Abstract
A growing number of studies indicate that coronavirus disease 2019 (COVID-19) is associated with inflammatory sequelae, but molecular signatures governing the normal versus pathologic convalescence process have not been well-delineated. Here, we characterized global immune and proteome responses in matched plasma and saliva samples obtained from COVID-19 patients collected between 20 and 90 days after initial clinical symptoms resolved. Convalescent subjects showed robust total IgA and IgG responses and positive antibody correlations in saliva and plasma samples. Shotgun proteomics revealed persistent inflammatory patterns in convalescent samples including dysfunction of salivary innate immune cells, such as neutrophil markers (e.g., myeloperoxidase), and clotting factors in plasma (e.g., fibrinogen), with positive correlations to acute COVID-19 disease severity. Saliva samples were characterized by higher concentrations of IgA, and proteomics showed altered myeloid-derived pathways that correlated positively with SARS-CoV-2 IgA levels. Beyond plasma, our study positions saliva as a viable fluid to monitor normal and aberrant immune responses including vascular, inflammatory, and coagulation-related sequelae.
Keywords: COVID-19, Saliva, Proteomics, Convalescent, Pathogenesis
Highlights
-
•
Significant IgA levels were detected in saliva, and IgG levels in plasma in COVID-19 cases, confirming convalescence.
-
•
Abnormal inflammatory and clotting responses were identified in both saliva and plasma fluids after SARS-CoV-2 infection.
-
•
Random forest machine learning identified plasma fibrinogen as a marker that differentiates COVID-19 vascular dysfunctions.
-
•
Sustained inflammation and active pathogenic pathways were detected in saliva of COVID-19 cases.
-
•
Saliva is an important and accessible biofluid to monitor immune pathways altered in COVID-19 in a populational scale.
1. Introduction
Patients infected with the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) driving the COVID-19 pandemic generally experience a course of acute illness that lasts for approximately 2 weeks. For example, Byrne et al. reported that the estimated mean time from COVID-19 symptom onset to two negative PCR tests was 13.4 days [1]. During this acute phase of illness, COVID-19 patients who do not experience further complications generally produce SARS-CoV-2 associated antibodies and enter the recovery or convalescent stage of the disease. However, despite SARS-CoV-2 antibody production and a decrease in clinical symptoms, it is possible that convalescent COVID-19 patients (20–90 days after initial illness) still experience immune or coagulation-related sequelae. Indeed, an increasing number of complications such as post-acute sequelae of COVID-19 (PASC), are being reported in individuals after acute COVID-19 [2,3]. One longitudinal study followed COVID-19 survivors for up to 6-, and 12-month after symptom onset. While most subjects returned to normal life and produced antibody levels, they exhibited a dynamic range of recovery levels [4]. Another longitudinal study described altered T cell responses associated with the disease severity, neutralizing antibody responses and development of PASC [5]. Thus, it is important to document molecular signatures, in addition to antibody levels, in convalescent COVID-19 subjects to better define the normal vs. pathologic convalescence process. This potentially can detect the initiation of aberrant innate immune activation, especially in body fluids that are in direct contact with SARS-CoV-2 [6].
We characterized global immune and proteome responses after SARS-CoV-2 infection in matched plasma and saliva obtained from convalescent COVID-19 subjects (saliva, n = 42; plasma, n = 42), with samples obtained from healthy individuals pre-COVID-19 era serving as healthy controls (saliva, n = 13; plasma = 13). We focused on the analysis of saliva in addition to plasma for the following reasons: 1) saliva is a practical and optimal body fluid to monitor for host and immune-inflammatory markers 2) salivary fluid is a direct surrogate for SARS-CoV-2 antibody responses derived from bronchial-alveolar lymphoid tissues (BALT) [7], 3) oral fluid can reflect systemic reactions to infection since more than 90% of body protein components are detected from saliva [[8], [9], [10]], 4) saliva contains oral microbiome commensals which shape the host immune profile [11,12], and 5) oral inflammation can influence the severity of systemic inflammatory responses [[13], [14], [15]]. A human salivary proteome database was initially developed to explore saliva as a source of mapping markers in health and disease and to further advance comparisons to other body fluids [16]. The current effort is now to share studies related to the composition and function of saliva biofluid (salivaryproteome.org), and make it publicaly available. To date, evidence is lacking to understand the immune and vascular responses present in fluids such as saliva and plasma on a global scale. We aimed to evaluate subjects that experienced and resolved SARS-CoV-2 infection to investigate pathological responses.
We measured and compared SARS-CoV-2 antibody responses in matched saliva and plasma samples with SARS-CoV-2 structural proteins and pseudoviruses bearing the spike (S) used to evaluate binding and virus neutralization, respectively. We then used shotgun proteomics to analyze early immune and host cell-mediated markers that may activate inflammation and endothelial damage during COVID-19 convalescence. Last, we identified altered host pathways potentially impaired by SARS-CoV-2 infection when comparing responses in convalescent COVID-19 subjects to that of healthy controls and by comparing responses in saliva to that of plasma, demonstrating convalescent phase provides signals for continuous damage to the host response.
2. Results
2.1. The salivary antibody repertoire towards SARS-CoV-2 antigens significantly correlated with matched plasma serology
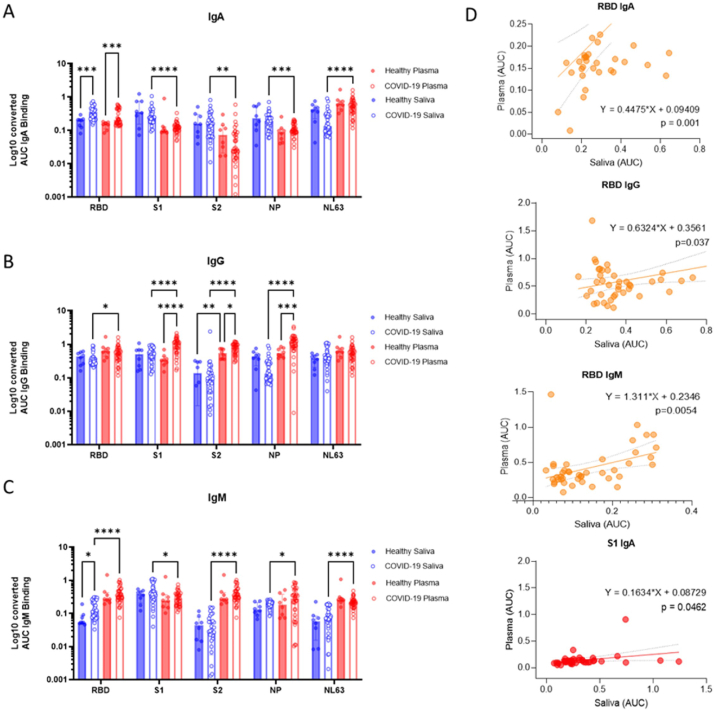
As body fluids are exposed to different antigens, we investigated how SARS-CoV-2 antibody responses in saliva compared to those in matched plasma. Paired saliva and plasma samples from COVID-19 donors in the convalescent phase (20–90 days) were collected and subjected to comparative analyses among demographic factors (age, gender, and initial COVID-19 disease severity), antibody, and proteomic responses (Fig. 1) (Table S1). Samples collected from healthy individuals collected pre-COVID-19 were included for comparison of health vs. COVID-19 (Fig. 1). We first evaluated antibody responses detected in saliva and plasma specific for the SARS-CoV-2 receptor-binding domain (RBD), subunit 1 (S1), and subunit 2 (S2) of the S, and the nucleoprotein (NP) (Fig. 2A–C). Antibodies to common cold coronaviruses were also evaluated by measuring antibodies that bind to the NL63 (NL63) S1 antigen. Our primary interest was in detecting antibodies specific to the RBD and S1 of SARS-CoV-2, the sites responsible for virus binding to cell receptors and major mutation sites. When comparing antibody titers in convalescent subjects versus healthy controls, significant increases were observed in IgA responses for RBD in saliva (p = 0.0001), and RBD in plasma (p = 0.0003). Increases were also found in IgG for S1 (p < 0.0001), S2 (p = 0.0373), and NP(p = 0.0002) antigens in plasma, whereas for IgM responses RBD elevated (p = 0.0118) (Fig. 2A–C). Supplementary Fig. 1A depicted each antibody titer of COVID-19 convalescent samples subtracted with the healthy cut-off (average healthy AUC titer – 2Xstandard deviation of the healthy AUC titer); which exhibits the increased salivary-, and plasma IgA for the SARS-CoV-2 RBD, plasma IgG for S1 and NP, and plasma IgM for the RBD, S2, and NP.
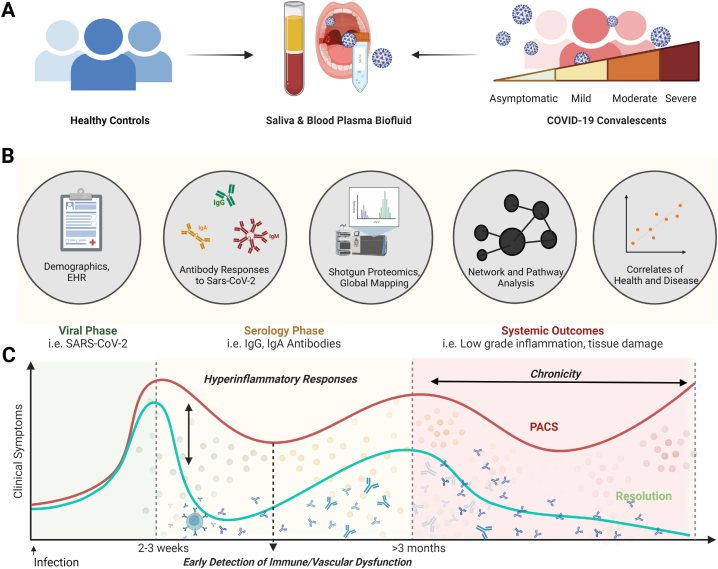
Fig. 1.
Study Design. (A) Saliva and plasma samples were collected from healthy and coronavirus disease (COVID-19) donors during the convalescent phase to investigate the viral-host axis in health versus disease. (B) The serology coupled with the global shotgun proteomic analysis of plasma and saliva samples was conducted in parallel, followed by correlation analyses to demographic factors, antibody-, and proteomic responses. (C) This study was designed to capture the inflammatory response (yellow/red dots) during the start of the convalescent phase (>20 days) after clinical symptom; antibody drawings) and investigate the correlation between biological, and demographic factors. Ultimately, our findings will be applied to discover early detection markers for the post-acute sequelae of SARS-CoV-2 infection. Total samples = 110 samples (Healthy Saliva = 13, Healthy Plasma n = 13, COVID19 Saliva n = 42, COVID19 Plasma = 42).
Fig. 2.
Compartmentalized antibody responses found in saliva and plasma collaborate in response to the SARS-CoV-2 infection. (A–C) The individual area under the curve (AUC) was plotted as blue or red hollow circles (saliva or plasma, respectively). Bars and whiskers represent median and standard deviation, respectively. Mixed-effect analysis with Tukey’s multiple comparisons test was used to measure statistical significance. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. (D) Five paired immunoglobulins showing significant correlation (p < 0.05) between plasma and saliva were depicted as simple linear regression models. The individual titer of saliva was plotted by paired plasma titers. The predicted regression line and deviations were depicted as a solid and dotted lines, respectively. Functions and p-value of regression analyses were indicated next to the regression lines. Correlations of immunoglobulins specific to the SARS-CoV-2 receptor binding site (RBD) or Spike protein 1 (S1) were colored yellow and red, respectively.
We further analyzed the antibody response by disease severity (Fig. S2A&B). In salivary samples, asymptomatic-to-mild individuals had significantly higher IgG for S1 (p = 0.00173), IgM for S1 (p = 0.0003), IgG for NP (p = 0.0115) and IgM for NL63 (p = 0.0297) compared to the healthy controls (pre-COVID-19). The IgG S1 was also increased in moderate and severe cases (p = 0.0007 and 0.0265, respectively). In plasma, significant increase was not observed after stratification by disease severity. In healthy samples, IgA antibodies from a few individuals showed high reactivity to the SARS-CoV-2 S1, resulting in higher median value of healthy groups than asymptomatic-mild groups (Fig. S2A). The same individuals showed high level of IgA titers which bind to the common cold strain (NL63), suggesting that the cross-reactivity between the IgA to the SARS-CoV-2 S1 and NL63 antigens.
While plasma and salivary antibody response showed distinct patterns, significant correlation was observed between paired saliva and plasma were also observed for SARS-CoV-2 RBD or S1 binding immunoglobulins (Fig. 2D). More significant correlations among immunoglobulin subclasses in plasma and saliva are summarized in Table S2. We also examined influence of sample collection timing (ranges between 20 and 90 days) on the pattern of antibody using simple linear regression analyses, but no significant pattern was observed (Fig. S3).
2.2. Immunoglobulin composition and neutralizing functions displayed unique patterns between saliva and plasma
We next investigated the antibody responses to confirm that our subjects were in the COVID-19 convalescent phase and produced protective antibody responses. In convalescent samples, antibody responses to the SARS-CoV-2 RBD antigens showed significant correlation between saliva and plasma; (RBD Saliva IgA vs. RBD plasma IgA (Pearson’s R = 0.701, p-value<0.0001); RBD Saliva IgG vs. RBD plasma IgG (Pearson’s R = 0.701, p-value = 0.037); RBD Saliva IgM vs. RBD plasma IgM (Pearson’s R = 0.422, p-value = 0.005) (Table S2B). Still, the overall antibody profile of saliva was different than plasma. For IgA response, convalescent saliva was significantly higher than convalescent plasma for the following SARS-CoV-2 antigens: S1, S2, NP, and NL63 (p < 0.0001, p = 0.0036 S1, p = 0.0009 S2, and p < 0.0001, respectively) (Fig. 2A). Yet, for IgG responses an opposite trend was found. The titers for plasma were significantly higher than in saliva for SARS-CoV-2 RBD, S1, S2, and NP (p = 0.0178, p < 0.0001, p < 0.0001, and p < 0.0001) (Fig. 2B). The IgM response was also significantly higher in plasma than saliva for all four SARS-CoV-2 and NL63 antigens (p < 0.0001 for RBD, p = 0.0117 for S1, p < 0.0001 for S2, p = 0.0338 for NP, and p < 0.0001 for NL63).
We also evaluated saliva and plasma for neutralizing activity against SARS-CoV-2 Spike bearing pseudovirus particles (rVSV-GFPΔG*Spike) to begin to confirm their functions. (Fig. S1B). Even though the donors in the healthy group were collected pre-COVID-19 era and never encountered the SARS-CoV-2 virus, more than half of their plasma displayed cross reactivity with a significant level of neutralizing activity (62.5%, median IC50 = 271.10). Convalescent COVID-19 subjects showed increased neutralizing activity in plasma with a positive rate and titer (92.16%, median IC50 = 317.30). In contrast, paired saliva samples were lower at neutralizing the pseudoviral particles despite the robust RBD S1-binding IgA antibody responses detected by ELISA (Fig. 2A). After further purification and concentration of the IgAs in saliva [17], neutralizing activity increased.
2.3. A global proteome analysis identified differentially expressed proteins
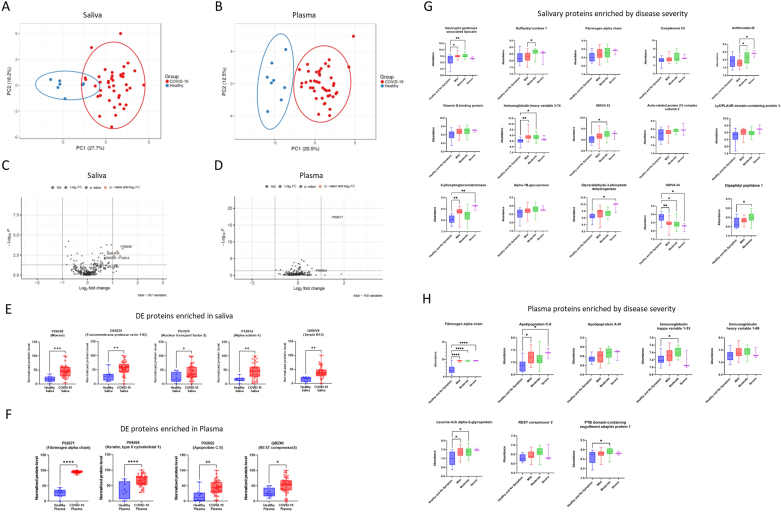
To characterize the oral mucosal and systemic host responses following SARS-CoV-2 infection more comprehensively, we profiled saliva and plasma samples with mass spectrometry and proteomics. Dimension reduction by principal component analysis (PCA) showed a separation of convalescent COVID-19 donors from healthy controls for both saliva and plasma samples (Fig. 3A&B). Differentially expressed (DE) proteins between convalescent versus healthy samples are displayed as in volcano plots (Fig. 3C&D) and all significant observations are summarized in Table S1. There were no DE proteins significantly enriched in healthy over disease (fold change < -2, p-value<0.05 in Fig. 3C&E). There were several DE proteins significantly enriching convalescent over healthy in saliva and plasma (fold change>2, p-value<0.05), are presented as in bar graphs (Fig. 3E&F). In saliva, convalescent COVID-19 samples showed a significant increase in expression of moesin (Uniprot accession number, AC=P26038, fold changes = 2.622, p-value<0.001), transmembrane protease serin D (AC=O60235, fold change = 2.109, p-value = 0.004), alpha-actin-1 (AC=P12814, fold changes = 2.450, p-value = 0.004), nuclear transport factor 2 (AC=P61970, fold changes = 2.251, p-value = 0.0498), and serpin B13 (AC = Q9UIV8, fold changes = 2.293, p-value = 0.001) (Fig. 3C, E and Table S2B). The significantly enriched DEs in plasma of healthy versus disease were the following biomarkers: fibrinogen alpha chain (AC=P0267, fold changes = 3.279, p-value<0.001), Keratin, type II cytoskeletal 1 (AC=P04264, fold change = 2.0231, p-value<0.001), apolipoprotein C-II (AC=P02655, fold changes = 3.688, p = 0.027), and REST corepressor2 (AC = Q8IZ40, fold changes = 2.247, p-value = 0.0122) (Fig. 3D, F, and Table S2A). We also discovered proteins enriched by the COVID-19 disease severity (No symptom (n = 2), mild (n = 18), moderate (n = 19) and severe (n = 2) (Fig. 3G&H) (Table S4B); including salivary neutrophil gelatinase-associated lipocalin, salivary sulfhydryl oxidase 1, salivary fibrinogen alpha chain, salivary complement C5, salivary antithrombin III, salivary and plasma fibrinogen alpha chain (Fig. 3G&H).
Fig. 3.
Comparative proteomic analyses revealed differentially expressed proteins (DEs) enriched in convalescent COVID-19 saliva and plasma. (A&B) Dimension reduction showed a separation of convalescent COVID-19 donors from healthy controls in saliva (A) and plasma (B). A nonparametric Wilcoxon rank sum test was applied to select subsets of proteins (p-value <0.01; 22 and 32 proteins were identified for saliva and plasma, respectively) before principal component analysis (PCA) was performed to visualize the samples. Circles indicate 95% confidence intervals of group memberships. Percentages along the axes indicate the degree of variance explained by that principal component. (C&D) Fold changes of protein expression of convalescent COVID-19 over healthy samples were plotted by negative log-transformed p-values in volcano plots. Dotted lines of the volcano plot represent thresholds for the fold changes (Log2 fold changes >1) and statistical significance (p < 0.05). (E&F) Differentially expressed proteins (Log2 fold changes>1, p < 0.05) in saliva and plasma, were depicted as in a relative abundance of five significantly up-regulated proteins. Individual dots represent individual values. The interquartile range, median, and min/max values were illustrated as box, middle line, and whiskers, respectively. Mixed-effect analysis with Tukey’s multiple comparisons test was used to measure statistical significance. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001. (G&H) Proteins differentially expressed (p < 0.05) by disease severity in saliva and plasma were depicted as in relative abundance. The interquartile range, median, and min/max values were illustrated as box, middle line, and whiskers, respectively. Mixed-effect analysis with Tukey’s multiple comparisons test was used to measure statistical significance. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001; ****p ≤ 0.0001.
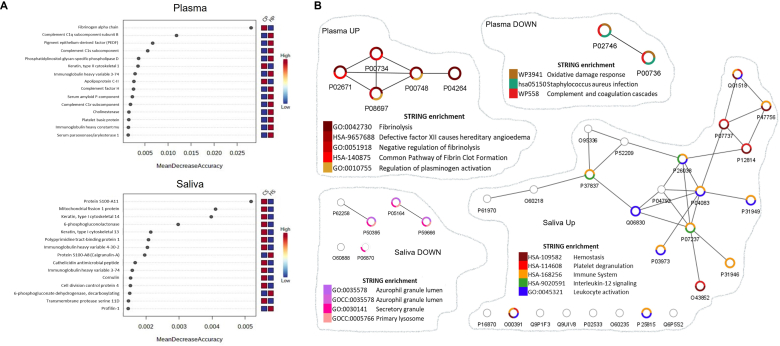
The proteomic signatures were confirmed by random forest (RF) machine learning and network enrichment analyses (STRING [citation]) (Fig. 4). The RF analysis showed that fibrinogen alpha chain and protein S100-A11 are the most important proteins that can differentiate healthy vs. convalescent COVID-19 in plasma and saliva samples, respectively (Fig. 4A). Network analyses performed based on differentially expressed proteins between convalescent COVID-19 and healthy samples showed that the convalescent plasma proteome displayed suppressed biological functions involved in oxidative damage response, antimicrobial properties against opportunistic infection (Staphylococcus aureus infection) and complement and coagulation cascades (Fig. 4B). In contrast, the pathways enriched in convalescent COVID-19 plasma were all associated with fibrin clot formation (Fig. 4B). Pathways enriched in convalescent samples included hemostasis, platelet degranulation, immune system, interleukin-12 signaling, and leukocyte activation (Fig. 4B). Pathways related to granule or lysozyme formation were suppressed in saliva from convalescent COVID-19 donors (Fig. 4B).
Fig. 4.
Network analyses depicted altered pathways and functions in convalescent plasma and saliva. (A) Random forest machine learning identified proteins contributing the most to the accuracy of COVID-19 classification. Top 15 DEs in plasma (top) and saliva (bottom) were depicted in rank order of mean decrease accuracy. (B) In parallel, interactions among DE proteins were predicted by the STRING enrichment analyses and depicted as network maps using Cytoscape. Pathways associated with each DE protein were depicted in a donut graph, color-coded based on terms discovered by the STRING enrichment assay. CP: COVID-19 plasma, HP: Healthy plasma, CS: COVID-19 Saliva, and HS: Healthy Saliva.
2.4. Altered proteomic functions in COVID-19 convalescent saliva directly correlated with the expression of RBD-binding IgA response in saliva
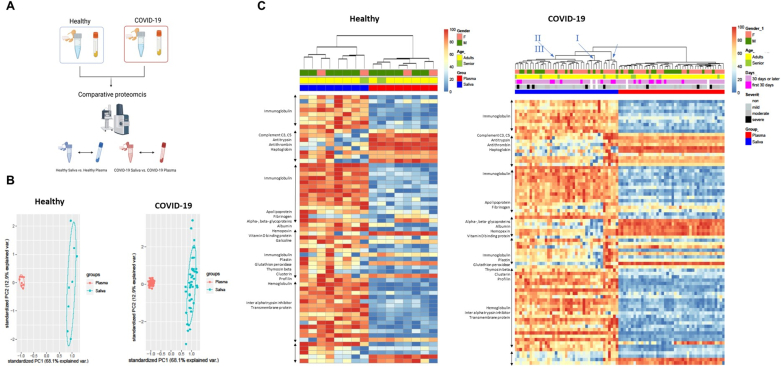
Comparative proteomic analyses between healthy vs. convalescent COVID-19 suggested that inflammatory markers induced by SARS-CoV-2 remained in both body fluids during the recovery phase. To better understand the inflammatory patterns that occur in the oral local mucosal and systemic immune system, we performed a separate set of analyses that compared the salivary and plasma proteome (Fig. 5A). The PCA analysis showed a clear separation between the saliva and plasma proteome for both healthy and convalescent COVID-19 samples (Fig. 5B). Hierarchical clustering heatmaps were clustered into two groups based on the origin of samples (saliva vs. plasma) while demographic factors (gender, age, and acute COVID-19 disease severity) did not contribute to the clustering. (Fig. 5C). In a healthy state, most DE proteins were higher in saliva, but proteins related to coagulation pathways (complement C3, C5, antithrombin) were higher in plasma (Fig. 5C). The convalescent COVID-19 heatmap had a similar pattern to the healthy heatmap, except that the expression pattern of apolipoprotein and fibrinogen was reversed (saliva > plasma in healthy; plasma > saliva in COVID-19, Fig. 5C). The cluster of saliva in the convalescent COVID-19 heatmap further diverged into three subclusters (Blue arrows and roman numerals on top of Fig. 5C), providing new clustering of proteins derived from the humoral immune response, clotting factors, and innate inflammatory responses. Each cluster was labeled as I, II, and III in increasing order of protein expression level.
Fig. 5.
Comparative proteomic analyses between saliva and plasma revealed the heterologous signatures of each biofluid and further divarication of convalescent COVID-19 saliva. (A) Proteomics data were further analyzed to compare proteomic composition between saliva vs. plasma. The data obtained from healthy and convalescent COVID-19 participants were separately analyzed. (B) Differentially expressed proteins (Log2 fold changes>1, p < 0.05) in saliva and plasma, were depicted as in a relative abundance of five significantly up-regulated proteins. The 95% confidence intervals of group memberships and degree of variance were indicated as circle and percentages on the axes, respectively. (C&D) The heterologous proteomic profile between saliva and plasma was further analyzed using clustered heatmap for both healthy and convalescent COVID-19. Relative abundance was calculated based on the proportion of normalized reads and displayed as color gradients. A dendrogram was constructed based on hierarchical clustering of relative abundances and color-coded demographic information of each participant was added to show their association with each clade. Blue Roman numerals and arrows indicate subclades in COVID-19 saliva. The numbering was in crescent with the expression level of DE proteins.
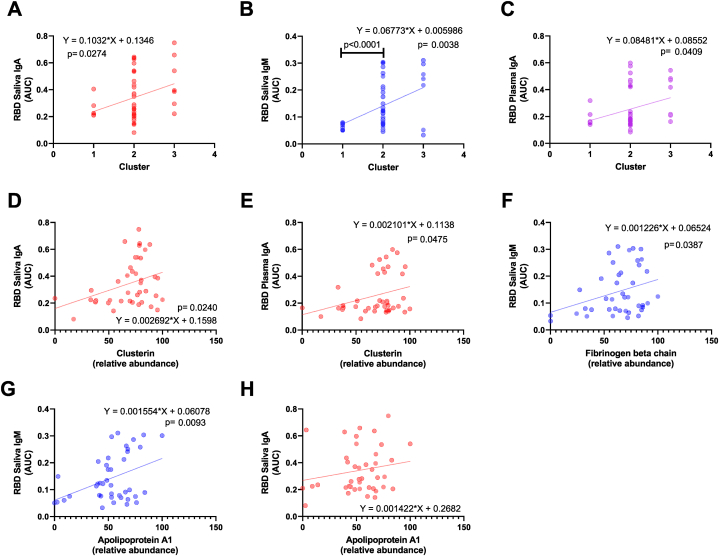
We then determined the influence of the immune subclusters in relation to serological results for each fluid by investigating correlations with immunoglobulin levels (Fig. 6A–C). Strikingly, significant correlations were observed with the RBD-binding saliva IgA, IgM, and plasma IgA titers, as in a linear increase by the order of subcluster numbers (p = 0.0274, p = 0.0038, and p = 0.0409, respectively, Fig. 6A–C). Since we observed an increasing trend in the expression level of DE protein, we also performed a correlation analysis between each RBD binding immunoglobulin with each DE protein involved in the convalescent COVID-19 saliva sub-clustering (Fig. 6D–H). The clustering showed significant positive correlations with RBD binding IgA in both saliva and plasma (Fig. 6D&E). Fibrinogen beta chain was significantly correlated with RBD binding IgA in saliva (Fig. 6F), and Apolipoprotein A1 was correlated with the RBD binding salivary IgM and plasma IgA (Fig. 6G&H).
Fig. 6.
Correlation analyses suggest that proteomic alterations in convalescent saliva are associated with antibody responses specific to the receptor binding site (RBD) of SARS-CoV-2. (A–C) A significant correlation (p < 0.05) between SARS-CoV-2 RBD specific immunoglobulins and convalescent COVID-19 salivary sub-clusters was depicted as simple linear regression models. The individual titer of immunoglobulin was plotted by subcluster numbers. The predicted regression line and deviations were depicted as solid and dotted lines, respectively. Functions and p-value of regression analyses were indicated next to the regression lines. (D–H) Three differentially expressed proteins responsible for the clustering were illustrated as simple regression models as described above.
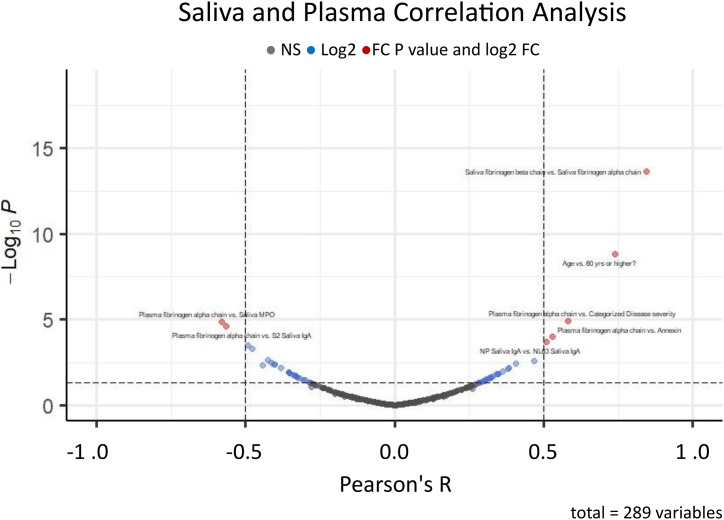
Lastly, we investigated correlations among parameters of interest (RBD binding immunoglobulins, fibrinogen proteins, and demographic factors) (Fig. 7). Significant positive correlations were observed between saliva fibrinogen beta chain and saliva fibrinogen alpha chain; plasma fibrinogen alpha chain and categorized disease severity (asymptomatic = 0, mild = 1, moderate = 2, and severe = 3), plasma fibrinogen alpha chain and salivary annexin; and NP Saliva IgA and NL63 saliva IgA. Significant positive correlations were between plasma fibrinogen alpha chain versus saliva myeloperoxidase (MPO); and plasma fibrinogen alpha chain and S2 saliva IgA.
Fig. 7.
Correlations among RBD binding immunoglobulins, fibrinogen proteins, and demographic factors. Pearson’s correlation R among RBD binding immunoglobulins, fibrinogen proteins, and demographic factors were plotted by negative log-transformed p-values in volcano plots. Dotted lines of the volcano plot represent thresholds for the R (0.5 and −0.5) and statistical significance (p < 0.05). Blue: values over R thresholds, Red: values over R thresholds with statistical significance.
Together, our results indicate that measurement of only antibody levels during the COVID-19 convalescent phase does not provide a full picture of the host response mounted after SARS-CoV-2 infection. Indeed, while we confirmed that our convalescent COVID-19 subjects had produced antibodies, our global proteomics analysis revealed novel aberrant immune signatures and clotting dynamics in plasma and saliva when compared to healthy controls. These biomarkers of inflammation and host response dysbiosis were unknown before. Overall, we demonstrated that population-based investigations of saliva can be used to map global host responses to local mucosal and systemic functions in addition to the characterization of antibody responses.
3. Discussion
Increasing evidence indicates that the immune and endothelial health of convalescent COVID-19 subjects may be compromised when compared to healthy controls [6]. Recovery from inflammatory responses to viral infection is mediated by multiple systems [18] and dependent on the overall host response. Molecular delineation of such components during physiological recovery versus pathologic transition is pivotal to develop host-directed strategies, aiming to sustain physiological function [19]. Our findings also demonstrate that abnormal inflammatory and clotting responses can be identified in both saliva and plasma fluids of convalescent COVID-19 subjects. This suggests that even when SARS-CoV-2 antibody responses are mounted, the COVID-19 convalescent phase can potentially reveal residual aberrant responses following an acute viral infection. Notably, we highlight saliva as an important and accessible fluid that can be harvested in a non-invasive manner. Salivary fluids are ideal to identify not just antibody responses, but also diverse immune pathways, including mucosal immunity, innate immune responses, neutrophil functions, and clotting pathways.
Carefully designed serosurveillance studies that investigate antibody responses rely solely on analyzing blood-derived fluids [20,21] and not saliva fluids. Here, we successfully detected SARS-CoV-2-specific antibodies during the early phase of the COVID-19 pandemic and clearly demonstrated that de novo generation of SARS-CoV-2 specific antibody responses by the first exposure to the virus. In accordance with current knowledge [22], the antibody response in plasma displayed distinct patterns from saliva. In plasma, there were significant increase of IgG responses in COVID-19 samples over healthy controls, such as S1 specific plasma IgG (p < 0.0001), S2 specific plasma IgG (p = 0.0373), and NP specific plasma IgG (p = 0.0002) (Fig. 2B). In the salivary antibody responses, we were able to observe a significant increase in RBD specific IgA response (p = 0.0001). Significant correlations between paired saliva and plasma showed positivity for SARS-CoV-2 RBD or S1 binding immunoglobulins, indicating that saliva is an available biofluid for monitoring the presence of protective antibody responses and immune responses. There were several similarities in antibody profiles detected between biofluid types; we also found unique patterns within saliva that distinguishes it from blood plasma. The IgA response in convalescence was significantly higher in saliva than in plasma, whereas the IgG response showed an opposite trend in that the titers in convalescent plasma were significantly higher than in saliva. This is expected as IgG is the dominant isotype in the blood [23], while IgA is found in mucosal tissues [24]. To date, however, evidence on antibody responses and neutralization levels to SARS-CoV-2 provided a limited range of information regarding the immune responses and pathogenesis of subjects that recovered, or not, from the natural infection.
Next-generation plasma profiling demonstrates a comprehensive overview of the immune response and has the potential to elucidate the impact of COVID-19 on the host. Zhong et al. showed that more than 200 proteins were found significantly different in plasma levels at the time of infection as compared to 14 days later [25]. In comparison to Zhong et al.’s findings, our plasma proteome appears to reflect a recovery process, displaying much fewer numbers of significantly enriched DE proteins (p-value<0.05, fold change>2) (Fig. 3D and Table S1). We speculate that the waning host response in the convalescent period contributed to less proteins detected. Yet, the participants of our cohort still displayed a significant enrichment of fibrinogen in plasma and in saliva. Random forest machine learning or correlation assay showed that the plasma fibrinogen is the most accurate biomarker to differentiate convalescent COVID-19 plasma (Fig. 4A) from the healthy group and correlated with salivary inflammatory markers (annexin or MPO), or salivary IgA responses (Fig. 7). A recent study also highlighted the role of fibrinogen in the pathogenesis of post-acute sequelae of COVID-19 [26] Taken together, plasma and saliva fibrinogen Levels are important target for understanding the resolution phase of COVID-19.
If not limited to the proteins upregulated by 2 fold or higher, convalescent plasma showed an increase in numerous proteins associated with neutrophil functions or migration, such as annexin 1 [27,28], antileukoproteinase [29], and Matrix metalloproteinase-9 [30,31] (Table S1A). Interestingly, salivary proteome appears to maintain activated inflammatory status longer than plasma, as significantly increased neutrophil activation markers, myeloperoxidase (MPO), annexin 1-2, alpha-actinin-1, and nuclear transport factor 2 are involved in the migration of neutrophils [[32], [33], [34]]. The convalescent saliva fluid also showed a significant increase in transmembrane protease serine 11D, which is known to activate the SARS-CoV-2 spike protein and facilitate the viral-cell fusion process [35,36], and serpin B13, known for regulating neutrophil serine proteases [37] and inflammatory caspases [38](Table S1B). Other interesting proteins found in our study are the inhibitors of cathepsin, which have been involved in viral cell entry and replication [39]. This was found to be significantly higher in plasma during acute infection versus convalescent COVID-19 cases and their analysis demonstrated that a group of patients display a “disease profile”, despite not having no symptoms of the disease [25]. In a recent study, the COVID-19 patients recovered from both non-severe and severe cases exhibited prolonged abnormality in plasma proteomic, and metabolomic profiles in 6 months after discharge [40]. A prolonged inflammatory status warrants hidden disease progress in seemingly recovered patients. Especially, long-term upregulation of neutrophil activation markers, such as MPO, is observed to trigger autoimmune activation which is associated with the development of Long COVID [[41], [42], [43], [44]]. It warrants the need of follow-up COVID-19 patients after recovery and discovery of early biomarkers. Dissecting proteomes of biofluids collected during the convalescence phase will be instrumental for a targeted approach in Long COVID research.
We further demonstrated that salivary IgA antibody responses to SARS-COV-2 could be involved in neutrophil-fibrinogen interactions at the oral mucosal surface. The plasma fibrinogen alpha chain displayed a significant correlation with salivary markers, such as salivary RBD IgA and salivary IgM (Table S2). Significant correlations were also observed between the salivary RBD IgM and salivary RBD IgA; salivary RBD IgM and salivary fibrinogen beta chain; salivary RBD IgM and severity of clinical illness during the acute disease phase. This suggests that salivary antibodies to the SARS-CoV-2 infection participate in the inflammatory response mediated via neutrophil extracellular traps [45] (NETs)-fibrin in oral mucosa and possibly contribute to the systemic inflammatory response represented by enhanced plasma fibrinogen. In severe COVID-19, neutrophil degradation and NETosis in blood and in the lung have been functionally linked to severe inflammation and thrombosis [[46], [47], [48], [49], [50], [51]]. Abnormal fibrinolysis is known to impact networks with neutrophil functions, including NET formation [47,52,53]. Of note, our correlation analysis between disease severity and protein abundance data revealed that markers associated with neutrophil activation (Neutrophil gelatinase-associated lipocalin) and clotting factors (salivary complement C5, antithrombin-III, and fibrinogen alpha chain in both saliva and plasma) (Fig. 3G&H and Table S4B). Indeed, excessive release of NETs, with a low resolution of inflammation, can lead to immune thrombosis in blood vessels, with NET-fibrin interactions contributing to the severity of tissue injury and pathogenesis [54]. Unique to the oral organ, the NET-fibrin axis also plays a unique role in regulating the constant deposition of fibrin produced by the commensal microbiome-triggered inflammation [55].
Other specific drivers of the abnormal inflammatory and clotting responses observed in our convalescent COVID-19 subjects require further study. Several research teams have detected SARS-CoV-2 or viral proteins as evidence of "viral reservoirs" or viral persistence collected from subjects’ months after acute COVID-19 [56,57]. For example, Gaebler et al. identified SARS-CoV-2 RNA and protein in 7 of 14 intestinal tissue samples obtained from asymptomatic COVID-19 patients with negative nasal-swab PCR at an average of 4 months after acute disease [58]. The SARS-CoV-2 S antigen S1 itself appears capable of directly interacting with platelets and fibrinogen to drive blood hypercoagulation [59]. This suggests that further studies of convalescent COVID-19 saliva and plasma would benefit from the measurement of SARS-CoV-2 RNA and spike antigen in addition to inflammatory and proteomic signatures. SARS-CoV-2 persistence in intestinal tissue or the oral mucosa, and possible shedding of spike antigen into saliva or blood, could also perpetuate chronic inflammatory and clotting sequelae.
The molecular mechanisms underlying higher concentrations of IgA but lower IgA neutralizing activity in convalescent saliva also require further exploration. It is possible that higher salivary IgA concentrations represent some form of extended antibody-mediated disease enhancement. Antibody-mediated disease enhancement has been reported in diverse RNA viral diseases, such as influenza, SARS-CoV-2, Dengue, and human immunodeficiency viruses infections [[60], [61], [62], [63]]. One team found that SARS-CoV-2 RBD-specific neutralizing dimeric IgAs isolated from nasal turbinate could facilitate viral infection, transmission, and injury in Syrian hamsters [64]. Aleyd et al. [65] demonstrated that IgA enhances NETosis as an effective defense mechanism to eliminate pathogens at mucosal surfaces. In contrast, neutrophil activation by IgA immune complex is also known to contribute to the immunopathogenesis of autoimmune diseases, such as IgA vasculitis, and nephropathy [[66], [67], [68]]. In respiratory viral disease models, such as influenza and SARS-CoV-2, the formation of an IgA-virus immune complex led to exacerbated NETosis of neutrophils isolated from peripheral blood mononuclear cells (PBMCs) ex vivo [69].
Our study has a limitation by subject enrollment, specifically, the types and numbers of volunteers during the early phase of the pandemics. We were not able to equalize the sample size of control (n = 13) with the COVID-19 group (n = 42) with a total of 110 samples; and for proteomics we had control (n = 8) with the COVID-19 group (n = 42), with a total of 100 samples . The samples were collected at UCSD as patients arrived in the clinic, which was not an ideal setting for a statistical power but still we were able to compare antibody and proteomic results between convalescent samples and healthy control. A few previous studies were also conducted in a setting with unequal healthy control sample size. Zoodsma et al. performed targeted proteomics using samples collected from 3 cohorts: 350 hospitalized COVID-19 patients, 186 post-COVID-19 individuals, and 61 healthy controls [70]. Despite the unequal sample size, they were able to discover biomarkers for differentiating disease severity and recovery rate (e.g., metalloproteinase inhibitor 1 (TIMP1), vascular endothelial growth factor A (VEGFA), and TRAIL-R2). Another case, Sharif-Askari used samples collected from 18 healthy controls and 62 patients (15 asymptomatic, 16 mild, 15 moderate and 16 severe) [71]. Similar to our study, and with limitation in that sample size, samples were collected at only a one-time point, as a cross-sectional study. After metabolomics they found biomarkers for severe cases (e.g., Sphingosine and 5-Aminolevulinic acid). Here, the antibody levels or proteomic responses were not adjusted by the different baseline of each individual intervariability. Since we did not have matching pre-pandemic controls from the same subjects, the comparisons here focus on relative numbers from health versus COVID-19 pandemics yet clear markers were found and further explorations will investigate them mechanistically. As shown in Fig. 2 and Supplementary Fig. 1A&B, the healthy controls already presented a considerable level of antibodies binding to the SARS-CoV-2 proteins and neutralizing pseudovirus particles. The pre-existing antibody response to the SARS-CoV-2 virus in pre-pandemic samples has also been described elsewhere [[72], [73], [74], [75], [76]], which is suspected to be the cross-reactive response from prior endemic corona virus infections. The high baseline titer might have reduced the power to detect increases of antibody responses to the infection. Therefore, our findings are focused on predictive correlations. While the study subjects were able to report the severity of their acute COVID-19 illness (asymptomatic, mild, moderate, and severe), their clinical symptom data after convalescent phase were not obtained when the saliva and plasma samples were collected. However, we had limited access to the crucial information, such as how long the symptoms last since the onset (and whether the symptoms existed during sample collection), exact viral titer at the moment of diagnosis and presence of other underlying conditions.
Most of our samples were from mild to moderate cases, which showed relatively less separation from healthy controls. A few of previous studies conducted metabolomics on salivary samples showed that simple comparison between healthy and COVID-19 cases showed less discriminating ability than comparison between severe and mild cases [71,77]. In PCA analyses, salivary samples were clustered into three subgroups (non-infected controls, severe, and non-severe patients) while plasma samples were clustered in to four subgroups (non-infected controls, asymptomatic, severe, and non-severe patients). Frampas et al. reported similar findings in that salivary metabolomic data were the most clearly separated for the severe COVID-19 cases from low-severity or negative cases [77]. Since our study included only 2 severe cases, we had to focus on comparison between healthy vs. COVID-19 cases. Despite that the analyses setting might have reduced the possibility to discover novel biomarkers, our analyses were able to present distinctive feature of convalescent salivary, and plasma samples at convalescent phase of the COVID-19; suggesting a potential underlying mechanism related with the development of chronic sequelae of the COVID-19.
Pre-existing neutralizing activity in the absence of prior viral exposure has been described in multiple studies. Wang et al. reported that such pre-existing immunity to the SARS-CoV-2 virus is mainly derived by cross-reactive immune cells elicited by other coronaviruses, such as OC43 and HKU1 [78]. Ng et al. identified functionally relevant antigenic epitopes conserved among coronaviruses: SARS-CoV-2 spike protein showed closer homology with alphacoronaviruses HCoV-NL63 and HCoV-229E [73]. The pre-existing immunity could provide partial protection and boost-effect to the immunizations. But it can also result in antibody-dependent disease enhancement and intervene vaccine effectiveness. Especially in serologic studies, the cross-reactive antibodies can increase the baseline titer and complicate defining seroreactivity thresholds. Also in our study, healthy control samples exhibited binding activity to the SARS-CoV-2 antigens and neutralizing activity to the pseudo-viral particles (Fig. 2 and Supplementary Fig. 1, respectively). We did detect significant differences between healthy control and COVID-19 groups for the SARS-CoV-2 specific IgG response (Fig. 2). To overcome such limitations, future studies will tract the antibody response in paired saliva/plasma samples performed in larger cohorts over an extended time. Our data will be instrumental to design future studies, particularly to perform power-analyses for determination of cohort size.
The 22 significantly different salivary proteins, as listed in the Supplementary Table S3, were mostly associated with inflammation/infection pathways based on the STING enrichment analysis, which can potentially be linked to cell types for innate immunity, and neutrophil pathways. Our findings confirmed that MPO and Annexin upregulated in the COVID-19 convalescent group, which are important regulators for myeloid cells [79]. Also, our analysis identified S100A [80], which is reported as a predictor for severe forms of COVID-19, and GRN, which is reported to increase in saliva of COVID-19 patients [81]. In traditional machine learning classification, feature selection is one of the most important data processing steps. Feeding all measured features into machine classification or prediction usually misleads the classifiers and destroys the small but important mechanistic patterns that can only be found by using a subset of highly relevant features such as critical protein pathways or gene modules. However, identifying these unknown subsets of proteins in a fully unsupervised way (i.e., without prior knowledge) is highly challenging due to the curse of dimensionality. We carefully designed and implemented a specific feature selection solution for our study. Given protein abundances measured across samples, statistical tests are commonly used to quantify the importance of the proteins for sample classification. The protein abundance data in the COVID-19 convalescent study have gone through preprocessing steps including listwise deletion amputation and log transformation. Because we cannot assume the preprocessed data still follow the original distribution, a nonparametric Wilcoxon Rank Sum test was applied to compare the abundance of each salivary protein between the healthy control samples (N = 13) and the COVID-19 convalescent samples (N = 42). We found COVID-19 convalescent samples and the healthy controls are in clearly different clusters when the top 22 proteins were used, while the samples could not be separated when all the proteins were used. Because the healthy group, the proteomics sampls had only 8 samples for controls, we could not use cross-validation to evaluate the accuracy of classification. However, we did a pathway enrichment analysis to interpret these 22 proteins. Not only we found that differences on many of the 22 proteins (PCA plot, Fig. 3 A and B) meet our previous understanding about the COVID-19, but also most of these proteins were from inflammation-associated pathways, indicating persistent immune changes towards Sars-CoV-2.
Future studies would benefit from requiring convalescent COVID-19 subjects to report possible chronic symptoms longitudinally. This is especially pressing since up to 30% of patients infected with SARS-CoV-2 are developing a wide range of persistent symptoms that do not resolve over months or years [82]. These patients are being given the diagnosis LongCovid or Post-Acute Sequelae of COVID-19 (PASC) [83]. Persistence of SARS-CoV-2 in tissue, aberrant immune signaling, and microclot formation have been documented in PASC [84,85], but early molecular signs indicating direct risk to chronic symptoms have been elusive. Our current study sets the stage for global immune and proteome analyses that characterize inflammatory and clotting processes in PACs saliva and plasma in a manner that may be able to elucidate key aspects of the disease process and contribute to the development of targeted therapeutics.
4. Materials & methods
The research reported in this manuscript complies with all relevant ethical regulations and informed consent was obtained from all human participants. Additional information was collected on donor demographics (age and gender).
Ethics approval
This study has been approved by the University of California, San Diego Institutional Review Board (IRB, no. 200236X) and the J. Craig Venter Institute Review Board (IRB, no. 2020-286).
4.1. Experimental study design
Blood and saliva samples were collected from convalescent COVID-19 donors who visited the COVID clinic at the University of California, San Diego between March 2020, and June 2020 Samples were collected in the convalescent phase (20–90 days post initial symptoms), when most symptoms were resolved (n = 42). Confirmed COVID-19 cases were defined either by PCR or antigen test as previously described [86]. Throughout the sample collection, the original SARS-CoV-2 strain circulating throughout the study was the original strain (USA-WA1/2020) and the vaccine against SARS-CoV-2 was not available. For comparison, we included healthy donors (n = 13) from the pre-pandemic era, and subjects recruited to the study signed the institutional review board (IRB)– approved consent form (# 2018-268) [87]. To equalize sample size and statistical comparison the following was done: 1) healthy (n = 13), asymptomatic to mild (n = 21), moderate (n = 19) and severe (n = 2). This leads to a total n = 110 (Healthy Saliva = 13, Healthy Plasma n = 13, COVID19 Saliva n = 42, COVID19 Plasma = 42). The study models are shown in Fig. 1. All COVID-19 subjects were included across the studies, yet for proteomics, healthy controls (n=8, saliva, n=8 plasma) were evaluated. Overall study design was summarized in Fig. 1 and Table S1.
Peripheral blood samples were collected by venipuncture and collected into BD vacutainer SST tubes (Vitality Medical, Salt Lake City, Utah). After 1hr., the collected blood sample was centrifuged for plasma separation. Saliva was collected by the “passive drool technique” using the Saliva Collection Aid (Salimetrics, Carlsbad, CA). All samples were aliquoted and stored at −80 °C for long-term storage.
The general experimental approach was summarized in Fig. 1. Briefly, all collected plasma and saliva samples were tested for SARS-CoV-2 specific antibodies by enzyme-linked immunosorbent assay (ELISA) and pseudovirus neutralization assay (Fig. 2 and Suppl. Fig. 1, respectively). Correlations among all immunoglobulins (Ig) were investigated with Pearson’s correlation and simple linear regression analysis using the GraphPad Prism version 8.3.1. In parallel, separate sets of samples were processed and used for mass spectrometry to detect host antiviral-, and microbial proteins and peptides (Fig. 3, Fig. 4, Fig. 5). In the end, all collected data were collectively analyzed to verify the interaction among systemic and oral mucosal immune responses to the SARS-CoV-2 infection (Fig. 6).
4.2. Antibody responses
SARS-CoV-2 binding ELISAs Plasma and saliva samples were tested for binding to recombinant SARS-CoV-2 using an Enzyme-linked immunosorbent assay (ELISA) according to the manufacturers with slight modifications and as previously described the recombinant spike protein from human coronavirus NL63 (NL63) was also included as a coating antigen to estimate the presence of cross-reactive antibodies to common cold coronaviruses. All procedures were repeated twice, once manually and once by using Hamilton Microlab STAR (Hamilton, Reno, NV). Briefly, ELISA plates (Nunc MaxiSorp™ flat-bottom, Thermo Fisher Scientific, Waltham, MA) were coated with antigen (10ng/50 μL) at 4 °C overnight. Four different coating antigens were included; SARS-CoV-2 Spike Glycoprotein (S1) RBD, His-Tag (HEK293) (NativeAntigen, Oxfordshore, United Kingdom), SARS-CoV-2 Spike Glycoprotein (S1), His-Tag (Insect Cells) (NativeAntigen, Oxfordshore, United Kingdom), SARS-CoV-2 Spike Glycoprotein (S2), His-Tag (Insect Cells) (NativeAntigen, Oxfordshore, United Kingdom), SARS-CoV-2 Nucleoprotein, His-Tag (E. coli) (NativeAntigen, Oxfordshore, United Kingdom), and Human Coronavirus NL63 Spike Glycoprotein (S1) His-Tag (HEK293) (NativeAntigen, Oxfordshore, United Kingdom). The next day, coated plates were washed three times with PBS-Tween (0.05%) and blocked with 200 μl of 5% milk blocking solution at room temperature for 30 min. During incubation, plasma and saliva samples were initially diluted 1:54 and 1:2, respectively, and three-fold serial dilution was performed. After blocking, diluted samples were added to the wells (50 μL/well) and incubated for 1 h at room temperature. After 4X washing, 100 μL of 1:5000 diluted Goat anti-human secondary antibody was added into each well and incubated at 37 °C for 1 h (Goat Anti-Human IgG ɣ Chain Specific HRP conjugated, species Adsorbed (Human IgM, IgD, and IgA) Polyclonal Antibody for IgG (Cat# AP504P, EMD Millipore, Burlington, MA), Goat Anti-Human IgA, a-chain specific Peroxidase conjugate for IgA (Cat# 401132-2 ML, Calbiochem, San Diego, CA) for IgA, and Goat Anti-Human IgM Fc5μ Fragment specific HRP conjugated secondary antibody for IgM (Cat# AP114P, EMD Millipore, Burlington, MA). After the incubation, plates were washed four times and 200 μL of the substrate (cat# P9187, Sigma, St. Louis, MO) was added for color development. After incubation in a dark room for 20 min, the reaction was stopped by the addition of 50 μL 3 M H2SO4, and plates were read at 492 nm [88]. A non-linear regression curve was first determined and then the area under the curve (AUC) was calculated using GraphPad Prism version 8.3.1 (GraphPad Software, Inc., San Diego, CA, USA).
4.3. Generation of pseudo-virus (rVSV-GFPΔG*Spike)
For pseudoviruses construction, spike genes from strain Wuhan-Hu-1 (GenBank: MN908947) were codon-optimized for human cells and cloned into eukaryotic expression plasmid pCAGGS to generate the envelope recombinant plasmids pCAGGS.S as described previously with slight modifications [89]. For this VSV pseudovirus system, the backbone was provided by VSV G pseudotyped virus (G*ΔG-VSV) that packages expression cassettes for firefly luciferase instead of VSV-G in the VSV genome. Briefly, 293T cells were transfected with pCAGGS.S (30 μg for a T75 flask) using Lipofectamine 3000 (Invitrogen, L3000015) following the manufacturer’s instructions. Twenty-four hours later, the transfected cells were infected with G*ΔG-VSV with a multiplicity of four. Two hours after infection, cells were washed with PBS three times, and then a new complete culture medium was added. Twenty-four hours post-infection, SARS-CoV-2 pseudoviruses containing culture supernatants were harvested, filtered (0.45-μm pore size, Millipore, SLHP033RB), and stored at −70 °C in 2-ml aliquots until use. The 50% tissue culture infectious dose (TCID50) of SARS-CoV-2 pseudovirus was determined using a single-use aliquot from the pseudovirus bank; all stocks were used only once to avoid inconsistencies that could have resulted from repeated freezing-thawing cycles. For titration of the SARS-CoV-2 pseudovirus, a 2-fold initial dilution was made in hexaplicate wells of 96-well culture plates followed by serial 3-fold dilutions (nine dilutions in total). The last column served as the cell control without the addition of pseudovirus. Then, the 96-well plates were seeded with trypsin-treated mammalian cells adjusted to a pre-defined concentration. After 24 h incubation in a 5% CO2 environment at 37 °C, the culture supernatant was aspirated gently to leave 100 μl in each well; then, 100 μl of luciferase substrate (Perkinelmer, 6066769) was added to each well. Two min after incubation at room temperature, 150 μl of lysate was transferred to white solid 96-well plates for the detection of luminescence using a microplate luminometer (PerkinElmer, Ensight). The positive well was determined as ten-fold relative luminescence unit (RLU) values higher than the cell background. The 50% tissue culture infectious dose (TCID50) was calculated using the Reed–Muench method, as described previously [90].
4.4. Pseudovirus neutralization assay
Neutralizing activity against rVSV-GFPΔG*Spike was determined as previously described with slight modification [91]. Briefly, Vero cells were seeded at a density of 2.5 × 104/50 μL in a Greiner Bio-One™ CellStar™ μClear™ 96-Well, Cell Culture-Treated, Flat-Bottom, Half-Area Microplate (Thermo Fisher Scientific, Waltham, MA). The next day, the cell monolayer was rinsed with 0.01 M PBS (Thermo Fisher Scientific, Waltham, MA). Due to the contamination by the commensal bacteria in saliva, total IgA was purified from saliva using Peptide M/agarose (InvivoGen Inc., San Diego, CA, USA) and used for the neutralization at low-dilution (1:2–1:10), as previously described [17]. Plasma and saliva IgAs were three-fold diluted (starting from 1:50 and 1:2 dilution, respectively) with infection media (DMEM medium (cat# 11995065, Thermo Fisher Scientific, Waltham, MA) containing 2% fetal bovine serum (FBS)). Twenty-five μL of diluted samples was incubated with the same volume of pseudovirus (rVSV-GFPΔG*Spike) at 37 °C for 1 h. The sample-virus mixture was added to the Vero Cell monolayer and incubated at 37 °C with 5% CO2. On the following day (12–16 h), the expression of GFP was visualized and quantified by Celigo Image Cytometer (Cyntellect Inc, San Diego, CA). The neutralizing activity [88,92,93] of the plasma sample was determined as pNT50 calculated from a transformed non-linear regression curve generated by GraphPad Prism version 8.3.1. (GraphPad Software, Inc., San Diego, CA, USA). Due to the low titer of salivary samples, the 50% inhibitory dilution (IC50) was determined by the reciprocal of the highest dilution of the sample corresponding to 50% reduction in GFP count compared with virus control minus sample control using the Reed-Muench method [94].
4.5. Proteomics sample preparation
Saliva and plasma specimens were deactivated for viral activity by temperature incubation [95] (60 °C for 60 mins), and saliva was preprocessed by centrifuging at 5,000g for 5 mins to remove large debris prior to storing at -80°C. Next, samples passed through 10-kDa cutoff filters (Microcon, Millipore) by centrifuging at 14,000 g for 15 min. The filtrates and the remaining materials on filters were subjected to peptidomics and proteomics analysis, respectively. For peptidomics analysis, the peptides in the filtrates were enriched using an Oasis HLB extraction cartridge (Waters, Milford, MA) following the manufacturer's procedure. The elution dried in SpeedVac and stored in −80C until further LC-MS/MS analysis. For proteomics analysis, the proteins remaining on filters (materials used for this study) were digested using the filter aided sample preparation (FASP) approach as described previously [87,96].
4.6. Liquid chromatography with tandem mass spectrometry (LC-MS/MS) analysis
For the LC-MS/MS analysis, the Ultimate 3000 nanoLC coupled to Q Exactive mass spectrometer (Thermo Scientific) was used as previously described [10]. Peptides were first loaded onto a trap column (PepMap C18, 2 cm × 100 mm x I.D.; Thermo Scientific), and they were separated using an in-house packed analytical column (C18 ReproSil, 3.0 mm, Dr. Maisch GmbH; 20 cm × 75 mm I.D.) and binary buffer system (buffer A: 0.1% formic acid in water; buffer B: 0.1% formic acid in acetonitrile) with a 150-min gradient (2–35% buffer B over 105min; 35–80% buffer B over 10min; back to 2% B in 5 min for equilibration after staying on 80% B for 5 min). For the MS data acquisition, a top-10 data-dependent acquisition (DDA) method was applied. The maximum injection time was set to 20 ms, and the scan range was set to 350–1800 m/z with an AGC target of 1e6. The MS/MS acquisition was performed with 30% HCD collision energy. The target value was set to 5e5, and the maximum injection time was set to 100 ms. Full MS and MS/MS scans were acquired at resolutions of 70,000 and 17,500, respectively. Dynamic exclusion was set to 20s. The mass to charge ratio (m/z [Da]) from mass spectrometry data was normalized and used for the calculation of fold changes of differentially expressed (DE) proteins (health vs. COVID-19; saliva vs. plasma).
4.7. Database search and bioinformatics analysis
For proteomics data analysis, protein identification and quantitation were performed using the MaxQuant-Andromeda software suite (version 1.6.3.4) as previously described [97]8. The majority of the default settings were taken, including trypsin as the enzyme, two missed cleavage sites, peptide length with minimum of seven amino acids, oxidation (M) as variable modification, and carbamidomethylation (C) as fixed modification. An UniProt human sequence database (20,376 sequences) was used for the protein database search. The false discovery rate (FDR) was set at 1% on both protein and peptide levels. Significantly enriched proteins in convalescent samples (p < 0.05) (Table S3) were subjected to the network analysis by STRING enrichment analysis (Cytoscape software v. 3.9.1) [98]. Protein abundances were also evaluated for their correlation with disease severity categories (healthy and asymptomatic = 0, mild = 1, and moderate = 2 and severe = 3) by simple linear regression and Pearson’s R (GraphPad Software, Inc., San Diego, CA, USA). The heatmap was created using the pheatmap package in R using a hierarchical distance matrix and clustering option [99]. The volcano plots were generated using the EnhancedVolcano package in R [99].
Data analytical steps for identifying protein biomarkers and classifying the COVID-19 convalescent and healthy controls (shown in Fig. 3) include data preprocessing, nonparametric statistical comparison, pathway enrichment interpretation, and result visualization. In the preprocessing step, the protein abundance values were log transformed without any normalization. We then calculated the feature saliency for classification using non-parametric statistical tests, including the popular Wilcoxon rank sum test and the Wald-Wolfowitz test. The proteins were ranked based on their p-values output by the statistical tests. Because of the limited number of healthy controls, cross validation was not applicable. For the same reason, we and there is also no need to correct the p-values for the purpose of ranking the proteins. For interpretation of the identified top-ranked proteins, they were compared with the STING network database in Cytoscape for pathway enrichment analysis, before the samples were visualized on principal component analysis (PCA) plots using these top-ranked proteins only, instead of using all proteins. We chose PCA over LDA (linear discriminative analysis) because of the small number of samples [100]. Also, PCA is unsupervised and less biased for unbalanced cohort sizes and LDA assumes the input data follows a Gaussian distribution. The PCA plots show that the COVID-19 convalescent samples can be clearly separated from healthy controls using features identified by either Based on the visualization, the Wilcoxon rank sum test or separated the COVID-19 convalescent from healthy controls in a better way than the Wald-Wolfowitz test. We further compared different p-value cutoffs for selecting the top-ranked proteins. The PCA plots showing that p-value = 0.01 led to should be used to generate the PCA plots, which produced an optimal classification of the two cohorts.
4.8. Statistics
Data was statistically analyzed using the R or Graphpad Prism-8 suites of software (GraphPad Software, Inc., San Diego, CA, USA). Representatives of a minimum of two independent experiments were presented as the median and standard deviation and are representative of a minimum of two independent experiments. Data points for quantitative in vitro experiments represent all technical repeats for experiments done in triplicate. Antibody titers were analyzed by mixed-effect analysis with Tukey’s multiple comparisons. The significance of fold changes in DE proteins were measured using the student’s t-test. Correlation among different parameters (antibody titers, proteomic marker expression levels, categorized demographic information, and salivary protein subgrouping) was evaluated by both Pearson’s R and simple linear regression analyses using Graphpad Prism-8 suites of software (GraphPad Software, Inc., San Diego, CA, USA).
Funding
The Conrad Prebys Foundation Grant (20-122) (MF, GT). U.S. Public Health Service Grants from the National Institute of Dental and Craniofacial Research grant #R21DE029625 (MF), andSupplement grant #R01DE016937-16S1 (MF).
Author contribution statement
Marcelo Freire: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Hyesun Jang and Saibyasachi Choudhury: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Gene S. Tan, Yanbao Yu, Stephen A. Rawlings, Davey Smith and Harinder Singh: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Wrote the paper.
Benjamin L. Sievers and Terri Gelbart: Performed the experiments; Analyzed and interpreted the data.
Amy Proal and Yu Qian: Analyzed and interpreted the data; Wrote the paper.
Data availability statement
Data associated with this study has been deposited at the source data file (ftp://MSV000086946@massive.ucsd.edu) at the Global Natural Products Social (GNPS) molecular networking depository via the Mass spectrometry Interactive Virtual Environment (MASSIVE) and available to Human Salivary Proteome Wiki, salivaryproteome.org.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Marcelo Freire reports financial support was provided by J Craig Venter Institute La Jolla. Marcelo Freire reports a relationship with Mars that includes: board membership.
Davey Smith has consulted for the following companies: Model Medicine, Linear Therapies, FluErgy, Bayer, Evidera and Pharma Holdings
Acknowledgments
We would like to thank Wan Choi for his technical support, and staff at the J. Craig Venter Institute and University of California San Diego for support during the sample collection and transferring.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2023.e17958.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Byrne A.W., McEvoy D., Collins A.B., Hunt K., Casey M., Barber A., et al. Inferred duration of infectious period of SARS-CoV-2: rapid scoping review and analysis of available evidence for asymptomatic and symptomatic COVID-19 cases. BMJ Open. 2020;10(8) doi: 10.1136/bmjopen-2020-039856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Davis H.E., Assaf G.S., McCorkell L., Wei H., Low R.J., Re’em Y., et al. Characterizing long COVID in an international cohort: 7 months of symptoms and their impact. EClinicalMedicine. 2021;38 doi: 10.1016/j.eclinm.2021.101019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perlis R.H., Santillana M., Ognyanova K., Safarpour A., Lunz Trujillo K., Simonson M.D., et al. Prevalence and correlates of long COVID symptoms among US adults. JAMA Netw. Open. 2022;5(10) doi: 10.1001/jamanetworkopen.2022.38804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang L., Yao Q., Gu X., Wang Q., Ren L., Wang Y., et al. 1-year outcomes in hospital survivors with COVID-19: a longitudinal cohort study. Lancet. 2021;398(10302):747–758. doi: 10.1016/S0140-6736(21)01755-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peluso M.J., Deitchman A.N., Torres L., Iyer N.S., Munter S.E., Nixon C.C., et al. Long-term SARS-CoV-2-specific immune and inflammatory responses in individuals recovering from COVID-19 with and without post-acute symptoms. Cell Rep. 2021;36(6) doi: 10.1016/j.celrep.2021.109518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Florence W.J.C., Siew-Wai F., Barnaby E.Y., Kan-Xing W., Anthony S., Shuba K., Yi-Hao C., Guillaume C., Louis L.Y.T., Fei G., Ru San T., Liang Z., Angela S.K., Seow-Yen T., Paul A.T., Laurent R., Lisa F.P.N., David C.L., Christine C. Convalescent COVID-19 patients are susceptible to endothelial dysfunction due to persistent immune activation. elife. 2021;10 doi: 10.7554/eLife.64909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brandtzaeg P. Secretory immunity with special reference to the oral cavity. Enferm Infecc Microbiol Clin. 2020;38(6):279–282. doi: 10.3402/jom.v5i0.20401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siqueira W.L., Salih E., Wan D.L., Helmerhorst E.J., Oppenheim F.G. Proteome of human minor salivary gland secretion. J. Dent. Res. 2008;87(5):445–450. doi: 10.1177/154405910808700508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei C.X., Yu Y., Aleti G., Torralba M., Edlund A., Nelson K.E., et al. In: Salivary Bioscience: Foundations of Interdisciplinary Saliva Research and Applications. Granger D.A., Taylor M.K., editors. Springer International Publishing; Cham: 2020. Salivary bioscience and periodontal medicine; pp. 419–447. [Google Scholar]
- 10.Lin Y.H., Eguez R.V., Torralba M.G., Singh H., Golusinski P., Golusinski W., et al. Self-assembled STrap for global proteomics and salivary biomarker discovery. J. Proteome Res. 2019;18(4):1907–1915. doi: 10.1021/acs.jproteome.9b00037. [DOI] [PubMed] [Google Scholar]
- 11.Zenobia C., Herpoldt K.L., Freire M. Is the oral microbiome a source to enhance mucosal immunity against infectious diseases? NPJ Vaccines. 2021;6(1):80. doi: 10.1038/s41541-021-00341-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Torralba M.G., Aleti G., Li W., et al. Oral Microbial Species and Virulence Factors Associated with Oral Squamous Cell Carcinoma. Microb Ecol. 2021;82:1030–1046. doi: 10.1007/s00248-020-01596-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brandini D.A., Takamiya A.S., Thakkar P., Schaller S., Rahat R., Naqvi A.R. Covid-19 and oral diseases: crosstalk, synergy or association? Rev. Med. Virol. 2021;31(6):e2226. doi: 10.1002/rmv.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasturk H., Kantarci A., Van Dyke T.E. Oral inflammatory diseases and systemic inflammation: role of the macrophage. Front. Immunol. 2012;3:118. doi: 10.3389/fimmu.2012.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heron S.E., Elahi S. HIV infection and compromised mucosal immunity: oral manifestations and systemic inflammation. Front. Immunol. 2017;8:241. doi: 10.3389/fimmu.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lau W.W., Hardt M., Zhang Y.H., Freire M., Ruhl S. The human salivary proteome Wiki: a community-driven research platform. J. Dent. Res. 2021;100(13):1510–1519. doi: 10.1177/00220345211014432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsunetsugu-Yokota Y., Ito S., Adachi Y., Onodera T., Kageyama T., Takahashi Y. Saliva as a useful tool for evaluating upper mucosal antibody response to influenza. PLoS One. 2022;17(2) doi: 10.1371/journal.pone.0263419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sugimoto M.A., Vago J.P., Perretti M., Teixeira M.M. Mediators of the resolution of the inflammatory response. Trends Immunol. 2019;40(3):212–227. doi: 10.1016/j.it.2019.01.007. [DOI] [PubMed] [Google Scholar]
- 19.Ayres J.S. Surviving COVID-19: a disease tolerance perspective. Sci. Adv. 2020;6(18) doi: 10.1126/sciadv.abc1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Metcalf C.J.E., Farrar J., Cutts F.T., Basta N.E., Graham A.L., Lessler J., et al. Use of serological surveys to generate key insights into the changing global landscape of infectious disease. Lancet. 2016;388(10045):728–730. doi: 10.1016/S0140-6736(16)30164-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Organization W.H., Others . WHO; Geneva: 2020. Unity Studies: Early Investigation Protocols. [Google Scholar]
- 22.Macpherson A.J., McCoy K.D., Johansen F.E., Brandtzaeg P. The immune geography of IgA induction and function. Mucosal Immunol. 2008;1(1):11–22. doi: 10.1038/mi.2007.6. [DOI] [PubMed] [Google Scholar]
- 23.Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexander R., Mestecky J. Neutralizing antibodies in mucosal secretions: IgG or IgA? Curr. HIV Res. 2007;5(6):588–593. doi: 10.2174/157016207782418452. [DOI] [PubMed] [Google Scholar]
- 25.Zhong W., Altay O., Arif M., Edfors F., Doganay L., Mardinoglu A., et al. Next generation plasma proteome profiling of COVID-19 patients with mild to moderate symptoms. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kell D.B., Laubscher G.J., Pretorius E. A central role for amyloid fibrin microclots in long COVID/PASC: origins and therapeutic implications [Internet] Biochem. J. 2022;479:537–559. doi: 10.1042/bcj20220016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perretti M., Flower R.J. Annexin 1 and the biology of the neutrophil. J. Leukoc. Biol. 2004;76(1):25–29. doi: 10.1189/jlb.1103552. [DOI] [PubMed] [Google Scholar]
- 28.Perretti M., Solito E. Annexin 1 and neutrophil apoptosis. Biochem. Soc. Trans. 2004;32(Pt3):507–510. doi: 10.1042/BST0320507. [DOI] [PubMed] [Google Scholar]
- 29.Sehnert B., Cavcic A., Böhm B., Kalden J.R., Nandakumar K.S., Holmdahl R., et al. Antileukoproteinase: modulation of neutrophil function and therapeutic effects on anti-type II collagen antibody-induced arthritis. Arthritis Rheum. 2004;50(7):2347–2359. doi: 10.1002/art.20339. [DOI] [PubMed] [Google Scholar]
- 30.Heissig B., Nishida C., Tashiro Y., Sato Y., Ishihara M., Ohki M., et al. Role of neutrophil-derived matrix metalloproteinase-9 in tissue regeneration. Histol. Histopathol. 2010;25(6):765–770. doi: 10.14670/HH-25.765. [DOI] [PubMed] [Google Scholar]
- 31.Gelzo M., Cacciapuoti S., Pinchera B., De Rosa A., Cernera G., Scialò F., et al. Matrix metalloproteinases (MMP) 3 and 9 as biomarkers of severity in COVID-19 patients. Sci. Rep. 2022;12(1):1212. doi: 10.1038/s41598-021-04677-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsumoto M., Hirata T. Moesin regulates neutrophil rolling velocity in vivo. Cell. Immunol. 2016;304–305:59–62. doi: 10.1016/j.cellimm.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Liu X., Yang T., Suzuki K., Tsukita S., Ishii M., Zhou S., et al. Moesin and myosin phosphatase confine neutrophil orientation in a chemotactic gradient. J. Exp. Med. 2015;212(2):267–280. doi: 10.1084/jem.20140508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yürüker B., Niggli V. Alpha-actinin and vinculin in human neutrophils: reorganization during adhesion and relation to the actin network. J. Cell Sci. 1992;101(Pt 2):403–414. doi: 10.1242/jcs.101.2.403. [DOI] [PubMed] [Google Scholar]
- 35.Lan T, Han L, Zhang L, Li H, Sahu SK, Zhu Y, et al. The functional prediction of transmembrane serine protease 2 (TMPRSS2) in priming S-protein of SARS-CoV-2 among vertebrates [Internet]. Available from: 10.22541/au.159225464.44600406. [DOI]
- 36.Berdowska I., Matusiewicz M. Cathepsin L, transmembrane peptidase/serine subfamily member 2/4, and other host proteases in COVID-19 pathogenesis - with impact on gastrointestinal tract. World J. Gastroenterol. 2021;27(39):6590–6600. doi: 10.3748/wjg.v27.i39.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang, N., Pérez, P., Kato, T. et al. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med27, 892–903 (2021). 10.1038/s41591-021-01296-8https://www.nature.com/articles/s41591-021-01296-8. [DOI] [PMC free article] [PubMed]
- 38.Choi Y.J., Kim S., Choi Y., Nielsen T.B., Yan J., Lu A., et al. SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 2019;20(3):276–287. doi: 10.1038/s41590-018-0303-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawase M., Shirato K., Matsuyama S., Taguchi F. Protease-mediated entry via the endosome of human coronavirus 229E. J. Virol. 2009;83(2):712–721. doi: 10.1128/JVI.01933-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H., Li X., Wu Q., Wang X., Qin Z., Wang Y., et al. Plasma proteomic and metabolomic characterization of COVID-19 survivors 6 months after discharge. Cell Death Dis. 2022;13(3):235. doi: 10.1038/s41419-022-04674-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Al-Hakeim HK, Al-Rubaye HT, Al-Hadrawi DS, Almulla AF, Maes M. Long-COVID post-viral chronic fatigue and affective symptoms are associated with oxidative damage, lowered antioxidant defenses and inflammation: a proof of concept and mechanism study [Internet]. Available from: 10.21203/rs.3.rs-1610917/v1. [DOI] [PMC free article] [PubMed]
- 42.Saheb Sharif-Askari N., Saheb Sharif-Askari F., Ahmed S.B.M., Hannawi S., Hamoudi R., Hamid Q., et al. Enhanced expression of autoantigens during SARS-CoV-2 viral infection. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.686462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sapkota H.R., Nune A. Long COVID from rheumatology perspective — a narrative review [Internet] Clin. Rheumatol. 2022;41:337–348. doi: 10.1007/s10067-021-06001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.George P.M., Reed A., Desai S.R., Devaraj A., Faiez T.S., Laverty S., et al. A persistent neutrophil-associated immune signature characterizes post–COVID-19 pulmonary sequelae. Sci. Transl. Med. 2022;14(671) doi: 10.1126/scitranslmed.abo5795. [DOI] [PubMed] [Google Scholar]
- 45.Scieszka D., Lin YH., Li W., et al. NETome: A model to Decode the Human Genome and Proteome of Neutrophil Extracellular Traps. Sci Data. 2022;9:702. doi: 10.1038/s41597-022-01798-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peñaloza H.F., Lee J.S., Ray P. Neutrophils and lymphopenia, an unknown axis in severe COVID-19 disease. PLoS Pathog. 2021;17(9) doi: 10.1371/journal.ppat.1009850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ackermann M., Anders H.J., Bilyy R., Bowlin G.L., Daniel C., De Lorenzo R., et al. Patients with COVID-19: in the dark-NETs of neutrophils. Cell Death Differ. 2021;28(11):3125–3139. doi: 10.1038/s41418-021-00805-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gillot C., Favresse J., Mullier F., Lecompte T., Dogné J.M., Douxfils J. NETosis and the immune system in COVID-19: mechanisms and potential treatments. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.708302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomar B., Anders H.J., Desai J., Mulay S.R. Neutrophils and neutrophil extracellular traps drive necroinflammation in COVID-19. Cells [Internet] 2020;9(6) doi: 10.3390/cells9061383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paludan S.R., Mogensen T.H. Innate immunological pathways in COVID-19 pathogenesis. Sci. Immunol. 2022;7(67) doi: 10.1126/sciimmunol.abm5505. [DOI] [PubMed] [Google Scholar]
- 51.Borges L., Pithon-Curi T.C., Curi R., Hatanaka E. COVID-19 and neutrophils: the relationship between hyperinflammation and neutrophil extracellular traps. Mediat. Inflamm. 2020 doi: 10.1155/2020/8829674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huckriede J., Anderberg S.B., Morales A., de Vries F., Hultström M., Bergqvist A., et al. Evolution of NETosis markers and DAMPs have prognostic value in critically ill COVID-19 patients. Sci. Rep. 2021;11(1):15701. doi: 10.1038/s41598-021-95209-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arcanjo A., Logullo J., Menezes C.C.B., de Souza Carvalho Giangiarulo T.C., Dos Reis M.C., de Castro G.M.M., et al. The emerging role of neutrophil extracellular traps in severe acute respiratory syndrome coronavirus 2 (COVID-19) Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-76781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varjú I., Kolev K. Networks that stop the flow: a fresh look at fibrin and neutrophil extracellular traps. Thromb. Res. 2019;182:1–11. doi: 10.1016/j.thromres.2019.08.003. [DOI] [PubMed] [Google Scholar]
- 55.Silva L.M., Doyle A.D., Tran C.L., Greenwell-Wild T., Dutzan N., Lum A.G., et al. Fibrin is a critical regulator of neutrophil effector function at mucosal barrier sites. bioRxiv. 2021 doi: 10.1101/2021.01.15.426743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cheung C.C.L., Goh D., Lim X., Tien T.Z., Lim J.C.T., Lee J.N., et al. Residual SARS-CoV-2 viral antigens detected in GI and hepatic tissues from five recovered patients with COVID-19. Gut. 2022;71(1):226–229. doi: 10.1136/gutjnl-2021-324280. [DOI] [PubMed] [Google Scholar]
- 57.Silva J., Lucas C., Sundaram M., Israelow B., Wong P., Klein J., et al. Saliva viral load is a dynamic unifying correlate of COVID-19 severity and mortality. medRxiv. 2021 doi: 10.1101/2021.01.04.21249236. [DOI] [Google Scholar]
- 58.Gaebler C., Wang Z., Lorenzi J.C.C., Muecksch F., Finkin S., Tokuyama M., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591(7851):639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Grobbelaar LM, Venter C, Vlok M, Ngoepe M, Laubscher GJ, Lourens PJ, et al. SARS-CoV-2 spike protein S1 induces fibrin(ogen) resistant to fibrinolysis: Implications for microclot formation in COVID-19 [Internet]. Available from: 10.1101/2021.03.05.21252960. [DOI] [PMC free article] [PubMed]
- 60.Winarski K.L., Tang J., Klenow L., Lee J., Coyle E.M., Manischewitz J., et al. Antibody-dependent enhancement of influenza disease promoted by increase in hemagglutinin stem flexibility and virus fusion kinetics. Proc. Natl. Acad. Sci. U. S. A. 2019;116(30):15194–15199. doi: 10.1073/pnas.1821317116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osiowy C., Horne D., Anderson R. Antibody-dependent enhancement of respiratory syncytial virus infection by sera from young infants. Clin. Diagn. Lab. Immunol. 1994;1(6):670–677. doi: 10.1128/cdli.1.6.670-677.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Alwis R., Williams K.L., Schmid M.A., Lai C.Y., Patel B., Smith S.A., et al. Dengue viruses are enhanced by distinct populations of serotype cross-reactive antibodies in human immune sera. PLoS Pathog. 2014;10(10) doi: 10.1371/journal.ppat.1004386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Füst G. Enhancing antibodies in HIV infection. Parasitology. 1997;115(Suppl):S127–S140. doi: 10.1017/s0031182097001819. [DOI] [PubMed] [Google Scholar]
- 64.Zhou B., Zhou R., Chan J.F.W., Zeng J., Zhang Q., Yuan S., et al. SARS-CoV-2 hijacks neutralizing dimeric IgA for enhanced nasal infection and injury. bioRxiv. 2021 doi: 10.1080/22221751.2023.2245921. https://www.biorxiv.org/content/10.1101/2021.10.05.463282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Aleyd E., van Hout M.W.M., Ganzevles S.H., Hoeben K.A., Everts V., Bakema J.E., et al. IgA enhances NETosis and release of neutrophil extracellular traps by polymorphonuclear cells via fcα receptor I [Internet] J. Immunol. 2014;192:2374–2383. doi: 10.4049/jimmunol.1300261. [DOI] [PubMed] [Google Scholar]
- 66.Chen X.Q., Tu L., Zou J.S., Zhu S.Q., Zhao Y.J., Qin Y.H. The Involvement of neutrophil extracellular traps in disease activity associated with IgA vasculitis. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.668974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heineke M.H., Ballering A.V., Jamin A., Ben Mkaddem S., Monteiro R.C., Van Egmond M. New insights in the pathogenesis of immunoglobulin A vasculitis (Henoch-Schönlein purpura) Autoimmun. Rev. 2017;16(12):1246–1253. doi: 10.1016/j.autrev.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 68.Li Q., Chen P., Shi S., Liu L., Lv J., Zhu L., et al. Neutrophil-to-lymphocyte ratio as an independent inflammatory indicator of poor prognosis in IgA nephropathy. Int. Immunopharm. 2020;87 doi: 10.1016/j.intimp.2020.106811. [DOI] [PubMed] [Google Scholar]
- 69.Stacey H.D., Golubeva D., Posca A., Ang J.C., Novakowski K.E., Zahoor M.A., et al. IgA potentiates NETosis in response to viral infection. Proc. Natl. Acad. Sci. U S A [Internet] 2021;118(27) doi: 10.1073/pnas.2101497118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zoodsma M., de Nooijer A.H., Grondman I., Gupta M.K., Bonifacius A., Koeken V.A.C.M., et al. Targeted proteomics identifies circulating biomarkers associated with active COVID-19 and post-COVID-19. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1027122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Saheb Sharif-Askari N., Soares N.C., Mohamed H.A., Saheb Sharif-Askari F., Alsayed H.A.H., Al-Hroub H., et al. Saliva metabolomic profile of COVID-19 patients associates with disease severity. Metabolomics. 2022;18(11):81. doi: 10.1007/s11306-022-01936-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nasrallah G.K. Do preexisting antibodies against seasonal coronaviruses have a protective role against SARS-CoV-2 infections and impact on COVID-19 severity? EBioMedicine. 2022;76 doi: 10.1016/j.ebiom.2022.103831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ng K.W., Faulkner N., Cornish G.H., Rosa A., Harvey R., Hussain S., et al. Preexisting and de novo humoral immunity to SARS-CoV-2 in humans. Science. 2020;370(6522):1339–1343. doi: 10.1126/science.abe1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li N., Li X., Wu J., Zhang S., Zhu L., Chen Q., et al. Pre-existing humoral immunity to low pathogenic human coronaviruses exhibits limited cross-reactive antibodies response against SARS-CoV-2 in children [Internet] Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1042406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science [Internet] 2020;(6520):370. doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sealy R.E., Hurwitz J.L. Cross-reactive immune responses toward the common cold human coronaviruses and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): mini-review and a murine study. Microorganisms. 2021;9(8) doi: 10.3390/microorganisms9081643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Frampas C.F., Longman K., Spick M., Lewis H.M., Costa C.D.S., Stewart A., et al. Untargeted saliva metabolomics by liquid chromatography—mass spectrometry reveals markers of COVID-19 severity. PLoS One. 2022;17(9) doi: 10.1371/journal.pone.0274967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang G., Xiang Z., Wang W., Chen Z. Seasonal coronaviruses and SARS-CoV-2: effects of preexisting immunity during the COVID-19 pandemic. J. Zhejiang Univ. - Sci. B. 2022;23(6):451–460. doi: 10.1631/jzus.B2200049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Saibyasachi N. Choudhury, Mark Novotny, Brian D Aevermann et al. A Protocol for Revealing Oral Neutrophil Heterogeneity by Single-Cell Immune Profiling in Human Saliva, 30 June 2020, PROTOCOL (Version 1) available at Protocol Exchange [ 10.21203/rs.3.pex-953/v1]. [DOI]
- 80.Bagheri-Hosseinabadi Z., Abbasi M., Kahnooji M., Ghorbani Z., Abbasifard M. The prognostic value of S100A calcium binding protein family members in predicting severe forms of COVID-19. Inflamm. Res. 2022;71(3):369–376. doi: 10.1007/s00011-022-01545-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Özgeris F.B., Koçak Ö.F., Kurt N., Parlak E., Yüce N., Keles M.S. High serum progranulin levels in COVID-19 patients: a pilot study. Biochemistry. 2022;87(3):207–214. doi: 10.1134/S0006297922030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Logue J.K., Franko N.M., McCulloch D.J., McDonald D., Magedson A., Wolf C.R., et al. Sequelae in adults at 6 Months after COVID-19 infection. JAMA Netw. Open. 2021;4 doi: 10.1001/jamanetworkopen.2021.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Proal A.D., VanElzakker M.B. Long COVID or post-acute sequelae of COVID-19 (PASC): an overview of biological factors that may contribute to persistent symptoms. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.698169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mehandru S., Merad M. Pathological sequelae of long-haul COVID. Nat. Immunol. 2022;23(2):194–202. doi: 10.1038/s41590-021-01104-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pretorius E., Vlok M., Venter C., Bezuidenhout J.A., Laubscher G.J., Steenkamp J., et al. Persistent clotting protein pathology in Long COVID/Post-Acute Sequelae of COVID-19 (PASC) is accompanied by increased levels of antiplasmin. Cardiovasc. Diabetol. 2021;20(1):172. doi: 10.1186/s12933-021-01359-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.El Alayli** Osama Altayar** Payal Patel** Yngve Falck-Ytter** Valé KEHAMCCAAMKHJAEMJLMLRPA, , **Methodologist December 23 2020 Version 2. 0. 0. Has Been Released, contains revisions to sample types (recommendation 2), rapid vs. non-rapid testing (recommendation 7), revised recommendations for testing, 16). May 6, 2020 Version 1. 0. 1 of the guideline has been released. IDSA guidelines on the diagnosis of COVID-19: Molecular diagnostic testing [Internet]. [cited 2023 Jan 25]. Available from: https://www.idsociety.org/practice-guideline/covid-19-guideline-diagnostics/.
- 87.Yu Y., Sikorski P., Smith M., Bowman-Gholston C., Cacciabeve N., Nelson K.E., et al. Comprehensive metaproteomic analyses of urine in the presence and absence of neutrophil-associated inflammation in the urinary tract. Theranostics. 2017;7(2):238–252. doi: 10.7150/thno.16086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chakraborty S., Gonzalez J.C., Sievers B.L., Mallajosyula V., Chakraborty S., Dubey M., et al. Early non-neutralizing, afucosylated antibody responses are associated with COVID-19 severity. Sci. Transl. Med. 2022;14(635) doi: 10.1126/scitranslmed.abm7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Whitt M.A. Generation of VSV pseudotypes using recombinant ΔG-VSV for studies on virus entry, identification of entry inhibitors, and immune responses to vaccines. J. Virol Methods. 2010;169(2):365–374. doi: 10.1016/j.jviromet.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lei C., Yang J., Hu J., Sun X. On the calculation of TCID50 for quantitation of virus Infectivity. Virol. Sin. 2021;36(1):141–144. doi: 10.1007/s12250-020-00230-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nie J., Li Q., Wu J., Zhao C., Hao H., Liu H., et al. Quantification of SARS-CoV-2 neutralizing antibody by a pseudotyped virus-based assay. Nat. Protoc. 2020;15(11):3699–3715. doi: 10.1038/s41596-020-0394-5. [DOI] [PubMed] [Google Scholar]
- 92.Sievers B.L., Chakraborty S., Xue Y., Gelbart T., Gonzalez J.C., Cassidy A.G., et al. Antibodies elicited by SARS-CoV-2 infection or mRNA vaccines have reduced neutralizing activity against Beta and Omicron pseudoviruses. Sci. Transl. Med. 2022;14(634) doi: 10.1126/scitranslmed.abn7842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sievers B.L., Gelbart T., Tan G.S. A high-throughput SARS-CoV-2 pseudovirus multiplex neutralization assay. STAR Protoc. 2022;3(4) doi: 10.1016/j.xpro.2022.101835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Reed L.J., Muench H. A simple method of estimating fifty per cent ENDPOINTS12 [Internet. Am. J. Epidemiol. 1938;27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 95.Amanat F., Stadlbauer D., Strohmeier S., et al. A serological assay to detect SARS-CoV-2 seroconversion in humans. Nat Med. 2020;26:1033–1036. doi: 10.1038/s41591-020-0913-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lin Y.-H., Eguez R.V., Torralba M.G., Singh H., Golusinski P., Golusinski W., Masternak M., Nelson K.E., Freire M., Yu Y. Self-assembled STrap for global proteomics and salivary biomarker discovery. J. Proteome Res. 2019;18:1907–1915. doi: 10.1021/acs.jproteome.9b00037. [DOI] [PubMed] [Google Scholar]
- 97.Cantwell A.M., Singh H., Platt M., Yu Y., Lin Y.H., Ikeno Y., et al. Kinetic multi-omic analysis of responses to SARS-CoV-2 infection in a model of severe COVID-19. J. Virol. 2021;95(20) doi: 10.1128/JVI.01010-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Team R.C. 2013. Others. R: A Language and Environment for Statistical Computing.http://r.meteo.uni.wroc.pl/web/packages/dplR/vignettes/intro-dplR.pdf Available from: [Google Scholar]
- 100.Martinez A., Kak A. vol. 23. 2001. PCA versus LDA IEEE Transactions on Pattern Analysis and Machine Intelligence. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data associated with this study has been deposited at the source data file (ftp://MSV000086946@massive.ucsd.edu) at the Global Natural Products Social (GNPS) molecular networking depository via the Mass spectrometry Interactive Virtual Environment (MASSIVE) and available to Human Salivary Proteome Wiki, salivaryproteome.org.