Abstract
Phenotypic analysis using heterologous host systems localized putative Bordetella pertussis ferric alcaligin transport genes and Fur-binding sequences to a 3.8-kb genetic region downstream from the alcR regulator gene. Nucleotide sequencing identified a TonB-dependent receptor family homolog gene, fauA, predicted to encode a polypeptide with high amino acid sequence similarity with known bacterial ferric siderophore receptors. In Escherichia coli, the fauA genes of both B. pertussis and Bordetella bronchiseptica directed the production of a 79-kDa polypeptide, approximating the predicted size of the mature FauA protein. B. bronchiseptica fauA insertion mutant BRM17 was unable to utilize ferric alcaligin, and in complementation analyses ferric alcaligin utilization was restored to this mutant by supplying the wild-type fauA gene in trans. Mutant BRM18, carrying a nonpolar in-frame fauA deletion mutation, was defective in ferric alcaligin utilization and 55Fe-ferric alcaligin uptake and no longer produced a 79-kDa iron-regulated outer membrane protein. In complementation analyses, BRM18 merodiploids bearing the wild-type fauA gene in trans regained ferric alcaligin siderophore transport and utilization functions and produced the 79-kDa protein. Analysis of a plasmid-borne fauA-lacZ operon fusion confirmed that fauA is subject to iron regulation at the transcriptional level and that cis-acting transcriptional control elements mediating fauA iron repressibility reside within the 3.8-kb PstI fauA DNA region. Moreover, expression of the fauA-lacZ fusion gene under iron starvation conditions was shown to be alcR dependent. FauA is a 79-kDa iron-regulated outer membrane receptor protein required for transport and utilization of ferric alcaligin siderophore complexes by Bordetella species.
Under iron-limiting growth conditions, bacteria express high-affinity transport systems to scavenge and assimilate nutritional iron. These transport processes often involve the synthesis and excretion of soluble siderophores that chelate ferric iron and are subsequently conveyed into the bacterial cell by ligand-specific cell surface receptors and permeases (34, 39). To exploit the availability of diverse iron sources that may be present in their environment, some microbes produce transport proteins that enable them to utilize siderophores produced by other microbial species (18, 39). For all gram-negative bacteria examined to date, siderophore-mediated iron uptake is dependent on the activities of TonB, ExbB, and ExbD, which work in conjunction with the cognate outer membrane receptor protein to translocate iron across the bacterial outer membrane against a concentration gradient (37). Further translocation of iron to the cytoplasm is accomplished by a periplasmic binding protein and an ATP binding cassette (ABC)-dependent permease in the cytoplasmic membrane.
Bordetella pertussis and Bordetella bronchiseptica are gram-negative pathogens that inhabit the respiratory mucosae of humans and nonhuman mammals. When nutritional iron is limiting in availability, these organisms produce and utilize the macrocyclic dihydroxamate siderophore alcaligin (16, 38). B. pertussis and B. bronchiseptica are also capable of utilizing iron complexed with the heterologous siderophores enterobactin (8), ferrichrome, and desferrioxamine B (9). In addition to siderophores, the mammalian host-derived molecules lactoferrin (1), transferrin (1, 26), hemin (9), and hemoglobin (40) have been reported as iron sources for these bacteria. The capacity of Bordetella cells to utilize these alternative iron sources implies that they are capable of producing the cognate transporters required for their utilization.
Biosynthesis of the alcaligin siderophore in B. pertussis and B. bronchiseptica requires an ornithine decarboxylase activity that yields the requisite alcaligin precursor putrescine from ornithine (14). Dedicated alcaligin biosynthesis activities are encoded within the iron-regulated alcABCDER operon (29, 30). AlcA, AlcB, and AlcC are required for alcaligin biosynthesis; mutations in alcA, alcB, or alcC render Bordetella cells unable to produce alcaligin (30). The alcA (25, 30), alcB, and alcC gene products have strong primary amino acid sequence similarity with the Escherichia coli aerobactin biosynthesis enzymes IucD, IucB, and IucC, respectively (19, 30). Based on these amino acid sequence similarities and the known structure of alcaligin, it is postulated that AlcA is an oxygenase that catalyzes the hydroxylation of putrescine, and AlcB functions in an acylation step involving succinate. AlcC is similar to IucC aerobactin synthetase and may function in one of the final reactions yielding alcaligin. AlcD was reported to have no significant amino acid sequence similarity with any known proteins, while AlcE has similarity to iron-sulfur-containing dioxygenases (43). Although their role in alcaligin biosynthesis has not been established, the location of these two genes within the alc biosynthesis operon and the prediction that AlcE may function as a dioxygenase suggest that alcD and alcE are also involved in the biosynthesis of alcaligin. The alcR gene is involved in the regulation of both alcaligin biosynthesis (11, 43) and transport of the ferric alcaligin complex (11). AlcR is an AraC-like transcriptional regulator similar to the Pseudomonas aeruginosa PchR pyochelin siderophore biosynthesis and transport regulator protein (27) and the YbtA yersiniabactin siderophore receptor gene regulator of Yersinia pestis (23). The alcR gene is transcribed primarily from the Fur-regulated promoter upstream from alcA as part of the alcABCDER operon and is also transcribed from a secondary Fur-regulated promoter located immediately upstream from alcR (11, 29). Adjacent to alcR is an open reading frame predicted to encode a multidrug efflux pump protein homolog (11, 43). This gene was named bcr in a previous study (43) on the basis of amino acid sequence similarity of the predicted product with the E. coli bicyclomycin resistance efflux protein, Bcr. However, since efflux pumps are generally not substrate specific (42) and the function of the Bordetella protein remains unknown, we presently refer to this gene as orfX.
In the present study, we localized a Bordetella genetic region downstream from orfX that is involved in ferric alcaligin transport and identified the fauA gene encoding the ferric alcaligin outer membrane receptor protein. FauA was localized in the outer membrane fraction prepared from iron-starved B. bronchiseptica cells and was produced in E. coli by using an inducible protein expression system. B. bronchiseptica fauA mutants were unable to transport and utilize ferric alcaligin, and these functions were restored to the mutants by genetic complementation using the cloned B. bronchiseptica or B. pertussis fauA gene. Analysis of a fauA-lacZ operon fusion confirmed that expression of fauA is regulated at the transcriptional level by iron and by the AlcR regulator protein.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
B. bronchiseptica strains used in this study are listed in Table 1. E. coli DH5α (Bethesda Research Laboratories, Gaithersburg, Md.) was used as the host for routine plasmid propagation and DNA cloning procedures and as the donor strain in triparental matings. E. coli SURE (Stratagene, La Jolla, Calif.) was used for propagation of M13 mp18 bacteriophages (41), and E. coli host strain CJ236 (28) was used to produce uracil-containing single-stranded M13 bacteriophage DNA for in vitro site-directed mutagenesis procedures. E. coli S17-1(λpir) (36, 48) was the donor strain for pUT mini-Tn5 lacZ1 suicide plasmids in transposon mutagenesis, and E. coli K38(pGP1-2) was the host strain in bacteriophage T7 RNA polymerase/promoter system protein expression studies (51). E. coli H1717 (fhuE-lacZ aroB) served as the indicator host strain in Fur repressor titration assays (50) used to identify potential Bordetella Fur-binding DNA sequences borne on multicopy plasmids. Pyochelin- and pyoverdin-deficient P. aeruginosa mutant strain IA1 (5) was used as a heterologous host for identification of genes encoding putative Bordetella ferric alcaligin transport and utilization functions.
TABLE 1.
Bordetella strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description | Reference or source |
|---|---|---|
| B. bronchiseptica strains | ||
| B013N | fauA+, nalidixic acid-resistant derivative of swine isolate strain B | 6 |
| BRM17 | fauA1::Kanr mutant derivative of B013N, with an insertion of a 1.2-kb BamHI kanamycin resistance cassette into the BglII restriction site in fauA | This study |
| BRM18 | ΔfauA2 mutant derivative of B013N, with a nonpolar, in-frame deletion of 255 bp of fauA coding sequences | This study |
| BRM11 | ΔalcR1 mutant derivative of B013N | 11 |
| Recombinant plasmids | ||
| pCP1.11 | B. pertussis strain UT25 fauA+ cosmid, source of sequenced fauA DNA, RK2 origin, RK2-Mob+, RK2-Tra−, Tcr | 30 |
| pBRM6 | 20-kb fauA+ SalI genomic DNA fragment of B. bronchiseptica alcC mutant BRM6 cloned in pGEM3Z, Apr, ColE1 origin | 6 |
| p3Z44 | 7-kb B. bronchiseptica fauA+ EcoRI-SalI DNA fragment subcloned from pBRM6 to pGEM3Z, Apr, ColE1 origin | This study |
| p3Z48 | 3.8-kb B. bronchiseptica fauA+ PstI DNA fragment subcloned from p3Z44 to pGEM3Z, Apr, ColE1 origin | This study |
| p3Z49 | As p3Z48 except opposite orientation of the 3.8-kb fauA+ PstI DNA insert fragment | This study |
| pBB24 | 3.8-kb B. bronchiseptica fauA+ PstI DNA fragment subcloned from p3Z44 to pBBR1MCS, Cmr | This study |
| pBB24ΔfauA | As pBB24 except carrying the B. bronchiseptica ΔfauA2 mutant allele on the PstI DNA insert fragment | This study |
| p3Z63 | 3.8-kb B. pertussis UT25 fauA+ PstI DNA fragment subcloned from pCP1.11 to pGEM3Z, Apr, ColE1 origin | This study |
| p3Z69 | As p3Z48 except carrying the B. bronchiseptica ΔfauA2 mutant allele on the PstI DNA insert fragment | This study |
| pBB24/mini-Tn5 lacZ1 | As pBB24 except carrying the B. bronchiseptica φ(fauA′-lacZ+)1 allele generated by mini-Tn5 lacZ1 transposon mutagenesis, Cmr, Kanr | This study |
B. bronchiseptica and E. coli strains were grown in Luria-Bertani (LB) broth or on LB agar plates (44). E. coli strains for bacteriophage T7 RNA polymerase/promoter protein expression experiments were cultured in M9 medium (44) supplemented with 0.2% glucose and 0.01% l-amino acids (minus cysteine and methionine). Defined broth culture medium for Bordetella strains was Stainer-Scholte medium (SS) (49) modified as described elsewhere (46); iron-replete SS contained 36 μM FeSO4, and iron-depleted SS culture medium was prepared by treatment of the medium with Chelex 100 (Bio-Rad, Hercules, Calif.) as described by Armstrong and Clements (6). Growth was monitored as optical density, using a spectrophotometer or a Klett-Summerson colorimeter equipped with a no. 54 filter (Klett Mfg. Co., Long Island City, N.Y.). Antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml; chloramphenicol, 30 μg/ml; gentamicin, 10 μg/ml; kanamycin, 50 μg/ml; and nalidixic acid, 35 μg/ml.
Plasmids and genetic methods.
Plasmid cloning vectors pGEM3Z (Promega, Madison, Wis.), pBBR1MCS (31), and pBluescript II KS+ (Stratagene) were used in the construction of recombinant plasmids. Recombinant Bordetella plasmids used in this study are listed in Table 1. Plasmid pRK2013 (24) was used to provide transfer functions in triparental matings. Plasmid pBSL15 (3) was the source of the kanamycin resistance gene used in the construction of B. bronchiseptica insertion mutant BRM17. Suicide plasmid vectors pEG7 and pEG18.3 (2) were used in allelic exchange procedures in the construction of B. bronchiseptica mutant BRM18, and suicide plasmid pUT mini-Tn5 lacZ1 (20) was used to generate fauA-lacZ transcriptional fusions.
General genetic techniques were performed essentially as described previously (44). Verification of allelic exchange in Bordetella mutants was by Southern hybridization analysis or PCR analysis of genomic DNA samples. DNA probes used in nucleic acid hybridizations were radiolabeled by the random priming method (22) using the Random Primers DNA labeling system (Gibco BRL, Gaithersburg, Md.) and [α-32P]dCTP (ICN Radiochemicals, Irvine, Calif.). The nucleotide sequence of fauA was determined for both DNA strands by using the dideoxy-chain termination method (45) and double-stranded plasmid DNA templates, with nucleotide sequencing services provided by the University of Tennessee Molecular Biology Resource Facility and by the University of Minnesota Microchemical Facility. Management and analysis of nucleotide sequence data used the Lasergene sequence analysis software system for the Macintosh PowerPC computer (DNASTAR, Inc., Madison, Wis.). Database searches and data retrievals were performed by using the BLAST (4) server provided by the National Center for Biotechnology Information at the National Library of Medicine. Multiple amino acid sequence alignments were performed by the CLUSTAL V method, using the MegAlign module of the Lasergene sequence analysis software system (DNASTAR). Putative Bordetella Fur-binding sequences were identified by using the MegAlign software to locate DNA regions of at least 50% identity over a 30-nucleotide search window with the dyad sequence, 5′-GATAATGATAATCATTATC-3′, representing the proposed consensus E. coli Fur-binding site (17, 21). Translated fauA DNA sequences were scanned for the occurrence of PROSITE protein patterns by using the ScanProsite tool of the ExPASy molecular biology server of the Swiss Institute of Bioinformatics. Stained protein gels and protein gel autoradiographs were imaged by using a Hewlett Packard ScanJet 4c color flatbed scanner and Adobe Photoshop LE software for the Power Macintosh computer.
Conjugal transfer of plasmids to Bordetella strains was accomplished as described previously (14). Transformation of Bordetella or E. coli strains by electroporation was performed by using standard methods (Bio-Rad).
The Fur repressor titration assay used to screen B. pertussis cosmid pCP1.11 fauA region subclones for functional Fur repressor-binding sequences was performed as described by Stojiljkovic and coworkers (50), using lactose MacConkey agar supplemented with 30 μM ferrous ammonium sulfate and appropriate antibiotics to select for plasmid maintenance.
Construction of a B. bronchiseptica fauA insertion mutant.
A fauA insertion mutation was constructed by ligation of the 1.2-kb BamHI kanamycin resistance cassette of plasmid pBSL15 into the unique BglII site of the B. bronchiseptica DNA insert fragment of p3Z48. Since this plasmid is a ColE1 replicon and unable to replicate in Bordetella strains, it was used as a suicide plasmid to deliver the mutation to the B. bronchiseptica chromosome by homologous recombination. The plasmid was introduced into B. bronchiseptica wild-type strain B013N by electroporation, and kanamycin-resistant clonal isolates were subsequently screened for loss of the ampicillin resistance plasmid marker by replicate plating on LB agar and LB agar containing ampicillin. Allelic exchange in B. bronchiseptica mutant BRM17 was confirmed by Southern hybridization using fauA-specific and plasmid vector DNA probes.
Construction of a B. bronchiseptica fauA deletion mutant.
A nonpolar fauA mutation was constructed by in vitro site-directed mutagenesis using the Kunkel method (32), and the mutated fauA gene was introduced into wild-type B. bronchiseptica strain B013N by allelic exchange. The purified 3.8-kb fauA PstI DNA fragment of plasmid p3Z49 was ligated with PstI-digested bacteriophage M13 mp18 replicative-form DNA, and the ligation mixture was used to transfect E. coli host strain SURE. Recombinant bacteriophages bearing the 3.8-kb PstI B. bronchiseptica DNA insert fragment were plaque purified and used to infect E. coli host strain CJ236. Bacteriophages were recovered from infected CJ236 culture supernatants by polyethylene glycol-NaCl precipitation, and single-stranded uracil-containing bacteriophage DNA was purified by using standard procedures. The mutagenic 42-mer oligonucleotide fau1 (5′-TTGCCCGAGCCTGTCATCAGGGATTGTGGTGTTTCGCGTGGC-3′) is complementary to two noncontiguous regions of the fauA sense strand, spanning the site of the desired deletion mutation. This primer was phosphorylated using T4 polynucleotide kinase and ATP, annealed to the single-stranded uracil-containing fauA template DNA, and extended by using T7 DNA polymerase and deoxyribonucleotides in the presence of T4 DNA ligase. A portion of the resulting reaction mixture was used to transfect E. coli SURE, and replicative-form DNA samples isolated from plaque-purified bacteriophage progeny were mapped by using the unique EcoRV and BglII restriction endonuclease sites in fauA that flank the desired mutation. Bacteriophage DNA samples from phage clones produced from two independent mutagenesis reactions were sequenced to confirm the desired deletion of fauA coding sequences.
The sacBR positive selection allelic exchange system of Akerley and coworkers (2) was used to cross the fauA deletion mutation carried on the 3.8-kb fauA PstI DNA fragment into the chromosome of wild-type B. bronchiseptica strain B013N. Deletion mutations were verified by PCR analysis of genomic DNA samples prepared from putative ΔfauA mutants, using oligonucleotide primers 5′-GACACTCTCGCCACGCGAAAC-3′ (upstream primer) and 5′-CGTGATGGGCACGGAGATGTC-3′ (downstream primer) flanking the chromosomal deletion mutation. Based on the known fauA nucleotide sequence, these primers were predicted to direct the synthesis of a 430-bp PCR product from the wild-type fauA region and a 175-bp PCR product from ΔfauA mutants. Agarose gel electrophoresis was used to determine the sizes of PCR reaction products corresponding to the presence of wild-type or ΔfauA mutant alleles in the bacterial chromosome.
Measurement of Bordetella siderophore activity.
The chrome azurol S universal siderophore detection assay (47) was used to quantitate alcaligin production by Bordetella cells grown in iron-depleted SS as described previously (6, 16) to monitor iron nutritional status of bacteria. Siderophore assays were performed in triplicate.
Ferric alcaligin utilization bioassays.
Growth stimulation of B. bronchiseptica strains and siderophore-deficient P. aeruginosa mutant strain IA1 by ferric alcaligin siderophore complexes was measured by quantitative ferric alcaligin utilization bioassays using iron-restricted agar plates containing ethylenediaminedi-[(o-hydroxyphenyl)acetic acid] (EDDA) at a concentration of 100 μg/ml, performed as described previously (11, 16). Alcaligin siderophore was purified from spent Bordetella culture supernatants as described by Brickman and coworkers (16) and used at aqueous concentrations of 250, 125, and 63 μg/ml.
Ferric alcaligin transport assays.
B. bronchiseptica cells were assayed for the ability to bind and transport ferric alcaligin by measuring 55Fe(III) accumulation by the method of Langman et al. (33) as modified by Brickman and coworkers (16), using 55Fe(III) supplied as 5 μM alcaligin–3.3 μM 55Fe(III) complexes (3:2 molar ratio, pH 8.0). Cell-associated 55Fe counts per minute were measured by scintillation counting.
Production of plasmid-encoded FauA proteins in E. coli.
Plasmid-encoded FauA proteins were conditionally produced in E. coli K38(pGP1-2) by using the bacteriophage T7 polymerase/promoter system of Tabor and Richardson (51). After induction of fauA expression at mid-logarithmic growth phase by incubation at 42°C, approximately 5 × 107 cells (0.1 units of optical density at 600 nm) were harvested by centrifugation, solubilized in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer at 100°C for 7 min and analyzed by SDS-PAGE on 10% polyacrylamide gels. Proteins were visualized by intrinsic radiolabeling using Tran35S-label (ICN Biochemicals, Inc.), SDS-PAGE, and autoradiography of dried electrophoretic gels.
Cell fractionation and SDS-PAGE.
Bordetella cells from iron-replete and iron-depleted SS cultures were harvested by centrifugation at late-logarithmic growth phase, resuspended in 50 mM HEPES buffer (pH 7.4), and lysed by passage through a French pressure cell (American Instrument Company, Silver Spring, Md.). The insoluble cell fraction sediment obtained by ultracentrifugation was extracted with Triton X-100 detergent as described previously (46), and the outer membrane protein-enriched detergent-insoluble fraction was resuspended in 50 mM HEPES buffer (pH 7.4) and solubilized in SDS-PAGE sample buffer at 100°C for 7 min. Proteins were resolved by SDS-PAGE on 7.5 or 10% polyacrylamide gels containing 0.5 M urea as described previously (46), with approximately 20 μg of protein applied to each lane. Protein bands were visualized by staining with Coomassie blue dye.
Construction and analysis of a fauA-lacZ transcriptional fusion plasmid.
Transposon mini-Tn5 lacZ1 (20) was delivered to B. bronchiseptica B013N carrying fauA plasmid pBB24 by conjugal transfer of suicide plasmid pUT mini-Tn5 lacZ1 from donor strain E. coli S17-1(λpir). After 5 h of mating on an LB agar plate surface, bacterial growth was harvested and total plasmid DNA was prepared by the alkaline lysis method (12). Plasmid DNA pools were used to transform E. coli DH5α to kanamycin and chloramphenicol resistance, and mutated pBB24 plasmids subsequently isolated from transformants were screened by restriction endonuclease mapping for insertions in fauA. Plasmids carrying fauA insertions in the fauA sense orientation were transferred to wild-type B. bronchiseptica strain B013N and alcR mutant strain BRM11 by conjugation. Transconjugants were assayed for β-galactosidase activity by the method of Miller (35) as modified by Brickman et al. (15) after culture in iron-replete or in iron-depleted SS medium.
Nucleotide sequence accession number.
The GenBank accession number assigned to the B. pertussis UT25 fauA gene is AF135154.
RESULTS
Preliminary identification of putative ferric alcaligin transport and utilization genes and Fur-binding sequences by phenotypic analysis using heterologous host systems.
Since microbial siderophore system genes are usually clustered on the chromosome, it was hypothesized that alcaligin transport genes may reside near known alcaligin biosynthesis genes on B. pertussis cosmid pCP1.11 or on the adjacent overlapping cosmids described previously (30). Prior alcaligin biosynthesis studies using a heterologous host system established that pyochelin- and pyoverdin-deficient P. aeruginosa mutant strain IA1 (5) carrying pCP1.11 produced authentic alcaligin (10). In parallel studies using mutant strain IA1 as a host system for expression of potential Bordetella alcaligin transport and utilization genes, analysis of subcloned segments of pCP1.11 identified an approximately 4-kb PstI genetic region downstream from alcR (Fig. 1) that conferred a positive growth stimulation phenotype to IA1 in bioassays using purified alcaligin as an iron source (data not shown). When cloned to high-copy-number plasmids, the same B. pertussis genetic region had strong in vivo Fur repressor-binding activity by the Fur repressor titration assay in E. coli (data not shown), consistent with the presence of previously unidentified functional Fur repressor-binding sequences. Based on these preliminary data suggesting the existence of previously unknown ferric alcaligin transport and utilization genes and functional Fur-binding sequences downstream from alcR, the corresponding DNA region was further analyzed by nucleotide sequencing.
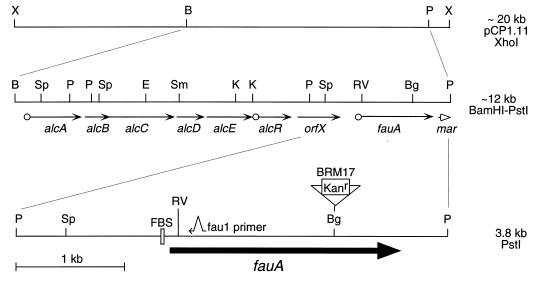
FIG. 1.
Spatial organization of known Bordetella alcaligin siderophore system genes. The 20-kb XhoI insert DNA fragment of B. pertussis cosmid pCP1.11 is depicted at the top. The enlarged map in the center represents an approximately 12-kb BamHI-PstI subregion of pCP1.11; arrows indicate the direction of transcription and genetic limits of all known genes within the Bordetella alcaligin gene cluster, and open circles represent known or hypothetical Fur-regulated promoter-operator regions. The detailed lowermost map is enlarged to represent the 3.8-kb PstI DNA subregion encoding fauA, and a 1-kb scale bar is shown. FBS indicates the position of the computer-predicted Fur-binding site immediately upstream from the start of the fauA coding sequences. The approximate position of sequences complementary to mutagenic primer fau1 used in the construction of ΔfauA mutant BRM18 is indicated by the small arrow. The triangle indicates the position of the BglII site of insertion of the kanamycin resistance cassette (Kanr) in fauA mutant BRM17. Abbreviations: X, XhoI; B, BamHI; E, EcoRI; Sp, SphI; P, PstI; Sm, SmaI; K, KpnI; RV, EcoRV; Bg, BglII.
Nucleotide sequencing of putative ferric alcaligin transport and utilization genes.
Complete nucleotide sequence analysis of an approximately 3-kb genetic region downstream from alcR identified a large open reading frame encoding a putative B. pertussis TonB-dependent receptor family homolog. In BLASTP searches, the GenBank database sequences producing the six highest-scoring alignments with the deduced Bordetella receptor homolog (GenBank accession no. AF135154) were ferric siderophore receptor proteins. These included the ferripyoverdin receptor FpvA of P. aeruginosa (GenBank accession no. U07359), the Pseudomonas putida ferric pseudobactin 358 receptor PupA (GenBank accession no. P25184), the E. coli FhuE outer membrane receptor for ferric coprogen, ferric ferrioxamine B, and ferric rhodotorulic acid (GenBank accession no. P16869), the P. putida PupB ferric pseudobactin BN7/BN8 receptor (GenBank accession no. P38047), P. aeruginosa ferric pyochelin receptor FptA (GenBank accession no. P42512), and PbuA, the ferric pseudobactin M114 receptor of Pseudomonas sp. strain M114 (GenBank accession no. Q08017). Overall percent similarity between the Bordetella ferric siderophore receptor homolog and these highest-scoring database retrievals ranged from 33.1% for FpvA to 27.0% for PbuA at the primary amino acid sequence level.
The Bordetella ferric siderophore receptor homolog gene was tentatively named fauA, for ferric alcaligin uptake gene. As deduced from the nucleotide sequence, the FauA precursor protein is a 734-amino-acid polypeptide with a calculated molecular mass of approximately 81.6 kDa. A PROSITE database search revealed that at C-terminal amino acid positions 717 to 734, FauA exhibits the TonB-dependent receptor protein signature 2 (PROSITE PDOC00354) that is characteristic of most other members of the TonB-dependent receptor family (Fig. 2). Consistent with the earlier identification of functional Fur-binding DNA sequences in the fauA genetic region by Fur repressor titration assay, a computer-predicted Fur repressor-binding site, 5′-AAAAAAGATAATTCCTATT-3′, was identified at nucleotide positions 35 to 50 upstream from the start of the fauA coding sequence (Fig. 1).
FIG. 2.
Partial amino acid sequence alignments of FauA with TonB-dependent bacterial siderophore receptor proteins of high similarity. (A) Multiple sequence alignments of the FauA C terminus with C termini of bacterial siderophore receptor proteins producing the six highest-scoring alignments with FauA in GenBank database searches (GenBank accession numbers: P. aeruginosa FpvA, U07359; P. putida PupA, P25184; E. coli FhuE, P16869; P. putida PupB, P38047; P. aeruginosa FptA, P42512; Pseudomonas sp. strain M114 PbuA, Q08017). With the exceptions of residues Q717 and Y734 of FauA and T812 of FpvA (boxed), all amino acid residues shown are consistent with the PROSITE database TonB-dependent receptor protein signature 2 (PROSITE PDOC00354) consensus pattern (B) shared by most members of the TonB-dependent receptor family.
Beginning approximately 300 bp downstream from the end of the fauA coding region and extending to the PstI terminus of the sequenced DNA region (Fig. 1) is a partial open reading frame encoding a polypeptide with primary amino acid sequence similarity to multiple antibiotic resistance protein MarC of E. coli (GenBank accession no. P31123) and Salmonella typhimurium (GenBank accession no. Q56068). MarC proteins are members of SwissProt uncharacterized protein family UPF0056 of integral membrane proteins. This Bordetella marC-like gene, tentatively named mar, is transcribed in the same polarity as fauA, but it is not yet known whether it is operonic with fauA or whether it encodes any function related to iron metabolism.
Construction and analysis of B. bronchiseptica fauA insertion mutant BRM17.
To preliminarily address the hypothesized role of fauA in transport and utilization of ferric alcaligin, fauA insertion mutant BRM17 was constructed by insertion of a kanamycin resistance cassette at the unique BglII site of fauA (Fig. 1). In growth stimulation bioassays, BRM17 was fully defective in utilization of ferric alcaligin as an iron source (Table 2). Genetic complementation of BRM17 by the wild-type 3.8-kb B. bronchiseptica PstI fauA DNA region as plasmid pBB24 resulted in restoration of ferric alcaligin utilization. In these complementation analyses, BRM17 carrying multicopy fauA plasmids reproducibly displayed larger growth haloes than the wild-type indicator strain, presumably because of FauA overproduction and enhanced ferric alcaligin utilization. Complementation of the BRM17 ferric alcaligin utilization defect by B. bronchiseptica plasmids carrying the 3.8-kb PstI fauA DNA region was independent of insert DNA orientation with respect to the plasmid cloning vector (data not shown), suggesting that fauA was expressed from a natural Bordetella promoter borne on that plasmid construct. In addition, a B. pertussis fauA plasmid carrying the 3.8-kb PstI fauA DNA region (Fig. 1) was equally capable of restoring ferric alcaligin utilization to BRM17 (data not shown), indicating that the B. pertussis fauA gene was fully functional in B. bronchiseptica by this growth stimulation bioassay method.
TABLE 2.
Utilization of ferric alcaligin by wild-type and mutant B. bronchiseptica strainsa
| Indicator strain | Growth zone (mm) surrounding wells containing purified alcaligin at concn of:
|
||
|---|---|---|---|
| 63 μg/ml | 125 μg/ml | 250 μg/ml | |
| B013N (wild type) | 16 | 20 | 24 |
| BRM17 | — | — | — |
| BRM17(pBBR1MCSb) | — | — | — |
| BRM17(pBB24ΔfauA) | — | — | — |
| BRM17(pBB24) | 17 | 22 | 27 |
| BRM18 | — | — | — |
| BRM18(pBBR1MCS) | — | — | — |
| BRM18(pBB24) | 18 | 23 | 28 |
Aqueous alcaligin solution (50 μl/well) at the indicated concentration was added to 6-mm-diameter wells in EDDA-containing iron-restricted agar medium seeded with indicator strains, and diameters of growth zones were measured after 18 h incubation at 37°C. No growth occurred in the absence of added alcaligin siderophore. —, no detectable growth stimulation.
Plasmid vector control.
Construction of a B. bronchiseptica fauA deletion mutant.
A specific fauA deletion mutation was constructed in vitro, and the mutated B. bronchiseptica fauA gene was crossed into wild-type B. bronchiseptica strain B013N by allelic exchange as described in Materials and Methods. The mutation was designed to generate an in-frame deletion removing 255 bp (85 codons) of fauA coding sequences that specify a FauA segment (amino acid residues 89 to 173 in the FauA precursor protein) that is highly conserved among known TonB-dependent outer membrane siderophore complex receptors; thus, the deleted sequences were hypothesized to encode a region of functional importance in the FauA protein. Further, an in-frame deletion was desired in order to avoid potential polarity effects of a mutation on unknown, potentially cotranscribed downstream genes that may also be involved in ferric alcaligin transport or utilization.
After in vitro mutagenesis of cloned fauA sequences, bacteriophage DNA from clones produced from two independent mutagenesis reactions was sequenced, and both were found to carry the desired deletion mutation. To preliminarily assess the phenotypic effect of the mutation, the mutated fauA PstI fragment was subcloned to the broad-host-range plasmid vector pBBR1MCS to produce pBB24ΔfauA, and this plasmid was introduced into B. bronchiseptica fauA insertion mutant BRM17, which is defective in ferric alcaligin utilization. Unlike the wild-type fauA plasmid equivalent pBB24, mutated plasmid pBB24ΔfauA was unable to complement the growth defect of BRM17 when supplied with ferric alcaligin in growth stimulation bioassays (Table 2). These data were consistent with the prediction that the fauA deletion mutation would ultimately result in a functionally defective FauA protein once transferred to the B. bronchiseptica chromosome by allelic exchange.
A positive selection allelic exchange system (2) was used to cross the fauA deletion mutation into the chromosome of wild-type B. bronchiseptica strain B013N, and the allelic exchange was confirmed by genomic PCR analysis. Electrophoretic analysis of PCR products generated from genomic DNA samples prepared from four putative B013N ΔfauA mutants by using oligonucleotide primers flanking the deletion junction revealed that three of the four putative mutants carried only the ΔfauA allele (data not shown). The fourth putative B013N ΔfauA mutant carried only a wild-type fauA allele, yielding a single PCR product indistinguishable from that produced from a wild-type control genomic DNA template.
FauA requirement for growth stimulation by ferric alcaligin siderophore.
The three B013N ΔfauA mutants identified by genomic PCR analysis were assayed for growth stimulation by ferric alcaligin under iron starvation culture conditions using bioassays conducted in parallel with the wild-type parent strain B013N. Consistent with the results of physical mapping of the chromosomal fauA regions by genomic PCR analysis, the deletion mutants examined in growth stimulation bioassays were fully defective in ferric alcaligin transport or utilization compared with the wild-type control strain. One B013N ΔfauA mutant chosen for further study was designated BRM18 (Table 1). In complementation analyses, growth stimulation by ferric alcaligin in bioassays was restored to BRM18 by the wild-type B. bronchiseptica fauA gene supplied as plasmid pBB24 (Table 2). As in BRM17 complementation analyses, growth stimulation zones for the complemented BRM18 ΔfauA mutant were consistently larger than those produced with wild-type indicator cells. BRM18 growth stimulation by ferric alcaligin was also restored by plasmids carrying the analogous wild-type 3.8-kb PstI fauA genetic region of B. pertussis strain UT25 (Fig. 1), indicating that the fauA alleles of the two different Bordetella species are interchangeable in this functional assay (data not shown). These growth stimulation bioassays confirmed the predicted correlation between the deletion mutation detected by genomic PCR mapping and a defective ferric alcaligin transport or utilization phenotype. Genetic complementation results confirmed that ferric alcaligin utilization could be fully restored to BRM18 by the wild-type fauA gene of either B. pertussis or B. bronchiseptica in trans.
FauA requirement for ferric alcaligin-mediated iron transport.
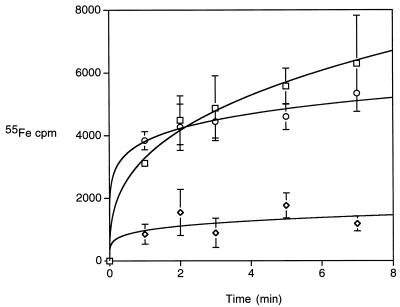
B. bronchiseptica ΔfauA mutant BRM18 and the wild-type parent strain B013N were assayed for the ability to transport iron supplied as 55Fe(III)-alcaligin complexes. Iron transport by iron-starved Bordetella cells was monitored as the accumulation of cell-associated 55Fe (Fig. 3). Compared with wild-type B013N iron transport activity, ΔfauA mutant strain BRM18 carrying the pBBR1MCS control plasmid vector was grossly defective in 55Fe uptake. Consistent with the results of the growth stimulation bioassays, B. bronchiseptica fauA plasmid pBB24 complemented the BRM18 ferric alcaligin transport defect, restoring accumulation of cell-associated 55Fe to wild-type levels. Although cell-associated 55Fe levels for the complemented mutant are not significantly different from the wild-type level, mathematical curve fitting indicated that the initial 55Fe uptake rate for the complemented mutant was greater than the wild-type rate, possibly reflecting a higher receptor density at the cell surface.
FIG. 3.
Bordetella 55Fe-ferric alcaligin transport assays. The rate of uptake of the 55Fe label of 55Fe-ferric alcaligin by iron-starved B. bronchiseptica wild-type strain B013N (squares), ΔfauA mutant BRM18 (diamonds), and ΔfauA mutant BRM18 carrying wild-type fauA plasmid pBB24 (circles), using an alcaligin concentration of 5 μM, together with 3.3 μM 55Fe(III) is shown over a 7-min period of time. Mean 55Fe counts per minute ± standard deviations (n = 3) are plotted as a function of time.
Bacteriophage T7 promoter-directed FauA protein production.
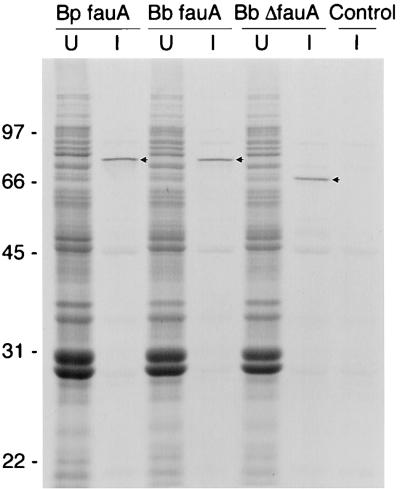
Protein production directed by the 3.8-kb Bordetella PstI fauA DNA region was examined by using a bacteriophage T7 RNA polymerase/promoter expression system in E. coli (Fig. 4). In the fauA sense orientation with respect to the plasmid vector T7 gene 10 promoter, both B. pertussis and B. bronchiseptica 3.8-kb PstI DNA regions directed the production of a single strongly radiolabeled polypeptide migrating at an apparent molecular mass of approximately 79 kDa. These products approximate the predicted size (ca. 77.7 kDa) of the mature FauA protein, based on amino acid sequence comparisons of FauA with processed forms of similar ferric siderophore receptor proteins. The cloned B. bronchiseptica ΔfauA region directed the production of a smaller polypeptide with an apparent molecular mass of approximately 69 kDa, consistent with the loss of an 85-amino-acid segment resulting from the ΔfauA in-frame deletion.
FIG. 4.
Production of Bordetella FauA polypeptides, using a bacteriophage T7 RNA polymerase/promoter expression system in E. coli. Intrinsically labeled plasmid-encoded FauA proteins were resolved by SDS-PAGE and visualized by autoradiography. Whole-cell lysates of E. coli K38(pGP1-2) containing p3Z63 (B. pertussis 3.8-kb PstI fauA fragment; Bp fauA), p3Z48 (B. bronchiseptica 3.8-kb PstI fauA fragment; Bb fauA), p3Z69 (B. bronchiseptica 3.8-kb PstI ΔfauA fragment deleted of the 255-bp internal fauA region; Bb ΔfauA), and pGEM3Z (plasmid vector control; Control) are shown. U, uninduced; I, induced for expression of T7 promoter-directed genes by temperature shift and in the presence of rifampin. Positions of marker proteins are given along with their apparent molecular masses in kilodaltons. The radiolabeled FauA products are indicated by arrows.
FauA protein production in B. bronchiseptica.
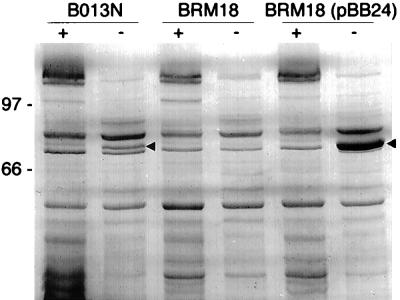
In this and previous SDS-PAGE analyses (11, 13), a polypeptide with an apparent molecular mass of approximately 79 kDa was detected in whole-cell lysates and outer membrane-enriched fractions of B013N cultured under iron-depleted conditions. The 79-kDa iron-regulated outer membrane protein was not present at detectable levels in samples prepared from B. bronchiseptica ΔfauA mutant BRM18 but was very strongly produced under iron-depleted culture conditions by BRM18 carrying wild-type fauA plasmid pBB24 (Fig. 5). Production of this 79-kDa iron-regulated outer membrane protein correlates with the ability of fauA plasmid pBB24 to restore ferric alcaligin growth stimulation and 55Fe-alcaligin uptake to mutant BRM18. The protein has an apparent molecular mass consistent with predictions based on the fauA nucleotide sequence and appears indistinguishable from wild-type FauA proteins produced in T7 protein expression experiments in E. coli. Elevated levels of FauA detected in outer membrane preparations of BRM18 carrying wild-type fauA plasmid pBB24 correlate with enhanced ferric alcaligin growth stimulation observed in bioassays for fauA mutants BRM17 and BRM18 complemented with this plasmid.
FIG. 5.
SDS-PAGE analysis of iron-regulated FauA outer membrane protein production in B. bronchiseptica. FauA outer membrane protein production by wild-type B. bronchiseptica B013N, ΔfauA mutant BRM18, and ΔfauA mutant BRM18 carrying wild-type fauA plasmid pBB24 was analyzed by SDS-PAGE. Outer membrane proteins were visualized by staining gels with Coomassie blue dye. +, outer membrane proteins from iron-replete cells; −, outer membrane proteins from iron-depleted cells. Arrowheads indicate positions of the 79-kDa FauA protein in the wild-type B013N and BRM18(pBB24) outer membrane samples. Positions of marker proteins are given along with their apparent molecular masses in kilodaltons.
Iron regulation and alcR-dependent expression of a fauA-lacZ transcriptional fusion gene.
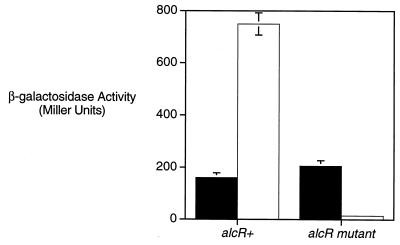
Fifteen mini-Tn5 lacZ1 transposon insertions were mapped to fauA sequences in plasmid pBB24 by restriction endonuclease mapping. One insertion chosen for further analysis mapped near the center of fauA, approximately 900 bp downstream from the start of the fauA coding sequence. The orientation of this transposon insertion in the fauA sense placed lacZ reporter gene transcription under the control of any putative fauA regulatory sequences present on the 3.8-kb PstI fauA insert DNA fragment of pBB24. The fusion plasmid, pBB24/mini-Tn5 lacZ1, was transferred to wild-type B. bronchiseptica strain B013N and isogenic alcR mutant strain BRM11 by conjugation, and fauA transcriptional activity was monitored as β-galactosidase reporter gene activity of cells cultured under iron-replete versus iron-depleted conditions in SS defined medium. In this plasmid context in wild-type B. bronchiseptica strain B013N, fauA transcriptional activity was elevated nearly fivefold by iron starvation (Fig. 6). Analysis of the fauA-lacZ fusion gene in alcR mutant strain BRM11 revealed that fauA transcription was dependent on the regulatory gene alcR; expression of the fauA fusion gene in the alcR mutant was drastically reduced. The apparent requirement of alcR for fauA expression is consistent with the previously observed loss of production of an iron-repressible 79-kDa membrane protein by B. bronchiseptica alcR mutants BRM10 and BRM11 (11). The alcR mutant BRM10, which is also defective in alcaligin production, was identified in a screening for ferric alcaligin transport mutants on the basis of a ferric alcaligin utilization-defective phenotype. These data indicate that fauA transcription is iron regulated and alcR dependent and that cis-acting transcriptional control elements are active within the pBB24 plasmid-borne fauA DNA region.
FIG. 6.
Iron regulation and alcR-dependent expression of a fauA-lacZ transcriptional fusion gene. Transcriptional activity of a fauA-lacZ fusion gene carried as plasmid pBB24/mini-Tn5 lacZ1 in wild-type B. bronchiseptica B013N (alcR+) or isogenic alcR mutant strain BRM11 (alcR mutant) was assessed by measuring β-galactosidase activity of bacteria cultured under iron-replete (■) and iron-depleted (□) conditions. Mean β-galactosidase activities ± standard deviations (n = 3) are shown in Miller units.
DISCUSSION
In our previous report of the purification and spectroscopic analysis of alcaligin produced by B. pertussis and B. bronchiseptica, evidence for biological activity of the purified alcaligin siderophore was obtained by using growth stimulation bioassays and quantitative ferric alcaligin transport assays with Bordetella indicator strains (16). Uptake rates and saturability of iron uptake observed in quantitative ferric alcaligin transport assays were consistent with the involvement of a high-affinity receptor-mediated ferric alcaligin transport system and provided the first direct evidence of iron delivery to Bordetella cells mediated by the native alcaligin siderophore. Despite the demonstration of rapid and saturable transport of iron supplied as ferric alcaligin, the specific outer membrane receptor for ferric alcaligin or other potential alcaligin transport proteins remained unidentified.
The first Bordetella ferric siderophore receptor identified was the B. pertussis BfeA receptor for the heterologous siderophore ferric enterobactin (8), found in a screen of E. coli to detect iron-repressible genes encoding exported proteins. BfeA was found to share a high degree of similarity with the ferric enterobactin receptors FepA of E. coli and PfeA of P. aeruginosa, and growth stimulation by ferric enterobactin was shown to be defective in B. pertussis and B. bronchiseptica bfeA mutants. Later, using the same E. coli screening approach, the putative ferric siderophore receptor gene bfrA was identified in B. bronchiseptica (9). The bfrA gene was repressible by Fur and iron and was predicted to encode an 80-kDa outer membrane protein with strong similarity with members of the TonB-dependent outer membrane receptor family, especially with Cir of E. coli and IrgA of Vibrio cholerae, and with three known ferric enterobactin receptors. In phenotypic analyses, Bordetella bfrA mutants were found to be unaffected in their ability to utilize ferric alcaligin, ferric enterobactin, and 2,3-dihydroxybenzoylserine. 2,3-Dihydroxybenzylserine utilization was shown to be dependent on the Bordetella ferric enterobactin receptor BfeA. Ferrichrome, desferrioxamine B, and hemin were shown to stimulate Bordetella growth, albeit by mechanisms functioning independently of both the BfeA and BfrA proteins in B. bronchiseptica and B. pertussis. The specificity of bfrA and its potential role in iron transport remain unknown. Two additional Fur-regulated ferric siderophore receptor homologs of unknown specificity, bfrB and bfrC, have since been identified in B. bronchiseptica (7). B. bronchiseptica and B. pertussis bfrB and bfrC mutants and a B. bronchiseptica bfrB bfrC double mutant had no phenotypic defects related to growth stimulation by any known Bordetella iron sources. Since phenotypic characterization of ferric siderophore receptor homolog mutants determined that BfrA, BfrB, and BfrC are not involved in the utilization of any known Bordetella iron sources, these data suggest that the iron-scavenging potential of Bordetella species may include additional, unidentified iron sources. With the exception of BfeA, it has proven difficult to predict the specificity of the putative Bordetella siderophore receptors based on nucleotide sequence information. Recent analysis of B. pertussis and B. bronchiseptica tonB mutants confirmed that as with other characterized gram-negative bacteria, TonB function is required for specific utilization of all known iron sources by these species (40).
In this study, the hypothesis that previously unknown Bordetella ferric alcaligin siderophore transport genes might be clustered near known alcaligin biosynthesis genes was tested by screening for B. pertussis genes encoding ferric alcaligin utilization functions in a heterologous host system. We identified a 3.8-kb PstI genetic region mapping immediately downstream from the known alcaligin system gene alcR that conferred a strong positive ferric alcaligin growth stimulation phenotype to siderophore-deficient P. aeruginosa IA1, an indicator strain that is normally incapable of ferric alcaligin utilization. The ability of the fauA gene alone to confer a ferric alcaligin utilization phenotype to IA1 implies that IA1 is capable of supplying any additional transport and utilization activities required for growth stimulation by ferric alcaligin. Nucleotide sequencing of the region identified a putative ferric siderophore receptor gene, fauA, with deduced primary amino acid sequence similarity with known members of the TonB-dependent receptor family. The fauA DNA regions of B. pertussis UT25 and B. bronchiseptica B013N each directed the production of a 79-kDa protein in E. coli, approximating the predicted size of the mature FauA protein.
Functional Fur repressor-binding sequences were detected on the 3.8-kb PstI DNA fragment by the Fur repressor titration assay in E. coli. Analysis of a plasmid-borne fauA-lacZ operon fusion in wild-type B. bronchiseptica provided evidence of iron regulation of fauA at the transcriptional level, mediated by cis-acting control elements residing within the 3.8-kb PstI fauA DNA region. By using an alcR mutant host strain, transcription of the fauA-lacZ fusion gene was also shown to be alcR dependent, an effect that is consistent with the previous observation of alcR-dependent production of an unidentified 79-kDa membrane protein, presumably FauA, in phenotypic analysis of B. bronchiseptica alcR mutants (11). A previous report also described a 79-kDa protein that was normally iron regulated but constitutively produced by a B. bronchiseptica fur mutant (13). Complete genetic characterization of fauA regulation will require further analysis.
Presumptive evidence for the requisite role of fauA in ferric alcaligin utilization was obtained by phenotypic analysis of B. bronchiseptica fauA insertion mutant BRM17. BRM17 was incapable of utilizing ferric alcaligin in growth stimulation bioassays, and in complementation analyses ferric alcaligin utilization was restored to BRM17 by the wild-type fauA gene of either B. pertussis or B. bronchiseptica supplied in trans. The ability of the fauA gene alone to complement the ferric alcaligin utilization defect of BRM17 when supplied in trans suggested that the BRM17 mutant phenotype was not due to polarity effects of the insertion mutation. However, to test the requirement for FauA function in ferric alcaligin transport and utilization in the absence of potential polarity effects of insertion mutations on any downstream genes, a nonpolar in-frame fauA deletion mutation was crossed into B. bronchiseptica B013N to produce ΔfauA mutant BRM18. BRM18 was defective in ferric alcaligin utilization in growth stimulation bioassays and in iron transport activity using quantitative 55Fe-ferric alcaligin uptake assays. BRM18 also failed to produce detectable quantities of a 79-kDa iron-regulated outer membrane protein produced by the wild-type parent strain B013N. In complementation studies, BRM18 merodiploids carrying a wild-type fauA gene in trans regained ferric alcaligin transport and utilization capacity, and production of the 79-kDa iron-regulated outer membrane protein was restored.
Genetic and biochemical evidence presented in this report corroborates and extends our previous observations of production of a 79-kDa outer membrane protein regulated by Fur and AlcR (11, 13), the ferric alcaligin utilization defect associated with alcR mutants (11), and the rapid and saturable transport of ferric alcaligin siderophore complexes by B. bronchiseptica (16). The fauA gene encodes the iron-regulated outer membrane receptor protein required for ferric alcaligin transport and utilization. Further studies will be necessary to identify additional gene products involved in transport and utilization of ferric alcaligin by Bordetella species.
ACKNOWLEDGMENTS
We thank Peggy Cotter, Jeff F. Miller, Igor Stojiljkovic, Bob Ankenbauer, and Victor de Lorenzo for bacterial strains and plasmids. We acknowledge Chantel Sabus and Jessica Boeldt for technical assistance. We are grateful to Cari Vanderpool for assistance with ferric alcaligin transport assays, FauA expression studies, and Bordetella cell fractionation and for helpful discussions and critical reading of the manuscript.
This work was supported by Public Health Service grant AI-31088 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Agiato L-A, Dyer D W. Siderophore production and membrane alterations by Bordetella pertussis in response to iron starvation. Infect Immun. 1992;60:117–123. doi: 10.1128/iai.60.1.117-123.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akerley B J, Cotter P A, Miller J F. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction. Cell. 1995;80:611–620. doi: 10.1016/0092-8674(95)90515-4. [DOI] [PubMed] [Google Scholar]
- 3.Alexeyev M F. Three kanamycin resistance gene cassettes with different polylinkers. BioTechniques. 1995;18:52–56. [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Ankenbauer R, Sriyosachati S, Cox C D. Effects of siderophores on the growth of Pseudomonas aeruginosa in human serum and transferrin. Infect Immun. 1985;49:132–140. doi: 10.1128/iai.49.1.132-140.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Armstrong S K, Clements M O. Isolation and characterization of Bordetella bronchiseptica mutants deficient in siderophore activity. J Bacteriol. 1993;175:1144–1152. doi: 10.1128/jb.175.4.1144-1152.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beall B. Two iron-regulated putative ferric siderophore receptor genes in Bordetella bronchiseptica and Bordetella pertussis. Res Microbiol. 1988;149:189–201. doi: 10.1016/s0923-2508(98)80079-x. [DOI] [PubMed] [Google Scholar]
- 8.Beall B, Sanden G N. A Bordetella pertussis fepA homologue required for utilization of exogenous ferric enterobactin. Microbiology. 1995;141:3193–3205. doi: 10.1099/13500872-141-12-3193. [DOI] [PubMed] [Google Scholar]
- 9.Beall B, Hoenes T. An iron-regulated outer-membrane protein specific to Bordetella bronchiseptica and homologous to ferric siderophore receptors. Microbiology. 1997;143:135–145. doi: 10.1099/00221287-143-1-135. [DOI] [PubMed] [Google Scholar]
- 10.Beaumont F C, Armstrong S K. Abstracts of the 95th General Meeting of the American Society for Microbiology 1995. Washington, D.C.: American Society for Microbiology; 1995. A heterologous expression system for the genetic analysis of Bordetella siderophore synthesis, abstr. B-361; p. 228. [Google Scholar]
- 11.Beaumont F C, Kang H Y, Brickman T J, Armstrong S K. Identification and characterization of alcR, a gene encoding an AraC-like regulator of alcaligin siderophore biosynthesis and transport genes in Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1998;180:862–870. doi: 10.1128/jb.180.4.862-870.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brickman T J, Armstrong S K. Bordetella pertussis fur gene restores iron-repressibility of siderophore and protein expression to deregulated Bordetella bronchiseptica mutants. J Bacteriol. 1995;177:268–270. doi: 10.1128/jb.177.1.268-270.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brickman T J, Armstrong S K. The ornithine decarboxylase gene odc is required for alcaligin siderophore biosynthesis in Bordetella spp.: putrescine is a precursor of alcaligin. J Bacteriol. 1996;178:54–60. doi: 10.1128/jb.178.1.54-60.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brickman T J, Ozenberger B A, McIntosh M A. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J Mol Biol. 1990;212:669–682. doi: 10.1016/0022-2836(90)90229-F. [DOI] [PubMed] [Google Scholar]
- 16.Brickman T J, Hansel J-G, Miller M J, Armstrong S K. Purification, spectroscopic analysis, and biological activity of the macrocyclic dihydroxamate siderophore alcaligin produced by Bordetella pertussis and Bordetella bronchiseptica. BioMetals. 1996;9:191–203. doi: 10.1007/BF00144625. [DOI] [PubMed] [Google Scholar]
- 17.Calderwood S B, Mekalanos J J. Iron regulation of Shiga-like toxin expression in Escherichia coli is mediated by the fur locus. J Bacteriol. 1987;169:4759–4764. doi: 10.1128/jb.169.10.4759-4764.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Lorenzo V, Bindereif A, Paw B H, Neilands J B. Aerobactin biosynthesis and transport genes of plasmid ColV-K30 in Escherichia coli K-12. J Bacteriol. 1986;165:570–578. doi: 10.1128/jb.165.2.570-578.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feinberg A P, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 23.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 24.Figurski D H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giardina P C, Foster L-A, Toth S I, Roe B A, Dyer D W. Identification of alcA, a Bordetella bronchiseptica gene necessary for alcaligin production. Gene. 1995;167:133–136. doi: 10.1016/0378-1119(95)00659-1. [DOI] [PubMed] [Google Scholar]
- 26.Gorringe A R, Woods G, Robinson A. Growth and siderophore production by Bordetella pertussis under iron-restricted conditions. FEMS Microbiol Lett. 1990;66:101–106. doi: 10.1016/0378-1097(90)90265-r. [DOI] [PubMed] [Google Scholar]
- 27.Heinrichs D E, Poole K. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J Bacteriol. 1993;175:5882–5889. doi: 10.1128/jb.175.18.5882-5889.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyce C M, Grindley N D F. Method for determining whether a gene of Escherichia coli is essential: application to the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang H Y, Armstrong S K. Transcriptional analysis of the Bordetella alcaligin siderophore biosynthesis operon. J Bacteriol. 1998;180:855–861. doi: 10.1128/jb.180.4.855-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang H Y, Beaumont F C, Brickman T J, Armstrong S K. Identification and characterization of iron-regulated Bordetella pertussis alcaligin siderophore biosynthesis genes. J Bacteriol. 1996;178:4877–4883. doi: 10.1128/jb.178.16.4877-4884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovach M E, Phillips R W, Elzer P H, Roop II R M, Peterson K M. pBBR1MCS: a broad-host-range cloning vector. BioTechniques. 1994;16:800–802. [PubMed] [Google Scholar]
- 32.Kunkel T A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Langman L, Young I G, Frost G E, Rosenberg H, Gibson F. Enterochelin system of iron transport in Escherichia coli: mutations affecting ferric-enterochelin esterase. J Bacteriol. 1972;112:1142–1149. doi: 10.1128/jb.112.3.1142-1149.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lankford C E. Bacterial assimilation of iron. Crit Rev Microbiol. 1973;2:273–331. [Google Scholar]
- 35.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 352–355. [Google Scholar]
- 36.Miller V L, Mekalanos J J. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 38.Moore C H, Foster L-A, Gerbig D G, Dyer D W, Gibson B W. Identification of alcaligin as the siderophore produced by Bordetella pertussis and Bordetella bronchiseptica. J Bacteriol. 1995;177:1116–1118. doi: 10.1128/jb.177.4.1116-1118.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Neilands J B. Microbial iron compounds. Annu Rev Biochem. 1981;50:715–731. doi: 10.1146/annurev.bi.50.070181.003435. [DOI] [PubMed] [Google Scholar]
- 40.Nicholson M L, Beall B. Disruption of tonB in Bordetella bronchiseptica and Bordetella pertussis prevents utilization of ferric siderophores, haemin and haemoglobin as iron sources. Microbiology. 1999;145:2453–2462. doi: 10.1099/00221287-145-9-2453. [DOI] [PubMed] [Google Scholar]
- 41.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 42.Paulsen I T, Brown M H, Skurray R A. Proton-dependent multidrug efflux systems. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pradel E, Guiso N, Locht C. Identification of AlcR, an AraC-type regulator of alcaligin siderophore synthesis in Bordetella bronchiseptica and Bordetella pertussis. J Bacteriol. 1998;180:871–880. doi: 10.1128/jb.180.4.871-880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider D R, Parker C D. Effect of pyridines on phenotypic properties of Bordetella pertussis. Infect Immun. 1982;38:548–553. doi: 10.1128/iai.38.2.548-553.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwyn B, Neilands J B. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 48.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 49.Stainer D W, Scholte M J. A simple chemically defined medium for the production of phase I Bordetella pertussis. J Gen Microbiol. 1970;63:211–220. doi: 10.1099/00221287-63-2-211. [DOI] [PubMed] [Google Scholar]
- 50.Stojiljkovic I, Baumler A J, Hantke K. Fur regulon in gram-negative bacteria. Identification and characterization of new iron-regulated Escherichia coli genes by a Fur titration assay. J Mol Biol. 1994;236:531–545. doi: 10.1006/jmbi.1994.1163. [DOI] [PubMed] [Google Scholar]
- 51.Tabor S, Richardson C C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82:1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]