Abstract
Purpose
Although there is an established role for microbiome dysbiosis in the pathobiology of colorectal cancer (CRC), CRC patients of various race/ethnicities demonstrate distinct clinical behaviors. Thus, we investigated microbiome dysbiosis in Egyptian, African American (AA), and European American (EA) CRC patients.
Patients and methods
CRCs and their corresponding normal tissues from Egyptian (n = 17) patients of the Alexandria University Hospital, Egypt, and tissues from AA (n = 18) and EA (n = 19) patients at the University of Alabama at Birmingham were collected. DNA was isolated from frozen tissues, and the microbiome composition was analyzed by 16S rRNA sequencing. Differential microbial abundance, diversity, and metabolic pathways were identified using linear discriminant analysis (LDA) effect size analyses. Additionally, we compared these profiles with our previously published microbiome data derived from Kenyan CRC patients.
Results
Differential microbiome analysis of CRCs across all racial/ethnic groups showed dysbiosis. There were high abundances of Herbaspirillum and Staphylococcus in CRCs of Egyptians, Leptotrichia in CRCs of AAs, Flexspiria and Streptococcus in CRCs of EAs, and Akkermansia muciniphila and Prevotella nigrescens in CRCs of Kenyans (LDA score >4, adj. p-value <0.05). Functional analyses showed distinct microbial metabolic pathways in CRCs compared to normal tissues within the racial/ethnic groups. Egyptian CRCs, compared to normal tissues, showed lower l-methionine biosynthesis and higher galactose degradation pathways.
Conclusions
Our findings showed altered mucosa-associated microbiome profiles of CRCs and their metabolic pathways across racial/ethnic groups. These findings provide a basis for future studies to link racial/ethnic microbiome differences with distinct clinical behaviors in CRC.
Keywords: Colorectal cancer, Microbiome, African American, European American, Egyptian
1. Introduction
Worldwide, the third most diagnosed cancer is colorectal cancer (CRC) [1]. Data from the Surveillance, Epidemiology, and End Results (SEER) program show that the overall incidence of CRCs for African Americans is 41.9 per 100,000 and 37.0 per 100,000 for European Americans [2]. For Egyptians, according to the Egyptian National Cancer Registry Program in 2014, CRC is the seventh most diagnosed cancer [3].
CRC has a multifactorial pathobiology, including genetic mutations; ethnicity; and age, as most CRC patients are older than 50 years (during the last decade, however, there has been an increased rate of early-onset CRC) [4]. Inflammatory intestinal conditions are also a risk factor for CRC [5]. Another risk factor is diet, as low-fiber, high-fat western diets are associated with a higher prevalence of CRC. For that reason, CRC has been called a westernized disease [6]. Westernized diets alter the microbiome symbiosis. Other well-established risk factors for colorectal neoplasia are diabetes, obesity, heavy alcohol consumption, and smoking [7,8]. Compared to European Americans (EAs), African Americans (AAs) have a high CRC incidence and mortality rate [4] and experience less treatment-related toxicity of adjuvant 5-fluorouracil-based chemotherapies [9]. A retrospective study shows that Egyptian CRC patients have clinical behaviors, epidemiologic profiles, and survival estimates that are distinctive from those of more developed countries [10]. In Egypt, the mean age of CRC diagnosis is 53 years, which is less than mean ages of CRC diagnosis in AAs (64 years) and in EAs (68 years) [10,11]. Some of these disparities can be attributed to genetics, which contribute to 35% of the overall CRC incidences [12]; metagenomics, socioeconomic disparities, and access to screening and therapy are also involved [13]. As determined by racial microbiome/diet studies, diet and microbiome influence the CRC incidence rate [14].
Advances in CRC research have improved our understanding of the pathobiology of CRC and CRC-associated microbiome dysbiosis. However, little work has been accomplished for other populations compared to those of the Western world (Europe/United States). Moreover, microbiome-profiling studies are needed to characterize the behavior of CRCs in other racial/ethnic groups. Thus, the present study assessed microbiome differential abundance (via 16S rRNA sequencing) and their metabolic pathway profiles (via computational functional analyses) in CRCs and their corresponding normal tissues derived from Egyptians, AAs, and EAs. These profiles were also compared with our recent microbiome findings for Kenyan CRC patients. Our findings aid in understanding the diversity in microbiome profiles of CRCs of these racial/ethnic groups and provide insights to the clinical behavior of CRCs.
2. Materials and methods
2.1. Patients and tissue collection
We used banked, frozen CRC and corresponding normal specimens of EA (n = 19) and AA (n = 18) patients from the University of Alabama at Birmingham (UAB) biorepository to conduct 16S rRNA sequencing. For the Egyptian cohort, we prospectively collected 17 CRCs and their matching normal mucosa specimens from Alexandria University Hospital, Alexandria, Egypt. Samples have been frozen until 16S rRNA sequencing analysis. The inclusion criteria of this study are: a) patients with a histopathologically confirmed diagnosis of CRC; and b) patients over 18 years old of both genders. The samples were de-identified for the study investigators until the data analyses. The Institutional Review Boards of UAB (IRB-060911009) and Alexandria University (IRB NO. 00007555-FWA NO. 00018699) approved this study. The microbiome data for Kenyan CRC patients were from our recently published study [15]. These data were used to compare with microbiome data for AA, CA, and Egyptian CRCs.
2.2. 16S rRNA sequencing for the mucosa-associated microbiome
DNA from CRCs and their corresponding normal specimens was extracted using Qiagen DNA isolation kits according to the manufacturer's instructions (Qiagen, USA). Amplification of V4 variable regions of the 16S rRNA gene was performed by using amplification primers as described earlier [15,16]. Illumina adaptor sequences and indexes were added to the amplicons. At the UAB Genomic Core, an Illumina MiSeq platform was used for sequencing.
2.3. Gut mucosal microbiome data processing and quality assessment
The raw paired-end 16S rRNA sequencing reads from CRCs and corresponding normal gut samples were analyzed using QIIME2 2019.10 (https://docs.qiime2.org/2019.10/install/) [17]. For taxonomic and functional profiling of microbiomes, we followed the previous procedure with minor modifications [18]. Briefly, the sequencing reads were imported into a QIIME2 environment and de-multiplexed into FASTQ files for individual samples. The QIIME2-DADA2 pipeline was used for sequence quality control analysis and to identify the amplicon sequence variants (ASVs) or features. The input reads to the QIIME2-DADA2 pipeline were filtered based on quality scores by adjusting the trim and truncate options. The QIIME2 and Naïve Bayes classifier trained on reference sequences from ‘Greengenes 13_8 99% operational taxonomic units (OTUs) from the V4 region’ [19] were used to classify the ASVs. Then, the feature tables were filtered to remove rare ASVs, contaminants, and unclassified ASVs using a QIIME2-feature-table filter-features tool, and low-depth samples were removed from the feature tables using QIIME2 feature-table filter-samples tool. Further, the feature tables were collapsed to species-level taxonomy, and microbial taxa with relative abundance >0.01% and prevalence >5% of the samples were used for downstream analyses. Similar microbiome data-processing methods were employed in analyzing the Kenyan CRC microbiome data [15].
2.4. Diversity analysis
Alpha (Shannon, ACE, Pielou's evenness and Chao1) and beta (unweighted UniFrac and weighted UniFrac) diversity indices of microbial communities were calculated on rarefied sequences using a QIIME2 diversity plugin. Alpha diversity indices between groups were assessed using Wilcoxson signed-rank tests and presented as box plots; unweighted and weighted UniFrac distances between microbial communities were presented as ordination plots based on principal coordination analysis (‘cmdscale’ function) using stats and ggplot2 packages in R (https://ggplot2.tidyverse.org/. The covariate effect of age and gender on species diversity (Shannon diversity) was tested using a multivariate linear regression method. Results were plotted using ggplot2 based on adjusted and unadjusted diversity coefficients. Beta diversity between groups was assessed based on adonis2 function (permutational multivariate analysis of variance, at 1000 permutations) using the Vegan package in R.
2.5. Functional analysis
By using PICRUSt2 v2.2 software (https://github.com/picrust/picrust2/releases), the metabolic pathway profiles of CRCs and normal samples of all datasets were computationally predicted [20]. The predicted MetaCyc pathways were filtered according to the criteria of relative abundance >0.1% and prevalence in >50% of the samples for downstream analysis.
2.6. Statistical analyses for differential abundances of microbial species and metabolic pathways
Differential abundances of microbial species and metabolic pathways between groups were identified using linear discriminant analysis (LDA) effect size (LEfSe) (https://bioconductor.org/packages/devel/bioc/manuals/lefser/man/lefser.pdf) [21] analysis. Other statistical analysis and visualizations were performed in R v 4.0.0 and above (https://cran.r-project.org/doc/manuals/r-patched/R-admin.html and https://github.com/rstudio/) [22]. A p-value less than 0.05 was considered significant.
3. Results
3.1. Characteristics of study participants
The study participants were 17 Egyptian CRC patients with an average age of 55.5 ± 8.9 years (males: 8, females: 9), 18 AA CRC patients with an average age of 57.3 ± 9 years (males: 7, females: 11), 19 EA CRC patients with an average age of 58 ± 8.5 years (males: 8, females: 11), and 18 Kenyan CRC patients with an average age of 49.6 ± 18.3 years (males: 12, females: 6). (Table 1).
Table 1.
Patient characteristics.
| Egyptian | African American | European American | Kenyan | |
|---|---|---|---|---|
| Number of Patients | 17 | 18 | 19 | 18 |
| Normal | 17 | 18 | 19 | – |
| CRCs | 17 | 18 | 19 | 18 |
| Gender | ||||
| Male | 8 | 7 | 8 | 12 |
| Female | 9 | 11 | 11 | 6 |
| Average Age (years) | 55.5 ± 8.9 | 57.3 ± 9 | 58 ± 8.5 | 49.6 ± 18.3 |
| Tumor Stage | ||||
| Stage I | 1 | – | – | – |
| Stage II | 1 | 2 | 1 | – |
| Stage III | 3 | 9 | 10 | 8 |
| Stage IV | 3 | 7 | 8 | 10 |
| NA | 9 | – | – | – |
NA denotes that information on stage is not available.
3.2. Gut microbial community structure and diversity
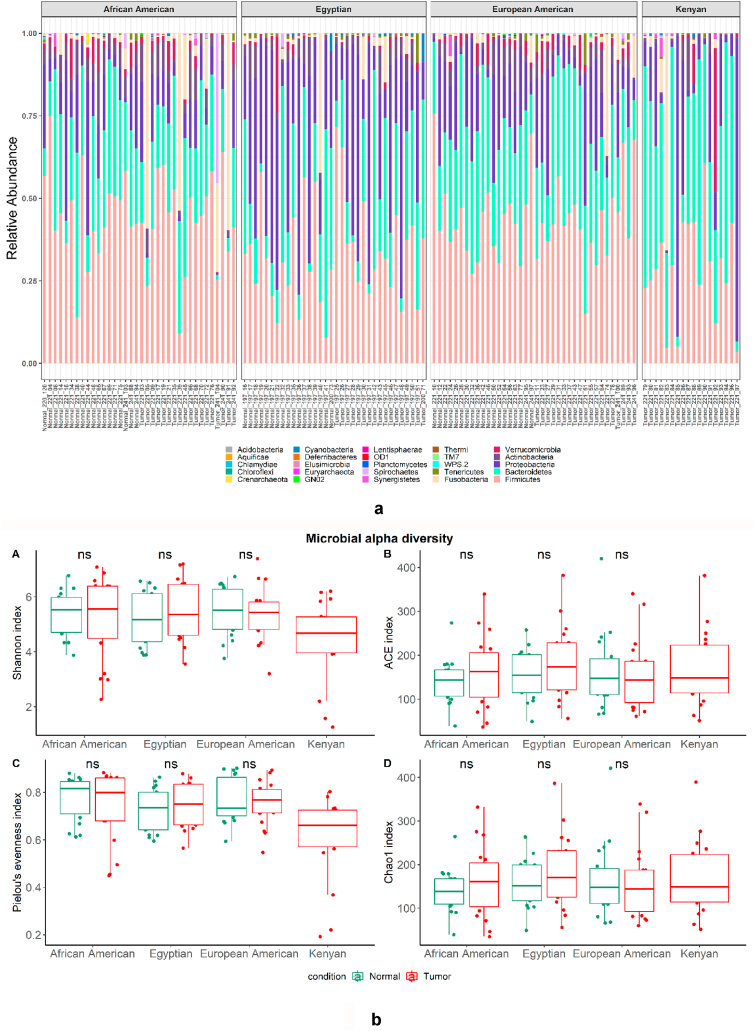
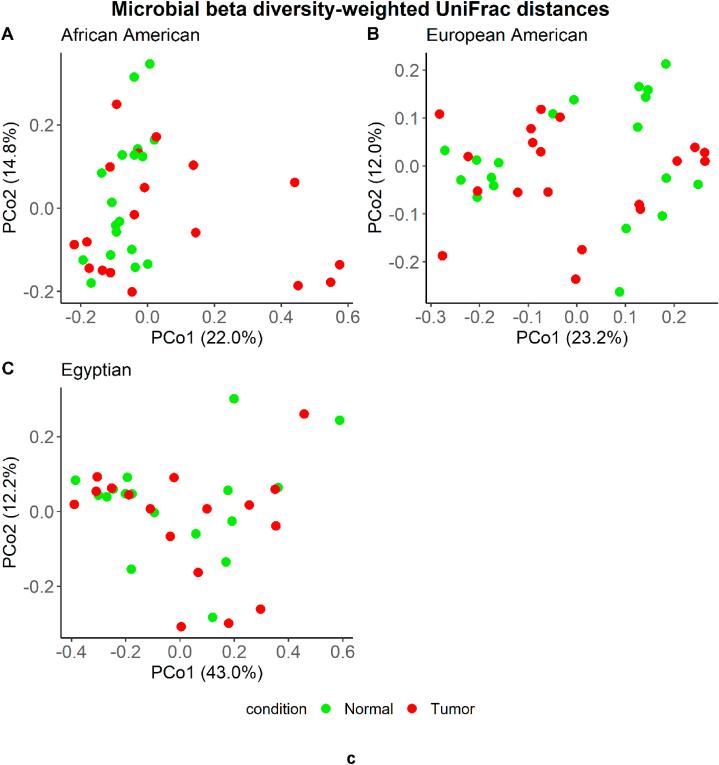
The results showed diverse microbiome profiles (Fig. 1Sa and 1Sb) and 16S rRNA sequences were clustered into 25 phyla (Fig. 1a). Across races, the species richness indices (Shannon, ACE, Pielou's evenness, and Chao1) showed no differences between CRCs and corresponding normal tissues (Fig. 1b [A-D]). In addition, within each race, beta diversity measures did not show significant differences between the CRC and normal microbial community structures (Fig. 1c [A-C]). Moreover, across racial/ethnic groups, beta diversity shows differences in CRCs as well as in Normal tissues (Fig. 2Sa and 2Sb).
Fig. 1.
a: Stacked bar plots depicting the relative abundance of microbiota at the phylum level in each sample of CRCs and normal tissues of different racial/ethnic groups.
b: Alpha diversity analysis of the microbiota of tumor (CRC) and normal samples of different racial/ethnic groups. Boxplot representing different alpha diversity indices of each group, (A) Shannon index, B) ACE index, C) Pielou's evenness index, and D) Chao1 index. ns indicates statistically non-significant differences within the alpha diversity indices between CRCs and normal samples (Wilcoxson signed-rank test, P > 0.05).
c: Principal coordinate analysis (PCoA) based on weighted UniFrac distances of microbial communities in each race/ethnic group. Differences between tumor (CRC) and normal samples are shown in A) African Americans (p-value 0.12087), B) European American (p-value 0.64035), and C). Egyptians (p-value 0.83716). p-Values are from the adonis2 test of vegan package R.
The species diversity was not affected by potential confounding factors such as age in AA, EA, and Kenyan groups. However, age showed a confounding effect on a few species in Egyptians, which include Herbaspirillum, Prevotella copri, Leptotrichia, Bacteroides, Prevotella stercorea, and Prevotella intermedia (Fig. 3S). Whereas in AAs and EAs, the species diversity was affected by sex differences. In AAs, Treponema socranskii, Alistipes onderdonkii, Selenomonas, Enterbacteriaceae, Akkermansia muciniphila, Bacteroides, Porphyromonas, and Gemellaceae and, in EAs, Schwartzia, Selenomonas, Enterobacteriaceae, Bacteroides, Flexispira, Porphyromonas, Faecalibacterium prausntitzii, and Bacteroides showed differences in diversity coefficients based on sex in multivariate analyses (Fig. 3S).
3.3. Differences in microbiome composition between tumor and normal gut mucosa for African American, European American, and Egyptian CRC patients
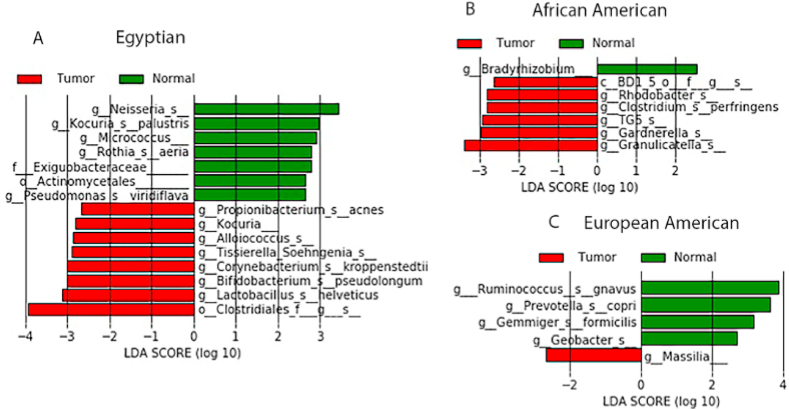
For CRCs of Egyptians, differential microbiome analysis showed significantly higher abundances of Lactobacillus helveticus, Bifidobacterium pseudolongum, Corynebacterium kroppenstedtii, Tissierella Soehngenia, Alloiococcus Kocuria, and Propionibacterium acnes relative to normal gut mucosa. In contrast, Neisseria, Kocuria palustris, Micrococcus, Rothia aeria, and Pseudomonas viridiflava showed lower abundances in tumor tissues relative to normal tissues (adj. p-value <0.05) (Fig. 2A). For CRCs of AAs, differential microbiome analysis showed significantly higher abundances of Granulicatella, Gardnerella, Clostridium perfringens, and Rhodobacter relative to their normal gut mucosa. However, in CRC tissues, Bradyrhizobium showed a lower abundance relative to normal tissues (adj. p-value <0.05) (Fig. 2B). For CRCs of EAs, differential microbiome analysis showed a significantly higher abundance of Massilia relative to normal gut mucosa. In contrast, Ruminococcus gnavus, Prevotella copri, Gemmiger formicilis, and Geobacter showed lower abundances in CRC tissues relative to normal tissues (adj. p-value <0.05) (Fig. 2C).
Fig. 2.
LEfSe analysis shows histograms of differential microbial taxa, at the species level, between tumor (CRC) and normal samples in each race/ethnic group. A) African Americans, B) European Americans, and C) Egyptians. Differential species with LDA score >2 at P < 0.05 are shown here.
3.4. Differences in microbiome profiles of CRCs between African American, European American, Egyptian, and Kenyan patients
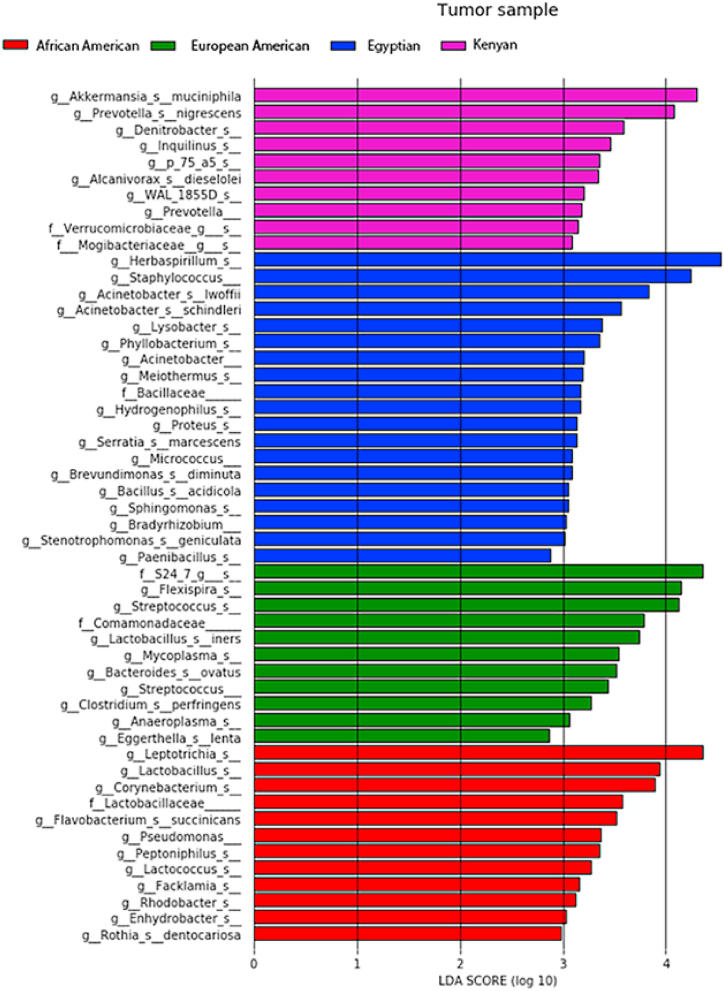
Differential microbiome analysis of CRCs across all racial/ethnic groups showed high abundances of Herbaspirillum and Staphylococcus in Egyptian CRCs, Leptotrichia in CRCs of AAs, Flexspiria and Streptococcus in CRCs of EAs, and Akkermansia muciniphila and Prevotella nigrescens in Kenyan CRCs (LDA score >4, adj. p-value <0.05) (Fig. 3).
Fig. 3.
LEfSe analysis shows histograms of differential microbial taxa between tumor (CRC) samples of different races at the species level. Differential species with LDA score >2 at P < 0.05 are shown here.
3.5. Functional analyses of microbiomes for African American, European American, and Egyptian patients
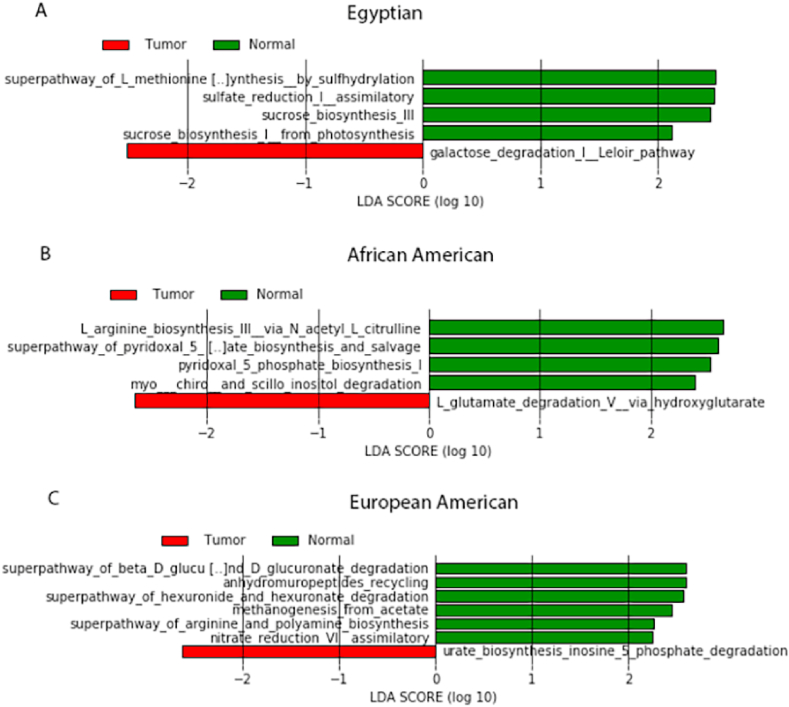
For CRCs from Egyptians, functional analysis showed a lower l-methionine biosynthesis pathway and a higher galactose degradation pathway compared to normal tissue (Fig. 4A). For CRCs of AAs, functional analysis showed a decreased l-arginine biosynthesis III pathway and an increased pathway for l-glutamate degradation V compared to normal tissues (Fig. 4B). For CRCs of EAs, functional analysis showed a decreased d-glucuronate degradation pathway and increased pathways of urate biosynthesis and inosine-5-phosphate degradation compared to normal tissues (Fig. 4C).
Fig. 4.
LEfSe analysis showing differential predicted metabolic pathways between tumors (CRC) and normal tissues in each racial/ethnic group. Differential pathways with LDA score (log10) >2 at P < 0.05 are shown in the histograms.
4. Discussion
Although multifactorial etiologies of CRC carcinogenesis are established, a dysbiosis of the gut microbiota contributes to CRC development. The findings of the present study show an altered mucosa-associated microbiome and their metabolic pathways between CRCs and their corresponding normal tissues as well as microbial disparities among racial/ethnic groups. Additionally, there were no differences in microbiome diversity based on age with an exception of a few bacterial species in Egyptian patients. However, sex has an influence on microbiome diversity in AA and EA patients.
Among several microbiota, the higher abundance of C. perfringens and the lower abundance of Bradyrhizobium, relative to normal gut mucosa, found in CRCs of AAs may contribute to increased risk of CRC. Indeed, C. perfringens septicemia is a risk factor of CRC [23,24]. APC-deficient mice show less carcinogenesis upon suppression of C. perfringens [25], and activation of the YAP gene is an underlying mechanism for the C. perfringens-induced CRC carcinogenesis [26]. In agreement with our results, a previous metagenomic study of CRC patients in India shows a depletion of Bradyrhizobium in CRC patients compared to healthy individuals [27]. In the context of the bacterial driver-passenger model [28], a potential driver bacterium for CRC is Bradyrhizobium, which transiently colonizes and initiates tumorigenesis, then get replaced by other microbiota [29].
Our results showed, in CRCs of EAs, a higher abundance of Massilia and lower abundances of R. gnavus, P. copri, G. formicilis, and Geobacter, relative to normal gut mucosa. In contrast to our findings, previous studies show a link between a high abundance of R. gnavus and the severity of inflammatory bowel diseases [30], which are established risk factors of CRC and elicit a pro-inflammatory cascade in colonic epithelial cells [31]. For CRCs of Moroccan patients, there is depletion of P. copri, a low-abundant bacterium in EAs [32], and in our recent study of CRCs of Kenyan patients [15]. Moreover, a meta-analysis shows that P. copri is highly abundant in healthy populations with non-westernized diets [33]. Additionally, a study that investigated the fecal microbiome of patients who had the immunochemical-based fecal occult blood test (iFOBT), a CRC screening assay, to determine the effect of blood in the stool on microbiome profiles, found a decreased abundance of P. copri and G. formicilis [34] in patients with blood in their stool. These findings suggest that depletions of these microbes are indicators of the presence of fecal blood, a known symptom of CRC. A previous study also shows a low abundance of G. formicilis in CRC patients compared to healthy individuals [35]. Additionally, preclinical and clinical studies show that CRC patients with a high abundance of G. formicilis have a better response to checkpoint blockade therapy [36]. For Chinese CRC patients, Geobacter, an additional lower abundant microbiota found in CRCs of EAs, is in low abundance compared to healthy controls [37]. Thus, the differential microbiome profiles of EAs observed in the present study suggest that they contribute to CRC development.
Among microbial alterations observed in CRCs of Egyptian patients, the lower abundance of Neisseria in CRCs, relative to normal tissues, is in agreement with a previous study showing that smoking, a risk factor for CRC, results in a low abundance of Neisseria [38] and in pathophysiological consequences of gut mucosa [38]. However, higher abundances of L. helveticus and B. pseudolongum, found in CRCs of Egyptians, are beneficial microbiota in CRC animal models [39,40]. However, some of these findings from experimental animals may not relate to human CRC pathophysiology.
CRC microbiome analyses based on race/ethnicity show high abundances of A. muciniphila and P. nigrescens in Kenyan CRCs; Herbaspirillum in Egyptian CRCs; Flexspiria in EA CRCs; and Leptotrichia in AA CRCs. An A. muciniphila supplementation improves the therapy response to FOLFOX, a first line therapy for CRC [41]. For a French population, P. nigrescens is also highly abundant in patients with CRC [42], and, in CRCs, Herbaspirillum is associated with NRAS mutations [43]. Our previous study of Kenyan CRC patients shows that Herbaspirillum is associated with early-onset of this disease [44]. Further, in CRCs of an Irish population, Leptotrichia is highly abundant [45].
Since a rural African diet is higher in fiber and lower in fat and meat, and an AA diet is higher in fat and meat and lower in fiber, a diet switch, within two weeks, has a profound effect on the microbiome and the fecal metabolome [46]. These variations in abundance of risk microbiota may be attributed to the dietary habits of various populations.
Functional analysis showed altered bacterial metabolic pathways in CRCs compared to normal tissues. For AAs, the l-arginine biosynthesis III pathway was lower in CRCs compared to normal tissues. In mouse models of colitis, dietary supplementation of arginine has a protective effect [47]. Further, l-arginine-producing bacteria modulate metabolism of the tumor microenvironment and enhance the efficacy of immunotherapies [48]. For EAs, the inosine-5-phosphate degradation pathway was higher in CRCs compared to normal tissues. Mice colonized with microbiota that enhance anti-tumor immunity have higher levels of inosine monophosphate [49], and inosine increases T cell proliferation and differentiation and improves the efficacy of immunotherapies [50], suggesting that combination of inosine supplements with immunotherapy is a promising intervention, especially for EA CRC patients. Since our finding that, for CRCs of Egyptians, a decreased l-methionine biosynthesis pathway and an increased galactose degradation pathway have not been reported previously in CRC, further investigations are needed to assess their utility in assessing the clinical behavior of CRCs.
5. Conclusion
The differential microbiome abundance and diversity in CRCs, compared to adjacent normal tissues, of various race/ethnicities, as reported in this study, provides insights about the clinical behavior of cancers. Nevertheless, further studies are needed to link the variations in microbiota and their metabolic pathways to clinical outcomes for CRCs.
Limitations of the study: Although our descriptive study reveals microbiome dysbiosis between CRCs and normal tissues, further studies with large sample sizes are needed to integrate the tumor differential gene expression and differential abundance of the microbiomes. Moreover, integration of host multiomics (microbiome, transcriptome, mutational profiles, and metabolomics) and CRC risk factors (e.g., high-fat diet, obesity, colonic inflammatory diseases, and smoking) is also necessary. This approach will improve our understanding of how race/ethnicity-specific microbiomes affect the development of CRCs and will aid in developing prevention strategies for these cancers. Moreover, multiple studies show a link between microbiome profiles and host genetics, diet, age, gender, lifestyle, and mode of birth/delivery [51]. However, our findings will provide a basis for future studies to identify these variations as biomarkers of clinical behaviors CRC.
Author contributions
Upender Manne: Conceived and designed the experiments; Wrote the paper.
Waleed Arafat; Hesham Saed; Amira Embaby; Mona Fouad: Conceived and designed the experiments.
Amr Elkholy; Nagavardhini Avuthu: Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Mohammad Abdalla; Mostafa Mohamed; Sarah Obuya; Prachi Bajpai; Nefertiti El-Nikhely: Performed the experiments.
Michael Behring: Analyzed and interpreted the data; Wrote the paper.
Chittibabu Guda; Mansoor Saleh; Sooryanarayana Varambally; Sejong Bae: Analyzed and interpreted the data.
Hyung-Gyoon Kim; Doaa Header; Reham AH. Abo Elwafa; Sarah Obuya; Ahmed Ashour Badawy; Ahmed Nawar; Farrukh Afaq; Laura Q. Rogers; James M. Shikany; Lori Brand Bateman; Mansoor Saleh; Temesgen Samuel: Contributed reagents, materials, analysis tools or data.
Funding statement
This work was funded by National Academy of Science, NAS 2000007148 and STDF-USC17-144 awarded to MF and WA, respectively. The effort was also partly supported by 5U54CA118948 and by institutional funds (Department of Pathology and Heersink School of Medicine of the University of Alabama at Birmingham, UAB) awarded to UM.
Declaration and Institutional Review Board
The study was conducted according to the guidelines of the Declaration of Helsinki, and approved by the Institutional Review Board and the Ethics Committee of University of Alabama at Birmingham (IRB NO. IRB-060911009) and Alexandria University (IRB NO.: 00007555-FWA NO.: 00018,699). The authors declare no competing interests.
Informed consent
Written informed consent was obtained from all subjects involved in the study to utilize their specimens.
Data availability
The data presented in this study has been uploaded to the Sequence Read Archive (SRA) under BioProject number PRJNA986115, and the previously published data on Kenyan patients will be available in the SRA under BioProject PRJNA986175. These data will be publicly available on 07-01-2024.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We acknowledge the Microbiome/Gnotobiotics Shared Facility of UAB Comprehensive Cancer Center (P30CA013148) for performing the 16s rRNA sequencing, and the Bioinformatics and Systems Biology Core (BSBC) Facility, of the University of Nebraska Medical Center for the bioinformStics analyses. BSBC receives support from the Nebraska Research Initiative and NIH awards (2P20GM103427, 5P30CA036727, and 2U54GM115458). We thank Dr. Donald Hill, a faculty member of the UAB O’Neal Comprehensive Cancer Center, for his editing the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e18035.
Contributor Information
Waleed Arafat, Email: waleed.arafat@alexmed.edu.eg.
Upender Manne, Email: upendermanne@uabmc.edu.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Ferlay J., et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136(5):E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Carethers J.M. Racial and ethnic disparities in colorectal cancer incidence and mortality. Adv. Cancer Res. 2021;151:197–229. doi: 10.1016/bs.acr.2021.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ibrahim A.S., et al. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014;2014 doi: 10.1155/2014/437971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel R.L., et al. Colorectal cancer statistics, 2020. Ca - Cancer J. Clin. 2020;70(3):145–164. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 5.Beaugerie L., Itzkowitz S.H. Cancers complicating inflammatory bowel disease. N. Engl. J. Med. 2015;372(15):1441–1452. doi: 10.1056/NEJMra1403718. [DOI] [PubMed] [Google Scholar]
- 6.Mármol I., et al. Colorectal carcinoma: a general overview and future perspectives in colorectal cancer. Int. J. Mol. Sci. 2017;18(1):197. doi: 10.3390/ijms18010197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kuipers E.J., et al. Colorectal cancer. Nat. Rev. Dis. Prim. 2015;1(1) doi: 10.1038/nrdp.2015.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davenport J.R., et al. Modifiable lifestyle factors associated with risk of sessile serrated polyps, conventional adenomas and hyperplastic polyps. Gut. 2018;67(3):456–465. doi: 10.1136/gutjnl-2016-312893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCollum A.D., et al. Outcomes and toxicity in african-american and caucasian patients in a randomized adjuvant chemotherapy trial for colon cancer. J. Natl. Cancer Inst. 2002;94(15):1160–1167. doi: 10.1093/jnci/94.15.1160. [DOI] [PubMed] [Google Scholar]
- 10.Metwally I.H., et al. Epidemiology and survival of colon cancer among Egyptians: a retrospective study. Journal of Coloproctology (Rio de Janeiro) 2018;38:24–29. [Google Scholar]
- 11.Krapcho M., et al. posted to the SEER website; 2019. SEER Cancer Statistics Review, 1975-2016. Based on November 2018 SEER Data Submission. [Google Scholar]
- 12.Lichtenstein P., et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000;343(2):78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 13.Alexander D.D., et al. African-American and Caucasian disparities in colorectal cancer mortality and survival by data source: an epidemiologic review. Cancer Biomarkers. 2007;3(6):301–313. doi: 10.3233/cbm-2007-3604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Royston K.J., Adedokun B., Olopade O.I. Race, the microbiome and colorectal cancer. World J. Gastrointest. Oncol. 2019;11(10):773–787. doi: 10.4251/wjgo.v11.i10.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Obuya S., et al. A signature of Prevotella copri and Faecalibacterium prausnitzii depletion, and a link with bacterial glutamate degradation in the Kenyan colorectal cancer patients. J. Gastrointest. Oncol. 2022;13(5):2282–2292. doi: 10.21037/jgo-22-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso J.G., et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bolyen E., et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019;37(8):852–857. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar S.R.P., et al. Role of small intestine and gut microbiome in plant-based oral tolerance for hemophilia. Front. Immunol. 2020;11:844. doi: 10.3389/fimmu.2020.00844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDonald D., et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6(3):610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douglas G.M., et al. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020;38(6):685–688. doi: 10.1038/s41587-020-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Segata N., et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12(6):R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Team, R.D.C. A language and environment for statistical computing. 2009. http://www.R-project.org
- 23.Pushpanathan P., et al. Gut microbiota and its mysteries. Indian J. Med. Microbiol. 2019;37(2):268–277. doi: 10.4103/ijmm.IJMM_19_373. [DOI] [PubMed] [Google Scholar]
- 24.Kwong T.N.Y., et al. Association between bacteremia from specific microbes and subsequent diagnosis of colorectal cancer. Gastroenterology. 2018;155(2):383–390.e8. doi: 10.1053/j.gastro.2018.04.028. [DOI] [PubMed] [Google Scholar]
- 25.Sasada T., et al. Chlorinated water modulates the development of colorectal tumors with chromosomal instability and gut microbiota in apc-deficient mice. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0132435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fujiwara-Tani R., et al. Role of Clostridium perfringens enterotoxin on YAP activation in colonic sessile serrated adenoma/polyps with dysplasia. Int. J. Mol. Sci. 2020;21(11) doi: 10.3390/ijms21113840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bamola V.D., et al. A metagenomic assessment of gut microbiota in Indian colon cancer patients. J. Cancer Res. Therapeut. 2022;18(1):96–102. doi: 10.4103/0973-1482.341139. [DOI] [PubMed] [Google Scholar]
- 28.Tjalsma H., et al. A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat. Rev. Microbiol. 2012;10(8):575–582. doi: 10.1038/nrmicro2819. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y., et al. Analyses of potential driver and passenger bacteria in human colorectal cancer. Cancer Manag. Res. 2020;12:11553–11561. doi: 10.2147/CMAR.S275316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Png C.W., et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am. J. Gastroenterol. 2010;105(11):2420–2428. doi: 10.1038/ajg.2010.281. [DOI] [PubMed] [Google Scholar]
- 31.Yu S., et al. Paneth cell-derived lysozyme defines the composition of mucolytic microbiota and the inflammatory tone of the intestine. Immunity. 2020;53(2):398–416.e8. doi: 10.1016/j.immuni.2020.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allali I., et al. Gut microbiome of Moroccan colorectal cancer patients. Med. Microbiol. Immunol. 2018;207(3–4):211–225. doi: 10.1007/s00430-018-0542-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tett A., et al. The Prevotella copri complex comprises four distinct clades underrepresented in westernized populations. Cell Host Microbe. 2019;26(5):666–679 e7. doi: 10.1016/j.chom.2019.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chénard T., et al. The influence of blood on the human gut microbiome. BMC Microbiol. 2020;20(1):44. doi: 10.1186/s12866-020-01724-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He T., Cheng X., Xing C. The gut microbial diversity of colon cancer patients and the clinical significance. Bioengineered. 2021;12(1):7046–7060. doi: 10.1080/21655979.2021.1972077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaput N., et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann. Oncol. 2017;28(6):1368–1379. doi: 10.1093/annonc/mdx108. [DOI] [PubMed] [Google Scholar]
- 37.Huang R., et al. Changes of intestinal microflora in colorectal cancer patients after surgical resection and chemotherapy. Comput. Math. Methods Med. 2022;2022 doi: 10.1155/2022/1940846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Kato I., et al. Oral microbiome and history of smoking and colorectal cancer. J Epidemiol Res. 2016;2(2):92–101. doi: 10.5430/jer.v2n2p92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zareie M., et al. Probiotics prevent bacterial translocation and improve intestinal barrier function in rats following chronic psychological stress. Gut. 2006;55(11):1553–1560. doi: 10.1136/gut.2005.080739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo W., et al. Protective effects of a novel probiotic Bifidobacterium pseudolongum on the intestinal barrier of colitis mice via modulating the pparγ/STAT3 pathway and intestinal microbiota. Foods. 2022;11(11) doi: 10.3390/foods11111551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou X., et al. Akkermansia muciniphila potentiates the antitumor efficacy of FOLFOX in colon cancer. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.725583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zeller G., et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014;10:766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sarhadi V., et al. Gut microbiota and host gene mutations in colorectal cancer patients and controls of Iranian and Finnish origin. Anticancer Res. 2020;40(3):1325–1334. doi: 10.21873/anticanres.14074. [DOI] [PubMed] [Google Scholar]
- 44.Obuya S., et al. A signature of Prevotella copri and Faecalibacterium prausnitzii depletion, and a link with bacterial glutamate degradation in the Kenyan colorectal cancer patients. J. Gastrointest. Oncol. 2022;13(5):2282–2292. doi: 10.21037/jgo-22-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flemer B., et al. The oral microbiota in colorectal cancer is distinctive and predictive. Gut. 2018;67(8):1454–1463. doi: 10.1136/gutjnl-2017-314814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Keefe S.J., et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Singh K., et al. Dietary arginine regulates severity of experimental colitis and affects the colonic microbiome. Front. Cell. Infect. Microbiol. 2019;9:66. doi: 10.3389/fcimb.2019.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Canale F.P., et al. Metabolic modulation of tumours with engineered bacteria for immunotherapy. Nature. 2021;598(7882):662–666. doi: 10.1038/s41586-021-04003-2. [DOI] [PubMed] [Google Scholar]
- 49.Tanoue T., et al. A defined commensal consortium elicits CD8 T cells and anti-cancer immunity. Nature. 2019;565(7741):600–605. doi: 10.1038/s41586-019-0878-z. [DOI] [PubMed] [Google Scholar]
- 50.Wang T., et al. Inosine is an alternative carbon source for CD8(+)-T-cell function under glucose restriction. Nat Metab. 2020;2(7):635–647. doi: 10.1038/s42255-020-0219-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hasan N., Yang H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ. 2019;7:e7502. doi: 10.7717/peerj.7502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data presented in this study has been uploaded to the Sequence Read Archive (SRA) under BioProject number PRJNA986115, and the previously published data on Kenyan patients will be available in the SRA under BioProject PRJNA986175. These data will be publicly available on 07-01-2024.