Summary
Coxiella burnetii is an obligate zoonotic bacterium that targets macrophages causing a disease called Q fever. It has a biphasic developmental life cycle where the extracellular and metabolically inactive small cell variant (SCV) transforms inside the host into the vegetative large cell variant (LCV). However, details about the morphological and structural changes of this transition are still lacking. Here, we used cryo-electron tomography to image both SCV and LCV variants grown either under axenic conditions or purified directly from host cells. We show that SCVs are characterized by equidistant stacks of inner membrane that presumably facilitate the transition to LCV, a transition coupled with the expression of the Dot/Icm type IVB secretion system (T4BSS). A class of T4BSS particles were associated with extracellular densities possibly involved in host infection. Also, SCVs contained spherical multilayered membrane structures of different sizes and locations suggesting no connection to sporulation as once assumed.
Subject areas: Microbiology, Structural biology
Graphical abstract
Highlights
-
•
Imaging developmental stages of Coxiella burnetii cells using cryo-electron tomography
-
•
Revealing the macromolecular structure of C. burnetii type IV secretion system
-
•
The ultrastructural features of the C. burnetii small and large cell variants
Microbiology; Structural biology
Introduction
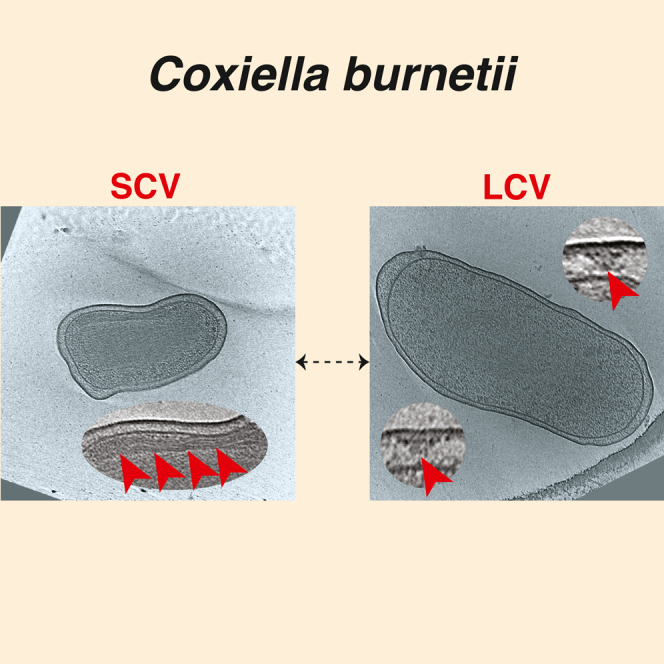
Coxiella burnetii is a highly infectious wide-ranging zoonotic pathogenic bacterium and the causative agent of human Q fever, a severe and debilitating form of influenza-like illness.1,2 C. burnetii exhibits a biphasic developmental cycle of metabolically active and replicating (exponential phase) large cell variant (LCV) present inside the host, to nonreplicating (stationary phase) dormant small cell variant (SCV) that can survive outside the host cell.3,4,5,6 Each variant has distinct morphological features; while SCVs are characterized by their rod-shaped, shorter length (0.2–0.6 μm), dense periplasmic space and condensed DNA, LCVs are pleomorphic in shape, with length exceeding 1 μm, and exhibit relatively sparse periplasmic space and dispersed DNA.3,4,5 The SCVs are highly stable in external environments (outside the host cell) and recalcitrant to physical and chemical stresses.3,7,8
The primary mode of human infection is inhalation of mainly SCVs through contaminated aerosol.9 C. burnetii SCVs predominantly target primary mononuclear phagocytes (e.g., alveolar macrophages) to begin the intracellular biphasic life cycle.10,11 Following internalization, C. burnetii establishes a specialized membrane-bound replicative niche known as the Coxiella containing vacuole (CCV) which matures into an autophagolysosome-like compartment with an acidified lumen (pH∼4.75), acid hydrolases, cationic peptides, and lysosomal markers such as LAMP-1 and CD63 (LAMP-3).12,13 The acidification of the CCV upregulates metabolic activity and gene expression in SCVs leading to a transition to the metabolically active LCVs.5,14,15 In addition, these developmental transitions can be mimicked in host cell-free conditions by growing C. burnetii in an axenic medium known as second generation acidified citrate cysteine medium (ACCM-2).16
The maintenance and maturation of the CCV is largely mediated by the activity of the C. burnetii Dot/Icm (defective in organelle trafficking/intracellular multiplication) type IV secretion system (T4SS) that delivers more than 130 unique effector proteins into the host cytosol.17,18 The bacterial T4SSs are multimegadalton molecular machines that span through the bacterial cell envelope and are involved in transport of nucleoprotein complexes as well as proteins across the cellular envelope.19 Based on genetic composition and phylogenetic analysis, they are classified into two major classes, T4ASS and T4BSS.20 The C. burnetii T4SS belongs to the T4BSS class and is phylogenetically very closely related to the Legionella pneumophila T4BSS.21,22
C. burnetii T4BSS effector proteins subvert multiple host cellular pathways such as autophagic, secretory, and endolysosomal trafficking and aid biogenesis of the CCV to facilitate intracellular replication of C. burnetii.23,24 Earlier studies suggested that the transition from SCV to LCV inside the CCV is correlated with the upregulation of T4BSS expression and activity.5,25,26,27 Intriguingly, under in vitro and axenic (host cell-free) growth conditions, C. burnetii slowly differentiates from LCVs to SCVs; this transition is evident after 10 days, and the majority of LCVs convert to SCVs after 21 days.16
Despite decades of research, the structural changes and remodeling that occur in C. burnetii during its biphasic cycle from LCV to SCV and vice versa remain elusive in part because of the lack of high-resolution in situ imaging of this obligate pathogen. While recent advances in single particle cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) methods have enabled the investigation of T4SSs in great detail in various bacterial species,28,29,30,31,32 the macromolecular architecture, localization and developmental regulation of the C. burnetii T4BSS with respect to its biphasic cycle remain poorly understood. Moreover, previous studies on the ultrastructure of C. burnetii, including their ability to form “spore-like” structures”, used conventional transmission electron microscopy (TEM) where sample preparation (fixation and dehydration) and staining may disrupt membranes.33,34,35
Here, we used cryo-ET to visualize C. burnetii LCVs and SCVs in axenic (host cell-free) growth conditions as well as host-derived C. burnetii variants at macromolecular resolution. Our comprehensive cryo-ET analyses captured changes in cellular morphology and intricate membrane dynamics associated with LCV to SCV transition. In addition, in situ structural analyses of the C. burnetii Dot/Icm T4BSS under different developmental conditions provided insights into the regulation of its expression and activity.
Results
Cryo-ET imaging of C. burnetii cells grown under axenic conditions
To reveal the morphological features of C. burnetii cells and the macromolecular architecture of their T4BSS and its regulation during different developmental stages, we used cryo-ET to image frozen hydrated C. burnetii cells (both SCVs and LCVs) grown in host cell-free second-generation acidified citrate cysteine media (ACCM-2). Earlier studies showed that the biphasic developmental transition and the growth kinetics and viability of C. burnetii can be recapitulated in ACCM-2 media.16 These transition and growth kinetic of C. burnetii are very similar to those occurring inside a host cell such as Vero (African green monkey kidney fibroblasts) cells.5,16 Hence, we decided to image C. burnetii grown in ACCM-2 medium for 5 and 14 days to capture the morphological and structural signatures of SCV to LCV transition and vice versa. While we cannot exclude that axenic growth might induce modifications not naturally occurring in the physiological biphasic growth, this approach is advantageous for cryo-ET imaging where the small size of C. burnetii makes them suitable for direct imaging.
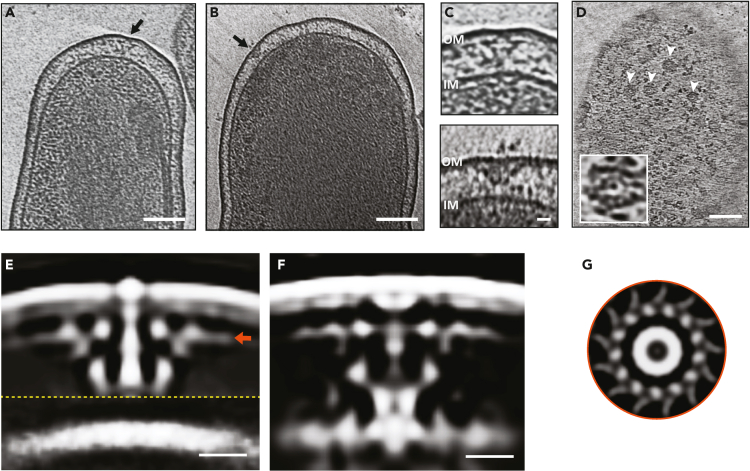
Our cryo-ET imaging of the day 5 axenic culture showed that ∼62% of the cells are LCVs (cell size >800 nm), ∼35% cells are in their transition state (transition state cell variant (TCV), 600–800 nm) and only 3% cells were SCVs (<600 nm) (Figure S1). We based our classification on the morphological and ultrastructural features of the cells like cell length and compact chromatin (for SCVs), as previously described.36 Similar to what is recently reported,37 the ribosomes appeared to be excluded from the compact nucleoid region in SCVs. Compared to SCVs, LCVs exhibited relatively sparse periplasmic space, dispersed DNA, and contained many “Wi-Fi” shaped structures spanning across the cell envelope (Figures 1A and 1B). These structures, with numbers ranging between 1 and 8 per cell (∼4 on average per cell), were composed of two major densities - an outer membrane (OM) associated layer and a lower periplasmic layer (Figure 1C). In partially lysed cells, we occasionally observed clear top views of these particles that appeared to have two concentric rings with the outer ring diameter being ∼40 nm (Figure 1D). Owing to their similarity to the T4BSS particles in L. pneumophila,38 we hypothesized that these particles are the T4BSS of C. burnetii. Accordingly, these particles were absent in 17 cryotomograms of a C. burnetii strain with a complete dot/icm knockout, confirming that they are T4BSS particles (Figure S2A). In contrast, cryo-ET imaging of the day 15 axenic culture contained a mixed population of SCVs (19%), LCVs (37%) and TCVs (44%), and here also T4BSS particles were present only in LCVs and TCVs but not in SCVs (Figure S2B). Our observation that the assembly and maintenance of the T4BSS apparatus is coupled to the developmental transition from SCV to LCV is consistent with previous studies.5,25,26,27
Figure 1.
In situ structure of the C. burnetii T4SS
(A and B) Slices through electron cryotomograms of LCV C. burnetii cells highlighting the presence of T4SS particles (black arrows). Scale bar 100 nm.
(C) Enlargements of the T4SS particles shown in A (top panel) and B (lower panel). Scale bar 10 nm.
(D) A slice thorough an electron cryotomogram of a lysed C. burnetii cell illustrating the presence of multiple top views of T4SS particles (white arrows). Scale bar 100 nm. An enlargement of a top view of a T4SS particle is shown in the white-boxed area.
(E and F) Slices through the subtomogram averages of T4SS of C. burnetii at pH 7.2 (left) and pH 4.75 (right). Dashed yellow line indicates a composite of two averages obtained with different masks (see STAR Methods) concatenated together. Scale bar 10 nm.
(G) A cross section through the T4SS OM complex (at the level indicated with the orange arrow in E) showing 13-fold symmetry. OM = outer membrane, IM = inner membrane.
While the majority of T4BSS particles of C. burnetii were located at the cell poles, there were also a few (7 from 825 particles in 138 tomograms) that were positioned away from the cell poles (Figure S3A). In our previous work on L. pneumophila, we observed only a few T4BSS particles located away from the cell pole in >3,500 cryotomograms.39,40 Similar to our previous observation in L. pneumophila, we identified T4BSS particles at the division plane in dividing C. burnetii, suggesting that they start assembling at the future new poles during the septation process (Figure S3B).
Macromolecular architecture of C. burnetii T4BSS
To decipher the macromolecular architecture of the C. burnetii T4BSS, we averaged 414 particles and generated a subtomogram average at a resolution of 2.5–4.5 nm (Figures 1E and S4A). This average was generated from 69 cryotomograms of cells grown in ACCM-2 for 5 days and resuspended in phosphate buffer saline pH 7.2. Our initial average nicely resolved densities associated with the OM and the periplasmic complex; however, the inner membrane (IM) associated densities were missing. A closer look at the individual particles revealed that the distance between the OM and IM varied significantly in individual particles with most of them lacking clear densities associated with the IM (Figure 1C and later in Figure 2). Consequently, we generated two separate averages with different masks, one with a mask on the OM and one on the IM and juxtaposed them together to produce a composite average with a local resolution between 2.5 and 4.5 nm (Figures 1E and S4A). At this resolution, this composite average showed marked structural similarity to the T4BSS of L. pneumophila, where both consisted of an OM complex, a periplasmic complex and a stem-like structure connecting the two (supplemental information Figures S5A and S5B). The OM complex showed a 13-fold symmetry and a maximum diameter of ∼40 nm (Figure 1G), and formed together with the periplasmic complex what appears to be a secretion chamber similar to the L. pneumophila T4BSS system. This structural similarity between the two systems is consistent with the homology between at least 23 genes of the 30 T4BSS genes in L. pneumophila.22
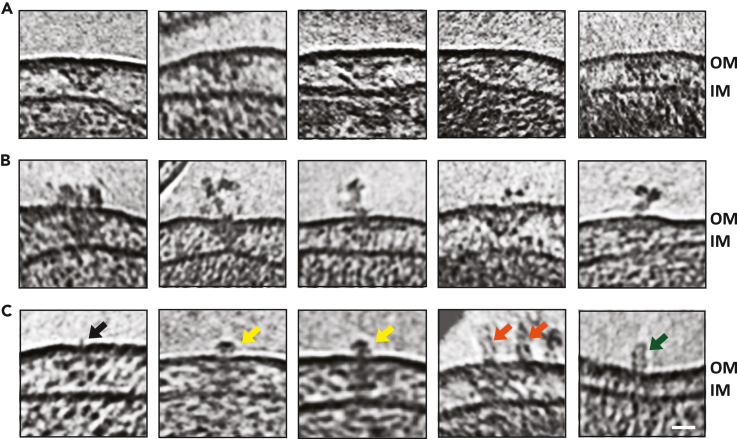
Figure 2.
T4SS-associated extracellular densities
(A) Slices through electron cryotomograms of C. burnetii cells indicating the presence of T4SS particles without any extracellular densities associated with them.
(B) Slices through electron cryotomograms of C. burnetii cells showing the presence of unstructured extracellular densities above the T4SS particles.
(C) Slices through electron cryotomograms of C. burnetii cells illustrating different types of extracellular densities associated with T4SS such as: short filament-like density (black arrow), crown-like density (yellow arrows), tubular densities (orange arrows). Green arrow point to tubular densities where no T4SS could be identified at their bases. Scale bar is 20 nm. OM = outer membrane, IM = inner membrane.
Subsequently, we docked the recently reported near-atomic structure of the L. pneumophila T4BSS core complex41 into our subtomogram average which allowed us to tentatively assign components that contribute to different densities in our structure. This fitting suggested that the OM core complex comprised DotC, DotD, DotF, DotH, DotG and DotK proteins while the periplasmic ring complex contains parts of DotG and DotH (supplemental information Figures S5C). Despite several data processing trials, we could not resolve the cytoplasmic ATPase complex. This is consistent with a contemporaneous study on C. burnetii T4BSS which used more than 7,000 particles to generate an in situ average,37 but it also lacked the cytoplasmic complex suggesting that it is either flexible or rarely associated.
Earlier studies have shown that acidification of the CCV inside the host triggers the differentiation of SCVs into LCVs. This developmental transition has been linked to enhanced metabolic activities, higher gene expression and biogenesis and secretion of the C. burnetii T4BSS.5,25,26,27 Based on this, we hypothesized that under autolysosomal pH conditions (pH ∼4.75), we might capture actively secreting T4BSSs in our cryotomograms. Therefore, we cultured C. burnetii cells in ACCM-2 media for 5 days and then resuspended them in a low pH media (citrate buffer saline pH ∼4.75). Unlike the particles observed in the previous tomograms of cells at pH 7.2, individual particles in these tomograms of cells at pH 4.75 showed less variability in the distance between the OM and IM. We extracted 411 T4BSS particles to produce a subtomogram average (with a single mask) at 2.5–5 nm resolution (Figures 1F and S4B) which revealed a few structural differences (as suggested at the resolution of our structures) compared to the average at pH 7.2 including the existence of what could be a secretion channel and density for the wing structures (as seen before in the L. pneumophila T4BSS, Figures S5 and S6). These observations are consistent with the fact that the T4BSS is active in the acidic autolysosome environment and therefore acidic pH might have ‘primed’ these complexes.
Of interest, ∼17% of individual T4BSS particles showed extracellular densities associated with them in day 5 (both pH 4.75 and 7.2) and day 15 cultures. We classified these extracellular densities into 4 major categories: (1) those with an amorphous shape, (2) thin filament-like, (3) crown-like structures, and (4) those with a rod or tubular-like form (Figures 2A–2C). However, some of these extracellular densities were also occasionally localized on the cell surfaces without an obvious T4BSS particle underneath them (Figure 2C, right most). This could be either because they are relics after the T4BSSs are disassembled or they were secreted and then drifted away from T4BSS. Alternatively, it is also possible that they were secreted in a T4BSS-independent manner. These extracellular densities were absent in 17 cryotomograms of a C. burnetii strain with a complete dot/icm knockout. The low number of particles with extracellular densities precluded the possibility of producing a decent subtomogram average to see if there are any structural changes in T4BSS associated with the presence of these extracellular densities.
Of the different types, the ‘tube-like’ densities were the most interesting as they are reminiscent of a T4ASS pilus filament. These tube-like filaments were ∼8 nm wide, ∼25 nm in length and showed flexibility. No extracellular features associated with T4SS were reported in previous studies of C. burnetii,37 or other species such as L. pneumophila.39,40 Furthermore, no pilin-like candidates have been identified for any of the T4BSSs to date. Therefore, what these different extracellular densities are and if they are related to released/secreted proteins (such as DotA or IcmX37) remain unknown.
Inner membrane stacking, a characteristic feature observed only in SCVs
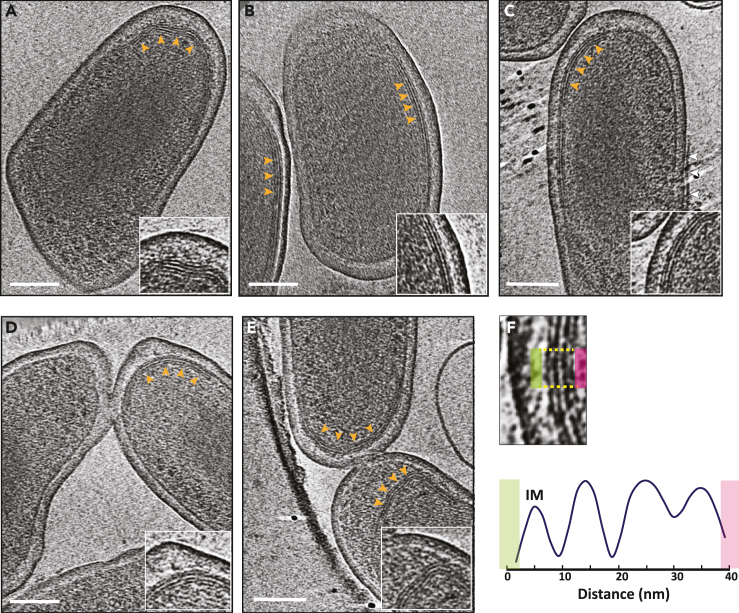
In the day 15 axenic culture, 19% of cells were SCVs, 37% were LCVs and 44% are in their transition state (Figure S1). The SCVs, which are 600 nm or shorter, exhibited relatively dense periplasmic space and condensed DNA compared to LCVs (Figure S2B). Another interesting feature in the cryotomograms of SCVs was the presence of a stack of tightly packed membranes in the cytoplasm (Figures 3A–3E, supplemental information Video S1). In 58 cryotomograms, we always observed a single membrane stack per SCV with each stack having 2–6 layers consistently spread ∼10 nm apart as revealed by density profile analysis (Figure 3F and supplemental information Video S1). This membrane stacking was generated by invaginating the IM toward the cytoplasm and was initiated in LCVs, but densely packed mature stacks were exclusively found in SCVs which suggests that maturation of these stacks is developmentally linked to the LCV to SCV differentiation.
Figure 3.
Multilayered cytoplasmic membrane invaginations in C. burnetii SCVs
(A–E) Slices through electron cryotomograms of C. burnetii cells (purified from infected vero cells 28 dpi) highlighting the presence of multilayered cytoplasmic membrane invaginations (orange arrows) with zoom-ins of these invaginations shown in the enlargements at the lower right corner of each panel. The white arrows in panel C point to a membrane of presumably the host vacuole still attached to the OM of C. burnetii cell. Scale bar is 100 nm.
(F) Lower panel: average density profile taken along the area indicated between the yellow lines in the upper panel highlighting the equidistant spacing of ∼10 nm between the different layers of the inner membrane invaginations.
To investigate if the membrane stacking changes during an infection condition, we imaged the 28 days post-infection host (Vero cells) -derived C. burnetii variants using cryo-ET. In this sample, ∼35% of the cells were SCVs, 41% were in transition state and 24% were LCVs. Similar to the day 15 axenic culture, only host-cell derived SCVs had densely packed membrane stacks while LCVs and transition state cells only revealed early stages of these membrane invaginations. Intriguingly, only in the host-cell-derived SCVs we also observed a dark linear density with an average length of few hundred nanometers (∼100–300 nm) nm and located ∼10 nm from the IM (Figure S7). This linear density appeared morphologically rather different from the IM stacks suggesting that it is of a different nature.
Cytoplasmic spore-like structures in SCVs and transitional cells
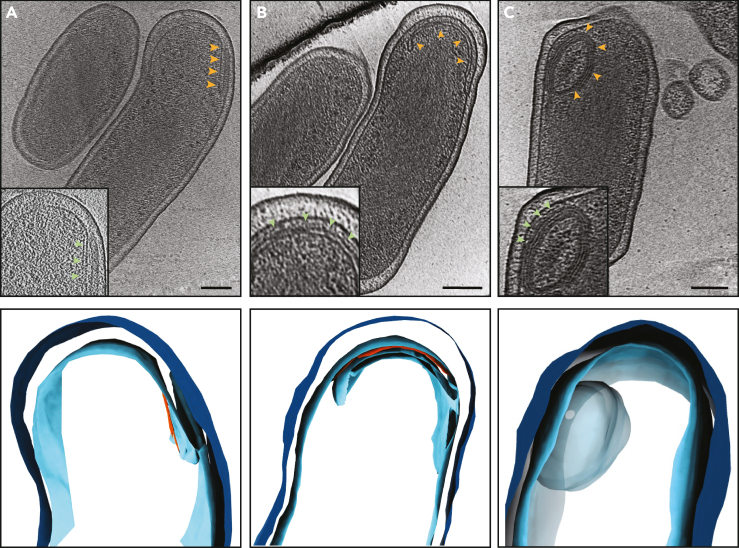
Another prominent feature in C. burnetii SCVs of the day 15 axenic culture is the presence of IM invaginations that extended and curved to form a multilayered spherical structure in the cytoplasm. Initially, the IM invaginated into the cytoplasm forming a septum with a protein-like layer visible at one side (Figure 4 and supplemental information Video S2). This protein-like layer was located ∼7–8 nm from the invaginated membrane. Subsequently, this septum elongated and curled with the protein-like layer still visible at one side to ultimately form a concentric multilamellar spherical cytoplasmic structure with an average diameter of ∼120–150 nm (Figures 4 and S8, Video S2). In total, we identified 16 such structures in 58 cryo-tomograms.
Figure 4.
Different stages of membrane invaginations in C. burnetii cells
(A–C) Top panels: Slices through electron cryotomograms of C. burnetii cells (grown in ACCM-2 for 15 days) illustrating different stages of inner membrane invaginations (orange arrows) with 3D segmentations of these stages shown in the lower panels. An early stage of IM invagination is shown in panel A where a protein-like density can be seen at one side of this invagination (red density in the segmentation). Panel B shows a long and curved IM invagination where a protein-like density can be seen between this invagination and the IM (red density in the segmentation, green arrow in inset). A whole multilayered vesicular structure can be seen in panel C. Note that it was difficult to trace the different layers in the tomogram hence it is shown as one large blob in the segmentation in the lower panel (See supplemental informationVideo S2). Scale bar is 100 nm.
It was difficult to determine how many layers this structure has at the resolution of our cryo-tomograms but the number of membrane layers in these structures appeared to be variable in different cells as they had different thickness. It was also not possible to determine whether they were connected to the IM or not. In 5 examples, this structure adopted an ellipsoidal shape instead of a spherical one. This structure had polar cellular localization in 12 examples, whereas the remaining 4 were located at the center of the cell and appeared darker than the polar one (Figure S9). In one cell, two such structures were present while the other examples had one per cell. Finally, the SCVs of the day 15 axenic culture were also associated with many outer membrane vesicles (OMVs, Figure S9). Both the cytoplasmic multilayered structures and the OMVs were lacking in SCVs of the other samples, namely, the 5 days axenic culture and the host-cell-derived SCVs.
Cytoplasmic tubes in SCVs
Another important morphological feature in the cells of the 5-day axenic culture, both at pH 7.2 and pH 4.75, was the presence of cytoplasmic tubes that were ∼10 nm wide and up to few hundreds of nanometers long (Figure S10). We identified these tubes in 43 cells in total (from 267 tomograms) and each cell contained many tubes, which were either single or stacked together as a bundle (Figure S10). We could only identify two cells with such tubes in the host-cell-derived SCVs, and none in the day 15 axenic culture SCVs. Of interest, these tubes were located at the center of the cell amidst the DNA density and were not found in areas outside the DNA where the ribosomes are present.
Discussion
Our cryo-ET imaging of C. burnetii cells enabled us to visualize the morphological changes associated with transition from SCV to LCV and vice versa at macromolecular resolution under native (hydrated, unfixed) conditions. In addition, it also allowed us to visualize the structural details of the T4BSS molecular machine and its regulation with respect to the biphasic cell cycle. We imaged mainly cells grown under axenic conditions which are believed to largely mimic the physiological biphasic cycle.42 However, the presence of certain morphological features only in host-derived cells (e.g., a linear density underneath the inner membrane), or only in axenically grown cells (e.g., cytoplasmic tubes and OM vesicles) suggest that there are differences between these two modes of growth that should be further investigated in the future.
C. burnetii T4BSS particles are primarily located at the cell poles, but we noticed some particles away from the cell pole suggesting the polar localization in C. burnetii is not as tightly controlled compared to L. pneumophila. The two key proteins, DotU and IcmF, that target the L. pneumophila T4BSS to the cell poles,40 have homologues in C. burnetii; however, icmF has a point mutation which leads to a premature stop,22 which could explain why some of the T4BSS particles were away from the cell pole in C. burnetii. The presence of T4BSS particles in C. burnetii near the division plane suggest that they start assembling near the future cell pole at the beginning of the septation process similar to L. pneumophila. Of interest, the T4BSS complexes were only observed in LCVs and never in SCVs, which is consistent with earlier studies suggesting that the expression and activity of T4BSS are tightly regulated with respect to developmental biphasic transitions.5,25,26,27 Moreover, the absence of T4BSS in SCVs implies that they are not essential for host-entry and early stages of infection.
The identity of the extracellular proteinaceous densities associated with some T4BSS particles remains unknown. One possibility is that they are related to the secretion of some of T4BSS components like DotA and IcmX in a T4BSS-dependent manner when grown in host free axenic media.43 Intriguingly, some of these extracellular densities appeared as short tubular structures; and while T4ASS is known to harbor extracellular pili containing VirB2 and VirB5 proteins,44 the T4BSS has no putative pilin candidates and there are no reports of this system producing a pilus. Previous studies proposed that the C. burnetii T4BSSs interact with the CCV membrane using short tethers that could facilitate translocation of T4BSS effectors,45 and a contemporaneous study also reported short filamentous densities between the bacterial OM and the CCV membrane.37 It is possible that the short tubular structures above some T4BSS particles are used to establish such an intimate contact between the bacterial OM and CCV membrane. Of interest, it has been shown that a population of the conjugation pilus (the F-pilus), which is also a product of the T4SS, consists only of the pilus docked in the outer membrane without any associated periplasmic density at its base.46
Our in situ structural analysis of the C. burnetii T4BSS using subtomogram averaging revealed that it is architecturally similar to that of the L. pneumophila T4BSS at our resolution.40,47 Recently, Sheedlo et al. used cryo-EM single particle analysis to resolve a near-atomic resolution structure of the L. pneumophila T4BSS core complex.41 Fitting their structure into our subtomogram average allowed us to tentatively assign certain components of the OM core complex and the periplasmic ring complex. In addition, while Sheedlo et al. identified multiple species-specific proteins for the L. pneumophila T4BSS - Dis1 (Lpg0657), Dis2 (Lpg0823), and Dis3 (Lpg2847),41 the counterparts of those proteins, if any, in the C. burnetii T4BSS remain unknown.
Another important morphological feature associated with the biphasic developmental cycle of C. burnetii is the presence of tightly packed membrane layers directly below the IM in the day 15 axenic culture and in host cell derived (28 dpi) SCVs. Similar membrane invaginations were reported by McCaul et al. nearly 4 decades ago using traditional TEM of chemically fixed SCV cells3 and recently by Park et al.37 These membrane stacks could be a way to store membranes for future rapid developmental transitions to LCVs. Of interest, the day 15 axenic culture cells also produced many OMVs, not seen in other samples, which could be a result of depleted nutrients in the medium.
A controversial point in the C. burnetii biology is related to whether it can produce spores. Conventional TEM experiments of fixed and negatively stained C. burnetii cells, performed since 1980s, indicated the presence of small multilayered structures in the periplasm of some cells.33 Based on morphological similarity between these structures and the spores of other bacterial species, it was hypothesized that LCVs could produce endogenous spores and ultimately release them into the environment with other SCV cells where the spores eventually also transform into SCV to invade new host cells.33 In addition, it was shown that these structures contain DNA, and a homologue of the sporulation gene SpoIIIE was identified in C. burnetii.34,48 Subsequent studies reported these supposed “endospores” also in C. burnetii cells purified from the heart valves of patients diagnosed with Q fever endocarditis.35
However, the nature of these structures remained controversial for a long time [see reference49], hence they are usually referred to as spore-like particles (SPL) [See ref.49,50 and references therein]. There are multiple reasons now to state that sporulation does not happen in C. burnetii including the absence of the biochemical spore marker, dipicolinic acid,3 and more importantly, the sequencing of C. burnetii genome revealed that it lacks sporulation genes.51 Our results here are also in accordance with this notion. First, these structures have different shapes, sizes, layers, and cellular localization which is inconsistent with a tightly regulated sporulation process. Second, we never detected these structures independently outside the bacterial cells and only observed them inside cells (grown in ACCM2 medium for 15 days). In addition, we do not know what the relationship between these structures and the membrane stacks seen in SCVs. It is possible that the inner membrane stacks mature into the spherical multilayered structures.
Finally, SCVs from the axenic culture day 5 (both at pH 4.75 and pH 7.2) contained bundles of cytoplasmic tubes ∼10 nm in diameter amidst the DNA region of the cell. Morphologically, they are reminiscent of filamentous bundles of unknown functions described in other bacterial species.52 The fact that these bundles are always present inside the DNA region in the cell suggests they could be related to other protein-DNA polymers such as those formed by RecA and MuB, which are also ∼9.5 nm in vitro.53,54 Another possibility is that these are a protective crystalline form of DNA and DNA-binding proteins known to form under stress conditions.55 The other two interesting uncharacterized morphological features include the linear density seen under the IM in host-derived SCVs C. burnetii, and the protein-like polymer seen on the side of early IM invaginations, which might play a role in the formation of the membrane stacks and/or spherical multilayered structures. Further studies are required to understand the biogenesis and actual function of the multilayered membranes stacks, ‘endo-spore’ like structures and other uncharacterized features described in this study.
Limitations of the study
One clear limitation of our study here is that we investigated C. burnetii cells either grown under axenic conditions or purified from host cells, and not directly inside the host. In addition, we could not unambiguously capture the T4SS in its active state, and we still do not know what the identity of the extracellular density observed on the surface of some cells are. Finally, the mechanism by which the membrane stacks form in SCV cells remains unknown. Further studies in the future should be performed to image all of the infection cycle, from host entry to host exit, in detail using cryo-ET can help to shed light on some of these points.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial strains | ||
| Coxiella burnetii Nine Mile phase II (clone 4, RSA 439) | Hoover et al.56 | N/A |
| Coxiella burnetii Nine Mile phase II Δdot/icm mutant | This study | N/A |
| Experimental models: Cell lines | ||
| African green monkey kidney (Vero) fibroblasts (CCL-81) | American Type Culture Collection | https://www.atcc.org/products/ccl-81 |
| Deposited data | ||
| Subtomogram average of the T4SS of Coxiella Burnetii at pH 4.75 | This study | EMD-28281 |
| Subtomogram average of the T4SS of Coxiella burnetii at pH 7 | This study | EMD-28282 |
| Subtomogram average of the T4SS of Coxiella burnetii at pH7 with an inner membrane mask | This study | EMD-28283 |
| Software and algorithms | ||
| UCSF Tomography | Zheng et al.58 | https://emcore.ucsf.edu/ucsf-software |
| Eman2 | Chen et al.60,62 | https://blake.bcm.edu/emanwiki/EMAN2 |
| UCSF ChimeraX | Goddard et al.66 | https://www.rbvi.ucsf.edu/chimerax/ |
| ImageJ | Schindelin et al.61 | https://imagej.net/ |
| IMOD | Kremer et al.59 | https://bio3d.colorado.edu/imod/ |
| Knossos | https://knossos.app | |
| Blender | https://www.blender.org/ | |
Resource availability
Lead contact
Further inquiries and request for data, strains and resources should be directed to the lead contact Debnath Ghosal, debnath.ghosal@unimelb.edu.au
Materials availability
C. burnetii stains and other reagents generated/used in this study (see key resources table) are available upon request from the lead contact.
Experimental model and study participant details
Coxiella burnetii strains
For all experiments, the Coxiella burnetii Nine Mile phase II (clone 4, RSA 439) strain56 or a mutant Nine Mile phase II strain lacking all dot/icm genes were used. For samples purified from eukaryotic host cells, C. burnetii was grown in African green monkey kidney (Vero) fibroblasts (CCL-81; American Type Culture Collection) which are cultured in RPMI medium (Invitrogen, Carlsbad, Calif.) supplemented with 2% fetal bovine serum as described in ref. 5.
Method details
Coxiella burnetii growth
C. burnetii (wild type Nine Mile phase II strain or an isogenic dot/icm mutant of this strain) was grown under axenic cultivation conditions as described in.36,42 In more details, C. burnetii was cultivated in ACCM-2, which is a modified ACCM medium.36 In this modified medium, 1 mg/ml methyl-β-cyclodextrin (Mβ-CD) is used to replace 1% FBS. Bacterial cells were cultured in this medium in T-75 flasks or 0.2-μm-pore-size-filter-capped 125-ml Erlenmeyer flasks with 20 ml of medium, T-25 flasks with 5 ml of medium, or 3 ml of medium in each well of a 6-well tissue culture plate. Bacterial cultures were kept in a CO-170 incubator (New Brunswick Scientific, NJ) with the following conditions: 37°C in a 2.5% O2 and 5% CO2. Subsequently, cells were washed and resuspended either in PBS pH 7.2 or in Citrate buffered saline pH 4.75 (PBS + 10 mM citrate) and then frozen at -80°C until they were thawed before plunge-freezing as described below.
For samples purified from eukaryotic host cells, African green monkey kidney (Vero) fibroblast host cells were infected in 150-cm2 flasks for 4 weeks without changing the growth medium. During the first week, cells were incubated at 37°C in 5% CO2 and subsequently the flasks containing the cells were kept for three weeks at room temperature with their caps tightened. Bacterial cells were then purified from the infected eukaryotic cells using Renografin density gradient centrifugation.57 Purified SCV cells were resuspended in K-36 buffer (0.1 M KCl, 0.015 M NaCl, 0.05 M potassium phosphate, pH 7.0) and stored at -80°C until they were thawed before plunge-freezing as described below.
Cryo-ET sample preparation and imaging
R2/2 carbon-coated 200 mesh copper Quantifoil grids (Quantifoil Micro Tools) were first glow-discharged for 60 seconds. 1 μL of BSA-treated 10-nm gold solution was added to 4 μL of cells thawed from a -80°C stock. The combination of cells and gold was then pipetted on the grids in a Vitrobot chamber (FEI) with 100% humidity. The extra fluid was blotted off using a Whatman filter paper and the grids were plunge-frozen in a liquid ethane/propane mixture. The samples were imaged using an FEI Polara 300 keV field emission gun electron microscope (FEI, Hillsboro, OR, USA) equipped with a Gatan image filter and K2 Summit direct electron detector in counting mode (Gatan, Pleasanton, CA, USA). Data were collected using the UCSF Tomography software58 with each tilt series ranging from −60° to 60° in 1°, 2° or 3° increments, an underfocus of ∼7 μm, an electron dose of 130 e-/Å2, and a pixel size of 3.9 Å.
Image processing and subtomogram averaging
Three-dimensional reconstructions of tilt-series were done with either IMOD software package59 or using EMAN2 software.60 In total, 88 tomograms of C. burnetii grown at pH 7 and 69 tomograms of C. burnetii grown at pH 4 were of sufficient quality for further sub-volume analysis. Particles were manually identified and selected from tomograms in each dataset. In the tomograms of C. burnetii grown at pH 7, 726 particles were identified, and 414 particles were identified in tomograms of C. burnetii grown at pH 4. Particles were extracted using a box size of 96 from tomograms at x2 binning (8.06 Å/pix).The EMAN2 program e2spt_sgd_new.py was used to generate initial models using a subset of 60 high-quality particles from each dataset at 4x binning. Initial models with C1 symmetry were generated with default parameters in the e2spt_sgd_new.py program. To generate initial models with C13 symmetry, the program e2symsearch.py with the “-- symmetrize” option was used to identify the C13 symmetry axis of the OMCC and rotate C1 models such that the C13 symmetry axis was parallel to the Z-axis.
Initial models with C1 or C13 were filtered to 50 Å resolution and used for subsequent sub-tomogram refinement. Extracted particles from each dataset were used for sub-tomogram averaging with either C1 or C13 symmetry. Particles with x2 binning were subjected to two rounds of 3D particle orientation refinement, 2D sub-tilt translation refinement, and sub-tilt translation & rotation refinement, followed by a sub-tilt defocus refinement. Finally, soft masks for either the Outer membrane core complex (OMCC) or Inner membrane were applied to focus the alignment of particles to each respective region.
The density projection profile of the cell envelope and membrane invaginations was calculated in ImageJ61 using a 50 nm wide linear section of the cell envelope. Density plots were generated using Microsoft excel software.
Segmentation of cryotomograms and visualization
Membranes, membrane invaginations, and spores were manually segmented using drawing tools in IMOD.59 Outer membrane vesicles were segmented with a convolutional neural network in EMAN2.62 Alternatively, deep convolutional neural networks (CNNs) were trained using the manually generated ground truth and fine-tuned until a good quality of segmentation was reached for automated segmentation in 3D. The segmentation has been proofread and cleaned-up by expert inspection using the open-source software Knossos (https://knossos.app/). For each segmentation class, meshes and binary Tiff masks were generated for subsequent rendering and visualization in Knossos and in the open-source 3days visualization tool Blender (https://www.blender.org/). Data visualization was also performed using ChimeraX.63
Acknowledgments
This project was funded by the National Institutes of Health (grant R01 AI127401 to G.J.J.), an NHMRC grant (APP1196924 to D.G.), and the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases (Grant number AI000931-20 to R.A.H. and C.L.L.). D.C.S. is supported by the Melbourne Research Scholarship. We are grateful to Prof. Elitza Tocheva for insightful discussions, and Somavally Dalvi for her help with figure preparation.
Author contributions
Conceptualization, D.G., G.J.J., and M.K.; Methodology, D.G., M.K., D.C.S., C.L.L., P.D., P.A.B., and E.K.; Investigation, M.K., D.G., D.C.S., C.L.L., P.D., and P.A.B.; Formal Analysis, M.K., D.G., D.C.S., C.L.L., P.D., P.A.B., K.W.K., N.V., E.K., and R.A.H.; Writing – Original Draft, D.G., M.K., and N.V.; Writing – Review and Editing, M.K., D.G., D.C.S., C.L.L., P.D., P.A.B., K.W.K., N.V., E.K., R.A.H., and G.J.J.; Visualization, M.K., D.G., and D.C.S.; Supervision, G.J.J. and D.G.; Funding Acquisition, G.J.J. and D.G.
Declaration of interests
The authors declare no competing interests.
Published: June 24, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107210.
Contributor Information
Mohammed Kaplan, Email: mohammedk@uchicago.edu.
Grant J. Jensen, Email: grant_jensen@byu.edu.
Debnath Ghosal, Email: debnath.ghosal@unimelb.edu.au.
Supplemental information
Data and code availability
-
•
The subtomogram averages have been deposited to EMBD (EMD-28281, EMD-28282 and EMD-28283) and will be publicly accessible. The raw data is available upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data presented in this study is available from the lead contact upon request.
References
- 1.Maurin M., Raoult D. Q Fever. Clin. Microbiol. Rev. 1999;12:518–553. doi: 10.1128/CMR.12.4.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brooke R.J., Kretzschmar M.E.E., Mutters N.T., Teunis P.F. Human dose response relation for airborne exposure to Coxiella burnetii. BMC Infect. Dis. 2013;13:488. doi: 10.1186/1471-2334-13-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCaul T.F., Williams J.C. Developmental cycle of Coxiella burnetii: structure and morphogenesis of vegetative and sporogenic differentiations. J. Bacteriol. 1981;147:1063–1076. doi: 10.1128/jb.147.3.1063-1076.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinzen R.A., Hackstadt T., Samuel J.E. Developmental biology of Coxiella burnetii. Trends Microbiol. 1999;7:149–154. doi: 10.1016/S0966-842X(99)01475-4. [DOI] [PubMed] [Google Scholar]
- 5.Coleman S.A., Fischer E.R., Howe D., Mead D.J., Heinzen R.A. Temporal Analysis of Coxiella burnetii Morphological Differentiation. J. Bacteriol. 2004;186:7344–7352. doi: 10.1128/JB.186.21.7344-7352.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voth D.E., Heinzen R.A. Lounging in a lysosome: the intracellular lifestyle of Coxiella burnetii. Cell Microbiol. 2007;9:829–840. doi: 10.1111/j.1462-5822.2007.00901.x. [DOI] [PubMed] [Google Scholar]
- 7.Amano K., Williams J.C. Sensitivity of Coxiella burnetii peptidoglycan to lysozyme hydrolysis and correlation of sacculus rigidity with peptidoglycan-associated proteins. J. Bacteriol. 1984;160:989–993. doi: 10.1128/jb.160.3.989-993.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amano K., Williams J.C., McCaul T.F., Peacock M.G. Biochemical and immunological properties of Coxiella burnetii cell wall and peptidoglycan-protein complex fractions. J. Bacteriol. 1984;160:982–988. doi: 10.1128/jb.160.3.982-988.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Angelakis E., Raoult D. Q fever. Vet. Microbiol. 2010;140:297–309. doi: 10.1016/j.vetmic.2009.07.016. [DOI] [PubMed] [Google Scholar]
- 10.Khavkin T., Tabibzadeh S.S. Histologic, immunofluorescence, and electron microscopic study of infectious process in mouse lung after intranasal challenge with Coxiella burnetii. Infect. Immun. 1988;56:1792–1799. doi: 10.1128/iai.56.7.1792-1799.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham J.G., MacDonald L.J., Hussain S.K., Sharma U.M., Kurten R.C., Voth D.E. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages: Coxiella infection of alveolar macrophages. Cell Microbiol. 2013;15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Howe D., Shannon J.G., Winfree S., Dorward D.W., Heinzen R.A. Coxiella burnetii Phase I and II Variants Replicate with Similar Kinetics in Degradative Phagolysosome-Like Compartments of Human Macrophages. Infect. Immun. 2010;78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maurin M., Benoliel A.M., Bongrand P., Raoult D. Phagolysosomes of Coxiella burnetii-infected cell lines maintain an acidic pH during persistent infection. Infect. Immun. 1992;60:5013–5016. doi: 10.1128/iai.60.12.5013-5016.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berón W., Gutierrez M.G., Rabinovitch M., Colombo M.I. Coxiella burnetii Localizes in a Rab7-Labeled Compartment with Autophagic Characteristics. Infect. Immun. 2002;70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howe D., Melnicákova J., Barák I., Heinzen R.A. Fusogenicity of the Coxiella burnetii Parasitophorous Vacuole. Ann. N. Y. Acad. Sci. 2003;990:556–562. doi: 10.1111/j.1749-6632.2003.tb07426.x. [DOI] [PubMed] [Google Scholar]
- 16.Sandoz K.M., Sturdevant D.E., Hansen B., Heinzen R.A. Developmental transitions of Coxiella burnetii grown in axenic media. J. Microbiol. Methods. 2014;96:104–110. doi: 10.1016/j.mimet.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Larson C.L., Beare P.A., Voth D.E., Howe D., Cockrell D.C., Bastidas R.J., Valdivia R.H., Heinzen R.A. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect. Immun. 2015;83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weber M.M., Chen C., Rowin K., Mertens K., Galvan G., Zhi H., Dealing C.M., Roman V.A., Banga S., Tan Y., et al. Identification of Coxiella burnetii Type IV Secretion Substrates Required for Intracellular Replication and Coxiella-Containing Vacuole Formation. J. Bacteriol. 2013;195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa T.R.D., Ilangovan A., Ukleja M., Redzej A., Santini J.M., Smith T.K., Egelman E.H., Waksman G. Structure of the Bacterial Sex F Pilus Reveals an Assembly of a Stoichiometric Protein-Phospholipid Complex. Cell. 2016;166:1436–1444.e10. doi: 10.1016/j.cell.2016.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christie P.J., Vogel J.P. Bacterial type IV secretion: conjugation systems adapted to deliver effector molecules to host cells. Trends Microbiol. 2000;8:354–360. doi: 10.1016/s0966-842x(00)01792-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sexton J.A., Vogel J.P. Type IVB Secretion by Intracellular Pathogens: Type IVB Secretion by Intracellular Pathogens. Traffic. 2002;3:178–185. doi: 10.1034/j.1600-0854.2002.030303.x. [DOI] [PubMed] [Google Scholar]
- 22.Vogel J.P. Turning a tiger into a house cat: using Legionella pneumophila to study Coxiella burnetii. Trends Microbiol. 2004;12:103–105. doi: 10.1016/j.tim.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Campoy E.M., Zoppino F.C.M., Colombo M.I. The Early Secretory Pathway Contributes to the Growth of the Coxiella -Replicative Niche. Infect. Immun. 2011;79:402–413. doi: 10.1128/IAI.00688-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonough J.A., Newton H.J., Klum S., Swiss R., Agaisse H., Roy C.R. Host Pathways Important for Coxiella burnetii Infection Revealed by Genome-Wide RNA Interference Screening. mBio. 2013;4:e00606–e00612. doi: 10.1128/mBio.00606-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandoz K.M., Popham D.L., Beare P.A., Sturdevant D.E., Hansen B., Nair V., Heinzen R.A. Transcriptional Profiling of Coxiella burnetii Reveals Extensive Cell Wall Remodeling in the Small Cell Variant Developmental Form. PLoS One. 2016;11:e0149957. doi: 10.1371/journal.pone.0149957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Newton H.J., McDonough J.A., Roy C.R. Effector Protein Translocation by the Coxiella burnetii Dot/Icm Type IV Secretion System Requires Endocytic Maturation of the Pathogen-Occupied Vacuole. PLoS One. 2013;8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton P., Thomas D.R., Reed S.C.O., Lau N., Xu B., Ong S.Y., Pasricha S., Madhamshettiwar P.B., Edgington-Mitchell L.E., Simpson K.J., et al. Lysosomal degradation products induce Coxiella burnetii virulence. Proc. Natl. Acad. Sci. USA. 2020;117:6801–6810. doi: 10.1073/pnas.1921344117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosal D., Kaplan M., Chang Y.-W., Jensen G.J. In Situ Imaging and Structure Determination of Bacterial Toxin Delivery Systems Using Electron Cryotomography. Methods Mol. Biol. 2019;1921:249–265. doi: 10.1007/978-1-4939-9048-1_16. [DOI] [PubMed] [Google Scholar]
- 29.Macé K., Vadakkepat A.K., Redzej A., Lukoyanova N., Oomen C., Braun N., Ukleja M., Lu F., Costa T.R.D., Orlova E.V., et al. Cryo-EM structure of a type IV secretion system. Nature. 2022;607:191–196. doi: 10.1038/s41586-022-04859-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang Y.-W., Shaffer C.L., Rettberg L.A., Ghosal D., Jensen G.J. In Vivo Structures of the Helicobacter pylori cag Type IV Secretion System. Cell Rep. 2018;23:673–681. doi: 10.1016/j.celrep.2018.03.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu B., Khara P., Song L., Lin A.S., Frick-Cheng A.E., Harvey M.L., Cover T.L., Christie P.J. In Situ Molecular Architecture of the Helicobacter pylori Cag Type IV Secretion System. mBio. 2019;10 doi: 10.1128/mBio.00849-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shepherd D.C., Dalvi S., Ghosal D. From cells to atoms: Cryo-EM as an essential tool to investigate pathogen biology, host–pathogen interaction, and drug discovery. Mol. Microbiol. 2022;117:610–617. doi: 10.1111/mmi.14820. [DOI] [PubMed] [Google Scholar]
- 33.McCaul T.F The developmental cycle of Coxiella burnetii. In Q Fever The Biology of Coxiella bumetii. In (J. C. Williams and H. A. Thompson, eds.) (CRC Press, Boca Raton.), pp. 223-258
- 34.McCAUL T.F., Williams J.C. Localization of DNA in Coxiella burnetii by Post-embedding Immunoelectron Microscopy. Ann. N. Y. Acad. Sci. 1990;590:136–147. doi: 10.1111/j.1749-6632.1990.tb42216.x. [DOI] [PubMed] [Google Scholar]
- 35.McCaul T.F., Dare A.J., Gannon J.P., Galbraith A.J. In vivo endogenous spore formation by Coxiella burnetii in Q fever endocarditis. J. Clin. Pathol. 1994;47:978–981. doi: 10.1136/jcp.47.11.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Omsland A., Cockrell D.C., Howe D., Fischer E.R., Virtaneva K., Sturdevant D.E., Porcella S.F., Heinzen R.A. Host cell-free growth of the Q fever bacterium Coxiella burnetii. Proc. Natl. Acad. Sci. USA. 2009;106:4430–4434. doi: 10.1073/pnas.0812074106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Park D., Steiner S., Shao M., Roy C.R., Liu J. Developmental Transitions Coordinate Assembly of the Coxiella burnetii Dot/Icm Type IV Secretion System. Infect. Immun. 2022;90:e0041022. doi: 10.1128/iai.00410-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosal D., Chang Y.-W., Jeong K.C., Vogel J.P., Jensen G.J. In situ structure of the Legionella Dot/Icm type IV secretion system by electron cryotomography. EMBO Rep. 2017;18:726–732. doi: 10.15252/embr.201643598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jeong K.C., Ghosal D., Chang Y.-W., Jensen G.J., Vogel J.P. Polar delivery of Legionella type IV secretion system substrates is essential for virulence. Proc. Natl. Acad. Sci. USA. 2017;114:8077–8082. doi: 10.1073/pnas.1621438114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosal D., Jeong K.C., Chang Y.-W., Gyore J., Teng L., Gardner A., Vogel J.P., Jensen G.J. Molecular architecture, polar targeting and biogenesis of the Legionella Dot/Icm T4SS. Nat. Microbiol. 2019;4:1173–1182. doi: 10.1038/s41564-019-0427-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheedlo M.J., Durie C.L., Chung J.M., Chang L., Roberts J., Swanson M., Lacy D.B., Ohi M.D. Cryo-EM reveals new species-specific proteins and symmetry elements in the Legionella pneumophila Dot/Icm T4SS. Elife. 2021;10:e70427. doi: 10.7554/eLife.70427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Omsland A., Beare P.A., Hill J., Cockrell D.C., Howe D., Hansen B., Samuel J.E., Heinzen R.A. Isolation from Animal Tissue and Genetic Transformation of Coxiella burnetii Are Facilitated by an Improved Axenic Growth Medium. Appl. Environ. Microbiol. 2011;77:3720–3725. doi: 10.1128/AEM.02826-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luedtke B.E., Mahapatra S., Lutter E.I., Shaw E.I. The Coxiella Burnetii type IVB secretion system (T4BSS) component DotA is released/secreted during infection of host cells and during in vitro growth in a T4BSS-dependent manner. Pathog. Dis. 2017;75:ftx047. doi: 10.1093/femspd/ftx047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Costa T.R.D., Felisberto-Rodrigues C., Meir A., Prevost M.S., Redzej A., Trokter M., Waksman G. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat. Rev. Microbiol. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- 45.Larson C.L., Martinez E., Beare P.A., Jeffrey B., Heinzen R.A., Bonazzi M. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol. 2016;11:919–939. doi: 10.2217/fmb-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu B., Khara P., Christie P.J. Structural bases for F plasmid conjugation and F pilus biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA. 2019;116:14222–14227. doi: 10.1073/pnas.1904428116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chetrit D., Hu B., Christie P.J., Roy C.R., Liu J. A unique cytoplasmic ATPase complex defines the Legionella pneumophila type IV secretion channel. Nat. Microbiol. 2018;3:678–686. doi: 10.1038/s41564-018-0165-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oswald W., Thiele D. A Sporulation Gene in Coxiella burnetii? J. Vet. Med. Ser. B. 1993;40:366–370. doi: 10.1111/j.1439-0450.1993.tb00151.x. [DOI] [PubMed] [Google Scholar]
- 49.Heinzen R.A. In: Rickettsial Infection and Immunity Infectious Agents and Pathogenesis. Anderson B., Friedman H., Bendinelli M., editors. Kluwer Academic Publishers; 2002. Intracellular Development of Coxiella burnetii; pp. 99–129. [DOI] [Google Scholar]
- 50.Gürtler L., Bauerfeind U., Blümel J., Burger R., Drosten C., Gröner A., Heiden M., Hildebrandt M., Jansen B., Offergeld R., et al. Coxiella burnetii - Pathogenic Agent of Q (Query) Fever (2014) Transfus. Med. Hemotherapy. 2014;41:60–72. doi: 10.1159/000357107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Seshadri R., Paulsen I.T., Eisen J.A., Read T.D., Nelson K.E., Nelson W.C., Ward N.L., Tettelin H., Davidsen T.M., Beanan M.J., et al. Complete genome sequence of the Q-fever pathogen Coxiella burnetii. Proc. Natl. Acad. Sci. USA. 2003;100:5455–5460. doi: 10.1073/pnas.0931379100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dobro M.J., Oikonomou C.M., Piper A., Cohen J., Guo K., Jensen T., Tadayon J., Donermeyer J., Park Y., Solis B.A., et al. Uncharacterized Bacterial Structures Revealed by Electron Cryotomography. J. Bacteriol. 2017;199:e00100-17. doi: 10.1128/JB.00100-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Egelman E.H., Stasiak A. Structure of helical RecA-DNA complexes. J. Mol. Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- 54.Mizuno N., Dramićanin M., Mizuuchi M., Adam J., Wang Y., Han Y.-W., Yang W., Steven A.C., Mizuuchi K., Ramón-Maiques S. MuB is an AAA+ ATPase that forms helical filaments to control target selection for DNA transposition. Proc. Natl. Acad. Sci. USA. 2013;110:E2441–E2450. doi: 10.1073/pnas.1309499110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wolf S.G., Frenkiel D., Arad T., Finkel S.E., Kolter R., Minsky A. DNA protection by stress-induced biocrystallization. Nature. 1999;400:83–85. doi: 10.1038/21918. [DOI] [PubMed] [Google Scholar]
- 56.Hoover T.A., Culp D.W., Vodkin M.H., Williams J.C., Thompson H.A. Chromosomal DNA Deletions Explain Phenotypic Characteristics of Two Antigenic Variants, Phase II and RSA 514 (Crazy), of the Coxiella burnetii Nine Mile Strain. Infect. Immun. 2002;70:6726–6733. doi: 10.1128/IAI.70.12.6726-2733.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hackstadt T., Messer R., Cieplak W., Peacock M.G. Evidence for proteolytic cleavage of the 120-kilodalton outer membrane protein of rickettsiae: identification of an avirulent mutant deficient in processing. Infect. Immun. 1992;60:159–165. doi: 10.1128/iai.60.1.159-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng S.Q., Keszthelyi B., Branlund E., Lyle J.M., Braunfeld M.B., Sedat J.W., Agard D.A. UCSF tomography: an integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J. Struct. Biol. 2007;157:138–147. doi: 10.1016/j.jsb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 60.Chen M., Bell J.M., Shi X., Sun S.Y., Wang Z., Ludtke S.J. A complete data processing workflow for cryo-ET and subtomogram averaging. Nat. Methods. 2019;16:1161–1168. doi: 10.1038/s41592-019-0591-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen M., Dai W., Sun S.Y., Jonasch D., He C.Y., Schmid M.F., Chiu W., Ludtke S.J. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nat. Methods. 2017;14:983–985. doi: 10.1038/nmeth.4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goddard T.D., Huang C.C., Meng E.C., Pettersen E.F., Couch G.S., Morris J.H., Ferrin T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis: UCSF ChimeraX Visualization System. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The subtomogram averages have been deposited to EMBD (EMD-28281, EMD-28282 and EMD-28283) and will be publicly accessible. The raw data is available upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data presented in this study is available from the lead contact upon request.