Abstract
The aim of this study is to explore the active components and potential molecular mechanism of action of Rubia cordifolia L. against nasopharyngeal carcinoma (NPC). We used network pharmacology, molecular docking, and bioinformatics analysis to identify the active components and their role against NPC. The experimental verification was detected by MTT, AnnexinV-FITC/PI double fluorescence staining and Western blotting method. Network pharmacology identified that mollugin is one of the most effective components inRubia cordifolia L. Important NPC targets included HSP90AA1, CDK1, EGFR, PIK3CA, MAPK14, and CDK2. Molecular docking revealed considerable binding activity of mollugin with either of the 6 important NPC targets. Bioinformatics analysis showed that these 6 important targets were mutated in NPC, and the expression of HSP90AA1, PIK3CA, and CDK2 in cancer tissues was significantly different from that in normal tissues. MTT detection and AnnexinV-FITC/PI double fluorescence staining showed that mollugin inhibited the proliferation and induced apoptosis of NPC cells. Western blotting indicated that the molecular mechanism of mollugin against NPC was related to the regulation of the expression of Survivin and XIAP. This study predicted and partially verified the pharmacological and molecular mechanism of action of Rubia cordifolia L. against NPC. Mollugin was identified as a potential active ingredient against NPC. These results prove the reliability of network pharmacology approaches and provide a basis for further research and application of Rubia cordifolia L. against NPC.
Keywords: Network pharmacology, Molecular docking, Bioinformatics, Rubia cordifolia L., Nasopharyngeal carcinoma, Molecular mechanism
1. Introduction
Nasopharyngeal carcinoma (NPC) is a malignant tumor found in the nasopharynx, with the highest incidence among head and neck malignant tumors (HNSC). Due to the characteristics of hidden lesion location and unobvious symptoms, most patients with NPC are in the middle and late stage when diagnosed, resulting in high rates of treatment failure and poor prognosis [1]. Platinum and pyrimidine compounds are often used in the clinical chemotherapy of NPC. Although NPC is sensitive to radiotherapy, its side effects, including radionecrosis, dysphagia and vomiting, are detrimental to patients. A number of patients also develop drug resistance. Despite the emergence of third generation platinum drugs, such as lobaplatin, grade 3–4 treatment-related adverse events (including mucositis, leukopenia, and neutropenia) are still affecting patients [2]. Therefore, there is an urgent need to find a combination of drugs that can enhance the efficacy of NPC therapy and protect normal cells from damage. Several studies have shown that supplementation with traditional Chinese medicine (TCM) has improved the efficacy of radiotherapy, chemotherapy, immunotherapy, and targeted therapy [3]. Concomitantly, Wang et al., also found that the overall risk of death in patients with NPC in the Chinese herbal treatment group was much lower than that in those in the control group, suggesting the potential therapeutic value of Chinese herbal medicine [4].
Rubia cordifolia L., which belongs to the Rubiaceae family, is a famous Ayurvedic medicinal herb and also a TCM [5]. Rubia cordifolia L. has been reported to exert a variety of pharmacological effects, including anti-inflammatory, antipyretic and analgesic, antioxidant, immunoregulatory, and antitumor effects [5,6]. Among them, Rubia cordifolia L. plays a significant role in anti-tumor. Recent studies showed that Rubia extract inhibited the proliferation of human laryngeal carcinoma Hep-2 cells in a dose-dependent manner and induced apopto-sis by increasing the levels of reactive oxygen species [7]; however, it is not clear which phytochemicals in the extract exerted this effect. At present, the known anticancer components of Rubia cordifolia L. mainly include Mollugin, β-sitosterol, dehydroalpha - raquat quinone and xylitone. Among them, Mollugin is considered as an important candidate for the treatment of cancer because of its good anti-tumor activity. Mollugin is a highly abundant naphthoquinone compound, which was first isolated from Rubia cordifolia L. by Itakawa et al., in 1983. Its molecular formula is C17H16O4 and its molecular weight is 284.31 [8]. Studies have shown that Mollugin can regulate multiple signal pathways, including PI3K/AKT/mTOR/p70S6K, ERK, JAK-STAT and HER2/Akt/SREBP-2c, and play an anti-tumor effect [6,8,9]. Interestingly, studies by Zheng-Guang et al. have shown that Mollugin may even be a natural JAK2 inhibitor, inhibiting the activation of JAK-STAT signaling pathway to inhibit LPS-induced inflammation [9]. However, only few studies have focused on the anticancer effects of active components of Rubia cordifolia L., especially against NPC. Therefore, a method is needed to scientifically and systematically explain the effect of Rubia cordifolia L. on cancer and its mechanism.
Network pharmacology is a promising research approach, which combines pharmacology, molecular biology, and bioinformatics to generate a network relationship among pharmacologic effective components, related targets, pathways, and diseases [10]. As such, it explains the development of disease from the perspective of systems biology, pharmacology, and biological networks. This method can not only predict the relationship between drugs and diseases from the network point of view but also visualize and analyze complex biological systems [[11], [12], [13]].
The aim of this study was to explore the potential molecular mechanism of action of Rubia cordifolia L. in the treatment of NPC using network pharmacology and molecular experiments, so as to provide reference for follow-up pharmacological research and future clinical therapeutic applications.
2. Results
2.1. Screening of active components of Rubia cordifolia L.
We searched for the active components of Rubia cordifolia L. in the TCMSP database. After removing the repeated components, we obtained a total of 19 bioactive components. The results are shown in Table 1.
Table 1.
Main active ingredients of Rubia cordifolia L.
| Mol ID | Molecule Name | OB (%) | DL |
|---|---|---|---|
| MOL003283 | (2R,3R,4S)-4-(4-hydroxy-3-methoxy-phenyl)-7-methoxy-2,3-dimethylol-tetralin-6-ol | 66.51 | 0.39 |
| MOL000358 | beta-sitosterol | 36.91 | 0.75 |
| MOL000359 | sitosterol | 36.91 | 0.75 |
| MOL005638 | Mollugin | 42.34 | 0.26 |
| MOL005869 | daucostero_qt | 36.91 | 0.75 |
| MOL006139 | 1,3-dimethoxy-2-carboxyanthraquinone | 102.89 | 0.33 |
| MOL006141 | 1,3-dihydroxy-2-hydroxymthylanthraquinone-3-O-xylosyl (1 → 6)-glucoside_qt | 71.27 | 0.27 |
| MOL006147 | Alizarin-2-methylether | 32.81 | 0.21 |
| MOL006149 | 7-hydroxy-8-methyl-4-vinyl-9,10-dihydrophenanthrene-1-carboxylic acid | 56.99 | 0.27 |
| MOL006150 | 1-acetoxy-6-hydroxy2-methylanthraquinone-3-O-α-rhamnosyl (1 → 4)-α-glucoside | 30.74 | 0.64 |
| MOL006153 | 2′-hydroxymollugin | 40.5 | 0.29 |
| MOL006155 | 4-hydroxy-9,10-dioxoanthracene-2-carboxylic acid | 45.98 | 0.25 |
| MOL006160 | Alizarin | 32.67 | 0.19 |
| MOL006162 | Nordamnacanthal | 53.97 | 0.24 |
| MOL006164 | Pallasone | 43.87 | 0.4 |
| MOL006167 | methyl 6-hydroxy-2,2-dimethyl-3,4-dihydrobenzo [h]chromene-5-carboxylate | 51.09 | 0.25 |
| MOL006170 | Henine | 77.12 | 0.24 |
| MOL006171 | rubiprasin B | 35.97 | 0.68 |
| MOL006174 | Xyloidone | 31.61 | 0.18 |
2.2. Screening of disease targets for NPC
We searched for disease targets of NPC in the OMIM, TTD, and GeneCards databases using “nasopharyngeal carcinoma” as the key word, and identified a total of 2383 disease targets.
2.3. Common target screening and interactive network construction
We also screened 413 drug target genes and 2383 NPC-related genes using the Venny 2.1.0 map and constructed the Wayne diagram (Fig. 1). We then inputted 143 common targets into the STRING data platform to build the PPI network (Fig. 2). We subsequently calculated the occurrence frequency of each protein target using the R software. We observed that the protein targets with the highest frequency included heat shock protein 90 alpha family class A member 1 (HSP90AA1), cyclin dependent kinase 1 (CDK1), epidermal growth factor receptor (EGFR), phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha (PIK3CA), mitogen-activated protein kinase 14 (MAPK14), and cyclin dependent kinase 2 (CDK2), all of which were suggested as potential targets of Rubia cordifolia L. in the treatment of NPC (Fig. 3).
Fig. 1.

Venn diagram of the action targets of active ingredients of Rubia cordifolia L. and the therapeutic targets of NPC.
Fig. 2.
The protein-protein interaction (PPI) network.
Fig. 3.
Frequency of common protein targets.
2.4. Construction of drug active components-NPC-target interaction network
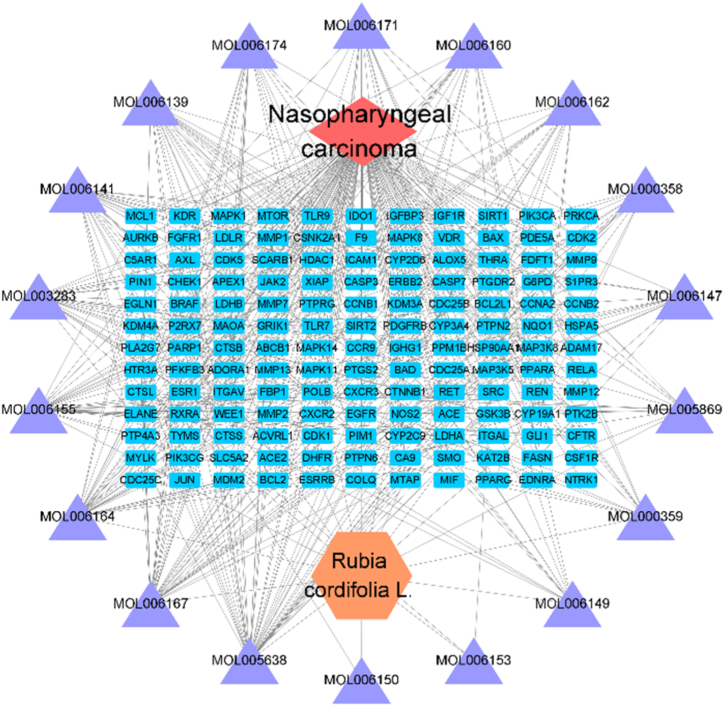
We also inputted the common drug-disease targets into the Cytoscape3.8.2 software and constructed the drug-NPC-target interaction network (Fig. 4). In Fig. 4, yellow represents Rubia cordifolia L., purple represents 18 active components (henine did not intersect with disease targets), blue represents 143 common targets, and red represents NPC. We found that the main active components of Rubia cordifolia L. in the active component-NPC-target interaction network (Fig. 4) included mollugin, pallasone, xyloidone, and rubiprasin B.
Fig. 4.
Active ingredients-NPC-target interaction network.
2.5. GO biological function analysis and core pathway screening of Rubia cordifolia L. in the treatment of NPC
Following GO enrichment analysis, we obtained the respective enriched biological processes, cell composition, and molecular functions, and generated the strip bubble diagram (Fig. 5). Our GO analysis identified the enrichment of 618 biological pathways, among which the negative regulation of apoptotic process, protein phosphorylation, response to drug, and response to xenobiotic stimulus were enriched in 89 cellular components, including the cytoplasm, cytosol, and receptor complexes, and in 128 molecular functional-related pathways, including protein tyrosine kinase activity, protein kinase activity, and enzyme binding. These results suggested that Rubia cordifolia L. might treat NPC by regulating multiple complex biological processes. Fig. 6 shows the results of the KEGG pathway enrichment analysis of active components of Rubia cordifolia L. The signal pathways of ≥10 tumor-related enriched targets are listed in Table 2. Fig. 7 shows the potential targets of mollugin on PI3K/AKT signaling pathway against NPC.
Fig. 5.
GO analysis bar chart.
Fig. 6.
Results of KEGG enrichment analysis.
Table 2.
The signaling pathway of KEGG enrichment target number ≥10.
| ID | Pathway | Number of genes | Genes |
|---|---|---|---|
| hsa04151 | PI3K-Akt signaling pathway | 25 | CSF1R, GSK3B, RELA, EGFR, PIK3CG, IGF1R, RXRA, ERBB2, KDR, MAPK1, ITGAV, JAK2, MCL1, PDGFRB, NTRK1, HSP90AA1, BAD, PRKCA, MTOR, PIK3CA, CDK2, BCL2, MDM2, FGFR1, BCL2L1 |
| hsa04066 | HIF-1 signaling pathway | 14 | EGLN1, PFKFB3, NOS2, PRKCA, EGFR, MTOR, RELA, IGF1R, LDHB, LDHA, PIK3CA, ERBB2, BCL2, MAPK1 |
| hsa04668 | TNF signaling pathway | 14 | JUN, PTGS2, MAPK14, MMP9, RELA, ICAM1, MAPK11, CASP7, MAPK8, PIK3CA, CASP3, MAPK1, MAP3K8, MAP3K5 |
| hsa04657 | IL-17 signaling pathway | 13 | GSK3B, JUN, HSP90AA1, MMP1, PTGS2, MAPK14, MMP9, RELA, MAPK11, MAPK8, MMP13, CASP3, MAPK1 |
| hsa04010 | MAPK signaling pathway | 21 | PDGFRB, NTRK1, CSF1R, JUN, BRAF, PRKCA, MAPK14, EGFR, RELA, CDC25B, IGF1R, MAPK11, PPM1B, MAPK8, CASP3, ERBB2, KDR, MAPK1, MAP3K8, FGFR1, MAP3K5 |
| hsa04012 | ErbB signaling pathway | 12 | GSK3B, JUN, MAPK8, PIK3CA, SRC, BAD, ERBB2, MAPK1, BRAF, PRKCA, EGFR, MTOR |
| hsa04115 | p53 signaling pathway | 11 | CCNB2, CCNB1, CASP3, IGFBP3, CHEK1, CDK2, MDM2, BCL2, CDK1, BAX, BCL2L1 |
| hsa05206 | MicroRNAs in cancer | 20 | PDGFRB, ABCB1, HDAC1, PRKCA, CDC25C, PTGS2, SIRT1, MMP9, CDC25A, EGFR, MTOR, CDC25B, PIK3CA, CASP3, ERBB2, MDM2, PIM1, BCL2, MAPK1, MCL1 |
| hsa04068 | FoxO signaling pathway | 13 | BRAF, MAPK14, SIRT1, EGFR, IGF1R, MAPK11, CCNB2, CCNB1, MAPK8, PIK3CA, CDK2, MDM2, MAPK1 |
| hsa04015 | Rap1 signaling pathway | 15 | PDGFRB, CSF1R, SRC, BRAF, PRKCA, ITGAL, MAPK14, EGFR, IGF1R, MAPK11, PIK3CA, KDR, MAPK1, CTNNB1, FGFR1 |
| hsa04912 | GnRH signaling pathway | 10 | MAPK11, JUN, MAPK8, SRC, MMP2, PTK2B, MAPK1, PRKCA, MAPK14, EGFR |
| hsa04625 | C-type lectin receptor signaling pathway | 10 | MAPK11, JUN, MAPK8, PIK3CA, SRC, MDM2, MAPK1, MAPK14, PTGS2, RELA |
| hsa04620 | Toll-like receptor signaling pathway | 10 | MAPK11, JUN, MAPK8, PIK3CA, TLR9, MAPK1, TLR7, MAP3K8, MAPK14, RELA |
| hsa04660 | T cell receptor signaling pathway | 10 | MAPK11, GSK3B, JUN, MAPK8, PIK3CA, MAPK1, PTPN6, MAP3K8, MAPK14, RELA |
| hsa04014 | Ras signaling pathway | 14 | PDGFRB, NTRK1, CSF1R, BAD, PRKCA, EGFR, RELA, IGF1R, MAPK8, PIK3CA, KDR, MAPK1, FGFR1, BCL2L1 |
| hsa04152 | AMPK signaling pathway | 10 | CCNA2, PFKFB3, PIK3CA, FASN, PPARG, SIRT1, FBP1, CFTR, MTOR, IGF1R |
| hsa04621 | NOD-like receptor signaling pathway | 12 | P2RX7, MAPK11, HSP90AA1, JUN, MAPK8, BCL2, MAPK1, XIAP, MAPK14, RELA, CTSB, BCL2L1 |
| hsa04630 | JAK-STAT signaling pathway | 11 | PDGFRB, PIK3CA, BCL2, PIM1, PTPN6, JAK2, EGFR, PTPN2, MTOR, BCL2L1, MCL1 |
| hsa04915 | Estrogen signaling pathway | 10 | HSP90AA1, JUN, PIK3CA, SRC, MMP2, BCL2, MAPK1, ESR1, MMP9, EGFR |
| hsa04024 | cAMP signaling pathway | 12 | EDNRA, JUN, MAPK8, PIK3CA, BAD, ADORA1, MAPK1, BRAF, PPARA, GLI1, RELA, CFTR |
Fig. 7.
Potential targets of Rubia cordifolia L. on PI3K/AKT signaling pathway.
2.6. Key genes docked with mollugin molecules
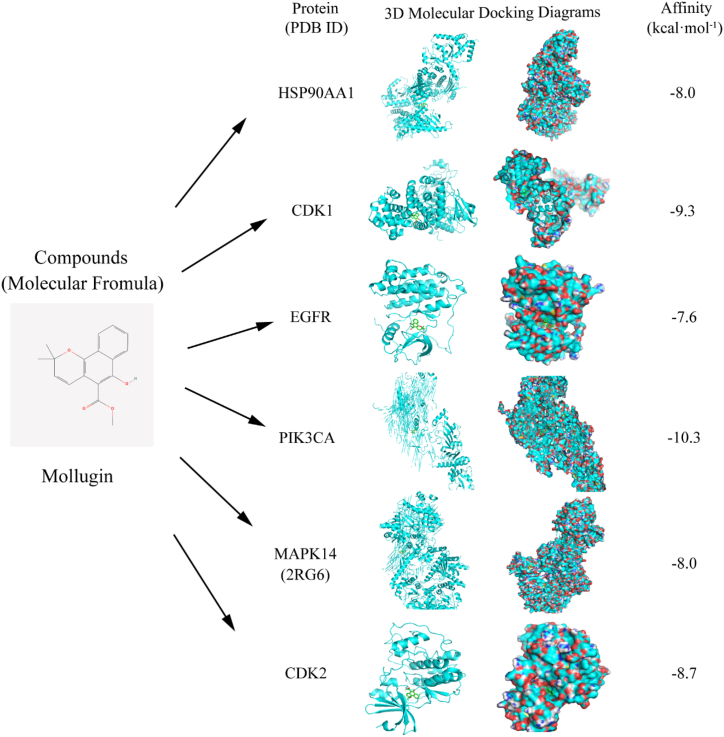
Based on the KEGG analysis results, we selected and docked the target protein, which ranks first in the PPI network and is closely related to NPC, with mollugin, which ranked first among the active compounds of Rubia cordifolia L. We downloaded the following protein data from PDB: HSP90AA1 (PDB ID: 7RY1), CDK1 (PDB ID: 4Y72), EGFR (PDB ID: 7TVD), PIK3CA (PDB ID: 2ENQ), MAPK14 (PDB ID: 1A9U), and CDK2 (PDB ID: 7UDU). The docking of 3D structures with their respective compounds was visualized using PYMOL, with lower binding energies representing high affinity interactions (Fig. 8).
Fig. 8.
Molecular docking of Mollugin with NPC targets.
2.7. Bioinformatics analysis
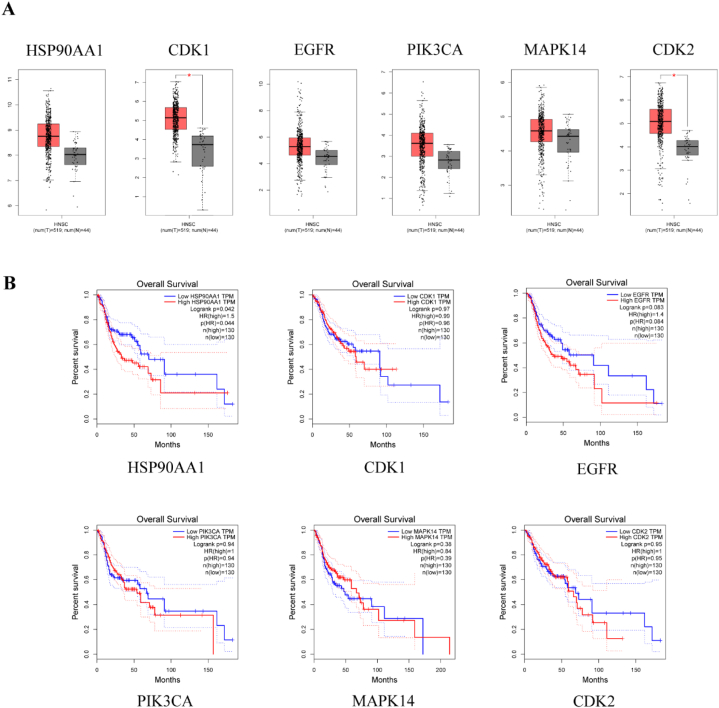
The results of the HPA database analysis (Fig. 9) showed that compared with normal nasopharyngeal tissues, the expression of HSP90AA1 was lower, whereas that of CDK1, PIK3CA, MAPK14, and CDK2 was higher in head and neck tumors. However, we did not detect any significant difference in the expression of EGFR. These findings suggested that the above 5 protein targets (HSP90AA1, CDK1, PIK3CA, MAPK14, and CDK2) might play an important role in the occurrence and development of NPC.
Fig. 9.
The protein expression levels in the HPA database.
The results of the GEPIA database analysis (Fig. 10A, B) showed that the mRNA levels of CDK1 and CDK2 were significantly higher in head and neck tumors (Fig. 10A) than those of CDK1 and CDK2 in normal nasopharyngeal tissues. Moreover, HUB target survival analysis (Fig. 10B) showed that there was a significant difference in the prognostic value between HSP90AA1 and EGFR.
Fig. 10.
Gene expression level and overall survival time (OS) in GEPIA database.
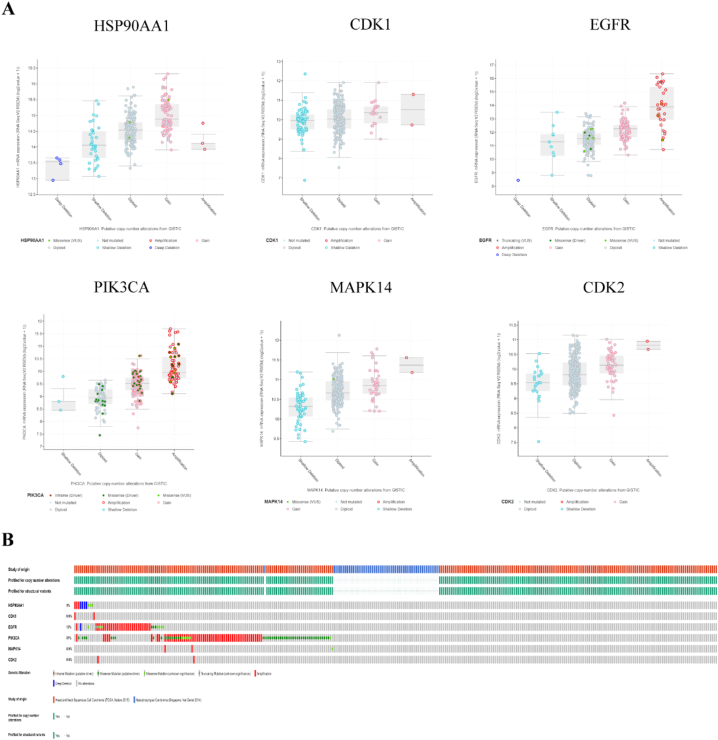
We then used the CBioPortal tool (Fig. 11A, B)to identify the number of gene mutations after selecting research data from HNSC (TCGA, Nature 2015) [14] and NPC (Singapore, Nat Genet2014) [15] (Fig. 11A). We found that the mutation rates of PIK3CA and EGFR genes were 31% and 12%, respectively (Fig. 11B). Interestingly, we observed that the mutation rate of PIK3CA was the highest among the 6 core targets.
Fig. 11.
Genetic information of hub targets.
2.8. Anti-NPC effect of mollugin
2.8.1. Mollugin inhibited the proliferation of NPC cells
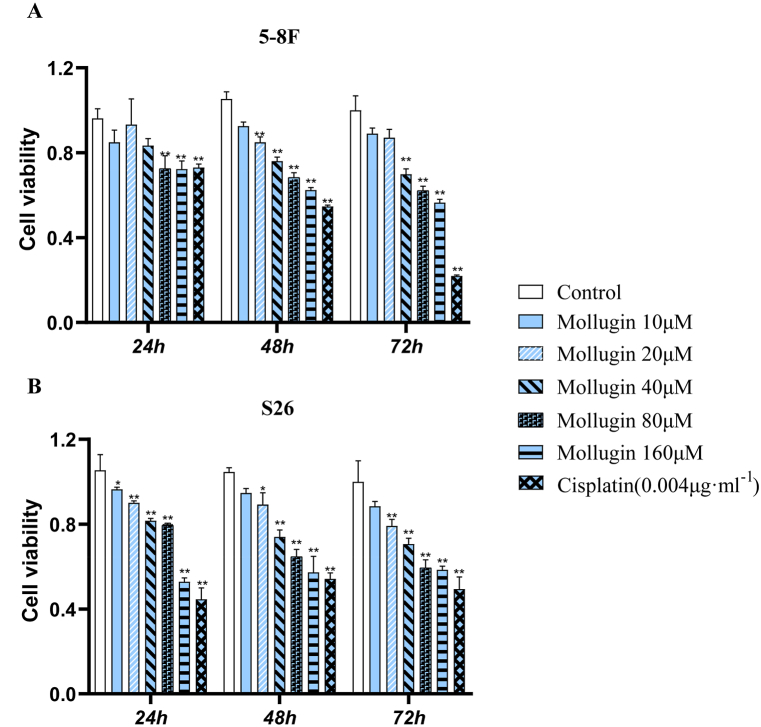
Based on the previous research and previous analysis, we selected mollugin for experimental verification to study its effect on the biological process of proliferation and apoptosis of nasopharyngeal carcinoma cells. Our MTT assay showed that mollugin significantly inhibited the proliferation of 5–8F and S26 cells after 24, 48, and 72 h (P < 0.01) (Fig. 12).
Fig. 12.
MTT assessment of the effect of Mollugin on the proliferation of 5–8F and S26 cells (vs. the control group: ∗P < 0.05 and ∗∗P < 0.01, ).
2.8.2. Mollugin induced apoptosis in NPC cells
To verify the results of the bioinformatics analysis and investigate the reason behind the growth inhibition induced by mollugin in 5–8F and S26 cells, apoptosis were examined by Annexin V-FITC/PI dual-fluorescence staining and Western blot, as a result, we found that mollugin promotes 5–8F and S26 cells apoptosis, and the expressions of Survivin and XIAP were decreased with increasing concentrations of mollugin (P < 0.05). Survivin and XIAP were apoptosis suppressor gene, and therefore, the decrease in their levels may suggest that mollugin may reverse the inhibition of apoptosis in nasopharyngeal carcinoma cells (Fig. 13).
Fig. 13.
Experimental verification: (A)Mollugin promotes 5–8F and S26 cells apoptosis. (B) WB analysis of the levels of the apoptosis-related proteins Survivin and XIAP in tumors treated with different concentrations of mollugin (vs. the control group: ∗P < 0.05 and ∗∗P < 0.01, ).
3. Discussion
There is an urgent need for alternative therapies to ensure the survival of patients with NPC while minimizing the side effects of chemotherapeutic drugs. TCM for the treatment of NPC may be an alternative. Rubia cordifolia L. is a potential natural medicine, and its pharmacological effects have been recently receiving increasing attention from researchers in the field. However, the effects and underlying mechanisms of action of the active components of Rubia cordifolia L. on NPC have not been fully explored. Recently, mollugin, one of the effective active components of Rubia cordifolia L., was reported to induce apoptosis and autophagy of tumor cells through the induction of the extracellular regulated protein kinase (ERK) signal pathway 6.
In this study, we used the network pharmacology approach to study the active components of Rubia cordifolia L. against NPC, in order to provide a theoretical and experimental basis for the clinical treatment of NPC. Network pharmacology can explain the role of TCM and its active components in a scientific and concrete manner. However, this approach has some limitations, i.e., mainly its oneness restricts its reliability.
At present, it has not been reported to analyze the anti-tumor mechanism of Rubia cordifolia L. by network pharmacology and bioinformatics. The novelty of this study was that it incorporated methods of molecular docking and bioinformatics into the network pharmacology approach to explore the possible molecular mechanisms of mollugin in the treatment of NPC. Furthermore, we performed in vitro experiments to verify our hypothesis that mollugin inhibits the proliferation and induces the apoptosis of NPC cells in a concentration-time-dependent manner.
Initially, our network pharmacology analysis identified 18 bioactive components, including mollugin, pallasone, xyloidone, and rubiprasin B, of Rubia cordifolia L. involved in the treatment of NPC. Among these, mollugin ranked first, and attracted our attention.
We constructed a PPI network to help us further identify the common targets of Rubia cordifolia L. and NPC. Among the identified targets, HSP90AA1, CDK1, EGFR, PIK3CA, MAPK14, and CDK2 appeared more frequently than the other targets. Our GO analysis suggested that Rubia might be involved in a variety of biological processes, including apoptosis and protein phosphorylation, while our KEGG analysis suggested that Rubia might exert its anti-NPC effect through the PI3K/AKT signal pathway. Interestingly, among the 6 identified targets, HSP90AA1, EGFR, PIK3CA, and CDK2 are involved in the PI3K/AKT signaling pathway, with some of them participating in tumor cell apoptosis by protein phosphorylation. Subsequently, mollugin was docked with each of the above 6 important protein targets using a molecular docking technique.
From the perspective of intracellular signal transduction, PI3K is in a central position, connecting upstream factors, such as EGFR. PIK3CA encodes the p110 catalytic subunit for the synthesis of class I PI3Ks. Pathogenic mutations in PIK3CA lead to the continuous activation of PI3K, which in turn leads to the uncontrollable proliferation of cells and finally the formation of tumor. The EGFR is known to be overexpressed in more than 90% of head and neck cancers, and PI3K inhibitors have been widely used in treatments targeting EGFR [16]. At present, the PI3K inhibitors approved by FDA include alpelisib, idelalisib, and umbralisib [17,18]; however, specific PI3K inhibitors for NPC are still lacking. Some studies have shown that mollugin induced apoptosis and autophagy by inhibiting the PI3K/AKT/mTOR/p70S6K signaling pathway in glioblastoma cells [6], suggesting the possibility of mollugin targeting PI3K in the treatment of cancer. MAPK14, a member of the mitogen-activated protein kinase family, has been shown to be upregulated in NPC [19]. HSP90AA1, or HSP90, is an important member of the heat shock protein family, the levels of which increase significantly under stress conditions, such as oxidation and carcinogenesis. A study reported significant differences in the levels of plasma Hsp90 α in patients with NPC at early stage (I + II), stage III and IV, and before and after treatment (P < 0.001). Significant differences were also reported in the levels of Hsp90 α between patients with NPC metastasis and patients without metastasis (P < 0.001). The plasma level of Hsp90 α in 196 patients with NPC was significantly higher than that of Hsp90 α in healthy control individuals and was related to the level of EBV DNA [20]. The above results suggested that the level of plasma HSP90 α might be closely related to the clinical stage, metastasis, and therapeutic effect of NPC. In addition, CDK1, which promotes the proliferation of NPC cells, has been shown to be highly expressed in NPC cells. Studies identified CDK1 as a direct target of microRNA-96-5p, the binding of which prevented the induction of apoptosis and cell cycle arrest in CNE-2Z cells [21]. CDK2, another member of the CDK family, plays a key role in the G1/S phase transition of cells. Zhaohai et al., demonstrated that cinobufagin, an active ingredient in a Chinese herbal medicine called VenenumBufonis, induced cell cycle arrest of HK-1 cells in the S phase by downregulating the levels of CDK2 and cyclin E28. In summary, we identified 6 very important targets involved in the occurrence and development of NPC. However, whether mollugin plays a role through the regulation of the complex biological network of these 6 targets warrants further exploration.
We used molecular docking to analyze the binding ability of these six targets (HSP90AA1, CDK1, EGFR, PIK3CA, MAPK14 and CDK2) with Mollugin, and bioinformatics techniques were used to analyze whether there were mutations and differences in the expression of these six targets in NPC. However, this paper also has some limitations. There is a lack of in vitro and in vivo experiments to further detect the effect of Mollugin on the expression of the above six targets. This cannot directly indicate the relationship between Mollugin and six target proteins. We expect more future studies to focus on the relationship between Mollugin and these targets, because mollugin may have the potential of molecular targeted inhibitors [9].
To the best of our knowledge for the first time, we have demonstrated that mollugin is a potential active ingredient against NPC. Concomitantly, we predicted and verified the possible targets of mollugin in the treatment of NPC, thus providing new biomarkers for the diagnosis and treatment of NPC. Meanwhile, the limitation of this study is that how mollugin exerts its anti-NPC effect through these well-combined key targets is still unknown, and whether this effect is related to the close degree of the combination of the two is also unknown. We look forward to further studying these issues in the future. Due to its characteristics, the diagnosis of NPC based on clinical symptoms is difficult. In the future, highly specific and high-performance biomarkers will improve the clinical diagnosis of NPC.
4. Materials and methods
4.1. Screening of bioactive components
The pharmacological database and analysis platform of the traditional Chinese medicine system (TCMSP) database (http://lsp.nwu.edu.cn/tcmsp.php) were used to search for active components of Rubia cordifolia L. The bioactive components and their structures were obtained by setting oral bioavailability (OB) ≥ 30% and drug-like drugs (DL) ≥ 0.18 [22]. The action targets were obtained using the TCMSP and Swiss Target Prediction databases (http://www.swisstargetprediction.ch/).
4.2. Screening of disease targets for NPC
We set “nasopharyngeal carcinoma” as the key word and searched for disease targets of NPC in the OMIM (https://www.omim.org/), TTD (http://db.idrblab.net/ttd/), and GeneCards (https://www.genecards.org/) databases. The results obtained from the screening of these 3 databases were combined, and disease targets of NPC were obtained after weight removal.
4.3. Screening of common targets of the active components and construction of protein-protein interaction network
The obtained disease targets were intersected with drug targets using the R software (https://www.r-project.org/), and a Venn diagram was drawn. The protein-protein interaction (PPI) network was constructed using the STRING plug-in (https://string-db.org/) [23]. The protein type was set to “Homo sapiens”, with 0.9 as the lowest interaction threshold. Isolated nodes were removed and the PPI network was constructed. Subsequently, the PPI network was imported into Cystoscap 3.8.2 [24], its topology was analyzed using the Network Analyzer tool, and the bar chart was drawn using R 4.0.5.
4.4. Construction of drug active component-NPC-target interaction network
The interaction network between active components of Rubia cordifolia L. and disease targets was constructed using the Cytoscape3.8.2 software. In addition, the interaction network diagram of drug active components-NPC targets was drawn, and the potential mechanism of action of Rubia cordifolia L. in the treatment of NPC was investigated.
4.5. Gene ontology (GO) biological functional analysis and kyoto encyclopedia of genes and genomes (KEGG) pathway enrichment analysis
The R language (https://www.r-project.org/) was used to analyze the common targets of the active components of Rubia cordifolia L. and NPC using GO and KEGG pathway enrichment analyses. GO analysis was mainly used to describe the functions of gene products, including cellular, molecular, and biological functions. KEGG pathway enrichment analysis was performed to analyze the enrichment degree of core pathways according to the value of the enrichment factor, and explore the possible biological functions and signal pathway mechanisms of Rubia cordifolia L. in the treatment of NPC.
4.6. Verification of compound-target interactions
Based on the results of network pharmacological analysis, the core targets were selected for molecular docking; we specifically tested the binding ability of mollugin to core targets. First, because the degree of mollugin ranks first in Rubia cordifolia L., mollugin was selected as the ligand, and the target “degree” in the PPI network was selected as the receptor for molecular docking. We downloaded the required protein structure from the PDB and SWISS-MODEL databases, docked it using the LibDock mode with the three-dimensional structural ligands downloaded from PubChem, and selected the molecular-protein docking map with the highest score (LibDock-Score > 100; the conformation was retained).
4.7. Bioinformatics analysis
Six core targets were inputted into the HPA database [25] (https://www.proteinatlas.org/) to study the expression of core target proteins in normal nasopharyngeal tissues and head and neck tumors. The levels of mRNA expression of these 6 targets and overall survival time of patients in TCGA-STAD were verified using the GEPIA database [26] (http://gepa.oma-pku.cn/index.html). The CBioPortalTool [27] (http://www.cbioportal.org/) was used to identify the correlation between genetic information and gene expression of core targets.
4.8. Experimental verification
NPC cells were treated with different concentrations (0, 10, 20, 40, 80, and 160 μM) of mollugin and control drug (cisplatin, 4 μg·mL-1). The proliferation of NPC cells was monitored using the MTT assay. The apoptotic rate was detected using AnnexinV-FITC/PI double fluorescence staining. Western blotting was performed to detect the expression of apoptosis-related proteins and the effect of mollugin on key proteins (Survivin and Xiap) of the phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) signal pathway in NPC cells.
4.9. Statistical analysis
All data were processed using the SPSS26.0 statistical software. The experimental data obeyed normal distribution and were expressed as the mean ± standard deviation. Single factor analysis of variance and the LSD method were used to compare the single factor design and multigroup measurement data, respectively. The analysis map was created using the GraphPad Prism 8.0 software.
5. Conclusions
This study predicted and partially verified the pharmacological and molecular mechanism of action of Rubia cordifolia L. against NPC. Mollugin was identified as a potential active ingredient against NPC. These results prove the reliability of network pharmacology approaches and provide a basis for further research and application of Rubia cordifolia L. against NPC, but this result needs to be futher studied in vivo experiments.
Author contribution statement
Ximing Yang: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Yangyang Tao, Runshi Xu, Wang Luo: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Ting Lin, Fangliang Zhou, Le Tang: Performed the experiments; Analyzed and interpreted the data.
Lan He, Yingchun He: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data will be made available on request.
Funding statement
This research was funded by the National Natural Science Foundation of China (82104941; 81874408), Hunan Provincial Department of Education-funded projects (21B0359), the Natural Science Foundation of Hunan Province (2022JJ30447; 2023JJ30449; 2023JJ40500), the Natural Science Foundation of Changsha city (kq2208206), Chinese Academy of Engineering Academician Liang Liu's Workstation of Hunan University of Chinese Medicine (22YS001), the Project of Hunan Provincial Health Commission (202207015049), the College Student Innovation Project of Hunan Province (S202210541143), the Domestic First-Class Discipline Construction Project of Chinese Medicine of Hunan University of Chinese Medicine (22JBZ011), and the Project of Hunan University of Chinese Medicine (2022XJJJ014).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We would like to thank Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17078.
Contributor Information
Lan He, Email: helan@hnucm.edu.cn.
Yingchun He, Email: heyingchun@hnucm.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Tian Y.M., Liu M.Z., Zeng L., et al. Long-term outcome and pattern of failure for patients with nasopharyngeal carcinoma treated with intensity-modulated radiotherapy. Head Neck. 2019;41(5):1246–1252. doi: 10.1002/hed.25545. [DOI] [PubMed] [Google Scholar]
- 2.Lv X., Cao X., Xia W.X., et al. Induction chemotherapy with lobaplatin and fluorouracil versus cisplatin and fluorouracil followed by chemoradiotherapy in patients with stage III-IVB nasopharyngeal carcinoma: an open-label, non-inferiority, randomised, controlled, phase 3 trial. Lancet Oncol. 2021;22(5):716–726. doi: 10.1016/S1470-2045(21)00075-9. [DOI] [PubMed] [Google Scholar]
- 3.Shen J., Yang T., Tang Y., et al. delta-Tocotrienol induces apoptosis and inhibits proliferation of nasopharyngeal carcinoma cells. Food Funct. 2021;12(14):6374–6388. doi: 10.1039/d1fo00461a. [DOI] [PubMed] [Google Scholar]
- 4.Wang C.Y., Wang T.C., Liang W.M., et al. Effect of Chinese herbal medicine therapy on overall and cancer related mortality in patients with advanced nasopharyngeal carcinoma in Taiwan. Front. Pharmacol. 2020;11 doi: 10.3389/fphar.2020.607413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Z., Li M.Y., Mi C., Wang K.S., Ma J., Jin X. Mollugin has an anti-cancer therapeutic effect by inhibiting TNF-alpha-Induced NF-kappaB activation. Int. J. Mol. Sci. 2017;18(8) doi: 10.3390/ijms18081619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang L., Wang H., Zhu J., Xu J., Ding K. Mollugin induces tumor cell apoptosis and autophagy via the PI3K/AKT/mTOR/p70S6K and ERK signaling pathways. Biochem. Biophys. Res. Commun. 2014;450(1):247–254. doi: 10.1016/j.bbrc.2014.05.101. [DOI] [PubMed] [Google Scholar]
- 7.Shilpa P.N., Sivaramakrishnan V., Niranjali Devaraj S. Induction of apoptosis by methanolic extract of Rubia cordifolia Linn in HEp-2 cell line is mediated by reactive oxygen species. Asian Pac. J. Cancer Prev. APJCP. 2012;13(6):2753–2758. doi: 10.7314/apjcp.2012.13.6.2753. [DOI] [PubMed] [Google Scholar]
- 8.Do M.T., Hwang Y.P., Kim H.G., Na M., Jeong H.G. Mollugin inhibits proliferation and induces apoptosis by suppressing fatty acid synthase in HER2-overexpressing cancer cells. J. Cell Physiol. 2013;228(5):1087–1097. doi: 10.1002/jcp.24258. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Z.G., Jin H., Yu P.J., Tian Y.X., Zhang J.J., Wu S.G. Mollugin inhibits the inflammatory response in lipopolysaccharide-stimulated RAW264.7 macrophages by blocking the Janus kinase-signal transducers and activators of transcription signaling pathway. Biol. Pharm. Bull. 2013;36(3):399–406. doi: 10.1248/bpb.b12-00804. [DOI] [PubMed] [Google Scholar]
- 10.Li S., Zhang B., Jiang D., Wei Y., Zhang N. Herb network construction and co-module analysis for uncovering the combination rule of traditional Chinese herbal formulae. BMC Bioinf. 2010;11(Suppl 11) doi: 10.1186/1471-2105-11-S11-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogales C., Mamdouh Z.M., List M., Kiel C., Casas A.I., Schmidt H. Network pharmacology: curing causal mechanisms instead of treating symptoms. Trends Pharmacol. Sci. 2022;43(2):136–150. doi: 10.1016/j.tips.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Ge H., Zhang B., Li T., et al. Potential targets and the action mechanism of food-derived dipeptides on colitis: network pharmacology and bioinformatics analysis. Food Funct. 2021;12(13):5989–6000. doi: 10.1039/d1fo00469g. [DOI] [PubMed] [Google Scholar]
- 13.Lv X., Xu Z., Xu G., et al. Investigation of the active components and mechanisms of Schisandra chinensis in the treatment of asthma based on a network pharmacology approach and experimental validation. Food Funct. 2020;11(4):3032–3042. doi: 10.1039/d0fo00087f. [DOI] [PubMed] [Google Scholar]
- 14.Cancer Genome Atlas N. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin D.C., Meng X., Hazawa M., et al. The genomic landscape of nasopharyngeal carcinoma. Nat Genet. Aug. 2014;46(8):866–871. doi: 10.1038/ng.3006. [DOI] [PubMed] [Google Scholar]
- 16.Xu M.J., Johnson D.E., Grandis J.R. EGFR-targeted therapies in the post-genomic era. Cancer Metastasis Rev. 2017;36(3):463–473. doi: 10.1007/s10555-017-9687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alzahrani A.S. PI3K/Akt/mTOR inhibitors in cancer: at the bench and bedside. Semin. Cancer Biol. 2019;59:125–132. doi: 10.1016/j.semcancer.2019.07.009. [DOI] [PubMed] [Google Scholar]
- 18.Vasan N., Toska E., Scaltriti M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann. Oncol. 2019;30(Suppl_10):x3–x11. doi: 10.1093/annonc/mdz281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pua L.J.W., Mai C.W., Chung F.F., et al. Functional roles of JNK and p38 MAPK signaling in nasopharyngeal carcinoma. Int. J. Mol. Sci. 2022;23(3) doi: 10.3390/ijms23031108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mao M., Wang X., Sheng H., et al. Heat shock protein 90alpha provides an effective and novel diagnosis strategy for nasopharyngeal carcinoma. Adv. Ther. 2021;38(1):413–422. doi: 10.1007/s12325-020-01518-4. [DOI] [PubMed] [Google Scholar]
- 21.Luo X., He X., Liu X., Zhong L., Hu W. miR-96-5p suppresses the progression of nasopharyngeal carcinoma by targeting CDK1. OncoTargets Ther. 2020;13:7467–7477. doi: 10.2147/OTT.S248338. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Xu X., Zhang W., Huang C., et al. A novel chemometric method for the prediction of human oral bioavailability. Int. J. Mol. Sci. 2012;13(6):6964–6982. doi: 10.3390/ijms13066964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Athanasios A., Charalampos V., Vasileios T., Ashraf G.M. Protein-protein interaction (PPI) network: recent advances in drug discovery. Curr. Drug Metabol. 2017;18(1):5–10. doi: 10.2174/138920021801170119204832. [DOI] [PubMed] [Google Scholar]
- 24.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Digre A., Lindskog C. The Human Protein Atlas-Spatial localization of the human proteome in health and disease. Protein Sci. 2021;30(1):218–233. doi: 10.1002/pro.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang Z., Li C., Kang B., Gao G., Li C., Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gao J., Aksoy B.A., Dogrusoz U., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.