Abstract
Classical cadherins are calcium dependent cell-cell adhesion proteins that play key roles in the formation and maintenance of tissues. Deficiencies in cadherin adhesion are hallmarks of numerous cancers. Here, we review recent biophysical studies on the regulation of cadherin structure and adhesion. We begin by reviewing distinct cadherin binding conformations, their biophysical properties, and their response to mechanical stimuli. We then describe design rules for engineering antibodies that can regulate adhesion by either stabilizing or destabilizing cadherin interactions. Finally, we review molecular mechanisms by which cytoplasmic proteins regulate the conformation of cadherin extracellular regions, from the inside-out.
Classical cadherins are essential, calcium dependent cell-cell adhesion proteins that physically connect cells in tissues and play key roles in tissue formation and in the maintenance of tissue integrity. Cadherin adhesions withstand mechanical stress and orchestrate complex cell movements during morphogenesis and wound healing (1, 2). Cadherins maintain the integrity of epithelial barriers, thereby preventing harmful agents from accessing underlying tissue. Cadherins are also expressed in a variety of leukocytes, including conventional dendritic cells, Langerhans cells, and macrophages (3). Dysregulation of cadherin adhesion results in a loss of contact inhibition and increased cell mobility, a hallmark of numerous cancers and immunodeficiencies (3, 4).
Recently, there has been much progress in understanding the biophysical mechanisms that underlie the regulation of the conformation and adhesion of cadherin extracellular domains (5). Here, we briefly review these finding. We begin by reviewing distinct cadherin adhesive conformations and their biomechanical properties. We then describe the molecular mechanisms by which antibodies can be engineered to stabilize/destabilize cadherin binding and modulate adhesion. Finally, we review the biophysical mechanisms by which cytoplasmic proteins regulate the conformation of the cadherin extracellular region from the inside-out. Our review focuses on the trans binding (i.e. binding from opposing cells) of classical cadherins such as E-cadherin, N-cadherin, P-cadherin and C-cadherin, which are among the most widely studied members of the cadherin superfamily.
Structure and energetics of cadherin binding:
Classical cadherins are transmembrane proteins and their extracellular regions (or ectodomains), comprised of five tandemly arranged domains (EC1–5), mediate cell-cell adhesion (Figure 1a). Ectodomains from opposing cells bind in two trans adhesive conformations: strand-swap dimers and X-dimers (6–12). Besides binding in distinct trans conformations, cadherins on the same cell surface oligomerize in cis orientations which cluster cadherins and form robust adherens junctions (13, 14). While this review focuses on cadherin trans binding, cadherin cis association has been reviewed elsewhere (15).
Figure 1: Structure and biomechanics of E-cadherin trans dimers.
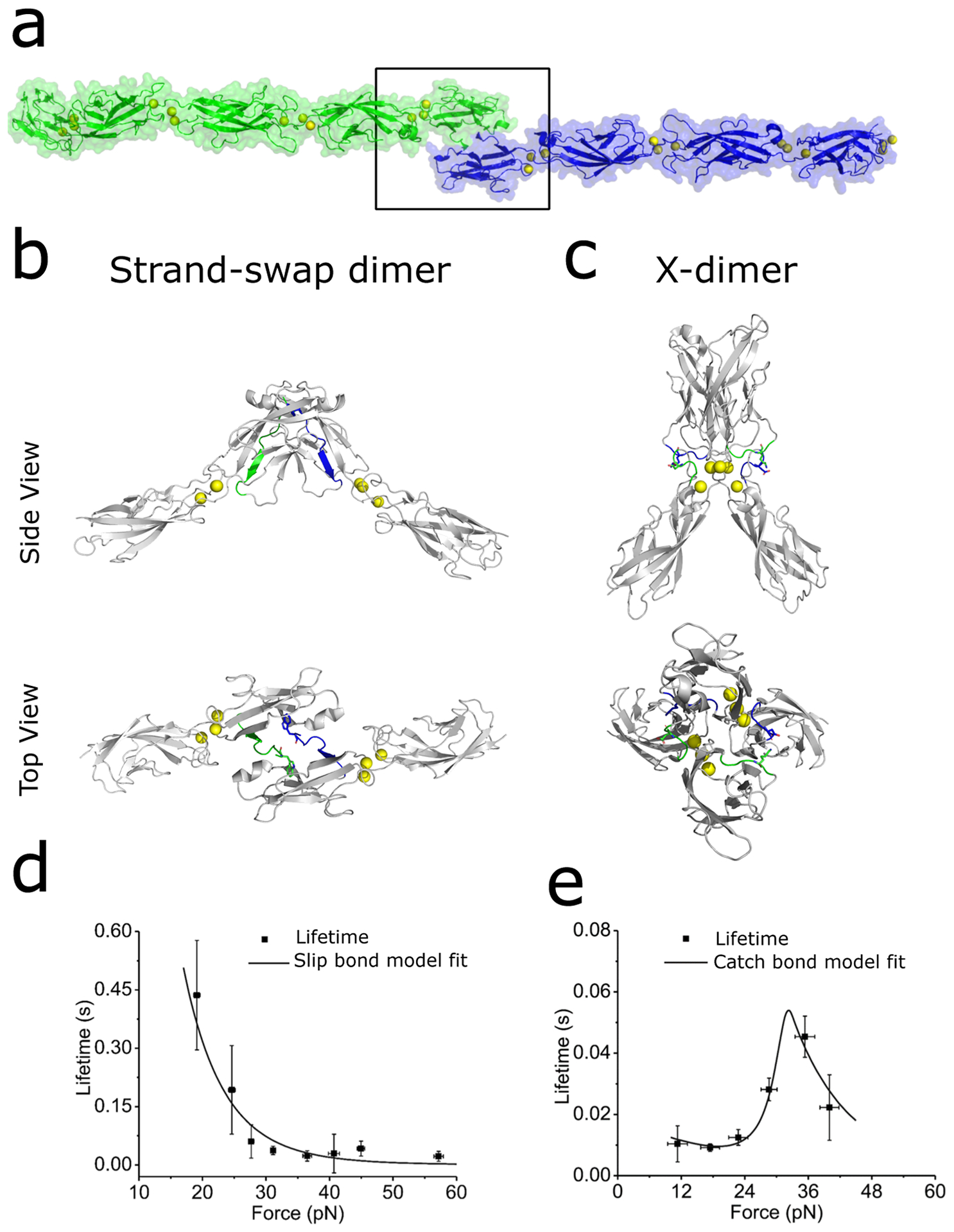
(a) Strand-swap dimers formed by the interaction of full length ectodomains of E-cadherin from opposing cell surfaces (green and blue, PDB: 3Q2V). (b) Structure of the outer two domains (EC1–2) forming a strand-swap dimer (PDB: 2O72). Strand-swap dimers are formed by exchanging the N terminal β-strands (residues 1–12 from opposing ectodomains are highlighted in green and blue). (c) X-dimers (PDB: 4ZT1) are formed due to interactions between EC1–2 domains of opposing cadherins. These interactions include hydrogen bonds and a key salt bridge (K14-D138) between loops 11–15 and 135–139 which are highlighted in green and blue. (d) Strand-swap dimers form slip bonds. The lifetime of a slip bond decreases with increasing in pulling force. (e) X-dimers form catch bonds. Catch bonds initially strengthen before weakening beyond a critical pulling force. Panel (d) is adapted from reference (29) while panel (e) is adapted from reference (42).
Strand-swap dimers are the primary trans adhesive conformation and are formed when the EC1 domains of opposing cadherins symmetrically exchange their N-terminal β-strands and insert a conserved Tryptophan at position 2 (W2) into hydrophobic pockets on their adhesive partners (Figure 1a, 1b) (16–20). Structural and computational studies show that strand swapping occurs because the β-strand in cadherin monomers is under a conformational strain which is relieved by exchange of β-strands (21).
Single molecule biophysics experiments first demonstrated that prior to strand-swapping, cadherin monomers interact in a non-swapped trans binding conformation (6). In these studies, E-cadherins were trapped in the non-swapped ‘initial encounter complex’ by mutating the W2 residue on the N-terminal β-strands to an Alanine (W2A mutation). Structural studies showed that the W2A mutants adopted an X-dimer structure due to extensive surface interactions between the base of the EC1 domain, EC1-EC2 inter-domain linker region and the apex of the EC2 domain (Figure 1c) (22). An X-dimer structure was also adopted when N-terminal β-strands were extended by a few amino acids (23), presumably because this reduced the conformational strain that drives the formation of strand-swap dimers (21). Key interactions that stabilize the X-dimer include salt-bridges between Lysine (K14) and Aspartic acid (D138) on opposing cadherins (9, 22). The affinity for X-dimer formation in solution was measured to be significantly weaker than the affinity for strand-swap dimerization: while the Kd for strand-swap dimerization ranged from 64 μM to 97 μM (24–26), the Kd for X-dimer formation was 916 μM (22).
X-dimers have been proposed to serve as an intermediate during the formation and rupture of strand-swap dimers (6, 22, 27). Mutating K14 in the cadherin X-dimer binding interface to Glutamic acid (K14E), which abolishes X-dimer formation, reduced both association and dissociation rates of strand-swap dimerization by factors of ~104, but did not change the affinity and the structure of the strand-swap dimer (22, 28). This suggests that X-dimers serve as non-obligatory, but crucial intermediates in the formation of strand-swap dimers. In epithelial cells, inactivation of X dimers result in extraordinarily stable cell-cell junctions suggesting that X-dimers are also an intermediate in the pathway to dissociation of strand-swap dimers (27). Computational studies and single molecule Atomic Force Microscopy (AFM) measurements also show that cadherins tune adhesion by interconverting between X-dimers and strand-swap dimer structures (29). Recent single molecule AFM measurements in live cells demonstrate that approximately 70% of E-cadherins on the surface of epithelial Madin-Darby canine kidney (MDCK) cells form strand-swap dimers while the remaining cadherins form X-dimers (30). This presumably provides cells with two cadherin pools with different adhesive properties. However, recent cryo-EM experiments (31), NMR (28) and high-speed AFM imaging experiments (32), show that X-dimers exist stably alongside monomers and strand-swap dimers, indicating that the X-dimer conformation is not just an intermediate, but is a stable adhesive structure.
Cadherins can interact in alternate structures:
While X-dimers and strand-swap dimers are the primary cadherin binding conformations, they are not the only trans adhesive structures that classical cadherins adopt. Biophysical studies show that wild type E-cadherin ectodomains in solution can interconvert between X-dimer and strand-swap dimer conformations via a metastable intermediate state (28, 29, 32). This intermediate conformation resembles an X-dimer, but with both W2 residues swapped (28, 29). Furthermore, recent biophysical experiments show that cadherins can also form trans dimers without the symmetric involvement of two W2s or of two K14-D138 salt-bridges (33). In this conformation, opposing cadherin ectodomains interact asymmetrically swapping just one W2, while simultaneously forming just one K14-D138 salt bridge (33).
Early biophysical studies showed the classical cadherins interact in three alternate conformations that involved interactions along the length of the ectodomain (34–36). In agreement with this finding, recent cryo-EM structures report a novel EC4-mediated E-cadherin trans dimer (31). Recently, high speed-AFM imaging of dimers formed by full-length ectodomains of E-cadherin revealed the existence of a novel S-shaped trans dimer structure formed by membrane-distal E-cadherin domains interacting via a broad binding interface. Imaging the conversion between X-dimers and strand-swap dimers showed that the formation of S-dimers precedes formation of X- and strand-swap dimers (32). Similar S-shaped dimers have previously been observed in the interaction of non-classical, desmosomal cadherins (37, 38). Taken together, these novel binding conformations, that are distinct from X-dimers and strand-swap dimers, indicate that the biophysics of classical cadherin interactions are still not completely understood and that cadherins can adopt a more diverse range of adhesive structures that need to be further explored.
Given the similarity in their sequence and structure, it is not surprising that different classical cadherins heterophilically interact with each other. Indeed, heterophilic binding between N-cadherin, E-cadherin and C-cadherin have been measured using ensemble force measurements (39). Kinetic measurements demonstrate that N-cadherin/E-cadherin heterophilic binding affinity lies in between the homophilic affinities of N-cadherin and E-cadherin respectively (25). Similarly, interactions between E-cadherin and P-cadherin have been detected using proximity labeling and AFM (40). However the structure of these classical cadherin heterotypic dimers are still unknown.
Biomechanics of cadherin trans dimers:
Besides their structural differences, X-dimers and strand-swap dimers can also be distinguished by their distinct responses to mechanical forces (29, 41, 42). Single molecule AFM measurements and computer simulations show that when X-dimers are pulled, the interacting protomers reorient which results in the formation of seven force-induced hydrogen bonds that lock the X-dimer into tighter contact (42). Consequently, the lifetime of an X-dimer initially increases with force before subsequently decreasing (Figure1 e) (41). These biphasic interactions, which are known as catch bonds, strengthen X-dimers in the presence of a pulling force (41). Biophysical experiments also show that X-dimer catch bonds are Ca2+ dependent (42). In contrast to X-dimers, single molecule AFM experiments show that cadherin strand-swap dimers form more conventional slip bonds that weaken upon pulling (Figure 1d) (41, 42).
It is possible that switching between X-dimer and strand-swap dimer conformations enable cells to tune their adhesive properties (29, 30). This ability to modulate adhesion may be important in phenomenon such as collective cell migration, which serves to keep tissue intact during morphogenesis, wound repair and cancer metastasis (43, 44). However, the biological roles of E-cadherin catch and slip bonds still remains to be determined.
Regulating cadherin adhesion using antibodies:
Classical cadherins are essential regulators of tissue homeostasis, and disruption of cadherin adhesion signals disease progression (45). E-cadherin acts as a tumor suppressor and deficiencies in E-cadherin adhesion are associated with the metastasis of breast cancer (46), colorectal cancer (47), gastric cancer (48), and lung cancer (49). Similarly, reduced expression of neuronal N-cadherin, strongly correlates with metastasis in neuroblastoma (50). Conversely, re-expression of E-cadherin in cadherin deficient cancer cells can prevent tumor progression and invasion (51). Due to their tumor suppressive properties, there is intense interest in developing antibodies that activate or strengthen E-cadherin adhesion for potential applications in reducing cancer metastasis.
Several monoclonal antibodies (mAbs) have been identified, which target the E-cadherin ectodomain and enhance cell-cell adhesion (52). These mAbs strongly activate adhesion in Colo 205 cells, a non-adhesive cell-line with a full but inactive complement of E-cadherin and its cytoplasmic binders. Activation of adhesion also induced dephosphorylation of specific residues in p120-catenin (52), an cadherin cytoplasmic binding partner that stabilizes the cadherin complex by preventing its internalization and degradation (53). Mutating the phosphorylation sites on p120-catenin to either phosphomimetic or non-phosphorylatable amino acids confirmed that p120-catenin phosphorylation regulates cadherin-dependent cell aggregation (52).
One of these mAbs, 19A11, was shown to prevent the metastatic invasion of mouse lung cancer cells expressing human E-cadherin (54, 55), enhance epithelial barrier function and limit progression of inflammatory bowel disease (56). Intercellular adhesion frequency measurements showed that either treating Colo 205 cells with 19A11, or dephosphorylating p120-catenin increased the homophilic binding affinity of E-cadherin (57). These results suggest that conformational changes in the E-cadherin ectodomain induced by 19A11 binding, allosterically correlate with p120-catenin associated changes across the cell membrane.
Recently, the structure of 19A11 bound to the EC1–2 domains of E-cadherin has been determined using X-ray crystallography and the molecular mechanism by which 19A11 strengthens adhesion has been identified using biophysical methods (58). This structure demonstrates that 19A11 binds to the EC1 domain of E-cadherin, between the swapped β-strand and the pocket region – two regions that are known to be key energetic determinants of strand-swap dimer stability (Figure 2) (21, 59). Computer simulations and single molecule AFM measurements show that 19A11 forms two key salt bridge interactions with E-cadherin which stabilize both the swapped β-strand and the pocket region which houses a W2 from its binding partner. To strengthen E-cadherin adhesion, at least one of these salt bridges needs to be formed between both E-cadherins in the trans dimer and their bound 19A11. Abolishing these salt-bridges eliminates adhesion strengthening (58).
Figure 2: Regulating classical cadherin adhesion.
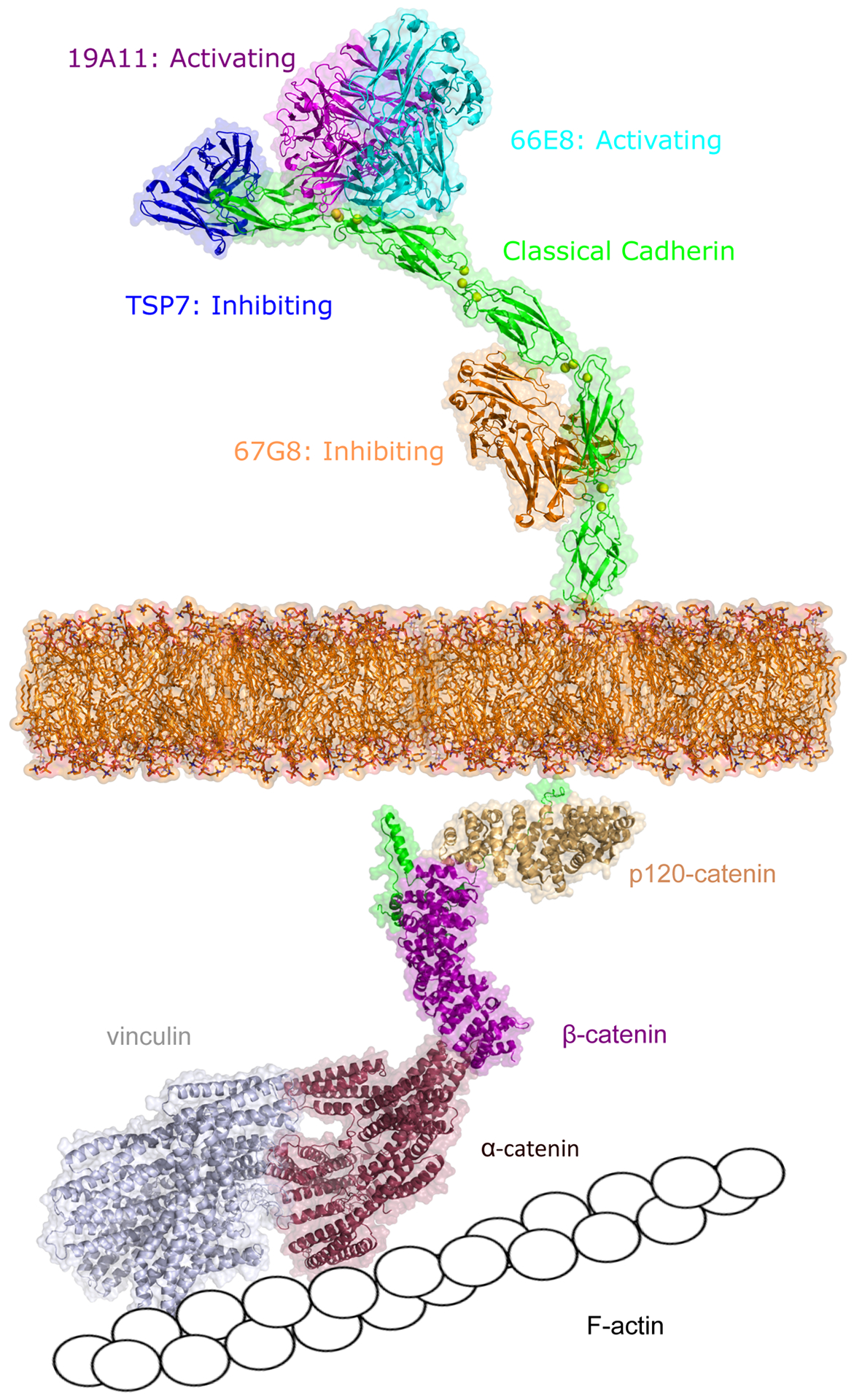
Cadherin adhesion can be regulating either via extracellular binding with antibodies or from the inside-out by cytoplasmic proteins. Activating antibodies 19A11 (magenta, PDB: 6CXY) and 66E8 (cyan, PDB:6VEL) recognize E-cadherin EC1 or EC2 domain (green, PDB: 3Q2V) near the cadherin trans binding sites. Antibody TSP7 (blue, PDB: 5JYL) inhibits P-cadherin adhesion by binding on the EC1 domain while 67G8 (orange) inhibits E-cadherin adhesion by binding on EC5 domain. The intracellular domain of cadherin (green) associates with various signaling molecules, including β-catenin (pink, PDB: 3L6X), p120-catenin (light yellow, PDB: 4R10), α-catenin (red), and vinculin (grey), which eventually link the cadherin cytoplasmic region to F-actin. The structures of α-catenin, vinculin, and E-cadherin intracellular domain were predicted using Alphafold (94). Unless apparent from the crystal structure, all interactions were predicted using Alphafold.
Simultaneously, structures of three activating mAbs (19A11, 66E8 and 59D2) bound to the full ectodomain of E-cadherin were resolved using X-ray crystallography and cryo-Electron Microscopy (Figure 2) (31). These structures revealed that mAb binding resulted in a distinctive twisted strand-swap dimer conformation caused by an outward shift in the N-terminal β-strands that may represent a strengthened adhesive state (31). All three activating antibodies bound at or near the anchor points of the swapped β-strands, again suggesting that stabilization of this strand is the dominant mechanism by which 19A11 strengthens E-cadherin adhesion (Figure 2) (31). These structural and biophysical studies outline mAb ‘design principles’ that can be exploited to strengthen strand-swap dimer adhesion. These studies demonstrate that selective stabilization of the swapped β-strand and its complementary binding pocket are sufficient to strengthen E-cadherin adhesion. Importantly, the activating mAbs do not inhibit access to the cis-binding interfaces (31), suggesting that these mAbs do not impact normal cadherin clustering. It remains to be seen if these design principles can be applied to develop new classes of mAbs that strengthen the adhesion of other classical cadherins as well.
Besides adhesion strengthening, inhibition of cadherin adhesion also has potential cancer therapeutic applications. Classical cadherins like P-cadherin act as tumor enhancers and overexpression of P-cadherin is associated with various types of cancer progression (60, 61). Consequently, there has been much recent effort in developing strategies to inhibit P-cadherin adhesion. An inhibiting antibody, TSP7, was developed that binds to the P-cadherin EC1 domain (Figure 2); steric hindrance between two bound TSP7s was shown to prevent the formation of X-dimers thus kinetically reducing strand-swap dimer formation and disrupting P-cadherin mediated cell adhesion (62). Similarly, a small chemical fragment that binds to a cavity in between the EC1 and EC2 domains of P-cadherin, was shown to prevent the formation of hydrogen bonds that are crucial for X-dimer formation (63). These experiments outline a plausible strategy for inhibiting cadherin adhesion by preventing X-dimer formation. However, this strategy may not be applicable to E-cadherin since E-cadherin strengthening mAbs 19A11, 66E8 and 59D2 all prevent X-dimer formation but still strengthen cell adhesion (31, 58). This suggests that X-dimers are necessary intermediates in P-cadherin strand-swap dimer formation, but may be non-obligatory in E-cadherin binding. These structural insights illustrate that differential biophysical targeting of each type of classical cadherin may be necessary for designing specific antibodies to tune adhesion.
Furthermore, studies indicate that there are several strategies to design antibodies that inhibit classical cadherin adhesion. E-cadherin inhibiting antibodies, namely 52F9, 67G8 and DECMA1, have been reported to recognize the EC5 domain and inhibit E-cadherin mediated cell adhesion (Figure 2) (31, 52, 64). Since the inhibiting antibody epitope is located distant from the E-cadherin trans binding site, it is unlikely that these inhibiting antibodies destabilize or inhibit trans binding conformations. Rather, binding to the EC5 domain may activate cytoplasmic signaling pathways which may in turn inhibit adhesion. Indeed, E-cadherin inhibition induced by the binding of DECMA1 has been reported to be associated with the activation of Epidermal Growth Factor Receptor 1/2 (HER1/2) and their downstream signaling pathways, which also has the potential to suppress the HER2 positive breast cancer (64). Recent data also suggests that DECMA1 functionally disrupts cell-cell adhesion by promoting proteolysis of E-cadherin (65).
Regulation of cadherin adhesion by cytoplasmic proteins:
The cytoplasmic region of classical cadherins associate with the catenin family of proteins: namely p120-catenin, β-catenin, γ-catenin/plakoglobin and α-catenin. The cadherin-catenin complex, in turn, links to filamentous actin (F-actin) either by the direct binding of α-catenin and F-actin or by the indirect association of α-catenin and F-actin via vinculin (Figure 2) (66). Additionally, recent studies show that β-catenin can directly bind to vinculin (67) and form an alternate bypass connection from cadherin to the actin cytoskeleton. These linkages between cadherin and the actin cytoskeleton are not static. Single molecule AFM and optical tweezer measurements in live cells reveal that on the apical region of MDCK cells, only ~50% of E-cadherin are linked to the cytoskeleton (30, 68).
Many of the cytoplasmic linkages between E-cadherin and the actin cytoskeleton are mechanosensitive – the binding of both α-catenin and vinculin to F-actin have been shown to display catch bond behaviors (69–71). Adhesive forces transmitted across intercellular junctions by cadherin, induce conformational changes in α-catenin (72, 73), strengthen F-actin binding (69) and recruit vinculin to the sites of force application (74, 75). These force-induced changes mediate cytoskeletal rearrangements and recruit myosin to cell-cell junctions (76–78). Cadherin coupling to α-catenin, vinculin, and the actin cytoskeleton is also regulated by the phosphorylation of p120-catenin (79) which decouples cadherin from vinculin and F-actin (80).
Studies show that α-catenin and vinculin play important roles in strengthening and stabilizing cadherin adhesion: bead twisting experiments show force-induced stiffening of E-cadherin-based junctions and cell doublet stretching experiments demonstrate reinforcement of cell-cell adhesion in vinculin and α-catenin dependent manners (81–83). AFM measurements with α-catenin knockdown cells also show that reducing the amount of cytoplasmic α-catenin decreases unbinding force of E-cadherin ectodomains (84, 85). Intercellular adhesion frequency measurements also provide biophysical evidence for the allosteric regulation of E-cadherin binding by the phosphorylation status of p120-catenin (57). However, the molecular mechanism for the ‘inside-out’ regulation of cadherin extracellular conformation and adhesion is only now beginning to be resolved.
Recent live-cell, single molecule AFM measurements show that the association of vinculin with the E-cadherin cytoplasmic tail allosterically drives the conversion of X-dimers to strand-swap dimers (30). These measurements demonstrate that while E-cadherins bound to vinculin form robust strand-swap dimers, E-cadherins are trapped in a weaker X-dimer conformation when vinculin is knocked-out or when vinculin binding to α-catenin is disrupted. AFM experiments and computer simulations show that vinculin binding to the E-cadherin cytoplasmic tail recruits myosin II to the sites of cell-cell contact. Forces due to actomyosin contractility propagates to the E-cadherin ectodomain and promotes the conversion of weak X-dimers to stronger strand-swap dimers (30).
However, the molecular mechanisms by which other cytoplasmic proteins (besides vinculin) regulate E-cadherin adhesion remain to be clarified. For instance, the biophysical mechanisms by which p120-catenin allosterically alters the adhesive properties of E-cadherin needs to be worked out. Similarly, it is unclear if α-catenin solely remodels cell-cell junctions or if it also allosterically alters the adhesive properties of individual E-cadherins.
Besides catenins, diverse signaling molecules are found at cell-cell contacts and many of these molecules are activated in a cadherin-dependent manner (86). For instance, cadherin binding sites are major locations for protein tyrosine phosphorylation, including both receptor tyrosine kinases and cytoplasmic kinases. One example is the Src-family kinases (SFKs), which are cytoplasmic tyrosine kinases, often found at cadherin-based cell–cell contacts (87, 88). Both E-cadherin and P-cadherin mediated cell adhesions have been shown to signal the activation of Src kinases at cell-cell contacts, and inhibiting Src kinases signaling is known to impair the functions of cadherins (89, 90). However, the molecular connections between Src kinases and cadherin adhesion is poorly understood and it is unclear if Src kinase activation affects cadherin ectodomain binding conformation.
Conclusions:
The distinct mechanical signatures of X-dimers and strand-swap dimers (41, 42) suggest different functional roles for these conformations in processes like collective cell migration, tissue formation and wound healing. However, the biological roles of X-dimer catch bonds and strand-swap dimer slip bonds remains to be elucidated. Determining this would likely require the development of conformation-specific antibodies that can distinguish between these structures. Similarly, new biophysical studies continue to reveal novel ectodomain binding conformations that are distinct from X-dimers and strand-swap dimers (29, 31–33). The biophysical properties of these conformations and their mechanistic roles in cadherin adhesion remain to be resolved.
Furthermore, how these distinct cadherin conformations are regulated in the context of cell function are unknown. Analogous to the case of integrins where adhesion is regulated from the inside-out (91), classical cadherin adhesion is also regulated by cytoplasmic proteins. However, the molecular mechanisms by which cytoplasmic effectors such as p120-catenin, α-catenin, and Src kinases regulate cadherin ectodomain conformation remains to be clarified.
While several mAbs that strengthen E-cadherin adhesion have been developed (52), only the biophysical mechanism by which 19A11 strengthens adhesion has been resolved in molecular detail (31, 57, 58). Determining the activating mechanisms for the remaining mAbs will provide a more holistic picture of how mAbs enhance cadherin adhesion. Additionally, although several antibodies have been found to inhibit cadherin function, the conclusive molecular mechanisms by which they act is unclear. Furthermore, several other classical cadherins are implicated in cancer metastasis including P-cadherin and N-cadherin (43, 60, 92, 93). The molecular mechanism of E-cadherin activating antibodies such as 19A11 (31, 58) may prove useful in designing mAbs that regulate the adhesion of other classical cadherins.
Grant support:
This research was supported in part by the National Institute of General Medical Sciences of the National Institutes of Health (R01GM121885 and R01GM133880).
References
- 1.Takeichi M 2022. Cell sorting in vitro and in vivo: How are cadherins involved? Seminars in Cell & Developmental Biology. [DOI] [PubMed] [Google Scholar]
- 2.Gumbiner BM 2005. Regulation of cadherin-mediated adhesion in morphogenesis. Nat. Rev. Mol. Cell Biol 6: 622–634. [DOI] [PubMed] [Google Scholar]
- 3.Nawijn MC, Hackett TL, Postma DS, van Oosterhout AJ, and Heijink IH. 2011. E-cadherin: gatekeeper of airway mucosa and allergic sensitization. Trends Immunol 32: 248–255. [DOI] [PubMed] [Google Scholar]
- 4.Mendonsa AM, Na T-Y, and Gumbiner BM. 2018. E-cadherin in contact inhibition and cancer. Oncogene 37: 4769–4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Priest AV, Koirala R, and Sivasankar S. 2019. Single molecule studies of classical and desmosomal cadherin adhesion. Current Opinion in Biomedical Engineering 12: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sivasankar S, Zhang Y, Nelson WJ, and Chu S. 2009. Characterizing the initial encounter complex in cadherin adhesion. Structure 17: 1075–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bayas MV, Leung A, Evans E, and Leckband D. 2006. Lifetime measurements reveal kinetic differences between homophilic cadherin bonds. Biophys. J 90: 1385–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chien YH, Jiang N, Li F, Zhang F, Zhu C, and Leckband D. 2008. Two stage cadherin kinetics require multiple extracellular domains but not the cytoplasmic region. J. Biol. Chem 283: 1848–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ciatto C, Bahna F, Zampieri N, VanSteenhouse HC, Katsamba PS, Ahlsen G, Harrison OJ, Brasch J, Jin XS, Posy S, Vendome J, Ranscht B, Jessell TM, Honig B, and Shapiro L. 2010. T-cadherin structures reveal a novel adhesive binding mechanism. Nat. Struct. Mol. Biol 17: 339–U110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perret E, Leung A, Feracci H, and Evans E. 2004. Trans-bonded pairs of E-cadherin exhibit a remarkable hierarchy of mechanical strengths. Proc. Natl. Acad. Sci. U. S. A 101: 16472–16477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prakasam A, Chien YH, Maruthamuthu V, and Leckband DE. 2006. Calcium site mutations in cadherin: Impact on adhesion and evidence of cooperativity. Biochemistry 45: 6930–6939. [DOI] [PubMed] [Google Scholar]
- 12.Shi QM, Maruthamuthu V, Li F, and Leckband D. 2010. Allosteric Cross Talk between Cadherin Extracellular Domains. Biophys. J 99: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu Y, Vendome J, Shapiro L, Ben-Shaul A, and Honig B. 2011. Transforming binding affinities from three dimensions to two with application to cadherin clustering. Nature 475: 510–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu Y, Honig B, and Ben-Shaul A. 2013. Theory and simulations of adhesion receptor dimerization on membrane surfaces. Biophys J 104: 1221–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Priest AV, Shafraz O, and Sivasankar S. 2017. Biophysical basis of cadherin mediated cell-cell adhesion. Exp. Cell Res 358: 10–13. [DOI] [PubMed] [Google Scholar]
- 16.Boggon TJ, Murray J, Chappuis-Flament S, Wong E, Gumbiner BM, and Shapiro L. 2002. C-cadherin ectodomain structure and implications for cell adhesion mechanisms. Science 296: 1308–1313. [DOI] [PubMed] [Google Scholar]
- 17.Haussinger D, Ahrens T, Aberle T, Engel J, Stetefeld J, and Grzesiek S. 2004. Proteolytic E-cadherin activation followed by solution NMR and X-ray crystallography. EMBO J. 23: 1699–1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Patel SD, Ciatto C, Chen CP, Bahna F, Rajebhosale M, Arkus N, Schieren I, Jessell TM, Honig B, Price SR, and Shapiro L. 2006. Type II cadherin ectodomain structures: Implications for classical cadherin specificity. Cell 124: 1255–1268. [DOI] [PubMed] [Google Scholar]
- 19.Parisini E, Higgins JMG, Liu JH, Brenner MB, and Wang JH. 2007. The crystal structure of human E-cadherin domains 1 and 2, and comparison with other cadherins in the context of adhesion mechanism. J. Mol. Biol 373: 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shapiro L, Fannon AM, Kwong PD, Thompson A, Lehmann MS, Grubel G, Legrand JF, Alsnielsen J, Colman DR, and Hendrickson WA. 1995. Structural Basis of Cell-Cell Adhesion by Cadherins. Nature 374: 327–337. [DOI] [PubMed] [Google Scholar]
- 21.Vendome J, Posy S, Jin X, Bahna F, Ahlsen G, Shapiro L, and Honig B. 2011. Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat. Struct. Mol. Biol 18: 693–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harrison OJ, Bahna F, Katsamba PS, Jin XS, Brasch J, Vendome J, Ahlsen G, Carroll KJ, Price SR, Honig B, and Shapiro L. 2010. Two-step adhesive binding by classical cadherins. Nat. Struct. Mol. Biol 17: 348–U121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagar B, Overduin M, Ikura M, and Rini JM. 1996. Structural basis of calcium-induced E-cadherin rigidification and dimerization. Nature 380: 360–364. [DOI] [PubMed] [Google Scholar]
- 24.Chappuis-Flament S, Wong E, Hicks LD, Kay CM, and Gumbiner BM. 2001. Multiple cadherin extracellular repeats mediate homophilic binding and adhesion. J. Cell Biol 154: 231–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, Shapiro L, and Honig BH. 2009. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc. Natl. Acad. Sci. U. S. A 106: 11594–11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koch AW, Pokutta S, Lustig A, and Engel J. 1997. Calcium binding and homoassociation of E-cadherin domains. Biochemistry 36: 7697–7705. [DOI] [PubMed] [Google Scholar]
- 27.Hong SJ, Troyanovsky RB, and Troyanovsky SM. 2011. Cadherin exits the junction by switching its adhesive bond. J. Cell Biol 192: 1073–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Altorelli N, Bahna F, Honig B, Shapiro L, and Palmer A. 2013. Mechanism of E-cadherin dimerization probed by NMR relaxation dispersion. P Natl Acad Sci USA 110: 16462–16467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manibog K, Sankar K, Kim S-A, Zhang Y, Jernigan RL, and Sivasankar S. 2016. Molecular determinants of cadherin ideal bond formation: Conformation-dependent unbinding on a multidimensional landscape. Proc. Natl. Acad. Sci. U. S. A 13: E5711–E5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koirala R, Priest AV, Yen C-F, Cheah JS, Pannekoek W-J, Gloerich M, Yamada S, and Sivasankar S. 2021. Inside-out regulation of E-cadherin conformation and adhesion. Proc. Natl. Acad. Sci. U. S. A 118: e2104090118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maker A, Bolejack M, Schecterson L, Hammerson B, Abendroth J, Edwards TE, Staker B, Myler PJ, and Gumbiner BM. 2022. Regulation of multiple dimeric states of E-cadherin by adhesion activating antibodies revealed through Cryo-EM and X-ray crystallography. PNAS Nexus 1: pgac163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishiguchi S, Furuta T, and Uchihashi T. 2022. Multiple dimeric structures and strand-swap dimerization of E-cadherin in solution visualized by high-speed atomic force microscopy. Proceedings of the National Academy of Sciences 119: e2208067119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Priest AV, Koirala R, and Sivasankar S. 2022. Cadherins can dimerize via asymmetric interactions. FEBS Lett. 596: 1639–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sivasankar S, Brieher W, Lavrik N, Gumbiner B, and Leckband D. 1999. Direct molecular force measurements of multiple adhesive interactions between cadherin ectodomains. Proc. Natl. Acad. Sci. U. S. A 96: 11820–11824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivasankar S, Gumbiner B, and Leckband D. 2001. Direct measurements of multiple adhesive alignments and unbinding trajectories between cadherin extracellular domains. Biophys. J 80: 1758–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu B, Chappuis-Flament S, Wong E, Jensen IE, Gumbiner BM, and Leckband D. 2003. Functional analysis of the structural basis of homophilic cadherin adhesion. Biophys. J 84: 4033–4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.He WZ, Cowin P, and Stokes DL. 2003. Untangling desmosomal knots with electron tomography. Science 302: 109–113. [DOI] [PubMed] [Google Scholar]
- 38.Sikora M, Ermel UH, Seybold A, Kunz M, Calloni G, Reitz J, Vabulas RM, Hummer G, and Frangakis AS. 2020. Desmosome architecture derived from molecular dynamics simulations and cryo-electron tomography. Proceedings of the National Academy of Sciences 117: 27132–27140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prakasam AK, Maruthamuthu V, and Leckband DE. 2006. Similarities between heterophilic and homophilic cadherin adhesion. Proc. Natl. Acad. Sci. U. S. A 103: 15434–15439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shafraz O, Xie B, Yamada S, and Sivasankar S. 2020. Mapping transmembrane binding partners for E-cadherin ectodomains. Proc Natl Acad Sci U S A 117: 31157–31165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rakshit S, Zhang Y, Manibog K, Shafraz O, and Sivasankar S. 2012. Ideal, catch, and slip bonds in cadherin adhesion. Proc. Natl. Acad. Sci. U. S. A 109: 18815–18820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Manibog K, Li H, Rakshit S, and Sivasankar S. 2014. Resolving the molecular mechanism of cadherin catch bond formation. Nat. Commun 5: 3941. [DOI] [PubMed] [Google Scholar]
- 43.Friedl P, and Gilmour D. 2009. Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol 10: 445–457. [DOI] [PubMed] [Google Scholar]
- 44.Rørth P 2009. Collective cell migration. Annu. Rev. Cell Dev. Biol 25: 407–429. [DOI] [PubMed] [Google Scholar]
- 45.Bruner HC, and Derksen PWB. 2018. Loss of E-Cadherin-Dependent Cell-Cell Adhesion and the Development and Progression of Cancer. Cold Spring Harb Perspect Biol 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alsaleem M, Toss MS, Joseph C, Aleskandarany M, Kurozumi S, Alshankyty I, Ogden A, Rida PCG, Ellis IO, Aneja R, Green AR, Mongan NP, and Rakha EA. 2019. The molecular mechanisms underlying reduced E-cadherin expression in invasive ductal carcinoma of the breast: high throughput analysis of large cohorts. Mod Pathol 32: 967–976. [DOI] [PubMed] [Google Scholar]
- 47.King LE, Zhang HH, Gould CM, Thomas DW, Whitehead LW, Simpson KJ, Burgess AW, and Faux MC. 2020. Genes regulating membrane-associated E-cadherin and proliferation in adenomatous polyposis coli mutant colon cancer cells: High content siRNA screen. PLoS One 15: e0240746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caldeira J, Figueiredo J, Bras-Pereira C, Carneiro P, Moreira AM, Pinto MT, Relvas JB, Carneiro F, Barbosa M, Casares F, Janody F, and Seruca R. 2015. E-cadherin-defective gastric cancer cells depend on Laminin to survive and invade. Hum Mol Genet 24: 5891–5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Asnaghi L, Vass WC, Quadri R, Day PM, Qian X, Braverman R, Papageorge AG, and Lowy DR. 2010. E-cadherin negatively regulates neoplastic growth in non-small cell lung cancer: role of Rho GTPases. Oncogene 29: 2760–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lammens T, Swerts K, Derycke L, De Craemer A, De Brouwer S, De Preter K, Van Roy N, Vandesompele J, Speleman F, Philippé J, Benoit Y, Beiske K, Bracke M, and Laureys G. 2012. N-cadherin in neuroblastoma disease: expression and clinical significance. Plos One 7: e31206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarro P, Gómez M, Pizarro A, Gamallo C, Quintanilla M, and Cano A. 1991. A role for the E-cadherin cell-cell adhesion molecule during tumor progression of mouse epidermal carcinogenesis. J Cell Biol 115: 517–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrova YI, Spano MM, and Gumbiner BM. 2012. Conformational epitopes at cadherin calcium-binding sites and p120-catenin phosphorylation regulate cell adhesion. Mol. Biol Cell 23: 2092–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, and Kowalczyk AP. 2003. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J Cell Biol 163: 535–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Na TY, Schecterson L, Mendonsa AM, and Gumbiner BM. 2020. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. P Natl Acad Sci USA 117: 5931–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Petrova YI, Schecterson L, and Gumbiner BM. 2016. Roles for E-cadherin cell surface regulation in cancer. Mol. Biol Cell 27: 3233–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bandyopadhyay C, Schecterson L, and Gumbiner BM. 2021. E-cadherin activating antibodies limit barrier dysfunction and inflammation in mouse inflammatory bowel disease. Tissue Barriers 9: 1940741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shashikanth N, Petrova YI, Park S, Chekan J, Maiden S, Spano M, Ha T, Gumbiner BM, and Leckband DE. 2015. Allosteric regulation of E-cadherin adhesion. J. Biol. Chem 290: 21749–21761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie B, Maker A, Priest AV, Dranow DM, Phan JN, Edwards TE, Staker BL, Myler PJ, Gumbiner BM, and Sivasankar S. 2022. Molecular mechanism for strengthening E-cadherin adhesion using a monoclonal antibody. Proceedings of the National Academy of Sciences 119: e2204473119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vendome J, Felsovalyi K, Song H, Yang Z, Jin X, Brasch J, Harrison OJ, Ahlsen G, Bahna F, and Kaczynska A. 2014. Structural and energetic determinants of adhesive binding specificity in type I cadherins. Proceedings of the National Academy of Sciences 111: E4175–E4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vieira AF, and Paredes J. 2015. P-cadherin and the journey to cancer metastasis. Mol. Cancer 14: 178–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim MA, Jung EJ, Lee HS, Lee HE, Yang HK, Oh DY, Bang YJ, and Kim WH. 2010. P-cadherin expression in gastric carcinoma: its regulation mechanism and prognostic significance. Hum Pathol 41: 877–885. [DOI] [PubMed] [Google Scholar]
- 62.Kudo S, Caaveiro JMM, Nagatoishi S, Miyafusa T, Matsuura T, Sudou Y, and Tsumoto K. 2017. Disruption of cell adhesion by an antibody targeting the cell-adhesive intermediate (X-dimer) of human P-cadherin. Sci Rep-Uk 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Senoo A, Ito S, Nagatoishi S, Saito Y, Ueno G, Kuroda D, Yoshida K, Tashima T, Kudo S, Sando S, and Tsumoto K. 2021. Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion. Commun Biol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brouxhon SM, Kyrkanides S, Teng X, Raja V, O’Banion MK, Clarke R, Byers S, Silberfeld A, Tornos C, and Ma L. 2013. Monoclonal antibody against the ectodomain of E-cadherin (DECMA-1) suppresses breast carcinogenesis: involvement of the HER/PI3K/Akt/mTOR and IAP pathways. Clin Cancer Res 19: 3234–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayward AN, Aird EJ, and Gordon WR. 2019. A toolkit for studying cell surface shedding of diverse transmembrane receptors. eLife 8: e46983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ratheesh A, and Yap AS. 2012. A bigger picture: classical cadherins and the dynamic actin cytoskeleton. Nat. Rev. Mol. Cell Biol 13: 673–679. [DOI] [PubMed] [Google Scholar]
- 67.Peng X, Cuff LE, Lawton CD, and DeMali KA. 2010. Vinculin regulates cell-surface E-cadherin expression by binding to β-catenin. Journal of Cell Science 123: 567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sako Y, Nagafuchi A, Tsukita S, Takeichi M, and Kusumi A. 1998. Cytoplasmic regulation of the movement of E-cadherin on the free cell surface as studied by optical tweezers and single particle tracking: corralling and tethering by the membrane skeleton. J. Cell Biol 140: 1227–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Buckley CD, Tan J, Anderson KL, Hanein D, Volkmann N, Weis WI, Nelson WJ, and Dunn AR. 2014. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346: 1254211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang DL, Bax NA, Buckley CD, Weis WI, and Dunn AR. 2017. Vinculin forms a directionally asymmetric catch bond with F-actin. Science 357: 703–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arbore C, Sergides M, Gardini L, Bianchi G, Kashchuk A, Pertici I, Bianco P, Pavone F, and Capitanio M. 2022. α-catenin switches between a slip and an asymmetric catch bond with F-actin to cooperatively regulate cell junction fluidity. Nat. Commun 13: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yonemura S, Wada Y, Watanabe T, Nagafuchi A, and Shibata M. 2010. alpha-Catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol 12: 533–U535. [DOI] [PubMed] [Google Scholar]
- 73.Kim T-J, Zheng S, Sun J, Muhamed I, Wu J, Lei L, Kong X, Leckband DE, and Wang Y. 2015. Dynamic visualization of α-catenin reveals rapid, reversible conformation switching between tension states. Curr. Biol 25: 218–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Choi H-J, Pokutta S, Cadwell GW, Bobkov AA, Bankston LA, Liddington RC, and Weis WI. 2012. αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. U. S. A 109: 8576–8581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yao M, Qiu W, Liu R, Efremov AK, Cong P, Seddiki R, Payre M, Lim CT, Ladoux B, and Mège R-M. 2014. Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun 5. [DOI] [PubMed] [Google Scholar]
- 76.Carisey A, and Ballestrem C. 2011. Vinculin, an adapter protein in control of cell adhesion signalling. Eur. J. Cell Biol 90: 157–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, and de Rooij J. 2010. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J. Cell Biol 189: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Leerberg JM, Gomez GA, Verma S, Moussa EJ, Wu SK, Priya R, Hoffman BD, Grashoff C, Schwartz MA, and Yap AS. 2014. Tension-sensitive actin assembly supports contractility at the epithelial zonula adherens. Curr. Biol 24: 1689–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Piedra J, Miravet S, Castaño J, Pálmer HG, Heisterkamp N, de Herreros AG, and Dunach M. 2003. p120 Catenin-associated Fer and Fyn tyrosine kinases regulate β-catenin Tyr-142 phosphorylation and β-catenin-α-catenin Interaction. Mol. Cell. Biol 23: 2287–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lilien J, Balsamo J, Arregui C, and Xu G. 2002. Turn‐off, drop‐out: functional state switching of cadherins. Developmental Dynamics 224: 18–29. [DOI] [PubMed] [Google Scholar]
- 81.le Duc Q, Shi Q, Blonk I, Sonnenberg A, Wang N, Leckband D, and de Rooij J. 2010. Vinculin potentiates E-cadherin mechanosensing and is recruited to actin-anchored sites within adherens junctions in a myosin II-dependent manner. J Cell Biol 189: 1107–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Thomas WA, Boscher C, Chu Y-S, Cuvelier D, Martinez-Rico C, Seddiki R, Heysch J, Ladoux B, Thiery JP, and Mege R-M. 2013. α-Catenin and vinculin cooperate to promote high E-cadherin-based adhesion strength. J. Biol. Chem 288: 4957–4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barry AK, Tabdili H, Muhamed I, Wu J, Shashikanth N, Gomez GA, Gottardi CJ, De Rooij J, Wang N, and Leckband DE. 2014. α-catenin cytomechanics–role in cadherin-dependent adhesion and mechanotransduction. J. Cell Sci 127: 1779–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bajpai S, Correia J, Feng Y, Figueiredo J, Sun SX, Longmore GD, Suriano G, and Wirtz D. 2008. {alpha}-Catenin mediates initial E-cadherin-dependent cell-cell recognition and subsequent bond strengthening. P Natl Acad Sci USA 105: 18331–18336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bajpai S, Feng Y, Krishnamurthy R, Longmore GD, and Wirtz D. 2009. Loss of alpha-catenin decreases the strength of single E-cadherin bonds between human cancer cells. The Journal of Biological Chemistry 284: 18252–18259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Yap AS, and Kovacs EM. 2003. Direct cadherin-activated cell signaling: a view from the plasma membrane. J Cell Biol 160: 11–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsukita S, Oishi K, Akiyama T, Yamanashi Y, Yamamoto T, and Tsukita S. 1991. Specific proto-oncogenic tyrosine kinases of src family are enriched in cell-to-cell adherens junctions where the level of tyrosine phosphorylation is elevated. J Cell Biol 113: 867–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thomas SM, and Brugge JS. 1997. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol 13: 513–609. [DOI] [PubMed] [Google Scholar]
- 89.McLachlan RW, Kraemer A, Helwani FM, Kovacs EM, and Yap AS. 2007. E-cadherin adhesion activates c-Src signaling at cell-cell contacts. Mol Biol Cell 18: 3214–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ribeiro AS, Nobre AR, Mendes N, Almeida J, Vieira AF, Sousa B, Carvalho FA, Monteiro J, Polonia A, Fonseca M, Sanches JM, Santos NC, Seruca R, and Paredes J. 2018. SRC inhibition prevents P-cadherin mediated signaling and function in basal-like breast cancer cells. Cell Commun Signal 16: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kim M, Carman CV, and Springer TA. 2003. Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301: 1720–1725. [DOI] [PubMed] [Google Scholar]
- 92.Berx G, and van Roy F. 2009. Involvement of Members of the Cadherin Superfamily in Cancer. Cold Spring Harbor Perspect. Biol 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jeanes A, Gottardi CJ, and Yap AS. 2008. Cadherins and cancer: how does cadherin dysfunction promote tumor progression? Oncogene 27: 6920–6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, and Hassabis D. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]


