Abstract
We present TrialsSummarizer, a system that aims to automatically summarize evidence presented in the set of randomized controlled trials most relevant to a given query. Building on prior work (Marshall et al., 2020), the system retrieves trial publications matching a query specifying a combination of condition, intervention(s), and outcome(s), and ranks these according to sample size and estimated study quality. The top-k such studies are passed through a neural multi-document summarization system, yielding a synopsis of these trials. We consider two architectures: A standard sequence-to-sequence model based on BART (Lewis et al., 2019), and a multi-headed architecture intended to provide greater transparency to end-users. Both models produce fluent and relevant summaries of evidence retrieved for queries, but their tendency to introduce unsupported statements render them inappropriate for use in this domain at present. The proposed architecture may help users verify outputs allowing users to trace generated tokens back to inputs. The demonstration video is available at: https://vimeo.com/735605060 The prototype, source code, and model weights are available at: https://sanjanaramprasad.github.io/trials-summarizer/.
1. Introduction
Patient treatment decisions would ideally be informed by all available relevant evidence. However, realizing this aim of evidence-based care has become increasingly difficult as the medical literature (already vast) has continued to rapidly expand (Bastian et al., 2010). Well over 100 new RCT reports are now published every day (Marshall et al., 2021). Language technologies — specifically automatic summarization methods — have the potential to provide concise overviews of all evidence relevant to a given clinical question, providing a kind of systematic review on demand (Wang et al., 2022; DeYoung et al., 2021; Wallace et al., 2021).
We describe a demonstration system, TrialsSummarizer, which combines retrieval over clinical trials literature with a summarization model to provide narrative overviews of current published evidence relevant to clinical questions. Figure 1 shows an illustrative query run in our system and the resultant output. A system capable of producing accurate summaries of the medical evidence on any given topic could dramatically improve the ability of caregivers to consult the whole of the evidence base to inform care.
Figure 1:

An example query (regarding use of statins to reduce risk of stroke) and output summary provided by the system. In this example, the summary accurately reflects the evidence, but this is not always the case.
However, current neural summarization systems are prone to inserting inaccuracies into outputs (Kryscinski et al., 2020; Maynez et al., 2020; Pagnoni et al., 2021; Ladhak et al., 2021; Choubey et al., 2021). This has been shown specifically to be a problem in the context of medical literature summarization (Wallace et al., 2021; Otmakhova et al., 2022), where there is a heightened need for factual accuracy. A system that produces plausible but often misleading summaries of comparative treatment efficacy is useless without an efficient means for users to assess the validity of outputs.
Motivated by this need for transparency when summarizing clinical trials, we implement a summarization architecture and interface designed to permit interactions that might instill trust in outputs. Specifically, the model associates each token in a generated summary with a particular source “aspect” extracted from inputs. This in turn allows one to trace output text back to (snippets of) inputs, permitting a form of verification. The architecture also provides functionality to “in-fill” pre-defined template summaries, providing a compromise between the control afforded by templates and the flexibility of abstractive summarization. We realize this functionality in our system demonstration.
2. Related Work
The (lack of) factuality of neural summarization systems is an active area of research (Chen et al., 2021; Cao et al., 2020; Dong et al., 2020; Liu et al., 2020; Goyal and Durrett, 2021; Zhang et al., 2021; Kryscinski et al., 2020; Xie et al., 2021). This demo paper considers this issue in the context of a specific domain and application. We also explored controllability to permit interaction, in part via templates. This follows prior work on hybrid template/neural summarization (Hua and Wang, 2020; Mishra et al., 2020; Wiseman et al., 2018).
We also note that this work draws upon prior work on visualizing summarization system outputs (Vig et al., 2021; Strobelt et al., 2018; Tenney et al., 2020) and biomedical literature summarization (Plaza and Carrillo-de Albornoz, 2013; Demner-Fushman and Lin, 2006; Mollá, 2010; Sarker et al., 2017; Wallace et al., 2021). However, to our knowledge this is the first working prototype to attempt to generate (draft) evidence reviews that are both interpretable and editable on demand.
3. System Overview
Our interface is built on top of Trialstreamer (Marshall et al., 2020), an automated system that identifies new reports of randomized controlled trials (RCTs) in humans and then extracts and stores salient information from these in a database of all published trial information. Our system works by identifying RCT reports relevant to a given query using a straightforward retrieval technique (Section 3.1), and then passing the top-k of these through a multi-document summarization model (Section 3.2). For the latter component we consider both a standard sequence-to-sequence approach and a aspect structured architecture (Section 3.3) intended to provide greater transparency.
3.1. Retrieving Articles
Trialstreamer (Marshall et al., 2020; Nye et al., 2020) monitors research databases — specifically, PubMed1 and the World Health Organization International Clinical Trials Registry Platform — to automatically identify newly published reports of RCTs in humans using a previously validated classifier (Marshall et al., 2018).
Articles describing RCTs are then passed through a suite of machine learning models which extract key elements from trial reports, including: sample sizes; descriptions of trial populations, interventions, and outcomes; key results; and the reliability of the evidence reported (via an approximate risk of bias score; Higgins et al. 2019). This extracted (semi-)structured information is stored in the Trialstreamer relational database.
Extracted free-text snippets describing study populations, interventions, and outcomes (PICO elements) are also mapped onto MeSH terms,2 using a re-implementation of MetaMap Lite (Demner-Fushman et al., 2017).
To facilitate search, users can enter MeSH terms for a subset of populations, interventions, and outcomes, which is used to search for matches over the articles and their corresponding extracted key data in the database. Matched studies are then ranked as a score function of sample size s and risk of bias score rob: score = s/rob; that is, we prioritize retrieval of large, high-quality trial reports.
The novelty on offer in this system demonstration is the inclusion of a summarization component, which consumes the top-k retrieved trials (we use k=5 here) and outputs a narrative summary of this evidence in the style of a systematic review abstract (Wallace et al., 2021). By combining this summarization module with the Trialstreamer database, we can provide real-time summarization of all trials that match a given query (Figure 1).
3.2. Summarizing Trials
We consider two realizations of the summarization module. We train both models on a dataset introduced in prior work which comprises collections of RCT reports (PICO elements extracted from abstracts) as inputs and Authors’ Conclusions sections of systematic review abstracts authored by members of the Cochrane Collaboration as targets (Wallace et al., 2021) (see Section 4).
As a first model, we adopt BART (Lewis et al., 2019) with a Longformer (Beltagy et al., 2020) encoder to accommodate the somewhat lengthy multi-document inputs. As inputs to the model we concatenate spans extracted from individual trials containing salient information, including populations, interventions, outcomes, and “punchlines.” The latter refers to extracted snippets which seem to provide the main results or findings, e.g., “There was a significant increase in mortality ...”; see (Lehman et al., 2019) for more details. We enclose these spans in special tags. e.g., <population>Participants were diabetics ... </population>. As additional supervision we run the same extraction models over the targets and also demarcate these using the same set of tags.
An issue with standard sequence-to-sequence models for this task is that they provide no natural means to assess the provenance of tokens in outputs, which makes it difficult to verify the trustworthiness of generated summaries. Next we discuss an alternative architecture which is intended to provide greater transparency and controllability.
3.3. Proposed Aspect Structured Architecture to Increase Transparency
We adopt a multi-headed architecture similar to (Goyal et al., 2021), which explicitly generates tokens corresponding to the respective aspects (Figure 2). We assume inputs are segmented into texts corresponding to a set of K fields or aspects. Here these are descriptions of trial populations, interventions, and outcomes, and “punchline” snippets reporting the main study findings. We will denote inputs for each of the K aspects by , where denotes the text for aspect k extracted from input x. Given that this is a multi-document setting (each input consists of multiple articles), is formed by concatenating aspect texts across all documents using special tokens to delineate individual articles.
Figure 2:
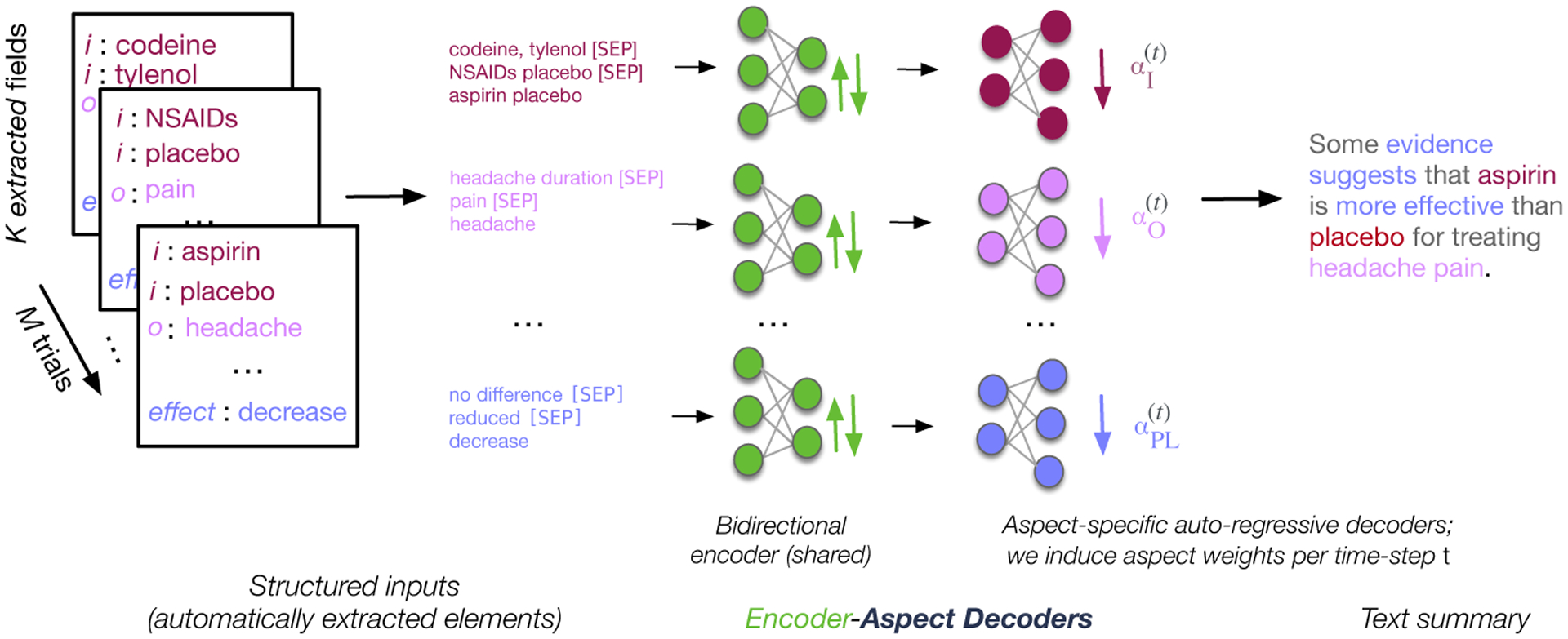
Our proposed structured summarization approach entails synthesizing individual aspects (automatically extracted in a pre-processing step), and conditionally generating text about each of these.
We encode aspect texts separately to obtain aspect-specific embeddings . We pass these (respectively) to aspect-specific decoders and a shared language model head to obtain vocabulary distributions . All model parameters are shared save for the last two decoder layers which comprise aspect-specific parameters. Importantly, the representation for a given aspect is only based on the text associated with this aspect .
We model the final output as a mixture over the respective aspect distributions: . Mixture weights encode a soft selection over aspects for timestep t and are obtained as a dot product between each penultimate representation of the decoder (prior to passing them through a language model head) and a learnable parameter, Wz ∈ RD. The K logits are then normalized via a Softmax before multiplying with the aspect-specific vocabulary distributions
Tracing outputs to inputs
This architecture permits one to inspect the mixture weights associated with individual tokens in a generated summary, which suggests which aspect (most) influenced the output. Further inspection of the corresponding snippets from studies for this aspect may facilitate verification of outputs, and/or help to resolve errors and where they may have been introduced.
Controlled generation
Neural summarization models often struggle to appropriately synthesize conflicting evidence to arrive at the correct overall determination concerning a particular intervention effectiveness. But while imperfect, summarization models may be useful nonetheless by providing a means to rapidly draft synopses of the evidence to be edited. The multi-headed architecture naturally permits template in-filling, because one can explicitly draw tokens from heads corresponding to aspects of interest. In our demo, we allow users to toggle between different templates which correspond to different conclusions regarding the overall effectiveness of the intervention in question. (It would be simple to extend this to allow users to specify their own templates to be in-filled.)
To in-fill templates we use template text preceding blanks as context and then generate text from the language head corresponding to the designated aspect. To determine span length dynamically we monitor the mixture distribution and stop when the it shifts to the another aspect (Figure 3).
Figure 3:
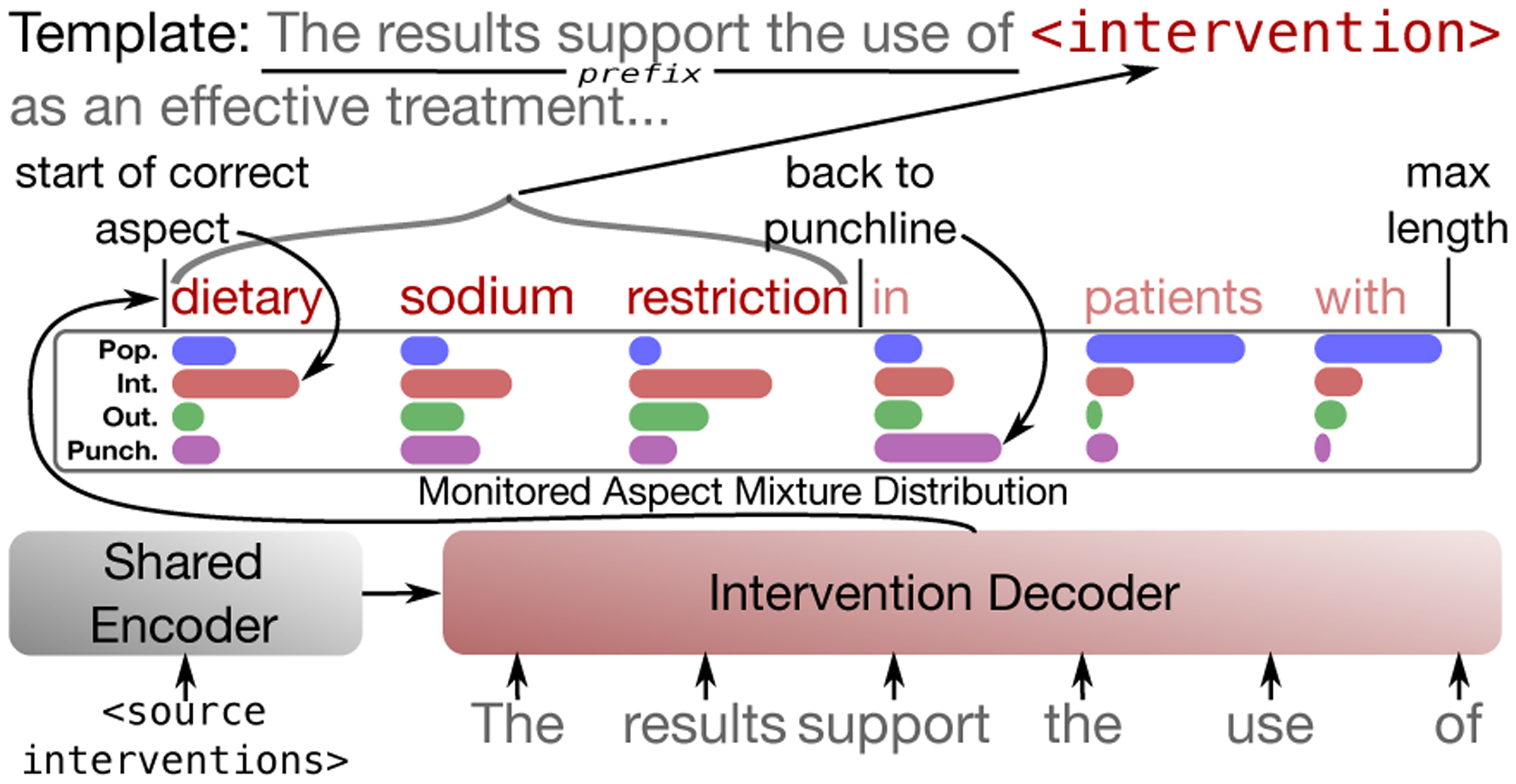
Template generation. To in-fill, we force generation from a specific head and monitor the model’s mixture distribution to decide when to stop.
3.4. User Interface
Figure 5 shows the interface we have built integrating the multi-headed architecture. Highlighted aspects in the summary provide a means of interpreting the source of output tokens by indicating the aspects that informed their production. One can in turn inspect the snippets associated with these aspects, which may help to identify unsupported content in the generated summary. To this end when users click on a token we display the subset of the input that most informed its production.
Figure 5:
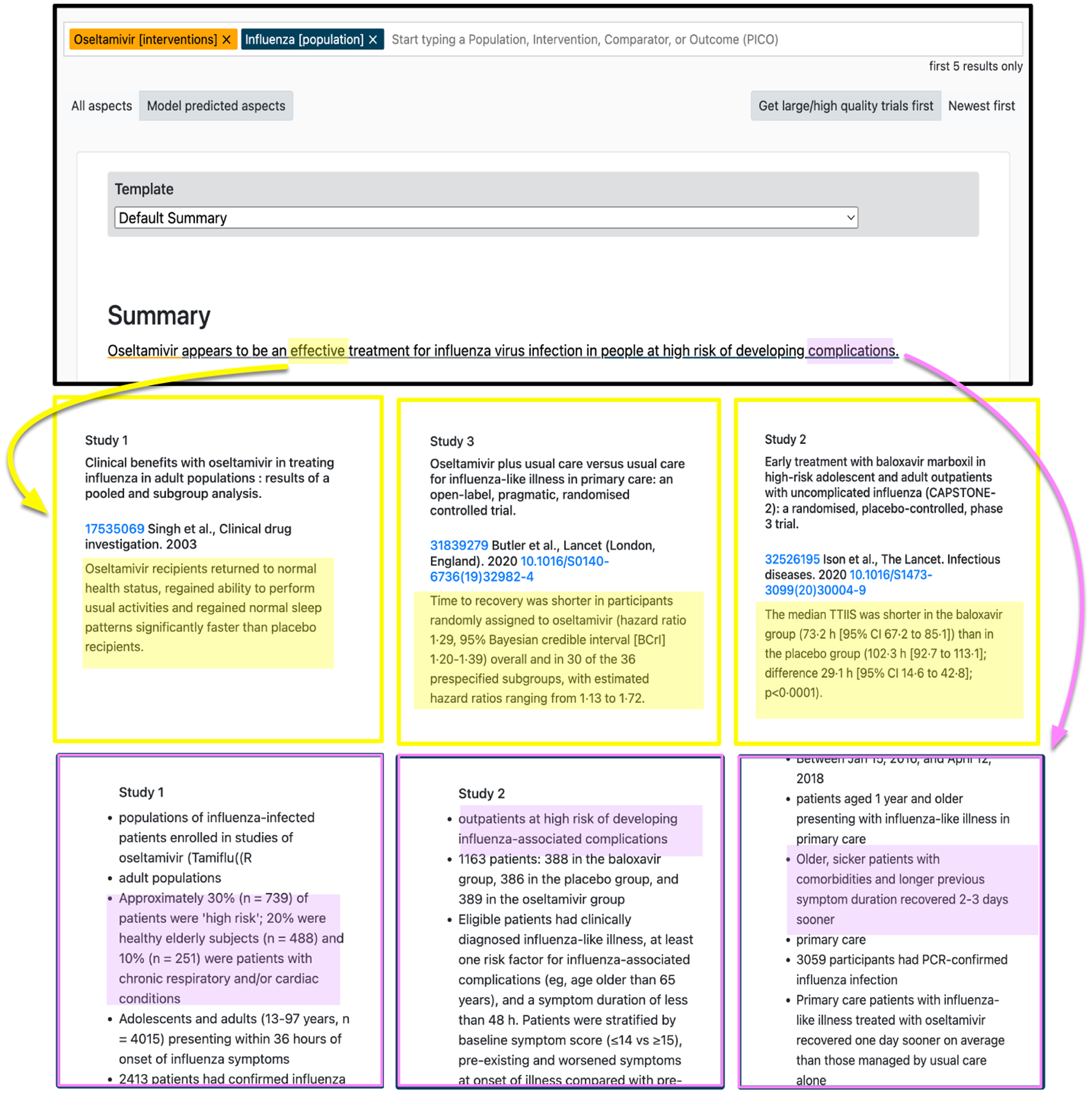
Qualitative example where the structured summarization model (and associated interface) permits token-level verification of the summary generated regarding the use of oseltamivir on influenza-infected patients. This approach readily indicates support for the claim that it is “effective” (top; yellow) and for the description of the population as individuals at risk of “complications” (bottom; purple).
We provide additional context by displaying overviews (i.e., “punchlines”) communicating the main findings of the trials. Because standard sequence-to-sequence models do not provide a mechanism to associate output tokens with input aspects, we display all aspects (and punchlines) for all trials alongside the summary for this model.
Capitalizing on the aforementioned in-filling abilities of our model, we also provide pre-defined templates for each possible “direction” of aggregate findings (significant vs. no effect). We discuss the interface along with examples in Section 5.
4. Dataset and Training Details
We aim to consume collections of titles and abstracts that describe RCTs addressing the same clinical question to abstractive summaries that synthesize the evidence presented in these. We train all models on an RCT summarization dataset (Wallace et al., 2021) where we extract clinically salient elements — i.e., our aspects — from each of the (unstructured) inputs as a pre-processing step using existing models (Marshall et al., 2020).
Training
We use the Huggingface Transformers library (Wolf et al., 2020) to implement both models. We initalize both models to bart-base (Lewis et al., 2019). We fine-tune the models with a batch size of 2 for 3 epochs, using the Adam optimizer (Kingma and Ba, 2014) with a learning rate of 3e-5.
Inference
We use beam search with a beam size of 3. We set the min and max length of generated text to be 10 and 300, respectively.
5. Case Study: Verification and Controllability
To demonstrate the potential usefulness of the interface (and the architecture which enables it), we walk through two case studies. We highlight the type of interpretability for verification our proposed approach provides, also demonstrate the ability to perform controllable summarization to show how this might be useful. The queries used in these case studies along with the investigation were performed by a co-author IJM, a medical doctor with substantial experience in evidence-based medicine. We also compare the models and report automatic scores for ROUGE and factuality in the Appendix section A and find that the two models perform comparably.
Model Interpretability
As an example to highlight the potential of the proposed architecture and interface to permit verification, we consider a query regarding the effect of Oseltamivir as an intervention for patients infected with influenza. The standard architecture produces a summary of the top most relevant RCTs to this query shown in Figure 4. This comprises two claims: (1) The intervention has been shown to reduce the risk of adverse events among adults and children, and, (2) There is no consensus as to the most effective dosage. One can inspect the inputs to attempt to verify these. Doing so, we find that reported results do tend to indicate a reduced risk of adverse events and that adolescents and adults were included in some of these studies, indicating that the first claim is accurate. The second claim is harder to verify on inspection; no such uncertainty regarding dosage is explicitly communicated in the inputs. Verifying these claims using the standard seq2seq architecture is onerous because the abstractive nature of such models makes it difficult to trace parts of the output back to inputs. Therefore, verification requires reading through entire inputs to verify different aspects.
Figure 4:
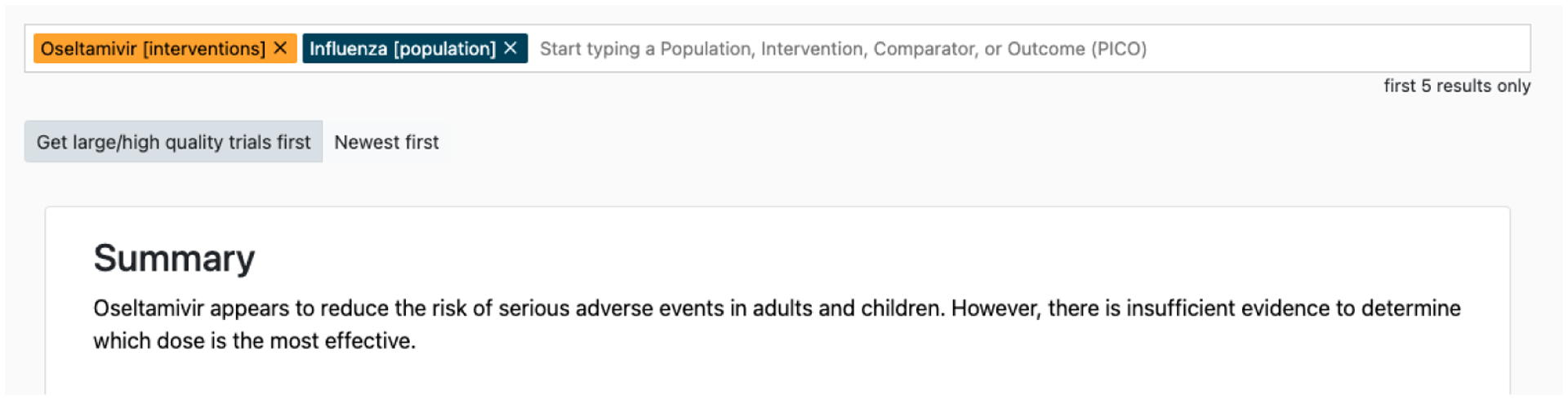
Example output and interface using a standard BART (Lewis et al., 2019) model.
The multi-headed architecture allows us to provide an interactive interface intended to permit easier verification. In particular, associating each output token with a particular aspect provides a natural mechanism for one to inspect snippets of the inputs that might support the generated text. Figure 5 illustrates this for the aforementioned Oseltamivir and flu example. Here we show how the “effective” token in the output can be clicked on to reveal the aspect that influenced its production (Figure 2), in this case tracing back to the extracted “punchlines” conveying main study findings. This readily reveals that the claim is supported. Similarly, we can verify the bit about the population being individuals at risk of complications by tracing back to the population snippets upon which this output was conditioned.
Controllability
As mentioned above, another potential benefit of the proposed architecture is the ability to “in-fill” templates to imbue neural generative models with controllability. In particular, given that the overall (aggregate) treatment efficacy is of primary importance in this context, we pre-define templates which convey an effect direction. The idea is that if upon verification one finds that the model came to the wrong aggregate effect direction, they can use a pre-defined template corresponding to the correct direction to generate a more accurate summary on-demand.
We show an example of a summary generated by the structured model in the top part of Figure 6. By using the interpretability features for verification discussed above, we find that the model inaccurately communicates that the intervention Chloroquine is effective for treating COVID-19. However, with the interactive interface we are able to immediately generate a new summary featuring the corrected synthesis result (direction), as depicted in the bottom of Figure 6, without need for manual drafting.
Figure 6:
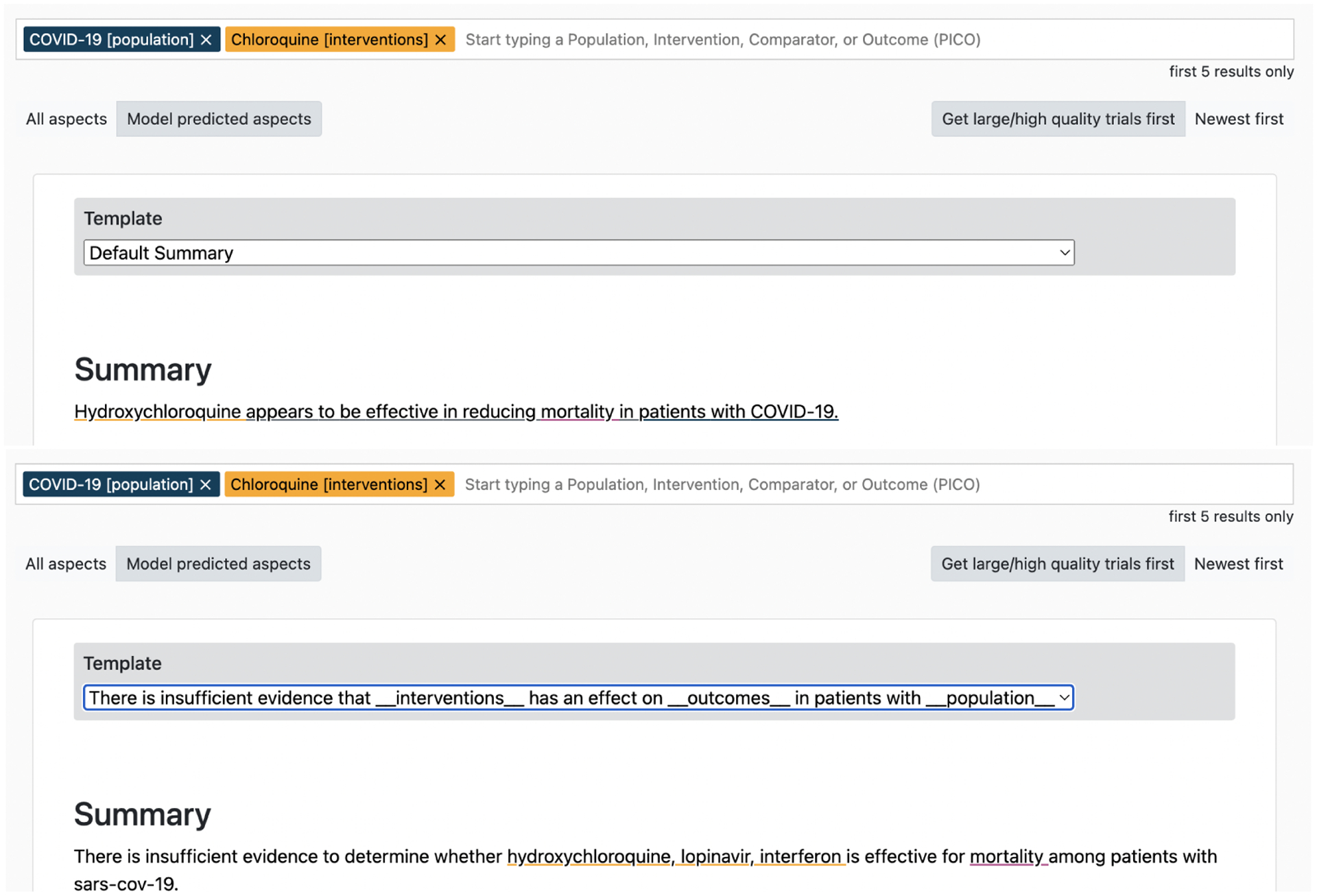
Inaccurate summaries generated by the structured model regarding the effect of Chloroquine on patients with COVID-19 (top). Template-controlled summary using the structured model (bottom).
We provide additional case studies in Appendix Section B.
6. Conclusions
We have described TrialsSummarizer, a prototype system for automatically summarizing RCTs relevant to a given query. Neural summarization models produce summaries that are readable and (mostly) relevant, but their tendency to introduce unsupported or incorrect information into outputs means they are not yet ready for use in this domain.
We implement a multi-headed architecture intended to provide greater transparency. We provided qualitative examples intended to highlight its potential to permit faster verification and controllable generation. Future work is needed to test the utility of this functionality in a user trial, and to inform new architectures that would further increase the accuracy and transparency of models for summarizing biomedical evidence.
Limitations and Ethical Issues
Limitations
This work has several limitations. First, as stated above, while the prospect of automatic summarization of biomedical evidence is tantalizing, existing models are not yet fit for the task due to their tendency to introduce factual errors. Our working prototype serves in part to highlight this and motivate work toward resolving issues of reliability and trusworthiness.
In this demo paper we have also attempted to make some progress in mitigating such issues by way of the proposed structured summarization model and accompanying interface and provided qualitative examples highlighting its potential, but really a formal user study should be conducted to assess the utility of this. This is complicated by the difficulty of the task: To evaluate the factuality of automatic summaries requires deep domain expertise and considerable time to read through constituent inputs and determine the veracity of a generated summary.
Another limitation of this work is that we have made some ad-hoc design decisions in our current prototype system. For example, at present we (arbitrarily) pass only the top-5 (based on trial sample size and estimated reliability) articles retrieved for a given query through the summarization system. Future work might address this by considering better motivated methods to select which and how many studies ought to be included.
Ethics
Accurate summaries of the biomedical evidence have the potential to ultimately improve patient care by supporting the practice of evidence-based medicine. However, at present such models bring inherent risks. In particular, one may be tempted to blindly trust model outputs; given the limitations of current summarization technologies, this would be ill-advised.
Our prototype demonstration system is designed in part to highlight existing challenges that must be solved in this space before any model might actually be adopted (and beyond this, we emphasize that need for verification of outputs, which has been the focus of the present effort). In the interface we indicate with a hard-to-miss warning message that this system should only be used for research purposes and these summaries are unreliable and not to be trusted.
Acknowledgements
This work was supported in part by the National Institutes of Health (NIH) under award R01LM012086, and by the National Science Foundation (NSF) awards 1901117 and 2211954. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH or the NSF.
Appendix
A. Automatic Evaluation
We report ROUGE scores with respect to the target (manually composed) Cochrane summaries, for both the development and test sets. We report scores for both the vanilla standard BART model along with our proposed multi-headed model intended to aid verifiability and controllability. The models perform about comparably with respect to this metric as can be seen in Table 1.
However ROUGE measures are based on (exact) n-gram overlap, and cannot measure the factuality of generated texts. Measuring factuality is in general an open problem, and evaluating the factual accuracy of biomedical reviews in particular is further complicated by the complexity of the domain and texts. Prior work has, however, proposed automated measures for this specific task (Wallace et al., 2021; DeYoung et al., 2021). These metrics are based on models which infer the reported directionality of the findings, e.g., whether or not a summary indicates that the treatment being described was effective. More specifically, we make binary predictions regarding whether generated and reference summaries report significant results (or not) and then calculate the F1 score of the former with respect to the latter.
Table 1:
ROUGE scores achieved by the standard BART model and our proposed multi-headed architecture on the dev and test sets.
| Model | ROUGE-L (dev) | ROUGE-L(test) |
|---|---|---|
| BART | 20.4 | 19.7 |
| Multi-head | 19.9 | 19.3 |
Table 2:
Directionality scores on the vanilla BART model and our proposed multi-headed architecture on the dev and test sets.
| Model | Direc (dev) | Direc(test) |
|---|---|---|
| BART | 49.6 | 51.8 |
| Multi-head | 49.3 | 52.7 |
B. Additional Case Studies
In this section we highlight a few more use cases that demonstrate the need for interpretability and controllability.
Interpretability
We first highlight a set of examples where verifying model generated summaries is difficult without an interface explicitly designed to provide interpretability capabilities. In Figure 7 (a) we show an example where the model generates a summary that accurately synthesized a summary on the effect of using Mirtazapine for patients with depression. However, the summary also includes a statement that states the need for adequate, well-designed trials. Because this statement is generic and does not point to discussing any of the PICO elements, it is unclear what element was responsible for the generation of the statement. A user would therefore need to review all (raw) input texts.
Figure 7:
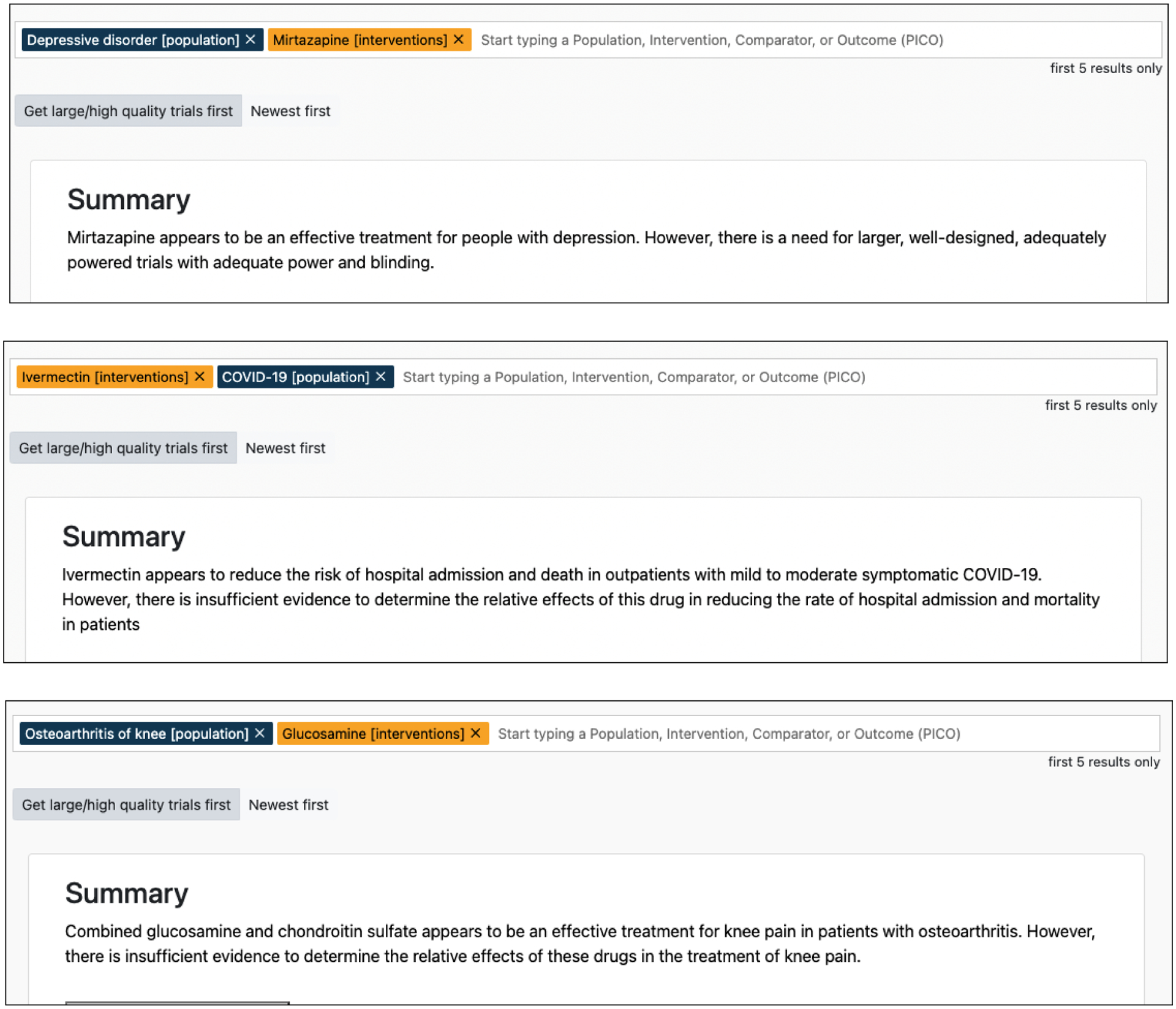
a) BART generated summary when queried about the use of Mirtazapine to treat depression b) BART generated summary when queried about the use of Ivermectin to treat COVID-19)
Figure 8:
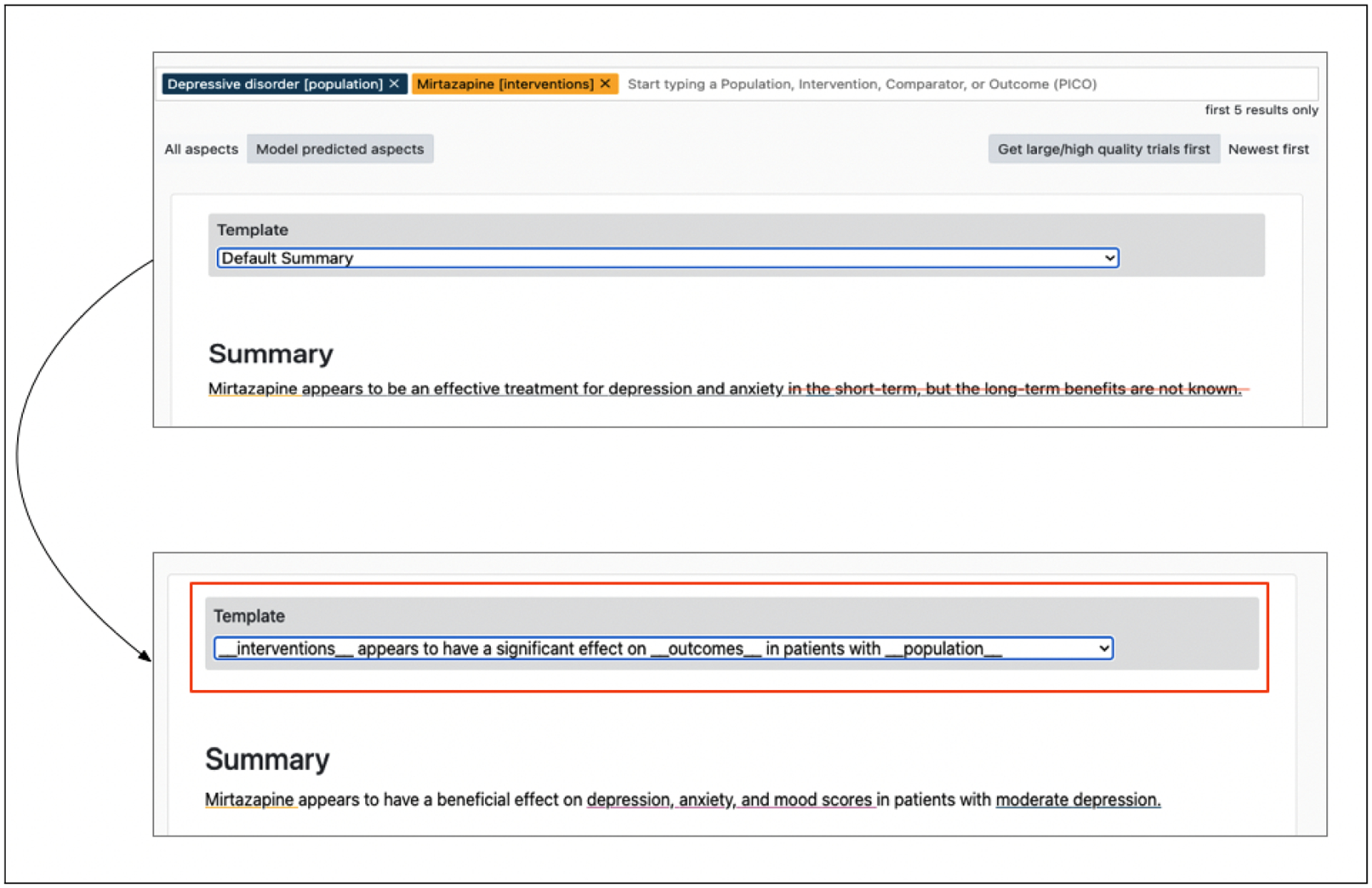
The summary on top shows the default summary generated by the multi-headed model when queried for the effect of Mirtazapine on depression. The bottom summary shows the controlled summary using a pre-defined template.
In the case of Figure 7 (b), the model generated summaries has two contradicting sentences. The first sentence indicates a reduction in hospital admission and death among COVID-19 patients when Ivermectin was used and the second sentence claims there is insufficient evidence for the same. However without interpretability capabilities it is not possible to debug and verify if the same set of elements were responsible for contradicting statements or not.
The example in Figure 7 (c) shows a case where the model first accurately synthesizes the findings in the studies of the effect of glucosamine in combination of chondroitin sulfate on knee pain. However, the following statement talks about the relative effects of the two. Again, in this case it is is not intuitive which element led to the generation of the statement and verification requires careful reviewing of all the text and their implication in all elements.
Controllability
We next highlight examples where one can effectively control the generation of summaries that would otherwise be incorrect using the template in-filling capabilities afforded by our model. While the interpretability features may permit efficient verification, models still struggle to consistently generate factual accurate summaries. We showcase instances where one can arrive at more accurate summaries quickly via the controllability (template in-filling) made possible by our model.
In the example shown in Figure 8 the default summary synthesizes the effect accurately. However, the model summary discusses the effect on short-term and long-term benefits generated from the punchlines of the studies. Reading through extracted ‘punchlines’, we find that the studies indicate issues upon withdrawal but do not necessarily provide information on long-term use of the medication. In-filling templates constrains the output, and can be used to produce more accurate summaries while still taking some advantage of the flexibility afforded by generation. For instance in this case we can see that the edited summary induced using the template is more accurate.
Similarly, in Figure 9 when the multi-headed model is queried for the effect of Glucosamine on Osteoarthritis of knee, we observe that the model on its own produces a summary conveying an incorrect aggregate effect of studies. We can verify this by inspecting the elements responsible for the generation, as discussed above. We then arrive at a more accurate summary using the template shown.
The example in Figure 10 is an interesting mistake made by the model. Because the outcomes can be presented with the same information but in a positive or negative direction (e.g., weight loss vs weight gain), the model has to accurately infer the effect of all studies. In this case, the model generates a summary with the right effect but views weight loss as an undesirable effect. Here again we select a template and allow the model quickly in-fill, yielding a more accurate summary.
Figure 9:
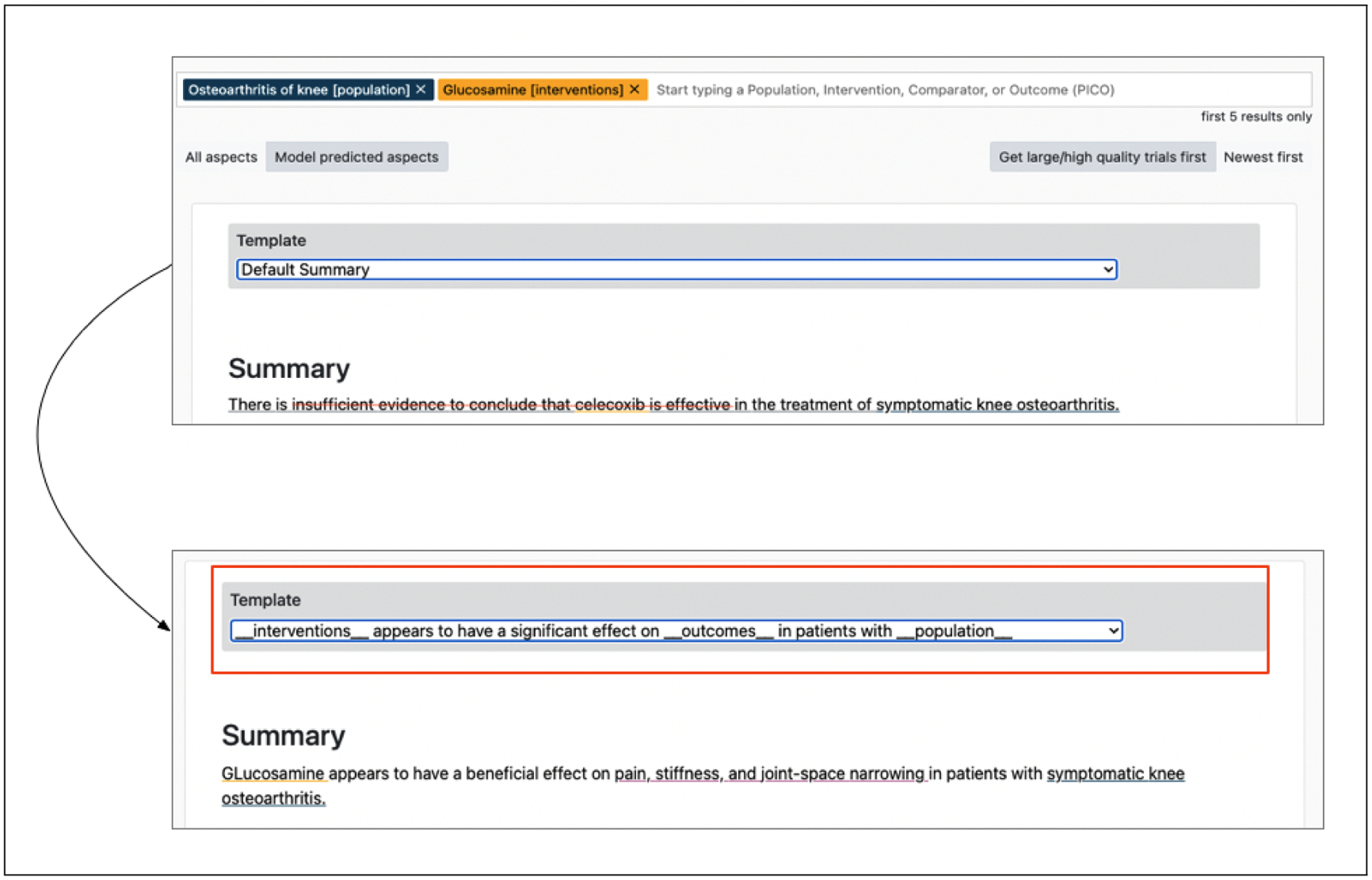
The summary on top shows the default summary generated by the multi-headed model when queried for the effect of Glucosamine on Osteoarthritis of knee. The bottom summary shows the edited summary using a pre-defined template
Figure 10:
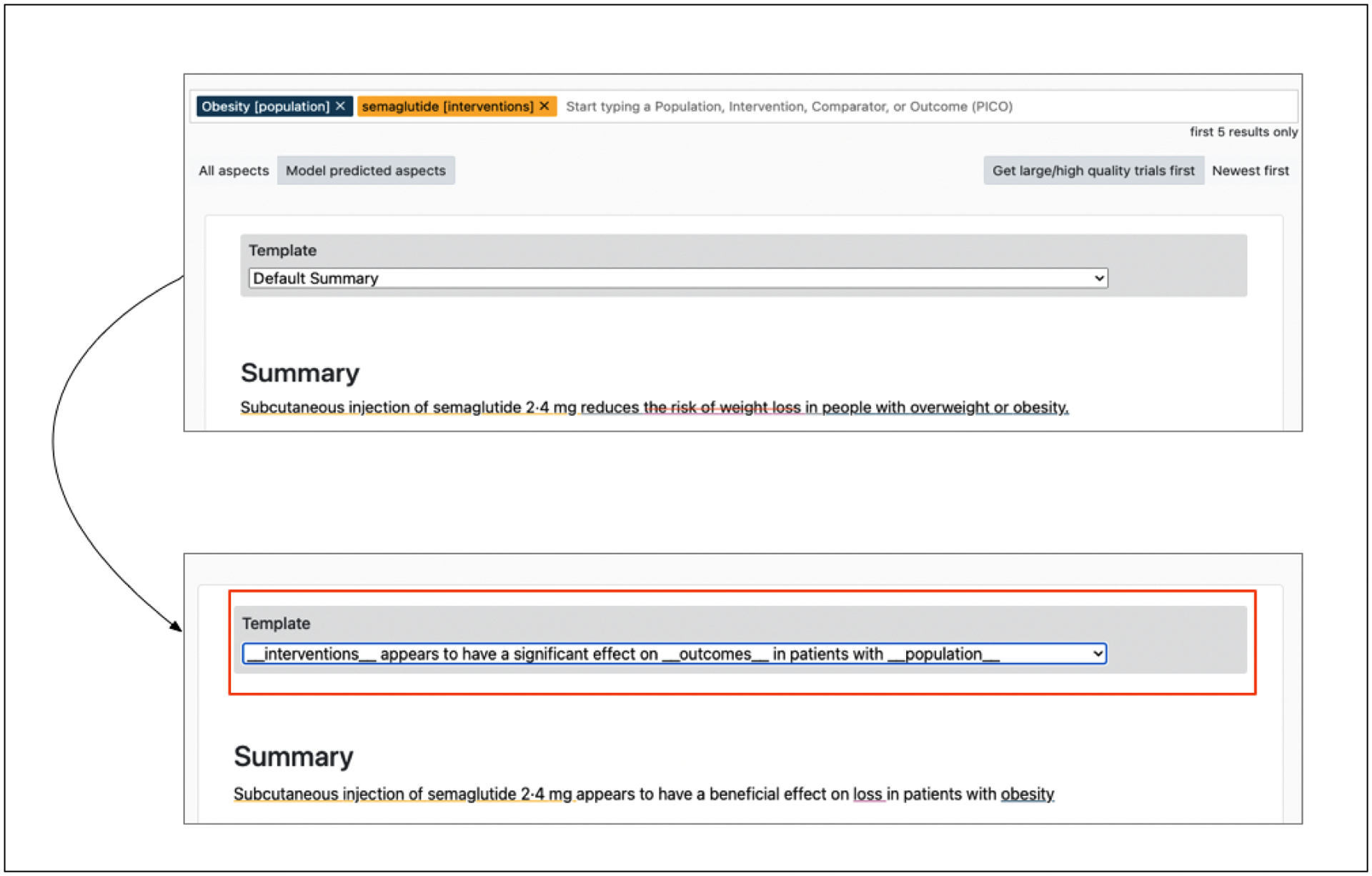
The summary on top shows the default summary generated by the multi-headed model when queried for the effect of Semaglutide on obese patients. The bottom summary shows the edited summary using a pre-defined template
Footnotes
MeSH — short for Medical Subject Headings — is a controlled vocabulary maintained by the National Library of Medicine (NLM).
Contributor Information
Sanjana Ramprasad, Northeastern University.
Iain J. Marshall, King’s College London
Denis Jered McInerney, Northeastern University.
Byron C. Wallace, Northeastern University
References
- Bastian H, Glasziou P, and Chalmers I. 2010. Seventy-five trials and eleven systematic reviews a day: how will we ever keep up? PLoS medicine, 7(9). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltagy Iz, Peters Matthew E, and Cohan Arman. 2020. Longformer: The long-document transformer. arXiv preprint arXiv:2004.05150.
- Cao Meng, Dong Yue, Wu Jiapeng, and Cheung Jackie Chi Kit. 2020. Factual error correction for abstractive summarization models. arXiv preprint arXiv:2010.08712.
- Chen Sihao, Zhang Fan, Sone Kazoo, and Roth Dan. 2021. Improving faithfulness in abstractive summarization with contrast candidate generation and selection. arXiv preprint arXiv:2104.09061.
- Prafulla Kumar Choubey Jesse Vig, Liu Wenhao, and Rajani Nazneen Fatema. 2021. Mofe: Mixture of factual experts for controlling hallucinations in abstractive summarization. arXiv preprint arXiv:2110.07166.
- Demner-Fushman Dina and Lin Jimmy. 2006. Answer extraction, semantic clustering, and extractive summarization for clinical question answering. In Proceedings of the 21st International Conference on Computational Linguistics and 44th Annual Meeting of the Association for Computational Linguistics, pages 841–848. [Google Scholar]
- Demner-Fushman Dina, Rogers Willie J, and Aronson Alan R. 2017. Metamap lite: an evaluation of a new java implementation of metamap. Journal of the American Medical Informatics Association, 24(4):841–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung Jay, Beltagy Iz, Zuylen Madeleine van, Kuehl Bailey, and Wang Lucy Lu. 2021. Ms2: Multi-document summarization of medical studies. arXiv preprint arXiv:2104.06486.
- Dong Yue, Wang Shuohang, Gan Zhe, Cheng Yu, Cheung Jackie Chi Kit, and Liu Jingjing. 2020. Multi-fact correction in abstractive text summarization. arXiv preprint arXiv:2010.02443.
- Goyal Tanya and Durrett Greg. 2021. Annotating and modeling fine-grained factuality in summarization. arXiv preprint arXiv:2104.04302.
- Goyal Tanya, Rajani Nazneen Fatema, Liu Wenhao, and Kryściński Wojciech. 2021. Hydrasum: Disentangling stylistic features in text summarization using multi-decoder models. arXiv preprint arXiv:2110.04400.
- Higgins Julian PT, Savović Jelena, Page Matthew J, Elbers Roy G, and Sterne Jonathan AC. 2019. Assessing risk of bias in a randomized trial. Cochrane handbook for systematic reviews of interventions, pages 205–228. [Google Scholar]
- Hua Xinyu and Wang Lu. 2020. Pair: Planning and iterative refinement in pre-trained transformers for long text generation. arXiv preprint arXiv:2010.02301.
- Kingma Diederik P and Ba Jimmy. 2014. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980.
- Kryscinski Wojciech, Bryan McCann Caiming Xiong, and Socher Richard. 2020. Evaluating the factual consistency of abstractive text summarization. In Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), pages 9332–9346, Online.Association for Computational Linguistics. [Google Scholar]
- Ladhak Faisal, Durmus Esin, He He, Cardie Claire, and McKeown Kathleen. 2021. Faithful or extractive? on mitigating the faithfulness-abstractiveness tradeoff in abstractive summarization. arXiv preprint arXiv:2108.13684.
- Lehman Eric, Jay DeYoung, Barzilay Regina, and Wallace Byron C. 2019. Inferring which medical treatments work from reports of clinical trials. arXiv preprint arXiv:1904.01606.
- Lewis Mike, Liu Yinhan, Goyal Naman, Ghazvininejad Marjan, Mohamed Abdelrahman, Levy Omer, Stoyanov Ves, and Zettlemoyer Luke. 2019. Bart: Denoising sequence-to-sequence pre-training for natural language generation, translation, and comprehension. arXiv preprint arXiv:1910.13461.
- Liu Zhenghao, Xiong Chenyan, Dai Zhuyun, Sun Si, Sun Maosong, and Liu Zhiyuan. 2020. Adapting open domain fact extraction and verification to covid-fact through in-domain language modeling. In Findings of the Association for Computational Linguistics: EMNLP 2020, pages 2395–2400. [Google Scholar]
- Marshall Iain J, Noel-Storr Anna, Kuiper Joël, Thomas James, and Wallace Byron C. 2018. Machine learning for identifying randomized controlled trials: an evaluation and practitioner’s guide. Research synthesis methods, 9(4):602–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Iain J, Nye Benjamin, Kuiper Joël, Noel-Storr Anna, Marshall Rachel, Maclean Rory, Soboczenski Frank, Nenkova Ani, Thomas James, and Wallace Byron C. 2020. Trialstreamer: A living, automatically updated database of clinical trial reports. Journal of the American Medical Informatics Association, 27(12):1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall Iain James, L’Esperance Veline, Marshall Rachel, Thomas James, Noel-Storr Anna, Soboczenski Frank, Nye Benjamin, Nenkova Ani, and Wallace Byron C. 2021. State of the evidence: a survey of global disparities in clinical trials. BMJ Global Health, 6(1):e004145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynez Joshua, Narayan Shashi, Bohnet Bernd, and McDonald Ryan. 2020. On faithfulness and factuality in abstractive summarization. arXiv preprint arXiv:2005.00661.
- Mishra Abhijit, Chowdhury Md Faisal Mahbub, Manohar Sagar, Gutfreund Dan, and Sankaranarayanan Karthik. 2020. Template controllable keywords-to-text generation. arXiv preprint arXiv:2011.03722.
- Diego Mollá. 2010. A corpus for evidence based medicine summarisation. In Proceedings of the Australasian Language Technology Association Workshop, pages 76–80. [Google Scholar]
- Nye Benjamin E, Nenkova Ani, Marshall Iain J, and Wallace Byron C. 2020. Trialstreamer: mapping and browsing medical evidence in real-time. In Proceedings of the conference. Association for Computational Linguistics. North American Chapter. Meeting, volume 2020, page 63. NIH Public Access. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otmakhova Yulia, Verspoor Karin, Baldwin Timothy, and Han Lau Jey. 2022. The patient is more dead than alive: exploring the current state of the multi-document summarisation of the biomedical literature. In Proceedings of the 60th Annual Meeting of the Association for Computational Linguistics (Volume 1: Long Papers), pages 5098–5111, Dublin, Ireland. Association for Computational Linguistics. [Google Scholar]
- Pagnoni Artidoro, Balachandran Vidhisha, and Tsvetkov Yulia. 2021. Understanding factuality in abstractive summarization with frank: A benchmark for factuality metrics. arXiv preprint arXiv:2104.13346.
- Plaza Laura and Carrillo-de Albornoz Jorge. 2013. Evaluating the use of different positional strategies for sentence selection in biomedical literature summarization. BMC bioinformatics, 14(1):1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker Abeed, Molla Diego, and Paris Cecile. 2017. Automated text summarisation and evidence-based medicine: A survey of two domains.
- Strobelt Hendrik, Gehrmann Sebastian, Behrisch Michael, Perer Adam, Pfister Hanspeter, and Rush Alexander M. 2018. S eq 2s eq-v is: A visual debugging tool for sequence-to-sequence models. IEEE transactions on visualization and computer graphics, 25(1):353–363. [DOI] [PubMed] [Google Scholar]
- Tenney Ian, Wexler James, Bastings Jasmijn, Bolukbasi Tolga, Coenen Andy, Gehrmann Sebastian, Jiang Ellen, Pushkarna Mahima, Radebaugh Carey, Reif Emily, et al. 2020. The language interpretability tool: Extensible, interactive visualizations and analysis for nlp models. arXiv preprint arXiv:2008.05122.
- Vig Jesse, Wojciech Kryściński Karan Goel, and Rajani Nazneen Fatema. 2021. Summvis: Interactive visual analysis of models, data, and evaluation for text summarization. arXiv preprint arXiv:2104.07605.
- Wallace Byron C, Sayantan Saha, Soboczenski Frank, and Marshall Iain J. 2021. Generating (factual?) narrative summaries of rcts: Experiments with neural multi-document summarization. In AMIA Annual Symposium Proceedings, volume 2021, page 605. American Medical Informatics Association. [PMC free article] [PubMed] [Google Scholar]
- Wang Lucy Lu, DeYoung Jay, and Wallace Byron. 2022. Overview of MSLR2022: A shared task on multi-document summarization for literature reviews. In Proceedings of the Third Workshop on Scholarly Document Processing, pages 175–180, Gyeongju, Republic of Korea. Association for Computational Linguistics. [Google Scholar]
- Wiseman Sam, Shieber Stuart M, and Rush Alexander M. 2018. Learning neural templates for text generation. arXiv preprint arXiv:1808.10122. [Google Scholar]
- Wolf Thomas, Debut Lysandre, Sanh Victor, Chaumond Julien, Delangue Clement, Moi Anthony, Cistac Pier-ric, Rault Tim, Louf Rémi, Funtowicz Morgan, et al. 2020. Transformers: State-of-the-art natural language processing. In Proceedings of the 2020 conference on empirical methods in natural language processing: system demonstrations, pages 38–45. [Google Scholar]
- Xie Yuexiang, Sun Fei, Deng Yang, Li Yaliang, and Ding Bolin. 2021. Factual consistency evaluation for text summarization via counterfactual estimation. arXiv preprint arXiv:2108.13134. [Google Scholar]
- Zhang Sen, Niu Jianwei, and Wei Chuyuan. 2021. Fine-grained factual consistency assessment for abstractive summarization models. In Proceedings of the 2021 Conference on Empirical Methods in Natural Language Processing, pages 107–116. [Google Scholar]


