Abstract
Objective
The AMP-activated protein kinase (AMPK) gets activated in response to energetic stress such as contractions and plays a vital role in regulating various metabolic processes such as insulin-independent glucose uptake in skeletal muscle. The main upstream kinase that activates AMPK through phosphorylation of α-AMPK Thr172 in skeletal muscle is LKB1, however some studies have suggested that Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) acts as an alternative kinase to activate AMPK. We aimed to establish whether CaMKK2 is involved in activation of AMPK and promotion of glucose uptake following contractions in skeletal muscle.
Methods
A recently developed CaMKK2 inhibitor (SGC-CAMKK2-1) alongside a structurally related but inactive compound (SGC-CAMKK2-1N), as well as CaMKK2 knock-out (KO) mice were used. In vitro kinase inhibition selectivity and efficacy assays, as well as cellular inhibition efficacy analyses of CaMKK inhibitors (STO-609 and SGC-CAMKK2-1) were performed. Phosphorylation and activity of AMPK following contractions (ex vivo) in mouse skeletal muscles treated with/without CaMKK inhibitors or isolated from wild-type (WT)/CaMKK2 KO mice were assessed. Camkk2 mRNA in mouse tissues was measured by qPCR. CaMKK2 protein expression was assessed by immunoblotting with or without prior enrichment of calmodulin-binding proteins from skeletal muscle extracts, as well as by mass spectrometry-based proteomics of mouse skeletal muscle and C2C12 myotubes.
Results
STO-609 and SGC-CAMKK2-1 were equally potent and effective in inhibiting CaMKK2 in cell-free and cell-based assays, but SGC-CAMKK2-1 was much more selective. Contraction-stimulated phosphorylation and activation of AMPK were not affected with CaMKK inhibitors or in CaMKK2 null muscles. Contraction-stimulated glucose uptake was comparable between WT and CaMKK2 KO muscle. Both CaMKK inhibitors (STO-609 and SGC-CAMKK2-1) and the inactive compound (SGC-CAMKK2-1N) significantly inhibited contraction-stimulated glucose uptake. SGC-CAMKK2-1 also inhibited glucose uptake induced by a pharmacological AMPK activator or insulin. Relatively low levels of Camkk2 mRNA were detected in mouse skeletal muscle, but neither CaMKK2 protein nor its derived peptides were detectable in mouse skeletal muscle tissue.
Conclusions
We demonstrate that pharmacological inhibition or genetic loss of CaMKK2 does not affect contraction-stimulated AMPK phosphorylation and activation, as well as glucose uptake in skeletal muscle. Previously observed inhibitory effect of STO-609 on AMPK activity and glucose uptake is likely due to off-target effects. CaMKK2 protein is either absent from adult murine skeletal muscle or below the detection limit of currently available methods.
Keywords: Ca2+/calmodulin dependent protein kinase kinase 2, AMP-activated protein kinase, SGC-CAMKK2-1, STO-609, Glucose uptake
Graphical abstract
Highlights
-
•
CaMKK2 is not an upstream regulator of AMPK in skeletal muscle.
-
•
SGC-CAMKK2-1 is a more selective CaMKK inhibitor compared to STO-609.
-
•
CaMKK2 inhibition reduces contraction-induced glucose uptake via off-target effects.
-
•
CaMKK2 protein is undetectable in skeletal muscle.
1. Introduction
AMP-activated protein kinase (AMPK) is a vital energy sensor which functions to maintain cellular homeostasis through coordinating metabolic pathways in response to energetic stresses (e.g., contractions, hypoxia, mitochondrial poisoning) [1,2]. AMPK exists as heterotrimeric complexes comprised of catalytic α-subunits, regulatory β-subunits, and γ-subunits. There are multiple genes encoding two α-isoforms (α1/α2), two β-isoforms (β1/β2), and three γ-isoforms (γ1/γ2/γ3). The expression of AMPK isoforms varies among different cell types and tissues, with α1, β1, and γ1 being the most ubiquitously expressed. Assays of immunoprecipitated AMPK isoforms in mouse skeletal muscle revealed that the α2-containing complexes (α2β2γ1, α2β2γ3, α2β1γ1) account for ∼90% and ∼70% of the total AMPK trimers in glycolytic extensor digitorum longus (EDL) and oxidative soleus muscle, respectively [3].
AMPK heterotrimers are active when a threonine residue (Thr172) within the activation loop of the α-subunit kinase domain is phosphorylated [4]. The α-Thr172-phosphorylated/activated form of AMPK can be maintained by the binding of AMP or ADP to the cystathionine β-synthase domains of the γ subunit [5,6]. Moreover, AMP can increase AMPK activity further through an allosteric mechanism [7]. In contrast, ATP antagonizes the effects of AMP and ADP, and this forms the basis by which AMPK can respond to cellular fluctuations of the AMP:ATP and ADP:ATP ratios during times of energy demand and maintain ATP at a constant level.
The major upstream kinases phosphorylating α-Thr172 are Liver kinase B 1 (LKB1) and Ca2+/calmodulin-dependent protein kinase kinase 2 (CaMKK2) [8]. In skeletal muscle, we and others have provided genetic evidence using muscle-specific LKB1 knock-out (mLKB1 KO) mouse models that LKB1 is the primary upstream kinase to activate AMPK [[9], [10], [11]]. We and others also showed that LKB1 is required for muscle glucose uptake in response to strenuous contractions or pharmacological treatments that increase intracellular levels of AMP or its mimetic ZMP [[11], [12], [13]]. Although activity of α2-AMPK is ablated [10,11,13], α1-AMPK activity was largely [10,13] or residually [11] detectable in skeletal muscle tissues from mLKB1 KO mice. Some studies reported that mLKB1 KO mice retained an ability to activate α1-AMPK in skeletal muscle in response to treatment with 5-aminoimidazole-4-carboxamide riboside (AICAR, a cellular ZMP-raising pro-drug) and electrically-stimulated contractions ex vivo [10] or treadmill exercise in vivo [13], suggesting that there might be an alternative kinase regulating α1-AMPK.
Both CaMKK1 and CaMKK2 isoforms were initially demonstrated to activate AMPK in cell-free assays [14]. However, subsequent studies revealed a predominant role for CaMKK2 as a physiological upstream kinase of AMPK in particular cell types (e.g., T cells, neuronal, and endothelial cells) that predominantly express α1-AMPK (reviewed in [8,15,16]). The pharmacological inhibition of CaMKK2 predominantly relies on the use of 7-Oxo-7h-benzimidazo-[2,1-a]benz[de]isoquinoline-7-one-3-carboxylic acid (known as STO-609), which inhibits CaMKK2 activity 5–10 fold more effectively than CaMKK1 activity in cell-free assays (IC50 value = ∼1 μM) [[17], [18], [19], [20], [21]]. In perfused rat hindlimb in vivo, STO-609 (5 μM) resulted in a significant inhibition of contraction-stimulated AMPK activation and glucose uptake in skeletal muscle [22]. However, in incubated mouse skeletal muscles ex vivo STO-609 (5 μM) inhibited contraction- stimulated AMPK activation and glucose uptake in one study [23] whereas in another study STO-609 (∼2.7 μM) had no effect on muscle glucose uptake or AMPKα (Thr172) phosphorylation [24]. While the on-target effects of STO-609 are compelling when using cells expressing STO-609-resistant CaMKK mutants [25], careful examinations have revealed that it also inhibits several other protein kinases with a similar potency to CaMKKs [19,20,26]. Notably, STO-609 inhibits AMPK with an IC50 of 1.7 μM in cell-free assays [19], which raises concerns regarding the interpretation of experiments studying the physiological roles of the CaMKK2-AMPK signaling pathway in intact cells/tissues. Furthermore, although the Ca2+/CaMKK signaling axis plays a key role in metabolic homeostasis [27], to date no study has provided genetic evidence that CaMKK2 plays a role in the regulation of AMPK and/or glucose uptake in skeletal muscle.
In the current study, we aimed to address these limitations and establish whether CaMKK2 functions as an upstream activator of AMPK in response to contractions and mediates contraction-stimulated glucose uptake in skeletal muscle. To this end, we employed a recently developed potent and selective CaMKK2 inhibitor alongside a structurally related but inactive compound [28], as well as CaMKK2 KO mice.
2. Materials and methods
2.1. Materials
Protein G Sepharose was purchased from Cytiva. Cation-exchange paper (P81) was obtained from SVI Phosphocellulose (https://www.svi.edu.au/resources/phosphocellulose_paper/). [γ-32P]-ATP was purchased from PerkinElmer. Anti-FLAG M2 agarose beads were obtained from MerckMilliporeSigma. The AMARA peptide and FLAG-tagged CaMKK1 (DU73944) and CaMKK2 (DU73349) constructs used for antibody specificity validation (shown in Figure 5) from MRC PPU Reagents and Services. Ionomycin and STO-609 were purchased from Tocris. SGC-CAMKK2-1 and SGC-CAMKK2-1 compounds were synthesized as described [28]. All other reagents were from MerckMilliporeSigma unless otherwise stated. List of primary and secondary antibodies are in the Supplementary Tables 1 and 2.
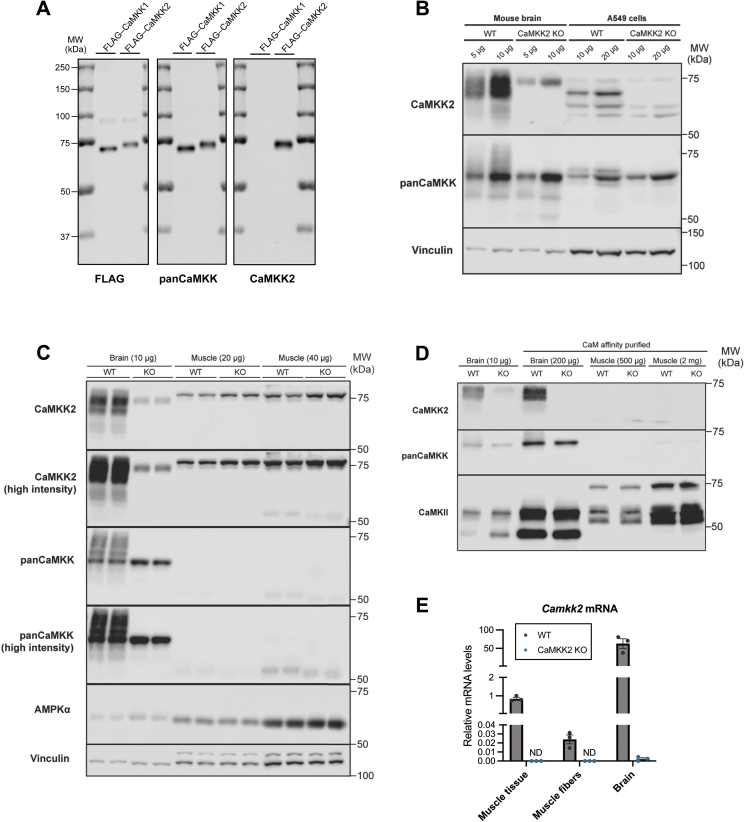
Figure 5.
CaMKK2proteinis undetectable in mouse skeletal muscle. (A) Validation of isoform-specificity of CaMKK antibodies. HEK293 cells were transiently transfected with FLAG-tagged CaMKK1 or CaMKK2. Affinity purification was performed using FLAG-agarose beads and the equal amount of purified FLAG-CaMKK1 or FLAG-CaMKK2 were subjected to immunoblot analysis using the indicated antibodies. (B) Immunoblot analysis of CaMKK isoforms in wild-type (WT) and CaMKK2 knock-out (KO) mouse brain and A549 cell lysates. (C) Different amounts of skeletal muscle (gastrocnemius) or brain lysates from WT and CaMKK2 KO mice were subjected to immunoblot analysis using the indicated antibodies. Immunoblots with pan-CaMKK and CaMKK2 antibodies were visualized at either lower or higher intensity. (D) Calmodulin (CaM) affinity purification was used to enrich for CaM-binding proteins which were subsequently subjected to immunoblot analysis using the indicated antibodies. Representative blots from three independent experiments are shown. Mouse samples used in panels (B–D) are from male, 17–18-week-old mice. (E) Levels of Camkk2 mRNA expression in WT or CaMKK2 KO skeletal muscle (gastrocnemius) tissue, isolated skeletal muscle (gastrocnemius) fibers and brain were determined by qPCR using Taqman probes (n = 3). Shown values are relative to WT skeletal muscle tissue. ND represents non-detected levels.
2.2. Animal ethics and models
Animal studies involving CaMKK2 knock-out (KO) and its littermate control wild-type (WT) mice were approved by Indiana University School of Medicine (IUSM) Institutional Animal Care and Use Committee (IACUC) and all experiments were performed in compliance with NIH guidelines on the use and care of laboratory and experimental animals. CaMKK2 KO mice (C57BL/6 background) were previously generated through the targeted deletion of exons 2–4 of the mouse Camkk2 gene [29]. CaMKK2 KO and WT littermates were housed in the IUSM Laboratory Animal Resource Center (Indianapolis, IN).
Other animal experiments were conducted in accordance with the European directive 2010/63/EU of the European Parliament and of the Council of the protection of animals used for scientific purposes. Ethical approval was given by the Danish Animal Experiments Inspectorate (license number #2021-15-0201-00884). WT C57BL/6NTac male mice were obtained from Taconic Biosciences and housed in the animal facility at the Faculty of Health and Medical Sciences (University of Copenhagen). All the animals were kept and maintained according to local regulations under a light/dark cycle of 12 h and had free access to a standard chow diet.
2.3. Protein kinase screen and IC50 determination of the selected kinases in vitro
All protein kinases in the kinase panel were expressed, purified, and assayed at the Protein Phosphorylation and Ubiquitylation Unit International Centre for Protein Kinase Profiling, University of Dundee, as previously described [20,30]. Assays were performed for 30 min (except Lck and PBK that were incubated for 15 min) at room temperature using Multidrop Micro reagent dispensers (Thermo Electron Corporation) in a 96-well format. The concentration of magnesium acetate in the assays was 10 mM and [γ-33P]ATP (∼800 cpm/pmol) was used at either 5, 20, or 50 μM in order to be at or below the Km for ATP for each enzyme (described under ‘kinase panel’ in the webpage http://www.kinase-screen.mrc.ac.uk/services/premier-screen). The half-maximal inhibitory concentration (IC50) analysis of LKB1 against STO-609 and SGC-CAMKK2-1 was performed by the International Centre for Kinase Profiling team (MRC Protein Phosphorylation and Ubiquitylation Unit, Dundee) and determined after carrying out assays at ten different concentrations of the compounds. Briefly, LKB1 activity was measured as following. Recombinant human LKB1 complex (LKB1 [1–433], MO25 [1–341], STRAD [1–431], 5–20 mU diluted in 50 mM Tris pH 7.5, 0.1 mM EGTA, 0.1% β-mercaptoethanol, 1 mg/ml BSA) is assayed against LSNLYHQGKFLQTFCGSPLYRRR in a final volume of 25.5 μl containing 50 mM Tris pH 7.5, 0.1 mM EGTA, 0.2 mM LSNLYHQGKFLQTFCGSPLYRRR, 10 mM magnesium acetate and 0.02 mM [33P-γ-ATP] (50–1000 cpm/pmole) and incubated for 30 min at room temperature. Assays are stopped by addition of 5 μl of 0.5 M (3%) orthophosphoric acid and then harvested onto P81 Unifilter plates with a wash buffer of 50 mM orthophosphoric acid.
2.4. Cell culture and lysates preparation
HEK293 cells and A549 cells were obtained from Invitrogen (Flp-In™-293 Cell Line, #R75007) and ATCC (#CRM-CCL-185) respectively and mouse embryonic fibroblasts (MEFs) were obtained as described [31]. Cells were cultured in DMEM (Invitrogen, #31966) supplemented with 10% (v/v) fetal bovine serum (Sigma, F7524) and 1% (v/v) penicillin/streptomycin in a 37 °C incubator with 5% CO2. C2C12 cells were obtained from ATCC (#CRL-1772). For myoblast experiments, C2C12 myoblasts were seeded at 10,000 cells/cm2 and permitted to proliferate in growth medium (4.5 mmol/L glucose DMEM, 10% fetal bovine serum, 1% Penicillin–Streptomycin) until approximately 90% confluent. For differentiation experiments, C2C12 myoblasts were seeded at 40,000 cells/cm2 in growth medium. Once confluent, differentiation media consisting of 4.5 mmol/L glucose DMEM supplemented with 2% horse serum was added and myotubes permitted to form for 5–6 days. For knocking down of CaMKK2 by siRNA, C2C12 myoblasts were seeded at 10,000 cells/cm2 in antibiotic-free growth medium, before being subjected to reverse transfection with an siRNA construct containing 10 nM ON-TARGETplus siCaMKK2 or non-targeting siControl (L-040625-00-0005 and D-001810-0X, respectively; Horizon Discovery) and lipofectamine RNAiMAX (Invitrogen). 24 h post transfection, media was refreshed, and cells permitted to grow to approximately 90% confluence, before harvesting for immunoblot analysis.
When appropriate, cells were lysed in QIAzol Lysis Reagent (Qiagen) or ice-cold lysis buffer (270 mM sucrose, 50 mM Tris–HCl (pH 7.5), 1 mM EDTA, 1 mM EGTA, 1% (v/v) Triton X-100, 20 mM glycerol-2-phosphate, 50 mM NaF, 5 mM Na4P2O7, 1 mM Na3VO4, 1 mM DTT, 0.1 mM PMSF, 1 mM benzamidine, 1 μg/ml microcystin-LR, 2 μg/ml leupeptin, and 2 μg/ml pepstatin A) for RNA and protein collection, respectively. All cells were routinely tested for the presence of mycoplasma and all of them tested negative.
2.5. Generation of CaMKK2 knockout (KO) cell line with CRISPR/Cas9
A549 cells (ATCC #CRM-CCL-185) were first transduced with the Cas9 expression vector FUCas9Cherry (Addgene #70182) and sorted for mCherry expression. Cells were subsequently transduced with lentiGuide-Puro-CaMKK2 gRNA (BRDN0001146379) (Addgene #76699) or lentiGuide-Puro-non-targeting control gRNA (BRDN0001149198) (Addgene #80248) followed by selection with 2 μg/ml puromycin. Lentivirus were produced by co-transfection of envelope plasmid pCMV-VSV-G (Addgene #8454) and packaging plasmid psPAX2 (Addgene #12260), as described previously [32]. A549 cells were transduced with 0.22 μm-filtered viral supernatant supplemented with polybrene. Genomic editing of CaMKK2 KO in A549 cell was validated with Sanger sequencing and analyzed using Synthego Performance Analysis, ICE Analysis. 2019. v3.0. Synthego. The absence of CaMKK2 protein expression was further confirmed by immunoblot.
2.6. In vitro CaMKK and AMPK activity assay
Heterotrimeric human AMPK FLAG-α2β1γ1 and FLAG-α2β2γ1, as well as human FLAG-CaMKK1 and FLAG-CaMKK2, were produced in mammalian cells as described [33,34]. For AMPK expression, the cells were triply transfected at 60% confluency using FuGene HD (Roche Applied Science) and 1 μg of pcDNA3 plasmid expression constructs for AMPK FLAG-α2, β1-Myc or β2-Myc, and HA-γ1. For CaMKK1 and CaMKK2 expression, the cells were transfected with either 1 μg of pcDNA3 FLAG-CaMKK1 or FLAG-CaMKK2 plasmid. After 48 h, the transfected cells were harvested by rinsing with ice-cold PBS, followed by rapid lysis using 500 μl of lysis buffer.
AMPK activity was determined by phosphorylation of the synthetic peptide substrate SAMS (HMRSAMSGLHLVKRR), whereas CaMKK1 and CaMKK2 activity was determined using CaMKKtide (LSNLYHQGKFLQTFCGAPLYRRR).
For the AMPK assays, recombinant AMPK was immunoprecipitated from 10 μg of transfected cultured cell lysate using 10 μl of anti-FLAG M2 agarose beads (50% (v/v)), washed and then added to a 25 μl reaction containing assay buffer (50 mM HEPES-NaOH, pH 7.4, 1 mM DTT, and 0.02% (v/v) Brij-35), 200 μM synthetic peptide substrate (SAMS), 200 μM [γ-32P]-ATP, 5 mM MgCl2, in the presence of SGC-CAMKK2-1 (0–100 μM) or STO-609 (0–100 μM).
For the CaMKK1 and CaMKK2 assays, recombinant kinase was immunoprecipitated from 10 μg of transfected cultured cell lysate using 10 μl of anti-FLAG M2 agarose beads (50% (v/v)), washed and then added to a 35 μl reaction containing assay buffer (50 mM HEPES-NaOH, pH 7.4, 1 mM DTT, and 0.02% (v/v) Brij-35), 200 μM synthetic peptide substrate (CaMKKtide), 200 μM [γ-32P]-ATP, 100 μM CaCl2, 1 μM calmodulin (MerckMilliporeSigma), 5 mM MgCl2, in the presence of SGC-CAMKK2-1 (0–10 μM) or STO-609 (0–10 μM). Reactions were performed at 30 °C and terminated after 10 min by spotting 15 μl onto P81 phosphocellulose paper. Radioactivity was quantified by liquid scintillation counting.
2.7. Ex vivo skeletal muscle incubation, electrically-stimulated contractions and measurement of glucose uptake
Animals were anesthetized via Avertin [stock of 1 g/ml tribromoethanol (#T48402, MerckMilliporeSigma) in 2-methyl-2-butanol (#152463, MerckMilliporeSigma)], diluted 1:20 in saline, and dosed at 10 μl/g body weight or sodium pentobarbital (80–90 g/kg body weight) via intraperitoneal injection, and EDL or soleus muscles were rapidly dissected and mounted in oxygenated (95% O2 and 5% CO2), and warmed (30 °C) Krebs–Ringer buffer (KRB) supplemented with 2 mM pyruvate in the presence of the indicated drug or vehicle for 50 min. The muscles were either further incubated for 10 min in glucose uptake buffer (KRB buffer containing 1.5 μCi/ml [3H]-2-deoxy-d-glucose, 1 mM 2-deoxy-d-glucose, 0.45 μCi/ml [14C]-mannitol, 7 mM mannitol) or electrically-stimulated contractions for 10 min followed by an additional 10 min in glucose uptake buffer. Muscle contractions were evoked by electrical stimulation every 15 s with 1-s trains of 0.2-ms impulses delivered at 100 Hz (∼30 V) [23]. Force measurements were performed at optimal length tension, which was determined via micromanipulations of muscle length and a series of single twitch contractions. At the end of incubation period, muscles were frozen in liquid nitrogen and subsequently processed for glucose uptake (as described in [11]) and immunoblot analysis.
2.8. Single fiber isolation
Flash-frozen dissected samples of gastrocnemius muscle from WT or CaMKK2 KO mice were first placed in a 1:1 solution of glycerol and relaxing buffer (4 mM Mg-ATP, 1 mM free Mg2+, 20 mM imidazole, 7 mM EGTA, 14.5 mM creatine phosphate, KCl to adjust the ionic strength to 180 mM and pH to 7.0) and incubated at −20 °C for 18 h. After this incubation samples were removed from this buffer and placed into a sterile filtered collagenase solution (2 mg/ml in serum-free Dulbecco's Modified Eagle Medium) for 90 min at 37 °C with constant agitation. Following this incubation samples were dissected at 4 °C in relaxing buffer under a light microscope. This dissection was to remove any residual connective tissue from the sample which had not previously been removed during chemical or enzymatic digestion. Once fibers had been isolated, they were placed in protein lysis buffer or RNAprotect (Qiagen) for RNA isolation and stored at −80 °C prior to processing.
2.9. Cell/tissue extracts preparation and immunoblotting
Mouse skeletal muscle tissue or isolated fibers were homogenized in ice-cold lysis buffer using a TissueLyser II (Qiagen) at 30 Hz for 1 min. Cell or tissue lysates were clarified by centrifugation at 6000×g for 10 min at 4 °C, and total protein concentration was determined using Bradford reagent (#23200, ThermoFisher) and BSA as standard. Proteins were separated by SDS-PAGE and transferred to nitrocellulose membrane. Membranes with proteins were blocked with 3% nonfat dry milk or BSA and incubated with primary antibodies diluted in Tris-buffered saline/0.1% (v/v) Tween 20 containing 4% BSA, overnight at 4 °C. Horseradish peroxidase-conjugated secondary antibodies were added to the membrane with proteins. After adding the enhanced chemiluminescence substrate (Millipore, WBKLS0500), the protein bands were visualized by Odyssey® XF imaging system. The volume densities of the protein bands were quantified with the ImageStudio Lite Ver 5.2 software. Specific blot/band signals in each lane alongside housekeeping vinculin or tubulin were obtained, and the background signal was subtracted in the software. The highest signal detected for the housekeeping protein was used to normalize the bands in each blot, with a normalization factor being calculated for each lane. To determine phospho/total ratios, measured signals from each blot were then divided by their respective normalization factor, yielding relative quantitative comparisons.
2.10. Immunoprecipitation of AMPK and in vitro activity assay
Lysates of skeletal muscle (100 μg) were incubated on an orbital shaker at 4 °C for 2 h with a mix of 5–7.5 μl packed Protein G Sepharose beads and either polyclonal rabbit α1-or α2-AMPK antibody or IgG (as control) (Supplementary Table 1). The beads were pelleted at 500×g for 1 min and initially washed twice with 0.5 ml lysis buffer containing 500 mM NaCl and subsequently washed twice with the same amount of buffer A [50 mM HEPES (pH 7.4), 150 mM NaCl, 1 mM EGTA and 1 mM DTT]. The AMPK complexes were subjected to in vitro activity assay as previously described using AMARA peptide (AMARAASAAALARRR) as substrate [35].
2.11. Calmodulin (CaM) affinity purification
CaM binding proteins were affinity purified from protein lysates with CaM agarose beads (MerckMilliporeSigma) in an overnight incubation at 4 °C in buffer containing 50 mM Tris–HCl (pH 7.4), 5 mM EGTA, 5 mM sodium pyrophosphate, 2 mM DTT, 4 μg/ml leupeptin, 100 μM benzamidine, 250 μM PMSF supplemented with 10 mM CaCl2. Agarose beads with captured CaM binding proteins were washed with the same buffer containing CaCl2. CaM binding proteins were eluted with 2× Laemmli buffer containing 100 mM DTT by incubation for 10 min at 65 °C and the eluates were subjected to immunoblot analysis.
2.12. RNA isolation and qPCR
Brain, whole gastrocnemius muscle tissue, and isolated muscle fibers from gastrocnemius muscle were homogenized in Qiazol Lysis reagent (Qiagen, #79309) using a TissueLyser II (Qiagen) at 30 Hz. Qiazol lysates from tissues and cell lines were cleared by centrifugation at 12,000×g for 10 min at 4 °C and RNA extracted by chloroform. The aqueous phase was used for RNA isolation using QIAwave RNA mini kit (Qiagen, #74536) with on-column DNA digestion (Qiagen RNase-Free DNase Set cat #79256) according to the manufacturer's instructions. 2 μg RNA were transcribed to cDNA using random primers (Roche, #11034731001), dNTPs (Invitrogen, #10297018), RNaseOUT (Invitrogen, #10777-019) and SuperScript III Reverse Transcriptase (Invitrogen, #18080-044) according to the manufacturer's instructions. qPCR was performed using Taqman probes and TaqMan Fast Advanced Master mix (ThermoFisher, #4444556) on a Roche Lightcycler480. Gene expression was calculated using the delta–delta Ct method and normalized to the house keeping gene Rn18S. The Taqman probes used are described in the Supplementary Table 3.
2.13. Statistical analysis
Data are reported as mean ± SEM. Statistical analyses were performed using GraphPad Prism software version 9.5.0. Differences between groups which involved a single variable or factor were analyzed using a one-way analysis of variance (ANOVA). In groups involving two or more factors, a two-way ANOVA test was performed instead. All analyses were subjected to Tukey's correction for multiple comparisons, unless otherwise indicated in the respective figure legends. Statistical significance was accepted at P < 0.05.
3. Results
3.1. Protein kinase screen reveals that SGC-CAMKK2-1 is a more selective CaMKK inhibitor compared to STO-609
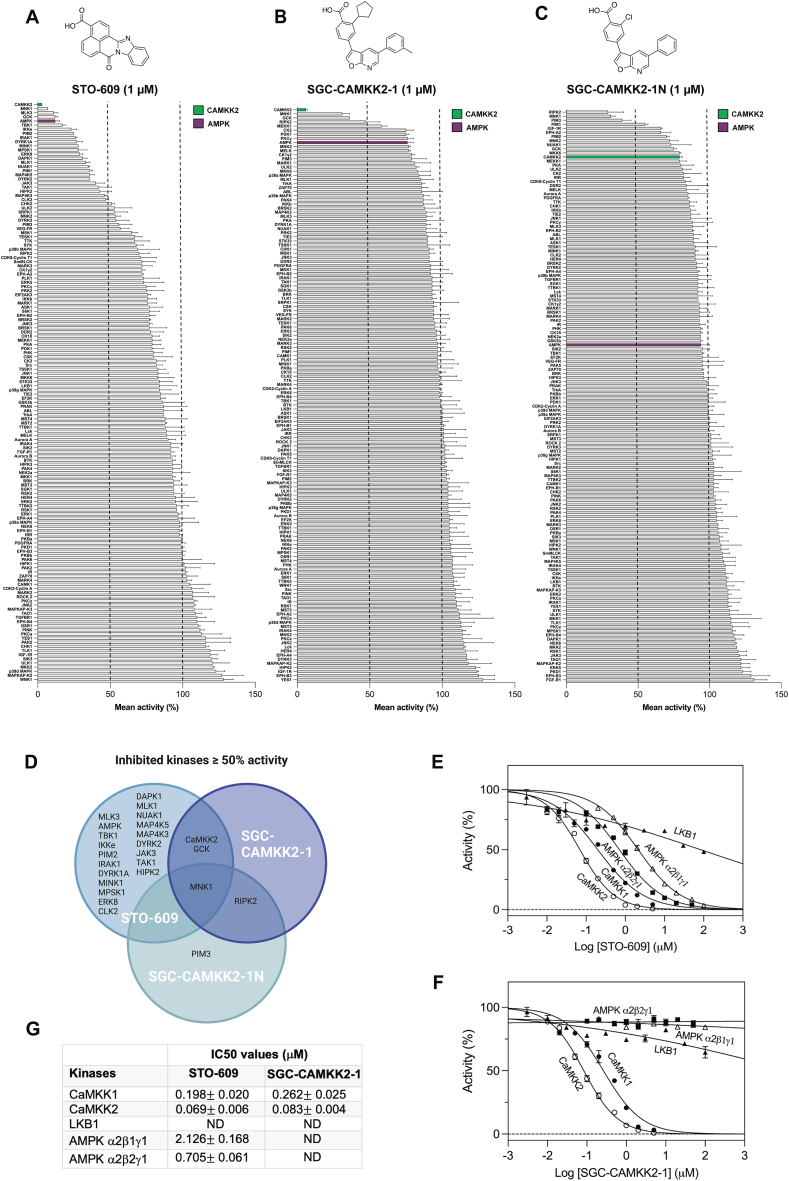
A new chemical probe, SGC-CAMKK2-1, which can be used as an additional pharmacological tool to explore CaMKK2 functions has recently been developed [28]. We took advantage of the crystal structure of UNC-YL-10, a close analog of the SGC-CAMKK2-1 [21], bound to the CaMKK2 kinase domain and overlayed SGC-CAMKK2-1 and STO-609 (Supplementary Figure 1A and B). Even though SGC-CAMKK2-1 and STO-609 share some of the same interactions, the compounds have different shapes, and thus occupy different portions of the active site. The cyclopentyl moiety that is present on the furopyridine compound UNC-YL-10, and by extension, the same group on the chemical probe SGC-CAMKK2-1, fills a space unoccupied by STO-609, likely contributing to its enhanced selectivity towards CaMKK2 (Supplementary Figure 1A and B). SGC-CAMKK2-1 was previously profiled against the DiscoverX KINOMEscan® assay panel and demonstrated significantly higher binding affinity for CaMKK1 and CaMKK2 compared to other >400 human protein kinases [28]. To further characterize and establish compound selectivity, we tested SGC-CAMKK2-1 and STO-609 in a cell-free kinase activity assay against a panel of 140 human protein kinases (Figure 1A and B). We also included SGC-CAMKK2-1N (Figure 1C), a structurally related but CaMKK2-inactive compound which had two minor modifications (i.e. removal of the methyl group from the 5-position aryl ring and replacement of the cyclopentyl moiety with a chloro) to SGC-CAMKK2-1 [28]. STO-609 and SGC-CAMKK2-1, but not SGC-CAMKK2-1N, most robustly inhibited CaMKK2 (>90% at 1 μM) among other kinases screened. Consistent with previous studies [19,20], STO-609 inhibited (>50%) several other kinases including AMPK, whereas SGC-CAMKK2-1 and its control SGC-CAMKK2-1N compound inhibited (≥50%) only a few kinases (Figure 1A–D). Notably, all three compounds commonly inhibited mitogen-activated protein kinase-interacting kinase 1 (MNK1) (Figure 1D).
Figure 1.
Kinase selectivity of STO-609, SGC-CAMKK2-1 and SGC-CAMKK2-1N. (A–C) Inhibition of a panel of 140 human protein kinases by 1 μM of CaMKK2 inhibitor STO-609 (A), SGC-CAMKK2-1 (B) and negative control SGC-CAMKK2-1N (C) with respective chemical structures. For clarity, CaMKK2 is highlighted in green and AMPK in purple. (D) Graphic showing the overlap in inhibited kinases ≥50% activity in the presence of the respective compounds (1 μM) from (A–C). Inhibition of CaMKK1, CaMKK2, AMPK α2β1γ1, AMPK α2β2γ1 and LKB1 by increasing concentrations of STO-609 (E) or SGC-CAMKK2-1 (F), presented as mean ± SEM, with LKB1 (n = 2) and all other kinases (n = 3). Data in (E and F) was subjected to a non-linear regression fitting using the model [Inhibitor] vs. normalized response – (Variable slope) performed in GraphPad Prism. (G) Estimated half-maximal inhibitory concentration (IC50) values (μM ± SEM) from kinase activity in the presence of inhibitors STO-609 or SGC-CAMKK2-1, represented in (E and F). Note that for some kinases IC50 values were not determined (ND).
We next performed a detailed in vitro enzyme kinetics analysis for the selected kinases and determined the compound concentrations giving 50% inhibition (IC50). In line with previous findings [18,19,28], inhibition of CaMKK2 by STO-609 and SGC-CAMKK2-1 was 2–3 fold more sensitive than CaMKK1, with comparable IC50 values of ∼70–80 nM for CaMKK2 and ∼200–260 nM for CaMKK1 (Figure 1E–G). However, it should be noted that because the inhibitors are competitive with ATP, the exact values depend on the ATP concentration used in the assay. STO-609, but not SGC-CAMKK2-1, inhibited AMPKα2-containing trimeric complexes at IC50 of ∼0.7–2 μM (Figure 1E–G). Both STO-609 and SGC-CAMKK2-1 did not significantly inhibit LKB1 up to high μM concentration range (Figure 1E and F).
3.2. STO-609 and SGC-CAMKK2-1 are equally potent in inhibiting ionomycin/Ca2+-stimulated AMPKα phosphorylation in LKB1-deficient A549 cells
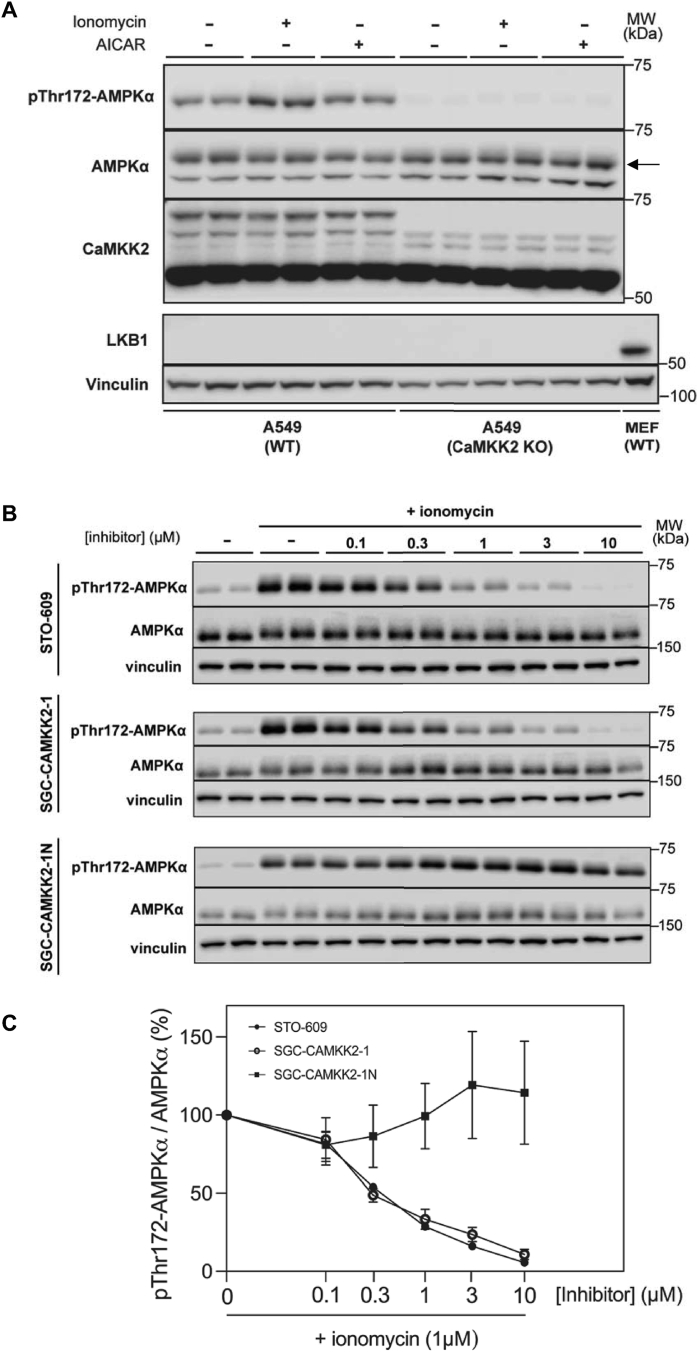
To specifically assess the efficacy of the CaMKK inhibitors (STO-609 and SGC-CAMKK2-1) in suppressing Ca2+/CaMKK2-dependent AMPKα T172 phosphorylation at cellular level, we used LKB1-deficient human lung adenocarcinoma A549 cells. Immunoblot analysis confirmed absence of LKB1 protein expression and a lack of AMP-mimetic compound (5-aminoimidazole-4-carboxamide-1-β-d-ribofuranoside)-stimulated AMPKα phosphorylation in A549 cells (Figure 2A). We also verified that both basal and ionomycin-stimulated AMPKα phosphorylation was ablated in CaMKK2 knock-out (KO) A549 cells, despite comparable expression of total AMPKα between parental/wild-type (WT) and CaMKK2 KO cells (Figure 2A). In the WT cells, ionomycin treatment of WT A549 cells increased AMPKα phosphorylation by 3–5 fold (Figure 2B). Treatment of the cells with either STO-609 or SGC-CAMKK2-1 resulted in a nearly identical dose-dependent inhibition kinetic of ionomycin-stimulated AMPKα phosphorylation (Figure 2B and C). In contrast, SGC-CAMKK2-1N, an inactive analogue of SGC-CAMKK2-1, had no inhibitory effect on AMPKα phosphorylation (Figure 2B and C). Collectively, STO-609 and SGC-CAMKK2-1 inhibit Ca2+-/CaMKK2-dependent AMPKα phosphorylation at similar potency and efficacy in A549 cells, and a complete inhibition of the AMPKα phosphorylation was achieved at 10 μM.
Figure 2.
Cellular efficacy of STO-609 and SGC-CAMKK2-1 in inhibiting Ca2+/CaMKK2-dependent phosphorylation of AMPK. (A) LKB1-deficient wild-type (WT) or CaMKK2 knock-out (KO) A549 cells were treated with either vehicle (0.1% DMSO), ionomycin (1 μM) or AICAR (2 mM) and cell extracts were subjected to immunoblot analysis with the indicated antibodies. Cell extracts from mouse embryonic fibroblasts (MEFs) were used as control for LKB1. Representative immunoblots from two independent experiments are shown. An arrow indicates the band of interest. (B) WT A549 cells were serum starved for 2 h and pre-incubated for 30 min with vehicle (−) or increasing doses of STO-609, SGC-CAMKK2-1 or SGC-CAMKK2-1N followed by treatment with or without ionomycin (1 μM) for 5 min. Cell extracts were subjected to immunoblot analysis with the indicated antibodies. Representative immunoblots from three independent experiments are shown. (C) Percent (%) changes of AMPKα Thr172 phosphorylation levels relative to the ionomycin-treated condition (100%, 0 inhibitor) shown in (B) with data points represented as mean ± SEM. n = 2 per treatment condition.
3.3. Neither STO-609 nor SGC-CAMKK2-1 inhibit AMPK phosphorylation, but both inhibit contraction-stimulated glucose uptake in mouse skeletal muscle
We assessed the effect of STO-609 or SGC-CAMKK2-1 (both at 10 μM) on AMPKα phosphorylation and glucose uptake in response to contractions (Figure 3A) evoked by electrical stimulation (10 min) in extensor digitorum longus (EDL), a glycolytic muscle, ex vivo. Incubation of isolated EDL muscle with either STO-609 or SGC-CAMKK2-1 had no significant effect on basal or contraction-stimulated phosphorylation of AMPKα and its substrate acetyl-CoA carboxylase-2 (ACC2) compared to respective vehicle-treated muscle (Figure 3B–D). In contrast, both STO-609 and SGC-CAMKK2-1 significantly inhibited (∼20%) contraction-stimulated glucose uptake in EDL muscle, but with no inhibition in rested muscle (Figure 3E). We confirmed that neither STO-609 nor SGC-CAMKK2-1 affected contractile function, as defined by peak force and fatigue curve (Figure 3F and G). The observed inhibitory effect of SGC-CAMKK2-1 was most likely off-target, as contraction-stimulated glucose uptake in EDL was also significantly inhibited with an inactive control compound SGC-CAMKK2-1N (Figure 3H) without affecting contractile function (Supplementary Figure S2A and B). The off-target effect of SGC-CAMKK2-1 on glucose uptake was further exemplified by the observations that it inhibited in response to both an allosteric AMPK activator MK-8722- and insulin-stimulated glucose uptake without affecting the phosphorylation of AMPKα or Akt, respectively (Figure 3I and J, Supplementary Figure S2C and D). Taken together, we have demonstrated that CaMKK inhibitors do not inhibit AMPKα phosphorylation but inhibit contraction-stimulated glucose uptake through a CaMKK-independent mechanism.
Figure 3.
Effect of CaMKK inhibitors on contraction-stimulated AMPK phosphorylation and glucose uptake in mouse skeletal muscles. (A) Schematic depicting the signaling pathways and experimental strategy to study the potential role for CaMKK2 in stimulating AMPK and glucose uptake in response to contractions in skeletal muscle. (B) Isolated extensor digitorum longus (EDL) muscles from wild-type C57BL/6 mice (male, an age range of 10–20 weeks) were incubated in Krebs–Ringer bicarbonate buffer ex vivo with vehicle (0.1% DMSO) or the indicated compounds (10 μM) for 1 h. During the last 10 min of incubation, muscle contractions were evoked by electrical stimulation. Muscle tissue extracts were subjected to immunoblot analysis with the indicated antibodies. Representative immunoblots are shown. (C and D) Quantification of AMPKα and ACC2 phosphorylation from (B). Data are n = 6 from two separate membranes, ∗P < 0.05 (Basal vs. contracted treatments) as indicated. (E) At the end of the 50 min incubation period with or without contractions, EDL muscles were further incubated for 10 min and [3H]-2-deoxy-glucose uptake was assessed. Shown data from n = 9–11 from two separate experiments. ∗P < 0.05 (Basal vs. indicated treatment) and #P < 0.05 (Vehicle contracted vs. indicated treatment) (F and G) Quantification of peak force production (n = 6–7) and force kinetics during the 10-min contractions in the presence of the indicated treatments. (H–J) Isolated EDL muscles were subjected to the indicated treatments and [3H]-2-deoxy-glucose uptake following the 50 min incubation period. Data are n = 5–6 (H), n = 11–12 from two separate experiments (I) and n = 6–7 (J). Data shown are ∗P < 0.05 (Basal vs. indicated treatment) and #P < 0.05 (Vehicle contracted vs. indicated treatment) (H) and #P < 0.05 (treatment vs. treatment + SGC-CAMKK2-1 (I and J). One-way ANOVA (F, I, J) and two-way ANOVA (C-E, H) tests were performed with Tukey's correction for multiple comparisons respectively. All data points shown are mean ± S.E.M. NS; not significant. PI3K; phosphoinositide-3-kinase, PDK1; phosphoinositide-dependent kinase 1, mTORC2; mTOR complex 2, GLUT4: glucose transporter 4.
3.4. CaMKK2 is not required for contraction-stimulated AMPK phosphorylation and glucose uptake in mouse skeletal muscle
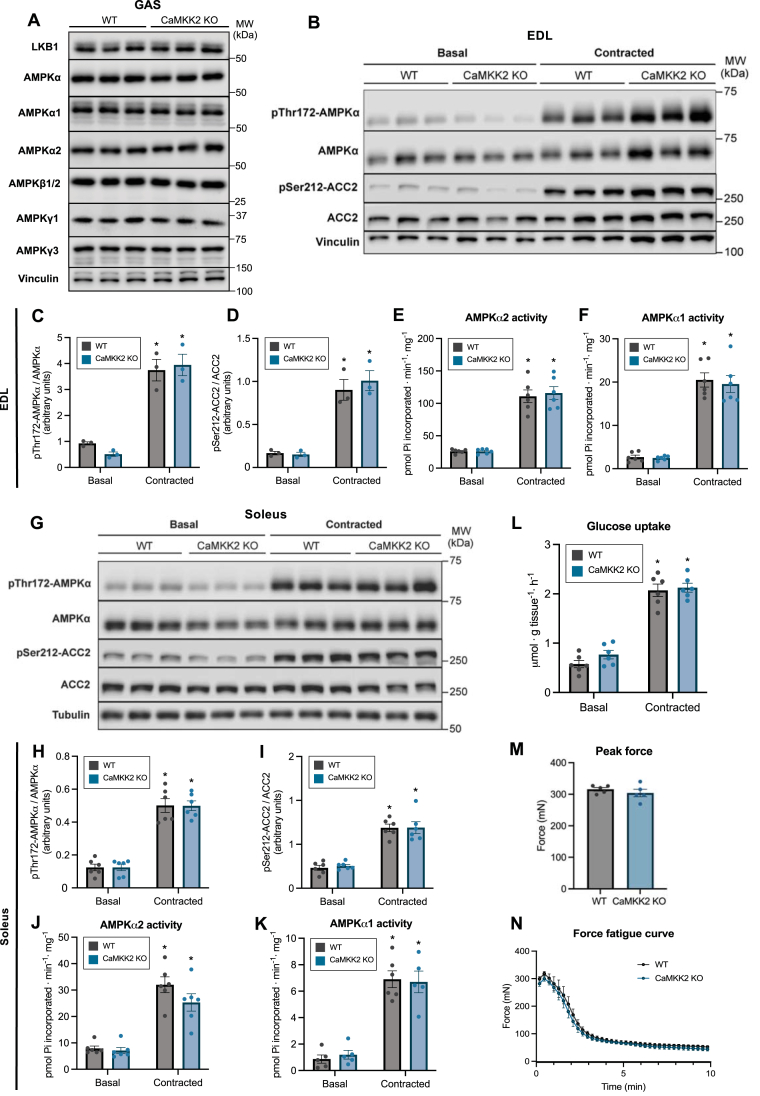
We next undertook a genetic approach and assessed if CaMKK2 is required for contraction-stimulated AMPK phosphorylation/activity and glucose uptake ex vivo employing CaMKK2 KO mouse skeletal muscles. Immunoblot analysis confirmed that protein expression of LKB1 and AMPK subunits/isoforms in skeletal muscle was comparable between WT and CaMKK2 KO mice (Figure 4A). Electrically-stimulated contractions ex vivo resulted in robust and comparable increases in phosphorylation of AMPKα and ACC2 in both EDL (glycolytic) and soleus (oxidative) muscles from WT and CaMKK2 KO mice (Figure 4B–D, G–I). Consistent with this, isoform-specific activity of AMPKα1 and α2 was robustly and similarly increased in both EDL and soleus muscles from the two genotypes (Figure 4E, F, J, K). Both basal and contraction-stimulated glucose uptake in EDL were comparable between WT and CaMKK2 KO mice (Figure 4L). There was no difference in peak force and fatigue curve of EDL muscle between WT and CaMKK2 KO mice (Figure 4M and N).
Figure 4.
CaMKK2 deficiency does not affect contraction-stimulated AMPK phosphorylation/activityand glucose uptake in mouse skeletal muscles. (A) Analysis of LKB1 and AMPK subunit/isoform expression in gastrocnemius muscles from wild-type (WT) or CaMKK2 knock-out (KO) mice (male, 17–18 weeks old). Muscle extracts were subjected to immunoblot analysis using the indicated antibodies. (B) Isolated extensor digitorum longus (EDL) muscles from WT or CaMKK2 KO mice (male, 14–15 weeks old) were incubated in Krebs–Ringer bicarbonate buffer ex vivo for 1 h. During the last 10 min of incubation, muscle contractions were evoked by electrical stimulation. Muscle tissue extracts were subjected to immunoblot analysis with the indicated antibodies. Representative immunoblots are shown. (C and D) Quantification of AMPKα and ACC2 phosphorylation shown in (B). (E and F) Isoform-specific AMPK activity in WT and CaMKK2 KO mice (male, 17 weeks old). (G) Soleus muscle from the indicated genotypes was analyzed using the same experimental setup as (B). (H and I) Quantification of AMPKα and ACC2 phosphorylation shown in (G) and (J and K) isoform-specific AMPK activity from WT and CaMKK2 KO mice (male, 17–18 weeks). (L) [3H]-2-deoxy-glucose uptake in isolated EDL muscle from the indicated genotypes in response to contractions ex vivo. (M and N) Quantification of peak force production and force kinetics during the 10-min contractions. Data are mean ± S.E.M. n = 3 for (C and D) and n = 5–6 (E and F, H–M). A two-way ANOVA test was performed with Tukey's correction for multiple comparisons. ∗P < 0.05 (Basal vs. contracted).
3.5. CaMKK2 protein is undetectable in mouse skeletal muscle
It has been controversial as to whether CaMKK2 protein is expressed in skeletal muscle fiber/tissue. We wanted to definitively address this and first characterized isoform-specificity of the CaMKK antibodies (CaMKK2 and pan-CaMKK1/2) used in the current study. FLAG-tagged recombinant CaMKK1 or CaMKK2 was ectopically expressed in HEK293 cells and immunopurified from the cell lysates using anti-FLAG antibody-conjugated agarose beads. Immunoblotting of the purified CaMKK1 and CaMKK2 with FLAG, pan-CaMKK and isoform-specific CaMKK2 antibody validated equal protein loading, isoform-cross reactivity, and isoform-specificity, respectively (Figure 5A). We next sought to validate specificity of the CaMKK antibodies for detection of endogenous CaMKK proteins in WT or CaMKK2 KO mouse brain and WT or CaMKK2 KO A549 cell lysates. Using the CaMKK2-specific antibody, we detected multiple variants ranging from ∼60 to 75 kDa in WT mouse brain and A549 cell extracts (Figure 5B). A few non-specific bands were detected in CaMKK2 KO brain/A549 cell lysates. In contrast, in mouse skeletal muscle we were not able to detect any signal/bands within the expected ranges (∼60–75 kDa) of CaMKK2 variants when we loaded up to 40 μg of tissue extracts (Figure 5C). Even though a distinct band was detected just above 75 kDa molecular weight indicator, it was present equally in both WT and CaMKK2 KO skeletal muscle lysates. A faint band was also detected at ∼75 kDa (just below the distinct band), but it was also present equally in both genotypes (Figure 5C). Pan-CaMKK antibody detected several variants/bands in WT brain lysates, however the majority of which, except a single distinct band, was undetectable in CaMKK2 KO lysates. In contrast, we were not able to detect any visible signal in either WT or CaMKK2 KO mouse skeletal muscle lysates (Figure 5C). We took an alternative approach to detect CaMKK2 through enrichment of calmodulin-binding proteins using the calmodulin-agarose affinity purification (Figure 5D). We were able to detect markedly higher amount of CaMKKs through calmodulin-binding protein enrichment from 200 μg brain lysates compared to direct immunoblotting of 10 μg brain lysates. Employing the calmodulin-agarose affinity purification method using up to 2,000 μg skeletal muscle lysates, we were able to detect CaMKII (a calmodulin binding protein), but not CaMKK2. To determine if CaMKK2 is expressed at transcript/mRNA level in mouse skeletal muscle tissue or fiber, we performed a quantitative RT-PCR analysis. Camkk2 mRNA was detectable in isolated skeletal muscle fibers, but its expression was much lower compared to brain and also much lower compared to whole skeletal muscle tissue (Figure 5E). We further investigated and evaluated CaMKK2 and CaMKK1 protein expression by mass spectrometry-based proteomics of mouse skeletal muscle and C2C12 myotubes. We obtained the raw mass spectrometry files from a previous study [36] and reprocessed them using the latest software (MaxQuant version 2.0.1.0) and updated FASTA file. The re-analysis yielded consistent results with the earlier findings [36], as CaMKK2 and CaMKK1 were identified exclusively in C2C12 myotubes, with 9 and 4 detected peptides, respectively. These peptides are not detected in mouse skeletal muscle (Supplementary Figure S4A and B).
Relatively low expression of Camkk1 mRNA was also detected in mouse skeletal muscle tissue at comparable levels between WT and CaMKK2 KO mice, but it was only marginally detected in isolated muscle fibers from both genotypes (Supplementary Figure S3A).
4. Discussion
Previous studies proposing the involvement of CaMKK2 in activating AMPK through α-Thr172 phosphorylation in response to contractions relied on the use of STO-609. While one study in isolated mouse muscles demonstrated significant inhibitory effects of STO-609 (5 μM) on contraction-stimulated AMPK activation and glucose uptake [23], another study demonstrated no significant effect of STO-609 (∼2.7 μM) on either AMPK (Thr172) phosphorylation or muscle glucose uptake [24].
Even though we used the same contraction protocol [23], higher concentration of STO-609 (10 μM) [23], and an additional structurally distinct CaMKK2 selective inhibitor (SGC-CAMKK2-1) [28], we did not observe inhibition of contraction-stimulated AMPK phosphorylation and activity in mouse skeletal muscle. This discrepancy could stem from purity of the STO-609 inhibitor between the studies. We thoroughly tested the effects of CaMKK inhibitors in cell-free and cell-based assays and confirmed their potency, selectivity, cellular efficacy [20,28] as well as non-significant effect on contractile functions during the ex vivo skeletal muscle incubation studies. Nonetheless, our results are further backed up using an alternative loss of function approach where we demonstrated that genetic ablation of CaMKK2 does not affect contraction-stimulated AMPK phosphorylation and isoform-specific activity in glycolytic and oxidative mouse skeletal muscle.
It is not uncommon that protein kinase inhibitors affect glucose uptake in skeletal muscle and adipocytes through off-target effects. The p38 MAPK inhibitor SB-203580, for example, has been shown to inhibit insulin-stimulated glucose uptake in skeletal muscle tissue/cells and adipocytes [37]. Using a SB-203580-resistant mutant and knockout of p38 MAPK it was unambiguously shown that the inhibitory effect of SB-203580 on glucose transport was independent of p38 MAPK [37,38]. More recently, we and others have shown that SBI-0206965, a selective inhibitor of AMPK and ULK1 [39,40], potently inhibits basal and insulin-stimulated glucose uptake independent of AMPK in skeletal muscle tissues/cells and adipocytes [30,41]. To mitigate off-target effects of a given compound, a structurally related but inactive compound, as well as genetic models (e.g., knockout or drug-insensitive mutant) should ideally be used alongside the compound to give additional confidence in its physiological effects. We took advantage of SGC-CAMKK2-1N, a structurally related, but inactive compound of SGC-CAMKK2-1 [28], to substantiate that the inhibitory effect of SGC-CAMKK2-1 on glucose uptake is CaMKK2-independent. Consistent with the previous studies [19,20,26], STO-609 (1 μM) inhibited an additional ∼20 kinases in cell-free assays (50% reduction), including Nuak1, GCK, MNK1, MLKs, DAMK, DYRKs, and RIPK2. In contrast, SGC-CAMKK2-1 (1 μM) only inhibited three kinases that were also inhibited by STO-609 (i.e., CaMKK2, GCK, and MNK1 as well as CaMKK1). Notably, we found that MNK1 was inhibited not only with STO-609 and SGC-CAMKK2-1, but also SGC-CAMKK2-1N in cell-free assay. We did not investigate if MNK1 could be inhibited in skeletal muscle tissue by CaMKK inhibitors, but pharmacological inhibition of ERK1/2, upstream kinases of MNK1, did not affect contraction-stimulated glucose uptake in skeletal muscle [42].
CaMKK2 is highly expressed in brain and distributed to many regions [[43], [44], [45]], while it is detected at lower levels in testis, spleen, and lung [44]. Further, protein expression of CaMKK2 has been confirmed in preadipocytes [46], embryonic fibroblasts, hepatocytes [47], mesenchymal stem cells, osteoblasts, osteocytes and osteoclasts [48,49] and in cells of the myeloid lineage, including bone marrow-derived and peritoneal macrophages [50,51], using respective cell types from CaMKK2 KO mice as control. It had been controversial whether CaMKK2 is expressed in skeletal muscle, mainly due to a lack of robust antibody validation along with the use of CaMKK2 KO muscle as negative control. It was previously reported that CaMKKs were difficult to detect in mouse skeletal muscle by direct immunoblotting. However, prior affinity-purification of calmodulin-binding proteins from mouse skeletal muscle extracts followed by immunoblotting with a pan-CaMKK antibody resulted in a detection of polypeptides of the expected size [23]. Even though we could robustly detect CaMKK2 and CaMKII in brain lysates and CaMKII in skeletal muscle lysates following the calmodulin-binding protein enrichment, we could not detect CaMKK2 in mouse muscle using either CaMKK2 isoform-specific or pan-CaMKK antibody. We have no clear explanation to reconcile this discrepancy, however, the previous study did not clarify the identity of the specific CaMKK isoform detected by the pan-CaMKK antibody [23]. Consistent with our current observation, a previous study reported an absence of CaMKK2 protein expression in mouse skeletal muscle [52]. Notably, however, studies examining skeletal muscle following one week of chronic mechanical overloading by ablation of synergistic muscle demonstrated a marked increase in the expression of both CaMKK1 [52,53] and CaMKK2 proteins in mouse plantaris muscle [52]. Mechanical overloading causes profound inflammatory responses, starting with immune cell invasion in the muscle interstitium and the number of macrophages progressively increases, reaching a peak at day 5–7 [54]. It is unknown whether the detected CaMKK proteins following the overloading originated from myofibers or non-muscle cells within skeletal muscle tissue. It would be interesting to perform single-cell analysis of CaMKK expression within skeletal muscle tissue following mechanical overloading. According to the current Human Protein Atlas (https://www.proteinatlas.org/), CaMKK2 proteins have been identified in skeletal muscle tissue, but not in myocyte. Taken together, even though we could detect relatively low levels of Camkk2 mRNA in isolated mouse skeletal muscle fibers by qPCR, we were unable to reliably detect CaMKK2 protein by immunoblotting with or without prior enrichment of calmodulin binding proteins in mouse skeletal muscle. We conclude that expression of CaMKK2 protein is below the detection limit of currently available reagents/methods we tested.
It has been suggested that CaMKK2 plays an inhibitory role in myoblast proliferation and differentiation through AMPK in the C2C12 muscle cell line, and that upon induction of myoblast differentiation the expression of CaMKK2 was downregulated [55]. We also observed that CaMKK2 is abundantly expressed in C2C12 myoblasts and that it was modestly reduced upon differentiation into myotubes (Supplementary Figure S3B and C), while expression of skeletal muscle selective AMPKγ3 isoform is markedly induced upon differentiation (Supplementary Figure S3B and D). Caution needs to be taken when studying skeletal muscle regulation and physiological role of CaMKK and AMPK pathways using C2C12 cells as a model system, as they are markedly different from intact skeletal muscle in terms of their isoform expression and abundance as well as expression profiles of their downstream metabolic regulators [36]. A deep proteomics analysis of mouse skeletal muscle and C2C12 myotubes revealed profound differences in expression profiles of AMPK isoforms and upstream kinases. C2C12 myotube predominantly expresses α1-AMPK in contrast to predominant expression of α2-AMPK in mouse skeletal muscle [36]. CaMKK2 was not detected in recently published studies of the human skeletal muscle proteome [[56], [57], [58], [59]]. This suggests that with the existing proteomics technology, there is no detectable expression of CaMKK2 in mouse or human skeletal muscle tissue.
5. Conclusions
We demonstrated that pharmacological inhibition or genetic loss of CaMKK2 does not affect contraction-stimulated AMPK α-Thr172 phosphorylation and activity, as well as glucose uptake in skeletal muscle. Previously observed inhibitory effect of STO-609 on AMPK activity and glucose uptake is likely due to off-target effects. CaMKK2 protein is either absent from adult skeletal muscle or below the detection limit of currently available methods.
Author contributions
Conceptualization: K.S., C.A.W. Experimental design: F.N., A.B.A., K.H., J.W.S., C.A.W., K.S. Experimental execution: F.N., A.B.A., K.H., C.F.B., P.S.H., T.J.S., M.A., C.T.A.L., D.A., J.W.S., C.A.W. Reagent/tool development and donation: N.X.Y.L., J.S.O., M.A.H., D.H.D., U.S. Data analysis: F.N., A.B.A., K.H., C.F.B., P.S.H., J.K.L., A.S.D., J.W.S., C.A.W., K.S. Data visualization and figure formatting: C.F.B. Supervision: K.S., C.A.W., J.O., J.J.B. Writing- Original draft preparation: K.S. Writing- Reviewing and Editing: All authors.
Grants
The work is supported by the Novo Nordisk Foundation (NNF21OC0070257) to K.S. Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center based at the University of Copenhagen, Denmark, and partially funded by an unconditional donation from the Novo Nordisk Foundation (Grant number NNF18CC0034900). This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases (R01DK103562) and National Institute of Arthritis and Musculoskeletal Diseases (R21AR081593) grants to C.A.W., the Carlsberg Foundation grant (CF20-0113) to J.O., National Cancer Institute of the National Institutes of Health (R01 CA218442) to D.H.D., National Institutes of Health (R01AR070200) to J.B and National Institute of Arthritis and Musculoskeletal Diseases (R01AR076477) to U.S. J.W.S is funded by the Australian Research Council (DP210102840) and National Medical and Health Research Council (GNT2001817). J.S.O. is funded by the Australian Research Council (DP220103700). The Structural Genomics Consortium (SGC) is a registered charity (number 1097737) that receives funds from Bayer AG, Boehringer Ingelheim, the Canada Foundation for Innovation, the Eshelman Institute for Innovation, Genentech, Genome Canada through Ontario Genomics Institute [OGI-196], EU/EFPIA/OICR/McGill/KTH/Diamond, Innovative Medicines Initiative 2 Joint Undertaking [EUbOPEN grant 875510], Janssen, Merck KGaA (aka EMD in Canada and USA),Pfizer, the Sao Paulo Research Foundation-FAPESP, and Takeda.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We thank Thomas E. Jensen and Erik A. Richter (University of Copenhagen) for their valuable input. We thank D. Grahame Hardie (University of Dundee) for his generous donation of CaMKK2 antibody. We thank Amy Ehrlich and LeAnna Phillips for their technical assistance. We thank Han Wee Ong for creation of supplementary Figure S1. Illustrations shown in Graphical Abstract, Figure 1D, and Figure 3A were created with BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101761.
Contributor Information
Carol A. Witczak, Email: cwitczak@iu.edu.
Kei Sakamoto, Email: kei.sakamoto@sund.ku.dk.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Hardie D.G., Sakamoto K. AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology. 2006;21:48–60. doi: 10.1152/physiol.00044.2005. [DOI] [PubMed] [Google Scholar]
- 2.Garcia D., Shaw R.J. AMPK: mechanisms of cellular energy sensing and restoration of metabolic balance. Mol Cell. 2017;66(6):789–800. doi: 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Treebak J.T., Birk J.B., Hansen B.F., Olsen G.S., Wojtaszewski J.F. A-769662 activates AMPK beta1-containing complexes but induces glucose uptake through a PI3-kinase-dependent pathway in mouse skeletal muscle. Am J Physiol Cell Physiol. 2009;297(4):C1041–C1052. doi: 10.1152/ajpcell.00051.2009. [DOI] [PubMed] [Google Scholar]
- 4.Hawley S.A., Davison M., Woods A., Davies S.P., Beri R.K., Carling D., et al. Characterization of the AMP-activated protein kinase kinase from rat liver and identification of threonine 172 as the major site at which it phosphorylates AMP-activated protein kinase. J Biol Chem. 1996;271(44):27879–27887. doi: 10.1074/jbc.271.44.27879. [DOI] [PubMed] [Google Scholar]
- 5.Xiao B., Sanders M.J., Underwood E., Heath R., Mayer F.V., Carmena D., et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472(7342):230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oakhill J.S., Steel R., Chen Z.P., Scott J.W., Ling N., Tam S., et al. AMPK is a direct adenylate charge-regulated protein kinase. Science. 2011;332(6036):1433–1435. doi: 10.1126/science.1200094. [DOI] [PubMed] [Google Scholar]
- 7.Gowans G.J., Hawley S.A., Ross F.A., Hardie D.G. AMP is a true physiological regulator of AMP-activated protein kinase by both allosteric activation and enhancing net phosphorylation. Cell Metab. 2013;18(4):556–566. doi: 10.1016/j.cmet.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hardie D.G., Ross F.A., Hawley S.A. AMPK: a nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thomson D.M., Porter B.B., Tall J.H., Kim H.J., Barrow J.R., Winder W.W. Skeletal muscle and heart LKB1 deficiency causes decreased voluntary running and reduced muscle mitochondrial marker enzyme expression in mice. Am J Physiol Endocrinol Metab. 2007;292(1):E196–E202. doi: 10.1152/ajpendo.00366.2006. [DOI] [PubMed] [Google Scholar]
- 10.Koh H.J., Arnolds D.E., Fujii N., Tran T.T., Rogers M.J., Jessen N., et al. Skeletal muscle-selective knockout of LKB1 increases insulin sensitivity, improves glucose homeostasis, and decreases TRB3. Mol Cell Biol. 2006;26(22):8217–8227. doi: 10.1128/MCB.00979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sakamoto K., McCarthy A., Smith D., Green K.A., Hardie D.G., Ashworth A., et al. Deficiency of LKB1 in skeletal muscle prevents AMPK activation and glucose uptake during contraction. EMBO J. 2005;24(10):1810–1820. doi: 10.1038/sj.emboj.7600667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koh H.J., Toyoda T., Fujii N., Jung M.M., Rathod A., Middelbeek R.J., et al. Sucrose nonfermenting AMPK-related kinase (SNARK) mediates contraction-stimulated glucose transport in mouse skeletal muscle. Proc Natl Acad Sci U S A. 2010;107(35):15541–15546. doi: 10.1073/pnas.1008131107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeppesen J., Maarbjerg S.J., Jordy A.B., Fritzen A.M., Pehmoller C., Sylow L., et al. LKB1 regulates lipid oxidation during exercise independently of AMPK. Diabetes. 2013;62(5):1490–1499. doi: 10.2337/db12-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawley S.A., Selbert M.A., Goldstein E.G., Edelman A.M., Carling D., Hardie D.G. 5'-AMP activates the AMP-activated protein kinase cascade, and Ca2+/calmodulin activates the calmodulin-dependent protein kinase I cascade, via three independent mechanisms. J Biol Chem. 1995;270(45):27186–27191. doi: 10.1074/jbc.270.45.27186. [DOI] [PubMed] [Google Scholar]
- 15.Steinberg G.R., Hardie D.G. New insights into activation and function of the AMPK. Nat Rev Mol Cell Biol. 2022;24(4):255–272. doi: 10.1038/s41580-022-00547-x. [DOI] [PubMed] [Google Scholar]
- 16.Means A.R. The Year in Basic Science: calmodulin kinase cascades. Mol Endocrinol. 2008;22(12):2759–2765. doi: 10.1210/me.2008-0312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokumitsu H., Sakagami H. Molecular mechanisms underlying Ca(2+)/calmodulin-dependent protein kinase kinase signal transduction. Int J Mol Sci. 2022;23(19) doi: 10.3390/ijms231911025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tokumitsu H., Inuzuka H., Ishikawa Y., Ikeda M., Saji I., Kobayashi R. STO-609, a specific inhibitor of the Ca(2+)/calmodulin-dependent protein kinase kinase. J Biol Chem. 2002;277(18):15813–15818. doi: 10.1074/jbc.M201075200. [DOI] [PubMed] [Google Scholar]
- 19.Hawley S.A., Pan D.A., Mustard K.J., Ross L., Bain J., Edelman A.M., et al. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2(1):9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 20.Bain J., Plater L., Elliott M., Shpiro N., Hastie C.J., McLauchlan H., et al. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408(3):297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eduful B.J., O'Byrne S.N., Temme L., Asquith C.R.M., Liang Y., Picado A., et al. Hinge binder scaffold hopping identifies potent calcium/calmodulin-dependent protein kinase kinase 2 (CAMKK2) inhibitor chemotypes. J Med Chem. 2021;64(15):10849–10877. doi: 10.1021/acs.jmedchem.0c02274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abbott M.J., Edelman A.M., Turcotte L.P. CaMKK is an upstream signal of AMP-activated protein kinase in regulation of substrate metabolism in contracting skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2009;297(6):R1724–R1732. doi: 10.1152/ajpregu.00179.2009. [DOI] [PubMed] [Google Scholar]
- 23.Jensen T.E., Rose A.J., Jorgensen S.B., Brandt N., Schjerling P., Wojtaszewski J.F., et al. Possible CaMKK-dependent regulation of AMPK phosphorylation and glucose uptake at the onset of mild tetanic skeletal muscle contraction. Am J Physiol Endocrinol Metab. 2007;292(5):E1308–E1317. doi: 10.1152/ajpendo.00456.2006. [DOI] [PubMed] [Google Scholar]
- 24.Witczak C.A., Fujii N., Hirshman M.F., Goodyear L.J. Ca2+/calmodulin-dependent protein kinase kinase-alpha regulates skeletal muscle glucose uptake independent of AMP-activated protein kinase and Akt activation. Diabetes. 2007;56(5):1403–1409. doi: 10.2337/db06-1230. [DOI] [PubMed] [Google Scholar]
- 25.Fujiwara Y., Hiraoka Y., Fujimoto T., Kanayama N., Magari M., Tokumitsu H. Analysis of distinct roles of CaMKK isoforms using STO-609-resistant mutants in living cells. Biochemistry. 2015;54(25):3969–3977. doi: 10.1021/acs.biochem.5b00149. [DOI] [PubMed] [Google Scholar]
- 26.O'Byrne S.N., Scott J.W., Pilotte J.R., Santiago A.D.S., Langendorf C.G., Oakhill J.S., et al. In depth analysis of kinase cross screening data to identify CAMKK2 inhibitory scaffolds. Molecules. 2020;25(2) doi: 10.3390/molecules25020325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcelo K.L., Means A.R., York B. The Ca(2+)/calmodulin/CaMKK2 axis: nature's metabolic CaMshaft. Trends Endocrinol Metab. 2016;27(10):706–718. doi: 10.1016/j.tem.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells C., Liang Y., Pulliam T.L., Lin C., Awad D., Eduful B., et al. SGC-CAMKK2-1: a chemical probe for CAMKK2. Cells. 2023;12(2) doi: 10.3390/cells12020287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson K.A., Ribar T.J., Lin F., Noeldner P.K., Green M.F., Muehlbauer M.J., et al. Hypothalamic CaMKK2 contributes to the regulation of energy balance. Cell Metab. 2008;7(5):377–388. doi: 10.1016/j.cmet.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Ahwazi D., Neopane K., Markby G.R., Kopietz F., Ovens A.J., Dall M., et al. Investigation of the specificity and mechanism of action of the ULK1/AMPK inhibitor SBI-0206965. Biochem J. 2021;478(15):2977–2997. doi: 10.1042/BCJ20210284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Collodet C., Foretz M., Deak M., Bultot L., Metairon S., Viollet B., et al. AMPK promotes induction of the tumor suppressor FLCN through activation of TFEB independently of mTOR. FASEB J. 2019;33(11):12374–12391. doi: 10.1096/fj.201900841R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dite T.A., Ling N.X.Y., Scott J.W., Hoque A., Galic S., Parker B.L., et al. The autophagy initiator ULK1 sensitizes AMPK to allosteric drugs. Nat Commun. 2017;8(1):571. doi: 10.1038/s41467-017-00628-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ngoei K.R.W., Langendorf C.G., Ling N.X.Y., Hoque A., Varghese S., Camerino M.A., et al. Structural determinants for small-molecule activation of skeletal muscle AMPK alpha2beta2gamma1 by the glucose importagog SC4. Cell Chem Biol. 2018;25(6):728–737 e729. doi: 10.1016/j.chembiol.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Scott J.W., Park E., Rodriguiz R.M., Oakhill J.S., Issa S.M., O'Brien M.T., et al. Autophosphorylation of CaMKK2 generates autonomous activity that is disrupted by a T85S mutation linked to anxiety and bipolar disorder. Sci Rep. 2015;5:14436. doi: 10.1038/srep14436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakamoto K., Goransson O., Hardie D.G., Alessi D.R. Activity of LKB1 and AMPK-related kinases in skeletal muscle: effects of contraction, phenformin, and AICAR. Am J Physiol Endocrinol Metab. 2004;287(2):E310–E317. doi: 10.1152/ajpendo.00074.2004. [DOI] [PubMed] [Google Scholar]
- 36.Deshmukh A.S., Murgia M., Nagaraj N., Treebak J.T., Cox J., Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol Cell Proteomics. 2015;14(4):841–853. doi: 10.1074/mcp.M114.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antonescu C.N., Huang C., Niu W., Liu Z., Eyers P.A., Heidenreich K.A., et al. Reduction of insulin-stimulated glucose uptake in L6 myotubes by the protein kinase inhibitor SB203580 is independent of p38MAPK activity. Endocrinology. 2005;146(9):3773–3781. doi: 10.1210/en.2005-0404. [DOI] [PubMed] [Google Scholar]
- 38.Turban S., Beardmore V.A., Carr J.M., Sakamoto K., Hajduch E., Arthur J.S., et al. Insulin-stimulated glucose uptake does not require p38 mitogen-activated protein kinase in adipose tissue or skeletal muscle. Diabetes. 2005;54(11):3161–3168. doi: 10.2337/diabetes.54.11.3161. [DOI] [PubMed] [Google Scholar]
- 39.Dite T.A., Langendorf C.G., Hoque A., Galic S., Rebello R.J., Ovens A.J., et al. AMP-activated protein kinase selectively inhibited by the type II inhibitor SBI-0206965. J Biol Chem. 2018;293(23):8874–8885. doi: 10.1074/jbc.RA118.003547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Egan D.F., Chun M.G., Vamos M., Zou H., Rong J., Miller C.J., et al. Small molecule inhibition of the autophagy kinase ULK1 and identification of ULK1 substrates. Mol Cell. 2015;59(2):285–297. doi: 10.1016/j.molcel.2015.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knudsen J.R., Madsen A.B., Persson K.W., Henriquez-Olguin C., Li Z., Jensen T.E. The ULK1/2 and AMPK inhibitor SBI-0206965 blocks AICAR and insulin-stimulated glucose transport. Int J Mol Sci. 2020;21(7) doi: 10.3390/ijms21072344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wojtaszewski J.F., Lynge J., Jakobsen A.B., Goodyear L.J., Richter E.A. Differential regulation of MAP kinase by contraction and insulin in skeletal muscle: metabolic implications. Am J Physiol. 1999;277(4):E724–E732. doi: 10.1152/ajpendo.1999.277.4.E724. [DOI] [PubMed] [Google Scholar]
- 43.Gong S., Zheng C., Doughty M.L., Losos K., Didkovsky N., Schambra U.B., et al. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425(6961):917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- 44.Anderson K.A., Means R.L., Huang Q.H., Kemp B.E., Goldstein E.G., Selbert M.A., et al. Components of a calmodulin-dependent protein kinase cascade. Molecular cloning, functional characterization and cellular localization of Ca2+/calmodulin-dependent protein kinase kinase beta. J Biol Chem. 1998;273(48):31880–31889. doi: 10.1074/jbc.273.48.31880. [DOI] [PubMed] [Google Scholar]
- 45.Ohmstede C.A., Jensen K.F., Sahyoun N.E. Ca2+/calmodulin-dependent protein kinase enriched in cerebellar granule cells. Identification of a novel neuronal calmodulin-dependent protein kinase. J Biol Chem. 1989;264(10):5866–5875. [PubMed] [Google Scholar]
- 46.Lin F., Ribar T.J., Means A.R. The Ca2+/calmodulin-dependent protein kinase kinase, CaMKK2, inhibits preadipocyte differentiation. Endocrinology. 2011;152(10):3668–3679. doi: 10.1210/en.2011-1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Anderson K.A., Lin F., Ribar T.J., Stevens R.D., Muehlbauer M.J., Newgard C.B., et al. Deletion of CaMKK2 from the liver lowers blood glucose and improves whole-body glucose tolerance in the mouse. Mol Endocrinol. 2012;26(2):281–291. doi: 10.1210/me.2011-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cary R.L., Waddell S., Racioppi L., Long F., Novack D.V., Voor M.J., et al. Inhibition of Ca(2)(+)/calmodulin-dependent protein kinase kinase 2 stimulates osteoblast formation and inhibits osteoclast differentiation. J Bone Miner Res. 2013;28(7):1599–1610. doi: 10.1002/jbmr.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams J.N., Irwin M., Li Y., Kambrath A.V., Mattingly B.T., Patel S., et al. Osteocyte-derived CaMKK2 regulates osteoclasts and bone mass in a sex-dependent manner through secreted calpastatin. Int J Mol Sci. 2023;24(5) doi: 10.3390/ijms24054718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Racioppi L., Noeldner P.K., Lin F., Arvai S., Means A.R. Calcium/calmodulin-dependent protein kinase kinase 2 regulates macrophage-mediated inflammatory responses. J Biol Chem. 2012;287(14):11579–11591. doi: 10.1074/jbc.M111.336032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Racioppi L., Nelson E.R., Huang W., Mukherjee D., Lawrence S.A., Lento W., et al. CaMKK2 in myeloid cells is a key regulator of the immune-suppressive microenvironment in breast cancer. Nat Commun. 2019;10(1):2450. doi: 10.1038/s41467-019-10424-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGee S.L., Mustard K.J., Hardie D.G., Baar K. Normal hypertrophy accompanied by phosphoryation and activation of AMP-activated protein kinase alpha1 following overload in LKB1 knockout mice. J Physiol. 2008;586(6):1731–1741. doi: 10.1113/jphysiol.2007.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ferey J.L., Brault J.J., Smith C.A., Witczak C.A. Constitutive activation of CaMKKalpha signaling is sufficient but not necessary for mTORC1 activation and growth in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2014;307(8):E686–E694. doi: 10.1152/ajpendo.00322.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bernard C., Zavoriti A., Pucelle Q., Chazaud B., Gondin J. Role of macrophages during skeletal muscle regeneration and hypertrophy-implications for immunomodulatory strategies. Physiol Rep. 2022;10(19) doi: 10.14814/phy2.15480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ye C., Zhang D., Zhao L., Li Y., Yao X., Wang H., et al. CaMKK2 suppresses muscle regeneration through the inhibition of myoblast proliferation and differentiation. Int J Mol Sci. 2016;17(10) doi: 10.3390/ijms17101695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hingst J.R., Bruhn L., Hansen M.B., Rosschou M.F., Birk J.B., Fentz J., et al. Exercise-induced molecular mechanisms promoting glycogen supercompensation in human skeletal muscle. Mol Metab. 2018;16:24–34. doi: 10.1016/j.molmet.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hostrup M., Lemminger A.K., Stocks B., Gonzalez-Franquesa A., Larsen J.K., Quesada J.P., et al. High-intensity interval training remodels the proteome and acetylome of human skeletal muscle. Elife. 2022;11 doi: 10.7554/eLife.69802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Karlsen A., Gonzalez-Franquesa A., Jakobsen J.R., Krogsgaard M.R., Koch M., Kjaer M., et al. The proteomic profile of the human myotendinous junction. iScience. 2022;25(2):103836. doi: 10.1016/j.isci.2022.103836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Blazev R., Carl C.S., Ng Y.K., Molendijk J., Voldstedlund C.T., Zhao Y., et al. Phosphoproteomics of three exercise modalities identifies canonical signaling and C18ORF25 as an AMPK substrate regulating skeletal muscle function. Cell Metab. 2022;34(10):1561–1577 e1569. doi: 10.1016/j.cmet.2022.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.