Abstract
Semiconductor-based biointerfaces are typically established either on the surface of the plasma membrane or within the cytoplasm. In Gram-negative bacteria, the periplasmic space, characterized by its confinement and the presence of numerous enzymes and peptidoglycans, offers additional opportunities for biomineralization, allowing for nongenetic modulation interfaces. We demonstrate semiconductor nanocluster precipitation containing single- and multiple-metal elements within the periplasm, as observed through various electron- and x-ray-based imaging techniques. The periplasmic semiconductors are metastable and display defect-dominant fluorescent properties. Unexpectedly, the defect-rich (i.e., the low-grade) semiconductor nanoclusters produced in situ can still increase adenosine triphosphate levels and malate production when coupled with photosensitization. We expand the sustainability levels of the biohybrid system to include reducing heavy metals at the primary level, building living bioreactors at the secondary level, and creating semi-artificial photosynthesis at the tertiary level. The biomineralization-enabled periplasmic biohybrids have the potential to serve as defect-tolerant platforms for diverse sustainable applications.
Periplasmic semiconductors yield optically-active biohybrids.
INTRODUCTION
Biomineralization involves the deposition of inorganic species around biological cells and tissues, leading to the formation of soft-hard composites (Fig. 1A) (1–4). These biologically produced composites typically exhibit multifunctionality, where both the hard and soft components have resilience and adaptability (5). Fish scales, for example, are macroscopic layers of the exoskeleton that are attached to the epidermis and provide mechanical protection for the soft interior and locomotion. Coccoliths are produced by unicellular plankton and serve as a protective exoskeleton for the microorganism. In natural biomineralized cells and tissues, inorganic ions can be extracted from the biological or chemical environment, resulting in the construction of multifunctional exoskeletons (Fig. 1A). Aside from fish and plankton, bacteria are another type of aquatic organism that can harvest metal ions from the environment and produce functional materials.
Fig. 1. Periplasmic biomineralization would provide biointerfaces for modulation and sustainability.
(A) An illustration of aquatic and biomineralized soft-hard composites with a wide range of length scales and a variety of structures and functions. Examples include fish scales, coccoliths, and the proposed nanostructured exoskeletons in the periplasm of Gram-negative bacteria. The scale bars are only representative. (B) Schematic illustrating the synthesis of semiconductor nanoclusters within the periplasm of Gram-negative bacteria. The periplasm is the space between the inner membrane (IM) and the outer membrane (OM). (C to E) Electron and x-ray microscopy images of the biohybrids. Pseudo-colored scanning electron microscope (SEM) image (C) showing E. coli (green) with extensive amounts of fimbriae (purple). STEM image (D) and energy dispersive x-ray spectroscopy (EDS) mapping (E) showing that bacterial cell can mineralize semiconductor clusters composed of multiple elements, including Cd, Pb, and Hg, within the periplasm in single bacterial cell. (F) We demonstrate that our biohybrid system with periplasmic biomineralization could enable metal removal, as well as semi-artificial photosynthesis, and construct a living reactor. The biomineralized semiconductor nanoclusters may couple the photoelectrons with the microbial metabolism to enhance the biosynthesis under light.
Materials-based microbial interfaces are generally established either on the cell surface or within the cytoplasm of the microbe. These interfaces have yielded innovations in diverse areas such as semi-artificial photosynthesis (6–15), biological modulation (16–24), microbial fuel cells (25, 26), and water disinfection (27). Within Gram-negative bacteria, the periplasmic space (Fig. 1B) is defined by its spatial restriction and the substantial presence of various enzymes and peptidoglycans (28, 29). These characteristics provide prospects for biomineralization, thereby permitting interfaces for nongenetic modulation (30–32). In addition, in Gram-negative bacteria, the electron transport chain is bound to the inner membrane (IM) and is in close proximity to the periplasm (33). We believe that building such a periplasmic biointerface might offer a worthy category of photosynthetic semiconductor-based biohybrids for solar-driven biocatalysis and sustainability (34–36). In addition, semiconductors could provide highly adjustable synthesis and, thus, controllable biocompatibility, which may effectively pair with bacterial components and serve as a source of electrons for metabolism (36). While there are reports of metallic nanoparticle synthesis in the periplasm, semiconductor-based biointerfaces in periplasm have been largely underexplored, especially for biological modulation and multilevel sustainability (37–40).
Here, we demonstrate a nongenetic method for building periplasmic biohybrids by mineralizing metastable semiconductors to create nanostructured “exoskeletons” and establish semiconductor-based periplasmic interfaces toward sustainable applications. We introduced metal ions and cysteine to Escherichia coli culture to mineralize semiconductors such as CdS, one of the most-studied optically active materials (36), in the periplasm. The selection of E. coli was based on the fact that it is one of the most studied organisms on earth. A number of favorable characteristics (41), including genetic tractability, favorable growth conditions, well-characterized biochemistry and physiology, and the availability of genetic manipulation tools, make E. coli an ideal platform for periplasm-based biointerface. The CdS nanoclusters exhibit low crystallinity and are stabilized by the peptidoglycan matrix of periplasms, thus providing a “softer” interface with bacterial cells (Fig. 1B). We also examined the ability of periplasmic semiconductor clusters to couple photosensitization with bacterial metabolism to enhance intracellular biosynthesis and solar-to-chemical production and explored the underneath mechanism with both material and biological characterization, including scanning transmission electron microscopy (STEM), three-dimensional (3D) x-ray fluorescence tomography, microspectrofluorometry, x-ray powder diffraction (XRD), inductively coupled plasma mass spectrometry (ICP-MS), transcriptome, knockout mutants, and others. The biohybrid displays a higher density of fimbriae following semiconductor cluster mineralization (Fig. 1C), and such a biohybrid is also capable of mineralizing multiple metal elements to form “high-entropy” semiconductor clusters (Fig. 1, D and E). Besides removing heavy metal in flask culture, the production of periplasmic hybrids can be performed in a continuous bioreactor, and the produced biohybrids can couple light for semi-artificial photosynthesis. The key discoveries and corresponding data are summarized in table S1. These results indicate that the periplasm, an underexplored subcellular space, is an attractive substrate for constructing semiconductor-based biointerfaces for a wide range of applications, including water purification, manufacturing of living bioreactors, and semi-artificial photosynthesis for biological modulation and sustainability (Fig. 1F).
RESULTS
Cysteine can facilitate biomineralization and form microbial biohybrids
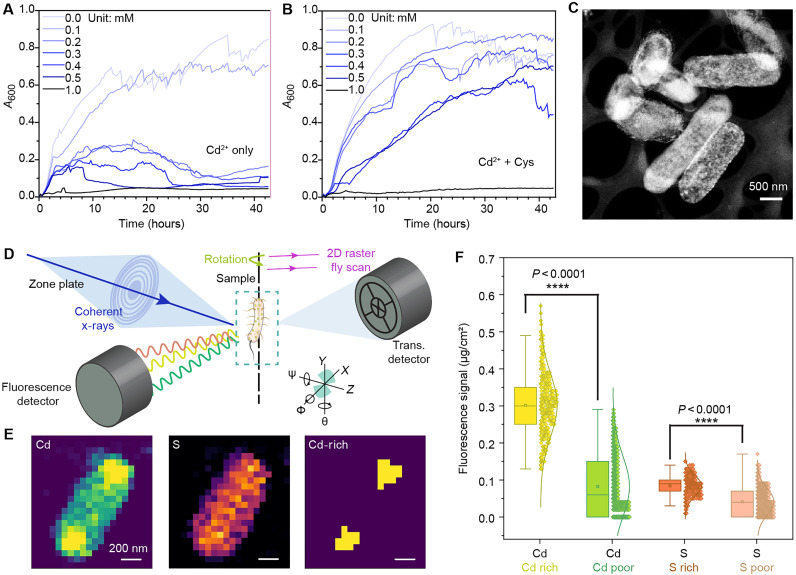
Heavy metal ions such as Cd2+ could be toxic to bacterial cells. We confirmed that E. coli growth in the minimal medium (M9) was notably suppressed in the absence of cysteine at Cd2+ concentrations greater than 0.1 mM (Fig. 2A). The M9 medium is chosen because of its definite components, very low autofluorescence, and low absorbance (42). The E. coli, however, were able to tolerate concentrations of Cd2+ as high as 0.3 mM in the presence of cysteine (1 mM; Fig. 2B). Using STEM, we examined images of E. coli cells grown in a medium containing both Cd2+ and cysteine, and it showed a high density of biomineralization (Fig. 2C). The E. coli also grew a substantial number of fimbriae over time, which is confirmed by scanning electron microscope (SEM) images (fig. S1). In the absence of cysteine or Cd2+, no biomineralization or fimbria formation was observed (figs. S2 and S3). Furthermore, we used ICP-MS to study the metal precipitation and found that Cd2+ cannot be removed without bacteria or cysteine, while in the presence of both cysteine and E. coli, 99.28% of Cd2+ were eliminated (fig. S4). Mixing Cd2+ and cysteine without the presence of bacterial cells cannot produce CdS as well (fig. S5). These results suggest that the metal ions were actively removed from the medium in the presence of cysteine and were used by the bacteria for biomineralization. Notably, metal ions, cysteine, and bacteria are all essential for this process.
Fig. 2. Cysteine can facilitate biomineralization and form microbial biohybrids.
(A) Optical density measurements showing that in the absence of Cys, E. coli growth is severely inhibited at Cd2+ concentrations >0.1 mM. (B) Optical density measurements showing that in the presence of Cys, E. coli cells can tolerate Cd2+ concentrations up to 0.3 mM with little adverse effect on growth. The cysteine concentration is fixed at 1 mM. (C) STEM image indicating the formation of biohybrids with mineralized aggregates. Scale bar, 500 nm. (D) Schematic of the synchrotron-based, 3D x-ray fluorescence tomography experiment. Using a zone plate, a monochromatized x-ray beam is focused on the sample. While the sample is raster-scanned, x-ray fluorescence spectra are recorded, forming 2D elemental maps and 3D reconstructions in tomography mode. Using a quadrant photodiode, differential phase contrast images are generated from transmission signals. (E) Selected slice of the 3D x-ray fluorescence tomography reconstruction showing the presence of both the Cd and S elements within an intact bacterial cell. In each virtual slice, Cd-rich regions are identified when pixel intensity is beyond two times of SD from the mean in each virtual slice. Scale bars, 200 nm. (F) Boxplot showing the correlation between the distributions of Cd and S in 3D reconstruction slices. The data are generated from multiple slices in one 3D x-ray fluorescence tomography (check fig. S6). Boxes bind interquartile range (IQR) divided by the median; whiskers extend 1.5 ± IQR. All data points are plotted (n ≥ 380). A two-tailed unpaired t test is used to determine P values. A600, absorbance at 600 nm.
While STEM suggested biomineralization in the aggregate form, we cannot exclude the possibility of metal ions being immobilized at the molecular level within the cells. Next, we used synchrotron-based, 3D x-ray fluorescence tomography (Fig. 2D) on the biohybrids, a method sensitive enough to establish position correlations between various elements (Fig. 2, D to F, and fig. S6). The following observations were noteworthy. First, the results confirmed the presence of Cd and S elements within the cell (Fig. 2E and figs. S7 and S8), indicating the efficient uptake of Cd2+ and likely the cysteine by E. coli. Second, after determining a threshold for Cd/S-rich regions (i.e., areas with pixel intensity beyond two times of SD from the mean in each virtual slice), we found a tendency for Cd/S-rich areas to be located in the outer regions of the bacteria (Fig. 2E, right), possibly associated with periplasm- or cell wall–bound aggregates. Third, the estimated molar ratio (Fig. 2F) of Cd/S in the Cd/S-rich region is 1.02, which happens to be the same stoichiometry of Cd/S in CdS nanoparticles. This might indicate that Cd may be primarily bonded to S to form CdS nanoparticles in the Cd-rich region.
Biomineralization takes place in the periplasm
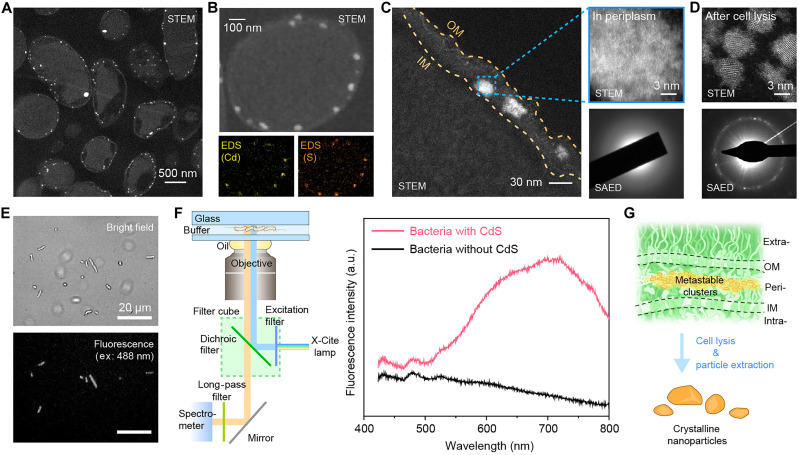
The x-ray fluorescence tomography indicates that the deposition location of CdS aggregates exhibits a preference. To gain a more comprehensive understanding of the localization of Cd/S aggregates within the bacterial cells, we prepared cross-sectional samples of biohybrids through microtoming. STEM images of cross-sectional samples (Fig. 3A and fig. S9A) revealed that the biomineralized Cd/S aggregates had an average size of ~29.3 nm (fig. S9B), and they were primarily located between the IMs and the outer membranes (OMs), which correspond to periplasmic space (fig. S9C). STEM–energy dispersive x-ray spectroscopy (STEM-EDS) mapping showed that the aggregates were composed of Cd and S elements (Fig. 3B and fig. S10), whose locations are highly correlated. STEM images and selected-area electron diffraction (SAED) of the individual Cd/S aggregates show low-crystallinity or almost amorphous structures (Fig. 3C), suggesting that the building blocks of biominerals are nanoclusters. Furthermore, after collecting the aggregates by cell lysis and centrifugation, we found that the biominerals dissociated into smaller particles with an average size of ~6.02 nm (Fig. 3D and figs. S11 and S12), suggesting that the associations between the nanoclusters in the periplasm are largely due to the peptidoglycan matrix (i.e., the biomineralization template). Notably, the removal of the periplasmic components from the aggregates increases the crystallinity and forms Zincblende-phase crystallites (Fig. 3D and fig. S13), likely through oriented attachment or other nonclassical crystallization pathways (43). These observations, in conjunction with the Cd/S ratio provided by x-ray fluorescent tomography (Fig. 2F), indicate that CdS is the primary material mineralized in the periplasm. The CdS nanoclusters in bacteria give the biohybrids fluorescence properties, as indicated by fluorescence imaging (Fig. 3E) and microspectrofluorometry measurements from a few cells (Fig. 3F). Biohybrids show a defect-dominant fluorescence spectrum, characterized by a broad distribution of emissions around ~700 nm, which differs notably from the band edge emission of bulk CdS (i.e., ~510 nm). Together, these results suggest that CdS nanoclusters mineralized in the periplasm are metastable and mainly disordered and defect-rich aggregates (Fig. 3G).
Fig. 3. Biomineralization occurs in the periplasm, and the nanoclusters are metastable.
(A) Cross-sectional STEM images showing that the aggregates are predominantly located between the IM and OM. Scale bar, 500 nm. (B) STEM image and EDS maps confirming the Cd and S composition for the aggregates. Scale bar, 100 nm. (C) Zoom-in STEM images and SAED showing that the aggregates display low crystallinity and are nanoclusters. Scale bars, 30 nm and 3 nm. (D) STEM image and SAED of the minerals (with TEM) after cell lysis and particle extraction, showing smaller particle sizes and improved crystallinity. Scale bar, 3 nm. (E) Bright-field and fluorescence optical microscopy images of the biohybrids (excitation wavelength, 488 nm). Scale bars, 20 μm. (F) Ultraviolet-induced microspectrofluorometry spectra showing that the bacteria with CdS nanoclusters have defect-dominant fluorescent properties with a broad emission peak. (G) A schematic showing the transformation of low-crystallinity nanoclusters to high-crystallinity nanoparticles upon periplasmic material removal, suggesting the metastable nature of the biomineralized nanoclusters. a.u. arbitrary units.
Biomineralization is mediated by an H2S-producing pathway
The bacteria must be alive for the CdS precipitation to occur (fig. S14), suggesting that both the active cellular behavior (e.g., enzymatic activities) and the passive physiochemical properties of the periplasm are essential for biomineralization. To probe the potential cellular processes, we performed a transcriptomic study on Cd2+/cysteine-treated E. coli samples (with CdS nanocluster formation) and the control Cd2+-treated E. coli samples (without CdS nanocluster formation; figs. S15 and S16). We found that several key genes related to H2S production, including cysK, dcyD, cysM, metB, metC, and sufS, were up-regulated in Cd2+/cysteine-treated samples compared to Cd2+-treated samples (Fig. 4A and fig. S17A). The cysK, dcyD, cysM, and metC genes are directly involved in different biological pathways that convert cysteine into hydrogen sulfide (H2S; Fig. 4B and fig. S18). The additional quantitative polymerase chain reaction (qPCR) experiments confirmed that cysteine alone is capable of up-regulating genes related to H2S production, and the presence of Cd2+ in conjunction with cysteine appears to have a synergistic effect, further enhancing the up-regulation of specific genes. (fig. S19). Notably, we conducted knockout experiments for several genes involved in H2S production, specifically targeting cysK, cysM, and metC. The mutants’ experiments suggest functional redundancy of multiple genes involved in H2S synthesis (fig. S20). The up-regulation of efflux-related genes in transcriptomic study, including tolC, cueO, and macA (Fig. 4A and fig. S17B), suggested the efflux of Cd2+ into the periplasmic space following uptake by the E. coli cells (Fig. 4B) (44–46). H2S is a highly lipophilic molecule that freely penetrates the cell membrane (47, 48). To validate the ability to generate H2S, we conducted experiments with multiple benchmark samples to measure H2S production following a reported methodology (49). We discovered that cysteine is essential for detectable H2S production over a 24-hour experiment (fig. S21). In the presence of cysteine, H2S became detectable after 1 hour, whereas it took more than 2 hours for the sample containing both Cd2+ and cysteine, and the group with both cysteine and Cd2+ produced less detectable H2S over a 24-hour experiment (fig. S22). The H2S produced in Cd2+ and cysteine might be used for the biomineralization of Cd2+ within the bacterial cells (50, 51), resulting in reduced detectable H2S within the microbial culture. Taking into account all the aforementioned data, it strongly indicates that the reaction between H2S and Cd2+ for the precipitation of CdS nanoclusters primarily occurs within the periplasmic space. (Fig. 4B); this is further supported by the up-regulation of multiple genes encoding proteins that are typically located in the periplasmic space (fig. S23).
Fig. 4. Biomineralization is driven by enzymatic activities, and it can process with multi-elements and flow-based systems.
(A) Heatmap of transcriptomic study confirming the up-regulation of genes related to H2S production and ion efflux pumps. E_1, E_2, and E_3 represent three independent experimental groups, while C_1, C_2, and C_3 are the control groups. The number of the color bar represents fragments per kilobase of exon per million mapped fragments (FPKM). (B) Schematic illustrating how E. coli synthesizes the CdS nanoclusters in the periplasm. (C) Schematic showing that E. coli can use different heavy metals (Mn+), including Cd2+, Pb2+, and Hg2+, from flask cultures to form metal sulfides. (D) Plot summarizing individual heavy metal ion removal upon biomineralization. The metal conversion rates of Cd2+, Pb2+, and Hg2+ (0.3 mM each) are 99.28%, 94.72%, and 95.68%, respectively. The cysteine concentration is fixed at 1 mM. The data points represent means ± SD (n = 4). (E) Plot illustrating the simultaneous removal of heavy metal ions upon multiplex biomineralization. The metal conversion rates of Cd2+, Pb2+, and Hg2+ (0.1 mM each) are 98.91%, 98.43%, and 97.10%, respectively. The cysteine concentration is fixed at 1 mM. The data points represent means ± SD (n = 4). (F) STEM image and EDS map showing that E. coli can biosynthesize CdxPbyHgzS (x + y + z ~ 1), as the locations for different elements are highly correlated in space. Scale bar, 200 nm. (G) Schematic showing a process of forming high-entropy nanoclusters through periplasm-supported biomineralization. (H) Schematic illustrating biomineralization over a replaceable living membrane, composed of E. coli and filter paper, in a continuous bioprocessing reactor. The solid or dashed arrows suggest the flow pattern throughout the bioreactor and its accessories. The green matrix beneath the bacteria is filter. (I) Plot showing the metal conversion of Cd2+, Pb2+, and Hg2+ in the continuous bioprocessing reactor. The semitransparent background shows the actual 3D design of the bioreactor. The data points represent means ± SD (n = 3).
H2S can react with Cd2+ to precipitate CdS particles in various biological systems, such as bacteria (50, 51), yeast (49), and fungi (52). Given the fact that bacteria must be metabolically active for the biomineralization to occur (fig. S14) and the transcriptomic results (Fig. 4A and figs. S15 to S18 and S23), we conclude in this case that (i) the cysteine can be converted into H2S and (ii) Cd2+ may enter the cell and even reach the cytoplasm, but they mainly interact with H2S to form CdS nanoclusters within the periplasmic space because of the efflux (Fig. 4B).
In addition, we discovered that the formation of CdS could induce high oxidative stress, as evidenced by the up-regulation of dps, gor, grxB, and oxyR genes under the OxyR regulon (fig. S17C) and multiple oxidative stress-responsive genes, including msrA, sodA, sodB, sodC, sufA, sufD, uspE, ygiW, and yodD (fig. S17D). The induced oxidative stress could induce the fimbria and pilus formation (53, 54), as confirmed with the major fimbrial subunit gene fimA (14.46 fold) and major curli subunit gene csgA (3.36 fold), corresponding to the fimbria formation (Fig. 1E and fig. S17E).
Biomineralization is compatible with multi-elements and flow-based systems
We used the flask culture to further extend the biomineralization using different metal ions (Fig. 4C). The bacteria can use most of Cd2+ (99.28%), Pb2+ (94.72%), and Hg2+ (95.68%) within 24 hours for biohybrids synthesis (Fig. 4D), and the concentrations of metals can be as high as 0.3 mM. Using cross-sectional STEM images and EDS maps, we confirmed the presence of PbS (fig. S24A) and HgS (fig. S24B) aggregates within the periplasm in bacterial cells. Similar to what we observed for metastable CdS (Fig. 3D and fig. S11), these biosynthetic PbS and HgS aggregates can also be extracted from the periplasm and transformed into crystalline and smaller nanocrystals (figs. S11 to S13 and S25).
Besides single–metal element sulfide formation, we also explored multi–metal element biomineralization simultaneously. At individual ion concentration of 0.1 mM, 98% of Cd2+, 98% of Pb2+, and 97% of Hg2+ can be simultaneously removed from the mixture within 24 hours (Fig. 4E). The slight difference in removal efficiency between Cd2+, Pb2+, and Hg2+ may be due to the different affinity of E. coli cells for each metal ion (55). STEM imaging (Fig. 4F), EDS mapping (Fig. 4F), and EDS spectrum analysis (fig. S26) confirm the findings. The different ions are mainly colocalized (i.e., precipitating as CdxPbyHgzS, with x + y + z ~ 1) in the periplasm (Fig. 4F), as opposed to forming separate sulfide nanoclusters (i.e., precipitating as CdS, PbS, and HgS in different periplasmic regions). The morphologies of bacterial cell are similar upon both single-metal and multiple-metal ion precipitations (Figs. 3B and 4F and fig. S27). This suggests that a single bacterium can simultaneously use Cd2+, Pb2+, and Hg2+ to form high-entropy (56) sulfide aggregates with multiple elements simultaneously (Fig. 4G).
Besides flask culture, we have also constructed replaceable membranes that contain living bacteria and filter membranes and mounted them onto a 3D printed bioreactor for continuous processing (Figs. 4, H and I, and fig. S28). The bioreactor was supplied with 200 ml of M9 medium containing Cd2+, Pb2+, and Hg2+ (0.1 mM each), and Cd2+ and Pb2+ had already reached 79.1% and 91.2% utilization efficiency after 2 hours, respectively. After 24 hours, the metal conversation rate of Cd2+, Pb2+, and Hg2+ reached 98.7%, 98.1%, and 87.3%, respectively (Fig. 4I). Without further optimization, this small living composite membrane (diameter, 25 mm) could process up to 200 ml of medium for biomineralization. The continuous system (Fig. 4H, left) may be further scaled up and integrated with programmable and self-regulating systems for application in modulation biointerface and synthetic biology (57).
Optically modulate the periplasm-based biointerfaces for semi-artificial photosynthesis
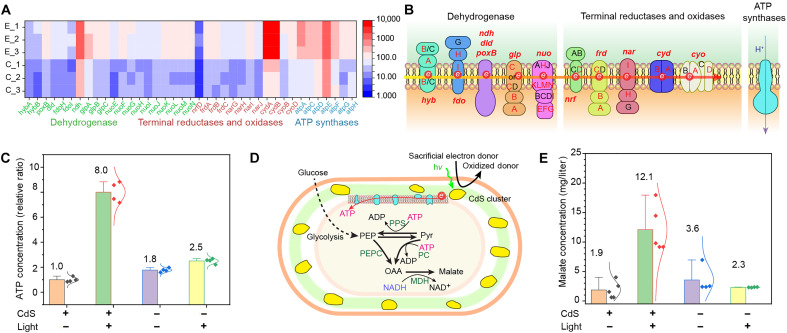
The transcriptomic studies, in addition to providing information related to H2S production and oxidative stress (Fig. 4A), show the up-regulation of dehydrogenase genes in the presence of periplasmic CdS, including expression of formate dehydrogenases, reduced form of nicotinamide adenine dinucleotide dehydrogenases, glyceraldehyde-3-phosphate dehydrogenases, pyruvate oxidase, d-lactate dehydrogenase, and glucose dehydrogenase (Fig. 5, A and B, and fig. S29A), as well as up-regulation of terminal reductase and oxidase genes (Fig. 5, A and B, and fig. S29B). Moreover, periplasmic CdS biomineralization also resulted in the up-regulation of seven of eight adenosine triphosphate (ATP) synthase subunit genes, including atpA, atpC, atpD, atpE, atpF, atpG, and atpH (Fig. 5, A and B, and fig. S29C). On the basis of these up-regulations, an accelerated bacterial respiratory efficiency and a higher rate of ATP synthesis may be possible. Nanocrystalline CdS is known to produce photoelectrons in bilayer lipid membranes (58). Given that the metastable CdS nanoclusters remain optically active (Fig. 3F) and are physically close in proximity to the electron transfer chain (which is located within the IM of E. coli; Fig. 5B), we next explored the possibility of optically modulating microbial processes using the periplasmic biointerfaces.
Fig. 5. Periplasmic semiconductor biointerface may couple with electron transport chain for semi-artificial photosynthesis.
(A) A heatmap from transcriptomic analysis indicates that CdS biomineralization up-regulates genes related to the electron transport chain, including dehydrogenases, terminal reductases and oxidases, and ATP synthases. (B) Schematic showing the electron transport chain with up-regulated genes highlighted in red letters. The red arrow indicates the direction of electron transport, while the purple arrow indicates proton transport. (C) Production of ATP in E. coli cells with biomineralized CdS nanoclusters under light is 8.1 times of the ATP production in cells in the dark. The data points represent means ± SD (n = 4). P values (<0.001) are determined by unpaired two-tailed t test. White light intensity, 6.25 mW/cm2; duration, 24 hours. (D) Schematic of an artificial power system derived from biohybrids to assist ATP production and speed up the production of high-value biochemicals (e.g., malate). PPS, phosphoenolpyruvate synthase; PEPC, PEP carboxylase; PC, pyruvate carboxylase; MDH, malate dehydrogenase. (E) Production of malate in E. coli with CdS nanoclusters increases from 1.9 mg/liter (in the dark) to 12.1 mg/liter (under light). The data points represent means ± SD (n = 5). P values (<0.001) are determined by unpaired two-tailed t test. Notably, E. coli without CdS nanoclusters produces no significant difference in malate production in light or in the dark. P values (0.3121) are determined by unpaired two-tailed t test. The data points represent means ± SD (n = 4). White light intensity, 6.25 mW/cm2; duration, 24 hours.
We incubated E. coli with in situ synthesized CdS nanoclusters under light illumination. We found that ATP level in E. coli under light was 8.1 times of that in the dark condition (Fig. 5C). Repeating the same experiment in E. coli without CdS nanocluster mineralization, the ATP level under light was only 1.4 times that of the dark condition (Fig. 5C). This benchmark experiment suggested that the CdS nanoclusters may contribute to accelerated bacterial respiration and ATP level.
Bioproduction is dependent on the intracellular supply of ATP. Hence, establishing a cell factory that enhances ATP level under artificially controlled conditions is a promising strategy for improving bioproduction yields. To this end, we use the production of malate as a test system to determine whether our biohybrids can enhance biosynthesis under light stimulation. Malate is a four-carbon dicarboxylic acid identified as an optimal building block that could be derived from biomass (59), and it is widely used in fine chemical synthesis precursors and food industry additive (60). The reductive tricarboxylic acid pathway uses ATP to generate malate (61). Specifically, increasing ATP levels may activate the enzymes phosphoenolpyruvate synthase and pyruvate carboxylase, which would then redirect carbon flux from pyruvate to malate (Fig. 5D). The CdS nanocluster biomineralization increases the malate production from 1.87 to 12.10 mg/liter under light stimulation (Fig. 5E), while the biomass of flask cultures [optical density of 600 nm (OD600)] is similar (fig. S30). The light-to-malate conversion efficiency (external quantum yield) is estimated to be about 1.7%. However, the control bacterial group without CdS nanocluster does not have statistically significant differences in malate production with and without light stimulation (Fig. 5E). We also performed benchmark experiments with malate production at various conditions, and it confirmed that cysteine alone cannot enhance malate bioproduction with light stimulation (fig. S31). The results suggest that semiconductor-based biointerfaces in periplasm could be used for semi-artificial photosynthesis. Notably, the fact that the semiconductor nanoclusters are defect-rich and metastable (Fig. 3) suggests that modulation biointerfaces may not necessarily need high-quality components if efficient coupling can be achieved (in this case, via the periplasmic in situ biomineralization), which hold promise for the biohybrid to be used for semi-artificial photosynthesis in a sustainable manner.
DISCUSSION
The intersection of materials science, semiconductor-enabled technologies, and synthetic biology is likely to become a major focus of research in the coming years, as it offers distinctive opportunities for addressing pressing sustainability challenges (62–65). In this work, we show that the periplasm, an underappreciated subcellular region in materials research, can offer a nongenetic pathway for biomineralization of metastable semiconductor nanoclusters. This process can be applied to the construction of a continuous bioreactor based on living materials for multi-element conversion. Under light stimulation, the metastable semiconductor nanoclusters could couple with the electron transport chain to increase ATP level and enhance malate production. By using the biohybrid for semi-artificial photosynthesis, we do not have to discard the defect-rich (i.e., low-grade) biomineralized semiconductors with their cellular hosts. The current demonstration of semiconductor-based biointerfaces in the periplasm and the use of hybrid living materials provide a valuable path toward programmable and adaptable systems where existing cellular chassis and a wide range of nanocluster-cell combinations can be used. Using the E. coli strain as a model, the current system to develop periplasmic biointerface for enhanced solar-to-chemical production might be extended to other bacterial cells to potentially endow additional sustainability level to the bioremediation applications (66). Incorporating semiconductors, additional genetic tools, and existing microbial models, periplasmic biohybrid platforms will enable the preparation of valuable targets in a cost-effective manner.
MATERIALS AND METHODS
Growth of E. coli with CdS nanoclusters
E. coli K12 MG1655 was cultured overnight in LB medium at 37°C. Then, 1% bacterial culture was inoculated in 50 ml fresh LB medium and grown at 37°C for ~3 hours to an OD600 of ~0.6. The cells were centrifuged and resuspended in modified M9 medium containing NH4Cl (1 g/liter), NaCl (0.5 g/liter), tris-HCl buffer (10.99 g/liter; pH 7.5), thiamine (340 mg/liter), 0.4% (v/v) glycerol, glucose (4 g/liter), 2 mM MgSO4, and 0.1 mM CaCl2. In a typical experiment, the E. coli cells were inoculated at 37°C, and 0.3 mM CdCl2 and 1 mM cysteine were added to the culture. The samples were collected for electron imaging, x-ray imaging, RNA sequencing (RNA-seq), and intracellular ATP measurement at specific time points.
Growth of E. coli with different concentrations of CdCl2
E. coli were cultured overnight in LB medium at 37°C. Then, 1% bacterial culture was transferred to 50 ml of fresh LB medium and grown at 37°C for ~3 hours. Cells were centrifuged and resuspended in a modified M9 medium to an OD600 of ~0.1. The E. coli cells were cultured in a 96-well plate reader with or without 1 mM cysteine and CdCl2 at concentrations of 0.0, 0.1, 0.2, 0.3, 0.4, 0.5, and 1.0 mM. The growth of E. coli cells was determined according to the absorbance at 600 nm.
Construction of E. coli mutants
The E. coli mutants were constructed via CRISPR-Cas9 system according to reported protocols (67). The E. coli competent cells containing pCas were prepared following previous literature (68). pTargetT series DNA (pTargetTcysK, pTargetTcysM, pTargetTdcyD, and pTargetTmetC) contains single guide RNA, and donor template DNA was constructed (67). Arabinose (10 mM) was added to the culture for λ-red induction. For electroporation, 50 μl of cells was mixed with 100 ng of pTargetT series DNA; electroporation was done in a 2-mm Gene Pulser cuvette (Bio-Rad) at 2.5 kV, and the product was suspended immediately in 1 ml of ice-cold LB medium. Cells were recovered at 30°C for 1 hour before being spread onto LB agar containing kanamycin (50 mg/liter) and incubated overnight at 30°C. Transformants were identified by colony PCR and DNA sequencing. The pTargetT and pCas plasmid were cured accordingly (67).
Critical point drying
To image the bacterial samples using an electron microscope or x-ray characterization in a vacuum environment, we first performed critical point drying to dehydrate the bacterial samples. The bacterial sample was first fixed using 3% glutaraldehyde in phosphate-buffered saline at 4°C overnight. These samples were then sequentially submerged into an ethanol series (30, 50, 70, 80, 90, 96, and 100%) at 25°C for 10 min each to remove the water content. A critical point dryer (Leica EM CPD300) was used to dry the samples with supercritical carbon dioxide (304.13 K and 73.8 bar). Carbon dioxide was injected at low speed with a delay of 120 s, and it exchanged ethanol at speed 3 for 14 cycles.
Scanning electron microscopy
Bacterial samples or biofilm samples were dried by critical point drying (CPD) and then imaged using SEM (Carl Zeiss, Merlin). All samples were coated with a conductive layer of 10-nm-thick platinum/palladium coating by sputter coating (Agar AGB7341) before SEM imaging at an acceleration voltage of 2 kV. For each sample, a minimum of 10 measurements were taken to confirm the repeatability of the experiments.
X-ray fluorescence microscopy and tomography
Synchrotron-based x-ray fluorescence microscopy and tomography were performed at the Bionanoprobe (69) located at beamline 9-ID-B of the Advanced Photon Source at Argonne National Laboratory to characterize the spatial distribution of CdS nanocluster in the bacterial cells. Bacterial cells with CdS nanocluster were first fixed with 3% glutaraldehyde and then immersed with a series of ethanol solutions (30, 50, 70, 80, 90, 96, and 100%). A critical point dryer (Leica EM CPD300) was used to fully dry the bacterial cells for x-ray imaging. X-ray photons of 10.5 keV were focused using zone plate optics onto a sample with a focal size of ~80 nm. The sample was raster-scanned through the focused x-ray beam in a fly-scan mode with a step size of 60 nm and 50-ms dwell time per pixel. A full spectrum was collected at each pixel to construct 2D elemental maps of a single bacterial cell. A series of such 2D projections were acquired while the sample rotated around a vertical axis from −68° to 63° with respect to the incident x-ray beam with a 1° increment to form a 3D tomography dataset. Per-pixel spectrum fitting and quantification were performed using MAPS software on each projection (70). An AXO standard thin film (AXO products, Dresden, Germany) was used for elemental concentration calibration. Before performing a 3D volume reconstruction, the quantitative 2D maps were further segmented using k-means clustering analysis to extract features of interest. Tomography reconstruction was done using the gridrec algorithm (71) in an open source software named TomoPy (72).
TEM and STEM
The particle suspension was dropped onto copper grids (Lacey Formvar/Carbon; 200 meshes; Ted Pella Inc.) and imaged on TEM FEI 30 with 300-kV acceleration voltage in the Advanced Electron Microscopy Facility at the University of Chicago after it was thoroughly dried. After CPD treatment, bacterial samples were dropped onto copper grids. The sample was imaged under FEI F30 with 300-kV acceleration voltage and JEOL JEM-ARM200CF with 200-kV acceleration voltage in the Electron Microscopy Core at the University of Illinois at Chicago in STEM mode. For bacterial cross-sectional samples, bacterial cells were collected and fixed with glutaraldehyde and paraformaldehyde. The sample was then stained with uranyl acetate, followed by dehydration, infiltration, and polymerization. Sections (thickness, 90 nm) were prepared with Leica EM UC6. Bacterial cross-sectional samples were also imaged under FEI F30 with 300-kV acceleration voltage and JEOL JEM-ARM200CF with 200-kV acceleration voltage in the Electron Microscopy Core at the University of Illinois at Chicago in STEM mode. EDS mapping was collected with JEOL JEM-ARM200C in STEM mode. For each sample, more than 10 imaging measurements were performed to confirm the repeatability of the experiments.
Fluorescence microscopy and microspectrofluorometry
Following bacterial cell culture with or without CdS, bacterial cells are centrifuged and resuspended in a fresh M9 solution to maintain the same fluorescent background. A drop of bacterial suspension was then transferred to the microscope slide and sealed with a glass coverslip and microscope nail polish (Fisher Scientific) to prevent the evaporation of buffer. The fluorescent images of bacteria with CdS were taken with a Nikon eclipse Ti2 inverted microscope with the excitation light at 488 nm. Microspectrofluorometry of bacterial culture was performed on an inverted microscope (Olympus IX71) using an UPlanFL100x/1.30 (Olympus) oil lens. A fluorescent X-Cite lamp (120PC Q, Lumen Dynamics) was used as a light source for excitation. A U-MNUA2 filter cube with BP360-370 excitation filter and DM400 dichroic, but without emission filter, was used for spectrum recording. Residual scattering of excitation wavelength has been filtered out using GG400 colored glass filter (400 nm, long-pass; FGL400S, Thorlabs) before analysis in the spectrometer. Spectra were recorded using a Fergie spectrometer and LightField software (Princeton Instruments). The spectra were corrected after measurement for detector quantum efficiency and dichroic mirror and long-pass filter transmissions using data obtained from technical specifications.
Biomineral extraction
E. coli were cultured with different metallic ions, and the bacterial cells were harvested after 24 hours. The cells were concentrated by centrifugation and resuspended in deionized (DI) water. Cell lysis was performed via sonication in an ice bath for 2 hours with a probe sonicator (Sonics Ultrasonic Vibra-Cell) under pulse mode. The particles were collected by centrifuging the sample at 13,000 rpm for 1 hour. Further surface chemical modification was applied to further disperse the biosynthesized nanoparticles. In a typical experiment, the particles were resuspended with Li2S/formamide solution (5 mg/ml) and sonicated for 15 min to obtain a uniform solution. Then, didodecyldimethylammonium bromide (DDAB)/toluene (50 mg/ml) was added to the top of the solution under continuous stirring. Following the overnight reaction, nanoparticles underwent modification with DDAB in the upper layer of the solution (oil layer). Subsequently, they were collected and subjected to three rounds of thorough washing with acetone and ethanol to remove any excess DDAB. The dispersion was then centrifuged at 10,000 rpm for 10 min to obtain the precipitant, which was later redispersed in toluene. The characterization of the nanoparticles involved the use of TEM, STEM, and XRD.
X-ray powder diffraction
The extracted nanoparticle solution was drop-cast on silica substrates as a thin film. Wide-angle powder x-ray diffraction patterns were collected using a Bruker D8 diffractometer with a Cu Kα x-ray source operating at 40 kV and 40 mA.
RNA extraction and RNA-seq analysis
Cells were cultured in 0.3 mM CdCl2 with or without 1 mM cysteine following the cultivation method described above and collected after 6 hours. The total RNA was extracted using the TRIzol Reagent (Invitrogen, catalog number 15596026), and the quality was determined using the Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). The ribosomal RNA was removed using the I Ribo-Zero Magnetic Kit (Epicenter). The RNA was sheared and reverse-transcribed using random primers to obtain cDNA, which was used for library construction. Libraries were run on a BGISEQ-500 (BGI). All RNA-seq experiments were performed in biological triplicates from distinct samples.
Real-time PCR (qPCR) analysis
The RNA extraction was the same as RNA-seq experiments. The product was quantified via real-time PCR using the CFX96 thermal cycler (Bio-Rad). The reaction mixture (20 μl) contained Power SYBR Green PCR Master Mix (Bio-Rad) and 0.4 mM gene-specific primers (table S1). The PCR parameters were 1 cycle of 95°C for 2 min, followed by 40 cycles of 95°C for 20 s, 60°C for 20 s, and 72°C for 15 s. The expression level of each gene was normalized with the value for the housekeeping gene 16S.
Detection of H2S
We adapted the measurement protocol from previous literature (49). Quantitative H2S gas was monitored using the IFU Hydrogen Sulfide Dräger-Tube (Dräger, Germany). The E. coli strain was cultured overnight in LB medium at 37°C. Then, 1% bacterial culture was further transferred to fresh LB medium and grown at 37°C for about 3 hours. Cells were centrifuged and resuspended with a modified M9 medium to an OD600 of ~0.6. The bacterial cultures were transfer into a seal-flask bottle and cultured under four different conditions, including (i) cysteine + Cd2+, (ii) cysteine only, (iii) Cd2+ only, and (iv) control group without cysteine and Cd2+. A made-to-measure rubber stopper fitted by Dräger-Tube was corked in 125-ml Erlenmeyer flasks containing 40 ml of cultures. To quantify H2S gas production, the numbers on the Dräger-Tube were recorded at specific time points.
Flask culture for multi-element sulfide biosynthesis
E. coli were cultured overnight in LB medium at 37°C. Then, 1% bacterial culture was further transferred to fresh LB medium and grown at 37°C for about 3 hours. Cells were centrifuged and resuspended with a modified M9 medium to an OD600 of ~0.6. The bacterial culture was grown at 37°C with various metal ions (CdCl2, HgCl2, and PbCl2) at a concentration of 0.1 mM with or without 1 mM cysteine. After 12 hours, the cells pellets were collected for nanoparticle analysis, and the supernatants were collected for inductively coupled plasma to evaluate the removal efficiency.
Inductively coupled plasma mass spectrometry
After incubating the bacteria in M9 minimal medium in the presence of heavy metal ions for different times, the heavy metal concentrations were characterized by inductively coupled plasma (Agilent 700). The bacterial suspension is centrifuged at 13,000 rpm for half an hour, and the supernatant is collected for inductively coupled plasma measurement. High-purity concentrated nitric acid (68 w/w%; TraceSELECT) was used to digest the sample and then further diluted using DI water to the proper concentrations. A multi-element standard solution (Sigma-Aldrich, 51844) was used as the standard for all analyses. Four standard samples were prepared, with concentrations of 0, 1, 5, and 25 parts per million (ppm) for each element (Cd and Pb) being analyzed. These standards have the same concentration of nitric acid as the samples. For the analyses of the Hg atoms, 12.5 v/v% of 400-ppm gold (III) chloride trihydrate solution was also added to both standard solution and samples to stabilize Hg atoms during the measurement. All the experiments were repeated at least three times to confirm the repeatability of the experiments.
Continuous bioprocessing reactor
Continuous bioprocessing systems are composed of a replaceable living material membrane, a tower bioreactor, and a cyclic pumping system. The living material membrane was composed of E. coli biofilm on the filter membrane. Specifically, E. coli were cultured overnight in LB medium at 37°C. Then, 1% bacterial culture was further transferred to a 50-ml fresh LB medium and grown at 37°C for about 3 hours. Cells were harvested from shake flask cultures at an OD600 of ~0.6. The cell culture was drop-cast on a filter membrane (0.22-μm mixed cellulose ester membrane; MF-Millipore) and gently vacuum-filtrated to leave a layer of cells on the membrane. The membrane was later transferred onto an LB agar plate and grown at 30°C for 48 hours. The biofilm, together with the filter membrane, served as the living material membrane. The tower bioreactor comprises three portions, and each of them was printed by 3D printing (Ultimaker 3). The living membrane was integrated into the bioreactor before assembling. The structure of the bioreactor is shown in fig. S21. The cyclic pump system was composed of a pump (Fisherbrand Variable-Flow Peristaltic Pumps, 13-876-2) and silicone tubes (Cole-Parmer, 06422-074) to cycle the fluids in the bioreactor. Solution samples containing Cd2+, Pb2+, and Hg2+ with 0.1 mM each were pumped inside the bioreactor and cycled at 37°C. We sampled the cyclic solution at different time points, followed by centrifugation to collect the supernatant for ICP-MS measurements to determine the metal conversion efficiency.
Intracellular ATP assay in E. coli cells
E. coli cells were cultured and resuspended with a modified M9 medium. CdCl2 (0.3 mM) and cysteine (1 mM) were added to the culture, and the culture was grown at 37°C in a shaker at 220 rpm with and without light stimulation (light intensity, 6.25 mW/cm2). The same experiment was repeated for cell cultures without the addition of CdCl2 and cysteine. Intracellular ATP levels in the cells were measured using the Promega ATP assay kit. Specifically, we collected 2 ml of bacterial culture medium and resuspended it in 0.2 ml of distilled water. Samples were boiled at 95°C for 5 min to deactivate ATPases, and the supernatant was collected for ATP measurement. ATP measurement was conducted by following the manufacturer’s instructions. Meanwhile, the intracellular protein concentrations of the collected bacterial samples were measured using a Thermo Fisher Scientific BCA assay kit. ATP reading values were adjusted by protein concentrations to avoid loading amount variance. The intracellular ATP levels were presented as in fold changes compared to samples without light stimulation.
Detection of malate
E. coli cells were cultured overnight in LB medium at 37°C. Then, 1% bacterial culture was inoculated in fresh LB medium and grown at 37°C for ~3 hours to an OD600 of ~0.6. Cells were centrifuged and resuspended in a modified M9 medium to an OD600 of ~0.5; then, 0.3 mM CdCl2 and 1 mM cysteine were added. The E. coli cells were cultured in a photoreaction flask at 37°C at 220 rpm with and without light stimulation (light intensity, 6.25 mW/cm2). After 24 hours of cultivation, the supernatant was filtered and detected by high-performance liquid chromatography (Agilent 1260 Infinity II). Malate was determined with an Aminex HPX-87H column (300 mm by 7.8 mm; Bio-Rad) at a wavelength of 210 nm. The mobile phase was 5 mM H2SO4, and the flow rate was 0.6 ml/min.
External quantum yield calculation
The emission spectrum of the light-emitting diode light was integrated to determine the photon flux. The measured power was 6.25 mW cm−2, and the estimated photon energy absorbed by hybrid cells was about 0.558 μmol hour−1 with consideration of light reflection and absorption by glass vials and cell concentrations (8). The external quantum efficiency may be calculated as
where the [Cmalate]light is the malate production in the medium from light, [Cmalate]dark is the amount in the dark under parallel conditions, Mmalate = 132 g mol−1 is the molar mass of malate, the factor of 3 accounts for 3e−1 molecules of pyruvate to convert to malate, V is the culture medium volume for producing malate, and t is the total illumination time.
Acknowledgments
We thank K. Watters for scientific editing of the manuscript and E. Maxey for beamline support at the Advanced Photon Source. B.T. acknowledges the support from the NSF QuBBE Quantum Leap Challenge Institute (NSF OMA-2121044), the Zhong Ziyi Educational Foundation Award, and the University of Chicago startup grant. This work made use of instruments in the Electron Microscopy Core, Research Resources Center at University of Illinois Chicago. D.V.T. acknowledges support as part of the Center for Advanced Materials for Energy Water Systems (AMEWS), an Energy Frontier Research Center funded by the U.S. Department of Energy (DOE), Office of Science, Basic Energy Sciences (BES). Use of the Advanced Photon Source and the Center for Nanoscale Materials, both U.S. Department of Energy Office of Science User Facilities, was supported by the U.S. Department of Energy, Office of Science, under contract no. DE-AC02-06CH11357. Synchrotron data analysis effort was partially supported by Argonne Laboratory Directed Research and Development (2020-0038).
Author contributions: B.T. supervised the research. Y. Lin, X.G. and B.T. designed the experiments. Y. Lin, X.G., J.S., W.F., J.Y., Y. Luo, S.C., Y.J., H.H., C.Z., F.S., A.P., and D.V.T. contributed to the biohybrid preparation, structural and chemical characterizations, and the biomineralization mechanism studies. Y. Lin, J.S., and J.Y. performed the ATP production experiment, with the results verified by W.F. and X.G. separately. W.F., B.Y., W.X., and X.G. contributed to the malate biosynthesis data. Y. Lin, X.G., J.S., J.Y., and B.T wrote the manuscript and received comments from all the other authors.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Source data are available at https://doi.org/10.5061/dryad.5hqbzkhbm. XRF tomography reconstruction was done using the gridrec algorithm in an open source software named TomoPy: https://tomopy.readthedocs.io/en/latest/about.html.
Supplementary Materials
This PDF file includes:
Table S1
Figs. S1 to S31
REFERENCES AND NOTES
- 1.Reznikov N., Steele J. A. M., Fratzl P., Stevens M. M., A materials science vision of extracellular matrix mineralization. Nat. Rev. Mater. 1, 16041 (2016). [Google Scholar]
- 2.Mann S., Self-assembly and transformation of hybrid nano-objects and nanostructures under equilibrium and non-equilibrium conditions. Nat. Mater. 8, 781–792 (2009). [DOI] [PubMed] [Google Scholar]
- 3.Lin Y., Fang Y., Yue J., Tian B., Soft–hard composites for bioelectric interfaces. Trends Chem. 2, 519–534 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barthelat F., Yin Z., Buehler M. J., Structure and mechanics of interfaces in biological materials. Nat. Rev. Mater. 1, 16007 (2016). [Google Scholar]
- 5.Fang Y., Yang X., Lin Y., Shi J., Prominski A., Clayton C., Ostroff E., Tian B., Dissecting biological and synthetic soft–hard interfaces for tissue-like systems. Chem. Rev. 122, 5233–5276 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakimoto K. K., Wong A. B., Yang P., Self-photosensitization of nonphotosynthetic bacteria for solar-to-chemical production. Science 351, 74–77 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Zhang H., Liu H., Tian Z., Lu D., Yu Y., Cestellos-Blanco S., Sakimoto K. K., Yang P., Bacteria photosensitized by intracellular gold nanoclusters for solar fuel production. Nat. Nanotechnol. 13, 900–905 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Guo J., Suástegui M., Sakimoto K. K., Moody V. M., Xiao G., Nocera D. G., Joshi N. S., Light-driven fine chemical production in yeast biohybrids. Science 362, 813–816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu C., Colón B. C., Ziesack M., Silver P. A., Nocera D. G., Water splitting–biosynthetic system with CO2 reduction efficiencies exceeding photosynthesis. Science 352, 1210–1213 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Su Y., Cestellos-Blanco S., Kim J. M., Shen Y., Kong Q., Lu D., Liu C., Zhang H., Cao Y., Yang P., Close-packed nanowire-bacteria hybrids for efficient solar-driven CO2 fixation. Joule 4, 800–811 (2020). [Google Scholar]
- 11.Kornienko N., Sakimoto K. K., Herlihy D. M., Nguyen S. C., Alivisatos A. P., Harris C. B., Schwartzberg A., Yang P., Spectroscopic elucidation of energy transfer in hybrid inorganic–biological organisms for solar-to-chemical production. Proc. Natl. Acad. Sci. U.S.A. 113, 11750–11755 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu C., Gallagher J. J., Sakimoto K. K., Nichols E. M., Chang C. J., Chang M. C. Y., Yang P., Nanowire–bacteria hybrids for unassisted solar carbon dioxide fixation to value-added chemicals. Nano Lett. 15, 3634–3639 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu C., Colón B. E., Silver P. A., Nocera D. G., Solar-powered CO2 reduction by a hybrid biological | inorganic system. J. Photochem. Photobiol. A. Chem. 358, 411–415 (2018). [Google Scholar]
- 14.Nangle S. N., Sakimoto K. K., Silver P. A., Nocera D. G., Biological-inorganic hybrid systems as a generalized platform for chemical production. Curr. Opin. Chem. Biol. 41, 107–113 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Torella J. P., Gagliardi C. J., Chen J. S., Bediako D. K., Colón B., Way J. C., Silver P. A., Nocera D. G., Efficient solar-to-fuels production from a hybrid microbial–water-splitting catalyst system. Proc. Natl. Acad. Sci. U.S.A. 112, 2337–2342 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song E., Li J., Won S. M., Bai W., Rogers J. A., Materials for flexible bioelectronic systems as chronic neural interfaces. Nat. Mater. 19, 590–603 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Dai Z., Lee A. J., Roberts S., Sysoeva T. A., Huang S., Dzuricky M., Yang X., Zhang X., Liu Z., Chilkoti A., You L., Versatile biomanufacturing through stimulus-responsive cell–material feedback. Nat. Chem. Biol. 15, 1017–1024 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Higgins S. G., Becce M., Belessiotis-Richards A., Seong H., Sero J. E., Stevens M. M., High-aspect-ratio nanostructured surfaces as biological metamaterials. Adv. Mater. 32, 1903862 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen R., Canales A., Anikeeva P., Neural recording and modulation technologies. Nat. Rev. Mater. 2, 16093 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Frank J. A., Antonini M.-J., Anikeeva P., Next-generation interfaces for studying neural function. Nat. Biotechnol. 37, 1013–1023 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maya-Vetencourt J. F., Manfredi G., Mete M., Colombo E., Bramini M., Di Marco S., Shmal D., Mantero G., Dipalo M., Rocchi A., DiFrancesco M. L., Papaleo E. D., Russo A., Barsotti J., Eleftheriou C., Di Maria F., Cossu V., Piazza F., Emionite L., Ticconi F., Marini C., Sambuceti G., Pertile G., Lanzani G., Benfenati F., Subretinally injected semiconducting polymer nanoparticles rescue vision in a rat model of retinal dystrophy. Nat. Nanotechnol. 15, 698–708 (2020). [DOI] [PubMed] [Google Scholar]
- 22.Jiang Y., Carvalho-de-Souza J. L., Wong R. C. S., Luo Z., Isheim D., Zuo X., Nicholls A. W., Jung I. W., Yue J., Liu D.-J., Wang Y., De Andrade V., Xiao X., Navrazhnykh L., Weiss D. E., Wu X., Seidman D. N., Bezanilla F., Tian B., Heterogeneous silicon mesostructures for lipid-supported bioelectric interfaces. Nat. Mater. 15, 1023–1030 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang Y., Li X., Liu B., Yi J., Fang Y., Shi F., Gao X., Sudzilovsky E., Parameswaran R., Koehler K., Nair V., Yue J., Guo K., Fang Y., Tsai H.-M., Freyermuth G., Wong R. C. S., Kao C.-M., Chen C.-T., Nicholls A. W., Wu X., Shepherd G. M. G., Tian B., Rational design of silicon structures for optically controlled multiscale biointerfaces. Nat. Biomed. Eng. 2, 508–521 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Becce M., Klöckner A., Higgins S. G., Penders J., Hachim D., Bashor C. J., Edwards A. M., Stevens M. M., Assessing the impact of silicon nanowires on bacterial transformation and viability of Escherichia coli. J. Mater. Chem. B 9, 4906–4914 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao B., Zhao Z., Peng L., Shiu H.-Y., Ding M., Song F., Guan X., Lee C. K., Huang J., Zhu D., Fu X., Wong G. C. L., Liu C., Nealson K., Weiss P. S., Duan X., Huang Y., Silver nanoparticles boost charge-extraction efficiency in Shewanella microbial fuel cells. Science 373, 1336–1340 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Ding M., Shiu H.-Y., Li S.-L., Lee C. K., Wang G., Wu H., Weiss N. O., Young T. D., Weiss P. S., Wong G. C. L., Nealson K. H., Huang Y., Duan X., Nanoelectronic investigation reveals the electrochemical basis of electrical conductivity in Shewanella and Geobacter. ACS Nano 10, 9919–9926 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Liu C., Kong D., Hsu P.-C., Yuan H., Lee H.-W., Liu Y., Wang H., Wang S., Yan K., Lin D., Maraccini P. A., Parker K. M., Boehm A. B., Cui Y., Rapid water disinfection using vertically aligned MoS2 nanofilms and visible light. Nat. Nanotechnol. 11, 1098–1104 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Prochnow H., Fetz V., Hotop S.-K., García-Rivera M. A., Heumann A., Brönstrup M., Subcellular quantification of uptake in gram-negative bacteria. Anal. Chem. 91, 1863–1872 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Stock J. B., Rauch B., Roseman S., Periplasmic space in Salmonella typhimurium and Escherichia coli. J. Biol. Chem. 252, 7850–7861 (1977). [PubMed] [Google Scholar]
- 30.Tang T.-C., An B., Huang Y., Vasikaran S., Wang Y., Jiang X., Lu T. K., Zhong C., Materials design by synthetic biology. Nat. Rev. Mater. 6, 332–351 (2021). [Google Scholar]
- 31.Tang T.-C., Tham E., Liu X., Yehl K., Rovner A. J., Yuk H., de la Fuente-Nunez C., Isaacs F. J., Zhao X., Lu T. K., Hydrogel-based biocontainment of bacteria for continuous sensing and computation. Nat. Chem. Biol. 17, 724–731 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu Y.-Y., Wang Y.-Z., Fang Z., Shi Y.-T., Cheng Q.-W., Chen Y.-X., Shi W., Yong Y.-C., Single cell electron collectors for highly efficient wiring-up electronic abiotic/biotic interfaces. Nat. Commun. 11, 4087 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaila V. R. I., Wikström M., Architecture of bacterial respiratory chains. Nat. Rev. Microbiol. 19, 319–330 (2021). [DOI] [PubMed] [Google Scholar]
- 34.Wang B., Zeng C., Chu K. H., Wu D., Yip H. Y., Ye L., Wong P. K., Enhanced biological hydrogen production from Escherichia coli with surface precipitated cadmium sulfide nanoparticles. Adv. Energy Mater. 7, 1700611 (2017). [Google Scholar]
- 35.Xiao K., Liang J., Wang X., Hou T., Ren X., Yin P., Ma Z., Zeng C., Gao X., Yu T., Si T., Wang B., Zhong C., Jiang Z., Lee C.-S., Yu J. C., Wong P. K., Panoramic insights into semi-artificial photosynthesis: Origin, development, and future perspective. Energy Environ. Sci. 15, 529–549 (2022). [Google Scholar]
- 36.Cestellos-Blanco S., Zhang H., Kim J. M., Shen Y., Yang P., Photosynthetic semiconductor biohybrids for solar-driven biocatalysis. Nat. Catal. 3, 245–255 (2020). [Google Scholar]
- 37.Konishi Y., Tsukiyama T., Ohno K., Saitoh N., Nomura T., Nagamine S., Intracellular recovery of gold by microbial reduction of AuCl4− ions using the anaerobic bacterium Shewanella algae. Hydrometallurgy 81, 24–29 (2006). [Google Scholar]
- 38.Pugazhenthiran N., Anandan S., Kathiravan G., Udaya Prakash N. K., Crawford S., Ashokkumar M., Microbial synthesis of silver nanoparticles by Bacillus sp. J. Nanopart. Res. 11, 1811–1815 (2009). [Google Scholar]
- 39.Riddin T. L., Govender Y., Gericke M., Whiteley C. G., Two different hydrogenase enzymes from sulphate-reducing bacteria are responsible for the bioreductive mechanism of platinum into nanoparticles. Enzyme Microb. Technol. 45, 267–273 (2009). [Google Scholar]
- 40.Wing-Shan Lin I., Lok C.-N., Che C.-M., Biosynthesis of silver nanoparticles from silver(i) reduction by the periplasmic nitrate reductase c-type cytochrome subunit NapC in a silver-resistant E. coli. Chem. Sci. 5, 3144–3150 (2014). [Google Scholar]
- 41.Pontrelli S., Chiu T.-Y., Lan E. I., Chen F. Y.-H., Chang P., Liao J. C., Escherichia coli as a host for metabolic engineering. Metab. Eng. 50, 16–46 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Pardee A. B., Jacob F., Monod J., The genetic control and cytoplasmic expression of “Inducibility” in the synthesis of β-galactosidase by E. coli. J. Mol. Biol. 1, 165–178 (1959). [Google Scholar]
- 43.De Yoreo J. J., Gilbert P. U. P. A., Sommerdijk N. A. J. M., Penn R. L., Whitelam S., Joester D., Zhang H., Rimer J. D., Navrotsky A., Banfield J. F., Wallace A. F., Michel F. M., Meldrum F. C., Cölfen H., Dove P. M., Crystallization by particle attachment in synthetic, biogenic, and geologic environments. Science 349, aaa6760 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Routh M. D., Zalucki Y., Su C.-C., Long F., Zhang Q., Shafer W. M., Yu E. W., Efflux pumps of the resistance-nodulation-division family: A perspective of their structure, function and regulation in gram-negative bacteria. Adv. Enzymol. Relat. Areas Mol. Biol. 77, 109–146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nies D. H., Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbiol. Rev. 27, 313–339 (2003). [DOI] [PubMed] [Google Scholar]
- 46.Lu S., Zgurskaya H. I., MacA, a periplasmic membrane fusion protein of the macrolide transporter MacAB-TolC, binds lipopolysaccharide core specifically and with high affinity. J. Bacteriol. 195, 4865–4872 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riahi S., Rowley C. N., Why can hydrogen sulfide permeate cell membranes? J. Am. Chem. Soc. 136, 15111–15113 (2014). [DOI] [PubMed] [Google Scholar]
- 48.Mathai J. C., Missner A., Kügler P., Saparov S. M., Zeidel M. L., Lee J. K., Pohl P., No facilitator required for membrane transport of hydrogen sulfide. Proc. Natl. Acad. Sci. U.S.A. 106, 16633–16638 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun G. L., Reynolds E. E., Belcher A. M., Using yeast to sustainably remediate and extract heavy metals from waste waters. Nat. Sustain. 3, 303–311 (2020). [Google Scholar]
- 50.Bang S. W., Clark D. S., Keasling J. D., Engineering hydrogen sulfide production and cadmium removal by expression of the thiosulfate reductase gene (phsABC) from Salmonella enterica serovar typhimurium in Escherichia coli. Appl. Environ. Microbiol. 66, 3939–3944 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakpirom J., Kantachote D., Siripattanakul-Ratpukdi S., McEvoy J., Khan E., Simultaneous bioprecipitation of cadmium to cadmium sulfide nanoparticles and nitrogen fixation by Rhodopseudomonas palustris TN110. Chemosphere 223, 455–464 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Reyes L. R., Gómez I., Garza M. T., Biosynthesis of cadmium sulfide nanoparticles by the fungi Fusarium sp. Int. J. Green Nanotechnol. Biomed. 1, B90–B95 (2009). [Google Scholar]
- 53.Kulkarni R., Antala S., Wang A., Amaral F. E., Rampersaud R., LaRussa S. J., Planet P. J., Ratner A. J., Cigarette smoke increases Staphylococcus aureus biofilm formation via oxidative stress. Infect. Immun. 80, 3804–3811 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geier H., Mostowy S., Cangelosi G. A., Behr M. A., Ford T. E., Autoinducer-2 triggers the oxidative stress response in mycobacterium avium, leading to biofilm formation. Appl. Environ. Microbiol. 74, 1798–1804 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hui C.-Y., Guo Y., Yang X.-Q., Zhang W., Huang X.-Q., Surface display of metal binding domain derived from PbrR on Escherichia coli specifically increases lead(II) adsorption. Biotechnol. Lett. 40, 837–845 (2018). [DOI] [PubMed] [Google Scholar]
- 56.George E. P., Raabe D., Ritchie R. O., High-entropy alloys. Nat. Rev. Mater. 4, 515–534 (2019). [Google Scholar]
- 57.Lu L., Guest J. S., Peters C. A., Zhu X., Rau G. H., Ren Z. J., Wastewater treatment for carbon capture and utilization. Nat. Sustain. 1, 750–758 (2018). [Google Scholar]
- 58.Baral S., Fendler J. H., Cadmium sulfide-mediated photoelectric effects in bilayer lipid membranes. J. Am. Chem. Soc. 111, 1604–1614 (1989). [Google Scholar]
- 59.T. Werpy, G. Petersen, Top Value Added Chemicals from Biomass: Volume I—Results of Screening for Potential Candidates from Sugars and Synthesis Gas (DOE/GO-102004-1992, National Renewable Energy Lab, 2004).
- 60.Chen X., Xu G., Xu N., Zou W., Zhu P., Liu L., Chen J., Metabolic engineering of Torulopsis glabrata for malate production. Metab. Eng. 19, 10–16 (2013). [DOI] [PubMed] [Google Scholar]
- 61.Jiang Y., Zheng T., Ye X., Xin F., Zhang W., Dong W., Ma J., Jiang M., Metabolic engineering of Escherichia coli for L-malate production anaerobically. Microb. Cell Fact. 19, 165 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen A. Y., Deng Z., Billings A. N., Seker U. O. S., Lu M. Y., Citorik R. J., Zakeri B., Lu T. K., Synthesis and patterning of tunable multiscale materials with engineered cells. Nat. Mater. 13, 515–523 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gilbert C., Tang T.-C., Ott W., Dorr B. A., Shaw W. M., Sun G. L., Lu T. K., Ellis T., Living materials with programmable functionalities grown from engineered microbial co-cultures. Nat. Mater. 20, 691–700 (2021). [DOI] [PubMed] [Google Scholar]
- 64.Cubillos-Ruiz A., Guo T., Sokolovska A., Miller P. F., Collins J. J., Lu T. K., Lora J. M., Engineering living therapeutics with synthetic biology. Nat. Rev. Drug Discov. 20, 941–960 (2021). [DOI] [PubMed] [Google Scholar]
- 65.Huang J., Liu S., Zhang C., Wang X., Pu J., Ba F., Xue S., Ye H., Zhao T., Li K., Wang Y., Zhang J., Wang L., Fan C., Lu T. K., Zhong C., Programmable and printable Bacillus subtilis biofilms as engineered living materials. Nat. Chem. Biol. 15, 34–41 (2019). [DOI] [PubMed] [Google Scholar]
- 66.Yin K., Wang Q., Lv M., Chen L., Microorganism remediation strategies towards heavy metals. Chem. Eng. J. 360, 1553–1563 (2019). [Google Scholar]
- 67.Jiang Y., Chen B., Duan C., Sun B., Yang J., Yang S., Multigene editing in the Escherichia coli genome via the CRISPR-Cas9 system. Appl. Environ. Microbiol. 81, 2506–2514 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sharan S. K., Thomason L. C., Kuznetsov S. G., Court D. L., Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S., Deng J., Yuan Y., Flachenecker C., Mak R., Hornberger B., Jin Q., Shu D., Lai B., Maser J., Roehrig C., Paunesku T., Gleber S. C., Vine D. J., Finney L., VonOsinski J., Bolbat M., Spink I., Chen Z., Steele J., Trapp D., Irwin J., Feser M., Snyder E., Brister K., Jacobsen C., Woloschak G., Vogt S., The Bionanoprobe: Hard X-ray fluorescence nanoprobe with cryogenic capabilities. J. Synchrotron Radiat. 21, 66–75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vogt S., MAPS: A set of software tools for analysis and visualization of 3D X-ray fluorescence data sets. J. Phys. IV Proc. 104, 635–638 (2003). [Google Scholar]
- 71.B. A. Dowd, G. H. Campbell, R. B. Marr, V. V. Nagarkar, S. V. Tipnis, L. Axe, D. P. Siddons, Developments in synchrotron x-ray computed microtomography at the National Synchrotron Light Source, in Developments in X-Ray Tomography II (SPIE, 1999), vol. 3772, pp. 224–236. [Google Scholar]
- 72.Gürsoy D., De Carlo F., Xiao X., Jacobsen C., TomoPy: A framework for the analysis of synchrotron tomographic data. J. Synchrotron Radiat. 21, 1188–1193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1
Figs. S1 to S31