Abstract
In response to the unprecedented global healthcare crisis of the COVID-19 pandemic, the scientific community has joined forces to tackle the challenges and prepare for future pandemics. Multiple modalities of data have been investigated to understand the nature of COVID-19. In this paper, MIDRC investigators present an overview of the state-of-the-art development of multimodal machine learning for COVID-19 and model assessment considerations for future studies. We begin with a discussion of the lessons learned from radiogenomic studies for cancer diagnosis. We then summarize the multi-modality COVID-19 data investigated in the literature including symptoms and other clinical data, laboratory tests, imaging, pathology, physiology, and other omics data. Publicly available multimodal COVID-19 data provided by MIDRC and other sources are summarized. After an overview of machine learning developments using multimodal data for COVID-19, we present our perspectives on the future development of multimodal machine learning models for COVID-19.
Keywords: COVID-19, Multimodal data, Machine learning
1. Introduction
The coronavirus disease 2019 (COVID-19) pandemic caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has claimed over 6 million lives with over 630 million confirmed cases around the world to date [1]. Besides the primary target organ, i.e., the respiratory system, the virus has exhibited deleterious impacts on other organs, including brain, heart, kidney, liver, and the endocrine system [2]. Moreover, while the majority of COVID-19 patients recover within weeks, some patients suffer from post-acute COVID-19 syndrome with long-term complications [3]. In response to this unprecedented global healthcare crisis, the scientific community has joined forces to tackle the challenges and prepare for future pandemics. The purpose of this paper is to present our perspective on the use of multimodal data and machine learning technologies for assessing infection and progression of COVID-19.
Multi-modality data are under investigation for a variety of COVID-19 clinical tasks, e.g., detection/diagnosis, prognostication, severity characterization, and prediction/monitoring of treatment response. The gold standard for detection of the SARS-CoV-2 virus is the Reverse Transcription Polymerase Chain Reaction (RT-PCR) test. Multiple imaging modalities, such as chest x-ray radiography (CXR), computed tomography (CT), magnetic resonance imaging (MRI), and ultrasound (US), are being used or investigated for diagnosis (especially when RT-PCR is not available), for characterization of disease severity, and for monitoring treatment response [4,5]. Plasma multi-omic profiles and clinical data are also being investigated for the characterization of COVID-19 severity [6]. Additionally, measurements of gas exchange, hemodynamics, and lung mechanics, as well as chest CT scans, are enabling the building of computational models on ventilation-perfusion inequality, and thus shedding light on the pathophysiology of the disease [7].
The availability of multimodal data could enable the comprehensive understanding of disease thereby potentially informing better patient care than might a single modality, which is especially beneficial when investigating a new disease such as COVID-19. Clinically, critical decisions are often made during multidisciplinary consultation meetings (e.g., tumor boards) with experts from multiple specialties, as interpretation of multimodal data may be beyond any single physician's capability. Machine learning (ML) holds great promise in the analysis and integration of large amounts of complex multimodal data for specified clinical tasks, as has been demonstrated in cancer diagnosis/prognosis such as radiogenomics [8,9], fusion of pathology and genomics data [10,11], among many others. However, despite the large number of publications on using ML techniques for COVID-19, a recent review [12] of prediction models using multimodal data for COVID-19 concluded that “almost all published prediction models are poorly reported and at high risk of bias.” Another review [13] of ML methods using CXR and CT concluded that “none of the models identified are of potential clinical use due to methodological flaws and/or underlying biases.” As more multimodal data become available and more ML models are developed for COVID-19, we believe that an overview and critical appraisal of the state-of-the-art techniques are crucial to inform better development strategies and assessment of study designs. We emphasize that our focus is on multimodal data and ML models that integrate different types of data. Reviews of general ML development in COVID-19 can be found in the literature [[12], [13], [14]]; however, these reviews either focus on imaging based studies [13,14] or include models based on different types of data but not integration of multimodal data [12].
In this paper, we begin with a discussion of lessons learned from radiogenomic studies for cancer diagnosis. This actively-researched field, although on a different clinical task, investigates techniques and assessment methods to study the correlation and integration of multimodal data, which can shed light on a new disease such as COVID-19. We then summarize the types of data and ML techniques relevant to COVID-19. Finally, we present our perspectives on future development of ML techniques using multimodal data for COVID-19.
1.1. Lessons learned from radiogenomics studies for cancer diagnosis
Radiogenomic studies in cancer have generally made use of multimodal data to better understand and potentially improve cancer diagnosis, prognosis, and treatment. Historically, radiogenomic studies have combined transcriptomic data from tissue or blood samples with features extracted from images (a) to explore relationships between the underlying cancer biology and the disease presentation on medical images, and (b) to combine image and molecular features for better diagnosis and treatment planning. While this approach has not yet been leveraged for COVID-19, we describe it below as it has many parallels that may be explored in the future.
Early studies of radiogenomics in cancer [8,15] focused on hepatocellular carcinoma [[16], [17], [18]], lung cancer [[19], [20], [21], [22], [23]], glioma [24,25], breast cancer [[26], [27], [28]] and head and neck squamous carcinoma [22,29]. Early work in lung cancer radiogenomics showed that lung cancer radiographic images reflect important molecular properties of lung cancer patients including EGFR mutation status [[30], [31], [32]] and relevant gene expression pathways [33]. For example, an initial radiogenomic map of lung cancer patients showed that 56 gene expression metagenes can be predicted from CT image features with mean accuracy of 72% [21]. Similarly, radiogenomics analysis of head and neck squamous carcinoma showed that radiomic patterns reflect mutation status, DNA methylation patterns, histopathological diagnosis, and clinical outcome [29,34].
Breast cancer researchers collaborated in the MRI mapping of tumors to various clinical, molecular, and genomics markers of prognosis and risk of recurrence, including gene expression profiles [[35], [36], [37], [38], [39]]. The MRI mapping had been conducted on de-identified datasets of invasive breast carcinomas on which radiomic algorithms yielded computer-extracted quantitative lesion features from dynamic contrast-enhanced breast MRIs from The Cancer Imaging Archive (TCIA) [35] and clinical and genomic data from The Cancer Genome Atlas (TCGA) [36]. The investigators showed some specific imaging-genomic associations, such as a positive correlation between the transcriptional activities of genetic pathways and a blurred tumor margin indicating potentially tumor invasion into surrounding tissue. From association studies, they showed that MicroRNA (miRNA) expressions were associated with breast cancer tumor size and enhancement texture; indicating that miRNAs might mediate tumor growth and tumor heterogeneity of angiogenesis [35].
While an abundance of literature in radiogenomics has shown promising results, it should be noted that many of the studies involve small numbers of subjects and lack external validation, thereby limiting the generalizability of the ML technologies to patient populations at large and warranting further research [8,40,41].
1.2. Types of data for COVID-19
Multiple types of data have been investigated and used for developing ML algorithms for COVID-19 diagnosis/prognosis, including symptoms and other clinical data, laboratory tests, imaging, omics data, and pathology/physiology (Fig. 1).
Fig. 1.
Multimodal data for COVID-19.
Typical COVID-19 symptoms as listed by the US Center of Disease Control (CDC) [42] include fever, cough, shortness of breath, fatigue, muscle or body aches, headache, new loss of taste or smell, sore throat, congestion or runny nose, nausea or vomiting, and diarrhea. While the symptoms may range from mild to severe, they are usually recorded as binary features (e.g., presence or absence) in ML models. Besides patient symptoms, other important clinical data include patient demographics (age/gender/race), comorbidities, admission to ICU, use of assisted ventilation, and final clinical outcomes such as discharge (recover) or death, which can be used as severity and prognosis endpoints in ML models. For Clinical Laboratory Tests (CLTs), RT-PCR is currently the gold standard for detection of COVID-19 and is often used as the clinical endpoint for ML training and prediction. Besides RT-PCR, CLTs usually include complete blood count, a comprehensive metabolic panel and a coagulation panel, and additional tests such testing for inflammatory markers such as ESR, C-reactive protein, D-dimer etc [43].
The role of imaging has evolved during the pandemic. Chest radiography and CT imaging were initially suggested in some regions as an alternative and possibly superior testing method especially when RT-PCR was not widely available. Then, imaging applications evolved into different roles, such as for characterization of disease severity, examining COVID-19 manifestations in other organs, and investigation of the long-term sequelae of COVID-19 [5]. Moreover, lung ultrasound (LUS) was more widely employed by emergency and intensive care physicians in the UK during the COVID-19 pandemic, despite lacking patient outcome data associated with LUS findings [44]. Clinical research approaches using pulmonary magnetic resonance imaging (MRI) have been reported to be concordant with chest CT in manifestation of typical features of COVID-19 pneumonia and thus have been suggested as potential alternatives for patients who should avoid exposure to ionizing radiation [4]. However, MRI applications remain limited in clinical practice. Multimodal brain imaging studies have shown evidence for brain-related abnormalities in COVID-19; e.g., a recent study with longitudinal MRI scans showed significant longitudinal effects including reductions in gray matter thickness and global brain size in COVID-19 positive patients [45].
Besides sequencing of the virus itself, identification of variants, and their evolution and differential risk, genomic analysis of blood draws is the most common strategy to generate molecular data for COVID-19 patients [6]. Both the plasma and Peripheral Blood Mononuclear Cells (PBMCs) harbor important signals about health and disease. The plasma can be analyzed for cell free DNA & cell free RNA using sequencing, and proteins can be analyzed using proteomic and metabolomic technologies. PBMCs include lymphocyte, monocyte, and dendritic cells that can be analyzed with single cell multi-omics technologies. More specifically, by using innovative sequencing techniques the whole transcriptome, surface protein levels and TCR sequences can be simultaneously analyzed from the same cell. A recent study performed a comprehensive analysis of both the plasma and PBCMs using these omics technologies in 139 COVID-19 patients representing the spectrum of infection severities, and showed that a major immunological shift is present between mild and moderate disease [6]. While a full multi-omics analysis of both plasma and PBMCs might be prohibitive, prior studies can inform new specific omics analyses that can be customized to the clinical question to be answered of COVID-19 patients.
The physiology and pathophysiology of COVID-19 is still poorly understood in its entirety. The SARS-CoV-2 virus binds to the Angiotensin Converting Enzyme 2 (ACE2) receptors present in the lung [46] (also in heart, blood vessels, brain, intestine, Kidney, testis [46]) and results in generalized vasoconstriction in the lung, impacting gas exchange [47]. ACE2 binding also impacts diuretic and anti-inflammatory pathways, and may contribute to a cascade of vascular and vascular endothelial effects, increased cytokine production, increasing vascular responsiveness to inflammatory cytokines across multiple organs and systems [46]. Consistently, the relevant pathophysiological data for COVID-19 spans multiple organs and multiple spatial and temporal scales and spans both the immediate post-infection period and the Post-Acute Sequelae of Covid-19 (PASC). Pathophysiological mechanistic understanding of PASC is even more incipient. PASC affects multiple organs and systems, with risk that seems independent of acute disease severity [48] resulting in lingering inflammation, increased cardiovascular risk, increased risk of clot formation, as well as changes in brain structure [45], erectile dysfunction [49], kidney damage [50], and long-term lung damage [51].
The combination of multimodal data, comprising clinical data, data from well controlled physiological experiments, data from electronic health records (EHR), pathology, real-world data (wearable sensors) with imaging and machine learning may prove essential to the understanding of the underlying physiology spanning multiple scales and organ systems, as well as explain the observed heterogeneity, and contribute to identifying therapeutic targets and interventions.
In Table 1, we list several publicly available datasets that contain both medical imaging and other types of data for COVID-19 studies. The number of patients in these datasets ranges from 100 to 3000. The most commonly used clinical imaging modalities in publicly available multimodal datasets for COVID-19 are chest CT and X-ray imaging. Non-imaging data in these datasets are mainly clinical data including patient demographics, symptoms, treatment(s) received, and clinical laboratory tests. Other types of data, while relevant to COVID-19, have rarely been found in public multimodal datasets. It is also worth noting that many of these multimodal datasets are not fully paired, i.e., missing data are common. For example, in the “integrative CT images and Clinical Features for COVID-19 (iCTCF)” dataset [52], out of the 1521 subjects, 1342 subjects had both CT and clinical data. In the Stony Brook University (SBU) dataset, out of 1384 subjects, for imaging data, only 458 subjects had CT scans, and 1365 subjects had X-ray images; for clinical measurements, 271 subjects had no records for any significant comorbidities or symptoms, and for different CLTs, the number of missing values ranges from 208 to 1170.
Table 1.
A list of Public COVID-19 datasets containing multimodal data (imaging and clinical data).
| Dataset | Hosts | Sample size (#subjects) | Phenotype | Imaging modality | Non-imaging data |
|---|---|---|---|---|---|
| BIMCV COVID-19+52 | https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/ | 1131 | COVID-19 | CT, CR, DX | demographic data, radiological reports, CLTs |
| iCTCF [51] | https://ngdc.cncb.ac.cn/ictcf/; | 1521 | COVID-19 and non-COVID-19 | CT | demographic data, medical history, CLTs |
| V2-COVID19-NII [53] | https://data.uni-hannover.de/dataset/cov-19-img | 243 | COVID-19 and non-COVID-19 | CR, DX | demographic data, treatment, CLTs |
| Available in MIDRC (https://www.midrc.org; https://data.midrc.org)(∼105,000 medical imaging studies, >44,000 patients, COVID-19+/−, CR, DX, CT and limited clinical data) | |||||
| PETAL RED CORAL54,* | https://data.midrc.org/explorer | 1480 | COVID-19+ | CR, DX, CT | Demographic data, medical history, clinical lab tests |
| N3C-MIDRC** | https://ncats.nih.gov/n3c | 3000 | COVID-19 | CR, DX | Electronic Health records |
| SBU dataset [54] | TCIA: https://doi.org/10.7937/TCIA.BBAG-2923 | 1384 | COVID-19 | CT, CR, DX, MRI, PET | demographic data, medical history, symptoms, CLTs |
| MIDRC: https://data.midrc.org/explorer | |||||
| COVID-19-AR [55] | TCIA: https://doi.org/10.7937/tcia.2020.py71-5978 | 105 | COVID-19 | CT, CR, DX | demographic data, medical history, treatment |
| MIDRC: https://data.midrc.org/explorer | |||||
*Interoperability of two platforms: MIDRC for medical images, and BioData Catalyst for Electronic Health Record (EHR) data; ** Interoperability of two platforms: MIDRC for medical images, and N3C for EHR data.
1.3. Overview of statistical and machine learning techniques
This section provides an overview of machine learning techniques utilizing multimodal data for COVID-19, which broadly include statistical analysis using semantic features, hand-crafted (human-engineered) features, and deep learning models. Table 2 summarizes the technological characteristics of these techniques with references that are overviewed in this section.
Table 2.
Overview of statistical and machine learning techniques for COVID-19 using multimodal data.
| Type of data | Technological characteristics | References |
|---|---|---|
| CT images, clinical measurements, laboratory tests | Traditional statistical methods, correlation analysis | Xiong et al. [56], Qin et al. [57], Sun et al. [58], Dane et al. [59] |
| Physiology measurements and physical activities | Random forest model for early detection of COVID-19 infection | Mason et al. [60] |
| demographics, medications, laboratory tests, CPT and ICD codes | Fusion machine learning model to predict severity of COVID-19 | Tariq et al. [61] |
| CT images and clinical features | Integrative deep learning model for prediction of COVID-19 morbidity and mortality | Ning et al. [51] |
Generally, machine learning using medical images relies on the ability to ascribe features to images. There are two basic categories of features: (1) semantic features refer to words or phrases, which may have numerical codes associated with them, that describe images or regions of interest (ROIs) within them, and (2) computational features are numerical variables that can be computed directly from the images. Semantic features can be ascribed to images by experts or assigned based on machine learning using images and curated associated features for training. Hand-crafted computational features can be defined in advance as mathematical combinations of image pixel locations and intensity values that relate to visual characteristics, e.g., object shape, margin sharpness, gray or color value distributions, and texture, or they can be discovered by machine learning algorithms. Finally, it is often desirable to limit observation to certain parts of the image (e.g., a tumor in cancer studies, the lungs in a chest CT), giving rise to the need for image segmentation. While this has shown to be extremely important in cancer applications, regions of the lungs impacted by COVID may be hard for humans and computers to segment. End-to-end deep learning models learn features and identify regions automatically in a seamless pipeline and thereby do not rely on segmentation and human-engineered features. However, this flexibility is associated with the need for large amount of training data and poor interpretability.
Several studies [[53], [54], [55], [62]] have focused on analyzing the statistical relationship between CT characteristics, clinical measurements, and laboratory findings in COVID-19 patients. In all of these, CT image features were extracted by multiple radiologists following clinical guidelines. Such manually-extracted CT features may include the following: lesion location (e.g., peripheral, central, both central and peripheral); lesion attenuation (e.g., ground glass opacities (GGOs), consolidations, and mixed GGOs); lesion pattern (e.g., patchy, oval); distribution of affected lobes; pleural effusion, air bronchogram, interstitial thickening or reticulation, tree-in-bud signs, etc. Statistical analyses have been applied to analyze the relationship between CT image features, clinical characteristics and COVID-19. Statistical analyses have, for example, included, chi-square or Fisher exact test for categorical features, Mann-Whitney U test or Student t-test for quantitative features, linear regression [53] and logistic regression [62], and Spearman or Pearson correlation analysis [53,54]. These analyses may shed light on the design of more complicated ML models for the integration of multimodal data or help with interpretation of ML models.
Mason et al. [56] developed random forest models for early detection of COVID-19 infection using multimodal data from a wearable device including photoplethysmography-based measurement of physiology parameters and accelerometry-based measurement of physical activities of patients. This unique effort yielded promising results with cross-validation on a limited dataset, thereby warranting further development in diverse populations. Tariq et al. [57] compared early, middle, and late fusion ML models using multi-modal data including demographics, medications, laboratory tests, CPT and ICD codes to predict the severity of COVID-19 at the time of testing and a COVID-19 positive patient's need for hospitalization. While promising results were reported, the authors admit that the study has limited data from a highly integrated academic healthcare system putting the generalizability of the models into question.
While most machine learning models use either Clinical Features (CFs) or imaging features as predictors, Ning et al. [52] developed an integrative CT images and CFs for COVID-19 (iCTCF) model that integrates the CFs with chest CT images for prediction of COVID-19 morbidity and mortality outcomes using a deep learning based algorithm trained with data from 1170 patients. In their model, a 13-layer Convolutional Neural Network (CNN) model was first used to classify individual CT slices into three types (non-informative, positive, negative) and the positive images were input to the second 13-layer CNN to predict patient outcomes. In the meantime, the CFs were combined by a 7-layer Deep Neural Network (DNN). Finally, the predictions using CT images and CFs were integrated using the penalized logistic regression model to output final predictions on morbidity or mortality outcomes of patients.
1.4. Perspective on future studies
We believe that development of machine learning models using multimodal data has great potential to help understand COVID-19 more comprehensively, provide clinical tools in the detection, diagnosis, and triage of patients as well as detection and treatment of long COVID complications. To inform future development of multimodal ML models for COVID-19, we present our perspective on several important aspects: data curation, model strategy, clinical relevance, and assessment.
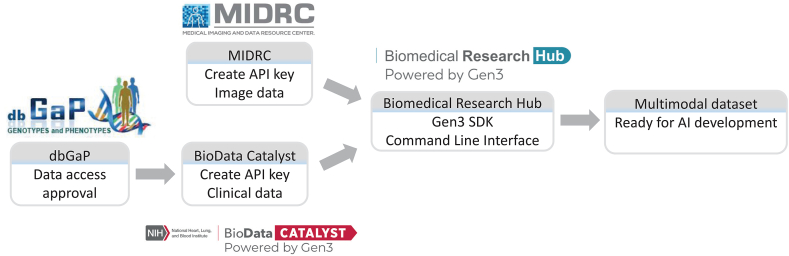
Curation of high-quality and diverse multimodal datasets plays a vital role. It is critically important that a dataset for ML development is diverse enough to represent the intended patient population and minimize bias. It is also crucial that a dataset is large enough to train a complex model such as deep neural networks to avoid overfitting. The curation of multimodal dataset is generally more challenging than a single-modality dataset due to its scarcity and the possible need for effort to match data from multiple sources. As an example, Fig. 2 shows the workflow of the curation of a multimodal dataset of US patients hospitalized with COVID-19 originally collected by the Prevention and Early Treatment of Acute Lung Injury (PETAL) Network [58]. The clinical data of 1480 patients hosted by BioData Catalyst (BDCat) [59] include patient demographics, medical history, symptoms, and laboratory results. While the dataset is publicly available, the access to these data needs an approval through dbGaP (https://www.ncbi.nlm.nih.gov/gap/). A subset of these patients have imaging data that are hosted by MIDRC including chest radiographs and CTs. Both data servers are powered by Gen3 (https://gen3.org/), which provides a command line interface using the Gen3 SDK to query/retrieve data. One can match the data from MIDRC and BDCat by using the API keys created from the two sources and a crosswalk service provided by the Biomedical Research Hub (BRH) (https://brh.data-commons.org/login).
Fig. 2.
Curation of the PETAL multimodal dataset involving two sources. The clinical data is hosted by Biodata Catalyst and access requires approval through dbGaP. The imaging data is hosted by MIDRC. Both data servers are powered by Gen3 and matching of the data from the two sources is achieved by a command line interface using the Gen3 Software Development Kit (SDK) on the Biomedical Research Hub, which requires Application Programming Interface (API) keys created on MIDRC and BioData catalyst.
Interoperability is also available between MIDRC and the National COVID Cohort Collaborative (N3C, https://ncats.nih.gov/n3c). MIDRC currently hosts imaging data from 3000 patients with the corresponding EHR data available in N3C. Medical imaging and EHR data can be associated using Privacy Preserving Record Linkage tokens through a linkage honest broker, and a crosswalk service provided by the Biomedical Research Hub (BRH). Access to the EHR data requires access to N3C, as set out in https://ncats.nih.gov/n3c/about/applying-for-access.
The availability of public datasets from multiple sources is beneficial to help improve data diversity. On the other hand, merging datasets from multiple sources may be challenging due to variations and biases in patient populations, data collection and image acquisition protocols. Great attention must be paid to the image quality differences across different sources that may lead to shortcut learning [60,61], i.e., models learning institution-specific markers or annotations instead of pathology in the image. Data harmonization techniques may help mitigate this issue. Representativeness of data is key, as this is a potentially significant source of bias. For COVID-19, different tasks will require different target population distributions; typically based on ‘reference’ distributions such as data from the US Census, COVID-19 diagnosis, hospitalization, and mortality present significant differences across age and self-reported racial groups, e.g., which need to be accounted for when creating training, tuning and test cohorts. Cross-repository cohort building, availability of desired data types and specific data may further limit sample size, or result in significant missing data. Indeed, missing data are common and oftentimes only a subset of patients in a multimodal dataset have data from all modalities. Moreover, even when multimodal data are available, they may be acquired at different times; such temporal misalignment must be considered for a disease such as COVID-19 that is evolving quickly. Multimodal machine learning model development must take these into consideration.
Multimodal machine learning has many design strategies to address medical problems, particularly for COVID-19. Representation refers to techniques for representing and summarizing multimodal data in a way that exploits the complementarity and redundancy of multiple modalities. Correlation studies investigate the quantitative correlation and relations between different modalities for a certain clinical task, which provides basis for translation or replacement of one modality to another (e.g., use of non-invasive testing to replace invasive ones). Fusion models aim to join information from multiple modalities thereby taking advantage of the complementarity to improve accuracy. Huang et al. [63] provide a systematic review and implementation guidelines on the fusion of medical imaging and EHR data using deep learning, in which, like earlier reviews in more general AI applications [64], pros and cons of different fusion strategies are discussed including early fusion (concatenating multimodal features), late fusion (integration decisions of models from individual models), and hybrid fusion models (allowing joint learning and optimizing features).
In addition to data driven associations, the ability to use established biology and physiology knowledge to create quantitative imaging or clinical features may contribute to improve model accuracy. Physiology driven mechanistic features and models can enrich phenomenological data-driven models through cohort normalization, missing data imputation, etc. [65]. Probably the most noteworthy example is the use of digital twins [66,67], where computational physiological models create constrained, plausible, individualized artificial data instances of data, including human physiology. Synthetic digital twins, the generation of which involves fusion of multimodal data, may be used to enrich datasets, probe for potential bias, or assess sensitivity to parameter changes in a multimodal machine learning model.
For a multimodal machine learning model to be clinically useful, it is crucial to target the right clinical question. In relation to the current stage of COVID-19, some of the most pressing questions are linked to PASC (long-COVID), especially who is likely to develop PASC; what mechanistic pathways may be common to multiple PASC presentations; and who is likely to respond to specific therapeutical interventions, e.g. from depression [68]. Considering the complexity of PASC and the large ongoing efforts to collect and curate imaging, omics, clinical and EHR data in long-COVID (the NIH Recover Initiative, MIDRC-N3C interoperability, for example), PASC may prove to be an important ground for the multimodal, multi-data type, machine learning approaches that are the focus of this manuscript.
Assessment methods play a critical role in the development of multimodal ML models to avoid pitfalls and enhance quality in the design, analysis, and reporting of ML studies. Statistical methods are essential for study design to appropriately collect and/or partition data for training, tuning, internal and external validations to avoid bias, for analysis of results to yield generalizable results with assessment of uncertainty. Tools, such as the PROBAST tool [69], are available to facilitate the development of ML models with minimized bias and better applicability. Many machine-learning-good-practice checklists/guidelines are also generally useful tools such as the checklist for artificial intelligence in medical imaging (CLAIM) [70] and the guidelines for publication of AI in medical physics [71].
2. Conclusion
Machine learning technologies using multimodal data have great potential to help better understand COVID-19 and provide clinical tools in the diagnosis, prognosis, and triage of patient as well as prediction and treatment of long COVID complications. Experience gained and lessons learned from radiogenomics and general multimodal machine learning technologies have and will continue to facilitate the development for COVID-19. Conversely, the experience gained through the development of multimodal COVID-19 machine learning will contribute to other efforts, such as cancer diagnosis and precision medicine [72]. For example, the MIDRC infrastructure and data curation techniques for data sharing, the interoperability of MIDRC with other data repositories as shown in this paper, and MIDRC developed tools such as the bias identification and mitigation tool [73] can be applied to ML models in other applications as well. We recognize that multimodal machine learning, whether for general medical applications or for COVID-19 specifically, is still in its infancy. A major bottleneck is the scarcity of high-quality multimodal data. Our MIDRC projects have put significant efforts into the curation of multimodal data. It can be expected that the availability of multimodal data and the development of ML models will greatly advance our understanding of COVID-19 and potentially lead to clinically impactful tools.
Author contribution statement
All authors listed have significantly contributed to the development and the writing of this article.
Data availability statement
This is a review paper and datasets under review were public and their sources were included in the paper.
Additional information
Supplementary content related to this article has been published online at [URL].
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Research reported is part of MIDRC and was made possible by the National Institute of Biomedical Imaging and Bioengineering (NIBIB) of the National Institutes of Health under contracts 75N92020C00008 and 75N92020C00021. This work is supported by NIH through the Data and Technology Advancement (DATA) National Service Scholar program.
References
- 1.World Health Organization . 2022. Weekly Epidemiological Update on COVID-19 - 30 November 2022. [Google Scholar]
- 2.Rana R., Tripathi A., Kumar N., Ganguly N.K. A comprehensive overview on COVID-19: future perspectives. Front. Cell. Infect. Microbiol. 2021;11 doi: 10.3389/fcimb.2021.744903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nalbandian A., et al. Post-acute COVID-19 syndrome. Nat. Med. 2021;27:601–615. doi: 10.1038/s41591-021-01283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fields B.K.K., Demirjian N.L., Dadgar H., Gholamrezanezhad A. Imaging of COVID-19: CT, MRI, and PET. Semin. Nucl. Med. 2021;51:312–320. doi: 10.1053/j.semnuclmed.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kanne J.P., et al. COVID-19 imaging: what we know now and what remains unknown. Radiology. 2021;299:E262–E279. doi: 10.1148/radiol.2021204522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Su Y., et al. Multi-omics resolves a sharp disease-state shift between mild and moderate COVID-19. Cell. 2020;183:1479–1495.e1420. doi: 10.1016/j.cell.2020.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busana M., et al. The impact of ventilation–perfusion inequality in COVID-19: a computational model. J. Appl. Physiol. 2021;130:865–876. doi: 10.1152/japplphysiol.00871.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Napel S., Giger M. Special section guest editorial:radiomics and imaging genomics: quantitative imaging for precision medicine. J. Med. Imaging. 2015;2 doi: 10.1117/1.JMI.2.4.041001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trivizakis E., et al. Artificial intelligence radiogenomics for advancing precision and effectiveness in oncologic care. Int. J. Oncol. 2020;57:43–53. doi: 10.3892/ijo.2020.5063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen R.J., et al. Pan-cancer integrative histology-genomic analysis via multimodal deep learning. Cancer Cell. 2022;40:865–878.e866. doi: 10.1016/j.ccell.2022.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheerla A., Gevaert O. Deep learning with multimodal representation for pancancer prognosis prediction. Bioinformatics. 2019;35:i446–i454. doi: 10.1093/bioinformatics/btz342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wynants L., et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ. 2020;369:m1328. doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roberts M., et al. Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat. Mach. Intell. 2021;3:199–217. [Google Scholar]
- 14.Komolafe T.E., et al. Diagnostic test accuracy of deep learning detection of COVID-19: a systematic review and meta-analysis. Acad. Radiol. 2021;28:1507–1523. doi: 10.1016/j.acra.2021.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colen R., et al. NCI workshop report: clinical and computational requirements for correlating imaging phenotypes with genomics signatures. Transl Oncol. 2014;7:556–569. doi: 10.1016/j.tranon.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuo M.D., Gollub J., Sirlin C.B., Ooi C., Chen X. Radiogenomic analysis to identify imaging phenotypes associated with drug response gene expression programs in hepatocellular carcinoma. J. Vasc. Intervent. Radiol. 2007;18:821–831. doi: 10.1016/j.jvir.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 17.Renzulli M., et al. Can current preoperative imaging Be used to detect microvascular invasion of hepatocellular carcinoma? Radiology. 2016;279:432–442. doi: 10.1148/radiol.2015150998. [DOI] [PubMed] [Google Scholar]
- 18.Sagir Kahraman A. Radiomics in hepatocellular carcinoma. J. Gastrointest. Cancer. 2020;51:1165–1168. doi: 10.1007/s12029-020-00493-x. [DOI] [PubMed] [Google Scholar]
- 19.Das A.K., Bell M.H., Nirodi C.S., Story M.D., Minna J.D. Radiogenomics predicting tumor responses to radiotherapy in lung cancer. Semin. Radiat. Oncol. 2010;20:149–155. doi: 10.1016/j.semradonc.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nair V.S., et al. Prognostic PET 18F-FDG uptake imaging features are associated with major oncogenomic alterations in patients with resected non-small cell lung cancer. Cancer Res. 2012;72:3725–3734. doi: 10.1158/0008-5472.CAN-11-3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gevaert O., et al. Non-small cell lung cancer: identifying prognostic imaging biomarkers by leveraging public gene expression microarray data--methods and preliminary results. Radiology. 2012;264:387–396. doi: 10.1148/radiol.12111607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aerts H.J., et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Napel S., Mu W., Jardim-Perassi B.V., Aerts H., Gillies R.J. Quantitative imaging of cancer in the postgenomic era: radio(geno)mics, deep learning, and habitats. Cancer. 2018;124:4633–4649. doi: 10.1002/cncr.31630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zinn P.O., et al. Radiogenomic mapping of edema/cellular invasion MRI-phenotypes in glioblastoma multiforme. PLoS One. 2011;6 doi: 10.1371/journal.pone.0025451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gevaert O., et al. Glioblastoma multiforme: exploratory radiogenomic analysis by using quantitative image features. Radiology. 2014;273:168–174. doi: 10.1148/radiol.14131731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto S., Maki D.D., Korn R.L., Kuo M.D. Radiogenomic analysis of breast cancer using MRI: a preliminary study to define the landscape. AJR Am. J. Roentgenol. 2012;199:654–663. doi: 10.2214/AJR.11.7824. [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto S., et al. Breast cancer: radiogenomic biomarker reveals associations among dynamic contrast-enhanced MR imaging, long noncoding RNA, and metastasis. Radiology. 2015;275:384–392. doi: 10.1148/radiol.15142698. [DOI] [PubMed] [Google Scholar]
- 28.Grimm L.J. Breast MRI radiogenomics: current status and research implications. J. Magn. Reson. Imag. 2016;43:1269–1278. doi: 10.1002/jmri.25116. [DOI] [PubMed] [Google Scholar]
- 29.Huang C., et al. Development and validation of radiomic signatures of head and neck squamous cell carcinoma molecular features and subtypes. EBioMedicine. 2019;45:70–80. doi: 10.1016/j.ebiom.2019.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang S., et al. Predicting EGFR mutation status in lung adenocarcinoma on computed tomography image using deep learning. Eur. Respir. J. 2019;53 doi: 10.1183/13993003.00986-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Minamimoto R., et al. Prediction of EGFR and KRAS mutation in non-small cell lung cancer using quantitative (18)F FDG-PET/CT metrics. Oncotarget. 2017;8:52792–52801. doi: 10.18632/oncotarget.17782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gevaert O., et al. Predictive radiogenomics modeling of EGFR mutation status in lung cancer. Sci. Rep. 2017;7 doi: 10.1038/srep41674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou M., et al. Non-small cell lung cancer radiogenomics map identifies relationships between molecular and imaging phenotypes with prognostic implications. Radiology. 2018;286:307–315. doi: 10.1148/radiol.2017161845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee P., et al. CT-Based radiomic signatures for predicting histopathologic features in head and neck squamous cell carcinoma. Radiol Imaging Cancer. 2020;2 doi: 10.1148/rycan.2020190039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu Y., et al. Deciphering genomic underpinnings of quantitative MRI-based radiomic phenotypes of invasive breast carcinoma. Sci. Rep. 2015;5 doi: 10.1038/srep17787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo W., et al. Prediction of clinical phenotypes in invasive breast carcinomas from the integration of radiomics and genomics data. J. Med. Imaging. 2015;2 doi: 10.1117/1.JMI.2.4.041007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H., et al. Quantitative MRI radiomics in the prediction of molecular classifications of breast cancer subtypes in the TCGA/TCIA data set. npj Breast Cancer. 2016;2 doi: 10.1038/npjbcancer.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H., et al. MR imaging radiomics signatures for predicting the risk of breast cancer recurrence as given by research versions of MammaPrint, oncotype DX, and PAM50 gene assays. Radiology. 2016;281:382–391. doi: 10.1148/radiol.2016152110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burnside E.S., et al. Using computer-extracted image phenotypes from tumors on breast magnetic resonance imaging to predict breast cancer pathologic stage. Cancer. 2016;122:748–757. doi: 10.1002/cncr.29791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bai H.X., et al. Imaging genomics in cancer research: limitations and promises. Br. J. Radiol. 2016;89 doi: 10.1259/bjr.20151030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pinker K., et al. Background, current role, and potential applications of radiogenomics. J. Magn. Reson. Imag. 2018;47:604–620. doi: 10.1002/jmri.25870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Centers for Disease Control and Prevention of USA (CDC). Symptoms of COVID-19. 2021. https://www.cdc.gov/coronavirus/2019-ncov/symptoms-testing/symptoms.html
- 43.Cascella M., Aleem A., et al. 2021. Features, Evaluation, and Treatment of Coronavirus (COVID-19)https://www.ncbi.nlm.nih.gov/books/NBK554776/ [PubMed] [Google Scholar]
- 44.Jackson K., Butler R., Aujayeb A. Lung ultrasound in the COVID-19 pandemic. Postgrad. Med. 2021;97:34. doi: 10.1136/postgradmedj-2020-138137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Douaud G., et al. SARS-CoV-2 is associated with changes in brain structure in UK Biobank. Nature. 2022 doi: 10.1038/s41586-022-04569-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bourgonje A.R., et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19) J. Pathol. 2020;251:228–248. doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karmouty-Quintana H., et al. Emerging mechanisms of pulmonary vasoconstriction in SARS-CoV-2-induced acute respiratory distress syndrome (ARDS) and potential therapeutic targets. Int. J. Mol. Sci. 2020;21:8081. doi: 10.3390/ijms21218081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A., et al. Symptoms and risk factors for long COVID in non-hospitalized adults. Nat. Med. 2022;28:1706–1714. doi: 10.1038/s41591-022-01909-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kaynar M., Gomes A.L.Q., Sokolakis I., Gül M. Tip of the iceberg: erectile dysfunction and COVID-19. Int. J. Impot. Res. 2022;34:152–157. doi: 10.1038/s41443-022-00540-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yende S., Parikh C.R. Long COVID and kidney disease. Nat. Rev. Nephrol. 2021;17:792–793. doi: 10.1038/s41581-021-00487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grist J.T., et al. Lung abnormalities detected with hyperpolarized 129Xe MRI in patients with long COVID. Radiology. 2022;305:709–717. doi: 10.1148/radiol.220069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ning W., et al. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nature biomedical engineering. 2020;4:1197–1207. doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiong Y., et al. Clinical and high-resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes. Invest. Radiol. 2020 doi: 10.1097/RLI.0000000000000674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun D., et al. CT quantitative analysis and its relationship with clinical features for assessing the severity of patients with COVID-19. Korean J. Radiol. 2020;21:859. doi: 10.3348/kjr.2020.0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dane B., Brusca-Augello G., Kim D., Katz D.S. Unexpected findings of coronavirus disease (COVID-19) at the lung bases on abdominopelvic CT. Am. J. Roentgenol. 2020;215:603–606. doi: 10.2214/AJR.20.23240. [DOI] [PubMed] [Google Scholar]
- 56.Mason A.E., et al. Detection of COVID-19 using multimodal data from a wearable device: results from the first TemPredict Study. Sci. Rep. 2022;12:3463. doi: 10.1038/s41598-022-07314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tariq A., et al. Patient-specific COVID-19 resource utilization prediction using fusion AI model. npj Digital Medicine. 2021;4:94. doi: 10.1038/s41746-021-00461-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Peltan I.D., et al. Characteristics and outcomes of US patients hospitalized with COVID-19. Am. J. Crit. Care. 2022;31:146–157. doi: 10.4037/ajcc2022549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.National Heart Lung, Blood Institute. National Institutes of Health, U.S . (National Heart, Lung, and Blood Institute, National Institutes of Health, U.S. Department of Health and Human Services. 2020. Department of health and human services. The NHLBI BioData catalyst. [Google Scholar]
- 60.López-Cabrera J.D., Orozco-Morales R., Portal-Díaz J.A., Lovelle-Enríquez O., Pérez-Díaz M. Current limitations to identify covid-19 using artificial intelligence with chest x-ray imaging (part ii). The shortcut learning problem. Health Technol. 2021:1–15. doi: 10.1007/s12553-021-00609-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.DeGrave A.J., Janizek J.D., Lee S.-I. AI for radiographic COVID-19 detection selects shortcuts over signal. Nat. Mach. Intell. 2021;3:610–619. [Google Scholar]
- 62.Qin L., et al. A predictive model and scoring system combining clinical and CT characteristics for the diagnosis of COVID-19. Eur. Radiol. 2020;30:6797–6807. doi: 10.1007/s00330-020-07022-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang S.C., Pareek A., Seyyedi S., Banerjee I., Lungren M.P. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ Digit Med. 2020;3:136. doi: 10.1038/s41746-020-00341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baltrusaitis T.a.A., Chaitanya, Morency Louis-Philippe. Multimodal machine learning: a survey and taxonomy. IEEE Trans. Pattern Anal. Mach. Intell. 2019;41:423–443. doi: 10.1109/TPAMI.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 65.Jones E., et al. Phenotyping heart failure using model-based analysis and physiology-informed machine learning. J. Physiol. 2021;599:4991–5013. doi: 10.1113/JP281845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Coorey G., et al. The health digital twin to tackle cardiovascular disease—a review of an emerging interdisciplinary field. npj Digital Medicine. 2022;5:126. doi: 10.1038/s41746-022-00640-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stahlberg E.A., et al. Exploring approaches for predictive cancer patient digital twins: opportunities for collaboration and innovation. Front Digit Health. 2022;4 doi: 10.3389/fdgth.2022.1007784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oakley T., Coskuner J., Cadwallader A., Ravan M., Hasey G. IEEE Trans Biomed Eng; 2022. EEG Biomarkers to Predict Response to Sertraline and Placebo Treatment in Major Depressive Disorder. [DOI] [PubMed] [Google Scholar]
- 69.Wolff R.F., et al. PROBAST: a tool to assess the risk of bias and applicability of prediction model studies. Ann. Intern. Med. 2019;170:51–58. doi: 10.7326/M18-1376. [DOI] [PubMed] [Google Scholar]
- 70.Mongan J., Moy L., Kahn Charles E. J. Checklist for artificial intelligence in medical imaging (CLAIM): a guide for authors and reviewers. Radiology: Artif. Intell. 2020;2 doi: 10.1148/ryai.2020200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.El Naqa I., et al. AI in medical physics: guidelines for publication. Med. Phys. 2021;48:4711–4714. doi: 10.1002/mp.15170. [DOI] [PubMed] [Google Scholar]
- 72.Boehm K.M., Khosravi P., Vanguri R., Gao J., Shah S.P. Harnessing multimodal data integration to advance precision oncology. Nat. Rev. Cancer. 2022;22:114–126. doi: 10.1038/s41568-021-00408-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Karen D., et al. Toward fairness in artificial intelligence for medical image analysis: identification and mitigation of potential biases in the roadmap from data collection to model deployment. J. Med. Imag. 2023;10 doi: 10.1117/1.JMI.10.6.061104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This is a review paper and datasets under review were public and their sources were included in the paper.