Abstract
Conserving species and their genetic variation are a global priority to safeguard evolutionary potential in a rapidly changing world. Species are fundamental units in research and nature management, but taxonomic work is increasingly undermined. Increasing knowledge on the species genetic diversity would aid in prioritizing conservation efforts. Sphagnum is a diverse, well‐known bryophyte genus, which makes the genus suited to study speciation and cryptic variation. The species share specific characteristics and can be difficult to separate in the field. By combining molecular data with thorough morphological examination, new species have recently been discovered. Still, there are taxonomic uncertainties, even for species assessed on the IUCN Red List of threatened species. Here, we use molecular data to examine three rare species within the subgenus Acutifolia described based on morphological characters. All species have narrow distributions and limited dispersability. First, we confirm the genetic origin of S. skyense. Second, we show that S. venustum is a haploid species genetically distinct from morphologically similar species. Lastly, S. nitidulum was found to have a distinct haplotype, but cannot be genetically separated from other red Acutifolia species. We also found high genetic variation within red Acutifolia specimens, indicating the need of further morphological examination and possibly taxonomic revision. Until then, our results have shown that genetic data can aid in prioritizing targets of conservation efforts when taxonomy is unresolved. All three taxa should be further searched for by field biologists to increase knowledge about their distribution ranges.
Keywords: genetic structure, microsatellites, molecular data, morphology, peatmosses, speciation, species identification
We used molecular data to resolve taxonomic uncertainty of three rare Sphagnum species described based on morhological characters. We confirm the parents of Sphagnum skyense and validate the species status of S. venustum. Sphagnum nitidulum has a distinct haplotype, but more data are needed to conclude whether it is a valid species.

1. INTRODUCTION
Recognizing and describing species is an essential task in biology, as species are the fundamental biodiversity units used in many research fields and nature management. Correct species identification is crucial to, for instance, obtain species distribution maps and biodiversity estimates, which are commonly used in a wide range of ecological studies (Stropp et al., 2022). Also, conservation efforts are usually targeting species, and global, regional, and national Red lists make assessments at the species level. Furthermore, the inclusion of genetic data in conservation management and prioritizations is becoming increasingly recognized, as maintaining genetic diversity can make species more resilient to rapid environmental changes (Andrello et al., 2022). In fact, maintaining genetic diversity within species to safeguard their adaptive potential is listed under Goal A in the “Kunming‐Montreal Biodiversity Framework” (CBD, 2022). Thus, increasing knowledge about diversity among and within species is highly important to make informed decisions about where conservation efforts should be applied.
The genus Sphagnum L. is species‐rich and one of the most studied bryophyte genera, partly because it is the main driver of peat accumulation in boreal peatlands. Consequently, Sphagnum stores more carbon than any other plant genera (Yu et al., 2011). Evolutionary, the genus is on a long branch separated from the rest of the mosses, but with relatively recent, rapid diversification (Shaw et al., 2010). A major driver of speciation in Sphagnum seems to be adaptation to niches along the water table gradient within the mire landscape (Johnson et al., 2015; Rydin, 1986). Another important speciation mechanism is allopolyploidization, a process where two species hybridize and give viable offspring. Around 20% of Sphagnum species have originated as a result of allopolyploidization (Meleshko et al., 2018). Allopatric speciation due to barriers like the Atlantic Ocean is much less prevalent in bryophytes than seed plants, as Europe and North America share the majority of moss species (Carter et al., 2016; Frahm & Vitt, 1993). This is mainly explained by effective dispersal of bryophyte spores (Heinrichs et al., 2009; Kyrkjeeide, Hassel, Flatberg, Shaw, Brochmann, & Stenoien, 2016; Szövenyi et al., 2008). High species diversity of Sphagnum is found in the boreal region, a paradox considering the short time since the last glacial maximum. High dispersibility leading to colonization may explain this, but the role of geographical isolation in driving speciation in bryophytes is not well‐understood (but see Yousefi et al., 2019).
Sphagnum species are morphologically similar and often display phenotypic plasticity (Stenøien et al., 2014), and closely related species may have overlapping niches (Hassel et al., 2018; Yousefi et al., 2017). Thus, species delineation has been a debated topic within Sphagnum biology (Cronberg, 1989; Flatberg, 1986). Lately, investigating genetic variation has aided the identification of new species, even within common and well‐studied species such as S. magellanicum Brid. sensu lato (Hassel et al., 2018; Kyrkjeeide, Hassel, Flatberg, Shaw, Yousefi, & Stenoien, 2016; Shaw et al., 2022; Yousefi et al., 2019). Despite the effort to describe Sphagnum diversity, there are still unresolved taxonomical issues, even for species assessed in the IUCN Red List of threatened species (Gabriel & Sim‐Sim, 2019).
Taxonomic work is time‐consuming, and becoming a species expert may take years of training in the field and/or identification of microscopical characters in the laboratory. Field trips enable experts to collect specimens and enroll them in scientific collections. These collections have value for scientific discoveries and as a historical record of biodiversity (Pyke & Ehrlich, 2010; Suarez & Tsutsui, 2004), but not if specimens stay hidden in storage. For example, S. beothuk Andrus, described from Newfoundland, Canada (Andrus, 2006), was not detected in Europe before it was discovered to be genetically the same as specimens collected for years as a “dark morph” of S. fuscum (Schimp.) H. Klinggr. (Kyrkjeeide et al., 2015). Also, within the widespread S. warnstorfii Russow, extensively used in ecological studies (Bengtsson et al., 2016; Granath et al., 2010; Hájek et al., 2021), European specimens belonging to different genotypes seem to hold morphological variation (Mikulášková et al., 2015).
Here, we use molecular data to explore genetic differentiation and to clarify the taxonomic status of three rare species from Sphagnum subgenus Acutifolia (Wilson) A.J. Shaw: S. skyense Flatberg, S. venustum Flatberg, and S. nitidulum Warnstorf. All three species are described solely based on morphological data (Figure 1).
FIGURE 1.

Sphagnum skyense (left), S. venustum (middle), and S. nitidulum (right) are three rare species described based on morphological data. Photos: K.I. Flatberg.
Sphagnum skyense was detected as a distinct red‐colored peat moss in the heathland on Isle of Skye, Scotland, and later described as a new species based on morphology (Flatberg, 1988a). The large size of S. skyense and its morphological resemblance to both S. quinquefarium (Lindb.) Warnst. and S. subnitens Russow & Warnst. lead to the hypothesis that this likely was an allopolyploid species. Genetic analysis including most species in subgenus Acutifolia (except S. quinquefarium) confirmed the allopolyploid origin, but suggested S. warnstorfii and S. subnitens as the likely progenitors of S. skyense (Shaw et al., 2005). No further attempt has been made in studying the ploidy level or the origin of S. skyense, with the latter remaining an open question for more than 30 years.
Sphagnum venustum is one of the most recent contributions of new species in Acutifolia. It was discovered and described based on collections from Labrador, eastern Canada (Flatberg, 2008), and later several new localities were reported from Quebec, Canada (Ayotte & Rochefort, 2019). In 2011, it was also discovered in Norway, but so far from one single site. Hill (2019) asked whether this single site occurrence of S. venustum could be a result of separate hybridization events on different continents. However, we hypothesis that it is a haploid species due to the small size, as allopolyploids tend to be larger than their progenitors (Flatberg, 1988a, 1988b).
Sphagnum nitidulum was described in 1911 and is still only known from one site on the Azorean island Terceira. It grows on soil in warm spots in the sulfurous fumaroles—a very rare substrate for Sphagnum species (Séneca & Söderström., 2009). The species has been seen as conspecific with S. rubellum (Andrews, 1941), but anyhow been included on several checklists (Gabriel & Sim‐Sim, 2019 and references therein). The type was assumed to be destructed, but it was photographed in 1991 (Gabriel & Sim‐Sim, 2019). Collections from the type locality in herbarium TRH were obtained in 2004 (four specimens) and 2021 (two specimens). Sphagnum nitidulum is one of the “red Acutifolia species.” Within this group, species delineation of S. capillifolium and S. rubellum has been debated and thoroughly studied, concluding that the two species may overlap morphologically (Cronberg, 1989), but are genetically distinct (Cronberg, 1996, 1998). Morphological overlap has also been observed between specimens identified as both S. capillifolium and S. warnstorfii (K. I. Flatberg, personal communication). Such specimens have been collected in mainland Europe the last two decades and tagged under different provisional names (specimens in TRH). We included these specimens (hereafter called conspecific specimens) to explore whether S. nitidulum is genetically distinct from other red Acutifolia species and whether the observed morphological similarities between S. nitidulum and deviating specimens of S. warnstorfii and S. capillifolium are reflected in molecular data.
We aim to identify the parental species of S. skyense, confirm the haploid level and the genetic distinctness of S. venustum, and evaluate whether S. nitidulum is genetically distinct from other “red Acutifolia specimens.”
2. MATERIALS AND METHODS
We used 72 Sphagnum specimens from the scientific collection (herbarium TRH) in Trondheim, Norway, collected in Europe and Eastern Canada. An overview of species and number of specimens are given in Table 1. DNA was extracted from the apical part of gametophytes, that is the middle part of the capitula, for each specimen using the NucleoSpin Plant II, Mini kit for plant DNA (Macherey‐Nagel) or E.Z.N.A.® HP Plant DNA Mini Kit (Omega Bio‐tek), following the manufacturers' protocols. The ploidy level of bryophytes is based on the gametophytic phase. Most Sphagnum species are haploid, whereas allopolyploid species have diploid or triploid gametophytes.
TABLE 1.
Voucher information for Sphagnum specimens used in the molecular analyses.
| DNA ID | TRH ID | Herb. assigned taxon | Country | Province/county | Longitude | Latitude | Year | Collector | Assigned taxon in analyses |
|---|---|---|---|---|---|---|---|---|---|
| ss1 | 728,102 | S. skyense | Scotland | Highland | 1987 | KIF | Skyense | ||
| ss3 | 728,099 | S. skyense | Scotland | Highland | 2004 | MOH | Skyense | ||
| ss4 | 741,748 | S. cf. skyense | Ireland | Tipperary | 2005 | NGH | Skyense | ||
| ss5 | 93,374 | S. subnitens ssp. subnitens | Norway | Vestland | 4,97883 | 60,59546 | 2017 | MJ, CP, MOK, KIF | Subnitens |
| ss6 | 93,414 | S. subnitens ssp. subnitens | Norway | Vestland | 5,10753 | 59,99331 | 2017 | MJ, CP, MOK, KIF | Subnitens |
| ss7 | 728,280 | S. subnitens ssp. subnitens | Scotland | Highland | 1987 | KIF | Subnitens | ||
| ss8 | 120,164 | S. subnitens ssp. subnitens | Ireland | Mayo | −9,51535 | 54,10317 | 2011 | MOK, KOK | Subnitens |
| ss9 | 728,298 | S. subnitens ssp. subnitens | Wales | Gwynedd | 1980 | KIF | Subnitens | ||
| ss10 | 93,420 | S. quinquefarium | Norway | Vestland | 5,20637 | 59,59257 | 2017 | MJ, CP, MOK, KIF | Quinquefarium |
| ss11 | 93,405 | S. quinquefarium | Norway | Vestland | 5,10615 | 59,99316 | 2017 | MJ, CP, MOK, KIF | Quinquefarium |
| ss12 | 120,123 | S. quinquefarium | Scotland | Highland | −6,06299 | 57,17806 | 2011 | MOK | Quinquefarium |
| ss13 | 4802 | S. quinquefarium | Scotland | Scottish Borders | −2,85016 | 55,19173 | 2014 | KH | Quinquefarium |
| ss68 | 109,503 | S. quinquefarium | Scotland | Highland | −6,24462 | 57,42046 | 2019 | NGH | Quinquefarium |
| ss23 | 93,432 | S. capillifolium | Norway | Vestland | 5,19754 | 59,61486 | 2017 | MJ, CP, MOK, KIF | Capillifolium |
| ss24 | 673,473 | S. capillifolium | Scotland | Highland | 1987 | KIF | Capillifolium | ||
| ss26 | 13,176 | S. capillifolium | Scotland | Highland | −5,01957 | 58,19938 | 2015 | KH | Capillifolium |
| ss28 | 94,003 | S. capillifolium | Austria | Salzburg | 13,78167 | 47,08359 | 2017 | KIF, CS | Capillifolium |
| ss29 | 38,148 | S. capillifolium | Slovakia | Prešov Region | 20,22992 | 49,2195 | 2016 | KH | Capillifolium |
| ss30 | 98,303 | S. capillifolium | Slovakia | Prešov Region | 20,22212 | 49,21094 | 2016 | KIF | Capillifolium |
| ss31 | 93,386 | S. capillifolium | Norway | Vestland | 5,01412 | 60,56181 | 2017 | MJ, CP, MOK, KIF | Capillifolium |
| ss36 | 93,415 | S. capillifolium | Norway | Vestland | 5,10753 | 59,99331 | 2017 | MJ, CP, MOK, KIF | Capillifolium |
| ss37 | 93,414 | S. capillifolium | Norway | Vestland | 5,10753 | 59,99331 | 2017 | MJ, CP, MOK, KIF | Capillifolium |
| ss39 | 740,734 | S. capillifolium | Norway | Trøndelag | 10,41161 | 63,20311 | 2003 | KIF | Capillifolium |
| 224 | 108,510 | S. rubellum | Norway | Trøndelag | 11,21917 | 64,23135 | 2019 | KH, OM, MOK, KIF | Conspecific |
| 230 | 115,591 | S. cf. capillifolium × warnstorfii | Norway | Trøndelag | 9,099786 | 63,06362 | 2016 | KIF | Conspecific |
| 241 | 148,557 | S. cf. capillifolium × warnstorfii | Norway | Trøndelag | 10,46162 | 63,38908 | 2019 | KIF | Conspecific |
| ss21 | 93,450 | S. cf. warnstorfii | Norway | Vestland | 5,19795 | 59,61641 | 2017 | MJ, CP, MOK, KIF | Conspecific |
| ss27 | 94,018 | S. capillifolium | Austria | Salzburg | 13,67968 | 47,08395 | 2017 | KIF, CS | Conspecific |
| ss34 | 94,041 | S. cf. capillifolium × warnstorfii | Austria | Oberosterreich | 12,95769 | 48,0541 | 2017 | KIF | Conspecific |
| ss35 | 94,020 | S. cf. capillifolium × warnstorfii | Austria | Salzburg | 13,67906 | 47,08405 | 2017 | KIF, CS | Conspecific |
| SW01 | 148,553 | S. cf. warnstorfii | Denmark | Jylland | 9,135816 | 56,37582 | 2021 | IG | Conspecific |
| SN11 | 727,588 | S. cf. nitidulum | Portugal | Azores | 2004 | KIF | Nitidulum | ||
| SN12* | 121,476 | S. nitidulum | Portugal | Azores | −27,2 | 38,73333 | 2021 | EFC | Nitidulum |
| SN13* | 121,475 | S. nitidulum | Portugal | Azores | −27,2167 | 38,73333 | 2021 | EFC | Nitidulum |
| SN14* | 727,587 | S. cf. nitidulum | Portugal | Azores | 2004 | KIF | Nitidulum | ||
| RB01** | 728,039 | S. cf. rubellum | Portugal | Azores | 2004 | KIF | Nitidulum | ||
| 221 | 725,290 | S. rubellum | Canada | Quebec | −57,1541 | 51,43078 | 2007 | BF, KIF | Rubellum |
| 222 | 725,305 | S. rubellum | Canada | Newfoundland and Labrador | −56,7302 | 51,57122 | 2007 | BF, KIF | Rubellum |
| 223 | 37,834 | S. rubellum | Norway | Trøndelag | 11,43851 | 64,70585 | 2016 | KH | Rubellum |
| 240 | 148,556 | S. rubellum | Norway | Trøndelag | 10,46162 | 63,38908 | 2019 | KIF | Rubellum |
| 242 | 148,558 | S. rubellum | Norway | Trøndelag | 10,46162 | 63,38908 | 2019 | KIF | Rubellum |
| ss15 | 11,185 | S. cf. warnstorfii | Norway | Vestland | 5,16446 | 59,65667 | 2015 | KH | Rubellum |
| ss25 | 120,124 | S. rubellum | Scotland | Highland | −6,37338 | 57,37548 | 2011 | MOK | Rubellum |
| ss32 | 98,281 | S. cf. capillifolium | Czech Republic | Pardubický kraj | 15,96429 | 49,73827 | 2016 | KIF | Rubellum |
| ss33* | 98,279 | S. cf. capillifolium | Czech Republic | Pardubický kraj | 15,96429 | 49,73827 | 2016 | KIF | Rubellum |
| ss61 | 158,861 | S. rubellum | Norway | Trøndelag | 10,06233 | 63,97804 | 2002 | KIF | Rubellum |
| ss63 | 725,288 | S. rubellum | Canada | British Columbia | −126 | 49,16667 | 2000 | KIF | Rubellum |
| 229 | 115,591 | S. warnstorfii | Norway | Trøndelag | 9,099786 | 63,06362 | 2016 | KIF | Warnstorfii |
| ss14 | 728,537 | S. warnstorfii | Scotland | Highland | 1987 | KIF | Warnstorfii | ||
| ss16 | 740,618 | S. warnstorfii | Norway | Vestland | 5,64284 | 60,41393 | 2008 | KIF | Warnstorfii |
| ss17 | 38,119 | S. warnstorfii | Czech Republic | Pardubický kraj | 15,96413 | 49,73867 | 2016 | KH | Warnstorfii |
| ss18 | 98,276 | S. warnstorfii | Czech Republic | Pardubický kraj | 15,93139 | 49,70528 | 2016 | KIF | Warnstorfii |
| ss19 | 93,992 | S. warnstorfii | Austria | Salzburg | 13,86189 | 47,18654 | 2017 | KIF, CS | Warnstorfii |
| ss20 | 94,022 | S. warnstorfii | Austria | Salzburg | 13,7101 | 47,10077 | 2017 | KIF, CS | Warnstorfii |
| ss65 | 109,505 | S. warnstorfii | Scotland | Highland | −6,22782 | 57,4338 | 2019 | NGH | Warnstorfii |
| ss66 | 109,504 | S. warnstorfii | Scotland | Highland | −6,22782 | 57,4338 | 2019 | NGH | Warnstorfii |
| ss67 | 109,506 | S. warnstorfii | Scotland | Highland | −6,22782 | 57,4338 | 2019 | NGH | Warnstorfii |
| ss64 | 728,492 | S. venustum | Canada | Newfoundland and Labrador | −56,3042 | 51,97892 | 2007 | BF, KIF | Venustum |
| ss70 | 741,059 | S. venustum | Norway | Trøndelag | 11,69203 | 63,93091 | 2011 | KIF | Venustum |
| ss71 | 728,496 | S. venustum | Canada | Quebec | −72,6333 | 53,85 | 2010 | MW | Venustum |
| 220 | 741,060 | S. venustum | Norway | Trøndelag | 11,69203 | 63,93091 | 2011 | KIF | Venustum |
| 217 | 724,524 | S. beothuk | Canada | Newfoundland and Labrador | −56,7053 | 51,62902 | 2007 | BF, KIF | Beothuk |
| 213 | 108,518 | S. beothuk | Norway | Trøndelag | 11,24113 | 64,57635 | 2019 | KH, OM, MOK, KIF | Beothuk |
| 214 | 108,530 | S. beothuk | Norway | Trøndelag | 11,07712 | 64,4292 | 2019 | KH, OM, MOK, KIF | Beothuk |
| 215 | 724,518 | S. beothuk | Canada | Newfoundland and Labrador | −55,6243 | 52,37197 | 2007 | BF, KIF | Beothuk |
| 219 | 115,819 | S. fuscum | Canada | Quebec | −72,2329 | 52,81405 | 2018 | LB, MFI | Fuscum |
| 210 | 724,520 | S. fuscum | Canada | Quebec | −57,2362 | 51,50422 | 2007 | BF, KIF | Fuscum |
| 212 | 724,523 | S. fuscum | Canada | Newfoundland and Labrador | −56,7088 | 51,66303 | 2007 | BF, KIF | Fuscum |
| 209 | 114,017 | S. fuscum | Norway | Trøndelag | 11,21456 | 64,26283 | 2020 | KH, KIF, OM | Fuscum |
| 211 | 676,317 | S. fuscum | Norway | Trøndelag | 11,6934 | 63,93074 | 2013 | TP | Fuscum |
| ss58 | 158,973 | S. fuscum | Norway | Trøndelag | 10,51795 | 64,09067 | 2002 | KIF | Fuscum |
| ss60 | 724,509 | S. fuscum | Canada | British Columbia | 2000 | KIF | Fuscum |
Note: The specimens were analyzed in three different datasets: the Actuifolia dataset included all specimens except S. skyense and samples marked with * (assumed to be clones sampled at the same site), the ‘red Acutifolia’ dataset further excluded samples marked as ** (identical haplotypes), and the S. skyense dataset that included all individuals in bold. The specimen order in the table is the same as the specimen order in the boxplots in Figures 2 and 3 from left to right. Collectors: BF, B. Flatberg; CP, C. Pötsch; CS, C. Schröck; EF, E. Feldmayer‐Christe; IG, I. Goldberg; KH, K. Hassel; KIF, K.I. Flatberg; KOK, K.O. Kyrkjeeide; LB, L. Bourgouin; MFI, M.‐F. Indorf; MJ, M. Jokerud; MOH, M.O. Hill; MOK, M.O. Kyrkjeeide; MW, M. White; NGH, N.G. Hodgetts; OM, O. Meleshko; TP, T. Prestø.
We used microsatellite markers to explore genetic diversity of the specimens. These markers have been frequently used as a mean of identifying new species (e.g., Karlin et al., 2008) and genetic structuring patterns (Kyrkjeeide, Hassel, Flatberg, Shaw, Yousefi, & Stenoien, 2016), and they allow identification of ploidy levels as allopolyploid species have more than one allele present at several loci (Kyrkjeeide et al., 2019; Ricca et al., 2008). Furthermore, patterns obtained by microsatellite data has been confirmed using much larger SNP datasets (Duffy et al., 2020; Shaw et al., 2022). Altogether, 15 microsatellite markers (1, 7, 9, 12, 17, 19, 20, 22, 29, 30, 56, 65, 68, 78, 93) were amplified for all specimens and genotyped in GeneMapper (Table 2, see e.g., Kyrkjeeide, Hassel, Flatberg, Shaw, Brochmann, & Stenoien, 2016 for methods).
TABLE 2.
Allele data of Acutifolia specimens from 15 microsatellite markers (ssr) developed for Sphagnum.
| DNA ID | Assigned name in analyses | ssr1 | ssr7 | ssr12 | ssr68 | ssr19 | ssr93 | ssr29 | ssr30 | ssr9 | ssr56 | ssr20 | ssr22 | ssr17 | ssr65 | ssr78 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ss1 | skyense | 245 | 189/198 | 117 | 218/225 | 264 | 221/253 | 198 | 127/139 | 187/189 | 200/206 | 277/282 | 94/103 | 158/0 | 190 | 201/213 |
| ss3 | skyense | 245 | 189/198 | 117 | 218/225 | 264 | 221/253 | 198 | 127/139 | 187/189 | 200/206 | 277/282 | 94/103 | 158/164 | 190 | 201/213 |
| ss4 | skyense | 245 | 189/200 | 117 | 218/225 | 264 | 221/250 | 198 | 127/139 | 187/187 | 200/206 | 277/282 | 94/103 | 158/164 | 190 | 201/207 |
| ss5 | subnitens | 245 | 189 | 117 | 225 | 264 | 221 | 198 | 127 | 187 | 209 | 282 | 94 | 164 | 190 | 207 |
| ss6 | subnitens | 245 | 189 | 117 | 225 | 264 | 221 | 198 | 127 | 187 | 203 | 282 | 94 | 164 | 190 | 207 |
| ss7 | subnitens | 245 | 189 | 117 | 225 | 264 | 221 | 198 | 127 | 187 | 203 | 282 | 94 | 164 | 190 | 207 |
| ss8 | subnitens | 245 | 197 | 117 | 225 | 264 | 221 | 198 | 127 | 187 | 197 | 282 | 94 | 164 | 190 | 213 |
| ss9 | subnitens | 245 | 189 | 117 | 225 | 264 | 221 | 198 | 127 | 187 | 197 | 282 | 94 | 164 | 190 | 207 |
| ss10 | quinquefarium | 245 | 198 | 117 | 218 | 256 | 0 | 201 | 139 | 186 | 206 | 277 | 103 | 158 | 190 | 201 |
| ss11 | quinquefarium | 245 | 198 | 117 | 218 | 256 | 250 | 198 | 139 | 189 | 206 | 277 | 103 | 156 | 190 | 201 |
| ss12 | quinquefarium | 245 | 198 | 117 | 218 | 256 | 253 | 201 | 139 | 189 | 206 | 277 | 103 | 156 | 190 | 201 |
| ss13 | quinquefarium | 245 | 198 | 117 | 218 | 256 | 0 | 198 | 139 | 186 | 206 | 277 | 103 | 158 | 190 | 201 |
| ss68 | quinquefarium | 245 | 198 | 117 | 218 | 256 | 253 | 198 | 139 | 186 | 206 | 277 | 103 | 156 | 190 | 201 |
| ss23 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 201 | 137 | 172 | 239 | 276 | 106 | 155 | 187 | 195 |
| ss24 | capillifolium | 245 | 179 | 119 | 222 | 256 | 229 | 201 | 137 | 185 | 239 | 276 | 106 | 161 | 187 | 213 |
| ss26 | capillifolium | 245 | 177 | 119 | 222 | 270 | 229 | 201 | 137 | 185 | 230 | 276 | 106 | 161 | 187 | 195 |
| ss28 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 202 | 137 | 185 | 221 | 277 | 106 | 161 | 190 | 195 |
| ss29 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 201 | 137 | 172 | 233 | 276 | 106 | 155 | 187 | 213 |
| ss30 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 200 | 137 | 168 | 233 | 277 | 106 | 155 | 187 | 195 |
| ss31 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 201 | 137 | 185 | 221 | 276 | 106 | 161 | 187 | 195 |
| ss36 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 200 | 137 | 169 | 230 | 276 | 106 | 161 | 187 | 195 |
| ss37** | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 200 | 137 | 169 | 230 | 276 | 106 | 161 | 187 | 195 |
| ss39 | capillifolium | 245 | 177 | 119 | 222 | 256 | 229 | 201 | 137 | 172 | 239 | 277 | 106 | 155 | 187 | 195 |
| 224 | conspecific | 245 | 177 | 119 | 225 | 258 | 229 | 198 | 137 | 185 | 215 | 274 | 109 | 155 | 184 | 210 |
| 230 | conspecific | 245 | 179 | 119 | 225 | 267 | 247 | 204 | 137 | 0 | 215 | 274 | 109 | 155 | 185 | 195 |
| 241 | conspecific | 245 | 186 | 123 | 223 | 256 | 229 | 198 | 137 | 184 | 215 | 274 | 112 | 157 | 190 | 210 |
| ss21 | conspecific | 245 | 177 | 123 | 225 | 256 | 229 | 195 | 143 | 169 | 215 | 274 | 109 | 157 | 190 | 195 |
| ss27** | conspecific | 245 | 0 | 119 | 223 | 264 | 229 | 198 | 137 | 162 | 224 | 274 | 112 | 157 | 184 | 195 |
| ss34 | conspecific | 245 | 179 | 123 | 225 | 0 | 229 | 195 | 137 | 184 | 215 | 274 | 109 | 155 | 190 | 207 |
| ss35 | conspecific | 245 | 0 | 119 | 223 | 264 | 229 | 198 | 137 | 162 | 224 | 274 | 112 | 157 | 184 | 195 |
| SW01 | conspecific | 245 | 179 | 119 | 225 | 267 | 229 | 195 | 137 | 184 | 215 | 300 | 109 | 152 | 184 | 207 |
| SN1*3* | nitidulum | 245 | 175 | 119 | 231 | 259 | 247 | 204 | 147 | 169 | 233 | 275 | 109 | 157 | 190 | 201 |
| SN11 | nitidulum | 245 | 175 | 119 | 231 | 259 | 247 | 204 | 147 | 169 | 233 | 275 | 109 | 157 | 190 | 201 |
| SN12* | nitidulum | 245 | 175 | 119 | 231 | 259 | 247 | 204 | 147 | 169 | 233 | 275 | 109 | 157 | 190 | 201 |
| SN14* | nitidulum | 245 | 175 | 119 | 231 | 259 | 247 | 204 | 147 | 169 | 233 | 275 | 109 | 157 | 190 | 201 |
| RB01** | nitidulum | 245 | 175 | 119 | 231 | 259 | 247 | 204 | 147 | 169 | 233 | 275 | 109 | 157 | 190 | 201 |
| 221 | rubellum | 245 | 177 | 119 | 231 | 264 | 232 | 198 | 137 | 195 | 221 | 277 | 112 | 155 | 0 | 216 |
| 222 | rubellum | 245 | 173 | 119 | 225 | 259 | 247 | 198 | 137 | 180 | 227 | 295 | 109 | 155 | 184 | 210 |
| 223 | rubellum | 245 | 0 | 119 | 225 | 264 | 241 | 195 | 137 | 0 | 230 | 295 | 112 | 157 | 196 | 207 |
| 240 | rubellum | 245 | 187 | 125 | 225 | 264 | 229 | 196 | 137 | 169 | 191 | 274 | 94 | 157 | 194 | 201 |
| 242 | rubellum | 245 | 179 | 125 | 225 | 263 | 241 | 200 | 147 | 179 | 224 | 298 | 109 | 157 | 190 | 210 |
| ss15 | rubellum | 245 | 177 | 123 | 225 | 259 | 229 | 204 | 139 | 169 | 233 | 295 | 109 | 152 | 184 | 210 |
| ss25 | rubellum | 245 | 179 | 119 | 225 | 266 | 244 | 201 | 143 | 169 | 224 | 274 | 106 | 152 | 184 | 204 |
| ss32 | rubellum | 245 | 179 | 123 | 228 | 264 | 244 | 196 | 137 | 169 | 224 | 295 | 109 | 157 | 190 | 195 |
| ss33* | rubellum | 245 | 179 | 123 | 228 | 264 | 244 | 196 | 137 | 169 | 224 | 295 | 109 | 157 | 190 | 195 |
| ss61 | rubellum | 245 | 173 | 125 | 225 | 264 | 232 | 204 | 137 | 169 | 224 | 298 | 90 | 157 | 196 | 195 |
| ss63 | rubellum | 245 | 181 | 123 | 225 | 274 | 232 | 196 | 147 | 172 | 218 | 274 | 106 | 152 | 194 | 201 |
| 229 | warnstorfii | 245 | 190 | 119 | 225 | 267 | 244 | 201 | 139 | 172 | 194 | 290 | 100 | 155 | 187 | 204 |
| ss14 | warnstorfii | 245 | 194 | 119 | 225 | 270 | 244 | 201 | 141 | 172 | 188 | 289 | 100 | 155 | 181 | 204 |
| ss16 | warnstorfii | 245 | 190 | 0 | 225 | 267 | 247 | 201 | 139 | 172 | 212 | 289 | 100 | 157 | 187 | 0 |
| ss17 | warnstorfii | 245 | 190 | 121 | 225 | 267 | 247 | 201 | 141 | 172 | 224 | 289 | 100 | 155 | 187 | 195 |
| ss18 | warnstorfii | 245 | 190 | 0 | 225 | 270 | 244 | 201 | 139 | 172 | 212 | 289 | 100 | 155 | 187 | 204 |
| ss19 | warnstorfii | 245 | 190 | 117 | 225 | 267 | 243 | 201 | 139 | 179 | 212 | 292 | 103 | 155 | 187 | 204 |
| ss20 | warnstorfii | 245 | 190 | 0 | 225 | 270 | 243 | 201 | 139 | 179 | 194 | 289 | 103 | 155 | 187 | 0 |
| ss65** | warnstorfii | 245 | 190 | 119 | 0 | 269 | 244 | 201 | 141 | 196 | 221 | 289 | 103 | 155 | 181 | 0 |
| ss66 | warnstorfii | 245 | 190 | 119 | 225 | 265 | 247 | 200 | 141 | 178 | 224 | 289 | 100 | 155 | 187 | 0 |
| ss67 | warnstorfii | 245 | 190 | 119 | 225 | 269 | 244 | 201 | 141 | 196 | 221 | 289 | 103 | 155 | 181 | 201 |
| 220 | venustum | 245 | 175 | 119 | 218 | 282 | 251 | 198 | 137 | 196 | 227 | 277 | 106 | 155 | 191 | 204 |
| ss64 | venustum | 245 | 175 | 119 | 218 | 279 | 251 | 201 | 147 | 196 | 227 | 277 | 106 | 155 | 191 | 195 |
| ss70** | venustum | 245 | 175 | 119 | 218 | 282 | 251 | 0 | 137 | 196 | 227 | 277 | 106 | 155 | 191 | 204 |
| ss71 | venustum | 245 | 175 | 119 | 218 | 273 | 251 | 195 | 143 | 196 | 227 | 277 | 106 | 155 | 191 | 204 |
| 217 | beothuk | 245 | 179 | 121 | 218 | 256 | 250 | 198 | 127 | 182 | 206 | 277 | 106 | 0 | 194 | 201 |
| 213 | beothuk | 245 | 179 | 121 | 218 | 256 | 238 | 198 | 127 | 182 | 206 | 277 | 106 | 157 | 194 | 201 |
| 214 | beothuk | 245 | 179 | 121 | 218 | 256 | 238 | 198 | 127 | 182 | 206 | 277 | 106 | 157 | 194 | 201 |
| 215 | beothuk | 245 | 179 | 121 | 218 | 0 | 247 | 198 | 127 | 182 | 206 | 277 | 106 | 157 | 194 | 201 |
| 219 | fuscum | 245 | 189 | 121 | 225 | 256 | 241 | 198 | 127 | 182 | 209 | 294 | 103 | 158 | 175 | 198 |
| 210 | fuscum | 245 | 189 | 121 | 225 | 256 | 241 | 198 | 130 | 186 | 212 | 288 | 103 | 158 | 176 | 198 |
| 212 | fuscum | 245 | 189 | 121 | 225 | 256 | 241 | 198 | 127 | 182 | 209 | 288 | 109 | 158 | 176 | 198 |
| 209 | fuscum | 245 | 193 | 121 | 225 | 256 | 241 | 198 | 130 | 182 | 212 | 288 | 97 | 158 | 176 | 198 |
| 211 | fuscum | 245 | 193 | 121 | 227 | 256 | 241 | 198 | 130 | 186 | 212 | 291 | 103 | 158 | 176 | 198 |
| ss58 | fuscum | 245 | 189 | 121 | 225 | 256 | 241 | 198 | 130 | 186 | 215 | 291 | 106 | 158 | 176 | 198 |
| ss60 | fuscum | 245 | 189 | 121 | 225 | 255 | 241 | 198 | 127 | 186 | 212 | 291 | 103 | 153 | 176 | 198 |
Note: The DNA ID correspond to the ID in Table 1 and Figure 4. Specimens were analyzed in three different datasets (see main text and Table 1). The specimen order in the table is the same as the specimen order in the boxplots in Figures 2 and 3 from left to right. Sphagnum skyense named skyense under Assigned name in analyses, is diploid and have two alleles at each microsatellite.
After visually studying the number of alleles identified for each morphological species, three datasets were prepared for statistical analyses. The first dataset included all specimens identified as haploid (one allele at each locus), this only excluded S. skyense. In addition, four samples were excluded as they were collected at the same site and had identical haplotypes, and hence were likely clones (see Table 2). A total of 65 specimens were included in the dataset and we hereafter call it the Acutifolia dataset. The second dataset, hereafter called the “red Acutifolia” dataset, included specimens of S. rubellum, S. capillifolium, S. warnstorfii, S. nitidulum and conspecific specimens, and also S. venustum, as analyses of the Acutifolia dataset show that this species cluster with “red Acutifolia” (see below). Specimens with identical haplotypes were removed from the dataset, and a total of 39 specimens were analyzed. The third dataset, hereafter called the S. skyense dataset, included S. skyense and the hypothesized parental species: S. subnitens, S. quinqefarium, and S. warnstorfii. In total, 17 specimens were included in this dataset and scored as diploid individuals, with the second allele scored as missing for the three latter species.
The software Structure (Falush et al., 2003, 2007; Pritchard et al., 2000) was used to identify the number of genetic clusters and potential admixture among individuals of morphologically described taxa (sensu van Hengstum et al., 2012). The analysis was run for the first dataset with the number of genetic clusters (K) ranging from 1 to 10, to explore clustering among all Acutifolia species. The second dataset was run using K = 2–6. The dataset included five morphologically described species and seven specimens that were collected as conspecific (Table 1). The third dataset was run for K = 3, as S. skyense is described as a hybrid species and the aim of the analyses were to detect potential admixture between the parental species. For each K, the analyses were replicated 10 times with a burnin of 100,000 steps followed by a Monte Carlo Markov chain of 200,000 steps. We ran Structure with different settings for the first dataset applying the admixture model and both the correlated and independent allele frequencies model. The correlated allele frequencies model is better at detecting distinct populations that are closely related (Porras‐Hurtado et al., 2013), and this model was used when analyzing the second and third datasets. The results were further assessed using Clumpak (Kopelman et al., 2015) and visualized with StructureSelector (Li & Liu, 2017).
The data were also explored in GenAlEx v6.501 (Peakall & Smouse, 2012). A genetic distance matrix was made of the Acutifolia dataset and the “red Acutifolia” dataset using the haploid (ssr) option and interpolating missing data. The distance matrices were used in PCoA analyses with the covariance‐standardized method in GenAlEx, but for the Acutifolia dataset also in SplitsTree v4.19.0 (Huson & Bryant, 2006) to make a Neighbor‐Net network (Bryant & Moulton, 2002). A PCoA was also made for the S. skyense dataset, but the distance matrix was made with the codominant option and the haploid specimens scored as homozygotes to avoid missing data at the second allele.
3. RESULTS
We used the number of alleles recognized for each locus to confirm the ploidy level of S. skyense and S. venustum (sensu Ricca et al., 2008). Our results confirm that S. venustum is a haploid species, as only one allele was identified for each locus, while S. skyense is diploid as 10 of 15 loci have two alleles. At the five loci with only one allele, four of them have the same allele in S. skyense, S. subnitens and S. quinquefarium (Table 2). Furthermore, a visual comparison of alleles across the S. skyense dataset, shows that the alleles in the heterozygote loci are mainly the same as alleles found in S. subnitens and S. quinquefarium. The Structure analysis of this dataset shows that S. skyense is indeed admixed between S. subnitens and S. quinquefarium, strongly supporting that they are the parental species of S. skyense (Figure 2).
FIGURE 2.
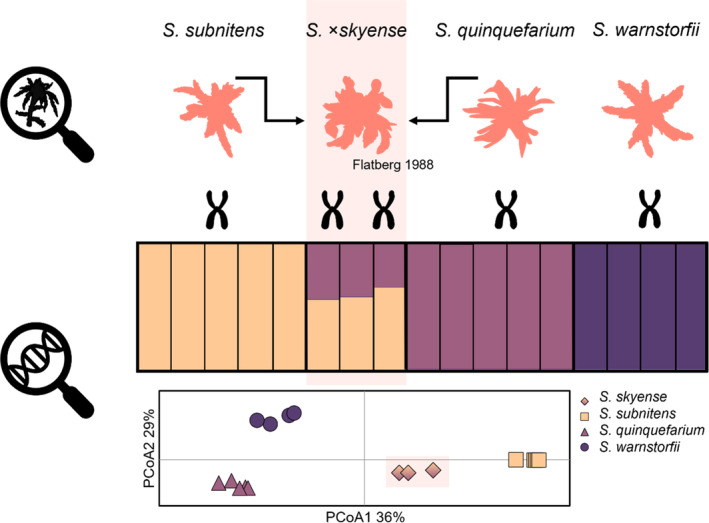
Microsatellite data of Sphagnum × skyense show that the species has two alleles at most of its loci, strongly indicating that it is a diploid species originating from hybridization. The barplot shows the result of STRUCTURE analysis of microsatellite data of S. × skyense and three hypothesized parental species. The result confirms that S. × skyense is an admixed species between S. subnitens and S. quinquefarium. This is also evident in the PCoA (below) which shows that S. × skyense is placed between its parental species. In the barplot, each bar represents one individual. The order of the bars, from left to right, follows the order of individuals listed in Table 1 (dataset S. × skyense).
Also, specimens of S. venustum had one allele at each loci, thus, the species is haploid. A sample collected as S. cf. rubellum in the Azores (Terceira) was genetically identical to the samples collected as S. nitidulum (Table 2). The Azores specimens share alleles with other Acutifolia species, but the same alleles overlap at maximum four loci with other specimens (Table 2). All the conspecific specimens had one allele at each locus; hence, they were all haploids, even though many of them were morphologically assigned as hybrids (Table 1).
The Structure analysis of the Acutifolia dataset shows that S. venustum mainly clusters with “red Acutifolia” species and that morphological species separate into different genetic clusters as the value of K increases. The Azorean specimens cluster with S. rubellum at K = 2–5, but as a separate cluster at K = 7–10. However, in analyses of the “red Acutifolia” dataset, where only unique haplotypes were included, the Azorean specimen cluster with S. rubellum at all K‐values (Figure 3). Furthermore, this analysis separates S. capillifolium, S. warnstorfii, S. rubellum and “conspecific” specimens, and S. venustum into different genetic clusters at K = 4, while the “conspecific” specimens form a fifth genetic cluster at K = 5 (Figure 3). Specimens of S. rubellum show varying extent of admixture with this cluster.
FIGURE 3.
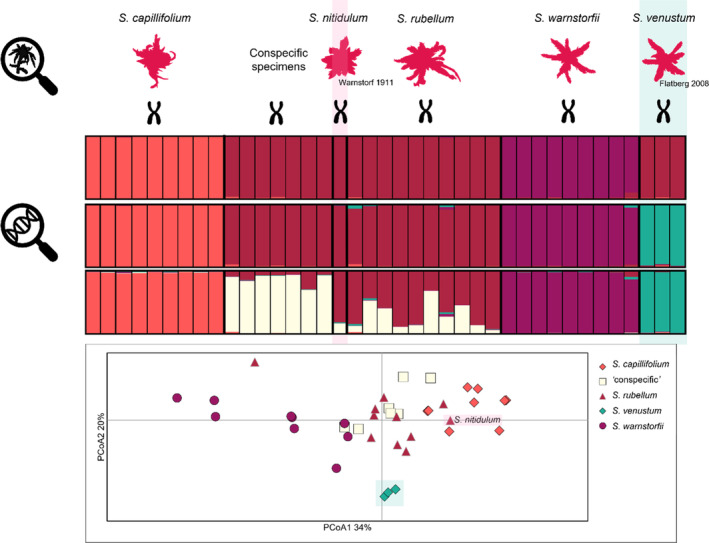
Barplots show the result of STRUCTURE analysis of 15 microsatellite data of five morphologically described Sphagnum sect. Acutifolia species and seven specimens assigned as conspecific. The order of the bars, from left to right, follows the order of individuals listed in Tables 1 and 2. The barplots show the results for different number of genetic clusters: 3 (results of 10/10 runs), 4 (results of 8/10) and 5 (10/10 runs). The PCoA (below) shows that S. venustum is separated from the other species, that are overlapping. The S. nitidulum sample (named and colored pink in the plot) was included as S. rubellum in the analysis and found within specimens of S. rubellum and S. capillifolium.
The results of the PCoA show that there is large variation within S. warnstorfii, S. rubellum, S. capillifolium, and the conspecific specimens, and that they overlap to some extent (Figures 3 and 4). Also, one individual of S. rubellum group with S. subnitens in the PCoA, but not in the Structure analysis (results not shown). Sphagnum venustum is clearly separated from the rest, and S. nitidulum is close to, but genetically distant from the bigger group of “red Acutifolia” (Figure 4). The distinctness of S. venustum and S. nitidulum is also evident the Neighbor‐Net network generated in SplitsTree (Figure 4). However, in the PCoA analysis of the “red Acutifolia” dataset, S. nitidulum is found within specimens of S. capillifolium and S. rubellum. In the network, specimens of S. subnitens, S. quinquefarium, S. beothuk, and S. fuscum group in their respective species, while S. warnstorfii, S. rubellum and S. capillifolium are not fully resolved, with occurrences in several places in the network. Also, the conspecific specimens are spread among S. rubellum and S capillifolium.
FIGURE 4.

PCoA (left) generated in GenAlEx and Neighbor‐Net network (right) constructed in SplitsTree4, both based on genetic distance matrix calculated in GenAlEx6.501 of nine haploid Sphagnum species in subgenus Acutifolia and a group named “conspecific” that consists of specimens deviating morphologically and also somewhat genetically (see Structure results in Figure 3) from describe “red Acutifolia” species. Specimen ID in the network are the same as in Table 1, where voucher information is given.
4. DISCUSSION
Our findings seem to point to different paths leading to speciation in the subgenus Acutifolia. Sphagnum skyense is an example of instant speciation by polyploidization, a common speciation mode in Sphagnum (reviewed by Meleshko et al., 2018). We were able to identify the parents, which is not always possible (Kyrkjeeide et al., 2019; Kyrkjeeide, Hassel, Flatberg, Shaw, Yousefi, & Stenoien, 2016) as they may, for example, have gone extinct (Kyrkjeeide et al., 2019). Our genetical examination of S. skyense concludes that S. subnitens and S. quinquefarium are indeed the parental species. Sphagnum subnitens has two close relatives not included in the analyses: S. subfulvum Sjörs and S. flavicomans (Cardot.) Warnst. However, since they have been found to be genetically distinct based on microsatellite data (Kyrkjeeide et al., 2018), and S. subnitens and S. skyense mainly overlap in alleles, it is unlikely that either S. subfulvum or S. flavicomans are the parental species. Another allopolyploid species that has both parental species identified, is S. troendelagicum Flatberg (Såstad et al., 2001). Stenøien, Shaw, Stengrundet, and Flatberg (2011) estimated the origin of this species to about 40,000 years before present, indicating that the young age of the species make it possible to trace the parents. Both S. troendelagicum and S. skyense have narrow distribution ranges within areas glaciated during the last glacial maximum. Thus, S. skyense could have a recent origin like S. troendelagicum.
Genetic data show that S. venustum is a haploid species. Furthermore, it is genetically distinct from other similar Acutifolia species sharing the same habitat and geographical distribution range. Thus, we hypothesize that speciation may have taken place by niche differentiation along the mire structure gradient known in other Sphagnum species (Johnson et al., 2015).
Our PCoA result shows that S. nitidulum clusters within other ‘red Acutifolia’ species, but the haplotype is genetically distinct. We cannot conclude that it is a valid species based on our results, as a bigger sample size is needed to further explore whether it rather belongs to S. rubellum. Sphagnum rubellum is not with certainty collected from Terceira, and the included specimen collection of S. cf. rubellum shares its haplotype with S. nitidulum. The specimen of S. cf. rubellum from Terceira was collected in moist, sloping heath. As it is genetically identical to S. nitidulum, the genetic distinctness of the latter is likely not a result of adaptation to the sulfuric springs S. nitidulum is described from. A detailed morphological examination of the specimens was outside the scope of this study, but preliminary analysis indicate that the Azorean specimens collected as S. nitidulum and S. cf. rubellum are morphologically similar, further supporting no adaptation to an uncommon habitat. However, additional samples from Azores should be examined both morphologically and genetically to confirm which taxa occur at Terceira. Sphagnum warnstorfii is not reported from Terceira, but S. capillifolium is. It is necessary to study the phylogenetic relationship of the species to conclude on the validity of S. nitidulum, but also the origin of this taxon on Terceira. A larger selection of specimens from North America should be included in future analyses, as the Atlantic Ocean is a weak barrier to dispersal (Kyrkjeeide, Hassel, Flatberg, Shaw, Brochmann, & Stenoien, 2016; Stenøien et al., 2011).
The three red species, S. warnstorfii, S. capillifolium, and S. rubellum, form three distinct genetic groups, as expected (Shaw et al., 2005). However, the specimens collected as conspecific, group with S. rubellum, but overlap also with S. capillifolium and group close to S. warnstorfii in the PCoA. They have been identified as morphologically deviating, but conspecific to S. warnstorfii or S. capillifolium. Four specimens were collected as S. cf. capillifolium × warnstorfii, one was collected as S. cf. warnstorfii, and three more specimens were originally identified as S. warnstorfii, S. capillifolium and S. rubellum. A brief re‐examination of the morphology indicates that the conspecific group of specimens share characters and differ from other red Acutifolia, but a more thorough morphological examination and more genetic data should be included in a taxonomic revision to further explore this potential new taxon.
All three investigated taxa can be considered rare, since even though the amphi‐Atlantic S. venustum produces spores and has a wide distribution range, it seems to form very small populations where it occurs. It is only found at one site in Europe and assumed overlooked (Hallingbäck, 2019), and is assessed as data deficient in the European Red List of bryophytes (Hodgetts, Calix, et al., 2019). Both S. skyense and S. nitidulum, for the time being, are narrow endemics without known spore production, seemingly dependent upon fragmentation for dispersal. While S. nitidulum is evaluated in the IUCN global Red List of species as critically endangered (Gabriel & Sim‐Sim, 2019; Hodgetts, Calix, et al., 2019) due to the very small population size restricted to one site with declining habitat area, S. skyense occurs at several sites with no signs of decline and is thus evaluated as of least concern (Hodgetts, Calix, et al., 2019; Hodgetts, Lockhart, et al., 2019).
A morphological approach to describing species in the genus Sphagnum has proven useful, but due to large morphological variation along ecological gradients, like hight above water table, nutrient availability and light conditions, the determination of specimens can be very difficult both in the field and in the laboratory. Also, there is usually a time lag from the description of a new species until scientists and field biologist are reporting the species from new locations. Sphagnum skyense was described from Isle of Skye in 1988 (Flatberg, 1988a, 1988b), and then recorded again 16 years later. However, several new records were made the following years (Hill, 2014). All three species, but especially S. venustum and potentially S. nitidulum, should be searched for in mainland Europe. It could be that S. venustum has successfully established in Europe only once, but the species is small, with a somewhat dull color, and is therefore easy to overlook. The species is characterized morphologically (Flatberg, 2008) and included in floras (Flatberg, 2013; Laine et al., 2018), aiding field biologists with identification. On the other hand, if S. nitidulum is indeed a valid species, a morphological revision is needed to make it easier to distinguish it from other red Acutifolia species. In addition, a wider range of conspecific specimens should be investigated in terms of morphology, genetics, distribution ranges, and habitat preferences.
We have provided genetic data of three rare Sphagnum species with limited occurrences and assessments in Red lists. We have not studied intraspecific variation within these species, but found high genetic variation among the “red Acutifolia” species. Even though we were not able to fully resolve if S. nitidulum and the conspecific samples are genetically differentiated from S. rubellum, S. capillifolium, and S. warnstorfii, our findings indicate that there is genetic diversity within this group of species that should be prioritized for conservation. Ensuring protection of genetic variation safeguard biodiversity even in species groups where taxonomy is uncertain (Andrello et al., 2022; Rosauer et al., 2018). This is often the case for bryophyte species, where genetic diversity is not always expressed in identifiable morphological characters (Hedenäs, 2016), but is valid for all organisms groups were taxonomic work is limited due to high species diversity, few morphological characters or even lack of experts and taxonomists.
AUTHOR CONTRIBUTIONS
Magni Olsen Kyrkjeeide: Conceptualization (equal); data curation (equal); formal analysis (lead); methodology (lead); visualization (lead); writing – original draft (lead); writing – review and editing (lead). Olena Meleshko: Conceptualization (equal); data curation (supporting); formal analysis (supporting); methodology (supporting); writing – review and editing (equal). Kjell Ivar Flatberg: Conceptualization (equal); data curation (equal); writing – review and editing (equal). Kristian Hassel: Conceptualization (equal); data curation (supporting); methodology (supporting); writing – original draft (supporting); writing – review and editing (equal).
CONFLICT OF INTEREST STATEMENT
All authors declare that they have no conflicts of interest.
ACKNOWLEDGMENTS
Thanks to Line Birkeland at NINAGen for laboratory assistance. Thanks to three anonymous reviewers for valuable input.
Kyrkjeeide, M. O. , Meleshko, O. , Flatberg, K. I. , & Hassel, K. (2023). Short stories from Sphagnum of rare species, taxonomy, and speciation. Ecology and Evolution, 13, e10356. 10.1002/ece3.10356
DATA AVAILABILITY STATEMENT
The data are available in Table 2.
REFERENCES
- Andrello, M. , D'Aloia, C. , Dalongeville, A. , Escalante, M. A. , Guerrero, J. , Perrier, C. , Torres‐Florez, J. P. , Xuereb, A. , & Manel, S. (2022). Evolving spatial conservation prioritization with intraspecific genetic data. Trends in Ecology & Evolution, 37, 553–564. [DOI] [PubMed] [Google Scholar]
- Andrews, A. L. (1941). Notes on the Warnstorf Sphagnum herbarium II. The section Malacosphagnum. The Bryologist, 44, 97–102. [Google Scholar]
- Andrus, R. E. (2006). Six new species of Sphagnum (Bryophyta; Sphagnaceae). Sida, 22, 959–972. [Google Scholar]
- Ayotte, G. , & Rochefort, L. (2019). Les sphaignes de l’Est du Canada ‐ Clé d’identification visuelle et cartes de répartition. Éditions JFD. [Google Scholar]
- Bengtsson, F. , Granath, G. , & Rydin, H. (2016). Photosynthesis, growth, and decay traits in Sphagnum – A multispecies comparison. Ecology and Evolution, 6, 3325–3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant, D. , & Moulton, V. (2002). NeighborNet: An agglomerative method for the construction of planar phylogenetic networks. In Guig'o R. & Gusfield D. (Eds.), Algorithms in bioinformatics (Vol. 2452, pp. 375–391). WABI, LNCS. [Google Scholar]
- Carter, B. E. , Shaw, B. , & Shaw, A. J. (2016). Endemism in the moss flora of North America. American Journal of Botany, 103, 769–779. [DOI] [PubMed] [Google Scholar]
- CBD Secretariat . (2022). COP15: Final text of Kunming‐Montreal global biodiversity framework | convention on biological diversity. CBD Secretariat. [Google Scholar]
- Cronberg, N. (1989). Patterns of variation in morphological characters and isozymes in populations of Sphagnum capillifolium (Erh.) Hedw. And S. rubellum Wils. From two bogs in southern Sweden. Journal of Bryology, 15, 683–696. [Google Scholar]
- Cronberg, N. (1996). Isozyme evidence of relationships within Sphagnum sect. Acutifolia (Sphagnaceae, Bryophyta). Plant Systematics and Evolution, 203, 41–64. [Google Scholar]
- Cronberg, N. (1998). Population structure and interspecific differentiation of the peat moss sister species Sphagnum rubellum and S. capillifolium (Sphagnaceae) in northern Europe. Plant Systematics and Evolution, 209, 139–158. [Google Scholar]
- Duffy, A. M. , Aguero, B. , Stenøien, H. K. , Flatberg, K. I. , Ignatov, M. S. , Hassel, K. , & Shaw, A. J. (2020). Phylogenetic structure in the Sphagnum recurvum complex (Bryophyta) in relation to taxonomy and geography. American Journal of Botany, 107, 1283–1295. [DOI] [PubMed] [Google Scholar]
- Falush, D. , Stephens, M. , & Pritchard, H. W. (2007). Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Molecular Ecology Notes, 1, 574–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falush, D. , Stephens, M. , & Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics, 164, 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flatberg, K. I. (1986). Taxonomy, morphovariation, distribution andecology of the Sphagnum imbricatum complex with main reference to Norway. Gunneria, 54, 1–118. [Google Scholar]
- Flatberg, K. I. (1988a). Sphagnum skyense sp. Nov. Journal of Bryology, 15, 101–107. [Google Scholar]
- Flatberg, K. I. (1988b). Sphagnum troendelagicum sp. Nov. (sect. Cuspidata). Lindbergia, 14, 33–39. [Google Scholar]
- Flatberg, K. I. (2008). Sphagnum venustum (Bryophyta), a noticeable new species in sect. Acutifolia from Labrador, Canada. Lindbergia, 33, 2–12. [Google Scholar]
- Flatberg, K. I. (2013). Norges torvmoser. Akademika Trondheim. [Google Scholar]
- Frahm, J.‐P. , & Vitt, D. H. (1993). Comparisons between the moss floras of North‐America and Europe. Nova Hedwigia, 56, 307–333. [Google Scholar]
- Gabriel, R. , & Sim‐Sim, M. (2019). Sphagnum nitidulum. The IUCN red list of threatened species. 10.2305/IUCN.UK.2019-2.RLTS.T87567839A87716016.en [DOI]
- Granath, G. , Strengbom, J. , & Rydin, H. (2010). Rapid ecosystem shifts in peatlands: Linking plant physiology and succession. Ecology, 91, 3047–3056. [DOI] [PubMed] [Google Scholar]
- Hájek, M. , Hájková, P. , Apostolova, I. , Sopotlieva, D. , Goia, I. , & Dítě, D. (2021). The vegetation of rich fens (Sphagno warnstorfii‐Tomentypnion nitentis) at the southeastern margins of their European range. Vegetation Classification and Survey, 2, 177–190. [Google Scholar]
- Hallingbäck, T. (2019). Sphagnum venustum. The IUCN red list of threatened species. 2019: e.T90103201A90103600.
- Hassel, K. , Kyrkjeeide, M. O. , Yousefi, N. , Prestø, T. , Stenøien, H. K. , Shaw, J. A. , & Flatberg, K. I. (2018). Sphagnum divinum (sp. Nov.) and S. medium Limpr. and their relationship to S. magellanicum brid. Journal of Bryology, 40, 197–222. [Google Scholar]
- Hedenäs, L. (2016). Intraspecific diversity matters in bryophyte conservation – Internal transcribed spacer and rpl16 G2 intron variation in some European mosses. Journal of Bryology, 38, 173–182. [Google Scholar]
- Heinrichs, J. , Hentschel, J. , Feldberg, K. , Bombosch, A. , & Schneider, H. (2009). Phylogenetic biogeography and taxonomy of disjunctly distributed bryophytes. Journal of Systematics and Evolution, 47, 497–508. [Google Scholar]
- Hill, M. O. (2014). Sphagnum skyense . In Blockeel T., Bosanquet M. O., Hill M. O., & Preston C. D. (Eds.), Atlas of British and Irish bryophytes (p. 377). Pisces Publications. [Google Scholar]
- Hill, M. O. (2019). Sphagnum mosses: The stars of European mires. Journal of Bryology, 41, 386–387. [Google Scholar]
- Hodgetts, N. , Calix, M. , Englefield, E. , Fettes, N. , Garcia Criado, M. , Patin, L. , Nieto, A. , Bergamini, A. , Bisang, I. , Baisheva, E. , Campisi, P. , Cogoni, A. , Hallingback, T. , Konstantinova, N. , Lockhart, N. , Sabovljevic, M. , Schnyder, N. , Schrock, C. , Sergio, C. , … Żarnowiec, J. (2019). A miniature world in decline: European red list of mosses, liverworts and hornworts. IUCN. [Google Scholar]
- Hodgetts, N. , Lockhart, N. , Rothero, G. , & Vanderpoorten, A. (2019). Sphagnum skyense. The IUCN red list of threatened species. 2019: e.T87569523A87826226. 10.2305/IUCN.UK.2019-2.RLTS.T87569523A87826226.en [DOI]
- Huson, D. H. , & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254–267. [DOI] [PubMed] [Google Scholar]
- Johnson, M. G. , Granath, G. , Tahvanainen, T. , Pouliot, R. , Stenøien, H. K. , Rochefort, L. , Rydin, H. , & Shaw, A. J. (2015). Evolution of niche preference in Sphagnum peat mosses. Evolution, 69, 90–103. [DOI] [PubMed] [Google Scholar]
- Karlin, E. F. , Boles, S. , & Shaw, A. J. (2008). Resolving boundaries between species in Sphagnum section Subsecunda using microsatellite markers. Taxon, 57, 1189–1200. [Google Scholar]
- Kopelman, N. M. , Mayzel, J. , Jakobsson, M. , Rosenberg, N. A. , & Mayrose, I. (2015). Clumpak: A program for identifying clustering modes and packaging population structure inferences across K. Molecular Ecology Resources, 15, 1179–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Aguero, B. , Shaw, A. J. , Temsch, E. M. , & Flatberg, K. I. (2018). Sphagnum incundum a new species in Sphagnum subg. acutifolia (Sphagnaceae) from boreal and arctic regions of North America. Phytotaxa, 333, 1–21. [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Aguero, B. , Temsch, E. M. , Afonina, O. M. , Shaw, A. J. , Stenøien, H. K. , & Flatberg, K. I. (2019). Sphagnum ×lydiae, the first allotriploid peatmoss in the northern hemisphere. The Bryologist, 122, 38–61. [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Flatberg, K. I. , Shaw, A. J. , Brochmann, C. , & Stenoien, H. K. (2016). Long‐distance dispersal and barriers shape genetic structure of peatmosses (Sphagnum) across the Northern Hemisphere. Journal of Biogeography, 43, 1215–1226. [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Flatberg, K. I. , Shaw, A. J. , Yousefi, N. , & Stenoien, H. K. (2016). Spatial genetic structure of the abundant and widespread peatmoss Sphagnum magellanicum brid. PLoS One, 11, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrkjeeide, M. O. , Hassel, K. , Stenøien, H. K. , Prestø, T. , Boström, E. , Shaw, A. J. , & Flatberg, K. I. (2015). The dark morph of Sphagnum fuscum in Europe is conspecific with the North American S. beothuk . Journal of Bryology, 37, 251–266. [Google Scholar]
- Laine, J. , Flatberg, K. I. , Harju, P. , Timonen, T. , Minkkinen, K. , Laine, A. , Tuittila, E. S. , & Vasander, H. (2018). Sphagnum mosses – The stars of European mires. Department of Forest Science, University of Helsinki, Sphagna Ky. [Google Scholar]
- Li, Y.‐L. , & Liu, J.‐X. (2017). STRUCTURESELECTOR: A web‐based software to select and visualize the optimal number of clusters using multiple methods. Molecular Ecology Resources, 18, 176–177. [DOI] [PubMed] [Google Scholar]
- Meleshko, O. , Stenøien, H. K. , Speed, J. D. M. , Flatberg, K. I. , Kyrkjeeide, M. O. , & Hassel, K. (2018). Is interspecific gene flow and speciation in peatmosses (Sphagnum) constrained by phylogenetic relationship and life‐history traits? Lindbergia, 1, linbg.01107. [Google Scholar]
- Mikulášková, E. , Hájek, M. , Veleba, A. , Johnson, M. G. , Hájek, T. , & Shaw, J. A. (2015). Local adaptations in bryophytes revisited: The genetic structure of the calcium‐tolerant peatmoss Sphagnum warnstorfii along geographic and pH gradients. Ecology and Evolution, 5, 229–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in excel. Population genetic software for teaching and research – An update. Bioinformatics, 28, 2537–2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porras‐Hurtado, L. , Ruiz, Y. , Santos, C. , Phillips, C. , Carracedo, Á. , & Lareu, M. V. (2013). An overview of STRUCTURE: Applications, parameter settings, and supporting software. Frontiers in Genetics, 4, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyke, G. H. , & Ehrlich, P. R. (2010). Biological collections and ecological/environmental research: A review, some observations and a look to the future. Biological Reviews of the Cambridge Philosophical Society, 85, 247–266. [DOI] [PubMed] [Google Scholar]
- Ricca, M. , Beecher, F. W. , Boles, S. B. , Temsch, E. , Greilhuber, J. , Karlin, E. F. , & Shaw, A. J. (2008). Cytotype variation and allopolyploidy in North American species of the Sphagnum subsecundum complex (Sphagnaceae). American Journal of Botany, 95, 1606–1620. [DOI] [PubMed] [Google Scholar]
- Rosauer, D. F. , Margaret, B. , Blom, M. P. K. , Coates, D. J. , Donnellan, S. , Doughty, P. , Keogh, J. S. , Kinloch, J. , Laver, R. J. , Myers, C. , Oliver, P. M. , Potter, S. , Rabosky, D. L. , Silva, A. C. A. , Smith, J. , & Moritz, C. (2018). Real‐world conservation planning for evolutionary diversity in the Kimberley, Australia, sidesteps uncertain taxonomy. Conservation Letters, 11, e12438. [Google Scholar]
- Rydin, H. (1986). Competition and niche separation in Sphagnum . Canadian Journal of Botany, 64, 1817–1824. [Google Scholar]
- Såstad, S. M. , Stenøien, H. K. , Flatberg, K. I. , & Bakken, S. (2001). The narrow endemic Sphagnum troendelagicum is an allopolyploid derivative of the widespread S. balticum and S. tenellum . Systematic Botany, 26, 66–74. [Google Scholar]
- Séneca, A. , & Söderström, L. J. J. o. B. (2009). Sphagnophyta of Europe and Macaronesia: A checklist with distribution data. Journal of Bryology, 31, 243–254. [Google Scholar]
- Shaw, A. J. , Cox, C. J. , & Boles, S. B. (2005). Phylogeny, species delimitation, and recombination in Sphagnum section Acutifolia . Systematic Botany, 30, 16–33. [Google Scholar]
- Shaw, A. J. , Devos, N. , Cox, C. J. , Boles, S. B. , Shaw, B. , Buchanan, A. M. , Cave, L. , & Seppelt, R. (2010). Peatmoss (Sphagnum) diversification associated with Miocene Northern Hemisphere climatic cooling? Molecular Phylogenetics and Evolution, 55, 1139–1145. [DOI] [PubMed] [Google Scholar]
- Shaw, A. J. , Piatkowski, B. , Duffy, A. M. , Aguero, B. , Imwattana, K. , Nieto‐Lugilde, M. , Healey, A. , Weston, D. J. , Patel, M. N. , Schmutz, J. , Grimwood, J. , Yavitt, J. B. , Hassel, K. , Stenøien, H. K. , Flatberg, K.‐I. , Bickford, C. P. , & Hicks, K. A. (2022). Phylogenomic structure and speciation in an emerging model: The Sphagnum magellanicum complex (Bryophyta). New Phytologist, 236, 1497–1511. [DOI] [PubMed] [Google Scholar]
- Stenøien, H. K. , Hassel, K. , Segreto, R. , Gabriel, R. , Karlin, E. F. , Shaw, A. J. , & Flatberg, K. I. (2014). High morphological diversity in remote Island populations of the peat moss Sphagnum palustre: Glacial refugium, adaptive radiation or just plasticity? The Bryologist, 117, 95–109. [Google Scholar]
- Stenøien, H. K. , Shaw, A. J. , Shaw, B. , Hassel, K. , & Gunnarsson, U. (2011). North American origin and recent European establishment of the amphi‐Atlantic peat moss Sphagnum angermanicum . Evolution, 65, 1181–1194. [DOI] [PubMed] [Google Scholar]
- Stenøien, H. K. , Shaw, A. J. , Stengrundet, K. , & Flatberg, K. I. (2011). The narrow endemic Norwegian peat moss Sphagnum troendelagicum originated before the last glacial maximum. Heredity, 106, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stropp, J. , Ladle, R. J. , Emilio, T. , Lessa, T. , & Hortal, J. (2022). Taxonomic uncertainty and the challenge of estimating global species richness. Journal of Biogeography, 49, 1654–1656. [Google Scholar]
- Suarez, A. V. , & Tsutsui, N. D. (2004). The value of museum collections for research and society. Bioscience, 54, 66–74. [Google Scholar]
- Szövenyi, P. , Terracciano, S. , Ricca, M. , Giordano, S. , & Shaw, A. J. (2008). Recent divergence, intercontinental dispersal and shared polymorphism are shaping the genetic structure of amphi‐Atlantic peatmoss populations. Molecular Ecology, 17, 5364–5377. [DOI] [PubMed] [Google Scholar]
- van Hengstum, T. , Lachmuth, S. , Oostermeijer, J. G. B. , den Nijs, H. C. M. , Meirmans, P. G. , & van Tienderen, P. H. (2012). Human‐induced hybridization among congeneric endemic plants on Tenerife, Canary Islands. Plant Systematics and Evolution, 298, 1119–1131. [Google Scholar]
- Yousefi, N. , Hassel, K. , Flatberg, K. I. , Kemppainen, P. , Trucchi, E. , Shaw, A. J. , Kyrkjeeide, M. O. , Szövényi, P. , & Stenøien, H. K. (2017). Divergent evolution and niche differentiation within the common peatmoss Sphagnum magellanicum . American Journal of Botany, 104, 1060–1072. [DOI] [PubMed] [Google Scholar]
- Yousefi, N. , Mikulášková, E. , Stenøien, H. K. , Flatberg, K. I. , Košuthová, A. , Hájek, M. , & Hassel, K. (2019). Genetic and morphological variation in the circumpolar distribution range of Sphagnum warnstorfii: Indications of vicariant divergence in a common peatmoss. Botanical Journal of the Linnean Society, 189, 408–423. [Google Scholar]
- Yu, Z. , Beilman, D. W. , Frolking, S. , MacDonald, G. M. , Roulet, N. T. , Camill, P. , & Charman, D. J. (2011). Peatlands and their role in the global carbon cycle. Eos, Transactions, American Geophysical Union, 92, 97–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data are available in Table 2.


