Abstract
Prophylactic efficacy of two different delivery platforms for vaccination against Mycobacterium avium (M. avium) were tested in this study; a subunit and an RNA-based vaccine. The vaccine antigen, ID91, includes four mycobacterial antigens: Rv3619, Rv2389, Rv3478, and Rv1886. We have shown that ID91+GLA-SE is effective against a clinical NTM isolate, M. avium 2-151 smt. Here, we extend these results and show that a heterologous prime/boost strategy with a repRNA-ID91 (replicon RNA) followed by protein ID91+GLA-SE boost is superior to the subunit protein vaccine given as a homologous prime/boost regimen. The repRNA-ID91/ID91+GLA-SE heterologous regimen elicited a higher polyfunctional CD4+ TH1 immune response when compared to the homologous protein prime/boost regimen. More significantly, among all the vaccine regimens tested only repRNA-ID91/ID91+GLA-SE induced IFN-γ and TNF-secreting CD8+ T cells. Furthermore, the repRNA-ID91/ID91+GLA-SE vaccine strategy elicited high systemic proinflammatory cytokine responses and induced strong ID91 and an Ag85B-specific humoral antibody response a pre- and post-challenge with M. avium 2-151 smt. Finally, while all prophylactic prime/boost vaccine regimens elicited a degree of protection in beige mice, the heterologous repRNA-ID91/ID91+GLA-SE vaccine regimen provided greater pulmonary protection than the homologous protein prime/boost regimen. These data indicate that a prophylactic heterologous repRNA-ID91/ID91+GLA-SE vaccine regimen augments immunogenicity and confers protection against M. avium.
Introduction
Pulmonary lung disease caused by nontuberculous mycobacteria (NTM), is an intensifying health threat. NTM infections can often lead to chronic disease as current treatment options are lengthy, induce toxic side effects, and can lead to incomplete treatment or reinfection[1]. One of our goals is to develop a vaccine for NTM, which could be used either as a prophylactic vaccine in people with specific known risks for pulmonary lung disease, or as an adjunct to drug treatment, enabling a reduction in treatment time and associated severe side-effects.
Despite garnering less attention than the more well-known mycobacteria, Mycobacterium tuberculosis (M.tb), NTM cause significant morbidity and financial health burden in certain regions of the world, including the United States[2]. It is not surprising that NTM, which are ubiquitous in water and soil[3, 4], along with physiologically- or immune-compromised hosts and perpetual exposures, are causing an increase in infection incidences globally[5]. In the U.S., NTM pulmonary disease (NTM-PD) is predominantly caused by Mycobacterium avium complex (MAC), however Mycobacterium abscessus and Mycobacterium kansasii also represent NTM strains that are identified in patients diagnosed with this disease[6–8].
Unfortunately, NTM infections are still hard to diagnose, which can lead to inadequate treatment regimens[9, 10]. Even when appropriately diagnosed, patients have to undergo lengthy, complex, and sometimes poorly tolerated drug regimens over many months to years. The current recommendation for treating pulmonary MAC infection is a 12 month regimen of rifampin, ethambutol and clarithromycin or azithromycin[11], which is only successful in about 50–70% of individuals[12]. Furthermore, individuals with emphysema, cystic fibrosis, bronchiectasis, and rheumatoid arthritis are predisposed to NTM pulmonary disease[8, 10, 13, 14] and treatments for inflammatory diseases including anti-TNFα therapies such as Infliximab, Etanercept, and Adalimumab are added risk factors for both NTM and M.tb[13, 15, 16]. Many bacterial factors also complicate the treatment of NTM infections, particularly with antibiotics, including the bacterial cell wall, multiple drug efflux mechanisms[17–20], and biofilm formation, all of which allow NTM to survive in the host[21–23]. Furthermore, recurrent infections with new strains of mycobacteria or relapse of infection caused by the original organism occur[24]. While new drug strategies, such as liposomal amikacin, have helped[25], they are not likely to eliminate NTM infection and disease[26]. Improved therapeutic options with long-term protection are needed for patients with treatment of refractory NTM lung disease caused by MAC and in those with inherited or acutely acquired host risk factors.
An alternative approach for improved therapies against NTM disease is host-directed therapy, through the design and development of candidate vaccines against NTM. By including a host-directed vaccine approach as an adjunct to drug treatment, this could lead to a reduction in the incidence of disease and a decrease in the severe side effects associated with prolonged drug treatment[27, 28]. The evaluation of new vaccine candidates for pulmonary NTM is hampered by the lack of defined correlates of protection and an incomplete understanding of immune responses required for induction of long-lasting immunity. We hypothesize that ID91 is a promising candidate for studying these mechanisms. The ID91 vaccine contains proteins that are virulence factors and are produced under different conditions in the host: of the four M.tb proteins include Rv3619 (esxV, EsX family), Rv2389 (produced under hypoxia, resuscitation factor D), Rv3478 (PE/PPE family member), and Rv1886 (Ag85B, mycolyl-transferase). The recombinant fusion protein, ID91, combined with a synthetic TLR4 agonist adjuvant [glucopyranosyl lipid adjuvant formulated in an oil-in-water stable nano-emulsion (GLA-SE)], affords robust protection against M. avium 2-151 smt infection in both wildtype C57BL/6 and immunocompromised Beige mice[29].
The current M.tb vaccine pipeline heavily relies on a host response that includes CD4+ T helper 1 (TH1) type responses. While the correlates of protection against either M.tb or NTM are not fully understood[30, 31], there is ample evidence shown from our group and others to suggest that IFNγ-producing CD4+ T cells alone are not sufficient for protective efficacy[32–35]. The vaccine development field targeting mycobacterial infections has also demonstrated the significance of CD8+ T cells, particularly in M.tb infection control[36–41], however this cell population remains an underappreciated target for vaccine-induced efficacy. Recently, Darrah et al. showed that intravenous immunization with BCG induced robust protection in 9 out of 10 macaques, and unlike the groups that received BCG through other routes of immunization, elicited both CD4+ and CD8+ T cell IFN-γ, TNF, IL-2, or IL-17 cytokine responses[42]. We hypothesize that both CD4+ and CD8+ T cells will be important drivers of protection against NTM, similar to that required for vaccine-mediated protection against M.tb.
Recently, during the COVID-19 pandemic, RNA-based vaccines have been employed to address the needs of a rapid turnaround for vaccine development as well as reducing costs, both of which are attractive for designing vaccines targeting global populations. RNA vaccine platforms known for stimulating CD8+ T cell response have not been leveraged for NTM vaccines. However, a recent study reported that that exhaustion of CD8+ T cells increased bacterial burden in patients with NTM lung disease[43], which prompted us to evaluate this approach. Previous studies have demonstrated robust humoral and cellular responses, including CD8+ T cell responses, induced by replicon RNA (repRNA) vaccines delivered via multiple modalities[44–47]. Additionally, a recent study from our group has shown that both CD4+ and CD8+ proliferative responses were generated, with a bias to a higher frequency of CD4+ T cells after protein vaccination and a CD8+ biased T cell response following an ID91-repRNA vaccination (https://doi.org/10.1101/2022.02.23.481669, bioRxiv). Furthermore, the subunit platform, or a combined platform approach with both RNA and protein, are protective against M.tb H37Rv, providing proof-of-concept that the repRNA-ID91 vaccine can contribute to generating a protective immune response against mycobacterial infections (https://doi.org/10.1101/2022.02.23.481669, bioRxiv).
While homologous prime/boost strategies, which are repeated administrations of the same vaccine, have been proven to be effective in augmenting humoral responses[48, 49], they appear to be relatively less efficient at enhancing cellular immunity, likely because prior immunity to the vaccine tends to prematurely sequester robust antigen presentation and the generation of diverse inflammatory signals for T cells. One strategy to overcome this limitation has been the heterologous prime/boost approach, which is the sequential administration of vaccines using different antigen delivery systems, like recombinant live vectors, DNA or RNA vaccines, or adjuvanted subunit vaccines. Indeed, previous studies have demonstrated improved effector T cell responses following heterologous prime/boost compared to homologous strategies for pathogens such as HIV, malaria, and even tuberculosis[50–55]. However, vaccine design against NTM has not yet capitalized on this coordinated effort to control bacterial burden and disease by engaging multiple arms of the innate and adaptive immune system. Therefore, in this study we evaluated repRNA-ID91 given as a homologous prime/boost or a heterologous prime/boost strategy with a protein/adjuvant vaccine candidate, to determine which regimen induces an optimal protective response against M. avium challenge in mice.
Material and Methods
Preclinical Animal Model
Male and female C57BL/6 bg/bg (beige) mice were bred in-house and used for experiments at 4–6 weeks of age. Mice were housed at the Seattle Children’s Research Institute (SCRI) biosafety level 3 (BSL-3) animal facility under pathogen-free conditions and were handled in accordance with experimental protocols that were approved by the SCRI Institutional Animal Care and Use Committee (IACUC). All methods were carried out in accordance with relevant guidelines and regulations with respect to animal welfare. Mice were infected using an aerosol challenge with 104 CFU of M. avium 2-151 smooth transparent (smt) (obtained from Dr. Diane Ordway at Colorado State University, Fort Collins, CO) using a Glas-Col aerosol infection chamber. Twenty-four hours post challenge the lungs of three mice were homogenized and plated on Middlebrook 7H10 agar (Fisher Scientific) to confirm and quantify the delivery of M. avium 2-151 smt.
Vaccines and adjuvants
Cohorts of mice were immunized intramuscularly (i.m.) two times four weeks apart in a homologous or heterologous prime/boost vaccine regimen, with the final immunization occurring 4 weeks before challenge with M. avium 2-151 smt.
Mice received either saline alone, or vaccinations containing 0.5 μg/dose of ID91 recombinant fusion protein combined with 5.0 μg/dose GLA-SE, as previously published[29, 56, 57]. A separate cohort of mice were immunized with 1.0 μg/dose of ID91 replicon RNA (repRNA-ID91) formulated in LION™ delivery system[45]. Homologous or heterologous vaccine regimens were administered using the doses and regimens outlined above. A separate cohort of mice were immunized once intradermally (i.d.) with 104 CFU of bacillus Calmette–Guérin (BCG) (Pasteur strain, AERAS) 8 weeks before challenge; mice received 50 μl at the base of the tail.
Bacterial burden
At 24 hours and 6-weeks post infection with M. avium 2-151 smt, 3 mice total or 7 mice per group, respectively, were euthanized and bacterial counts were enumerated as described previously[29]. Briefly, mice were euthanized with CO2, and lung, spleen, and liver tissues from infected animals were isolated and homogenized in 5 mL of either RPMI + FBS (lung) or PBS + 10% Tween-80 (spleen and liver) (Sigma-Aldrich, St. Louis, Missouri, USA) using an Omni tissue homogenizer (Omni International, Kennesaw, GA, USA). Serial dilutions of homogenate were prepared in PBS + 10% Tween-80 and aliquots were plated on Middlebrook 7H10 agar plates and subsequently incubated at 37 °C and 5% CO2 for 2–3 weeks before colonies were counted. Bacterial burden, indicated as colony forming units/mL (CFU/mL), was calculated per organ and is presented as Log10 values. Reduction in the bacterial burden was calculated as the difference in mean Log10 values between the groups assessed as shown.
Histopathology and image analysis
Lung accessory lobes were collected from infected mice 6 weeks after M. avium 2-151 smt challenge and were perfused and stored in 10% normal buffered formalin. Fixed lungs were embedded in paraffin at the University of Washington Histology and Imaging (Seattle, WA, USA) as previously described[29]. Paraffin embedded lungs were sent to Dr. Brendan Podell, a boardcertified veterinary pathologist at Colorado State University. Lungs were processed and sections were H&E-stained and scanned at 20X magnification using a Vectra Polaris slide scanner (Akoya Biosciences, Marlborough, MA). Visiopharm software (Horsholm, Denmark) was used for image analysis as previously published[58]. For each tissue section, a region of interest (ROI) was generated at a low magnification with a custom tissue detecting algorithm using decision forest training and classification to differentiate tissue versus background based on color and area. Lesions were identified within tissue ROI’s at a high magnification with an additional custom-made algorithm using decision forest training and classification based on staining intensity, color normalization and deconvolution, area, and morphological features. Percent lesion calculations were integrated into the same algorithm and calculated from tissue area and lesion area as designated by the ROI and lesions detected.
Multiplex Cytokine analysis
Serum samples were collected from terminal bleeds immediately following euthanasia 1-week after the boost immunization and 6-weeks after infection with M. avium. Circulating cytokines were measured using the Meso Scale Discovery (MSD) mouse V-PLEX Proinflammatory Panel (Meso Scale Discovery, Rockville, MD) that included the following analytes: IFN-γ, IL-1β, IL-2, IL-4, IL-5, IL-6, IL-10, IL-12p70, TNF-α, and KC/GRO (a neutrophil-activating protein that shows hematopoietic activity). Serum was added in a 1:2 dilution to plates pre-coated with cytokine-specific antibodies following manufacturer’s instructions. Concentrations of each of the cytokines were determined for all experimental samples using the MSD Discovery Workbench analysis software (Meso Scale Discovery, Rockville, MD). Values below the limit of detection of the assay were assigned a value half that of the lowest standard. The levels of IL-4 and IL-12p70 were below limit of detection for all animals at both time points. The results are reported in pg/ml as the means from duplicate wells.
Antibody ELISAs
Serum samples were diluted 1:100 and added to plates coated with 2 μg/mL of ID91 or Ag85B as previously described[29]. After an overnight incubation, plates were exposed to HRP-conjugated Total IgG, IgG1, or IgG2c antibodies (Southern Biotech, Birmingham, AL). Plates were then developed with tetramethylbenzidine (TMB) substrate (KPL/Seracare, Milford, MA) and stopped with 1 N H2SO4 (Fisher). Plates were read on a SpectraMax iD3 microplate reader (Molecular Devices, San Jose, CA) at 450nm with 570nm background subtraction. Reciprocal dilutions corresponding to endpoint titers (EPT) were determined with GraphPad Prism 9.4.1 (GraphPad Software, San Diego, CA) with a cutoff value of saline treated serum control wells +2SD.
Flow Cytometry
Intracellular flow cytometry (ICS) was performed on splenocytes 1-week post boost immunization. Samples were lysed, incubated, washed, and stimulated as previously described[29, 58]. Briefly, cells were stimulated with media alone, 10 μg/mL of recombinant ID91, 10 μg/mL of ID91 component proteins (Rv1886, Rv3619, Rv3478, Rv2389), or 1 μg/mL phorbol myristate acetate (PMA) (Calbiochem) + 1 μg/mL ionomycin (Sigma-Aldrich, St. Louis, Missouri, USA) and incubated at 37 °C. Culture media stimulations also contained fluorescently labeled anti-mouse CD107a (1D4B, BioLegend). After two hours of incubation, 1 μg/μL of GolgiPlug (BD Biosciences) was added and samples were incubated at 37 °C for an additional 10 h. Samples then remained at 4°C until staining. After stimulation, samples were stained for markers of interest using fluorescent conjugated antibodies. Primary surface staining included: anti-mouse CD4 (clone RM4–5, BioLegend), CD8 (clone 53–6.7, BioLegend), CD44 (clone IM7, eBiosciences), and 1 μg/mL of Fc receptor block anti-CD16/CD32 (clone 93, eBioscience) in PBS with 1% bovine serum albumin (BSA). Cells were then washed with PBS/BSA and fixed using BD Biosciences Fix/Perm reagent for 20 min at RT. Intracellular staining was performed in Perm/Wash (BD Biosciences) reagent with anti-mouse IFN-γ (clone XMG1.2, Invitrogen), IL-2 (clone JES6-5H4, eBioscience), TNF-α (clone MP6-XT22, eBioscience), and CD154 (PerCP-710, clone MR7, eBioscience) for 15 min at RT. All antibodies were used at 1:100 dilution. Samples were acquired on a LSRII flow cytometer (BD Biosciences) with FACSDiva software (BD Biosciences). Gating strategies were based on our previous publications[29, 56, 59] (and shown in Supplementary Fig 1), and analysis was performed using FlowJo v10.6.1 (BD Biosciences) and SPICE (National Institutes of Health, http://exon.niaid.nih.gov/spice).
Statistical Analysis
Circulating cytokine levels and humoral immune responses at both post-boost and post-challenge timepoints were compared between groups using two-way ANOVA with Dunnett’s multiple comparisons test. Bacterial burden, single-cytokine-producing T cells and polyfunctional-cytokine-producing T cells were assessed using a one-way ANOVA with Dunnett’s multiple comparisons test between vaccinated groups and saline control. All the above analyses were performed using GraphPad Prism 9.4.1 (GraphPad Software, San Diego CA, United States).
Significant differences are labeled accordingly in the figures where *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001, as previously described by our group[29, 58].
Results
Study design
In order to evaluate the efficacy of both the homologous and heterologous prime/boost vaccine strategies with our repRNA-ID91 vaccine candidate and adjuvanted ID91+GLA-SE subunit protein vaccine candidate against M. avium challenge, we employed the use of the beige mouse model that has been previously established in our group[29]. Since NTM infection and disease burden are more pronounced in immunocompromised populations, the beige mouse strain was used as the representative susceptible mouse model. Indeed, beige mice have been considered the standard mouse model for slow-growing NTM, and are extremely susceptible to MAC infection due to reduced bactericidal activity of granulocytes, decreased neutrophil recruitment, deficiency in NK cells, and minor defect/delay in humoral responses[60–66]. In this study we used our preclinical vaccine candidate, ID91, a fusion of four M.tb proteins: Rv3619 (esxV; ESAT6-like protein), Rv2389 (RpfD), Rv3478 (PPE60) and Rv1886 (Ag85B), as (1) it has already been proven to be effective against an NTM clinical isolate, (2) ID91 antigens formulated in GLA-SE were verified to be immunogenic in beige mice [29], and (3) thorough protein homology analysis indicated that protein percent identity ranged between 88% and 85% for both Rv3619/esxV and Rv1886/Ag85B, respectively, while Rv2389 (RpfD) and Rv3478 (PPE60) have 49% and 47% protein percent identity, respectively, when compared to the M.avium homolog[29]. Also, using the IEDB database to predict CD4+ T cell epitopes, we have previously shown an overlap in the predicted MHCII binding peptides between M.tb and M. avium peptides that can be seen in mice with an H2-1AB allelic background and in humans, particularly with Rv3619 and Rv1886, as expected based on the high sequence identity with M.avium[29, 67]. Therefore, ID91 was included as a fusion protein formulated with GLA-SE (ID91+GLA-SE), and a replicon-RNA (repRNA) [44] formulated in LION™, which is an RNA delivery technology based on HDT Bio’s proprietary LION™ formulation (repRNA-ID91), that was designed for use in this study (Fig 1A). Since anti-vector immunity has been detrimental for promising RNA vaccine candidates, partnering the repRNA with lipid-based delivery formulations provides stability and protection from RNAses and removes the requirement for viral delivery. To evaluate the performance of ID91 fusion protein + GLA-SE and repRNA-ID91, beige mice were immunized with both vaccine candidates in homologous or heterologous prophylactic prime/boost strategies two times, four weeks apart (Fig 1B). Vaccine induced preliminary immunogenicity was assessed one week post boost immunization. Four weeks post boost, mice were aerosol challenged with clinical isolate M. avium 2-151 smt and vaccine induced protection was assessed six weeks post infection (Fig 1C).
Figure 1. Overview of vaccine strategies and experimental design.
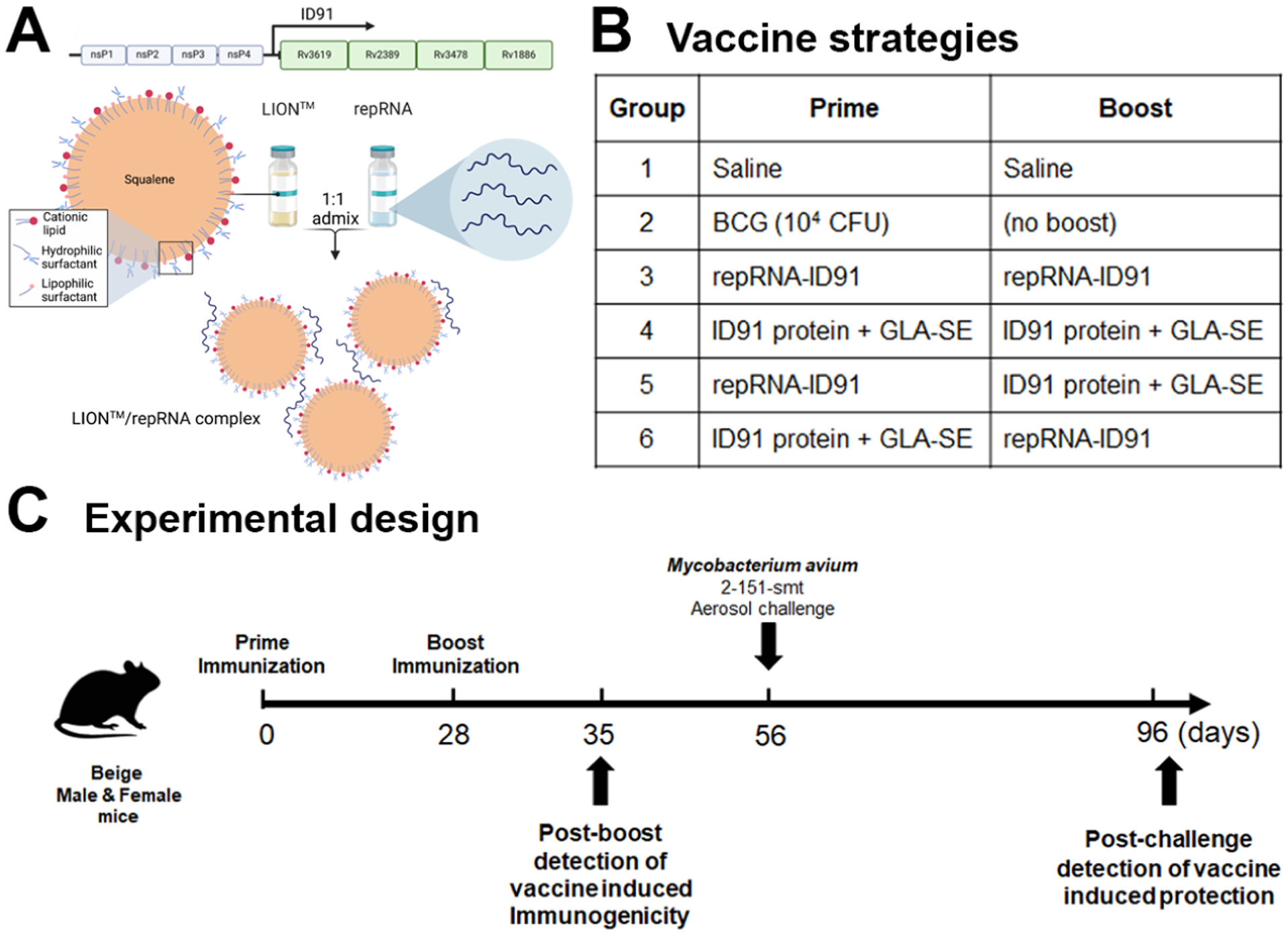
(A) Schematic of ID91 replicon RNA (repRNA) formulated in a Lipid InOrganic Nanoparticle (LION™) delivery system. (B) Prime/boost vaccine strategies received by each group. (C) Timeline of immunizations and M. avium 2-151 smt aerosol challenge. Experimental samples were collected at two time-points: D35 (one week post boost immunization) and D96 (six weeks post M. avium challenge).
Prophylactic Immunogenicity and Protective Efficacy
Systemic Proinflammatory Response
Prior reports have shown that infection with NTM leads to infection of macrophages, monocytes, and dendritic cells and a subsequent decrease in TNF-α, IFN-γ, and other proinflammatory cytokines[68–71]. Conversely, there is an increase in IL-10 production which promotes intracellular NTM survival within the host[72]. Therefore, we first evaluated vaccine-induced circulating IFN-γ, TNF-α, IL-1β, IL-2, IL-6, KC/GRO, IL-10, and IL-5 levels in serum samples collected 1-week post boost immunization and 6-weeks post M. avium challenge from cohorts of mice that were immunized with the homologous or heterologous prophylactic prime/boost vaccine strategies and then challenged with a LDA of M. avium 2-151 smt (Fig 2A–H). One week after the second immunization, we observed a significant increase in circulating IL1-β and IL-6 levels in beige mice immunized with both the homologous ID91+GLA-SE (green dots) and heterologous repRNA-ID91/ID91+GLA-SE (orange dots) vaccine strategies (Fig 2C, E). Interestingly, we also observed a significant increase in circulating IL-10 levels in the serum of both the BCG (gray dots) and homologous ID91+GLA-SE immunized groups (Fig 2G). Six weeks post challenge with M. avium, we observed a significant increase in circulating IFN-γ, TNF-α, IL-2, and KC/GRO levels in beige mice immunized with both the homologous ID91+GLA-SE and heterologous repRNA-ID91/ID91+GLA-SE vaccine strategies (Fig 2A, B, D, and F). However, the heterologous vaccine regimen elicited 1 to 2-fold higher circulating levels of proinflammatory cytokines (IFN-γ, TNF-α, and IL-2) than the homologous regimen. Furthermore, only the heterologous repRNA-ID91/ID91+GLA-SE vaccine strategy led to a significant increase in circulating IL-6 levels post M. avium challenge. No significant changes were observed in circulating IL-5 levels post boost or post challenge in any of the groups. These data indicate that the heterologous RNA prime/protein boost vaccine strategy elicits a strong ex vivo systemic proinflammatory response both post-boost and post-challenge with M. avium.
Figure 2. Heterologous RNA prime/protein boost vaccine strategy drives a robust systemic proinflammatory response both post-boost and post- M. avium challenge.
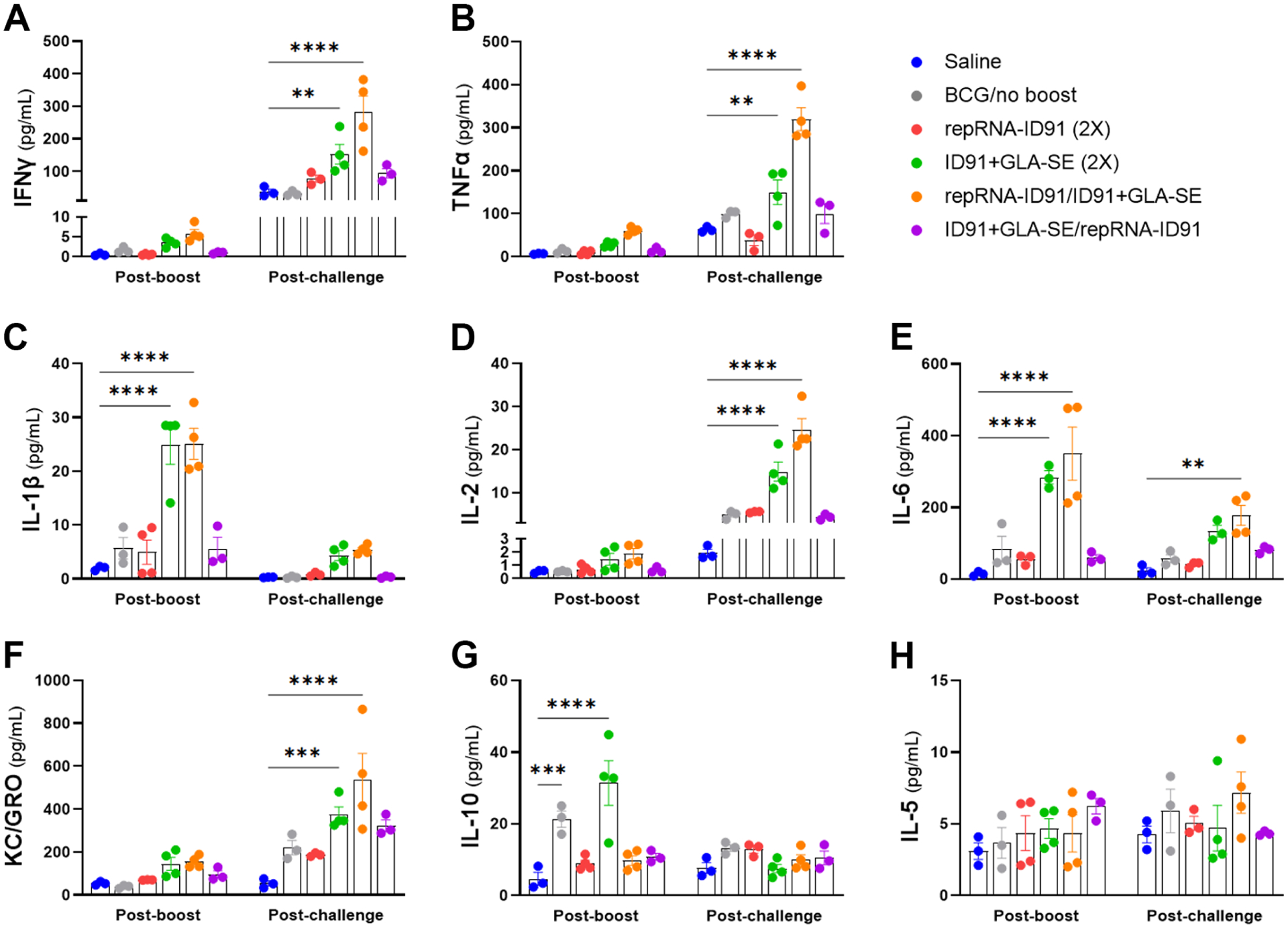
Circulating proinflammatory cytokine levels were measured in serum samples collected one week post boost immunization and six weeks post M. avium challenge. (A-H) Concentration of IFN-γ, TNF-α, IL-1β, IL-2, IL-6, KC/GRO, IL-10, and IL-5 (measured in pg/mL) at both time points. Bars show mean ± SEM, dots represent individual mice, n = 3–4/group. Asterisks indicate statistical significance compared to the saline alone group, where **p<0.01, ***p < 0.001, and ****p < 0.0001 using two-way ANOVA with Dunnett’s multiple comparisons test.
Humoral Immune Response
As antibody-mediated responses could play a role in protection against M. avium, we next evaluated humoral vaccine immunogenicity and protection in cohorts of mice that were immunized with the homologous or heterologous prophylactic prime/boost vaccine strategies and then challenged with M. avium 2-151 smt (Fig 3). Serum samples were collected 1-week post boost immunization and 6-weeks post M. avium challenge and assessed for ID91 -specific and Ag85B-specific humoral immune response (Ag85B is included as a component of the ID91-based vaccine platform and is expressed by BCG). At both timepoints, beige mice immunized with the homologous ID91+GLA-SE (green dots) or heterologous repRNA-ID91/ID91+GLA-SE (orange dots) vaccine strategies demonstrated the most robust ID91 -specific (Fig 3A–C) and Ag85B-specific (Fig 3D–F) total IgG, IgG1, and IgG2c antibody responses, whereas the other vaccine regimens induced much weaker or non-existent ID91- and Ag85B-specific humoral responses. Interestingly however, the heterologous vaccine regimen induced stronger trending IgG1 responses to both antigens post M. avium challenge. These data indicate that the heterologous RNA prime/protein boost strategy induces a higher magnitude ID91- and Ag85B-specific humoral response both post-boost and post-challenge with M. avium.
Figure 3. Heterologous RNA prime/protein boost vaccine strategy induces ID91 and Ag85B antigen-specific humoral immune responses post-boost and post M. avium challenge.
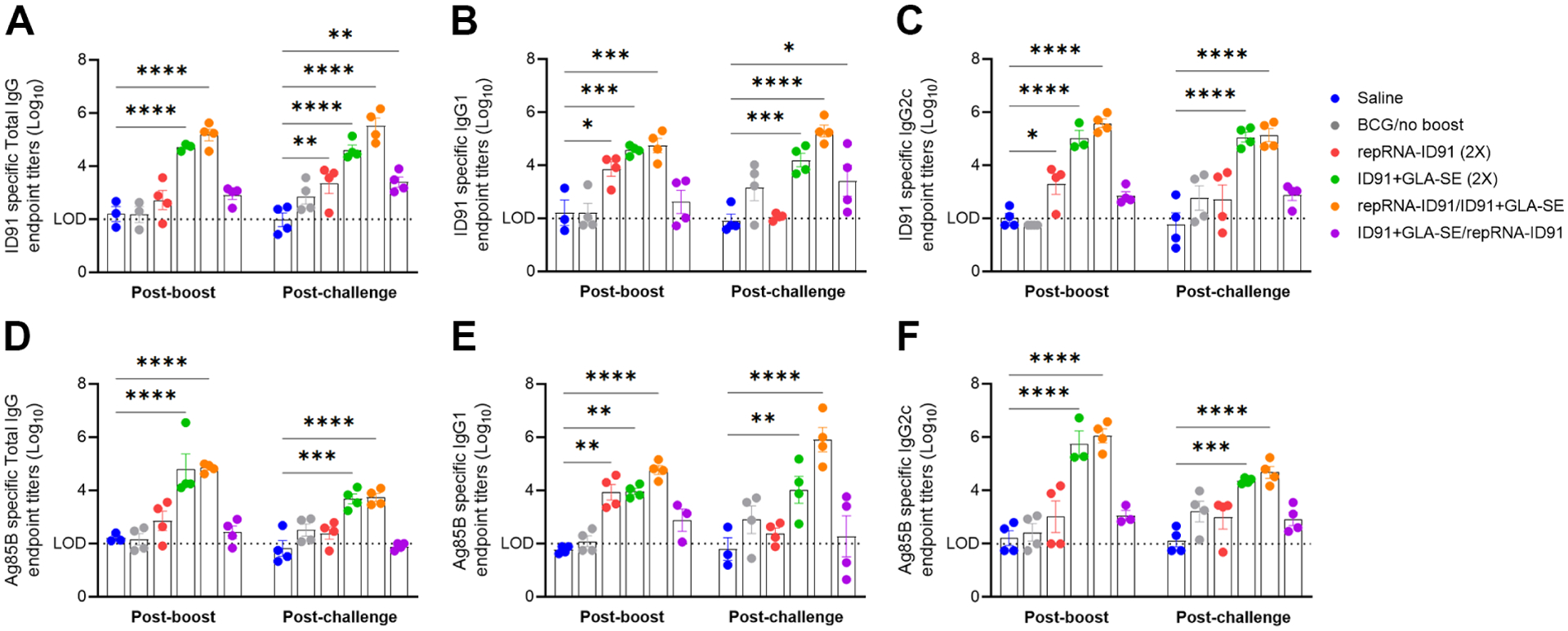
Serum samples were collected one week post boost immunization and six weeks post M. avium challenge and were evaluated for: (A-C) ID91 antigen specific Total IgG (A), IgG1 (B), and IgG2c (C) responses; (D-F) Ag85B-specific Total IgG (D), IgG1 (E), and IgG2c (F) responses. Log10 endpoint titer (EPT) is shown (LOD = limit of detection). Bars show mean ± SEM, dots represent individual mice, n = 3–4/group. Asterisks indicate statistical significance compared to saline alone cohort, where **p < 0.01, ***p < 0.001, and ****p < 0.0001 using two-way ANOVA with Dunnett’s multiple comparisons test.
Cellular Immune Responses
Next, to determine vaccine mediated ID91-specific CD4+ TH1 immune responses, splenocytes were isolated from immunized cohorts one week post boost immunization and restimulated ex vivo with ID91 fusion antigen or ID91 component antigens (Rv1886, Rv3619, Rv3478, Rv2389). These cells were then stained and evaluated by flow cytometry for CD4+ TH1 responses (Fig 4). In response to ID91 stimulation, we observe that the activation marker CD154 is significantly elevated in mice immunized with both the homologous ID91+GLA-SE (green dots) and heterologous repRNA-ID91/ID91+GLA-SE (orange dots) vaccine strategies compared to the saline (blue dots) group (Fig 4A). When assessing TH1 cellular immunity, both vaccine regimens also elicited a robust CD4+ TH1 response, including production of IFN-γ and TNF-α cytokines. However, only the heterologous vaccine regimen induced a significantly higher frequency of IL-2 producing CD4+CD44+ cells compared to the saline group (Fig 4A). Furthermore, polyfunctional ID91-specific CD4+ T cells expressing two or more cytokines (which included the CD154 activation marker and/or three cytokines including IFN-γ, IL-2 and TNF-α) were significantly increased in both the homologous ID91+GLA-SE (green dots) and heterologous repRNA-ID91/ID91+GLA-SE (orange dots) immunized groups compared to the other vaccine strategies (Fig 4B), although only the heterologous vaccine regimen induced triple TH1 cytokine-expressing (IFN-γ, IL-2 and TNF-α) CD4+ T cells (Fig 4B). Interestingly, when stimulated with ID91 component antigens, the homologous protein prime/boost vaccine regimen only induced CD4+ TH1 responses to Rv1886 and Rv3619 (Fig 4C–D), while the heterologous repRNA-ID91/ID91+GLA-SE vaccine regimen elicited robust CD4+ TH1 immune responses to all four ID91 component proteins (Fig 4C–F).
Figure 4. Heterologous RNA prime/protein boost vaccine strategy elicits robust ID91-specific CD4+ TH1 immune responses post-boost.
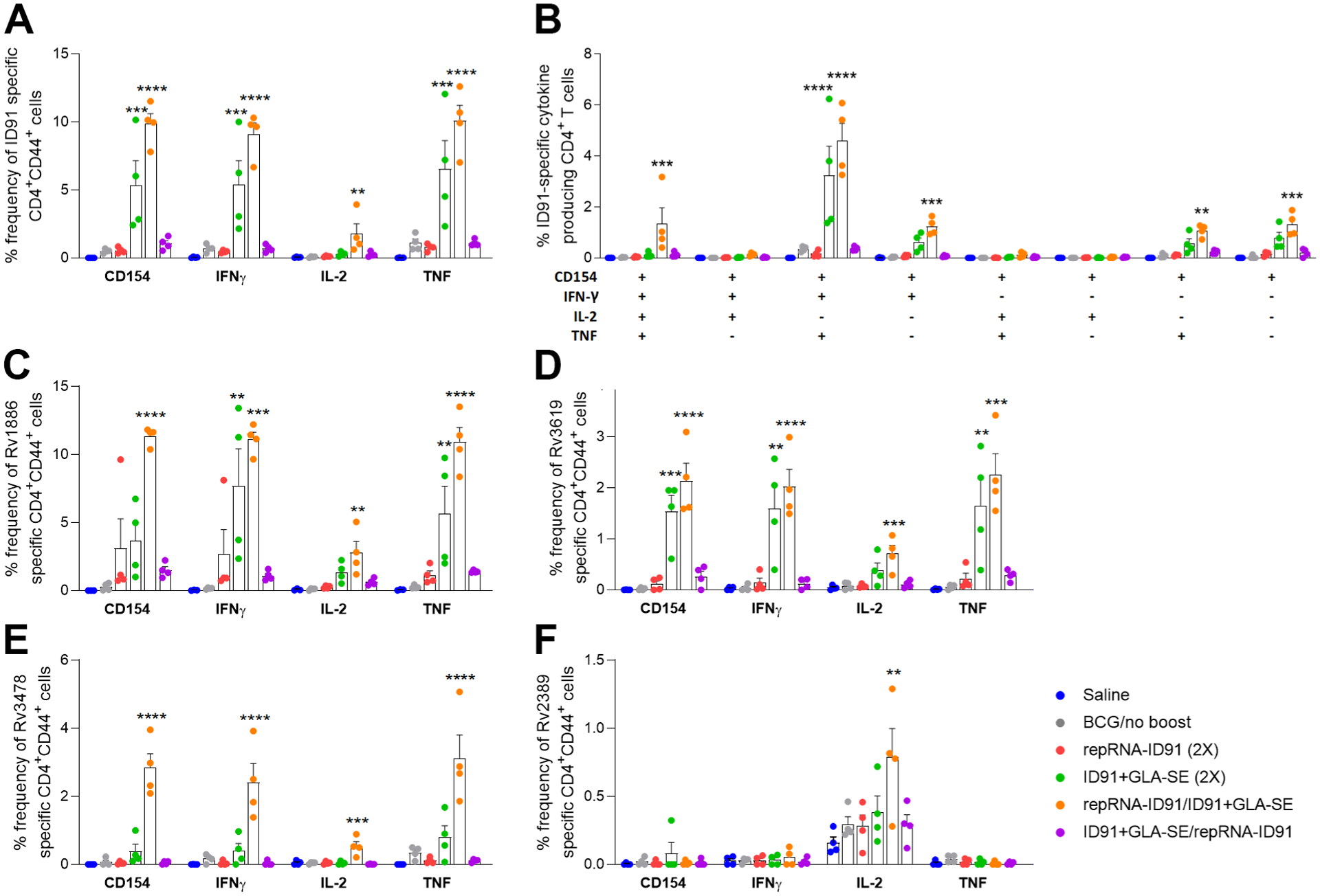
Splenocytes were cultured from all six groups of mice one week post boost immunization and stimulated with ID91 or ID91 components (Rv1886, Rv3619, Rv3478, Rv2389) ex vivo and evaluated for CD4+ T cell responses by intracellular cytokine staining flow cytometry including: (A) Percent frequency of CD4+CD44+ ID91-specific single-cytokine-producing cells; (B) ID91-specific CD4+CD44+ polyfunctional TH1 cytokine-producing cells; (C-F) Percent frequency of CD4+CD44+ Rv1886 (C), Rv3619 (D), Rv3478 (E), and Rv2389 (F) specific single-cytokine-producing cells. Bars show mean ± SEM, dots represent individual mice, n = 4/group. Asterisks indicate statistical significance, where **p < 0.01, ***p < 0.001, and ****p < 0.0001 using one-way ANOVA with Dunnett’s multiple comparisons test.
Since CD8+ T cells represent an underappreciated target for mycobacterial vaccine-induced efficacy endpoints, and the repRNA vaccine platform has been shown to induce CD8+ T cell responses, we next assessed vaccine-induced ID91-specific CD8+ immune responses in splenocytes isolated from immunized cohorts one week post boost immunization following ex vivo stimulation with ID91 (Fig 5). Significantly, only the heterologous repRNA-ID91/ID91+GLA-SE vaccine strategy (orange dots) elicited robust CD8+ cytokine responses, including production of IFN-γ and TNF-α cytokines (Fig 5A). Furthermore, polyfunctional ID91-specific CD8+ T cells expressing two or more cytokines (including CD154 and/or three cytokines including IFN-γ, IL-2 and TNF-α) were also significantly increased in the heterologous repRNA-ID91/ID91+GLA-SE (orange dots) immunized group compared to the other vaccine strategies (Fig 5B). Interestingly, none of the vaccine strategies induced triple cytokine-expressing (IFN-γ, IL-2 and TNF-α) CD8+ T cells. We also did not observe any changes in CD4+ or CD8+ CD107a expression, a marker of CD8+ T cell degranulation, between groups (data not shown). Although we did not observe significant CD8+ cytotoxicity measured by CD107a, the significant CD8+ T cell cytokine data indicates that the heterologous RNA prime/protein boost strategy is the most effective regimen for generating both CD4+ and CD8+ immune responses.
Figure 5. Heterologous RNA prime/protein boost vaccine strategy is the most effective regimen for CD8+ T cell responses post-boost.
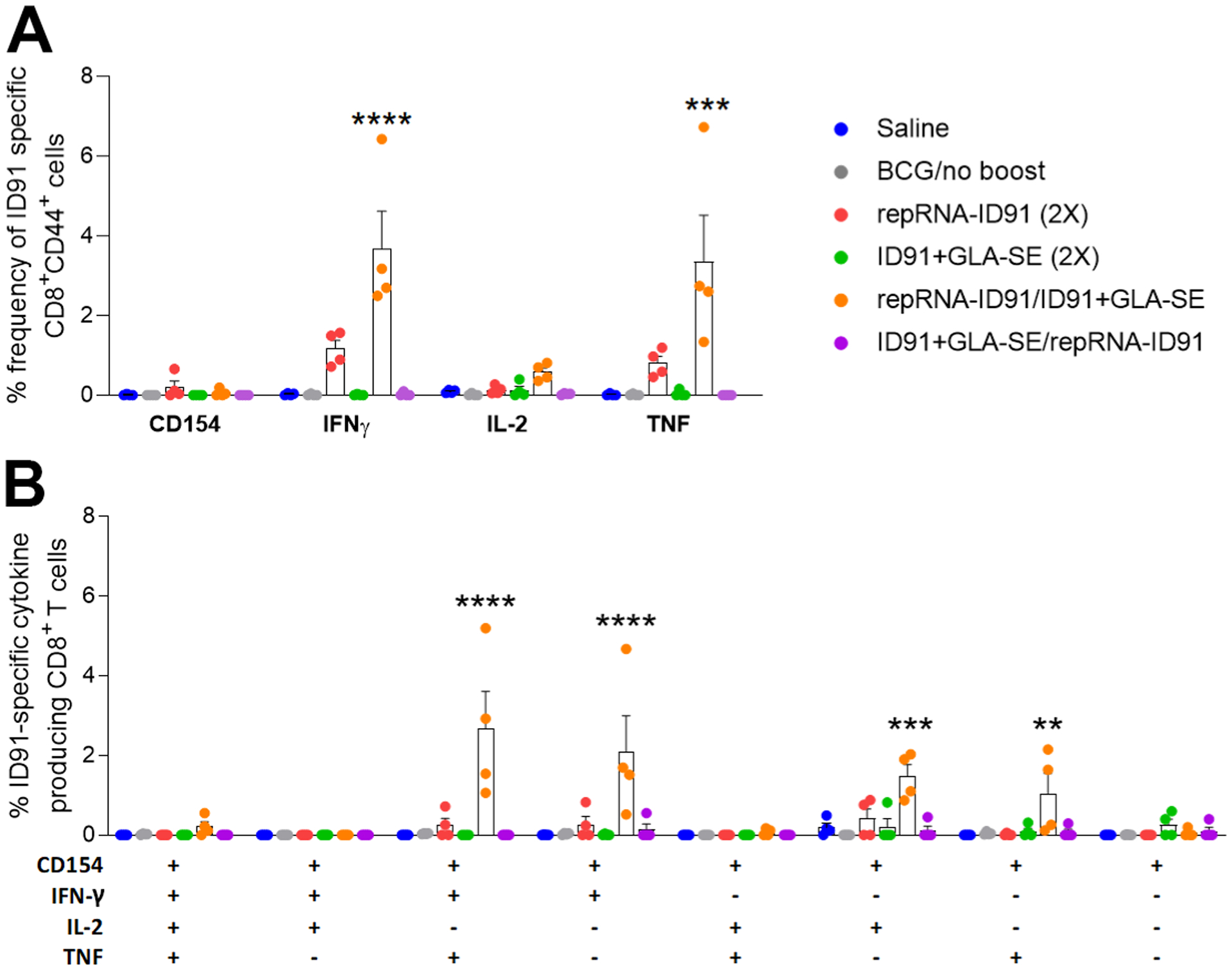
Splenocytes were cultured one week post boost immunization and stimulated with ID91 ex vivo and evaluated for CD8+ T cell responses by intracellular cytokine staining flow cytometry including: (A) Percent frequency of CD8+CD44+ ID91-specific single-cytokine-producing cells; (B) ID91-specific CD8+CD44+ polyfunctional cytokine-producing cells. Bars show mean ± SEM, dots represent individual mice, n = 4/group. Asterisks indicate statistical significance, where **p < 0.01, ***p < 0.001, and ****p < 0.0001 using one-way ANOVA with Dunnett’s multiple comparisons test.
Prophylactic Pulmonary Protection
Six-weeks following aerosol M. avium 2-151 smt infection, a cohort of seven mice per group were assessed for bacterial burden within the lung, spleen, and liver as the primary endpoints for protective efficacy. Interestingly, while all vaccine strategies induced significant prophylactic protection as observed by a reduction of bacterial burden in the lung, spleen, and liver of beige mice compared to the saline group (Fig 6A–C), the heterologous repRNA-ID91/ID91+GLA-SE vaccine regimen provided greater pulmonary protection in beige mice compared to the homologous protein prime/boost regimen, as shown by a significant decrease in bacterial load in the lung (7.550 Log10 CFU vs. 8.038 Log10 CFU, respectively; Fig 6A).
Figure 6. All prime/boost vaccine strategies provide prophylactic pulmonary protection against M. avium in beige mice.
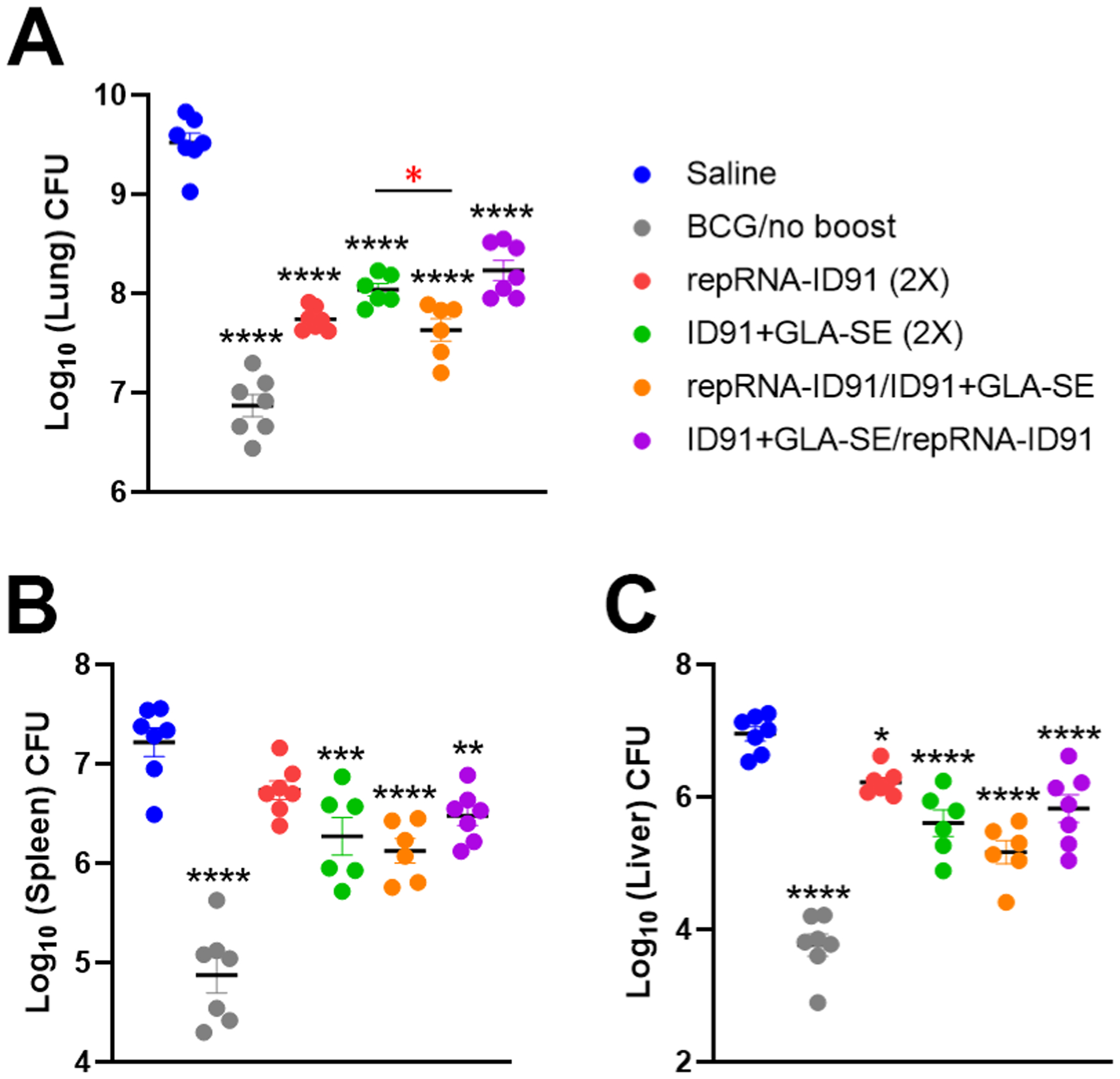
Beige mice were infected with M. avium 2-151 smt by aerosol route four weeks post final immunization. (A-C) Bacterial burden was assessed by colony forming unit (CFU) in lung (A), spleen (B), and liver (C) organ homogenates six weeks post challenge. CFU means were compared between each group using one-way ANOVA with Dunnett’s multiple comparisons test. Black line and error bars show mean ± SEM, dots represent individual mice, n = 6–7/group. Asterisks indicate statistical significance, where *p < 0.05, **p < 0.01, ***p < 0.001 and ****p < 0.0001.
We next evaluated pulmonary immunopathology six-weeks post M. avium 2-151 smt infection, including the assessment of lung lesions, using Visiopharm software and a novel, custom-made algorithm using decision forest training and classification (at Colorado State University) to detect areas of inflammation on H&E stained sections from the accessory lung lobes (Fig 7A–F). This high throughput workflow incorporates the entire accessory lobe in the unbiased analysis for a more progressive and reproducible scoring system. Consistent with the reduction in bacterial burden, all prophylactic prime/boost vaccine strategies led to decreased lung immunopathology as shown by a significantly lower percentage of pulmonary lesions in all vaccinated groups compared to saline (Fig 7G).
Figure 7. All prophylactic prime/boost vaccine strategies decrease lung immunopathology in Beige mice post challenge with M. avium.
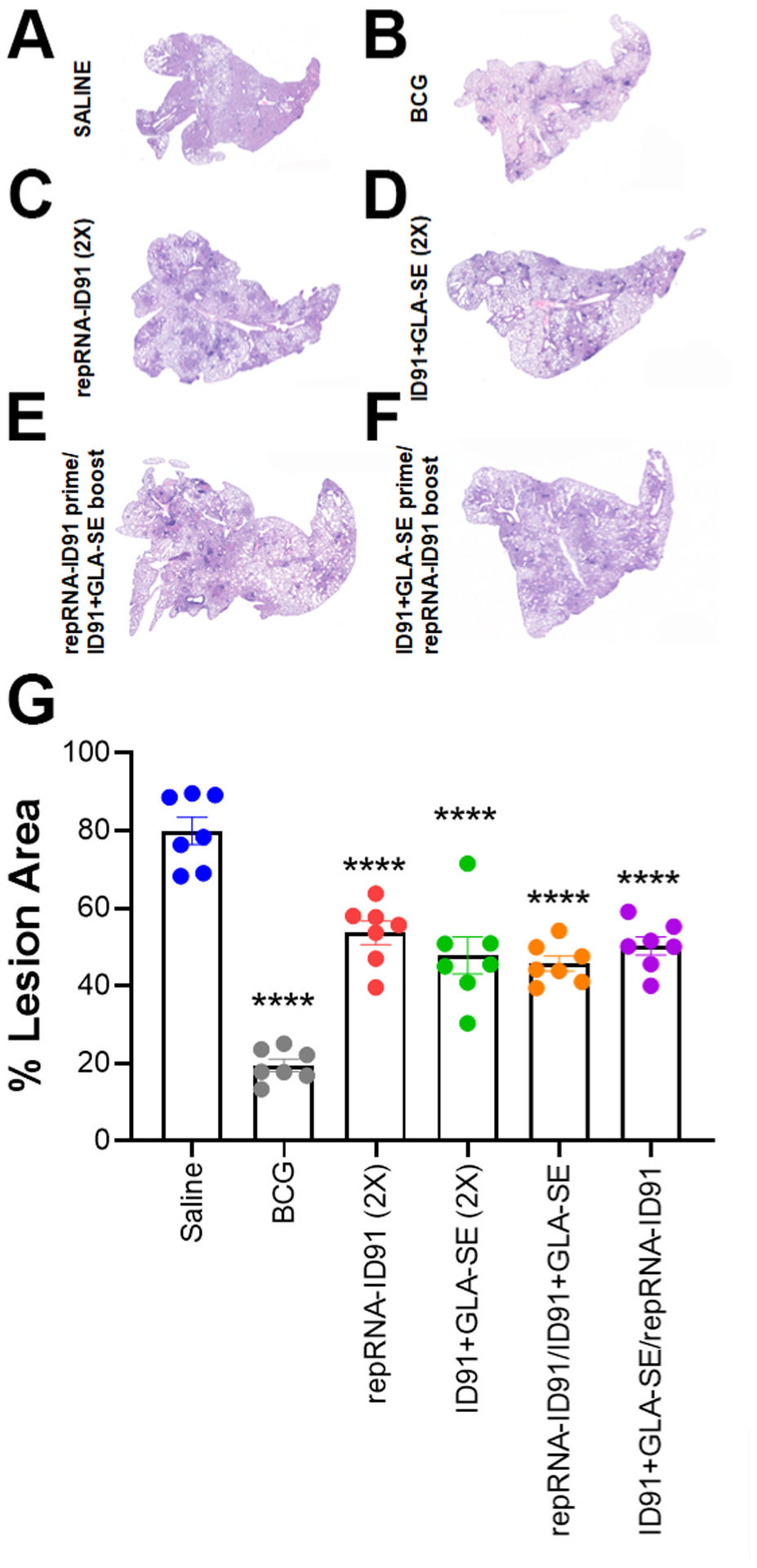
(A-F) Representative images of accessory lung lobes from beige mice immunized with saline (A), BCG (B)v, repRNA-ID91 (C), ID91+GLA-SE (D), repRNA-ID91 prime/ID91+GLA-SE boost (E), or ID91+GLA-SE prime/repRNA-ID91 boost (F) after H&E staining for pulmonary lesions (dark purple). (G) Bars show mean ± SEM percent lesion area, dots represent individual mice, n = 7/group. Asterisks indicate statistical significance, where ****p < 0.0001 using one-way ANOVA with Dunnett’s multiple comparisons test.
To summarize, these data indicate that while the homologous ID91+GLA-SE prime/boost strategy demonstrates prophylactic protection against M. avium 2-151 smt aerosol challenge as published previously[29], the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine strategy further augments prophylactic immunogenicity and confers pulmonary protection post M. avium 2-151 infection by triggering a robust systemic proinflammatory response, inducing strong antigen-specific humoral immune responses, eliciting robust ID91-specific TH1 CD4+ and CD8+ immune responses, and reducing M. avium bacterial burden in the lung.
Discussion
NTM including M. avium can cause pulmonary and extra-pulmonary infections, and since these bacteria exist in the environment it makes contraction of the disease prevalent in both immunocompetent and immunocompromised individuals[24, 73]. Once the host is infected, NTM promote their survival through the reduction of inflammatory cytokines necessary for inducing appropriate innate and adaptive immunity, such as IL-12, while also promoting immunosuppressive cytokines, such as IL-10, which enhances intracellular survival[68, 74, 75]. Not surprisingly, immunocompromised individuals tend to develop a more severe form of the diseaseand while most NTM infections can be treated with antibiotics, the treatment is often complicated by adverse side effects[76], antibiotic resistance, and lack of new drug candidates[77]. The development of a therapeutic vaccineused as an adjunct to drug treatment , would be advantageous to patients. Previous studies have shown that BCG and the ID91+GLA-SE vaccine provide partial immunity to M. avium infections through cross-reactive immunity, resulting in a robust immune response via an increase in production of proinflammatory cytokines and adaptive T cell immunity[29, 78, 79]. However, new vaccine strategies and platforms are required for the induction of an optimal cellular immune response against M. avium infection, ultimately leading to protection against pulmonary lung disease.
In this study, we evaluated both a homologous and a heterologous prime/boost strategy with our repRNA-ID91 vaccine candidate and adjuvanted ID91+GLA-SE subunit protein vaccine candidate against M. avium 2-151 smt challenge in immunocompromised Beige mice. The M. avium 2-151 smt strain was used due to its ability to establish a persistent pulmonary infection in the mouse model and importantly since it is a clinical isolate. The mouse model was able to provide resolution for deciphering vaccine-induced protection against pulmonary disease. Furthermore, the vaccine-induced mechanisms of control/resistance can be pursued in this mouse model due to its C57BL/6 genetic background[62, 80].
The data presented here provides the first report of the evaluation of a replicon RNA-based vaccine strategy in the context of NTM infection and disease, and the efficacy of this platform when used in a heterologous prime/boost regimen. A significant advantage of RNA vaccine platforms is their ability to stimulate antigen-specific CD8+ T cells and antibody responses, which both aid in the control of intracellular infections. As a result, a heterologous strategy combining protein and nucleic acid vaccines is a promising approach for inducing protective immunity through CD4+ and CD8+ T cell-mediated immune responses[81, 82]. We observed that the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine strategy significantly reduced pulmonary bacterial burden in Beige mice and all the single immunological endpoints (proinflammatory cytokine levels, total IgG, IgG1, IgG2c antibody titers, TH1 CD4+ or CD8+ T cells) trended with this protective efficacy. It is also of significant importance to note that of the two heterologous vaccine strategies evaluated, the repRNA-ID91 prime/ID91+GLA-SE boost vaccine strategy was the only successful regimen to induce these robust cellular and humoral immune responses both pre- and post M. avium infection. This indicates that the order in which the vaccines are administered play a key role in establishing immunogenicity and ensuing protection. Interestingly, a similar phenomenon was previously observed with heterologous immunization using a RNA prime and subunit vaccine boost against Leishmania donovani (L. donovani) wherein this vaccine regimen protected mice by significantly reducing L. donovani parasites in the liver compared to the saline injected group[83]. While the mechanism of protective efficacy of the repRNA-ID91 prime and ID91+GLA-SE boost regimen is currently unknown, we hypothesize that there is early generation of vaccine-specific CD8+ T cells following a repRNA-ID91 prime immunization which are subsequently maintained with robust ID91-specific TH1 CD4+ T cells after boosting with ID91-GLA-SE.
Inflammatory cytokines influence the outcome of mycobacterial infection by affecting the macrophage bactericidal capacity (IFN-γ, TNF-α), granuloma formation and maintenance (TNF-α, IL-1β), differentiation of T cells (IL-2), increased (IL-6) and decreased (IL-10) effector responses in target T cells and macrophages[84–88]. Indeed, the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine strategy elicited strong circulating proinflammatory cytokine responses both pre- (IL1-β and IL-6) and post-challenge with M. avium (IFN-γ, TNF-α, IL-2, IL-6). Interestingly, while IL-10 levels increased after BCG and homologous ID91+GLA-SE immunization, no significant changes were observed with IL-10 in the heterologous repRNA-ID91/ID91+GLA-SE immunized group. Neutrophils have also been implicated in the control of mycobacterial infections as they may play an important role in the transition from innate to adaptive immune responses by producing critical cytokines and chemokines[31, 89–93]. For example, neutrophils from M. avium infected mice have been shown to produce TNF-α, IL-12, and IL1-β, and have a putative role in early host responses[94]. To this end, we observed that the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine strategy led to a significant increase in circulating KC/GRO levels, a chemokine responsible for activating and recruiting neutrophils during inflammation, following M. avium infection. It is unclear at this point whether neutrophils play a protective role as a result of this strategy.
Several functional attributes of antibodies could potentially play a role in protection against mycobacteria infection, including antibody-dependent cellular cytotoxicity (ADCC) and antibody-dependent cellular phagocytosis (ADCP)[31, 95, 96]. A recent study showed that M.tb specific IgM, IgA, and IgG1 antibodies in the bronchoalveolar lavage fluid (BAL) negatively correlated with mycobacteria bacterial burden in macaques[97]. In our study, we observed robust ID91- and Ag85B-specific total IgG, IgG1, and IgG2c antibody responses in the serum of heterologous repRNA-ID91/ID91+GLA-SE immunized mice both pre- and post-challenge with M. avium. The IgG1 antibody subclass is able to neutralize bacterial toxins, opsonize capsular polysaccharides for phagocytosis, and activate the complement membrane-attack complex for bacterial lysis[98–103]. The IgG2c subclass, alternatively, has an added advantage due to their high capacity to bind to all activating Fcγ-receptor (FcγRs), leading to a maximal activation of innate immune effector cells, such as monocytes, macrophages, and neutrophils[104–108]. The heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine regimen induced the strongest magnitude of IgG1 and IgG2c responses to both ID91 and Ag85B antigens following M. avium infection. This knowledge highlights the need to understand how vaccine-mediated humoral changes can be leveraged both prophylactically and therapeutically in humans and preclinical models[109, 110].
Protective mycobacterial immunity includes the induction of TH1 CD4+ T cells. Depletion of CD4+ T cells may contribute to disseminated extrapulmonary NTM infections, as has been shown with M.tb[111, 112] and furthermore, in a recent study characterizing the lung cellular composition of patients with NTM lung disease, M. avium-induced CD4+ T cell dysfunction was observed with elevated expression of PD-1, CTLA-4, and TIM-3[113, 114]. Elevated M. avium induced IL-4+ CD4+ T cells has also been shown to skew immune responses towards a TH2 type response, which may lead to a dampened ability to clear the mycobacteria[115]. IFN-γ-secreting CD4+ T cells play a pivotal role in immune defense against mycobacteria which has been shown to activate neutrophils and macrophages to phagocytose and/or kill intracellular pathogens such as NTM[111, 112]. TNF-α is another prominent cytokine that has been clinically shown to help control NTM infections, and if inhibited can result in increased risk of infection[13, 15, 16, 116]. In our study, the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine strategy elicited the greatest magnitude of ID91-specific CD4+ TH1 responses, including production of IFN-γ, TNF-α, and IL-2 cytokines one week post boost immunization. This vaccine regimen was also the only one to elicit robust CD4+ TH1 immune responses to all four ID91 component proteins. Lastly, the heterologous vaccine strategy also induced significant polyfunctional CD4+CD44+CD154+ TH1 responses (IFN-γ, TNF-α, and/or IL-2). Although we did not actively look for cytotoxic CD4+ T cells in this study (except for CD107a responses), our group has previously shown that the GLA-SE adjuvant induces CD4+ cytolytic T lymphocytes (CTLs) with the ability to kill autologous B cells that present a cognate MHC class II peptide complex. Interestingly, this activity did not rely on any of the canonical CTL mechanisms, including perforin, granzymes, Fas-FasL, or TRAIL-DR5, but rather the CD4+ CTLs induced cell death by ligating CD40 on target cells via expression of CD154+[117]. These data indicate that the role of CD4+ CTLs in protective immunity should be further explored in the context of different vaccine strategies against other mycobacteria, including M. avium.
CD8+ T cells also likely contribute to limiting mycobacterial infections by producing effector cytokines or recognizing and directly lysing infected cells. Leveraging the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine strategy, or RNA-based platform alone, to induce robust CD8+ T cell responses enabled the investigation of different cellular outcomes, including the elicitation of CD8-biased responses and/or CD8+ and CD4+ T cell responses with combined RNA and protein platform strategies. Furthermore, including a subunit only approach presented a CD4-biased approach as a comparator. Multiple studies conducted in both humans and preclinical models (mice and NHP) have delineated a role for CD8+ T cells in the control of mycobacterial infections[118–120]. CD8+ T cells have been shown to migrate to the site of mycobacteria infection and can play a unique role in host defense[121–123]. Cytolytic CD8+ T cells produce IFN-γ and cytolytic granules such as perforin and granzyme B, which can help lyse infected macrophages[124–127]. This ability to distinguish infected cells suggests that the magnitude of the CD8+ T cell response may represent a sensor of intracellular burden[128, 129]. CD8+ T cells are also able to inhibit mycobacterial growth and induce apoptosis in infected cells[39]. Interestingly, only the heterologous repRNA-ID91/ID91+GLA-SE vaccine strategy elicited significant ID91-specific CD8+ cytokine responses, including production of IFN-γ and TNF-α cytokines. Furthermore, it was also the only vaccine strategy to induce robust polyfunctional CD8+CD44+ CD154+ IFN-γ and TNF-α cytokine responses.
The primary endpoints for protective efficacy in this study were reduction of bacterial burden and pulmonary pathology. While all vaccine strategies demonstrated prophylactic protection against M. avium 2-151 smt aerosol challenge, the heterologous repRNA-ID91/ID91+GLA-SE vaccine regimen provided greater pulmonary protection in beige mice compared to the homologous ID91+GLA-SE prime/boost regimen. The main difference between the two regimens included the induction of enhanced ID91-specific TH1 CD4+ and CD8+ cellular responses in the heterologous repRNA-ID91/ID91+GLA-SE prime/boost vaccine group. Surprisingly, despite inducing little to no immune responses in the assays measured, BCG immunization conferred the greatest protection compared to all other vaccine strategies, which emphasizes the need for more work in NTM specific antigen design and optimization for future vaccine candidates and platforms. Similar to bacterial burden, all prophylactic prime/boost vaccine strategies led to decreased pulmonary lesion scores suggesting that the mechanism of protection may be related to maintaining pulmonary architecture. Although BCG is given safety to infants in many countries, live-attenuated vaccines are generally not advisable for immunocompromised individuals, which is the population most significantly impacted by NTM disease. Furthermore, therapeutic use of BCG vaccination post NTM exposure didn’t seem to protect from NTM lung disease[130] which makes it a less than ideal approach for treatment against M. avium. Therefore, we believe that a vaccine platform selection and design against NTM and specifically M. avium needs to be developed as a safe strategy against this opportunistic infection.
These studies show that a prophylactic heterologous prime/boost vaccine strategy with repRNA-ID91 prime followed by protein ID91+GLA-SE boost is superior to the subunit-protein + adjuvant given as a homologous prime/boost regimen. Compared to the saline control, the heterologous vaccine strategy elicited higher circulating levels of proinflammatory cytokines, induced stronger ID91 and Ag85B antigen-specific IgG1 and IgG2c humoral antibody responses, elicited stronger CD4+ TH1 immune responses to ID91 and all four ID91 component antigens (Rv1886, Rv3619, Rv3478, Rv2389), induced IFN-γ and TNF-secreting CD8+ T cells, and furthermore, provided greater pulmonary protection in beige mice post M. avium 2-151 smt infection than the homologous ID91+GLA-SE regimen. The work detailed here demonstrates that two ID91 vaccine delivery platforms administered in a heterologous prime/boost strategy is a viable approach for novel interventions against M. avium infection.
Future work will focus on further optimizing the repRNA vaccine platform to determine whether longer intervals between RNA immunizations can enhance protection, and to test the efficacy of different routes of vaccine administration. Research focused on designing and developing vaccine candidates against M. avium and other NTM using NTM-specific antigens that are immunogenic in humans should also be prioritized to (1) determine whether these regimens induce long-lived memory immune responses or are protective against other NTM, (2) test these vaccines in the context of therapy as an adjunct to drug treatment, and (3) determine if these vaccines can enhance BCG protection in BCG primed animals, as BCG protection has been known to wane over time. These vaccine strategies can not only significantly help reduce the NTM disease burden globally, but can also help in the development of novel vaccine strategies against other mycobacteria.
Supplementary Material
Supplementary Figure 1. Intracellular cytokine staining (ICS) gating scheme for CD44+, CD4+, and CD8+ T cells expressing TH1 (IFN-γ, TNF-α, IL-2), TH2 (IL-5), and TH17 cytokines (IL-17A). ID91 stimulation shown.
Acknowledgements
The authors would like to express their gratitude to Seattle Children’s Research Institute Leadership for their guidance, compassion, and mentorship. The datasets generated and/or analyzed during the study presented here are available from the corresponding authors on reasonable request.
Funding
Research reported here was supported by the National Institute of Allergy and Infectious Diseases (NIAID) of the National Institutes of Health (NIH) under R21AI142267, R01AI125160, IMPAcTB 75N93021C00029, and by BMGF (INV-025713 to DF). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Competing interests
The authors declare no competing interests.
References
- [1].Baldwin SL, Larsen SE, Ordway D, Cassell G, and Coler RN, “The complexities and challenges of preventing and treating nontuberculous mycobacterial diseases,” PLoS Negl Trop Dis, vol. 13, no. 2, p. e0007083, Feb 2019, doi: 10.1371/journal.pntd.0007083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Strollo SE, Adjemian J, Adjemian MK, and Prevots DR, “The Burden of Pulmonary Nontuberculous Mycobacterial Disease in the United States,” Ann Am Thorac Soc, vol. 12, no. 10, pp. 1458–64, Oct 2015, doi: 10.1513/AnnalsATS.201503-173OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dirac MA et al. , “Environment or host?: A case-control study of risk factors for Mycobacterium avium complex lung disease,” Am J Respir Crit Care Med, vol. 186, no. 7, pp. 684–91, Oct 1 2012, doi: 10.1164/rccm.201205-0825OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Falkinham JO 3rd, “Environmental sources of nontuberculous mycobacteria,” Clin Chest Med, vol. 36, no. 1, pp. 35–41, Mar 2015, doi: 10.1016/j.ccm.2014.10.003. [DOI] [PubMed] [Google Scholar]
- [5].Brode SK, Daley CL, and Marras TK, “The epidemiologic relationship between tuberculosis and non-tuberculous mycobacterial disease: a systematic review,” Int J Tuberc Lung Dis, vol. 18, no. 11, pp. 1370–7, Nov 2014, doi: 10.5588/ijtld.14.0120. [DOI] [PubMed] [Google Scholar]
- [6].Orme IM and Ordway DJ, “Host response to nontuberculous mycobacterial infections of current clinical importance,” Infect Immun, vol. 82, no. 9, pp. 3516–22, Sep 2014, doi: 10.1128/IAI.01606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Spaulding AB et al. , “Geographic Distribution of Nontuberculous Mycobacterial Species Identified among Clinical Isolates in the United States, 2009–2013,” Ann Am Thorac Soc, vol. 14, no. 11, pp. 1655–1661, Nov 2017, doi: 10.1513/AnnalsATS.201611-860OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Abdelaal HFM, Chan ED, Young L, Baldwin SL, and Coler RN, “Mycobacterium abscessus: It’s Complex,” Microorganisms, vol. 10, no. 7, Jul 19 2022, doi: 10.3390/microorganisms10071454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ryu YJ, Koh WJ, and Daley CL, “Diagnosis and Treatment of Nontuberculous Mycobacterial Lung Disease: Clinicians’ Perspectives,” Tuberc Respir Dis (Seoul), vol. 79, no. 2, pp. 74–84, Apr 2016, doi: 10.4046/trd.2016.79.2.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Griffith DE et al. , “An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases,” Am J Respir Crit Care Med, vol. 175, no. 4, pp. 367–416, Feb 15 2007, doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- [11].Kwon YS, Koh WJ, and Daley CL, “Treatment of Mycobacterium avium Complex Pulmonary Disease,” Tuberc Respir Dis (Seoul), vol. 82, no. 1, pp. 15–26, Jan 2019, doi: 10.4046/trd.2018.0060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Falkinham JO 3rd, “Epidemiology of infection by nontuberculous mycobacteria,” Clin Microbiol Rev, vol. 9, no. 2, pp. 177–215, Apr 1996, doi: 10.1128/CMR.9.2.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Winthrop KL, Chang E, Yamashita S, Iademarco MF, and LoBue PA, “Nontuberculous mycobacteria infections and anti-tumor necrosis factor-alpha therapy,” Emerg Infect Dis, vol. 15, no. 10, pp. 1556–61, Oct 2009, doi: 10.3201/eid1510.090310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Bar-On O et al. , “Increasing nontuberculous mycobacteria infection in cystic fibrosis,” J Cyst Fibros, vol. 14, no. 1, pp. 53–62, Jan 2015, doi: 10.1016/j.jcf.2014.05.008. [DOI] [PubMed] [Google Scholar]
- [15].Winthrop KL et al. , “Mycobacterial diseases and antitumour necrosis factor therapy in USA,” Ann Rheum Dis, vol. 72, no. 1, pp. 37–42, Jan 2013, doi: 10.1136/annrheumdis-2011-200690. [DOI] [PubMed] [Google Scholar]
- [16].Yoo JW et al. , “Mycobacterial diseases developed during anti-tumour necrosis factor-alpha therapy,” Eur Respir J, vol. 44, no. 5, pp. 1289–95, Nov 2014, doi: 10.1183/09031936.00063514. [DOI] [PubMed] [Google Scholar]
- [17].Brown-Elliott BA, Nash KA, and Wallace RJ Jr., “Antimicrobial susceptibility testing, drug resistance mechanisms, and therapy of infections with nontuberculous mycobacteria,” Clin Microbiol Rev, vol. 25, no. 3, pp. 545–82, Jul 2012, doi: 10.1128/CMR.05030-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Brown-Elliott BA and Wallace RJ Jr., “Clinical and taxonomic status of pathogenic nonpigmented or late-pigmenting rapidly growing mycobacteria,” Clin Microbiol Rev, vol. 15, no. 4, pp. 716–46, Oct 2002, doi: 10.1128/CMR.15.4.716-746.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Maurer FP, Bruderer VL, Ritter C, Castelberg C, Bloemberg GV, and Bottger EC, “Lack of antimicrobial bactericidal activity in Mycobacterium abscessus,” Antimicrob Agents Chemother, vol. 58, no. 7, pp. 3828–36, Jul 2014, doi: 10.1128/AAC.02448-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Petrini B, “Mycobacterium abscessus: an emerging rapid-growing potential pathogen,” APMIS, vol. 114, no. 5, pp. 319–28, May 2006, doi: 10.1111/j.1600-0463.2006.apm_390.x. [DOI] [PubMed] [Google Scholar]
- [21].Greendyke R and Byrd TF, “Differential antibiotic susceptibility of Mycobacterium abscessus variants in biofilms and macrophages compared to that of planktonic bacteria,” Antimicrob Agents Chemother, vol. 52, no. 6, pp. 2019–26, Jun 2008, doi: 10.1128/AAC.00986-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rhoades ER, Archambault AS, Greendyke R, Hsu FF, Streeter C, and Byrd TF, “Mycobacterium abscessus Glycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-alpha by preventing interaction with TLR2,” J Immunol, vol. 183, no. 3, pp. 1997–2007, Aug 1 2009, doi: 10.4049/jimmunol.0802181. [DOI] [PubMed] [Google Scholar]
- [23].van Ingen J, Boeree MJ, van Soolingen D, and Mouton JW, “Resistance mechanisms and drug susceptibility testing of nontuberculous mycobacteria,” Drug Resist Updat, vol. 15, no. 3, pp. 149–61, Jun 2012, doi: 10.1016/j.drup.2012.04.001. [DOI] [PubMed] [Google Scholar]
- [24].Johnson MM and Odell JA, “Nontuberculous mycobacterial pulmonary infections,” J Thorac Dis, vol. 6, no. 3, pp. 210–20, Mar 2014, doi: 10.3978/j.issn.2072-1439.2013.12.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Griffith DE et al. , “Amikacin Liposome Inhalation Suspension for Treatment-Refractory Lung Disease Caused by Mycobacterium avium Complex (CONVERT). A Prospective, Open-Label, Randomized Study,” Am J Respir Crit Care Med, vol. 198, no. 12, pp. 1559–1569, Dec 15 2018, doi: 10.1164/rccm.201807-1318OC. [DOI] [PubMed] [Google Scholar]
- [26].Stout JE, Koh WJ, and Yew WW, “Update on pulmonary disease due to non-tuberculous mycobacteria,” Int J Infect Dis, vol. 45, pp. 123–34, Apr 2016, doi: 10.1016/j.ijid.2016.03.006. [DOI] [PubMed] [Google Scholar]
- [27].Daniel-Wayman S et al. , “Advancing Translational Science for Pulmonary Nontuberculous Mycobacterial Infections. A Road Map for Research,” Am J Respir Crit Care Med, vol. 199, no. 8, pp. 947–951, Apr 15 2019, doi: 10.1164/rccm.201807-1273PP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].van Ingen J et al. , “Management of Drug Toxicity in Mycobacterium avium Complex Pulmonary Disease: An Expert Panel Survey,” Clin Infect Dis, vol. 73, no. 1, pp. e256–e259, Jul 1 2021, doi: 10.1093/cid/ciaa1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Larsen SE et al. , “Subunit vaccine protects against a clinical isolate of Mycobacterium avium in wild type and immunocompromised mouse models,” Sci Rep, vol. 11, no. 1, p. 9040, Apr 27 2021, doi: 10.1038/s41598-021-88291-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Martin C, Aguilo N, Marinova D, and Gonzalo-Asensio J, “Update on TB Vaccine Pipeline,” Applied Sciences, vol. 10, no. 7, p. 2632, 2020. [Online]. Available: https://www.mdpi.com/2076-3417/10/7/2632. [Google Scholar]
- [31].Larsen SE, Williams BD, Rais M, Coler RN, and Baldwin SL, “It Takes a Village: The Multifaceted Immune Response to Mycobacterium tuberculosis Infection and Vaccine-Induced Immunity,” Front Immunol, vol. 13, p. 840225, 2022, doi: 10.3389/fimmu.2022.840225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lewinsohn DA, Lewinsohn DM, and Scriba TJ, “Polyfunctional CD4(+) T Cells As Targets for Tuberculosis Vaccination,” Front Immunol, vol. 8, p. 1262, 2017, doi: 10.3389/fimmu.2017.01262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tameris MD et al. , “Safety and efficacy of MVA85A, a new tuberculosis vaccine, in infants previously vaccinated with BCG: a randomised, placebo-controlled phase 2b trial,” Lancet, vol. 381, no. 9871, pp. 1021–8, Mar 23 2013, doi: 10.1016/S0140-6736(13)60177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lu LL et al. , “IFN-gamma-independent immune markers of Mycobacterium tuberculosis exposure,” Nat Med, vol. 25, no. 6, pp. 977–987, Jun 2019, doi: 10.1038/s41591-019-0441-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Smith CM et al. , “Functionally Overlapping Variants Control Tuberculosis Susceptibility in Collaborative Cross Mice,” mBio, vol. 10, no. 6, Nov 26 2019, doi: 10.1128/mBio.02791-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Behar SM, “Antigen-specific CD8(+) T cells and protective immunity to tuberculosis,” Adv Exp Med Biol, vol. 783, pp. 141–63, 2013, doi: 10.1007/978-1-4614-6111-1_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Behar SM, Woodworth JS, and Wu Y, “Next generation: tuberculosis vaccines that elicit protective CD8+ T cells,” Expert Rev Vaccines, vol. 6, no. 3, pp. 441–56, Jun 2007, doi: 10.1586/14760584.6.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Boom WH, “New TB vaccines: is there a requirement for CD8 T cells?,” J Clin Invest, vol. 117, no. 8, pp. 2092–4, Aug 2007, doi: 10.1172/JCI32933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lazarevic V and Flynn J, “CD8+ T cells in tuberculosis,” Am J Respir Crit Care Med, vol. 166, no. 8, pp. 1116–21, Oct 15 2002, doi: 10.1164/rccm.2204027. [DOI] [PubMed] [Google Scholar]
- [40].Lewinsohn DA et al. , “Comprehensive definition of human immunodominant CD8 antigens in tuberculosis,” NPJ Vaccines, vol. 2, 2017, doi: 10.1038/s41541-017-0008-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Lin PL and Flynn JL, “CD8 T cells and Mycobacterium tuberculosis infection,” Semin Immunopathol, vol. 37, no. 3, pp. 239–49, May 2015, doi: 10.1007/s00281-015-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Darrah PA et al. , “Prevention of tuberculosis in macaques after intravenous BCG immunization,” Nature, vol. 577, no. 7788, pp. 95–102, Jan 2020, doi: 10.1038/s41586-019-1817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang PH, Wu MF, Hsu CY, Pan SW, Shu CC, and Cheng SL, “The Trend of TIM3 Expression on T Cells in Patients With Nontuberculous Mycobacterial Lung Disease: From Immune Cell Dysfunction to Clinical Severity,” Front Immunol, vol. 12, p. 738056, 2021, doi: 10.3389/fimmu.2021.738056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Erasmus JH et al. , “A Nanostructured Lipid Carrier for Delivery of a Replicating Viral RNA Provides Single, Low-Dose Protection against Zika,” Mol Ther, vol. 26, no. 10, pp. 2507–2522, Oct 3 2018, doi: 10.1016/j.ymthe.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Erasmus JH et al. , “An Alphavirus-derived replicon RNA vaccine induces SARS-CoV-2 neutralizing antibody and T cell responses in mice and nonhuman primates,” Sci Transl Med, vol. 12, no. 555, Aug 5 2020, doi: 10.1126/scitranslmed.abc9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ljungberg K and Liljestrom P, “Self-replicating alphavirus RNA vaccines,” Expert Rev Vaccines, vol. 14, no. 2, pp. 177–94, Feb 2015, doi: 10.1586/14760584.2015.965690. [DOI] [PubMed] [Google Scholar]
- [47].Leitner WW et al. , “Alphavirus-based DNA vaccine breaks immunological tolerance by activating innate antiviral pathways,” Nat Med, vol. 9, no. 1, pp. 33–9, Jan 2003, doi: 10.1038/nm813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].McShane H and Hill A, “Prime-boost immunisation strategies for tuberculosis,” Microbes Infect, vol. 7, no. 5–6, pp. 962–7, May 2005, doi: 10.1016/j.micinf.2005.03.009. [DOI] [PubMed] [Google Scholar]
- [49].Ramshaw IA and Ramsay AJ, “The prime-boost strategy: exciting prospects for improved vaccination,” Immunol Today, vol. 21, no. 4, pp. 163–5, Apr 2000, doi: 10.1016/s0167-5699(00)01612-1. [DOI] [PubMed] [Google Scholar]
- [50].Schell JB et al. , “Significant protection against high-dose simian immunodeficiency virus challenge conferred by a new prime-boost vaccine regimen,” J Virol, vol. 85, no. 12, pp. 5764–72, Jun 2011, doi: 10.1128/JVI.00342-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Knudsen ML et al. , “Kinetic and phenotypic analysis of CD8+ T cell responses after priming with alphavirus replicons and homologous or heterologous booster immunizations,” J Virol, vol. 88, no. 21, pp. 12438–51, Nov 2014, doi: 10.1128/JVI.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Jiang G et al. , “Sterile protection against Plasmodium knowlesi in rhesus monkeys from a malaria vaccine: comparison of heterologous prime boost strategies,” PLoS One, vol. 4, no. 8, p. e6559, Aug 10 2009, doi: 10.1371/journal.pone.0006559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Barefoot B et al. , “Comparison of multiple vaccine vectors in a single heterologous prime-boost trial,” Vaccine, vol. 26, no. 48, pp. 6108–18, Nov 11 2008, doi: 10.1016/j.vaccine.2008.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Walczak M et al. , “Heterologous prime-boost immunizations with a virosomal and an alphavirus replicon vaccine,” Mol Pharm, vol. 8, no. 1, pp. 65–77, Feb 7 2011, doi: 10.1021/mp1002043. [DOI] [PubMed] [Google Scholar]
- [55].Hu Z et al. , “Heterologous prime-boost vaccination against tuberculosis with recombinant Sendai virus and DNA vaccines,” J Mol Med (Berl), vol. 97, no. 12, pp. 1685–1694, Dec 2019, doi: 10.1007/s00109-019-01844-3. [DOI] [PubMed] [Google Scholar]
- [56].Larsen SE et al. , “Enhanced Anti-Mycobacterium tuberculosis Immunity over Time with Combined Drug and Immunotherapy Treatment,” Vaccines (Basel), vol. 6, no. 2, May 24 2018, doi: 10.3390/vaccines6020030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Coler RN et al. , “Development and characterization of synthetic glucopyranosyl lipid adjuvant system as a vaccine adjuvant,” PLoS One, vol. 6, no. 1, p. e16333, Jan 26 2011, doi: 10.1371/journal.pone.0016333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Baldwin SL et al. , “Therapeutic efficacy against Mycobacterium tuberculosis using ID93 and liposomal adjuvant formulations,” Front Microbiol, vol. 13, p. 935444, 2022, doi: 10.3389/fmicb.2022.935444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Baldwin SL et al. , “Prophylactic efficacy against Mycobacterium tuberculosis using ID93 and lipid-based adjuvant formulations in the mouse model,” PLoS One, vol. 16, no. 3, p. e0247990, 2021, doi: 10.1371/journal.pone.0247990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gangadharam PR, “Beige mouse model for Mycobacterium avium complex disease,” Antimicrob Agents Chemother, vol. 39, no. 8, pp. 1647–54, Aug 1995, doi: 10.1128/AAC.39.8.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Gangadharam PR, Perumal VK, Farhi DC, and LaBrecque J, “The beige mouse model for Mycobacterium avium complex (MAC) disease: optimal conditions for the host and parasite,” Tubercle, vol. 70, no. 4, pp. 257–71, Dec 1989, doi: 10.1016/0041-3879(89)90020-2. [DOI] [PubMed] [Google Scholar]
- [62].Appelberg R, Castro AG, Gomes S, Pedrosa J, and Silva MT, “Susceptibility of beige mice to Mycobacterium avium: role of neutrophils,” Infect Immun, vol. 63, no. 9, pp. 3381–7, Sep 1995, doi: 10.1128/iai.63.9.3381-3387.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Saunders BM and Cheers C, “Intranasal infection of beige mice with Mycobacterium avium complex: role of neutrophils and natural killer cells,” Infect Immun, vol. 64, no. 10, pp. 4236–41, Oct 1996, doi: 10.1128/iai.64.10.4236-4241.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bannai M et al. , “Disparate effect of beige mutation on cytotoxic function between natural killer and natural killer T cells,” Immunology, vol. 100, no. 2, pp. 165–9, Jun 2000, doi: 10.1046/j.1365-2567.2000.00040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Roder JC, Lohmann-Matthes ML, Domzig W, and Wigzell H, “The beige mutation in the mouse. II. Selectivity of the natural killer (NK) cell defect,” J Immunol, vol. 123, no. 5, pp. 2174–81, Nov 1979. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/158612. [PubMed] [Google Scholar]
- [66].Pflumio F, Fonteneau P, and Loor F, “Impaired antibody response of C57BL/6 beige mutant mice to a thymus-independent type 2 antigen,” Immunol Lett, vol. 23, no. 4, pp. 269–74, Feb 1990, doi: 10.1016/0165-2478(90)90071-w. [DOI] [PubMed] [Google Scholar]
- [67].Dhanda SK et al. , “Predicting HLA CD4 Immunogenicity in Human Populations,” Front Immunol, vol. 9, p. 1369, 2018, doi: 10.3389/fimmu.2018.01369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Orujyan D et al. , “Protective Efficacy of BCG Vaccine against Mycobacterium leprae and Non-Tuberculous Mycobacterial Infections,” Vaccines (Basel), vol. 10, no. 3, Mar 3 2022, doi: 10.3390/vaccines10030390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Greinert U, Schlaak M, Rusch-Gerdes S, Flad HD, and Ernst M, “Low in vitro production of interferon-gamma and tumor necrosis factor-alpha in HIV-seronegative patients with pulmonary disease caused by nontuberculous mycobacteria,” J Clin Immunol, vol. 20, no. 6, pp. 445–52, Nov 2000, doi: 10.1023/a:1026407815946. [DOI] [PubMed] [Google Scholar]
- [70].Safdar A, White DA, Stover D, Armstrong D, and Murray HW, “Profound interferon gamma deficiency in patients with chronic pulmonary nontuberculous mycobacteriosis,” Am J Med, vol. 113, no. 9, pp. 756–9, Dec 15 2002, doi: 10.1016/s0002-9343(02)01313-x. [DOI] [PubMed] [Google Scholar]
- [71].Vankayalapati R et al. , “Cytokine profiles in immunocompetent persons infected with Mycobacterium avium complex,” J Infect Dis, vol. 183, no. 3, pp. 478–84, Feb 1 2001, doi: 10.1086/318087. [DOI] [PubMed] [Google Scholar]
- [72].Lim A, Allison C, Price P, and Waterer G, “Susceptibility to pulmonary disease due to Mycobacterium avium-intracellulare complex may reflect low IL-17 and high IL-10 responses rather than Th1 deficiency,” Clin Immunol, vol. 137, no. 2, pp. 296–302, Nov 2010, doi: 10.1016/j.clim.2010.07.011. [DOI] [PubMed] [Google Scholar]
- [73].Akram SM and Attia FN, “Mycobacterium Avium Intracellulare,” in StatPearls. Treasure Island (FL), 2022. [Google Scholar]
- [74].Wagner D, Sangari FJ, Kim S, Petrofsky M, and Bermudez LE, “Mycobacterium avium infection of macrophages results in progressive suppression of interleukin-12 production in vitro and in vivo,” J Leukoc Biol, vol. 71, no. 1, pp. 80–8, Jan 2002. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/11781383. [PubMed] [Google Scholar]
- [75].Sampaio EP et al. , “Mycobacterium abscessus and M. avium trigger Toll-like receptor 2 and distinct cytokine response in human cells,” Am J Respir Cell Mol Biol, vol. 39, no. 4, pp. 431–9, Oct 2008, doi: 10.1165/rcmb.2007-0413OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Al-Anazi KA, Al-Jasser AM, and Al-Anazi WK, “Infections caused by non-tuberculous mycobacteria in recipients of hematopoietic stem cell transplantation,” Front Oncol, vol. 4, p. 311, 2014, doi: 10.3389/fonc.2014.00311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Ratnatunga CN et al. , “The Rise of Non-Tuberculosis Mycobacterial Lung Disease,” Front Immunol, vol. 11, p. 303, 2020, doi: 10.3389/fimmu.2020.00303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Zimmermann P, Finn A, and Curtis N, “Does BCG Vaccination Protect Against Nontuberculous Mycobacterial Infection? A Systematic Review and Meta-Analysis,” J Infect Dis, vol. 218, no. 5, pp. 679–687, Jul 24 2018, doi: 10.1093/infdis/jiy207. [DOI] [PubMed] [Google Scholar]
- [79].Abate G, Hamzabegovic F, Eickhoff CS, and Hoft DF, “BCG Vaccination Induces M. avium and M. abscessus Cross-Protective Immunity,” Front Immunol, vol. 10, p. 234, 2019, doi: 10.3389/fimmu.2019.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Florido M, Appelberg R, Orme IM, and Cooper AM, “Evidence for a reduced chemokine response in the lungs of beige mice infected with Mycobacterium avium,” Immunology, vol. 90, no. 4, pp. 600–6, Apr 1997, doi: 10.1046/j.1365-2567.1997.00206.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Baldwin SL et al. , “Protection against tuberculosis with homologous or heterologous protein/vector vaccine approaches is not dependent on CD8+ T cells,” J Immunol, vol. 191, no. 5, pp. 2514–2525, Sep 1 2013, doi: 10.4049/jimmunol.1301161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Lu YJ, Barreira-Silva P, Boyce S, Powers J, Cavallo K, and Behar SM, “CD4 T cell help prevents CD8 T cell exhaustion and promotes control of Mycobacterium tuberculosis infection,” Cell Rep, vol. 36, no. 11, p. 109696, Sep 14 2021, doi: 10.1016/j.celrep.2021.109696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Duthie MS et al. , “Heterologous Immunization With Defined RNA and Subunit Vaccines Enhances T Cell Responses That Protect Against Leishmania donovani,” Front Immunol, vol. 9, p. 2420, 2018, doi: 10.3389/fimmu.2018.02420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Appelberg R, “Pathogenesis of Mycobacterium avium infection: typical responses to an atypical mycobacterium?,” Immunol Res, vol. 35, no. 3, pp. 179–90, 2006, doi: 10.1385/IR:35:3:179. [DOI] [PubMed] [Google Scholar]
- [85].Berrington WR and Hawn TR, “Mycobacterium tuberculosis, macrophages, and the innate immune response: does common variation matter?,” Immunol Rev, vol. 219, pp. 167–86, Oct 2007, doi: 10.1111/j.1600-065X.2007.00545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Haug M et al. , “Dynamics of immune effector mechanisms during infection with Mycobacterium avium in C57BL/6 mice,” Immunology, vol. 140, no. 2, pp. 232–43, Oct 2013, doi: 10.1111/imm.12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Cooper AM, Mayer-Barber KD, and Sher A, “Role of innate cytokines in mycobacterial infection,” Mucosal Immunol, vol. 4, no. 3, pp. 252–60, May 2011, doi: 10.1038/mi.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Liu X et al. , “IL-2 Restores T-Cell Dysfunction Induced by Persistent Mycobacterium tuberculosis Antigen Stimulation,” Front Immunol, vol. 10, p. 2350, 2019, doi: 10.3389/fimmu.2019.02350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Eruslanov EB et al. , “Neutrophil responses to Mycobacterium tuberculosis infection in genetically susceptible and resistant mice,” Infect Immun, vol. 73, no. 3, pp. 1744–53, Mar 2005, doi: 10.1128/IAI.73.3.1744-1753.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Fulton SA, Reba SM, Martin TD, and Boom WH, “Neutrophil-mediated mycobacteriocidal immunity in the lung during Mycobacterium bovis BCG infection in C57BL/6 mice,” Infect Immun, vol. 70, no. 9, pp. 5322–7, Sep 2002, doi: 10.1128/IAI.70.9.5322-5327.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Pedrosa J, Saunders BM, Appelberg R, Orme IM, Silva MT, and Cooper AM, “Neutrophils play a protective nonphagocytic role in systemic Mycobacterium tuberculosis infection of mice,” Infect Immun, vol. 68, no. 2, pp. 577–83, Feb 2000, doi: 10.1128/IAI.68.2.577-583.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Cassatella MA, “The production of cytokines by polymorphonuclear neutrophils,” Immunol Today, vol. 16, no. 1, pp. 21–6, Jan 1995, doi: 10.1016/0167-5699(95)80066-2. [DOI] [PubMed] [Google Scholar]
- [93].Scapini P, Lapinet-Vera JA, Gasperini S, Calzetti F, Bazzoni F, and Cassatella MA, “The neutrophil as a cellular source of chemokines,” Immunol Rev, vol. 177, pp. 195–203, Oct 2000, doi: 10.1034/j.1600-065x.2000.17706.x. [DOI] [PubMed] [Google Scholar]
- [94].Petrofsky M and Bermudez LE, “Neutrophils from Mycobacterium avium-infected mice produce TNF-alpha, IL-12, and IL-1 beta and have a putative role in early host response,” Clin Immunol, vol. 91, no. 3, pp. 354–8, Jun 1999, doi: 10.1006/clim.1999.4709. [DOI] [PubMed] [Google Scholar]
- [95].Achkar JM, Chan J, and Casadevall A, “B cells and antibodies in the defense against Mycobacterium tuberculosis infection,” Immunol Rev, vol. 264, no. 1, pp. 167–81, Mar 2015, doi: 10.1111/imr.12276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Lu LL, Suscovich TJ, Fortune SM, and Alter G, “Beyond binding: antibody effector functions in infectious diseases,” Nat Rev Immunol, vol. 18, no. 1, pp. 46–61, Jan 2018, doi: 10.1038/nri.2017.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Irvine EB et al. , “Robust IgM responses following intravenous vaccination with Bacille Calmette-Guerin associate with prevention of Mycobacterium tuberculosis infection in macaques,” Nat Immunol, vol. 22, no. 12, pp. 1515–1523, Dec 2021, doi: 10.1038/s41590-021-01066-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Tarr PI, Hosea SW, Brown EJ, Schneerson R, Sutton A, and Frank MM, “The requirement of specific anticapsular IgG for killing of Haemophilus influenzae by the alternative pathway of complement activation,” J Immunol, vol. 128, no. 4, pp. 1772–5, Apr 1982. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/6977569. [PubMed] [Google Scholar]
- [99].Frank MM, Joiner K, and Hammer C, “The function of antibody and complement in the lysis of bacteria,” Rev Infect Dis, vol. 9 Suppl 5, pp. S537–45, Sep-Oct 1987, doi: 10.1093/clinids/9.supplement_5.s537. [DOI] [PubMed] [Google Scholar]
- [100].Shackelford PG, Granoff DM, Nelson SJ, Scott MG, Smith DS, and Nahm MH, “Subclass distribution of human antibodies to Haemophilus influenzae type b capsular polysaccharide,” J Immunol, vol. 138, no. 2, pp. 587–92, Jan 15 1987. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/3491852. [PubMed] [Google Scholar]
- [101].Ferrante A, Beard LJ, and Feldman RG, “IgG subclass distribution of antibodies to bacterial and viral antigens,” Pediatr Infect Dis J, vol. 9, no. 8 Suppl, pp. S16–24, Aug 1990. [Online]. Available: https://www.ncbi.nlm.nih.gov/pubmed/2216603. [PubMed] [Google Scholar]
- [102].Schechter MC, Satola SW, and Stephens DS, “Host defenses to extracellular bacteria,” in Clinical Immunology: Elsevier, 2019, pp. 391–402. e1. [Google Scholar]
- [103].McLean MR, Lu LL, Kent SJ, and Chung AW, “An Inflammatory Story: Antibodies in Tuberculosis Comorbidities,” Front Immunol, vol. 10, p. 2846, 2019, doi: 10.3389/fimmu.2019.02846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Hamaguchi Y, Xiu Y, Komura K, Nimmerjahn F, and Tedder TF, “Antibody isotype-specific engagement of Fcgamma receptors regulates B lymphocyte depletion during CD20 immunotherapy,” J Exp Med, vol. 203, no. 3, pp. 743–53, Mar 20 2006, doi: 10.1084/jem.20052283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Kao D, Lux A, Schaffert A, Lang R, Altmann F, and Nimmerjahn F, “IgG subclass and vaccination stimulus determine changes in antigen specific antibody glycosylation in mice,” Eur J Immunol, vol. 47, no. 12, pp. 2070–2079, Dec 2017, doi: 10.1002/eji.201747208. [DOI] [PubMed] [Google Scholar]
- [106].Kao D et al. , “A Monosaccharide Residue Is Sufficient to Maintain Mouse and Human IgG Subclass Activity and Directs IgG Effector Functions to Cellular Fc Receptors,” Cell Rep, vol. 13, no. 11, pp. 2376–2385, Dec 22 2015, doi: 10.1016/j.celrep.2015.11.027. [DOI] [PubMed] [Google Scholar]
- [107].Nimmerjahn F and Ravetch JV, “Divergent immunoglobulin g subclass activity through selective Fc receptor binding,” Science, vol. 310, no. 5753, pp. 1510–2, Dec 2 2005, doi: 10.1126/science.1118948. [DOI] [PubMed] [Google Scholar]
- [108].Nimmerjahn F, Bruhns P, Horiuchi K, and Ravetch JV, “FcgammaRIV: a novel FcR with distinct IgG subclass specificity,” Immunity, vol. 23, no. 1, pp. 41–51, Jul 2005, doi: 10.1016/j.immuni.2005.05.010. [DOI] [PubMed] [Google Scholar]
- [109].Nziza N et al. , “Defining Discriminatory Antibody Fingerprints in Active and Latent Tuberculosis,” Front Immunol, vol. 13, p. 856906, 2022, doi: 10.3389/fimmu.2022.856906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Coler RN et al. , “The TLR-4 agonist adjuvant, GLA-SE, improves magnitude and quality of immune responses elicited by the ID93 tuberculosis vaccine: first-in-human trial,” NPJ Vaccines, vol. 3, p. 34, 2018, doi: 10.1038/s41541-018-0057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Yao S, Huang D, Chen CY, Halliday L, Wang RC, and Chen ZW, “CD4+ T cells contain early extrapulmonary tuberculosis (TB) dissemination and rapid TB progression and sustain multieffector functions of CD8+ T and CD3-lymphocytes: mechanisms of CD4+ T cell immunity,” J Immunol, vol. 192, no. 5, pp. 2120–32, Mar 1 2014, doi: 10.4049/jimmunol.1301373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lin PL et al. , “CD4 T cell depletion exacerbates acute Mycobacterium tuberculosis while reactivation of latent infection is dependent on severity of tissue depletion in cynomolgus macaques,” AIDS Res Hum Retroviruses, vol. 28, no. 12, pp. 1693–702, Dec 2012, doi: 10.1089/AID.2012.0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Han SA, Ko Y, Shin SJ, and Jhun BW, “Characteristics of Circulating CD4(+) T Cell Subsets in Patients with Mycobacterium avium Complex Pulmonary Disease,” J Clin Med, vol. 9, no. 5, May 3 2020, doi: 10.3390/jcm9051331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Schenkel AR et al. , “Characterization of Immune Cells From the Lungs of Patients With Chronic Non-Tuberculous Mycobacteria or Pseudomonas aeruginosa Infection,” Immune Netw, vol. 22, no. 3, p. e27, Jun 2022, doi: 10.4110/in.2022.22.e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Pfeffer PE, Hopkins S, Cropley I, Lowe DM, and Lipman M, “An association between pulmonary Mycobacterium avium-intracellulare complex infections and biomarkers of Th2-type inflammation,” Respir Res, vol. 18, no. 1, p. 93, May 15 2017, doi: 10.1186/2931-017-0579-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Brode SK et al. , “Increased risk of mycobacterial infections associated with anti-rheumatic medications,” Thorax, vol. 70, no. 7, pp. 677–82, Jul 2015, doi: 10.1136/thoraxjnl-2014-206470. [DOI] [PubMed] [Google Scholar]
- [117].Coler RN, Hudson T, Hughes S, Huang PW, Beebe EA, and Orr MT, “Vaccination Produces CD4 T Cells with a Novel CD154-CD40-Dependent Cytolytic Mechanism,” J Immunol, vol. 195, no. 7, pp. 3190–7, Oct 1 2015, doi: 10.4049/jimmunol.1501118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Brighenti S and Andersson J, “Induction and regulation of CD8+ cytolytic T cells in human tuberculosis and HIV infection,” Biochem Biophys Res Commun, vol. 396, no. 1, pp. 50–7, May 21 2010, doi: 10.1016/j.bbrc.2010.02.141. [DOI] [PubMed] [Google Scholar]
- [119].Chen CY et al. , “A critical role for CD8 T cells in a nonhuman primate model of tuberculosis,” PLoS Pathog, vol. 5, no. 4, p. e1000392, Apr 2009, doi: 10.1371/journal.ppat.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Mazzaccaro RJ et al. , “Cytotoxic T lymphocytes in resistance to tuberculosis,” Adv Exp Med Biol, vol. 452, pp. 85–101, 1998, doi: 10.1007/978-1-4615-5355-7_11. [DOI] [PubMed] [Google Scholar]
- [121].Kamath AB, Woodworth J, Xiong X, Taylor C, Weng Y, and Behar SM, “Cytolytic CD8+ T cells recognizing CFP10 are recruited to the lung after Mycobacterium tuberculosis infection,” J Exp Med, vol. 200, no. 11, pp. 1479–89, Dec 6 2004, doi: 10.1084/jem.20041690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Jeyanathan M et al. , “Murine airway luminal antituberculosis memory CD8 T cells by mucosal immunization are maintained via antigen-driven in situ proliferation, independent of peripheral T cell recruitment,” Am J Respir Crit Care Med, vol. 181, no. 8, pp. 862–72, Apr 15 2010, doi: 10.1164/rccm.200910-1583OC. [DOI] [PubMed] [Google Scholar]
- [123].Smith SM et al. , “Human CD8(+) T cells specific for Mycobacterium tuberculosis secreted antigens in tuberculosis patients and healthy BCG-vaccinated controls in The Gambia,” Infect Immun, vol. 68, no. 12, pp. 7144–8, Dec 2000, doi: 10.1128/IAI.68.12.7144-7148.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Tascon RE, Stavropoulos E, Lukacs KV, and Colston MJ, “Protection against Mycobacterium tuberculosis infection by CD8+ T cells requires the production of gamma interferon,” Infect Immun, vol. 66, no. 2, pp. 830–4, Feb 1998, doi: 10.1128/IAI.66.2.830-834.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Serbina NV and Flynn JL, “CD8(+) T cells participate in the memory immune response to Mycobacterium tuberculosis,” Infect Immun, vol. 69, no. 7, pp. 4320–8, Jul 2001, doi: 10.1128/IAI.69.7.4320-4328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Serbina NV, Liu CC, Scanga CA, and Flynn JL, “CD8+ CTL from lungs of Mycobacterium tuberculosis-infected mice express perforin in vivo and lyse infected macrophages,” J Immunol, vol. 165, no. 1, pp. 353–63, Jul 1 2000, doi: 10.4049/jimmunol.165.1.353. [DOI] [PubMed] [Google Scholar]
- [127].Brookes RH, Pathan AA, McShane H, Hensmann M, Price DA, and Hill AV, “CD8+ T cell-mediated suppression of intracellular Mycobacterium tuberculosis growth in activated human macrophages,” Eur J Immunol, vol. 33, no. 12, pp. 3293–302, Dec 2003, doi: 10.1002/eji.200324109. [DOI] [PubMed] [Google Scholar]
- [128].Nyendak MR et al. , “Mycobacterium tuberculosis specific CD8(+) T cells rapidly decline with antituberculosis treatment,” PLoS One, vol. 8, no. 12, p. e81564, 2013, doi: 10.1371/journal.pone.0081564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Lancioni C et al. , “CD8+ T cells provide an immunologic signature of tuberculosis in young children,” Am J Respir Crit Care Med, vol. 185, no. 2, pp. 206–12, Jan 15 2012, doi: 10.1164/rccm.201107-1355OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Orme IM, Roberts AR, and Collins FM, “Lack of evidence for a reduction in the efficacy of subcutaneous BCG vaccination in mice infected with nontuberculous mycobacteria,” Tubercle, vol. 67, no. 1, pp. 41–6, Mar 1986, doi: 10.1016/0041-3879(86)90030-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Intracellular cytokine staining (ICS) gating scheme for CD44+, CD4+, and CD8+ T cells expressing TH1 (IFN-γ, TNF-α, IL-2), TH2 (IL-5), and TH17 cytokines (IL-17A). ID91 stimulation shown.


