Abstract
The monomer units in the Escherichia coli and Staphylococcus aureus cell wall peptidoglycans differ in the nature of the third amino acid in the l-alanyl-γ-d-glutamyl-X-d-alanyl-d-alanine side chain, where X is meso-diaminopimelic acid or l-lysine, respectively. The murE gene from S. aureus encoding the UDP-N-acetylmuramoyl-l-alanyl-d-glutamate: l-lysine ligase was identified and cloned into plasmid vectors. Induction of its overexpression in E. coli rapidly results in abnormal morphological changes and subsequent cell lysis. A reduction of 28% in the peptidoglycan content was observed in induced cells, and analysis of the peptidoglycan composition and structure showed that ca. 50% of the meso-diaminopimelic acid residues were replaced by l-lysine. Lysine was detected in both monomer and dimer fragments, but the acceptor units from the latter contained exclusively meso-diaminopimelic acid, suggesting that no transpeptidation could occur between the ɛ-amino group of l-lysine and the α-carboxyl group of d-alanine. The overall cross-linking of the macromolecule was only slightly decreased. Detection and analysis of meso-diaminopimelic acid- and l-lysine-containing peptidoglycan precursors confirmed the presence of l-lysine in precursors containing amino acids added after the reaction catalyzed by the MurE ligase and provided additional information about the specificity of the enzymes involved in these latter processes.
Bacterial-cell-wall peptidoglycan (murein) is a giant macromolecule of periodic structure whose basic unit, a disaccharide-pentapeptide, is polymerized linearly via the disaccharide motif and cross-linked laterally via the peptide motif (for a review, see reference 15). Any alteration of the basic unit thus results in a global change of peptidoglycan structure and properties. Such global variations are encountered in nature as conserved variations along phyletic lines (30) but have sometimes been acquired as a mechanism of resistance against cell-wall-targeted antibiotics (5, 6, 8). The amino acid residue located at the third position in the peptide chain plays a key role in the integrity of the sacculus since it is directly involved in peptide cross-linkages. This vital function is fulfilled by meso-diaminopimelic acid (meso-A2pm) in Escherichia coli and l-lysine in Staphylococcus aureus.
In bacteria, free endogenous meso-A2pm is either irreversibly decarboxylated into l-lysine (27) or used to form the peptidoglycan precursor UDP-N-acetylmuramoyl-l-alanyl-γ-d-glutamyl-meso-A2pm, the latter reaction being catalyzed by the murE gene product (14, 24, 26). E. coli mutants altered in the A2pm pathway require exogenous A2pm for growth and lyse if lysine but not A2pm is supplied (18, 27). However, the A2pm auxotrophy can be suppressed in some cases by endogenous metabolic modifications (28) or, in the presence of lysine, by the addition of certain A2pm analogs (7, 18). The replacement of A2pm by an analog thus appeared to be a very useful tool for analyzing the specificity of the different enzymes involved in its insertion into peptidoglycan metabolism and the complexity of the transpeptidation reactions.
The E. coli UDP-MurNAc-l-Ala-d-Glu:meso-A2pm ligase (also named meso-A2pm-adding enzyme, EC 6.3.2.13) has been previously purified, and its kinetic properties have been investigated in detail (24, 26). The specificity of this enzyme for both its nucleotide and amino acid substrates is very high but not absolutely strict. Considering in particular the amino acid site, ll-A2pm and many analogs of A2pm are substrates of the reaction (3, 18, 21), but l-lysine is not (18). The same was observed with the A2pm-adding enzyme from other bacteria (14). Less information is available on the UDP-MurNAc-l-Ala-d-Glu–l-lysine ligase (l-lysine-adding enzyme, EC 6.3.2.7), but Ito and Strominger showed that the enzyme from S. aureus (13) and other bacterial species (14) does not accept meso-A2pm as an alternative substrate. Since pools of lysine and A2pm coexist in bacteria, the high (and inverse) specificities of the MurE enzymes from E. coli and S. aureus clearly prevent these strains from incorporating these nonspecific compounds into cell wall peptidoglycan. It was thus tempting to speculate that the expression of the E. coli murE gene in S. aureus or inversely the S. aureus murE gene in E. coli could have dramatic effects on peptidoglycan metabolism and cell growth. In the present study we describe the cloning of the murE gene from S. aureus and show that its overexpression in E. coli results in a large and toxic recruitment of l-lysine in the pathway for peptidoglycan synthesis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli BL21(DE3)/pLysS (Promega) was used as the host for the plasmids as well as for the overproduction of the MurE enzyme. 2YT medium (25) was used for growing cells, and growth was monitored at 600 nm with a Shimadzu UV-1601 spectrophotometer. Antibiotics were used at the following concentrations: ampicillin (100 μg · ml−1), kanamycin (40 μg · ml−1), and chloramphenicol (30 μg · ml−1).
Cloning of the S. aureus murE gene and plasmid construction.
Standard procedures for molecular cloning were used (29). S. aureus murE gene was PCR amplified from strain RN4220 by using primers containing the start and the stop codons of the gene (5′-TTGGATGCAAGTACGTTGTTT-3′ and 5′-TTATTGATCAACAGGGCCACC-3′) (sequence data supplied by SmithKline Beecham Pharmaceuticals). A 1,485-bp product was amplified, cloned into pGEM-T Easy (Promega), and confirmed as S. aureus murE by sequencing. The resulting construct (pMuSa1) was then digested with EcoRI, and the excised S. aureus gene was ligated into EcoRI-digested pET30b (Novagen). The orientation of murE was determined by EcoRV digestion, and constructs containing the gene in the correct orientation (pMuSa2) were subsequently transformed into E. coli BL21(DE3)/pLysS for expression.
Extraction and quantitation of peptidoglycan precursors.
Cells of BL21(DE3)/pLysS/pMuSa2 (1-liter cultures) were grown exponentially at 37°C in 2YT medium. When an optical density (OD) of 0.4 (600 nm) was reached (approximately 2.5 × 108 cells · ml−1), IPTG (isopropyl-β-d-thiogalactopyranoside) was added to one culture at a final concentration of 1 mM. As soon as the first effects on cell growth were observed in induced cells (ca. 1 h later at a final OD of 0.8), cultures were stopped by rapid chilling to 0 to 4°C, and cells were harvested in the cold. The extraction of peptidoglycan nucleotide precursors, as well as the analytical procedures used for their quantitation, were as described previously (9, 19, 20).
Isolation of sacculi and quantitation of peptidoglycan.
Cells of BL21(DE3)/pLysS/pMuSa2 (0.5-liter cultures) were grown and induced with IPTG as described above. Harvested cells were washed with cold 0.85% NaCl solution and centrifuged again. Bacteria were then rapidly suspended under vigorous stirring in a hot (95 to 100°C) aqueous 4% sodium dodecyl sulfate (SDS) solution (20 ml) for 30 min. After being allowed to stand overnight at room temperature, the suspensions were centrifuged for 30 min at 200,000 × g in a Beckman TL100 centrifuge, and the pellets were washed several times with water. Final suspensions were made in 2 ml of water, and aliquots (100 μl) were hydrolyzed and analyzed with a Biotronik model LC2000 amino acid analyzer. The peptidoglycan content of the sacculi was expressed in terms of its muramic acid content (19, 22).
Purification of peptidoglycan and structure analysis.
First, the crude preparations of E. coli sacculi were subjected to successive treatments with pancreatin, pronase, and trypsin to eliminate peptidoglycan-associated proteins (4, 19). After several washings with water, hydrolysis of an aliquot of this material showed that it contained only peptidoglycan constituents: muramic acid, glucosamine, alanine, glutamic acid, and A2pm (or A2pm plus lysine in induced cells) in the expected molar ratios, i.e., approximately 1/1/2/1/1.
The structural analysis of the purified peptidoglycan material was then carried out by the method of Glauner et al. (10, 11) in slightly modified form. Purified peptidoglycan was in all cases digested to 90 to 95% by a mixture of lysozyme and cellosyl (Streptomyces coelicolor muramidase). The resulting soluble fragments were reduced with sodium borohydride in 0.25 M borate buffer (pH 9) for 30 min at room temperature. After the pH was adjusted to 4 with phosphoric acid, the reduced compounds were separated by reversed-phase high-pressure liquid chromatography (HPLC) on a LiChrosorb RP-18 column (4 by 250 mm) by using a gradient of methanol in sodium phosphate buffer. Peptidoglycan fragments from A2pm-containing sacculi were identified by their retention times compared to previously purified muropeptides (18, 21) and were designated according to the method of Glauner (10). The main monomers and dimers from lysine-containing sacculi were recognized by amino acid and hexosamine analyses, both before and after dinitrophenylation of their recovered reduced forms. The amounts of monomer and dimer fragments were quantified either by integration of the peaks recorded during HPLC or by determination of their amino acid composition after isolation, both methods yielding similar results (18).
Preparation of crude protein extracts.
Cells (0.5-liter cultures) grown as described above were harvested in the cold and washed with 40 ml of cold 20 mM potassium phosphate buffer (pH 7.4) containing 0.5 mM MgCl2 and 0.1% 2-mercaptoethanol. The wet cell pellet was suspended in 7.5 ml of the same buffer and disrupted by sonication in the cold (Bioblock Vibracell sonicator), and the resulting suspension was centrifuged at 4°C for 30 min at 200,000 × g. The supernatant was dialyzed overnight at 4°C against 100 volumes of the same phosphate buffer, and the resulting solution (10 mg of protein · ml−1) designated as crude enzyme was stored at −20°C. SDS-polyacrylamide gel electrophoresis (PAGE) analysis of proteins was performed as previously described (16) by using 12% polyacrylamide gels. Protein concentrations were determined by the method of Lowry et al. (17) with bovine serum albumin as the standard.
Enzymatic assays. (i) meso-A2pm-adding activity.
The standard assay mixture contained 100 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 100 mM MgCl2, 0.1 mM meso-[14C]A2pm (500 Bq), 0.2 mM UDP-MurNAc-l-Ala-d-Glu, and crude enzyme (5 μg of protein) in a final volume of 100 μl.
(ii) l-Lysine-adding activity.
The standard assay mixture contained 100 mM Tris-HCl buffer (pH 8.6), 5 mM ATP, 100 mM MgCl2, 0.2 mM UDP-MurNAc-l-Ala-d-[14C]Glu (500 Bq), 0.5 mM l-lysine, and crude enzyme (1 to 125 μg of protein, depending on overexpression factor) in a final volume of 100 μl.
In both cases, mixtures were incubated at 37°C for 30 min, and reactions were stopped by the addition of 10 μl of acetic acid. Reaction products were separated by high-voltage electrophoresis on Schleicher & Schuell 3469 paper in 2% formic acid (pH 1.9) for 45 min at 40 V · cm−1 by using an LT36 apparatus (Savant Instruments). The radioactive spots corresponding to substrate and reaction product were detected by overnight autoradiography with type R2 films (3M, St. Paul, Minn.) or with a radioactivity scanner (Multi-Tracermaster LB285; Berthold France, Elancourt, France). The spots were cut out and counted in an Intertechnique SL30 liquid scintillation spectrophotometer with a solvent system consisting of 2 ml of water and 13 ml of Aqualyte mixture (J.T. Baker Chemicals, Deventer, The Netherlands). One unit of enzyme activity was defined as the amount which catalyzed the synthesis of 1 μmol of UDP-MurNAc-tripeptide in 1 min.
Chemicals.
The preparation of UDP-MurNAc-peptides and meso-A2pm was previously described (9, 33). UDP-MurNAc-l-Ala-d-[14C]Glu was synthesized as described earlier (21) by using purified UDP-MurNAc-l-Ala:d-Glu ligase (2), and meso-[14C]A2pm was purchased from the CEA (Saclay, France). IPTG was obtained from Eurogentec (Seraing, Belgium). Lysozyme was from Sigma, and cellosyl was a gift from Hoechst Marion Roussel.
RESULTS AND DISCUSSION
Effect of overexpression of S. aureus murE in E. coli on cell survival.
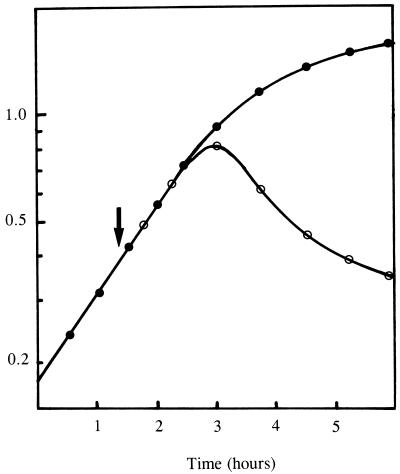
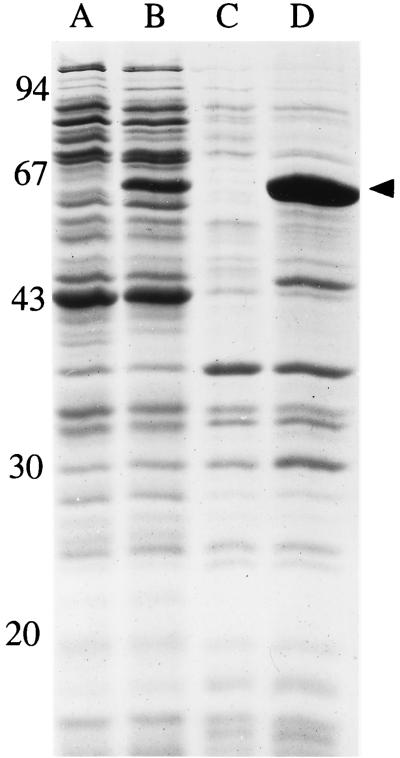
When the expression of the S. aureus murE gene was induced with IPTG in E. coli cells carrying the pMuSa2 plasmid, abnormal morphological changes of cell shape and size rapidly occurred, which were followed ca. 1 h later by an arrest of growth and finally by cell lysis (Fig. 1). Gram staining of the induced cells 2 h after induction revealed almost all cells to be lysed (data not shown), suggesting defective or greatly altered cell wall peptidoglycan biosynthesis. SDS-PAGE analysis of crude cell extracts showed that induced cells had greatly accumulated the MurE protein (Fig. 2). The latter was found in both the soluble and particulate fractions (Fig. 2) due to the formation of aggregates at such a high level of expression (inclusion bodies were effectively observed in induced cells by optical microscopy). Appropriate enzymatic assays confirmed the expression of the l-lysine-adding enzyme (S. aureus MurE) in pMuSa2 harboring cells (Table 1). A low but detectable activity observed in the absence of IPTG was due to a basal expression from the plasmid pMuSa2 since this activity was not detected in BL21(DE3)/pLysS control cells (data not shown). The specific activity of the l-lysine-adding enzyme was increased by a factor of 330 after IPTG induction, while that of the meso-A2pm-adding enzyme (E. coli MurE) was similar in noninduced and induced cells (Table 1). The ratio of S. aureus to E. coli MurE enzyme activities varied from 0.033 to 11 upon induction with IPTG.
FIG. 1.
Lytic effect of the expression of the S. aureus murE gene in E. coli cells. Cells of BL21(DE3)/pLysS/pMuSA2 were grown exponentially at 37°C in 2YT-ampicillin medium. At the time indicated by the arrow (OD = 0.4), IPTG was added at a final concentration of 1 mM. Growth of cells induced (○) or not induced (●) with IPTG was monitored at 600 nm.
FIG. 2.
Overproduction of S. aureus MurE enzyme in E. coli cells. Cells of BL21(DE3)/pLysS/pMuSa2 were grown and induced for 1 h with IPTG as described in the legend to Fig. 1. Cells were harvested at an OD of 0.8 and were disrupted by sonication. The protein contents from both soluble and membrane fractions obtained after high-speed centrifugation of the crude extracts were analyzed by SDS-PAGE. Molecular weight standards (in thousands) indicated on the left are as follows: phosphorylase b, 94; bovine serum albumin, 67; ovalbumin 43; carbonic anhydrase, 30; and soybean trypsin, 20. Lanes: A and B, analysis of the soluble fractions from noninduced and IPTG-induced cells, respectively; C and D, analysis of the membrane fractions from noninduced and IPTG-induced cells, respectively. The arrow points to the overproduced S. aureus MurE enzyme.
TABLE 1.
Pool levels of peptidoglycan precursors, peptidoglycan content, and specific activities of MurE enzymes in E. coli cells harboring the pMuSa2 plasmida
| Peptidoglycan precursor, peptidoglycan, or enzyme | Pool level or sp actb in:
|
|
|---|---|---|
| Noninduced cells | Induced cells | |
| Peptidoglycan precursors | ||
| UDP-GlcNAc | 160 | 370 |
| UDP-MurNAc | 90 | 90 |
| UDP-MurNAc-pentapeptide(A2pm) | 1,230 | 400 |
| UDP-MurNAc-pentapeptide(lysine) | 150 | 1,710 |
| Peptidoglycanc | 8,370 | 6,120 |
| Enzymes | ||
| A2pm-adding enzyme | 2.1 | 2.1 |
| Lysine-adding enzyme | 0.07 | 23.4 |
Cells of BL21(DE3)/pLysS(pMuSa2) were induced or noninduced with 1 mM IPTG for 1 h, as described in Materials and Methods. In each case, all of the parameters were tested from samples of the same culture.
The pool level (for peptidoglycan precursors and peptidoglycan) is given in nanomoles per gram (dry weight) of bacteria. The specific activity (enzymes) is given in units per milligram of protein.
Effects of the expression of the S. aureus MurE enzyme on peptidoglycan metabolism.
Cells of BL21(DE3)/pLysS/pMuSa2 induced with IPTG were harvested just before the first effects on cell growth were observed, and their peptidoglycan was extracted and quantified. In induced cells the peptidoglycan content was 28% lower than in noninduced cells (Table 1), suggesting that dysfunctioning of one (or more) step(s) in the pathway had occurred after overproduction of the S. aureus enzyme. The most likely explanation was a toxic recruitment of lysine by the flow of metabolites going to the cell wall peptidoglycan. The presence of lysine-containing peptidoglycan precursors was demonstrated (Table 1). In particular, both lysine- and A2pm-containing UDP-MurNAc-pentapeptides were detected in a ratio which paralleled the relative abundances of the two enzyme activities in vivo (Table 1). The UDP-MurNAc-pentapeptide(lysine) present in noninduced cells resulted from the basal expression of the S. aureus murE gene from plasmid pMuSa2, as discussed above. The total amount of UDP-MurNAc-pentapeptide was slightly higher in induced cells, suggesting some limitation in their in vivo utilization by membrane steps catalyzed by the mraY and murG gene products. The finding that the pool of UDP-N-acetylglucosamine, the other nucleotide substrate of the membrane steps, was also increased in induced cells was consistent with this hypothesis. The pool level of UDP-MurNAc-tripeptide(A2pm) is known to be very low in E. coli (19, 22). UDP-MurNAc-tripeptide(lysine) was detected in induced cells at a very low concentration (at most a few nanomoles per gram of bacterial dry weight), suggesting that this compound was efficiently utilized by the enzyme MurF, which catalyzes the subsequent step of addition of d-alanyl-d-alanine in the pathway (23, 32).
Incorporation of lysine into peptidoglycan.
To determine whether lysine was eventually incorporated at the place of A2pm in the macromolecule, peptidoglycan preparations were first made free of all traces of covalently associated proteins by successive treatments with proteases. Analyses showed that the material purified from noninduced cells contained only peptidoglycan constituents: muramic acid, glucosamine, alanine, glutamic acid, A2pm, and lysine in a ratio of 1/1/2.2/1/0.95/0.1, respectively. It was earlier established that some A2pm residues from E. coli peptidoglycan were covalently linked to C-terminal lysine residues of outer-membrane lipoprotein (4). Since these A2pm-lysine links (α-carboxyl-ɛ-amino amide bond) are not cleaved by proteases, the 10% of lysine found in the peptidoglycan purified from noninduced cells could consist of these residues but might also consist of lysine which had effectively replaced A2pm in the peptide chains. The latter was likely as it was shown above that a basal expression from pMuSa2 plasmid resulted in a small synthesis of lysine-containing peptidoglycan precursors. A similar analysis performed on the peptidoglycan from induced cells gave for the same constituents the following relative abundances: 1/1/1.7/1/0.51/0.6. It showed that a large incorporation of lysine had occurred in the macromolecule, half of A2pm residues in cell-wall peptidoglycan being now replaced by lysine.
HPLC analysis of peptidoglycan structure.
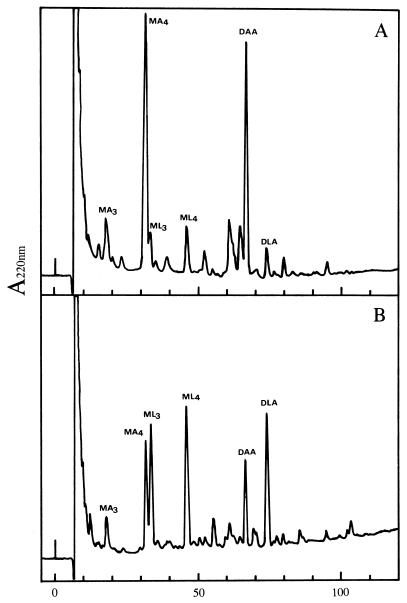
The purified peptidoglycan preparations were subjected to prolonged digestion with specific N-acetylmuramidases, leading to the breakdown of glycan strands into monomer, dimer, and trimer fragments that could be separated by HPLC after reduction with NaBH4 (10, 11). The main monomer (tetra) and dimer (tetra-tetra), as well as the less-abundant monomer (tri), encountered in the solubilized material from noninduced cells were those classically detected during analyses of peptidoglycan from wild-type E. coli cells (Fig. 3 and 4) (11, 18, 21). The nature of the compound in each peak was confirmed by analysis of its amino acid and hexosamine contents after acid hydrolysis (data shown in the legend to Fig. 4). Small additional peaks observed on the elution profile were identified as tri, tetra, and tetra-tetra fragments in which A2pm was replaced by lysine. As shown in Fig. 3, the retention time of these compounds was significantly higher than that of their A2pm counterparts, due to a great difference of polarity between lysine and A2pm residues. When the peptidoglycan from induced cells was analyzed in this way, the main difference was the large increase of the three peaks corresponding to lysine-containing monomers (tri and tetra) and dimer (tetra-tetra) (Fig. 3 and 4). Analysis of the latter dimer showed that it contained equimolar amounts of A2pm and lysine, and dinitrophenylation experiments further indicated that the ɛ-amino group of lysine was free (Fig. 4). This demonstrated that lysine was restricted to the donor unit in this dimer (designated DLA in Fig. 3 and 4) and that cross-linking was thus supported by A2pm. No other peaks of significant importance were observed in the elution profile that could consist of a hetero-dimer with lysine in the acceptor unit or a dimer containing exclusively lysine, suggesting that no transpeptidation could occur between the ɛ-amino group of lysine and the α-carboxyl group of d-alanine. However, a possibility exists that such dimers were formed but were too poorly represented to be detected by the technique employed here. It was noteworthy that the overall cross-linking of the macromolecule (as defined by the following ratio: Σ dimers/ [Σ monomers + 2 × Σ dimers] [see reference 10]) was not significantly modified (Table 2).
FIG. 3.
Separation of muropeptides by reversed-phase HPLC. Cells of BL21(DE3)/pLysS/pMuSa2 were grown and induced with IPTG as described in the legend to Fig. 1. The peptidoglycan from noninduced cells (A) and induced cells (B) was extracted and digested with muramidases, and the resulting fragments were reduced with NaBH4 and separated by reversed-phase HPLC on a LiChrosorb RP-18 column (4 by 250 mm). Elution was performed at 0.5 ml · min−1 with 50 mM sodium phosphate buffer (pH 4.5) and a linear gradient of methanol (0 to 15% from 0 to 100 min). Eluted compounds were detected at 220 nm at a sensitivity of 0.04 absorbance unit (full scale). MA3, monomer tri(A2pm); MA4, monomer tetra(A2pm); DAA, dimer tetra(A2pm)-tetra(A2pm); ML3, monomer tri(lysine); ML4, monomer tetra(lysine); DLA, dimer tetra(lysine)-tetra(A2pm).
FIG. 4.
Structures of the main muropeptides as separated in Fig. 3, based on amino acid and hexosamine composition. Glucosamine, Ala, Glu, A2pm, and Lys were detected after acid hydrolysis of HPLC-purified muropeptides in the following ratios (taking Glu as reference): MA3, 0.95/0.92/1/0.95/0; ML3, 1.07/1.02/1/0/1.03; MA4, 0.97/1.95/1/1.05/0; ML4, 0.97/2.1/1/0/0.97; DAA, 0.95/1.97/1/1/0; and DLA, 1.04/1.9/1/0.48/0.45 (abbreviations are as defined for Fig. 3). When the two latter muropeptides were dinitrophenylated before acid hydrolysis, half of the A2pm from DAA and all of the lysine, but not the A2pm, from DLA were lost.
TABLE 2.
Muropeptide composition and cross-linking of peptidoglycan in E. coli cellsa
| Cell type | Relative abundance (mol%) of:
|
% Cross-linkageb | |||||
|---|---|---|---|---|---|---|---|
| Tri(A2pm) | Tri(lysine) | Tetra(A2pm) | Tetra(lysine) | Tetra(A2pm)-tetra(A2pm) | Tetra(lysine)-tetra(A2pm) | ||
| Noninduced | 6.8 | 6.7 | 40.1 | 8.0 | 33 | 4.9 | 27.5 |
| Induced | 6.4 | 21.4 | 18.8 | 22.3 | 12.1 | 19.0 | 26.0 |
Conclusions.
Some bacteria contain meso-A2pm and others lysine at the third position of the peptide side chain in cell wall peptidoglycan (30). In each case, the MurE enzymes efficiently discriminate between the two amino acids in vitro, since they are only able to catalyze the addition of either meso-A2pm or lysine to UDP-MurNAc-l-Ala-d-Glu (13, 14, 18). As these two amino acids effectively coexist in bacterial cells (27), the high specificities of the MurE enzymes act as gatekeepers to ensure that only the specific substrate is incorporated in the peptidoglycan precursor. However, this specificity is not absolute since other A2pm analogs (lanthionine, cystathionine, 3-hydroxy-A2pm, and diaminosuberic acid) were earlier shown to complement a A2pm auxotrophic strain and to totally replace meso-A2pm in E. coli peptidoglycan (7, 18). Enzymes catalyzing subsequent steps, from cytoplasmic synthetase MurF to membrane transglycosylases, are clearly less selective enzymes since they can accept a broader range of substrates (1, 12, 14, 31). In fact, the critical step after the incorporation of analogs of A2pm into E. coli peptidoglycan always appeared to be the final stage of transpeptidation, in which A2pm is directly involved by its free amino group (15, 18, 21, 28). We observed that the pool levels of lysine-containing precursors (UDP-MurNAc-tripeptide and UDP-MurNAc-pentapeptide) in E. coli expressing S. aureus murE were quite similar to those of their A2pm analogs in cells not expressing the staphylococcal enzyme. This suggested that in vivo the replacement of A2pm by lysine in these precursors had little effect on their immediate subsequent use in the formation of peptidoglycan lipid intermediates. As demonstrated earlier with in vitro assays, the MurF, MraY, and MurG enzymes which catalyze these reactions utilized these alternative substrates with comparable kinetics (1, 12, 31). The rapid recruitment of lysine into the macromolecule (50% of A2pm residues were replaced by lysine within one generation time) was consistent with this finding. Since the internal production of free A2pm was in theory not altered, the extent of incorporation of lysine into the macromolecule should be determined by the relative abundances of the two MurE enzymes, the Km values and the respective pool levels of the two substrates A2pm and lysine. As previously shown for some A2pm analogs which poorly supported the growth of A2pm auxotrophs and lead to morphological alterations and lysis (7), the penicillin-sensitive transpeptidation reactions involved in septation were clearly the critical step for proper growth with an A2pm analog. Lysine can only utilize the E. coli pathway if the appropriate MurE ligase is supplied, but it is unable to fulfill the final essential role of A2pm to ensure peptidoglycan cross-linking. This explains the toxic effect of a large incorporation of this amino acid in the macromolecule. Most likely a lower expression of S. aureus murE gene (a reduced level of lysine incorporated) could be tolerated by E. coli cells, and there is probably a critical ratio between A2pm and lysine that is compatible with cell integrity.
ACKNOWLEDGMENTS
This work was supported by a grant from the Centre National de la Recherche Scientifique (EP1088) and grant “Biotechnologies” from the Ministère de l’Education Nationale de la Recherche et de la Technologie (97.C.0177) to the laboratory in Orsay and by a grant to I. Chopra from SmithKline Beecham Pharmaceuticals.
REFERENCES
- 1.Anderson M S, Eveland S S, Onishi H R, Pompliano D L. Kinetic mechanism of the Escherichia coli UDP-MurNAc-tripeptide d-alanyl-d-alanine-adding enzyme: use of a glutathione S-transferase fusion. Biochemistry. 1996;35:16264–16269. doi: 10.1021/bi961872+. [DOI] [PubMed] [Google Scholar]
- 2.Auger A, Martin L, Bertrand J, Ferrari P, Fanchon E, Vaganay S, Pétillot Y, van Heijenoort J, Blanot D, Dideberg O. Large-scale preparation, purification, and cristallization of UDP-N-acetylmuramoyl-l-alanine:d-glutamate ligase from Escherichia coli. Protein Expr Purif. 1998;13:23–29. doi: 10.1006/prep.1997.0850. [DOI] [PubMed] [Google Scholar]
- 3.Auger G, van Heijenoort J, Vederas J C, Blanot D. Effect of analogs of diaminopimelic acid on the meso-diaminopimelate-adding enzyme from Escherichia coli. FEBS Lett. 1996;391:171–174. doi: 10.1016/0014-5793(96)00619-9. [DOI] [PubMed] [Google Scholar]
- 4.Braun V, Rehn K. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem. 1969;10:426–438. doi: 10.1111/j.1432-1033.1969.tb00707.x. [DOI] [PubMed] [Google Scholar]
- 5.Bugg T D H, Dutka-Malen S, Arthur M, Courvalin P, Walsh C T. Identification of vancomycin resistance protein VanA as a d-alanine:d-alanine ligase of altered substrate specificity. Biochemistry. 1991;30:2017–2021. doi: 10.1021/bi00222a002. [DOI] [PubMed] [Google Scholar]
- 6.Caparrós M, Pisabarro A G, de Pedro M A. Effect of d-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J Bacteriol. 1992;174:5549–5559. doi: 10.1128/jb.174.17.5549-5559.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaloupka J, Strnadová M, Caslavská J, Vereš K. Growth and cell division of Escherichia coli 173-25 in the presence of some analogs of diaminopimelic acid. Z Allg Mikrobiol. 1974;14:283–296. doi: 10.1002/jobm.3630140403. [DOI] [PubMed] [Google Scholar]
- 8.de Jonge B L M, Chang Y-S, Gage D, Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. J Biol Chem. 1992;267:11248–11254. [PubMed] [Google Scholar]
- 9.Flouret B, Mengin-Lecreulx D, van Heijenoort J. Reverse-phase high pressure liquid chromatography of uridine diphosphate N-acetylmuramyl peptide precursors of bacterial cell wall peptidoglycan. Anal Biochem. 1981;114:59–63. doi: 10.1016/0003-2697(81)90451-6. [DOI] [PubMed] [Google Scholar]
- 10.Glauner B. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal Biochem. 1988;172:451–464. doi: 10.1016/0003-2697(88)90468-x. [DOI] [PubMed] [Google Scholar]
- 11.Glauner B, Höltje J-V, Schwarz U. The composition of the murein from Escherichia coli. J Biol Chem. 1988;263:10088–10095. [PubMed] [Google Scholar]
- 12.Hammes W P, Neuhaus F C. On the specificity of phospho-N-acetylmuramyl-pentapeptide translocase. The peptide subunit of uridine diphosphate-N-acetylmuramyl-pentapeptide. J Biol Chem. 1974;249:3140–3150. [PubMed] [Google Scholar]
- 13.Ito E, Strominger J L. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. III. Purification and properties of l-lysine-adding enzyme. J Biol Chem. 1964;239:210–214. [PubMed] [Google Scholar]
- 14.Ito E, Strominger J L. Enzymatic synthesis of the peptide in bacterial uridine nucleotides. VII. Comparative biochemistry. J Biol Chem. 1973;248:3131–3136. [PubMed] [Google Scholar]
- 15.Labischinski H, Maidhof H. Bacterial peptidoglycan: overview and evolving concepts. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. New comprehensive biochemistry. Vol. 27. Amsterdam, The Netherlands: Elsevier Science B.V.; 1994. pp. 23–38. [Google Scholar]
- 16.Laemmli U K, Favre M. Maturation of the head of bacteriophage T4. J Mol Biol. 1973;80:575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- 17.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 18.Mengin-Lecreulx D, Blanot D, van Heijenoort J. Replacement of diaminopimelic acid by cystathionine or lanthionine in the peptidoglycan of Escherichia coli. J Bacteriol. 1994;176:4321–4327. doi: 10.1128/jb.176.14.4321-4327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Cytoplasmic steps of peptidoglycan synthesis in Escherichia coli. J Bacteriol. 1982;151:1109–1117. doi: 10.1128/jb.151.3.1109-1117.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mengin-Lecreulx D, Flouret B, van Heijenoort J. Pool levels of UDP-N-acetylglucosamine and UDP-N-acetylglucosamine-enolpyruvate in Escherichia coli and correlation with peptidoglycan synthesis. J Bacteriol. 1983;154:1284–1290. doi: 10.1128/jb.154.3.1284-1290.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mengin-Lecreulx D, Michaud C, Richaud C, Blanot D, van Heijenoort J. Incorporation of ll-diaminopimelic acid into peptidoglycan of Escherichia coli mutants lacking diaminopimelate epimerase encoded by dapF. J Bacteriol. 1988;170:2031–2039. doi: 10.1128/jb.170.5.2031-2039.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mengin-Lecreulx D, van Heijenoort J. Effect of growth conditions on peptidoglycan content and cytoplasmic steps of its biosynthesis in Escherichia coli. J Bacteriol. 1985;163:208–212. doi: 10.1128/jb.163.1.208-212.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michaud C, Blanot D, Flouret B, van Heijenoort J. Partial purification and specificity studies of the d-glutamate-adding and d-alanyl-d-alanine-adding enzymes from Escherichia coli. Eur J Biochem. 1987;166:631–637. doi: 10.1111/j.1432-1033.1987.tb13560.x. [DOI] [PubMed] [Google Scholar]
- 24.Michaud C, Mengin-Lecreulx D, van Heijenoort J, Blanot D. Over-production, purification and properties of the uridine-diphosphate-N-acetylmuramoyl-l-alanyl-d-glutamate: meso-2,6-diaminopimelate ligase from Escherichia coli. Eur J Biochem. 1990;194:853–861. doi: 10.1111/j.1432-1033.1990.tb19479.x. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Mizuno Y, Ito E. Purification and properties of uridine diphosphate N-acetylmuramyl-l-alanyl-d-glutamate: meso-2,6-diaminopimelate ligase. J Biol Chem. 1968;243:2665–2672. [PubMed] [Google Scholar]
- 27.Patte J-C. Diaminopimelate and lysine. In: Herrmann K M, Somerville R L, editors. Amino acids: biosynthesis and genetic regulation. Reading, Mass: Addison-Wesley Publishing Co.; 1983. pp. 213–228. [Google Scholar]
- 28.Richaud C, Mengin-Lecreulx D, Pochet S, Johnson E J, Cohen G N, Marlière P. Directed evolution of biosynthetic pathways. Recruitment of cysteine thioethers for constructing the cell wall of Escherichia coli. J Biol Chem. 1993;268:26827–26835. [PubMed] [Google Scholar]
- 29.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 30.Schleifer K H, Kandler O. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev. 1972;36:407–477. doi: 10.1128/br.36.4.407-477.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taku A, Fan D P. Purification and properties of a protein factor stimulating peptidoglycan synthesis in toluene- and LiCl-treated Bacillus megaterium cells. J Biol Chem. 1976;251:1889–1895. [PubMed] [Google Scholar]
- 32.van Heijenoort J. Murein synthesis. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1025–1034. [Google Scholar]
- 33.van Heijenoort J, Bricas E. Contribution à l’étude des isomères de l’acide α,α′-diaminopimélique. Bull Soc Chim Fr. 1968;7:2828–2831. [Google Scholar]