., Hada M., Kizu A., Kitada K., Eguchi-Kasai K., Kokubo T., Teramura T., Yano S., Suzuki H.H., Watanabe H., Kondoh G. Comparison of biological measurement and physical estimates of space radiation in the International Space Station. Heliyon. 2022;8(8) doi: 10.1016/j.heliyon.2022.e10266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenech M. The cytokinesis-block micronucleus technique: a detailed description of the method and its application to genotoxicity studies in human populations. Mutat. Res. Fund Mol. Mech. Mutagen. 1993;285(1):35–44. doi: 10.1016/0027-5107(93)90049-l. [DOI] [PubMed] [Google Scholar]
- 12.Fenech M. Environmental Genomics. Humana Press; 2008. The micronucleus assay determination of chromosomal level DNA damage; pp. 185–216. [DOI] [PubMed] [Google Scholar]
- 13.Vral A., Fenech M., Thierens H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis. 2011;26(1):11–17. doi: 10.1093/mutage/geq078. [DOI] [PubMed] [Google Scholar]
- 14.Fenech M. The cytokinesis-block micronucleus technique and its application to genotoxicity studies in human populations. Environ. Health Perspect. 1993;101(suppl 3):101–107. doi: 10.1289/ehp.93101s3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lusk C.P., King M.C. Rotten to the core: why micronuclei rupture. Dev. Cell. 2018;47(3):265–266. doi: 10.1016/j.devcel.2018.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Luzhna L., Kathiria P., Kovalchuk O. Micronuclei in genotoxicity assessment: from genetics to epigenetics and beyond. Front. Genet. 2013;4:131. doi: 10.3389/fgene.2013.00131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Cao J., Wang Z.Q., Bai Y.S., Lü Y.M., Huang Q.L., Zhao W.Z., Li J., Jiang L.P., Tang W.S., Fu B.H. Dose estimation by chromosome aberration analysis and micronucleus assays in victims accidently exposed to 60Co radiation. Br. J. Radiol. 2009;82(984):1027–1032. doi: 10.1259/bjr/62484075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pajic J., Rakic B., Rovcanin B., Jovicic D., Novakovic I., Milovanovic A., Pajic V. Inter-individual variability in the response of human peripheral blood lymphocytes to ionizing radiation: comparison of the dicentric and micronucleus assays. Radiat. Environ. Biophys. 2015;54(3):317–325. doi: 10.1007/s00411-015-0596-3. [DOI] [PubMed] [Google Scholar]
- 19.Müller W.U., Rode A. The micronucleus assay in human lymphocytes after high radiation doses (5–15 Gy) Mutat. Res. Fund Mol. Mech. Mutagen. 2002;502(1–2):47–51. doi: 10.1016/s0027-5107(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 20.Yao B., Li Y., Liu G., Guo M., Bai J., Man Q., Qiu L., Ai H. Estimation of the biological dose received by five victims of a radiation accident using three different cytogenetic tools. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2013;751(1):66–72. doi: 10.1016/j.mrgentox.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 21.Da Cruz A.D., McArthur A.G., Silva C.C., Curado M.P., Glickman B.W. Human micronucleus counts are correlated with age, smoking, and cesium-137 dose in the Goiania (Brazil) radiological accident. Mutat. Res. Environ. Mutagen Relat. Subj. 1994;313(1):57–68. doi: 10.1016/0165-1161(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 22.Beninson D., Lloyd D.C., Natarajan A.T., Obe G., Preston R.J., Sasaki M.S. vol. 260. Technical Reports Series; 1986. (Biological Dosimetry: Chromosomal Aberration Analysis for Dose Assessment). [Google Scholar]
- 23.Hatch E.M., Hetzer M.W. Linking micronuclei to chromosome fragmentation. Cell. 2015;161(7):1502–1504. doi: 10.1016/j.cell.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 24.Wilhelm T., Said M., Naim V. DNA replication stress and chromosomal instability: dangerous liaisons. Genes. 2020;11(6):642. doi: 10.3390/genes11060642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachs R.K., Hlatky L.R., Trask B.J. Radiation-produced chromosome aberrations: colourful clues. Trends Genet. 2000;16(4):143–146. doi: 10.1016/s0168-9525(99)01960-5. [DOI] [PubMed] [Google Scholar]
- 26.Ainsbury E.A., Bakhanova E., Barquinero J.F., Brai M., Chumak V., Correcher V., Darroudi F., Fattibene P., Gruel G., Guclu I., Horn S. Review of retrospective dosimetry techniques for external ionising radiation exposures. Radiat. Protect. Dosim. 2011;147(4):573–592. doi: 10.1093/rpd/ncq499. [DOI] [PubMed] [Google Scholar]
- 27.Amundson S.A., Bittner M., Meltzer P., Trent J., Fornance A.J., Jr. Biological indicators for the identification of ionizing radiation exposure in humans. Expert Rev. Mol. Diagn. 2001;1(2):211–219. doi: 10.1586/14737159.1.2.211. [DOI] [PubMed] [Google Scholar]
- 28.Horn S., Barnard S., Rothkamm K. Gamma-H2AX-based dose estimation for whole and partial body radiation exposure. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0025113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Belyaev I.Y. Radiation-induced DNA repair foci: spatio-temporal aspects of formation, application for assessment of radiosensitivity and biological dosimetry. Mutat. Res. Rev. Mutat. Res. 2010;704(1–3):132–141. doi: 10.1016/j.mrrev.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Rothkamm K., Barnard S., Moquet J., Ellender M., Rana Z., Burdak-Rothkamm S. DNA damage foci: Meaning and significance. Environ. Mol. Mutagen. 2015;56(6):491–504. doi: 10.1002/em.21944. [DOI] [PubMed] [Google Scholar]
- 31.Redon C.E., Weyemi U., Parekh P.R., Huang D., Burrell A.S., Bonner W.M. γ-H2AX and other histone post-translational modifications in the clinic. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2012;1819(7):743–756. doi: 10.1016/j.bbagrm.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rizzo A., Brandi G. Radiofrequency ablation, radiation therapy, transarterial chemoembolization, and yttrium 90: no differences for local treatment of liver cancer? Acta Oncol. 2022;61(6) doi: 10.1080/0284186X.2022.2046849. 730-730. [DOI] [PubMed] [Google Scholar]
- 33.Mollica V., Rizzo A., Rosellini M., Marchetti A., Ricci A.D., Cimadamore A., Scarpelli M., Bonucci C., Andrini E., Errani C., Santoni M. Bone targeting agents in patients with metastatic prostate cancer: state of the art. Cancers. 2021;13(3):546. doi: 10.3390/cancers13030546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Federico A., Rizzo A., Carloni R., De Giglio A., Bruno R., Ricci D., Brandi G. Atezolizumab-bevacizumab plus Y-90 TARE for the treatment of hepatocellular carcinoma: preclinical rationale and ongoing clinical trials. Expet Opin. Invest. Drugs. 2022;31(4):361–369. doi: 10.1080/13543784.2022.2009455. [DOI] [PubMed] [Google Scholar]
- 35.Ricci A.D., Rizzo A., Bonucci C., Tavolari S., Palloni A., Frega G., Mollica V., Tober N., Mazzotta E., Felicani C., Serra C. The (Eternal) debate on microwave ablation versus radiofrequency ablation in BCLC-A hepatocellular carcinoma. In Vivo. 2020;34(6):3421–3429. doi: 10.21873/invivo.12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudprasert W., Belyakov O.V., Tashiro S. Biological and internal dosimetry for radiation medicine: current status and future perspectives. J. Radiat. Res. 2022;63(2):247–254. doi: 10.1093/jrr/rrab119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alsbeih G.A., Al-Hadyan K.S., Al-Harbi N.M., Bin Judia S.S., Moftah B.A. Establishing a reference dose–response calibration curve for dicentric chromosome aberrations to assess accidental radiation exposure in Saudi arabia. Front. Public Health. 2020;8 doi: 10.3389/fpubh.2020.599194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkins R.C., Romm H., Kao T.C., Awa A.A., Yoshida M.A., Livingston G.K., Jenkins M.S., Oestreicher U., Pellmar T.C., Prasanna P.G. Interlaboratory comparison of the dicentric chromosome assay for radiation biodosimetry in mass casualty events. Radiat. Res. 2008;169(5):551–560. doi: 10.1667/RR1272.1. [DOI] [PubMed] [Google Scholar]
- 39.Pajic J., Rakic B., Jovicic D., Milovanovic A. Construction of dose response calibration curves for dicentrics and micronuclei for X radiation in a Serbian population. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014;773:23–28. doi: 10.1016/j.mrgentox.2014.07.009. [DOI] [PubMed] [Google Scholar]
- 40.Lusiyanti Y., Syaifudin M., Budiantari T., Purnami S., Ramadhani D. Development of dose-response calibration curve for dicentric chromosome induced by X-rays. Genome Integr. 2019;10 doi: 10.4103/genint.genint_1_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hunter N., Muirhead C.R. Review of relative biological effectiveness dependence on linear energy transfer for low-LET radiations. J. Radiol. Prot. 2009;29(1):5. doi: 10.1088/0952-4746/29/1/R01. [DOI] [PubMed] [Google Scholar]
- 42.Hill M.A. The variation in biological effectiveness of X-rays and gamma rays with energy. Radiat. Protect. Dosim. 2004;112(4):471–481. doi: 10.1093/rpd/nch091. [DOI] [PubMed] [Google Scholar]
- 43.Pardini B., Viberti C., Naccarati A., Allione A., Oderda M., Critelli R., Preto M., Zijno A., Cucchiarale G., Gontero P., Vineis P. Increased micronucleus frequency in peripheral blood lymphocytes predicts the risk of bladder cancer. Br. J. Cancer. 2017;116(2):202–210. doi: 10.1038/bjc.2016.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barquinero J.F., Barrios L., Caballín M.R., Miró R., Ribas M., Subias A., Egozcue J. Establishment and validation of a dose-effect curve for γ-rays by Cytogenetic analysis. Mutat. Res. Fund Mol. Mech. Mutagen. 1995;326(1):65–69. doi: 10.1016/0027-5107(94)00150-4. [DOI] [PubMed] [Google Scholar]
- 45.Dainiak N., Albanese J., Kaushik M., Balajee A.S., Romanyukha A., Sharp T.J., Blakely W.F. Concepts of operations for a us dosimetry and biodosimetry network. Radiat. Protect. Dosim. 2019;186(1):130–138. doi: 10.1093/rpd/ncy294. [DOI] [PubMed] [Google Scholar]
- 46.Kulka U., Ainsbury L., Atkinson M., Barquinero J.F., Barrios L., Beinke C., Bognar G., Cucu A., Darroudi F., Fattibene P., Gil O. Realising the European network of biodosimetry (RENEB) Radiat. Protect. Dosim. 2012;151(4):621–625. doi: 10.1093/rpd/ncs157. [DOI] [PubMed] [Google Scholar]
- 47.Blakely W.F., Carr Z., Chu M.C.M., Dayal-Drager R., Fujimoto K., Hopmeir M., Kulka U., Lillis-Hearne P., Livingston G.K., Lloyd D.C., Maznyk N. WHO 1st consultation on the development of a global biodosimetry laboratories network for radiation emergencies (BioDoseNet) Radiat. Res. 2009;171(1):127–139. doi: 10.1667/RR1549.1. [DOI] [PubMed] [Google Scholar]
- 48.Chaurasia R.K., Shirsath K.B., Desai U.N., Bhat N.N., Sapra B.K. Establishment of in vitro calibration curve for 60Co-γ-rays induced phospho-53BP1 foci, rapid biodosimetry and initial triage, and comparative evaluations with γH2AX and cytogenetic assays. Front. Public Health. 2022;10 doi: 10.3389/fpubh.2022.845200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chaurasia R.K., Bhat N.N., Gaur N., Shirsath K.B., Desai U.N., Sapra B.K. Establishment and multiparametric-cytogenetic validation of 60Co-gamma-ray induced, phospho-gamma-H2AX calibration curve for rapid biodosimetry and triage management during radiological emergencies. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2021;866 doi: 10.1016/j.mrgentox.2021.503354. [DOI] [PubMed] [Google Scholar]
- 50.Narayanasamy G., Saenz D., Cruz W., Ha C.S., Papanikolaou N., Stathakis S. Commissioning an Elekta versa HD linear accelerator. J. Appl. Clin. Med. Phys. 2016;17(1):179–191. doi: 10.1120/jacmp.v17i1.5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Greinert R., Detzler E., Volkmer B., Harder D. Kinetics of the formation of chromosome aberrations in X-irradiated human lymphocytes: analysis by premature chromosome condensation with delayed fusion. Radiat. Res. 1995;144(2):190–197. [PubMed] [Google Scholar]
- 52.Darroudi F., Fomina J., Meijers M., Natarajan A.T. Kinetics of the formation of chromosome aberrations in X-irradiated human lymphocytes, using PCC and FISH. Mutat. Res. Fund Mol. Mech. Mutagen. 1998;404(1–2):55–65. doi: 10.1016/s0027-5107(98)00095-5. [DOI] [PubMed] [Google Scholar]
- 53.International Organization for Standardization (ISO) ISO; Geneva: 2004. Radiation Protection–Performance Criteria for Service Laboratories Performing Biological Dosimetry by Cytogenetics; p. 2014. 19238. [Google Scholar]
- 54.Chaurasia R.K., Shirsath K.B., Sapra B.K. Protocol for one-step selective lysis of red blood cells and platelets with long-term preservation of white blood cells (human) at ambient temperature. STAR protocols. 2021;2(4) doi: 10.1016/j.xpro.2021.100834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chaurasia R.K., Balakrishnan S., Kunwar A., Yadav U., Bhat N., Anjaria K., Nairy R., Sapra B.K., Jain V.K., Priyadarsini K.I. Cyto-genotoxicity assessment of potential radioprotector, 3, 3′-diselenodipropionic acid (DSePA) in Chinese Hamster Ovary (CHO) cells and human peripheral blood lymphocytes. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2014;774:8–16. doi: 10.1016/j.mrgentox.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 56.Ainsbury E.A., Lloyd D.C. Dose estimation software for radiation biodosimetry. Health Phys. 2010;98(2):290–295. doi: 10.1097/01.HP.0000346305.84577.b4. [DOI] [PubMed] [Google Scholar]
- 57.Szłuińska M., Edwards A.A., Lloyd D.C. Health Protection Agency, Centre for Radiation, Chemical and Environmental Hazards, Radiation Protection Division; 2005. Statistical Methods for Biological Dosimetry. [Google Scholar]
- 58.Fenech M., Kirsch-Volders M., Rossnerova A., Sram R., Romm H., Bolognesi C., Ramakumar A., Soussaline F., Schunck C., Elhajouji A., Anwar W. HUMN project initiative and review of validation, quality control and prospects for further development of automated micronucleus assays using image cytometry systems. Int. J. Hyg Environ. Health. 2013;216(5):541–552. doi: 10.1016/j.ijheh.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 59.Jones C.G. A review of the history of US radiation protection regulations, recommendations, and standards. Health Phys. 2005;88(6):697–716. [PubMed] [Google Scholar]
- 60.Gianfaldoni S., Gianfaldoni R., Wollina U., Lotti J., Tchernev G., Lotti T. An overview on radiotherapy: from its history to its current applications in dermatology. Open access Macedonian journal of medical sciences. 2017;5(4):521. doi: 10.3889/oamjms.2017.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Reisz J.A., Bansal N., Qian J., Zhao W., Furdui C.M. Effects of ionizing radiation on biological molecules—mechanisms of damage and emerging methods of detection. Antioxidants Redox Signal. 2014;21(2):260–292. doi: 10.1089/ars.2013.5489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Evans H.J., Buckton K.E., Hamilton G.E., Carothers A. Radiation-induced chromosome aberrations in nuclear-dockyard workers. Nature. 1979;277(5697):531–534. doi: 10.1038/277531a0. [DOI] [PubMed] [Google Scholar]
- 63.Swartz H.M., Williams B.B., Flood A.B. Overview of the principles and practice of biodosimetry. Radiat. Environ. Biophys. 2014;53(2):221–232. doi: 10.1007/s00411-014-0522-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Blakely W.F., Port M., Abend M. Early-response multiple-parameter biodosimetry and dosimetry: risk predictions. J. Radiol. Prot. 2021;41(4):R152. doi: 10.1088/1361-6498/ac15df. [DOI] [PubMed] [Google Scholar]
- 65.Swartz H.M., Flood A.B., Gougelet R.M., Rea M.E., Nicolalde R.J., Williams B.B. A critical assessment of biodosimetry methods for large-scale incidents. Health Phys. 2010;98(2):95. doi: 10.1097/HP.0b013e3181b8cffd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satyamitra M.M., DiCarlo A.L., Hollingsworth B.A., Winters T.A., Taliaferro L.P. 2022. Development of Biomarkers for Radiation Biodosimetry and Medical Countermeasures Research: Current Status, Utility, and Regulatory Pathways. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vorobiev A.I. Acute radiation disease and biological dosimetry in 1993. Stem Cell. 1997;15(S1):269–274. doi: 10.1002/stem.5530150736. [DOI] [PubMed] [Google Scholar]
- 68.Romm H., Oestreicher U., Kulka U. Cytogenetic damage analysed by the dicentric assay. Ann. Ist. Super Sanita. 2009;45(3):251–259. [PubMed] [Google Scholar]
- 69.Bauchinger M. Quantification of low-level radiation exposure by conventional chromosome aberration analysis. Mutat. Res. Rev. Genet. Toxicol. 1995;339(3):177–189. doi: 10.1016/0165-1110(95)90010-1. [DOI] [PubMed] [Google Scholar]
- 70.Shi L., Fujioka K., Sakurai-Ozato N., Fukumoto W., Satoh K., Sun J., Awazu A., Tanaka K., Ishida M., Ishida T., Nakano Y. Chromosomal abnormalities in human lymphocytes after computed tomography scan procedure. Radiat. Res. 2018;190(4):424–432. doi: 10.1667/RR14976.1. [DOI] [PubMed] [Google Scholar]
- 71.Vaurijoux A., Gruel G., Pouzoulet F., Grégoire E., Martin C., Roch-Lefèvre S., Voisin P., Voisin P., Roy L. Strategy for population triage based on dicentric analysis. Radiat. Res. 2009;171(5):541–548. doi: 10.1667/RR1664.1. [DOI] [PubMed] [Google Scholar]
- 72.Beinke C., Braselmann H., Meineke V. Establishment of an x-ray standard calibration curve by conventional dicentric analysis as prerequisite for accurate radiation dose assessment. Health Phys. 2010;98(2):261–268. doi: 10.1097/HP.0b013e3181b35a53. [DOI] [PubMed] [Google Scholar]
- 73.Oliveira M., Einbeck J., Higueras M., Ainsbury E., Puig P., Rothkamm K. Zero‐inflated regression models for radiation‐induced chromosome aberration data: a comparative study. Biom. J. 2016;58(2):259–279. doi: 10.1002/bimj.201400233. [DOI] [PubMed] [Google Scholar]
- 74.Baeyens A., Swanson R., Herd O., Ainsbury E., Mabhengu T., Willem P., Thierens H., Slabbert J.P., Vral A. A semi-automated micronucleus-centromere assay to assess low-dose radiation exposure in human lymphocytes. Int. J. Radiat. Biol. 2011;87(9):923–931. doi: 10.3109/09553002.2011.577508. [DOI] [PubMed] [Google Scholar]
- 75.Lloyd D.C., Purrott R.J., Dolphin G.W., Bolton D., Edwards A.A., Corp M.J. The relationship between chromosome aberrations and low LET radiation dose to human lymphocytes. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 1975;28(1):75–90. doi: 10.1080/09553007514550781. [DOI] [PubMed] [Google Scholar]
- 76.Lloyd D.C., Edwards A.A., Prosser J.S. Chromosome aberrations induced in human lymphocytes by in vitro acute X and gamma radiation. Radiat. Protect. Dosim. 1986;15(2):83–88. [Google Scholar]
- 77.Prasanna P.G.S., Loats H., Gerstenberg H.M., Torres B.N., Shehata C.W. ARMED FORCES RADIOBIOLOGY RESEARCH INST; BETHESDA MD: 2002. AFRRI's Gamma-Ray, X-Ray, and Fission-Neutron Calibration Curves for the Lymphocyte Dicentric Assay: Application of a Metaphase Finder System. [Google Scholar]
- 78.Rawojć K., Tarnawska D.M., Miszczyk J.U., Swakoń J., Stolarczyk L., Rydygier M. Application of the micronucleus assay performed by different scorers in case of large-scale radiation accidents. Nukleonika. 2015;60(3):643–649. part 2) [Google Scholar]
- 79.Thierens H., Vral A. The micronucleus assay in radiation accidents. Annali Dell Istituto Superiore Di Sanita. 2009;45(3):260–264. [PubMed] [Google Scholar]
- 80.Bonassi S., El-Zein R., Bolognesi C., Fenech M. Micronuclei frequency in peripheral blood lymphocytes and cancer risk: evidence from human studies. Mutagenesis. 2011;26(1):93–100. doi: 10.1093/mutage/geq075. [DOI] [PubMed] [Google Scholar]
- 81.Sullivan J.M., Prasanna P.G., Grace M.B., Wathen L., Wallace R.L., Koerner J.F., Coleman C.N. Assessment of biodosimetry methods for a mass-casualty radiological incident: medical response and management considerations. Health Phys. 2013;105(6) doi: 10.1097/HP.0b013e31829cf221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Carvalho Cavalcante D.N., do Amaral Crispim B., Grisolia B.B., Viana L.F., Maran N.H., Solórzano J.C.J., de Oliveira K.M.P., Barufatti A. Effects of age, sex, medication, and environmental conditions on genetic alterations in oral mucosa cells. Heliyon. 2019;5(6) doi: 10.1016/j.heliyon.2019.e01840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bonassi S., Coskun E., Ceppi M., Lando C., Bolognesi C., Burgaz S., Holland N., Kirsh-Volders M., Knasmueller S., Zeiger E., Carnesoltas D. The HUman MicroNucleus project on eXfoLiated buccal cells (HUMNXL): the role of life-style, host factors, occupational exposures, health status, and assay protocol. Mutat. Res. Rev. Mutat. Res. 2011;728(3):88–97. doi: 10.1016/j.mrrev.2011.06.005. [DOI] [PubMed] [Google Scholar]
- 84.Rehm J., Marmet S., Anderson P., Gual A., Kraus L., Nutt D.J., Room R., Samokhvalov A.V., Scafato E., Trapencieris M., Wiers R.W. Defining substance use disorders: do we really need more than heavy use? Alcohol Alcohol. 2013;48(6):633–640. doi: 10.1093/alcalc/agt127. [DOI] [PubMed] [Google Scholar]
- 85.Wang Q., Rodrigues M.A., Repin M., Pampou S., Beaton-Green L.A., Perrier J., Garty G., Brenner D.J., Turner H.C., Wilkins R.C. Automated triage radiation biodosimetry: integrating imaging flow cytometry with high-throughput robotics to perform the cytokinesis-block micronucleus assay. Radiat. Res. 2019;191(4):342–351. doi: 10.1667/RR15243.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alexander G.A., Swartz H.M., Amundson S.A., Blakely W.F., Buddemeier B., Gallez B., Dainiak N., Goans R.E., Hayes R.B., Lowry P.C., Noska M.A. BiodosEPR-2006 Meeting: acute dosimetry consensus committee recommendations on biodosimetry applications in events involving uses of radiation by terrorists and radiation accidents. Radiat. Meas. 2007;42(6–7):972–996. [Google Scholar]
- 87.Abend M., Blakely W.F., Ostheim P., Schüle S., Port M. Early molecular markers for retrospective biodosimetry and prediction of acute health effects. J. Radiol. Prot. 2022;42(1) doi: 10.1088/1361-6498/ac2434. [DOI] [PubMed] [Google Scholar]
- 88.González A.J. An international perspective on radiological threats and the need for retrospective biological dosimetry of acute radiation overexposures. Radiat. Meas. 2007;42(6–7):1053–1062. [Google Scholar]
- 89.Howard M., Beltran C., Sarkaria J., Herman M.G. Characterization of relative biological effectiveness for conventional radiation therapy: a comparison of clinical 6 MV X-rays and 137Cs. J. Radiat. Res. 2017;58(5):608–613. doi: 10.1093/jrr/rrx018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang Q., Wang X., Sun Q., Jin Y., Li Y., Li Z., Sun T., Wang L. Investigation and application of high megavoltage X-ray imaging mode in radiotherapy. Int. J. Med. Phys. Clin. Eng. Radiat. Oncol. 2016;5(1):42–50. [Google Scholar]
- 91.Ferlay J., Ervik M., Lam F., Colombet M., Mery L., Piñeros M., Znaor A., Soerjomataram I., Bray F. Lyon International Agency Res. Cancer; 2020. Observatory:“Cancer Today”. [Google Scholar]
- 92.de Martel C., Georges D., Bray F., Ferlay J., Clifford G.M. Global burden of cancer attributable to infections in 2018: a worldwide incidence analysis. Lancet Global Health. 2020;8(2):e180–e190. doi: 10.1016/S2214-109X(19)30488-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. material/referenced in article.