Abstract
Background
Hepatocellular carcinoma (HCC), the main type of liver cancer, is the second most lethal tumor worldwide, with a 5-year survival rate of only 18%. Driver genes facilitate cancer cell growth and spread in the tumor microenvironment. Here, a comprehensive driver gene signature for the prognosis of HCC was developed.
Methods
HCC driver genes were analyzed comprehensively to develop a better prognostic signature. The dataset of HCC patients included mRNA sequencing data and clinical information from the TCGA, the ICGC, and the Guangxi Medical University Cancer Hospital cohorts. First, LASSO was performed to develop a prognostic signature for differentially expressed driver genes in the TCGA cohort. Then, the robustness of the signature was assessed using survival and time-dependent ROC curves. Furthermore, independent predictors were determined using univariate and multivariate Cox regression analyses. Stepwise multi-Cox regression analysis was employed to identify significant variables for the construction of a nomogram that predicts survival rates. Functional analysis by Spearman correlation analysis, enrichment analysis (GO, KEGG, and GSEA), and immunoassay (ssGSEA and xCell) were performed.
Result
A 4-driver gene signature (CLTC, DNMT3A, GMPS, and NRAS) was successfully constructed and showed excellent predictive efficiency in three cohorts. The nomogram indicated high predictive accuracy for the 1-, 3-, and 5-year prognoses of HCC patients, which included clinical information and risk score. Enrichment analysis revealed that driver genes were involved in regulating oncogenic processes, including the cell cycle and metabolic pathways, which were associated with the progression of HCC. ssGSEA and xCell showed differences in immune infiltration and the immune microenvironment between the two risk groups.
Conclusion
The 4-driver gene signature is closely associated with the survival prediction of HCC and is expected to provide new insights into targeted therapy for HCC patients.
Keywords: Driver genes, Prognosis, Hepatocellular carcinoma, Nomogram, Immune
1. Introduction
Primary liver cancer is the sixth most diagnosed cancer and the third most common cause of cancer mortality worldwide as of 2020, leading to more than 1.3 million deaths yearly [1]. Its survival rate is only 18%, being the lowest of all cancer types [2]. HCC accounts for approximately 90% of primary liver cancers [3]. The survival rate of HCC patients remains low after surgical resection and treatment because of the relapse that follows [4].
Thus, it is important to continuously seek advances in screening, diagnosis, and treatment strategies to improve the prognosis of this malignancy [5]. Early diagnosis is a promising strategy to reduce HCC mortality, as it can greatly contribute to detecting HCC at the early stage and applying effective treatments [6]. However, there is still no good method for the early diagnosis and accurate prognosis prediction of HCC [7]. Therefore, predicting the prognosis of patients with HCC can be another way to raise the survival rate because it can help clinicians choose more suitable treatment options for patients [8].
Nevertheless, prognostic prediction of HCC patients has high complexity [9]. On the one hand, the complex etiological landscape contributes to the dilemma created by the molecular and biological heterogeneity of cancer cells [10]. On the other hand, the accurate prognostication of HCC patients necessitates a comprehensive understanding of the entire natural history of the disease, coupled with the identification of optimal survival predictors at each stage. Regrettably, these data are only partially attainable [11].
Specific biomarkers for auxiliary examination are tremendously helpful for the prognostic evaluation of HCC patients in clinical practice [12]. In recent years, many biomarkers for the prognosis of HCC have been reported. For example, CTNNB1 can serve as a prognostic biomarker for HCC, which is related to metabolic reprogramming [13]. CDT1 is an essential factor in the initiation of DNA replication, playing a pivotal role in regulating eukaryotic cell cycle progression and replication. Its expression in HCC was associated with potential diagnostic and prognostic significance [14]. APEX1 has also been shown to be a potential diagnostic and prognostic biomarker in HCC by analyzing multiple databases [15]. Despite the recent noteworthy improvements in the development of new biomarkers for HCC, it still lacks biomarkers able to predict the prognosis or identify subgroups of patients who would benefit from clinical management [16,17]. Therefore, it is urgent to find out specific new biomarkers to evaluate the prognosis and help clinical management of HCC.
In some recent studies, it has been found that driver genes may serve as important genomic biomarkers for diagnosis and prognosis [18,19]. Various studies have shown that driver genes play a crucial role in tumorigenesis, including HCC, lung adenocarcinoma, bladder cancer, prostate cancer, and other malignant cancers [[20], [21], [22], [23]]. Cancer driver genes are a set of mutational genes that possess the ability to drive tumorigenesis [24]. Driver mutations occurring in driver genes are causally implicated in oncogenesis, which confers a growth advantage on cancer cells and affects the homeostatic development of a set of key cellular functions [25,26]. With the clonal expansion, these mutations will be present in all subsequent cancer cells [27]. Although researchers have confirmed the presence of driver genes in HCC by bioinformatics, the significance of all driver genes for the prognosis of liver cancer is still unclear [26,28].
In this study, the overall survival (OS) rate of HCC patients was systematically associated with driver genes, and a signature was established. The robustness of the signature was externally validated by the ICGC cohort and Guangxi Medical University Cancer Hospital cohort. The signature was constructed to help clarify the underlying mechanism of HCC and accurately predict the prognosis for patients with HCC. At the same time, the association between the prognostic signature and the immune cell infiltration of HCC was explored, which makes a valuable contribution to treatment strategies.
2. Materials and methods
2.1. RNA-seq data collection from public databases
The transcriptome data and related clinical details of HCC were downloaded from the TCGA-LIHC project using The Cancer Genome Atlas (TCGA; v31.0) transfer portal GDC (Genomic Data Commons) Data Portal (GDC (cancer.gov)). Additional mRNA expression data and information on 230 clinical HCC samples, the ICGC-LIRI-JP cohort, were obtained from the International Cancer Genome Consortium (ICGC) portal (http://dcc.icgc.org). Simple nucleotide variation data (SNV) were collected by the R package “TCGAbiolinks” [29].
2.2. Clinical information collection from Guangxi Medical University Cancer Hospital
We collected paired HCC and normal tissues from 116 HCC patients after surgical procedures at Guangxi Medical University Cancer Hospital in recent years. Transcriptome expression data for each sample were obtained by tissue sequencing. Apart from the collection of tissue samples, comprehensive clinical and pathological data were obtained from patients, encompassing age, gender, tumor quantity, and size, Barcelona Clinic Liver Cancer (BCLC) stage, Edmondson grade, recurrence status, and survival outcome. All experimental procedures adhered to relevant guidelines and regulations.
2.3. Construction of driver genes sets of HCC
To construct a comprehensive set of HCC driver genes, HCC driver genes were extracted from the IntOGen platform (IntOGen - Cancer driver mutations in Hepatic cancer) and a previous study [26,28].
2.4. Identification of differentially expressed driver genes
The expression of 78 driver genes in cancer and normal tissues was analyzed by the Wilcoxon test in the TCGA database (n = 424). Meanwhile, the degree of differential expression of each driver gene was presented in box plots. The driver genes with significant differential expression were selected as candidate driver genes.
2.5. Development of the 4-driver gene signature by LASSO cox regression analysis
Survival-associated candidate driver genes were identified by univariate Cox regression analysis of OS, and P values were adjusted using Benjamini and Hochberg (BH) correction.
Thereafter, the least absolute shrinkage and selection operator (LASSO) Cox regression was used to select prognosis-related driver genes through the R package “glmnet” with normalized expression data considered independent variables, and the corresponding OS time and patient survival states (dead or alive) were viewed as response variables. The penalty parameter (λ) in the prognostic signature was determined using Tenfold cross-validation and followed minimum criteria. The normalized expression value of each prognostic driver gene was combined with its LASSO Cox regression coefficient to calculate the risk score for each patient:
represents the expression value of driver genes within the signature, while denoting the regression coefficient values of differential genes obtained through Cox regression analysis. After that, the signature was evaluated in the TCGA cohort, Kaplan‒Meier (K-M) survival analysis was carried out by using the “ggsurvplot” function in the R “survminer” package, and time-dependent receiver operating characteristic curves (ROC) were generated by the “survivalROC” package. The efficacy of the signature can be effectively estimated on prognosis.
2.6. Independent validation of the 4-driver gene signature
To exclude the differences between data samples and clinical variations, 230 HCC samples from the ICGC cohort and 116 HCC patients undergoing long-term follow-up at Guangxi Medical University Cancer Hospital cohort were used as the validation cohort. To evaluate the robustness of the signature, two independent validation cohorts were analyzed through K-M survival curves and time-dependent ROC curves. K–M survival curves were undertaken on high- and low-risk groups to compare the survival rate. The area under the curve (AUC) of the time-dependent ROC curve was used to estimate the signature prediction efficiency.
2.7. Construction and validation of a predictive nomogram
Nomograms are widely utilized in predicting the prognoses of HCC patients, primarily due to their ability to condense statistical prediction models into a singular numerical evaluation of the probability of overall survival that is customized to an individual patient's profile. Stepwise multi-Cox regression analysis was used to filter nomogram variables that included clinical information and risk score. The calibration plots and the C-index were utilized to assess the predictive accuracy of the nomogram through the R-related package and “Nomogram” function. The consistency of the model can be assessed by the overlap degree of the correction curve and the reference line.
2.8. Differentially expressed genes analysis in high- and low-risk groups
HCC patients were divided into high- and low-risk groups by median risk score, after which “DESeq2” was used to analyze two clusters of differentially expressed genes (DEGs) in the TCGA cohort. The threshold values were |log2FoldChange | >1.5 and adjusted P value < 0.05. It was considered to be statistically significant for the determination of DEGs.
2.9. Functional analysis based on the prognostic signature
Spearman correlation analysis was used to compare the correlation between the expression of 4 driver genes, and the obtained coefficient represented the degree of correlation between the linear changes of two continuous variables. Enrichment analysis was performed to find important biological pathways between high- and low-risk group DEGs, which involved the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO). In addition, gene set enrichment analysis (GSEA)was used with the “clusterProfiler” package to determine molecular pathways and consistent heterogeneities.
2.10. Single nucleotide variant analysis and immune infiltration assessment
SNV data, which were stored in the Mutation Annotation Format (MAF) of the TCGA database, depicted the HCC patients' mutation burden landscape according to waterfall plots through the R package “maftools”. The ESTIMATE algorithm was utilized to compute the immune score, stromal score, ESTIMATE score, and tumor purity based on the relative proportions of immune cells and stromal cells. Single-sample GSEA (ssGSEA) and xCell were utilized to analyze the immune landscape and calculate the activity of immune cells and immune infiltration between the high- and low-risk groups. The marker genes of different immune cells were obtained from previous studies of 28 immune cell types in the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/). P < 0.05 was considered statistically significant.
2.11. Statistical analysis
All statistical analyses were conducted by R (4.1.2), and corresponding figures were generated. Box plots were analyzed with the Wilcoxon test. Spearman's coefficient was utilized to compute the correlations among the four driver genes Univariate and stepwise multivariate Cox analysis were used to evaluate the survival status of patients in the TCGA cohort and the clinical characteristics affecting the cohort. Survival curves were constructed by the log-rank test using the K–M method. The ssGSEA scores were subjected to the Mann-Whitney test, and an adjusted p-value was calculated using the BH method. All hypothetical tests were two-sided, and a significant p value was defined as <0.05.
3. Result
3.1. Identification of DEGs of driver genes in HCC
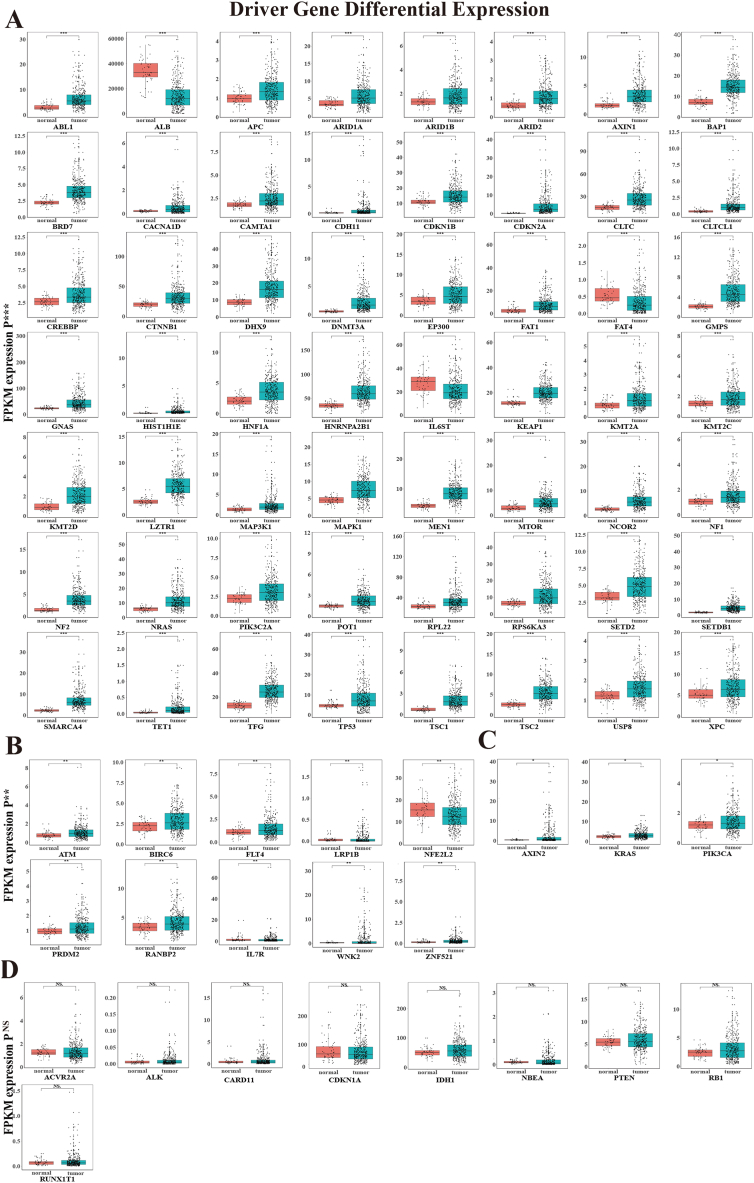
A gene set consisting of 78 HCC driver genes was constructed (Supplementary Table 1). Based on the TCGA database, the driver gene mRNA expression between normal and tumor samples of HCC was analyzed using Wilcoxon tests. The analysis showed that there were 69 differentially expressed driver genes: 56 driver genes (p < 0.001, Fig. 1A), 10 driver genes (0.001 < p < 0.01, Fig. 1B), and 3 driver genes (0.01 < p < 0.05, Fig. 1C). Additionally, 9 driver genes showed no differential expression (Fig. 1D).
Fig. 1.
| The differentially expressed profiles of 78 HCC driver genes. The box of them was shown by the Wilcoxon test between HCC and normal tissues (A) 56 driver genes (***P < 0.001), (B) 10 driver genes (0.001<**P < 0.01), (C) 3 driver genes (0.01< *P < 0.05), (D) 9 driver genes NS (no significance).
3.2. Development of a prognostic-related risk signature
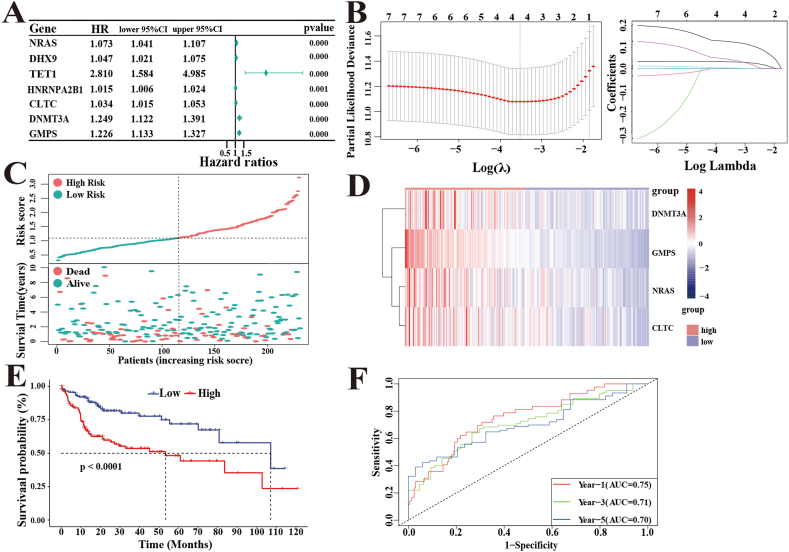
Among 56 (p < 0.001) DEGs, 7 candidate driver genes were significantly related to the OS rate of HCC patients in the TCGA cohort by univariate Cox analysis (using a cutoff threshold of Cox P < 0.001) (Fig. 2A). Then, the above 7 genes were utilized to build a prognostic signature by LASSO Cox regression analysis. LASSO-penalized Cox analysis with penalty parameter tuning performed via 10-fold cross-validation was established to further narrow the candidate driver genes, whose confidence interval was under each lambda. Then, a 4-driver gene signature involving NARS, DNMT3A, CLTC, and GMPS was successfully identified. The risk score of each HCC patient was calculated in the TCGA cohort (n = 231) using the following formula: risk score = 0.0317 × exp (NRAS)+0.0028 × exp (CLTC)+0.0426 × exp (DNMT3A) +
Fig. 2.
LASSO-Cox regression analysis develops a 4-driver genes signature in the TCGA cohort. (A)Univariate Cox analysis identified prognosis-related 7 driver genes. (B)Determination of the number of factors by LASSO analysis. (C)The distribution of risk scores and survival status. (D)Heatmap of 4-driver gene expression. (E)K–M survival analysis of the OS between the risk groups. (F)The time-dependent ROC curves of the risk signature predict OS of 1-, 3-, and 5- years.
0.1256 × exp (GMPS) (Fig. 2B). The patients in the TCGA cohort were divided into high-risk (n = 115) and low-risk groups (n = 116) according to the median risk score (Fig. 2C). The heatmap showed high expression of DNMT3A, GMPS, NARS, and CLTC in the high-risk group, while the other group was in the opposite situation (Fig. 2D). At the same time, the K-M survival curves indicated that the high-risk group had a significantly poorer OS rate than the low-risk group (p < 0.0001) (Fig. 2E). In addition, time-dependent ROC curves were used to evaluate the prediction efficiency of the 4-driver gene signature. The areas under the ROC curve (AUCs) for 1-, 3-, and 5-year survival were 0.75, 0.71, and 0.70, respectively (Fig. 2F). This supported the accurate prediction of the 4-driver gene signature of OS in HCC patients.
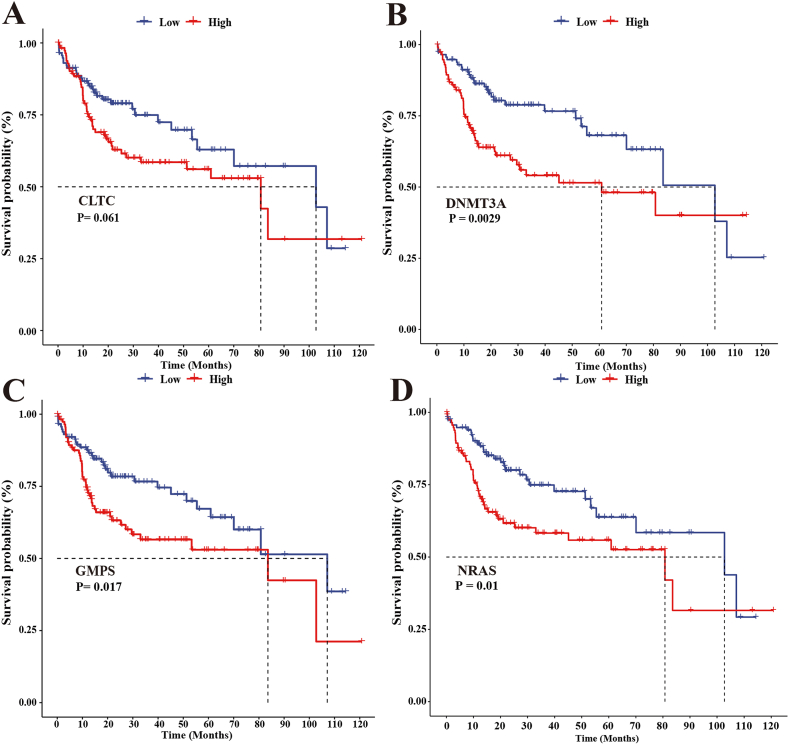
Further analysis of the K-M survival curves in the TCGA cohort showed that the high-expression groups of CLTC, DNMT3A, GMPS, and NRAS had worse OS than the low-expression group (Fig. 3A, B, C, and D). The 4-driver gene signature (Fig. 2E) was able to distinguish the survival probability of HCC patients more significantly than the respective 4 genes (Fig. 3 A, B, C, and D).
Fig. 3.
The 4 driver genes predict HCC patients' survival. K-M survival curves showed the correlation between the expression of driver genes and OS in the TCGA cohort. (A)CLTC, (B)DNMT3A, (C)GMPS, (D)NRAS.
3.3. Validation of the 4-driver gene signature in different independent cohorts
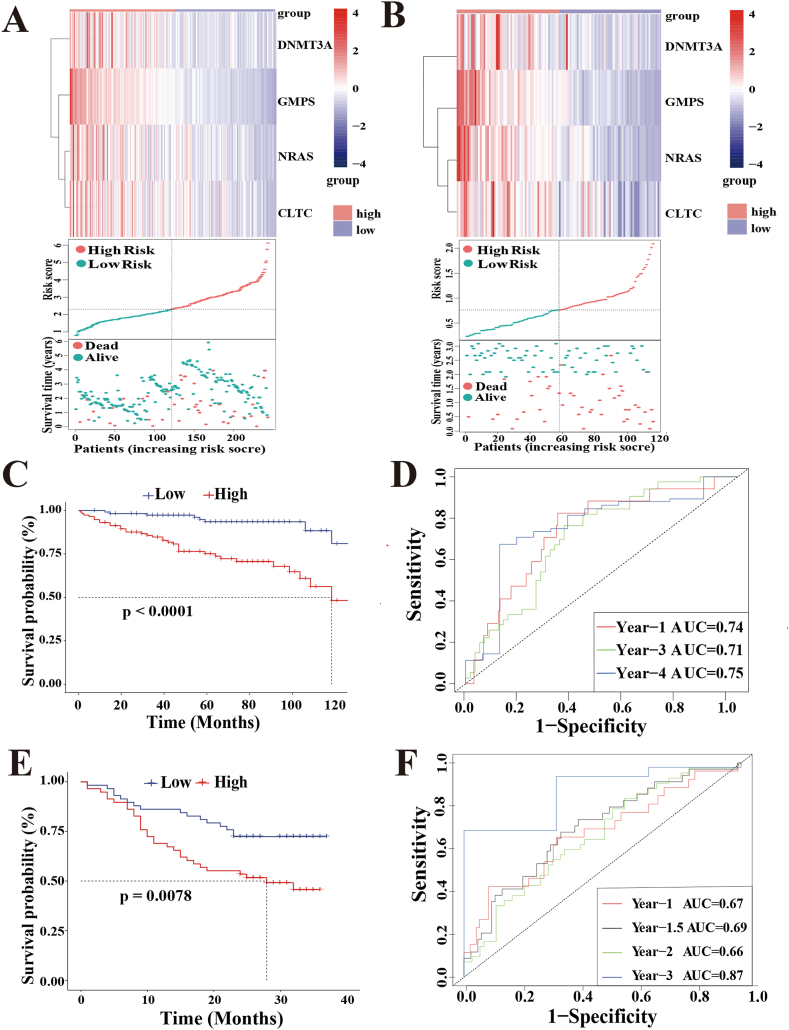
The median risk score of the two independent cohorts (Supplementary Table 2) was calculated according to the same formula as the TCGA cohort. According to the median risk score, these patients were also classified into high- or low-risk groups (Fig. 4A and B). Two cohorts, ICGC, and Guangxi Medical University Cancer Hospital, had worse survival outcomes, as illustrated by the K-M survival curves in the high-risk group. (p < 0.0001 and p = 0.0078) (Fig. 4C, E).
Fig. 4.
External validation of 4-driver genes signature. (A)The distribution of signature 4-driver genes expression, risk scores, and survival statuses in the ICGC cohort. (B) The distribution of signature 4-driver genes expression, risk scores, and survival statuses in Guangxi Medical University Cancer Hospital cohort. (C)K-M assessed the 4-driver genes signature in the ICGC cohort. (D)Time-dependent ROC forecasted in the ICGC cohort. (E)K-M assessed the 4-driver genes signature in Guangxi Medical University Cancer Hospital cohort. (F)Time-dependent ROC forecasted in Guangxi Medical University Cancer Hospital cohort.
Additionally, the AUC values of the ROC curves at 1, 3, and 5 years in the ICGC cohort were 0.74, 0.71, and 0.75, respectively (Fig. 4D). The AUC values for the 1-, 1.5-, 2-, and 3-year prognoses were 0.67, 0.69, 0.66, and 0.87, respectively, in Guangxi Medical University Cancer Hospital cohort (Fig. 4F).
3.4. Construction and validation of a nomogram integrating independent predictive factors
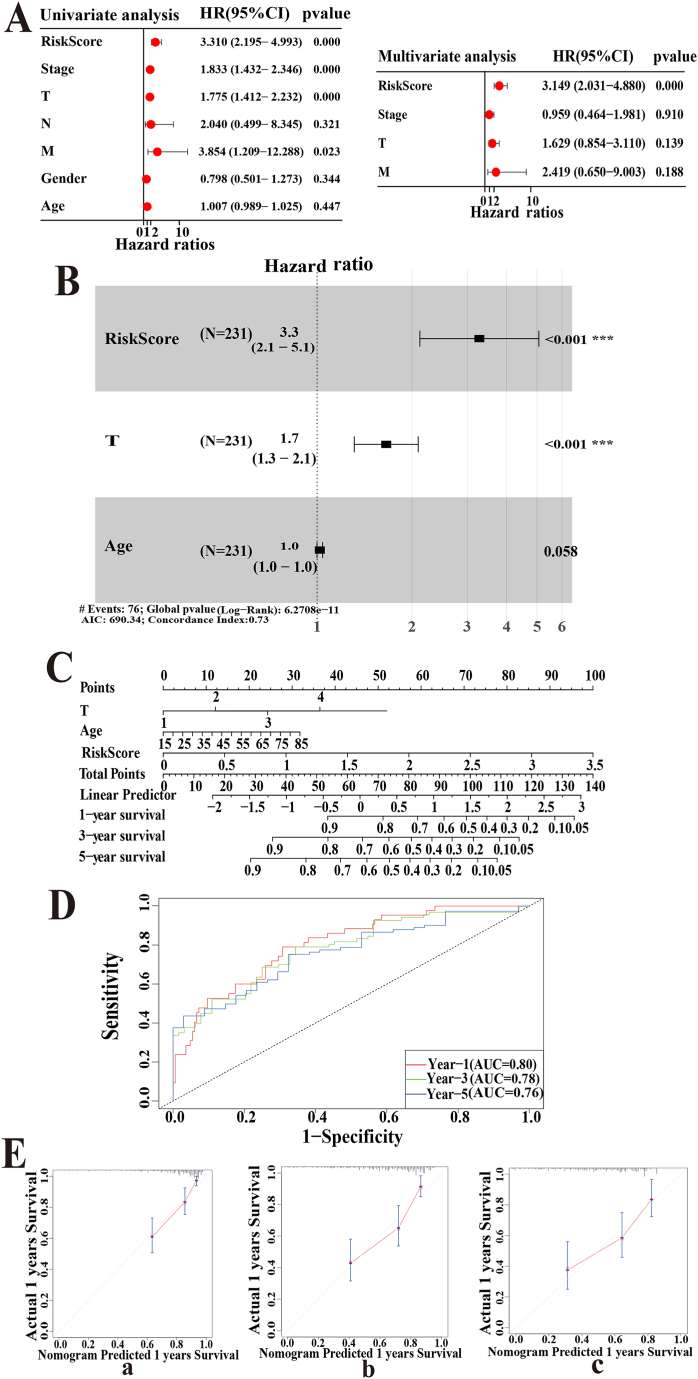
Univariate and multivariate Cox analyses showed that the risk score could be an independent prognostic factor for HCC (Fig. 5A).
Fig. 5.
Construction and assessment of a nomogram integrating clinical factors in the TCGA cohort. (A)Univariate and multi-Cox regression associated the risk score and clinicopathological characteristics with OS. (B)Stepwise multi-Cox regression analysis constructed a nomogram model to predict OS for HCC patients. (C) Nomograms for quantitatively predicting the survival probability in HCC patients. (D) Time-dependent ROC curve to judge the predictive efficacy of the model. (E)The calibration plots for the internal validation of the nomogram predicting 1- (a),3- (b), and 5- years (c) OS.
The variables for the nomogram were chosen using stepwise Cox regression based on the risk score and the clinical information of 231 HCC samples from TCGA. Ultimately, risk score (P < 0.01, HR = 1.905), T stage (P < 0.001 H R = 2.218), and age (P = 0.58, HR = 1.735) were included in the nomogram prognosis prediction (Fig. 5B).
A nomogram incorporating these predictive factors has been developed to accurately quantify the probability of survival in HCC patients (Fig. 5C). To understand the performance of the model, its discrimination and calibration ability were evaluated. The time-dependent ROC curves in terms of 1-, 3-, and 5-year prognoses were analyzed, and the AUCs were 0.80, 0.78, and 0.76, respectively, in the TCGA cohort (Fig. 5D). This model showed better discriminative ability than the 4-driver gene signature (1-year AUC: 0.80 vs. 0.75, 3-year AUC: 0.78 vs. 0.71, 5-year AUC: 0.76 vs. 0.70). The nomogram's calibration curves closely followed the 45° lines (Fig. 5E), with a C-index of 0.73 indicating high consistency between predicted and actual results.
3.5. Functional analysis of subgroups in the TCGA cohort
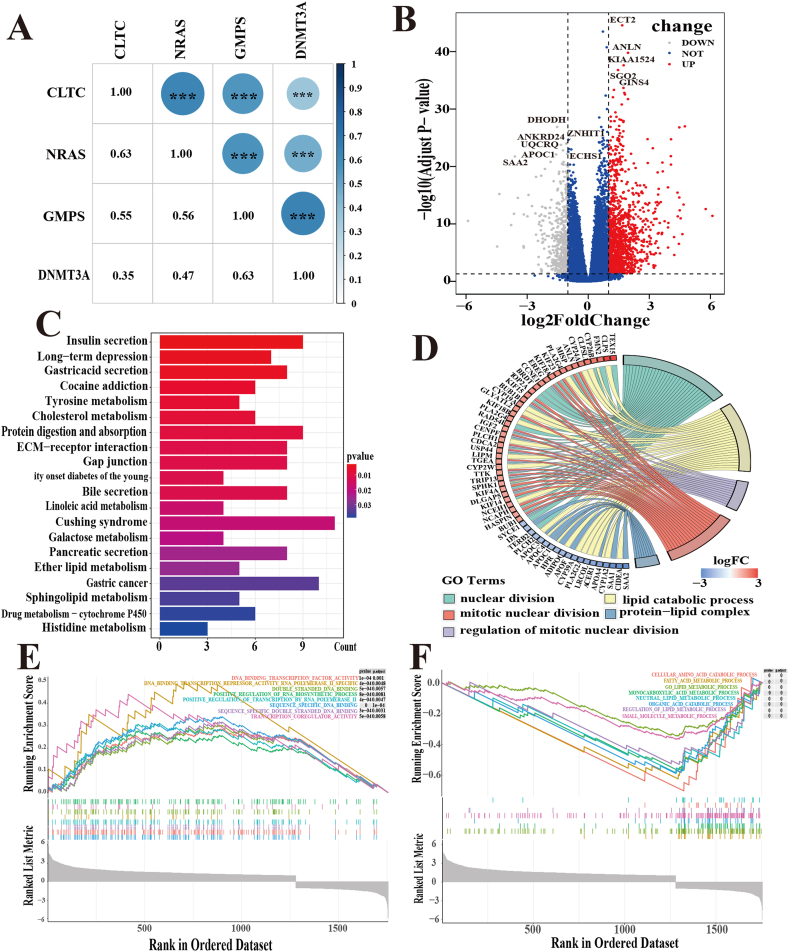
Above all, the relationship between the 4 driver genes was investigated and a positive correlation was observed in their expression levels (Fig. 6A).
Fig. 6.
Functional analysis in the TCGA cohort. (A) The correlation between the expression of 4-driver genes by Spearman analysis. (B)Volcano plot of DEGs in HCC samples. (C)The top 20 most enriched KEGG pathways of DEGs. (D)The chord diagram shows the pathway of GO enrichment analysis. (E、F)GSEA results showed gene sets were differentially enriched signaling pathways in the two risk groups.
Then, the volcano plots displayed 727 DEGs between the high- and low-risk groups with thresholds of p < 0.05 and | log2 (fold change) | >1.5 (Fig. 6B). The biological characteristics of the DEGs were further investigated through function and pathway annotations. KEGG analysis showed that the key pathways correlated with tyrosine metabolism, cholesterol metabolism, protein digestion, absorption, and sphingolipid metabolism (P < 0.05) (Fig. 6C). GO analysis indicated that these DEGs were mainly involved in the regulation of cell cycle processes and lipid catabolic processes (Fig. 6D), suggesting that the differences in survival outcomes between subgroups may be related to the patient's metabolic status. GESA analysis implied that the high-risk group was involved in pathways such as DNA-binding transcription factor activity, double-stranded DNA binding, and other cell cycle processes (Fig. 6E). Interestingly, compared to the outcomes of GO enrichment, the low-risk group was involved in material metabolic processes such as cellular amino acid catabolic processes and fatty acid metabolic processes (Fig. 6F).
3.6. Mutation landscape and differences in immune cell infiltration
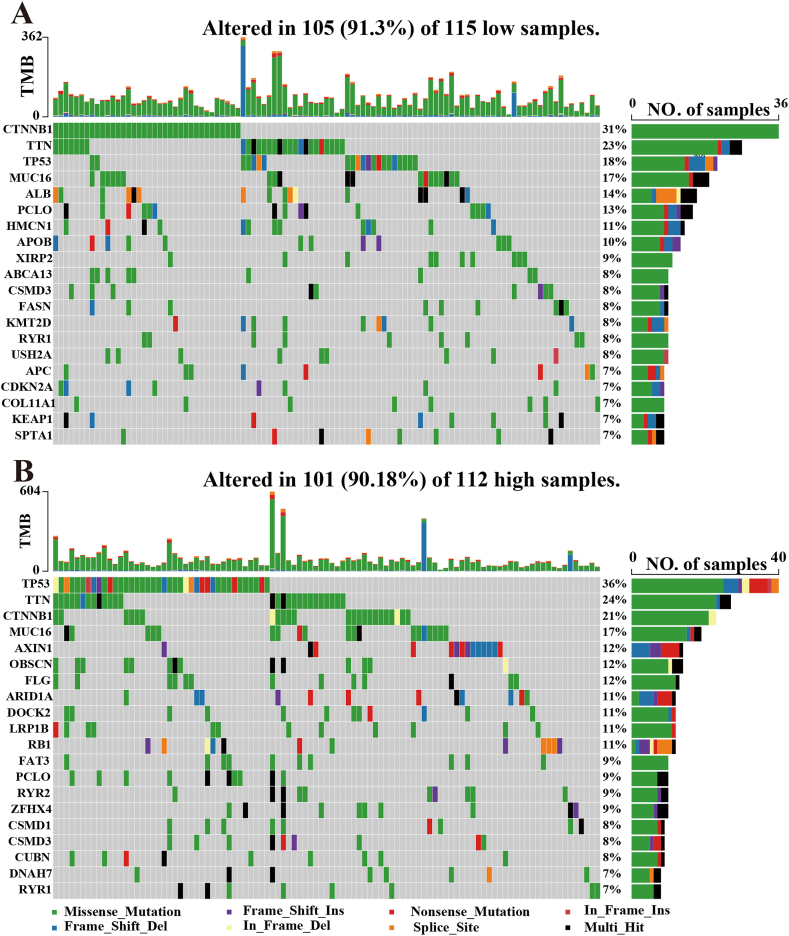
Genomic landscape single nucleotide variants for the two risk groups were visualized by waterfall plots (Fig. 7A and B). The top 20 mutated genes between the high- and low-risk groups were shown for SNV dates in the TCGA cohort. Missense mutations were the most common somatic mutational types. TP53 had a high mutation rate and more abundant mutation forms in the high-risk group, while CTNNB1 had a high mutation rate in the other group.
Fig. 7.
Mutation analysis of TCGA cohort based on risk scores. Mutation status analysis between (A)low- and (B) high-risk groups of the HCC patient's samples. The barplot showed the proportion of each variant type.
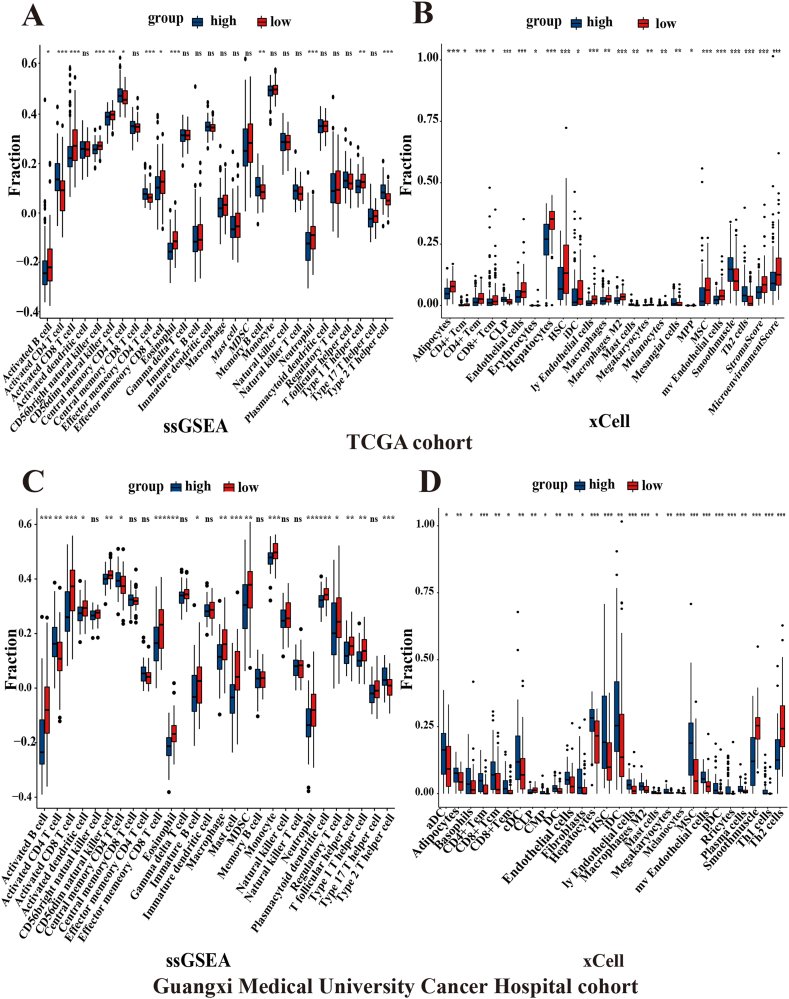
ssGSEA and xCell were used to determine the level of immune cell infiltration in the TCGA cohort and Guangxi Medical University Cancer Hospital cohort, and a median risk score was adopted to classify HCC patients into high- and low-risk groups. ssGSEA results showed significant differences in activated B cells, activated CD4 T cells, activated CD8 T cells, CD56dim natural killer cells, central memory CD4 T cells, effector memory CD8 T cells, eosinophils, neutrophils, and type 1 T helper cells in the TCGA and Guangxi Medical University Cancer Hospital cohorts (all P < 0.05, Fig. 8A, C). Interestingly, the xCell results indicated evident differences in CD4+ Tem cells, hepatocytes, HSCs, ly endothelial cells, M2 macrophages, melanocytes, MSCs, mv endothelial cells, and smooth muscle cells in the two cohorts (all P < 0.05, Fig. 8B, D).
Fig. 8.
ssGSEA and xCell analysis in TCGA cohort and Guangxi Medical University Cancer Hospital cohort. The box plot showed the levels of immune cell infiltration for the high- and low-risk groups in the TCGA cohort (A, B) and Guangxi Medical University Cancer Hospital cohort (C, D) through ssGESA analysis and xCell analysis, respectively. (***P < 0.001, **P < 0.01, *P < 0.05, NS (no significance)).
4. Discussion
To date, it is generally acknowledged that mutational cancer driver genes confer selective advantages on cells relative to the surrounding environment [30]. Different driver gene mutations bring heterogeneity to tumors [31]. Cooperation among driver genes leads to a hepatocellular immune landscape with unique histology. Combinations of expression levels and specific changes in driver genes can shape tumor phenotypes and result in intratumor heterogeneity [32]. HCC is highly heterogeneous, which makes its diagnosis and prognosis difficult. However, proper molecular biomarkers can assist in predicting the prognosis of HCC patients and improve clinical decisions. Hence, there is no doubt that driver genes, as key factors in tumorigenesis and development, can be considered markers for personalized tumor therapy [33].
The 78 driver genes of HCC were comprehensively analyzed and 7 differentially expressed driver genes significantly associated with the survival of HCC patients were found by univariate Cox regression analysis. Furthermore, LASSO Cox regression analysis was used to construct a 4-driver gene signature (CLTC, DNMT3A, GMPS, and NRAS). Ingeniously, the 4-driver gene signature showed excellent predictive efficacy in both the public database and Guangxi Medical University Cancer Hospital cohort. Meanwhile, the risk score was an independent predictor, shown by univariate and multivariate Cox regression analyses. Stepwise multi-Cox regression analysis ascertained that T stage, age, and risk score were nomogram variables for HCC patients in the TCGA cohort. The construction of the nomogram model combining the risk score and clinical features can provide a more accurate prediction measure of HCC prognosis. Compared to the recently constructed prognostic signature of HCC driver genes [34], only HCC driver genes were specifically used to construct the prognostic model. In addition, fewer detection targets were incorporated into this study, thus reducing the clinical detection burden and equipping the model with higher specificity.
This study focused on differentially expressed driver genes in HCC that were associated with OS in HCC patients. As a result, four driver genes were selected to develop the prognostic signature. CLTC encodes the clathrin heavy chain (CHC), which interacts with ATG16L1 and is involved in the autophagic process of endocytosis and degradation of cytoplasmic contents in the transport of various macromolecules [35]. Clathrin can inhibit apoptosis by impairing NOX4 upregulation and ROS production by TGF-β. High expression of TGF-β1 and CLTC is related to worse prognosis and lower OS in HCC patients [36]. Additionally, alterations in CHC prolong mitosis, leading to destabilization of mitotic fibers, defective chromosome binding to the midpalatal, and sustained activation of the spindle checkpoint [37]. The DNMT3A (DNA methyltransferase 3 alpha) gene plays an essential role in encoding enzymes that catalyze DNA methylation [38]. Research has confirmed that upregulation of the DNMT3A gene promotes cancer development. During HCC tumorigenesis, the upregulated mRNA level of DNMT3A is an early event and contributes to the progression of HCC, which can predict patient survival of HCC [39,40]. GMPS (guanine monophosphate synthase) is an important bifunctional dual-domain enzyme. According to previous findings, when it enters the nucleus, the GMPS-USP7 complex is formed with ubiquitin-specific protease 7 (USP7), which can promote the degradation of the p53 protein by deubiquitinating the negative p53 regulatory protein MDM2 [41,42]. Thereafter, it leads to a drop in the intracellular level of p53 and helps tumors evade attack. Meanwhile, GMPS plays a key role in infection, pathogenicity, and axonal transmission [43,44]. NRAS belongs to the RAS superfamily of proteins with physiological functions that regulate cell proliferation, differentiation, and survival [45]. In addition, when NRAS is activated by promoting MAPK and PI3K signaling under normal physiological conditions, NRAS mutation activates MAPK signaling indefinitely, leading to dysregulated cell cycle and cell proliferation signaling, which induces tumor formation [46,47].
Immunotherapy is a promising stratagem for treating cancers by harnessing the cytotoxic potentiality of human immune system [48]. Immune checkpoint blockade (ICB) immunotherapy based on the programmed death-1/ligand-1 (PD-1/PD-L1) axis, which can augment tumor-directed T-cell responses, has reshaped oncology [49]. Some immune checkpoint inhibitors (ICIs), such as atezolizumab, pembrolizumab and nivolumab, have now become the first-line systemic therapy drugs for some advanced cancers, significantly increasing the rate of long-term survival, whereas some other tumors do not respond to ICI monotherapy [50,51]. The expression of PD-L1 has been used to forecast the response to ICB in some cancers [48,52]. However, the predictive role of PD-L1 expression in HCC patients receiving immunotherapy remains controversial [51]. There is significant heterogeneity of PD-L1 expression. Only approximately one-tenth of tumor cells express PD-L1 [53,54]. Therefore, it is necessary to find new biomarkers to identify priori patients who will respond to ICI treatment. A series of researches in some different cancers have revealed connectivity between the tumor immune infiltrating cell square measure and immune checkpoint therapy response [55,56]. In this study, activated B cells, activated CD4 T cells and activated CD8 T cells showed marked differences between the low-risk and high-risk groups. A previous study showed that B-cell markers were the most differentially expressed genes in the tumors of patients who responded to ICB versus nonresponding patients. Significantly higher expression of B-cell-related genes was observed in responders versus nonresponders [57]. The presence of polyclonal CD8 T cells in the tumor is also related to effective anti-PD-1 immunotherapy [58]. Different CD8 T-cell populations can be used as relevant biomarkers of HCC outcomes during immunotherapy studies [59]. ICB therapy can increase the percentage of CD4 T cells [60]. These results imply that grouping based on the risk score may screen potential patients who may benefit from immunotherapy.
Admittedly, there were also certain limitations in the current study. First, since median cutoff values retrospectives data were used in each cohort, more prospective data are needed for accurate validation. Second, considering that the association between the prognostic risk score and immune cell infiltration was based on estimated tumor characteristics, this association may be incomplete. Therefore, their association remains to be experimentally resolved. Third, the signature was not combined with widely used HCC markers, such as AFP, which limits its accuracy in further improving prognosis.
Although it had some deficiencies, the prognosis signature in three different cohorts all showed a better prediction performance. With the advent of the era of precision medicine, clinical trials can be designed using a risk score based on the 4-driver gene signature to select patients who are most likely to develop poor prognoses. This can provide potential translational value for the clinical management of HCC patients. Therefore, novel or more intensive postoperative therapies can be developed in the future. Consequently, it is of great value for future tumor treatment to focus more on precision medicine and molecular markers.
5. Conclusion
A 4-driver genes signature was successfully built to predict prognostic viability. The signature has resulted in being of significant predictive value in the ICGC cohort and Guangxi Medical University Cancer Hospital cohort. This indicates that driver genes play a considerable function in the development of HCC, thus inspiring a novel research direction for future targeted therapy of liver cancer.
Author contribution
Houtian Guo, Fei Lu, Meiqi Huang, Rongqi Lu: Performed the experiments; Analyzed and interpreted the data; Contributed analysis tools and data; Wrote the paper.
Xuejing Li: Analyzed and interpreted the data; Wrote the paper.
Jianhui Yuan: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed analysis tools and data: Wrote the paper.
Feng Wang: Conceived and designed the experiments; Performed the experiments, Analyzed and interpreted the data; Contributed analysis tools and data; Wrote the paper.
Funding statement
This work is supported by the Natural Science Foundation of China (Grant No. 81660510), the Natural Science Foundation of Guangxi in China (Grant No. 2014GXNSFBA118206), the Project of Improving the Basic Research Ability of Young and Middle-aged Teachers in Guangxi Universities (Grant No. 2022KY0103) and National College Students’ Innovation and Entrepreneurship Training Program (Grant No. 202210598006).
Data availability statement
Data included in article/supp. Material/referenced in article.
Ethical approval
TCGA cohort and ICGC cohort data that we analyzed were available to the public in the TCGA and ICGC databases and all processes followed the relevant guidelines and policies of the present study. Informed consent of each patient in Guangxi Medical University Cancer Hospital cohort data was obtained, which was approved by the Ethics Committee from Guangxi Medical University Cancer Hospital (Ethical Number: 20,200,137).
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17054.
Contributor Information
Jianhui Yuan, Email: yuanjianhui@gxmu.edu.cn.
Feng Wang, Email: wangf@sr.gxmu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Ca - Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Yang X., Ou Q., Yang W., Shi Y., Liu G. Diagnosis of liver cancer by ftir spectra of serum. Spectrochim. Acta, Part A. 2021;263 doi: 10.1016/j.saa.2021.120181. [DOI] [PubMed] [Google Scholar]
- 3.European Association for the Study of the Liver Easl clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 4.Wang M.D., Li C., Liang L., Xing H., Sun L.Y., Quan B., Wu H., Xu X.F., Wu M.C., Pawlik T.M., Lau W.Y., Shen F., Yang T. Early and late recurrence of hepatitis B virus-associated hepatocellular carcinoma. Oncol. 2020;25(10):e1541–e1551. doi: 10.1634/theoncologist.2019-0944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinero F., Dirchwolf M., Pessoa M.G. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6) doi: 10.3390/cells9061370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bruix J., Sherman M., Llovet J.M., Beaugrand M., Lencioni R., Burroughs A.K., Christensen E., Pagliaro L., Colombo M., Rodes J., Hcc E.P.o.E.o. Clinical management of hepatocellular carcinoma. Conclusions of the barcelona-2000 easl conference. European association for the study of the liver. J. Hepatol. 2001;35(3):421–430. doi: 10.1016/s0168-8278(01)00130-1. [DOI] [PubMed] [Google Scholar]
- 7.Yang J.D., Heimbach J.K. New advances in the diagnosis and management of hepatocellular carcinoma. BMJ. 2020;371:m3544. doi: 10.1136/bmj.m3544. [DOI] [PubMed] [Google Scholar]
- 8.Pallozzi M., Di Tommaso N., Maccauro V., Santopaolo F., Gasbarrini A., Ponziani F.R., Pompili M. Non-invasive biomarkers for immunotherapy in patients with hepatocellular carcinoma: current knowledge and future perspectives. Cancers. 2022;14(19):4631. doi: 10.3390/cancers14194631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serper M., Taddei T.H., Mehta R., D'addeo K., Dai F., Aytaman A., Baytarian M., Fox R., Hunt K., Goldberg D.S., Valderrama A., Kaplan D.E., Group V.S. Association of provider specialty and multidisciplinary care with hepatocellular carcinoma treatment and mortality. Gastroenterology. 2017;152(8):1954–1964. doi: 10.1053/j.gastro.2017.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J., Tang W., Budhu A., Forgues M., Hernandez M.O., Candia J., Kim Y., Bowman E.D., Ambs S., Zhao Y., Tran B., Wu X., Koh C., Surana P., Liang T.J., Guarnera M., Mann D., Rajaure M., Greten T.F., Wang Z., Yu H., Wang X.W. A viral exposure signature defines early onset of hepatocellular carcinoma. Cell. 2020;182(2):317–328 e10. doi: 10.1016/j.cell.2020.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Llovet J.M., Bru C., Bruix J. Prognosis of hepatocellular carcinoma: the bclc staging classification. Semin. Liver Dis. 1999;19(3):329–338. doi: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 12.Li N., Li L., Chen Y. The identification of core gene expression signature in hepatocellular carcinoma. Oxid. Med. Cell. Longev. 2018 doi: 10.1155/2018/3478305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huo J., Wu L., Zang Y. Development and validation of a ctnnb1-associated metabolic prognostic model for hepatocellular carcinoma. J. Cell Mol. Med. 2021;25(2):1151–1165. doi: 10.1111/jcmm.16181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cai C., Zhang Y., Hu X., Hu W., Yang S., Qiu H., Chu T. Cdt1 is a novel prognostic and predictive biomarkers for hepatocellular carcinoma. Front. Oncol. 2021;11 doi: 10.3389/fonc.2021.721644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cao L., Cheng H.W., Jiang Q.X., Li H., Wu Z.X. Apex1 is a novel diagnostic and prognostic biomarker for hepatocellular carcinoma. Aging-Us. 2020;12(5):4573–4591. doi: 10.18632/aging.102913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen V.L., Xu D., Wicha M.S., Lok A.S., Parikh N.D. Utility of liquid biopsy analysis in detection of hepatocellular carcinoma, determination of prognosis, and disease monitoring: a systematic review. Clin. Gastroenterol. Hepatol. 2020;18(13):2879–28902 e9. doi: 10.1016/j.cgh.2020.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cerrito L., Ainora M.E., Mosoni C., Borriello R., Gasbarrini A., Zocco M.A. Prognostic role of molecular and imaging biomarkers for predicting advanced hepatocellular carcinoma treatment efficacy. Cancers. 2022;14(19):4647. doi: 10.3390/cancers14194647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen J., Zhou C., Liu Y. Establishing a cancer driver gene signature-based risk model for predicting the prognoses of gastric cancer patients. Aging (Albany NY) 2022;14(5):2383–2399. doi: 10.18632/aging.203948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sun X., Yi J., Yang J., Han Y., Qian X., Liu Y., Li J., Lu B., Zhang J., Pan X., Liu Y., Liang M., Chen E., Liu P., Lu Y. An integrated epigenomic-transcriptomic landscape of lung cancer reveals novel methylation driver genes of diagnostic and therapeutic relevance. Theranostics. 2021;11(11):5346–5364. doi: 10.7150/thno.58385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang S., Yim S., Park H. The cancer driver genes idh1/2, Jarid1c/Kdm5c, and utx/Kdm6a: crosstalk between histone demethylation and hypoxic reprogramming in cancer metabolism. Exp. Mol. Med. 2019;51(6):1–17. doi: 10.1038/s12276-019-0230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inamura K. Clinicopathological characteristics and mutations driving development of early lung adenocarcinoma: tumor initiation and progression. Int. J. Mol. Sci. 2018;19(4) doi: 10.3390/ijms19041259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tran L., Xiao J.F., Agarwal N., Duex J.E., Theodorescu D. Advances in bladder cancer biology and therapy. Nat. Rev. Cancer. 2021;21(2):104–121. doi: 10.1038/s41568-020-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao X., Lei Y., Li G., Cheng Y., Yang H., Xie L., Long H., Jiang R. Integrative analysis of cancer driver genes in prostate adenocarcinoma. Mol. Med. Rep. 2019;19(4):2707–2715. doi: 10.3892/mmr.2019.9902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4(3):177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., Edkins S., O'meara S., Vastrik I., Schmidt E.E., Avis T., Barthorpe S., Bhamra G., Buck G., Choudhury B., Clements J., Cole J., Dicks E., Forbes S., Gray K., Halliday K., Harrison R., Hills K., Hinton J., Jenkinson A., Jones D., Menzies A., Mironenko T., Perry J., Raine K., Richardson D., Shepherd R., Small A., Tofts C., Varian J., Webb T., West S., Widaa S., Yates A., Cahill D.P., Louis D.N., Goldstraw P., Nicholson A.G., Brasseur F., Looijenga L., Weber B.L., Chiew Y.E., Defazio A., Greaves M.F., Green A.R., Campbell P., Birney E., Easton D.F., Chenevix-Trench G., Tan M.H., Khoo S.K., Teh B.T., Yuen S.T., Leung S.Y., Wooster R., Futreal P.A., Stratton M.R. Patterns of somatic mutation in human cancer Genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez-Jimenez F., Muinos F., Sentis I., Deu-Pons J., Reyes-Salazar I., Arnedo-Pac C., Mularoni L., Pich O., Bonet J., Kranas H., Gonzalez-Perez A., Lopez-Bigas N. A compendium of mutational cancer driver genes. Nat. Rev. Cancer. 2020;20(10):555–572. doi: 10.1038/s41568-020-0290-x. [DOI] [PubMed] [Google Scholar]
- 27.Stratton M.R., Campbell P.J., Futreal P.A. The cancer Genome. Nature. 2009;458(7239):719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey M.H., Tokheim C., Porta-Pardo E., Sengupta S., Bertrand D., Weerasinghe A., Colaprico A., Wendl M.C., Kim J., Reardon B., Ng P.K., Jeong K.J., Cao S., Wang Z., Gao J., Gao Q., Wang F., Liu E.M., Mularoni L., Rubio-Perez C., Nagarajan N., Cortes-Ciriano I., Zhou D.C., Liang W.W., Hess J.M., Yellapantula V.D., Tamborero D., Gonzalez-Perez A., Suphavilai C., Ko J.Y., Khurana E., Park P.J., Van Allen E.M., Liang H., Group M.C.W., Cancer Genome Atlas Research N., Lawrence M.S., Godzik A., Lopez-Bigas N., Stuart J., Wheeler D., Getz G., Chen K., Lazar A.J., Mills G.B., Karchin R., Ding L. Comprehensive characterization of cancer driver genes and mutations. Cell. 2018;173(2):371–385 e18. doi: 10.1016/j.cell.2018.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Colaprico A., Silva T.C., Olsen C., Garofano L., Cava C., Garolini D., Sabedot T.S., Malta T.M., Pagnotta S.M., Castiglioni I., Ceccarelli M., Bontempi G., Noushmehr H. Tcgabiolinks: an R/bioconductor package for integrative analysis of tcga data. Nucleic Acids Res. 2016;44(8):e71. doi: 10.1093/nar/gkv1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vogelstein B., Kinzler K.W. The path to cancer --Three strikes and you're out. N. Engl. J. Med. 2015;373(20):1895–1898. doi: 10.1056/NEJMp1508811. [DOI] [PubMed] [Google Scholar]
- 31.Colaprico A., Olsen C., Bailey M.H., Odom G.J., Terkelsen T., Silva T.C., Olsen A.V., Cantini L., Zinovyev A., Barillot E., Noushmehr H., Bertoli G., Castiglioni I., Cava C., Bontempi G., Chen X.S., Papaleo E. Interpreting pathways to discover cancer driver genes with moonlight. Nat. Commun. 2020;11(1):69. doi: 10.1038/s41467-019-13803-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Molina-Sanchez P., Ruiz De Galarreta M., Yao M.A., Lindblad K.E., Bresnahan E., Bitterman E., Martin T.C., Rubenstein T., Nie K., Golas J., Choudhary S., Barcena-Varela M., Elmas A., Miguela V., Ding Y., Kan Z., Grinspan L.T., Huang K.L., Parsons R.E., Shields D.J., Rollins R.A., Lujambio A. Cooperation between distinct cancer driver genes underlies intertumor heterogeneity in hepatocellular carcinoma. Gastroenterology. 2020;159(6):2203–2220. doi: 10.1053/j.gastro.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata T. Genomic landscape of hepatocarcinogenesis. J. Hum. Genet. 2021;66(9):845–851. doi: 10.1038/s10038-021-00928-8. [DOI] [PubMed] [Google Scholar]
- 34.Zou J., Qin W. Comprehensive analysis of the cancer driver genes constructs a seven-gene signature for prediction of survival and tumor immunity in hepatocellular carcinoma. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.937948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravikumar B., Moreau K., Jahreiss L., Puri C., Rubinsztein D.C. Plasma membrane contributes to the formation of pre-autophagosomal structures. Nat. Cell Biol. 2010;12(8):747–757. doi: 10.1038/ncb2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caballero-Diaz D., Bertran E., Penuelas-Haro I., Moreno-Caceres J., Malfettone A., Lopez-Luque J., Addante A., Herrera B., Sanchez A., Alay A., Sole X., Serrano T., Ramos E., Fabregat I. Clathrin switches transforming growth factor-beta role to pro-tumorigenic in liver cancer. J. Hepatol. 2020;72(1):125–134. doi: 10.1016/j.jhep.2019.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Royle S.J., Bright N.A., Lagnado L. Clathrin is required for the function of the mitotic spindle. Nature. 2005;434(7037):1152–1157. doi: 10.1038/nature03502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lyko F. The DNA methyltransferase family: a versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018;19(2):81–92. doi: 10.1038/nrg.2017.80. [DOI] [PubMed] [Google Scholar]
- 39.Dong Y., Wang A. Aberrant DNA methylation in hepatocellular carcinoma tumor suppression (review) Oncol. Lett. 2014;8(3):963–968. doi: 10.3892/ol.2014.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito Y., Kanai Y., Sakamoto M., Saito H., Ishii H., Hirohashi S. Expression of mrna for DNA methyltransferases and methyl-cpg-binding proteins and DNA methylation status on cpg islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology. 2001;33(3):561–568. doi: 10.1053/jhep.2001.22507. [DOI] [PubMed] [Google Scholar]
- 41.Reddy B.A., Van Der Knaap J.A., Bot A.G., Mohd-Sarip A., Dekkers D.H., Timmermans M.A., Martens J.W., Demmers J.A., Verrijzer C.P. Nucleotide biosynthetic enzyme gmp synthase is a trim21-controlled relay of P53 stabilization. Mol. Cell. 2014;53(3):458–470. doi: 10.1016/j.molcel.2013.12.017. [DOI] [PubMed] [Google Scholar]
- 42.Pei Y., Fu J., Shi Y., Zhang M., Luo G., Luo X., Song N., Mi T., Yang Y., Li J., Zhou Y., Zhou B. Discovery of a potent and selective degrader for Usp7. Angew Chem. Int. Ed. Engl. 2022;61(33) doi: 10.1002/anie.202204395. [DOI] [PubMed] [Google Scholar]
- 43.Iglesias-Gato D., Martin-Marcos P., Santos M.A., Hinnebusch A.G., Tamame M. Guanine nucleotide pool imbalance impairs multiple steps of protein synthesis and disrupts Gcn4 translational control in Saccharomyces cerevisiae. Genetics. 2011;187(1):105–122. doi: 10.1534/genetics.110.122135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ipe D.S., Sullivan M.J., Goh K.G.K., Hashimi S.M., Munn A.L., Ulett G.C. Conserved bacterial de novo guanine biosynthesis pathway enables microbial survival and colonization in the environmental niche of the urinary tract. ISME J. 2021;15(7):2158–2162. doi: 10.1038/s41396-021-00934-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin C., Zhu B., Zhang T., Liu T., Chen S., Liu Y., Li X., Miao X., Li S., Mi X., Zhang J., Li L., Wei G., Xu Z.X., Gao X., Huang C., Wei Z., Goding C.R., Wang P., Deng X., Cui R. Pharmacological targeting of Stk19 inhibits oncogenic nras-driven melanomagenesis. Cell. 2019;176(5):1113–1127 e16. doi: 10.1016/j.cell.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 46.Garcia-Alvarez A., Ortiz C., Munoz-Couselo E. Current perspectives and novel strategies of nras-mutant melanoma. OncoTargets Ther. 2021;14:3709–3719. doi: 10.2147/OTT.S278095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Randic T., Kozar I., Margue C., Utikal J., Kreis S. Nras mutant melanoma: towards better therapies. Cancer Treat Rev. 2021;99 doi: 10.1016/j.ctrv.2021.102238. [DOI] [PubMed] [Google Scholar]
- 48.Kalbasi A., Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20(1):25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rahim M.K., Okholm T.L.H., Jones K.B., Mccarthy E.E., Liu C.C., Yee J.L., Tamaki S.J., Marquez D.M., Tenvooren I., Wai K., Cheung A., Davidson B.R., Johri V., Samad B., O'gorman W.E., Krummel M.F., Van Zante A., Combes A.J., Angelo M., Fong L., Algazi A.P., Ha P., Spitzer M.H. Dynamic Cd8(+) T cell responses to cancer immunotherapy in human regional lymph nodes are disrupted in metastatic lymph nodes. Cell. 2023;186(6):1127–1143. doi: 10.1016/j.cell.2023.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petitprez F., De Reynies A., Keung E.Z., Chen T.W., Sun C.M., Calderaro J., Jeng Y.M., Hsiao L.P., Lacroix L., Bougouin A., Moreira M., Lacroix G., Natario I., Adam J., Lucchesi C., Laizet Y.H., Toulmonde M., Burgess M.A., Bolejack V., Reinke D., Wani K.M., Wang W.L., Lazar A.J., Roland C.L., Wargo J.A., Italiano A., Sautes-Fridman C., Tawbi H.A., Fridman W.H. B cells are associated with survival and immunotherapy response in sarcoma. Nature. 2020;577(7791):556–560. doi: 10.1038/s41586-019-1906-8. [DOI] [PubMed] [Google Scholar]
- 51.Rizzo A., Cusmai A., Gadaleta-Caldarola G., Palmiotti G. Which role for predictors of response to immune checkpoint inhibitors in hepatocellular carcinoma? Expet Rev. Gastroenterol. Hepatol. 2022;16(4):333–339. doi: 10.1080/17474124.2022.2064273. [DOI] [PubMed] [Google Scholar]
- 52.Oh S.A., Wu D.C., Cheung J., Navarro A., Xiong H., Cubas R., Totpal K., Chiu H., Wu Y., Comps-Agrar L., Leader A.M., Merad M., Roose-Germa M., Warming S., Yan M., Kim J.M., Rutz S., Mellman I. Pd-L1 expression by dendritic cells is a key regulator of T-cell immunity in cancer. Nat. Can. (Que.) 2020;1(7):681–691. doi: 10.1038/s43018-020-0075-x. [DOI] [PubMed] [Google Scholar]
- 53.Lei J., Zhang D., Yao C., Ding S., Lu Z. Development of a predictive immune-related gene signature associated with hepatocellular carcinoma patient prognosis. Cancer Control. 2020;27(1) doi: 10.1177/1073274820977114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Viscardi G., Tralongo A.C., Massari F., Lambertini M., Mollica V., Rizzo A., Comito F., Di Liello R., Alfieri S., Imbimbo M., Della Corte C.M., Morgillo F., Simeon V., Lo Russo G., Proto C., Prelaj A., De Toma A., Galli G., Signorelli D., Ciardiello F., Remon J., Chaput N., Besse B., De Braud F., Garassino M.C., Torri V., Cinquini M., Ferrara R. Comparative assessment of early mortality risk upon immune checkpoint inhibitors alone or in combination with other agents across solid malignancies: a systematic review and meta-analysis. Eur. J. Cancer. 2022;177:175–185. doi: 10.1016/j.ejca.2022.09.031. [DOI] [PubMed] [Google Scholar]
- 55.Mao X., Xu J., Wang W., Liang C., Hua J., Liu J., Zhang B., Meng Q., Yu X., Shi S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: new findings and future perspectives. Mol. Cancer. 2021;20(1):131. doi: 10.1186/s12943-021-01428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Bao X., Hu M., Chang H., Jiao M., Cheng J., Xie L., Huang Q., Li F., Li C.Y. Inhibition of Pcsk9 potentiates immune checkpoint therapy for cancer. Nature. 2020;588(7839):693–698. doi: 10.1038/s41586-020-2911-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Helmink B.A., Reddy S.M., Gao J., Zhang S., Basar R., Thakur R., Yizhak K., Sade-Feldman M., Blando J., Han G., Gopalakrishnan V., Xi Y., Zhao H., Amaria R.N., Tawbi H.A., Cogdill A.P., Liu W., Lebleu V.S., Kugeratski F.G., Patel S., Davies M.A., Hwu P., Lee J.E., Gershenwald J.E., Lucci A., Arora R., Woodman S., Keung E.Z., Gaudreau P.O., Reuben A., Spencer C.N., Burton E.M., Haydu L.E., Lazar A.J., Zapassodi R., Hudgens C.W., Ledesma D.A., Ong S., Bailey M., Warren S., Rao D., Krijgsman O., Rozeman E.A., Peeper D., Blank C.U., Schumacher T.N., Butterfield L.H., Zelazowska M.A., Mcbride K.M., Kalluri R., Allison J., Petitprez F., Fridman W.H., Sautes-Fridman C., Hacohen N., Rezvani K., Sharma P., Tetzlaff M.T., Wang L., Wargo J.A. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puig-Saus C., Sennino B., Peng S., Wang C.L., Pan Z., Yuen B., Purandare B., An D., Quach B.B., Nguyen D., Xia H., Jilani S., Shao K., Mchugh C., Greer J., Peabody P., Nayak S., Hoover J., Said S., Jacoby K., Dalmas O., Foy S.P., Conroy A., Yi M.C., Shieh C., Lu W., Heeringa K., Ma Y., Chizari S., Pilling M.J., Ting M., Tunuguntla R., Sandoval S., Moot R., Hunter T., Zhao S., Saco J.D., Perez-Garcilazo I., Medina E., Vega-Crespo A., Baselga-Carretero I., Abril-Rodriguez G., Cherry G., Wong D.J., Hundal J., Chmielowski B., Speiser D.E., Bethune M.T., Bao X.R., Gros A., Griffith O.L., Griffith M., Heath J.R., Franzusoff A., Mandl S.J., Ribas A. Neoantigen-targeted Cd8(+) T cell responses with Pd-1 blockade therapy. Nature. 2023;615:697–704. doi: 10.1038/s41586-023-05787-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barsch M., Salie H., Schlaak A.E., Zhang Z., Hess M., Mayer L.S., Tauber C., Otto-Mora P., Ohtani T., Nilsson T., Wischer L., Winkler F., Manne S., Rech A., Schmitt-Graeff A., Bronsert P., Hofmann M., Neumann-Haefelin C., Boettler T., Fichtner-Feigl S., Van Boemmel F., Berg T., Rimassa L., Di Tommaso L., Saeed A., D'alessio A., Pinato D.J., Bettinger D., Binder H., John Wherry E., Schultheiss M., Thimme R., Bengsch B. T-cell exhaustion and residency dynamics inform clinical outcomes in hepatocellular carcinoma. J. Hepatol. 2022;77(2):397–409. doi: 10.1016/j.jhep.2022.02.032. [DOI] [PubMed] [Google Scholar]
- 60.Luoma A.M., Suo S., Wang Y., Gunasti L., Porter C.B.M., Nabilsi N., Tadros J., Ferretti A.P., Liao S., Gurer C., Chen Y.H., Criscitiello S., Ricker C.A., Dionne D., Rozenblatt-Rosen O., Uppaluri R., Haddad R.I., Ashenberg O., Regev A., Van Allen E.M., Macbeath G., Schoenfeld J.D., Wucherpfennig K.W. Tissue-resident memory and circulating T cells are early responders to pre-surgical cancer immunotherapy. Cell. 2022;185(16):2918–2935. doi: 10.1016/j.cell.2022.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. Material/referenced in article.