Summary
Distant metastasis remains the leading cause of high mortality in patients with non-small-cell lung cancer (NSCLC). DIRAS3 is a candidate tumor suppressor protein that is decreased in various tumors. However, the regulatory mechanism of DIRAS3 on metastasis of NSCLC remains unclear. Here, we found that DIRAS3 suppressed the migration of NSCLC cells. Besides, DIRAS3 stimulated the polyubiquitination of RAC1 and suppressed its protein expression. Furthermore, RNF19B, a member of the RBR E3 ubiquitin ligase family, was observed to be the E3 ligase involved in the DIRAS3-induced polyubiquitination of RAC1. DIRAS3 could promote the binding of RAC1 and RNF19B, thus enhancing the degradation of RAC1 by the ubiquitin-proteasome pathway. Finally, the DIRAS3-RNF19B-RAC1 axis was confirmed to be associated with the malignant progression of NSCLC. These findings may be beneficial for developing potential prognostic markers of NSCLC and may provide an effective treatment strategy.
Subject areas: Molecular biology, Cell biology, Biotechnology
Graphical abstract

Highlights
-
•
DIRAS3 inhibits the migration of NSCLC cells by downregulating RAC1
-
•
DIRAS3 stimulates RAC1 polyubiquitination
-
•
RNF19B is a potential E3 ubiquitin ligase of RAC1
-
•
The DIRAS3-RNF19B-RAC1 axis is associated with NSCLC malignant progression
Molecular biology; Cell biology; Biotechnology
Introduction
Lung cancer is one of the most common diseases threatening human health and the first leading cause of cancer-related death worldwide.1 As the most frequent type of lung cancer, non-small-cell lung cancer (NSCLC) accounts for more than 80% of all lung cancer diagnoses.2 Presently, distant metastasis remains the leading cause of high mortality, about 80% in patients with NSCLC.3
As a small GTPase of the RHO family, RAC1, similar to other small GTPases, is activated or inactivated by cycling between the GTP- and GDP-bound states.4,5 The most well-known function of RAC1 is to regulate cytoskeleton and gene expression, which is involved in processes such as cell adhesion, proliferation, and inflammatory responses.6,7 Therefore, overexpression or mutation of RAC1 is observed in a series of cancers.8,9,10,11 Currently, accumulating evidence has implicated that RAC1 plays a critical role in many aspects of cancer development, particularly in cancer cell invasion and metastasis.12,13,14 Based on this, RAC1 provides an attractive therapeutic target for combating several human cancers. In order to effectively target RAC1 clinically, an in-depth understanding of factors which modulate its expression is necessary. Various post-translational modifications have recently been shown to regulate the activity and function of RAC1 and other RHO GTPases, including lipidation, ubiquitination, phosphorylation, and adenylylation,15 and ubiquitination is the most crucial way to mediate RAC1 degradation among them.
DIRAS3 is also a small GTPase that belongs to the RAS superfamily sharing 60% amino acid homology with RAS.16 However, unlike RAS which can stimulate cell growth, DIRAS3 was observed to suppress cell proliferation. This growth-inhibitory function is attributed to the unique 34 amino acids N-terminal extension of DIRAS3, which is distinct from RAS.17 Notably, DIRAS3 is present in humans, primates, cattle, and pigs, but absent in mice or rats due to lineage divergence.18 As a tumor suppressor protein, DIRAS3 has been reported to inhibit cancer development in many aspects, such as inducing cytotoxicity, inhibiting cell growth and migration, as well as leading to autophagy and tumor dormancy.17,19,20,21 Consequently, the downregulation of DIRAS3 has been confirmed in breast, lung, thyroid, prostate, pancreatic, and hepatocellular carcinoma.22,23,24,25,26 It is worth noting that, like other members of the RAS family, DIRAS3 can attach to the inner leaflet of the plasma membrane to regulate the expression or activity of some small G proteins, such as RHOA and RAS.19,27
In this study, we found the mechanism of DIRAS3-induced downregulation of RAC1 protein and the effects of DIRAS3 on NSCLC cell migration. This will be beneficial in identifying the role of DIRAS3 as a potential prognostic biomarker as well as in modulating DIRAS3 as a therapeutic approach.
Results
The expression of DIRAS3 is clinically correlated with the malignant progression in patients with lung cancer
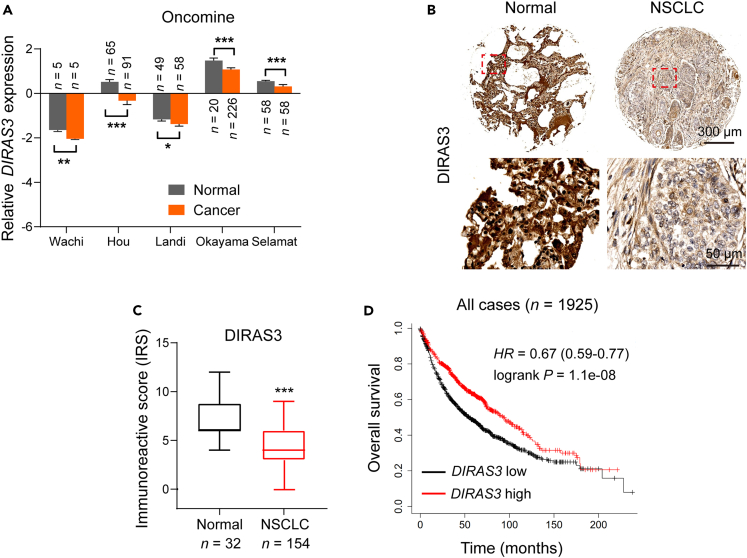
To explore the expression of DIRAS3 in patients with lung cancer, we analyzed 5 Oncomine datasets, namely Wachi, Hou, Landi, Okayama, and Selamat (Figure 1A). The data show that the mRNA levels of DIRAS3 were signiFIcantly lower in lung cancer tissues than that in normal tissues (Figure 1A). Furthermore, high-density tissue microarrays (TMAs) containing 186 normal and patients with NSCLC were used to examine the expression of DIRAS3 protein (Figures 1B and 1C). Similarly, the DIRAS3 protein level was also lower in the NSCLC cases compared with the normal cases (Figures 1B and 1C, and Table S1). Using the Kaplan-Meier method followed by the log rank test, we further conFIrmed that higher expression of DIRAS3 was correlated with higher overall survival (OS) in all cases (Figure 1D). Collectively, these data reveal that DIRAS3, a potential tumor suppressor protein, is clinically correlated with the malignant progression in patients with lung cancer.
Figure 1.
The expression of DIRAS3 is clinically correlated with the malignant progression in patients with lung cancer
(A) Boxplots of DIRAS3 mRNA levels determined from 5 Oncomine datasets, namely Wachi, Hou, Landi, Okayama, and Selamat (∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001; p-values were obtained using two-tailed Student’s t-tests).
(B) Representative IHC images showing the DIRAS3 expression in 32 normal and 154 NSCLC samples.
(C) Immunoreactivity scores (IRS) of DIRAS3 in 32 normal and 154 NSCLC samples were shown as a histogram (∗∗∗p < 0.001; p-value was calculated via two-tailed Student’s t test).
(D) Kaplan-Meier plots of the overall survival (OS) of all patients stratiFIed by DIRAS3 expression. The data were acquired from the Kaplan-Meier plotter database (p-value was obtained using the log rank test).
DIRAS3 inhibits the migration of NSCLC cells and downregulates the expression of RAC1
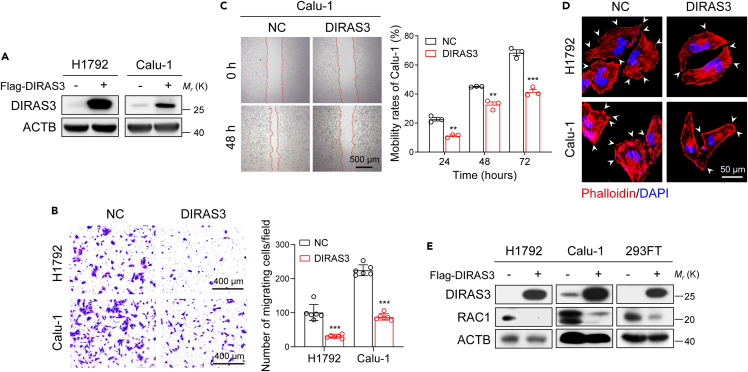
Next, DIRAS3 was overexpressed in NSCLC cells to investigate whether it would reduce the proliferation or migration ability of cells. Western blot assays confirmed the overexpression of DIRAS3 in H1792 and Calu-1 cells (Figure 2A). Figures 2B and 2C show that DIRAS3 overexpression restrained migration of H1792 and Calu-1 cells using transwell and wound-healing scratch assays. However, CCK-8 proliferation assays verified that cell proliferation abilities were not suppressed after DIRAS3 overexpression (Figures S1A and S1B). Then, we performed TRITC-phalloidin staining to label filamentous actin of H1792 and Calu-1 cells and found that the cell protrusions were reduced after DIRAS3 overexpression (Figure 2D). It has been reported that the small G protein RAC1-deficient cells were unable to form cellular lamellipodia.28,29 Indeed, as shown in Figures 2E, S1C, and S1D, overexpression of DIRAS3 markedly downregulated RAC1 protein levels without decreasing RNA levels in H1792, Calu-1, and 293FT cells. Taken together, these findings indicate that DIRAS3 inhibits the migration of NSCLC cells and downregulates the expression of the RAC1 protein.
Figure 2.
DIRAS3 inhibits migration of NSCLC cells and downregulates the expression of RAC1
(A) Overexpression of DIRAS3 in H1792 and Calu-1 cells. Cell lysates were analyzed by western blotting with antibodies against DIRAS3 and ACTB.
(B) Transwell assays of H1792 and Calu-1 cells that overexpressed DIRAS3. The statistics were performed using two-tailed Student’s t-tests, ∗∗∗p < 0.001.
(C) Wound-healing scratch assays of Calu-1 cells that overexpressed DIRAS3. Quantification of the data from three independent experiments using the ImageJ software (∗∗p < 0.01, ∗∗∗p < 0.001; p-values were obtained using two-tailed Student’s t-tests).
(D) TRITC-phalloidin staining of H1792 and Calu-1 cells that overexpressed DIRAS3. The white arrows denote the typical protrusions.
(E) Overexpression of DIRAS3 in H1792, Calu-1, and 293FT cells. Cell lysates were analyzed by western blotting with antibodies against DIRAS3, RAC1, and ACTB.
DIRAS3 interacts with RAC1 and stimulates its polyubiquitination
To identify the mechanism underlying the DIRAS3-induced reduction of RAC1 protein, the proteasome inhibitor MG132 and the lysosomal inhibitor E64D were used to treat Calu-1 cells that overexpressed DIRAS3 (Figure 3A). Western blot assays demonstrated that DIRAS3-mediated negative regulation of RAC1 could be significantly rescued by MG132 (Figure 3A). Furthermore, we observed that DIRAS3 increased the ubiquitination of RAC1 in 293FT cells using co-immunoprecipitation analysis (Figure 3B). Next, co-immunoprecipitation analysis indicated that DIRAS3 could bind to both exogenous and endogenous RAC1 protein (Figures 3C and 3D). Immunofluorescence further revealed the colocalization of endogenous DIRAS3 and RAC1 in Calu-1 cells (Figure 3E). Therefore, these results indicate that RAC1 was degraded by DIRAS3 through ubiquitin-proteasome pathway.
Figure 3.
DIRAS3 interacts with RAC1 and stimulates its polyubiquitination
(A) Calu-1 cells were transfected with Flag-DIRAS3 or control plasmid and cultured for 24 h before further incubating with MG132 (20 μM) or E64D (15 μM) for 6 h. Cell lysates were analyzed by western blotting with antibodies against Flag, RAC1, and ACTB.
(B) Co-immunoprecipitation analysis of the ubiquitination of RAC1 in 293FT cells co-transfected with Flag-DIRAS3 plasmid, Myc-RAC1 plasmid, and HA-Ub plasmid.
(C) 293FT cells were transfected with Myc-RAC1 and Flag-DIRAS3 plasmids and were cultured for 24 h before being further incubated with MG132 (20 μM) for 6 h. Then, the co-immunoprecipitation assays were carried out with Flag antibody and the co-eluted proteins were detected by western blot assays with Flag and Myc antibodies.
(D) Co-immunoprecipitation of endogenous RAC1 with DIRAS3 in H1299 cells.
(E) Immunofluorescence staining for detecting endogenous DIRAS3 and RAC1 expression in Calu-1 cells. The intensity profiles of DIRAS3 and RAC1 along the white line are plotted in the right panel.
RNF19B reduces the abundance of RAC1 and binds to RAC1 directly
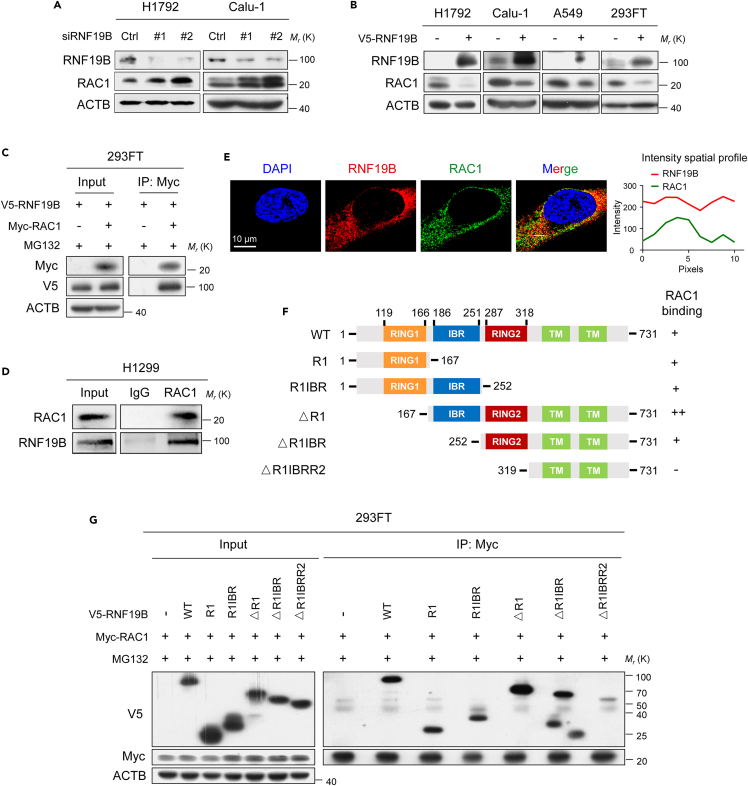
Given that DIRAS3 cannot catalyze ubiquitinylation, we considered the possible involvement of other E3 ubiquitin ligases. Mass spectrometry data from the BioGRID database revealed that DIRAS3 might interact with the E3 ubiquitin ligase RNF19B.30 We therefore speculate that RNF19B may mediate DIRAS3-induced polyubiquitination of RAC1. Various RNF E3 ubiquitin ligases, such as RNF5, RNF43, RNF47, RNF103, RNF130, RNF139, RNF149, RNF170, RNF180, and RNF19B, were knocked down with siRNAs (Figures S2A and 4A). Indeed, we found that RAC1 protein levels were increased after knocking down RNF19B rather than knocking down of other RNF proteins in H1792 and Calu-1 cells using western blot assays (Figures S2A and 4A). In contrast, overexpression of RNF19B markedly reduced RAC1 levels in H1792, Calu-1, A549, and 293FT cells (Figure 4B). Next, co-immunoprecipitation analysis indicated that RNF19B could bind to both exogenous and endogenous RAC1 protein (Figures 4C and 4D). Immunofluorescence further revealed the colocalization of endogenous RNF19B and RAC1 in Calu-1 cells (Figure 4E). To define which domain(s) of RNF19B is important for RAC1 binding, we constructed different truncated plasmids of RNF19B which sequences were predicted from the UniProt database (Figure 4F). 293FT cells were co-transfected with vectors for RAC1 and various regions of RNF19B, followed by co-immunoprecipitation assays (Figure 4G). As shown in Figures 4F and 4G, RAC1 bound to RING1, IBR, and RING2 domains of RNF19B to varying degrees, but it did not bind to RNF19B lacking these three domains. Furthermore, we observed that RNF19B overexpression inhibited the migration of Calu-1 cells using wound-healing assays (Figures S3A and S3B), while RNF19B knockdown promoted the migration of Calu-1 cells (Figures S3C and S3D). In summary, these data described previously demonstrate the binding of RAC1 to the RBR region (RING1, IBR, and RING2 domains) of RNF19B as well as the function of RNF19B to suppress NSCLC cell migration.
Figure 4.
RNF19B reduces the abundance of RAC1 and binds to RAC1 directly
(A) Knockdown of RNF19B expression by siRNAs in H1792 and Calu-1 cells. Cell lysates were analyzed by western blotting with antibodies against RNF19B, RAC1, and ACTB.
(B) Overexpression of RNF19B in H1792, Calu-1, A549, and 293FT cells. Cell lysates were analyzed by western blotting with antibodies against RNF19B, RAC1, and ACTB.
(C) 293FT cells were transfected with V5-RNF19B and Myc-RAC1 plasmids and were cultured for 24 h before being further incubated with MG132 (20 μM) for 6 h. Then, the co-immunoprecipitation assays were carried out with Myc antibody and the co-eluted proteins were detected by western blot assays with Myc and V5 antibodies.
(D) Co-immunoprecipitation of endogenous RAC1 with RNF19B in H1299 cells.
(E) Immunofluorescence staining for detecting endogenous RNF19B and RAC1 expression in Calu-1 cells. The intensity profiles of RNF19B and RAC1 along the white line are plotted in the right panel.
(F) The schematic representation of RNF19B domain structure and the summary of the relative binding affinity with RAC1 from 5 experimental sets.
(G) Co-immunoprecipitation analysis of the interaction between RAC1 and RNF19B or RNF19B truncation mutants in 293FT cells co-transfected with V5-RNF19B-WT plasmid or V5-RNF19B truncation mutant plasmids together with Myc-RAC1 plasmid.
RNF19B catalyzes K48-linked polyubiquitination of RAC1
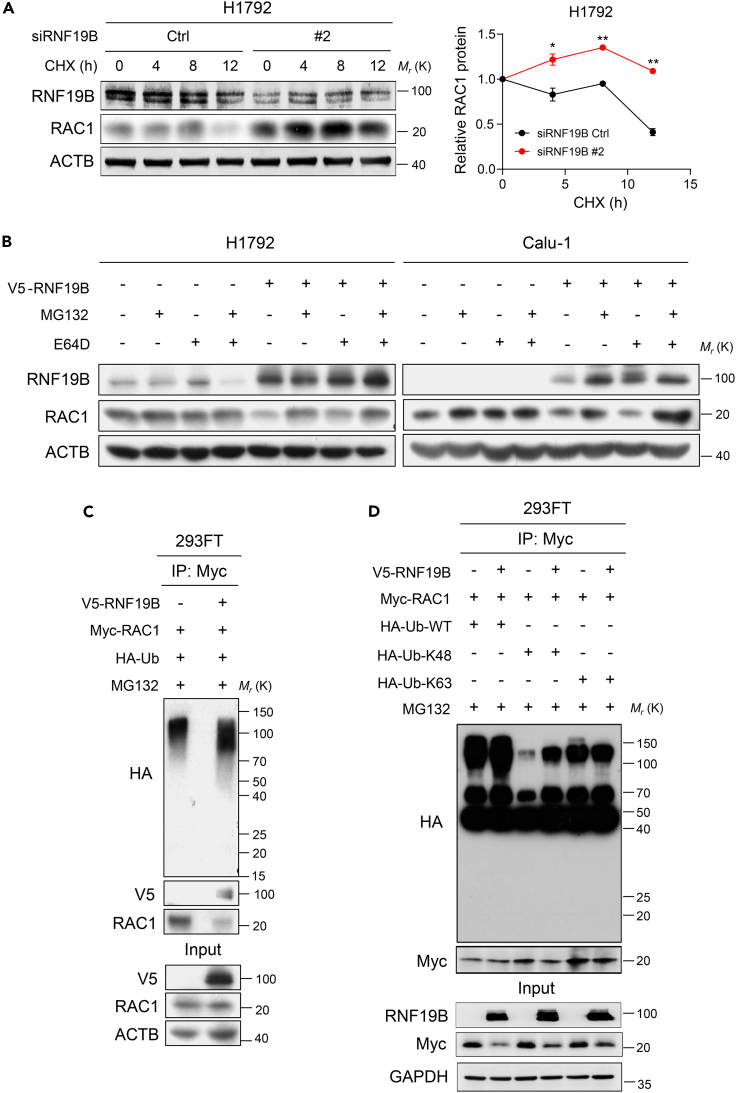
Since we identified a direct interaction between RNF19B and RAC1, we were further interested in defining whether RNF19B regulated the stability of RAC1 protein. H1792 cells were treated with 10 μg/mL cycloheximide at various times to inhibit new protein translation and examine the turnover of RAC1 protein (Figure 5A). Figure 5A shows increased stability of RAC1 protein after RNF19B knockdown compared with controls in H1792 cells. Then, MG132 and E64D were used to treat H1792 and Calu-1 cells that overexpressed RNF19B (Figure 5B). Western blot assays demonstrated that RNF19B-mediated reduction of RAC1 protein could be significantly rescued by MG132 (Figure 5B). Besides, we observed that RNF19B promoted the ubiquitination of RAC1 in 293FT cells (Figure 5C). Notably, co-immunoprecipitation analysis further confirmed that RNF19B catalyzed K48-linked polyubiquitination of RAC1, whereas it did not catalyze K6, K11, K27, K29, and K33-linked polyubiquitination of RAC1 (Figures 5D, S2B, and S2C). Together, these data suggest that RNF19B serves as a potential E3 ligase of RAC1 and catalyzes its K48-linked polyubiquitination.
Figure 5.
RNF19B catalyzes K48-linked polyubiquitination of RAC1
(A) Left: Knockdown of RNF19B expression by RNF19B-2 siRNA in H1792 cells in the presence or absence of cycloheximide (CHX) at 10 μg/mL for the indicated times (0, 4, 8, and 12 h). Cell lysates were analyzed by western blotting with antibodies against RNF19B, RAC1, and ACTB. Right: The band intensity of RAC1 was quantified by ImageJ software and plotted. This experiment was repeated three times independently with similar results (mean ± SEM, n = 3 independent experiments; ∗p < 0.05, ∗∗p < 0.01; p-values were obtained using two-tailed Student’s t-tests).
(B) H1792 and Calu-1 cells were transfected with V5-RNF19B or control plasmid and were cultured for 24 h before being further incubated with MG132 (20 μM) or E64D (15 μM) for 6 h. Cell lysates were analyzed by western blotting with antibodies against RNF19B, RAC1, and ACTB.
(C) Co-immunoprecipitation analysis of the ubiquitination of RAC1 in 293FT cells co-transfected with V5-RNF19B plasmid, Myc-RAC1 plasmid, and HA-Ub plasmid.
(D) Co-immunoprecipitation analysis of the ubiquitination of RAC1 in 293FT cells co-transfected with V5-RNF19B plasmid, Myc-RAC1 plasmid, and HA-Ub-WT, HA-Ub-K48, or HA-Ub-K63 plasmid.
The DIRAS3-RNF19B-RAC1 axis modulates migration of NSCLC cells
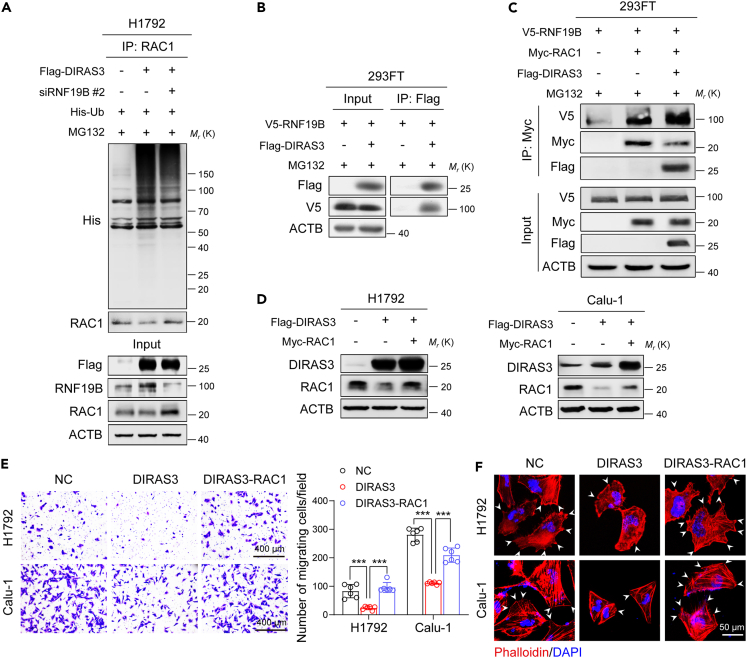
Considering the E3 ligase catalytic activity of RNF19B, we speculated that RNF19B might be involved in DIRAS3-induced RAC1 degradation. Figure 6A shows that the DIRAS3-induced upregulation of RAC1 ubiquitination was reversed after RNF19B knockdown by siRNA using co-immunoprecipitation assays in H1792 cells. Furthermore, DIRAS3 was confirmed to interact with RNF19B in 293FT cells (Figure 6B). Interestingly, co-immunoprecipitation analysis revealed that DIRAS3 promoted the interaction between RAC1 and RNF19B in 293FT cells compared with the interaction in control cells (Figure 6C), which might be the reason why DIRAS3 could induce ubiquitination and degradation of RAC1. Next, both DIRAS3 and RAC1 were overexpressed in H1792 and Calu-1 cells (Figure 6D). As shown in Figure 6E, the addition of RAC1 restored DIRAS3-induced suppression of cell migration in H1792 and Calu-1 cells using transwell assays. Besides, TRITC-phalloidin staining indicated that the cell protrusions were increased after DIRAS3 and RAC1 co-overexpression compared with that overexpressing DIRAS3 only (Figure 6F). In summary, these results reveal that the DIRAS3-RNF19B-RAC1 axis plays a vital role in modulating migration of NSCLC cells.
Figure 6.
The DIRAS3-RNF19B-RAC1 axis modulates migration of NSCLC cells
(A) Co-immunoprecipitation analysis of the ubiquitination of RAC1 in H1792 cells co-transfected with Flag-DIRAS3 plasmid, RNF19B-2 siRNA, and His-Ub plasmid.
(B) 293FT cells were transfected with V5-RNF19B and Flag-DIRAS3 plasmids and were cultured for 24 h before being further incubated with MG132 (20 μM) for 6 h. Then, the co-immunoprecipitation assays were carried out with Flag antibody and the co-eluted proteins were detected by western blot assays with Flag and V5 antibodies.
(C) 293FT cells were transfected with V5-RNF19B, Myc-RAC1, and Flag-DIRAS3 plasmids and were cultured for 24 h before being further incubated with MG132 (20 μM) for 6 h. Then, the co-immunoprecipitation assays were carried out with Myc antibody and the co-eluted proteins were detected by western blot assays with V5, Myc, and Flag antibodies.
(D) Overexpression of DIRAS3 or RAC1 in H1792 and Calu-1 cells. Cell lysates were analyzed by western blotting with antibodies against DIRAS3, RAC1, and ACTB.
(E) Transwell assays of H1792 and Calu-1 cells that overexpressed DIRAS3 and RAC1. The statistics were performed using two-tailed Student’s t-tests, ∗∗∗p < 0.001. (F) TRITC-phalloidin staining of H1792 and Calu-1 cells that overexpressed DIRAS3 and RAC1. The white arrows denote the typical protrusions.
The DIRAS3-RNF19B-RAC1 axis is associated with malignant progression in patients with lung cancer
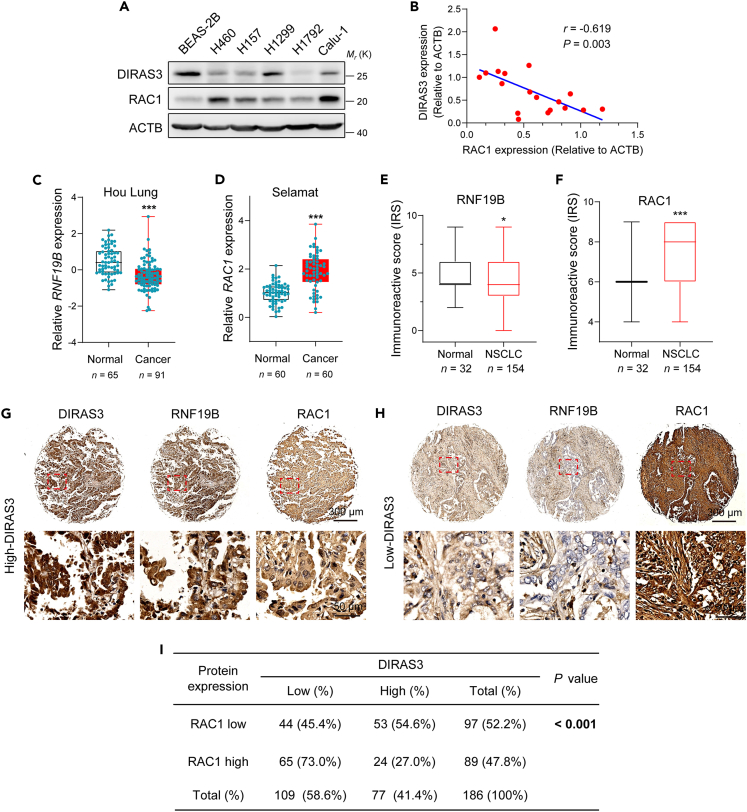
Next, western blot assays were performed to test the expression of DIRAS3 and RAC1 proteins in human bronchial epithelial cell BEAS-2B and a series of NSCLC cells, such as H460, H157, H1299, H1792, and Calu-1 (Figure 7A). Figure 7A shows that DIRAS3 expression was higher in BEAS-2B cell than that in NSCLC cells. In contrast, the expression of RAC1 was increased in NSCLC cells compared with that in BEAS-2B cells (Figure 7A). In addition, we observed that DIRAS3 expression tended to be negatively associated with the expression of RAC1 through the quantitative analysis of data from Figure 7A (Figure 7B). To determine whether our findings are clinically relevant, two Oncomine datasets, namely Hou Lung and Selamat, were used to examine the mRNA expression of RNF19B or RAC1 (Figures 7C and 7D). The data show that the mRNA levels of RNF19B were significantly lower in lung cancer tissues than that in normal tissues, while RAC1 mRNA levels were higher in lung cancer tissues compared with normal cases (Figures 7C and 7D). Using the Kaplan-Meier method followed by the log rank test, we further confirmed that higher expression of DIRAS3 was correlated with higher first-progression survival (Figure S4A) and post-progression survival (Figure S4B) in patients with lung cancer. However, higher expression of RAC1 was correlated with lower OS in patients with lung cancer (Figure S4C). Finally, TMAs that containing 186 normal and patients with NSCLC were used to examine the expression of DIRAS3, RNF19B, and RAC1 proteins (Figures 7E, 7F, 7G, and 7H). As shown in Figures 1C, 7E, and 7F, DIRAS3 and RNF19B protein levels were lower in the NSCLC cases compared with the normal cases, while RAC1 expression was higher in NSCLC tissues than in normal tissues. Besides, we found that the patients with high DIRAS3 and RNF19B levels often displayed low RAC1 expression and vice versa (Figures 7G and 7H). Chi-squared tests were then performed to determine whether the observed differences in the expression of the 3 proteins had statistical significance (Table S1). Indeed, there was an inverse correlation between DIRAS3 and RAC1 levels (Figure 7I). Collectively, these data suggest that the DIRAS3-RNF19B-RAC1 axis is associated with malignant progression in patients with lung cancer.
Figure 7.
The DIRAS3-RNF19B-RAC1 axis is associated with malignant progression in patients with lung cancer
(A) Western blot analysis of endogenous DIRAS3 and RAC1 protein expression in BEAS-2B, H460, H157, H1299, H1792, and Calu-1 cells.
(B) Scatterplots showing the correlation of DIRAS3 expression with RAC1 expression in cells in (A). The R value was calculated via Spearman’s rank correlation coefficient analysis.
(C) Boxplots of RNF19B mRNA levels were determined from 1 Oncomine dataset, namely Hou Lung (∗∗∗p < 0.001; p-value was obtained using two-tailed Student’s t-tests).
(D) Boxplots of RAC1 mRNA levels were determined from 1 Oncomine dataset, namely Selamat (∗∗∗p < 0.001; p-value was obtained using two-tailed Student’s t-tests).
(E–F) Immunoreactivity scores (IRS) of RNF19B (E) and RAC1 (F) in 32 normal and 154 NSCLC samples shown as a histogram (∗p < 0.05, ∗∗∗p < 0.001; p-values were calculated via two-tailed Student’s t-tests).
(G–H) Representative IHC images of DIRAS3, RNF19B, and RAC1 expression in 186 normal and NSCLC samples.
(I) The correlation between DIRAS3 and RAC1 expression in 186 normal and NSCLC samples. p-value was calculated via the chi-square test.
Discussion
The tumor suppressor protein DIRAS3 has been reported to play a critical role in the development of various tumors.31 Here, we observed that the expression of DIRAS3 in lung cancer tissues was significantly lower than that in benign tissues. Its levels were clinically correlated with the malignant progression in patients with lung cancer. Loss of DIRAS3 function in tumors may owe to the genetic, epigenetic, transcriptional, and posttranscriptional mechanisms.31,32,33 DIRAS3 overexpression was reported to suppress the malignant progression of tumor cells via modulating apoptosis, autophagy, proliferation, or migration.16,26,34 As a tumor suppressor protein of the RAS family, DIRAS3 directly bound to RAS through its unique N-terminal extension, inhibiting RAS clustering and suppressing the RAF kinase and downstream MAPK signaling pathway.27 In addition, re-expression of DIRAS3 blocked FAK-mediated RHOA signaling, resulting in decreased levels of GTP-RHOA in epidermal growth factor-stimulated ovarian cancer cells.19 Notably, a meta-analysis of more than 1200 cases of metastatic or recurrent breast cancer showed that the allelic loss and methylation of the DIRAS3 and 1p31 chromosomal region were associated with an increased risk of metastasis.35 Therefore, exploring the role of DIRAS3 in NSCLC metastasis is worth attention.
In this study, we found that DIRAS3 downregulated the level of RAC1 protein which is best known for its role in regulating the cytoskeleton and metastasis of cells.4,5,36 Mechanistically, DIRAS3 induces the ubiquitin-proteasome degradation of RAC1, thus decreasing RAC1 expression. Given that DIRAS3 does not have the function of catalyzing ubiquitination, there might be an E3 ligase involved in the DIRAS3-mediated RAC1 degradation. Finally, we discovered that RNF19B acts as the E3 ligase of RAC1, which was initially predicted by the BioGRID database.
RNF19B, as a member of RBR E3 ubiquitin ligases, has a conserved catalytic RING1-IBR-RING2 module and was initially found to be significantly upregulated in IFN-β/IL-2-stimulated natural killer (NK) cells and antigen-specific-induced CD8+ cytotoxic T lymphocytes.37,38 Subsequent studies have further revealed that RNF19B increased granule exocytosis in NK cells and nitric oxide synthesis in macrophages, thereby enhancing their target-killing ability.39,40 Moreover, RNF19B could also modulate the expression of inflammatory factors such as IL-6, IFN-γ, MCP-1, MIP-1α, and RANTES, which are considered to play important roles in leukocyte recruitment to inflammatory lesions.41,42 Some reports claimed that RNF19B improved the survival rate of animals infected with the pathogenic avian influenza virus, Sendai virus, Streptococcus pneumoniae, and bacteria based on its role in innate immunity.41,42,43 Furthermore, RNF19B was also found to influence cell autophagy by regulating the overall expression of LC3 and the conversion of LC3I to LC3II.42 Rnf19b knockout mice showed higher metastasis rates in melanoma, T cell lymphoma, and breast cancer compared to wild-type mice.44,45 Similarly, our research also confirmed the inhibitory role of RNF19B in NSCLC cell migration. As an E3 ligase, RNF19B has been revealed to promote the ubiquitination and transcriptional activity of STAT1.46 Furthermore, it was also observed to interact with uridine-cytidine kinase-like-1 (UCKL-1), inducing UCKL-1 degradation through ubiquitination and tumor apoptosis.47,48
In the work reported in this manuscript, we found that RNF19B bound to RAC1 through its RBR domain, and catalyzed K48-linked ubiquitination of RAC1 to decline RAC1 expression, thereby inhibiting NSCLC metastasis. Further study confirmed the inhibitory function of RNF19B in NSCLC cell migration. DIRAS3 could enhance the connection between RNF19B and RAC1, thus promoting RAC1 ubiquitin-proteasome-dependent degradation and inhibiting NSCLC cell metastasis. Of course, this study still has some limitations, such as the need for in vivo animal experiments to confirm the functional role of the DIRAS3-RNF19B-RAC1 axis in NSCLC metastasis. We plan to address this topic in our future research as well.
In conclusion, our study demonstrates that the DIRAS3-RNF19B-RAC1 axis plays a crucial role in NSCLC cell metastasis and is associated with malignant development in clinical lung cancer patients. Thus, we can predict the malignancy of NSCLC by detecting the protein levels of DIRAS3, RNF19B, and RAC1 in NSCLC patient specimens. Alternatively, upregulating the expression of DIRAS3 through specific DIRAS3 activators may represent a promising way to suppress NSCLC metastasis.
Limitations of the study
The current research still has some limitations. Firstly, only overexpressing cell lines were used to explore the role of DIRAS3 in NSCLC cell migration, without constructing knocked-out or knocked-down cell lines. In addition, no related in vivo experiments support the current conclusions. The Diras3 gene is absent in mice due to lineage divergence, which prevented traditional studies of tumor suppression in genetically engineered mice, suggesting that the mouse model may not be the optimal model for studying processes that rely on DIRAS3. Moreover, further exploration is needed on how to apply the DIRAS3-RNF19B-RAC1 axis as a biological marker in clinical practice.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-DIRAS3 | Thermo Fisher Scientific | Cat# PA5-25785; RRID: AB_2543285 |
| Anti-RNF19B | Sigma-Aldrich | Cat# HPA049587; RRID: AB_2680826 |
| Anti-RNF19B | Sigma-Aldrich | Cat# SAB1301675 |
| Anti-RAC1 | Proteintech Group | Cat# 66122-1-Ig; RRID: AB_2881521 |
| Anti-RAC1 | Cell Signaling Technology | Cat# 2465S |
| Anti-Flag | Sigma-Aldrich | Cat# F7425; RRID: AB_439687 |
| Anti-His | Sigma-Aldrich | Cat# SAB1306085 |
| Anti-Myc | Sigma-Aldrich | Cat# C3956; RRID: AB_439680 |
| Anti-V5 | MBL Life Science | Cat# M215-3 |
| Anti-HA | Sigma-Aldrich | Cat# H3663 |
| Anti-ACTB | Sigma-Aldrich | Cat# A1978; RRID: AB_476692 |
| Anti-GAPDH | Sigma-Aldrich | Cat# G8795; RRID: AB_1078991 |
| Biological samples | ||
| Fetal bovine serum | Sigma-Aldrich | F0193 |
| Lung cancer TMA | Avilabio | DC-Lun01087 |
| Chemicals, peptides, and recombinant proteins | ||
| CHX | MedChem Express | HY-12320 |
| MG132 | MedChem Express | HY-13259 |
| E64D | MedChem Express | HY-100229 |
| TRITC-phalloidin | Sigma-Aldrich | P1951 |
| CCK8 | Dojindo | CK04 |
| DAPI | Beyotime Biotechnology | C1002 |
| Experimental models: Cell lines | ||
| H1792 | ATCC | CRL-5895 |
| Calu-1 | ATCC | HTB-54 |
| H1299 | ATCC | CRL-5803 |
| A549 | ATCC | CRM-CCL-185 |
| H460 | ATCC | HTB-177 |
| H157 | ATCC | CRL-5802 |
| BEAS-2B | National Collection of Authenticated Cell Cultures (China) | SCSP-5067 |
| 293FT | Invitrogen™ | R70007 |
| Oligonucleotides | ||
| siRNA targeting sequence: RNF19B #1: CCGGGAGGUGGCCCUGAAUTT | This paper | N/A |
| siRNA targeting sequence: RNF19B #2: CUCAGCCCCUCUGGCUGUATT | This paper | N/A |
| Primer: DIRAS3 Forward: GTACCTGCCGACCATTGAAAA Reverse: GGGTTTCCTTCTTGGTGACTG |
This paper | N/A |
| Primer: RAC1 Forward: ATGTCCGTGCAAAGTGGTATC Reverse: CTCGGATCGCTTCGTCAAACA |
This paper | N/A |
| Primer: GAPDH Forward: ACGGATTTGGTCGTATTGGG Reverse: CGCTCCTGGAAGATGGTGAT |
This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pcDNA3.1-Flag-DIRAS3 | This paper | N/A |
| Plasmid: pcDNA3.1-Myc-RAC1 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K48 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K63 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K6 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K11 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K27 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K29 | This paper | N/A |
| Plasmid: pcDNA3.1-HA-Ub-K33 | This paper | N/A |
| Plasmid: pcDNA3.1-V5-RNF19B | This paper | N/A |
| Plasmid: pcDNA3.1-V5-RNF19B-R1 | This paper | N/A |
| Plasmid: pcDNA3.1-V5-RNF19B-R1IBR | This paper | N/A |
| Plasmid: pcDNA3.1-V5-RNF19B-△R1 | This paper | N/A |
| Plasmid: pcDNA3.1-V5-RNF19B-△R1IBR | This paper | N/A |
| Plasmid: pcDNA3.1-V5-RNF19B-△R1IBRR2 | This paper | N/A |
| Software and algorithms | ||
| SPSS for Windows version 13.0 | IBM SPSS | https://www.ibm.com/docs/zh/spss-statistics/ |
| GraphPad Prism 8 | GraphPad | https://www.graphpad.com/ |
| Image J | National Institutes of Health | https://imagej.nih.gov/ij/ |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Xiangguo Liu (xgliu@sdu.edu.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Cell lines and cell culture
H1792, Calu-1, H1299, A549, H460, H157 human NSCLC cell lines, BEAS-2B human bronchial epithelial cells, and 293FT human embryonic kidney cells were initially obtained from the American Type Culture Collection (ATCC). The NSCLC cells were cultured in RPMI 1640 medium with 10% FBS. BEAS-2B and 293FT cells were cultured in DMEM medium with 10% FBS. All of the cells were cultured at 37°C in a humidified atmosphere consisting of 5% CO2 and tested for no mycoplasma contamination.
Method details
Immunofluorescence (IF) staining
Calu-1 cells were fixed with PHEMO buffer for 15 min at room temperature before washing with PBS 3 times. Then, cells were incubated with blocking buffer (2% BSA) for 30 min at room temperature, followed by primary antibodies against DIRAS3 (Cat. no. PA5-25785; Thermo Fisher Scientific), RNF19B (Cat. no. HPA049587; Sigma-Aldrich) or RAC1 (Cat. no. 66122-1-Ig; Proteintech Group) for 1 h at room temperature. After washing with PBS 3 times, cells were incubated with Alex Fluor-488 and Alex Fluor-555-conjugated secondary antibodies (Invitrogen) at room temperature for 1 h. For the F-actin staining assay, cells were blocked in PBS containing 3% BSA for 30 min and then incubated with TRITC-phalloidin (Sigma-Aldrich, 400 ng/ml) for 40 min instead incubation with secondary antibodies. In the end, cells were mounted with 1μg/ml DAPI (Beyotime Biotechnology) to stain the nucleus, and examined with a Zeiss LSM900 confocal microscope.
Tissue microarrays (TMA) and immunohistochemistry (IHC) staining
High-density tissue microarrays (TMA) of human NSCLC clinical samples (Cat. no. DC-Lun01087) were obtained from a cohort of 186 patients and constructed by Avilabio Inc. (Xian, China). For immunohistochemistry (IHC), tissue sections were baked at 60 °C for 2 h, then de-paraffinized by three 10-min extractions in 100% xylene, followed by 5-min each of descending grade of alcohol (100%, 95%, 80%, and 70%). Samples were washed briefly with phosphate-buffered saline (PBS) before transferring to boiling 10 mM sodium citrate buffer (pH 6.0) for 30 min. Sections were then pre-treated with 3% hydrogen peroxide for 10 min before blocking. Blocking was performed with 5% normal goat serum in PBS for 30 min at room temperature followed by primary antibody incubation overnight at 4°C. After incubation with horse radish peroxidase-conjugated anti-rabbit polyclonal antibody, sections were visualized with a DAB substrate (Cat. no. ZLI-9018; ZSGB-BIO) according to the manufacturer’s instruction.
After IHC staining, the TMA chips of human NSCLC clinical samples were digitally scanned by the automated slide scanner (VS100, Nanjing Jiangnan Novel Optics Co., Ltd., China), and the whole field of each tissue spot was obtained for IHC evaluation. The expression levels of DIRAS3, RNF19B, and RAC1 were scored semiquantitatively based on staining intensity and distribution using the immunoreactive score (IRS). Briefly, immunoreactive score (IRS) = SI (staining intensity) × PP (percentage of positive cells). SI was assigned as: 0 = negative; 1 = weak; 2 = moderate; 3 = strong. PP is defined as 0 = 0%; 1 = 0-25%; 2 = 25-50%; 3 = 50-75%; 4 = 75-100%. For categorization of the continuous IRS values into low and high, we chose a cutoff point for the measurements (range 0-12, cut point ≤ 4 versus > 4 for DIRAS3 and RNF19B; cut point ≤ 6 versus > 6 for RAC1).
Data mining
The DIRAS3, RNF19B, and RAC1 mRNA expression of normal or lung cancer tissues were obtained by Oncomine Cancer Microarray database analysis (http://www.oncomine.org). Then data were retrieved from the website and reanalyzed in GraphPad software. For analysis of the relationship between DIRAS3 or RAC1 mRNA expression and the OS, FPS, and PPS of lung cancer patients, data were obtained from the Kaplan–Meier plotter database (http://kmplot.com/analysis).
Reagents and antibodies
CHX, MG132, and E64D were purchased from MedChem Express. The primary antibodies used in IF and IHC stainings were as follows: anti-DIRAS3 (Cat. no. PA5-25785; Thermo Fisher Scientific), RNF19B (Cat. no. HPA049587; Sigma-Aldrich), and RAC1 (Cat. no. 66122-1-Ig; Proteintech Group). The primary antibodies used in Western blot assays and co-immunoprecipitation were as follows: anti-DIRAS3 (Cat. no. PA5-25785; Thermo Fisher Scientific), RNF19B (Cat. no. SAB1301675; Sigma-Aldrich), RAC1 (Cat. no. 2465S; CST), RAC1 (Cat. no. 66122-1-Ig; Proteintech Group), Flag (Cat. no. F7425; Sigma-Aldrich), His (Cat. no. SAB1306085; Sigma-Aldrich), Myc (Cat. no. C3956; Sigma-Aldrich), V5 (Cat. no. M215-3; MBL Life Science), HA (Cat. no. H3663; Sigma-Aldrich), and ACTB (Cat. no. A1978; Sigma-Aldrich).
Western blot analysis
Cells were harvested and rinsed with pre-chilled PBS on ice. They were lysed in lysis buffer on ice for 30 min and then purified via centrifugation for 13 min at 4°C. Protein extracts were resolved through 8%–15% SDS-PAGE, transferred to PVDF membranes, and probed with primary antibodies. Peroxidase-conjugated anti-mouse or rabbit antibody (Bio-Rad Laboratories) was used as a secondary antibody, and the antigen-antibody reaction was visualized by enhanced chemiluminescence assay.49
Cell migration assay
BD Falcon cell culture inserts (8 μm) were used for the cell migration assay (Cat. no. 353097, BD). Cells were detached from the plates, and the cell suspension was placed into the upper chamber in 0.2 ml of DMEM serum-free medium (1.5x104 cells per filter). DMEM medium supplemented with 10% FBS was placed in the lower chamber as a chemoattractant. Migration was scored following 12h. Cells at the lower surface of the inserts were then fixed in 4% formaldehyde for 30 min at room temperature, stained using crystal violet for 30 min, visualized, and counted. Values for cell migration were expressed as the mean number of cells per microscopic field over 6 fields per one insert for triplicate experiments. Experiments were repeated at least three times.
Immunoprecipitation
Cells were lysed in lysis buffer (20 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1 mM Na2EDTA; 1 mM EGTA; 2.5 mM sodium pyrophosphate; 1 mM β-glycerophosphate; 1 mM Na3VO4; 0.5% Triton) on ice for 30 min then purified via centrifugation for 15 min at 4°C. The supernatants were incubated with the antibody at 4°C for 4 h. Then the mixture was incubated with protein A/G beads (Invitrogen) at 4°C for 2 h. The beads were washed twice with 1 ml of lysis buffer. 20 μl 2× SDS buffer were added for elution (100°C, 10 min). Samples were centrifuged for Western blot analysis.
siRNA and plasmid transfection
The cells were transfected with jetPRIME transfection reagent (Polyplus transfection) or LipoMax reagent (Sudgen Biotechnology) in serum-free Opti-MEM (Gibco) according to the instruction manual. All siRNAs were synthesized from GenePharma (Shanghai, China). The sense and anti-sense strands of siRNAs were as follows:
RNF19B siRNA-1 sense: 5′-CCGGGAGGUGGCCCUGAAUTT-3′
RNF19B siRNA-1 antisense: 5′-AUUCAGGGCCACCUCCCGGTT-3′
RNF19B siRNA-2 sense: 5′-CUCAGCCCCUCUGGCUGUATT-3′
RNF19B siRNA-2 antisense: 5′-UACAGCCAGAGGGGCUGAGTT-3′
RT-qPCR
Reverse transcription-quantitative real-time PCR (RT-qPCR) was performed with a LightCycler 480 System (Roche Diagnostics), using Real-Time PCR Super Mix (Mei5Bio, China) according to the manufacturer’s instructions. All reactions were done in a 20 μl reaction volume in triplicate. Primers were obtained from Sangon Biotech. Following an initial denaturation at 95°C for 30 s, 40 cycles of PCR amplification were performed at 95°C for 5 s and 60°C for 30 s. Standard curves were generated, and the relative amount of target gene mRNA was normalized to GAPDH. The primer sequences are as follows:
DIRAS3 forward: 5′-GTACCTGCCGACCATTGAAAA-3′
DIRAS3 reverse: 5′-GGGTTTCCTTCTTGGTGACTG-3′
RAC1 forward: 5′-ATGTCCGTGCAAAGTGGTATC-3′
RAC1 reverse: 5′-CTCGGATCGCTTCGTCAAACA-3′
GAPDH forward: 5′-ACGGATTTGGTCGTATTGGG-3′
GAPDH reverse: 5′-CGCTCCTGGAAGATGGTGAT-3′
Wound-healing scratch assay
The Calu-1 cells were seeded in 6-well plates and then transfected with plasmids or siRNAs. They were treated with mitomycin-C (proliferation inhibitors, 10 μg/ml) for 1 h before performing the scratch assay. The scratches were made with a p-200 pipette tip across the bottom of the 6-well plates, and the suspended cells were washed gently with PBS. Then cells were cultured with medium including 1% FBS for 36-48 h at 37°C in a humidified incubator with 5% CO2. The sites of the scratch wounds were imaged using a microscope equipped with a digital camera. Images were obtained at the indicated time points and the migration rate was calculated.
Cell growth
Cells were seeded into 96-well plates at 5000 cells/well overnight, and each well was subsequently replaced with mixtures of 10 μl of CCK8 (Dojindo, Japan) and 90 μl of culture medium. Two hours later, absorbance was measured at an OD value of 450 nm using an enzyme microplate reader (Molecular Devices, US). The proliferation level of each cell was calculated by GraphPad Prism 8 (GraphPad Software, San Diego, CA).
Quantification and statistical analysis
All statistical analyses were performed using SPSS for Windows version 13.0 (SPSS). Two-tailed Student’s t-tests were used for comparisons between two groups, and one-way ANOVA followed by Bonferroni’s posthoc test was used for multiple comparisons (three or more groups). The Kaplan-Meier curves for survival analyses were determined using the log-rank test. Spearman’s rank correlation coefficient analysis was performed to assess the correlation of DIRAS3 expression with RAC1 expression in Western blot assays. A chi-square test was performed to determine the relationship between DIRAS3 and RAC1 protein in the NSCLC tissue microarrays. All experiments for cell cultures were performed independently at least three times and in triplicate each time. In all cases, P values < 0.05 were considered statistically significant.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (81902994, 82172970, 31771526) and The Future Plan for Young Scholars of Shandong University. We thank Yuyu Guo and Haiyan Yu from SKLMT (State Key Laboratory of Microbial Technology, Shandong University) for their assistance in confocal microscopy analyses.
Author contributions
X.L., L.S., and Y.W. conceived and designed the study. Y.W., M.W., and M.S. acquired and analyzed the data. Z.D., J.D., Y.W., X.L., and J.D. supervised the study. Y.W. and X.L. wrote and reviewed the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 16, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107157.
Contributor Information
Ling Su, Email: suling@sdu.edu.cn.
Xiangguo Liu, Email: xgliu@sdu.edu.cn.
Supplemental information
Data and code availability
-
•
All data has been included in main figures or supplemental information. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.
References
- 1.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. Ca - Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 2.Salehi M., Movahedpour A., Tayarani A., Shabaninejad Z., Pourhanifeh M.H., Mortezapour E., Nickdasti A., Mottaghi R., Davoodabadi A., Khan H., et al. Therapeutic potentials of curcumin in the treatment of non-small-cell lung carcinoma. Phytother Res. 2020;34:2557–2576. doi: 10.1002/ptr.6704. [DOI] [PubMed] [Google Scholar]
- 3.Arbour K.C., Mezquita L., Long N., Rizvi H., Auclin E., Ni A., Martínez-Bernal G., Ferrara R., Lai W.V., Hendriks L.E.L., et al. Impact of Baseline Steroids on Efficacy of Programmed Cell Death-1 and Programmed Death-Ligand 1 Blockade in Patients With Non-Small-Cell Lung Cancer. J. Clin. Oncol. 2018;36:2872–2878. doi: 10.1200/jco.2018.79.0006. [DOI] [PubMed] [Google Scholar]
- 4.Marei H., Malliri A. Rac1 in human diseases: The therapeutic potential of targeting Rac1 signaling regulatory mechanisms. Small GTPases. 2017;8:139–163. doi: 10.1080/21541248.2016.1211398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kotelevets L., Chastre E. Rac1 Signaling: From Intestinal Homeostasis to Colorectal Cancer Metastasis. Cancers. 2020;12:665. doi: 10.3390/cancers12030665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 7.Bosco E.E., Mulloy J.C., Zheng Y. Rac1 GTPase: a "Rac" of all trades. Cell. Mol. Life Sci. 2009;66:370–374. doi: 10.1007/s00018-008-8552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan Y., Bi F., Liu N., Xue Y., Yao X., Zheng Y., Fan D. Expression of seven main Rho family members in gastric carcinoma. Biochem. Biophys. Res. Commun. 2004;315:686–691. doi: 10.1016/j.bbrc.2004.01.108. [DOI] [PubMed] [Google Scholar]
- 9.Kamai T., Yamanishi T., Shirataki H., Takagi K., Asami H., Ito Y., Yoshida K.I. Overexpression of RhoA, Rac1, and Cdc42 GTPases is associated with progression in testicular cancer. Clin. Cancer Res. 2004;10:4799–4805. doi: 10.1158/1078-0432.ccr-0436-03. [DOI] [PubMed] [Google Scholar]
- 10.Gastonguay A., Berg T., Hauser A.D., Schuld N., Lorimer E., Williams C.L. The role of Rac1 in the regulation of NF-kappaB activity, cell proliferation, and cell migration in non-small cell lung carcinoma. Cancer Biol. Ther. 2012;13:647–656. doi: 10.4161/cbt.20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Yang M., Chen R., Su W., Li P., Chen S., Chen Z., Chen A., Li S., Hu C. IBP regulates epithelial-to-mesenchymal transition and the motility of breast cancer cells via Rac1, RhoA and Cdc42 signaling pathways. Oncogene. 2014;33:3374–3382. doi: 10.1038/onc.2013.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M., Niu J., Gao L., Gao Y., Gao S. Zerumbone inhibits migration in ESCC via promoting Rac1 ubiquitination. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2019;109:2447–2455. doi: 10.1016/j.biopha.2018.11.134. [DOI] [PubMed] [Google Scholar]
- 13.Liu L., Cui J., Zhao Y., Liu X., Chen L., Xia Y., Wang Y., Chen S., Sun S., Shi B., Zou Y. KDM6A-ARHGDIB axis blocks metastasis of bladder cancer by inhibiting Rac1. Mol. Cancer. 2021;20:77. doi: 10.1186/s12943-021-01369-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ni J., Zhang X., Li J., Zheng Z., Zhang J., Zhao W., Liu L. Tumour-derived exosomal lncRNA-SOX2OT promotes bone metastasis of non-small cell lung cancer by targeting the miRNA-194-5p/RAC1 signalling axis in osteoclasts. Cell Death Dis. 2021;12:662. doi: 10.1038/s41419-021-03928-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson M.F. Rho GTPases, their post-translational modifications, disease-associated mutations and pharmacological inhibitors. Small GTPases. 2018;9:203–215. doi: 10.1080/21541248.2016.1218407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reexpression of the Tumor Suppressor Gene ARHI Induces Apoptosis in Ovarian and Breast Cancer Cells through a Caspase-independent Calpain-dependent Pathway. Cancer research; 2002. [PubMed] [Google Scholar]
- 17.Luo R.Z., Fang X., Marquez R., Liu S.Y., Mills G.B., Liao W.S.L., Yu Y., Bast R.C. ARHI is a Ras-related small G-protein with a novel N-terminal extension that inhibits growth of ovarian and breast cancers. Oncogene. 2003;22:2897–2909. doi: 10.1038/sj.onc.1206380. [DOI] [PubMed] [Google Scholar]
- 18.Fitzgerald J., Bateman J.F. Why mice have lost genes for COL21A1, STK17A, GPR145 and AHRI: evidence for gene deletion at evolutionary breakpoints in the rodent lineage. Trends Genet. 2004;20:408–412. doi: 10.1016/j.tig.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Badgwell D.B., Lu Z., Le K., Gao F., Yang M., Suh G.K., Bao J.J., Das P., Andreeff M., Chen W., et al. The tumor-suppressor gene ARHI (DIRAS3) suppresses ovarian cancer cell migration through inhibition of the Stat3 and FAK/Rho signaling pathways. Oncogene. 2012;31:68–79. doi: 10.1038/onc.2011.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Z., Luo R.Z., Lu Y., Zhang X., Yu Q., Khare S., Kondo S., Kondo Y., Yu Y., Mills G.B., et al. The tumor suppressor gene ARHI regulates autophagy and tumor dormancy in human ovarian cancer cells. J. Clin. Invest. 2008;118:3917–3929. doi: 10.1172/jci35512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu Y., Xu F., Peng H., Fang X., Zhao S., Li Y., Cuevas B., Kuo W.L., Gray J.W., Siciliano M., et al. NOEY2 (ARHI), an imprinted putative tumor suppressor gene in ovarian and breast carcinomas. Proc. Natl. Acad. Sci. USAUSA. 1999;96:214–219. doi: 10.1073/pnas.96.1.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalai I., Missiaglia E., Barbi S., Butturini G., Doglioni C., Falconi M., Scarpa A. Low expression of ARHI is associated with shorter progression-free survival in pancreatic endocrine tumors. Neoplasia. 2007;9:181–183. doi: 10.1593/neo.06838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang J., Lin Y., Li L., Qing D., Teng X.M., Zhang Y.L., Hu X., Hu Y., Yang P., Han Z.G. ARHI, as a novel suppressor of cell growth and downregulated in human hepatocellular carcinoma, could contribute to hepatocarcinogenesis. Mol. Carcinog. 2009;48:130–140. doi: 10.1002/mc.20461. [DOI] [PubMed] [Google Scholar]
- 24.Lin D., Cui F., Bu Q., Yan C. The expression and clinical significance of GTP-binding RAS-like 3 (ARHI) and microRNA 221 and 222 in prostate cancer. J. Int. Med. Res. 2011;39:1870–1875. doi: 10.1177/147323001103900530. [DOI] [PubMed] [Google Scholar]
- 25.Wu X., Liang L., Dong L., Yu Z., Fu X. Effect of ARHI on lung cancer cell proliferation, apoptosis and invasion in vitro. Mol. Biol. Rep. 2013;40:2671–2678. doi: 10.1007/s11033-012-2353-x. [DOI] [PubMed] [Google Scholar]
- 26.Kuang P., Xie A., Deng J., Tang J., Wang P., Yu F. GTP-binding protein Di-RAS3 diminishes the migration and invasion of non-small cell lung cancer by inhibiting the RAS/extracellular-regulated kinase pathway. Bioengineered. 2022;13:5663–5674. doi: 10.1080/21655979.2022.2031671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sutton M.N., Lu Z., Li Y.C., Zhou Y., Huang T., Reger A.S., Hurwitz A.M., Palzkill T., Logsdon C., Liang X., et al. DIRAS3 (ARHI) Blocks RAS/MAPK Signaling by Binding Directly to RAS and Disrupting RAS Clusters. Cell Rep. 2019;29:3448–3459.e6. doi: 10.1016/j.celrep.2019.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fiedler L.R. Rac1 regulates cardiovascular development and postnatal function of endothelium. Cell Adhes. Migrat. 2009;3:143–145. doi: 10.4161/cam.3.2.8279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kunschmann T., Puder S., Fischer T., Steffen A., Rottner K., Mierke C.T. The Small GTPase Rac1 Increases Cell Surface Stiffness and Enhances 3D Migration Into Extracellular Matrices. Sci. Rep. 2019;9:7675. doi: 10.1038/s41598-019-43975-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huttlin E.L., Bruckner R.J., Navarrete-Perea J., Cannon J.R., Baltier K., Gebreab F., Gygi M.P., Thornock A., Zarraga G., Tam S., et al. Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell. 2021;184:3022–3040.e28. doi: 10.1016/j.cell.2021.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bildik G., Liang X., Sutton M.N., Bast R.C., Jr., Lu Z. DIRAS3: An Imprinted Tumor Suppressor Gene that Regulates RAS and PI3K-driven Cancer Growth, Motility, Autophagy, and Tumor Dormancy. Mol. Cancer Therapeut. 2022;21:25–37. doi: 10.1158/1535-7163.MCT-21-0331. [DOI] [PubMed] [Google Scholar]
- 32.Aberrant Methylation and Silencing of ARHI, an Imprinted Tumor Suppressor Gene in Which the Function Is Lost in Breast Cancers. Cancer research; 2003. [PubMed] [Google Scholar]
- 33.Fu Y., Chen J., Pang B., Li C., Zhao J., Shen K. EZH2-induced H3K27me3 is associated with epigenetic repression of the ARHI tumor-suppressor gene in ovarian cancer. Cell Biochem. Biophys. 2015;71:105–112. doi: 10.1007/s12013-014-0168-1. [DOI] [PubMed] [Google Scholar]
- 34.Lu Z., Baquero M.T., Yang H., Yang M., Reger A.S., Kim C., Levine D.A., Clarke C.H., Liao W.S.L., Bast R.C., Jr. DIRAS3 regulates the autophagosome initiation complex in dormant ovarian cancer cells. Autophagy. 2014;10:1071–1092. doi: 10.4161/auto.28577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomassen M., Tan Q., Kruse T.A. Gene expression meta-analysis identifies chromosomal regions and candidate genes involved in breast cancer metastasis. Breast Cancer Res. Treat. 2009;113:239–249. doi: 10.1007/s10549-008-9927-2. [DOI] [PubMed] [Google Scholar]
- 36.Payapilly A., Malliri A. Compartmentalisation of RAC1 signalling. Curr. Opin. Cell Biol. 2018;54:50–56. doi: 10.1016/j.ceb.2018.04.009. [DOI] [PubMed] [Google Scholar]
- 37.Dove K.K., Klevit R.E. RING-Between-RING E3 Ligases: Emerging Themes amid the Variations. J. Mol. Biol. 2017;429:3363–3375. doi: 10.1016/j.jmb.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence D.W., Willard P.A., Cochran A.M., Matchett E.C., Kornbluth J. Natural Killer Lytic-Associated Molecule (NKLAM): An E3 Ubiquitin Ligase With an Integral Role in Innate Immunity. Front. Physiol. 2020;11:573372. doi: 10.3389/fphys.2020.573372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lawrence D.W., Gullickson G., Kornbluth J. E3 ubiquitin ligase NKLAM positively regulates macrophage inducible nitric oxide synthase expression. Immunobiology. 2015;220:83–92. doi: 10.1016/j.imbio.2014.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawrence D.W., Kornbluth J. E3 ubiquitin ligase NKLAM is a macrophage phagosome protein and plays a role in bacterial killing. Cell. Immunol. 2012;279:46–52. doi: 10.1016/j.cellimm.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lawrence D.W., Kornbluth J. Reduced inflammation and cytokine production in NKLAM deficient mice during Streptococcus pneumoniae infection. PLoS One. 2018;13:e0194202. doi: 10.1371/journal.pone.0194202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence D.W., Shornick L.P., Kornbluth J. Mice deficient in NKLAM have attenuated inflammatory cytokine production in a Sendai virus pneumonia model. PLoS One. 2019;14:e0222802. doi: 10.1371/journal.pone.0222802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uchida Y., Watanabe C., Takemae N., Hayashi T., Oka T., Ito T., Saito T. Identification of host genes linked with the survivability of chickens infected with recombinant viruses possessing H5N1 surface antigens from a highly pathogenic avian influenza virus. J. Virol. 2012;86:2686–2695. doi: 10.1128/JVI.06374-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoover R.G., Gullickson G., Kornbluth J. Natural killer lytic-associated molecule plays a role in controlling tumor dissemination and metastasis. Front. Immunol. 2012;3:393. doi: 10.3389/fimmu.2012.00393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoover R.G., Gullickson G., Kornbluth J. Impaired NK cytolytic activity and enhanced tumor growth in NK lytic-associated molecule-deficient mice. J. Immunol. 2009;183:6913–6921. doi: 10.4049/jimmunol.0901679. [DOI] [PubMed] [Google Scholar]
- 46.Lawrence D.W., Kornbluth J. E3 ubiquitin ligase NKLAM ubiquitinates STAT1 and positively regulates STAT1-mediated transcriptional activity. Cell. Signal. 2016;28:1833–1841. doi: 10.1016/j.cellsig.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fortier J.M., Kornbluth J. NK lytic-associated molecule, involved in NK cytotoxic function, is an E3 ligase. J. Immunol. 2006;176:6454–6463. doi: 10.4049/jimmunol.176.11.6454. [DOI] [PubMed] [Google Scholar]
- 48.Geiger T., Madden S.F., Gallagher W.M., Cox J., Mann M. Proteomic portrait of human breast cancer progression identifies novel prognostic markers. Cancer Res. 2012;72:2428–2439. doi: 10.1158/0008-5472.CAN-11-3711. [DOI] [PubMed] [Google Scholar]
- 49.Liu X., Yue P., Zhou Z., Khuri F.R., Sun S.Y. Death receptor regulation and celecoxib-induced apoptosis in human lung cancer cells. J. Natl. Cancer Inst. 2004;96:1769–1780. doi: 10.1093/jnci/djh322. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All data has been included in main figures or supplemental information. All data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon reasonable request.