Abstract
The main virulence factors of the phytopathogenic bacterium Erwinia chrysanthemi are pectinases which attack pectin, the major constituent of the plant cell wall. Of these enzymes, the alkaline isoenzyme named PelD in strain 3937 and PelE in strain EC16 has been described as being particularly important, based on virulence studies of plants. Expression of the pelD and pelE genes is tightly modulated by various regulators, including the KdgR repressor and the cyclic AMP-cyclic AMP receptor protein (CRP) activator complex. The use of a lacZ reporter gene allowed us to quantify the repression of E. chrysanthemi 3937 pelD expression exerted by PecS, another repressor of pectinase synthesis. In vitro DNA-protein interaction experiments, centered on the pelD and pelE wild-type or pelE mutated promoter regions, allowed us to define precisely the sequences involved in the binding of these three regulators and of RNA polymerase (RNAP). These studies revealed an unusual binding of the KdgR repressor and suggested the presence of a UP (upstream) element in the pelD and pelE genes. Investigation of the simultaneous binding of CRP, KdgR, PecS, and the RNAP to the regulatory region of the pelD and pelE genes showed that (i) CRP and RNAP bind cooperatively, (ii) PecS partially inhibits binding of the CRP activator and of the CRP-RNAP complex, and (iii) KdgR stabilizes the binding of PecS and prevents transcriptional initiation by RNAP. Taken together, our data suggest that PecS attenuates pelD and pelE expression rather than acting as a true repressor like KdgR. Overall, control of the pelD and pelE genes of E. chrysanthemi appears to be both complex and novel.
The phytopathogenicity of the pectinolytic erwiniae is mainly due to their capacity to synthesize and secrete depolymerizing enzymes which macerate the major component of plant cell walls. Among these enzymes, pectate lyases (Pels) play a major role since, when purified, they are able to mimic symptoms of the bacterial infection (11).
Erwinia chrysanthemi 3937 produces five major pectate lyases encoded by pelA, pelB, pelC, pelD, and pelE (13). PelB and PelC have moderately basic pIs (7.5 to 8.5), PelA is acidic (pI 4.5), and PelD and PelE are strongly basic (pI 9.5 to 10.5) (13). In the other well-studied E. chrysanthemi strain, EC16, only four major Pel isoenzymes are present (29). DNA sequence analysis in this latter strain revealed a deletion event that removed most of the coding region for the gene corresponding to pelE in strain 3937. Thus, the major basic isoenzyme in strain EC16, which corresponds to that encoded by the 3937 pelD gene, was named PelE (13).
Production of the Pels by E. chrysanthemi is tightly regulated and responds to various physiological controls, including growth phase-dependent induction, catabolic repression and variations in environmental conditions such as the presence of pectin or plant extract, temperature, or nitrogen starvation (13). Several mechanisms modulating the expression of the pel genes in E. chrysanthemi 3937 have been elucidated. It has been demonstrated that the full synthesis of Pels requires the presence of the cyclic AMP (cAMP)-cAMP receptor protein (CRP) complex (23), CRP being proposed to act as the primary activator of the pelB, pelC, pelD, and pelE promoters (20). The KdgR repressor essentially mediates the induction of pel gene expression by pectic compounds (24), whereas two other loci involved in the negative regulation of pel gene expression, pecS-pecM and pecT, have also been characterized, but the signals to which these regulators respond remain unknown (9, 25, 28). Although in vivo deletion and mutation analyses conducted on the strain EC16 pelE regulatory region revealed the existence of various regulatory sequences (12), only one regulatory gene, pir (plant-inducible gene), was formally identified (21). It was shown that this gene directs induction of the pel genes by plant extracts.
In this study, we established that the specific regulators of pectinolysis identified in strain 3937, PecS and KdgR, are also present in strain EC16 and display DNA binding activity. Moreover, the elements directing CRP, KdgR, and PecS binding were identified in the EC16 pelE regulatory region, and their occupancy by these three proteins as well as by RNA polymerase (RNAP) was investigated in parallel to the corresponding 3937 pelD promoter. Finally, we propose a mechanism that incorporates the new data and earlier observations to explain the regulation of these two allelic loci.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids are described in Table 1. E. chrysanthemi and Escherichia coli cells were grown at 30 and 37°C, respectively, in Luria-Bertani (LB) medium, synthetic M63 medium, or 2YT medium (18) supplemented, when required, with the antibiotics ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (50 μg/ml). Carbon source was added at 2 g/liter with the exception of polygalacturonate (PGA) (grade II; Sigma Chemical Co.), which was added at 4 g/liter.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or descriptiona | Source or reference |
|---|---|---|
| E. coli NM522 | Δ(lac-proAB) thi hsd-5 supE [F′ proAB+ lacIqlacZΔM15] | Stratagene |
| E. chrysanthemi | ||
| EC16 | Wild-type strain | A. Chatterjee, University of Missouri |
| 3937 | Wild-type strain isolated from Saintpaulia ionanthia | Laboratory collection |
| A350 | lmrT(Con) lacZ2 | Laboratory collection |
| A1510 | A350, kdgK arg10 pelD::lacZ Cmr | Laboratory collection |
| A837 | A350, kdgR | Laboratory collection |
| A2011 | A350, pecS::uidA Kanr | Laboratory collection |
| A2348 | A837, pecS::uidA Kanr | Laboratory collection |
| A3485 | A350, pelD::lacZ Cmr | This work |
| A3486 | A2011, pelD::lacZ Cmr | This work |
| A3487 | A837, pelD::lacZ Cmr | This work |
| A3489 | A2348, pelD::lacZ Cmr | This work |
| Plasmids | ||
| pUC18 | Cloning plasmid | 31 |
| pBluescript | Apr, lacZ′ | Stratagene |
| pSR2159 | pUC18 with the 495-bp BstEII-HpaI fragment containing the pelA regulatory region from strain 3937 inserted between the SmaI and XbaI sites | 19 |
| pN1272 | pBluescript with 725-bp HindIII-BglII fragment contained the pelD regulatory region from strain 3937 inserted into the HindIII-BamHI sites | N. Hugouvieux-Cotte-Pattat |
| pWN2481 | pBluescript with 318-bp HpaI-HindIII fragment contained the pelD regulatory region from strain 3937 inserted into the EcoRV-HindIII sites | This work |
| pPEL743 | pUC18 carrying pelE gene from strain EC16 and 5′ sequences | 29 |
| pElux781 | Oligonucleotide mutant in which bases 34 to 37 (AAAC) of the EC16 pelE promoter were deleted:ΔOP2 | 12 |
| pElux782 | Oligonucleotide mutant in which bases 19 to 22 (ATTT) of the EC16 pelE promoter were deleted:ΔOP1 | 12 |
| pElux783 | Oligonucleotide mutant in which bases −41 to −43 (TGA) of the EC16 pelE promoter were deleted: ΔCRP1 | 12 |
| pElux784 | Oligonucleotide mutant in which the deletions in both pElux781 and pElux782 were introduced into the EC16 pelE promoter: ΔOP1 + OP2 | 12 |
Genotype designations are according to reference 2. lmrT(Con) indicates that the transport system encoded by the gene lmrT, which mediates entry of lactose, melibiose, and raffinose into cells, is constitutively expressed. lacZ′ indicates that the 3′ end of the lacZ gene is truncated.
Proteins.
The KdgR, CRP, and PecS of strain 3937 used in this work were purified as described previously (19, 20, 22). Protein concentrations were determined by Bradford’s method (5). E. coli RNAP was purchased from TEBU (distributor for Epicentre Technologies).
Preparation of operator fragments for binding studies.
The regulatory regions from the E. chrysanthemi EC16 and 3937 pel genes were cloned in pUC18 and pBluescript vectors, respectively (Table 1). The EC16 pelE operator was labeled at the top strand by incorporation of [α-32P]dCTP (3,000 Ci/mmol−1; Amersham) with the Klenow fragment of DNA polymerase at the MluI end of the NdeI-MluI fragment (260 bp). For the bottom strand, the region between −259 to +139 was amplified by PCR using pPEL743 as the template and two primers (5′ AGGGGCTTTCAAGCTTTAATAAGGCAC 3′ and 5′ GTAAACTTTTAGTTCCTCGAGAACGTAC 3′) overlapping the extremities of this region and containing HindIII and XhoI cutting sites, respectively. The bottom strand was labeled by incorporating [α-32P]dATP (3,000 Ci/mmol−1; Amersham) with the Klenow fragment of DNA polymerase at the HindIII end. For the 3937 pelD gene, plasmid pN1272 was digested by HpaI and HindIII. The top strand was labeled at the HindIII end by incubation in the presence of [α-32P]dCTP (3,000 Ci/mmol−1) and Klenow fragment of DNA polymerase. For labeling the bottom strand, the HpaI-HindIII fragment was cloned into the EcoRV-HindIII sites of pBluescript KS+, (Apr) giving rise to plasmid pWN2481. This recombinant plasmid was digested by HindII and EcoRI and further labeled at the EcoRI end by incorporation of [α-32P]dATP (3,000 Ci/mmol−1; Amersham) with the Klenow fragment of DNA polymerase. These labeled fragments were further purified by the DEAE-cellulose paper procedure (1) or with a Qiagen quick extraction kit.
Gel retardation assay.
Band shift assays were conducted as described by Nasser et al. (20) and Praillet et al. (22). Cobinding studies were performed with a buffer allowing correct fixation for each of the various regulatory proteins, consisting of 10 mM Tris-HCl (pH 7.8 or 7), 75 mM KCl, 1 mM dithiothreitol, 100 μM cAMP, 4 μg of acetylated bovine serum albumin (BSA), and 1 μg of poly(dI-dC) · (dI-dC) (Pharmacia LKB) as bulk carrier DNA. After addition of the DNA probe (50,000 cpm) and of various amounts of the purified CRP, PecS, RNAP, or KdgR, the reaction mixtures were incubated for 30 min at 30°C, then loaded onto a 4% nondenaturing polyacrylamide gel, and electrophoresed in 10 mM Tris-HCl (pH 7.8 or 7) containing 100 μM cAMP. Gels were then dried and exposed to Amersham MP film.
The apparent dissociation constants (Kd app) were determined as described by Carey (8), with minor modifications. Band shift assays were performed as described above, with a large dilution scale of the three different regulators (CRP, PecS, and KdgR). Autoradiograms were subjected to densitometric analysis using BIOPROFIL software (Vilber Loumat). For each dilution of the regulatory proteins, the ratio of free probe to total DNA was then calculated and plotted. The Kd app is the regulatory protein concentration for which half of the DNA probe is complexed to the protein.
Interference experiments.
Base removal was achieved by using formic acid as a depurinating reagent and hydrazine as a depyrimidinating reagent (6). Interference experiments were performed by incubating modified DNA (50,000 cpm) with KdgR (50 nM) as described for the gel retardation assays. Reaction mixtures were analyzed by a preparative gel retardation assay. Bands corresponding to free and complexed DNA were visualized by autoradiography on the wet gel after 3 h exposure to Amersham MP film at 4°C. Labeled DNA was cut out of the gel, eluted for subsequent piperidine cleavage, and analyzed in a sequencing gel.
Shift-Western blotting.
After the gel shift, Western blotting was done by the semidry blotting method using a multiphor II NovaBlot electrophoretic transfer unit (Pharmacia) at a fixed current of 0.8 mA per square centimeter of gel surface area for 90 min. Tris (48 mM)-glycine (39 mM) containing 0.0375% sodium dodecyl sulfate and 20% (vol/vol) methanol was used as the buffer, and a nitrocellulose filter was used as the membrane. After electrotransfer, the membranes were saturated for 2 h at 37°C with 30 g of BSA/liter in Tris-buffered saline (TBS; 137 mM NaCl, 20 mM Tris-HCl [pH 7.5]), rinsed extensively with TBS containing 0.1% (vol/vol) Tween 20 (T-TBS), and then incubated for at least 4 h at room temperature with a 1/400 dilution of the primary antibodies in T-TBS containing 0.5% BSA. The primary antibodies used in this work were purified from a rabbit antiserum as described by Sakakibara et al. (27). Finally, enhanced chemiluminescence protein detection was carried out as described by Amersham, with a goat anti-rabbit immunoglobulin G conjugated to peroxidase.
Footprinting with DNase I.
DNase I footprinting was performed as described by Nasser et al. (20), with minor modifications. About 100,000 cpm of DNA probe, labeled at one end, was incubated for 30 min at 30°C with the purified protein(s) in the buffer used for mobility shift assay, and the reaction mixtures were adjusted to 10 mM MgCl2 and 5 mM CaCl2 just before the addition of DNase I. DNase I (5 × 10−3 U; Boehringer Mannheim) was added, and the mixture was incubated at 30°C for 1 min. Digestion was blocked by the addition of 25 μl of stop solution (100 mM EDTA [pH 8.0], 0.4 mg of yeast tRNA/ml), and 50 μl of ice-cold Tris-EDTA (pH 8.0) was added to increase the volume of the mixture. After phenol-chloroform extraction, DNA fragments were ethanol precipitated, resuspended in 10 μl of formamide-dye mixture (1), and separated by electrophoresis on a 6% polyacrylamide sequencing gel. Bands were detected by autoradiography.
RESULTS AND DISCUSSION
Occurrence of active KdgR and PecS proteins in E. chrysanthemi EC16.
The presence of EC16 proteins reacting with anti-KdgR and anti-PecS in immunoblotting experiments was previously reported. These results, as well as the observation of increased expression of the pel genes in the presence of pectic derivatives, suggested that homologues of KdgR and PecS are present in E. chrysanthemi EC16 (12, 22, 29a). However, no direct evidence supporting this hypothesis has been obtained.
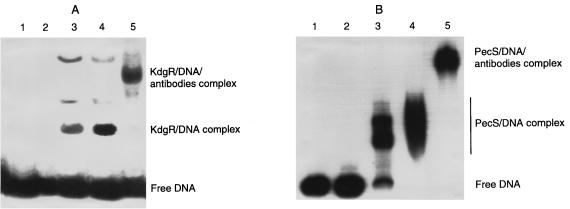
To investigate the existence of active KdgR and PecS proteins in E. chrysanthemi EC16, we performed band shift experiments using protein extracts from E. chrysanthemi EC16 cells grown in LB medium and the regulatory regions of pectinolysis genes from strain EC16 (pelE) or 3937 (pelD and pelA). The protein extracts were enriched in KdgR or PecS by fractional precipitation with ammonium sulfate. Fractions containing KdgR and PecS were identified by Western blot analysis (fractions corresponding to 20 to 40% and 55 to 70% ammonium sulfate saturation, respectively). Gel retardation assays revealed in each case the formation of DNA protein complexes with both EC16 pelE and 3937 pelD or pelA. The complexes obtained with the fraction containing KdgR and PecS could be displaced by addition of specific KdgR and PecS antibodies, respectively (Fig. 1). Using shift-Western blotting experiments in the presence of the specific KdgR or PecS antibody, we detected a band in the major complexes obtained with the fraction corresponding to 20 to 40 or 55 to 70% ammonium sulfate saturation, respectively (data not shown). Thus, E. chrysanthemi EC16 contains active PecS and KdgR proteins. Moreover, binding of the EC16 PecS and KdgR proteins on the 3937 pelA and pelD regulatory regions demonstrated that the regulators PecS and KdgR from E. chrysanthemi EC16 and 3937 are interchangeable.
FIG. 1.
Detection of the E. chrysanthemi EC16 KdgR and PecS proteins by electrophoresis mobility shift assays. The E. chrysanthemi 3937 pelD promoter-operator region (A) was incubated with 0, 2, 10, and 30 μg of E. chrysanthemi EC16 proteins contained in the 20 to 40% ammonium sulfate-saturated fraction (lane 1 to 4) or with 30 μg of E. chrysanthemi EC16 proteins contained in the 20 to 40% ammonium sulfate-saturated fraction followed by the addition of KdgR antibodies (lane 5). The E. chrysanthemi EC16 pelE promoter-operator region (B) was incubated with 0, 2, 10, and 30 μg of E. chrysanthemi EC16 proteins contained in the 55 to 70% ammonium sulfate-saturated fraction (lane 1 to 4) or with 30 μg followed by the addition of PecS antibodies (lane 5).
Interaction of KdgR and CRP with the EC16 pelE regulatory region.
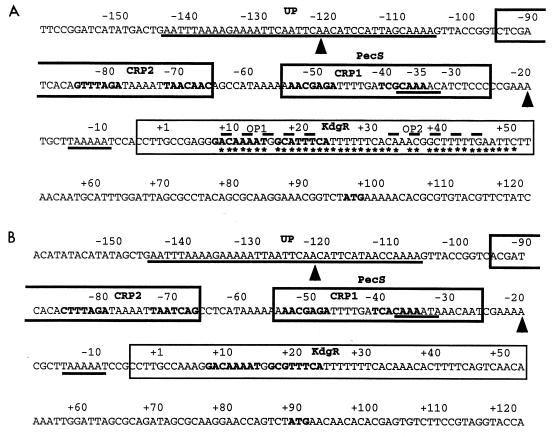
Previous deletion and mutation analyses of the E. chrysanthemi EC16 pelE gene allowed identification of two putative negative regulatory sequences, named operator 1 (OP1) and operator 2 (OP2), and a putative positive operator which was proposed as being a CRP binding site centered at position −43.5 (12) (Fig. 2). Further in vitro DNA-protein interaction studies conducted with the E. chrysanthemi 3937 pelD promoter, which is similar in organization of regulatory sequences to the EC16 pelE gene (13) (Fig. 2), and the purified KdgR or CRP established that the positive operator corresponds to a CRP binding site. Moreover, the region protected by KdgR, as revealed by DNase I footprinting, encompassed OP1 as well as OP2 (20). Noticeable was the fact that only OP1 contained the consensus sequence recognized by KdgR, called a KdgR box (19) (Fig. 2). Therefore, it was of interest to confirm the involvement of the different operators identified in the EC16 pelE gene in the binding of CRP and KdgR. For this purpose, the modified promoters previously used for in vivo analyses (12) were submitted to band shift and footprinting experiments in the presence of the purified regulatory proteins.
FIG. 2.
Organization of the promoter-operator region of the E. chrysanthemi EC16 pelE (A) and 3937 pelD (B) genes. The sequences are numbered from the transcription start site (+1, G base). The regions protected by the various regulatory proteins are delineated based on the examination of both DNA strands. Regions corresponding to the −10/−35 promoter sites and the AT-rich region protected by RNAP, and which include the sequence showing homology with the consensus (AAA[A/T][A/T]T[A/T]TTTT--AAAA) proposed by Ross et al. (26) (putative UP elements), are underlined; arrowheads indicate the ends of the region protected by PecS in DNase I footprinting experiments; the sequences boxed with bold and solid lines indicate the binding sites for cAMP-CRP and KdgR, as defined by DNase I footprinting experiments, respectively; the nucleotides in bold, located in the boxed regions, correspond to the sequences that show homology with the consensus binding site for cAMP-CRP (AATGTGAN6TCACATT [15]) and KdgR ([AATAGAAATC]N[NCATGTTTTCA] [19]), respectively; the regions corresponding to the previously proposed OP1 and OP2 by Gold et al. (12) are overlined by dashed lines; the ATG translation initiation codons are indicated in bold; on the EC16 pelE operator, asterisks indicate the nucleotides contacting the KdgR repressor, as deduced from base removal experiments.
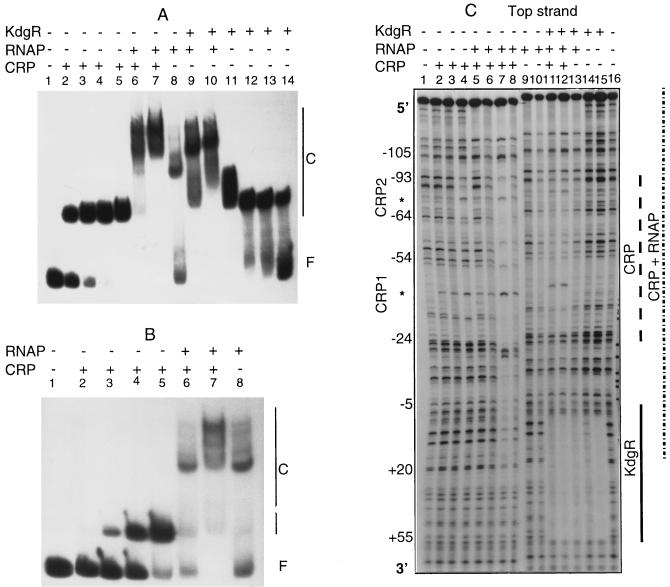
In vitro analysis of the CRP binding on the EC16 pelE promoter revealed that there are in fact two distinct binding sites of different affinities. The major one, centered at position −43.5, corresponds to that identified in the in vivo experiments. The second one, displaying a high degree of degeneration with regard to the consensus proposed for the binding site of the E. coli CRP (15), is centered at position −74.5 (Fig. 2). Binding experiments performed with the pelE promoter modified in the CRP high-affinity site (i.e., deleted of the TGA residues at position −43) showed that CRP is able to bind to the upstream site, but with an eightfold-decreased affinity (Fig. 3A and B). Moreover, DNase I footprinting experiments using this mutated OP1 revealed that protection of the low-affinity CRP binding site requires a concentration of CRP higher than that necessary with the wild-type promoter (400 nM versus 100 nM) (data not shown). Based on these two observations and taking into account the proximity of the two protected regions (Fig. 2) gradually occupied by CRP (Fig. 3C), it is reasonable to assume a cooperative binding of CRP at these two adjacent sites. This cooperativity could compensate for the relatively high degenerency of the two CRP binding sites and thus explain why pelD expression is more highly CRP regulated in vivo than expression of the other pectinolysis genes (23).
FIG. 3.
Binding of CRP, RNAP, and KdgR on the EC16 pelE promoter-operator region. (A) Band shift assays on the parental operator. Lanes 1 to 5, incubation with 0, 10, 20, 50, and 200 nM purified CRP, respectively; lane 6, incubation with 20 nM CRP and 70 nM RNAP; lane 7, incubation with 200 nM CRP and 250 nM RNAP; lane 8, incubation with 250 nM RNAP; lane 9, incubation with 250 nM RNAP and 50 nM KdgR; lane 10, incubation with 250 nM RNAP and 200 nM KdgR; lanes 11 to 14, incubation with 200, 100, 50 and 10 nM purified KdgR, respectively. (B) Band shift assays for the binding of CRP and RNAP on the operator modified in the CRP high-affinity site (i.e., deleted of the ATG residue at position −43). Lanes 1 to 5, incubation with 0, 10, 20, 50, and 200 nM purified CRP, respectively; lane 6, incubation with 20 nM CRP and 70 nM RNAP; lane 7, incubation with 200 nM CRP and 250 nM RNAP; lane 8, incubation with 250 nM RNAP. (C) DNase I footprinting digestions on the parental operator. Lanes 1 and 16, control digestions; lanes 2 to 4, digestion in the presence of 10, 50, and 200 nM purified CRP, respectively; lane 5, digestion in the presence of 10 nM CRP and 70 nM RNAP; lane 6, digestion in the presence of 50 nM CRP and 70 nM RNAP; lane 7, digestion in the presence of 200 nM CRP and RNAP; lane 8, reaction in the presence of 200 nM CRP and 70 nM RNAP; lanes 9 and 10, reaction in the presence of 70 or 250 nM RNAP, respectively; lane 11, digestion in the presence of 250 nM RNAP, 50 nM CRP, and 50 nM KdgR; lane 12, reaction in the presence of 250 nM RNAP, 100 nM CRP, and 100 nM KdgR; lane 13, digestion in the presence of 250 nM RNAP and 50 nM KdgR; lanes 14 and 15, reaction in the presence of 10 and 50 nM KdgR, respectively. The asterisks indicate the DNase I-hypersensitive sites induced by CRP binding, the CRP1 and CRP2 sites are indicated on the left.
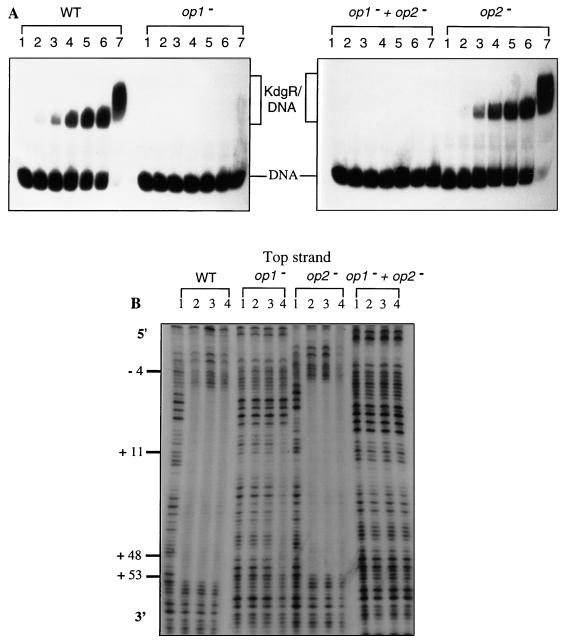
A unique KdgR complex, displaying an affinity (Kd = 0.9 nM) similar to that obtained for the 3937 pelD gene (20), was observed with the native pelE promoter. However, at a high KdgR concentration (200 nM), an overshift of the KdgR-DNA complex was obtained (Fig. 4A). This overshift could result from the binding on this DNA fragment of an additional KdgR dimer. The KdgR binding capacity was strongly decreased when OP1, containing the KdgR consensus, was partially deleted, clearly demonstrating the recognition of this site by KdgR (Fig. 4A). In contrast, a mutation within OP2 did not seem to significantly affect KdgR binding. A double mutation in both operators resulted in the absence of complex formation (Fig. 4A). Accordingly, DNase I footprinting analysis revealed that KdgR protects the region spanning −4 to +52 in the parental operator and in the OP2-modified operator, whereas no protected region could be observed with the operator modified in both OP2 and OP1 sequences. A more limited region (+11 to +47) was protected on the OP1-modified operator when high KdgR concentrations were used (200 nM versus 20 nM for the parental operator) (Fig. 4B). These results suggest that although OP2 is not essential for the efficient binding of KdgR on the pelE promoter, it could favor the binding of this repressor in particular conditions, for example, during the elongation step when the transcriptional machinery overlaps a part of the KdgR box.
FIG. 4.
Interactions between the KdgR repressor and the wild-type E. chrysanthemi EC16 pelE operator (WT) or its derivatives modified in OP1 (op1−), in OP2 (op2−), or in both (op1− + op2−). (A) Band shift assay. The DNA fragment (about 10 fmol), isolated and labeled as described in Materials and Methods, was incubated with 0, 1, 5, 10, 15, 20, or 200 nM purified KdgR (lane 1, 2, 3, 4, 5, 6, or 7, respectively). (B) DNase I footprinting experiments. The purified KdgR protein was added to a final concentration of 20, 75, or 200 nM (lane 2, 3, or 4, respectively). Lanes 1, control digestion. The sequences are numbered with respect to the transcription start site (+1).
To assess if the KdgR protein indeed interacts with nucleotides of OP2, we used the missing-contact chemical approach, whereby individual bases within the DNA helix can be removed by treatment with formic acid or hydrazine (6). The corresponding electrophoresis patterns are shown in Fig. 5A. By comparing the intensity of bands corresponding to complexed DNA (lanes C) and free DNA (lanes F), significant alterations affecting 43 bases essentially distributed in OP1 and OP2 were observed. Quantitative analysis of the involvement of all of these nucleotides in KdgR binding (Fig. 5B) revealed that most of the bases belonging to OP1, particularly those which constitute the AAAA (+11 to +14) and ATTT (+19 to +22) motifs previously proposed as key elements in the KdgR box (19), strongly interact with the KdgR repressor. In addition to the nucleotides of OP1, the motif TTT (+42 to +44), probably belonging to the right half-site of OP2, also strongly interacts with KdgR. In contrast, only a slight interaction could be detected between KdgR and the bases of the OP2 left half-site (+34 to +37). This result, which differs from the model proposed for KdgR binding consisting of a symmetrical interaction with the two half-sites of a KdgR box (19), as observed with OP1, could explain the preferential binding of KdgR on OP1 versus OP2 revealed by band shift and DNase I footprinting experiments. Finally, as previously mentioned for the 3937 pelE operator (19), removal of the thymines (+25 to +30) juxtaposing the OP1 right half-site interfered with KdgR binding. Furthermore, of particular interest was the detection of increased KdgR affinity for the EC16 operator when the guanine bases at position +6, +7, and +16 were modified (Fig. 5). This could result from a higher flexibility of the operator. This finding suggests that KdgR affinity for a DNA fragment depends not only on the homology of its sequence with the KdgR box but also on its relative AT content, which is crucial for flexibility.
FIG. 5.
Analysis of base removal on the coding strand of the E. chrysanthemi EC16 pelE promoter-operator region. (A) DNA molecules were treated with either formic acid to remove G+A or hydrazine to remove C+T and then incubated with KdgR before electrophoresis. DNA isolated from repressor-DNA complexes (lanes C) and DNA that was free of complexes (lanes F) were cleaved by piperidine, electrophoresed, and autoradiographed. (B) Summary of the data obtained by the interference method for the EC16 pelE gene. The magnitude of the effect observed upon removal of a given base is indicated by the size of the bar above (binding interference) or below (enhanced binding) this base; the regions deleted in the modified operators, OP1 and OP2, are underlined.
Based on in vitro DNA-protein interactions, it appears that (i) signals required for the binding of KdgR are contained in the pelE OP1 identified by Gold et al. (12), which perfectly matches the consensus previously determined for the KdgR binding site (19), and (ii) the efficient binding of KdgR on OP2 (12) probably requires prebinding of the KdgR dimer on OP1. This new insight into the KdgR binding on the pelD/E promoter correlates with in vivo observations (12), especially the fact that a double OP1-OP2 mutation displays the same phenotype as a single OP1 mutation.
Two hypotheses could explain the partial constitutivity resulting from a single OP2 mutation. (i) At high KdgR concentrations, OP2 allows for the efficient binding of an additional dimer, giving rise to a high-order KdgR complex responsible for improved repression. Similar examples have been reported for various repressors, including E. coli TyrR and ArgR (10). (ii) Alternatively, OP2 could allow the binding of an unidentified regulator protein acting synergically with KdgR and whose action would strictly depend on the simultaneous presence of KdgR. This isorepressor may be the regulator found by Tsuyumu et al. (30) in E. chrysanthemi extracts and able to bind to the OP2 operator.
Overall, the particular organization of the KdgR binding sites revealed by this work and the possible involvement of a corepressor offer the first explanation for the fact that pelD is more tightly regulated by the KdgR repressor than any other of the genes encoding pectinases in E. chrysanthemi 3937 (20, 24).
The CRP and PecS high-affinity binding sites are superimposed on the pelD/pelE promoter.
Although gel retardation experiments showed that PecS is able to specifically interact with the regulatory region of some virulence genes (i.e., genes encoding pectate lyases, the EGZ cellulase, or OutC, belonging to the specific machinery responsible for pectinase and cellulase secretion), no clear mechanism for PecS activity has yet been proposed (22). Indeed, in most cases (pelA, pelB, pelC, pelE, and pelL of strain 3937), it was impossible to footprint the precise location of the PecS binding site on target genes. In other cases (celZ and outC genes), the transcriptional starts of the regulated genes were not available. To provide further information on PecS control of the pel genes, in vitro interaction experiments were performed with the regulatory regions of the 3937 pelD and EC16 pelE genes.
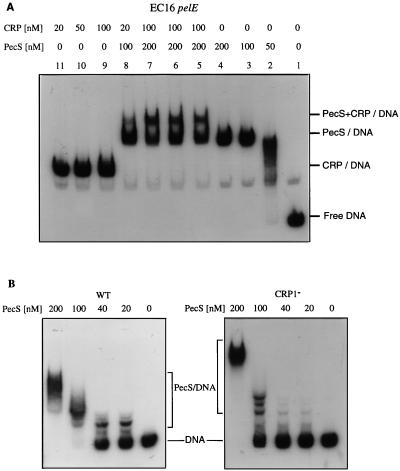
The PecS protein displayed similar affinities (Kd of about 200 nM) for the 3937 pelD and EC16 pelE genes. In cobinding experiments using the concentration determined as saturating for the CRP activator and the PecS repressor, only a slight overshift corresponding to the binding of both proteins was observed (Fig. 6). Most of the probe gives a band migrating at the position of the PecS-DNA complex, suggesting a preferential binding of PecS (Fig. 6A). When the EC16 pelE operator was modified in its high-affinity CRP binding site, its affinity for PecS decreased about fivefold (Fig. 6B). On the contrary, modifications in the regulatory sequences OP1 and OP2 had no significant effect on PecS binding (data not shown). Thus, it appears that the high-affinity binding sites for PecS and CRP are either superimposed or at least overlapping. Moreover, DNase I footprinting analysis revealed a single highly protected region by PecS (−70 to −20) which entirely encompasses the CRP high-affinity binding site (CRP1) (Fig. 7). The partial formation of a ternary complex (CRP-PecS-DNA), detected in the band shift assays (Fig. 6A), could then result from the binding of the PecS and CRP to the sites corresponding to CRP1 and CRP2, respectively. The repressor activity of PecS could then essentially result from its capacity to inhibit the binding of the CRP activator at its high-affinity site, CRP1.
FIG. 6.
Cobinding of PecS and CRP on the EC16 pelE promoter-operator region. (A) Gel shift assays with the wild-type EC16 regulatory region. The concentration of the proteins used are indicated at the top. In lanes 6 and 8, the two proteins were added simultaneously; in lane 5, CRP was incubated 30 min before addition of PecS; in lane 7, PecS was incubated 30 min before addition of CRP. (B) Analysis of the effect of the modification in the CRP binding site 1 (CRP1−) on the PecS binding capacity by electrophoresis gel shift assays. WT, wild type.
FIG. 7.
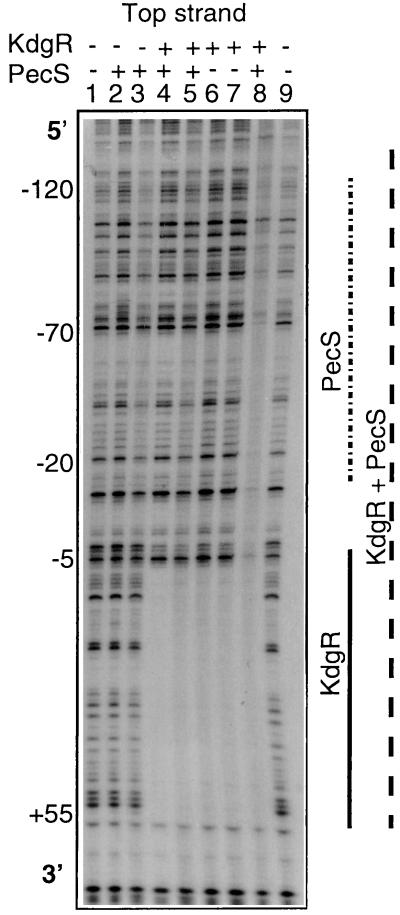
Analysis of the cobinding of KdgR and PecS on the E. chrysanthemi 3937 pelD promoter-operator region by DNase I footprinting. Lanes 1 and 9, control digestions; lanes 2 and 3, reaction in the presence of 50 and 200 nM purified PecS, respectively; lane 4, digestion in the presence of 25 nM PecS and 20 nM KdgR; lane 5, reaction in the presence of 50 nM PecS and 20 nM KdgR; lanes 6 and 7, digestion in the presence of 20 and 200 nM KdgR, respectively; lane 8, reaction in the presence of PecS 200 nM and 100 nM KdgR.
The E. chrysanthemi EC16 pelE and 3937 pelD genes contain a putative UP element.
The synergistic binding of CRP and RNAP has already been described (7). We therefore conducted band shift and DNase I protection assays with the 3937 pelD and the EC16 pelE genes with RNAP in the presence or absence of CRP. Similar results were obtained for both genes. Thus, only data for the EC16 pelE gene are presented (Fig. 3). In the presence of 70 nM RNAP, which corresponds to a subsaturating concentration based on band shift experiments, only the protected region spanning −142 to −104 was detectable. At a saturating concentration of RNAP (250 nM), a second, weaker protected region spanning −50 to +15 appeared (Fig. 3C). This second protected region, which encompasses the predicted ς70 RNAP consensus, became clearer in the presence of both CRP and RNAP, parallel to the protection of the two CRP binding sites (Fig. 3C), thus confirming the synergistic binding of these two proteins. Similar experiments carried out with the EC16 pelE operator containing a mutation in the CRP high-affinity binding site showed that the synergistic binding of CRP and RNAP is conserved but a higher CRP concentration is needed (200 nM versus 20 nM for the parental operator) (Fig. 3A and B). Of particular interest is the detection of the RNAP-protected region spanning −142 to −104. This area, which is particularly rich in A+T residues (up to 80%), could act as a UP (upstream) element. Because of the very low level of pelD/E expression obtained in the absence of the activator protein CRP (12, 23), the involvement of this sequence in pelD/E transcription could not be supported by in vitro evidence. However, the protection of this region by RNAP observed in DNase I footprinting experiments and the remarkable homology of the middle part of the sequence (positions −132 to −116, AAAATTCAATTCAACAT for EC16 pelE and AAAATTAATTCAACATT for 3937 pelD) (Fig. 2) with the consensus (AAA[A/T][A/T]T[A/T]TTTT--AAAA) proposed by Ross et al. (26) argue in favor of the existence of an UP element. Accordingly, previous in vivo deletion studies have shown that the removal of the EC16 pelE region located upstream of position −90 decreased EC16 pelE gene expression by about 30-fold (12). This is comparable to the variation reported when the UP element identified in the E. coli rrnBP1 promoter was deleted (26). Remarkable is the presence of similar AT-rich sequences in the distal promoter regions of the three other major pectinase genes whose transcription is also strictly CRP dependent (E. chrysanthemi 3937 pelB, pelC, and pelE). The UP element could thus be a new feature common to the major pel genes.
Simultaneous binding of the transcriptional machinery with the PecS or KdgR repressor on the EC16 pelE and 3937 pelD genes.
Although the mechanism of action of the KdgR and PecS repressors on pel gene expression was individually investigated in vivo, no detailed description of the simultaneous action of these two regulatory proteins on pel gene expression has been provided. To assess this question, we used a double in vivo and in vitro approach. Quantification of pelD-lacZ transcriptional fusion expression in the parental strain and in kdgR, pecS, and kdgR-pecS mutants revealed that the derepression ratio observed in the two single mutants was slightly lower than that obtained in the double pecS-kdgR mutant (Table 2). This result suggests that the effects of the kdgR and pecS mutations on pelD gene expression are cumulative.
TABLE 2.
Expression of the pelD::lacZ fusion in several backgroundsa
| Strain | Genotype | β-Galactosidase sp act (nmol of product liberated min−1 mg of bacterial dry wt−1) | Derepression ratio |
|---|---|---|---|
| A3485 | Wild type | 13 | |
| A3487 | kdgR | 644 | 50 |
| A3486 | pecS | 42 | 3 |
| A3489 | kdgR pecS | 2,581 | 200 |
Cultures were grown at 30°C for 14 h in M63 minimum medium supplemented with glycerol (2 g liter−1). β-Galactosidase activity reflects expression of the pelD::lacZ fusion. The results reported are averages of five independent experiments, each with a standard deviation of less than 20%.
In vitro band shift assays revealed that both KdgR and PecS could simultaneously interact with the EC16 pelE and 3937 pelD genes (data not shown). To analyze whether PecS and KdgR can interact in a cooperative, independent, or antagonistic way, we performed band shift assays in the presence of these two regulators. The mutual influence of PecS and KdgR on their binding ability was estimated by using control reactions containing only one of the two proteins. Addition of a subsaturating quantity of KdgR and PecS to a solution containing the 3937 pelD or EC16 pelE promoter fragment resulted in three protein-DNA complexes: two corresponding to the KdgR-DNA and PecS-DNA individual complexes and one corresponding to the KdgR-PecS-DNA complex. At a saturating concentration of PecS and KdgR proteins, only a ternary complex was observed (data not shown). Simultaneous binding of both regulators did not modify their respective affinities for 3937 pelD and EC16 pelE promoters. Thus, these results suggest that there is neither cooperativity nor competition in the binding of the two repressors. Besides, DNase I footprinting experiments showed that the protected area observed after the simultaneous incubation of PecS and KdgR roughly corresponds to the addition of the areas protected by PecS and KdgR proteins alone (Fig. 7). However, in the presence of KdgR, the region corresponding to PecS binding is more strongly protected (Fig. 7). This could reflect stabilization of the PecS-DNA interaction by the KdgR regulator and could explain the low basal level of pelD expression due to more effective repression by the KdgR-PecS complex. Furthermore, the dependency between these two repressors which respond to different signals could allow for a gradual but coordinate derepression of pelD/E expression.
As previously shown for the 3937 pelD gene (20), CRP and KdgR are able to bind simultaneously and independently to the EC16 pelE regulatory region. To elucidate the mechanism directing the repression of transcription of these two genes by KdgR and PecS, we performed cobinding experiments involving the proteins of the transcription initiation machinery, CRP and RNAP, and each of the two repressors.
Band shift assays clearly showed that KdgR and RNAP are able to bind simultaneously to the regulatory regions of EC16 pelE (Fig. 3) and 3937 pelD (data not shown). DNase I footprinting experiments revealed that the presence of KdgR does not prevent occupancy of the putative UP region by the RNAP or occupancy of the −142 to −24 region (encompassing the putative UP region and the two CRP binding sites) by the RNAP-CRP complex. However, in the presence of KdgR, the −24 to −5 region encompassing the −10 promoter sequence of the EC16 pelE and 3937 pelD genes is no longer protected (Fig. 3). Thus, KdgR function could prevent positioning of the RNAP on the −10 sequence rather than inhibit binding of transcription complex initiation. This particular role of KdgR would allow for a rapid transcription initiation when derepression occurs since the CRP-RNAP complex already correctly contacts the promoter. In the promoter regions of other pel genes of strain 3937, the region protected by KdgR encompasses the whole RNAP binding sites, i.e., the −10 and −35 boxes (20). In these cases, the function of KdgR seems to be a classical prevention of RNAP binding. This hypothesis is supported by previously reported results (16, 17) which showed that among the E. chrysanthemi 3937 pel genes, pelD is the gene first expressed during plant infection. This particular effect of KdgR on the pelD promoter could contribute to the observed dominance of the PelD isoenzyme in E. chrysanthemi pathogenicity (3, 4).
Results of band shift experiments suggest that PecS and the RNAP can bind simultaneously to the promoter-operator region of the EC16 pelE and 3937 pelD genes, respectively (data not shown). DNase I footprinting assays have revealed that the presence of PecS results in a partial inhibition of CRP-RNAP complex binding to the two CRP binding sites and the RNAP −10/−35 promoter sequences (data not shown). This inhibition could in fact result from the inhibition by PecS of CRP binding, mentioned above. The PecS protein could then act as a moderator of 3937 pelD and EC16 pelE transcription rather than as a strong repressor like KdgR. This hypothesis is supported by the slight effect of a pecS mutation on 3937 pelD expression (3-fold increase) observed in vivo in comparison with that obtained in a kdgR mutant (50-fold increase) (Table 2).
In accordance with its major role in E. chrysanthemi pathogenicity, the pelD-pelE gene was shown to be the most tightly regulated by environmental conditions (14) and the most highly and quickly expressed during plant infection (16, 17). However, the first in vitro data obtained with KdgR and CRP (20) could not be integrated in a coherent model explaining the high-level regulation of pelD. The results of this study with respect to the involvement of KdgR, CRP, PecS, and the RNAP reveal multicomponent control of pelD-pelE transcription. Into this complex model should be further integrated the Pir activator (21), possibly the PecT repressor (9), and putative additional regulators.
ACKNOWLEDGMENTS
This work was supported by grants from the CNRS, the DRED, and the Actions Concertées Coordonnées-Sciences du Vivant 6 from the Ministère de l’Education Nationale, de l’Enseignement Supérieur, de la Recherche et de la Formation Professionnelle, and the Japan Society for Promotion of Sciences.
We are indebted to N. Hugouvieux-Cotte-Pattat, G. Condemine, and A. Buchet for their critical opinions and V. James for reading the manuscript. We thank Y. Rhabé for photograph printing. We also thank J. Robert-Baudouy for her interest and support during this work.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley-Interscience; 1987. [Google Scholar]
- 2.Bachmann B S. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990;54:130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaulieu C, Boccara M, Van Gijsegem F. Pathogenic behavior of pectinase-defective Erwinia chrysanthemi mutants on different plants. Mol Plant-Microbe Interact. 1993;6:197–202. [Google Scholar]
- 4.Boccara M, Diolez A, Rouve M, Kotoujansky A. The role of individual pectate lyases of Erwinia chrysanthemi strain 3937 in pathogenicity on Saintpaulia plants. Physiol Mol Plant Pathol. 1988;33:95–104. [Google Scholar]
- 5.Bradford M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Brunelle A, Schleif R M. Missing contact probing of DNA-protein interactions. Proc Natl Acad Sci USA. 1987;84:6673–6676. doi: 10.1073/pnas.84.19.6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Busby S, Ebright R H. Transcription activation at class II CAP-dependent promoters. Mol Microbiol. 1997;23:853–859. doi: 10.1046/j.1365-2958.1997.2771641.x. [DOI] [PubMed] [Google Scholar]
- 8.Carey J. Gel retardation at low pH resolves trp repressor-DNA complexes for quantitative study. Proc Natl Acad Sci USA. 1988;85:975–979. doi: 10.1073/pnas.85.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castillo A, Nasser W, Condemine G, Reverchon S. The PecT repressor interacts with regulatory regions of pectate lyase genes in Erwinia chrysanthemi. Biochim Biophys Acta. 1998;1442:148–160. doi: 10.1016/s0167-4781(98)00158-4. [DOI] [PubMed] [Google Scholar]
- 10.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collmer A, Keen N T. The role of pectic enzymes in plant pathogenesis. Annu Rev Phytopathol. 1986;24:383–409. [Google Scholar]
- 12.Gold S, Nishio S, Tsuyumu S, Keen N T. Analysis of the pelE promoter in Erwinia chrysanthemi EC16. Mol Plant-Microbe Interact. 1992;5:170–178. [PubMed] [Google Scholar]
- 13.Hugouvieux-Cotte-Pattat N, Condemine G, Nasser W, Reverchon S. Regulation of pectinolysis in Erwinia chrysanthemi. Annu Rev Microbiol. 1996;50:213–257. doi: 10.1146/annurev.micro.50.1.213. [DOI] [PubMed] [Google Scholar]
- 14.Hugouvieux-Cotte-Pattat N, Dominguez H, Robert-Baudouy J. Environmental conditions affect the transcription of the pectinase genes of Erwinia chrysanthemi 3937. J Bacteriol. 1992;174:7807–7818. doi: 10.1128/jb.174.23.7807-7818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kolb A, Busby S, Buc H, Garges S, Adhya S. Transcriptional regulation by cAMP and its receptor protein. Annu Rev Biochem. 1993;62:749–795. doi: 10.1146/annurev.bi.62.070193.003533. [DOI] [PubMed] [Google Scholar]
- 16.Lojkowska E, Dorel C, Reignault P, Hugouvieux-Cotte-Pattat N, Robert-Baudouy J. Use of GUS fusions to study the expression of Erwinia chrysanthemi pectinase genes during infection of potato tubers. Mol Plant-Microbe Interact. 1993;6:488–494. [Google Scholar]
- 17.Masclaux C, Hugouvieux-Cotte-Pattat N, Expert D. Iron is a triggering factor for differential expression of Erwinia chrysanthemi strain 3937 pectate lyases in pathogenesis of African violets. Mol Plant-Microbe Interact. 1996;9:198–205. [Google Scholar]
- 18.Miller J H. Experiment in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 19.Nasser W, Reverchon S, Condemine G, Robert-Baudouy J. Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. J Mol Biol. 1994;236:427–440. doi: 10.1006/jmbi.1994.1155. [DOI] [PubMed] [Google Scholar]
- 20.Nasser W, Robert-Baudouy J, Reverchon S. Antagonistic effect of CRP and KdgR in the transcription control of the Erwinia chrysanthemi pectinolysis genes. Mol Microbiol. 1997;26:1071–1082. doi: 10.1046/j.1365-2958.1997.6472020.x. [DOI] [PubMed] [Google Scholar]
- 21.Nomura K, Nasser W, Kawagishi H, Tsuyumu S. The pir gene of Erwinia chrysanthemi EC16 regulates hyperinduction of pectate lyase virulence genes in response to plant signals. Proc Natl Acad Sci USA. 1998;95:14034–14039. doi: 10.1073/pnas.95.24.14034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Praillet T, Nasser W, Robert-Baudouy J, Reverchon S. Purification and functional characterization of PecS: a regulator of virulence factor synthesis in Erwinia chrysanthemi. Mol Microbiol. 1996;20:391–402. doi: 10.1111/j.1365-2958.1996.tb02626.x. [DOI] [PubMed] [Google Scholar]
- 23.Reverchon S, Expert D, Robert-Baudouy J, Nasser W. The cyclic AMP receptor protein is the main activator of the pectinolysis genes in Erwinia chrysanthemi. J Bacteriol. 1997;179:3500–3508. doi: 10.1128/jb.179.11.3500-3508.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reverchon S, Nasser W, Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol. 1991;5:2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- 25.Reverchon S, Nasser W, Robert-Baudouy J. pecS: a locus controlling pectinase, cellulase and blue pigment production in Erwinia chrysanthemi. Mol Microbiol. 1994;11:1127–1139. doi: 10.1111/j.1365-2958.1994.tb00389.x. [DOI] [PubMed] [Google Scholar]
- 26.Ross W, Aiyar S E, Salomon J, Gourse R L. Escherichia coli promoters with UP elements of different strengths: modular structure of bacterial promoters. J Bacteriol. 1998;180:5375–5383. doi: 10.1128/jb.180.20.5375-5383.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sakakibara H, Watanabe M, Hase T, Sugiyama T. Molecular cloning and characterization of complementary DNA encoding for ferrodoxin-dependent glutamate synthetase in maize leaf. J Biol Chem. 1991;226:2028–2035. [PubMed] [Google Scholar]
- 28.Surgey N, Robert-Baudouy J, Condemine G. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J Bacteriol. 1996;178:1593–1599. doi: 10.1128/jb.178.6.1593-1599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tamaki S J, Gold S, Robeson M, Manulis S, Keen N T. Structure and organization of the pel genes from Erwinia chrysanthemi EC16. J Bacteriol. 1988;170:3468–3478. doi: 10.1128/jb.170.8.3468-3478.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Thomson, N., et al. Unpublished results.
- 30.Tsuyumu S, Miura M, Nishio S. Distinct induction of pectinases as a factor determining host specificity of soft-rotting Erwinia. In: Patil S, Ouchi S, Mills D, Vance C, editors. Molecular strategies of pathogens and host plants. New York, N.Y: Springer-Verlag; 1991. pp. 31–43. [Google Scholar]
- 31.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]