Abstract
The propanediol utilization (pdu) operon of Salmonella enterica serovar Typhimurium LT2 contains genes needed for the coenzyme B12-dependent catabolism of 1,2-propanediol. Here the completed DNA sequence of the pdu operon is presented. Analyses of previously unpublished pdu DNA sequence substantiated previous studies indicating that the pdu operon was acquired by horizontal gene transfer and allowed the identification of 16 hypothetical genes. This brings the total number of genes in the pdu operon to 21 and the total number of genes at the pdu locus to 23. Of these, six encode proteins of unknown function and are not closely related to sequences of known function found in GenBank. Two encode proteins involved in transport and regulation. Six probably encode enzymes needed for the pathway of 1,2-propanediol degradation. Two encode proteins related to those used for the reactivation of adenosylcobalamin (AdoCbl)-dependent diol dehydratase. Five encode proteins related to those involved in the formation of polyhedral organelles known as carboxysomes, and two encode proteins that appear distantly related to those involved in carboxysome formation. In addition, it is shown that S. enterica forms polyhedral bodies that are involved in the degradation of 1,2-propanediol. Polyhedra are formed during either aerobic or anaerobic growth on propanediol, but not during growth on other carbon sources. Genetic tests demonstrate that genes of the pdu operon are required for polyhedral body formation, and immunoelectron microscopy shows that AdoCbl-dependent diol dehydratase is associated with these polyhedra. This is the first evidence for a B12-dependent enzyme associated with a polyhedral body. It is proposed that the polyhedra consist of AdoCbl-dependent diol dehydratase (and perhaps other proteins) encased within a protein shell that is related to the shell of carboxysomes. The specific function of these unusual polyhedral bodies was not determined, but some possibilities are discussed.
Salmonella enterica serovar Typhimurium LT2 degrades 1,2-propanediol by a pathway that requires coenzyme B12, adenosylcobalamin (AdoCbl) (29). Several lines of evidence indicate the importance of this process to the Salmonella lifestyle. 1,2-Propanediol is produced by the fermentation of the common plant sugars rhamnose and fucose (31, 34). Fucose is also found in the glycoconjugates of intestinal cells, where it is involved in host-parasite interactions (12). In vivo expression technology has indicated that 1,2-propanediol utilization (pdu) genes may be important for growth in host tissues, and competitive index studies with mice have shown that pdu mutations confer a virulence defect (14, 25). The pdu genes are contiguous and coregulated with the cobalamin (cob) (B12) biosynthetic genes, indicating that propanediol catabolism is the primary reason for de novo B12 synthesis in S. enterica (2, 8, 37, 41). If one includes the cob genes, S. enterica maintains 40 to 50 genes primarily for the transformation of propanediol. Moreover, nearly all natural isolates of Salmonella tested synthesized B12 de novo and degraded propanediol (30). Some of these aspects of Salmonella biology have been reviewed recently (40).
The pathway of 1,2-propanediol degradation has been investigated (34, 54). It initiates with the conversion of 1,2-propanediol to propionaldehyde by an AdoCbl-dependent diol dehydratase (1). Subsequently, propionaldehyde is catabolized to propionic acid and propanol, presumably by coenzyme A (CoA)-dependent aldehyde dehydrogenase, phosphotransacylase, propionate kinase, and alcohol dehydrogenase. This pathway provides a source of ATP, an electron sink, and carbon compounds that can be diverted to central metabolism via known pathways (27, 56). In addition, S. enterica can carry out the anaerobic respiration of 1,2-propanediol with tetrathionate as terminal electron acceptor (9). However, 1,2-propanediol respiration is not supported by the more common anaerobic electron acceptors, nitrate, fumarate, trimethylamine-N-oxide (TMAO), or dimethyl sulfoxide (DMSO) (9).
The genes required for 1,2-propanediol degradation cluster at the pdu locus on centisome 44 of the S. enterica chromosome (29). This locus includes the pocR and pduF genes, as well as the genes of the adjacent and divergently transcribed pdu operon (10, 13). The pocR and pduF genes encode a positive transcriptional regulatory protein and a 1,2-propanediol diffusion facilitator, respectively (10, 13, 37). The pdu operon is estimated to include about 20 genes (9). Those identified thus far are pduABCDEGHJ (10, 13, 58). The pduA and pduB genes encode a close and a distant relative of carboxysome shell proteins (13). The pduCDE genes encode an AdoCbl-dependent diol dehydratase, and the pduG gene encodes a putative cobalamin adenosyltransferase (10, 58). The pduHJ genes were identified by genetic tests, but neither their DNA sequence nor their specific function is known (58). The DNA sequences of the pocR, pduF, and pduABCDE genes and a portion of the pduG genes were determined, and analyses of these sequences indicated that the pdu locus was acquired by a horizontal gene transfer (10, 13, 41). The regulation of the pdu operon has also been investigated. It is coinduced with the adjacent cob operon in response to 1,2-propanediol, and its induction is influenced by cyclic AMP levels, the redox state of the cell, iron, magnesium, pH, and perhaps the growth phase (2, 8, 25, 37–39). In addition, recent electron microscopy (EM) studies have shown that S. enterica forms polyhedral bodies similar in size and appearance to carboxysomes during anaerobic growth on 1,2-propanediol (46). However, neither the composition of these polyhedra nor their role in 1,2-propanediol degradation has been investigated.
Here the completed DNA sequence of the pdu operon and its analysis are presented. In addition, results establish that polyhedral bodies are involved in AdoCbl-dependent 1,2-propanediol degradation. The first evidence for the association of a coenzyme B12-dependent enzyme with a polyhedral organelle is presented, and we propose that these organelles consist of AdoCbl-dependent diol dehydratase (and perhaps other proteins) encased within a protein shell that is related to the shell of carboxysomes. The specific function of these unusual organelles is not determined, but some possibilities are discussed.
MATERIALS AND METHODS
Chemicals and reagents.
Fumaric acid, tetrathionate, vitamin B12, MgSO4, CaCl2 · 2H2O, Na2SeO4, MnSO4 · H2O, FeSO4 · 7H2O, and sodium cacodylate were from Sigma Chemical Company, St. Louis, Mo. Formaldehyde, (r,s) 1,2-propanediol, pyruvic acid, Na2MoO4 · 2H2O, and CoCl2 were from Fisher Scientific, Pittsburgh, Pa. Glutaraldehyde was from Tousimis, Rockville, Md. Uranyl acetate was from E.M. Sciences, Ft. Washington, Pa. Osmium tetroxide and LR White resin were from Ted Pella, Inc., Redding, Calif. Yeast extract and Luria-Bertani (LB) broth were from Difco Laboratories, Detroit, Mich. Powdered milk was from Nestle, Glendale, Calif.
Bacterial strains, media, and growth conditions.
The bacterial strains used in this study are derivatives of S. enterica serovar Typhimurium LT2, formerly S. typhimurium LT2 (Table 1). The minimal medium used was NCE (6, 57) supplemented with 0.01% yeast extract and DB minerals, which consist of 4.5 μM CaCl2, 2 μM Na2MoO4, 2 μM Na2SeO4, 2 μM MnSO4, 2 μM FeSO4, and 5 μM CoCl2. LB medium was the rich medium used (32). 1,2-Propanediol was used at 82 mM, tetrathionate was used at 10 mM, pyruvate was used at 40 mM, fumarate was used at 20 mM, and vitamin B12 (CN-Cbl) was used at 0.15 μM. Cultures were grown for about 24 h at 37°C in 5 ml of minimal medium supplemented with the appropriate growth substrates and inoculated with 0.1 ml of rich medium culture that had been grown about 16 h at 37°C.
TABLE 1.
Genotypes of the strains used in this study
| Straina | Genotype |
|---|---|
| RT818 | pdu-8::mudA |
| RT822 | pdu-12::mudA |
| BE22 | cobD24::mudJ |
| BE25 | pdu-12::mudJ |
| BE26 | hut+ galE542 muHP1 (muCts62 hP1-1)/pEG5005 |
| BE27 | pdu-12::mudA/pEM55 |
| BE28 | pdu-12::mudA/pTA417 |
| BE43 | cbiA-700::Camr |
| TT18117 | DEL1077 (metE) ara-9 DEL1715 [(cobD24)*mudJ*(zea-3666)] |
Strains are derivatives of S. enterica serovar Typhimurium LT2 (formerly S. typhimurium LT2).
Cloning and DNA sequencing.
Two pdu clones, EM55 and TA417, were used as a template for completing the DNA sequence of the pdu operon. These were obtained by screening for clones that complemented a pdu mutation as follows. An S. enterica gene library was prepared by using strain BE26 (vector pEG5005) and the in vivo cloning method of Groisman and Casadaban (24). A transducing lysate was prepared from this library by using P22HT105/1int-201 and cells that were grown at 30°C on LB medium supplemented with 0.2% glucose and 0.02% galactose (18, 43). Plasmid clones were transferred to S. enterica RT822 (pdu-12::mudA) by transduction with all incubations carried out at 30°C. Kanamycin resistance was selected and transductants were screened for their Pdu phenotype on MacConkey–1,2-propanediol–B12 indicator medium (29). A red color indicated complementation of the pdu-12::mudA mutation by the plasmid clone. The procedure was performed twice. About 5,000 plasmid-containing transductants were screened, and two complementing clones were identified, pTA417 and pEM55.
Plasmid DNA was purified from strains containing plasmids pTA417 and pEM55 by using Qiagen tip 100 columns (Qiagen, Inc., Chatsworth, Calif.). Purified DNA was used as template for obtaining new pdu DNA sequence by primer walking. DNA sequencing was carried out by the University of Florida Interdisciplinary Center for Biotechnology Research DNA Sequencing Core Facility by using Applied Biosystems, Inc., automated sequencing equipment (Perkin-Elmer, Norwalk, Conn.).
DNA sequence analysis.
Several DNA sequence analysis programs were employed, and, except where noted, default parameters were used. Genes were identified by using Genemark software with the S. typhimurium species option selected (11). BlastP and Ψ-Blast softwares were used to search the nonredundant (nr) database of the National Center for Biotechnology Information (NCBI) for protein sequences related to those encoded by the pdu genes (4, 5). ProDom was used for the identification of homologous protein domains (15). ClustalW and Blast2 were used for sequence alignments (4, 50). Phylogenetic trees were constructed by using the PHYLIP package of Felsenstein (21). Codon usage bias and G+C composition were analyzed as previously described (45). Edited multiple sequence alignments were obtained as follows: whenever a gap character was present in one or more sequences, that region of the alignment was deleted. In addition, overhangs at the ends of alignments were deleted. This editing procedure removes nonhomologous protein regions and generally improves phylogenetic comparisons (19).
EM.
Two fixation protocols were used. In the standard protocol, cells were fixed in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2) for 30 min at room temperature and then in 1% osmium tetroxide in the same buffer for 1 h at 4°C. The samples were then dehydrated through a graded ethanol series followed by absolute acetone and embedded in Spurr’s low-viscosity resin. The modified protocol differed only in that samples were held at room temperature overnight in 75% ethanol containing 1% uranyl acetate during the alcohol dehydration series; this modification imparted extra contrast to the polyhedral bodies.
Specimens were thin sectioned on an LKB Nova or an RMC MT-6000-XL ultramicrotome, collected on Formvar-coated copper grids, post-stained with lead citrate, and observed and photographed with a Zeiss EM-10CA transmission electron microscope.
For immunogold localization of diol dehydratase, cells were fixed in 0.5% glutaraldehyde–4% formaldehyde on ice for 20 min (3). They were then dehydrated in a graded series of ethanol to absolute ethanol. Samples were embedded in LR White resin and polymerized at 50°C for 5 days (3). Thin sections were placed on Formvar-coated nickel grids, blocked for 20 min on 1% powdered milk in phosphate-buffered saline (PBS) at pH 7.2, and floated overnight at 4°C on rabbit polyclonal antibody to diol dehydratase from Klebsiella oxytoca diluted 1:1,000 with PBS. After washing on high-salt Tris-Tween buffer (3), grids were floated for 1 h on goat anti-rabbit antibody conjugated with 12-nm-particle-diameter colloidal gold (Jackson ImmunoResearch Laboratories, Inc., West Grove, Pa.). Samples were washed with buffer and deionized water, and then sections were poststained with 0.5% uranyl acetate and lead citrate.
Nucleotide sequence accession number.
The sequence reported here has been assigned GenBank accession no. af026270.
RESULTS
Cloning and sequencing of pdu DNA.
Plasmids pTA417 and pEM55 were needed to complete the DNA sequence of the pdu operon, (see Materials and Methods). Plasmid pTA417 was used to determine the pdu DNA sequence through bp 18,781, and plasmid pEM55 was used to complete the DNA sequence of the pdu operon (bp 19,215). DNA sequence was determined for both strands, and all ambiguities were resolved by additional sequencing reactions with different primers. A total of 11,624 bp of previously unreported pdu DNA sequence was determined.
Identification of pdu genes.
Analysis of previously unreported pdu DNA sequence with GeneMark software allowed the identification of 16 hypothetical genes, pduGHJKLMNOPQSTUVWX. The pduG sequence completed a previously reported partial open reading frame (ORF), ORF1 (10). In addition, the codon adaptation indices and the G+C contents for each codon position of the proposed pdu genes were consistent with those of expressed sequences (Table 2).
TABLE 2.
Codon usage bias and G+C content of the S. enterica pduABCDEGHJKLMNOPQSTUVWX coding regions
| ORF | CAIa | % G+C contentb | % G+C content by positionc
|
||
|---|---|---|---|---|---|
| 1st | 2nd | 3rd | |||
| pduA | 0.297 | 53 | 67 | 41 | 52 |
| pduB | 0.131 | 60 | 63 | 48 | 70 |
| pduC | 0.463 | 57 | 61 | 41 | 69 |
| pduD | 0.372 | 57 | 63 | 42 | 66 |
| pduE | 0.432 | 56 | 63 | 43 | 63 |
| pduG | 0.332 | 60 | 71 | 44 | 64 |
| pduH | 0.280 | 58 | 68 | 42 | 63 |
| pduJ | 0.421 | 56 | 69 | 44 | 57 |
| pduK | 0.309 | 56 | 64 | 48 | 58 |
| pduL | 0.290 | 60 | 70 | 42 | 69 |
| pduM | 0.367 | 62 | 73 | 41 | 71 |
| pduN | 0.213 | 62 | 68 | 50 | 68 |
| pduO | 0.344 | 62 | 66 | 49 | 69 |
| pduP | 0.386 | 59 | 59 | 46 | 71 |
| pduQ | 0.349 | 60 | 63 | 45 | 73 |
| pduS | 0.316 | 62 | 70 | 46 | 69 |
| pduT | 0.302 | 59 | 61 | 43 | 72 |
| pduU | 0.316 | 56 | 57 | 44 | 67 |
| pduV | 0.318 | 58 | 57 | 46 | 71 |
| pduW | 0.276 | 53 | 60 | 41 | 58 |
| pduX | 0.264 | 56 | 60 | 45 | 63 |
| cob operon | 56 | 63 | 44 | 61 | |
| Typical gene of S. enterica | 0.2–0.8 | 54 | 58 | 44 | 58 |
The codon adaptation index (CAI) was determined as described previously (45).
Percent G+C content of the indicated coding sequence.
Percent G+C content of the first, second, or third codon position of the indicated coding sequence.
The S. enterica pdu locus on centisome 44 now includes 23 genes, pocR, pduF, and pduABCDEGHJKLMNOPQSTUVWX. We propose that the pdu operon ends at bp 19,215 and that the terminal gene of the operon is pduX. Seventy-eight base pairs downstream of the pduX gene is the end of a long ORF transcribed in the opposite direction. It is related to an Escherichia coli gene of unknown function, yeeX. However, experimental evidence that the pduX gene is the terminal gene of the pdu operon has not yet been obtained.
With one exception, the pdu genes previously identified by genetic tests, but of unknown DNA sequence, can be correlated to the pdu genes identified here. The pduG genes identified here and previously were shown to be the same by complementation analysis (7). The previously identified pduH gene represented a region in the pdu operon identified by deletion endpoints (58), and hence can correspond to the pduH gene identified here. However, the pduJ gene previously identified was not correlated with the pduJ gene identified here, and this possible lack of correspondence will need to be addressed in the future.
Analyses of previously unpublished pdu DNA sequence supported previous work indicating that S. enterica acquired the pdu locus and the adjacent cob operon by a single horizontal gene transfer (10, 30, 41). The G+C content of the 16 putative coding sequences identified here averaged about 59%, which is significantly higher than the typical 54% average for native S. enterica coding sequences (Table 2). In addition, the G+C contents of the first, second, and third codon positions varied from that of native S. enterica coding sequences. On the other hand, the G+C composition of pdu genes is similar to that of the adjacent cob genes, for which there is considerable evidence of horizontal gene transfer (41).
Sequences similar to those of the PduGHJKLMNOPQSTUVWX proteins.
BlastP software and Ψ-Blast software were used to search the nr database of the NCBI for protein sequences related to those encoded by the pduGHJKLMNOPQSTUVWX genes. Table 3 summarizes the results of those analyses and also includes information on previously sequenced pdu genes (10, 13, 41).
TABLE 3.
Pdu proteins and proteins of related amino acid sequences
| Pdu protein
|
Related protein sequencesa
|
||
|---|---|---|---|
| Name | Function (reference or references) | Name(s) | Function (reference or references) |
| PocR | Transcriptional regulator (8, 37) | AraC | Transcriptional activator protein (8, 37) |
| PduF | Propanediol diffusion facilitator (13) | GlpF | Glycerol facilitator (13) |
| PduA | Polyhedral bodies (13) | See Table 4 | Carboxysomes, CO2 concentration (36, 48) |
| PduB | Polyhedral bodies (13) | CsoS1A (distantly related, 22% identical over 85 amino acids) | Carboxysomes, CO2 concentration (36, 48) |
| PduC | B12-dependent diol dehydratase large subunit (10) | PddA, DhaB, GldA | B12-dependent glycerol and diol dehydratases large subunit (16, 44, 51, 52) |
| PduD | B12-dependent diol dehydratase medium subunit (10) | PddB, DhaC, GldB | B12-dependent glycerol and diol dehydratases medium subunit (16, 44, 51, 52) |
| PduE | B12-dependent diol dehydratase small subunit (10) | PddC, DhaE, GldC | B12-dependent glycerol and diol dehydratases small subunit (16, 44, 51, 52) |
| PduG | Diol dehydratase reactivation (10) | DdrA, ORFZ | Diol dehydratase reactivation factor large subunit, cobalamin adenosyltransferase (33, 55) |
| PduH | Diol dehydratase reactivation (this study) | DdrB | Diol dehydratase reactivating factor small subunit (33, 55) |
| PduJ | Polyhedral bodies (this study) | See Table 4 | Carboxysomes, CO2 concentration (36, 48) |
| PduK | Polyhedral bodies (this study) | See Table 4 | Carboxysomes, CO2 concentration (36, 48) |
| PduL | Unknown (this study) | CpcE (distantly related, 25% identical over 93 amino acids) | Phycocyanobilin lyase, required for chromophorylation of CpcA (20) |
| PduM | Unknown (this study) | None | None |
| PduN | Polyhedral bodies (this study) | CchB, CcmL | Carboxysomes, CO2 concentration (36, 48) |
| PduO | B12 related (this study) | ORFW, ORFY | Hypothetical genes grouped with genes for B12-dependent glycerol dehydratase (16, 44) |
| PduP | CoA-dependent propionaldehyde dehydrogenase (this study) | EutE, Adh | CoA-dependent aldehyde dehydrogenase (49) |
| PduQ | Propanol dehydrogenase (this study) | EutG, AdhE | Alcohol dehydrogenase (23, 49) |
| PduS | Unknown (this study) | RnfC (distantly related, 28% identical [90 of 318 amino acids]) | Membrane-associated oxidoreductases (42) |
| PduT | Polyhedral bodies (this study) | See Table 4 | Carboxysomes, CO2 concentration (36, 48) |
| PduU | Polyhedral bodies (this study) | EutS | Carboxysomes, CO2 concentration (36, 48) |
| PduV | Unknown (this study) | EutP | None |
| PduW | Propionate kinase (this study) | Ack | Acetate kinase |
| PduX | Unknown (this study) | None | None |
Except where sequences are stated to be distantly related, all sequences were ≥35% identical and had BlastP Expect values of ≤7 × 10−6.
1,2-Propanediol utilization pathway genes.
The hypothetical PduP protein identified here is related to a number of CoA-dependent aldehyde dehydrogenases. It is most closely related to the EutE protein of S. enterica. These two proteins are 45% identical in sequence over 465 amino acids. The EutE protein is a putative CoA-dependent aldehyde dehydrogenase proposed to function in the AdoCbl-dependent pathway of ethanolamine degradation (49). Accordingly, it seems likely that the PduP protein is a CoA-dependent aldehyde dehydrogenase used in the pdu pathway for the conversion of propionaldehyde to propionyl-CoA.
The sequence of the PduQ protein was found to be related to those of many alcohol dehydrogenases. PduQ is most closely related to the AdhE enzyme of E. coli (23); these proteins are 35% identical in sequence over 330 amino acids. The PduQ protein aligns with the carboxy-terminal portion of the bifunctional AdhE enzyme, which has both alcohol dehydrogenase and aldehyde dehydrogenase activity. In addition, the PduQ protein is closely related to a number of monofunctional alcohol dehydrogenases, including the hypothetical alcohol dehydrogenase involved in ethanolamine degradation in S. enterica, the EutG protein (49). Thus, we propose that the PduQ protein functions as a propanol dehydrogenase in the pathway of 1,2-propanediol degradation.
The PduW protein was found to be related to acetate kinases. It is 87% identical in sequence, over 400 amino acids, to the ack gene product of S. enterica serovar Typhimurium LT2. Thus, the pduW protein is likely a propionate kinase whose role in 1,2-propanediol degradation is the conversion of propionyl-phosphate to propionate.
Genes for the reactivation of AdoCbl-dependent diol dehydratase.
The PduGH proteins appear to be involved in the reactivation of diol dehydratase, and the PduG protein may also be involved in the adenosylation of B12. The PduG protein is 92% identical in sequence, over 611 amino acids, to the DdrA protein K. oxytoca, and the PduH protein is 87% identical in sequence (99 of 124 amino acids) to the DdrB protein of K. oxytoca. The DdrAB proteins are involved in the reactivation of AdoCbl-dependent diol dehydratase (33, 55). Inactivation of diol dehydratase occurs due to the breakdown of AdoCbl to an inactive form that has an upper ligand other than an adenosyl group (26, 53). The DdrAB proteins are proposed to reactivate diol dehydratase by removing the inactive cofactor and replacing it with AdoCbl (33, 55). The PduGH proteins may have a similar function. In addition, the PduG protein has been proposed to be an adenosyltransferase on the basis of genetic tests (58), and it has high sequence similarity to a proposed cobalamin adenosyltransferase from Citrobacter freundii (17, 44). Hence, the PduG protein may be bifunctional.
Pdu proteins of unknown function.
The PduO protein may have resulted from the fusion of two genes. The sequence of the N-terminal 170 amino acids of this protein is 35% identical to that of ORFW of C. freundii and is also similar to those of unknown ORFs from Clostridium, Pyrococcus, Bacillus, and Mycobacterium. The sequence of the C-terminal 121 amino acids of PduO is 37% identical to that of ORFY from C. freundii and also has similarity to ORFs from Clostridium, Klebsiella, and Pseudomonas. ORFW and ORFY are arranged with genes involved in the AdoCbl-dependent degradation of glycerol by C. freundii (17, 44). Thus, the PduO protein appears to have a function that is common to AdoCbl-dependent degradation of 1,2-propanediol and glycerol.
The PduS protein was found to be 30% identical in sequence over 72% of its length to the glucose repression mediator protein of E. coli. However, the sequence of this E. coli protein is unpublished. It was submitted to GenBank by the E. coli genome sequencing project being conducted in Japan, and it is unclear how this function was assigned. The PduS protein was also found to be distantly related to a number of membrane oxidoreductases, and of these, it was most closely related to the RnfC protein of Rhodobacter capsulatus (42). The PduS and RnfC proteins are 28% identical (90 of 318 amino acids).
The PduL protein is distantly related to phycocyanobilin lyase (CpcE). Ψ-Blast reiteration 1 gave an Expect value of 2 × 10−24. Phycocyanobilin lyase catalyzes the covalent attachment of phycocyanobilin (a linear tetrapyrrole) to phycocyanin via a thioether linkage (20). Since cobalamins are tetrapyrroles, perhaps the PduL protein includes a pyrrole binding site.
The PduV protein was compared to the Prosite database and was found to have a domain closely related to the ATP and GTP binding motifs. Analyses of the other Pdu proteins of unknown function did not allow the identification of motifs similar to those of the Prosite dictionary.
Pdu proteins related to carboxysome proteins.
Protein sequence similarity analyses showed that five Pdu proteins (AJKNT) are clearly related to those involved in the formation of polyhedral organelles known as carboxysomes and that two Pdu proteins (BU) are tentatively related to carboxysome proteins.
The PduAJKT proteins are clearly related to a group of proteins that includes the major shell proteins of carboxysomes (Table 4). The percent identities shown in Table 4 were determined from an edited multiple sequence alignment (see Materials and Methods). The edited alignment consisted of sequences 85 amino acids in length that aligned to the following PduA amino acids: ALGMVETKGLTAAIEAADAMVKSANVMLVGYEKIGSGLVTVIVRGDVGAVKAATDAGAAAARNVKAVHVIPRPHTDVEKILPKEL.
TABLE 4.
Amino acid sequence identities between the PduAJKT proteins and homologous regions of carboxysome shell proteins and their relatives
| Protein | Gi | Organism | % Identity to indicated Pdu proteina
|
|||
|---|---|---|---|---|---|---|
| PduA | PduJ | PduK | PduT | |||
| PduA | 5069450 | S. enterica LT2 | ||||
| PduJ | 5069453 | S. enterica LT2 | 82 | |||
| PduK | 5069454 | S. enterica LT2 | 35 | 36 | ||
| PduT | 5069462 | S. enterica LT2 | 30 | 31 | 24 | |
| EutM | 3885916 | S. enterica LT2 | 64 | 63 | 29 | 30 |
| CchA | 1788799 | E. coli | 64 | 62 | 29 | 30 |
| CsoS1C | 4105524 | T. intermedius | 56 | 55 | 27 | 27 |
| CsoS1A | 4105525 | T. intermedius | 56 | 56 | 27 | 27 |
| CsoS1B | 4105526 | T. intermedius | 56 | 54 | 28 | 28 |
| CsoS1C | 3449372 | T. neapolitanus | 58 | 58 | 27 | 29 |
| CsoS1A | 3449373 | T. neapolitanus | 57 | 58 | 27 | 29 |
| CsoS1B | 3449374 | T. neapolitanus | 58 | 58 | 27 | 28 |
| CsoS1C | 3282390 | T. denitrificans | 56 | 56 | 27 | 28 |
| CsoS1A | 3282391 | T. denitrificans | 56 | 56 | 27 | 29 |
| CsoS1B | 3282392 | T. denitrificans | 57 | 56 | 27 | 24 |
| CcmK | 3182944 | Synechococcus sp. strain WH7803 | 56 | 56 | 30 | 24 |
| b2438 | 1788779 | E. coli | 42 | 42 | 30 | 29 |
| YffI | 3183436 | E. coli | 42 | 42 | 30 | 29 |
| EutK | 3885926 | S. enterica LT2 | 40 | 42 | 29 | 29 |
| CcmK | 541311 | Synechococcus sp. strain PCC7942 | 57 | 55 | 27 | 24 |
| CcmK | 3182943 | Synechococcus sp. strain PCC7002 | 55 | 54 | 25 | 23 |
| Cck1 | 2493552 | Synechocystis sp. strain PCC6803 | 55 | 51 | 27 | 21 |
| Cck2 | 2493553 | Synechocystis sp. strain PCC6803 | 56 | 54 | 27 | 22 |
| Cck3 | 2493554 | Synechocystis sp. strain PCC6803 | 43 | 45 | 25 | 17 |
| Cck4 | 2493555 | Synechocystis sp. strain PCC6803 | 35 | 35 | 21 | 21 |
| Yrb2 | 141357 | Synechococcus sp. strain PCC6301 | 51 | 50 | 27 | 25 |
| Yrb2 | 1176828 | Synechococcus sp. strain PCC7942 | 51 | 50 | 27 | 25 |
| Yrb1 | 141354 | Synechococcus sp. strain PCC6301 | 43 | 42 | 28 | 23 |
| Y436 | 3183228 | Synechocystis sp. strain PCC6803 | 47 | 44 | 30 | 30 |
Amino acid identities were determined from the edited multiple sequence alignment described in the text.
The PduN protein is closely related to the CcmL-CchB family of proteins, which are needed for the proper assembly and function of carboxysomes (36, 48). A phylogenetic tree constructed from an edited multiple sequence alignment indicated that the PduN protein is more closely related to homologous proteins from Synechococcus, Synechocystis, and E. coli than to homologous proteins from Thiobacillus. The percent identities between the PduN amino acid sequence and sequences derived from other CcmL-CchB family members ranged from 28 to 53%. The tree was constructed from 1,000 bootstrap replicates, and the high bootstrap values (626 to 999) indicated a statistically significant branching order. The edited multiple sequence alignment (see Materials and Methods) used for tree construction consisted of a group of sequences 77 amino acids in length that aligned to the following PduN amino acid sequence: MHLARVTGAVVSTQKSPSLIGKKLLLVRGDEV AVDSVGAGVGELVLLSGGSSARHVFSGPNEAIDLAVV GIVDTLSC.
The PduB and PduU proteins were found to be distantly related to proteins involved in polyhedral body formation. The PduB protein was shown to be 29% identical in sequence (53 of 178 amino acids) to the EutL protein of S. enterica and to the Eut b2439 protein of E. coli by BlastP analysis. The PduB protein is also distantly related to the carboxysome shell proteins shown in Table 4. When a Ψ-Blast search was conducted starting with Yrb2 protein (accession no. P46205), the 20 most closely related proteins identified were carboxysome shell proteins listed in Table 4. Reiteration with these 20 shell proteins indicated that they are related to the PduB protein, with an Expect value of 2 × 10−4.
The PduU protein was determined to be 57% identical to the EutS protein of S. enterica and 56% identical to the Eut b2462 protein of E. coli by BlastP analysis. The EutS protein is a putative carboxysome structural protein distantly related to the carboxysome proteins listed in Table 4.
Formation of polyhedral bodies by S. enterica.
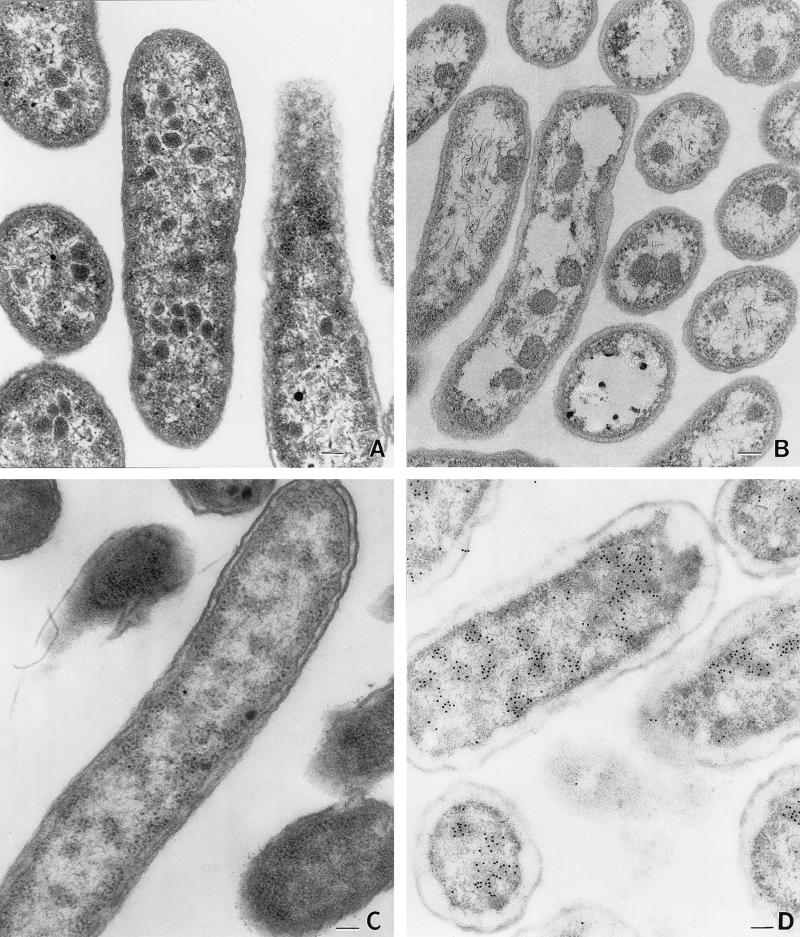
S. enterica formed polyhedral structures similar in size and appearance to carboxysomes during aerobic growth on minimal 1,2-propanediol–B12 medium (Fig. 1A), but not during aerobic growth on glucose (Fig. 1C). Polyhedra were 100 to 200 nm in cross-section and appeared to consist of a proteinaceous shell and interior. The bodies formed by S. enterica are somewhat irregular in shape compared to the carboxysomes of Thiobacillus neapolitanus, suggesting some differences between these structures (Fig. 1A and B).
FIG. 1.
Electron micrographs of S. enterica grown aerobically on minimal, 1,2-propanediol, vitamin B12 medium (A), Thiobacillus neapolitanus grown autotrophically (B), S. enterica grown aerobically on minimal glucose medium (C), and immunogold localization of the S. enterica AdoCbl-dependent diol dehydratase (D). Bars, 100 nm.
Similar polyhedra were also formed during anaerobic growth on 1,2-propanediol–tetrathionate, but not during anaerobic growth on glucose or pyruvate-tetrathionate, indicating that polyhedra are specifically involved in the catabolism of 1,2-propanediol. It was expected that polyhedra would be formed during anaerobic growth on glycerol-tetrathionate. Although S. enterica does not degrade glycerol in a B12-dependent manner, glycerol induces expression of the pdu operon to high levels (8, 37). However, polyhedra were observed in very few cells grown anaerobically on glycerol-tetrathionate. This suggests that 1,2-propanediol plays a role in formation of polyhedra in addition to induction of the pdu operon. Tetrathionate was used as a terminal electron acceptor for these experiments, because other terminal electron acceptors, such as fumarate, nitrate, DMSO, and TMAO, do not support anaerobic growth on 1,2-propanediol. Vitamin B12 supplementation did not affect polyhedral body formation either aerobically or anaerobically.
Role of pdu genes in polyhedral body formation.
EM studies showed that pdu mutants either failed to produce polyhedral bodies or produced aberrantly shaped structures. During either aerobic or anaerobic growth in the presence of 1,2-propanediol, strain BE25 (pdu-12::mudJ) failed to produce polyhedra, whereas an otherwise isogenic strain formed these structures under similar growth conditions. These results show that genes of the pdu operon are necessary for polyhedral body formation, and strongly indicate that polyhedra have a role in the catabolism of 1,2-propanediol. The pdu-12::mudJ mutation is a polar mutation that was previously shown to be located in either the pduD or pduE genes, both of which encode subunits of diol dehydratase (58).
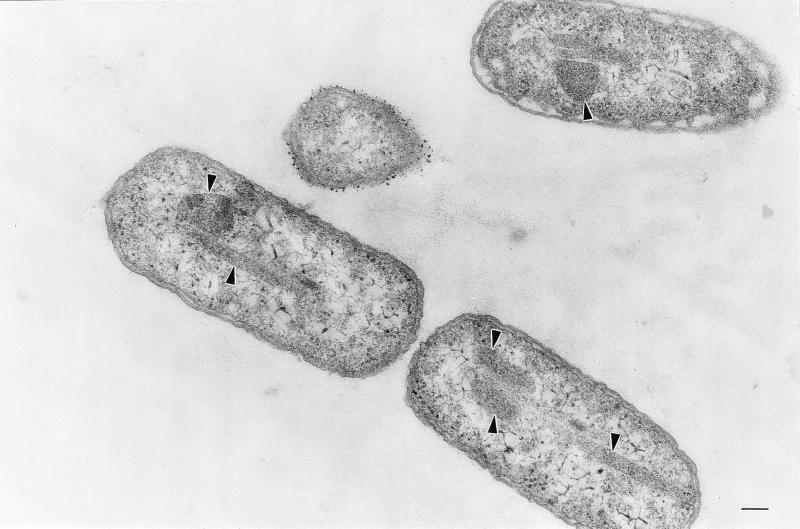
An additional pdu mutant (strain RT818, pdu-8::mudA) was found to produce bodies that appeared to have a proteinaceous shell and interior, but were aberrantly shaped (Fig. 2). Many were highly elongated and spanned the entire length of the cell. This shows that genes on either side of the pdu-8::mudA insertion are required for polyhedral body formation. Thus, multiple pdu genes are required for the formation of polyhedra.
FIG. 2.
Electron micrograph of the aberrant polyhedra (arrowheads) formed by S. enterica pdu mutant RT818 (pdu-8::mudJ). Bar, 100 nm.
Neither the catabolism of propanediol nor genes of the cob operon are needed for polyhedral body formation.
Strains unable to synthesize AdoCbl de novo (Cob−) produced polyhedral bodies in the presence of 1,2-propanediol both aerobically and anaerobically regardless of vitamin B12 supplementation. This showed that B12-dependent catabolism of 1,2-propanediol is not required for formation of polyhedra. Results also showed that the genes of the cob operon (which are coregulated with those of the pdu operon) are not needed for polyhedral body formation. Both strains BE43 and TT18117 formed polyhedra. BE43 contains a polar mutation in the first gene of the cob operon that prevents expression of the first 17 of 20 cob genes, and TT18117 contains a deletion of the 17 terminal genes of the cob operon. In the above experiments, pyruvate or succinate was used as a carbon and energy source because expression of the pdu operon is relatively high during growth on these compounds (2, 8).
Association of B12-dependent diol dehydratase with polyhedral bodies.
Immuno-EM indicated that the AdoCbl-dependent diol dehydratase of S. enterica is associated with polyhedral bodies (Fig. 1D). In the micrograph, antibody-conjugated gold particles (solid black circles) indicate the location of diol dehydratase. Clustering of the gold particles in the interior of the polyhedral bodies shows that the AdoCbl-dependent diol dehydratase is associated with the polyhedra and is consistent with the encasement of this enzyme within a protein shell. Because of differences in the fixation procedures, polyhedra are somewhat less distinct in the immuno-EM pictures than in the standard thin sections (Fig. 1). Immunogold labeling, similar to that described above, was also carried out with pdu mutants unable to express diol dehydratase. Little to no labeling occurred; the very small amount of labeling observed probably resulted from nonspecific antibody binding.
Visualization of polyhedral bodies by EM.
The polyhedral structures were easiest to observe in older cells that had begun to lyse. Early-log-phase cells contained the structures, but they were obscured by the cytoplasmic contents of the cell. The modified EM fixation procedure with an overnight stain of uranyl acetate in alcohol enhanced the visibility of the structures. The cytoplasm of the fixed cells in the modified fixation process was less dense, and it was easier to visualize the sharp edges of the more darkly stained polyhedral bodies.
DISCUSSION
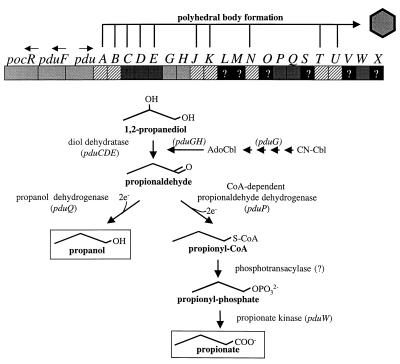
To better understand the physiology and molecular biology of AdoCbl-dependent processes, 1,2-propanediol degradation by S. enterica serovar Typhimurium LT2 was investigated. The DNA sequence of the pdu operon was completed and analyzed, and evidence was presented that polyhedral organelles are involved in AdoCbl-dependent 1,2 propanediol degradation. Analyses of previously unpublished pdu DNA sequence substantiated previous studies indicating that the pdu and cob operons were acquired by a single horizontal gene transfer event (30) and allowed the identification of 16 hypothetical genes. In all, 23 pdu genes are proposed, and these genes fall into six classes: pathway, diol dehydratase reactivation, unknown, polyhedral body formation, transport, and regulation (Fig. 3 and Table 3).
FIG. 3.
The pdu locus of S. enterica with a summary of the encoded functions. Genes thought to be involved in the formation of polyhedra are indicated above the genetic map. Genes thought to be involved in the pathway of 1,2-propanediol degradation and in the conversion of vitamin B12 (CN-Cbl) to coenzyme B12 (AdoCbl) are indicated below the genetic map. Arrows above pocR, pduF, and pdu indicate the direction of transcription.
With one exception, pdu genes corresponding to each enzyme of the proposed 1,2-propanediol degradative pathway have now been identified (Fig. 3). A pdu gene corresponding to a proposed phosphotransacylase was not identified. This enzyme might be encoded by one of the six pdu genes of unknown function, or perhaps the proposed CoA-dependent aldehyde dehydrogenase (PduP) is bifunctional or a kinase; a Blast-ProDom search showed that the PduP protein shares a domain with ProA proteins, enzymes that catalyze the reduction of glutamate-5-semialdehyde to gamma-glutamyl-5-phosphate.
Six hypothetical Pdu proteins of unknown function (PduLMOSVX) were also identified. Genetic tests have shown that the pdu operon encodes functions for the conversion of vitamin B12 (CN-Cbl) to AdoCbl, the active cofactor of diol dehydratase. The PduG protein has been implicated in this process, but additional Pdu proteins might also be involved. Studies with several different systems have indicated that a decyanase, one or two cobalt reductases, and an adenosyltransferase are needed (22, 28). The PduL protein is a possibility, since related proteins are found in an operon that encodes an AdoCbl-dependent glycerol dehydratase (17, 44). Other possible functions for Pdu proteins are suggested by physiological studies. S. enterica grows via the anaerobic respiration of 1,2-propanediol with tetrathionate as a terminal electron acceptor (9). Hence, some Pdu proteins might be specific to the 1,2-propanediol–tetrathionate respiration. The PduS protein is a possibility, since it is related in amino acid sequence to several membrane-bound oxidoreductases. In addition, previous studies have indicated that the pdu operon encodes a protein involved in regulation of the prpBCDE operon (56). Hence, one of the Pdu proteins of unknown function may fulfill this regulatory role.
The polyhedral bodies formed by S. enterica during growth on 1,2-propanediol were also investigated. Based on results reported here and previous results, we propose that S. enterica forms polyhedral organelles involved in 1,2-propanediol degradation that consist of AdoCbl-dependent diol dehydratase (and perhaps other proteins) encased within a protein shell related to the shell of carboxysomes. During growth on 1,2-propanediol, S. enterica forms polyhedra that are proteinaceous in nature and that have sharp edges indicative of a shell (this study and reference 46). Immunogold labeling indicated that diol dehydratase is associated with the polyhedra and that it is localized to the interior of these structures. DNA sequence analyses showed that the pdu operon encodes five to seven proteins that are related to those involved in the formation of carboxysomes (this study and reference 13), and genetic tests showed that genes of the pdu operon are required for polyhedral body formation. In addition, a pdu mutant was found that produced aberrantly shaped polyhedra. Thus, some pdu genes are required for proper organelle shape, while others apparently encode its basic structural components. Analogous Synechococcus mutants (that produce aberrantly shaped carboxysomes) were previously identified (35).
Although the polyhedral bodies involved in 1,2-propanediol degradation are apparently related to carboxysomes structurally, a functional relationship is uncertain. Carboxysomes are proposed to play a role in concentrating CO2 for RuBisCo, since mutations in shell genes result in strains that require high CO2 for autotrophic growth (36, 47, 48). On the other hand, the polyhedra of S. enterica function in AdoCbl-dependent catabolism of 1,2-propanediol, and this process has no known association with CO2. Previous reports, which identified the carboxysome shell protein gene homologues in the eut and pdu operons of S. enterica, discussed some possible functions for polyhedral bodies in the AdoCbl-dependent catabolism of ethanolamine and 1,2-propanediol (13, 40, 49). It was suggested that polyhedral bodies could be used to sequester toxic aldehydes formed both during 1,2-propanediol and ethanolamine degradation and channel them to subsequent pathway enzymes. It was also suggested that polyhedra might be used to protect diol dehydratase and ethanolamine ammonia-lyase from oxygen, a molecule to which both are sensitive (13). The finding reported here, that AdoCbl-dependent diol dehydratase is associated with polyhedra, is consistent with both of these hypotheses.
Although their precise function is unknown, the size of the polyhedral organelles and the number of genes involved attest to the substantial resources devoted to AdoCbl-dependent 1,2-propanediol degradation. Formation of these bodies must play an important role in S. enterica survival and niche establishment among the competitive flora of natural environments.
ACKNOWLEDGMENTS
This work was supported by grant GM59486 from the National Institutes of Health, by the Florida Agricultural Experiment Station, and by a Research Project Enhancement Award from the Experiment Station.
We thank Tetsuo Toraya for providing the antibody used in the immunogold labeling studies.
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-07046.
REFERENCES
- 1.Abeles R H, Lee H A., Jr An intramolecular oxidation-reduction requiring a cobamide coenzyme. J Biol Chem. 1961;236:2347–2350. [PubMed] [Google Scholar]
- 2.Ailion M, Bobik T A, Roth J R. Two global regulatory systems (Crp and Arc) control the cobalamin/propanediol regulon of Salmonella typhimurium. J Bacteriol. 1993;175:7200–7208. doi: 10.1128/jb.175.22.7200-7208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aldrich H C, McDowell L, Barbosa M de F S, Yomano L P, Scopes R K, Ingram L O. Immunocytochemical localization of glycolytic and fermentative enzymes in Zymomonas mobilis. J Bacteriol. 1992;174:4504–4508. doi: 10.1128/jb.174.13.4504-4508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. A basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 5.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkowitz D, Hushon J M, Whitfield H J, Jr, Roth J, Ames B N. Procedure for identifying nonsense mutations. J Bacteriol. 1968;96:215–220. doi: 10.1128/jb.96.1.215-220.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bobik, T. A. Unpublished results.
- 8.Bobik T A, Ailion M, Roth J R. A single regulatory gene integrates control of vitamin B12 synthesis and propanediol degradation. J Bacteriol. 1992;174:2253–2266. doi: 10.1128/jb.174.7.2253-2266.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bobik, T. A., and J. R. Roth. Unpublished results.
- 10.Bobik T A, Xu Y, Jeter R M, Otto K E, Roth J R. Propanediol utilization genes (pdu) of Salmonella typhimurium: three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. doi: 10.1128/jb.179.21.6633-6639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Borodovsky M, McIninch J D. GeneMark: parallel gene recognition for both DNA strands. Comp Chem. 1993;17:123–133. [Google Scholar]
- 12.Bry L, Falk P G, Midtvedt T, Gordon J I. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273:1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 13.Chen P, Andersson D, Roth J R. The control region of the pdu/cob regulon in Salmonella typhimurium. J Bacteriol. 1994;176:5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conner C P, Heithoff D M, Julio S M, Sinsheimer R L, Mahan M J. Differential patterns of acquired virulence genes distinguish Salmonella strains. Proc Natl Acad Sci USA. 1998;95:4641–4645. doi: 10.1073/pnas.95.8.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Corpet F, Gouzy J, Kahn D. Recent improvements of the ProDom database of protein domain families. Nucleic Acids Res. 1999;27:263–267. doi: 10.1093/nar/27.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daniel R, Bobik T A, Gottschalk G. Biochemistry of coenzyme B12-dependent glycerol and diol dehydratases and organization of the encoding genes. FEMS Microbiol Rev. 1999;22:553–566. doi: 10.1111/j.1574-6976.1998.tb00387.x. [DOI] [PubMed] [Google Scholar]
- 17.Daniel R, Gottschalk G. Growth temperature-dependent activity of glycerol dehydratase in Escherichia coli expressing the Citrobacter freundii dha regulon. FEMS Microbiol Lett. 1995;100:281–286. doi: 10.1111/j.1574-6968.1992.tb14053.x. [DOI] [PubMed] [Google Scholar]
- 18.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 19.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fairchild C D, Zhao J, Zhou J, Colson S E, Bryant D A, Glazer A N. Phycocyanin alpha-subunit phycocyanobilin lyase. Proc Natl Acad Sci USA. 1992;89:7017–7021. doi: 10.1073/pnas.89.15.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Felsenstein J. revision date. [Online.] PHYLIP (Phylogeny Inference Package) version 3.5c. Seattle: Department of Genetics, University of Washington; 21 January 1999. http://evolution.genetics.washington.edu/phylip.html . [22 August 1999, last date accessed.] [Google Scholar]
- 22.Friedmann H C. Biosynthesis of corrinoids. In: Babior B M, editor. Cobalamin. New York, N.Y: John Wiley and Sons; 1975. pp. 75–103. [Google Scholar]
- 23.Goodlove P E, Cunningham P R, Parker J, Clark D P. Cloning and sequence analysis of the fermentative alcohol-dehydrogenase-encoding gene of Escherichia coli. Gene. 1989;85:209–214. doi: 10.1016/0378-1119(89)90483-6. [DOI] [PubMed] [Google Scholar]
- 24.Groisman E A, Casadaban M J. Mini-Mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusions. J Bacteriol. 1986;168:357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Heithoff D M, Conner C P, Hentschel U, Govantes F, Hanna P C, Mahan M J. Coordinate intracellular expression of Salmonella genes induced during infection. J Bacteriol. 1999;181:799–807. doi: 10.1128/jb.181.3.799-807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda S, Toraya T, Fukui S. In situ reactivation of glycerol-inactivated coenzyme B12-dependent enzymes, glycerol dehydratase and diol dehydratase. J Bacteriol. 1980;143:1458–1465. doi: 10.1128/jb.143.3.1458-1465.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horswill R A, Escalante-Semerena J C. Propionate catabolism in Salmonella typhimurium LT2: two divergently transcribed units comprise the prp locus at 8.5 centisomes, prpR encodes a member of the sigma-54 family of activators, and the prpBCDE genes constitute an operon. J Bacteriol. 1997;179:928–940. doi: 10.1128/jb.179.3.928-940.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huennekens F M, Viktols K S, Fujii K, Jacobsen D W. Biosynthesis of the cobalamin coenzymes. In: Dolphin D, editor. B12. New York, N.Y: John Wiley and Sons; 1982. pp. 145–168. [Google Scholar]
- 29.Jeter R M. Cobalamin-dependent 1,2-propanediol utilization by Salmonella typhimurium. J Gen Microbiol. 1990;136:887–896. doi: 10.1099/00221287-136-5-887. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence J G, Roth J R. Evolution of coenzyme B12 synthesis among enteric bacteria: evidence for loss and reacquisition of a multigene complex. Genetics. 1996;142:11–24. doi: 10.1093/genetics/142.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 1. Washington, D.C.: American Society for Microbiology; 1987. pp. 244–284. [Google Scholar]
- 32.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 33.Mori K, Tobimatsu T, Hara T, Toraya T. Characterization, sequencing, and expression of the genes encoding a reactivating factor for glycerol-inactivated adenosylcobalamin-dependent diol dehydratase. J Biol Chem. 1997;272:32034–32041. doi: 10.1074/jbc.272.51.32034. [DOI] [PubMed] [Google Scholar]
- 34.Obradors N, Badía J, Baldomà L, Aguilar J. Anaerobic metabolism of the l-rhamnose fermentation product 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1988;170:2159–2162. doi: 10.1128/jb.170.5.2159-2162.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Price G D, Gadger M R. Isolation and characterization of high CO2-requiring-mutants of the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1989;91:514–525. doi: 10.1104/pp.91.2.514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Price G D, Sultemeyer D, Klughammer B, Ludwig M, Badger M R. The functioning of the CO2 concentrating mechanism in several cyanobacterial strains: a review of general physiological characteristics, genes, proteins, and recent advances. Can J Bot. 1998;76:973–1002. [Google Scholar]
- 37.Rondon M R, Escalante-Semerena J. The poc locus is required for 1,2-propanediol-dependent transcription of the cobalamin biosynthetic (cob) and propanediol utilization (pdu) genes of Salmonella typhimurium. J Bacteriol. 1992;174:2267–2272. doi: 10.1128/jb.174.7.2267-2272.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rondon M R, Escalante-Semerena J C. Integration host factor is required for 1,2-propanediol-dependent transcription of the cob/pdu regulon in Salmonella typhimurium LT2. J Bacteriol. 1997;179:3797–3800. doi: 10.1128/jb.179.11.3797-3800.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rondon M R, Kazmierczak R, Escalante-Semerena J C. Glutathione is required for maximal transcription of the cobalamin biosynthetic and 1,2-propanediol utilization (cob/pdu) regulon and for the catabolism of ethanolamine, 1,2-propanediol, and propionate in Salmonella typhimurium LT2. J Bacteriol. 1995;177:5434–5439. doi: 10.1128/jb.177.19.5434-5439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roth J R, Bobik T A, Lawrence J G. Cobalamin (coenzyme B12): synthesis and biological significance. Annu Rev Microbiol. 1996;50:137–181. doi: 10.1146/annurev.micro.50.1.137. [DOI] [PubMed] [Google Scholar]
- 41.Roth J R, Lawrence J G, Rubenfield M, Kieffer-Higgins S, Church G M. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J Bacteriol. 1993;175:3303–3316. doi: 10.1128/jb.175.11.3303-3316.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmehl M, Jahn A, Meyer zu Vilsendorf A, Hennecke S, Masepohl B, Schuppler M, Marxer M, Oelze J, Klipp W. Identification of a new class of nitrogen fixation genes in Rhodobacter capsulatus: a putative membrane complex involved in electron transport to nitrogenase. Mol Gen Genet. 1993;241:602–615. doi: 10.1007/BF00279903. [DOI] [PubMed] [Google Scholar]
- 43.Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110:378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- 44.Seyfried M, Daniel R, Gottschalk G. Cloning, sequencing, and overexpression of the genes encoding coenzyme B12-dependent glycerol dehydratase of Citrobacter freundii. J Bacteriol. 1996;178:5793–5796. doi: 10.1128/jb.178.19.5793-5796.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharp P M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 46.Shively J M, Bradburne C E, Aldrich H C, Bobik T A, Mehlman J L, Jin S, Baker S H. Sequence homologs of the carboxysomal polypeptide CsoS1 of the thiobacilli are present in cyanobacteria and enteric bacteria that form carboxysomes-polyhedral bodies. Can J Bot. 1998;76:906–916. [Google Scholar]
- 47.Shively J M, English R S. The carboxysome, a prokaryotic organelle: a mini-review. Can J Bot. 1991;69:957–962. [Google Scholar]
- 48.Shively J M, van Keulen G, Meijer W G. Something from almost nothing: carbon dioxide fixation in chemoautotrophs. Annu Rev Microbiol. 1998;52:191–230. doi: 10.1146/annurev.micro.52.1.191. [DOI] [PubMed] [Google Scholar]
- 49.Stojiljkovic I, Bäumler A J, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. J Bacteriol. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thompson J D, Higgins D G, Gibson T J. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobimatsu T, Azuma M, Matsubara H, Takatori H, Niida T, Nishimoto K, Satoh H, Hayashi R, Toraya T. Cloning, sequencing, and high level expression of the genes encoding adenosylcobalamin-dependent glycerol dehydrase of Klebsiella pneumoniae. J Biol Chem. 1996;271:22352–22357. doi: 10.1074/jbc.271.37.22352. [DOI] [PubMed] [Google Scholar]
- 52.Tobimatsu T, Hara T, Sakaguchi M, Kishimoto Y, Wada Y, Isoda M, Sakai T, Toraya T. Molecular cloning, sequencing, and expression of the genes encoding adenosylcobalamin-dependent diol dehydrase of Klebsiella oxytoca. J Biol Chem. 1995;270:7142–7148. doi: 10.1074/jbc.270.13.7142. [DOI] [PubMed] [Google Scholar]
- 53.Toraya T, Fukui S. Diol dehydratase. In: Dolphin D, editor. B12. New York, N.Y: John Wiley & Sons; 1982. pp. 234–259. [Google Scholar]
- 54.Toraya T, Honda S, Fukui S. Fermentation of 1,2-propanediol and 1,2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J Bacteriol. 1979;139:39–47. doi: 10.1128/jb.139.1.39-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Toraya T, Mori K. A reactivating factor for coenzyme B12-dependent diol dehydratase. J Biol Chem. 1999;274:3372–3377. doi: 10.1074/jbc.274.6.3372. [DOI] [PubMed] [Google Scholar]
- 56.Tsang A W, Horswill A R, Escalante-Semerena J C. Studies of regulation of expression of the propionate (prpBCDE) operon provide insights into how Salmonella typhimurium LT2 integrates its 1,2-propanediol and propionate catabolic pathways. J Bacteriol. 1998;180:6511–6518. doi: 10.1128/jb.180.24.6511-6518.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogel H J, Bonner D M. Acetylornithase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- 58.Walter D, Ailion M, Roth J. Genetic characterization of the pdu operon: use of 1,2-propanediol in Salmonella typhimurium. J Bacteriol. 1997;179:1013–1022. doi: 10.1128/jb.179.4.1013-1022.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]