Abstract
Success in the treatment of infants with spinal muscular atrophy (SMA) underscores the potential of vectors based on adeno-associated virus (AAV). However, a major obstacle to the full realization of this potential is pre-existing natural and therapy-induced anti-capsid humoral immunity. Structure-guided capsid engineering is one possible approach to surmounting this challenge but necessitates an understanding of capsid-antibody interactions at high molecular resolution. Currently, only mouse-derived monoclonal antibodies (mAbs) are available to structurally map these interactions, which presupposes that mouse and human-derived antibodies are functionally equivalent. In this study, we have characterized the polyclonal antibody responses of infants following AAV9-mediated gene therapy for SMA and recovered 35 anti-capsid mAbs from the abundance of switched-memory B (smB) cells present in these infants. For 21 of these mAbs, seven from each of three infants, we have undertaken functional and structural analysis measuring neutralization, affinities, and binding patterns by cryoelectron microscopy (cryo-EM). Four distinct patterns were observed akin to those reported for mouse-derived mAbs, but with early evidence of differing binding pattern preference and underlying molecular interactions. This is the first human and largest series of anti-capsid mAbs to have been comprehensively characterized and will prove to be powerful tools for basic discovery and applied purposes.
Keywords: adeno-associated virus vector, anti-vector immunity, monoclonal antibody, capsid, neutralization, Zolgensma, spinal muscular atrophy, capsid structure, affinity, recombinant AAV
Graphical abstract
Logan and colleagues show that Zolgensma-treated infants mount robust and sustained anti-vector antibody responses. They also isolate the single largest collection of human anti-AAV monoclonal antibodies and functionally and structurally characterize their capsid-binding properties to provide valuable insights into host-vector responses as well as a powerful toolkit for scientific applications.
Introduction
Recombinant viral vectors based on adeno-associated virus (AAV) are delivering transformative therapeutic impact across an increasing spectrum of diseases. The successful treatment of spinal muscular atrophy (SMA), a devastating neurodegenerative disease of infancy caused by progressive motor neuron die-off, is a leading example.1,2 An important feature of the AAV vector system is the ability to package recombinant viral genomes into different capsid serotypes depending on the gene transfer properties required.3 For instance, the ability to cross the blood-brain barrier and transduce motor neurons in the CNS following systemic delivery underpinned the selection of the AAV9 capsid for the treatment of SMA.4 The tight link between capsid biology and therapeutic utility has led to an intense interest in the engineering of novel AAV capsids with desired biological properties using directed evolution of variants from complex randomly diversified libraries or structure-guided approaches.3,5
The structure-guided engineering approach requires a comprehensive understanding of the capsid surface at high molecular resolution. AAV capsids are highly organized structures that assemble from 60 viral proteins (VPs) into T = 1 icosahedral capsids via 2-, 3-, and 5-fold symmetry-related VP interactions to form ∼260 Å diameter particles.6 Although the amino acid sequence identity of AAV serotypes can vary by ∼50%, the virions conserve general surface features, including cylindrical channels at the 5-fold axes, protrusions surrounding the 3-fold axes, depressions located at the 2-fold axes, and raised regions between the 2- and 5-fold axes that are termed 2/5-fold walls.6
An important constraint in the use of AAV-based vectors is the presence of pre-existing neutralizing antibodies (NAbs) that target the AAV capsid surface in a proportion of patients and diminish the efficacy of therapy.7 This pre-existing anti-capsid humoral immunity is induced by exposure to circulating wild-type AAVs, most commonly AAV2, which are endemic in the human population and share reactive epitopes with the diversity of AAV capsid serotypes being used in human therapy8,9 Treatment-induced anti-capsid humoral immunity is also problematic, as it has the potential to preclude retreatment of patients with an inadequate initial therapeutic response. Addressing the challenges imposed by anti-capsid humoral immunity is therefore important to realization of the full therapeutic potential of AAV-based vector technology.
Strategies being explored to ameliorate the detrimental impact on therapy of anti-capsid immunity can be broadly divided into those focused on modulation of host immune responses and those focused on modifying the vector. Examples of the former include depletion of the B cell compartment10 and antibody removal by physical11 or enzymatic means,12 while examples of the latter include masking the capsid from the immune system by exosome enclosure13 and by antibody evasion through structure-guided evolution of antigenically distinct AAV capsids.14
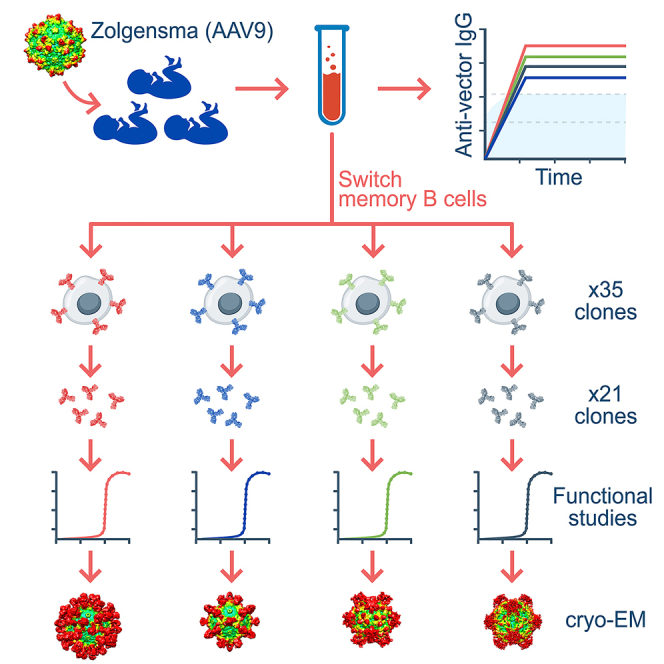
Capsid bioengineering approaches are well established for the modification of tropism in different cell types and tissues3 but are relatively less well developed for immune evasion. The latter demands an intimate understanding of antibody-capsid interactions. Previous studies have analyzed these interactions with various AAV serotypes using mouse monoclonal antibodies (mAbs).15,16,17,18,19,20 As mouse antibodies may not faithfully recapitulate human-derived antibodies, understanding the antigenic interactions have been constrained by a lack of human mAbs raised against clinically relevant AAV capsid serotypes. To this end, we have exploited privileged access to sera and fresh whole blood from a series of infants identified with SMA through a newborn screening program and treated using an AAV9 vector (Zolgensma). We have used these samples to study the anti-capsid serological responses and to clone and characterize the anti-AAV9 capsid mAbs involved.
All infants studied mounted robust IgG anti-capsid immune responses that not only reacted to the AAV9 capsid, but also with a hierarchy of cross-reactivity to other clinically relevant AAV capsid serotypes. This robust anti-capsid seroreactivity correlated with an abundance of switched-memory B (smB) cells that powerfully facilitated the recovery of sequences encoding light- and heavy-chain variable regions for reconstitution of 35 anti-AAV9 mAbs from three infants. The neutralization properties of 21 of these antibodies were determined and cryoelectron microscopy (cryo-EM) performed with the Fab fragments to resolve patterns of antibody interaction with the capsid surface. All tested mAbs were found to be capable of neutralizing an AAV vector pseudo-serotyped with the AAV9 capsid in an in vitro transduction assay, and a proportion also demonstrated cross-reactive neutralization of vectors pseudo-serotyped with other AAV capsids. Cryo-EM analysis of the individual Fabs revealed four distinct binding regions of the capsid surface involving either the 2-fold, 3-fold, and 5-fold axes of symmetry or the 2/5-fold wall. Although this directly correlates with the binding patterns observed with anti-capsid mAbs raised in mice,16,18,19 the series of human mAbs reported here show strong early evidence of differing capsid binding behavior, both in binding pattern preference and in the molecular architecture of the mAb-capsid interactions underlying these higher order patterns. Binding affinities ranged from 0.05 to 200 nM, without displaying a trend in binding strength and preferred binding region.
Collectively, this report not only offers insight into the amplitude, duration, and complexity of the human humoral response to AAV capsid exposure in the context of gene therapy but also provides the first human and single largest set of structurally and functionally characterized mAbs directed against any single AAV capsid. Specifically, these novel reagents provide an unprecedented opportunity to map human mAb-AAV9 capsid interactions for both basic discovery and applied purposes.
Results
Infants with SMA mount robust anti-AAV9 capsid humoral immune responses
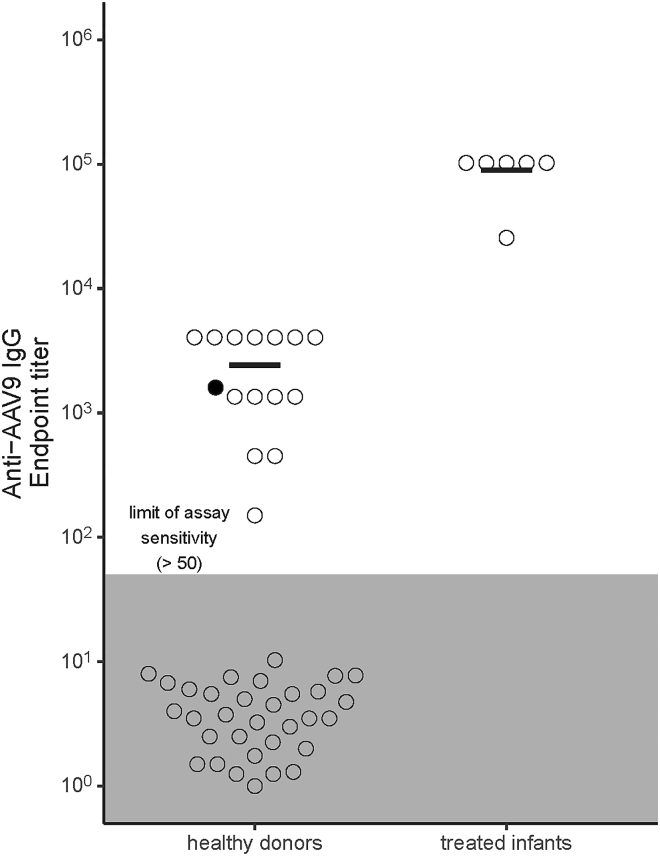
Sera collected from six infants treated for SMA with Zolgensma, an AAV gene therapy vector bearing the serotype 9 capsid, were assayed for anti-AAV9 IgG endpoint titers using a sensitive in-house ELISA and compared with titers measured in healthy donors (n = 51; Figure 1). The endpoint titers in the treated infants were universally high relative to those detected in healthy donors. Furthermore, anti-AAV9 IgG was not detected in more than half of healthy donors resulting in a bimodal pattern of anti-AAV9 IgG in this group.
Figure 1.
AAV9-based gene therapy induces high anti-AAV9 antibody endpoint titers in treated infant sera
Sera from AAV-treated SMA infants (n = 6) and healthy donors (n = 51) were analyzed to determine anti-AAV9 IgG endpoint titers. Black bars in each group depicted in the bee swarm plot indicate the mean of samples with a detectable titer. Samples from treated infants were collected between 169 and 335 days post-treatment. The donor indicated in black is a woman whose child was excluded from access to AAV9 gene therapy because of passively acquired antibodies in utero. The gray zone indicates levels below the limit of assay sensitivity. Results are representative of three independent assays.
The differences in serological profiles between the two groups can almost certainly be attributed to the treated infants’ exposure to a high AAV9 capsid antigen load (>1014 vector genome [vg] per kilogram), while seroreactivity observed in healthy donors is likely to reflect cross-reactivity to natural infection, most commonly with AAV2.8 The difference may also be influenced by the time interval between capsid antigen exposure and serological testing. Of particular interest, one of the healthy donor samples was from a mother whose infant was diagnosed with SMA by newborn screening but excluded from Zolgensma therapy because of anti-AAV capsid antibodies passively acquired in utero (Figure 1, black dot). This infant was subsequently treated after seroreactivity subsided.
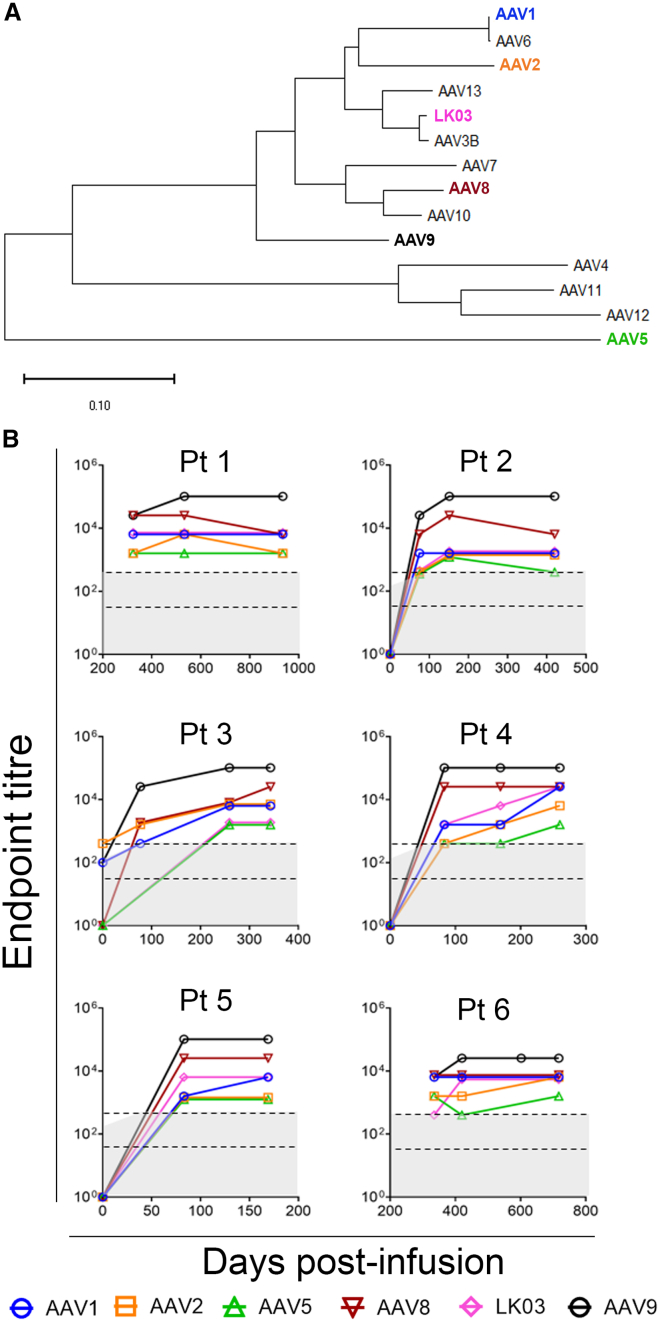
The sera collected from the AAV9-treated infants presented a unique opportunity to examine IgG responses mounted by the naive human immune system after AAV capsid exposure. Anti-AAV9 capsid reactivity and cross-reactivity with other capsid serotypes, selected on the basis of clinical use, were evaluated (Figures 2A and 2B). Pre-treatment samples were available in 4 of 6 infants and, in one of these (patient 3), pre-existing anti-AAV9 capsid seroreactivity was detected in the pre-treatment serum but was below the cut-off for exclusion from treatment (on the basis of the trial reference laboratory result). Maximal levels of AAV9 seroreactivity were stably maintained in all infants out to the most contemporary sera samples available, up to 42 months post-treatment (Figure S1), and a hierarchy of cross-reactivity to other AAV capsid serotypes was observed that correlates with phylogenetic relatedness (Figure 2B). This included reactivity to AAV5, the most phylogenetically distant capsid tested, that consistently gave an endpoint titer 10- to 100-fold lower than AAV9, which consistently exhibited the greatest seroreactivity.
Figure 2.
Anti-AAV9 IgG responses are sustained and cross-reactive to distinct AAV capsid serotypes
(A) Dendrogram showing the phylogenetic relatedness of AAV VP1 amino acid sequences from native capsid serotypes 1–13 and the engineered capsid LK03. (B) Anti-AAV IgG endpoint titers to AAV capsids 1, 2, 5, 8, 9, and LK03 in treated infants were measured using ELISA. The x axis indicates day of serum collection post-AAV infusion. Gray zones indicate levels below the limit of assay sensitivity with a lower sensitivity for pre-infusion samples. Area between dashed black lines indicates the cut-off range to preclude gene therapy treatment, accommodating different levels of assay sensitivity. Pre-infusion sera were not available for patients 1 and 6.
Gene therapy induced a high frequency of anti-AAV9 reactive switched-memory B cells
Switched-memory B cells are B lymphocytes that have undergone antigen stimulation, somatic hypermutation, and antibody class switching. They can mount a rapid response after secondary antigen stimulation, and their presence in the circulatory system makes them easily accessible to clone heavy and light chain sequences to permit monoclonal antibody reconstitution.21 The frequency of smB cells that can secrete anti-AAV9 IgG was surveyed in post-infusion peripheral blood mononuclear cells (PBMCs) collected from 3 of the 6 infants that were sero-surveyed (patients 1, 2, and 3) as well as in the earlier mentioned mother of patient 5. In treated infant samples, the frequency of smB cells reactive to the AAV9 capsid comprised between approximately 2% and 10% of the total circulating smB cell population, values up to 760-fold higher than those observed in the naturally exposed mother (Table 1). We took advantage of this rich source of AAV9-reactive smB cells in these infants to recover light- and heavy-chain antibody variable sequences for mAb reconstitution (Figure S2).
Table 1.
Frequency of smB cells expressing AAV9-reactive antibody
| PBMC source | Days post-antigen stimulus | Wells seeded with smB cells | IgG+ wells | IgG+ wells, % | AAV9 reactive clones | Anti-AAV9 reactive smB clones (%) |
|---|---|---|---|---|---|---|
| Patient 1 | 324 | 2,464 | 714 | 29 | 39 | 5.5 |
| Patient 2 | 151 | 1,232 | 221 | 18 | 22 | 9.9 |
| Patient 3 | 259 | 616 | 363 | 59 | 8 | 2.2 |
| Mother of patient 5 | unknown | 9,856 | 7,491 | 76 | 1 | <0.1 |
Reconstitution and sequence characterization of anti-AAV9 capsid monoclonal antibodies
The smB cell clones showing reactivity to AAV9 capsid antigen were subjected to RT-PCR to recover either the kappa or lambda light-chain variable region sequences and the IgG heavy-chain variable region sequence. After sequence validation, the amplicons were subcloned into their respective constant region scaffolds in plasmid constructs. Paired light- and heavy-chain encoding constructs derived from individual smB cell clones were then used to transfect HEK293 cells for mAb reconstitution. In total, 35 anti-AAV9 reactive mAbs were reconstituted, 15 from patient 1, 13 from patient 2, and 7 from patient 3. There was no consistent bias for kappa or lambda light-chain use across the mAbs recovered (Figure S3A). Mean CDR3 region lengths of the light chains for the three patients fell within the typical range of 9–10 amino acids, but heavy chain CDR3 were longer than the 15 amino acids typically found in healthy humans (patient 1, 19; patient 2, 22; patient 3, 20; Figure S3B22,23). Aligning variable region sequences against germline sequences showed no dominant V, D, or J gene family use for any patient (Table S1). As expected, relative to germline sequences, multiple nucleotide differences were observed throughout variable regions with a preference for non-synonymous changes (Figure S2C). Furthermore, these changes occurred more frequently in the CDR1 and CDR2 regions than the framework regions (Figure S3D). All antibody amino acid sequences were unique across the 35 mAbs reconstituted, although two clearly related mAbs isolated from patient 2 encoded identical lambda chain sequences and differed by only a single amino acid in the heavy chain CDR3 region (data not shown). The IgG subclass distribution, determined by examination of sequences located downstream of the heavy-chain variable domains, showed that most mAbs were derived from smB cell clones that had switched to the IgG1 subclass (n = 29), a smaller proportion switched to IgG3 (n = 3), and the remainder were indeterminate (n = 3). Importantly, this subclass distribution directly recapitulated what was observed in sera, with IgG1 being the most dominant subclass of anti-AAV9 IgG, with substantially lower levels of IgG3 and IgG4 (Figure S4). This subclass hierarchy is typical of the response seen to virus infection.24 Collectively, these observations indicate that the smB cell clones showing anti-AAV9 reactivity have arisen through functional selection involving somatic hypermutation and affinity maturation.
Anti-AAV9 capsid monoclonal antibodies block transduction and show variable cross-reactivity
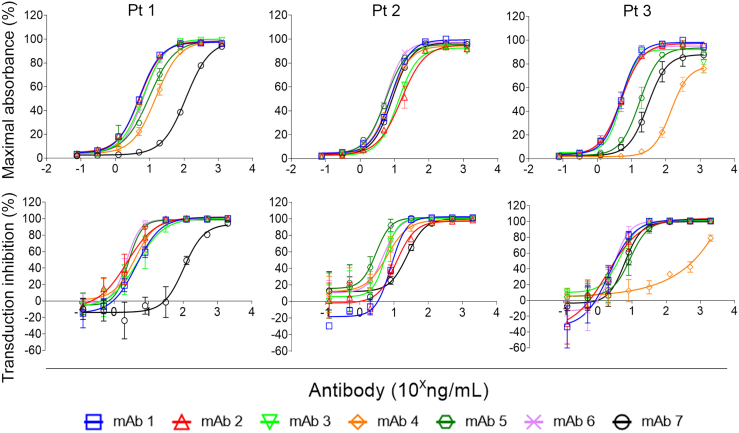
For each of the treated infants a subset of seven mAbs were selected, on the basis of the chronological order of isolation, and purified mAb stocks prepared for characterization. Titration of the mAbs and assay by ELISA revealed an approximately 30-fold range of anti-AAV9 reactivity determined by the mAb concentration required to produce 50% maximal absorbance (Figure 3, top panel; half-maximal effective concentration [EC50]; Table S2). Titration and assessment by neutralization assay showed that all mAbs were capable of blocking vector transduction; however, some also produced a modest but reliable enhancement of AAV9-mediated transduction at low concentrations (Figure 3, bottom panel). There was a modest correlation using the Spearman rho test (rho = 0.657; Figure S5) between EC50 (Table S2) and the concentration of mAb required to inhibit 50% of vector transduction (half-maximal inhibitory concentration [IC50], Table S3).
Figure 3.
Anti-AAV9 mAbs neutralize functional transduction
Anti-AAV9 reactivity of twenty-one mAbs recovered from three treated infants were serially diluted and tested for anti-AAV9 reactivity by ELISA (top panel) or AAV9 transduction neutralization assay (bottom panel). Data are pooled from duplicate and triplicate independent experiments for the ELISA and neutralization assays, respectively. Error bars indicate SEM.
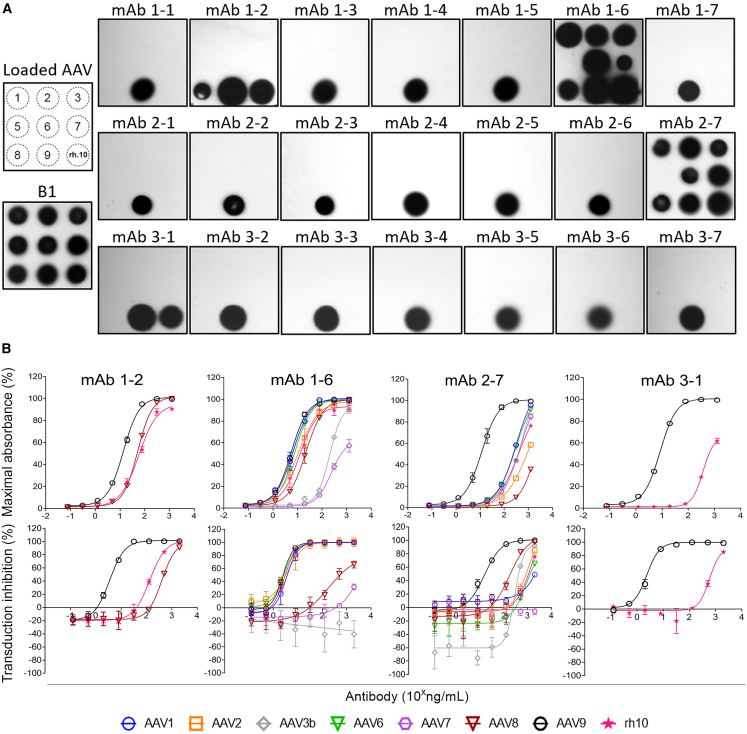
The mAbs were also screened by native dot immunoblot to explore cross-reactivity to other natural AAV serotypes. The assay was performed against serotypes 1, 2, 3, 5, 6, 7, 8, 9, and rh10, with the majority of mAbs (17 of 21) being AAV9 specific (Figure 4A). Four mAbs showed cross-reactivity to other capsid serotypes, with mAb1-2 cross-reacting with AAV8 and AAVrh10; mAb3-1 cross-reacting with AAVrh10; and two mAbs, mAb1-6 and mAb2-7, cross-reacting with all serotypes except the phylogenetically distant AAV5 capsid. These cross-reactive mAbs were further tested by ELISA and neutralization assays against the capsid serotypes to which they showed reactivity in the dot-blot assay (Figure 4B). The ELISA data proved consistent with the dot-blot data, but more clearly revealed a hierarchy of cross-reactivity spanning up to a 100-fold mAb concentration. With exceptions, the neutralization assay also correlated with the dot-blot data. For the broadly cross-reactive mAb1-6, neutralization of AAV8- and AAV3-mediated transduction was weak or absent, respectively, relative to the ELISA signal. For mAb2-7, another interesting observation was enhancement of transduction at low mAb concentrations, particularly for AAV3B, in the single cell line (2v6.11) tested.
Figure 4.
A subset of anti-AAV9 mAbs cross-react with other capsid serotypes
(A) Immuno-dot blot of the twenty-one mAbs, obtained from three treated infants, against the indicated AAV serotypes arrayed as per the template to the left of the panel. Equivalent antigen loading was confirmed using the B1 mAb. (B) Cross-reactive mAbs identified by immune dot blot were tested for anti-capsid reactivity by ELISA (upper row) and neutralization in a functional transduction assay (lower row). The immune dot blot is representative of three replicates. Data are pooled from duplicate and triplicate independent experiments for the ELISA and neutralization assays, respectively. Error bars indicate SEM.
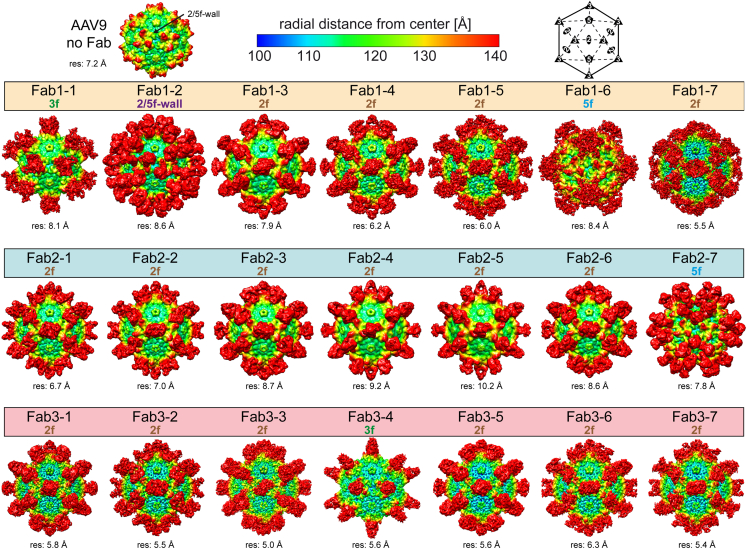
Cryo-EM mapping reveals the antigenically dominant 2-fold region of the AAV9 capsid
Fabs were prepared for the 21 mAbs and mixed with AAV9 to map their binding sites to the surface of the capsids. Cryo-EM data were collected of the complexed samples and three-dimensional (3D) image reconstruction was used to generate ∼5–10 Å resolution complex maps from ∼2,000 to 10,000 individual particles. Fabs were found to bind either the 2-, 3-, and 5-fold axis of symmetry or the 2/5-fold wall of the AAV9 capsid (Figure 5). However, although the Fabs showed binding to all four topological regions, Fabs binding near or across the 2-fold symmetry axis were the most prevalent, as ∼76% (n = 16) of Fabs bound to this region. This dominant binding pattern was observed for all three patients (patient 1, 57%; patient 2, 86%; patient 3, 86%). Among the 2f-binding Fabs, different interacting conformations were observed (e.g., Fab1-3 vs. Fab1-7 vs. Fab2-1). For the non-2f-binding Fabs, two were shown to bind to the center of the 3-fold axis of symmetry (Fab1-1 and Fab3-4), one to the 2/5-fold wall (Fab1-2), and two to the 5-fold region (Fab1-6 and Fab2-7) with different binding conformations. In summary, the Fabs derived from the 21 human mAbs bound either of the four topological regions of the capsid surface but showed antigenic dominance to the 2-fold region of the AAV9 capsid.
Figure 5.
Fab binding sites on the AAV9 capsid
Three-dimensional reconstruction maps of AAV9 (without Fab) and in complex with Fab1-7 from patients 1, 2, and 3. The maps are radially colored according to distance to the center of the capsid blue to red, as indicated by the scale at the top of the panel. The icosahedral 2-, 3-, and 5-fold axes and the 2/5-fold wall are indicated on the schematic and “AAV9 no Fab” capsid map, and Fab-binding sites are given above each map. The estimated resolutions, determined at a Fourier shell correlation (FSC) threshold value of 0.143, is shown below each map.
Fab binding affinity correlates with IC50 but not Fab binding site
The Fabs were further characterized by determining their binding kinetics to the AAV9 capsid. Using immobilized AAV9 capsids, the kD value for each Fab was determined by biolayer interferometry (BLI). The Fabs display high binding affinities, with kD values ranging from approximately 50 pM (Fab3-3) to 200 nM (Fab1-7) (Table 2). The Fabs binding the 2/5-fold wall (Fab1-2, 0.23 nM) and the 3-fold region (Fab1-1, 0.30 nM) showed comparable sub-nanomolar binding affinity. The two 5-fold binding Fabs (Fab1-6, 0.25 nM; Fab2-7, 14.46 nM) showed a ∼60-fold difference in affinity. Similarly, the large group of 2-fold binding Fabs contains both the strongest and weakest binding antibodies (Fab3-3, 50 pM; Fab1-7, 200 nM). However, there is a modest correlation between Fab binding affinity and the IC50 values for the corresponding mAb (rho = 0.693; Figure S6). For comparison, the binding affinities of the mouse anti-AAV9 mAbs, ADK9, HL2368, and HL2372,25 were also determined. ADK9 binding across the 2-fold axis showed a kD of 0.21 nM, HL2368, a 2/5-fold wall binder, 1.87 nM, and HL2372, a 5-fold binder, 0.29 nM. In summary, identifying the Fab binding site on the AAV capsid is not predictive of the binding kinetics of an antibody, and no significant difference of the antibody affinity was observed between mouse and human mAbs.
Table 2.
Binding kinetics of the Fabs (average of n = 3 experiments)
| Fab | kD, nM | Fab | kD, nM | Fab | kD, nM |
|---|---|---|---|---|---|
| 1-1 | 0.30 | 2-1 | 0.22 | 3-1 | 0.24 |
| 1-2 | 0.23 | 2-2 | 1.29 | 3-2 | 0.18 |
| 1-3 | 0.28 | 2-3 | 24.80 | 3-3 | 0.05 |
| 1-4 | 2.30 | 2-4 | 2.57 | 3-4 | 6.12 |
| 1-5 | 0.15 | 2-5 | 0.26 | 3-5 | 26.40 |
| 1-6 | 0.25 | 2-6 | 2.94 | 3-6 | 0.18 |
| 1-7 | 199.50 | 2-7 | 14.46 | 3-7 | 30.60 |
| ADK9 | 0.21 | HL2368 | 1.87 | HL2372 | 0.29 |
Discussion
The AAV vector system is the clinically dominant platform for direct in vivo gene therapy applications and has significant therapeutic potential. Full realization of this potential is dependent upon ongoing technological development and an increased understanding of the complex host-vector interactions that influence the efficacy and safety of AAV-based therapeutic interventions. Anti-AAV capsid humoral immunity, both pre-existing and gene therapy induced, remain major challenges to the field, limiting the number of patients eligible for treatment and precluding retreatment, respectively. Here we report the first study to examine the serological responses of the naive immune system in human infants following exposure to high-dose AAV9 vector for the treatment of SMA. We not only show that infants mount a vigorous and durable anti-capsid humoral immune response but have dissected the polyclonal architecture of this response by cloning and structural characterization of the AAV capsid-binding properties of 21 mAbs recovered from three treated infants. Remarkably, all were neutralizing, showed an IgG subclass distribution reflecting that present in patient plasma, and fell into one of four distinct capsid surface binding patterns, with the dominant pattern being binding in the region of the 2-fold axis of symmetry. The majority were specific for the AAV9 capsid, but several exhibited cross-reactivity with other phylogenetically related capsid serotypes, most notably those binding the 5-fold axis of symmetry. This unprecedented collection of patient-derived mAbs, induced by AAV vector exposure, provides a powerful toolkit to explore host-capsid interactions and exploit the insights gained to engineer a new generation of AAV capsids with reduced seroreactivity. Future studies recovering human mAbs from the pediatric and adult population following natural AAV infection will further strengthen this toolkit.
Although it has been long appreciated that treatment-induced immunity is likely to preclude the readministration of AAV vectors, the amplitude, durability, and cross-reactivity of the polyclonal response to high-dose AAV9 exposure reported here underscore the challenge posed by anti-capsid humoral immunity. Moreover, the robust nature of the responses observed occurred despite the use of steroids during the period in which the immune response was being induced. In all treated infants examined, the induced antibody response against the AAV9 capsid was in the order of 500- to 2,000-fold above the levels considered preclusive of treatment26 and was stably maintained. Furthermore, patient serum showed broad cross-reactivity to other clinically relevant capsid serotypes, in a loose hierarchy correlating with phylogenic relatedness. Even the most phylogenetically distant capsid, AAV5, would likely be precluded from subsequent use. Importantly, this degree of cross-reactivity induced by therapeutic exposure to the AAV9 capsid is consistent with the frequency with which natural exposure to endemic AAV serotypes, most commonly AAV2, can induce sufficient anti-AAV9 cross-reactivity to preclude initial treatment. To understand the capsid binding interactions that underlie both direct reactivity and cross-reactivity, it is necessary to examine the properties of individual mAbs that make up the complex architecture of the polyclonal immune response.
In the present study, dissection of the polyclonal immune response was facilitated by the abundance of smB cells producing anti-AAV9 antibody in the PBMC fraction obtained from treated infants. Moreover, this abundance of smB cells was maintained for at least many months. The frequencies observed were dramatically higher than those observed in the mother of an infant with SMA initially precluded from treatment through passive acquisition of maternal antibody, and consistent with or higher than the frequencies of antigen-specific memory B cells reported following vaccination.27,28,29 The commonality of the post-gene therapy and vaccination contexts is that both involve a known antigen with known timing of exposure and elevated levels of antigenic stimulation by either high antigen load or concurrent use of adjuvant. Our success in reliably obtaining multiple anti-AAV9 mAbs from three different infants, selected simply on the basis of blood sample availability, demonstrates the scientific and translational value of such samples as a largely untapped resource and provides a template for recovering mAbs against the multiple different AAV capsid serotypes in clinical use.30
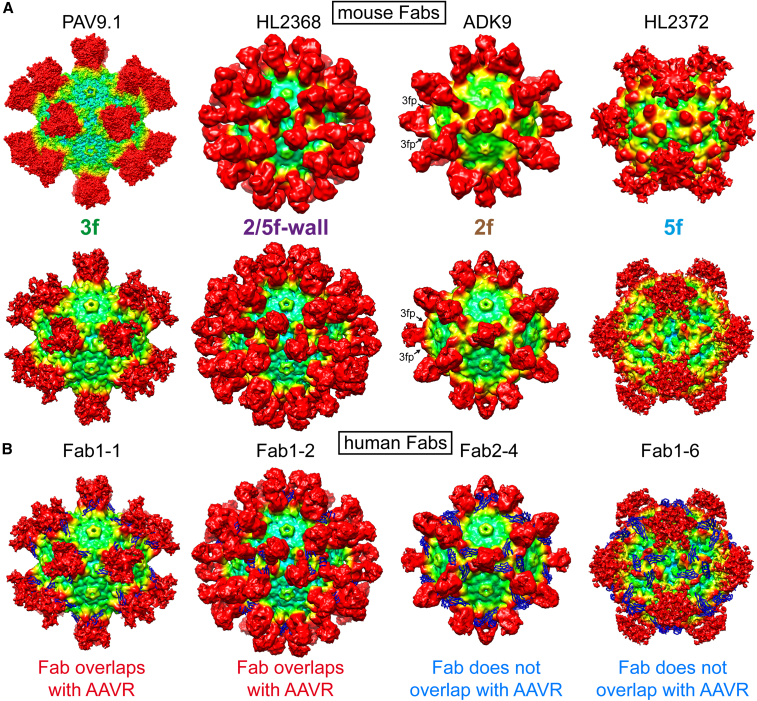
To date, a total of 28 individual AAV capsid-antibody complex structures have been published, using 22 different mAbs, all generated in mice, for the capsid serotypes AAV1, AAV2, AAV4, AAV5, AAV6, AAV8, AAV9, and AAVrh.10.16,17,18 Six of these 22 murine mAbs are generated against the AAV9 capsid.17,18 Thus, the series of 21 mAbs from three donors reported here are not only of human origin but represent the single largest collection to have had their capsid-binding properties structurally characterized using cryo-EM. Recently, a series of human mAbs from a single donor with broad naturally induced anti-capsid seroreactivity has been reported.31 However, attempts to map binding sites by cryo-EM were unsuccessful, and these mAbs did not show neutralization activity. The use of anti-capsid mouse mAbs to simulate the human anti-capsid immune response has faced criticism, as mouse antibodies may not faithfully recapitulate the capsid binding behavior of human-derived antibodies. Accordingly, our study has allowed the first structural comparison of mouse and human antibodies binding to any AAV capsid. Overall, the antibodies from humans and mice bind the same regions on the capsids (Figure 6A). In fact, some antibody-complex structures appear almost identical (e.g., Fab1-2 vs. HL2368), whereas others show slight variations (e.g., Fab1-6 vs. HL2372). In the case of mouse ADK9-derived Fab, while its binding site is across the 2-fold axis, the Fab does not enter the depression and instead binds the top of the 3-fold protrusion and bridges the 2-fold depression. This binding mode was not observed among any of the recovered human mAbs. Furthermore, two additional mouse mAbs, HL2370 and HL2374 (not shown), which exclusively bind the top of the 3-fold protrusions,15 did not have a corresponding human mAb counterpart in our series. Overall, the mouse anti-AAV9 mAbs appear to preferentially bind the 3-fold regions (4 of 6 mAbs, two-thirds) whereas for the human mAbs, the 2-fold depression including the side of the 3-fold protrusions, appear to be antigenically dominant (16 of 21 mAbs, approximately three-quarters). This difference has led prior studies to focus primarily on amino acids changes in the 3-fold protrusion which, although yet to be demonstrated, may have resulted in the engineering of capsids that would escape only a small portion of human NAbs, but not the preponderance binding near the 2-fold region.15,16,19,20 A shared property of the mouse and human mAbs is that binding of the derived Fabs near the 5-fold region of the capsid confers cross-reactivity to other AAV serotypes. However, although the previously reported mouse mAb, HL2372, showed only cross-reactivity to AAV8 and AAV9,25 the human mAb1-6 and mAb2-7 showed cross-reactivity to many AAV serotypes, with the exception of AAV5. With these characteristics, the human mAbs are akin to the camelid AVB nanobody that also binds most AAV serotypes including AAV5, but not AAV9.32 This is due to generally higher sequence and structural conservation in this region between different AAVs compared with other regions of the capsid.33
Figure 6.
Human and murine anti-AAV9 Fabs bind the four topological regions of the AAV capsid
(A) Top row: mouse Fabs binding to the 3-fold, 2/5-fold wall, 2-fold, and 5-fold regions are shown. In the absence of a density map for PAV9.1,17 a surface map from a pseudoatomic model was generated. The maps are radially colored according to distance to the center of the capsid (blue to red, as in Figure 6). Lower row: a selection of the human mAbs that bind regions equivalent to the murine mAbs. For the ADK9 and Fab2-4 density maps, the location of the 3-fold protrusions (3fp) are indicated. (B) The atomic model of AAV9-AAVR (PDB: 7WJX) was fitted into the density maps to determine whether the human Fabs overlap with AAVR (blue ribbon).
An additional notable feature is that all 21 human mAbs neutralized AAV9 transduction. Despite some of the mAbs cross-reacting with other serotypes, neutralizing capacity for these AAV serotypes were generally lower and/or required higher antibody concentrations. One outlier is mAb1-6 that neutralizes AAV1, AAV2, AAV6, and AAVrh.10 equally to AAV9, whereas neutralization of AAV7 and AAV8 is reduced, and no neutralization was observed for AAV3B. This could be due to lower affinities of the mAbs toward certain AAVs or the use of different receptors not affected by the antibodies. A similar observation was made for the AVB nanobody, which was shown to neutralize AAV1 and AAV5 but not AAV2.32
Currently, the neutralization mechanisms of the 21 human mAbs are unknown. For the AAV9 capsid the binding sites of two receptors have been mapped. The galactose binding pocket is located at the base of the 3-fold protrusion near the 2/5-fold wall,34 which overlaps with the binding site of mAb1-2. Binding to AAVR was shown to be essential for efficient AAV9 transduction.35 Recently, the complex structure of AAV9 and AAVR was determined.36 In this structure the PKD domain 2 of AAVR binds to the AAV9 capsid on top of the 2/5-fold wall along the base of the 3-fold protrusion toward the 3-fold symmetry axis. Because of its size, the PKD domain directly overlaps with 15 of the 21 human mAbs except for the 5-fold binding mAbs (mAb1-6 and mAb2-7) and a few 2-fold binding mAbs (mAb2-3, mAb2-4, mAb3-3, and mAb3-5) (Figure 6B). However, as AAVR is a multi-domain protein, it is possible that the adjacent domains clash with these mAbs as well. Furthermore, the mAbs binding near the 5-fold region might interfere with the externalization of the VP1u region from the interior of the capsid. The neutralization mechanisms of the human mAbs will be further investigated in future studies.
This is also the first study to determine the affinity of mAbs to AAV capsids. Most of the human antibodies showed kD values in the picomolar to low nanomolar range irrespective of the binding site on the capsid surface. Similarly, the kD values of the mouse anti-AAV9 mAbs ADK9, HL2368, and HL2372 fell into the same range. For the 5-fold binding mAbs, high concentrations of Fabs resulted in binding curves that fell below the base line during dissociation (data not shown). The reason for this observation is likely that the Fabs compete with the AAVX nanobody coupled to the Gator Bio BLI probe used to immobilize the AAV9 capsid, which also binds to the 5-fold region of the capsid (manuscript in preparation). Overall, antibody capsid binding affinity did not correlate with capsid binding site, but there was a correlation between affinity and the efficiency of AAV9 vector neutralization in an in vitro transduction assay. The in vivo implications of these observations are yet to be resolved.
Beyond these structure-function insights obtained from ∼5–10 Å resolution structures, higher resolution mapping of capsid-antibody binding interactions at 2–3 Å resolution will allow precise localization of the capsid surface amino acid residues involved in antibody recognition and neutralization. This applies not only to binding sites unique to the AAV9 capsid but also to those that underlie cross-reactivity with other capsid serotypes. The former provides a direct basis for structure-guided engineering of AAV9 escape variants that might allow retreatment following AAV9-based therapy, while the latter provides insights into how the AAV9 capsid might be engineered to escape antibody responses driven by natural exposure to other serotypes, most notably AAV2. Such variants could potentially prove useful in the treatment of patients currently precluded of AAV9-based therapies by pre-existing cross-reactive immunity. Importantly, the feasibility of engineering capsids to evade polyclonal anti-capsid responses has previously been established.14 The series of human mAbs reported could also be used in orthogonal approaches to provide selection pressure on randomly diversified capsid libraries14,37 to similarly identify antibody-escape variants with clinical utility. Other translational applications for these reagents include use in immune detection and purification methodologies.
Materials and methods
Study design
Serum and PBMCs were collected from a series of infants detected with SMA through newborn screening and subsequently treated with Zolgensma. Serum anti-AAV IgG responses to the AAV9 capsid were measured using ELISA and cross-reactivity to other capsid serotypes was similarly measured. SmB cells in PBMC samples obtained from three treated infants were isolated by negative selection and individual cells clonally expanded. Clones that secreted anti-AAV9 IgG were identified using ELISA. The variable regions from these clones encoding light or heavy chain variable sequences were recovered by nested RT-PCR amplification and ligated into expression plasmids containing the required antibody scaffold sequences. Co-transfection of paired light- and heavy-chain antibody plasmid constructs and testing of culture supernatants for anti-AAV9 reactivity by ELISA confirmed successful reconstitution of 35 anti-AAV9 mAb from three patients. A subset of 21 mAb (seven from each patient) were further titrated and characterized by ELISA. The mAb were also tested for the capacity to inhibit AAV9 vector transduction in vitro and used to generate Fabs for capsid binding studies using cryo-EM and biolayer interferometry.
Blood samples
Human ethics approval (HREC/18/SCHN/373) permitted collection of blood from SMA patients before and after treatment with Zolgensma at the age of 30 days (patient 1), 27 days (patient 2), 22 days (patient 3), 6 months (patient 4), 12 months (patient 5) and 35 days (patient 6). Serum was removed from clotted blood after centrifugation (1,000 × g), while PBMCs were isolated from lithium heparin-treated blood using centrifugation through Ficoll-Paque (GE Healthcare) according to the manufacturer’s instructions. PBMCs were resuspended in media containing 90% heat-inactivated fetal bovine serum (HI-FBS) and 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen before use.
Tissue culture
HEK293 (CRL-1573; American Type Culture Collection) and 2v6.1138 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM; Gibco) supplemented with glutamine (10 mM) and 10% heat-inactivated FBS (Complete media) in a humidified incubator at 37°C/5% CO2 and passaged using trypsin (Gibco). To generate an HEK-293 cell line expressing the CD154 ligand, the CD154 cDNA coding sequence was RT-PCR amplified from RNA isolated from PBMCs using forward primer: GCCACCATGATCGAAACATACAACCAAACTTCT and reverse primer: TCAGAGTTTGAGTAAGCCAAAGGAC and subcloned into pTARGET Mammalian Plasmid Expression System (Promega). A plasmid clone encoding human CD154 was validated using Sanger sequencing and transfected into HEK-293 cells by calcium-phosphate transfection. At 48 h post-transfection, cell selection was performed in complete media supplemented with 600 μg/mL G418 (Roche) until control HEK-293 cells (i.e., no plasmid control cells) were eliminated. G418-selected HEK-293 were enriched for CD154 by antibody labeling (TRAP-1 clone; BD Biosciences) and fluorescence-activated cell sorting (FACS) using the FACSAria III (BD Biosciences). The sorted HEK293-CD154 cell line was cultured, harvested, resuspended, and stored in liquid nitrogen in media containing DMEM supplemented with 20% HI-FBS and 10% DMSO.
AAV construct and vector production
The AAV vector genome construct contained the inverted terminal repeat (ITR) from AAV serotype 2 flanking an expression cassette composed of a promoter from respiratory syncytial virus, an enhanced GFP coding sequence, a 2A viral element, and a luciferase reporter coding sequence followed by a polyadenylation sequence. The construct was packaged into capsid serotype 1,39 serotype 2-like natural isolate, hu.li.02,40 AAV3,39 AAV5,39 AAV6,41 AAV7,42 AAV8,42 AAV9,43 or rh10 capsids,43 as previously described,44 except that transfections were performed using calcium transfection or polyethylenimine. AAV vectors were purified45 and titered by qPCR using primers eGFP-forward: 5′-TCAAGATCCGCCACAACATC-3′ and eGFP-reverse: 5′-TTCTCGTTGGGGTCTTTGCT-3′.46
Isolation and culture of switched-memory B cells from PBMCs
PBMCs for patients 1, 2, and 3 were collected and stored at days 324, 151, and 259 post-vector treatment, respectively, then later thawed at 37°C, resuspended in complete medium, and pelleted by centrifugation at 300 × g for 10 min at 4°C. smB cells were isolated from the PBMCs by negative selection using a switched-memory B cell isolation kit (Miltenyi Biotech) according to the manufacturer’s instructions. The purity of isolated smB cells was always found to be >80%, as determined by the proportion of CD19+CD27+ double-positive cells after immunolabeling (anti-CD19 mAb, HIB19 clone [BD Biosciences]; anti-CD27 mAb, M-T271 clone [BD Biosciences]) and FACS analysis. Purified smB cells were cloned and cultured as previously described.21 Briefly, smB cells were mixed with irradiated HEK293-CD154 (7.5 Gy) in Iscove’s modified DMEM and seeded in 50 μL to wells of 384-culture plates so that each well contained 1.3 smB cells and 7 × 103 HEK293-CD154. The media was also supplemented with final concentrations of Glutamax, penicillin-streptomycin, and Normocin (InvivoGen) (all at 1×) 10% heat-inactivated FBS, interleukin-2 (100 U/mL; Roche) and interleukin-21 (100 μg/mL; Peprotech). The outer wells of each plate contained 100 μL PBS to minimize evaporation and plate effects. Plates were incubated at 37°C/5% CO2 for 12 days, when one representative plate was assessed for IgG production by ELISA (see protocol below). On day 13, 40 μL of the supernatants from the remainder of plates were transferred to new 384-well plate while cells that smB cell clones that remained in the wells were lysed by adding 20 μL of cell lysis buffer (150 mM Tris-HCl [pH 8.0], 500 U/mL RNase inhibitor (New England Biolabs, prepared in DEPC-treated water) to each well. Supernatants and cell lysates were stored at −80°C before use.
Enzyme-linked immunosorbent assay to detect total or anti-AAV IgG
Sera were assayed for reactivity to AAV by ELISA. Ninety-six-well polystyrene Maxisorp ELISA plates (Nunc) were coated overnight at 4°C with 50 μL per well of AAV vector stocks (2.5 × 1010 vg/mL) diluted in coating buffer (carbonate-bicarbonate buffer; Sigma-Aldrich). Plates were washed 3 times with wash buffer (phosphate-buffered saline [PBS]; Sigma-Aldrich) + 0.05% Tween 20 (Sigma-Aldrich) and then received 100 μL per well of blocking buffer (PBS + 5% skim milk + 0.05% Tween 20). After incubation at room temperature (RT) for 2 h, plates were washed 3 times in wash buffer and received 50 μL per well of sera (serially diluted in blocking buffer as indicated with duplicate wells for each dilution). Plates were incubated for 2 h at RT and washed 3 times with wash buffer before receiving 50 μL per well of horseradish peroxidase (HRP)-conjugated goat anti-human IgG (AP309P; Sigma-Aldrich) diluted 1:10,000 in blocking buffer. Plates were incubated for 1 h at RT and washed 4 times using wash buffer before receiving 75 μL per well of 3,3′,5,5′-tetramethylbenzidine (TMB; Sigma-Aldrich). Plates were incubated in the dark for 30 min at RT before reactions were stopped using 75 μL per well 1 M H2SO4. The absorbance of each well was measured at 450 nm wavelength using a VersaMax microplate reader (Molecular Devices). Duplicate wells containing no AAV served as background controls for each sera dilution. The mean value for each sample dilution was calculated for wells both with (foreground) and without coated vector (background) and the endpoint titer was determined as the lowest dilution where this ratio was >8.3. The limit of sensitivity for each assay is indicated in the graphs. ELISA to detect anti-AAV9 IgG subclasses in patient sera was performed as above using a 1:100 dilution of patient sera and substituting HRP-conjugated secondary antibody for subclass specific secondary antibodies for IgG1 (1:200; A1068; Thermo Fisher Scientific), IgG2 (1:400; MH1722; Invitrogen), IgG3 (1:800; 053620; Invitrogen), and IgG4 (1:400; A10656; Thermo Fisher Scientific)
An ELISA was similarly conducted to assess anti-AAV reactivity by mAbs and in smB cell culture supernatants, except that the wells were initially coated with AAV vector stocks diluted to 1 × 1010 vg/mL, and supernatants were diluted 1:55 in blocking buffer. The smB cell culture supernatants were regarded as reactive when the 450 nm absorbance level of test samples was more than 2-fold higher than the mean absorbance of the blanks.
A similar ELISA protocol was followed to detect human IgG in smB cell culture by ELISA, with the exception that Maxisorp ELISA plates were coated overnight with 50 μL per well of anti-human Fc-specific IgG capture antibody (I2136; Sigma-Aldrich; diluted 1:6,000 in carbonate-bicarbonate buffer) and 50 μL per well horseradish peroxidase conjugated anti-human Fc-specific IgG (CST32935; Cell Signaling Technologies; diluted 1:6,000 in blocking buffer) was used as a detection antibody.
Recovery of antibody variable regions from switched-memory B cell clones and reconstitution and purification of mAbs and Fab fragments
Three hundred eighty-four-well plates containing stored smB cell lysates were thawed on ice and well contents recovered by pipetting, taking care to avoid any contamination from adjacent wells. Total RNA was isolated from the lysates using the RNeasy Micro Kit (Qiagen), according to the manufacturer’s instructions. The purification included treatment using an RNase-free DNase reagent (Qiagen). The RNA was eluted from the Qiagen column in 60 μL nuclease-free dH2O.
The cDNA encoding the variable regions of light and heavy antibody chains were amplified by reverse transcription (RT-PCR) followed by nested PCR using a previously reported method with some modifications.47 Note that all reagents were kept on ice for preparation of RT-PCR and nested PCR. Briefly, 5′ primers for each of the kappa, lambda, or IgG heavy-chain variable regions were synthesized and mixed in equal proportions for each group. This was repeated for the 3′ primers. Primers for each group were then added to RT-PCR master mixes (OneStep RT-PCR kit; Qiagen) to bring the final forward or reverse primer concentration to 0.6 μM in a 22.5 μL volume. RT-PCR was performed on each lysate to separately amplify kappa, lambda, or IgG heavy-chain variable regions. Purified RNA (2.5 μL) was added to each reaction, and tubes were thermocycled using the following program: 50°C for 30 min for reverse transcription, 95°C for 15 min to deactivate RT and activate thermal Taq, then 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 1 min, followed by 1 cycle of 72°C for 10 min.
A 25 μL nested PCR was then performed on each RT-PCR using a cocktail of forward and reverse nested primers and Taq DNA polymerase (M0267L; New England Biolabs), with the following reaction conditions: 95°C for 4 min then 40 cycles of 95°C for 1 min, 55°C for 1 min, 72°C for 1.5 min, and then 1 cycle of 72°C for 5 min. Five microliters of the nested PCR products were analyzed on a 1.5% Agarose gel. Amplicons at 400–500 bp (depending on light- and heavy-chain use) were recovered, purified (Wizard SV 96 PCR Clean-Up Kit, A6792; Promega), and Sanger sequenced using a mixture of the nested primers for either light or heavy chains. Sequences were analyzed for each PCR product using the default settings in IMGTV-Quest (http://www.imgt.org/IMGT_vquest/analysis) to confirm capture of a productive immunoglobulin V(D)J rearrangement.48 The framework regions and the complementary determining regions (CDR) in each sequence were identified, and total nucleotide and amino acid substitutions from germline were determined by alignment to the most likely germline locus for each chain (excluding CDR3). The mutation frequency was determined by dividing the number of mutations over the length of the given sequence.
Variable region sequences were synthesized as g-Blocks (Integrated DNA Technologies) and subcloned into their respective constant region scaffolds in plasmid constructs.49 MAbs and respective Fab fragments were reconstituted by co-transfection of corresponding light and heavy chain constructs in HEK293T cells using established protocols.49
Transduction inhibition assays
2V6.11 cells were seeded at 1 × 104 cells per well in 96-well tissue culture plates (black walls and flat transparent bottoms; CLS3603; Sigma-Aldrich) in 70 μL complete media supplemented with Ponasterone (1.0 μg/mL; Sigma-Aldrich) and incubated overnight at 37°C/5% CO2. The next day, 60 μL AAV vector (4 × 108 vg) was mixed with 60 μL titrated mAb (final mAb concentration indicated on graphs) and incubated at 37°C for 30 min. Vector-antibody solutions were then added (30 μL) to each of triplicate wells containing the 2V6.11 cells (multiplicity of infection [MOI] = 10,000). After 48 h, incubation at 37°C/5% CO2, luciferase was measured in each well using a Pierce Firefly Luc One-Step Glow Assay Kit (Life Technologies), per the manufacturer’s instructions. The levels of activity in each well were quantitated on the Victor 3 multi-label plate reader (PerkinElmer). The values for triplicate wells were averaged, and the percentage inhibition to vector transduction was calculated relative to vector similarly treated using a non-reactive control mAb, which had been titrated across the indicated range. Inhibition curves were generated in GraphPad Prism using non-linear regression model with least squares of regression. Outliers with Q values of 30%–40% were eliminated.
Native dot immunoblot analysis
AAV capsids were adsorbed onto nitrocellulose membranes (Bio-Rad, Hercules, CA) in a dot-blot manifold (Schleicher and Schuell, Dassel, Germany). Excess fluid was drawn through the membrane by vacuum filtration. The membrane was removed from the manifold and blocked with 6% milk in PBS (pH 7.4) for 1 h. Primary antibody in the form of hybridoma supernatant was applied to the membrane at a 1:50 dilution in PBS with 6% milk, 0.1% Tween 20 and incubated for 1 h. The membrane was then washed with PBS, and horseradish peroxidase-linked secondary antibody (GE Healthcare) was applied at a dilution of 1:3,000 in PBS and incubated for 1 h. The membrane was washed with PBS, and then Immobilon Chemiluminescent Substrate (Millipore, Darmstadt, Germany) was applied to the membrane and the signal detected on X-ray film.
Fab generation and purification
For the generation of Fabs from the purified MAbs, immobilized papain was used according to the manufacturer’s instructions (Thermo Fisher Scientific) and mixed with 1 mg purified IgG. The slurry was incubated under constant rotation at 37°C overnight. Following the reaction, additional sample buffer (1.5 mL, 10 mM Tris-HCl [pH 7.5]) was added and the mixture gently centrifuged (200 × g, 5 min) to pellet the immobilized papain-agarose beads. The aqueous supernatant was transferred and diluted in 20 mM sodium phosphate buffer (pH 8.5). To separate the Fab from the Fc fragments and undigested IgGs, the sample was applied to a Hi-Trap protein A column (GE Healthcare, Uppsala, Sweden) using a peristaltic pump at a rate of 1 mL/min. The Fabs were collected in the flowthrough and concentrated on Apollo concentrators (Orbital Biosciences). The success of IgG cleavage, their concentration, and purity were determined by SDS-PAGE.
Cryo-EM sample preparation, data collection, and 3D image reconstruction
Purified AAV9 capsids were mixed with Fabs at a molecular ratio of ∼2 Fabs per potential VP binding site in the 60-meric capsid, giving a final ratio of ∼1:120 (capsid to Fab). Aliquots (3.5 μL) of the purified samples either with or without Fabs were applied to glow-discharged Quantifoil copper grids with 2 nm continuous carbon support over holes (Quantifoil R 2/2 400 mesh), blotted, and vitrified using a Vitrobot Mark 4 (FEI) at 95% humidity and 4°C. The grids were imaged using a FEI Tecnai G2 F20-TWIN microscope (FEI) operated under low-dose conditions (200 kV, ∼20e−/Å2). Images were collected on a GatanUltraScan 4000 charge-coupled device (CCD) camera (Gatan) at a pixel size of 1.82 Å. Data for the samples using the Fabs from patient 3 (Fab3-1 to Fab3-7) and Fab1-7 were collected at the Stanford-SLAC Cryo-EM Center (S2C2) using a Titan Krios (FEI) electron microscope operated at 300 kV equipped with a Falcon 4 direct electron detector (Thermo Fisher Scientific). Fifty movie frames were collected per micrograph at a total electron dose of ∼50 e−/Å2 and a pixel size of 0.95 Å. Motion corrected micrographs were obtained by aligning the movie frames using MotionCor2 with dose weighting. To obtain ∼5–6 Å structures, the micrographs were binned using a bin factor of 3. For 3D image reconstruction the cisTEM software package was used, and the data processed as described previously.32 The resolution of the cryo-reconstructed density maps was estimated on the basis of a Fourier shell correlation of 0.143. The density maps were analyzed using Chimera.50
Biolayer interferometry
For the determination of the binding kinetics of the Fabs and IgGs toward the AAV9 capsid the GatorPrime instrument with AAVX probes (Gator Bio) was used. For this purpose, the purified AAV9 capsids were diluted in Q-buffer (1× PBS with 0.02% Tween 20 and 0.2% BSA) to a titer of 1 × 1011 capsids/mL. Similarly, the Fabs or IgGs were diluted in Q-buffer in a 2-fold dilution series ranging from 200 to 3.125 nM. In the first step, the AAVX probes were incubated in Q-buffer for 120 s to establish a baseline prior to loading AAV9 capsids. The capsids were loaded until a wavelength shift of ∼1 nm was achieved. The loading step was followed by another incubation in Q-buffer (150 s) to establish the new baseline. Subsequently, the Fabs or IgGs were associated with the capsids for 600 s followed by a 600 s dissociation step using Q-buffer. Probes without loaded AAV9 capsids were used as controls to account for any unspecific binding of the Fabs. The results of the assay were analyzed using the Gator software to determine the kinetic parameters.
Data and code availability
All data and materials reported in this study are available to any researcher for purposes of reproducing or extending the findings. The antibodies are freely available to academic interests subject to the execution of a Materials Transfer Agreement (MTA).
Acknowledgments
The authors would like to thank the late Dr. Mavis Agbandje-McKenna for her pioneering studies of AAV capsid structures. We thank Suzanne Scott for writing script to permit data visualization and Katherine Jackson for providing sequences to identify the subclass of IgG expressed by switched-memory B cell clones. The authors also thank the University of Florida Interdisciplinary Center for Biotechnology Research (UF-ICBR) electron microscopy core for access to electron microscopes used for cryoelectron micrograph screening. Some of this work was performed at the Stanford-SLAC Cryo-EM Center (S2C2), which is supported by the National Institutes of Health Common Fund Transformative High-Resolution Cryo-Electron Microscopy program (grant U24 GM129541). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The study was funded by National Health and Medical Research Council of Australia grants (grant APP2004320 to I.E.A. and G.J.L. and grant APP1194940 to M.A.F.), the Rebecca Cooper Foundation (grant PG2019449 to G.J.L.), and NIH grant R01 NIH GM082946 (to R.M.).
Author contributions
G.J.L., M. Mietzsch, N.K., C.C.G., J.H.R., R.M., and I.E.A. devised the methodology and guided experiments. M.A.F. and A.D. cared for patients and acquired blood samples. G.J.L., M. Mietzsch, N.K., D.A., M. Mandwie, J.H., and A.R.N. performed the experiments. J.J., P.S., and D.C. synthesized and purified the Fabs and monoclonal antibodies. P.C. provided the resources for cryo-EM analysis. G.J.L., M. Mietzsch, R.M., and I.E.A. designed the study and wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.03.032.
Supplemental information
References
- 1.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K., et al. Single-dose gene-replacement therapy for spinal muscular atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 2.Strauss K.A., Farrar M.A., Muntoni F., Saito K., Mendell J.R., Servais L., McMillan H.J., Finkel R.S., Swoboda K.J., Kwon J.M., et al. Onasemnogene abeparvovec for presymptomatic infants with two copies of SMN2 at risk for spinal muscular atrophy type 1: the Phase III SPR1NT trial. Nat. Med. 2022;28:1381–1389. doi: 10.1038/s41591-022-01866-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lisowski L., Tay S.S., Alexander I.E. Adeno-associated virus serotypes for gene therapeutics. Curr. Opin. Pharmacol. 2015;24:59–67. doi: 10.1016/j.coph.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 4.Merkel S.F., Andrews A.M., Lutton E.M., Mu D., Hudry E., Hyman B.T., Maguire C.A., Ramirez S.H. Trafficking of adeno-associated virus vectors across a model of the blood-brain barrier; a comparative study of transcytosis and transduction using primary human brain endothelial cells. J. Neurochem. 2017;140:216–230. doi: 10.1111/jnc.13861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barnes C., Scheideler O., Schaffer D. Engineering the AAV capsid to evade immune responses. Curr. Opin. Biotechnol. 2019;60:99–103. doi: 10.1016/j.copbio.2019.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mietzsch M., Pénzes J.J., Agbandje-McKenna M. Twenty-five years of structural parvovirology. Viruses. 2019;11 doi: 10.3390/v11040362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronzitti G., Gross D.A., Mingozzi F. Human immune responses to adeno-associated virus (AAV) vectors. Front. Immunol. 2020;11:670. doi: 10.3389/fimmu.2020.00670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boutin S., Monteilhet V., Veron P., Leborgne C., Benveniste O., Montus M.F., Masurier C. Prevalence of serum IgG and neutralizing factors against adeno-associated virus (AAV) types 1, 2, 5, 6, 8, and 9 in the healthy population: implications for gene therapy using AAV vectors. Hum. Gene Ther. 2010;21:704–712. doi: 10.1089/hum.2009.182. [DOI] [PubMed] [Google Scholar]
- 9.Calcedo R., Vandenberghe L.H., Gao G., Lin J., Wilson J.M. Worldwide epidemiology of neutralizing antibodies to adeno-associated viruses. J. Infect. Dis. 2009;199:381–390. doi: 10.1086/595830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corti M., Elder M., Falk D., Lawson L., Smith B., Nayak S., Conlon T., Clément N., Erger K., Lavassani E., et al. B-cell depletion is protective against anti-AAV capsid immune response: a human subject case study. Molecular therapy. Mol. Ther. Methods Clin. Dev. 2014;1 doi: 10.1038/mtm.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bertin B., Veron P., Leborgne C., Deschamps J.Y., Moullec S., Fromes Y., Collaud F., Boutin S., Latournerie V., van Wittenberghe L., et al. Capsid-specific removal of circulating antibodies to adeno-associated virus vectors. Sci. Rep. 2020;10:864. doi: 10.1038/s41598-020-57893-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leborgne C., Barbon E., Alexander J.M., Hanby H., Delignat S., Cohen D.M., Collaud F., Muraleetharan S., Lupo D., Silverberg J., et al. IgG-cleaving endopeptidase enables in vivo gene therapy in the presence of anti-AAV neutralizing antibodies. Nat. Med. 2020;26:1096–1101. doi: 10.1038/s41591-020-0911-7. [DOI] [PubMed] [Google Scholar]
- 13.Hudry E., Martin C., Gandhi S., György B., Scheffer D.I., Mu D., Merkel S.F., Mingozzi F., Fitzpatrick Z., Dimant H., et al. Exosome-associated AAV vector as a robust and convenient neuroscience tool. Gene Ther. 2016;23:380–392. doi: 10.1038/gt.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tse L.V., Klinc K.A., Madigan V.J., Castellanos Rivera R.M., Wells L.F., Havlik L.P., Smith J.K., Agbandje-McKenna M., Asokan A. Structure-guided evolution of antigenically distinct adeno-associated virus variants for immune evasion. Proc. Natl. Acad. Sci. USA. 2017;114 doi: 10.1073/pnas.1704766114. E4812-e4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Emmanuel S.N., Smith J.K., Hsi J., Tseng Y.S., Kaplan M., Mietzsch M., Chipman P., Asokan A., McKenna R., Agbandje-McKenna M. Structurally mapping antigenic epitopes of adeno-associated virus 9: development of antibody escape variants. J. Virol. 2022;96 doi: 10.1128/jvi.01251-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mietzsch M., Yu J.C., Hsi J., Chipman P., Broecker F., Fuming Z., Linhardt R.J., Seeberger P.H., Heilbronn R., McKenna R., Agbandje-McKenna M. Structural study of Aavrh.10 receptor and antibody interactions. J. Virol. 2021;95 doi: 10.1128/jvi.01249-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giles A.R., Govindasamy L., Somanathan S., Wilson J.M. Mapping an adeno-associated virus 9-specific neutralizing epitope to develop next-generation gene delivery vectors. J. Virol. 2018;92 doi: 10.1128/jvi.01011-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emmanuel S.N., Mietzsch M., Tseng Y.S., Smith J.K., Agbandje-McKenna M. Parvovirus capsid-antibody complex structures reveal conservation of antigenic epitopes across the family. Viral Immunol. 2021;34:3–17. doi: 10.1089/vim.2020.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Havlik L.P., Simon K.E., Smith J.K., Klinc K.A., Tse L.V., Oh D.K., Fanous M.M., Meganck R.M., Mietzsch M., Kleinschmidt J., et al. Coevolution of adeno-associated virus capsid antigenicity and tropism through a structure-guided approach. J. Virol. 2020;94 doi: 10.1128/jvi.00976-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jose A., Mietzsch M., Smith J.K., Kurian J., Chipman P., McKenna R., Chiorini J., Agbandje-McKenna M. High-resolution structural characterization of a new adeno-associated virus serotype 5 antibody epitope toward engineering antibody-resistant recombinant gene delivery vectors. J. Virol. 2019;93 doi: 10.1128/jvi.01394-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang J., Doria-Rose N.A., Longo N.S., Laub L., Lin C.L., Turk E., Kang B.H., Migueles S.A., Bailer R.T., Mascola J.R., Connors M. Isolation of human monoclonal antibodies from peripheral blood B cells. Nat. Protoc. 2013;8:1907–1915. doi: 10.1038/nprot.2013.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.IJspeert H., van Schouwenburg P.A., van Zessen D., Pico-Knijnenburg I., Driessen G.J., Stubbs A.P., van der Burg M. Evaluation of the antigen-experienced B-cell receptor repertoire in healthy children and adults. Front. Immunol. 2016;7:410. doi: 10.3389/fimmu.2016.00410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vigdorovich V., Oliver B.G., Carbonetti S., Dambrauskas N., Lange M.D., Yacoob C., Leahy W., Callahan J., Stamatatos L., Sather D.N. Repertoire comparison of the B-cell receptor-encoding loci in humans and rhesus macaques by next-generation sequencing. Clin. Transl. Immunol. 2016;5:e93. doi: 10.1038/cti.2016.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vidarsson G., Dekkers G., Rispens T. IgG subclasses and allotypes: from structure to effector functions. Front. Immunol. 2014;5:520. doi: 10.3389/fimmu.2014.00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tseng Y.S., Vliet K.V., Rao L., McKenna R., Byrne B.J., Asokan A., Agbandje-McKenna M. Generation and characterization of anti-Adeno-associated virus serotype 8 (AAV8) and anti-AAV9 monoclonal antibodies. J. Virol. Methods. 2016;236:105–110. doi: 10.1016/j.jviromet.2016.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Day J.W., Finkel R.S., Mercuri E., Swoboda K.J., Menier M., van Olden R., Tauscher-Wisniewski S., Mendell J.R. Adeno-associated virus serotype 9 antibodies in patients screened for treatment with onasemnogene abeparvovec. Molecular therapy. Mol. Ther. Methods Clin. Dev. 2021;21:76–82. doi: 10.1016/j.omtm.2021.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Crotty S., Felgner P., Davies H., Glidewell J., Villarreal L., Ahmed R. Cutting edge: long-term B cell memory in humans after smallpox vaccination. J. Immunol. 2003;171:4969–4973. doi: 10.4049/jimmunol.171.10.4969. [DOI] [PubMed] [Google Scholar]
- 28.Wrammert J., Smith K., Miller J., Langley W.A., Kokko K., Larsen C., Zheng N.Y., Mays I., Garman L., Helms C., et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goel R.R., Painter M.M., Apostolidis S.A., Mathew D., Meng W., Rosenfeld A.M., Lundgreen K.A., Reynaldi A., Khoury D.S., Pattekar A., et al. mRNA vaccines induce durable immune memory to SARS-CoV-2 and variants of concern. Science. 2021;374:abm0829. doi: 10.1126/science.abm0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Au H.K.E., Isalan M., Mielcarek M. Gene therapy advances: a meta-analysis of AAV usage in clinical settings. Front. Med. 2021;8 doi: 10.3389/fmed.2021.809118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Giles A.R., Calcedo R., Tretiakova A.P., Wilson J.M. Isolating human monoclonal antibodies against adeno-associated virus from donors with pre-existing immunity. Front. Immunol. 2020;11:1135. doi: 10.3389/fimmu.2020.01135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mietzsch M., Smith J.K., Yu J.C., Banala V., Emmanuel S.N., Jose A., Chipman P., Bhattacharya N., McKenna R., Agbandje-McKenna M. Characterization of AAV-specific affinity ligands: consequences for vector purification and development strategies. Molecular therapy. Mol. Ther. Methods Clin. Dev. 2020;19:362–373. doi: 10.1016/j.omtm.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mietzsch M., Hull J.A., Makal V.E., Jimenez Ybargollin A., Yu J.C., McKissock K., Bennett A., Penzes J., Lins-Austin B., Yu Q., et al. Characterization of the serpentine adeno-associated virus (SAAV) capsid structure: receptor interactions and antigenicity. J. Virol. 2022;96 doi: 10.1128/jvi.00335-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bell C.L., Gurda B.L., Van Vliet K., Agbandje-McKenna M., Wilson J.M. Identification of the galactose binding domain of the adeno-associated virus serotype 9 capsid. J. Virol. 2012;86:7326–7333. doi: 10.1128/jvi.00448-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dudek A.M., Pillay S., Puschnik A.S., Nagamine C.M., Cheng F., Qiu J., Carette J.E., Vandenberghe L.H. An alternate route for adeno-associated virus (AAV) entry independent of AAV receptor. J. Virol. 2018;92 doi: 10.1128/jvi.02213-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu G., Zhang R., Li H., Yin K., Ma X., Lou Z. Structural basis for the neurotropic AAV9 and the engineered AAVPHP.eB recognition with cellular receptors. Mol. Ther. Methods Clin. Dev. 2022;26:52–60. doi: 10.1016/j.omtm.2022.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rapti K., Grimm D. Adeno-associated viruses (AAV) and host immunity - a race between the hare and the hedgehog. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.753467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mohammadi E.S., Ketner E.A., Johns D.C., Ketner G. Expression of the adenovirus E4 34k oncoprotein inhibits repair of double strand breaks in the cellular genome of a 293-based inducible cell line. Nucleic Acids Res. 2004;32:2652–2659. doi: 10.1093/nar/gkh593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rabinowitz J.E., Rolling F., Li C., Conrath H., Xiao W., Xiao X., Samulski R.J. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 2002;76:791–801. doi: 10.1128/jvi.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabanes-Creus M., Hallwirth C.V., Westhaus A., Ng B.H., Liao S.H.Y., Zhu E., Navarro R.G., Baltazar G., Drouyer M., Scott S., et al. Restoring the natural tropism of AAV2 vectors for human liver. Sci. Transl. Med. 2020;12 doi: 10.1126/scitranslmed.aba3312. [DOI] [PubMed] [Google Scholar]
- 41.Rutledge E.A., Halbert C.L., Russell D.W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao G., Vandenberghe L.H., Alvira M.R., Lu Y., Calcedo R., Zhou X., Wilson J.M. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004;78:6381–6388. doi: 10.1128/jvi.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xiao X., Li J., Samulski R.J. Production of high-titer recombinant adeno-associated virus vectors in the absence of helper adenovirus. J. Virol. 1998;72:2224–2232. doi: 10.1128/jvi.72.3.2224-2232.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ayuso E., Mingozzi F., Montane J., Leon X., Anguela X.M., Haurigot V., Edmonson S.A., Africa L., Zhou S., High K.A., et al. High AAV vector purity results in serotype- and tissue-independent enhancement of transduction efficiency. Gene Ther. 2010;17:503–510. doi: 10.1038/gt.2009.157. [DOI] [PubMed] [Google Scholar]
- 46.Dane A.P., Wowro S.J., Cunningham S.C., Alexander I.E. Comparison of gene transfer to the murine liver following intraperitoneal and intraportal delivery of hepatotropic AAV pseudo-serotypes. Gene Ther. 2013;20:460–464. doi: 10.1038/gt.2012.67. [DOI] [PubMed] [Google Scholar]
- 47.Schofield P., Vazquez-Lombardi R., Abdelatti M., Nevoltris D., Goodnow C.C., Christ D., Reed J.H. Sequencing and affinity determination of antigen-specific B lymphocytes from peripheral blood. Methods Mol. Biol. 2018;1827:287–309. doi: 10.1007/978-1-4939-8648-4_15. [DOI] [PubMed] [Google Scholar]
- 48.Giudicelli V., Brochet X., Lefranc M.P. IMGT/V-QUEST: IMGT standardized analysis of the immunoglobulin (IG) and T cell receptor (TR) nucleotide sequences. Cold Spring Harb. Protoc. 2011;2011:695–715. doi: 10.1101/pdb.prot5633. [DOI] [PubMed] [Google Scholar]
- 49.Vazquez-Lombardi R., Nevoltris D., Luthra A., Schofield P., Zimmermann C., Christ D. Transient expression of human antibodies in mammalian cells. Nat. Protoc. 2018;13:99–117. doi: 10.1038/nprot.2017.126. [DOI] [PubMed] [Google Scholar]
- 50.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and materials reported in this study are available to any researcher for purposes of reproducing or extending the findings. The antibodies are freely available to academic interests subject to the execution of a Materials Transfer Agreement (MTA).