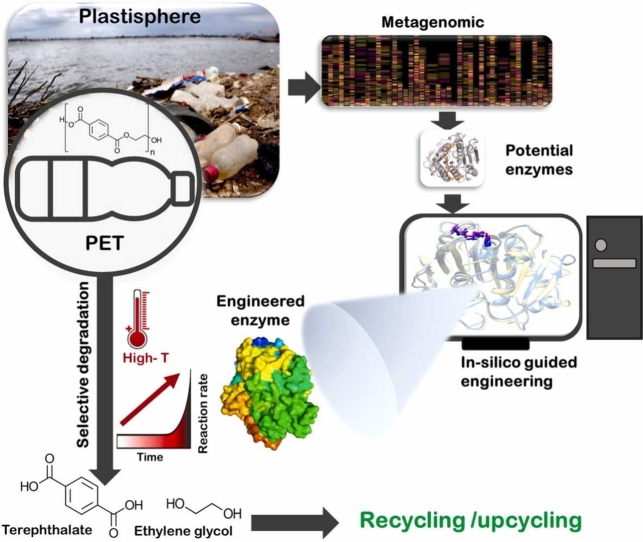
Abstract
Polyethylene terephthalate (PET) is the largest produced polyester globally, and less than 30% of all the PET produced globally (∼6 billion pounds annually) is currently recycled into lower-quality products. The major drawbacks in current recycling methods (mechanical and chemical), have inspired the exploration of potentially efficient and sustainable PET depolymerization using biological approaches. Researchers have discovered efficient PET hydrolyzing enzymes in the plastisphere and have demonstrated the selective degradation of PET to original monomers thus enabling biological recycling or upcycling. However, several significant hurdles such as the less efficiency of the hydrolytic reaction, low thermostability of the enzymes, and the inability of the enzyme to depolymerize crystalline PET must be addressed in order to establish techno-economically feasible commercial-scale biological PET recycling or upcycling processes. Researchers leverage a synthetic biology-based design; build, test, and learn (DBTL) methodology to develop commercially applicable efficient PET hydrolyzing enzymes through 1) high-throughput metagenomic and proteomic approaches to discover new PET hydrolyzing enzymes with superior properties: and, 2) enzyme engineering approaches to modify and optimize PET hydrolyzing properties. Recently, in-silico platforms including molecular mechanics and machine learning concepts are emerging as innovative tools for the development of more efficient and effective PET recycling through the exploration of novel mutations in PET hydrolyzing enzymes. In-silico-guided PET hydrolyzing enzyme engineering with DBTL cycles enables the rapid development of efficient variants of enzymes over tedious conventional enzyme engineering methods such as random or directed evolution. This review highlights the potential of in-silico-guided PET degrading enzyme engineering to create more efficient variants, including Ideonella sakaiensis PETase (IsPETase) and leaf-branch compost cutinases (LCC). Furthermore, future research prospects are discussed to enable a sustainable circular economy through the bioconversion of PET to original or high-value platform chemicals.
Keywords: PET hydrolases, Mutagenesis, PET bio-recycling, Molecular mechanics, Machine learning
Graphical Abstract
Highlights
-
•
Multiscale modeling with rational enzyme engineering for better PET hydrolases.
-
•
This approach is used to produce efficient variants of PETase and LCC enzymes.
-
•
The engineered enzymes efficiently depolymerize PET into its monomers.
-
•
The engineered enzymes are expected to pave the path for a sustainable circular economy for PET.
1. Introduction
1.1. PET recycling and upcycling
Poly (Ethylene Terephthalate) (PET) is a synthetic polymer composed of terephthalic acid (TPA) and ethylene glycol (EG), which are derived from non-renewable petroleum-based sources. It is the third most widely utilized plastic commodity worldwide. PET is mainly designed for single-use food and beverage packaging applications, and for textile fabric production[1]. For instance, the USA, the second-largest PET-packaging market with 20.5% of the global share, in 2020 reached a production of 22 million tons with a value of ∼$44 billion, and it is predicted to have a compound annual growth rate (CAGR) of 3.7% pushing demand to 27 million tons by 2025 [2]. PET is a classic linear “take–make–waste” economy model. Landfills or incineration are no longer viable options for end-of-life treatment of PET in a circular economy [3]. Eco-friendly PET recycling for material recovery is necessary for reducing greenhouse gas (GHG) emissions and plastic pollution. The current physical and/or chemical limitations of PET recycling make it challenging to establish a fully closed-loop (PET-to-PET) economy. These limitations decrease the quality of recycled PET and increase the need for new PET to be produced. For instance, mechanical PET recycling, which dramatically deteriorates the material properties, is generally a “downcycling’ process, and has end products with lower-quality PET. Chemical recycling deconstructs plastic into its intermediates or original monomers that permit repolymerization [4]. However, the chemical process is also economically unfeasible due to the extensive energy it consumes and harsh conditions it requires [4]. Hence, developing innovative, eco-friendly techniques to valorize plastic is urgent for establishing a circular PET economy.
Researchers leverage the biological processes in nature to develop cost-effective, sustainable plastic bio-deconstruction and bioconversion techniques [5]. Recent research efforts have made significant advances with the discovery of plastic-degrading enzymes, such as PET hydrolases, from certain microorganisms evolved in plastisphere that can selectively degrade PET into its constituent monomers thus offering more sustainable and eco-friendly bio-based PET recycling and waste management [6]. Notably, those microbes employ a two-enzyme system for PET deconstruction, in which one enzyme, PETase, converts the polymer into soluble intermediates: and, the other, MHETase, produces the constituent PET monomers [7]. Bio-based processes can remarkably reduce energy consumption and GHG emission compared to traditional recycling processes, as well as produce virgin PET from petroleum-based sources. Simultaneously, revolutionary plastic bio-upcycling technologies are emerging that involve breaking down post-consumer or post-industrial PET waste and feedstock to synthesize high-value products such as fuels, platform chemicals, and polymers using engineered PET hydrolyzing enzymes and microbes (i.e., biocatalysts) [8]. The high-value precursors obtained via biocatalytic approaches offer a path to establish the open-loop (PET-to-X) economy for PET [9], and a promising solution to address the global plastic waste problem. For instance, in bio-upcycling, several high-value advanced chemicals, such as muconate, 2-pyrone-4,6-dicarboxylic acid, and β-ketoadipate can be obtained through bio-funneling PET-derived monomers via engineered microbes [10]. Those monomers can be used to produce advanced-performance materials. In sum, the PET hydrolyzing enzymes play a key role in developing the bio-based circular economy for PET.
1.2. Mining PET-hydrolyzing enzymes and their limitations in industrial applications
PET hydrolyzing enzymes are carboxylic ester hydrolase enzymes such as cutinases, lipases, and esterases produced by microorganisms such as bacteria and fungi [11], [12]. Researchers have developed high-throughput, culture-based screening methods to identify environmental plastic-degrading microbes from the plastisphere [13]. Briefly, microorganisms expressing the PET enzymes are first enriched and isolated under proper cultivation conditions, and molecular biological (e.g., sub cloning and enzyme purification) or computational approaches (e.g., in silico protein homology search) are used to identify potent PET hydrolyzing enzymes. Indeed, multi-omics analyses, including genomic, proteomics, transcriptomic, and metabolomics, of those microbes enable the identification of novel PET hydrolyzing enzymes [14], [15]. The main drawback of conventional culture-dependent methods is that not all microorganisms in the plastisphere are culturable, or they require unique culturing conditions (e.g., micronutrients), making it challenging to find potent plastic-degrading enzymes through this approach [8].
Metagenomic is a culture-independent powerful tool to discover the potent PET hydrolyzing enzymes from the plastisphere. Advancement in high-throughput next-generation DNA sequencing technology, in in silico bioinformatics tools, and in metagenomic-library screening technologies enable efficient mining of promising PET-degrading enzymes via a metagenomics approach. Culture-independent metagenomics studies follow two approaches: 1) homology-based sequence screening; and, 2) functional-based screening [16], [17]. Homology-based screens use in silico similarity comparison of functional genes of known plastic degrading enzymes. It is an inexpensive and rapid method. Several PET hydrolyzing enzymes have been discovered by this method [18], [19], [20]. Homology-based screens give the structure and function of unknown enzymes based on their sequence similarity with known enzymes in the databases, under the assumption that similar sequences have similar functions. There are several approaches to identify homologous sequencing, including Basic Local Alignment Search Tool (BLAST), Hidden Markov Models (HMMs), and profile-based methods. For instance, Danso and coworkers demonstrated the successful application of HMMs for in-silico screening of PET hydrolyzing enzymes [21]. Despite the popularity of metagenomic methods for mining potential plastic degrading enzymes, there are some drawbacks to this approach such as limited comparison to current known plastic degrading enzymes in databases, missing potential plastic degrading enzymes due to low sequence similarity, and the need of further study to validate the functionality of the enzyme [17]. The other approach of metagenomics uses activity assays to find potential PET-degraders. This method does not require prior knowledge of the sequence. It is considered a more effective method than homology-based screening and it uncovers completely novel groups of enzymes for which the sequences are divergent from existing homologous ones. However, it is more costly, and has limitations and challenges associated with the heterologous expression of genes, such as host compatibility, which damper its effectiveness [8], [22].
In addition to metagenomics, researchers leverage a proteomic-based approach to discover PET hydrolyzing enzymes. Generally, PET depolymerization occurs extracellularly, using enzymes secreted by microbes. Hence, the exoproteome is the principal target for identifying potential plastic-degrading enzymes. Researchers mostly use comparative proteomics, which relies on differential induction of microbes to express plastic hydrolyzing enzymes when presented with a plastic surface [23]. Although this approach can directly detect and quantify protein expression, proteomic studies have only been conducted pure cultures and not in complex environmental samples, revealing the complex nature of high-quality protein extraction and the lack of high-throughput bioinformatic analysis for metaproteomics [24].
In general, the inferior catalytic performance of natural plastic-degrading enzymes is a key technical challenge to establishing the feasible biocatalytic-based PET-recycling or -upcycling technology. Indeed, an industrially applicable PET hydrolyzing enzyme needs to have higher conversion efficiency, improved robustness (thermal and chemical), expanded half-life, and reusability with prolonged functional properties. Hence, the engineering of efficient PET-hydrolyzing enzymes is necessary for a realistic application of biocatalytic-based PET depolymerization on an industrial scale. PET-degrading enzymes can be optimized either through a rational engineering approach or by directed evolution. The latter involves screening huge libraries of PET enzyme variants, which is laborious and time-consuming. The need for efficient high-throughput screening techniques is a crucial technical barrier to the directed evolution [25]. By contrast, rational engineering primarily leverages precise modifications of enzymes based on a deep understanding of their structure and function using computational simulation and modeling of proteins coupled with experimental techniques such as X-ray crystallography, NMR, and enzyme kinetics to enhance desired PET-degrading enzyme characteristics. The available structural and mechanistic information for many identified PET-hydrolyzing enzymes enables the efficient engineering of plastic-degrading enzymes via a rational approach. Researchers leverage systems and synthetic biology-based design, build, test, and learn (DBTL) cycles to develop commercially applicable, efficient PET hydrolyzing enzymes through rational engineering. This review highlights the trends of using an in-silico-guided PET hydrolyzing enzyme engineering approach to develop efficient PET hydrolyzing enzymes.
2. Overview of developing in-silico platform to study the plastic hydrolyzing enzymes
In-silico enzyme engineering is primarily twofold: rational and combinatorial [26]. In rational engineering, hot spots for mutations and potential substituent residues are identified through the results of previous experiments or through binding surface analysis via molecular docking. Combinatorial approaches involve generating and screening larger libraries of enzymes to identify potential variants for specific properties such as increased thermal stability or enhanced catalytic activity. Machine Learning (ML) algorithms have recently been utilized to effectively screen a large sequence space. However, in practical ML approaches, rational engineering inputs are incorporated to guide the algorithms search for a specific enzyme activity, or to truncate the sequence space. Once the potential variants are identified, their kinetics are studied through multi-scale computational modeling [27].
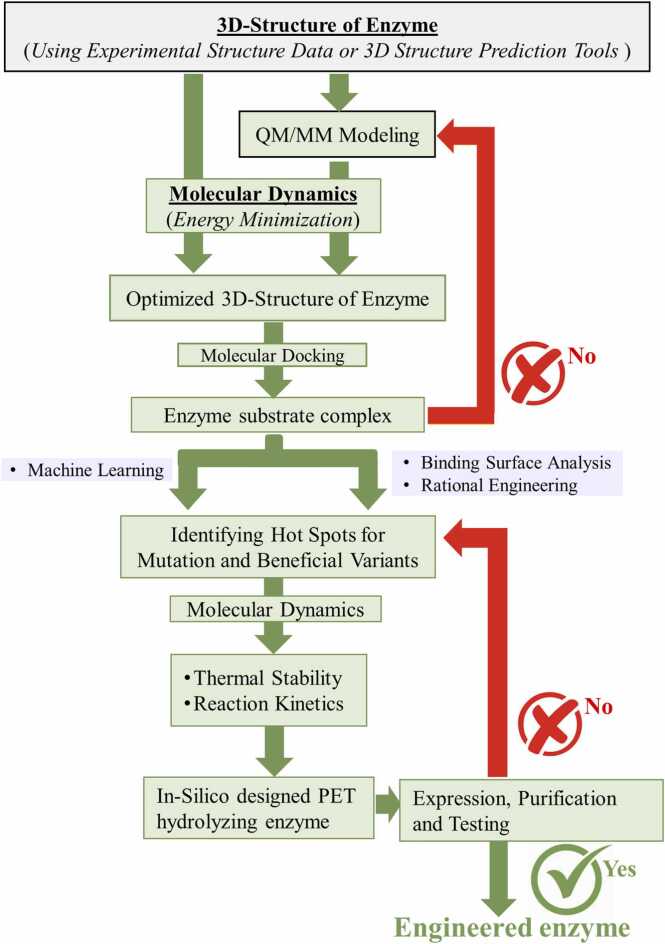
As shown in Fig. 1, the modeling process initiates with an atomic configuration of the PET hydrolyzing enzyme obtained from X-ray crystallography or NMR spectroscopy [28]. After optimizing the enzyme’s atomic configuration, an initial configuration of the enzyme/substrate (e.g., MHET or BHET) complex with the lowest binding energy is made using molecular docking programs by sampling the conformational space with various algorithms [29], [30], [31]. The docked configurations can further be analyzed to visualize the active sites of the binding pockets, and to identify the residues which are crucial for the PET degradation process. The residues of the enzyme in the active site play a critical role in determining the size, shape as well as hydrophobicity of the binding site [32]. These key residues can be considered for further mutation (i.e., mutagenesis).
Fig. 1.
Flowchart of the in-silico multiscale modeling process for enzyme engineering.
A deeper understanding of enzyme/substrate interactions and their dynamical behavior is crucial in evaluating the effect of mutagenesis, which is possible with Molecular Dynamics (MD) simulations. MD simulate the dynamics of atoms in force fields originating from their surroundings as described by classical Newtonian physics [29], [30], [33]. The accuracy or reliability of the MD depends significantly on the quality of the force field, which typically describes the potential energy of the systems with parameters related to bonded and non-bonded interactions. In the bonded model, force fields are described by structural parameters such as bond lengths, bond angles, and dihedral angles, which can be extracted from quantum mechanical calculations [34]. Once the force fields are generated, the time evolution of the system’s energy as a function of its structural details is simulated via Newtonian mechanics using molecular dynamic simulations. MD programs typically include various analysis tools for studying the configurations and trajectories that were calculated throughout the simulation such as energy-optimized atomic configurations, binding energies, flexibility of residues, hydrogen bonding analysis, and root mean square deviation (RMSD). RMSD compares the atomic coordinates of two molecular configurations. In enzyme engineering, RMSD can be used to analyze the relative stability of a mutant enzyme with respect to that of wild type. However, MD does not provide sub-atomic details such as electronic charge transfer.
Electronic properties of a system are described by the laws of Quantum Mechanics, which can be obtained by solving the Schrodinger equation. However, solving Schrodinger’s equation for a many-electron system is challenging and only possible using approximations [35], [36]. Ab-initio Density Function Theory (DFT) is one of the mathematical approximations that has proven crucial to studying biological processes, but it is still a challenging process that requires high performance computing. Hybrid Quantum Mechanics/Molecular Mechanics (QM/MM) methods further help to overcome this problem [37], [38], [39]. In QM/MM simulations, a small portion of the system, usually the active site, is treated using QM, while the remainder of the system is handled using MM. For biological systems such as the PET/IsPETase complex, we can identify regions that are known to play a crucial role in catalysis and truncate the larger system to a few hundred atoms around the area of interest to perform DFT calculations. In the case of PET/IsPETase system, electronic details will be important around the catalytic triad and the metal site. We can investigate the fine geometric details, charge transfer details, and energy barriers from the output of QM/MM calculations.
While more efficient variants were reported through rational engineering, the optimum enzyme configurations may be hidden in the intractable chemical configuration space, which can be explored through Machine Learning (ML) algorithms. Recently, Pirillo and coworkers presented a semi-rational protein evolution workflow, where hotspots for potential mutations were identified by bioinformatics, and site-saturated mutagenesis was applied to identify high-stability variants. The stable variants chosen from a MD study were then tested for PET hydrolysis under various conditions [6].
Further, ML is a promising tool that can assist in-silico modeling of enzyme engineering in various ways [40]. ML models can be trained to understand the correlations between the characteristics and enzyme sequences to predict variants with targeted properties. Various protein predictors such as distances between amino acids, torsion angles, and active site residues have been used in this process [41]. ML models can also be used to develop more accurate force fields to achieve better results from MD simulations [42].
It is evident that information at various time/length scales must be investigated to achieve a deeper insight into the PET - hydrolysis process. Therefore, combined application of several in silico platforms is essential to pave the path for developing industrial-scale PET hydrolysis.
2.1. Current knowledge of engineering LCC and IsPETase
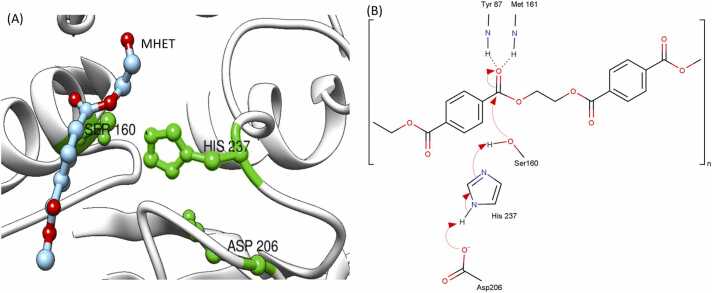
The PET hydrolyzing enzymes belong to the hydrolase family with a Nucleophile-His-Acid catalytic triad made of serine, histidine, and aspartic acid. Most of these hydrolases have the GX1 SX2G motif in the active site, where central Serine is a part of the catalytic triad [32]. As the PET substrate binds to the enzyme, its carbonyl bond attached to the first benzene ring must be harbored close to Serine in the catalytic triad (Fig. 2). Serine gets polarized by histidine, which is then stabilized by aspartic acid. Polarized Serine will attack the carbonyl bond (C-O) of the polyester, resulting in a tetrahedral intermediate, which is then stabilized by the oxyanion hole. The hydrolysis procedure is completed by a second nucleophilic attack mediated through a water molecule [43], [44]. With the results from a QM/MM approach combined with statistical sampling, Zheng and coworkers found a unified PET hydrolysis mechanism for LCC and IsPETase [45]. Various other enzyme-specific residues support (or inhibit) the hydrolysis reaction. The activity can be enhanced by mutagenesis. A summary of recently found variants is described below and summarized in Table 1, Fig. 3, and Fig. 4.
Fig. 2.
A mechanistic overview of mutants to enhance PETase activity. Panel (A) shows the catalytic triad of MHET-docked PETase (from 5xjh.pdb). Amino acids of catalytic triad, Ser, His, and Asp are shown in green. The MHET molecule is shown in ball-and-stick model. The carbonyl bond of MHET is closely harbored to Ser 160 to initiate the hydrolysis reaction. Panel (B) shows initial steps of the PET hydrolysis mechanism.
Table 1.
Enzymes developed through in silico-based engineering.
| Enzyme | Development platform/Software used | Specific mutations incorporated into the enzyme | Type of PET used as a substrate for enzyme assay | In vitro/in vivo conversion efficiency | Ref. |
|---|---|---|---|---|---|
| FAST-PETase: functional, active, stable, and tolerant PETase) | MutCompute three-dimensional CNN (3DCNN) model. A structure-based machine learning algorithm |
Three from ML algorithm prediction (N233K/R224Q/S121E) and two (D186H/R280A) from parental scaffold) Active in ambient temperature from 30 ℃ To 50 ℃ | untreated, postconsumer-PET from 51 different thermoformed products (Marked thermostability and reactivity toward amorphous and less crystalline PET (1.2–11.7% crystallinity) at elevated temperatures (e.g., 50 ℃)) |
98.4% of TPA From digestion solution. Complete degradation of untreated, post-consumer-PET from 51 different thermoformed products in 1 week. Depolymerization of untreated, amorphous portions of a commercial water bottle and an entire thermally pretreated water bottle at 50 ºC |
[54] |
| Dura-PETase | Greedy accumulated strategy for protein engineering, GRAPE. A systematic clustering analysis combined with greedy accumulation of beneficial mutations in a computationally derived. library |
melting temperature increased by 31 °C | 1. semicrystalline poly (ethylene terephthalate) (PET) films (30%)2. microplastics | 1. 30% enhancement of PET degradation at mild temperatures (over 300-fold). 2. Complete biodegradation of 2 g/L microplastics 2. Complete biodegradation of 2 g/L microplastics |
[52] |
| LCC-ICCG | M.Do. Binding analysis to 2- HE(MHET)3 | F243I/D238C/S283C/Y127G | post-consumer colored-flake PET waste (PcW-PET) | 90% of PET depolymerization into monomers over 10 h, Productivity is 16.7 g of TPA/L/h (200 g/kg of PET suspension, with an enzyme concentration of 3 mg/g of PET) | [56] |
| Thermo-PETase |
IsPETaseS121E/D186H/R280A variant, have a stabilized β6-β7 connecting loop and extended subsite IIc, Thermostable IsPETase with Tm 56.8 ℃) that harbors three mutations, S121E, D186H, and R280A |
Tm value increased by 8.81 °C, and PET degradation activity was enhanced by 14-fold at 40 °C compared with IsPETaseWT. | [49] | ||
| IsPETaseS121E/D186H/S242T/N246D | Structural bioinformatics-based protein engineering | integrating the S242 T and N246D mutations into the previously reported IsPETaseS121E/D186H/R208A variant | A 58-fold increase in activity compared with IsPETaseWT. | [50] | |
| IsPETase W159H/F229Y variant | the mutation design tool, Premuse | two mutations (W159H and F229Y) | p-NPP, amorphous PET, and PET bottle | Its Tm and catalytic efficiency values (kcat/Km) increased by 10.4 °C and 2.0-fold using p-NPP as the substrate compared with the wild type. The degradation activity for amorphous PET was increased by almost 40-fold compared with the wild type at 40 °C in 24 h. biodegradation of PET bottles at a mean rate of 23.4 mgPET/h/mg enzyme. | [53] |
| L92F/Q94Y variant of PES-H1 | Structural analyses and computational modeling using MD simulation | Amorphous Goodfellow PET (Gf-PET) films and pretreated postconsumer (‘real-world’) PET waste | 2.3-fold and 3.4-fold improved hydrolytic activity against amorphous PET films and pretreated real-world PET waste, respectively. hydrolyzed low-crystallinity PET materials 2.2-fold more efficiently | [59] | |
| Ple629 polyester hydrolase variant | D226A/S279A mutations improved activity and thermo-stability | PET nanoparticles | 5.5-fold improved activity | [60] | |
| Cut190* | S226P/R228S increased activity and higher thermostability. The mutant of the cutinases-like enzyme, Cut190, from Saccharomonospora viridis AHK190 | Model substrate poly (butylene succinate-co-adipate). | [61] | ||
| IsPETase double mutant | Homology modeling and g-induced fit docking (IFD) | Two mutations are introduced at S238F/W159H to make the PETase-active site more like cutinase enzyme’s | polyethylene-2,5-furan dicarboxylate (PEF), PET coupons with an initial crystallinity of 14.8 ± 0.2% |
[51] |
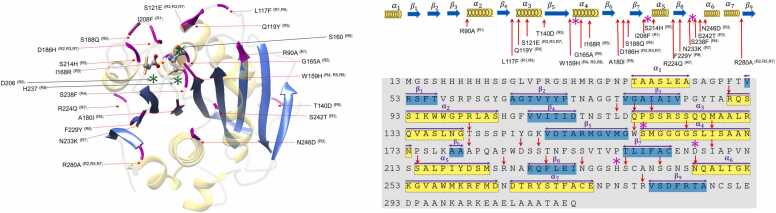
Fig. 3.
Overview of in-silico engineering of IsPETase (a) is the structure of IsPETase from 5xjh.pdb [44]. The catalytic triads are shown in the ball-stick model, α helices are shown in shaded yellow, β strands are shown in blue, and the residue spots considered in various studies for mutagenesis are marked in magenta. (b) shows the sequence of IsPETase. helices and strands are marked in yellow and blue. Catalytic triads (S160, H237, D206) are marked with stars, and red arrows point to the residue spots considered for mutation. (Rx) denotes the references as R1-[32], R2-[49], R3-[50], R4-[51], R5-[52], R6-[53], R7-[54], R8-[44].
Fig. 4.
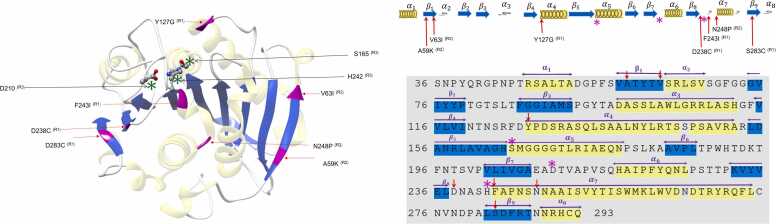
Overview of in-silico engineering of LCC: (a) The structure of LCC from 4eb0.pdb [28]. The catalytic triad is shown in the ball-stick model, α helices are shown in shaded yellow, β strands are shown in blue, and the residue spots considered in various studies for mutagenesis are marked in magenta. (b) The sequence of LCC (from 4eb0.pdb). helics and strands are marked in yellow and blue. Catalytic triad (S165, H242, D210) is marked with stars and red arrows point to the residue spots considered for mutation. (Rx) denotes the references as R1-[56], R2-[62], R3-[37].
2.1.1. Rational engineering of IsPETase
IsPETase secreted from Ideonella sakaiensis 201-F6 gained much attention due to its PET hydrolysis activity at mild temperature [46]. It shows 5–120-fold increase in PET hydrolysis activity compared to other mesophilic PET-hydrolyzing enzymes [43], [44]. IsPETase has 51% of amino acid similarity to the cutinase from Thermobifida fusca [46]. It has a few notable features that further support the hydrolyzing process [44]. It gains stability with two disulfide bonds, while only one conserved disulfide bond exists in other homologs. Conserved disulfide bond C273-C289 connects the last loop to the C-terminal helix. The IsPETase-specific disulfide bond C203-C239 harbors the catalytic acid and the base [43], and has been shown to result in a lower energy barrier/higher efficiency for PET hydrolysis [47]. Trp185 is found to be wobbling due to the presence of Serine, i. e. specific to IsPETase, and forms T-stacking with the benzene ring of the substrate, which also plays a vital role in the process of cleavage in PET hydrolysis. A. Crnjar and coworkers confirmed the flexibility of wobbling Trp by S214H and I218F with MD simulations [48].
Using covalent docking calculations followed by experiments, Joo and co-workers reported a 22.4% increase of PET degradation activity in IsPETase-R280A. Even though R280 is far from the catalytic triad, its replacement with a smaller hydrophobic residue enhances the substrate binding [44]. Melting of this variant is reported to increase by 8.81 ℃ with the addition of S121E/D186H [49]. Thermal stability of this variant (S121E/D186H/R280A) is achieved by a newly introduced water-mediated hydrogen bond between S121E and Asn172. The new variant named ThermoPETase, IsPETase-S121E/D186H/R280A gives a 14-fold increase in hydrolysis activity at 40 ℃ [49]. In a later study, ThermoPETase was further engineered to IsPETase S121E/D186H/N246D/S242T, which has a melting temperature of 37 ℃ and a 58-fold higher activation rate compared to wildtype. N246D was found to increase the activity and S242H to improve the durability [50].
Ma and coworkers identified the potential mutation sites that could increase the active site space or hydrophobicity to increase PET binding affinity by docking IsPETase with 2PET substrate [32]. Out of the six mutation sites chosen from a molecular docking study and tested through experiments, R90A, L117F and I208F (R61A, L88F, and I179F in the original paper) have shown significant effects with a 1.4-, 2.1- and 2.5-fold increases in activity, respectively. Electrostatic surface potential analysis suggests that alanine is more hydrophobic and less massive compared to arginine, thus it increases activity. Additionally, R61A enhances the cleavage of products due to its smaller size, L88F improves the effect of oxyanion hole, and I179F increases the substrate localization, with a similar effect as W156. Surprisingly, narrowing down the binding cleft with double mutant IsPETase-S238F/W159H has shown to improve the PET hydrolysis activity [51].
To further understand the PET hydrolysis mechanism, Boneta and coworkers and Jerves and co-workers performed a DFT calculation on a smaller region of the IsPETase /MHET complex and identified a 4-step mechanism [37], [38]. The energy barrier for the rate limiting acylation step is 20.0 kJ/mol. While DFT calculations are time-consuming and require high-performance computing resources, it is crucial to identify the role of each residue at the electronic level. However, this approach is limited to small systems due to computational cost. It is hard to understand the effect of residues far from the QM region, but it can be accessed through QM/MM approach.
2.1.2. Rational engineering of LCC
The leaf and branch compost cutinase (LCC), first derived in 2012, shows a high potential for industrial application mainly due to its activity at a relatively high melting temperature, 65 ℃[55]. LCC and the IsPETase enzymes have a similar PET-hydrolyzing mechanism. The two enzymes are structurally similar with an identity of 49.5% with the same catalytic triad made of Serine, Histidine, and Aspartic Acid [37]. By binding surface analysis through Molecular Docking studies, Tournier and coworkers considered 11 residues in the contact shell for potential mutations. Of these the F243 substitution with Isoleucine (F243I) is the most promising, probably because it expands the binding cleft for PET [56], [57]. They also confirmed the melting temperature increase in LCC by Ca2+ ions, which was also reported in previous studies [55]. Key residues that were on the metal site of known homolog enzymes were identified as D238 and S283. They mutated by cysteine to introduce a disulfide bonding to increase melting temperature instead of metal ion stabilization. The engineered LCC version named LCC-ICCG (with D238C/S283C along with F243I and Y127G) is reported to acieve 90% of PET depolymerization. Recent study by Zeng and coworkers reported an increase in the melting temperature of LCC-ICCG to 98.9 ℃ by addition of A59K, V63I, and N248P mutations. However, the optimal hydrolyzing temperature was found to be 74 ℃ [58]. Using DFT, Boneta and coworkers analyzed the reaction mechanism of PET hydrolysis through LCC in 4 major steps [37]. Zheng and coworkers analyzed the correlations between structural details, such as bond lengths and dihedral angles, and activation energies using a combined Quantum Mechanics (DFT) and Molecular Dynamics simulations. More work is needed to clearly understand the correlations between the atomistic details and the hydrolysis performance [58].
2.1.3. Leverage machine learning with MD simulation for engineering the enzymes
We discussed above changes in PET hydrolysis by IsPETase and LCC because of targeted single/multiple point mutations chosen by rational protein engineering. An intuitive next step is to investigate their synergistic performance. However, searching for combinatorial mutations to identify optimal variants is beyond the capacity for conventional in-silico approaches. Such a problem can only be tackled by in-silico machine learning (ML) approaches.
Recently, Pirillo and coworkers presented a semi-rational protein evolution workflow, where hotspots for potential mutations were identified by bioinformatics, and site saturated mutagenesis was applied to identify highly stable variants [6]. The stable variants chosen from a MD study were then tested for PET hydrolysis under various conditions.
Cui and co-workers reported a new variant of IsPETase, named DURA-PETase with a 31 ℃ increase in melting temperature, and with a remarkable 300-fold increase of degradation activity on semi-crystalline PET [52]. GRAPE algorithm was used on 21 single point mutations, which were chosen by other energy-based algorithms, and verified through experiments. These 21 single-point mutants were clustered into three groups, and the most promising member of each group was chosen as the parent. The multi-hierarchical GRAPE algorithm continues accepting the next off-spring of the system only if the performance is increased because of the addition. This study resulted in DURA-PETase with mutations S214H-I168R-W159H-S188Q-R280A-A180I-G165A-Q119Y-L117F-T140D. MD was used to verify the activity of each alteration.
Meng and coworkers published a web tool named Premuse to design enzyme variants by comparing to their homologs [53]. Premuse integrates pairwise alignment of residues between the target protein and its homologs, position specific amino acid probabilities (PSAP), and preferred mutation selections to design new variants. Double-mutant IsPETase W159H/F229Y, which was derived by Premuse after comparing it to 1486 homologs was found to have a 10.4 ℃ increase in melting temperature and a 2.0 -fold increase in degrading activity for p-NPP substrate. MD study on this new variant on 2HE(MHET)4 found new hydrogen bonds in the active site compared to those in the wildtype enzyme.
Lu and coworkers used a ML platform, MuteCompute, which employs a CNN analysis, to identify the unstable regions of the WT IsPETase [54]. Based on the information on the chemical environment of 19000 stable protein structures from protein data banks, 159 mutations were predicted to have better stability than the wild type.
Out of the mutations, the four with higher melting temperature and higher degradation rate were filtered. Twenty-Nine combinatorial configurations of those four mutations were added to three enzymes, WT-PETase, Thermo-PETase, and Dura-PETase. The highest melting temperatures were observed for WT-PETaseT140D/N233K (58.1 ℃), ThermoPETaseN233K (67.2 ℃), and DuraPETaseN233K (83.5 ℃). They show melting temperature increases of 10, 9 and 5 ℃ respectively, relative to their host scaffolds, IsPETase, ThermoPetase and DuraPETase. The best PET degradation rates of 3.4 and 29- fold compared to WT at 30 ℃ and 40 ℃ was achieved with variant IsPETase-S121E/D186H/R224Q/N233K/R280A, which was named FAST-PETase (Functional, Active, Stable and Tolerant PETase). The overall PET degrading performance of FAST PETase was found to be superior to those IsPETase, Dura-PETase, Thermo-PPETase, LCC, as well as LCC-ICCM variants. The increased thermal stability of FAST-PETase can be ascribed to the newly formed salt bridge between N233K and E204 and the hydrogen bond between R224Q and S192. Crystal structure analysis shows further water-mediated hydrogen bonding between H186 and N172.
3. Summary and outlook
3.1. Outlook
Due to the enormous size of sequence space, the result of its combinatorial nature, combinatorial sequence space, rational protein engineering may fail to identify highly beneficial variants of already discovered enzymes for PET hydrolysis. Recent development of Machine Learning approaches plays a crucial role in searching for the beneficial variants in the enormity of this sequence space. That was demonstrated by the engineering of IsPETase into the more efficient FAST-PETase [54]. However, ignoring the fundamental laws of physics (such as Quantum Mechanics as well as Classical Newtonian Mechanics) that primarily govern the process of biodegradation in machine learning approaches may guide us towards incorrect directions. Physics-learned ML models are expected to be successful in paving the path for protein engineering towards desired activities [63].
There are several ways in which ML approaches can be trained with Physics-based models. Recently, there has been significant research on ML-modeled force fields based on input from Quantum Mechanics.
Another approach is to use Physics-based descriptors to train ML modeling [64]. The performance of a ML algorithm depends on the selection of the features used for describing a desired property. On the other hand, understanding reaction pathways with the details of electron transfer is possible through Quantum Mechanical DFT approaches, which are computationally expensive. It is, therefore, a challenge to use Machine Learning algorithms to effectively predict reaction pathways. There have been significant efforts to identify suitable descriptors, and more accurate Machine Learning models in order to incorporate quantum mechanical effects in enzyme engineering. For example, Song and coworkers calculated the minimum energy pathways (MEP) using a multiscale modeling approach [65]. Many initial configurations were obtained using the Molecular Dynamics trajectory. A region of interest with a ∼150 atoms was selected to study with QM. QM features of that active region were investigated, while the rest of the system was considered with Molecular Dynamics. Features related to atomistic details such as bond lengths of specific heavy atoms, hydrogen bond distances, and dihedral angles of intermediate states can be analyzed. ML algorithms can extract the important features of minimum energy pathways.
Physics-learned Machine Learning algorithms based on features evaluated from a multi-scale modeling approach including Molecular Docking, Molecular Dynamics, and Quantum Mechanics are expected to accelerate the engineering of PET Hydrolysis with biocatalysts.
The expression and purification of in-silico-designed PET hydrolyzing enzymes is one of the key challenges. Overexpression of recombinant protein often leads to severe burdens on the physiology of host strain. The novel enzymes can be less soluble and tends to make inclusion bodies, folding defects, and become toxic to traditionally employed model systems such as E. coli. Furthermore, not all suggested mutations can be tested in the invitro reactions. Different tags such as GST, Fh8, SUMO, His, TRX, and MBP at the N- or C-terminal enhance the solubility of PET hydrolyzing enzymes and help in affinity purification. Researchers implement protein expression control (i.e., regulatory promotor) by lowering the temperature after culture induction to promote soluble PET hydrolyzing enzyme production. This strategy increases protein stability and proper folding. Further, novel promoters and glycoengineering E. coli cells also lead to increased recombinant protein expression [66]. It was also revealed that enhanced recombinant protein yields were obtained in the E. coli periplasm by combining signal peptide and production rate screening. One study established a scale-up of a type I secretion system in E. coli using a defined mineral medium, paving the way for industrial application [67]. Recently, Deng and coworkers demonstrated enhancement of activity and thermostability of IsPETase through glycosylation engineering [62]. Other robust protein-expressing host strains such as yeast (e.g., Pichia pastoris) need to be used to produce the targeted PET hydrolyzing enzymes with the desired post translation modifications (i.e., glycosylation).
3.2. Summary
In conclusion, we discussed the current efforts on identifying more efficient variants of IsPETase and LCC for PET degradation. In-silico multiscale modeling approaches, which includes quantum mechanical density functional theory, molecular docking, molecular dynamics, and machine learning algorithms have the potential to uncover more beneficial variants. Engineered enzymes provide a promising path to develop a sustainable enzyme or microbial cell-based circular economy through the bioconversion of PET to original or high-value platform chemicals. We discussed the potential of physics-learned machine learning algorithms for future research in enzyme engineering and the challenges of testing those in-silico predicted varians in in-vitro techniques.
Funding
This work was supported by the grants from Advanced Energy Institute [Grant numbers: 2625575] and Green Core LCC, Japan [Grant numbers C-22-0027].
CRediT authorship contribution statement
LNJ, TJ, and PS conceptualized and led the writing and editing of the overall manuscript. SKJ, HDD, BJ, LD, CM, MMSS, and RM collected the literature write the draft, and edit the manuscript; all authors reviewed and edited the whole manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this manuscript.
Acknowledgements
The author thanks Prof. Aldo Migone, an Emeritus Professor of Physics at Southern Illinois University Carbondale, for proofreading and language editing of the manuscript. LNJ, TJ, and SP acknowledge funding from Advanced Energy Institute, SIU-Carbondale to support the study. LNJ also acknowledges funding from Green Core LCC, Japan [Grant numbers C-22–0027].
References
- 1.Nisticò R. Polyethylene terephthalate (PET) in the packaging industry. Polym Test. 2020;90(106707) doi: 10.1016/j.polymertesting.2020.106707. [DOI] [Google Scholar]
- 2.Smithers.(2023) Global PET packaging demand to reach $44.1 billion in 2020 says Smithers report. Available from: https://www.smithers.com/en-gb/resources/2020/sept/global-pet-packaging-demand-to-reach-$44-1-billion
- 3.Anonymous.(2023) Plastics and the circular economy Deep dive. [cited 2023 03.03.2023]; Available from: https://ellenmacarthurfoundation.org/plastics-and-the-circular-economy-deep-dive
- 4.Jehanno C., Alty J.W., Roosen M., De Meester S., Dove A.P., et al. Critical advances and future opportunities in upcycling commodity polymers. Nature. 2022;603(7903):803–814. doi: 10.1038/s41586-021-04350-0. 〈https://www.ncbi.nlm.nih.gov/pubmed/35354997〉 [DOI] [PubMed] [Google Scholar]
- 5.Qi X., Yan W., Cao Z., Ding M., Yuan Y. Current advances in the biodegradation and bioconversion of polyethylene terephthalate. Microorganisms. 2021;10(1) doi: 10.3390/microorganisms10010039. 〈https://www.ncbi.nlm.nih.gov/pubmed/35056486〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pirillo V., Orlando M., Tessaro D., Pollegioni L., Molla G. An efficient protein evolution workflow for the improvement of bacterial PET hydrolyzing enzymes. Int J Mol Sci. 2021;23(1) doi: 10.3390/ijms23010264. 〈https://www.ncbi.nlm.nih.gov/pubmed/35008691〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knott Erickson, Allen Gado, Graham, et al. Characterization and engineering of a two-enzyme system for plastics depolymerization. Proc Natl Acad Sci. 2020;117(41):25476–25485. doi: 10.1073/pnas.2006753117. 〈https://www.ncbi.nlm.nih.gov/pubmed/32989159〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu B., Wang D., Wei N. Enzyme discovery and engineering for sustainable plastic recycling. Trends Biotechnol. 2022;40(1):22–37. doi: 10.1016/j.tibtech.2021.02.008. 〈https://www.ncbi.nlm.nih.gov/pubmed/33676748〉 [DOI] [PubMed] [Google Scholar]
- 9.AnonymousPlastic upcycling Nat Catal 2 11 2019 945 946.(Published online)〈https://doi.org/10.1038/s41929-019-0391-7〉.
- 10.Dissanayake L., Jayakody L.N. Engineering microbes to bio-upcycle polyethylene terephthalate. Front Bioeng Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.656465. 〈https://www.ncbi.nlm.nih.gov/pubmed/34124018〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kawai Kawabata, Oda Current state and perspectives related to the polyethylene terephthalate hydrolases available for biorecycling. ACS Sustain Chem Eng. 2020;8(24):8894–8908. doi: 10.1021/acssuschemeng.0c01638. [DOI] [Google Scholar]
- 12.Maurya A., Bhattacharya A., Khare S.K. Enzymatic remediation of polyethylene terephthalate (PET)-based polymers for effective management of plastic wastes: an overview. Front Bioeng Biotechnol. 2020;8 doi: 10.3389/fbioe.2020.602325. 〈https://www.ncbi.nlm.nih.gov/pubmed/33330434〉 doi: 10.3389/fbioe.2020.602325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viljakainen V.R., Hug L.A. New approaches for the characterization of plastic-associated microbial communities and the discovery of plastic-degrading microorganisms and enzymes. Comput Struct Biotechnol J. 2021;19:6191–6200. doi: 10.1016/j.csbj.2021.11.023. 〈https://www.ncbi.nlm.nih.gov/pubmed/34900132〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dey S., Rout A.K., Behera B.K., Ghosh K. Plastisphere community assemblage of aquatic environment: plastic-microbe interaction, role in degradation and characterization technologies. Environ Micro. 2022;17(1):32. doi: 10.1186/s40793-022-00430-4. 〈https://www.ncbi.nlm.nih.gov/pubmed/35739580〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purohit J., Chattopadhyay A., Teli B. Metagenomic exploration of plastic degrading microbes for biotechnological application. Curr Genom. 2020;21(4):253–270. doi: 10.2174/1389202921999200525155711. 〈https://www.ncbi.nlm.nih.gov/pubmed/33071619〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankara Subramanian S.H., Balachandran K.R.S., Rangamaran V.R., Gopal D. RemeDB: tool for rapid prediction of enzymes involved in bioremediation from high-throughput metagenome data sets. J Comput Biol. 2020;27(7):1020–1029. doi: 10.1089/cmb.2019.0345. [DOI] [PubMed] [Google Scholar]
- 17.Ufarté L., Laville É., Duquesne S., Potocki-Veronese G. Metagenomics for the discovery of pollutant degrading enzymes. Biotechnol Adv. 2015;33(8):1845–1854. doi: 10.1016/j.biotechadv.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 18.Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., et al. New insights into the function and global distribution of polyethylene terephthalate (PET)-Degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl Environ Microbiol. 2018;84(8) doi: 10.1128/AEM.02773-17. 〈https://www.ncbi.nlm.nih.gov/pubmed/29427431〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim D.-W., Ahn J.-H., Cha C.-J. Biodegradation of plastics: mining of plastic-degrading microorganisms and enzymes using metagenomics approaches. J Microbiol. 2022;60:969–976. doi: 10.1007/s12275-022-2313-7. [DOI] [PubMed] [Google Scholar]
- 20.Zrimec J., Kokina M., Jonasson S., Zorrilla F., Zelezniak A. Plastic-degrading potential across the global microbiome correlates with recent pollution trends. mBio. 2020 doi: 10.1128/mBio.02155-21. DOI: https://doi.org/10.1101/2020.12.13.422558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Danso D., Schmeisser C., Chow J., Zimmermann W., Wei R., et al. New insights into the function and global distribution of polyethylene terephthalate (PET)-Degrading bacteria and enzymes in marine and terrestrial metagenomes. Appl Environ Microbiol. 2018;84(8):e02773–02717. doi: 10.1128/AEM.02773-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lam K.N., Cheng J., Engel K., Neufeld J.D., Charles T.C. Current and future resources for functional metagenomics. Front Microbiol. 2015;6:1196. doi: 10.3389/fmicb.2015.01196. 〈https://www.ncbi.nlm.nih.gov/pubmed/26579102〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weinberger S., Beyer R., Schuller C., Strauss J., Pellis A., et al. High throughput screening for new fungal polyester hydrolyzing enzymes. Front Microbiol. 2020;11:554. doi: 10.3389/fmicb.2020.00554. 〈https://www.ncbi.nlm.nih.gov/pubmed/32390956〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas R., Sarkar A. In: ‘Omics’ Tools in Soil Microbiology: The State of the Art, in Advances in Soil Microbiology: Recent Trends and Future Prospects. Adhya T.K., et al., editors. Springer Singapore; singapore: 2018. pp. 35–64. [Google Scholar]
- 25.Sheludko Y.V., Fessner W.-D. Winning the numbers game in enzyme evolution – fast screening methods for improved biotechnology proteins. Curr Opin Struct Biol. 2020;63:123–133. doi: 10.1016/j.sbi.2020.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Korendovych I.V. In: Protein Engineering Methods in Molecular Biology. Bornscheuer U., Höhne M., editors. Humana Press; New York, NY: 2018. Rational and Semirational Protein Design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhai C., Li T., Shi H., Yeo J. Discovery and design of soft polymeric bio-inspired materials with multiscale simulations and artificial intelligence. J Mater Chem B. 2020;8:6562–6587. doi: 10.1039/D0TB00896F. [DOI] [PubMed] [Google Scholar]
- 28.Anonymous,(2023) RCSB Protein Data Bank (RCSB PDB).
- 29.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Comput Chem. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 30.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E., III, et al. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comput Phys Commun. 1995;91(1–3):1–41. doi: 10.1016/0010-4655(95)00041-D. [DOI] [Google Scholar]
- 31.Phillips J.C., Hardy D.J., Maia J.D.C., Stone J.E., Ribeiro J.V., et al. Scalable molecular dynamics on CPU and GPU architectures with NAMD. J Chem Phys. 2020;153(4) doi: 10.1063/5.0014475. 〈https://www.ncbi.nlm.nih.gov/pubmed/32752662〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ma Y., Yao M., Li B., Ding M., He B., et al. Enhanced Poly(ethylene terephthalate) hydrolase activity by protein engineering. Engineering. 2018;4(6):888–893. doi: 10.1016/j.eng.2018.09.007. [DOI] [Google Scholar]
- 33.Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., et al. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. 〈https://www.ncbi.nlm.nih.gov/pubmed/19399780〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li, Merz MCPB.py: a python based metal center parameter builder. J Chem Inf Model. 2016;56(4):599–604. doi: 10.1021/acs.jcim.5b00674. 〈https://www.ncbi.nlm.nih.gov/pubmed/26913476〉 [DOI] [PubMed] [Google Scholar]
- 35.Barca G.M.J., Bertoni C., Carrington L., Datta D., De Silva N., et al. Recent developments in the general atomic and molecular electronic structure system. J Chem Phys. 2020;152(15) doi: 10.1063/5.0005188. 〈https://www.ncbi.nlm.nih.gov/pubmed/32321259〉 [DOI] [PubMed] [Google Scholar]
- 36.Fox S.J., Pittock C., Fox T., Tautermann C.S., Malcolm N., et al. Electrostatic embedding in large-scale first principles quantum mechanical calculations on biomolecules. J Chem Phys. 2011;135(22) doi: 10.1063/1.3665893. 〈https://www.ncbi.nlm.nih.gov/pubmed/22168680〉 [DOI] [PubMed] [Google Scholar]
- 37.Boneta S., Arafet K., Moliner V. QM/MM study of the enzymatic biodegradation mechanism of polyethylene terephthalate. J Chem Inf Model. 2021;61(6):3041–3051. doi: 10.1021/acs.jcim.1c00394. 〈https://www.ncbi.nlm.nih.gov/pubmed/34085821〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jerves C., Neves R.P.P., Ramos M.J., Da Silva S., Fernandes P.A. Reaction Mechanism of the PET Degrading Enzyme PETase Studied with DFT/MM Molecular Dynamics Simulations. ACS Catal. 2021;11(18):11626–11638. doi: 10.1021/acscatal.1c03700. [DOI] [Google Scholar]
- 39.Shrimpton-Phoenix E., Mitchell J.B.O., Buhl M. Computational insights into the catalytic mechanism of is-petase: an enzyme capable of degrading poly(ethylene) terephthalate. Chemistry. 2022;28(70) doi: 10.1002/chem.202201728. 〈https://www.ncbi.nlm.nih.gov/pubmed/36112344〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang X.-F., Gao P., Liu Yi-F., Li H.-F., Lu F. Predicting thermophilic proteins by machine learning. Curr Bioinforma. 2020;15(5):493–502. DOI: http://dx.doi.org/10.2174/1574893615666200207094357. [Google Scholar]
- 41.Song Z., Trozzi F., Palzkill T., Tao P. QM/MM modeling of class A β-lactamases reveals distinct acylation pathways for ampicillin and cefalexin. Org Biomol Chem. 2021;19:9182–9189. doi: 10.1039/D1OB01593A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Han Y., Wang Z., Wei Z., Liu J., Li J. Machine learning builds full-QM precision protein force fields in seconds. Brief Bioinforma. 2021;22(6) doi: 10.1093/bib/bbab158. [DOI] [PubMed] [Google Scholar]
- 43.Han X., Liu W., Huang J.W., Ma J., Zheng Y., et al. Structural insight into catalytic mechanism of PET hydrolase. Nat Commun. 2017;8(1):2106. doi: 10.1038/s41467-017-02255-z. 〈https://www.ncbi.nlm.nih.gov/pubmed/29235460〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Joo S., Cho I.J., Seo H., Son H.F., Sagong H.Y., et al. Structural insight into molecular mechanism of poly(ethylene terephthalate) degradation. Nat Commun. 2018;9(1):382. doi: 10.1038/s41467-018-02881-1. 〈https://www.ncbi.nlm.nih.gov/pubmed/29374183〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng M., Li Y., Dong W., Zhang W., Feng S., et al. Depolymerase-catalyzed polyethylene terephthalate hydrolysis: a unified mechanism revealed by quantum mechanics/molecular mechanics analysis. ACS Sustain Chem Eng. 2022;10(22):7341–7348. doi: 10.1021/acssuschemeng.2c01093. [DOI] [Google Scholar]
- 46.Yoshida S., Hiraga K., Takehana T., Taniguchi I., Hironao Y., et al. A bacterium that degrades and assimilate poly(ethylene terephthalate) Science. 2016;351(6278):1196–1199. doi: 10.1126/science.aad6359. [DOI] [PubMed] [Google Scholar]
- 47.Aboelnga M.M. and Kalyaanamoorthy S.,(2022) QM/MM Investigation to Identify the Hallmarks of Superior PET Biodegradation Activity of PETase over Cutinase.10(48): p. 15857–15868. DOI: 10.1021/acssuschemeng.2c04913 [DOI]
- 48.Crnjar A., Griñen A., Kamerlin S.C.L., Ramírez-Sarmiento César A. Conformational selection of a tryptophan side chain drives the generalized increase in activity of PET Hydrolases through a Ser/Ile Double Mutation. ACS Org Inorg Au. 2023 doi: 10.1021/acsorginorgau.2c00054. DOI: 10.1021/acsorginorgau.2c00054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Son H.F., Cho IJ S., Seo H., Sagong H.-Y., et al. Rational protein engineering of thermo-stable PETase from Ideonella sakaiensis for Highly Efficient PET Degradation. ACS Catal. 2019;9(4):3519–3526. doi: 10.1021/acscatal.9b00568. [DOI] [Google Scholar]
- 50.Son H.F., Joo S., Seo H., Sagong H.Y., Lee S.H., et al. Structural bioinformatics-based protein engineering of thermo-stable PETase from Ideonella sakaiensis. Enzym Micro Technol. 2020;141 doi: 10.1016/j.enzmictec.2020.109656. 〈https://www.ncbi.nlm.nih.gov/pubmed/33051015〉 [DOI] [PubMed] [Google Scholar]
- 51.Austin H.P., Allen M.D., Donohoe B.S., Rorrer N.A., Kearns F.L., et al. Characterization and engineering of a plastic-degrading aromatic polyesterase. Proc Natl Acad Sci. 2018;115(19):E4350–E4357. doi: 10.1073/pnas.1718804115. 〈https://www.ncbi.nlm.nih.gov/pubmed/29666242〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cui Y., Chen Y., Liu X., Dong S., Tian Y., et al. Computational Redesign of a PETase for Plastic Biodegradation under Ambient Condition by the GRAPE Strategy. ACS Catal. 2021;11(3):1340–1350. doi: 10.1021/acscatal.0c05126. [DOI] [Google Scholar]
- 53.Meng X., Yang L., Liu H., Li Q., Xu G., et al. Protein engineering of stable IsPETase for PET plastic degradation by Premuse. Int J Biol Macromol. 2021;180:667–676. doi: 10.1016/j.ijbiomac.2021.03.058. 〈https://www.ncbi.nlm.nih.gov/pubmed/33753197〉 [DOI] [PubMed] [Google Scholar]
- 54.Lu H., Diaz D.J., Czarnecki N.J., Zhu C., Kim W., et al. Machine learning-aided engineering of hydrolases for PET depolymerization. Nature. 2022;604(7907):662–667. doi: 10.1038/s41586-022-04599-z. 〈https://www.ncbi.nlm.nih.gov/pubmed/35478237〉 [DOI] [PubMed] [Google Scholar]
- 55.Sulaiman S., Yamato S., Kanaya E., Kim J.J., Koga Y., et al. Isolation of a novel cutinase homolog with polyethylene terephthalate-degrading activity from leaf-branch compost by using a metagenomic approach. Appl Environ Microbiol. 2012;78(5):1556–1562. doi: 10.1128/AEM.06725-11. 〈https://www.ncbi.nlm.nih.gov/pubmed/22194294〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tournier V., Topham C.M., Gilles A., David B., Folgoas C., et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature. 2020;580(7802):216–219. doi: 10.1038/s41586-020-2149-4. 〈https://www.ncbi.nlm.nih.gov/pubmed/32269349〉 [DOI] [PubMed] [Google Scholar]
- 57.Wei R., von Haugwitz G., Pfaff L., Mican J., Badenhorst C.P.S., et al. Mechanism-based design of efficient PET hydrolases. ACS Catal. 2022;12(6):3382–3396. doi: 10.1021/acscatal.1c05856. 〈https://www.ncbi.nlm.nih.gov/pubmed/35368328〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng M., Li Y., Dong W., Feng S., Zhang Q., et al. Computational biotransformation of polyethylene terephthalate by depolymerase: a QM/MM approach. J Hazard Mater. 2022;423(Pt A) doi: 10.1016/j.jhazmat.2021.127017. 〈https://www.ncbi.nlm.nih.gov/pubmed/34464862〉 [DOI] [PubMed] [Google Scholar]
- 59.Pfaff L., Gao J., Li Z., Jackering A., Weber G., et al. Multiple substrate binding mode-guided engineering of a thermophilic PET hydrolase. ACS Catal. 2022;12(15):9790–9800. doi: 10.1021/acscatal.2c02275. 〈https://www.ncbi.nlm.nih.gov/pubmed/35966606〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li Z., Zhao Y., Wu P., Wang H., Li Q., et al. Structural insight and engineering of a plastic degrading hydrolase Ple629. Biochem Biophys Res Commun. 2022;626:100–106. doi: 10.1016/j.bbrc.2022.07.103. 〈https://www.ncbi.nlm.nih.gov/pubmed/35981419〉 [DOI] [PubMed] [Google Scholar]
- 61.Kawabata T., Oda M., Kawai F. Mutational analysis of cutinase-like enzyme, Cut190, based on the 3D docking structure with model compounds of polyethylene terephthalate. J Biosci Bioeng. 2017;124(1):28–35. doi: 10.1016/j.jbiosc.2017.02.007. [DOI] [PubMed] [Google Scholar]
- 62.Zeng W., Li X., Yang Y., Min J., Huang J.-W., et al. Substrate-binding mode of a thermophilic PET hydrolase and engineering the enzyme to enhance the hydrolytic efficacy. ACS Catal. 2022;12(5):3033–3040. doi: 10.1021/acscatal.1c05800. [DOI] [Google Scholar]
- 63.Khan A.Z., Bilal M., Rasheed T., Iqbal H.M. Advancements in biocatalysis: from computational to metabolic engineering. Chin J Catal. 2018;39(12):1861–1868. doi: 10.1016/s1872-2067(18)63144-4. [DOI] [Google Scholar]
- 64.Song Z., Zhou H., Tian H., Wang X., Tao P. Unraveling the energetic significance of chemical events in enzyme catalysis via machine-learning based regression approach. Commun Chem. 2020;3(1):134. doi: 10.1038/s42004-020-00379-w. 〈https://www.ncbi.nlm.nih.gov/pubmed/36703376〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Song Z., Trozzi F., Tian H., Yin C., Tao P. Mechanistic insights into enzyme catalysis from explaining machine-learned quantum mechanical and molecular mechanical minimum energy pathways. ACS Phys Chem Au. 2022;2(4):316–330. doi: 10.1021/acsphyschemau.2c00005. 〈https://www.ncbi.nlm.nih.gov/pubmed/35936506〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shilling P.J., Mirzadeh K., Cumming A.J., Widesheim M., Kock Z., et al. Improved designs for pET expression plasmids increase protein production yield in Escherichia coli. Commun Biol. 2020;3(1):214. doi: 10.1038/s42003-020-0939-8. 〈https://www.ncbi.nlm.nih.gov/pubmed/32382055〉 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ihling N., Uhde A., Scholz R., Schwarz C., Schmitt L., et al. Scale-up of a Type I secretion system in E. coli using a defined mineral medium. Biotechnol Prog. 2020;36(2) doi: 10.1002/btpr.2911. 〈https://www.ncbi.nlm.nih.gov/pubmed/31513739〉 [DOI] [PubMed] [Google Scholar]