Abstract
ParB is one of two P1-encoded proteins that are required for active partition of the P1 prophage in Escherichia coli. To probe the native domain structure of ParB, we performed limited proteolytic digestions of full-length ParB, as well as of several N-terminal and C-terminal deletion fragments of ParB. The C-terminal 140 amino acids of ParB form a very trypsin-resistant domain. In contrast, the N terminus is more susceptible to proteolysis, suggesting that it forms a less stably folded domain or domains. Because native ParB is a dimer in solution, we analyzed the ability of ParB fragments to dimerize, using both the yeast two-hybrid system and in vitro chemical cross-linking of purified proteins. These studies revealed that the C-terminal 59 amino acids of ParB, a region within the protease-resistant domain, are sufficient for dimerization. Cross-linking and yeast two-hybrid experiments also revealed the presence of a second self-association domain within the N-terminal half of ParB. The cross-linking data also suggest that the C terminus is inhibitory to multimerization through the N-terminal domain in vitro. We propose that the two multimerization domains play distinct roles in partition complex formation.
Bacterial chromosome and low-copy-number plasmid segregation, or partition, is an essential process, but its mechanism(s) is not well understood. It is an active positioning reaction that ensures that each daughter cell receives an intact genome at cell division. Active partition systems have been identified in several low-copy-number plasmids (58). The P1 prophage exists as a unit-copy-number plasmid whose stability requires the action of its partition system. The latter consists of two proteins, ParA and ParB, and a cis-acting site, parS (1). Genes encoding homologous proteins have also been identified in the chromosomes of several bacterial species including Bacillus subtilis and Caulobacter crescentus (31, 40). This suggests that an analogous mechanism of segregation operates in these diverse systems.
ParA and ParB are multifunctional proteins. Both are essential for partition and play roles in the regulation of their own genes. The ParA protein belongs to a large family of ATP binding proteins (37, 41), and it binds and hydrolyzes ATP (11, 13). ParA binds to a DNA site next to the par promoter, thereby repressing expression of both parA and parB genes (13, 29). Repression is enhanced by the presence of ParB, although ParB does not repress transcription on its own (20). ParA’s ATPase and site-specific DNA binding activities are both stimulated by ParB (11, 13). Although ParA’s role in partition is still not defined, ParA has been shown to interact directly with ParB bound to parS in an ATP-dependent manner (3).
ParB binds specifically to the parS site (12, 22). The Escherichia coli protein integration host factor (IHF) and ParB bind cooperatively to parS to form the partition complex, with each greatly enhancing the other’s affinity for the site (23–25). ParB recognizes two distinct sequences within parS, the heptameric box A sequence and the hexameric box B sequence (12, 24), which are asymmetrically arranged on either side of an IHF recognition sequence (22, 23). IHF is a DNA bending protein (49, 50, 56). The IHF-directed bend at parS is thought to facilitate ParB contacts with its recognition sequences across this bend, resulting in a partition complex in which parS DNA is wrapped around a ParB-IHF protein core (22, 23). It is this complex that directs the plasmid to its specific location(s) inside the cell.
The hydrodynamic and cross-linking properties of ParB indicate that it is an asymmetric dimer in solution (23). However, there are no canonical dimerization motifs, such as leucine zippers, in ParB, and therefore the regions involved in dimerization are not obvious. Point mutations within the C-terminal region of ParB disrupt the protein’s cross-linking activity, indicating that this region is important for dimerization (39). These mutations also eliminated DNA binding activity, and it was suggested that this was a result of the dimerization defect. In addition, previous studies have indicated that ParB is involved in various types of self-association interactions. These include the observations that excess ParB destabilizes plasmids containing parS (21) and that ParB can silence the expression of genes that are located near parS (51). In the former case, the evidence suggests that excess ParB self-associates and forms ParB-ParB-plasmid aggregates that can no longer be properly partitioned and are quickly lost from a population of cells. The silencing data indicate that when ParB binds the DNA at parS, it polymerizes and forms a nucleoprotein filament that extends beyond par. Finally, it has been proposed that plasmids pair during partition, mediated by ParB-ParB association (2, 43). A pairing interaction has been observed in an analogous plasmid partition system, that of the R1 plasmid. ParR, the ParB equivalent, binds DNA at parC and has an intrinsic pairing activity. This activity is stimulated by ParM, the ParA analogue (33). Therefore, multimers of P1 ParB, including dimers and higher-order oligomers, form and contribute to ParB function in vivo.
In this study we have used limited proteolytic digestion of ParB to identify the resistant structural domains of the protein and begin to correlate these domains with function, specifically ParB’s ability to dimerize. Limited proteolysis is a classical way to isolate and define such functional domains (reviewed in reference 36). Typically, at low protease concentrations, the more flexible linker regions of proteins are accessible to the protease and are cleaved while the more stably structured regions are left intact. Recent examples of this approach include domain analysis of Thermus thermophilus UvrB protein (42), T4 intron-encoded I-TevI endonuclease (14), the Saccharomyces cerevisiae transcription factor Swi6 (53), and the human apurinic/apyrimidinic endonuclease (54). SopB, a ParB homologue encoded by the F plasmid, has also been examined by limited proteolytic digestion (27), and the C terminus of the protein was determined to be required for DNA binding activity in vitro. Here we show that the C terminus of ParB forms a domain that is highly resistant to protease.
The yeast two-hybrid system, a genetic assay for protein-protein interactions performed in S. cerevisiae (18), has been successful in identifying interacting partners in many eukaryotic systems (reviewed in reference 19) and in several prokaryotic systems (30, 34, 46, 57). A direct interaction between ParM and ParR of the R1 plasmid was initially demonstrated with the yeast two-hybrid system (32). Self-association domains have also been identified by two-hybrid analysis (9, 59). In this study, we have used the yeast two-hybrid system as well as chemical cross-linking to examine the self-association interactions of ParB.
By assaying a series of N-terminal and C-terminal fragments of ParB, we found that the last 59 amino acids contained a dimerization domain. Interestingly, this domain is located within the region of ParB that is highly resistant to proteolysis. These studies also revealed that a second self-association activity was present in the N-terminal half of ParB. Together, these multimerization domains likely regulate the oligomeric structure of ParB and contribute to its stability and activity throughout partition.
MATERIALS AND METHODS
Bacterial and yeast strains.
E. coli DH5 [F− endA1 hsdR17 (rK− mK+) supE44 thi-1 recA1 gyrA96 relA1] was used for all plasmid constructions. E. coli BL21 [F− ompT hsdSB(rB− mB−) dcm gal](λDE3, pLysS) (55) and BB101 [ara Δ(lac pro) halA argE(Am) rif thi-1 ΔslyDF′ lacIq lacZ::Tn5 pro+] (λDE3) were used for fusion protein expression and purification.
S. cerevisiae Y153 (MATa gal4 gal80 his3 trp1-901 ade2-101 ura3-52 leu2-3,112 URA3::GALlacZ LYS2::GALHIS3) (16) was used for the yeast two-hybrid analysis.
Media and antibiotics.
All bacterial cells were grown in Luria-Bertani (LB) medium or on LB plates (52). The antibiotics and concentrations used were as follows: ampicillin, 100 μg/ml; chloramphenicol, 25 μg/ml; and kanamycin, 25 μg/ml. Yeast cells were grown in YPD medium (1% yeast extract, 2% Bacto Peptone, 2% glucose). Plasmid-containing yeast strains were grown in SD broth (0.67% yeast nitrogen base, 2.5% glucose) supplemented with the appropriate nutrients (tryptophan, 40 mg/liter; leucine, 100 mg/liter; histidine, 20 mg/liter; adenine, 40 mg/liter; uracil, 20 mg/liter). Agar was added to a concentration of 2% for plates. To detect expression of the HIS3 reporter gene, SD plates lacked histidine and contained 25 mM 3-amino-1,2,4-triazole (3-AT) (16).
Reagents and buffers.
Sources for reagents were as follows: 3-AT, amino acids, dithiobis[succinimidyl propionate] (DSP), bovine serum albumin (BSA), and guanidine hydrochloride, Sigma; trypsin and chymotrypsin, Worthington Biochemical; X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactopyranoside), Jersey Supply Lab; imidazole, Research Organics Inc.; yeast nitrogen base, Difco; Bradford reagent, Bio-Rad. Enzymes for cloning were purchased from New England Biolabs or Boehringer Mannheim. Resins used were Ni-nitrilotriacetic acid resin (Qiagen) or chelating Sepharose Fast-Flow (Pharmacia). The latter was preequilibrated with Ni2+ by washing twice with 4 volumes of sterile water, mixing with 2 volumes of 0.1 M NiSO4 for at least 10 min and then washing with 5 volumes of water. The Ni2+-agarose resins were equilibrated with 10 volumes of the appropriate purification buffer before use. Sonication buffer was 50 mM sodium phosphate (pH 8.0)–300 mM NaCl–7 mM β-mercaptoethanol. Wash buffer was sonication buffer with 10% glycerol. Buffer A was 100 mM NaH2PO4–10 mM Tris–6 M guanidine-HCl (pH 8.0). Buffer F was 0.2 M acetic acid–6 M guanidine-HCl.
Plasmid construction.
The DNA encoding full-length ParA (FL-ParA) and FL-ParB and the DNA encoding ParB fragments were cloned into one of four vectors for analysis in this work (Tables 1 and 2). The vectors for the yeast two-hybrid system were pAS1 and pACTII (16), and the vector for purification was either pET19b-HMK (carrying a heart muscle kinase [HMK] fragment) (8) or pJS124 (Table 1). To create pJS124, pET19b-HMK was digested with BamHI and treated with Klenow DNA polymerase and deoxynucleoside triphosphates. Two complementary linkers, 5′GGATCCATGAGTGAGTGA and 5′TCACTCACTCATGGATCC, were then ligated into this site, creating a new BamHI site and inserting stop codons in all three frames downstream of the BamHI site. The stop codons provide translational stop signals for the 3′ deletions of parB. The sequence of the linkers was designed to avoid hydrophobic residues at the C terminus of the fusion proteins, to avoid targeting them for intracellular proteolysis (4, 45).
TABLE 1.
Vectors and P1 plasmids
| Vector | Description | Marker(s)a | Reference |
|---|---|---|---|
| pAS1 | GAL4 DNA binding domain vector | TRP1, Apr | 16 |
| pACTII | GAL4 activation domain vector | LEU2, Apr | 16 |
| pET19b-HMK | Protein expression vector; N-terminal 10-His tag; HMK phosphorylation site | Apr | 8 |
| pJS124 | pET19b-HMK derivative; stop codons inserted in all 3 frames at BamHI site | Apr | This study |
| pJS9 | parB in pBluescript SK+ | Apr | This study |
| pJS10 | parB in pBluescript SK+; opposite orientation of pJS9 | Apr | This study |
| pJS49 | pJS10 with BglII site immediately upstream of parB | Apr | This study |
Apr indicates ampicillin resistance in E. coli. TRP1 and LEU2 are nutritional markers in S. cerevisiae.
TABLE 2.
Plasmids for yeast two-hybrid analysis and for protein expression
| Plasmid | Vector | ParB fragmenta |
|---|---|---|
| For yeast two-hybrid analysis | ||
| pJS50 | pAS1 | FL-ParB |
| pJS51 | pACTII | FL-ParB |
| pMR2 | pAS1 | 30-333 ParB |
| pMR4 | pACTII | 30-333 ParB |
| pJS37 | pACTII | 47-333 ParB |
| pJS38 | pACTII | 67-333 ParB |
| pJS39 | pACTII | 87-333 ParB |
| pJS40 | pACTII | 93-333 ParB |
| pJS41 | pACTII | 187-333 ParB |
| pJS44 | pACTII | 275-333 ParB |
| pJS45 | pAS1 | 275-333 ParB |
| pJS141 | pACTII | 1-312 ParB |
| pMBP1 | pACTII | 1-293 ParB |
| pJS136 | pACTII | 1-277 ParB |
| pJS139 | pACTII | 1-234 ParB |
| pJS140 | pACTII | 1-189 ParB |
| pJS138 | pACTII | 1-177 ParB |
| pJS137 | pACTII | 1-128 ParB |
| pJS182 | pACTII | 1-61 ParB |
| pJS181 | pAS1 | 1-61 ParB |
| pBEF217 | pACTII | FL-ParA |
| For protein expression | ||
| pJS12 | pET19b-HMK | FL-ParB (tag A1) |
| pJS117 | pET19b-HMK | 47-333 ParB (tag B) |
| pJS118 | pET19b-HMK | 67-333 ParB (tag B) |
| pJS119 | pET19b-HMK | 87-333 ParB (tag B) |
| pJS120 | pET19b-HMK | 93-333 ParB (tag B) |
| pMD11 | pET19b-HMK | 152-333 ParB (tag A2) |
| pMD12 | pET19b-HMK | 175-333 ParB (tag A2) |
| pJS121 | pJS124 | 187-333 ParB (tag A3) |
| pMD13 | pET19b-HMK | 267-333 ParB (tag A2) |
| pJS123 | pJS124 | 275-333 ParB (tag A3) |
| pJS172 | pJS124 | 1-312 ParB (tag A4) |
| pJS151 | pJS124 | 1-293 ParB (tag A4) |
| pJS128 | pJS124 | 1-277 ParB (tag A4) |
| pJS189 | pJS124 | 1-274 ParB (tag A4) |
| pJS146 | pJS124 | 1-245 ParB (tag A4) |
| pJS164 | pJS124 | 1-234 ParB (tag A4) |
| pJS143 | pJS124 | 1-189 ParB (tag A4) |
| pJS144 | pJS124 | 1-188 ParB (tag A4) |
| pJS145 | pJS124 | 1-182 ParB (tag A4) |
| pJS129 | pJS124 | 1-177 ParB (tag A4) |
| pJS166 | pJS124 | 1-128 ParB (tag A4) |
| pJS149 | pJS124 | 1-114 ParB (tag A4) |
| pJS148 | pJS124 | 1-61 ParB (tag A4) |
| pJS198 | pJS124 | 47-177 ParB (tag A3) |
Sequences of the N-terminal tags are shown in Fig. 1A.
Parent parB plasmids were generated for subsequent constructions. First, the DraI site in P1 DNA downstream of parB was changed to a BamHI site, and the resulting P1 BglII-BamHI parB fragment was cloned into the BamHI site of pBluescriptII SK+ (Stratagene), creating pJS9 and pJS10 (opposite orientations of the complete parB gene). Site-directed mutagenesis (38) of pJS10 introduced a new BglII site upstream of the parB ATG, creating pJS49. The new BglII site allowed in-frame fusion of the parB ATG to the reading frames of the two-hybrid vectors and of pJS124. In a separate mutagenesis, a BglII site was introduced in a different location upstream of parB to allow in-frame cloning into pET19b-HMK.
The parB deletion plasmids were created by exonuclease III digestion of parB, using a modified protocol from the New England Biolabs Biolabs Exo-Size kit. For 5′ deletions, pJS9 was digested with KpnI and EcoRI and then treated with exonuclease III. Exonuclease products were treated with mung bean nuclease and then Klenow DNA polymerase and deoxynucleoside triphosphates to ensure blunt DNA ends. Following ligation to 12-bp BamHI linkers, the DNA was recircularized and used to transform E. coli DH5. The 3′ deletions of parB were constructed in a similar manner, except that the starting plasmid was pJS49 (see above). The endpoint of each parB deletion was determined by dideoxy DNA sequencing (Pharmacia T7 sequencing kit) (Table 2). This process produced parB gene fragments flanked by BamHI and/or BglII sites. Fragments in which the deletion was in the proper reading frame for insertion into the BamHI sites of pAS1 and pACTII were chosen for the yeast two-hybrid experiments.
ParB fragments covering a wide range of sizes were selected for in vitro analyses. In most cases, the corresponding parB gene fragments were cloned as BamHI or BamHI/BglII fragments into the BamHI site of pJS124 or pET19b-HMK for protein purification. This created fragments fused to histidine tag A (Fig. 1A). Several of the 5′ deletions of parB were digested with BamHI and then treated with Klenow DNA polymerase. This DNA fragment was ligated into pET19b-HMK that had been digested with XhoI and treated with Klenow DNA polymerase. This cloning strategy removed the HMK sequence from the tag, resulting in tag B (Fig. 1A).
FIG. 1.
(A) Sequences of the N-terminal tags of the purified fusion proteins used in this study. Tags A1 to A4 are encoded by pET19b-HMK and pJS124. Slightly different versions of the tag were generated as a result of cloning from different sources. In tag B, the HMK sequence has been eliminated in the cloning protocol (see Materials and Methods). The HMK recognition sequence is a phosphorylation site for the catalytic subunit of bovine HMK. (B) Sequence of ParB. The arrows indicate the N terminus of each proteolytic fragment identified in this work. Bands A to E were generated by trypsin digestion. Band F was produced by chymotrypsin digestion.
To create the 47–177 ParB fragment, pJS18 (parB deleted for the nucleotides encoding amino acids 1 to 46 cloned into pBluescript SK+) was used as a substrate for PCR, and the region encoding up to amino acid 177 was amplified. The upstream primer was the M13 reverse primer. The downstream primer (5′GCGCAGATCTTACAGCCCTTCTTTGGCTGC) changed amino acid 178 to a stop codon and created a BglII site to facilitate cloning. The PCR product was purified from an agarose gel, digested with BamHI and BglII, and cloned into the BamHI site of pJS124.
To construct pBEF217, the P1 parA gene was modified so that it was flanked by two NdeI sites. Creation of the upstream NdeI site, which overlaps the parA ATG start codon, is described in reference 10. The PvuI site in the beginning of the parB gene was altered with synthetic linkers to create an NdeI site downstream of parA. The resulting parA NdeI fragment was then inserted into pACTII.
Yeast two-hybrid analysis.
Y153 cells were transformed by two different plasmids by using an adaptation of a high-efficiency transformation protocol (26). To test for expression of the lacZ reporter gene in yeast, transformants were grown as 1- to 2-cm patches on minimal plates. Cell patches were replica plated onto new plates overlaid with a no. 50 Whatman filter circle and incubated overnight at 30°C. Next, a no. 3 Whatman filter circle was immersed in 2.5 ml of X-Gal solution (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 0.038 mM β-mercaptoethanol, 0.2% X-Gal) in a sterile petri plate. The no. 50 replica filter was frozen in liquid N2, warmed to room temperature, and then overlaid colony side up on the saturated no. 3 filter circle. The plate was closed, wrapped in Parafilm, and incubated at 30°C. The time required for color development ranged from 1 h to overnight. Each fusion protein was tested for activation of the reporter genes in the presence of its partner GAL4 domain alone. None exhibited more than low, background levels of lacZ expression, and none promoted growth in the absence of histidine.
Protein purification.
For native protein purification, a 500-ml culture of BL21(λDE3, pLysS) cells transformed by a plasmid encoding a histidine-tagged ParB fusion protein was grown at 37°C in LB medium containing ampicillin and chloramphenicol to an A600 of approximately 0.5. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to 1 mM, and the culture was incubated for an additional 2 h at 37°C. The cells were collected by centrifugation, washed and resuspended in sonication buffer, and frozen in liquid nitrogen. The cells were thawed on ice, lysed by sonication bursts, and centrifuged at 25,000 × g for 1 h at 0°C. The remaining steps were performed at 4°C. The supernatant was collected and loaded onto a small (0.5 to 1 ml) Ni2+-agarose column. The column was washed with 10 column volumes of sonication buffer and then with 20 column volumes of wash buffer. The fusion protein was then eluted in steps of increasing imidazole concentration. Most of the fusions eluted at about 400 mM imidazole, and the majority of the ParB fragments purified well with this method. However, some fragments, particularly the small N-terminal fragments (1-33, 1-61, and 1-114 ParB), were much cleaner when purified with a denaturing protocol. This method did not affect the cross-linking of the proteins, as determined by examination of several proteins (His-ParB, 47-333 ParB, and 1-293 ParB) that were purified in parallel by both methods.
For denaturing purification, a 25-ml culture of BB101 cells transformed by a plasmid encoding a ParB fusion protein was grown at 37°C in LB medium containing ampicillin and kanamycin to an A600 of approximately 0.8. IPTG was added to 1 mM, and the culture was incubated at 37°C for another 2 h. The cells were collected by centrifugation, resuspended in 0.75 ml of buffer A, and mixed gently by slow rotation at room temperature for at least 1 h. The lysate was centrifuged at 14,000 rpm for 15 min in a microcentrifuge (Heraeus Biofuge). The supernatant was removed and mixed gently with 75 μl of Ni2+-agarose resin at room temperature for 15 min. The resin was collected by centrifugation and washed three times with 1 ml of buffer A. To elute protein, the resin was mixed with 750 μl of buffer F and recentrifuged. The supernatant was collected and dialyzed against decreasing concentrations of guanidine-HCl until the protein was in wash buffer.
ParB (with no His tag) was purified as described previously (10, 23). Protein concentrations were determined by the Bradford protein assay (5).
DSP cross-linking.
Protein samples were diluted to between 5 and 20 μg/ml in 50 mM HEPES-KOH (pH 7.5)–150 mM NaCl–0.1 mM EDTA. DSP (20 mg/ml in dimethylformamide) was added to 0.1 mg/ml, and the mixtures were incubated at room temperature. To stop the cross-linking reaction and to precipitate the protein; 750-μl samples were removed, mixed with an equal volume of 30% trichloroacetic acid, and incubated on ice for 20 min. The precipitate was collected by centrifugation at 4°C, washed with acetone, and resuspended in 30 μl of 62.5 mM Tris-HCl (pH 6.8)–2% sodium dodecyl sulfate (SDS)–10% glycerol–0.025% bromophenol blue. The samples were incubated at 90°C for 3 min and then analyzed by SDS-polyacrylamide gel electrophoresis (PAGE).
Proteolysis.
ParB or ParB fragments were incubated with trypsin or chymotrypsin in 50 mM Tris-HCl (pH 7.5)–100 mM NaCl–0.1 mM EDTA–20% glycerol at room temperature. The protein/protease ratios used ranged from 100:1 to 5,000:1 (wt/wt) and are indicated in the figure legends. Digestion was stopped by the addition of acetic acid to 1%. For sequencing of the N termini, proteolytic digestions were flash frozen on dry ice and stored at −20°C.
Protein sequencing.
The protein products were separated by electrophoresis in a 15% SDS-polyacrylamide gel and then were transferred to a polyvinylidene difluoride membrane, using a Multiphor II electrophoresis system (Pharmacia). The filter was rinsed several times in distilled water and stained with 0.2% Coomassie blue in 50% methanol for 5 min. The filter was destained with several washes of 50% methanol, air dried, wrapped in plastic, and stored at −20°C. Sequencing was then performed at the HSC Biotechnology Service Centre, Toronto, Ontario, Canada, and at the Alberta Peptide Institute, Edmonton, Alberta, Canada.
Nucleotide sequence accession number.
The GenBank accession number for ParB is 215655.
RESULTS
ParB is a multifunctional protein with several different biochemical activities. These include its ability to dimerize, its specific DNA binding to sequences within parS, and its interaction with ParA. It may also be involved in interactions with host cell components required for partition. In this work, we took two different approaches to identify and map the structural and functional domains in ParB. One approach was to use proteolytic digestion to analyze the domain structure of the protein. We also assayed fragments of ParB for dimerization activity to define functional domains.
Proteolytic digestion of ParB.
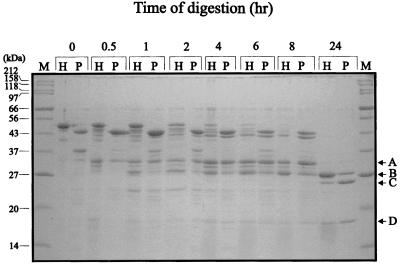
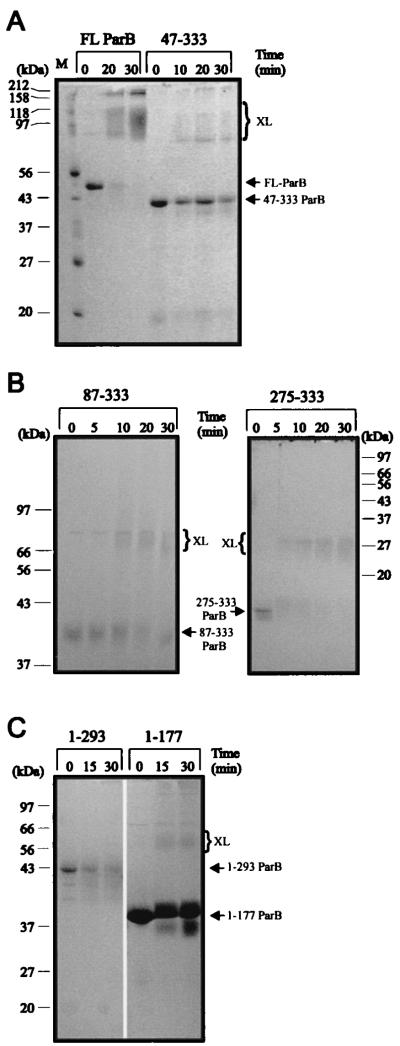
We first treated ParB with low concentrations of trypsin, a protease that cleaves on the carboxy side of lysine and arginine residues (36), and examined the digestion patterns by gel electrophoresis (Fig. 2). We compared this pattern to that produced from a version of ParB that was tagged at its N terminus with a polyhistidine sequence (Fig. 2). ParB and His-ParB contain 53 and 56 arginine plus lysine residues, respectively. After tryptic digestion of ParB and His-ParB, a discrete pattern of proteolytic products was seen. The major proteolytic fragments were identified by sequencing their N termini (Fig. 1 and Table 3). The first major fragment that appeared migrated at about 32 kDa (Fig. 2, band A) and started at amino acid 83. At later time points, a smaller band with an apparent size of 30 kDa (band B), starting at residue 124, was produced. With more extensive digestion, two fragments (bands C and D) with apparent sizes of 25 and 18 kDa, respectively, were generated. The latter two fragments began at residues 142 and 185, respectively, and persisted, even after very long periods of digestion. A smaller fragment of about 10 kDa (starting at residue 263) would occasionally be seen, especially in digestions performed at 30°C (Fig. 3), but in general bands C and D were particularly resistant to further proteolysis. Note that the sizes cited here are only estimates because ParB runs anomalously on SDS-PAGE. The calculated molecular masses of ParB and His-ParB are 37.4 and 41.7 kDa, respectively, but they migrate at about 44 and 50 kDa, respectively.
FIG. 2.
Tryptic digestion of ParB and His-ParB. ParB (lanes P) and His-ParB (lanes H) were treated with trypsin at a protein/protease ratio of 1,000:1 (wt/wt) at 20°C for the indicated times. Digestion was stopped with 1% acetic acid. Proteolytic fragments were separated by electrophoresis in SDS–15% polyacrylamide gels and were visualized with Coomassie blue. Undigested ParB and His-ParB migrate with 44- and 50-kDa proteins, respectively. The arrows at the right indicate the major tryptic fragments identified in Table 3. Lane M, size markers.
TABLE 3.
N-terminal sequences of proteolytic fragments generated by trypsin and chymotrypsin
| Banda | N-terminal sequenceb | Starts at position: | Molecular mass (kDa)c
|
|
|---|---|---|---|---|
| Apparent (on gel) | Predicted (to C terminus) | |||
| A | ?-Thr-Ile-Lys-His-Gln | 83 | 32.2 | 28.4 |
| B | Val-Leu-Val-Thr-Asp | 124 | 27.9 | 23.9 |
| C | Asp-Val-Gln-Thr-Ala | 142 | 26.8 | 21.9 |
| D | Ala-Leu-Gln-Ala-Ala | 185 | 17.6 | 17.1 |
| E | Glu/Gly-Ala-Ser-Leu-Leu | 263 | 9.1 | 8.4 |
| F | Lys-?-Ile-Arg-Ser-Thr | 79 | 32.2 | 28.9 |
The sequences were determined by Edman degradation, and the first five or six residues are shown. Despite blanks in some sequences, each was consistent with only one position in ParB, assuming that each N-terminal residue was preceded by either an arginine or a lysine for tryptic fragments and by a hydrophobic residue for chymotryptic fragments.
The apparent molecular masses were determined by linear regression analysis (Multi-Analyst software) of the protein gels, and the predicted masses were calculated on the assumption that the fragments extend to the C terminus of the protein.
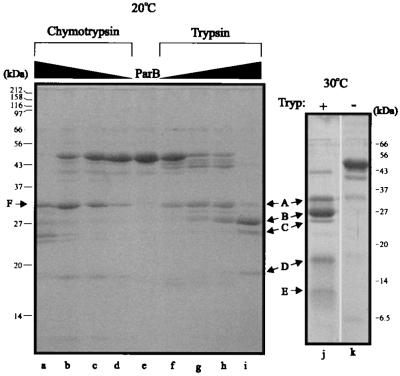
FIG. 3.
Chymotrypsin and trypsin digestion of His-ParB. (Left) Five micrograms of protein was incubated with increasing amounts of protease for 2 h at room temperature. Protein-to-protease ratios (wt/wt) were 100:1 (lanes a and i), 500:1 (lanes b and h), 1,000:1 (lanes c and g), and 5,000:1 (lanes d and f). Lane e, no protease. Arrows indicate the fragments whose N termini were sequenced. (Right) Tryptic digest (protein/protease ratio of 1,000:1) performed at 30°C for 7.5 h, illustrating an additional band (E) that was also sequenced. The positions of size markers are indicated beside each gel. Tryp, trypsin.
The cleavage patterns of the two proteins were very similar. The main difference was that His-ParB digestion generated a number of fragments that migrated slightly faster than the intact protein, which likely represent removal of the His tag from His-ParB (these fragments no longer bound to Ni2+ affinity resin [data not shown]). The similarity in rate and pattern of cleavage for both native and His-tagged ParB indicated that the N-terminal tag did not significantly alter the conformation of ParB. Consequently, to simplify purification of the truncated versions of ParB, we have used the His-tagged versions of ParB and ParB fragments for domain analysis.
Next we examined whether chymotrypsin, which cleaves on the carboxy side of hydrophobic residues (36), detects similar domains in His-ParB (Fig. 3). Several fragments with sizes similar to those of fragments produced by trypsin were observed. Specifically, band F (Fig. 3) appeared only slightly larger than band A and was found to begin at amino acid 79, which was very close to the initial tryptic cleavage. The proteolytic data indicate that the N-terminal approximately 80 amino acids of ParB are relatively accessible to protease and are rapidly digested, suggesting that this region is less stably folded under these conditions.
Bands B, C, D, and E represent increasingly C-terminal portions of ParB. Since these fragments, particularly band D (starting at amino acid 185) and band E (starting at amino acid 263), were observed only at the later time points, we concluded that they must be derived from the larger fragments that are subsequently reduced or disappear. All of these proteolytic fragments migrated more slowly than predicted for fragments that extend to the C terminus (Fig. 3 and Table 3), suggesting that the C terminus is included in these fragments.
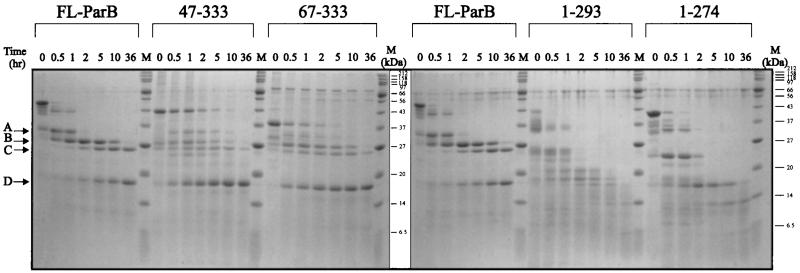
We compared the proteolytic patterns of digestion of FL-ParB with those of two C-terminal fragments and two N-terminal fragments of ParB (Fig. 4). The two C-terminal fragments, His-47-333 ParB and His-67-333 ParB, both generated a cleavage pattern similar to that of full-length His-ParB, including the resistant domains (especially band D) that remained at the later time points. The digestion of two N-terminal fragments, His-1-274 ParB and His-1-293 ParB, produced a series of fragments that were similar to each other but were all smaller than those produced from full-length protein at comparable times. Therefore, removal of the C terminus, but not of the N terminus, of ParB altered the pattern of resistant regions. These results, along with the N-terminal sequences and the sizes (Table 3) of the proteolytic fragments, suggest that the tryptic fragments likely extend to, or very close to, the extreme C terminus of ParB.
FIG. 4.
Tryptic digestion of His-ParB, two C-terminal fragments (His-47-333 ParB and His-67-333 ParB), and two N-terminal fragments (His-1-293 ParB and His-1-274 ParB). For each time course, the protein-to-protease ratio was 1,000:1 (wt/wt), and the digestions were performed at room temperature. Arrows at the left indicate the major proteolytic fragments of His-ParB. M, size markers.
We attempted to confirm the identity of the tryptic fragments by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectroscopy. However, because His-ParB contains 56 arginine plus lysine residues, there was a very large number of potential tryptic fragments. This complicated the analysis, particularly of fragments with a mass of less than 10 kDa. Also, the technique was unable to reproducibly detect fragments of His-ParB larger than about 21 kDa. Despite these problems, one peak of average mass of 17,106 ± 30 Da was seen consistently by mass spectroscopy (data not shown). This mass corresponds only to a fragment consisting of amino acids 185 to 333, whose N terminus is identical to that of band D and whose C terminus corresponds to the C terminus of ParB. Since it is likely that band D is derived from the larger proteolytic fragments, we expect bands A, B, C, and F also extend to amino acid 333. Given the mobility and apparent molecular weight of band E, it likely also extends to or close to the extreme C terminus of ParB.
The data strongly suggest that the region from residues 185 to 333 forms a resistant core structure, within which lies a further resistant domain, from amino acid 263 to 333. Within this structural domain lies a region that has previously been implicated in ParB’s dimerization activity (39). The proteolytic resistance of the C-terminal half of ParB may also be the result of a core structure centered around a strong dimerization interface at the extreme C terminus.
Definition of dimerization domains by yeast two-hybrid and in vitro cross-linking analysis.
We used deletion analysis to define regions of ParB that are required for dimerization. Our first approach was the yeast two-hybrid system, which provided the advantage that both homologous and heterologous interactions could be examined. In the system that we used, ParB or ParB fragments were fused to the DNA binding domain and/or to the activation domain of the S. cerevisiae GAL4 protein. The interaction of the DNA binding fusion with an activation domain fusion activated a lacZ reporter gene, detected by filter tests with X-Gal as a substrate, and a HIS3 reporter gene, detected as growth on minimal plates lacking histidine and containing 3-AT. 3-AT inhibits imidazole glycerol phosphate dehydratase, thus reducing basal histidine biosynthesis (35). An interaction was considered positive only if both reporter genes were activated.
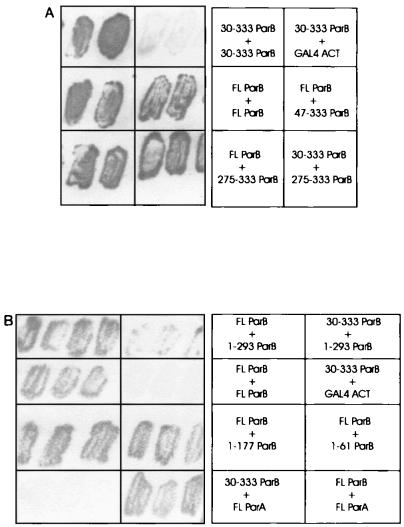
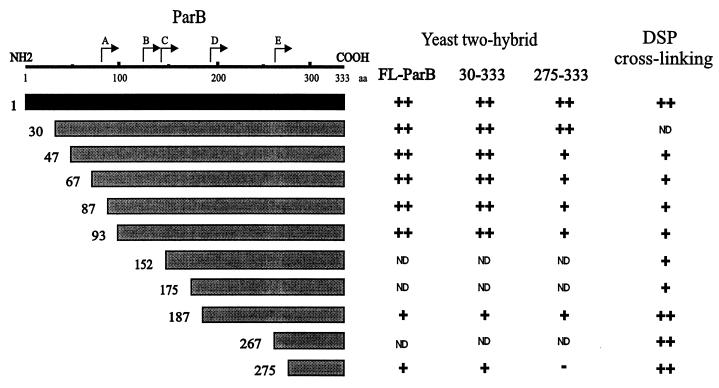
Examples of the activation of lacZ, as detected by X-Gal filter tests, are shown in Fig. 5. When FL-ParB was tested for a self-association interaction (homodimerization) the yeast cells turned blue within a few hours in X-Gal filter tests (Fig. 5A). These transformants also grew well in the absence of histidine. We concluded that ParB dimerization activity was detectable in yeast. Seven different C-terminal fragments of ParB, when fused to the GAL4 activation domain, interacted with FL-ParB (Fig. 6). Two of these fragments, 87-333 ParB and 187-333 ParB, correspond closely to tryptic fragments A and D, respectively. The shortest fragment consisted of only the last 59 amino acids of ParB, indicating that this region of ParB is sufficient to mediate a dimerization interaction with longer ParB fragments. All of the deletions, including 275-333 ParB, were also able to interact with 30-333 ParB when this ParB fragment was fused with the GAL4 DNA binding domain (Fig. 6). Western blotting of cell lysates indicated that in each instance the recombinant proteins were expressed in yeast (data not shown).
FIG. 5.
Examples of filter tests to determine β-galactosidase activity in the yeast two-hybrid system. The light patches were white and the grey patches were blue on the filters. Each panel (A and B) is a separate filter, but the patches within each panel are from the same filter. The particular interactions tested are indicated at the right of each filter. Various ParB and ParB fragment interactions (A and B) and ParA-ParB interactions (B) are shown. GAL4-ACT is the GAL4 activation domain alone encoded by pACTII and represents one of the negative controls for these assays.
FIG. 6.
Summary of dimerization assays with C-terminal fragments of ParB. In yeast two-hybrid analysis, the ParB fragments shown in the diagram were fused to the GAL4 activation domain and were tested against FL-ParB, 30-333 ParB, and 275-333 ParB (in the columns) that were fused to the GAL4 DNA binding domain. For the cross-linking experiments, ParB fragments fused to a polyhistidine tag (Table 2) were purified and examined in vitro (Materials and Methods). The results from the yeast two-hybrid experiments were categorized as follows: −, no color development on filter tests or no growth on plates without histidine; +, moderate color development and moderate growth in the absence of histidine; ++, dark blue color and good growth in the absence of histidine. ND, not determined. The DSP cross-linking results were similarly categorized: −, no cross-linking; +, some cross-linking activity; ++, strong cross-linking, often to completion. Neither set of categories is intended to imply relative strengths of the interactions, which are presumably dependent on the assay. The N termini of the tryptic proteolytic fragments are indicated above the schematic of ParB.
The 275-333 ParB fragment was then fused to the GAL4 DNA binding domain in order to test it against itself and all other C-terminal fragments (Fig. 6). While it interacted with all larger fragments, 275-333 ParB did not interact with itself in yeast. This suggested that a more N-terminal region, between amino acids 187 and 274, was required in at least one monomer for dimerization to be detectable in yeast. To measure dimerization independently of the yeast system, we turned to an in vitro cross-linking assay.
Full-length and truncated versions of ParB, fused to a polyhistidine sequence, were purified and then treated with DSP, a thiol-cleavable cross-linker that interacts with lysines. As has been shown previously (23), FL-ParB cross-linked efficiently to a dimer-sized smear following this treatment (Fig. 7). Because ParB contains 29 lysines, both inter- and intramolecular cross-links occur, resulting in smeary rather than discrete bands on SDS-polyacrylamide gels. All of the C-terminal fragments, including His-275-333 ParB, cross-linked to dimer in this assay, although cross-linking was not always complete. We conclude that the most C-terminal 59 amino acids of ParB define a region that is sufficient for dimerization.
FIG. 7.
Cross-linking of His-ParB and His-ParB fragments with DSP. Ten micrograms of protein was incubated with DSP at room temperature for the indicated times and analyzed by electrophoresis as described in Materials and Methods. Arrows indicate the positions of monomers and the brackets indicate the positions of cross-linked products (XL). (A) Cross-linking of His-ParB compared with that of His-47-333 ParB on an SDS–10% gel. His-ParB cross-links efficiently to a dimer-size and possibly a tetramer-size smear. Lane M, size markers. (B) Cross-linking of two smaller C-terminal fragments in the presence of DSP. His-87-333 ParB and His-275-333 were treated with DSP and analyzed on a 10% polyacrylamide gel (left) and a 12% polyacrylamide gel (right), respectively. (C) Cross-linking reactions of two N-terminal fragments of ParB, His-1-293 ParB, and His-1-177 ParB, in a 12% polyacrylamide gel.
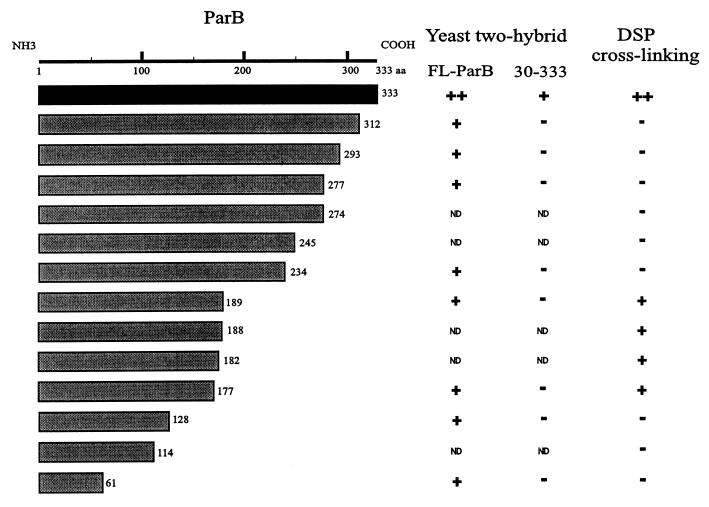
A second self-association domain of ParB.
We next tested various N-terminal fragments (i.e., C-terminal truncations) for the ability to cross-link in vitro. Fragments lacking the extreme C terminus (e.g., His-1-274 ParB) did not cross-link, which was expected since the C-terminal dimerization domain defined above was removed. Surprisingly, as more of the C terminus of ParB was removed, the protein fragments recovered the ability to cross-link with DSP (e.g., His-1-177 ParB). This indicated a second multimerization domain within the N-terminal half of ParB. The data also suggested the presence of an inhibitory region within the C-terminal half of ParB. Fusions consisting of only the first 114 amino acids or less no longer cross-linked with DSP (Fig. 8), but this may be explained by the fact that only a few lysines remain in these fragments.
FIG. 8.
Summary of dimerization assays with N-terminal fragments of ParB. In yeast two-hybrid analysis, the ParB fragments shown in the diagram were fused to the GAL4 activation domain and were tested against FL-ParB and 30-333 ParB (in the columns) fused to the GAL4 DNA binding domain. For the cross-linking experiments, polyhistidine-tagged protein fusions were purified and tested in vitro. The categories are described in the legend to Fig. 6. ND, not determined.
A possible trivial explanation for cross-linking of 1-177 ParB and 1-189 ParB is that the deletion created sticky ends, leading to nonspecific hydrophobic interactions. Two experiments suggested that this was not the case. When an equal concentration of BSA was included in the assay, no interaction between 1-177 ParB and BSA was observed (data not shown). We also removed the N-terminal 46 amino acids from the His-1-177 construct. His-47-177 did not cross-link under the conditions in which His-1-177 did cross-link (data not shown). Both experiments imply that the interaction of His-1-177 with itself is specific. This suggestion was further supported by yeast two-hybrid analysis (see below). It also appears that deletion of the N-terminal 46 amino acids of ParB is sufficient to disrupt the N-terminal self-association domain.
A series of these N-terminal ParB fragments was tested for interactions with full-length ParB and ParB lacking the first 29 amino acids (30-333 ParB) in yeast (Fig. 8). All interacted with FL-ParB, including those that did not cross-link in vitro, indicating that only the first 61 amino acids of ParB are required for this interaction. However, none of the C-terminally truncated proteins interacted with 30-333 ParB-DB. Finally, as with cross-linking of 47-177 ParB, the removal of both the C terminus and the N terminus eliminated self-association of ParB. The inhibition that the C terminus exerted on cross-linking of the N-terminal self-association domain was, however, not seen in yeast. All N-terminal fragments could interact with FL-ParB (Fig. 8). Unfortunately, it was not possible to test for homodimerization of these proteins in yeast because the DNA binding fusions of 1-312, 1-293, 1-277, and 1-234 ParB activated both reporter genes in the absence of an interacting partner. Nevertheless, these results support the presence of a second multimerization domain within the N-terminal region of ParB.
ParA-ParB interactions.
ParA fused to the GAL4 activation domain was able to interact with FL-ParB in the yeast two-hybrid system (Fig. 5B). However, when only the first 29 amino acids were removed from ParB, the interaction was eliminated. These data indicate that the extreme N terminus of ParB is required for an interaction with ParA and are consistent with recent results from experiments using P1-P7 hybrid partition proteins to probe the specificity of ParA-ParB interactions (48). The latter showed that the first 28 amino acids of ParB are necessary for P1 ParB to recognize P1 ParA.
DISCUSSION
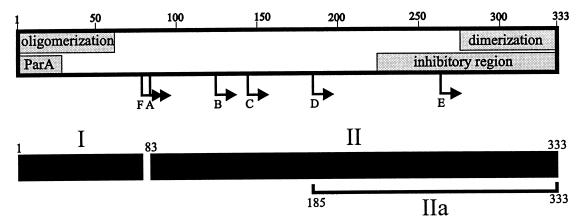
We have probed the domain structure of ParB by partial proteolysis and by analysis of ParB fragments for self-association interactions. Our model of ParB from these experiments is shown in Fig. 9. The major proteolytic fragments were identified and shown to be C terminal, extending to or very close to the extreme C terminus of ParB (Table 3). Our proteolysis results suggest that an approximately 80-amino-acid region at the N terminus of ParB forms an unstable domain (or domains), that is easily accessible and rapidly digested by protease (Fig. 9, region I). The remaining approximately 250-amino-acid region is more structured (Fig. 9, region II). In particular, the 185-333 fragment (band D in Fig. 2 to 4) is very resistant to protease, although further digestion to the 263-333 fragment was also observed. Therefore, the last approximately 140 residues of ParB form an inaccessible, folded structure (Fig. 9, region IIa), within which is a smaller resistant core of 70 amino acid residues. This core structure contains the C-terminal dimerization domain that is defined by chemical cross-linking and yeast two-hybrid analyses. The dimerization interface may contribute to the protection of these C-terminal residues from proteolytic digestion. Consistent with this possibility is the observation that point mutations within the C terminus of ParB that disrupt dimerization result in proteins that are much more susceptible to the OmpT protease (39).
FIG. 9.
Model of ParB’s functional and structural domains. The shaded boxes represent regions of the protein involved in ParB-ParB and ParA-ParB interactions. Arrows indicate the N termini of the proteolytic fragments (A to F) identified in this study. We propose that the C-terminal self-association domain is required for ParB dimerization and the N-terminal self-association domain mediates oligomerization. The inhibitory region affects the self-association of ParB that is mediated by the N-terminal oligomerization domain, as measured by cross-linking assays. The ParA box indicates a region of ParB that is necessary for interactions with ParA. The lower black boxes predict the general structural domains of ParB, from proteolytic assays. Region I represents the protease-accessible N-terminal region, which may mean that it is less structured in solution. Region II is more stable and more protease resistant. Region IIa represents the smallest highly protease resistant region of ParB.
ParB’s self-association domains.
In vitro cross-linking and the yeast two-hybrid system have provided complementary information regarding the dimerization activities of ParB. The last 59 amino acids of ParB were sufficient to interact with full-length ParB and with the other C-terminal fragments in the yeast two-hybrid system (Fig. 5A and 6). This fragment was also cross-linked by DSP in vitro. These data indicate that the C-terminal 59 amino acids contain a dimerization domain. Further, the yeast data indicate that removal of only 21 amino acids from the C terminus disrupts this dimerization interaction (Fig. 8). We also discovered that a fragment consisting of the first 177 amino acids of ParB was cross-linked by DSP in the absence of the C-terminal dimerization domain, providing evidence of an additional multimerization determinant within the N-terminal half of ParB. Deletion of the first 46 amino acids from 1-177 ParB disrupted this determinant. Similarly, while all the N-terminal fragments interacted with FL-ParB in the yeast two-hybrid system, none interacted with 30-333 ParB (Fig. 8). These results strongly suggest that an intact N terminus in both partners is crucial for an interaction in the absence of the C terminus in even one partner. We favor a model in which the C terminus of ParB dimerizes the protein, while the N terminus is involved in forming tetramer or higher oligomeric complexes (see below).
A C-terminal region of ParB inhibits cross-linking in the absence of the C terminus.
ParB fragments missing only a portion of the C terminus (21 to 100 residues) did not cross-link in the presence of DSP, but removal of an additional 54 residues (from residue 190) restored cross-linking activity. Therefore, a region at the C terminus (Fig. 9) is inhibitory to dimerization via the N terminus in a cross-linking assay. This result may explain why point mutations in the C terminus of ParB destroyed its cross-linking ability (39) and failed to reveal the N-terminal self-association domain. On the other hand, this inhibition was not apparent in the yeast two-hybrid experiments. ParB fragments lacking the C terminus, when fused to the GAL4 activation domain, could interact with FL-ParB that was fused to the GAL4 DNA binding domain (Fig. 8). If an inhibition occurred, it was not sufficient to completely destroy the interaction. Alternatively, the yeast two-hybrid system may provide a distinct context that allows these interactions to occur. For example, the full-length partner may prevent the free C terminus of its partner from occluding its N-terminal domain, binding of ParB to DNA in yeast (presumably nonspecifically) may alter the conformation of one or both partners, or the addition of a large GAL4 fusion at the N terminus of the C-terminal deletions may expose the N-terminal self-association domain.
Whether the C terminus of FL-ParB normally prevents self-association of the N-terminal domain is unknown, but this possibility has interesting implications for partition. The C terminus in the intact protein may physically block oligomerization through the N terminus until a specific point in partition, for example, until ParB binds to parS or until ParB binds to ParA. A similar situation exists with the E. coli regulatory protein NtrC (17), which contains two multimerization domains. The first mediates constitutive dimerization and is located at the C terminus of the protein. The second is near the N terminus, and the central domain of the protein sterically inhibits its oligomerization activity, until it is phosphorylated by NtrB. That no resistant region corresponding to the N terminus of ParB was detected following proteolytic digestion may also indicate that the N terminus is not oligomerized in solution.
Possible roles for dimerization in ParB activity in partition.
In this work, we have shown that ParB contains a distinct domain structure, within which exist two self-association determinants that can function independently. Do these two regions have distinct roles in partition? Experiments with hybrid P1-P7 ParB proteins indicate that the C terminus of ParB likely contains more than one function (28, 47). Bacteriophage P7 encodes a partition system that is very similar to that of P1, with homologous ParA and ParB proteins and a similar cis-acting parS site. Species specificity appears to be mediated through recognition of the parS box B sequences, since the parS box A sequences in P1 and P7 are identical. The C terminus of a hybrid ParB protein is responsible for recognizing its cognate box B sequence. The dimerized C terminus may bind directly to the box B sites, and dimerization at the C terminus may be required for box B binding.
It has been proposed that a putative helix-turn-helix (HTH) motif in the center of ParB (amino acids 166 to 189) (15) is responsible for box A binding (47). Classically, HTH DNA binding proteins, such as the Trp repressor, λ Cro, and E. coli catabolite gene activator protein, must be dimeric in order to efficiently bind the DNA, with each partner contributing a DNA binding half site (44). ParB dimerization may bring two HTH motifs together to form a stable DNA binding domain, and one or both multimerization domains may be required to ensure that the HTH domain is intact (Fig. 9).
In vitro studies have shown that ParB is a dimer under all conditions tested (23). We have shown that ParB fragments missing the extreme C terminus do not dimerize in vitro. Similarly, point mutations within the C terminus disrupt dimerization (39). We therefore propose that the C-terminal domain promotes ParB dimerization and that this is a very strong interaction (Fig. 9). We suggest that the N-terminal self-association domain mediates dimer-dimer interactions. This interaction is weaker and occurs only under certain conditions such as when ParB is bound to parS. It leads to the formation of tetramers and higher-order oligomers that are inferred from the observation of plasmid pairing of R1 ParR (33) and gene silencing by P1 ParB (51). It is also of interest that the N terminus of the F-encoded ParB homologue SopB is involved in gene silencing, suggesting that it may also be involved in protein multimerization (27). The lac repressor similarly has two multimerization domains, one that allows dimerization and a second at its extreme C terminus that promotes tetramerization through dimer-dimer interactions (6, 7).
The 29 amino acids at the extreme N terminus of ParB are required for a ParA-ParB interaction (48) (Fig. 5B). Therefore, the N-terminal domain of ParB contains both a ParA and a ParB (self) association function. ParA-ParB interactions occur in at least two aspects of partition. ParB acts as a corepressor to stimulate the repressor activity of ParA (20), and ParA assembles on the ParB-IHF partition complex at parS in an ATP-dependent reaction (3). In the latter case, it is interesting that at high ParA-to-ParB ratios, ParA prevents or inhibits ParB binding to parS. Perhaps the ParA-ParB interaction (when in excess) interferes with the N-terminal ParB-ParB association. We do not yet know whether such ParA-ParB and ParB-ParB interactions both occur in the context of the partition complex or whether they are mutually exclusive. In light of pairing proposals, we favor the former possibility but this has still to be determined. The next step is to establish how the self-association domains contribute to ParB’s activities in partition.
ACKNOWLEDGMENTS
We are grateful to Don Awrey for performing the MALDI-TOF experiments and for help in analyzing the results. We also thank Alan Davidson and Marc Perry for critical reading of the manuscript.
This work was supported by a University of Toronto Open Fellowship (to J.A.S.) and a grant from the Medical Research Council of Canada (to B.E.F.).
REFERENCES
- 1.Abeles A L, Friedman S A, Austin S J. Partition of unit-copy miniplasmids to daughter cells. III. The DNA sequence and functional organization of the P1 partition region. J Mol Biol. 1985;185:261–272. doi: 10.1016/0022-2836(85)90402-4. [DOI] [PubMed] [Google Scholar]
- 2.Austin S, Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983;169:373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- 3.Bouet J-Y, Funnell B E. P1 ParA interacts with the P1 partition complex at parS and an ATP-ADP switch controls ParA activities. EMBO J. 1999;18:1415–1424. doi: 10.1093/emboj/18.5.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowie J U, Sauer R T. Identification of C-terminal extensions that protect proteins from intracellular proteolysis. J Biol Chem. 1989;264:7596–7602. [PubMed] [Google Scholar]
- 5.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Matthews K S. Subunit dissociation affects DNA binding in a dimeric Lac repressor produced by C-terminal deletion. Biochemistry. 1994;33:8728–8735. doi: 10.1021/bi00195a014. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Suredran R, Lee J C, Matthews K S. Construction of a dimeric repressor: dissection of subunit interfaces in Lac repressor. Biochemistry. 1994;33:1234–1241. doi: 10.1021/bi00171a025. [DOI] [PubMed] [Google Scholar]
- 8.Copeland J W, Nasaidka A, Dietrich B H, Krause H M. Patterning of the Drosophila embryo by a homeodomain-deleted Ftz polypeptide. Nature. 1996;379:162–165. doi: 10.1038/379162a0. [DOI] [PubMed] [Google Scholar]
- 9.Creasy C L, Ambrose D M, Chernoff J. The Ste20-like protein kinase, Mst1, dimerizes and contains an inhibitory domain. J Biol Chem. 1996;271:21049–21053. doi: 10.1074/jbc.271.35.21049. [DOI] [PubMed] [Google Scholar]
- 10.Davey M J, Funnell B E. The P1 plasmid partition protein ParA. A role for ATP in site-specific DNA binding. J Biol Chem. 1994;269:29908–29913. [PubMed] [Google Scholar]
- 11.Davey M J, Funnell B E. Modulation of the P1 plasmid partition protein ParA by ATP, ADP and P1 ParB. J Biol Chem. 1997;272:15286–15292. doi: 10.1074/jbc.272.24.15286. [DOI] [PubMed] [Google Scholar]
- 12.Davis M A, Austin S J. Recognition of the P1 plasmid centromere analog involves binding of the ParB protein and is modified by a specific host factor. EMBO J. 1988;7:1881–1888. doi: 10.1002/j.1460-2075.1988.tb03021.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis M A, Martin K A, Austin S J. Biochemical activities of the ParA partition protein of the P1 plasmid. Mol Microbiol. 1992;6:1141–1147. doi: 10.1111/j.1365-2958.1992.tb01552.x. [DOI] [PubMed] [Google Scholar]
- 14.Derbyshire V, Kowalski J C, Dansereau J T, Hauer C R, Belfort M. Two-domain structure of the td intron-encoded endonuclease I-TevI correlates with the two-domain configuration of the homing site. J Mol Biol. 1997;265:494–506. doi: 10.1006/jmbi.1996.0754. [DOI] [PubMed] [Google Scholar]
- 15.Dodd I B, Egan J B. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res. 1990;18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durfee T, Becherer K, Chen P-L, Yeh S-H, Yang Y, Kilburn A E, Lee W-H, Elledge S J. The retinoblastoma protein associates with the protein phosphatase type 1 catalytic subunit. Genes Dev. 1993;7:555–569. doi: 10.1101/gad.7.4.555. [DOI] [PubMed] [Google Scholar]
- 17.Fiedler U, Weiss V. A common switch in activation of the response regulators NtrC and PhoB: phosphorylation induces dimerization of the receiver modules. EMBO J. 1995;14:3696–3705. doi: 10.1002/j.1460-2075.1995.tb00039.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fields S, Song O-K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 19.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- 20.Friedman S A, Austin S J. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988;19:103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- 21.Funnell B E. Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J Bacteriol. 1988;170:954–958. doi: 10.1128/jb.170.2.954-960.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Funnell B E. Participation of Escherichia coli integration host factor in the P1 plasmid partition system. Proc Natl Acad Sci USA. 1988;85:6657–6661. doi: 10.1073/pnas.85.18.6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Funnell B E. The P1 partition complex at parS: the influence of Escherichia coli integration host factor and of substrate topology. J Biol Chem. 1991;266:14328–14337. [PubMed] [Google Scholar]
- 24.Funnell B E, Gagnier L. The P1 plasmid partition complex at parS. II. Analysis of ParB protein binding activity and specificity. J Biol Chem. 1993;268:3616–3624. [PubMed] [Google Scholar]
- 25.Funnell B E, Gagnier L. P1 plasmid partition: binding of P1 ParB protein and Escherichia coli integration host factor to altered parS sites. Biochimie. 1994;76:924–932. doi: 10.1016/0300-9084(94)90017-5. [DOI] [PubMed] [Google Scholar]
- 26.Geitz D, Jean A S, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hanai R, Liu R P, Benedetti P, Caron P R, Lynch A S, Wang J C. Molecular dissection of a protein SopB essential for Escherichia coli F plasmid partition. J Biol Chem. 1996;271:17469–17475. doi: 10.1074/jbc.271.29.17469. [DOI] [PubMed] [Google Scholar]
- 28.Hayes F, Austin S J. Specificity determinants of the P1 and P7 plasmid centromere analogs. Proc Natl Acad Sci USA. 1993;90:9228–9232. doi: 10.1073/pnas.90.19.9228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayes F, Radnedge L, Davis M A, Austin S J. The homologous operons for P1 and P7 plasmid partition are autoregulated from dissimilar operator sites. Mol Microbiol. 1994;11:249–260. doi: 10.1111/j.1365-2958.1994.tb00305.x. [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Cao C, Lutkenhaus J. Interaction between FtsZ and inhibitors of cell division. J Bacteriol. 1996;178:5080–5085. doi: 10.1128/jb.178.17.5080-5085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ireton K, Gunther N W, Grossman A D. spoOJ is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J Bacteriol. 1994;176:5320–5329. doi: 10.1128/jb.176.17.5320-5329.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen R B, Gerdes K. Partitioning of plasmid R1. The ParM protein exhibits ATPase activity and interacts with the centromere-like ParR-parC complex. J Mol Biol. 1997;269:505–513. doi: 10.1006/jmbi.1997.1061. [DOI] [PubMed] [Google Scholar]
- 33.Jensen R B, Lurz R, Gerdes K. Mechanism of DNA segregation in prokaryotes: replicon pairing by parC of plasmid R1. Proc Natl Acad Sci USA. 1998;95:8550–8555. doi: 10.1073/pnas.95.15.8550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiesau P, Eikmanns U, Gutowski-Eckel Z, Weber S, Hammelmann M, Entian K-D. Evidence for a multimeric subtilin synthase complex. J Bacteriol. 1997;179:1475–1481. doi: 10.1128/jb.179.5.1475-1481.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kishore G M, Shah D M. Amino acid biosynthesis inhibitors as herbicides. Annu Rev Biochem. 1988;57:627–663. doi: 10.1146/annurev.bi.57.070188.003211. [DOI] [PubMed] [Google Scholar]
- 36.Konigsberg W H. Limited proteolysis of DNA polymerases as probe of functional domains. Methods Enzymol. 1995;262:331–346. doi: 10.1016/0076-6879(95)62028-1. [DOI] [PubMed] [Google Scholar]
- 37.Koonin E V. A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J Mol Biol. 1993;229:1165–1174. doi: 10.1006/jmbi.1993.1115. [DOI] [PubMed] [Google Scholar]
- 38.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 39.Lobocka M, Yarmolinsky M. P1 plasmid partition: a mutational analysis of ParB. J Mol Biol. 1996;259:366–382. doi: 10.1006/jmbi.1996.0326. [DOI] [PubMed] [Google Scholar]
- 40.Mohl D A, Gober J W. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 41.Motallebi-Veshareh M, Rouch D A, Thomas C M. A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol Microbiol. 1990;4:1455–1463. doi: 10.1111/j.1365-2958.1990.tb02056.x. [DOI] [PubMed] [Google Scholar]
- 42.Nakagawa N, Masui R, Kato R, Kuramitsu S. Domain structure of Thermus thermophilus UvrB protein. J Biol Chem. 1997;272:22703–22713. doi: 10.1074/jbc.272.36.22703. [DOI] [PubMed] [Google Scholar]
- 43.Nordström K, Austin S J. Mechanisms that contribute to the stable segregation of plasmids. Annu Rev Genet. 1989;23:37–69. doi: 10.1146/annurev.ge.23.120189.000345. [DOI] [PubMed] [Google Scholar]
- 44.Pabo C O, Sauer R T. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 45.Parsell D A, Silber K R, Sauer R T. Carboxy-terminal determinants of intracellular protein degradation. Genes Dev. 1990;4:277–286. doi: 10.1101/gad.4.2.277. [DOI] [PubMed] [Google Scholar]
- 46.Pichoff S, Benedikt V, Touriol C, Bouche J-P. Deletion analysis of gene minE which encodes the topological specificity factor of cell division in Escherichia coli. Mol Microbiol. 1995;18:321–329. doi: 10.1111/j.1365-2958.1995.mmi_18020321.x. [DOI] [PubMed] [Google Scholar]
- 47.Radnedge L, Davis M A, Austin S J. P1 and P7 plasmid partition: ParB protein bound to its partition site makes a separate discriminator contact with the DNA that determines species specificity. EMBO J. 1996;15:1155–1162. [PMC free article] [PubMed] [Google Scholar]
- 48.Radnedge L, Youngren B, Davis M, Austin S. Probing the structure of complex macromolecular interactions by homolog specificity scanning: the P1 and P7 plasmid partition systems. EMBO J. 1998;17:6076–6085. doi: 10.1093/emboj/17.20.6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rice P A, Yang S W, Mizuuchi K, Nash H A. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 50.Robertson C A, Nash H A. Bending of the bacteriophage λ attachment site by Escherichia coli integration host factor. J Biol Chem. 1988;263:3554–3557. [PubMed] [Google Scholar]
- 51.Rodionov O, Lobocka M, Yarmolinsky M. Silencing of genes flanking the P1 plasmid centromere. Science. 1999;283:546–549. doi: 10.1126/science.283.5401.546. [DOI] [PubMed] [Google Scholar]
- 52.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 53.Sedgewick S G, Taylor I A, Adam A C, Spanos A, Howell S, Morgan B A, Treiber M K, Kanuga N, Banks G R, Foord R, Smerdon S J. Structural and functional architecture of the yeast cell-cycle transcription factor Swi6. J Mol Biol. 1998;281:763–775. doi: 10.1006/jmbi.1998.1996. [DOI] [PubMed] [Google Scholar]
- 54.Strauss P R, Holt C M. Domain mapping of human apurinic/apyrimidinic endonuclease. J Biol Chem. 1998;273:14435–14441. doi: 10.1074/jbc.273.23.14435. [DOI] [PubMed] [Google Scholar]
- 55.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 56.Thompson J F, Landy A. Empirical estimation of protein-induced DNA bending angles: applications to λ site-specific recombination complexes. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang X, Hiand J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP, and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Williams D R, Thomas C M. Active partitioning of bacterial chromosomes. J Gen Microbiol. 1992;138:1–16. doi: 10.1099/00221287-138-1-1. [DOI] [PubMed] [Google Scholar]
- 59.Zelicof A, Protopopov V, David D, Lin X-Y, Lustgarten V, Gerst J E. Two separate functions are encoded by the carboxyl-terminal domains of the yeast cyclase-associated protein and its mammalian homologs. J Biol Chem. 1996;271:18243–18252. doi: 10.1074/jbc.271.30.18243. [DOI] [PubMed] [Google Scholar]