ABSTRACT
The ability of bacteria to thrive in diverse habitats and to adapt to ever-changing environmental conditions relies on the rapid and stringent modulation of gene expression. It has become evident in the past decade that small regulatory RNAs (sRNAs) are central components of networks controlling the bacterial responses to stress. Functioning at the posttranscriptional level, sRNAs base-pair with cognate mRNAs to alter translation, stability, or both to either repress or activate the targeted transcripts; the RNA chaperone Hfq participates in stabilizing sRNAs and in promoting pairing between target and sRNA. In particular, sRNAs act at the heart of crucial stress responses, including those dedicated to overcoming membrane damage and oxidative stress, discussed here. The bacterial cell envelope is the outermost protective barrier against the environment and thus is constantly monitored and remodeled. Here, we review the integration of sRNAs into the complex networks of several major envelope stress responses of Gram-negative bacteria, including the RpoE (σE), Cpx, and Rcs regulons. Oxidative stress, caused by bacterial respiratory activity or induced by toxic molecules, can lead to significant damage of cellular components. In Escherichia coli and related bacteria, sRNAs also contribute significantly to the function of the RpoS (σS)-dependent general stress response as well as the specific OxyR- and SoxR/S-mediated responses to oxidative damage. Their activities in gene regulation and crosstalk to other stress-induced regulons are highlighted.
INTRODUCTION
One major paradigm for RNA-based regulation in both eukaryotes and prokaryotes is small regulatory RNAs (sRNAs) that pair with mRNAs, leading to changes in translation and mRNA stability. In bacteria, rather than the highly processed very short microRNAs found in eukaryotes, these sRNAs are generally on the order of 50 to 200 nucleotides (nt) long, and in the Gram-negative organisms that are the major focus of this review, annealing of sRNAs to their target mRNAs is usually dependent on the RNA chaperone, Hfq. Annealing can lead to positive regulation of translation, by remodeling inhibitory RNA structures or blocking access of negative regulators (for instance, RNases or the Rho transcription termination factor), or negative regulation, by inhibiting translation, recruiting RNases, or both. A given sRNA can have multiple targets, and can carry out both negative and positive regulation (1–3).
A member of the conserved family of Sm and Sm-like (LSm) proteins, Hfq assembles as a stable, homohexameric ring that offers three principal binding sites for RNA: the proximal and distal surfaces of the ring, as well as the lateral rim. Hfq binds sRNAs on the proximal face, recognizing the uridine stretch at the 3′ end of the sRNA’s Rho-independent terminator sequence. In the absence of the chaperone, almost all of these Hfq-binding sRNAs become quite unstable, and that may be sufficient to explain the loss of sRNA function in hfq mutants (see, for instance, reference 4). In addition, Hfq binds to mRNAs, frequently but not always via its distal surface. In vitro, Hfq promotes pairing of sRNAs and mRNAs, suggesting that it is likely to do that as well in vivo (5–7). Overall, for the discussion here, the phenotypes of hfq mutants serve as a starting point for understanding the role of sRNA-based regulation in Escherichia coli and Salmonella. Many but not all of these phenotypes are now understood and point to major roles for sRNAs in the use of alternative sigma factors and the response to stress—including envelope and oxidative damage—in bacteria.
Loss of Hfq-Dependent Regulation Leads to Low Levels of RpoS
The first descriptions of phenotypes of a mutation in hfq in E. coli (8), including osmosensitivity and elongated cell shape, were noted as consistent with the phenotypes of mutants of rpoS (encoding the alternative sigma subunit of RNA polymerase, RpoS [also called σS]). We now know that at least three sRNAs, DsrA, RprA, and ArcZ, each produced under different conditions, interact with Hfq to positively regulate RpoS translation (9) (Fig. 1A). Being a major stress sigma factor, RpoS controls >10% of all protein-coding genes in E. coli (10–12); during stationary phase, the RpoS-mediated general stress response provides resistance to a variety of cell-damaging conditions, including, for example, oxidative stress, low pH, and high osmolarity (9). The demonstration that the hfq mutant was defective in the production of RpoS (13, 14) thus explained many of its phenotypes, including its sensitivity to high osmolarity and low pH.
FIGURE 1.
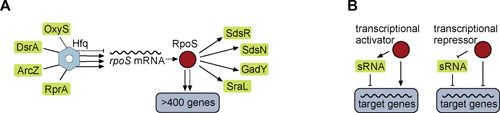
Activity of sRNAs in the general stress response. (A) Together with Hfq, the sRNAs DsrA, ArcZ, and RprA activate translation of the rpoS transcript by alleviating a self-inhibitory structure within the 5′ UTR of the mRNA. The sRNA OxyS functions as an indirect, negative regulator of rpoS expression. The major alternative sigma factor RpoS governs the general stress response and controls >400 genes in E. coli and related enterobacteria, including at least four sRNAs (SdsR, SdsN, GadY, and SraL). (B) Transcription factors which are restricted to function as either activators or repressors utilize sRNAs to facilitate opposite regulation.
In addition to the direct effect of sRNAs and Hfq on RpoS synthesis, a set of sRNAs are RpoS-dependent (see Fig. 1A), and thus any phenotypes associated with those RNAs will also be Hfq-dependent, both for their expression and for their own function and stability. For instance, GadY, an Hfq-dependent sRNA whose synthesis is controlled by RpoS in E. coli and Shigella, positively regulates GadX and GadW, transcriptional activators of genes for glutamate-dependent acid resistance (15, 16), and the E. coli-specific SdsN represses genes involved in the metabolism of oxidized nitrogen compounds (17). More widely conserved are the RpoS-controlled sRNAs SraL and SdsR. A single target has been identified for SraL (tig mRNA, encoding the chaperone trigger factor [18]), while SdsR interferes with translation of >20 transcripts in E. coli and Salmonella and has been implicated in the response to antibiotics and in mismatch repair upon DNA damage (19–22).
Loss of Hfq-Dependent Regulation Leads to High Levels of RpoE
A second major phenotype of hfq mutants was first highlighted in studies examining the transcriptional changes when Hfq was absent (23). These experiments were carried out specifically under conditions under which RpoS is not abundant, i.e., early exponential phase. Transcripts encoding outer membrane (OM) β-barrel proteins were significantly overrepresented and generally were upregulated in the hfq mutant, while RpoE (or σE, encoded by the rpoE gene), the alternative sigma factor that regulates OM stress responses, was induced. Combined with studies by others in Salmonella (24), and studies of specific sRNAs and their targets (25), these observations have led to evidence that loss of sRNA downregulation of OM protein synthesis leads to an RpoE-inducing stress. Thus, hfq mutants express RpoE-dependent genes at a high level, discussed further below.
Specialized Sigma Factors and Hfq: Changing the Sign of Regulation
Worth noting here is that the two major phenotypes of hfq mutants discussed above are due to effects on the levels of two specialized sigma factors. Sigma factors act with core RNA polymerase to direct it to particular promoters, and thus, other than competing for core, are not themselves capable of carrying out negative regulation. Therefore, any negative regulation dependent upon a specialized sigma factor is likely indirect, by positive regulation of a negative regulator, including, in many cases, sRNAs (Fig. 1B). Thus, in the initial studies of hfq mutants in the RpoE response (23), many upregulated genes have now been shown to be negatively regulated by RpoE-dependent sRNAs (26). This switch in the sign of regulation is also seen for repressors (for instance, positive regulation by the Fur repressor [27]; see Fig. 1B), and thus an unexpected direction of regulation should lead to examining the possible involvement of Hfq and sRNAs.
CONSTRUCTING sRNA REGULATORY NETWORKS: GENERAL PRINCIPLES AND EXPECTATIONS
For any stress, one can consider the roles of sRNAs and what they tell us about how the stress is sensed and responded to. As shown in Fig. 1A for the RpoS general stress response, sRNAs can act upstream, to regulate expression of a transcriptional regulator, or downstream, as part of the regulon. In some cases, sRNAs do both, providing feedback regulatory loops. Possibly because sRNAs oftentimes work in a stoichiometric fashion (28), i.e., the regulatory RNA is degraded together with the mRNA it is pairing with, the promoters of sRNAs are frequently among the best regulated and most robust in a given stress regulon (see, for instance, reference 29). Thus, they can also serve as excellent reporters for the stress response. The general expectations discussed here are primarily relevant to sRNAs expressed as part of a stress response (i.e., those whose expression is dependent on the transcriptional signals for the response) and are outlined here to provide some guidelines for considering the role of sRNAs within regulons.
sRNAs as Guides to a Stress Response
What sRNAs are expressed in response to the stress (and/or are regulated by the known transcriptional regulators for that stress)? Presumably these sRNAs have effects that help in repairing or avoiding the damage from the stress. Can that contribution to avoiding or overcoming stress be demonstrated? If not, do the sRNA targets suggest novel components of the stress response, not previously appreciated?
Are there sRNAs expressed as part of other regulons that contribute, positively or negatively, to the stress under consideration? sRNAs can provide interactions between different regulons, modulating or setting hierarchies for regulon expression.
A few clear examples of the types of sRNA functions in stress responses are noted here; some of these are discussed in more detail elsewhere.
Reinforcing or Helping Implement the Stress Response (Positive Feedback Loops)
Spot 42 sRNA synthesis is negatively regulated by cyclic AMP (cAMP)/cAMP receptor protein (CRP) and negatively regulates many operons involved in alternative carbon source use that are dependent upon cAMP (30). Thus, Spot 42 contributes to reducing the basal levels of these operons when cAMP is low (favoring efficient use of glucose/favored carbon sources).
Minimizing Stress Signals (Negative Feedback Loops)
The negative feedback loop in which RpoE-dependent sRNAs (as well as others) downregulate translation of many OM proteins is discussed further below. In another, more indirect example of negative feedback, RyhB, made when iron is limiting, inhibits synthesis of nonessential iron-binding proteins, thus helping to overcome the stress by increasing the availability of iron (31).
Connecting Regulons/Stress Responses/Setting Hierarchies
sRNAs connect different regulons, for purposes that are not always yet well understood. In two cases noted here, sRNAs modulate the interaction between specific regulatory responses and a specialized sigma factor. For instance, the Hfq-dependent OxyS sRNA, synthesized under the control of OxyR, negatively regulates RpoS (32). Given that OxyR regulates genes involved in the response to oxidative stress, which RpoS does as well, it would seem that OxyS helps the cell to use the specific (OxyR-dependent) response rather than the general stress response under some conditions. PhoQ-PhoP, a two-component system (TCS) that is activated at low Mg2+ concentrations and leads to the synthesis of genes regulating Mg2+ homeostasis as well as lipopolysaccharide (LPS) modifications, is negatively regulated by the RpoE-dependent MicA sRNA (33).
INVOLVEMENT OF sRNAs IN THE ENVELOPE STRESS RESPONSE
Cell Envelope Structure and Function in Gram-Negative Bacteria
The cell envelope represents a barrier shielding the bacterium from its environment and allowing the selective passage of both harmful and beneficial molecules (34). In Gram-negative bacteria, the envelope is composed of two concentric membrane layers that enclose the periplasmic space containing a thin peptidoglycan (PG) cell wall (35). The inner membrane (IM) separating the cytoplasm from the periplasm is a phospholipid bilayer, and the proteins associated with or integrated in the IM are frequently involved in key cellular processes including energy generation, signal transduction, metabolism, transport, and cell division (35). The periplasm is an aqueous cellular compartment densely packed with proteins and harbors the mesh-like PG layer, which is formed from linear amino-sugar polymers cross-connected via oligopeptide chains (36). Structural integrity of the cell envelope is ensured by the tight linkage of the PG layer to the cellular OM via the lipoprotein Lpp (also referred to as Braun’s lipoprotein) (37). Lpp is the most abundant protein in E. coli (>500,000 copies/cell) (38), and lipids attached to the N-terminus of Lpp embed it into the OM. Concomitantly, Lpp can be covalently attached to the peptide cross-bridges of the PG layer via its carboxy-terminal end (39). In contrast to the IM, the OM is an asymmetric bilayer consisting of phospholipids in the inner and LPS in the surface-exposed leaflet (35). LPS is a complex glycolipid composed of lipid A (a glucosamine disaccharide decorated with fatty acids anchoring LPS to the membrane), an oligosaccharide core, and an extended polysaccharide chain commonly referred to as O-antigen (40). Tightly packed LPS serves as an effective permeability barrier to hydrophobic substances (41), but certain other molecules are able to cross the OM through protein transporters. Small, hydrophilic compounds can diffuse through the lumen of porins, which are highly abundant β-barrel OM proteins (OMPs) that only discriminate their substrates by size (34). Gated, high-affinity uptake of ligands including siderophores, vitamins, and carbohydrates is mediated by an additional class of larger, integral β-barrel OMPs. Active transport via so-called TonB-dependent receptors into the periplasmic space involves coupling to a protein complex localized in the IM (42).
The integrity of the cellular envelope is essential for bacterial survival, and consequently, its architecture and composition are tightly regulated. Bacteria have evolved a suite of stress responses that function in monitoring impairment or deficiencies of the envelope and govern adaptation of the bacterial gene expression to alleviate stress (43). In E. coli and related enterobacteria, at least five envelope stress responses coordinately function to maintain membrane homeostasis (44). While no sRNAs have yet been associated with the minor membrane stress responses coordinated via the BaeS/R or the Psp system, the three major pathways (regulated through RpoE and the Cpx and Rcs systems, respectively) all rely on the activity of regulatory RNAs. Intriguingly, sRNAs function not only as effectors regulating individual target genes but also in mediating the extensive crosstalk and adaptation of individual stress responses.
sRNAs in the RpoE-Mediated Envelope Stress Response
Maintenance of OM homeostasis in E. coli and related bacteria relies on the dedicated activity of the alternative sigma factor RpoE, which is encoded by the rpoE gene and cotranscribed with rseABC from a RpoD-dependent promoter (45). Under nonstress conditions, RpoE is only expressed at a low basal level (38) and sequestered to the plasma membrane in an inactive state by its cognate anti-sigma factor, RseA (46) (Fig. 2A). The activity of RpoE is under complex control and tuned to perturbations of OMP folding, the status of LPS, as well as nutrient availability and growth phase (47). The accumulation of misfolded OMPs within the periplasmic space is the most thoroughly characterized signal triggering RpoE activation (47) (Fig. 2A). The C-termini of unfolded OMPs—which are inaccessible in properly assembled porins—are recognized by the periplasmic PDZ domain of a serine protease, DegS (48). The resulting conformational change initiates a protease cascade resulting in degradation of RseA and consequent release of RpoE into the cytoplasm (47). Maximal induction of RpoE requires the integration of a second activating signal. In response to mislocalized LPS (49), the RseB protein, which protects the RseA anti-sigma factor from cleavage by DegS, is displaced.
FIGURE 2.
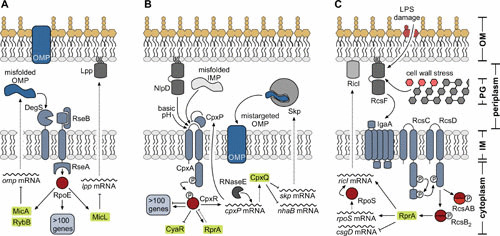
The role of sRNAs in the major envelope stress responses. Gram-negative bacteria are diderm, with the OM and IM being separated by the periplasmic space containing the PG cell wall. (A) OM homeostasis is regulated by the RpoE response. A series of proteolysis steps results in the degradation of the anti-sigma factor RseA and concomitant release of RpoE. The large regulon of the alternative sigma factor also comprises at least three sRNAs: MicA and RybB function to downregulate the transcripts of all major OMPs to reduce the accumulation of misfolded porins within the periplasm. MicL specifically represses translation of the lpp mRNA. (B) Maintenance of the IM relies on the CpxA-CpxR TCS, which amongst other targets controls expression of at least three sRNAs, CyaR, RprA, and CpxQ. CpxQ is a stable fragment released by RNase E processing from the 3′ end of the cpxP mRNA. In association with Hfq, CpxQ functions to repress translation of several transcripts including skp mRNA, which encodes a periplasmic chaperone promoting the mistargeting of OMPs into the IM. (C) The IM-associated histidine kinase RcsC, phosphotransfer protein RcsD, and response regulator RcsB constitute the core of the Rcs system. The sRNA RprA is one highly induced component of the Rcs response, which is activated by LPS damage and perturbations of the cell wall. While acting as a negative regulator of the csgD mRNA, RprA also promotes translation of both the rpoS and the ricI messages. As transcription of ricI (encoding an inhibitor of the conjugation machinery) is dependent on RpoS, RprA functions at the heart of a posttranscriptional feedforward loop for RicI activity.
The RpoE-orchestrated envelope stress response possesses two parallel branches, one acting through proteins and one mediated by sRNAs. In E. coli, RpoE drives transcription of ∼100 genes, including genes encoding all components required to assemble and transport OMPs and LPS to the OM (29, 50, 51). However, the rate at which new OM components are synthesized may easily exceed the capacity of chaperones and transporters. While RpoE, as for all sigma factors, is intrinsically restricted to function as a transcriptional activator, it employs regulatory RNAs to function as repressors of gene expression at the posttranscriptional level.
The strongest promoters within the RpoE regulon control RpoE itself and two Hfq-dependent sRNAs, RybB and MicA (29, 52, 53). Both sRNAs act as global regulators of the envelope stress response and together govern expression of >30 targets in E. coli. Most prominently, RybB and/or MicA repress all major OMPs, as well as several lipoproteins and transporters (26). Rapid decay of these target transcripts results in an immediate relief for the periplasmic folding machinery. In addition, the target suites of RybB and MicA also encompass genes involved in production of OM vesicles and the response regulator phoP (26, 33) (see below).
Mutants of rpoE in E. coli are not viable, but cells can be depleted for RpoE by overexpression of its antagonists, RseA and RseB (54, 55). The important contribution of the two regulatory RNAs, RybB and MicA, to cell homeostasis is reflected by their ability to counteract the growth and viability phenotypes associated with loss of RpoE (26).
Mechanistically, both RybB and MicA exert their regulatory activity by a variety of means. RybB employs a conserved seed region of 16 nt located at the very 5′ end of the molecule to interact with its target genes (56). Depending on the location of the base-pairing site on the target mRNA, RybB is able to interfere with ribosome association at the translational start site (57), to promote RNase cleavage in the coding sequence of target mRNAs, or to interrupt structural elements within the 5′ untranslated region (5′ UTR) (56). In the latter case, base-pairing can abrogate the protective effect of 5′-terminal structures, which have been shown to stabilize transcripts by restricting access of nucleases, including RNase E (58, 59). Similar to RybB, the first 24 nt of MicA are hyperconserved and involved in regulation of its target genes. In contrast to RybB, the seed region of MicA is less stringently defined, and the first 7 nt of the molecule are dispensable for regulation of approximately half of its targets (26). In the majority of cases, predicted interaction sites of MicA overlap the translation initiation region of its target transcripts; base-pairing of MicA close to the start codon for one noteworthy target, phoP (also see below), has been confirmed (26, 33). The regulatory mechanisms underlying the observed repression of other targets are yet to be experimentally validated (26).
Both RybB and MicA are highly conserved in numerous enterobacteria and likely contribute to OMP homeostasis in these species (60). Albeit not conserved at the sequence level, the human pathogen Vibrio cholerae encodes a functional homolog of these E. coli sRNAs. The 140-nt VrrA is under strict control of RpoE and represses translation of the major OMPs OmpA and OmpT as well as the biofilm matrix component RbmC and the ribosome binding protein Vrp in response to perturbations of the OM (61). Of note, accumulation of the VrrA sRNA is not affected in an hfq deletion mutant of V. cholerae, and possibly for this reason, the RNA chaperone is not strictly required for regulation of all targets (61–64).
In E. coli, a third sRNA is transcribed from an RpoE-dependent promoter positioned within the cutC coding sequence. The 308-nt MicL (also known as RyeF [53] and SlrA [65]) is processed by a currently unknown cleavage mechanism to a smaller, ∼80-nt transcript (MicL-S), and both isoforms associate with the Hfq chaperone (66). In stark contrast to the many targets of RybB and MicA, MicL appears to interact with only few transcripts (6), and the only experimentally confirmed target of this sRNA is currently the lpp gene (66). By binding to a target sequence located within the beginning of the coding sequence, MicL inhibits translation of lpp mRNA and triggers accelerated degradation of the transcript. Why is repression of Lpp synthesis beneficial to the RpoE-dependent envelope stress response? As an OM lipoprotein, folding and transport of Lpp are dependent on the LolAB system. The same machinery is, however, also required for the installation of other lipoproteins, including BamD and LptE, which are essential to chaperone OM insertion of OMPs and LPS, respectively (35). Consequently, MicL-mediated downregulation of Lpp may relieve envelope stress by reducing the demand on the Lol machinery, indirectly promoting the correct assembly and localization of other OM components.
sRNAs in the Cpx-Mediated Envelope Stress Response
Maintenance of OM integrity via the RpoE response is complemented by another major regulon primarily controlling homeostasis of the periplasm and the IM. The central hub of the Cpx envelope stress response is a TCS consisting of the IM-localized histidine kinase CpxA and its cognate DNA-binding response regulator, CpxR (67). While the molecular characteristics of the stimulus perceived by CpxA remain to be determined, numerous environmental cues triggering the signaling cascade have been identified, including alterations in IM composition (68), alkaline pH (69), and overexpression of the OM lipoprotein NlpE (70). Recent work also reports that the Cpx system may respond to perturbations of the PG cell wall (71). Activation of CpxA results in autophosphorylation and phosphotransfer to CpxR, which consequently enables transcriptional control of the Cpx regulon, comprising >100 genes (69) (Fig. 2B). In the absence of an inducing signal, CpxA acts as a phosphatase on CpxR∼P to rapidly inactivate the response regulator. Different from sigma factors, CpxR functions as both an activator and a repressor (69). It downregulates the expression of envelope-associated, macromolecular complexes, including cellular appendages (72) and respiratory complexes (73), and at the same time fosters transcription of periplasmic proteases and chaperones to alleviate the burden of protein folding (67). Another highly upregulated gene controlled by CpxR is cpxP, encoding a periplasmic inhibitor of the Cpx pathway that likely exerts its negative feedback control by masking the CpxA sensory domain (74, 75).
Processing of the 3′ UTR of the cpxP transcript by the major endonuclease RNase E liberates a stable, ∼60-nt long mRNA fragment, termed CpxQ, which associates with the Hfq chaperone (52, 76). A transcriptomic approach in Salmonella revealed that CpxQ acts as a trans-encoded sRNA negatively regulating multiple targets (76). Strikingly, the CpxQ regulon is enriched for proteins localizing to the IM, including those controlling the proton motive force (PMF). Employing one of two seed regions, CpxQ also represses translation of skp (encoding a periplasmic chaperone) and nhaB mRNAs (encoding a proton/sodium antiporter). Different from other chaperones, Skp is able to mistarget OMPs into the IM if the OM insertion machinery is compromised. By downregulating skp, CpxQ prevents the unrestricted flux over the IM via ion-permeable pores that would result from OMPs within the IM, and thus protects cells from collapse of the PMF. CpxQ addresses a similar problem by repression of nhaB, as overexpression of the protein, and concomitant increase in proton uptake, results in a loss of membrane polarization. In protecting the integrity of the IM and the PMF, CpxQ appears to play a major function as a repressive arm of the Cpx response (76, 77).
Additional sRNAs have also been shown to be integrated into this complex network. For example, CyaR and RprA sRNAs appear to be both directly and indirectly controlled by the Cpx response, with some differences depending on whether enteropathogenic E. coli or E. coli K-12 was examined (78).
Expression of CyaR (formerly RyeE [79]), a conserved, Hfq-associated sRNA, is under complex regulation by both the Crp and Cpx regulons. Crp induces CyaR under conditions when cAMP levels are high (80–82). CpxR functions as a transcriptional repressor of the cyaR promoter (72); CyaR in turn functions as a posttranscriptional repressor of several target genes, including the yqaE transcript (80). As expression of yqaE, encoding an IM protein of currently unknown function, is at the same time induced by CpxR∼P at the transcriptional level, repression of cyaR by CpxR integrates the sRNA into a coherent feedforward loop within the Cpx regulon (72, 80). The physiological importance of this regulation has not been examined; presumably, the CpxR effect on the cyaR promoter may not be significant under conditions when cAMP levels are high (i.e., growth in poorer carbon sources), and repression of yqaE may not be important under those conditions.
The promoter of rprA, a conserved enterobacterial sRNA, is activated under conditions of high CpxR∼P and is directly bound by CpxR∼P. Overexpression of RprA in turn feeds back to repress the Cpx response, indirectly via a currently undefined target (78). However, rprA expression is primarily controlled by an additional envelope stress response, the Rcs pathway, discussed below.
Integration of the Cpx and Rcs Envelope Stress Responses via RprA sRNA
Damage of the surface-exposed LPS, mutations in genes required for disulfide bond formation in the periplasm, and perturbations of PG cell wall biosynthesis are all cues triggering the Rcs phosphorelay (83, 84) (Fig. 2C). Signal transduction in this stress response system is more complex than in conventional TCSs: under inducing conditions, the surface-exposed lipoprotein, RcsF, likely inactivates the periplasmic IgaA repressor to trigger a phosphorelay. Upon activation, RcsC is autophosphorylated, and phosphotransfer via RcsD relays the signal to the cognate response regulator, RcsB. RcsB in turn controls the transcription of the Rcs stress response by binding to target promoters either as a heterodimer in cooperation with an additional DNA-binding protein (RcsA in the case of activation of colanic acid production), or as a homodimer (as is the case for the rprA gene). Originally identified in E. coli as one of the sRNAs activating rpoS translation (85), RprA has recently been demonstrated to posttranscriptionally modulate expression of >60 additional genes in Salmonella (86), although how many of these are direct targets remains to be explored. RprA is a substrate of RNase E, and both the full-length (107 nt) and the cleaved versions of the sRNA (∼50 nt) lacking the 5′ end are present in the cell (86, 87). Interestingly, both variants associate with Hfq (52) and control different sets of mRNAs (86).
One of the mRNA targets activated by RprA is the Salmonella-specific ricI transcript (86). Similar to the positive regulation of rpoS, RprA also promotes translation of ricI by interference with a self-inhibitory structure within the 5′ UTR of the mRNA. Of note, while only full-length RprA harbors the site required for base-pairing with rpoS mRNA, the ricI transcript is recognized via a conserved sequence stretch located downstream of the cleavage site of RprA. As expression of ricI is controlled by RpoS at the transcriptional level, RprA functions as the centerpiece of a posttranscriptional feedforward loop. RicI acts as an inhibitor for conjugative transfer of the Salmonella virulence plasmid pSLT by binding to the conjugation apparatus at the cytoplasmic membrane. With regard to envelope homeostasis, the Rcs regulon might employ RprA to prevent assembly of the complex conjugation machinery when membrane integrity is compromised.
In E. coli, RprA is one of six currently known sRNAs (together with OmrA/B, McaS, GcvB, and RydC [88–91, 142]) to repress translation of csgD, encoding a transcriptional regulator of curli fimbriae and cellulose production (92). In addition, RprA also downregulates expression of ydaM, which encodes a diguanylate cyclase involved in activating csgD transcription. While curli are required for efficient adhesion of bacterial cells in growing biofilms, massive synthesis of surface-exposed curli fimbriae may be detrimental to cells experiencing envelope stress (90). In addition, it has been speculated that RprA may function to balance expression of curli/cellulose and colanic acid, an additional biofilm matrix component directly controlled by the Rcs pathway (93).
Although none of the signaling components of the Cpx system has been shown to be directly controlled by RprA, overexpression of the sRNA exerts negative feedback onto the stress response in a CpxR-dependent manner (78). Further investigation regarding the integration of the sRNA into the Cpx regulon is required, but two tempting hypotheses may explain the observed phenotype. First, a yet-to-be-identified auxiliary factor modulating CpxR activity could be under control of RprA (43). Alternatively, RprA could indirectly reduce induction of the Cpx response by contributing to stress relief. Intriguingly, one of the most upregulated genes following pulse overexpression of RprA in Salmonella is dsbG. Together with the CpxR-controlled dsbB, dsbG functions in disulfide bond formation within the periplasm (86, 94), and upregulation of the gene would be consistent with the induction of the Rcs phosphorelay upon loss of DsbA (83).
Additional Pathways Mediating Envelope Homeostasis
The activity of the major envelope stress response is complemented by several additional regulatory pathways modulating membrane homeostasis and modifications. In many cases, sRNAs constitute central nodes of these systems.
The EnvZ/OmpR system is one of the most thoroughly studied TCSs and contributes to the maintenance of the OM by controlling expression of multiple OMPs. The environmental cues triggering activation of the sensor kinase, EnvZ, include increased growth temperature, acidic pH, and, most importantly, increased osmolarity (95). Phosphotransfer from activated EnvZ to its cognate response regulator, OmpR, in conditions of high osmolarity controls (among other genes) the ratio of the major OMPs OmpC and OmpF (96). The opposite regulation of the two OMP genes by OmpR∼P, i.e., induction of ompC and repression of ompF transcription, respectively, is reinforced at the posttranscriptional level. While the promoter of the MicC sRNA (which represses ompC mRNA) is repressed by OmpR∼P, the transcription factor activates production of MicF sRNA (which represses ompF mRNA) (97, 98).
In addition, the two homologous sRNAs OmrA and OmrB, encoded in tandem orientation on the E. coli chromosome, are under positive control of OmpR∼P (99). Together, OmrA/OmrB repress the synthesis of the TonB-dependent receptors CirA, FecA, and FepA; the OM protease OmpT; as well as the transcription factor CsgD (88, 99). In addition, OmrA/B autoregulates its own transcription by negatively regulating the ompR-envZ mRNA (100).
Involvement of sRNAs in Modification of LPS
In pathogenic bacteria, the cell surface provides numerous exposed epitopes recognized by the host’s immune response after infection (101). Moreover, given its essential functions, the bacterial cell envelope is an effective target for antimicrobial peptides (AMPs). To evade the response of the host immune system, bacteria are able to tune the composition of OMPs within the OM. In addition, modifications of the LPS also contribute to the bacterial survival strategy in the presence of the host immune defense and AMPs (41).
Several sRNAs are integrated into the regulatory loops governing LPS modifications, either directly by controlling expression of modifying enzymes or indirectly by influencing the activity of transcriptional regulators (Fig. 3).
FIGURE 3.

Posttranscriptional regulation of LPS modification. The PhoQ-PhoP TCS, a major determinant of LPS modifications, is activated in response to Mg2+ starvation as well as by AMPs. Translation of the phoPQ bicistronic transcript is repressed by two sRNAs, MicA and GcvB. PhoQ-PhoP controls expression of MgrR, which, together with ArcZ, inhibits phosphoethanolamine (PEA) addition to the LPS oligosaccharide core by EptB. Both GcvB and MgrR are regulated at the posttranscriptional level by the sRNA SroC, which acts as a sponge and induces decay of its target sRNAs. Downregulation of lpxR mRNA by MicF decreases lipid A deacylation.
The LPS component lipid A is a common site for modifications and can, for example, be subject to dephosphorylation, deacylation, or hydroxylation (102). One of the enzymes responsible for lipid A deacylation, encoded by lpxR in Salmonella and some other bacteria, including the pathogenic E. coli O157:H7, is posttranscriptionally repressed by MicF (103). Of note, MicF, which itself is controlled by the EnvZ-OmpR TCS, uses two RNA stretches to form base-pairing interactions both at the translation initiation site and within the coding sequence of lpxR mRNA.
A major regulon controlling the physiology of LPS is the PhoQ-PhoP TCS, which is activated by low levels of Mg2+ ions, as well as by AMPs (104). The PhoQ-PhoP system has been extensively studied in the enteric pathogen Salmonella, where its integrity is essential for the infection process (105). The TCS is, however, widely conserved in several enterobacterial species, where it is involved in the adaptation to low-Mg2+ environments and/or the regulation of virulence factors (104). One of the several dozen genes directly controlled via PhoQ-PhoP encodes an sRNA, MgrR (termed Stnc560 in Salmonella), which was originally identified in Hfq coimmunoprecipitation experiments (53, 106). At the posttranscriptional level, the activity of MgrR is counteracted by yet another sRNA, SroC, which acts as a sponge RNA to sequester and trigger the decay of MgrR (107). SroC, processed from the gltIJKL mRNA, encoding a glutamate/aspartate ABC transporter, was first described to directly base-pair and repress GcvB sRNA (108), where it provides a feedforward loop regulating amino acid transport.
MgrR represses at least two mRNA targets, ygdQ (encoding an IM protein of unknown function) and eptB (pmrC in Salmonella) (109). The eptB transcript is one of the most highly deregulated genes in hfq mutant strains of Salmonella (24) and encodes a phosphoethanolamine transferase modifying the outer Kdo (3-deoxy-d-manno-octulosonic acid) unit of the LPS core (110). Thus, while directly activating other LPS-modifying enzymes, the PhoQ-PhoP TCS employs the sRNA MgrR to negatively act on eptB expression. EptB is barely expressed under standard laboratory growth conditions but is transcriptionally activated by the RpoE response, hinting at the benefit of the EptB-mediated LPS modification during OM stress. The deletion of mgrR from the E. coli chromosome, and the consequent expression of eptB, result in increased resistance to the AMP polymyxin B due to modification of the LPS structure (109). In addition, eptB mRNA is also repressed by the sRNA ArcZ, further specifying timing and degree of EptB-mediated LPS modification (111). Since ArcZ is preferentially expressed under aerobic conditions (112), the cooperative activity of both ArcZ and MgrR allows the expression of EptB only when cells encounter an Mg2+/Ca2+-rich, anaerobic (or microaerobic) environment (111).
Additional sRNAs mediate the crosstalk between the PhoQ-PhoP TCS and other regulons. The phoQ and phoP genes are encoded in a bicistronic operon, and the phoQP mRNA has been shown to be posttranscriptionally controlled by both MicA and GcvB sRNAs. However, given that phoQP mRNA levels are elevated in the absence of hfq even when micA and gcvB have been deleted from the genome, additional, yet-to-be-identified sRNAs might contribute to the complex regulation of the TCS (113), or possibly Hfq alone can repress this mRNA (114).
MicA, induced by the RpoE response, represses translation of the TCS by base-pairing within the translation initiation site of phoP (33). Repression of PhoP activity by MicA is consistent with and reinforces the activation of eptB (see above) under RpoE-inducing conditions. Similarly, GcvB sRNA inhibits translation initiation of phoP by binding a region in close proximity to the MicA pairing site (113). The posttranscriptional activity of GcvB links the PhoQ-PhoP regulon to cell metabolism, as the sRNA is mainly involved in limiting amino acid and peptide uptake under nutrient-rich conditions (115). It is intriguing, given the negative regulation of phoP by GcvB, that SroC provides another link between GcvB and the PhoP-dependent MgrR sRNA. With the SroC sponge acting as a competitor for mRNA targets of the different sRNAs, gene expression will depend not only on the binding affinities and relative concentrations of cognate sRNA/mRNA pairs but also critically on the expression level of the sponge RNA. Consequently, SroC might serve to fine-tune the coordination of posttranscriptional control via the MgrR and GcvB networks.
sRNAs AND THE RESPONSE TO OXIDATIVE STRESS AND DNA DAMAGE
Under aerobic growth conditions, reactive oxygen species (ROS), including, for example, superoxide and hydrogen peroxide, are generated as natural by-products of bacterial metabolic activity (116). ROS are able to harm the cell by damaging DNA, iron-sulfur (Fe-S) clusters, and other enzymes (117). Exploiting these toxic effects, one of the host defense mechanisms to counteract infection with Salmonella and other intracellular bacteria is the production of ROS (118). In response, bacteria have evolved mechanisms to detoxify ROS and to respond to and help the cell repair oxidative damage. As is the case for many major stress responses of Gram-negative bacteria, sRNAs are embedded in these networks (Fig. 4). However, exactly what these sRNAs do is not entirely clear.
FIGURE 4.

The OxyS and MicF sRNAs are integrated into the enterobacterial response to oxidative stress. OxyS, induced by the hydrogen peroxide-responsive OxyR, downregulates fhlA mRNA (encoding a transcription factor regulating formate metabolism) and indirectly represses rpoS expression. In addition, OxyS-mediated repression of nusG results in increased expression of kilR, encoded in the cryptic Rac prophage. KilR sequesters FtsZ, thereby leading to inhibition of cell division and growth arrest, which allows the cell to facilitate DNA damage repair. MicF contributes to increased bacterial resistance against antibiotics of different classes by repressing the major porin OmpF. Additional targets of MicF include lpxR mRNA (encoding an LPS modification enzyme), as well as lrp mRNA (encoding a transcriptional regulator of amino acid metabolism and transport). Expression of MicF is positively controlled by the transcription factors OmpR, MarA, Rob, and SoxS, with the last being induced in the presence of superoxide.
One major contributor to resistance to oxidative stress in E. coli and Salmonella, as well as other bacteria, is the general stress sigma factor RpoS and the genes under its control, including catalase (encoded by katE), required for the detoxification of hydrogen peroxide. As noted above, RpoS levels are low in hfq mutants because sRNAs are required to activate its translation (Fig. 1A). To what extent increased sensitivity to oxidative stress of hfq mutants is due to the RpoS defect has not been investigated. However, ArcZ, RprA, and DsrA, the sRNAs that promote RpoS translation, certainly should contribute to resistance to oxidative stress by inducing RpoS.
Aside from RpoS, microbes use additional, distinct mechanisms to detect different forms of oxidative stress. For example, the SoxR/S and OxyR regulons mediate the response to superoxide and hydrogen peroxide stresses, respectively, in E. coli and related enterobacteria, and each of the systems also involves the activity of sRNAs.
sRNAs Induced by Oxidative Stress
MicF, one of the first-described chromosomally encoded antisense sRNAs (98), is not only part of the OmpR-mediated envelope stress response (see above) but is also integrated into the cellular program to defeat oxidative damage. Expression of MicF is induced by three homologs of the AraC family of transcription factors with overlapping activity, SoxS, Rob, and MarA, all of which help the bacteria respond to a range of toxic molecules, including antibiotics, as well as free radicals such as superoxide and nitric oxide (119). SoxS is activated upon oxidation of an Fe-S cluster in its regulator, SoxR, in response to treating cells with the superoxide-generating drug paraquat; repression of MarA by MarR is relieved in the presence of certain phenolic compounds (120); and the activity of Rob is posttranscriptionally increased when bacteria encounter the iron chelators 2,2′- or 4,4′-dipyridyl (121). Under these conditions, downregulation of OmpF by MicF contributes to increased bacterial resistance against antibiotics entering the cell through this porin. In addition, the SoxR/S regulon, and consequently MicF, is activated by nitric oxide, produced by activated macrophages during the infection process. Deletion of micF, and thus loss of MicF-mediated repression of OmpF, results in similar hypersensitivity to killing of E. coli by murine macrophages, as observed for soxR/S mutants (122). Combined with its activity in repressing the LPS modification enzyme LpxR (see above) and the transcriptional regulator Lrp (103, 123), MicF can be considered a bacterial virulence factor, acting by restricting the entry of a variety of harmful molecules, some of which cause oxidative stress.
One of the first Hfq-dependent sRNAs described in E. coli was OxyS, which is induced as part of the OxyR regulon. Initial studies found no evidence that OxyS directly affects the resistance of bacteria to hydrogen peroxide; however, expression of the sRNA confers a protective effect against spontaneous and hydrogen peroxide-induced mutagenesis (32).
A major effect of OxyS overproduction is reduced expression of RpoS (32, 124). As noted above, RpoS contributes significantly to resistance to oxidative stress in stationary phase, e.g., via synthesis of the katE-encoded catalase or the gor-encoded glutathione oxidoreductase (125, 126). However, RpoS can also compete with RpoD for RNA polymerase, and one can imagine that the OxyS-mediated reduction of RpoS might help favor the RpoD-dependent OxyR response to oxidative stress. Repression of RpoS once OxyR is activated might avoid redundant activation of stress genes. No direct pairing of OxyS with the rpoS mRNA has been detected. The current interpretation is that OxyS might compete with other sRNAs and/or the rpoS leader itself for binding to Hfq, thereby blocking the sRNA-dependent activation of RpoS translation (127, 128). Deletion of oxyS from the E. coli chromosome results in significantly higher intracellular levels of both hydrogen peroxide and superoxide compared to the wild type (129). This phenotype was suggested to result from the ability of OxyS to restrict cellular respiration, and thus to reduce the burden of ROS produced during metabolic activities (129). A model in which OxyS helps reduce the endogenous sources of the inducing stress may be characteristic of many of the sRNA arms of stress regulons. Consistent with this idea, the best-characterized OxyS target is fhlA, which is repressed by the sRNA through the formation of a kissing-loop interaction targeting sites in both the 5′ UTR and the coding sequence of the transcript (130). FhlA is a transcriptional activator of complexes involved in formate metabolism (131), whose metal cofactors likely increase cellular damage under oxidative stress conditions (132). In addition, confirmed targets of OxyS also include wrbA, encoding an NAD(P)H:quinone oxidoreductase (133).
The ability of OxyS to function as an antimutator is the subject of a very recent study (134). One of the transcripts downregulated by OxyS is nusG mRNA, coding for a highly conserved regulator of RNA polymerase (Fig. 4). Together with Rho, NusG aids termination at a subset of sites and plays an important role in silencing horizontally acquired genetic elements by inhibiting their transcription (135). One of the loci silenced by NusG and Rho is the E. coli cryptic prophage rac, encoding amongst others the kilR gene (136). When NusG levels are low, i.e., when OxyS is active, rac genes are transcribed and KilR is produced. Interfering with assembly of the cell division machinery, KilR inhibits cell cycle progression, which allows the cell to facilitate damage repair. In line with this model, cells expressing OxyS and KilR appear elongated, and the decreased rate of mutations observed in the presence of OxyS is likewise dependent on KilR (134). Repression of nusG by OxyS, and thus loss of silencing of cryptic prophages and other horizontally acquired elements, is reminiscent of prophage induction during the SOS response. In both cases, DNA damage may act as a signal for repressed prophages to jump ship and seek new, undamaged hosts.
RNA-Based Regulation of DNA Mutagenesis
OxyS is not the only known posttranscriptional regulator of DNA damage in bacteria. Cells strictly control the rates at which mutations can occur, presumably to balance beneficial and detrimental changes of the genome. One system to limit the integration of mutations upon DNA damage is the mismatch repair (MMR) system, which is dedicated to the recognition and repair of mismatches in the genome. One central component of the E. coli MMR system is MutS, which detects and binds to DNA mismatches, thus initiating the repair process (137). While it had long been known that the cellular levels of MutS decrease in an Hfq-dependent manner when E. coli enters stationary growth (138, 139), the molecular principles underlying this regulation were only recently discovered. Overexpression of two sRNAs, the RpoS-dependent SdsR and the RpoS-activating ArcZ (see Fig. 1A), results in significant repression of a posttranscriptional mutS reporter (114). More importantly, however, Hfq itself binds to the 5′ UTR of mutS mRNA independently of base-pairing sRNAs, and thereby inhibits translation of the transcript. In turn, the cell is able to react to changing environments by limiting mutS expression and thus raising the mutagenesis rate under specific stress conditions (through induction of ArcZ or SdsR), as well as by titration of Hfq (through high levels of sRNAs or transcripts competing for binding).
RpoS contributes to stationary-phase mutagenesis and break repair by different mechanisms, including the induction of error-prone polymerases and the SdsR sRNA (9, 21, 22, 140). Cells lacking the sRNA GcvB were found to limit mutagenic break repair and mutagenesis in stationary phase; surprisingly, this phenotype of GcvB was entirely suppressed by loss of RpoE (141). The authors find a modest induction of RpoE in cells devoid of GcvB and suggest that the higher level of RpoE may compete with RpoS access to core polymerase. While the mode of action of GcvB and the basis for loss of RpoS activity will need further investigation, the results do point out the degree to which stress responses and the roles of sRNAs in regulating them are entangled in organisms like E. coli.
CONCLUSIONS AND FUTURE DIRECTIONS
sRNAs exist in essentially all bacterial stress responses, and their major roles within the regulons of specialized sigma factors have been studied extensively. In many other cases, the physiological functions of the sRNAs within a given regulon are only partially understood but can provide new insight into what cells perceive as stress and the pathways they can use to overcome stress or minimize intrinsic sources of stress. The recent discovery of many sRNAs encoded within the 3′ UTRs of genes has expanded the identification of sRNA-dependent arms for stress responses. The sRNAs also play critical roles in crosstalk between regulons, in setting regulatory hierarchies, and in providing feedback loops that allow rapid response to stress and rapid return to equilibrium when stresses are dealt with. Although we highlight here many examples of sRNA-mediated control that we understand, the sRNAs frequently have multiple targets for which the physiological role of the regulation remains to be understood, necessary for a full understanding of the role of the regulon.
ACKNOWLEDGMENTS
We thank members of our laboratories for comments on the manuscript and thank S. Altuvia for sharing results prior to publication and for comments on the manuscript. Preparation of this review was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (S.G.), and the BioMentoring Program of the LMU Faculty of Biology (K.S.F.).
REFERENCES
- 1.Papenfort K, Vanderpool CK. 2015. Target activation by regulatory RNAs in bacteria. FEMS Microbiol Rev 39:362–378. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Waters LS, Storz G. 2009. Regulatory RNAs in bacteria. Cell 136:615–628. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Storz G, Vogel J, Wassarman KM. 2011. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43:880–891. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. 2013. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J Mol Biol 425:3678–3697. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kawamoto H, Koide Y, Morita T, Aiba H. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol Microbiol 61:1013–1022. [PubMed] [DOI] [PubMed] [Google Scholar]
- 6.Melamed S, Peer A, Faigenbaum-Romm R, Gatt YE, Reiss N, Bar A, Altuvia Y, Argaman L, Margalit H. 2016. Global mapping of small RNA-target interactions in bacteria. Mol Cell 63:884–897. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soper TJ, Woodson SA. 2008. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA 14:1907–1917. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tsui HC, Leung HC, Winkler ME. 1994. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol 13:35–49. [PubMed] [DOI] [PubMed] [Google Scholar]
- 9.Battesti A, Majdalani N, Gottesman S. 2011. The RpoS-mediated general stress response in Escherichia coli. Annu Rev Microbiol 65:189–213. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber H, Polen T, Heuveling J, Wendisch VF, Hengge R. 2005. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters, and sigma factor selectivity. J Bacteriol 187:1591–1603. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dong T, Schellhorn HE. 2009. Control of RpoS in global gene expression of Escherichia coli in minimal media. Mol Genet Genomics 281:19–33. [PubMed] [DOI] [PubMed] [Google Scholar]
- 12.Patten CL, Kirchhof MG, Schertzberg MR, Morton RA, Schellhorn HE. 2004. Microarray analysis of RpoS-mediated gene expression in Escherichia coli K-12. Mol Genet Genomics 272:580–591. [PubMed] [DOI] [PubMed] [Google Scholar]
- 13.Muffler A, Fischer D, Hengge-Aronis R. 1996. The RNA-binding protein HF-I, known as a host factor for phage Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev 10:1143–1151. [PubMed] [DOI] [PubMed] [Google Scholar]
- 14.Muffler A, Traulsen DD, Fischer D, Lange R, Hengge-Aronis R. 1997. The RNA-binding protein HF-I plays a global regulatory role which is largely, but not exclusively, due to its role in expression of the σS subunit of RNA polymerase in Escherichia coli. J Bacteriol 179:297–300. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Opdyke JA, Fozo EM, Hemm MR, Storz G. 2011. RNase III participates in GadY-dependent cleavage of the gadX-gadW mRNA. J Mol Biol 406:29–43. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Opdyke JA, Kang JG, Storz G. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J Bacteriol 186:6698–6705. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hao Y, Updegrove TB, Livingston NN, Storz G. 2016. Protection against deleterious nitrogen compounds: role of σS-dependent small RNAs encoded adjacent to sdiA. Nucleic Acids Res 44:6935–6948. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva IJ, Ortega AD, Viegas SC, García-Del Portillo F, Arraiano CM. 2013. An RpoS-dependent sRNA regulates the expression of a chaperone involved in protein folding. RNA 19:1253–1265. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parker A, Gottesman S. 2016. Small RNA regulation of TolC, the outer membrane component of bacterial multidrug transporters. J Bacteriol 198:1101–1113. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fröhlich KS, Haneke K, Papenfort K, Vogel J. 2016. The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res 44:10406–10422. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fröhlich KS, Papenfort K, Berger AA, Vogel J. 2012. A conserved RpoS-dependent small RNA controls the synthesis of major porin OmpD. Nucleic Acids Res 40:3623–3640. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gutierrez A, Laureti L, Crussard S, Abida H, Rodríguez-Rojas A, Blázquez J, Baharoglu Z, Mazel D, Darfeuille F, Vogel J, Matic I. 2013. β-Lactam antibiotics promote bacterial mutagenesis via an RpoS-mediated reduction in replication fidelity. Nat Commun 4:1610. 10.1038/ncomms2607. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guisbert E, Rhodius VA, Ahuja N, Witkin E, Gross CA. 2007. Hfq modulates the σE-mediated envelope stress response and the σ32-mediated cytoplasmic stress response in Escherichia coli. J Bacteriol 189:1963–1973. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Figueroa-Bossi N, Lemire S, Maloriol D, Balbontín R, Casadesús J, Bossi L. 2006. Loss of Hfq activates the σE-dependent envelope stress response in Salmonella enterica. Mol Microbiol 62:838–852. [PubMed] [DOI] [PubMed] [Google Scholar]
- 25.Vogel J, Papenfort K. 2006. Small non-coding RNAs and the bacterial outer membrane. Curr Opin Microbiol 9:605–611. [PubMed] [DOI] [PubMed] [Google Scholar]
- 26.Gogol EB, Rhodius VA, Papenfort K, Vogel J, Gross CA. 2011. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc Natl Acad Sci U S A 108:12875–12880. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Massé E, Gottesman S. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc Natl Acad Sci U S A 99:4620–4625. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schu DJ, Zhang A, Gottesman S, Storz G. 2015. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J 34:2557–2573. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mutalik VK, Nonaka G, Ades SE, Rhodius VA, Gross CA. 2009. Promoter strength properties of the complete σE regulon of Escherichia coli and Salmonella enterica. J Bacteriol 191:7279–7287. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beisel CL, Storz G. 2011. The base-pairing RNA Spot 42 participates in a multioutput feedforward loop to help enact catabolite repression in Escherichia coli. Mol Cell 41:286–297. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massé E, Salvail H, Desnoyers G, Arguin M. 2007. Small RNAs controlling iron metabolism. Curr Opin Microbiol 10:140–145. [PubMed] [DOI] [PubMed] [Google Scholar]
- 32.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. 1997. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell 90:43–53. [PubMed] [DOI] [PubMed] [Google Scholar]
- 33.Coornaert A, Lu A, Mandin P, Springer M, Gottesman S, Guillier M. 2010. MicA sRNA links the PhoP regulon to cell envelope stress. Mol Microbiol 76:467–479. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nikaido H. 2003. Molecular basis of bacterial outer membrane permeability revisited. Microbiol Mol Biol Rev 67:593–656. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silhavy TJ, Kahne D, Walker S. 2010. The bacterial cell envelope. Cold Spring Harb Perspect Biol 2:a000414. 10.1101/cshperspect.a000414. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollmer W, Blanot D, de Pedro MA. 2008. Peptidoglycan structure and architecture. FEMS Microbiol Rev 32:149–167. [PubMed] [DOI] [PubMed] [Google Scholar]
- 37.Braun V. 1975. Covalent lipoprotein from the outer membrane of Escherichia coli. Biochim Biophys Acta 415:335–377. [PubMed] [DOI] [PubMed] [Google Scholar]
- 38.Li GW, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157:624–635. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Inouye M, Shaw J, Shen C. 1972. The assembly of a structural lipoprotein in the envelope of Escherichia coli. J Biol Chem 247:8154–8159. [PubMed] [PubMed] [Google Scholar]
- 40.Whitfield C, Trent MS. 2014. Biosynthesis and export of bacterial lipopolysaccharides. Annu Rev Biochem 83:99–128. [PubMed] [DOI] [PubMed] [Google Scholar]
- 41.Maldonado RF, Sá-Correia I, Valvano MA. 2016. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol Rev 40:480–493. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Braun V, Endriss F. 2007. Energy-coupled outer membrane transport proteins and regulatory proteins. Biometals 20:219–231. [PubMed] [DOI] [PubMed] [Google Scholar]
- 43.Grabowicz M, Silhavy TJ. 2017. Envelope stress responses: an interconnected safety net. Trends Biochem Sci 42:232–242. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bury-Moné S, Nomane Y, Reymond N, Barbet R, Jacquet E, Imbeaud S, Jacq A, Bouloc P. 2009. Global analysis of extracytoplasmic stress signaling in Escherichia coli. PLoS Genet 5:e1000651. 10.1371/journal.pgen.1000651. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raina S, Missiakas D, Georgopoulos C. 1995. The rpoE gene encoding the σE (σ24) heat shock sigma factor of Escherichia coli. EMBO J 14:1043–1055. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ades SE, Grigorova IL, Gross CA. 2003. Regulation of the alternative sigma factor σE during initiation, adaptation, and shutoff of the extracytoplasmic heat shock response in Escherichia coli. J Bacteriol 185:2512–2519. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ades SE. 2004. Control of the alternative sigma factor σE in Escherichia coli. Curr Opin Microbiol 7:157–162. [PubMed] [DOI] [PubMed] [Google Scholar]
- 48.Walsh NP, Alba BM, Bose B, Gross CA, Sauer RT. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61–71. [PubMed] [DOI] [PubMed] [Google Scholar]
- 49.Lima S, Guo MS, Chaba R, Gross CA, Sauer RT. 2013. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 340:837–841. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Skovierova H, Rowley G, Rezuchova B, Homerova D, Lewis C, Roberts M, Kormanec J. 2006. Identification of the σE regulon of Salmonella enterica serovar Typhimurium. Microbiology 152:1347–1359. [PubMed] [DOI] [PubMed] [Google Scholar]
- 51.Rhodius VA, Suh WC, Nonaka G, West J, Gross CA. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol 4:e2. 10.1371/journal.pbio.0040002. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. 2012. An atlas of Hfq-bound transcripts reveals 3′ UTRs as a genomic reservoir of regulatory small RNAs. EMBO J 31:4005–4019. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. 2003. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol 50:1111–1124. [PubMed] [DOI] [PubMed] [Google Scholar]
- 54.Hayden JD, Ades SE. 2008. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. 10.1371/journal.pone.0001573. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Las Peñas A, Connolly L, Gross CA. 1997. σE is an essential sigma factor in Escherichia coli. J Bacteriol 179:6862–6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Papenfort K, Bouvier M, Mika F, Sharma CM, Vogel J. 2010. Evidence for an autonomous 5′ target recognition domain in an Hfq-associated small RNA. Proc Natl Acad Sci U S A 107:20435–20440. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. 2008. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell 32:827–837. [PubMed] [DOI] [PubMed] [Google Scholar]
- 58.Celesnik H, Deana A, Belasco JG. 2007. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell 27:79–90. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emory SA, Bouvet P, Belasco JG. 1992. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev 6:135–148. [PubMed] [DOI] [PubMed] [Google Scholar]
- 60.Papenfort K, Pfeiffer V, Mika F, Lucchini S, Hinton JC, Vogel J. 2006. σE-Dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol Microbiol 62:1674–1688. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Song T, Mika F, Lindmark B, Liu Z, Schild S, Bishop A, Zhu J, Camilli A, Johansson J, Vogel J, Wai SN. 2008. A new Vibrio cholerae sRNA modulates colonization and affects release of outer membrane vesicles. Mol Microbiol 70:100–111. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Song T, Sabharwal D, Wai SN. 2010. VrrA mediates Hfq-dependent regulation of OmpT synthesis in Vibrio cholerae. J Mol Biol 400:682–688. [PubMed] [DOI] [PubMed] [Google Scholar]
- 63.Song T, Sabharwal D, Gurung JM, Cheng AT, Sjöström AE, Yildiz FH, Uhlin BE, Wai SN. 2014. Vibrio cholerae utilizes direct sRNA regulation in expression of a biofilm matrix protein. PLoS One 9:e101280. 10.1371/journal.pone.0101280. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabharwal D, Song T, Papenfort K, Wai SN. 2015. The VrrA sRNA controls a stationary phase survival factor Vrp of Vibrio cholerae. RNA Biol 12:186–196. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Klein G, Kobylak N, Lindner B, Stupak A, Raina S. 2014. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J Biol Chem 289:14829–14853. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Guo MS, Updegrove TB, Gogol EB, Shabalina SA, Gross CA, Storz G. 2014. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev 28:1620–1634. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Raivio TL. 2014. Everything old is new again: an update on current research on the Cpx envelope stress response. Biochim Biophys Acta 1843:1529–1541. [PubMed] [DOI] [PubMed] [Google Scholar]
- 68.Mileykovskaya E, Dowhan W. 1997. The Cpx two-component signal transduction pathway is activated in Escherichia coli mutant strains lacking phosphatidylethanolamine. J Bacteriol 179:1029–1034. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raivio TL, Silhavy TJ. 1997. Transduction of envelope stress in Escherichia coli by the Cpx two-component system. J Bacteriol 179:7724–7733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Snyder WB, Davis LJ, Danese PN, Cosma CL, Silhavy TJ. 1995. Overproduction of NlpE, a new outer membrane lipoprotein, suppresses the toxicity of periplasmic LacZ by activation of the Cpx signal transduction pathway. J Bacteriol 177:4216–4223. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Delhaye A, Collet JF, Laloux G. 2016. Fine-tuning of the Cpx envelope stress response is required for cell wall homeostasis in Escherichia coli. mBio 7:e00047-e16. 10.1128/mBio.00047-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogt SL, Nevesinjac AZ, Humphries RM, Donnenberg MS, Armstrong GD, Raivio TL. 2010. The Cpx envelope stress response both facilitates and inhibits elaboration of the enteropathogenic Escherichia coli bundle-forming pilus. Mol Microbiol 76:1095–1110. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guest RL, Wang J, Wong JL, Raivio TL. 2017. A bacterial stress response regulates respiratory protein complexes to control envelope stress adaptation. J Bacteriol 199:e00153-17. 10.1128/JB.00153-17. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danese PN, Silhavy TJ. 1998. CpxP, a stress-combative member of the Cpx regulon. J Bacteriol 180:831–839. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Raivio TL, Popkin DL, Silhavy TJ. 1999. The Cpx envelope stress response is controlled by amplification and feedback inhibition. J Bacteriol 181:5263–5272. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chao Y, Vogel J. 2016. A 3′ UTR-derived small RNA provides the regulatory noncoding arm of the inner membrane stress response. Mol Cell 61:352–363. [PubMed] [DOI] [PubMed] [Google Scholar]
- 77.Grabowicz M, Koren D, Silhavy TJ. 2016. The CpxQ sRNA negatively regulates Skp to prevent mistargeting of β-barrel outer membrane proteins into the cytoplasmic membrane. mBio 7:e00312-16. 10.1128/mBio.00312-16. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vogt SL, Evans AD, Guest RL, Raivio TL. 2014. The Cpx envelope stress response regulates and is regulated by small noncoding RNAs. J Bacteriol 196:4229–4238. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev 15:1637–1651. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.De Lay N, Gottesman S. 2009. The Crp-activated small noncoding regulatory RNA CyaR (RyeE) links nutritional status to group behavior. J Bacteriol 191:461–476. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Johansen J, Eriksen M, Kallipolitis B, Valentin-Hansen P. 2008. Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP-CRP- and σE-dependent CyaR-ompX regulatory case. J Mol Biol 383:1–9. [PubMed] [DOI] [PubMed] [Google Scholar]
- 82.Papenfort K, Pfeiffer V, Lucchini S, Sonawane A, Hinton JC, Vogel J. 2008. Systematic deletion of Salmonella small RNA genes identifies CyaR, a conserved CRP-dependent riboregulator of OmpX synthesis. Mol Microbiol 68:890–906. [PubMed] [DOI] [PubMed] [Google Scholar]
- 83.Majdalani N, Gottesman S. 2005. The Rcs phosphorelay: a complex signal transduction system. Annu Rev Microbiol 59:379–405. [PubMed] [DOI] [PubMed] [Google Scholar]
- 84.Laubacher ME, Ades SE. 2008. The Rcs phosphorelay is a cell envelope stress response activated by peptidoglycan stress and contributes to intrinsic antibiotic resistance. J Bacteriol 190:2065–2074. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Majdalani N, Chen S, Murrow J, St John K, Gottesman S. 2001. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol 39:1382–1394. [PubMed] [DOI] [PubMed] [Google Scholar]
- 86.Papenfort K, Espinosa E, Casadesús J, Vogel J. 2015. Small RNA-based feedforward loop with AND-gate logic regulates extrachromosomal DNA transfer in Salmonella. Proc Natl Acad Sci U S A 112:E4772–E4781. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madhugiri R, Basineni SR, Klug G. 2010. Turn-over of the small non-coding RNA RprA in E. coli is influenced by osmolarity. Mol Genet Genomics 284:307–318. [PubMed] [DOI] [PubMed] [Google Scholar]
- 88.Holmqvist E, Reimegård J, Sterk M, Grantcharova N, Römling U, Wagner EG. 2010. Two antisense RNAs target the transcriptional regulator CsgD to inhibit curli synthesis. EMBO J 29:1840–1850. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Thomason MK, Fontaine F, De Lay N, Storz G. 2012. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol 84:17–35. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mika F, Busse S, Possling A, Berkholz J, Tschowri N, Sommerfeldt N, Pruteanu M, Hengge R. 2012. Targeting of csgD by the small regulatory RNA RprA links stationary phase, biofilm formation and cell envelope stress in Escherichia coli. Mol Microbiol 84:51–65. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jørgensen MG, Nielsen JS, Boysen A, Franch T, Møller-Jensen J, Valentin-Hansen P. 2012. Small regulatory RNAs control the multi-cellular adhesive lifestyle of Escherichia coli. Mol Microbiol 84:36–50. [PubMed] [DOI] [PubMed] [Google Scholar]
- 92.Ogasawara H, Yamamoto K, Ishihama A. 2011. Role of the biofilm master regulator CsgD in cross-regulation between biofilm formation and flagellar synthesis. J Bacteriol 193:2587–2597. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Boehm A, Vogel J. 2012. The csgD mRNA as a hub for signal integration via multiple small RNAs. Mol Microbiol 84:1–5. [PubMed] [DOI] [PubMed] [Google Scholar]
- 94.Pogliano J, Lynch AS, Belin D, Lin EC, Beckwith J. 1997. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev 11:1169–1182. [PubMed] [DOI] [PubMed] [Google Scholar]
- 95.Pratt LA, Hsing W, Gibson KE, Silhavy TJ. 1996. From acids to osmZ: multiple factors influence synthesis of the OmpF and OmpC porins in Escherichia coli. Mol Microbiol 20:911–917. [PubMed] [DOI] [PubMed] [Google Scholar]
- 96.Alphen WV, Lugtenberg B. 1977. Influence of osmolarity of the growth medium on the outer membrane protein pattern of Escherichia coli. J Bacteriol 131:623–630. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Chen S, Zhang A, Blyn LB, Storz G. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J Bacteriol 186:6689–6697. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mizuno T, Chou MY, Inouye M. 1984. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA). Proc Natl Acad Sci U S A 81:1966–1970. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guillier M, Gottesman S. 2006. Remodelling of the Escherichia coli outer membrane by two small regulatory RNAs. Mol Microbiol 59:231–247. [PubMed] [DOI] [PubMed] [Google Scholar]
- 100.Brosse A, Korobeinikova A, Gottesman S, Guillier M. 2016. Unexpected properties of sRNA promoters allow feedback control via regulation of a two-component system. Nucleic Acids Res 44:9650–9666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh SP, Williams YU, Klebba PE, Macchia P, Miller S. 2000. Immune recognition of porin and lipopolysaccharide epitopes of Salmonella typhimurium in mice. Microb Pathog 28:157–167. [PubMed] [DOI] [PubMed] [Google Scholar]
- 102.Needham BD, Trent MS. 2013. Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nat Rev Microbiol 11:467–481. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Corcoran CP, Podkaminski D, Papenfort K, Urban JH, Hinton JC, Vogel J. 2012. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol Microbiol 84:428–445. [PubMed] [DOI] [PubMed] [Google Scholar]
- 104.Groisman EA. 2001. The pleiotropic two-component regulatory system PhoP-PhoQ. J Bacteriol 183:1835–1842. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miller SI, Kukral AM, Mekalanos JJ. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc Natl Acad Sci U S A 86:5054–5058. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sittka A, Lucchini S, Papenfort K, Sharma CM, Rolle K, Binnewies TT, Hinton JC, Vogel J. 2008. Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 4:e1000163. 10.1371/journal.pgen.1000163. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Acuña LG, Barros MJ, Peñaloza D, Rodas PI, Paredes-Sabja D, Fuentes JA, Gil F, Calderón IL. 2016. A feed-forward loop between SroC and MgrR small RNAs modulates the expression of eptB and the susceptibility to polymyxin B in Salmonella Typhimurium. Microbiology 162:1996–2004. [PubMed] [DOI] [PubMed] [Google Scholar]
- 108.Miyakoshi M, Chao Y, Vogel J. 2015. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J 34:1478–1492. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moon K, Gottesman S. 2009. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol Microbiol 74:1314–1330. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Reynolds CM, Kalb SR, Cotter RJ, Raetz CR. 2005. A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J Biol Chem 280:21202–21211. [PubMed] [DOI] [PubMed] [Google Scholar]
- 111.Moon K, Six DA, Lee HJ, Raetz CR, Gottesman S. 2013. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol Microbiol 89:52–64. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mandin P, Gottesman S. 2010. Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. EMBO J 29:3094–3107. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Coornaert A, Chiaruttini C, Springer M, Guillier M. 2013. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet 9:e1003156. 10.1371/journal.pgen.1003156. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen J, Gottesman S. 2017. Hfq links translation repression to stress-induced mutagenesis in E. coli. Genes Dev 31:1382–1395. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sharma CM, Papenfort K, Pernitzsch SR, Mollenkopf HJ, Hinton JC, Vogel J. 2011. Pervasive post-transcriptional control of genes involved in amino acid metabolism by the Hfq-dependent GcvB small RNA. Mol Microbiol 81:1144–1165. [PubMed] [DOI] [PubMed] [Google Scholar]
- 116.Storz G, Imlay JA. 1999. Oxidative stress. Curr Opin Microbiol 2:188–194. [DOI] [PubMed] [Google Scholar]
- 117.Imlay JA. 2013. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11:443–454. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Frawley ER, Fang FC. 2014. The ins and outs of bacterial iron metabolism. Mol Microbiol 93:609–616. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alekshun MN, Levy SB. 1997. Regulation of chromosomally mediated multiple antibiotic resistance: the mar regulon. Antimicrob Agents Chemother 41:2067–2075. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Demple B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53–57. [DOI] [PubMed] [Google Scholar]
- 121.Rosner JL, Dangi B, Gronenborn AM, Martin RG. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J Bacteriol 184:1407–1416. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nunoshiba T, deRojas-Walker T, Wishnok JS, Tannenbaum SR, Demple B. 1993. Activation by nitric oxide of an oxidative-stress response that defends Escherichia coli against activated macrophages. Proc Natl Acad Sci U S A 90:9993–9997. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee HJ, Gottesman S. 2016. sRNA roles in regulating transcriptional regulators: Lrp and SoxS regulation by sRNAs. Nucleic Acids Res 44:6907–6923. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. 1998. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-I) protein. EMBO J 17:6061–6068. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Schellhorn HE, Hassan HM. 1988. Transcriptional regulation of katE in Escherichia coli K-12. J Bacteriol 170:4286–4292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Becker-Hapak M, Eisenstark A. 1995. Role of rpoS in the regulation of glutathione oxidoreductase (gor) in Escherichia coli. FEMS Microbiol Lett 134:39–44. [DOI] [PubMed] [Google Scholar]
- 127.Moon K, Gottesman S. 2011. Competition among Hfq-binding small RNAs in Escherichia coli. Mol Microbiol 82:1545–1562. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hämmerle H, Večerek B, Resch A, Bläsi U. 2013. Duplex formation between the sRNA DsrA and rpoS mRNA is not sufficient for efficient RpoS synthesis at low temperature. RNA Biol 10:1834–1841. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.González-Flecha B, Demple B. 1999. Role for the oxyS gene in regulation of intracellular hydrogen peroxide in Escherichia coli. J Bacteriol 181:3833–3836. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Argaman L, Altuvia S. 2000. fhlA repression by OxyS RNA: kissing complex formation at two sites results in a stable antisense-target RNA complex. J Mol Biol 300:1101–1112. [PubMed] [DOI] [PubMed] [Google Scholar]
- 131.Schlensog V, Böck A. 1990. Identification and sequence analysis of the gene encoding the transcriptional activator of the formate hydrogenlyase system of Escherichia coli. Mol Microbiol 4:1319–1327. [PubMed] [DOI] [PubMed] [Google Scholar]
- 132.Altuvia S, Zhang A, Argaman L, Tiwari A, Storz G. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J 17:6069–6075. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. 2006. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res 34:2791–2802. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Barshishat S, Elgrably-Weiss M, Edelstein J, Georg J, Govindarajan S, Haviv M, Wright PR, Hess WR, Altuvia S. 2017. OxyS small RNA induces cell cycle arrest to allow DNA damage repair. EMBO J 37:413–426. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ray-Soni A, Bellecourt MJ, Landick R. 2016. Mechanisms of bacterial transcription termination: all good things must end. Annu Rev Biochem 85:319–347. [PubMed] [DOI] [PubMed] [Google Scholar]
- 136.Conter A, Bouché JP, Dassain M. 1996. Identification of a new inhibitor of essential division gene ftsZ as the kil gene of defective prophage Rac. J Bacteriol 178:5100–5104. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Li GM. 2008. Mechanisms and functions of DNA mismatch repair. Cell Res 18:85–98. [PubMed] [DOI] [PubMed] [Google Scholar]
- 138.Feng G, Tsui HC, Winkler ME. 1996. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. J Bacteriol 178:2388–2396. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tsui HC, Feng G, Winkler ME. 1997. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol 179:7476–7487. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Layton JC, Foster PL. 2003. Error-prone DNA polymerase IV is controlled by the stress-response σ factor, RpoS, in Escherichia coli. Mol Microbiol 50:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Barreto B, Rogers E, Xia J, Frisch RL, Richters M, Fitzgerald DM, Rosenberg SM. 2016. The small RNA GcvB promotes mutagenic break repair by opposing the membrane stress response. J Bacteriol 198:3296–3308. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bordeau V, Felden B. 2014. Curli synthesis and biofilm formation in enteric bacteria are controlled by a dynamic small RNA module made up of a pseudoknot assisted by an RNA chaperone. Nucleic Acids Res 42:4682–4696. [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]


