Abstract
Bacteriocins from lactic acid bacteria (LAB) have attracted widespread attention as a new type of biological preservative due to their safety, high efficiency, and non-toxic characteristics. In this study, we focused on Sakacin ZFM225, a novel bacteriocin produced by Lactobacillus sakei ZFM225, which was isolated from raw milk. It was purified by a strategy including precipitation with 70% ammonium sulfate, cation exchange chromatography and reverse-phase high performance liquid chromatography (RP-HPLC). The predicted molecular weight of Sakacin ZFM225 was 14950.92 Da. Sakacin ZFM225 exhibited resistance to high temperatures, strong activity under acidic conditions, and sensitivity to trypsin and pepsin. Bacteriocins from Lactobacillus sakei mainly inhibited the growth of Listeria monocytogenes. The bacteriocin possessed a broad-spectrum inhibition which could kill many foodborne pathogens such as Pseudomonas aeruginosa, Micrococcus luteus, and Staphylococcus aureus. We further demonstrated that the mode of action of Sakacin ZFM225 was related to the formation of cell membrane porosity, and excluded Lipid Ⅱ as its target. These results suggest that this new bacteriocin has great potential in food industry as a biological preservative and even medical field.
Keywords: Bacteriocin, Sakacin ZFM225, Purification, Characteristic, Mode of action
Highlights
-
•
a novel bacteriocin Sakacin ZFM225 produced by Lactobacillus sakei ZFM225 who isolated from raw milk was purified.
-
•
Sakacin ZFM225 was active under high temperature and acidic conditions, sensitive to trypsin and pepsin.
-
•
Sakacin ZFM225 can kill foodborne pathogens such as Pseudomonas aeruginosa and Staphylococcus aureus, which possessed a broad-spectrum inhibition.
-
•
We further demonstrated that the mode of action of Sakacin ZFM225 was related to formation of cell membrane porosity, and excludes Lipid Ⅱ as its target.
1. Introduction
Food spoilage occurs regularly in our daily life, especially for food with high water activity, such as fish, meat, and vegetables. Rapid deterioration of these perishable foods leads to significant wastage. Microbial contamination is the main cause of food spoilage [1]. Although food preservatives effectively alleviate the problem of food spoilage caused by microorganisms, some food additives may have adverse effects on the most fragile population such as the elderly and infants [2]. Consequently, the development of new non-toxic and harmless food preservatives is the focus of research in the food bioengineer [3].
Bacteriocins are extracellular products synthesized by ribosomes during the cell growth of some bacteria, which exert high antibacterial effects by forming channels on the plasma membrane of target bacteria or directly inhibiting crucial functions in target bacteria cells to inhibit or kill them [4]. Due to their non-toxic, non-resistant, non-residual, and environmental friendly characteristics, the development of bacteriocins as novel antimicrobial compounds capable of effective defense against food-borne pathogens has opened a new field of research [[5], [6], [7]]. However, most bacteriocins remain unexplored or underexploited, with nisin being the only commercially available bacteriocins in high-purity form [8]. The limited application of bacteriocin is primarily attributed to the difficulties in establishing purification methods, low yield, high cost, complex purification process, and unclear antibacterial mechanism [8]. Therefore, the purification technology of a new high-performance bacteriocin is still a challenging task to promote bacteriocin as a widely used biological preservative. The separation and purification of new bacteriocins is still a research hotspot at present.
Lactic acid bacteria (LAB) are high-performing producers of bacteriocins [9]. The LAB Lactobacillus sakei (L. sakei) is primarily found in meat products and plants. They play an important role in fermentation and preservation primarily by producing organic acids or by producing compounds that inhibit spoilage bacteria growth [10].
In Western Europe, it is widely used as a starter for fermented sausage together with Micrococcus and yeasts [10]. Many strains of L. sakei were found to produce one or more bacteriocins which described previously are sakacin A [11], sakacin P [12], sakacin G [13], sakacin D98 [14], sakacin X and sakacin T [15], and these bacteriocins mainly inhibited the growth of Listeria monocytogenes (L. monocytogenes). Understanding the action modes of bacteriocins is an important prerequisite for optimizing their use and development [16]. Bacteriocins have various modes action, including pore formation, membrane permeability, cell wall damage, blocking of metabolic pathways, and reducing the risk of cross-resistance development [17]. These mechanisms were more precisely called antibacterial models. Down to the molecular level, there were some relevant reports showed that bacteriocins need a target molecule at the surface of a sensitive cell to be active [18], but they didn't propose a target molecule of L. sakei bacteriocin. In the present study, a bacteriocin produced from Lactobacillus sakei ZFM225, isolated from fresh milk, was partially purified and characterized. Micrococcus luteus 10209 as an indicator, the antibacterial action mode, and even the target of Sakacin ZFM225 were studied, which is of great value for application in the food industry as a natural preservative.
2. Materials and methods
2.1. Strain cultivation
L. sakei ZFM225, the bacteriocin-producing strain, was isolated from raw milk at Hangzhou dairy farm, Zhejiang province, China. It was stored in China Center for Type Culture Collection (CCTCC, Wuhan, China) with the collection number of CCTCC: M 2016669. L. sakei was inoculated in MRS broth (2% v/v) with pH 6.5. Incubation was carried out at 37 °C for 24 h in stationary conditions. Cell-free supernatant (CFS) was separated from the fermentation broth at 8000 rpm for 30 min at 4 °C. Determination of antimicrobial activity was tested by agar well diffusion assay [19]. The indicator strain M. luteus 10209 came from China Center for Industrial Culture Collection (CICC, Beijing, China) with the collection number of CICC 10209 and was incubated at 30 °C for 12 h.
2.2. Purification of Sakacin ZFM225
The CFS produced by L. sakei ZFM225 was obtained by the method described above.
Ammonium sulfate powder was slowly added ammonium to 1 L CFS to the saturation of 20, 30, 40, 50, 60, 70, 80, 90, and 100% (25 °C, 707g ammonium sulfate was contained in 1L saturated ammonium sulfate solution at 25 °C). The salt-protein system was left at 4 °C with constant stirring for 12 h. The system was subsequently centrifuged and then dissolved precipitates in 30 mL of pure water. Crude protein extract (CPE) was obtained. After desalting treatment by Sephadex G-10, the antimicrobial activity of CPE was assayed. The best gradient precipitation was selected for subsequent experiments.
The desalted CPE was filtered through a 0.22-μm filter, then injected into a fast protein liquid chromatography (FPLC) system at 1 mL/min with SP Sepharose Fast Flow cation-exchange column (HiPrepTMSP XL 16/10, GE Healthcare, Sweden). The crude bacteriocin was eluted with a NaCl gradient (0–100% of 1 M) in sodium acetate buffer (20 mM, pH 4.0) at the same flow rate. The eluted protein peaks were monitored at 280 nm with a UV detector and collected one tube every 5 mL. The protein fractions were desalinated and rotary evaporated to dryness and then dissolved in water for activity testing by using M. luteus 10209 as indicator bacteria.
The crude bacteriocin obtained after cation-exchange chromatography was further purified by RP-HPLC with a SunFire C18 Prep column (5 μm, 10 mm × 100 mm, Waters, USA) at 5 mL/min. Diluting with ultrapure water to the appropriate concentration (i.e. 0.5–1 mg/mL), the crude bacteriocin was eluted with a linear gradient of acetonitrile with 0.05% TFA from 5 to 95% in 30 min. Fractions were monitored at 280 nm with a UV detector and collected manually. The fractions were rotary evaporated to dryness and then dissolved in water for activity testing.
2.3. Determination of protein concentration
Determination of protein concentration used the BCA protein quantification kit (Beyotime, Product number: P0012, Jiangsu Kaiji Biotechnology) according to the instructions. The kit was based on the Lowry method using bovine serum albumin(BSA) as a standard [20]. The same volume of Sample and BCA reagent were mixed and then placed at 37 °C for 30 min. The protein concentration of the mixture was measured at 562 nm by comparing with BSA.
2.4. Molecular weight estimation
Sodium dodecyl sulfate-polyacrylamide gel (15%) electrophoresis (SDS-PAGE) was performed to estimate the molecular mass of Sakacin ZFM225. The target protein band was cut from the lanes, then washed with water. Samples stored at −20 °C were sent to Shanghai Applied Protein Technology Company (Shanghai, China) for LC-MS/MS analysis.
Protein digestion was performed with the FASP procedure (Wiśniewski et al., 2009). LC-MS/MS analysis was performed on a Q Exactive mass spectrometer (Thermo Scientific, America) and operated in positive ion mode. The following experimental conditions were used: Peptide mass tolerance, 20 ppm, MS/MS tolerance, 0.1 Da, Enzyme, Trypsin, Missed cleavage, 2, Fixed modification, Carbamidomethyl (C), Variable modification, Oxidation(M).
2.5. Characteristics of Sakacin ZFM225
The Sakacin ZFM225 solution (about 2 mg/mL) was treated at 4-37-50-60-80-100 °C for 30 min, respectively to evaluate stability at different temperatures. The Sakacin ZFM225 solution was adjusted to different pH values (2.0–10.0) with 1 M HCl and NaOH respectively to maintain a consistent final concentration of bacteriocin. The enzymes used in the experiment to test the sensitivity of bacteriocin to enzymes included pepsin (pH 2.0) prepared in glycine HCl buffer, papain (pH 7.0) prepared in PBS buffer, trypsin (pH 5.4) prepared in PBS -citric acid buffer and protease K prepared in PBS buffer (pH 7.6). Enzymes were dissolved in buffer at a concentration of 1 mg/mL and added to the same amount of lyophilized bacteriocin powder to each (2 mg). Samples were left at 37 °C for 4 h to fully react and then treated at 100 °C for 5 min to inactivate the enzyme. The pH was adjusted back to the initial value with 1 M HCl and NaOH solution respectively; the untreated bacteriocin solution was taken as the blank control. All the above samples were tested for bacteriocin activity using M. luteus 10209 as indicator bacteria.
2.6. Antibacterial spectrum and minimum inhibitory concentration of Sakacin ZFM225
In this experiment, 17 strains of microorganisms including Gram-positive bacteria, Gram-negative bacteria, and molds were selected as indicator bacteria. The antimicrobial activity of Sakacin ZFM225 was tested by agar well diffusion assay performed in triplicates.
Using M. luteus 10209 and Staphylococcus aureus D48 (Sa. D48) as indicator bacteria, the 96-well titer plate method was used to measure the minimum inhibitory concentrations (MIC) of bacteriocins. S. aureus D48 was screened by Zhejiang Provincial Center for Disease Control and Prevention (Zhejiang CDC, Hangzhou, China) and preserved in Zhejiang CDC. M. luteus 10209 and Sa. D48 were inoculated in LB broth (1% v/v) pH 7. Incubation was carried out at 30 °C (M. luteus 10209) /37 °C (Sa. D48) for 12 h in a stationary condition. The OD600 value of the activated indicator bacteria culture medium was adjusted to 0.05. The lyophilized bacteriocin powder was redissolved in 0.05% acetic acid and serially diluted to 2, 1, 0.5, 0.25, 0.125, 0.0625, 0.031, 0.015, and 0.007 mg/mL. After being diluted to different concentration gradients, Sakacin ZFM225 was mixed with the indicator bacteria, cultured at the optimum growth temperature of the indicator bacteria for 24 h, and the absorbance value at 600 nm was measured by microplate reader Spectra Max 190 (Molecular Devices, USA). The bacterial solution without bacteriocin was used as the control. If the OD600 value did not increase after 24 h, the corresponding bacteriocin concentration under this condition was the MIC.
2.7. Mode of action
2.7.1. Effects on ATP levels in M. luteus
Single colony of M. luteus 10209 was picked out to 10 mL LB liquid medium and cultured overnight, then, subcultured at 30 °C until OD600 reached 0.5. The cell pellet washed twice with 5 mm HEPES buffer by centrifugation was resuspended in 5 mM HEPES buffer (1 mL). Glucose solution (50 μL,10 mM), the processed bacterial suspension (50 μL), and Sakacin ZFM225 (1 × MIC) were mixed without sequence requirements. Negative control (Ultra-pure water) was treated in the same way. Immediately, the suspensions treated with Sakacin ZFM225 or control reagent were mixed with 25 μL ATP lysate which could effectively lyse and release ATP in common cultured cells and tissues, and then 100 μL ATP reagent. ATP lysate was one of the reagents in the ATP detection kit of Beyotime. The multifunctional micrometer was used to measure the fluorescence intensity of the samples in a black 96-well plate (VICTOR Nivo™, America).
2.7.2. Effects on potential difference across the membrane (ΔΨ)
The test strain M. luteus 10209 was obtained as above. Fluorescence was detected using the Kinetics software of a Cary Eclipse fluorescence spectrophotometer (Agilent, USA). Test parameters: excitation wavelength (Ex) of 650 nm with a slit of 5 nm, emission wavelength (Em) of 670 nm with a slit of 5 nm, scanning time with 20 min. 2 mL of special buffer for fluorescence leakage (4.5% glucose, 0.05 mM magnesium sulfate, 25 mM potassium chloride, 10% PBS) and 20 μL bacterial suspension were successively added into a cuvette. After zero adjustments, a 3 μL 0.1 mM DISC2(5) probe was added into the cuvette, fully mixing. Starting immediately for fluorescence detection. After the fluorescence value tends to be stable, Sakacin ZFM225 was quickly added with the final concentration of twice MIC in the experimental group. In parallel, 0.05% acetic acid of the same volume was added as the negative control and tested according to the same method above.
2.7.3. Effects on pH difference inside and outside the membrane (ΔpH)
First, we added HEPES buffer (5 mM, 20 mL) to the cell pellet obtained as above. After 30 min, added with 0.5 μL BCECF AM pH fluorescent probe (Beyotime, China), and the bacterial suspension system was in the dark for 60 min. A fluorescent cuvette which was a supporting instrument of the Cary Eclipse fluorescence spectrophotometer (Agilent, USA) was added with HEPES buffer (5 mM, 2 mL) and bacterial suspension system (20 μL), then quickly added Sakacin ZFM225 with the final concentration of 1 × MIC in the experimental group, added 0.05% acetic acid of the same volume as the negative control. Test parameters: excitation wavelength (Ex) of 488 nm, emission wavelength (Em) of 585 nm, scanning in sequence every 1 min for recording the fluorescence value.
2.7.4. Lipid Ⅱ binding experiment with Sakacin ZFM225
Sakacin ZFM225 solution (2 mg/mL) was added to Lipid II to give a final concentration of Lipid II of 1 mM. Nisin and Lipid Ⅱ were mixed so that the final concentration of Nisin in the solution was 10 μM and the final concentration of Lipid Ⅱ was 1 mM; the inhibition activity of the mixture against M. luteus 10209 was tested and the size of the inhibition circle was observed. Lipid II was presented by Professor Eefjan Breukink of Utrecht University in the Netherlands. Nisin was purchased from Zhejiang Changqing Chemical Corporation (China).
3. Results and discussion
3.1. Purification of Sakacin ZFM225
The CFS fermented by L. sakei ZFM225 was purified by a three-step purification project.
The results showed that the crude protein had the best antimicrobial effect under the saturation of 70% ammonium sulfate. Therefore, the crude protein through the treatment of ammonium sulfate with the saturation of 70% was desalted and subsequently use. The concentration was approximately 38.15-fold, and the recovery was 25.45%.
Next step, the active crude protein was processed by a strong cation exchange chromatography column. As shown in Fig. 1A, the penetration peak (peak after buffer flushing) and elution peak (peak after eluent elution) were concentrated to 2 mg/mL by rotary evaporation. Fig. 1B shows the eluted protein peak fractions had good bacteriostatic activity, indicating that bacteriocin was concentrated. The concentration was approximately 53.31-fold, and the recovery was 9.33%.
Fig. 1.
Purification of Sakacin ZFM225: (A) Chromatogram of purification on SP-Sepharose. the first four absorption peaks were “penetration peaks”; the absorption peaks 5 was “eluted protein peak”; the ConcB means a NaCl gradient (0–100% of 1 M) in sodium acetate buffer (20 mM, pH 4.0); (B) Antibacterial activity of elution parts from cation exchange chromatography; (C) Chromatogram of antibacterial eluents by preparative HPLC.
Finally, the eluted active protein was purified by RP-HPLC shown in Fig. 1C. It can be seen that there are two single peaks after purification by high performance liquid chromatography. After collecting every single peak for the bacteriostatic test, it is found that peak 2, with a retention time of 12 min, has good bacteriostatic activity against M. luteus 10209. The concentration was approximately 73.68-fold, and the recovery was 1.47%.
In Table 1, we summarized the increase in the specific activity and purification obtained step by step.
Table 1.
Purification yield rate of bacteriocin of L. sakei ZFM225.
| Purification step | Volume (mL) | Total activity (IU) | Total protein (mg) | Specific activity (IU/mg) | Purification fold | Recovery (%) |
|---|---|---|---|---|---|---|
| CFS | 2000 | 532000 | 50000 | 10.64 | 1.00 | 100.00 |
| ammonium sulfate precipitation | 60 | 135420 | 334 | 405.45 | 38.11 | 25.45 |
| SP cation exchange | 45 | 49680 | 91 | 545.93 | 51.31 | 9.33 |
| HPLC | 10 | 7840 | 10 | 784.00 | 73.68 | 1.47 |
3.2. Molecular weight estimation
SDS-PAGE electrophoretic map, Fig. 2A, showed a single band which was about 14 kDa. The specific molecular mass of Sakacin ZFM225 was detected by LC-MS/MS. The polypeptide fragments centered on 1584.78 Da were manually selected (Fig. 2B) and searched with two kinds of frequently used databases: Public Library (UniProtKB was mostly used) and self-built library (protein database translated from transgenome database). The predicted protein molecular weight was 14950.92 Da.
Fig. 2.
Molecular weight estimation: (A) The SDS-PAGE map of Sakacin ZFM225; (B) Mass spectrum of Sakacin ZFM225.
Although the molecular weight varies largely between bacteriocins from L. sakei, it is mostly small, around 3500 Da. For example, sakacin A was produced by L. sakei Lb706 with a molecular weight of 4308.7 Da [11,21]. Sakacin P, produced by L. sake LTH 673 was isolated from fermented sausages with a molecular weight of about 2560 Da [22]. Sakacin G (3834.32 Da) produced by L. sakei 2512 was isolated from the Rhodia Food [13]. Sakacin D98 (3509.9 Da) produced by L. sakei D98 was isolated from a kind of rice malt called Shubo [14]. Interestingly, the predicted molecular weight of Sakacin ZFM225 is about one order of magnitude larger than that of already found sakacin. Based on the above results, Sakacin ZFM225 was a new bacteriocin.
3.3. Characterization of bacteriocin activity
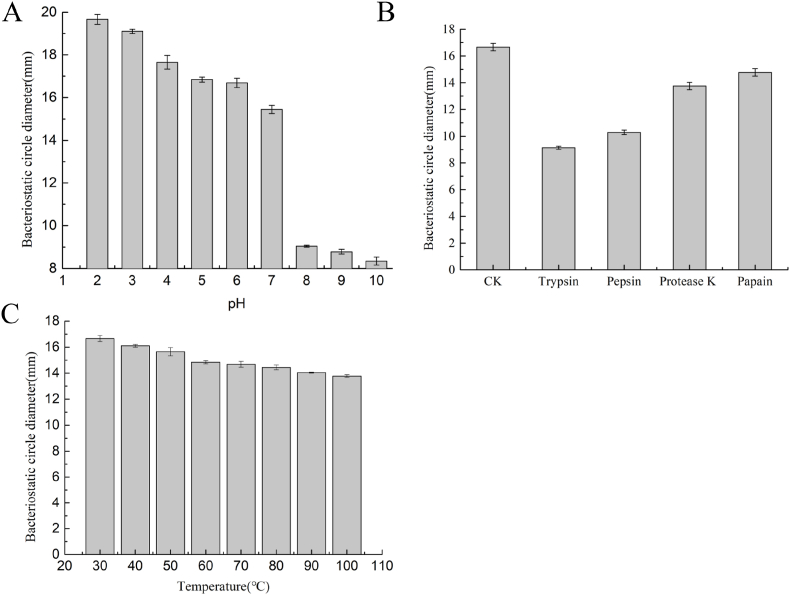
The bacteriostatic activity of bacteriocin was measured by the diameter of the bacteriostatic ring. The larger the diameter of the bacteriostatic ring is, the better the bacteriostatic activity is. The effects of pH on antimicrobial activity are shown in Fig. 3A. Under alkaline conditions, the characteristic of Sakacin ZFM225 activity decreased significantly or was lost. It was reported that many bacteriocins produced by lactic acid bacteria were stable within a certain pH range. Some of them could play a better role under acidic conditions but be irreversibly inactivated after alkali treatment. The Sakacin ZFM225 suitable pH range of bacteriocin is 2-7. The effects of temperature on antimicrobial activity are shown in Fig. 3C. Although the diameter of the bacteriostatic circle slightly decreased with the increase in temperature, more than 95% of antibacterial activity remained. This characteristic of Sakacin ZFM225 established a cornerstone for application in food preservation and applied to the occasion when some foods need to be heated in the production process.
Fig. 3.
Characterization of bacteriocin activity: (A) Effect on antibacterial activity of Sakacin ZFM225 at different pH values; (B) Effect of different protein enzymes on the antibacterial activity of Sakacin ZFM225; (C) Heat stability of Sakacin ZFM225. *CK means control.
The effects of enzymes are shown in Fig. 3B. The results revealed that the antibacterial activity of Sakacin ZFM225 was reduced in all cases, but it was most sensitive to trypsin and pepsin. After treatment with these two enzymes, the activity was almost completely lost.
When bacteriocin is added to food as a biological preservative, it is necessary to understand the sensitivity of target organisms. Sakacin ZFM225 is a thermostable protein that is stable in acidic environments and suitable for use in food. It is generally believed that the smaller the molecular weight of bacteriocin is, the more stable it is to heat. For example, bacteriocin with a molecular weight of 6 Da-17 kDa was generally not inactivated after treatment at 100 °C for 30 min, while bacteriocin with molecular weight less than 6 kDa could often maintain good activity after treatment at 121 °C for 30 min [23]. Sakacin ZFM225 is a kind of protein that remains active after heating at 100 °C for 30 min that is heat-stable for conventional heating of food. Similar to nisin, it shows strong antibacterial activity under acidic conditions and is inactivated at pH 8–10 [24]. Therefore, Sakacin ZFM225 is most active in acidic conditions like nisin. Sakacin ZFM225 can be inactivated by the proteolytic enzyme trypsin and pepsin which could prevent their accumulation in the body. This feature ensures its safety and makes it suitable for application in the food industry. Trypsin specifically cleaves peptide bonds of lysine and arginine carboxyl side chains [25]. Pepsin is generally able to hydrolyze peptide bonds containing any 20 amino acids at the P1 residue position [26]. So it can be predicted that lysine and arginine are two of the amino acid components of Sakacin ZFM225. However, there is a need for further structural characterization by Edman degradation.
3.4. Characterization of bacteriocin activity
The results showed in Table 2 revealed that Sakacin ZFM225 was a bacteriocin with a broad spectrum antibacterial activity. It had good antibacterial activity against most Gram-positive bacteria, such as M. luteus 10209 with MIC values of 0.125 mg/mL, S. aureus D48 with MIC values of 0.500 mg/mL. At the same time, it had certain antibacterial activity on Escherichia coli DH5 α, S. paratyphi A CMCC 50093, S. cholerae swine ATCC 13312, and Pseudomonas aeruginosa ATCC 47085, but had no antibacterial effect on molds. We found that the bacteriocin has a good antibacterial effect on some common foodborne pathogenic bacteria, so it has potential application as a food biological preservative.
Table 2.
Antibacterial spectrum of Sakacin ZFM225.
| Indicator strain | IZD (mm) | |
|---|---|---|
| G+ | Micrococcus luteus CICC 10209 | 15.70 ± 0.45 |
| Staphylococcus aureus D48 | 13.40 ± 0.37 | |
| Listeria monocytogenes LM1 | 13.50 ± 0.25 | |
| Staphylococcus warneri | / | |
| Staphylococcus muscae | 15.98 ± 0.56 | |
| Staphylococcus carnosus pCA 44 | 17.36 ± 0.33 | |
| Staphylococcus carnosus pet 20 | 17.56 ± 0.45 | |
| Bacillus subtilis BAS2 | / | |
| G− | Escherichia coli DH5α | 13.44 ± 0.43 |
| Salmonella paratyphi B CMCC50094 | / | |
| Salmonella paratyphi A CMCC 50093 | 11.43 ± 0.15 | |
| Salmonella typhimurium CMCC 50015 | / | |
| Salmonella choleraesuis ATCC 13312 | 11.97 ± 0.65 | |
| Salmonella enterica subsp. Arizonae CMCC(B) 47001 | / | |
| Pseudomonas aeruginosa ATCC 47085 | 13.76 ± 0.44 | |
| Fungus | Candida albicans | / |
| Aspergillus niger ATCC 13073 | / |
* "/" indicates no antibacterial effect. IZD means Inhibitory zone diameter (IZD). G+ means Gram positive bacteria. G− means Gram negative bacteria. Inhibitory zone diameter was measured with vernier caliper.
As mentioned above, bacteriocins from L. sakei mainly inhibited the growth of Listeria monocytogenes which is a Gram-positive pathogen but couldn't inhibit Gram-negative bacteria. Sakacin ZFM225 remedies this defect because it is effective against Gram-negative pathogenic bacteria. Therefore, it has the potential as a natural and effective therapeutic biomolecule. Pseudomonas aeruginosa (P. aeruginosa) is the most prevalent cause of serious hospital-acquired infections, including nosocomial bloodstream infections (BSI) are associated with high mortality rates and increased healthcare costs. Moreover, limited sensitivity to antibiotics and drug resistance found during the treatment of P. aeruginosa make treatment more difficult [27]. Sakacin ZFM225 had a strong inhibitory effect on P. aeruginosa ATCC 47085. Besides, it also had a good effect on E. coli and some types of Salmonella. Therefore, as a biological preservative, Sakacin ZFM225 had potential application value in the field of food safety and even in medical treatment.
3.5. Mode of action
3.5.1. Effects on ATP levels in M. luteus
As a coupling agent of energy metabolism in biological cells, ATP plays a very important role in cells. ATP is the central substance for the release, storage, and utilization of energy in cells. Therefore, the change in intracellular ATP level is closely related to the driving of cell function. Generally, when cells are in the state of necrosis or apoptosis, the intracellular ATP content will decrease to a certain extent. Therefore, the change in intracellular ATP content can reflect the state of cells to a certain extent. In addition, for cells, the dissipation of the proton electromotive force of the cell membrane will affect the synthesis of ATP, so detecting the level of intracellular ATP is also a verification of the change of the proton electromotive force of cells.
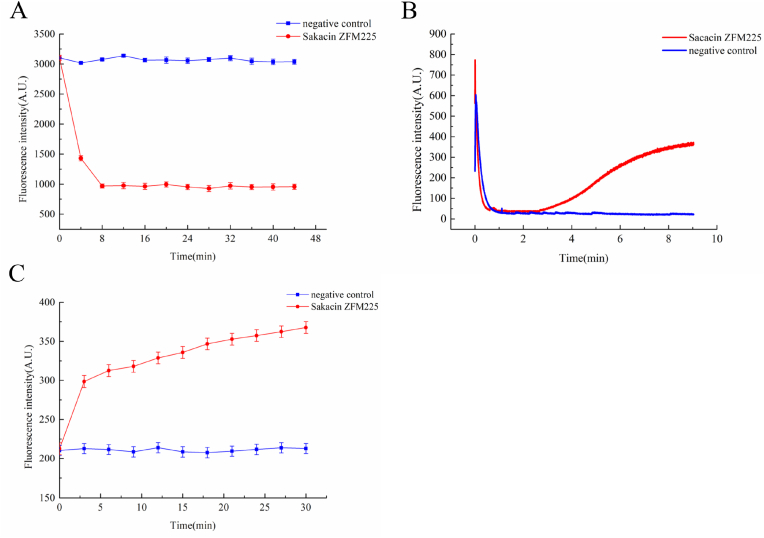
It can be seen from Fig. 4A that the intracellular ATP level of the indicator bacteria treated with 0.05% acetic acid didn't change, while the intracellular ATP level of the experimental group treated with Sakacin ZFM225 decreased and tended to be stable within a certain time, indicating that the intracellular ATP synthesis of the indicator bacteria was inhibited after bacteriocin treatment, This result was consistent with the effect of Sakacin ZFM225 on the proton electromotive force of indicator bacteria.
Fig. 4.
Mode of action: (A) Analysis of intracellular ATP of M. luteus cells treated by Sakacin ZFM225. (B) Analysis of ΔΨ of M. luteus cells treated by Sakacin ZFM225 with different concentrations. (C). Analysis of ΔpH of M. luteus cells treated by Sakacin ZFM225.
3.5.2. Effects on ΔΨ
If bacteriocin can destroy the cell membrane of indicator bacteria and let them die, this process is bound to cause the dissipation of bacterial cell membrane potential difference. Therefore, we used a fluorescence leakage experiment to observe the change in cell membrane potential difference after bacteriocin treatment. DISC2(5) is a lipophilic cationic fluorescent material sensitive to membrane potential. It can be combined with the phospholipid bilayer of the cell membrane and enter the fluorescent probe cell to produce fluorescence quenching. When the cell membrane structure was damaged, DISC2(5), which was originally located in the phospholipid bilayer of the cell, would be discharged out of the cell, thus enhancing the fluorescence detection signal. Therefore, we could infer whether the cell membrane was damaged according to the change in fluorescence response value after the addition of bacteriocin. The results are shown in Fig. 4B.
It was found that after adding DISC2(5) to the treated bacterial suspension, the fluorescence intensity decreased rapidly and gradually stabilized. Then, it was found that the fluorescence value rose gradually and stabilized finally after adding Sakacin ZFM225 to bring the final concentration to the MIC of the indicator bacteria. However, in the negative control group, the fluorescence did not increase and remained stable. Therefore, it can be speculated that Sakacin ZFM225 could destroy the structure of the cell membrane, break the membrane potential difference, lead to the leakage of DISC2(5), and strengthen the fluorescence signal.
3.5.3. Effects on ΔpH
The proton concentration gradient of the cell membrane is also an important factor affecting the proton dynamic potential of the cell membrane. The fluorescent dye BCECF AM can be used to detect the changes in the proton concentration gradient of the cell membrane. BCECF AM fluorescent dye itself does not produce fluorescence, but it can penetrate the cell membrane and be cut into BCECF by esterase in the cell after entering the cell membrane. BCECF can be excited into green fluorescence at an appropriate pH. Therefore, when it penetrates into the cell membrane, the protons in the cell flow out, resulting in the increase of the pH value in the cell, so the corresponding fluorescence intensity will also change. Generally, the excitation wavelength (Ex) 488 nm and emission wavelength (EM) 535 nm are used to detect the change in fluorescence intensity.
As shown in Fig. 4C, in the negative control group where 0.05% acetic acid was added fluorescence intensity value basically remained unchanged. When adding Sakacin ZFM225 with a final concentration of MIC, it was found that the extracellular fluorescence response value was gradually increasing and tends to be stable, indicating that Sakacin ZFM225 could affect the cell membrane's permeability and increase the intracellular pH, to enhance the corresponding fluorescence intensity.
3.5.4. Lipid Ⅱ binding experiment with Sakacin ZFM225
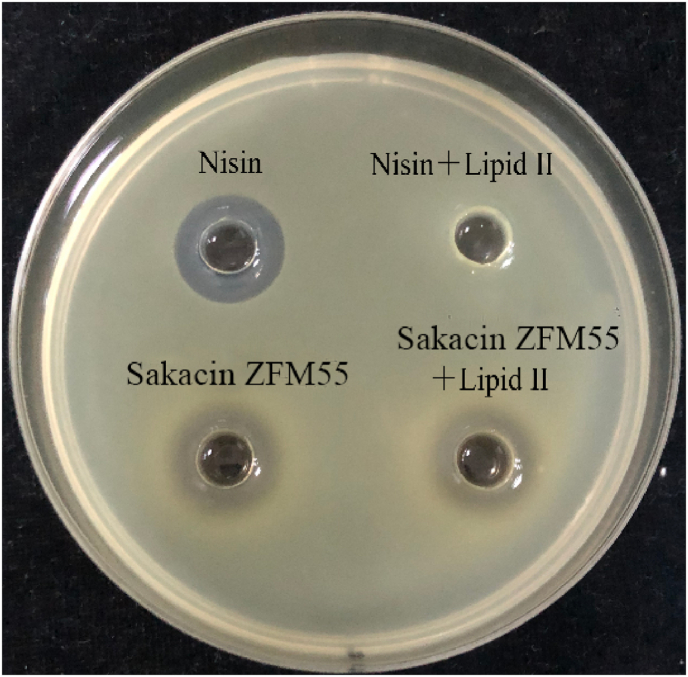
Lipid Ⅱ is the target of nisin acting on sensitive cell membranes. Therefore, take nisin as the positive control to explore whether the target of Sakacin ZFM225 on the cell membrane is lipid Ⅱ or not. The results were shown in Fig. 5. When nisin was mixed with excess lipid Ⅱ, lipid Ⅱ competed with lipid Ⅱ on the cell membrane of sensitive bacteria to bind nisin. Therefore, nisin could not bind with lipid Ⅱ on the cell membrane and lost the antibacterial effect. After Sakacin ZFM225 was mixed with lipid Ⅱ, the antimicrobial effect basically was analogous to the original, indicating that lipid Ⅱ could not compete with the binding sites on the cell membrane of sensitive bacteria. Therefore, it could be inferred that lipid Ⅱ wasn't the target of Sakacin ZFM225 acting on the cell membrane of sensitive bacteria.
Fig. 5.
Binding experiment of Sakacin ZFM225 with Lipid Ⅱ.
* Indicator strain used M. luteus 10209.
Studies on the antimicrobial mode of action of bacteriocin produced by L. sakei have rarely been reported. The study by Trinetta et al. [28]mentioned the antimicrobial mechanism of Sakacin A. Sakacin A is a class Ⅱa bacteriocin which can inhibit the growth of several LAB and L. monocytogenes. What we already know is that the highly conserved N-terminal domain with the consensus motif YGNGV is characteristic of class Ⅱa bacteriocin which makes the responsible for activity against L. monocytogenes [28]. After contacting with these bacteriocins, leakage of ions and small molecules in sensitive cells is accompanied by the dissipation of proton power and the consumption of intracellular ATP [29]. The antimicrobial effects of Sakacin A were characterized by the same method as ours to measure the fluorescence to study the mechanism of Sakacin A against L. monocytogenes cells. The results suggested that Sakacin A acts rapidly by altering the charge distribution across the membrane to dissipate the proton motive force, as does Sakacin ZFM225. The difference was that they did a cell wall breakdown assay and showed another mechanism of action that Sakacin A slowly breaks down the cell walls of sensitive bacteria while acting on the polysaccharide and peptide components of the cell walls peptidoglycan. But they still hadn't clarified the specific target at the molecular level. Bacteriocins need a target molecule on the surface of sensitive cells to be active. It is now clear that the antibacterial mechanism of nisin is due to Lipid Ⅱ, which mediates the formation of cell membrane pores while preventing the formation of the cell wall of sensitive bacteria to reach antibacterial effects [30], so do Gallidermin and Mutacin 1140 [31]. According to the combined test, we have known that the specific target of Sakacin ZFM225 in the cell membrane wasn't Lipid Ⅱ, which promoted the development of an action mechanism at the molecular level.
4. Conclusion
L.sakei ZFM225 was isolated from raw milk. In this study, Sakacin ZFM225 produced by this strain was purified and its biological characteristics were examined. The purification process involved a three-step method that included ammonium sulfate gradient purification, strong cation exchange chromatography, and RP-HPLC. The broad-spectrum antimicrobial activity of sakacin ZFM225 is not common among known sakacins. Moreover, Sakacin ZFM225 could be applied to thermally processed foods. Thus, it could be used as a potential bio preservative to deter foodborne pathogens and foodborne bacteria, which can improve food safety and extend their shelf life. The antibacterial mechanism of Sakacin ZFM225 was explored which resulted in the damage of the cell membrane and the leakage of intracellular electrolytes, resulting in bacterial death and excluding Lipid Ⅱ as its target. The purification method of Sakacin ZFM225 established in this paper requires only three steps, to avoid a complex purification process. However, the purification yield, purity, and cost-effectiveness of Sakacin ZFM225 still needs further improvement.
Author contributions
Methodology, Investigation, Writing - Original Draft, Huifei Shentu; Methodology, Investigation, Pengxin Ye. Investigation, Qingqing Zhou. Investigation, Ping Li. Conceptualization, Writing - Review & Editing, Funding acquisition, Qing Gu.
Funding
This project was funded by the Chinese Academy of Engineering Academy-Locality Cooperation Project (No. 2019- ZJ-JS-02), the Key Research and Development Program of Zhejiang Province (No.2020C04002), and the Joint Funds of the National Natural Science Foundation of China (No. U20A2066).
Declaration of competing interest
All authors declare no conflict of interest.
Contributor Information
Ping Li, Email: ping-biology@outlook.com.
Qing Gu, Email: guqing2002@hotmail.com.
Data availability
Data will be made available on request.
References
- 1.Thakali A., MacRae J.D. A review of chemical and microbial contamination in food: what are the threats to a circular food system? Environ. Res. 2021;194 doi: 10.1016/j.envres.2020.110635. [DOI] [PubMed] [Google Scholar]
- 2.Trasande L., Shaffer R.M., Sathyanarayana S. COUNCIL on environmental health, food additives and child health. Pediatrics. 2018;142 doi: 10.1542/peds.2018-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pisoschi A.M., Pop A., Georgescu C., Turcuş V., Olah N.K., Mathe E. An overview of natural antimicrobials role in food. Eur. J. Med. Chem. 2018;143:922–935. doi: 10.1016/j.ejmech.2017.11.095. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Y., Zhou Q., Li P., Gu Q. Purification, characterization, and mode of action of Paracin 54, a novel bacteriocin against Staphylococci. Appl. Microbiol. Biotechnol. 2021;105:6735–6748. doi: 10.1007/s00253-021-11505-6. [DOI] [PubMed] [Google Scholar]
- 5.Gálvez A., Abriouel H., López R.L., Ben Omar N. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 2007;120:51–70. doi: 10.1016/j.ijfoodmicro.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 6.Ołdak A., Zielińska D. Bacteriocins from lactic acid bacteria as an alternative to antibiotics. Postepy Hig. Med. Dosw. 2017;71:328–338. doi: 10.5604/01.3001.0010.3817. [DOI] [PubMed] [Google Scholar]
- 7.Kumariya R., Garsa A.K., Rajput Y.S., Sood S.K., Akhtar N., Patel S. Bacteriocins: classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019;128:171–177. doi: 10.1016/j.micpath.2019.01.002. [DOI] [PubMed] [Google Scholar]
- 8.Peng Z., Xiong T., Huang T., Xu X., Fan P., Qiao B., Xie M. Factors affecting production and effectiveness, performance improvement and mechanisms of action of bacteriocins as food preservative. Crit. Rev. Food Sci. Nutr. 2022:1–14. doi: 10.1080/10408398.2022.2100874. [DOI] [PubMed] [Google Scholar]
- 9.Mokoena M.P. Lactic acid bacteria and their bacteriocins: classification, biosynthesis and applications against uropathogens: a mini-review. Molecules. 2017;22:E1255. doi: 10.3390/molecules22081255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Champomier-Vergès M.C., Chaillou S., Cornet M., Zagorec M. Lactobacillus sakei: recent developments and future prospects. Res. Microbiol. 2001;152:839–848. doi: 10.1016/s0923-2508(01)01267-0. [DOI] [PubMed] [Google Scholar]
- 11.Holck A., Axelsson L., Birkeland S.-E., Aukrust T., Blom H. Purification and amino acid sequence of sakacin A, a bacteriocin from Lactobacillus sake Lb706. J. Gen. Microbiol. 1992;138:2715–2720. doi: 10.1099/00221287-138-12-2715. [DOI] [PubMed] [Google Scholar]
- 12.Tichaczek P.S., Vogel R.F., Hammes W.P. Cloning and sequencing of sakP encoding sakacin P, the bacteriocin produced by Lactobacillus sake LTH 673. Microbiology. 1994;140:361–367. doi: 10.1099/13500872-140-2-361. [DOI] [PubMed] [Google Scholar]
- 13.Simon L., Fremaux C., Cenatiempo Y., Berjeaud J.M., Sakacin g. A new type of antilisterial bacteriocin. Appl. Environ. Microbiol. 2002;68:6416–6420. doi: 10.1128/AEM.68.12.6416-6420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sawa N., Koga S., Okamura K., Ishibashi N., Zendo T., Sonomoto K. Identification and characterization of novel multiple bacteriocins produced by Lactobacillus sakei D98. J. Appl. Microbiol. 2013;115:61–69. doi: 10.1111/jam.12226. [DOI] [PubMed] [Google Scholar]
- 15.Vaughan A., Eijsink V.G., O'Sullivan T.F., O'Hanlon K., van Sinderen D. An analysis of bacteriocins produced by lactic acid bacteria isolated from malted barley. J. Appl. Microbiol. 2001;91:131–138. doi: 10.1046/j.1365-2672.2001.01365.x. [DOI] [PubMed] [Google Scholar]
- 16.Li Z., Velkov T. Polymyxins: mode of action. Adv. Exp. Med. Biol. 2019;1145:37–54. doi: 10.1007/978-3-030-16373-0_4. [DOI] [PubMed] [Google Scholar]
- 17.Wang Y., Haqmal M.A., Liang Y., Muhammad I., Zhao X., Elken E.M., Gao Y., Jia Y., He C., Wang Y., Kong L., Ma H. Antibacterial activity and cytotoxicity of a novel bacteriocin isolated from Pseudomonas sp. strain 166. Microb. Biotechnol. 2022:1751–7915. doi: 10.1111/1751-7915.14096. 14096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Katla T., Naterstad K., Vancanneyt M., Swings J., Axelsson L. Differences in susceptibility of Listeria monocytogenes strains to sakacin P, sakacin A, pediocin PA-1, and nisin. Appl. Environ. Microbiol. 2003;69:4431–4437. doi: 10.1128/AEM.69.8.4431-4437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallynck Th, Pijck J. An agar well punching-sucking device for diffusion assays. J. Antimicrob. Chemother. 1978;4:94–95. doi: 10.1093/jac/4.1.94. [DOI] [PubMed] [Google Scholar]
- 20.Lowry O.H., Rosebrough N.J., Farr A.L., Randall R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 21.Holck A.L., Axelsson L., Hühne K., Kröckel L. Purification and cloning of sakacin 674, a bacteriocin from Lactobacillus sake Lb674. FEMS Microbiol. Lett. 1994;115:143–149. doi: 10.1111/j.1574-6968.1994.tb06629.x. [DOI] [PubMed] [Google Scholar]
- 22.Tichaczek P.S., Nissen-Meyer J., Nes I.F., Vogel R.F., Hammes W.P. Characterization of the bacteriocins curvacin A from Lactobacillus curvatus LTH1174 and sakacin P from L. sake LTH673. Syst. Appl. Microbiol. 1992;15:460–468. doi: 10.1016/S0723-2020(11)80223-7. [DOI] [Google Scholar]
- 23.Zamfir M., Callewaert R., Cornea P.C., Savu L., Vatafu I., De Vuyst L. Purification and characterization of a bacteriocin produced by Lactobacillus acidophilus IBB 801. J. Appl. Microbiol. 1999;87:923–931. doi: 10.1046/j.1365-2672.1999.00950.x. [DOI] [PubMed] [Google Scholar]
- 24.Tafreshi S.-Y.H., Mirdamadi S., Khatami S. Comparison of different nisin separation and concentration methods: industrial and cost-effective perspectives. Probiotics Antimicrob Proteins. 2020;12:1226–1234. doi: 10.1007/s12602-019-09607-9. [DOI] [PubMed] [Google Scholar]
- 25.Bhugaloo-Vial P., Douliez J.P., Moll D., Dousset X., Boyaval P., Marion D. Delineation of key amino acid side chains and peptide domains for antimicrobial properties of divercin V41, a pediocin-like bacteriocin secreted by Carnobacterium divergens V41. Appl. Environ. Microbiol. 1999;65:2895–2900. doi: 10.1128/AEM.65.7.2895-2900.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S., Bech Thoegersen J., Kragh K.M. Comparative study of protease hydrolysis reaction demonstrating normalized peptide bond cleavage frequency and protease substrate broadness index. PLoS One. 2020;15 doi: 10.1371/journal.pone.0239080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Six A., Mosbahi K., Barge M., Kleanthous C., Evans T., Walker D. Pyocin efficacy in a murine model of Pseudomonas aeruginosa sepsis. J. Antimicrob. Chemother. 2021;76:2317–2324. doi: 10.1093/jac/dkab199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trinetta V., Morleo A., Sessa F., Iametti S., Bonomi F., Ferranti P. Purified sakacin A shows a dual mechanism of action against Listeria spp: proton motive force dissipation and cell wall breakdown. FEMS Microbiol. Lett. 2012;334:143–149. doi: 10.1111/j.1574-6968.2012.02630.x. [DOI] [PubMed] [Google Scholar]
- 29.Drider D., Fimland G., Héchard Y., McMullen L.M., Prévost H. The continuing story of class IIa bacteriocins. Microbiol. Mol. Biol. Rev. 2006;70:564–582. doi: 10.1128/MMBR.00016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Panina I., Krylov N., Nolde D., Efremov R., Chugunov A. Environmental and dynamic effects explain how nisin captures membrane-bound lipid II. Sci. Rep. 2020;10:8821. doi: 10.1038/s41598-020-65522-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pokhrel R., Bhattarai N., Baral P., Gerstman B.S., Park J.H., Handfield M., Chapagain P.P. Lipid II binding and transmembrane properties of various antimicrobial lanthipeptides. J. Chem. Theor. Comput. 2022;18:516–525. doi: 10.1021/acs.jctc.1c00666. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.