Abstract
Background and aims
Amplification of S100A8 occurs in 10–30% of all breast cancers and has been linked to poorer prognosis. Similarly, the protein S100A8 is overexpressed in a roughly comparable proportion of breast cancers and is also found in infiltrating myeloid-lineage cells, again linked to poorer prognosis. We explore the relationship between these findings.
Methods
We examined S100A8 copy number (CN) alterations using fluorescence in situ hybridization in 475 primary breast cancers and 117 corresponding lymph nodes. In addition, we studied S100A8 protein expression using immunohistochemistry in 498 primary breast cancers from the same cohort.
Results
We found increased S100A8 CN (≥ 4) in tumor epithelial cells in 20% of the tumors, increased S100A8 protein expression in 15%, and ≥ 10 infiltrating S100A8 + polymorphonuclear cells in 19%. Both increased S100A8 CN and protein expression in cancer cells were associated with high Ki67 status, high mitotic count and high histopathological grade. We observed no association between increased S100A8 CN and S100A8 protein expression, and only a weak association (p = 0.09) between increased CN and number of infiltrating S100A8 + immune cells. Only S100A8 protein expression in cancer cells was associated with significantly worse prognosis.
Conclusions
Amplification of S100A8 does not appear to be associated with S100A8 protein expression in breast cancer. S100A8 protein expression in tumor epithelial cells identifies a subgroup of predominantly non-luminal tumors with a high mean age at diagnosis and significantly worse prognosis. Finally, S100A8 alone is not a sufficient marker to identify infiltrating immune cells linked to worse prognosis.
Keywords: S100A8, Breast cancer, Copy number, Amplification, MDSC, Protein expression
Introduction
Despite significant advances in our understanding of breast cancer (BC) and its treatment, there is still an urgent need for new prognostic and predictive markers to reduce overall mortality due to the disease and morbidity associated with its treatment.
S100A8 and S100A9, in addition to their multitudinous other functions [1], are two of the most abundant alarmins in the human body and can be found in significantly increased quantities across almost all inflammatory conditions [2–4]. Increasingly, they are also recognized for their role in the initiation and progression of cancer [5]. In BC, the gene S100A8 is found to be amplified in about 10–30% of patients [6–8], and amplification of its chromosomal neighborhood, 1q21.3, has been linked to poorer prognosis in BC patients [8]. Similarly, the protein S100A8 is overexpressed in many common cancers, particularly in BC [9, 10]. Overexpression of S100A8 in cancer cells has been studied in several different populations of BC patients and has consistently been found to be associated with poor clinicopathological features and reduced survival [11–14].
In addition to the pro-tumor effects of S100A8 when expressed in cancer cells which, in BC, so far have mostly been linked to transformation, invasion and migration [15–17], it is thought to contribute further to cancer progression through its association with a loosely defined class of immune-suppressive myeloid cells. These are commonly referred to as Myeloid Derived Suppressor Cells (MDSCs) and can be found in elevated levels in most forms of cancer [18, 19]. S100A8, along with its partner S100A9, has been found to induce the accumulation and differentiation of these cells [20–23]. This has specifically been shown in mouse models of BC, where they also appear to have a role in preparing pre-metastatic niches [21, 24]. Both Drews-Elger et al. and, more recently, Woo et al. have independently reported finding infiltrating S100A8 + immune cells in human BCs, which are associated with reduced survival [13, 25].
While both amplification of S100A8 and S100A8 expression in BC cells have been studied separately, the relationship between the two remains to be elucidated. Furthermore, the relationship between infiltrating S100A8 + immune cells and increased S100A8 copy number (CN) or S100A8 expression in tumor cells has not been described in detail. Finally, there is little data on the histopathological features of tumors with increased S100A8 CN.
To address these points, we used fluorescence in situ hybridization (FISH) to study S100A8 CN in formalin-fixed, paraffin-embedded tissue (FFPE) from primary BCs and their corresponding axillary lymph node metastases. We also performed immunohistochemistry (IHC), staining for S100A8 in the same primary BCs.
Materials and methods
Study population
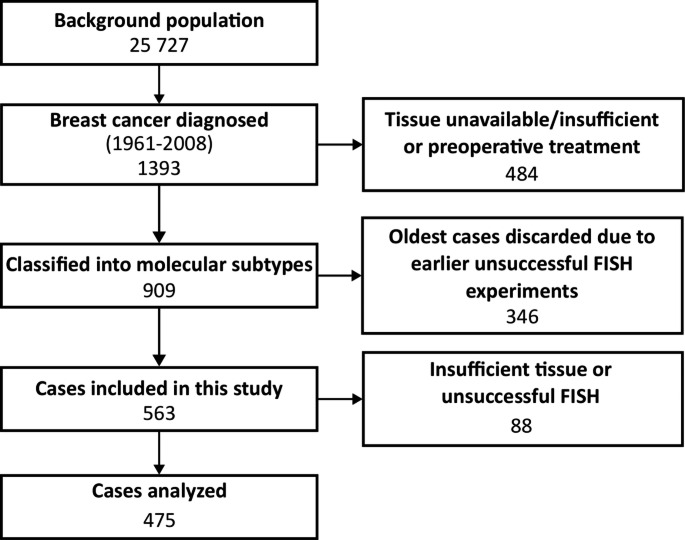
Between 1956 and 1959, a cohort of 25 727 women from Nord-Trøndelag, Norway, were invited to participate in a population-based program for the early detection of BC [26]. Using information from the Cancer Registry of Norway and the Norwegian Cause of Death Registry, a total of 1396 BC cases were identified in this cohort during the follow-up period from 1961 to 2008 [Fig. 1]. 909 of these were again reclassified into molecular subtypes [27] and followed from time of diagnosis until either death from BC, death from other causes, or the 31st of December 2015.
Fig. 1.
Overview of the study population. Inclusion and exclusion criteria
Specimen characteristics
Full-face sections from the primary BCs had previously been reclassified into histopathological type and grade in accordance with established guidelines [28, 29]. After that, tissue microarray (TMA) blocks were constructed by extracting three 1 mm tissue cores from the periphery of the FFPE primary BCs and axillary lymph node metastases using a Tissue Arrayer MiniCore with TMA designer2 software (Alphelys). These blocks were then cut into 4 μm thick sections and kept at -20 °C until use. Using IHC and chromogenic in-situ hybridization applied to TMAs in place of gene expression analysis, all cases were then classified into molecular subtypes as previously described (Table 1) [27].
Table 1.
Classification and distribution of molecular subtypes
| ER + and/or PR+ | HER2 | Ki67 | CK5 + and/or EGFR+ | Number included | ||||
|---|---|---|---|---|---|---|---|---|
| Luminal A | + | - | < 15% | 268/498 | ||||
| Luminal B (HER2−) | + | - | ≥ 15% | 116/498 | ||||
| Luminal B (HER2+) | + | + | 36/498 | |||||
| HER2 type | - | + | 28/498 | |||||
| 5NP | - | - | - | 12/498 | ||||
| BP | - | - | + | 35/498 | ||||
ER estrogen receptor, PR progesterone receptor, HER2 human epidermal growth factor receptor 2, 5NP 5 negative phenotype, BP basal phenotype, CK5 cytokeratin 5, EGFR epidermal growth factor receptor 1
Only BCs of more recent date, from the 1980s onwards, were included in this study, leaving 563 BCs available for analysis. Of these 563 cases, 169 had axillary lymph node metastases at the time of diagnosis and from which tissue was available.
Fluorescence in situ hybridization
Following manufacturer instructions, FISH was carried out using a DAKO Histology FISH accessory Kit (Code K 5799). TMA slides were first heated at 60 °C for 1–2 h before dewaxing and rehydrating. Subsequently, they were boiled for 15 min in a pre-treatment solution (Dako) before cooling to room temperature and washing in Dako buffer (2 × 3 min). The slides were then immersed in pepsin solution (37 °C, 30 min) for protein digestion, washed in Dako buffer (2 × 3 min), dehydrated (2 min each, 70%, 85%, and 96% ethanol), and allowed to air-dry. Then, 3 µL of a custom S100A8 FISH-probe (Empire Genomics) was mixed with 12 µL of hybridization buffer (Empire Genomics) and applied to the slides, which were coverslipped and sealed with coverslip sealant (Dako). They were then placed in a Dako Hybridizer and denatured (83 °C, 3 min) before they were renatured at 37 °C overnight. After this, the slides were rinsed in Stringent Wash Buffer (Dako) (72 °C, 2 min) and Dako wash buffer (2 × 3 min) at room temperature. Next, they were dehydrated (2 min each, 70%, 85%, and 96% ethanol) and dried at 37 °C for 15 min. Finally, 20 µL DAPI (VYSIS Abbott no 06J50-001) was applied to the slides, which were again coverslipped.
Immunohistochemistry
Immunohistochemistry was performed on 498 of the original 563 primary BCs using a Dako EnVision + Dual Link System-HRP (DAB+) (Cat #4065) following the manufacturer’s protocol. A total of 65 BCs were excluded due to insufficient tissue. TMA slides were first deparaffinized and rehydrated. Epitope retrieval was achieved by boiling the slides for 15 min in citrate buffer pH 6 (Sigma, Cat#C9999) and letting them air-dry before rehydrating and washing them in PBS. The slides were then covered with Dual Endogen Enzyme block (Dako) for 5–10 min at room temperature and washed in PBS (2 × 5 min). After this, anti-human monoclonal mouse IgG1 S100A8 antibody (R&D systems, Cat #MAB4570, 1:8000) was applied, and the slides were incubated at 4 °C overnight. The next morning the slides were washed in PBS (3 × 5 min) before the secondary antibody and enzyme were added (Dako Labelled polymer-HRP, 30 min at room temperature) followed by another wash in PBS (3 × 5 min). Subsequently, the slides were covered with a mixture of 20 µL DAB chromogen (Dako) and 1 mL Substrate buffer (Dako) for 5–10 min at room temperature, rinsed with water and contrast stained with hematoxylin. Finally, the slides were dehydrated (2 min each, 70%, 85% and 96% ethanol), cleared using xylene, and coverslips were mounted.
Scoring and reporting
S100A8 FISH
All samples were analyzed using a Nikon Eclipse 90i fluorescence microscope. The number of fluorescent signals within 20 well-preserved, non-overlapping cancer cell nuclei from a single TMA core was counted for each case. Where multiple tissue cores were available, the one with the best signal quality was chosen. In cases with no noticeable difference in signal quality, the core with the highest apparent count number was selected. Where that again failed to distinguish the cores, one was chosen arbitrarily. Similarly, where cancer cell populations within a single core were heterogeneous with respect to CN, nuclei with the highest CN of S100A8 were counted. The means S100A8 CN for each tumor and each metastasis were then calculated. Finally, the cases were divided into two categories according to mean CN: <4 and ≥ 4 CN of S100A8. The REMARK criteria for tumor marker studies were followed [30].
S100A8 immunohistochemistry
Two different types of S100A8 staining were assessed using a bright-field microscope. Staining within tumor cells was recorded as an estimate of the percentage of positively staining cells among all epithelial tumor cells within a single TMA core. The cases were divided into three categories based on this percentage: < 1%, ≥ 1% < 10%, and ≥ 10%. In addition, within a single TMA core, the absolute number of positively stained infiltrating polymorphonuclear cells was recorded. Cells were regarded as infiltrating if found within the tumor, either in the epithelial or stromal compartment, but not visibly contained in a vessel. This data was used to define two categories: < 10 and ≥ 10 infiltrating S100A8 + polymorphonuclear cells. Again, where more than one TMA core was available, the one with the highest number of staining cells was chosen. The cut-off of 10 cells was chosen arbitrarily.
Statistical analyses
The differences in tumor characteristics between the defined categories were examined using Pearson’s χ2 test. A log-normal accelerated failure time was chosen for the survival analysis because the proportional hazards assumption was inappropriate for some of the included covariates. The log-normal distribution was chosen after fitting the model using several different distributions (log-logistic, log-normal, Weibull, exponential, generalized gamma) and comparing the goodness of fit of each model using both the Akaike and the Bayesian information criteria. In the final model, the impact of the included variables on survival is given as a time ratio with a 95% confidence interval.
Results
Fluorescence in situ hybridization
FISH was successful in 475 of the primary tumors and 117 of the axillary lymph node metastases. Eighty-three primary tumors and 52 lymph node metastases were excluded either due to an insufficient amount of tissue available or unsatisfactory FISH.
Table 2 shows patient and tumor characteristics of the 475 cases included in the FISH study, further divided into two categories: those with a mean S100A8 CN per cancer cell < 4, and those with CN ≥ 4. The mean age at diagnosis in the study population was 76.1 years, and the mean follow-up after diagnosis was 8.7 years. By the end of follow-up 171 (36%) women had died from BC while 255 (54%) had died from other causes.
Table 2.
Characteristics of the study population according to mean S100A8 copy number
| Study Population | Mean S100A8 CN in the primary tumor | |||
|---|---|---|---|---|
| < 4 | ≥ 4 | p-value (χ2) | ||
| N (%) | 475 | 379 (80) | 96 (20) | |
| Mean age at diagnosis, years (SD) | 76.1 (8.2) | 76.4 (8.3) | 75.1 (7.6) | |
| Mean follow-up after diagnosis, years (SD) | 8.7 (6.8) | 8.9 (6.8) | 8.1 (6.7) | |
| Deaths from BC (%) | 171 (36) | 131 (35) | 40 (42) | |
| Deaths from other causes (%) | 255 (54) | 206 (54) | 49 (51) | |
| Grade (%) | ||||
| 1 | 48 (10) | 42 (11) | 6 (6) | < 0.001 |
| 2 | 267 (56) | 233 (61) | 34 (35) | |
| 3 | 160 (34) | 104 (27) | 56 (58) | |
| Lymph node metastasis (%) | ||||
| Yes | 165 (35) | 134 (35) | 31 (32) | 0.85 |
| No | 200 (42) | 158 (42) | 42 (44) | |
| Unknown histology | 110 (23) | 87 (23) | 23 (24) | |
| Mean S100A8 CN in lymph node metastasis (%) | ||||
| < 4 | 70 (74) | 60 (86) | 14 (58) | 0.005 |
| ≥ 4 | 24 (26) | 10 (14) | 10 (42) | |
| Molecular subtype (%) | ||||
| Luminal A | 251 (53) | 212 (56) | 39 (41) | 0.002 |
| Luminal B (HER2-) | 114 (24) | 92 (24) | 22 (23) | |
| Luminal B (HER2+) | 37 (8) | 26 (7) | 11 (11) | |
| HER2 type | 25 (5) | 17 (5) | 8 (8) | |
| 5NP | 11 (2) | 10 (3) | 1 (1) | |
| BP | 36 (8) | 21 (6) | 15 (16) | |
| Histologic subtype (%) | ||||
| Ductal (NOS) | 324 (68) | 258 (68) | 66 (69) | 0.3 |
| Lobular | 64 (13) | 55 (15) | 9 (9) | |
| Other | 87 (18) | 66 (17) | 21 (22) | |
| Ki67 high / low (%) | ||||
| Ki67 < 15% | 286 (60) | 239 (63) | 47 (49) | 0.03 |
| Ki67 ≥ 15% | 188 (40) | 139 (37) | 49 (51) | |
| Mitoses / 10 HPF, median (IQR p25, p75) | 5 (1, 12) | 4 (1, 11) | 9 (3, 17) | |
| Mitoses / 10 HPF, quartiles (%) | ||||
| ≤ 1 | 114 (27) | 100 (29) | 14 (18) | 0.003 |
| > 1, ≤ 5 | 110 (26) | 97 (28) | 13 (16) | |
| > 5, ≤ 12 | 102 (24) | 79 (23) | 23 (29) | |
| > 12 | 102 (24) | 73 (21) | 29 (37) | |
N number of patients, SD standard deviation, BC breast cancer, HER2 human epidermal growth factor receptor 2, 5NP 5 negative phenotype, BP basal phenotype, HPF high-power field, IQR interquartile range
S100A8 was found to be amplified exclusively within cancer cells [Fig. 2], where it was found in a pattern consistent with extrachromosomal amplification.
Fig. 2.
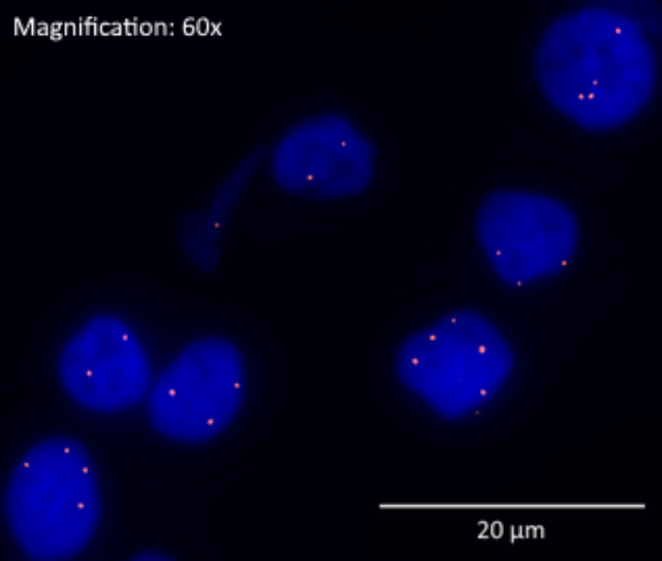
Fluorescence in situ hybridization for S100A8 at 60x magnification, scale bars: 20 μm, showing increased copy number of S100A8 (red dots) in tumor cell nuclei stained with Dapi (blue)
S100A8 CN and molecular subtypes
Increased S100A8 CN was found across all molecular subtypes. Among luminal tumors increased S100A8 CN was found in 39/251 (16%) Luminal A, 22/114 (18%) Luminal B, and 11/37 (30%) Luminal B (HER2+) tumors. Among the non-luminal tumors, it was found in 8/25 (32%) HER2 tumors, 1/11 (9%) 5NP, and 15/36 (42%) BP tumors (p = 0.002).
S100A8 CN, proliferation and histopathological grade
Tumors with increased S100A8 CN tended to be of higher histopathological grade. Of these tumors 6/96 (6%) were grade 1, 34/96 (35%) were grade 2, and 56/96 (58%) were grade 3 (p < 0.001). S100A8 CN increase was also significantly associated with increased proliferation. Forty-nine of 96 (51%) of the tumors with S100A8 CN ≥ 4 were Ki67 high (p = 0.03).
S100A8 CN in primary tumors and lymph node metastases
Of the 475 cases with successful FISH, 165 (35%) had confirmed metastases to axillary lymph-nodes at the time of diagnosis. S100A8 CN ≥ 4 was found in 96/475 (20%) cases. Copy number data for cases with lymph node metastases was available for 94 cases and 24/94 (25%) showed S100A8 CN ≥ 4 in the primary tumor. Of these, 10/24 (42%) also had increased S100A8 CN in the lymph-node metastasis while 14/24 (58%) did not (p = 0.005).
Immunohistochemistry
Table 3 shows patient and tumor characteristics of the 498 cases from the same study population, with successful IHC-staining for S100A8, divided into three categories (< 1%, ≥ 1% < 10%, ≥ 10%) based on the proportion of epithelial tumor cells staining positively for the protein S100A8. The mean age at diagnosis was 75.9 years while the mean follow-up time was 9.2 years. By end of follow-up 167/498 (34%) of the women in the study population had died of BC while 279/498 (56%) had died from other causes.
Table 3.
Characteristics of the study population according to the proportion of S100A8 staining tumor epithelial cells
| Study Population | Cytoplasmic S100A8 staining | |||||
|---|---|---|---|---|---|---|
| < 1% | ≥ 1%, < 10% | ≥ 10% | p-value (χ2) | |||
| N (%) | 498 | 420 (84) | 47 (9) | 31 (6) | ||
| Mean age at diagnosis, years (SD) | 75.9 (8.5) | 75.5 (8.4) | 76.3 (8.4) | 79.9 (9.5) | ||
| Mean follow-up after diagnosis, years (SD) | 9.2 (7.1) | 9.6 (7.1) | 8.3 (6.7) | 4.7 (4.9) | ||
| Deaths from BC (%) | 167 (34) | 134 (32) | 14 (30) | 19 (61) | ||
| Deaths from other causes (%) | 279 (56) | 241 (58) | 26 (55) | 12 (39) | ||
| Grade (%) | ||||||
| 1 | 54 (11) | 52 (12) | 2 (4) | 0 (0) | < 0.001 | |
| 2 | 287 (58) | 261 (62) | 19 (40) | 7 (23) | ||
| 3 | 155 (31) | 105 (25) | 26 (55) | 24 (77) | ||
| Lymph node metastasis (%) | ||||||
| Yes | 169 (34) | 134 (32) | 22 (47) | 9 (29) | 0.08 | |
| No | 226 (45) | 198 (47) | 19 (40) | 13 (42) | ||
| Unknown histology | 103 (21) | 88 (21) | 6 (13) | 9 (29) | ||
| Molecular subtype (%) | ||||||
| Luminal A | 268 (54) | 247 (59) | 17 (36) | 4 (13) | < 0.001 | |
| Luminal B (HER2-) | 116 (23) | 106 (25) | 8 (17) | 2 (7) | ||
| Luminal B (HER2+) | 36 (7) | 30 (7) | 5 (11) | 1 (3) | ||
| HER2 type | 28 (6) | 12 (3) | 8 (17) | 8 (27) | ||
| 5NP | 12 (2) | 4 (1) | 5 (11) | 3 (10) | ||
| BP | 35 (7) | 19 (5) | 4 (9) | 12 (40) | ||
| Histologic subtype (%) | ||||||
| Ductal (NOS) | 351 (70) | 294 (70) | 35 (75) | 22 (71) | 0.4 | |
| Lobular | 65 (13) | 60 (14) | 3 (6) | 2 (6) | ||
| Other | 82 (16) | 66 (16) | 9 (19) | 7 (23) | ||
| Ki67 high / low (%) | ||||||
| Ki67 < 15% | 301 (60) | 268 (64) | 25 (53) | 8 (26) | < 0.001 | |
| Ki67 ≥ 15% | 194 (39) | 150 (36) | 22 (47) | 22 (71) | ||
| Mitoses / 10 HPF, median (IQR p25, p75) | 5 (1, 12) | 4 (1, 9) | 9 (2, 16) | 14 (8, 21) | ||
| Mitoses / 10 HPF, quartiles (%) | ||||||
| ≤ 1 | 136 (27) | 127 (30) | 7 (15) | 2 (6) | < 0.001 | |
| > 1, ≤ 5 | 134 (27) | 121 (29) | 9 (19) | 4 (13) | ||
| > 5, ≤ 12 | 116 (23) | 94 (22) | 13 (28) | 9 (29) | ||
| > 12 | 112 (22) | 78 (19) | 18 (38) | 15 (48) | ||
| Number of infiltrating S100A8 + PMN cells (%) | ||||||
| < 10 | 405 (81) | 342 (81) | 38 (81) | 25 (81) | 0.99 | |
| ≥ 10 | 93 (19) | 78 (19) | 9 (19) | 6 (19) | ||
N number of patients, SD standard deviation, BC breast cancer, HER2 human epidermal growth factor receptor 2, 5NP 5 negative phenotype, BP basal phenotype, HPF high-power field, IQR interquartile range
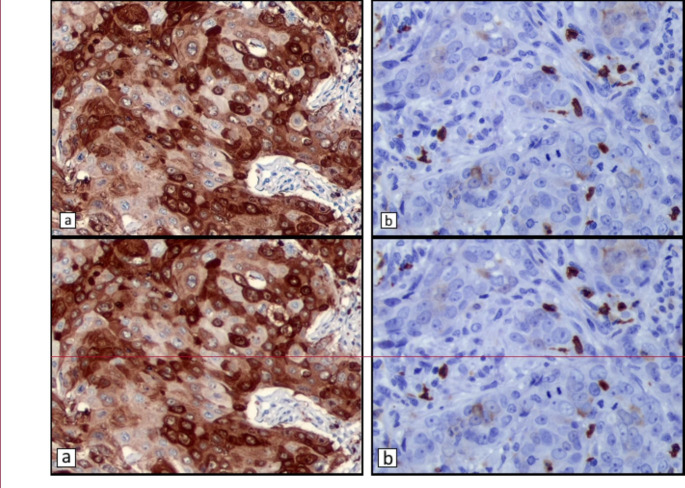
Positive S100A8 staining was observed both in epithelial tumor cells and in polymorphonuclear (PMN) cells. In both cell types, staining was diffusely cytoplasmic. [Figure 3] shows representative images of this type of staining. Patient and tumor characteristics according to the proportion of S100A8 + tumor epithelial cells are presented in Table 3 and explained in this section. Results for S100A8 in PMNs are presented in Table 4.
Fig. 3.
Immunohistochemical staining showing positive cytoplasmic staining at 400x magnification in (a) cancer cells, and (b) infiltrating polymorphonuclear cells
Table 4.
Characteristics of the study population according to the presence of ≥ 10 infiltrating S100A8 + PMN cells
| Study Population | Infiltration of S100A8 + cells | |||
|---|---|---|---|---|
| < 10 | ≥ 10 | p-value (χ2) | ||
| N (%) | 498 | 405 (81) | 93 (19) | |
| Mean age at diagnosis, years (SD) | 75.9 (8.5) | 76.0 (8.5) | 75.1 (8.5) | |
| Mean follow-up after diagnosis, years (SD) | 9.2 (7.1) | 9.1 (7.1) | 9.6 (7.1) | |
| Deaths from BC (%) | 167 (34) | 133 (33) | 34 (37) | |
| Deaths from other causes (%) | 279 (56) | 233 (58) | 46 (49) | |
| Grade (%) | ||||
| 1 | 54 (11) | 49 (12) | 5 (5) | < 0.001 |
| 2 | 287 (58) | 247 (61) | 40 (43) | |
| 3 | 155 (31) | 107 (27) | 48 (52) | |
| Lymph node metastasis (%) | ||||
| Yes | 169 (34) | 134 (33) | 35 (38) | 0.7 |
| No | 226 (45) | 186 (46) | 40 (43) | |
| Unknown histology | 103 (21) | 85 (21) | 18 (19) | |
| Molecular subtype (%) | ||||
| Luminal A | 268 (54) | 234 (58) | 34 (37) | < 0.001 |
| Luminal B (HER2-) | 116 (23) | 94 (23) | 22 (24) | |
| Luminal B (HER2+) | 36 (7) | 25 (6) | 11 (12) | |
| HER2 type | 28 (7) | 18 (5) | 10 (11) | |
| 5NP | 12 (2) | 9 (2) | 3 (3) | |
| BP | 35 (7) | 22 (5) | 13 (14) | |
| Histologic subtype (%) | ||||
| Ductal (NOS) | 351 (70) | 289 (71) | 62 (67) | 0.04 |
| Lobular | 65 (13) | 57 (14) | 8 (9) | |
| Other | 82 (16) | 59 (15) | 23 (25) | |
| Ki67 high / low (%) | ||||
| Ki67 < 15% | 301 (60) | 264 (65) | 37 (40) | < 0.001 |
| Ki67 ≥ 15% | 194 (39) | 138 (34) | 56 (60) | |
| Mitoses / 10 HPF, median (IQR p25, p75) | 5 (1, 12) | 4 (1, 9) | 9 (4, 20) | |
| Mitoses / 10 HPF, quartiles (%) | ||||
| ≤ 1 | 136 (27) | 123 (30) | 13 (14) | < 0.001 |
| > 1, ≤ 5 | 134 (27) | 116 (29) | 18 (19) | |
| > 5, ≤ 12 | 116 (23) | 89 (22) | 27 (29) | |
| > 12 | 111 (22) | 76 (19) | 35 (38) | |
N number of patients, SD standard deviation, BC breast cancer, HER2 human epidermal growth factor receptor 2, 5NP 5 negative phenotype, BP basal phenotype, HPF high-power field, IQR interquartile range
S100A8 expression in primary tumors
S100A8 + staining in ≥ 1% of tumor cells was observed in 78/498 (16%) of cases. Of these, 47/498 (9%) had between ≥ 1% <10% positive-staining tumor epithelial cells, while 31/498 (6%) had ≥ 10%. The mean age at diagnosis among the latter group of patients was 79.9 years, 4 years higher than that of the study population as a whole and 19/31 (61%), had died from BC by the end of follow-up.
S100A8 and molecular subtypes
Tumors with S100A8 + tumor epithelial cells were found among all molecular subtypes. There was a gradual shift away from luminal types in favor of non-luminal types with increasing levels of S100A8 staining. Among tumors with ≥ 10% S100A8 + tumor epithelial cells, we found 4/268 (1%) Luminal A, 2/116 (2%) Luminal B, and 1/36 (3%) Luminal B (HER2+) tumors, whereas we found 8/28 (29%) HER2, 3/12 (25%) 5NP, and 12/35 (34%) BP tumors expressed S100A8 in ≥ 10% of tumor epithelial cells. Notably, of all tumors with S100A8 + ≥ 10%, 23/31 (74%) were of non-luminal types.
S100A8, proliferation and histopathological grade
Higher proportions of S100A8 + tumor epithelial cells were significantly associated both with increased proliferation and with higher histopathological grade. Of tumors with ≥ 10% S100A8 + cancer cells 22/31 (71%) were Ki67 high. In the same group there were no grade 1 tumors, 7/31 (23%) grade 2 tumors, while the remaining 24/31 (77%) were grade 3.
S100A8 and lymph node metastases
A weak association between S100A8 staining in tumor epithelial cells and axillary lymph-node status at the time of diagnosis was observed (p = 0.08). Metastases to axillary lymph-nodes were more frequent in the intermediate group (≥ 1% <10%), 22/47 (47%). Only 9/31 (29%) of tumors with ≥ 10% positive staining cells had metastases to axillary lymph-nodes at time of diagnosis.
S100A8 in cancer cells and in infiltrating PMN cells
No association was observed between the proportion S100A8 + tumor epithelial cells and the number of infiltrating S100A8 + PMN cells.
S100A8 in infiltrating PMN cells
Ten or more infiltrating S100A8 + PMN cells were found in 93/498 (19%) cases. These tumors were found among all molecular subtypes: 67/93 (72%) among luminal types and 26/93 (28%) among non-luminal types. They tended to be of high histopathological grade with 48/93 (52%) tumors being grade 3 and 40/93 (40%) being grade 2. No significant associations between the presence of these cells and metastases to axillary lymph-nodes were observed (Table 4).
S100A8 CN and protein expression
As shown in Table 5, no significant association was observed between mean S100A8 CN/tumor epithelial cell and the proportion of S100A8 + cells. There was, however, a weak association between mean S100A8 CN and the number of infiltrating S100A8 + PMN cells (p = 0.09). A somewhat higher proportion of tumors with increased S100A8 CN also had ≥ 10 S100A8 + PMN cells.
Table 5.
Relationship between mean S100A8 copy number (CN) and protein expression
| Mean S100A8 CN | |||||
|---|---|---|---|---|---|
| < 4 | ≥ 4 | p-value (χ2) | |||
| Proportion of S100A8 + cancer cells (%) | |||||
| < 1% | 356 (83) | 292 (84) | 64 (81) | 0.5 | |
| ≥ 1% < 10% | 42 (10) | 35 (10) | 7 (9) | ||
| ≥ 10% | 30 (7) | 22 (6) | 8 (10) | ||
| Number of infiltrating S100A8 + PMN cells (%) | |||||
| < 10 | 348 (81) | 289 (83) | 60 (75) | 0.09 | |
| ≥ 10 | 80 (19) | 59 (17) | 20 (25) | ||
Survival analysis
Figure 4. Relative prognosis was estimated by using a log-normal accelerated failure time model. The results of such an analysis are shown in Table 6 for four different variables, each previously described. A plot for one of these variables is shown in Fig. 4. No significant differences in survival were observed between cases with and without increased S100A8 CN, neither in the primary tumor nor in axillary lymph-node metastases. Neither was the presence of ≥ 10 S100A8 + PMN cells in the tumor associated with significantly different patient prognosis. However, patients with tumors with ≥ 10% S100A8 + tumor epithelial cells did have significantly poorer survival (p < 0.001). The time ratio for this variable was estimated to be 0.2 (95% CI 0.10–0.42), meaning that the average estimated lifespan of patients in this group, from time of diagnosis, was 20% of that of cases with less than 1% of S100A8 + tumor epithelial cells. As is shown in Table 7 this time ratio remained significantly different from 1 when adjusted for age, grade, histologic subtype, molecular subtype and Ki67 status.
Fig. 4.
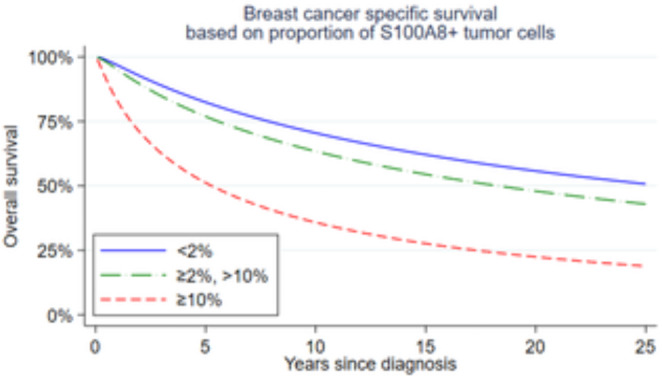
Breast cancer specific survival based on proportion of S100A8 + tumor epithelial cells using a log-normal AFT model. Time ratios as well as p-values are given in Table 6
Table 6.
Parameter estimates and 95% confidence intervals for log-normal accelerated failure time model
| Variable | Time ratio (95% CI) | P |
|---|---|---|
| Mean S100A8 CN (primary tumor) | ||
| < 4 | Reference | |
| ≥ 4 | 0.77 (0.47–1.24) | 0.28 |
| Mean S100A8 CN (lymph node metastases) | ||
| < 4 | Reference | |
| ≥ 4 | 0.60 (0.30–1.20) | 0.15 |
| Proportion of S100A8 + tumor epithelial cells | ||
| < 1% | Reference | |
| ≥ 1%, < 10% | 0.71 (0.37–1.36) | 0.30 |
| ≥ 10% | 0.20 (0.10–0.42) | < 0.001 |
| Number of infiltrating S100A8 + PMN cells | ||
| < 10 | Reference | |
| ≥ 10 | 0.83 (0.51–1.37) | 0.46 |
TR time ratio, CI confidence interval, CN copy number, PMN polymorphonuclear
Table 7.
Parameter estimates for different log-normal AFT models with proportion of S100A8 + tumor epithelial cells as the main covariate, unadjusted and adjusted for age, grade, histologic subtype, molecular subtype and Ki67 status
| Proportion of S100A8 + tumor cells | |||
|---|---|---|---|
| < 1% | ≥ 1%, < 10% | ≥ 10% | |
| TR unadjusted (95% CI) | Reference | 0.71 (0.37–1.36) | 0.20 (0.10–0.42) |
| TR adjusted for age (95% CI) | Reference | 0.78 (0.43–1.39) | 0.23 (0.12–0.43) |
| TR adjusted for grade (95% CI) | Reference | 0.91 (0.47–1.77) | 0.30 (0.14–0.61) |
| TR adjusted for histological subtype (95% CI) | Reference | 0.66 (0.34–1.27) | 0.20 (0.10–0.41) |
| TR adjusted for molecular subtype (95% CI) | Reference | 1.05 (0.54–2.05) | 0.39 (0.18–0.86) |
| TR adjusted for Ki67 status (95% CI) | Reference | 0.78 (0.41–1.49) | 0.25 (0.12–0.51) |
TR time ratio, CI confidence interval
Discussion
The aim of this study was to examine the relationship between S100A8 CN increase and S100A8 protein expression in BC epithelial cells and infiltrating PMN cells, as well as to explore associations between each of these with proliferation, histopathological grade, molecular subtypes and prognosis. We found that both S100A8 protein expression in tumor cells and, to a lesser extent, S100A8 CN increase, were associated with high tumor grade and increased proliferation. S100A8 protein expression was strongly associated with non-luminal tumors, and in particular with HER2-type and BP tumors. While S100A8 protein expression in at least 10% of epithelial tumor cells was associated with poor survival, CN increase of S100A8 was not. We observed no association between S100A8 CN increase and S100A8 protein expression in tumor cells and only a weak association (p = 0.09) between increased S100A8 CN and the presence of 10 or more S100A8 + infiltrating PMN cells.
The incidence of S100A8 CN increase we report here is congruent with the incidence of S100A8 amplification found in other data sets. Using cBioPortal [31, 32] to explore the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) data set reveals that S100A8 is amplified in 21% of the tumors included therein [6, 7]. Similarly, Goh et al. found S100A8 amplification in between 10 and 30% of tumors in the TCGA breast cancer data set depending on molecular subtype [8].
In the same paper Goh et al. also found that amplification of a profile of 17 genes on chromosome 1q21.3, S100A8 included, was associated with worse survival and proposed a mechanism involving a reciprocal feedback loop between S100A7, S100A8, S100A9 and IRAK1. A possible reason we did not observe such an association in our population might be that the presence of this amplification alters sensitivity to adjuvant therapy, which the women included in our study for the most part would not have received. Of note, Lang et al. has reported that targeted silencing of the S100A8 gene by miR-24 increases chemotherapy sensitivity of endometrial carcinoma cells to paclitaxel [33], while Li et al. found that the level of S100A8 expression was superior to molecular subtyping in predicting chemo responses in 120 cases of BC patients [34].
It should also be noted that the amplifications involved in these studies encompass more genes than simply S100A8. The FISH probe used in the present study, for example, is ~ 168Kb long and covers six genes (PGLYRP3, PGLYRP4, S100A9, S100A12, S100A7A) in addition to S100A8, making it difficult to attribute specific findings directly to any one gene.
Our findings regarding S100A8 protein expression in BC cells are well in concordance with previous studies [11–13]. Like our findings, Woo et al. reported on S100A8 staining both in BC cells as well as in infiltrating immune cells. However, unlike us, they found that both types of staining were linked to poorer prognosis. In a different study on the same population, we examined another potential marker for MDSCs, arginase-1 (ARG1), and found that 27/92 (29%) of the tumors with significant infiltration of S100A8 + PMNs also had significant infiltration of ARG1 + PMNs. The latter, however, were significantly associated with poorer prognosis (unpublished data). We therefore propose that S100A8 alone may not be the most appropriate surrogate marker for MDSCs due to its abundant expression in different neutrophil and monocyte subpopulations.
Interestingly, all these studies found S100A8 staining exclusively in the cytoplasm of tumor cells. This may be a limitation of the methods applied given that S100A8 has both known extracellular and intranuclear functions associated with cancer [5, 16]. Furthermore, the function and stability of S100A8 is highly dependent on S100A9, and while the genes are generally coamplified, the proteins are not always coexpressed [12]. In future studies it would therefore be an advantage to look at the expression of both proteins.
We observed that women in the group with the largest proportion of S100A8 + tumor epithelial cells had a mean age at diagnosis of 79.9 years, 4 years higher than the mean age at diagnosis of the study population, and that this group had a worse prognosis even after adjusting for age. This is remarkable given that 74% of these tumors were of non-luminal types which are relatively more common in younger women. S100A8 expression may be a response to increasing levels of reactive oxygen and nitrogen species in these tumors, of which both S100A8 and S100A9 are excellent scavengers [35]. In addition, S100A8 and S100A9 are age associated damage-associated molecular patterns (DAMPs) and they, along with MDSCs, have been connected to chronic inflammation during aging [36, 37].
The 563 tumors included in this study came from a well-described, and relatively homogeneous cohort of Norwegian breast cancer patients who were in most cases followed until time of death [26, 27]. Data on patient outcome was taken from high quality national registries [38, 39]. Due to either advanced age at diagnosis, or the time period in which they were diagnosed, most of these women would not have been offered the adjuvant therapy, which is common today, allowing us to study a near-natural course of the disease after surgery. The tumors were collected over a period of more than 40 years and were thus subject to varying pre-analytical conditions. This makes it difficult to study mRNA expression in these BCs. The findings should also be verified in a larger number of non-luminal BCs.
In conclusion, though both the amplification of S100A8 and the expression of the protein in BC cells may identify BCs with more malignant characteristics, they appear to be unrelated phenomena.
Acknowledgements
The authors thank the Department of Pathology, St. Olav’s Hospital, Trondheim University Hospital, for making the archives available for the study and the Cancer Registry of Norway for providing patient data.
Funding
The Medical Student Research Program, NTNU, the Tromsø Research Foundation, and the Trond Mohn Foundation (180°N project; MSG), have given the study financial support.
Open access funding provided by NTNU Norwegian University of Science and Technology (incl St. Olavs Hospital - Trondheim University Hospital)
Data availability
Data are available upon reasonable request. The datasets included in the current study are not publicly available but are available from the corresponding author on reasonable request.
Declarations
Ethics statement
This study was approved by the Regional Committee for Medical and Health Research Ethics, Central Norway. Approval number: REK 836-09. The approval of this study includes dispensation from the general requirement of patient consent.
Conflict of interest
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Pruenster M, et al. S100A8/A9: from basic science to clinical application. Pharmacol Ther. 2016;167:120–131. doi: 10.1016/j.pharmthera.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 2.Ehrchen JM, et al. The endogenous toll–like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 3.Wang S et al (2018) S100A8/A9 in inflammation. Front Immunol 9. 10.3389/fimmu.2018.01298
- 4.Yang D, Han Z. Oppenheim Alarmins and immunity. Immunol Rev. 2017;280:41–56. doi: 10.1111/imr.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shabani F, et al. Calprotectin (S100A8/S100A9): a key protein between inflammation and cancer. Inflamm Res. 2018;67:801–812. doi: 10.1007/s00011-018-1173-4. [DOI] [PubMed] [Google Scholar]
- 6.Curtis C, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pereira B, et al. The somatic mutation profiles of 2,433 breast cancers refine their genomic and transcriptomic landscapes. Nat Commun. 2016;7:1–16. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goh JY, et al. Chromosome 1q21. 3 amplification is a trackable biomarker and actionable target for breast cancer recurrence. Nat Med. 2017;23:1319–1330. doi: 10.1038/nm.4405. [DOI] [PubMed] [Google Scholar]
- 9.Cross S, et al. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46:256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 10.Huang A et al (2020) Prognostic role of S100A8 in human solid cancers: a systematic review and validation. Front Oncol 10. 10.3389/fonc.2020.564248 [DOI] [PMC free article] [PubMed]
- 11.Wang D et al (2018) Clinical significance of elevated S100A8 expression in breast cancer patients. Front Oncol 8. 10.3389/fonc.2018.00496 [DOI] [PMC free article] [PubMed]
- 12.Arai K, et al. S100A8 and S100A9 overexpression is associated with poor pathological parameters in invasive ductal carcinoma of the breast. Curr Cancer Drug Targets. 2008;8:243–252. doi: 10.2174/156800908784533445. [DOI] [PubMed] [Google Scholar]
- 13.Woo JW, et al. Prognostic significance of S100A8-positive immune cells in relation to other immune cell infiltration in pre-invasive and invasive breast cancers. Cancer Immunol Immunother. 2021;70:1365–1378. doi: 10.1007/s00262-020-02776-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller P, et al. Elevated S100A8 protein expression in breast cancer cells and breast tumor stroma is prognostic of poor disease outcome. Breast Cancer Res Treat. 2017;166:85–94. doi: 10.1007/s10549-017-4366-6. [DOI] [PubMed] [Google Scholar]
- 15.Bao Y, Wang A, Mo J. A8/A9 is associated with estrogen receptor loss in breast cancer. Oncol Lett. 2016;100:1936–1942. doi: 10.3892/ol.2016.4134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song R, Struhl K. S100A8/S100A9 cytokine acts as a transcriptional coactivator during breast cellular transformation. Sci Adv. 2021;7:eabe5357. doi: 10.1126/sciadv.abe5357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin C, et al. RAGE-binding S100A8/A9 promotes the migration and invasion of human breast cancer cells through actin polymerization and epithelial–mesenchymal transition. Breast Cancer Res Treat. 2013;142:297–309. doi: 10.1007/s10549-013-2737-1. [DOI] [PubMed] [Google Scholar]
- 18.Ostrand-Rosenberg S, et al. Cross-talk between myeloid-derived suppressor cells (MDSC), macrophages, and dendritic cells enhances tumor-induced immune suppression. Sem Cancer Biol. 2012;22:275–281. doi: 10.1016/j.semcancer.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umansky V et al (2016) The role of myeloid-derived suppressor cells (MDSC) in cancer progression. Vaccines 4. 10.3390/vaccines4040036 [DOI] [PMC free article] [PubMed]
- 20.Cheng P, et al. Inhibition of dendritic cell differentiation and accumulation of myeloid-derived suppressor cells in cancer is regulated by S100A9 protein. J Exp Med. 2008;205:2235–2249. doi: 10.1084/jem.20080132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vrakas CN, et al. The measure of DAMPs and a role for S100A8 in recruiting suppressor cells in breast cancer lung metastasis. Immunol Investig. 2015;44:174–188. doi: 10.3109/08820139.2014.952818. [DOI] [PubMed] [Google Scholar]
- 22.Sinha P, et al. Proinflammatory S100 proteins regulate the accumulation of myeloid-derived suppressor cells. J Immunol. 2008;181:4666–4675. doi: 10.4049/jimmunol.181.7.4666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ortiz ML, et al. Myeloid-derived suppressor cells in the development of lung CancerMDSC and the development of Lung Cancer. Cancer Immunol Res. 2014;2:50–58. doi: 10.1158/2326-6066.CIR-13-0129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eisenblaetter M, et al. Visualization of tumor-immune interaction-target-specific imaging of S100A8/A9 reveals pre-metastatic niche establishment. Theranostics. 2017;7:2392. doi: 10.7150/thno.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drews-Elger K, et al. Infiltrating S100A8 + myeloid cells promote metastatic spread of human breast cancer and predict poor clinical outcome. Breast Cancer Res Treat. 2014;148:41–59. doi: 10.1007/s10549-014-3122-4. [DOI] [PubMed] [Google Scholar]
- 26.Kvale G, HEUCH I. EIDE A prospective study of reproductive factors and breast cancer: I. Parity. Am J Epidemiol. 1987;126:831–841. doi: 10.1093/oxfordjournals.aje.a114720. [DOI] [PubMed] [Google Scholar]
- 27.Engstrøm MJ, et al. Molecular subtypes, histopathological grade and survival in a historic cohort of breast cancer patients. Breast Cancer Res Treat. 2013;140:463–473. doi: 10.1007/s10549-013-2647-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lakhani S et al (2012) WHO classification of Tumours of the breast. Lyon, France: International Agency for Research on Cancer (IARC); 2012. World Health Organization Classification of Tumours, p 4
- 29.Elston CW. Ellis pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow‐up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- 30.Altman DG, et al. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med. 2012;10:1–39. doi: 10.1007/s10549-006-9242-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1–pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lang B, Shang C (2016) Meng targeted silencing of S100A8 gene by miR-24 to increase chemotherapy sensitivity of endometrial carcinoma cells to paclitaxel. Med Sci Monitor: Int Med J Experimental Clin Res 22. 10.12659/MSM.899179 [DOI] [PMC free article] [PubMed]
- 34.Li Y-h, et al. The value of detection of S100A8 and ASAH1 in predicting the chemotherapy response for breast cancer patients. Hum Pathol. 2018;74:156–163. doi: 10.1016/j.humpath.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 35.Gomes LH, et al. S100A8 and S100A9—oxidant scavengers in inflammation. Free Radic Biol Med. 2013;58:170–186. doi: 10.1016/j.freeradbiomed.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 36.Swindell WR, et al. Robust shifts in S100a9 expression with aging: a novel mechanism for chronic inflammation. Sci Rep. 2013;3:1–13. doi: 10.1038/srep01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pawelec G et al (2021) MDSCs, ageing and inflammageing. Cell Immunol 362. 10.1016/j.cellimm.2021.104297 [DOI] [PubMed]
- 38.Larsen IK, et al. Data quality at the Cancer Registry of Norway: an overview of comparability, completeness, validity and timeliness. Eur J Cancer. 2009;45:1218–1231. doi: 10.1016/j.ejca.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 39.Pedersen AG, Ellingsen CL. Data quality in the Causes of Death Registry. Tidsskrift for Den norske legeforening. 2015 doi: 10.4045/tidsskr.14.1065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are available upon reasonable request. The datasets included in the current study are not publicly available but are available from the corresponding author on reasonable request.