Abstract
Introduction
In light of the clinically meaningful results of the PI3K inhibitors in PIK3CA-mutated metastatic breast cancer (BC) patients, the reliable identification of PIK3CA mutations is of outmost importance. However, lack of evidence on the optimal site and timing of assessment, presence of temporal heterogeneity and analytical factors pose several challenges in clinical routine. We aimed to study the discordance rates of PIK3CA mutational status between primary and matched metastatic tumors.
Methods
A systematic literature search was performed in three different databases (Embase, Pubmed, Web of Science) and—upon screening—a total of 25 studies reporting PIK3CA mutational status both on primary breast tumors and their matched metastases were included in this meta-analysis. The random-effects model was used for pooled analyses of discordance of PIK3CA mutational status.
Results
The overall discordance rate of PIK3CA mutational status was 9.8% (95% CI, 7.0–13.0; n = 1425) and did not significantly differ within BC subtypes or metastatic sites. The change was bi-directional, more commonly observed from PIK3CA mutated to wild-type status (14.9%, 95% CI 11.8–18.2; n tumor pairs = 453) rather than the opposite direction (8.9%, 95% CI 6.1–12.1; n tumor pairs = 943).
Conclusions
Our results indicate the need of obtaining metastatic biopsies for PIK3CA-mutation analysis and the possibility of testing of the primary tumor, in case a re-biopsy deemed non-feasible.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10549-023-07010-1.
Keywords: PIK3CA, Breast cancer, Mutation, Primary, Metastasis
Introduction
Despite the therapeutic advances in the management of breast cancer (BC), metastatic disease still remains incurable. However, the identification of molecular alterations has led to the development of novel targeted treatments which substantially prolong survival outcomes in the advanced setting.
The phosphatidylinositol 3-kinase (PI3K)/protein kinase B (AKT)/mammalian target of rapamycin (mTOR) represents one of the signaling pathways which plays crucial role in cell proliferation, growth and other cellular processes in several cancer types, including BC and more prominently the hormone receptor positive (HR +)/human epidermal growth factor receptor-2 negative (HER2-) subtype [1]. The pathway hyperactivation occurs mainly due to oncogenic mutations in PIK3CA gene encoding the p110a catalytic subunit of the PI3Kα heterodimeric protein complex, observed in approximately 40% of HR + /HER2- BC patients [2]. PIK3CA mutations display differential frequency and prognostic value in the early and metastatic setting whereas they have been associated with resistance to endocrine and HER2-targeted treatment [3–5]. Of note, inhibition of PI3K/Akt/mTOR pathway has provided clinically meaningful improved outcomes, mostly in patients with HR + /HER2- metastatic disease who have developed endocrine resistance [6, 7]. Based on randomized evidence showing significant progression-free and numerical clinically meaningful overall survival benefit when the oral PI3K—selective inhibitor alpelisib is combined with fulvestrant in patients with PIK3CA-mutated HR + /HER2- advanced or metastatic BC who progressed during or after endocrine therapy, this treatment combination has been approved from regulatory authorities [8, 9].
The predictive role of PIK3CA mutational status for the treatment with alpelisib poses some challenges on how evidence from randomized trials is implemented into clinical practice. The detection of PIK3CA mutations in archival or fresh tumor tissue and plasma-derived circulating tumor DNA (ctDNA) is dependent on analytical and methodological factors, thus possibly affecting clinical validity and utility [10]. Furthermore, the presence of tumor heterogeneity and clonal evolution over time could potentially influence the mutational status and any discordance between primary and metastatic disease should be acknowledged and managed based on available evidence. Following the paradigm of other common BC biomarkers (i.e. ER, PR, HER2, PD-L1), little is known about how PIK3CA mutational status would change during metastatic progression and potentially drive treatment selection in BC patients.
Given the treatment option of PI3K inhibitors in patients with HR + /HER2- metastatic breast cancer, the temporal heterogeneity within breast tumors and the diagnostic challenges associated with identification of mutational status, reliable identification of PIK3CA-mutated patients who will benefit from treatment with PI3K inhibitors remains of outmost importance. The aim of this systematic review and study-level meta-analysis was to evaluate the discordance rates of PIK3CA mutational status between primary and matched metastatic tumors in BC patients.
Methods
Search algorithm and study selection criteria
The protocol of the current systematic review and meta-analysis has been published on the PROSPERO database (CRD42023398005).
The literature search was performed in the following three databases: PubMed and Web of Science (November 2022) and Embase (February 2023). The detailed search strategy is presented in the Supplementary Material and included terms “PIK3CA”, “breast cancer” and “primary or metastatic disease” (MeSH terms) in the title or abstract. These terms were also adapted according to the corresponding Embase controlled vocabulary.
Studies were included in the meta-analysis based on the following criteria: (i) studies reporting PIK3CA mutational status both on primary breast tumors and their matched metastases, using tumor tissue, irrespective of the detection method; (ii) studies that included at least 10 patients. Studies including circulating tumor cells or ctDNA analyses, in vitro and/or in vivo experiments, reviews, case reports or previous meta-analyses or written in language other than English were excluded.
Data extraction and quality assessment
The initial study selection on the basis of abstract and title screening and the full text screening were performed independently by two investigators (JR, ES), a third investigator (AV) resolved any discrepancies and consensus was reached for all eligible studies. Two investigators (JR, ES) performed the data extraction using a predefined form and a third (IZ) resolved any discrepancies by comparing the databases.
The following data were collected from each study: first author, journal, year of publication, country, if a study was multicentric or if only a single center was involved, study type (prospective/retrospective), number of patients with paired samples, if the study focused on specific metastatic sites and if it concerned a specific breast cancer subtype, overall number of discordant cases, direction of discordance (from PIK3CA-mutated in primary tumor to PIK3CA-wild-type in metastasis or vice versa) and mutation detection method.
The reporting quality of all studies included in the meta-analysis were assessed by two investigators (JR, ES) and a third investigator resolved any discrepancies, according to REporting recommendations for tumour MARKer prognostic studies (REMARK) checklist, consisting of 20 items, as previously described [11]. Each item (involving the reporting of study aims, methods, results and their contextual discussion) was scored on a scale from 0 to 2 depending on how adequately each of the 20 items was defined in the study, thus generating a maximum total quality assessment score of 40.
Outcomes and definitions
The outcomes of interests included: i) overall discordance (n pairs with discordance / total n of pairs analyzed), ii) overall discordance rate from PIK3CA-mutated to PIK3CA-wild-type (n samples changed from mutated to wild-type in metastatic tumor / n samples with PIK3CA-mutated in primary tumor), iii) overall discordance rate from PIK3CA-wild-type to PIK3CA-mutated (n samples changed from wild-type to mutated in metastatic tumor / n samples with PIK3CA-wild-type in primary tumor).
Statistical analysis
The discordance rates were calculated for the overall population and when feasible, within breast cancer subtypes (HR + /HER2-negative, HER2-positive, triple negative breast cancer), according to the site of recurrence (locoregional / distant) or per metastatic site (brain / liver / other).
The random-effects model was used for pooled analyses of discordance and corresponding 95% confidence interval (CI). Chi-square test was used to compare potential differences among the pooled discordant rates. Sensitivity analysis was performed within subgroups of interest, by excluding studies either without HER2 status or using gene expression profiling data for defining subtypes. Publication bias was evaluated based on both the visual inspection/qualitative assessment of the asymmetry on funnel plots and Egger’s test. Statistical heterogeneity was assessed using I2 statistic for each pooled analysis, with a 50% cut off for considerable heterogeneity. Each reported p-value was two-sided with significance being set at 0.05. All statistical analyzes were performed with StatsDirect (StatsDirect Ltd. UK, 2013).
Results
Study characteristics
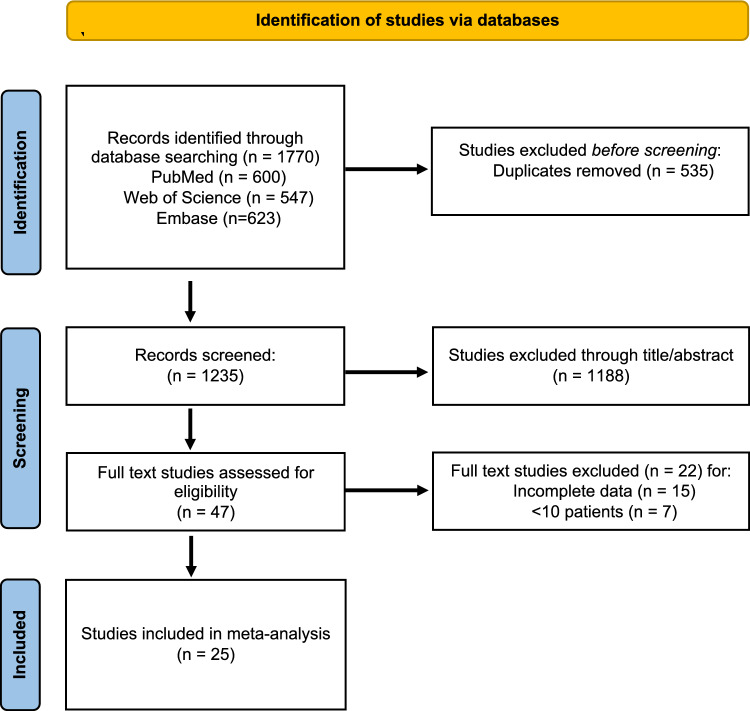
The Preferred Reporting Items for Systematic Reviews and Meta-analyses (PRISMA) study flowchart is depicted in Fig. 1. The initial search generated a total of 1770 records, 600 from PubMed, 547 from Web of Science and 623 from Embase. Upon deduplication, 1235 studies were screened for abstract and title 47 were assessed in full text.; 25 studies fulfilled the criteria and included in the meta-analysis.
Fig. 1.
PRISMA flowchart of search and study selection
The studies were published between 2010 and 2022 and the study characteristics are summarized in Table 1 [12–36]. The number of paired cases ranged between 10 and 242 patients. Twenty-one out of the 25 studies (84%) included in the meta-analysis were retrospective whereas the rest included patient cohorts collected within the scope of a prospective study. None of the studies presented results on treatment outcomes with alpelisib or other PI3K inhibitors. PIK3CA mutations were detected using targeted next-generation sequencing [21] methodology in the majority of the studies (14 of 25; 56%). A total of 19 studies (76%) evaluated the PIK3CA mutational status according to the site of recurrence (locoregional and/or distant), while 13 studies (52%) focused on specific metastatic sites including brain, liver or other localization (Table 2).
Table 1.
Characteristics of the studies included in the meta-analysis
| Author (reference) | Year | Country | Study type | Multi-center study | Metastatic site | Number of paired patient samples | Detection method | Quality assessment score |
|---|---|---|---|---|---|---|---|---|
| Aftimos (12) | 2021 | Belgium | Prospective | Yes | All | 242 | tNGS | 34 |
| Agahozo (13) | 2019 | Netherlands | Retrospective | No | All | 26 | SNaPshot, dPCR | 17 |
| Akahane (14) | 2020 | Japan | Retrospective | Yes | All | 11 | tNGS | 19 |
| Akcakanat (15) | 2021 | USA | Retrospective | No | All | 10 | tNGS | 23 |
| Arthur (16) | 2014 | UK | Retrospective | No | All | 89 | PCR | 28 |
| Basho (17) | 2016 | USA | Retrospective | Yes | All | 89 | tNGS | 29 |
| Bertucci (18) | 2016 | France | Retrospective | No | All | 23 | aCGH, tNGS | 20 |
| Callens (19) | 2021 | France | Prospective | Yes | All | 67 | tNGS, WES | 30 |
| Chen (20) | 2021 | China | Retrospective | No | Lymph nodes | 131 | tNGS | 22 |
| Da Silva (21) | 2010 | Australia | Retrospective | Yes | Brain | 12 | PCR combined with MALDI-TOF MS | 18 |
| Drury (22) | 2011 | UK | Retrospective | Yes | All | 21 | PCR | 22 |
| Dupont Jensen (23) | 2011 | Denmark | Retrospective | No | All | 100 | SNaPshot, RT-PCR | 29 |
| Fumagalli C (24) | 2020 | Italy | Retrospective | No | All | 61 | tNGS | 29 |
| Fumagalli D (25) | 2016 | Belgium | Prospective | No | All | 68 | PCR-based MUT-MAP | 31 |
| Giannoudis (26) | 2021 | UK | Retrospective | No | Brain | 32 | PCR-based UltraSEEK® panel | 19 |
| Gonzalez-Angulo (27) | 2011 | USA | Retrospective | Yes | All | 47 | PCR- and mass spectometry based | 21 |
| Gonzales-Martinez (28) | 2022 | Spain | Retrospective | Yes | Skin | 33 | tNGS | 25 |
| Kim (29) | 2019 | South Korea | Retrospective | Yes | All | 19 | PCR combined with MALDI-TOF MS | 36 |
| Lee (30) | 2015 | South Korea | Retrospective | No | Brain | 15 | tNGS | 20 |
| Meric-Bernstam (31) | 2014 | USA | Retrospective | Yes | All | 33 | tNGS | 20 |
| Park (32) | 2022 | South Korea | Retrospective | No | All | 49 | ddPCR | 23 |
| Roy-Chowduri (33) | 2015 | USA | Prospective | No | All | 31 | tNGS | 23 |
| Schleifman (34) | 2014 | USA | Retrospective | No | All | 73 | PCR-based MUT-MAP, SNP | 21 |
| Thulin (35) | 2021 | Sweden | Retrospective | Yes | Brain | 37 | tNGS | 24 |
| van Geelen (36) | 2020 | Australia | Prospective | No | All | 76 | tNGS | 30 |
tNGS targeted next-generation sequencing, dPCR digital polymerase chain reaction, RT-PCR real-time polymerase chain reaction, aCGH array-based comparative genomic hybridization, WES whole-exome sequencing; SNaPshot genotyping primerextension or minisequencing, ddPCR droplet digital polymerase chain reaction, MUT-MAP mutation multi-analyte panel, SNP single nucleotide polymorphism genotyping, MALDI-TOF MS matrix-assisted laser desorption/ionization coupled to time-of-flight mass spectrometry
Table 2.
Pooled discordance rates according to direction of PIK3CA mutational status change and within subgroups of interest
| Parameters | N studies (n pairs) | Pooled discordance, % | 95% Confidence Interval, % | Statistical heterogeneity (I2) | p-value for comparison |
|---|---|---|---|---|---|
| Direction of change | 0.003 | ||||
| Mut to wild-type | 24 (453) | 14.9 | 11.8–18.2 | 35.3 | |
| Wild type to mut | 24 (943) | 8.9 | 6.1–12.1 | 56.4 | |
| Breast cancer subtype | 0.577 | ||||
| HR + /HER2-negative | 13 (583) | 10.2 | 6.4–14.8 | 56.6 | |
| HER2-positive | 10 (149) | 8.7 | 4.8–13.7 | 0.0 | |
| TNBC | 11 (151) | 8.8 | 4.9–13.7 | 0.0 | |
| Site of recurrence | 0.839 | ||||
| Locoregional | 8 (306) | 9.6 | 6.6–13.1 | 29.0 | |
| Distant (any) | 11 (301) | 9.9 | 6.8–13.5 | 49.4 | |
| Metastatic site | 0.330 | ||||
| Brain | 5 (106) | 9.6 | 1.1–24.9 | 77.2 | |
| Liver | 3 (59) | 6.4 | 1.7–13.9 | 0.0 | |
| Other | 5 (67) | 11.2 | 4.9–19.6 | 7.2 | |
Quality of the eligible studies, magnitude of heterogeneity and publication bias
The median reporting quality assessment score was 23 (range: 17–36) according to REMARK guidelines. Substantial statistical heterogeneity among eligible studies (I2 > 50%) was observed in most of the analyses, except for the overall discordance rate of PIK3CA mutational status from mutated to wild-type and the HER2 + and triple-negative breast cancer (TNBC) subgroup analyses. Furthermore, no convincing evidence of publication bias was observed in pooled analyses (Supplementary Fig. 1).
Pooled discordance rates of PIK3CA mutational status and direction of change in paired primary and metastatic tumors
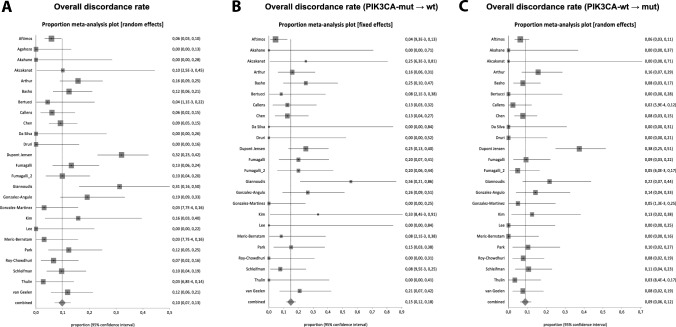
The overall discordance rate of PIK3CA mutational status between matched primary and metastatic breast cancer, regardless of subtype was 9.8% (95% CI, 7.0–13.0; I2 = 70.0%), including 1425 patients from 25 studies (Fig. 2A). The direction of change was more commonly observed from PIK3CA mutated to wild-type status (14.9%, 95% CI 11.8–18.2; n studies = 24, n tumor pairs = 453; I2 = 35.3%) (Fig. 2B) rather than from PIK3CA wild-type to mutated status (8.9%, 95% CI 6.1–12.1; n studies = 24; n tumor pairs = 943; I2 = 56.4%) (Fig. 2C). The difference between the directions of PIK3CA mutational status change was statistically significant (p = 0.003) (Table 2).
Fig. 2.
Forest plots for PIK3CA mutational status pooled discordance rates. A Overall discordance rate. B Overall discordance rate from PIK3CA-mutated to PIK3CA-wild-type. C Overall discordance rate from PIK3CA-wild-type to PIK3CA-mutated
Pooled discordance rates of PIK3CA mutational status in paired primary and metastatic lesions in subgroups of interest
Pooled results of PIK3CA mutational status discordance between matched primary and metastatic breast tumors within the different breast cancer subtypes of interest are presented in Table 2. Data from 13 studies including 583 HR + /HER2- BC patients demonstrated an overall discordance of 10.2% (95% CI 6.4–14.8; I2 = 56.6%). A sensitivity analysis was performed after exclusion of studies without available HER2 status or with subtyping based on gene expression profiling data and showed similar pooled discrepancy rates (9.7%, 95% CI 5.1 – 15.6%, n = 442 and 10.3%, 95% CI 5.8 – 15.8%, n = 515, respectively). A comparable overall discordance rate was noted between HER2-positive (8.7% 95% CI 4.8–13.7, n = 149, I2 = 0%) and TNBC (8.8%, 95% CI 4.9–13.7, n = 151, I2 = 0%) breast cancer patients (p = 0.557 for the comparison among the three subtypes). Similar results on the discrepancy rates were obtained though sensitivity analyses, upon exclusion of gene expression-based studies for both subtypes (8.8%, 95% CI: 4.6–14.0%, n = 133 for HER2 + and 8.2%, 95% CI: 4.0–13.8%, n = 117 for the TNBC).
Further subgroup analyses were performed according to the site of recurrence and metastatic site. Comparable pooled discordance rates were noted between the primary tumors and matched locoregional (9.6%, 95% CI 6.6–13.1) or distant (9.9%, 95% CI 6.6–13.1) recurrences (p = 0.839 for the comparison). When discordance rates were pooled based on distant metastatic site, a numerical but not statistically significant difference was observed between patients with matched metastatic lesion from brain compared to liver (9.6% versus 6.4%) (Table 2).
Discussion
This is, to the best of our knowledge, the first meta-analysis providing data on the discordance rates of PIK3CA-mutations between primary breast tumors and their matched metastases. Given the clinically meaningful results of the PI3K inhibitors in PIK3CA-mutated metastatic BC patients [8, 9], the reliable detection of PIK3CA mutations remains of outmost importance as it could guide physicians’ choices (level of evidence I-A according to ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) score) [37]. The results of our meta-analysis indicate that the pooled discordance rate of PIK3CA mutations between primary tumors and paired metastases was relatively low, observed in approximately 1 out of 10 patients. Of note, we also showed that the discordance of PIK3CA mutational status was bi-directional, though more commonly observed from PIK3CA-mutated in primary tumor to wild-type status in the metastases rather than in the opposite direction. This finding could help clinicians in deciding how to proceed with PIK3CA-mutation analysis in clinical practice to ensure reliable results and thus facilitating decision-making process.
The prevalence and clinical implications of PIK3CA mutations in BC varies according to the disease setting and also within subtypes. In early BC, PIK3CA mutations have been detected in 37%, 22% and 18% of ER + /HER2-, HER2 + and ER-/HER2- subtypes, respectively and associated with improved invasive disease-free survival but also with resistance to HER2-targeted treatments [3, 5]. In contrast, PIK3CA mutations have been detected in 28% of HR + /HER2- metastatic BC patients and correlated with worse overall survival as well as resistance to chemo- and endocrine therapy [4]. However, none of the aforementioned studies have assessed PIK3CA mutations in matched primary and metastatic tumors. When evaluating the discordance rates in paired samples in the present study, we demonstrated comparable results among all different BC subtypes.
In the pivotal phase III randomized trial SOLAR-1 that confirmed the predictive value of PIK3CA-mutation status on the clinical benefit of the PI3K-α-selective inhibitor alpelisib in PIK3CA-mutated patients with HR + /HER2- advanced or metastatic BC [8, 9, 38], the vast majority of PIK3CA mutations were determined at the primary tumor (77%) rather than at the metastatic sites (22%) [39], whereas no matched tumors have been evaluated. Nonetheless, the presence of temporal tumor heterogeneity during metastatic progression should not be disregarded, since it could influence patient prognosis and drive treatment selection. Following the paradigm of other common BC biomarkers (i.e. ER, PR, HER2, PD-L1) and their discordance over time [40–42], the magnitude of discordance on PIK3CA mutational status between primary and metastatic lesions could have important clinical implications. The results of this meta-analysis indicate a fairly substantial change in the PIK3CA-mutational status—although at a lower level compared to immunohistochemistry-based biomarkers—further motivating metastatic biopsies, despite the anatomical, technical and analytical challenges [43]. On the other hand, one could argue that an approximately 10% discordance rate might be acceptable under certain circumstances and, as a result, PIK3CA-mutational analysis of the primary tumor could serve as a suitable option in patients where metastatic biopsies are deemed inappropriate or technically infeasibly.
Although the results of subgroup analyses based on metastatic sites are limited by the small study numbers, a numerically higher discordance rate in brain compared to liver lesions was observed. These findings should be confirmed by subsequent studies but the different discordance rates among different metastatic sites seem to be in accordance with emerging data on specific genomic alterations linked to specific organotropisms in breast cancer [44, 45].
The evaluation process of the PIK3CA mutational status could be influenced by several factors as the source of testing material and the detection method used. Regarding the former, the introduction of liquid biopsy and ctDNA analysis has gained interest as an appealing, non-invasive alternative method to tissue (re)biopsy [46]. Despite the fact that in most studies a benefit for PI3K inhibitors was demonstrated in ctDNA-detected PIK3CA-mutated patients [47], few studies have investigated the concordance of ctDNA- versus tissue-based approaches. These studies report a modest concordance (70–83%) between the two methods [48–50], indicating the risk of false negative result due to low- or non-tumor shedding, technical challenges and/or tumor heterogeneity. Of note, although the positive ctDNA PIK3CA-mutated patients received similar magnitude of benefit from alpelisib as for the tissue-based detection (HR = 0.55) in the SOLAR-1 study [50], negative ctDNA result did not preclude the presence of a PIK3CA mutation [51], imposing the analysis on tumor tissue and the need for obtaining a metastatic biopsy in patients with ctDNA-negative result and reflex testing in the primary tumor when biopsy from metastatic lesion is not feasible. Investigating the concordance between tissue-based and liquid biopsy-based PIK3CA analysis was beyond the scope of this systematic review and meta-analysis. Considering the mutation detection methods, the regulatory FDA approval of alpelisib included the use of therascreen PIK3CA RGQ PCR Kit, the FoundationOne® CDx and FoundationOne® Liquid CDx assays as companion diagnostics for the detection of PIK3CA mutational status. However, a post-hoc targeted NGS analysis of the SOLAR-1 tissue samples (initially tested with PCR-based assays aimed to detect 12 mutations in exons 7, 9, and 20) revealed that in 12% of patients with PIK3CA-altered status, a PIK3CA mutation was not previously detected by PCR and that these patients had a favorable outcome when treated with alpelisib [52]. Therefore, the analytical performance of the assays needs to be refined and standardized in order to reliably detect PIK3CA mutations. In our analysis, the majority of the studies included tissue samples tested with targeted NGS, without any further reported direct method comparison.
The present study suffers from limitations that need to be addressed. First, this is a study-level meta-analysis not including individual patient data that would enable a deeper analysis of patient subgroups, thus reflecting a substantial between-the-study heterogeneity. Second, substantial clinical differences in patient cohorts, treatment strategies, metastatic sites, detection methods used and determination of the PIK3CA mutational status among eligible studies were observed, thus reflecting a substantial statistical heterogeneity in almost all pooled analyses. In an effort to reduce the risk of bias due to between-study heterogeneity, we used random-effects model for pooled analyses. Furthermore, the small numbers of studies and patients resulted in limited number of tumor pairs for certain subgroups analyses including IHC-based subtypes and metastatic sites. Finally, no study included information on treatment with alpelisib or other PI3K inhibitors among eligible patients and no information about the effectiveness of PI3K inhibitors in discordant cases was reported.
In conclusion, this meta-analysis provides information on the overall discordance rates of PIK3CA-mutational status between primary and matched metastatic breast tumors, the direction of change as well as the impact of different subtypes and metastatic sites on discordance. Given the clinical benefit of PI3KCA inhibitors in PIK3CA-mutated metastatic BC, the analytical challenges of ctDNA testing and the observation that PIK3CA status could change in 1 out of 10 patients, our results indicate the need of obtaining metastatic biopsies for PIK3CA-mutation analysis but also the possibility of testing of the primary tumor, in case a re-biopsy deemed non-feasible.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Ioannis Zerdes is supported by the Region Stockholm (clinical postdoctorial appointment, FoUI-977295), the Swedish Society of Oncology postdoctoral grant and Iris, Stig och Gerry Castenbäcks foundation. This study received no funding.
Author contributions
Conceptualization: A.V.; methodology: J.R., E.S, A.V; data extraction: J.R., E.S, A.V., I.Z.; formal analysis: A.V; data interpretation: I.Z, A.V.; writing—original draft preparation: J.R., E.S., I.Z.; writing—review and editing: A.V., I.Z; visualization: I.Z, A.V.; supervision, A.V.; resources and funding acquisition: A.V., I.Z. All authors have read and agreed to the published version of the manuscript.
Funding
Open access funding provided by Karolinska Institute.
Data availability
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Conflict of interest
The authors of this manuscript have no conflicts of interest to disclose.
Data sharing
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Justus Rosin and Ella Svegrup have contributed equally to the work.
Antonios Valachis and Ioannis Zerdes have jointly supervised the work.
References
- 1.Fruman DA, et al. The PI3K Pathway in Human Disease. Cell. 2017;170:605–635. doi: 10.1016/j.cell.2017.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 3.Zardavas D, et al. Tumor PIK3CA genotype and prognosis in early-stage breast cancer: a pooled analysis of individual patient data. J Clin Oncol. 2018;36:981–990. doi: 10.1200/jco.2017.74.8301. [DOI] [PubMed] [Google Scholar]
- 4.Mosele F, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020;31:377–386. doi: 10.1016/j.annonc.2019.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Rasti AR, et al. PIK3CA mutations drive therapeutic resistance in human epidermal growth factor receptor 2-positive breast cancer. JCO Precis Oncol. 2022;6:e2100370. doi: 10.1200/po.21.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Araki K, Miyoshi Y. Mechanism of resistance to endocrine therapy in breast cancer: the important role of PI3K/Akt/mTOR in estrogen receptor-positive, HER2-negative breast cancer. Breast Cancer. 2018;25:392–401. doi: 10.1007/s12282-017-0812-x. [DOI] [PubMed] [Google Scholar]
- 7.Vitale SR, et al. PI3K inhibition in breast cancer: Identifying and overcoming different flavors of resistance. Crit Rev Oncol Hematol. 2021;162:103334. doi: 10.1016/j.critrevonc.2021.103334. [DOI] [PubMed] [Google Scholar]
- 8.André F, et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N Engl J Med. 2019;380:1929–1940. doi: 10.1056/NEJMoa1813904. [DOI] [PubMed] [Google Scholar]
- 9.André F, et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: final overall survival results from SOLAR-1. Ann Oncol. 2021;32:208–217. doi: 10.1016/j.annonc.2020.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Fusco N, et al. PIK3CA mutations as a molecular target for hormone receptor-positive, HER2-negative metastatic breast cancer. Front Oncol. 2021;11:644737. doi: 10.3389/fonc.2021.644737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. PLoS Med. 2012;9:e1001216. doi: 10.1371/journal.pmed.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aftimos P, et al. Genomic and transcriptomic analyses of breast cancer primaries and matched metastases in AURORA, the Breast International Group (BIG) molecular screening initiative. Cancer Discov. 2021;11:2796–2811. doi: 10.1158/2159-8290.Cd-20-1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agahozo MC, et al. PIK3CA mutations in ductal carcinoma in situ and adjacent invasive breast cancer. Endocr Relat Cancer. 2019;26:471–482. doi: 10.1530/erc-19-0019. [DOI] [PubMed] [Google Scholar]
- 14.Akahane T, et al. Targeted next-generation sequencing assays using triplet samples of normal breast tissue, primary breast cancer, and recurrent/metastatic lesions. BMC Cancer. 2020;20:944. doi: 10.1186/s12885-020-07432-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Akcakanat A, et al. Genomic, transcriptomic, and proteomic profiling of metastatic breast cancer. Clin Cancer Res. 2021;27:3243–3252. doi: 10.1158/1078-0432.Ccr-20-4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arthur LM, et al. Changes in PIK3CA mutation status are not associated with recurrence, metastatic disease or progression in endocrine-treated breast cancer. Breast Cancer Res Treat. 2014;147:211–219. doi: 10.1007/s10549-014-3080-x. [DOI] [PubMed] [Google Scholar]
- 17.Basho RK, et al. Clinical outcomes based on multigene profiling in metastatic breast cancer patients. Oncotarget. 2016;7:76362–76373. doi: 10.18632/oncotarget.12987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bertucci F, et al. Comparative genomic analysis of primary tumors and metastases in breast cancer. Oncotarget. 2016;7:27208–27219. doi: 10.18632/oncotarget.8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Callens C, et al. Molecular features of untreated breast cancer and initial metastatic event inform clinical decision-making and predict outcome: long-term results of ESOPE, a single-arm prospective multicenter study. Genome Med. 2021;13:44. doi: 10.1186/s13073-021-00862-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen B, et al. Genetic and immune characteristics of sentinel lymph node metastases and multiple lymph node metastases compared to their matched primary breast tumours. EBioMedicine. 2021;71:103542. doi: 10.1016/j.ebiom.2021.103542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Da Silva L, et al. HER3 and downstream pathways are involved in colonization of brain metastases from breast cancer. Breast Cancer Res. 2010;12:R46. doi: 10.1186/bcr2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Drury SC, et al. Changes in breast cancer biomarkers in the IGF1R/PI3K pathway in recurrent breast cancer after tamoxifen treatment. Endocr Relat Cancer. 2011;18:565–577. doi: 10.1530/erc-10-0046. [DOI] [PubMed] [Google Scholar]
- 23.Dupont Jensen J, et al. PIK3CA mutations may be discordant between primary and corresponding metastatic disease in breast cancer. Clin Cancer Res. 2011;17:667–677. doi: 10.1158/1078-0432.Ccr-10-1133. [DOI] [PubMed] [Google Scholar]
- 24.Fumagalli C, et al. Inter-tumor genomic heterogeneity of breast cancers: comprehensive genomic profile of primary early breast cancers and relapses. Breast Cancer Res. 2020;22:107. doi: 10.1186/s13058-020-01345-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fumagalli D, et al. Somatic mutation, copy number and transcriptomic profiles of primary and matched metastatic estrogen receptor-positive breast cancers. Ann Oncol. 2016;27:1860–1866. doi: 10.1093/annonc/mdw286. [DOI] [PubMed] [Google Scholar]
- 26.Giannoudis A, et al. Genomic profiling using the UltraSEEK panel identifies discordancy between paired primary and breast cancer brain metastases and an association with brain metastasis-free survival. Breast Cancer Res Treat. 2021;190:241–253. doi: 10.1007/s10549-021-06364-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonzalez-Angulo AM, et al. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther. 2011;10:1093–1101. doi: 10.1158/1535-7163.Mct-10-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.González-Martínez S, et al. Differences in the molecular profile between primary breast carcinomas and their cutaneous metastases. Cancers (Basel) 2022 doi: 10.3390/cancers14051151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim SB, et al. BioPATH: a biomarker study in asian patients with HER2+ advanced breast cancer treated with lapatinib and other anti-HER2 therapy. Cancer Res Treat. 2019;51:1527–1539. doi: 10.4143/crt.2018.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JY, et al. Mutational profiling of brain metastasis from breast cancer: matched pair analysis of targeted sequencing between brain metastasis and primary breast cancer. Oncotarget. 2015;6:43731–43742. doi: 10.18632/oncotarget.6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meric-Bernstam F, et al. Concordance of genomic alterations between primary and recurrent breast cancer. Mol Cancer Ther. 2014;13:1382–1389. doi: 10.1158/1535-7163.Mct-13-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Park J, et al. Analysis of PIK3CA Mutation Concordance and Frequency in Primary and Different Distant Metastatic Sites in Breast Cancer. Cancer Res Treat. 2023;55:145–154. doi: 10.4143/crt.2022.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roy-Chowdhuri S, et al. Multigene clinical mutational profiling of breast carcinoma using next-generation sequencing. Am J Clin Pathol. 2015;144:713–721. doi: 10.1309/ajcpwdeqycyc92jq. [DOI] [PubMed] [Google Scholar]
- 34.Schleifman EB, et al. Targeted biomarker profiling of matched primary and metastatic estrogen receptor positive breast cancers. PLoS One. 2014;9:e88401. doi: 10.1371/journal.pone.0088401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thulin A, et al. Discordance of PIK3CA and TP53 mutations between breast cancer brain metastases and matched primary tumors. Sci Rep. 2021;11:23548. doi: 10.1038/s41598-021-02903-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Geelen CT, et al. Clinical implications of prospective genomic profiling of metastatic breast cancer patients. Breast Cancer Res. 2020;22:91. doi: 10.1186/s13058-020-01328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gennari A, et al. ESMO Clinical Practice Guideline for the diagnosis, staging and treatment of patients with metastatic breast cancer. Ann Oncol. 2021;32:1475–1495. doi: 10.1016/j.annonc.2021.09.019. [DOI] [PubMed] [Google Scholar]
- 38.Matikas A, Foukakis T. SOLAR1s: alpelisib returns to earth? Ann Oncol. 2021;32:129–132. doi: 10.1016/j.annonc.2020.12.003. [DOI] [PubMed] [Google Scholar]
- 39.Rugo, HS et al. (2019) Abstract CT142: Prevalence of PIK3CAmutations in patients with hormone receptor-positive, human epidermal growth factor-2-negative advanced breast cancer from the SOLAR-1 trial. Cancer Research79, CT142 . doi:10.1158/1538-7445.Am2019-ct142
- 40.Schrijver W, et al. Receptor conversion in distant breast cancer metastases: a systematic review and meta-analysis. J Natl Cancer Inst. 2018;110:568–580. doi: 10.1093/jnci/djx273. [DOI] [PubMed] [Google Scholar]
- 41.Boman, C. et al. Discordance of PD-L1 status between primary and metastatic breast cancer: A systematic review and meta-analysis. Cancer Treat Rev99, 102257, (2021). doi:10.1016/j.ctrv.2021.102257 [DOI] [PubMed]
- 42.Lindström LS, et al. Clinically used breast cancer markers such as estrogen receptor, progesterone receptor, and human epidermal growth factor receptor 2 are unstable throughout tumor progression. J Clin Oncol. 2012;30:2601–2608. doi: 10.1200/jco.2011.37.2482. [DOI] [PubMed] [Google Scholar]
- 43.Foukakis T, Åström G, Lindström L, Hatschek T, Bergh J. When to order a biopsy to characterise a metastatic relapse in breast cancer. Ann Oncol. 2012;23(Suppl 10):x349–353. doi: 10.1093/annonc/mds297. [DOI] [PubMed] [Google Scholar]
- 44.Gerratana L, et al. Understanding the organ tropism of metastatic breast cancer through the combination of liquid biopsy tools. Eur J Cancer. 2021;143:147–157. doi: 10.1016/j.ejca.2020.11.005. [DOI] [PubMed] [Google Scholar]
- 45.Nguyen B, et al. Genomic characterization of metastatic patterns from prospective clinical sequencing of 25,000 patients. Cell. 2022;185:563–575.e511. doi: 10.1016/j.cell.2022.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ignatiadis M, Sledge GW, Jeffrey SS. Liquid biopsy enters the clinic - implementation issues and future challenges. Nat Rev Clin Oncol. 2021;18:297–312. doi: 10.1038/s41571-020-00457-x. [DOI] [PubMed] [Google Scholar]
- 47.Criscitiello C, Marra A, Curigliano G. PIK3CA mutation assessment in HR+/HER2− metastatic breast cancer: overview for oncology clinical practice. J Mol Pathol. 2021;2:42–54. doi: 10.3390/jmp2010005. [DOI] [Google Scholar]
- 48.Chae YK, et al. Concordance of Genomic Alterations by Next-Generation Sequencing in Tumor Tissue versus Circulating Tumor DNA in Breast Cancer. Mol Cancer Ther. 2017;16:1412–1420. doi: 10.1158/1535-7163.Mct-17-0061. [DOI] [PubMed] [Google Scholar]
- 49.Kodahl AR, et al. Correlation between circulating cell-free PIK3CA tumor DNA levels and treatment response in patients with PIK3CA-mutated metastatic breast cancer. Mol Oncol. 2018;12:925–935. doi: 10.1002/1878-0261.12305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Juric D et al. (2019) Abstract GS3-08: Alpelisib + fulvestrant for advanced breast cancer: subgroup analyses from the phase III SOLAR-1 trial. Cancer Res 79, GS3-08. doi:10.1158/1538-7445.Sabcs18-gs3-08
- 51.Ciruelos EM et al. (2021) Abstract PD2-06: Clinical outcomes of alpelisib plus fulvestrant in hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer with PIK3CA alterations detected in plasma ctDNA by next-generation sequencing: Biomarker analysis from the SOLAR-1 study. Cancer Res 81: PD2-06. doi:10.1158/1538-7445.
- 52.Juric D, et al. Abstract P4–10-04: Clinical outcomes of alpelisib in hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer by next-generation sequencing-detected PIK3CA alteration status and phosphatase and tensin homolog loss: Biomarker analysis from the SOLAR-1 study. Can Res. 2020;80:P4–10. doi: 10.1158/1538-7445. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.