Abstract
As it is well known, the gut is one of the primary sites in any host for xenobiotics, and the many microbial metabolites responsible for the interactions between the gut microbiome and the host. However, there is a growing concern about the negative impacts on human health induced by toxic xenobiotics. Metabolomics, broadly including lipidomics, is an emerging approach to studying thousands of metabolites in parallel. In this review, we summarized recent advancements in mass spectrometry (MS) technologies in metabolomics. In addition, we reviewed recent applications of MS-based metabolomics for the investigation of toxic effects of xenobiotics on microbial and host metabolism. It was demonstrated that metabolomics, gut microbiome profiling, and their combination have a high potential to identify metabolic and microbial markers of xenobiotic exposure and determine its mechanism. Further, there is increasing evidence supporting that reprogramming the gut microbiome could be a promising approach to the intervention of xenobiotic toxicity.
Keywords: Gut Microbiome, Xenobiotic Exposure, Mass Spectrometry, Metabolomics, Lipidomics, Metabolic Flux Analysis
1. Introduction
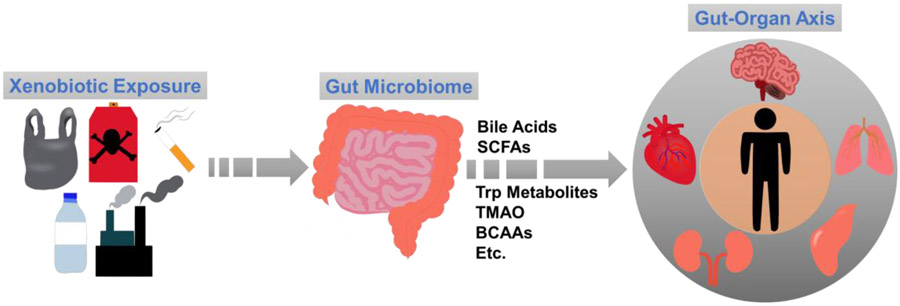
The gut microbiome in the human gastrointestinal tract is a complex and dynamic microorganism community that plays an important role in microbial metabolism and maintenance of the host health [1-5]. In humans, gut microbiome has hundreds of bacterial species and nearly two million microbial genes that are around a hundred times as many as the host [6]. Gut microbiota was identified as an “organ” in the human body in the past decade, and it co-evolves with the host and participates in numerous local physiological functions [7, 8], such as maintaining the intestinal membrane, mucus layer, and epithelium, as well as metabolizing xenobiotics and interacting with host immunity [9-11]. Remotely, gut microbiome can crosstalk with multiple host organs throughout the “gut-organ axis” (Fig. 1) [12-14]. There is increasing evidence showing that the gut microbiome is vital to a host’s health; e.g., if the homeostatic state of the gut microbiome is disrupted (dysbiosis), the crosstalk between the microbes and mucosal immune system will be compromised, which can cause various inflammatory responses and immune diseases [15, 16]. Numerous factors, including diet, environment, host health conditions, etc., are connected to potential gut microbiota disturbances. One of the most common factors is exposure to xenobiotics [17-19].
Fig. 1.
The gut serves as a crucial site for several xenobiotic exposures, which can impact a variety of essential microbial metabolites, including bile acids, short-chain fatty acids (SCFAs), tryptophan (Trp) metabolites, trimethylamine N-oxide (TMAO), and branched-chain amino acids (BCAA). In addition, the gut can remotely communicate with other organs within the host, leading to toxic effects through the gut-organ axis.
The gut serves as a major location within the host for diverse xenobiotics that are foreign chemical substances not naturally present within an organism [20-23]. Exposure to xenobiotics is unavoidable, since these substances are present in drugs, dietary supplements, industrial chemicals, food additives, pesticides, and various other environmental pollutants [24-27]. Exposure to these xenobiotics can occur in a variety of ways such as ingestion, inhalation, and dermal absorption [28]. Many of these xenobiotics are first accumulated and absorbed in the gut, and they undergo metabolic processes of reducing toxicity, increasing hydrophilicity, and becoming readily excretable [29, 30]. Gut microbiome plays a significant role in modulating the bioavailability and metabolism of xenobiotics. Not only can gut microbiome affect both the kinetics and dynamics of xenobiotic metabolism, but also the gut-derived molecules can enter the host’s circulation and directly affect the biological functions of host organs [31, 32]. As a result, multiple gut-organ axis, such as gut-liver axis (GLA), gut-brain axis (GBA), gut-pancreas axis (GPA), and gut-kidney axis (GKA), represent bidirectional signal pathways from the gut to various organs in the host (Fig. 1) [33-37]. These interactions are possible through means of metabolic, neural, endocrine, immune, and humoral connections [33, 38]. Chronic or acute exposure to toxic xenobiotics may alter the gut microbial composition and even the location of certain gut microbes, which eventually impacts host health [21]. The alarming issue of xenobiotic pollution, characterized by its high toxicity, long-lasting persistence, and limited biodegradability, has caused severe environmental harm on a global scale. Microorganisms have the potential to use xenobiotic compounds as a source of carbon or nitrogen to support their growth and metabolic functions. As a result, microbial-assisted degradation of xenobiotics is widely acknowledged as an effective and environmentally friendly approach. For example, various microbial strains such as Alcaligenes and Cellulosimicrobium have been identified and isolated for their exceptional biodegradation potential against diverse xenobiotic pollutants in soil and water environments [39]. In addition, growing evidence shows that reprogramming the gut microbiota through the administration of probiotics, prebiotics, and microbiome transplants can promote a healthy gut environment in the host, establishing a beneficial bacterial community. This approach can be utilized to alleviate the toxicity of xenobiotics, enhance immune function, and potentially aid in targeted disease (such as cancer) therapy [40-42].
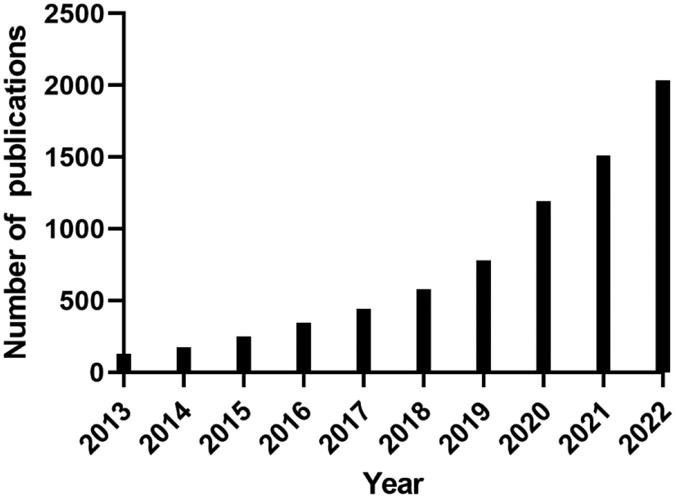
The interaction between the gut microbiome and the host is critically impacted by metabolism [43-47]. Over the past 10 years, there has been a dramatic increase in the number of publications investigating the association between gut microbiome and metabolome, as well as their connections to the host’s health/disease (Fig. 2). Through producing numerous essential microbial metabolites during food digestion and xenobiotic metabolism, the gut microbiota plays a crucial role in preserving the host's health and homeostasis, as supported by numerous pieces of evidence (Fig. 1) [16, 48]. For example, bile acids (BAs) are well-known pleiotropic signaling molecules mechanistically involved in gut-liver crosstalk [49-52]. BAs are a group of steroids produced in the liver from cholesterol [53]. These primary BAs are secreted into the lumen of the duodenum and metabolized by the gut microbiota into more lipophilic secondary BAs, the majority of which are absorbed and recirculated to the liver [50, 54]. In addition, short chain fatty acids (SCFAs) are mechanistically involved along the gut-brain axis [55, 56]. SCFAs, such as acetic acid, propionic acid, and butyric acid, are produced through the gut bacterial fermentation of dietary carbohydrates, mainly including fiber and starch, by bacteria in the gut [57, 58]. SCFAs have immunomodulatory properties and can interact with nerve cells by stimulating the sympathetic and autonomic nervous systems via G-protein-coupled receptor 41 (GPR41) and 43 (GPR43) [59]. Gut-derived SCFAs can enter the circulatory system and cross the blood-brain barrier (BBB), which makes it possible for them to directly modulate brain development and behavior [60, 61]. It is interesting to note that introducing Akkermansia muciniphila, which is a major producer of SCFAs, from an external source can have positive effects on neurological disorders like Alzheimer’s disease and seizures. The proposed ways in which this works include improving gene expression by enhancing histone acetylation, regulating amino acids and γ-aminobutyric acid (GABA) signaling in the central nervous system, and reducing inflammation [62-66]. Further, microbiota-dependent metabolites, including those in tryptophan (Trp) metabolism, play an important role in the gut-heart axis [67, 68]. Indole-3-propionic acid (IPA) is a microbial tryptophan derivative and a mitochondrial modulator in cardiomyocytes, and can directly impact cardiac functions [69]. Gut microbiota facilitates mammalian host nicotinamide adenine dinucleotide (NAD+) biosynthesis through a microbial nicotinamidase (PncA) [70]. Earlier studies showed that NAD+-dependent acetylation contributes to pressure overload-induced heart failure. While, activation of the NAD+ biosynthetic pathways can elevate NAD+ levels and alleviate cardiac dysfunction [71, 72].
Fig. 2.
The number of publications in the past 10 years, using the searching key words of “metabolome” and “microbiome” in PubMed (https://pubmed.ncbi.nlm.nih.gov/).
In this review, we aim to elucidate the key steps involved in the applications of omics, mainly including metabolomics and microbiomics, in recent selected xenobiotic exposure studies. In brief, we first outlined the general study design. In addition, we reviewed targeted and untargeted mass spectrometry (MS)-based metabolomics, metabolic flux analysis, metagenomic sequencing, as well as recent methods development related to these technologies. Further, we performed a comprehensive review of recent studies investigating xenobiotic exposures of wide interest and their effects on the gut microbiome and metabolome.
2. General Study Design
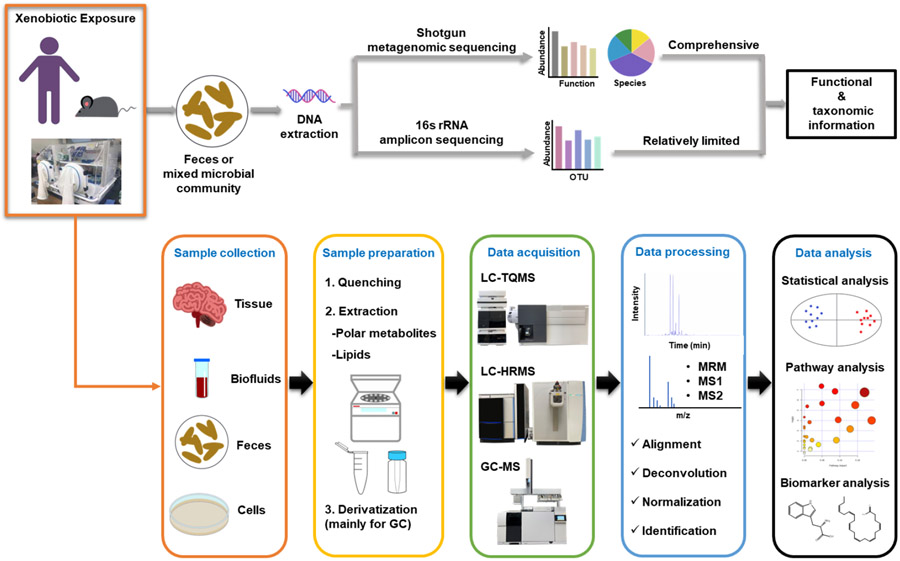
The general workflow is displayed in Fig. 3. Xenobiotic exposure can be conducted on humans, rodents, and cell models. To assess the diversity and functions of gut microbiota, samples of feces and/or intestinal content are collected. In general, the analysis of microbial communities begins with the sequencing of DNA to generate a catalog of genes and/or organisms [73]. Metagenomic sequencing analyzes genetic materials from mixed microbial populations through targeted or shotgun sequencing. Shotgun sequencing performs a more comprehensive analysis of the microbiome's composition and functions, providing an integrated view of both taxonomic and functional information without targeting specific genes or regions. This approach provides a more in-depth view of the microbiome, allowing for the identification of both known and unknown microorganisms, as well as their potential functions. 16S rRNA analysis is a targeted approach that focuses on a specific gene, the 16S ribosomal RNA gene, which is present in bacteria and archaea. The gene is conserved across these microbial groups but contains variable regions that can be used to identify and classify different bacterial and archaeal species. In contrast, whole shotgun metagenomic sequencing involves the sequencing of all DNA present in a sample, including the DNA from bacteria, archaea, viruses, and other microorganisms. Both approaches have their advantages and disadvantages, 16S rRNA analysis is often used as a cost-effective and rapid method to assess the microbial community composition in a sample. It can be particularly useful for large-scale surveys of microbial diversity or when the focus is on bacterial or archaeal communities. Whole shotgun metagenomic sequencing, on the other hand, provides a more detailed view of the microbiome but is often more expensive and computationally more intensive [74-77].
Fig. 3.
General flowchart for identifying metabolite biomarkers and investigating metabolic mechanisms, connecting gut microbiome, xenobiotic exposure, and host health.
Metabolomics, broadly including lipidomics, promises novel avenues for the detection of thousands of microbial and host metabolites associated with xenobiotic exposure and understanding the metabolic mechanisms that involve dysbiosis of gut microbiome [78-89]. Metabolomics focuses on detecting alterations at the metabolite level using analytical chemistry techniques and multivariate statistical analysis. To profile microbial metabolites, feces and intestine content samples are often collected. Host metabolic profiles will be measured from tissue, biofluid, and cell samples. In general, samples are first subjected to quenching, followed by metabolite extraction, and sometimes derivatization if necessary. The appropriate solvent systems are utilized to extract metabolites from the biological samples, and they are optimized based on the specific classes of metabolites under investigation. Typically, aqueous solvents are used to extract polar metabolites like amino acids and sugars, while relatively nonpolar metabolites such as lipids and fatty acids are extracted using a high portion of organic solvents. Optimal sample preparation methods and MS procedures may vary depending on the specific applications. Previous studies have established various workflows. For example, Courant et al. presented a tutorial that includes key steps from study design, sample preparation, MS data acquisition, and data analysis of the MS-based metabolomics flow, which could help beginners understand the concept [90]. Miggiels et al. focused on sample preparation methods, such as solid-phase extraction, liquid-liquid microextraction, and electro-driven extractions, as well as the introduction of NMR, liquid chromatography-mass spectrometry (LC-MS), gas chromatography-mass spectrometry (GC-MS), and capillary electrophoresis-MS [91]. Gong et al. and Hemmati et al. focused on sample preparation methods used in previous metabolomics studies, such as dry blood spot extraction, headspace-solid phase microextraction, microwave-assisted extraction, ultrasound-assisted extraction, and enzyme-assisted extraction techniques [92, 93]. Potential metabolite biomarkers will be selected after compound identification and statistical analysis, and metabolic pathway analysis is used to gain a better understanding of their potential roles in the biological processes. Importantly, further validation studies are typically needed to confirm the significance of identified biomarkers for specific applications. This workflow in Fig. 3 enables the integration of metabolic profiles and microbial community analysis results to yield metabolite and microbial biomarkers, along with a mechanistic understanding of the associated xenobiotic exposure.
Investigation of both microbial and host metabolites is a promising approach to accurately evaluate the toxicity of xenobiotics and develop therapeutic targets. In this review, we will summarize the recent advancements in metabolomics, especially those studies related to toxic xenobiotics using in vivo and in vitro biological models. Notably, we will focus on a few selected xenobiotics of wide interest in recent years. We are aware that there are many more xenobiotics currently under investigation, for more comprehensive and in-depth reviews, please see [21, 28, 94-96].
3. Recent Advancements of Metabolic Profiling Approaches
Currently, MS is the most commonly used analytical detection technology in metabolomics due to its high sensitivity and specificity [91, 97-100]. LC-MS and GC-MS are mainly used to detect metabolites and lipids, both widely used in targeted and untargeted metabolomics [101, 102]. The pace of method development in MS-based metabolomics has been rapid, in this review, a few highlights from the past few years will be reintroduced.
3.1. Untargeted metabolomics
Untargeted metabolomics, broadly including untargeted lipidomics, is widely used to generate hypotheses by identifying metabolites and pathways that are altered under certain conditions. Untargeted metabolomics is a powerful approach that allows for the comprehensive analysis of metabolites in a biological sample without prior knowledge of which metabolites are of interest. This contrasts with targeted metabolomics, which focuses on the measurement of specific metabolites. Untargeted metabolomics can be used to identify biomarkers of diseases or to gain a more comprehensive understanding of metabolic pathways and their regulation in various biological contexts. The choice between targeted and untargeted metabolomics depends on the specific research question and the available preliminary results [103-107]. Untargeted metabolomics is used to acquire unbiased information of the metabolites and their variations in the host and gut microbiome. High-sensitivity and high-resolution MS (HRMS) analyzers, such as time-of-flight (Tof) and orbitrap, are predominantly applied in untargeted metabolomics approaches, since mass accuracy is very informative for structure identification of the detected metabolites [97]. Typically, the mass range for aqueous metabolites is <1,000 Da, while for lipids it is < 2,000 Da [108-111]. However, the actual range used in specific studies may vary depending on the research questions and specific metabolites of interest. These metabolic results can be further associated with metagenomic sequencing or 16S rRNA analysis to investigate the microbial functions and their interactions with the host [98].
Once data is collected, specialized software packages are utilized for various tasks such as peak deconvolution, peak picking, data integration, structure annotation, statistical analysis, and pathway analysis [112-117]. To date, open-source software for small molecule discovery in untargeted metabolomics primarily includes MZmine, mzMatch, MS-DIAL, Ideom, and XCMS. For example, Fiehn’s group developed MS-DIAL that is a software pipeline for data-independent acquisition (DIA)-based identification and quantification of small molecules in comprehensive untargeted acquisition of metabolic data [113]. Zhu’s group established the Metabolite identification and Dysregulated Network Analysis (MetDNA) approach for identification of metabolites in LC-MS-based untargeted metabolomics [117]. Commercial software can also be used, such as SIEVE, MassHunter, MassProfiler Professional, Progenesis CoMet, Compound Discoverer, LipidSearch, MarkerLynx, and Progenesis Q. The databases for compound identification and annotation mainly include Human Metabolome Database (HMDB), METLIN, Chemspider, BioCyc, Cayman Chemical Compounds Database, KEGG, LipidMAPS, and mzCloud. Siuzdak’s group established the METLIN Metabolite and Chemical Entity Database that is the largest repository of MS/MS and neutral loss data acquired from standards to assist in metabolite and chemical entity identification [118, 119]. Xia’s and Wishart’s groups developed MetaboAnalyst and HMDB, which are incredibly useful for statistics analysis and metabolic pathways analysis in metabolomics. MetaboAnalyst is a free web-based platform for comprehensive metabolomics data analysis, supporting raw MS spectra processing, data normalization, statistical analysis, functional analysis, meta-analysis, and integrative analysis with other omics data [120, 121]. HMDB is a freely available web database containing comprehensive and detailed information of small molecules in the human body [122, 123]. Notably, >8,610 proteins, including enzymes and transporters, are also linked to these metabolites and lipids. Many entries are hyperlinked to other databases, such as KEGG and GenBank. There is extensive literature available that evaluates the performance of different software packages in various metabolomics analyses. The listed software packages are widely used and have been well organized in many previous reviews and metabolomics studies [124-127].
Table 1 summarizes the key parameters of the selected studies related to the impact of xenobiotic exposures on the gut microbiome. Methanol (MeOH) and acetonitrile (ACN) are the most commonly used solvents for protein precipitation and efficient extraction of metabolites in biological samples [86, 87, 128-130]. In lipid extraction, chloroform and MTBE are two commonly used solvents [131-133]. For the separation of hydrophilic compounds, the recommended separation mode is hydrophilic interaction liquid chromatography (HILIC), with popular options like ACQUITY UPLC BEH Amide and XBridge BEH Amide columns [87, 134]. Reverse phase columns are widely employed for hydrophobic compounds, mainly including XSelect HSS T3, ZORBAX Eclipse Plus C8/C18, ACQUITY UPLC BEH C18, ACQUITY UPLC HSS T3, Hypersil Gold C18, etc. [86, 128, 131, 134, 135] Untargeted metabolomics often utilizes both negative and positive ionization modes to gather comprehensive metabolic profiling [136]. Chromatographic conditions may differ between these modes, such as the use of distinct columns or mobile phase conditions. [137]. For GC separation, helium is often used as the carrier gas, and the metabolite extracts of the samples are usually separated on a capillary column, such as HP-5 and DB-5, after derivatization [129, 130]. Detailed sample preparation protocols for extracting microbial metabolites are well documented in previous reviews, including sample collection, quenching, and extraction for different types of samples (tissue, medium, cells, etc.) [93, 98, 99].
Table 1.
Summary of Recent Publications Related to Metabolomics, Gut Microbiome and Xenobiotic Exposure
| Xenobiotics, dose |
(Animal strain) Sample type for metabolomics |
Sample preparation (extraction solvent; ISs; derivatization reagent for GC) |
Column for analytical platform |
Nontargeted metabolomics | Targeted metabolomics | Transcriptomics | Genomics | References | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Analytical platform |
Listed differential metabolites |
Analytical platform |
Target analytes |
|||||||
| BDE-3, 0.0015-30 (mg/kg) | (C57BL/6J) Mouse testis, urine, serum | 80% MeOH (testis), MeOH (urine, serum); 2-Chloro-L-phenylalanine | XSelect HSS T3 | UPLC-Q-TOF/MS | 76, 38, and 31 metabolites in testis, urine, and serum | - | - | - | - | Wei et al. (2018) [128] |
| BDE-47, 10.0 (mg/kg) | (SD) Rat serum | MeOH; N/A; BSTFA | HP-5MS | GC-MS | 26 metabolites | - | - | - | Fecal 16S rRNA Seq | Gao et al. (2021) [129] |
| 18 PBDE congeners, N/A | Human placenta | 80% MeOH; tridecanoate and 3 deuterated ISs; MSTFA | DB-5MS | GC-MS | 66 metabolites | - | - | - | - | Wang et al. (2022) [130] |
| BDE-47, 48.5; BDE-99, 56.5 (mg/kg) | (C57BL/6J) Mouse serum, liver, small and large intestine content | MeOH (serum), ACN-MeOH (liver, small and large intestine content); 5 deuterated ISs | XSelect HSS T3 | - | - | UPLC-QQQ-MS | 56 BAs | Liver RNA-Seq | Fecal 16S rRNA Seq | Li et al. (2018) [86] |
| BDE-47, 48.5; BDE-99, 56.5 (mg/kg) | (C57BL/6J) Mouse serum, liver, small and large intestine content | MeOH (serum), 80% MeOH (liver, small intestine content, large intestine content); 2 13C-labeled ISs | Xbridge BEH Amide | - | - | UPLC-QQQ-MS | >300 aqueous metabolites | Liver RNA-Seq | Fecal 16S rRNA Seq | Scoville et al. (2019) [87] |
| BDE-47, 0.2; TBBPA, 0.2; BPS, 0.2 (mg/kg) | (CD-1) Mouse liver, fecal; fecal (SCFAs) | BAs: ACN-MeOH; 5 deuterated ISs SCFAs: MeOH-H2O; hexanoic acid-6,6,6-d3; MTBSTFA | ZORBAX Eclipse Plus C18; HP-5MS | - | - | UPLC-QQQ-MS; GC-MS | 56 BAs, 14 SCFAs | BA-processing related genes and cytokines in liver | Fecal 16S rRNA Seq | Gomez et al. (2021) [135] |
| PCB 126, 1.0 (μmol/kg) | (Ldlr −/−) Mouse liver; fecal (SCFAs) | SCFAs: MTBE; N/A; MTBSTFA | ACQUITY UPLC BEH C18, UPLC BEH Amide | - | - | UPLC-Q-Exactive; GC-MS | 25 Hepatic Metabolism metabolites; 5 SCFAs | Inflammatory, metabolic, and gut health markers in jejunum and colon | Fecal 16S rRNA Seq | Petriello et al. (2018) [134] |
| Fox River PCB mixture, 6-30 (mg/kg) | (C57BL/6J) Mouse liver, serum | MeOH (serum), 80% MeOH (liver); 2 13C-labeled ISs | XBridge BEH Amide | - | - | UPLC-QQQ-MS | >300 aqueous metabolites | Liver RNA-Seq | Fecal 16S rRNA Seq | Lim et al. (2020) [182] |
| PCB 126, 1.0 (μg/L) | (D.rerio) Zebrafish intestine, serum | 50% ACN (Metabolomics) MeOH and MTBE (lipidomics) ; N/A | ACQUITY UPLC BEH Amide, ACQUITY UPLC HSS T3 | UPLC-Q-Exactive (metabolomics, lipidomics) | - | - | - | BA metabolism related genes in liver | Fecal Metagenomic Seq | Hu et al. (2021) [137] |
| Aroclor 1260 PCB mixture, 20 (ng/kg) | Pig serum | MeOH-H2O-chloroform; 4 deuterated ISs for metabolomics, LPC (15:0), PC (15:0/15:0), and TG (17:0/17:0/17:0 for lipidomics | Hypersil Gold C18, ACQUITY CSH C18 | UPLC-Q-Exactive (metabolomics, lipidomics) | 33 polar metabolites, 39 lipids | - | - | - | - | Hernandez-Mesa et al. (2022) [131] |
| PCB 126, 24 (μg/kg) | (C57BL/6J) Mouse urine, cecal content, liver, fecal | Metabolomics: 80% MeOH BAs: MeOH; deuterated ISs | XSelect HSS T3 (metabolomics); ACQUITY C8 BEH (BAs quantification) | 1H NMR, UPLC-Q-Exactive (metabolomics, lipidomics) | - | UPLC-QQQ-MS | BAs | BA and FA metabolism related genes in liver and intestine | Cecal content 16S rRNA Seq, Cecal content metagenomic Seq | Tian et al. (2022) [230] |
| PCB 126, PCB 153, TCDF, 0.6-6 (μM); TCDD, 0.06-6 (μM) | (C57BL/6J) Mouse cecal microbiota (In vitro) | MeOH-H2O-chloroform; chlorpropamide and 15:0-18:1-d7-PC | Hydro-RP C18, CSH C18 column | 1H NMR, UPLC-Q-Exactive (metabolomics, lipidomics) | - | - | - | Cecal bacterial mixtures RNA-Seq | Cecal bacterial mixtures 16S rRNA Seq | Tian et al. (2020) [231] |
| PFOS, 0.3-30 (μg/g) | (C57BL/6J) Mouse fecal | Diethyl ether; N/A | Rtx-Wax | - | - | GC-MS | 5 SCFAs | Inflammation related genes in colon | Fecal 16S rRNA Seq | Wang et al. (2020) [240] |
| PFOS, 0.003-0.012% for mouse 5-50 (μg/g) for bacterial | (C57BL/6J) Mouse liver, cecal bacterial suspension | FAs: MeOH-chloroform; C15:0 free FA, methyl ester of C17:0 | HP-5MS | 1H NMR | - | GC-MS | 5 FAs | Inflammation and other metabolism related genes in liver | Cecal content 16S rRNA Seq | Zhang et al. (2020) [132] |
| PFOA, 2.5-30 (mg/kg) | (C57BL/6J) Mouse fecal | Diethyl ether; N/A | N/A | - | - | GC-MS | 5 SCFAs | - | Cecal content 16S rRNA Seq | Wang et al. (2021) [241] |
| PFOA, 0.5-3 (mg/kg) | (C57BL/6J) Mouse fecal | Ether; N/A | N/A | - | - | GC-MS | 6 SCFAs | Inflammation related genes in colon | Fecal 16S rRNA Seq | Shi et al. (2020) [242] |
| PFOA, 300 (mg/kg) | (C57BL/6J) Mouse fecal | Ether; N/A | Rtx-Wax | - | - | GC-MS | SCFAs | - | Fecal 16S rRNA Seq | Shi et al. (2021) [243] |
| PFAS, 32.7±1.1 (μg/L) | (Emydura macquarii macquarii) Turtle fecal | N/A; 2 13C labeled ISs | ZORBAX Extend C18 for targeted analysis | UPLC-Q-TOF/MS | - | UPLC-QQQ-MS | Central carbon metabolism metabolites | - | Fecal 16S rRNA Seq | Beale et al. (2022) [244] |
| As, 0.5-10 (ppm) | (SD) Rat serum | MeOH; N/A | Kinetex C18 | UPLC-Orbitrap | 18 metabolites in lipid, amino acid, and nucleotide metabolism | - | - | Lipids and AA metabolism related genes in liver | - | Wang et al. (2015) [259] |
| As, N/A | Human urine | Water (no extraction); N/A | ACQUITY UPLC HSS T3 | UPLC-Q-Exactive | 38 metabolites | - | - | - | - | Khanam et al. (2022) [260] |
| As, 10 (ppm) | (C57BL/6J) Mouse fecal, urine, plasma | MeOH (urine, plasma), 50% MeOH (fecal) ; N/A | Waters C18 T3 | UPLC-Q-TOF/MS | 370 features in fecal, 40 and 10 metabolites in urine and plasma | - | - | - | Fecal 16S rRNA Seq | Lu et al. (2014) [261] |
| As, 10 (ppm) | (C57BL/6J) Mouse serum | MeOH; N/A | Poroshell 120 EC-C18 | UPLC-Q-TOF/MS | 33 metabolites | - | - | - | - | Xue et al. (2019) [262] |
| As, 30 (mg/kg) | (C57BL/6J) Mouse brain | Metabolomics: MeOH-ACN-H2O Lipidomics: MeOH and MTBE; 3 deuterated ISs | ACQUITY UPLC HSS T3, Kinetex C18 | UPLC-Q-TOF-MS/MS (metabolomics), UPLC-Q-Exactive (lipidomics) | 118 polar metabolites, 17 lipids | - | - | - | Fecal 16S rRNA Seq | Wang et al. (2021) [263] |
| As, 30 (mg/kg) | (C57BL/6J) Mouse fecal, plasma, cortex | MeOH-ACN-H2O (fecal, brain); MTBE (plasma) ; N/A | N/A | UPLC-Q-Exactive | 31, 1, and 17 metabolites in fecal, cortex, and plasma | - | - | - | Fecal 16S rRNA Seq | Luo et al. (2023) [133] |
| Cd, 1-5 (μmol/L) | Human urine, Pancreatic beta cells (In vitro) | Urine: No extraction, L-2-chlorophenylalanine Cells: MeOH-ACN | HILIC column | UPLC-TOF/MS | 76 metabolites in cells, 53 metabolites in human urine | - | - | Mitochondrial TCA cycle and fatty acid oxidation related genes in pancreatic beta cells and mouse pancreas | - | Hong et al. (2022) [266] |
| Cd, 1-50 (ppm) | (C57BL/6J) Mouse brain | MeOH-H2O; 13C3-lactate | ACQUITY UPLC BEH C18 | UPLC-Q-Exactive | 101 altered metabolites | UPLC-Orbitrap ID-X | Lactate | Brain RNA-Seq | - | Hudson et al. (2021) [267] |
| Cd 0.6-3 (mg/L) | (Humanized ApoE3-KI and ApoE4-K1) Mouse plasma | MeOH-H20; hexanoic acid-6,6,6-d3; MTBSTFA | HP-5MS | - | - | GC-MS | SCFAs | Liver RNA-Seq and inflammation related genes in liver | Large intestinal content 16S rRNA Seq | Zhang et al. (2021) [268] |
| Cd, As, 50 (ppm) | (C57BL/6J) Mouse fecal | ACN-H2O; N/A | self-packed 15 cm C18 column | UPLC-Q-Exactive | 33 and 2 features in Cd and As treated group | - | - | - | Fecal 16S rRNA Seq | Li et al. (2019) [269] |
| Cd, 0.24-0.46; Se, 0.6-6 (mg/L) | Honey bee abdomen | MeOH-ACN-H2O-isopropanol; N/A | CSH phenyl-hexyl column | UPLC-Q-TOF/MS | 2 Cd-induced and 7 Se-induced metabolites | - | - | - | Whole-bee 16S rRNA Seq | Rothman et al. (2019) [270] |
| Mn, 20 (mg/kg) | (C57BL/6J) Mouse fecal | MeOH-H2O-chloroform; N/A; BSTFA | N/A | GC-MS | 36 and 32 metabolites in female and male mice | - | - | - | Fecal 16S rRNA Seq, Fecal metagenomic Seq | Chi et al. (2017) [275] |
| Mn, 200 (mg/L) | (SD) Rat plasma | MeOH-ACN; N/A | ACQUITY UPLC HSS T3 | UPLC-Q-TOF/MS | 17 metabolites | - | - | - | Fecal 16S rRNA Seq, Fecal metagenomics Seq | Wang et al. (2020) [276] |
| Pb, N/A | Human fecal | MeOH-H2O-chloroform; 11 ISs; BSTFA | HP-5MS | GC-MS | 19 metabolites | - | - | - | Fecal 16S rRNA Seq | Zeng et al. (2022) [277] |
| PM2.5, 198.52 ± 21.56 (μg/m3) | (Balb/C) Mouse ileum | MeOH-H2O-chloroform (nontargeted metabolomics), MeOH (targeted analysis); 2-chlorobenzalanine | ACQUITY UPLC HSS T3 C18 | UPLC-Orbitrap | 31 metabolites | UPLC-QQQ-MS | 17 metabolites in arachidonic acid metabolism | Inflammation related genes in ileum | Fecal 16S rRNA Seq | Dai et al. (2022) [292] |
| TRAP, N/A | Human plasma, saliva | ACN; N/A | HILIC column (pos ionization mode), C18 column (negionization mode) | UPLC-Q-Exactive | 1291 associated features | - | - | - | - | Liang et al. (2018) [293] |
| TRAP, N/A | Human plasma | ACN; 14 isotopic labeled ISs | C18 column | UPLC-Q-Exactive | 5112 associated features | - | - | - | - | Li et al. (2021) [294] |
N/A: no information provided.
In gut microbiome metabolic studies, there are several approaches that could be employed to increase the stability of sample preparation, reduce data variation, and increase data quality. To prevent/correct analytical drift and provide data consistency including intensity monitoring, either a quality control (QC) sample is applied, and/or stable isotope labeled internal standards (SIL-ISs) are used. QC samples are useful for assessing the overall quality of the data and detecting any systematic errors that may have occurred during sample preparation or measurement. In addition, SIL-ISs can provide quantification of the analytes of interest and improve the precision and reproducibility of the data. For example, He et al. spiked a SIL-IS mixture that includes 14 deuterium labeled lipids to serum samples during methyl tert-butyl ether (MTBE) extraction, and ~1,500 lipids were detected with high reproducibility within 60 min gradient length by Orbitrap-based MS2 [138].
3.2. Targeted metabolomics, pseudotargeted metabolomics, and beyond
In targeted metabolomics, the focus is on a specific set of metabolites that are relevant to the hypothesis being tested, while untargeted metabolomics is used where all metabolites are measured without prior knowledge of which ones may be of interest [139-142]. Triple quadrupole mass spectrometry (TQMS) based on selective ion monitoring mode (SIM) or multiple reaction monitoring (MRM) analysis [143-145], and HRMS-based parallel reaction monitoring (PRM) analysis [146, 147] are commonly used to perform targeted metabolomics. To date, various essential metabolites involved in microbial metabolism have been identified in previous studies, such as SCFAs, BAs, tryptophan metabolites, amino acids, and those in central carbon metabolism [148-150]. For these metabolites of strong interest, LC-MS was used for quantification/semi-quantification of BAs [151-153], SCFAs [154], tryptophan catabolites [155, 156], and methylamines [157, 158]. Nicholson’s group described an LC-MS procedure for sensitive and quantitative targeted analysis of 145 primary, secondary, and tertiary bile acids [153]. The assay had a great linearity with a lower limit of quantification (LLOQ) of 0.25-10 nM and an upper limit of quantification (ULOQ) of 2.5-5 μM. The group also confirmed precision (≈6.5%), accuracy (81.2-118.9% on inter- and intraday analysis), and recovery (serum/plasma 88% and urine 93%). Notably, SCFAs were also often measured by GC-MS after derivatization [159-163].
Large-scale targeted metabolomics was developed for the detection of over 300 aqueous metabolites and >280 lipids from >35 different metabolic pathways of strong biological significance [141, 164-167]. Munjoma et al. have developed a high-throughput HILIC-based LC-MS method for the semi-quantitative screening of more than 2,000 lipids using over 4,000 MRM transitions [168]. In another study, Zhou et al. developed a PRM assay to monitor 237 polar metabolites [169]. In addition, Zhang et al. applied PRM to analyze 20 amino acids and 40 derivatives in targeted analysis [170]. Our own assay of this type has been successfully used in a growing number of studies, including metabolic reprogramming during Myc-regulated tumorigenesis [171], cardiac metabolic shifts induced by rapamycin [172], metabolic characterization of mammalian skeletal muscles [173], and metabolome regulation during naive-to-primed human embryonic stem cell transition [174]. Chromatographic separations are observed by using HILIC and reverse phase modes for aqueous metabolites and lipids, respectively. LC-MS/MS system is optimized using metabolite and lipid standards, including precursor ion, product ion, collision voltage, and retention times (RTs). Targeted MS data acquisition is performed in the MRM mode, due to significant advantages of great selectivity and excellent quantitation. Samples are often run with a set of internal or external standards such that data variation will be minimized.
Xu’s group established pseudotargeted metabolomics, in order to combine the advantages of untargeted and targeted metabolomics [175-179]. Pseudotargeted methods acquire MS/MS fragmentation information from pooled biological samples in the full scan untargeted mode and collect as many ion pairs as possible under different collision energy (CE) voltages by multiple parallel injections. After removing the redundant features, the remaining interested ion pairs are utilized for CE optimization and MRM transitions in TQMS for measuring the samples. Zheng et al. provided a detailed protocol that illustrates the entire timeline of pseudotargeted metabolomics development. The process involves various steps, including sample preparation, data acquisition, MRM transition definition, parameter optimization, and method evaluation, which typically takes ~5 days to complete, allowing for the analysis of 800-1,300 metabolites [175]. Notably, the actual duration may vary depending on the number of samples analyzed in practical scenarios. This method was first introduced in 2012 using GC-MS, then in 2013 with LC-QTOF and LC-QQQ-MS platforms, and in 2015 with an upgraded procedure. The review by Liu et al. detailed pseudotargeted metabolomics analytical platforms [97].
Similarly, we developed a new analytical approach, globally optimized targeted (GOT)-MS, which has the capability to detect unknowns, broad metabolite coverage, and excellent quantitation [180]. From a serum sample, we obtained 595 precursor ions and 1,890 MRM transitions. For many MRMs/metabolites, the analytical performance of GOT-MS is better than or at least comparable to untargeted metabolomics. To significantly improve identification in GOT-MS, Shi et al. recently established a database-assisted globally optimized targeted mass spectrometry (dGOT-MS) approach that can provide a strong coverage of identified aqueous metabolites and lipids (310 confirmed with pure chemicals) [181]. Lim et al. applied dGOT-MS to investigate the metabolite changes due to polychlorinated biphenyls (PCB, a persistent organic pollutant) exposure in mouse liver and serum samples. The results showed that NADP and arginine varied with drug-metabolizing enzymes, which were highly correlated with Ruminiclostridium and Roseburia alterations [182].
3.3. Metabolic flux analysis (MFA)
MFA using stable isotope labeled tracers is extremely important for understanding metabolic mechanisms of various biological processes [183-188]. The whole metabolome can be viewed as an extensive map, in which metabolites (cities) and metabolic pathways (roads) comprise the metabolic reaction network. Metabolite levels provide “snapshots” of biochemical abundances; however, cellular metabolism in a living system is not static. In addition, many metabolites are involved in multiple pathways; therefore, concentration changes of a particular metabolite can result from perturbations in several pathways, which often confuses the analysis of metabolite level data. In contrast, MFA investigates the rate of metabolite turnover in biochemical reactions, which enables a dynamic model of cellular phenotype through tracing metabolite conversions taking place in the metabolic network. Furthermore, MFA can be easily connected to other important areas in systems biology, such as proteomics and genomics since the flux rate is regulated by enzymes. Therefore, MFA providing pathway-specific information is often required to fully understand the biological process [189-191]. In isotope tracing experiments, a biological system will receive isotopically labeled substrates, e.g., 13C, 15N, and 2H. GC/LC-MS data are collected and processed to detect metabolic features and align retention times. Enrichment of isotope labeling is processed to compare labeling patterns between different conditions. MetaboAnalyst, IsoCor, X13CMS, MZmine, and XCMS are software tools for processing and analyzing isotope labeling data. They provide normalization, statistical analysis, pathway analysis, and data visualization. Isotope labeling experiments and advanced software tools enable the investigation of metabolic pathways and fluxes in biological systems, providing a dynamic and deeper understanding of cellular metabolism and its response to perturbations.
Fan’s group developed stable isotope-resolved metabolomics (SIRM) that can analyze the fate of individual atoms from stable isotope-enriched precursors to deduce metabolic pathways and networks [184, 191]. Deng et al. developed an untargeted SIRM method for metabolic profiling of isotopic microbial metabolites after treating the fecal microbes with 13C-labeled dietary fibers (inulin or cellulose) [192]. Their results showed that 13C enrichment was successfully examined in both microbial cells and the culture medium. In addition, Patti’s group developed X13CMS for the identification of compounds that have incorporated the isotopic tracer, in an untargeted way [183].
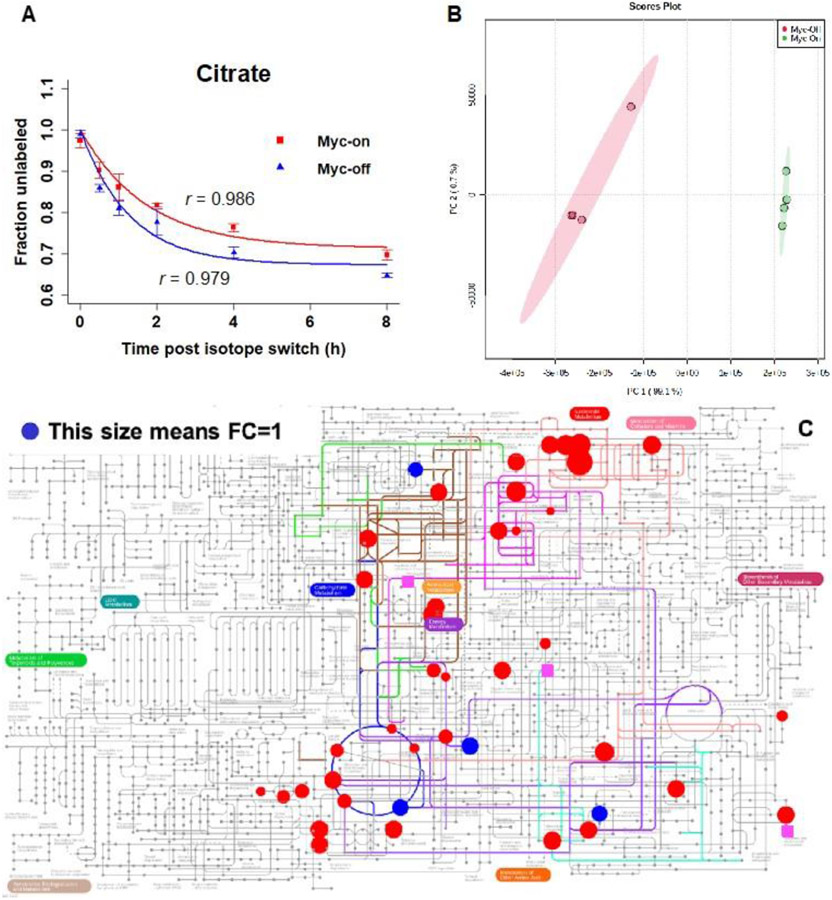
Recently, Shi et al. established a TQMS-based comprehensive isotopic targeted mass spectrometry method in MFA [171]. This method was used to track 13C-enriched metabolites in Myc(oncogene)-On and Myc-Off Tet21N human neuroblastoma cells after being cultured in U-13C6-glucose medium. A LC-MS/MS assay was developed to analyze 310 metabolites across 35 metabolic pathways. For each metabolite isotopologue, an isotopic MRM was generated based on the number of carbon atoms. The precursor ion and product ion m/z values were determined using the chemical formula. Isotopic MRMs were utilized in the LC-MS/MS measurements, and IsoCor was used to compute the mass isotopomer distribution and mean enrichment for each identified metabolite using the isotopic intensities. As a result, out of the 310 metabolites analyzed, 142 were detected with a high level of reliability, and 46 of these were found to be kinetically enriched. In addition, to investigate the time-course study under the pseudosteady state, the rate constant of decay and relative flux were calculated using the relative quantitation data. Citrate was examined as an example, and the results demonstrated an exponential decay pattern (as shown in Fig. 4A). The flux data allowed for clear differentiation between Myc-On and Myc-Off cells in the principal component analysis (PCA) score plot (as depicted in Fig. 4B). Further, Fig. 4C shows the metabolites with significant relative flux differences between Myc-On and Myc-Off cells in Ipath Pathways [171].
Fig.4.
A) Exponential fitting of the time-course data of citrate, B) the PCA score plot, and C) pathway view of metabolites with relative flux derived, for the comparison of Myc-On and Myc-Off Tet21N cells under pseudosteady state following U-13C6-glucose labeling: red circles have p values < 0.05, blue circles have p values > 0.05, pink squares indicate metabolites present only in the Myc-Off group. This figure is reproduced from [171] with permission.
3.4. Novel MS techniques with highly promising applications in gut microbiome and xenobiotics studies
Metabolome within a living cell is highly dynamic and undergoes rapid shifts in response to the intracellular environment. Therefore, measuring metabolites within individual cells as they advance through metabolic pathways may provide a better understanding of cell metabolic activities [193, 194]. As such, MS-based single-cell metabolomics serves as a valuable tool to explore the metabolic capabilities and functions of individual microbial cells within this intricate ecosystem [195]. In single-cell metabolomics, various techniques, such as secondary ion mass spectrometry (SIMS), matrix-assisted laser desorption (MALDI), and laser ablation electrospray ionization (LAESI), can be employed for sampling and ionizing metabolites from individual cells [196]. In addition, the applications of nano-electrospray ionization (ESI) in single-cell metabolomics have been reviewed previously [197, 198]. Notably, challenges also exist in single-cell metabolomics [195], such as the small size of bacterial cells (approximately 1 μm in size) and metabolite identification (e.g., generation of large molecular fragments in SIMS high-energy ion beams). Additional limitations include difficulties in accurately measuring and quantifying metabolites within individual cells due to low abundance, potential leakage, and enzymatic degradation [199].
Another method with high potential in gut microbiome and xenobiotics studies is mass spectrometry imaging (MSI). MSI is a technique that provides information about the distribution and localization of molecules (focusing on metabolites in this review) in different regions of tissues or organisms [200]. MSI provides complementary information to that from histological methods [201]. MSI uses different ionization techniques such as MALDI, SIMS, and desorption electrospray ionization (DESI) [202]. Previous studies have utilized MSI to enhance the understanding of metabolic functions in different parts of the intestine. Hulme et al. utilized MSI to investigate alterations in neurotransmitters and spatial patterns in both the gut and brain in mice [203]. Their study showed that many metabolic changes were induced by gut microbes in the microbiome-gut-brain axis following an antibiotic treatment. Watrous et al. utilized two-dimensional MALDI-TOF MSI to three-dimensionally visualize metabolic exchange during microbial interactions between different microorganisms [204]. Similarly, MSI has limitations, such as difficulties in quantitation, sophisticated instrumentation, and annotating microbial metabolites [205].
Interestingly, MasSpec Pen, another novel MS-based technique, has emerged as a handheld device with immense clinical applications [206]. MasSpec Pen was specifically designed to enable rapid, non-destructive, and in situ analysis of the molecular composition of tissue samples during surgical procedures. The device can directly touch the tissue surface and release a droplet of water to extract metabolites from the tissue, particularly for patients under surgery. These extracted metabolites are subsequently transferred to MS, enabling the detection and characterization of the molecular composition associated with disease pathology (i.e., cancer type, tumor margin, and other disease-related features) [207, 208]. MasSpec Pen shows promises as a real-time, intraoperative tissue diagnostic tool, providing surgeons with valuable information leading to faster, safer, and more effective surgeries for patients. Notably, while we introduced single-cell metabolomics, MSI, and MasSpec Pen herein and they do have shown high potential for biological and clinical studies, to date, the applications of these technologies in gut microbiome and xenobiotic studies are very rare.
4. Persistent Organic Pollutant (POPs) Exposure, Gut Microbiome, and Metabolomics
POPs are highly toxic organic chemicals causing worldwide concerns due to their persistence in the environment and resistance to biodegradation [209-211]. POPs can easily remain in the environment for prolonged periods of time and accumulate in the food chain [209]. Exposure to POPs increases risks of cancer, autoimmune diseases, diabetes, obesity, and reproductive disorders [212-214]. There is an increasing number of studies published on metabolomics-based POPs exposure both in vivo and in vitro. Table 1 presents selected studies with essential parameters, such as animal model utilized, exposure levels of xenobiotics, sample preparation conditions, and analytical instruments along with the columns used. In this review, we will focus on a few POPs with predominant existence in the environment, including polybrominated diphenyl ethers (PBDEs), tetrabromobisphenol A (TBBPA), and polychlorinated biphenyls (PCBs).
4.1. Polybrominated diphenyl ethers (PBDEs) and tetrabromobisphenol A (TBBPA)
PBDEs are an important class of POPs, widely used in a variety of consumer products such as flame retardants, electronics, and textiles [215, 216]. Most PBDEs have been banned due to their neurotoxicity, but they still have managed to become widespread worldwide and accumulate as severe environmental pollutants. PBDEs can be found in seafood, meat, dietary products, and even indoor dust [217]. Nowadays, these compounds are found in human serum and breast milk samples in many areas and countries around the world [218]. Previous studies showed that acute exposure to BDE-47 and BDE-99 (the topmost enriched PBDE congeners) decreased the anti-diabetic tryptophan microbial metabolite, 3-Indolepropionic acid (IPA), in a gut microbiome-dependent manner [87]. Since PBDEs’ ban, TBBPA is currently the most widely used brominated flame retardant, and it constitutes approximately 60% of the worldwide demand [219, 220]. Like PBDEs, TBBPA can cross the placenta and can be passed from mother to fetus [221]. TBBPA has also been detected in 44-50% of breast milk samples from postpartum mothers, posing significant health risks to offspring during pregnancy and lactation [222]. TBBPA is glucuronidated and sulfated by the host and deconjugated by the gut microbiota before being excreted in feces as the parent compound [223].
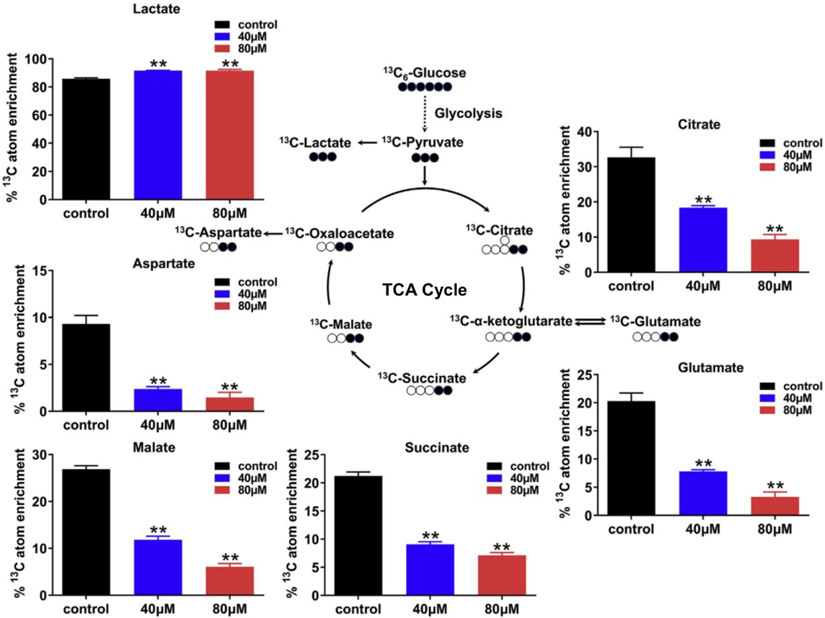
In an in vitro study, He et al. observed that BDE-47 (a PBDE congener) exposure induced significant attenuation of mitochondrial respiration and glycolysis elevation [224]. To conduct GC-MS-based MFA, isotopically labeled glucose was substituted for unlabeled glucose in the cell culture medium. The presence of elevated isotopically labeled lactate in the BDE-47 treatment group confirmed an increase in glycolysis. Conversely, isotopically labeled metabolites involved in the TCA cycle, such as citrate, α-ketoglutarate, succinate, malate, and oxaloacetate, exhibited a significant decrease after BDE-47 treatment, indicating a reduction in mitochondrial respiration activity (Fig. 5). In addition, they found that multiple metabolic pathways including glycine, serine, threonine, and glutathione metabolism were disturbed via large-scale metabolomics, and eventually 17 metabolites were selected as the potential biomarkers of BDE-47 exposure.
Fig. 5.
Incorporation of glucose-derived carbon atoms into the TCA cycle and glycolysis metabolites. Bar charts left to right: control (black), 40 μM BDE-47 (blue), and 80 μM BDE-47 (red) groups; the Y-axis depicts the mean of isotopic enrichment of 13C in the metabolites. ** indicate a highly significant difference (p < 0.01). This figure is reproduced form Ref. [224] with permission.
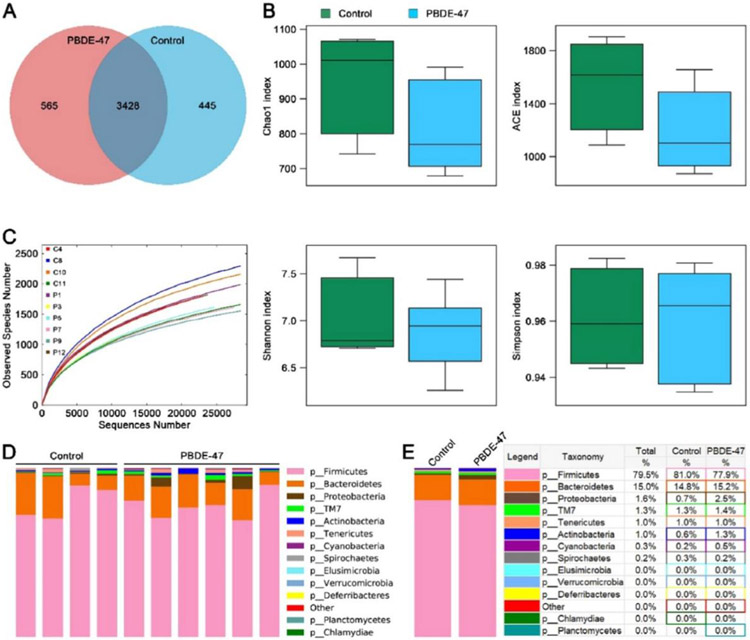
Untargeted metabolomics approaches were applied to investigate the impacts of PBDE exposure on the gut microbiome-host metabolite profiles. Wei et al. used UHPLC-Q-TOF-MS in untargeted metabolomics, and they found that male mice presented a significant reproductive toxicity after BDE-3 exposure [128]. As a results, 76, 38, and 31 metabolites involved in nucleotides, lipids, tyrosine, purine, and riboflavin metabolism were significantly changed in testis, urine, and serum samples, respectively. Gao et al. studied BDE-47 exposure-induced impacts and association between the rat serum metabolites and gut microbes before, during, and after pregnancy [129]. Through GC-MS untargeted metabolomics, a total of 26 metabolites in amino acid, lipid, carbohydrate, and energy metabolism were found to have significant responses to BDE-47. The exposure to BDE-47 resulted in maternal gut dysbiosis, as displayed in Fig. 6. The operational taxonomic unit (OTU) clustering of 10 fecal samples yielded 577,315 effective sequences, revealing 3,428 shared OTUs between healthy controls and BDE-47-treated rats. Control animals had 445 unique OTUs, while BDE-47-treated rats had 565 unique OTUs. The rarefaction curve suggested that sufficient sampling had been conducted, while the α-diversity indexes revealed a slight decline in the BDE-47 group, although not statistically significant compared to the control group. The Bacteroidetes, Proteobacteria, and Cyanobacteria exhibited a decrease in relative abundance, while Firmicutes increased, although not significantly. At the phylum level, the 16S rRNA results indicated that BDE-47 exposure reduced the relative abundance of Actinobacteria. Recently, Wang et al. investigated the impacts of environmental PBDE exposure on prenatal metabolites in two different areas in China [130]. They confirmed prenatal exposure to multiple types of PBDEs by GC-MS/MS in the MRM mode, and they observed 66 PBDEs-associated prenatal metabolites using GC-MS in the full scan mode. The disrupted metabolic pathways included pentose phosphate pathway, arginine biosynthesis, ascorbate, threonine, butanoate, and lipid metabolism.
Fig. 6.
Impacts of BDE-47 exposure on the richness and pattern of maternal intestinal microbiota. A) Venn diagram of shared and unique OUT numbers of BDE-47-treated rats compared to controls. B) Alpha diversity indexes of gut microbiota in the control (n = 4) and BDE-47-treated rats (n = 6). The top and bottom boundaries of each box indicate the 75th and 25th percentiles, respectively; the line within each box represents the median; the bottom and top edges show the minimum and maximum, respectively. C) The observed species number. D-E) Gut microbial pattern at the phylum level within D) and between E) the control (n = 4) and the BDE-47 groups (n = 6) were assessed using 16S high throughput sequencing. This figure is reproduced from [129] with permission.
Microbial metabolites, such as BAs, SCFAs, and tryptophan catabolism metabolites, were targeted in xenobiotic metabolomics studies. Li et al. orally administrated BDE-47 and BDE-99 to male conventional and germ-free mice [86]. Both congeners decreased bacterial diversity but increased the abundances of Akkermansia muciniphila and Erysipelotrichaceae Allobaculum spp., as well as microbial 7α-dehydroxylation enzymes for secondary BA synthesis. Targeted metabolomics showed that the exposure also caused increased unconjugated BAs in multiple biospecimen. In addition, targeted proteomics revealed that PBDEs increased the expression of host intestinal transporters for BA absorption, while they downregulated host BA-synthesizing enzymes and transporters in livers of conventional mice. Scoville et al. found that PBDEs altered a majority number of aqueous metabolites in the large intestine content of germ-free mice [87]. PBDEs also regulated the correlation between microbial taxa composition and serum metabolites in the conventional mice. Gomez et al. investigated the effects of maternal exposure to BDE-47 on the metabolite profiles and gut microbiome composition in adult male mice [135]. Consistent up-regulation of BAs in 12α hydroxylation pathway was observed in fecal and liver samples after BDE-47 exposure, corresponding to an up-regulation of the hepatic BA synthetic enzyme Cyp7a1. Consistent with previous results, they showed that PBDE exposure modified the gut-liver axis, through altering gut microbiota, as well as multiple hosts and microbial metabolic pathways.
Related to TBBPA, previous studies showed that cells exposed to TBBPA exhibited mitochondrial dysfunction [225, 226]. In A549 cells, TBBPA caused morphological and ultrastructural changes, such as dilated smooth endoplasmic reticulum and extensively injured mitochondria, as well as excessive reactive oxygen species (ROS) generation [225]. Recently, Yu et al. observed that TBBPA significantly reduced the viability of A549 cells and attenuated mitochondrial respiration [227]. LC-MS data showed significant reductions in TCA cycle metabolites, and MFA indicated reduced oxidative capacity in mitochondrial metabolism following TBBPA exposure. These findings indicate that the cytotoxicity of TBBPA was at least partially due to the perturbed TCA cycle metabolism and mitochondrial respiration. In vivo, maternal exposure to TBBPA persistently modified the gut-liver axis, which may produce an immune-suppressive and dyslipidemia-prone signature later in life [135]. The most distinct microbial biomarker was Rikenellaceae for TBBPA exposure. For metabolites, fecal BAs were persistently up regulated by TBBPA exposure, and TBBPA also increased propionic acid and succinate.
4.2. Polychlorinated biphenyls (PCBs)
PCBs are a large group of chemicals possessing unique chemical stability and fire-resistant properties [228]. Although they are no longer widely used due to their toxicity, the bioaccumulation of PCBs has continued to increase in the past decades much like PBDEs [229]. Toxicity of PCB 126 on mouse gut microbiome and metabolic homeostasis was investigated by Petriello et al., and 16S rRNA results showed an obvious alteration of cecal microbial diversity and bacterial genera [134]. Using untargeted metabolomics, hepatic metabolism was found to be significantly changed, including glycolysis and lipogenesis. A significantly decreased level of formic acid was discovered in fecal samples. Lim et al. investigated the correlation between the aqueous metabolites in serum and liver with the hepatic transcriptome in PCBs-exposed mice, as well as the metabolism differences between conventional and germ-free mice [182]. PCBs induced upregulation of aryl hydrocarbon receptor and pregnane X receptor. Constitutive androstane receptor expression was increased in conventional mice compared to germ-free mice, indicating that PCBs impact the hepatic transcriptome at least partially through the gut microbiome. Hu et al. discovered that PCBs induced the blockage of bacterial primary BAs conversion and intestinal accumulation of BAs, through decreased microbial activities related to primary BA metabolism in zebrafish intestines [137]. A systematic deficiency of essential vitamins was detected due to PCBs. Hernandez-Mesa et al. performed a PCBs exposure study on pigs, where untargeted metabolomics and lipidomics results showed that 33 polar metabolites and 39 lipids involved in fatty acid metabolism, glycerophospholipid metabolism, and tryptophan-kynurenine pathway were significantly changed in serum [131]. Tian et al. found similar results in mouse models, and they confirmed that PCB 126 exposure induced abnormal liver amino acid, BA, and nucleotide metabolism, as well as enhanced hepatic lipogenesis [230]. Interestingly, another Tian’s study showed significantly lower unsaturated fatty acids (UFA) and monounsaturated fatty acids (MUFA) but higher saturated fatty acids (SFA) due to PCB 153 exposure, while the opposite results were observed in PCB 126 exposure [231]. They also found that PCBs induced higher levels of lipids in the bacterial membrane, including fatty acids, phosphatidylcholine (PC), phosphatidylethanolamine (PE), etc.
4.3. Per- and polyfluoroalkyl substances (PFASs)
PFASs are a class of synthetic chemicals used to make fluoropolymer coatings and products produced since the 1940s and used in various consumer and industrial products [232]. Although PFASs are now widespread in our environment and have been linked to severe health impacts such as hypercholesterolemia, ulcerative colitis, and liver cell damage, there are no enforceable regulatory standards to safeguard public health and drinking waters from this serious hazard [233]. While some nanoparticle-based methods have been developed to remove PFASs, their extensive use and persistence can cause reproductive issues, immunodeficiencies, and hormonal disruptions in both humans and wildlife [234-236]. Exposure to PFASs can directly impact the gut microbiome, as evidenced by the ability of the human fecal microbiome to transform the PFAS surfactant 8:2 monosubstituted polyfluoroalkyl phosphate ester [237]. PFASs exposure can occur in infants through human milk, potentially increasing their gut microbiome diversity and increasing the relative abundance of Bacteroides vulgatus [238]. A study tracked 124 individuals from birth to age 28, finding weak links between PFASs exposure and fastidious anaerobes. Several individual microbes were associated with certain compounds, including Bilophila wadsworthia, Faecalibacterium prautzii, Dorea longicatena, and Sutterella wadsworthensis [239]. Perfluorooctane sulfonate (PFOS) is one of the most abundant PFASs in the environment, extensively researched, and has been found to affect inflammatory reactions in the liver and colon, promoting the development of metabolic disorders. This includes altering adipocytokine signaling pathway, steroid hormone biosynthesis, flavonoid biosynthesis, lipid metabolism, oxidative stress, inflammation, TCA cycle, glucose, and amino acid metabolism. Exposure to PFOS in mice has been shown to dysregulate the levels of genera such as Firmicutes, Bacteroides, Proteobacteria, Clostridium, Streptococcus, and Blautia, and induce a loss of gut barrier integrity by reducing production of SCFAs and expression of intestinal tight junction proteins [132, 240]. In addition, Wang et al. demonstrated that perfluorooctanoic acid (PFOA) exposure resulted in liver inflammation, antioxidative homeostasis disruption, SCFAs level reduction, and liver histological abnormalities with hepatomegaly and injury. Subacute exposure altered the abundances of Dehalobacterium and Bacteroides genera, which contribute to liver inflammation and oxidative stress, while subchronic exposure decreased the abundances of potentially beneficial Lactobacillus and Bifidobacterium genera [241]. According to a publication by Shi et al., exposure to PFOA resulted in cognitive deficits in mice through changes in the composition of gut microbiota, impairment of the intestinal barrier, and an increase in inflammation in both the gut and brain. However, the symptoms were alleviated by FMT treatment [242]. In addition, the use of lactic acid bacteria (Lactobacillus strains) reduced liver inflammation induced by PFOA by providing antioxidants and biosorption, alleviating dysbiosis, and preventing decreased production of SCFAs [243]. Research conducted on wild freshwater turtles, where PFOS levels were elevated in their serum, demonstrated that the exposure to this substance affected the metabolism of amino acids, butanoate, purine, and pyrimidine in these turtles. Furthermore, the metagenomic analysis of fecal samples from PFAS-exposed turtles revealed a greater prevalence of Firmicutes and a decreased prevalence of Bacteroidota, compared to the control group of turtles [244].
5. Heavy Metal Exposure, Gut Microbiome, and Metabolomics
Heavy metal exposure is considered a global public health concern, including the exposure of arsenic (As), cadmium (Cd), manganese (Mn), lead (Pb), copper (Cu), lithium (Li), tungsten (W), etc. [245, 246]. Exposure to heavy metals may cause a shift in the gut microbiome composition; and eventually severe damage from genome to metabolome such as DNA breakdown, irreversible impairment to the nervous system, immune system disruption, and tumorigenesis [247, 248]. Heavy metals are omnipresent in the environment, and the human population is at risk for daily exposure [249]. There is a growing evidence based on metabolomics approaches, supporting the important role of gut microbiome participating in adverse consequences induced by heavy metal exposure (Table 1) [250-252].
5.1. Arsenic (As)
Arsenic is well known as a poison, and this chemical is present in the environment in both organic (oAs) and inorganic (iAs) forms [253, 254]. As has been found in some water sources in the USA with high levels (>10mg/L), which leads to a frequent route of exposure through dietary uptake of seafood [255, 256]. A correlation has been found between arsenic exposure and tumors in the kidneys, lungs, liver, and bladder [257, 258].
HRMS-based metabolic profiling showed that chronic As exposure through drinking water caused disruptions in lipids and amino acid metabolism in rat serum, which had a positive correlation with up-regulation of the hepatic genes including cpt2, lcat, cact, crot, and mtr [259]. A recent untargeted metabolomics study performed by Khanam et al. investigated the toxic impacts of As exposure on Pakistani male urinary metabolome [260]. The results showed that As caused oxidative stress, disrupted one-carbon, purine, and caffeine metabolism. These tests showed 38 potential metabolite biomarkers of As exposure, including xanthines, purines, and testosterone. Lu et al. found that gut microbiome was a key player participating in the energy metabolism of As-treated mice, through investigating the association between gut microbial composition alterations and the typical gut microflora–related metabolites (indole-containing compounds, isoflavone metabolites, and BAs [261]. Xue et al. discovered a high correlation between serum metabolic profiles and gut microbiome perturbation in As-exposed Helicobacter-free mice [262]. Dramatic changes in numerous pathways were observed, including fatty acid, lipid, and tryptophan metabolism, and the serum metabolic disorder was attenuated in mice with healthy gut microbiota. Using untargeted metabolomics and lipidomics approaches, Wang et al. confirmed the important role of gut microbiome in As-inflicted neurodegeneration in the mouse brain [263]. Potential metabolite biomarkers of neurodegeneration, including 118 polar metabolites and 17 lipids involved in 30 metabolic pathways (fatty acid, lipid, amino acid, glycolysis, and nucleic acid metabolism), were significantly changed after the exposure, which was correlated with 12 kinds of gut microbes. In another recent publication from Luo et al., a significant microbial detoxification was discovered in situ in As-exposed mice after fecal microbiome transplantation (FMT) [133]. After the gut transplant, As accumulation was lower in fecal, liver, and plasma. Metabolites potentially beneficial to the host were higher in feces, plasma, and cerebral cortex in the FMT recipients than non-FMT mice. This suggests that FMT could be a promising treatment for As exposure.
5.2. Cadmium (Cd)
Cd is a metal element in the earth crust and exists in many industrial products such as batteries, alloys, electroplates, solar cells, plastic stabilizers, and pigments [264, 265]. Cd pollution also happens in industrial zones in developing countries [24]. The toxicity of Cd involves depletion of reduced glutathione, binding sulfhydryl groups with proteins, causing production of ROS, and potential genetic mutation and chromosomal deletions [248, 257, 265].
Recently, an untargeted metabolomics study found 76 altered metabolites mainly involved in organic acid, nucleoside, and lipid metabolism in vitro, and 14 metabolites co-existed in the urine of Cd-exposed workers were selected as potential biomarkers for Cd-exposure [266]. Using a mouse model, Hudson et al. employed a multi-omics approach to investigate the connections among maternal Cd exposure, impaired neurodevelopment in newborn brains, and behavior alterations at adulthood [267]. Maternal Cd exposure reduced mitochondrial DNA levels in newborn brains and disturbed hypoxic responses, cellular energy pathways, and retinoic acid signaling. MS measurement of 101 identified compounds showed that the metabolites in cellular energy pathways and hypoxia, such as retinoids, were significantly altered after the exposure. Zhang et al. investigated how Cd exposure impacts the gut microbiome and the host in an AD model (ApoE4-KI mice) [268]. They examined liver transcriptome (liver RNA-seq), gut microbiome (fecal 16S rRNA), and sera SCFAs concentration (GC-MS targeted quantification). The results showed that Cd exposure indeed increased microbial AD biomarkers while down-regulating energy supply-related pathways in the gut. Cd induced inflammation and disturbed xenobiotic biotransformation in the host liver. Cd exposure changed SCFAs concentrations in the mouse serum, which was correlated with gut microbiome composition. Li et al. combined metabolic profiles with 16S rRNA sequencing results, and they confirmed that Cd exposure caused significant changes in BA and amino acid metabolism and induced decreased microbial species, including Blautia, Eisenbergiella, Clostridium_XlVa, etc. [269]. Rothman et al. identified that metabolites involved in detoxification, proteolysis, and lipolysis were increased in Cd-exposed honeybees, and these alterations may cause oxidative damage to lipids, proteins, and detoxification genes [270]. In addition, they discovered seven kinds of bee-associated strains (such as L. micheneri, L. quenuiae, and L. kunkeei) that could be used to protect bees from Cd exposure.
5.3. Other heavy metals
Manganese (Mn) is a naturally occurring mineral metal that can exist in nuts and vegetables [271]. Mn is an essential micronutrient element for human health, but it is toxic at high physiological levels [272]. Overexposure of Mn occurs via contaminated water, welding fumes, or pesticides [273]. Mn toxicity includes neurodegeneration, neuroinflammation, spasticity, tremor, dystonia, and bradykinesia [274]. Mn-exposed mice were investigated by Chi et al., and they found that the gut microbiome composition and bacterial genes related to tryptophan, GABA metabolism, and LPS synthesis where DNA repair was significantly altered after the exposure [275]. GC-MS metabolic profiling showed that neurotransmitters and their precursors such as phenylalanine, glycine, and glutamic acid were significantly changed. Wang et al. found that bioaccumulated Mn increased β-amyloid (Aβ), receptor-interacting protein kinase 3 (RIP3), and caspase-3 production in the brain [276]. Mn significantly altered the metabolites involved in β-hydroxypyruvic acid, tryptamine, taurodeoxycholic acid, and urocanic acid metabolism. Mn exposure also reduced the gut microbiota richness, especially the composition of Prevotellaceae, Fusobacteriaceae, and Lactobacillaceae. Interestingly, Mn-induced neurotoxicity was alleviated with the recovery of gut microbiome composition by FMT from normal rats, which supplies a novel therapeutic strategy for the treatment of Mn toxicity by remodeling the gut microbiota.
Lead (Pb) is also classified as a heavy metal toxicant, which is found all throughout the environment in the water, soil, and air [271, 277]. This heavy metal is a known pollutant and can also be found in dietary and marine products, and most known to be found in old paint cans [278, 279]. Exposure to Pb can lead to multiple illnesses, such as gut microbiome dysbiosis, central nervous system disorders, inflammation, and liver toxicity [257, 277, 280]. Zeng et al. showed a high blood and urinary Pb level in the children living in electronic-waste areas [277]. Pb exposure reduced pediatric development parameters and altered 58 kinds of gut microbial genera. Through GC-MS metabolic profiling, 19 metabolites showed significant correlation with Pb exposure.
6. Inhalation Toxicant Exposure, Gut Microbiome, and Metabolomics
Inhalation xenobiotics are a public health emergency, including wildfire smoke (WFS), ambient air pollution (AAP) such as particulate matters with the aerodynamic diameter < 2.5 μm (PM2.5) or < 10 μm (PM10), traffic-related air pollution (TRAP), and third hand smoking (THS) [281, 282]. These inhalation exposures may be linked to a series of health problems, including neurodegeneration, hypertension, diabetes, asthma, and pulmonary diseases [283-285]. Cigarette smoking and THS exposure have a strong capability to shift the cecal microbial taxa diversity, and interestingly, they also significantly increased glycolysis and pyruvate decarboxylation, as well as significantly decreased coenzyme A biosynthesis and pyrimidine deoxyribonucleoside salvage [286-289]. WFS is an increasing global concern, although there are limited studies about WFS-exposed gut microbiome and metabolic profiles. To determine the effects of WFS on neuroinflammation and neuro-metabolomics in mice, Scieszka et al. performed a multi-omics study in a mobile laboratory, >300 km away from massive wildfires burning in California [290]. As a result, anti-aging metabolites, such as NAD+ and taurine, were decreased and the amyloid-beta protein (a key protein in neurodegeneration) was up-regulated by WFS exposure.
Multiple epidemiological studies have shown that exposure to AAP is associated with dysregulated composition of the gut microbiome. Bailey et al. found that AAP induced gut microbiota alterations in 6-month infants using 16S rRNA amplicon sequencing [291]. PM2.5 increased Actinomyces, which was negatively correlated with Alistipes. PM10 exposure was positively correlated with systemic inflammation related Dialister and Dorea. NO2 exposure was positively associated with Actinomyces, Enterococcus, Clostridium, and Eubacterium. In addition, a recent study showed that PM2.5 induced body weight loss, liver injury, and intestinal barrier disorder in adult mice [292]. The ratio of Bacteroidetes/Firmicutes of gut flora was associated with arachidonic acid metabolism and linoleic acid metabolism in the mice ileum. Freeway TRAP exposure has negative impacts on gut microbial taxa in overweight and obese adolescents. Liang et al. and Li et al. discovered that over thousands of metabolites and metabolic features were significantly changed in the serum samples of TRAP-exposed participants [293, 294]. The influenced metabolites were mainly involved in the oxidative stress-related pathways, nutrient metabolism (e.g., fatty acid metabolism), acute inflammation (e.g., histidine metabolism and tyrosine metabolism), leukotriene and vitamin E metabolism, and nucleic acids damage/repair (e.g., pyrimidine metabolism).
7. Conclusion
Toxic xenobiotic exposure is an increasing global health concern. Gut is a primary site for many xenobiotics in the host, and it was hypothesized that gut microbiota plays a vital role in xenobiotic toxicological effects. Metabolites can be signaling molecules responsible for the interaction between the gut microbiome and host; therefore, metabolomics is a promising approach for the investigation of metabolic effects due to xenobiotic exposure. This review summarized recent advancements in ultramodern MS-based metabolomics platforms and recent applications of metabolomics in xenobiotic exposure and gut microbiome studies. Based on the literature in the past few years, research into gut microbiomes, metabolomics, and their combination are effective to identify metabolic and microbial markers of xenobiotic exposure and determine its mechanisms. Importantly, there is increasing evidence showing that reprogramming the gut microbiome through probiotics, prebiotics, and/or FMT could be a promising approach to treat or reduce xenobiotic toxicity.
Highlights.
Applications of metabolomics combined with other omics platforms in xenobiotic metabolism and gut microbiome are reviewed.
The central hypothesis is that the interaction between gut microbiome and the host is mainly through metabolites.
Recent advancements in mass spectrometry-based metabolomics and metabolic flux analysis are summarized.
The toxicity of a wide variety of xenobiotics and potential therapeutic approaches through reprograming the gut microbiome are discussed.
Acknowledgments
This work was supported by the NIH (1R01ES030197, 1P01HL146369-01A1). In addition, we thank Freeman Lewis for reviewing and improving this paper.
Abbreviations:
- MS
mass spectrometry
- GLA
gut-liver axis
- GBA
gut-brain axis
- GPA
gut-pancreas axis
- GKA
gut-kidney axis
- BAs
bile acids
- SCFAs
short chain fatty acids
- Trp
tryptophan
- TMAO
trimethylamine N-oxide
- BCAA
branched-chain amino acids
- BAs
bile acids
- GPR
G-protein-coupled receptor
- BBB
blood-brain barrier
- GABA
γ-aminobutyric acid
- IPA
Indole-3-propionic acid
- NAD+
nicotinamide adenine dinucleotide
- LC-MS
liquid chromatography-mass spectrometry
- GC-MS
gas chromatography-mass spectrometry
- HRMS
high-resolution MS detectors
- Tof
time-of-flight
- QC
quality control
- SIL-ISs
stable isotope labeled internal standards
- MeOH
methanol
- ACN
acetonitrile
- ESI
electrospray ionization
- MTBE
methyl tert-butyl ether
- TQMS
quadrupole mass spectrometry
- SIM
selective ion monitoring mode
- MRM
multiple reaction monitoring
- PRM
parallel reaction monitoring
- LLOQ
lower limit of quantification
- ULOQ
upper limit of quantification
- HILIC
hydrophilic interaction chromatography
- RT
retention time
- CE
collision energy
- GOT-MS
globally optimized targeted mass spectrometry
- dGOT-MS
database-assisted globally optimized targeted mass spectrometry
- PCB
polychlorinated biphenyls
- MFA
Metabolic flux analysis
- SIRM
stable isotope-resolved metabolomics
- PCA
principal component analysis
- POP
Persistent Organic Pollutant
- PBDEs
polybrominated diphenyl ethers
- TBBPA
tetrabromobisphenol A
- OUT
operational taxonomic unit
- PCBs
polychlorinated biphenyls
- UFA
unsaturated fatty acids
- MUFA
monounsaturated fatty acids
- SFA
saturated fatty acids
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PFASs
Per- and polyfluoroalkyl substances
- PFOS
perfluorooctane sulfonate
- PFOA
perfluorooctanoic acid
- As
arsenic
- Cd
cadmium
- Mn
manganese
- Pb
lead
- FMT
fecal microbiome transplantation
- WFS
wildfire smoke
- AAP
ambient air pollution
- TRAP
traffic-related air pollution
- THS
third hand smoking
Footnotes
Conflict of Interest Disclosure
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Sekirov I, Russell SL, Antunes LC, Finlay BB, Gut microbiota in health and disease, Physiol. Rep 90 (2010) 859–904 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- [2].Santoro A, Ostan R, Candela M, Biagi E, Brigidi P, Capri M, Franceschi C, Gut microbiota changes in the extreme decades of human life: a focus on centenarians, Cell. Mol. Life Sci 75 (2018) 129–148 10.1007/s00018-017-2674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Heintz-Buschart A, Wilmes P, Human gut microbiome: function matters, Trends Microbiol. 26 (2018) 563–574 10.1016/j.tim.2017.11.002. [DOI] [PubMed] [Google Scholar]
- [4].Cani PD, Human gut microbiome: hopes, threats and promises, Gut 67 (2018) 1716–1725 10.1136/gutjnl-2018-316723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Durack J, Lynch SV, The gut microbiome: relationships with disease and opportunities for therapy, J. Exp. Med 216 (2019) 20–40 10.1084/jem.20180448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Zhu B, Wang X, Li L, Human gut microbiome: the second genome of human body, Protein Cell 1 (2010) 718–725 10.1007/s13238-010-0093-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Woting A, Blaut M, The intestinal microbiota in metabolic disease, Nutrients 8 (2016) 202 10.3390/nu8040202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Thursby E, Juge N, Introduction to the human gut microbiota, Biochem. J 474 (2017) 1823–1836 10.1042/BCJ20160510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Nageshwar Reddy D, Role of the normal gut microbiota, World J. Gastroenterol 21 (2015) 8787–8803 10.3748/wjg.v21.i29.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sommer F, Backhed F, The gut microbiota--masters of host development and physiology, Nat. Rev. Microbiol 11 (2013) 227–238 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- [11].Antonini M, Lo Conte M, Sorini C, Falcone M, How the interplay between the commensal microbiota, gut barrier integrity, and mucosal immunity regulates brain autoimmunity, Front. Immunol 10 (2019) 1937 10.3389/fimmu.2019.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Stacchiotti V, Rezzi S, Eggersdorfer M, Galli F, Metabolic and functional interplay between gut microbiota and fat-soluble vitamins, Crit. Rev. Food Sci. Nutr 61 (2021) 3211–3232 10.1080/10408398.2020.1793728. [DOI] [PubMed] [Google Scholar]
- [13].Jones ML, Martoni CJ, Ganopolsky JG, Labbe A, Prakash S, The human microbiome and bile acid metabolism: dysbiosis, dysmetabolism, disease and intervention, Expert Opin. Biol. Ther 14 (2014) 467–482 10.1517/14712598.2014.880420. [DOI] [PubMed] [Google Scholar]
- [14].de Vos WM, Tilg H, Van Hul M, Cani PD, Gut microbiome and health: mechanistic insights, Gut 71 (2022) 1020–1032 10.1136/gutjnl-2021-326789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Levy M, Kolodziejczyk AA, Thaiss CA, Elinav E, Dysbiosis and the immune system, Nat. Rev. Immunol 17 (2017) 219–232 10.1038/nri.2017.7. [DOI] [PubMed] [Google Scholar]
- [16].Fan Y, Pedersen O, Gut microbiota in human metabolic health and disease, Nat. Rev. Microbiol 19 (2021) 55–71 10.1038/s41579-020-0433-9. [DOI] [PubMed] [Google Scholar]
- [17].Illiano P, Brambilla R, Parolini C, The mutual interplay of gut microbiota, diet and human disease, FEBS J. 287 (2020) 833–855 10.1111/febs.15217. [DOI] [PubMed] [Google Scholar]
- [18].Flandroy L, Poutahidis T, Berg G, Clarke G, Dao MC, Decaestecker E, Furman E, Haahtela T, Massart S, Plovier H, Sanz Y, Rook G, The impact of human activities and lifestyles on the interlinked microbiota and health of humans and of ecosystems, Sci. Total Environ 627 (2018) 1018–1038 10.1016/j.scitotenv.2018.01.288. [DOI] [PubMed] [Google Scholar]
- [19].Zeng H, Umar S, Rust B, Lazarova D, Bordonaro M, Secondary bile acids and short chain fatty acids in the colon: a focus on colonic microbiome, cell proliferation, inflammation, and cancer, Int. J. Mol. Sci 20 (2019) 1214 10.3390/ijms20051214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Omiecinski CJ, Vanden Heuvel JP, Perdew GH, Peters JM, Xenobiotic metabolism, disposition, and regulation by receptors: from biochemical phenomenon to predictors of major toxicities, Toxicol. Sci 120 (2011) S49–S75 10.1093/toxsci/kfq338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Collins SL, Patterson AD, The gut microbiome: an orchestrator of xenobiotic metabolism, Acta. Pharm. Sin. B 10 (2020) 19–32 10.1016/j.apsb.2019.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Maurice CF, Haiser HJ, Turnbaugh PJ, Xenobiotics shape the physiology and gene expression of the active human gut microbiome, Cell 152 (2013) 39–50 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Claus SP, Guillou H, Ellero-Simatos S, The gut microbiota: a major player in the toxicity of environmental pollutants?, NPJ Biofilms Microbiomes 2 (2016) 1–11 10.1038/npjbiofilms.2016.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Shi XM, Liu S, Song L, Wu CS, Yang B, Lu HZ, Wang X, Zakari S, Contamination and source-specific risk analysis of soil heavy metals in a typical coal industrial city, central China, Sci. Total Environ 836 (2022) 155694 10.1016/j.scitotenv.2022.155694. [DOI] [PubMed] [Google Scholar]
- [25].Al-Gubory KH, Environmental pollutants and lifestyle factors induce oxidative stress and poor prenatal development, Reprod. Biomed. Online 29 (2014) 17–31 10.1016/j.rbmo.2014.03.002. [DOI] [PubMed] [Google Scholar]
- [26].Trasande L, Shaffer RM, Sathyanarayana S, H. Council On Environmental, Food additives and child health, Pediatrics 142 (2018) 10.1542/peds.2018-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Katsikantami I, Colosio C, Alegakis A, Tzatzarakis MN, Vakonaki E, Rizos AK, Sarigiannis DA, Tsatsakis AM, Estimation of daily intake and risk assessment of organophosphorus pesticides based on biomonitoring data - The internal exposure approach, Food Chem. Toxicol 123 (2019) 57–71 10.1016/j.fct.2018.10.047. [DOI] [PubMed] [Google Scholar]
- [28].Tsiaoussis J, Antoniou MN, Koliarakis I, Mesnage R, Vardavas CI, Izotov BN, Psaroulaki A, Tsatsakis A, Effects of single and combined toxic exposures on the gut microbiome: Current knowledge and future directions, Toxicol. Lett 312 (2019) 72–97 10.1016/j.toxlet.2019.04.014. [DOI] [PubMed] [Google Scholar]
- [29].Nakov R, Velikova T, Chemical metabolism of xenobiotics by gut microbiota, Curr. Drug Metab 21 (2020) 260–269 10.2174/1389200221666200303113830. [DOI] [PubMed] [Google Scholar]
- [30].Croom E, Metabolism of xenobiotics of human environments, Prog. Mol. Biol. Transl. Sci 112 (2012) 31–88 10.1016/B978-0-12-415813-9.00003-9. [DOI] [PubMed] [Google Scholar]
- [31].Turroni S, Brigidi P, Cavalli A, Candela M, Microbiota-host transgenomic metabolism, bioactive molecules from the inside, J. Med. Chem 61 (2018) 47–61 10.1021/acs.jmedchem.7b00244. [DOI] [PubMed] [Google Scholar]
- [32].Tsunoda SM, Gonzales C, Jarmusch AK, Momper JD, Ma JD, Contribution of the gut microbiome to drug disposition, pharmacokinetic and pharmacodynamic variability, Clin. Pharmacokinet 60 (2021) 971–984 10.1007/s40262-021-01032-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Ahlawat S, Asha KK Sharma, Gut-organ axis: a microbial outreach and networking, Lett. Appl. Microbiol 72 (2021) 636–668 10.1111/lam.13333. [DOI] [PubMed] [Google Scholar]
- [34].Lim JJ, Dutta M, Dempsey JL, Lehmler HJ, MacDonald J, Bammler T, Walker C, Kavanagh TJ, Gu H, Mani S, Cui JY, Neonatal exposure to BPA, BDE-99, and PCB produces persistent changes in hepatic transcriptome associated with gut dysbiosis in adult mouse livers, Toxicol. Sci 184 (2021) 83–103 10.1093/toxsci/kfab104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Yang T, Richards EM, Pepine CJ, Raizada MK, The gut microbiota and the brain-gut-kidney axis in hypertension and chronic kidney disease, Nat. Rev. Nephrol 14 (2018) 442–456 10.1038/s41581-018-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Milosevic I, Vujovic A, Barac A, Djelic M, Korac M, Radovanovic Spurnic A, Gmizic I, Stevanovic O, Djordjevic V, Lekic N, Russo E, Amedei A, Gut-liver axis, gut microbiota, and its modulation in the management of liver diseases: a review of the literature, Int. J. Mol. Sci 20 (2019) 10.3390/ijms20020395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Mohr AE, Jasbi P, Vander Wyst KB, van Woerden I, Shi X, Gu H, Whisner CM, Bruening M, Association of food insecurity on gut microbiome and metabolome profiles in a diverse college-based sample, Sci. Rep 12 (2022) 14358 10.1038/s41598-022-18515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Gao K, Mu CL, Farzi A, Zhu WY, Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain, Adv. Nutr 11 (2020) 709–723 10.1093/advances/nmz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Mishra S, Lin Z, Pang S, Zhang W, Bhatt P, Chen S, Recent Advanced Technologies for the Characterization of Xenobiotic-Degrading Microorganisms and Microbial Communities, Front. Bioeng. Biotechnol 9 (2021) 632059 10.3389/fbioe.2021.632059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Akatsu H, Exploring the effect of probiotics, prebiotics, and postbiotics in strengthening immune activity in the elderly, Vaccines (Basel) 9 (2021) 10.3390/vaccines9020136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lee KA, Shaw HM, Bataille V, Nathan P, Spector TD, Role of the gut microbiome for cancer patients receiving immunotherapy: Dietary and treatment implications, Eur. J. Cancer 138 (2020) 149–155 10.1016/j.ejca.2020.07.026. [DOI] [PubMed] [Google Scholar]
- [42].Sieow BF, Wun KS, Yong WP, Hwang IY, Chang MW, Tweak to Treat: Reprograming Bacteria for Cancer Treatment, Trends Cancer 7 (2021) 447–464 https://www.ncbi.nlm.nih.gov/pubmed/33303401. [DOI] [PubMed] [Google Scholar]
- [43].Tremaroli V, Backhed F, Functional interactions between the gut microbiota and host metabolism, Nature 489 (2012) 242–249 10.1038/nature11552. [DOI] [PubMed] [Google Scholar]
- [44].Visconti A, Le Roy CI, Rosa F, Rossi N, Martin TC, Mohney RP, Li W, de Rinaldis E, Bell JT, Venter JC, Nelson KE, Spector TD, Falchi M, Interplay between the human gut microbiome and host metabolism, Nat. Commun 10 (2019) 4505 10.1038/s41467-019-12476-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang C, Zhang M, Wang S, Han R, Cao Y, Hua W, Mao Y, Zhang X, Pang X, Wei C, Zhao G, Chen Y, Zhao L, Interactions between gut microbiota, host genetics and diet relevant to development of metabolic syndromes in mice, ISME J. 4 (2010) 232–241 10.1038/ismej.2009.112. [DOI] [PubMed] [Google Scholar]
- [46].Nicholson JK, Holmes E, Kinross J, Burcelin R, Gibson G, Jia W, Pettersson S, Host-gut microbiota metabolic interactions, Science 336 (2012) 1262–1267 10.1126/science.1223813. [DOI] [PubMed] [Google Scholar]
- [47].Ussar S, Griffin NW, Bezy O, Fujisaka S, Vienberg S, Softic S, Deng L, Bry L, Gordon JI, Kahn CR, Interactions between gut microbiota, host genetics and diet modulate the predisposition to obesity and metabolic syndrome, Cell Metab. 22 (2015) 516–530 10.1016/j.cmet.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Peredo-Lovillo A, Romero-Luna HE, Jimenez-Fernandez M, Health promoting microbial metabolites produced by gut microbiota after prebiotics metabolism, Food Res. Int 136 (2020) 109473 10.1016/j.foodres.2020.109473. [DOI] [PubMed] [Google Scholar]
- [49].Klaassen CD, Cui JY, Review: mechanisms of how the intestinal microbiota alters the effects of drugs and bile acids, Drug Metab. Dispos 43 (2015) 1505–1521 10.1124/dmd.115.065698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ridlon JM, Kang DJ, Hylemon PB, Bile salt biotransformations by human intestinal bacteria, J. Lipid Res 47 (2006) 241–259 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- [51].Stofan M, Guo GL, Bile acids and FXR: novel targets for liver diseases, Front. Med 7 (2020) 544 10.3389/fmed.2020.00544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chiang JY, Bile acid regulation of hepatic physiology: III. Bile acids and nuclear receptors, Am. J. Physiol. Gastrointest Liver Physiol 284 (2003) G349–G356 10.1152/ajpgi.00417.2002. [DOI] [PubMed] [Google Scholar]
- [53].Ethanic M, Stanimirov B, Pavlovic N, Golocorbin-Kon S, Al-Salami H, Stankov K, Mikov M, Pharmacological applications of bile acids and their derivatives in the treatment of metabolic syndrome, Front. Pharmacol 9 (2018) 1382 10.3389/fphar.2018.01382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sberna AL, Assem M, Gautier T, Grober J, Guiu B, Jeannin A, Pais de Barros JP, Athias A, Lagrost L, Masson D, Constitutive androstane receptor activation stimulates faecal bile acid excretion and reverse cholesterol transport in mice, J. Hepatol 55 (2011) 154–161 10.1016/j.jhep.2010.10.029. [DOI] [PubMed] [Google Scholar]
- [55].Dalile B, Van Oudenhove L, Vervliet B, Verbeke K, The role of short-chain fatty acids in microbiota-gut-brain communication, Nat. Rev. Gastroenterol. Hepatol 16 (2019) 461–478 10.1038/s41575-019-0157-3. [DOI] [PubMed] [Google Scholar]
- [56].O'Riordan KJ, Collins MK, Moloney GM, Knox EG, Aburto MR, Fulling C, Morley SJ, Clarke G, Schellekens H, Cryan JF, Short chain fatty acids: microbial metabolites for gut-brain axis signalling, Mol. Cell Endocrinol 546 (2022) 111572 10.1016/j.mce.2022.111572. [DOI] [PubMed] [Google Scholar]
- [57].Stumpff F, A look at the smelly side of physiology: transport of short chain fatty acids, Pflugers Arch. 470 (2018) 571–598 10.1007/s00424-017-2105-9. [DOI] [PubMed] [Google Scholar]
- [58].Koh A, De Vadder F, Kovatcheva-Datchary P, Backhed F, From dietary fiber to host physiology: short-chain fatty acids as key bacterial metabolites, Cell 165 (2016) 1332–1345 10.1016/j.cell.2016.05.041. [DOI] [PubMed] [Google Scholar]
- [59].Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH, Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice, Gastroenterology 145 (2013) 396–406 10.1053/j.gastro.2013.04.056. [DOI] [PubMed] [Google Scholar]
- [60].Parker A, Fonseca S, Carding SR, Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health, Gut Microbes 11 (2020) 135–157 10.1080/19490976.2019.1638722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Martin CR, Osadchiy V, Kalani A, Mayer EA, The brain-gut-microbiome axis, Cell. Mol. Gastroenterol. Hepatol 6 (2018) 133–148 10.1016/j.jcmgh.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].MacFabe DF, Cain DP, Rodriguez-Capote K, Franklin AE, Hoffman JE, Boon F, Taylor AR, Kavaliers M, Ossenkopp KP, Neurobiological effects of intraventricular propionic acid in rats: possible role of short chain fatty acids on the pathogenesis and characteristics of autism spectrum disorders, Behav. Brain Res 176 (2007) 149–169 10.1016/j.bbr.2006.07.025. [DOI] [PubMed] [Google Scholar]
- [63].Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A, Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression, J. Alzheimers. Dis 26 (2011) 187–197 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- [64].Olson CA, Vuong HE, Yano JM, Liang QY, Nusbaum DJ, Hsiao EY, The gut microbiota mediates the anti-seizure effects of the ketogenic diet, Cell 173 (2018) 1728–1741 e1713 10.1016/j.cell.2018.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bian X, Wu W, Yang L, Lv L, Wang Q, Li Y, Ye J, Fang D, Wu J, Jiang X, Shi D, Li L, Administration of Akkermansia muciniphila ameliorates dextran sulfate sodium-induced ulcerative colitis in mice, Front. Microbiol 10 (2019) 2259 10.3389/fmicb.2019.02259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Sun M, Wu W, Chen L, Yang W, Huang X, Ma C, Chen F, Xiao Y, Zhao Y, Ma C, Yao S, Carpio VH, Dann SM, Zhao Q, Liu Z, Cong Y, Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis, Nat. Commun 9 (2018) 3555 10.1038/s41467-018-05901-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liu G, Chen S, Zhong J, Teng K, Yin Y, Crosstalk between tryptophan metabolism and cardiovascular disease, mechanisms, and therapeutic implications, Oxid. Med. Cell Longev 2017 (2017) 1602074 10.1155/2017/1602074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Guarner F, Malagelada JR, Gut flora in health and disease, Lancet 361 (2003) 512–519 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- [69].Gesper M, Nonnast ABH, Kumowski N, Stoehr R, Schuett K, Marx N, Kappel BA, Gut-derived metabolite indole-3-propionic acid modulates mitochondrial function in cardiomyocytes and alters cardiac function, Front. Med 8 (2021) 648259 10.3389/fmed.2021.648259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Shats I, Williams JG, Liu J, Makarov MV, Wu X, Lih FB, Deterding LJ, Lim C, Xu X, Randall TA, Lee E, Li W, Fan W, Li JL, Sokolsky M, Kabanov AV, Li L, Migaud ME, Locasale JW, Li X, Bacteria boost mammalian host NAD metabolism by engaging the deamidated biosynthesis pathway, Cell Metab. 31 (2020) 564–579 10.1016/j.cmet.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Lee CF, Chavez JD, Garcia-Menendez L, Choi Y, Roe ND, Chiao YA, Edgar JS, Goo YA, Goodlett DR, Bruce JE, Tian R, Normalization of NAD+ Redox Balance as a Therapy for Heart Failure, Circulation 134 (2016) 883–894 10.1161/CIRCULATIONAHA.116.022495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Karamanlidis G, Lee CF, Garcia-Menendez L, Kolwicz SC Jr., Suthammarak W, Gong G, Sedensky MM, Morgan PG, Wang W, Tian R, Mitochondrial complex I deficiency increases protein acetylation and accelerates heart failure, Cell Metab. 18 (2013) 239–250 10.1016/j.cmet.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK, Dorrestein PC, Mass spectrometry-based metabolomics in microbiome investigations, Nat. Rev. Microbiol 20 (2022) 143–160 10.1038/s41579-021-00621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Poretsky R, Rodriguez RL, Luo C, Tsementzi D, Konstantinidis KT, Strengths and limitations of 16S rRNA gene amplicon sequencing in revealing temporal microbial community dynamics, PLoS One 9 (2014) e93827 10.1371/journal.pone.0093827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Tringe SG, Hugenholtz P, A renaissance for the pioneering 16S rRNA gene, Curr. Opin. Microbiol 11 (2008) 442–446 10.1016/j.mib.2008.09.011. [DOI] [PubMed] [Google Scholar]
- [76].Ranjan R, Rani A, Metwally A, McGee HS, Perkins DL, Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing, Biochem. Biophys. Res. Commun 469 (2016) 967–977 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Lugli GA, Ventura M, A breath of fresh air in microbiome science: shallow shotgun metagenomics for a reliable disentangling of microbial ecosystems, Microbiome Res. Rep 1 (2022) 8 10.20517/mrr.2021.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Dumas M-E, Barton RH, Toye A, Cloarec O, Blancher C, Rothwell A, Fearnside J, Tatoud R, Blanc V, Lindon JC, Metabolic profiling reveals a contribution of gut microbiota to fatty liver phenotype in insulin-resistant mice, Proc. Natl. Acad. Sci. U.S.A 103 (2006) 12511–12516 10.1073/pnas.0601056103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hartman AL, Lough DM, Barupal DK, Fiehn O, Fishbein T, Zasloff M, Eisen JA, Human gut microbiome adopts an alternative state following small bowel transplantation, Proc. Natl. Acad. Sci. U.S.A 106 (2009) 17187–17192 10.1073/pnas.0904847106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Marshall DD, Powers R, Beyond the paradigm: combining mass spectrometry and nuclear magnetic resonance for metabolomics, Prog. Nucl. Magn. Reson. Spectrosc 100 (2017) 1–16 10.1016/j.pnmrs.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Kirpich IA, Petrosino J, Ajami N, Feng W, Wang Y, Liu Y, Beier JI, Barve SS, Yin X, Wei X, Zhang X, McClain CJ, Saturated and unsaturated dietary fats differentially modulate ethanol-induced changes in gut microbiome and metabolome in a mouse model of alcoholic liver disease, Am. J. Pathol 186 (2016) 765–776 10.1016/j.ajpath.2015.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Dey P, Sasaki GY, Wei P, Li J, Wang L, Zhu J, McTigue D, Yu Z, Bruno RS, Green tea extract prevents obesity in male mice by alleviating gut dysbiosis in association with improved intestinal barrier function that limits endotoxin translocation and adipose inflammation, J. Nutr. Biochem 67 (2019) 78–89 10.1016/j.jnutbio.2019.01.017. [DOI] [PubMed] [Google Scholar]
- [83].Li H, Xu M, Zhu J, Headspace gas monitoring of gut microbiota using targeted and globally optimized targeted secondary electrospray ionization mass spectrometry, Anal. Chem 91 (2019) 854–863 10.1021/acs.analchem.8b03517. [DOI] [PubMed] [Google Scholar]
- [84].Moran MA, Kujawinski EB, Schroer WF, Amin SA, Bates NR, Bertrand EM, Braakman R, Brown CT, Covert MW, Doney SC, Dyhrman ST, Edison AS, Eren AM, Levine NM, Li L, Ross AC, Saito MA, Santoro AE, Segre D, Shade A, Sullivan MB, Vardi A, Microbial metabolites in the marine carbon cycle, Nat. Microbiol 7 (2022) 508–523 10.1038/s41564-022-01090-3. [DOI] [PubMed] [Google Scholar]
- [85].Wu L, Han Y, Zheng Z, Peng G, Liu P, Yue S, Zhu S, Chen J, Lv H, Shao L, Sheng Y, Wang Y, Li L, Li L, Wang B, Altered gut microbial metabolites in amnestic mild cognitive impairment and Alzheimer's disease: signals in host–microbe interplay, Nutrients 13 (2021) 10.3390/nu13010228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Li CY, Dempsey JL, Wang D, Lee S, Weigel KM, Fei Q, Bhatt DK, Prasad B, Raftery D, Gu H, Cui JY, PBDEs altered gut microbiome and bile acid homeostasis in male C57BL/6 mice, Drug Metab. Dispos 46 (2018) 1226–1240 10.1124/dmd.118.081547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Scoville DK, Li CY, Wang D, Dempsey JL, Raftery D, Mani S, Gu H, Cui JY, Polybrominated diphenyl ethers and gut microbiome modulate metabolic syndrome–related aqueous metabolites in mice, Drug Metab. Dispos 47 (2019) 928–940 10.1124/dmd.119.086538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Cirstea MS, Yu AC, Golz E, Sundvick K, Kliger D, Radisavljevic N, Foulger LH, Mackenzie M, Huan T, Finlay BB, Appel-Cresswell S, Microbiota composition and metabolism are associated with gut function in Parkinson's disease, Mov. Disord 35 (2020) 1208–1217 10.1002/mds.28052. [DOI] [PubMed] [Google Scholar]
- [89].Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, Singh S, Madamba EV, Bettencourt R, Richards L, Yu RT, Atkins AR, Huan T, Brenner DA, Sirlin CB, Downes M, Evans RM, Loomba R, A universal gut-microbiome-derived signature predicts cirrhosis, Cell Metab. 32 (2020) 878–888 10.1016/j.cmet.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Courant F, Antignac JP, Dervilly-Pinel G, Le Bizec B, Basics of mass spectrometry based metabolomics, Proteomics 14 (2014) 2369–2388 10.1002/pmic.201400255. [DOI] [PubMed] [Google Scholar]
- [91].Miggiels P, Wouters B, van Westen GJ, Dubbelman A-C, Hankemeier T, Novel technologies for metabolomics: more for less, Trends Anal. Chem 120 (2019) 115323 10.1016/j.trac.2018.11.021. [DOI] [Google Scholar]
- [92].Gong ZG, Hu J, Wu X, Xu YJ, The Recent Developments in Sample Preparation for Mass Spectrometry-Based Metabolomics, Crit. Rev. Anal. Chem 47 (2017) 325–331 10.1080/10408347.2017.1289836. [DOI] [PubMed] [Google Scholar]
- [93].Hemmati M, Nix C, Crommen J, Servais A-C, Fillet M, Benefits of microsampling and microextraction for metabolomics studies, Trends Anal. Chem 127 (2020) 115899 10.1016/j.trac.2020.115899. [DOI] [Google Scholar]
- [94].Spanogiannopoulos P, Bess EN, Carmody RN, Turnbaugh PJ, The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism, Nat. Rev. Microbiol 14 (2016) 273–287 10.1038/nrmicro.2016.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Koppel N, Maini Rekdal V, Balskus EP, Chemical transformation of xenobiotics by the human gut microbiota, Science 356 (2017) 10.1126/science.aag2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Clarke G, Sandhu KV, Griffin BT, Dinan TG, Cryan JF, Hyland NP, Gut reactions: breaking down xenobiotic–microbiome interactions, Pharmacol. Rev 71 (2019) 198–224 10.1124/pr.118.015768. [DOI] [PubMed] [Google Scholar]
- [97].Liu X, Zhou L, Shi X, Xu G, New advances in analytical methods for mass spectrometry-based large-scale metabolomics study, Trends Anal. Chem 121 (2019) 115665 10.1016/j.trac.2019.115665. [DOI] [Google Scholar]
- [98].Zhou L, Yu D, Zheng S, Ouyang R, Wang Y, Xu G, Gut microbiota-related metabolome analysis based on chromatography-mass spectrometry, Trends Anal. Chem 143 (2021) 116375 10.1016/j.trac.2021.116375. [DOI] [Google Scholar]
- [99].Ye D, Li X, Shen J, Xia X, Microbial metabolomics: from novel technologies to diversified applications, Trends Anal. Chem (2022) 116540 10.1016/j.trac.2022.116540. [DOI] [Google Scholar]
- [100].Xu Y-J, Wang C, Ho WE, Ong CN, Recent developments and applications of metabolomics in microbiological investigations, Trends Anal. Chem 56 (2014) 37–48 10.1016/j.trac.2013.12.009. [DOI] [Google Scholar]
- [101].Harrieder E-M, Kretschmer F, Böcker S, Witting M, Current state-of-the-art of separation methods used in LC-MS based metabolomics and lipidomics, J. Chromatogr. B 1188 (2022) 123069 10.1016/j.jchromb.2021.123069. [DOI] [PubMed] [Google Scholar]
- [102].Wang R, Yin Y, Zhu Z-J, Advancing untargeted metabolomics using data-independent acquisition mass spectrometry technology, Anal. Bioanal. Chem 411 (2019) 4349–4357 10.1007/s00216-019-01709-1. [DOI] [PubMed] [Google Scholar]
- [103].Lacalle-Bergeron L, Izquierdo-Sandoval D, Sancho JV, López FJ, Hernández F, Portolés T, Chromatography hyphenated to high resolution mass spectrometry in untargeted metabolomics for investigation of food (bio) markers, Trends Anal. Chem 135 (2021) 116161 10.1016/j.trac.2020.116161. [DOI] [Google Scholar]
- [104].Cui L, Lu H, Lee YH, Challenges and emergent solutions for LC-MS/MS based untargeted metabolomics in diseases, Mass. Spectrom. Rev 37 (2018) 772–792 10.1002/mas.21562. [DOI] [PubMed] [Google Scholar]
- [105].Ivanisevic J, Want EJ, From samples to insights into metabolism: uncovering biologically relevant information in LC-HRMS metabolomics data, Metabolites 9 (2019) 10.3390/metabo9120308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Cajka T, Fiehn O, Toward merging untargeted and targeted methods in mass spectrometry-based metabolomics and lipidomics, Anal. Chem 88 (2016) 524–545 10.1021/acs.analchem.5b04491. [DOI] [PubMed] [Google Scholar]
- [107].Vinayavekhin N, Homan EA, Saghatelian A, Exploring disease through metabolomics, ACS Chem. Biol 5 (2010) 91–103 10.1021/cb900271r. [DOI] [PubMed] [Google Scholar]
- [108].Bird SS, Marur VR, Sniatynski MJ, Greenberg HK, Kristal BS, Lipidomics profiling by high resolution LC-MS and HCD fragmentation: focus on characterization of mitochondrial cardiolipins and monolysocardiolipins, Anal. Chem 83 (2011) 940 10.1021/ac102598u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Knittelfelder OL, Weberhofer BP, Eichmann TO, Kohlwein SD, Rechberger GN, A versatile ultra-high performance LC-MS method for lipid profiling, J. Chromatogr. B 951 (2014) 119–128 10.1016/j.jchromb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Holcčapek M, Liebisch G, Ekroos K, Lipidomic analysis, Anal. Chem (2018) 10.1021/acs.analchem.7b05395. [DOI] [PubMed] [Google Scholar]
- [111].Wang J, Wang C, Han X, Tutorial on lipidomics, Anal Chim Acta 1061 (2019) 28–41 10.1016/j.aca.2019.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Nash WJ, Dunn WB, From mass to metabolite in human untargeted metabolomics: Recent advances in annotation of metabolites applying liquid chromatography-mass spectrometry data, Trends Anal. Chem 120 (2019) 115324 10.1016/j.trac.2018.11.022. [DOI] [Google Scholar]
- [113].Tsugawa H, Cajka T, Kind T, Ma Y, Higgins B, Ikeda K, Kanazawa M, VanderGheynst J, Fiehn O, Arita M, MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis, Nat. Methods 12 (2015) 523–526 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Gorrochategui E, Jaumot J, Lacorte S, Tauler R, Data analysis strategies for targeted and untargeted LC-MS metabolomic studies: Overview and workflow, Trends. Anal. Chem 82 (2016) 425–442 10.1016/j.trac.2016.07.004. [DOI] [Google Scholar]
- [115].Blazenovic I, Kind T, Ji J, Fiehn O, Software tools and approaches for compound identification of LC-MS/MS data in metabolomics, Metabolites 8 (2018) 10.3390/metabo8020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Kind T, Liu KH, Lee DY, DeFelice B, Meissen JK, Fiehn O, LipidBlast in silico tandem mass spectrometry database for lipid identification, Nat. Methods 10 (2013) 755–758 10.1038/nmeth.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Shen X, Wang R, Xiong X, Yin Y, Cai Y, Ma Z, Liu N, Zhu ZJ, Metabolic reaction network-based recursive metabolite annotation for untargeted metabolomics, Nat. Commun 10 (2019) 1516 10.1038/s41467-019-09550-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhu ZJ, Schultz AW, Wang J, Johnson CH, Yannone SM, Patti GJ, Siuzdak G, Liquid chromatography quadrupole time-of-flight mass spectrometry characterization of metabolites guided by the METLIN database, Nat. Protoc 8 (2013) 451–460 10.1038/nprot.2013.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G, XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification, Anal. Chem 78 (2006) 779–787 10.1021/ac051437y. [DOI] [PubMed] [Google Scholar]
- [120].Pang Z, Zhou G, Ewald J, Chang L, Hacariz O, Basu N, Xia J, Using MetaboAnalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data, Nat. Protoc 17 (2022) 1735–1761 10.1038/s41596-022-00710-w. [DOI] [PubMed] [Google Scholar]
- [121].Xia J, Wishart DS, Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst, Nat. Protoc 6 (2011) 743–760 10.1038/nprot.2011.319. [DOI] [PubMed] [Google Scholar]
- [122].Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, Querengesser L, HMDB: the human metabolome database, Nucleic Acids Res. 35 (2007) D521–D526 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Wishart DS, Guo A, Oler E, Wang F, Anjum A, Peters H, Dizon R, Sayeeda Z, Tian S, Lee BL, Berjanskii M, Mah R, Yamamoto M, Jovel J, Torres-Calzada C, Hiebert-Giesbrecht M, Lui VW, Varshavi D, Varshavi D, Allen D, Arndt D, Khetarpal N, Sivakumaran A, Harford K, Sanford S, Yee K, Cao X, Budinski Z, Liigand J, Zhang L, Zheng J, Mandal R, Karu N, Dambrova M, Schioth HB, Greiner R, Gautam V, HMDB 5.0: the human metabolome database for 2022, Nucleic Acids Res. 50 (2022) D622–D631 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Misra BB, New software tools, databases, and resources in metabolomics: updates from 2020, Metabolomics 17 (2021) 49 https://www.ncbi.nlm.nih.gov/pubmed/33977389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chang HY, Colby SM, Du X, Gomez JD, Helf MJ, Kechris K, Kirkpatrick CR, Li S, Patti GJ, Renslow RS, Subramaniam S, Verma M, Xia J, Young JD, A Practical Guide to Metabolomics Software Development, Anal. Chem 93 (2021) 1912–1923 10.1021/acs.analchem.0c03581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Spicer R, Salek RM, Moreno P, Canueto D, Steinbeck C, Navigating freely-available software tools for metabolomics analysis, Metabolomics 13 (2017) 106 10.1007/s11306-017-1242-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Chen Y, Li EM, Xu LY, Guide to Metabolomics Analysis: A Bioinformatics Workflow, Metabolites 12 (2022) 10.3390/metabo12040357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Wei Z, Xi J, Gao S, You X, Li N, Cao Y, Wang L, Luan Y, Dong X, Metabolomics coupled with pathway analysis characterizes metabolic changes in response to BDE-3 induced reproductive toxicity in mice, Sci. Rep 8 (2018) 5423 10.1038/s41598-018-23484-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Gao H, Wan X, Xiao B, Yang K, Wang Y, Zhang C, Li P, Liu L, Xia T, Wang A, Zhang S, Impacts of PBDE-47 exposure before, during and after pregnancy on the maternal gut microbiome and its association with host metabolism, Ecotoxicol. Environ. Saf 222 (2021) 112530 10.1016/j.ecoenv.2021.112530. [DOI] [PubMed] [Google Scholar]
- [130].Wang Y, Wang Q, Zhou L, Zeng Z, Zhao C, You L, Lu X, Liu X, Ouyang R, Wang Y, Xu X, Tian X, Guo Y, Huo X, Xu G, Metabolomics insights into the prenatal exposure effects of polybrominated diphenyl ethers on neonatal birth outcomes, Sci. Total Environ 836 (2022) 155601 10.1016/j.scitotenv.2022.155601. [DOI] [PubMed] [Google Scholar]
- [131].Hernandez-Mesa M, Narduzzi L, Ouzia S, Soetart N, Jaillardon L, Guitton Y, Le Bizec B, Dervilly G, Metabolomics and lipidomics to identify biomarkers of effect related to exposure to non-dioxin-like polychlorinated biphenyls in pigs, Chemosphere 296 (2022) 133957 10.1016/j.chemosphere.2022.133957. [DOI] [PubMed] [Google Scholar]
- [132].Zhang L, Rimal B, Nichols RG, Tian Y, Smith PB, Hatzakis E, Chang SC, Butenhoff JL, Peters JM, Patterson AD, Perfluorooctane sulfonate alters gut microbiota-host metabolic homeostasis in mice, Toxicology 431 (2020) 152365 10.1016/j.tox.2020.152365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Luo Y, Wang J, Wang C, Wang D, Li C, Zhang B, Zhong X, Chen L, Li H, Su H, Zheng Q, Zhu D, Tang H, Guo L, The fecal arsenic excretion, tissue arsenic accumulation, and metabolomics analysis in sub-chronic arsenic-exposed mice after in situ arsenic-induced fecal microbiota transplantation, Sci. Total Environ (2022) 158583 10.1016/j.scitotenv.2022.158583. [DOI] [PubMed] [Google Scholar]
- [134].Petriello MC, Hoffman JB, Vsevolozhskaya O, Morris AJ, Hennig B, Dioxin-like PCB 126 increases intestinal inflammation and disrupts gut microbiota and metabolic homeostasis, Environ. Pollut 242 (2018) 1022–1032 10.1016/j.envpol.2018.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Gomez MV, Dutta M, Suvorov A, Shi X, Gu H, Mani S, Yue Cui J, Early life exposure to environmental contaminants (BDE-47, TBBPA, and BPS) produced persistent alterations in fecal microbiome in adult male mice, Toxicol. Sci 179 (2021) 14–30 10.1093/toxsci/kfaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Banerjee S, Mazumdar S, Electrospray ionization mass spectrometry: a technique to access the information beyond the molecular weight of the analyte, Int. J. Anal. Chem 2012 (2012) 282574 10.1155/2012/282574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Hu C, Liu M, Wan T, Tang L, Sun B, Zhou B, Lam JCW, Lam PKS, Chen L, Disturbances in microbial and metabolic communication across the gut-liver axis induced by a dioxin-like pollutant: an integrated metagenomics and metabolomics analysis, Environ. Sci. Technol 55 (2021) 529–537 10.1021/acs.est.0c06884. [DOI] [PubMed] [Google Scholar]
- [138].He Y, Brademan DR, Hutchins PD, Overmyer KA, Coon JJ, Maximizing MS/MS Acquisition for Lipidomics Using Capillary Separation and Orbitrap Tribrid Mass Spectrometer, Anal. Chem 94 (2022) 3394–3399 10.1021/acs.analchem.1c05552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Kapoore RV, Vaidyanathan S, Towards quantitative mass spectrometry-based metabolomics in microbial and mammalian systems, Philos. Trans. Royal Soc. A 374 (2016) 10.1098/rsta.2015.0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Reveglia P, Paolillo C, Ferretti G, De Carlo A, Angiolillo A, Nasso R, Caputo M, Matrone C, Di Costanzo A, Corso G, Challenges in LC-MS-based metabolomics for Alzheimer's disease early detection: targeted approaches versus untargeted approaches, Metabolomics 17 (2021) 78 10.1007/s11306-021-01828-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Roberts LD, Souza AL, Gerszten RE, Clish CB, Targeted metabolomics, Curr. Protoc. Mol. Biol Chapter 30 (2012) Unit 30 32 31–24 10.1002/0471142727.mb3002s98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Griffiths WJ, Koal T, Wang Y, Kohl M, Enot DP, Deigner HP, Targeted metabolomics for biomarker discovery, Angew. Chem. Int. Ed. Engl 49 (2010) 5426–5445 10.1002/anie.200905579. [DOI] [PubMed] [Google Scholar]
- [143].Stachniuk A, Fornal E, Liquid chromatography-mass spectrometry in the analysis of pesticide residues in food, Food Anal. Methods 9 (2016) 1654–1665 10.1007/s12161-015-0342-0. [DOI] [Google Scholar]
- [144].Cajka T, Fiehn O, Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry, Trends Anal. Chem 61 (2014) 192–206 10.1016/j.trac.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Gianazza E, Banfi C, Post-translational quantitation by SRM/MRM: applications in cardiology, Expert Rev. Proteomics 15 (2018) 477–502 10.1080/14789450.2018.1484283. [DOI] [PubMed] [Google Scholar]
- [146].Zhou J, Yin Y, Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry, Analyst 141 (2016) 6362–6373 10.1039/c6an01753c. [DOI] [PubMed] [Google Scholar]
- [147].Zhou J, Liu C, Si D, Jia B, Zhong L, Yin Y, Workflow development for targeted lipidomic quantification using parallel reaction monitoring on a quadrupole-time of flight mass spectrometry, Anal Chim Acta 972 (2017) 62–72 10.1016/j.aca.2017.04.008. [DOI] [PubMed] [Google Scholar]
- [148].Skonieczna-Zydecka K, Jakubczyk K, Maciejewska-Markiewicz D, Janda K, Kazmierczak-Siedlecka K, Kaczmarczyk M, Loniewski I, Marlicz W, Gut biofactory-neurocompetent metabolites within the gastrointestinal tract. a scoping review, Nutrients 12 (2020) 10.3390/nu12113369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Gasaly N, de Vos P, Hermoso MA, Impact of bacterial metabolites on gut barrier function and host immunity: a focus on bacterial metabolism and its relevance for intestinal inflammation, Front. Immunol 12 (2021) 658354 10.3389/fimmu.2021.658354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Michaudel C, Sokol H, The gut microbiota at the service of immunometabolism, Cell Metab. 32 (2020) 514–523 10.1016/j.cmet.2020.09.004. [DOI] [PubMed] [Google Scholar]
- [151].Choucair I, Nemet I, Li L, Cole MA, Skye SM, Kirsop JD, Fischbach MA, Gogonea V, Brown JM, Tang WHW, Hazen SL, Quantification of bile acids: a mass spectrometry platform for studying gut microbe connection to metabolic diseases, J. Lipid Res 61 (2020) 159–177 10.1194/jlr.RA119000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Garcia-Canaveras JC, Donato MT, Castell JV, Lahoz A, Targeted profiling of circulating and hepatic bile acids in human, mouse, and rat using a UPLC-MRM-MS-validated method, J. Lipid Res 53 (2012) 2231–2241 10.1194/jlr.D028803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Sarafian MH, Lewis MR, Pechlivanis A, Ralphs S, McPhail MJ, Patel VC, Dumas ME, Holmes E, Nicholson JK, Bile acid profiling and quantification in biofluids using ultra-performance liquid chromatography tandem mass spectrometry, Anal. Chem 87 (2015) 9662–9670 10.1021/acs.analchem.5b01556. [DOI] [PubMed] [Google Scholar]
- [154].Han J, Lin K, Sequeira C, Borchers CH, An isotope-labeled chemical derivatization method for the quantitation of short-chain fatty acids in human feces by liquid chromatography-tandem mass spectrometry, Anal. Chim. Acta 854 (2015) 86–94 10.1016/j.aca.2014.11.015. [DOI] [PubMed] [Google Scholar]
- [155].Chen GY, Zhong W, Zhou Z, Zhang Q, Simultaneous determination of tryptophan and its 31 catabolites in mouse tissues by polarity switching UHPLC-SRM-MS, Anal. Chim. Acta 1037 (2018) 200–210 10.1016/j.aca.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Whiley L, Nye LC, Grant I, Andreas N, Chappell KE, Sarafian MH, Misra R, Plumb RS, Lewis MR, Nicholson JK, Holmes E, Swann JR, Wilson ID, Ultrahigh-performance liquid chromatography tandem mass spectrometry with electrospray ionization quantification of tryptophan metabolites and markers of gut health in serum and plasma-application to clinical and epidemiology cohorts, Anal. Chem 91 (2019) 5207–5216 10.1021/acs.analchem.8b05884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Liu J, Zhao M, Zhou J, Liu C, Zheng L, Yin Y, Simultaneous targeted analysis of trimethylamine-N-oxide, choline, betaine, and carnitine by high performance liquid chromatography tandem mass spectrometry, J. Chromatogr. B 1035 (2016) 42–48 10.1016/j.jchromb.2016.09.026. [DOI] [PubMed] [Google Scholar]
- [158].Lu WH, Chiu HH, Kuo HC, Chen GY, Chepyala D, Kuo CH, Using matrix-induced ion suppression combined with LC-MS/MS for quantification of trimethylamine-N-oxide, choline, carnitine and acetylcarnitine in dried blood spot samples, Anal. Chim. Acta 1149 (2021) 338214 10.1016/j.aca.2021.338214. [DOI] [PubMed] [Google Scholar]
- [159].Gu H, Jasbi P, Patterson J, Jin Y, Enhanced detection of short-chain fatty acids using gas chromatography mass spectrometry, Curr. Protoc 1 (2021) e177 10.1002/cpz1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Rotondo-Trivette S, Wang B, Luan Y, Fiehn O, Sun F, Michail S, Reduced fecal short-chain fatty acids in hispanic children with ulcerative colitis, Physiol. Rep 9 (2021) e14918 10.14814/phy2.14918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Lotti C, Rubert J, Fava F, Tuohy K, Mattivi F, Vrhovsek U, Development of a fast and cost-effective gas chromatography-mass spectrometry method for the quantification of short-chain and medium-chain fatty acids in human biofluids, Anal. Bioanal. Chem 409 (2017) 5555–5567 10.1007/s00216-017-0493-5. [DOI] [PubMed] [Google Scholar]
- [162].He L, Prodhan MAI, Yuan F, Yin X, Lorkiewicz PK, Wei X, Feng W, McClain C, Zhang X, Simultaneous quantification of straight-chain and branched-chain short chain fatty acids by gas chromatography mass spectrometry, J. Chromatogr. B 1092 (2018) 359–367 10.1016/j.jchromb.2018.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].He L, Li F, Yin X, Bohman P, Kim S, McClain CJ, Feng W, Zhang X, Profiling of polar metabolites in mouse feces using four analytical platforms to study the effects of cathelicidin-related antimicrobial peptide in alcoholic liver disease, J. Proteome Res 18 (2019) 2875–2884 10.1021/acs.jproteome.9b00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Eghlimi R, Shi X, Hrovat J, Xi B, Gu H, Triple negative breast cancer detection using LC-MS/MS lipidomic profiling, J. Proteome Res 19 (2020) 2367–2378 10.1021/acs.jproteome.0c00038. [DOI] [PubMed] [Google Scholar]
- [165].Birsoy K, Wang T, Possemato R, Yilmaz OH, Koch CE, Chen WW, Hutchins AW, Gultekin Y, Peterson TR, Carette JE, Brummelkamp TR, Clish CB, Sabatini DM, MCT1-mediated transport of a toxic molecule is an effective strategy for targeting glycolytic tumors, Nat. Genet 45 (2013) 104–108 10.1038/ng.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Zhu J, Djukovic D, Deng L, Gu H, Himmati F, Chiorean EG, Raftery D, Colorectal cancer detection using targeted serum metabolic profiling, J. Proteome Res 13 (2014) 4120–4130 10.1021/pr500494u. [DOI] [PubMed] [Google Scholar]
- [167].Wang J, Ferruzzi MG, Ho L, Blount J, Janle EM, Gong B, Pan Y, Gowda GA, Raftery D, Arrieta-Cruz I, Sharma V, Cooper B, Lobo J, Simon JE, Zhang C, Cheng A, Qian X, Ono K, Teplow DB, Pavlides C, Dixon RA, Pasinetti GM, Brain-targeted proanthocyanidin metabolites for Alzheimer's disease treatment, J. Neurosci 32 (2012) 5144–5150 10.1523/JNEUROSCI.6437-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Munjoma N, Isaac G, Muazzam A, Cexus O, Azhar F, Pandha H, Whetton AD, Townsend PA, Wilson ID, Gethings LA, Plumb RS, High Throughput LC-MS Platform for Large Scale Screening of Bioactive Polar Lipids in Human Plasma and Serum, J. Proteome Res 21 (2022) 2596–2608 10.1021/acs.jproteome.2c00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Zhou J, Liu H, Liu Y, Liu J, Zhao X, Yin Y, Development and Evaluation of a Parallel Reaction Monitoring Strategy for Large-Scale Targeted Metabolomics Quantification, Anal. Chem 88 (2016) 4478–4486 10.1021/acs.analchem.6b00355. [DOI] [PubMed] [Google Scholar]
- [170].Zhang L, Zheng W, Li X, Wang S, Xiao M, Xiao R, Zhang D, Ke N, Cai H, Cheng J, Chen X, Gong M, A merged method for targeted analysis of amino acids and derivatives using parallel reaction monitoring combined with untargeted profiling by HILIC-Q-Orbitrap HRMS, J. Pharm. Biomed. Anal 203 (2021) 114208 10.1016/j.jpba.2021.114208. [DOI] [PubMed] [Google Scholar]
- [171].Shi X, Xi B, Jasbi P, Turner C, Jin Y, Gu H, Comprehensive isotopic targeted mass spectrometry: reliable metabolic flux analysis with broad coverage, Anal. Chem 92 (2020) 11728–11738 10.1021/acs.analchem.0c01767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Dai DF, Karunadharma PP, Chiao YA, Basisty N, Crispin D, Hsieh EJ, Chen T, Gu H, Djukovic D, Raftery D, Beyer RP, MacCoss MJ, Rabinovitch PS, Altered proteome turnover and remodeling by short-term caloric restriction or rapamycin rejuvenate the aging heart, Aging Cell 13 (2014) 529–539 10.1111/acel.12203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Reyes NL, Banks GB, Tsang M, Margineantu D, Gu H, Djukovic D, Chan J, Torres M, Liggitt HD, Hirenallur SD, Hockenbery DM, Raftery D, Iritani BM, Fnip1 regulates skeletal muscle fiber type specification, fatigue resistance, and susceptibility to muscular dystrophy, Proc. Natl. Acad. Sci. U.S.A 112 (2015) 424–429 10.1073/pnas.1413021112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Sperber H, Mathieu J, Wang Y, Ferreccio A, Hesson J, Xu Z, Fischer KA, Devi A, Detraux D, Gu H, Battle SL, Showalter M, Valensisi C, Bielas JH, Ericson NG, Margaretha L, Robitaille AM, Margineantu D, Fiehn O, Hockenbery D, Blau CA, Raftery D, Margolin AA, Hawkins RD, Moon RT, Ware CB, Ruohola-Baker H, The metabolome regulates the epigenetic landscape during naive-to-primed human embryonic stem cell transition, Nat. Cell Biol 17 (2015) 1523–1535 10.1038/ncb3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Zheng F, Zhao X, Zeng Z, Wang L, Lv W, Wang Q, Xu G, Development of a plasma pseudotargeted metabolomics method based on ultra-high-performance liquid chromatography–mass spectrometry, Nat. Protoc 15 (2020) 2519–2537 10.1038/s41596-020-0341-5. [DOI] [PubMed] [Google Scholar]
- [176].Luo P, Yin P, Zhang W, Zhou L, Lu X, Lin X, Xu G, Optimization of large-scale pseudotargeted metabolomics method based on liquid chromatography-mass spectrometry, J. Chromatogr. A 1437 (2016) 127–136 10.1016/j.chroma.2016.01.078. [DOI] [PubMed] [Google Scholar]
- [177].Chen S, Kong H, Lu X, Li Y, Yin P, Zeng Z, Xu G, Pseudotargeted metabolomics method and its application in serum biomarker discovery for hepatocellular carcinoma based on ultra high-performance liquid chromatography/triple quadrupole mass spectrometry, Anal. Chem 85 (2013) 8326–8333 10.1021/ac4016787. [DOI] [PubMed] [Google Scholar]
- [178].Luo P, Dai W, Yin P, Zeng Z, Kong H, Zhou L, Wang X, Chen S, Lu X, Xu G, Multiple reaction monitoring-ion pair finder: a systematic approach to transform nontargeted mode to pseudotargeted mode for metabolomics study based on liquid chromatography-mass spectrometry, Anal. Chem 87 (2015) 5050–5055 10.1021/acs.analchem.5b00615. [DOI] [PubMed] [Google Scholar]
- [179].Li Y, Ruan Q, Li Y, Ye G, Lu X, Lin X, Xu G, A novel approach to transforming a non-targeted metabolic profiling method to a pseudo-targeted method using the retention time locking gas chromatography/mass spectrometry-selected ions monitoring, J. Chromatogr. A 1255 (2012) 228–236 10.1016/j.chroma.2012.01.076. [DOI] [PubMed] [Google Scholar]
- [180].Gu H, Zhang P, Zhu J, Raftery D, Globally optimized targeted mass spectrometry: reliable metabolomics analysis with broad coverage, Anal. Chem 87 (2015) 12355–12362 10.1021/acs.analchem.5b03812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Shi X, Wang S, Jasbi P, Turner C, Hrovat J, Wei Y, Liu J, Gu H, Database-assisted globally optimized targeted mass spectrometry (dGOT-MS): Broad and reliable metabolomics analysis with enhanced identification, Anal. Chem 91 (2019) 13737–13745 10.1021/acs.analchem.9b03107. [DOI] [PubMed] [Google Scholar]
- [182].Lim JJ, Li X, Lehmler HJ, Wang D, Gu H, Cui JY, Gut microbiome critically impacts PCB-induced changes in metabolic fingerprints and the hepatic transcriptome in mice, Toxicol. Sci 177 (2020) 168–187 10.1093/toxsci/kfaa090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Huang X, Chen YJ, Cho K, Nikolskiy I, Crawford PA, Patti GJ, X13CMS: global tracking of isotopic labels in untargeted metabolomics, Anal. Chem 86 (2014) 1632–1639 10.1021/ac403384n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Fan TW, Lorkiewicz PK, Sellers K, Moseley HN, Higashi RM, Lane AN, Stable isotope-resolved metabolomics and applications for drug development, Pharmacol. Ther 133 (2012) 366–391 10.1016/j.pharmthera.2011.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Hackett SR, Zanotelli VR, Xu W, Goya J, Park JO, Perlman DH, Gibney PA, Botstein D, Storey JD, Rabinowitz JD, Systems-level analysis of mechanisms regulating yeast metabolic flux, Science 354 (2016) 10.1126/science.aaf2786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Davis RJ, Gonen M, Margineantu DH, Handeli S, Swanger J, Hoellerbauer P, Paddison PJ, Gu H, Raftery D, Grim JE, Hockenbery DM, Margolin AA, Clurman BE, Pan-cancer transcriptional signatures predictive of oncogenic mutations reveal that Fbw7 regulates cancer cell oxidative metabolism, Proc. Natl. Acad. Sci. U.S.A 115 (2018) 5462–5467 10.1073/pnas.1718338115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Antoniewicz MR, A guide to metabolic flux analysis in metabolic engineering: Methods, tools and applications, Metab. Eng 63 (2021) 2–12 10.1016/j.ymben.2020.11.002. [DOI] [PubMed] [Google Scholar]
- [188].Zhang H, Badur MG, Divakaruni AS, Parker SJ, Jager C, Hiller K, Murphy AN, Metallo CM, Distinct metabolic states can support self-renewal and lipogenesis in human pluripotent stem cells under different culture conditions, Cell Rep. 16 (2016) 1536–1547 10.1016/j.celrep.2016.06.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Chokkathukalam A, Kim DH, Barrett MP, Breitling R, Creek DJ, Stable isotope-labeling studies in metabolomics: new insights into structure and dynamics of metabolic networks, Bioanalysis 6 (2014) 511–524 10.4155/bio.13.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Hui S, Cowan AJ, Zeng X, Yang L, TeSlaa T, Li X, Bartman C, Zhang Z, Jang C, Wang L, Lu W, Rojas J, Baur J, Rabinowitz JD, Quantitative fluxomics of circulating metabolites, Cell Metab. 32 (2020) 676–688 10.1016/j.cmet.2020.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Bruntz RC, Lane AN, Higashi RM, Fan TW, Exploring cancer metabolism using stable isotope-resolved metabolomics (SIRM), J. Biol. Chem 292 (2017) 11601–11609 10.1074/jbc.R117.776054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Deng P, Valentino T, Flythe MD, Moseley HNB, Leachman JR, Morris AJ, Hennig B, Untargeted stable isotope probing of the gut microbiota metabolome using (13)C-labeled dietary fibers, J. Proteome Res 20 (2021) 2904–2913 10.1021/acs.jproteome.1c00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Liu R, Yang Z, Single cell metabolomics using mass spectrometry: Techniques and data analysis, Anal. Chim. Acta 1143 (2021) 124–134 10.1016/j.aca.2020.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Zenobi R, Single-cell metabolomics: analytical and biological perspectives, Science 342 (2013) 1243259 10.1126/science.1243259. [DOI] [PubMed] [Google Scholar]
- [195].Lanekoff I, Sharma VV, Marques C, Single-cell metabolomics: where are we and where are we going?, Curr. Opin. Biotechnol 75 (2022) 102693 10.1016/j.copbio.2022.102693. [DOI] [PubMed] [Google Scholar]
- [196].Liu Q, Martínez-Jarquín S, Zenobi R, Recent Advances in Single-Cell Metabolomics Based on Mass Spectrometry, CCS Chem. 5 (2023) 310–324 10.31635/ccschem.022.202202333. [DOI] [Google Scholar]
- [197].Zhan L, Liu H, Hou Z, Gao Y, Chu B, Huang G, Recent advances in single bacterium metabolic analysis techniques, Trends Anal. Chem (2023) 117076 10.1016/j.trac.2023.117076. [DOI] [Google Scholar]
- [198].Duncan KD, Fyrestam J, Lanekoff I, Advances in mass spectrometry based single-cell metabolomics, Analyst 144 (2019) 782–793 10.1039/C8AN01581C. [DOI] [PubMed] [Google Scholar]
- [199].Guo S, Zhang C, Le A, The limitless applications of single-cell metabolomics, Curr. Opin. Biotechnol 71 (2021) 115–122 10.1016/j.copbio.2021.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Buchberger AR, DeLaney K, Johnson J, Li L, Mass spectrometry imaging: a review of emerging advancements and future insights, Anal. Chem 90 (2018) 240 https://pubs.acs.org/doi/10.1021/acs.analchem.7b04733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Lin B-J, Kuo T-C, Chung H-H, Huang Y-C, Wang M-Y, Hsu C-C, Yao P-Y, Tseng YJ, MSIr: Automatic Registration Service for Mass Spectrometry Imaging and Histology, Anal. Chem 95 (2023) 3317–3324 10.1021/acs.analchem.2c04360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Soudah T, Zoabi A, Margulis K, Desorption electrospray ionization mass spectrometry imaging in discovery and development of novel therapies, Mass. Spectrom. Rev 42 (2023) 751–778 10.1002/mas.21736. [DOI] [PubMed] [Google Scholar]
- [203].Hulme H, Meikle LM, Strittmatter N, Swales J, Hamm G, Brown SL, Milling S, MacDonald AS, Goodwin RJ, Burchmore R, Mapping the influence of the gut microbiota on small molecules across the microbiome gut brain axis, J. Am. Soc. Mass. Spectrom 33 (2022) 649–659 10.1021/jasms.1c00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Watrous JD, Phelan VV, Hsu C-C, Moree WJ, Duggan BM, Alexandrov T, Dorrestein PC, Microbial metabolic exchange in 3D, ISME J. 7 (2013) 770–780 10.1038/ismej.2012.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Dunham SJ, Ellis JF, Li B, Sweedler JV, Mass spectrometry imaging of complex microbial communities, Acc. Chem. Res 50 (2016) 96–104 10.1021/acs.accounts.6b00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Zhang J, Rector J, Lin JQ, Young JH, Sans M, Katta N, Giese N, Yu W, Nagi C, Suliburk J, Nondestructive tissue analysis for ex vivo and in vivo cancer diagnosis using a handheld mass spectrometry system, Sci. Transl. Med 9 (2017) eaan3968 10.1126/scitranslmed.aan3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Sans M, Zhang J, Lin JQ, Feider CL, Giese N, Breen MT, Sebastian K, Liu J, Sood AK, Eberlin LS, Performance of the MasSpec Pen for rapid diagnosis of ovarian cancer, Clin. Chem 65 (2019) 674–683 https://academic.oup.com/clinchem/article/65/5/674/5608042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].King ME, Zhang J, Lin JQ, Garza KY, DeHoog RJ, Feider CL, Bensussan A, Sans M, Krieger A, Badal S, Rapid diagnosis and tumor margin assessment during pancreatic cancer surgery with the MasSpec Pen technology, Proc. Natl. Acad. Sci. U.S.A 118 (2021) e2104411118 10.1073/pnas.2104411118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Ashraf MA, Persistent organic pollutants (POPs): a global issue, a global challenge, Environ. Sci. Pollut. Res. Int 24 (2017) 4223–4227 10.1007/s11356-015-5225-9. [DOI] [PubMed] [Google Scholar]
- [210].El-Shahawi MS, Hamza A, Bashammakh AS, Al-Saggaf WT, An overview on the accumulation, distribution, transformations, toxicity and analytical methods for the monitoring of persistent organic pollutants, Talanta 80 (2010) 1587–1597 10.1016/j.talanta.2009.09.055. [DOI] [PubMed] [Google Scholar]
- [211].Pariatamby A, Kee YL, Persistent organic pollutants management and remediation, Procedia Environ. Sci 31 (2016) 842–848 10.1016/j.proenv.2016.02.093. [DOI] [Google Scholar]
- [212].Maqbool F, Mostafalou S, Bahadar H, Abdollahi M, Review of endocrine disorders associated with environmental toxicants and possible involved mechanisms, Life Sci 145 (2016) 265–273 10.1016/j.lfs.2015.10.022. [DOI] [PubMed] [Google Scholar]
- [213].Alharbi OM, Khattab RA, Ali I, Health and environmental effects of persistent organic pollutants, J. Mol. Liq 263 (2018) 442–453 10.1016/j.molliq.2018.05.029. [DOI] [Google Scholar]
- [214].Yuan J, Liu Y, Wang J, Zhao Y, Li K, Jing Y, Zhang X, Liu Q, Geng X, Li G, Wang F, Long-term persistent organic pollutants exposure induced telomere dysfunction and senescence-associated secretary phenotype, J. Gerontol. A Biol. Sci. Med. Sci 73 (2018) 1027–1035 10.1093/gerona/gly002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Dodson RE, Perovich LJ, Covaci A, Van den Eede N, Ionas AC, Dirtu AC, Brody JG, Rudel RA, After the PBDE phase-out: a broad suite of flame retardants in repeat house dust samples from California, Environ. Sci. Technol 46 (2012) 13056–13066 10.1021/es303879n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Talsness CE, Overview of toxicological aspects of polybrominated diphenyl ethers: a flame-retardant additive in several consumer products, Environ. Res 108 (2008) 158–167 10.1016/j.envres.2008.08.008. [DOI] [PubMed] [Google Scholar]
- [217].Law RJ, Covaci A, Harrad S, Herzke D, Abdallah MA, Fernie K, Toms LM, Takigami H, Levels and trends of PBDEs and HBCDs in the global environment: status at the end of 2012, Environ. Int 65 (2014) 147–158 10.1016/j.envint.2014.01.006. [DOI] [PubMed] [Google Scholar]
- [218].Jones-Otazo HA, Clarke JP, Diamond ML, Archbold JA, Ferguson G, Harner T, Richardson GM, Ryan JJ, Wilford B, Is house dust the missing exposure pathway for PBDEs? An analysis of the urban fate and human exposure to PBDEs, Environ. Sci. Technol 39 (2005) 5121–5130 10.1021/es048267b. [DOI] [PubMed] [Google Scholar]
- [219].Eriksson P, Jakobsson E, Fredriksson A, Brominated flame retardants: a novel class of developmental neurotoxicants in our environment?, Environ. Health Perspect 109 (2001) 903–908 10.1289/ehp.01109903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Jarosiewicz M, Milowska K, Krokosz A, Bukowska B, Evaluation of the effect of selected brominated flame retardants on human serum albumin and human erythrocyte membrane proteins, Int. J. Mol. Sci 21 (2020) 10.3390/ijms21113926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Sheller-Miller S, Radnaa E, Arita Y, Getahun D, Jones RJ, Peltier MR, Menon R, Environmental pollutant induced cellular injury is reflected in exosomes from placental explants, Placenta 89 (2020) 42–49 10.1016/j.placenta.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Zhou H, Yin N, Faiola F, Tetrabromobisphenol A (TBBPA): a controversial environmental pollutant, J. Environ. Sci. (China) 97 (2020) 54–66 10.1016/j.jes.2020.04.039. [DOI] [PubMed] [Google Scholar]
- [223].Borghoff SJ, Wikoff D, Harvey S, Haws L, Dose- and time-dependent changes in tissue levels of tetrabromobisphenol A (TBBPA) and its sulfate and glucuronide conjugates following repeated administration to female Wistar Han Rats, Toxicol. Rep 3 (2016) 190–201 10.1016/j.toxrep.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].He H, Shi X, Lawrence A, Hrovat J, Turner C, Cui JY, Gu H, 2,2',4,4'-tetrabromodiphenyl ether (BDE-47) induces wide metabolic changes including attenuated mitochondrial function and enhanced glycolysis in PC12 cells, Ecotoxicol. Environ. Saf 201 (2020) 110849 10.1016/j.ecoenv.2020.110849. [DOI] [PubMed] [Google Scholar]
- [225].Cho J-H, Lee S, Jeon H, Kim AH, Lee W, Lee Y, Yang S, Yun J, Jung Y-S, Lee J, Tetrabromobisphenol A-induced apoptosis in neural stem cells through oxidative stress and mitochondrial dysfunction, Neurotox. Res 38 (2020) 74–85 10.1007/s12640-020-00179-z. [DOI] [PubMed] [Google Scholar]
- [226].Zhang Y, Wang X, Chen C, An J, Shang Y, Li H, Xia H, Yu J, Wang C, Liu Y, Guo S, Regulation of TBBPA-induced oxidative stress on mitochondrial apoptosis in L0 cells through the Nrf2 signaling pathway, Chemosphere 226 (2019) 463–471 10.1016/j.chemosphere.2019.03.167. [DOI] [PubMed] [Google Scholar]
- [227].Yu X, Yin H, Peng H, Lu G, Liu Z, Dang Z, OPFRs and BFRs induced A54 cell apoptosis by caspase-dependent mitochondrial pathway, Chemosphere 221 (2019) 693–702 10.1016/j.chemosphere.2019.01.074. [DOI] [PubMed] [Google Scholar]
- [228].Erickson MD, Kaley RG 2nd, Applications of polychlorinated biphenyls, Environ. Sci. Pollut. Res. Int 18 (2011) 135–151 10.1007/s11356-010-0392-1. [DOI] [PubMed] [Google Scholar]
- [229].Wu JP, Luo XJ, Zhang Y, Luo Y, Chen SJ, Mai BX, Yang ZY, Bioaccumulation of polybrominated diphenyl ethers (PBDEs) and polychlorinated biphenyls (PCBs) in wild aquatic species from an electronic waste (e-waste) recycling site in South China, Environ. Int 34 (2008) 1109–1113 10.1016/j.envint.2008.04.001. [DOI] [PubMed] [Google Scholar]
- [230].Tian Y, Rimal B, Gui W, Koo I, Yokoyama S, Perdew GH, Patterson AD, Early life short-term exposure to polychlorinated biphenyl 126 in mice leads to metabolic dysfunction and microbiota changes in adulthood, Int. J. Mol. Sci 23 (2022) 10.3390/ijms23158220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Tian Y, Gui W, Rimal B, Koo I, Smith PB, Nichols RG, Cai J, Liu Q, Patterson AD, Metabolic impact of persistent organic pollutants on gut microbiota, Gut Microbes 12 (2020) 1–16 10.1080/19490976.2020.1848209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Gluge J, Scheringer M, Cousins IT, DeWitt JC, Goldenman G, Herzke D, Lohmann R, Ng CA, Trier X, Wang Z, An overview of the uses of per- and polyfluoroalkyl substances (PFAS), Environ. Sci. Process. Impacts 22 (2020) 2345–2373 10.1039/D0EM00291G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Brennan NM, Evans AT, Fritz MK, Peak SA, von Holst HE, Trends in the Regulation of Per- and Polyfluoroalkyl Substances (PFAS): A Scoping Review, Int. J. Environ. Res. Public Health 18 (2021) 10.3390/ijerph182010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Ankley GT, Cureton P, Hoke RA, Houde M, Kumar A, Kurias J, Lanno R, McCarthy C, Newsted J, Salice CJ, Sample BE, Sepulveda MS, Steevens J, Valsecchi S, Assessing the Ecological Risks of Per- and Polyfluoroalkyl Substances: Current State-of-the Science and a Proposed Path Forward, Environ. Toxicol. Chem 40 (2021) 564–605 10.1002/etc.4869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Long M, Donoso J, Bhati M, Elias WC, Heck KN, Luo YH, Lai YS, Gu H, Senftle TP, Zhou C, Wong MS, Rittmann BE, Adsorption and Reductive Defluorination of Perfluorooctanoic Acid over Palladium Nanoparticles, Environ. Sci. Technol 55 (2021) 14836–14843 10.1021/acs.est.1c03134. [DOI] [PubMed] [Google Scholar]
- [236].Long M, Elias WC, Heck KN, Luo Y-H, Lai YS, Jin Y, Gu H, Donoso J, Senftle TP, Zhou C, Hydrodefluorination of Perfluorooctanoic Acid in the H2-Based Membrane Catalyst-Film Reactor with Platinum Group Metal Nanoparticles: Pathways and Optimal Conditions, Environ. Sci. Technol 55 (2021) 16699–16707 10.1021/acs.est.1c06528. [DOI] [PubMed] [Google Scholar]
- [237].Peskett ST, Rand AA, The human fecal microbiome contributes to the biotransformation of the PFAS surfactant 8:2 monosubstituted polyfluoroalkyl phosphate ester, Environ. Sci. Process. Impacts 24 (2022) 1758–1768 10.1039/D2EM00225F. [DOI] [PubMed] [Google Scholar]
- [238].Laue HE, Moroishi Y, Palys TJ, Christensen BC, Criswell RL, Peterson LA, Huset CA, Baker ER, Karagas MR, Madan JC, Romano ME, Early-life exposure to per- and polyfluoroalkyl substances and infant gut microbial composition, Environ. Epidemiol 7 (2023) e238 https://www.ncbi.nlm.nih.gov/pubmed/36777525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Thompson KN, Oulhote Y, Weihe P, Wilkinson JE, Ma S, Zhong H, Li J, Kristiansen K, Huttenhower C, Grandjean P, Effects of Lifetime Exposures to Environmental Contaminants on the Adult Gut Microbiome, Environ. Sci. Technol 56 (2022) 16985–16995 10.1021/acs.est.2c03185. [DOI] [PubMed] [Google Scholar]
- [240].Wang G, Sun S, Wu X, Yang S, Wu Y, Zhao J, Zhang H, Chen W, Intestinal environmental disorders associate with the tissue damages induced by perfluorooctane sulfonate exposure, Ecotoxicol. Environ. Saf 197 (2020) 110590 https://www.ncbi.nlm.nih.gov/pubmed/32283409. [DOI] [PubMed] [Google Scholar]
- [241].Wang G, Pan R, Liang X, Wu X, Wu Y, Zhang H, Zhao J, Chen W, Perfluorooctanoic acid-induced liver injury is potentially associated with gut microbiota dysbiosis, Chemosphere 266 (2021) 129004 10.1016/j.chemosphere.2020.129004. [DOI] [PubMed] [Google Scholar]
- [242].Shi L, Zheng J, Yan S, Li Y, Wang Y, Liu X, Xiao C, Exposure to Perfluorooctanoic Acid Induces Cognitive Deficits via Altering Gut Microbiota Composition, Impairing Intestinal Barrier Integrity, and Causing Inflammation in Gut and Brain, J. Agric. Food Chem 68 (2020) 13916–13928 10.1021/acs.jafc.0c05834. [DOI] [PubMed] [Google Scholar]
- [243].Shi L, Pan R, Lin G, Liang X, Zhao J, Zhang H, Chen W, Wang G, Lactic acid bacteria alleviate liver damage caused by perfluorooctanoic acid exposure via antioxidant capacity, biosorption capacity and gut microbiota regulation, Ecotoxicol. Environ. Saf 222 (2021) 112515 10.1016/j.ecoenv.2021.112515. [DOI] [PubMed] [Google Scholar]
- [244].Beale DJ, Nguyen TV, Shah RM, Bissett A, Nahar A, Smith M, Gonzalez-Astudillo V, Braun C, Baddiley B, Vardy S, Host-Gut Microbiome Metabolic Interactions in PFAS-Impacted Freshwater Turtles (Emydura macquarii macquarii), Metabolites 12 (2022) 10.3390/metabo12080747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Planchart A, Green A, Hoyo C, Mattingly CJ, Heavy metal exposure and metabolic syndrome: evidence from human and model system studies, Curr. Environ. Health Rep 5 (2018) 110–124 10.1007/s40572-018-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Song Q, Li J, A review on human health consequences of metals exposure to e-waste in China, Environ. Pollut 196 (2015) 450–461 10.1016/j.envpol.2014.11.004. [DOI] [PubMed] [Google Scholar]
- [247].Rehman K, Fatima F, Waheed I, Akash MSH, Prevalence of exposure of heavy metals and their impact on health consequences, J. Cell Biochem 119 (2018) 157–184 10.1002/jcb.26234. [DOI] [PubMed] [Google Scholar]
- [248].Wirth JJ, Mijal RS, Adverse effects of low level heavy metal exposure on male reproductive function, Syst. Biol. Reprod. Med 56 (2010) 147–167 10.3109/19396360903582216. [DOI] [PubMed] [Google Scholar]
- [249].Rai PK, Lee SS, Zhang M, Tsang YF, Kim KH, Heavy metals in food crops health risks, fate, mechanisms, and management, Environ. Int 125 (2019) 365–385 10.1016/j.envint.2019.01.067. [DOI] [PubMed] [Google Scholar]
- [250].Duan H, Yu L, Tian F, Zhai Q, Fan L, Chen W, Gut microbiota: A target for heavy metal toxicity and a probiotic protective strategy, Sci. Total Environ 742 (2020) 140429 10.1016/j.scitotenv.2020.140429. [DOI] [PubMed] [Google Scholar]
- [251].Arun KB, Madhavan A, Sindhu R, Emmanual S, Binod P, Pugazhendhi A, Sirohi R, Reshmy R, Awasthi MK, Gnansounou E, Pandey A, Probiotics and gut microbiome - Prospects and challenges in remediating heavy metal toxicity, J. Hazard. Mater 420 (2021) 126676 10.1016/j.jhazmat.2021.126676. [DOI] [PubMed] [Google Scholar]
- [252].Breton J, Daniel C, Dewulf J, Pothion S, Froux N, Sauty M, Thomas P, Pot B, Foligne B, Gut microbiota limits heavy metals burden caused by chronic oral exposure, Toxicol. Lett 222 (2013) 132–138 10.1016/j.toxlet.2013.07.021. [DOI] [PubMed] [Google Scholar]
- [253].Ben Issa N, Rajakovic-Ognjanovic VN, Marinkovic AD, Rajakovic LV, Separation and determination of arsenic species in water by selective exchange and hybrid resins, Anal. Chim. Acta 706 (2011) 191–198 10.1016/j.aca.2011.08.015. [DOI] [PubMed] [Google Scholar]
- [254].de Almeida Rodrigues P, Ferrari RG, Kato LS, Hauser-Davis RA, Conte-Junior CA, A systematic review on metal dynamics and marine toxicity risk assessment using crustaceans as bioindicators, Biol. Trace. Elem. Res 200 (2022) 881–903 10.1007/s12011-021-02685-3. [DOI] [PubMed] [Google Scholar]
- [255].Kato LS, Ferrari RG, Leite JVM, Conte-Junior CA, Arsenic in shellfish: a systematic review of its dynamics and potential health risks, Mar. Pollut. Bull 161 (2020) 111693 10.1016/j.marpolbul.2020.111693. [DOI] [PubMed] [Google Scholar]
- [256].Byeon E, Kang HM, Yoon C, Lee JS, Toxicity mechanisms of arsenic compounds in aquatic organisms, Aquat. Toxicol 237 (2021) 105901 10.1016/j.aquatox.2021.105901. [DOI] [PubMed] [Google Scholar]
- [257].Balali-Mood M, Naseri K, Tahergorabi Z, Khazdair MR, Sadeghi M, Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic, Front. Pharmacol 12 (2021) 643972 10.3389/fphar.2021.643972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Chung JY, Yu SD, Hong YS, Environmental source of arsenic exposure, J. Prev. Med. Public Health 47 (2014) 253–257 10.3961/jpmph.14.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Wang X, Mu X, Zhang J, Huang Q, Alamdar A, Tian M, Liu L, Shen H, Serum metabolomics reveals that arsenic exposure disrupted lipid and amino acid metabolism in rats: a step forward in understanding chronic arsenic toxicity, Metallomics 7 (2015) 544–552 10.1039/c5mt00002e. [DOI] [PubMed] [Google Scholar]
- [260].Khanam T, Liang S, Xu S, Musstjab Akber Shah Eqani SA, Shafqat MN, Rasheed H, Bibi N, Shen H, Zhang J, Arsenic exposure induces urinary metabolome disruption in Pakistani male population, Chemosphere 312 (2022) 137228 10.1016/j.chemosphere.2022.137228. [DOI] [PubMed] [Google Scholar]
- [261].Lu K, Abo RP, Schlieper KA, Graffam ME, Levine S, Wishnok JS, Swenberg JA, Tannenbaum SR, Fox JG, Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis, Environ. Health. Perspect 122 (2014) 284–291 10.1289/ehp.1307429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [262].Xue J, Lai Y, Chi L, Tu P, Leng J, Liu CW, Ru H, Lu K, Serum metabolomics reveals that gut microbiome perturbation mediates metabolic disruption induced by arsenic exposure in mice, J. Proteome Res 18 (2019) 1006–1018 10.1021/acs.jproteome.8b00697. [DOI] [PubMed] [Google Scholar]
- [263].Wang C, Deng H, Wang D, Wang J, Huang H, Qiu J, Li Y, Zou T, Guo L, Changes in metabolomics and lipidomics in brain tissue and their correlations with the gut microbiome after chronic food-derived arsenic exposure in mice, Ecotoxicol. Environ. Saf 228 (2021) 112935 10.1016/j.ecoenv.2021.112935. [DOI] [PubMed] [Google Scholar]
- [264].Turner A, Cadmium pigments in consumer products and their health risks, Sci. Total Environ 657 (2019) 1409–1418 10.1016/j.scitotenv.2018.12.096. [DOI] [PubMed] [Google Scholar]
- [265].Genchi G, Sinicropi MS, Lauria G, Carocci A, Catalano A, The effects of cadmium toxicity, Int. J. Environ. Res. Public Health 17 (2020) 3782 10.3390/ijerph17113782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Hong H, Xu J, He H, Wang X, Yang L, Deng P, Yang L, Tan M, Zhang J, Xu Y, Tong T, Lin X, Pi H, Lu Y, Zhou Z, Cadmium perturbed metabolomic signature in pancreatic beta cells correlates with disturbed metabolite profile in human urine, Environ. Int 161 (2022) 107139 10.1016/j.envint.2022.107139. [DOI] [PubMed] [Google Scholar]
- [267].Hudson KM, Shiver E, Yu J, Mehta S, Jima DD, Kane MA, Patisaul HB, Cowley M, Transcriptomic, proteomic, and metabolomic analyses identify candidate pathways linking maternal cadmium exposure to altered neurodevelopment and behavior, Sci. Rep 11 (2021) 16302 10.1038/s41598-021-95630-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Zhang A, Matsushita M, Zhang L, Wang H, Shi X, Gu H, Xia Z, Cui JY, Cadmium exposure modulates the gut-liver axis in an Alzheimer's disease mouse model, Commun. Biol 4 (2021) 1398 10.1038/s42003-021-02898-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Li X, Brejnrod AD, Ernst M, Rykaer M, Herschend J, Olsen NMC, Dorrestein PC, Rensing C, Sorensen SJ, Heavy metal exposure causes changes in the metabolic health-associated gut microbiome and metabolites, Environ. Int 126 (2019) 454–467 10.1016/j.envint.2019.02.048. [DOI] [PubMed] [Google Scholar]
- [270].Rothman JA, Leger L, Kirkwood JS, McFrederick QS, Cadmium and selenate exposure affects the honey bee microbiome and metabolome, and bee-associated bacteria show potential for bioaccumulation, Appl. Environ. Microbiol 85 (2019) 10.1128/AEM.01411-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].He ZL, Yang XE, Stoffella PJ, Trace elements in agroecosystems and impacts on the environment, J. Trace Elem. Med. Biol 19 (2005) 125–140 10.1016/j.jtemb.2005.02.010. [DOI] [PubMed] [Google Scholar]
- [272].Fraga CG, Relevance, essentiality and toxicity of trace elements in human health, Mol. Aspects. Med 26 (2005) 235–244 10.1016/j.mam.2005.07.013. [DOI] [PubMed] [Google Scholar]
- [273].Crossgrove J, Zheng W, Manganese toxicity upon overexposure, NMR Biomed. 17 (2004) 544–553 10.1002/nbm.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Harischandra DS, Ghaisas S, Zenitsky G, Jin H, Kanthasamy A, Anantharam V, Kanthasamy AG, Manganese-induced neurotoxicity: new insights into the triad of protein misfolding, mitochondrial impairment, and neuroinflammation, Front. Neurosci 13 (2019) 654 10.3389/fnins.2019.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Chi L, Gao B, Bian X, Tu P, Ru H, Lu K, Manganese-induced sex-specific gut microbiome perturbations in C57BL/6 mice, Toxicol. Appl. Pharmacol 331 (2017) 142–153 10.1016/j.taap.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Wang H, Zhang S, Yang F, Xin R, Wang S, Cui D, Sun Y, The gut microbiota confers protection in the CNS against neurodegeneration induced by manganism, Biomed. Pharmacother 127 (2020) 110150 10.1016/j.biopha.2020.110150. [DOI] [PubMed] [Google Scholar]
- [277].Zeng X, Zeng Z, Wang Q, Liang W, Guo Y, Huo X, Alterations of the gut microbiota and metabolomics in children with e-waste lead exposure, J. Hazard. Mater 434 (2022) 128842 10.1016/j.jhazmat.2022.128842. [DOI] [PubMed] [Google Scholar]
- [278].Njati SY, Maguta MM, Lead-based paints and children's PVC toys are potential sources of domestic lead poisoning - A review, Environ. Pollut 249 (2019) 1091–1105 10.1016/j.envpol.2019.03.062. [DOI] [PubMed] [Google Scholar]
- [279].Meyer PA, Brown MJ, Falk H, Global approach to reducing lead exposure and poisoning, Mutat. Res 659 (2008) 166–175 10.1016/j.mrrev.2008.03.003. [DOI] [PubMed] [Google Scholar]
- [280].Tchounwou PB, Yedjou CG, Patlolla AK, Sutton DJ, Heavy metal toxicity and the environment, Exp. Suppl 101 (2012) 133–164 10.1007/978-3-7643-8340-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Dujardin CE, Mars RAT, Manemann SM, Kashyap PC, Clements NS, Hassett LC, Roger VL, Impact of air quality on the gastrointestinal microbiome: A review, Environ. Res 186 (2020) 109485 10.1016/j.envres.2020.109485. [DOI] [PubMed] [Google Scholar]
- [282].Celebi Sozener Z, Cevhertas L, Nadeau K, Akdis M, Akdis CA, Environmental factors in epithelial barrier dysfunction, J. Allergy Clin. Immunol 145 (2020) 1517–1528 10.1016/j.jaci.2020.04.024. [DOI] [PubMed] [Google Scholar]
- [283].Qin P, Luo X, Zeng Y, Zhang Y, Li Y, Wu Y, Han M, Qie R, Wu X, Liu D, Huang S, Zhao Y, Feng Y, Yang X, Hu F, Sun X, Hu D, Zhang M, Long-term association of ambient air pollution and hypertension in adults and in children: A systematic review and meta-analysis, Sci. Total Environ 796 (2021) 148620 10.1016/j.scitotenv.2021.148620. [DOI] [PubMed] [Google Scholar]
- [284].Landrigan PJ, Air pollution and health, Lancet Public Health 2 (2017) e4–e5 10.1016/S2468-2667(16)30023-8. [DOI] [PubMed] [Google Scholar]
- [285].Poh TY, Ali N, Mac Aogain M, Kathawala MH, Setyawati MI, Ng KW, Chotirmall SH, Inhaled nanomaterials and the respiratory microbiome: clinical, immunological and toxicological perspectives, Part. Fibre Toxicol 15 (2018) 46 10.1186/s12989-018-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].He L, Zhou YX, Zhang Y, Hang B, Chang H, Schick SF, Celniker SE, Xia Y, Snijders AM, Mao JH, Thirdhand cigarette smoke leads to age-dependent and persistent alterations in the cecal microbiome of mice, Microbiologyopen 10 (2021) e1198 10.1002/mbo3.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Savin Z, Kivity S, Yonath H, Yehuda S, Smoking and the intestinal microbiome, Arch. Microbiol 200 (2018) 677–684 10.1007/s00203-018-1506-2. [DOI] [PubMed] [Google Scholar]
- [288].Fluhr L, Mor U, Kolodziejczyk AA, Dori-Bachash M, Leshem A, Itav S, Cohen Y, Suez J, Zmora N, Moresi C, Gut microbiota modulates weight gain in mice after discontinued smoke exposure, Nature 600 (2021) 713–719 10.1038/s41586-021-04194-8. [DOI] [PubMed] [Google Scholar]
- [289].Stewart CJ, Auchtung TA, Ajami NJ, Velasquez K, Smith DP, De La Garza R 2nd, Salas R, Petrosino JF, Effects of tobacco smoke and electronic cigarette vapor exposure on the oral and gut microbiota in humans: a pilot study, PeerJ 6 (2018) e4693 10.7717/peerj.4693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Scieszka D, Hunter R, Begay J, Bitsui M, Lin Y, Galewsky J, Morishita M, Klaver Z, Wagner J, Harkema JR, Herbert G, Lucas S, McVeigh C, Bolt A, Bleske B, Canal CG, Mostovenko E, Ottens AK, Gu H, Campen MJ, Noor S, Neuroinflammatory and neurometabolomic consequences from inhaled wildfire smoke-derived particulate matter in the western United States, Toxicol. Sci 186 (2022) 149–162 10.1093/toxsci/kfab147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Bailey MJ, Holzhausen EA, Morgan ZEM, Naik N, Shaffer JP, Liang D, Chang HH, Sarnat J, Sun S, Berger PK, Schmidt KA, Lurmann F, Goran MI, Alderete TL, Postnatal exposure to ambient air pollutants is associated with the composition of the infant gut microbiota at 6-months of age, Gut Microbes 14 (2022) 2105096 10.1080/19490976.2022.2105096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Dai S, Wang Z, Yang Y, Du P, Li X, PM2.5 induced weight loss of mice through altering the intestinal microenvironment: Mucus barrier, gut microbiota, and metabolic profiling, J. Hazard. Mater 431 (2022) 128653 10.1016/j.jhazmat.2022.128653. [DOI] [PubMed] [Google Scholar]
- [293].Liang D, Moutinho JL, Golan R, Yu T, Ladva CN, Niedzwiecki M, Walker DI, Sarnat SE, Chang HH, Greenwald R, Jones DP, Russell AG, Sarnat JA, Use of high-resolution metabolomics for the identification of metabolic signals associated with traffic-related air pollution, Environ. Int 120 (2018) 145–154 10.1016/j.envint.2018.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Li Z, Liang D, Ye D, Chang HH, Ziegler TR, Jones DP, Ebelt ST, Application of high-resolution metabolomics to identify biological pathways perturbed by traffic-related air pollution, Environ. Res 193 (2021) 110506 10.1016/j.envres.2020.110506. [DOI] [PMC free article] [PubMed] [Google Scholar]