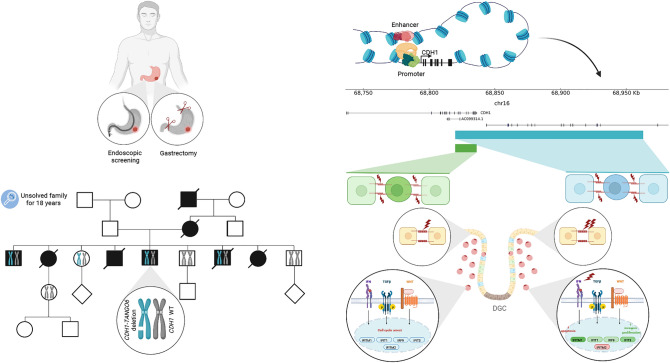
Abstract
Background
Germline CDH1 pathogenic or likely pathogenic variants cause hereditary diffuse gastric cancer (HDGC). Once a genetic cause is identified, stomachs’ and breasts’ surveillance and/or prophylactic surgery is offered to asymptomatic CDH1 carriers, which is life-saving. Herein, we characterized an inherited mechanism responsible for extremely early-onset gastric cancer and atypical HDGC high penetrance.
Methods
Whole-exome sequencing (WES) re-analysis was performed in an unsolved HDGC family. Accessible chromatin and CDH1 promoter interactors were evaluated in normal stomach by ATAC-seq and 4C-seq, and functional analysis was performed using CRISPR-Cas9, RNA-seq and pathway analysis.
Results
We identified a germline heterozygous 23 Kb CDH1-TANGO6 deletion in a family with eight diffuse gastric cancers, six before age 30. Atypical HDGC high penetrance and young cancer-onset argued towards a role for the deleted region downstream of CDH1, which we proved to present accessible chromatin, and CDH1 promoter interactors in normal stomach. CRISPR-Cas9 edited cells mimicking the CDH1-TANGO6 deletion display the strongest CDH1 mRNA downregulation, more impacted adhesion-associated, type-I interferon immune-associated and oncogenic signalling pathways, compared to wild-type or CDH1-deleted cells. This finding solved an 18-year family odyssey and engaged carrier family members in a cancer prevention pathway of care.
Conclusion
In this work, we demonstrated that regulatory elements lying down-stream of CDH1 are part of a chromatin network that control CDH1 expression and influence cell transcriptome and associated signalling pathways, likely explaining high disease penetrance and very young cancer-onset. This study highlights the importance of incorporating scientific–technological updates and clinical guidelines in routine diagnosis, given their impact in timely genetic diagnosis and disease prevention.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s10120-023-01395-0.
Keywords: CDH1, Hereditary diffuse gastric cancer, Regulatory elements, Copy number variants, Deletion, Type-I interferon immune response
Introduction
Pathogenic and likely pathogenic germline truncating single-nucleotide (SNVs) and copy number variants (CNVs) affecting the CDH1 coding sequence [1–3], as well as, truncating CTNNA1 SNVs [4, 5] cause hereditary diffuse gastric cancer (HDGC) syndrome [3, 5, 6]. CDH1 and CTNNA1 genes encode epithelial adherens junctions proteins, E-cadherin and αE-catenin respectively [7, 8], whose loss of function reduces cell–cell adhesion and increases invasiveness and cell survival, amongst other consequences [7, 9]. HDGC is autosomal-dominant and predisposes to diffuse gastric cancer (DGC) and lobular breast cancer (LBC) [1, 3].
In CDH1-associated HDGC, the cumulative DGC incidence ranges from 42 to 72% in males [10, 11], and from 33 to 55% in females, whilst the risk of LBC ranges from 39 to 55% in females, by 80 years of age [10–13]. Even taking into account that estimates are likely to be biassed, either due to patient geographical origin or due to clinical ascertainment [11], the risk of developing DGC or LBC remains high across different cohorts and studies [3]. Individuals with suspected HDGC are identified and referred for genetic testing, according to guidelines and clinical criteria, which have been evolving since 1999 (supplementary Fig. 1) [14–17], based on technological advances and recognition of novel HDGC-related clinical presentations [3]. Identification of a CDH1 variant in the proband, classified as pathogenic or likely pathogenic, according to the American College of Medical Genetics and Genomics and the Association for Molecular Pathology (ACMG/AMP) variant curation guidelines for CDH1 germline variants [18], leads to cascade genetic testing in other family members [17]. Once carrier status is confirmed in asymptomatic relatives, endoscopic surveillance and/or prophylactic surgery are offered according to guidelines to prevent DGC in both genders and/or LBC in females [17].
Many families fulfilling the 2020 HDGC clinical criteria remain genetically unexplained, despite the use of candidate-gene, whole-exome (WES), and whole-genome (WGS) sequencing approaches [6, 19]. Missing hereditability in such families may derive from screening approaches excluding CNV analysis or by technical limitations still existing in next-generation sequencing (NGS) protocols and analyses pipelines [20, 21]. In addition, missing hereditability may be hidden in regulatory elements or unscreened genomic regions [22]. The 3D chromatin architecture, and enhancers, may drive gene expression across large genomic distances through physical proximity to the promoter [23]. These mechanisms are often cell-type specific and control overall gene expression, including that of important disease-causing genes [24, 25]. In fact, the gene expression profile of some disease-causing genes is controlled by regulatory elements located in gene bodies, intergenic regions in their vicinity, and other regions of the causal-gene topological-associating domain [26]. CNVs or other type of aberrations affecting these regulatory regions, which consequently interfere with the normal 3D chromatin architecture, may lead to misexpression patterns, causing or aggravating disease [26].
Given the advances in knowledge on all above-described layers, retesting families who tested negative prior to a syndrome-specific breakthrough, implementation of relevant technological improvements and integration of well-validated novel disease-causative mechanisms in routine testing are important aspects in diagnosis and management [27–29].
Herein, we wish to characterize a novel inherited mechanism, that is simultaneously disease-causing and likely responsible for extremely early-onset gastric cancer and atypical high penetrance of HDGC.
Materials/subjects and methods
Subjects
A HDGC-suspected family from Mexico was enrolled for meeting the 1999 HDGC clinical criteria [14]. The family presents eight DGC with signet-ring cells. WES of individuals III-1, III-3, III-5, III-6, III-7 and III-9 was available for CNVs re-analysis. Multiplex ligation-dependent probe amplification (MLPA) validation was performed for individuals III-1, III-5, III-6, III-9, IV-1 and IV-3.
Whole-exome sequencing (WES) re-analysis
WES from HDGC-suspected family [19] was re-analyzed and screened for CNVs using three variant callers (ExomeDepth [30], ClinCNV [31] and Manta [32]) in tumour risk syndromes-associated genes (supplementary table 1) and annotated using AnnotSV [33]. Candidate calls were prioritized considering quality scores, calling by more than one caller and visual evaluation using Integrated Genome Viewer (IGV).
Multiplex ligation-dependent probe amplification (MLPA)
Specific CDH1 probes (SALSA MLPA-Probemix P083 CDH1, MRC-Holland) were used to identify gene dosage alterations, as previously described [2, 34] and following manufacturer’s recommendations. Probemix includes 20 CDH1-specific probes, 2 probes in CDH1 flanking genes and 13 reference probes in relatively copy number stable regions. Data analysis was performed using Coffalyser.Net (MRC-Holland).
ATAC-seq and 4C-seq
ATAC-seq data was collected from phase 3 ENCODE project [35]. For 4C-seq, gastric mucosa cells from bariatric surgeries were scraped and enzymatically dissociated. 4C-seq libraries were generated, as previously described [36, 37], digested with DnpII (New England Biolabs) and Csp6I (New England Biolabs) and re-circularized with T4 DNA Ligase (Thermo Fisher Scientific). 4C-seq libraries were PCR-amplified (supplementary Table 2) and sequenced on Illumina HiSeqX technology.
CDH1 interactions on a genome scale were mapped, based on Pipe4C [37] and PeakC [38] with window size 2, alpha fdr 0.1 and minimal distance 500.
Cell culture
MKN74 human gastric cancer cell line was purchased from the Japanese collection of research bioresources cell bank. MKN74 and isogenic clones were cultured in RPMI medium (Gibco), supplemented with 10% foetal bovine serum (Biowest) and 1% penicillin streptomycin (Gibco) and maintained at 37 °C under 5% CO2 humidified atmosphere.
CRISPR-Cas9 editing
sgRNAs were designed to target the family CNV or the CDH1 portion of that CNV using Benchling online platform (supplementary table 3). Individual sgRNAs (Invitrogen) were cloned in LentiCRISPRv2GFP (addgene 82416) or LentiCRISPRv2-mCherry (addgene 99154) vectors using BsmBIv2 (New England Biolabs). Plasmids were transformed into Stbl3 competent cells and colonies were sequenced (supplementary Table 4). Lentiviral particles were produced resourcing to HEK293T cell line with pMD2.G (addgene 12259) and pCMV-dR8.91 (addgene) vectors, following Lipofectamine 3000 manufacture’s protocol (Invitrogen) and collected at 48 h. MKN74 was infected with pairs of lentivirus particles in medium supplemented with 10 μg/μl hexadimethrine bromide (Merck Life Science S.L.U.) for 48 h. Transduced cells were selected for GFP and mCherry positive expression at 7 days post-infection using FACS ARIA (BD Biosciences).
Genotyping of edited clones
gDNA was extracted using NZY Tissue gDNA isolation kit (NZYTech), according to the manufacturers’ protocol. gDNA was amplified using primers flanking the edition sites (supplementary table 4) and Multiplex PCR kit (Qiagen). PCR products were analysed in gel electrophoresis and Sanger sequenced on an ABI-3130 Genetic Analyzer (Applied Biosystems).
CDH1/E-cadherin expression analysis
CDH1 mRNA expression was assessed by qPCR in triplicates. RNA was extracted using mirVana RNA Isolation Kit (Invitrogen), according to manufacturers’ protocol. cDNA was synthesized using SuperScriptII reverse transcriptase (Invitrogen), according to the manufacturers’ protocol. CDH1 mRNA expression was analyzed by qPCR with KAPA PROBE FAST qPCR Master Mix (2X) Kit (Sigma-Aldrich) and probes for CDH1 (Hs.PT.58.3324071, TaqMan) and 18S (custom assay, IDT). Reactions were sequenced on a 7500 Real-Time PCR System (Applied Biosystems). Relative expression was normalized for the endogenous 18S control and quantified using the 2−∆∆Ct method.
E-cadherin expression was assessed by flow cytometry in triplicates. Cells were detached with Versene (Gibco) and blocked with 3% bovine serum albumin–phosphatase-buffered saline. Cells were incubated with primary mouse monoclonal antibody HECD-1 (Invitrogen), washed and incubated with secondary antibody anti-mouse Alexa Fluor 647 (Invitrogen). Fluorescence was measured using FACS ARIA (BD Biosciences) and Flow Jo version 10 software was used to analyse the data.
Statistical analysis
Statistical analysis was performed using GraphPad Prism version 7.00 software (GraphPad Software Inc.). A Student’s t test was used for comparison analysis, assuming equal variance between clones and WT samples. Differences were considered significant when p value < 0.05.
RNA-seq and whole transcriptome sequencing bioinformatics analysis
Duplicated RNA samples collected from control and CRISPR-cas9-edited cell lines were sequenced with TruSeq stranded total RNA with Ribo-Zero Gold library in NovaSeq6000. Unique reads were mapped to GRCh38 human genome using STAR and RSeQC and annotated with USCS GRCh38 (Genecode v36). Batch effect was assessed using a PCA and corrected with ComBat_seq from sba (v.3.44) R package for replicates and samples. Differential expression analysis was performed using Deseq2 (v.1.36.0) R package and canonical transcripts were selected considering |log2fold-change|≥ 1 and corrected p value < 0.05. ClusterProfiler (v.4.4.4) R package was used for enrichment GO terms analysis (q value < 0.05). Statistics were performed using R (v.4.2.0).
Results
The genetic odyssey of a HDGC-suspected family ends with the finding of a CDH1-TANGO6 deletion
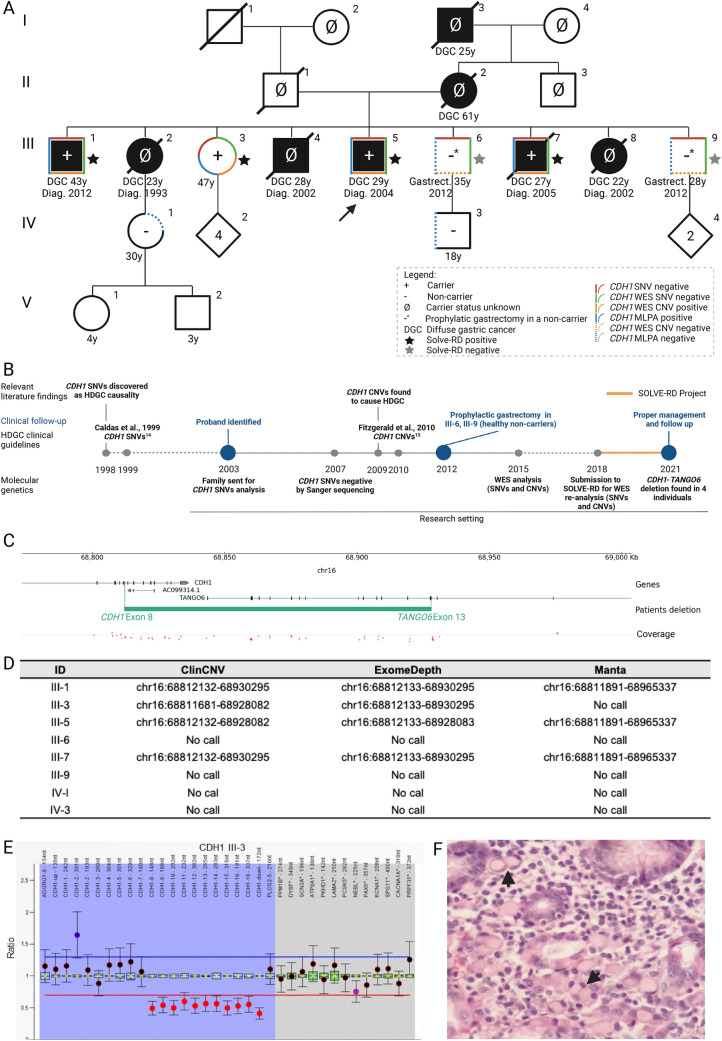
A family of Mexican origin, meeting the 1999 HDGC clinical criteria [14], was identified in 2003 showing five DGC in three consecutive generations, three occurring in the same generation at ages 23 (III-2), 28 (III-4) and 22 (III-8) (Fig. 1A). In addition, individuals III-5 and III-7 also developed DGC at age 29 and 27, in 2004 and 2005, respectively. Given the highly suggestive HDGC pedigree, samples from family members that remained alive (III-1, III-3, III-5, III-6, III-7 and III-9), were all sent out for CDH1 Sanger sequencing, under a research protocol (Fig. 1A, B). The genetic test was negative for the presence of CDH1 SNVs in 2007. Meanwhile, all asymptomatic family members were integrated into a gastric surveillance protocol, similar to that offered to genetically unsolved HDGC families [14]. In 2012, at age 43, individual III-1 developed DGC and underwent gastric surgery with curative intent. Due to the absence of a genetic diagnosis to find individuals at higher risk of DGC, the clinical team decided to offer (presumably) prophylactic total gastrectomy to asymptomatic individuals III-6 and III-9 (Fig. 1A).
Fig. 1.
Diagnostic odyssey, pedigree, variant calling details and cancer histology of a HDGC family carrying a causative CDH1 CNV. A Current family pedigree. Full black symbols: individuals with confirmed DGC; Red outline: negative for SNVs (Sanger sequencing); green outline: negative for SNVs (WES); orange outline: positive for CNVs (WES); blue outline: MLPA-positive for the CDH1 deletion; traced orange outline: negative for CNVs (WES); traced blue outline: MLPA-negative for the CDH1 deletion. All family members, submitted to multiple genetic tests herein described, signed informed consents. The research work has been approved by the Ethics Committee of Centro Hospitalar Universitário São João, in Porto, Portugal, in the frame of the Solve-RD project with the reference CHUSJ_445/2020; B Timeline of events and diagnostic odyssey of the family; C CDH1 CNV found by WES, encompassing part of the CDH1 and TANGO6 genes (represented in green) and IGV coverage; Green text represents first and last edited exons; D Detected CDH1 CNV in ClinCNV [31], ExomeDepth [30] and Manta [32]; E MLPA analysis performed using SALSA MLPA-Probemix P083 CDH1 (MRC Holland) in patient III-3; Ratio above blue line indicates increased copy number, whilst ratio below red line indicates reduced copy number; Blue highlight represents CDH1 probes and 2 neighbour genes; Grey highlight represents reference probes; F Haematoxylin and eosin staining of DGC depicting signet-ring cells (arrow heads) (100 × magnification) from the proband III-5
Although, CDH1 CNVs were first described as a cause of HDGC in 2009 [2], and integrated in HDGC clinical guidelines in 2010 [15], samples from this family were not tested by MLPA. Samples were rather submitted to WES in 2015, again as part of a research protocol, and analysed for SNVs and CNVs which result turned out negative (Fig. 1B), likely due to technical limitations in WES analysis or sample/sequencing quality [39].
WES re-analysis led to the identification of a heterozygous deletion involving half of the CDH1 gene (starting downstream of CDH1 exon 7) and the downstream TANGO6 gene (ending upstream of TANGO6 exon 14) (Fig. 1B–D). This CNV was identified by several variant callers, namely ‘ClinCNV’ [31], ‘Manta’ [32] and ‘ExomeDepth’ [30] (Fig. 1C,D), and supported by Integrative Genomics Viewer (IGV) visual inspection [40], despite the low quality of the sequencing data (Fig. 1C). The CNV was after validated by MLPA (Fig. 1E) in individuals III-1, III-3, III-5 and III-7 (Fig. 1A). Individuals III-6 and III-9, who had opted for risk-reducing gastrectomy, were actually non-carriers of the CNV (Fig. 1A).
As the pedigree evidences (Fig. 1A), three out of four carriers in generation III, developed DGC with signet-ring cells at ages 27, 29 and 43 (Fig. 1F). Individual III-3 is the only carrier to our knowledge that remains asymptomatic at the age of 47. Besides these family members, there are five not-tested DGC patients, two (I-3 and II-2) died from DGC at ages 25 and 61, being in principle obligated carriers. There are three other relatives (III-2, III-4 and III-8) who died of DGC at age 23, 28 and 22 and were likely also carriers of this predisposing CNV (Fig. 1A). After the genetic diagnosis, a blood sample was obtained from individuals IV-1 and IV-3, who were old enough to be tested, and the CNV screening performed by MLPA returned a negative result for both (Fig. 1A).
Chromatin conformation analysis highlights physical interactions between the CDH1 promoter and sequences within the deleted region in normal stomach
Although highly penetrant, HDGC caused by CDH1 pathogenic or likely pathogenic variants is rarely as penetrant as in this family, which presents an uncommonly high number of individuals in a single generation affected by DGC (six out of seven carriers/presumed carriers). Additionally, five out of six developed DGC before age 29, and average age of cancer onset was 29 ± 7.55 years in six carriers/presumed carriers from generation III. We therefore hypothesised that sequences within the deleted region, and extending into the downstream intergenic region, and the neighbouring TANGO6 gene sequence, would affect CDH1 expression and the locus 3D chromatin structure more strongly, than deletion of the CDH1 alone.
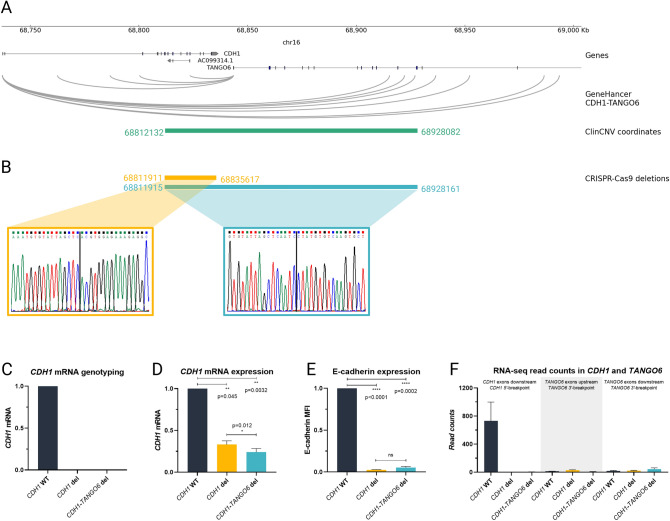
To address this hypothesis, we mined the GeneHancer database [41] and found that several enhancers within the deleted region, interact with either TANGO6 or CDH1 promoters (Fig. 2A). Additionally, we used normal epithelial cells from bariatric surgeries to profile CDH1 promoter interactions using 4C-seq, and analysed accessible chromatin regions using ATAC-seq from the ENCODE project [35]. We found that the CDH1 promoter interacts not only with downstream sequences within CDH1, but also TANGO6 and the intergenic region, all deleted by the family CNV, and displaying regions of accessible chromatin (Fig. 2A). These results suggest a role of CDH1/TANGO6 co-regulation in stomach, besides gene physical proximity.
Fig. 2.
CDH1 and TANGO6 regulatory network and characterization of CRISPR-cas9-edited clones. A CDH1/TANGO6 potential enhancers depicted in GeneHancer, accessible chromatin and CDH1 promotor interactors in normal stomach tissue; B Sanger sequencing genotyping of the CNV breakpoints, and deletion coordinates in CDH1 and CDH1-TANGO6 CRISPR-cas9-edited clones; C mRNA genotyping of CDH1 and CDH1-TANGO6-edited clones resourcing to a probe located in the deleted CDH1 region; D CDH1 mRNA expression of CDH1- and CDH1-TANGO6-edited clones resourcing to a probe located in exons 6–7 (Hs.PT.58.3324071, TaqMan); E E-cadherin protein expression of CDH1- and CDH1-TANGO6-edited clones measured by flow cytometry (monoclonal antibody HECD-1, Invitrogen, 1:100 dilution; and mouse Alexa Fluor 647, Invitrogen, 1:500 dilution); F RNA-seq expression read counts of CDH1 and TANGO6 genes. Data are represented as the mean ± SEM, MFI: median fluorescence intensity. Experiments depicted in panels C–E were performed in triplicates and differences considered statistically significant if p value < 0.05 in a t-test (details in Materials and Methods)
Deletion of CDH1 and downstream regulatory sequences, present within the HDGC causal CNV, strongly impact CDH1 expression and specific signalling pathways
To understand the impact of deleting several CDH1 promoter-interacting sequences, beyond the CDH1 locus, we CRISPR-Cas9 deleted either the CDH1 portion of the CNV, or mimicked the family CNV. This was performed in a human gastric cancer cell line bearing normal CDH1/E-cadherin expression and function [42]. We used sgRNAs flanking target regions (supplementary Table 3), and successful editing was confirmed by Sanger sequencing and qRT-PCR (Fig. 2B,C). A clone bearing a 23.705 bp homozygous deletion spanning from CDH1 intron 7 and extending till exon 16 (chr16:68,811,911–68,835,617, hg38) and a clone bearing a 116.245 bp homozygous deletion starting in CDH1 intron 7 and ending after TANGO6 intron 13 (chr16:68,811,915–68,928,161, hg38) were produced (Fig. 2B).
We found that both CDH1 and CDH1-TANGO6 deletions led to a significant decrease in CDH1 mRNA compared to the wild-type (WT) control (p value = 0.045 and p value = 0.0032, respectively), being this downregulation stronger for the CDH1-TANGO6 deletion as compared to the CDH1 deletion alone (p value = 0.012) (Fig. 2D). The level of protein downregulation in relation to the WT control was equivalent in both deletion clones, as assessed by flow cytometry (p value < 0.0001 and p value = 0.0002, respectively) (Fig. 2E).
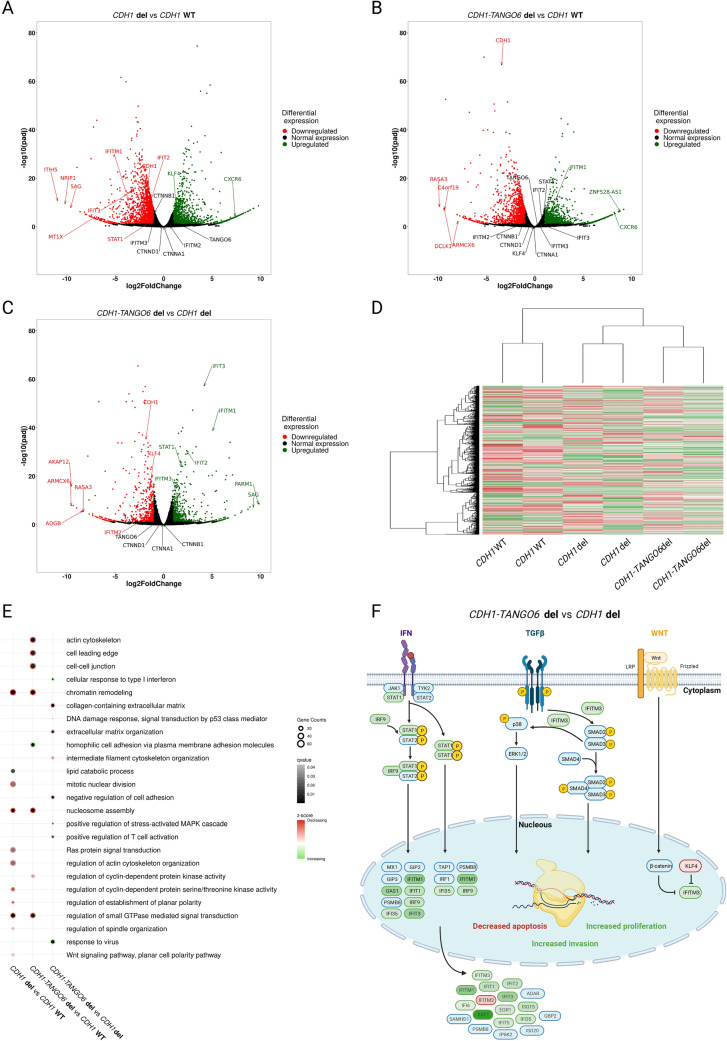
We next explored the effect of both deletions in the cells’ transcriptomes. For this, we evaluated the differential transcriptome between CDH1 or CDH1-TANGO6 deletion clones and WT cells, and between CDH1-TANGO6 and CDH1 deletion clones. We confirmed that CDH1 mRNA expression was more strongly downregulated (3.4-fold) in the CDH1-TANGO6 deletion than in the CDH1 deletion alone (1.6-fold), when each was compared to CDH1 mRNA expression in WT cells (p value < 0.0001) (Figs. 2F, 3A,B). We also verified that TANGO6 is not expressed in WT cells, and its expression remains, as expected, unchanged after the 5’-deletion of the TANGO6 gene (Fig. 2F).
Fig. 3.
Genome-wide transcriptome regulation of CDH1 CNV and CDH1-TANGO6 CNV. A) Volcano-plot illustrating CDH1 CNV differentially expressed genes with CDH1 WT; B Volcano-plot illustrating CDH1-TANGO6 CNV differentially expressed genes with CDH1 WT; C Volcano-plot illustrating CDH1-TANGO6 CNV differentially expressed genes with CDH1 CNV; D Heatmap illustrating genome-wide differentially expressed genes; E Selected genome-wide transcriptomic misregulated pathways of CDH1-TANGO6 CNV and CDH1 CNV. Grey scale represents q value and red–green scale represents z-score (red for pathways with mainly downregulated genes, white for equally downregulated and upregulated genes and green for pathways with mainly upregulated genes); F Response to interferon, TGF-β and Wnt signalling pathways misregulated in CDH1-TANGO6 CNV vs CDH1 CNV, created with BioRender.com. Red–green scale represents down- to upregulated genes and blue represents normal expressed genes
We also observed that the global patterns of differentially expressed genes clearly differ between clones bearing the CDH1 deletion alone, the CDH1-TANGO6 deletion and the CDH1 WT cells (Fig. 3D). There were 3,272 differentially expressed genes between the CDH1-deleted clone and CDH1 WT cells; 2 492 differentially expressed genes between the CDH1-TANGO6-deleted clone and CDH1 WT cells; and 1,102 differentially expressed genes between the CDH1-TANGO6 and the CDH1-deleted clones (supplementary Tables 5,6,7). Gene ontology analysis further revealed that ‘nucleosome assembly’, ‘chromatin remodelling’, ‘regulation of small GTPase-mediated signal transduction’ as well as pathways related to cyclin-dependent protein kinase activity and actin cytoskeleton were impacted in both in CDH1 alone and in the CDH1-TANGO6 deletion clones (Fig. 3E, supplementary Table 8).
CDH1 deletion exclusive events encompass downregulation of ‘mitotic nuclear division’, ‘regulation of spindle organization’ and planar polarity-related pathways, as well as upregulation of ‘lipid catabolic processes’, amongst others (Fig. 3E). Events exclusively altered in the CDH1-TANGO6 deletion clone include mainly impairment of adhesion- and cytoskeleton-associated pathways (Fig. 3E).
The direct comparison between the transcriptional profile of clones bearing the CDH1-TANGO6 deletion and the CDH1 deletion alone emphasises the negative impact in adhesion-, p53-mediated DNA damage response and cytoskeleton-associated pathways and the positive impact in MAPK cascade-associated proliferation, presented by the CDH1-TANGO6 deletion clone (Fig. 3E). Additional positive and negative impacts were found in several immune-associated pathways, namely related to ‘cellular response to type I interferon’ and ‘response to virus’ in the CDH1-TANGO6 deletion clone (Fig. 3E,F). The expression of IFITM genes (IFITM1, IFITM3 and IFITM2) and some of their upstream regulators (STAT1 and KLF4) was particularly affected in the CDH1-TANGO6 deletion clone (Fig. 3C,F).
Altogether, and upon data integration, cancer cells arising in patients bearing the CDH1-TANGO6 deletion are likely to present decreased apoptosis, increased proliferation and invasion, and no cell cycle arrest (Fig. 3F).
Discussion
In this work, we demonstrated that regulatory elements, lying within the CDH1-TANGO6 deletion, are part of a chromatin network that, not only control CDH1 expression, but also influence the cell transcriptome and associated signalling pathways. We believe this finding explains the high penetrance and extremely young age of cancer-onset in patients from this family. In addition, we describe the diagnostic odyssey of a HDGC-suspected family, that remained genetically unsolved for 18 years, to highlight the importance of incorporating scientific–technological updates and clinical guidelines in routine diagnosis. This is expected to ultimately improve clinical management and care of hereditary cancer patients and families.
A distinctive feature of this family, in comparison with other CDH1-positive HDGC families, is the high disease penetrance reflected in generation III, with six out of seven individuals being CNV carriers or presumed carriers and developing DGC. The other is the very early age of cancer-onset (29 ± 7.55 years), compared to the average age reported for DGC in most HDGC families (46,7 ± 16,6 years) [3, 11, 43]. In accordance with our findings, an additional HDGC family has been reported in the literature, bearing a 275 kb CDH1 (exon 7) and TANGO6 (full gene) deletion, with four patients in four consecutive generations affected by gastric cancer, from which two were confirmed with signet ring cell carcinoma at ages 30 and 34 years old [44]. These features prompted the next question of this study, which was related to a potential role for the downstream intergenic region and the neighbouring TANGO6 gene sequence, in modulating CDH1 expression, the CDH1 locus 3D chromatin structure and the transcriptome, in aggravating the clinical presentation in this family.
CRISPR-Cas9 clones mimicking the CDH1 deletion alone or the family full CNV, demonstrated that the full CNV was more efficient in promoting downregulation of CDH1 mRNA than the CDH1 deletion alone. This finding supports the existence of regulatory DNA elements controlling CDH1 expression, downstream of the CDH1 locus, and enclosed in the CNV, which we have also proved to interact with the CDH1 promoter. Similar findings have been previously reported, as for instance those related to neighbour murine Shh and Lmbr1 genes, in which the ZRS enhancer located within the Lmbr1 gene, regulates Shh gene expression [45].
Our data on differential expression and gene ontology analysis of RNA-seq data from clones mimicking the CDH1 deletion alone or the family full CNV in comparison to CDH1 WT, highlighted shared biological terms between these clones related to cell division and spindle orientation, and involving chromatin remodelling. These terms have been previously associated with E-cadherin function [46–49], and likely represent the contribution of the CDH1 locus deletion itself to the differential transcriptome. The fact that adhesion-related (‘negative regulation of cell adhesion’) and actin-related pathways (‘actin cytoskeleton’) were found specifically downregulated upon CDH1-TANGO6 deletion, and in comparison, to CDH1-deleted cells, suggests that the perturbations induced by the CDH1-TANGO6 deletion are greater than those induced by the CDH1 deletion alone in these pathways. Although it is widely known that E-cadherin deficiency leads to weaker adhesion and abnormal microtubule organization [50, 51], which may be involved in migration upon malignant transformation [52], our data specifically link homophilic cell adhesion to the longer CDH1-TANGO6 deletion. Indeed, this clone presents upregulation of the pathway ‘homophilic cell adhesion via plasma membrane adhesion molecules’, which may reflect a yet to prove adhesion-compensatory mechanism, promoted by upregulation of protocadherins in the absence of E-cadherin, seen in our data. A similar switch has been previously demonstrated in lobular breast cancer, whereby E-cadherin is replaced by P-cadherin in cancer cells [53].
Immune-associated genes and pathways were particularly altered in the CDH1-TANGO6 deletion clone, comparing to the clone bearing the CDH1 deletion alone. IFITM1 gene was particularly overexpressed in the CDH1-TANGO6 deletion, a feature previously observed in several aggressive tumours, and shown to enhance tumour proliferation and invasion [42, 54]. In gastroesophageal tumours, for instance, IFITM1 was found overexpressed comparing to normal adjacent tissues [55–57]. Additionally, IFITM3 overexpression has been shown to promote gastric cancer growth, through the activation of several pathways, resulting in increased proliferation, invasion, and metastasis [58, 59]. IFITM3 has been shown to promote SMAD2/3/4 phosphorylation and interact with STAT1/2 and PIP3/PI3K. This results in activation of downstream proliferation and oncogenic signalling pathways, such as p38/MAPK [60], JAK/STAT [61–63] and PI3K [64]. IFITM3 plays an additional role in the Wnt signalling pathway, being regulated by β-catenin [58] and KLF4 [65]. Indeed, KLF4, known to promote IFITM3 transcriptional inhibition [65], is downregulated in the CDH1-TANGO6 deletion clone providing a rationale for the IFITM3 upregulation seen in this clone. Finally, IFITM2, a pro-apoptotic gene, encoding a protein which has been linked to protection against tumour proliferation [66], was found downregulated in the CDH1-TANGO6 deletion clone. Most abovementioned observations support a relationship between IFITM proteins misregulation and the severe phenotype, observed in this HDGC family. Increased proliferation associated with MAPK cascade [67, 68] and decreased DNA damage response mediated by p53 signalling [69–71], also found in cells bearing the family CNV, may hint to more aggressive DGC, than the commonly described for HDGC patients bearing CDH1-coding variants, who generally die by dissemination to the peritoneum [43]. Supporting this, is the fact that individual III-4 died at age 28 with DGC metastases in the lung and bone, a very unusual event in patients with coding CDH1 variants.
Our data also seem to indicate that whilst a role has not been found for the TANGO6 mRNA itself, the genomic sequences downstream of the CDH1 locus, and within the sequence deleted in this family, host positive regulators of CDH1 expression. This would explain not only the lower CDH1 mRNA expression, but also the wider transcriptional and signalling impact, which likely contribute to the clinical presentation in this family. Only two HDGC families have been reported to carry large deletions involving specifically CDH1 and TANGO6, both presenting early-onset DGC and gastric cancer in consecutive generations [44]. Therefore, evidence for the role of regulatory CDH1 downstream regions in early-onset DGC is still limited. Yet, this study provides the first evidence on the role of CDH1 downstream sequences in CDH1 expression control, which may apply to other HDGC families harbouring similar deletions.
This HDGC-suspected family with a severe clinical presentation was somehow lost in genetic follow-up, as clinical standardized diagnostic tests were never used in their clinical path. Negative tests for CDH1-coding SNVs and CNVs, both were obtained from research projects, although management guidelines proposed formal genetic testing with Sanger sequencing and MLPA since 2010 in families fulfilling HDGC clinical criteria [2, 15]. This highlights the risk associated with molecular diagnosis being performed in the frame of research studies, rather than in proper diagnostic and clinically approved laboratories, as part of patients’ and families’ pathways of care. In this case, the most obvious disease gene/causing mechanism was overlooked, as for research purposes, these should have been, in principle, excluded. The result of a WES data re-analysis 18 years after clinical diagnosis, revealed a CNV involving the most obvious HDGC-causing gene, CDH1 [1, 3, 6, 17]. This case highlights the importance of revisiting, re-contacting and retesting families fulfilling clinical criteria but lacking molecular diagnosis, in diagnostic and clinically approved laboratories, as novel and consolidated genetic susceptibility causes of hereditary syndromes arise, and both technologies and management guidelines evolve.
This family odyssey also illustrates the importance of timely genetic diagnosis. For this family, the greater consequences included two carriers developing cancer, two and nine years after the family was recognized clinically as an HDGC-suspected family; and two non-carrier siblings undergoing unnecessary and presumably prophylactic total gastrectomy, nine years after clinical diagnosis, based on family history only. The Solve-RD project (https://solve-rd.eu/) emerged in this case, as a solution to overcome an unfortunate diagnostic odyssey. The finding of a causal CDH1 CNV was reported to the clinical team at the hospital of origin in 2021, which promoted re-engagement with the family and effective genetic counselling and disease risk management.
Taken together, our data suggest that patients bearing the CDH1-TANGO6 deletion develop high penetrance, earlier-onset and possibly more aggressive DGC, than patients with pathogenic variants confined to the CDH1, likely due to the impairment of regulatory elements able to activate oncogenic signalling pathways. It also illustrates the inequities and complexities of genetic diagnosis and integrated care in rare diseases, in a rapidly evolving field, which can impact timely genetic diagnosis and disease prevention.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
This work was supported by: (1) the European Reference Network on Genetic Tumour Risk Syndromes (ERN GENTURIS) – Project ID No. 739547. ERN GENTURIS is partly co-funded by the European Union within the framework of the Third Health Programme “ERN-2016 – Framework Partnership Agreement 2017–2021”; (2) Doctoral Programme in Biomedicine—Faculty of Medicine, University of Porto, to JG-P and CSJ; (3) Doctoral Programme in Computational Sciences, Faculty of Sciences, University of Porto, to MF; (4) Masters in Molecular Oncology, Faculty of Medicine, University of Porto, to NM. The team would also like to thank Nuno Alves from i3S for fruitful discussions on immuno-oncology; Sónia Sousa, Cátia Pereira and Luis Cirnes from Ipatimup Diagnostics, Porto-Portugal for MLPA analysis.
Solve-RD DITF-GENTURIS: Stefan Aretz1,2, Gabriel Capella3, Sérgio Castedo4-7, Richarda M. de Voer8,9, Gareth Evans10, Susana Fernandes11, José Garcia-Pelaez4,5,12, Luzia Garrido7, Elke Holinski-Feder13,14, Nicoline Hoogerbrugge8,9, David Huntsman15,16, Arne Jahn17,18, C. Marleen Kets8,9, Andreas Laner14, Marjolijn Ligtenberg8,9,19, Andrea Meinhardt17, Arjen Mensenkamp8,9, Carla Oliveira4-6, Sophia Peters1, Isabel Quintana3, Evelin Schröck17,18, Anna Sommer1, Isabel Spier1,2, Liesbeth Spruijt8,9, Verena Steinke-Lange13,14, Iris te Paske8,9, Marc Tischkowitz20, Laura Valle3, Rachel van der Post9,19, Yasmijn van Herwaarden9,21, Wendy van Zelst-Stams8,9, Doreen William17,18. Affiliations: 1.Institute of Human Genetics, Medical Faculty, University of Bonn, Bonn, Germany. 2.Center for Hereditary Tumor Syndromes, University Hospital Bonn, Bonn, Germany. 3.Bellvitge Biomedical Research Institute (IDIBELL), Barcelona, Spain. 4.IPATIMUP - Institute of Molecular Pathology and Immunology of the University of Porto, Portugal. 5.i3S - Instituto de Investigação e Inovação em Saúde, Universidade do Porto, Portugal. 6.Faculty of Medicine, University of Porto, Portugal. 7.CHUSJ, Centro Hospitalar e Universitário de São João, Porto, Portugal. 8.Department of Human Genetics, Radboud University Medical Center, Nijmegen, The Netherlands. 9.Radboud Institute for Molecular Life Sciences, Nijmegen, The Netherlands. 10.Division of Evolution, Infection and Genomics, School of Biological Sciences, Faculty of Biology, Medicine and Health, University of Manchester, Manchester M13 9WL, UK. 11.Departament of Genetics, Faculty of Medicine, University of Porto, Portugal. 12.Doctoral Programme in Biomedicine, Faculty of Medicine, University of Porto, Portugal. 13.Medizinische Klinik und Poliklinik IV – Campus Innenstadt, Klinikum der Universität München, Munich, Germany. 14.MGZ - Medical Genetics Center, Munich, Germany. 15.Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada. 16.British Columbia Cancer Agency, Vancouver, British Columbia, Canada. 17.Institute of Clinical Genetics, University Hospital Carl Gustav Carus, Technical University Dresden, Dresden, Germany. 18.National Center for Tumor Diseases (NCT), Dresden, Germany. 19.Department of Pathology, Radboud University Medical Center, Nijmegen, the Netherlands. 20.Academic Department of Medical Genetics, Cambridge University, Cambridge, UK. 21.Department of Gastroenterology, Radboud University Medical Center, Nijmegen, the Netherlands.
Author contributions
CSJ and JGP contributed equally to the work. CO: supervision, project administration, funding acquisition; CSJ, JGP and CO: conceptualization, formal analysis; MTT, OA, SS, BM-B, MLO-S and MR-T: acquisition of clinical and pathological data; CSJ, JGP, AA and NM: experimental work; MF, GD and SL: bioinformatic analysis; Solve-RD DITF-GENTURIS, AS, IteP, CC, MT, NH and RMdeV: integration of genetics and clinical data; all authors: writing, review, editing and approval of the manuscript.
Funding
This work received funding from: (1) the European Union’s Horizon 2020 research and innovation programme under grant agreement No 779257 (Solve‐RD: H2020-SC1-2017-Single-Stage-RTD), which provided Ph.D. salaries to JG-P, AS, ItP; (2) The European Regional Development Fund (ERDF) through the COMPETE 2020-Operacional Programme for Competitiveness and Internationalisation (POCI), Portugal 2020, and by Portuguese funds through Fundação para a Ciência e Tecnologia (FCT)/Ministério da Ciência, Tecnologia e Inovação (LEGOH: PTDC/BTM-TEC/6706/2020); (3) FCT with salary support for Ph.D. studentships to CSJ (Ref. SFRH/BD/140796/2018), JG-P (Ref. 2022.11952.BD) and MF (Ref. 2020.05763.BD); (4) MT is supported by the NIHR Cambridge Biomedical Research Centre (BRC-1215-20014). Open access funding provided by FCT|FCCN (b-on).
Data availability
Raw data supporting the findings of this study are available upon request from the corresponding author. Processed data are available in supplementary materials.
Declarations
Conflcit of interest
The authors disclose no conflict of interest.
Footnotes
The collaborators names associated with Solve-RD DITF-GENTURIS are given in acknowledgements sections.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Celina São José and José Garcia-Pelaez contributed equally to this work.
Contributor Information
Carla Oliveira, Email: carlaol@i3s.up.pt.
Solve-RD DITF-GENTURIS:
Stefan Aretz, Gabriel Capella, Sérgio Castedo, Richarda M. de Voer, Gareth Evans, Susana Fernandes, José Garcia-Pelaez, Luzia Garrido, Elke Holinski-Feder, Nicoline Hoogerbrugge, David Huntsman, Arne Jahn, C. Marleen Kets, Andreas Laner, Marjolijn Ligtenberg, Andrea Meinhardt, Arjen Mensenkamp, Carla Oliveira, Sophia Peters, Isabel Quintana, Evelin Schröck, Anna Sommer, Isabel Spier, Liesbeth Spruijt, Verena Steinke-Lange, Iris te Paske, Marc Tischkowitz, Laura Valle, Rachel van der Post, Yasmijn van Herwaarden, Wendy van Zelst-Stams, and Doreen William
References
- 1.Guilford P, Hopkins J, Harraway J, McLeod M, McLeod N, Harawira P, et al. E-cadherin germline mutations in familial gastric cancer. Nature. 1998;392(6674):402–405. doi: 10.1038/32918. [DOI] [PubMed] [Google Scholar]
- 2.Oliveira C, Senz J, Kaurah P, Pinheiro H, Sanges R, Haegert A, et al. Germline CDH1 deletions in hereditary diffuse gastric cancer families. Hum Mol Genet. 2009;18(9):1545–1555. doi: 10.1093/hmg/ddp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garcia-Pelaez J, Barbosa-Matos R, Lobo S, Dias A, Garrido L, Castedo S, et al. Genotype-first approach to identify associations between CDH1 germline variants and cancer phenotypes: a multicentre study by the European reference network on genetic tumour risk syndromes. Lancet Oncol. 2022 doi: 10.1016/s1470-2045(22)00643-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Majewski IJ, Kluijt I, Cats A, Scerri TS, de Jong D, Kluin RJ, et al. An α-E-catenin (CTNNA1) mutation in hereditary diffuse gastric cancer. J Pathol. 2013;229(4):621–629. doi: 10.1002/path.4152. [DOI] [PubMed] [Google Scholar]
- 5.Lobo S, Benusiglio PR, Coulet F, Boussemart L, Golmard L, Spier I, et al. Cancer predisposition and germline CTNNA1 variants. Eur J Med Genet. 2021;64(10):104316. doi: 10.1016/j.ejmg.2021.104316. [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Pelaez J, Barbosa-Matos R, São José C, Sousa S, Gullo I, Hoogerbrugge N, et al. Gastric cancer genetic predisposition and clinical presentations: Established heritable causes and potential candidate genes. Eur J Med Genet. 2022;65(1):104401. doi: 10.1016/j.ejmg.2021.104401. [DOI] [PubMed] [Google Scholar]
- 7.Harris TJ, Tepass U. Adherens junctions: from molecules to morphogenesis. Nat Rev Mol Cell Biol. 2010;11(7):502–514. doi: 10.1038/nrm2927. [DOI] [PubMed] [Google Scholar]
- 8.Takeichi M. Multiple functions of α-catenin beyond cell adhesion regulation. Curr Opin Cell Biol. 2018;54:24–29. doi: 10.1016/j.ceb.2018.02.014. [DOI] [PubMed] [Google Scholar]
- 9.Figueiredo J, Mercadillo F, Melo S, Barroso A, Gonçalves M, Díaz-Tasende J, et al. Germline CDH1 G212E missense variant: combining clinical, in vitro and in vivo strategies to unravel disease burden. Cancers (Basel). 2021 doi: 10.3390/cancers13174359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, et al. Hereditary diffuse gastric cancer syndrome: CDH1 mutations and beyond. JAMA Oncol. 2015;1(1):23–32. doi: 10.1001/jamaoncol.2014.168. [DOI] [PubMed] [Google Scholar]
- 11.Roberts ME, Ranola JMO, Marshall ML, Susswein LR, Graceffo S, Bohnert K, et al. Comparison of CDH1 penetrance estimates in clinically ascertained families vs families ascertained for multiple gastric cancers. JAMA Oncol. 2019;5(9):1325–1331. doi: 10.1001/jamaoncol.2019.1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pharoah PD, Guilford P, Caldas C. Incidence of gastric cancer and breast cancer in CDH1 (E-cadherin) mutation carriers from hereditary diffuse gastric cancer families. Gastroenterology. 2001;121(6):1348–1353. doi: 10.1053/gast.2001. [DOI] [PubMed] [Google Scholar]
- 13.Xicola RM, Li S, Rodriguez N, Reinecke P, Karam R, Speare V, et al. Clinical features and cancer risk in families with pathogenic CDH1 variants irrespective of clinical criteria. J Med Genet. 2019;56(12):838–843. doi: 10.1136/jmedgenet-2019-105991. [DOI] [PubMed] [Google Scholar]
- 14.Caldas C, Carneiro F, Lynch HT, Yokota J, Wiesner GL, Powell SM, et al. Familial gastric cancer: overview and guidelines for management. J Med Genet. 1999;36(12):873–880. [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald RC, Hardwick R, Huntsman D, Carneiro F, Guilford P, Blair V, et al. Hereditary diffuse gastric cancer: updated consensus guidelines for clinical management and directions for future research. J Med Genet. 2010;47(7):436–444. doi: 10.1136/jmg.2009.074237.PubMedPMID:20591882;PubMedCentralPMCID:PMC2991043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Post RS, Vogelaar IP, Carneiro F, Guilford P, Huntsman D, Hoogerbrugge N, et al. Hereditary diffuse gastric cancer: updated clinical guidelines with an emphasis on germline CDH1 mutation carriers. J Med Genet. 2015;52(6):361–374. doi: 10.1136/jmedgenet-2015-103094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blair VR, McLeod M, Carneiro F, Coit DG, D'Addario JL, van Dieren JM, et al. Hereditary diffuse gastric cancer: updated clinical practice guidelines. Lancet Oncol. 2020;21(8):e386–e397. doi: 10.1016/s1470-2045(20)30219-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee K, Krempely K, Roberts ME, Anderson MJ, Carneiro F, Chao E, et al. Specifications of the ACMG/AMP variant curation guidelines for the analysis of germline CDH1 sequence variants. Hum Mutat. 2018;39(11):1553–1568. doi: 10.1002/humu.23650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sommer AK, Te Paske I, Garcia-Pelaez J, Laner A, Holinski-Feder E, Steinke-Lange V, et al. Solving the genetic aetiology of hereditary gastrointestinal tumour syndromes- a collaborative multicentre endeavour within the project Solve-RD. Eur J Med Genet. 2022;65(5):104475. doi: 10.1016/j.ejmg.2022.104475. [DOI] [PubMed] [Google Scholar]
- 20.Kamps R, Brandão RD, Bosch BJ, Paulussen AD, Xanthoulea S, Blok MJ, et al. Next-generation sequencing in oncology: genetic diagnosis, risk prediction and cancer classification. Int J Mol Sci. 2017 doi: 10.3390/ijms18020308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vos JR, Giepmans L, Röhl C, Geverink N, Hoogerbrugge N. Boosting care and knowledge about hereditary cancer: European reference network on genetic tumour risk syndromes. Fam Cancer. 2019;18(2):281–284. doi: 10.1007/s10689-018-0110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Allou L, Balzano S, Magg A, Quinodoz M, Royer-Bertrand B, Schöpflin R, et al. Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature. 2021;592(7852):93–98. doi: 10.1038/s41586-021-03208-9. [DOI] [PubMed] [Google Scholar]
- 23.Andrey G, Mundlos S. The three-dimensional genome: regulating gene expression during pluripotency and development. Development. 2017;144(20):3646–3658. doi: 10.1242/dev.148304. [DOI] [PubMed] [Google Scholar]
- 24.Cannavò E, Khoueiry P, Garfield DA, Geeleher P, Zichner T, Gustafson EH, et al. Shadow enhancers are pervasive features of developmental regulatory networks. Curr Biol. 2016;26(1):38–51. doi: 10.1016/j.cub.2015.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kragesteen BK, Spielmann M, Paliou C, Heinrich V, Schöpflin R, Esposito A, et al. Dynamic 3D chromatin architecture contributes to enhancer specificity and limb morphogenesis. Nat Genet. 2018;50(10):1463–1473. doi: 10.1038/s41588-018-0221-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Will AJ, Cova G, Osterwalder M, Chan WL, Wittler L, Brieske N, et al. Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog) Nat Genet. 2017;49(10):1539–1545. doi: 10.1038/ng.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deignan JL, Chung WK, Kearney HM, Monaghan KG, Rehder CW, Chao EC, et al. Points to consider in the reevaluation and reanalysis of genomictest results: a statement of the American College of Medical Genetics and Genomics(ACMG) Genet Med. 2019;21(6):1267–1270. doi: 10.1038/s41436-019-0478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robertson AJ, Tan NB, Spurdle AB, Metke-Jimenez A, Sullivan C, Waddell N. Re-analysis of genomic data: an overview of the mechanisms and complexities of clinical adoption. Genet Med. 2022;24(4):798–810. doi: 10.1016/j.gim.2021.12.011. [DOI] [PubMed] [Google Scholar]
- 29.Dettwyler SA, Koeppe ES, Jacobs MF, Stoffel EM. Outcomes of retesting in patients with previously uninformative cancer genetics evaluations. Fam Cancer. 2022;21(3):375–385. doi: 10.1007/s10689-021-00276-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plagnol V, Curtis J, Epstein M, Mok KY, Stebbings E, Grigoriadou S, et al. A robust model for read count data in exome sequencing experiments and implications for copy number variant calling. Bioinformatics. 2012;28(21):2747–2754. doi: 10.1093/bioinformatics/bts526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Demidov G, Sturm M, Ossowski S. ClinCNV: multi-sample germline CNV detection in NGS data. bioRxiv. 2022 doi: 10.1101/2022.06.10.495642. [DOI] [Google Scholar]
- 32.Chen X, Schulz-Trieglaff O, Shaw R, Barnes B, Schlesinger F, Källberg M, et al. Manta: rapid detection of structural variants and indels for germline and cancer sequencing applications. Bioinformatics. 2016;32(8):1220–1222. doi: 10.1093/bioinformatics/btv710. [DOI] [PubMed] [Google Scholar]
- 33.Geoffroy V, Herenger Y, Kress A, Stoetzel C, Piton A, Dollfus H, et al. AnnotSV: an integrated tool for structural variations annotation. Bioinformatics. 2018;34(20):3572–3574. doi: 10.1093/bioinformatics/bty304. [DOI] [PubMed] [Google Scholar]
- 34.Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30(12):e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moore JE, Purcaro MJ, Pratt HE, Epstein CB, Shoresh N, Adrian J, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583(7818):699–710. doi: 10.1038/s41586-020-2493-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Werken HJ, de Vree PJ, Splinter E, Holwerda SJ, Klous P, de Wit E, et al. 4C technology: protocols and data analysis. Methods Enzymol. 2012;513:89–112. doi: 10.1016/b978-0-12-391938-0.00004-5. [DOI] [PubMed] [Google Scholar]
- 37.Krijger PHL, Geeven G, Bianchi V, Hilvering CRE, de Laat W. 4C-seq from beginning to end: a detailed protocol for sample preparation and data analysis. Methods. 2020;170:17–32. doi: 10.1016/j.ymeth.2019.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Geeven G, Teunissen H, de Laat W, de Wit E. peakC: a flexible, non-parametric peak calling package for 4C and capture-C data. Nucleic Acids Res. 2018;46(15):e91. doi: 10.1093/nar/gky443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fewings E, Larionov A, Redman J, Goldgraben MA, Scarth J, Richardson S, et al. Germline pathogenic variants in PALB2 and other cancer-predisposing genes in families with hereditary diffuse gastric cancer without CDH1 mutation: a whole-exome sequencing study. Lancet Gastroenterol Hepatol. 2018;3(7):489–498. doi: 10.1016/s2468-1253(18)30079-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754.PubMedPMID:21221095;PubMedCentralPMCID:PMC3346182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fishilevich S, Nudel R, Rappaport N, Hadar R, Plaschkes I, Iny Stein T, et al. GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017 doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pinheiro H, Carvalho J, Oliveira P, Ferreira D, Pinto MT, Osório H, et al. Transcription initiation arising from E-cadherin/CDH1 intron2: a novel protein isoform that increases gastric cancer cell invasion and angiogenesis. Hum Mol Genet. 2012;21(19):4253–4269. doi: 10.1093/hmg/dds248. [DOI] [PubMed] [Google Scholar]
- 43.Oliveira C, Pinheiro H, Figueiredo J, Seruca R, Carneiro F. Familial gastric cancer: genetic susceptibility, pathology, and implications for management. Lancet Oncol. 2015;16(2):e60–70. doi: 10.1016/s1470-2045(14)71016-2. [DOI] [PubMed] [Google Scholar]
- 44.Yamada M, Fukagawa T, Nakajima T, Asada K, Sekine S, Yamashita S, et al. Hereditary diffuse gastric cancer in a Japanese family with a large deletion involving CDH1. Gastric Cancer. 2014;17(4):750–756. doi: 10.1007/s10120-013-0298-y. [DOI] [PubMed] [Google Scholar]
- 45.Paliou C, Guckelberger P, Schöpflin R, Heinrich V, Esposito A, Chiariello AM, et al. Preformed chromatin topology assists transcriptional robustness of Shh during limb development. Proc Natl Acad Sci USA. 2019;116(25):12390–12399. doi: 10.1073/pnas.1900672116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gloerich M, Bianchini JM, Siemers KA, Cohen DJ, Nelson WJ. Cell division orientation is coupled to cell-cell adhesion by the E-cadherin/LGN complex. Nat Commun. 2017;8:13996. doi: 10.1038/ncomms13996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Dong B, Zhang K, Ji Z, Cheng C, Zhao H, et al. E-cadherin bridges cell polarity and spindle orientation to ensure prostate epithelial integrity and prevent carcinogenesis in vivo. PLoS Genet. 2018;14(8):e1007609. doi: 10.1371/journal.pgen.1007609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan HB, Wang XF, Zhang Q, Tang ZQ, Jiang YH, Fan HZ, et al. Reduced expression of the chromatin remodeling gene ARID1A enhances gastric cancer cell migration and invasion via downregulation of E-cadherin transcription. Carcinogenesis. 2014;35(4):867–876. doi: 10.1093/carcin/bgt398. [DOI] [PubMed] [Google Scholar]
- 49.Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 50.Chen A, Beetham H, Black MA, Priya R, Telford BJ, Guest J, et al. E-cadherin loss alters cytoskeletal organization and adhesion in non-malignant breast cells but is insufficient to induce an epithelial-mesenchymal transition. BMC Cancer. 2014;14:552. doi: 10.1186/1471-2407-14-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mestre T, Figueiredo J, Ribeiro AS, Paredes J, Seruca R, Sanches JM. Quantification of topological features in cell meshes to explore E-cadherin dysfunction. Sci Rep. 2016;6:25101. doi: 10.1038/srep25101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Elisha Y, Kalchenko V, Kuznetsov Y, Geiger B. Dual role of E-cadherin in the regulation of invasive collective migration of mammary carcinoma cells. Sci Rep. 2018;8(1):4986. doi: 10.1038/s41598-018-22940-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Christgen M, Bartels S, van Luttikhuizen JL, Bublitz J, Rieger LU, Christgen H, et al. E-cadherin to P-cadherin switching in lobular breast cancer with tubular elements. Mod Pathol. 2020;33(12):2483–2498. doi: 10.1038/s41379-020-0591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lui AJ, Geanes ES, Ogony J, Behbod F, Marquess J, Valdez K, et al. IFITM1 suppression blocks proliferation and invasion of aromatase inhibitor-resistant breast cancer in vivo by JAK/STAT-mediated induction of p21. Cancer Lett. 2017;399:29–43. doi: 10.1016/j.canlet.2017.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borg D, Hedner C, Gaber A, Nodin B, Fristedt R, Jirström K, et al. Expression of IFITM1 as a prognostic biomarker in resected gastric and esophageal adenocarcinoma. Biomark Res. 2016;4:10. doi: 10.1186/s40364-016-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee J, Goh SH, Song N, Hwang JA, Nam S, Choi IJ, et al. Overexpression of IFITM1 has clinicopathologic effects on gastric cancer and is regulated by an epigenetic mechanism. Am J Pathol. 2012;181(1):43–52. doi: 10.1016/j.ajpath.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Lee JH, Kim KY, Song HK, Kim JK, Yoon SR, et al. The interferon-inducible 9–27 gene modulates the susceptibility to natural killer cells and the invasiveness of gastric cancer cells. Cancer Lett. 2005;221(2):191–200. doi: 10.1016/j.canlet.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 58.Hu J, Wang S, Zhao Y, Guo Q, Zhang D, Chen J, et al. Mechanism and biological significance of the overexpression of IFITM3 in gastric cancer. Oncol Rep. 2014;32(6):2648–2656. doi: 10.3892/or.2014.3522. [DOI] [PubMed] [Google Scholar]
- 59.Rajapaksa US, Jin C, Dong T. Malignancy and IFITM3: Friend or Foe? Front Oncol. 2020;10:593245. doi: 10.3389/fonc.2020.593245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Min J, Feng Q, Liao W, Liang Y, Gong C, Li E, et al. IFITM3 promotes hepatocellular carcinoma invasion and metastasis by regulating MMP9 through p38/MAPK signaling. FEBS Open Bio. 2018;8(8):1299–1311. doi: 10.1002/2211-5463.12479. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 61.Liu X, Chen L, Fan Y, Hong Y, Yang X, Li Y, et al. IFITM3 promotes bone metastasis of prostate cancer cells by mediating activation of the TGF-β signaling pathway. Cell Death Dis. 2019;10(7):517. doi: 10.1038/s41419-019-1750-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, Wu C, Zhang C, Li Z, Zhu T, Chen J, et al. TGF-β-induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR-30a interactions. Cancer Lett. 2018;436:52–62. doi: 10.1016/j.canlet.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 63.Wang H, Tang F, Bian E, Zhang Y, Ji X, Yang Z, et al. IFITM3/STAT3 axis promotes glioma cells invasion and is modulated by TGF-β. Mol Biol Rep. 2020;47(1):433–441. doi: 10.1007/s11033-019-05146-2. [DOI] [PubMed] [Google Scholar]
- 64.Lee J, Robinson ME, Ma N, Artadji D, Ahmed MA, Xiao G, et al. Nature. 2020;588(7838):491–497. doi: 10.1038/s41586-020-2884-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li D, Peng Z, Tang H, Wei P, Kong X, Yan D, et al. KLF4-mediated negative regulation of IFITM3 expression plays a critical role in colon cancer pathogenesis. Clin Cancer Res. 2011;17(11):3558–3568. doi: 10.1158/1078-0432.Ccr-10-2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Daniel-Carmi V, Makovitzki-Avraham E, Reuven EM, Goldstein I, Zilkha N, Rotter V, et al. The human 1–8D gene (IFITM2) is a novel p53 independent pro-apoptotic gene. Int J Cancer. 2009;125(12):2810–2819. doi: 10.1002/ijc.24669. [DOI] [PubMed] [Google Scholar]
- 67.Bonni A, Brunet A, West AE, Datta SR, Takasu MA, Greenberg ME. Cell survival promoted by the Ras-MAPK signaling pathway by transcription-dependent and -independent mechanisms. Science. 1999;286(5443):1358–1362. doi: 10.1126/science.286.5443.1358. [DOI] [PubMed] [Google Scholar]
- 68.Hou F, Shi DB, Chen YQ, Gao P. Human epidermal growth factor receptor-2 promotes invasion and metastasis in gastric cancer by activating mitogen-activated protein kinase signaling. Appl Immunohistochem Mol Morphol. 2019;27(7):529–534. doi: 10.1097/pai.0000000000000672. [DOI] [PubMed] [Google Scholar]
- 69.Ohtsuka J, Oshima H, Ezawa I, Abe R, Oshima M, Ohki R. Functional loss of p53 cooperates with the in vivo microenvironment to promote malignant progression of gastric cancers. Sci Rep. 2018;8(1):2291. doi: 10.1038/s41598-018-20572-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Busuttil RA, Zapparoli GV, Haupt S, Fennell C, Wong SQ, Pang JM, et al. Role of p53 in the progression of gastric cancer. Oncotarget. 2014;5(23):12016–12026. doi: 10.18632/oncotarget.2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51(23 Pt 1):6304–6311. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data supporting the findings of this study are available upon request from the corresponding author. Processed data are available in supplementary materials.