Abstract
To understand the mechanism of acquired resistance to poly(ADP-ribose) polymerase inhibitors (PARPi) olaparib, we induced the formation of polyploid giant cancer cells (PGCCs) in ovarian and breast cancer cell lines, high-grade serous cancer (HGSC)–derived organoids, and patient-derived xenografts (PDXs). Time-lapse tracking of ovarian cancer cells revealed that PGCCs primarily developed from endoreplication after exposure to sublethal concentrations of olaparib. PGCCs exhibited features of senescent cells but, after olaparib withdrawal, can escape senescence via restitutional multipolar endomitosis and other noncanonical modes of cell division to generate mitotically competent resistant daughter cells. The contraceptive drug mifepristone blocked PGCC formation and daughter cell formation. Mifepristone/olaparib combination therapy substantially reduced tumor growth in PDX models without previous olaparib exposure, while mifepristone alone decreased tumor growth in PDX models with acquired olaparib resistance. Thus, targeting PGCCs may represent a promising approach to potentiate the therapeutic response to PARPi and overcome PARPi-induced resistance.
PARPi activates a pregnant-like process via forming polyploid giant cancer cells and is blocked by contraceptive drug RU-486.
INTRODUCTION
Approximately 50% of high-grade serous carcinomas (HGSCs) exhibit defective homologous recombination (HR) DNA repair caused by genetic or epigenetic alterations of HR pathway genes, most commonly the tumor-suppressor genes BRCA1 and BRCA2 (1). HR-deficient tumors with BRCA1/2 alterations are susceptible to poly(ADP-ribose) polymerase inhibitors (PARPi), which selectively kill BRCA1/2-deficient cells while sparing BRCA1/2-proficient normal cells (2, 3). Currently, three PARPi—olaparib, rucaparib, and niraparib—have been granted regulatory approval as maintenance therapy for platinum-sensitive recurrent ovarian cancer on the basis of substantial improvements in progression-free survival in three randomized phase 3 trials (4–7).
Despite the remarkable clinical benefit of PARPi, acquired drug resistance is observed in most patients with advanced HGSC and other cancers (8). Multiple potential mechanisms of PARPi resistance have been proposed in preclinical models, including overexpression of drug-efflux transporter genes (9), decreased PARP trapping (10), stabilization of stalled replication forks (11), or reactivation of HR (12). However, most of these mechanisms have only been studied in vitro, and it is unclear whether they apply to treated tumors in patients. Moreover, 50% of HGSC patients are HR-proficient and do not substantially benefit from PARPi treatment (12), highlighting the importance of identifying alternative clinically relevant mechanisms that potentiate the therapeutic effect of PARPi and the mechanisms of acquired resistance in patients regardless of HR status.
Emerging evidence suggests that moderate, clinically relevant doses of anticancer drugs can trigger senescence of cancer cells in addition to apoptosis in solid tumors and tumor-derived cell lines (13, 14). These senescent cells contain either a highly enlarged nucleus or multiple nuclei, often accompanied by markedly increased cell size and genomic content, and are now referred to as polyploid giant cancer cells (PGCCs) (15, 16). Although PGCCs were often overlooked or mispresented as “dead cells” in the past owing to their inability to execute mitosis, they are now known to generate therapy-resistant daughter cells via nuclear budding or bursting (16–22), forming transient cell-in-cell structures called fecundity cells (22, 23). It has been recently shown that PGCCs recapitulate properties of the blastomere-stage embryonic program of dedifferentiation (16, 20, 22, 24, 25). PGCCs are also known to contribute to therapy resistance in various types of solid tumors, including ovarian (16), prostate (19, 26–28), colon (29), and breast cancers (30) and melanoma (31). However, whether PGCCs account for resistance to PARPi in HGSC remains unknown.
This work used several different preclinical ovarian cancer models, including multiple ovarian cell lines with or without p53 mutation and one breast cancer cell line, HGSC organoids, and patient-derived xenograft (PDX) models to extensively investigate the essential role of PGCCs in the therapeutic response as well as PARPi resistance in ovarian cancer. We analyzed the phenotype of PGCCs and used time-lapse photography to describe how PGCCs evolve and generate daughter cells. We further evaluated the possibility of potentiating the therapeutic effect of PARPi and overcoming PARPi resistance by eliminating PGCCs.
RESULTS
PGCCs accumulate in PARPi-resistant HGSC models with acquired resistance to olaparib
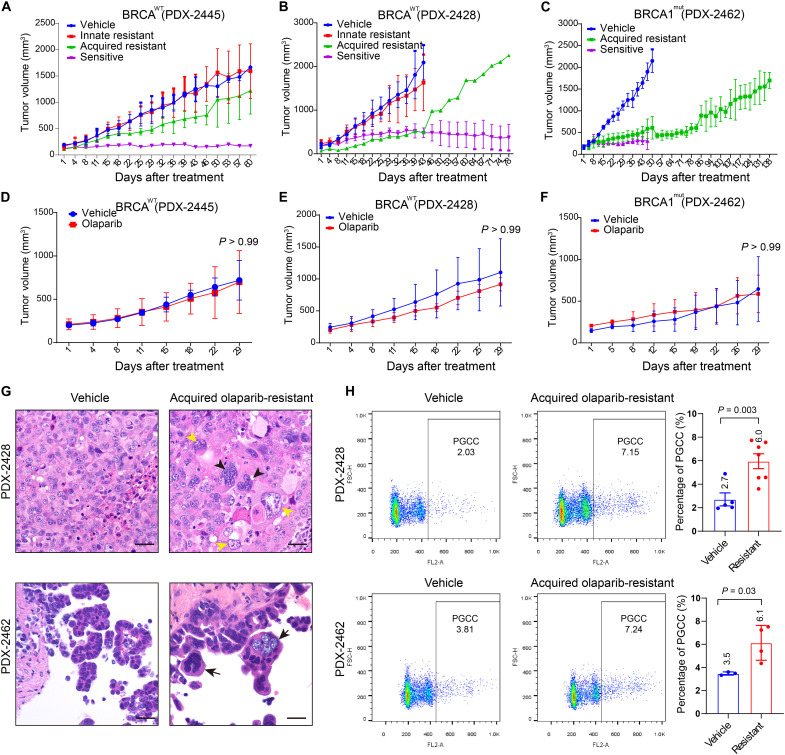
PDX models recapitulate the original tumor’s heterogeneity and preserve its three-dimensional histologic and architectural characteristics of cancer tissue (32). To establish an olaparib-resistant PDX model, we transplanted HGSC PDX tumors [two BRCA1/2 wild type (BRCAWT) and one BRCA1 mutant (BRCA1MUT)] subcutaneously into the left flanks of female nude mice. When the tumor volume reached about 200 mm3, mice were given olaparib (50 mg/kg) or vehicle intraperitoneally daily for at least 8 weeks. The response of BRCAWT and BRCA1MUT tumors to olaparib displayed great intratumor and intertumor heterogeneity. Among the BRCAWT PDX-2445 xenografts treated with olaparib, 7 of 10 xenografts were innately resistant, 1 was sensitive, and 2 gradually developed resistance (Fig. 1A). Similarly, four of the eight BRCAWT PDX-2428 xenografts were innately resistant to olaparib, three were sensitive, and one was initially sensitive but acquired olaparib resistance after 6 weeks of treatment (Fig. 1B). In contrast, all of the BRCA1MUT PDX-2462 xenografts were initially sensitive to olaparib, and three of seven developed resistance over time (Fig. 1C).
Fig. 1. Establishment of olaparib-resistant PDX models of BRCAWT and BRCA1MUT ovarian HGSC.
(A to C) Tumor growth curves of HGSC xenografts. Mice were treated with vehicle or olaparib (50 mg/kg per day, 5 days/week) for 60 days. Data are shown as mean ± SD. (A) BRCAWT PDX-2445: vehicle (n = 3), olaparib (n = 10). (B) BRCAWT PDX-2428: vehicle (n = 3), olaparib (n = 8). (C) BRCA1MUT PDX-2462: vehicle (n = 3), olaparib (n = 7). (D to F) Validation of acquired olaparib resistance in xenograft tumors. Olaparib-treated PDX tumors were harvested, retransplanted into different mice, and expanded. PDX-bearing mice were then treated with vehicle or olaparib (50 mg/kg per day, 5 days/week). (D) PDX-2445: vehicle (n = 3), olaparib (n = 5). (E) PDX-2428: vehicle (n = 3), olaparib (n = 6). (F) PDX-2462: vehicle (n = 3), olaparib (n = 3). Data are shown as mean ± SD [two-way analysis of variance (ANOVA)]. (G) Representative hematoxylin and eosin (H&E)–stained sections of PDX-2428 and PDX-2462 xenografts. The vehicle-treated tumors mostly consist of relatively uniform tumor cells. In contrast, the tumors with acquired olaparib resistance exhibit enriched PGCCs in the forms of multinucleated giant cells (black arrowheads) or mononucleated giant cells (yellow arrowheads). Scale bars, 50 μm. (H) Propidium iodide (PI) flow cytometry quantification of polyploidy in PDX-2428 and PDX-2462 xenografts. PDX-2428: vehicle (n = 5), resistant (n = 7). PDX-2462: vehicle (n = 3), resistant (n = 4). Cells with DNA content > 4C were defined as PGCCs. Data are shown as mean ± SD. The exact P values are shown on the graph (Welch’s t test).
To evaluate whether the xenografts that acquired olaparib resistance were bona fide resistant tumors, they were harvested, retransplanted, and expanded in different mice, and the mice were treated with olaparib or vehicle as above. As expected, these xenografts displayed resistance to olaparib, and their tumor growth was similar to that of vehicle-treated xenografts regardless of their BRCA status (Fig. 1, D to F). Histologically, the olaparib-sensitive tumors were mainly composed of residual tumor cells on a background of fibrotic stromal cells. PGCCs constituted one of the major components of the residual tumor cells, especially in the germline BRCA1MUT tumors (fig. S1). PGCCs were also highly enriched in both the BRCAWT PDX-2428 and the BRCA1MUT PDX-2462 tumors with acquired olaparib resistance (Fig. 1G). Quantitative analysis using flow cytometry shows that there was approximately twofold enrichment of PGCCs in tumors with acquired resistance as compared with vehicle-treated controls (Fig. 1H). Thus, our PDX experiments showed that PGCCs survived the antitumor effect of olaparib and were accumulated in HGSC tumors with acquired olaparib resistance.
A sublethal concentration of olaparib leads to the development of PGCCs in ovarian and breast cancer cell lines
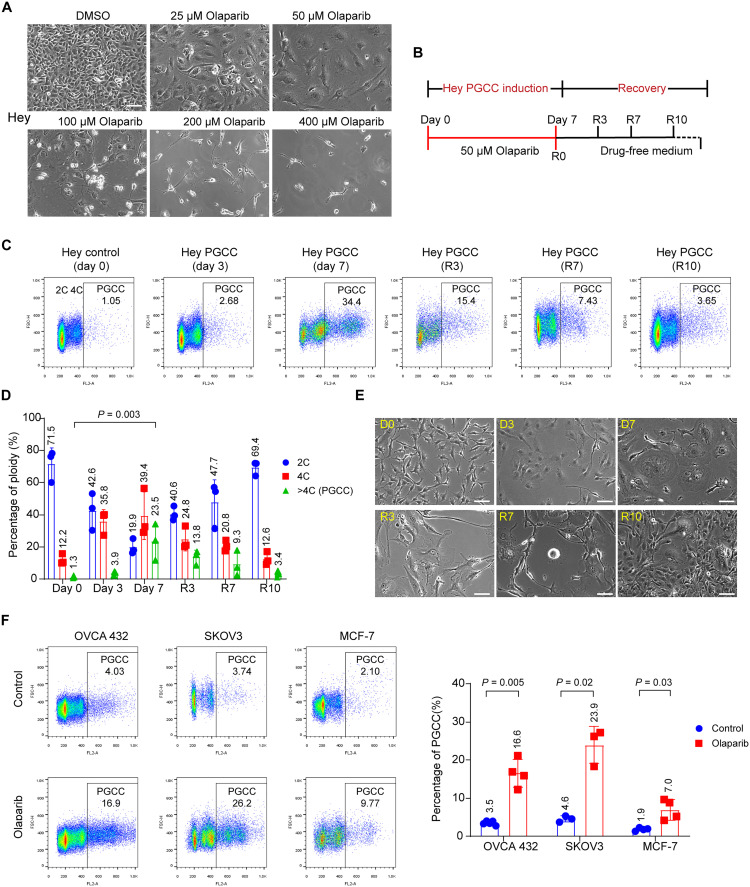
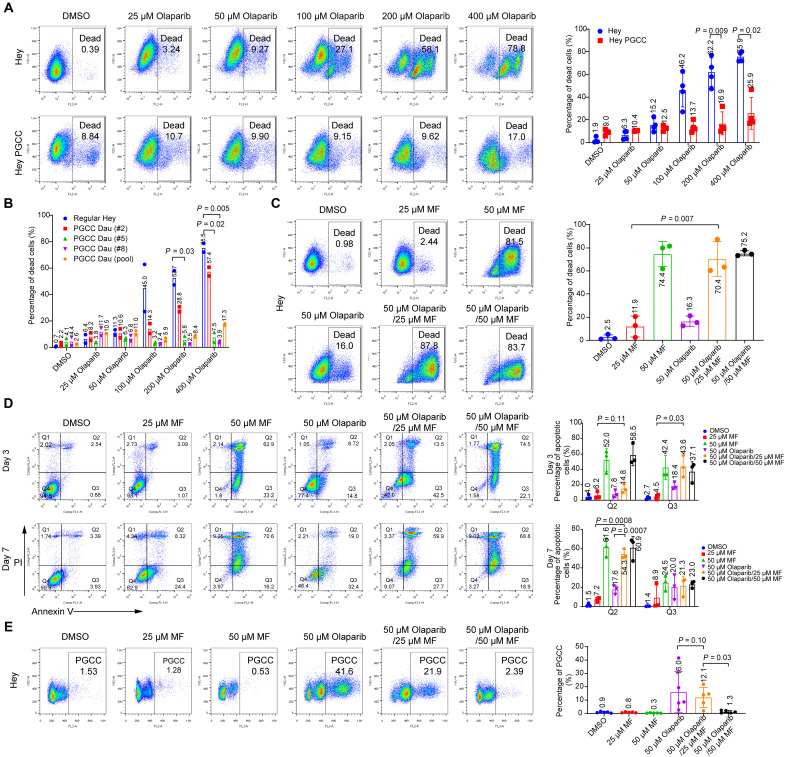
To investigate the mechanism underlying the increased proportion of PGCCs in tumors with acquired olaparib resistance, we treated Hey ovarian cancer cells with various concentrations of olaparib for 1 week. We found that a high concentration of olaparib (>100 μM) induced massive cell death. However, lower concentrations of olaparib (25 and 50 μM) resulted in the enlargement of the cytoplasm and nucleus (Fig. 2A). Therefore, 50 μM olaparib was selected as the optimal concentration for inducing Hey PGCCs for the following experiments. In general, Hey PGCCs were induced by treating Hey cells with 50 μM olaparib for 7 days. Olaparib was then withdrawn from culture on day 7, and PGCCs were allowed to recover in drug-free culture medium and to generate daughter cells (Fig. 2B).
Fig. 2. Olaparib induces the formation of PGCCs.
(A) Representative phase-contrast microscopy images of Hey HGSC cells exposed to the indicated concentrations of olaparib or vehicle (0.1% DMSO) for 7 days. The sublethal concentration of olaparib (50 μM) led to formation of PGCCs, characterized by enlarged cytoplasm and nuclei. Scale bar, 100 μm. (B) Schematic illustration of induction of PGCCs and PGCC-derived daughter cells. Hey cells were treated with 50 μM olaparib for 7 days. Cells were then allowed to recover in drug-free culture medium for up to 10 days to generate daughter cells. R0 refers to the day on which olaparib was withdrawn. (C and D) PI flow cytometry quantification of polyploidy in Hey cells exposed to 50 μM olaparib at the indicated times. The exact P values are shown on the graph (one-way ANOVA). (E) Representative phase-contrast microscopy images showing the morphological changes in Hey cells exposed to olaparib at the indicated times. Freshly seeded Hey cells are slender. Once exposed to 50 μM of olaparib, cells gradually became flattened with enlarged cytoplasm and nuclei. PGCCs proliferated and produced daughter cells during recovery (R) days 3 to 10. Scale bars, 100 μm. (F) PI flow cytometry quantification of polyploidy (left) and PGCCs as a percentage of total tumor cells (right) in human ovarian and breast cancer cell lines. OVCA-432 PGCCs were induced by exposure to 400 μM olaparib for 72 hours and then allowed to recover for another 72 hours. SKOV3 and MCF-7 PGCCs were induced by exposure to 50 μM olaparib for 7 days. Each data point corresponds to one biological replicate in (C), (D), and (F), and data are shown as mean ± SD. The exact P values are shown on the graph (Welch’s t test).
To determine how PGCCs change over time, Hey cells were exposed to 50 μM olaparib for 7 days and allowed to recover for another 10 days. The percentage of PGCCs (defined by DNA content > 4C) in untreated Hey cells was only 1.05%, but it markedly increased, to 34.4%, after 7 days of treatment. During recovery, the proportion of PGCCs in Hey cells decreased over time (Fig. 2, C and D). The morphological characteristics of PGCCs observed under a light microscope closely corresponded with flow cytometry analyses of these cells (Fig. 2E). Freshly seeded Hey cells mainly consisted of diploid cells (day 0). By day 3 of exposure to olaparib, the cells became flat, and the cytoplasm gradually enlarged. At day 7, olaparib exposure had induced the enlargement of the nuclei and cytoplasm of Hey cells. Once olaparib was withdrawn from the culture, PGCCs proliferated and produced progeny cells [recovery (R) days 3 to 10].
Next, we examined whether olaparib could facilitate the formation of PGCCs in five other ovarian cancer cell lines and one breast cancer cell line as compared with the Hey cell line. First, we determined the olaparib sensitivity in these cell lines (fig. S2A). Then, olaparib concentrations were optimized to induce PGCCs in different cell lines. Olaparib induced a remarkable increase in the proportion of PGCCs in all cell lines tested (Fig. 2F, fig. S2B, and table S1). Last, we also tested whether other PARPi could also induce PGCCs. Niraparib induced PGCCs at an even lower concentration in all cell lines tested (fig. S2C). These results suggest that the emergence of PGCCs may be a general biologic response to PARPi treatment.
PGCCs exhibit hallmarks of cellular senescence
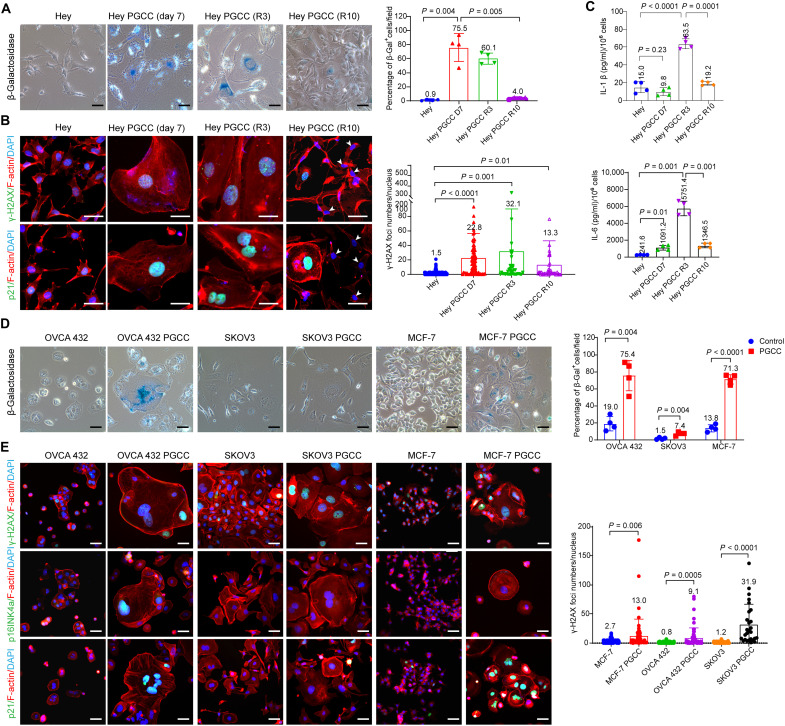
Their enlarged size and flattened morphology suggest that PGCCs may be senescent. The senescence phenotype is often characterized by the induction of γ-H2A histone family member X (γ-H2AX) nuclear foci (33), cell cycle arrest regulated by the cyclin-dependent kinase inhibitors p16INK4a and p21 (33, 34), an increase of senescence-associated β-galactosidase (SA-β-gal) activity (35), and enhanced expression of cytokines [e.g., interleukin-1 (IL-1), IL-6, and IL-8] (36). We therefore evaluated a panel of markers that are commonly used for senescence detection. We first measured acidic β-gal staining. The untreated Hey cells were barely stained with β-gal. However, Hey PGCCs exhibited remarkable blue-green–positive staining in the cytoplasm (Fig. 3A). The PGCC progeny cells at R10 expressed less β-gal than did the parental PGCCs. We then assessed the expression of γ-H2AX foci and p21 in Hey cells by immunofluorescence staining. The untreated Hey cells showed minimal staining for γ-H2AX and p21. Conversely, γ-H2AX foci and p21 expression were highly enhanced in the nuclei of PGCCs and dropped to an undetectable level in the progeny cells (Fig. 3B). Another senescence marker, p16INK4a, was not detected in any of the samples. Last, we analyzed the levels of IL-1β and IL-6, two components of the senescence-associated secretory phenotype (SASP), in the culture medium of Hey cells by the enzyme-linked immunosorbent assay (ELISA). The secretion of IL-1β and IL-6 was significantly higher in PGCCs that had recovered in olaparib-free medium for 3 days than in control Hey cells or Hey PGCCs that had not been allowed to recover (Fig. 3C).
Fig. 3. PGCCs exhibit multiple characteristics of senescent cells.
(A) Representative images (left) and quantification (right) of β-galactosidase (β-gal) staining of control Hey cells and Hey PGCCs. The β-gal–positive cells exhibit dark blue staining in the cytoplasm. Scale bars, 50 μm. Four randomly selected fields per group (10× objective lenses) were used for quantification analysis. (B) Immunofluorescence images of γ-H2AX foci and p21 expression (left) and quantification (right) of γ-H2AX foci number in control Hey cells and Hey PGCCs at the indicated times. Both γ-H2AX foci and p21 expression were highly elevated in the nuclei of PGCCs on treatment day 7 and gradually decreased during the recovery period in the PGCC-derived daughter cells (white arrowheads). Scale bars, 50 μm. Filamentous actin (F-actin) was visualized by phalloidin staining. At least 50 cells per group were counted and used for quantification. (C) Hey PGCCs were induced by exposure of Hey cells to 50 μM olaparib for 7 days and allowed to recover for 3 or 10 days. Supernatants were collected and analyzed for IL-1β and IL-6 secretion by ELISA. Each data point corresponds to one biological replicate. (D) Phase-contrast images (left) and quantification (right) of β-gal staining of control OVCA-432, SKOV3, and MCF-7 cells and their corresponding PGCCs. Scale bars, 50 μm. Four randomly selected fields per group (10× objective lenses) were used for quantification analysis. (E) Immunofluorescence images (left) of γ-H2AX foci, p16INK4a, and p21 expression and quantification (right) of γ-H2AX foci in the indicated cell lines and their corresponding PGCCs. Scale bars, 50 μm. At least 50 cells per group were counted and used for quantification (27 cells for SKOV3 PGCCs). Data are shown as mean ± SD in (A) to (E). The exact P values are shown on the graph (Welch’s t test).
We next evaluated whether the PGCCs induced by olaparib in other cancer cell lines were also senescent. PGCCs derived from OVCA-432 ovarian cancer and MCF-7 breast cancer cell lines were strongly stained with β-gal, but β-gal staining in SKOV3 ovarian cancer PGCCs was almost negligible (Fig. 3D). Furthermore, the expression of senescence marker proteins varied among the cell lines. For example, PGCCs derived from OVCA-432 or MCF-7 cells expressed abundant γ-H2AX foci and p21 protein. OVCA-432 PGCCs were also positively stained with p16INK4a (Fig. 3E). However, SKOV3 PGCCs were only positively stained for γ-H2AX foci. These results suggest that PGCCs do not necessarily express all markers involving senescence, depending on their genetic background.
To identify the potential biomarkers of PGCCs, we performed RNA sequencing analysis of whole transcriptomes in the above-used cell lines and three ovarian cancer organoids treated with olaparib (fig. S3A and table S2). Gene set enrichment analysis indicated that multiple pathways were enriched in the PGCCs and PGCC-derived daughter cells. The most prominent enriched pathways involved cytokines and chemokines associated with the SASP phenotype such as increased tumor necrosis factor–α signaling and cytokine activity. Enrichment of other gene sets, including methylation and MYC signaling (fig. S3B), demonstrated the down-regulation of major cell proliferation–related pathways in PGCCs. We also evaluated the protein level changes of core senescence-determining genes such as GATA binding protein 4 (GATA4) and p21 from the PGCCs to subsequent cell division. The expression of GATA4 and p21 were up-regulated in PGCCs (day 7) and gradually down-regulated in PGCC-derived daughter cells. Together, these data demonstrated that PGCCs display several major hallmarks of cellular senescence.
PGCCs escape from senescence and generate mitotic competent daughter cells
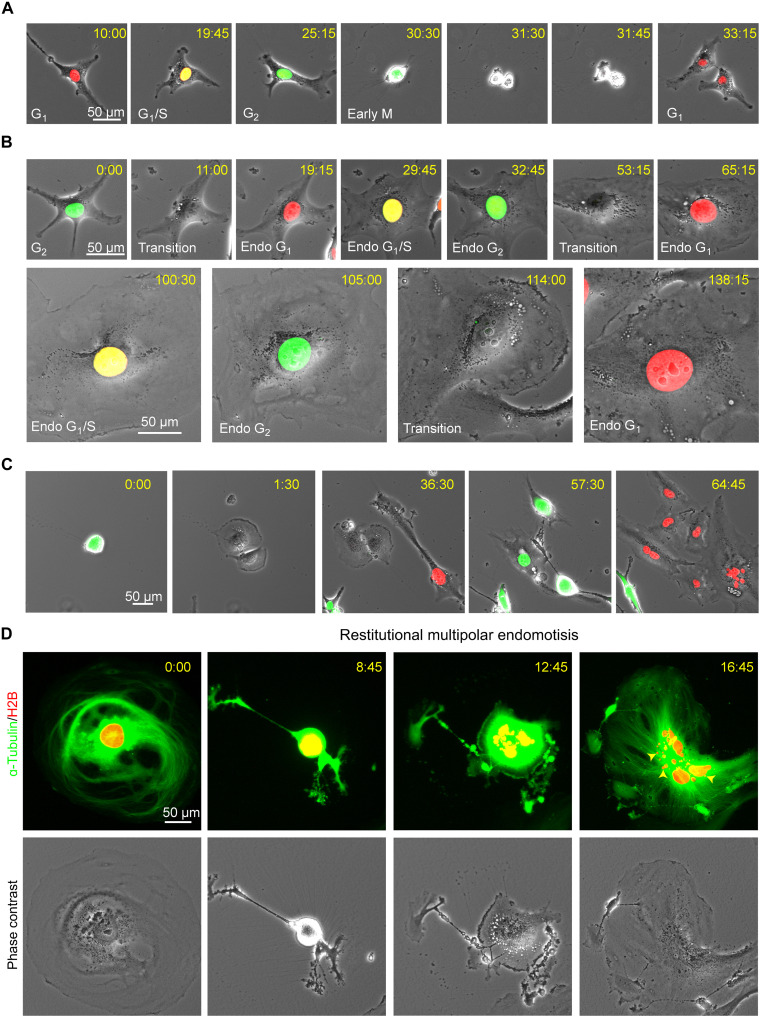
Senescent cells are traditionally considered to be nondividing cells because they lack the ability to undergo mitosis. One of the major unsolved questions about PGCCs is how they survive PARPi-induced therapeutic stress and whether they can escape senescence and resume proliferation. To answer these questions, we labeled Hey cells with the fluorescent ubiquitination–based cell cycle indicator (FUCCI) system and tracked cell cycle changes with time-lapse photography. The mitotic cell cycle consists of two main stages: interphase (G1 phase, S phase, and G2 phase) and M phase (mitosis and cytokinesis). The FUCCI system labels nuclei in red at the G1 phase, yellow at the G1/S transition phase, and green at the S/G2/M phases (37). Consistent with our earlier finding with paclitaxel-induced PGCCs (20), we found that untreated Hey cells divided via the canonical mitotic cell cycle and produced two identical daughter cells once they completed cytokinesis (Fig. 4A and movie S1). However, olaparib (50 μM) prevented Hey cells from undergoing mitosis. Instead, in olaparib-treated Hey cells, the cell cycle consisted of alternating S and G phases without cell division. Eventually, the cells became flattened and their nuclei enlarged due to the accumulation of genomic DNA (Fig. 4B and movie S2). This process is termed endoreplication or endocycling (38).
Fig. 4. Olaparib induces the formation of PGCCs through endoreplication.
Time-lapse monitoring of Hey cells labeled with FUCCI (A to C) or histone H2B-mCherry and EGFP-ɑ-tubulin (D). The FUCCI system labels nuclei in red at the G1 phase, yellow at the G1/S transition phase, and green at the S/G2/M phases. (A) The typical mitotic cell cycle consists of interphase (G1, S, and G2) and mitosis (M). (B) Tracking of cell cycle changes in a Hey cell exposed to 50 μM olaparib. A diploid Hey cell gradually becomes a mononucleated PGCC after undergoing multiple cycles of endoreplication without cell division under olaparib treatment. (C) A Hey PGCC re-enters the mitosis cycle to generate daughter cells with strong self-renewal capacity. (D) A mononucleated PGCC generates a multinucleated PGCC via restitutional multipolar endomotisis. The yellow arrowheads indicate micronuclei.
We next investigated whether PGCCs can exit the endoreplication cycle and re-enter mitosis when cultured in the drug-free recovery medium. Tripolar mitosis occurred in some cases; fig. S4A and movie S3 show that PGCC can generate three daughter cells (28:15), two of which re-fused (36:00). All daughter cells then continued endocycling. PGCCs also gave rise to daughter cells via bipolar mitosis. In the example shown in Fig. 4C and movie S4, the first cell division occurred at 1:30, and the resulting daughter cells underwent the second and third rounds of cell division at 36:30 and 64:45, respectively.
To visualize the dynamic changes of chromosomes and the spindle movement in PGCCs during the division process, we labeled Hey cells’ chromosomes with histone H2B-mCherry and microtubules with enhanced green fluorescent protein (EGFP)–ɑ-tubulin. Untreated Hey cells divided via canonical bipolar mitosis (fig. S4B and movie S5). By contrast, olaparib-induced Hey PGCCs exhibited diverse modes of division. In some cases, PGCCs produced two separate daughter cells via bipolar mitosis (fig. S4C and movie S6). In other cases, PGCCs underwent tripolar mitosis. As shown in fig. S4D, a mononucleated PGCC gave rise to three daughter cells connected at the midbody (white arrow at 2:45), two of which re-fused and formed a binucleated PGCC (movie S7). In another case, a PGCC divided into three multinucleated daughter cells via tripolar mitosis (fig. S4E and movie S8). In addition, during division, PGCCs can undergo restitution multipolar endomitosis (RMEM) (22), resulting in a massively fragmented multinucleated PGCC (Fig. 4D, fig. S4F, and movies S9 and S10) with multiple micronuclei (yellow arrows in Fig. 4D and fig. S4F). These results collectively indicated that a sublethal concentration of olaparib resulted in a switch from the mitotic cell cycle to various modes of endoreplication with defective nuclear division or cytokinesis. PGCCs then divided in various ways to produce mononucleated or multinucleated daughter cells with massively altered genomes via creating genomic chaos (21, 39), a subset of daughter cells can survive and acquire therapeutic resistance.
PGCCs and daughter cells are resistant to cell death triggered by olaparib
To test whether PGCCs and PGCC-derived daughter cells confer olaparib resistance, we compared olaparib sensitivity between control Hey cells, Hey PGCCs, and PGCC-derived daughter cells as shown in Fig. 5A, and Hey and Hey PGCCs (pretreated with 50 μM olaparib for 1 week) were exposed to concentrations of olaparib ranging from 25 to 400 μM for 7 days. As also shown in Fig. 5A, olaparib induced the death of control Hey cells in a dose-dependent manner. In contrast, Hey PGCCs showed extreme resistance to olaparib, especially to high concentrations (>100 μM). To test whether PGCC-derived progeny cells confer drug resistance, we subcloned the Hey PGCCs and established four daughter cell lines derived from a single PGCC. Compared with control Hey cells, all the tested daughter cell lines exhibited varying degrees of resistance to olaparib-induced cell death (Fig. 5B and fig. S5A). To determine whether the varying sensitivity of PGCC subclones is related to the proliferation of the clones, we compared the proliferative capacity of these daughter cell clones and found no difference (fig. S5B). We also assessed the sensitivity of the PGCC subclones to common chemotherapy drugs such as carboplatin and paclitaxel (fig. S5C). Although the dose-response of PGCC subclones to carboplatin was consistent with that of Hey cells, they were more resistant to paclitaxel, especially PGCC subclone #10. These data demonstrate that daughter cells derived from PGCCs maintain the PGCCs’ acquired resistance to olaparib.
Fig. 5. Mifepristone inhibits olaparib-mediated PGCC formation by promoting apoptosis.
(A) Hey cells and Hey PGCCs were treated with vehicle (0.1% DMSO) or olaparib at the indicated concentrations for 7 days, stained with PI, and analyzed with flow cytometry. Representative percentages of dead cells are shown on the left, and statistical analysis results are shown on the right. (B) Quantitative analysis of olaparib sensitivity in Hey and Hey PGCC daughter cells. Hey cells and Hey PGCC-derived daughter (Dau) cells (derived from a single PGCC or pooled PGCCs) were exposed to the indicated increasing concentrations of olaparib for 7 days and assayed by PI staining. Data represent mean ± SD from two independent experiments. (C) Hey cells were treated with the indicated concentrations of mifepristone (MF) alone, olaparib alone, or a combination of both drugs for 7 days. Cell viability was determined by PI flow cytometry. Percentages of dead cells are shown on the left, and statistical analysis results are shown on the right. (D) Hey cells were treated with the indicated concentrations of MF or olaparib alone or with a combination of both drugs for 3 or 7 days. Apoptotic cells were identified by annexin V–PI staining. Q2 and Q3 represent the late apoptotic cells and early apoptotic cells, respectively. Statistical analysis results are shown on the right. (E) Hey cells were exposed to 50 μM olaparib with or without MF for 7 days to induce PGCC formation. Polyploidy was measured by PI flow cytometry analysis, and the percentage of PGCCs is shown. Data are shown as mean ± SD in (A) to (E). Each data point corresponds to one biological replicate in (A), (C), (D), and (E). The exact P values are shown on the graph (Welch’s t test).
PGCCs exhibit sensitivity to the combination of olaparib and mifepristone
Our previous studies showed that PGCCs dedifferentiate from mature somatic cells by recapitulating a blastomere-stage cleavage program that augments the nucleus and gives rise to embryonic life (16, 20, 22). Because they mimic early embryonic development, we propose that PGCCs may represent “somatic blastomeres” (28, 40–42). Therefore, we reasoned that certain contraceptive drugs may be able to block the life cycle of PGCCs to block development of somatic blastomeres. One candidate drug is mifepristone, which has been used as an emergency contraceptive drug for decades (43). Mifepristone has also been reported to inhibit repopulation of ovarian cancer cells after cisplatin/paclitaxel combination therapy and delay the growth of ovarian carcinoma xenografts (44, 45). Therefore, we hypothesized that mifepristone might decrease tumor recurrence by inhibiting olaparib-induced PGCC formation. To test this hypothesis, we first determined the cytotoxic effect of mifepristone on Hey cells. A lower concentration (25 μM) of mifepristone had minimal effect on the viability of Hey cells, and olaparib alone resulted in only 16.0% cell death. However, the combination of mifepristone and olaparib led to 87.8% cell death (Fig. 5C). The synergy between mifepristone and olaparib disappeared when a higher concentration of mifepristone (50 μM) was used, probably because it alone was sufficient to kill most cells. Analysis of the apoptotic response of cells to the compounds showed that treatment with mifepristone (25 μM) in combination with olaparib for 3 days accelerated the production of early apoptotic cells (42.5%, Q3) compared to cells treated with mifepristone (1.1%) or olaparib alone (14.8%) (Fig. 5D). The combination treatment also led to the emergence of more late apoptotic cells (59.9%, Q2) than did treatment with mifepristone (8.3%) or olaparib (19.0%) alone for 7 days. Cell cycle analysis further revealed that mifepristone alone did not significantly reduce the proportion of preexisting PGCCs among Hey cells. However, the combined use of mifepristone and olaparib markedly inhibited the olaparib-mediated development of PGCCs in a concentration-dependent manner (Fig. 5E).
We further validated the above findings in OVCA-432, MCF-7, SKOV3, and PEO-1 cells. In these cells, too, the combination of mifepristone and olaparib resulted in more early and late apoptotic cells than did mifepristone or olaparib alone (fig. S6). Together, these data demonstrate that the mifepristone/olaparib combination blocked olaparib-induced PGCC development by promoting apoptosis.
Olaparib enhances the frequency of PGCCs in HGSC-derived organoids
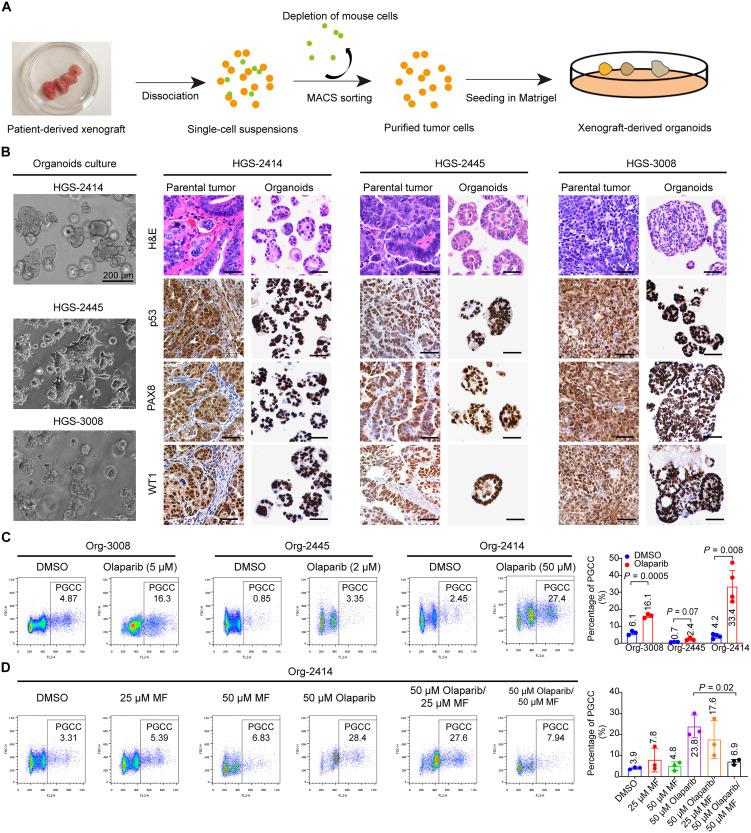
Organoids can faithfully maintain the heterogeneity and the histomorphological characteristics of the parental tumor and can better predict drug response than cell lines (46, 47). To develop an ex vivo platform of organoid models, we established several organoids from HGSC PDXs (Fig. 6A and table S3). Hematoxylin and eosin (H&E) staining of these HGSC PDX-derived organoids revealed that the organoids harbored multiple histologic and characteristics of their parental tumors, such as the presence of histologic architectures, papillary (HGSC-2414 and HGSC-2445) or solid (HGSC-3008) patterns, and nuclear and cellular atypia (Fig. 6B). We also compared the expression of HGSC protein biomarkers between organoids and parental tumors. HGSC parental tumors were characterized by intense nuclear staining of paired box gene 8 (PAX8), a marker of the serous subtype and Wilms tumor 1 (WT1), as well as either positive nuclear staining or completely absent expression of tumor protein p53 (48, 49). As shown in Fig. 6B, the organoids retained the p53, PAX8, and WT1 expression status of their corresponding parental tumors.
Fig. 6. Olaparib enhances polyploidy in patient-derived ovarian cancer organoids.
(A) Flowchart of procedures for establishing organoids from PDXs. MACS, magnetic-activated cell sorting. (B) Phase-contrast images of 3 HGSC organoids (left) together with H&E and immunohistochemical staining (right) in organoids and corresponding parental tumors. Tumors and organoids showed robust expression of p53, PAX8 (a marker of serous subtype), and WT1 in the nucleus. Scale bars, 100 μm (right). (C) PI flow cytometry analysis (left) and quantification (right) of polyploidy in human ovarian cancer-derived organoids. Organoids (Org) were exposed to vehicle (DMSO) or olaparib at the indicated concentrations for 7 days and then collected and dissociated into single cells. Polyploidy was determined by PI flow cytometry analysis. (D) PI flow cytometry analysis (left) and quantification (right) of polyploidy in Org-2414. Org-2414 was exposed to the indicated concentrations of mifepristone (MF), 50 μM olaparib, or a combination of both drugs for 7 days. Data are shown as mean ± SD in (C) and (D). Each data point corresponds to one biological replicate. The exact P values are shown on the graph (Welch’s t test).
We next investigated whether olaparib induces the development of PGCCs in organoids. Olaparib treatment remarkably increased the number of PGCCs in organoid (Org)–3008 and Org-2445 when used at a sublethal concentration (Fig. 6C and fig. S7A). Similarly, exposure to 50 μM olaparib induced the highest proportion of PGCCs in Org-2414. PGCCs induced by olaparib in the organoids exhibited the phenotypes of senescent cells (fig. S7, B and C). γ-H2AX foci, p16INK4a, and p21 were highly expressed in Org-2414 PGCCs (fig. S7C).
Consistent with our observations in Hey cells, mifepristone alone did not change the percentage of PGCCs in Org-2414. However, the proportion of PGCCs in this organoid shrank from 28.4% with olaparib monotherapy to 7.94% with combined mifepristone and olaparib (50 μM each) (Fig. 6D). We also found that a combination of mifepristone and olaparib enhanced apoptosis in Org-2414, Org-2445, and Org-3008 (fig. S7D). These data suggest that mifepristone blockage of olaparib-induced PGCC formation can be applied to HGSC organoid models.
Mifepristone suppresses tumor growth in olaparib-naïve and olaparib-resistant HGSC PDX models
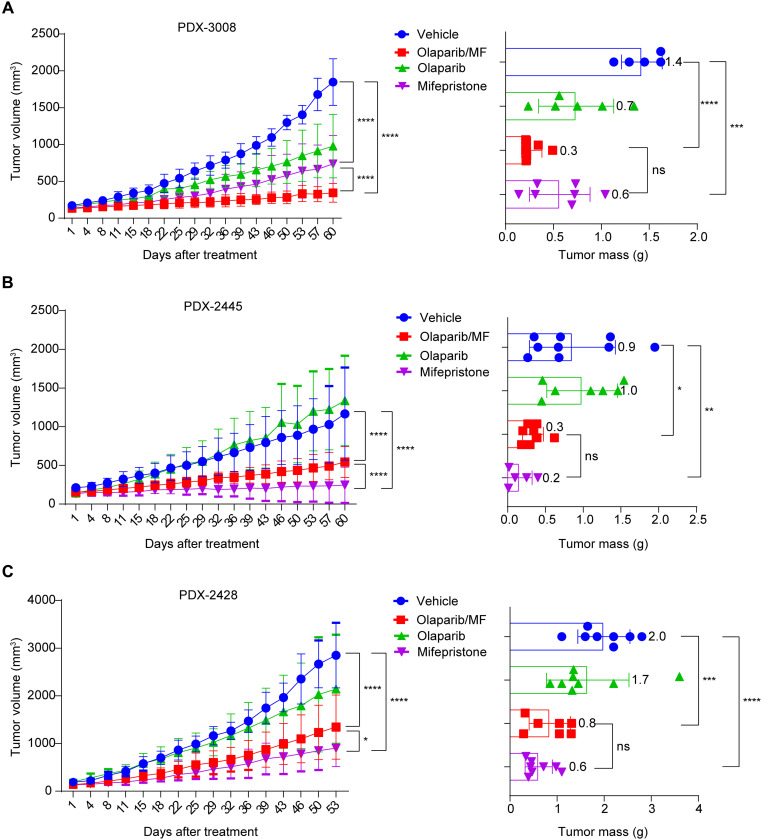
To assess whether blockage of PGCC development could suppress tumor growth, we tested the in vivo efficacy of mifepristone in mouse PDX models. We first compared the effects of mifepristone monotherapy and a mifepristone/olaparib combination using an olaparib-naïve PDX model (i.e., one without prior exposure to olaparib). Mifepristone monotherapy induced mild tumor growth inhibition in BRCAWT PDX-3008 compared to vehicle treatment, with increased tumor necrosis and a reduction in the number of proliferating cells as shown by Ki-67 staining (Fig. 7A and fig. S8A). By contrast, the combination of mifepristone with olaparib was more effective at reducing tumor growth than either single agent. Furthermore, both mifepristone monotherapy and the mifepristone/olaparib combination significantly decreased the tumor mass compared to vehicle-treated tumors (P = 0.0002 and P < 0.0001, respectively).
Fig. 7. Mifepristone mitigates tumor growth in ovarian HGSC PDX models.
(A) Ovarian HGSC PDX-3008 xenografts (BRCAWT, olaparib-naive tumors) were treated with olaparib (n = 6), mifepristone (n = 7), vehicle (n = 5), or olaparib in combination with mifepristone (n = 7) for 60 days. The left shows the mean tumor volume in the xenograft-bearing mice at the indicated times; the right shows the tumor mass after 60 days of treatment. Mifepristone monotherapy and mifepristone/olaparib treatment significantly suppressed tumor growth compared with vehicle (two-way ANOVA, ****P < 0.0001). Tumors treated with mifepristone alone (Welch’s t test, ***P = 0.0002) and those treated with mifepristone/olaparib (****P < 0.0001) had significantly smaller masses at harvest than did vehicle-treated tumors. Error bars indicate SD. (B) Olaparib-resistant BRCAWT ovarian HGSC PDX-2445 xenografts were treated with olaparib (n = 7), mifepristone (n = 5), vehicle (n = 9), or mifepristone/olaparib (n = 8) for 60 days. Both mifepristone monotherapy (two-way ANOVA, ****P < 0.0001) and mifepristone/olaparib combination therapy (****P < 0.0001) significantly suppressed tumor growth compared with vehicle treatment. Tumors treated with mifepristone monotherapy (Welch’s t test, **P = 0.0061) and mifepristone/olaparib combination therapy (*P = 0.0257) had significantly smaller masses at harvest (60 days after treatment) than did vehicle-treated tumors. Error bars indicate SD. (C) Olaparib-resistant BRCAWT ovarian HGSC PDX-2428 xenografts were treated with olaparib (n = 8), mifepristone (n = 8), vehicle (n = 8), or mifepristone/olaparib (n = 7) for 53 days. Both mifepristone monotherapy (two-way ANOVA, ****P < 0.0001) and mifepristone/olaparib combination treatment (****P < 0.0001) significantly suppressed tumor growth compared with vehicle treatment. Tumors treated with mifepristone monotherapy (Welch’s t test, ****P < 0.0001) and mifepristone/olaparib combination therapy (***P = 0.0006) had significantly smaller masses at harvest than did vehicle-treated tumors. Error bars indicate SD. ns, not significant.
Next, we tested the role of mifepristone in the acquired olaparib resistance models PDX-2445 and PDX-2428. The olaparib-resistant tumors were expanded in mice, which were then treated with vehicle, olaparib only, mifepristone only, or olaparib and mifepristone. The olaparib-treated tumors had a similar growth pattern to that of vehicle-treated tumors, demonstrating that they were bona fide olaparib-resistant tumors. Moreover, mifepristone monotherapy and the mifepristone/olaparib combination therapy significantly inhibited tumor growth compared to vehicle treatment (Fig. 7B). Mifepristone monotherapy showed significantly better suppression of tumor growth than did mifepristone/olaparib combination therapy (P < 0.0001), and mifepristone monotherapy and the mifepristone/olaparib combination also led to massive tumor necrosis and reduced cell proliferation, as indicated by Ki-67 staining (fig. S8B). Similar results were obtained in PDX-2428 (Fig. 7C and fig. S8C). In addition, in a subset of tumors that responded to mifepristone, there is a marked increase in fibrosis and macrophages (fig. S8B), suggesting that mifepristone may attenuate the tumor growth in patient tumors with acquired resistance by targeting embryonic properties of PGCCs toward benign lineages. Inclusion of olaparib may in fact interfere with the differentiation ability of mifepristone in PDX models that have already acquired resistance. Together, our data suggest that combination therapy with olaparib/mifepristone may be a better choice for olaparib-naïve tumors, while mifepristone monotherapy may be more effective for treating olaparib-resistant tumors.
DISCUSSION
The inevitable tumor recurrence in both patients with BRCA mutations and those with WT BRCA following olaparib therapy suggests that the underlying mechanism of PARPi resistance may be independent of the BRCA status of the tumor. Multiple mechanisms of resistance to PARPi have been described, including up-regulation of drug efflux via overexpression of adenosine triphosphate–binding cassette, restoration of HR or WT BRCA sequence, target specific resistance, or restoration of stalled replication fork protection (3, 50). These mechanisms remain important ones to understand how resistance occurs to PARPi in different subtypes of tumor; however, it is unlikely that they represent all of the mechanisms involved in resistance generation, which prompts us to look into PGCCs, an emerging new field of cancer biology that may provide a generalized resistant mechanism by activating our previously described giant cell life cycle that leads to whole genome reprograming in response to catastrophic stress (15, 21, 42).
To address this issue, we developed PDX models with acquired olaparib resistance from both BRCA1MUT and BRCAWT HGSC patient tumors. We found that PGCCs were more common in the olaparib-treated PDXs, suggesting that PGCCs, not BRCA status, are associated with acquired resistance to olaparib in patients with relapsed ovarian cancer. This result is consistent with the previous report that increased ploidy were found in cellular and clinical ovarian cancer tumor samples with PARPi resistance (51). To understand how PGCCs are associated with PARPi resistance and tumor recurrence, we evaluated the effect of olaparib on the development of PGCCs in six human ovarian cancer cell lines and one breast cancer cell line. Long-term (1-week) exposure to sublethal concentrations of olaparib markedly increased the proportion of PGCCs in these cancer cell lines. Using time-lapse photography of FUCCI-labeled Hey cells, we demonstrated that olaparib led to a transition of the cell cycle from mitosis to endoreplication, a common cell cycle variant during which cells increase their genomic DNA content without dividing. The endoreplication cycle eventually resulted in the development of PGCCs (mostly mononucleated PGCCs). Here, we found that PGCCs divided into daughter cells through diverse variants of mitosis, such as multipolar mitosis and RMEM as we recently reported (22). These aberrant mitoses generated multiple micronuclei and inevitably increased chromosomal instability, genomic chaos, and cancer macroevolution (22, 39, 52, 53), which is known to contribute to chemotherapy resistance (54, 55). Thus, PGCCs are not “dead,” as has been assumed in the past but rather undergo a variety of aberrant cell cycles that may underlie their ability to promote cancer recurrence and therapy resistance via whole genomic duplication-mediated reprogramming.
Consistent with our early observations with paclitaxel-induced PGCCs (20, 56), our data demonstrate that olaparib-induced PGCCs exhibit most of the phenotypes of senescent cells. Most of the Hey-derived PGCCs showed enhanced SA-β-gal activity and p21 expression, greater numbers of γ-H2AX foci, and increased secretion of SASP factors, whereas the daughter cells of PGCCs minimally expressed β-gal, p21, and γ-H2AX. This finding suggested that PGCC progeny cells can escape senescence to facilitate recurrence, consistent with early observations made by our group (15). The expression of senescence markers in PGCCs varied in different cell types. For instance, SA-β-gal activity was elevated in the PGCCs of all but one cell line, which only exhibited increased γ-H2AX nuclear foci. These differences could be attributable to the different genetic backgrounds of the parental cell lines.
To better model tumor’s therapy response, we developed organoids from PDXs from patients with ovarian HGSC. We formulated a relatively simple medium to foster the HGSC PDX-derived organoids modified from the media used in previous reports (47, 57). These HGSC organoids recapitulated the histomorphological characteristics of the parental tumor and were able to proliferate in vitro over the long term (>40 passages). The organoid-derived PGCCs induced by olaparib also exhibited the phenotypes of senescent cells. These data provide further evidence that sustained DNA damage results in a senescent cell-like phenotype in PGCCs and that this conserved mechanism allows these cells to resist apoptosis.
Mutation in the TP53 gene was found in 96% of HGSC (1). In this study, we used both p53 WT and multiple p53 mutant ovarian cancer cell lines. We have observed the formation of PGCCs in all cell lines regardless of TP53 mutations. These data support our early conclusion that PGCCs represent a general genomic response to high level stresses via an evolutionarily conserved polyploidization program (15, 27, 58). Although mutation or loss of p53 can sensitize the cancer cell to polyploidization due to defective cell checkpoints, however, the mutation in p53 per se is not required. Depending on the types of stressors, the level of polyploidization program could respond at the different levels by varying speed or number of genomic copy to generate newly programmed daughter cells for resistance.
We have recently described how PGCCs confer acquired therapeutic resistance and dormancy by de-repressing an embryonic program that is suppressed during human growth and development. PGCCs may therefore represent somatic blastomeres in tumor initiation, resistance, dormancy, and metastasis (16, 20, 22, 40). The different stages of the PGCC life cycle may offer vulnerabilities for potential therapeutic intervention (42) . Our data here provide additional proof of principle for this hypothesis. By using the antiprogestin contraceptive drug mifepristone to block the initiation of PGCCs by olaparib, we found that mifepristone can notably block the formation and survival of PGCCs induced by olaparib. Mifepristone synergistically acts with olaparib to promote apoptosis of cells that are undergoing endoreplication, resulting in the inability to form PGCCs. This mechanism suggests that mifepristone may be more effective in killing newly formed PGCCs induced by therapeutic stress than preexisting PGCCs in patient tumors.
Mifepristone can directly decrease tumor growth with acquired resistance to PARPi. One possibility is that mifepristone could also promote the differentiation of PGCCs and daughter cells toward differentiation into benign lineages, as our early studies demonstrated that PGCCs acquired blastomere-like stemness and are prone to adipose or fibrous cell differentiation (16, 20, 24). We observed massive fibrosis in mifepristone-treated tumors (fig. S8B), although such observation remains to be validated in a large cohort of PDX models. In addition, mifepristone is also known to block the function of the glucocorticoid receptor, and it remains to be determined if dysfunctional glucocorticoid receptors play any role in resistance formation and tumor recurrence following PARPi treatment (59).
This mechanistic understanding may explain our finding that the combined use of olaparib and mifepristone achieved the most potent tumor inhibition in olaparib-naïve PDXs, while mifepristone monotherapy showed better tumor inhibition effect in PDXs with acquired olaparib resistance. The above results suggest that olaparib can interfere tumor growth inhibition effect of mifepristone if the patient’s tumor has already acquired PARPi-induced resistance. This finding has important clinical implications: Addition of mifepristone to a PARPi could potentiate therapeutic effects of PARPi for patients who have not been previously exposed to PARPi, while mifepristone monotherapy may block tumor growth in patients who have acquired resistance to PARPi.
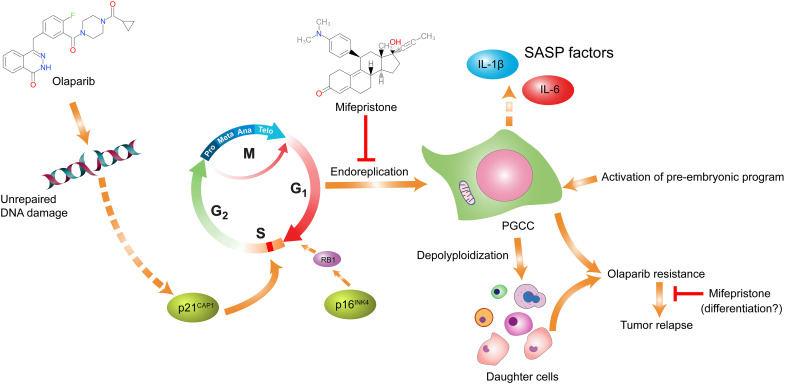
On the basis of the above data, we propose a model to elucidate the mechanism of acquired resistance to olaparib in HGSC (Fig. 8). In tumors that have not been exposed to olaparib, the administration of olaparib induces DNA damage, shuts down mitosis, and leads to activation of the life cycle of PGCCs via de-repressing a pre-embryonic program that is suppressed in cancer growth and progression to generate somatic blastomeres, which leads to whole genomic reorganization at both genomic and epigenetic levels and produces genomically reprogramed and mitotically competent progeny cells with acquired resistance, causing tumor recurrence. Mifepristone promotes apoptosis of cells undergoing endoreplication, thereby blocking the formation of PGCCs, preventing the development of somatic cell “pregnancy,” and suppressing tumor growth. The inclusion of mifepristone with olaparib can synergistically block the endoreplication and survival of PGCCs, increasing the antitumor efficacy compared to either drug alone. In addition, mifepristone can attenuate tumor growth with acquired resistance to PARPi, as these tumors may have acquired high level of embryonic stemness and prone to differentiation toward benign lineages as demonstrated in our previous studies (16, 21). Thus, our data provide proof of principle that targeting the life cycle of PGCCs represents a promising approach to potentiate a therapeutic effect of PARPi and to overcome acquired resistance in ovarian cancer.
Fig. 8. Schematic model on how mifepristone potentiates olaparib-induced therapeutic response and blocks acquired resistance to PARPi in ovarian cancer.
Continuous exposure to olaparib results in unrepaired DNA damage, causing the activation of the aberrant endoreplication cell cycle. Cells that undergo endoreplication develop into senescent PGCCs, which give rise to daughter cells via a variety of modes of depolyploidization. The daughter cells acquired resistance via the life cycle of PGCCs and escaped senescence, eventually leading to tumor recurrence. The combined use of mifepristone and olaparib could block the development of PGCCs, thereby suppressing tumor growth. In tumors with acquired resistance, mifepristone can directly attenuate tumor growth, possibly by inducing differentiation toward benign lineages.
MATERIALS AND METHODS
Study design
We used several preclinical ovarian cancer models, including cell lines, organoids, and PDXs models, to investigate the essential role of PGCCs in PARPi resistance in ovarian cancer. A panel of markers of senescent cells were used to identify PGCCs in the cell lines and organoids. Time-lapse photography was used to monitor how PGCCs evolved. The in vivo efficacy of mifepristone was evaluated in mouse PDX models.
Cell lines and organoids
The human ovarian cancer cell lines Hey, SKOV3, OVCA-432, OVCAR8, OVCAR5, and PEO-1 and the human breast cancer cell line MCF-7 were obtained from our laboratory stocks. The p53 status of the cell lines was as follows: [WT p53: Hey (60) and MCF-7 (61); p53-null: SKOV3 (62); mutant p53: OVCA 432 (63), OVCAR8, OVCAR5 (64), and PEO-1 (65)]. Hey, OVCA-432, and MCF-7 cells were cultured in Eagle’s minimum essential medium (30-2003, American Type Culture Collection) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. SKOV3 cells were grown in modified McCoy’s 5A media (16600082, Gibco) with 10% FBS and 1% penicillin/streptomycin. OVCAR5 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (10-013-CV, Corning) supplemented with 10% FBS and 1% penicillin/streptomycin. OVCAR8 and PEO-1 cells were cultured in RPMI 1640 medium (10-040-CV, Corning) with 10% FBS and 1% penicillin/streptomycin. Organoids (2414, 2445, and 3008) were established from xenografts derived from patients with ovarian HGSC. MD Anderson Characterized Cell Line Core facility confirmed the authenticity of the cell lines and organoids via short tandem repeat sequence analysis.
Human specimens
HGSC tissues were obtained from patients undergoing resection of ovarian tumors at The University of Texas MD Anderson Cancer Center. The Institutional Review Board at MD Anderson approved the use of these samples. In addition, all patients gave informed consent to their tissue’s use for scientific research. The clinicopathological and genomic characterization data of the patients participating in the study are summarized in table S4.
Ovarian cancer PDX tissue processing
Under sterile conditions, xenografts were removed from tumor-bearing mice and transferred onto a 60-mm petri dish. The necrotic tissue was removed with a scalpel and forceps, and well-formed tumor tissue was rinsed with ice-cold phosphate-buffered saline (PBS) at least three times. The tumor tissue was then minced and transferred to a 15-ml conical tube containing 10-ml prewarmed basal medium [Advanced DMEM/F12 with 1× Glutamax, 10 mM Hepes, (Gibco), and Primocin antimicrobial agent (100 μg/ml; Invivogen)] supplemented with dispase II (0.6 to 2.4 U/ml; 17105041, Gibco) and 10 μM Rock inhibitor (Y-27632, Selleckchem). The tube was incubated in a 37°C water bath for 15 min to dissociate the tissue, and the cell slurry was manually agitated every 5 min. The digested cell suspension was sheared using a 5-ml serological pipette and transferred onto a cell strainer (100-μm mesh) placed on top of a 50-ml conical tube. Next, 2% fetal calf serum was added to the strained cell suspension, and the mixture was centrifuged at 300 relative centrifugal force (rcf) for 5 min. The cell pellet was then dissociated with 1 ml of TrypLE Express enzyme (Invitrogen) containing 10 μM Y-27632 at room temperature for 5 min. The dissociated cell clusters were sheared into a single-cell suspension by using a P1000 pipette with a P20 tip without a filter, resuspended with 10 ml of basal medium supplemented with 5% FBS and 10 μM Y-27632, and passed through a 40-μm cell strainer. Human tumor cells were enriched by using a mouse cell depletion kit (130-104-694, Miltenyi Biotec) according to the manufacturer’s instructions.
Organoid culture
The purified human tumor cell pellet was resuspended in a small volume of ice-cold growth factor–reduced Matrigel (354230, Corning). Up to four 50-μl drops of the Matrigel cell suspension were plated into a prewarmed six-well cell suspension culture plate (M9062, Greiner) at a density of about 15,000 to 20,000 cells per drop. The Matrigel was solidified for 15 min at 37°C, and 3 ml of prewarmed organoid culture medium (table S5A) was added to each well. In addition, 10 μM Y-27632 was added upon plating to supplement the culture medium for 3 days. Organoids were cultured at 37°C in an atmosphere of 5% CO2 in a humidified incubator. The medium was changed every 3 to 4 days, and organoids were passaged at a ratio of 1:2 to 1:4 every 2 weeks. For passaging, dispase II (1 mg/ml) was added to the culture medium and incubated at 37°C for 1 hour. The Matrigel was then mechanically disrupted, and organoids were transferred into a 15-ml conical tube and centrifuged at 300 rcf for 5 min. Subsequently, organoids were dissociated by resuspension in 1 ml of TrypLE Express enzyme containing 10 μM Y-27632. Organoids were then incubated at room temperature for 3 min and mechanically sheared into small cell clusters with a P1000 pipette connected to a P20 tip without a filter. Organoid fragments were then washed with 3 ml of basal medium, spun down, and reseeded as described above. Organoids were frozen in 90% FBS and 10% dimethyl sulfoxide (DMSO) to make stocks and stored in liquid nitrogen.
Drug-response assay
Cell viability of cancer cell lines was determined with a Cell Counting Kit-8 (CCK-8; CK04-13, Dojindo). Cells were seeded on 96-well plates (3595, Corning) at a density of 2000 cells per well in quadruplicate and incubated overnight. The cells were then exposed to olaparib (LC Laboratories), niraparib (LC Laboratories), carboplatin (Sigma-Aldrich), or paclitaxel (Sigma-Aldrich) at from five to seven different concentrations for 5 days. After drug exposure, CCK-8 solution was added to each well, and the plates were incubated for 2 hours at 37°C. The absorbance was measured at 450 nm using a microplate reader (BMG Labtech CLARIOstar).
Cell viability of organoids was assayed with a CellTiter-Glo 3D kit (G9683, Promega). Organoids were collected 3 days after passaging and strained with a 100-μm cell strainer (431752, Corning) to remove large organoids. Organoids were then plated in a 4-μl organoid culture media and Matrigel mix (v/v 1:3) in a 96-well white plate (655083, Greiner Bio-One) at a density of 100 to 200 organoids per well in quadruplicate. At 24 hours after plating, organoids were exposed to olaparib at six to seven different concentrations for 5 days. Cell viability was determined according to the manufacturer’s instructions, and results were normalized to vehicle controls.
Data were analyzed using the GraphPad Prism 8.0.2 software, and the values of median inhibitory concentration were calculated by applying nonlinear regression (curve fit) and the equation (inhibitors) versus response--variable slope (four parameters).
Cell viability, cell cycle, apoptosis, and immunofluorescence assays
Target cells were plated in a six-well culture plate at 1 × 105 cells per well. The next day, the cells were treated with olaparib, mifepristone (Sigma-Aldrich), or both for the times indicated in the figure legends. Cell survival was determined by measuring propidium iodide (PI)–stained cells by flow cytometry as described previously (66). The DNA content of tumor cells was detected by PI staining and flow cytometry as described previously (67). For apoptosis assays, cells were incubated with the drugs for the times indicated in the figure legends and evaluated with a fluorescein isothiocyanate (FITC) Annexin V Apoptosis Detection Kit (556547, BD Biosciences). All flow cytometry experiments were conducted on a FACSCalibur flow cytometer (BD Biosciences). Data were analyzed with FlowJo software (Tree Star Inc.).
For immunofluorescence, cells or organoids were seeded onto a coverslip, treated as described in the figure legends, fixed, and stained with primary antibodies (table S6) overnight at 4°C. Samples were then incubated with a secondary antibody labeled with FITC (Invitrogen) at room temperature for 1 hour. After F-actin staining with phalloidin, samples were mounted with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories). Images were acquired with an Axio Imager A2 microscope.
RNA sequencing and data analysis
RNA samples were retrieved from cell lines and organoids that had been treated with olaparib or vehicle (table S2). Total RNA was extracted using RNeasy Mini Kits (Qiagen). The RNA quality was determined by the RNA integrity number (RIN) value with an Agilent 2100 Bioanalyzer. Only specimens with RIN values ≥ 7.0 and a 28S/18S ratio ≥ 1.0 were used in this study. RNA library preparation and transcriptome resequencing (20M reads per sample, DNA sequencing 100 paired ends) were performed by BGI Genomics on an Illumina HiSeq 1000 sequencing system. Differentially expressed genes (DEGs) between sample pairs (PGCC versus control) were obtained by identifying genes with more than twofold changes in transcripts per kilobase of exon model per million mapped reads (TPM), with more highly expressed genes having TPM more than 0.5. DEG was collected only if it showed changes in a scenario shown in fig. S2: changed in PGCCs on day 3 only, changed in PGCCs on day 3/7, changed in PGCCs on day 3/7 and recovery day 7, changed in PGCCs on day 7 and recovery day 7, changed in PGCCs on day 7 only, and changed on recovery day 7 only.
Western blotting
Cells were lysed in radioimmunoprecipitation assay buffer (89900, Thermo Fisher Scientific) supplemented with a protease and phosphatase inhibitor cocktail (78442, Thermo Fisher Scientific) for 30 min on ice, followed by centrifugation at 15,000g for 15 min at 4°C. Proteins were subjected to SDS–polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane. The membranes were blocked with 5% bovine serum albumin and probed with antibodies against GATA4 (1:4000; ab124265, Abcam), p21(1:1000; 2947S, Cell Signaling Technology), or β-actin (1:4000; A1978, Sigma-Aldrich). Detection was performed with a chemiluminescent substrate (32132, Thermo Fisher Scientific) followed by exposure to a ChemiDoc MP Imaging System (Bio-Rad).
Histology and immunohistochemistry
Tissue and organoids were processed for paraffin sectioning, and H&E staining was performed on 5-μm paraffin sections using standard protocols. Briefly, organoids were harvested with dispase II, washed in PBS, fixed in 4% paraformaldehyde at 4°C overnight, and centrifuged at 300 rcf for 5 min. The cell pellet was resuspended in 30 μl of HistoGel specimen processing gel (HG-4000-012, Thermo Fisher Scientific) before processing and embedding. For immunohistochemical staining, 5-μm paraffin sections were deparaffinized in xylene, rehydrated with a graded series of ethanol, and treated with a heat retrieval solution (RV1000M, Biocare Medical) in a digital electric pressure cooker (Decloaking Chamber, Biocare Medical). Slides were incubated with Ki-67 antibody (1:200 dilution; ab16667, Abcam) diluted in Da Vinci Green diluent (PD900M, Biocare Medical) overnight at 4°C. Slides were then incubated with a Polink-2 HRP Plus Rabbit DAB Detection System (D39-18, GBI Labs) according to the manufacturer’s instructions. Immunohistochemical staining of p53, PAX-8, and WT1 was performed by the Research Histology Core Laboratory at MD Anderson. Images were obtained with an Axio Imager A2 microscope (Carl Zeiss).
Plasmids, lentiviral manipulation, and time-lapse cell cycle imaging
For production of lentiviral particles, human embryonic kidney–293T cells were transfected using fuGENE transfection reagent (Promega) with a mixture of the following plasmids: 10 μg of a lentiviral plasmid (table S7), 5 μg of the packaging plasmid psPAX2 (12260, Addgene), and 2.5 μg of the pMD2.G envelope-expressing plasmid (12259, Addgene). The supernatant containing the lentiviral particles was collected 48 and 72 hours after transfection, pooled, filtered (0.45 μm), and concentrated with PEG-it Virus Precipitation Solution (LV810A-1, SBI). Hey cells were subsequently infected with the lentiviral vectors pLenti6-H2B-mCherry and L304-EGFP-Tubulin-WT to visualize the chromosomes and cytoskeleton, respectively (68, 69). In addition, the FUCCI system was used to record cell cycle changes (37). To label cells with FUCCI, Hey cells were infected with the lentiviral vectors mKO2-hCdt1 (30/120) and mAG-hGeminin (1/110).
For time-lapse imaging, fluorescently labeled Hey cells or Hey PGCCs were plated on a glass-bottomed six-well plate (P06G-1.0-20-F, MatTek) at a density of 1 × 103 cells per well. The following day, cells were maintained in the imaging medium (table S5B) and imaged with a Lionheart FX automated microscope (BioTek) in a humidified chamber kept at 37°C with 5% CO2. Cells were imaged every 15 min using a 10× objective lens for up to 1 week. The imaging program includes (1) stabilize incubation temperature to 37°C, (2) laser autofocus, (3) phase-contrast image acquisition, (4) fluorescence image acquisition, (5) move to the next beacon; loop steps 2 to 5. The raw images were imported into Fiji ImageJ software (RRID: SCR_002285) (version 1.52p) to generate image stacks with pseudocolor rendering procedures. The time-lapse videos were further edited in Adobe Premiere Pro (RRID: SCR_021315) and coded in H.264 format.
Enzyme-linked immunosorbent assay
Supernatants were collected from control Hey cells, Hey PGCCs, or Hey PGCC-derived daughter cells. ELISA was performed according to the instructions included with commercial kits (IL-1β: #DLB50; IL-6: #D6050, R&D Systems). The data were normalized to the number of cells in each sample and presented as picograms per milliliter protein per 106 cells.
SA-β-gal staining
Cells were stained for SA-β-gal activity at pH 6.0 as described previously (70). For quantification, cells were counterstained with DAPI to determine the total cell number. Four randomly selected fields (10× objective lenses) were photographed, and the number of cells blue with x-gal was divided by the total cell number.
Xenograft studies
We purchased 6- to 8-week-old athymic nude (nu/nu) female mice from Envigo/Harlan Labs. All mouse experiments were performed according to protocols approved by the Institutional Animal Care and Use Committee at MD Anderson Cancer Center (protocol number: 00001249-RN03). To establish the PDX model, fresh ovarian cancer tumor tissue obtained from debulking surgeries conducted at MD Anderson Cancer Center was sectioned into chunks (4 mm by 4 mm by 4 mm) and engrafted in both flanks of the mice subcutaneously. Once the tumor volume reached approximately 700 to 1000 mm3, tumors were harvested, expanded, and banked for future use.
To develop PDXs of tumors with acquired olaparib resistance, cryopreserved tumor tissue was thawed, washed twice with PBS, and engrafted subcutaneously to one flank of each nude mouse. Tumor length (L) and width (W) were measured with calipers. Tumor volume [V = (L * W2)/2] was calculated as previously described (9). Once tumor volume reached around 200 mm3, mice were randomly assigned to treatment with vehicle (10% 2-hydroxy-propyl-beta-cyclodextrin/PBS) or olaparib [50 mg/ml solubilized in DMSO and diluted to 5 mg/ml with vehicle, 50 mg/kg per day, intraperitoneally (i.p.), 5 days/week] for at least 8 weeks until tumors grew back. The recurring tumors were expanded in different mice and treated according to the same regimen. Acquired resistance to olaparib was defined as a comparable growth rate in olaparib-treated recurring tumors and vehicle-treated recurring tumors.
To investigate the effect of mifepristone on tumor growth, olaparib-naïve tumors or tumors with acquired olaparib resistance were transplanted subcutaneously into nude mice. Once tumor volume reached 150 to 200 mm3, mice were randomized to the following treatment arms: vehicle (10% 2-hydroxy-propyl-beta-cyclodextrin/PBS), olaparib (50 mg/kg per day, i.p., 5 days/week), mifepristone (50 mg/ml solubilized in DMSO and diluted to 5/3 mg/ml with vehicle, 25 mg/kg per day, i.p., 5 days/week), and olaparib/mifepristone (olaparib 50 mg/kg per day + mifepristone 25 mg/kg per day, i.p., 5 days/week). Tumor volume and body weight were monitored twice weekly. Animals were euthanized by inhalation of CO2 followed by cervical dislocation after 6 to 8 weeks of treatment. Tumors were collected and snap-frozen for protein analysis and immunohistochemistry.
Statistical analyses
Statistical methods are described in the respective figure legends. All statistical analyses were performed with GraphPad Prism 8.0.2 software. Data from multiple experiments are presented as mean ± SD. Statistical differences were determined by unpaired two-tailed Welch’s t test or two-way analysis of variance (ANOVA). P < 0.05 was considered statistically significant.
Acknowledgments
We especially thank A. Ninetto, Scientific Editor, Research Medical Library, MD Anderson Cancer Center, for editing the manuscript.
Funding: This work was supported by The University of Texas MD Anderson Ovarian Cancer Moon Shot Program, the National Institutes of Health (SPORE in Ovarian Cancer, grant number P50CA217685), the American Cancer Society, the Ovarian Cancer Research Alliance, the Dunwoody Fund, the NIH/NCI under award number P30CA016672, and used the Research Animal Support Facility, the Research Histology Core Laboratory, and the Cytogenetics and Cell Authentication Core.
Author contributions: X.Z. developed the culture protocol for PDX-derived HGSC organoids and conducted most of the experiments. J.Y. analyzed the RNA data. X.L. helped with flow cytometry and time-lapse experiments, performed the γ-H2AX foci quantification and participated in the design of experiments. N.N. conducted histologic analyses, and Y.L. assisted with PDX models. G.P., S.W., and R.A.H. helped with data analysis. X.Z., A.K.S., and J.L. designed the experiments, analyzed and interpreted the data, and wrote the paper. A.K.S. and J.L. supervised the study and revised the manuscript.
Competing interests: A.K.S. is a consultant for AstraZeneca, GSK/Tesaro, KIYATEC, ImmunoGen, Iylon, Merck, and Onxeo and is a shareholder of BioPath. S.W. is a consultant for AstraZeneca, Caris, Clovis Oncology, Eisai, EQRX, GSK, ImmunoGen, Lilly, Merck, Mereo, Mersana, NGM Bio, Nuvectis, Roche/Genentech, SeaGen, Verastem, Vincerx, and Zentalis. J.L., A.K.S., X.Z., J.Y., X.L., N.N., and Y.L. have a pending U.S. patent application on progestin analog for cancer therapy. The authors declare that they have no other competing interests.
Data and materials availability: RNA sequencing data have been deposited in NCBI’s Gene Expression Omnibus (GEO) and are accessible through accession number GSE229119. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S8
Tables S1 to S7
Legends for movies S1 to S10
Other Supplementary Material for this manuscript includes the following:
Movies S1 to S10
REFERENCES AND NOTES
- 1.Cancer Genomic Atlas Research Network , Integrated genomic analyses of ovarian carcinoma. Nature 474, 609–615 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.H. E. Bryant, N. Schultz, H. D. Thomas, K. M. Parker, D. Flower, E. Lopez, S. Kyle, M. Meuth, N. J. Curtin, T. Helleday, Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature 434, 913–917 (2005). [DOI] [PubMed] [Google Scholar]
- 3.M. P. Dias, S. C. Moser, S. Ganesan, J. Jonkers, Understanding and overcoming resistance to PARP inhibitors in cancer therapy. Nat. Rev. Clin. Oncol. 18, 773–791 (2021). [DOI] [PubMed] [Google Scholar]
- 4.R. L. Coleman, A. M. Oza, D. Lorusso, C. Aghajanian, A. Oaknin, A. Dean, N. Colombo, J. I. Weberpals, A. Clamp, G. Scambia, A. Leary, R. W. Holloway, M. A. Gancedo, P. C. Fong, J. C. Goh, D. M. O'Malley, D. K. Armstrong, J. Garcia-Donas, E. M. Swisher, A. Floquet, G. E. Konecny, I. A. McNeish, C. L. Scott, T. Cameron, L. Maloney, J. Isaacson, S. Goble, C. Grace, T. C. Harding, M. Raponi, J. Sun, K. K. Lin, H. Giordano, J. A. Ledermann; ARIEL3 investigators , Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 390, 1949–1961 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.K. A. Gelmon, M. Tischkowitz, H. Mackay, K. Swenerton, A. Robidoux, K. Tonkin, H. Hirte, D. Huntsman, M. Clemons, B. Gilks, R. Yerushalmi, E. Macpherson, J. Carmichael, A. Oza, Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: A phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 12, 852–861 (2011). [DOI] [PubMed] [Google Scholar]
- 6.E. Pujade-Lauraine, J. A. Ledermann, F. Selle, V. Gebski, R. T. Penson, A. M. Oza, J. Korach, T. Huzarski, A. Poveda, S. Pignata, M. Friedlander, N. Colombo, P. Harter, K. Fujiwara, I. Ray-Coquard, S. Banerjee, J. Liu, E. S. Lowe, R. Bloomfield, P. Pautier; SOLO2/ENGOT-Ov21 investigators , Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): A double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 18, 1274–1284 (2017). [DOI] [PubMed] [Google Scholar]
- 7.M. R. Mirza, B. J. Monk, J. Herrstedt, A. M. Oza, S. Mahner, A. Redondo, M. Fabbro, J. A. Ledermann, D. Lorusso, I. Vergote, N. E. Ben-Baruch, C. Marth, R. Madry, R. D. Christensen, J. S. Berek, A. Dorum, A. V. Tinker, A. du Bois, A. Gonzalez-Martin, P. Follana, B. Benigno, P. Rosenberg, L. Gilbert, B. J. Rimel, J. Buscema, J. P. Balser, S. Agarwal, U. A. Matulonis, Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N. Engl. J. Med. 375, 2154–2164 (2016). [DOI] [PubMed] [Google Scholar]
- 8.C. J. Lord, A. Ashworth, PARP inhibitors: Synthetic lethality in the clinic. Science 355, 1152–1158 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. Rottenberg, J. E. Jaspers, A. Kersbergen, E. van der Burg, A. O. Nygren, S. A. Zander, P. W. Derksen, M. de Bruin, J. Zevenhoven, A. Lau, R. Boulter, A. Cranston, M. J. O'Connor, N. M. Martin, P. Borst, J. Jonkers, High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc. Natl. Acad. Sci. U.S.A. 105, 17079–17084 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.S. J. Pettitt, D. B. Krastev, I. Brandsma, A. Drean, F. Song, R. Aleksandrov, M. I. Harrell, M. Menon, R. Brough, J. Campbell, J. Frankum, M. Ranes, H. N. Pemberton, R. Rafiq, K. Fenwick, A. Swain, S. Guettler, J. M. Lee, E. M. Swisher, S. Stoynov, K. Yusa, A. Ashworth, C. J. Lord, Genome-wide and high-density CRISPR-Cas9 screens identify point mutations in PARP1 causing PARP inhibitor resistance. Nat. Commun. 9, 1849 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Ray Chaudhuri, E. Callen, X. Ding, E. Gogola, A. A. Duarte, J. E. Lee, N. Wong, V. Lafarga, J. A. Calvo, N. J. Panzarino, S. John, A. Day, A. V. Crespo, B. Shen, L. M. Starnes, J. R. de Ruiter, J. A. Daniel, P. A. Konstantinopoulos, D. Cortez, S. B. Cantor, O. Fernandez-Capetillo, K. Ge, J. Jonkers, S. Rottenberg, S. K. Sharan, A. Nussenzweig, Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature 535, 382–387 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.P. A. Konstantinopoulos, R. Ceccaldi, G. I. Shapiro, A. D. D'Andrea, Homologous recombination deficiency: Exploiting the fundamental vulnerability of ovarian cancer. Cancer Discov. 5, 1137–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.I. B. Roninson, Tumor cell senescence in cancer treatment. Cancer Res. 63, 2705–2715 (2003). [PubMed] [Google Scholar]
- 14.R. H. te Poele, A. L. Okorokov, L. Jardine, J. Cummings, S. P. Joel, DNA damage is able to induce senescence in tumor cells in vitro and in vivo. Cancer Res. 62, 1876–1883 (2002). [PubMed] [Google Scholar]
- 15.J. Liu, J. Erenpreisa, E. Sikora, Polyploid giant cancer cells: An emerging new field of cancer biology. Semin. Cancer Biol. 81, 1–4 (2022). [DOI] [PubMed] [Google Scholar]
- 16.S. Zhang, I. Mercado-Uribe, Z. Xing, B. Sun, J. Kuang, J. Liu, Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene 33, 116–128 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.M. Sundaram, D. L. Guernsey, M. M. Rajaraman, R. Rajaraman, Neosis: A novel type of cell division in cancer. Cancer Biol. Ther. 3, 207–218 (2004). [DOI] [PubMed] [Google Scholar]
- 18.J. A. Erenpreisa, M. S. Cragg, B. Fringes, I. Sharakhov, T. M. Illidge, Release of mitotic descendants by giant cells from irradiated Burkitt's lymphoma cell line. Cell Biol. Int. 24, 635–648 (2000). [DOI] [PubMed] [Google Scholar]
- 19.K. Mittal, S. Donthamsetty, R. Kaur, C. Yang, M. V. Gupta, M. D. Reid, D. H. Choi, P. C. G. Rida, R. Aneja, Multinucleated polyploidy drives resistance to Docetaxel chemotherapy in prostate cancer. Br. J. Cancer 116, 1186–1194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.N. Niu, I. Mercado-Uribe, J. Liu, Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene 36, 4887–4900 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.N. Niu, J. Zhang, N. Zhang, I. Mercado-Uribe, F. Tao, Z. Han, S. Pathak, A. S. Multani, J. Kuang, J. Yao, R. C. Bast, A. K. Sood, M. C. Hung, J. Liu, Linking genomic reorganization to tumor initiation via the giant cell cycle. Oncogenesis 5, e281 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.X. Li, Y. Zhong, X. Zhang, A. K. Sood, J. Liu, Spatiotemporal view of malignant histogenesis and macroevolution via formation of polyploid giant cancer cells. Oncogene 42, 665–678 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D. Diaz-Carballo, S. Saka, J. Klein, T. Rennkamp, A. H. Acikelli, S. Malak, H. Jastrow, G. Wennemuth, C. Tempfer, I. Schmitz, A. Tannapfel, D. Strumberg, A distinct oncogenerative multinucleated cancer cell serves as a source of stemness and tumor heterogeneity. Cancer Res. 78, 2318–2331 (2018). [DOI] [PubMed] [Google Scholar]
- 24.S. Zhang, I. Mercado-Uribe, J. Liu, Tumor stroma and differentiated cancer cells can be originated directly from polyploid giant cancer cells induced by paclitaxel. Int. J. Cancer 134, 508–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.S. Zhang, I. Mercado-Uribe, A. Sood, R. C. Bast, J. Liu, Coevolution of neoplastic epithelial cells and multilineage stroma via polyploid giant cells during immortalization and transformation of mullerian epithelial cells. Genes Cancer 7, 60–72 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.K. C. Lin, G. Torga, Y. Sun, R. Axelrod, K. J. Pienta, J. C. Sturm, R. H. Austin, The role of heterogeneous environment and docetaxel gradient in the emergence of polyploid, mesenchymal and resistant prostate cancer cells. Clin. Exp. Metastasis 36, 97–108 (2019). [DOI] [PubMed] [Google Scholar]
- 27.J. Chen, N. Niu, J. Zhang, L. Qi, W. Shen, K. V. Donkena, Z. Feng, J. Liu, Polyploid giant cancer cells (PGCCs): The evil roots of cancer. Curr. Cancer Drug Targets 19, 360–367 (2019). [DOI] [PubMed] [Google Scholar]
- 28.J. Liu, The "life code": A theory that unifies the human life cycle and the origin of human tumors. Semin. Cancer Biol. 60, 380–397 (2020). [DOI] [PubMed] [Google Scholar]
- 29.L. M. Lopez-Sanchez, C. Jimenez, A. Valverde, V. Hernandez, J. Penarando, A. Martinez, C. Lopez-Pedrera, J. R. Munoz-Castaneda, J. R. De la Haba-Rodriguez, E. Aranda, A. Rodriguez-Ariza, CoCl2, a mimic of hypoxia, induces formation of polyploid giant cells with stem characteristics in colon cancer. PLOS ONE 9, e99143 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.I. Sirois, A. Aguilar-Mahecha, J. Lafleur, E. Fowler, V. Vu, M. Scriver, M. Buchanan, C. Chabot, A. Ramanathan, B. Balachandran, S. Legare, E. Przybytkowski, C. Lan, U. Krzemien, L. Cavallone, O. Aleynikova, C. Ferrario, M. C. Guilbert, N. Benlimame, A. Saad, M. Alaoui-Jamali, H. U. Saragovi, S. Josephy, C. O'Flanagan, S. D. Hursting, V. R. Richard, R. P. Zahedi, C. H. Borchers, E. Bareke, S. Nabavi, P. Tonellato, J. A. Roy, A. Robidoux, E. A. Marcus, C. Mihalcioiu, J. Majewski, M. Basik, A unique morphological phenotype in chemoresistant triple-negative breast cancer reveals metabolic reprogramming and PLIN4 expression as a molecular vulnerability. Mol. Cancer Res. 17, 2492–2507 (2019). [DOI] [PubMed] [Google Scholar]
- 31.S. Meierjohann, Effect of stress-induced polyploidy on melanoma reprogramming and therapy resistance. Semin. Cancer Biol. 81, 232–240 (2022). [DOI] [PubMed] [Google Scholar]
- 32.F. Ricci, F. Bizzaro, M. Cesca, F. Guffanti, M. Ganzinelli, A. Decio, C. Ghilardi, P. Perego, R. Fruscio, A. Buda, R. Milani, P. Ostano, G. Chiorino, M. R. Bani, G. Damia, R. Giavazzi, Patient-derived ovarian tumor xenografts recapitulate human clinicopathology and genetic alterations. Cancer Res. 74, 6980–6990 (2014). [DOI] [PubMed] [Google Scholar]
- 33.N. E. Sharpless, C. J. Sherr, Forging a signature of in vivo senescence. Nat. Rev. Cancer 15, 397–408 (2015). [DOI] [PubMed] [Google Scholar]
- 34.C. D. Wiley, J. M. Flynn, C. Morrissey, R. Lebofsky, J. Shuga, X. Dong, M. A. Unger, J. Vijg, S. Melov, J. Campisi, Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence. Aging Cell 16, 1043–1050 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.B. Y. Lee, J. A. Han, J. S. Im, A. Morrone, K. Johung, E. C. Goodwin, W. J. Kleijer, D. DiMaio, E. S. Hwang, Senescence-associated β-galactosidase is lysosomal β-galactosidase. Aging Cell 5, 187–195 (2006). [DOI] [PubMed] [Google Scholar]
- 36.H. Chen, P. D. Ruiz, W. M. McKimpson, L. Novikov, R. N. Kitsis, M. J. Gamble, MacroH2A1 and ATM play opposing roles in paracrine senescence and the senescence-associated secretory phenotype. Mol. Cell 59, 719–731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.A. Sakaue-Sawano, H. Kurokawa, T. Morimura, A. Hanyu, H. Hama, H. Osawa, S. Kashiwagi, K. Fukami, T. Miyata, H. Miyoshi, T. Imamura, M. Ogawa, H. Masai, A. Miyawaki, Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 132, 487–498 (2008). [DOI] [PubMed] [Google Scholar]
- 38.B. A. Edgar, T. L. Orr-Weaver, Endoreplication cell cycles. Cell 105, 297–306 (2001). [DOI] [PubMed] [Google Scholar]
- 39.J. Heng, H. H. Heng, Genome chaos: Creating new genomic information essential for cancer macroevolution. Semin. Cancer Biol. 81, 160–175 (2022). [DOI] [PubMed] [Google Scholar]
- 40.J. Liu, The dualistic origin of human tumors. Semin. Cancer Biol. 53, 1–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.J. Liu, Giant cells: Linking McClintock's heredity to early embryogenesis and tumor origin throughout millennia of evolution on Earth. Semin. Cancer Biol. 81, 176–192 (2022). [DOI] [PubMed] [Google Scholar]
- 42.J. Liu, N. Niu, X. Li, X. Zhang, A. K. Sood, The life cycle of polyploid giant cancer cells and dormancy in cancer: Opportunities for novel therapeutic interventions. Semin. Cancer Biol. 81, 132–144 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.I. M. Spitz, Mifepristone: Where do we come from and where are we going? Clinical development over a quarter of a century. Contraception 82, 442–452 (2010). [DOI] [PubMed] [Google Scholar]
- 44.C. D. Gamarra-Luques, A. A. Goyeneche, M. B. Hapon, C. M. Telleria, Mifepristone prevents repopulation of ovarian cancer cells escaping cisplatin-paclitaxel therapy. BMC Cancer 12, 200 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.A. A. Goyeneche, R. W. Caron, C. M. Telleria, Mifepristone inhibits ovarian cancer cell growth in vitro and in vivo. Clin. Cancer Res. 13, 3370–3379 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.N. Sachs, J. de Ligt, O. Kopper, E. Gogola, G. Bounova, F. Weeber, A. V. Balgobind, K. Wind, A. Gracanin, H. Begthel, J. Korving, R. van Boxtel, A. A. Duarte, D. Lelieveld, A. van Hoeck, R. F. Ernst, F. Blokzijl, I. J. Nijman, M. Hoogstraat, M. van de Ven, D. A. Egan, V. Zinzalla, J. Moll, S. F. Boj, E. E. Voest, L. Wessels, P. J. van Diest, S. Rottenberg, R. G. J. Vries, E. Cuppen, H. Clevers, A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386.e10 (2018). [DOI] [PubMed] [Google Scholar]
- 47.O. Kopper, C. J. de Witte, K. Lohmussaar, J. E. Valle-Inclan, N. Hami, L. Kester, A. V. Balgobind, J. Korving, N. Proost, H. Begthel, L. M. van Wijk, S. A. Revilla, R. Theeuwsen, M. van de Ven, M. J. van Roosmalen, B. Ponsioen, V. W. H. Ho, B. G. Neel, T. Bosse, K. N. Gaarenstroom, H. Vrieling, M. P. G. Vreeswijk, P. J. van Diest, P. O. Witteveen, T. Jonges, J. L. Bos, A. van Oudenaarden, R. P. Zweemer, H. J. G. Snippert, W. P. Kloosterman, H. Clevers, An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity. Nat. Med. 25, 838–849 (2019). [DOI] [PubMed] [Google Scholar]
- 48.M. Kobel, S. E. Kalloger, N. Boyd, S. McKinney, E. Mehl, C. Palmer, S. Leung, N. J. Bowen, D. N. Ionescu, A. Rajput, L. M. Prentice, D. Miller, J. Santos, K. Swenerton, C. B. Gilks, D. Huntsman, Ovarian carcinoma subtypes are different diseases: Implications for biomarker studies. PLOS Med. 5, e232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.M. Kobel, A. M. Piskorz, S. Lee, S. Lui, C. LePage, F. Marass, N. Rosenfeld, A. M. Mes Masson, J. D. Brenton, Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J. Pathol. Clin. Res. 2, 247–258 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.E. Gogola, S. Rottenberg, J. Jonkers, Resistance to PARP inhibitors: Lessons from preclinical models of BRCA-associated cancer. Annu. Rev. Cancer Biol. 3, 235–254 (2019). [Google Scholar]
- 51.A. Farkkila, A. Rodriguez, J. Oikkonen, D. C. Gulhan, H. Nguyen, J. Dominguez, S. Ramos, C. E. Mills, F. Perez-Villatoro, J. B. Lazaro, J. Zhou, C. S. Clairmont, L. A. Moreau, P. J. Park, P. K. Sorger, S. Hautaniemi, S. Frias, A. D. D'Andrea, Heterogeneity and clonal evolution of acquired PARP inhibitor resistance in TP53- and BRCA1-deficient cells. Cancer Res. 81, 2774–2787 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Y. Stewenius, L. Gorunova, T. Jonson, N. Larsson, M. Hoglund, N. Mandahl, F. Mertens, F. Mitelman, D. Gisselsson, Structural and numerical chromosome changes in colon cancer develop through telomere-mediated anaphase bridges, not through mitotic multipolarity. Proc. Natl. Acad. Sci. U.S.A. 102, 5541–5546 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.H. H. Heng, Genomic chaos: Rethinking genetics, evolution, and molecular medicine. Academic Press; (2019).
- 54.S. F. Bakhoum, O. V. Danilova, P. Kaur, N. B. Levy, D. A. Compton, Chromosomal instability substantiates poor prognosis in patients with diffuse large B-cell lymphoma. Clin. Cancer Res. 17, 7704–7711 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.A. J. X. Lee, D. Endesfelder, A. J. Rowan, A. Walther, N. J. Birkbak, P. A. Futreal, J. Downward, Z. Szallasi, I. P. M. Tomlinson, M. Howell, M. Kschischo, C. Swanton, Chromosomal instability confers intrinsic multidrug resistance. Cancer Res. 71, 1858–1870 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.N. Niu, J. Yao, R. C. Bast, A. K. Sood, J. Liu, IL-6 promotes drug resistance through formation of polyploid giant cancer cells and stromal fibroblast reprogramming. Oncogenesis 10, 65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.S. J. Hill, B. Decker, E. A. Roberts, N. S. Horowitz, M. G. Muto, M. J. Worley Jr., C. M. Feltmate, M. R. Nucci, E. M. Swisher, H. Nguyen, C. Yang, R. Morizane, B. S. Kochupurakkal, K. T. Do, P. A. Konstantinopoulos, J. F. Liu, J. V. Bonventre, U. A. Matulonis, G. I. Shapiro, R. S. Berkowitz, C. P. Crum, A. D. D'Andrea, Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov. 8, 1404–1421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.S. D. Horne, S. K. Chowdhury, H. H. Heng, Stress, genomic adaptation, and the evolutionary trade-off. Front. Genet. 5, 92 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.F. Cadepond, A. Ulmann, E. E. Baulieu, RU486 (mifepristone): Mechanisms of action and clinical uses. Annu. Rev. Med. 48, 129–156 (1997). [DOI] [PubMed] [Google Scholar]
- 60.J. L. Watson, A. Greenshields, R. Hill, A. Hilchie, P. W. Lee, C. A. Giacomantonio, D. W. Hoskin, Curcumin-induced apoptosis in ovarian carcinoma cells is p53-independent and involves p38 mitogen-activated protein kinase activation and downregulation of Bcl-2 and survivin expression and Akt signaling. Mol. Carcinog. 49, 13–24 (2010). [DOI] [PubMed] [Google Scholar]
- 61.M. A. Troester, J. I. Herschkowitz, D. S. Oh, X. He, K. A. Hoadley, C. S. Barbier, C. M. Perou, Gene expression patterns associated with p53 status in breast cancer. BMC Cancer 6, 276 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Y. Yaginuma, H. Westphal, Abnormal structure and expression of the p53 gene in human ovarian carcinoma cell lines. Cancer Res. 52, 4196–4199 (1992). [PubMed] [Google Scholar]
- 63.C. S. Muenyi, A. R. Pinhas, T. W. Fan, G. N. Brock, C. W. Helm, J. C. States, Sodium arsenite +/− hyperthermia sensitizes p53-expressing human ovarian cancer cells to cisplatin by modulating platinum-DNA damage responses. Toxicol. Sci. 127, 139–149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.P. De Feudis, D. Debernardis, P. Beccaglia, M. Valenti, E. Graniela Sire, D. Arzani, S. Stanzione, S. Parodi, M. D'Incalci, P. Russo, M. Broggini, DDP-induced cytotoxicity is not influenced by p53 in nine human ovarian cancer cell lines with different p53 status. Br. J. Cancer 76, 474–479 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.S. L. Cooke, C. K. Ng, N. Melnyk, M. J. Garcia, T. Hardcastle, J. Temple, S. Langdon, D. Huntsman, J. D. Brenton, Genomic analysis of genetic heterogeneity and evolution in high-grade serous ovarian carcinoma. Oncogene 29, 4905–4913 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.L. C. Crowley, A. P. Scott, B. J. Marfell, J. A. Boughaba, G. Chojnowski, N. J. Waterhouse, Measuring cell death by propidium iodide uptake and flow cytometry. Cold Spring Harb. Protoc. 2016, pdb.prot087163 (2016). [DOI] [PubMed] [Google Scholar]
- 67.L. C. Crowley, G. Chojnowski, N. J. Waterhouse, Measuring the DNA content of cells in apoptosis and at different cell-cycle stages by propidium iodide staining and flow cytometry. Cold Spring Harb. Protoc. 2016, pdb.prot087247 (2016). [DOI] [PubMed] [Google Scholar]
- 68.H. Pemble, P. Kumar, J. van Haren, T. Wittmann, GSK3-mediated CLASP2 phosphorylation modulates kinetochore dynamics. J. Cell Sci. 130, 1404–1412 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.W. Yang, X. Guo, S. Thein, F. Xu, S. Sugii, P. W. Baas, G. K. Radda, W. Han, Regulation of adipogenesis by cytoskeleton remodelling is facilitated by acetyltransferase MEC-17-dependent acetylation of α-tubulin. Biochem. J. 449, 605–612 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.G. P. Dimri, X. H. Lee, G. Basile, M. Acosta, C. Scott, C. Roskelley, E. E. Medrano, M. Linskens, I. Rubelj, O. Pereira-Smith, A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S8
Tables S1 to S7
Legends for movies S1 to S10
Movies S1 to S10