Summary
The impact of the peptide amino acids side-chain modifications on the immunological recognition has been scarcely explored. We investigate here the effect of methionine oxidation on the antigenicity of the melanoma immunodominant peptide 369-YMDGTMSQV-377 (YMD). Using CD8+ T cell activation assays, we found that the antigenicity of the sulfoxide form is higher when compared to the YMD peptide. This is consistent with free energy computations performed on HLA-A∗02:01/YMD/TCR complex showing that this is lowered upon oxidation, paired with a steep increase in order at atomic level. Oxidized YMD forms were identified at the melanoma cell surface by LC-MS/MS analysis. These results demonstrate that methionine oxidation in the antigenic peptides may generate altered peptide ligands with increased antigenicity, and that this oxidation may occur in vivo, opening up the possibility that high-affinity CD8+ T cells might be naturally primed in the course of melanoma progression, as a result of immunosurveillance.
Subject areas: Immunology, Biochemistry, Cancer
Graphical abstract

Highlights
-
•
Oxidation of the Met residues in YMD tyrosinase yields sulfoxides/sulfone peptides
-
•
LC-MS analysis detects oxidized YMD-HLA∗02:01 complexes at the cell surface
-
•
YMD-restricted CD8+ T cell clones assays reveal that oxidation increases activation
-
•
ΔG computations indicate tighter binding upon oxidation in HLA-YMD-TCR complex
Immunology; Biochemistry; Cancer
Introduction
The increasing incidence of malignant melanoma1 and the high mortality due to malignant skin tumors2 require therapeutic options with improved efficiency and lower toxicity. Although in the last years immune checkpoint therapy revealed an improved clinical efficacy,3,4,5,6 still a large proportion of patients do not respond to it and significant immune-related toxicity may occur.7 Recent studies suggest enhanced immunogenic response when combinatorial immunotherapy is used.8,9,10 Therapeutic vaccines have shown efficacy in preclinical studies. This strategy is based on the ability of cytotoxic T cells (CTL) to recognize through their T cell receptor (TCR) different human leukocyte antigen (HLA)-peptide complexes exposed at the cancer cell surface to specifically kill tumor cells.
One promising cancer antigen for translational research in immunotherapy is tyrosinase.11 The maturation and glycosylation of this antigen depends upon the endoplasmic reticulum (ER)-associated quality control.12,13 In melanoma, tyrosinase is frequently retained in the ER as a misfolded protein and sent to degradation, resulting in the production of peptides presented at the cell surface by HLA molecules.14,15,16 While these have shown to induce detectable specific T cell responses in patients, peptide-based vaccination proved limited patient benefit in clinical trials.17 One strategy to circumvent this problem is to design antigens with increased immunogenicity, by the modification of the peptide’s amino acid sequence in order to provide a better immune response.18 Although initially this approach showed promising results in vitro,19 it did not deliver the expected clinical response in patients with melanoma.20 Another alternative is to chemically modify the amino acid side chain of the HLA-associated peptides. The finding that the tyrosinase-derived epitope Tyr369-377 (YMNGTMSQV) is presented at the cell surface of melanoma cells in the deamidated form by the conversion of asparagine-Asn to aspartic acid-Asp (N371D resulting YMDGTMSQV - YMD peptide) suggested that chemical modifications of HLA-associated peptides can have an impact on the immunological scale.21 This was further confirmed by functional T cell assays, which revealed the existence of distinct CD8+ T cell clones for the Asn and Asp containing sequence in patients with melanoma.22 However, this epitope contains other amino acids susceptible to potential modifications and given its use in clinical trials for melanoma,23 these could also alter its immunological response.
It is well known that peptides and proteins containing methionine (Met) or cysteine residues can become oxidized under different conditions leading to the generation of multiple chemical species with distinct physicochemical and even biological properties.24 However, little is known about the immunological response of such modifications and previous reports in this field were inconclusive. Initial results pointed to the hypothesis that oxidized HLA-associated peptides could boost the immune response in patients with cancer, as it was found that the oxidation of an ovarian cancer cell line enhanced its immunogenic response by increasing antigen processing and cross-presentation by dendritic cells.25 Though CD8+ T cell clones specific for the human immunogenic peptide CMV495-503 displayed reduced antigen avidity for the sulfoxide version of the peptide, the oxidized version and the native peptide showed similar HLA bindings.26 On the other hand, both, the sulfoxide- and the sulfone-modified forms of the tyrosinase reversed spliced peptide IYMDGTADFSF were similarly recognized by specific CTL clones, compared with the non-modified version of the peptide.27 Moreover, recent studies suggest that peptide chemical modifications are involved in regulation of MHCII-specific T cells activation.28,29 In a similar approach, during our initial experiments in which we aimed to study the impact of N-glycosylation on the presentation of YMD peptide [16], we observed the spontaneous oxidation of this epitope using liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis.
Here, we address several important features of this oxidation in which we show that the peptide can be oxidized on both of its Met residues, yielding distinct oxidation intermediates (sulfoxides) and final products (sulfone derivatives). We describe the kinetics of the oxidation, study the HLA-binding properties of the oxidized forms, and show that the Met oxidation products trigger a better cytotoxic activity of CD8+ T cell clones isolated from peripheral blood mononuclear cells (PBMC) of patients with melanoma. We also investigate the potential HLA presentation of such oxidized forms by melanoma cells. To our knowledge, this is the first report clearly showing that the oxidized products of an epitope can trigger a better immune response ex vivo, addressing important questions from a biochemical and immunological point of view. These findings pave the road for a new window of opportunity in peptide-based vaccination immunotherapy of melanoma.
Results
Analysis of the tyrosinase 369-377 epitope oxidation
The immunogenic epitope Tyr369-377 (YMD peptide) is one of the most abundant epitopes derived from tyrosinase, estimated in the order of thousands of presented copies on melanoma cells.30 This contains two methionine residues, which can undergo oxidation21,31 to different forms resulting in multiple chemical derivatives (Figure 1A). While previous investigations have not described in detail the oxidized forms, the LC-MS analysis of the standard YMD peptide revealed besides the native form, three additional peaks, designated as M2SOM6SO, M2SO, and M6SO (Figure 1B, upper panel), corresponding to a mass shift increase of 32 and respective 16 Da each. Analysis of the CID fragmentation pattern of these precursors confirmed the presence of oxidized forms of the YMD peptide, pinpointing the location of the oxidation for each peak (Figures S1A–S1D). Thus, the peptide can become spontaneously oxidized to one or both methionine residues from its sequence by the addition of an oxygen atom. A similar pattern of oxidation was observed when analyzing the YMD peptide isolated from the surface of a melanoma cell line modified to express high level of tyrosinase molecules32 (Figure 1B, lower panel). However, we did not detect oxidation derivatives with sulfone forms of the peptide in biological samples. These results indicated that YMD peptide processed from tyrosinase could be oxidized either during the LC-MS analysis, as found for other peptides,24 or endogenously in a subcellular compartment and presented in complex with HLA-A2 at the cell surface. We further scrutinized the significance of the YMD oxidation in antigen presentation.
Figure 1.
Identification of the YMD-oxidized derivatives using mass spectrometry and HPLC profile of its oxidation potential
(A) Schematic representation of the theoretical Met-oxidized forms of the YMD peptide. ∗ designates experimentally identified peptide variants in this work.
(B) LC-MS identification of the YMD peptide Met-oxidized derivatives from the standard peptide (upper panel) and from an acidic cell surface elution of A375 melanoma cells, stably expressing tyrosinase (A375+Tyr): extracted ion chromatograms for the native (N – m/z 516.21226), Met2 sulfoxide (M2SO – m/z 524.20973), Met6 sulfoxide (M6SO – m/z 524.20973), and the sulfoxide at Met2 and Met6 (M2SOM6SO – m/z 532.20717) of the YMD peptide. Retention time (min) and identity are assigned to each peak. The calculated mass of each peak corresponds to an increase of 16 and respective 32 Da compared to the native form.
(C) The standard peptide was subject to a series of oxidation reactions with several concentrations of hydrogen peroxide as described under STAR methods. An aliquot of 1 μg of peptide was analyzed using reversed phase high-performance liquid chromatography (RP-HPLC) for the separation of the oxidized products (left panel). Peak integration was performed for the annotated peaks N, M2SO, M6SO, and M2SOM6SO at each peroxide concentration. At 120 mM, ∼15% of the peptide is still detected as native, while at concentrations of 600 mM and higher, the peptide is almost completely oxidized (right panel). Points are mean of triplicate replicates and error bars are SEM.
(D) Similar to C, but the oxidation reaction was monitored in time (left panel) to derive the kinetics of the reaction (right panel). It can be observed that after 60 min of incubation with hydrogen peroxide, less than 10% native form was detected. Inset: a linear correlation (R2: 0.9766) was found between the log of the native peptide remained fraction and the reaction time suggesting a pseudo-first-order reaction ([N]0 and [N]t are the concentrations of the reduced Met at the initial t0 time and time t).
Since peptides containing methionine residues can be oxidized spontaneously during storage and/or manipulation,24,33 we examined the susceptibility to oxidation of the peptide following incubation with hydrogen peroxide. We tested various concentrations of oxidant and separated the oxidized forms of the peptide using reversed phase high-performance liquid chromatography coupled with UV-visible detection. Incubation of the peptide with different concentrations of hydrogen peroxide resulted in a concentration-dependent decrease of retention on a reversed phase C18 column. This was observed by the appearance of three earlier eluting peaks, designated as M2SOM6SO, M2SO, and M6SO (Figure 1C, left panel). The observed shift to more hydrophilic species suggests that each peak represents a different oxidation form of the peptide. Since no apparent change in 275 nm absorbance for the native and oxidized forms of the peptide was observed, the peaks were quantified by integration of the absorbance, highlighting the transition from native (N) to partial sulfoxide forms (M2SO and M6SO) and to complete sulfoxide derivative M2SOM6SO, (Figure 1C, right panel). Without peroxide treatment, minor amounts of monosulfoxide derivatives can be observed in the standard (M6SO), reflecting the natural tendency of the peptide to incorporate oxygen atoms in aqueous solutions. Low amounts of hydrogen peroxide induced the appearance of both monosulfoxide forms M2SO, M6SO. Starting at 24 mM, the complete sulfoxide form was detected. There is a significant change at 120 mM hydrogen peroxide, when only roughly 15% is still detected as native (Figure 1C, right panel). At concentrations of 600 mM or higher, almost complete transformation of the peptide to sulfoxide forms was observed after 30 min incubation at 4°C. Thus, the peptide can become oxidized starting at millimolar hydrogen peroxide concentrations. To obtain higher peptide quantities from each sulfoxide variant, we selected the concentration of 120 mM hydrogen peroxide. As seen in Figure 1D, right panel after 60 min incubation of the peptide with 120 mM hydrogen peroxide at 4°C less than 10% was detected as a native form (Figure 1D, right panel). The oxidation reaction follows a pseudo-first-order kinetics (Figure 1D, inset from right panel), with similar characteristics of the protein-bound Met residues oxidation.34 Using these conditions, we determined that 50% should oxidize at 120 mM hydrogen peroxide in almost 20 min. It was previously reported that the spliced peptide IYMDGTADFSF from tyrosinase spontaneously oxidizes to sulfoxide and under strong oxidative conditions, to sulfone, gaining one and two oxygen atoms, respectively, at its Met side chain.27 This is an important aspect, since the spliced peptide shares the amino acids from position 2 to position 6 with the YMD peptide, so one could expect that at least the Met from the second position of the YMD sequence could be additionally oxidized.
To test this hypothesis, we performed a time-dependent oxidation assay under strong oxidative conditions. As expected, a high-performance liquid chromatography (HPLC) injection immediately after the addition of 3 M hydrogen peroxide resulted in complete oxidation of the native peptide to the M2SOM6SO form (Figure S1E). However, after 24 h, two additional peaks were observed, concomitant with a drop in the signal for the M2SOM6SO peak, indicating partial sulfone derivatives: M2SO2M6SO and M2SOM6SO2. After 48 h incubation of the peptide in 3 M hydrogen peroxide, an additional, third late eluting peak was observed (Figure S1E) indicating complete sulfone-oxidized derivative M2SO2M6SO2. Thus, under strong oxidative conditions, the peptide can be completely oxidized to sulfone forms. However, peak quantification analysis revealed that the kinetics of the reaction is rather slow compared to the oxidation to sulfoxide forms. After 10 days, almost 66% of the total was found as complete oxidized to sulfone forms (Figure S1E, right panel).
Characterization of the YMD-oxidized HPLC fractions
Thus, after establishing the oxidation kinetics conditions, we upscaled the HPLC method to obtain each oxidative form of the YMD peptide. The specificity of the oxidation reaction was confirmed by MS/MS analysis of the HPLC-purified fractions. A targeted-like method was used, in which all masses of the native peptide and peptides corresponding to different oxygen molecules addition +16, +32, +48, and +64 Da were included for fragmentation along the ∼60 min gradient. We focused on the analysis of the double-charged precursors (Table S1) to capture as many fragment ions as possible (+1 or +2), which would confirm the peptide sequence and its modifications.
To increase the confidence of our result, we devised the method with Orbitrap acquisition for its full-range CID spectra, thus employing a parallel reaction monitoring-like approach in our data sample acquisition. Our results confirmed oxidation of the peptide-Met residues as observed in Figures 2 and S2.
Figure 2.
Identification of YMD oxidative modifications from the HPLC fractions and their purity estimation
(A) Theoretical b and y ions detectable following CID fragmentation of the std. YMD peptide. The diagnostic ions, flanking the two Met residues, used in our analysis are labeled in green.
(B) Schematic reaction of the Met sulfoxide side-chain neutral loss during CID fragmentation. Sulfoxide peptides can lose methanesulfenic acid following CID fragmentation rendering −64 Da neutral loss species.
(C) MS/MS fragmentation pattern obtained by CID fragmentation of the mass corresponding to the Met6 sulfoxide version (M6SO – upper panel) and the native peptide (N – lower panel). Pink lines correspond to the theoretical b and y ions obtained by in silico fragmentation of M6SO, respective native peptide. Black lines denote the m/z of the ions detected following fragmentation and blue and red peaks map the b and y ions. Green peaks denote the ions flanking the two Met residues (Met2 and Met6). The green arrows above the MS/MS spectra also indicate these ions.
(D) Extracted ion chromatograms (EIC) corresponding to the precursor ions (left panel) and product ions flanking the two Met residues for the M6SO version. For both precursor and product ions, the mass accuracy is indicated and the corresponding retention time.
(E) Similar as in C, but for the Met2 sulfoxide version (M2SO).
(F) Similar as in D, but for the M2SO.
(G) Similar as in C, but for sulfoxide version of both Met residues (M2SOM6SO).
(H) Similar as in D but for M2SOM6SO.
(I) UV-VIS HPLC trace at 275 nm assesed on a Hypersil Gold preparative column cat no 25005-109070, ThermoFisher Scientifc of the fractions corresponding to the native (N-first panel), Met2 sulfoxide (M2SO - second panel), Met6 sulfoxide (M6SO – third panel), Met2 and Met6 sulfoxide versions (M2SOM6SO – fourth panel).
More specific, following careful analysis of the CID spectrum obtained for the stdYMD native peptide, we noticed the presence of b5+, b6+ and y3+, y4+ ions neighboring the Met residue from the sixth position and b2+, y7+, and y82+ for the second Met residue (Figure 2C, lower panel). Since we aimed to use two pairs of ions for each of the Met residues and the ion corresponding to b1 was detected at a very low abundance, we looked for alternative ions in the MS/MS spectra. Guan et al. reported the formation of characteristic neutral loss (NL) ions following CID fragmentation of sulfoxide methionine peptides,35 resulting in methanesulfenic acid (CH3SOH, 64 Da) loss (Figure 2B), which is a unique characteristic of sulfoxide peptides, as the authors suggest using these NL ions to discriminate between isobaric ions. Thus, we also included the 64 Da NL of the y82+ in our analysis (y#82+) to investigate two pairs of ions flanking the two Met residues (y3+, y4+ and b5+, b6+ for M6 and respectively y7+, y82+, b2+, y#82+ for M2, these are mapped in green and indicated by green arrows above the MS/MS spectra in Figures 2 and S2). The MS/MS spectra of the M6SO confirm the oxidative modification and its position to the sixth Met residue as the ion b6+ displays a shift of 16 Da compared with the same ion in the native spectra (Figure 2C). At the same time, y3+ shows the same m/z thus confirming that no amino acid is oxidized in the C terminus of the peptide. Moreover, y4+ shows a similar 16 Da shift while the ion b2+ displays no increase of the mass thus confirming the oxidation and its position on the sixth Met. These observations are also confirmed by the transitions of the Met flanking ions obtained following extraction of their ion current as these confirmed the oxidation and its position (Figure 2D).
Similarly, for the M2SO, we observed a 16 Da increase for the b2+ and y82+ ions while no modification for the y3+, y4+, or y7+ ions suggesting the oxidation of the second Met residue (Figure 2E). The y#82+ ion was found as modified. The extracted ion chromatograms confirmed the modification for both the precursors and product ions (Figure 2F, left and right panels).
Furthermore, we analyzed the CID MS/MS spectra corresponding to the M2SOM6SO (Figure 2G). We observed a net increase of 16 Da for the mass corresponding to the ion b2+, compared with the native (Figure 2G). At the same time, the ion b5+ displays a similar 16 Da higher mass compared with the native while the ion b6+ shows a +32 Da mass increase compared with the native strongly suggesting that M6 is also sulfoxide (Figure 2G). Furthermore, the oxidative modifications were also confirmed by the y ions series as y3+ displayed no mass increase compared with the native, suggesting no oxidation toward C terminus and y4+ indeed displayed a mass shift of 16 Da. Moreover, y82+ was found at a 32 Da higher mass compared with the native, while the y7+ displayed only a 16 Da higher mass, thus also confirming the oxidation of the second Met residue to the sulfoxide variant. These were also evidenced by analysis of the transition for the M2SOM6SO (Figure 2H).
Using a similar strategy, we have analyzed the MS/MS CID spectra corresponding to the partial sulfone M2SO2M6SO (Figures S2A and S2B), M2SOM6SO2 (Figures S2C and S2D), and sulfone M2SO2M6SO2 (Figures S2E and S2F), described in detail in Data S1.
Thus, we have identified and mapped the oxidative modifications for each peptide variant of each of its Met residues. We next focused our attention for the purity estimation of our fractions. We thus re-injected fractions corresponding to the baseline-resolved peaks and confirmed that indeed the 275 nm detection chromatogram displayed a single eluting peak with different retention times according to the oxidative variants hydrophobicity (Figure 2I). Moreover, we had taken advantage of our dedicated method to also analyze the purity of the HPLC fractions using nanoLC-MS/MS data discussed in detail in Data S1 and displayed the results in Figure S2H, confirming our results.
The oxidized peptide binds to HLA-A∗02:01 with similar affinity as the native peptide
We next evaluated the relative binding affinities of the native and sulfoxide forms of the peptide. We performed a competition-binding assay based on the ability of YMD antigenic peptide forms (competitor peptides) to compete binding of a fluorescent, Cy5-labeled HBVc, peptide to MHC class I from the surface of T2 cells.36 The positive and negative controls (ELAGIGILTV (Melan-A26-35) and EAAGIGILTI) used displayed the expected trend, validating our assay (Figure S3A). The results did not reveal any significant differences between the native and the bulk of the oxidized YMD (Figure 3A, left and right panels), although a slightly increase was observed for IC50 specific for the oxidized form. This suggests a weaker binding potential of YMD-oxidized derivatives compared to native form for the HLA-A∗02:01.
Figure 3.
HLA-A∗02:01 recognition profile of the YMD peptide-oxidized derivatives
(A and B) Binding affinity of the YMD native (N) and a desalted mix of the oxidized derivative YMD OX (∼54% M2SOM6SO, ∼37% partial sulfone, and ∼7% complete sulfone, HPLC assessed) in A. and HPLC purified YMD sulfoxide derivatives (≥90% purity HPLC assessed) in B. for HLA-A∗02:01, using competition binding assay. Left panel: A representative graph (n = 2 biological replicates) depicting non-linear regression binding-competitive curves for YMD derivatives, error bars are SD of n = 2 technical replicates. Right panel: A violin plot of the corresponding IC50 values estimated from one site-Fit log IC50 equation in Prism software (GraphPad V.9.4.1).
(C) Binding affinity of the native YMD and M2SOM6SO derivative determined by an alternative in vitro binding assay. Left panel: representative graph (n = 2 biological replicates) depicting non-linear regression binding-competitive curves for YMD derivatives, error bars are SD of n = 2 technical replicates. Right panel: A violin plot of the corresponding IC50 values estimated from one site-Fit log IC50 equation in Prism software (GraphPad V.9.4.1).
(D) The HLA-A∗02:01-YMD peptide derivatives stability kinetics across a 72-h time course, error bars are SD of n = 2 technical replicates.
We further tested each sulfoxide form separated by HPLC and did not find any significant differences in the ability to displace the fluorescent peptide between the native and any sulfoxide derivative (Figure 3B, left and right panels). As in the case of bulk sulfoxides, each oxidative derivative showed a slightly increase of IC50, more evident for sulfoxide form, M2SOM6SO, suggesting a weaker binding potential for HLA-A∗02:01 of all oxidized derivatives. Using the same assay, we also tested the sulfone forms of the peptide and did not observe significant differences compared with the native peptide (Figure S3B), only the trend of a weak binding potential of sulfone forms for HLA-A∗02:01.
Furthermore, this was strengthened by the results obtained in a direct (in vitro) competition experiment for the HLA-A∗02:01 between the native and M2SOM6SO variant, our results suggesting that both the native and the sulfoxide derivative display similar affinity for the same HLA-I (Figure 3C, left and right panels). We observed an increased IC50 for the M2SOM6SO as in the competition-binding assay, thus these confirmed our initial observations.
We also assessed the stability of the two complexes in a 72 h time course (Figure 3D) and observed that although the HLA-A∗02:01-M2SOM6SO complex is more stable at t0, the stability decreases more abruptly compared with the one of the HLA-A∗02:01-N complex along the 72 h tested. During this time, the peptides derivatives keep their integrity as assessed by mass spectrometry evaluation (Figure S3C). This would suggest that although the formation of the complex with the M2SOM6SO form is favored compared with the native one (Figure 3D), this does not necessarily translate to a more stable complex in time.
The methionine sulfoxide derivatives of the YMD peptide are more efficiently recognized by cytotoxic Tyr369-377-specific CD8+ T cell clones compared with the native antigenic peptide
We next evaluated the potential of the Tyr369-377-restricted CD8+ T cells, isolated from PBMC of patients with melanoma, to recognize the peptide oxidation forms in complex with HLA-A∗02:01. We used a standard 4-h 51Chromium release assay, where serial dilutions of the native and oxidized peptides are used to pulse the T2 51Cr-labeled cells during their co-culture with tyrosinase-specific CD8+ T cell clones.
We considered first CD8+ YMD-specific T cell activation by the native YMD peptide (YMD N) and the oxidized bulk, a mix of various sulfoxide and sulfone derivatives, purified by C18 stage-tip desalting (Figure 4A). Both Tyr369-377-specific CD8+ T cell clones revealed an increased lysis when the target cells were pulsed with the oxidized peptide compared with the native one. The EC50 values indicated that the oxidized peptide(s) was nearly 100-fold more potent as antigen than the native antigenic peptide (p ≤ 0.01, Figure 4B).
Figure 4.
Activation of cytotoxic CD8+ T cells from patients with melanoma by YMD native and oxidized forms
Recognition efficiency of YMD-specific CD8+ T cell clones for the native peptide (YMD N) and a mix of oxidized form YMD OX (∼54% M2SOM6SO, ∼37% partial sulfone, and ∼7% complete sulfone, HPLC assessed) in A. and the native peptide (YMD N) and HPLC-purified YMD derivatives (≥90% purity HPLC assessed) in C., by a standard 4-h 51Chromium release assay.
(A) Shown are non-linear dose-response regression curves with points denoting average of specific lysis duplicates of T2 pulsed with YMD N and OX, co-cultured with YMD-specific CD8+ T cell clones 3 and 26.
(B) EC50 of YMD OX, for n = 3 YMD-specific CD8+ T cells, applied statistics-unpaired two-sample two-tailed t-test.
(C) Shown are non-linear dose-response regression curves with points denoting the mean of specific lysis duplicates of T2 pulsed with YMD derivatives, co-cultured with YMD-specific CD8+ T cell clones 3, 5, and 21.
(D) EC50 of YMD derivatives for n = 4 YMD-specific CD8+ T cell clones, applied statistics-non-parametric one-way Kruskal-Wallis test with Dunn’s correction (p ≤ 0.05 for N vs. M6SO, p ≤ 0.01 for N vs. M2SOM6SO).
(E) IFNγ secretion assay for the native and sulfoxide derivatives of the YMD peptide. Shown are mean of duplicates for YMD-specific CD8+ T cell clones 58, 5, and 21.
We further tested each sulfoxide derivative obtained by HPLC, using three distinct CD8+ T cell clones. All the clones revealed an increased sensitivity, particularly for the M6 sulfoxide variants (M2SOM6SO and M6SO), as observed in Figure 4C. M2SO displayed a relative similar recognition profile to native across all the peptide dilutions considered (Figure 4C). The EC50 values across all the clones, for each peptide variant showed that at least M2SOM6SO and M6SO forms, are characterized by significant lower values of EC50 (p ≤ 0.05 for M6SO, p ≤ 0.01 for M2SOM6SO, Figure 4D).
Moreover, when we evaluated the sulfone derivatives of the YMD peptide, obtained by HPLC separation, in the same recognition assays, we observed that M6 sulfoxide derivatives also display an increased antigenicity for three distinct CD8+ T cell clones (Figure S4A). However, the complete sulfone variant of the peptide showed variable responses compared with native one, depending on the clone used (Figure S4A). Furthermore, the estimated EC50 values revealed that M6SO derivatives have significant lower values compared with the native (Figure S4B, p ≤ 0.001).
Thus, these results suggest that at least the M6 sulfoxide peptide derivatives are characterized by a relative increased activation of Tyr369-377-restricted CD8+ T cell clones. This observation is further strengthened by the results obtained in IFNγ secretion assay, an independent functional T cell assay, which revealed that at least two distinct CTL clones released more IFNγ when co-cultured with M2SOM6SO and M6SO pulsed T2 cells. In addition, these clones showed no significant differences regarding IFNγ concentration released in the presence of N and M2SO (Figure 4E).
Together, these results indicate that the sulfoxide M2SOM6SO and M6SO variants of the YMD peptide trigger an increased activation of Tyr369-377-restricted CD8+ T cell clones. This was also evidenced by the assay performed using the complete and partial sulfone derivatives, which suggest that at least one of the Met residues should acquire the sulfoxide form to enhance CTL recognition, compared with the native one (Figures S4A–S4C), suggesting that the M6 sulfoxide is mainly responsible for the increased peptide’s antigenicity and that any variant with M6 sulfoxide (M6SO, M2SOM6SO, and M2SO2M6SO) can induce a stronger activation.
Assessment of HLA-A∗02:01-YMD oxidation forms presentation at the cell surface of melanoma cells
We next evaluated whether the oxidized species could be presented at the surface of melanoma cells. To assess this, we used a mild acid elution protocol (MAE) as previously reported.32 This approach allows the direct analysis of the HLA-A∗02:01/YMD complexes and does not require multiple sample processing steps as in the standard HLA-I immunoprecipitation, which could favor oxidation during sample handling. To quantify the extent of the oxidation, we spiked-in, in each sample, a synthetic heavy-labeled version of the YMD peptide (13C15N-Val, +6 Da) at the start of each experiment (Figure 5A). Samples were then analyzed by a custom LC-MS/MS-targeted method screening for the YMD heavy (H) and light (L) native and oxidative variants.
Figure 5.
Assessment of HLA-A∗02:01-YMD sulfoxide complex presentation by melanoma cells
(A) Schematic diagram of the workflow. HLA-A2 and tyrosinase-positive melanoma cells were subjected to the mild acid-elution (MAE) protocol for the YMD peptide sulfoxide estimation. These were compared to the abundance of the same sulfoxide variants of a heavy-labeled YMD peptide (13C15N-Val +6 Da) to assess their presentation.
(B) Flow cytometry analysis for cell surface HLA-A2 detection of melanoma cell line considered. Error bars are SEM of biological triplicates.
(C) Western blot detection of total tyrosinase (left panel) and EndoH-sensitive fraction (right panel) for each melanoma cell line considered.
(D) Results in C were quantified and the relative total tyrosinase expression level (left panel) and the ratio of EndoH resistant/sensitive (right panel) were calculated. Error bars are SEM of biological triplicates.
(E) Adjusted mean ratios for each Met residue of the YMD peptide.
(F) Estimation of YMD copy numbers for each cell line, using only the native form of the light and heavy versions. Error bars are SEM of four-five biological replicates.
We selected four cell lines positive for HLA-A2, as assessed by flow cytometry (Figures 5B and S5A), with various tyrosinase expression levels: Me319, Me275, Me290, and A375 overexpressing human tyrosinase (A375+Tyr) (Figures 5C and 5D left panels). Since previous reports showed a positive correlation between HLA-I cell surface complexes and the ER-retained protein fraction targeted for proteasomal degradation,15,32,37,38 we evaluated tyrosinase maturation in the selected cell lines (Figures 5C and 5D right panels). The results revealed that all the cell lines have a major fraction of misfolded tyrosinase, retained in the ER, with Me290 and A375+Tyr also containing a considerable fraction of mature tyrosinase.
These cell lines were subjected further to the MAE protocol for the detection of possible YMD peptide oxidation forms. We first evaluated the transitions for both, native and oxidized forms, regarding the light and heavy versions of the peptide and these were found to co-elute, as expected (Figures S5B–S5E). We then extracted the L/H ratios of each YMD-peptide derivative and compared these ratios between the native and oxidative forms by dividing the aggregated ratios of each Met-sulfoxide derivative to the L/H ratios of the native form. If the oxidation level would be similar between them, a ratio close to 1 would be expected. However, if the light version would be more oxidized than the heavy version, then the ratios would be > 1 and vice versa if ratios <1 are found. We thus obtained aggregated mean oxidative/native ratios from the L/H ratios extracted from Skyline (Figure S5H). However, we should also consider the oxidation found initially in the heavy peptide and adjust the ratios accordingly (Figure S5G). We thus evaluated this level of oxidation (Figure S5I) and calculated the adjusted ratios (Figure 5E). Moreover, we assessed the variability of oxidation estimation by injection of different peptide amounts (Figure S5J). Our results did not reveal any statistically significant differences regarding the oxidation extent between the exogenous and endogenous forms of the peptide. However, we found slightly higher values of the light peptide oxidation for the YMD presented at the cell surface of some cell lines, particularly for the oxidation of Met6 from its sequence (Figure 5E). We thus conclude that some melanoma cell lines could present at the cell surface oxidized YMD-HLA∗02:01 complexes, but in lower abundance relative to the total number of YMD-presented complexes.
Since we used an external standard, spiked-in in known concentrations in our samples, we were able to compare the melanoma cell lines regarding the number of HLA-A∗02:01-YMD complexes, by considering the native peptide version. As seen in Figure 5F, all melanoma cells presented the HLA-A∗02:01-YMD complexes at the cell surface and besides the expression level or the processing of tyrosinase, there are also other alterations within the melanoma cell influencing the number of presented complexes. Interestingly, as shown before,30 there is an unexpectedly high number of HLA-A∗02:01-YMD complexes in all analyzed tyrosinase-positive cells, ranging from 1,500 to 50,000 copies per cell, even though tyrosinase has a relatively low expression in melanoma cells.
Overall, we conclude that melanoma cells could present at their surface HLA-A∗02:01-oxidized YMD complexes, considering that various oxidative variants are able to form stable complexes with HLA-A∗02:01. These are recognized by tumor-infiltrating Tyr369-377-specific CD8+ T cells and the oxidation levels are slightly upregulated compared with the ones found for the exogenous standard, particularly regarding Met6.
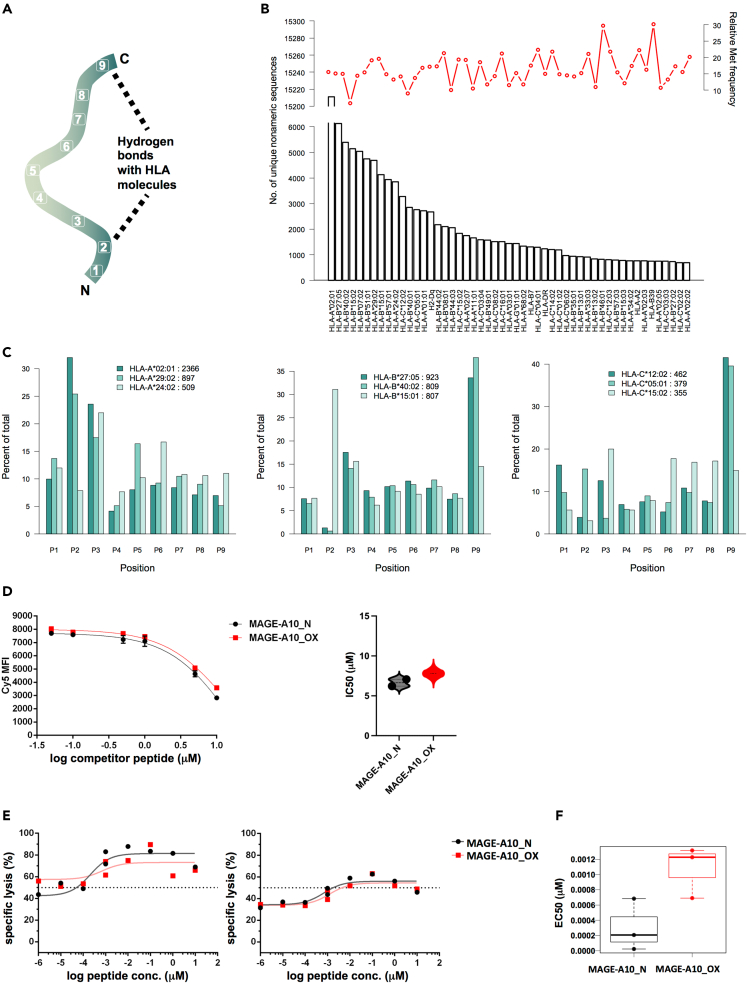
Met is favored in key positions for HLA-A∗02:01 but CD8+ T cell avidity is not generally enhanced for oxidized Met-containing peptides
We next wondered if our observations regarding T cell recognition could be valid for other Met-containing peptides associated with HLA-I considering peptide amino acid positions involved in interactions with HLA-I (Figure 6A). For this, we first interrogated the IEDB database (Immune Epitope Database and Analysis Resource) for epitopes containing Met residues. We observed that roughly over 20% of the presented epitopes have at least one Met residue in their sequence (Figure 6B), although there was a higher variation for the understudied alleles. Compared with Met abundance in UniProtKB database (Figure S6), this is more abundant, suggesting that peptides containing Met residues are favored for HLA class I presentation. We also studied the relative frequency of Met positions in nonameric peptides and observed that in general most of Met residues are found for P2 and P9 positions, although this is also allele dependent (Figure 6C). This could explain why M2SO sulfoxide does not reveal a similar T cell response as M6SO, as this position would require contacts with HLA-I.
Figure 6.
Analysis of Met distribution in IEDB database and the immunogenicity of other peptide suggest a YMD-related structural-based feature responsible for increased recognition
(A) Schematic representation of the hydrogen bonds between the nonameric epitope and the HLA molecule.
(B) Allele-specific analysis of the Met frequency in IEDB epitope database. Only linear human nonameric entries were considered for analysis. For each allele, the number of unique entries is displayed and the corresponding estimated Met frequency.
(C) Analysis of Met relative positional frequency for top three most represented alleles from IEDB database for each HLA class I gene (A – left panel, B – middle panel and C – right panel).
(D) Binding affinity of native MAGE-A10 (MAGE-A10_N) and oxidized form (MAGE-A10_OX) to the HLA-A∗02:01 from T2 cells surface, estimated by competition binding assay. Left panel: representative graph (n = 2 biological replicates) depicting non-linear regression binding-competitive curves for MAGE-A10 derivatives (error bars are SD of n = 2 technical replicates). Right panel: A violin plot of the corresponding IC50 values estimated from one site-Fit log IC50 equation in Prism (GraphPad V.9.4.1).
(E) CTL assay of native and oxidized form of MAGE-A10 peptide. Shown are non-linear dose-response regression curves with points denoting the mean of specific lysis duplicates of T2 pulsed with MAGE-A10 derivatives, co-cultured with MAGE-A10-specific CD8+ T cell clones 2 and 16.
(F) EC50 of MAGE-A10, N, and OX, for n = 3 MAGE-A10-specific CD8+ T cells.
We next examined another HLA-I-associated peptide containing Met residue, the Melanoma Antigen Family A-10 (MAGE-A10) 254–262 peptide (GLYDGMEHL). First, we inspected the binding affinity of oxidized version compared with native and observed that the differences are not significant (Figure 6D, left and right panels). As in the case of oxidized YMD derivatives, a slightly increase for IC50 corresponding to oxidized form was observed, indicating a slight alteration in the potential of oxidized form to bind HLA-A∗02:01.
We further tested antigenic capacity of MAGE-A10254-262-oxidized variant, and we did not find an increase in MAGE-A10-specific CD8+ T cells activation (Figures 6E and 6F, right and left panels) as for Tyr369-377. Unfortunately, these experiments were not very conclusive as the clones were quite sensitive and there was no titration of the antigenic peptides. Even though, estimated EC50 value for oxidized MAGE-A10 254-262 is slightly increased, suggesting a slow decrease in avidity of CD8+ T cells against T2 cells pulsed with oxidized forms. This finding is in agreement with a previous report which has shown that CD8+ T cells stimulated for two weeks with the native peptide version have a decreased avidity toward the HLA-I-CMV495-503 (NLVPMVATV)-oxidized peptide complexes compared with the native version.26
However, these results suggest that the increased CTL response of the sulfoxide variants of the YMD peptide could be more related toward some structural aspects of the TCR-HLA-YMD peptide ternary complex, rather than a more general feature of Met-containing peptides, although this does not rule out the possibility that other Met sulfoxide-containing antigens would show a similar response in CTL assays.
Free energy calculations indicate increased stability of the TCR-HLA-oxidized YMD peptide compared with the native version
To evaluate computationally the effect of YMD oxidation on antigenicity, we generated four structural models of the HLA–YMD–TCRc3 ternary complex with the YMD peptide in oxidized (M2SOM6SO) and N forms, and further calculated the absolute free energies binding of YMD peptide to the HLA-TCRc3 assembly by an alchemical transformation between fully coupled and decoupled ligand states. As described under STAR methods, the work involved the following stages: 1) Modeling the HLA-YMD complexes in N and M2SOM6SO forms, 2) Building the HLA–YMD–TCRc3 models, and 3) Computing the ternary complex absolute binding free energies for YMD-N and M2SOM6SO for two YMD starting conformations as shown in the thermodynamic cycle from Figure 7A. In addition, the fully coupled receptor-ligand simulations were retained for structural analysis.
Figure 7.
Molecular modeling of HLA-A∗02:01/YMD peptide forms/cognate TCR
(A) The thermodynamic cycle on which the estimation of oxidation effect was based.
(B) 3D Model of the two binary complex, in the two conformations (“Up” – Left half; “Down” – Right Half). HLA-A2, β-microglobulin, and both chains of the TCR are shown in gray transparent cartoon representation, the YMD peptide is shown in licorice representation, along with its wireframe surface (1 & 3) and opaque cartoon representation (2 & 4). The methionine residues are shown in orange and their respective sulfur atoms is represented with a yellow sphere, while the rest of the peptide is shown in dark gray.
(C) The overall HLA-A∗02:01-YMD-TCR ternary complex model. HLA-A∗02:01 & β-microglobulin are shown in blue shades; TCR α- and β-chains are shown in magenta shades; YMD peptide is shown in red with M2 and M6 in yellow-mesh/ribbon. 1 – “Up” conformation; 2 – “Down” conformation.
The YMD native and oxidized forms in the two starting conformations were docked into the HLA-I binding groove. The selection of YMD templates performed as described under STAR methods yielded YLSPIASPL and MLIYSMWGK structures as the most appropriate templates for modeling the YMD peptide: YMDGTMSQV. The first template Y9L from Toxoplasma gondii found in the pdb structure 5F9J is bound to the experimental HLA-A∗02:01. This adequately matches the hydrophobicity and volumetric profiles of YMD and similarly, it contains tyrosine in the first position in stacking interaction with HLA-A2. The second peptide derived from Vaccinia virus virion membrane protein A14 (vmpA14), found in the pdb structure 4N8V was chosen because the methionine in position 6 even though the HLA-I type, HLA-A∗11, is different from the experimental one. Both these YMD templates have hydrophobic amino acids in positions 2 and 6, with the side chains pointing toward HLA-I. However, their configuration in the HLA-I groove differs significantly, with side chain in position 6 pointing directly toward the bottom of the groove in the case of Y9L template and pointing almost parallel to the surface in the case of vmpA14 template—allowing in this way to assess the effects induced by the starting configuration of the YMD model on free energy estimates (Figures 7B1 and 7B2).
After docking the TCRc3 models on top of the YMD- HLA-A∗02:01 models, the YMD-M6 side chain showed two stable poses one toward HLA-A∗02:01 (↓) and the second toward TCR (↑) (Figure 7C). The two oxidation states and two poses gave a total of four YMD structures: N↓, N↑, M2SOM6SO↓, and M2SOM6SO↑. These were used as starting structures in absolute binding free energies calculation of native and oxidized epitopes. For the sake of saving computing power, as the flipping between the poses may be assumed as a rare event, calculations were run separately for each pose leading to a total of four experiments. The 4 free energies were then used to compute ΔΔGs between the native and oxidized state in the two poses. Results are shown in Table 1.
Table 1.
Absolute binding free energies of the HRE simulation (reported in kcal/mole)
| ΔG N ↑ | ΔG M2SOM6SO ↑ | ΔG N ↓ | ΔG M2SOM6SO ↓ |
|---|---|---|---|
| −43.070 ± 1.085 | −43.277 ± 0.595 | −17.097 ± 0.669 | −21.05 ± 0.557 |
| ΔΔG↑ | ΔΔG↓ | ||
| ∼0.0 kcal/mol | ∼ −4.0 kcal/mol |
As can be seen, only the difference between free energy variations in the “down” conformation ΔΔG↓ is statistically significant, suggesting that the equilibrium toward the oxidized form seen experimentally in the ternary complex is shifted primarily due to the “down” pose. The fact that the higher affinity for the oxidized form is basically driven by the “down” pose of YMD seems somehow surprising given that one might expect that side-chain oxidation, especially in position 6, i.e., M6SO, should affect “directly” the interaction with TCR. However, care should be taken in this regard as free energy variations are rather a reflection of complex thermodynamic equilibria implying both enthalpic and entropic terms resulting from a very large number of local interactions displayed at room temperature by myriad of molecular copies in various conformations.
To understand the underlying mechanism, we looked more closely at what happens within a 5 Å region around the oxidized methionines in the YMD↓ “down” conformation during fully coupled N↓ vs. M2SOM6SO↓ simulations of ΔG computations (Data S4 and Figure S7). Analysis of the mean residence time of water molecules in these 5 Å M2/M6 regions indicates a significant decrease in water dynamics upon oxidation (Figure S7F). While around 650 water molecules are transiting these regions in the native form during simulations, less than half were transiting the same region in the oxidized M2SOM6SO form. Hence, sulfoxidation reduces the local water mobility in the system. Moreover, during fully coupled ternary complex simulations, stability-related properties at the ternary complex interface greatly increase upon oxidation. This is reflected in a lower RMSF of both YMD↓ peptide and TCRc3-CDR loops in the oxidized form (Figures S7C and S7D). This result was further confirmed by pairwise RMSD and Cluster Count Analysis (Figure S7E). At a clustering cutoff of 1.3 Å, the number of clusters in the native “down” N↓ conformation is significantly higher than in the M2SOM6SO↓ form and comes as strong evidence that the oxidized peptide is also more tightly bound to HLA-A∗02:01 in the “down” conformation. An in-depth analysis of all contacts along the molecular dynamics trajectories indicating further a freezing effect and stronger bonding induced by oxidation in the YMD↓- HLA-A∗02:01 complex formation is presented in Datas S2–S4 and Figure S7.
All these data indicate that the main factor that drives the higher affinity of the oxidized YMD↓- HLA-A∗02:01 for TCRc3 is the enthalpic contribution indicated by a significant increase in local order of the system upon oxidation.
Discussion
In this work, we show that YMD peptide, one of the immunodominant tyrosinase epitopes used in immunotherapy, can become oxidized at both of its methionine residues. The susceptibility of the peptide to form sulfoxide intermediates, and complete sulfoxide products, is clearly higher than the potential for transformation to sulfone derivatives. This observation is in agreement with the findings for the tyrosinase spliced peptide IYMDGTADFSF in which the methionine from the third position converts easily to sulfoxide and only in strong oxidative conditions to sulfone.27 The native and oxidized versions of the YMD peptide show similar HLA-A2 binding affinity (Figures 3A and 3B), including both, sulfoxide and sulfone versions. However, unlike previous reports for other epitopes,26,27 our data show that the oxidized derivatives of the YMD peptide result in increased cytotoxic activity of Tyr369-377-specific CD8+ T cells, compared with the native sequence (Figure 4). This could have a major impact for peptide-based vaccination of patients with melanoma and demonstrates that novel altered peptide ligands could be obtained by in vitro modification. Additionally, we show that such an altered peptide ligand is also generated naturally in melanoma cells, although it is hard to assess the exact contribution of the spontaneous oxidation in the ion current, or where in the cell is the peptide oxidized. However, considering its oxidation potential, it is hard to believe that given different cellular pro-oxidative environments, the peptide would never be oxidized as we observed oxidation even by incubating the peptide at 37°C.
The finding that a T cell clone isolated from a melanoma patient could be primed by the oxidized YMD peptide could suggest that T cells have previously encountered this oxidized species. Specifically, our results suggest that melanoma cells are able to present the HLA-A2-oxidized YMD complex versions to T cells (Figure 5). Several independent observations are in agreement and support this conclusion. First, the existence of specific enzymes MSRs (methionine sulfoxide reductases), which can recognize stereospecifically the oxidized Met residues in proteins and peptides39 reducing them to the native state. Second, the pro-oxidative environment of ER40 where, frequently, misfolded tyrosinase is retained and sent to degradation37 and where the HLA-I-peptide complexes are assembled. And third, the finding that Met374 from tyrosinase is oxidized by intracellular hydrogen peroxide found in millimolar concentrations,41 similar with the ones used in our experiments. Met374 from tyrosinase corresponds to the sixth residue in the amino acid sequence of the YMD antigenic peptide, which we found to trigger an increased lysis of the tumor cells by several HLA-A2-restricted, tyrosinase-specific independent CD8+ T cell clones.
We also investigated the possibility of obtaining similar results for another Met-containing peptide and found that although Met residues are found in anchor positions more frequently (Figure 6C), the oxidative modification does not impose a difference regarding TCR recognition of HLA-I-peptide complexes using a different model-peptide and its corresponding clones: MAGE-A10254-262 peptides. This did not reveal any differences in the avidity of CD8+ T cell clones between the native and oxidized versions of this peptide, suggesting that the results obtained for the YMD peptide are rather due to some specific structural restrictions between the TCR and HLA-A2-sulfoxide peptide complex. This hypothesis was also confirmed by the in silico free energy calculations between the native and oxidized state as shown by Table 1. Nevertheless, these data have to be taken only qualitatively given the many assumptions made in building the model. First, computations were not performed starting from a crystal structure but rather from homology models of the interacting molecular objects with a high degree of uncertainty along the alignment concentrated in the interacting loops of the TCR, which is the very region responsible for the interaction. Moreover, a water molecule was placed in the model at the interface between YMD and HLA-A2 given that this is present in 80% of the analyzed ternary crystal structures. Moreover, the CD8 co-receptor and other stabilizing factors that may impose on the real value of the free energy—such as CD3 or membrane lipid rafts—were not included in the model given the lack of structural information. However, some of the model assumptions were confirmed by computations.
Interestingly, computational experiments indicate that, at room temperature, the YMD peptide may be found in stable HLA-A2-YMD-TCRc3 complexes in more than one conformation. In addition, by oxidation, only some of these YMD conformations generate more stable ternary complexes. Hence, computational data suggest that the underlying mechanism by which YMD oxidation might increase the TCR recognition relies on increasing the local order in the system rather than due to a direct involvement of sulfoxides in the direct interaction with the TCR. This is somehow consistent with the fact that more than two-thirds of the interface with TCR is contributed by HLA-A2 and only one-third to the peptide, and therefore the direct contribution of the peptide atoms in ternary complex formation is far lower than that of HLA-A2. A possible mechanism for increased efficiency of recognition of the oxidized peptide by specific CD8+ T cells relies on the fact that the local HLA-A2 structure is modified by the oxidized YMD, resulting in an increased avidity of the TCR for the HLA-A2-YMD oxidative complex.
Overall, the present results clearly show how oxidative stress could modulate the cellular immune response, in line with some other reports.25,26,28,29 These results demonstrate that oxidation of the side chain of a residue in the antigenic peptide may generate altered peptide ligands with increased antigenicity, and that this oxidation may occur in vivo, opening the possibility that high-affinity CD8+ T cells might be naturally primed in the course of melanoma progression, as a result of immunosurveillance.
Limitations of the study
To assess the level of intracellular oxidation of HLA-presented epitopes, we simultaneously measured by LC-MS the spontaneous oxidation of a control isotope-labeled peptide that could only partially overcome the variations found in the level of HLA-peptide presented by the range of melanoma cells. The computational evaluation of the YMD oxidation started from a homology model in the absence of an available crystal structure; therefore, the model has a level of uncertainty at the TCR-interacting loops level.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Tyrosinase T311 | Santa Cruz Biotechnology | Cat#sc-20035; R RID:AB_628420 |
| Calnexin | Abcam | Cat#ab-22595; R RID:AB_2069006 |
| FITC Mouse Anti-Human HLA-A2 Clone BB7.2 | BD Pharmingen™ | Cat# 551285; R RID:AB_394130 |
| FITC Mouse Anti-Human CD8 Clone RPA-T8 | BD Pharmingen™ | Cat#561948; R RID: AB_11154582 |
| anti-human MHC Class I (HLA-A, HLA-B, HLA-C) W6/32 | Department of Oncology, UNIL-CHUV, University of Lausanne, Epalinges, Switzerland | N/A |
| Donkey anti-mouse | JacksonImmunoResearch | Cat#715-035-150; RRID: AB_2340770 |
| Donkey anti-rabbit | JacksonImmunoResearch | Cat# 711-035-152; RRID: AB_10015282 |
| Chemicals, peptides, and recombinant proteins | ||
| Hydrogen peroxide | Sigma-Aldrich | 16911 |
| Methanol | Sigma-Aldrich | 34966 |
| LC-MS water | Sigma-Aldrich | 39253 |
| Formic acid – FA | Sigma-Aldrich | 56302 |
| Acetonitrile – ACN | Sigma-Aldrich | 34967 |
| MgCl2 | Sigma Aldrich | M4880 |
| NaCl | Santa Cruz Biotechnology | sc-203274 |
| EDTA | Santa Cruz Biotechnology | sc-29092 |
| Citric acid | Merck Millipore | 100241 |
| Na2HPO4 | Santa Cruz Biotechnology | sc-203277 |
| Tris | Santa Cruz Biotechnology | sc-3715 |
| Triton X-100 | Sigma Aldrich | T-9284 |
| Peptide: YM2SODGTM6SOSQV (>98 % purity), | Gene Script | N/A |
| Peptides: YMDGTMSQV (Tyr369-377) light, heavy, YM2SODGTM6SOSQV (>98 % purity), | Gene Script | N/A |
| Peptide: GLYDGMEHL (MAGE-A10254-262) | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Peptides: GILGFVFTL (Influenza matrix58-66) labelled or unlabelled with biotin, | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Peptide: ELAGIGILTV (Melan-A26-35) | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Peptide: EAAGIGILTI | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Peptide: Cy5-labeled HBVc | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Tetramer: APC-labeled HLA-A2/YMDGTMSQV | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Tetramer: APC-labeled HLA-A2/GLYDGMEHL | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| E coli purified HLA-A∗02:01 | Peptide and Tetramer Core Facility, UNIL-CHUV, Epalinges, Switzerland | N/A |
| Human β2-microglobulin | Proteintech | Ag27803 |
| IL-2 | PeproTech | 200-02 |
| Critical commercial assays | ||
| IFNγ -ELISA kit | MyBiosource | MBS161542 |
| Pierce™ BCA Protein Assay Kit | Thermo Fisher Scientific | 23225 |
| Immobilon Crescendo Western HRP substrate | Merck Millipore | WBLUR0500 |
| Deposited data | ||
| Raw and analyzed data | This paper | PXD039318 |
| Experimental models: Cell lines | ||
| Tyrosinase369-377(YMD) and MAGE-A10 specific CD8+ T cells | Ludwig Institute for Cancer Research, Lausanne Branch; Lausanne; Switzerland. | N/A |
| Human T2 cells, CEMx721.174.T2 | Ludwig Institute for Cancer Research, Lausanne Branch; Lausanne; Switzerland. | RRID: CVCL_2211 |
| A375 | European Collection of Animal Cell Cultures | RRID: CVCL_0132 |
| A375-Tyr | Chiritoiu G. N. et al. (2016)32 | 10.1002/elps.201500449 |
| Me 290 | Ludwig Institute for Cancer Research, Lausanne Branch; Lausanne; Switzerland. | RRID: CVCL_S598 |
| Me 275 | Ludwig Institute for Cancer Research, Lausanne Branch; Lausanne; Switzerland. | RRID: CVCL_S597 |
| Me 319 | Ludwig Institute for Cancer Research, Lausanne Branch; Lausanne; Switzerland. | N/A |
| Software and algorithms | ||
| XCalibur v2.2/3.0 | Thermo Fisher Scientific | https://thermo.flexnetoperations.com |
| LTQ Tune v2.70.112SP2 | Thermo Fisher Scientific | https://thermo.flexnetoperations.com |
| Skyline software v2.6.0 | MacCoss Lab Software | https://skyline.ms/project/home/software/Skyline/begin.view |
| CellQuest | BD Bioscience | https://www.bdbiosciences.com/content/dam/bdb/marketing-documents/14_cellquest_prosoft_acquisit.pdf |
| FlowJo | Beckman Coulter | https://www.flowjo.com/solutions/flowjo/downloads |
| Cytobank | Beckman Coulter | https://community.cytobank.org |
| Generalized Amber Force Field - GAFF2 | Wang, J. et al. (2004).42 | https://ambermd.org/AmberModels_organic.php |
| Antechamber from the AmberTools suite | Case, D.A. et al. (2020).43 | http://ambermd.org/antechamber/ac.html |
| OpenMM v.7.4.1 | Eastman, P. et al. (2017).44 | https://github.com/openmm/openmm/releases |
| Modeller v.9.21 | Webb, B. et al. (2016).45 | https://salilab.org/modeller/9.21/manual/ |
| YANK v.0.25.2 | Wang, K. et al. (2013).46 | N/A |
| PyMBAR package | Shirts, M.R. et al. (2008).47 | https://pypi.org/project/pymbar/ |
| MDTraj 1.9.3 | McGibbon, R. T. et al. (2015).48 | https://mdtraj.org/1.9.3/index.html |
| FreeSASA 2.0.3 | Mitternacht, S. (2016).49 | https://pypi.org/project/freesasa/2.0.3/ |
| Scipy v. 1.3.2 | Virtanen, P. et al. (2020).50 | https://pypi.org/project/scipy/1.3.2/ |
| PyMOL v.2.2.3 | Schrödinger, L. (2018).51 | https://pymol.org/2/ |
| Molprobity server | Davis, I.W. et al. (2007).52 | http://molprobity.biochem.duke.edu |
| R v3.6.3 | R Core Team | https://www.r-project.org |
| Prism software (GraphPad v.6.0). | GraphPad | https://www.graphpad.com |
| Prism software (GraphPad v.9.4.1) | GraphPad | https://www.graphpad.com |
| Image J | Image J | https://imagej.nih.gov/ij/download.html |
| Other | ||
| DMEM | Gibco | 31966047 |
| RPMI | Gibco | 61870044 |
| FBS | Gibco | 10270106 |
| Glutamine | Gibco | 25030081 |
| HEPES | Gibco | 15630056 |
| Sodium pyruvate | Gibco | 11360039 |
| Penicillin/Streptomycin | Gibco | 15140122 |
| Geneticin | Invivogen | ant-gn-5 |
| Na251CrO4 (51Cr) | PerkinElmer | NEZ030S |
| EndoH | Neb | P0702 |
| Protease inhibitors | Roche | 11697498001 |
| LumaPlate | PerkinElmer | 6005630 |
| Terasaki multiwell plates | Greiner Bio-One | 654180 |
| v-bottom 96-well plate | Greiner Bio-One | 651101 |
| Analytical column (100 mm or 150 mm x 75 μm ID) packed with 3 μm ReproSil-Pur C18-AQ resin | Dr. Maisch GmbH | https://www.thermofisher.com/ |
| Reversed-phase (RP) C18, 1.9 μm, Hypersil GOLD column (50 mm x 2.1 ID) | Thermo Fisher Scientific (Cat. No.: 25002-052130 | https://www.thermofisher.com/ |
| Preparative RP column C18 (100 x 10 mm, 5 μm) | Thermo Fischer Scientific (Cat. No.: 25005-109070) | https://www.thermofisher.com/ |
| Dionex AFC 3000 fraction collector | Thermo Fisher Scientific | https://www.thermofisher.com/ |
| Easy nanoLC II | Proxeon Biosystems | https://www.thermofisher.com/ |
| LTQ-Orbitrap Velos Pro | Thermo Fisher Scientific | https://www.thermofisher.com/ |
| Nanospray Flex Ion Source | Thermo Fisher Scientific | https://www.thermofisher.com/ |
| HPLC Accela 600 | Thermo Fisher Scientific | https://www.thermofisher.com/ |
| FACS Calibur | BD Bioscience | https://www.bdbiosciences.com/en-eu/products/instruments/flow-cytometers/clinical-cell-analyzers/calibur-discontinuation |
| Gallios | Beckman Coulter | https://www.beckman.com/resources/videos/products/gallios-overview |
| FACS Verse | BD Bioscience | https://www.bdbiosciences.com/en-eu/products/instruments/flow-cytometers/research-cell-analyzers/bd-facsverse |
| TopCount NXT Microplate Scintillation | PerkinElmer | https://resources.perkinelmer.com/corporate/cmsresources/images/44-73884spc_topcountnxtmicropltscint.pdf |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact: Stefana M. Petrescu (stefana_petrescu@yahoo.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Cell lines and TIL cultures
Melanoma cell lines used for assessment of cell surface presentation of HLA-A∗02:01-YMD/YMD oxidised derivatives complexes, were grown as detailed below.
Amelanotic melanoma cell line A37554 (RRID:CVCL_0132), isolated from skin of a 54 years female, from European Collection of Animal Cell Cultures (Porton Down, UK) and A375 modified to overexpress tyrosinase (A375-Tyr),32 were grown in DMEM supplemented with 10% FBS, with or without 400μg/mL Geneticin at 37°C with 5% CO2. Metastatic melanoma cell lines: Me290 (RRID: CVCL_S598), Me275 (RRID:CVCL_S597), Me319 obtained from the adherent fraction of lymph node (LN) surgical resected from patients: LAU203 (female), LAU50 (male) respectively LAU25755,56,57,58,59 were grown in RPMI supplemented with 10% FBS, 1% Glutamine, 1% HEPES, 1% sodium pyruvate and 1% Penicillin/Streptomycin at 37°C with 5% CO2.
Human T2, a hybrid cell line, HLA-A2+, deficient in TAP, (CEMx721.174.T2)60 (RRID:CVCL_2211) used as antigen presenter cells61 were grown in RPMI supplemented with 10% FBS, 1% Glutamine, 1% HEPES, 1% sodium pyruvate and 1% Penicillin/Streptomycin at 37°C with 5% CO2.
Tyrosinase specific CD8+ T cells, were isolated from PBMCs of HLA-A2+ melanoma patients (LAU 1015-clones 5, 21, 26 and LAU 156-clones 3, 58) after positive selection of CD8 expressing cells by Magnetic Activated Cell Sorting (MACS) (Miltenyi Biotec Inc., Sunnyvale,CA). Selected cells were stained with specific conjugated antibodies and multimers: anti-CD8 FITC and allophycocyanin (APC)-labelled HLA-A2/YMDGTMSQV tetrameric complex, to assess the frequency and sort YMD specific CD8+ lymphocytes with Cell Sorter (FACS Aria, ).55,62,63 Positive cells were cloned by limiting dilutions in Terasaki multiwell plates. The obtained clones were expanded and cultivated in RPMI supplemented with 8% pooled Human Serum, 150 IU/ml recombinant human IL-2 1% Glutamine, 1% HEPES, 1% sodium pyruvate and 1% Penicillin/Streptomycin at 37°C with 5% CO2. CD8+ T cells specific for MAGE-A10254-26264,65 were isolated using the same protocol, with specific labelling of CD8+ selected T cells with APC-labelled HLA-A2/GLYDGMEHL tetrameric complex.
Multimers were loaded with the native peptide, and reactivity of growing YMD-specific CD8+ T cell clones and MAGE-A10-specific CD8+ T cell clones were tested against native or oxidized peptide forms.
Method details
HLA peptide extraction
To isolate cell surface peptides, melanoma cells grown to semi-confluence were detached with 0.02% EDTA, counted, and ∼15-20x106 cells were pelleted. The cell pellets were resuspended in citrate-phosphate buffer, pH 3.3-3.4 (131.5 mM citric acid, 61.5 mM Na2HPO4) and incubated for 2 minutes on ice, followed by inactivation with 1M Tris buffer.66,67 For the estimation of Met oxidation, a heavy-labelled version (13C15N, Val +6Da) of the YMD peptide was spiked-in in the acidic elution buffer (∼ 1 ng). The supernatant was recovered by centrifugation at 1500 RPM for 3 min at 4°C. The samples were filtered through a 10 kDa ultrafiltration membrane, desalted and eluted with acetonitrile (ACN), vacuum-dried and kept at -20°C until further analysis.
HPLC oxidation profiling of the YMD peptide using hydrogen peroxide
The synthetic YMD peptide (98 % HPLC assay) was oxidized with varying concentrations of hydrogen peroxide and kept for 30 min at 4°C. For each concentration, the reaction was performed in 25 μl of solution (LC-MS water) at a concentration of ∼ 155 μM peptide. For the time dependent oxidation assay a similar concentration of peptide was used at the indicated concentration of hydrogen peroxide, in 150 μl solution of solvent A (0.1 % formic acid – FA + 2 % acetonitrile – ACN). For each point of the assay 1 μg of peptide was injected on a reversed-phase (RP) C18, 1.9 μm, Hypersil GOLD column (50 mm x 2.1 ID, ThermoFisher Scientific), equilibrated in 95 % solvent A and 5 % solvent B (0.1 % FA + 50 % ACN). The oxidized peptide species were separated using a linear gradient from 5 % B to 100 % B in 50 min at a flow rate of 0.5 ml/min with detection at 275 nm. For offline MS/MS validation of each chemical derivative, fractions were collected automatically using a Dionex AFC 3000 fraction collector and the resulting fractions were analysed further. For the kinetic analysis the concentration of hydrogen peroxide was considered constant following the course of the reaction as the assay was performed with an excess of hydrogen peroxide compared with the peptide concentration (120 mM vs. 310 μM peptide-bound Met). Large scale peptide purification and purity assesement was performed using a preparative RP C18 column (100 x 10 mm, 5 μm, ThermoFisher Scientific).
Mass spectrometry analysis
All the samples were reconstituted in solvent A and analyzed using an Easy nanoLC II (Proxeon Biosystems) connected online to an Orbitrap Velos Pro (Thermo Fisher Scientific) using a Nanospray Flex Ion Source (Thermo Fisher Scientific). The peptides were separated by a 60-min or 90-min gradient elution (2-30% B: 0.1 % formic acid + 98 % ACN) on an analytical column (100 mm or 150 mm x 75 μm ID) packed with 3 μm ReproSil-Pur C18-AQ resin. (Dr. Maisch GmbH). Light and heavy versions of the standard YMD peptide were injected before analysis to assess retention time and m/z values. The eluted peptides were analyzed using a targeted MS method involving a full Orbitrap scan at 60 000 resolution (m/z 400), followed by a SIM scan (500-600 m/z) at 60 000 resolution and a subsequent scan for peptide MS/MS spectra acquisition. Peptides were selected for CID fragmentation based on a custom inclusion list containing the m/z values detected for both, the light and heavy version of the standard peptide [16].
Mass spectrometry data analysis
For peptide identification, SIM scans and MS/MS scans were manually processed with XCalibur v2.2 (Thermo Fisher Scientific) using 7 ppm mass accuracy for Orbitrap scans and 0.5 Da for linear ion trap scans. For peptide quantification Skyline software v2.6.068,69 was used in which the raw files were imported and parent and fragment ion chromatograms were extracted and integrated for quantification. The following settings were used: precursor and b, y as ion types, precursor charges of +2, fragment ion charges +1, +2 and five-eight product ions for each precursor m/z. For MS1 filtering settings isotope peaks included were set to count, the precursor mass analyser was selected as Orbitrap with resolving power of 60 000 at m/z 400 and for MS2 filtering the acquisition method was set to targeted, product mass analyser as QIT with resolution 0.7 m/z for ion trap data and Orbitrap product mass analyser with 7 500 resolution at 400 m/z for MS/MS spectra detected in the orbital trap. The retention times filtering was based only on scans within five minutes of MS/MS IDs. For the analysis of the HPLC collected fractions eight product ions were used (two b/y ion pairs flanking each Met residue), besides the precursors peaks (M, M+1 and M+2). Moreover, the MS/MS spectra were compared to the native and the in silico obtained ones to validate the oxidation position (Figures 2 and S2). For oxidation level estimation of the light and heavy versions of the YMD peptide, all the sulfoxide variants were considered by extracting the Skyline reported heavy/light total area (Figures S5F and S5H). These are calculated by the software following manual/automatic integration of the heavy and light peptide versions weighted mean of the transition ratios, where the weight is the area of the internal standard, or heavy area. For this analysis, the three double charged precursors (M, M+1 and M+2 peaks) were considered along five product ions from the y ions series covering the two Met residues. We choose only y ions since the heavy peptide is labelled to the C-terminus Val residue and thus y ions can discriminate between the light and heavy peptide versions. For heavy peptide initial oxidation the Skyline reported peak areas were considered for all the selected transitions. These were summed and divided by the total ion current signal of the peptide (including the native form) to obtain the relative oxidation level (ROS) (Figure S5F). These were calculated from injections of the standard heavy YMD peptide and were estimated separately for each of the two Met residues. These were further used to adjust the aggregated ratios obtained for each of the two Met residues (Figure S5G). Copy-numbers were estimated using only the native peptide from the light and heavy, by considering the concentration of the heavy peptide added in each sample.
Western Blot
The pelleted cells were lysed in TritonX-100 containing buffer (50 mM Hepes, 1.5 mM MgCl2, 50 mM NaCl, 1 mM EDTA, 1% Triton X-100, and pH 7.4), supplemented with protease inhibitors. Endo H digestion was performed following manufacturer's instructions. An equal amount of protein from each sample, estimated by BCA assay, was separated by SDS-PAGE and transferred to nitrocellulose membranes. These were subsequently probed with the indicated primary antibodies, tyrosinase-T311 and calnexin for loading control and corresponding HRP coupled secondary antibodies, donkey anti-mouse and donkey anti-rabbit, detected with ECL substrate. Bands from three independent experiments were quantified by Image J and the obtained results were represented as bar plots using Prism software (GraphPad v.6.0).
Flow cytometry analysis of the HLA-A2 level of melanoma cell lines
Cells were detached with EDTA 0.02%, counted and 1x105 cells from each cell line were labelled for 30 min, at 4°C with anti-human HLA-A2-FITC antibody. The cells were washed three times before acquisition on a FACS Calibur (BD Bioscience). Samples were analyzed with the CellQuest software and values of Median Fluorescence Intensity (MFI) of three independent experiments with three technical replicates each were considered. The MFI values were graphically represented as bar plots Using Prism software (GraphPad v.6.0).
Competition binding assay
The competition binding assay was performed according to the method of P. Baumgaertner and colleagues.36 More specific, T2 cells were incubated for 1 h at 37°C with 1μg/ mL W6/32 anti-MHC class I antibody for MHCI complex stabilization. Subsequent, the cells were washed and incubated for 4 h at 37°C with 6 dilutions (10,5,1,0.5,0.1,0.05 μM) of antigenic competitor peptide, native and each HPLC purified form, the reference HBVc-Cy5 fluorescently labelled peptide in fixed concentration-0.2 μM and 1,5 μg/mL β2-microglobulin, all diluted in complete RPMI. The fluorescence intensity of Cy5-labelled HBVc bound to the T2 cells surface after competing with titrated dose of peptide derivatives was measured at Flow Cytometers (Gallios, Beckman Coulter and FACS Verse, BD Bioscience). Samples were analyzed with FlowJo and Cytobank (Beckman Coulter) software’s and Median Fluorescence Intensity (MFI) of technical duplicates were represented as nonlinear regression, binding-competitive curves using Prism software (GraphPad v.6.0). Each experiment was replicated twice and a representative graph is shown in the coressponding figure. For each individual biological replicate the IC50 was interpolated using One Site-Fit logIC50 from Prism Software (GraphPad v.9.4.1) and represented as violin plots in Prism software (Graph Pad v.9.4.1).
In vitro binding assay and stability of pMHC
The competition assays were performed following a previously published method.70 More specifically, 1 μM HLA-A∗02:01 and 2 μM human β2 microglobulin, purified from Escherichia coli were refolded in a v-bottom 96-well plate in 150 μl refolding buffer (100 mM Tris HCl pH 8, 400 mM L-arginine HCl, 2 mM EDTA, 5 mM reduced glutathione, 0.5 mM oxidized glutathione, 0.5 mM phenylmethylsulfonyl fluoride) with biotinylated-Influenza matrix 58-66 peptide (GILGFVFTL) at fixed concentration (2 μM) and candidate peptide to each well to a final concentration of 0, 0.09, 0.19, 0.39, 0.78, 1.56, 3.12, 6.25, 12.5, 25, 50 and 100 μM. As control, unbiotinylated-Influenza matrix 58-66 peptide (GILGFVFTL) was used at the same concentrations as the candidate peptide. The mixture was incubated 72 hours at 4°C. The binding of the biotin tagged Influenza matrix 58-66 peptide (GILGFVFTL) to HLA-A∗02:01 molecule was measured by ELISA. The obtained values were represented as non-linear regression curves, binding-competitive in Prism software (GraphPad v6.0) and a representative experiment is shown in the manuscript. The experiments were replicated twice and for each individual replicate the IC50 was interpolated using One Site-Fit logIC50 from Prism Software (GraphPad v.9.4.1) and represented as violin plot.
For the stability assay refolded HLA-A∗02:01 complexes with Influenza matrix58-66 peptide (GILGFVFTL), Tyr369-377 peptide (YMDGTMSQV) native (N), Tyr369-377 peptide (YM2SODGTM6SOSQV) oxidised version (M2SOM6SO) or without peptide as negative control (-) were incubated at 37 °C at different time points 0, 8h, 24h, 48 and 72h. The integrity of each complex was measured by ELISA.
CD8+ T cell activation assays
Antigenicity of YMD oxidized derivatives was assessed by a standard 4h 51Chromium-release assay and IFNγ secretion assay.
Chromium release assay
This method determines the lytic activity of different Tyr369-377 and MAGE-A10254-262, specific CD8+ T cells activated by native or oxidized peptide derivatives presented by T2 cells. More specific, different clones of YMD-specific CD8+ T cells were incubated together with 51Chromium labelled T2 cells, pulsed with serial dilutions of peptides in native or oxidized forms, for 4-hours at 37°C. For each condition duplicate samples were performed. After 4 h incubation, 50 μl of supernatant were transferred in a LumaPlate containing solid scintillant and let to dry overnight (ON). Radioactive signal from dried plates was counted with a TopCount NXT Microplate Scintillation (PerkinElmer) and the percentage of specific lysis was calculated using the standard equation: [(experimental - spontaneous release) / (maximum load - spontaneous release)] x 100. Specific lysis was represented as the nonlinear log [Agonist] vs. response curves for each clone in Prism software (GraphPad v.6.0) and representative clones were shown. Using the equation describing the nonlinear [Agonist] vs. response curve, the functional avidity (EC50-peptide concentration giving 50% maximal killing) for each Tyr369-377 specific CD8+ T cell clone was estimated. Calculated EC50 values were graphically represented as box plots and the statistical significance was assessed using an unpaired t test for YMD oxidized mix forms and the non-parametric option of one-way ANOVA with Dunn’s correction for each YMD derivative. MAGE-A10254-262 peptide variants were processed in the same way.
IFNγ secretion assay
Tyrosinase specific CD8+ T cells were incubated at 1/1 ratio with the T2 cells pulsed with serial dilutions of native YMD peptide or oxidized forms, each condition in duplicate, for IFNγ quantification by ELISA after 24 h of co-culture. The concentration of IFNγ secreted in each condition was determined by integration in an exponential IFNγ standard curve equation. Dilutions of standard IFNγ were processed simultaneously with the samples according to ELISA manufacturer indications. The obtained IFNγ concentration were graphically represented in Prism software (GraphPad v.6.0), as shown in the figure.
Analysis of Met relative frequency and position in IEDB database
The full IEDB database was downloaded from https://www.iedb.org (4537458 entries as of 08/2022) and the corresponding data was further analysed. Only human sequences from linear peptide epitopes of nonamers were further kept. To reduce redundancy only unique sequences were considered for the next steps of the analysis. The relative Met frequency was calculated by dividing the entries containing at least one Met residue to all entries available in the database. These were calculated for each allele from IEDB. For Met abundance in the corresponding proteome, the human version of the UniProtKB database was retrieved from the UniProtKB web-site https://www.uniprot.org/ (79740 entries as of 09/2022). Only unique sequences of at least 100 amino acids long were further kept for analysis. For Met frequency positional analysis the top three most represented alleles from IEDB for each major HLA class I genes were selected and the relative positional Met frequency was assessed by calculating the number of entries with Met residues for each of the available nine possibilities.
Modeling work and binding free energy computation
The computational work involved the following three main stages:
-
1.
Modeling the HLA-A∗02:01 - YMD binary complex in native (N) and oxidized form (M2SOM6SO);
-
2.
Building the HLA-A∗02:01 - YMD TCR ternary complex for the N and M2SOM6SO form; and
-
3.
Computing the ternary complex absolute binding free energies for YMD-N and M2SOM6SO using alchemical transition from the unbound to the bound state and estimating the difference between them using a thermodynamic cycle.
Modeling of HLA-A∗02:01 - YMD binary complex
To select the best possible templates for homology modelling HLA-YMD complex, all available HLA-peptide (HP) and HLA-peptide-TCR (HPT) structures were retrieved from the Protein Data Bank71 using BLAST with HLA-A∗02:01 sequence as query. In this way a structural database was compiled, HP/HTP-DB containing 319 HP binary complexes and 105 HPT ternary complexes in which HLA structures include the β-microglobulin besides the HLA-α chain. Identification and elimination of redundant low resolution and disordered structures and selection of the best representative reduced the set to 237 HP and 28 HPT structures. The nonredundant antigenic peptides were subjected to structural and statistical analysis with in-house tools aiming to select the closest YMD templates (Data S2).
Given that in the HPLC experiments M2SOM6SO was found to be dominant amongst the 8 possible oxidized YMD forms - the two peptide templates were used to build only 4 starting YMD-HLA-A∗02:01 binary complex models corresponding to the Native and M2SOM6SO states in each of the two conformations. In building the oxidized form models the missing force field parameters for Methionine sulfoxide were taken from the Generalized Amber Force Field - GAFF2.42 For the new residue, partial charges were calculated using the Antechamber from the AmberTools suite.43
Modeling the ternary complex HLA-A∗02:01 -TCR
Models of the ternary HLA-A∗02:01 -TCR complex were built in two steps: (1) the TCR build-up step of models with patient specific, clone 3 CDR loops, TCRc3, by joint fragment homology modelling, and (2) docking step of the TCR-c3 model to the four HP binary complexes generated in stage 1.
In order to generate the TCR-c3 model, the HP/HTP-DB database mentioned above was scanned for closest candidates for clone 3 CDR regions. Sequence analysis revealed that each CDR loop and each constant region had different best candidates in terms of sequence similarity. Thus, a joint fragment homology modelling procedure72,73 was used starting from four reference structures (PDB codes: 5HHM, 3PWP, 2PYE and 2BNQ). The 5HHM and 3PWP structures were used to build the α- and β-chains conserved scaffolds respectively, while 2PYE and 2BNQ CDR templates were used to build the CDR-TCRc3 loops, as shown in Data S3 multiple sequence alignment.
In step 2, the ternary complexes were built by docking the TCRc3 on top of the previously built HLA-A∗02:01 -YMD binary complex using 2PYE ternary complex crystal structure as an overall scaffold. In addition, a water molecule was placed by coordinate transfer at the HLA-A∗02:01 -YMD interface near position M6 as 3 out of the 4 templates present this water in the mentioned location.
Conformation of these models were further iteratively optimized by repeated rounds of simulated annealing and minimization using the L-BFGS algorithm, with a tolerance of 10 kJ/mole, with OpenMM v.7.4.1.44 The stability of all ternary complex models was assessed by 100 ns molecular dynamics (MD) simulations each, performed with OpenMM (v.7.4.1) using explicit solvent, at 300K temperature and 1 bar constant pressure. Trajectory integration was rendered by Langevin integrator at 2 fs integration timestep and 1ps-1 friction coefficient.
As during these ternary complex simulations, the M6 side-chain of the first starting conformation remained firmly oriented toward HLA, while in the second starting conformation M6 side-chain drifted slowly toward TCR (Figures 7B3 and 7B4), we called these two alternative peptide conformations - 'down' (↓) and 'up' (↑) respectively. All HLA-A∗02:01 and TCR-c3 models built in steps 1 and 2 were raised with Modeller v.9.21.45 Briefly, variable stretches connecting the sequence-conserved regions SCR were generated iteratively using the Modeller loop optimization protocol relying on the distance-dependent statistical potential of mean force, developed by Melo and Feytmans.74 The sequence variable regions were optimized by repeated alternative rounds of simulated annealing and energy minimization. Models were validated using the Molprobity server52 resulting in 0.59 and 1.00 scores for HLA-A∗02:01 and TCR respectively.
Free energy estimation
In stage 3, the Absolute Binding Free Energy estimates were calculated using Hamiltonian Replica Exchange (HRE) with thermodynamic states scaled between a coupled and a decoupled ligand state. Briefly, this entails gradual reduction of the interactions exerted by HLA-A∗02:01 and TCR onto the YMD till extinction endpoint. The two HLA-A∗02:01 -YMD- TCR ternary complex models - with YMD in native form and with methionine residues M2 & M6 sulfoxide forms (M2SOM6SO) - were subjected to a total of 472.5 ns simulation time across 135 replicas. Calculations were performed with YANK v.0.25.246 which uses Hamiltonian Replica Exchange (HRE) as sampling method and Multistate Bennett Acceptance Ratio implemented in the PyMBAR package,47 a method widely used for ΔG estimation in general75 that has recently proven useful in investigating protein-RNA interactions76 and evaluating the impact of point mutations in the barnase:barstar system.77 The number of replicas and their corresponding coupling parameters were set using automated replica detection protocol with default settings. In order to ensure that enough replicas were used replica mixing was computed using YANK’s automatic scripts. The ligand was kept within the binding site using harmonic restraints. The HREs were ran for 3.5 ns ∗135 replicas with a replica swap being attempted every 10-3 ns. The fully coupled trajectories were further used for differential structural analysis including stability, cluster count and solvent accessibility analysis. To this end, RMSD and RMSF were calculated using MDTraj 1.9.3.48 Solvent Accessible Surface Area was computed using FreeSASA 2.0.3.49 Cluster count analysis was performed with Scipy v. 1.3.2,50 using RMSD as distance. PyMOL v.2.2.3 was used to generate all figures containing 3D structures of proteins.51
Quantification and statistical analysis
Experimental design and statistical analysis
For Chromium release assay, distinct CD8+ T cell clones were used for experiments in duplicates. To assess the relevance of the YMD derivative immunogenicity’s, we calculated the concentration giving 50% maximal killing (EC50) for all YMD specific CD8+ T cell clones tested, illustrated in box plot graphics. Finally, the statistical significance of differences in the efficiency of oxidative variants of YMD recognition by Tyr369-377 restricted CD8+ T cells was assessed by unpaired two-sample two-tailed t-test or by non-parametric one-way ANOVA with Dunn’s correction for each YMD derivative using Prism software (GraphPad v.6.0).
Experiments using IFNγ secretion readouts were performed in duplicates for each YMD-specific CD8+ T cell clone used.
For the competition binding assay, the experiments were performed in biological duplicates and one representative experiment was shown as nonlinear regression curves in the figure. For each experiment the IC50 was calculated and represented as box plot using Prism software (GraphPad v.9.4.1).
For HLA-A2 detection by flow cytometry, experiments were performed in biological triplicates and mean of triplicates were represented as bar plots with SEM as error bars, using Prism software (Graph Pad v.6.0).
Western Blot experiments were performed in three biological replicates and values obtained from bands quantification using ImageJ were represented as bar plots of mean values with error bars denoting SEM.
The experiments for endogenous oxidation were performed in three or four biological replicates.
For all figures we show mean with error bars denoting SEM, if not otherwise denoted.
Acknowledgments
We would like to thank Dr. Philippe Guillaume for performing in vitro binding and stability assays and Dr. Livia Sima for technical assistance in using FACS Verse and Cytobank data analysis guidance. This work was funded by the Romanian Ministry of Research and Innovation, CNCS- UEFISCDI grants, PN-III-P1-1.1-PD-2016-1528 (Project No.: PD62/2018), PN-III-P1-1.1-PD-2019-1242 (Project No.: PD176/2020), PN-III-P1-1.1-PD-2019-1278 (Project No.: PD183/2020), grant PN-III-P4-ID-PCE-2020-2444 (Project No, PCE148/2021), RO-SWISS Project IZERZO No. 142216 and the Romanian Academy programs 1 & 4 of IBAR.
Author contributions
G.C. and C.V.A.M.: conceptualization, methodology, investigation, writing-original draft-review and funding, T.A.Ș. and L.S.: methodology, computational investigation, writing-original draft, funding, A.J.P.: conceptualization, supervision, writing-review and editing, C.J. and P.R.: conceptualization, writing-review, editing supervision, funding, S.M.P.: conceptualization, supervision, writing-review, editing and funding.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 25, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107205.
Supplemental information
Data and code availability
-
•
Data have been deposited at the ProteomeXchange Consortium via the PRIDE53 partner repository with the dataset identifier PXD039318 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyse the data reported in this paper is available from the lead contact upon request.
References
- 1.Erdei E., Torres S.M. A new understanding in the epidemiology of melanoma. Expert Rev. Anticancer Ther. 2010;10:1811–1823. doi: 10.1586/era.10.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Radović-Kovacević V., Pekmezović T., Adanja B., Jarebinski M., Marinković J., Tomin R. Survival analysis in patients with cutaneous malignant melanoma. Srp. Arh. Celok. Lek. 1997;125:132–137. [PubMed] [Google Scholar]
- 3.Topalian S.L., Hodi F.S., Brahmer J.R., Gettinger S.N., Smith D.C., McDermott D.F., Powderly J.D., Carvajal R.D., Sosman J.A., Atkins M.B., et al. Safety, Activity, and Immune Correlates of Anti–PD-1 Antibody in Cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brahmer J.R., Tykodi S.S., Chow L.Q.M., Hwu W.-J., Topalian S.L., Hwu P., Drake C.G., Camacho L.H., Kauh J., Odunsi K., et al. Safety and Activity of Anti–PD-L1 Antibody in Patients with Advanced Cancer. N. Engl. J. Med. 2012;366:2455–2465. doi: 10.1056/NEJMoa1200694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raedler L.A. Keytruda (Pembrolizumab): First PD-1 Inhibitor Approved for Previously Treated Unresectable or Metastatic Melanoma. Am. Health Drug Benefits. 2015;8:96–100. [PMC free article] [PubMed] [Google Scholar]
- 6.Twomey J.D., Zhang B. Cancer Immunotherapy Update: FDA-Approved Checkpoint Inhibitors and Companion Diagnostics. AAPS J. 2021;23:39. doi: 10.1208/s12248-021-00574-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martins F., Sofiya L., Sykiotis G.P., Lamine F., Maillard M., Fraga M., Shabafrouz K., Ribi C., Cairoli A., Guex-Crosier Y., et al. Adverse effects of immune-checkpoint inhibitors: epidemiology, management and surveillance. Nat. Rev. Clin. Oncol. 2019;16:563–580. doi: 10.1038/s41571-019-0218-0. [DOI] [PubMed] [Google Scholar]
- 8.Kjeldsen J.W., Andersen M.H., Svane I.M. Combination therapy with nivolumab and PD-L1/IDO peptide vaccine to patients with metastatic melanoma. A clinical trial in progress. Ann. Oncol. 2017;28:xi27–xi28. doi: 10.1093/annonc/mdx711.075. [DOI] [Google Scholar]
- 9.Tagliamonte M., Ciliberto G. In: Cancer Vaccines as Immunotherapy of Cancer. Buonaguro L., Van Der Burg S., editors. Academic Press; 2022. Chapter 8 - Combinatorial immunotherapy strategies for cancer vaccines; pp. 137–154. [DOI] [Google Scholar]
- 10.George D.D., Armenio V.A., Katz S.C. Combinatorial immunotherapy for melanoma. Cancer Gene Ther. 2017;24:141–147. doi: 10.1038/cgt.2016.56. [DOI] [PubMed] [Google Scholar]
- 11.Cheever M.A., Allison J.P., Ferris A.S., Finn O.J., Hastings B.M., Hecht T.T., Mellman I., Prindiville S.A., Viner J.L., Weiner L.M., Matrisian L.M. The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin. Cancer Res. 2009;15:5323–5337. doi: 10.1158/1078-0432.CCR-09-0737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Branza-Nichita N., Negroiu G., Petrescu A.J., Garman E.F., Platt F.M., Wormald M.R., Dwek R.A., Petrescu S.M. Mutations at Critical N-Glycosylation Sites Reduce Tyrosinase Activity by Altering Folding and Quality Control. J. Biol. Chem. 2000;275:8169–8175. doi: 10.1074/jbc.275.11.8169. [DOI] [PubMed] [Google Scholar]
- 13.Petrescu S.M., Petrescu A.-J., Titu H.N., Dwek R.A., Platt F.M. Inhibition of N-Glycan Processing in B16 Melanoma Cells Results in Inactivation of Tyrosinase but Does Not Prevent Its Transport to the Melanosome. J. Biol. Chem. 1997;272:15796–15803. doi: 10.1074/jbc.272.25.15796. [DOI] [PubMed] [Google Scholar]
- 14.Petrescu S.M., Branza-Nichita N., Negroiu G., Petrescu A.J., Dwek R.A. Tyrosinase and Glycoprotein Folding: Roles of Chaperones That Recognize Glycans. Biochemistry. 2000;39:5229–5237. doi: 10.1021/bi000107z. [DOI] [PubMed] [Google Scholar]
- 15.Ostankovitch M., Robila V., Engelhard V.H. Regulated folding of tyrosinase in the endoplasmic reticulum demonstrates that misfolded full-length proteins are efficient substrates for class I processing and presentation. J. Immunol. 2005;174:2544–2551. doi: 10.4049/jimmunol.174.5.2544. [DOI] [PubMed] [Google Scholar]
- 16.Slingluff C.L., Jr. Tumor antigens and tumor vaccines: peptides as immunogens. Semin. Surg. Oncol. 1996;12:446–453. doi: 10.1002/(SICI)1098-2388(199611/12)12:6<446::AID-SSU10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 17.Marchand M., Weynants P., Rankin E., Arienti F., Belli F., Parmiani G., Cascinelli N., Bourlond A., Vanwijck R., Humblet Y., et al. Tumor regression responses in melanoma patients treated with a peptide encoded by gene MAGE-3. Int. J. Cancer. 1995;63:883–885. doi: 10.1002/ijc.2910630622. [DOI] [PubMed] [Google Scholar]
- 18.Tourdot S., Scardino A., Saloustrou E., Gross D.A., Pascolo S., Cordopatis P., Lemonnier F.A., Kosmatopoulos K. A general strategy to enhance immunogenicity of low-affinity HLA-A2. 1-associated peptides: implication in the identification of cryptic tumor epitopes. Eur. J. Immunol. 2000;30:3411–3421. doi: 10.1002/1521-4141(2000012)30:12<3411::AID-IMMU3411>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 19.Valmori D., Fonteneau J.F., Lizana C.M., Gervois N., Liénard D., Rimoldi D., Jongeneel V., Jotereau F., Cerottini J.C., Romero P. Enhanced generation of specific tumor-reactive CTL in vitro by selected Melan-A/MART-1 immunodominant peptide analogues. J. Immunol. 1998;160:1750–1758. [PubMed] [Google Scholar]
- 20.Filipazzi P., Pilla L., Mariani L., Patuzzo R., Castelli C., Camisaschi C., Maurichi A., Cova A., Rigamonti G., Giardino F., et al. Limited induction of tumor cross-reactive T cells without a measurable clinical benefit in early melanoma patients vaccinated with human leukocyte antigen class I-modified peptides. Clin. Cancer Res. 2012;18:6485–6496. doi: 10.1158/1078-0432.CCR-12-1516. [DOI] [PubMed] [Google Scholar]
- 21.Skipper J.C., Hendrickson R.C., Gulden P.H., Brichard V., Van Pel A., Chen Y., Shabanowitz J., Wolfel T., Slingluff C.L., Jr., Boon T., et al. An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J. Exp. Med. 1996;183:527–534. doi: 10.1084/jem.183.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Altrich-VanLith M.L., Ostankovitch M., Polefrone J.M., Mosse C.A., Shabanowitz J., Hunt D.F., Engelhard V.H. Processing of a class I-restricted epitope from tyrosinase requires peptide N-glycanase and the cooperative action of endoplasmic reticulum aminopeptidase 1 and cytosolic proteases. J. Immunol. 2006;177:5440–5450. doi: 10.4049/jimmunol.177.8.5440. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg S.A., Sherry R.M., Morton K.E., Yang J.C., Topalian S.L., Royal R.E., Kammula U.S., Restifo N.P., Hughes M.S., Schwarz S.L., et al. Altered CD8(+) T-cell responses when immunizing with multiepitope peptide vaccines. J. Immunother. 2006;29:224–231. doi: 10.1097/01.cji.0000190399.98802.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Potgieter H.C., Ubbink J.B., Bissbort S., Bester M.J., Spies J.H., Vermaak W.J. Spontaneous oxidation of methionine: effect on the quantification of plasma methionine levels. Anal. Biochem. 1997;248:86–93. doi: 10.1006/abio.1997.2075. [DOI] [PubMed] [Google Scholar]
- 25.Chiang C.L.L., Ledermann J.A., Aitkens E., Benjamin E., Katz D.R., Chain B.M. Oxidation of ovarian epithelial cancer cells by hypochlorous acid enhances immunogenicity and stimulates T cells that recognize autologous primary tumor. Clin. Cancer Res. 2008;14:4898–4907. doi: 10.1158/1078-0432.CCR-07-4899. [DOI] [PubMed] [Google Scholar]
- 26.Weiskopf D., Schwanninger A., Weinberger B., Almanzar G., Parson W., Buus S., Lindner H., Grubeck-Loebenstein B. Oxidative stress can alter the antigenicity of immunodominant peptides. J. Leukoc. Biol. 2010;87:165–172. doi: 10.1189/jlb.0209065. [DOI] [PubMed] [Google Scholar]
- 27.Dalet A., Robbins P.F., Stroobant V., Vigneron N., Li Y.F., El-Gamil M., Hanada K.i., Yang J.C., Rosenberg S.A., Van den Eynde B.J. An antigenic peptide produced by reverse splicing and double asparagine deamidation. Proc. Natl. Acad. Sci. USA. 2011;108:E323–E331. doi: 10.1073/pnas.1101892108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clement C.C., Moncrieffe H., Lele A., Janow G., Becerra A., Bauli F., Saad F.A., Perino G., Montagna C., Cobelli N., et al. Autoimmune response to transthyretin in juvenile idiopathic arthritis. JCI Insight. 2016;1 doi: 10.1172/jci.insight.85633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clement C.C., Nanaware P.P., Yamazaki T., Negroni M.P., Ramesh K., Morozova K., Thangaswamy S., Graves A., Kim H.J., Li T.W., et al. Pleiotropic consequences of metabolic stress for the major histocompatibility complex class II molecule antigen processing and presentation machinery. Immunity. 2021;54:721–736.e10. doi: 10.1016/j.immuni.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Michaeli Y., Denkberg G., Sinik K., Lantzy L., Chih-Sheng C., Beauverd C., Ziv T., Romero P., Reiter Y. Expression hierarchy of T cell epitopes from melanoma differentiation antigens: unexpected high level presentation of tyrosinase-HLA-A2 Complexes revealed by peptide-specific, MHC-restricted, TCR-like antibodies. J. Immunol. 2009;182:6328–6341. doi: 10.4049/jimmunol.0801898. [DOI] [PubMed] [Google Scholar]
- 31.Pan H., Chen K., Chu L., Kinderman F., Apostol I., Huang G. Methionine oxidation in human IgG2 Fc decreases binding affinities to protein A and FcRn. Protein Sci. 2009;18:424–433. doi: 10.1002/pro.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiritoiu G.N., Jandus C., Munteanu C.V.A., Ghenea S., Gannon P.O., Romero P., Petrescu S.M. Epitope located N-glycans impair the MHC-I epitope generation and presentation. Electrophoresis. 2016;37:1448–1460. doi: 10.1002/elps.201500449. [DOI] [PubMed] [Google Scholar]
- 33.Morand K., Talbo G., Mann M. Oxidation of peptides during electrospray ionization. Rapid Commun. Mass Spectrom. 1993;7:738–743. doi: 10.1002/rcm.1290070811. [DOI] [PubMed] [Google Scholar]
- 34.Hui A., Lam X.M., Kuehl C., Grauschopf U., Wang Y.J. Kinetic Modeling of Methionine Oxidation in Monoclonal Antibodies from Hydrogen Peroxide Spiking Studies. PDA J. Pharm. Sci. Technol. 2015;69:511–525. doi: 10.5731/pdajpst.2015.01059. [DOI] [PubMed] [Google Scholar]
- 35.Guan Z., Yates N.A., Bakhtiar R. Detection and characterization of methionine oxidation in peptides by collision-induced dissociation and electron capture dissociation. J. Am. Soc. Mass Spectrom. 2003;14:605–613. doi: 10.1016/S1044-0305(03)00201-0. [DOI] [PubMed] [Google Scholar]
- 36.Baumgaertner P., Schmidt J., Costa-Nunes C.-M., Bordry N., Guillaume P., Luescher I., Speiser D.E., Rufer N., Hebeisen M. CD8 T cell function and cross-reactivity explored by stepwise increased peptide-HLA versus TCR affinity. Front. Immunol. 2022;13:973986. doi: 10.3389/fimmu.2022.973986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halaban R., Cheng E., Zhang Y., Moellmann G., Hanlon D., Michalak M., Setaluri V., Hebert D.N. Aberrant retention of tyrosinase in the endoplasmic reticulum mediates accelerated degradation of the enzyme and contributes to the dedifferentiated phenotype of amelanotic melanoma cells. Proc. Natl. Acad. Sci. USA. 1997;94:6210–6215. doi: 10.1073/pnas.94.12.6210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michaeli Y., Sinik K., Haus-Cohen M., Reiter Y. Melanoma cells present high levels of HLA-A2-tyrosinase in association with instability and aberrant intracellular processing of tyrosinase. Eur. J. Immunol. 2012;42:842–850. doi: 10.1002/eji.201141511. [DOI] [PubMed] [Google Scholar]
- 39.Moskovitz J. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 40.Hwang C., Sinskey A.J., Lodish H.F. Oxidized redox state of glutathione in the endoplasmic reticulum. Science. 1992;257:1496–1502. doi: 10.1126/science.1523409. [DOI] [PubMed] [Google Scholar]
- 41.Wood J.M., Decker H., Hartmann H., Chavan B., Rokos H., Spencer J.D., Hasse S., Thornton M.J., Shalbaf M., Paus R., Schallreuter K.U. Senile hair graying: H2O2-mediated oxidative stress affects human hair color by blunting methionine sulfoxide repair. Faseb. J. 2009;23:2065–2075. doi: 10.1096/fj.08-125435. [DOI] [PubMed] [Google Scholar]
- 42.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 43.Case D.A., K.B., Ben-Shalom I.Y., Brozell S.R., Cerutti D.S., Cheatham T.E., III, Cruzeiro V.W.D., Darden T.A., Duke R.E., Giambasu G., et al. 2020. AMBER. [Google Scholar]
- 44.Eastman P., Swails J., Chodera J.D., McGibbon R.T., Zhao Y., Beauchamp K.A., Wang L.-P., Simmonett A.C., Harrigan M.P., Stern C.D., et al. OpenMM 7: Rapid development of high performance algorithms for molecular dynamics. PLoS Comput. Biol. 2017;13 doi: 10.1371/journal.pcbi.1005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webb B., Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Bioinformatics. 2016;54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang K., Chodera J.D., Yang Y., Shirts M.R. Identifying ligand binding sites and poses using GPU-accelerated Hamiltonian replica exchange molecular dynamics. J. Comput. Aided Mol. Des. 2013;27:989–1007. doi: 10.1007/s10822-013-9689-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shirts M.R., Chodera J.D. Statistically optimal analysis of samples from multiple equilibrium states. J. Chem. Phys. 2008;129 doi: 10.1063/1.2978177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McGibbon R.T., Beauchamp K.A., Harrigan M.P., Klein C., Swails J.M., Hernández C.X., Schwantes C.R., Wang L.-P., Lane T.J., Pande V.S. MDTraj: A Modern Open Library for the Analysis of Molecular Dynamics Trajectories. Biophys. J. 2015;109:1528–1532. doi: 10.1016/j.bpj.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mitternacht S. FreeSASA: An open source C library for solvent accessible surface area calculations. F1000Res. 2016;5:189. doi: 10.12688/f1000research.7931.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Virtanen P., Gommers R., Oliphant T.E., Haberland M., Reddy T., Cournapeau D., Burovski E., Peterson P., Weckesser W., Bright J., et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat. Methods. 2020;17:261–272. doi: 10.1038/s41592-019-0686-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schrödinger L. 2018. The PyMOL Molecular Graphics System. [Google Scholar]
- 52.Davis I.W., Leaver-Fay A., Chen V.B., Block J.N., Kapral G.J., Wang X., Murray L.W., Arendall W.B., III, Snoeyink J., Richardson J.S., Richardson D.C. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perez-Riverol Y., Bai J., Bandla C., García-Seisdedos D., Hewapathirana S., Kamatchinathan S., Kundu D.J., Prakash A., Frericks-Zipper A., Eisenacher M., et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kozlowski J.M., Hart I.R., Fidler I.J., Hanna N. A Human Melanoma Line Heterogeneous With Respect to Metastatic Capacity in Athymic Nude Mice234. JNCI. J. Natl. Cancer Inst. 1984;72:913–917. doi: 10.1093/jnci/72.4.913. [DOI] [PubMed] [Google Scholar]
- 55.Romero P., Dunbar P.R., Valmori D., Pittet M., Ogg G.S., Rimoldi D., Chen J.-L., Liénard D., Cerottini J.-C., Cerundolo V. Ex Vivo Staining of Metastatic Lymph Nodes by Class I Major Histocompatibility Complex Tetramers Reveals High Numbers of Antigen-experienced Tumor-specific Cytolytic T Lymphocytes. J. Exp. Med. 1998;188:1641–1650. doi: 10.1084/jem.188.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rimoldi D., Rubio-Godoy V., Dutoit V., Lienard D., Salvi S., Guillaume P., Speiser D., Stockert E., Spagnoli G., Servis C., et al. Efficient Simultaneous Presentation of NY-ESO-1/LAGE-1 Primary and Nonprimary Open Reading Frame-Derived CTL Epitopes in Melanoma. J. Immunol. 2000;165:7253–7261. doi: 10.4049/jimmunol.165.12.7253. [DOI] [PubMed] [Google Scholar]
- 57.Giard D.J., Aaronson S.A., Todaro G.J., Arnstein P., Kersey J.H., Dosik H., Parks W.P. In Vitro Cultivation of Human Tumors: Establishment of Cell Lines Derived From a Series of Solid Tumors2. J. Natl. Cancer Inst. 1973;51:1417–1423. doi: 10.1093/jnci/51.5.1417. [DOI] [PubMed] [Google Scholar]
- 58.Derré L., Bruyninx M., Baumgaertner P., Devevre E., Corthesy P., Touvrey C., Mahnke Y.D., Pircher H., Voelter V., Romero P., et al. In Vivo Persistence of Codominant Human CD8+ T Cell Clonotypes Is Not Limited by Replicative Senescence or Functional Alteration. J. Immunol. 2007;179:2368–2379. doi: 10.4049/jimmunol.179.4.2368. [DOI] [PubMed] [Google Scholar]
- 59.Derré L., Ferber M., Touvrey C., Devevre E., Zoete V., Leimgruber A., Romero P., Michielin O., Lévy F., Speiser D.E. A Novel Population of Human Melanoma-Specific CD8 T Cells Recognizes Melan-AMART-1 Immunodominant Nonapeptide but Not the Corresponding Decapeptide. J. Immunol. 2007;179:7635–7645. doi: 10.4049/jimmunol.179.11.7635. [DOI] [PubMed] [Google Scholar]
- 60.Salter R.D., Cresswell P. Impaired assembly and transport of HLA-A and -B antigens in a mutant TxB cell hybrid. EMBO J. 1986;5:943–949. doi: 10.1002/j.1460-2075.1986.tb04307.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luft T., Rizkalla M., Tai T.Y., Chen Q., MacFarlan R.I., Davis I.D., Maraskovsky E., Cebon J. Exogenous Peptides Presented by Transporter Associated with Antigen Processing (TAP)-Deficient and TAP-Competent Cells: Intracellular Loading and Kinetics of Presentation. J. Immunol. 2001;167:2529–2537. doi: 10.4049/jimmunol.167.5.2529. [DOI] [PubMed] [Google Scholar]
- 62.Valmori D., Fonteneau J.F., Valitutti S., Gervois N., Dunbar R., Liénard D., Rimoldi D., Cerundolo V., Jotereau F., Cerottini J.C., et al. Optimal activation of tumor-reactive T cells by selected antigenic peptide analogues. Int. Immunol. 1999;11:1971–1980. doi: 10.1093/intimm/11.12.1971. [DOI] [PubMed] [Google Scholar]
- 63.Pittet M.J., Valmori D., Dunbar P.R., Speiser D.E., Liénard D., Lejeune F., Fleischhauer K., Cerundolo V., Cerottini J.C., Romero P. High frequencies of naive Melan-A/MART-1-specific CD8(+) T cells in a large proportion of human histocompatibility leukocyte antigen (HLA)-A2 individuals. J. Exp. Med. 1999;190:705–715. doi: 10.1084/jem.190.5.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Valmori D., Dutoit V., Rubio-Godoy V., Chambaz C., Liénard D., Guillaume P., Romero P., Cerottini J.-C., Rimoldi D. Frequent Cytolytic T-Cell Responses to Peptide MAGE-A10254–262 in Melanoma1. Cancer Res. 2001;61:509–512. [PubMed] [Google Scholar]
- 65.Dutoit V.r., Rubio-Godoy V., Dietrich P.-Y., Quiqueres A.-L., Schnuriger V.r., Rimoldi D., Liénard D., Speiser D., Guillaume P., Batard P., et al. Heterogeneous T-Cell Response to MAGE-A10254–262: High Avidity-specific Cytolytic T Lymphocytes Show Superior Antitumor Activity1. Cancer Res. 2001;61:5850–5856. [PubMed] [Google Scholar]
- 66.Sugawara S., Abo T., Kumagai K. A simple method to eliminate the antigenicity of surface class I MHC molecules from the membrane of viable cells by acid treatment at pH 3. J. Immunol. Methods. 1987;100:83–90. doi: 10.1016/0022-1759(87)90175-x. [DOI] [PubMed] [Google Scholar]
- 67.Storkus W.J., Zeh H.J., 3rd, Salter R.D., Lotze M.T. Identification of T-cell epitopes: rapid isolation of class I-presented peptides from viable cells by mild acid elution. J. Immunother. Emphasis Tumor Immunol. 1993;14:94–103. [PubMed] [Google Scholar]
- 68.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schilling B., Rardin M.J., MacLean B.X., Zawadzka A.M., Frewen B.E., Cusack M.P., Sorensen D.J., Bereman M.S., Jing E., Wu C.C., et al. Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol. Cell. Proteomics. 2012;11:202–214. doi: 10.1074/mcp.M112.017707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garboczi D.N., Hung D.T., Wiley D.C. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc. Natl. Acad. Sci. USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Slootweg E.J., Spiridon L.N., Roosien J., Butterbach P., Pomp R., Westerhof L., Wilbers R., Bakker E., Bakker J., Petrescu A.J., et al. Structural determinants at the interface of the ARC2 and leucine-rich repeat domains control the activation of the plant immune receptors Rx1 and Gpa2. Plant Physiol. 2013;162:1510–1528. doi: 10.1104/pp.113.218842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rajaraman J., Douchkov D., Hensel G., Stefanato F.L., Gordon A., Ereful N., Caldararu O.F., Petrescu A.J., Kumlehn J., Boyd L.A., Schweizer P. An LRR/Malectin Receptor-Like Kinase Mediates Resistance to Non-adapted and Adapted Powdery Mildew Fungi in Barley and Wheat. Front. Plant Sci. 2016;7:1836. doi: 10.3389/fpls.2016.01836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Melo F., Feytmans E. Novel knowledge-based mean force potential at atomic level. J. Mol. Biol. 1997;267:207–222. doi: 10.1006/jmbi.1996.0868. [DOI] [PubMed] [Google Scholar]
- 75.Matsunaga Y., Kamiya M., Oshima H., Jung J., Ito S., Sugita Y. Use of multistate Bennett acceptance ratio method for free-energy calculations from enhanced sampling and free-energy perturbation. Biophys. Rev. 2022;14:1503–1512. doi: 10.1007/s12551-022-01030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Minuesa G., Albanese S.K., Xie W., Kazansky Y., Worroll D., Chow A., Schurer A., Park S.-M., Rotsides C.Z., Taggart J., et al. Small-molecule targeting of MUSASHI RNA-binding activity in acute myeloid leukemia. Nat. Commun. 2019;10:2691. doi: 10.1038/s41467-019-10523-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang I., Rufa D.A., Iván P., Henry M.M., Rosen L.E., Hauser K., Singh S., Chodera J.D. Identifying and overcoming the sampling challenges in relative binding free energy calculations of a model protein:protein complex. bioRxiv. 2023 doi: 10.1101/2023.03.07.530278. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data have been deposited at the ProteomeXchange Consortium via the PRIDE53 partner repository with the dataset identifier PXD039318 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to re-analyse the data reported in this paper is available from the lead contact upon request.