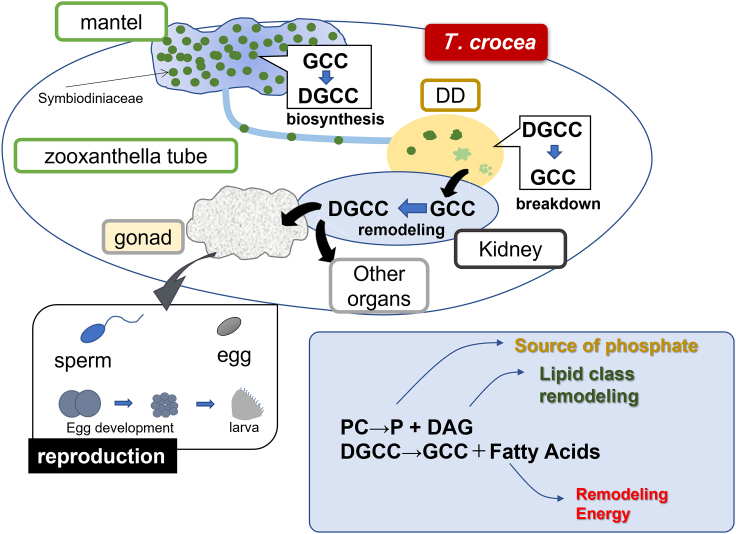
Summary
The giant clam Tridacna crocea thrives in poorly nourished coral reef water by forming a holobiont with zooxanthellae and utilizing photosynthetic products of the symbiont. However, detailed metabolic crosstalk between clams and symbionts is elusive. Here, we discovered that the nonphosphorous microalgal betaine lipid DGCC (diacylglycerylcarboxy-hydroxymethylcholine) and its deacylated derivative GCC are present in all tissues and organs, including algae-free sperm and eggs, and are metabolized. Colocalization of DGCC and PC (phosphatidylcholine) evidenced by MS imaging suggested that DGCC functions as a PC substitute. The high content of GCC in digestive diverticula (DD) suggests that the algal DGCC was digested in DD for further utilization. Lipidomics analysis showing the organ-specific distribution pattern of DGCC species suggests active utilization of DGCC as membrane lipids in the clam. Thus, the utilization of zooxanthellal DGCC in animal cells is a unique evolutionary outcome in phosphorous-deficient coral reef waters.
Subject areas: Marine organism, Biological sciences, Biochemistry
Graphical abstract
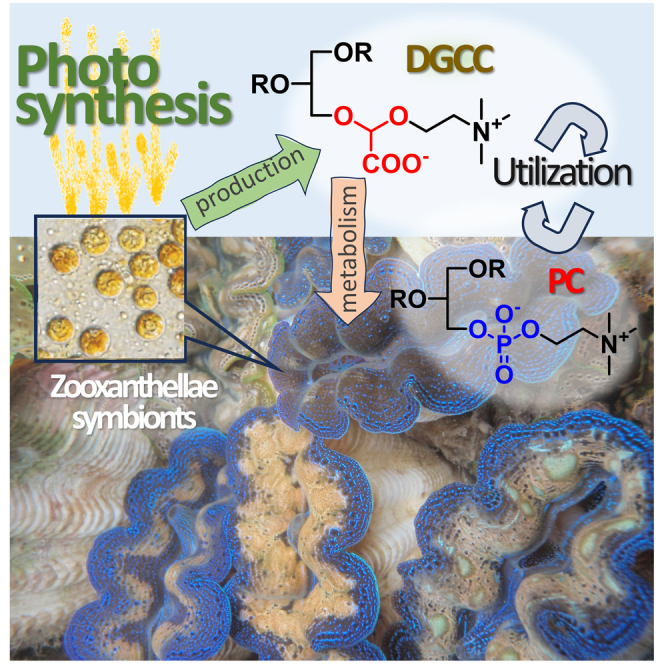
Highlights
-
•
Giant clams rely on symbiosis with zooxanthellae dinoflagellates for nutrients
-
•
Giant clams contain DGCC, a nonphosphorus membrane lipid of zooxanthellae origin
-
•
Giant clams metabolize and utilize DGCC in all tissues and organs
Marine organism; Biological sciences; Biochemistry
Introduction
The boring giant clam Tridacna crocea is a tridacnid bivalve mollusk and is found in shallow coral reefs. Giant clams are known to engage in symbiosis with the photosynthetic dinoflagellate family Symbiodiniaceae, also known as “zooxanthellae”.1,2 The symbiodiniacean cells in giant clams are harbored within a specialized tubular system, “zooxanthella tubes”, which extend from the stomach of the clam and branch out in their mantle region.3
This symbiotic relationship between giant clams and algal cells has previously been observed from an energy flow viewpoint. For example, algal symbionts within the T. crocea mantle release glucose, and 46%–80% of fixed carbon is translocated from the symbionts to the host tissues.4 Although these values can vary among studies and clam species, symbiont algae generally contribute more than half of the carbon requirements of giant clams.1,5 This implies that giant clams are largely dependent on the symbiont algae for survival, and the symbiotic algae are protected from harmful UV radiation by the host clam.6 Recently, we reported the isolation of ten different natural sunscreen compounds, mycosporines, from the mantle tissue of a giant clam.7 Mass spectrometry and UV imaging studies have indicated that the distribution of mycosporines within mantle tissues differs among compounds. This is thought to be related to their UV absorbing function and biosynthetic stage, suggesting that mycosporines can first be biosynthesized by clams or symbiotic algae and then translocated to the area where they can function best with appropriate structural modification.7,8
These findings revealed close and complex relationships between the host animal and photosynthetic microalgae, whereby the functional metabolites in the symbiotic system are not just “utilized” as nutrients but are fine-tuned to optimize their functions to thrive in shallow tropical waters. This “smart utilization” of limited metabolites may have positively contributed to the coevolution of coral reef invertebrates and their symbiont algae. Symbiotic relationships between invertebrates and microalgae are widely observed in coral reef environments.9,10 Each case of symbiosis is thought to have independently evolved, reflecting their ecological and physiological characteristics. Therefore, many interesting examples of the “smart utilization” of metabolites are expected in each symbiotic relationship.
Although numerous studies on the symbiotic relationship between invertebrate hosts and dinoflagellates have been conducted, these studies have mainly focused on the chemical aspects of carbon and nitrogen flow.11,12 Limited studies have investigated the metabolic relationship in the symbiotic consortium by identifying individual compounds, including betaine lipids diacylglycerylcarboxy-hydroxymethylcholine (DGCC), present in the host and symbiont.13 Interestingly, the lipid component in DGCC is tightly related to the breaching history of corals, suggesting some fundamental functions of DGCC in coral symbionts.14,15 This study, however, did not suggest a metabolic relationship of DGCC between the symbionts and the hosts. Therefore, further insights into the unique metabolic exchange between the host and symbiont are essential to explain the paradoxical high production in coral reefs. Structural identification of key metabolites by advanced liquid chromatography‒mass spectrometry (LC‒MS) in combination with the determination of loci by MS imaging may provide a great deal of information regarding the flow and functions of metabolites that govern symbiotic relationships between algae and invertebrates.7
In the present study, we focused on the lipid component in the giant clam-Symbiodiniaceae system, as lipids can be useful marker molecules for revealing the smart utilization of metabolites, from a metabolic flow perspective, between the host and symbiont. Here, we show the first experimental evidence of the utilization of algal lipids in giant clams. This finding may facilitate further insight into the coevolution of coral reef organisms on a molecular level.
Results
Anatomical analysis of T. crocea and distribution of Symbiodiniaceae cells
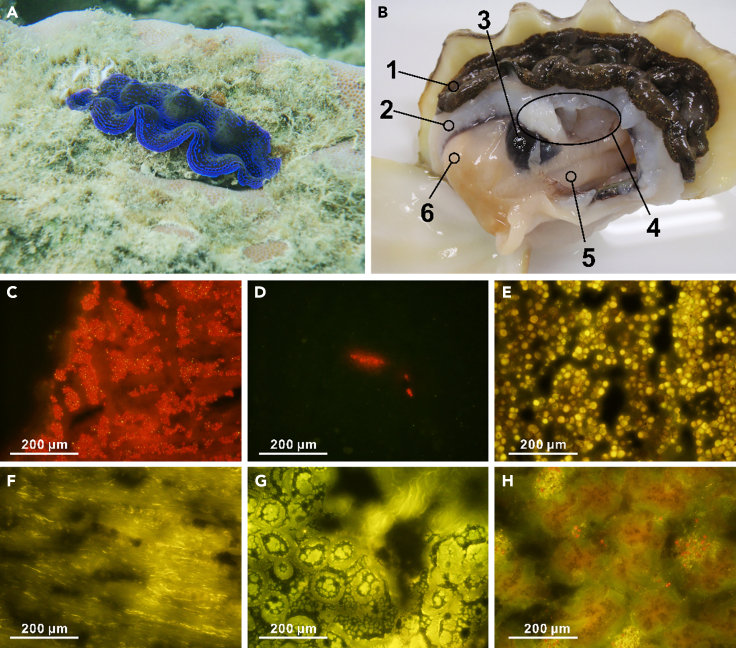
An anatomical overview of T. crocea is shown in Figure 1. The outermost mantle tissue (epidermal layer; EL) of the clam densely harbors Symbiodiniaceae cells. Conversely, the pale white mantle tissue (inner layer; IL) contains algal cells at a lower density than that observed in the EL. The kidney is a large and dark-colored organ. The adductor muscle and posterior byssal/pedal retractor muscles are located in the middle of the shell. Ctenidia (gill) can be recognized as a comb-like structure. The digestive diverticula (DD) are surrounded by off-white gonads. To avoid cross-contamination as much as possible, each of these parts was carefully separated (Figure S1). However, DD are embedded in the gonads; thus, a small fraction of the components from interconnected organs was inevitable. To confirm whether algal cells were present, we observed algal chlorophyll fluorescence in both the EL and IL of the mantle, muscle, gonad, DD, and kidney in thin sections (Figures 1C–1H). The algal cells shown as red fluorescent dots under blue excitation light were most densely localized in the EL (Figure 1C), followed by the IL (Figure 1D). Although the DD was completely bound by gonads,16 the DD and gonads could be easily distinguished by color (Figure S1). Algal cells (including digested cells) were also found in the DD region (Figure 1H). Although the gonad region was occupied by eggs and sperm (Figure 1G), small brown patches were observed in rare instances and often contained a few algal cells (Figure S2). No algal cells were observed in the muscle (Figure 1F). Although a characteristic granular structure, the nephrolith,17 was observed in the kidneys (Figure 1E), no algal population was found in this organ. Unfortunately, ctenidia could not be seen in the observed section.
Figure 1.
Anatomical analysis of T. crocea and distribution of Symbiodiniaceae cells
(A) Underwater photograph of T. crocea found at – 1 m in Ishigaki, Okinawa.
(B) Dissected specimen (shell length 80.0 mm). 1; mantle (outer epidermal layer: EL), 2; mantle (inner tissue layer: IL), 3; kidney, 4; muscle (adductor muscle and posterior byssal/pedal retractor muscles), 5; ctenidia (gill), 6; gonads and digestive diverticula (DD; inside).
(C–H) Fluorescence micrographs of visceral parts of T. crocea under blue (460–490 nm) excitation light. (C) Mantle EL, (D) Mantle IL, (E) kidney, (F) muscle, (G) gonad, (H) digestive diverticula. The red dots in fluorescence micrographs (C, D, and H) indicate symbiont algal chlorophyll autofluorescence.
Lipidomic analysis of T. crocea organs
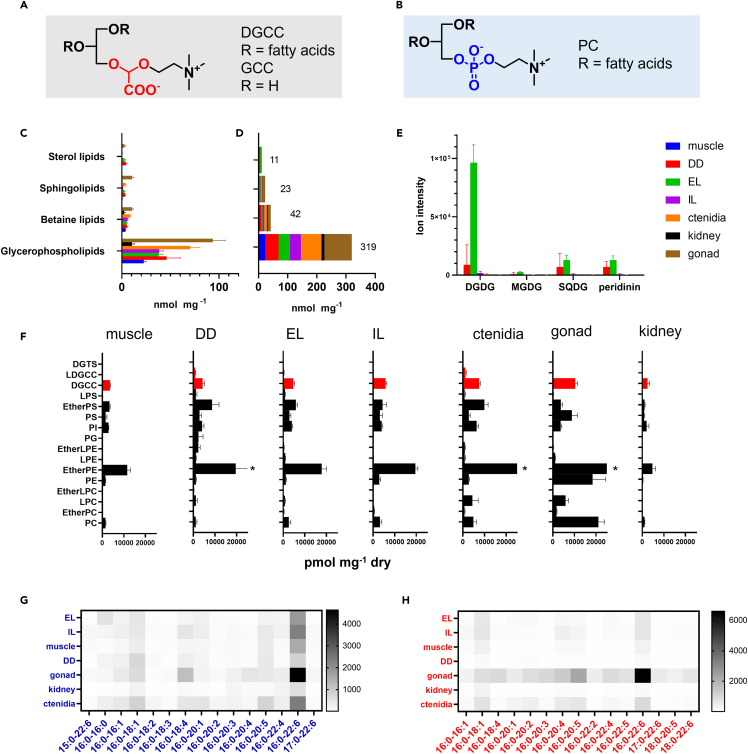
We first conducted LC‒MS-based lipidomic analyses of extracts from each of the seven organs, including mantle (EL and IL), adductor muscle, kidney, DD, gonads, and ctenidia, to yield a total of 566 lipids (Table S1). These could be divided into six categories: glycerophospholipids (12 classes, 167 species), betaine lipids (3 classes, 76 species), glycerolipids (9 classes, 209 species), sterol lipids (2 classes, 32 species), sphingolipids (3 classes, 60 species), and free fatty acids (22 species) (Table S1). Semiquantitative data of the representative lipid classes are summarized in Figure 2. In all organs, glycerophospholipids were predominant among the membrane-associated lipids (Figure 2C). Betaine lipids were the second most abundant class and were distributed in all the tissues analyzed, including those that were free of symbiotic algae. The total amounts of glycerophospholipids, betaine lipids, sphingolipids, and sterol lipids were 319, 42, 23, and 11 nmol mg−1 dry tissue, respectively (Figure 2D). To confirm the presence of algal cells in each giant clam body part, we further analyzed algal lipids, namely, peridinin and galactosyl lipids, using LC‒MS. As expected, peridinin, a dinoflagellate-borne carotenoid, was found at a substantial ion intensity in EL and DD, followed by IL (Figure 2E, Table S2). Only trace ions for peridinin were found in other body parts (Table S2). Moreover, we found three galactosyl lipids: digalactosyldiacylglycerol (DGDG), sulfoquinovosyl diacylglycerols (SQDG), and monogalactosyldiacylglycerol (MGDG) (Tables S1 and S2). Among them, DGDG was the most abundant class, followed by SQDG; MGDG was detected only at low intensities (Figure 2E, Table S2). Other tissues exhibited negligible peak intensities (Table S2). Subsequently, we semiquantitatively compared the amounts of phospholipids and betaine lipids at the subclass level in each organ (Figure 2F). The lipidomic profile of each body part differed substantially (Figure 2F). Glycerol phosphatidyl ethanolamines (PE) with ether lipids (ether-PE) were predominant among the glycerophospholipid subclasses in all tissues; ctenidia and gonads were rich in this subclass (Figure 2F). The membrane lipid profiles among muscle, IL, and EL were comparable. Gonads contained considerable amounts of PC and PE in addition to ether-PE. The storage lipid triacylglycerol (TG) was especially prevalent in the gonads (Figure S3). The kidney was the leanest organ with the lowest phospholipid diversity. Among the three known subclasses of betaine lipids, DGCC was predominant. Only trace amounts of diacylglyceryltrimethylhomoserine (DGTS)/diacylglycerylhydroxymethylalanine (DGTA) were found in all body parts (10–100 pmol mg−1 dry tissue) except for the kidney, where less than 10 pmol mg−1 was detected. A trace amount of lyso-DGCCs was detected in the ctenidia, DD, EL, gonad, and IL (Table S1).
Figure 2.
Lipidomic analysis of T. crocea organs
Structures of (A) DGCC and (B) PC.
(C) Semiquantitative data for four representative classes of membrane-associated lipids in each organ.
(D) Sum of each lipid class. Number on the histogram is the total amount in nmol. Error bars were omitted for clarity.
(E) Ion intensity of galactosyl lipids and peridinin in each organ. The color label in the upper light panel is applicable to all graphs in (C–E).
(F) Semiquantitative analysis of the membrane lipids in T. crocea. The total amount of each class of lipids in pmol mg−1 of dry tissue is shown. Glycerol phosphatidyl lipids, black bars; and betaine lipids, red bars at fixed scale. Some data for ether-PE are clipped∗ at the given scale (Figure S3). Comparisons of the 15 most abundant fatty acid species of DGCC (G) and PC (H) in each organ of T. crocea. All data here were collected from 5 specimens independently (Tables S3 and S4). In histograms, the average ±SD is indicated.
See also Figure S13.
We next compared the fatty acid composition of DGCC and phosphatidylcholine (PC) because both lipids contain choline as a polar group, and thus, they are structurally compatible (Figures 2A and 2B). We detected 18 PC and 69 DGCC species, of which the most abundant 15 species were compared (Figures 2G and 2H; Tables S3 and S4), and found that 16:0-22:6 was the major fatty acid in both PC and DGCC. Both classes comprised mostly 16:0-containing species (e.g., 16:0-22:5, 16:0-18:1, 16:0-20:5, 16:0-16:1, 16:0-20:1, and 16:0-22:4). Some odd numbered species 17:0-22:6 in both DGCC and PC and 15:0-22:6 in PC were found in T. crocea (Figures 2G and 2H). Additionally, we analyzed two Symbiodiniaceae culture strains isolated from giant clams (TsIS-H4 and TsIS-G10, Figure S4) and found that, as in the case of T. crocea, 16:0-22:6 DGCC was the most abundant species in the tested strains, as reported previously.14,18,19 However, the overall composition of minor species differed greatly from that of T. crocea (Figure S4). The profile for PC had the same tendency, where clams had a more diverse profile than the alga.
To yield further chemical evidences, we actually separated DGCC16:0-22:6 and characterized it by spectroscopic and spectrometric methods. The 1H NMR data showing a characteristic acetal proton at δ4.78 (Figure S5 and Table S5), and mass spectral data with a diagnostic fragment ions at m/z 104 and 132 in electrospray ionization mass spectrometry (ESI-MS) (Figure S6) agreed well with those reported previously from Pavlova lutheri.20 We finally confirmed the relative amount of DGCC in each organ by high-performance liquid chromatography (HPLC) analysis using charge aerosol detector that allows the detection of molecule independent of ionization efficiency and found that the data obtained by LC‒MS were supported by this detention method (Figure S7).
Identification of glycerylcarboxy-hydroxymethylcholine in T. crocea and cultured Symbiodiniaceae cells
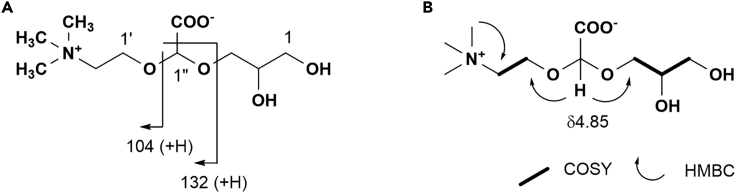
When the aqueous extract of T. crocea was analyzed using LC‒MS, we found a novel ion at m/z 252. This ion was absent from the organic extract, suggesting that this compound is a polar water-soluble substance. The molecular formula C10H21NO6 suggested that the molecule responsible for this ion was the deacylated derivative of DGCC. To obtain pure glycerylcarboxy-hydroxymethylcholine (GCC), we searched for the same ion in cultured Symbiodiniaceae strains; the compound was separated using gel filtration chromatography followed by HPLC. As the proposed structure for GCC has not been previously reported as a free molecule, the planar structure of GCC was thus determined based on spectral data analyses (Figures S8–S10, Table S5). The molecular formula for GCC, C10H21NO6 was established by the high-resolution ESI-MS molecular ion at m/z 252.1442 (M + H)+, Δ 2 ppm. 1H NMR data (Figure S6, Table S5) indicated a presence of trimethyl amino group, and a characteristic singlet appeared at δ4.85. In the 13C NMR data, only 6 resonances (Figure S7) accounting for 8 carbons appeared, while resonances for C-1″ and carboxylate were missing. Two-dimensional NMR data, however, assigned most 1H and 13C resonances as shown in Table S5. The heteronuclear single quantum coherence spectrum was particularly useful to detect acetal carbon 1″ which was missing in 13C NMR data. Thus, these data constructed the planar structure of GCC mostly, except for a carboxylate group. ESI-MS/MS data showing diagnostic ions for DGCC at m/z 104 and 132 (Scheme 1, Figures S8 and S9), however, supported the proposed structure with the carboxylate group at C-1″. Thus, the planar structure of GCC was assigned. LC‒MS/MS analysis of GCC from Symbiodiniaceae culture strains confirmed the identity between the isolate and that from the clam (Figure S11).
Scheme 1.
Identification of glycerylcarboxy-hydroxymethylcholine (GCC)
(A) Diagnostic fragment ions for GCC in ESIMS.
(B) NMR connectivity observed for GCC. Spectral data are shown in Figures S8–S10. LC-MS analysis for GCC of Symbiodiniaceae and T. crocea origin is shown in Figure S11.
Lipidomic analysis of T. crocea sperm, fertilized eggs, and larvae
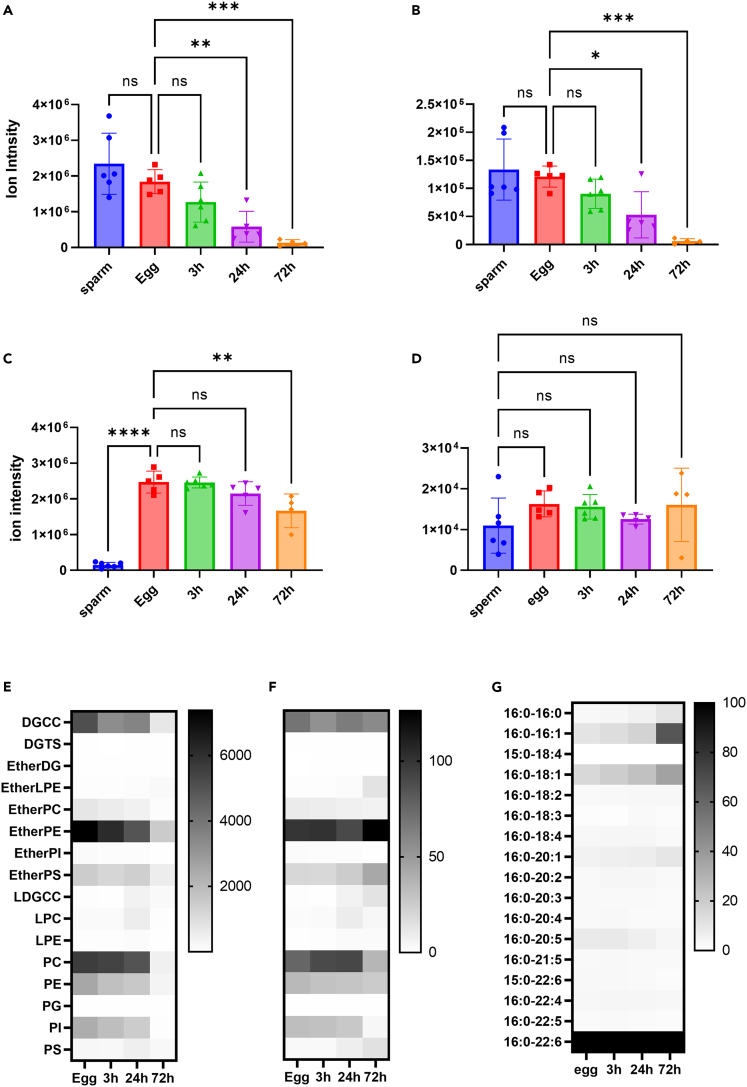
Symbiodiniacean cells are essential for giant clams; however, fertilized eggs and trochophore stage larvae are free of the symbiont;21 thus, the larvae or juveniles must acquire the symbionts from the ambient environment.22 A subtle decrease in TG ion intensity was observed only after 72 h, while those of DGCC and PC decreased from 24 h and markedly at 72 h after fertilization (Figures 3A–3C). These observations are in line with progress in fertilization, as eggs started to divide 3 h after fertilization, while veliger larvae and D-shaped larvae with thin shells were observed at 24 and 72 h after fertilization, respectively (Figure S12). Notably, GCC was found at a stable level in all sperm and egg stages (Figure 3D).
Figure 3.
Lipidomic analysis of T. crocea sperm, fertilized eggs, and larvae
Relative abundance of (A) DGCC ∗∗p value < 0.006, ∗∗∗∗p value <0.0006, ns: not significant.
(B) PC, ∗p value <0.0215, ∗∗∗p value < 0.0004, (C) TG, ∗∗p value < 0.0011, ∗∗∗∗p value <0.0001, and (D) GCC. Each histogram indicates the average value from four to six independent specimens ±SD. One-way ANOVA followed by Dunnett’s test for multiple group comparisons, ns: not significant.
(E) Semiquantitative analysis of lipid classes at each developmental stage (pmol mg−1).
(F) Relative concentration of lipids in each stage.
(G) Representative fatty acid composition of DGCC in each stage. In F and G, data in each column were normalized so that the highest value becomes 100 and the lowest value is equal to 0.
See also Figure S12.
Next, we semiquantitatively compared the membrane lipid composition of eggs and larvae during development. Similar to the gonad of adult clams, the amounts of ether-PE, PC, and DGCC were predominant over other lipids in eggs and larvae up to 24 h. The original compositions of all classes were maintained at 3 h after fertilization (Figures 3E and 3F). Lipid compositions started to change at 24 h and then changed drastically at 72 h (Figure 3F). Because DGCC was consumed rapidly, we asked if the fatty acid species in DGCC change over developmental stages. The relative composition of fatty acids in DGCC at each developmental stage showed acyl group changes at the developmental stages, in that fatty acids including 16:0-16:0, 16:0-16:1, 16:0-18:1, and 16:0-20:1 increased but 16:0-20:5 decreased over time.
Lipidomic analysis of other giant clams and bivalves
The above results suggested that DGCC is an indispensable class of fatty acids in T. crocea. We thus investigated whether DGCC was present in other giant clams and nonsymbiotic clams. To confirm this, we compared DGCC in the adductor muscles of Tridacna squamosa, Tridacna derasa, and Hippopus hippopus along with two other Symbiodiniaceae-free bivalves, Atactodea striata and Donax faba, which inhabit the sandy shore in Okinawa. The LC‒MS data indicated that the giant clams contained DGCC, whereas those in the nonsymbiotic clams did not have detectable amounts of DGCC (Figure S13).
MS imaging analyses
Next, we investigated the localization of lipids in T. crocea tissues to assess whether DGCC and PC, structurally compatible lipids, share distribution patterns. The imaging data were analyzed for DGCC and PC with two different fatty acid compositions, 16:0-22:6 and 16:0-18:1, which were predominant molecules in T. crocea tissues (Figure 2). As expected, 16:0-22:6 DGCC and PC showed similar distribution patterns; that is, they were dense in the EL and gonads, but only trace levels were visible in the DD, kidney, and muscle (Figure 4). In the case of 16:0-18:1 species, DGCC was distributed throughout the mantle tissues (Figure 4B) and visceral parts, including DD (Figure 4A), while PC showed a similar profile to that of the 16:0-22:6 species, except for some signals that were observed in the IL region of the mantle tissue.
Figure 4.
Mass imaging data for cryosections of T. crocea
(A) Atlas of the HE-stained cryo-visceral-section recovered after MS imaging. M, muscle; K, kidney; G, gonad; and DD, digestive diverticula. Heatmaps (0–100%) show relative intensities for two representative ion species (16:0-22:6) and (16:0-18:1) of PC and DGCC, and lyso-PC (16:0), and lyso-DGCC (16:0) in positive mode, (M + Na)+ for PC and (M + H)+ for DGCC.
(B) HE-stained cryo-mantle-section and heatmaps.
(C) MS imaging data for the detection of GCC.
(D) LC‒MS analysis of GCC in each organ. Each histogram indicates the average value from five independent specimens ±SD. Detailed results of multiple comparisons are given in Figure S14.
We then analyzed the distribution of lyso-PC and lyso-DGCC, which can be enzymatically transformed from PC and DGCC, respectively. The distribution pattern of lyso-PC (16:0) was similar to that of PC (16:0-18:1); faint signals were found in the DD, kidney, and muscle. Notably, in DD, signals for lyso-DGCC (16:0) were clearly observed as opposed to those of lyso-PC (16:0) and 16:0-22:6 DGCC; however, the distribution pattern was somewhat similar to that of 16:0-18:1 DGCC.
Further analysis of the distribution of GCC, a deacylated form of DGCC, revealed that its ion distribution was different from that of 16:0-22:6 DGCC, showing dense signals in the DD and kidney but low intensity in gonads (Figure 4D). Overall, these observations supported those of the LC‒MS analysis of the dissected organs (Figures 2 and S7).
Discussion
Presence of betaine lipid DGCC and its deacylated derivative in giant clams and Symbiodiniaceae
In the present study, we performed LC‒MS lipidomic and metabolomic analyses of the giant clam T. crocea and cultured giant clam-associated Symbiodiniaceae to identify the unique metabolic relationships between symbiotic dinoflagellates and their host clams. Strikingly, we found a betaine lipid, DGCC, and its polar head group, GCC, not only in the algal cells but also in the clam tissue extracts at concentrations comparable to those of PC. To date, three different classes of betaine lipids, DGTS, DGTA, and DGCC, have been identified. DGTS and DGTA are structural isomers found in a wide variety of lower plants, such as algae, pteridophyta, bryophyta, lichens, and fungi.23,24 However, DGCC is known only in limited taxa of microalgae, including Haptophyceae, Bacillariophyceae, and Dinophyceae,18,25 and thus, it is most likely biosynthesized in microalgae.26 DGCC, however, was detected in the corals Montipora capitata15 and Palythoa sp.27, likely due to symbiotic algae contained in these animal species affecting the total lipidome profiles.28
Localization of DGCC suggests its metabolism in T. crocea
The true origin of DGCC in these invertebrates is elusive since it is difficult to separate symbiont algae from the host to precisely analyze its origin. We combined lipidomics and MS imaging approaches on large dissectible giant clams to overcome this problem. As expected, DGCC was detected in the Symbiodiniaceae-rich mantle because the zooxanthellae are known to contain DGCC;18 however, the presence of DGCC in the muscle, kidney, sperm, and eggs where no symbiotic algae exist was unexpected. This result led to the hypothesis that T. crocea takes up algal DGCC and metabolites in their own cells and tissues. Our data, along with previous observations showing that algal DGCC is localized in plasma membranes,24,29 strongly suggest that DGCC is incorporated in the lipid bilayer of clam cells, up to a level equivalent to that of PC.23,24,30 The MS imaging data for two major fatty acid species illustrated that the most abundant species 16:0-22:6 of PC and DGCC share similar distribution profiles, suggesting that these lipids are physiologically compatible. However, the second most abundant 16:0-18:1 species of PC and DGCC were distributed somewhat differently (Figure 4A). This suggests that lipid molecules at the class and even species levels have defined loci to present and roles to play.31,32
Our results clearly show that T. crocea accumulates DGCC in all tissues, which strongly suggests that giant clams have evolved to utilize DGCC as a phospholipid substitute to thrive in oligotrophic coral reef waters.
The presence of degraded forms of DGCC, lyso-DGCC, and GCC was found in the clams and warrants further discussion. Lyso-DGCC was found in the DD and ctenidia in substantially higher amounts than in the EL, indicating that lyso-one is probably generated by the enzymatic digestion of DGCC in DD. Notably, significant quantities of lyso-DGCCs were found in ctenidia, which may suggest that they have specific functions in the loci. One of the notable observations here is the presence and distribution of GCC, which can be both a precursor and catabolite of DGCC. GCC was distributed in the DD and kidney, which is a complementary pattern to DGCC (Figure 4D). This observation supports that the clam digests its own symbiont algae in the DD, and algal DGCC can be degraded to form GCC (Figure 5). The high GCC signal observed in the DD in the MS imaging data supports this deduction. In fact, degraded algal cells were observed in the DD (Figure 1H). Fankboner’s detailed microscopic observation of aged algal cells that were culled and digested by amoebocytes and conveyed to DD33,34 further supports our finding. Therefore, GCC can be reacylated to produce DGCC or lyso-DGCC, which is redistributed and utilized in other organs and tissues as a component of the lipid membrane (Figure 5).
Figure 5.
Possible scenario for utilization of DGCC in giant clams
A schematic diagram of metabolic flow in T. crocea. Symbiotic algae fix carbons and biosynthesize GCC followed by DGCC. The algal cells are culled, transported, and digested in DD. Algal DGCC can be hydrolyzed to form GCC, which is reacylated to form DGCC with new acyl substituents (remodeling). Newly formed DGCCs are redistributed to each organ to be incorporated into the plasma membrane. In phosphorus (P)-deficient conditions, the use of DGCC allows consumption of PC as an extra source of P.
In addition to DD, the kidney can be a key organ for the metabolism of DGCC because a high intensity of GCC was found in the kidney tissue. This indicates that digested GCC was excreted as urine or stored as waste. However, it is thought that the kidney in giant clams associates closely with symbiosis with zooxanthella.17 Although the function of the kidneys in giant clams is not well understood, these organs are disproportionally large, and high enzymatic activity involving proteases is evident.17,35 The body plan connecting the DD to other parts, such as the gonads, gastrointestinal system, and ctenidia, suggests that the kidneys may play a central role in the use of Symbiodiniaceae metabolites in giant clams.17,36
Can DGCC be remodeled in the clam?
We found that the variation in fatty acid species in DGCC from T. crocea was highly diverse (49 species), while the diversity in cultured Symbiodiniaceae strains, which were isolated from giant clams, was lower (18 species); however, their profiles differed greatly, apart from 16:0-22:6 being the major species in both organisms (Figures 3F and S4). This differential fatty acid diversity suggests that the algal DGCC could be remodeled37 in the clam to constitute lipids that meet the physiological needs of the animal. This could also be due to other factors, such as differential metabolism between cultured and symbiotic algae or the presence of diverse Symbiodiniaceae species in the clam; however, our observation in the strictly algae-free life stage of clam discussed below supports the metabolism of DGCC in T. crocea.
DGCCs were present and metabolized in algae-free eggs, sperm, and larvae
The distribution of DGCC in completely algae-free eggs and sperm suggests maternal transfer of these molecules. DGCC was consumed rapidly as development progressed, similar to PC, while the storage lipid TG was consumed relatively slowly (Figures 3A–3C). It was also evident that lipid class remodeling occurred at approximately 24 h after fertilization, suggesting that de novo lipid biosynthesis occurs throughout development. Moreover, some fatty acid compositions in DGCC were shown to be changed over developmental periods, suggesting that lipid remodeling occurs at the fatty acid species level in T. crocea. The fact that GCC was found in all egg and sperm stages suggests that not only DGCC but also GCC was supplied maternally. The level of GCC in sperm, eggs, and larvae was consistent throughout the stages, suggesting that free GCC can be pooled at certain levels. Although the biosynthesis of DGCC is not understood to date,26,38 the presence of DGCC and GCC in both Symbiodiniaceae and the clam may provide some clues for investigating the metabolism of DGCC.
Possible metabolic role of DGCC
Phytoplankton in oligotrophic waters with low phosphate concentrations can compensate for this phosphorous requirement by breaking down PC and concomitantly fulfill their cellular phosphorus requirements by substituting nonphosphorus membrane lipids for phospholipids.30,39 Some marine phytoplankton employ betaine lipids for this purpose.30,39 It has been reported that DGTS plays the role of PC in phosphorous-deficient conditions and can function as a hub of glycerolipid remodeling in some stramenopile microalgae.40,41 Although little is known about DGCC, in P. lutheri, cytoplasmic DGCC was shown to act as a carrier of fatty acids to plastid MGDG,29 suggesting the presence of acyltransferase that utilizes DGCC as a substrate. Taken together, our data suggested that DGCC acts to serve fatty acids in the lipid biosynthesis of T. crocea, analogous to other betaine lipids in lower plants.
Possible scenario for utilization of DGCC in giant clams
The lipidomic study combined with MS imaging techniques uncovered an unusual metabolic flow in the giant clam-algae consortium. First, our observation that DGCC was distributed not only within the symbiont-rich sites in the clam but also in the symbiont-free tissues, including eggs and sperm, suggests that the clam uses this lipid as a plasma membrane component. Second, DGCC is most likely enzymatically digested, remodeled, and then redistributed to organs within the clam. These findings have identified a novel scenario in the metabolic flow between symbiotic algae and clams. Namely, GCC is biosynthesized within symbiotic algae and acylated to form DGCC. The algae are trafficked and digested by clams in the DD to generate GCC, which can then be used as a substrate for acyltransferase for remodeling. This transformation may take place in either the DD or kidney. The newly formed DGCC can be redistributed to other organs as a building block of plasma membranes. Giant clams thus utilize DGCC as a membrane component, which in turn allows the fast breakdown of PC to supply phosphate for other indispensable phosphorus-containing biomolecules (Figure 5). In the present study, we detected DGCC and GCC from giant clam species closely associated with symbiont algae, while the nonsymbiotic two clam species tested here did not contain these lipids. It is possible, however, that there might be bivalve species that digest planktonic microalgae containing DGCC/GCC and utilize its lipids. Nevertheless, our results demonstrated that animal cells can utilize betaine lipids, possibly with unique coupling with phospholipid metabolism, thereby showing that “smart utilization” of novel metabolites helps giant clams thrive in the oligotrophic milieu.30 These results provide a basis for future investigations into the paradoxical productivity and biodiversity of coral reef ecosystems.
Limitations of the study
In the present study, we used several species of giant clams and two nonsymbiotic bivalves for analysis to show that Symbiodiniaceae-harboring species acquired DGCC. However, this result cannot be generalized to all giant clams and to all other Symbiodiniaceae-harboring clams, or nonsymbiotic clams. Similarly, this result alone cannot predict lipid metabolism in other invertebrates with algal symbionts, including corals, foraminifera, jellyfish, or sea anemone. The detailed course of the biosynthesis, metabolism, and catabolism of DGCC in the giant clam-Symbiodiniaceae holobiont is yet to be understood and awaits further studies.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Tridacna crocea | Field collected in Okinawa | Permission from Okinawa Prefectural Government for research use (No. 30-82, 2-60) |
| Tridacna squamosa, | Field collected in Okinawa | Purchased from local fisherman |
| Tridacna derasa | Commercially cultivated in Okinawa | Purchased from local fisherman |
| Hippopus hippopus | Field collected in Okinawa | Purchased from local fisherman |
| Atactodea striata | Field collected in Okinawa | |
| Donax faba | Field collected in Okinawa | |
| Chemicals, peptides, and recombinant proteins | ||
| EquiSPLASH | Avanti | 330731-1EA |
| diacyl-N,N,N-trimethylhomoserine 16:0–16:0(d9) (DGTS) | Avanti | 857464P |
| Software and algorithms | ||
| Prism | GraphPad | https://www.graphpad.com/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Ryuichi Sakai (ryu.sakai@fish.hokudai.ac.jp).
Materials availability
This study did not generate unique reagents.
Experimental model and subject details
Collection of clam specimens
Specimens of T. crocea were collected in the coastal waters of Ishigaki Island, Okinawa, Japan under permission from the Okinawa Prefectural Government for research use (No. 30-82, 2-60). T. crocea specimens were dissected and each organ including the mantle, adductor muscle, kidney, digestive diverticula, and ctenidia (gill) was identified based on a previous report.16 The mantles were further divided into EL and IL. Other species of giant clams, T. derasa, T. squamosa, and H. hippopus were purchased from local fishers in Okinawa, and Symbiodiniaceae-free clams, Atactodea striata and Donax faba, were also collected in coastal waters of Ishigaki Island.
Preparation of sperm, eggs, and larvae
The gonad cutting method was employed to obtain Symbiodiniaceae-free T. crocea reproductive cells.41 The gonads were collected from three different T. crocea individuals. Giant clams are simultaneous hermaphrodites; thus, the gonads consist of both eggs and sperm. Eggs and sperm were separated using a 5 μm mesh filter. Fertilization was conducted using sperm from the other two individuals, under an egg:sperm ratio of 1:50.42 After 3 h of fertilization, eggs were washed with 0.2 μm filtered seawater, and were collected. The remaining fertilized eggs were kept in a 27°C incubator and the water was replaced once a day. The larvae at 24 and 72 h after fertilization were then collected. During the experiments, the larvae were not supplied food source microalgae or Symbiodiniaceae cells. We repeated this process with another three individuals. Thus, we used a total of 6 sperm, egg, and larvae samples. Each sample was lyophilized and extracted as described below for LC-MS analysis.
Method details
LC-MS/MS-based lipidomics
Approximately 5 mg of lyophilized samples were homogenized with a mortar and pestle and weighed in 1.5 mL tubes. Then, ice-cold extraction solvent (water∶methanol∶chloroform 0.8:2:1, v/v/v) was add (200 μL/mg dry sample). To this extraction mixture EquiSPLASH (Avanti) [a mixture of deuterium labeled phosphatidylcholine 15:0–18:1(d7) (PC), lysophosphatidylcholine 18:1(d7) (LPC), phosphatidylethanolamine 15:0–18:1(d7) (PE), lysophosphatidylethanolamine 18:1(d7) (LPE), phosphatidylglycerol 15:0–18:1(d7) (PG), phosphatidylinositol 15:0–18:1(d7) (PI), phosphatidylserine 15:0–18:1(d7) (PS), triacylglycerol 15:0–18:1(d7)-15:0 (TG), diacylglycerol 15:0–18:1(d7) (DG), monoacylglycerol 18:1(d7) (MG), cholesteryl ester 18:1(d7) (CE), sphingomyelin 18:1(d9) (SM)], and diacyl-N,N,N-trimethylhomoserine 16:0–16:0(d9) (DGTS, Avanti) were added to final concentration of 1 μg/mL lipid standards. The mixture was homogenized on ice for 2 min by an ultrasonic homogenizer. The extraction mixture was allowed to stand for 10 min, vortexed, and then centrifuged at 16,000×g at 4°C for 3 min. Next, the supernatant (760 μL) was collected to another vial and each 200 μL of chloroform and distilled water was added followed by vortex for 30 s. Centrifugation at 16,000×g at 4°C for 3 min phase-separated the mixture and the lower layer (250 μL) was transferred to new 1.5 mL glass vials and evaporated to dryness in an SPEED VAC (Thermo Scientific). The dried extracts were resuspended in MeOH (250 μL) and diluted 10-fold with MeOH for LC-MS analysis.
High-performance LC-tandem MS (HPLC-MS/MS) was performed on a quadrupole time-of-flight (TOF) mass spectrometer (TripleTOF 5600+; Sciex, Framingham, MA, USA) with a BEH C8 column (2.1x150, S 2.5 μm; Waters, Milford, MA, USA) and solvents A (0.1% formate +10 mM ammonium formate in water) and B (0.1% formate +10 mM ammonium formate in MeOH: 2-propanol (85:15, v/v)). The flow rate was 0.3 mL/min with the following time program: B conc 75%, 0–2 min; 75%–99%, 2–18 min; 99%, 18–24 min; 99%–75%, 24–25 min; 30 min stop. The column oven temperature was 50°C, and 3 (positive) and 5 μL (negative) of the samples were injected. The following MS parameters were set based on the lipidomic analysis reported previously:43 MS range 100–1,250 Da, ion spray voltage floating +5.5 kV (positive) and −4,5 kV (negative), gas temperature 350°C, declustering potential 80V, and accumulation time 250 ms. MS2 was measured using the high sensitivity mode for information dependent analysis (IDA), which acquires 15 times MS2 per cycle. MS2 parameters were MS range 100–1,250 Da, gas temperature 350°C, collision energy 40–70 V (positive) and -30–-60 V (negative). The collision energy in the positive ion mode was stronger than that used by Tsugawa43 for clearer acquisition of the fatty acid fragment of diacylglycerylcarboxy-hydroxymethylcholine (DGCC).
We used a mixture of all samples for quality control (QC). QC data were acquired for every five samples during the analysis to monitor the intensity drift of peaks detected by LC-MS and reduce the number of failed MS2 acquisitions of peaks in the IDA mode.
Lipidomic data analysis
LC-MS/MS data were analyzed using MS-DIAL version 4.24, and lipid annotation was automatically conducted by matching with an in silico MS/MS library available on the RIKEN PRIMe website (http://prime.psc.riken.jp/); the results were manually curated with the confirmation of the diagnostic product ion and neutral losses in addition to the fatty acid fragmentation of each lipid species. We used the table reported by Tsugawa to characterize each lipid species.44
The parameters below were used for the MS-DIAL analysis: retention time begin, 0.5 min; retention time end, 28 min; mass range begin, 100 Da; mass range end, 2000 Da; accurate mass tolerance (MS1), 0.01 Da; MS2 tolerance, 0.025 Da; maximum change number, 2; smoothing level, 3; minimum peak width, 5 scans; minimum peak height 1000; mass slice width, 0.1 Da; sigma window value, 0.5; exclude after precursor ion, true; keep the isotopic ions until 0.5 Da; retention time tolerance for identification, 100 min; MS1 for identification, 0.01 Da; accurate mass tolerance (MS2) for identification, 0.05 Da; identification score cut-off, 70%; using retention time for scoring, true; relative abundance cut-off, 0; top candidate report, true; retention time tolerance for alignment, 0.5 min; MS1 tolerance for alignment, 0.015 Da; peak count filter, 0; remove feature based on blank information, true; sample max/blank average, 5; keep identified metabolites, true; keep removable features and assign the tag, true; and gap filling by compulsion, true.
Isolation of GCC from cultured Symbiodiniaceae cells
To analyze betaine lipid of Symbiodiniaceae cells, we used two Symbiodiniaceae culture strains isolated from giant clams. Giant clam species are known to harbor clades A, C, and D Symbiodiniaceae45 (they are now classified into genus Symbiodinium, Cladocopium, and Durusdinium, respectively). The culture strains (TsIS-H4 and TsIS-G10) used in this study are Symbiodinium tridacnidorum (ITS2 type A6) and Durusdinium sp. (ITS2 type D4-5), respectively. The cell pellets from culture strains were extracted with water on ice using an ultrasonic homogenizer (Smurt NR 50M; Microtec, Funabashi, Chiba, Japan). The water extract was separated by dialysis and the small molecular fraction was subjected to Sephadex LH-20 gel filtration chromatography (GE Healthcare, Chicago, IL, USA). The glycerylcarboxy-hydroxymethylcholine (GCC)-containing fraction was further separated using a Hillic HPLC column (Develosil ANIDIUS, NOMURA CHEMICAL CO., LTD, Japan) with an acetonitrile–water gradient, and pure GCC (IH2-78-7, 1.00 mg) was obtained ([α]D 20= -110˚ (H2O, c 0.007), HRESIMS (m/z 252.1442 C10H21NO6, Δ 2 ppm). 1H NMR, see Table S5. Detailed structure determination is described in Supplementary results.
Isolation of DGCC of T. crocea
Cultured T. crocea (8 specimens) were dissected to yield EL (28g), IL (23g), ctenidia (5g), kidney (4g), DD (2g), foot (10g), muscle (6g). The muscle was used to isolate DGCC. Samples were first extracted with 27 mL of extraction solvent system (water:methanol:chloroform = 2 : 5 : 2) and water (12mL) was added after homogenization. The solvent was partitioned by centrifugation and both upper layer and lower layer were concentrated to dryness. The organic extract (lower layer, 11 mg) was separated by counter current partition chromatography (CPC MODEL LLB; SIC Japan, Tokyo, Japan) using a solvent system; chloroform: n-heptane: n-butanol: methanol: acetic acid (60%) at 3:5:3:3:5 ratio. The lower phase was used as the mobile phase at a rotor speed of 1000 rpm. The sample was suspended in the upper and lower layers of the solvent system (3 mL) and injected in the descending mode at a flow of 9.0 mL/min. Elution was initially performed in the descending mode at 2 mL/min with a column pressure of 35 kg/cm2 and then switched to the ascending mode (9 mL/min, 22 kg/cm2). The column was then flushed with methanol, after which the eluents (31 tubes, 10 mL/tube) was collected and combined into 10 fractions (IH9-28-1∼10). Each fraction was analyzed by thin layer chromatography and LC-MS/MS to characterize its lipid composition. Fraction 3 (IH9-28-3, 4 mg) containing DGCC 22:6-16:0 ([α]D 20= -30˚ (MeOH, c 0.064), HRESIMS m/z 800.5954, M + H+, C10H21NO6) as a major lipid was used for spectral analysis.
Analysis of DGCC on HPLC-CAD
As DGCC does not offer strong and characteristic UV absorption, charge aerosol detector (CAD, CORONA Ultra RS; Thermo Fisher Scientific, Waltham, MA, USA) was used to quantify DGCC to further support our LC/MS analysis data. We used the lipophilic portion of the dissected samples described above. Each of the extract was dissolved in MeOH (1mg/mL), and POPG was added as an internal standard at final concentration of 1mg/mL. InertSustain C8-5 (4.6 x 250 mm, GL Sciences) was used with a gradient of acetonitrile (0.05% TFA) and water (0.05% TFA) at oven temperature at 50°C. Amount of DGCC was calculated relative to that of POPG (Figure S5).
MS imaging
Consecutive 10-μm sections were cut directly from the frozen samples using a cryostat (CM 1950; Leica Microsystems, Wetzlar, Germany). Serial sections were mounted onto glass slides for with/without hematoxylin and eosin (HE) staining and onto indium tin oxide-coated glass slides (Bruker Daltonics, Billerica, MA, USA) for MS imaging. The section without HE stain was observed under epifluorescent microscope (BX50, Olympus, Tokyo, Japan) to confirm the distribution/localization of Symbiodiniaceae cells in the section. After MS imaging, the sections were subjected to HE staining for morphological observation. Samples were prepared as previously described.46,47 Briefly, a matrix solution containing 50 mg/mL 2,5-dihydroxybenzoic acid in methanol: water (8:2, v/v) was used, with 1–2 mL prepared before use and sprayed uniformly over the frozen sections using an airbrush with a 0.2-mm nozzle (Procon Boy FWA Platinum; Mr. Hobby, Tokyo, Japan). MS analysis was performed using TOF/TOF 5800 (AB Sciex, Framingham, MA, USA) and SolariX Fourier-transform ion cyclotron resonance (FT-ICR) (Bruker Daltonics) mass spectrometers. To optimize FT-ICR MS, we set the mass range from m/z 400–1200 for DGCC/PC, and m/z 100-500 for GCC and the spatial resolution to 150 μm for mantle tissue and 220 μm for the frozen viscera of the animal.
Statistical analysis
Data were obtained from randomly collected specimens. Number of specimens is indicated in figure legends. Datapoints representing mean value and error bars ±SD were analyzed using one-way analysis of variance (ANOVA) and Dunnett’s multiple comparison test using GraphPad Prism 8.0.3.
Acknowledgments
The authors thank the staff at the Yeyama field station, Fisheries Technology Institute, Japan Fisheries Research and Education Agency for their support during clam sampling. Hiroki Ikeda at Hokkaido University assisted in the initial data collection. We are grateful to Mr. Teruo Yamashita who provided cultivated and wild collected giant clams. We would like to thank Editage (www.editage.com) for English language editing. This study was supported by the Japan Society for the Promotion of Science KAKENHI grant no. 21H04742 to H.Y. Some research equipment for the MEXT Project for promoting public utilization of advanced research infrastructure (Program for advanced research equipment platforms, grant number JPMXS0450200120) was used in this study.
Author contributions
Conceptualization, R.S., H.Y., and N.G.I.; Methodology, R.S., H.Y., and N.G.I.; Investigation, N.A., Y.K., H.Y., and N.G.I.; Writing – Original Draft, R.S.; Writing –Review & Editing, R.S., H.Y., T.M., and N.G.I.; Funding Acquisition, H.Y.; Resources, H.Y. and R.S.; Supervision, R.S., H.Y., and T.M.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 28, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107250.
Contributor Information
Ryuichi Sakai, Email: ryu.sakai@fish.hokudai.ac.jp.
Hiroshi Yamashita, Email: yamashita_hiroshi21@fra.go.jp.
Supplemental information
Data and code availability
The lipidomics data generated in this study were deposited at https://doi.org/10.17632/93hgsjz5sv.1 and are publicly available as of the date of publication.
The paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Trench R.K., Wethey D.S., Porter J.W. Observations on the symbiosis with zooxanthellae among the Tridacnidae (Mollusca, Bivalvia) Biol. Bull. 1981;161:180–198. doi: 10.2307/1541117. [DOI] [Google Scholar]
- 2.Ikeda S., Yamashita H., Kondo S.-n., Inoue K., Morishima S.-y., Koike K. Zooxanthellal genetic varieties in giant clams are partially determined by species-intrinsic and growth-related characteristics. PLoS One. 2017;12:e0172285. doi: 10.1371/journal.pone.0172285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Norton J.H., Shepherd M.A., Long H.M., Fitt W.K. The zooxanthellal tubular system in the giant clam. Biol. Bull. 1992;183:503–506. doi: 10.2307/1542028. [DOI] [PubMed] [Google Scholar]
- 4.Ishikura M., Adachi K., Maruyama T. Zooxanthellae release glucose in the tissue of a giant clam, Tridacna crocea. Mar. Biol. 1999;133:665–673. doi: 10.1007/s002270050507. [DOI] [Google Scholar]
- 5.Klumpp D., Lucas J. Nutritional ecology of the giant clams Tridacna tevoroa and T. derasa from Tonga: influence of light on filter-feeding and photosynthesis. Mar. Ecol. Prog. Ser. 1994;107:147–156. doi: 10.3354/meps107147. [DOI] [Google Scholar]
- 6.Ishikura M., Kato C., Maruyama T. UV-absorbing substances in zooxanthellate and azooxanthellate clams. Mar. Biol. 1997;128:649–655. doi: 10.1007/s002270050131. [DOI] [Google Scholar]
- 7.Goto-Inoue N., Sato T., Morisasa M., Yamashita H., Maruyama T., Ikeda H., Sakai R. Mass spectrometry imaging reveals differential localization of natural sunscreens in the mantle of the giant clam Tridacna crocea. Sci. Rep. 2020;10:656. doi: 10.1038/s41598-019-57296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shick J.M. The continuity and intensity of ultraviolet irradiation affect the kinetics of biosynthesis, accumulation, and conversion of mycosporine-like amino acids (MAAs) in the coral Stylophora pistillata. Limnol. Oceanogr. 2004;49:442–458. doi: 10.4319/lo.2004.49.2.0442. [DOI] [Google Scholar]
- 9.Trench R.K. Microalgal-invertebrate symbiosis—a review. Endocytobiosis Cell Res. 1993;9:135–175. [Google Scholar]
- 10.Baker A.C. Flexibility and specificity in coral-algal symbiosis: diversity, ecology, and biogeography of Symbiodinium. Annu. Rev. Ecol. Evol. Syst. 2003;34:661–689. doi: 10.1146/annurev.ecolsys.34.011802.132417. [DOI] [Google Scholar]
- 11.Wild C., Huettel M., Klueter A., Kremb S.G., Rasheed M.Y.M., Jørgensen B.B. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature. 2004;428:66–70. doi: 10.1038/nature02344. [DOI] [PubMed] [Google Scholar]
- 12.De Goeij J.M., Van Oevelen D., Vermeij M.J.A., Osinga R., Middelburg J.J., de Goeij A.F.P.M., Admiraal W. Surviving in a marine desert: the sponge loop retains resources within coral reefs. Science. 2013;342:108–110. doi: 10.1126/science.1241981. [DOI] [PubMed] [Google Scholar]
- 13.Gordon B.R., Leggat W. Symbiodinium—invertebrate symbioses and the role of metabolomics. Mar. Drugs. 2010;8:2546–2568. doi: 10.3390/md8102546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosset S., Koster G., Brandsma J., Hunt A.N., Postle A.D., D’Angelo C. Lipidome analysis of Symbiodiniaceae reveals possible mechanisms of heat stress tolerance in reef coral symbionts. Coral Reefs. 2019;38:1241–1253. doi: 10.1007/s00338-019-01865-x. [DOI] [Google Scholar]
- 15.Roach T.N.F., Dilworth J., Jones A.D., Jones A.D., Quinn R.A., Drury C. Metabolomic signatures of coral bleaching history. Nat. Ecol. Evol. 2021;5:495–503. doi: 10.1038/s41559-020-01388-7. [DOI] [PubMed] [Google Scholar]
- 16.Norton J.H., Jones G.W. Australian Centre for International Agricultural Research; 1992. The Giant Clam: An Anatomical and Histological Atlas. Monographs. number 118700. [DOI] [Google Scholar]
- 17.Reid R., Fanklboner P., Brand D. Studies of the physiology of the giant clam Tridacna gigas linné—II. Kidney function. Comp. Biochem. Physiol. Part A: Physiol. 1984;78:103–108. doi: 10.1016/0300-9629(84)90100-2. [DOI] [Google Scholar]
- 18.Kato M., Sakai M., Adachi K., Ikemoto H., Sano H. Distribution of betaine lipids in marine algae. Phytochemistry. 1996;42:1341–1345. doi: 10.1016/0031-9422(96)00115-X. [DOI] [Google Scholar]
- 19.Leblond J.D., Khadka M., Duong L., Dahmen J.L. Squishy lipids: Temperature effects on the betaine and galactolipid profiles of a C18/C18 peridinin-containing dinoflagellate, Symbiodinium microadriaticum (Dinophyceae), isolated from the mangrove jellyfish, Cassiopea xamachana. Phycol. Res. 2015;63:219–230. doi: 10.1111/pre.12093. [DOI] [Google Scholar]
- 20.Kato M., Adachi K., Hajiro-Nakanishi K., Ishigaki E., Sano H., Miyachi S. A betaine lipid from Pavlova lutheri. Phytochemistry. 1994;37:279–280. doi: 10.1016/0031-9422(94)85041-0. [DOI] [Google Scholar]
- 21.Fitt W.K., Trench R.K. Spawning, development, and acquisition of zooxanthellae by Tridacna squamosa (Mollusca, Bivalvia) Biol. Bull. 1981;161:213–235. doi: 10.2307/1540800. [DOI] [Google Scholar]
- 22.Fitt W.K. The role of chemosensory behavior of Symbiodinium microadriaticum, intermediate hosts, and host behavior in the infection of coelenterates and molluscs with zooxanthellae. Mar. Biol. 1984;81:9–17. doi: 10.1007/BF00397620. [DOI] [Google Scholar]
- 23.Cañavate J.P., Armada I., Ríos J.L., Hachero-Cruzado I. Exploring occurrence and molecular diversity of betaine lipids across taxonomy of marine microalgae. Phytochemistry. 2016;124:68–78. doi: 10.1016/j.phytochem.2016.02.007. [DOI] [PubMed] [Google Scholar]
- 24.Künzler K., Eichenberger W. Betaine lipids and zwitterionic phospholipids in plants and fungi. Phytochemistry. 1997;46:883–892. doi: 10.1515/znc-1997-7-811. [DOI] [PubMed] [Google Scholar]
- 25.Aveiro S.S., Melo T., Figueiredo A., Domingues P., Pereira H., Maia I.B., Silva J., Domingues M.R., Nunes C., Moreira A.S.P. The polar lipidome of cultured Emiliania huxleyi: a source of bioactive lipids with relevance for biotechnological applications. Biomolecules. 2020;10:1434. doi: 10.3390/biom10101434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato M., Kobayashi Y., Torii A., Yamada M. Advanced research on plant lipids. Springer; 2003. Betaine lipids in marine algae; pp. 19–22. [DOI] [Google Scholar]
- 27.Sikorskaya T.V., Efimova K.V., Imbs A.B. Lipidomes of phylogenetically different symbiotic dinoflagellates of corals. Phytochemistry. 2021;181:112579. doi: 10.1016/j.phytochem.2020.112579. [DOI] [PubMed] [Google Scholar]
- 28.Imbs A.B., Ermolenko E.V., Grigorchuk V.P., Sikorskaya T.V., Velansky P.V. Current progress in lipidomics of marine invertebrates. Mar. Drugs. 2021;19:660. doi: 10.3390/md19120660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eichenberger W., Gribi C. Lipids of Pavlova lutheri: cellular site and metabolic role of DGCC. Phytochemistry. 1997;45:1561–1567. doi: 10.1016/S0031-9422(97)00201-X. [DOI] [Google Scholar]
- 30.Van Mooy B.A.S., Fredricks H.F., Pedler B.E., Dyhrman S.T., Karl D.M., Koblízek M., Lomas M.W., Mincer T.J., Moore L.R., Moutin T., et al. Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature. 2009;458:69–72. doi: 10.1038/nature07659. [DOI] [PubMed] [Google Scholar]
- 31.Bourceau P., Michellod D., Geier B., Liebeke M. Spatial metabolomics shows contrasting phosphonolipid distributions in tissues of marine bivalves. PeerJ Anal. Chem. 2022;4:e21. doi: 10.7717/peerj-achem.21. [DOI] [Google Scholar]
- 32.Murphy R.C., Hankin J.A., Barkley R.M. Imaging of lipid species by MALDI mass spectrometry. J. Lipid Res. 2009;50:S317–S322. doi: 10.1194/jlr.R800051-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fankboner P.V. Intracellular digestion of symbiontic zooxanthellae by host amoebocytes in giant clams (Bivalvia: Tridacnidae), with a note on the nutritional role of the hypertrophied siphonal epidermis. Biol. Bull. 1971;141:222–234. doi: 10.2307/1540113. [DOI] [Google Scholar]
- 34.Morton B. The diurnal rhythm and the processes of feeding and digestion in Tridacna crocea (BivaMa: Tridacnidae) J. Zool. 1978;185:371–387. doi: 10.1111/j.1469-7998.1978.tb03339.x. [DOI] [Google Scholar]
- 35.Rees T.A.V., Fitt W.K., Yellowlees D. Host glutamine synthetase activities in the giant clam—zooxanthellae symbiosis: effects of clam size, elevated ammonia and continuous darkness. Mar. Biol. 1994;118:681–685. doi: 10.1007/BF00347516. [DOI] [Google Scholar]
- 36.Fankboner P.V., Reid R.G. In: Proceedings of a memorial symposium in honour of Sir Charles Maurice Yonge. Morton B., editor. Hong Kong University Press; 1990. Nutrition in giant clams (Tridacnidae). The bivalvia; pp. 195–209. [Google Scholar]
- 37.Huang B., Marchand J., Thiriet-Rupert S., Carrier G., Saint-Jean B., Lukomska E., Moreau B., Morant-Manceau A., Bougaran G., Mimouni V. Betaine lipid and neutral lipid production under nitrogen or phosphorus limitation in the marine microalga Tisochrysis lutea (Haptophyta) Algal Res. 2019;40:101506. doi: 10.1016/j.algal.2019.101506. [DOI] [Google Scholar]
- 38.Hunter J.E., Brandsma J., Dymond M.K., Koster G., Moore C.M., Postle A.D., Mills R.A., Attard G.S. Lipidomics of Thalassiosira pseudonana under phosphorus stress reveal underlying phospholipid substitution dynamics and novel diglycosylceramide substitutes. Appl. Environ. Microbiol. 2018;84:e02034-17. doi: 10.1128/AEM.02034-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin P., Van Mooy B.A.S., Heithoff A., Dyhrman S.T. Phosphorus supply drives rapid turnover of membrane phospholipids in the diatom Thalassiosira pseudonana. ISME J. 2011;5:1057–1060. doi: 10.1038/ismej.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cañavate J.P., Armada I., Hachero-Cruzado I. Interspecific variability in phosphorus-induced lipid remodelling among marine eukaryotic phytoplankton. New Phytol. 2017;213:700–713. doi: 10.1111/nph.14179. [DOI] [PubMed] [Google Scholar]
- 41.Murakoshi M., Hirata H. Self-fertilization in four species of giant clam. Nippon Suisan Gakkaishi. 1993;59:581–587. doi: 10.2331/suisan.59.581. [DOI] [Google Scholar]
- 42.Neo M., Todd P., Chou L., Teo S. Spawning induction and larval development in the fluted giant clam, Tridacna squamosa (Bivalvia: Tridacnidae) Nat. Singap. 2011;4:157–161. [Google Scholar]
- 43.Tsugawa H., Cajka T., Kind T., Ma Y., Higgins B., Ikeda K., Kanazawa M., VanderGheynst J., Fiehn O., Arita M. MS-DIAL: data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods. 2015;12:523–526. doi: 10.1038/nmeth.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsugawa H., Satoh A., Uchino H., Cajka T., Arita M., Arita M. Mass spectrometry data repository enhances novel metabolite discoveries with advances in computational metabolomics. Metabolites. 2019;9:119. doi: 10.3390/metabo9060119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.DeBoer T.S., Baker A.C., Erdmann M.V., Jones P.R., Jones P., Barber P. Patterns of Symbiodinium distribution in three giant clam species across the biodiverse Bird’s Head region of Indonesia. Mar. Ecol. Prog. Ser. 2012;444:117–132. doi: 10.3354/meps09413. [DOI] [Google Scholar]
- 46.Goto-Inoue N., Kashiwagi A., Kashiwagi K., Mori T. Metabolomic approach for identifying and visualizing molecular tissue markers in tadpoles of Xenopus tropicalis by mass spectrometry imaging. Biol. Open. 2016;5:1252–1259. doi: 10.1007/s00216-012-5809-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Goto-Inoue N., Manabe Y., Miyatake S., Ogino S., Morishita A., Hayasaka T., Masaki N., Setou M., Fujii N.L. Visualization of dynamic change in contraction-induced lipid composition in mouse skeletal muscle by matrix-assisted laser desorption/ionization imaging mass spectrometry. Anal. Bioanal. Chem. 2012;403:1863–1871. doi: 10.1242/bio.019646. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The lipidomics data generated in this study were deposited at https://doi.org/10.17632/93hgsjz5sv.1 and are publicly available as of the date of publication.
The paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.