Abstract
The organoids represent one of the greatest revolutions in the biomedical field in the past decade. This three-dimensional (3D) micro-organ cultured in vitro has a structure highly similar to that of the tissue and organ. Using the regeneration ability of stem cells, a 3D organ-like structure called intestinal organoids is established, which can mimic the characteristics of real intestinal organs, including morphology, function, and personalized response to specific stimuli. Here, we discuss current stem cell-based organ-like 3D intestinal models, including understanding the molecular pathophysiology, high-throughput screening drugs, drug efficacy testing, toxicological evaluation, and organ-based regeneration of inflammatory bowel disease (IBD). We summarize the advances and limitations of the state-of-the-art reconstruction platforms for intestinal organoids. The challenges, advantages, and prospects of intestinal organs as an in vitro model system for precision medicine are also discussed.

Key applications of stem cell-derived intestinal organoids. Intestinal organoids can be used to model infectious diseases, develop new treatments, drug screens, precision medicine, and regenerative medicine.
Subject terms: Haematopoietic stem cells, Inflammatory bowel disease
Facts
ISCs in the intestinal crypts maintain the integrity of the intestinal epithelium.
ISCs or PSCs can self-organize and differentiate into “intestinal organoids” by specific 3D culture.
SCs-derived intestinal organoids are ideal models of the intestinal tract and have obvious advantages in the study of intestinal physiology and intestinal diseases.
SCs-derived intestinal organoids have been successfully transplanted into the intestinal mucosa and have been shown to have a repairing effect on the intestinal epithelium, which holds great prospects for the treatment of intestinal diseases.
Open questions
How to optimize the culture system and culture process of intestinal organoids?
How to improve the simulation and homogeneity of intestinal organoid models?
Can new therapeutic targets be identified using intestinal organoids?
Can intestinal organoid transplantation be a method to repair intestinal epithelial damage?
Introduction
Inflammatory bowel disease (IBD) is a chronic disease with an increasing incidence worldwide [1]. The intestinal epithelial barrier plays a major role in IBD [2, 3]. However, the lack of techniques for long-term culture of human primary epithelial cells in vitro hinders the study of the role of intestinal epithelium in IBD. In recent years, intestinal stem cells (ISCs) have become the focus of research on intestinal injury and regeneration, colorectal cancer, and other intestinal diseases [4]. At present, leucine rich repeat containing G protein coupled receptor 5+ (Lgr5+) stem cells (SCs) play an important role in maintaining normal intestinal epithelial structure by renewing the intestinal epithelium by generating billions of cells at an alarming rate every day [5]. Studies have shown that ISCs and crypts in vitro can form hollow spheres with intact intestinal epithelial-like structures, known as “intestinal organoids”, through 3D culture mode in matrix glue (Matrigel) under the action of appropriate growth factors [6]. In addition, pluripotent SCs (PSC) or induced pluripotent SCs (iPSC) derived from patients with IBD are also capable of forming intestinal organoids under in vitro induction. These organs are hollow globules with complete intestinal epithelioid structures. Intestinal epithelial organs cultured in vitro for several months retain their cellular characteristics and functions because they are fully integrated into the recipient epithelial cells [7–9]. They retain their original cell characteristics and functions even after several months of in vitro culture, making them ideal physiological models of the intestinal epithelium and unique disease models, and can be transplanted intact into the recipient intestinal mucosa to achieve repair of intestinal epithelial damage [10, 11]. Cellular sources and methods for the generation of intestinal organoid have illustrated in Fig. 1.
Fig. 1. Schematic diagram of intestinal tissue engineering by organoid technology.
Intestinal organoid is generated from the intestinal crypts of the small intestine, and Lgr5+ stem cells were isolated from small intestinal tissues are then embedded in Matrigel with a culture medium. Clonal fibroblast subclones and clonal iPSCs were established in the parental fibroblast population and then co-generated with fragmented intestinal tissue on 3D scaffold material to generate intestinal organoids.
This review discusses the progress of research on the formation of intestinal organoids by SCs in culture. And discusses the research progress and current limitations of intestinal organoids in establishing intestinal epithelial models and treating patients with IBD. Finally, we outline the urgent need to technically standardize laboratory procedures for intestinal organs in order to make them more widely used in clinical IBD studies.
IBD intestinal model
Clinical studies and applications of IBD are subject to strict technical and ethical restrictions, and high-quality preclinical studies are needed to ensure the efficacy and safety of clinical studies [12, 13]. The main preclinical research models for IBD are animal models and cellular models. For a long time, animal models have been the mainstay of intestinal research due to the lack of representativeness of cellular models [14]. However, animal models have long time cycles and racial differences from humans with being expensive and subject to animal ethics [14]. Therefore, it requires effective, safe, and feasible cellular models to avoid unnecessary animal experiments as much as possible. Besides, animal models are difficult to be widely carried out in disease research [15, 16]. The intestinal cell model is limited by the technology of intestinal epithelial cell culture, and the mechanism of intestinal epithelial damage for IBD patients is under-researched and the treatment methods are limited. In the past decade or so, SCs-derived intestinal organoid models have brought a leap forward in IBD research and can provide new directions and methods for IBD mechanism research and treatment development [17, 18].
2D cellular model
The in vitro cell model is to isolate primary cells from in vivo tissues, perform cell culture and generate different types of cell lines [19, 20]. Until the ISCs were isolated, the intestinal cell model could only be replaced by intestinal tumor cells. The developed Caco-2, HT-29, and T84 cell lines were derived from human colorectal adenocarcinoma [21–24].
The 2D cell culture environment is far from the complex cellular environment in vivo, which affects the structure and function of cells [21]. 2D culture cell lines have few cell types and confusing tissue structure, and the cell lines will gradually flatten, abnormally differentiate and lose their differentiation phenotype, which cannot reflect the interaction between different cells and matrix, and cannot form similar cellular tissue structure in vivo. It cannot reflect the interactions between different cells and matrix, and cannot form similar cellular tissue structures in vivo [20, 25]. It is generally accepted that 2D cell models are difficult to reflect the in vivo situation, have little reference value, and the experimental results are not widely accepted [26–28].
3D cellular model
3D cell culture is an in vitro co-culture of carriers with 3D structures and different materials with different cell types so that the cells can migrate and grow in the carriers with 3D spatial structure, forming 3D cell-carrier complexes [28, 29]. A 3D cell model optimizes the limitations of a 2D cell model. 3D culture environment can better simulate the in vivo cell growth environment, mimic the in vivo cell growth environment, regulate cell proliferation and differentiation, and cultivate more cell types to form tissue structures similar to in vivo [30]. Currently, 3D cell models are widely used in various fields such as organoid, microtumor, and microcarrier [31, 32].
Lgr5(+) ISCs were successfully isolated in 2007, and have become the most important cell source for intestinal cell models [33]. In vivo, Lgr5+ crypt base columnar cells (CBC) are located at the base of the intestinal mucosal crypt and differentiate to form different intestinal mucosal cells by continuously proliferating and migrating toward the top of the crypt [34]. In vitro, Lgr5(+) CBC has a short survival time and cannot be cultured in a 2D culture environment, but can survive for a long time in a 3D culture environment [35]. In 3D culture, Lgr5+ CBC was found to divide once every 24 h and could differentiate into all intestinal epithelial cells (IEC) except mesenchymal cells and immune cells [36].
Intestinal organoid model
Intestinal organoid is a miniature hollow sphere with an intestinal epithelial crypt structure formed by SCs in 3D culture, which mimics the ecological niche of ISCs, and precisely regulates the proliferation and differentiation of SCs [17, 37–48]. These spheres contain most types of IEC, and when differentiated mature, they have physiological functions such as absorption, secretion, mucus production, and material transport [49, 50]. Intestinal organoids have short culture cycles, can be stored frozen for long periods of time, and can also simulate complex environments in vivo or be adapted to the culture environment. Intestinal organoids are able to preserve the intestinal epithelial crypt structure and maintain stable phenotypic and genetic characteristics [7, 43, 51–53]. Compared with cell line models, organoids have similar tissue structure and function to those in vivo, improving the realism and reliability of the study [18, 27]. In addition, the culture cycle is shorter than that of animal models, the process is easy to manipulate, and there are no animal ethical issues involved, allowing for a wider range of studies [16, 29, 54, 55]. Table 1 Summary of current intestine models. Figure 2 Intestinal Organoids as models for IBD research.
Table 1.
Summary of current intestine models.
| Intestinal models | Advantage | Limitation | Ref. |
|---|---|---|---|
| Intestine animal models | Traditional model; Technology maturity; Approaching the stage of clinical studies. | long time cycles; expensive; racial differences from human; subject to animal ethics. | [15, 16] |
| 2D intestine cellular model | The culture is simple, the technology is mature, the cost is low, and it is adapted to simple preclinical studies. | Planar culture environment, monolayer cell culture, gradually planarizing and losing differentiation phenotype, inability to form intestinal epithelial structures. | [168–173] |
| 3D intestine organoid model | 3D culture environment that mimics the growth environment of ISCs in vivo, forms intestinal epithelial structures, and maintains stable phenotypic and genetic characteristics. | Culture process and technology is relatively complex, and cost is relatively high. There are still limitations that hinder clinical translation. | [174–178] |
| Intestine Organoid microarray | More precise regulation of the cultivation environment, simulating the physical and microbial environment in the body. | The culture system and process lack standards, the culture technology needs to be improved, and the cost is high. | [179–184] |
Fig. 2. Intestinal Organoids as models for IBD research.
Intestinal organoids can be used for intestinal development and IBD disease modeling, drug/toxicity testing, and host-pathogen interaction studies. In cultures of intestinal organoids from IBD patients, we can add multiple growth factors, cytokines, or drug molecules to modulate the culture environment, add autoimmune cells for co-culture, or digest into single cells. The functions can evaluate by phenotypic analysis, drug screening, qRT-PCR, single cell RNA sequencing, flow cytometry, imaging, ELISA, and other kinds of indicators.
The cell sources of intestinal organoids include ISCs from intestinal crypts, PSCs, embryonic SCs (ESCs), and iPSCs [18, 56]. In 2009, Hans et al. reported for the first time in Nature the formation of murine intestinal organoids with intestinal epithelial crypt structure using Lgr5(+) ISCs from mouse intestinal crypt in vitro [6]. In 2011, Hans et al. cultured human intestinal organoids with intestinal epithelial crypt structure using Lgr5(+) ISCs from human intestinal crypt [47]. After more than a decade of development, ISCs-derived intestinal organoid cultures have matured with high success rates, and are currently the main source of intestinal organoid cultures [57, 58]. However, the cultures do not contain MSCs and immune cells [42, 59]. In 2011, Spence et al. reported the use of PSCs or embryonic SCs to culture human intestinal organoids [10]. In 2017, Miura and Suzuki reported the formation of intestinal organoids using mouse and human iPSCs cultures, respectively [60]. PSC, ESC, and iPSC cultures form intestinal organoids with crypt structures of the intestinal epithelium and contain various types of IEC, but the differentiated IEC is not sufficiently mature [61, 62]. There is a potential risk of genetic and epigenetic variation during induction, with some differences in structure, function, and genetic characteristics from the IEC in vivo [60, 62]. Currently, the success rate of various PSCs cultured to form intestinal organoids is low and the technique is still flawed, pending subsequent improvements [10, 43, 61].
Intestinal organoid culture technology
Organoid culture technology was named one of the "Top Ten Breakthroughs" by Science in 2013 and Nature Methods in 2017, and an excellent preclinical disease model by The New England Journal of Medicine in 2019 [63]. The organoid culture technology includes core technologies such as 3D culture, detection, and identification, as well as basic technologies such as stem cell isolation, organoid extraction, and organoid preservation [58, 64]. In addition, emerging technologies such as 3D bioprinting, organoid microarrays, and gene editing technologies can be combined to increase environmental simulation conditions or expand the scope of research [53, 65, 66]. To better simulate the intestinal epithelial growth environment in vivo, organoid culture is systematized and a 3D culture system for intestinal organoids is established [47, 67–69]. The current 3D organoid culture system can be divided into scaffold-free culture system and scaffold-based culture system according to the presence or absence of scaffold deriving from natural components or artificially synthesized [70].
scaffold-free culture system
The scaffold-free culture system has no support structures for cell adhesion, growth, and spreading [64]. The 3D culture environment is formed by various physicochemical principles, and the cells aggregate in the medium to form microtissue spheroids similar to the source tissue [55, 64]. Hanging drops, which have no plane of attachment, can be prepared as a variety of hanging drop plates allowing cells to self-assemble under the influence of gravity to form microtissue spheroids [71–73]. The cells can be magnetized and suspended in a magnetic field for 3D culture [74–76]. Special synthetic polymer materials can be used to form microplate structures with ultra-low adhesion surfaces on which the cells migrate and adhere to each other to form microtissue spheroids [64, 77]. Alternatively, the agar interface can be used to reduce the stiffness of the culture surface to create a 3D Petri Dish in which cells can migrate and spread to form microtissue spheroids [43, 78].
Scaffold-based culture system
Scaffold-based culture systems allow cells to attach to a scaffold composed of solid particles or liquid gels, which are made to float in the culture medium by gentle agitation [38]. With the development of technology, scaffold appears in more and more forms to solve different problems [67]. According to the source of scaffold, it can be divided into scaffold supported by natural extracellular matrix (ECM) and scaffold supported by synthetic materials [38, 67, 79].
Natural ECM-supported scaffold
Natural ECM is used as the support material to optimize the 3D culture matrix formulation according to the cultured cell types meeting the culture needs of different tissue cells [80]. Matrix gel is a natural ECM extracted from the basement membrane of mouse sarcoma cells in liquid gel form, which is a natural scaffold with good compatibility for both human and mouse cell cultures [80, 81]. The main components of matrix gel are laminin, type IV collagen, nestin, heparan sulfate glycoprotein, and also contains growth factors and matrix metalloproteinases, providing rich nutrition [43]. However, natural ECM has some immunogenicity and risk of pathogenic infection and is less stable [82]. Natural ECM may have the disadvantage of batch variability due to different matrices and extraction techniques [81, 83].
Synthesized material-supported scaffold
The types of synthetic scaffold materials are quite diverse. Synthetic materials have consistency, stability, and biocompatibility bias, and cannot provide growth factors and small molecule compounds needed for cell culture. The synthetic scaffold can be divided into hydrogel and solid scaffold according to the attachment method of cells [67, 79].
Hydrogels
The hydrogel is mimics of ECM and can be made from natural polymers or synthetic polymers [64]. SCs are dispersed in liquid hydrogels and then cross-linked to achieve 3D culture [38, 84]. The hydrogel scaffold has good stability and adjustability [29, 38, 64]. Hydrogels can alter cell signaling by increasing or decreasing protein concentration and changing the density of cell adhesion ligands [39, 66]. Polymeric gelling agents can be pre-designed, and functional additives can be adjusted as required, allowing the hydrogel to obtain specific properties such as temperature, magnetic properties, and pH, and meet a wider range of research requirements [39, 40, 67].
Solid scaffold
Solid scaffold uses a variety of porous materials to prepare a solid, porous microcarrier structure [85, 86]. SCs are "seeded" in these solid scaffold porous microcarriers to achieve a 3D culture environment [86, 87]. Sponges or foams have high porosity and homogeneous connectivity structures, and the prepared porous microcarrier scaffolds are widely used in tissue engineering [85, 88]. Solid scaffolds prepared by applying biodegradable polymers, such as PLA, have been extensively studied for their good biocompatibility and proper porous structure [89, 90]. Enables efficient 3D cell attachment for superior cell viability [67, 89, 91].
Organoid detection and identification techniques
Intestinal organoids are microtissue spheroids with intestinal epithelial structure, and their detection and identification techniques are different from traditional 2D cell lines [58]. It is necessary to evaluate the quality of intestinal spheroids in culture, as well as to evaluate the research results of intestinal spheroid models. The development of various characterization techniques and assays has enriched the identification and evaluation of organoids. The application and promotion of intestinal organoid technology in the study of intestinal diseases have been promoted [29, 47, 58, 92]. The tests that can be performed now include morphological structure, proliferative activity, intestinal epithelial barrier function, and genetics [93–95].
Organoid microarray technology
Stationary 3D cell culture is performed in culture dishes or culture plates, and the culture environment that can be regulated is very limited [96]. The intestinal epithelial cells are regulated by multiple systems of the organism and are also influenced by the intestinal microecology and microenvironment [97, 98]. Organoid microarrays are microfluidic microarray technology combined with 3D organoid culture technology to form a dynamic 3D cell culture environment in vitro by more rigorously mimicking the functional units of human tissues and organs in a miniature organoid culture vehicle [41, 99]. The organoid chip can simulate the physical environment such as in vivo mechanics, and magnetic and electric fields by dynamically regulating the culture medium and additives through microfluidics [100]. It can add different cells or microorganisms to mediate cell-cell and cell-microbe interactions [100–102]. Microarray technology can also be combined with imaging instruments to monitor cell biological changes in real time, record behavioral changes of cells in disease states, and record the whole process of cellular response to drugs [103–105]. Microarray technology enables more systematic and mechanized organoid culture, increases throughput, expands the scope of research, and takes a firm step toward clinical translation [69, 100, 106–108].
Bio-3D printing technology
Bio-3D printing technology has broken through cell printing and manipulated cell-containing bio-ink to construct active structures [109, 110]. The hybrid printing of SCs with biological scaffolds through multi-jet 3D printing technology to form organoid structures could be the next generation of organoid construction methods [111]. It has been shown that ‘assembloids’ assembled by cell-based 3D printing technology go beyond Organoid and are closer to human tissues and organs in terms of structure and function [112]. At present, bio-3D printing is mainly used for in vitro research models, for forming 3D culture systems, organoid chips, and printing biomimetic materials, such as gelatin, alginate, and creating conditions for in vitro culture of SCs into desirable organoids [113]. Currently, although it is possible to print cell-containing structures that resemble in vivo tissues and organs in shape and structure, there is still a big gap between them and the complex physiological functions of real organs. There is still quite a long way to go before active organs can be printed using for transplantation [68, 77, 114].
Intestinal organoid applications
The applications of Intestinal organoids are focused on both research models and clinical treatments. The intestinal organoid has the structural characteristics and physiological functions of the intestinal epithelium, which is an ideal model of intestinal epithelial physiology and intestinal diseases [55]. The organoid is able to maintain stable phenotypic and genetic characteristics, and has obvious advantages in individualized disease modeling and precision medicine [7, 115]. As an intestinal epithelial physiological model, the growth and development, tissue structure, and physiological function of intestinal epithelium can be studied. As a model of IBD disease, it can explore the pathogenesis of IBD and test the treatment effect of IBD. Intestinal organoids can be used in IBD treatment to repair intestinal epithelial damage through organoid transplantation and to improve genetic susceptibility and gene expression through gene therapy [17, 50, 53, 116].
Studying intestinal physiology
Intestinal organoids maintain the structure and function of the intestinal epithelium [29]. By observing the formation process of the organoid, the complex environmental changes in vivo are simulated, thus understanding the structure and physiological functions of the intestinal epithelium. It also allows in-depth study of the proteomics, lipidomics, genomics, and transcriptomics of intestinal epithelial cells [29, 36, 63, 117, 118].
It was found that mature absorptive cells, cupped cells, and various intestinal endocrine cells could be detected in intestinal epithelial organoids derived from Lgr5(+) CBC [36, 117]. These cells have corresponding physiological functions such as transporting substances, absorbing nutrients, and synthesizing secretion [57, 59]. By comparing colonic epithelial organoids from healthy humans with those from patients with ulcerative colitis (UC), Dotti et al. found differences in DNA methylation profiles and gene expression profiles. suggesting that the UC colonic organoid model retains a disease-specific genetic and expression profile [7, 9, 115].
Exploring the pathogenesis of IBD
Currently, the etiology and pathogenesis of IBD are unclear. In order to explore the pathogenesis of IBD and find the etiology of intestinal injury, colonic organoids are ideal models for disease research. Being able to establish an individualized disease model, preserving the morphological structure and genetic characteristics of diseased tissue cells, can lead to more realistic and reliable research results [9, 29].
In terms of genetics
A study comparing colonic epithelial organoids of terminal ileal origin from UC patients and Crohn’s disease (CD) patients found that they differ in DNA methylation profiles and gene expression profiles, although they have similar morphological structures [9]. It indicates that there are different disease-related genetic genes and expression profiles in each of UC and CD, which can provide a basis for subsequent differential diagnosis and gene therapy for IBD [7, 9, 93].
In terms of mucosal inflammatory damage
A study found that transcript levels of IL-1β and γ-IFN were lower in colonic organoid cultures than in their source mucosal biopsy specimens. The investigators concluded that the levels of inflammatory factors in mucosal specimens do not carry over to colonic organoids and that overexpression of these inflammatory factors requires re-stimulation [52]. In 2018, Biton et al. studied Th and its inflammatory factors in intestinal organoids on the proliferation and biochemistry of Lgr5(+) CBC [119]. Lgr5(+) CBC expressing major histocompatibility complex II (MHC II) was found to be an atypical antigen-presenting cell that interacts with Th via MHC II. Th1, Th2, Th17 and their secreted pro-inflammatory factors IFN-γ, IL-13 and IL-17A promote Lgr5(+) CBC differentiation [119, 120]. And regulatory Th (Treg) and the base-secreted cytokine IL-10 promote Lgr5(+) CBC regeneration [119]. A study found that the regenerative capacity of ISCs and the rate of organoid formation were reduced after short-term intervention of intestinal organoid Interleukin-22 (IL-22), but the number of transiently expanded progenitor cells increased and differentiated to various types of epithelial cells, resulting in an increase in the volume of formed organoids [121, 122]. It is hypothesized that IL-22 contributes to the differentiation of transiently expanded progenitor cells to various types of epitheliums, and promotes intestinal epithelial repair during acute injury of intestinal epithelium [121, 123]. However, IL-22 may be detrimental to epithelial regeneration and repair by causing damage to ISCs when chronic injury hampering the intestinal epithelium [123, 124]. The clinical application of IL-22 to promote mucosal repair in IBD patients should take into account its long-term chronic damage to ISCs [121].
In terms of intestinal flora
Wang et al. combined 3D differentiation technology at the air-liquid interface to construct an intestinal epithelial organoid and cell co-culture system to simulate pseudomembranous colitis caused by C. difficile infection of the intestine and to explore the pathogenic mechanism of C. difficile [125]. Hou et al. established a co-culture system of Lactobacillus, intestinal lamina propria lymphocytes, and intestinal organoid, and found Lactobacillus stimulated the proliferation and repair of intestinal epithelium of Intestinal organoids by promoting the secretion of IL-22 and TNF-α from lymphocytes in the lamina propria of the intestine [126].
In terms of fibrosis and cancerization
One study isolated the crypt from the mouse colon and cultured it into intestinal organoids. Tumor necrosis factor-α (TNF-α) stimulation and Transforming growth factor-β1 (TGF-β1) stimulation were successively administered to intestinal organoids to induce IEC mesenchymal cell transformation, providing a convenient and effective in vitro model for studying intestinal fibrosis in IBD [127]. In another study, cytokines (TNF-α, IL-1β, IL-6) and bacterial components (bacterial lipopolysaccharide, flagellin) were mixed and added to mouse colonic organoid medium on alternate days to simulate chronic inflammatory stimuli for more than 1 year of continuous intervention [128]. The results revealed that colonic organoid underwent a cellular transformation that may be associated with UC carcinogenesis after chronic inflammatory stimulation due to activation of the nuclear factor kappa-B (NF-κB) signaling pathway and impaired cell differentiation [128].
Testing the effect of IBD treatment
IBD is mainly treated with drugs, and the application of colonic organoid models for drug testing is ideal. Colonic epithelial organoids can be cultured to simulate a more reviewed in vivo environment, and have a very high structural and functional similarity to the in vivo intestinal epithelium. It is highly realistic and reliable as a research model to study drug responses to intestinal epithelial structures. There is a significant increase in the success rate of clinical translation of IBD therapeutic agents or treatments that pass the test in the colonic organoid model [12, 118, 129, 130]. In precision medicine, colonic organoids derived from ISCs or iPSCs established from IBD patients are mostly used for drug testing [118]. Testing conventional therapeutic drugs and exploring new drugs for IBD treatment can help identify more optimal options for IBD patients [118, 130, 131].
Glucocorticoids are commonly used in the treatment of IBD [132]. Xu et al. observed the penetration of FITC-Dextran 4 (FD4) markers in the lumen of intestinal organoids using coaggregation microscopy [133]. It was found that exposure of the organoid to the glucocorticoid prednisolone significantly reduced intraluminal FD4 infiltration, and decreased inflammatory cytokine expression in the culture medium, indicating that glucocorticoids are well suited to suppress inflammation and reduce intestinal epithelial permeability in IBD treatment [133]. One study added azathioprine and 5-aminosalicylic acid to TNF-α-treated intestinal organoid medium, respectively [134]. The results revealed that TNF-α-treated intestinal organoids showed internalization and abnormal disruption of E-calmodulin and reduced bridging granule core protein-2 levels. The addition of 5-aminosalicylic acid or azathioprine treatment restored E-calmodulin levels on cell membranes. Addition of 5-aminosalicylic acid restored bridging granule core protein-2 levels [134]. Studies have shown that azathioprine and 5-aminosalicylic acid can repair the integrity of the intestinal barrier [134, 135]. Infliximab is an anti-tumor necrosis factor agent that can be used in the treatment of IBD [136]. One study examined the effect of infliximab on IEC using intestinal organoids from UC patients. The results found that concomitant treatment of UC patient’s organoids with infliximab and TNF-α had no significant effect on their viability or morphology, but resulted in a significant decrease in ubiquitin D (UBD) mRNA expression [136, 137]. UBD is a ubiquitin-like modifier involved in protein degradation and is upregulated in inflamed intestinal tissues, suggesting an anti-inflammatory effect of infliximab [136]. Lloyd et al. used healthy human-derived colonic organoids organs incorporating the macrolide clarithromycin and found that clarithromycin exerted anti-inflammatory effects in the intestinal epithelium [138]. This suggests that in addition to its antibacterial effects, clarithromycin has the potential to inhibit the inflammatory response of the intestinal epithelium [138]. Table 2 Novel molecular targets and therapy approach identified using intestinal organoids.
Table 2.
Novel molecular targets and therapy approach identified using intestinal organoids.
| molecular targets | effect | Drug | Species | Ref. |
|---|---|---|---|---|
| Molecular target identified by treatment of intestinal organoids with current therapies for IBD | ||||
| E-cadherin | Re-distribution of protein on intestinal surface restored correct permeability | 5-aminosalycilic acid, azathioprine | Mouse (IL-10−/−) | [134] |
| Desmoglein-2 | Restored physiological desmoglein-2 expression levels | 5-aminosalycilic acid | Mouse (IL-10−/−) | [134] |
| UBD | Restored physiological UBD expression levels | Infliximab | Human (UC patients) | [136] |
| CLDN-2 | Restored physiological CLDN-2 expression levels | Prednisolone, tofacitinib | Human (CD and CRC patients) | [133] |
| ZO-1 | Re-distribution of protein on intestinal surface restored correct | Permeability, Tofacitinib | Human (CRC patients) | [185] |
| Novel potential molecular targets identified using intestinal organoids | ||||
| LRH-1 | Improved resistance to pro-inflammatory mediators and induced mucosal healing | — | Humanized mouse (Lrh-1−/−) LRH-1(+/+) and Human | [186] |
| PXR | Reduced NF-kB activity | — | Human (IBD patients) | [187] |
| IL-22-pSTAT3 SP | Restored tissue damage and intestinal homeostasis | — | Mouse (ATF3−/−) | [188] |
| TGF-β SP | Arrested inflammatory signals | — | Mouse | [189] |
| SIRT2 | Regulated Wnt/β-catenin SP | — | Mouse (Sirt2−/−) | [190] |
| Potential therapeutic approaches for IBD treatment identified using intestinal organoids | ||||
| Sex hormones | Decreased expression of ER stress markers | — | Human (UC female patients) | [191] |
| Naltrexone | Reduced ER stress levels, increased the expression of endogenous encephalins and endorphins | — | Human (IBD patients) | [192] |
| Bacillus subtilis (RZ001) | Promoted intestinal mucosa repair | — | Mouse | [193] |
| Bacterial indoleacrylic acid | Promoted anti-inflammatory cytokines secretion while inducing goblet cells differentiation | — | Mouse | [194] |
| Hyaluronan 35 kDa | Promoted epithelial wound healing | — | Mouse | [195] |
ATF3 Activating transcription factor 3, CD Crohn’s disease, CLDN-2 Claudin-2, CRC Colorectal cancer, ER Endoplasmic reticulum, IBD Inflammatory bowel disease, IL Interleukin, IL-10 Interleukin-10, LRH-1 Liver receptor homolog 1, NF-kB nuclear factor kappa-B, PXR Pregnane X receptor, SIRT2 Human sirtuin protein 2, SP Signaling pathway, STAT Signal transducer and activator of transcription, TGF-βTransforming growth factor-β, UBD Ubiquitin D, UC Ulcerative colitis, ZO-1 Zonula occludens.
Repair of intestinal epithelial injury
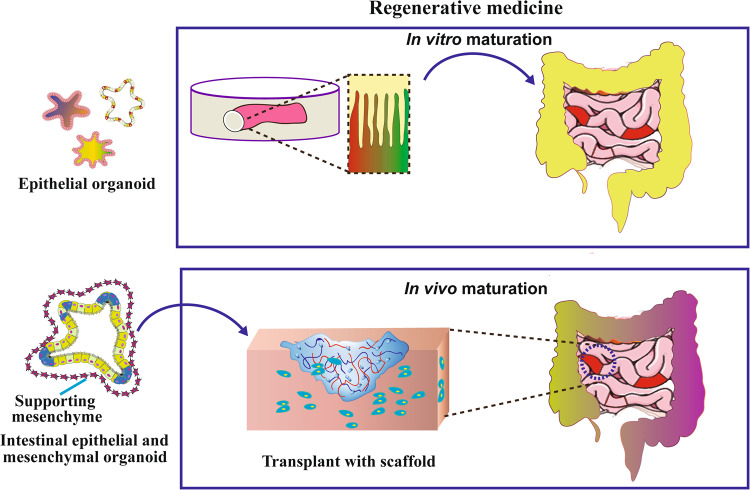
IBD presents as chronic inflammatory colonic injury, most notably colonic epithelial injury. In non-surgical resection therapy, there is a lack of effective treatment to repair chronic colonic injury and maintain the integrity of the intestinal epithelial barrier [2]. SCs therapy has been shown to be effective in the treatment of IBD, maintaining prolonged remission, repairing intestinal damage, and achieving mucosal healing (MH) [139, 140]. Intestinal organoid transplantation has significant advantages over stem cell transplantation [141]. Colonic organoids are derived from colonic SCs, which have colonic epithelial-like structural features and physiological functions, and retain genetic characteristics [11]. Transplantation into the body is more likely to integrate into the damaged colonic epithelial tissue and achieve the therapeutic goal of MH by regenerating and renewing intestinal epithelial cells and repairing colonic epithelial damage [140, 142, 143]. Figure 3 shows regenerative medicine for intestinal diseases. The development of direct repair of injured epithelial cells or partial replacement of abnormal intestinal epithelial cells with normal epithelial cells would be promising new therapeutic approaches for IBD. Organoid cells can also be transplanted in vivo, which provides a pre-clinical tool for regenerative medicine. For example, one study described in detail how epithelial organoids were transplanted into the colon of a mouse model of inflammatory enteritis. In this experiment, they injected the organoids into the luminal space at the anus. The injected organoids then attach to the injured area and reconstruct the donor-derived epithelium. This method has been successfully applied to epithelial cell-derived organoid tissue from adult colon and small intestine epithelium as well as fetal small intestine [144]. It has been shown that intestinal organoids are implanted through the anus into the dextran sodium sulfate(DSS)-induced colitis model, the clinical activity score can be significantly improved, and the donor cells can be accurately targeted to localize in the colitis-induced ulcer surface, and begin to reconstruct and repair the crypt structure [145–147]. In 2012, Yui et al. reported that in vitro cultured intestinal organoids were able to repair colitis through anal enucleation in mice, and the donor cells were able to target the colonic ulcer surface and start to reconstruct and repair the crypt structure [142]. In 2022, the same mouse-derived intestinal organoids were injected anally into UC mice by Tokyo Medical and Dental University (TMDU), Japan, to repair colitis, and these cultured organoids could precisely reach the location in the damaged intestinal epithelium and repair the damaged intestinal epithelium [148]. Subsequently, the TMDU research team announced the first successful transplantation of "organoids" into the intestinal mucosa of a patient with refractory UC [149]. The "organoid" is derived from the patient’s normal intestinal mucosa (including ISCs), and cultured in vitro in 3D to form 0.1–0.2 mm diameter "organoids", which are then endoscopically transplanted to the colon lesion site [148, 149]. Periodic endoscopic observation of mucosal repair and improvement of colonic lesions was performed. In addition, ISCs secrete a large number of extracellular vesicles in the culture medium during the formation of intestinal organoids [150–152]. The extracellular vesicles extracted from the culture medium have regulatory repair functions similar to those of mother cells [153–155]. It can be used as cell-free therapy and applied in IBD treatment, which can achieve the effects of repairing intestinal epithelium, promoting mucosal healing, and regulating immune dysfunction [152, 155, 156].
Fig. 3. Transplantation of intestinal organoids for the regenerative medicine of ulcerative colitis tissues.
Combining organoids with 3D scaffold material can induce the maturation of intestinal organs into functional intestines by implantation into the host. Also in vitro maturation of intestinal organs could repair the original intestinal structure and function.
Gene therapy
Organoid culture technology can be combined with gene editing technology. The gene editing technology represented by clustered regularly interspaced short palindromic repeats-associated protein 9 (CRISPR/Cas9) is becoming mature [157–160]. Gene editing of SCs or organoids using CRISPR/Cas9 for genetic modification of intestinal organoids [157, 161]. Genetic correction is performed in the case of genetic defects to achieve improvement of genetic susceptibility factors for IBD, prevention of carcinogenesis, or inhibition of tumor growth [94, 162, 163]. Alternatively, gene expression in intestinal organoids can be altered by DNA or RNA transfection, lentiviral infection, and other methods [164, 165]. Clevers et al. removed a small number of ISCs from patients with Cystic fibrosis (CF), modified the genes of these SCs using CRISPR/Cas9 technology, inserted normal cystic fibrosis transmembrane conductance regulator (CFTR) genes, and made them develop into organoid. These organoids, if reintroduced into the source patient, may partially cure CF disease [166].
Limitations of intestinal organoids technology
The organoid has more than 10 years of development and fruitful research results, but there are still certain limitations [13, 29, 167].
Potential hazards
In the process of 3D culture and gene editing, the differentiation of SCs and genetic modification needs to be precisely regulated, and there is a possibility of abnormal differentiation and gene mutation of SCs, and the chance of tumor formation increases. Allogeneic organ transplantation requires consideration of immunogenicity, which poses certain safety risks [13, 167]. Regarding culture systems and culture additives, it is important to guard against microbial contamination. It is important to consider the toxic components contained in synthetic materials and the immunogenicity of natural biological materials [67].
Tissue differences
Current organoid culture systems do not completely mimic the complex growth environment of the in vivo intestine. In comparison with the in vivo intestine, intestinal organoids also have certain tissue differences and functional defects. The intestinal organoid is dominated by intestinal epithelium and lacks connective, muscular, and neural tissues. There are no blood vessels, lymphatic vessels, and neurogenesis, and a lack of immune cells and smooth muscle cells [54].
Defective culture system
Materials and techniques are the basis for establishing culture systems. With the development and improvement of materials, diverse culture systems can be established, but all have different degrees of defects. It is still technically difficult to completely simulate the complex environment in vivo and monitor the culture process in real time, and there is still a long way to go before the integrated culture and commercial application of intestinal organoids [47, 61, 70].
Organoid heterogeneity
Both cell source and culture conditions affect the structure and function of organoids. At present, the sources of SCs are diverse, the cultural techniques are not uniform, and the research results are obviously individual. There are certain obstacles to the sharing of results and technology diffusion [43].
Summary
Currently, organoids have a wide range of applications in organ development, precision medicine, regenerative medicine, drug screening, gene editing, and disease modeling. SCs-derived intestinal organoid is an ideal intestinal epithelial physiological model and intestinal disease model, which has obvious advantages in individualized disease modeling and precision medicine. As a model of IBD disease, it can explore the pathogenesis of IBD, test the therapeutic effect of IBD, and find new therapeutic targets again. Intestinal organoids transplanted into IBD patients can repair intestinal epithelial damage. After gene therapy, intestinal organoid transplants can also improve IBD genetic susceptibility and improve gene expression. Intestinal organoid-derived extracellular vesicles have also demonstrated a role in repairing intestinal damage and regulating immune disorders.
However, the limitations of the intestinal organoid limit its clinical translation, and continuous research is still needed to break through its limitations. First of all, we need to expand the scope of research, conduct more extensive model studies, explore new therapeutic approaches, and prove the effectiveness, safety, and feasibility of organoid applications. Within the scope of ethics, we should optimize organoid transplantation and gene therapy technology, and conduct more animal or clinical trials to promote the clinical translation of organoids. Secondly, we need to optimize the culture system. Through material and technical improvements, we can better simulate the in vivo intestinal physicochemical and microbial environment, homogenize culture, and narrow the differences between organoid and in vivo organs. To simulate in vivo cellular interactions and establish a multicellular co-culture system in vitro. Table 3 Overview of the currently established organoid–cell co-culture systems. With the breakthrough of technical and ethical limitations, the clinical translation of intestinal organoids can be widely carried out. The future of intestinal organoids can bring a bright future to the research and treatment of IBD.
Table 3.
Overview of the currently established organoid–cell co-culture systems.
| Intestinal cells | Co-culture | Device material | Technique | Major observations | Ref. |
|---|---|---|---|---|---|
| Caco-2 | L. rhamnosus | PDMS, type I collagen, and Matrigel mix | Soft lithography | Fluid flow accelerated intestinal epithelial differentiation and organization into villi-like structures, mechanical stimulation enhanced specific differentiation features, sustained long-term co-culture with commensal bacteria | [196] |
| Human duodenal organoids (pediatric donors) | HIMECs | PDMS, type I collagen, and Matrigel mix | Soft lithography | Transcriptome of the intestinal tissue-on-chip more closely resembled that of the duodenum in vivo than the initial organoid culture from which it was derived, co-culture with endothelial cells accelerated the formation of the epithelial monolayer | [184] |
| Human duodenal organoids (adult donors) | HIMECs | PDMS, type IV collagen, and Matrigel mix | Soft lithography | Showed culture system suitability for studying intestinal metabolism and drug transport | [197] |
| Human jejunal organoids (adult donors) | HUVECs | PDMS, type IV collagen | Soft lithography | Shear stress generated by luminal and basolateral flow produced a model of continuous intestinal differentiation, no villi-like structures observed with stem cell expansion media on the luminal side | [198] |
| Mouse colon tissue explant | Intestinal submucosal and muscular layers, microbiota |
Cyclin olefin polymer and polyurethane no ECM |
Injection molding | Dual-flow microfluidics allowed for the culture of full thickness explants over 3 days, recapitulated the in vivo oxygen gradient across the epithelial layer | [199] |
| Mouse proximal small intestine organoids | Cryptosporidium parvum | PDMS, Type I collagen, and Matrigel mix-coated 3D scaffold | Soft lithography and laser ablation | Established a long-lived and tube-shaped intestinal epithelial culture system by using crypt-like microcavities under flow, induced topography-guided self-organization of a functional epithelium with crypt- and villus-like domains similar to that observed in vivo, the culture system showed self-regeneration capacity and response to bacterial infection | [200] |
| Caco-2 | L. rhamnosus GG and Bacteroides caccae | Polycarbonate, type I collagen; porcine gastric mucin | Computer-controlled milling, laser cutting, and bolting | Engineered a modular architecture consisting of 3 microchambers to facilitate human and microbial cell interface, allowed measuring individual transcriptional responses in different infectious contexts and real-time monitoring of oxygen concentrations | [201] |
| Caco-2 | Human gut microbiota, E. coli, human PBMCs, human microvascular endothelial cells, and human lymphatic microvascular endothelial cells | PDMS, type I collagen, and Matrigel mix | Soft lithography | Established a stable long-term co-culture system of commensal and pathogenic microbes with intestinal epithelial cells, lack of mechanical stimulation induced bacterial overgrowth, similar to what is observed in IBD patients, emulated intestinal infection and inflammatory responses | [180] |
| Caco-2 | Human gut microbiota, E. coli, PBMCs | PDMS, type I collagen, and Matrigel mix | Soft lithography | Re-created a dextran sodium sulfate–induced epithelial inflammatory response, described intestinal barrier dysfunction as a critical trigger of inflammation onset in the gut | [202] |
| Caco-2 | S. flexneri | PDMS, type I collagen, and Matrigel mix | Soft lithography | Enabled the replication of Shigella infection hallmarks, Shigella invaded directly via the luminal side of the epithelium composed solely of enterocytes, 3D crypt-like structures provided a safe harbor for bacteria against luminal washout | [203] |
| Human colon organoids (pediatric and adult donors) | HIMECs, EHEC | PDMS, type I collagen, and Matrigel mix | Soft lithography | Observed that infectious activity of EHEC is promoted by human gut microbiome metabolites, when compared with those derived from mouse, recapitulated the proinflammatory and anti-inflammatory cytokine profiles induced by EHEC infection | [204] |
| Caco-2 | Bifidobacterium adolescentis and Eubacterium hallii | PDMS, type I collagen and Matrigel mix | Soft lithography | Simulated a steady-state vertical oxygen gradient, the transepithelial anoxic interface allowed co-culture with obligate anaerobes | [205] |
| Caco-2 and human ileal organoids (pediatric donors) | Bacteroides fragilis, human gut microbiota, HIMECs | PDMS, type I collagen, and Matrigel mix | Soft lithography | Established an oxygen gradient compatible with co-culture of a complex community of anaerobic commensal microorganisms | [183] |
| Caco-2 | HUVECs, PBMCs, mucosal macrophages, dendritic cells, L. rhamnosus, Candida albicans | Polystyrol, PET | Injection molding | Characterized immunologic responses to luminal lipopolysaccharide and endotoxemia, addressed the role of probiotics in protecting from opportunistic infections | [206] |
| Caco-2 | Neutrophils, monocytic THP1 cells |
Glass, polystyrene, and proprietary polymers Membrane-free (Phase Guide) type I collagen |
— | Simulated acute intestinal inflammatory responses by enabling neutrophil recruitment to the parenchymal compartment | [38] |
| Human colon organoids (pediatric and adult donors) | PBMCs |
Glass, polystyrene, and proprietary polymers type I collagen |
— | Studied IBD-associated inflammatory responses | [207] |
| Human ileal organoids (pediatric donors) | Endothelial colony forming cell derived endothelial cells, intestinal subepithelial myofibroblasts | PDMS, Polycarbonate type IV collagen | Soft lithography | Characterized ISEMF-induced angiogenesis and its physiological parameters, evaluated the effect of perfused vasculature on intestinal epithelial cell culture | [208] |
| Mouse small intestine | Lamina propria lymphocytes and ILC3s | recombinant mouse IL-2, IL-7, IL-15 and IL-23 | — | IL-22-dependent stem cell proliferation | [123] |
| Mouse small intestine | Pre-stimulated CD4+ splenocytes | Organoid medium | — | Intestinal stem cell differentiation | [119] |
| Human fetal intestine | Pre-stimulated fetal lamina propria T cells | p38 MAPK inhibitor, recombinant human IL-2 | — | Organoid outgrowth | [209] |
| Mouse small intestine | αβ and γδ IELs | recombinant mouse IL-2, IL-7, IL-15 | — | IEL survival, proliferation, and incorporation in the epithelium | [210] |
| Mouse small intestine | OT-I CD8+ splenocytes | EGF, noggin, R-spondin1 or R-spondin3 | — | T cell proliferation and IEL phenotype | [211] |
| Mouse Lgr5(+) ISCs at single-cell level | OT-II splenocytes (naive) | Organoid medium | — | T cell proliferation | [119] |
CAR chimeric antigen receptor, CDK cyclin-dependent kinase, CRC colorectal cancer, DC dendritic cell, DKK1 dickkopf Wnt signaling pathway inhibitor 1, ECM extracellular matrix, EGF epidermal growth factor, EHEC enterohemorrhagic E coli, FZD9 frizzled class receptor 9, HIMEC human intestinal microvascular endothelial cell, HUVEC human umbilical vein endothelial cell, IEL intraepithelial lymphocyte, ILC3 group 3 innate lymphoid cell, ISEMF intestinal subepithelial myofibroblast, MAPK mitogen-activated protein kinase, MDOTS mouse-derived organotypic tumor spheroid, MUC2 mucin 2, NSCLC non-small-cell lung cancer, PD1 programmed cell death 1, PDMS polydimethylsiloxane, PDOTS patient-derived organotypic tumor spheroid, PET polyethylene terephthalate, RSV respiratory syncytial virus, TCR T cell receptor, THP1 Tohoku Hospital Pediatrics-1 cell line, WNT3A WNT family member 3A.
Author contributions
YL contributed towards the conceiving and illustration of this manuscript. CT, DL, MY was responsible for the writing of this review. RS, NY, YLQT, HX JF, and MZ were responsible for assisting in collecting the literature and revising this review. YZ, YN, and JW provided some suggestions. JY and LW contributed to the discussion. All authors read and approved the final manuscript.
Funding
This work was supported by Science and Technology Innovation Committee of Shenzhen (JCYJ20190807145617113, JCYJ20210324113802006, JCYJ20200109150700942, and JCYJ2022053015180024), Guangdong Basic and Applied Basic Research Foundation (Grant 2021A1515010985, 2023A1515011936), Shenzhen Fund for Guangdong Provincial High Level Clinical Key Specialties (Grant SZGSP013), and the Shenzhen Key Medical Discipline Construction Fund (Grant SZXK042).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Cheng-mei Tian, Mei-feng Yang, Hao-ming Xu, Min-zheng Zhu.
Contributor Information
Rui-yue Shi, Email: ruiyueshi@126.com.
Jun Yao, Email: yao.jun@szhospital.com.
Li-sheng Wang, Email: wanglsszrmyy@163.com.
Yu-jie Liang, Email: liangyjie@126.com.
De-feng Li, Email: ldf830712@163.com.
References
- 1.Ng SC, Shi HY, Hamidi N, Underwood FE, Tang W, Benchimol EI, et al. Worldwide incidence and prevalence of inflammatory bowel disease in the 21st century: a systematic review of population-based studies. Lancet (Lond, Engl) 2017;390:2769–78. doi: 10.1016/s0140-6736(17)32448-0. [DOI] [PubMed] [Google Scholar]
- 2.Ramos GP, Papadakis KA. Mechanisms of disease: inflammatory bowel diseases. Mayo Clin Proc. 2019;94:155–65. doi: 10.1016/j.mayocp.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu Q, Yang MF, Liang YJ, Xu J, Xu HM, Nie YQ, et al. Immunology of inflammatory bowel disease: molecular mechanisms and therapeutics. J Inflamm Res. 2022;15:1825–44. doi: 10.2147/jir.S353038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rahman S, Ghiboub M, Donkers JM, van de Steeg E, van Tol EAF, Hakvoort TBM et al. The progress of intestinal epithelial models from cell lines to gut-on-chip. Int J Mol Sci. 2021;22 10.3390/ijms222413472 [DOI] [PMC free article] [PubMed]
- 5.Leung C, Tan SH, Barker N. Recent advances in Lgr5(+) stem cell research. Trends Cell Biol. 2018;28:380–91. doi: 10.1016/j.tcb.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–5. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 7.Dotti I, Mora-Buch R, Ferrer-Picón E, Planell N, Jung P, Masamunt MC, et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut. 2017;66:2069–79. doi: 10.1136/gutjnl-2016-312609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Wu J, Chen J, Dou W, Zhao Q, Han J, et al. Advances in reconstructing intestinal functionalities in vitro: from two/three dimensional-cell culture platforms to human intestine-on-a-chip. Talanta. 2021;226:122097. doi: 10.1016/j.talanta.2021.122097. [DOI] [PubMed] [Google Scholar]
- 9.Sarvestani SK, Signs S, Hu B, Yeu Y, Feng H, Ni Y, et al. Induced organoids derived from patients with ulcerative colitis recapitulate colitic reactivity. Nat Commun. 2021;12:262. doi: 10.1038/s41467-020-20351-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature. 2011;470:105–9. doi: 10.1038/nature09691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oda M, Hatano Y, Sato T. Intestinal epithelial organoids: regeneration and maintenance of the intestinal epithelium. Curr Opin Genet Dev. 2022;76:101977. doi: 10.1016/j.gde.2022.101977. [DOI] [PubMed] [Google Scholar]
- 12.Baker EJ, Beck NA, Berg EL, Clayton-Jeter HD, Chandrasekera PC, Curley JL, et al. Advancing nonclinical innovation and safety in pharmaceutical testing. Drug Discov Today. 2019;24:624–8. doi: 10.1016/j.drudis.2018.11.011. [DOI] [PubMed] [Google Scholar]
- 13.Mollaki V. Ethical challenges in organoid use. BioTech (Basel). 2021;10 10.3390/biotech10030012 [DOI] [PMC free article] [PubMed]
- 14.Chen MK, Beierle EA. Animal models for intestinal tissue engineering. Biomaterials. 2004;25:1675–81. doi: 10.1016/s0142-9612(03)00517-9. [DOI] [PubMed] [Google Scholar]
- 15.Franco R, Cedazo-Minguez A. Successful therapies for Alzheimer’s disease: why so many in animal models and none in humans? Front Pharm. 2014;5:146. doi: 10.3389/fphar.2014.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Justice MJ, Dhillon P. Using the mouse to model human disease: increasing validity and reproducibility. Dis Model Mech. 2016;9:101–3. doi: 10.1242/dmm.024547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wakisaka Y, Sugimoto S, Sato T. Organoid medicine for inflammatory bowel disease. Stem Cells. 2022;40:123–32. doi: 10.1093/stmcls/sxab020. [DOI] [PubMed] [Google Scholar]
- 18.Rossi G, Manfrin A, Lutolf MP. Progress and potential in organoid research. Nat Rev Genet. 2018;19:671–87. doi: 10.1038/s41576-018-0051-9. [DOI] [PubMed] [Google Scholar]
- 19.Wells JM, Spence JR. How to make an intestine. Development. 2014;141:752–60. doi: 10.1242/dev.097386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duval K, Grover H, Han LH, Mou Y, Pegoraro AF, Fredberg J, et al. Modeling physiological events in 2D vs. 3D cell culture. Physiol (Bethesda) 2017;32:266–77. doi: 10.1152/physiol.00036.2016. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 21.Sun H, Chow EC, Liu S, Du Y, Pang KS. The Caco-2 cell monolayer: usefulness and limitations. Expert Opin Drug Metab Toxicol. 2008;4:395–411. doi: 10.1517/17425255.4.4.395. [DOI] [PubMed] [Google Scholar]
- 22.Lea T. in The Impact of Food Bioactives on Health: in vitro and ex vivo models (eds K Verhoeckx et al.) 103–11 (Springer Copyright 2015, The Author(s). 2015) [PubMed]
- 23.Rousset M. The human colon carcinoma cell lines HT-29 and Caco-2: two in vitro models for the study of intestinal differentiation. Biochimie. 1986;68:1035–40. doi: 10.1016/s0300-9084(86)80177-8. [DOI] [PubMed] [Google Scholar]
- 24.Murakami H, Masui H. Hormonal control of human colon carcinoma cell growth in serum-free medium. Proc Natl Acad Sci USA. 1980;77:3464–8. doi: 10.1073/pnas.77.6.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Evans GS, Flint N, Somers AS, Eyden B, Potten CS. The development of a method for the preparation of rat intestinal epithelial cell primary cultures. J Cell Sci. 1992;101:219–31. doi: 10.1242/jcs.101.1.219. [DOI] [PubMed] [Google Scholar]
- 26.Huch M, Knoblich JA, Lutolf MP, Martinez-Arias A. The hope and the hype of organoid research. Development. 2017;144:938–41. doi: 10.1242/dev.150201. [DOI] [PubMed] [Google Scholar]
- 27.Li Y, Kilian KA. Bridging the gap: from 2D cell culture to 3D microengineered extracellular matrices. Adv Health Mater. 2015;4:2780–96. doi: 10.1002/adhm.201500427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ylostalo JH. 3D stem cell culture. Cells. 2020;9 10.3390/cells9102178 [DOI] [PMC free article] [PubMed]
- 29.Zachos NC, Kovbasnjuk O, Foulke-Abel J, In J, Blutt SE, de Jonge HR, et al. Human enteroids/colonoids and intestinal organoids functionally recapitulate normal intestinal physiology and pathophysiology. J Biol Chem. 2016;291:3759–66. doi: 10.1074/jbc.R114.635995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Su J, Wei J, Jiang N, Ge X. Recent advances in three-dimensional stem cell culture systems and applications. Stem Cells Int. 2021;2021:9477332. doi: 10.1155/2021/9477332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen Q, Wang Y. The application of three-dimensional cell culture in clinical medicine. Biotechnol Lett. 2020;42:2071–82. doi: 10.1007/s10529-020-03003-y. [DOI] [PubMed] [Google Scholar]
- 32.McKee C, Chaudhry GR. Advances and challenges in stem cell culture. Colloids Surf B Biointerfaces. 2017;159:62–77. doi: 10.1016/j.colsurfb.2017.07.051. [DOI] [PubMed] [Google Scholar]
- 33.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–7. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 34.Yan KS, Janda CY, Chang J, Zheng GXY, Larkin KA, Luca VC, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5(+) intestinal stem-cell self-renewal. Nature. 2017;545:238–42. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Browning TH, Trier JS. Organ culture of mucosal biopsies of human small intestine. J Clin Invest. 1969;48:1423–32. doi: 10.1172/jci106108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–84. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 37.van der Flier LG, Clevers H. Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. doi: 10.1146/annurev.physiol.010908.163145. [DOI] [PubMed] [Google Scholar]
- 38.Gjorevski N, Sachs N, Manfrin A, Giger S, Bragina ME, Ordóñez-Morán P, et al. Designer matrices for intestinal stem cell and organoid culture. Nature. 2016;539:560–4. doi: 10.1038/nature20168. [DOI] [PubMed] [Google Scholar]
- 39.Urbischek M, Rannikmae H, Foets T, Ravn K, Hyvönen M, de la Roche M. Organoid culture media formulated with growth factors of defined cellular activity. Sci Rep. 2019;9:6193. doi: 10.1038/s41598-019-42604-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malik M, Yang Y, Fathi P, Mahler GJ, Esch MB. Critical considerations for the design of multi-organ microphysiological systems (MPS) Front Cell Dev Biol. 2021;9:721338. doi: 10.3389/fcell.2021.721338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koyilot MC, Natarajan P, Hunt CR, Sivarajkumar S, Roy R, Joglekar S et al. Breakthroughs and applications of organ-on-a-chip technology. Cells. 2022;11 10.3390/cells11111828 [DOI] [PMC free article] [PubMed]
- 42.Mizutani T, Clevers H. Primary intestinal epithelial organoid culture. Methods Mol Biol. 2020;2171:185–200. doi: 10.1007/978-1-0716-0747-3_11. [DOI] [PubMed] [Google Scholar]
- 43.Miura S, Suzuki A. Brief summary of the current protocols for generating intestinal organoids. Dev Growth Differ. 2018;60:387–92. doi: 10.1111/dgd.12559. [DOI] [PubMed] [Google Scholar]
- 44.Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell. 2012;149:1192–205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 45.Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, et al. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–6. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 46.Stanger BZ, Datar R, Murtaugh LC, Melton DA. Direct regulation of intestinal fate by Notch. Proc Natl Acad Sci USA. 2005;102:12443–8. doi: 10.1073/pnas.0505690102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sato T, Stange DE, Ferrante M, Vries RG, Van Es JH, Van den Brink S, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–72. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 48.Shin YC, Shin W, Koh D, Wu A, Ambrosini YM, Min S et al. Three-dimensional regeneration of patient-derived intestinal organoid epithelium in a physiodynamic mucosal interface-on-a-chip. Micromachines (Basel). 2020;11 10.3390/mi11070663 [DOI] [PMC free article] [PubMed]
- 49.Fair KL, Colquhoun J & Hannan NRF. Intestinal organoids for modelling intestinal development and disease. Philos Trans R Soc Lond B Biol Sci. 2018;373 10.1098/rstb.2017.0217 [DOI] [PMC free article] [PubMed]
- 50.Yoo JH, Donowitz M. Intestinal enteroids/organoids: a novel platform for drug discovery in inflammatory bowel diseases. World J Gastroenterol. 2019;25:4125–47. doi: 10.3748/wjg.v25.i30.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tsai YH, Czerwinski M, Wu A, Dame MK, Attili D, Hill E, et al. A method for cryogenic preservation of human biopsy specimens and subsequent organoid culture. Cell Mol Gastroenterol Hepatol. 2018;6:218–222.e217. doi: 10.1016/j.jcmgh.2018.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noben M, Verstockt B, de Bruyn M, Hendriks N, Van Assche G, Vermeire S, et al. Epithelial organoid cultures from patients with ulcerative colitis and Crohn’s disease: a truly long-term model to study the molecular basis for inflammatory bowel disease? Gut. 2017;66:2193–5. doi: 10.1136/gutjnl-2016-313667. [DOI] [PubMed] [Google Scholar]
- 53.Lucafò M, Muzzo A, Marcuzzi M, Giorio L, Decorti G, Stocco G. Patient-derived organoids for therapy personalization in inflammatory bowel diseases. World J Gastroenterol. 2022;28:2636–53. doi: 10.3748/wjg.v28.i24.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dutta D, Heo I, Clevers H. Disease modeling in stem cell-derived 3D organoid systems. Trends Mol Med. 2017;23:393–410. doi: 10.1016/j.molmed.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 55.Lancaster MA, Knoblich JA. Organogenesis in a dish: modeling development and disease using organoid technologies. Science. 2014;345:1247125. doi: 10.1126/science.1247125. [DOI] [PubMed] [Google Scholar]
- 56.Goldrick C, Guri I, Herrera-Oropeza G, O’Brien-Gore C, Roy E, Wojtynska M, et al. 3D multicellular systems in disease modelling: From organoids to organ-on-chip. Front Cell Dev Biol. 2023;11:1083175. doi: 10.3389/fcell.2023.1083175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jung P, Sato T, Merlos-Suárez A, Barriga FM, Iglesias M, Rossell D, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17:1225–7. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 58.Pleguezuelos-Manzano C, Puschhof J, van den Brink S, Geurts V, Beumer J, Clevers H. Establishment and culture of human intestinal organoids derived from adult stem cells. Curr Protoc Immunol. 2020;130:e106. doi: 10.1002/cpim.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sato T, Clevers H. Growing self-organizing mini-guts from a single intestinal stem cell: mechanism and applications. Science. 2013;340:1190–4. doi: 10.1126/science.1234852. [DOI] [PubMed] [Google Scholar]
- 60.Miura S, Suzuki A. Generation of mouse and human organoid-forming intestinal progenitor cells by direct lineage reprogramming. Cell Stem Cell. 2017;21:456–471.e455. doi: 10.1016/j.stem.2017.08.020. [DOI] [PubMed] [Google Scholar]
- 61.Takahashi Y, Sato S, Kurashima Y, Yamamoto T, Kurokawa S, Yuki Y, et al. A refined culture system for human induced pluripotent stem cell-derived intestinal epithelial organoids. Stem Cell Rep. 2018;10:314–28. doi: 10.1016/j.stemcr.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McCracken KW, Howell JC, Wells JM, Spence JR. Generating human intestinal tissue from pluripotent stem cells in vitro. Nat Protoc. 2011;6:1920–8. doi: 10.1038/nprot.2011.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li M, Izpisua Belmonte JC. Organoids - preclinical models of human disease. N Engl J Med. 2019;380:569–79. doi: 10.1056/NEJMra1806175. [DOI] [PubMed] [Google Scholar]
- 64.Sugimoto S, Sato T. Establishment of 3D intestinal organoid cultures from intestinal stem cells. Methods Mol Biol. 2017;1612:97–105. doi: 10.1007/978-1-4939-7021-6_7. [DOI] [PubMed] [Google Scholar]
- 65.Okamoto R, Shimizu H, Suzuki K, Kawamoto A, Takahashi J, Kawai M, et al. Organoid-based regenerative medicine for inflammatory bowel disease. Regen Ther. 2020;13:1–6. doi: 10.1016/j.reth.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hofer M, Lutolf MP. Engineering organoids. Nat Rev Mater. 2021;6:402–20. doi: 10.1038/s41578-021-00279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Farahani M, Carthew J, Bhowmik S, Shard C, Nunez-Nescolarde A, Gomez GA, et al. Emerging biomaterials and technologies to control stem cell fate and patterning in engineered 3D tissues and organoids. Biointerphases. 2022;17:060801. doi: 10.1116/6.0002034. [DOI] [PubMed] [Google Scholar]
- 68.Zheng F, Xiao Y, Liu H, Fan Y, Dao M. Patient-specific organoid and organ-on-a-Chip: 3D cell-culture meets 3D printing and numerical simulation. Adv Biol (Weinh) 2021;5:e2000024. doi: 10.1002/adbi.202000024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sasserath T, Rumsey JW, McAleer CW, Bridges LR, Long CJ, Elbrecht D, et al. Differential monocyte actuation in a three-organ functional innate immune system-on-a-chip. Adv Sci (Weinh) 2020;7:2000323. doi: 10.1002/advs.202000323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choudhury D, Ashok A, Naing MW. Commercialization of organoids. Trends Mol Med. 2020;26:245–9. doi: 10.1016/j.molmed.2019.12.002. [DOI] [PubMed] [Google Scholar]
- 71.Takahashi J, Mizutani T, Sugihara HY, Nagata S, Kato S, Hiraguri Y, et al. Suspension culture in a rotating bioreactor for efficient generation of human intestinal organoids. Cell Rep. Methods. 2022;2:100337. doi: 10.1016/j.crmeth.2022.100337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Giandomenico SL, Mierau SB, Gibbons GM, Wenger LMD, Masullo L, Sit T, et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat Neurosci. 2019;22:669–79. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu XY, Jiang W, Ma D, Ge LP, Yang YS, Gou ZC, et al. SYTL4 downregulates microtubule stability and confers paclitaxel resistance in triple-negative breast cancer. Theranostics. 2020;10:10940–56. doi: 10.7150/thno.45207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jaganathan H, Gage J, Leonard F, Srinivasan S, Souza GR, Dave B, et al. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci Rep. 2014;4:6468. doi: 10.1038/srep06468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Urbanczyk M, Zbinden A, Layland SL, Duffy G, Schenke-Layland K. Controlled heterotypic pseudo-islet assembly of human β-cells and human umbilical vein endothelial cells using magnetic levitation. Tissue Eng Part A. 2020;26:387–99. doi: 10.1089/ten.TEA.2019.0158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tseng H, Balaoing LR, Grigoryan B, Raphael RM, Killian TC, Souza GR, et al. A three-dimensional co-culture model of the aortic valve using magnetic levitation. Acta Biomater. 2014;10:173–82. doi: 10.1016/j.actbio.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garreta E, Kamm RD, Chuva de Sousa Lopes SM, Lancaster MA, Weiss R, Trepat X, et al. Rethinking organoid technology through bioengineering. Nat Mater. 2021;20:145–55. doi: 10.1038/s41563-020-00804-4. [DOI] [PubMed] [Google Scholar]
- 78.Dergham Y, Sanchez-Vizuete P, Le Coq D, Deschamps J, Bridier A, Hamze K et al. Comparison of the genetic features involved in bacillus subtilis biofilm formation using multi-culturing approaches. Microorganisms. 2021;9. 10.3390/microorganisms9030633 [DOI] [PMC free article] [PubMed]
- 79.Kaur S, Kaur I, Rawal P, Tripathi DM, Vasudevan A. Non-matrigel scaffolds for organoid cultures. Cancer Lett. 2021;504:58–66. doi: 10.1016/j.canlet.2021.01.025. [DOI] [PubMed] [Google Scholar]
- 80.Berrier AL, Yamada KM. Cell-matrix adhesion. J Cell Physiol. 2007;213:565–73. doi: 10.1002/jcp.21237. [DOI] [PubMed] [Google Scholar]
- 81.Asano Y, Nishiguchi A, Matsusaki M, Okano D, Saito E, Akashi M, et al. Ultrastructure of blood and lymphatic vascular networks in three-dimensional cultured tissues fabricated by extracellular matrix nanofilm-based cell accumulation technique. Microsc (Oxf) 2014;63:219–26. doi: 10.1093/jmicro/dfu005. [DOI] [PubMed] [Google Scholar]
- 82.Saldin LT, Cramer MC, Velankar SS, White LJ, Badylak SF. Extracellular matrix hydrogels from decellularized tissues: structure and function. Acta Biomater. 2017;49:1–15. doi: 10.1016/j.actbio.2016.11.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang Y, Gunasekara DB, Reed MI, DiSalvo M, Bultman SJ, Sims CE, et al. A microengineered collagen scaffold for generating a polarized crypt-villus architecture of human small intestinal epithelium. Biomaterials. 2017;128:44–55. doi: 10.1016/j.biomaterials.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim S, Min S, Choi YS, Jo SH, Jung JH, Han K, et al. Tissue extracellular matrix hydrogels as alternatives to Matrigel for culturing gastrointestinal organoids. Nat Commun. 2022;13:1692. doi: 10.1038/s41467-022-29279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang L, Xiao L, Jung Poudel A, Li J, Zhou P, Gauthier M, et al. Porous chitosan microspheres as microcarriers for 3D cell culture. Carbohydr Polym. 2018;202:611–20. doi: 10.1016/j.carbpol.2018.09.021. [DOI] [PubMed] [Google Scholar]
- 86.Ratheesh G, Venugopal JR, Chinappan A, Ezhilarasu H, Sadiq A, Ramakrishna S. 3D fabrication of polymeric scaffolds for regenerative therapy. ACS Biomater Sci Eng. 2017;3:1175–94. doi: 10.1021/acsbiomaterials.6b00370. [DOI] [PubMed] [Google Scholar]
- 87.Bhuptani RS, Patravale VB. Porous microscaffolds for 3D culture of dental pulp mesenchymal stem cells. Int J Pharm. 2016;515:555–64. doi: 10.1016/j.ijpharm.2016.10.040. [DOI] [PubMed] [Google Scholar]
- 88.Fong ELS, Toh TB, Lin QXX, Liu Z, Hooi L, Mohd Abdul Rashid MB, et al. Generation of matched patient-derived xenograft in vitro-in vivo models using 3D macroporous hydrogels for the study of liver cancer. Biomaterials. 2018;159:229–40. doi: 10.1016/j.biomaterials.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 89.Singhvi MS, Zinjarde SS, Gokhale DV. Polylactic acid: synthesis and biomedical applications. J Appl Microbiol. 2019;127:1612–26. doi: 10.1111/jam.14290. [DOI] [PubMed] [Google Scholar]
- 90.He J, Zhang X, Xia X, Han M, Li F, Li C, et al. Organoid technology for tissue engineering. J Mol Cell Biol. 2020;12:569–79. doi: 10.1093/jmcb/mjaa012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Paşca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat Methods. 2015;12:671–8. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Takebe T, Wells JM. Organoids by design. Science. 2019;364:956–9. doi: 10.1126/science.aaw7567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horita N, Tsuchiya K, Hayashi R, Fukushima K, Hibiya S, Fukuda M, et al. Fluorescent labelling of intestinal epithelial cells reveals independent long-lived intestinal stem cells in a crypt. Biochem Biophys Res Commun. 2014;454:493–9. doi: 10.1016/j.bbrc.2014.10.091. [DOI] [PubMed] [Google Scholar]
- 94.Artegiani B, Hendriks D, Beumer J, Kok R, Zheng X, Joore I, et al. Fast and efficient generation of knock-in human organoids using homology-independent CRISPR-Cas9 precision genome editing. Nat Cell Biol. 2020;22:321–31. doi: 10.1038/s41556-020-0472-5. [DOI] [PubMed] [Google Scholar]
- 95.Michels BE, Mosa MH, Streibl BI, Zhan T, Menche C, Abou-El-Ardat K, et al. Pooled In Vitro and In Vivo CRISPR-Cas9 screening identifies tumor suppressors in human colon organoids. Cell Stem Cell. 2020;26:782–e787. doi: 10.1016/j.stem.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 96.Nahak BK, Mishra A, Preetam S, Tiwari A. Advances in organ-on-a-chip materials and devices. ACS Appl Bio Mater. 2022;5:3576–607. doi: 10.1021/acsabm.2c00041. [DOI] [PubMed] [Google Scholar]
- 97.Allaire JM, Crowley SM, Law HT, Chang SY, Ko HJ, Vallance BA. The intestinal epithelium: central coordinator of mucosal immunity. Trends Immunol. 2018;39:677–96. doi: 10.1016/j.it.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 98.Guan Q. A comprehensive review and update on the pathogenesis of inflammatory bowel disease. J Immunol Res. 2019;2019:7247238. doi: 10.1155/2019/7247238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang L, Li Z, Xu C, Qin J. Bioinspired engineering of organ-on-chip devices. Adv Exp Med Biol. 2019;1174:401–40. doi: 10.1007/978-81-13-9791-2_13. [DOI] [PubMed] [Google Scholar]
- 100.Low LA, Mummery C, Berridge BR, Austin CP, Tagle DA. Organs-on-chips: into the next decade. Nat Rev Drug Discov. 2021;20:345–61. doi: 10.1038/s41573-020-0079-3. [DOI] [PubMed] [Google Scholar]
- 101.Luni C, Serena E, Elvassore N. Human-on-chip for therapy development and fundamental science. Curr Opin Biotechnol. 2014;25:45–50. doi: 10.1016/j.copbio.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 102.Takebe T, Zhang B, Radisic M. Synergistic engineering: organoids meet organs-on-a-chip. Cell Stem Cell. 2017;21:297–300. doi: 10.1016/j.stem.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 103.Brandenberg N, Hoehnel S, Kuttler F, Homicsko K, Ceroni C, Ringel T, et al. High-throughput automated organoid culture via stem-cell aggregation in microcavity arrays. Nat Biomed Eng. 2020;4:863–74. doi: 10.1038/s41551-020-0565-2. [DOI] [PubMed] [Google Scholar]
- 104.Omerzu M, Fenderico N, de Barbanson B, Sprangers J, de Ridder J & Maurice MM. Three-dimensional analysis of single molecule FISH in human colon organoids. Biol Open. 2019;8. 10.1242/bio.042812 [DOI] [PMC free article] [PubMed]
- 105.Title AC, Karsai M, Mir-Coll J, Grining ÖY, Rufer C, Sonntag S, et al. Evaluation of the effects of harmine on β-cell function and proliferation in standardized human islets using 3D high-content confocal imaging and automated analysis. Front Endocrinol (Lausanne) 2022;13:854094. doi: 10.3389/fendo.2022.854094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Vunjak-Novakovic G, Ronaldson-Bouchard K, Radisic M. Organs-on-a-chip models for biological research. Cell. 2021;184:4597–611. doi: 10.1016/j.cell.2021.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Skardal A, Shupe T, Atala A. Organoid-on-a-chip and body-on-a-chip systems for drug screening and disease modeling. Drug Discov Today. 2016;21:1399–411. doi: 10.1016/j.drudis.2016.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.van de Stolpe A, den Toonder J. Workshop meeting report Organs-on-Chips: human disease models. Lab Chip. 2013;13:3449–70. doi: 10.1039/c3lc50248a. [DOI] [PubMed] [Google Scholar]
- 109.Jang J, Park JY, Gao G, Cho DW. Biomaterials-based 3D cell printing for next-generation therapeutics and diagnostics. Biomaterials. 2018;156:88–106. doi: 10.1016/j.biomaterials.2017.11.030. [DOI] [PubMed] [Google Scholar]
- 110.Huang J, Xiong J, Wang D, Zhang J, Yang L, Sun S et al. 3D Bioprinting of hydrogels for cartilage tissue engineering. Gels (Basel, Switzerland). 2021;7 10.3390/gels7030144 [DOI] [PMC free article] [PubMed]
- 111.Carberry BJ, Hergert JE, Yavitt FM, Hernandez JJ, Speckl KF, Bowman CN et al. 3D printing of sacrificial thioester elastomers using digital light processing for templating 3D organoid structures in soft biomatrices. Biofabrication. 2021;13 10.1088/1758-5090/ac1c98 [DOI] [PMC free article] [PubMed]
- 112.Kim E, Choi S, Kang B, Kong J, Kim Y, Yoon WH, et al. Creation of bladder assembloids mimicking tissue regeneration and cancer. Nature. 2020;588:664–9. doi: 10.1038/s41586-020-3034-x. [DOI] [PubMed] [Google Scholar]
- 113.Brassard JA, Nikolaev M, Hübscher T, Hofer M, Lutolf MP. Recapitulating macro-scale tissue self-organization through organoid bioprinting. Nat Mater. 2021;20:22–29. doi: 10.1038/s41563-020-00803-5. [DOI] [PubMed] [Google Scholar]
- 114.Ren Y, Yang X, Ma Z, Sun X, Zhang Y, Li W, et al. Developments and opportunities for 3D bioprinted organoids. Int J Bioprint. 2021;7:364. doi: 10.18063/ijb.v7i3.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Howell KJ, Kraiczy J, Nayak KM, Gasparetto M, Ross A, Lee C, et al. DNA methylation and transcription patterns in intestinal epithelial cells from pediatric patients with inflammatory bowel diseases differentiate disease subtypes and associate with outcome. Gastroenterology. 2018;154:585–98. doi: 10.1053/j.gastro.2017.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen Y, Lin Y, Davis KM, Wang Q, Rnjak-Kovacina J, Li C, et al. Robust bioengineered 3D functional human intestinal epithelium. Sci Rep. 2015;5:13708. doi: 10.1038/srep13708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15:19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 118.Mochel JP, Jergens AE, Kingsbury D, Kim HJ, Martín MG, Allenspach K. Intestinal stem cells to advance drug development, precision, and regenerative medicine: a paradigm shift in translational research. Aaps J. 2017;20:17. doi: 10.1208/s12248-017-0178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Biton M, Haber AL, Rogel N, Burgin G, Beyaz S, Schnell A et al. T Helper cell cytokines modulate intestinal stem cell renewal and differentiation. Cell. 2018;175:1307–20.e2, 10.1016/j.cell.2018.10.008 (2018) [DOI] [PMC free article] [PubMed]
- 120.Asfaha S. Intestinal stem cells and inflammation. Curr Opin Pharm. 2015;25:62–6. doi: 10.1016/j.coph.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 121.Zwarycz B, Gracz AD, Rivera KR, Williamson IA, Samsa LA, Starmer J, et al. IL22 inhibits epithelial stem cell expansion in an ileal organoid model. Cell Mol Gastroenterol Hepatol. 2019;7:1–17. doi: 10.1016/j.jcmgh.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Abo H, Denning TL. Epithelial traffic control: IL22 gives TA cells the green light. Cell Mol Gastroenterol Hepatol. 2019;7:409–10. doi: 10.1016/j.jcmgh.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lindemans CA, Calafiore M, Mertelsmann AM, O’Connor MH, Dudakov JA, Jenq RR, et al. Interleukin-22 promotes intestinal-stem-cell-mediated epithelial regeneration. Nature. 2015;528:560–4. doi: 10.1038/nature16460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ray K. Stem cells: IL-22 activates ISCs for intestinal regeneration. Nat Rev Gastroenterol Hepatol. 2016;13:64. doi: 10.1038/nrgastro.2015.221. [DOI] [PubMed] [Google Scholar]
- 125.Engevik MA, Engevik KA, Yacyshyn MB, Wang J, Hassett DJ, Darien B, et al. Human Clostridium difficile infection: inhibition of NHE3 and microbiota profile. Am J Physiol Gastrointest Liver Physiol. 2015;308:G497–509. doi: 10.1152/ajpgi.00090.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hou Q, Ye L, Liu H, Huang L, Yang Q, Turner JR, et al. Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 2018;25:1657–70. doi: 10.1038/s41418-018-0070-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Hahn S, Nam MO, Noh JH, Lee DH, Han HW, Kim DH, et al. Organoid-based epithelial to mesenchymal transition (OEMT) model: from an intestinal fibrosis perspective. Sci Rep. 2017;7:2435. doi: 10.1038/s41598-017-02190-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Hibiya S, Tsuchiya K, Hayashi R, Fukushima K, Horita N, Watanabe S, et al. Long-term inflammation transforms intestinal epithelial cells of colonic organoids. J Crohns Colitis. 2017;11:621–30. doi: 10.1093/ecco-jcc/jjw186. [DOI] [PubMed] [Google Scholar]
- 129.Jekunen A. Decision-making in product portfolios of pharmaceutical research and development–managing streams of innovation in highly regulated markets. Drug Des Dev Ther. 2014;8:2009–16. doi: 10.2147/dddt.S68579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kopper JJ, Iennarella-Servantez C, Jergens AE, Sahoo DK, Guillot E, Bourgois-Mochel A, et al. Harnessing the biology of canine intestinal organoids to heighten understanding of inflammatory bowel disease pathogenesis and accelerate drug discovery: a one health approach. Front Toxicol. 2021;3:773953. doi: 10.3389/ftox.2021.773953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lacombe J, Soldevila M, Zenhausern F. From organ-on-chip to body-on-chip: the next generation of microfluidics platforms for in vitro drug efficacy and toxicity testing. Prog Mol Biol Transl Sci. 2022;187:41–91. doi: 10.1016/bs.pmbts.2021.07.019. [DOI] [PubMed] [Google Scholar]
- 132.Pithadia AB, Jain S. Treatment of inflammatory bowel disease (IBD) Pharmacol Rep. 2011;63:629–42. doi: 10.1016/s1734-1140(11)70575-8. [DOI] [PubMed] [Google Scholar]
- 133.Xu P, Elizalde M, Masclee A, Pierik M, Jonkers D. Corticosteroid enhances epithelial barrier function in intestinal organoids derived from patients with Crohn’s disease. J Mol Med (Berl) 2021;99:805–15. doi: 10.1007/s00109-021-02045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Khare V, Krnjic A, Frick A, Gmainer C, Asboth M, Jimenez K, et al. Mesalamine and azathioprine modulate junctional complexes and restore epithelial barrier function in intestinal inflammation. Sci Rep. 2019;9:2842. doi: 10.1038/s41598-019-39401-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Swidsinski A, Loening-Baucke V, Bengmark S, Lochs H, Dörffel Y. Azathioprine and mesalazine-induced effects on the mucosal flora in patients with IBD colitis. Inflamm Bowel Dis. 2007;13:51–56. doi: 10.1002/ibd.20003. [DOI] [PubMed] [Google Scholar]
- 136.Kawamoto A, Nagata S, Anzai S, Takahashi J, Kawai M, Hama M, et al. Ubiquitin D is upregulated by synergy of notch signalling and TNF-α in the inflamed intestinal epithelia of IBD patients. J Crohns Colitis. 2019;13:495–509. doi: 10.1093/ecco-jcc/jjy180. [DOI] [PubMed] [Google Scholar]
- 137.Tytgat KM, van der Wal JW, Einerhand AW, Büller HA, Dekker J. Quantitative analysis of MUC2 synthesis in ulcerative colitis. Biochem Biophys Res Commun. 1996;224:397–405. doi: 10.1006/bbrc.1996.1039. [DOI] [PubMed] [Google Scholar]
- 138.Lloyd K, Papoutsopoulou S, Smith E, Stegmaier P, Bergey F, Morris L et al. Using systems medicine to identify a therapeutic agent with potential for repurposing in inflammatory bowel disease. Dis Model Mech13, 10.1242/dmm.044040 (2020) [DOI] [PMC free article] [PubMed]
- 139.Zhang HM, Yuan S, Meng H, Hou XT, Li J, Xue JC et al. Stem cell-based therapies for inflammatory bowel disease. Int J Mol Sci. 2022;23. 10.3390/ijms23158494 [DOI] [PMC free article] [PubMed]
- 140.Sugimoto S, Kobayashi E, Fujii M, Ohta Y, Arai K, Matano M, et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature. 2021;592:99–104. doi: 10.1038/s41586-021-03247-2. [DOI] [PubMed] [Google Scholar]
- 141.Mishra R, Dhawan P, Srivastava AS, Singh AB. Inflammatory bowel disease: therapeutic limitations and prospective of the stem cell therapy. World J Stem Cells. 2020;12:1050–66. doi: 10.4252/wjsc.v12.i10.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, Zheng X, et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat Med. 2012;18:618–23. doi: 10.1038/nm.2695. [DOI] [PubMed] [Google Scholar]
- 143.Fordham RP, Yui S, Hannan NR, Soendergaard C, Madgwick A, Schweiger PJ, et al. Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury. Cell Stem Cell. 2013;13:734–44. doi: 10.1016/j.stem.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Watanabe S, Kobayashi S, Ogasawara N, Okamoto R. Transplantation of intestinal organoids into a mouse model of colitis. Nat Protoc. 2022;17:649–71. doi: 10.1038/s41596-021-00658-3. [DOI] [PubMed] [Google Scholar]
- 145.Date S, Sato T. Mini-gut organoids: reconstitution of the stem cell niche. Annu Rev Cell Dev Biol. 2015;31:269–89. doi: 10.1146/annurev-cellbio-100814-125218. [DOI] [PubMed] [Google Scholar]
- 146.Shaffiey SA, Jia H, Keane T, Costello C, Wasserman D, Quidgley M, et al. Intestinal stem cell growth and differentiation on a tubular scaffold with evaluation in small and large animals. Regen Med. 2016;11:45–61. doi: 10.2217/rme.15.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sugimoto S, Ohta Y, Fujii M, Matano M, Shimokawa M, Nanki K, et al. Reconstruction of the human colon epithelium In Vivo. Cell Stem Cell. 2018;22:171–176.e175. doi: 10.1016/j.stem.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 148.Watanabe S, Kobayashi S, Ogasawara N, Okamoto R, Nakamura T, Watanabe M, et al. Transplantation of intestinal organoids into a mouse model of colitis. Nat Protoc. 2022;17:649–71. doi: 10.1038/s41596-021-00658-3. [DOI] [PubMed] [Google Scholar]
- 149.Sugimoto S, Kobayashi E, Kanai T, Sato T. In Vivo intestinal research using organoid transplantation. Keio J Med. 2022;71:73–81. doi: 10.2302/kjm.2022-0019-IR. [DOI] [PubMed] [Google Scholar]
- 150.Nakanishi A, Toyama S, Onozato D, Watanabe C, Hashita T, Iwao T, et al. Effects of human induced pluripotent stem cell-derived intestinal organoids on colitis-model mice. Regen Ther. 2022;21:351–61. doi: 10.1016/j.reth.2022.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Li DF, Yang MF, Xu J, Xu HM, Zhu MZ, Liang YJ, et al. Extracellular vesicles: the next generation theranostic nanomedicine for inflammatory bowel disease. Int J Nanomed. 2022;17:3893–911. doi: 10.2147/ijn.S370784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Tian CM, Yang MF, Xu HM, Zhu MZ, Zhang Y, Yao J, et al. Emerging role of bacterial outer membrane vesicle in gastrointestinal tract. Gut Pathog. 2023;15:20. doi: 10.1186/s13099-023-00543-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Liang Y, Iqbal Z, Lu J, Wang J, Zhang H, Chen X, et al. Cell-derived nanovesicle-mediated drug delivery to the brain: principles and strategies for vesicle engineering. Mol Ther : J Am Soc Gene Ther. 2023;31:1207–24. doi: 10.1016/j.ymthe.2022.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Xu L, Xu X, Liang Y, Wen C, Ouyang K, Huang J, et al. Osteoclast-targeted delivery of anti-miRNA oligonucleotides by red blood cell extracellular vesicles. J Control Release : Off J Control Release Soc. 2023;358:259–72. doi: 10.1016/j.jconrel.2023.04.043. [DOI] [PubMed] [Google Scholar]
- 155.Tian CM, Yang MF, Xu HM, Zhu MZ, Zhang Y, Yao J, et al. Mesenchymal stem cell-derived exosomes: novel therapeutic approach for inflammatory bowel diseases. Stem Cells Int. 2023;2023:4245704. doi: 10.1155/2023/4245704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li DF, Yang MF, Xu HM, Zhu MZ, Zhang Y, Tian CM, et al. Nanoparticles for oral delivery: targeted therapy for inflammatory bowel disease. J Mater Chem B. 2022;10:5853–72. doi: 10.1039/d2tb01190e. [DOI] [PubMed] [Google Scholar]
- 157.Liang Y, Iqbal Z, Wang J, Xu L, Xu X, Ouyang K, et al. Cell-derived extracellular vesicles for CRISPR/Cas9 delivery: engineering strategies for cargo packaging and loading. Biomater Sci. 2022;10:4095–106. doi: 10.1039/d2bm00480a. [DOI] [PubMed] [Google Scholar]
- 158.Liang Y, Xu X, Xu L, Iqbal Z, Ouyang K, Zhang H, et al. Chondrocyte-specific genomic editing enabled by hybrid exosomes for osteoarthritis treatment. Theranostics. 2022;12:4866–78. doi: 10.7150/thno.69368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Iqbal Z, Rehman K, Xia J, Shabbir M, Zaman M, Liang Y, et al. Biomaterial-assisted targeted and controlled delivery of CRISPR/Cas9 for precise gene editing. Biomater Sci. 2023;11:3762–83. doi: 10.1039/d2bm01636b. [DOI] [PubMed] [Google Scholar]
- 160.Duan L, Xu L, Xu X, Qin Z, Zhou X, Xiao Y, et al. Exosome-mediated delivery of gene vectors for gene therapy. Nanoscale. 2021;13:1387–97. doi: 10.1039/d0nr07622h. [DOI] [PubMed] [Google Scholar]
- 161.Duan L, Ouyang K, Wang J, Xu L, Xu X, Wen C, et al. Exosomes as targeted delivery platform of CRISPR/Cas9 for therapeutic genome editing. Chembiochem: a Eur J Chem Biol. 2021;22:3360–8. doi: 10.1002/cbic.202100359. [DOI] [PubMed] [Google Scholar]
- 162.Fujii M, Clevers H, Sato T. Modeling human digestive diseases with CRISPR-Cas9-modified organoids. Gastroenterology. 2019;156:562–76. doi: 10.1053/j.gastro.2018.11.048. [DOI] [PubMed] [Google Scholar]
- 163.Li T, Yang Y, Qi H, Cui W, Zhang L, Fu X, et al. CRISPR/Cas9 therapeutics: progress and prospects. Signal Transduct Target Ther. 2023;8:36. doi: 10.1038/s41392-023-01309-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Koo BK, Stange DE, Sato T, Karthaus W, Farin HF, Huch M, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 2011;9:81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 165.Koo BK, Sasselli V, Clevers H. Retroviral gene expression control in primary organoid cultures. Curr Protoc Stem Cell Biol. 2013;27:5a.6.1–5a.6.8. doi: 10.1002/9780470151808.sc05a06s27. [DOI] [PubMed] [Google Scholar]
- 166.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–8. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 167.Wang X, Yamamoto Y, Wilson LH, Zhang T, Howitt BE, Farrow MA, et al. Cloning and variation of ground state intestinal stem cells. Nature. 2015;522:173–8. doi: 10.1038/nature14484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Borchardt RT, Hidalgo IJ, Raub TJ, Borchardt RT. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability, Gastroenterology, 96, 736-749, 1989–the backstory. Aaps J. 2011;13:323–7. doi: 10.1208/s12248-011-9283-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ma’ayeh SY, Knörr L, Sköld K, Garnham A, Ansell BRE, Jex AR, et al. Responses of the differentiated intestinal epithelial cell line Caco-2 to infection with the giardia intestinalis GS isolate. Front Cell Infect Microbiol. 2018;8:244. doi: 10.3389/fcimb.2018.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Bakhshi B, Barzelighi HM, Daraei B. The anti-adhesive and anti-invasive effects of recombinant azurin on the interaction between enteric pathogens (invasive/non-invasive) and Caco-2 cells. Micro Pathog. 2020;147:104246. doi: 10.1016/j.micpath.2020.104246. [DOI] [PubMed] [Google Scholar]
- 171.VanDussen KL, Marinshaw JM, Shaikh N, Miyoshi H, Moon C, Tarr PI, et al. Development of an enhanced human gastrointestinal epithelial culture system to facilitate patient-based assays. Gut. 2015;64:911–20. doi: 10.1136/gutjnl-2013-306651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Mitra A, Mishra L, Li S. Technologies for deriving primary tumor cells for use in personalized cancer therapy. Trends Biotechnol. 2013;31:347–54. doi: 10.1016/j.tibtech.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Tian CM, Zhang Y, Yang MF, Xu HM, Zhu MZ, Yao J, et al. Stem cell therapy in inflammatory bowel disease: a review of achievements and challenges. J Inflamm Res. 2023;16:2089–119. doi: 10.2147/jir.S400447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li XG, Zhu M, Chen MX, Fan HB, Fu HL, Zhou JY, et al. Acute exposure to deoxynivalenol inhibits porcine enteroid activity via suppression of the Wnt/β-catenin pathway. Toxicol Lett. 2019;305:19–31. doi: 10.1016/j.toxlet.2019.01.008. [DOI] [PubMed] [Google Scholar]
- 175.Zhou JY, Huang DG, Gao CQ, Yan HC, Zou SG, Wang XQ. Heat-stable enterotoxin inhibits intestinal stem cell expansion to disrupt the intestinal integrity by downregulating the Wnt/β-catenin pathway. Stem Cells. 2021;39:482–96. doi: 10.1002/stem.3324. [DOI] [PubMed] [Google Scholar]
- 176.Co JY, Margalef-Català M, Li X, Mah AT, Kuo CJ, Monack DM, et al. Controlling epithelial polarity: a human enteroid model for host-pathogen interactions. Cell Rep. 2019;26:2509–2520.e2504. doi: 10.1016/j.celrep.2019.01.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Roodsant T, Navis M, Aknouch I, Renes IB, van Elburg RM, Pajkrt D, et al. A human 2D primary organoid-derived epithelial monolayer model to study host-pathogen interaction in the small intestine. Front Cell Infect Microbiol. 2020;10:272. doi: 10.3389/fcimb.2020.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Braverman J, Yilmaz ÖH. From 3D organoids back to 2D enteroids. Dev Cell. 2018;44:533–4. doi: 10.1016/j.devcel.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 179.Delon LC, Guo Z, Oszmiana A, Chien CC, Gibson R, Prestidge C, et al. A systematic investigation of the effect of the fluid shear stress on Caco-2 cells towards the optimization of epithelial organ-on-chip models. Biomaterials. 2019;225:119521. doi: 10.1016/j.biomaterials.2019.119521. [DOI] [PubMed] [Google Scholar]
- 180.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. 2016;113:E7–15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Bu XD, Li N, Tian XQ, Huang PL. Caco-2 and LS174T cell lines provide different models for studying mucin expression in colon cancer. Tissue Cell. 2011;43:201–6. doi: 10.1016/j.tice.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 182.Damiano S, Sasso A, De Felice B, Di Gregorio I, La Rosa G, Lupoli GA, et al. Quercetin increases MUC2 and MUC5AC gene expression and secretion in intestinal goblet cell-like LS174T via PLC/PKCα/ERK1-2 pathway. Front Physiol. 2018;9:357. doi: 10.3389/fphys.2018.00357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jalili-Firoozinezhad S, Gazzaniga FS, Calamari EL, Camacho DM, Fadel CW, Bein A, et al. A complex human gut microbiome cultured in an anaerobic intestine-on-a-chip. Nat Biomed Eng. 2019;3:520–31. doi: 10.1038/s41551-019-0397-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kasendra M, Tovaglieri A, Sontheimer-Phelps A, Jalili-Firoozinezhad S, Bein A, Chalkiadaki A, et al. Development of a primary human Small Intestine-on-a-Chip using biopsy-derived organoids. Sci Rep. 2018;8:2871. doi: 10.1038/s41598-018-21201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lei H, Crawford MS & McCole DF. JAK-STAT Pathway regulation of intestinal permeability: pathogenic roles and therapeutic opportunities in inflammatory bowel disease. Pharmaceuticals (Basel). 2021;14. 10.3390/ph14090840 [DOI] [PMC free article] [PubMed]
- 186.Bayrer JR, Wang H, Nattiv R, Suzawa M, Escusa HS, Fletterick RJ, et al. LRH-1 mitigates intestinal inflammatory disease by maintaining epithelial homeostasis and cell survival. Nat Commun. 2018;9:4055. doi: 10.1038/s41467-018-06137-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Deuring JJ, Li M, Cao W, Chen S, Wang W, de Haar C, et al. Pregnane X receptor activation constrains mucosal NF-κB activity in active inflammatory bowel disease. PLoS One. 2019;14:e0221924. doi: 10.1371/journal.pone.0221924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Glal D, Sudhakar JN, Lu HH, Liu MC, Chiang HY, Liu YC, et al. ATF3 sustains IL-22-induced STAT3 phosphorylation to maintain mucosal immunity through inhibiting phosphatases. Front Immunol. 2018;9:2522. doi: 10.3389/fimmu.2018.02522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Means AL, Freeman TJ, Zhu J, Woodbury LG, Marincola-Smith P, Wu C, et al. Epithelial Smad4 deletion up-regulates inflammation and promotes inflammation-associated cancer. Cell Mol Gastroenterol Hepatol. 2018;6:257–76. doi: 10.1016/j.jcmgh.2018.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Li C, Zhou Y, Rychahou P, Weiss HL, Lee EY, Perry CL, et al. SIRT2 contributes to the regulation of intestinal cell proliferation and differentiation. Cell Mol Gastroenterol Hepatol. 2020;10:43–57. doi: 10.1016/j.jcmgh.2020.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.van der Giessen J, van der Woude CJ, Peppelenbosch MP & Fuhler GM. A direct effect of sex hormones on epithelial barrier function in inflammatory bowel disease models. Cells. 2019;8. 10.3390/cells8030261 [DOI] [PMC free article] [PubMed]
- 192.Lie M, van der Giessen J, Fuhler GM, de Lima A, Peppelenbosch MP, van der Ent C, et al. Low dose Naltrexone for induction of remission in inflammatory bowel disease patients. J Transl Med. 2018;16:55. doi: 10.1186/s12967-018-1427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Li Y, Zhang T, Guo C, Geng M, Gai S, Qi W et al. Bacillus subtilis RZ001 improves intestinal integrity and alleviates colitis by inhibiting the Notch signalling pathway and activating ATOH-1. Pathog Dis. 2020;78. 10.1093/femspd/ftaa016 [DOI] [PubMed]
- 194.Wlodarska M, Luo C, Kolde R, d’Hennezel E, Annand JW, Heim CE, et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe. 2017;22:25–37.e26. doi: 10.1016/j.chom.2017.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kim Y, West GA, Ray G, Kessler SP, Petrey AC, Fiocchi C, et al. Layilin is critical for mediating hyaluronan 35kDa-induced intestinal epithelial tight junction protein ZO-1 in vitro and in vivo. Matrix Biol. 2018;66:93–109. doi: 10.1016/j.matbio.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–74. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 197.Kasendra M, Luc R, Yin J, Manatakis DV, Kulkarni G, Lucchesi C et al. Duodenum Intestine-Chip for preclinical drug assessment in a human relevant model. Elife. 2020;9. 10.7554/eLife.50135 [DOI] [PMC free article] [PubMed]
- 198.Yin J, Sunuwar L, Kasendra M, Yu H, Tse CM, Talbot CC, Jr., et al. Fluid shear stress enhances differentiation of jejunal human enteroids in Intestine-Chip. Am J Physiol Gastrointest Liver Physiol. 2021;320:G258–g271. doi: 10.1152/ajpgi.00282.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Bersini S, Moretti M. 3D functional and perfusable microvascular networks for organotypic microfluidic models. J Mater Sci Mater Med. 2015;26:180. doi: 10.1007/s10856-015-5520-5. [DOI] [PubMed] [Google Scholar]
- 200.Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature. 2020;585:574–8. doi: 10.1038/s41586-020-2724-8. [DOI] [PubMed] [Google Scholar]
- 201.Shah P, Fritz JV, Glaab E, Desai MS, Greenhalgh K, Frachet A, et al. A microfluidics-based in vitro model of the gastrointestinal human-microbe interface. Nat Commun. 2016;7:11535. doi: 10.1038/ncomms11535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shin W, Kim HJ. Intestinal barrier dysfunction orchestrates the onset of inflammatory host-microbiome cross-talk in a human gut inflammation-on-a-chip. Proc Natl Acad Sci USA. 2018;115:E10539–e10547. doi: 10.1073/pnas.1810819115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Grassart A, Malardé V, Gobaa S, Sartori-Rupp A, Kerns J, Karalis K, et al. Bioengineered human organ-on-chip reveals intestinal microenvironment and mechanical forces impacting shigella infection. Cell Host Microbe. 2019;26:565. doi: 10.1016/j.chom.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 204.Tovaglieri A, Sontheimer-Phelps A, Geirnaert A, Prantil-Baun R, Camacho DM, Chou DB, et al. Species-specific enhancement of enterohemorrhagic E. coli pathogenesis mediated by. Microbiome Metab Microbiome. 2019;7:43. doi: 10.1186/s40168-019-0650-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Shin W, Wu A, Massidda MW, Foster C, Thomas N, Lee DW, et al. A robust longitudinal co-culture of obligate anaerobic gut microbiome with human intestinal epithelium in an anoxic-oxic interface-on-a-chip. Front Bioeng Biotechnol. 2019;7:13. doi: 10.3389/fbioe.2019.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Maurer M, Gresnigt MS, Last A, Wollny T, Berlinghof F, Pospich R, et al. A three-dimensional immunocompetent intestine-on-chip model as in vitro platform for functional and microbial interaction studies. Biomaterials. 2019;220:119396. doi: 10.1016/j.biomaterials.2019.119396. [DOI] [PubMed] [Google Scholar]
- 207.Beaurivage C, Kanapeckaite A, Loomans C, Erdmann KS, Stallen J, Janssen RAJ. Development of a human primary gut-on-a-chip to model inflammatory processes. Sci Rep. 2020;10:21475. doi: 10.1038/s41598-020-78359-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Seiler KM, Bajinting A, Alvarado DM, Traore MA, Binkley MM, Goo WH, et al. Patient-derived small intestinal myofibroblasts direct perfused, physiologically responsive capillary development in a microfluidic Gut-on-a-Chip Model. Sci Rep. 2020;10:3842. doi: 10.1038/s41598-020-60672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Schreurs R, Baumdick ME, Sagebiel AF, Kaufmann M, Mokry M, Klarenbeek PL, et al. Human fetal TNF-α-Cytokine-producing CD4(+) effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity. 2019;50:462–476.e468. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 210.Nozaki K, Mochizuki W, Matsumoto Y, Matsumoto T, Fukuda M, Mizutani T, et al. Co-culture with intestinal epithelial organoids allows efficient expansion and motility analysis of intraepithelial lymphocytes. J Gastroenterol. 2016;51:206–13. doi: 10.1007/s00535-016-1170-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Rogoz A, Reis BS, Karssemeijer RA, Mucida D. A 3-D enteroid-based model to study T-cell and epithelial cell interaction. J Immunol Methods. 2015;421:89–95. doi: 10.1016/j.jim.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]