Abstract
Previous characterization of Bacillus subtilis hemN, encoding a protein involved in oxygen-independent coproporphyrinogen III decarboxylation, indicated the presence of a second hemN-like gene (B. Hippler, G. Homuth, T. Hoffmann, C. Hungerer, W. Schumann, and D. Jahn, J. Bacteriol. 179:7181–7185, 1997). The corresponding hemZ gene was found to be split into the two potential open reading frames yhaV and yhaW by a sequencing error of the genome sequencing project. The hemZ gene, encoding a 501-amino-acid protein with a calculated molecular mass of 57,533 Da, complemented a Salmonella typhimurium hemF hemN double mutant under aerobic and anaerobic growth conditions. A B. subtilis hemZ mutant accumulated coproporphyrinogen III under anaerobic growth conditions. A hemN hemZ double mutant exhibited normal aerobic and anaerobic growth, indicating the presence of a third alternative oxygen-independent enzymatic system for coproporphyrinogen III oxidation. The hemY gene, encoding oxygen-dependent protoporphyrinogen IX oxidase with coproporphyrinogen III oxidase side activity, did not significantly contribute to this newly identified system. Growth behavior of hemY mutants revealed the presence of an oxygen-independent protoporphyrinogen IX oxidase in B. subtilis. A monocistronic hemZ mRNA, starting 31 bp upstream of the translational start codon, was detected. Reporter gene fusions of hemZ and hemN demonstrated a fivefold anaerobic induction of both genes under nitrate ammonifying growth conditions. No anaerobic induction was observed for fermentatively growing B. subtilis. The B. subtilis redox regulatory systems encoded by resDE, fnr, and ywiD were indispensable for the observed transcriptional induction. A redox regulation cascade proceeding from an unknown sensor via resDE, through fnr and ywiD to hemN/hemZ, is suggested for the observed coregulation of heme biosynthesis and the anaerobic respiratory energy metabolism. Finally, only hemZ was found to be fivefold induced by the presence of H2O2, indicating further coregulation of heme biosynthesis with the formation of the tetrapyrrole enzyme catalase.
During bacterial heme biosynthesis, coproporphyrinogen III is oxidatively decarboxylated to protoporphyrinogen IX (3, 22, 24). In bacteria, two structurally unrelated enzymatic systems catalyzing this reaction were identified (37, 40, 41, 47, 48). One type of coproporphyrinogen III oxidase (HemF) requires molecular oxygen (42, 43, 47). Since it obviously cannot function in the absence of molecular oxygen, a structurally different oxygen-independent enzyme (designated HemN and HemZ) is required for anaerobic growth and heme biosynthesis (42, 43, 47, 48). Up to now, genes coding for oxygen-independent enzymes (hemN and hemZ) were found only in bacteria (13, 34). Expression of the Rhodobacter sphaeroides hemN gene in Escherichia coli led to an increase in coproporphyrinogen III oxidase activity which was dependent on the presence of methionine, ATP, NADP+, NADH, and Mg2+ (4).
In Bacillus subtilis, the anaerobic energy metabolism via nitrate ammonification requires oxygen-independent heme biosynthesis (14, 15, 26, 30, 31). The anaerobic enzymatic systems are induced via a regulatory cascade. Low oxygen tension is sensed by an unknown system, and the signal generated is transferred to the pleiotropic two-component regulatory system ResDE. ResDE is responsible, directly or indirectly, for induction of the gene encoding the redox regulator Fnr (32). Fnr in turn induces genes of nitrate respiration and the regulatory gene ywiD (5, 27). Subsequently, ywiD activates anaerobic respiratory and fermentative loci (27). Oxygen tension-dependent coregulation of energy metabolism and heme biosynthesis has been described for various bacteria (6, 18–21, 33). The genes encoding coproporphyrinogen III oxidases were found to be key redox regulatory targets in Pseudomonas aeruginosa (34). Fnr binding sites were located in the promoter sequences of coproporphyrinogen III oxidase genes from Alcaligenes eutrophus, P. aeruginosa, Pseudomonas stutzeri, and R. sphaeroides (25, 34, 49).
A hemF gene encoding the oxygen-dependent enzyme has not been found in the genome sequence of B. subtilis (13, 23). Recently, we described the identification and functional characterization of the B. subtilis hemN gene located upstream of the B. subtilis dnaK operon (13, 16). It is not part of the two hem operons in B. subtilis, hemAXCDBL at 244° and hemEHY operon at 94° of the genomic map, encoding almost all enzymes required for the formation of protoheme from glutamyl-tRNA (9, 11). However, mutation of B. subtilis hemN had no obvious consequences for aerobic and anaerobic growth, suggesting the presence of a second hemN type gene (13). Here, we describe the identification and functional characterization of the second hemN-type gene, hemZ. Both, hemN and hemZ are subject to redox regulation mediated by resDE, fnr, and ywiD. Peroxide stress regulation is limited to hemZ.
MATERIALS AND METHODS
Bacterial strains, plasmids and DNA manipulations.
Bacterial strains and plasmids used in this study are listed in Table 1. B. subtilis and E. coli strains were grown aerobically and anaerobically in complex and minimal media with additions as detailed before (14, 30, 36). Cloning procedures were carried out by standard protocols (36). PCR products were generated with Deep Vent DNA polymerase (New England Biolabs, Schwalbach, Germany). PCR primers were obtained from ARK Scientific GmbH Biosystems (Darmstadt, Germany). For DNA sequence determination of the hemZ locus from B. subtilis 1012, four different plasmids were constructed by PCR. The PCR products generated with primers containing artificial BamHI restriction sites were inserted into the BamHI site of pBluescript SK+II. Cloning of a DNA fragment corresponding to the former yhaW gene (nucleotides 180 to 681 in Fig. 1), generated by PCR using primers YHAW5 (5′-GGCCATGGATCCTTGCAAATTAAAATAGAAGGCATA-3′; BamHI restriction site underlined) and YHAW3 (5′-GGCCATGGATCCTTACTCGGTACAAATCCGGCACTG-3′), resulted in plasmid phemZ1RPT3. The former open reading frame (ORF) yhaV carrying plasmid (nucleotides 813 to 1685 in Fig. 1), generated via PCR using primers YHAV5 (5′-GGCCATGGATCCATGCAGAAAATCGGTGAATGGCTG-3′) and YHAV3 (5′-GGCCATGGATCCTCAGTGCTGCTTTGTCGTTTTTTC-3′), was designated phemZ2RPT3. The third construct, pALF01, contains the complete former yhaW gene, the 5′ end of postulated yhaV, and the region between these potential ORFs (nucleotides 180 to 962), generated by PCR using primers YHAW5 (see above) and ALF (5′-GGCCATGGATCCAATGTTTTTCACATCCGGGAAGGA-3′). Using the same primers (YHAWS and ALF), PCR products were generated from genomic DNA isolated from B. subtilis 168 and JH642 and cloned as outlined above. The fourth construct, phemZ-PEX, containing the promoter region upstream of hemZ and the complete former yhaW (nucleotides 4 to 681 in Fig. 1), was generated via PCR using primers HEMZPEX5 (5′-GGCCATGGATCCAGGAATATTTCCGAATGCAGCAGC-3′) and YHAW3 (see above). Complete DNA sequence determination of all cloned fragments was performed with an ALFexpress automatic DNA sequencer (Amersham Pharmacia, Freiburg, Germany).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| Strains | ||
| B. subtilis | ||
| 1A594 | trpC2 hemG321 | BGSCa |
| 168 | Wild type | BGSC |
| 3G18Δ2 | trpC2 met ade ΔhemY::ble | 9 |
| 1012 | leuA8 metB5 trpC2 hsrM1 | 35 |
| AM01 | 1012 with amyE::cat | 28 |
| AM7 | Promoterless bgaB inserted at amyE | 28 |
| AM10 | lepA::bgaB transcriptional fusion at amyE | 28 |
| ARB1 | HZ04 with resDE mutant from MH5081 | This work |
| ARB2 | HZ04 with fnr mutant from THB2 | This work |
| ARB3 | HZ04 with ywiD mutant from THB99 | This work |
| ARB4 | AM10 with resDE mutant from MH5081 | This work |
| ARB5 | AM10 with fnr mutant from THB2 | This work |
| ARB6 | AM10 with ywiD mutant from THB99 | This work |
| ARB7 | AM7 with resDE mutant from MH5081 | This work |
| ARB8 | AM7 with fnr mutant from THB2 | This work |
| ARB9 | AM7 with ywiD mutant from THB99 | This work |
| ARB10 | HZ03 with hemY mutation from ARB11, hemN hemZ hemY triple mutant | This work |
| ARB11 | 1012 carry hemY mutant introduced by using pMUTIN-hemY | This work |
| HZ01 | 1012 with 501-bp in-frame deletion in hemN, previously described as hemNΔ | 17 |
| HZ02 | 1012 with replacement of a part of hemZ by a cat cassette | This work |
| HZ03 | HZ01 with part of hemZ replaced by a cat cassette, hemN hemZ double mutant | This work |
| HZ04 | 1012 with hemZ::bgaB transcriptional fusion inserted at amyE | This work |
| JH642 | pheA1 trpC2 | BGSC |
| MH5081 | trpC2 pheA1 ΔresDE::tet | 39 |
| THB2 | trpC2 pheA1 fnr::spec | 14 |
| THB99 | trpC2 pheA1 ywiD::spec | 27 |
| E. coli | ||
| DH10B | Host in cloning experiments | Gibco BRL |
| DH5α | hsdR recA1 lacZYA f80 lacZDM15 gyrA96 | 36 |
| S. typhimurium TE3006 | env-53 hemN704::Mud-J(b) hemF707::Tn10d-Tet | 47 |
| Plasmids | ||
| pALF01 | pBluescript SK+II with 783-bp PCR product carrying the complete former yhaW and the 5′ end of former yhaV | This work |
| pBgaB | Promoter-test vector containing the bgaB gene | 28 |
| pBluehemZ | pBluescript SK+II with 1,706-bp PCR product carrying hemZ and its promoter | This work |
| pBlueSalISmaIΔ | pBluescript SK+II with deletion of the polylinker region between and including SalI and SmaI | 17 |
| pBluescript SK+II | Cloning vector | Stratagene |
| pBluescript KS+II | Cloning vector | Stratagene |
| pGH02 | 1,256-bp ClaI fragment in pBluescript KS+ containing lepA and its 5′ region | 17 |
| phemNΔ | PCR product of the mutated hemN gene from B. subtilis hemNΔ in pBluescript KS+II | This work |
| phemZ-PEX | pBluescript SK+ carrying 677-bp PCR product of former yhaW and its 5′ region | This work |
| phemZ1RPT3 | pBluescript SK+II carrying 500-bp PCR product of former yhaW | This work |
| phemZ2RPT3 | pBluescript SK+II carrying 877-bp PCR product of former yhaV | This work |
| phemZΔ01 | Fusion of a 149-bp PCR product carrying the 5′ region of former yhaV with a 153-bp fragment of the 3′ end of hemZ in pBluescript SK+II | This work |
| phemZΔ02 | 316-bp BamHI fragment from phemZΔ01 in pBlueSalISmaIΔ01 | This work |
| phemZ::cat | cat cassette integrated in EcoRV site of the phemZΔ02 insert | This work |
| pBluehemN | B. subtilis hemN in pBluescript SK+II | 13 |
| pBluehemZ::cat | PCR product of the tagged hemZ gene from B. subtilis hemZ::cat in pBluescript KS+II | This work |
| pHZ01 | 175-bp hemZ promoter fused to bgaB in pBgaB | This work |
| pMUTIN-hemY | pMUTIN4 with 324-bp fragment of hemY | This work |
| pMUTIN4 | Integration vector | 44 |
| pOKBShemN | 1,302-bp fragment containing the B. subtilis hemN gene in pOK12 | 13 |
| pSKCAT | cat cassette cloned into EcoRV site of pBluescript SK+II | This work |
BGSC, Bacillus Genetic Stock Center.
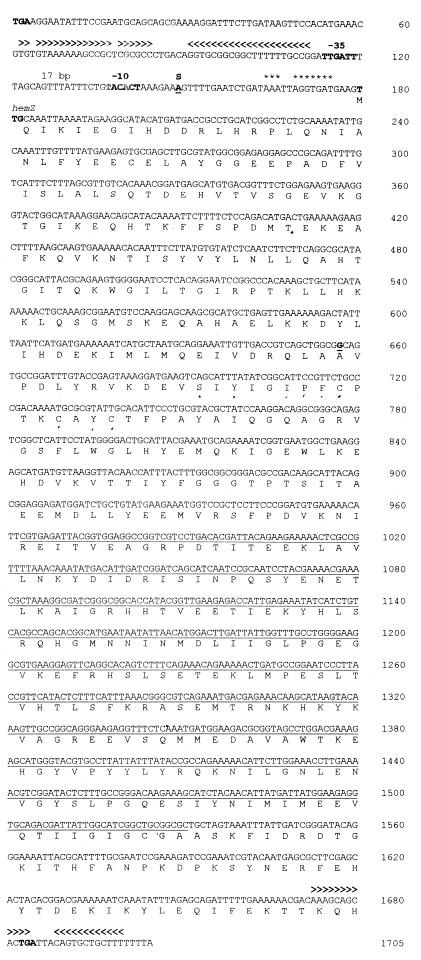
FIG. 1.
Nucleotide sequence of hemZ and deduced protein sequence. The putative translational start and stop codon of hemZ and the stop codon of the upstream-located yhaX are shown in boldface. The stem-loop structures upstream of hemZ and at its 3′ end are indicated by arrowheads above the sequence. The potential hemZ promoter is shown in boldface, and the −35 and −10 regions of the potential ςA promoter are indicated above the sequence. The mapped transcriptional start point (S) and the G residue at position 657, that is absent in the published genome sequence are underlined and in boldface. The hemZ region replaced by a cat cassette in various mutants is underlined.
RNA manipulations.
Preparation of total RNA of B. subtilis and Northern blot analysis were performed as described before (17). Hybridizations specific for hemZ were carried out with digoxigenin (DIG)-labeled RNAs synthesized in vitro with T3 RNA polymerase from EcoRI-linearized plasmids phemZ1RPT3 and phemZ2RPT3. In vitro RNA labeling was accomplished according to the manufacturer’s instructions (DIG-RNA-labeling kit; Boehringer, Mannheim, Germany). Primer extension experiments were carried out with the 32P-labeled primer hemZ-PEX (5′-CTGCGGGCTCCTCTCCGCCA-3′) as outlined before (46). DNA sequencing reactions utilizing the same primer and plasmid phemZ-PEX as template were performed, and the sequencing products were separated on the same gel.
Construction of reporter gene fusions.
The intercistronic region between yhaX and hemZ carrying the hemZ promoter (nucleotides 4 to 179 in Fig. 1) was amplified by PCR using primers HEMZPEX5 (see above) and HEMZTF3 (5′-GGCCATGAATTCCTTCATCACCTAATTTATCAGATT-3′). The hemZ-distal primer carried a BamHI restriction site at its 5′ end; the hemZ-proximal primer carried an EcoRI site. The PCR product was inserted in BamHI-EcoRI-digested pBgaB vector (28). The reporter gene bgaB encodes a thermostable β-galactosidase from Bacillus stearotheromophilus. In the resulting plasmid, pHZ01, the bgaB reporter gene is under control of the hemZ promoter region. B. subtilis AM01 carrying a cat cassette in amyE (28) was transformed with pHZ01 (28). Double-crossover integration of the fusion at amyE was selected by neomycin resistance and screened for the loss of chloramphenicol resistance of B. subtilis (named HZ04). Reporter gene fusions were tested under the indicated growth conditions as described earlier (28). The construction of the PlepA-bgaB transcriptional fusion (AM10) has been described before (28). Mutations in the regulatory loci resDE, fnr, and ywiD were introduced into HZ04 (hemZ::bgaB fusion), AM10 (hemN::bgaB fusion), and AM7 (bgaB) via transformation using genomic DNA prepared from MH5081 (resDE), THB2 (fnr), and THB99 (ywiD), respectively, as described previously (14, 27, 39).
Construction of a B. subtilis hemZ single mutant and a hemN hemZ double mutant.
A PCR product containing the 5′-terminal region of former yhaV was generated with primers YHAV5 (see above) and YHAV5EV (5′-GGCCATGATATCAATGTTTTTCACATCCGGGAAGGA-3′) (nucleotides 813 to 962 in Fig. 1). This fragment was designed to carry a promoter-proximal BamHI site and a promoter-distal EcoRV site. A second PCR product enclosing the 3′ end of hemZ (nucleotides 1532 to 1685 in Fig. 1), generated with primers YHAVEV (5′-GGCCATGATATCTGCTAGTAAATTTATTGATCGGGA-3′) and YHAV3 (see above) carried an EcoRV site at its promoter-proximal end and a BamHI site at its promoter-distal end. Both fragments were fused and ligated into the BamHI site of pBluescript SK+II. The resulting plasmid phemZΔ01 was digested with BamHI, and the 316-bp fragment obtained was subcloned into the BamHI site of pBlueSalISmaIΔ, a derivative of pBluescript SK+II in which the polylinker region between the SalI and SmaI site was deleted, to generate phemZΔ02 (17). With this strategy, phemZΔ02 carried only one EcoRV site, the one between the two hemZ fragments. A cat cassette was liberated from pSKCAT (cat cassette cloned into the EcoRV site of pBluescript SK+II) by using EcoRV and inserted into phemZΔ02 after linearization with EcoRV, generating phemZ::cat. In phemZ::cat, transcription of cat occurs in the same direction as the hemZ reading frame. phemZ::cat was linearized with ScaI and transformed into the chromosome of the B. subtilis hemN mutant HZ01 (16) and wild-type B. subtilis 1012. After selection on plates containing neomycin and 5 μg of hemin per ml, several transformants were obtained and checked by PCR for correct double-crossover integration of the cat cassette into the chromosomal copy of hemZ. The resulting mutant strains were designated B. subtilis HZ03 (hemN hemZ double mutant) and B. subtilis HZ02 (hemZ mutant). In both cases, the part of hemZ underlined in Fig. 1 is replaced by the cat cassette.
Construction of a hemY and a hemN hemZ hemY triple mutant.
An internal fragment of the B. subtilis hemY gene corresponding to positions 1088210 to 1088509 of the B. subtilis genome sequence (GenBank accession no. A1009126) was amplified by PCR using primers HEMY5 (5′-GGCCATGAATTCATTGACAAGCTCAGCCTGATGTCG-3′), carrying a BamHI restriction site at its 5′ end, and HEMY3 (5′-GGCCATGGATCCTGAATCAGCATCAAGTGTGACGCC-3′), carrying an EcoRI restriction site at its 5′ end. The 324-bp PCR product was digested with BamHI and EcoRI and inserted into BamHI-EcoRI-restricted pMUTIN4 (44). The resulting plasmid, pMUTIN-hemY, was used to transform B. subtilis 1012 and the hemN hemZ double mutant HZ03. Transformants were selected on plates containing erythromycin plus 5 μg of hemin per ml and checked via PCR as described above. The obtained hemY and the hemN hemZ hemY triple mutant were designated B. subtilis ARB11 and ARB10, respectively.
Complementation experiments.
To test for the hemN-like function, we cloned the B. subtilis hemZ gene into pBluescript SK+II and performed complementation experiments with a heme-auxotrophic S. typhimurium hemF hemN double mutant TE3006 (47). For this purpose, primers HEMZPEX5 (see above) and YHAV3 (see above), corresponding to positions 1056984 to 1057007 and 1058641 to 1058664 of the recently cloned B. subtilis genomic region containing hemZ (GenBank accession no. AL009126), were used in a PCR to generate a 1,706-bp fragment containing the complete 1,506-bp B. subtilis hemZ and 188 bp of its 5′ region including its promoter. The fragment containing BamHI restriction sites introduced by the primer sequences was cut with BamHI, and the resulting 1,692-bp fragment was cloned into pBluescript SK+II to generate pBluehemZ. S. typhimurium TE3006 lacking the intact genes for the oxygen-dependent coproporphyrinogen III oxidase (hemF) and a component of the oxygen-independent enzyme (hemN) was transformed via electroporation with the newly constructed pBluehemZ containing B. subtilis hemZ, pBluehemN containing B. subtilis hemN as positive control, and the cloned PCR products of the inactivated hemN gene from B. subtilis HZ01(phemNΔ) and the tagged B. subtilis hemZ from HZ02(phemZ::cat) as negative controls. Transformants were subsequently screened aerobically and anaerobically for the recovery of heme sufficiency by plating on Luria-Bertani medium supplemented with 100 μg of ampicillin per ml and 10 μg of tetracycline per ml but without further addition of hemin. For comparison of the complementation efficiency of B. subtilis hemN and hemZ, growth experiments with the indicated strains in liquid medium under aerobic and anaerobic conditions followed by optical density measurements were performed.
High-performance liquid chromatography analysis of porphyrins.
Porphyrins were extracted, modified, and separated as described before (13, 38).
RESULTS AND DISCUSSION
Identification and sequence analysis of a second hemN-like gene hemZ containing the previously proposed ORFs yhaV and yhaW.
Recently, we provided experimental evidence for the existence of a second HemN-type enzyme in B. subtilis (13). To identify the corresponding gene, we analyzed the completed and published DNA sequence of the B. subtilis genome with the program SubtiList and the BLAST algorithm, using the protein sequence deduced from B. subtilis hemN (16, 23, 29). One ORF encoding a protein with significant amino acid sequence identity to HemN on the protein level (26% identity and 51% similarity) was identified. This ORF, designated yhaV, encoded a putative protein of 290 amino acids with a calculated molecular mass of 33,556 Da. YhaV revealed a high degree of amino acid sequence similarity to numerous HemN proteins in the database, the strongest to HemN of B. subtilis. However, it was significantly smaller than other known HemN proteins. Initial inspection of the published yhaV sequence first led to the assumption that yhaV forms a bicistronic operon with the promoter-distal gene yhaW. The potential orf yhaW encoded a protein of 166 amino acids with a calculated molecular mass of 19,024 Da. This hypothetical protein showed no homology to any other protein in the SwissProt database. No obvious ribosome binding site was present upstream of the predicted yhaV coding sequence. Moreover, the 130-bp intercistronic sequence between yhaW and yhaV seemed to be unusually large. These observations suggested a sequencing error in the published genome sequence (23). To substantiate this hypothesis, we resequenced the whole yhaV locus of B. subtilis 1012, using four independently generated PCR products from this locus (phemZ1RPT3, phemZ2RP3, pALF01, and phemZ-PEX). The four products covered the complete region of the postulated bicistronic operon. Three of the cloned fragments contained the 3′ end of yhaW, and they all contained a G residue 25 bp upstream of the 3′ end of the postulated yhaW coding sequence (Fig. 1). This G was lacking in the published genome sequence (23). No additional sequencing errors were detected. Identical results were obtained for a PCR-based analysis of the same genomic region of B. subtilis 168 and B. subtilis JH642. After correction of the DNA sequence, the coding region of the former reading frames yhaW and yhaV were fused to one large ORF of 1,506 bp, which was designated hemZ (Fig. 1). The newly identified hemZ gene encodes a protein of 501 amino acids with a calculated molecular mass of 57,526 Da. The molecular mass was confirmed by in vivo expression experiments in E. coli using T7 RNA polymerase-driven transcription and radioactive protein labeling (data not shown). The protein deduced from hemZ showed significant homology to numerous HemN proteins in the database, the strongest to B. subtilis HemN (28% identity and 51% similarity) and to the Aquifex aeolicus HemN (28% identity and 50% similarity). However, alignment of the currently known approximately 30 HemN and HemZ proteins revealed the phylogenetically most distant position for B. subtilis HemZ in the HemN/HemZ protein family tree (data not shown). B. subtilis HemZ carries a unique approximately 100-amino-acid residue N-terminal extension, while 50 amino acid residues present in all other HemN/HemZ proteins are missing close to the C terminus. However, all highly conserved amino acid residues of HemN/HemZ proteins, like the potential iron-sulfur cluster signature sequence (CXXXCXXC) or the glycine-rich box (GGGIP), are present. A biochemical characterization of recombinant B. subtilis HemZ is under way.
The hemZ gene is preceded by a distinct ribosome binding site (AAAUUAGGUGAU) with a high degree of identity to the consensus ribosome binding sequence of B. subtilis (AAAGGAGGUGAU). Further upstream of hemZ, a potential vegetative ςA-dependent promoter (TTGATT-17 bp-TACACT) with sequence similarity to the consensus sequence of vegetative B. subtilis promoters (TTGACA-17 bp-TATAAT) was identified; 2 bp upstream of this potential promoter, a strong secondary structure (ΔG = −29 kcal/mol) can be predicted at the RNA level. The observed structure could serve as transcriptional terminator for the upstream located yhaX gene. Immediately downstream of hemZ and partially overlapping with the 3′ end of its coding sequence a second potential RNA secondary structure (ΔG = −18 kcal/mol) was located followed by a 7-U stretch. We assume that hemZ transcription is terminated here (Fig. 1).
B. subtilis hemZ complemented a Salmonella typhimurium hemF hemN double mutant under aerobic and anaerobic conditions.
S. typhimurium TE3006 (hemZ hemN double mutant) is unable to grow in the absence of heme both under aerobic and anaerobic conditions (47). To determine whether the B. subtilis hemZ gene is able to complement the mutant strain TE3006 for growth without added heme, it was transformed with plasmid pBluehemZ. Transformants were able to grow both under aerobic and anaerobic conditions, indicating that the HemZ protein is involved in the oxygen-independent coproporphyrinogen III decarboxylation. It had already been shown that the B. subtilis hemN gene was able to complement strain TE3006 in a similar way (13). In contrast, plasmids carrying inactivated hemZ (phemZ::cat) or hemN (phemNΔ) failed to complement TE3006 under these growth conditions. Comparison of the growth behavior of Salmonella mutants complemented with B. subtilis hemN and hemZ revealed significant differences. Strain TE3006 carrying either hemZ (doubling time [Td] = 2.5 h) or hemN (Td = 2 h) grew faster under anaerobic ammonifying growth conditions than under aerobic conditions. Under aerobic growth conditions, mutants complemented with hemN (Td = 3.2 h) grew significantly faster than their hemZ (Td = 11.0 h)-complemented counterparts. The amounts of hemN and hemZ mRNA isolated from the complemented Salmonella mutants and analyzed via RNA dot blot experiments were found to be approximately identical under all analyzed conditions (data not shown).
A B. subtilis hemZ mutant accumulates coproporphyrinogen III under anaerobic conditions.
To demonstrate the involvement of B. subtilis hemZ in the metabolism of coproporphyrinogen III, the cellular porphyrin profiles of wild-type B. subtilis, the hemZ mutant HZ02 (construction described in Materials and Methods), and the previously described hemN mutant HZ01 were compared. All three strains were grown aerobically and anaerobically in the presence of 10 mM nitrate. The porphyrins were extracted, modified, and separated via high-pressure liquid chromatography as described before (13, 38). B. subtilis wild-type cells did not accumulate any significant amounts of porphyrins. In agreement with our previous findings, porphyrins extracted from the hemN mutant HZ01 grown under anaerobic conditions exhibited a clear peak of coproporphyrin III. A coproporphyrin III accumulation of approximately 40 nmol/g (dry weight) was deduced. Similar to these results, the hemZ mutant HZ02 accumulated coproporphyrin III only under anaerobic conditions. However, only 25% of the coproporphyrin III amount (10 nmol/g [dry weight]) detected with the hemN mutant were observed. The amount of coproporphyrin III (55 nmol/g [dry weight]) accumulated by the hemN hemZ double mutant almost exactly corresponded to the sum of precursors detected in the two single mutants. These results demonstrated the involvement of the B. subtilis hemZ gene product in the oxygen-independent metabolism of coproporphyrinogen III.
B. subtilis hemZ and hemN hemZ double mutants have no obvious growth phenotype.
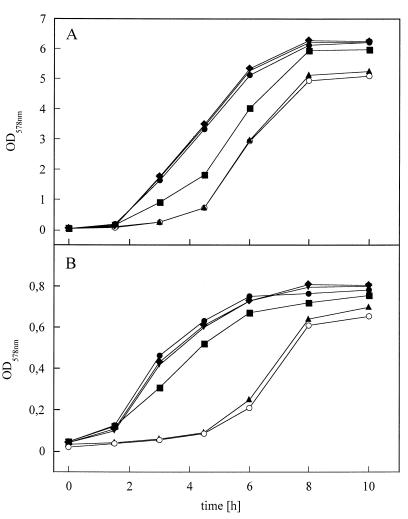
The consequences of hemZ inactivation for the growth of B. subtilis were studied. No obvious growth phenotype under aerobic and anaerobic nitrate respiratory or fermentative conditions was observed for the B. subtilis hemZ mutant (Fig. 2). To investigate whether hemN supplemented for hemZ inactivation, a B. subtilis hemN hemZ double mutant was constructed. For this purpose, the hemZ mutation was introduced into the hemN deletion strain B. subtilis HZ01 (16). B. subtilis HZ01 contains in the hemN gene an in-frame deletion reducing the size of the HemN protein from 363 to 167 amino acid residues. As described above, the knockout phenotype of this deletion was verified by the absence of complementation of the S. typhimurium hemN hemF double mutant with the cloned, partially deleted hemN gene (phemNΔ). The newly constructed hemN hemZ double mutant was designated B. subtilis HZ03. Surprisingly, mutation of both genes did not abolish aerobic and anaerobic respiratory growth. However, reduction of aerobic and anaerobic growth was observed (Fig. 2). These results point to the existence of an additional third oxygen-independent enzymatic system for the conversion of coproporphyrinogen III to protoporphyrinogen IX. One potential candidate, at least for aerobic heme formation, was the product of the hemY gene in combination with a yet unknown enzyme.
FIG. 2.
Growth behavior of B. subtilis 1012 (●), the hemN mutant HZ01 (⧫), the hemZ mutant HZ02 (▾), the hemY mutant ARB11 (▴), the hemN hemZ double mutant HZ03 (■), and the hemN hemZ hemY triple mutant ARB10 (○) in minimal medium under aerobic (A) and anaerobic nitrate ammonifying (B) conditions. OD, optical density.
Analysis of B. subtilis hemZ hemN hemY triple mutant.
Hansson et al. described the oxidation of coproporphyrinogen III to coproporphyrin III as a side activity of the oxygen-dependent protoporphyrinogen oxidase encoded by hemY (8). It was postulated that in combination with a new type of enzyme, a coproporphyrin III decarboxylase which converts coproporphyrin III into protoporphyrin IX, HemY could convert coproporphyrinogen III into protoporphyrin (8, 10). However, due to the oxygen dependence of HemY, this pathway would be limited to aerobic conditions. First, the effect of hemY mutation on aerobic and anaerobic growth was tested in the newly constructed hemY mutant ARB11. Surprisingly, reduced growth of the B. subtilis hemY mutant under aerobic and anaerobic conditions was observed (Fig. 2). To reconfirm this observation, the B. subtilis hemY deletion mutants 3G18Δ2 and 1A594 were subjected to similar growth experiments (9, 10). Almost identical growth behaviors to ARB11 were observed (data not shown). The differences of these observations from the previously detected clear detrimental effects of the hemY mutation are possibly caused by differences in the growth media used (9, 10). We concluded from our results that a still unknown oxygen-independent protoporphyrinogen IX oxidase partially compensated under aerobic and anaerobic conditions for hemY mutation. In S. typhimurium and P. aeruginosa, oxygen-independent HemN compensated for the loss of the oxygen-dependent HemF under aerobic and anaerobic growth conditions (34, 48). Due to the anaerobic respiratory energy metabolism of B. subtilis, an oxygen-independent protoporphyrinogen IX oxidase is required at least for anaerobic heme biosynthesis.
Moreover, the growth experiment indicated an unexpected anaerobic role for HemY. Similar observations were made for P. aeruginosa hemF, encoding the oxygen-dependent coproporphyrinogen III oxidase. Chromosomal deletion of hemF resulted in reduced aerobic and anaerobic growth of P. aeruginosa (34). In agreement with this observation, a strong anaerobic induction of hemF transcription was observed (34). The potential anaerobic roles for both oxygen-dependent enzymes remain to be elucidated.
To test for the hemY function in in vivo conversion of coproporphyrinogen III into protoporphyrin IX a hemN hemZ hemY triple mutant (ARB10) was constructed and tested for aerobic and anaerobic growth. The aerobic and anaerobic growth phenotype of this mutant was identical to the phenotype of the hemY mutant ARB10 (Fig. 2). These experiments indicated that hemY is not directly responsible for the viability of the hemN hemZ double mutant.
From our results, we concluded the existence of a third oxygen-independent enzymatic system for coproporphyrinogen III oxidation and an oxygen-independent system for protoporphyrinogen IX oxidation in B. subtilis.
Analysis of hemZ transcriptional unit.
The existence of two genes (hemN and hemZ) encoding two structurally highly related proteins with similar functions could provide the cell with a regulatory tool to differentially respond to variations in the cellular heme requirement as a consequence of changing environmental conditions. Regulated gene expression in response to changes in oxygen tension was previously observed for hemN and hemF from P. aeruginosa and for E. coli hemN (34, 42). Moreover, highly conserved oxygen tension regulator Fnr binding sites have been identified in the 5′ region of the A. eutrophus hemN and R. capsulatus hemZ (25, 49).
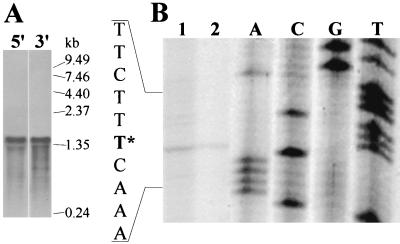
We started our transcriptional analysis of B. subtilis hemZ with the definition of the transcriptional unit by Northern blot hybridization. Computer analysis of the hemZ locus suggested a monocistronic transcriptional organization of hemZ (see above). Total RNA was isolated from exponentially growing B. subtilis 1012 cultivated under aerobic conditions. We used two different riboprobes in the Northern blot hybridizations, one with specificity to the 5′-terminal part of hemZ (the former yhaW) and the second complementary to the 3′-terminal part (the former yhaV). Northern blot analysis detected one transcript of around 1.6 kb with both probes, indicating a monocistronic transcriptional unit for hemZ (Fig. 3A). To determine the 5′ end of the hemZ transcript, we performed a primer extension analysis. Total RNA isolated from exponentially aerobically growing B. subtilis 1012 was hybridized to a 32P-labeled oligonucleotide primer (hemZ-PEX) complementary to the 5′ region of the hemZ mRNA and was extended by using avian myeloblastosis virus reverse transcriptase. The resulting autoradiograph revealed one single transcriptional start site which corresponded to an A on the RNA level located 31 bp upstream the translational start of hemZ (Fig. 3B). This putative transcriptional start site was within the appropriate distance (7 bases) from the postulated ςA-dependent promoter mentioned above (Fig. 1). No obvious differences in transcript length and transcriptional start site were detected with RNA prepared from anaerobically grown cells (data not shown).
FIG. 3.
(A) Characterization of hemZ transcriptional unit by Northern blot analyses. RNA was isolated from B. subtilis wild-type strain 1012 grown under aerobic conditions. After electrophoresis in 1.2% denaturing agarose gels and transfer to nylon membranes, hybridization was performed with riboprobes directed against the 5′ end (5′) and 3′ end (3′) of hemZ. We used 5 μg of RNA for the assay shown in lane 5′ and 10 μg of RNA for the assay shown in lane 3′. The positions of RNA size markers are indicated. (B) Mapping of the 5′ end of the hemZ mRNA. Primer extension analyses were carried out with the oligonucleotide hemZ-PEX and 20 μg (lane 1) or 5 μg (lane 2) of RNA prepared from exponentially growing B. subtilis 1012 cells under aerobic conditions. DNA sequencing reactions utilizing the same primer and plasmid phemZPEX were performed in parallel, and the reaction products were separated on the same gel (lanes A, C, G, and T). The mapped 5′ end of hemZ mRNA is denoted by an asterisk.
Regulation of hemZ and hemN expression by oxygen tension, nitrate, and H2O2.
After identification of the hemZ promoter region, we analyzed the regulation of B. subtilis hemZ and hemN expression by reporter gene fusions using the bgaB system (28). For this purpose, the promoter regions of hemZ and hemN were amplified by PCR and cloned into plasmid pBgaB immediately upstream of the promoterless bgaB gene (28). Since hemN forms an operon with the upstream-located lepA, the promoter region of lepA was fused to bgaB (13, 16). The bgaB gene encodes a thermostable β-galactosidase of B. stearotheromophilus. Using this construct, the transcriptional fusions PhemZ-bgaB and PhemN-bgaB were integrated into the chromosome at the amyE locus by double crossover to generate B. subtilis HZ04 and AM10. Both strains were grown under the indicated conditions into mid-exponential growth phase, and β-galactosidase activity was assayed as described before (28). As a control, B. subtilis AM07 carrying the bgaB gene without any promoter in amyE was analyzed in parallel. The results of the various BgaB assays are summarized in Table 2. A clear fivefold induction of hemZ and hemN transcription was observed under anaerobic growth conditions in the presence of the alternative electron acceptors nitrate and nitrite compared to aerobic growth conditions. Fermentative growth conditions failed to induce hemZ and hemN transcription, in agreement with the greatly reduced heme requirement for nonrespiratory fermentative growth. The observed anaerobic induction was visible only during growth on well-defined minimal medium. In our previous investigation of hemN expression via RNA slot blot experiments using RNA prepared from B. subtilis grown aerobically and anaerobically on complex medium, we failed to detect significant redox regulation (13). Similar results were obtained in this study for the hemZ-bgaB and hemN-bgaB fusion strains grown aerobically and anaerobically on complex medium (data not shown). The exact nature of the inhibitory compound contained in the complex medium remains to be elucidated.
TABLE 2.
Regulation of B. subtilis hemN and hemZ
| Strain | β-Galactosidase activity (U/mg of protein)a
|
|||
|---|---|---|---|---|
| +O2 | +H2O2b | −O2 | −O2 + 10 mM nitrate | |
| HZ01(hemZ::bgaB) | 9 | 52 | 4 | 47 |
| ARB1(hemZ::bgaB resDE) | 10 | NT | 3 | 10 |
| ARB2(hemZ::bgaB fnr) | 9 | NT | 2 | 4 |
| ARB3(hemZ::bgaB ywiD) | 14 | NT | 1 | 2 |
| AM10(hemN::bgaB) | 7 | 6 | 4 | 34 |
| ARB4(hemN::bgaB resDE) | 8 | NT | 4 | 8 |
| ARB5(hemN::bgaB fnr) | 9 | NT | 2 | 5 |
| ARB6(hemN::bgaB ywiD) | 9 | NT | 2 | 3 |
| AM7(bgaB) | 2 | 2 | 2 | 3 |
| ARB7(bgaB resDE) | 2 | NT | 2 | 2 |
| ARB8(bgaB fnr) | 3 | NT | 2 | 2 |
| ARB9(bgaB ywiD) | 2 | NT | 2 | 3 |
Strains were grown aerobically or anaerobically as described in Materials and Methods. All cell extracts were prepared and assayed for β-galactosidase activity as described before (28). The results represent averages of three different experiments performed in triplicate. A standard deviation of approximately 1.5 U/mg of protein was observed. NT, not tested.
Growing cultures were treated for 15 min with 0.1 mM H2O2 before assaying for reporter gene expression.
A second regulatory factor, peroxide stress, was specific to hemZ expression. An approximately fivefold induction of the hemZ-bgaB fusion was observed in the presence of H2O2, while the expression of the hemN-bgaB fusion remained unchanged. The H2O2-neutralizing enzyme catalase possesses a heme cofactor. The catalase gene katA is also under the control of H2O2 (1, 7). A catalase-overproducing B. subtilis mutant was found to overproduce tetrapyrroles, too (7). A consensus sequence called the per box was detected upstream the promoter of katA and a second H2O2-regulated gene, mrgA (2). The same per box was present at two positions upstream of the hemAXCDBL operon, encoding the enzyme for initial steps of heme biosynthesis in B. subtilis, indicating peroxide stress regulation for this operon (2). However, no obvious DNA sequence matching the per box was found upstream of hemZ. Other examples of H2O2-regulated genes without a per box are known (7, 12, 45). The alternative sigma factor ςB is responsible for the general stress response of B. subtilis, including the expression of the second catalase gene, katE, induced by oxidative stress (12). However, no obvious promoter sequence for ςB was found in the 5′ region of hemZ (Fig. 1).
Anaerobic induction of hemN and hemZ is dependent on resDE, fnr, and ywiD.
To investigate the molecular basis for the observed transcriptional activation of hemN and hemZ, we analyzed expression of the reporter gene constructs in mutants of the previously identified redox regulatory loci resDE, fnr, and ywiD (Table 2). While aerobic expression of hemN and hemZ remained mainly unaffected by the resDE, fnr, and ywiD mutants, anaerobic expression was found to be drastically reduced, indicating the importance of all three loci for anaerobic hemN and hemZ induction. Mutation in ywiD resulted in the most severe reduction of hemN and hemZ expression compared to the fnr and resDE mutation (Table 2). The observed low expression under anaerobic fermentative conditions remained unchanged. Previously, the requirement of resDE for efficient fnr expression was established (32). Furthermore, Fnr mediates the anaerobic induction of ywiD via a highly conserved consensus binding site (Fnr box) (27). Finally, YwiD activates anaerobic transcription of genes encoding nitrite reductase (nasDE), a potential flavohemoglobin (hmp), lactate dehydrogenase (lctE), and enzymes of acetoin fermentation (alsSD), all involved in anaerobic metabolism (27). A similar redox regulatory cascade is proposed for the regulation of hemN and hemZ. However, from our results a direct involvement of resDE in anaerobic hemN and hemZ cannot be excluded. A direct activation mediated by fnr is very unlikely since appropriate Fnr boxes are missing in the hemN and hemZ promoter regions.
Since heme requirements drastically vary between aerobic respiratory, anaerobic respiratory, and anaerobic fermentative conditions, heme biosynthesis is obviously coregulated via hemN and hemZ expression (33). The known regulators employed by B. subtilis for its general redox adaptation encoded by resDE, fnr, and ywiD are also used for the coordination of heme biosynthesis with the anaerobic energy metabolism. Moreover, the resDE genes are also involved in the regulation of qcrABC (encoding subunits of the cytochrome bc complex), ctaA (required for heme A synthesis), and genes important for cytochrome c biogenesis, loci all related to tetrapyrrole-associated processes (39).
ACKNOWLEDGMENTS
This work was supported by grants of the Deutsche Forschungsgemeinschaft and the Fonds der Chemischen Industrie to D.J. and W.S., the Max-Planck-Gesellschaft, the Sonderforschungsbereich 388, and the Graduiertenkolleg “Biochemie der Enzyme” of the Albert-Ludwigs-Universität Freiburg to D.J. We are indebted to T. Elliott (West Virginia University, Morgantown) for the gift of several S. typhimurium strains and to M. Nakano (Louisiana State University, Shreveport), H. Cruz Ramos, and P. Glaser (Pasteur Institute, Paris, France) for the gift of B. subtilis strains. We thank T. Hoffmann (Max-Planck-Institut, Marburg, Germany) for helpful discussions and Antonio Espin for technical assistance. We thank R. K. Thauer (Max-Planck-Institut) for helpful discussions and continuous support. We are indebted to L. Hederstedt and M. Hansson (University of Lund, Lund, Sweden) for the gift of B. subtilis strains and helpful discussions and suggestions.
REFERENCES
- 1.Bol D K, Yasbin R E. Analysis of the dual regulatory mechanisms controlling expression of the vegetative catalase gene of Bacillus subtilis. J Bacteriol. 1994;176:6744–6748. doi: 10.1128/jb.176.21.6744-6748.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L, Keramati L, Helmann J D. Coordinate regulation of Bacillus subtilis peroxide stress genes by hydrogen peroxide and metal ions. Proc Natl Acad Sci USA. 1995;92:8190–8194. doi: 10.1073/pnas.92.18.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ciba Foundation Symposia Staff. The biosynthesis of the tetrapyrrole pigments. Ciba Foundation Symposia 180. New York, N.Y: John Wiley & Sons; 1994. [Google Scholar]
- 4.Coomber S A, Jones R M, Jordan P M, Hunter C N. A putative anaerobic coproporphyrinogen III oxidase in Rhodobacter sphaeroides. I. Molecular cloning, transposon mutagenesis and sequence analysis of the gene. Mol Microbiol. 1992;6:3155–3169. doi: 10.1111/j.1365-2958.1992.tb01772.x. [DOI] [PubMed] [Google Scholar]
- 5.Cruz Ramos H, Boursier L, Moszer I, Kunst F, Danchin A, Glaser P. Anaerobic transcription activation in Bacillus subtilis: identification of distinct FNR-dependent and -independent regulatory mechanisms. EMBO J. 1995;14:5984–5994. doi: 10.1002/j.1460-2075.1995.tb00287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doss M O, Philipp-Dornston W K. Porphyrin and heme biosynthesis from endogenous and exogenous δ-aminolevulinic acid in Escherichia coli, Pseudomonas aeruginosa, and Achromobacter metalcaligenes. Hoppe-Seyler’s Z Physiol Chem. 1971;352:725–733. doi: 10.1515/bchm2.1971.352.1.725. [DOI] [PubMed] [Google Scholar]
- 7.Dowds B C A. The oxidative stress response in Bacillus subtilis. FEMS Microbiol Lett. 1994;124:255–264. doi: 10.1111/j.1574-6968.1994.tb07294.x. [DOI] [PubMed] [Google Scholar]
- 8.Hansson M, Gustafsson M C, Kannangara C G, Hederstedt L. Isolated Bacillus subtilis HemY has coproporphyrinogen III to coproporphyrin III oxidase activity. Biochim Biophys Acta. 1997;20:97–104. doi: 10.1016/s0167-4838(97)00030-7. [DOI] [PubMed] [Google Scholar]
- 9.Hansson M, Hederstedt L. Cloning and characterization of the Bacillus subtilis hemEHY gene cluster, which encodes protoheme IX biosynthetic enzymes. J Bacteriol. 1992;174:8081–8093. doi: 10.1128/jb.174.24.8081-8093.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansson M, Hederstedt L. Bacillus subtilis HemY is a peripheral membrane protein essential for protoheme IX synthesis which can oxidize coproporphyrinogen III and protoporphyrinogen IX. J Bacteriol. 1994;176:5962–5970. doi: 10.1128/jb.176.19.5962-5970.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hansson M, Rutberg L, Schröder I, Hederstedt L. The Bacillus subtilis hemAXCDBL gene cluster, which encodes enzymes of the biosynthetic pathway from glutamate to uroporphyrinogen III. J Bacteriol. 1991;173:2590–2599. doi: 10.1128/jb.173.8.2590-2599.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hecker M, Völker U. Non-specific, general and multiple stress resistance of growth-restricted Bacillus subtilis cells by the expression of the δB regulon. Mol Microbiol. 1998;29:1129–1136. doi: 10.1046/j.1365-2958.1998.00977.x. [DOI] [PubMed] [Google Scholar]
- 13.Hippler B, Homuth G, Hoffmann T, Schumann W, Jahn D. Characterization of Bacillus subtilis hemN. J Bacteriol. 1997;179:7181–7185. doi: 10.1128/jb.179.22.7181-7185.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann T, Frankenberg N, Marino M, Jahn D. Ammonification in Bacillus subtilis utilizing dissimilatory nitrite reductase is dependent on resDE. J Bacteriol. 1998;180:186–189. doi: 10.1128/jb.180.1.186-189.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann T, Troup B, Szabo A, Hungerer C, Jahn D. The anaerobic life of Bacillus subtilis. Cloning and characterization of the genes encoding the respiratory nitrate reductase system. FEMS Microbiol Lett. 1995;131:219–225. doi: 10.1111/j.1574-6968.1995.tb07780.x. [DOI] [PubMed] [Google Scholar]
- 16.Homuth G, Heinemann M, Zuber U, Schumann W. The genes lepA and hemN form a bicistronic operon in B. subtilis. Microbiology. 1996;142:1641–1649. doi: 10.1099/13500872-142-7-1641. [DOI] [PubMed] [Google Scholar]
- 17.Homuth G, Masuda S, Mogk A, Kobayashi Y, Schumann W. The dnaK operon of Bacillus subtilis is heptacistronic. J Bacteriol. 1997;179:1153–1164. doi: 10.1128/jb.179.4.1153-1164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs N J, Jacobs J M. Nitrate, fumarate, and oxygen as electron acceptors for a late step in microbial heme synthesis. Biochim Biophys Acta. 1976;13:1–9. doi: 10.1016/0005-2728(76)90002-5. [DOI] [PubMed] [Google Scholar]
- 19.Jacobs N J, Jacobs J M, Mills B A. Role of oxygen in the late steps of heme synthesis in Pseudomonads and Escherichia coli. Enzyme. 1973;16:50–56. doi: 10.1159/000459361. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs N J, Jacobs J M, Morgan J R. Comparative effect of oxygen and nitrate on protoporphyrin and heme synthesis from d-aminolevulinic acid in bacterial cultures. J Bacteriol. 1972;112:1444–1445. doi: 10.1128/jb.112.3.1444-1445.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahn D, Verkamp E, Söll D. Glutamyl-transfer RNA: a precursor of heme and chlorophyll biosynthesis. Trends Biochem Sci. 1992;17:215–218. doi: 10.1016/0968-0004(92)90380-r. [DOI] [PubMed] [Google Scholar]
- 22.Jordan P M. Biosynthesis of tetrapyrroles. In: Neuberger A, van Deenen L L M, editors. New comprehensive biochemistry. Vol. 19. Amsterdam, The Netherlands: Elsevier; 1991. [Google Scholar]
- 23.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, Bolotin A, Borchert S, Borriss R, Boursier L, Brans A, Braun M, Briginell S C, Bron S, Brouillet S, Bruschi C V, Caldwell B, Capuano V, Carter N M, Choi S K, Codani J J, Connerton I F, Cummings N J, Danie R A, Denizot F, Devine K M, Dusterhoft A, Ehrlich S D, Emmerson P T, Entian K D, Errington J, Fabret C, Ferrari E, Foulger D, Fritz C, Fujita M, Fujita Y, Fuma S, Galizzi A, Galleron N, Ghim S Y, Glaser P, Goffeau A, Golightly E J, Grandi G, Guiseppi G, Guy B J, Haga K, Haiech J, Harwood C R, Hasahara Y, Henaut A, Hilbert H, Holsappel S, Hoson S, Hullo M F, Itaya M, Jones L, Joris B, Karamata D, Klaerr-Blanchard M, Klein C, Kobayashi Y, Koetter P, Koningstein G, Krogh S, Kumano M, Kurita K, Lapidus A, Lardinois S, Lauer J, Lazarevic V, Lee S M, Levine A, Liu H, Masuda S, Mauel C, Medigue C, Medina N, Medallo R P, Mizuno M, Moestl D, Nakai S, Noback M, Noone D, O’Reilly M, Ogawa K, Ogiwara A, Oudega B, Park S H, Parro V, Pohl T M, Portetelle D, Porwollik S, Prescott A M, Presecan E, Pujic P, Purnelle B, Rapoport G, Rey M, Reynolds S, Rieger M, Rivolta C, Rocha E, Roche B, Rose M, Sadaie Y, Sato T, Scanlan E, Schleich S, Schroeter R, Scoffone F, Sekiguchi J, Sekowska A, Seror S J, Serror P, Shin B S, Soldo B, Sorokin A, Tacconi E, Takagi T, Takahashi H, Takemaru K, Takeuchi M, Tamakoshi A, Tanaka T, Terpstra P, Tognini A, Tosato V, Uchiyama S, Vandenbol M, Vannier F, Vassarotti A, Viari A, Wambutt R, Wedler E, Wedler H, Weitzenegger T, Winters P, Wipat A, Yamamoto H, Yamane K, Yasumoto K, Yata K, Yoshida K, Yoshikawa H, Yoshikawa H F, Zumstein E, Danchin A. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1998;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 24.Leeper F. Intermediate steps in the biosynthesis of chlorophylls. In: Scheer H, editor. Chlorophylls. London, England: CRC Press; 1991. pp. 407–432. [Google Scholar]
- 25.Lieb C, Siddiqui R A, Hippler B, Jahn D, Friedrich B. The Alcaligenes eutrophus hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase is required for heme biosynthesis during anaerobic growth. Arch Microbiol. 1998;169:52–60. doi: 10.1007/s002030050540. [DOI] [PubMed] [Google Scholar]
- 26.Mach H, Hecker M, Mach F. Physiological studies on cAMP synthesis in Bacillus subtilis. FEMS Microbiol Lett. 1988;52:189–192. [Google Scholar]
- 27.Marino, M., H. Cruz Ramos, T. Hoffmann, P. Glaser, and D. Jahn. 1999. Unpublished results.
- 28.Mogk A, Hayward R, Schumann W. Integrative vectors for constructing single-copy transcriptional fusions between Bacillus subtilis promoters and various reporter genes encoding heat-stable enzymes. Gene. 1996;182:33–36. doi: 10.1016/s0378-1119(96)00447-7. [DOI] [PubMed] [Google Scholar]
- 29.Moszer I, Glaser P, Danchin A. SubtiList: a relational database for the Bacillus subtilis genome. Microbiology. 1995;141:261–268. doi: 10.1099/13500872-141-2-261. [DOI] [PubMed] [Google Scholar]
- 30.Nakano M M, Hoffmann T, Zhu Y, Jahn D. Nitrogen and oxygen regulation of Bacillus subtilis nasDEF encoding NADH-dependent nitrite reductase by TnrA and ResDE. J Bacteriol. 1998;180:5344–5350. doi: 10.1128/jb.180.20.5344-5350.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nakano M M, Zuber P. Anaerobic growth of a “strict aerobe” (Bacillus subtilis) Annu Rev Microbiol. 1998;52:165–190. doi: 10.1146/annurev.micro.52.1.165. [DOI] [PubMed] [Google Scholar]
- 32.Nakano M M, Zuber P, Glaser P, Danchin A, Hulett F M. Two-component regulatory proteins ResD-ResE are required for transcriptional activation of fnr upon oxygen limitation in Bacillus subtilis. J Bacteriol. 1996;178:3796–3802. doi: 10.1128/jb.178.13.3796-3802.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Philipp-Dornston W K, Doss M O. Comparison of porphyrin and heme in various heterotrophic bacteria. Enzyme. 1973;16:57–64. doi: 10.1159/000459362. [DOI] [PubMed] [Google Scholar]
- 34.Rompf A, Hungerer C, Hoffmann T, Lindenmeyer M, Römling U, Groß U, Doss M O, Arai H, Igarashi Y, Jahn D. Regulation of Pseudomonas aeruginosa hemN and hemF by the dual action of the redox response regulators Anr and Dnr. Mol Microbiol. 1998;29:985–997. doi: 10.1046/j.1365-2958.1998.00980.x. [DOI] [PubMed] [Google Scholar]
- 35.Saito H, Shibata T, Ando T. Mapping of genes determining nonpermissiveness and host-specific restriction to bacteriophages in Bacillus subtilis Marburg. Mol Gen Genet. 1979;170:117–122. doi: 10.1007/BF00337785. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Seehra J S, Jordan P M, Akhtar M. Anaerobic and aerobic copro-porphyrinogen III oxidases of Rhodopseudomonas spheroides. Biochem J. 1982;269:709–718. doi: 10.1042/bj2090709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straka J G. High-performance liquid chromatography of porphyrin methylesters. Methods Enzymol. 1986;123:352–363. doi: 10.1016/s0076-6879(86)23042-6. [DOI] [PubMed] [Google Scholar]
- 39.Sun G, Sharkova E, Chesnut R, Birkey S, Duggan M F, Sorokin A, Pujic P, Ehrlich S D, Hulett F M. Regulators of aerobic and anaerobic respiration in Bacillus subtilis. J Bacteriol. 1996;178:1374–1385. doi: 10.1128/jb.178.5.1374-1385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tait G H. Coproporphyrinogenase activity in extracts from Rhodopseudomonas spheroides. Biochem Biophys Res Commun. 1969;37:116–122. doi: 10.1016/0006-291x(69)90888-2. [DOI] [PubMed] [Google Scholar]
- 41.Tait G H. Coproporphyrinogenase activities in extracts of Rhodopseudomonas spheroides and Chromatium strain D. Biochem J. 1972;128:1159–1169. doi: 10.1042/bj1281159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Troup B, Hungerer C, Jahn D. Cloning and characterization of the Escherichia coli hemN gene encoding the oxygen-independent coproporphyrinogen III oxidase. J Bacteriol. 1995;177:3326–3331. doi: 10.1128/jb.177.11.3326-3331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Troup B, Jahn M, Hungerer C, Jahn D. Isolation of the hemF operon containing the gene for the Escherichia coli aerobic coproporphyrinogen III oxidase by in vivo complementation of a yeast HEM13 mutant. J Bacteriol. 1994;176:673–680. doi: 10.1128/jb.176.3.673-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 45.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 46.Wetzstein M, Völker U, Dedio S, Löbau U, Zuber U, Schiesswohl M, Herget C, Hecker M, Schumann W. Cloning, sequencing, and molecular analysis of the dnaK locus from Bacillus subtilis. J Bacteriol. 1992;174:3300–3310. doi: 10.1128/jb.174.10.3300-3310.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu K, Delling J, Elliott T. The genes required for heme synthesis in Salmonella typhimurium include those encoding alternative functions for aerobic and anaerobic coproporphyrinogen oxidation. J Bacteriol. 1992;174:3953–3963. doi: 10.1128/jb.174.12.3953-3963.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu K, Elliott T. Cloning, DNA sequence, and complementation analysis of the Salmonella typhimurium hemN gene encoding a putative oxygen-independent coproporphyrinogen III oxidase. J Bacteriol. 1994;176:3196–3203. doi: 10.1128/jb.176.11.3196-3203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zeilstra-Ryalls J H, Kaplan S. Regulation of the 5-aminolevulinic acid synthesis in Rhodobacter sphaeroides 2.4.1: genetic basis of mutant H-5 auxotrophy. J Bacteriol. 1995;177:2760–2768. doi: 10.1128/jb.177.10.2760-2768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]