Abstract
Affinity-based biosensing can enable point-of-care diagnostics and continuous health monitoring, which commonly follows bottom-up approaches and is inherently constrained by bioprobes’ intrinsic properties, batch-to-batch consistency, and stability in biofluids. We present a biomimetic top-down platform to circumvent such difficulties by combining a “dual-monolayer” biorecognition construct with graphene-based field-effect-transistor arrays. The construct adopts redesigned water-soluble membrane receptors as specific sensing units, positioned by two-dimensional crystalline S-layer proteins as dense antifouling linkers guiding their orientations. Hundreds of transistors provide statistical significance from transduced signals. System feasibility was demonstrated with rSbpA-ZZ/CXCR4QTY-Fc combination. Nature-like specific interactions were achieved toward CXCL12 ligand and HIV coat glycoprotein in physiologically relevant concentrations, without notable sensitivity loss in 100% human serum. The construct is regeneratable by acidic buffer, allowing device reuse and functional tuning. The modular and generalizable architecture behaves similarly to natural systems but gives electrical outputs, which enables fabrication of multiplex sensors with tailored receptor panels for designated diagnostic purposes.
A modular membrane protein design based biomimetic sensing architecture behaves as biological systems with electrical outputs.
INTRODUCTION
Affinity-based electrical biosensors provide sensitive and specific detection of analytes in complex biofluids at low cost with real-time readouts, which can revolutionize portable point-of-care disease diagnostics, continuous health monitoring, and precision medicine (1, 2). Almost all current systems adopt the bottom-up approach by identifying analytes of interest and selectively detecting them with antibodies or aptamers, which is intrinsically constrained by the availability and properties of probes (3–5). Although both types of probes exhibit high sensitivity and specificity, antibodies require immunogenicity, are expensive to produce, and lack batch-to-batch consistency, while aptamers are sensitive to nuclease degradation and unstable in biofluids such as human serum. Their detection range can also fall out of clinical relevance (6). An alternative type of bioprobes without these issues is natural receptor proteins, which are cells’ input/output machineries and mostly reside on plasma membranes, including G protein–coupled receptors (GPCRs) that comprise the largest protein family in human proteome (7). They specifically recognize diverse stimuli ranging from ions and odorants to biomolecules including peptides, amino acids, and many pathogens. However, membrane receptors suffer from poor water solubility that requires arduous and nontransferable efforts for synthesis and stabilization in aqueous environments, while additives like detergents may further decrease the signal-to-noise ratio during electrical sensing (8, 9). Despite a few reports on receptor integrations in biosensing (10–13), the lack of generality hinders their widespread use beyond lab benches.
Here, we take a top-down approach and present a modular biomimetic architecture to overcome the bottleneck in previous biosensing systems. Instead of searching for probes to detect target analytes, we identify critical receptors in human physiological processes as well as redesign and integrate them with electronic devices to generate electrical outputs with biophysical significance. The architecture combines a protein design–based “dual-monolayer” construct consisting of (i) water-soluble membrane receptors as specific sensing units and (ii) two-dimensional (2D) crystalline S-layer protein intermediates forcing receptor orientation, with (iii) graphene-based field-effect transistor (GFET) array for electrical signal transduction (14). The challenges in the membrane receptor optimization are circumvented via our design tool named QTY code to produce their water-soluble functional equivalents in low-cost Escherichia coli system (Fig. 1A and fig. S1). Redesigned receptors exhibit native ligand selectivity and high thermostability, as have been demonstrated in our previous works (fig. S2) (15, 16). S-layer proteins, which form 2D crystalline arrays by self-assembly (17), are adopted to connect receptors and signal transducing devices (17–21). They outperform common linking approaches such as π stacking, His-tag binding, and N-hydroxysuccinimide (NHS)–1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC) conjugation by (i) providing high-density anchoring sites at ~1012 U/cm2 and forcing receptors to expose their functional domains (fig. S3), (ii) not requiring invasive modifications on substrates that negatively affect electrical properties, and (iii) offering antifouling against nonspecific bindings to the signal transducing layer underneath.
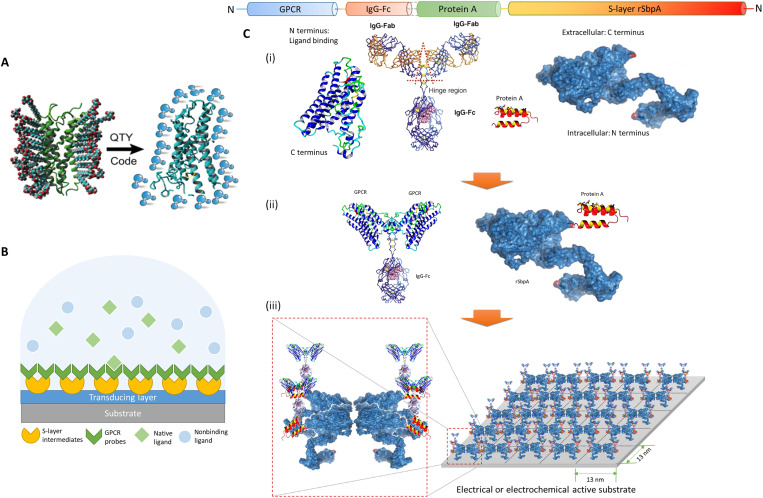
Fig. 1. Schematic illustration of the architecture.
(A) The QTY code transforms hydrophobic membrane receptors to water-soluble functional equivalents that do not require detergents to stabilize in aqueous solutions. (B) S-layer proteins and solubilized receptors are sequentially coated onto designated sensing devices. Target analytes are specifically recognized in aqueous media, while the detection signal is transduced for electrical responses. GPCR, G protein–coupled receptor. (C) Sequence layout of the dual-monolayer construct. (i) The four elements of the construct: redesigned water-soluble membrane receptor, heavy chain Fc region of immunoglobulin G (IgG) protein, Fc-binding fragment of protein A, and two-dimensional (2D) crystal-forming S-layer protein. (ii) Solubilized receptors are fused to IgG-Fc to serve as the biospecific probes, while S-layer proteins are fused with protein A fragments to connect probes to the device. (iii) Sequentially, S-layer/protein A fusion proteins reproduce its native nanoscale 2D crystalline lattice on the substrate, after which the receptor probes with Fc fusions were applied to bind the intermediate layer in high affinity and capacity. The specific construct presented here (rSbpA-ZZ/CXCR4QTY-Fc) has p4 symmetry, a lattice parameter of ~13 nm2, and a receptor density in the order of ~1012 U/cm2.
In mammalian sensory systems, native receptors’ redundancy in both their types and numbers plays an important role in ensuring reliable behavioral sensitivity (22–24). We fabricate an integrated GFET array with more than 200 functionalized sensors to mimic such redundancy, providing an ensemble of sensing units with certain degree of performance variations, similar to a nature sensory system. Signals are transduced in the electrical domain from large numbers of sensory inputs (Fig. 1B), which encloses statistically significant and machine learning computable readouts from rapid measurements with a custom-built portable interface (25). The built-in statistical significance provides a realistic computational model of a nature system for high sensitivity and exquisite discriminatory capacity to ligands (26). The FET device geometry has been widely adopted in biosensors to provide high sensitivity detections toward analytes of interests (27). Guo et al. (28) showed single-molecule detection of coronavirus disease 2019 antigens by functionalizing devices with commercial nanobodies, whereas another antibodies-based FET biosensor was used for bladder cancer diagnostics and classification (29). Graphene is chosen as the transducing material in our setup due to its large surface-to-volume ratio, high carrier mobility, chemical stability, and potential scalability (30, 31). The modular design allows freedom for specificity tuning and multiplexing while being adaptive to alternative targets with simple surface receptor exchanges. With prospective integration of a wide variety of human-sourced receptors trained by deep learning–based analytical algorithms, this highly scalable top-down approach may provide a biomimetic sensing platform that behaves like a living organism, with similar recognition powers, and gives real-time electrical readouts.
RESULTS
Fabrication of CXCR4QTY-Fc/rSbpA-ZZ dual-monolayer construct
To demonstrate the feasibility and biomimicry of the architecture, CXCR4QTY receptor was selected as an example probe, which is an inflammatory chemokine receptor within the GPCR family. Together with its native ligand CXCL12, the axis plays major roles in multiple sclerosis (32); in the growth, invasion, and metastasis of 23 types of cancers; (33); and in germinal center responses that are critical in humoral immunity (34). Monitoring the CXCL12 level in biofluids from specific organs can provide valuable information about human immune responses in inflammatory diseases and can predict metastasis and prognosis in patients with cancer. CXCR4 is also one of the co-receptors for HIV entry into T cells, which enables HIV diagnostics and monitoring for clinical managements (35). S-layer protein SbpA from Geobacillus sphaericus CCM2177 was selected as linkers, which can form tetramers to recrystallize on solid and flexible surfaces, with p4 symmetry, a lattice parameter of ~13 nm, and a height of ~7 nm (36).
A direct gene fusion of CXCR4 to SbpA was not possible, as the N terminus of SbpA is needed to recrystallize on solid supports, while the N terminus of CXCR4 is involved in the ligand binding and required for selective biorecognition. Thus, we installed the components onto functional devices by a dual-monolayer architecture, the layout of which is shown in Fig. 1C. The four elements in the construct are (i) CXCR4QTY receptors, the fragment crystallizable (Fc) region of immunoglobulin G (IgG) proteins, Fc-binding fragments from protein A, and SbpA S-layer proteins, where (ii) the receptor is fused to the Fc region of IgG at the C terminus (denoted as CXCR4QTY-Fc) and SbpA is fused with two Fc-binding fragments of protein A also at the C terminus as described in a previous report (denoted as rSbpA-ZZ; Supplementary Materials) (37). The two fusion protein components are then sequentially coated, where (iii) rSbpA-ZZ first reproduces its 2D crystalline lattice in nanoscale on the electronic substrate, followed by the application of CXCR4QTY-Fc that bound the intermediate monolayer in high affinity and capacity. The sequence construct of CXCR4QTY-Fc is shown in table S1. Rabbit IgG-Fc was selected for its strong binding with protein A, with hinge region retained and linkers added to enable enough flexibility against steric collisions between two components.
Biological behavior and solution test
Several biological and spectroscopic characterizations were conducted to confirm the biological functionality of each component. We first examined whether the Fc fusion will interfere with CXCR4QTY. On the basis of the AlphaFold2 prediction (fig. S4A), the fusion protein exhibited a distinct two-domain structure with high confidence. Superimpositions of each segment with reported crystal structures from Protein Data Bank (CXCR4: 4RWS and rabbit IgG-Fc: 2VUO) revealed closely matched conformations with RMSDs (root mean square deviations between equivalent atomic positions) between 1 and 2 Å (fig. S4, B to D). Respective RMSDs increased by 0.3 to 0.4 Å when superimpositions were conducted on the fusion protein model, indicating additional spatial constraints on subunits when they were connected.
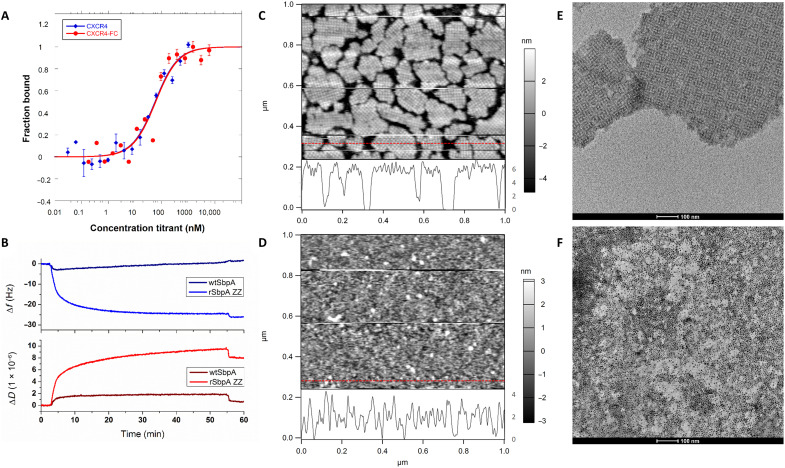
CXCR4QTY-Fc was then expressed in E. coli and purified with yield between 5 and 10 mg/liter in Luria-Bertani (LB) media (fig. S5A). Far ultraviolet (UV) circular dichroism (CD) spectra showed typical α-helical structures with signature valleys at 208 and 222 nm. The helical content is contributed by CXCR4QTY and is inversely proportional to the molecular weight (MW) of each protein, while reduced propensity of the α-helix was observed for CXCR4QTY-Fc (fig. S5B). As shown in Fig. 2A, the ligand binding of CXCR4QTY-Fc toward CXCL12 was determined by microscale electrophoresis (MST), where overlapping profiles were observed compared to that from CXCR4QTY, suggesting no discernable impact from Fc fusion on the receptor function.
Fig. 2. Biological verification of the construct.
(A) Comparison of microscale electrophoresis (MST) binding profiles between CXCR4QTY and CXCR4QTY-Fc toward their native ligand CXCL12. (B) CXCR4QTY-Fc is bound to rSbpA-ZZ at basic buffer condition (pH = 9) and can be reversibly dissociated from the complex at acidic buffer condition (pH = 3). No association is observed between CXCR4QTY-Fc and wild-type (wt) SbpA. (C) Atomic force microscopy (AFM) topography of rSbpA-ZZ–coated silicon wafer. Height profile extracted from the red line is shown at the bottom of the image. (D) AFM topography of CXCR4QTY-Fc/rSbpA-ZZ–coated silicon wafer. Height profile extracted from the red line is shown at the bottom of the image. (E) Transmission electron microscopy (TEM) graph of the standalone rSbpA-ZZ 2D crystalline sheets in solution (scale bar, 100 nm). (F) TEM graph of CXCR4QTY-Fc–coated standalone rSbpA-ZZ 2D crystalline sheets in solution and visualized by His-binding Au nanoparticles (scale bar, 100 nm).
The reversible construction of dual-monolayers is confirmed using quartz crystal microbalance (QCM). Specific association was observed only between CXCR4QTY-Fc and rSbpA-ZZ but not with wild-type (wt) SbpA (Fig. 2B). Applying acidic (pH = 3) buffer to the construct caused receptor dissociation. Atomic force microscopy (AFM) inspection in deionized (DI) water confirmed the sequential formation of each monolayer on a silicon wafer. While rSbpA-ZZ formed well-ordered waffle-like patches, subsequent CXCR4QTY-Fc coating did not exhibit 2D crystalline lattice, since there are not predefined lateral interactions between helical regions of adjacent receptors (Fig. 2, C and D). Height profiles corresponded well with monolayers of either rSbpA-ZZ 2D lattice or folded CXCR4QTY-Fc (~3 to 4 nm). The construct is applicable in other substrates (fig. S6A) and can be expanded with alternative receptors (CCR10QTY-Fc and CXCR5QTY-Fc), whereas similar profiles also dependent on receptor properties were obtained (fig. S6, B and C) (19). Trace-retrace scanning profiles revealed a nonrigid body association between CXCR4QTY-Fc and rSbpA-ZZ, which can be moved by AFM probes with the slightest touch using real-time setpoint adjustments at low scanning speed (fig. S7). It is possible to further cross-link the biomolecular coatings with dimethyl pimelimidate for enhanced rigidity and durability at the sacrifice of device reusability, which was not conducted in the current study (37, 38). The distribution of CXCR4QTY-Fc was visualized by transmission electron microscopy (TEM) in solution on stand-alone rSbpA-ZZ sheets and His-binding gold nanoparticles (Fig. 2, E and F). High-density coverage was observed without significant clustering, indicating near-homogenous receptor monolayer without apparent aggregates.
The initial electrochemical verification was conducted in solution. An open-circuit voltage (OCV) setup was used to evaluate the redox potential changes during receptor-ligand binding (fig. S8). CCL5, the native ligand for chemokine receptor CCR5 that is similar in MW and biophysical properties to CXCL12 (table S2) but does not interact with CXCR4, was selected as the negative control to interrogate the system. The binding assays showed potential changes in the order of tens of millivolts (mV), which was not observed in the nonbinding set, demonstrating biological specificity. The fusion protein exhibited a similar profile to the receptor alone but smaller in magnitude of voltage change, probably due to their higher MW and relatively less conformational changes in binding. An alternative combination of CCR10-CCL28 was also test as shown in fig. S8D.
Ligand detection in buffer
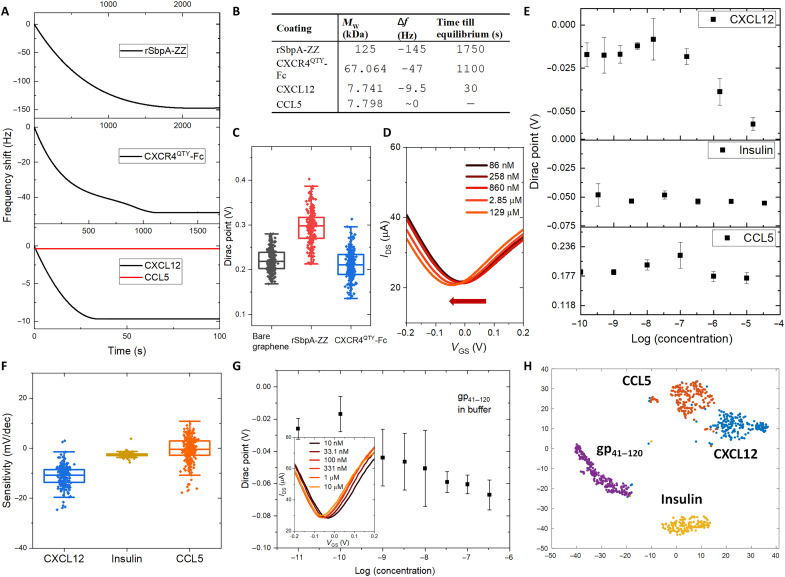
The dual-monolayer construct was first tested on a biologically derived QCM impedance assay with gold sensor chips. Analyte binding with the chip would induce a frequency shift (Δf) in the readout. The Δf was recorded as a function of time for sequential coatings of rSbpA-ZZ and CXCR4QTY-Fc and detections of CXCL12 and CCL5 in the nanomolar concentration (Fig. 3, A and B). The Δf and time till equilibrium are correlated to the MW of bound proteins in each step as summarized in Fig. 3B. No discernible Δf was observed when the control sample CCL5 was added, indicating detection specificity. Notably, the Δf profile equilibrated within ~30 s during CXCL12 detection by a fully constructed sensing chip, which agreed with the ligand-binding kinetics determined by surface plasmon resonance assays in previous reports, and enabled ultrafast detection of the target analyte in real time (39).
Fig. 3. Ligand detection in buffer.
(A) Frequency shift profile in each step of protein coatings and ligand detections as a function of time. (B) Table summarizes the frequency shift (Δf), time till equilibrium, and molecular weight (MW) of bound proteins for sequential coatings of rSbpA-ZZ, CXCR4QTY-Fc, and detections of CXCL12 and CCL5 in the nanomolar concentration. (C) Average Dirac points (DPs) of graphene-based field-effect transistor (GFET) arrays for bare graphene, rSbpA-ZZ coating, and CXCR4QTY-Fc coating. (D) The left shift in averaged drain current with respect to gate-to-source voltage(IDS-VGS) curves from the fully constructed array with respect to a gradient concentration of CXCL12. (E) The averaged DP shifts from the fully constructed array with respect to a gradient concentration of CXCL12, CCL5, and insulin. Error bar indicates the standard deviation of three repeat measurements. (F) Distribution of sensor sensitivities toward the native ligand and the negative controls. (G) The plot of averaged concentration-dependent response and IDS-VGS curve (insert) from the fully constructed device array toward recombinant HIV coat glycoprotein gp41–120. Error bar indicates the standard deviation of three repeat measurements. (H) Well-defined nonoverlapping clusters via t-distributed stochastic neighbor embedding (t-SNE) dimension reduction processing of data from experimental sets in buffer.
The GFET array was then fabricated to fulfill the overall architecture providing an electrical assay (25, 30). The device geometry is shown in fig. S9A, where more than 200 sensing units were integrated to provide statistical significance for the readouts in sensor responses. The rSbpA-ZZ and CXCR4QTY-Fc were sequentially coated onto the graphene channel. A low-profile Ag/AgCl reference electrode was used as a global top gate for the sensor array. The drain current with respect to gate-source voltage [referred as “IDS-VGS” (current-voltage relation) characteristics] of the chip before and after each stage of functionalization are shown in fig. S9B. Device-to-device variation was present in the commonly observed range for advanced material–based devices due to material synthesis, device fabrication, and biomolecule deposition–induced uncertainties (31, 40). By averaging data from all devices on the highly integrated sensing chip, we were able to reduce the noise and capture the universal trend through statistical analysis and machine learning algorithms to obtain biologically relevant results. The IDS-VGS characteristics shifted during coating, which were quantified by tracking the location of minimum conduction in graphene, i.e., Dirac point (DP; fig. S10). Hysteresis of the dual-monolayer–coated graphene sensors is mitigated by applying a slow voltage sweeping rate of 20 mV/s. We obtained stable and reproducible measurements and extrapolation of DP values as shown in fig. S11. The sweeping window was kept constant during gradient measurements against individual analyte in each testing environment to eliminate its potential impact on DP shift. As shown in Fig. 3C, the average DP shifted (cumulatively determined from fig. S9C) in opposite directions when rSbpA-ZZ and CXCR4QTY-Fc were coated. The functionalized array was tested against different concentrations of CXCL12 in deionized (DI) water spanning six orders of magnitude. A left shift of the average IDS-VGS characteristics was observed as shown in Fig. 3D, indicating an n-doping effect in binding. By fitting with Langmuir adsorption isotherm, we extrapolated the surface dissociation constant (Kd) from the gradient concentration responses in hundreds of sensors (fig. S12A). The calculated Kd is 20.1 ± 9.1 nM, which is similar to the Kd obtained from biological assays. This result suggests that the electrical biomimetic architecture reacts similarly to the native ligand as in biological systems. The Langmuir adsorption isotherm was chosen over the law of mass action-based model, as the ligand detection on devices can be modeled as a surface adsorption process, different from in-solution reactions with predefined molecular concentrations of binding partners (15, 16). The limit of detection (LOD) was estimated by finding the intercept between normalized baseline and the linear fit of the logarithmic dynamic range that determined to be ~18 nM, as shown in fig. S13 (41). The average sensitivity of CXCR4QTY-Fc–functionalized sensors is −11.4 ± 6.1 mV/decade. The CXCL12 in the range of 10−9 to 10−7 M but not >1 μM (10−6 M) were present in different organs and found to mediate multiple biological and pathological processes (42–44); therefore, the performance of the system is accurately suitable in applications like continuously health monitoring and cancer metastasis evaluation.
To verify sensing specificity, we again used CCL5 to interrogate the system. Human insulin was introduced to be another negative control as in previous biological assays (15, 16). The averaged DP shifts with respect to different ligand concentrations were plotted in Fig. 3E. The analytes were tested between 10−10 and 10−5 M to fully cover the clinically relevant ligand concentrations while following the manufacturer’s reconstitution protocols. CXCR4QTY-Fc–functionalized sensors showed notable responses to CXCL12 with concentrations greater than 10 nM, while negative controls induced negligible change even with concentrations as high as 0.1 mM. Figure 3F shows the distribution of sensor sensitivities toward CXCL12 and negative controls. This result highlights our claim of sensor selectivity against nonspecific binding.
The shift of DP was proposed to be induced by the isoelectric point (pI) of each protein (table S2). Because tests were conducted in the neutral pH environment, the rSbpA-ZZ has an acidic pI, carries a net negative charge at pH = 7, and serves as electron donors, while CXCR4QTY-Fc has a basic pI, carries a net positive charge at pH = 7, and serves as electron acceptors. This was reinstated by ligand-binding tests, which also induced a negative shift on the DP with positively charged CXCL12. However, despite that insulin and CCL5 have pIs on the opposite sides of pH = 7, loading them onto the array did not induce any DP shift, as they did not interact with receptors. The current density change profiles with zero gate were plotted against gradient concentrations of CXCL12, CCL5, and insulin in fig. S14, which cross-referenced well with DP changes. Because current response is different when gated at left or right side of the DP for graphene-based FETs (fig. S15), the change of DP will be used as the primary feature for evaluating the sensor performance.
To evaluate biomimicry of the architecture compared to a natural system, we further tested the constructed devices with HIV coat glycoprotein. CXCR4 is one of receptors for HIV human infection by binding with coat protein gp41 and gp120 (45). Their recombinant fusion named gp41–120 was obtained from Prospec, which did not contain virus RNA but had necessary machineries for docking. A negative DP shift was observed similar to CXCL12. Extracted average DP with respect to gp41–120 concentrations is shown in Fig. 3G, with shift of averaged IDS-VGS curves shown in the inset. The array showed concentration-dependent responses toward gp41–120 with a Kd of 5.8 ± 3.1 nM, similar to that obtained from biological assays. The LOD for gp41–120 in buffer is estimated to be 95 pM. Thanks to the large number of available sensors, we are able to perform statistical analysis to further distinguish different types of analytes. T-distributed stochastic neighbor embedding (t-SNE) is used as a dimension reduction technique to visualize the data collected. As shown in Fig. 3H, clusters of different analytes are well separated. This result demonstrates the potential of the sensing platform to distinguish different biomolecules and to be used for rapid HIV diagnostics.
Analyte detection in serum and antifouling test
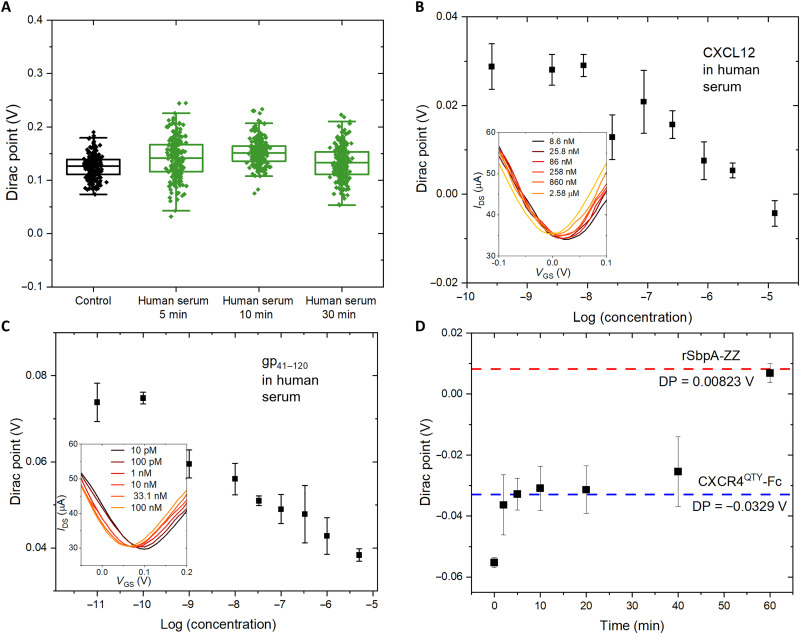
One major obstacle against affinity-based biosensors is their lack of resistance to nonspecific bindings and device degradation in complex biofluids (46). Here, we test the usability of our system in human serum, since the dense crystalline layer of rSbpA-ZZ should provide efficient antifouling against unspecific adsorptions of nontargeting biomolecules by the graphene channel underneath (17, 47, 48). As shown in Fig. 4A, incubating fully constructed devices in heat-inactivated human serum for extended period of time beyond the testing requirement did not induce notable DP shift. The current density change during incubation was also unobservable (fig. S16), suggesting negligible nonspecific associations of undesired biomolecules.
Fig. 4. Analyte detection in serum and device regeneration.
(A) Device DP shift after incubation in the human serum for extended period of time. (B) Concentration-dependent response of the fully constructed array toward a gradient of CXCL12 dissolved in human serum. Insert shows the change of IDS-VGS curves. Error bar indicates the standard deviation of three repeat measurements. (C) Concentration-dependent response of the fully constructed array toward a gradient of gp41–120 dissolved in human serum. Insert shows the change of IDS-VGS curves. Error bar indicates the standard deviation of three repeat measurements. (D) DP regeneration as a function of time by immersing the sensing array after ligand testing in glycine buffer at pH = 3.
Tests with gradient concentrations of CXCL12 and gp41–120 were conducted using human serum as buffers, where similarly well-behaved binding profiles were obtained compared to assays in DI water, demonstrating that minimal interference was induced in our concentration-dependent DP measurements in human serum (Fig. 4, B and C). The two-way analysis of variance (ANOVA) multiple comparison test with Tukey’s post hoc test procedure was performed on different analyte concentrations for gp41–120 tests both in buffer and in human serum. Calculated P values indicate strong confidence in statistically distinguishable DP measurements with various gp41–120 concentrations (tables S3 and S4). The Kd was 24.8 ± 13.0 and 19.1 ± 6.1 nM for CXCL12 and gp41–120 in serum, compared to 20.1 ± 9.1 and 5.8 ± 3.1 nM in DI water, respectively. Slightly decreased Kd values agreed with biological assays in 50% human serum (compared to 100% human serum in device tests) and were attributed to reduced performances from receptors in complex biofluids rather than devices. The results are summarized in Table 1 (16). The similarity between device-derived Kd values and those from biological assays for both solubilized and native proteins reinstate our claim about biomimicry of the architecture, proving their compatibility with complex biofluids that does not interfere with the specific detection of target analytes. The LODs for CXCL12 and gp41–120 in human serum are estimated to be 7.2 nM and 26 pM, respectively. All estimated LOD values in the current testing sets are summarized in table S5.
Table 1. Comparison of Kds.
The dissociation constant (Kd) between CXCR4QTY and its binding partners obtained from device models agree with those from biological assays. GFET, graphene-based field-effect transistor.
| Ligand | Biological assay (nM)* | GFET device test (nM) | |
|---|---|---|---|
| Native | QTY | Langmuir fitting model | |
| CXCL12 buffer | ∼5 | 17.3 ± 4.2 | 20.1 ± 9.1 |
| CXCL12 serum† | 31.4 ± 12.2 | 24.8 ± 13.0 | |
| gp41–120 buffer | ∼200‡ | 7.0 ± 1.9 | 5.8 ± 3.1 |
| gp41–120 serum† | 12.7 ± 2.6 | 19.1 ± 6.1 | |
*Kd values from biological assays were obtained from our previously published paper (16).
†Because of the technical implementation, biological assays were conducted in 50% human serum, while the device tests were conducted in 100% human serum.
‡Kd between native CXCR4 and HIV coat glycoprotein was measured by a cell-based assay.
Because reversible association-dissociation between rSbpA-ZZ and CXCR4QTY-Fc was observed in the QCM assay (Fig. 2B), we determine whether it allows the regeneration of GFET devices. After sensing experiments with the highest concentration of ligands (10−5 M), we immersed the whole chip in a glycine-based buffer at pH = 3. The DP was monitored with incremental time intervals, whereas the full restoration to the original position corresponding to rSbpA-ZZ coating (fig. S17) was observed after 1 hour of cumulative immersion (Fig. 4D). No further DP change was observed because of the higher durability of crystalline S-layer coating compared to affinity-based receptor associations. The capability of regenerating sensing devices enables their reuse with convenient exchanges of surface receptors. Despite that our system can mitigated the device-to-device variability and low reproducibility issue by providing a large number of devices, not all 200 devices are needed to obtain statistically significant results. We analyzed the DP distributions using gp41–120 testing sets with randomly selected groups of devices and found almost identical DP distributions from only 100 devices compared to the full array (fig. S18). Fifty devices still resulted in statistically confident responses, whereas less than 20 units are more susceptible to device variations. Potentially more than one type of receptor probes can be integrated in the same GFET array to achieve multiplexing and realize accurate disease diagnostics (25).
DISCUSSION
In summary, we have devised a modular and scalable biomimetic sensing platform using a dual-monolayer construct to transform biospecific interactions into readable electrical outputs. The construct adopted water-soluble membrane receptors as specific bioprobes (15, 16) and 2D crystalline S-layer protein intermediates (17). The water-soluble receptor design was achieved by a widely applicable design tool named QTY code, which enables cost-efficient production of previously inaccessible membrane proteins in a scale-up quantity. S-layer proteins guided receptor orientations and forced exposure of their functional sites to enhance signal-to-noise ratio as well as provided antifouling for the device underneath. The construct is naturally multimodal (49–51), which overcomes one of the fundamental challenges to ultrasensitive bioelectronic detections in obtaining well-defined sensing structures on device surfaces (4). The dual monolayer was then introduced to a GFET array integrated with more than 200 transistors to mimic the redundancy in natural systems and provide statistically significant electrical readouts (25).
We demonstrated the feasibility and biomimicry of this system by rSbpA-ZZ/CXCR4QTY-Fc combination. Specific detections were observed toward the native ligand and binding partner of CXCR4 but not negative controls. High sensitivity and specificity were observed also in 100% human serum, suggesting sufficient antifouling capability for fully constructed GFET arrays in complex biofluids. The architecture can be regenerated in acidic buffer, which enables the reuse of devices and functional tuning by surface receptor exchanges. Comparison of performances for our platform with other works is summarized in table S6. The current system exhibited excellent specificity, good stability in biofluids, excellent sensitivity and suitable detection range, as well as device reusability.
The design approach is easily applicable to a wider range of receptor probes including another 11 cytokine receptors in full-length or truncated forms already experimentally verified, which can be conveniently integrated into the system for targets of interest (15, 16, 52, 53). Combinations of receptors can be tailored to fabricate multiplex sensors on the GFET array for designated diagnostic purposes. The sensor array analyzes the collective response from multichannel inputs instead of assuming absolute specificity and sensitivity of one device. Microfluidic channels can be further integrated onto the system for precise sample manipulation during the tests. With properly calibrated and well-trained disease models using large sets of real-world biological samples in future studies, data from individual patients in clinical trials can be analyzed by deep learning algorithms for highly accurate analyte predictions, disease diagnostics, and continuously health monitoring, which enable precision medicine.
On the other hand, the biomimicry enables the use of our platform as physical simulators of a nature system, beyond the function of a traditional biosensor for health care. Potential applications include membrane receptor deorphanization, drug screening including antibodies and aptamers, and specificity/off-target screening for cell therapies like chimeric antigen receptor–T, using statistically significant electrical signals as a quantification criterion. Such applications cannot be achieved in traditional bottom-up designed biosensors with antibody or aptamer probes.
While conception of the architecture was demonstrated here, its significance lies beyond the current stage of research. Compared to common bottom-up approaches, where researchers identify target analytes and find bioprobes to detect them, we take a top-down route to build a biomimetic system with important receptors in proteome, redesigning and installing them onto bioelectronic devices to generate electrical signals that can be analyzed from a biophysical aspect. The “plug-and-play” regime in combination with multiplex GFET arrays opens up the pathway to fabricate integrated bioelectronic systems with a panel of receptors that behaves in a synergistic manner across different functional molecules like the living organism but through real-time electrical communications. The approach may be especially advantageous in electronic nose applications where state-of-the-art methods inherently lack specificity in complex environments, which also provides new routes for reemerging interests in disease diagnostics by patients’ exhaled breath, tear drops, and sweat (54).
MATERIALS AND METHODS
Bioinformatics of the QTY variants
Protein properties including MW and pI were calculated on the basis of its primary sequence via the open-access web-based tool ExPASy (https://web.expasy.org/protparam/).
Structure prediction and superimposition
Structure prediction for QTY code designed CXCR4, and its Fc fusion were conducted via ColabFold (55). Google Colaboratory notebook called AlphaFold2_advanced was used, since it provides more accurate predictions. The best model was analyzed on the basis of pLDDT (predicted local distance difference test) values of each prediction with PyMOL (version 2.4.1). Structural superpositions were made by align command against reported crystal structures obtained from the Protein Data Bank, and visualizations were made in PyMOL. The parameters are summarized in Table 2.
Table 2. Parameters for structure predictions.
| Parameter | Value |
|---|---|
| homooligomer | 1 |
| msa_method | mmseq2 |
| msa_format | fas |
| pair_mode | unpaired |
| pair_cov | 50 |
| pair_qid | 20 |
| rank_by | pLDDT |
| max_msa | True |
| num_models | 512:1024 |
| use_ptm | True |
| num_ensemble | 5 |
| max_recycles | 3 |
| tol | 0 |
| num_samples | 1 |
| subsample_msa | True |
| num_relax | None |
E. coli synthesis of CXCR4QTY-Fc
Genes of CXCR4QTY-Fc were codon-optimized for E. coli expression and obtained from GenScript. The genes were cloned into pET20b expression vector with carbenicillin resistance. The plasmids were reconstituted and transformed into E. coli BL21(DE3) strain. Transformants were selected on LB medium plates with carbenicillin (100 μg/ml). E. coli cultures were grown at 37°C until the optical density at 600 nm (OD600) reached 0.4 to 0.8, after which IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 1 mM, followed by 4 hours of expression. Cells were lysed by sonication in B-PER protein extraction agent (Thermo Fisher Scientific) and centrifuged (at 23,000g for 40 min at 4°C) to collect the inclusion body. The biomass was then subsequently washed twice in buffer 1 [50 mM tris-HCl (pH 7.4), 50 mM NaCl, 10 mM CaCl2, 0.1% (v/v) Triton X-100, and 2 M urea, 0.2 μm filtered], once in buffer 2 [50 mM tris-HCl (pH 7.4), 1 M NaCl, 10 mM CaCl2, 0.1% (v/v) Triton X-100, and 2 M urea, 0.2 μm filtered], and again in buffer 1. Pellets from each washing step were collected by centrifugation (at 23,000g for 25 min at 4°C).
Washed inclusion bodies were fully solubilized in denaturation buffer (6 M guanidine hydrochloride, 1× phosphate-buffered saline, and 10 mM dithiothreitol, 0.2 μm filtered) at room temperature for 1.5 hours with magnetic stirring. The solution was centrifuged at 23,000g for 40 min at 4°C. The supernatant with proteins was then purified by QIAGEN Ni-NTA (nitrilotriacetic acid) beads (His-tag), followed by size exclusion chromatography (SEC) using an ÄKTA Purifier Protein Purification System and a GE Healthcare Superdex 200 gel filtration column. Purified protein was collected and dialyzed twice against renaturation buffer [50 mM tris-HCl (pH 9.0), 3 mM reduced glutathione, 1 mM oxidized glutathione, 5 mM ethylenediaminetetraacetic acid, and 0.5 M l-arginine]. Following an overnight refolding process, the renatured protein solution was dialyzed against 50 mM tris-HCl (pH 9.0) with various arginine content and filtered through a 0.2-μm syringe filter to remove aggregates.
E. coli synthesis of rSbpA-ZZ
The plasmid of rSbpA-ZZ was constructed as has been reported in the previous publication (37). It was transformed into E. coli BL21(DE3) strain and selected on LB medium plates with kanamycin (50 μg/ml). Cultures were grown at 37°C until the OD600 reached 0.4 to 0.8 and induced by 1 mM IPTG for 4 hours. Cells were collected and lysed in lysis buffer [0.05 M Hepes (pH 7.4), 0.5 M NaCl, 0.01 M imidazole, 15% glycerol, 0.5% Triton X-100, 1× protease inhibitor cocktail, and 1 × 10−4 M phenylmethylsulfonyl fluoride]. Inclusion bodies were collected by centrifugation (at 23,000g for 40 min at 4°C), washed twice in buffer S1 [50 mM tris-HCl (pH 7.4), 50 mM NaCl, and 10 mM CaCl2], once in buffer S2 [50 mM tris-HCl (pH 7.4), 1 M NaCl, and 10 mM CaCl2], and again in buffer S1. The pellet was denatured using extraction buffer [50 mM tris-HCl (pH 7.4), 150 mM NaCl, and 5 M GHCl] and centrifuged, and the supernatant was renatured against SEC running buffer [50 mM tris-HCl (pH 7.4) and 150 mM NaCl] overnight. Supernatant filtered through a 0.2-μm syringe filter was purified by ÄKTA Purifier system with GE Superdex 200 column and checked via SDS–polyacrylamide gel electrophoresis. Protein concentration was determined using Implen NanoPhotometer NP80 NanoDrop system using an extinction coefficient of 1.64. Purified proteins can be stored in aqueous solution at 4°C for several months. Recrystallization can be induced by applying a buffer system containing 10 mM CaCl2.
MST measurement
MST detects changes in thermophoretic movement and temperature-related intensity changes of the protein-attached fluorophore upon ligand binding. Because synthesized receptors were labeled with Monolith NT Protein Labeling Kit RED-NHS (NanoTemper Technologies) in storage buffer to obtain unique fluorescent signals. Receptor at 5 nM was tested against 0.0916 to 3000 nM the ligand at 80% MST power and 15% light-emitting diode power in premium capillaries on a Monolith NT.115 pico instrument at 25°C. MST time traces were recorded and analyzed to obtain highest possible signal to noise levels and amplitudes >5 Fnorm units. The recorded fluorescence was plotted against the concentration of ligand, and curve fitting was performed with KaleidaGraph 4.5 using the Kd fit formula derived from the law of mass action. For clarity, binding graphs of each independent experiment were normalized to fraction bound (0 = unbound and 1 = bound). MST measurements were performed at 2bind GmbH, Regensburg, Germany.
Transmission electron microscopy
The ability of the rSbpA-ZZ to form standalone 2D crystalline self-assembly was demonstrated with an FEI Tecnai T20 transmission electron microscope operated at 160 kV (Thermo Fisher Scientific, Eindhoven, The Netherlands) via negative staining. CaCl2 was added to the rSbpA-ZZ proteins to induce recrystallization in solution [1 mg/ml protein, 10 mM CaCl2, and 0.5 mM tris-HCl (pH 9.0)]. After overnight recrystallization, the formed self-assemblies were adsorbed on 300 mesh copper grids (Christine Gröpl Elektronenmikroskopie, Tulln, Austria) coated with a Formvar support film and a thin carbon layer. A chemical fixation of the protein adsorbed on the copper grids was done with 2.5% glutaraldehyde in cacodylate buffer (pH 7.4) for 10 min. For negative staining, samples were placed on 2% uranium acetate drops for 4 min. All steps were performed at room temperature. To demonstrate the accessibility and the binding of biological components, CXCR4QTY-Fc was applied to the self-assembly products, followed by an incubation with His-binding nanogold particles (Ni-NTA nanogold, d = 10 nm; nanoprobe) at pH 9.0 for 60 min. The binding events were visualized in TEM after negative staining as described above.
Atomic force microscopy
The surface morphology for each layer of the construct was examined on an ultraflat silicon wafer (<100>; Ted Pella). The wafer was cut into 1 cm–by–1 cm squares and cleaned sequentially in acetone, isopropanol, ethanol, and DI water for 10 min each using a sonication bath. Recrystallization solution of rSbpA-ZZ was prepared and applied onto the clean silicon chip, which was placed in a closed humid environment, and incubated overnight at 4°C before removal. Coated surfaces were rinsed repeatedly and stored in DI water for 1 day to remove excessive rSbpA-ZZ to ensure the formation of the monolayer before AFM inspection or CXCR4QTY-Fc coating. For CXCR4QTY-Fc coating, the protein was renatured following the procedure above to a temporary storage buffer [50 mM tris-HCl (pH 9.0) and 50 mM arginine] and diluted by 0.1 M glycine-NaOH (pH 9.0) at a ratio of 1:9 with extensive mixing. The solution was kept at room temperature for 3 min, centrifuged at 16,000g for 5 min to remove aggregates, and applied to rSbpA-ZZ–coated wafer for 1.5 hours at room temperature. The sample was immersed and stored in DI water before AFM characterizations. The Asylum Cypher ES Environmental AFM system in the Center for Nanoscale Systems at Harvard was used for the inspection. Examinations were conducted by contact mode in liquid with NanoWorld Pyrex-Nitride triangular AFM probes. The resolution is 384 points per line, and the scanning rate is 1 Hz in an area of 1 μm × 1 μm. Real-time adjustment on the setpoint was performed to ensure minimal contact force between the probe tip and proteins to obtain optimal image reflexing surface geometry.
Circular dichroism
CD spectra were recorded using JASCO Model J-1500 CD Spectrometer in Biophysical Instrumentation Facility at Massachusetts Institute of Technology. CXCR4QTY and CXCR4QTY-Fc were dialyzed and refolded into CD buffer [0.05% (v/v) trifluoroacetic acid and 1 mM tris (2-carboxyethyl)phosphine]. For far UV CD, spectra between 195 and 250 nm were collected with a step size of 0.5 nm, a bandwidth of 1 nm, and a scanning speed of 50 nm/min in a 0.1-cm path length cuvette. Baselines were established using dialysis buffer without protein and subtracted from the measurements. The baseline-subtracted spectra were scaled to obtain mean residue ellipticity and normalized by protein concentrations, which were 1 to 4 μM as determined by NanoDrop with calculated extinction coefficient from ExPASy.
OCV test
The device for OCV measurement was made in-house using a Thermo Fisher Scientific Slide-A-Lyzer MINI dialysis tube [0.5 ml, 3.5K MWCO (molecular weight cut off)]. Copper tape was attached to the inner wall of the lower and upper chambers to serve as electrodes. Receptors were predialyzed overnight in a parallel tube without the electrodes to reach environmental equilibrium in the buffer. Final concentrations of the receptors were ~0.11 to 0.16 mg/ml in the upper chamber. After transferring the equilibrated solutions in both chambers to the measurement device, analytes were added to the upper chambers in 1:1 molar ratio to the receptor. The relative changes of OCV between the two chambers were recorded at fixed time intervals using a Hewlett Packard 34401A Multimeter. The reported values in fig. S8 were the average of three measurements at each time point. Parallel control tests were conducted to establish a baseline to eliminate interferences from the buffer-electrode interactions, whereas changes in OCV readings were subtracted from those obtained in the testing sets and normalized to the readings at 0 min.
QCM test
The initial QCM verifications were performed with a QSense E4 instrument (QSense, Biolin Scientific, Västra Frölunda, Sweden). Gold-coated quartz sensors (QSX 301, QSense, Biolin Scientific) were cleaned by sonification in 2% (w/w) SDS solution for 20 min and subsequently rinsed with ultrapure water and ethanol, followed by a drying step using nitrogen. Subsequently, the cleaned sensors were treated with UV/ozone (BioForce Nanosciences, Ames, IA, USA) for 30 min and coated with 1 mM solution of octanethiol in ethanol to obtain a more hydrophobic surface. The octanethiol-coated sensors were then sonicated in ethanol and lastly mounted into the QCM-D (dissipation) chamber. Injection of wtSbpA or rSbpA-ZZ (50 μg/ml), all washing steps, and the addition of CXCR4QTY-Fc were performed by means of a peristaltic pump (Ismatec, Cole-Parmer GmbH, Wertheim, Germany) operating at a flow rate of 0.3 ml/min. Experiments were performed at 25°C.
The QCM sensor chip was obtained from the Gamry company (USA). The fundamental frequency of the chip was 5 MHz with a double-side gold electrode surface. CXCR4QTY-Fc tests were performed by QCM-І (impedance) (Gamry, USA). The quartz chips were sonicated in piranha solution (98% H2SO4:30% H2O2 = 7:3) for 10 min to remove organic pollutants remaining on the crystals. Subsequently, the quartz chips were incubated in 25 ml of 3-aminopropyltriethoxysilane (100 μl) and ethanol solution for 12 hours at room temperature. The quartz chips were rinsed with ethanol and dried with N2 gas. The dried quartz chips were incubated in the recrystallization solution of rSbpA-ZZ (0.5 ml) for 12 hours. The treated quartz chips were rinsed with ethanol, dried with N2 gas, and subjected to CXCR4QTY-Fc coating and analyte detections.
Graphene-based sensor array fabrication
Similar fabrication method was reported by our team before (25). The graphene-based sensor arrays were fabricated on a 4-inch, 200-μm-thick Corning willow glass wafer (MTI Corporation). After solvent cleaning, 25 nm of aluminum oxide was deposited on the wafer using atomic layer deposition (ALD) to enhance adhesion in subsequent photolithography steps. Ti/Au (5/100 nm) was deposited using electron beam deposition as the row contacts of the sensor array. A 30-nm layer of aluminum oxide was then deposited as the interlayer dielectric using ALD. A second metal layer of Ti/Au (5 or 100 nm) was then deposited using electron beam deposition. Openings were etched into the interlayer dielectric using a BCl3 plasma to expose the first metal layer for contacts. The sensing chips were diced from the wafer using a diesaw machine before the graphene transfer step. Graphene coated with poly(methyl methacrylate) (PMMA) [1 cm by 1 cm; ACS (Advanced Chemicals Supplier) Materials Trivial Transfer Graphene] was transferred on the substrate to cover the entirety of the array. The chip was baked at 60°C for 30 min and 130°C for 10 min. The sensor array chip was then immersed in acetone for several hours to remove the PMMA. The chip was subsequently annealed at 350°C in 400 standard cubic centimeter per minute (SCCM) of Ar and 7000 SCCM of H2 to reduce PMMA residue and further enhance adhesion between the graphene and substrate. Graphene sensors were isolated from each other using oxygen plasma and a patterned PMGI/SPR700 resist stack as a mask. The resist stack was then removed by immersion in N-methyl-2-pyrrolidone for several hours. The chip was spin coated with SU-8, and openings were defined over the graphene channel regions and contact leads.
Dual-monolayer coating on graphene-based sensor array
Sequential coatings of rSbpA-ZZ and CXCR4QTY-Fc were conducted on the as-fabricated GFET arrays. Recrystallization solution of rSbpA-ZZ was applied onto the device. The devices were placed in a closed humid environment and incubated overnight at 4°C before removing the coating buffer. Coated surfaces were rinsed repeatedly and stored in DI water for 1 day to remove excessive rSbpA-ZZ and ensure the formation of the monolayer. Simultaneously, CXCR4QTY-Fc were renatured following the procedure above to a temporary storage buffer [50 mM tris-HCl (pH 9.0) and 50 mM arginine] and diluted by 0.1 M glycine-NaOH (pH 9.0) at a ratio of 1:9 with extensive mixing. The coating solution was kept at room temperature for 3 min and centrifuged at 16,000g for 5 min to remove aggregates. The fully coated devices were rinsed repeatedly and stored in DI water till testing. Drying out of the devices was carefully avoided during handling to prevent the denaturation of surface proteins which might cause the loss of function.
Graphene-based sensor array measurement
The analytes and human serum were obtained directly from commercial sources [CXCL12 (Sino Biological, 13511-HNCE), CCL5 (Sino Biological, 10900-HNAE), insulin (MilliporeSigma, 91077C), gp41–120 (Prospec, HIV-158), and human serum (MilliporeSigma, H5667)]. All electrical measurements were performed using the custom-built measurement system that is previously reported by our team at room temperature (25). The drain-source voltage VDS was held constant, and the gate-source voltage VGS was swept from −0.6 to 0.6 or 0.9 V in 10-mV increments. A 10-s hold time was used before the gate-source voltage VGS was swept at a rate of 10 mV/500 ms. This provided adequate time for charged species to migrate and reach steady-state before measurement. Analytes were reconstituted and diluted in DI water (manufacture recommended) or human serum with the exception of insulin (0.25 mM HEPES, pH 8.0) due to its solubility issue. The analytes were applied onto the devices and kept for 5 min to allow sufficient binding and rinsed off by repeated mild pipetting, followed by I-V measurements in DI water. The experiments started with the lowest concentration to reduce the potential for altering solution concentrations due to cross contamination. The glycine regeneration test was done by immersing the whole device in 0.1 M glycine-NaOH (pH 3.0) and periodically removing the device for testing the DP shift in DI water.
DP fitting
Minimum conduction points, or DPs, were calculated by fitting the experimental I-V characteristic with a polynomial and finding the minimum of the polynomial fit (25). This ensures the most accurate possible DP interpolation. I-V curves were fitted using a sixth-order polynomial. Experimental data were discretized using a step size of 10 mV, which could lead to some additional error if the DP was estimated simply by taking the minimum of the discretized dataset.
Kd calculation–based on Langmuir adsorption isotherm and LOD estimation
The Kd value was calculated using Langmuir model. DPs were extracted using the estimation method mentioned above. The device response S is calculated by finding the change in the DP upon binding with different concentrations of analytes with respect to the baseline. A linear fit was performed with the x value being the concentration of the analyte (C), while the y value being the ratio between concentration of the analyte and device response (C/S) as shown in fig. S12A. Kd can be extrapolated as y intercept/slope. Devices with poor fitting accuracy were filtered out. All calculation is done in MATLAB.
The LODs of each analyte testing set were estimated by finding the intercept between normalized baselines and the linear fit of the logarithmic dynamic range, following the equation of: y = a × log (x) + b, where y is the DP shift, x is the analyte concentration, and a and b are the fitting parameters. LOD is extrapolated from the x values at the intercept.
T-distributed stochastic neighbor embedding
We applied t-SNE to visualize the clusters of different analytes. We first standardized the sensor response by finding the change in the DP upon binding with six different concentrations of analytes with respect to the baseline (DI water). The standardized DP values of each device were used as features for clustering. The dimensionality of our sensor response was the number of different concentrations tested and was restricted to six. The data from multiple chips were combined for analysis with a matrix size of 813 × 6. Each data point represents a single sensor’s response toward one concentration of analyte. The t-SNE function in MATLAB’s Statistics and Machine Learning Toolbox was used to produce the final 2D projection.
Acknowledgments
We thank J. Song and F. Yan from Hong Kong Polytechnic University for their support in the initiation stage of this project.
Funding: This work was supported by the National Science Foundation grant DMR-1231319 and W. Chu, Defond Group, Hong Kong.
Author contributions: Conceptualization: R.Q., M.X., J.Z., G.A., U.B.S., and S.Z. Experimental: R.Q., M.X., J.Z., L.W., A.B., E.S., and T.S. Data analysis: R.Q., M.X., L.W., and A.B. Supervision: R.Q., J.K., T.P., U.B.S., and S.Z. Writing: R.Q., M.X., L.W., A.B., E.S., D.J., T.P., U.B.S., and S.Z.
Competing interests: Massachusetts Institute of Technology (MIT) filed the following patent relating to the work described here: S.Z., R.Q., U.B.S., and A.B. U.S. Patent 11293923, active: 5 April 2022. S.Z. is an inventor of the QTY code and has a minor equity of OH2 Laboratories. S.Z. founded a startup 511 Therapeutics to generate therapeutic monoclonal antibodies against solute carrier transporters to treat pancreatic cancer. S.Z. has majority equity in 511 Therapeutics. However, this article does not work on solute carrier transporters to treat pancreatic cancer at all. Defond Co Ltd. in Hong Kong partially sponsored the study but had no influence and interference in the design of the study, in the data collection, analyses, or interpretation of data and in the writing of the manuscript as well as in the decision to publish the results. S.Z. is also a cofounder and scientific advisor for 3D Matrix Co Ltd. in Japan that produces self-assembling peptide hydrogels for accelerating wound healing for surgical and dental wound healing applications. S.Z. is a coinventor on the following patents related to this work: 8,637,452, filed 28 January 2014 (licensed to OH2 Laboratories 2014); 9,309,302, filed 12 April 2016 (licensed to OH2 Laboratories 2014); 10,035,837, filed 31 July 2018 (licensed to OH2 Laboratories 2014); and WO2019046699A1, filed 7 March 2019. S.Z., R.Q., D.J, and U.B.S. are coinventors on a patent application filed at MIT (US11,401,320 B2, filed 2 August 2022). The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Supplementary Text
Figs. S1 to S18
Tables S1 to S6
References
REFERENCES AND NOTES
- 1.P. Mehrotra, Biosensors and their applications–A review. J. Oral Biol. Craniofac. Res. 6, 153–159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.M. X. Ouyang, D. D. Tu, L. Tong, M. Sarwar, A. Bhimaraj, C. Z. Li, G. L. Cote, D. Di Carlo, A review of biosensor technologies for blood biomarkers toward monitoring cardiovascular diseases at the point-of-care. Biosens. Bioelectron. 171, 112621 (2021). [DOI] [PubMed] [Google Scholar]
- 3.J. B. Tu, R. M. Torrente-Rodriguez, M. Q. Wang, W. Gao, The era of digital health: A review of portable and wearable affinity biosensors. Adv. Funct. Mater. 30, 1906713 (2020). [Google Scholar]
- 4.A. Q. Zhang, C. M. Lieber, Nano-bioelectronics. Chem. Rev. 116, 215–257 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.N. Gao, T. Gao, X. Yang, X. C. Dai, W. Zhou, A. Q. Zhang, C. M. Lieber, Specific detection of biomolecules in physiological solutions using graphene transistor biosensors. Proc. Natl. Acad. Sci. U.S.A. 113, 14633–14638 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A. Adesina, P. Mashazi, Oriented antibody covalent immobilization for label-free impedimetric detection of c-reactive protein via direct and sandwich immunoassays. Front. Chem. 9, 587142 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O. S. Kwon, H. S. Song, T. H. Park, J. Jang, Conducting nanomaterial sensor using natural receptors. Chem. Rev. 119, 36–93 (2019). [DOI] [PubMed] [Google Scholar]
- 8.A. E. Rawlings, Membrane proteins: Always an insoluble problem? Biochem. Soc. Trans. 44, 790–795 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R. Qing, S. Hao, E. Smorodina, D. Jin, A. Zalevsky, S. Zhang, Protein design: From the aspect of water solubility and stability. Chem. Rev. 122, 14085–14179 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. De-Eknamkul, X. W. Zhang, M. Q. Zhao, W. Z. Huang, R. Y. Liu, A. T. C. Johnson, E. Cubukcu, MoS2-enabled dual-mode optoelectronic biosensor using a water soluble variant of mu-opioid receptor for opioid peptide detection. 2d Mater 7, 014004 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.M. B. Lerner, F. Matsunaga, G. H. Han, S. J. Hong, J. Xi, A. Crook, J. M. Perez-Aguilar, Y. W. Park, J. G. Saven, R. Liu, A. T. Johnson, Scalable production of highly sensitive nanosensors based on graphene functionalized with a designed G protein-coupled receptor. Nano Lett. 14, 2709–2714 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.B. R. Goldsmith, J. J. Mitala Jr., J. Josue, A. Castro, M. B. Lerner, T. H. Bayburt, S. M. Khamis, R. A. Jones, J. G. Brand, S. G. Sligar, Biomimetic chemical sensors using nanoelectronic readout of olfactory receptor proteins. ACS Nano 5, 5408–5416 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D. M. Goodwin, F. Walters, M. M. Ali, E. Daghigh Ahmadi, O. J. Guy, Graphene bioelectronic nose for the detection of odorants with human olfactory receptor 2AG1. Chem 9, 174 (2021). [Google Scholar]
- 14.S. Zhang, R. Qing, A. Breitwieser, U. Sleytr, S-Layer protein 2D lattice coupled detergent-free GPCR bioelectronic interfaces, devices, and methods for the use thereof, US Patent 11,293,923 (2022).
- 15.S. Zhang, F. Tao, R. Qing, H. Tang, M. Skuhersky, K. Corin, L. Tegler, A. Wassie, B. Wassie, Y. Kwon, B. Suter, C. Entzian, T. Schubert, G. Yang, J. Labahn, J. Kubicek, B. Maertens, QTY code enables design of detergent-free chemokine receptors that retain ligand-binding activities. Proc. Natl. Acad. Sci. U.S.A. 115, E8652–E8659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.R. Qing, Q. Han, M. Skuhersky, H. Chung, M. Badr, T. Schubert, S. Zhang, QTY code designed thermostable and water-soluble chimeric chemokine receptors with tunable ligand affinity. Proc. Natl. Acad. Sci. U.S.A. 116, 25668–25676 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.U. B. Sleytr, B. Schuster, E. M. Egelseer, D. Pum, S-layers: Principles and applications. FEMS Microbiol. Rev. 38, 823–864 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.U. B. Sleytr, B. Schuster, E. M. Egelseer, D. Pum, C. M. Horejs, R. Tscheliessnig, N. Ilk, Nanobiotechnology with S-layer proteins as building blocks. Prog. Mol. Biol. Transl. 103, 277–352 (2011). [DOI] [PubMed] [Google Scholar]
- 19.R. Qing, Self-assembling 2D nano-crystalline of recombinant surface layer proteins (S-layer) on solid substrates and electrical responses. MRS Adv. 2, 349–355 (2017). [Google Scholar]
- 20.U. B. Sleytr, P. Messner, D. Pum, M. Sara, Crystalline bacterial cell surface layers (s layers): From supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. Engl. 38, 1034–1054 (1999). [DOI] [PubMed] [Google Scholar]
- 21.M. H. Ucisik, S. Küpcü, A. Breitwieser, N. Gelbmann, B. Schuster, U. B. Sleytr, S-layer fusion protein as a tool functionalizing emulsomes and CurcuEmulsomes for antibody binding and targeting. Colloids Surf. B Biointerfaces 128, 132–139 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.A. E. Proudfoot, Chemokine receptors: Multifaceted therapeutic targets. Nat. Rev. Immunol. 2, 106–115 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E. K. Jenkins, M. T. DeChant, E. B. Perry, When the nose doesn’t know: Canine olfactory function associated with health, management, and potential links to microbiota. Front Vet. Sci. 5, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C. Sarafoleanu, C. Mella, M. Georgescu, C. Perederco, The importance of the olfactory sense in the human behavior and evolution. J. Med. Life 2, 196–198 (2009). [PMC free article] [PubMed] [Google Scholar]
- 25.M. Xue, C. Mackin, W.-H. Weng, J. Zhu, Y. Luo, S.-X. L. Luo, A.-Y. Lu, M. Hempel, E. McVay, J. Kong, T. Palacios, Integrated biosensor platform based on graphene transistor arrays for real-time high-accuracy ion sensing. Nat. Commun. 13, 5064 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.M. Bernabei, S. Pantalei, K. C. Persaud, Description and characterisation of a large array of sensors mimicking an artificial olfactory epithelium. Procedia Eng. 87, 863–866 (2014). [Google Scholar]
- 27.C. A. Vu, W. Y. Chen, Field-effect transistor biosensors for biomedical applications: Recent advances and future prospects. Sensors (Basel) 19, 4214 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.K. Guo, S. Wustoni, A. Koklu, E. Diaz-Galicia, M. Moser, A. Hama, A. A. Alqahtani, A. N. Ahmad, F. S. Alhamlan, M. Shuaib, A. Pain, I. McCulloch, S. T. Arold, R. Grunberg, S. Inal, Rapid single-molecule detection of COVID-19 and MERS antigens via nanobody-functionalized organic electrochemical transistors. Nat. Biomed. Eng. 5, 666–677 (2021). [DOI] [PubMed] [Google Scholar]
- 29.Y. Yang, J. Wang, W. Huang, G. Wan, M. Xia, D. Chen, Y. Zhang, Y. Wang, F. Guo, J. Tan, H. Liang, B. Du, L. Yu, W. Tan, X. Duan, Q. Yuan, Integrated urinalysis devices based on interface-engineered field-effect transistor biosensors incorporated with electronic circuits. Adv. Mater. 34, e2203224 (2022). [DOI] [PubMed] [Google Scholar]
- 30.C. Mackin, V. Schroeder, A. Zurutuza, C. Su, J. Kong, T. M. Swager, T. Palacios, Chemiresistive graphene sensors for ammonia detection. ACS Appl. Mater. Interfaces 10, 16169–16176 (2018). [DOI] [PubMed] [Google Scholar]
- 31.M. Lanza, Q. Smets, C. Huyghebaert, L. J. Li, Yield, variability, reliability, and stability of two-dimensional materials based solid-state electronic devices. Nat. Commun. 11, 5689 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.K. Shi, H. Li, T. Chang, W. He, Y. Kong, C. Qi, R. Li, H. Huang, Z. Zhu, P. Zheng, Bone marrow hematopoiesis drives multiple sclerosis progression. Cell 185, 2234–2247.e17 (2022). [DOI] [PubMed] [Google Scholar]
- 33.S. Chatterjee, B. Behnam Azad, S. Nimmagadda, The intricate role of CXCR4 in cancer. Adv. Cancer Res. 124, 31–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.C. D. Allen, K. M. Ansel, C. Low, R. Lesley, H. Tamamura, N. Fujii, J. G. Cyster, Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 5, 943–952 (2004). [DOI] [PubMed] [Google Scholar]
- 35.L. Lopalco, CCR5: From natural resistance to a new anti-HIV strategy. Viruses-Basel 2, 574–600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.N. Ilk, C. Vollenkle, E. M. Egelseer, A. Breitwieser, U. B. Sleytr, M. Sara, Molecular characterization of the S-layer gene, sbpA, of Bacillus sphaericus CCM 2177 and production of a functional S-layer fusion protein with the ability to recrystallize in a defined orientation while presenting the fused allergen. Appl. Environ. Microbiol. 68, 3251–3260 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.C. Vollenkle, S. Weigert, N. Ilk, E. Egelseer, V. Weber, F. Loth, D. Falkenhagen, U. B. Sleytr, M. Sara, Construction of a functional S-layer fusion protein comprising an immunoglobulin G-binding domain for development of specific adsorbents for extracorporeal blood purification. Appl. Environ. Microbiol. 70, 1514–1521 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.J. DeCaprio, T. O. Kohl, Cross-linking antibodies to beads using dimethyl pimelimidate (DMP). Cold Spring Harbor. Protoc. 2019, pdb. prot098624 (2019). [DOI] [PubMed] [Google Scholar]
- 39.L. Tegler, K. Corin, H. Pick, J. Brookes, M. Skuhersky, H. Vogel, S. Zhang, The G protein coupled receptor CXCR4 designed by the QTY code becomes more hydrophilic and retains cell signaling activity. Sci. Rep. 10, 21371 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.C. Mackin, T. Palacios, Large-scale sensor systems based on graphene electrolyte-gated field-effect transistors. Analyst 141, 2704–2711 (2016). [DOI] [PubMed] [Google Scholar]
- 41.B. D. Wilson, H. T. Soh, Re-evaluating the conventional wisdom about binding assays. Trends Biochem. Sci. 45, 639–649 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. Zlatopolskiy, J. Laurence, “Reverse gear” cellular movement mediated by chemokines. Immunol. Cell Biol. 79, 340–344 (2001). [DOI] [PubMed] [Google Scholar]
- 43.X. Cheng, H. Wang, X. Zhang, S. Zhao, Z. Zhou, X. Mu, C. Zhao, W. Teng, The role of SDF-1/CXCR4/CXCR7 in neuronal regeneration after cerebral ischemia. Front. Neurosci. 11, 590 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.A. Muller, B. Homey, H. Soto, N. Ge, D. Catron, M. E. Buchanan, T. McClanahan, E. Murphy, W. Yuan, S. N. Wagner, J. L. Barrera, A. Mohar, E. Verastegui, A. Zlotnik, Involvement of chemokine receptors in breast cancer metastasis. Nature 410, 50–56 (2001). [DOI] [PubMed] [Google Scholar]
- 45.T. D'huys, S. Claes, T. Van Loy, D. Schols, CXCR7/ACKR3-targeting ligands interfere with X7 HIV-1 and HIV-2 entry and replication in human host cells. Heliyon 4, e00557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.J. S. del Rio, O. Y. F. Henry, P. Jolly, D. E. Ingber, An antifouling coating that enables affinity-based electrochemical biosensing in complex biological fluids. Nat. Nanotechnol. 14, 1143–1149 (2019). [DOI] [PubMed] [Google Scholar]
- 47.M. M. Picher, S. Kupcu, C. J. Huang, J. Dostalek, D. Pum, U. B. Sleytr, P. Ertl, Nanobiotechnology advanced antifouling surfaces for the continuous electrochemical monitoring of glucose in whole blood using a lab-on-a-chip. Lab Chip 13, 1780–1789 (2013). [DOI] [PubMed] [Google Scholar]
- 48.D. Moll, C. Huber, B. Schlegel, D. Pum, U. B. Sleytr, M. Sára, S-layer-streptavidin fusion proteins as template for nanopatterned molecular arrays. Proc. Natl. Acad. Sci. U.S.A. 99, 14646–14651 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.A. Breitwieser, P. Siedlaczek, H. Lichtenegger, U. B. Sleytr, D. Pum, S-layer protein coated carbon nanotubes. Coatings 9, 492 (2019). [Google Scholar]
- 50.A. Breitwieser, D. Pum, J. Toca-Herrera, U. Sleytr, Magnetic beads functionalized with recombinant S-layer protein exhibit high human IgG-binding and anti-fouling properties. Curr. Top. Pept. Protein Res. 17, 45–55 (2016). [Google Scholar]
- 51.M. Pleschberger, D. Saerens, S. Weigert, U. B. Sleytr, S. Muyldermans, M. Sara, E. M. Egelseer, An S-layer heavy chain camel antibody fusion protein for generation of a nanopatterned sensing layer to detect the prostate-specific antigen by surface plasmon resonance technology. Bioconjug. Chem. 15, 664–671 (2004). [DOI] [PubMed] [Google Scholar]
- 52.M. A. Skuhersky, F. Tao, R. Qing, E. Smorodina, D. Jin, S. Zhang, Comparing native crystal structures and alphafold2 predicted water-soluble g protein-coupled receptor QTY variants. Life (Basel) 11, 1285 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.S. Hao, D. Jin, S. Zhang, R. Qing, QTY code-designed water-soluble Fc-fusion cytokine receptors bind to their respective ligands. QRB Discov. 1, e4 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.E. Kabir, N. Raza, V. Kumar, J. Singh, Y. F. Tsang, D. K. Lim, J. E. Szulejko, K.-H. Kim, Recent advances in nanomaterial-based human breath analytical technology for clinical diagnosis and the way forward. Chem 5, 3020–3057 (2019). [Google Scholar]
- 55.M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov, M. Steinegger, ColabFold: Making protein folding accessible to all. Nat. Methods 19, 679–682 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.N. Ilk, E. M. Egelseer, U. B. Sleytr, S-layer fusion proteins–Construction principles and applications. Curr. Opin. Biotechnol. 22, 824–831 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.U. B. Sleytr, P. Messner, D. Pum, M. Sára, Crystalline bacterial cell surface layers (S-layers): From supramolecular cell structure to biomimetics and nanotechnology. Angew. Chem. Int. Ed. 38, 1034–1054 (1999). [DOI] [PubMed] [Google Scholar]
- 58.B. Schuster, U. B. Sleytr, Nanotechnology with S-layer proteins. Methods Mol. Biol. 2073, 195–218 (2020). [DOI] [PubMed] [Google Scholar]
- 59.C. H. Naylor, N. J. Kybert, C. Schneier, J. Xi, G. Romero, J. G. Saven, R. Liu, A. T. Johnson, Scalable production of molybdenum disulfide based biosensors. ACS Nano 10, 6173–6179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.H. Liu, A. Yang, J. Song, N. Wang, P. Lam, Y. Li, H. K. Law, F. Yan, Ultrafast, sensitive, and portable detection of COVID-19 IgG using flexible organic electrochemical transistors. Sci. Adv. 7, eabg8387 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.R. Desplantes, C. Leveque, B. Muller, M. Lotierzo, G. Ferracci, M. Popoff, M. Seagar, R. Mamoun, O. El Far, Affinity biosensors using recombinant native membrane proteins displayed on exosomes: Application to botulinum neurotoxin B receptor. Sci. Rep. 7, 1032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text
Figs. S1 to S18
Tables S1 to S6
References