Abstract
Corticosteroids regulate vital processes, including stress responses, systemic metabolism, and blood pressure. Here, we show that corticosteroid synthesis is related to the polyunsaturated fatty acid (PUFA) content of mitochondrial phospholipids in adrenocortical cells. Inhibition of the rate-limiting enzyme of PUFA synthesis, fatty acid desaturase 2 (FADS2), leads to perturbations in the mitochondrial lipidome and diminishes steroidogenesis. Consistently, the adrenocortical mitochondria of Fads2−/− mice fed a diet with low PUFA concentration are structurally impaired and corticoid levels are decreased. On the contrary, FADS2 expression is elevated in the adrenal cortex of obese mice, and plasma corticosterone is increased, which can be counteracted by dietary supplementation with the FADS2 inhibitor SC-26192 or icosapent ethyl, an eicosapentaenoic acid ethyl ester. In humans, FADS2 expression is elevated in aldosterone-producing adenomas compared to non-active adenomas or nontumorous adrenocortical tissue and correlates with expression of steroidogenic genes. Our data demonstrate that FADS2-mediated PUFA synthesis determines adrenocortical steroidogenesis in health and disease.
FADS2 determines the lipidome and steroidogenic function of the adrenal gland.
INTRODUCTION
The corticosteroids aldosterone and cortisol are produced in the zona glomerulosa and zona fasciculata of the adrenal cortex, respectively. Aldosterone increases the blood volume and pressure, while cortisol mediates stress responses, including regulation of glucose and lipid metabolism (1, 2). Excess production of aldosterone in primary hyperaldosteronism can cause resistant hypertension and hypokalemia (1). Hypercortisolismus leads to severe comorbidities, including central obesity, dyslipidemia, and hypertension collectively termed Cushing syndrome (3). Obesity is also associated with increased aldosterone and glucocorticoid production, which exacerbates central obesity, hyperlipidemia, hypertension, and risk for cardiovascular disease (4–8). However, the underlying mechanisms of the increased corticoid production in obesity are not fully understood.
Aldosterone production is induced by angiotensin II through calcium signaling in the steroidogenic cells of the zona glomerulosa (9). Adrenocorticotropic hormone (ACTH), released by the pituitary, triggers cyclic adenosine 3′,5′-monophosphate signaling and induces glucocorticoid synthesis in the steroidogenic cells of the zona fasciculata. Corticosteroid synthesis requires the release of cholesterol from lipid droplets where it is stored in the form of cholesterol esters (CE) and its translocation into mitochondria via the steroidogenic acute regulatory (StAR) protein, which is the rate-limiting step of steroidogenesis. Once inside the mitochondria, cholesterol is used for steroidogenesis through a series of synthetic steps. CYP11A1 (P450scc)-mediated side-chain cleavage of cholesterol produces pregnenolone which is then converted to progesterone by the nicotinamide adenine dinucleotide–dependent 3β-hydroxysteroid dehydrogenase (3β-HSD). CYP21 catalyzes the NADPH-dependent hydroxylation of progesterone and 17a-hydroxyprogesterone to 11-deoxycorticosterone and 11-deoxycortisol, respectively. In the zona glomerulosa, CYP11B2 catalyzes the production of aldosterone from 11-deoxycorticosterone. In the zona fasciculata, CYP11B1 catalyzes the conversion of 11-deoxycortisol to cortisol in humans and 11-deoxycorticosterone to corticosterone in mice (2, 10).
We here investigated the impact of mitochondrial membrane lipids on steroidogenesis in adrenocortical cells. We show that inhibition of fatty acid desaturase 2 (FADS2), the rate-limiting enzyme of polyunsaturated fatty acid (PUFA) synthesis, transforms the mitochondrial lipidome, inhibits cholesterol import, and diminishes steroidogenesis. Accordingly, FADS2 deficiency in mice impairs mitochondrial structure in adrenocortical cells and reduces corticosterone and aldosterone production. Conversely, FADS2 expression is up-regulated in the adrenal glands of obese mice and in aldosterone-producing adenomas compared to non-active adenomas (producing low amounts of aldosterone) and nontumorous adrenocortical tissue of patients, while FADS2 inhibition reduces corticoid levels in obese mice. These findings demonstrate that FADS2 determines mitochondrial structure and function and highlight the crucial role of FADS2 in adrenocortical steroidogenesis.
RESULTS
FADS2 determines the mitochondrial lipidome and is required for steroidogenesis in adrenocortical cells
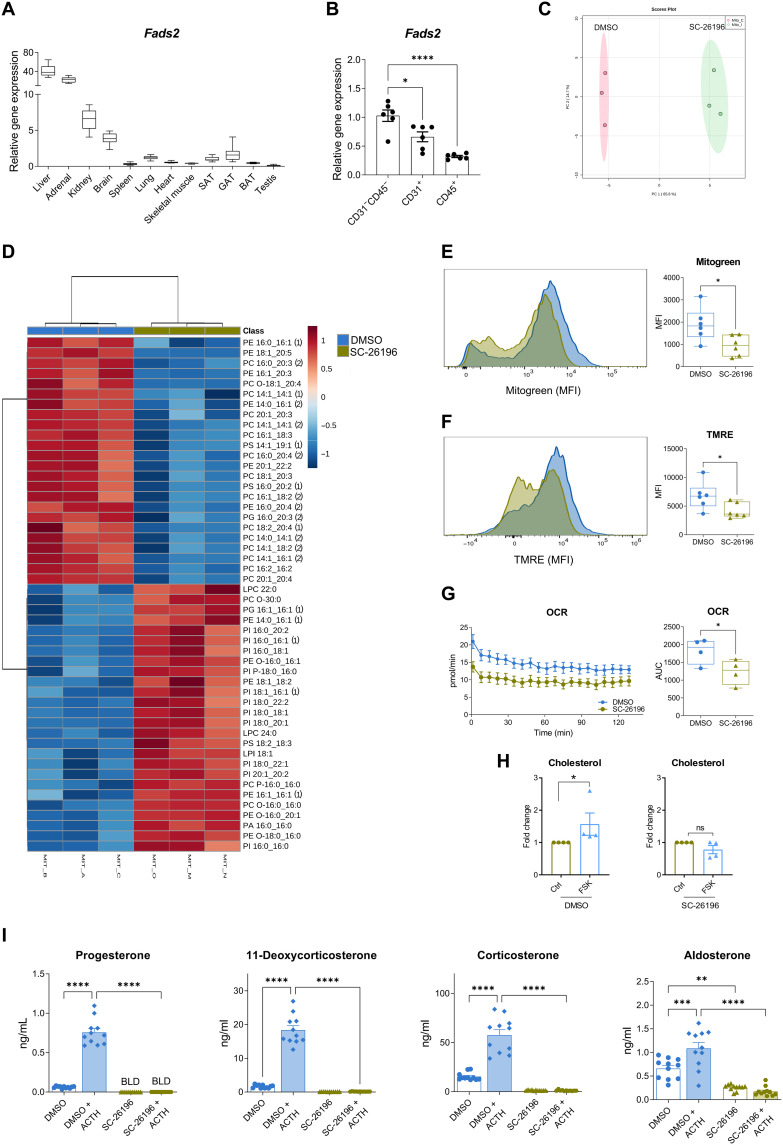
As key steps of steroidogenesis take place in mitochondria (10), we first interrogated to which extent the lipid composition of mitochondria affects steroidogenesis in adrenocortical cells. We chose Δ-6-desaturase FADS2 as a target to modulate mitochondrial lipid composition in adrenocortical cells, since it is highly expressed in the adrenal gland compared to other tissues and organs (Fig. 1A and fig. S1A), and its expression is higher in steroidogenic cells (CD31−CD45−) compared to endothelial cells (CD31+) and leukocytes (CD45+) of the adrenal cortex (Fig. 1B). The adrenal gland and the liver were the organs with the highest FADS2 mRNA and protein expression among many different tested organs and tissues (Fig.1A and fig. S1A). Moreover, FADS2 is the rate-limiting enzyme in the synthesis of PUFAs, which are important components of functional and storage lipids (11, 12). FADS2 catalyzes the conversion of the omega-6 linoleic acid (18:2 n-6, LA) to γ-linolenic acid (18:3 n-6, GLA) and the omega-3 α-linolenic acid (18:3 n-3, ALA) to stearidonic acid (18:4 n-3) (fig. S1B) (13).
Fig. 1. FADS2 determines the mitochondrial lipidome and is required for steroidogenesis in adrenocortical cells.
(A) Relative Fads2 expression in different tissues of 8-week-old WT C57BL6J mice determined by qPCR using Tbp as a housekeeping gene. Data are shown as mean 2ΔCt (n = 8). BAT, brown adipose tissue; SAT, subcutaneous adipose tissue; GAT, gonadal adipose tissue. (B) Relative Fads2 expression in CD31−CD45−, CD31+, and CD45+ cell populations of the mouse adrenal cortex. Expression of Fads2 in CD31−CD45− cells was set to 1. 18S was used as an internal control (n = 6). (C and D) PCA of the phospholipidome (C) and heatmap showing the differentially regulated lipid species (D) of mitochondrial fractions of NCI-H295R cells treated for 18 hours with SC-26196 (10 μM) or equal amount of DMSO (n = 3, one of two experiments). (E and F) Mitogreen (E) and TMRE (F) staining in primary adrenocortical cells treated for 18 hours with SC-26196 (10 μM) or DMSO (n = 6). MFI, mean fluorescence intensity. (G) OCR measurement of primary adrenocortical cells treated for 18 hours with SC-26196 or DMSO (n = 4). AUC, area under curve (H) Cholesterol levels determined by LC-MS/MS in mitochondrial fractions of NCI-H295R cells treated for 18 hours with SC-26196 or DMSO and stimulated or not with forskolin (FSK; 1 μM) for 15 min. Results of three independent experiments are shown here. (I) Progesterone, 11-deoxycorticosterone, corticosterone, and aldosterone levels were determined by LC-MS/MS in supernatants of primary adrenal cell cultures treated for 18 hours with SC-26196 or DMSO and then stimulated or not for 1 hour with ACTH (10 ng/ml) in the presence of SC-26196 or DMSO. The cell culture medium was changed before ACTH treatment (n = 12). Data in (B), (H), and (I) are shown as mean ± SEM, *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
We treated NCI-H295R adrenocortical carcinoma cells for 18 hours with the FADS2 inhibitor SC-26196 and analyzed the lipidome of mitochondrial fractions [identified by succinate dehydrogenase B (SDHB) expression] by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (fig. S1C). FADS2 inhibition reprogramed the mitochondrial lipidome, as shown by principal components analysis (PCA) (Fig. 1C) and strongly altered the mitochondrial lipid composition at individual species level (Fig. 1D). The abundance of phosphatidylcholines (PC) and phosphatidylethanolamines (PE) containing PUFA acyl chains, such as PC 16:0_20:4, PC 20:1_20:3, PC 20:1_20:4, PE 16:0_20:4, PE 18:1_20:5, and PE 16:1_20:3, was reduced in SC-26196–treated cells, while conversely lipids with saturated or monounsaturated acyl chains, such as PE 14:0_16:1, PE 16:1_16:1, phosphatidylinositol (PI) PI 16:0_16:0, PI 16:0_16:1, PI 16:0_18:1, PI 18:1_16:1, PI 18:0_18:1, PI 18:0_20:1, PI 18:0_22:1, and phosphatidylglycerol (PG) PG 16:1_16:1, were increased (Fig. 1D). Cardiolipins, a unique component of the inner mitochondrial membrane (IMM), remained unaffected (not shown).
PC and PE are major lipid components of mitochondrial membranes, and therefore, changes in their consistency are likely to influence mitochondrial physiology (14, 15). FADS2 inhibition decreased the mitochondrial load, mitochondrial membrane potential, and oxygen consumption rate (OCR) in primary adrenocortical cells (Fig. 1, E to G). Electron transport chain–dependent mitochondrial bioenergetics are intimately linked with and required for steroidogenesis (10, 16–19). Hence, the observed reduction of the mitochondrial membrane potential (Fig. 1F) and OCR (Fig. 1G) is expected to lead to reduced steroidogenesis. Along this line, we asked whether cholesterol transport into mitochondria, which is the rate-limiting step of steroidogenesis, is affected by FADS2 inhibition. Cholesterol import into mitochondria involves the interaction between StAR, voltage-dependent anion channel (VDAC), and translocator protein, 18 kDa (TSPO) (10). StAR shuttles from the cytoplasm to the outer mitochondrial membrane (OMM), transferring cholesterol to a complex, which is assembled by VDAC and TSPO at the OMM-IMM contact sites, called transduceosome (10, 19). In adrenocortical steroidogenic cells, cholesterol taken up from circulating lipoproteins or synthesized in the endoplasmic reticulum (ER), is mainly stored in the form of CE in lipid droplets, from where it is transferred by StAR and the transduceosome into the mitochondria for pregnenolone synthesis, the first steroid to be synthesized in the steroidogenic cascade (19). To examine whether FADS2 inhibition affects cholesterol uptake in the mitochondria, we analyzed the cholesterol content of mitochondrial (SDHB positive) fractions of NCI-H295R cells treated or not with SC-26196 upon stimulation with forskolin (fig. S1C). Forskolin treatment triggered a strong increase in mitochondrial cholesterol levels in control but not SC-26196–treated cells (Fig. 1H). In accordance, cholesterol and CE accumulated in the lipid droplet [hormone-sensitive lipase (HSL) positive] fractions in SC-26196–treated cells (fig. S1, C and D).
Reduced mitochondrial bioenergetics (Fig. 1, F and G) and cholesterol mitochondrial import (Fig. 1H) in SC-26196–treated cells are expected to lead to impaired steroidogenesis. To verify this, we analyzed steroid hormone production by LC-MS/MS in primary adrenal cell cultures treated with SC-26196 and subsequently stimulated with ACTH. ACTH efficiently increased progesterone, 11-deoxycorticosterone, corticosterone, and aldosterone production, while FADS2 inhibition abolished their production (Fig. 1I), without affecting cell viability (fig. S1E). FADS2 inhibition also reduced progesterone and 11-deoxycortisol production in forskolin-treated NCI-H295R cells (fig. S1F). The effect of FADS2 inhibition on steroidogenesis was not due to decreased mRNA expression of StAR or steroidogenic enzymes, Cytochrome P450 Family 11 Subfamily A Member 1 (Cyp11a1, cholesterol side-chain cleavage enzyme), 3β-Hsd2, Cyp11b1, or Cyp11b2; their expression was increased upon FADS2 inhibition, perhaps as a compensatory response to low steroid hormone synthesis (fig. S1G). These data indicate that increased expression of steroidogenic enzymes is not sufficient for successful steroidogenesis when FADS2 is blocked, thereby underscoring the importance of FADS2-dependent PUFA synthesis for steroidogenesis. In accordance, FADS2 overexpression (fig. S1, H and I) increased 11-deoxycorticosterone, cortisol, and aldosterone production in forskolin-treated NCI-H295R cells (fig. S1J) substantiating the promoting effect of FADS2 on steroidogenesis.
High-fat diet reprograms the adrenal lipidome and increases glucocorticoid production
Next, we asked whether FADS2 could drive increased adrenocortical hormone production in vivo. As an experimental model we used high-fat diet (HFD)–induced obesity in mice, which was previously shown to lead to increased corticosterone and aldosterone production (5). Wild-type (WT) C57BL6/J mice were fed for 20 weeks a low-fat diet (LFD; 10% kcal deriving from fat) or a HFD (60% kcal deriving from fat). HFD feeding substantially increased the body weight of the animals (fig. S4A), as previously reported in a large number of studies (20, 21). As expected, progesterone and corticosterone plasma levels were elevated in HFD compared to LFD mice (Fig. 2A). ACTH plasma levels were also increased in obese mice (fig. S2A), standing in accordance with the notion that chronic inflammation and dysmetabolism may lead to chronic activation of the hypothalamic-pituitary-adrenal (HPA) axis (22, 23). Adrenal gland weights did not change with obesity (fig. S2B). We next interrogated the lipidomic changes in the adrenal glands of obese mice, and whether these changes may contribute to the increased steroidogenesis in an autonomous fashion.
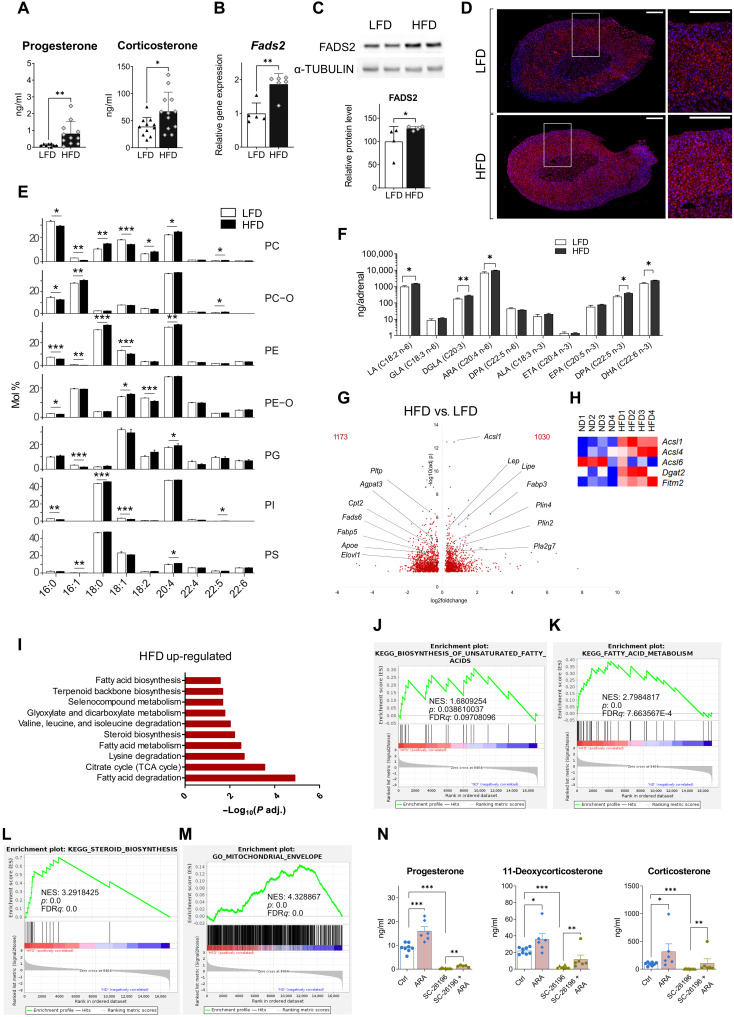
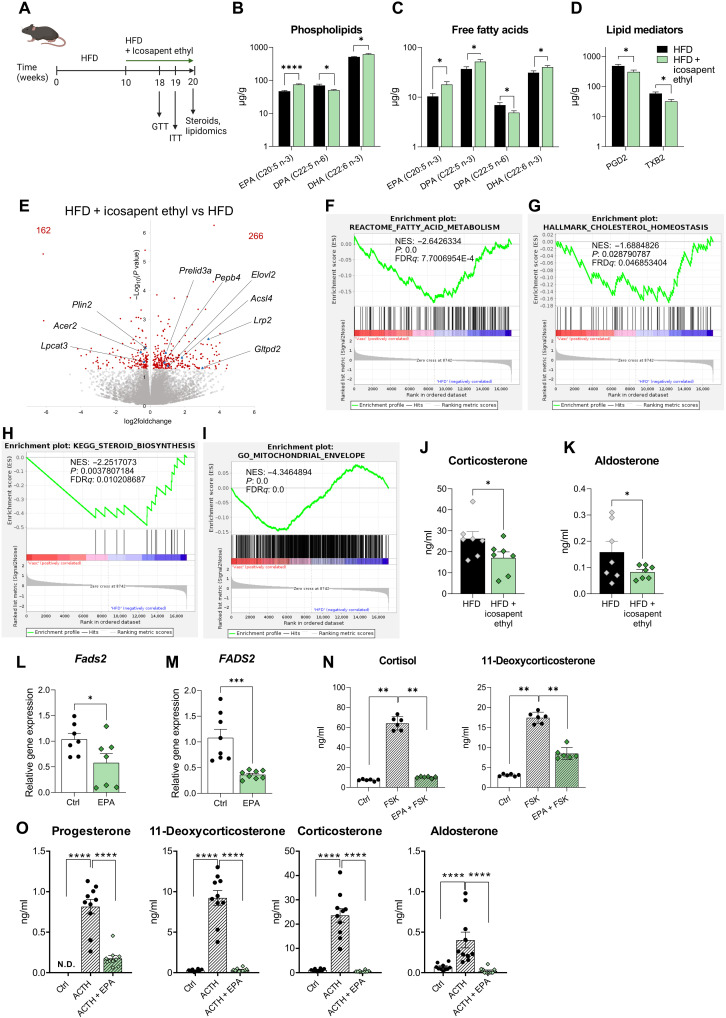
Fig. 2. HFD reprograms the adrenal lipidome and increases glucocorticoid production.
(A) Progesterone and corticosterone plasma levels in mice fed for 20 weeks a LFD or HFD (n = 8 to 12). (B and C) Fads2 mRNA expression (B) and protein expression (C) in adrenal glands of LFD and HFD mice (n = 4 to 6). (D) Immunofluorescence for FADS2 (red) and 4′,6-diamidino-2-phenylindole (DAPI; blue) in adrenal glands of LFD- and HFD mice (1 of 5 mice per condition). Scale bars, 200 μm, zoomed-in inserts are shown at the right. (E) Acyl chain profile of nonstorage lipids analyzed by shotgun lipidomics. Lipids in adrenal glands from lean and obese mice were grouped according to the acyl chain within each lipid class (mol % is relative to the lipid class). Only the features with a mean abundance >5 mol % are shown, mean mol % ± SD is shown (n = 7 to 8). (F) Acyl chain isomers in phospholipids in adrenal glands of LFD and HFD mice determined by HPLC-MS (n = 5 to 6). (G to M) RNA-seq in the adrenal cortex of LFD and HFD mice (n = 4 mice per group). Volcano plot showing differentially expressed genes in HFD versus LFD mice (G). Heatmap of differentially expressed genes involved in fatty acid processing (P adjusted < 0.05) (H). EGSEA of RNA-seq data in the adrenal cortex of HFD versus LFD mice showing 10 among most up-regulated metabolic pathways (KEGG database) (I). GSEA for genes involved in biosynthesis of unsaturated fatty acids (J), fatty acid metabolism (K), steroid biosynthesis (L), and mitochondrial envelope formation (M). NES, normalized enrichment score; FDR, false discovery rate. (N) Progesterone, 11-deoxycorticosterone, and corticosterone levels in supernatants of primary adrenal cell cultures treated for 18 hours with SC-26196 (10 μM) or DMSO (Ctrl) in the presence or absence of ARA (150 μM) (n = 6 to 8). Data in (A) to (C) are shown as mean ± SD and in (F) and (N) as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001.
FADS2 expression was elevated at mRNA and protein level in the adrenal glands of HFD-fed mice (Fig. 2, B to D), while its expression did not change in other organs, such as the liver or the brain (fig. S2C). To understand the role of FADS2 in regulating the adrenocortical lipidome, we investigated the adrenal gland lipidome by shotgun lipidomics and revealed reprograming of the whole lipidome, i.e., both nonstorage lipids and storage lipids upon HFD feeding (fig. S2D). Nonstorage, i.e., membrane lipids include phospholipids and their ether-linked versions [PC, PE, PC O-, PE O-, PI, PG, phosphatidylserine (PS), and phosphatidate (PA)], lysolipids [lyso-phosphatidate (LPA), lyso-phosphatidylcholine (LPC), phosphatidylethanolamine (LPE), LPE O-, lyso-phosphatidylglycerol (LPG), lyso-phosphatidylinositol (LPI), and lyso-phosphatidylserine (LPS)], sphingomyelin (SM), diacylglycerol (DAG), cardiolipins (CL), ceramides, and hexosylceramides (HexCer). Storage lipids comprise CE and triacylglycerol (TAG) (24). Whole and nonstorage lipidomes of adrenal glands from lean and obese mice segregated along the first principal component (PC1), and the topology of the samples was notably similar, while storage lipids regrouped with statistical significance along the second principal (PC2) (fig. S2D), suggesting that nonstorage lipids were mainly responsible for the grouping of the whole lipidome according to the diet.
Yet, storage lipids (TAG and CE) constituted the largest portion of the identified lipids in adrenal glands (86.98 ± 5.43% in LFD mice and 90.12 ± 1.93% in HFD mice), while nonstorage lipids accounted for 13.02 ± 5.22% and 9.87 ± 1.89% of all identified lipids in the adrenal glands of mice under LFD and HFD, respectively (table S1). The total amounts of storage and nonstorage lipids did not substantially differ in the adrenal glands of lean and obese mice (table S1). Likewise, lipid classes did not significantly differ between the adrenal glands of lean and obese mice (table S2). CE as well as non-esterified cholesterol were also not significantly increased in the adrenal glands of obese mice (table S2).
Our data show that it is not the lipid amount but the total acyl chain length and degree of unsaturation of storage and nonstorage lipids that substantially changed with obesity. TAG species with 50 to 54 carbon atoms (fig. S2E) and two to four acyl chain double bonds (fig. S2F) were highly abundant (mol % > 10). Adrenal glands of HFD mice exhibited a significant increase in long (with a total acyl chain length of 54 carbon atoms) and unsaturated (with four, five, and more than six double bonds) TAG species. In contrast, adrenal glands from LFD-fed mice showed a larger amount of TAG species containing shorter acyl chains (with 48 and 50 carbon atoms) with one or two double bonds (fig. S2, E and F). To determine whether the diet affected the overall mean weighted length and unsaturation level of lipids, we calculated the double bond index (DBI) and the length index (LI). Storage lipids in the adrenal glands of obese mice showed a significantly higher DBI compared to lean mice (table S3). Hence, upon HFD feeding, TAG in the adrenal glands became overall more unsaturated.
Most nonstorage lipids contained a cumulative of 34 to 38 carbon atoms (fig. S2G) and four double bonds in their acyl chains (fig. S2H). Similarly to what was found for TAG, adrenal glands from HFD mice showed a significant increase in longer and more unsaturated (with four double bonds) nonstorage lipids, while shorter and mono-unsaturated nonstorage lipid species were more abundant in the adrenal glands of LFD-fed mice (fig. S2, G and H). In accordance, membrane lipids in the adrenal glands of HFD-fed mice had significantly higher DBI and LI compared to LFD-fed mice (table S3), indicating that nonstorage (membrane) lipids were longer and more unsaturated in the adrenal glands of obese mice.
Notably, 20:4 was one of the most abundant acyl chains in PC, PC O-, PE, PE O-, PG, and PI, and the amount of PC, PE, and PG lipids containing 20:4 was higher in the adrenal glands of obese compared to lean mice (Fig. 2E). In contrast, long-chain PUFAs other than 20:4, such as 22:4, 22:5, and 22:6, were less abundant (Fig. 2E). Accordingly, the phospholipids PC 18:0_20:4 and PE 18:0_20:4 were highly abundant and higher in the adrenal glands of HFD mice (fig. S2, I and J).
Isomer analysis showed that arachidonic acid (ARA; 20:4 n-6, all-cis-5,8,11,14-eicosatetraenoic acid) but not eicosatetraenoic acid (ETA; 20:4 n-3, all-cis-8,11,14,17-eicosatetraenoic acid) was the abundant 20:4 isomer form (Fig. 2F), in agreement with previous reports showing very little ETA in the tissue lipidome (25). Furthermore, docosapentaenoic acid (DPA; 22:5 n-3) and docosahexaenoic acid (DHA 22:6 n-3) were more abundant in adrenal gland phospholipids of obese mice, while eicosapentaenoic acid (EPA; 20:5 n-3) was not (Fig. 2F). Given that FADS2 is the rate-limiting enzyme for ARA, DPA, and DHA synthesis (fig. S1B), the increased amounts of these phospholipidic acyl chains in the adrenal glands of HFD mice could be the result of an enhanced adrenal FADS2 expression in obese mice (Fig. 2, B to F).
To correlate the observed shift at the lipidome level with transcriptional changes, we performed RNA sequencing (RNA-seq) in the adrenal cortex of LFD and HFD mice that revealed 2203 differentially expressed genes, of which 1030 were up-regulated and 1173 were down-regulated by HFD compared to LFD feeding (Fig. 2G). Several genes involved in lipid metabolism were highly up-regulated, such as Acyl-coenzyme A synthetase long-chain family member 1 (Acsl1), Fatty acid binding protein 3 (Fabp3), Lipase e (Lipe), and Perilipin 4 (Plin4), or down-regulated, such as Phospholipid transfer protein (Pltp), Fads6, 1-Acylglycerol-3-phosphate O-acyltransferase 3 (Agpat3), or Carnitine palmitoyltransferase (Cpt2) in the adrenal cortex of HFD mice (Fig. 2G). Fads2 was also up-regulated in the adrenal cortex of HFD- compared to LFD-fed mice (log2 fold change = 1.244, adjusted P value = 0.0117), as were several other genes involved in fatty acid processing, such as Acsl4, the most highly expressed Acsl isoform in the adrenal gland, Diacylglycerol O-acyltransferase 2 (Dgat2), and Fat storage–inducing transmembrane protein 2 (Fitm2) (Fig. 2H). Ensemble of Gene Set Enrichment Analysis (EGSEA) showed that fatty acid metabolism, steroid biosynthesis, and tricarboxylic acid (TCA) cycle pathways were among the most up-regulated pathways in the adrenal cortex of HFD mice (Fig. 2I). Similarly, Gene Set Enrichment Analysis (GSEA) also demonstrated significant positive enrichment of genes related to biosynthesis of unsaturated fatty acids, lipid metabolism, and steroid biosynthesis in the adrenal cortex of obese mice (Fig. 2, J to L, and fig. S2K). Also genes associated with mitochondrial biogenesis (fig. S2L), formation of the mitochondrial envelope (Fig. 2M), and mitochondrial function, including respiratory electron transport, oxidative phosphorylation, TCA cycle, and beta-oxidation (fig. S2, M to P), were all positively enriched in the adrenal cortex of HFD mice.
In summary, these findings demonstrate that HFD-induced obesity is associated with lipidomic transformation of the adrenal gland, particularly enhanced PUFA synthesis, increased FADS2 expression, and transcriptional changes promoting increased PUFA metabolism and mitochondrial bioenergetics in the adrenal cortex. A particularly abundant PUFA in adrenal PC and PE is ARA (Fig. 2, E and F). ARA was previously shown to promote steroidogenesis and potentially be required for cholesterol import into mitochondria (26, 27). We found that FADS2 is required for the incorporation of ARA acyl chains in mitochondrial PC and PE (Fig. 1D). Hence, we tested whether ARA supplementation can counteract the inhibitory effect of FADS2 inhibition on steroidogenesis. ARA increased progesterone, 11-deoxycorticosterone, and corticosterone production in primary adrenocortical cells treated with SC-26196 (Fig. 2N). The fact that ARA only partially restored steroidogenesis could be due to insufficient lipidomic remodeling achieved by ARA supplementation under the used experimental conditions or may indicate the requirement of additional PUFAs for full restorage of steroidogenesis under FADS2 inhibition. Overall, these data imply that intra-adrenal FADS2-dependent ARA synthesis is required for adrenocortical steroidogenesis.
Medulla-derived PUFAs promote adrenocortical steroidogenesis
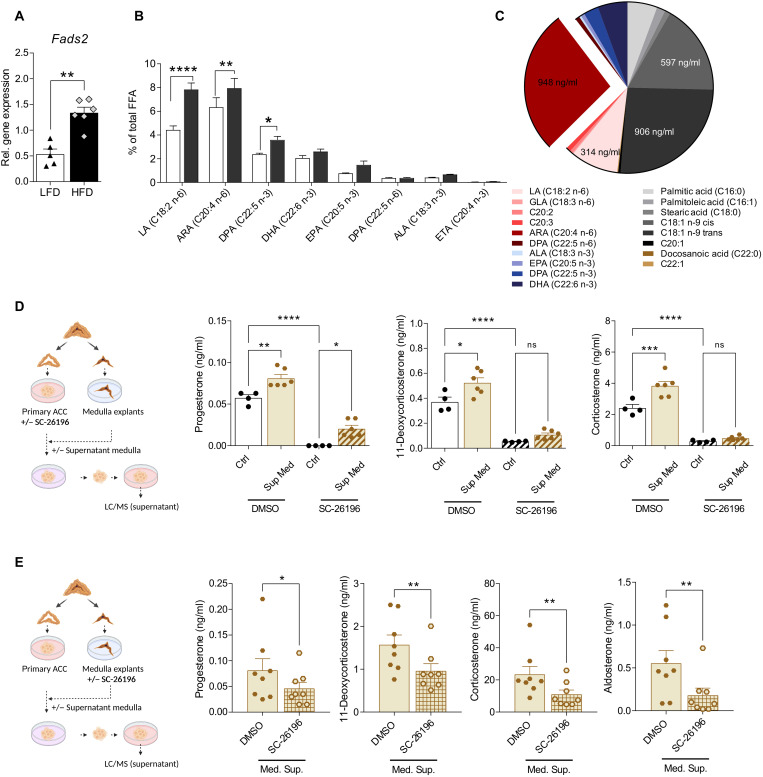
The adrenal gland consists of the medulla and the cortex. Early reports showed that chromaffin (medullary) cells may release ARA (28). We asked whether FADS2-mediated PUFA synthesis in the medulla could influence adrenocortical steroidogenesis. Fads2 expression in the adrenal medulla increased with obesity (Figs. 2D and 3A). In addition, free LA (C18:2 n-6) and ARA were increased in the medulla of obese compared to lean animals (Fig. 3B). In addition, free ARA was the most abundant free fatty acid (FFA) in the supernatant of medulla organ cultures (Fig. 3C). Together with our finding that ARA supplementation increased steroidogenesis in adrenocortical cells (Fig. 2N), these data suggest that ARA may mediate a medulla-cortex interaction promoting adrenocortical steroidogenesis.
Fig. 3. Medulla-derived PUFAs promote adrenocortical steroidogenesis.
(A) Relative Fads2 expression in the adrenal medulla of mice fed for 20 weeks a LFD or HFD (n = 5 to 6 mice per group). Tbp was used as a housekeeping gene. (B) FFA were measured in the adrenal medulla of mice fed for 20 weeks a LFD or HFD by HPLC-MS (n = 3 mice per group). Results are presented as % of total FFA. (C) FFA measured in the supernatants of adrenal medulla explants kept for 18 hours in 100 μl of medium. Results are presented as % of total FFAs (n = 4). (D) Adrenocortical cells were treated for 6 hours with SC-26196 (10 μM) or DMSO and then received medulla explant–conditioned or control medium for another 18 hours. Steroids were measured in the adrenocortical cell supernatant by LC-MS/MS (n = 4 to 6). (E) Adrenal medulla explants were treated for 6 hours with SC-26196 (10 μM) or DMSO, washed thoroughly, and left another 18 hours in culture. Their supernatant was then applied on adrenocortical cell cultures, and 8 hours later, adrenocortical cell supernatants were collected and analyzed by LC-MS/MS (n = 8). Data in (A), (B), (D), and (E) are shown as mean ± SEM. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. ns, not significant.
To investigate how PUFAs from chromaffin cells may affect steroidogenesis in adrenocortical cells, we applied supernatants from medulla explants onto primary adrenocortical cell cultures. Medulla-conditioned supernatants increased progesterone, 11-dehydrocorticosterone, and corticosterone production in primary adrenocortical cultures compared to control medium (Fig. 3D). This effect was attenuated when medulla explants were treated with the FADS2 inhibitor SC-26196, underscoring the importance of medullar PUFA synthesis in the medulla-cortex interaction (Fig. 3E). However, treatment of adrenocortical cells with SC-26196 diminished steroid synthesis in cells receiving medulla-conditioned medium (Fig. 3D), suggesting that despite the fact that adrenocortical cells may take up medulla-deriving PUFA, it is mainly endogenous synthesis of PUFA in adrenocortical cells, which is required for steroidogenesis.
FADS2 inhibition reduces corticosteroid levels in obese mice
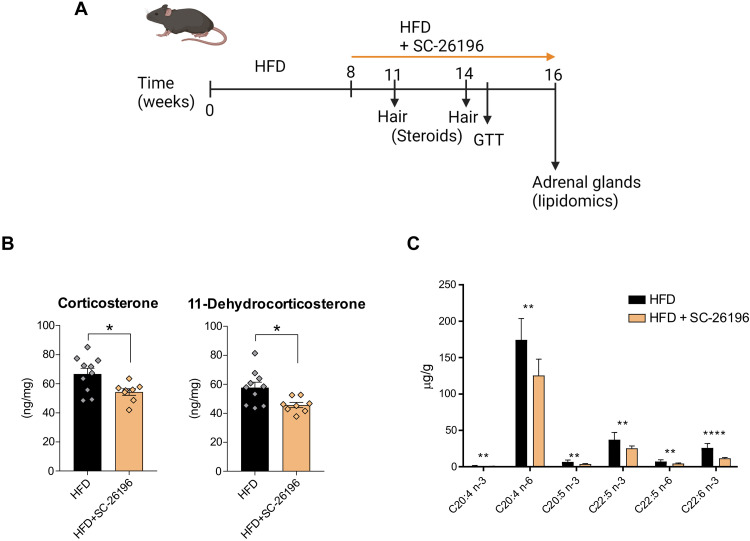
To validate the role of FADS2 in adrenocortical steroidogenesis in obesity, we treated HFD mice with the FADS2 inhibitor SC-26196 and assessed corticosteroid production. WT C57BL/6J mice were fed for 8 weeks a HFD and another 8 weeks with HFD containing or not SC-26196 (0.625 g/kg; corresponding to approximately 100 mg/kg of animal weight) (Fig. 4A). We decided to determine corticosteroid levels at two different time points, i.e., 3 and 6 weeks after SC-26196 treatment start, during the course of the HFD feeding in hair, instead of plasma, since hair cutting causes minimal stress to mice compared to blood retrieval (Fig. 4A). Notably, corticosteroid levels in newly grown hair (2 to 3 mm above the skin) reflect the cumulative corticosteroid production over a period of 3 to 4 weeks (29, 30). Already at 3 weeks after SC-26196 treatment start, levels of 11-dehydrocorticosterone in newly grown hair were reduced in HFD mice receiving the inhibitor compared to control mice (fig. S3A). 11-Dehydrocorticosterone is the inactive form of corticosterone, which is converted to the latter by 11β-HSD1 in tissues (31). Hence, reduced availability of 11-dehydrocorticosterone can lead to lower (active) corticosterone levels in tissues (31). At 6 weeks after SC-26196 treatment start, both 11-dehydrocorticosterone and corticosterone levels were reduced in the hair of mice receiving the FADS2 inhibitor compared to control mice (Fig. 4B). Lower corticosteroid production associated with reduced free PUFA, including ARA, levels in the adrenal glands of SC-26196 treated mice (Fig. 4C). Glucose tolerance was not affected by treatment of mice with SC-26196 (fig. S3B). These findings demonstrate that FADS2 inhibition in established obesity can alter the PUFA content of the adrenal glands and down-regulate adrenocortical steroidogenesis.
Fig. 4. FADS2 inhibition reduces corticosteroid levels in obese mice.
(A) Schematic representation of the experimental setup. (B) Corticosterone and 11-dehydrocorticosterone levels were determined by LC-MS/MS at feeding week 14 in hair of HFD mice receiving or not for 6 weeks SC-26196. Steroid concentrations were normalized to hair weight (B). (C) FFA were measured by HPLC-MS in the adrenal glands of mice fed a HFD with or without SC-26196 (n = 8 to 10 mice per group). Data are presented as mean ± SEM, *P < 0.05; **P < 0.01; ****P < 0.0001.
Dietary supplementation with icosapent ethyl reduces corticoid levels in obese mice
To further elaborate on the requirement of FADS2-dependent PUFA synthesis for adrenocortical steroidogenesis, we asked whether enhanced activation of the FADS2-ARA axis can be regulated by dietary interventions. In general, increased intake or elevated blood levels of omega-3 lipids are associated with health benefits, while an omega-6/omega-3 imbalance is a risk factor for chronic disease (32, 33). Icosapent ethyl, a highly purified and stable EPA ethyl ester, is approved as an adjunct to diet to reduce triglyceride, non–high-density lipoprotein (HDL) and apolipoprotein B levels and to lower the risk of cardiovascular events in hypertriglyceridemic patients, as shown by the recent Reduction of Cardiovascular Events with Icosapent Ethyl–Intervention Trial and other clinical studies (34–36). We therefore asked whether dietary supplementation with icosapent ethyl can lower corticosterone and aldosterone levels in mice with diet-induced obesity. To this end, WT C57BL/6J mice were fed for 10 weeks a HFD and another 10 weeks with HFD containing or not icosapent ethyl (1.125 g/kg; corresponding to approximately 180 mg/kg of animal weight) (Fig. 5A). Icosapent ethyl treatment did not alter the body weight gain, glucose, or insulin tolerance of HFD mice (fig. S4, A to C). Lipidomic analysis in the adrenal glands of these mice showed increased EPA incorporation in the phospholipids of icosapent ethyl–treated mice, as well as higher DHA and lower omega-6 DPA levels in adrenal phospholipids (Fig. 5B). Accordingly, free EPA, omega-3 DPA, and DHA were increased, and free omega-6 DPA was decreased in the adrenal glands of mice receiving icosapent ethyl (Fig. 5C). Moreover, analysis of lipid mediators (oxylipins) showed reduced prostaglandin D2 and thromboxane B2 levels in the adrenal glands of icosapent ethyl–treated mice (Fig. 5D). Hence, dietary supplementation with icosapent ethyl in obesity transforms the adrenal lipidome toward increased omega-3 lipid content and reduced levels of proinflammatory lipid mediators.
Fig. 5. Dietary supplementation with icosapent ethyl reduces corticoid levels in obese mice.
(A) Schematic representation of the experimental setup. (B to D) Acyl chains of phospholipids (B), FFA (C), and lipid mediators (D) were measured by HPLC-MS in the adrenal glands of mice fed a HFD with or without icosapent ethyl (n = 7 mice per group). (E) Volcano plot based on RNA-seq data in the adrenal cortex of mice, which received HFD supplemented with icosapent ethyl versus mice fed a HFD without icosapent ethyl (n = 4 mice per group). (F to I) GSEA analysis for genes related to fatty acid metabolism, cholesterol homeostasis, steroid biosynthesis, and mitochondrial envelope formation (n = 4 mice per group). (J and K) Corticosterone and aldosterone plasma levels in mice, which received HFD without and with icosapent ethyl, measured by LC-MS/MS (n = 7 mice per group). (L and M) Primary CD31−CD45− adrenocortical cells (L) and NCI-H295R cells (M) were treated with 66 μM EPA for 18 or 48 hours, respectively, and Fads2 expression was determined by qPCR using 18S expression as a housekeeping gene [n = 7 in (L) and n = 8 in (M)]. (N) NCI-H295R cells were treated for 48 hours with 66 μM EPA and stimulated or not with forskolin (FSK;1 μM) in the last 24 hours. Steroids were measured in the culture supernatant by LC-MS/MS (n = 6). (O) Primary adrenocortical cells were treated for 18 hours with 66 μM EPA and stimulated or not with ACTH (10 ng/ml). Steroids were measured in the culture supernatant by LC-MS/MS (n = 10). Data in (B) to (D) and (L) and (M) are presented as mean ± SEM, data in (N) and (O) as mean ± SD. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. N.D., not determined.
The lipidomic alterations caused by icosapent ethyl dietary supplementation were accompanied by transcriptional changes in the adrenal cortex, as shown by RNA-seq, with 266 genes being up- and 162 genes being down-regulated (Fig. 5E). Among the differentially expressed genes, several lipid metabolism–associated genes, such as Acsl4 and Elovl2, were up-regulated, while others, such as Plin2 and lysophosphatidylcholine acyltransferase 3 (Lpcat3), were down-regulated (Fig. 5E). GSEA analysis showed negative enrichment of genes related to fatty acid metabolism, cholesterol homeostasis, and steroid biosynthesis in the adrenal cortex of icosapent ethyl–treated mice (Fig. 5, F to H), pathways which were up-regulated in the adrenal cortex of HFD compared to LFD mice (Fig. 2, J to L). In addition, genes related to mitochondrial envelope formation, respiratory electron transport, oxidative phosphorylation, and TCA cycle were negatively enriched in the adrenal cortex of icosapent ethyl–receiving mice (Fig. 5I and fig. S4, D to F), again standing in stark contrast to the up-regulation of these pathways in the adrenal glands of HFD versus LFD mice (Fig. 2M and fig. S2, M to P). Icosapent ethyl dietary supplementation reduced corticosterone and aldosterone plasma levels in obese mice (Fig. 5, J and K) without altering ACTH plasma levels (fig. S4G). These data collectively demonstrate that dietary administration of icosapent ethyl in obesity reduces adrenocortical steroidogenesis in an HPA axis–independent fashion. Instead, it is associated with lipidomic and transcriptional reprograming of the adrenal cortex, suggesting an intra-adrenal regulation of corticosteroid production. In support of these findings, EPA treatment down-regulated FADS2 (Fig. 5, L and M) and reduced steroidogenesis in forskolin-stimulated NCI-H295R cells (Fig. 5N) and ACTH-stimulated primary adrenocortical cells (Fig. 5O), further substantiating that FADS2 determines adrenocortical hormone production.
FADS2 deficiency perturbs adrenal gland function
To further confirm the role of the FADS2-dependent PUFA synthesis in adrenocortical function, we used Fads2 knockout (Fads−/−) mice, which were kept for 10 weeks under a diet with low or high PUFA concentration (Fig. 6A and table S4). FADS2 deficiency was verified in the adrenal gland at mRNA and protein level (Fig. 6, B and C). Adrenal glands from Fads2−/− mice did not show any obvious abnormalities in size or macroscopic structure compared to littermate WT mice fed a PUFA-rich or low-PUFA diet (fig. S5A). However, progesterone, corticosterone, and aldosterone plasma concentrations were reduced in Fads2−/− mice fed the low-PUFA diet but not in mice fed the PUFA-rich diet (Fig. 6D). Reduced corticosteroid production was not due to reduced ACTH levels, i.e., WT and Fads2−/− mice fed with either diet displayed similar ACTH plasma levels (fig. S5B). In accordance, StAR expression was reduced in Fads2−/− mice, which were kept under low-PUFA diet but not in mice fed the PUFA-rich diet (Fig. 6E). Expression of other steroidogenic genes (Cyp11a1, 3β-Hsd2, Cyp11b1, and Cyp11b2) was not affected by FADS2 deficiency (fig. S5C).
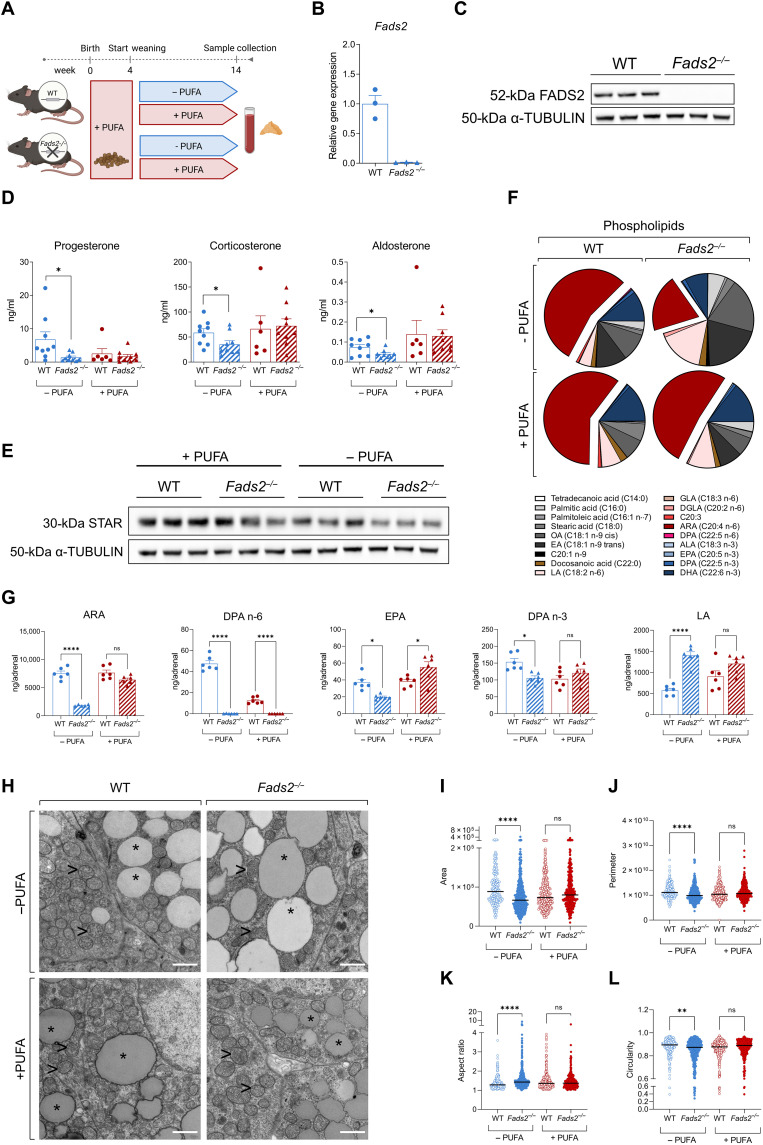
Fig. 6. FADS2 deficiency perturbs adrenal gland function.
(A) Scheme demonstrating the experimental setup. (B and C) FADS2 deletion efficiency in Fads2−/− mice. FADS2 expression was determined in adrenal glands of WT and Fads2−/− mice by qPCR using Tbp expression as a housekeeping gene (B) and Western blot using α-Tubulin as a loading control (C) (n = 3 mice per group). (D) Progesterone, corticosterone, and aldosterone levels in the plasma of WT and Fads2−/− mice fed a low-PUFA or PUFA-rich diet (n = 6 to 9 mice per group). (E) Western blot for StAR in adrenal glands of WT and Fads2−/− mice fed a low-PUFA or PUFA-rich diet. α-Tubulin was used as a loading control (n = 3 mice per group). (F and G) Acyl chain composition of phospholipids in the adrenal gland of WT and Fads2−/− mice fed a low-PUFA or PUFA-rich diet (n = 6 mice per group). Results are presented as % of all phospholipidic acyl chains (F) and as ng per adrenal gland (G). OA, oleic acid; EA, elaidic acid. (H) Representative electron microscopy images of adrenocortical cells; adrenal glands of two WT and two Fads2−/− mice fed a low-PUFA and 1 WT and 1 Fads2−/− mice fed a PUFA-rich diet were imaged (magnification 6,800x). Scale bars, 1 μM. Asterisks (*) depict lipid droplets, and arrowheads (>) depict mitochondria. (I to L) Quantification of mitochondrial area (I), perimeter (J), aspect ratio (K), and circularity (L) in WT and Fads2−/− mice fed a low-PUFA or PUFA-rich diet (in total, 198 to 429 mitochondria were quantified in two WT and two Fads2−/− mice fed a low-PUFA and one WT and one Fads2−/− mice fed a PUFA-rich diet). Data in (B), (D), and (G) are shown as means ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001.
As outlined above, FADS2-mediated ARA synthesis is crucial for adrenal steroidogenesis. Lipidomic analysis showed that ARA abundance in adrenal phospholipids of WT mice was not affected by feeding with the low-PUFA diet (Fig. 6, F and G), suggesting that the ARA content in adrenal phospholipids is not affected by dietary ARA uptake when FADS2 is expressed. However, in Fads2−/− mice fed the low-PUFA diet the ARA content in adrenal phospholipids was strongly reduced compared to WT mice fed the same diet but not in mice fed a PUFA-rich diet (Fig. 6, F and G, and table S5). DPA (C22:5 n-6) levels in phospholipids were also diminished in the adrenal glands of Fads2−/− mice independently of the diet (Fig. 6, F and G, and table S5). The levels of EPA and DPA (C22:5 n-3) were reduced in the adrenal glands of FADS2-deficient mice receiving a low-PUFA diet (Fig. 6, F and G), although to a lesser extent than the omega-6 fatty acids ARA and DPA (C22:5 n-6), suggesting that FADS2 may rather promote the synthesis of omega-6 fatty acids. In accordance, the PUFA precursor LA increased in the adrenal glands of Fads2−/− mice fed the low-PUFA diet presumably due to its diminished use for PUFA synthesis (Fig. 6, F and G).
Moreover, adrenocortical cells of Fads2−/− mice fed the low-PUFA diet had larger lipid droplets compared to WT mice receiving the same diet, as shown by electron microscopy (Fig. 6H and fig. S5D). This stands in accordance with the observed reduced steroid hormone production (Fig. 6D); hence, the attenuated use of CE for steroidogenesis, which are thus accumulating in cells forming larger lipid droplets. Adrenocortical cells of Fads2−/− mice fed the low-PUFA diet also displayed fewer, smaller, and less circular mitochondria (Fig. 6, H to L), indicating perturbed mitochondrial function, thus standing in agreement with impaired steroidogenesis. Together, these data further indicate that FADS2-dependent PUFA synthesis determines the lipidomic landscape, mitochondrial structure, lipid droplet load, and steroidogenesis in adrenocortical cells.
FADS2 expression correlates with the expression of steroidogenic genes in the human adrenal gland and is increased in aldosterone-producing adenomas
To validate the significance of our findings in the human adrenal gland, we mined the public GEPIA2 database (http://gepia2.cancer-pku.cn/#index) (37) for steroidogenic genes and analyzed their expression in correlation with the expression of FADS2. FADS2 expression positively correlated with the expression of STAR, VDAC1, CYP11A1, Steroidogenic Factor-1 (SF-1), and protein kinase A (PKA) catalytic subunit A (PRKACA) in the human adrenal gland (fig. S6, A to E). VDAC1 interacts with StAR at the OMM, facilitating cholesterol transport into mitochondria (38). CYP11A1 catalyzes the side-chain cleavage reaction, which is the first step of steroidogenesis generating pregnenolone (39). PRKACA mediates PKA activation, which triggers steroidogenesis, and is involved in adrenocortical pathologies with ACTH-independent hypercortisolism, such as adrenocortical adenomas, carcinomas, or bilateral adrenocortical hyperplasias (40). SF-1 is the major regulator of steroidogenesis-related genes (41). The correlation of the expression of FADS2 with the expression of genes that play a critical role in the initiation of steroidogenesis in the human adrenal gland underscores the relevance of our findings. On the basis of GEPIA2 data, FADS2 also positively correlates with SF-1 and PRKACA expression in adrenocortical carcinoma (fig. S6, F and G).
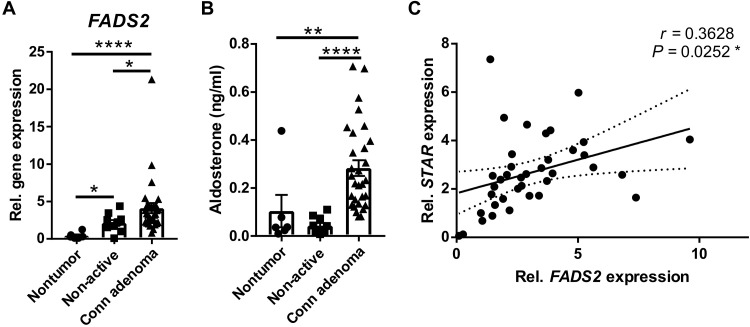
Along this line, we investigated the changes in FADS2 expression in human adrenal tumors. FADS2 expression was analyzed in nontumorous adrenocortical tissue, nonfunctioning adrenocortical adenomas (hormonally inactive), and aldosterone-producing adenomas (Conn adenomas) and was found to be significantly higher in aldosterone-producing adenomas compared to both, nontumorous adrenocortical tissue and nonfunctioning adenomas (Fig. 7A). Notably, FADS2 expression was also elevated in hormonally inactive tumors compared to nontumorous adrenocortical tissue (Fig. 7A), suggesting a potential role of FADS2 in adrenocortical tumorigenesis. Increased FADS2 expression in Conn adenomas was associated with enhanced pre-operative aldosterone levels in these patients compared to patients of the two other groups (Fig. 7B). In addition, FADS2 expression positively correlated with STAR expression in aldosterone- and cortisol-producing adrenocortical adenomas (Fig. 7C). These data collectively support that FADS2 is not only required for steroidogenesis in the human adrenal gland but may also promote increased steroid production in adrenocortical adenomas.
Fig. 7. FADS2 expression is increased in aldosterone-producing adenomas.
(A) FADS2 expression was determined in nontumorous adrenocortical tissue from pheochromocytoma patients (n = 6 patients), non-active adenomas (n = 10 patients), and aldosterone-producing adenomas (Conn adenomas) (n = 30 patients) by qPCR using 18S as a housekeeping gene. (B) Pre-operative aldosterone plasma levels in the same patients as in (A) were measured by LC-MS/MS. (C) Correlation of FADS2 with STAR expression in aldosterone- and cortisol-producing adrenocortical adenomas from 30 and 7 patients, respectively. FADS2 and STAR expression was determined by qPCR using 18S as a housekeeping gene. Data in (A) and (B) are presented as mean ± SEM. *P < 0.05; **P < 0.01; ****P < 0.0001
DISCUSSION
Chronically elevated cortisol levels can have deleterious consequences, including disturbed lipid and glucose metabolism, immunodeficiency, and neurological disorders (3, 22). Increased aldosterone levels promote hypertension and increase the risk of cardiovascular disease (1). Moderately elevated corticoid levels in obese subjects contribute to complications such as insulin resistance and hypertension and can thus substantially affect life quality but are rarely treated (4, 7, 8). The mechanisms underlying derailed adrenocortical hormone production in obesity are incompletely understood. Here, we investigated to which extent the lipidomic landscape of the adrenal gland affects its steroidogenic function. We analyzed the adrenal lipidome of lean and obese mice and found that a high content of phospholipids in longer and more unsaturated lipids is associated with increased steroidogenesis in obese mice. ARA is a particularly abundant acyl chain in adrenal phospholipids and is increased with obesity. We identified FADS2, the rate-limiting enzyme for PUFA synthesis, as a key determinant for the adrenal lipidome and steroidogenic function. FADS2 is highly expressed in the adrenal gland compared to other tissues and increases in the adrenal gland of obese animals. Moreover, we propose that not only adrenocortical FADS2-mediated ARA synthesis but also medulla-derived ARA may affect adrenocortical steroidogenesis.
The importance of the FADS2-ARA axis for steroidogenesis was validated in Fads2−/− mice, which displayed lower corticosterone and aldosterone plasma levels compared to WT littermates when receiving for 10 weeks a low-PUFA diet, while corticoid levels were restored by a PUFA-rich diet. In accordance, adrenocortical cells in Fads2−/− mice fed a low-PUFA diet had larger lipid droplets and disturbed mitochondria. Along the same line, in vitro treatment of adrenocortical cells with SC-26196, a FADS2 inhibitor, potently reduced the abundance of PUFA, including ARA acyl chains of mitochondrial phospholipids, mainly PC and PE. These are the most abundant phospholipids in all cellular membranes including mitochondrial membranes (14, 15) and important stores of ARA residues (42–44). FADS2 inhibition also reduced the mitochondrial membrane potential, oxygen consumption, and mitochondrial cholesterol uptake and diminished steroidogenesis, which was partially reversed by ARA supplementation. ARA was previously reported to be required for steroidogenesis and high amounts of arachidonic cholesterylesters are found in rat adrenal glands (27, 45). However, our study suggests that the phospholipidomic landscape and the ARA content of mitochondria specifically determines steroidogenesis. On the basis of our findings, we hypothesize that the PUFA content of phospholipids assembling the mitochondrial membrane is critical for proper mitochondrial membrane potential and bioenergetics, which is pivotal for cholesterol transport into mitochondria and steroidogenesis (10, 16–19).
Abundance of long-chain PUFA in phospholipids constructing phospholipid layers increases membrane fluidity, in contrast to saturated fatty acids, which make membranes more rigid (46, 47). In turn, membrane fluidity can determine enzymatic activity and secretory functions (46–49). Steroid hormones are rapidly synthesized from cholesterol through a cascade of enzymatic reactions taking place in the mitochondria and the ER (10). The activity of the enzymes involved in steroid biosynthesis depends on the membrane fluidity of these organelles (50, 51). Along this line, our data suggest that FADS2 providing PUFA residues may increase membrane fluidity in adrenocortical cells thereby promoting steroidogenesis. In agreement with this hypothesis, FADS2-deficient female and male mice are sterile due to changes in the membrane fluidity of gonadal cells encompassing less PUFA acyl groups (11, 12). Fertility of these mice is restored by dietary supplementation with ARA, EPA, and DHA (11, 52). On the other hand, FADS2 deficiency was reported to lead to a noncanonical conversion of linoleate to eicosatrienoic acid (20:3 n-6), which substitutes ARA in phospholipids thereby critically changing the structure and function of ER and Golgi membranes (53). However, the precise mechanisms linking the mitochondrial lipidome with steroidogenesis in the adrenal gland remain to be elucidated.
Having found that the adrenal lipidome is integral to the regulation of steroidogenesis, we next asked whether it can be modulated in established obesity by FADS2 inhibition. Along this line, we demonstrate that administration of SC-26196 through the food reduced the PUFA content of the adrenal gland and down-regulated corticosteroid production in obese mice. We also asked whether the adrenal lipidome and corticoid production can be modulated by other dietary interventions. To this end, EPA was selected as a candidate dietary component. EPA together with DHA, i.e., the two main fish oil components used for food supplementation, was previously shown to reduce corticosterone levels and anxiety in rats subjected to restrained stress test (54). EPA and DHA supplementation also reduced cortisol saliva levels and perceived stress in abstinent alcoholics (55). In addition, EPA decreased serum cortisol levels in patients with major depression disorder (56). Hence, the concept of omega-3 fatty acid–mediated regulation of glucocorticoid levels is not new, but the underlying mechanisms remained little understood. Here, we fed mice with HFD for 10 weeks and another 10 weeks with HFD containing or not icosapent ethyl, a highly purified EPA ethyl ester (36). We chose to administrate icosapent ethyl instead of EPA or EPA with DHA, based on the findings of large clinical studies, which showed that icosapent ethyl alone lowered the risk of cardiovascular events, non-HDL cholesterol, and triglycerides in patients with established cardiovascular disease, hypertriglyceridemia, or diabetes (34–36). We demonstrate that icosapent ethyl food supplementation increased the omega-3 fatty acid content of the adrenal lipidome, instigated transcriptional changes in the adrenal cortex opposite to the ones evoked by HFD, which were associated to lipid metabolism, mitochondrial function and steroid biosynthesis, and reduced corticosterone and aldosterone plasma levels. Accordingly, EPA treatment of cultured adrenocortical cells reduced FADS2 expression and diminished forskolin-stimulated corticoid production, indicating that icosapent ethyl dietary supplementation may directly regulate the steroidogenic function of adrenocortical cells. In agreement with these results, EPA was previously shown to down-regulate FADS2 expression in THP-1 cells (57), 3T3-L1 adipocytes (58), and HepG2 hepatocytes (59). Hence, icosapent ethyl is likely to influence the lipidome of not only the adrenal gland but also several other tissues.
Last, to substantiate the role of FADS2 in the human adrenal gland, we analyzed its expression in adrenal nontumorous and tumorous tissue. We demonstrate that FADS2 expression is higher in aldosterone-producing adenomas compared to nonfunctioning adenomas or nontumorous adrenocortical tissue and correlates with the expression of STAR in aldosterone- and cortisol-producing adenomas.
These data collectively demonstrate that FADS2 is a major regulator of steroidogenesis in the adrenal gland due to its key role in shaping the lipidomic landscape of adrenocortical cells. Moreover, our findings indicate that the adrenal lipidome can be modulated by icosapent ethyl dietary supplementation, which consequently influences corticoid production, thereby endorsing the evaluation of icosapent ethyl dietary supplementation as a means of regulation of cortisol and aldosterone levels in obese subjects. Last, our data indicate a clear correlation between FADS2 expression and steroidogenic capacity in the human adrenal gland and suggest that the role of FADS2–mediated lipidomic changes in adrenocortical tumorigenesis merits further investigation.
MATERIALS AND METHODS
Mice and in vivo experiments
The Fads2−/− mice were developed by Stroud et al. (12). The first 4 postnatal weeks female WT and Fads2−/− littermate mice were kept with their mothers and fed a PUFA-rich diet. After weaning at 4 weeks, they were kept for another 10 weeks on a PUFA-rich or low-PUFA diet. Composition of the diets is shown in table S4.
Six-weeks-old male WT C57BL/6J mice were fed for 20 weeks a LFD (D12450B) or a HFD (D12492) both from Research Diets, NJ, USA. In some experiments, WT C57BL/6J mice were fed for 8 weeks a HFD and for another 8 weeks a HFD supplemented with SC-26196 (Biorbyt Ltd.). SC-26196 was mixed in the HFD at a concentration of 625 mg/kg of diet corresponding to approximately 100 mg/kg of animal weight. In other experiments, WT mice were fed for 10 weeks a HFD and for another 10 weeks a HFD supplemented with icosapent ethyl (Vascepa). The content of Vascepa capsules was mixed by Research Diets with the diet at a concentration of 1.125 g/kg icosapent ethyl (corresponding to approximately 180 mg/kg of animal weight). Mice were weighed every week. Glucose and insulin tolerance tests were performed as previously described (20, 21). Three to 5 mg of hair (2 to 3 mm close to the skin on the back of the mice) was cut while gently holding the mice without anesthesia.
At the end of the experiment, blood was collected retroorbitally in EDTA tubes, the mice were sacrificed by cervical dislocation, and organs were excised. Tissues were immediately snap-frozen or processed for cell isolation. For electron microscopy, mice were anesthetized with isoflurane and systemically perfused with 4% paraformaldehyde (PFA), 0.1% glutaraldehyde (GA), 0.1 M sodium phosphate buffer, or phosphate-buffered saline (PBS) (pH 7.4). Experiments were approved by the Landesdirektion Sachsen, Germany and the institutional animal care and use committee of Azabu University.
Human samples
The study included retrospective data and biosamples from 53 patients who underwent surgery because of adrenal tumors, 10 patients with nonfunctional adenomas, 30 patients with aldosterone-producing adenomas (Conn adenomas), 7 with cortisol-producing adenomas, and 6 patients with pheochromocytoma. Patients were enrolled in the ENSAT and the NeoExNet registry studies and the prospective clinical trials, Prospheo/Muppet (https://muppet.eu/) and PMT-trial (pmt-study.pressor.org) in three tertiary centers in Germany: University Hospital Carl Gustav Carus, TU Dresden, University Hospital Wuerzburg, and University Hospital Munich, Ludwig-Maximilians-Universität München. The studies were approved by the Ethics Committees at the Medical Faculty of each University. All patients provided written informed consent. Measurements of steroids in plasma were performed using LC-MS/MS (60). Potential contamination with medulla tissue was excluded in all samples by examination of tyrosine hydroxylase (TH) and phenylethanolamine N-methyltransferase (PNMT) expression by quantitative polymerase chain reaction (qPCR). Among the limitations of the study design is that possible influences from antihypertensive medication interfering with the renin-angiotensin-aldosteron system were not considered in the analysis.
Sorting of adrenocortical cell populations
Adrenal glands were isolated and cleaned from the surrounding fat tissue, and the adrenal cortex was separated from the medulla under a dissecting microscope. The cortex was collected in a digestion buffer, consisting of collagenase I and bovine serum albumin (BSA; both at 1.6 mg/ml concentration; Sigma-Aldrich) dissolved in PBS, and digested for 25 min at 37°C while shaking at 900 rpm. Dissociated cells were passed through a 22-G needle and then a 100-μm cell strainer and centrifuged at 300g for 5 min at 4°C. Cells were then washed in MACS buffer (0.5% BSA and 2 mM EDTA in PBS), and endothelial cells (CD31+) and leukocytes (CD45+) were sequentially positively selected using CD31 and CD45 MicroBeads (Miltenyi Biotec), respectively, according to manufacturer’s instructions. Briefly, pelleted cells were resuspended in 190-μl MACS buffer, mixed with 10-μl CD31 MicroBeads, incubated for 15 min at 4°C, washed with 2-ml MACS buffer, and centrifuged at 300g for 10 min. The cell pellet was resuspended in 500-μl MACS buffer and applied onto MS Column placed on a MACS Separator. The columns were washed and the flow-through was collected. CD31+ cells were positively sorted. The flow-through was centrifuged at 300g for 5 min, and the pelleted cells were subjected to the same procedure using CD45 MicroBeads. The flow-through containing CD31−CD45− adrenocortical cells was centrifuged at 300g for 5 min, and the pelleted cells were collected. CD45+ cells were positively sorted. Collected cell populations were kept for transcriptional analysis (61).
Cell and explant culture and treatments
Adrenal glands or adrenal corteces were isolated from WT C57BL/6J mice, cleaned from the surrounding fat tissue, and incubated in collagenase buffer [collagenase type I (1.6 mg/ml) and BSA (1.6 mg/ml)] for 45 min at 37°C while shaking at 900 rpm. Dissociated cells were passed through 22-G needle and a 100-μm cell strainer and centrifuged at 300g for 5 min at 4°C. Cells of a pool from two adrenals of each mouse were distributed in four 0.2% gelatin-precoated wells of a 96-well plate in DMEM/F12 supplemented with 1% fetal bovine serum, penicillin (50 U/ml), and streptomycin (50 μg/ml). Two hours after seeding, 10 μM SC-26196 (Tocris) or the same amount of carrier [dimethyl sulfoxide (DMSO)] was added without changing the medium. In some experiments, medium was additionally supplemented with 150 μM ARA (Sigma-Aldrich) or same amount of solvent (endotoxin-free water). When induction of steroidogenesis was required, the medium was changed 18 hours after treatment, and the cells were incubated with or without ACTH (10 ng/ml) for 60 min in the presence of either 10 μM SC-26196 or DMSO. Supernatants were kept for steroid hormone measurement, and cell lysates were collected for transcriptional analysis. In some experiments, primary CD31−CD45− adrenocortical cells were treated or not for 18 hours with 66 μM EPA (Sigma-Aldrich) followed or not by ACTH stimulation (61).
Medullas were isolated from WT C57BL/6J mice and transferred into DMEM/F12 medium. Six hours later, the medulla explant supernatants were collected and applied onto adrenocortical cell cultures. These were previously treated for 6 hours with SC-26196 (10 μM) or DMSO. SC-26196 or DMSO was re-added, and adrenocortical cells were incubated for another 18 hours until supernatants were collected for steroid measurement. In other experiments, medulla explants were treated for 6 hours with SC-26196 (10 μM) or DMSO, washed thoroughly, and left another 18 hours in culture. Their supernatant was then applied on adrenocortical cell cultures, and 8 hours later, supernatants of the latter were collected and analyzed by LC-MS/MS.
NCI-H295R cells (obtained from American Type Culture Collection catalog no. CRL-2128) were maintained in DMEM/F12 medium supplemented with 2.5% Nu serum type I (Corning), 1% insulin transferrin selenium (Gibco), penicillin (50 U/ml), and streptomycin (50 μg/ml). They were treated for 18 hours with 10 μM SC-26196 or DMSO. In some experiments, NCI-H295R cells were treated or not for 48 hours 66 μM EPA, and in the last 24 hours, they were stimulated or not with forskolin (10 μM; Sigma-Aldrich) (61).
Cell fractionations
Cells were trypsinized and pelleted by centrifugation (500g, 5 min, 4°C). The cells were gently resuspended in homogenization buffer [200 mM mannitol, 50 mM sucrose, 10 mM KCl, 1 mM EDTA, 25 mM Hepes (pH 7.4), 1 μM aprotinin, 10 μM leupeptin, and 20 μM phenylmethylsulfonyl fluoride] and homogenized on ice with 20 strokes with a Dounce homogenizer. The samples were then centrifuged (800g, 10 min, 4°C) to pellet the nuclei. The supernatant was mixed with iodixanol solution (Optiprep, Sigma-Aldrich) to a final concentration of 20%. The samples were placed at the bottom of a tube and overlaid with 500 μl of iodixanol solutions (18, 16, 14, 12, 10, and 5%) and lastly with 500 μl of Hepes-buffered saline (HBS) [50 mM Hepes and 150 mM NaCl (pH 7.4)]. The samples were centrifuged in a Beckman SW60Ti swing-out rotor (45,000 rpm, 5 hours, 4°C). After centrifugation, 15 280-μl aliquots were collected from top of the tubes.
Fractions 10 to 12 (SDHB-positive) and 4 to 5 (HSL-positive) were considered as mitochondrial and lipid droplet fractions, respectively. Protein concentration was measured in the mitochondrial and lipid droplet pools with the BCA Protein Assay Kit (Thermo Fisher Scientific), and fractions were analyzed for cholesterol and CE concentrations.
Lipidomic analysis of isolated mitochondria
Fractions containing mitochondria were combined and extracted with three volumes of ice-cold chloroform/methanol [10:1, v/v, both containing 0.1% butylated hydroxytoluene (Merck, ≥99%)]. Samples were incubated on ice for 15 min, before the organic phase was collected after centrifugation (5 min, 5,000g). The aqueous phase was re-extracted with two volumes chloroform/methanol as described above. Combined organic phases were dried in vacuo. Methanol (ULC-MS grade, Biosolve B.V.) and chloroform (EMSURE ACS, ISO, Reag. Ph Eur; Supleco) were of highest purity.
Lipid extracts were dissolved in 50 μl of 2-propanol (ULC/MS-CC/SFC grade, >99.95%; Biosolve B.V.) and analyzed by LC-MS/MS. Lipids (2.5 μl loaded in positive mode, 5 μl in negative mode) were separated by reverse phase chromatography (Accucore C30 column; 150 mm by 2.1 mm, 2.6 μM, 150 Å; Thermo Fisher Scientific) using a Vanquish Horizon ultrahigh-performance liquid chromatography (UHPLC) system (Thermo Fisher Scientific) coupled on-line to a Q Exactive Plus Hybrid Quadrupole Orbitrap mass spectrometer (Thermo Fisher Scientific) equipped with a heated electrospray ionization (HESI) source. Column was operated at 50°C and a flow rate of 0.3 ml/min. Chromatographic gradient included following steps: 0 to 20 min, 10 to 86% B; 20 to 22 min, 86 to 95% B; 22 to 26 min, 95% isocratic; 26 to 26.1 min, 95 to 10% B, followed by 5 min re-equilibration at 10% B. Eluent A consisted of acetonitrile/water (50:50, v/v, both ULC/MS-CC/SFC grade, Biosolve B.V.) and Eluent B of 2-propanol/acetonitrile/water (85:10:5, v/v/v), both containing 5 mM ammonium formate (MS grade, Sigma-Aldrich) and 0.1% formic acid (ULC/MS-CC/SFC grade, Biosolve B.V.). HESI parameters are as follows: sheath gas, 40 arbitrary units; auxiliary gas, 10 liters/min; sweep gas, 1 liter/min; spray voltage, 3.5 kV (positive ion mode) and 2.5 kV (negative ion mode); ion transfer temperature, 300°C; S-lens radio frequency (RF) level, 35%; aux gas heater temperature, 370°C. Data were acquired using data-dependent acquisition: full scan resolution 140,000 at mass/charge ratio (m/z) 200, AGC target 1 × 106, maximum IT 100 ms, scan range m/z 350 to 1200; data-dependent MS/MS: resolution 17,500 at m/z 200, AGC target 1 × 105, maximum IT 60 ms, loop count 15, isolation window 1.2 m/z, stepped normalized collision energies of 10, 20, and 30%, AGC target of 2 × 102, dynamic exclusion 10 s, isotope exclusion on, default charge state 1 (62).
Lipid identification and relative quantification was performed using Lipostar2 (63). Briefly, supersample filter considering only lipids with isotopic pattern and with an MS/MS spectrum were kept. Features were searched against the LIPID MAPS structural database (downloaded November 2021) and proposed identifications obtained by automatic approval considering three and four stars. Data were normalized using the Sum/Average normalization (area sum). Statistical analysis was performed in MetaboAnalyst 5.0 (sample normalization by median, data log transformed, pareto scaled) (64).
Product ion scan for cholesterol and cholesteryl esters
For quantification of cholesterol and cholesteryl esters, equal volumes of mitochondria fractions (180 μl) were combined, cholesterol-2,3,4-13C (5 nmol; Sigma-Aldrich) and SPLASHLipidomix (3 μl; Avanti Polar Lipids Inc) spiked, and samples were incubated for 15 min on ice. Lipids were extracted as described above.
Lipid extracts were reconstituted in 55 μl of 2-propanol, and 10 μl was loaded onto a Accucore C30 column (150 mm by 2.1 mm, 2.6 μM, 150 Å; Thermo Fisher Scientific) installed on a Vanquish Horizon UHPLC (Thermo Fisher Scientific) coupled on-line to a Orbitrap Exploris 240 (Thermo Fisher Scientific) mass spectrometer equipped with a HESI source. Lipids were separated at a flow rate of 0.3 ml/min and a column temperature of 50°C by a gradient starting from 10% B (0 to 20 min 10 to 80% B, 20 to 37 min 80 to 95% B, 37 to 41 min 95 to 100% B, 41 to 49 min 100% B, and 49.1 to 57 min 10% B). The column was re-equilibrated for 7.9 min at 10% eluent B. Eluent A consisted of acetonitrile/water (50:50, v/v, both ULC/MS-CC/SFC grade Biosolve B.V.) and eluent B of 2-propanol/acetonitrile/water (85:10:5, v/v/v), both containing 5 mM Ammonium formate (MS grade, Sigma-Aldrich) and 0.1% formic acid (ULC/MS-CC/SFC grade, Biosolve B.V.).
For cholesterol and CE, a scheduled product ion scan method with the following parameters was used: spray voltage, 3500 V; sheath gas, 40 arbitrary units; aux gas, 10 arbitrary units; sweep gas, 1 arbitrary units; vaporizer, 370°C; ion transfer tube, 300°C; polarity, positive; default charge state, 1; isolation window, 1 m/z; resolution at m/z 200 to 45,000; HCD collision energy (%), 17, 27, 37; fixed first mass, 70 m/z; AGC target, standard; maximum injection time, auto; EASY-IC, start run. Quantification was performed in Skyline (version 22.2.0.255, MacCoss Lab, University of Washington) using cholesten ion (m/z 369.35) as a quantifier (65). CE was quantified relative to the 18:1-d7-cholesteryl ester present in the SPLASHLipidomix mix. Signal of cholesterol-2,3,4-13C was used to quantify endogenous cholesterol.
Shotgun lipidomics and analysis
Shotgun lipidomics were performed as previously described (61). Adrenal glands were homogenized in ammonium-bicarbonate buffer (150 mM ammonium bicarbonate, pH 7) with TissueLyser (Qiagen). Protein content was assessed using the BCA Protein Assay Kit (Thermo Fisher Scientific). Equivalents of 20 μg of protein were taken for MS analysis. MS-based lipid analysis was performed as described (24). Lipids were extracted using a two-step chloroform (Sigma-Aldrich)/methanol (Thermo Fisher Scientific) procedure (66). Samples were spiked with internal lipid standard mixture containing: CL 16:1/15:0/15:0/15:0, Cer 18:1;2/17:0, HexCer 18:1;2/12:0, LPA 17:0, LPC 12:0, LPE 17:1, LPG 17:1, LPI 17:1, LPS 17:1, PA 17:0/17:0, PC 17:0/17:0, PE 17:0/17:0, PG 17:0/17:0, PI 16:0/16:0, PS 17:0/17:0, CE 20:0, SM 18:1;2/12:0;0, cholesterol D6 (all Avanti Polar Lipids), TAG 17:0/17:0/17:0, and DAG 17:0/17:0 (both Larodan Fine Chemicals). Synthetic lipid standards were purchased from Avanti Polar Lipids, Larodan Fine Chemicals, and Sigma-Aldrich, and all chemicals were analytical grade. After extraction, the organic phase was transferred to an infusion plate and dried in a speed vacuum concentrator (Martin Christ). The dry extract from the first step was resuspended in 7.5 mM ammonium acetate (Sigma-Aldrich) in chloroform/methanol/propan-2-ol (Thermo Fisher Scientific) (1:2:4 V:V:V) and the second-step dry extract in 33% ethanol of methylamine (Sigma-Aldrich)/chloroform/methanol (0.003:5:1 V:V:V) solution. All liquid handling steps were performed using Hamilton Robotics STARlet robotic platform with the Anti Droplet Control feature for organic solvents pipetting.
Samples were analyzed by direct infusion on a QExactive mass spectrometer (Thermo Fisher Scientific) equipped with a TriVersa NanoMate ion source (Advion Biosciences, Ithaca, NY). Samples were analyzed in both positive and negative ion modes with a resolution of R(m/z=200) = 28,0000 for MS and R(m/z=200) = 17,500 for MS/MS experiments, in a single acquisition. MS/MS was triggered by an inclusion list encompassing corresponding MS mass ranges scanned in 1-Da increments (67). Both MS and MS/MS data were combined to monitor CE, DAG, and TAG ions as ammonium adducts; PC, PC O-, as acetate adducts; and CL, PA, PE, PE O-, PG, PI, and PS as deprotonated anions. MS only was used to monitor LPA, LPE, LPE O-, LPI, and LPS as deprotonated anions; Cer, HexCer, SM, LPC, and LPC O- as acetate adduct and cholesterol as ammonium adduct of an acetylated derivative (68).
Data were analyzed with an in-house developed lipid identification software based on LipidXplorer (69, 70). Data postprocessing and normalization were performed using an in-house developed data management system. Only lipid identifications with a signal-to-noise ratio > 5 and a signal intensity fivefold higher than in corresponding blank samples were considered for further data analysis. The dataset was filtered by applying an occupational threshold of 50%, meaning that lipids that were present in less than 50% of the samples in a group were set to zero in that group. Dataset was log2-scaled before PCA. The descriptive analysis was performed on the mol %–transformed dataset, i.e., picomol quantities were divided by the sum of the lipids detected in the respective sample and multiplied by 100. Total carbon chain length and double bonds plots result from grouping together all the lipids that present the same number of carbon atoms (total length) or the same number of double bonds (degree of unsaturation) and calculating the mean and standard deviation in each group of samples (LFD and HFD). To further inspect general trends in the degree of unsaturation and length of storage and membrane lipids under different perturbations, the average weighted number of double bonds (DBI) and length (LI), as previously described (71). Statistical tests were performed on the mol %–transformed data. First, normality was assessed by means of the Shapiro-Wilkinson test, and according to its result, either the Welch t test or the Wilcoxon test was used. P values were then adjusted following the Benjamini-Hochberg correction. Analysis was done in R (72). Plots were generated using ggplot2 (73).
Lipid isomer and oxylipin measurement
Frozen adrenal glands were homogenized in 200 μl of PBS buffer. Lipids were extracted from adrenal lysates or supernatants in chloroform/methanol (Folch’s protocol). The extract was evaporated to dryness under stream of nitrogen at 40°C. The residue was dissolved in 100 μl of ethanol. For FFA, 50 μl were used. HPLC measurement of FFA was performed using an Agilent 1290 HPLC system with binary pump, autosampler, and column thermostat equipped with a Phenomenex Kinetex-C18 column 2.6 μm, 2.1 by 150 mm column (Phenomenex) using a solvent system of acetic acid (0.05%) and acetonitrile. All solvents and buffers in LC-MS–grade were purchased from VWR Germany. The HPLC was coupled with an Agilent 6470 triplequad mass spectrometer with electrospray ionisation source operated in negative multiple reaction monitoring. The quantification was performed with individual calibration curves using deuterated standards (Cayman Chemical).
To release fatty acids and lipid mediators from phospholipids, 50 μl lipid extract was dissolved with 500 μl of phospholipase A2 (PLA2) buffer consisting of 80 mM Hepes, 300 mM NaCl, 20 mM CaCl2, and BSA (2 mg/ml) in 60% glycerol (pH 7.4). This solution was spiked with 5 μl of butylhydroxytoluen and 2.2 U/26 μl of PLA2 from honey bee (Merck) and incubated 1 hour at 25°C. One hundred microliters of hydrolysate was analyzed as described above. The results were the sum of FFAs and fatty acids from phospholipids; hence, the amount of fatty acyl chains of phospholipids was calculated by subtracting the amount of FFA from phospholipidic fatty acyl chains.
For measurement of lipid mediators, 400 μl of hydrolysate was spiked with 100 pg of d4-PGE2-13 as an internal standard (Cayman Chemical). Acetonitrile (400 μl) was added for protein precipitation. After centrifugation and pH adjustment at 6.0, the obtained supernatant was added to Bond Elute Certify II columns (Agilent Technologies) for solid phase extraction. The eluate was evaporated on a heating block at 40°C under a stream of nitrogen to obtain solid residues, which were dissolved in 100 μl of methanol/water (60/40). The residues were analyzed using an Agilent 1290 HPLC system with binary pump, multisampler, and column thermostat with a Zorbax Eclipse plus C-18, 2.1 by 150 mm, 1.8-μm column using a gradient solvent system of aqueous acetic acid (0.05%) and acetonitrile/methanol 50:50. The flow rate was set at 0.3 ml/min, and the injection volume was 20 μl. The HPLC was coupled with an Agilent 6495 Triplequad mass spectrometer (Agilent Technologies) with electrospray ionisation source. Analysis was performed with multiple reaction monitoring in negative mode, at least two mass transitions for each compound.
Steroid hormone measurement
Steroid hormones in plasma and cell culture supernatants were analyzed by LC-MS/MS as previously described (60, 61). Fifty to 100 μl of plasma or cell culture supernatants were extracted by solid phase extraction using positive pressure, followed by a dry-down under gentle stream of nitrogen. Residues were reconstituted in 100 μl of the initial LC mobile phase, and 10 μl were injected for detection by the triple quadrupole mass spectrometer in multiple reaction-monitoring scan mode using positive electrospray ionization. Quantification of steroid concentrations was done by comparisons of ratios of analyte peak areas to respective peak areas of stable isotope-labeled internal standards obtained in samples to those of calibrators.
Corticosterone and 11-dehydrocorticosterone in hair were extracted with methanol as described elsewhere (74). Briefly, hair (3 to 5 mg) was washed by addition of 2.5 ml of isopropanol. After 3 min, the isopropanol was decanted, and hair samples were dried in a fume hood. After drying, hair samples were extracted in 1.8 ml of methanol that contained the internal standard mixtures for 18 hours at room temperature (RT). The methanolic extract was dried down, followed by reconstitution in respective mobile phase and analysis by LC-MS/MS as previously described (60) with minor modifications. Briefly, a QTRAP 6500+ (Sciex, Darmstadt) coupled to an Aquity i-class ultra-performance liquid chromatography system (Waters, Eschborn) was used. Chromatographic separation was achieved as described before (60). Corticosterone and 11-dehydrocorticosterone were detected by using the multi reaction monitoring scan mode with respective quantifier ions of 347.1->329.2 and 345.1->121.1 and qualifier ions of 347.1->90.9 and 345.1->242.3. An automatic flow injection analysis revealed optimal ion source settings of curtain gas (40), ion spray voltage (5500), temperature (500), gas 1 (70), and gas 2 (50). Quantification of steroid hormones was achieved by comparisons of ratios of analyte peak areas to internal standard peak areas observed in samples to those in calibrators. Measured steroid concentrations were normalized to sample weights.
ACTH measurement
ACTH was measured in EDTA mouse plasma 4× diluted in PBS with an enzyme-linked immunosorbent assay (ELISA) kit (Abnova, KA0917) according to the manufacturer’s instructions.
RNA sequencing
For transcriptome mapping, strand-specific paired-end sequencing libraries from total RNA were constructed using a TruSeq stranded Total RNA kit (Illumina Inc). Sequencing was performed on an Illumina HiSeq3000 (1 × 75 base pairs). Low-quality nucleotides were removed with the Illumina fastq filter, and reads were further subjected to adaptor trimming using cutadapt (75). Alignment of the reads to the mouse genome was done using STAR Aligner (76) using the parameters: “–runMode alignReads –outSAMstrandField intronMotif –outSAMtype BAM SortedByCoordinate --readFilesCommand zcat”. Mouse Genome version GRCm38 (release M12 GENCODE) was used for the alignment. The parameters: “htseq-count -f bam -s reverse -m union -a 20,” HTSeq-0.6.1p1 (77) were used to count the reads that map to the genes in the aligned sample files. The GTF file (gencode.vM12.annotation.gtf) used for read quantification was downloaded from Gencode (https://gencodegenes.org/mouse/release_M12.html). Gene centric differential expression analysis was performed using DESeq2_1.8.1 (78). The raw read counts for the genes across the samples were normalized using “rlog” command of DESeq2, and subsequently, these values were used to render a PCA plot using ggplot2_1.0.1 (79).
Pathway and functional analyses were performed using GSEA (79) and EGSEA (80). GSEA is a standalone software with a graphical user interface. To run GSEA, a ranked list of all the genes from DESeq2-based calculations was created usingthe −log10 of the P value. This ranked list was then queried against Molecular Signatures Database–, Kyoto Encyclopedia of Genes and Genomes (KEGG)–, Gene Ontology–, Reactome-, and Hallmark-based repositories. EGSEA is an R/Bioconductor-based command line package. For doing functional analyses using EGSEA, a differentially expressed list of genes with parameters log2 foldchange > 0.3 and P adjusted <0.05 was used. The KEGG database repository was used for performing the functional analyses.
For constructing heatmaps, the “rlog-normalized” expression values of the significantly expressed genes (P adjusted < 0.05) were scaled using z-transformation. The resulting matrices were visually rendered using MORPHEUS.
Electron microscopy
After animal perfusion with PFA/GA, adrenal glands were immediately transferred into 4% PFA, 2% GA, 0.1 M sodium phosphate buffer, or PBS (pH 7.4) for 1 hour at RT and stored in 0.1% GA and 1% PFA in PBS at 4°C until processing. After washing with PBS, adrenal glands were further dissected into three pieces. They were contrasted with 1% osmium tetroxide/1.5% potassium cyanoferrate for an hour at RT. Contrast was further enhanced by incubation with 0.5% uranyl acetate overnight in the cold. Dehydration with a graded series of ethanol followed (10 min each step, three steps of pure ethanol for 20 min). Samples were gradually infiltrated and embedded in Epon resin and cured at 60°C for 48 hours. Ultrathin sections (70 nm) were cut on a Leica UC7 ultramicrotome and poststained with uranyl and lead. Samples were evaluated and images acquired on a Tecnai Biotwin T12 transmission electron microscope (Philips/FEI/Thermo Fisher Scientific) with a digital F416 charge-coupled device camera (Tietz Video and Image Processing Systems).
Quantification of mitochondrial structural features
Mitochondrial features (area, perimeter, circularity, and aspect ratio) were analyzed in electron microscope images using the Fiji software (81) and the “particle analysis” plugin. Outlines of mitochondria were marked by hand using the “freehand selection” tool. At least 16 images obtained from different areas were analyzed per adrenal gland.
Transcriptional analysis
RNA was isolated from frozen tissues with the TRI Reagent (Molecular Research Center, Inc.) after mechanical tissue disruption or from cells with the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions. RNA obtained with TRI Reagent was subsequently extracted with Chloroform and the NucleoSpin RNA, Maxi kit (Macherey-Nagel). Reverse transcription was performed with the iScript cDNA Synthesis kit (Bio-Rad), and cDNA was analyzed by qPCR using the SsoFast Eva Green Supermix (Bio-Rad), a CFX384 real-time System C1000 Thermal Cycler (Bio-Rad) and the Bio-Rad CFX Manager 3.1 software (61). Primers used are listed in table S6.
Plasmid transfection
NCI-H295R cells were transfected with a FADS2-overexpressing plasmid (Origene NM_004265) or control pCMV plasmid. Briefly, cells were plated in 24-well plates at 70% confluence, and the next day, they were transfected with 0.3 μg of DNA using Lipofectamine LTX with Plus Reagent (Thermo Fisher Scientific), according to the manufacturer’s instructions.
Western blot analysis
Cells or tissues were lysed with 10 mM tris-HCl (pH 7.4) + 1% SDS + 1 mM sodium vanadate, cell lysates were centrifuged at 16,000g for 5 min at 4°C, and supernatants were collected. Total protein concentration was measured using the BCA Protein Assay Kit (Thermo Fisher Scientific) in cell lysates or cell fractions. Protein samples were prepared with 5× Reducing Laemmli buffer containing same amount of protein, denatured at 95°C for 5 min, and loaded on a 10% acrylamide gel (Invitrogen) for SDS–polyacrylamide gel electrophoresis. PageRuler Prestained Protein Ladder (Thermo Fisher Scientific) was used as a protein size ladder. The separated proteins were transferred on Amersham Protran nitrocellulose membrane (GE Healthcare Lifescience). After blocking with 5% skimmed milk in Tris-buffered saline with Tween20 (TBS-T) [0.1% Tween 20 (Sigma-Aldrich) in 1× tris-buffered saline] for 1 hour at RT, membranes were incubated overnight at 4°C with anti-FADS2 (1:2000; Invitrogen, PA5-87765), anti-StAR (1:300; Santa Cruz, SC-25806), anti-SDHB (1:1000; Sigma-Aldrich, HPA002868), anti-HSL (1:1000, Cell Signaling, #4107), anti-tubulin (1:3,000; Sigma-Aldrich, T5168) or anti-vinculin (1:3000; Cell Signaling #4650) and diluted in 5% BSA in TBS-T. After washing, membranes were incubated for 1 hour at RT with secondary antibodies: goat anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP)–conjugated (1:3000; Jackson ImmunoResearch) or goat anti-mouse IgG HRP-conjugated (1:3000; Jackson ImmunoResearch), diluted in 5% skimmed milk in TBS-T. The signal was detected using the Western blot Ultra-Sensitive HRP Substrate (Takara) and imaged using the Fusion FX Imaging system (PeqLab Biotechnologie).
Immunofluorescence
Immunofluerescent stainings were performed as previously described (82). Adrenal glands cleaned from surrounding fat tissue were fixed in 4% PFA in PBS, washed overnight in PBS, cryopreserved in 30% sucrose (AppliChem GmbH) in PBS overnight at 4°C, embedded in Optimal Cutting Temperature (O.C.T.) compound (Tissue-Tek) and frozen at −80°C. Each adrenal gland was cut into 8-μm-thick serial sections. Before staining, adrenal sections were prewarmed at RT for 30 min. Adrenal sections were washed twice with PBS, permeabilized with 0.5% Triton X-100 in PBS for 15 min, treated with TrueBlack Lipofuscin Quencher (1:40 in 70% ethanol; Biotium) for 30 s to reduce autofluorescence, and blocked in Dako Protein Block, serum-free buffer for 1 hour at RT. Then, sections were incubated overnight at 4°C with anti-FADS2 (1:250, Invitrogen, PA5-87765), washed with PBS, and incubated for 2 hours at RT with the secondary antibody Alexa Fluor 555 donkey anti-rabbit (1:300; Thermo Fisher Scientific, #A-31572) together with 4′,6-diamidino-2-phenylindole (DAPI; 1:5000; Roche), both diluted in Dako Antibody Diluent. After washing with PBS, cryosections were mounted with Fluoromount (Sigma-Aldrich), covered with 0.17-mm cover glass, fixed with nail polish, and kept at 4°C until imaging. Adrenal cross sections were imaged as tile sections taken with an Axio Observer.Z1 (Zeiss) and a Plan-Apochromat 20× objective.
Histology
Adrenal glands were fixed in 4% PFA and paraffin-embedded. Sections were stained with hematoxylin and eosin. Images were obtained using an Axio Observer.Z1 (Zeiss) and a Plan-Apochromat 20× objective.
Flow cytometry
Adrenocortical cells (CD31−CD45−) were treated for 18 hours with DMSO or SC-26196 (10 μM) and were then incubated with Mitotracker Green (0.25 μM; Invitrogen), tetramethylrhodamine, ethyl ester (TMRE) (2.5 μM; Thermo Fisher Scientific), for 30 min at 37°C in FACS buffer (0.5% BSA and 2 mM EDTA in PBS) in dark. Live cells were selected by DAPI staining. FACS was performed using LSR Fortessa X20 flow cytometer, and data were analyzed with the FlowJo software.
Seahorse analysis
Primary adrenocortical cells were plated in 0.2% gelatin-precoated XF96 cell culture microplate (Agilent) (8*104 cells per well) and treated for 18 hours with DMSO or SC-26196 (10 μM). OCR was measured with a Seahorse XF96 Analyzer (Agilent Technologies). The experimental medium used was XF Base Medium supplemented with glucose (10 mM), pyruvate (1 mM), and glutamine (2 mM).
Statistical analysis
Data were analyzed with Mann-Whitney U test, Students t test, one-way analysis of variance (ANOVA) with post hoc Tukey’s test for multiple comparisons or Pearson analysis with P < 0.05 set as a significance level using the GraphPrism 7 software.
Acknowledgments
We gratefully acknowledge M. T. Nakamura (University of Illinois at Urbana-Champaign, Urbana, IL, USA), who generated the Fads2−/− mice and contributed to the establishment of the experimental setup used with these mice. We also thank D. Kaden and C. Mund for technical assistance. Last, we thank the Electron Microscopy Facility of the Max Planck Institute of Molecular Cell Biology and Genetics (MPI-CBG), especially W. Leng, for services.
Funding: This work was supported by funds from the Deutsche Forschungsgemeinschaft (SFB-TRR 205 to V.I.A., N.B., C.P., B.W., S.R.B., A.R., and M.K. and 465137452; INST 269/910-1 FUGG to M.P.).
Author contributions: A.W.: Methodology, validation, formal analysis, investigation, data curation, and writing—original draft; I.M.: Methodology, validation, investigation, and editing; A.P.: Software, formal analysis, and data curation; A.S.: Software, formal analysis, and data curation; M.W.: Methodology and investigation; A.H.: Investigation; N.B.: Human samples; C.P.: Human samples and clinical data; A.D.: Software and formal analysis; M.R.: Methodology, validation, and formal analysis; I.K.: Resources; C.A.: Human samples; A.R.: Human samples; B.W.: Resources and editing; S.R.B.: Editing; M.K.: Human samples and clinical data; M.P.: Formal analysis and data curation; T.M.: Methodology, validation, investigation, and resources; M.F.: Methodology, formal analysis, investigation, and data curation; M.G.: Methodology, formal analysis, investigation, and data curation; T.C.: Resources, editing, and funding acquisition; P.M.: Conceptualization; V.I.A.: Conceptualization, validation, resources, data curation, writing—original draft, editing, supervision, project administration, and funding acquisition.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. RNA-seq data are available through Gene Expression Omnibus (GEO) with accession number GSE216327.
Supplementary Materials
This PDF file includes:
Tables S1 to S6
Figs. S1 to S6
REFERENCES AND NOTES
- 1.R. D. Feldman, Aldosterone and blood pressure Regulation. Hypertension 63, 19–21 (2014). [DOI] [PubMed] [Google Scholar]
- 2.S. L. Lightman, M. T. Birnie, B. L. Conway-Campbell, Dynamics of ACTH and cortisol secretion and implications for disease. Endocr. Rev. 41, bnaa002 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.C. A. Koch, Cushing syndrome and glucocorticoid excess, in Disorders of Blood Pressure Regulation: Phenotypes, mechanisms, therapeutic options, A. Berbari, G. Mancia, Eds, (Springer, 2018), pp. 481–512. [Google Scholar]
- 4.D. A. Calhoun, K. Sharma, The role of aldosteronism in causing obesity-related cardiovascular risk. Cardiol. Clin. 28, 517–527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. M. Swierczynska, I. Mateska, M. Peitzsch, S. R. Bornstein, T. Chavakis, G. Eisenhofer, V. Lamounier-Zepter, S. Eaton, Changes in morphology and function of adrenal cortex in mice fed a high-fat diet. Int. J. Obes. (Lond) 39, 321–330 (2015). [DOI] [PubMed] [Google Scholar]
- 6.A. Hofmann, M. Peitzsch, C. Brunssen, J. Mittag, A. Jannasch, A. Frenzel, N. Brown, S. M. Weldon, G. Eisenhofer, S. R. Bornstein, H. Morawietz, Elevated steroid hormone production in the db/db mouse model of obesity and type 2 diabetes. Horm. Metab. Res. 49, 43–49 (2017). [DOI] [PubMed] [Google Scholar]
- 7.J. Galitzky, A. Bouloumie, Human visceral-fat-specific glucocorticoid tuning of adipogenesis. Cell Metab. 18, 3–5 (2013). [DOI] [PubMed] [Google Scholar]
- 8.C. Roberge, A. C. Carpentier, M. F. Langlois, J. P. Baillargeon, J. L. Ardilouze, P. Maheux, N. Gallo-Payet, Adrenocortical dysregulation as a major player in insulin resistance and onset of obesity. Am. J. Physiol. Endocrinol. Metab. 293, E1465–E1478 (2007). [DOI] [PubMed] [Google Scholar]
- 9.P. Q. Barrett, N. A. Guagliardo, P. M. Klein, C. Hu, D. T. Breault, M. P. Beenhakker, Role of voltage-gated calcium channels in the regulation of aldosterone production from zona glomerulosa cells of the adrenal cortex. J. Physiol. 594, 5851–5860 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.A. Midzak, V. Papadopoulos, Adrenal mitochondria and steroidogenesis: From individual proteins to functional protein assemblies. Front. Endocrinol. (Lausanne) 7, 106 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.W. Stoffel, B. Holz, B. Jenke, E. Binczek, R. H. Gunter, C. Kiss, I. Karakesisoglou, M. Thevis, A. A. Weber, S. Arnhold, K. Addicks, Delta6-desaturase (FADS2) deficiency unveils the role of omega3- and omega6-polyunsaturated fatty acids. EMBO J. 27, 2281–2292 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.C. K. Stroud, T. Y. Nara, M. Roqueta-Rivera, E. C. Radlowski, P. Lawrence, Y. Zhang, B. H. Cho, M. Segre, R. A. Hess, J. T. Brenna, W. M. Haschek, M. T. Nakamura, Disruption of FADS2 gene in mice impairs male reproduction and causes dermal and intestinal ulceration. J. Lipid Res. 50, 1870–1880 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.W. J. Park, K. S. Kothapalli, P. Lawrence, C. Tyburczy, J. T. Brenna, An alternate pathway to long-chain polyunsaturates: The FADS2 gene product Delta8-desaturates 20:2n-6 and 20:3n-3. J. Lipid Res. 50, 1195–1202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.J. E. Vance, Historical perspective: Phosphatidylserine and phosphatidylethanolamine from the 1800s to the present. J. Lipid Res. 59, 923–944 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.A. Poulaki, S. Giannouli, Mitochondrial lipids: From membrane organization to apoptotic facilitation. Int. J. Mol. Sci. 23, 3738 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.S. R. King, Z. Liu, J. Soh, S. Eimerl, J. Orly, D. M. Stocco, Effects of disruption of the mitochondrial electrochemical gradient on steroidogenesis and the Steroidogenic Acute Regulatory (StAR) protein. J. Steroid Biochem. Mol. Biol. 69, 143–154 (1999). [DOI] [PubMed] [Google Scholar]
- 17.D. B. Hales, J. A. Allen, T. Shankara, P. Janus, S. Buck, T. Diemer, K. H. Hales, Mitochondrial function in Leydig cell steroidogenesis. Ann. N. Y. Acad. Sci. 1061, 120–134 (2005). [DOI] [PubMed] [Google Scholar]
- 18.I. Mateska, A. Witt, E. Haga, A. Sinha, C. Yilmaz, E. Thanou, N. Sun, O. Kolliniati, M. Patschin, H. Abdelmegeed, H. Henneicke, W. Kanczkowski, B. Wielockx, C. Tsatsanis, A. Dahl, A. Walch, K. Li, M. Peitzsch, T. Chavakis, V. Alexaki, Inflammation-induced downregulation of succinate dehydrogenase regulates adrenocortical function. BioRxiv 2022.04.29.490066 (2022). 10.1101/2022.04.29.490066 [DOI]
- 19.L. Issop, M. B. Rone, V. Papadopoulos, Organelle plasticity and interactions in cholesterol transport and steroid biosynthesis. Mol. Cell. Endocrinol. 371, 34–46 (2013). [DOI] [PubMed] [Google Scholar]
- 20.R. Garcia-Martin, V. I. Alexaki, N. Qin, M. F. Rubin de Celis, M. Economopoulou, A. Ziogas, B. Gercken, K. Kotlabova, J. Phieler, M. Ehrhart-Bornstein, S. R. Bornstein, G. Eisenhofer, G. Breier, M. Bluher, J. Hampe, A. El-Armouche, A. Chatzigeorgiou, K. J. Chung, T. Chavakis, Adipocyte-specific HIF2α deficiency exacerbates obesity-induced brown adipose tissue dysfunction and metabolic dysregulation. Mol. Cell. Biol. 36, 376–393 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.K. J. Chung, A. Chatzigeorgiou, M. Economopoulou, R. Garcia-Martin, V. I. Alexaki, I. Mitroulis, M. Nati, J. Gebler, T. Ziemssen, S. E. Goelz, J. Phieler, J. H. Lim, K. P. Karalis, T. Papayannopoulou, M. Bluher, G. Hajishengallis, T. Chavakis, A self-sustained loop of inflammation-driven inhibition of beige adipogenesis in obesity. Nat. Immunol. 18, 654–664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.V. I. Alexaki, The impact of obesity on microglial function: Immune, metabolic and endocrine perspectives. Cells 10, 1584 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. Dionysopoulou, E. Charmandari, A. Bargiota, N. Vlahos, G. Mastorakos, G. Valsamakis, The role of hypothalamic inflammation in diet-induced obesity and its association with cognitive and mood disorders. Nutrients 13, 498 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.M. Grzybek, A. Palladini, V. I. Alexaki, M. A. Surma, K. Simons, T. Chavakis, C. Klose, U. Coskun, Comprehensive and quantitative analysis of white and brown adipose tissue by shotgun lipidomics. Mol. Metab. 22, 12–20 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. R. Pergande, F. Serna-Perez, S. B. Mohsin, J. Hanek, S. M. Cologna, Lipidomic analysis reveals altered fatty acid metabolism in the liver of the symptomatic niemann-pick, type C1 mouse model. Proteomics 19, e1800285 (2019). [DOI] [PubMed] [Google Scholar]
- 26.X. Wang, L. P. Walsh, A. J. Reinhart, D. M. Stocco, The role of arachidonic acid in steroidogenesis and steroidogenic acute regulatory (StAR) gene and protein expression. J. Biol. Chem. 275, 20204–20209 (2000). [DOI] [PubMed] [Google Scholar]
- 27.A. Duarte, A. F. Castillo, R. Castilla, P. Maloberti, C. Paz, E. J. Podesta, F. Cornejo Maciel, An arachidonic acid generation/export system involved in the regulation of cholesterol transport in mitochondria of steroidogenic cells. FEBS Lett. 581, 4023–4028 (2007). [DOI] [PubMed] [Google Scholar]
- 28.R. A. Frye, R. W. Holz, The relationship between arachidonic acid release and catecholamine secretion from cultured bovine adrenal chromaffin cells. J. Neurochem. 43, 146–150 (1984). [DOI] [PubMed] [Google Scholar]
- 29.Y. Li, M. Han, P. Lin, Y. He, J. Yu, R. Zhao, Hair growth promotion activity and its mechanism of polygonum multiflorum. Evid. Based Complement. Alternat. Med. 2015, 517901 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R. L. Erickson, C. A. Browne, I. Lucki, Hair corticosterone measurement in mouse models of type 1 and type 2 diabetes mellitus. Physiol. Behav. 178, 166–171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.K. Chapman, M. Holmes, J. Seckl, 11β-hydroxysteroid dehydrogenases: Intracellular gate-keepers of tissue glucocorticoid action. Physiol. Rev. 93, 1139–1206 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R. K. Saini, Y. S. Keum, Omega-3 and omega-6 polyunsaturated fatty acids: Dietary sources, metabolism, and significance - A review. Life Sci. 203, 255–267 (2018). [DOI] [PubMed] [Google Scholar]
- 33.K. Kaliannan, X. Y. Li, B. Wang, Q. Pan, C. Y. Chen, L. Hao, S. Xie, J. X. Kang, Multi-omic analysis in transgenic mice implicates omega-6/omega-3 fatty acid imbalance as a risk factor for chronic disease. Commun. Biol. 2, 276 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.H. E. Bays, C. M. Ballantyne, J. J. Kastelein, J. L. Isaacsohn, R. A. Braeckman, P. N. Soni, Eicosapentaenoic acid ethyl ester (AMR101) therapy in patients with very high triglyceride levels (from the Multi-center, plAcebo-controlled, Randomized, double-blINd, 12-week study with an open-label Extension [MARINE] trial). Am. J. Cardiol. 108, 682–690 (2011). [DOI] [PubMed] [Google Scholar]
- 35.C. M. Ballantyne, H. E. Bays, J. J. Kastelein, E. Stein, J. L. Isaacsohn, R. A. Braeckman, P. N. Soni, Efficacy and safety of eicosapentaenoic acid ethyl ester (AMR101) therapy in statin-treated patients with persistent high triglycerides (from the ANCHOR study). Am. J. Cardiol. 110, 984–992 (2012). [DOI] [PubMed] [Google Scholar]
- 36.D. L. Bhatt, P. G. Steg, M. Miller, E. A. Brinton, T. A. Jacobson, S. B. Ketchum, R. T. Doyle Jr., R. A. Juliano, L. Jiao, C. Granowitz, J. C. Tardif, C. M. Ballantyne, R.-I. Investigators, Cardiovascular risk reduction with icosapent ethyl for hypertriglyceridemia. N. Engl. J. Med. 380, 11–22 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Z. Tang, B. Kang, C. Li, T. Chen, Z. Zhang, GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 47, W556–W560 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Bose, R. M. Whittal, W. L. Miller, H. S. Bose, Steroidogenic activity of StAR requires contact with mitochondrial VDAC1 and phosphate carrier protein. J. Biol. Chem. 283, 8837–8845 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.N. Strushkevich, F. MacKenzie, T. Cherkesova, I. Grabovec, S. Usanov, H. W. Park, Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system. Proc. Natl. Acad. Sci. U.S.A. 108, 10139–10143 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.A. S. Berthon, E. Szarek, C. A. Stratakis, PRKACA: The catalytic subunit of protein kinase A and adrenocortical tumors. Front. Cell Dev. Biol. 3, 26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. P. Schimmer, P. C. White, Minireview: Steroidogenic factor 1: Its roles in differentiation, development, and disease. Mol. Endocrinol. 24, 1322–1337 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. R. Brash, Arachidonic acid as a bioactive molecule. J. Clin. Invest. 107, 1339–1345 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.M. K. Montgomery, S. H. J. Brown, T. W. Mitchell, A. C. F. Coster, G. J. Cooney, N. Turner, Association of muscle lipidomic profile with high-fat diet-induced insulin resistance across five mouse strains. Sci. Rep. 7, 13914 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.T. Hashidate-Yoshida, T. Harayama, D. Hishikawa, R. Morimoto, F. Hamano, S. M. Tokuoka, M. Eto, M. Tamura-Nakano, R. Yanobu-Takanashi, Y. Mukumoto, H. Kiyonari, T. Okamura, Y. Kita, H. Shindou, T. Shimizu, Fatty acid remodeling by LPCAT3 enriches arachidonate in phospholipid membranes and regulates triglyceride transport. eLife 4, e06328 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.B. Cheng, J. Kowal, Analysis of adrenal cholesteryl esters by reversed phase high performance liquid chromatography. J. Lipid Res. 35, 1115–1121 (1994). [PubMed] [Google Scholar]
- 46.W. Fan, R. M. Evans, Turning up the heat on membrane fluidity. Cell 161, 962–963 (2015). [DOI] [PubMed] [Google Scholar]
- 47.K. Hac-Wydro, P. Wydro, The influence of fatty acids on model cholesterol/phospholipid membranes. Chem. Phys. Lipids 150, 66–81 (2007). [DOI] [PubMed] [Google Scholar]
- 48.H. Sunshine, M. L. Iruela-Arispe, Membrane lipids and cell signaling. Curr. Opin. Lipidol. 28, 408–413 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.J. C. Holthuis, A. K. Menon, Lipid landscapes and pipelines in membrane homeostasis. Nature 510, 48–57 (2014). [DOI] [PubMed] [Google Scholar]
- 50.J. Gallay, M. Vincent, C. de Paillerets, M. Rogard, A. Alfsen, Relationship between the activity of the 3 beta-hydroxysteroid dehydrogenase from bovine adrenal cortex microsomes and membrane structure. Influence of proteins and steroid substrates on lipid "microviscosity". J. Biol. Chem. 256, 1235–1241 (1981). [PubMed] [Google Scholar]
- 51.S. Narasimhulu, Thermotropic transitions in fluidity of bovine adrenocortical microsomal membrane and substrate-cytochrome P-450 binding reaction. Biochim. Biophys. Acta 487, 378–387 (1977). [DOI] [PubMed] [Google Scholar]
- 52.M. Roqueta-Rivera, C. K. Stroud, W. M. Haschek, S. J. Akare, M. Segre, R. S. Brush, M. P. Agbaga, R. E. Anderson, R. A. Hess, M. T. Nakamura, Docosahexaenoic acid supplementation fully restores fertility and spermatogenesis in male delta-6 desaturase-null mice. J. Lipid Res. 51, 360–367 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.W. Stoffel, I. Hammels, B. Jenke, E. Binczek, I. Schmidt-Soltau, S. Brodesser, M. Odenthal, M. Thevis, Obesity resistance and deregulation of lipogenesis in Δ6-fatty acid desaturase (FADS2) deficiency. EMBO Rep. 15, 110–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.M. A. Perez, G. Terreros, A. Dagnino-Subiabre, Long-term ω-3 fatty acid supplementation induces anti-stress effects and improves learning in rats. Behav. Brain Funct. 9, 25 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.P. Barbadoro, I. Annino, E. Ponzio, R. M. Romanelli, M. M. D'Errico, E. Prospero, A. Minelli, Fish oil supplementation reduces cortisol basal levels and perceived stress: A randomized, placebo-controlled trial in abstinent alcoholics. Mol. Nutr. Food Res. 57, 1110–1114 (2013). [DOI] [PubMed] [Google Scholar]
- 56.S. Jazayeri, S. A. Keshavarz, M. Tehrani-Doost, M. Djalali, M. Hosseini, H. Amini, M. Chamari, A. Djazayery, Effects of eicosapentaenoic acid and fluoxetine on plasma cortisol, serum interleukin-1beta and interleukin-6 concentrations in patients with major depressive disorder. Psychiatry Res. 178, 112–115 (2010). [DOI] [PubMed] [Google Scholar]
- 57.P. J. Gillies, S. K. Bhatia, L. A. Belcher, D. B. Hannon, J. T. Thompson, J. P. Vanden Heuvel, Regulation of inflammatory and lipid metabolism genes by eicosapentaenoic acid-rich oil. J. Lipid Res. 53, 1679–1689 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.J. C. Ralston, S. Matravadia, N. Gaudio, G. P. Holloway, D. M. Mutch, Polyunsaturated fatty acid regulation of adipocyte FADS1 and FADS2 expression and function. Obesity (Silver Spring) 23, 725–728 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.S. Tanaka, N. Ishihara, S. Suzuki, Y. Watanabe, D. Nagayama, T. Yamaguchi, M. Ohira, A. Saiki, T. Tanaka, I. Tatsuno, Fatty acid desaturase 2 is up-regulated by the treatment with statin through geranylgeranyl pyrophosphate-dependent Rho kinase pathway in HepG2 cells. Sci. Rep. 9, 10009 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.M. Peitzsch, T. Dekkers, M. Haase, F. C. Sweep, I. Quack, G. Antoch, G. Siegert, J. W. Lenders, J. Deinum, H. S. Willenberg, G. Eisenhofer, An LC-MS/MS method for steroid profiling during adrenal venous sampling for investigation of primary aldosteronism. J. Steroid Biochem. Mol. Biol. 145, 75–84 (2015). [DOI] [PubMed] [Google Scholar]
- 61.A. Witt, P. Mirtschink, A. Palladini, I. Mateska, H. Abdelmegeed, M. Grzybek, B. Wielockx, M. Peitzsch, U. Coskun, T. Chavakis, V. I. Alexaki, Obesity-associated lipidomic remodeling of the adrenal gland indicates an important role of the FADS2-arachidonic acid axis in adrenocortical hormone production. BioRxiv 2020.09.04.282905 (2020). 10.1101/2020.09.04.282905 [DOI]
- 62.M. Lange, G. Angelidou, Z. Ni, A. Criscuolo, J. Schiller, M. Bluher, M. Fedorova, AdipoAtlas: A reference lipidome for human white adipose tissue. Cell Rep. Med. 2, 100407 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.L. Goracci, S. Tortorella, P. Tiberi, R. M. Pellegrino, A. Di Veroli, A. Valeri, G. Cruciani, Lipostar, a comprehensive platform-neutral cheminformatics tool for lipidomics. Anal. Chem. 89, 6257–6264 (2017). [DOI] [PubMed] [Google Scholar]
- 64.Z. Pang, G. Zhou, J. Ewald, L. Chang, O. Hacariz, N. Basu, J. Xia, Using metaboanalyst 5.0 for LC-HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 17, 1735–1761 (2022). [DOI] [PubMed] [Google Scholar]
- 65.K. J. Adams, B. Pratt, N. Bose, L. G. Dubois, L. St John-Williams, K. M. Perrott, K. Ky, P. Kapahi, V. Sharma, M. J. MacCoss, M. A. Moseley, C. A. Colton, B. X. MacLean, B. Schilling, J. W. Thompson; Alzheimer’s Disease Metabolomics Consortium , Skyline for small molecules: A unifying software package for quantitative metabolomics. J. Proteome Res. 19, 1447–1458 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.C. S. Ejsing, J. L. Sampaio, V. Surendranath, E. Duchoslav, K. Ekroos, R. W. Klemm, K. Simons, A. Shevchenko, Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 2136–2141 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.M. A. Surma, R. Herzog, A. Vasilj, C. Klose, N. Christinat, D. Morin-Rivron, K. Simons, M. Masoodi, J. L. Sampaio, An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 117, 1540–1549 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.G. Liebisch, M. Binder, R. Schifferer, T. Langmann, B. Schulz, G. Schmitz, High throughput quantification of cholesterol and cholesteryl ester by electrospray ionization tandem mass spectrometry (ESI-MS/MS). Biochim. Biophys. Acta 1761, 121–128 (2006). [DOI] [PubMed] [Google Scholar]
- 69.R. Herzog, K. Schuhmann, D. Schwudke, J. L. Sampaio, S. R. Bornstein, M. Schroeder, A. Shevchenko, LipidXplorer: A software for consensual cross-platform lipidomics. PLOS ONE 7, e29851 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.R. Herzog, D. Schwudke, K. Schuhmann, J. L. Sampaio, S. R. Bornstein, M. Schroeder, A. Shevchenko, A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 12, R8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.R. Pamplona, M. Portero-Otin, D. Riba, C. Ruiz, J. Prat, M. J. Bellmunt, G. Barja, Mitochondrial membrane peroxidizability index is inversely related to maximum life span in mammals. J. Lipid Res. 39, 1989–1994 (1998). [PubMed] [Google Scholar]
- 72.R. C. Team, R: A language and environment for statistical computing. https://r-project.org/ (2018).
- 73.H. Wickham, ggplot2: Elegant graphics for data analysis. Springer-Verlag New York https://ggplot2.tidyverse.org (2016).
- 74.W. Gao, T. Stalder, P. Foley, M. Rauh, H. Deng, C. Kirschbaum, Quantitative analysis of steroid hormones in human hair using a column-switching LC-APCI-MS/MS assay. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 928, 1–8 (2013). [DOI] [PubMed] [Google Scholar]
- 75.M. Martin, Cutadept removes adapter sequences from high-throughput sequncing reads. EMBnet.J. 7, 2803–2809 (2011). [Google Scholar]
- 76.A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski, S. Jha, P. Batut, M. Chaisson, T. R. Gingeras, STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.S. Anders, P. T. Pyl, W. Huber, HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.S. Anders, W. Huber, Differential expression analysis for sequence count data. Genome Biol. 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.A. Subramanian, P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, J. P. Mesirov, Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.M. Alhamdoosh, M. Ng, N. J. Wilson, J. M. Sheridan, H. Huynh, M. J. Wilson, M. E. Ritchie, Combining multiple tools outperforms individual methods in gene set enrichment analyses. Bioinformatics 33, 414–424 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.I. Mateska, A. Witt, E. Hagag, A. Sinha, C. Yilmaz, E. Thanou, N. Sun, O. Kolliniati, M. Patschin, H. Abdelmegeed, H. Henneicke, W. Kanczkowski, B. Wielockx, C. Tsatsanis, A. Dahl, A. Walch, K. Li, M. Peitzsch, T. Chavakis, V. I. Alexaki, Succinate mediates inflammation-induced adrenocortical dysfunction. BioRxiv 2022.04.29.490066 (2022). 10.1101/2022.04.29.490066 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S6
Figs. S1 to S6