Abstract
Objective
N4-acetylcytidine (ac4C) acetylation can promote target gene expression through improved mRNA stability. To explore the role of ac4C acetylation in osteosarcoma, U2OS and MG63 cell lines were treated with the N-acetyltransferase 10 (NAT10) inhibitor Remodelin. Reverse transcription–polymerase chain reaction (RT-PCR) and Western blot were used to test the gene and protein expression efficiency.
Methods
The proliferation rate of osteosarcoma cells was measured by a cell counting kit-8 (CCK8) assay. The cell cycle and apoptosis were analyzed by flow cytometry. The invasiveness of osteosarcoma cells was detected by a transwell invasion assay. The ac4C acetylation of target genes was screened by acetylated RNA immunoprecipitation and sequencing (acRIP-seq).
Results
We found that when osteosarcoma cells were treated with Remodelin at the optimal concentration, their NAT10 expression and the cell proliferation was inhibited, the cells in the G1 phase increased (P < 0.05) but those in the S phase decreased, the apoptotic cells in the early and late stages increased, and the cells invasiveness decreased (P < 0.05).
Conclusions
The farnesyltransferase subunit beta gene (FNTB) was identified by acRIP-seq as one of the target genes of ac4C acetylation and was further verified by RT-PCR and Western blot analyses. Remodelin was demonstrated to reduce the stability and protein translation efficiency of target gene mRNA in osteosarcoma cells. In conclusion, inhibition of ac4C acetylation in osteosarcoma can block proliferation and metastasis as well as promote apoptosis and cell cycle arrest. Ac4C acetylation contributes to the stability and protein translation efficiency of the downstream target gene mRNA.
Keywords: ac4C acetylation, NAT10, Osteosarcoma, acRIP-seq, FNTB gene
1. Introduction
As one of the most common primary bone tumors, osteosarcoma is highly malignant. Its drug resistance, rapid invasion, and other biological characteristics will ultimately lead to a poor prognosis. When surgery combined with chemotherapy is used as the primary treatment plan, the five-year survival rate of patients with osteosarcoma is only 50–60% [1]. At present, chemotherapy incorporating cisplatin, doxorubicin, methotrexate, and cyclophosphamide often cause serious side effects and leads to chemotherapy resistance [2]. In the current era of tumor-targeted therapy, there is still no ideal tumor-specific target for osteosarcoma, which greatly hinders its disease management. Furthermore, after most patients with osteosarcoma receiving neoadjuvant chemotherapy, the resected tumors frequently undergo transformation from severe necrosis to ossification [3]. Hence, the discovery of alternative treatment options is critical. Recent research has found that noncoding RNA is closely involved in regulating the occurrence and development of osteosarcoma; therefore, it has become an important marker and target for the diagnosis and treatment of osteosarcoma [[4], [5], [6]]. Further research exploring the molecular mechanism of osteosarcoma regulation is thus essential for developing a new targeted therapy strategy.
One of the most active research areas in biology in recent years has involved the elucidation of the structure and function of RNA via post-transcriptional modification, including N6-methyladenosine (m6A), 5-methylcytosine (m5C), N1-methyladenosine (m1A), and 7-methylguanosine (m7G) in eukaryotic mRNA. In addition, an increase of number studies have reported that post-transcriptional acetylation of mRNA exhibits dynamic characteristics and takes part in a variety of RNA regulatory processes [7,8]. At present, N4-acetylcytidine (ac4C) RNA acetylation, which is a new type of RNA acetylation, is another hot spot of epigenomics research after m6A methylation [9,10]. Ac4C RNA acetylation is a conservative chemical modification that is under the action of an RNA ac4C-modified enzyme. Early studies have revealed that this acetylation occurs on the serine and leucine residues of tRNA and 18SrRNA in eukaryotes, resulting in increased thermal stability of Watson–Crick base coordination with guanosine and coding accuracy of regulation in protein synthesis. Recent studies have demonstrated that ac4C is widely distributed in the human transcriptome, with the majority of instances occurring in the coding sequence (CDS), and promotes target gene expression through improved mRNA stability and translation [11,12]. The latest research shows that ac4C acetylation is closely related to a variety of tumors [13,14]. However, its role in osteosarcoma has not been studied. N-acetyltransferase 10 (NAT10) is a member of the N-terminal acetyltransferase superfamily associated with the acetyltransferase general control nonrepressed 5 protein. Studies have confirmed that NAT10-catalyzed nonhistone acetylation is widely involved in physiological and pathophysiological processes, including telomerase activity, rRNA synthesis, DNA damage repair, cell cycle, maintenance of mRNA stability, and regulation of translation efficiency, and is closely related to cancer and nuclear laminopathy. Therefore, the identification of new targets for understanding the pathogenesis and treatment of the aforementioned conditions [[15], [16], [17]] has become an important research topic. The structure of NAT10 mainly includes an acetyltransferase domain, tRNA-binding domain, and RNA helicase domain [18,19].
Recent studies have found that ac4C, including tRNA, rRNA, and mRNA, are all catalyzed by NAT10, which requires acetyl-coenzyme A to provide acetyl and ATP/GTP hydrolysis to provide energy [20,21]. In the present study, we found that downregulation of NAT10 in osteosarcoma cells can inhibit the acetylation of ac4C in osteosarcoma and further affect the biological behavior of osteosarcoma cells. To the best of our knowledge, this is the first study to investigate the mechanism and target of ac4C acetylation in osteosarcoma cells and to provide a new theoretical basis for the mechanism underlying the occurrence and progression of osteosarcoma and the development of targeted therapies.
2. Materials and methods
Cell line and culture. The human osteosarcoma cell lines MG63 and U2OS were purchased from the China Center for Type Culture Collection (Wuhan, China). Cells were cultured in RPMI 1640 medium (KeyGEN Biotech, China) supplemented with 10% fetal bovine serum (Gibco, USA). Cells were maintained at 37 °C in a humidified incubator containing 5% CO2.
Main reagents. NAT10 antibodies, Remodelin hydrobromide, actinomycin D (ActD), and MG132 were purchased from (MCE, China); related target gene antibodies were provided by Shanghai Jikai Genetics Co., Ltd. The routine reagents were provided by our laboratory. Matrigel and Transwell chambers were bought from BD, Inc. (Franklin, USA). Acetylated RNA sequencing was provided by Shanghai Jikai Genetics Co., Ltd.
Drug sensitivity test and concentration gradient test of the NAT10 inhibitor Remodelin. Our pilot study showed that the expression levels of the NAT10 gene in the MG63 and U2OS osteosarcoma cell lines were high and consistent. Remodelin is an effective and specific NAT10 inhibitor [22]. In accordance with the 48-h half maximal inhibitory concentration (IC50) cytotoxicity test and the cell counting kit-8 (CCK8) assay with the NAT10-specific inhibitor Remodelin, three concentrations of Remodelin were chosen to pretreat MG63 and U2OS cells, respectively. Western blot analysis was used to verify the inhibition efficiency of different concentrations of Remodelin on NAT10 in osteosarcoma in order to determine the optimal inhibition concentration.
Western blot. MG63 and U2OS cells were treated with the optimal concentration of Remodelin for 48 h, and the expression of NAT10 was detected by Western blot. MG63 and U2OS cells were lysed in cold radioimmunoprecipitation assay lysis buffer containing protease and phosphatase inhibitors. The concentration of extracted protein was measured by a BCA Protein Assay Kit. Cell lysates were separated by Bis-Tris protein gels and transferred to methanol-soaked polyvinylidene difluoride membranes, which were blocked by 5% nonfat milk, and immunoblotted with the appropriate primary antibodies and horseradish peroxidase-conjugated secondary antibodies. An enhanced chemiluminescence detection system was used for protein band visualization. Tris-buffered saline with 0.1% Tween-20 was used as the washing buffer for the entire procedure.
CCK8 assay. The two groups of cells in the logarithmic phase of growth were digested with trypsin, and then the cells were resuspended in complete culture medium and counted. The cell density was determined by the rate of cell growth (usually 4000 cells/well and 100 μL per well). The assay was repeated in 3–5 wells for each group, and the number of plates was determined according to the experimental design. In the experimental group, 500 μM Remodelin was added to the cells. There was no need to change the liquid after the addition of 10 μL of CCK8 reagent to the well at 2–4 h before culture termination. The cell inhibition rate was calculated after incubation for 48 h.
Cell cycle and apoptosis analyses. An apoptosis assay was performed. According to the manufacturer's instructions, apoptotic cells in each group were analyzed using an Annexin V-FITC/propidium iodide (PI) apoptosis double staining kit. The DNA content of the cells was analyzed by flow cytometry, and the sub-G1 population was considered to represent apoptotic cells. The effect of NAT10 on the proliferation of MG63 and U2OS cells was studied using cell cycle analysis. Cells in each group were washed twice with phosphate-buffered saline (PBS) and fixed with 75% cold ethanol before being stored at 4 °C for 24 h. The cells were washed with PBS after removing the ethanol, followed by incubation for 30 min at 37 °C in PBS containing 0.5 mL of 50 mg/mL PI, 1 mL EDTA, 0.1% Triton X-100, and 1 mg/mL RNase A. The stained cells were finally analyzed by flow cytometry. The experiment was repeated in triplicate.
Transwell invasion assay to detect the effect of ac4C acetylation on the invasiveness of osteosarcoma cells. The necessary number of chambers were transferred to a new 24-well plate and placed in an aseptic console to return them to room temperature. The Matrigel matrix layer was rehydrated by filling the upper and lower chambers with 500 μL of serum-free medium and placing it in an incubator at 37 °C for 2 h. The serum-free cell suspension was prepared. Following rehydration of the Matrigel matrix layer, all chambers were transferred to a new orifice plate, the upper chamber culture medium was carefully removed, 500 μL of cell suspension was added, and the cubicle was turned upside down on absorbent paper to remove the culture medium. The noninvasive cells in the chamber were gently removed with a cotton swab. The chamber was soaked and washed several times before being air-dried after dripping 2–3 drops of Giemsa staining solution onto the lower surface of the membrane, and the cells were stained for 35 min. For microscopic imaging, the field of view was randomly selected in each chamber, and four photos at 100 × magnification and nine photos at 200 × magnification were taken. The cells in the photos at 200 × magnification were counted, and the data were analyzed to compare the difference in cell invasion ability between the experimental and the control groups. The experiment was repeated in triplicate.
RNA-seq analysis. Total RNA from osteosarcoma cells with or without Remodelin treatment was isolated using TRIzol reagent. Poly(A)+ mRNA was subsequently purified from total RNA via a Dynabeads mRNA Purification Kit (61,006, Invitrogen) and then sequenced on an Illumina NovaSeq 6000 platform by Epibiotek (Guangzhou, China). The reads were mapped to human genome version 38 (GRCh38) with HISAT2 (v2.1.0) and then calculated using HT-seq (v0.7.2). Genes with a P value < 0.05 and fold change >1.2 were considered to be differentially expressed. The online tool DAVID (www.david.niaid.nih.gov) was used to perform GO enrichment and KEGG pathway analyses [23].
AcRIP-seq analysis. AcRIP-seq and subsequent data analyses were mainly supported by Epibiotek (Guangzhou, China). Total RNA from osteosarcoma cells with Remodelin treatment or control osteosarcoma cells was extracted using TRIzol reagent, digested with DNase I and fragmented into 100–200 nt oligonucleotides by RNA fragmentation reagents (ab252215, Ambion). After saving 50 ng of fragmented RNA as the input, the remainder (150 μg) was used for RNA immunoprecipitation with ac4C antibody (Abcam). Ac4C RNAs were immunoprecipitated with Dynabeads Protein G (10004D, Invitrogen) and recovered with HiPure cell miRNA (R4311-03, Magen, Guangzhou, China). Ribosomal RNA was removed from input RNA and ac4C-enriched RNA samples. RNA sequencing libraries for input RNA (RNA-seq) and ac4C-enriched RNA (acRIP-seq) were simultaneously constructed with the EpiTM mini longRNA-seq kit (E1802, Epibiotek) and then deep sequenced on the Illumina NovaSeq 6000 using PE150 strategy with two independent biological replicates. Reads were aligned to the human genome GRCh38/hg38 with HISAT2 software (v2.1.0). Then, ac4C peaks were identified using the ExomePeak R package (v2.13.2). The differential ac4C peaks with fold change >2.0 and P value < 0.05 were selected [24].
Effect of ac4C acetylation on the stability of the target gene mRNA. ActD is an inhibitor of RNA synthesis that can be used to judge the stability of the synthesized RNA by inhibiting the synthesis of new RNA. To explore the effect of ac4C acetylation on the stability of mRNA, we treated two types of osteosarcoma cells with ActD for 10 min, 30 min, and 60 min. The relative expression of target gene mRNA was measured by RT-PCR to reflect the effect of ac4C acetylation on the mRNA stability.
Effect of ac4C acetylation on target gene mRNA translation efficiency. MG132 is an inhibitor of proteasome degradation that can be used to inhibit the degradation of translated proteins by blocking the proteasome degradation pathway and to judge the efficiency of new protein translation synthesis. MG132 was incubated with osteosarcoma cells for 6 h and 18 h, respectively, after Remodelin treatment. The protein expression of target gene mRNA was measured by Western blot, and the expression efficiency of the target gene protein in the control group and experimental group was calculated.
Statistical analysis. SPSS version 13.0 for Windows was utilized for data analysis. All results were presented as the mean ± standard deviation. One-way analysis of variance, followed by Dunnett's t-test and Student's t-test, was used to examine the statistical difference in the experimental data between groups. P < 0.05 was considered statistically significant.
3. Results
Screening the optimal concentration of Remodelin for NAT10 inhibition by the 48-h chemosensitivity test and concentration gradient experiment. Based on the IC50 test and CCK8 results, we chose 3.96025 μM, 31.25 μM, and 500 μM as the concentrations of the NAT10-specific inhibitor Remodelin for the 48-h chemosensitivity test in tumor cells (Fig. 1A and B). In U2OS and MG63 osteosarcoma cells, the concentration of Remodelin with the greatest inhibitory effect on NAT10 was 500 μM (P < 0.05) (Fig. 1C and D).
Fig. 1.
Screening the best concentration of Remodelin for NAT10 inhibition by the 48-h chemosensitivity test and concentration gradient experiment. (A) The 48-h drug sensitivity test of Remodelin in U2OS osteosarcoma cells. (B) The 48-h drug sensitivity test of Remodelin in MG63 osteosarcoma cells. (C) At a concentration of 500 μM, the inhibition rate of U2OS osteosarcoma cells was the highest (P < 0.05). (D) At a concentration of 500 μM, the inhibition rate of MG63 osteosarcoma cells was the highest (P < 0.05).
Western blot analysis verifies the inhibitory effect of Remodelin on NAT10 in osteosarcoma cells. U2OS and MG63 osteosarcoma cells were grown in the presence of 500 g/mL Remodelin. Compared to the control group, the expression of NAT10 protein was significantly reduced in the experimental group (P < 0.05) (Fig. 2A and B).
Fig. 2.
Western blot analysis verified the inhibitory effect of Remodelin on NAT10 in osteosarcoma cells. (A) The expression of NAT10 protein in U2OS osteosarcoma cells decreased significantly after the addition of 500 μM Remodelin (P < 0.05). (B) The expression of NAT10 protein in MG63 osteosarcoma cells decreased significantly after the addition of 500 μM Remodelin (P < 0.05).
Effects of ac4C acetylation on the proliferation of osteosarcoma cells. A CCK8 proliferation assay was used to determine the effect of Remodelin on the proliferation of U2OS and MG63 osteosarcoma cells. After the addition of CCK8 reagents, the proliferation of U2OS and MG63 osteosarcoma cells was inhibited by 33.9% and 29.8%, respectively. As a result, compared with the control group, the proliferation of cells in the Remodelin treatment group was significantly inhibited (P < 0.05) (Fig. 3A and B).
Fig. 3.
Effect of ac4C acetylation on osteosarcoma cell proliferation. (A) The proliferation of U2OS osteosarcoma cells was significantly inhibited by the addition of Remodelin. (B) The proliferation of MG63 osteosarcoma cells was significantly inhibited by the addition of Remodelin.
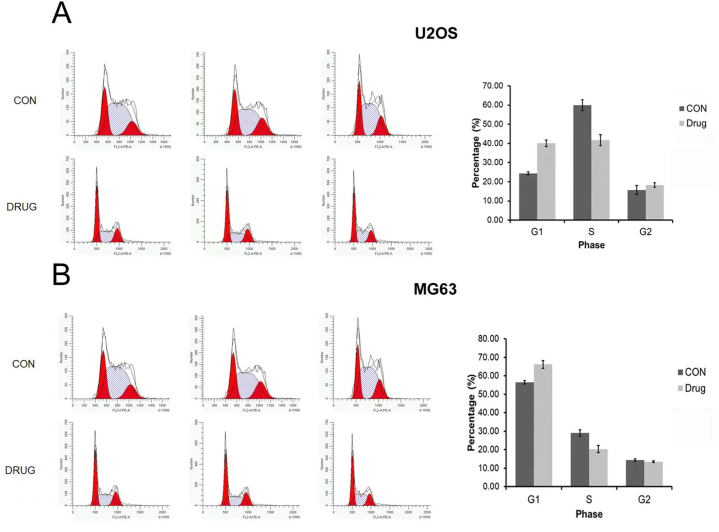
Effects of ac4C acetylation on the cell cycle and apoptosis of osteosarcoma cells. To better understand the effect of ac4C acetylation on osteosarcoma, we measured the cell cycle of osteosarcoma cells in the control and experimental groups. The results showed that compared with the control group, in the treatment group, the proportions of U2OS and MG63 cells in the G1 phase were increased, had no significant change in the G2 phase, and were decreased in the S phase, suggesting that the main reason for the inhibition of osteosarcoma cell proliferation after ac4C acetylation inhibition is that the cell cycle stagnated in the G1 phase (Fig. 4A and B). The results of the Annexin V-FITC/PI apoptosis double staining assay revealed that the early apoptosis and late apoptosis of osteosarcoma cells were significantly increased following the inhibition of ac4C acetylation in the treatment group (P < 0.05) (Fig. 5A and B).
Fig. 4.
Effect of ac4C acetylation on the cell cycle of osteosarcoma cells. (A) In the treatment group, the number of U2OS cells in the G1 phase was increased, the number of cells in the G2 phase had no significant change, and the number of cells in the S phase was decreased (P < 0.05). (B) In the treatment group, the number of MG63 cells in the G1 phase was increased, the number of cells in the G2 phase had no significant change, and the number of cells in the S phase was decreased (P < 0.05).
Fig. 5.
Effect of ac4C acetylation on the apoptosis of osteosarcoma cells. (A) The early apoptosis and late apoptosis of U2OS osteosarcoma cells were significantly increased after inhibition of ac4C acetylation in the treatment group. (B) The early apoptosis and late apoptosis of MG63 osteosarcoma cells were significantly increased after inhibition of ac4C acetylation in the treatment group.
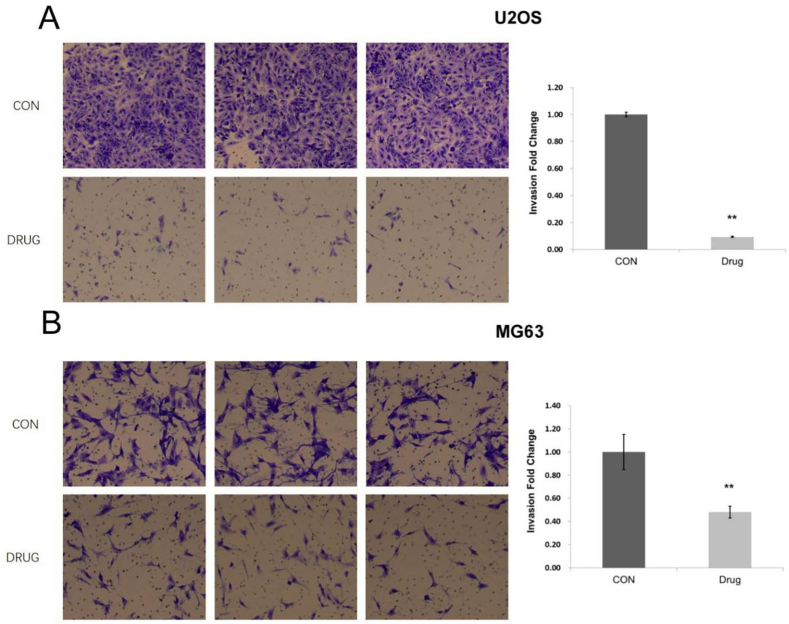
Effect of ac4C acetylation on the invasiveness of osteosarcoma cells. The invasiveness of cells in each group was detected by a Transwell invasion assay. According to the findings, the number of U2OS cells entering the invasion chamber in the treatment group was about 9% of that in the control group, while the number of MG63 cells entering the invasion chamber in the treatment group was about 48% of that in the control group. Compared with the control group, the invasiveness of osteosarcoma cells decreased significantly after ac4C acetylation inhibition in the treatment group (P < 0.05) (Fig. 6A and B).
Fig. 6.
Ac4C acetylation increases the invasiveness of osteosarcoma cells. (A) The invasiveness of U2OS osteosarcoma cells was significantly decreased after the inhibition of ac4C acetylation in the treatment group (P < 0.05). (B) The invasiveness of MG63 osteosarcoma cells was significantly decreased after the inhibition of ac4C acetylation in the treatment group (P < 0.05).
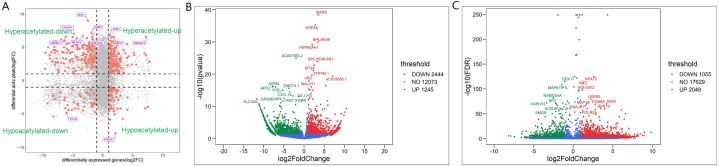
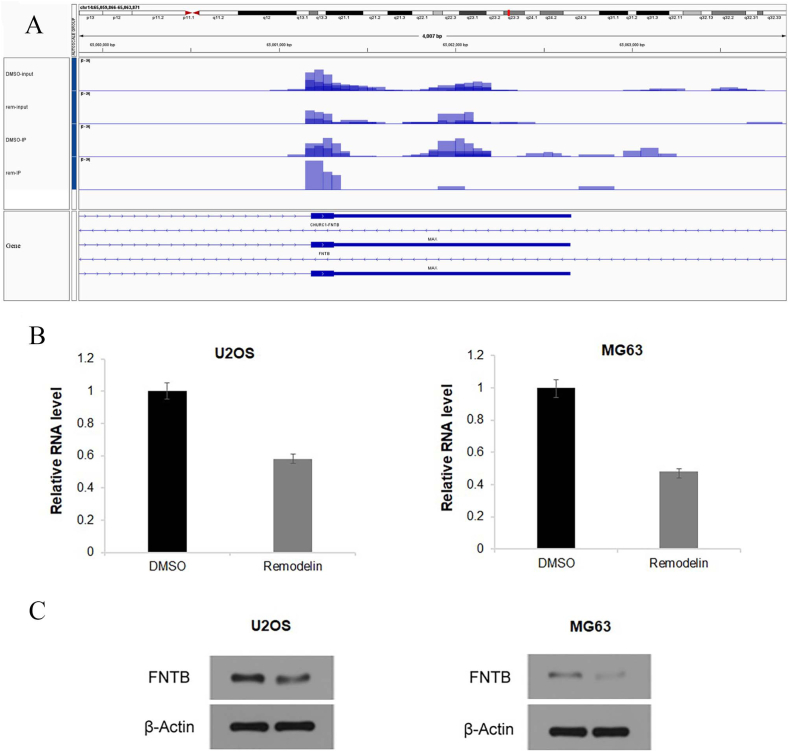
Screening of target genes related to ac4C acetylation by acRIP-seq and bioinformatics analyses. AcRIP-seq of osteosarcoma cells showed that there were 3621 different ac4C peaks between with and without Remodelin treatment, with the ac4C peaks primarily distributed in the mRNA CDS region (Fig. 7A and B). The ac4C-modified genes and genes with altered RNA expression levels were analyzed to identify key genes associated with disease phenotypes or experimental treatment conditions for future research. The association analysis was divided into four categories: hyperacetylated-down (downregulation of hyperacetylated RNA levels); hyperacetylated-up (upregulation of hyperacetylated RNA levels); hypoacetylated-down (downregulation of deacetylated RNA levels); and hypoacetylated-up (upregulation of deacetylated RNA levels). 10 most up regulated and 10 most down regulated genes were labeled (Fig. 8A–C). We identified five genes strongly associated with the downregulated deacetylated RNA levels: trafficking protein particle complex subunit 3 (TRAPPC3), B-TFIID TATA-box binding protein associated factor 1 (BTAF1), Rap guanine nucleotide exchange factor 6 (RAPGEF6), RAD9-HUS1-RAD1 interacting nuclear orphan 1 (RHNO1), and farnesyltransferase subunit beta (FNTB). RT-PCR and Western blot analyses revealed that FNTB is one of the ac4C acetylation target genes (Fig. 9A–C).
Fig. 7.
The acetylated RNA immunoprecipitation and sequencing results show that the ac4C peaks are mainly distributed in the CDS region of mRNA. (A) Metageneplot diagram of the ac4C peak distribution according to the mRNA structure. (B) Pie diagram of the ac4C peak distribution according to the RNA structure.
Fig. 8.
Analysis of genes related to ac4C acetylation. (A) There are four types of association analysis: hyperacetylated-down (downregulation of hyperacetylated RNA); hyperacetylated-up (upregulation of hyperacetylated RNA); hypoacetylated-down (downregulation of deacetylated RNA); and hypoacetylated-up (upregulation of deacetylated RNA). (B) Differential volcano diagram of ac4C acetylation. (C) Differential gene expression volcano map.
Fig. 9.
Screening and verification of the target gene FNTB for ac4C acetylation. (A) Integrative Genomics Viewer diagram showing that the peak value of ac4C acetylation of the FNTB gene in osteosarcoma cells decreased significantly after the addition of Remodelin. (B) RT-PCR results demonstrating that the expression of FNTB in U2OS and MG63 osteosarcoma cells decreased significantly after the addition of Remodelin (P < 0.05). (C) Western blots indicating that the expression of FNTB protein in U2OS and MG63 osteosarcoma cells decreased significantly after the addition of Remodelin (P < 0.05).
Effect of ac4C acetylation on the stability of FNTB mRNA in osteosarcoma cells. After the cells in the control group and experimental group were treated with ActD for 10 min, 30 min, and 60 min, respectively, we found that the mRNA level of FNTB in the control group remained stable. The mRNA decline rate of FNTB, however, significantly increased after Remodelin treatment, indicating that the mRNA stability of FNTB was compromised. As a result, inhibiting ac4C acetylation may significantly reduce the stability of FNTB mRNA in osteosarcoma cells (Fig. 10A and B).
Fig. 10.
Effect of ac4C acetylation on the stability of FNTB mRNA in osteosarcoma cells. (A) After treatment with Remodelin and ActD, the decrement rate of FNTB mRNA in U2OS osteosarcoma cells increased significantly. (B) After treatment with Remodelin and ActD, the decrement rate of FNTB mRNA in MG63 osteosarcoma cells increased significantly.
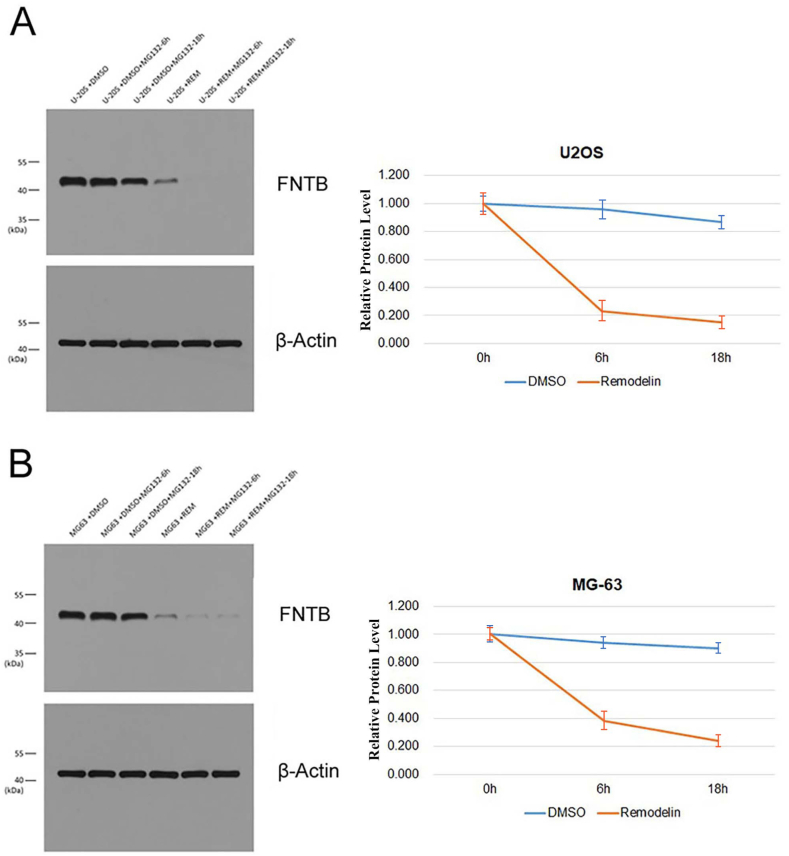
Effect of ac4C acetylation on FNTB translation efficiency in osteosarcoma cells. The cells in both the control and experimental groups were treated with MG132 to observe the effect of ac4C acetylation on the translation efficiency of FNTB. The FNTB protein in U2OS and MG63 cells was slowly decreased in the control group, but it was significantly faster in the Remodelin treatment group, indicating that the efficiency of new protein translation and synthesis was seriously compromised. Therefore, inhibition of ac4C acetylation could significantly inhibit the translation efficiency of the target gene FNTB (Fig. 11A and B).
Fig. 11.
Effect of ac4C acetylation on the FNTB translation efficiency in osteosarcoma cells. (A) After treatment with Remodelin and MG132, the degradation rate of FNTB protein in U2OS osteosarcoma cells increased significantly, indicating that the efficiency of protein synthesis decreased. (B) After treatment with Remodelin and MG132, the degradation rate of FNTB protein in MG63 osteosarcoma cells increased significantly, indicating that the efficiency of protein synthesis decreased.
4. Discussion
RNA acetylation is a new addition to the expanding list of RNA chemical modifications that can regulate the structure and function of RNA [25]. The presence of ac4C on tRNAs helps to improve the high fidelity of protein translation and preserve the heat tolerance of organisms. The presence of ac4C on rRNA is a trait of thermophilic organisms and is also essential for precise protein translation. Furthermore, ac4C acetylation of mRNA has different effects on the RNA structure and increases the mRNA stability and protein translation efficiency; thus, it has biological functions in normal development and disease progression. The detection of ac4C acetylation has clinical utility in tumor diagnosis, surgical treatment, and follow-up, and it is anticipated to become a new marker and a new research focus in the field of cancer treatment [[26], [27], [28], [29]]. However, there is a lack of research worldwide on its effect on the biological behavior of osteosarcoma.
For the first time, we investigated the effect of ac4C acetylation on the oncological properties of osteosarcoma cells in this study. The level of ac4C acetylation was downregulated using Remodelin to inhibit NAT10 expression, and the proliferation, migration, and metastasis of osteosarcoma cells were inhibited, indicating that ac4C acetylation can affect the occurrence and development of osteosarcoma. Furthermore, using acRIP-seq and bioinformatics analysis, we identified five gene fragments and found that ac4C acetylation mainly occurred in the CDS region of the downstream target mRNA. In our study, osteosarcoma cell U20S was analyzed by acRIP-seq, combined with RNA-seq and differential acetylation data analysis, including differential Peak analysis and differential ac4C gene GO/KEGG analysis and a series of bioinformatics correlation analysis, and the downstream target genes closely related to ac4C acetylation were screened [[30], [31], [32]]. Results of RNA-seq were combined our own results with those reported by some previous researchers. The FNTB gene was confirmed by RT-PCR and Western blot to be the downstream target gene of ac4C acetylation. Additionally, it was demonstrated that the mechanism of ac4C acetylation in osteosarcoma involves improvement of the stability and protein translation efficiency of the target gene's mRNA in osteosarcoma cells, which is consistent with the most recent literature reports from both Chinese and international sources [33,34].
An increasing number of studies have examined how ac4C and NAT10 regulate and affect the intrinsic and extrinsic characteristics of tumors. For example, Zhang et al. have discovered that NAT10 is essential for the processes of gastric cancer metastasis and epithelial-to-mesenchymal transition [35]. They also identified ac4C of the collagen type V alpha 1 chain (COL5A1) gene as a biomarker of gastric cancer metastasis promotion. In another study, NAT10 was found to be highly expressed in bladder cancer. In addition, acRIP-seq has demonstrated that ac4C modification on the target stimulates tumor proliferation, metastasis, and stemness in bladder cancer. NAT10 also has been shown to contribute to proliferation and tumorigenesis in vivo through its effect on the acetylation of downstream targets [36]. This study demonstrated that NAT10-mediated ac4C modification of target genes increases gene stability and translation during the progression of bladder cancer. To investigate the influence of ac4C-related mRNA modification patterns on the prognosis of hepatocellular carcinoma, researchers have developed an ac4C score model to represent ac4C-related mRNA modification patterns and stratified patients into high- and low-prognosis groups [37]. The high-prognosis group was associated with an advanced tumor stage, a higher TP53 mutation rate, a higher tumor stemness, more activated DNA-repair system pathways, a lower stromal score, a higher immune score, and higher infiltration of regulatory T cells. While patients in the low-prognosis group were associated with an abundance of memory CD4 T cells, a less-aggressive immune subtype, and a long-lasting therapeutic advantage. In a different bioinformatics study, Yang et al. examined the pan-cancer expression and correlations of NAT10 using Oncomine, PrognoScan, GEPIA2, and Kaplan–Meier Plotter [38]. They revealed that a high NAT10 expression level was a significant predictor of a poor prognosis for patients with adrenocortical carcinoma, head and neck squamous cell carcinoma, liver hepatocellular carcinoma, kidney renal papillary cell carcinoma, pheochromocytoma, or paraganglioma. In addition, significant positive correlations were observed between NAT10 expression and immune infiltrates. Collectively, these results confirm the central roles of ac4C mRNA modification in the progression of cancer.
In summary, a thorough understanding of the relationships between osteosarcoma characteristics and ac4C modification patterns will aid disease management and treatment. Increased levels of ac4C in urine, for example, have been linked to several different types of cancer [[39], [40], [41]]. Whether the ac4C variant of FNTB might be used to predict the progression of osteosarcoma is an intriguing question. On the other hand, our findings suggest that small-molecule inhibitors like Remodelin, which specifically targets NAT10, might one day be used to treat osteosarcoma.
5. Conclusion
In conclusion, this study confirmed that inhibiting ac4C acetylation reduces osteosarcoma proliferation and metastasis as well as promotes apoptosis and cell cycle arrest. Ac4C acetylation mainly occurs in the CDS region of the downstream target mRNA and plays a role by affecting the stability and protein translation efficiency of the downstream target gene mRNA.
Author contribution statement
Lei Fan conceived and designed the experiments.
Wenjie Zhang, Jia Gao, Xiaotong Zhang and Hui Mao performed the experiments.
Juan Wang and Lei Fan analyzed and interpreted the data.
Bin He and Yunhua Wang contributed reagents, materials, analysis tools or data.
Wenjie Zhang, Jia Gao and Lei Fan wrote the paper.
Wenjie Zhang drafted the article.
Jia Gao critically revised its important intellectual content.
Lei Fan made final approval of the version submitted.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Funding statement
The present study was supported by the Natural Science Foundation of Jiangsu Provincial Health Commission [grant No. Project M2020059 and No. Project LKM2022007].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Faisham W.I., Mat Saad A.Z., Alsaigh L.N., et al. Prognostic factors and survival rate of osteosarcoma: a single-institution study. Asia Pac. J. Clin. Oncol. 2017;13(2):e104–e110. doi: 10.1111/ajco.12346. [DOI] [PubMed] [Google Scholar]
- 2.Harrison D.J., Geller D.S., Gill J.D., et al. Current and future therapeutic approaches for osteosarcoma. Expert Rev. Anticancer Ther.: Janus. 2018;18(1):39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 3.Li B., Wang Z., Wu H., et al. Epigenetic regulation of CXCL12 plays a critical role in mediating tumor progression and the immune response in osteosarcoma. Cancer Res. 2018;78(14):3938–3953. doi: 10.1158/0008-5472.CAN-17-3801. [DOI] [PubMed] [Google Scholar]
- 4.Yuan T.Z., Wang J., Wei X.C., et al. MicroRNA-30a inhibits proliferation and metastasis of osteosarcoma cells by modulating autophagy. J. Biobased Mater. Bioenergy. 2016;10(4):265–271. [Google Scholar]
- 5.Fan L., Wu Q., Xing X., et al. MicroRNA-145 targets vascular endothelial growth factor and inhibits invasion and metastasis of osteosarcoma cells. Acta Biochim. Biophys. Sin. 2012;44(5):407–414. doi: 10.1093/abbs/gms019. [DOI] [PubMed] [Google Scholar]
- 6.Lin S., Shao N.N., Fan L., et al. Effect of MicroRNA-101 on proliferation and apoptosis of human osteosarcoma cells by targeting mTOR. J. Huazhong Univ. Sci. Technol. 2014;34(6):889–895. doi: 10.1007/s11596-014-1369-y. [DOI] [PubMed] [Google Scholar]
- 7.Boccaletto Pietro, Machnicka Magdalena A., Purta Elzbieta, et al. MODOMICS: a database of RNA acetylation pathways.2017 update. Nucleic Acids Res. 2018;46(D1):D303–D307. doi: 10.1093/nar/gkx1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haruehanroengra P., Zheng Y.Y., Zhou Y., et al. RNA acetylations and cancer. RNA Biol. 2020;17(11):1560–1575. doi: 10.1080/15476286.2020.1722449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karthiya R., Wasil S.M., Khandelia P. Emerging role of N4-acetylcytidine acetylation of RNA in gene regulation and cellular functions. Mol. Biol. Rep. 2020;47(11):9189–9199. doi: 10.1007/s11033-020-05963-w. [DOI] [PubMed] [Google Scholar]
- 10.Yang C., Wu T., Zhang J., et al. Prognostic and immunological role of mRNA AC4C regulator NAT10 in pan-cancer: new territory for cancer research? Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taniguchi Takaaki, Miyauchi Kenjyo, Sakaguchi Yuriko, et al. Acetate-dependent tRNA acetylation required for decoding fidelity in protein synthesis. Nat. Chem. Biol. 2018;14(11):1010–1020. doi: 10.1038/s41589-018-0119-z. [DOI] [PubMed] [Google Scholar]
- 12.Jin G., Xu M., Zou M., et al. The processing, gene regulation, biological functions, and clinical relevance of N4-acetylcytidine on RNA: a systematic review. Mol. Ther. Nucleic Acids. 2020;20:13–24. doi: 10.1016/j.omtn.2020.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arango D., Sturgill D., Alhusaini N., et al. Acetylation of cytidine in mRNA promotes translation efficiency. Cell. 2018;175(7):1872–1886. doi: 10.1016/j.cell.2018.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thalalla Gamage S., Sas-Chen A., Schwartz S., et al. Quantitative nucleotide resolution profiling of RNA cytidine acetylation by AC4C-seq. Nat. Protoc. 2021;16(4):2286–2307. doi: 10.1038/s41596-021-00501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Q., Liu X., Jin K., et al. NAT10 is upregulated in hepatocellular carcinoma and enhances mutant p53 activity. BMC Cancer. 2017;17(1):605. doi: 10.1186/s12885-017-3570-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liang P., Hu R., Liu Z., et al. NAT10 upregulation indicates a poor prognosis in acute myeloid leukemia. Curr. Probl. Cancer. 2020;44(2) doi: 10.1016/j.currproblcancer.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 17.Zhang Y., Jing Y., Wang Y., et al. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct. Targeted Ther. 2021;6(1):173. doi: 10.1038/s41392-021-00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sleiman S., Dragon F. Recent advances on the structure and function of RNA acetyltransferase Kre33/NAT10. Cells. 2019;8(9):1305. doi: 10.3390/cells8091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thalalla Gamage S., Sas-Chen A., Schwartz S., et al. Quantitative nucleotide resolution profiling of RNA cytidine acetylation by ac4C-seq. Nat. Protoc. 2021;16(4):2286–2307. doi: 10.1038/s41596-021-00501-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu Z., Xing X., Huang S., et al. NAT10 promotes osteogenic differentiation of mesenchymal stem cells by mediating N4-acetylcytidine acetylation of gremlin 1. Stem Cell. Int. 2021;2021 doi: 10.1155/2021/8833527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ikeuchi Y., Kitahara K., Suzuki T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008;27(16):2194–2203. doi: 10.1038/emboj.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shrimp J.H., Jing Y., Gamage S.T., et al. Remodelin is a cryptic assay interference chemotype that does not inhibit NAT10-dependent cytidine acetylation. ACS Med. Chem. Lett. 2020;12(6):887–892. doi: 10.1021/acsmedchemlett.0c00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Smyth G.K., Shi W. Feature Counts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Ashburner M., Ball C.A., Blake J.A., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ito S., Horikawa S., Suzuki T., et al. Human NAT10 is an ATP-dependent RNA acetyltransferase responsible for N4-acetylcytidine formation in 18 S ribosomal RNA (rRNA) J. Biol. Chem. 2014;289(52):35724–35730. doi: 10.1074/jbc.C114.602698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawai G., Hashizume T., Miyazawa T., et al. Conformational characteristics of 4-acetylcytidine found in tRNA. Nucleic Acids Symp. Ser. 1989;21:61–62. [PubMed] [Google Scholar]
- 27.Kumbhar B.V., Kamble A.D., Sonawane K.D. Conformational preferences of modified nucleoside N(4)-acetylcytidine, ac4C occur at "wobble" 34th position in the anticodon loop of tRNA. Cell Biochem. Biophys. 2013;66(3):797–816. doi: 10.1007/s12013-013-9525-8. [DOI] [PubMed] [Google Scholar]
- 28.Sharma S., Langhendries J.L., Watzinger P., et al. Yeast Kre33 and human NAT10 are conserved 18S rRNA cytosine acetyltransferases that modify tRNAs assisted by the adaptor Tan1/THUMPD1. Nucleic Acids Res. 2015;43(4):2242–2258. doi: 10.1093/nar/gkv075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orita I., Futatsuishi R., Adachi K., et al. Random mutagenesis of a hyperthermophilic archaeon identified tRNA modifications associated with cellular hyperthermotolerance. Nucleic Acids Res. 2019;47(4):1964–1976. doi: 10.1093/nar/gky1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho X.D., Phung P., Le V Q., et al. Whole transcriptome analysis identifies differentially regulated networks between osteosarcoma and normal bone samples. Exp. Biol. Med. (Maywood, NJ, U. S.) 2017;242(18):1802–1811. doi: 10.1177/1535370217736512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimann E., Kõks S., Ho X.D., et al. Whole exome sequencing of a single osteosarcoma case--integrative analysis with whole transcriptome RNA-seq data. Hum. Genom. 2014;8(1):20. doi: 10.1186/s40246-014-0020-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ho X.D., Nguyen H.G., Trinh L.H., et al. Analysis of the expression of repetitive DNA elements in osteosarcoma. Front. Genet. 2017;8:193. doi: 10.3389/fgene.2017.00193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartee D., Nance K.D., Meier J.L. Site-specific synthesis of N4-acetylcytidine in RNA reveals physiological duplex stabilization. J. Am. Chem. Soc. 2022;144(8):3487–3496. doi: 10.1021/jacs.1c11985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsai K., Jaguva Vasudevan A.A., Martinez Campos C., et al. Acetylation of cytidine residues boosts HIV-1 gene expression by increasing viral RNA stability. Cell Host Microbe. 2020;28(2):306–312. doi: 10.1016/j.chom.2020.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Jing Y., Wang Y., et al. NAT10 promotes gastric cancer metastasis via N4-acetylated COL5A1. Signal Transduct. Targeted Ther. 2021;6(1):173. doi: 10.1038/s41392-021-00489-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang G., Zhang M., Zhang Y., et al. NAT10-mediated mRNA N4-acetylcytidine modification promotes bladder cancer progression. Clin. Transl. Med. 2022;12(5):e738. doi: 10.1002/ctm2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S., Zhang Y., Qiu L., et al. Uncovering N4-acetylcytidine-related mRNA modification pattern and landscape of stemness and immunity in hepatocellular carcinoma. Front. Cell Dev. Biol. 2022;10 doi: 10.3389/fcell.2022.861000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang C., Wu T., Zhang J., et al. Prognostic and immunological role of mRNA ac4C regulator NAT10 in pan-cancer: new territory for cancer research? Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.630417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szymanska E., Markuszewski M.J., Markuszewski M., et al. Altered levels of nucleoside metabolite profiles in urogenital tract cancer measured by capillary electrophoresis. J. Pharm. Biomed. Anal. 2010;53(5):1305–1312. doi: 10.1016/j.jpba.2010.07.031. [DOI] [PubMed] [Google Scholar]
- 40.Zhang T., Wu X., Ke C., et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J. Proteome Res. 2013 Jan 4;12(1):505–512. doi: 10.1021/pr3009572. [DOI] [PubMed] [Google Scholar]
- 41.Li H., Qin Q., Shi X., et al. Modified metabolites mapping by liquid chromatography-high resolution mass spectrometry using full scan/all ion fragmentation/neutral loss acquisition. J. Chromatogr. A. 2019;1583:80–87. doi: 10.1016/j.chroma.2018.11.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.