Abstract
Brevetoxins (BTX) are a group of marine neurotoxins produced by the harmful alga Karenia brevis. Numerous studies have shown that BTX are rapidly accumulated and metabolized in shellfish and mammals. However, there are only limited data on BTX metabolism in fish, despite growing evidence that fish serve as vectors for BTX transfer in marine food webs. In this study, we aimed to investigate the in vitro biotransformation of BTX-2, the major constituent of BTX profiles in K. brevis, in several species of northern Gulf of Mexico fish. Metabolism assays were performed using hepatic microsomes prepared in-house as well as commercially available human microsomes for comparison, focusing on phase I reactions mediated by cytochrome P450 monooxygenase (CYP) enzymes. Samples were analyzed by UHPLC-HRMS(/MS) to monitor BTX-2 depletion and characterize BTX metabolites based on MS/MS fragmentation pathways. Our results showed that both fish and human liver microsomes rapidly depleted BTX-2, resulting in a 72–99% reduction within 1 h of incubation. We observed the simultaneous production of 22 metabolites functionalized by reductions, oxidations, and other phase I reactions. We were able to identify the previously described congeners BTX-3 and BTX-B5, and tentatively identified BTX-9, 41,43-dihydro-BTX-2, several A-ring hydrolysis products, as well as several novel metabolites. Our results confirmed that fish are capable of similar BTX biotransformation reactions as reported for shellfish and mammals, but comparison of metabolite formation across the tested species suggested considerable interspecific variation in BTX-2 metabolism potentially leading to divergent BTX profiles. We additionally observed non-enzymatic formation of BTX-2 and BTX-3 glutathione conjugates. Collectively, these findings have important implications for determining the ecotoxicological fate of BTX in marine food webs.
Keywords: Brevetoxin, High-resolution mass spectrometry, Metabolite identification, Fish, Phase I biotransformation, Liver microsomes
Graphical abstract
Highlights
-
•
BTX-2 is rapidly depleted by northern Gulf of Mexico fish liver microsomes within 1 h.
-
•
22 metabolites resulting from phase I reactions mediated by CYP enzymes were tentatively identified using UHPLC-HRMS/MS.
-
•
Observed glutathione conjugates of BTX-2 and BTX-3 likely formed by chemical rather than enzymatic reactions.
-
•
BTX-2 metabolite production varied across microsomes from different species and/or taxa.
-
•
Differences in metabolite formation between fish and human liver microsomes suggest taxa-specific BTX-2 metabolism pathways.
1. Introduction
Brevetoxins (BTX) are lipophilic marine neurotoxins originating from the harmful alga Karenia brevis (formerly Gymnodinium breve or Ptychodiscus brevis), which is responsible for recurrent “Florida red tide” blooms along the Gulf of Mexico and western Atlantic U.S. coast (Flewelling et al., 2005). BTX accumulate in a variety of marine organisms and undergo metabolic biotransformations leading to a diversity of chemical structures. Humans may be exposed to BTX through contaminated shellfish, which has been associated with neurotoxic shellfish poisoning (Ishida et al., 2004a; Abraham et al., 2008; Watkins et al., 2008). K. brevis blooms have also been linked to several mass mortality events of fish, marine mammals, sea turtles, and seabirds (Landsberg, 2002; Flewelling et al., 2005; Twiner et al., 2012; Van Deventer et al., 2012; Walker et al., 2018). Despite their apparent sensitivity to BTX following lethal exposure, there is evidence that fish may also serve as important vectors of BTX in marine foods webs. Fish exposed to chronic, sublethal concentrations of BTX can accumulate the toxin in their flesh and viscera, sometimes reaching levels beyond the regulatory guidance threshold for safe consumption of shellfish (Tester et al., 2000; Woofter et al., 2005; Naar et al., 2007; Flewelling et al., 2010). Findings of BTX-contaminated fish in the stomachs of dead dolphins (Fire et al., 2008, 2015; Twiner et al., 2012) and scavenging seabirds (Van Deventer et al., 2012) illustrate the ecological impact of BTX transferred through fish vectors and raise concern regarding the risk of human exposure to BTX via contaminated finfish (Fleming et al., 2011). This underscores the need to better assess the role that fish play in BTX transfer, including their capacity for BTX metabolism.
BTX congeners are classified based on their backbone structures (A- and B-type) and are further differentiated by modifications of their terminal side chain (Baden, 1989, Fig. 1). The B-type toxins are considered more prevalent than the A-type toxins, with BTX-2 representing the most prevalent algal congener in K. brevis in both bloom and culture settings (Twiner et al., 2007; Pierce et al., 2008; Errera et al., 2010; Lekan and Tomas, 2010). Aside from algal congeners, much of the present knowledge on BTX derivatives comes from studies with humans and rodents (Poli et al., 2000; Wang et al., 2005; Radwan and Ramsdell, 2006; Abraham et al., 2008; Guo et al., 2010), as well as with shellfish (Morohashi et al., 1995, 1999; Murata et al., 1998; Poli et al., 2000; Plakas et al., 2002, 2004; Ishida et al., 2004b; Wang et al., 2004). These studies have shown that BTX-2 is rapidly absorbed, distributed, and eliminated primarily as metabolites, indicating that biotransformation plays a major role in the post-exposure processing and excretion of BTX (Poli et al., 1990; Radwan et al., 2005). At least 70 BTX derivatives have been reported to date, although only about half of these have been structurally characterized (ANSES, 2021; Hort et al., 2021). Phase I BTX-2 metabolites result mainly from cytochrome P450 monooxygenase (CYP)-dependent functionalization to several reduction and hydrolysis products (e.g., BTX-3, -9, the carboxylic acid derivative BTX-B5, as well as epoxide, diol, and A-ring hydrolysis products) (Ishida et al., 2004a; Radwan and Ramsdell, 2006; Guo et al., 2010). A variety of phase II metabolites have additionally been identified in mammals and shellfish, including cysteine and cysteine-sulfoxide conjugates arising from glutathione S-transferase (GST) pathways involving intermediate glutathione (GSH) metabolites (Plakas et al., 2002, 2004; Wang et al., 2004; Radwan and Ramsdell, 2006).
Fig. 1.
Structure of BTX compounds. Backbone structures of A-type and B-type BTX compounds, as well as terminal side chains (R group) of major BTX congeners produced by K. brevis, including BTX-2, BTX-3, and BTX-B5 used as reference materials in this study.
Compared to advances in understanding BTX metabolism in mammals and shellfish, the metabolic fate of BTX in fish remains largely unexplored. Several phase I metabolites and conjugated products have been identified in fish using targeted analytical approaches, including BTX-2, -3, -6, -9, and several hydrolysis and cysteine conjugates (Naar et al., 2007; Fire et al., 2008; Flewelling et al., 2010). Some studies have also shown that exposure to BTX leads to activation of CYP- and GST-related enzyme activities (Washburn et al., 1994, 1996), indicating that biotransformation may be a similarly important pathway for BTX elimination in fish. However, there are few detailed studies describing BTX metabolites or biotransformation pathways in fish comparable to those available for mammals and shellfish.
The aim of this study was to investigate the in vitro biotransformation of BTX-2 using liver microsomes prepared from fish collected from the northern Gulf of Mexico, where K. brevis blooms regularly occur. We additionally explored in vitro BTX-2 biotransformation in commercial human liver microsomes (HLM) to further compare BTX-2 metabolism across taxa using a microsomal source for which information on BTX metabolism is available through previous studies (e.g., Guo et al., 2010). Through this research, we hope to characterize relevant BTX-2 metabolites in fish to better understand the role they play in the ecotoxicological fate and transfer of BTX.
2. Experimental
2.1. Materials and reagents
Optima LC-MS grade acetonitrile (MeCN), methanol (MeOH), and water were obtained from Thermo Fisher Scientific (Waltham, MA, USA). Formic acid (>98%) was purchased from Merck (Darmstadt, Germany). Reagents used for the in vitro phase I metabolism experiments, including β-nicotinamide adenine dinucleotide 2′-phosphate reduced tetrasodium salt hydrate (NADPH), β-nicotinamide adenine dinucleotide phosphate sodium salt hydrate (NADP+), HEPES sodium salt, d-glucose 6-phosphate sodium salt, and d-glucose 6-phosphate dehydrogenase from baker's yeast (Saccharomyces cerevisiae), were purchased from Sigma-Aldrich (St. Louis, MO, USA). Magnesium chloride hexahydrate (MgCl2 × 6H2O) was obtained from Honeywell Fluka (Bucharest, Romania).
The in vitro metabolic stability, biotransformation, and HRMS(/MS) experiments with brevetoxin-2 (BTX-2) were performed using synthetic BTX-2 (>95%) obtained from Abcam (Cambridge, UK; No. ab143469; Lots GR3275745–1 and GR3325074-2). Additionally, HRMS(/MS) experiments were performed using brevetoxin-3 (BTX-3; >95%) from Ptychodiscus brevis, which was also obtained from Abcam (No. ab143470, Lot GR3275242-3), and semi-synthetic brevetoxin B5 (BTX-B5; >95%) purchased from MARBIONC (Durham, NC, USA). A-ring hydrolyzed derivatives of BTX-2, BTX-3, and BTX-B5 were produced through base hydrolysis by reacting the standards (dissolved in 80% MeOH, v/v) with 0.1 M NaOH (1:1, v/v), prepared from a 10 M stock solution (BioUltra grade, Merck), at room temperature for 1 h, after which the parent toxins were no longer detectable. Reduced glutathione (GSH, ≥98%), used in chemical conjugation assays with BTX-2, was obtained from Merck.
2.2. Microsomal sources
The in vitro metabolism assays were performed using commercially available liver microsomes from humans (Homo sapiens) as well as liver microsomes prepared in-house from several species of northern Gulf of Mexico fish, which had been collected and prepared as part of a previous investigation (Gwinn et al., 2021). The human liver microsomes (HLM) consisted of a mixed gender pool of 50 donors and were obtained from Bioreclamation IVT (Westbury, NY, USA; No. X008067, Lots SBM, YAO, and IHG). The total protein concentrations, total CYP contents, and specific CYP enzyme activities of HLM were provided by the manufacturer. The HLM were stored in liquid N2 until use. Microsomes were prepared from fish representing five commercially and recreationally important species with ecological relevance throughout the northern Gulf of Mexico, and spanned several taxonomic groups and trophic levels. These included Lutjanus campechanus (red snapper; RSN), L. griseus (gray snapper; GSN), Pterois volitans (red lionfish; LNF), Nicholsina usta (emerald parrotfish; PRT), and Archosargus probatocephalus (sheepshead; SHP). The fish were collected during October and November of 2019 from several northern Gulf of Mexico sites. Aside from PRT, the specimens were obtained opportunistically with other research being conducted at the Dauphin Island Sea Lab (DISL), so collection methods varied depending on the species. RSN were caught by longlining from artificial reef structures offshore of Biloxi, MS, while GSN were collected by divers on SCUBA from natural gas platforms offshore from Gulf Shores, AL. Both snapper species were collected under federal collection permit F/SER24:RM and euthanized according to IACUC protocol 1562086. LNF were collected from artificial reef structures by recreational fishermen on SCUBA. SHP were captured live on hook-and-line from Mobile Bay, AL and Mississippi Sound, AL and donated by researchers after temporary holding at the DISL wet-lab facility with livers harvested immediately following euthanasia (IACUC protocols 1442738 and 1380906). PRT were collected using shallow seagrass trawls from St. Andrew Bay, FL according to a Special Activities License SAL-18-1230-SR and IACUC 1384468. In all cases, livers were dissected from fish immediately after euthanasia and flash-frozen in liquid N2. All livers were stored at −80 °C until microsome preparation.
2.3. Preparation of microsomes from northern Gulf of Mexico fish
Hepatic microsomes were prepared according to Ivanova et al. (2014). All preparation steps were carried out on ice, unless otherwise stated. Frozen liver tissue was added to approximately 5 vol (v/w) of ice-cold preparation buffer (0.1 M KPO4, pH 7.5) and manually homogenized using a Potter-Elvehjem tissue grinder (Sigma Aldrich, St. Louis, MO, USA). Homogenates were separated using ultracentrifugation (SW41Ti; Beckman Instruments, Palo Alto, CA, USA) at 4 °C, with steps at 16,000 g for 30 min and then at 100,000 g for 1 h to remove cellular debris and isolate the microsomal fractions, respectively. Microsomes were resolubilized in preparation buffer and stored at −80 °C until use in characterization and metabolism assays.
The prepared fish liver microsomes were characterized with respect to their microsomal protein concentrations according to the Lowry method (Bio-Rad, DC Protein Kit, Hercules, CA, USA), recovery indices, as well as enzyme activities related to major human CYP enzyme families (CYP1A, CYP2C, CYP2D, CYP2E, and CYP3A), as previously reported (Gwinn et al., 2021). Duplicate batches of RSN and GSN microsomes (denoted as RSN1 and 2 and GSN1 and 2, respectively) were prepared to assess potential inter-batch variability arising from the microsome preparation process. Information on the number and sex of specimens and combined liver mass included in each batch, the microsomal protein concentrations, and preparation yields is provided in Table S1, while details on the CYP characterizations may be found in Gwinn et al. (2021).
2.4. In vitro phase I metabolism of BTX-2
The in vitro phase I metabolism of BTX-2 was investigated in HLM and the prepared fish liver microsomes in incubations with CYP-specific cofactors. The reaction mixture included a NADPH regeneration system (0.91 mM NADPH, 0.83 mM NADP+, 19.4 mM glucose 6-phosphate, 9 mM MgCl2 × 6H2O dissolved in 0.05 M HEPES buffer, adjusted to pH 7.4), 1 U mL−1 glucose-6-phosphate dehydrogenase, and 2 mg mL−1 microsomal protein in a total volume of 0.5 mL. While previous studies have advised microsomal protein concentrations around 0.5 mg mL−1 (Jia and Liu, 2007), the authors have successfully used concentrations of 2 mg mL−1 in similar biotransformation assays to study the metabolism of other biotoxins in microsomes from a range of species, including humans, rodents, birds, and fish (e.g., Fæste et al., 2011; Ivanova et al., 2011; Ivanova et al., 2014; Johny et al., 2020), including the fish microsomes used in this study (Gwinn et al., 2021). The reaction mixture was pre-incubated for 2 min in a shaking water bath maintained at 37 °C for HLM or 25 °C for fish microsomes, as previously described (Gwinn et al., 2021). The metabolism reactions were initiated by adding 1.75 μL of a BTX-2 stock solution dissolved in 80% MeOH (v/v) at a concentration of 1.29 μg μL−1 (1.43 mM), resulting in a start concentration of 5 μM BTX-2 after dilution in the assay. After the addition of BTX-2, samples were capped and incubated for up to 1 h. Aliquots (70 μL) were withdrawn at 0, 5, 10, 15, 30, and 60 min, and reactions were terminated by immediately adding equal volumes of ice cold 100% MeCN. All samples were kept on ice until centrifugation at 20,000×g for 10 min at 4 °C to precipitate proteins, after which the supernatant was transferred to a 0.22 μm nylon Costar Spin-X filter (Corning Life Sciences, Corning, NY, USA) and centrifuged at 20,000×g for 1 min at 4 °C. The filtrates were collected in 300 μL fixed-insert HPLC vials (Thermo Fisher Scientific) and stored at −20 °C until analysis by ultra-high-performance liquid-chromatography high-resolution mass spectrometry (UHPLC-HRMS), as described below. Incubations were performed in duplicate for each batch of microsomes except for those with HLM and L. griseus (GSN2), for which a total of six replicates were performed each, as well as L. campechanus (RSN1), for which five replicates were performed.
Concentration optimization experiments were performed using both HLM and RSN1 to compare assay performance and linearity with start concentrations of 1, 5, and 10 μM BTX-2. Based on the results (Fig. S1), a start concentration of 5 μM BTX-2 was selected to maintain the ecological relevance of our results while compromising between substrate depletion and detectability of potential BTX metabolites. Assay conditions were further optimized during single-replicate incubations with HLM, in which reactions were initiated by the addition of microsomes rather than by substrate addition or by extending the incubation time to 90 min with aliquot withdrawal at 0, 15, 30, 60, and 90 min. However, since no relevant differences with respect to BTX-2 depletion or formation of the metabolites BTX-3, BTX-9, or BTX-B5 were observed (Fig. S2), a standard 60 min-assay was chosen for all subsequent incubations. Finally, duplicate control incubations were performed using HLM and RSN1 by replacing either BTX-2 or the NADPH regeneration system with equivalent volumes of HEPES buffer (Fig. S3). Controls without microsomes were evaluated to determine BTX-2 stability in the incubation buffer by preliminary deactivation of the reaction mixture in 50% MeCN, which was found suitable for testing the microsomal dependency of BTX-2 metabolism (Fig. S4).
2.5. UHPLC-HRMS(/MS) analysis of BTX-2 and potential phase I metabolites
UHPLC-HRMS(/MS) was applied to aid in the untargeted detection and structural characterization of known and novel phase I BTX-2 metabolites. Compared with methods previously used to study BTX metabolites in fish, the high mass accuracy of UHPLC-HRMS offers the advantage of unequaled resolving power and enables unambiguous determination of elemental compositions. The UHPLC-HRMS(/MS) analyses were performed using a Vanquish Horizon UHPLC instrument (Thermo Fisher Scientific) connected to a Q-Exactive mass spectrometer (Thermo Fisher Scientific), equipped with a HESI-II heated electrospray interface (ESI). Incubation sample aliquots were maintained at 20 °C in the UHPLC autosampler and chromatographed on a 100 mm × 2.1 mm i.d. Kinetex F5 column (1.7 μm, Phenomenex, Torrance, CA, USA) maintained at 20 °C. The capillary and probe heater temperatures were maintained at 270 °C and 300 °C, respectively. Compounds were eluted by a linear gradient using the mobile phases A (0.1% formic acid in MeCN/water, 5:95, v/v) and B (0.1% formic acid in MeCN/water, 95:5, v/v), where B was increased from 30% to 60% over 15 min, followed by flushing of the column at 99% B for 2 min, and then re-equilibration at 30% B for 3 min. The total run time was 20 min. For full scan data acquisition (FullMS), the mass spectrometer was set to scan mode in the mass range 750–1250 m/z with a mass resolution of 70,000 (at 200 m/z) in positive ionization mode, the AGC target was set to 1 × 106, and the maximum inject time was set to 256 ms. Important interface parameters included a spray voltage of 3.5 kV, a probe temperature of 300 °C, a sheath gas flow of 35 units, an auxiliary gas flow of 10 units and a transfer capillary temperature of 270 °C. Basic instrument maintenance, including mass calibration, cleaning of UHPLC–HRMS interface components and test of instrument response was performed at least on a weekly basis using Thermo Scientific Pierce calibration solutions.
For unambiguous identification and characterization, BTX standards (BTX-2, BTX-3, and BTX-B5) and known BTX metabolites with reported molecular masses and elemental formulae were analyzed by HRMS/MS in parallel reaction monitoring (PRM) mode using an isolation width of 1.5 m/z and a collision energy for higher-energy collision dissociation (HCD) of 35 eV. The mass resolution during PRM was set to 70,000 (at m/z 200), the automatic gain control target was 3 × 105, and the maximum injection time was 100 ms. All other interface settings were identical to FullMS. The UHPLC-HRMS(/MS) data were analyzed using Xcalibur software (v4.2; Thermo Fisher Scientific). Raw peak areas (Tables S2–S3) were calculated from extracted ion chromatograms (XIC) using the exact mass of individual compounds within a mass accuracy (Δm) of ±5 ppm in an expected retention time (RT) window of 30 s. The chemical formulae of compounds and fragments reported in the results are listed with the corresponding Δm (in ppm) of observed m/z values.
To search for potential novel phase I metabolites of BTX-2, FullMS data from replicate RSN1 and HLM incubations were processed using MZmine 2 (v2.53, downloaded from http://mzmine.github.io/). The methods and parameters used for MZmine2 analyses are detailed in Table S4. Peak lists generated during MZmine2 analysis were refined by considering only those peaks which met the following criteria: (1) showed at least 10-fold growth between discrete time points during the incubation (0–10, 0–30, 30–60, and/or 0–60 min); (2) the tenths place value of the observed m/z was either 4 or 5, consistent with minor modification of the BTX-2 substrate ([M+H]+, C50H71O14+, exact m/z 895.4838); (3) the observed m/z could be used to calculate a molecular formula within a mass accuracy (Δm) of 5 ppm with the elemental allowances near that of BTX-2 (C40–60, H60–80, O10–20); and (4) the RDBE was a half-integer value, consistent with the majority of ions produced by ESI. The FullMS data were also manually searched for potential metabolites with mass shifts expected for common phase I biotransformation reactions (e.g., oxidation, reduction, and hydrolysis reactions).
2.6. Statistical analyses and quality criteria
Substrate depletion rates were determined by exponential regression of BTX-2 peak area versus time curves of mean data from each incubation, where depletion was expressed as percent BTX-2 peak area relative to that at the incubation start (0 min). Metabolite formation data were likewise referred to the BTX-2 peak area at 0 min, which enabled comparison of production rates across species. The normalized metabolite formation curves were fitted using quadratic regression. For both substrate depletion and metabolite formation data, only regression curves producing R2 > 0.95 and including at least four time points were considered for subsequent comparisons across microsomes. Note that two of the incubation replicates were omitted from HLM (rep5) and RSN1 (rep1), because the BTX-2 depletion curves indicated insufficient mixing at the incubation start (e.g., higher peak areas at 5 min relative to 0 min time point) (Table S2). One GSN2 replicate (rep6) was omitted since it showed relatively low raw BTX-2 peak areas (Table S2) that suggested insufficient BTX-2 addition to the incubation mixture. Information on the substrate depletion and metabolite formation regression analyses is listed in Tables S2 and S3.
Total metabolite production throughout the incubation period was estimated as the area under the curve (AUC) of the peak area-to-incubation time graph. The AUC calculations were performed in R (v4.0.2) and were determined by integrating the fitted regression of each curve using the “integrate” function, where the number of subdivisions was set to 100 (v3.6.2; https://www.rdocumentation.org/packages/stats/versions/3.6.2/topics/integrate). Metabolite AUCs are shown in Table S3. For microsomal incubations with three or more replicates (HLM, RSN1, and GSN4), we tested for outlier AUC values using the interquartile range (IQR) method, where values more than 1.5 × IQR outside of Q1 and Q3 were considered outliers. Note that limits of detection (LOD) and quantification (LOQ) were not determined since we did not have standards for several of the compounds identified in this study or, especially in the case of novel metabolites, standards were unavailable.
2.7. GSH conjugation of BTX-2
Conjugation experiments were performed to verify GSH-conjugated metabolites observed during several microsomal incubations and to investigate whether they were formed by chemical or enzyme-catalyzed reaction. The conjugation experiments were performed by incubating 1.75 μL of BTX-2 stock solution in a total volume of 0.5 mL (resulting in a start concentration of 5 μM BTX-2) with either: (i) active RSN2 supplemented with 100 μM GSH; (ii) RSN2 deactivated by addition of 50% MeCN, supplemented with 100 μM GSH; or (iii) 100 μM GSH dissolved in 50% MeCN. For incubations including microsomes (i and ii), the RSN2 were adjusted to a total protein concentration of 2 mg mL−1 as in the metabolism assays. To prevent phase I biotransformation of BTX-2 during the GSH conjugation experiments, NADPH regeneration system was not included in the reaction mixture. Samples were collected from single-replicate incubations after 0, 5, 15, and 30 min, prepared for analysis, and analyzed by UHPLC-HRMS(/MS) as described above to monitor the depletion of BTX-2 and formation of BTX-2-GSH. Data were sum-normalized by expressing the peak areas of BTX-2 and BTX-2-GSH as percent of the summarized peak areas of the two compounds for each time point. Rates of BTX-2 depletion and BTX-2-GSH formation were then determined from normalized data by linear regression and compared across treatments.
3. Results
3.1. UHPLC-HRMS/MS of BTX standards
Detailed UHPLC-HRMS(/MS) analyses were performed on the BTX-2, BTX-3, and BTX-B5 standards to characterize their chromatographic properties, identify diagnostic product ions, and determine major fragmentation pathways for B-type BTXs. BTX-2 eluted at 11.4 min, compared with 8.7 and 9.4 min for BTX-3 and BTX-B5, respectively (Fig. S5), and the corresponding HRMS spectra for each peak were dominated by protonated ([M+H]+) molecules with nominal m/z 895 (C50H71O14+; Δm = 0.75), 897 (C50H73O14+; Δm = −0.20), and 911 (C50H71O15+; Δm = −0.07) (Fig. S5). Dehydrated ([M + H–H2O]+), sodium adducted ([M+Na]+), and potassium adducted ([M+K]+) ions were also observed (Δm ± 5 ppm), albeit with much lower relative intensities (Fig. S5). While previous studies have reported an abundance of sodiated molecules in the MS spectra of BTX compounds, these were reported to provide less reliable fragmentation spectra than their protonated counterparts (Hua and Cole, 2000), and these were not abundant in the HRMS spectra in this study. Therefore, the protonated molecules were targeted for each compound in the subsequent HRMS/MS analyses.
To characterize the structures of the BTX standards, HRMS/MS product ions were assigned in the same way as previously described for Caribbean ciguatoxins, another group of ladder-like polyether phycotoxins (Kryuchkov et al., 2020). Following this nomenclature, product ions were assigned both a letter and a number, where the letters p, q, and s are used to indicate the C–C or C–O bond cleavages during fragmentation, and numbers describe the number of intact or fragmented rings contained in the product ion (shown in inlay of Fig. 2). A prime symbol (‘) is used to differentiate fragments arising from the “head” (C-1 terminus) and “tail” sides of the molecule. Major head and tail fragments were confirmed by comparison of fragmentation spectra obtained from standards of BTX-2, -3, and -B5, which are distinguishable by modifications of the K-ring side chain by +2H and +O, respectively (Fig. 1).
Fig. 2.
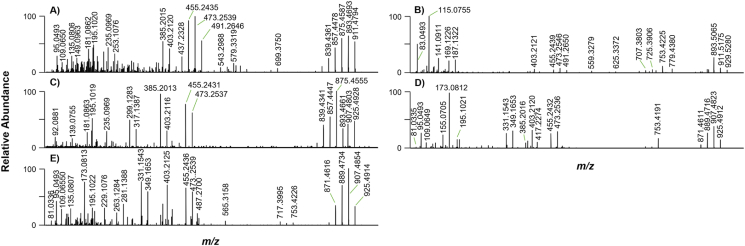
Proposed fragmentation scheme of BTX-2 (top panel) showing major product ions and associated water loss ions identified by comparison of HRMS/MS spectra (m/z 63–955) for BTX-2 (A), BTX-3 (B), and BTX-B5 (C) standards. Fragmentation was achieved using a collision energy of 35 eV. Spectra represent the average of multiple scans for each standard. Product ions are color-coded to indicate origin from either the head (blue) or tail (red) of the molecule. General nomenclature for MS/MS product ions of ladder-like polyether compounds used to predict BTX fragmentation pathways (Kryuchkov et al., 2020) is shown in the inlay.
A proposed fragmentation scheme for BTX-2 is provided in the upper panel of Fig. 2, based on the HRMS/MS spectra for the three BTX standards shown in Fig. 2A–C. While the molecular ions were relatively prominent in the HRMS/MS spectra of all three compounds, HCD resulted in substantial fragmentation yielding primary product ions derived from both the head and tail of each molecule. Additional product ions appeared to be derived secondarily by sequential loss of up to three water molecules (−H2O, Δ m/z = 18.0106) from primary fragments. In general, the most diagnostically valuable product ions appeared in the middle mass range between m/z 300 and 700, below which there was substantial noise in the spectra that made it more difficult to annotate fragments with diagnostic value.
3.1.1. Diagnostic fragments related to A-ring side (“head fragments”)
Several major head fragments associated with p8, p6, p5, q6, and s10 cleavages were detected across the HRMS/MS spectra for all three standards (Fig. 2). The m/z 491/473/455 and 421/403/385 ions were prominent in BTX-2 and -B5, showing relative intensities >30% (Fig. 2A–C). These were determined to be derived from the p6 and p5 series, respectively, with base ion formulae of C27H39O8+ (Δm = −1.5 to 0.25) and C23H33O7+ (Δm = −1.7 to 0.13). These fragments were previously reported as diagnostic for B-type BTXs (Hua and Cole, 2000; Wang et al., 2004; Abraham et al., 2006, 2012; Radwan and Ramsdell, 2006; Bottein et al., 2010; Shen et al., 2021). The spectrum of BTX-3 was dominated by m/z 643/625/607 ions arising from p8 (C36H51O10+; Δm = −1.5 to 0.20) as well as m/z 725/707 ions arising from s10 (C41H57O11+; Δm = −1.29 to −0.11) cleavage (Fig. 2B), with the latter being characteristic of BTX-3 fragmentation in previous studies (Abraham et al., 2012; Radwan and Ramsdell, 2006; Shen et al., 2021). The m/z 417/399 fragments derived from q6 (C24H35O7+; Δm = −1.83 to −0.16) were also detectable across all three standards (Fig. 2A–C), but with lower relative intensities.
Head fragments were also detected as product ions with nominal m/z 525, 333, and 249, which are visible in published MS/MS spectra of BTX-2 and -3, but were not previously annotated (Radwan and Ramsdell, 2006; Shen et al., 2021). The m/z 525 fragment was observed across the spectra for all three standards and identified as C31H41O7+ (Δm = −1.7 to 0.23), which we determined to arise from cleavage across the H-ring associated with the q6 series (Fig. 2). The m/z 333 fragment determined as C19H25O5+ (Δm = −1.4 to −0.45) is consistent with a BTX-2 tail fragment predicted to arise from the q′4 series opposite to q6 (Fig. 2A–C). However, observation of this product ion in the spectra of all three standards suggested that it could instead be related to the head side of the molecule, as shown in the fragmentation scheme in Fig. 2. The m/z 249 fragment was identified as C14H17O4+ (Δm = −0.95 to 0.66) and was similarly localized to the head of the molecules based on its occurrence in the HRMS/MS spectra of the three standards, although it was most prominent in BTX-2 and -B5; Fig. 2A–C). This fragment could arise by cleavage of the D-ring between the oxygen and methyl group followed by water loss to produce a fragment containing rings A–C (Fig. 2).
Additional head fragments were observed that did not correspond to the p, q, s nomenclature, including those with nominal m/z 779 and 559. The m/z 779 fragment was determined as C45H63O11+ (Δm = −1.1 to 3.7) arising from cleavage across the K-ring between C-38/-39 and the C–O bond adjacent to C-35 (Fig. 2). Its detection across the HRMS/MS spectra of all three standards (Fig. 2A–C) confirmed that it could be utilized as a diagnostic ion for B-type BTXs as previously suggested (Hua and Cole, 2000). The m/z 559 fragment identified as C32H47O8+ (Δm = −1.5 to 0.10) was seen across spectra (Fig. 2A–C) and is observable in previously published low-resolution MS/MS spectra without annotation (Hua and Cole, 2000; Radwan and Ramsdell, 2006; Abraham et al., 2012). A product ion with this composition could possibly be derived from breakage across the H-ring between C-26/-27 and the C–O bond adjacent to C-24, as shown in the Fig. 2 fragmentation scheme.
3.1.2. Diagnostic fragments related to K-ring side and C-39 side chain (“tail fragments”)
Consistent with their respective modifications in the K-ring side chain, the tail fragments of BTX-3 and -B5 were discernible by shifts of +2H and +O relative to BTX-2, respectively, although the latter was typically observed as –2H due to water loss. The most prominent tail fragments corresponded to the m/z 559 head fragment, but with the charge on the opposite side of the molecule (Fig. 2). For BTX-2, this tail fragment had a base ion formula of C18H25O6+, although it was predominantly detected as dehydration products with m/z 319/301(Δm = (m/z 321/303 and 317/299 for BTX-3 and -B5, respectively; Δm = −1.42 to 0.31) (Fig. 2A–C). This cleavage type was previously reported for 41,43-dihydro-BTX-B5, identified by Abraham et al. (2008) as a +2H product of BTX-B5 with m/z 913, in which the reduction was localized to the K-ring side chain.
The tail fragments associated with removal of the K-ring side chain between C-38/-39 and the C–O bond adjacent to C-39 (as shown in Fig. 2 and previously described by Hua and Cole, 2000), were relatively prominent in the HRMS/MS spectra of all three standards. This fragment had a base ion formula of C5H7O+ (Δm = −0.50) for BTX-2 and was observed at nominal m/z 83, compared with m/z 85 and 81 for BTX-3 and -B5, respectively (Fig. 2A–C). A product ion with nominal m/z 109 (C7H9O+; Δm = 0.08) was also observed in the spectrum of BTX-2 (Fig. 2A), which could arise from cleavage across the K-ring following the loss of the C-37 hydroxyl as water. This fragment showed an expected +2H shift to m/z 111 in BTX-3 (Fig. 2B), but was seen as m/z 109 in BTX-B5, suggesting that a second molecule of water had been lost from the carboxylic acid group prior to cleavage from the BTX-B5 skeleton.
Several dehydrated products arising from the p′2 and p′5 series were observed in the spectrum for BTX-2. These had base ion formulas of C13H17O4+ and C26H35O7+, respectively, and were predominantly seen as fragments with m/z 237/219 and 441/423 (Δm = 0.13–0.69) (Fig. 2A). Comparable +2H and –2H fragments were detectable in the spectra for BTX-3 and -B5, respectively, but these occurred at much lower relative intensities and were barely distinguishable from noise. Other previously reported tail fragments with nominal m/z 95, 125, 179, and 153/135 (Hua and Cole, 2000) were likewise observed in our analyses (Fig. 2A–C). However, we were unable to confirm their structural origin due to the substantial background noise in the lower m/z ranges of the spectra.
3.2. Identification and structural elucidation of phase I BTX-2 metabolites
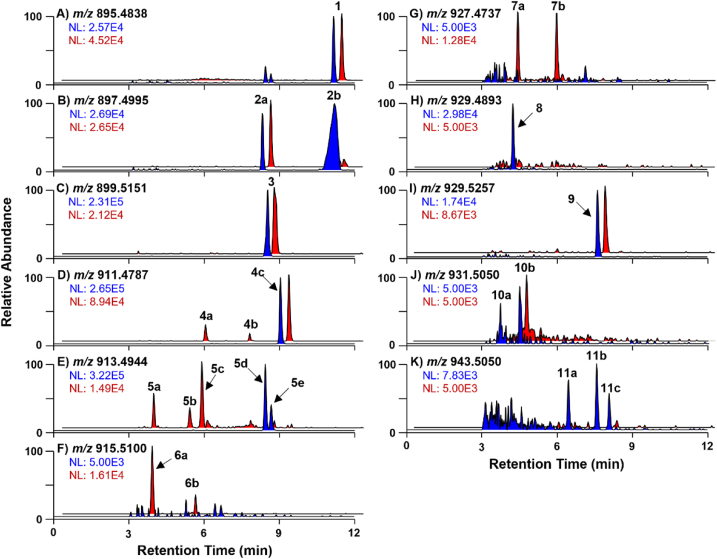
Potential biotransformation products of BTX-2 were determined by an untargeted approach using MZmine2 data processing software. The generated peak lists contained between 618 and 732 features, depending on the microsomal source from either HLM or RSN1. Full peak lists were refined to focus on m/z features with apparent relevance as BTX-2 metabolites based on the predicted structural formulae and relative formation during the incubation period. We thus obtained a final peak list consisting of six potential BTX-2 metabolites (Table S5) with m/z ratios corresponding to protonated molecules arising from phase I oxidation, reduction, and hydrolysis reactions. These included products shifted relative to BTX-2 (hereafter 1) by +H2 (m/z 897; 2), +H4 (m/z 899; 3), +O (m/z 911; 4), +H2O (m/z 913; 5), +H2O2 (m/z 929; 8), and +CH6O (m/z 929; 9). Structural isomers of these products, which showed identical m/z ratios (i.e., Δm within ±5 ppm) but eluted at different RT, were observed during manual inspection of FullMS data and were all considered as potential metabolites (2a and 2b; 4a to 4c; 5a to 5e). Additional biotransformation products, including +O2 (m/z 927; 7a and 7b), +H4O2 (m/z 931; 10a and 10b), and +CH4O2 (m/z 943; 11a to 11c), were identified by manual inspection of the FullMS data. Data were also searched for products shifted by +H4O relative to BTX-2 (m/z 915), since we had detected a corresponding compound after performing NaOH-catalyzed hydrolysis of the BTX-3 standard (Fig. S6), which resulted in observation of metabolites 6a and 6b. In total, we obtained a list of 22 potential phase I biotransformation products of BTX-2 (Fig. 4; Table 1). In subsequent HRMS/MS analyses, these compounds were investigated using PRM to aid in their tentative assignment and structural characterization by comparison of fragmentation patterns (Fig. 5, Fig. 6) with BTX-2, -3, and -B5 standards.
Fig. 4.
HRMS/MS spectra of protonated metabolites that were shifted relative to BTX-2 by: +H2(m/z897), leading to 2a (A) and 2b (B); +H4(m/z899), 3 (C); +O (m/z911), 4a (D), 4b (E), and 4c (F); +H2O (m/z913), 5b (G), 5c (H), 5d (I), and 5e (J). Data were obtained from samples collected after 60 min incubation of BTX-2 (5 μM, v/v, after dilution in the assay) with L. campechanus (RSN1) or human (HLM) liver microsomes. The m/z ranges are scaled to m/z 62–935 for m/z 897 and 899, and m/z 65–950 for m/z 911 and 913.
Table 1.
Molecular formulae, elemental changes relative to BTX-2,m/zof protonated ions, ring double bond equivalents (RDBE), retention times (RT), and tentative assignments for relevant compounds in this study. Listed accurate m/z values and RT for each compound were obtained from a sample collected after 60 min incubation of BTX-2 (5 μM, v/v, after dilution in the assay) with L. campechanus (RSN1), A. probatocephalus (SHP), or human (HLM) liver microsomes.
| Compound No. | Neutral Formula | Change relative to BTX-2 | Accurate [M+H]+m/z (Δm in ppm) | Ion RDBE | RTa (min) | Tentative Assignment |
|---|---|---|---|---|---|---|
| 1 | C50H70O14 | n/a | 895.4864 (+2.9) | 15.5 | 11.2 | BTX-2b |
| 2a | C50H72O14 | +H2 | 897.5030 (+3.9) | 14.5 | 8.3 | BTX-3b |
| 2b | C50H72O14 | +H2 | 897.5030 (+3.9) | 14.5 | 11.2 | 41,43-dihydro-BTX-2 |
| 3 | C50H74O14 | +H4 | 899.5183 (+3.5) | 13.5 | 8.5 | BTX-9 |
| 4a | C50H70O15 | +O | 911.4820 (+3.6) | 15.5 | 5.7 | Hydroxyl BTX-2 (C-9 or D-ring) |
| 4b | C50H70O15 | +O | 911.4828 (+4.4) | 15.5 | 7.5 | Hydroxyl BTX-2 (rings A-E) |
| 4c | C50H70O15 | +O | 911.4822 (+3.8) | 15.5 | 9.1 | BTX-B5b |
| 5a | C50H72O15 | +H2O | 913.4974 (+3.3) | 14.5 | 3.7 | |
| 5b | C50H72O15 | +H2O | 913.4980 (+3.9) | 14.5 | 5.1 | |
| 5c | C50H72O15 | +H2O | 913.4979 (+3.8) | 14.5 | 5.6 | A-ring hydrolyzed BTX-2b |
| 5d | C50H72O15 | +H2O | 913.4970 (+2.8) | 14.5 | 8.5 | 41,43-dihydro-BTX-B5c |
| 5e | C50H72O15 | +H2O | 913.4974 (+3.3) | 14.5 | 8.7 | 41,43-dihydro-BTX-B5c |
| 6a | C50H74O15 | +H4O | 915.5139 (+4.3) | 13.5 | 3.6 | A-ring hydrolyzed BTX-3b |
| 6b | C50H74O15 | +H4O | 915.5141 (+4.5) | 13.5 | 5.3 | |
| 7a | C50H70O16 | +O2 | 927.4769 (+3.5) | 15.5 | 4.1 | |
| 7b | C50H70O16 | +O2 | 927.4768 (+3.4) | 15.5 | 5.7 | |
| 8 | C50H72O16 | +H2O2 | 929.4920 (+2.9) | 14.5 | 4.2 | A-ring hydrolyzed BTX-B5b |
| 9 | C51H76O15 | +CH6O | 929.5295 (+4.1) | 13.5 | 7.6 | O-methylated BTX-2 |
| 10a | C50H74O16 | +H4O2 | 931.5090 (+4.3) | 13.5 | 3.7 | |
| 10b | C50H74O16 | +H4O2 | 931.5091 (+4.5) | 13.5 | 4.5 | |
| 11a | C51H74O16 | +CH4O2 | 943.5083 (+3.5) | 14.5 | 6.5 | |
| 11b | C51H74O16 | +CH4O2 | 943.5081 (+3.3) | 14.5 | 7.6 | |
| 11c | C51H74O16 | +CH4O2 | 943.5082 (+3.4) | 14.5 | 8.1 | |
| 12 | C60H87N3 O20S | +C10H17N3O6S | 1202.5654 (−1.9) | 18.5 | 4.2 | BTX-2-GSH |
| 13 | C60H89N3 O20S | +C10H19N3O6S | 1204.5820 (−1.1) | 17.5 | 3.5 | BTX-3-GSH |
Note that the RT of standards in UHPLC-HRMS(/MS) standard characterizations (Fig. S5) and those observed in incubation samples are shifted slightly due to instrumental variability across analyses as well as potential effects of the different matrices of incubation samples and standard solutions.
Identities confirmed by comparison with BTX-2, BTX-3, BTX-B5 reference standards and corresponding A-ring hydrolyzed standards.
Metabolites 5d and 5e likely represent a pair of diastereoisomers based on nearly identical RT and HRMS/MS spectra.
Fig. 5.
HRMS/MS spectra for the protonated metabolites that were shifted +H2O2(relative to BTX-2) withm/z929, 8 (A); +CH6O metabolites, also withm/z929, 9 (B); and +CH4O2metabolites withm/z943, 11a (C), 11b (D), and 11c (E). Data were obtained from samples collected after 60 min incubation of BTX-2 (5 μM, v/v, after dilution in the assay) with L. campechanus (RSN1) or human (HLM) liver microsomes. The m/z ranges (x-axes) are scaled to m/z 65–965 and 65–980 for m/z 929 and 943 metabolites, respectively.
Fig. 6.
BTX-2 substrate depletion rates (min−1) measured for HLM and northern Gulf of Mexico fish microsomes. Depletion rates were determined by exponential regression of the substrate depletion versus time curves, where BTX-2 peak areas were expressed as percent relative to that at the incubation start (0 min). Data are shown for N. usta (PRT), L. campechanus (RSN 1,2), L. griseus (GSN 1,2), A. probatocephalus (SHP), P. volitans (LNF), and human (HLM) liver microsomes. Error bars show standard deviation of replicate incubations.
3.2.1. Reduced metabolites: BTX-3, 41,43-dihydro-BTX-2 and BTX-9
Metabolites 2a and 2b were both observed with nominal m/z 897 (C50H73O14+; Δ m = 3.9), resulting from a +H2 shift relative to 1 (Fig. 3B; Table 1). The observed m/z, RT, and HRMS/MS spectrum of 2a (Fig. 3, Fig. 4A) were identical with those of the BTX-3 standard (Figs. S5 and 2B), which supported its identification as BTX-3. The other +H2 metabolite 2b co-eluted as a broad peak with 1 and was observed at high signal intensity in HLM relative to RSN1 (Fig. 3B). Comparing the HRMS/MS spectrum of metabolite 2b with that of BTX-2 traced the hydrogenation to the C-39 sidechain of the molecule via a corresponding shift for several prominent product ions. For example, the major m/z 83 fragment related to the C-39 sidechain in 1 was observed at m/z 85 in 2b (Fig. 4B). Additional product ions supported the observation, such as the m/z 111 and 321/303 fragments from K- and H-ring cleavages, respectively (Fig. 4B), which showed the same +H2 shift relative to corresponding fragments of 1 (Fig. 2A). This evidence, together with a RT that was nearly identical to that of 1, allowed the annotation of 2b as 41,43-dihydro-BTX-2, which had been previously identified by Guo et al. (2010). The broad and poorly shaped chromatographic peak could be explained by possible on-column equilibration of the aldehyde and the corresponding vinyl alcohol.
Fig. 3.
Extracted ion UHPLC-HRMS chromatograms for protonated molecules (± 5 ppm), of BTX-2 substrate (A) and phase I metabolites with elemental change of +H2(B), +H4(C), +O (D), +H2O (E), +H4O (F), +O2(G), +H2O2(H), +CH6O (I), +H4O2(J), and +CH4O2(K). Data were obtained from aliquots taken after 60 min incubation with liver microsomes from human (HLM; blue) or L. campechanus (RSN1; red, vertically and horizontally offset) microsomes, except for m/z 915.5100, for which the fish data (red trace) were taken from incubation with A. probatocephalus (SHP) microsomes. Normalization levels (NL) (arbitrary units) for the chromatograms are indicated in the panel for each species and were constrained to a minimum value of 5.00E3 to reduce noise. Peaks are labelled with the compound ID numbers shown in Table 1.
Both HLM and RSN1 produced metabolite 3 detected at nominal m/z 899 (C50H75O14+; Δm = 3.5), corresponding to the addition of +H4 relative to 1 (Fig. 3C; Table 1). The HRMS/MS spectrum of 3 contained several diagnostic head fragments that were identical to those observed for 1, such as m/z 779, 559, 473/455, and 403 (Fig. 4C). By contrast, a pair of ions was observed with m/z 305/287 (the parent ion and water loss fragment of C18H25O4+; Δm = 0.87–0.80) (Fig. 4C), which are shifted by +H4 in comparison to corresponding fragments of 1 produced by cleavage across the H-ring, which supported that 3 was generated by hydrogenation of the C-39 sidechain. The C-39 sidechain fragment was observed at nominal m/z 85 (Fig. 4C), which corresponded to a +H2 mass shift relative to the same fragment in 1. However, the mechanism of the K-ring side chain cleavage is reportedly influenced by the reduction state of both the aldehyde moiety and 41,43-double bond (Hua and Cole, 2000). A major m/z 85 product ion is thus expected for BTX-9, in which the aldehyde functionality is reduced to an alcohol and the 41,43-double bond is saturated (Poli et al., 1986). We therefore concluded that the m/z 899 metabolite 3 was equivalent to BTX-9.
3.2.2. Metabolites from mono-oxidation, oxidation/reduction, or water addition
The metabolites 4a to 4c were observed at nominal m/z 911 (C50H71O15+; Δm = 3.6–4.4) (Fig. 3D; Table 1), corresponding to mono-oxygenation (+O) of 1. The most prominent of these in the HLM and RSN1 incubations was metabolite 4c, which was identified as BTX-B5 by comparison of its observed m/z, RT, and HRMS/MS spectrum (Fig. 4F) to that of the BTX-B5 standard (Figs. S5 and 2C). In 4c, the aldehyde functionality in 1 is oxidized to the corresponding carboxylic acid. For this molecule, the C-39 sidechain fragment was observed at m/z 81 (Fig. 4F), indicating that the product ion was generated by the same fragmentation mechanism as in 1 and 2a after an initial water loss from the carboxylic acid moiety. Observation of prominent p6 head fragments (m/z 491/473/455) and BTX-B5-like product ions (m/z 317/299), originating from cleavage of the H-ring and related to the higher-order rings including the C-39 sidechain, confirmed the assumed structure of 4c as BTX-B5.
While metabolite 4c was the only prominent +O product in HLM, incubations with RSN1 (as well as other fish microsome preparations, as described in Section 3.4) resulted in the production of two additional +O metabolites, 4a and 4b, which eluted at RT 5.7 and 7.5 min, respectively (Fig. 3D; Table 1). Compared with 4c, the lack of a prominent m/z 81 fragment in the HRMS/MS spectrum of 4a (Fig. 4D) suggested that cleavage of the C-39 sidechain was not a preferred fragmentation mechanism in this molecule. The spectrum was instead dominated by a fragment with m/z 221 determined as C12H13O4+ (Δm = 0.75), which could arise from an s3 cleavage following water loss from a nearby hydroxyl group, e.g., at C-9. Hydroxylation of the head-part of the molecule was also supported by the presence of ions related to p5 and p6 observed at m/z 401/383 and 489.2481 from the base ions C23H29O6+ (Δm = −0.91) and C27H37O8+ (Δm = −0.40), respectively, that were shifted –H2 relative to 1 due to additional water loss prior to ring opening and cleavage.
The HRMS/MS spectrum of metabolite 4b (Fig. 4E) showed the C-39 sidechain product ion at m/z 83, which was identical to the fragment observed for 1. Furthermore, the m/z 319/301 ions associated with cleavage of the H-ring were prominent, which assigned the oxygenation in 4b to rings A–I. Observation of p6 fragments shifted by –H2 relative to 1 (m/z 489/471) further localized the site of oxygenation to rings A–F, while the presence of the p5 head fragment (m/z 401) limited it to rings A–E. Additional diagnostic ions that could further identify the site of oxygenation were not observed. It is worth noting that BTX-6, the H-ring epoxide of BTX-2 originally identified by Chou et al. (1985) and reported in some BTX metabolism and fish studies (Radwan and Ramsdell, 2006; Naar et al., 2007), has an identical structural formula and mass as 4a to 4c. However, localization of the oxygenation in these metabolites to either the head rings anterior to the H-ring or to the sidechain tail excluded their identification as BTX-6. No additional metabolites with m/z 911 that could be identified as BTX-6 were observed in this study and, to our knowledge, other isomers of BTX-B5 and BTX-6 that could aid in identifying 4a and 4b have not been reported, indicating that these represent novel metabolites.
We observed five products (5a to 5e) with nominal m/z 913 (C50H73O15+; Δm = 2.8–3.9) (Fig. 3E; Table 1) corresponding to an elemental shift of +H2O relative to 1, resulting either from addition of water or a combination of oxidation and reduction reactions. Interestingly, the three earlier-eluting metabolites 5a–5c (RT 3.7–5.6 min) were primarily observed in RSN1 incubations, whereas the two later-eluting isomers 5d and 5e (RT 8.5 and 8.7 min) were only produced by HLM (Fig. 3E). Metabolite 5a occurred with relatively low abundance that was insufficient to obtain HRMS/MS spectra to aid in its identification. Metabolite 5b was identified as a BTX-3-type molecule based on the presence of a m/z 85 fragment in its HRMS/MS spectrum (Fig. 4G), indicating a –H2 reduction in the C-39 sidechain. This was further supported by the presence of a dominant m/z 741 product ion identified as C41H57O12+ (Δm = −2.0) corresponding to the m/z 725 fragment originating from s10 cleavage in 2a, but shifted by +O. However, the HRMS/MS spectrum of 5b did not reveal additional ions to aid in identifying the oxidation site. Metabolite 5c had a RT and HRMS/MS spectrum (Fig. 3, Fig. 4H) identical to A-ring hydrolyzed BTX-2 generated by treating 1 with base (Figs. S6 and S7), and was thus determined to result from hydrolysis of the A-ring lactone. The HRMS/MS spectra of 5c (and other A-ring hydrolysis products, as described below) were dominated by head fragments from p5 (m/z 403/385) and p6 (m/z 473/455) cleavages (Fig. S7).
The HRMS/MS spectra of the other two m/z 913 metabolites observed in HLM incubations, 5d and 5e, were practically identical (Fig. 4I and J), indicating a pair of stereoisomers. Their fragmentation patterns were also highly similar to that of 1. The only notable difference occurred in the tail fragment, observed at m/z 81 for both 5d and 5e, which was equivalent to 4c and indicated a BTX-B5-like compound. An MS/MS spectrum with high similarity to that of 1 could be expected if the 41,43-double bond in BTX-B5 was reduced. The reduction of the 41,43-double bond would also introduce a stereogenic center into the molecule, which could explain the observation of two closely-eluting metabolites with nearly identical HRMS/MS spectra. A metabolite fitting this description was previously identified in human urine as 41,43-dihydro-BTX-B5 (Abraham et al., 2008) and has also been reported as a major metabolite produced by human CYP enzymes (Guo et al., 2010).
Metabolites 6a and 6b were observed with nominal m/z 915 (C50H75O15+; Δm = 4.2–4.4) (Fig. 3F; Table 1), corresponding to addition of +H4O relative to 1. While 6a was prominent and only observed in RSN1 incubations, traces of 6b were visible in both RSN1 and HLM (Fig. 4F). Metabolite 6a was identified as A-ring hydrolyzed BTX-3 based on its observed m/z and RT, which corresponded to that of base-treated 2a (Fig. S6). However, the signal intensities of 6a were insufficient to obtain HRMS/MS spectra of reasonable quality to compare to that of the A-ring hydrolyzed BTX-3 reference material (Fig. S7; also shown in previously published spectra by Radwan and Ramsdell, 2006). Similarly, the signal intensities of 6b were too low to allow further structural characterization via confirmation of diagnostic fragments. However, as other m/z 915 isomers have not been previously reported, it is possible that this compound may also represent a novel metabolite.
3.2.3. Metabolites from multiple biotransformation reactions
Two +O2 metabolites, 7a and 7b, were observed in RSN1 incubations with nominal m/z 927 (C50H71O16+; Δm = 3.4–3.5) (Fig. 3G; Table 1). However, as these were of relatively low abundance and were practically absent in HLM, the signal intensities were not sufficient to obtain HRMS/MS spectra of good quality. To our knowledge, BTX congeners consistent with m/z 927 have not yet been identified in previous studies, indicating that these products may represent novel metabolites. Another product observed in HLM, metabolite 8, was observed at nominal m/z 929 (C50H73O16+; Δm = 2.9), corresponding to +H2O2 relative to 1 (Fig. 3H; Table 1). This molecule was found to be equivalent to A-ring hydrolyzed BTX-B5 by comparison of its observed m/z, RT, and HRMS/MS spectrum (Fig. 5A) with that of the base-treated reference material (Figs. S6 and S7). As with 5c, the HRMS/MS spectrum of 8 was dominated by p5 (m/z 403/385; Δm = −1.9 to −0.78) and p6 (m/z 473/455; Δm = −2.0 to −1.6) fragments. It is worth noting that 27,28-diol-BTX-2, a previously reported hydrolysis product generated using purified CYP enzymes, has an identical chemical composition and its HRMS/MS spectrum contained similar product ions to those shown in published low-resolution spectra (Radwan and Ramsdell, 2006). However, the relative ion abundances differed slightly and appeared more similar to those in the spectrum of the base-hydrolyzed BTX-B5 standard (Fig. S7), which helped support our proposed identity of 8.
Metabolite 9 was detected in both HLM and RSN1 at nominal m/z 929 (C51H77O15+; Δm = 4.1) (Fig. 3I; Table 1), indicating a +CH6O shift relative to 1. The additional carbon atom in 9 was supported by the composition of the corresponding sodiated ion in the FullMS spectrum (data not shown). The HRMS/MS spectrum of 9 (Fig. 5B) was very similar to that of 1 and merely exhibited relative differences in the intensities of the different ions. The prominent m/z 83 (Δm = −2.9) ion suggested the presence of a BTX-2-like C-39 sidechain, while the RT (7.6 min) suggested an intact A-ring. As methylations are relatively uncommon phase I biotransformation reactions (in contrast to demethylation reactions), we initially hypothesized that 9 could be an artefact. However, there was no evidence for such impunities in the BTX-2 standard (dissolved in 80% MeOH), nor did the base hydrolysis of 1 (which was performed in aqueous MeOH) result in methylated products. Thus, we believe that 9 was indeed the result of O-methylation catalyzed by S-adenosyl-l-methionine-dependent methyltransferases such as catechol-O-transferase. This annotation was additionally supported by the fact that we did not observe oxidized +CH2 shifted metabolites of 1. While methylated products have not yet been reported for BTX, indicating a potentially novel transferase-mediated pathway for these toxins, O-methylation is known to be involved in the metabolism of catechol estrogens, dietary bioflavonoids, and tea polyphenols (Zhu, 2002).
At least two early-eluting +H4O2 metabolites, 10a and 10b, were observed with nominal m/z 931 (C50H75O16+; Δm = 4.3–4.5) (Fig. 3J; Table 1). While both 10a and 10b were detected in HLM samples, only 10b was observed in RSN1 incubations. A metabolite with m/z 931 had previously been reported as 27,28-diol-BTX-3, a hydrolysis product forming in incubations with rat microsomes (Radwan and Ramsdell, 2006). However, as the two metabolites observed in our study were of low signal intensities that were not sufficient to obtain informative HRMS/MS data, their identities could not be confirmed further.
Metabolites 11a to 11c were observed with m/z 943 (C51H75O16+; Δm = 3.3–3.5), equivalent to the addition of +CH4O2 (Fig. 3K; Table 1), and were primarily observed in incubations with HLM. The spectrum of 11a was dominated by ions from the p5 (m/z 403/385) and p6 (m/z 473/455) series (Fig. 5C). Fragments with m/z 317/299 (Δm = 1.1–1.7) were also observed, corresponding to identical fragments in 4c arising from cleavage across the H-ring. This indicated 11a as being a BTX-5-type compound in which the aldehyde function was oxidized to a carboxylic acid. However, no other product ions were present that could support the site of this suspected modification. The spectra of the later-eluting metabolites 11b and 11c (Fig. 5D and E) were similar to each other, with 11c showing generally higher product ion intensities. A BTX-5-type structure was also most likely for these compounds given that the characteristic tail fragment was observed at nominal m/z 81 (Δm = 0.11–1.3), albeit with a low relative intensity in both spectra. Interestingly, the diagnostic H-ring tail fragments (m/z 319/301 in 1) were observed at m/z 349/331 from the base ion C19H25O6+ (Δm = 0.91–2.1) for 11b and 11c, which corresponded to a +CH2O shift relative to 1. Yet, several head fragments in 11b and 11c were identical to those observed for 1, including those from the p5 (m/z 491/473/455) and p6 (m/z 491/473/455) series, indicating that the O-methyl functionality may be located somewhere in rings H–K for these metabolites. The spectra of 11b and 11c were otherwise dominated by an ion with m/z 173 identified as C8H13O4+ (Δm = −2.6 to −2.1), although the origin of this fragment could not be determined. To our knowledge, BTX metabolites with m/z 943 have not been previously reported.
3.2.4. GSH conjugation of BTX-2
In addition to the phase I metabolites identified during the in vitro biotransformation experiments, we also observed potential GSH conjugates of 1 and 2a. The formation of these products was first detected during the control experiments. Incubation of RSN1 microsomes in the absence of the NADPH regeneration system resulted in substantial depletion of the BTX-2 substrate, with depletion rates similar to those observed in incubations replete with the cofactor (Fig. S3, panel A). Metabolites 2a, 3, and 4c were not formed in incubations where the NADPH regeneration system was excluded, indicating that they were likely produced by NADPH-dependent CYP enzymes (Fig. S3, panels B–D). Exploration of the FullMS data from these incubations revealed peaks eluting at RT 4.2 and 3.5 min which were comprised of ions with nominal m/z 1202 (C60H88O20N3S+; Δm = −1.9) and 1204 (C60H88O20N3S+; Δm = −1.9), respectively (Fig. S8). These were putatively identified as the protonated ions of BTX-2-GSH (12) and BTX-3-GSH (13) (Table 1), respectively, each consistent with the addition of GSH (+C10H17N3O6S). Using different MS conditions and instrumentation, the BTX-2-GSH ion was previously reported with m/z 1204 (Wang et al., 2004; Radwan and Ramsdell, 2006), similar to what we report for BTX-3-GSH here, so our data provide a minor correction for these BTX-GSH metabolites. We also searched FullMS data of HLM and RSN1 incubations for BTX-2-cysteine and its sulfoxide oxidation product (m/z 1018 and 1034, respectively), but these were not detected.
The observation of GSH-conjugated BTX metabolites was unexpected, considering that GSH had not been added as a cofactor in the reaction mixture. Moreover, microsome preparations typically do not contain cytosolic GST enzymes, and the distinct microsomal forms have a limited metabolic capacity relative to predominant CYP enzymes (Morgenstern et al., 1982; Song et al., 2015, 2018). We therefore assume that 12 was formed non-enzymatically by chemical reaction with endogenous GSH present in the microsome preparations, but monitoring the reaction kinetics indicated a CYP-mediation formation mechanism for 13. In the controls without NADPH regeneration system, 12 increased continually from the incubation start, with comparatively minimal production of 13 (Fig. S9). In NADPH replete incubations, however, the formation of 12 and 13 initially proceeded at similar rates during the first 5 min, after which 12 declined while 13 continued to increase (Fig. S9). Thus, we conclude that 12 was produced directly from 1 by chemical conjugation of the α,β-unsaturated aldehyde in the molecule tail and that 13 arose secondarily through a CYP-dependent reduction of 12.
The chemical GSH conjugation of BTX-2 was further tested by incubating 1 with an excess of GSH (100 μM) and comparing conjugate formation in incubations without microsomes or with active or deactivated RSN2. Without microsomes, the reaction of 1 with GSH resulted in the formation of 12, which confirmed the identity of the conjugated product observed in the control incubations. Interestingly, the formation rate of 12 in incubations without microsomes or with deactivated RSN2 was considerably higher (0.46 min−1) than that observed with active RSN2 (0.18 min−1) (Fig. S10). This indicated a primarily chemical GSH conjugation, since otherwise the formation rate would have been higher using the active microsomes.
3.3. Depletion of BTX-2 in vitro and comparison across species
Following the optimization of experimental BTX-2 concentrations and confirmation of first-order depletion kinetics (Fig. S1), the metabolic stability of 1 was determined in the liver microsomes of the northern Gulf of Mexico fish and in HLM. The BTX-2 substrate was depleted by 71.9–98.5% within 60 min in all microsomal preparations (Fig. S11), indicating widespread capacity for BTX-2 biotransformation among the species tested. Comparison of substrate depletion rates revealed substantial variability in microsomal capacities for BTX-2 metabolism, which were highest in HLM and PRT (−0.111 ± 0.020 min−1 and –0.102 ± 0.018 min −1, mean ± standard deviation, respectively) (Fig. 6). The other fish microsomes showed about three-fold lower depletion rates, averaging −0.035 ± 0.002 min−1 across species (Fig. 6). No outliers in BTX-2 depletion rates across replicate incubations were detected using the IQR method.
3.4. Comparison of in vitro BTX-2 phase I metabolite formation across species
After the (tentative) structure identification of BTX-2 metabolites, we aimed to compare metabolite production across the prepared northern Gulf of Mexico fish microsomes and HLM. To standardize metabolite data across species and replicate incubations, all metabolite peak areas were normalized to that of 1 at the incubation start (0 min) in the respective assay (Figs. S12 and S13). The resultant metabolite formation curves were used to determine AUC as a measure for the total metabolite production in individual microsomal incubations. We chose to use AUC in place of other approaches (e.g., initial formation rates) since AUC are less influenced by differences in the metabolite formation curve progression (e.g., linear, quadratic, asymptotic, delayed). In this respect, we felt that the AUC estimates provided a more reliable and comprehensive measure of the overall capacity for BTX-2 metabolite production in the microsomes tested. We acknowledge the possibility that the presumed absence of a metabolite in a given incubation may be caused by values below the LOD. However, due to the lack of standards for individual BTX metabolites identified in this study, it was impossible to characterize potential differences in ionization efficiencies or other properties that may affect compound detectability. Therefore, we did not attempt to extrapolate metabolite concentrations from the known amount of BTX-2 substrate. AUC values are intended only to compare relative formation of a given metabolite across microsomes, rather than across different metabolites.
Comparing metabolite AUC across the fish and human microsomes tested indicated an extensive capacity for the biotransformation of 1 with some notable differences in the formation of specific metabolites among species and taxa (Figs. S14 and S15; summarized in Fig. 7). For example, there was considerably higher production of the +H2 metabolite 2a in PRT incubations as compared to other microsomes (Fig. 7), with AUC values ranging between approx. three to 18-fold lower in other fish microsome incubations (Fig. S14). Metabolite 2b, however, showed substantially greater production by HLM than the fish microsomes (Fig. 7 and S14). Notably, while 2b grew asymptotically in most fish preparations, in HLM the metabolite reached a maximum after about 15 min before decreasing considerably throughout the remainder of the incubation (Fig. S12). This time course, coupled with similarly high production of 3 by HLM, support previous work identifying 41,43-dihydro-BTX-2 as an important intermediate in the formation of BTX-9 in humans (Guo et al., 2010). Mean AUC values of 4c were also higher in HLM than in fish microsomes (Fig. 7), but the data were biased by a disproportionally high estimate in one replicate, while other replicates showed AUC values more similar to those in fish microsomes (Fig. S14). It should be noted that, as with the BTX-2 depletion rates, no outliers were detected in metabolite AUC across replicate incubations using the IQR method.
Fig. 7.
Cross-species comparison of phase I BTX-2 metabolite production. Heatmaps (left) show relative mean AUC of each metabolite across northern Gulf of Mexico fish microsomes and HLM. Data are shown for N. usta (PRT), L. campechanus (RSN 1,2), L. griseus (GSN 1,2), A. probatocephalus (SHP), P. volitans (LNF), and human (HLM) liver microsomes. The range of mean AUC values is shown for each metabolite (right). Incubations for which metabolite AUC could not be determined are indicated in panels with an "X".
The +H4O, +CH6O, and +H4O2 metabolites (6a to 6b, 9, and 10b, respectively) were also produced in all fish microsomes and in HLM (Fig. 7), but showed no obvious differences in relative formation across species (Fig. S14 and S15). The GSH conjugates were also detected across species (Fig. S13), with BTX-2-GSH (12) peaking at 5 min in all incubations and thereafter declining to give rise to BTX-3-GSH (13). While AUC values were not estimated given the focus of this investigation on phase I BTX-2 metabolism, we did observe differences in the relative peak areas of 12 and the formation of 13 across species (Fig. S13 and S14), although this seemed to be driven by relatively high variability between replicates (data not shown).
The later-eluting +H2O products (5c to 5e), as well as the +H2O2, +H4O2, and +CH4O2 metabolites (8, 10a, and 11a to 11c, respectively), occurred almost exclusively in HLM incubations (Fig. 7, S14, and S15), except 5c that was also relatively prominent in RSN incubations. While traces of these metabolites were produced in some RSN and GSN incubations, in some cases only at the last time point (60 min), mean AUC values were between one and four orders of magnitude higher in HLM and they were not found at all in other fish incubations. Metabolite 10a was only detected in HLM, albeit with relatively low AUC values, suggesting that it could be a relatively minor metabolite compared with the other HLM-specific products.
By contrast, several BTX-2 metabolites (4a and 4b, 5a and 5b, and 7a and 7b) were more prevalent or detected only in incubations with the northern Gulf of Mexico fish microsomes, with minimal or no production by HLM (Fig. 7, S14, and S15). There was, however, considerable variation in the occurrence or relative formation of these metabolites across fish species. For example, the +O metabolites 4a and 4b occurred mainly in the two Lutjanid species, with higher production by red snapper microsomes (RSN1,2) than those from gray snapper (GSN1,2) (Fig. 7 and S14). Yet, the consistency of the data for replicate microsome preparations from these two species supported that the observed differences in metabolite production represented differences in metabolic capacities across species rather than experimental bias. The + H2O metabolite 5a was only detected in RSN1 and RSN2 incubations, although with considerably lower intensity than 4a and 4b, while its isomer 5b was formed by all the northern Gulf of Mexico fish microsomes (Fig. 7 and S14). As with 5a, the +O2 metabolites 7a and 7b were also only found in RSN1 and RSN2 incubations (Fig. 7 and S15), indicating that these products may be specific to red snapper, or that 7a and 7b could be formed secondarily from 5a by a combination of oxidation and reduction reactions. The absence of 4a and 4b, 5a and 5b, and 7a and 7b in HLM may suggest that they were produced by fish specific enzymes or more specifically for Lutjanids (excluding 5b).
4. Discussion
The recurrent blooms of Karenia spp. and prevalence of BTX-2 throughout the northern Gulf of Mexico pose a threat to marine food webs and people that may consume BTX-contaminated seafood (Flewelling et al., 2005; Watkins et al., 2008). While fish can be affected by the ichthyotoxic properties of BTX, they can also accumulate sublethal levels of the lipophilic toxins, which may be detected in fish for weeks following water-borne K. brevis exposure (Woofter et al., 2005). The composition of BTX congeners retained in fish tissues following exposure likely depends on the species-specific enzymatic biotransformation reactions of the K. brevis toxins, yet few studies to date have compared enzymatic processes and metabolite profiles across fish species. In this study, we used microsomes from relevant fish species and humans, which have seen more research effort, in a comparative approach to investigate the metabolic fate of BTX-2.
For these experiments, we selected BTX-2 as the major substrate since it is considered the most prevalent algal congener in K. brevis, and targeted a range of fish species crossing multiple trophic levels in the northern Gulf of Mexico where these blooms occur. We aimed to use ecologically relevant levels of BTX-2 exposure based on previously reported concentrations of BTX in seawater and fish during and following K. brevis events. For instance, BTX-2 and BTX-3 levels in seawater from Sarasota Bay, FL collected during a 1994 K. brevis bloom were reported at 60 μg/L (0.07 μM) and 5.7 μg/L (6.4 nM), respectively (Hua et al., 1996). Between 1998 and 2005, total BTX-3 equivalents of 0.1 μM or less were detected in seawater during the annual algal bloom in the same area (Pierce et al., 2005, 2008; Abraham et al., 2006). In contrast, livers of fish that had died from BTX exposure showed levels as high as 106.9 μM BTX-3 equivalents (Van Deventer et al., 2012), while fish experimentally exposed to BTX demonstrated no adverse effects upon accumulation of up to 2.5 μM BTX-3 equivalents (Naar et al., 2007). By comparison, the livers of healthy, wild-caught fish collected from a region with frequent Karenia blooms contained an average 1.8 μM BTX-3 equivalents (Naar et al., 2007). Considering these varied reports in prior studies, we tested BTX-2 concentrations of 1 to 10 μM (Fig. S1) in our in vitro metabolism assays, which were lower but in the same order of magnitude as in previous studies with human and rat microsomes, where starting concentrations ranged from 10 to 50 μM (Radwan and Ramsdell, 2006; Guo et al., 2010).
Following rapid depletion of BTX-2 substrate by all the fish and human microsomes tested, we detected a total of 22 phase I BTX-2 metabolites in fish microsomes and HLM (Fig. 3; Table 1), including products of reduction, oxidation, methylation, or a combination of biotransformation reactions as well as glutathione conjugates. Some of the observed metabolites had been previously observed in fish through studies using targeted analytical methods. For example, a previous study on the prey fish of dolphins (Tursiops truncatus) including Lagodon rhomboides (pinfish), Orthopristis chrysoptera (pigfish), Mugil cephalus (striped mullet), and Leiostomus xanthurus (spot) and another study on sharks and rays showed specific detection of BTX-2, -3, several hydrolysis products, and cysteine conjugates (Fire et al., 2008; Flewelling et al., 2010). Experimental exposure of planktivorous M. cephalus to K. brevis cultures led to the accumulation of BTX-2 > BTX-3 > BTX -6 > BTX -9, while L. rhomboides and Micropogonias undulates (Atlantic croaker) fed toxic shellfish accumulated greater proportions BTX-2 cysteine conjugates (Naar et al., 2007). This same study found a similar prevalence of cysteine conjugated metabolites in six species of wild-caught fish collected from St. Joseph Bay, Florida as well as fish recovered from dolphin stomach contents. However, based on the toxin profiles of the shellfish and K. brevis cultures fed during the experimental exposures, the authors suggested that the observed metabolites in fish were likely accumulated directly from their diets. Our data thus provide some of the first direct evidence that fish are capable of BTX metabolism, potentially leading to the production of toxins previously reported in fish tissues. However, as our experiments relied on in vitro metabolism in fish microsomes, more work is needed to determine the relative role of BTXs obtained through diet compared with those produced through biotransformation in natural settings.
Several of the metabolites observed in our study were produced across fish microsomes and HLM, indicating widespread capacity for their production across species and taxa (Fig. 7). Yet, there were some clear differences in the presence or relative production of metabolites between fish and human microsomes, as well as among different fish species. For example, incubation of BTX-2 with microsomes from L. campechanus (RSN) and L. griseus (GSN) led to greater production of metabolites 4a and 4b, 5a, and 7a and 7b (Fig. 7, S14, and S15), while the N. usta (PRT) showed comparatively high depletion of the BTX-2 substrate coupled with notably greater production of 2a (Fig. 7 and S14). These results suggest that herbivorous fish that have closer ecological proximity to toxigenic Karenia spp. may exhibit different metabolic adaptations for coping with BTX-2 exposure than fish occupying higher trophic levels. Previous studies have shown that experimental exposure of immature Scianops ocellatus (red drum) and mature Morone saxatilis (striped bass) to BTX-3 resulted in activation of CYP1A- and CYP2B-related enzyme activities as well as of GST pathways (Washburn et al., 1994, 1996). Radwan and Ramsdell (2006) previously used recombinantly expressed CYP proteins to show that CYP2D1 and CYP3A1 showed substantially higher rates of BTX-2 depletion than other CYP enzymes tested. Thus, our previous findings that the PRT and RSN1 had higher CYP2D6 and CYP3A4 than the other northern Gulf of Mexico fish tested (Gwinn et al., 2021) may help to explain the unique patterns of metabolite production observed in these species (Fig. 7). To further elucidate the role of these enzymes in BTX biotransformation leading to specific metabolites in fish, we plan to perform more targeted experiments aimed at reaction phenotyping, e.g., through use of recombinant CYP enzymes or specific enzyme inhibitors.
To our knowledge, there are only a couple of studies comparing BTX metabolite profiles across species. In addition to the study by Naar et al. (2007) comparing relative toxin compositions across planktivorous or omnivorous fish species described above, other studies have found differing levels and time courses of BTX-2, -3, -B1, and -B5 across wild-caught and experimentally-exposed shellfish (e.g., Austrovenus stutchburyi, New Zealand cockle; Perna canaliculus, greenshell mussel; Crassostrea gigas, Pacific oyster) (Ishida et al., 2004a, 2004b) Otherwise, much of the available information on BTX in fish and comparisons across species is limited to composite toxicity measurements – which have ranged from below detection limits to > 95,000 ng BTX equivalents g−1 tissue depending on the species, tissue type, and location/timing with K. brevis bloom events (Naar et al., 2007; Fire et al., 2008; Flewelling et al., 2010; Van Deventer et al., 2012) – but without determination of toxin profiles. Considering that some of the novel metabolites reported here appeared to be unique to fish, our data help to complete the complex picture of the metabolic fate of BTX-2 in marine ecosystems, including the biotransformation mechanisms potentially underlying differences in toxicity across species. Consistent with previous work on other xenobiotics (de la Torre and Farré, 2004; Turesky, 2005; Gwinn et al., 2021), the results presented here also suggest that extrapolation of BTX biotransformation data across taxa may not be possible and highlights the need for species- or taxa-specific characterization of toxin metabolism pathways.
The occurrence of GSH-conjugated metabolites (12 and 13) in all species tested (Fig. 7 and S13) pointed at the importance of chemical modifications in the metabolic fate of BTX-2 in marine ecosystems. Previous work has suggested that BTX occurred primarily in conjugated forms in fish, with relatively high concentrations of cysteine adducts and their sulfoxide products (Fire et al., 2008). Importantly, the conjugation of BTX-2 with GSH and cysteine has been shown to increase its polarity and facilitate renal elimination and lead to lower intrinsic potency (Radwan et al., 2005; Dechraoui et al., 2007). Although we did not detect cysteine conjugates in the present work, the GSH conjugates observed in our study could potentially serve as intermediates leading to further biotransformation (Radwan et al., 2005; Radwan and Ramsdell, 2006) and identification of a non-enzymatic pathway for BTX conjugation underscores the interplay of chemical and enzymatic processes potentially affecting toxin fate. Follow-up studies with competent primary hepatocytes or S9 liver fractions of the different fish species are planned to elucidate the full extent of GSH conjugation in northern Gulf of Mexico fish.
Identification of BTX biotransformation pathways and products is not only critical for biomonitoring, the structural modifications imparted during biotransformation can alter the toxin's physicochemical and toxicological properties. Due to a paucity of commercially available BTX standards, data on the acute oral toxicity and elimination are limited for the known suite of BTX congeners, with previous work focusing primarily on BTX-2 and BTX-3. However, prior studies in rodents have shown that BTX-3 has a 10-fold higher oral toxicity than BTX-2 (Baden and Mende, 1982). It has also been demonstrated that BTX-2 and BTX-3 follow different elimination kinetics, with BTX-2 being more rapidly eliminated than BTX-3 due to conjugation with GSH and formation of a cysteine conjugate that is more readily excreted (Radwan et al., 2005). Thus, the differences in relative metabolic capabilities for transforming BTX observed here (Fig. 7), which may potentially lead to divergent toxin profiles in fish, could confer differences in overall toxicity and BTX bioaccumulation.
5. Conclusion
In this study, we investigated the in vitro biotransformation of BTX-2 using liver microsomes of five ecologically relevant fish species collected from the northern Gulf of Mexico, as well as commercially available human liver microsomes to evaluate taxonomic differences. Our goal was to contribute to the identification and characterization of known and novel phase I metabolites of BTX-2, and specifically to expand the available data on BTX metabolites in fish. The data presented here confirm that fish have the capacity to produce a variety of metabolites resulting from phase I reactions (e.g., reductions, oxidations, methylations). While about half of the compounds observed in this study have been previously reported in contaminated fish and/or humans, the others represented novel compounds that, to our knowledge, have not previously been identified. The observed differences in metabolite production across the microsomes tested, including six of the 22 metabolites that occurred uniquely in fish compared with eight in humans, suggest that BTX-2 biotransformation varies across species and taxa. Formation of GSH conjugates also highlights the role of nonenzymatic reactions in BTX-2 fate. Some important next steps will include confirming the relevance and predictability of the in vitro BTX metabolite profiles in naturally-exposed fish. Future work focused on characterizing the toxic equivalencies and kinetic properties of BTX compounds, including the novel metabolites identified in this study, are also needed to improve risk analysis, policy, and guidance that protect marine wildlife and seafood consumers.
Funding
This work was funded in part by the National Science Foundation (NSF) Partnerships in International Research and Education (PIRE) Program (CiguaPIRE; 1743802 to A.R.) and contributes to the NSF and National Institute of Environmental Health Sciences (NIEHS) Centers for Oceans and Human Health (COHH): Greater Caribbean Center for Ciguatera Research (NSF: 1841811 and NIH: 1P01ES028949–01, A.R.). The Norwegian Veterinary Institute contributions were supported by a supplemental PIRE grant through the Research Council of Norway (Grant No. 279247 to S.U.). Partial travel support for J.K.G. was provided through a Student Training Exchange Opportunity award from the Society of Environmental Toxicology and Chemistry.
Ethical statement
Fish microsomes used in this study were sourced from animals that had been collected with approved scientific collection permits and were humanely euthanized following approved IACUC protocols from the University of South Alabama. Red and gray snapper were collected under federal collection permit F/SER24:RM and euthanized following methods outlines in IACUC protocol 1562086. Emerald parrotfish were collected under Florida Special Activities License SAL-18-1230-SR and euthanized according to IACUC protocol 1384468. Sheepshead were humanely euthanized according to IACUC protocols 1442738 and 1380906. The lionfish were collected and donated by recreational fishers.
CRediT authorship contribution statement
Jessica Kay Gwinn: Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Data curation, Conceptualization. Alison Robertson: Writing – review & editing, Visualization, Validation, Supervision, Resources, Project administration, Funding acquisition, Conceptualization. Lada Ivanova: Methodology, Investigation. Christiane Kruse Fæste: Writing – review & editing, Validation. Fedor Kryuchkov: Methodology, Investigation. Silvio Uhlig: Writing – review & editing, Validation, Supervision, Resources, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors would like to thank Clay Bennett, Sean Collins, Crystal Hightower, Edward Kim, Merritt McCall, Sean P. Powers (from the University of South Alabama), and Alex Rodriguez, Grant Lockridge, Capt. Tom Guoba Dottie Byron David, and Kenneth L. Heck, Jr. (Dauphin Island Sea Lab) for assistance with fish collections.
Handling Editor: Dr. Denise Tambourgi
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.toxcx.2023.100168.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abraham A., Plakas S.M., Wang Z., Jester E.L.E., El Said K.R., Granade H.R., Henry M.S., Blum P.C., Pierce R.H., Dickey R.W. Characterization of polar brevetoxin derivatives isolated from Karenia brevis cultures and natural blooms. Toxicon. 2006;48(1):104–115. doi: 10.1016/j.toxicon.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Abraham A., Plakas S.M., Flewelling L.J., El Said K.R., Jester E.L.E., Granade H.R., White K.D., Dickey R.W. Biomarkers of neurotoxic shellfish poisoning. Toxicon. 2008;52(2):237–245. doi: 10.1016/j.toxicon.2008.04.175. [DOI] [PubMed] [Google Scholar]
- Abraham A., Wang Y., El Said K.R., Plakas S.M. Characterization of brevetoxin metabolism in Karenia brevis bloom-exposed clams (Mercenaria sp.) by LC-MS/MS. Toxicon. 2012;60(6):1030–1040. doi: 10.1016/j.toxicon.2012.06.016. [DOI] [PubMed] [Google Scholar]
- ANSES . 2021. Opinion of the French Agency for Food, Environmental and Occupational Health & Safety of 2 March 2021 on the State of Knowledge on Brevetoxins in Shellfish, Data on Toxicity, Occurrence and Brevetoxin-Producing Microalgae. Request No 2020-SA-0020) [Google Scholar]
- Baden D.G., Mende T.J. Toxicity of two toxins from the Florida red tide marine dinoflagellate, Ptychodiscus brevis. Toxicon. 1982;20(2):457–461. doi: 10.1016/0041-0101(82)90009-5. [DOI] [PubMed] [Google Scholar]
- Baden D.G. Brevetoxins: unique polyether dinoflagellate toxins. Faseb. J. 1989;3(7):1807–1817. doi: 10.1096/fasebj.3.7.2565840. [DOI] [PubMed] [Google Scholar]
- Bottein M.Y.D., Fuquay J.M., Munday R., Selwood A.I., van Ginkel R., Miles C.O., Loader J.I., Wilkins A.L., Ramsdell J.S. Bioassay methods for detection of N-palmitoylbrevetoxin-B2 (BTX-B4) Toxicon. 2010;55(2–3):497–506. doi: 10.1016/j.toxicon.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Chou H.N., Shimizu Y., Van Duyne G., Clardy J. Isolation and structures of two new polycyclic ethers from Gymnodinium breve Davis (= Ptychodiscus brevis) Tetrahedron Lett. 1985;26(24):2865–2868. doi: 10.1016/s0040-4039(00)98857-9. [DOI] [Google Scholar]
- Dechraoui M.Y.B., Wang Z., Ramsdell J.S. Intrinsic potency of synthetically prepared brevetoxin cysteine metabolites BTX-B2 and desoxyBTX-B2. Toxicon. 2007;50(6):825–834. doi: 10.1016/j.toxicon.2007.06.013. [DOI] [PubMed] [Google Scholar]
- de la Torre R., Farré M. Neurotoxicity of MDMA (ecstasy): the limitations of scaling from animals to humans. Trends Pharmacol. Sci. 2004;25(10):505–508. doi: 10.1016/j.tips.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Errera R.M., Bourdelais A., Drennan M.A., Dodd E.B., Henrichs D.W., Campbell L. Variation in brevetoxin and brevenal content among clonal cultures of Karenia brevis may influence bloom toxicity. Toxicon. 2010;55(2–3):195–203. doi: 10.1016/j.toxicon.2009.07.013. [DOI] [PubMed] [Google Scholar]
- Fæste C.K., Ivanova L., Uhlig S. In vitro metabolism of the mycotoxin enniatin B in different species and cytochrome p450 enzyme phenotyping by chemical inhibitors. Drug Metab. Dispos. 2011;39(9):1768–1776. doi: 10.1124/dmd.111.039529. [DOI] [PubMed] [Google Scholar]
- Fire S.E., Flewelling L.J., Naar J., Twiner M.J., Henry M.S., Pierce R.H., Gannon D.P., Wang Z., Davidson L., Wells R.S. Prevalence of brevetoxins in prey fish of bottlenose dolphins in Sarasota Bay, Florida. Mar. Ecol. Prog. Ser. 2008;368:283–294. doi: 10.3354/meps07643. [DOI] [Google Scholar]
- Fire S.E., Flewelling L.J., Stolen M., Durden W.N., De Wit M., Spellman A.C., Wang Z. Brevetoxin-associated mass mortality event of bottlenose dolphins and manatees along the east coast of Florida, USA. Mar. Ecol. Prog. Ser. 2015;526:241–251. doi: 10.3354/meps11225. [DOI] [Google Scholar]
- Fleming L.E., Kirkpatrick B., Backer L.C., Walsh C.J., Clark J., Reich A., Hollenbeck J., Benson J., Sung Y., Naar J., Pierce R., Bourdelais A.J., Abraham W.M., Kirkpatrick G., Zaias J., Wanner A., Mendes E., Shalat S., Stephan W., Bean J., Watkins S., Clarke T., Byrne M., Baden D.G. Review of Florida red tide and human health effects. Harmful Algae. 2011;10(2):224–233. doi: 10.1016/j.hal.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flewelling L.J., Naar J.P., Abbott J.P., Baden D.G., Barros N., Bossart G.D., Bottein M.-Y.D., Hammond D.G., Haubold E.M., Heil C.A., Henry M.S., Jacocks H.M., Leighfield T.A., Pierce R.H., Pitchford T.D., Rommetl S.A., Scott P.S., Steidinger K.A., Truby E.W., Van Dolah F.M., Landsberg J.H. Red tides and marine mammal mortalities. Nature. 2005;435(7043):755–756. doi: 10.1038/nature435755a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flewelling L.J., Adams D.H., Naar J.P., Atwood K.E., Granholm A.A., O'Dea S.N., Landsberg J.H. Brevetoxins in sharks and rays (Chondrichthyes, Elasmobranchii) from Florida coastal waters. Mar. Biol. 2010;157:1937–1953. doi: 10.1007/s00227-010-1463-z. [DOI] [Google Scholar]
- Guo F., An T., Rein K.S. Human metabolites of brevetoxin PbTx-2: identification and confirmation of structure. Toxicon. 2010;56(4):648–651. doi: 10.1016/j.toxicon.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn J.K., Uhlig S., Ivanova L., Fæste C.K., Kryuchkov F., Robertson A. In vitro glucuronidation of Caribbean ciguatoxins in fish: first report of conjugative ciguatoxin metabolites. Chem. Res. Toxicol. 2021;34(8):1910–1925. doi: 10.1021/acs.chemrestox.1c00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hort V., Abadie E., Arnich N., Dechraoui Bottein M.Y., Amzil Z. Chemodiversity of brevetoxins and other potentially toxic metabolites produced by Karenia spp. and their metabolic products in marine organisms. Mar. Drugs. 2021;19(12):656. doi: 10.3390/md19120656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y., Lu W., Henry M.S., Pierce R.H., Cole R.B. On-line liquid chromatography-electrospray ionization mass spectrometry for determination of the brevetoxin profile in natural “red tide” algae blooms. J. Chromatogr. A. 1996;750(1–2):115–125. doi: 10.1016/0021-9673(96)00470-0. [DOI] [Google Scholar]
- Hua Y., Cole R.B. Electrospray ionization tandem mass spectrometry for structural elucidation of protonated brevetoxins in red tide algae. Anal. Chem. 2000;72(2):376–383. doi: 10.1021/ac990433o. [DOI] [PubMed] [Google Scholar]
- Ishida H., Nozawa A., Nukaya H., Rhodes L., McNabb P., Holland P.T., Tsuji K. Confirmation of brevetoxin metabolism in cockle, Austrovenus stutchburyi, and greenshell mussel, Perna canaliculus, associated with New Zealand neurotoxic shellfish poisoning, by controlled exposure to Karenia brevis culture. Toxicon. 2004;43(6):701–712. doi: 10.1016/j.toxicon.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Ishida H., Nozawa A., Nukaya H., Tsuji K. Comparative concentrations of brevetoxins PbTx-2, PbTx-3, BTX-B1 and BTX-B5 in cockle, Austrovenus stutchburyi, greenshell mussel, Perna canaliculus, and Pacific oyster, Crassostrea gigas, involved neurotoxic shellfish poisoning in New Zealand. Toxicon. 2004;43(7):779–789. doi: 10.1016/j.toxicon.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Ivanova L., Fæste C.K., Uhlig S. In vitro phase I metabolism of the depsipeptide enniatin B. Anal. Bioanal. Chem. 2011;400:2889–2901. doi: 10.1007/s00216-011-4964-9. [DOI] [PubMed] [Google Scholar]
- Ivanova L., Fæste C.K., Van Pamel E., Daeseleire E., Callebaut A., Uhlig S. Presence of enniatin B and its hepatic metabolites in plasma and liver samples from broilers and eggs from laying hens. World Mycotoxin J. 2014;7(2):167–175. doi: 10.3920/WMJ2013.1609. [DOI] [Google Scholar]
- Jia L., Liu X. The conduct of drug metabolism studies considered good practice (II): in vitro experiments. Curr. Drug Metabol. 2007;8(8):822–829. doi: 10.2174/138920007782798207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johny A., Ivanova L., Østbye T.K.K., Fæste C.K. Biotransformation of phytoestrogens from soy in enzymatically characterized liver microsomes and primary hepatocytes of Atlantic salmon. Ecotoxicol. Environ. Saf. 2020;197 doi: 10.1016/j.ecoenv.2020.110611. [DOI] [PubMed] [Google Scholar]
- Kryuchkov F., Robertson A., Miles C.O., Mudge E.M., Uhlig S. LC–HRMS and chemical derivatization strategies for the structure elucidation of Caribbean ciguatoxins: identification of C-CTX-3 and -4. Mar. Drugs. 2020;18(4):182. doi: 10.3390/md18040182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg J.H. The effects of harmful algal blooms on aquatic organisms. Rev. Fish. Sci. 2002;10(2):113–390. doi: 10.1080/20026491051695. [DOI] [Google Scholar]
- Lekan D.K., Tomas C.R. The brevetoxin and brevenal composition of three Karenia brevis clones at different salinities and nutrient conditions. Harmful Algae. 2010;9(1):39–47. doi: 10.1016/j.hal.2009.07.004. [DOI] [Google Scholar]
- Morgenstern R., Guthenberg C., Depierre J.W. Microsomal glutathione S-transferase: purification, initial characterization and demonstration that it is not identical to the cytosolic glutathione S-transferases A, B and C. Eur. J. Biochem. 1982;128(1):243–248. doi: 10.1111/j.1432-1033.1982.tb06958.x. [DOI] [PubMed] [Google Scholar]
- Morohashi A., Satake M., Murata K., Naoki H., Kaspar H.F., Yasumoto T. Brevetoxin B3, a new brevetoxin analog isolated from the greenshell mussel Perna canaliculus involved in neurotoxic shellfish poisoning in New Zealand. Tetrahedron Lett. 1995;36(49):8995–8998. doi: 10.1016/0040-4039(95)01969-O. [DOI] [Google Scholar]
- Morohashi A., Satake M., Naoki H., Kaspar H.F., Oshima Y., Yasumoto T. Brevetoxin B4 isolated from greenshell mussels Perna canaliculus, the major toxin involved in neurotoxic shellfish poisoning in New Zealand. Nat. Toxins. 1999;7(2):45–48. doi: 10.1002/(SICI)1522-7189(199903/04)7:2<45::AID-NT34>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Murata K., Satake M., Naoki H., Kaspar H.F., Yasumoto T. Isolation and structure of a new brevetoxin analog, brevetoxin B2, from greenshell mussels from New Zealand. Tetrahedron. 1998;54(5–6):735–742. doi: 10.1016/S0040-4020(97)10336-2. [DOI] [Google Scholar]
- Naar J.P., Flewelling L.J., Lenzi A., Abbott J.P., Granholm A., Jacocks H.M., Gannon D., Henry M., Pierce R., Baden D.G., Wolny J., Landsberg J.H. Brevetoxins, like ciguatoxins, are potent ichthyotoxic neurotoxins that accumulate in fish. Toxicon. 2007;50(5):707–723. doi: 10.1016/j.toxicon.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce R.H., Henry M.S., Blum P.C., Hamel S.L., Kirkpatrick B., Cheng Y.S., Zhou Y., Irvin C.M., Naar J., Weidner A., Fleming L.E., Backer L.C., Baden D.G. Brevetoxin composition in water and marine aerosol along a Florida beach: assessing potential human exposure to marine biotoxins. Harmful Algae. 2005;4(6):965–972. doi: 10.1016/j.hal.2004.11.004. [DOI] [Google Scholar]
- Pierce R., Henry M., Blum P. Brevetoxin abundance and composition during ECOHAB-Florida field monitoring cruises in the Gulf of Mexico. Continent. Shelf Res. 2008;28(1):45–58. doi: 10.1016/j.csr.2007.04.012. [DOI] [Google Scholar]
- Plakas S.M., El Said K.R., Jester E.L.E., Ray Granade H., Musser S.M., Dickey R.W. Confirmation of brevetoxin metabolism in the Eastern oyster (Crassostrea virginica) by controlled exposures to pure toxins and to Karenia brevis cultures. Toxicon. 2002;40(6):721–729. doi: 10.1016/S0041-0101(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Plakas S.M., Wang Z., El Said K.R., Jester E.L.E., Granade H.R., Flewelling L., Scott P., Dickey R.W. Brevetoxin metabolism and elimination in the Eastern oyster (Crassostrea virginica) after controlled exposures to Karenia brevis. Toxicon. 2004;44(6):677–685. doi: 10.1016/j.toxicon.2004.07.027. [DOI] [PubMed] [Google Scholar]
- Poli M.A., Mende T.J., Baden D.G. Brevetoxins, unique activators of voltage-sensitive sodium channels, bind to specific sites in rat brain synaptosomes. Mol. Pharmacol. 1986;30(2):129–135. [PubMed] [Google Scholar]
- Poli M.A., Templeton C.B., Thompson W.L., Hewetson J.F. Distribution and elimination of brevetoxin PbTx-3 in rats. Toxicon. 1990;28:903–910. doi: 10.1016/0041-0101(90)90020-8. [DOI] [PubMed] [Google Scholar]
- Poli M.A., Musser S.M., Dickey R.W., Eilers P.P., Hall S. Neurotoxic shellfish poisoning and brevetoxin metabolites: a case study from Florida. Toxicon. 2000;38(7):981–993. doi: 10.1016/S0041-0101(99)00191-9. [DOI] [PubMed] [Google Scholar]
- Radwan F.F.Y., Wang Z., Ramsdell J.S. Identification of a rapid detoxification mechanism for brevetoxin in rats. Toxicol. Sci. 2005;85(2):839–846. doi: 10.1093/toxsci/kfi138. [DOI] [PubMed] [Google Scholar]
- Radwan F.F.Y., Ramsdell J.S. Characterization of in vitro oxidative and conjugative metabolic pathways for brevetoxin (PbTx-2) Toxicol. Sci. 2006;89(1):57–65. doi: 10.1093/toxsci/kfj013. [DOI] [PubMed] [Google Scholar]
- Shen H., Song X., Zhang Y., Zhang P., Li J., Song W., Yu Z. Profiling of brevetoxin metabolites produced by Karenia brevis 165 based on liquid chromatography-mass spectrometry. Toxins. 2021;13(5):354. doi: 10.3390/toxins13050354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W., Yu L., Peng Z. Targeted label-free approach for quantification of epoxide hydrolase and glutathione transferases in microsomes. Anal. Biochem. 2015;478:8–13. doi: 10.1016/j.ab.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Song W., Yu L., Peng Z. Dataset from proteomic analysis of human liver, lung, kidney and intestine microsomes. Data Brief. 2018;18:831–834. doi: 10.1016/j.dib.2018.03.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tester P.A., Turner J.T., Shea D. Vectorial transport of toxins from the dinoflagellate Gymnodinium breve through copepods to fish. J. Plankton Res. 2000;22(1):47–62. doi: 10.1093/plankt/22.1.47. [DOI] [Google Scholar]
- Turesky R.J. Interspecies metabolism of heterocyclic aromatic amines and the uncertainties in extrapolation of animal toxicity data for human risk assessment. Mol. Nutr. Food Res. 2005;49(2):101–117. doi: 10.1002/mnfr.200400076. [DOI] [PubMed] [Google Scholar]
- Twiner M.J., Bottein Dechraoui M.Y., Wang Z., Mikulski C.M., Henry M.S., Pierce R.H., Doucette G.J. Extraction and analysis of lipophilic brevetoxins from the red tide dinoflagellate Karenia brevis. Anal. Biochem. 2007;369(1):128–135. doi: 10.1016/j.ab.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Twiner M.J., Flewelling L.J., Fire S.E., Bowen-Stevens S.R., Gaydos J.K., Johnson C.K., Landsberg J.H., Leighfield T.A., Mase-Guthrie B., Schwacke L., van Dolah F.M., Wang Z., Rowles T.K. Comparative analysis of three brevetoxin-associated bottlenose dolphin (Tursiops truncatus) mortality events in the Florida Panhandle region (USA) PLoS One. 2012;7(8) doi: 10.1371/journal.pone.0042974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deventer M., Atwood K., Vargo G.A., Flewelling L.J., Landsberg J.H., Naar J.P., Stanek D. Karenia brevis red tides and brevetoxin-contaminated fish: a high risk factor for Florida’s scavenging shorebirds? Bot. Mar. 2012;55(1):31–37. doi: 10.1515/BOT.2011.122. [DOI] [Google Scholar]
- Walker J.S., Shaver D.J., Stacy B.A., Flewelling L.J., Broadwater M.H., Wang Z. Brevetoxin exposure in sea turtles in south Texas (USA) during Karenia brevis red tide. Dis. Aquat. Org. 2018;127(2):145–150. doi: 10.3354/dao03194. [DOI] [PubMed] [Google Scholar]
- Wang Z., Plakas S.M., El Said K.R., Jester E.L.E., Granade H.R., Dickey R.W. LC/MS analysis of brevetoxin metabolites in the Eastern oyster (Crassostrea virginica) Toxicon. 2004;43(4):455–465. doi: 10.1016/j.toxicon.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Wang W., Hua Y., Wang G., Cole R.B. Characterization of rat liver microsomal and hepatocytal metabolites of brevetoxins by liquid chromatography-electrospray tandem mass spectrometry. Anal. Bioanal. Chem. 2005;383(1):67–75. doi: 10.1007/s00216-005-3323-0. [DOI] [PubMed] [Google Scholar]
- Washburn B.S., Baden D.G., Gassman N.J., Walsh P.J. Brevetoxin: tissue distribution and effect on cytochrome P450 enzymes in fish. Toxicon. 1994;32(7):799–805. doi: 10.1016/0041-0101(94)90005-1. [DOI] [PubMed] [Google Scholar]
- Washburn B.S., Vines C.A., Baden D.G., Hinton D.E., Walsh P.J. Differential effects of brevetoxin and β-naphthoflavone on xenobiotic metabolizing enzymes in striped bass (Morone saxatilis) Aquat. Toxicol. 1996;35(1):1–10. doi: 10.1016/0166–445X(95)00050-E. [DOI] [Google Scholar]
- Watkins S.M., Reich A., Fleming L.E., Hammond R. Neurotoxic shellfish poisoning. Mar. Drugs. 2008;6(3):431–455. doi: 10.3390/md20080021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woofter R.T., Brendtro K., Ramsdell J.S. Uptake and elimination of brevetoxin in blood of striped mullet (Mugil cephalus) after aqueous exposure to Karenia brevis. Environ. Health Perspect. 2005;113(1):11–16. doi: 10.1289/ehp.7274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B. Catechol-O-Methyltransferase (COMT)-mediated methylation metabolism of endogenous bioactive catechols and modulation by endobiotics and xenobiotics: importance in pathophysiology and pathogenesis. Curr. Drug Metabol. 2002;3(3):321–349. doi: 10.2174/1389200023337586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.