Abstract
The activity of alginic acid as a cytotoxic agent was improved by structure modification using 4-aminophenol (4-AP) through condensation and polymerization processes. Then, silver nanoparticles were employed through doping to further enhance the cytotoxic activity of the modified polymer. The structure of the prepared materials was characterized by FT-IR, 1HNMR, UV spectroscopy, X-ray diffraction, and electron microscopy, and the thermal behavior of all synthesized materials was intensively studied. The cytotoxicity of the prepared compounds against cell lines of human hepatocellular (HepG-2) and lung (A-549) carcinomas was investigated. Alginic acid modified with 4-AP (Alg-4-AP3) showed the highest activity against HepG-2 and A-549 among all tested materials with IC50 values of 3.0 ± 0.19 μg/mL and 3.63 ± 0.23 μg/mL, respectively. Multitargeted molecular docking was employed to explore the binding modes of our compounds with the receptors EGFR, HER2, and VEGFR 2. The results revealed the inhibitory activity of our tested compounds against the proposed protein receptors, findings coincided with the in vitro results. In conclusion, the modification of alginic acid with 4-AP improved its cytotoxic activity against HepG-2 and A-549 cancer cells. In addition, doping the new materials with silver nanoparticles (AgNPs) further enhanced the cytotoxic activity.
Keywords: Alginic acid, 4-Aminophenol, Silver nanoparticles, Cytotoxicity
Graphical abstract
1. Introduction
During the last decade, silver nanoparticles (AgNPs) have offered extraordinary potential for applications in the catalytic, electrical, optical, biological, and medicinal fields because of their distinctive physical and chemical characteristics [[1], [2], [3], [4], [5], [6]]. Owing to their broad-spectrum therapeutic applications, AgNPs have recently attracted much attention [6,7]. AgNPs are generally obtained through the controlled reduction of silver ions by employing strong reducing agents, including sodium borohydride in chemical synthesis pathways [8]. However, AgNPs possess high tendency to aggregate and their shape is altered throughout catalytic reactions as a result of the high surface energy nascent from the high surface-to-volume ratio [9]. The aggregations might lead to considerable undesired depression in the catalytic activity and selectivity [10], and polymers could prevent aggregation when utilized as capping and stabilizing agents [11]. The production of AgNPs by conventional chemical routes are likely to involve harmful reagents and toxic byproducts that are hazardous to the environment. Therefore, the use of polymers for capping the nanoparticle surface is of significant value not only to prevent aggregation but also to protect the environment [12].
The synthesis of AgNPs is carried out using physical and chemical methods, biological or green methods, and photochemical methods [13,14]. The physical strategies include spark discharging, evaporation–condensation, and transformation [15]. The green synthesis approach in which environmentally friendly raw materials and/or sustainable synthetic methods is preferred [13]. The use of non-toxic solvents, reagents and separation products, such as water and plant extracts, minimization of energy usage, and the use of chemicals that are safe for both humans and the environment should all be incorporated into the syntheses whenever possible [16]. Green synthesis based on chemical or physical techniques such as ultrasound- or microwave-based synthesis, hydro- or solvothermal synthesis, photocatalysis, etc., and biological techniques could be used to prepare AgNPs [17]. Photophysical (top to bottom) and photochemical (bottom to up) are two strategies are commonly recognized for the photochemical synthesis of AgNPs, [15]. The former employs metals in their metallic state and the latter utilizes their ionic precursor [18]. The direct photoreduction technique is also employed for the synthesis of NPs of various metals [19]. Photosensitization is used to prepare AgNPs, where in metallic ion reduction occurs with excited species generated photochemically [15]. The growing interest to environmental problems such as pollution and waste disposal along with the subedited rules and regulations to these issues are leading factors to produce more eco-friendly materials for industrial and medicinal applications. Alginic acid (Alg), algin, or alginate are widely used natural anionic polysaccharide extracted from several brown seaweed, and could also obtained by the microbial fermentation [20,21]. Alginate could be prepared as a biobased fiber by mixing wet-spun sodium alginate with a coagulating bath including aqueous calcium chloride (CaCl2) solution [21,22]. The large length-to-diameter ratio of the produced alginate fibers suggest them as an advantageous supporting material for stabilizing AgNPs with easily accessible high surface area [2] The wide range of environmental and biological applications of alginate is mainly attributed to its cost-effective, biodegradable, non-toxic and biocompatible characteristics [21].
Dysregulation of the expression of growth factor receptors (GFRs) promoted by genetic, epigenetic and somatic changes can lead to cancer initiation and progression [23]. Vascular endothelial growth factor receptor (VEGFR), epidermal growth factor receptor (EGFR), human epidermal growth factor receptor 2 (HER2) are upregulated in many different types of malignancies [23,24]. Thus, suppression of EGFR, HER2 and VEGFR singling routes represents an efficient strategy for the production of novel antitumor agents [25]. Two essential routes for chemotherapeutic treatment were proposed. These include the production of active tyrosine kinase inhibitors and targeting the binding pocket domain EGFR and HER2 by means of monoclonal antibodies that suppress the dimerization and consequently the biochemical cascade [26]. The suppression of EGFR family inhibits the course of the cell cycle and increases apoptosis in a plethora of tumor types [27]. Drugs displaying inhibitory activity against HER2 by inhibiting tyrosine kinase domain of the receptors were generally accepted for the chemotherapy of various types of cancers [27]. VEGFR is believed to be one of the most effective pro-angiogenic signaling molecules responsible for angiogenesis in many different malignancies [28]. Accordingly, drugs exhibiting inhibitory activity against EGFR and VEGFR-2 are clinically approved for the treatment of many types of tumors [29]. However, drug resistance may be induced by administering the same treatments repeatedly, necessitating the development of new candidate drugs. The in silico approaches have offered an effective route for the identification of novel therapeutic agents against several diseases [[30], [31], [32]]. This work proposed the employment of three stages synthesized Alg. modified with 4-aminophenol (4-AP)_doped with AgNPs as potent cytotoxic agents against hepatocellular and lung cancer cell lines. The binding affinity of the tested agents against EGFR, HER2 and VEGFR-2 was investigated by molecular docking (MD) assessments.
2. Materials and methods
2.1. Materials
Alg (Mwt. 80,000) was purchased from Alfa-Aesar (USA) and 4-AP was supplied by Across-Organics (USA). Ammonium persulfate was purchased silver nitrate (AgNO3), xylene, and sodium citrate from Sigma (USA).
2.2. Methods
2.2.1. Synthesis of Alg. condensed with 4-AP
Alg. (2 g) and 4-AP (0.183 mol) were added to a round bottom flask with 100 mL of xylene and refluxed for 7 h at 200 °C using Dean–Stark apparatus. The solid product (Alg-4-AP1) was separated by filtration and dried at room temperature. Ammonium persulfate (0.0438 mol) dissolved in the least amount of water was added to 50 mL distilled water containing 1 g Alg-4-AP1 in a flask and adjusted at room temperature then 0.183 mol of 4-AP was added to the reaction mixture dropwise with stirring for 3 h. The solid product (Alg-4-AP2) was separated, washed with water and methanol then dried under a vacuum at 60 °C (Scheme 1).
Scheme 1.
Alginic acid modified with 4-Aminophenol.
2.2.2. Preparation of AgNPs
Chemical reduction using a reducing and capping agent has been followed as previously reported [33,34]. 0.000941 mol of AgNO3 was added to 100 mL distilled water and heated till boiling then 0.0968 mol of trisodium citrate dissolved in 10 mL of distilled water added drop wisely with stirring at room temperature till the solution turned yellow. The reaction mixture was cooled down and stirred for 4 h [35].
2.2.3. Assembling of Alg-4-AP2 on AgNPs (Alg-4-AP3)
20 mL of colloidal AgNPs was added to Alg-4-AP2 solution with stirring for 24 h till the solution color changed. The solid product (Alg-4-AP3) was separated by filtration, washed, and dried under vacuum at 60 C.
2.2.4. Cytotoxicity assay
HepG-2 and A-549 cell lines (American Type Culture Collection (ATCC, Rockville, MD)) were cultured in RPMI-1640 (Lonza, Belgium) supplemented with 10% fetal bovine serum (Sigma, USA), 1% l-glutamine, HEPES buffer, and antibiotics, and maintained at 37 °C and 5% CO2. The cells were seeded in 96-well plates (1 × 104 cells/well) in 100 μl medium and incubated for 24 h. A fresh medium containing different concentrations of the test agents or doxorubicin followed by incubation for 24 h. The MTT assay was employed to determine cell viability. In brief, MTT (0.5 mg/mL final concentration) was added, and the plate was incubated at 37 °C for 4 h and then 50 μl/well of DMSO. After 10 min, the absorbance was measured at 590 nm using a microplate reader (TECAN, USA). The 50% inhibitory concentration (IC50) was calculated using GraphPad Prism 8.
2.2.5. In silico molecular docking (MD) analysis
The binding profiles of the synthesized drug (Alg-4-AP3) against EGFR, HER2, and VEGFR were examined by MD analysis. The geometrical structure of the tested drug was fully optimized at the B3LYP exchange-correlation functional level of theory without constraints [36,37] using the 6-311G (d, p) basis set [38]. Density functional theory (DFT) investigation was utilized in this work and was executed using Gaussian 16 software [39]. Frequency calculations were implemented at the same level of theory to assure that the tested drug is in the ground state and to confirm the absence of imaginary frequencies. The 3D crystal structure of the three targets was downloaded from the Protein Data Bank (PDB). The pdb identification codes (ids) for the studied receptors are EGFR (PDB ID: 2J6M), HER2 (PDB ID: 3PP0), and VEGFR (PDB ID: 3B8Q). The MD run was performed using Autodock Tools (ADT) v1.5.6 and AutoDock Vina programs [40]. Different drug-receptor complexes were optimized for docking by removal of nonstandard amino acids residues, stripping out solvent molecules, adding polar hydrogens, and adjusting the grid box to the active site amino acids residues [30,31]. These optimization processes were carried out through ADT v1.5.6. UCSF Chimera software was used to eliminate original drugs from the original pdb structure and for clean original pdb generation [41]. PyMOL v2.4 was employed for high-resolution figures generation and binding interactions molecular visualizations.
2.2.6. Capacity of Alg-4-AP2 grafting
The number of 4-AP units in the grafting side chain of Alg-4-AP1 was calculated from the mole ratio between Alg-4-AP1 and 4-AP in the last step of graft as the chain length of 4-AP on Alg-4-AP1 is found to be 4.8 ≈ 5.
2.3. Instrumentations
UV–Vis spectra were recorded on Shimadzu 2040 instrument. Fourier transforms infrared (FTIR) spectrometer (VERTEX 70 F T-IR) spectra were measured in ATR discs at room temperature within the wavenumber range of 400–4000 cm−1. Bruker high-performance digital FT-NMR spectrometer Avance III 400 MHZ was employed to perform routine 1H/13C high–resolution spectra along with common 2D experiments (COSY, HMBC, HSQC, …). The crystalline structure of prepared polymers was checked by X-ray diffraction (2,020,964 PA Analytical Empyrean). A scanning electron microscope (SEM; JEOL (JSM-5200)) was employed to investigate the morphologies of the prepared polymers. Samples were prepared by placing a slight part of a film on a carbon tube on a stub, which was coated with a gold thin layer. Micrographs of the prepared new polymeric materials using a JEM-100S Transmission Electron Microscope (TEM, Jeol, Tokyo, Japan) were taken. The different area pictures were captured at adjustable magnifications that identify the particle size and morphology. TGA-50H thermogravimetric analyzer was used to study the thermal stability of synthesized polymeric materials. Samples were heated from 10 to 600 °C in a platinum pan with a heating rate of 10 °C min−1 under an N2 atmosphere with a flow rate of 25 mL min−1.
3. Results and discussion
3.1. Characterization
3.1.1. IR
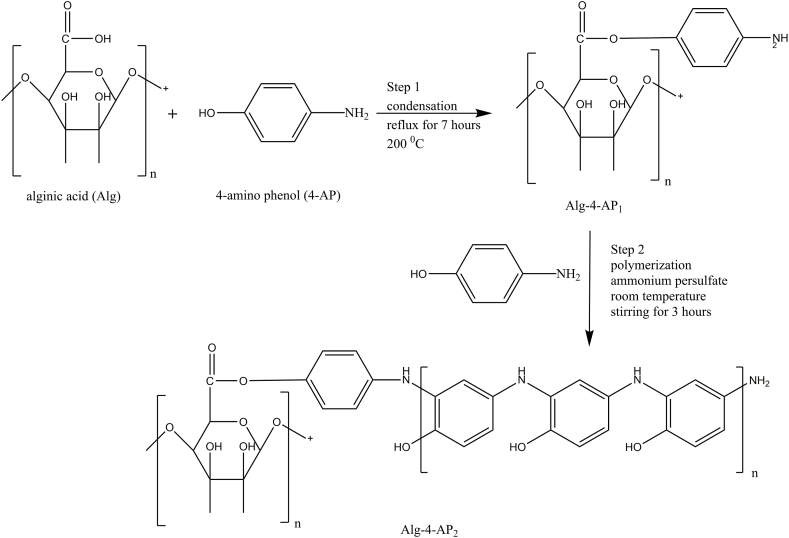
FT-IR was employed to figure out the functional groups present in Alg by means of typical vibration frequencies [42]. The following vibrational frequencies were detected in Alg sample by means of FTIR spectra (Fig. 1). The obtained broadband in the range of 3700–3000 cm−1 was detected for the OH stretching frequency and that in the range 3000–2300, cm−1 was assigned for C–H stretching vibration [43]. Alg is known to be the building unit of mannuronic and guluronic acid residues comprising one –COO group [44]. The bands near 1650 cm−1 and 1400 cm−1 in the obtained spectrum revealed an asymmetric and symmetric vibrations of carboxylate O–C–O stretching, respectively [45]. The deformation of O–H and C–O stretching frequencies were noticed at 1395 cm−1 and 1333 cm−1, respectively. The bands around 1100 cm−1 -1027 cm−1 were assigned to the anti-symmetric stretch of C–O–C [45]. The band at 1150 cm−1 showed the presence of o-acetyl ester in the bacterial alginates [46]. The first modification of alginic acid is condensation with 4-amino phenol (Alg-4-AP1) which is confirmed by the IR (Fig. 1). It is noticed that the bands appear in the range of 3341–3284 cm−1 which is assigned to the N–H stretching of primary amines. 1604 cm−1 was due to the N–H bending of aromatic amine and C-C group stretching. 1512 cm−1 was due to C–C in-ring stretching vibration peak. The vibration peaks for C–O were observed at 1031-1085 cm−1 and C–N stretching peaks were observed at 1269-1282 cm−1 which indicates the successful condensation between Alg through the –COO group and 4-AP through the OH group. The second modification is a polymerization process of Alg-4-AP1 with an excess of 4-aminophenol to give Alg-4-AP2 in polyaminophenol (Fig. 1). After the incorporation of AgNPs into the grafting process, all the peaks have been slightly shifted indicating the assembling of Alg-4-AP2 onto AgNPs by a chemical which we can observe a peak at 3191 cm−1 related to the bonding of Ag0 with N and/or O-atoms (Alg-4-AP3) (Fig. 1) [47]. The variations of the OH and –COO groups have been reported in the previous study on the synthesis of AgNPs with another polysaccharide [48].
Fig. 1.
FTIR spectrum of alginic acid (Alg), alginic acid condensed with 4- aminophenol (Alg-4-AP1), alginic acid condensed with p-aminophenol polymer (Alg-4-AP2), and alginic acid condensed with p-aminophenol dopped with silver nanoparticles (Alg-4-AP3).
3.1.2. 1H NMR
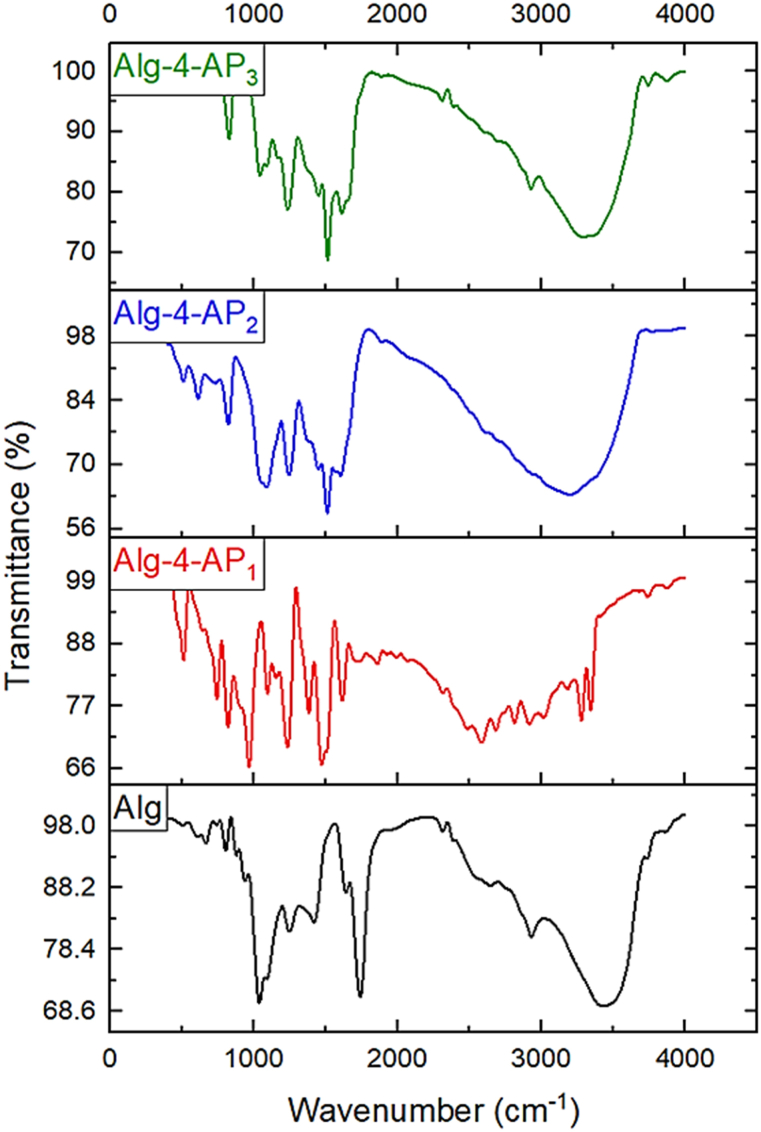
The 1H NMR spectra of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3 are represented in Fig. 2. Based on the 1H NMR spectrum resonances, the region between 3.2 and 4.4 ppm up-taking polysaccharides are sugar ring protons and the region from 4.4 to 5.6 up-taking polysaccharides are anomeric protons [49]. In addition to Alg peaks, a new peak appeared related to 4-AP at 4.7 ppm for H of an amino group and at 6.9–7 ppm for aromatic protons of the benzene ring. The disappearance of the shift related to the H for phenolic OH confirms the successful condensation between Alg and 4-AP. The very low-intensity peak related to aromatic protons of the benzene ring (6.9–7 ppm) which can be related to the presence of silver metals which share with polymer chains between the benzene rings with π-electrons of benzoid structure to form Sandwich composite was observed. Also, folded chains caused by the silver restrict the functionality of H protons which causes the low intensity of benzene ring protons.
Fig. 2.
Characterization by 1H NMR spectroscopy of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3.
3.1.3. X-ray diffraction (XRD)
XRD of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3 are illustrated in Fig. 3. From the XRD pattern of Alg, 3 characteristic peaks were observed at 2θ = 13.07°, 21.5°, and 41.15°, suggesting long-range disorder along with the amorphous feature exists in the Alg [[50], [51], [52]]. For Alg-4-AP, some peaks appeared to overlap with Alg peaks at around 2θ = 17.6°, 19.4°, 24°and 25°, revealing the crystalline nature of prepared 4-AP. For polymerized Alg-4-AP, the crystalline form of the polymer was different as compared with Alg and Alg-4-AP, where sharp peaks appeared at around 2θ = 20° and small peaks appeared around 2θ = 30. This observation suggested the successful reaction between Alg and 4-AP. For loaded silver nanoparticles with polymerized Alg-4-AP, there are main peaks at 20.36°, 28.04°, 32.4°, 38.3°. These peaks are due to the crystalline and amorphous organic phases, accompanying crystallized AgNPs. Also, the crystallite size after AgNPs loading became larger than before 918.453600 Å, wherein the case of Alg-4-AP1 was 55.132810 Å, polymerized Alg-4-AP2 became 48.164050 Å. These all observations confirmed the reaction.
Fig. 3.
XRD spectrum of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3.
3.1.4. UV–vis spectroscopy
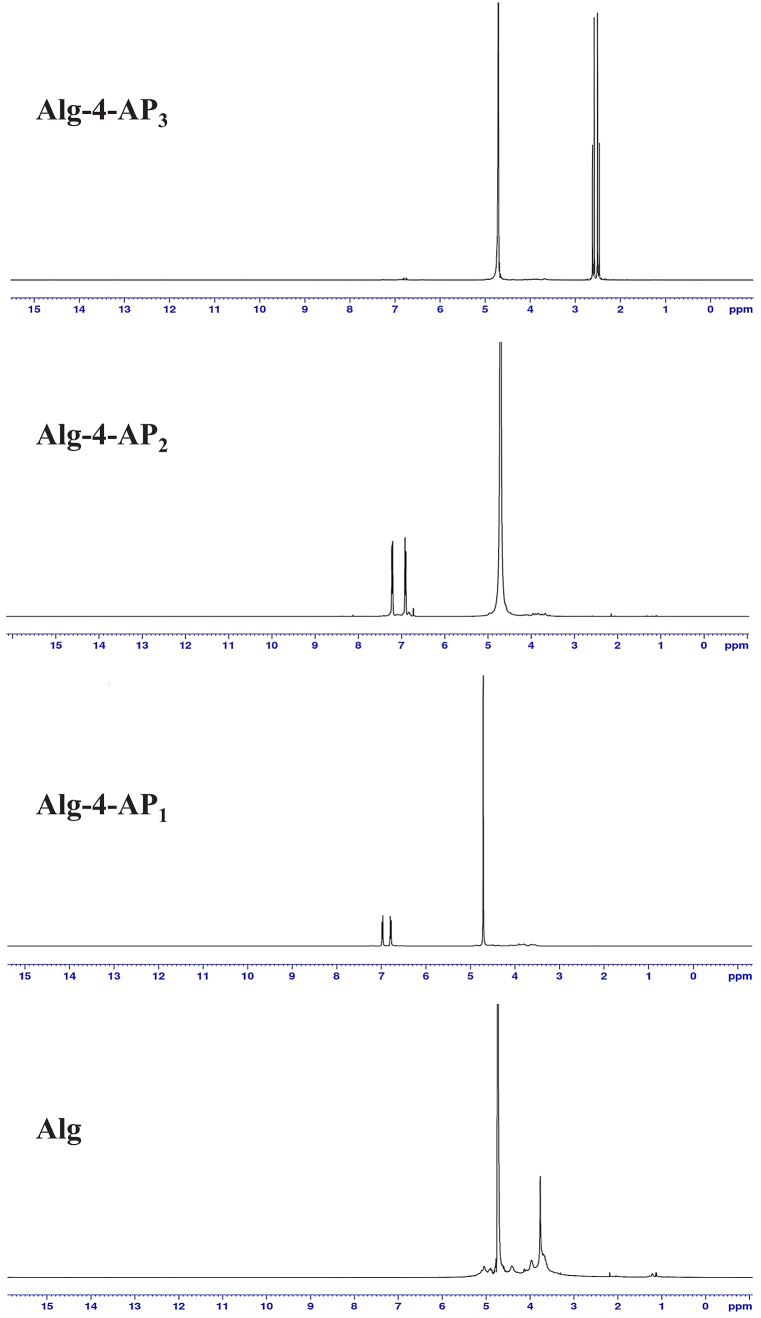
UV–Vis spectroscopy has been used to determine the optical properties of the prepared materials (Fig. 4). The strong absorption band at 290 nm for the Alg could be related to the n-π* transitions of carbonyl in the –COO group [52]. The presence of a new peak at around 238 nm indicates the presence of 4-AP [37]. The peak for AgNPs is shifted to 339 nm due to the presence of prepared polymer. The shift AgNPs peak in the UV–vis spectrum is mainly attributed to the doping of AgNPs to the newly synthesized polymer. Thus, the absorption of AgNPs is strongly influenced by the tendency of polymeric materials to adsorb Ag+ and AgNPs particles. Also, an increase in the absorbance intensity with the introduction AgNPs because of the interaction of the nanosized particles with the polymeric system was observed.
Fig. 4.
UV spectrum of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3.
3.1.5. Thermal analysis
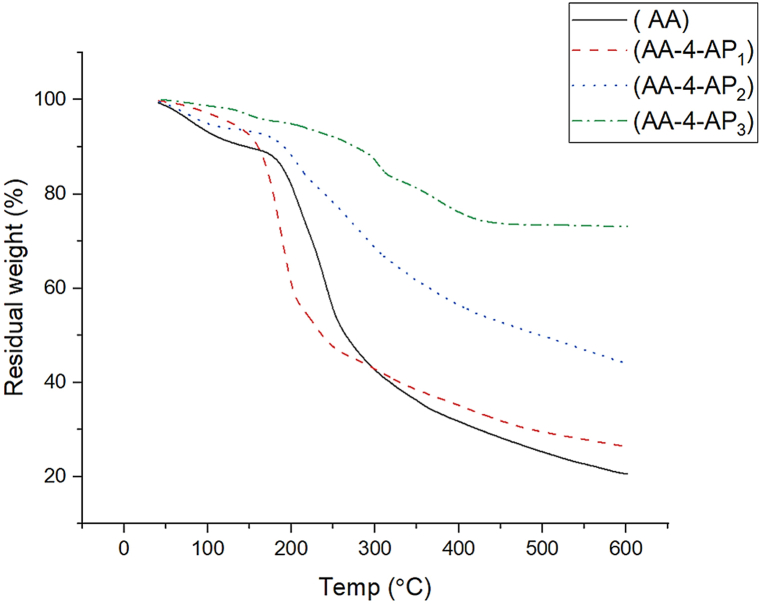
TGA analysis of alginic acid, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3 is shown in Fig. 5 and Table 1. The thermogram of Alg revealed weight loss in four succeeding steps. The 1st step (25–100 °C) a loss of moisture. The 2nd step (100–190 °C) decomposition of bonded water and degradation of –COO groups and consequent loss of CO2 (decarboxylation). The weight loss in the 3rd step (190–270 °C) could be ascribed to the fragmentation of the biopolymer backbone. Complete disintegration was observed in the 4th step (300–600 °C) due to the absolute decomposition of the polysaccharide skeleton [53] with a residual weight of about 20% at 600 °C. For Alg-4-AP1, three decomposition steps ((25–150 °C), (150–250 °C) and (250–600 °C)) with total residual weight 26% at 600 °C could be observed. For Alg-4-AP2, three decomposition steps first one at (25–100 °C), the second one (100–200 °C) with a residual weight of 90%, and the third step started at 200 °C–600 °C with residual weight 44% were observed. For Alg-4-AP3, we observed a great increase in thermal stability of the prepared materials as we can notice a slight decrease in (Alg-4-AP4) weight with an increase in temperature from 25 to 600 °C with residual weight 73% which can be related to the doping of AgNPs.
Fig. 5.
TGA of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3.
Table 1.
TGA fragment and assignments of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3.
| Temperature range (°C) | Polymeric sample weight loss % |
Assignment | Reference(s) | |||
|---|---|---|---|---|---|---|
| Alg | Alg-4-AP1 | Alg-4-AP2 | Alg-4-AP3 | |||
| 25–100 | 93 | 90 | 92 | 98 | Loss of moisture | [53,65] |
| 100–190 | 85 | – | – | – | Decarboxylation of bonded water | [53] |
| 100–200 | – | – | 87 | – | Decarboxylation of bonded water | – |
| 150–250 | – | 45 | – | – | Decarboxylation of bonded water | – |
| 190–270 | 48 | – | – | – | fragmentation of biopolymer backbone | [53] |
| 250–600 | – | 26 | – | – | Complete disintegration | – |
| 200–600 | – | – | 44 | – | Complete disintegration | – |
| 300–600 | 20 | – | – | 73 | Complete disintegration | [53,65] |
| Residual weight at 600 °C | 20 | 26 | 44 | 73 | – | – |
3.1.6. SEM
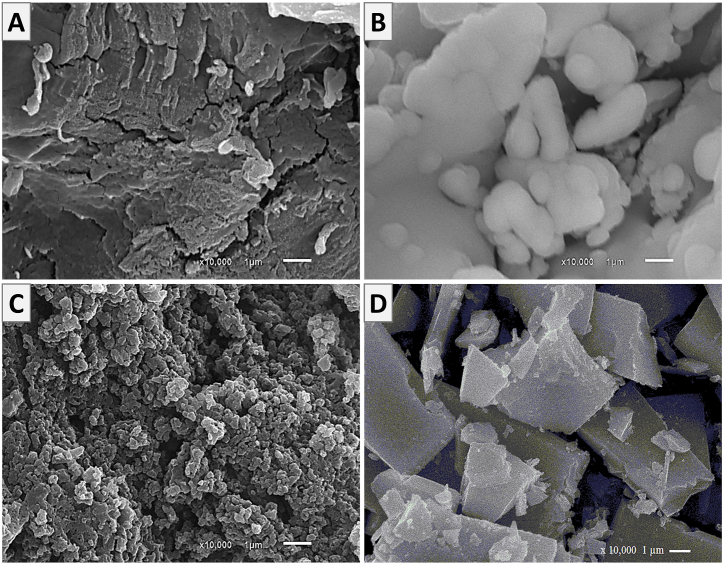
SEM of Alg, Alg-4-AP1, Alg-4-AP2, and Alg-4-AP3 are shown in Fig. 6 (A, B, C and D, respectively). The images showed a difference in surface morphology between all samples confirming the synthesis of new products. The smooth surface of Alg has been entirely distorted and changed to different structures. It seems that the particles looked bulky and the backbone of Alg is covered with 4-AP. SEM can be used as evidence of the modification process.
Fig. 6.
SEM of (A) Alg, (B) Alg-4-AP1, (C) Alg-4-AP2, and (D) Alg-4-AP3.
3.1.7. TEM
Fig. 7 shows TEM images of Alg (Fig. 7A), Alg-4-AP1 (Fig. 7B), Alg-4-AP2 (Fig. 7C), and Alg-4-AP3 (Fig. 7D). TEM images also confirmed the modification of Alg with 4-AP in the two modification steps (condensation and polymerization) TEM images of AgNPs show the spherical shape of the prepared particles in the range of 6.8–7.7 nm in addition to aggregated particles with larger size [54] (Fig. 7D).
Fig. 7.
TEM of (A) Alg, (B) Alg-4-AP1, (C) Alg-4-AP2, (D) Alg-4-AP3.
3.2. Cytotoxic activity
IC50 is the most widely employed tool to investigate the anti-proliferative efficacy of a specific drug. It reflects the amount of a drug required to inhibit a biological process by half, thus offering an effective tool to estimate the potency of an antagonist drug in pharmaceutical studies [55]. The potent cytotoxicity of whole synthesized compounds towards HepG-2 and A-549 cells was evaluated in vitro. The dose-response curves are introduced within Fig. 8, Fig. 9 for HepG-2 and A-549 cells, respectively.
Fig. 8.
Cytotoxic activity of Alg (A), Alg-4-AP1 (B), Alg-4-AP2 (C), Alg-4-AP3 (D), and doxorubicin (E) on HePG-2 cells.
Fig. 9.
Cytotoxic activity of Alg (A), Alg-4-AP1 (B), Alg-4-AP2 (C), Alg-4-AP3 (D), and doxorubicin (E) on A-549 cells.
All tested compounds exerted a reduction influence on HepG-2 (Fig. 8A–E) and A-549 (Fig. 9A–E) cell viability. The outputs revealed that increasing the concentration of the drugs led to a significant reduction of cell viability of the two tested cell lines. For HepG-2, Alg showed the lowest anti-proliferative activity (IC50 = 46.1 μg/mL) while Alg-4-AP3 was the most active compound (IC50 = 3.08 μg/mL). The same results also appeared for A-549 which showed that Alg has the lowest activity (IC50 = 87.5 μg/mL) while Alg-4-AP3 has the highest activity (IC50 = 3.63 μg/mL) (Fig. 9). Alg-4-AP1 showed a higher cytotoxic activity against HepG-2 and A-549 cells than Alg-4-AP2. This higher activity of Alg-4-AP1 could be attributed to the formation of a newly polymeric structure (Sandwich structure) which block the activity of free amino groups and restrict the activity of benzene rings in the polymeric chain. In addition, the prepared materials exhibited higher cytotoxic activity against HepG-2 than A-549 cells with the following order Alg-4-AP3 > Alg-4-AP1 > Alg-4-AP2 > Alg. By comparing the results with the positive control (Doxorubicin with IC50 = 0.95 μg/mL for A-549 cell line and IC50 = 2.62 μg/mL for HepG-2 cell line) we noticed that the prepared new materials exhibit a promising cytotoxic activity.
The efficacy of our newly synthesized compounds is mainly due to the introduction of amino groups and the cellular influence of AgNPs in the new chain. Consequently, the prepared drug exerts its cytotoxicity by three main routes, including increasing reactive oxygen species (ROS) production, depolarization of mitochondrial membrane and mitochondrial dysfunction [13]. Nevertheless, the influence of AgNPs in tumors and normal cells is compared, the latter showed lower or no toxicity. These meager toxicities in normal cells occurred due to the inclusion of natural compounds as surface capping for AgNPs or because of specific physical features of the NPs. Besides, this effect is likely because of the highest uptake of AgNPs by tumor cells, an outcome of their anomalous metabolic behavior and the increased proliferation rate which makes them more susceptible to AgNPs cell penetration.
3.3. MD analysis
Herein, we carried out a multitargeted MD to explore the potential binding profile of our newly synthesized compounds with three target receptors namely, EGFR, HER2, and VEGFR. The binding energies, hydrogen bonding, and probable hydrophobic interactions are represented in Table 2.
Table 2.
Binding affinities, polar bonding, and hydrophobic interactions of Alg-4-AP3 against the proposed target receptors.
| Target | Binding energy (kcal/mol) | Polar bonding | Hydrophobic interactions |
|---|---|---|---|
| EGFR | −6.9 | Met793 | Leu 718, Val 726, Ala 743, Lys 745, Glu 762, Leu 788, Thr790, Leu 792, Gly 796 and Leu 844 |
| HER2 | −7.4 | Pro 761, Lys 765, Glu 764, Arg 966 and Glu 975 | Lys 957, Cys 965, Pro 967 and Glu 971 |
| VEGFR 2 | −6.5 | Thr859, Thr861, Glu 917 and Glu 1038 | Leu 836, Gly 837, Lys 838, Pro 839, Asp 857, Arg 863, Ser 1037 and Lys 1043 |
Drug-receptor polar and hydrophobic interactions are considered the main factors controlling the stabilities of biological macromolecules. Polar interactions are believed to be the leading factor affecting the binding of various drugs in the active site of the target protein [56,57]. Thus, these interactions play a crucial role in maintaining ligands in the binding site of the protein, molecular recognition, and conformational alignment in the binding cavity [58,59]. The binding affinity of the ligand to the binding site is the second major factor affecting the stability of the drug-target complex. This energy is significantly influenced by the hydrophobic binding of the ligand's lipophilic surface to the hydrophobic residues in the active site [60,61]. The number of formed hydrogen bonds was shown to affect the binding energy values considerably [58]. Thus, for a thermodynamically favorable drug-protein interaction, a suitable geometrical alignment of the drug into the protein's binding pocket is essential. The coincidence between the results of MD and the biochemical outputs represents a chance to evaluate the efficacy of a specific drug as a potent cytotoxic agent.
The outputs of MD revealed the activity of our drug against EGFR as represented in Fig. 10A–C, a finding mainly attributed to the obtained low binding affinity of −6.9 kcal/mol. The proposed drug was shown to dock into the main central channel of the receptor surrounded by a dense network of hydrophobic interacting residues. Although the extent of polar interactions in EGFR bindings is not very common, one polar bond was detected with a significant key residue Met793 in the binding site of this complex. Thus, our results suggested a high probability for this drug to exhibit activity against tumor cells by targeting EGFR.
Fig. 10.
Binding site of Alg-4-AP3 with EFGR. Residues are shown as stick model (A), surface representation of the binding cavity of EFGR occupied by the drug (B), and residues exhibiting polar and hydrophobic interactions with the drug (C).
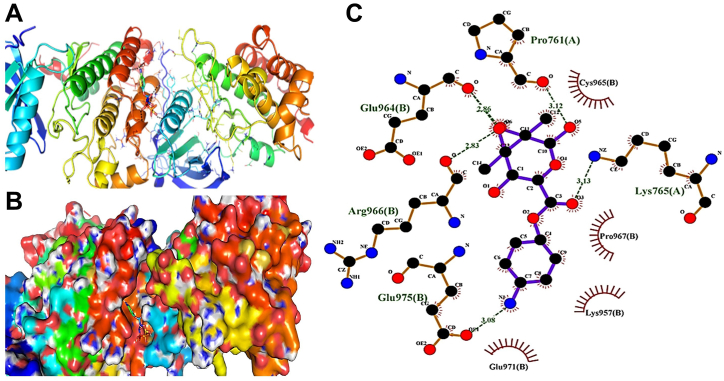
The binding affinity of the tested agent towards the kinase domain of HER2 was carried out using MD (Fig. 11A–C). This domain is thought to be a potential target for chemotherapeutic remedies for different tumors in humans [62]. The compatibility of the synthesized agent to the active site of HER2 is estimated from the obtained low binding free energy (−7.4 kcal/mol). Interestingly, six polar bonds were detected in the binding site of this complex. This inference along with the location of the drug on the surface of the main binding site suggests the formation of a stable drug-receptor complex. The low binding free energy and the involvement of significant key residues in the binding interactions of this complex suggested the inhibitory activity of the tested agent against HER2 [63].
Fig. 11.
Binding site of Alg-4-AP3 with HER2. Residues are shown as stick model (A), surface representation of the binding cavity of EFGR occupied by the drug (B), and residues exhibiting polar and hydrophobic interactions with the drug (C).
Because of the bioavailability of VEGFR as an essential receptor protein tyrosine kinase in charge of propagating signaling pathways, an MD analysis was executed to find out the activity of the proposed drug as an inhibitor (Fig. 12A–C). The results displayed the high coexistence of polar and hydrophobic interactions, including active recognized residues in VEGFR inhibition pathways [64]. The prepared drug was shown to occupy the main active site in the protein skeleton with a relatively low binding affinity (−6.5 kcal/mol).
Fig. 12.
Binding site of Alg-4-AP3 with VEGFR. Residues are shown as stick model (A), surface representation of the binding cavity of EFGR occupied by the drug (B), and residues exhibiting polar and hydrophobic interactions with the drug (C).
4. Conclusion
Alginic acid was successfully modified with 4-AP, and the structure of the newly synthesized compound was confirmed by FT-IR, 1HNMR, XRD, UV, spectroscopy, and electron microscopy. The obtained results showed that this modification improved its activity as a cytotoxic agent against HePG-2 and A-549 cancer cells. Doping the new materials with AgNPs further enhanced the cytotoxic activity. According to the obtained binding affinities, hydrogen bonds, and probable hydrophobic interactions of the tested drug against the proposed protein receptors EGFR, HER2, and VEGFR, the results pointed to the involvement of these receptors in the observed cytotoxic activity. However, further in vitro and in vivo studies are needed to explore the mechanism(s) underlying the anti-proliferative and anti-tumor activity of the tested agent.
Author contribution statement
Haneen A. Mahmoud: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Emadeldin M. Kamel, Ayman M. Mahmoud: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Reem S. Alruhaimi, Ali M. El-Zanaty, Hanafy M. Abd El-Salam: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Omayma F. Abdel-Gawad: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Data availability statement
Data included in article/supp. Material/referenced in article.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2023R381), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Contributor Information
Emadeldin M. Kamel, Email: emad.abdelhameed@science.bsu.edu.eg.
Omayma F. Abdel-Gawad, Email: omayma2013@hotmail.com.
References
- 1.Chook S.W., et al. A porous aerogel nanocomposite of silver nanoparticles-functionalized cellulose nanofibrils for SERS detection and catalytic degradation. rhodamine B. 2015;5(108):88915–88920. [Google Scholar]
- 2.Zhao X., et al. Alginate fibers embedded with silver nanoparticles as efficient catalysts for reduction. 4-nitrophenol. 2015;5(61):49534–49540. [Google Scholar]
- 3.Zayed M.F., et al. Malva parviflora extract assisted green synthesis. silver nanoparticles. 2012;98:423–428. doi: 10.1016/j.saa.2012.08.072. [DOI] [PubMed] [Google Scholar]
- 4.Kareem M., et al. IOP Conference Series: Materials Science and Engineering. IOP Publishing; 2020. Green synthesis of silver nanoparticles (AgNPs) for optical and photocatalytic applications: a review. [Google Scholar]
- 5.Arif R., Uddin R.J.M.D., Sensors A review on recent developments in the biosynthesis. Silver Nanoparticles Biomed. Appl. and its biomedical applications. 2021;4(1) [Google Scholar]
- 6.Mikhailova E., O J J o f b. Silver nanoparticles. Mech. Action Probable Bio-application. 2020;11(4):84. doi: 10.3390/jfb11040084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Afkhami F., et al. Silver Nanoparticles and Their Therapeutic Applications in Endodontics. Narrative Rev. 2023;15(3):715. doi: 10.3390/pharmaceutics15030715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Urzedo A.L., et al. vol. 6. 2020. pp. 2117–2134. (Cytotoxicity and Antibacterial Activity of Alginate Hydrogel Containing Nitric Oxide Donor and Silver Nanoparticles for Topical Applications). [DOI] [PubMed] [Google Scholar]
- 9.Levard C., et al. Environmental transformations of silver nanoparticles. Impact Stabil. Toxic. 2012;46(13):6900–6914. doi: 10.1021/es2037405. [DOI] [PubMed] [Google Scholar]
- 10.Gawande M.B., et al. Microwave-assisted chemistry: synthetic applications for rapid assembly. Nanomater. Org. 2014;47(4):1338–1348. doi: 10.1021/ar400309b. [DOI] [PubMed] [Google Scholar]
- 11.Jorge de Souza T.A., Rosa Souza L.R., Franchi L.P. Silver nanoparticles: an integrated view of green synthesis methods, transformation in the environment, and toxicity. Ecotoxicol. Environ. Saf. 2019;171:691–700. doi: 10.1016/j.ecoenv.2018.12.095. [DOI] [PubMed] [Google Scholar]
- 12.Rolim W.R., et al. Antimicrobial activity and cytotoxicity to tumor cells of nitric oxide donor and silver nanoparticles containing PVA/PEG films for topical applications. ACS Appl. Mater. Interfaces. 2019;11(6):6589–6604. doi: 10.1021/acsami.8b19021. [DOI] [PubMed] [Google Scholar]
- 13.Morais M., et al. Cytotoxic effect of silver nanoparticles synthesized by green methods in cancer. J. Med. Chem. 2020;63(23):14308–14335. doi: 10.1021/acs.jmedchem.0c01055. [DOI] [PubMed] [Google Scholar]
- 14.Dutta T., et al. Green synthesis of antibacterial and antifungal silver nanoparticles using Citrus limetta peel extract: experimental and theoretical studies. J. Environ. Chem. Eng. 2020;8(4) [Google Scholar]
- 15.Zahoor M., et al. A review on silver nanoparticles. Classif. Var. Methods Synth. Potential Roles Biomed. Appl. Water Treat. 2021;13(16):2216. [Google Scholar]
- 16.Prakash M., et al. Green synthesis of bismuth based nanoparticles and its applications. Rev. 2022;25 [Google Scholar]
- 17.Kharissova O.V., et al. Greener synthesis. Chem. Compd. Mater. 2019;6(11) [Google Scholar]
- 18.Bashir S.M., et al. Chitosan nanoparticles. Versatile Platform Biomed. Appl. 2022;15(19):6521. doi: 10.3390/ma15196521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y.-J., et al. vol. 197. 2021. (Direct Synthesis of Monolayer Gold Nanoparticles on Epoxy Based Photoresist by Photoreduction and Application to Surface-Enhanced Raman Sensing). [Google Scholar]
- 20.Grządka E., Matusiak J., Godek E.J.C.p. vol. 246. 2020. (Alginic Acid as a Stabilizer of Zirconia Suspensions in the Presence of Cationic Surfactants). [DOI] [PubMed] [Google Scholar]
- 21.Arafa E.G., et al. vol. 291. 2022. (Eco-friendly and Biodegradable Sodium Alginate/quaternized Chitosan Hydrogel for Controlled Release of Urea and its Antimicrobial Activity). [DOI] [PubMed] [Google Scholar]
- 22.Qin Y.J.P.i. Alginate fibres. Overv. Prod. Appl. Wound Manag. 2008;57(2):171–180. [Google Scholar]
- 23.Tiash S., Chowdhury E.H. Growth factor receptors: promising drug targets in cancer. J. Cancer Metastasis Treat. 2015;1:190–200. [Google Scholar]
- 24.Mourad A.A.E., et al. EGFR/VEGFR-2 dual inhibitor and apoptotic inducer: design, synthesis, anticancer activity and docking study of new 2-thioxoimidazolidin-4one derivatives. Life Sci. 2021;277 doi: 10.1016/j.lfs.2021.119531. [DOI] [PubMed] [Google Scholar]
- 25.Li J., et al. Discovery of a potential HER2 inhibitor from natural products for the treatment of HER2-positive breast cancer. Int. J. Mol. Sci. 2016;17(7) doi: 10.3390/ijms17071055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harari P.M. Epidermal growth factor receptor inhibition strategies in oncology. Endocr. Relat. Cancer. 2004;11(4):689–708. doi: 10.1677/erc.1.00600. [DOI] [PubMed] [Google Scholar]
- 27.Chae Y.S., et al. RIPK1 and CASP7 polymorphism as prognostic markers for survival in patients with colorectal cancer after complete resection. J. Cancer Res. Clin. Oncol. 2011;137(4):705–713. doi: 10.1007/s00432-010-0929-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontanella C., et al. Clinical advances in the development. Nov. VEGFR2 Inhibitors. 2014;2(12) doi: 10.3978/j.issn.2305-5839.2014.08.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Le Y., et al. Design, synthesis and in vitro biological evaluation of quinazolinone derivatives as EGFR inhibitors. Antitumor Treat. 2020;35(1):555–564. doi: 10.1080/14756366.2020.1715389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamel E.M., Lamsabhi A.M. The quasi-irreversible inactivation of cytochrome P450 enzymes by paroxetine: a computational approach. Org. Biomol. Chem. 2020;18(17):3334–3345. doi: 10.1039/d0ob00529k. [DOI] [PubMed] [Google Scholar]
- 31.Kamel E.M., Lamsabhi A.M. Water biocatalytic effect attenuates cytochrome P450-mediated carcinogenicity of diethylnitrosamine: a computational insight. Org. Biomol. Chem. 2021;19(41):9031–9042. doi: 10.1039/d1ob01439k. [DOI] [PubMed] [Google Scholar]
- 32.Kamel E.M., et al. Xanthine oxidase inhibitory activity of Euphorbia peplus L. Phenolics. 2022;25(8):1336–1344. doi: 10.2174/1386207324666210609104456. [DOI] [PubMed] [Google Scholar]
- 33.Begum I., et al. Facile fabrication of malonic acid capped silver nanoparticles and their antibacterial activity. J. King Saud Univ. Sci. 2021;33(1) [Google Scholar]
- 34.Ahari H., et al. Proc. 4th World Congress Mech. Chem. Mater. Eng. MCM, Madrid, Spain – August 16 – 18. 2018. Synthesis of the silver nanoparticle by chemical reduction method and preparation of nanocomposite based on AgNPS. [Google Scholar]
- 35.Abd El-Salam H., et al. Facile polyacrylamide graft based on poly (2-chloroaniline) silver nano. Compos. Antimicrob. 2019;68(5):278–286. [Google Scholar]
- 36.Lee C., Yang W., Parr R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B. 1988;37(2):785. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 37.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. 1988;38(6):3098. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 38.Hehre W.J., et al. vol. 67. Wiley; New York et al: 1986. (Ab Initio Molecular Orbital Theory). [Google Scholar]
- 39.Frisch M.J., et al. C.01; Wallingford, CT: 2016. Gaussian 16 Rev. [Google Scholar]
- 40.Trott O., A J J J o c c, Olson, Vina AutoDock. Vol. 31. 2010. pp. 455–461. (improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pettersen E.F., et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Chandıa N., Matsuhiro B., Vásquez A.J.C.P. Alginic acids in Lessonia trabeculata. Char. Formic Acid Hydrolysis FT-IR Spectrosc. 2001;46(1):81–87. [Google Scholar]
- 43.Jayawardena T.U., et al. Alginic acid from padina boryana Abate particulate matter-induced inflammatory responses. Keratinocytes Dermal Fibroblasts. 2020;25(23):5746. doi: 10.3390/molecules25235746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandía N.P., et al. Vol. 16. 2004. pp. 127–133. (Alginic acids in Lessonia vadosa: partial hydrolysis and elicitor properties of the polymannuronic acid fraction). [Google Scholar]
- 45.Nasrollahzadeh M., et al. vol. 1. 2021. pp. 317–478. (Physicochemical Characterization of Biopolymer-Based Metal Nanoparticles). [Google Scholar]
- 46.Meena S., Tripathi A.D., Ts R.L.J.B.R. vol. 27. 2020. (Optimization and Characterization of Alginic Acid Synthesized from a Novel Strain of Pseudomonas Stutzeri). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng A., et al. vol. 235. 2023. (Functional Bacterial Cellulose Nanofibrils with Silver Nanoparticles and its Antibacterial Application). [DOI] [PubMed] [Google Scholar]
- 48.Ma Z., et al. vol. 166. 2021. pp. 567–577. (Green Synthesis of Silver Nanoparticles Using Soluble Soybean Polysaccharide and Their Application in Antibacterial Coatings). [DOI] [PubMed] [Google Scholar]
- 49.Gonzalez-Gil G., et al. Vol. 5. 2015. pp. 1–12. (NMR and MALDI-TOF MS based characterization of exopolysaccharides in anaerobic microbial aggregates from full-scale reactors). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Srivastava M., et al. Synthesis of superparamagnetic bare Fe3O4 nanostructures and core/shell (Fe3O4/alginate) Nanocomposites. 2012;89(3):821–829. doi: 10.1016/j.carbpol.2012.04.016. [DOI] [PubMed] [Google Scholar]
- 51.Daemi H., Barikani M., Barmar M.J.C.p. vol. 92. 2013. pp. 490–496. (Compatible Compositions Based on Aqueous Polyurethane Dispersions and Sodium Alginate). [DOI] [PubMed] [Google Scholar]
- 52.Zare E.N., et al. Multilayered electromagnetic bionanocomposite based on alginic acid. Char. Biol. Activit. 2015;130:372–380. doi: 10.1016/j.carbpol.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 53.Rani P., et al. Vol. 27. 2019. pp. 2769–2783. (Alginic acid derivatives: synthesis, characterization and application in wastewater treatment). [Google Scholar]
- 54.Huang Y.-C., et al. Vol. 89. 2014. pp. 137–142. (Direct preparation of silver nanoparticles and thin films in CO2-expanded hexane). [Google Scholar]
- 55.Aykul S., Martinez-Hackert E.J.A.b. Vol. 508. 2016. pp. 97–103. (Determination of half-maximal inhibitory concentration using biosensor-based protein interaction analysis). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamel E.M., et al. Xanthine oxidase inhibitory activity of Euphorbia peplus L. phenolics. J. Comb. Chem. High Throughput Screening. 2022;25(8):1336–1344. doi: 10.2174/1386207324666210609104456. [DOI] [PubMed] [Google Scholar]
- 57.Baildya N., et al. Screening of potential drug from Azadirachta Indica (Neem) extracts for SARS-CoV-2: an insight from molecular docking and MD-simulation studies. J. Mol. Struct. 2021;1227 doi: 10.1016/j.molstruc.2020.129390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Abukhalil M.H., et al. Farnesol attenuates oxidative stress and liver injury and modulates fatty acid synthase and acetyl-CoA carboxylase in high cholesterol-fed rats. Environ. Sci. Pollut. Res. Int. 2020;27(24):30118–30132. doi: 10.1007/s11356-020-09296-w. [DOI] [PubMed] [Google Scholar]
- 59.Dutta T., et al. Inhibitory effect of anti-HIV compounds extracted from Indian medicinal plants to retard the replication and transcription process of SARS-CoV-2: an insight from molecular docking and MD-simulation studies. Netw. Model. Anal. Health Inf. Bioinf. 2021;10(1):32. doi: 10.1007/s13721-021-00309-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Alrashdi B.M., et al. A flavonoid-rich fraction of Monolluma quadrangula inhibits xanthine oxidase and ameliorates potassium oxonate-induced hyperuricemia in rats. Environ. Sci. Pollut. Res. Int. 2022;29(42):63520–63532. doi: 10.1007/s11356-022-20274-2. [DOI] [PubMed] [Google Scholar]
- 61.Khan A.A., et al. Inhibitory efficiency of potential drugs against SARS-CoV-2 by blocking human angiotensin converting enzyme-2: virtual screening and molecular dynamics study. Microb. Pathog. 2021;152 doi: 10.1016/j.micpath.2021.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmed M., et al. Assessment of new anti-HER2 ligands using combined docking, QM/MM scoring and MD simulation. J. Mol. Graph. Model. 2013;40:91–98. doi: 10.1016/j.jmgm.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 63.Sait K.H.W., et al. Molecular docking analysis of HER-2 inhibitor from the ZINC database as anticancer agents. Bioinformation. 2020;16(11):882–887. doi: 10.6026/97320630016882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sadremomtaz A., et al. Molecular docking, synthesis and biological evaluation of Vascular Endothelial Growth Factor (VEGF) B based peptide as antiangiogenic agent targeting the second domain of the Vascular Endothelial Growth Factor Receptor 1 (VEGFR1D2) for anticancer application. Signal Transduct. Targeted Ther. 2020;5(1):76. doi: 10.1038/s41392-020-0177-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Majeed Khan M.A., et al. vol. 6. 2011. pp. 1–8. (Structural and Thermal Studies of Silver Nanoparticles and Electrical Transport Study of Their Thin Films). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data included in article/supp. Material/referenced in article.