Abstract
Immune checkpoint blockade (ICB) treatment has demonstrated excellent medical effects in oncology, and it is one of the most sought after immunotherapies for tumors. However, there are several issues with ICB therapy, including low response rates and a lack of effective efficacy predictors. Gasdermin-mediated pyroptosis is a typical inflammatory death mode. We discovered that increased expression of gasdermin protein was linked to a favorable tumor immune microenvironment and prognosis in head and neck squamous cell carcinoma (HNSCC). We used the mouse HNSCC cell lines 4MOSC1 (responsive to CTLA-4 blockade) and 4MOSC2 (resistant to CTLA-4 blockade) orthotopic models and demonstrated that CTLA-4 blockade treatment induced gasdermin-mediated pyroptosis of tumor cells, and gasdermin expression positively correlated to the effectiveness of CTLA-4 blockade treatment. We found that CTLA-4 blockade activated CD8+ T cells and increased the levels of interferon γ (IFN-γ) and tumor necrosis factor α (TNF-α) cytokines in the tumor microenvironment. These cytokines synergistically activated the STAT1/IRF1 axis to trigger tumor cell pyroptosis and the release of large amounts of inflammatory substances and chemokines. Collectively, our findings revealed that CTLA-4 blockade triggered tumor cells pyroptosis via the release of IFN-γ and TNF-α from activated CD8+ T cells, providing a new perspective of ICB.
Keywords: pyroptosis, immune checkpoint blockade, CTLA-4 blockade, CD8+ T cells, gasdermins, IFN-γ, TNF-α, head and neck squamous cell carcinoma
Graphical abstract
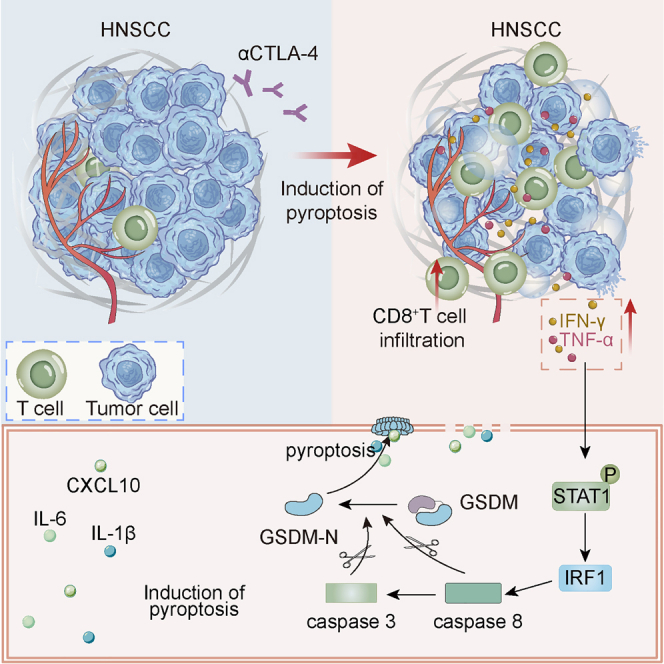
Sun and colleagues found that CTLA-4 blockade treatment induced gasdermin-mediated pyroptosis, which revealed an antitumor mechanism of immune checkpoint blockade treatment and suggested that targeting this pathway in combination with immune checkpoint blockade treatment is a therapeutic approach.
Introduction
Head and neck squamous cell carcinoma (HNSCC) is one of the most common cancers, and its incidence has risen recently due to an increase in the number of cases of human papillomavirus (HPV)-related cancer.1,2 Immune checkpoint blockade (ICB) therapies that target the programmed cell death protein 1 pathway (PD-1/PD-L1) and cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) have changed the therapies of many cancers and have resulted in excellent survival in some oncological patients in recent years.3,4,5 However, ICB response rate in HNSCC is only approximately 20%, which highlights the significance of elucidating the underlying mechanisms of initial resistance to immunotherapies.6 A critical objective in targeting ICB-resistant (e.g., “cold,” immune-excluded, or immune-desert) tumors is the induction of inflammation in the tumor microenvironment, particularly in tumor nests, to convert “cold” tumors into “hot” tumors that respond to ICB.7
Pyroptosis is an inflammatory cell death process mediated by gasdermins that plays a vital role in innate immunity. Pyroptosis is distinguished by cell swelling and rupture.8,9,10 Gasdermins A–E and DFNB59 are members of the gasdermin family.11 All of these proteins (with the exception of DFNB59) share the same two-domain structure, and their N-domain induces pyroptosis.12,13,14,15 The inflammatory consequences of gasdermin-mediated pyroptosis in the tumor environment may boost the performance of ICB by increasing lymphocyte infiltration and activation, which converts “cold” tumors into “hot” tumors.16,17,18 Recent evidence has directly linked gasdermins with tumor-infiltrating lymphocytes, and increasing gasdermin E (GSDME) expression in tumors enhanced the function and number of tumor-infiltrating natural killer and CD8+ T cells.19,20 Notably, recent research has shown that natural killer cells and cytotoxic T lymphocytes trigger pyroptosis in target cells9,21 and that tumor cell pyroptosis is induced during chimeric antigen receptor (CAR) T cell treatment.22 The objective of the present study was to determine the association between ICB therapy and the GSDM family in HNSCC.
We used The Cancer Genome Atlas (TCGA) database and human HNSCC specimens to demonstrate that overexpression of GSDMs in HNSCC was associated with a favorable immune microenvironment and better prognosis. We used the mouse HNSCC cell lines 4MOSC1 (responsive to CTLA-4 blockade) and 4MOSC2 (resistant to CTLA-4 blockade) as orthotopic models23 to provide evidence for the critical function of GSDM-mediated pyroptosis mediated in CTLA-4 blockade therapy. We discovered that blockade of CTLA-4 increased tumor-infiltrating CD8+ T lymphocytes, which activated the STAT1/IRF1 signaling axis in tumor cells by increasing interferon γ (IFN-γ) and tumor necrosis factor α (TNF-α) in the tumor microenvironment and induced robust pyroptosis of tumor cells to play an antitumor role.
Results
Overexpression of GSDMs in HNSCC indicates favorable prognosis and “hot” tumors
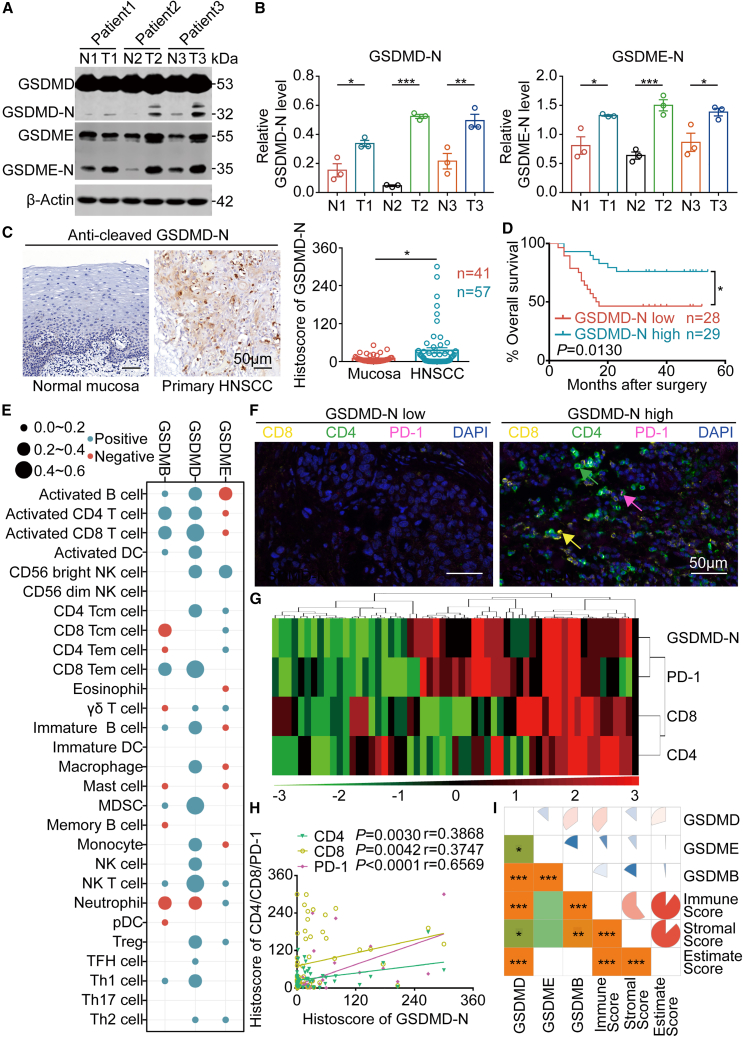
The present study determined the expression of GSDMD and other GSDM family proteins and their functions in the HNSCC microenvironment. We observed that GSDMB, GSDMD, and GSDME were overexpressed in HNSCC tissue compared with normal mucosa according to the GEPIA database (Figure S1). GSDM proteins only exert pyroptosis-inducing activity after cleavage, which results in the formation of an N-terminal fragment (GSDM-N) and a C-terminal fragment (GSDM-C), with GSDM-N causing pyroptosis.11 Western blotting has revealed that GSDMD-N and GSDME-N production was higher in HNSCC tissue than in the adjacent oral mucosa (Figures 1A and 1B). We used immunohistochemistry to stain HNSCC tissue for GSDMD-N as a representative marker to detect the relationship between activated GSDM proteins (pyroptosis) and prognosis since there are currently no antibodies against other GSDM protein N-terminal fragments.24 The results indicated that GSDMD-N production was higher in HNSCC tissue than in adjacent oral mucosa (Figure 1C), and higher production of GSDMD-N indicated a favorable prognosis (Figure 1D).
Figure 1.
Expression of GSDM family proteins in HNSCC
(A and B) The production of GSDMD-N and GSDME-N in paired normal oral mucosal tissue (N) and HNSCC (T) was measured using western blotting (left) and corresponding quantitative analysis (right). (C) Representative immunochemistry images of GSDMD-N production in primary HNSCC and normal mucosal tissues (left). Histoscores of GSDMD-N determined from GSDMD-N production levels in normal mucosal (n = 41) and primary HNSCC (n = 57) tissue samples (right). Scale bars, 50 μm. (D) Kaplan-Meier (K-M) survival analysis of the HNSCC patient cohort divided into high and low GSDMD-N production groups. (E) Bubble chart of the correlations between GSDM family proteins (GSDMB, GSDMD, and GSDME) and ssGSEA of 28 immune cell types in HNSCC samples from TCGA. (F) Representative graphs of multiplex immunohistochemistry (IHC; CD8, CD4, PD-1, and DAPI) in HNSCC patient cohort sections (high GSDMD-N production vs. low GSDMD-N production). CD8: yellow; CD4: green; PD-1: pink; DAPI: blue. Scale bars, 50 μm. (G) Positive correlations among CD8, CD4, PD-1, and GSDMD-N in the HNSCC microenvironment are displayed by hierarchical clustering. The color scale represents the histoscore. (H) The correlations between CD8, CD4, PD-1, and GSDMD-N in the HNSCC microenvironment. (I) Spearman’s correlation analysis between GSDM family proteins (GSDMB, GSDMD, and GSDME) and the immune score, stromal score, and ESTIMATE score. Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Tumor cells with increased GSDM expression are more vulnerable to endogenous stresses, such as hypoxia and endoplasmic reticulum (ER) stress; this stress predisposes these cells to pyroptosis, which then affects the tumor immune microenvironment.19 Therefore, we obtained data from the TCGA database to perform further analyses. We investigated how immune-associated molecules, such as major histocompatibility complex (MHC), immunostimulatory molecules, immune-inhibitory molecules, chemokines, and chemokine receptors, were linked with three markers directly associated with pyroptosis: GSDMB, GSDMD, and GSDME (Figures S2–S4). GSDMD was positively correlated with most immune-related molecules, including MHC molecules, PD-1, CD80, CD86, and CCL5 (Figure S2). Similarly, GSDMB and GSDME expression was also positively correlated with many immune-related molecules (Figures S3 and S4). Most immune-related molecules are associated with antitumor responses.25,26 For example, MHC molecules are involved in antigen presentation, while CD80 and CD86 are markers of activated dendritic cells (DCs).27 PD-1 is also a hallmark of activated T cells, and increased CCL5 expression increases T cell infiltration into solid tumors.28,29 Therefore, these results suggest that pyroptosis mediated by GSDMB, GSDMD, and GSDME contributes to a good antitumor response in HNSCC.
We performed single-sample gene set enrichment analysis (ssGSEA) and measured the infiltration levels of 28 immune cell types in the HNSCC microenvironment.25,30 The results indicated that GSDMB and GSDMD overexpression was strongly related to the high infiltration of activated CD8+ T cells, effector memory CD8+ T cells, activated CD4+ T cells, natural killer (NK) cells, myeloid-derived suppressor cells (MDSCs), and activated DCs (Figure 1E); moreover, patients with HNSCC who had more infiltrating activated CD8+ T cells had a favorable prognosis (Figure S5). Similar to the results from the TCGA database, CD8+ and CD4+ T cells were highly infiltrated in the HNSCC microenvironment where GSDMD-N production was high (Figure 1F). GSDMD-N production was significantly linked with CD8, CD4, and PD-1 expression (Figures 1G and 1H). GSDMD overexpression in TCGA HNSCC samples was linked to high expression of activated CD8+ T cell markers, such as interleukin-2 (IL-2), TNF-α, IFN-γ, and granzyme B (Figure S6). DCs were also of interest. The KEGG pathway analysis from TCGA data indicated that the antigen processing and presentation pathway was enriched in the HNSCC tissues with high GSDMB and GSDMD expression (Figures S7–S9). These results clearly showed that the HNSCC immune microenvironment with high expression of GSDMB and GSDMD was highly complex and varied (Figure 1I). Although the TCGA database found no relationship between GSDME and antigen presentation (Figure S10), a plethora of evidence suggests that GSDME-mediated pyroptosis activates considerable antitumor immune functions.18,19
CTLA-4 blockade induces pyroptosis in HNSCC
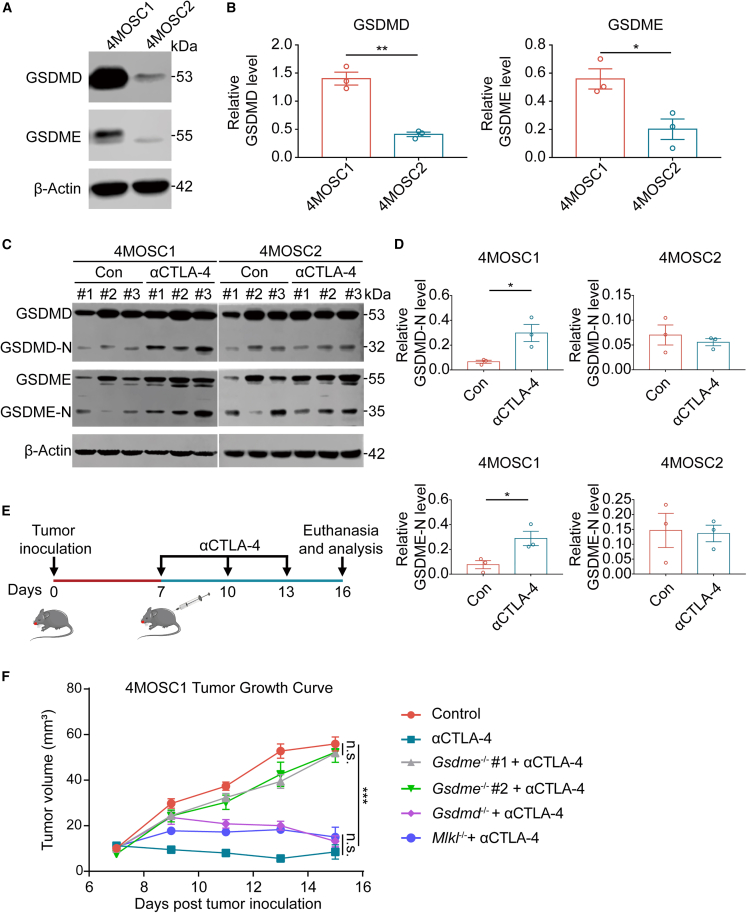
Our findings revealed that GSDM protein expression was adversely related to the immunological microenvironment of the tumor. Interestingly, we discovered that GSDMD and GSDME expression was significantly higher in the murine HNSCC 4MOSC1 cell line (responsive to CTLA-4 blockade) than in the 4MOSC2 cell line (resistant to CTLA-4 blockade) (Figures 2A and 2B), which prompted us to investigate the relationship between ICB and GSDM-mediated pyroptosis. 4MOSC1 and 4MOSC2 cells were transplanted into the dorsum linguae of C57Bl/6 mice, after which the mice were given an isotype control or an anti-CTLA-4 antibody intraperitoneally every 3 days (Figures S11A–S11C). GSDMD-N and GSDME-N production was significantly elevated in 4MOSC1 tissues after CTLA-4 blockade treatment (Figures 2C and 2D), but GSDMD-N and GSDME-N production did not change significantly in 4MOSC2 tissues after CTLA-4 blockade treatment (Figures 2C and 2D). The western blotting results indicated that pyroptosis is involved in the antitumor effect of CTLA-4 blocking treatment. To further validate the effect of GSDM-mediated pyroptosis on CTLA-4 blockade treatment, we constructed cell lines with stably reduced GSDME expression using a lentivirus and validated the reduction using western blotting and real-time PCR (Figures S12A–S12C). We discovered that decreased GSDME protein in 4MOSC1 cells greatly decreased the antitumor impact of CTLA-4 blockade (Figures S12D and S12E), which indicates a role for GSDM-mediated pyroptosis in the antitumor effect of CTLA-4 blockade. We also used a lentivirus to enhance the expression of GSDME in 4MOSC2 cells to analyze whether increased GSDM protein expression would improve the antitumor effect of ICB (Figure S13A). We demonstrated that increased GSDME expression significantly reduced the size of some tumors after CTLA-4 blockade (Figures S13B and S13C). In addition, we knocked out Gsdmd, Gsdme, and Mlkl by CRISPR-Cas9 and used western blotting to validate the knockout efficiency (Figures S14A–S14C). Our in vivo observations were consistent with the aforementioned findings that GSDME-mediated pyroptosis exhibited a more substantial influence on CTLA-4 blockade treatment than GSDMD-mediated pyroptosis and MLKL-mediated necroptosis (Figures 2E and 2F). Taken together, our findings indicate that GSDM expression in tumor cells may affect the efficacy of ICB, which supports the use of GSDM as a possible therapeutic target for future research into methods for increasing ICB response rates and overcoming ICB resistance.
Figure 2.
CTLA-4 blockade induces pyroptosis in HNSCC
(A and B) The expression of GSDMD and GSDME in the 4MOSC1 cell line and 4MOSC2 cell line was evaluated using western blotting (left) and corresponding quantitative analyses (right). (C and D) The production of GSDMD-N and GSDME-N was evaluated using western blotting in the control group (n = 3) and anti-CTLA-4 group (n = 3) for 4MOSC1 and 4MOSC2 tumors (left) and corresponding quantitative analyses (right). (E) Experimental schedule for tumor inoculation and CTLA-4 blockade. (F) Tumor volume of 4MOSC1 tumor-bearing mouse with CTLA-4 blockade. Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Intratumoral CD8+ T cells regulate tumor pyroptosis during CTLA-4 blockade treatment
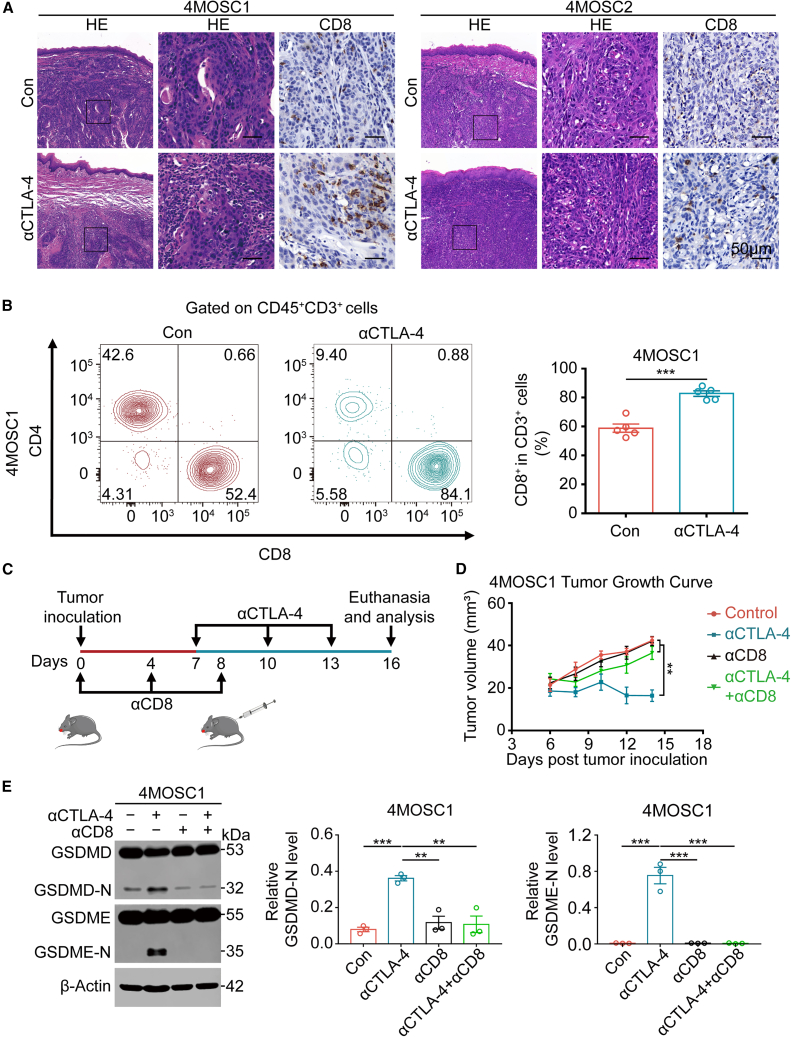
Cancer immunotherapy restores or increases the effector function of CD8+ T cells in the tumor immune microenvironment,31 and evidence has revealed that CD8+ T cells triggered by cancer immunotherapy cause tumor cell ferroptosis by releasing IFN-γ.32 Immunohistochemistry and flow cytometry revealed an increase in the number of CD8+ T cells in the 4MOSC1 tumor after CTLA-4 blockade therapy (Figures 3A and 3B). However, anti-CTLA4 therapy had little effect on the CD8+ T cell population in the 4MOSC2 tumor (Figure 3A), which suggests that activation of CD8+ T cells is required for anti-CTLA-4 therapy to exert its antitumor effects. We used a CD8 blocking antibody to reduce the number of CD8+ T cells in the 4MOSC1 tumor and demonstrated that the antitumor effect of CTLA-4 blockade therapy was decreased when fewer CD8+ T cells were present (Figures 3C and 3D). To exclude the effect of other cells, we also depleted CD4+ T cells and NK cells and demonstrated that intratumoral CD8+ T cells activated by ICB therapy are likely to destroy tumor cells via GSDM-mediated pyroptosis (Figures S15A and S15B). Consistent with this hypothesis, depletion of CD8+ T cells led to a considerable reduction in GSDMD-N and GSDME-N compared with anti-CTLA-4 treatment alone (Figure 3E), which indicates that activated CD8+ T cells exert their antitumor effects via pyroptosis.
Figure 3.
CD8+ T cells regulate tumor pyroptosis during CTLA-4 blockade treatment
(A) Representative images of H&E staining and CD8 expression in the control group (Con) and anti-CTLA-4 group of 4MOSC1 tumors (left) and 4MOSC2 tumors (right). Scale bars, 50 μm. (B) CD8+ T cell populations in the tumors (left) were measured using flow cytometry, and quantitative analyses were performed (right). (C) Experimental schedule for tumor inoculation and CD8 blockade or CTLA-4 blockade. (D) Tumor volume of 4MOSC1 tumor-bearing mice with different treatments. (E) The production of GSDMD-N and GSDME-N was evaluated using western blotting in different treatment groups for 4MOSC1 tumors (left) and corresponding quantitative analyses (right). Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Synergism of IFN-γ and TNF-α triggers tumor cell pyroptosis
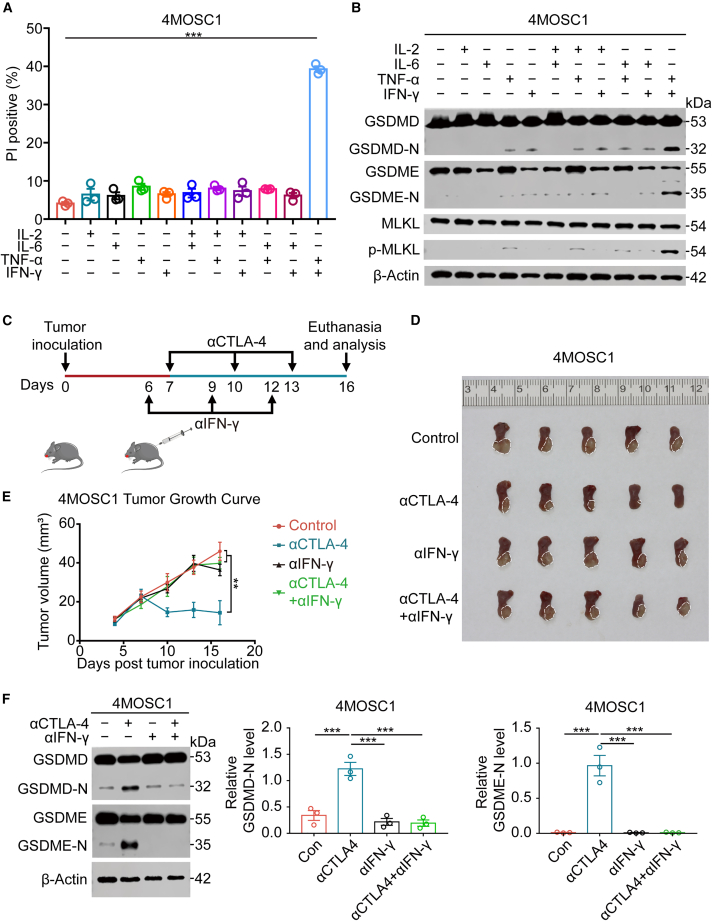
The mechanism by which intratumorally activated CD8+ T lymphocytes induce pyroptosis of 4MOSC1 cells upon ICB is a critical subject. CD8+ T lymphocytes trigger ferroptosis in tumor cells by secreting IFN-γ.32 Therefore, we hypothesized that CD8+ T lymphocytes induce tumor cells to undergo pyroptosis by releasing specific cytokines. We used real-time PCR to examine 6 common activating cytokines closely associated with CD8+ T cells and demonstrated that IL-2, IL-6, IFN-γ, and TNF-α were considerably enhanced in the 4MOSC1 tumor microenvironment during anti-CTLA-4 treatment (Figure S16). We tested the hypothesis that these elevated cytokines in the tumor microenvironment caused tumor cell pyroptosis by treating 4MOSC1 cells with these cytokines in vitro. We found that only the combination of IFN-γ and TNF-α significantly induced 4MOSC1 cell pyroptosis (Figures 4A and 4B, S17, and S18). ELISA and flow cytometry showed a link between the cytokines IFN-γ and TNF-α and CD8+ T cells (Figures S19A and S19B). Consistent with the in vitro findings, the antitumor effect of anti-CTLA-4 treatment was weakened when intratumoral IFN-γ was neutralized (Figures 4C–4E), and the capacity of anti-CTLA-4 therapies to trigger pyroptosis was diminished when intratumoral IFN-γ or TNF-α or IFN-γ and TNF-α together were neutralized (Figures 4F and S20). In summary, the synergy of intratumoral IFN-γ and TNF-α after ICB triggers tumor cell death via pyroptosis, which provides a new perspective for the mechanism of CD8+T lymphocytes attacking tumor cells in immunotherapy.
Figure 4.
Synergism of IFN-γ and TNF-α triggers tumor cell pyroptosis
(A) Apoptosis measured using flow cytometry of 4MOSC1 cells 48 h after different treatments. (B) The production of GSDMD-N, GSDME-N, and p-MLKL was evaluated using western blotting in different treatment groups of 4MOSC1 cells. (C) Experimental schedule for tumor inoculation and treatment with IFN-γ blockade or CTLA-4 blockade. (D) Tumor images of 4MOSC1 tumor-bearing mice with different treatments. (E) Tumor volume of 4MOSC1 tumor-bearing mice with different treatments. (F) The production of GSDMD-N and GSDME-N was evaluated using western blotting in different treatment groups for 4MOSC1 tumors (left) and corresponding quantitative analysis (right). Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Synergism of IFN-γ and TNF-α initiates pyroptosis in tumor cells via the STAT1/IRF1 axis
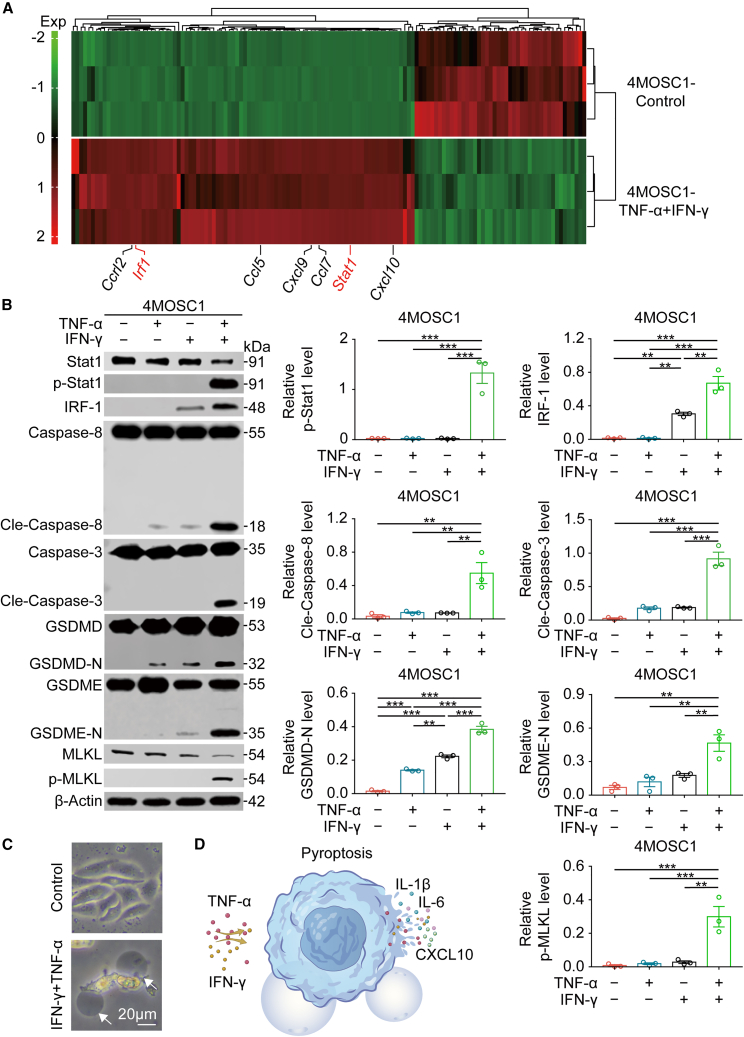
We performed RNA sequencing (RNA-seq) to further investigate the mechanism of the induction of tumor cell pyroptosis via IFN-γ and TNF-α. Our RNA-seq results and previous studies33,34 suggested that activation of the STAT1/IRF1 axis by IFN-γ and TNF-α result in tumor cell pyroptosis (Figures 5A, S21A, S21B, and S22A–S22C). We demonstrated that the addition of TNF-α and IFN-γ to 4MOSC1 cells led to the activation of a substantial number of genes associated with inflammation and immunity (Figure 5A). Western blotting analysis revealed that the addition of IFN-γ and TNF-α to 4MOSC1 cells significantly increased STAT1 phosphorylation and IRF1 expression as well as the activation of caspase-8 and caspase-3, which resulted in the activation of GSDMD and GSDME and tumor cell pyroptosis (Figures 5B–5D). When STAT1 expression was inhibited by small interfering RNA (siRNA) followed by the addition of IFN-γ and TNF-α, western blotting analysis revealed that neither STAT1 phosphorylation nor IRF-1 expression was significantly increased. Caspase-8 and caspase-3 activation was also reduced, which did not result in the onset of pyroptosis (Figures S23 and S24). Similarly, the siRNA-mediated suppression of IRF-1 expression followed by the addition of IFN-γ and TNF-α had no effect on STAT1 phosphorylation, but rather this suppression caused dramatic inhibition of caspase-8 and caspase-3 activation, and the initiation of pyroptosis was not significantly induced (Figure S25). These results indicated that synergism of IFN-γ and TNF-α in the tumor microenvironment activated the STAT1/IRF1 axis to trigger tumor cell pyroptosis and the release of inflammatory chemokines.
Figure 5.
Synergism of IFN-γ and TNF-α triggers tumor cell pyroptosis via the STAT1/IRF1 Axis
(A) Heatmap depicting the expression levels of genes in 4MOSC1 cells co-treated with TNF-α and IFN-γ for 48 h relative to their expression in untreated 4MOSC1 cells. (B) The expression of p-Stat1, IRF-1, Cle-caspase-8, Cle-caspase-3, and p-MLKL in 4MOSC1 cells and the production of GSDMD-N and GSDME-N in 4MOSC1 cells were evaluated using western blotting in different treatment groups (left) and corresponding quantitative analyses (right). (C) Representative bright-field microscopy images of 4MOSC1 cells with different treatments. The arrows indicate pyroptotic cells. Scale bars, 20 μm. (D) Schematic overview of TNF-α- and IFN-γ-induced tumor cell pyroptosis and inflammatory factor and chemokine release. Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
IFN-γ and TNF-α exert synergistic effects on pyroptosis in human HNSCC tissues
We evaluated the synergistic effects of recombinant human IFN-γ and TNF-α on human HNSCC cells and tissues. The expression levels of GSDMB, GSDMD, GSDME, and MLKL were quantified in several human HNSCC cell lines, including Cal27, SCC4, SCC9, and SCC25. GSDMB, GSDMD, GSDME, and MLKL expression levels were relatively high in the SCC9 cell line (Figures 6A and S26A). The GSDMD-N and GSDME-N fragments resulted from proteolytic cleavage by caspase-8 and caspase-3, respectively. Consistent with the results observed in 4MOSC1 cells, the synergistic stimulation of SCC9 cells with IFN-γ and TNF-α also eventually induced the onset of GSDMD- and GSDME-mediated pyroptosis (Figures 6B–6D and S26B) as well as a considerable increase in cell death (Figures S27A and S27B). We collected 3 human HNSCC specimens from the tongue or cheek and stimulated these human HNSCC tissues with IFN-γ and TNF-α. Consistent with the in vitro data, the synergistic effect of IFN-γ and TNF-α greatly increased GSDMD-N and GSDME-N production in human HNSCC tissue (Figures 6B, 6E, and S28). In conclusion, these results showed that human IFN-γ and TNF-α synergistically triggered HNSCC tumor pyroptosis, which provides a concept and mechanism for immune therapy.
Figure 6.
Synergism of IFN-γ and TNF-α triggers pyroptosis in human HNSCC cells and tissues
(A) The expression of GSDMD and GSDME was evaluated using western blotting in Cal27, SCC4, SCC9, and SCC25 human HNSCC cell lines (left) and corresponding quantitative analyses (right). (B) Schematic overview of SCC9 cells and fresh HNSCC tissue stimulated with the cytokines IFN-γ and TNF-α. (C) The production of GSDMD-N and GSDME-N was evaluated using western blotting in different treatment groups of SCC9 cells (left) and corresponding quantitative analyses (right). (D) Representative bright-field microscopy images of SCC9 cells with different treatments. The arrows indicate pyroptotic cells. Scale bars, 20 μm. (E) Quantitative analyses of GSDMD-N and GSDME-N production in different treatment groups of HNSCC tumors as assessed using western blotting. Data, mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Discussion
Several studies on GSDM-mediated pyroptosis in HNSCC have been published.35,36,37,38 It is reported that GSDM-mediated pyroptosis plays a critical role in antitumor therapy, and different treatments, including chemotherapy and targeted therapies, exert antitumor effects by causing GSDM-mediated pyroptosis.39,40,41,42 Pyroptosis of 15% of tumor cells is sufficient to eradicate an entire tumor,17 which demonstrates that the strong antitumor immunity induced by pyroptosis plays a significant role in tumor cell death. Pyroptosis may also be used in conjunction with PD-1 blockade therapy to increase the effectiveness of both treatments.17 GSDM-mediated pyroptosis modifies the tumor immune microenvironment and the inflow of tumor-infiltrating lymphocytes (TILs) to transform immunologically “cold” tumors into “hot” tumors.18 According to these discoveries, a tight association exists between GSDM-mediated pyroptosis and ICB therapy. However, whether ICB therapy promotes GSDM-mediated pyroptosis to exert antitumor effects is uncertain. To determine whether pyroptosis was involved in the mechanism of ICB therapy, we used a 4MOSC1 HNSCC orthotopic xenograft model that responds to CTLA-4 blockade and a 4MOSC2 HNSCC orthotopic xenograft model that does not respond to CTLA-4 blockade. The 4MOSC1 orthotopic xenograft model showed a significant increase in pyroptosis after CTLA-4 blockade treatment, but the 4MOSC2 orthotopic xenograft model showed no significant change in pyroptosis after CTLA-4 blockade treatment, which suggests that GSDM-mediated pyroptosis plays a role in the antitumor effects of CTLA-4 blockade treatment.
Although the relationship between GSDM expression and the response to ICB therapy is not clear, previous research has demonstrated that tumor cells with high levels of GSDMs, such as GSDME, are more susceptible to endogenous stress or extrinsic challenges that cause them to undergo pyroptosis and release large amounts of inflammatory substances into the tumor microenvironment. This release of inflammatory mediators increases the number and function of TILs in the tumor microenvironment and promotes phagocytosis by macrophages, which leads to antitumor effects.19 Our findings revealed that GSDMB, GSDMD, and GSDME were strongly expressed in HNSCC and favorably associated with many immune-related molecules and that high GSDMD-N production was associated with a favorable prognosis. Notably, GSDMD and GSDME expression was significantly higher in 4MOSC1 cells than in 4MOSC2 cells, which strongly suggests that the expression of GSDMs affects the effect of ICB treatment. Our subsequent experimental results also confirmed our hypothesis that the downregulation of GSDME expression in 4MOSC1 cells was associated with poorer efficacy of CTLA-4 blockade treatment and that the upregulation of GSDME expression in 4MOSC2 cells was associated with better efficacy of CTLA-4 blockade treatment.
The activation of cytotoxic T cells is critical for the efficacy of ICB therapy, as these cells destroy tumor cells either directly via the release of cytotoxic perforins and granzymes or indirectly via the release of potent proinflammatory cytokines.43 Insights into the mechanisms of cytokine-mediated cancer cell killing are critical because cancer cells disrupt granzyme- and perforin-mediated cell death.44 IFN-γ secreted by CD8+ T cells triggers cancer cell ferroptosis,32 which has an antitumor effect. Therefore, we hypothesized that CD8+ T cells induce cancer cell pyroptosis via cytokine secretion. Our findings support the hypothesis that TNF-α and IFN-γ work synergistically to cause cancer cell pyroptosis following CTLA-4 blockade therapy. Recent investigations have demonstrated that TNF-α and IFN-γ synergistically induce pyroptosis and the release of large amounts of inflammatory substances,33,34,45,46 and these results are consistent with our experimental findings. We used RNA-seq to investigate the mechanism by which TNF-α and IFN-γ synergistically promoted tumor cell pyroptosis. Taken together, these data offer new perspectives on methods to increase ICB therapy response rates.
In conclusion, we evaluated the expression of GSDMs in HNSCC and systematically investigated the relationship between GSDMs in HNSCC and the tumor immune microenvironment using the TCGA database, multiplexed immunohistochemistry, and other methods. We used the 4MOSC1 HNSCC orthotopic xenograft model that responds to CTLA-4 blockade and the 4MOSC2 HNSCC orthotopic xenograft model that does not respond to CTLA-4 blockade to demonstrate the occurrence of GSDM-mediated pyroptosis in ICB treatment and to further validate the effects of GSDM-mediated pyroptosis in ICB treatment by knocking down and overexpressing GSDM proteins. We demonstrated that CD8+ T cells were required for the regulation of CTLA-4-induced pyroptosis and increased the levels of IFN-γ and TNF-α cytokines in the tumor microenvironment. These cytokines synergistically activated the STAT1/IRF1 axis to trigger tumor cell pyroptosis and the release of large amounts of inflammatory substances and chemokines. Future studies should delineate the underlying mechanisms by which GSDM proteins influence the efficacy of ICB therapy in greater depth.
Materials and methods
Ethical statement, HNSCC tissue samples, and HNSCC patient cohort
The Medical Ethics Committee of School and Hospital of Stomatology, Wuhan University, approved this study (2014LUNSHENZI06), which was performed in accordance with the institutional guidelines. All participants provided written informed permission. Two pathologists performed clinical staging and histological grading in accordance with the appropriate guidelines, the International Alliance for Cancer Control (eighth edition) protocol,47 and the World Health Organization system.48 Protein extraction and in vitro cytokine stimulation assays were performed on HNSCC tissue specimens. The HNSCC tissue cohort contained 57 primary HNSCC tissue samples and 41 adjacent mucosal tissue samples.
Cell lines, animal models, and treatments
SCC4, SCC9, and SCC25 (human HNSCC cell lines from the American Type Culture Collection) were cultured in F12/DMEM plus 400 ng/mL hydrocortisone. Cal27 (the human HNSCC cell line from the American Type Culture Collection) was cultured in DMEM/high glucose.49 4MOSC1 and 4MOSC2 cells were gifts from Prof. J. Silvio Gutkind at the University of California San Diego via a materials transfer agreement (SD2017-202).23,50 4MOSC1 and 4MOSC2 cells were cultured in keratinocyte serum-free medium (K-SFM; Gibco-BRL). To construct orthotopic HNSCC mouse models, 4MOSC1 or 4MOSC2 cells (1 × 106/per mouse) were separately transplanted into the dorsum linguae of female C57Bl/6 mice aged 6–8 weeks and weighing 18–20 g.23 The mice were fed in compliance with the recommendations of the Institutional Animal Care and Use Committee of School and Hospital of Stomatology, Wuhan University (S07921090A, S07920090H, and S07921020D), and the ARRIVE Guidelines.51 The mice were given an isotype control antibody (10 mg/kg; BE0086; Bio X Cell) or CTLA-4 antibody (10 mg/kg; BE0164; Bio X Cell) intraperitoneally every 3 days after the tumors formed. The mice were subjected to strict observation. Tumor sizes were measured every 2 days. Tumor volume was calculated using the equation tumor length × tumor width2/2. When the treatment experiment was complete, the mice were euthanized and dissected to obtain the tumors. On days 0, 4, and 8, mice were given NK1.1 (10 mg/kg; BE0036; Bio X Cell), CD4 (10 mg/kg; BE0119; Bio X Cell), and CD8 (10 mg/kg; BE0004-1; Bio X Cell) target antibodies for in vivo NK cell, CD4+ T cell, and CD8+ T cell depletion. For in vivo cytokine neutralization, mice received IL-2 (10 mg/kg; BE0043; Bio X Cell), IL-6 (10 mg/kg; BE0046; Bio X Cell), TNF-α (10 mg/kg; BE0058; Bio X Cell), or IFN-γ (10 mg/kg; BE0055; Bio X Cell) neutralizing antibodies the day before each CTLA-4-targeting antibody treatment was given.
In vitro cytokine stimulation assay
For 4MOSC1 cells, IL-6 (25 ng/mL, Peprotech, 212-16), IL-2 (25 ng/mL, Peprotech, 212-12), TNF-α (25 ng/mL, Peprotech, 315-01A), and IFN-γ (25 ng/mL, Peprotech, 315-05) were used for 48 h. For SCC9 cells, 25 ng/mL TNF-α (Peprotech, AF-300-01A) and IFN-γ (Peprotech, 300-02) were used for 48 h.33,34 Within 30 min of resection, freshly resected HNSCC tissues were cleaned and split into various sections (1–5 mm3) using a scalpel, followed by injection of 5% glucose control, TNF-α (50 ng/mL, Peprotech, AF-300-01A), and IFN-γ (100 ng/mL, Peprotech, 300-02) in a 5% glucose solution at multiple sites. Each segment was cultured in 24-well plates for 24 h in 0.5 mL RPMI 1640 media.52
Western blotting
Western blotting was performed as previously described.49 The protein was detected using an ECL kit (Advansta), and images were obtained using an Odyssey system (LI-COR Biosciences). ImageJ was used to calculate the amount of protein in each sample (National Institutes of Health, Bethesda, MD, USA). The experiment was repeated at least 3 times. Detailed steps and a list of primary antibodies used for western blotting are provided in the supplemental information.
Real-time PCR
Total RNA was obtained from tumor tissues and cells as directed by the manufacturer (Axygen, Corning, NY, USA), and the RNA concentration was measured using a spectrometer (Shimadzu UV-2401PC, Kyoto, Japan). 1 μg total RNA was reverse transcribed using HiScript II Reverse Transcriptase (Vazyme Biotech, Nanjing, China) according to the manufacturer’s instructions, and approximately 1% of the cDNA was used as a template in each real-time PCR with SYBR master mix (Vazyme Biotech). Three steps of PCR were used. Detailed steps and a list of the primer sequences used for real-time PCR are provided in the supplemental information.
Immunohistochemistry
Paraffin-embedded tissues (4 μm) were sectioned consecutively, dewaxed in xylene, rehydrated in a gradient alcohol series, heated for antigen repair in a microwave oven, and incubated with endogenous peroxidase blocking agent and goat serum at 37°C for 20 min. The sectioned tissues were incubated with primary antibody overnight at 4°C. A list of primary antibodies used for immunohistochemistry is provided in the supplemental information. The secondary biotinylated immunoglobulin G (IgG) antibody and the antibiotinyl-peroxidase reagent were incubated in an incubator for 20 min. DAB reagent (Mxb Biotechnologies) was used for staining, and the nuclei were stained with hematoxylin. The entire process was based on a horseradish peroxidase (HRP) color system. The sections were scanned and analyzed using a Pannoramic Midi (3DHISTECH).
Multiplexed immunohistochemistry
The Opal serial immunostaining handbook was used to achieve multiplexed immunohistochemistry, as previously described.53 Formalin-fixed and paraffin-embedded HNSCC patient tissues were marked using an Opal 7-ColorManual immunohistochemistry Kit (NEL811001KT; PerkinElmer, Hopkinton, MA, USA). After deparaffinization and hydration, the sections were placed in AR buffer (pH = 6.0; PerkinElmer), sequentially incubated with blocking buffer (PerkinElmer) and primary antibodies, and subjected to tyramide signal amplification (TSA; PerkinElmer Opal Kit). Primary antibodies included CD4 (ab133616; Abcam, Boston, MA, USA), CD8 (85,336; Cell Signaling Technology), and PD-1 (86,163; Cell Signaling Technology). The sections were repeatedly subjected to these steps until incubation with the final primary antibody. Nuclear staining was performed with DAPI before mounting. PerkinElmer Vectra software was used to scan the slides (PerkinElmer).
Flow cytometry
Tumors were collected and processed into a single-cell suspension (gentleMACS Dissociator, Miltenyi Biotec), followed by staining with antibodies and T cell analysis. FlowJo 10 software (v.10.0.6, Tree Star) was used to analyze and show the research results. Flow cytometry analysis strategy for T cell gating is shown in Figure S29. Fluorescence-labeled antibodies included Fixable Viability Dye (eBioscience, eFluor 506); anti-CD45 (APC-CY7, BD Biosciences); anti-CD3 (FITC, BD Biosciences); anti-CD4 (eFluor450, Invitrogen); anti-CD8 (PerCP-Cyanine 5.5, BioLegend); anti-IFN-γ (PE, BioLegend); and anti-TNF-α (APC, BioLegend). The supplemental information contains detailed instructions for detecting apoptosis.
Stable cell line generation
Single guide RNA (sgRNA) targeting Gsdmd, Gsdme, and Mlkl was constructed as part of a Lenti-CRISPRv2 vector.54 Lentivirus was generated in 293T cells, after which 4MOSC1 cells were infected with lentivirus containing sgRNA. After 36 h, 4MOSC1 cells were selected with 5 μg/mL puromycin for 2 days. Cells were maintained in culture medium containing puromycin (Sigma Aldrich). The sequences are as follows: Gsdmd sgRNA 1: 5′-CACCGCAACAGCTTCGGAGTCGTG-3′; Gsdme sgRNA 1: 5′-CACCgACAGGCTGTATTGGCAATGG-3′; Gsdme sgRNA 2: 5′-CACCgATGGTGGfdb 9.1.450/W UnicodeCTTCGAGCATGAG-3′; and Mlkl sgRNA 1: 5′-CACCGCCCTCCCAACGGCCCTCTG-3′.
The short hairpin RNA (shRNA) targeting sequence for mouse Gsdme was purchased from Ubigene Biosciences (Guangzhou, China). The specific targeting sequences were as follows: shGsdme1(mouse): CCGTCAAGAGAACAGTTAATA; shGsdme2(mouse): TGGAGTCAGACTTCGTGAAAT; and shGsdme3(mouse): GAGCGTTGTCAATGGGTTATT.
The 4MOSC1 cells were infected with 8 μg/mL polybrene (Sigma Aldrich) and lentivirus supernatant then selected with 5 μg/mL puromycin (Sigma Aldrich).49 Protein and mRNA levels were verified using real-time PCR and western blotting.
To generate lentiviruses, EF-1aF/GFP-Puro empty vector (negative control) or EF-1aF-Gsdme was transfected into HEK293T cells with PG-P1-VSVG, PG-P2-REV, and PG-P3-RRE. The 4MOSC2 cells were infected with 8 μg/mL polybrene (Sigma Aldrich) and lentivirus supernatant then selected with 5 μg/mL puromycin (Sigma Aldrich). The mRNA level was verified using real-time PCR.
RNA interference
The transfection of 4MOSC1 cells with siRNA was performed using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA). The detailed transfection procedure was discussed previously.55 GenePharma (Shanghai, China) designed and produced siRNAs with the following sequences: Stat1 siRNA: 5′-AAGGAAAAGCAAGCGTAATCT-3′, and Irf1 siRNA: 5′-CCGAUACAAAGCAGGAGAATT-3'.
ELISA
Fresh mouse tumor homogenates (100 mg tissue/mL PBS) were centrifuged for 20 min at 3,000 RPM. Supernatants were obtained, and protein concentrations were adjusted using a BCA Protein Kit (Beyotime). TNF-α and IFN-γ concentrations in supernatants were measured using commercial ELISA kits (4A Biotech) in accordance with the manufacturer’s instructions.
RNA-seq and bioinformatics analysis
Beijing Genomics Institution (Shenzhen, China) created the RNA-seq library and performed RNA-seq construction. RNA-seq data have been deposited to GEO under the accession number GEO: GSE201367. To obtain more convincing results, patient RNA-seq data from the TCGA-HNSCC cohort were grouped based on the median values of IFN-γ and TNF. DEGs between the IFN-γHiTNFHi and IFN-γLoTNFLo groups were identified using the Deseq2 R package with significance cutoff parameters of p <0.05 (adjusted) and absolute fold change >2. Differentially expressed genes (DEGs) overlapping our RNA-seq data with TCGA-HNSCC RNA-seq data were extracted for further analysis. To examine the expression of GSDM family proteins and their relationship to the HNSCC microenvironment, we acquired RNA-seq data and the corresponding clinical features of patients with HNSCC from GEPIA (http://gepia.cancer-pku.cn/) and TCGA (https://portal.gdc.cancer.gov/). The enrichment scores for patients with HNSCC in the TCGA database were calculated and used to perform ssGSEA and differential enrichment analysis using GSEA and gene set variation analysis (GSVA).25 ssGSEA was used to determine the tumor-infiltrating levels of 28 immune cell types, which are represented as the normalized enrichment score (NES) of the tumor microenvironment.25 To further analyze the complexity and variations in the HNSCC microenvironment, we used the R package “estimate” to obtain an immune score, a stromal score, and an ESTIMATE score for HNSCC.56
Statistical analysis
For statistical analysis, GraphPad Prism 7.0 was used, with Student’s t test for two-group comparisons or ANOVA analysis for multigroup comparisons. Spearman’s correlation coefficient was used for correlation analyses. For survival analysis, a Mantel-Cox log rank test was performed to evaluate statistical significance in Kaplan-Meier analysis. p <0.05 was regarded as statistically significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). The results are presented as mean ± SEM.
Acknowledgments
We acknowledge Shuyan Liang and Zhixin Qiu from Wuhan Biobank Co., Ltd., for fluorescence-activated cell sorting (FACS) assistance. This work was financially supported by the National Natural Science Foundation of China 82273202 (Z.-J.S.), 82072996 (Z.-J.S.), and 81874131 (Z.-J.S.); the Fundamental Research Funds for the Central Universities (2042021kf0216); the Hubei Province International Science and Technology Cooperation Project 2021EHB027 (Z.-J.S.); and the National Key Research and Development Program 2022YFC2504200 (Z.-J.S.).
Author contributions
Data curation, S.W., Z.-Z.W., S.-W.Z., S.-C.W., M.-J.Z., B.-X.Z., and H.L.; conceptualization, S.W., Z.-Z.W., and Z.-J.S.; methodology, S.W., Z.-Z.W., M.-J.Z., B.-X.Z., and H.L.; formal analysis, S.W., Z.-Z.W., B.-X.Z., and H.L.; investigation, S.W., Z.-Z.W., S.-W.Z., S.-C.W., and L.M.; writing – original draft, S.W., Z.-Z.W., and Z.-J.S.; writing – review & editing, Z.-Y.W. and Z.-J.S.; supervision, Q.-C.Y., Y.X., and Z.-J.S.; resources, J.S.G. and Z.-J.S.; funding acquisition, Z.-J.S.; project administration, Z.-J.S.
Declaration of interests
No potential conflicts of interest were disclosed.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.02.023.
Supplemental information
Data availability
RNA-seq data will be shared publicly in an online depository prior to publication. All other data generated for this study will be made available upon reasonable request to the corresponding author.
References
- 1.Chow L.Q.M. Head and neck cancer. N. Engl. J. Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715. [DOI] [PubMed] [Google Scholar]
- 2.Johnson D.E., Burtness B., Leemans C.R., Lui V.W.Y., Bauman J.E., Grandis J.R. Head and neck squamous cell carcinoma. Nat. Rev. Dis. Primers. 2020;6:92. doi: 10.1038/s41572-020-00224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nör J.E., Gutkind J.S. Head and neck cancer in the new era of precision medicine. J. Dent. Res. 2018;97:601–602. doi: 10.1177/0022034518772278. [DOI] [PubMed] [Google Scholar]
- 4.Postow M.A., Callahan M.K., Wolchok J.D. Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalbasi A., Ribas A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020;20:25–39. doi: 10.1038/s41577-019-0218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferris R.L., Blumenschein G., Jr., Fayette J., Guigay J., Colevas A.D., Licitra L., Harrington K., Kasper S., Vokes E.E., Even C., et al. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2016;375:1856–1867. doi: 10.1056/NEJMoa1602252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ochoa de Olza M., Navarro Rodrigo B., Zimmermann S., Coukos G. Turning up the heat on non-immunoreactive tumours: opportunities for clinical development. Lancet Oncol. 2020;21:e419–e430. doi: 10.1016/S1470-2045(20)30234-5. [DOI] [PubMed] [Google Scholar]
- 8.Shi J., Gao W., Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem. Sci. 2017;42:245–254. doi: 10.1016/j.tibs.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Z., He H., Wang K., Shi X., Wang Y., Su Y., Wang Y., Li D., Liu W., Zhang Y., et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368 doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 10.Fink S.L., Bergsbaken T., Cookson B.T. Anthrax lethal toxin and Salmonella elicit the common cell death pathway of caspase-1-dependent pyroptosis via distinct mechanisms. Proc. Natl. Acad. Sci. USA. 2008;105:4312–4317. doi: 10.1073/pnas.0707370105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Broz P., Pelegrín P., Shao F. The gasdermins, a protein family executing cell death and inflammation. Nat. Rev. Immunol. 2020;20:143–157. doi: 10.1038/s41577-019-0228-2. [DOI] [PubMed] [Google Scholar]
- 12.Shi J., Zhao Y., Wang K., Shi X., Wang Y., Huang H., Zhuang Y., Cai T., Wang F., Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 13.Kayagaki N., Stowe I.B., Lee B.L., O'Rourke K., Anderson K., Warming S., Cuellar T., Haley B., Roose-Girma M., Phung Q.T., et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature. 2015;526:666–671. doi: 10.1038/nature15541. [DOI] [PubMed] [Google Scholar]
- 14.Ding J., Wang K., Liu W., She Y., Sun Q., Shi J., Sun H., Wang D.C., Shao F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 15.Liu X., Zhang Z., Ruan J., Pan Y., Magupalli V.G., Wu H., Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Minton K. Pyroptosis heats tumour immunity. Nat. Rev. Drug Discov. 2020;19:309. doi: 10.1038/d41573-020-00062-8. [DOI] [PubMed] [Google Scholar]
- 17.Wang Q., Wang Y., Ding J., Wang C., Zhou X., Gao W., Huang H., Shao F., Liu Z. A bioorthogonal system reveals antitumour immune function of pyroptosis. Nature. 2020;579:421–426. doi: 10.1038/s41586-020-2079-1. [DOI] [PubMed] [Google Scholar]
- 18.Rosenbaum S.R., Wilski N.A., Aplin A.E. Fueling the fire: inflammatory forms of cell death and implications for cancer immunotherapy. Cancer Discov. 2021;11:266–281. doi: 10.1158/2159-8290.CD-20-0805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang Z., Zhang Y., Xia S., Kong Q., Li S., Liu X., Junqueira C., Meza-Sosa K.F., Mok T.M.Y., Ansara J., et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang S., Zhang M.J., Wu Z.Z., Zhu S.W., Wan S.C., Zhang B.X., Yang Q.C., Xiao Y., Chen L., Sun Z.J. GSDME is related to prognosis and response to chemotherapy in oral cancer. J. Dent. Res. 2022;101:848–858. doi: 10.1177/00220345211073072. [DOI] [PubMed] [Google Scholar]
- 21.Nicolai C.J., Raulet D.H. Killer cells add fire to fuel immunotherapy. Science. 2020;368:943–944. doi: 10.1126/science.abc2502. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y., Fang Y., Chen X., Wang Z., Liang X., Zhang T., Liu M., Zhou N., Lv J., Tang K., et al. Gasdermin E-mediated target cell pyroptosis by CAR T cells triggers cytokine release syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.aax7969. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z., Wu V.H., Allevato M.M., Gilardi M., He Y., Luis Callejas-Valera J., Vitale-Cross L., Martin D., Amornphimoltham P., McDermott J., et al. Syngeneic animal models of tobacco-associated oral cancer reveal the activity of in situ anti-CTLA-4. Nat. Commun. 2019;10:5546. doi: 10.1038/s41467-019-13471-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang K., Sun Q., Zhong X., Zeng M., Zeng H., Shi X., Li Z., Wang Y., Zhao Q., Shao F., Ding J. Structural mechanism for GSDMD targeting by autoprocessed caspases in pyroptosis. Cell. 2020;180:941–955.e20. doi: 10.1016/j.cell.2020.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Charoentong P., Finotello F., Angelova M., Mayer C., Efremova M., Rieder D., Hackl H., Trajanoski Z. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 2017;18:248–262. doi: 10.1016/j.celrep.2016.12.019. [DOI] [PubMed] [Google Scholar]
- 26.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.H., Porta-Pardo E., Gao G.F., Plaisier C.L., Eddy J.A., et al. The immune landscape of cancer. Immunity. 2018;48:812–830.e14. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moutaftsi M., Mehl A.M., Borysiewicz L.K., Tabi Z. Human cytomegalovirus inhibits maturation and impairs function of monocyte-derived dendritic cells. Blood. 2002;99:2913–2921. doi: 10.1182/blood.v99.8.2913. [DOI] [PubMed] [Google Scholar]
- 28.Davidson T.B., Lee A., Hsu M., Sedighim S., Orpilla J., Treger J., Mastall M., Roesch S., Rapp C., Galvez M., et al. Expression of PD-1 by T cells in malignant glioma patients reflects exhaustion and activation. Clin. Cancer Res. 2019;25:1913–1922. doi: 10.1158/1078-0432.CCR-18-1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dangaj D., Bruand M., Grimm A.J., Ronet C., Barras D., Duttagupta P.A., Lanitis E., Duraiswamy J., Tanyi J.L., Benencia F., et al. Cooperation between constitutive and inducible chemokines enables T cell engraftment and immune attack in solid tumors. Cancer Cell. 2019;35:885–900.e10. doi: 10.1016/j.ccell.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C., et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou W., Wolchok J.D., Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016;8:328rv4. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang W., Green M., Choi J.E., Gijón M., Kennedy P.D., Johnson J.K., Liao P., Lang X., Kryczek I., Sell A., et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karki R., Sharma B.R., Tuladhar S., Williams E.P., Zalduondo L., Samir P., Zheng M., Sundaram B., Banoth B., Malireddi R.K.S., et al. Synergism of TNF-alpha and IFN-gamma triggers inflammatory cell death, tissue damage, and mortality in SARS-CoV-2 infection and cytokine shock syndromes. Cell. 2021;184:149–168.e17. doi: 10.1016/j.cell.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Malireddi R.K.S., Karki R., Sundaram B., Kancharana B., Lee S., Samir P., Kanneganti T.D. Inflammatory cell death, PANoptosis, mediated by cytokines in diverse cancer lineages inhibits tumor growth. Immunohorizons. 2021;5:568–580. doi: 10.4049/immunohorizons.2100059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu W., Ye Z., Chen L., Liang H., Cai Q. A pyroptosis-related lncRNA signature predicts prognosis and immune microenvironment in head and neck squamous cell carcinoma. Int. Immunopharmacol. 2021;101 doi: 10.1016/j.intimp.2021.108268. [DOI] [PubMed] [Google Scholar]
- 36.Zhang M.J., Gao W., Liu S., Siu S.P.K., Yin M., Ng J.C.W., Chow V.L.Y., Chan J.Y.W., Wong T.S. CD38 triggers inflammasome-mediated pyroptotic cell death in head and neck squamous cell carcinoma. Am. J. Cancer Res. 2020;10:2895–2908. [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Z., Liu H., Dong Q., Li H., Zhang B., Liu Y., Zhong L., Tang H. Prognostic role of DFNA5 in head and neck squamous cell carcinoma revealed by systematic expression analysis. BMC Cancer. 2021;21:951. doi: 10.1186/s12885-021-08692-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rioja-Blanco E., Arroyo-Solera I., Álamo P., Casanova I., Gallardo A., Unzueta U., Serna N., Sánchez-García L., Quer M., Villaverde A., et al. CXCR4-targeted nanotoxins induce GSDME-dependent pyroptosis in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2022;41:49. doi: 10.1186/s13046-022-02267-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Y., Gao W., Shi X., Ding J., Liu W., He H., Wang K., Shao F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 40.Erkes D.A., Cai W., Sanchez I.M., Purwin T.J., Rogers C., Field C.O., Berger A.C., Hartsough E.J., Rodeck U., Alnemri E.S., Aplin A.E. Mutant BRAF and MEK inhibitors regulate the tumor immune microenvironment via pyroptosis. Cancer Discov. 2020;10:254–269. doi: 10.1158/2159-8290.CD-19-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smalley K.S.M. Two worlds collide: unraveling the role of the immune system in BRAF-MEK inhibitor responses. Cancer Discov. 2020;10:176–178. doi: 10.1158/2159-8290.CD-19-1441. [DOI] [PubMed] [Google Scholar]
- 42.Xiao Y., Zhang T., Ma X., Yang Q.C., Yang L.L., Yang S.C., Liang M., Xu Z., Sun Z.J. Microenvironment-responsive prodrug-induced pyroptosis boosts cancer immunotherapy. Adv. Sci. 2021;8 doi: 10.1002/advs.202101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearney C.J., Vervoort S.J., Hogg S.J., Ramsbottom K.M., Freeman A.J., Lalaoui N., Pijpers L., Michie J., Brown K.K., Knight D.A., et al. Tumor immune evasion arises through loss of TNF sensitivity. Sci. Immunol. 2018;3 doi: 10.1126/sciimmunol.aar3451. [DOI] [PubMed] [Google Scholar]
- 44.Baginska J., Viry E., Berchem G., Poli A., Noman M.Z., van Moer K., Medves S., Zimmer J., Oudin A., Niclou S.P., et al. Granzyme B degradation by autophagy decreases tumor cell susceptibility to natural killer-mediated lysis under hypoxia. Proc. Natl. Acad. Sci. USA. 2013;110:17450–17455. doi: 10.1073/pnas.1304790110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Taft J., Markson M., Legarda D., Patel R., Chan M., Malle L., Richardson A., Gruber C., Martín-Fernández M., Mancini G.M.S., et al. Human TBK1 deficiency leads to autoinflammation driven by TNF-induced cell death. Cell. 2021;184:4447–4463.e20. doi: 10.1016/j.cell.2021.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gomez A., Serrano A., Salero E., Tovar A., Amescua G., Galor A., Keane R.W., de Rivero Vaccari J.P., Sabater A.L. Tumor necrosis factor-alpha and interferon-gamma induce inflammasome-mediated corneal endothelial cell death. Exp. Eye Res. 2021;207 doi: 10.1016/j.exer.2021.108574. [DOI] [PubMed] [Google Scholar]
- 47.O'Sullivan B., Brierley J., Byrd D., Bosman F., Kehoe S., Kossary C., Piñeros M., Van Eycken E., Weir H.K., Gospodarowicz M. The TNM classification of malignant tumours-towards common understanding and reasonable expectations. Lancet Oncol. 2017;18:849–851. doi: 10.1016/S1470-2045(17)30438-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reichart P.A., Philipsen H.P., Sciubba J.J. The new classification of Head and Neck Tumours (WHO)--any changes? Oral Oncol. 2006;42:757–758. doi: 10.1016/j.oraloncology.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 49.Chen L., Yang Q.C., Li Y.C., Yang L.L., Liu J.F., Li H., Xiao Y., Bu L.L., Zhang W.F., Sun Z.J. Targeting CMTM6 suppresses stem cell-like properties and enhances antitumor immunity in head and neck squamous cell carcinoma. Cancer Immunol. Res. 2020;8:179–191. doi: 10.1158/2326-6066.CIR-19-0394. [DOI] [PubMed] [Google Scholar]
- 50.Zheng D.W., Deng W.W., Song W.F., Wu C.C., Liu J., Hong S., Zhuang Z.N., Cheng H., Sun Z.J., Zhang X.Z. Biomaterial-mediated modulation of oral microbiota synergizes with PD-1 blockade in mice with oral squamous cell carcinoma. Nat. Biomed. Eng. 2022;6:32–43. doi: 10.1038/s41551-021-00807-9. [DOI] [PubMed] [Google Scholar]
- 51.Percie du Sert N., Hurst V., Ahluwalia A., Alam S., Avey M.T., Baker M., Browne W.J., Clark A., Cuthill I.C., Dirnagl U., et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. BMJ Open Sci. 2020;4 doi: 10.1136/bmjos-2020-100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S., Luo M., Wang Z., Feng Q., Wilhelm J., Wang X., Li W., Wang J., Cholka A., Fu Y.X., et al. Prolonged activation of innate immune pathways by a polyvalent STING agonist. Nat. Biomed. Eng. 2021;5:455–466. doi: 10.1038/s41551-020-00675-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xiao Y., Li H., Mao L., Yang Q.C., Fu L.Q., Wu C.C., Liu B., Sun Z.J. CD103(+) T and dendritic cells indicate a favorable prognosis in oral cancer. J. Dent. Res. 2019;98:1480–1487. doi: 10.1177/0022034519882618. [DOI] [PubMed] [Google Scholar]
- 54.Sun X., Liu T., Zhao J., Xia H., Xie J., Guo Y., Zhong L., Li M., Yang Q., Peng C., et al. DNA-PK deficiency potentiates cGAS-mediated antiviral innate immunity. Nat. Commun. 2020;11:6182. doi: 10.1038/s41467-020-19941-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S., Wang X.L., Wu Z.Z., Yang Q.C., Xiong H.G., Xiao Y., Li H., Sun Z.J. Overexpression of RRM2 is related to poor prognosis in oral squamous cell carcinoma. Oral Dis. 2021;27:204–214. doi: 10.1111/odi.13540. [DOI] [PubMed] [Google Scholar]
- 56.Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data will be shared publicly in an online depository prior to publication. All other data generated for this study will be made available upon reasonable request to the corresponding author.